Abstract
Drosophila melanogaster olfactory neurons have long been thought to express only one chemosensory receptor gene family. There are two main olfactory receptor gene families in Drosophila, the odorant receptors (ORs) and the ionotropic receptors (IRs). The dozens of odorant-binding receptors in each family require at least one co-receptor gene in order to function: Orco for ORs, and Ir25a, Ir8a, and Ir76b for IRs. Using a new genetic knock-in strategy, we targeted the four co-receptors representing the main chemosensory families in D. melanogaster (Orco, Ir8a, Ir76b, Ir25a). Co-receptor knock-in expression patterns were verified as accurate representations of endogenous expression. We find extensive overlap in expression among the different co-receptors. As defined by innervation into antennal lobe glomeruli, Ir25a is broadly expressed in 88% of all olfactory sensory neuron classes and is co-expressed in 82% of Orco+ neuron classes, including all neuron classes in the maxillary palp. Orco, Ir8a, and Ir76b expression patterns are also more expansive than previously assumed. Single sensillum recordings from Orco-expressing Ir25a mutant antennal and palpal neurons identify changes in olfactory responses. We also find co-expression of Orco and Ir25a in Drosophila sechellia and Anopheles coluzzii olfactory neurons. These results suggest that co-expression of chemosensory receptors is common in insect olfactory neurons. Together, our data present the first comprehensive map of chemosensory co-receptor expression and reveal their unexpected widespread co-expression in the fly olfactory system.
Research organism: D. melanogaster, Anopheles coluzzii, D. sechellia
Introduction
The sense of smell is crucial for many animal behaviors, from conspecific recognition and mate choice (Dweck et al., 2015; Stengl, 2010), to location of a food source (Auer et al., 2020; Hansson and Stensmyr, 2011), to avoidance of predators (Ebrahim et al., 2015; Kondoh et al., 2016; Papes et al., 2010) and environmental dangers (Mansourian et al., 2016; Stensmyr et al., 2012). Peripheral sensory organs detect odors in the environment using a variety of chemosensory receptors (Carey and Carlson, 2011; Su et al., 2009). The molecular repertoire of chemosensory receptors expressed by the animal, and the particular receptor expressed by any individual olfactory neuron, define the rules by which an animal interfaces with its odor environment. Investigating this initial step in odor detection is critical to understanding how odor signals first enter the brain to guide behaviors.
The olfactory system of the vinegar fly, Drosophila melanogaster, is one of the most extensively studied and well understood (Depetris-Chauvin et al., 2015). D. melanogaster is an attractive model for studying olfaction due to its genetic tractability, numerically simpler nervous system (compared to mammals), complex olfactory-driven behaviors, and similar organizational principles to vertebrate olfactory systems (Ache and Young, 2005; Wilson, 2013). Over 60 years of research have elucidated many of the anatomical, molecular, and genetic principles underpinning fly olfactory behaviors (Gomez-Diaz et al., 2018; Harris, 1972; Pask and Ray, 2016; Siddiqi, 1987; Stocker, 2001; Venkatesh and Naresh Singh, 1984; Vosshall and Stocker, 2007; Yan et al., 2020). Recent advances in electron microscopy and connectomics are revealing higher brain circuits involved in the processing of olfactory information (Bates et al., 2020; Berck et al., 2016; Frechter et al., 2019; Horne et al., 2018; Marin et al., 2020; Zheng et al., 2018); such endeavors will aid the full mapping of neuronal circuits from sensory inputs to behavioral outputs.
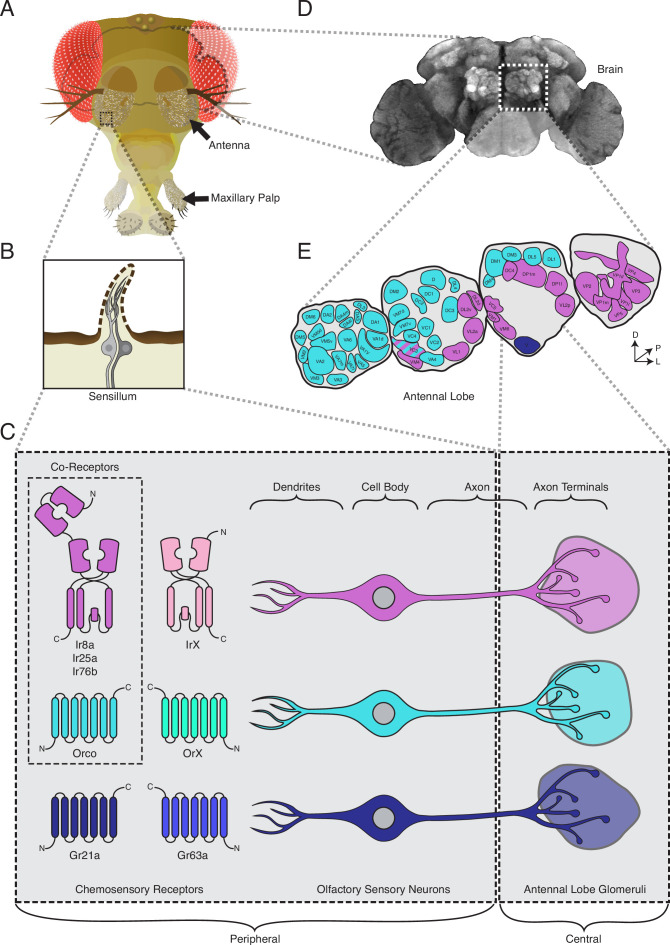
The fly uses two olfactory appendages to detect odorants: the antennae and maxillary palps (Figure 1A; Stocker, 1994). Each of these is covered by sensory hairs called sensilla, and each sensillum houses between one and four olfactory sensory neurons (OSNs) (Figure 1B; de Bruyne et al., 2001; Venkatesh and Naresh Singh, 1984). The dendrites of these neurons are found within the sensillar lymph, and they express chemosensory receptors from three gene families: odorant receptors (ORs), ionotropic receptors (IRs), and gustatory receptors (GRs) (Figure 1C, left; Benton et al., 2009; Clyne et al., 1999; Gao and Chess, 1999; Jones et al., 2007; Kwon et al., 2007; Vosshall et al., 1999; Vosshall et al., 2000). These receptors bind odorant molecules that enter the sensilla from the environment, leading to the activation of the OSNs, which then send this olfactory information to the fly brain (Figure 1D), to the first olfactory processing center – the antennal lobes (ALs) (Figure 1E; reviewed in Depetris-Chauvin et al., 2015; Gomez-Diaz et al., 2018; Pask and Ray, 2016). The standard view regarding the organization of the olfactory system in D. melanogaster is that olfactory neurons express receptors from only one of the chemosensory gene families (either ORs, IRs, or GRs), and all neurons expressing the same receptor (which can be considered an OSN class) project their axons to one specific region in the AL called a glomerulus (Figure 1C, right; Couto et al., 2005; Fishilevich and Vosshall, 2005; Gao et al., 2000; Laissue et al., 1999; Pinto et al., 1988; Vosshall et al., 2000). This pattern of projections creates a map in which the OR+ (Figure 1E, teal), IR+ (Figure 1E, purple), and GR+ (Figure 1E, dark blue) domains are segregated from each other in the AL. The OR+ domains innervate 38 anterior glomeruli, while the IR+ (19 glomeruli) and GR+ (1 glomerulus) domains occupy more posterior portions of the AL. One exception is the Or35a+ OSN class, which expresses an IR (Ir76b) in addition to the OR and Orco, and innervates the VC3 glomerulus (Figure 1E, striped glomerulus; Benton et al., 2009; Couto et al., 2005; Fishilevich and Vosshall, 2005). Different OSN classes send their information to different glomeruli, and the specific combination of OSN classes and glomeruli that are activated by a given smell (usually a blend of different odorants) constitutes an olfactory ‘code’ that the fly brain translates into an appropriate behavior (Grabe and Sachse, 2018; Haverkamp et al., 2018; Seki et al., 2017).
Figure 1. The standard view of olfactory receptor expression in Drosophila melanogaster.
(A) The adult fly head (left) has two olfactory organs: the antennae and the maxillary palps (arrows). Olfactory neurons from these organs project to the fly brain (D), to the first center involved in processing of olfactory information, the antennal lobes (E). (B) The olfactory organs are covered by sensory hairs called sensilla (left). Each sensillum contains between one and four olfactory sensory neurons (two example neurons are shown in gray). The dendrites of these neurons extend into the sensilla, and the axons target discrete regions of the antennal lobes called glomeruli (E). Neuronal compartments (dendrites, cell body, axon, axon terminals) are labeled in (C). ( C) Left: in the periphery, each olfactory sensory neuron is traditionally thought to express chemosensory receptors from only one of three gene families on its dendrites: ionotropic receptors (IRs, pink and purple), odorant receptors (ORs, teal and green), or gustatory receptors (GRs, light and dark blue). IRs and ORs require obligate co-receptors (dotted box outline) to form functional ion channels. All ORs utilize a single co-receptor, Orco (teal), while IRs can utilize one (or a combination) of three possible co-receptors (purple): Ir8a, Ir25a, or Ir76b. The two GRs form a functional carbon dioxide detecting channel expressed in only one class of neurons. All other olfactory neurons express one of the four co-receptors. Right: olfactory sensory neurons expressing ORs, IRs, and GRs are thought to project to mutually exclusive glomeruli in the antennal lobe (AL) of the central brain, forming the olfactory map shown in (E). (D) Fly brain stained with anti-brp synaptic marker (nc82), with left AL outlined by the dotted white box. (E) AL map with glomeruli color-coded by the chemosensory receptors (ORs, IRs, or GRs) expressed in the olfactory sensory neurons projecting to them. Only one glomerulus (VC3, striped) receives inputs from neurons expressing chemoreceptors from multiple gene families (ORs and IRs). Compass: D = dorsal, L = lateral, P = posterior.
The receptors within each chemosensory gene family form heteromeric ion channels (receptor complexes) (Abuin et al., 2011; Butterwick et al., 2018; Sato et al., 2008). The ORs require a single co-receptor, Orco, to function (Figure 1C, middle row; Benton et al., 2006; Larsson et al., 2004; Vosshall and Hansson, 2011). The ligand-binding OrX confers odorant specificity upon the receptor complex, while the co-receptor Orco is necessary for trafficking of the OrX to the dendritic membrane and formation of a functional ion channel (Benton et al., 2006; Larsson et al., 2004). Likewise, the ligand-binding IrXs require one or more IR co-receptors: Ir8a, Ir25a, and/or Ir76b (Figure 1C, top row). The IR co-receptors (IrCos) are similarly required for trafficking and ion channel function (Abuin et al., 2011; Abuin et al., 2019; Ai et al., 2013; Vulpe and Menuz, 2021). The GR gene family generally encodes receptors involved in taste, which are typically expressed outside the olfactory system (such as in the labella or the legs) (Dunipace et al., 2001; Park and Kwon, 2011; Scott, 2018; Scott et al., 2001); however, Gr21a and Gr63a are expressed in one antennal OSN neuron class and form a complex sensitive to carbon dioxide (Figure 1C, bottom row; Jones et al., 2007; Kwon et al., 2007).
The majority of receptors have been mapped to their corresponding OSNs, sensilla, and glomeruli in the fly brain (Bhalerao et al., 2003; Couto et al., 2005; Fishilevich and Vosshall, 2005; Frank et al., 2017; Grabe et al., 2016; Hallem and Carlson, 2006; Hallem et al., 2004; Knecht et al., 2017; Marin et al., 2020; Ray et al., 2008; Silbering et al., 2011). This detailed map has allowed for exquisite investigations into the developmental, molecular, electrophysiological, and circuit/computational bases of olfactory neurobiology. This work has relied on transgenic lines to identify and manipulate OSN classes (Ai et al., 2013; Brand and Perrimon, 1993; Couto et al., 2005; Fishilevich and Vosshall, 2005; Kwon et al., 2007; Lai and Lee, 2006; Larsson et al., 2004; Menuz et al., 2014; Potter et al., 2010; Silbering et al., 2011). These transgenic lines use regions of DNA upstream of the chemosensory genes that are assumed to reflect the enhancers and promoters driving expression of these genes. While a powerful tool, transgenic lines may not contain all of the necessary regulatory elements to faithfully recapitulate the expression patterns of the endogenous genes. In addition, the genomic insertional location of the transgene might affect expression patterns (positional effects). Some transgenic lines label a subset of the cells of a given olfactory class, while others label additional cells: for example, the transgenic Ir25a-Gal4 line is known to label only a portion of cells expressing Ir25a protein (as revealed by antibody staining) (Abuin et al., 2011); conversely, Or67d-Gal4 transgenes incorrectly label two glomeruli, whereas a Gal4 knock-in at the Or67d genetic locus labels a single glomerulus (Couto et al., 2005; Fishilevich and Vosshall, 2005; Kurtovic et al., 2007). While knock-ins provide a faithful method to capture a gene’s expression pattern, generating these lines has traditionally been cumbersome.
In this paper, we implement an efficient knock-in strategy to target the four main chemosensory co-receptor genes in D. melanogaster (Orco, Ir8a, Ir76b, Ir25a). We find broad co-expression of these co-receptor genes in various combinations in olfactory neurons, challenging the current view of segregated olfactory families in the fly. In particular, Ir25a is expressed in the majority of olfactory neurons, including most Orco+ OSNs. In addition, the Ir8a and Ir25a knock-in lines help to distinguish two new OSN classes in the sacculus that target previously unidentified glomerular subdivisions in the posterior AL. Recordings in Ir25a mutant sensilla in Orco+ neurons reveal subtle changes in odor responses, suggesting that multiple chemoreceptor gene families could be involved in the signaling or development of a given OSN class. We further extend our findings of co-receptor co-expression to two additional insect species, Drosophila sechellia and Anopheles coluzzii. These data invite a re-examination of odor coding in D. melanogaster and other insects. We present a comprehensive model of co-receptor expression in D. melanogaster, which will inform future investigations of combinatorial chemosensory processing.
Results
Generation and validation of co-receptor knock-in lines
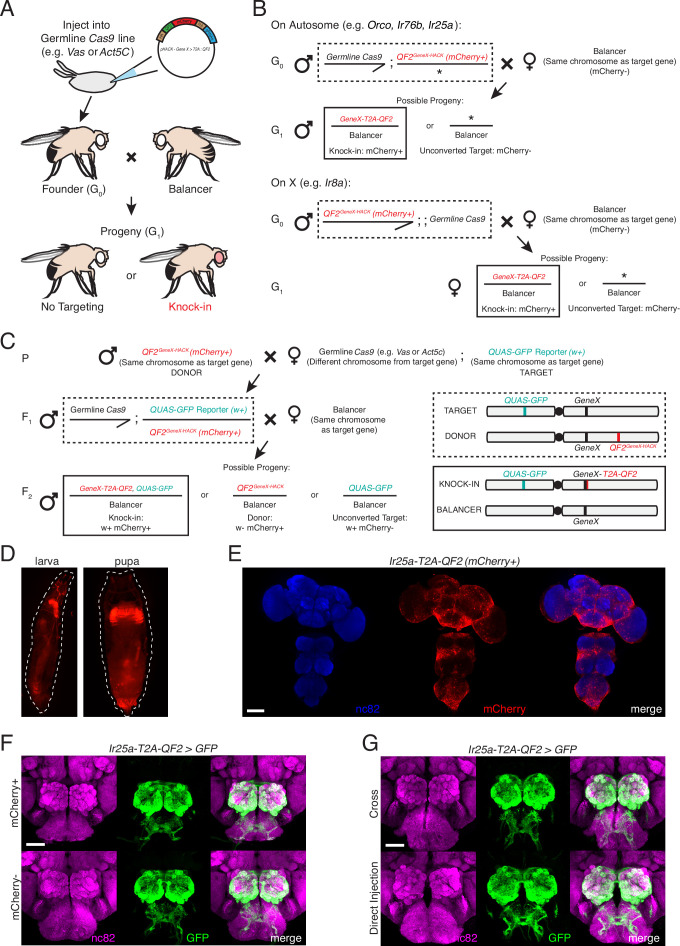
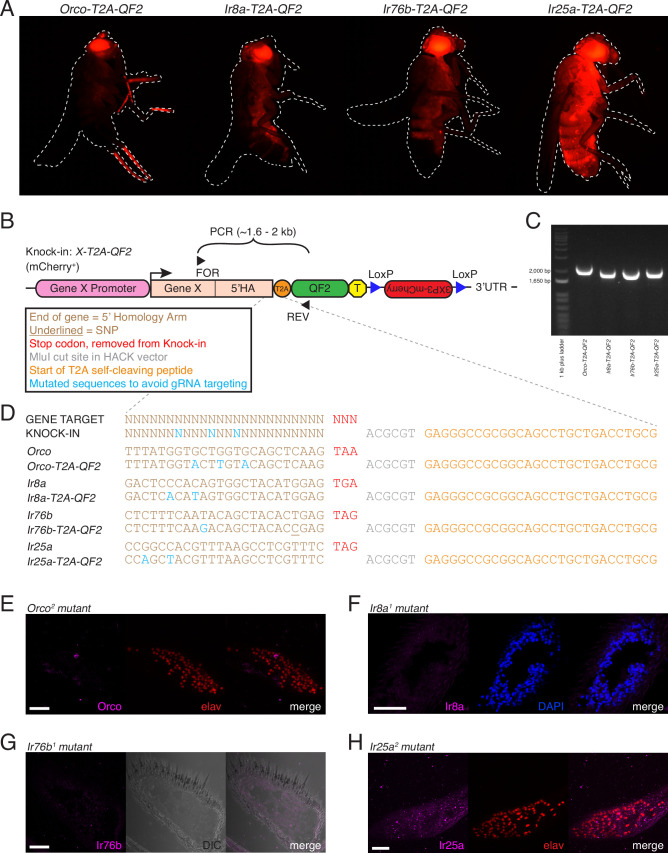
We previously developed the HACK technique for CRISPR/Cas9-mediated in vivo gene conversion of binary expression system components, such as the conversion of transgenic Gal4 to QF2 (Brand and Perrimon, 1993; Jinek et al., 2012; Lin and Potter, 2016a; Lin and Potter, 2016b; Potter et al., 2010; Riabinina et al., 2015; Xie et al., 2018). Here, we adapt this strategy for the efficient generation of targeted knock-ins (see Table 1 and Table 1—source data 1 for details). We chose to target the four chemosensory co-receptor genes to examine unmapped patterns of co-receptor expression in D. melanogaster. We inserted a T2A-QF2 cassette and mCherry selection marker before the stop codon of the four genes of interest (Figure 2A, Figure 2—figure supplement 1). By introducing the T2A ribosomal skipping peptide, the knock-in will produce the full-length protein of the gene being targeted as well as a functional QF2 transcription factor (Figure 2A, protein products). This approach should capture the endogenous expression pattern of the gene under the control of the gene’s native regulatory elements while retaining the gene’s normal function (Baena-Lopez et al., 2013; Bosch et al., 2020; Chen et al., 2020; Diao et al., 2015; Diao and White, 2012; Du et al., 2018; Gnerer et al., 2015; Gratz et al., 2014; Kanca et al., 2019; Lee et al., 2018; Li-Kroeger et al., 2018; Lin and Potter, 2016a; Vilain et al., 2014; Xue et al., 2014). We found that T2A-QF2 knock-ins were functional with some exceptions (see Figure 2—figure supplement 2 and Figure 2—source data 1). For example, Orco-T2A-QF2 knock-in physiology was normal, while a homozygous Ir25a-T2A-QF2 knock-in exhibited a mutant phenotype. This suggests that the addition of the T2A peptide onto the C-terminus of Ir25a might interfere with its co-receptor function.
Table 1. Summary of HACK knock-in efficiency (related to Figure 2).
| Gene | Approach | mCherry+ | mCherry- | Total | Efficiency (%) | Founders producing knock-in (#/total) | Knock-ins sampled | False positives | Confirmed | Correct (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| Orco | Direct injection | 180 | 365 | 545 | 33 | 43% (3/7) | 30 | 0 | 30 | 100 |
| Ir8a | Direct injection | 53 | 609 | 662 | 8 | 20% (4/20) | 5 | 0 | 5 | 100 |
| Ir76b | Direct injection | 79 | 184 | 263 | 30 | 100% (2/2) | 10 | 0 | 10 | 100 |
| Ir25a | Direct injection | 82 | 268 | 350 | 23 | 40% (2/5) | 6 | 0 | 6 | 100 |
| Orco | Cross | 37 | 96 | 133 | 28 | 100% (3/3) | 2 | 1 | 1 | 50 |
| Ir25a | Cross | 30 | 95 | 125 | 24 | 100% (2/2) | 30 | 5 | 25 | 83 |
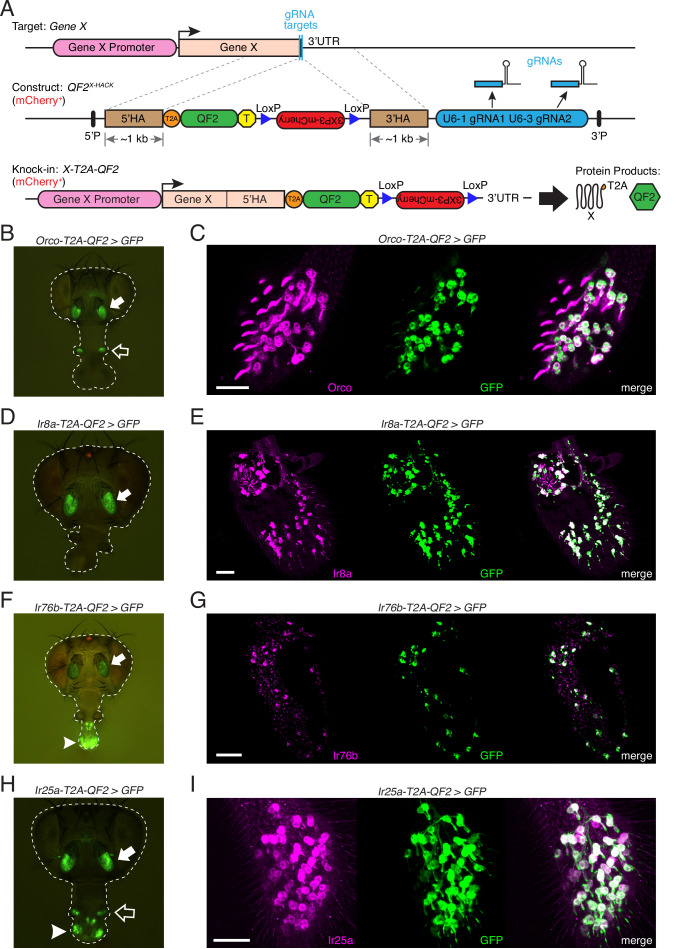
Figure 2. Generation and validation of chemosensory co-receptor knock-in lines.
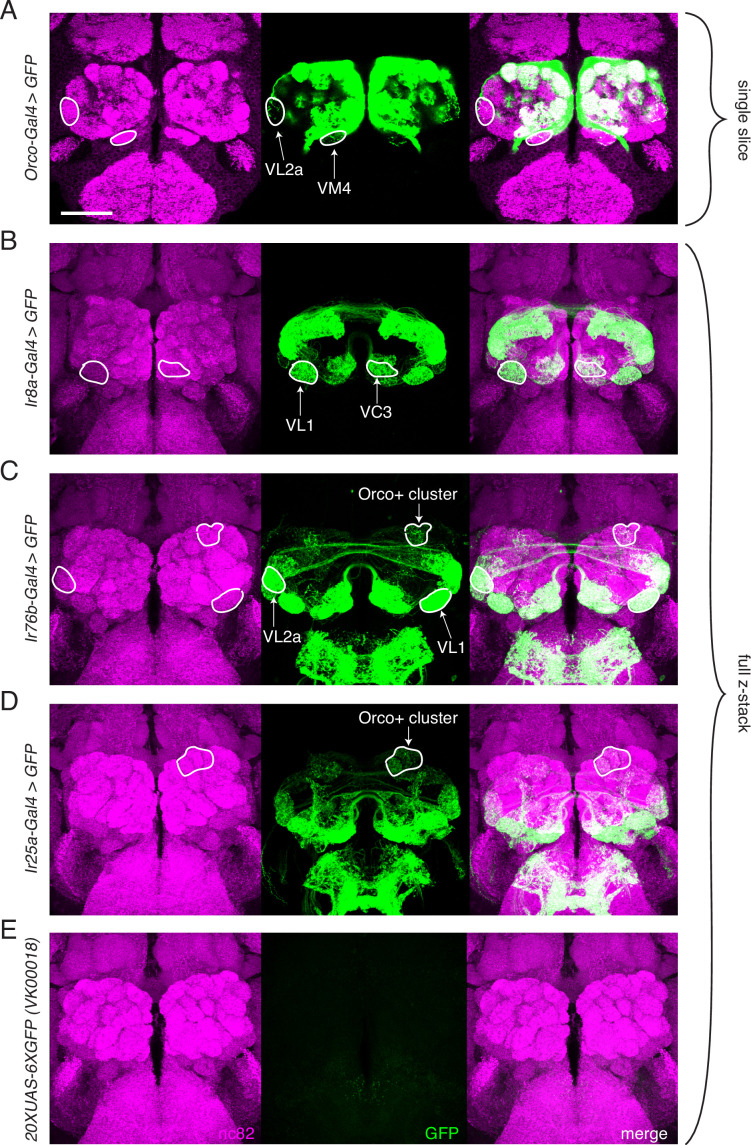
(A) Schematic of HACK knock-in approach. Top: two double-stranded breaks are induced on either side of the target gene stop codon with gRNAs (blue) expressed from the QF2X-HACK construct (middle) in the presence of Cas9. The construct includes T2A-QF2 and a floxed 3XP3-mCherry marker. The knock-in introduces a transcriptional stop (yellow T) after QF2. Bottom: the knock-in produces two protein products (right) from the targeted mRNA: target X and the QF2 transcription factor (Diao and White, 2012). The X-T2A-QF2 knock-in can be crossed to a reporter (e.g., QUAS-GFP) to examine the endogenous expression pattern of the target gene. (B) Orco-T2A-QF2 driving QUAS-GFP in adult fly head. GFP expression is found in the antennae (filled arrow) and maxillary palps (hollow arrow), as previously reported (Larsson et al., 2004). (C) Whole-mount anti-Orco antibody staining in Orco-T2A-QF2>GFP maxillary palps reveals a high degree of overlap of Orco+ and GFP+ cells. N = 3. (D) Ir8a-T2A-QF2 drives GFP in the antennae (arrow), as previously reported (Abuin et al., 2011). (E) Anti-Ir8a antibody staining of Ir8a-T2A-QF2>GFP antennal cryosections shows high correspondence between Ir8a+ and GFP+ cells. N = 7. (F) Ir76b-T2A-QF2 drives GFP expression in the antennae (filled arrow) and labella (hollow arrow), reflecting Ir76b’s role in olfaction and gustation, respectively (Benton et al., 2009; Zhang et al., 2013). (G) In situs on Ir76b-T2A-QF2>GFP antennal cryosections to validate that the knock-in faithfully recapitulates the endogenous expression pattern. N = 3. (H) Ir25a-T2A-QF2 drives GFP in the antennae (filled arrow) and labella (hollow arrow), which has been reported previously (Benton et al., 2009; Croset et al., 2010). Expression in the maxillary palps (arrowhead) has not been previously reported. (I) Whole-mount maxillary palp staining with an anti-Ir25a antibody in Ir25a-T2A-QF2>GFP flies. The knock-in and Ir25a antibody co-labeled the majority of olfactory neurons in the palps. N = 5. Scale bars = 25 µm. In (D) and (F), the 3XP3-mCherry knock-in marker can be weakly detected in the eyes and ocelli (red spot) of both Ir8a-T2A-QF2 and Ir76b-T2A-QF2. See also Figure 2—figure supplements 1–4, Tables 1 and 2, Table 1—source data 1, Figure 2—source data 1, and Materials and methods.
Figure 2—figure supplement 1. HACK crossing schematics, marker expression, and approach comparison.
Figure 2—figure supplement 2. T2A-QF2 HACK knock-in effects on target gene function.
Figure 2—figure supplement 3. Additional validation of co-receptor knock-in lines.
Figure 2—figure supplement 4. Knock-in expression in the larva.
We examined the expression of the co-receptor knock-in lines in the adult olfactory organs by crossing each line to the same 10XQUAS-6XGFP reporter (Figure 2B–I). Orco-T2A-QF2-driven GFP expression was detected in the adult antennae and maxillary palps (Figure 2B), as previously described (Larsson et al., 2004). We validated the Orco-T2A-QF2 knock-in line with whole-mount antibody staining of maxillary palps (Figure 2C) and found a high degree of correspondence between anti-Orco antibody staining and knock-in driven GFP in palpal olfactory neurons (quantified in Table 2; see also Figure 2—figure supplement 3A–D for PCR and sequencing validation of all knock-in lines). We confirmed the specificity of the anti-Orco antibody by staining Orco2 mutant palps and found no labeling of olfactory neurons (Figure 2—figure supplement 3E).
Table 2. Validation of T2A-QF2 knock-in expression in the antennae and maxillary palps (related to Figure 2).
To verify that the knock-in lines recapitulate the endogenous expression patterns of the target genes, antennae or maxillary palps of flies containing the knock-ins driving GFP expression were co-stained with the corresponding antibody (Ab) (anti-Orco, anti-Ir8a, or anti-Ir25a). The overlap of Ab+ and GFP+ cells was examined, and a high correspondence between antibody staining and knock-in driven GFP was found. WM: whole-mount; cryo: cryosection. See also Figure 2—figure supplement 3.
| Knock-in | Sample | Antibody (Ab) | Ab+ cells | GFP+ cells | Double-labeled cells | Total cells |
|---|---|---|---|---|---|---|
| Orco | Palp 1 (WM) | Anti-Orco | 125 | 127 | 125 | 127 |
| Orco | Palp 2 (WM) | Anti-Orco | 112 | 111 | 108 | 115 |
| Orco | Palp 6 (WM) | Anti-Orco | 125 | 126 | 123 | 128 |
| Total across samples: | 362 | 364 | 356 | 370 | ||
| Proportion of Ab+ cells that are GFP+: | Proportion of GFP+ cells that are Ab+: | Proportion of all cells that are double labeled: | ||||
| 0.98 | 0.98 | 0.96 | ||||
| Ir8a | Antenna 1 (cryo) | Anti-Ir8a | 20 | 21 | 20 | 21 |
| Ir8a | Antenna 2 (cryo) | Anti-Ir8a | 24 | 24 | 24 | 24 |
| Ir8a | Antenna 6 (cryo) | Anti-Ir8a | 40 | 43 | 40 | 43 |
| Ir8a | Antenna 7 (cryo) | Anti-Ir8a | 12 | 13 | 12 | 13 |
| Ir8a | Antenna 8 (cryo) | Anti-Ir8a | 16 | 16 | 16 | 16 |
| Ir8a | Antenna 9 (cryo) | Anti-Ir8a | 42 | 42 | 41 | 43 |
| Ir8a | Antenna 10 (cryo) | Anti-Ir8a | 41 | 40 | 40 | 41 |
| Total across samples: | 195 | 199 | 193 | 201 | ||
| Proportion of Ab+ cells that are GFP+: | Proportion of GFP+ cells that are Ab+: | Proportion of all cells that are double labeled: | ||||
| 0.99 | 0.97 | 0.96 | ||||
| Ir25a | Palp 1 (WM) | Anti-Ir25a | 107 | 105 | 104 | 108 |
| Ir25a | Palp 2 (WM) | Anti-Ir25a | 86 | 85 | 85 | 86 |
| Ir25a | Palp 3 (WM) | Anti-Ir25a | 111 | 111 | 110 | 112 |
| Ir25a | Palp 4 (WM) | Anti-Ir25a | 94 | 94 | 94 | 94 |
| Ir25a | Palp 5 (WM) | Anti-Ir25a | 83 | 83 | 81 | 85 |
| Total across samples: | 481 | 478 | 474 | 485 | ||
| Proportion of Ab+ cells that are GFP+: | Proportion of GFP+ cells that are Ab+: | Proportion of all cells that are double labeled: | ||||
| 0.99 | 0.99 | 0.98 |
Unlike Orco, Ir8a expression has previously been localized only to the antenna, to olfactory neurons found in coeloconic sensilla and in the sacculus (Abuin et al., 2011). As expected, the knock-in line drove GFP expression only in the antenna (Figure 2D). To validate the Ir8a-T2A-QF2 knock-in line, we performed antibody staining on antennal cryosections and found the majority of cells to be double labeled (Figure 2E, Table 2). There was no anti-Ir8a staining in control Ir8a1 mutant antennae (Figure 2—figure supplement 3F).
The Ir76b gene has previously been implicated in both olfaction and gustation and has been shown to be expressed in adult fly antennae, labella (mouthparts), legs, and wings (Abuin et al., 2011; Chen and Amrein, 2017; Croset et al., 2010; Ganguly et al., 2017; Hussain et al., 2016; Sánchez-Alcañiz et al., 2018; Zhang et al., 2013). We examined the Ir76b-T2A-QF2 knock-in line and found a similar pattern of expression in the periphery, with GFP expression in the antennae and labella (Figure 2F). Because an anti-Ir76b antibody has not previously been tested in fly antennae, we performed in situs on Ir76b-T2A-QF2>GFP antennal cryosections to validate knock-in expression (Figure 2G) and confirmed the specificity of the probe in Ir76b1 mutant antennae (Figure 2—figure supplement 3G).
Of the four D. melanogaster co-receptor genes, Ir25a has been implicated in the broadest array of cellular and sensory functions, from olfaction (Abuin et al., 2011; Benton et al., 2009; Silbering et al., 2011) and gustation (Chen and Amrein, 2017; Chen and Dahanukar, 2017; Jaeger et al., 2018), to thermo- and hygro-sensation (Budelli et al., 2019; Enjin et al., 2016; Knecht et al., 2017; Knecht et al., 2016), to circadian rhythm modulation (Chen et al., 2015). In the adult olfactory system, Ir25a expression has previously been reported in three types of structures in the antenna: coeloconic sensilla, the arista, and the sacculus (Abuin et al., 2011; Benton et al., 2009). We examined the Ir25a-T2A-QF2 knock-in line and found GFP expression in the adult antennae, labella, and maxillary palps (Figure 2H). This was surprising because no IR expression has previously been reported in fly palps. To verify Ir25a protein expression in the maxillary palps, we performed whole-mount anti-Ir25a antibody staining in Ir25a-T2A-QF2>GFP flies. We found broad Ir25a expression in palpal olfactory neurons (Figure 2I) and a high degree of overlap between knock-in driven GFP expression and antibody staining (Table 2). As expected, there was no anti-Ir25a staining in Ir25a2 mutant palps (Figure 2—figure supplement 3H).
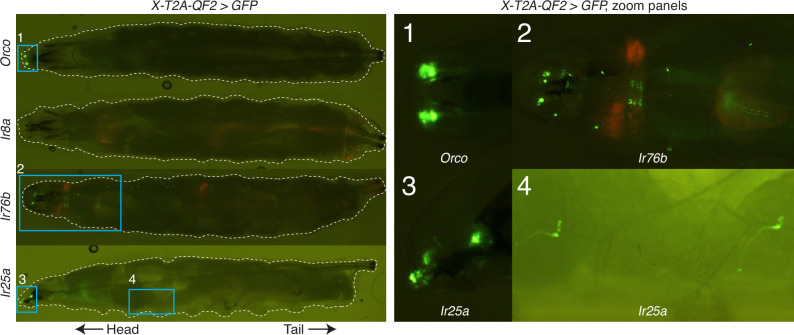
We also examined co-receptor knock-in expression in D. melanogaster larvae. As in the adult stage, larval GFP expression was broadest in the Ir25a-T2A-QF2 and Ir76b-T2A-QF2 knock-in lines, with GFP labeling of neurons in the head and throughout the body wall (Figure 2—figure supplement 4). The Orco-T2A-QF2 knock-in line labeled only the olfactory dorsal organs in the larva, while the Ir8a-T2A-QF2 knock-in line did not have obvious expression in the larval stage (Figure 2—figure supplement 4). All subsequent analyses focused on the adult olfactory system.
Expanded expression of olfactory co-receptors
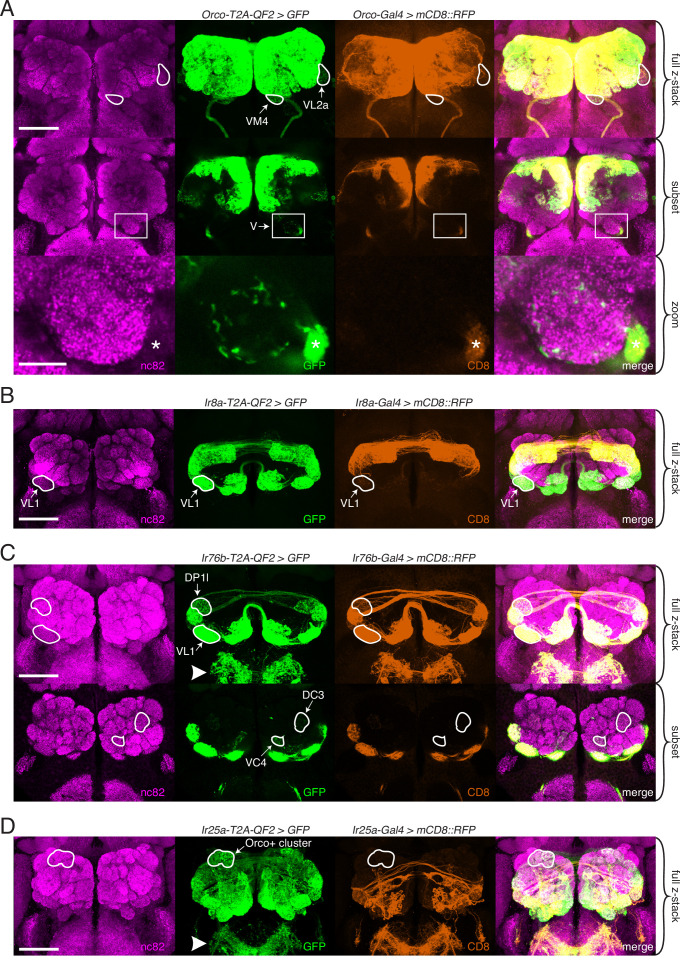
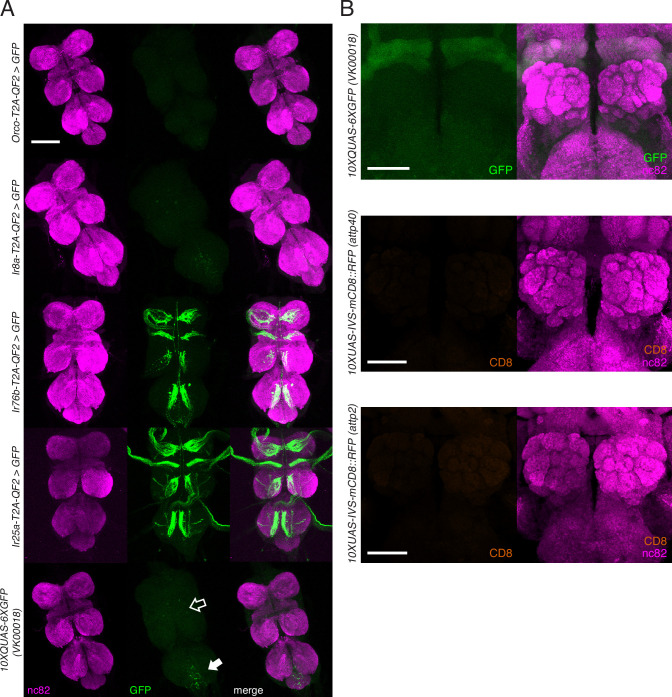
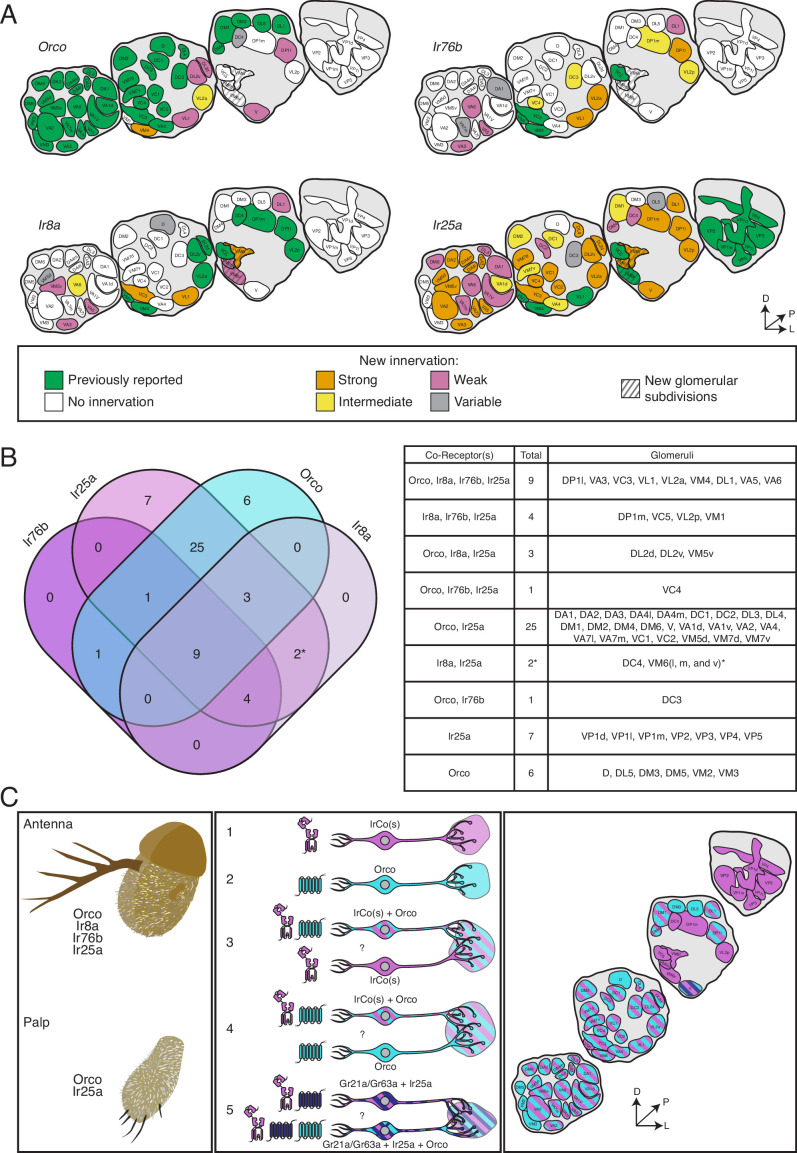
We next examined the innervation patterns of the four co-receptor knock-in lines in the adult central nervous system: the brain and ventral nerve cord (VNC) (Figure 3). Only two of the four lines (Ir25a and Ir76b) showed innervation in the VNC, consistent with the role of these genes in gustation in addition to olfaction (Figure 3—figure supplement 1A). In the brain, we compared the expression of each knock-in line (Figure 3A–D, green) to the corresponding transgenic Gal4 line (Figure 3A–D, orange) to examine the differences in expression to what has previously been reported. Reporter-alone controls for these experiments are shown in Figure 3—figure supplement 1B. All four knock-in lines innervated the ALs, and the Ir25a-T2A-QF2 and Ir76b-T2A-QF2 lines additionally labeled the subesophageal zone (SEZ), corresponding to gustatory axons from the labella (Figure 3C and D, arrowheads; Hussain et al., 2016; Zhang et al., 2013). The co-labeling experiments revealed that all four knock-ins label more glomeruli than previously reported (see Figure 3—source data 1 for AL analyses, Figure 3—source data 2 for traced examples of newly identified glomeruli in each knock-in line, and Table 3 for a summary of glomerular expression across all knock-in lines). Some glomeruli were not labeled consistently in all flies, which we define as variable expression (found in <50% of brains examined).
Figure 3. Expanded expression of olfactory co-receptors.
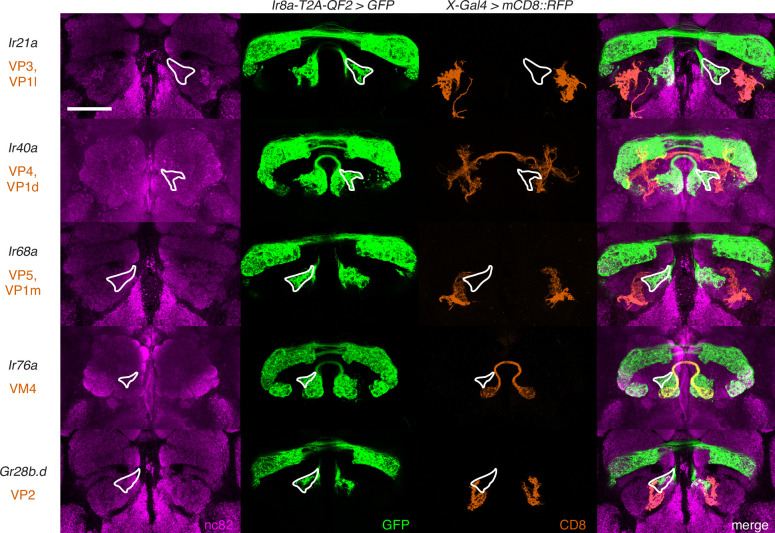
(A–D) Comparing knock-in innervation patterns of the antennal lobe (AL) with what has previously been reported for each co-receptor. Co-labeling experiments with each co-receptor knock-in line driving QUAS-GFP (green) and the corresponding transgenic co-receptor Gal4 line driving UAS-mCD8::RFP (anti-CD8, orange). The nc82 antibody labels synapses (magenta) and is used as a brain counterstain in these and all subsequent brain images. (A) The Orco-T2A-QF2 knock-in labels more glomeruli than the Orco-Gal4 line. Top: maximum intensity projection of full z-stack showing two additional glomeruli labeled by the knock-in, VM4 (Ir8a+/Ir76b+/Ir25a+) and VL2a (Ir8a+). Middle: subset of z-stack with a box around the V glomerulus. Bottom: zoom of boxed region showing sparse innervation of the V glomerulus (Gr21a+/Gr63a+) by the knock-in but not the Gal4 line. Asterisk indicates antennal nerve that is outside the V glomerulus. In the sub z-stack and zoom panel, gain has been increased in the GFP channel to visualize weak labeling more clearly. (B) The Ir8a-T2A-QF2 knock-in also drives GFP expression in more glomeruli than previously reported, including the outlined VL1 glomerulus (Ir25a+). (C) In the brain, Ir76b-T2A-QF2>GFP olfactory neurons innervate the ALs, while gustatory neurons from the labella innervate the subesophageal zone (SEZ, arrowhead). Top: both the Ir76b knock-in and transgenic Gal4 line label more glomeruli than previously reported, including VL1 (Ir25a+) and DP1l (Ir8a+). Bottom: the Ir76b-T2A-QF2 knock-in labels several Orco+ glomeruli, such as DC3 and VC4 (outlined). In the subset, gain has been increased in the GFP channel to visualize weakly labeled glomeruli more clearly. (D) The Ir25a-T2A-QF2 knock-in drives GFP expression broadly in the antennal lobes and SEZ (arrowhead). Ir25a+ neurons innervate many Orco+ glomeruli, such as those outlined. The transgenic Ir25a-Gal4 line labels a subset of the knock-in expression pattern. N = 3–10 for co-labeling experiments, N = 5–15 for additional analyses of the knock-in lines alone. Scale bars = 25 µm, except zoom panel scale bar = 10 µm. See also Figure 3—figure supplements 1 and 2, Table 3, and Figure 3—source data 1 and Figure 3—source data 2.
Figure 3—figure supplement 1. Knock-in expression in the adult ventral nerve cord (VNC) and reporter expression in the brain.
Figure 3—figure supplement 2. Transgenic co-receptor Gal4 lines do not fully recapitulate knock-in expression.
Table 3. Summary of expression patterns for all knock-in lines (related to Figures 3—5).
Summarized here are all of the olfactory sensory neuron (OSN) classes innervating the 58 antennal lobe glomeruli†; their corresponding sensilla and tuning receptors; the previously reported (original) co-receptors they express; and whether or not each of the co-receptor knock-in lines labels those glomeruli. Variable indicates that the glomerulus was labeled in <50% of brains examined in the given knock-in line. Sensilla or glomeruli that have been renamed or reclassified have their former nomenclature listed in parentheses. Question marks indicate expression that has been reported but not functionally validated. * See also Figure 3—source data 1 and Figure 3—source data 2.
*VM6l was initially named VC6 in version 1 of our pre-print (Task et al., 2020) but was reclassified using additional data from EM reconstructions in the antennal lobe (AL) and immunohistochemical experiments in the periphery (see Figure 5).
†The VM6 subdivisions (VM6v, VM6m, VM6l) are separated in this table for clarity but counted together as one glomerulus in accordance with Schlegel et al., 2021.
Orco-T2A-QF2 labels seven ‘non-canonical’ glomeruli consistently, and one sporadically. These include VM4 and VL2a, which correspond to Ir76b+ and Ir8a+ OSN populations, respectively (Figure 3A, outlines). We also found that the Orco knock-in sparsely but consistently labels the V glomerulus, which is innervated by Gr21a+/Gr63a+ neurons (Figure 3A, box and zoom panel). Orco-T2A-QF2 also labels one Ir25a+ glomerulus consistently (VL1), three additional Ir8a+ glomeruli consistently (DL2d, DL2v, DP1l), and one variably (DC4). Surprisingly, when we crossed the transgenic Orco-Gal4 line (Larsson et al., 2004) to a stronger reporter (Shearin et al., 2014), we found that several of these additional glomeruli were weakly labeled by the transgenic line (Figure 3—figure supplement 2A). This suggests that there are OSN populations in which Orco is expressed either at low levels or in few cells, which might be why this expression was previously missed. We found this to be the case with the IrCo knock-ins, as well (described below).
There has been some inconsistency in the literature as to which glomeruli are innervated by Ir8a-expressing OSNs. For example, Silbering et al., 2011 note that their Ir8a-Gal4 line labels approximately 10 glomeruli, 6 of which are identified (DL2, DP1l, VL2a, VL2p, DP1m, DC4). An Ir8a-Gal4 line generated by Ai et al., 2013 also labels about 10 glomeruli, only 2 of which are identified (DC4 and DP1m) and which correspond to 2 glomeruli in Silbering et al., 2011. Finally, Min et al., 2013 identify three additional glomeruli innervated by an Ir8a-Gal4 line (VM1, VM4, and VC5) but not reported in the other two papers. DL2 was later subdivided into two glomeruli (Prieto-Godino et al., 2017), bringing the total number of identified Ir8a+ glomeruli to 10. However, we found that Ir8a-T2A-QF2 consistently labels twice as many glomeruli as previously reported. These additional glomeruli include an Ir25a+ glomerulus (VL1, Figure 3B), numerous Orco+ glomeruli (such as VA3 and VA5), and an Orco+/Ir76b+ glomerulus (VC3) (see Figure 3—source data 1 for a full list of new glomeruli and Figure 3—source data 2 for outlined examples). Some of these additional glomeruli are weakly labeled by an Ir8a-Gal4 line (Figure 3—figure supplement 2B), but this innervation is only apparent when examined with a strong reporter.
Of the four chemosensory co-receptor genes, the previously reported expression of Ir76b is the narrowest, with only four identified glomeruli (VM1, VM4, VC3, VC5) (Silbering et al., 2011). The Ir76b-T2A-QF2 knock-in labels more than three times this number, including several Orco+ glomeruli (such as DC3 and VC4), most Ir8a+ glomeruli (including DP1l), and one additional Ir25a+ glomerulus (VL1) (Figure 3C). As with Orco and Ir8a, some but not all of these glomeruli can be identified by crossing the transgenic Ir76b-Gal4 line to a strong reporter (Figure 3—figure supplement 2C). However, the Ir76b-Gal4 line labels additional glomeruli not seen in the knock-in (Figure 3—figure supplement 2C, Orco+ cluster). In total, the Ir76b-T2A-QF2 knock-in labels 15 glomeruli consistently and two variably (Figure 3—source data 1 and Figure 3—source data 2).
Ir25a-T2A-QF2 innervation of the AL was the most expanded compared to what has previously been reported. In addition to the novel expression we identified in the palps (Figure 2H), we found that the Ir25a knock-in innervates many Orco+ glomeruli receiving inputs from the antennae (Figure 3D). The extensive, dense innervation of the AL by Ir25a+ processes made identification of individual glomeruli difficult and necessitated further experiments to fully characterize this expression pattern (described in greater detail below). While it was previously reported that the transgenic Ir25a-Gal4 line labels only a subset of Ir25a+ neurons (compared to anti-Ir25a antibody staining), it was assumed that neurons not captured by the transgenic line would reside in coeloconic sensilla, the arista, or sacculus (the original locations for all IR+ OSNs) (Abuin et al., 2011). When we crossed Ir25a-Gal4 to a strong reporter, we found labeling of a few Orco+ glomeruli (Figure 3—figure supplement 2D), but this was a small fraction of those labeled by the knock-in. To further examine Ir25a expression and the potential co-expression of multiple co-receptors in greater detail, we employed a combination of approaches, including single-nucleus RNAseq (snRNAseq), immunohistochemistry, and optogenetics.
Confirmation of co-receptor co-expression
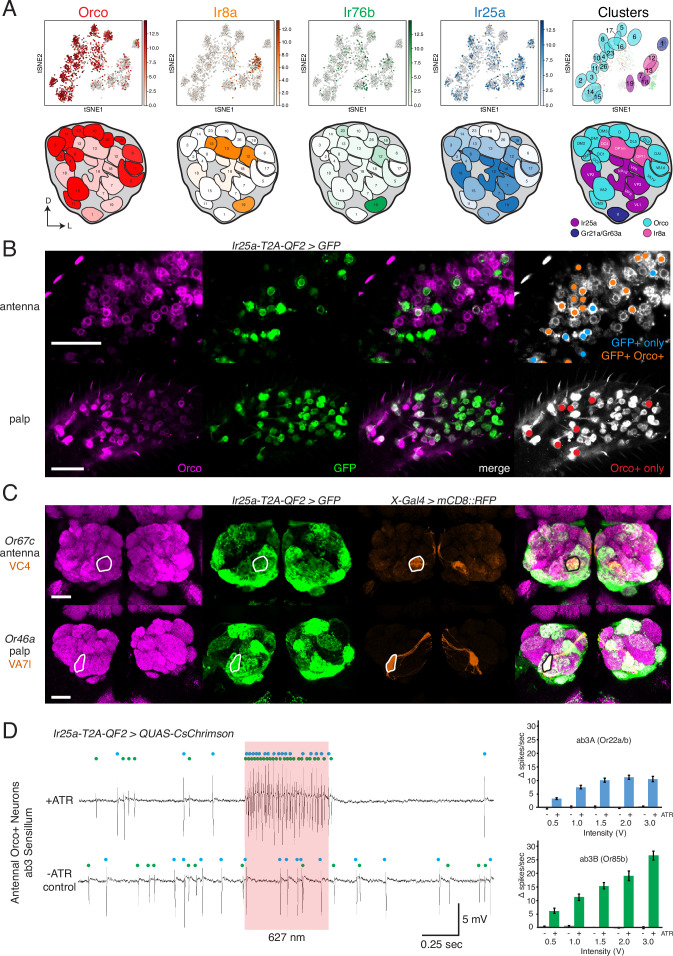
The innervation of the same glomeruli by multiple co-receptor knock-in lines challenges the previous view of segregated chemosensory receptor expression in D. melanogaster and suggests two possible explanations: either the same olfactory neurons express multiple co-receptors (co-expression) or different populations of olfactory neurons expressing different receptors converge upon the same glomeruli (co-convergence). These scenarios are not necessarily mutually exclusive. To examine these possibilities in a comprehensive, unbiased way, we analyzed snRNAseq data from adult fly antennae (McLaughlin et al., 2021). Figure 4A shows the expression levels of the four co-receptor genes in 20 transcriptomic clusters (tSNE plots [Van der Maaten and Hinton, 2008], top row), which were mapped to 24 glomerular targets in the brain (AL maps, bottom row). The proportion of cells in each cluster expressing the given co-receptor gene is indicated by the opacity of the glomerular fill color, normalized to maximum expression for that gene (see Materials and methods and Figure 4—source data 1 for details on expression normalization). The OSN classes to which these clusters map include Orco+ neurons (Figure 4A, right column, teal), Ir25a+ neurons (Figure 4A, right column, purple), Ir8a+ neurons (Figure 4A, right column, pink), and GR+neurons (Figure 4A, right column, dark blue). They also include example OSNs from all sensillar types (basiconic, intermediate, trichoid, coeloconic) as well as from the arista and sacculus. The snRNAseq analyses confirmed expanded expression of all four co-receptor genes into OSN classes not traditionally assigned to them. For example, Orco and Ir25a are expressed in cluster 1, which maps to the V glomerulus (Gr21a+/Gr63a+). Similarly, Ir8a and Ir76b are expressed in cluster 19 (VL1 glomerulus, Ir25a+), and Ir25a is expressed in multiple Orco+ clusters (such as 15/VA2, 16/DL3, and 8/DC1).
Figure 4. Confirmation of co-receptor co-expression.
(A) snRNAseq of adult fly antennae (McLaughlin et al., 2021) confirms expanded expression of olfactory co-receptors. Top: tSNE plots show expression of each co-receptor in 20 decoded olfactory sensory neuron (OSN) clusters. Bottom: clusters were mapped to 24 glomeruli. Opacity of fill in each glomerulus indicates the proportion of cells in that cluster expressing the given co-receptor, normalized to total expression for that co-receptor gene (see Figure 4—source data 1). Right column: clusters color-coded according to original chemoreceptor gene family. Compass: D = dorsal; L = lateral. (B) Anti-Orco antibody staining in antennal cryosections (top) and whole-mount palps (bottom) confirms co-expression of Orco and Ir25a in the periphery (genotype: Ir25a-T2A-QF2>GFP). Right panels show cells pseudo-colored gray with specific single- or double-labeled cells indicated by colored cell markers (GFP+ only in blue, GFP+Orco+ in orange, Orco+ only in red). (C) Co-labeling experiments with various transgenic Gal4 lines driving mCD8::RFP (orange) and the Ir25a-T2A-QF2 knock-in driving GFP (green). Ir25a-T2A-QF2 labels glomeruli innervated by both antennal (top) and palpal (bottom) OSNs. (D) Verification of Ir25a expression in antennal ab3 sensilla using optogenetics. Single sensillum recordings (SSR) from ab3 Orco+ neurons in Ir25a-T2A-QF2>QUAS-CsChrimson flies. Representative traces from ab3 using 1.5 V of 627 nm LED light (red box) to activate CsChrimson. Bottom trace is control animal, which has the same genotype as the experimental animal but was not fed the required all-trans retinal cofactor (-ATR). Spikes from the ab3A and ab3B neurons are indicated by blue and green dots, respectively. Right: quantification of neuronal activity in response to light at various LED intensities (N = 7–12). These optogenetic experiments support Ir25a expression in both ab3A neurons (Or22a/b, top; corresponding to DM2 glomerulus) and ab3B neurons (Or85b, bottom; corresponding to VM5d glomerulus). Scale bars = 25 µm. See also Figure 4—figure supplement 1, Table 3, Figure 4—source data 1, Figure 4—source data 2, and Figure 4—source data 3.
Figure 4—figure supplement 1. Optogenetic experiments to examine Ir25a expression in Orco+ neurons.
The snRNAseq analyses confirm transcript co-expression in olfactory neurons in the periphery. To demonstrate protein co-expression in OSNs, we performed anti-Orco antibody staining on Ir25a-T2A-QF2>GFP antennae and palps (Figure 4B). In the antennae, we found examples of Orco+ GFP+ double-labeled cells, as well as many cells that were either GFP+ or Orco+ (Figure 4B, top-right panel). Interestingly, in the palps the vast majority of cells were double labeled. We found a small population of palpal neurons that were only Orco+, and no neurons that were only GFP+ (Figure 4B, bottom-right panel). These results are consistent with our anti-Ir25a staining experiments in the palps (Figure 2I), which showed that most of the ~120 palpal OSNs express Ir25a protein.
The snRNAseq data from the antennae and peripheral immunohistochemical experiments in the palps helped to identify some of the novel OSN populations expressing Ir25a. We extended these analyses with co-labeling experiments in which we combined transgenic OrX-, IrX-, or GrX-Gal4 lines labeling individual glomeruli with the Ir25a knock-in to verify the glomerular identity of Ir25a+ axonal targets in the AL. Two examples are shown in Figure 4C (one antennal and one palpal OSN population), and the full list of OSN classes checked can be found in Figure 4—source data 2.
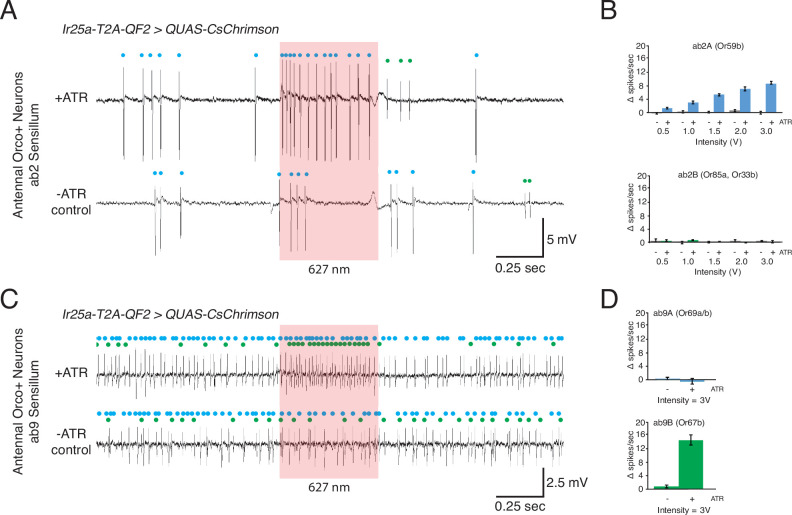
For some OSN classes not included in the snRNAseq dataset for which co-labeling experiments yielded ambiguous results, we employed an optogenetic approach. We used the Ir25a-T2A-QF2 knock-in to drive expression of QUAS-CsChrimson, a red-shifted channelrhodopsin (Klapoetke et al., 2014), and performed single sensillum recordings (SSR) from sensilla previously known to house only Orco+ neurons. If these neurons do express Ir25a, then stimulation with red light should induce neuronal firing. We recorded from ab3 sensilla, which have two olfactory neurons (A and B; indicated with blue and green dots, respectively, in Figure 4D). Ab3A neurons innervate DM2 and ab3B neurons innervate VM5d. Both neurons responded to pulses of 627 nm light at various intensities in a dose-dependent manner, confirming Ir25a expression in these neurons. No light-induced responses were found in control flies, which had the same genotype as experimental flies but were not fed all-trans retinal (-ATR), a necessary co-factor for channelrhodopsin function (see Materials and methods). We used similar optogenetic experiments to examine Ir25a expression in OSN classes innervating DM4 (ab2A, Or59b+) and DM5 (ab2B, Or85a/Or33b+) (Figure 4—figure supplement 1A and B), as well as D (ab9A, Or69aA/aB+) and VA3 (ab9B, Or67b+) (Figure 4—figure supplement 1C and D). These experiments indicated that Ir25a is expressed in ab2A (DM4) and ab9B (VA3) neurons, but not ab2B (DM5) or ab9A (D) neurons (see also Figure 4—source data 2 and Figure 4—source data 3). Results of these experiments are summarized in Table 3.
Identification of new OSN classes
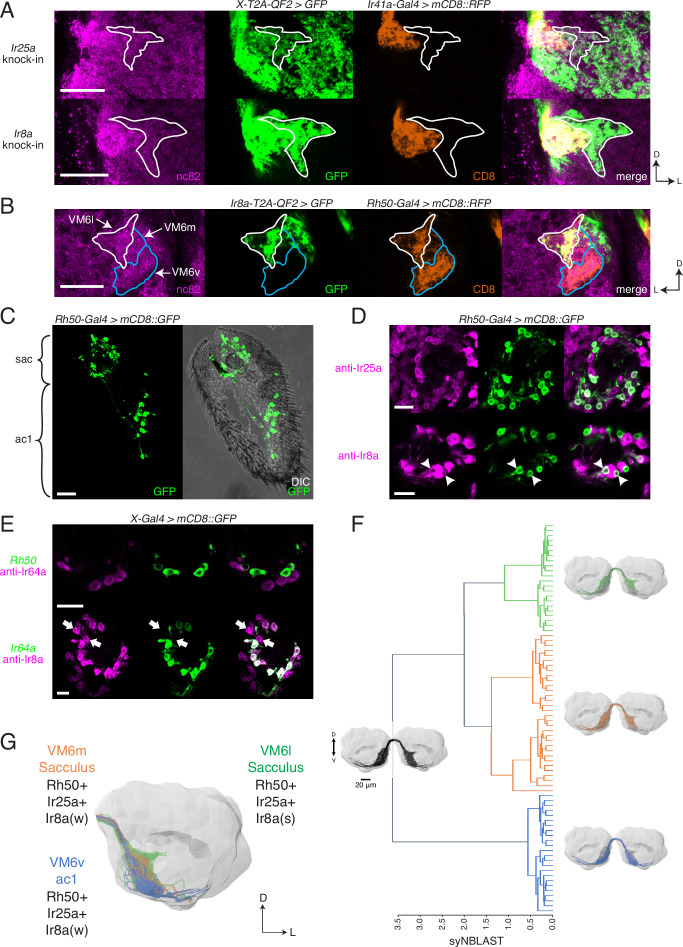
The co-receptor knock-ins allowed us to analyze the olfactory neuron innervation patterns for all AL glomeruli. Interestingly, the Ir8a-T2A-QF2 and Ir25a-T2A-QF2 knock-ins strongly labeled a previously uncharacterized posterior region of the AL. By performing a co-labeling experiment with Ir41a-Gal4, which labels the VC5 glomerulus, we narrowed down the anatomical location of this region and ruled out VC5 as the target of these axons (Figure 5A). While both knock-ins clearly labeled VC5, they also labeled a region lateral and slightly posterior to it (Figure 5A, outline). We performed additional co-labeling experiments with Ir8a-T2A-QF2 and various Gal4 lines labeling all known posterior glomeruli to confirm that this AL region did not match the innervation regions for other previously described OSN populations (Figure 5—figure supplement 1). We recognized that this novel innervation pattern appeared similar to a portion of the recently identified Rh50+ ammonia-sensing olfactory neurons (Vulpe et al., 2021). Co-labeling experiments with Rh50-Gal4 and Ir8a-T2A-QF2 confirmed that they indeed partially overlapped (Figure 5B). We determined that these Rh50+ olfactory neurons mapped to a portion of the VM6 glomerulus, with the strongly Ir8a+ region innervating the ‘horn’ of this glomerulus. The difference in innervation patterns between Ir8a+ and Rh50+ neurons in this AL region suggested at least two different subdivisions or OSN populations within this VM6 glomerulus. In fact, in between the main body of VM6 and the Ir8a+ horn there appeared to be a third region (Figure 5B, horn outlined in white, other two regions outlined in blue). We designated these subdivisions VM6l, VM6m, and VM6v (for lateral, medial, and ventral). We coordinated the naming of this glomerulus with recent connectomics analyses of the entire fly AL (Schlegel et al., 2021). In this connectomics study, dendrites of olfactory projection neurons were found to innervate the entire region described here as VM6l, VM6m, and VM6v. No projection neurons were identified to innervate only a subdomain. As such, the new VM6 nomenclature reflects this unique subdivision of a glomerulus by OSNs but not second-order projection neurons.
Figure 5. Identification of new olfactory sensory neuron (OSN) classes.
(A) Co-labeling experiments with Ir41a-Gal4 show that both Ir25a-T2A-QF2 and Ir8a-T2A-QF2 label the VC5 glomerulus (orange), and also a previously unidentified antennal lobe (AL) region (outline). (B) The new innervation pattern corresponds to the ‘horn’ (white outline) of the VM6 glomerulus labeled by Rh50+ neurons (orange). One portion of VM6 is strongly Ir8a+ (VM6l), while two other portions show little to no Ir8a expression (VM6m and VM6v, blue outlines). (C) Rh50-Gal4>GFP labels neurons in the sacculus (sac) and antennal coeloconic ac1 sensilla. (D) In the sacculus, all Rh50+ neurons appear to be Ir25a+ (top), and a subset are Ir8a+ (bottom, arrowheads). (E) Top: Rh50+ neurons in the sacculus do not overlap with Ir64a+ neurons. Bottom: there are two distinct populations of Ir8a+ neurons in the sacculus – those that are Ir64a+ and those that are Ir64a- (arrows). The latter likely correspond to Rh50+ neurons. (F) EM reconstructions of VM6 OSNs in a full brain volume (Dorkenwald et al., 2020) reveal three distinct subpopulations. (G) Model of OSN innervation of the VM6 region. VM6 can be subdivided into three OSN populations based on anatomical location in the periphery and chemoreceptor expression: VM6v (blue) OSNs originate in ac1, strongly (s) express Rh50 and Ir25a, and weakly (w) or infrequently express Ir8a; VM6m (orange) neurons originate in the sacculus and have a similar chemoreceptor expression profile to VM6v; VM6l (green) OSNs originate in the sacculus but strongly express Ir8a in addition to Rh50 and Ir25a. Compass: D = dorsal, L = lateral. Scale bars: 20 µm in (A–C) and (F), 10 µm in (D, E). N = 9–11 for (C–E). See also Figure 5—figure supplement 1 and Tables 3 and 4.
Figure 5—figure supplement 1. The new glomerular region labeled by the Ir8a knock-in does not correspond to previously identified posterior glomeruli.
We sought to determine the identity of the olfactory neurons that might be innervating these three VM6 subdivisions. Rh50+ neurons can be found in two regions of the antenna: ac1 coeloconic sensilla and the sacculus (Figure 5C; Vulpe et al., 2021). The shape of the VM6v subdomain most closely matches the glomerulus described as VM6 by previous groups (e.g., Couto et al., 2005; Endo et al., 2007), which had been suggested to be innervated by coeloconic sensilla (Chai et al., 2019; Li et al., 2016). In addition, antibody staining had previously shown that Rh50+ ac1 neurons broadly co-express Ir25a but generally not Ir8a (Vulpe et al., 2021). This suggested that the other VM6 subdomains might be innervated by the Rh50+ sacculus olfactory neurons. Antibody staining in Rh50-Gal4>GFP antennae confirmed co-expression with both Ir25a protein (broad overlap) and Ir8a protein (narrow overlap) in the third chamber of the sacculus (Figure 5D; quantified in Table 4). Most sacculus neurons appear to be Ir25a+, and in contrast to the Ir8a knock-in, the three VM6 subdivisions are all strongly innervated by the Ir25a knock-in (Figure 5A). Two previously described OSN populations in the third chamber of the sacculus had been characterized to express Ir8a along with Ir64a and innervate the DP1m and DC4 glomeruli (Ai et al., 2013; Ai et al., 2010). To demonstrate that the Rh50+ Ir8a+ sacculus neurons represented a distinct olfactory neuron population, we performed immunohistochemistry experiments in Rh50-Gal4>GFP antennae with an anti-Ir64a antibody (Figure 5E, top), and in Ir64a-Gal4>GFP antennae with an anti-Ir8a antibody (Figure 5E, bottom). These experiments confirmed a new, distinct population of Ir8a+ Ir64a- cells in the sacculus.
Table 4. Co-expression of Rh50 and Ir8a in the sacculus (related to Figure 5).
Antennal cryosections of Rh50-Gal4>GFP flies were stained with an anti-Ir8a antibody, and the overlap of Ir8a+ and GFP+ cells was quantified in the sacculus. 22% of Ir8a+ cells expressed Rh50, 35% of Rh50+ cells expressed Ir8a, and 16% of all cells were double labeled. N = 11.
| Genotype | Sample | Ir8a+ cells | GFP+ cells | Double-labeled cells | Total cells |
|---|---|---|---|---|---|
| Rh50-Gal4>GFP | 20210226 a1 | 18 | 9 | 2 | 25 |
| Rh50-Gal4>GFP | 20210226 a2 | 22 | 15 | 4 | 33 |
| Rh50-Gal4>GFP | 20210226 a3 | 41 | 22 | 7 | 56 |
| Rh50-Gal4>GFP | 20210226 a4 | 41 | 14 | 5 | 50 |
| Rh50-Gal4>GFP | 20210129 a1 | 26 | 20 | 9 | 37 |
| Rh50-Gal4>GFP | 20210129 a2 | 32 | 24 | 7 | 49 |
| Rh50-Gal4>GFP | 20210129 a3 | 29 | 19 | 7 | 41 |
| Rh50-Gal4>GFP | 20210216 a1 | 26 | 21 | 8 | 39 |
| Rh50-Gal4>GFP | 20210216 a2 | 30 | 18 | 7 | 41 |
| Rh50-Gal4>GFP | 20210216 a3 | 34 | 23 | 8 | 49 |
| Rh50-Gal4>GFP | 20210216 a4 | 34 | 23 | 9 | 48 |
| Total across samples: | 333 | 208 | 73 | 468 | |
| Proportion of Ir8a+ cells that are GFP+: | Proportion of GFP+ cells that are Ir8a+: | Proportion of all cells that are double labeled: | |||
| 0.22 | 0.35 | 0.16 |
The VM6l olfactory projections are difficult to identify in the hemibrain connectome (Scheffer et al., 2020) due to the medial truncation of the AL in that dataset (see Schlegel et al., 2021 for additional details). Here, we used FlyWire (Dorkenwald et al., 2020), a recent segmentation of a full adult fly brain (FAFB) (Zheng et al., 2018), to reconstruct the VM6 OSN projections in both left and right ALs. Synapse-based hierarchical clustering (syNBLAST) (Buhmann et al., 2021) of the VM6 OSNs demonstrated the anatomical segregation into three distinct subpopulations: VM6l, VM6m, and VM6v (Figure 5F). This subdivision was subsequently confirmed in a reanalysis of the VM6 glomerulus in the hemibrain dataset (Schlegel et al., 2021). Olfactory neurons innervating VM6l were strongly Ir8a+, while olfactory neurons innervating VM6m and VM6v were weakly and sparsely Ir8a+ (see Figure 3—source data 2, page 3). This pattern may be due to Ir8a expression in only one or a few cells.
Based on the EM reconstructions, genetic AL analyses, and peripheral staining experiments, we propose a model of the anatomical locations and molecular identities of the olfactory neurons innervating the VM6 subdivisions (Figure 5F). All VM6 subdivisions broadly express Rh50 and Ir25a; the VM6v OSNs are housed in ac1 sensilla and express Ir8a either weakly or only in a small subset of neurons; both the VM6m and VM6l OSNs are found in the sacculus and can be distinguished by their levels or extent of Ir8a expression, with VM6l neurons being strongly Ir8a+. Because all three VM6 subdivisions share the same downstream projection neurons, this AL region has been classified as a single glomerulus (Schlegel et al., 2021). We maintain this convention here, for a total of 58 AL glomeruli. It is possible that this number may need to be re-evaluated in the future, and the three VM6 subdivisions reconsidered as bona fide separate glomeruli (bringing the OSN glomerular total to 60). Such a separation might be warranted if it is found that these OSN populations express different tuning receptors, and those receptors respond to different odorants.
Table 3 summarizes the chemosensory receptor expression patterns for all four co-receptor knock-in lines across all OSNs, sensillar types, and glomeruli. For clarity, this summary considers the newly identified OSN populations described here separately. We find that Orco-T2A-QF2 consistently labels 45 total glomeruli out of 58 (7 more than previously reported); Ir8a-T2A-QF2 consistently labels 18 glomeruli (8 more than previously identified); Ir76b-T2A-QF2 consistently labels 15 glomeruli (11 more than previously identified); and Ir25a-T2A-QF2 consistently labels 51 glomeruli (39 more than previously identified).
Co-receptor contributions to olfactory neuron physiology
How might the broad, combinatorial co-expression of various chemosensory families affect olfactory neuron function? To begin to address this question, we examined olfactory responses in neuronal populations co-expressing just two of the four chemosensory receptor families (Orco and Ir25a). We chose to test eight OSN classes previously assigned to the Orco+ domain that we found to have strong or intermediate Ir25a expression – two in the antennae and six in the maxillary palps. The two antennal OSN classes are found in the same ab3 sensillum (ab3A, Or22a/b+, DM2 glomerulus; and ab3B, Or85b+, VM5d glomerulus). The six palpal OSN classes represent the entire known olfactory neuron population of the maxillary palps (pb1A, Or42a+, VM7d; pb1B, Or71a+, VC2; pb2A, Or33c/Or85e+, VC1; pb2B, Or46a+, VA7l; pb3A, Or59c+, VM7v; pb3B, Or85d+, VA4). In both the antennae and the palps, we compared the olfactory responses of OSNs to a panel of 13 odorants in three genotypes: wildtype, Ir25a2 mutant, and Orco2 mutant flies. This panel included odorants typically detected by ORs, such as esters and aromatics, and odorants typically detected by IRs, such as acids and amines (Silbering et al., 2011). In the previously accepted view of olfaction in D. melanogaster, Orco+ neurons express only Orco/OrX receptors, and all olfactory responses in the neurons can be attributed to these receptors. Thus, in an Ir25a2 mutant background, there should be no difference in olfactory responses from wildtype if either (a) Ir25a is not expressed in these neurons or (b) Ir25a is expressed, but is not playing a functional role in these neurons. In an Orco2 mutant background, there would be no trafficking of Orco/OrX receptors to the dendritic membrane, and no formation of functional ion channels (Benton et al., 2006; Larsson et al., 2004). Thus, in the traditional view of insect olfaction, Orco2 mutant neurons should have no odor-evoked activity. However, in the new co-receptor co-expression model of olfaction, if Ir25a is contributing to olfactory responses in Orco+ neurons, then mutating this co-receptor might affect the response profiles of these neurons. Similarly, Orco2 mutant neurons that co-express Ir25a might retain some odor-evoked activity.
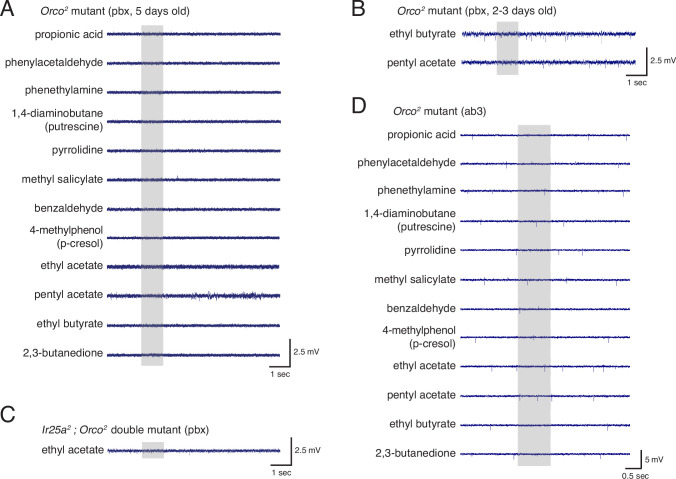
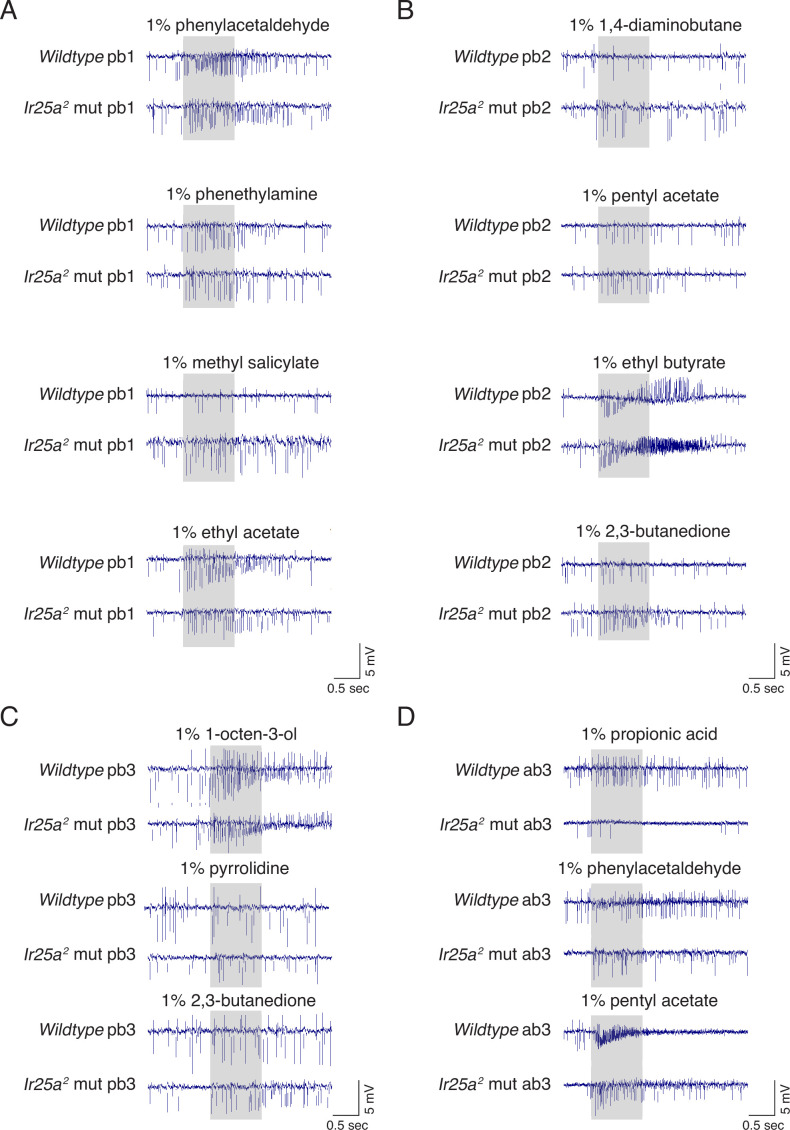
We first examined olfactory responses in palp basiconic sensilla. In the palps, three types of basiconic sensilla (pb1, pb2, and pb3) contain two neurons each (A and B) (Figure 6A), for a total of six OSN classes (Couto et al., 2005; de Bruyne et al., 1999; Fishilevich and Vosshall, 2005; Goldman et al., 2005; Ray et al., 2008; Ray et al., 2007). We found robust responses to several odorants in our panel in both the wildtype and Ir25a2 mutant flies, including odorants like 1-octen-3-ol typically considered as an OR ligand (Figure 6B), and IR ligands like pyrrolidine. Neither odor-evoked nor spontaneous activity was detected in the Orco2 mutant (Figure 6B, bottom row; see also Figure 6—figure supplement 1A). This was true of all sensilla tested in the palps. The SSR experiments in Figure 6A–D were performed at 4–8 DPE. We recently discovered that neurodegeneration of Orco2 mutant olfactory neurons occurs in the palps by ~6 DPE (Task and Potter, 2021), which could potentially confound our interpretation. We repeated the experiments in young (1–3 DPE) flies but similarly detected neither odor-evoked activity nor spontaneous activity in Orco2 mutant palpal neurons (Figure 6—figure supplement 1B). There was also no spontaneous or odor-evoked activity in an Ir25a2; Orco2 double mutant (Figure 6—figure supplement 1C). This suggests one of three possibilities: first, Orco2 mutant neurons in the palps could already be dysfunctional at this early stage, despite not yet showing cell loss, and Ir25a-dependent activity is not sufficient to maintain either baseline or stimulus-induced activity; second, Ir25a function may be Orco-dependent in these cells, or act downstream of Orco, such that loss of Orco function affects Ir25a function; third, we did not stimulate neurons with an Ir25a-dependent odorant. The latter possibility would not, however, explain why there is no spontaneous activity in these cells. Future experiments will be needed to address these possibilities. Given the lack of neuronal activity in the Orco2 mutant, we focused subsequent analyses in the palps on the two other genotypes: wildtype and Ir25a2.
Figure 6. Co-receptor contributions to olfactory neuron physiology.
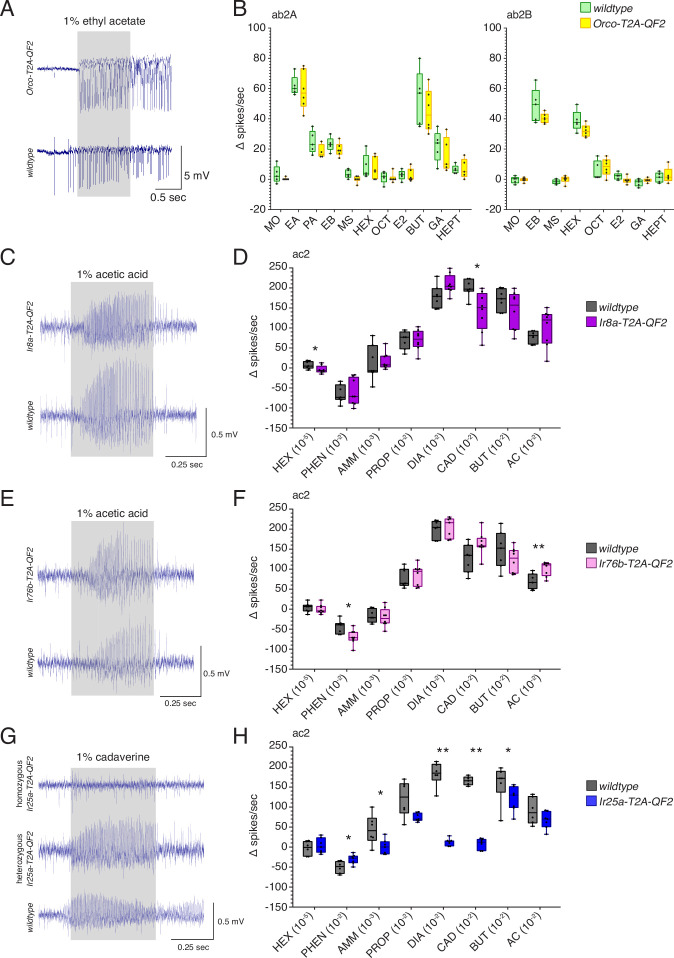
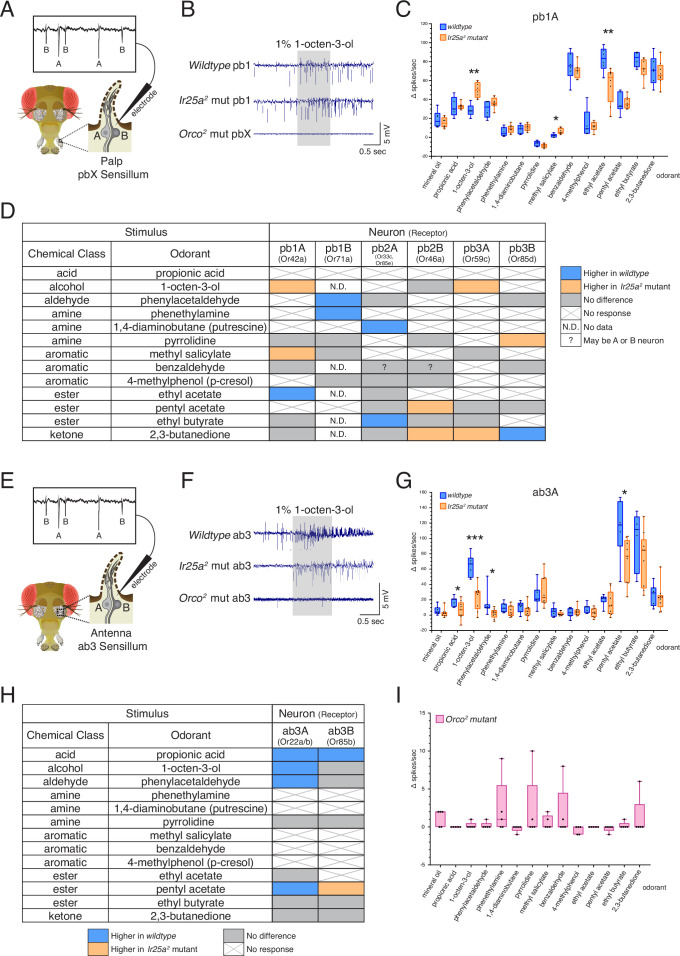
(A–I) Single sensillum recording (SSR) experiments were performed in three genetic backgrounds: wildtype, Ir25a2 mutant, and Orco2 mutant flies. A panel of 13 odorants was tested. In all box plots, *p<0.05, **p<0.01, and ***p<0.001. (A) Cartoon of a fly head, zooming in on a single sensillum in the palp. Each palpal sensillum (pbX) contains two neurons, A and B. An electrode is inserted into the sensillum, and neuronal activity is recorded in response to odorants. Activity of the A and B neurons can be distinguished based on their spike amplitudes (top). (B) Representative traces from recordings in palp basiconic pb1 sensilla in the three genotypes in response to 1% 1-octen-3-ol. Sensilla were identified based on responses to reference odorants (de Bruyne et al., 1999; see Materials and methods). The Orco2 mutant did not exhibit odor-evoked activity nor spontaneous activity, making it difficult to determine the identity of the recorded sensillum. Orco2 mutant sensilla are thus denoted pbX. (C) Quantification of responses to the panel of odorants in wildtype (blue; N = 5–9 flies) and Ir25a2 mutant (orange; N = 6–10 flies) pb1A neurons. Responses were higher in the Ir25a2 mutant than in the wildtype for 1-octen-3-ol and methyl salicylate, and lower in the Ir25a2 mutant for ethyl acetate. Mann–Whitney U tests indicated these differences were statistically significant: 1-octen-3-ol: MdnIr25amut = 50, Mdnwildtype = 28, U(NIr25amut = 8, Nwildtype = 5) = 0, p=0.0016; methyl salicylate: MdnIr25amut = 5, Mdnwildtype = 2, U(NIr25amut = 7, Nwildtype = 5) = 3, p=0.0177; ethyl acetate: MdnIr25amut = 63.5, Mdnwildtype = 83.5, U(NIr25amut = 8, Nwildtype = 6) = 4, p=0.008. (D) Summary of differences in responses across all six neuron classes in the palps between wildtype and Ir25a2 mutant flies. Comparisons were made using Mann–Whitney U tests. Orange indicates higher response in Ir25a2 mutant, blue indicates higher response in wildtype. Gray is no difference between genotypes, X indicates no response to the given stimulus, and N.D. is no data (strong A neuron response obscured B neuron spikes preventing quantification). In the wildtype, for one sensillum-odorant combination (pb2 and benzaldehyde), it could not be distinguished if responses arose from the A or B neuron or both (indicated by a question mark). (E) Fly head cartoon, zooming in on a single sensillum in the antenna. We recorded from antennal ab3 sensilla, each of which contains two neurons, A and B. As in the palps, responses from these neurons can be distinguished based upon their spike amplitude (top). (F) Representative traces from recordings in antennal basiconic ab3 sensilla in the three genotypes in response to 1% 1-octen-3-ol. In Orco2 mutant ab3 sensilla spontaneous activity was observed, but there was no significant odor-evoked activity. Wildtype N = 7 sensilla from five flies; Ir25a2 mutant N = 10 sensilla from five flies. (G) Quantification of responses in wildtype (blue; N = 7) and Ir25a2 mutant (orange; N = 9) ab3A neurons. Responses were significantly higher in wildtype compared to Ir25a2 mutant ab3A neurons for four odorants (Mann–Whitney U results in parentheses; all Nwildtype = 7 and NIr25amut = 9): propionic acid (Mdnwildtype = 21, MdnIr25amut = 7, U = 12.5, p=0.0441); 1-octen-3-ol (Mdnwildtype = 67, MdnIr25amut = 29, U = 1.5, p=0.0004); phenylacetaldehyde (Mdnwildtype = 10, MdnIr25amut = 3, U = 9, p=0.015); and pentyl acetate (Mdnwildtype = 118, MdnIr25amut = 77, U = 9, p=0.0164). Difference between wildtype and Ir25a2 mutant to phenylacetaldehyde is significant even with the large wildtype outlier removed (p=0.0336). (H) Summary of differences in responses in the two neuron classes in ab3 between wildtype and Ir25a2 mutant flies. Comparisons were made using Mann–Whitney U tests. Orange indicates higher response in Ir25a2 mutant, blue indicates higher response in wildtype, gray is no difference between genotypes, and X is no response to the given stimulus. One Ir25a2 mutant fly was excluded from analyses as it had high responses to the mineral oil control (40–53 Δ spikes/s), not seen in any other animal of any genotype. (I) Weak responses in Orco2 mutant flies to certain stimuli (≤10 Δ spikes/s) were occasionally detected. While there were some statistically significant differences from mineral oil control (pentyl acetate p=0.0109, propionic acid p=0.0434, ethyl acetate p=0.0434, 1,4-diaminobutane p=0.0109, p-cresol p=0.0021), these were not deemed biologically significant due to very small Δ spike values relative to zero. For more details, see Materials and methods. N = 5 flies. See also Figure 6—figure supplements 1–3 and Figure 6—source data 1.
Figure 6—figure supplement 1. Electrophysiological experiments to examine Ir25a function in Orco+ neurons.
Figure 6—figure supplement 2. No IrX expression of the top candidates in the maxillary palps.
Figure 6—figure supplement 3. Example traces for odorants eliciting differences between wildtype and Ir25a mutant sensilla.
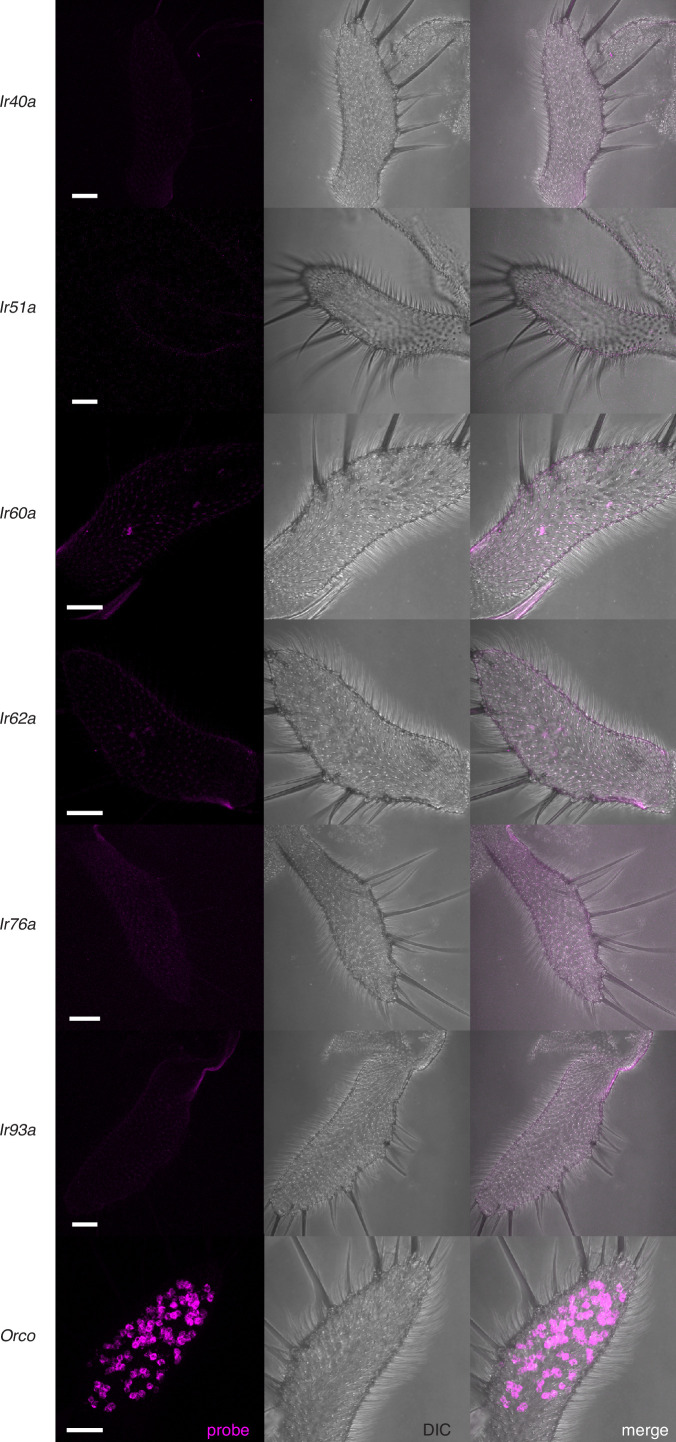
The response in the pb1A neuron to 1-octen-3-ol was significantly higher in the Ir25a2 mutant compared to the wildtype (Mann–Whitney U test, p=0.0016), as was the response to methyl salicylate (p=0.0177), while the response to ethyl acetate (EA) was higher in wildtype (p=0.008) (Figure 6C; see Figure 6—source data 1 for results of all statistical analyses). The differences in responses across all six OSN classes in the palps between wildtype and Ir25a2 mutant flies are summarized in Figure 6D. In each neuron class, we found 1–3 odorants whose response profiles differed between the two genotypes. However, the specific stimuli eliciting different responses, and the directionality of those responses, varied. For example, 2,3-butanedione elicited higher responses in the Ir25a2 mutant in both pb2B and pb3A neurons, but lower responses in the mutant (higher in the wildtype) in pb3B. Interestingly, when we examined a list of candidate IrX tuning receptors (Li et al., 2021) in the palps using in situs, we did not find expression (see Figure 6—figure supplement 2 and Appendix 1—key resources table). This suggests that Ir25a may not be functioning as a traditional co-receptor in Orco+ olfactory neurons in the palps (an expanded role for Ir25a beyond co-reception has previously been suggested; see Budelli et al., 2019; Chen et al., 2015).
We next examined olfactory responses in antennal basiconic ab3 sensilla in wildtype, Ir25a2 mutant, and Orco2 mutant flies (Figure 6E–I). As in the palps, ab3 sensilla contain two neurons, A and B (Figure 6E). In contrast to the palps, Orco2 mutant ab3 sensilla did occasionally show spontaneous activity (Figure 6F, bottom row; see Figure 6—figure supplement 1D and Figure 6—figure supplement 3 for additional example traces). Although there are two Orco+ neurons in this sensillum, we consistently observed only a single spike amplitude in the Orco2 mutant. Thus, we cannot determine at this time whether this activity arises from the A or B neuron. We occasionally observed small responses (≤10 Δ spikes/s) in the Orco2 mutant; however, across all flies tested, these responses were not significantly different from the mineral oil control (Figure 6I; statistical analyses can be found in Figure 6—source data 1). For these reasons, Orco2 mutant flies were excluded from the analyses in Figure 6G and H.
As in the palps, we found significant differences in the responses of both ab3A and ab3B neurons to some odorants between the two genotypes. A comparison of all ab3A responses between the wildtype and Ir25a2 mutant genotypes is shown in Figure 6G, and results from both the A and B neurons are summarized in Figure 6H (Mann–Whitney U, as in Figure 6A–C; see Figure 6—source data 1 for all analyses). In the ab3A neuron, the wildtype showed higher responses to propionic acid (p=0.0441), 1-octen-3-ol (p=0.0004), phenylacetaldehyde (p=0.015), and pentyl acetate (p=0.0164). Interestingly, two of these four odorants are typically associated with IRs (propionic acid and phenylacetaldehyde). In the ab3B neuron, only two odorants elicited significantly different responses between the wildtype and Ir25a2 mutant: propionic acid (response higher in wildtype, as with ab3A; p=0.0388), and pentyl acetate (response higher in mutant, in contrast to ab3A; p=0.0385). While responses to propionic acid are small in both ab3 neurons, they are abolished in the Ir25a2 mutant background (Kruskal–Wallis with uncorrected Dunn’s comparing odorant responses to mineral oil control; ab3A p=0.3957; ab3B p=0.5184), suggesting that propionic acid detection in ab3 may be Ir25a-dependent.
Co-receptor co-expression in other insect olfactory organs
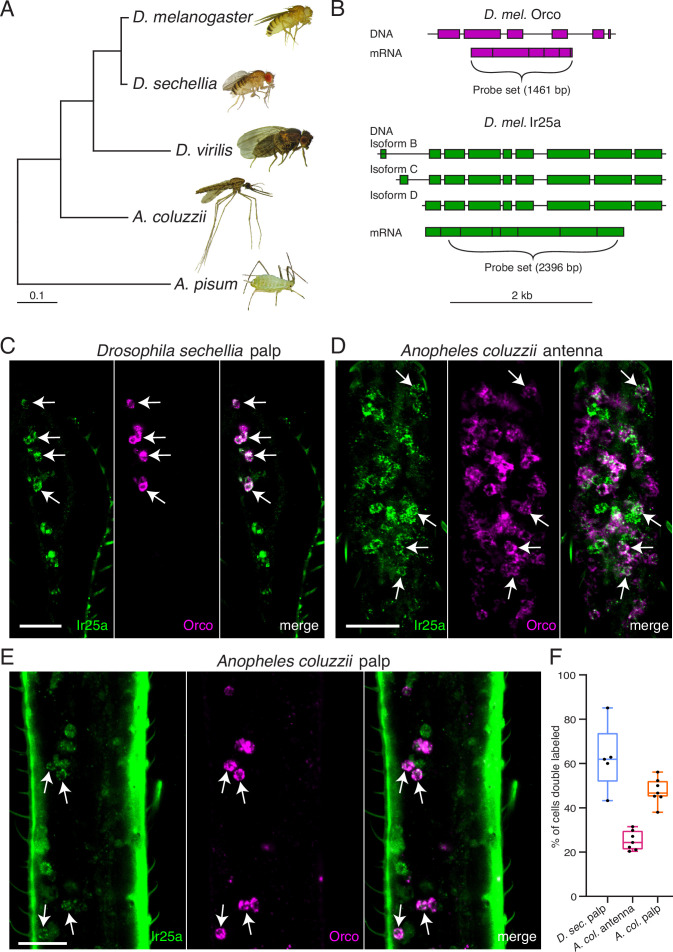
To determine if co-receptor co-expression might exist in other insects besides D. melanogaster, we used RNA in situ hybridization to examine expression of Orco and Ir25a orthologues in the fly D. sechellia and in the mosquito A. coluzzii (Figure 7). D. melanogaster and D. sechellia diverged approximately 5 million years ago (Hahn et al., 2007), while the Drosophila and Anopheles lineages diverged nearly 260 million years ago (Gaunt and Miles, 2002; Figure 7A). Because co-receptor sequences are highly conserved, we could use our D. mel. Orco and Ir25a in situ probes (Figure 7B) to examine the expression of these genes in the maxillary palps of D. sechellia. We found widespread co-expression of Orco and Ir25a (63% of all cells were double labeled), consistent with our findings in D. melanogaster (Figure 7C). For A. coluzzii mosquitoes, we designed Anopheles-specific Orco and Ir25a probes, and examined co-receptor co-expression in antennae (Figure 7D) and maxillary palps (Figure 7E). We observed broad co-expression of AcOrco and AcIr25a in the maxillary palp capitate peg sensilla (47% of all cells were double labeled), and narrower co-expression in the antennae (25% double labeled). Co-expression results for all tissues examined are summarized in Figure 7F, and cell counts can be found in Table 5. These results suggest that Orco and Ir25a co-receptor co-expression extends to other Drosophilid species as well as mosquitoes (see also Ye et al., 2021; Younger et al., 2020).
Figure 7. Orco and Ir25a are co-expressed in Drosophila sechellia and Anopheles coluzzii olfactory organs.
(A) Phylogenetic tree based on the Orco sequences from the five insects shown (D. = Drosophila, A. coluzzii = Anopheles coluzzii, A. pisum = Acyrthosiphon pisum). Evolutionary history was inferred using the Maximum Likelihood method and Tamura–Nei model (Tamura and Nei, 1993). Pea aphid image A was reproduced from PLoS Biology Issue Image (2010). (B) The Drosophila melanogaster Orco in situ probe set, which covers the entire Orco coding sequence (top, magenta), was used to examine Orco expression in the maxillary palps of other Drosophila fly species. We designed a new probe set covering the most conserved portion of D. mel. Ir25a (bottom) as determined by analyzing the Ir25a sequences from multiple fly species and comparing them to the various Drosophila melanogaster Ir25a isoforms (three of which are illustrated in green). (C) Many olfactory sensory neurons (OSNs) in the Drosophila sechellia maxillary palps co-express both Orco and Ir25a, as revealed by in situ experiments (four example cells indicated with arrows). N = 5. (D) In situs in Anopheles coluzzii antennae reveal a small proportion of cells expressing both co-receptors (arrows). N = 7. (E) In situs in Anopheles coluzzii maxillary palps show many cells express both Orco and Ir25a (four examples indicated with arrows). N = 7. (F) Summary of co-expression analyses in (C–E). For each olfactory organ examined, we divided the number of Orco+ Ir25a+ double-labeled cells by the total number of cells labeled by either probe. We found that 63% of D. sec. palpal OSNs express both Orco and Ir25a (blue), 25% of A. col. antennal OSNs express both co-receptors (pink), and 47% of A. col. palpal OSNs are double labeled (orange). (C–E) are maximum intensity projections of partial z-stacks. See also Table 5, and Appendix 1—key resources table.
Table 5. Co-expression of Orco and Ir25a in non-melanogaster insect olfactory organs (related to Figure 7).
Whole-mount palps from Drosophila sechellia flies, and whole-mount antennae and palps from Anopheles coluzzii mosquitoes, were examined using fluorescence in situ hybridization with probe sets against Orco and Ir25a. Co-expression between Orco and Ir25a co-receptors was observed in both insects, with D. sec. palps having the highest degree of co-expression (63% of cells double labeled) and A. col. antennae having the lowest (25% of cells double labeled). N = 5 for D. sec. and 7 for A. col.
| Species | Sample | Orco+ cells | Ir25a+ cells | Double-labeled cells | Total cells |
|---|---|---|---|---|---|
| Drosophila sechellia | Palp 1 | 70 | 44 | 44 | 70 |
| Drosophila sechellia | Palp 2 | 64 | 48 | 42 | 70 |
| Drosophila sechellia | Palp 3 | 63 | 39 | 39 | 63 |
| Drosophila sechellia | Palp 4 | 78 | 38 | 35 | 81 |
| Drosophila sechellia | Palp 5 | 86 | 75 | 74 | 87 |
| Total across samples: | 361 | 244 | 234 | 371 | |
| Proportion of Orco+ cells that are Ir25a+: | Proportion of Ir25a+ cells that are Orco+: | Proportion of all cells that are double labeled: | |||
| 0.65 | 0.96 | 0.63 | |||
| Anopheles coluzzii | Antenna 1 | 52 | 17 | 12 | 57 |
| Anopheles coluzzii | Antenna 2 | 47 | 24 | 17 | 54 |
| Anopheles coluzzii | Antenna 3 | 57 | 20 | 13 | 64 |
| Anopheles coluzzii | Antenna 4 | 50 | 27 | 14 | 63 |
| Anopheles coluzzii | Antenna 5 | 62 | 30 | 18 | 74 |
| Anopheles coluzzii | Antenna 6 | 53 | 21 | 17 | 57 |
| Anopheles coluzzii | Antenna 7 | 49 | 26 | 16 | 59 |
| Total across samples: | 370 | 165 | 107 | 428 | |
| Proportion of Orco+ cells that are Ir25a+: | Proportion of Ir25a+ cells that are Orco+: | Proportion of all cells that are double labeled: | |||
| 0.29 | 0.65 | 0.25 | |||
| Anopheles coluzzii | Palp 1 | 34 | 30 | 23 | 41 |
| Anopheles coluzzii | Palp 2 | 30 | 36 | 22 | 44 |
| Anopheles coluzzii | Palp 3 | 26 | 35 | 19 | 42 |
| Anopheles coluzzii | Palp 4 | 35 | 34 | 19 | 50 |
| Anopheles coluzzii | Palp 5 | 32 | 39 | 22 | 49 |
| Anopheles coluzzii | Palp 6 | 31 | 35 | 21 | 45 |
| Anopheles coluzzii | Palp 7 | 30 | 37 | 23 | 44 |
| Total across samples: | 218 | 246 | 149 | 315 | |
| Proportion of Orco+ cells that are Ir25a+: | Proportion of Ir25a+ cells that are Orco+: | Proportion of all cells that are double labeled: | |||
| 0.68 | 0.61 | 0.47 |
The co-receptor co-expression map of olfaction in D. melanogaster
Co-receptor co-expression of insect chemosensory receptors suggests that multiple receptors may influence the response properties of an olfactory neuron, as we have shown in ab3 and palpal sensilla. To aid future investigations of co-receptor co-expression signaling, we synthesized our results (Table 3) into a comprehensive new map of the AL. Figure 8 summarizes the expression patterns of all the co-receptor knock-in lines and presents a new model for chemosensory receptor expression in D. melanogaster. In Figure 8A, the expression pattern of each knock-in line is presented separately (see also Figure 3—source data 1). The new AL map is updated with the recent reclassification of VP1 into three glomeruli (Marin et al., 2020) and indicates the new VM6 subdivisions. In Figure 8A, the original glomerular innervation pattern for each co-receptor is shown in green, with new innervation revealed by the T2A-QF2 knock-in lines color coded by intensity: strongly labeled glomeruli are in orange, intermediate glomeruli in yellow, and weakly labeled glomeruli are in pink. Glomeruli labeled in <50% of brains examined are designated variable (gray), and glomeruli not labeled by the given knock-in are in white. The new VM6v, VM6m, and VM6l subdivisions are labeled with gray stripes.
Figure 8. The co-receptor co-expression map of olfaction in Drosophila melanogaster.
(A) Summary of antennal lobe (AL) expression for all co-receptor knock-in lines (from all brains examined in Figures 3—5; Orco N = 8, Ir8a N = 15, Ir76b N = 11, Ir25a N = 15). The previously reported innervation pattern for each co-receptor is shown in green; new innervation reported here is color-coded according to strength of glomerular labeling, from strong (orange), to intermediate (yellow), to weak (pink). Glomeruli labeled in <50% of brains examined for a given knock-in line are designated variable (gray); glomeruli not labeled are white. The novel VM6 glomerular subdivisions reported here are indicated by gray stripes. (B) Overlap of chemosensory modalities in the AL. In the Venn diagram (left), IR co-receptors are color-coded in shades of purple, while Orco is in teal, as in Figure 1. Numbers indicate how many glomeruli are found in the given intersection of co-receptors out of 58 total glomeruli. Variably labeled glomeruli were excluded from these analyses. The table lists the names of the glomeruli in each section of the Venn diagram. The new glomerular subdivisions are indicated with an asterisk. (C) New view of olfaction in Drosophila. Left: in the periphery, all four co-receptors are expressed in the antenna (top), while palpal neurons express Orco and Ir25a (bottom). Middle: many different classes of olfactory sensory neurons (OSNs) express various combinations of chemosensory receptors and co-receptors. While some neurons express only IrCos (purple, #1) or Orco (teal, #2), many neurons co-express these chemoreceptors (indicated with striped fill, #3 and 4). Within the latter group, there may be OSN populations in which IRs are the dominant receptors, and OR expression is sparse (#3), and other populations where ORs are the primary receptors and IR expression is infrequent (#4). GR+ neurons (dark blue) also express Ir25a (#5, dark blue and purple striped fill), and some of these neurons additionally express Orco (#5, dark blue, purple, and teal striped fill). Question marks indicate potential instances of co-convergence of different subtypes of OSNs onto the same glomeruli. Right: a comprehensive map of the antennal lobe shows that most glomeruli are innervated by OSNs that co-express multiple chemoreceptors. Compass in (A) and (C): D = dorsal, L = lateral, P = posterior. See also Table 3 and Figure 3—source data 1 and Figure 3—source data 2.
In the previous model of olfaction in Drosophila, the Orco/OR domain primarily occupied the anterior AL, while the IR domains innervated more posterior glomeruli. While the former is still, for the most part, accurate (Figure 8A, Orco), the latter is not: both Ir8a-T2A-QF2 and Ir76b-T2A-QF2 label several more anterior glomeruli (such as VA3 or VA6), and Ir25a-T2A-QF2 labels the majority of glomeruli throughout the anterior to posterior axis (Figure 8A, Ir25a). The expansion of the Ir25a+ domain is the most dramatic of the four co-receptors: previously, Ir25a+ glomeruli accounted for 21% of the AL (12/58 glomeruli) (Enjin et al., 2016; Frank et al., 2017; Marin et al., 2020; Silbering et al., 2011); the Ir25a-T2A-QF2 knock-in consistently labels 88% of the AL (51/58 glomeruli, excluding variable). This represents a greater than fourfold expansion. Similarly, the number of Ir76b+ glomeruli increased more than threefold, from 7% of the AL (4/58 glomeruli) (Silbering et al., 2011) to 26% (15/58, excluding variable). The Ir8a+ domain has nearly doubled, from 17% of the AL originally (10/58 glomeruli) (Silbering et al., 2011) to 31% (18/58 glomeruli, excluding variable). The most modest increase in reported expression is in the Orco+ domain: from 66% of the AL (38/58 glomeruli) (Couto et al., 2005; Fishilevich and Vosshall, 2005) to 78% (45/58, excluding variable).
The expression overlap in the AL of the four co-receptor families is summarized in the Venn diagram shown in Figure 8B (excluding the variably labeled glomeruli from Figure 8A). The table at the right lists the names of the glomeruli that correspond to the sections of the Venn diagram. This analysis reveals nine glomeruli labeled by all four knock-in lines; furthermore, it shows that the Ir8a+ and Ir76b+ domains do not have glomeruli unique to them. Most of the AL is innervated by Orco+ Ir25a+ neurons (25 glomeruli that are only Orco+ Ir25a+, plus an additional 13 that have Orco, Ir25a, and one or both other co-receptors). The Orco+ and Ir25a+ domains reveal glomeruli unique to them (six glomeruli that are only Orco+, seven glomeruli that are only Ir25a+). Expression analyses also reveal that Ir8a does not co-express with Orco alone or Ir76b alone.
A unified AL map organized by chemosensory gene families (ORs, IRs, and GRs) is shown in Figure 8C (right panel), and the left two panels extend this information into the periphery. Here, we include the GR+innervation of the V glomerulus. However, a knock-in line for either Gr21a or Gr63a does not currently exist; thus, it is possible these receptors (as well as other poorly characterized antennal GRs) might also be more broadly expressed than previous transgenic lines indicate (Fujii et al., 2015; Menuz et al., 2014). All four OR and IR co-receptors are expressed in the antenna, while olfactory neurons in the palps express Orco and Ir25a (Figure 8C, left panel). In the antennae, there are many different classes of OSNs expressing various combinations of chemosensory receptors and co-receptors: there are Orco+ only neurons (Figure 8C, middle panel, #2), such as those innervating the VM2 and VM3 glomeruli (teal); IrCo+ only neurons (purple), which include neurons expressing one, two, or all three IR co-receptors (such as VP2, VM6v, or DP1m, respectively) (Figure 8C, middle panel, #1); and neurons expressing both Orco and IrCo(s) (teal and purple stripe) (Figure 8C, middle panel, #3 and 4).
The expression data suggest that different subpopulations of olfactory neurons might be targeting a shared glomerulus. Our data indicate that both Orco+ and Ir25a+ neurons innervate the GR+ V glomerulus (dark blue; see also Figure 8A). Based on the sparse innervation of the V glomerulus by the Orco-T2A-QF2 knock-in (Figure 3A) and the lower expression levels in the snRNAseq data (Figure 4A), we hypothesize that Orco may be expressed in only a subset of Gr21a/Gr63a+ neurons. This contrasts with the Ir25a-T2A-QF2 knock-in, which appears to label most Gr21a/Gr63a+ neurons. Thus, two subpopulations of neurons may be co-converging upon the same V glomerulus: neurons that express Gr21a/Gr63a and Ir25a (dark blue and purple stripes), and neurons that express Gr21a/Gr63a, Ir25a, and Orco (dark blue, purple, and teal stripes) (Figure 8C, middle panel, #5). Such co-convergence has recently been shown in the olfactory system of Aedes aegypti mosquitoes (Younger et al., 2020). Similarly, the sparse Orco-T2A-QF2 knock-in innervation of the DP1l glomerulus suggests that there are OSN populations expressing mostly IRs, but with a subset of neurons that additionally express Orco (Figure 8C, middle panel, #3). The converse may also be possible (Figure 8C, middle panel, #4): OSN populations that have some neurons expressing only Orco, and a subset expressing both Orco and IrCo(s) co-converging onto the same glomerulus. There is some evidence for this in the palps, based on our anti-Orco and anti-Ir25a antibody staining (Figure 2C and I, Figure 4B, Table 2). The snRNAseq data suggest that this may also be the case in the antennae (see Figure 4—source data 1).
Discussion
Here, we present evidence of widespread chemosensory co-receptor co-expression in the olfactory system of D. melanogaster, contrasting a previously accepted view of segregated, mutually exclusive olfactory domains. By generating targeted knock-ins of the four main chemosensory co-receptor genes (Orco, Ir8a, Ir76b, Ir25a), we demonstrate that all four co-receptors have broader olfactory neuron expression than previously appreciated. The Ir25a co-receptor was previously thought to be expressed only in a small subset of olfactory neurons (coeloconic, sacculus, and arista neurons), but we present evidence that it is expressed in subsets of all sensilla classes and in OSNs that innervate the majority of the fly’s AL glomeruli. We further find that the Ir25a co-receptor may be involved in modulating the activity of some Orco+ OSNs, both in the antennae and maxillary palps. We present a new AL map that will aid future inquiries into the role that specific chemoreceptor co-expression plays in distinct OSN populations.
Based on the co-receptor innervation patterns in the ALs, we identified a glomerulus, VM6, that is uniquely partitioned by different OSNs (Figure 5; also see Schlegel et al., 2021). The co-receptor expression patterns allowed us to pinpoint the likely origin of the innervating OSNs. Since the VM6 glomerulus was labeled by both the Ir25a-T2A-QF2 and Ir8a-T2A-QF2 knock-in lines, the cell bodies of these neurons had to reside in the antenna; furthermore, since we did not find Ir8a-T2A-QF2 labeling of the arista, these neurons were likely to be either in coeloconic sensilla or in the sacculus. Indeed, we determined the VM6 glomerulus to be innervated by the newly discovered Rh50+ Amt+ olfactory neurons that reside in the sacculus and ac1 sensilla (Vulpe et al., 2021). Based on our results, Rh50+ Amt+ sacculus neurons are further subdivided into those that strongly express Ir8a, which innervate the VM6l region, and those that weakly or infrequently express Ir8a, which innervate VM6m and VM6v. The functional consequences of this unusual subdivision by olfactory neurons for a glomerulus, and how this relates to the fly’s olfactory perception of ammonia or other odorants, remain to be determined. These results also highlight the value of exploring chemosensory receptor expression patterns using knock-in lines even in the era of connectomics as the VM6 glomerulus and its subdivisions were not easily identifiable in prior electron microscopy reconstructions of the entire AL (Bates et al., 2020; Scheffer et al., 2020; Schlegel et al., 2021).
A model for OR/IR segregation was initially supported by developmental evidence. Two pro-neural genes specify the development of sensory structures on the antennae: amos and atonal. Amos mutants lack basiconic and trichoid sensilla, while atonal mutants do not develop coeloconic sensilla, the arista, or the sacculus (Goulding et al., 2000; Gupta and Rodrigues, 1997; Jhaveri et al., 2000a; Jhaveri et al., 2000b). It was observed that amos mutants lose Orco expression while retaining Ir25a expression (Benton et al., 2009). Our results generally do not conflict with this view. In the traditional segregated model of the fly olfactory system, it was presumed that atonal mutant antennae would show the reverse pattern: loss of IrCo expression but not Orco expression. However, our co-receptor knock-in expression results suggest that atonal mutants should have significant IrCo expression, particularly of Ir25a. This was indeed found to be the case in RNAseq analyses performed on atonal mutant antennae, which showed that both Ir25a and Ir76b expression, but not Ir8a expression, remained (Menuz et al., 2014). Based upon the strength of the corresponding glomerular innervations, it does appear that the previously reported Ir25a+ neurons have stronger or more consistent Ir25a expression, while the new Ir25a+ olfactory neurons in the antennae reported here (e.g., OR-expressing OSNs) are often weakly or stochastically labeled. This might also explain why Ir25a expression was initially overlooked in these Orco+ neural populations. The developmental pattern is different in the maxillary palps, where it is atonal and not amos, which is required for the development of the basiconic sensilla (Gupta and Rodrigues, 1997). Interestingly, the Ir25a knock-in expression corresponds well with the atonal developmental program across the olfactory appendages: the strongest expression of Ir25a is in coeloconic sensilla, but outside of these sensilla the strongest and most consistent Ir25a expression is in palp basiconic sensilla.
Chemosensory co-receptor co-expression may help clarify previously confounding observations regarding D. melanogaster odor coding. For example, olfactory neurons in the sacculus that express the Ir64a tuning receptor along with the Ir8a co-receptor project to two different glomeruli: DC4 and DP1m (Ai et al., 2013; Ai et al., 2010). These two glomeruli exhibit different olfactory response profiles, with DC4 being narrowly tuned to acids or protons, while DP1m is more broadly tuned. The molecular mechanism for these differences was previously unclear. However, the co-receptor knock-in data presented here reveals that the Ir64a+ OSN subpopulations express different combinations of co-receptors: in addition to the Ir8a co-receptor, neurons innervating DC4 express Ir25a (and occasionally Orco) (see Figure 8). In contrast, neurons innervating DP1m express Ir8a, Ir25a, and Ir76b. Thus, perhaps it is Ir76b expression in DP1m-targeting Ir64a+ neurons that makes them olfactory generalists. This idea is supported by experiments in which Ir64a was misexpressed in neurons targeting the VM4 glomerulus, conferring a DP1m-like, rather than a DC4-like, response profile to VM4 (Ai et al., 2010). We show here that VM4-targeting neurons express Ir8a, in addition to Ir25a and Ir76b (as well as Orco): thus, molecularly, the VM4 neuron profile is more similar to DP1m (co-expressing all IrCos) than DC4 (co-expressing two IrCos), and the key distinguishing component appears to be Ir76b. It would be interesting to repeat such misexpression experiments in an Ir76b- mutant background to test this hypothesis.
While we demonstrate here that multiple chemosensory co-receptors can be co-expressed in the same olfactory neurons, it remains to be determined if this also applies to tuning (odor-binding) receptors. Previous studies suggest that OrX tuning receptors are generally limited to a single class of olfactory neurons (Couto et al., 2005; Fishilevich and Vosshall, 2005). However, many IrX tuning receptors remain to be fully characterized and could be co-expressed in multiple olfactory neurons. For example, recordings from ab1, ab3, and ab6 sensilla indicate responses to the typical IR odors 1,4-diaminobutane, ammonia, and butyric acid, respectively (de Bruyne et al., 2001), suggesting that tuning IrXs may be involved. We show that Ir25a plays a functional role in Orco+ neurons in the antennae and palps; this suggests that these Orco+ neurons could also express as yet unidentified ligand binding IrXs. The recent release of the whole fly single-cell atlas, which includes RNAseq data from maxillary palps, allowed us to identify six IRs that might be expressed in palpal OSNs (Ir40a, Ir51a, Ir60a, Ir62a, Ir76a, Ir93a) (Li et al., 2021). However, in situ analyses for these six IRs in the maxillary palps did not detect a signal (see Figure 6—figure supplement 2 and Appendix 1—key resources table). This suggests that Ir25a in the palps may be playing a role independent of its role as a co-receptor, as discussed further below, or that a tuning IrX was missed by the RNAseq analyses. In antennal ab3 sensilla, we did find one odorant (propionic acid) that elicited a small response in wildtype neurons and no response in Ir25a2 mutant neurons. It is possible that other antennal Orco+ OSNs might utilize IR chemoreceptors for signaling. For example, the ac3B neuron, which expresses Or35a/Orco and all IR co-receptors, has recently been suggested to utilize an unidentified IrX to mediate responses to phenethylamine (Vulpe and Menuz, 2021). The chemoreceptor expression patterns revealed in this work will help the search for olfactory neurons that may utilize multiple chemosensory families for odor detection.
The widespread expression of Ir25a in the fly olfactory system raises the possibility that it might have roles in addition to its function as an IrX co-receptor. For example, Ir25a has been found to play a developmental role in forming the unique structure of Cold Cells in the arista (Budelli et al., 2019). Evolutionary studies also suggest that Ir25a is the most ancient of all the insect chemosensory receptors (Croset et al., 2010), and the currently broad expression might reflect its previous ubiquitous role in chemosensory signaling.
Co-expression of chemosensory co-receptors might function to increase the signaling capabilities of an olfactory neuron. For example, the signaling of an Orco+ olfactory neuron may be guided primarily by the tuning OrX, and the sensitivity range extended to include odors detectable by an IrX. Co-expression might also allow synergism, such that weak activation of a co-expressed receptor could increase neuronal activity to levels sufficient to drive behavior. This might be useful in tuning behavioral response to complex odors, such that certain combinations of odors lead to stronger olfactory neuron responses. Alternatively, a co-expressed receptor inhibited by odorants might be able to attenuate a neuron’s response to odor mixtures. The observed broad Ir25a co-expression might allow an Orco-positive olfactory neuron to be primed to express a functional IrX/Ir25a receptor complex. As suggested above, this could be an evolutionary advantage if the co-expressed IrX receptor improved olfactory responses to a complex but crucial biologically relevant odor, such as host-seeking cues as observed in the A. aegypti mosquito olfactory system (Younger et al., 2020). Co-expression of chemosensory receptors could thereby be a mechanism to increase the functional flexibility of a numerically limited olfactory system.
Ir25a expression might further modulate chemosensory neuron activity levels driven by Orco/OrX signaling by altering membrane resistance. This might explain the modest activity changes we observed in Ir25a mutant Orco-expressing neurons (Figure 6). In this manner, altering Ir25a expression levels could be a neuronal mechanism to adjust Orco/OrX activity. Alternatively, Ir25a may contribute to olfactory signal transduction or amplification as has recently been shown for a pickpocket ion channel (Ng et al., 2019). Experiments addressing potentially expanded roles for Ir25a in olfactory neurons will be aided by the new chemosensory co-receptor map presented here.
D. melanogaster often serves as a model for many other insect olfactory systems, and information gleaned from vinegar flies is frequently extrapolated to other insects (e.g., DeGennaro et al., 2013; Fandino et al., 2019; Riabinina et al., 2016; Trible et al., 2017; Yan et al., 2017). Indeed, prompted by our findings of Orco and Ir25a co-expression in D. melanogaster, we extended our observations to two additional insect species. Using in situ hybridization, we found that olfactory neurons in the palps of D. sechellia flies, and in the antennae and palps of A. coluzzii mosquitoes, also co-express Orco and Ir25a co-receptors. The work presented here raises the possibility that many insects may also exhibit co-expression of chemosensory co-receptors. Recent work in A. aegypti mosquitoes suggests this is indeed the case: A. aegypti mosquito olfactory neurons can co-express Orco/IrCo/Gr receptors (Younger et al., 2020). Furthermore, A. coluzzii mosquitoes have recently been shown to co-express Orco and Ir76b co-receptors in their olfactory organs (Ye et al., 2021). This suggests that co-expression of chemosensory co-receptors may be an important feature of insect olfactory neurons.
Materials and methods
Key resources table
See Appendices 1 and 2.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Christopher J. Potter (cpotter@jhmi.edu).
Materials availability
Fly lines generated in this study have been deposited to the Bloomington Drosophila Stock Center.
Data and code availability
The snRNAseq dataset analyzed in this paper is published in McLaughlin et al., 2021. Sequencing reads and preprocessed sequencing data are available in the NCBI Gene Expression Omnibus (GSE162121). Python code for generating figures is publicly available on GitHub (https://github.com/colleen-mclaughlin/ORN_seq/). VM6 reconstructions using FlyWire can be viewed at https://flywire.ai/#links/Task2021a/all. Raw data used to generate each figure is available at Johns Hopkins Research Data Repository (https://doi.org/10.7281/T1/9VJGPI ).
Experimental model and subject details
Fly husbandry
Fly stocks were maintained at 20–25°C on standard cornmeal-agar food. Male and female flies used for experiments were 3–11 days old, unless otherwise noted.
Fly stocks
Fly lines used in this paper can be found in the Appendix 1—key resources table.
While performing co-labeling experiments, we discovered that several OrX-Gal4 lines label multiple glomeruli, and thus do not accurately represent single OSN classes. These lines were excluded from analyses and should be used with caution: Or33c-Gal4 (BDSC# 9966), Or42a-Gal4 (BDSC# 9970), Or43b-Gal4 (BDSC# 23894), Or59b-Gal4 (BDSC# 23897), Or65a-Gal4 (BDSC# 9994), Or85a-Gal4 (BDSC# 23133), and Or85b-Gal4 (BDSC# 23911). We also found that the following Or35a lines label the newly identified VM6l glomerulus in addition to VC3: Or35a-Gal4 (BDSC# 9967), Or35a-Gal4 (BDSC# 9968), Or35a-mCD8.GFP (BDSC# 52624), as well as an Or35a-Gal4 line from the Carlson lab (Yao et al., 2005).
D. sechellia flies (strain: Cousin Island, Seychelles; SKU: 14021-0248.25) were obtained from the National Drosophila Species Stock Center (Cornell College of Agriculture and Life Sciences) and reared according to our D. melanogaster protocol.
Mosquito husbandry
A. gambiae mosquitoes were reared as previously described (Riabinina et al., 2016). The wildtype N’Gousso strain was a gift from the Insect Transformation Facility in Rockville, Maryland.
Generation of QUAS-CsChrimson
The sequence of CsChrimson.Venus was PCR amplified from the genomic DNA of UAS-CsChrimson.mVenus flies (Klapoetke et al., 2014) and cloned into the 10XQUAS vector (Addgene #163629). A fly line was established through random P-element insertion. Cloning was confirmed with Sanger sequencing (Genewiz) before being sent for injection (Rainbow Transgenic Flies, Inc). Primers used for PCR amplification and In-Fusion cloning:
IVS-FOR:
TGGGTTGGACTCAGGGAATAGATCTAAAAGGTAGGTTCAACCACT
EcoRI-SV40-REV:
GCTTACGTCAGAATTCAGATCGATCCAGACATGATAAGA
Generation of HACK knock-in lines
The HACK knock-in approach requires two components: a donor construct and Cas9 (Lin and Potter, 2016a). The donor includes gRNAs specific to the target gene, as well as the template for HDR-mediated insertion of T2A-QF2 into the genome (Figure 2A, middle row). This template includes ~1 kb homology arms directly up- and downstream of the gene’s stop codon flanking a cassette containing T2A-QF2 and a 3XP3-mCherry fluorescent eye marker (see Figure 2—figure supplement 1D and E and Figure 2—figure supplement 3A and B). Outside of these homology arms, the construct has two RNA polymerase III U6 promoters driving independent expression of two gRNAs specific to the region around the target gene’s stop codon (Port et al., 2014). Two gRNAs were used to increase the probability of successfully inducing double-stranded breaks in the target (Port et al., 2014). The knock-in construct replaces the target gene’s stop codon (Figure 2A, bottom row) and introduces a transcriptional stop at the end of QF2.
The donor construct can be supplied in one of two ways (Figure 2—figure supplement 1). The first is to inject the HACK construct directly into embryos expressing Cas9 in their germline (direct injection method) (Figure 2—figure supplement 1A and B). The second approach is to establish transgenic donor lines through random P-element insertion or ΦC31 integration (Bischof et al., 2007; Gloor et al., 1991; Groth et al., 2004) of the construct into the genome, followed by genetic crosses with germline Cas9 flies for the generation of the knock-in (cross method) (Figure 2—figure supplement 1C). Only one (direct injection method) or two (cross method) generations of crosses are required for the creation of a knock-in line (Figure 2—figure supplement 1B and C). The HACK 3XP3-mCherry selection marker is bright but shows positional effects (Figure 2—figure supplement 3A). Potential knock-in flies can be screened at the adult stage (Figure 2—figure supplement 3A), or at the larval or pupal stages (Figure 2—figure supplement 1D). We generated T2A-QF2 knock-in lines for all four co-receptor genes using the direct injection method. Additionally, we tested the feasibility of the cross approach with two genes: Orco and Ir25a. Knock-ins were confirmed by PCR genotyping and sequencing (Figure 2—figure supplement 3B–D), and by crosses to a QUAS-GFP reporter to check for expression in the brain (QUAS-mCD8::GFP was used only to establish the Orco-T2A-QF2 knock-in line, after which the reporter was removed via genetic crosses; for all AL analyses, we used the 10XQUAS-6XGFP reporter line). We found no difference in expression pattern in the brain between these two approaches (Figure 2—figure supplement 1G). After establishing a knock-in line, the 3XP3-mCherry marker can be removed via Cre recombination (Siegal and Hartl, 1996). This can be useful as 3XP3-mCherry is expressed broadly throughout the fly nervous system and can interfere with red fluorescent reporters (Figure 2—figure supplement 1E). We produced two unmarked knock-in lines (for Orco and Ir25a) and confirmed no difference in brain GFP expression between marked and unmarked lines (Figure 2—figure supplement 1F).
Both approaches produced knock-ins at high rates (Table 1). Efficiency was calculated as the number of potentially HACKed knock-in flies (mCherry+), divided by the total number of flies from the given cross (G1 or F2 progeny; see Figure 2—figure supplement 1A–C). We further calculated the percentage of founders producing knock-in lines as this gives an indication of effort (how many initial crosses need to be set up to produce a knock-in). The aggregate efficiency rates for a given target locus ranged from 8% for Ir8a to 33% for Orco (Table 1); however, for individual crosses, efficiency rates were as high as 100% (see Table 1—source data 1), meaning that all progeny were potential mCherry+ knock-ins. For the two genes for which we created knock-in lines via both direct injection and genetic cross (Orco and Ir25a), we found efficiency rates comparable between approaches (Orco: 33% for direct injection, 28% for cross; Ir25a: 23% for direct injection, 24% for cross). For the direct injection approach, we tested 51 independent knock-in lines across the four target genes and found 100% to be correctly targeted events (Table 1). However, for the genetic cross approach, of the 32 independent knock-in lines tested for the two target genes, 6 (~19%) had the HACK mCherry eye marker but did not have QF2-driven GFP expression in the brain.
Information on plasmid construction can be found in the ‘Method details’ section. All D. melanogaster embryo injections were performed by Rainbow Transgenic Flies, Inc (Camarillo, CA). For HACKing via genetic cross, Orco-T2A-QF2 and Ir25a-T2A-QF2 constructs were injected into w1118 flies for P-element insertion, and donor lines were established on the second or third chromosomes by crossing to double balancers (see Appendix 1—key resources table). Donor lines were then crossed to Vas-Cas9 (BDSC# 51323). Knock-in lines were established from cis-chromosomal HACK (donor line on same chromosome as target gene) (Lin and Potter, 2016a). For HACKing via direct injection, knock-in constructs were injected into the following lines: Vas-Cas9 (BDSC# 51324) for Ir8a; Act5C-Cas9 (BDSC# 54590) for Orco, Ir76b, and Ir25a. The following lines were used to verify knock-in expression: QUAS-mCD8::GFP (BDSC# 30003), 10XQUAS-6XGFP (BDSC# 52264). Knock-in lines were confirmed by PCR genotyping (Phusion, NEB) and Sanger sequencing (Genewiz). Unmarked Orco-T2A-QF2 and Ir25a-T2A-QF2 knock-in lines were generated by crossing mCherry+ knock-in flies to the Crey+ 1B fly line (see Appendix 1—key resources table; Siegal and Hartl, 1996).
To investigate the effect of T2A-QF2 knock-in on gene function, we performed SSR on homozygous flies for each co-receptor knock-in (Figure 2—figure supplement 2), comparing their responses to wildtype flies to panels of Orco- and Ir-dependent odorants (Abuin et al., 2011; de Bruyne et al., 2001; Lin and Potter, 2015). In ab2 basiconic sensilla, Orco-T2A-QF2 knock-in flies had slightly lower baseline activity as compared to wildtype (Figure 2—figure supplement 2A); however, there were no significant differences in odor-evoked activity between these two genotypes across all stimuli tested (Figure 2—figure supplement 2B). In ac2 coeloconic sensilla, responses of Ir8a-T2A-QF2 knock-in flies to hexanol and cadaverine were slightly lower than wildtype (Figure 2—figure supplement 2D); however, these are not typically considered Ir8a-dependent odorants. Responses of the Ir8a-T2A-QF2 knock-in to Ir8a-dependent odorants (Abuin et al., 2011) were similar to wildtype controls (example trace in Figure 2—figure supplement 2C, quantification in Figure 2—figure supplement 2D). Responses of Ir76b-T2A-QF2 knock-in ac2 neurons to phenethylamine and acetic acid differed slightly from wildtype controls (Figure 2—figure supplement 2E and F). The reasons for this are unclear. The largest difference in responses between a knock-in and wildtype were for Ir25a-T2A-QF2 (Figure 2—figure supplement 2G and H); the knock-in has significantly reduced or abolished responses to Ir25a-dependent odorants, recapitulating an Ir25a mutant phenotype (Abuin et al., 2011; Silbering et al., 2011; see also Figure 2—source data 1).
Method details
Plasmid construction
The construction of QF2X-HACK knock-in plasmids requires three steps of cloning, as previously described (Lin and Potter, 2016a). All knock-in constructs were created using the pHACK-QF2 plasmid (Addgene #80274) as the backbone. The backbone was digested with the following enzymes for cloning: MluI for the 5′ homology arms; SpeI for the 3′ homology arms; and BbsI for the gRNAs. All cloning was performed using In-Fusion Cloning (Clontech #639645). The homology arms were PCR-amplified from genomic DNA extracted from wildtype flies using the DNeasy Blood and Tissue Kit (QIAGEN #69506), while the gRNAs were PCR-amplified using the pHACK-QF2 backbone as a template, with the primers themselves containing the gRNA target sequences. All homology arms were approximately 1 kb (Orco: 5HA = 1012 bp, 3HA = 1027 bp; Ir8a: 5HA = 1027 bp, 3HA = 1079 bp; Ir76b: 5HA = 997 bp, 3HA = 956 bp; Ir25a: 5HA = 1119 bp, 3HA = 990 bp). gRNAs were selected by analyzing the region around the stop codon of each gene using an online tool (https://flycrispr.org/ Gratz et al., 2014). When possible, gRNAs were chosen to minimize potential off-target cleavage sites (zero predicted for Orco, Ir8a, and Ir76b; one predicted for Ir25a, discussed below). They were selected such that one gRNA targeted upstream of the stop codon, within the last exon of the gene; the second gRNA targeted downstream of the stop codon, within the 3′UTR; and the two gRNAs were <100 bp apart. In order to prevent the gRNAs from targeting the homology arms, three synonymous nucleotide substitutions were made in each homology arm. The final knock-in lines did not always have all three substitutions (see Figure 2—figure supplement 3D), possibly due to PCR or HDR error. Note that due to the way the primers are designed, each targeted gene loses a small portion of its native 3′UTR (Orco = 72 bp, Ir8a = 31 bp, Ir76b = 27 bp, Ir25a = 24 bp). Cloning was confirmed with Sanger sequencing (Genewiz) before being sent for injection (Rainbow Transgenic Flies, Inc). Below are the gRNAs used for each gene, with the PAM sequence in parentheses.
Orco:
CTGCACCAGCACCATAAAGT (AGG)
GCACAGTGCGGAGGGGGCAA (GGG)
Ir8a:
GTTTGTTTGTTCGGCCATGT (TGG)
GGTGCCTCTGACTCCCACAG (TGG)
Ir76b:
GCAGTGATGCGAACTTCATA (TGG)
GTATTGAAAGAGGGCCGCCG (AGG)
Ir25a:
GCCGGATACTGATTAAAGCG (CGG)
ATTATGGTAAAATGAGCACT (CGG)
Primers
Italics = In-Fusion Cloning 15 bp overhang; bold = gRNA; lowercase = adding back restriction site; underline = synonymous substitution to prevent Cas9 targeting of donor construct.
PCR primers for cloning
Orco_gRNA_FOR:
TCCGGGTGAACTTCGCACAGTGCGGAGGGGGCAAGTTTTAGAGCTAGAAATAGCAAGTTA
Orco_gRNA_REV:
TTCTAGCTCTAAAACACTTTATGGTGCTGGTGCAGCGACGTTAAATTGAAAATAGGTC
Orco_5HA_FOR:
CCCTTACGTAACGCGtCAGCTTGTTTGACTTACTTGATTAC
Orco_5HA_REV:
CGCGGCCCTCACGCGtCTTGAGCTGTACAAGTACCATAAAGT
Orco_3HA_FOR:
GTTATAGATCACTAGtCTCAGTACTATGCAACCAGCAATA
Orco_3HA_REV:
AATTCAGATCACTAGtGTTTTATGAAAGCTGCAAGAAATAA
Ir8a_gRNA_FOR:
TCCGGGTGAACTTCGTTTGTTTGTTCGGCCATGTGTTTTAGAGCTAGAAATAGCAAGTTA
Ir8a_gRNA_REV:
TTCTAGCTCTAAAACCTGTGGGAGTCAGAGGCACCGACGTTAAATTGAAAATAGGTC
Ir8a_5HA_FOR:
CCCTTACGTAACGCGtCTATTGGCTATTCGTCGTACTCATGC
Ir8a_5HA_REV:
CGCGGCCCTCACGCGtCTCCATGTAGCCACTATGTGAGTCAGAT
Ir8a_3HA_FOR:
GTTATAGATCACTAGtGTTTCTTGTCGCACCTAATTAACAAGTG
Ir8a_3HA_REV:
AATTCAGATCACTAGtCATACTTAAGCTCCTTGAGGTCCAGC
Ir76b_gRNA_FOR:
TCCGGGTGAACTTCGCAGTGATGCGAACTTCATAGTTTTAGAGCTAGAAATAGCAAGTTA
Ir76b_gRNA_REV:
TTCTAGCTCTAAAACCGGCGGCCCTCTTTCAATACGACGTTAAATTGAAAATAGGTC
Ir76b_5HA_FOR:
CCCTTACGTAACGCGtACCAATGAATCCTTGTCCATGCTAAA
Ir76b_5HA_REV:
CGCGGCCCTCACGCGtCTCGGTGTAGCTGTCTTGAAGGAA
Ir76b_3HA_FOR:
GTTATAGATCACTAGtGCCTAATTGGAATACCTTCTACATAATGGA
Ir76b_3HA_REV:
AATTCAGATCACTAGtGGCAAGGCACAAAATAAAACGAAG
Ir25a_gRNA_FOR:
TCCGGGTGAACTTCGCCGGATACTGATTAAAGCGGTTTTAGAGCTAGAAATAGCAAGTTA
Ir25a_gRNA_REV:
TTCTAGCTCTAAAACAGTGCTCATTTTACCATAATCGACGTTAAATTGAAAATAGGTC
Ir25a_5HA_FOR:
CCCTTACGTAACGCGtTGCATGACTTCATTGACACCTCAAG
Ir25a_5HA_REV:
CGCGGCCCTCACGCGtGAAACGAGGCTTAAACGTAGCTGGATATT
Ir25a_3HA_FOR:
GTTATAGATCACTAGtAATATTATGGTTAAGTGAGCTCTCGG
Ir25a_3HA_REV:
AATTCAGATCACTAGtCAAAGCTAAGTTCATCGTCATAGAGAC
Genotyping and sequencing primers (PCR fragment size):
Orco_Seq_FOR (~2 kb):
GATGTTCTGCTCTTGGCTGATATTC
Ir8a_Seq_FOR (~1.9 kb):
CATCGACTTCATCATCAGGCTTTCG
Ir76b_Seq_FOR (~1.9 kb):
CAACGATATCCTCACGAAGAACAAGC
Ir25a_Seq_FOR (~1.9 kb):
CGAAAGGATACAAAGGATACTGCAT
HACK_Seq_REV (same for all):
TGTATTCCGTCGCATTTCTCTC
Checking for off-target effects
One of the gRNAs for QF2Ir25a-HACK had one predicted potential off-target cut site in the genome, in the tetraspanin 42ej (Tsp42Ej) gene. We sequenced this locus in the Ir25a-T2A-QF2 knock-in line and compared the sequence to our wildtype lab stock. We found no evidence of indels in the knock-in line. Primers used:
Tsp42Ej_FOR:
GAGAAGTCGTTTCCCATAACACCCT
Tsp42Ej_REV:
GAGGAGCAGTTTTCGGAGTCGCCTTC
HACK marker screening
Adult flies were anesthetized on a CO2 pad and screened in one of two ways: either with a Nightsea Stereo Microscope Fluorescence Adapter with the green SFA-GR LED light source (Nightsea LLC, Lexington, MA) and viewed with a Zeiss Stemi SV6 stereo microscope; or illuminated with an X-Cite 120Q excitation light source and viewed with a Zeiss SteREO Discovery V8 microscope equipped with a ds-Red filter.
Whole-animal imaging
Whole adults were anesthetized on ice before imaging. Whole larvae, pupae, or freshly dissected adult heads were affixed to slides with clear nail polish before imaging. All animals were imaged on an Olympus SZX7 microscope equipped with GFP and RFP filters. Animals were illuminated with an X-Cite Series 120Q light source. Images were acquired using a QImaging QIClick Cooled digital CCD camera and Q-Capture Pro 7 software. Multiple images were taken at different focal planes and then merged in Photoshop (CS6). Gain was adjusted in Fiji. Images appear in the following figures/panels: Figure 2B, D, F and H; Figure 2—figure supplement 1D; Figure 2—figure supplement 3A; and Figure 2—figure supplement 4. For Figure 7, D. melanogaster, D. sechellia, D. virilis, and A. coluzzii animals were immobilized with clear nail polish and imaged on a Zeiss SteREO Discovery V8 microscope. Images were acquired with a smartphone camera attached to the microscope ocular and processed in Photoshop (CS6) to remove background.
Immunohistochemistry
All flies were used at 3–11 days old. Apart from the cryosection protocols and portions of the antennal whole-mount protocol, all immunostaining steps were done on a nutator. All steps involving or following the addition of fluorescently conjugated secondary antibodies were done in the dark.
Brain and VNC staining was performed as in Xie et al., 2018. The tissue was dissected in PBS and then fixed for 20 min at room temperature (RT) in 4% paraformaldehyde in PBT (1× PBS with 0.3% Triton X-100). After fixation, the tissue was quickly rinsed three times with PBT, then put through three longer washes in PBT at RT (at least 15 min each). The tissue was blocked in PBT + 5% normal goat serum (NGS) at RT for at least 30 min, then transferred to block + primary antibody solution, and incubated at 4°C in primary antibodies for 1–2 days. The tissue was then washed three times with PBT at RT (at least 15 min per wash) and incubated in a secondary antibody solution in block at 4°C for 1 day. The tissue was washed three final times with PBT at RT for 15 min each, and then mounted in SlowFade Gold (Thermo Fisher Scientific #S36936). For experiments in which the 10XQUAS-6XGFP or 20XUAS-6XGFP reporters were used, the endogenous, unstained GFP signal was visualized, and no secondary green antibodies were used. Primary antibodies used: mouse anti-nc82 (DSHB, 1:25) and rat anti-mCD8 (Thermo Fisher #14-0081-82, 1:100 or 1:200). Secondary antibodies used: Cy3 goat anti-rat (Jackson ImmunoResearch #112-165-167, 1:200), Cy3 goat anti-mouse (Jackson ImmunoResearch #115-165-166, 1:200), Alexa 647 goat anti-rat (Jackson ImmunoResearch #112-605-167, 1:200), and Alexa 647 goat anti-mouse (Jackson ImmunoResearch #115-605-166, 1:200).
Whole-mount staining of maxillary palps was performed according to the brain staining protocol above, with the exception of a shorter fixation step (15 min). Primary antibodies used: rabbit anti-Ir25a (gift from Richard Benton, University of Lausanne, 1:100), rabbit anti-Orco (gift from Leslie Vosshall, Rockefeller University, 1:100), and rat anti-elav (DSHB, 1:100). Secondary antibodies used: Cy3 goat anti-rabbit (Jackson ImmunoResearch #111-165-144, 1:200), Alexa 647 goat anti-rabbit (Jackson ImmunoResearch #111-605-144, 1:200), and Alexa 647 goat anti-rat (Jackson ImmunoResearch #112-605-167, 1:200). For whole-mount staining of Orco2 mutant palps, 3-day-old flies were used to check for Orco expression before neurons degenerate (Task and Potter, 2021).
The protocol for whole-mount staining of antennae was adapted from Karim et al., 2014; Saina and Benton, 2013; Younger et al., 2020. Fly heads were dissected into CCD buffer (50 units chitinase, 1000 units chymotrypsin [25 mg of 40 units/mg], 10 mL HEPES larval buffer [119 mM NaCl, 48 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 25 mM HEPES], 100 µL DMSO) on ice, then warmed on a 37°C heat block for 10 min. Heads were incubated in CCD buffer at 37°C while rotating for 1 hr 20 min. Antennae were subsequently dissected off heads into fixative solution (4% PFA in PBT). All subsequent steps were done without rotation to prevent antennae from sticking to the walls or lids of the tubes. The antennae were fixed at RT for 40 min, then washed with PBT three times at RT, at least 15 min each time, and blocked in PBT plus 5% NGS for at least 1 hr at RT. The antennae were incubated in primary antibodies in blocking solution at 4°C for 4 days, washed three times for 15 min each at RT, and incubated in a secondary antibody solution at 4°C for 3 days. The antennae were then washed three times for 15 min each time at RT and mounted in SlowFade Gold. Primary antibody: rabbit anti-Orco (gift from Leslie Vosshall, 1:100). Secondary antibody: Cy3 goat anti-rabbit (Jackson ImmunoResearch #111-165-144, 1:200). The endogenous GFP signal was visualized.
The cryosection protocol was adapted from Spletter et al., 2007. Fly heads were dissected and lined up in cryomolds (Tissue-Tek #4565), covered with OCT compound (Tissue-Tek #4583), and frozen at –80°C. The samples were sectioned at ~12 µm on a Microm HM 500 cryostat (Microm International GmbH, Walldorf, Germany) and collected on SuperFrost Plus slides (Fisher #12-550-15). Slides were stored at –80°C until further processing. The slides were fixed at RT for 15 min in 4% paraformaldehyde in PBT (1× PBS with 0.3% Triton X-100), washed three times in PBT at RT (15 min each), blocked at RT for at least 30 min in PBT + 2.5% NGS + 2.5% normal donkey serum (NDS), then incubated overnight at 4°C in primary antibodies in fresh block solution in a special humidified chamber. On the next day, the slides were washed three times (15 min each) with PBT at RT and then incubated in secondary antibodies in block at 4°C overnight in the same humidified chamber covered in foil. Finally, the slides were washed three times (15 min each) with PBT at RT. DAPI (1:10,000) was included in the first wash as a nuclear counterstain. After washes, the slides were mounted in SlowFade Gold (Thermo Fisher Scientific #S36936). Primary antibody: guinea pig anti-Ir8a (gift from Richard Benton, University of Lausanne, 1:1000). Secondary antibody: Cy3 donkey anti-guinea pig (Jackson ImmunoResearch #706-165-148, 1:200).
For sacculus staining, 7- to 10-day-old flies were placed in an alignment collar. Their heads were encased in OCT (Tissue-Plus Fisher) in a silicone mold, frozen on dry ice, and snapped off. The head blocks were stored in centrifuge tubes at –80°C. A Leica cryostat was used to collect 20 µm sections of antennae. Immunohistochemical staining was carried out by fixing tissue in 4% paraformaldehyde for 10 min, followed by three 5 min washes in 1× PBS. The tissue was washed in 1× PBS containing 0.2% Triton-X (PBST) for 30 min to permeabilize the cuticle. Lastly, the tissue was washed in PBST containing 1% bovine serum albumin (BSA) to block nonspecific antibody binding. Primary antibody solution was made in PBST + 1% BSA, 200 µL was pipetted onto each slide under bridged coverslips, and slides were placed at 4°C overnight to incubate. The following day, the primary antibody was removed, and the slides were washed three times for 10 min each in PBST. Secondary antibody solution was made in PBST + 1% BSA, 200 µL was pipetted onto each slide under bridged coverslips, and left at RT in a dark box to incubate for 2 hr. After the 2 hr incubation, the slides were washed three times for 5 min each in PBST. After the last wash, the slides were allowed to dry in the dark staining box for ~30 min before being mounted in Vectashield, coverslipped, and stored at 4°C. Primary antibodies: rabbit anti-Ir25a (gift from Richard Benton, University of Lausanne, 1:100), guinea pig anti-Ir8a (gift from Richard Benton, University of Lausanne, 1:100), and rabbit anti-Ir64a (gift from Greg Suh, NYU/KAIST, 1:100). Secondary antibodies: Jackson Immuno Cy3 conjugated AffiniPure 568 goat anti-rabbit (111-165-144, 1:500) and Alexa Fluor 568 goat anti-guinea pig (A11075, 1:500).
In situ HCR
Cryosectioning for antennal in situs was performed as described above. The HCR protocol was adapted from Molecular Instruments HCR v3.0 protocol for fresh frozen or fixed frozen tissue sections (Choi et al., 2018). Slides were fixed in ice-cold 4% PFA in PBT for 15 min at 4°C, dehydrated in an ethanol series (50% EtOH, 70% EtOH, 100% EtOH, 100% EtOH, 5 min each step), and air dried for 5 min at RT. The slides were then incubated in proteinase K solution in a humidified chamber for 10 min at 37°C, rinsed twice with PBS and dried, then pre-hybridized for 10 min at 37°C in a humidified chamber. The slides were then incubated in probe solution (0.4 pmol Ir76b probe) overnight in the 37°C humidified chamber. On day 2, the slides were washed with a probe wash buffer/SSCT series (75% buffer/25% SSCT, 50% buffer/50% SSCT, 25% buffer/75% SSCT, 100% SSCT, 15 min each) at 37°C, then washed for 5 min at RT with SSCT and dried. The slides were pre-amplified for 30 min at RT in the humidified chamber while hairpins were snap cooled (6 pmol concentration). The slides were incubated in fresh amplification buffer with hairpins overnight in a dark humidified chamber at RT. On day 3, the slides were rinsed in SSCT at RT (2 × 30 min, 1 × 10 min with 1:10,000 DAPI, 1 × 5 min) and mounted in SlowFade Diamond (Thermo Fisher S36972). For the overnight steps, the slides were covered with HybriSlips (Electron Microscopy Sciences 70329-62) to prevent solution evaporation.
The whole-mount palp in situ protocol for all Drosophila species tested in this paper was adapted from a combination of Prieto-Godino et al., 2020; Saina and Benton, 2013; Younger et al., 2020 and the D. melanogaster whole-mount embryo protocol from Molecular Instruments (Choi et al., 2016). All steps after dissection were performed while rotating unless otherwise noted. Fly mouthparts (palps and proboscises) were dissected into CCD buffer (same as for whole-mount IHC on antennae above), incubated for 20–65 min in CCD at 37°C (5 min on heat block, 15 min to 1 hr rotating), then pre-fixed in 4% PFA in PBT for 20 min at RT. Tissue was washed with 0.1% PBS-Tween on ice (4 × 5 min), incubated for 1 hr at RT in 80% methanol/20% DMSO, and washed for 10 min in PBS-Tween at RT. The tissue was incubated in Proteinase K solution (1:1000) in PBS-Tween at RT for 30 min, then washed in PBS-Tween at RT (2 × 10 min) and post-fixed in 4% PFA in PBS-Tween at RT for 20 min. After post-fixation, the tissue was washed in PBS-Tween at RT (3 × 15 min), then pre-hybridized in a pre-heated probe hybridization buffer at 37°C for 30 min. The tissue was incubated in probe solution (2–30 pmol in hybridization buffer) at 37°C for 2–3 nights. After probe incubation, the tissue was washed in a pre-heated probe wash buffer at 37°C (5 × 10 min), then washed in SSCT (1× SSC plus 1% Tween) at RT (2 × 5 min). The tissue was pre-amplified with RT-equilibrated amplification buffer at RT for 10 min, then incubated in hairpin mixture (6–30 pmol snap-cooled hairpins in amplification buffer) in the dark at RT for 1–2 nights. After hairpin incubation, the tissue was washed at RT with SSCT (2 × 5 min, 2 × 30 min, 1 × 5 min), then mounted in SlowFade Diamond (Thermo Fisher S36972). Sequences for all in situ probes can be found in Appendix 2. In addition to D. sechellia (Cousin Island, Seychelles genome line, SKU: 14021-0248.25; Figure 7), we also tested the same in situ probes for six other species of Drosophila (apart from D. virilis, all species were ordered from the National Drosophila Species Stock Center): Drosophila simulans (genome line w[501], SKU: 14021-0251.195), Drosophila erecta (wild-type genome line, SKU: 14021-0224.01), Drosophila ananassae (wildtype line, Queensland, Australia, SKU: 14024-0371.11), Drosophila pseudoobscura (genome line, Anderson, Mesa Verde, CO, SKU: 14011-0121.94), Drosophila mojavensis (wildtype line, Catalina Island, CA [2002], SKU: 15081-1352.22), and Drosophila virilis (wildtype, Carolina Biological Supply, item# 172890). While we could detect a clean signal for the D. mel. Orco probe in these other fly species, the high background and poor signal-to-noise ratio for D. mel. Ir25a prevented co-localization analyses for all species but D. sechellia.
Anopheles in situs were performed essentially as described above, with the following modifications: olfactory appendages were dissected from 5- to 6-day-old female mosquitoes and incubated in CCD buffer for either 20 min (antennae) or 1.5 hr (maxillary palps) at 37°C while rotating. The tissue was then pre-fixed for 24 hr at 4°C. After the PBS-Tween and methanol/DMSO washes, the tissue was incubated overnight at –20°C in absolute methanol. The next day, the tissue was rehydrated in a graded methanol/PBS-Tween series. Subsequent steps follow the Drosophila whole-mount palp protocol. Probe concentration was 8 pmol in hybridization buffer (two night incubation), and hairpin concentration was 18 pmol in amplification buffer (one night incubation). The tissue was rinsed three times in SlowFade Diamond before being mounted.
For two-color in situs, we used the Molecular Instruments B2 amplifier conjugated to Alexa 647 for Orco and the B4 amplifier conjugated to Alexa 488 for Ir25a. For single-color in situs (Orco, Ir76b or tuning IrXs), we used the B2 amplifier conjugated to Alexa 647.
Confocal imaging and analysis
Brains, VNCs, antennae, maxillary palps, and antennal cryosections were imaged on a Zeiss LSM 700 confocal microscope equipped with Fluar 10×/0.50 air M27, LCI Plan-Neofluar 25×/0.8 water Korr DIC M27, Plan-Apochromat 40×/1.3 Oil DIC M27, and C-Apochromat 63×/1.2 water Korr M27 objectives. Images were acquired at either 512 × 512 or 1024 × 1024-pixel resolution with 0.43, 0.58, 2.37, or 6.54 µm z-steps. For illustration purposes, confocal images were processed in Fiji/ImageJ to collapse Z-stacks into a single image using maximum intensity projection. Where noted, single slices or partial z projections were used as opposed to full stacks. For co-labeling experiments, Fiji was used to convert red Look-Up Tables (LUTs) to orange for a colorblind-friendly palette. Similarly, in Figure 2—figure supplement 1E, Fiji was used to convert magenta LUT to blue for clarity. For Figure 4B, Fiji was used to convert the two-channel maximum intensity projection to a gray LUT, and the cell-counting plug-in was used in separate channels to identify single- and double-labeled cells. Fiji was also used to adjust the gain in separate channels in all figures/images; no other image processing was performed on the confocal data. For Figure 8A, glomeruli were assigned to the categories strong, intermediate, and weak by visual inspection of the strength of their innervation compared to the previously reported glomeruli for each respective knock-in line. Strong glomeruli generally have similar brightness/intensity of GFP signal as most of the originally reported glomeruli for the given knock-in line.
For sacculus staining (Figure 5C–E), slides were imaged on a Nikon A1R confocal microscope in the UConn Advanced Light Microscopy Facility with a 40× oil immersion objective at 1024 × 1024-pixel resolution. Stacks of images (0.5 µm z-step size) were gathered and analyzed with ImageJ/Fiji software. Image processing was performed as described above.
Magnification used:
10×: Figure 1D (brain), Figure 2—figure supplement 1E
25×: Figure 2—figure supplement 1F and G, Figure 3—figure supplement 1A and B
40×: Figures 5C–E–7D and E
63×: Figure 2C, E, G, and I, Figure 3A–D, Figure 4B and C, Figure 5A and B, Figure 2—figure supplement 3E–H, Figure 3—figure supplement 2A–E, Figure 5—figure supplement 1, Figure 6—figure supplement 2, Figure 7C
Note regarding Ir8a knock-in expression: in the ALs, we found that the sparse Ir8a+ expression in olfactory neurons targeting VM6m and VM6v could potentially be sexually dimorphic. Male brains generally had stronger and more frequent Ir8a+ innervation in these two glomeruli compared to female brains, as shown in Figure 3—source data 2 (see Figure 3—source data 1 for a summary of AL analyses). However, we did not find corresponding evidence for sexual dimorphism in Ir8a+ expression in the periphery. The reason for this discrepancy is currently unclear, and future work will be needed to determine whether there are functional male/female differences in Ir8a+ neurons.
Basiconic single sensillum recordings
Flies were immobilized and visualized as previously described (Lin and Potter, 2015). Basiconic sensilla were identified either using fluorescent-guided SSR (for ab3 sensilla) or using the strength of the A and B neuron responses to the reference odorants 1% ethyl acetate (EA) (Sigma #270989) and 1% pentyl acetate (PA) (Sigma #109584) (for ab2 and pb1-3 sensilla) (de Bruyne et al., 1999; Lin and Potter, 2015). For example, pb1A has strong responses to both odorants, while pb3A does not respond to EA. Similarly, ab2 sensilla were distinguished from ab3 based on the A neuron responses: ab2A responds strongly to EA and weakly to PA, while ab3A neurons have the reverse response (weak EA and strong PA response). The glass recording electrode was filled with Beadle–Ephrussi Ringer’s solution (7.5 g of NaCl + 0.35 g of KCl + 0.279 g of CaCl2-2H2O in 1 L H2O). Extracellular activity was recorded by inserting the glass electrode into the shaft or base of the sensillum of 3- to 10-day-old flies (unless otherwise specified in the young Orco2 mutant experiments). A tungsten reference electrode was inserted into the fly eye. Signals were amplified 100× (USB-IDAC System; Syntech, Hilversum, the Netherlands), input into a computer via a 16-bit analog-digital converter, and analyzed offline with AUTOSPIKE software (USB-IDAC System; Syntech). The low cutoff filter setting was 50 Hz, and the high cutoff was 5 kHz. Stimuli consisted of 1000 ms air pulses passed over odorant sources. The Δ spikes/s was calculated by counting the spikes in a 1000 ms window from ~500 ms after odorant stimuli were triggered, subtracting the spikes in a 1000 ms window prior to each stimulation. For ab3 recordings from wildtype, Orco2 mutant, and Ir25a2 mutant flies, spikes were counted in a 500 ms window from the start of the response and multiplied by 2. Then, the spikes in the 1000 ms window prior to stimulation were subtracted from this to calculate the Δ spikes/s. Stimuli used: mineral oil (Sigma CAS# 8042-47-5), EA (Sigma CAS# 141-78-6), PA (Sigma CAS# 628-63-7), benzaldehyde (Sigma CAS# 100-52-7), ethyl butyrate (Sigma CAS# 105-54-4), hexanol (Sigma CAS# 111-27-3), e2-hexenal (Sigma CAS# 6728-26-3), geranyl acetate (Sigma CAS# 105-87-3), 2-heptanone (Sigma CAS# 110-43-0), 1-octen-3-ol (Sigma CAS#3391-86-4), 2,3-butanedione (Sigma CAS#431-03-8), phenylacetaldehyde (Sigma CAS# 122-78-1), phenethylamine (Sigma CAS# 64-04-0), propionic acid (Sigma CAS# 79-09-4), 1,4-diaminobutane (Sigma CAS# 110-60-1), pyrrolidine (Sigma CAS# 123-75-1), p-cresol (Sigma CAS# 106-44-5), and methyl salicylate (Sigma CAS# 119-36-8). Odorants were dissolved in mineral oil at a concentration of 1%, and 20 µL of solution was pipetted onto filter paper in a glass Pasteur pipette. Stimuli were delivered by placing the tip of the Pasteur pipette through a hole in a plastic pipette (Denville Scientific Inc, 10 mL pipette) that carried a purified continuous air stream (8.3 mL/s) directed at the antenna or maxillary palp. A solenoid valve (Syntech) diverted delivery of a 1000 ms pulse of charcoal-filtered air (5 mL/s) to the Pasteur pipette containing the odorant dissolved on filter paper. Fresh odorant pipettes were used for no more than five odorant presentations. Ir25a2 and Orco2 mutant fly lines were outcrossed into the w1118 wildtype genetic background for at least five generations. Full genotypes for ab3 fgSSR were Pin/CyO;Or22a-Gal4,15XUAS-IVS-mcd8GFP/TM6B (wildtype), Ir25a2;Or22a-Gal4,15XUAS-IVS-mcd8GFP/TM6B (Ir25a2 mutant), and Or22a-Gal4/10XUAS-IVS-mcd8GFP (attp40);Orco2 (Orco2 mutant). These stocks were made from the following Bloomington Stocks (outcrossed to the Potter lab w1118 genetic background): BDSC# 9951, 9952, 23130, 32186, 32193, and 41737.
Coeloconic single sensillum recordings
Coeloconic SSR was performed similarly as for basiconic sensilla. Three- to five-day-old female flies were wedged in the tip of a 200 µL pipette, with the antennae and half the head exposed. A tapered glass electrode was used to stabilize the antenna against a coverslip. A BX51WI microscope (Olympus) was used to visualize the prep, which was kept under a 2000 mL/min humidified and purified air stream. A borosilicate glass electrode was filled with sensillum recording solution (Kaissling and Thorson, 1980) and inserted into the eye as a reference electrode. An aluminosilicate glass electrode was filled with the same recording solution and inserted into individual sensilla. Different classes of coeloconic sensilla were identified by their known location on the antenna and confirmed with their responses to a small panel of diagnostic odorants: in wildtype flies, ac2 sensilla were identified by their strong responses to 1,4-diaminobutane and 2,3-butanedione. The absence of a strong response to ammonia was used to rule out ac1 sensilla, the absence of a hexanol response was used to rule out ac3 sensilla, and the absence of a phenethylamine response was used to rule out ac4 sensilla. In Ir25a mutant flies in which amine responses were largely abolished, ac2 and ac4 sensilla were distinguished based on anatomical location, as well as the strong response of ac2 to 2,3-butanedione and the moderate response to propanal (both absent in ac4). ac1 and ac3 sensilla were excluded similarly in the mutant and wildtype flies. No more than four sensilla per fly were recorded. Each sensillum was tested with multiple odorants, with a rest time of at least 10 s between applications. The odorants used were acetic acid (Fisher, 1%, CAS# 64-19-7), ammonium hydroxide (Fisher, 0.1%, CAS# 7664-41-7), cadaverine (Sigma-Aldrich, 1%, CAS# 462-94-2), hexanol (ACROS Organics, 0.001%, CAS# 111-27-3), 2,3-butanedione (ACROS Organics, 1%, CAS# 431-03-8), phenethylamine (ACROS Organics, 1%, CAS# 64-04-0), propanal (ACROS Organics, 1%, CAS# 123-38-6), and 1,4-diaminobutane (ACROS Organics, 1%, CAS# 110-60-1). Odorants were diluted in water or paraffin oil. Odorant cartridges were made by placing a 13 mm antibiotic assay disc (Whatman) into a Pasteur pipette, pipetting 50 µL odorant onto the disc, and closing the end with a 1 mL plastic pipette tip. Each odorant cartridge was used a maximum of four times. The tip of the cartridge was inserted into a hole in the main airflow tube, and odorants were applied at 500 mL/min for 500 ms. Delivery was controlled via LabChart Pro v8 software (ADInstruments), which directed the opening and closing of a Lee valve (02-21-08i) linked to a ValveBank 4 controller (AutoMate Scientific). Extracellular action potentials were collected with an EXT-02F amplifier (NPI) with a custom 10X head stage. Data were acquired and AC filtered (300–1700 Hz) at 10 kHz with a PowerLab 4/35 digitizer and LabChart Pro v8 software. Spikes were summed in coeloconic recordings due to their similar sizes, and they were counted over a 500 ms window, starting at 100 ms after stimulus onset.
Optogenetics
Ir25a-T2A-QF2 was crossed to QUAS-CsChrimson #11C and double balanced to establish a stable stock (Ir25a-T2A-QF2/CyO; QUAS-CsChrimson #11C/TM6B). Newly eclosed flies (age <1 day old) were transferred to fly vials containing 0.4 mM all trans-retinal in fly food (Sigma-Aldrich #R2500, dissolved in pure DMSO with stock concentration of 0.4 M). Vials with flies were kept in the dark for at least 4 days before experiments. 627 nm LED light source (1-up LED Lighting Kit, Part# ALK-1UP-EH-KIT) powered by an Arduino Uno (https://docs.arduino.cc/hardware/uno-rev3/) was used to activate CsChrimson. By setting the voltage to 2 V and the distance of the light source to 20 cm between the LED and the fly antenna, the light intensity was equivalent to 1.13 W/m2. The antenna was stimulated for 500 ms followed by 5 s of recovery period for the total recording length of 20 s (three stimulations). The identity of ab2 and ab3 sensilla were first verified with 1% EA (Sigma #270989) and 1% PA (Sigma #109584) before optogenetic experiments. Identification of ab9 sensilla was assisted by fluorescence-guided SSR (fgSSR) (Lin and Potter, 2015) using Or67b-Gal4 (BDSC #9995) recombined with 15XUAS-IVS-mCD8::GFP (BDSC #32193). The Δ spikes/s was calculated as for other basiconic SSR. For all optogenetic experiments, the control flies were of the same genotype as experimental flies but had not been fed all-trans retinal.
Single-nucleus RNA-sequencing analyses
Dataset analyzed in this paper was published in McLaughlin et al., 2021. The expression levels for the Ir co-receptors across all OSNs were lower than for Orco, even for their corresponding ‘canonical’ glomeruli. To account for these differences and facilitate comparisons, we performed within-gene normalization in Figure 4—source data 1 and used the normalized values to generate the AL maps in Figure 4A. The normalization was performed as follows: first, we determined the fraction of cells within each cluster expressing the given co-receptor (read counts per million, CPM threshold ≥3). The cluster with the highest fraction value was taken as the maximum. Then, the fraction for each cluster was divided by this maximum value. The normalized value shows the relative strength of expression within each cluster for the given co-receptor gene.
EM neuron reconstruction
VM6 OSNs (Figure 5F and G) were reconstructed in the FAFB EM volume (Zheng et al., 2018) using FlyWire (https://flywire.ai/; Dorkenwald et al., 2020). Initial candidates were selected based on either being upstream of the VM6 (previously called VC5 in Bates et al., 2020) projection neurons or based on co-fasciculation with already identified VM6 OSNs. Analyses were performed in Python using the open-source packages navis (https://github.com/schlegelp/navis) and fafbseq (https://github.com/flyconnectome/fafbseg-py). OSNs were clustered using FAFB synapse predictions (Buhmann et al., 2021) for a synapse-based NBLAST (‘syNBLAST,’ implemented in navis). The reconstructions can be viewed at https://flywire.ai/#links/Task2021a/all.
Phylogenetic analysis
D. melanogaster, D. sechellia, and D. virilis Orco sequences were compared using the FlyBase (https://flybase.org/) BLAST tool (reference sequences examined: XM_002038370.1, XM_032721743.1, XM_002056720.3). The A. coluzzii Orco sequence was downloaded from VectorBase (https://vectorbase.org/) (sequence reference UniProtKB/TrEMBL;Acc:A0A182LER8). The pea aphid Orco sequence was acquired from the European Nucleotide Archive (https://www.ebi.ac.uk/ena/browser/view/AQS60741) (sequence reference ENA|AQS60741|AQS60741.1). Sequences were aligned using MUSCLE in MEGA11 software. This alignment was used to generate the phylogenetic tree shown in Figure 7A. The tree with the highest log likelihood (–6037.19) was used. Initial trees for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Tamura–Nei model, and then selecting the topology with superior log likelihood value. The tree in Figure 7A is drawn to sale, with branch lengths measured in the number of substitutions per site (scale bar = 0.1). Codon positions included were 1st + 2nd + 3rd + Noncoding. There were a total of 1488 positions in the final dataset. Evolutionary analyses were conducted in MEGA11 (Tamura et al., 2021). The tree is rooted to the pea aphid (Acyrthosiphon pisum) outgroup, thought to represent one of the most evolutionarily ancient examples of functional Orco/Or complexes (Missbach et al., 2014; Smadja et al., 2009; Soffan et al., 2018).
Quantification and statistical analysis
Cell counting
To quantify knock-in co-expression with the corresponding antibodies (Figure 2), the 3D reconstruction software Amira (FEI, OR) was used to manually mark individual cell bodies throughout the z-stack in each channel (antibody in far red channel, knock-in in green channel), and the cell markers between channels were compared. We also used Amira for the D. sechellia palp cell counts (Figure 7C). Cells were first marked in the far red (Orco) channel, then subsequently in the green (Ir25a) channel.
For sacculus cell counts (Figure 5C–E), cells were counted within ImageJ/Fiji using the cell counter tool. Counts were done manually by going through each stack within an image and using different colored markers for each cell type.
For Anopheles cell counts (Figure 7D and E), cells were counted in the Zeiss software (Zen Black) with the help of a manual cell counter.
All cell count data were gathered in Excel and analyzed for percent colocalization.
Statistics
Statistical analyses on SSR data were done in GraphPad Prism (version 8), except for optogenetic experiments, which were analyzed in Microsoft Excel. Box plots were made using GraphPad Prism; bar graphs were made in Excel. For all analyses, significance level α = 0.05. The following analyses were performed on all SSR data (excluding optogenetics): within genotype, Kruskal–Wallis test with uncorrected Dunn’s to determine which odorant responses were significantly different from mineral oil or paraffin oil control; between genotype, Mann–Whitney U to compare responses of two genotypes to the same odorant (e.g., wildtype vs. Ir25a2 mutant, or wildtype vs. Orco-T2A-QF2 knock-in). Summary tables in Figure 6 are filled in based on the following criteria: no response means neither wildtype nor Ir25a2 mutant odor-evoked activity for given odorant was significantly different from its respective mineral oil control, nor was the difference between the genotypes statistically significant; no difference means that either wildtype or mutant or both had a significantly different odor-evoked response to the stimulus compared to mineral oil control, but the difference between the two genotypes was not statistically significant; higher response (in either wildtype or mutant) means that there was a statistically significant difference between genotypes for the given odorant. This could mean that (a) one genotype did not have a response, while the other did; (b) both genotypes had a response, and one was higher; (c) responses are different from each other, but not from their respective mineral oil controls; or (d) neural activity was inhibited by the odorant in one genotype compared to mineral oil control, and either not inhibited in the other genotype or inhibited to a lesser degree. Nonparametric tests were chosen due to small sample sizes and/or data that were not normally distributed.
In Figure 6I, stimulus responses that were statistically significantly different from mineral oil control were those whose Δ spike values were zero due to the fact that the mineral oil control Δ spike value was nonzero (median = 1.2, range = 0–2). Because of this, we did not deem these differences as biologically relevant. Nevertheless, p-values are reported in the figure legend of Figure 6, and detailed information can be found in Figure 6—source data 1.
Acknowledgements
We thank E Marr and K Robinson for splinkerette genetic mapping, Y-T Chang for cloning of pHACKIr8a components, O Riabinina for initial QUAS-CsChrimson characterization, S Maguire for preliminary SSR experiments, P Mohapatra for RNAseq insights, D Baktash for help with figures, J Konopka for advice on statistical and phylogenetic analyses, S Shankar for discussion of AL mapping, and R Mann for providing lab resources. We would like to thank EC Marin and M Costa for discussions regarding posterior AL glomeruli. We are also grateful to the Seung and Murthy labs for access to the flywire.ai reconstruction community. Many thanks to the following labs for sharing antibody and fly stock reagents: Leslie B Vosshall (Rockefeller), Richard Benton (UNIL), Paul Garrity (Brandeis), Marco Gallio (Northwestern), Greg Suh (NYU/KAIST), Andrew Gordus (JHU), and Thomas R Clandinin (Stanford). We are grateful for in situ advice from Richard Benton, Steeve Cruchet, and Margo Herre. We thank the Center for Sensory Biology Imaging Facility (NIH P30DC005211) for use of the LSM700 confocal microscope, and the Johns Hopkins School of Public Health Malaria Research Institute for use of the Olympus SZX7 microscope equipped with QImaging QIClick Cooled digital CCD camera. We thank S Maguire and J Konopka for comments on the manuscript and members of the Potter and Menuz labs for discussion. We thank the Vosshall lab for sharing their Aedes findings before publication. This work was supported by a Shelanski award to C-CL; a Wellcome Trust Collaborative Award (203261/Z/16/Z) and an NIH BRAIN Initiative grant (1RF1MH120679-01) to GSXEJ; grants from the National Institutes of Health to HL (R00 AG062746); grants from the National Institutes of Health to KM (NIGMS R35GM133209; NIDCD 1R21DC017868); grants from the Department of Defense to CJP (W81XWH-17-PRMRP) and from the National Institutes of Health to CJP (NIAID R01Al137078; NIDCD R01DC013070). Portions of this work appear in Chapter 3 of DT’s doctoral dissertation.
Appendix 1
Appendix 1—key resources table.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Anti-nc82 (mouse monoclonal) | DSHB | Cat# nc82; RRID:AB_2314866 |
IHC (1:25) |
| Antibody | Anti-cd8 (rat monoclonal) | Thermo Fisher Scientific | Cat# 14-0081-82; RRID:AB_467087 |
IHC (1:100 or 1:200) |
| Antibody | Anti-elav (rat monoclonal) | DSHB | Cat# Rat-Elav-7E8A10; RRID:AB_528218 |
IHC (1:100) |
| Antibody | Anti-Orco (rabbit polyclonal) | Gift from Leslie Vosshall Larsson et al., 2004 | IHC (1:100) | |
| Antibody | Anti-Ir25a (rabbit polyclonal) | Gift from Benton et al., 2009 | RRID:AB_2567027 | IHC (1:100) |
| Antibody | Anti-Ir8a (guinea pig polyclonal) | Gift from Richard Benton Abuin et al., 2011 | RRID:AB_2566833 | IHC (1:100 for whole-mount, 1:1000 for cryosections) |
| Antibody | Anti-Ir64a (rabbit polyclonal) | Gift from Greg Suh Ai et al., 2010 | RRID:AB_2566854 | IHC (1:100) |
| Antibody | Anti-guinea pig Alexa 568 (goat polyclonal) | Thermo Fisher Scientific | Cat# A11075; RRID:AB_141954 |
IHC (1:500) |
| Antibody | Anti-rabbit Cy3 conjugated AffiniPure 568 (goat polyclonal) |
Jackson ImmunoResearch | Cat# 111-165-144; RRID:AB_2338006 |
IHC (1:500) |
| Antibody | Anti-mouse Cy3 (goat polyclonal) | Jackson ImmunoResearch | Cat# 115-165-166; RRID:AB_2338692 |
IHC (1:200) |
| Antibody | Anti-mouse Alexa 647 (goat polyclonal) | Jackson ImmunoResearch | Cat# 115-605-166; RRID:AB_2338914 |
IHC (1:200) |
| Antibody | Anti-guinea pig Cy3 (donkey polyclonal) | Jackson ImmunoResearch | Cat# 706-165-148; RRID:AB_2340460 |
IHC (1:200) |
| Antibody | Anti-rat Cy3 (goat polyclonal) | Jackson ImmunoResearch | Cat# 112-165-167; RRID:AB_2338251 |
IHC (1:200) |
| Antibody | Anti-rat Alexa 647 (goat polyclonal) | Jackson ImmunoResearch | Cat# 112-605-167; RRID:AB_2338404 |
IHC (1:200) |
| Antibody | Anti-rabbit Cy3 (goat polyclonal) | Jackson ImmunoResearch | Cat# 111-165-144; RRID:AB_2338006 |
IHC (1:200) |
| Antibody | Anti-rabbit Alexa 647 (goat polyclonal) | Jackson ImmunoResearch | Cat# 111-605-144; RRID:AB_2338078 |
IHC (1:200) |
| Recombinant DNA reagent | pHACK-QF2 (plasmid) | Addgene Lin and Potter, 2016a | Plasmid# 80274; RRID:Addgene_80274 |
QF2 HACK backbone Contains QF2-hsp70, but no gRNAs |
| Recombinant DNA reagent | p10XQUAS-CsChrimson-SV40 (plasmid) | This paper | Plasmid# 163629; RRID:Addgene_163629 |
For red-shifted optogenetic activation of neurons under control of the Q-system; see ’Fly stocks’ |
| Recombinant DNA reagent | pHACK-QF2Orco (plasmid) | This paper | HACK construct targeting the Orco gene; see ‘Plasmid construction’ | |
| Recombinant DNA reagent | pHACK-QF2Ir8a (plasmid) | This paper | HACK construct targeting the Ir8a gene; see ‘Plasmid construction’ | |
| Recombinant DNA reagent | pHACK-QF2Ir76b (plasmid) | This paper | HACK construct targeting the Ir76b gene; see ‘Plasmid construction’ | |
| Recombinant DNA reagent | pHACK-QF2Ir25a (plasmid) | This paper | HACK construct targeting the Ir25a gene; see ‘Plasmid construction’ | |
| Genetic reagent (Drosophila melanogaster) | Orco-T2A-QF2 knock-in | This paper | BDSC 92400, 92401, 92402 | See ‘Generation of HACK knock-in lines’ |
| Genetic reagent (D. melanogaster) | Ir8a-T2A-QF2 knock-in | This paper | BDSC 92398, 92399 | See ‘Generation of HACK knock-in lines’ |
| Genetic reagent (D. melanogaster) | Ir76b-T2A-QF2 knock-in | This paper | BDSC 92396, 92397 | See ‘Generation of HACK knock-in lines’ |
| Genetic reagent (D. melanogaster) | Ir25a-T2A-QF2 knock-in | This paper | BDSC 92392, 92393, 92394, 92395 |
See ‘Generation of HACK knock-in lines’ |
| Genetic reagent (D. melanogaster) | 10XQUAS-CsChrimson (y[1] w[*]; Pin[1]/CyO; P(w[+mC] = 10XQUAS-CsChrimson.mVenus)11c) | This paper | BDSC 91996, FlyBase FBst0091996 |
See ‘Fly stocks’ |
| Genetic reagent (D. melanogaster) | Ir21a-T2A-Gal4 knock-in | Gift from Paul Garrity Marin et al., 2020 | ||
| Genetic reagent (D. melanogaster) | Ir68a-T2A-Gal4 knock-in | Gift from Paul Garrity Marin et al., 2020 | ||
| Genetic reagent (D. melanogaster) | Or7a-Gal4 knock-in (y[1] w[*] TI(GAL4)Or7a[KI-GAL4.w-]) | Potter lab | BDSC 91991, FlyBase FBti0214362 |
|
| Genetic reagent (D. melanogaster) | Ir64a-Gal4 | Gift from Greg Suh lab Ai et al., 2010 | ||
| Genetic reagent (D. melanogaster) | Rh50-Gal4 | Menuz lab Vulpe et al., 2021 | ||
| Genetic reagent (D. melanogaster) | Amt-Gal4 | Menuz lab Menuz et al., 2014 | ||
| Genetic reagent (D. melanogaster) | Repo-Gal80 | Awasaki et al., 2011 | FlyBase FBtp0067904 | |
| Genetic reagent (D. melanogaster) | wCS (Cantonized w1118) | Koh et al., 2014 | FlyBase FBrf0226011 | |
| Genetic reagent (D. melanogaster) | Or35a-Gal4 | Yao et al., 2005 | ||
| Genetic reagent (D. melanogaster) | Crey +1B (y[1] w[67c23] P(y[+mDint2] = Crey)1b; sna[Sco]/CyO) | Bloomington Drosophila Stock Center | BDSC 766, FlyBase FBti0012692 |
|
| Genetic reagent (D. melanogaster) | Gr21a-GAL4 (w[*]; P(w[ + mC] = Gr21a-GAL4.9.323)2/CyO; Dr[1]/TM3, Sb[+]) | Bloomington Drosophila Stock Center | BDSC 57600, FlyBase FBti0162643 |
|
| Genetic reagent (D. melanogaster) | Gr28b.d-GAL4 (w[*]; P(w[+mC] = Gr28b.d-GAL4)B27; Dr[1]/TM3, Sb[1]) | Bloomington Drosophila Stock Center | BDSC 57620, FlyBase FBst0057620 |
|
| Genetic reagent (D. melanogaster) | Ir25a-GAL4 (w[*]; P(w[ + mC] = Ir25a-GAL4.A)236.1; TM2/TM6B, Tb[1]) | Bloomington Drosophila Stock Center | BDSC 41728, FlyBase FBti0148895 |
|
| Genetic reagent (D. melanogaster) | Ir40a-GAL4 (w[*]; P(w[ + mC] = Ir40a-GAL4.3011)214.1; TM2/TM6B, Tb[1]) | Bloomington Drosophila Stock Center | BDSC 41727, FlyBase FBst0041727 |
|
| Genetic reagent (D. melanogaster) | Ir41a-GAL4 (w[*]; P(y[+ t7.7] w[+mC] = Ir41a-GAL4.2474)attP40; TM2/TM6B, Tb[1]) | Bloomington Drosophila Stock Center | BDSC 41749, FlyBase FBst0041749 | |
| Genetic reagent (D. melanogaster) | Ir64a-GAL4 (w[*]; P(w[+mC] = Ir64a-GAL4.A)183.8; TM2/TM6B, Tb[1]) | Bloomington Drosophila Stock Center | BDSC 41732, FlyBase FBti0148898 | |
| Genetic reagent (D. melanogaster) | Ir76a-GAL4 (w[*]; P(w[+ mC] = Ir76a-GAL4.PB)292.3B; TM2/TM6B, Tb[1]) | Bloomington Drosophila Stock Center | BDSC 41735, FlyBase FBst0041735 | |
| Genetic reagent (D. melanogaster) | Ir76b-GAL4 (w[*]; P(w[ + mC] = Ir76b-GAL4.916)226.8; TM2/TM6B, Tb[+]) | Bloomington Drosophila Stock Center | BDSC 41730, FlyBase FBti0153291 | |
| Genetic reagent (D. melanogaster) | Ir8a-GAL4 (w[*]; P(w[ + mC] = Ir8a-GAL4.A)204.8; TM2/TM6B, Tb[1]) | Bloomington Drosophila Stock Center | BDSC 41731, FlyBase FBti0148897 | |
| Genetic reagent (D. melanogaster) | Or10a-Gal4 (w[*]; l(2)*[*]/CyO; P(w[ + mC] = Or10a-GAL4.C)134t1.3) | Bloomington Drosophila Stock Center | BDSC 23885, FlyBase FBti0102042 | |
| Genetic reagent (D. melanogaster) | Or13a-Gal4 (w[*]; P(w[ + mC] = Or13a-GAL4.C)229t56.2/TM3, Sb[1]) | Bloomington Drosophila Stock Center | BDSC 23886, FlyBase FBti0102056 | |
| Genetic reagent (D. melanogaster) | Or22a-Gal4 (w[*]; P(w[ + mC] = Or22a-GAL4.7.717)14.2) | Bloomington Drosophila Stock Center | BDSC 9951, FlyBase FBti0101805 | |
| Genetic reagent (D. melanogaster) | Or22a-Gal4 (w[*]; P(w[ + mC] = Or22a-GAL4.7.717)14.21) | Bloomington Drosophila Stock Center | BDSC 9952, FlyBase FBti0101805 | |
| Genetic reagent (D. melanogaster) | Or33c-Gal4 (w[*]; P(w[ + mC] = Or33c-GAL4.F)78.3) | Bloomington Drosophila Stock Center | BDSC 9966, FlyBase FBti0101843 | |
| Genetic reagent (D. melanogaster) | Or35a-cd8GFP (w[*]; P(w[ + mC] = Or35a-Mmus\Cd8a.GFP)3/TM3, Sb[1]) | Bloomington Drosophila Stock Center | BDSC 52624, FlyBase FBti0156834 | |
| Genetic reagent (D. melanogaster) | Or35a-Gal4 (w[*]; P(w[ + mC] = Or35a-GAL4.F)109.2A) | Bloomington Drosophila Stock Center | BDSC 9967, FlyBase FBti0101810 | |
| Genetic reagent (D. melanogaster) | Or35a-Gal4 (w[*]; P(w[ + mC] = Or35a-GAL4.F)109.3) | Bloomington Drosophila Stock Center | BDSC 9968, FlyBase FBti0101844 | |
| Genetic reagent (D. melanogaster) | Or42a-Gal4 (w[*]; P(w[ + mC] = Or42a-GAL4.F)48.3B) | Bloomington Drosophila Stock Center | BDSC 9970, FlyBase FBti0101811 | |
| Genetic reagent (D. melanogaster) | Or42b-Gal4 (w[*]; P(w[ + mC] = Or42b-GAL4.F)64.3) | Bloomington Drosophila Stock Center | BDSC 9971, FlyBase FBti0101812 | |
| Genetic reagent (D. melanogaster) | Or43b-Gal4 (w[*]; P(w[ + mC] = Or43b-GAL4.C)110t8.1) | Bloomington Drosophila Stock Center | BDSC 23894, FlyBase FBti0102047 | |
| Genetic reagent (D. melanogaster) | Or46a-Gal4 (w[1,118]; P(w[ + mC] = Or46a-GAL4.G)32.1.y) | Bloomington Drosophila Stock Center | BDSC 23291, FlyBase FBti0076800 | |
| Genetic reagent (D. melanogaster) | Or47a-Gal4 (w[*]; P(w[ + mC] = Or47a-GAL4.8.239)15.4A) | Bloomington Drosophila Stock Center | BDSC 9982, FlyBase FBti0101851 | |
| Genetic reagent (D. melanogaster) | Or49b-Gal4 (w[*]; P(w[ + mC] = Or49b-GAL4.F)80.1) | Bloomington Drosophila Stock Center | BDSC 9986, FlyBase FBti0101853 | |
| Genetic reagent (D. melanogaster) | Or59b-Gal4 (w[*]; P(w[ + mC] = Or59b-GAL4.C)114t2.2) | Bloomington Drosophila Stock Center | BDSC 23897, FlyBase FBti0102060 | |
| Genetic reagent (D. melanogaster) | Or59c-Gal4 (w[*]; P(w[ + mC] = Or59c-GAL4.C)129t1.1) | Bloomington Drosophila Stock Center | BDSC 23899, FlyBase FBti0102061 | |
| Genetic reagent (D. melanogaster) | Or65a-Gal4 (w[*]; Bl[1]/SM1; P(w[ + mC] = Or65a-GAL4.F)72.1) | Bloomington Drosophila Stock Center | BDSC 9994, FlyBase FBti0101857 | |
| Genetic reagent (D. melanogaster) | Or67a-Gal4 (w[*]; P(w[ + mC] = Or67a-GAL4.C)137t3.3) | Bloomington Drosophila Stock Center | BDSC 23904, FlyBase FBti0102049 | |
| Genetic reagent (D. melanogaster) | Or67b-Gal4 (w[*]; P(w[ + mC] = Or67b-GAL4.F)68.3/TM6B, Tb[1]) | Bloomington Drosophila Stock Center | BDSC 9995, FlyBase FBti0101858 | |
| Genetic reagent (D. melanogaster) | Or67c-Gal4 (w[*]; P(w[ + mC] = Or67c-GAL4.C)116t3.2/CyO) | Bloomington Drosophila Stock Center | BDSC 23905, FlyBase FBti0102050 | |
| Genetic reagent (D. melanogaster) | Or71a-Gal4 (w[*]; P(w[ + mC] = Or71a-GAL4.F)30.4) | Bloomington Drosophila Stock Center | BDSC 23122, FlyBase FBti0101860 | |
| Genetic reagent (D. melanogaster) | Or83c-Gal4 (w[*]; P(w[ + mC] = Or83c-GAL4.F)73.3B) | Bloomington Drosophila Stock Center | BDSC 23131, FlyBase FBti0101829 | |
| Genetic reagent (D. melanogaster) | Or85a-Gal4 (w[*]; P(w[ + mC] = Or85a-GAL4.F)67.2) | Bloomington Drosophila Stock Center | BDSC 23133, FlyBase FBti0101830 | |
| Genetic reagent (D. melanogaster) | Or85b-Gal4 (w[*]; P(w[ + mC] = Or85b-GAL4.C)179t5.1) | Bloomington Drosophila Stock Center | BDSC 23911, FlyBase FBti0102053 | |
| Genetic reagent (D. melanogaster) | Or85d-Gal4 (w[*]; P(w[ + mC] = Or85d-GAL4.C)143t2.1) | Bloomington Drosophila Stock Center | BDSC 24148, FlyBase FBti0102066 | |
| Genetic reagent (D. melanogaster) | Or92a-Gal4 (w[*]; P(w[ + mC] = Or92a-GAL4.F)62.1) | Bloomington Drosophila Stock Center | BDSC 23139, FlyBase FBti0101867 | |
| Genetic reagent (D. melanogaster) | Or98a-Gal4 (w[*]; P(w[ + mC] = Or98a-GAL4.F)115.1) | Bloomington Drosophila Stock Center | BDSC 23141, FlyBase FBti0101868 | |
| Genetic reagent (D. melanogaster) | Orco-Gal4 (w[*]; P(w[ + mC] = Orco-GAL4.W)11.17; TM2/TM6B, Tb[1]) | Bloomington Drosophila Stock Center | BDSC 26818, FlyBase FBti0101150 | |
| Genetic reagent (D. melanogaster) | QUAS reporter (y[1] w[1,118]; P(w[ + mC] = QUAS-mCD8::GFP.P)5B/TM6B, Tb[1]) | Bloomington Drosophila Stock Center | BDSC 30003, FlyBase FBti0129937 | |
| Genetic reagent (D. melanogaster) | QUAS reporter (y[1] w[*]; PBac(y[ + mDint2] w[ + mC] = 10XQUAS-6XGFP)VK00018/CyO, P(Wee-P.ph0)Bacc[Wee-P20]) | Bloomington Drosophila Stock Center | BDSC 52264, FlyBase FBti0162759 | |
| Genetic reagent (D. melanogaster) | UAS reporter (w[*]; P(y[ + t7.7] w[ + mC] = 10XUAS-IVS-mCD8::GFP)attP40) | Bloomington Drosophila Stock Center | BDSC 32186, FlyBase FBti0131963 | |
| Genetic reagent (D. melanogaster) | UAS reporter (w[*]; P(y[ + t7.7] w[ + mC] = 15XUAS-IVS-mCD8::GFP)attP2) | Bloomington Drosophila Stock Center | BDSC 32193, FlyBase FBti0131935 | |
| Genetic reagent (D. melanogaster) | UAS reporter (w[*]; P(y[ + t7.7] w[ + mC] = 10XUAS-IVS-mCD8::RFP)attP2) | Bloomington Drosophila Stock Center | BDSC 32218, FlyBase FBti0131950 | |
| Genetic reagent (D. melanogaster) | UAS reporter (w[*]; P(y[ + t7.7] w[ + mC] = 10XUAS-IVS-mCD8::RFP)attP40) | Bloomington Drosophila Stock Center | BDSC 32219, FlyBase FBti0131967 | |
| Genetic reagent (D. melanogaster) | UAS reporter (y[1] w[*]; PBac(y[ + mDint2] w[ + mC] = 20XUAS-6XGFP)VK00018/CyO, P(Wee-P.ph0)Bacc[Wee-P20]) | Bloomington Drosophila Stock Center | BDSC 52261, FlyBase FBti0162758 | |
| Genetic reagent (D. melanogaster) | UAS reporter, sacculus experiments (UAS-mCD8::GFP (2nd)) | Lee and Luo, 1999 | FlyBase FBti0012685 | |
| Genetic reagent (D. melanogaster) | UAS reporter, sacculus experiments (UAS-mCD8::GFP (3rd)) | Lee and Luo, 1999 | FlyBase FBti0012686 | |
| Genetic reagent (D. melanogaster) | Orco2 mutant (w[*]; TI(w[ + m*] = TI)Orco[2]) | Bloomington Drosophila Stock Center | BDSC 23130, FlyBase FBti0168777 | |
| Genetic reagent (D. melanogaster) | Ir25a2 mutant (w[*]; TI(w[ + m*] = TI)Ir25a[2]/CyO) | Bloomington Drosophila Stock Center | BDSC 41737, FlyBase FBti0168524 | |
| Genetic reagent (D. melanogaster) | Ir8a1 mutant (w[*] TI(w[ + mW.hs] = TI)Ir8a[1]; Bl[1] L[2]/CyO) | Bloomington Drosophila Stock Center | BDSC 41744, FlyBase FBst0041744 | |
| Genetic reagent (D. melanogaster) | Ir76b1 mutant (w[*]; Ir76b[1]) | Bloomington Drosophila Stock Center | BDSC 51309, FlyBase FBst0051309 | |
| Genetic reagent (D. melanogaster) | Germline Cas9 (y[1] M(RFP[3xP3.PB] GFP[E.3xP3] = vas-Cas9)ZH-2A w[1,118]/FM7c) | Bloomington Drosophila Stock Center | BDSC 51323, FlyBase FBti0154823 | |
| Genetic reagent (D. melanogaster) | Germline Cas9 (w[1,118]; PBac(y[ + mDint2] = vas-Cas9)VK00027) | Bloomington Drosophila Stock Center | BDSC 51324, FlyBase FBti0154822 | |
| Genetic reagent (D. melanogaster) | Germline Cas9 (y[1] M(w[ + mC] = Act5C-Cas9.P)ZH-2A w[*]) | Bloomington Drosophila Stock Center | BDSC 54590, FlyBase FBti0159182 | |
| Genetic reagent (D. melanogaster) | Double balancer (19ADrok/FM7c; Pin/CyO) | Potter lab stock | Derived from BDSC 6666, FBba0000009, FBal0013831, FBba0000025 |
|
| Genetic reagent (D. melanogaster) | Double balancer (y,w; Pin/CyO; Dh/TM6B) | Potter lab stock | Derived from FBal0013831, FBba0000025, FBti0004009, FBba0000057, FBal0016730 |
|
| Genetic reagent (D. melanogaster) | Double balancer (y,w; S/CyO; Pr/TM6B) | Potter lab stock | Derived from FBal0015108, FBba0000025, FBal0013944, FBba0000057, FBal0016730 |
|
| Genetic reagent (D. melanogaster) | Single balancer (y,w; +/+; Pr/TM6B) | Potter lab stock | Derived from FBal0013944, FBba0000057, FBal0016730 |
|
| Genetic reagent (D. melanogaster) | Single balancer (y,w; S/CyO; +/+) | Potter lab stock | Derived from FBal0015108, FBba0000025 |
|
| Genetic reagent (D. melanogaster) | Wildtype (w1118 IsoD1) | Gift from Thomas R. Clandinin | Derived from FBal0018186 | |
| Genetic reagent (Drosophila sechellia) | Wildtype, genome Cousin Island, Seychelles | National Drosophila Species Stock Center | SKU:14021-0248.25 | |
| Genetic reagent (Anopheles coluzzii) | Wildtype, N’Gousso strain | Gift from Insect Transformation Facility (Rockville, MD) Riabinina et al., 2016 | ||
| Chemical compound, drug | Mineral oil | Sigma-Aldrich | CAS# 8042-47-5, Cat# 330779-1L | |
| Chemical compound, drug | Ethyl acetate | Sigma-Aldrich | CAS# 141-78-6, Cat# 650528-1L, Cat# 270989-100ML |
|
| Chemical compound, drug | Pentyl acetate | Sigma-Aldrich | CAS# 628-63-7, Cat# 109584-250ML |
|
| Chemical compound, drug | Benzaldehyde | Sigma-Aldrich | CAS# 100-52-7, Cat# 418099-100ML, Cat# B1334-100G |
|
| Chemical compound, drug | Ethyl butyrate | Sigma-Aldrich | CAS# 105-54-4, Cat# E15701-500M, Cat# E15701-25ML |
|
| Chemical compound, drug | Hexanol | Sigma-Aldrich | CAS# 111-27-3, Cat# H13303-100ML |
|
| Chemical compound, drug | E2-hexenal | Sigma-Aldrich | CAS# 6728-26-3, Cat# W256005-1KG-K |
|
| Chemical compound, drug | Geranyl acetate | Sigma-Aldrich | CAS# 105-87-3, Cat# 173495-25G, Cat# 45896-1ML-F |
|
| Chemical compound, drug | 2-Heptanone | Sigma-Aldrich | CAS# 110-43-0, Cat# 537683-100ML |
|
| Chemical compound, drug | 1-Octen-3-ol | Sigma-Aldrich | CAS# 3391-86-4, Cat# O5284-25G, Cat# W280518-SAMPLE-K |
|
| Chemical compound, drug | 2,3-Butanedione | Sigma-Aldrich | CAS# 431-03-8, Cat# 11038-1ML-F |
|
| Chemical compound, drug | Phenylacetaldehyde | Sigma-Aldrich | CAS# 122-78-1, Cat# 107395-100ML |
|
| Chemical compound, drug | Phenethylamine | Sigma-Aldrich | CAS# 64-04-0, Cat# 241008-50ML |
|
| Chemical compound, drug | Propionic acid | Sigma-Aldrich | CAS# 79-09-4, Cat# 402907-100ML |
|
| Chemical compound, drug | 1,4-Diaminobutane | Sigma-Aldrich | CAS# 110-60-1, Cat# D13208-100G |
|
| Chemical compound, drug | Pyrrolidine | Sigma-Aldrich | CAS# 123-75-1, Cat# P73803-100ML, Cat# P73803-5ML |
|
| Chemical compound, drug | P-cresol | Sigma-Aldrich | CAS# 106-44-5, Cat# 42429-5G-F, Cat# C85751-5G |
|
| Chemical compound, drug | Methyl salicylate | Sigma-Aldrich | CAS# 119-36-8, Cat# M6752-250ML |
|
| Chemical compound, drug | Paraffin Oil | ACROS Organics | CAS# 8012-95-1, Cat# 171400010 | |
| Chemical compound, drug | Water | SIGMA Life Science | CAS# 7732-18-5, Cat# W3500 |
|
| Chemical compound, drug | Acetic acid | Fisher Scientific | CAS# 64-19-7, Cat# A38S | |
| Chemical compound, drug | Ammonium hydroxide | Fisher Scientific | CAS# 7664-41-7, Cat# A669S | |
| Chemical compound, drug | Cadaverine | Sigma-Aldrich | CAS# 462-94-2, Cat# 33211-10ML-F | |
| Chemical compound, drug | Hexanol | ACROS Organics | CAS# 111-27-3, Cat# AC43386 | |
| Chemical compound, drug | 2,3-Butanedione | ACROS Organics | CAS# 431-03-8, Cat# AC10765 | |
| Chemical compound, drug | Phenethylamine | ACROS Organics | CAS# 64-04-0, Cat# AC156491000 | |
| Chemical compound, drug | Propanal | ACROS Organics | CAS# 123-38-6, Cat# AC220511000 | |
| Chemical compound, drug | 1,4-Diaminobutane | ACROS Organics | CAS# 110-60-1, Cat# AC11212-250 | |
| Chemical compound, drug | All trans-retinal | Sigma-Aldrich | CAS# 116-31-4, Cat# R2500 | |
| Commercial assay or kit | HCR v3.0 | Molecular Instruments | ||
| Commercial assay or kit | In-Fusion Cloning | Clontech Labs | Cat# 639645 | |
| Commercial assay or kit | DNeasy Blood and Tissue Kit | QIAGEN | Cat# 69506 | |
| Software, algorithm | Fiji (ImageJ) | Schindelin et al., 2012 | https://imagej.net/Fiji | |
| Software, algorithm | GraphPad Prism 8.0 | GraphPad Software | http://www.graphpad.com/ | |
| Software, algorithm | AutoSpike | Syntech | http://www.ockenfels-syntech.com/products/signal-acquisition-systems-2/ | |
| Software, algorithm | LabChart Pro v8 | ADInstruments | https://www.adinstruments.com/support/downloads/windows/labchart | |
| Software, algorithm | Adobe Illustrator CS6 | Adobe, Inc | https://www.adobe.com/products/illustrator.html | |
| Software, algorithm | Adobe Photoshop CS6 | Adobe, Inc | https://www.adobe.com/products/photoshop.html | |
| Software, algorithm | MacVector 16.0 | MacVector, Inc | https://macvector.com/ | |
| Software, algorithm | MEGA11 | Tamura et al., 2021 | https://www.megasoftware.net/ | |
| Software, algorithm | Zen Black | Carl Zeiss Microscopy | https://www.zeiss.com/microscopy/us/products/microscope-software/zen.html | |
| Software, algorithm | Venn Diagram web tool | VIB/UGent Bioinformatics & Evolutionary Genomics | http://bioinformatics.psb.ugent.be/webtools/Venn/ | |
| Software, algorithm | Amira | Thermo Fisher Scientific | https://www.thermofisher.com/us/en/home/industrial/electron-microscopy/electron-microscopy-instruments-workflow-solutions/3d-visualization-analysis-software/amira-life-sciences-biomedical.html | |
| Software, algorithm | Python packages | Schlegel et al., 2021 | https://github.com/schlegelp/navis, https://github.com/flyconnectome/fafbseg-py | |
| Software, algorithm | Q-Capture Pro 7 | Teledyne QImaging | https://www.qimaging.com/home | |
| Other | DAPI stain | Invitrogen | D1306 | IHC (1:10,000) |
| Sequence-based reagent | Orco_gRNA_FOR | This paper, Integrated DNA Technologies | PCR primers | TCCGGGTGAACT TCGCACAGTGCG GAGGGGGCAAG TTTTAGAGCT AGAAATAGCAAGTTA |
| Sequence-based reagent | Orco_gRNA_REV | This paper, Integrated DNA Technologies | PCR primers | TTCTAGCTCTAAAAC ACTTTATGGTGCT GGTGCAGCGACGTTA AATTGAAAATAGGTC |
| Sequence-based reagent | Orco_5HA_FOR | This paper, Integrated DNA Technologies | PCR primers | CCCTTACGTAACGCGTCAGCTT GTTTGACTTACTTGATTAC |
| Sequence-based reagent | Orco_5HA_REV | This paper, Integrated DNA Technologies | PCR primers | CGCGGCCCTCACGCGTCTTGA GCTGTACAAGTACCATAAAGT |
| Sequence-based reagent | Orco_3HA_FOR | This paper, Integrated DNA Technologies | PCR primers | GTTATAGATCACTAGTCTCAG TACTATGCAACCAGCAATA |
| Sequence-based reagent | Orco_3HA_REV | This paper, Integrated DNA Technologies | PCR primers | AATTCAGATCACTAGTGTTTT ATGAAAGCTGCAAGAAATAA |
| Sequence-based reagent | Ir8a_gRNA_FOR | This paper, Integrated DNA Technologies | PCR primers | TCCGGGTGAACTTCGTTTGT TTGTTCGGCCATGTGTTTTAG AGCTAGAAATAGCAAGTTA |
| Sequence-based reagent | Ir8a_gRNA_REV | This paper, Integrated DNA Technologies | PCR primers | TTCTAGCTCTAAAACCTGTGG GAGTCAGAGGCACCGACGT TAAATTGAAAATAGGTC |
| Sequence-based reagent | Ir8a_5HA_FOR | This paper, Integrated DNA Technologies | PCR primers | CCCTTACGTAACGCGtCTATT GGCTATTCGTCGTACTCATGC |
| Sequence-based reagent | Ir8a_5HA_REV | This paper, Integrated DNA Technologies | PCR primers | CGCGGCCCTCACGCGtCTCCA TGTAGCCACTATGTGAGTCAGAT |
| Sequence-based reagent | Ir8a_3HA_FOR | This paper, Integrated DNA Technologies | PCR primers | GTTATAGATCACTAGtGTTTCTT GTCGCACCTAATTAACAAGTG |
| Sequence-based reagent | Ir8a_3HA_REV | This paper, Integrated DNA Technologies | PCR primers | AATTCAGATCACTAGtCATAC TTAAGCTCCTTGAGGTCCAGC |
| Sequence-based reagent | Ir76b_gRNA_FOR | This paper, Integrated DNA Technologies | PCR primers | TCCGGGTGAACTTCGCAGT GATGCGAACTTCATAGTTTT AGAGCTAGAAATAGCAAGTTA |
| Sequence-based reagent | Ir76b_gRNA_REV | This paper, Integrated DNA Technologies | PCR primers | TTCTAGCTCTAAAACCGGCG GCCCTCTTTCAATACGACG TTAAATTGAAAATAGGTC |
| Sequence-based reagent | Ir76b_5HA_FOR | This paper, Integrated DNA Technologies | PCR primers | CCCTTACGTAACGCGtACCAATG AATCCTTGTCCATGCTAAA |
| Sequence-based reagent | Ir76b_5HA_REV | This paper, Integrated DNA Technologies | PCR primers | CGCGGCCCTCACGCGtCTC GGTGTAGCTGTCTTGAAGGAA |
| Sequence-based reagent | Ir76b_3HA_FOR | This paper, Integrated DNA Technologies | PCR primers | GTTATAGATC ACTAGtGCCTA ATTGGAATACC TTCTACATAATGGA |
| Sequence-based reagent | Ir76b_3HA_REV | This paper, Integrated DNA Technologies | PCR primers | AATTCAGATCACTAGtGGCAA GGCACAAAATAAAACGAAG |
| Sequence-based reagent | Ir25a_gRNA_FOR | This paper, Integrated DNA Technologies | PCR primers | TCCGGGTGAACTTCGCCGGA TACTGATTAAAGCGGTTTTA GAGCTAGAAATAGCAAGTTA |
| Sequence-based reagent | Ir25a_gRNA_REV | This paper, Integrated DNA Technologies | PCR primers | TTCTAGCTCTAAAACAGTG CTCATTTTACCATAATCGA CGTTAAATTGAAAATAGGTC |
| Sequence-based reagent | Ir25a_5HA_FOR | This paper, Integrated DNA Technologies | PCR primers | CCCTTACGTAACGCGtTGC ATGACTTCATTGACACCTCAAG |
| Sequence-based reagent | Ir25a_5HA_REV | This paper, Integrated DNA Technologies | PCR primers | CGCGGCCCTCACGCGtGAA ACGAGGCTTAAACGTAGCTGGATATT |
| Sequence-based reagent | Ir25a_3HA_FOR | This paper, Integrated DNA Technologies | PCR primers | GTTATAGATCACTAGtAATAT TATGGTTAAGTGAGCTCTCGG |
| Sequence-based reagent | Ir25a_3HA_REV | This paper, Integrated DNA Technologies | PCR primers | AATTCAGATCACTAGtCAAA GCTAAGTTCATCGTCATAGAGAC |
| Sequence-based reagent | Orco_Seq_FOR | This paper, Integrated DNA Technologies | PCR primers | GATGTTCTGCTCTTGGCTGATATTC |
| Sequence-based reagent | Ir8a_Seq_FOR | This paper, Integrated DNA Technologies | PCR primers | CATCGACTTCATCATCAGGCTTTCG |
| Sequence-based reagent | Ir76b_Seq_FOR | This paper, Integrated DNA Technologies | PCR primers | CAACGATATCCTCACGAAGAACAAGC |
| Sequence-based reagent | Ir25a_Seq_FOR | This paper, Integrated DNA Technologies | PCR primers | CGAAAGGATACAAAGGATACTGCAT |
| Sequence-based reagent | HACK_Seq_REV | This paper, Integrated DNA Technologies | PCR primers | TGTATTCCGTCGCATTTCTCTC |
| Sequence-based reagent | IVS-FOR | This paper, Integrated DNA Technologies | PCR primers | TGGGTTGGACTCAGGGAATA GATCTAAAAGGTAGGTTCAACCACT |
| Sequence-based reagent | EcoRI-SV40-REV | This paper, Integrated DNA Technologies | PCR primers | GCTTACGTCAGAATTCAGA TCGATCCAGACATGATAAGA |
| Sequence-based reagent | Tsp42Ej_FOR | This paper, Integrated DNA Technologies | PCR primers | GAGAAGTCGTTTCCCATAACACCCT |
| Sequence-based reagent | Tsp42Ej_REV | This paper, Integrated DNA Technologies | PCR primers | GAGGAGCAGTTTTCGGAGTCGCCTTC |
| Sequence-based reagent | D. melanogaster Orco | This paper, Molecular Instruments | RNA in situ HCR probe set | See Appendix 2 ‘In situ probe sequences’ |
| Sequence-based reagent | D. melanogaster Ir25a | This paper, Molecular Instruments | RNA in situ HCR probe set | See Appendix 2 ‘In situ probe sequences’ |
| Sequence-based reagent | D. melanogaster Ir40a | This paper, Molecular Instruments | RNA in situ HCR probe set | See Appendix 2 ‘In situ probe sequences’ |
| Sequence-based reagent | D. melanogaster Ir51a | This paper, Molecular Instruments | RNA in situ HCR probe set | See Appendix 2 ‘In situ probe sequences’ |
| Sequence-based reagent | D. melanogaster Ir60a | This paper, Molecular Instruments | RNA in situ HCR probe set | See Appendix 2 ‘In situ probe sequences’ |
| Sequence-based reagent | D. melanogaster Ir62a | This paper, Molecular Instruments | RNA in situ HCR probe set | See Appendix 2 ‘In situ probe sequences’ |
| Sequence-based reagent | D. melanogaster Ir76a | This paper, Molecular Instruments | RNA in situ HCR probe set | See Appendix 2 ‘In situ probe sequences’ |
| Sequence-based reagent | D. melanogaster Ir76b | This paper, Molecular Instruments | RNA in situ HCR probe set | See Appendix 2 ‘In situ probe sequences’ |
| Sequence-based reagent | D. melanogaster Ir93a | This paper, Molecular Instruments | RNA in situ HCR probe set | See Appendix 2 ‘In situ probe sequences’ |
| Sequence-based reagent | Anopheles coluzzii Orco | This paper, Molecular Instruments | RNA in situ HCR probe set | See Appendix 2 ‘In situ probe sequences’ |
| Sequence-based reagent | A. coluzzii Ir25a | This paper, Molecular Instruments | RNA in situ HCR probe set | See Appendix 2 ‘In situ probe sequences’ |
Appendix 2
In situ probe sequences
The following coding sequences (no introns or UTRs) from FlyBase were used by Molecular Instruments, Inc (Los Angeles, California) to produce custom probe sets:
Antenna
Ir76b (1911 bp; Probe set ID PRH997)
AUGGCCACUGGCAUCGAGCUGCUGGUGGCCGCCGCCCUCUGUGUCGCCUGUCCGCCGCUGAACGAUUCCCCGCCGACGAACCUAAUCCAAAUGGGCGAGAAUGGCACUCUUUCCCCGGUCACCGAGCUGCCCAUGGAUGUGGACGCAUCGGAAGCUGGAUUCGAUGCGGAUGCCCCCGUGGAGACGCUGGAGACCAUUAACAGGAAGAAGCCGAAGCUGCGGGAGAUGCUCGAUUGGAUCGGCGGCAAGCACCUGCGCAUCGCUACCCUGGAGGACUUUCCGCUCAGCUACACCGAGGUCCUGGAGAACGGCACCCGUGUGGGGCACGGAGUCUCCUUUCAGAUCAUCGACUUCCUCAAGAAGAAGUUCAACUUCACCUAUGAAGUGGUCGUGCCCCAAGAUAACAUCAUCGGCUCGCCUAGCGACUUUGAUCGCAGUCUCAUCGAGAUGGUAAACAGCAGUACGGUGGACUUGGCGGCGGCCUUCAUACCCUCGCUCUCUGACCAGCGCAGCUUCGUCUACUACUCCACAACGACGCUGGACGAGGGCGAAUGGAUAAUGGUGAUGCAGCGUCCCCGCGAGUCGGCUAGUGGGUCCGGACUGCUUGCGCCCUUCGAGUUCUGGGUGUGGAUCCUGAUCCUCGUCUCGCUGCUGGCCGUGGGGCCGAUCAUCUACGCGCUGAUCAUCCUGCGAAAUCGGCUGACCGGCGACGGCCAGCAGACGCCCUACUCCCUGGGCCACUGCGCUUGGUUCGUCUACGGAGCGCUGAUGAAGCAGGGCAGCACCCUGUCGCCCAUUGCAGACUCGACGCGGCUGCUCUUUGCCACCUGGUGGAUUUUCAUCACGAUACUGACGUCCUUCUACACGGCCAACUUGACCGCCUUCCUGACCCUUUCCAAGUUCACGCUGCCGUACAACACGGUCAACGAUAUCCUCACGAAGAACAAGCACUUUGUGUCCAUGCGGGGCGGUGGAGUGGAGUACGCCAUUCGAACGACCAAUGAAUCCUUGUCCAUGCUAAACCGAAUGAUCCAGAACAACUACGCCGUAUUCUCGGACGAGACCAACGACACCUACAAUCUGCAGAACUACGUGGAAAAGAAUGGCUAUGUUUUUGUGAGGGAUCGGCCGGCGAUAAACAUAAUGUUGUACAGGGACUACCUGUACCGCAAAACCGUGAGCUUUAGCGACGAGAAGGUCCACUGUCCGUUUGCCAUGGCCAAGGAGCCGUUCCUGAAGAAGAAGAGGACCUUUGCCUAUCCCAUCGGAUCGAAUUUGAGCCAAUUAUUUGACCCGGAGCUGCUACACCUGGUGGAAUCUGGAAUCGUGAAGCACCUGUCUAAGAGAAAUCUGCCCAGUGCCGAGAUCUGUCCGCAGGAUCUCGGCGGAACGGAGCGGCAGCUGAGGAACGGCGACCUAAUGAUGACCUACUACAUCAUGCUUGCCGGUUUCGCCACCGCACUGGCCGUCUUCAGCACGGAGCUAAUGUUCCGGUACGUCAAUAGUCGCCAGGAGGCGAAUAAGUGGGCGCGCCACGGAAUCGGACGAACGCCCAACGGCCAGUCGGUGGCUCCAUCCCGGUGGCUCCGUGGCUGGAGGCGAUUGAACAGUGGACAUGGGCAGCUCCUGGGCGCCUCCACCCACGGCCAAAAUGUCACUCCUCCGCCGCCGUACCAGAGCAUCUUCAACGGCGGCAGUCACGGAGAUCCACUGAAUCGCUGGCGACGUCCCCUCGCAAACGGAAACGCCCUUGGCAAUGGUGUCCUCCUGGGCGGCGAUUCUGAAGGUGGUGUACGGCGCCUGAUCAACGGACGCGACUACAUGGUAUUCCGCAAUCCAAAUGGUCAAAGCCAGCUCGUGCCGGUUAGAUCGCCCUCGGCGGCCCUCUUUCAAUACAGCUACACUGAGUAG
PALP
Ir40a (2203 bp; Probe set ID PRH239)
AUGCAUAAGUUUCUGGCAUUGGGCCUGCUGCCCUACCUUUUGGGAUUGCUAAACAGCACAAGGCUGACUUUUAUUGGUAACGAUGAGUCAGACACUGCAAUAGCGCUCACCCAAAUUGUAAGAGGCUUGCAACAAUCUUCUCUUGCCAUAUUGGCGCUACCAAGCCUCGCUCUAUCUGAUGGAGUUUGCCAGAAAGAGCGCAACGUUUAUCUUGACGAUUUUCUGCAGCGUCUUCAUCGCAGUAACUACAAGUCGGUGGUAUUCAGCCAGACGGAGCUCUUUUUUCAACACAUUGAGGAAAACCUUCAAGGUGCAAACGAGUGCAUCAGCCUGAUUUUGGACGAGCCCAACCAGCUGUUGAAUAGCCUCCACGAUCGACAUCUCGGACAUCGCUUAAGCCUAUUUAUUUUCUAUUGGGGAGCACGCUGGCCACCCAGCUCCCGUGUAAUUCGUUUUAGAGAGCCGCUUCGAGUGGUAGUCGUAACUCGUCCUCGCAAGAAGGCCUUCCGCAUUUACUACAACCAGGCUAGACCCUGUAGCGACAGUCAGCUACAGUUGGUUAAUUGGUACGACGGCGAUAACCUUGGUCUGCAACGAAUUCCCCUCCUCCCGACUGCAUUAUCCGUGUACGCCAACUUUAAAGGUCGUACCUUUCGGGUGCCCGUAUUUCAUUCUCCGCCGUGGUUUUGGGUAACGUAUUGCAAUAACAGCUUCGAAGAGGACGAGGAGUUUAACAGCCUAGACAGCAUAGAGAAGAGAAAGGUUCGGGUCACUGGUGGCCGCGAUCACCGCCUACUCAUGCUGCUAUCUAAGCAUAUGAACUUUCGGUUUAAGUAUAUCGAAGCACCCGGUCGAACCCAGGGCUCAAUGAGGUCAGAAGAUGGCAAGGAUUCGAACGACAGUUUCACAGGGGGCAUUGGAUUGCUGCAAAGUGGACAAGCUGACUUUUUUUUGGGAGAUGUCGGUCUAAGCUGGGAACGGCGGAAGGCCAUCGAGUUCUCUUUUUUCACACUGGCUGAUUCAGGAGCGUUUGCUACACACGCUCCCAGACGCCUUAAUGAGGCCCUGGCGAUUAUGCGCCCGUUUAAGCAAGACAUCUGGCCCCAUCUAAUCCUUACGAUAAUUUUCUCCGGCCCUAUUUUUUAUGGCAUUAUUGCCCUGCCUUAUAUUUGGCGUCGACGAUGGGCGAACUCAGAUGUUGAACAUCUCGGAGAAUUAUAUAUCCAUAUGACGUACUUAAAAGAGAUAACCCCACGCUUAUUAAAGCUCAAACCCAGAACUGUGCUGUCUGCCCACCAGAUGCCCCAUCAACUUUUUCAGAAGUGCAUAUGGUUCACUUUACGUCUGUUUUUAAAACAAUAUGGCAUGCAAUGAACUACAUAACGGAUACCGAGCCAAGUUUUUGACCAUAGUGUAUUGGAUAGCAGCGACCUAUGUUUUGGCCGAUGUAUAUUCAGCUCAACUGACCAGCCAAUUUGCACGUCCAGCUCGCGAGCCACCAAUCAAUACUCUUCAGCGCCUGCAAGCAGCGAUGAUUCAUGACGGUUACCGGCUAUAUGUGGAGAAGGAAAGCAGUUCAUUGGAGAUGUUGGAGAAUGGGACAGAACUGUUUCGUCAGCUUUAUGCUCUGAUGAGGCAGCAGGUGAUCAAUGACCCUCAAGGAUUUUUUAUUGACUCUGUGGAAGCGGGAAUUAAACUAAUUGCAGAGGGCGGCGAGGACAAGGCAGUACUCGGAGGGCGUGAAACACUGUUUUUCAACGUUCAGCAAUACGGAUCAAACAACUUUCAGCUCAGUCAAAAACUUUACACUCGUUAUUCGGCUGUGGCUGUUCAAAUCGGAUGUCCCUUUCUAGGUAGCCUCAAUAAUGUCUUGAUGCAGUUGUUUGAGAGCGGAAUCCUAGAUAAGAUGACCGCUGCCGAAUACGCAAAGCAGUACCAGGAGGUAGAAGCCACGAGAAUAUACAAGGGCAGCGUGCAGGCGAAAAACAGUGAGGCUUACAGUCGAACCGAAAGCUAUGACAGCACGGUUAUCAGUCCGCUUAAUCUACGAAUGCUGCAGGGCGCUUUUAUCGCUCUCGGAGUUGGUUCAUUGGCUGCAGGUGUAAUUUUGCUGUUAGAGAUAGUAUUUAUAAAACUGGAUCAAGCGCGAUUGUGGAUGCUGUGCUCACGGCUGCAAUGGAUUAGAUAUGACAGGAAAGUGUAA
Ir51a (1830 bp; Probe set ID PRD951) – sequence derived from Potter lab wildtype strain, which has no predicted premature stop codons.
AUGCAAGGAUUUCAAGAAGCCAAUGCACAGUUAACCACCAUGUACAACGUUCUGGUAUUGUUCCUAUUGCUUUUCACUCGUGCCCAGAUGGAACCCCAUAGAAGAGGUCACAACAUGACUUUGCUGAGAUCGGUGCUGACAGUCAUCCGCGGCAGGGAGAAUUGGAAAAAUACCCCCAUCUUCUUAGGCGGACAUUGUAAUUCGGAUGACCUGAAUAACUUGAUGAGUUGGCUUCAAAAUACGAUGGAAGUAACUUGUCAUACGGUGGAUACAUCUACUUCAGCCAAAAACGAAAACGCUCUGGGUCACUUUAACAUAAAUGCGGAUAAUUCGCUUGGUUUGUUGUUUUGCCAAAGCUCCCAUGAAUUGAUUUGGUUUAAUAUGGAUAAGAGACUUCGGCGAUUACGUGGUAUUCGCCUUAUUGUGAUUCUAUCAGACAAACGAAGUUCAUCCAGCAAAGCUAUAAUGAGCACGUUUAAGAGACUGUGGCACUUUCAGUUCCUCAGAGUUCUUGUUCUGCACAGAGAUCAGAUUUAUUCGUAUACUCCUUAUCCAGUCAUUCGAUUCUUUAAGCUUGAUAGUGACGUCUAUCCGCUUUUCCCACCGAGUGCAAAAAAUUUCCAAGGCUAUGUGGUGUCUACCCCAGUGGAAAACGACAUUCCGCGAGUGUUUUUCGUGAAGGACAAGAAAACCGGACGCAAGCAGAUCAGAGGAUUUGGAUAUCGCACAUUUGUGGAGUACCUGCAUCGCUACAAUGCCUCUUUACAUGUCAGUAAUUCUCAGCAGGAGCAUGCCAUCAACAGCAGCGUGAAUAUGGGUCGGAUAAUUAAUCAGAUUGUGGAUGGCCAAUUAGAAAUCUCCCUGCAUCCGUAUGUAGACGUUCCGGAAAAUAUGGGAGAUAACAGUUAUCCCCUUCUAAUAGCUAGCAAUUGUCUUAUUGUUCCGGUCAGGAACGAGAUAUCUCGCUACAUGUAUCUACUAUUGCCUCUCAACCAAUCAAGCUGGAUACUACUGCUCGGUUCUGUAAUUUAUAUCAGCGGAGUGCUCUACUACAUUCAGCCUGGUCUGCUGCACCGCACCUGGGAUCAGCGGAUUGGCCUCAACAUCCUAGAUAGCAUCAGCCGAAUAAUUAAUAUAUGCUCUCCCUCCAGGAUUUACAACCCAUCCCUGAGGUAUUUUAUAGUUUCGGUGCACCUUAGUAUUCUGGGCUUUGUGGUGACCAACCUCUACAGCAUUAUGUUGGGCAGCUUCUUCACCACCUUGGUAGUGGGCGAGCAGGUGGACAGCAUGCAGCAGCUCAUCCAGCAACAGCAAAAGGUACUUGUAAAAUAUUACGAAGUCAGUACAUUCUUACGGCAUGUGGAACCAGAUUUGGUGGACGGAGUAGCGCAACUAUUAGUGGGCGUGAAUGCCAGUGAGCAGGUGUCCGCUCUUCUGGGAUUCAAUCGGAGCUAUGCCUACCCCUUCACCCUGGAGAGAUGGGAGUUCUUCUCACUACAACAGCAAUACGCCUUCAAGCCAAUCUUCAGAUUCUCAUCGGCAUGCCUGGGCUCCCCGAUUAUAGGCUAUCCCAUGAAAAGUGACUGCCACCUGCAGUCGUCAUUGAACAUGUUCAUCAUGCGGAUCCAGGCCGCAGGACUUCUGCGGCACUGGGUAGUAUCCGAUUUUAACGAUGCAAUGCGCGCUGGCUAUGUUCGACUUCUGGAAAACUUCCUAGGAUUUCACUCGUUAGAUGUCGAUUCCUUGCGCCUGGGGUGGGCAGUACUCCUAUGUGGGUGGCUGCUAUCCACCUUGAUUUUUCUUUGCGAGCGCUGGCGCUUUUACCACUAA
Ir60a (2151 bp; Probe set ID PRD952)
AUGUGGUGUAAUAAUCCGGGUCUCAUCAUCAUCAUCUUUCUGGGGCAGAUUUUAAACCUGUGCCAGGGGAUAGUGAAUCUUUCGAAUGAGACGGCCAACACGGUCAUCUUCAUGCUGCCCGAAAAGGACUUGGGUCCCGAUGUGUGGAAGGCUGGAGUGGGCUGCCUCGAUAGCUUCGCCCAAAUCUUCUUCUUUCGCAACCCGAAGGAGCGCUUCACCAGAGCCUAUAACUUGAUGCUGGUGCACGCCUUUCAUCUGUCCAGUCCGGCGGAUCAGAUCCAGGAAGGAUUCAGCAAACUGAUCAACGAAGCAGUCACGAAUCCAGGGCCGCCGGACAGGGAGGAACUCUUCCAAAUGCGCGUGGCCAGUGAUUACAACAUCACGAAUGGGACCGAAGACAAGGGAGAGCUGAUCCUAGCGGACAAUUAUGUGAUAGUGGUGGACUCGGUGGAUCGAUUAAAAGAGCUAAUGAAGAAAAAAAUUGUCGAAAUGCGAUCCUGGAAUCCAGGAGCGCGUUUUCUGGUGCUCUUUCAUAAUGCGACUUGUCGGAAUCGACCGCUUGGCGUAGCCUCCAAUAUUUUCAAGGACCUCAUGGAAAUGUUUUACGUGCACCGAGUUGCUCUUCUCUAUGCCAACUCCACCAUGAACUACAAUCUGCUGGUCAAUGAUUACUACAGCAAUGUAAACUGCAGGAUUCUGAAUGUCCAGAGCGUGGGCCAGUGCCACGAUGGCAAACUUUACCCCAAUAAUGCUGUCGUUAAGGCCUCCAUGCAGGACUACGUAUCAGGAUUCAGUCCCAGGAACUGCACCUUUUUUGCCUGUUCCUCCAUCUCUGCUCCCUUUGUGGAGGCCGACUGUAUCCUGGGACUAGAGAUGAGGAUCCUGGGGUUCAUGAAAAAUCGACUGAAAUUCGAUGUAAACCAAACCUGCAGCCUGGAGUCACGUGGUGAAAUGGAUGGCCCAGCUAACUGGACUGGAUUACUGGGGAAAGUCCAGAACAACGAGUGCGACUUUGUCUUCGGCGGCUAUUAUCCGGACAACGAGGUGGCGGACCAUUUUUGGGGAUCCGAUACCUAUCUGCAGGAUGCGCACACGUGGUACAUAAAAAUGGCCGACAGAAGACCCGCCUGGCAGGCGUUGGUGGGUAUUUUCGAAGCCUACACCUGGAUUGGAUUCAUCCUGAUUCUAAUAAUCAGCUGGCUGUUCUGGUUCACCCUGGUUAUGAUUCUUCCGGAGCCGAAGUAUUACCAACAGUUGAGUCUUACGGCCAUUAAUGCCCUGGCCGUCACCAUAUCGAUAGCCGUCCAGGAACGACCCAUUUGCGAGACGACGAGGCUGUUCUUCAUGGCCCUGACUUUGUAUGGCCUGAACGUGGUGGCUACAUACACGUCCAAGAUGAUAGCCACCUUCCAGGAUCCCGGCUACCUUCACCAGCUGGACGAGCUGACGGAAGUGGUGGCCGCGGGUAUUCCGUUUGGCGGCCACGAGGAGAGUCGCGACUGGUUCGAGAAUGACGACGACAUGUGGAUCUUUAACGGAUACAACAUUUCGCCGGAGUUCAUUCCGCAAUCGAAGAACCUGGAGGCGGUGAAGUGGGGCCAGCGGUGCAUCCUGAGCAACCGGAUGUACACGAUGCAGAGUCCCCUGGCGGACGUCAUAUACGCCUUUCCCAACAACGUUUUCAGCAGUCCGGUGCAGAUGAUCAUGAAGGCGGGAUUCCCCUUUCUCUUCGAGAUGAACAGCAUCAUCCGCCUCAUGCGCGACGUGGGCAUUUUUCAGAAGAUCGACGCGGACUUUAGGUACAACAACACGUACCUCAACAGGAUCAACAAGAUGCGCCCCCAGUUCCCGGAAACGGCUAUCGUCCUGACCACGGAGCACCUGAAGGGACCCUUCUUCAUCCUGGUGGUGGGCAGCUGCUGGGCCGCCCUCACGUUCAUUGGCGAGCUAAUAAUCCACAGAUGGCGAACCCAGCUGGUCAGCACGAGCGAGCAGCAGGACAGGAGGAGUGACAAGAGGAGGAGGAGGAGGAGGAGGAGGAAGCCAGAAAAGGAUAACCGCUGGCAGCGACAAGUUCAAGUGGCUCCAGUCGUUCGAUUUACGCCAGUCAAACGACGCAAAGUUUUCCAGGGACAAACAAGUCAAAAGUAA
Ir62a (1821 bp; Probe set ID PRD953)
AUGUAUCUUCAGUUUUUGUUCGCGCUGUUUUUGUCGCGCUACCAAAUCGUGGCCACCGAAAACUUUGACCGCGCUUUCGAGCUGGCUCUGUUUCUGGACAGGAUCGGCCGCGUGCAUCGCUUGCAUGCCAUCACUAUAGUGAACAGCCUGGGAUCUGUUGAUCCCAGCUAUCUGGACGAUCUGCAUCGCGGCCUGAUGUGCAACAGCAGCAAUCACUUCUACAUGCUGCCGCAGAUGACGGCCACGGACAAGGAUUCCUCGCACGUCCACUUCAGCAGCCUGCAGGAUGAGGAGACCAUAUAUUUGGUCUUCGCCAGGGAUUCCAAGGAUGCGGUGAUUUACUUGCAGGCCGAAAGAGCCAGAGGAAGGCGUUACACGAGGACUAUGUUCCUGCUAAGAAAGCAGGAGUCGCAAAAGGACAUUAAAUACUUCUUUGAACUCCUGUGGAAACUGCAGUUCCGGAGCGCUUUGGUAGUGGUGGCCGCCAGAAACUUCUACCAAAUGGAUCCAUAUCCCACUGUUAGGGUGAUACGCAUGCGAAGAUUAUCCUCCUAUGAUCCGCAUCAUGUUUUUCCGCCUGCAAAUCGAAAAAAUUUCAGAGGCUACAGGAUGCGAUUGCCCGUACAGCAGGAUGUGCCCAAUACUUUUUGGUACAAGAAUCGCAGGACGAAGGCCUGGGAACUGGCCGGAUUGGGCGGGAUCUUGAUCAAUCAACUGAUGAUGCACCUGAACGUGACCAUGGAUUUGUUUAGAUUUGAAGUAAAUGGCUCUUCAUUGCUGAAUAUGGCUGCGCUCACGGAUCUGAUUGUAAAGGGCAAGGUGGAGCUGAGUCCGCACUUGUACGACACACUGCAAUCAAACACCAGUGUGGACUACUCAUAUCCCACGCAGGUGGCACCGCGCUGCUUCAUGAUCCCAUUGGACAAUGAAAUUUCGAGGAGCCUCUAUGUGUUCCUGCCAUUUAGUUUAACCAUGUGGCUGUGCUUGCUGUUUGUUCUGCUCGUAGUGCACUUCGUGUAUGUGCGCCGCCUGAUUCCAGACGGACAUUUUUGGGCCAUACUAGGAGUGCCUGGUGCUGGUCAAGUCAGAUAUGGCAACCGCAAGCCGGUCAGGCGCUUUAGUACCUUUCUGAUCCUCUUCGGAAUAUUCAUUCUUGGGCAAACGUACAGCACCAAACUAACUUCAUCUCUGACCGUGACUCUGAUCCGAAGACCAGACAACAGCUUGGAGGAGCUAUUCCUGCUGCCGUAUCGCAUUCUAGUGCUGCCCACGGAUGUGUACGCCAUCGUUGAUUCCCUGGGUCAUGCCGAGCAGUUUAGCACAAAGUUUAGCUGCACGGAUGCCGAGAACUUCAGCCAGAAGAGAAUCUCCAUGCAUCCGGAGUAUAUUUACCCCAUCAGUACAAUUCGUUGGCGUUUCUUCGAUAUGCAGCAGCGCUUUCUGAGAAAGAAACGCUUCUAUUUCUCCAAGAUUUGCCACGGCUCCUUUCCCUAUCAGUACCAGCUCAGGGUAGACUCACAUCUCAAGGAUGCCCUCCAUCGAUUCCUGCUGCAUGUGCAGCAAGCCGGGCUGCAUGAUCUUUGGCUGGAUACGUGCUAUCGCAAGGCCCAUCGCAUGGGCUAUCUCAAGGAUUUCUCCACACUGGCGGAGCUGGAGGAGAAACUGAGGCUCCGCCCACUGGCGCUCAAUCUGCUGGUGCCCGCGUUCAGUCUGUUUCUGUGCGGAAUGUUGGGCAGCGGAAUCGCCUUCCUGGUGGAGAUCCGUCACAGCUUCGGGUGCAGACAAAAGCCACCGUCCAUUAACCGCAACCCCGGGGAUUAA
Ir76a (3281 bp; Probe set ID PRH240)
AUGGAAAACUUGCUGGUAGAGUCAUAUUACUUUAGCACUGUGCUUAGUUUUUUUGCUCAGCAGUUCUUCGCGGACUCGCAUGCCACUUGUAUUUUUUGGCACCCGGCGUUUGAUUUUCGUCUGGAAACGGUUCAUCCAAUGCCAUUAAUAAUCAUGGACUGGCACAGAUGGGCCAAUCGUUCCGAUCAAGAUGUUUAUGAUUACAAAAUAAAGGAGGAUGAGUUCGAGGGGAAGGGAAUCCCCUACAACGAUUGGACUCUUAGACUGACUGUUGCCAUCGAAAGGUCGCACUGUGAGCUAGUCAAAGAUAGUAUCACACUUAGAGUAGGUAUCCCGCUGACAGUGAUGACAUGUGGCUCCAUCAAAACUCACUUGCAAGUGUUUAUCAUUCCUGAACUUAUCUGUGAACUUUUUCCGAGUAUCCGGUGUCUUAAUGAAAUCGAGCGUUAGCAAAAGCCGGUCUUUCCGAUCCGCUCCUGCAUCAGCCACAUUGUCCAAAAAUUGAUUUACAUCCUCGAUAGGCAUCUUCCAAUGUUUUCCCAAUCUCAGAUGAUUUAGGUUAUAGCAAUUUUGUACUAGCAACAUAAGUUGGCGACUUGGGUUUGCAGGUGAGUCAAUGCAAUCCAGGGCCAGGCACUCGAGAUCUUUCAACUGGCCCAACUGGGACACCGAUUGAGCUGGCCAGUUAUCACAGUCCAGAGCGUUGAGUUGCCUAAUCGCCAGGAUGUGAUGCACUUGGUCACCUCUAACGCGGACGGAUAUGAGCUGUAGCUUUUGGAGGUUACUAACUCUUUUGGCCACCGACCUAUAAAAGUCGUUGUCCACUUCUGCUGUCCAACGUGAGAUUAGGCGCAGGGAUCGGAGACCUUUAAAUUCACCCAACUCCGGGAAGCUCAUAUCAAAGUUUUCGAGAGUAAGGUGAUCCAAGUUGGGAAACUUAUCUGCCAACAACGGGACCUGGUGAGAUCUCCUGCGAUCCGGUCCGAAAUUGAGAAGCAGUCGCUUCAGGGAAGCCAUACUUUCAAACAUUUGUUGGAAGGAGGCGUUGCUGAGGUUUGGGUCGAUUCCAUCCAAGUCCAAGGCCUCCAGUUUACUGAAGUGCCGCAAUUGAUCUAGCUGUGCAGAUCUGGCAUCGAUCAGAGUGAGAUUGGUUAGACUGGGUAGCUCCAGGAGGAGCUGCAUGAUGUGGCCCCUGUCCCCACUUUUCGGCGGCGACUCUGUAACGGCGUGCAUAAAUAUGAUGACUGCCUGUCGCAGUUUGGGGCAGUGCCGUCCCAGGAGCCCCAGAAAGGGGUAGGUAAACGGCUCAUCCCAGCUACCAUGGGGCACUUCGCACCUGGACACUUCGGAACCGCACAGCUGCAGCAGGAACUCCCAGUCCGGAAUGGUUUUCAGAAGCCGCACAUUGAUUUUUUUGUGGCGCGUUCGCAAAAUGUCCGUCAGUACAACCUGGAAUAGUGGAUGGGCCCGGCCCAGAUUUAGCUGGUCCUCCAGCACAUUUAUAUAUGAAAAUAUCUGAUGGUAGCAGUCGUAGUUGAGAUCCAGGAAACCGGGUCCCCCACUCAUACUCUGCGGUCUUCCGACCAGGAAGGAGAAGGGGCAGCCUAAUCCCAGUUUGUCCAUACCUUCAUAGCAUUUCAGGAGCAGAUUCCCGAAUUCGCCAGAUAUUUUUAUCACGCCAGCAUCUACUCCAUUUGGAGAUCUCUGCGAAAUCGUUUUAUGUUCGUUUACACUAAAGAGUUUGAGGACAAAAAAGAUAGUUAUCUGUCCGGUUAUAUAUUCCAAGAUCAACCUAACAUCUUGGUAAUUACUUCACAAUACCUGAACUCAAGCACUUUUGAAAUUAAAACGAACCGAUUUGUCGGUCCGCGAAACUUUAACAAAAACCCGGAGCCUGUUGAGUUUUACAUACUCCAGCGUUUCGAUGCCAAGGGUACGAAAGCUACUUGGGAAACCCAGAGUGCAAUGUCCAGCAAGAUGCGAAACCUCAAGGGACGCGAGGUAGUCAUAGGCAUCUUCGACUACAAGCCUUUUAUGCUAUUGGAUUAUUUAUGUGAGUUAGGAAAAGCCACCAUUAUAUUAUGAUCGUUUUAUGAACACGACGGAUGUAACCAUUGAUGGAACAGAUAUUCAACUAAUGCUCAUUUUCUGCGAGUUGUACAACUGCACCAUUCAGGUGGACACAUCUGAACCAUACGACUGGGGUGACAUCUACUUGAAUGCCUCGGGCUAUGGUCUGGUUGGAAUGAUCCUCGAUAGACGAAACGAUUACGGAGUAGGGGGCAUGUAUUUGUGGUACGAGGCGUACGAGUAUAUGGACAUGACUCAUUUUCUGGGACGAUCUGGAGUAACCUGUCUGGUGCCCGCUCCGAACCGUUUAAUCAGCUGGACGCUCCUGCUCCGACCAUUCCAGUUUGUCCUCUGGAUGUGCGUGAUGCUCUGCCUUCUGCUCGAGAGCUUAGCUCUUGGUAUAACCCGUCGCUGGGAACACUCGUCGGUUGCGGCAGGCAAUUCCUGGAUCAGUAGUCUGCGCUUCGGUUGCAUCAGUACGCUGAAACUUUUCGUAAAUCAGAGCACCAAUUAUGUGACCAGCUCGUAUGCUCUAAGAACCGUCCUAGUGGCCAGCUACAUGAUCGACAUUAUAUUGACCACUGUGUACAGUGGCGGUCUGGCGGCCAUCCUCACUUUGCCCACUUUAGAGGAAGCGGCGGACUCUCGCCAACGUCUCUUCGACCAUAAGCUCAUUUGGACGGGGACUUCACAGGCCUGGAUUACCACCAUUGACGAGCGAUCGGCCGACGCCAGUUCUUCUUGGUUUAAUGGAGCAUUACCGAGUUUACGAUGCCAAUUUAAUAUCCGCCUUCUCGCACACGGAGCAAAUGGGAUUUGUCGUCGAGCGCCUUCAGUUUGGUCAUUUGGGCAACACCGAGCUAAUAGAAAACGAUGCCCUAAAGCGGCUGAAACUUAUGGUGGAUGAUAUUUACUUUGCAUUUACGGUGGCGUUUGUGCCGCGACUGUGGCCGCACUUAAAUGCCUACAACGACUUCAUUCUGGCUUGGCAUUCCUCGGGCUUUGACAAAUUCUGGGAAUGGAAGAUCGCCGCCGAAUACAUGAAUGCGCACCGCCAAAAUCGCAUCGUGGCAUCUGAGAAAACGAACCUAGAUAUAGGACCUGUUAAACUUGGCAUUGAUAAUUUUAUUGGCCUAAUCCUGCUUUGGUGCUUCGGCAUGAUUUGUAGCCUUCUGACAUUUCUCGGAGAACUCUGGAGGGGACAGGGGUAG
Ir93a (2607 bp; Probe set ID PRH241)
AUGAAUCCUGGCGAAAUGCGGCCUUCGGCUUGCCUUCUGCUCCUGGCUGGACUGCAGCUCUCUAUCCUGGUACCCACUGAGGCCAAUGACUUUUCGUCCUUCCUGAGCGCCAAUGCAUCGCUGGCCGUUGUGGUGGAUCACGAGUAUAUGACGGUUCAUGGCGAGAAUAUAUUGGCUCAUUUCGAGAAAAUCCUGAGCGACGUAAUACGGGAGAAUCUAAGGAACGGUGGCAUAAACGUAAAAUAUUUUAGCUGGAAUGCAGUGCGAUUGAAGAAGGAUUUUUUGGCUGCCAUAACUGUUACGGAUUGCGAGAAUACAUGGAACUUUUACAAGAACACUCAGGAAACUUCAAUUCUACUGAUCGCCAUUACGGAUUCCGACUGUCCCAGGCUGCCCCUAAAUAGAGCUCUAAUGGUACCCAUCGUUGAGAACGGCGAUGAAUUCCCCCAACUUAUUCUGGAUGCCAAGGUCCAGCAGAUUCUAAAUUGGAAGACCGCCGUUGUUUUUGUGGAUCAAACCAUAUUGGAGGAGAACGCACUUCUGGUAAAAUCGAUUGUGCACGAAAGUAUAACCAACCACAUCACCCCAAUCUCCCUGAUCCUUUACGAGAUCAACGACUCCCUGAGGGGCCAACAGAAGCGAGUUGCUCUGCGCCAAGCUCUGUCUCAAUUCGCUCCCAAAAAGCACGAGGAGAUGCGCCAGCAGUUCCUGGUCAUAUCUGCCUUUCACGAGGACAUCAUCGAAAUAGCCGAGACCCUGAACAUGUUUCACGUGGGCAAUCAGUGGAUGAUUUUCGUGCUGGACAUGGUGGCUCGGGACUUCGAUGCCGGCACUGUGACCAUAAACCUGGACGAGGGAGCCAACAUAGCCUUCGCCCUCAACGAAACGGAUCCCAACUGCCAGGACUCGCUAAACUGCACGAUCUCGGAAAUUAGUCUCGCUCUGGUCAACGCUAUUUCCAAAAUUACCGUCGAGGAGGAGUCCAUAUAUGGUGAGAUCUCCGAUGAGGAAUGGGAGGCCAUCCGCUUUACCAAGCAGGAAAAGCAGGCCGAGAUUCUGGAGUACAUGAAGGAAUUCCUGAAGACCAAUGCCAAGUGCUCCAGCUGCGCGAGAUGGCGCGUGGAGACGGCCAUUACCUGGGGCAAAAGCCAGGAGAAUCGCAAGUUUCGCUCAACUCCCCAACGCGACGCUAAGAACCGAAAUUUUGAGUUCAUCAACAUUGGCUAUUGGACACCCGUGCUGGGAUUCGUCUGCCAGGAGCUCGCCUUUCCGCACAUCGAGCACCACUUCCGCAACAUAACCAUGGACAUUCUGACCGUGCACAAUCCACCCUGGCAAAUCCUUACCAAGAACAGCAAUGGGGUCAUCGUGGAGCACAAGGGCAUUGUUAUGGAGAUCGUCAAGGAGCUGAGUCGCGCCCUAAACUUCAGCUACUACCUUCACGAAGCCUCCGCAUGGAAGGAAGAAGAUUCACUCAGCACAUCAGCGGGCGGAAAUGAAAGCGACGAGCUAGUUGGUUCCAUGACCUUUCGUAUACCCUAUCGAGUGGUGGAGAUGGUGCAGGGCAAUCAGUUUUUCAUCGCUGCCGUGGCAGCCACCGUUGAGGAUCCCGACCAAAAGCCCUUCAAUUAUACCCAGCCCAUCAGUGUGCAGAAGUACUCCUUCAUCACCCGCAAGCCGGAUGAGGUGUCCCGCAUUUACUUGUUCACGGCACCCUUCACCGUGGAGACUUGGUUCUGCCUAAUGGGCAUCAUUCUGCUGACUGCUCCCACGCUGUACGCCAUUAAUCGCCUAGCUCCUCUGAAGGAGAUGCGAAUCGUGGGCCUGUCCACAGUUAAGAGCUGUUUUUGGUAUAUAUUCGGGGCUUUGUUACAACAGGGAGGCAUGUACUUGCCCACAGCAGACAGUGGGCGCCUAGUGGUCGGCUUUUGGUGGAUCGUGGUUAUCGUGCUGGUGACCACCUAUUGCGGCAACCUUGUGGCCUUCCUCACGUUCCCCAAAUUUCAACCGGGCGUGGACUAUUUGAAUCAACUAGAGGACCACAAGGACAUUGUACAGUAUGGAUUGCGAAACGGCACCUUCUUCGAGCGGUACGUUCAGUCGACAACGCGGGAGGACUUCAAACACUACCUGGAACGGGCGAAAAUCUACGGCAGCGCCCAAGAGGAGGACAUCGAGGCGGUGAAGCGUGGCGAGCGCAUCAACAUCGAUUGGCGGAUCAAUCUGCAGUUGAUUGUUCAGCGGCACUUCGAGCGGGAGAAGGAGUGCCACUUUGCUUUGGGCAGGGAGAGCUUCGUGGACGAGCAGAUUGCCAUGAUUGUGCCGGCCCAGAGUGCGUAUCUGCACCUGGUAAACCGCCACAUCAAGAGCAUGUUCCGGAUGGGCUUCAUCGAGCGCUGGCACCAGAUGAACUUACCCAGCGCGGGCAAGUGCAACGGGAAGAGCGCCCAGCGCCAGGUUACCAACCACAAGGUGAACAUGGACGACAUGCAAGGGUGCUUUCUGGUCCUGCUCUUGGGCUUCACGUUGGCUCUUUUAAUAGUGUGCGGCGAGUUCUGGUAUCGUCGCUUUCGGGCCAGUCGAAAACGGCGUCAGUUCACCAACUGA.
Orco (1461 bp; Probe set ID PRD954) – positive control
AUGACAACCUCGAUGCAGCCGAGCAAGUACACGGGCCUGGUCGCCGACCUGAUGCCCAACAUCCGGGCGAUGAAGUACUCCGGCCUGUUCAUGCACAACUUCACGGGCGGCAGUGCCUUCAUGAAGAAGGUGUACUCCUCCGUGCACCUGGUGUUCCUCCUCAUGCAGUUCACCUUCAUCCUGGUCAACAUGGCCCUGAACGCCGAGGAGGUCAACGAGCUGUCGGGCAACACGAUCACGACCCUCUUCUUCACCCACUGCAUCACGAAGUUUAUCUACCUGGCUGUUAACCAGAAGAAUUUCUACAGAACAUUGAAUAUAUGGAACCAGGUGAACACGCAUCCCUUGUUCGCCGAGUCGGAUGCUCGUUACCAUUCGAUCGCACUGGCGAAGAUGAGGAAGCUGUUCUUUCUGGUGAUGCUGACCACAGUCGCCUCGGCCACCGCCUGGACCACGAUCACCUUCUUUGGCGACAGCGUAAAAAUGGUGGUGGACCAUGAGACGAACUCCAGCAUCCCGGUGGAGAUACCCCGGCUGCCGAUUAAGUCCUUCUACCCGUGGAACGCCAGCCACGGCAUGUUCUACAUGAUCAGCUUUGCCUUUCAGAUCUACUACGUGCUCUUCUCGAUGAUCCACUCCAAUCUAUGCGACGUGAUGUUCUGCUCUUGGCUGAUAUUCGCCUGCGAGCAGCUGCAGCACUUGAAGGGCAUCAUGAAGCCGCUGAUGGAGCUGUCCGCCUCGCUGGACACCUACAGGCCCAACUCGGCGGCCCUCUUCAGGUCCCUGUCGGCCAACUCCAAGUCGGAGCUAAUUCAUAAUGAAGAAAAGGAUCCCGGCACCGACAUGGACAUGUCGGGCAUCUACAGCUCGAAAGCGGAUUGGGGCGCUCAGUUUCGAGCACCCUCGACACUGCAGUCCUUUGGCGGGAACGGGGGCGGAGGCAACGGGUUGGUGAACGGCGCUAAUCCCAACGGGCUGACCAAAAAGCAGGAGAUGAUGGUGCGCAGUGCCAUCAAGUACUGGGUCGAGCGGCACAAGCACGUGGUGCGACUGGUGGCUGCCAUCGGCGAUACUUACGGAGCCGCCCUCCUCCUCCACAUGCUGACCUCGACCAUCAAGCUGACCCUGCUGGCAUACCAGGCCACCAAAAUCAACGGAGUGAAUGUCUACGCCUUCACAGUCGUCGGAUACCUAGGAUACGCGCUGGCCCAGGUGUUCCACUUUUGCAUCUUUGGCAAUCGUCUGAUUGAAGAGAGUUCAUCCGUCAUGGAGGCCGCCUACUCGUGCCACUGGUACGAUGGCUCCGAGGAGGCCAAGACCUUCGUCCAGAUCGUGUGCCAGCAGUGCCAGAAGGCGAUGAGCAUAUCGGGAGCGAAAUUCUUCACCGUCUCCCUGGAUUUGUUUGCUUCGGUUCUGGGUGCCGUCGUCACCUACUUUAUGGUGCUGGUGCAGCUCAAGUAA.
Drosophila sechellia experiments (using D. melanogaster sequence)
Orco – see above
Ir25a (2396 bp; Probe set ID PRL623)
CAAACGCCCCAUCUGAUCCUGGACACCACCAAAUCGGGCAUAGCCUCGGAAACGGUGAAGAGCUUCACCCAGGCUCUGGGUCUGCCCACCAUUAGUGCCUCCUAUGGCCAGCAGGGCGACUUGAGGCAGUGGCGCGACUUGGAUGAGGCGAAGCAGAAGUAUUUGCUGCAGGUGAUGCCGCCGGCGGAUAUUAUUCCCGAGGCCAUUCGAAGUAUAGUGAUUCACAUGAACAUCACGAAUGCUGCCAUUCUGUACGAUGAUUCCUUUGUCAUGGACCACAAGUACAAGUCCCUGCUGCAGAAUAUACAAACCCGUCAUGUGAUCACCGCCAUAGCCAAGGAUGGUAAGCGGGAGCGCGAGGAGCAAAUCGAAAAGCUGAGGAACUUGGACAUCAAUAACUUCUUUAUUCUGGGCACCCUGCAAUCGAUCCGCAUGGUCCUGGAGUCGGUGAAGCCAGCGUAUUUCGAGCGCAACUUCGCCUGGCACGCCAUCACUCAGAACGAAGGAGAGAUUAGCAGUCAGCGGGACAAUGCGACCAUUAUGUUUAUGAAACCCAUGGCGUAUACGCAAUAUCGAGAUCGCUUGGGAUUACUGCGAACCACUUACAAUCUGAACGAGGAGCCGCAGUUGUCAUCCGCGUUUUACUUCGAUCUGGCACUUAGGAGUUUCCUUACCAUCAAAGAAAUGUUACAAUCGGGCGCCUGGCCAAAGGAUAUGGAGUAUCUGAAUUGUGACGAUUUCCAAGGUGGCAACACACCCCAAAGGAACUUGGAUCUUCGAGAUUACUUCACCAAGAUUACCGAACCGACUUCGUAUGGAACCUUUGAUCUCGUCACGCAAUCCACUCAGCCAUUCAAUGGGCAUAGCUUCAUGAAAUUCGAAAUGGAUAUAAAUGUGCUGCAGAUUCGUGGUGGCAGUUCCGUGAACAGCAAGUCCAUUGGCAAAUGGAUAUCGGGUCUGAACUCGGAGCUCAUCGUCAAAGACGAGGAGCAGAUGAAGAAUCUCACUGCAGACACUGUUUAUCGAAUCUUUACUGUAGUGCAAGCUCCUUUCAUAAUGCGCGAUGAAACGGCUCCGAAAGGAUACAAAGGAUACUGCAUUGAUCUGAUCAACGAGAUAGCCGCAAUUGUCCACUUCGAUUACACCAUCCAGGAGGUGGAGGACGGCAAGUUUGGCAACAUGGACGAGAAUGGGCAAUGGAAUGGCAUUGUGAAGAAGCUGAUGGACAAACAGGCGGACAUUGGCCUUGGCAGCAUGUCGGUGAUGGCCGAACGGGAGAUAGUCAUUGACUUCACCGUUCCGUACUACGAUCUGGUCGGGAUUACGAUCAUGAUGCAGCGACCCAGUUCGCCAAGCUCGCUGUUCAAGUUCCUUACCGUGCUGGAAACGAACGUGUGGCUUUGCAUCCUGGCUGCCUACUUCUUUACCAGCUUUCUCAUGUGGAUCUUCGAUCGCUGGAGUCCCUAUAGCUAUCAGAACAAUCGAGAGAAGUACAAGGACGACGAGGAGAAGCGCGAGUUCAAUCUGAAGGAGUGCCUCUGGUUCUGCAUGACUUCAUUGACACCUCAAGGCGGUGGCGAGGCUCCAAAGAAUCUGUCUGGCCGUUUAGUGGCCGCCACCUGGUGGCUAUUCGGUUUUAUCAUUAUUGCUUCGUACACGGCCAAUUUGGCUGCCUUCUUGACCGUAUCACGUUUGGAUACGCCCGUUGAAAGCUUGGAUGACCUGGCGAAGCAGUACAAGAUCCUAUACGCUCCAUUGAAUGGCUCAUCUGCGAUGACAUAUUUCGAGCGUAUGUCCAACAUAGAGCAGAUGUUUUACGAGAUUUGGAAGGAUCUGUCGCUGAACGACUCCCUGACCGCCGUGGAGCGCUCCAAGCUGGCUGUUUGGGAUUAUCCAGUGAGCGACAAGUAUACCAAGAUGUGGCAGGCCAUGCAGGAGGCGAAGCUACCGGCCACCCUCGACGAAGCGGUGGCCCGGGUUAGAAAUUCGACAGCUGCCACGGGUUUUGCCUUUCUGGGCGAUGCCACCGAUAUACGCUACCUGCAGUUGACCAACUGUGAUUUGCAGGUGGUGGGCGAGGAGUUCUCCCGGAAACCCUAUGCCAUAGCUGUUCAGCAGGGAUCGCAUCUCAAGGAUCAGUUUAAUAAUGCAAUCCUGACCCUGCUCAACAAACGACAGCUGGAGAAGCUCAAGGAGAAGUGGUGGAAGAACGACGAAGCUCUGGCCAAGUGCGAUAAGCCGGAGGAUCAAUCGGAUGGCAUCUCGAUCCAGAACAUUGGCGGCGUCUUCAUUGUCAUAUUCGUGGGCAUUGGAAUGGCCUGCAUCACGCUGGUCUUUGAGUACUGGUGGUACAGGUACCGCAAGAAUCCGCGGAUCAUCGAUGUGGCCGAAGCCAA.
The following genomic sequences from VectorBase were used by Molecular Instruments, Inc (Los Angeles, California) to produce custom probe sets targeting coding sequences:
Anopheles coluzzii experiments
Orco (10,821 bp; Probe set ID PRL382) – AGAP002560
CGTCGAGCGGAAAGGACGTATCAAGTCGATTCGTCTATCAGTGTCGGAACGATAGTGATAGAAATCTACAGGCGCCAATCATAAACCGTTTACTGCTCGCGATAGGACACGCTTTGCTAACGTCTTGTGCATCGCGAAAACTAGTGATTGGAGTGTGGTTTTGTAGCTGTTTGTCGTTCTGTGCTCCACGTTGACTCTGTGTGTGTGTGCGCTGAATTCAACAAGACTTTTCTGCAACAGCATCATTTGCAAAGAATAACCGGCGCGACTTACGCGGTCTGACTTGCTGGTGCGCTGCTTTGTACGGCAAACGGCTACACAAGCGAATCGAATTATTTTCCTATCACGCTGCGCTTACCAGCGCCTGCTGGTAGGCAAAGAATGTGCAAAGTTTCATTTGGCTTGGTTCGTCTGCTTTGCTGTGAACGTGTGCACGGTTGCATCGCTAAGGTTTCGGTGTGAGCCGAGAAGTTGCAGATCGAAACCCCTTTGTGTGTGTGTGTGCCGTGGGAAGCATTGTGTTTAGTGAGAAGTGAAAAGAAAAGTGCTGAAAAGTAAGTATTCGATGTAGCTTCAACCTTTACACTTGACAAGGCGGAGATCTCAACCATTTTTCTGGTTCGAACGTTAAGCGGAACATTTGATCCTTTCAAAACAGTGTGACTGTGTGTGTGTGTATGTACAATCGCCCCCCATGTATTCCAAATATTTGCTTCTCGCAAATCAGGTCACAAAGCTGTGCTCAATGTTGGCACAATCAAAGCTTTCCCCATTCGTCTGGGCCAAGCCTAGCTCAATTGTTTGTACGAGAGATTTTACCGTTTGGCTCCAAGCATTTGCAATTGTCCTTTCCTGTACGCTTGCCGCCGCTTCACTGCGCCCTTCTAAGTACACAGAGCACACACAGCTTAACTCTTCCCTGTAGCTGTACGGCTTGGTGTCTGTTACCTAAAAATCGATAAATCAATTAGTCTCTTGCCGACCACACCAAACGACCACTACAAACGAGCCCGACTCGACCCGTTCACTTCCCGTTCGCAGATGCAAGTCCAGCCGACCAAGTACGTCGGCCTCGTTGCCGACCTGATGCCGAACATTCGGCTGATGCAGGCCAGCGGTCACTTTCTGTTCCGCTACGTCACCGGCCCGATACTGATCCGCAAGGTGTACTCCTGGTGGACGCTCGCCATGGTGCTGATCCAGTTCTTCGCCATCCTCGGCAACCTGGCGACGAACGCGGACGACGTGAACGAGCTGACCGCCAACACGATCACGACCCTGTTCTTCACGCACTCGGTCACCAAGTTCATCTACTTTGCGGTCAACTCGGAGAACTTCTACCGGACGCTCGCCATCTGGAACCAGACCAACACGCACCCGCTGTTTGCCGAATCGGACGCCCGGTACCATTCGATTGCGCTCGCCAAGATGCGGAAGCTGCTGGTGCTGGTGATGGCCACCACCGTCCTGTCGGTTGTCGGTATGTGTGTATGTGTGTGGCCGTTTGGGAAAGTGTCTTTGCGGCAGAACCCCAATCTACTGTTACGCTTGACTGGGTTTTTGTTTTTTTCTCGGTGGAGGGACGGGATAAAATATCTGAAAGAATAATTGAGTCAACCCACAGGGGGATGCAAGACATCGCAGGCAGAGAGTTTGGGTTTGATTTATCACCGCACACCGAATATCTTCACGGTTCATAAGCTTCACCGCGGTGAAAAGGGAACTCCCCATTTCCCTGTTTTCTTTTTTTTCTTCCTCTCGATAAATTACTCATCGCTTTTCGTTTTTTTTTTTTTGTTGTTGCTTCTTTCTTCTTTCATCCCTACTAGCCTGGGTTACGATAACATTTTTCGGCGAGAGCGTCAAGACTGTGCTCGATAAGGCAACCAACGAGACGTACACGGTGGATATACCCCGGCTGCCCATCAAGTCCTGGTATCCGTGGAATGCAATGAGCGGACCGGCGTACATTTTCTCTTTCATCTACCAGGTACGTTGGCGGAATGTCCTGCGCGTCACAGTTGGCAGTCAGTGAGCGGCAACACGGCGAAAAAATGGGACTAAAACCGGTCTTCACAGAGCCAACACATTCCTACAGCAATTGCATACCTTCGGGCGGTCGGGACTGGGCAATGCAGCTACAACATCCTCGCCTAAAGTTATGCAATTCGAGCGACAAATGTTGCCGTGTTAGGGCTTTTTGTGATAATAGTCGTTTTTTTGTCCTCTCGCTTATCAAACTCTATCAACGGAGGAAATCCATTTTCGCTACAATGCCTACAGCTCAAGTTTCAAGGTCAATCGAGCGGGTGGGGATCAACTTTTTTATTCATTTTGCTAACGCCCCATCAACAAATTCTATGTTCTCAATGGCAAAGATTACTGCCCGCACCAATCGCCCAACGAAACGGCAAAAGAAAAGCGACGATTATGAAGATGTCCAAACCATTGCCCGCCCGACGCTTTATCTGATGATTTGCGGGATGGCTTTTACTTGTCTGCTACTTTCAGGCACAAAAGGAAATGAAACCAGCGCAGGCTCGTTTGCCGGCTTGCGGAGGTTCTTCAGGCACTGAGGCTGAGTACTTAAATCGAACGATTTTTACGATTCTGGATCCAGTTTTATGATGTGGCCTGCATTACAGTGGCAATTATACCCTGATGTTCATTTCATTGCATTTTGTAAGTTTGTGCTGGTAACGCCCGTAACGATTAATTCTTTTCAAAGAGATTCTTTCAAAGAGATTCAAAATGTGTATAACAAATGCTAACGAATGGACCGTACTTGGAGGGTTGCGGAAAGTAACGTTTTAAAATATTCATCACAATCCTCTGCAAACTTGTGCTTAATTAATTGGTGCACAATAAGTTTAAACTGTGGCGGCAGATGTGTCGCTGTCCGCTTCCTTCCTTCCCAGCAAGCTCGTGCGAAATAATTTATTCCATCATTTTAATACAGCCGTTTGTGCATTTTAATTAGCAAAGCAATATAAAAAGCAGCTAACCATCCCCATTAAAACAAAGTGCTTCCGGGCCCAATTGTTATGGCGGTGGAAGTAATGGTTTTACCAGTGGAAGTGTCCTTTCCCATCGTGGGTACTTCGCGATATTCTTGTCTTATAACAAGTGCATACAGAAAAAAGGAACAAATCCTCCTTGCTATGGTCTAAGGGCCAGCTTCGGTACCGCTTCCGCTTCGGGATGTCATAAAGTTTGATGGGTGTTTTTAACATTACTTCCGCTCTTAACCACCTAATGGACTTTTCATGCTTGAGCTAAAGCTAAACCAGCCACCAGCGGTACGCACCGAGCCACGGTTGATTTCGGCGGCGGCCTCATCCCCAGTTTTGCGCCACCAATATTGCCTTCATTAATCTGTACCCTCGGAGCGTTAGGGCCCGCGGACGAGTCCTCGTTGTAATGCACCGCCATGCCACGGGACGGGATAATCCGTTGGGACGGCGCGAAAGCGACTATCGCGGACGGATTGGTTCGACCGTGCTACAACACATTTTATGCTTCACAGATTTACTTCCTGCTGTTTTCGATGGTCCAGAGCAACCTCGCGGATGTCATGTTCTGCTCCTGGTTGCTGCTAGCCTGCGAGCAGCTGCAACACTTGAAGGTAGGTACGGTAGCAAACGTGGTTGTCTTTACATCCGCGTGCAGCATTATCCTTATCGACGTGTAGTGTTAACGGTAAAAGAGGAAGCGATAAAAAAGCAACATTCTCTCACACCCTCGATCTCTCTTTATTTTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCCATCTCCCTCGGGCAGGGTATTATGCGATCGTTGATGGAGCTTTCGGCCTCGCTGGACACCTACCGGCCCAACTCTTCGCAACTGTTCCGAGCAATTTCAGCCGGTTCCAAATCGGAGCTGATCATCAACGAAGGTATGTGAAACGTGTGCTCGTGGCAGACGGACTCAAAGAGAGCATAACACAATCCCCTGGTAGTTCATTTCAATGACCTTAACACTCGGCAAGCTAAGCGAGACAGTGGGGACAGTGAGAAAGAGAGAACAAGAAAAAAAACCATCATCCGTACGACATCATCGCTACGTACCGGTATTTCAGGATGAGGAAATAAAACGCTAGGGGAATGAAAGTGCGACAGAATGATAAAACAATCCCCACCCAGGCCCCCAGCCTGGACGAACGGATGTAGTGTGCGAAGCGAGCAAAAAAAGTCAAATAAATTGAAGTTTAAAAATAGATTTTCCCCGTCCATCCGTGGTGGAGCGTAAAGCCCGGCGGACAACTTCGAGCACGGCGACCGTGCACAGTACTGTGCCACAGTTGTAGGGACGGATAAGCTCCGTTCCTTTTTTATCCTTTTTTTTTGGAGATTTGTTTGCGTTCGCATCGTTAGACGAGCTTAGTGCCGTGTTGCTCTAATTGCTATTTATTATAAAGCGCTTCCAAATAGAAGATCGGTTCTCTCCATTTAATCTATCGCGCCTGTACGCCTGAAACTATGCACTGTGCTGTGAAACCGTCAAGCTCGAGCACGACGAATGGCCCACCGTACCACGCCCGTGGTGCCCAAAGCGCAACGCGAATTGCATGTTAACAAACCTTTGCCTACCATCCAATCCGTGTGAAATTGCCCGCTCTCTTTCTCTCTTTTGCGCTTTCGGTGTATCGAACGGTTTTGTCCCTTTTTTTTACTTTGCTCTTGATCTCTTGCTGTGCTCACTTTCATCTCATGTTTTGCCTGACGGTGGTGGGTTTTCGAAAAAAGAGCGATTTCTTCTGCGTGTGTGTGTGGTTTTTTTAAATAACCGCTCCAGGTCGTGTTGAACGCTGCAGGACCGATCGGAGCTAGTTTATTATCAGCTTTAGTGTTTATCCCACCCATGCCCCACATCACGTCTGTGGAGAGTGGGGGAAGCTTAAGTCCAATGTAATTTACCGTGTTTCTGTCGTTCGTCACCTTCTTCGTCGATGGAGATTGGTGCGGTTGGCACGATAAAAGCCCACTGCACGTTACGGACCGAGGGAAAGGTCTTTTTGTAGGCCTAGCAACGGTCCTCATTCACCGCATGGGGGTGTAGCTCAGATGGTAGAGCGCTCGCTTAGCATGTGAGAGGTACCGGGATCGATACCCGGCATCTCCAACCCACACAAAACGTTTTTTAAGAAGATTTTTAGGGAAGATATTAACGCGGGTACACTGTGCTCCTCTAAGTTGGAAGAGTAGATGAGATGATGACAAGGGAGAAGGAACATGTGTACGTGTTTGATAGCAAACACACAAACAACAATATCATCTCTGATAATAATCTGATGTGTGATGTGTGTGTATTGTTGTTATGCTGCCTTTGCCATCTTGTCCCTCTCTCTCCTGTTCAACTCCTAAAAGAATTGTTTGGAGTCCTCTCAGTTCCTCGTAAAGATCCTTTCGAGATTCTTCTTTCCTTTTTATTATTTATTCCACGAGCCTCTGACATAAGTAGCCTTCCGCTTATTTCCTTCTCCTTGCACTTGTCAGTTCCGTGTAGAGCGTCATTTTGAGGTTTACACATTTCCCACCGACGCCTGATTGTTACATTGTCATCTACATTGCTTTCCGTTTACCGTTCCGCCCTTTTTTTTTAACGCTACCACAGAAAAGGATCCGGACGTTAAGGACTTTGATCTGAGCGGCATCTACAGCTCGAAGGCGGACTGGGGCGCCCAGTTCCGTGCGCCGTCGACGCTGCAAACGTTCGACGAGAATGGCAGGAACGGAAATCCGAACGGGCTTACCCGGAAGCAGGAAATGATGGTGCGCAGCGCCATCAAGTACTGGGTCGAGCGGCACAAGCACGTTGTACGGTAGGTATGGTAATTTCTAAGGTGTGGTGTAAAGCCTCCAGGTTCCATGAAAAAGGGATACTTTACCACAGTAAGAGTTTGTTTTGCTGGACTTACATTCTTTGGAGCATTGTTTGGTGTTGTGCTGAAACCGGTTGCAATATCGTTTTGCGAAGAAATTATGTGTAAAGCGTATTACAATCTCATTCCTCTGTTAATCTGTACCAATTGTGTCAGCCCCGACCGAAAGCAGGCCTAATTCGTACCAGAAAAACCACAAGCTGTTTGTAAGCATCGATACGCCCGAAGCTTTCAATCCAGCCAAGGCGCCACCTACTATTGACGTGACTTTTTGCACGTTCACACTCTCCCTCTCCCATTCTTTCTATAACCAATCGTCGCTCAGCCAGCATCGCCCGGAGTGAAGTTTTTATTTGAACGATATCACCCGTATCGATTTTCCACTAAACATGCTTAAATCGTTTCACAAAGCTCCCCCAAAATCCCATTTCACCAATCCACCAATTTGAAGTCCGTCGTCCTTTGTGTCCTTGTGTTTGTGTGTTTGTGTGAGCTGGAGACATGGGGGAGTGAGTAACCGAACAACCTCTTGCCGCTGCTTCACGATATCGAACAGCACCAAGATAAGCATCCCTTTTTCCCTAGCCGATGTCTCCGATATCTCGATTCCGCTTCCAGCGAGGCAAAGAAAAAGGCGAACTGGCTGACCTCACCCGGGGCGAGGAAAAAGCGTAGGGATTACGTCGAGCAGCACGAGTTGTGATTTCTTCTTCTTCTGGTTCCATAAATCGCTGACGGTTTCCATTACCGCCTGCGGAGTGCACACACGTGAAGGGAAAGCGAAAACGTTTAGATTCCAGCAGCAACGGCAGCACCAGAAGCAGCAGCAGCGCGGCAAATTGAATCATCCTGACGCGATGAGTTGTCTGGGTTTTCGGGTCGGTGGCTTACAGCACCACACCATCTGCTGCAGCTAATACAGCTGTAAATTTCGTTAGACATAGACTTGATTTTACAATATTACACACACACTTACACACACAGCTATAGATTTGTCGCTTGGCGTATGGCTCTGTACGGCGTGCCGTACATGCCGCGAGCCGTGTTGCTGCTGGTTGCGATACGGATCACGTCCGATTCGATTCAGCCTGCGTGTTTTTGGTGAAGATCCTTATCGGTGACCCACTTTCAGTGTGTCGAGAGCGAGGGTCACTATGGCGCCTGTCAGTTGGAAAGCTAGGCTCGATTCAAAGGGCCATTGTGCCAGTGTTCTTTTTAAGATAGCGATAAGCTTTTGATCGAAATAGTAAATCAAACATTGTTTCTTTTTTCCTATTCCAAACTGTTGCCAACCTCATTATTACGTTTTTGCAGCGGGTGTATAGTAAATTGCATACTTTAAGGCGTGATTTTCAAATGTAGCGTTCCGTATGCAGAAACGCCATGGATTATGCAATTTAAACAATGCTGCTTCCTTAACATTCAAATAACGGCTTATTAAGGAACTTTTTGTGCAATTTGTTTTTAACAGCAAATAGTTAGCTCAGAACGATCACATTTAGTATCGCTTCAACAAAGAACTCTTTTAAACACACAATTTGTAATGCCATTCCCTCGAGAAAGTTTCTTGTCAGTCCTCCTCTGCATCACAGCAACAACCAAACCTGCTCATGTTTCCTGCTCGTTTCCTAGCTGTTTTGAACGTTATTTCCGATTCCTGTGCTTGCCCGCTTTTCTTACAATCAACCACAATGGTTCAGATTTCGCTCTTATTTTATTGACCCACTGCTTTCGTGCTGAAGCCCGTGGAAACAATGCGCCAAGCTCAGCATCCAGCCATGCATGTAAAATGAGCCACGCGACAGATTTTAGACATCGCTTTCGCTCTGCACCGGAGGTGGTTTTATTCTTGTTTCCGATTCCCACGTCCATTCGTCCTGGGTCCGTCCGCCGGGCCCGAAACCGTAAGCCGTGCGGGGAATTACGCAATCGAAACGAGCCAGAAAATGAGCACGCCAAATGCAAAGAAAATCCCCTTTTGAGTGGTGCTCCTGCCACCACTCATCTCCCCAACTGGTGGGTGAAAAACCTTGTGCGCCCCTTCTCTTTCCAGAAAAAAAACGCCTCGCTCGCACAAAAACATGCTCGCCCGGTGAAGCTGCGTATGTCGCAGAAGCTCAAACCAACGCCGCCAGCAAGCATCAACAATTTCTATTCAAACACCCAACGCAGCGCCCAAACCGGGTGCACTGTACTCAGTAGCGAAGATGCTCAGATTGTCCCGTGCGCTGCTTTCGATGCCCGTTTCGGAGCGGGAAGCCATCGCTTGCCAACGTTGGCGATGTCTTTTAGCCGTGGATTTGAATTTTCTGAATATCACAGGCGGGCGCGGTTTGCCTGCAAGGTTGTTGCTTCCCACACGAGCATTGCTTTCCGTACCGCGGTGGGGCGAGTTTTCAACGCAACCTTCTACAAGCAACGCCACAACGCCTGGGAGCGATATTTAACAGAAACAAGAACATCCCGAACTTCAGCACATGCCGTGATTTGCCTGTTGGAAAAGCTTTTGTGAGCGTGTGAGTTGAACGAGCTCTATTTTCCCAGCGATGGGTGGCATTTGTGTGGCATGCTATCGTCAGCTTTTCTTGAATCTTTACCTCTCCATTCGCCTCCATTAGTACACGCGTATGGAAAATGGGTGCAACGGATCAGAACGGATTTTCCGCGACAGACTTAATAAAGGGAAAGCAACGCGTTTTTTGCATGTGTAGTGTTTATGAGCTTTATGCCGTTACTTTGCAATTAAAAATAGCAAAAAATAACAGTTTTTTTTTGTAAGCGGATTACAAAGAATGTATCAGAATATTACGTGAAACATTCATTTCATGCTGTTAACGCTCAAATAGAATAGTTTTGTAACACGGATTGCATACCTTGCCGGTATCGGTTACATTTTCGCCTAACAGTATGCAATCTGTTTAGCTTTGTTGTTTAATGACTGCGTTGGTAGTACAATATTTATTTACACCGCGTAATTTATCTCACAAATTGCAAAAAAATGTCAATCTGTATCGATTATTCACACAAATCAGATCCCGGAACCAGTGTAGCCCAATGTGCTCTTTATTGAATTACCACGAACAAATCAACCTGATGCCCGGGTCCGTTGGCAAACAGCTTGCGCCGAAGCCGCTCAGTGTTTCGTGCACTACCGTGCTGCCATTTTGCTGCCCTCATCGAACAGATAAACAGAAGGGCAACTCTTGTGAGCATCGCAATGCCCGTCTGAAGTTCCGTCGAAAATGGGCCTAAATTCAATTTGACGCATTTACCCGCGAACAATTGCGCGAAGGCTGTCAAGTGTGTTCCACGAACTGCGACAACAAGCACACACACAAACACAAATGTTATCGTTTCGGCATGTTTCTCGGTACAAAGCGTGTGGCGCTATGTGGCATGCCGATTCCCAGACAGAGTGATCGATAGTAAATGTAGCCTATCCGGTAGCATTCAATTTCCTTTTCTATCCTCGCAAACAAAGCCCATTCTGGGGAGGCGTGGTGAAGCTTTCAAAGGCATTGTGAAACAAATGTCCTGGTTCGGAGGGATGCTGGGGAAAGCAAACACGGTGCCGCCATCGCTGCTACCGTCAATCGATCATGCATGATGTGATTAATATTTGTGTTATTCACCTGCGTATCTATGCGTCCGTCGTGTCGTTCGATTTCCGGAGTCAAGGAAAAAGCGACTCCATTTGGGATTGGTTTTTGCAGCGAAAAATCAAAACATTCGCACAAAACCGTCCTCCATTTCAAATGCCTACACTTGTCACTGTATATCTCTCTTTCTCTCGTTTTGCCACGTTGCAGTCTCGTTTCAGCAATCGGAGATACGTACGGTCCTGCCCTGCTGCTACACATGCTGACCTCCACCATCAAGCTGACGCTGCTCGCCTACCAGGCAACGAAAATCGACGGTGTCAACGTGTACGGATTGACCGTAATCGGATATTTGTGCTACGCGTTGGCTCAGGTTTTCCTGTTTTGCATCTTTGGCAATCGGCTCATCGAGGAGGTACGTGCGCTCGGCGTGTTGCCGTGGGAAAGCATTCTCCCTGCCCCATATCGCTTCATTCTCCCAGATCACACATTTGCATCACAAAGCCAGCACACTTTTGCTTCGCCGCTGCCATCTCGGCTTCTGAATGTTTTCACTCTCCCATACCTCTCCCGTGCAGAGCTCATCCGTGATGGAGGCGGCCTATTCCTGCCACTGGTACGACGGGTCCGAGGAGGCAAAAACCTTCGTCCAGATCGTTTGTCAGCAGTGCCAGAAGGCGATGACTATTTCCGGAGCCAAGTTTTTCACCGTTTCGCTCGATCTGTTTGCTTCGGTAAGTGTAGCCTGGTGGCTGGCACAGAACAGGCTGGCAAAACAGGGACTTTGGCTCTAGCCTGATGGGTGGTATATGTGTGTCTATTTTTTGCTACCATTCTCGCATCCCTTCCTTTCCAGGTTCTTGGAGCCGTTGTCACCTACTTCATGGTGCTGGTGCAGCTGAAGTAAACAGCCGTGGCCCGGAAGGATGTGTTTTTTTTCGCTCGTTCGGTTGTTTGTTTGTGCACACTTTCTCTTGGACATTTTCTCTACTGCAAAGGTTTAACAAACAGCAACAACAAATAATCCCAAGTTTTCTTTTACAGATCTTTGCAAAATGATTAGATTTTAATAGATTAACAGTGCTTGATTATCTGTCCTGTAGCAACCGGGGCTGAAGAACGTTGATTTGGTAAAAGTACAAAAGGGACGTTGGAAATTGAAACAACAGAAGAGTGATATTTATGCAAAGCTCACCAAGGGAAATCTATGTATGTGTGATTTGCGCTCATCAAGCACTGTATGTGCCTTTCAACTAGTGCAGCAATAAAGAGTACAAATGTTTCTTAG.
Ir25a (4005 bp; Probe set ID PRK149) – AGAP010272
CCGTCGCTTATAGACCGATTCCGCTATACTGTTCGGCTTGGGTATATGTGTGTATTACAAAAATATCATGGATCCTAAGAACGGAAGGCGTTGGCTGGTTTTAATCCCTATTCAGCTTGCATCATATGCAATTATAGCGATCATGGGACAAACTACCCAAAATATTAATATATGTAAGCATTTTCATTAATTAACTCCTTTTGCACTGGAAGTAGAGATTGTTGTGAACATTAGAAAAAGCTCAAATTATTTTAAAACGTAAATTTCACTTTATTACCAGTGTTTGTCAATGAGGTCGATAACAATTTGGCTAATGTTGCTGTTGAAGTCGCACTGAACTATGTTAAAAAGAATCCTCAACTGGGGCTGTCCGTTGATATGATGTACGTAGAAGGGAACCGCACCGACTCCAAGGATCTGCTACAAGCGCGTAAGAACTATGAACACTTATGTCGTTGTTGTTATCTCTCTGGTGTTACTGCTTAAGGTTACAAAACGTACTATTTAATTGTGTGTGATTTTTACCTTTTCAATGTCTAATTTGGTAAATGAGAAAGTGTGCTCGAAGTATGGCCAGTCTTTGAGCGAAAATCGACCGCCACACTTACTACTGGATACCACGCTGACTGGAGTATCATCGGAAACGGTGAAATCGTTCAGTCTGGCACTTGGCATACCAACCGTATCGGCTTCGTTTGGACAGGAAGGAGATTTACGCCAGTGGAGAGACTTGACGCCAACCAAACGCGGCTATTTGCTTCAGGTAAATCATTATGTCTTCTGCAGGGTGCGGTTGCAATGGTAGTTTTCAGTGCCTTGCTTAGGCTGCAATCAAAATCGTTATGAAATGAAGCCAGAACGTTATGATTCAACATTCTTTATCCATGAAGGTTATGCCCCCGGCTGATATGATTCCGCAAGTGATTCGATCTATCATCATTTACATGAACATAACGAATGCTGCAATTCTGTACGACAACACATTCGTAATGGATCACAAATATAAAGCTTTACTACAAAATATACCCACGCGCCACGTCATCACCACCATTGCTGACGATCGCGATAGAGCCAGCCAGATTGAAAAGCTTCGCAATTTGGACATAAACAATTTCTTTATACTAGGTTCTTTAGCATCGATCAAGCAAGTATTGGGTAAGTGGGTTAGTAGTTGTCGTCGGAGGTGCGCGTTTTAATTAACGCTATTCTTGTTTTGGTCACTTGTTTTCACAGAATCGGCTAAAAATGAGTATTTTGAACGTAACTTTGCCTGGCATGTAATAACGCAAGAGCAAAAGGATCTAACGTGCAATGTTGAAAATGCCACTATCATGTTTCTTCGTCCGATGTCTGATAGTTCAAGCAAAGATCGATTGGGCAGTATACGCACAACATACAATCTTAAGCAGGAGCCTCAAATTACCGGATTTTTCTACTTCGACCTGACACTGCGCGCGTTAATTGCAATTAAGTAATATATCAAGCTTTATATGGTTGGCTAGCATTAGATGCTGAACTGAGTTGTTTTTTTAAATTATTTTCTTTCATTTGCTAGAAATATTTTGCAGTCCGGATCCTGGCCATCAAACATGAAATACATCACGTGTGAGGATTACGACGGGACAAACACACCAAATCACACAATCGACCTTAAAACGGCTTTCATTGAGGTGACCGAGCCAACCACTTTCGGACCGTTTGAAATACCAAAAGGCGGAAAAATGCAATTCAACGGTAACACTTACATGAAGTTTGATATGGACATTAACGCCGTTTCCATCCGTAGCGGCGCATCCGTTAATACGCGCAGTCTCGGAACATGGGAAGCGAGCTTAAATGCACCGATAAATGTAGCAAATGAGGCGGAAATAAAAAATCTTACTGCCGATGTTGTTTATCGTGTCTATACGGTTGTGGTAAGTGCGAGTGGATAAGATAATATATTGTAAGCGACAACAATGTTAACAAGCGTAAACCCCGTTTATTTCAGCAAGCGCCATTCATAATGCGAGACCCAACGGCACCAAAGGGCTTCAAGGGATACTGCATCGATCTGCTCAACAAAATTGCCGAGATCGTCGAATTTGACTATGAAATACGCGAAGTGGAGGACGGAAAGTTTGGCAACATGAATGAAAACGGCGAGTGGAATGGTATCGTACGGAAGCTGATCGACAAGCAAGCAGATATCGGACTCGGCTCGATGTCTGTAATGGCCGAGCGGGAAACAGTTATAGACTTTACCGTCCCGTACTATGATCTAGTTGGGATTAGTATTATGATGCAATTGCCAAGCACGCCGAGTTCGCTGTTTAAATTTTTAACCGTGCTGGAAACGAATGTGTGGCTCTGCATTTTGGCTGCCTACTTCTTTACCAGCTTTCTGATGTGGATTTTTGACCGCTATAGTCCATACAGCTACCAGAATAATCGAGAGAAGTACAAGAACGACGACGAAAAACGGGAGTTCAATATTAAAGAATGTCTGTGGTTCTGCATGACATCGTTGACACCGCAAGGTGGCGGAGAAGCACCCAAAAATTTGTCCGGTCGCTTAGTCGCTGCCACATGGTGGTTGTTTGGGTAAGTAGTAGCAGTATTGATAGAAGGAAAAATATGTTTCCATTGTGTGCCGTTTCCACCCGCGTGGTTACACGGTAGGTGTAATTATCATTGCATTTCTATCATTATGTCATTATTCAACAGATTTATCATCATTGCCTCGTATACGGCTAATTTGGCAGCTTTCTTGACTGTATCCCGACTAGACACGCCGGTTGAATCGCTGGATGATTTATCAAAGCAGTACAAAATTCTGTATGCTCCACTGAATGGATCATCGGCTATGACGTACTTCCAGCGAATGGCTGACATTGAAGCTAAATTCTACGAGTAAGAAGTGCCATATACCGGCGCAAGTATCTAAGGATATGTTGCTTACTTATTTTACTACTTTCTGGCAAACGCAGGATTTGGAAGGAGATGTCGCTCAATGACTCTCTGACGGCGGTTGAGCGATCGAAGCTAGCTGTCTGGGACTATCCGGTCAGCGATAAATACACAAAGATGTGGCAAGCCATGCTTGAGGCAGGTTTACCGAACAGTCTCGAAGAAGCCGTACAGCGCATACGAAATTCGACATCTGCCTCTGGATTCGCATTTCTGGGCGACGCAACCGACATACGCTACCAAGTGTTGACAAATTGCGATTTACAGATGGTTGGCGAAGAGTTTTCTCGCAAACCGTACGCGATCGCCGTCCAGCAAGGATCACCGCTGAAGGATCAATTTAACAATGCGTACGTATTAATTTCATTATTCCTTTTTGCGTTTTCTAACCTACACTTCTCTTCGCTGTTTAGTATACTGATGCTACTGAACCGACGCGAGCTGGAAAAGTTGAAAGAACAATGGTGGAAGAATGATGACGTACAAAACAAGTGCGAAAAGCCAGATGACCAGTCGGATGGCATCTCGATACAAAACATCGGAGGCGTATTCATCGTAATATTTGTGGGGATAGGGATGGCGTGCATTACGTTATTGTTTGAGTTTTGGTATTACAAGTATCGAAACAACTCCAAAGTGATCGATGTTGCCGAATCAACGGACCAGCAACACGGTGGAACAATAGTAAAAAATGTCCGTCCCGCTGGTAAGCTTATGAAGCAAGATTCGTTAAAAGACTCAACAAAAGGTCACAATTATCAAAACCTCCGAACACGTACCTTGATGCCGAATTTGAGCAAGTTTCAACCTCGCTTCTAAAGTAATGCGTTTTTGACTGCGCTTTGGAAGTACAACGAAAACATACTAATGCGTCGTGAGATTTACGAGCATTAGTACGACTACGGATAGTTATGAACTGTATTTTTTTAATTTTAACGTATGATATGAATTTAATGGGGATATATATGTAACTATCTTTACCAATCTTATATAACGATTTGTGCATTTTGAGCAATTCTTTGAATTATGCAACAAAACGCTAAAAATTAAATGAAA.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Christopher J Potter, Email: cpotter@jhmi.edu.
Ilona C Grunwald Kadow, Technical University of Munich, Germany.
K VijayRaghavan, National Centre for Biological Sciences, Tata Institute of Fundamental Research, India.
Funding Information
This paper was supported by the following grants:
National Institute of Allergy and Infectious Diseases R01Al137078 to Christopher J Potter.
National Institute on Deafness and Other Communication Disorders R01DC013070 to Christopher J Potter.
U.S. Department of Defense W81XWH-17-PRMRP to Christopher J Potter.
Shelanski Research Innovation Award in Pathology 2018 to Chun-Chieh Lin.
Wellcome Trust 203261/Z/16/Z to Gregory SXE Jefferis.
National Institutes of Health BRAIN Initiative to Gregory SXE Jefferis.
National Institutes of Health R00 AG062746 to Hongjie Li.
National Institute of General Medical Sciences R35GM133209 to Karen Menuz.
National Institute on Deafness and Other Communication Disorders R21DC017868 to Karen Menuz.
Additional information
Competing interests
No competing interests declared.
Author contributions
Conceptualization, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review and editing.
Conceptualization, Formal analysis, Investigation, Methodology, Visualization, Writing – review and editing.
Conceptualization, Formal analysis, Investigation, Methodology, Writing – review and editing.
Formal analysis, Investigation, Methodology, Writing – review and editing.
Formal analysis, Investigation, Methodology, Writing – review and editing.
Conceptualization, Formal analysis, Investigation, Methodology, Visualization, Writing – review and editing.
Conceptualization, Formal analysis, Investigation, Methodology, Visualization, Writing – review and editing.
Formal analysis, Investigation, Methodology, Writing – review and editing.
Supervision, Writing – review and editing.
Conceptualization, Formal analysis, Investigation, Methodology, Visualization, Writing – review and editing.
Conceptualization, Methodology, Supervision, Writing – review and editing.
Conceptualization, Funding acquisition, Methodology, Supervision, Visualization, Writing – original draft, Writing – review and editing.
Additional files
Data availability
All data generated or analyzed during this study are included in the manuscript and supporting files; Source Data files have been provided for Table 1, Figure 2, Figure 3, Figure 4, Figure 6, and Figure 7. Raw data used to generate each figure is available at Johns Hopkins Research Data Repository (https://doi.org/10.7281/T1/9VJGPI).
The following dataset was generated:
Task D, Lin CC, Vulpe A, Afify A, Ballou S, Brbić M, Schlegel P, Raji J, Jefferis GSXE, Li H, Menuz K, Potter CJ. 2023. Data associated with the publication: Chemoreceptor co-expression in Drosophila melanogaster olfactory neurons. Johns Hopkins Research Data Repository.
The following previously published datasets were used:
McLaughlin CN, Brbić M, Xie Q, Li T, Horns F, Kolluru SS, Kebschull JM, Vacek D, Xie A, Li J, Jones RC, Leskovec J, Quake SR, Luo L Li H. 2021. Single-cell transcriptomes of developing and adult olfactory receptor neurons in Drosophila. NCBI Gene Expression Omnibus. GSE162121
Schlegal P, Jefferis GSXE. 2021. EM Reconstructions of VM6 OSNs. FAFB EM volume. FAFB
References
- Abuin L, Bargeton B, Ulbrich MH, Isacoff EY, Kellenberger S, Benton R. Functional architecture of olfactory ionotropic glutamate receptors. Neuron. 2011;69:44–60. doi: 10.1016/j.neuron.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abuin L, Prieto-Godino LL, Pan H, Gutierrez C, Huang L, Jin R, Benton R. In vivo assembly and trafficking of olfactory Ionotropic Receptors. BMC Biology. 2019;17:0651-7. doi: 10.1186/s12915-019-0651-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ache BW, Young JM. Olfaction: diverse species, conserved principles. Neuron. 2005;48:417–430. doi: 10.1016/j.neuron.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Ai M, Min S, Grosjean Y, Leblanc C, Bell R, Benton R, Suh GSB. Acid sensing by the Drosophila olfactory system. Nature. 2010;468:691–695. doi: 10.1038/nature09537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai M, Blais S, Park JY, Min S, Neubert TA, Suh GSB. Ionotropic glutamate receptors IR64a and IR8a form a functional odorant receptor complex in vivo in Drosophila. The Journal of Neuroscience. 2013;33:10741–10749. doi: 10.1523/JNEUROSCI.5419-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer TO, Khallaf MA, Silbering AF, Zappia G, Ellis K, Álvarez-Ocaña R, Arguello JR, Hansson BS, Jefferis G, Caron SJC, Knaden M, Benton R. Olfactory receptor and circuit evolution promote host specialization. Nature. 2020;579:402–408. doi: 10.1038/s41586-020-2073-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasaki T, Huang Y, O’Connor MB, Lee T. Glia instruct developmental neuronal remodeling through TGF-β signaling. Nature Neuroscience. 2011;14:821–823. doi: 10.1038/nn.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-Lopez LA, Alexandre C, Mitchell A, Pasakarnis L, Vincent JP. Accelerated homologous recombination and subsequent genome modification in Drosophila. Development. 2013;140:4818–4825. doi: 10.1242/dev.100933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates AS, Schlegel P, Roberts RJV, Drummond N, Tamimi IFM, Turnbull R, Zhao X, Marin EC, Popovici PD, Dhawan S, Jamasb A, Javier A, Serratosa Capdevila L, Li F, Rubin GM, Waddell S, Bock DD, Costa M, Jefferis G. Complete Connectomic Reconstruction of Olfactory Projection Neurons in the Fly Brain. Current Biology. 2020;30:3183–3199. doi: 10.1016/j.cub.2020.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLOS Biology. 2006;4:e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136:149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berck ME, Khandelwal A, Claus L, Hernandez-Nunez L, Si G, Tabone CJ, Li F, Truman JW, Fetter RD, Louis M, Samuel AD, Cardona A. The wiring diagram of a glomerular olfactory system. eLife. 2016;5:e14859. doi: 10.7554/eLife.14859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao S, Sen A, Stocker R, Rodrigues V. Olfactory neurons expressing identified receptor genes project to subsets of glomeruli within the antennal lobe of Drosophila melanogaster. Journal of Neurobiology. 2003;54:577–592. doi: 10.1002/neu.10175. [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. PNAS. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch JA, Knight S, Kanca O, Zirin J, Yang-Zhou D, Hu Y, Rodiger J, Amador G, Bellen HJ, Perrimon N, Mohr SE. Use of the CRISPR-Cas9 System in Drosophila Cultured Cells to Introduce Fluorescent Tags into Endogenous Genes. Current Protocols in Molecular Biology. 2020;130:e112. doi: 10.1002/cpmb.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Budelli G, Ni L, Berciu C, van Giesen L, Knecht ZA, Chang EC, Kaminski B, Silbering AF, Samuel A, Klein M, Benton R, Nicastro D, Garrity PA. Ionotropic receptors specify the morphogenesis of phasic sensors controlling rapid thermal preference in Drosophila. Neuron. 2019;101:738–747. doi: 10.1016/j.neuron.2018.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhmann J, Sheridan A, Malin-Mayor C, Schlegel P, Gerhard S, Kazimiers T, Krause R, Nguyen TM, Heinrich L, Lee W-CA, Wilson R, Saalfeld S, Jefferis GSXE, Bock DD, Turaga SC, Cook M, Funke J. Automatic detection of synaptic partners in a whole-brain Drosophila electron microscopy data set. Nature Methods. 2021;18:771–774. doi: 10.1038/s41592-021-01183-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterwick JA, Del Mármol J, Kim KH, Kahlson MA, Rogow JA, Walz T, Ruta V. Cryo-EM structure of the insect olfactory receptor Orco. Nature. 2018;560:447–452. doi: 10.1038/s41586-018-0420-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey AF, Carlson JR. Insect olfaction from model systems to disease control. PNAS. 2011;108:12987–12995. doi: 10.1073/pnas.1103472108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai PC, Cruchet S, Wigger L, Benton R. Sensory neuron lineage mapping and manipulation in the Drosophila olfactory system. Nature Communications. 2019;10:643. doi: 10.1038/s41467-019-08345-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Buhl E, Xu M, Croset V, Rees JS, Lilley KS, Benton R, Hodge JJL, Stanewsky R. Drosophila Ionotropic Receptor 25a mediates circadian clock resetting by temperature. Nature. 2015;527:516–520. doi: 10.1038/nature16148. [DOI] [PubMed] [Google Scholar]
- Chen Y, Amrein H. Ionotropic receptors mediate Drosophila oviposition preference through sour gustatory receptor neurons. Current Biology. 2017;27:2741–2750. doi: 10.1016/j.cub.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-CD, Dahanukar A. Molecular and Cellular Organization of Taste Neurons in Adult Drosophila Pharynx. Cell Reports. 2017;21:2978–2991. doi: 10.1016/j.celrep.2017.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HM, Yao X, Ren Q, Chang CC, Liu LY, Miyares RL, Lee T. Enhanced Golic+: highly effective CRISPR gene targeting and transgene HACKing in Drosophila. Development. 2020;147:dev181974. doi: 10.1242/dev.181974. [DOI] [PubMed] [Google Scholar]
- Choi HMT, Calvert CR, Husain N, Huss D, Barsi JC, Deverman BE, Hunter RC, Kato M, Lee SM, Abelin ACT, Rosenthal AZ, Akbari OS, Li Y, Hay BA, Sternberg PW, Patterson PH, Davidson EH, Mazmanian SK, Prober DA, van de Rijn M, Leadbetter JR, Newman DK, Readhead C, Bronner ME, Wold B, Lansford R, Sauka-Spengler T, Fraser SE, Pierce NA. Mapping a multiplexed zoo of mRNA expression. Development. 2016;143:3632–3637. doi: 10.1242/dev.140137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HMT, Schwarzkopf M, Fornace ME, Acharya A, Artavanis G, Stegmaier J, Cunha A, Pierce NA. Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development. 2018;145:dev165753. doi: 10.1242/dev.165753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron. 1999;22:327–338. doi: 10.1016/s0896-6273(00)81093-4. [DOI] [PubMed] [Google Scholar]
- Couto A, Alenius M, Dickson BJ. Molecular, anatomical, and functional organization of the Drosophila olfactory system. Current Biology. 2005;15:1535–1547. doi: 10.1016/j.cub.2005.07.034. [DOI] [PubMed] [Google Scholar]
- Croset V, Rytz R, Cummins SF, Budd A, Brawand D, Kaessmann H, Gibson TJ, Benton R. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLOS Genetics. 2010;6:e1001064. doi: 10.1371/journal.pgen.1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruyne M, Clyne PJ, Carlson JR. Odor Coding in a Model Olfactory Organ: The Drosophila Maxillary Palp. The Journal of Neuroscience. 1999;19:4520–4532. doi: 10.1523/JNEUROSCI.19-11-04520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruyne M, Foster K, Carlson JR. Odor Coding in the Drosophila Antenna. Neuron. 2001;30:537–552. doi: 10.1016/s0896-6273(01)00289-6. [DOI] [PubMed] [Google Scholar]
- DeGennaro M, McBride CS, Seeholzer L, Nakagawa T, Dennis EJ, Goldman C, Jasinskiene N, James AA, Vosshall LB. orco mutant mosquitoes lose strong preference for humans and are not repelled by volatile DEET. Nature. 2013;498:487–491. doi: 10.1038/nature12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depetris-Chauvin A, Galagovsky D, Grosjean Y. Chemicals and chemoreceptors: ecologically relevant signals driving behavior in Drosophila. Frontiers in Ecology and Evolution. 2015;3:41. doi: 10.3389/fevo.2015.00041. [DOI] [Google Scholar]
- Diao F, White BH. A novel approach for directing transgene expression in Drosophila: T2A-Gal4 in-frame fusion. Genetics. 2012;190:1139–1144. doi: 10.1534/genetics.111.136291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao F, Ironfield H, Luan H, Diao F, Shropshire WC, Ewer J, Marr E, Potter CJ, Landgraf M, White BH. Plug-and-play genetic access to Drosophila cell types using exchangeable exon cassettes. Cell Reports. 2015;10:1410–1421. doi: 10.1016/j.celrep.2015.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorkenwald S, McKellar C, Macrina T, Kemnitz N, Lee K, Lu R, Wu J, Popovych S, Mitchell E, Nehoran B, Jia Z, Bae JA, Mu S, Ih D, Castro M, Ogedengbe O, Halageri A, Ashwood Z, Zung J, Brittain D, Collman F, Schneider-Mizell C, Jordan C, Silversmith W, Baker C, Deutsch D, Encarnacion-Rivera L, Kumar S, Burke A, Gager J, Hebditch J, Koolman S, Moore M, Morejohn S, Silverman B, Willie K, Willie R, Yu S, Murthy M, Seung HS. FlyWire: Online Community for Whole-Brain Connectomics. bioRxiv. 2020 doi: 10.1101/2020.08.30.274225. [DOI] [PMC free article] [PubMed]
- Du L, Zhou A, Sohr A, Roy S. An efficient strategy for generating tissue-specific binary transcription systems in Drosophila by genome editing. Journal of Visualized Experiments. 2018;10:58268. doi: 10.3791/58268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunipace L, Meister S, McNealy C, Amrein H. Spatially restricted expression of candidate taste receptors in the Drosophila gustatory system. Current Biology. 2001;11:822–835. doi: 10.1016/s0960-9822(01)00258-5. [DOI] [PubMed] [Google Scholar]
- Dweck HKM, Ebrahim SAM, Thoma M, Mohamed AAM, Keesey IW, Trona F, Lavista-Llanos S, Svatoš A, Sachse S, Knaden M, Hansson BS. Pheromones mediating copulation and attraction in Drosophila. PNAS. 2015;112:E2829. doi: 10.1073/pnas.1504527112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahim SAM, Dweck HKM, Stökl J, Hofferberth JE, Trona F, Weniger K, Rybak J, Seki Y, Stensmyr MC, Sachse S, Hansson BS, Knaden M. Drosophila avoids parasitoids by sensing their semiochemicals via a dedicated olfactory circuit. PLOS Biology. 2015;13:1002318. doi: 10.1371/journal.pbio.1002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo K, Aoki T, Yoda Y, Kimura K, Hama C. Notch signal organizes the Drosophila olfactory circuitry by diversifying the sensory neuronal lineages. Nature Neuroscience. 2007;10:153–160. doi: 10.1038/nn1832. [DOI] [PubMed] [Google Scholar]
- Enjin A, Zaharieva EE, Frank DD, Mansourian S, Suh GSB, Gallio M, Stensmyr MC. Humidity Sensing in Drosophila. Current Biology. 2016;26:1352–1358. doi: 10.1016/j.cub.2016.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fandino RA, Haverkamp A, Bisch-Knaden S, Zhang J, Bucks S, Nguyen TAT, Schröder K, Werckenthin A, Rybak J, Stengl M, Knaden M, Hansson BS, Große-Wilde E. Mutagenesis of odorant coreceptor Orco fully disrupts foraging but not oviposition behaviors in the hawkmoth Manduca sexta. PNAS. 2019;116:15677–15685. doi: 10.1073/pnas.1902089116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishilevich E, Vosshall LB. Genetic and functional subdivision of the Drosophila antennal lobe. Current Biology. 2005;15:1548–1553. doi: 10.1016/j.cub.2005.07.066. [DOI] [PubMed] [Google Scholar]
- Frank DD, Enjin A, Jouandet GC, Zaharieva EE, Para A, Stensmyr MC, Gallio M. Early integration of temperature and humidity stimuli in the Drosophila brain. Current Biology. 2017;27:2381–2388. doi: 10.1016/j.cub.2017.06.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frechter S, Bates AS, Tootoonian S, Dolan MJ, Manton J, Jamasb AR, Kohl J, Bock D, Jefferis G. Functional and anatomical specificity in a higher olfactory centre. eLife. 2019;8:e44590. doi: 10.7554/eLife.44590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Yavuz A, Slone J, Jagge C, Song X, Amrein H. Drosophila sugar receptors in sweet taste perception, olfaction, and internal nutrient sensing. Current Biology. 2015;25:621–627. doi: 10.1016/j.cub.2014.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly A, Pang L, Duong VK, Lee A, Schoniger H, Varady E, Dahanukar A. A Molecular and Cellular Context-Dependent Role for Ir76b in Detection of Amino Acid Taste. Cell Reports. 2017;18:737–750. doi: 10.1016/j.celrep.2016.12.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Chess A. Identification of candidate Drosophila olfactory receptors from genomic DNA sequence. Genomics. 1999;60:31–39. doi: 10.1006/geno.1999.5894. [DOI] [PubMed] [Google Scholar]
- Gao Q, Yuan B, Chess A. Convergent projections of Drosophila olfactory neurons to specific glomeruli in the antennal lobe. Nature Neuroscience. 2000;3:780–785. doi: 10.1038/77680. [DOI] [PubMed] [Google Scholar]
- Gaunt MW, Miles MA. An Insect Molecular Clock Dates the Origin of the Insects and Accords with Palaeontological and Biogeographic Landmarks. Molecular Biology and Evolution. 2002;19:748–761. doi: 10.1093/oxfordjournals.molbev.a004133. [DOI] [PubMed] [Google Scholar]
- Gloor GB, Nassif NA, Johnson-Schlitz DM, Preston CR, Engels WR. Targeted gene replacement in Drosophila via P element-induced gap repair. Science. 1991;253:1110–1117. doi: 10.1126/science.1653452. [DOI] [PubMed] [Google Scholar]
- Gnerer JP, Venken KJT, Dierick HA. Gene-specific cell labeling using MiMIC transposons. Nucleic Acids Research. 2015;43:gkv113. doi: 10.1093/nar/gkv113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman AL, Van der Goes W, Lessing D, Warr CG, Carlson JR. Coexpression of two functional odor receptors in one neuron. Neuron. 2005;45:661–666. doi: 10.1016/j.neuron.2005.01.025. [DOI] [PubMed] [Google Scholar]
- Gomez-Diaz C, Martin F, Garcia-Fernandez JM, Alcorta E. The two main olfactory receptor families in Drosophila, ORs and IRs: A comparative approach. Frontiers in Cellular Neuroscience. 2018;12:253. doi: 10.3389/fncel.2018.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulding SE, zur Lage P, Jarman AP. amos, a Proneural Gene for Drosophila Olfactory Sense Organs that Is Regulated by lozenge. Neuron. 2000;25:69–78. doi: 10.1016/s0896-6273(00)80872-7. [DOI] [PubMed] [Google Scholar]
- Grabe V, Baschwitz A, Dweck HKM, Lavista-Llanos S, Hansson BS, Sachse S. Elucidating the Neuronal Architecture of Olfactory Glomeruli in the Drosophila Antennal Lobe. Cell Reports. 2016;16:3401–3413. doi: 10.1016/j.celrep.2016.08.063. [DOI] [PubMed] [Google Scholar]
- Grabe V, Sachse S. Fundamental principles of the olfactory code. Bio Systems. 2018;164:94–101. doi: 10.1016/j.biosystems.2017.10.010. [DOI] [PubMed] [Google Scholar]
- Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, O’Connor-Giles KM. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics. 2014;196:961–971. doi: 10.1534/genetics.113.160713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta BP, Rodrigues V. Atonal is a proneural gene for a subset of olfactory sense organs in Drosophila. Genes to Cells. 1997;2:225–233. doi: 10.1046/j.1365-2443.1997.d01-312.x. [DOI] [PubMed] [Google Scholar]
- Hahn MW, Han MV, Han SG. Gene family evolution across 12 Drosophila genomes. PLOoS Genetics. 2007;3:e197. doi: 10.1371/journal.pgen.0030197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallem EA, Ho MG, Carlson JR. The molecular basis of odor coding in the Drosophila antenna. Cell. 2004;117:965–979. doi: 10.1016/j.cell.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Carlson JR. Coding of odors by a receptor repertoire. Cell. 2006;125:143–160. doi: 10.1016/j.cell.2006.01.050. [DOI] [PubMed] [Google Scholar]
- Hansson BS, Stensmyr MC. Evolution of insect olfaction. Neuron. 2011;72:698–711. doi: 10.1016/j.neuron.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Harris WA. The maxillae of Drosophila melanogaster as revealed by scanning electron microscopy. Journal of Morphology. 1972;138:451–456. doi: 10.1002/jmor.1051380405. [DOI] [PubMed] [Google Scholar]
- Haverkamp A, Hansson BS, Knaden M. Combinatorial codes and labeled lines: How insects use olfactory cues to find and judge food, mates, and oviposition sites in complex environments. Frontiers in Physiology. 2018;9:49. doi: 10.3389/fphys.2018.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne JA, Langille C, McLin S, Wiederman M, Lu Z, Xu CS, Plaza SM, Scheffer LK, Hess HF, Meinertzhagen IA. A resource for the Drosophila antennal lobe provided by the connectome of glomerulus VA1v. eLife. 2018;7:e37550. doi: 10.7554/eLife.37550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain A, Zhang M, Üçpunar HK, Svensson T, Quillery E, Gompel N, Ignell R, Grunwald Kadow IC. Ionotropic Chemosensory Receptors Mediate the Taste and Smell of Polyamines. PLOS Biology. 2016;14:1002454. doi: 10.1371/journal.pbio.1002454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger AH, Stanley M, Weiss ZF, Musso PY, Chan RC, Zhang H, Feldman-Kiss D, Gordon MD. A complex peripheral code for salt taste in Drosophila. eLife. 2018;7:e37167. doi: 10.7554/eLife.37167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhaveri D, Sen A, Reddy GV, Rodrigues V. Sense organ identity in the Drosophila antenna is specified by the expression of the proneural gene atonal. Mechanisms of Development. 2000a;99:101–111. doi: 10.1016/s0925-4773(00)00487-1. [DOI] [PubMed] [Google Scholar]
- Jhaveri D, Sen A, Rodrigues V. Mechanisms underlying olfactory neuronal connectivity in Drosophila-the atonal lineage organizes the periphery while sensory neurons and glia pattern the olfactory lobe. Developmental Biology. 2000b;226:73–87. doi: 10.1006/dbio.2000.9855. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones WD, Cayirlioglu P, Kadow IG, Vosshall LB. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature. 2007;445:86–90. doi: 10.1038/nature05466. [DOI] [PubMed] [Google Scholar]
- Kaissling KE, Thorson J. In: In Receptors for Neurotransmitters, Hormones and Pheromones in Insects. Kaissling KE, editor. Elsevier/North-Holland Biomedical Press; 1980. Insect olfactory sensilla: structural, chemical and electrical aspects of the functional organisation; pp. 261–282. [Google Scholar]
- Kanca O, Zirin J, Garcia-Marques J, Knight SM, Yang-Zhou D, Amador G, Chung H, Zuo Z, Ma L, He Y, Lin WW, Fang Y, Ge M, Yamamoto S, Schulze KL, Hu Y, Spradling AC, Mohr SE, Perrimon N, Bellen HJ. An efficient CRISPR-based strategy to insert small and large fragments of DNA using short homology arms. eLife. 2019;8:e51539. doi: 10.7554/eLife.51539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim MR, Endo K, Moore AW, Taniguchi H. Whole mount immunolabeling of olfactory receptor neurons in the Drosophila antenna. Journal of Visualized Experiments. 2014;1:51245. doi: 10.3791/51245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapoetke NC, Murata Y, Kim SS, Pulver SR, Birdsey-Benson A, Cho YK, Morimoto TK, Chuong AS, Carpenter EJ, Tian Z, Wang J, Xie Y, Yan Z, Zhang Y, Chow BY, Surek B, Melkonian M, Jayaraman V, Constantine-Paton M, Wong GK-S, Boyden ES. Independent optical excitation of distinct neural populations. Nature Methods. 2014;11:338–346. doi: 10.1038/nmeth.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knecht ZA, Silbering AF, Ni L, Klein M, Budelli G, Bell R, Abuin L, Ferrer AJ, Samuel AD, Benton R, Garrity PA. Distinct combinations of variant ionotropic glutamate receptors mediate thermosensation and hygrosensation in Drosophila. eLife. 2016;5:e17879. doi: 10.7554/eLife.17879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knecht ZA, Silbering AF, Cruz J, Yang L, Croset V, Benton R, Garrity PA. Ionotropic Receptor-dependent moist and dry cells control hygrosensation in Drosophila. eLife. 2017;6:e26654. doi: 10.7554/eLife.26654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh TW, He Z, Gorur-Shandilya S, Menuz K, Larter NK, Stewart S, Carlson JR. The Drosophila IR20a clade of ionotropic receptors are candidate taste and pheromone receptors. Neuron. 2014;83:850–865. doi: 10.1016/j.neuron.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondoh K, Lu Z, Ye X, Olson DP, Lowell BB, Buck LB. A specific area of olfactory cortex involved in stress hormone responses to predator odours. Nature. 2016;532:103–106. doi: 10.1038/nature17156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtovic A, Widmer A, Dickson BJ. A single class of olfactory neurons mediates behavioural responses to a Drosophila sex pheromone. Nature. 2007;446:542–546. doi: 10.1038/nature05672. [DOI] [PubMed] [Google Scholar]
- Kwon JY, Dahanukar A, Weiss LA, Carlson JR. The molecular basis of CO2 reception in Drosophila. PNAS. 2007;104:3574–3578. doi: 10.1073/pnas.0700079104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai SL, Lee T. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nature Neuroscience. 2006;9:703–709. doi: 10.1038/nn1681. [DOI] [PubMed] [Google Scholar]
- Laissue PP, Reiter CH, Hiesinger PR, Halter S, Fischbach KF, Stocker RF. Three‐dimensional reconstruction of the antennal lobe in Drosophila melanogaster. The Journal of Comparative Neurology. 1999;405:543–552. [PubMed] [Google Scholar]
- Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, Vosshall LB. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43:703–714. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lee P-T, Zirin J, Kanca O, Lin W-W, Schulze KL, Li-Kroeger D, Tao R, Devereaux C, Hu Y, Chung V, Fang Y, He Y, Pan H, Ge M, Zuo Z, Housden BE, Mohr SE, Yamamoto S, Levis RW, Spradling AC, Perrimon N, Bellen HJ. A gene-specific T2A-GAL4 library for Drosophila. eLife. 2018;7:e35574. doi: 10.7554/eLife.35574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Barish S, Okuwa S, Maciejewski A, Brandt AT, Reinhold D, Jones CD, Volkan PC. A functionally conserved gene regulatory network module governing olfactory neuron diversity. PLOS Genetics. 2016;12:e1005780. doi: 10.1371/journal.pgen.1005780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Janssens J, De Waegeneer M, Kolluru SS, Davie K, Gardeux V, Saelens W, David F, Brbić M, Leskovec J, McLaughlin CN, Xie Q, Jones RC, Brueckner K, Shim J, Tattikota SG, Schnorrer F, Rust K, Nystul TG, Carvalho-Santos Z, Ribeiro C, Pal S, Przytycka TM, Allen AM, Goodwin SF, Berry CW, Fuller MT, White-Cooper H, Matunis EL, DiNardo S, Galenza A, O’Brien LE, Dow JAT, Jasper H, Oliver B, Perrimon N, Deplancke B, Quake SR, Luo L, Aerts S, FCA Consortium Fly Cell Atlas: A Single-Cell Transcriptomic Atlas of the Adult Fruit Fly. bioRxiv. 2021 doi: 10.1101/2021.07.04.451050. [DOI] [PMC free article] [PubMed]
- Li-Kroeger D, Kanca O, Lee PT, Cowan S, Lee MT, Jaiswal M, Salazar JL, He Y, Zuo Z, Bellen HJ. An expanded toolkit for gene tagging based on MiMIC and scarless CRISPR tagging in Drosophila. eLife. 2018;7:e38709. doi: 10.7554/eLife.38709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CC, Potter CJ. Re-Classification of Drosophila melanogaster Trichoid and Intermediate Sensilla Using Fluorescence-Guided Single Sensillum Recording. PLOS ONE. 2015;10:e0139675. doi: 10.1371/journal.pone.0139675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CC, Potter CJ. Editing transgenic DNA components by inducible gene replacement in drosophila melanogaster. Genetics. 2016a;203:1613–1628. doi: 10.1534/genetics.116.191783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CC, Potter CJ. Non-mendelian dominant maternal effects caused by CRISPR/Cas9 transgenic components in Drosophila melanogaster. G3: Genes, Genomes, Genetics. 2016b;6:3685–3691. doi: 10.1534/g3.116.034884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansourian S, Corcoran J, Enjin A, Löfstedt C, Dacke M, Stensmyr MC. Fecal-Derived Phenol Induces Egg-Laying Aversion in Drosophila. Current Biology. 2016;26:2762–2769. doi: 10.1016/j.cub.2016.07.065. [DOI] [PubMed] [Google Scholar]
- Marin EC, Büld L, Theiss M, Sarkissian T, Roberts RJV, Turnbull R, Tamimi IFM, Pleijzier MW, Laursen WJ, Drummond N, Schlegel P, Bates AS, Li F, Landgraf M, Costa M, Bock DD, Garrity PA, Jefferis GSXE. Connectomics Analysis Reveals First-, Second-, and Third-Order Thermosensory and Hygrosensory Neurons in the Adult Drosophila Brain. Current Biology. 2020;30:3167–3182. doi: 10.1016/j.cub.2020.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin CN, Brbić M, Xie Q, Li T, Horns F, Kolluru SS, Kebschull JM, Vacek D, Xie A, Li J, Jones RC, Leskovec J, Quake SR, Luo L, Li H. Single-cell transcriptomes of developing and adult olfactory receptor neurons in Drosophila. eLife. 2021;10:e63856. doi: 10.7554/eLife.63856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menuz K, Larter NK, Park J, Carlson JR. An RNA-seq screen of the Drosophila antenna identifies a transporter necessary for ammonia detection. PLOS Genetics. 2014;10:e1004810. doi: 10.1371/journal.pgen.1004810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min S, Ai M, Shin SA, Suh GSB. Dedicated olfactory neurons mediating attraction behavior to ammonia and amines in Drosophila. PNAS. 2013;110:E1321–E1329. doi: 10.1073/pnas.1215680110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missbach C, Dweck HK, Vogel H, Vilcinskas A, Stensmyr MC, Hansson BS, Grosse-Wilde E. Evolution of insect olfactory receptors. eLife. 2014;3:e02115. doi: 10.7554/eLife.02115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa Y, Koganezawa M, Yamamoto D. Antennae sense heat stress to inhibit mating and promote escaping in Drosophila females. Journal of Neurogenetics. 2018;32:353–363. doi: 10.1080/01677063.2018.1513507. [DOI] [PubMed] [Google Scholar]
- Ng R, Salem SS, Wu ST, Wu M, Lin HH, Shepherd AK, Joiner WJ, Wang JW, Su CY. Amplification of Drosophila Olfactory Responses by a DEG/ENaC Channel. Neuron. 2019;104:947–959. doi: 10.1016/j.neuron.2019.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L, Bronk P, Chang EC, Lowell AM, Flam JO, Panzano VC, Theobald DL, Griffith LC, Garrity PA. A gustatory receptor paralogue controls rapid warmth avoidance in Drosophila. Nature. 2013;500:580–584. doi: 10.1038/nature12390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papes F, Logan DW, Stowers L. The Vomeronasal Organ Mediates Interspecies Defensive Behaviors through Detection of Protein Pheromone Homologs. Cell. 2010;141:692–703. doi: 10.1016/j.cell.2010.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Kwon JY. A systematic analysis of Drosophila gustatory receptor gene expression in abdominal neurons which project to the central nervous system. Molecules and Cells. 2011;32:375–381. doi: 10.1007/s10059-011-0128-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pask GM, Ray A. In: Chemosensory Transduction. Akabas MH, editor. Elsevier; 2016. Insect olfactory receptors: an interface between chemistry and biology; pp. 101–122. [DOI] [Google Scholar]
- Pinto L, Stocker RF, Rodrigues V. Anatomical and neurochemical classification of the antennal glomeruli in Drosophila melanogaster meigen (Diptera: Drosophilidae) International Journal of Insect Morphology and Embryology. 1988;17:335–344. doi: 10.1016/0020-7322(88)90014-1. [DOI] [Google Scholar]
- Port F, Chen HM, Lee T, Bullock SL. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. PNAS. 2014;111:E2967–E2976. doi: 10.1073/pnas.1405500111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CJ, Tasic B, Russler EV, Liang L, Luo L. The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell. 2010;141:536–548. doi: 10.1016/j.cell.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto-Godino LL, Rytz R, Cruchet S, Bargeton B, Abuin L, Silbering AF, Ruta V, Dal Peraro M, Benton R. Evolution of Acid-Sensing Olfactory Circuits in Drosophilids. Neuron. 2017;93:661–676. doi: 10.1016/j.neuron.2016.12.024. [DOI] [PubMed] [Google Scholar]
- Prieto-Godino LL, Silbering AF, Khallaf MA, Cruchet S, Bojkowska K, Pradervand S, Hansson BS, Knaden M, Benton R. Functional integration of “undead” neurons in the olfactory system. Science Advances. 2020;6:eaaz7238. doi: 10.1126/sciadv.aaz7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, van Naters W, Shiraiwa T, Carlson JR. Mechanisms of odor receptor gene choice in Drosophila. Neuron. 2007;53:353–369. doi: 10.1016/j.neuron.2006.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, van der Goes van Naters W, Carlson JR. A regulatory code for neuron-specific odor receptor expression. PLOS Biology. 2008;6:0060125. doi: 10.1371/journal.pbio.0060125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riabinina O, Luginbuhl D, Marr E, Liu S, Wu MN, Luo L, Potter CJ. Improved and expanded Q-system reagents for genetic manipulations. Nature Methods. 2015;12:219–222. doi: 10.1038/nmeth.3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riabinina O, Task D, Marr E, Lin C-C, Alford R, O’Brochta DA, Potter CJ. Organization of olfactory centres in the malaria mosquito Anopheles gambiae. Nature Communications. 2016;7:13010. doi: 10.1038/ncomms13010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saina M, Benton R. Visualizing olfactory receptor expression and localization in Drosophila. Methods in Molecular Biology. 2013;1003:211–228. doi: 10.1007/978-1-62703-377-0_16. [DOI] [PubMed] [Google Scholar]
- Sánchez-Alcañiz JA, Silbering AF, Croset V, Zappia G, Sivasubramaniam AK, Abuin L, Sahai SY, Münch D, Steck K, Auer TO, Cruchet S, Neagu-Maier GL, Sprecher SG, Ribeiro C, Yapici N, Benton R. An expression atlas of variant ionotropic glutamate receptors identifies a molecular basis of carbonation sensing. Nature Communications. 2018;9:4252. doi: 10.1038/s41467-018-06453-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Pellegrino M, Nakagawa T, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452:1002–1006. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- Scheffer LK, Xu CS, Januszewski M, Lu Z, Takemura S-Y, Hayworth KJ, Huang GB, Shinomiya K, Maitlin-Shepard J, Berg S, Clements J, Hubbard PM, Katz WT, Umayam L, Zhao T, Ackerman D, Blakely T, Bogovic J, Dolafi T, Kainmueller D, Kawase T, Khairy KA, Leavitt L, Li PH, Lindsey L, Neubarth N, Olbris DJ, Otsuna H, Trautman ET, Ito M, Bates AS, Goldammer J, Wolff T, Svirskas R, Schlegel P, Neace E, Knecht CJ, Alvarado CX, Bailey DA, Ballinger S, Borycz JA, Canino BS, Cheatham N, Cook M, Dreher M, Duclos O, Eubanks B, Fairbanks K, Finley S, Forknall N, Francis A, Hopkins GP, Joyce EM, Kim S, Kirk NA, Kovalyak J, Lauchie SA, Lohff A, Maldonado C, Manley EA, McLin S, Mooney C, Ndama M, Ogundeyi O, Okeoma N, Ordish C, Padilla N, Patrick CM, Paterson T, Phillips EE, Phillips EM, Rampally N, Ribeiro C, Robertson MK, Rymer JT, Ryan SM, Sammons M, Scott AK, Scott AL, Shinomiya A, Smith C, Smith K, Smith NL, Sobeski MA, Suleiman A, Swift J, Takemura S, Talebi I, Tarnogorska D, Tenshaw E, Tokhi T, Walsh JJ, Yang T, Horne JA, Li F, Parekh R, Rivlin PK, Jayaraman V, Costa M, Jefferis GS, Ito K, Saalfeld S, George R, Meinertzhagen IA, Rubin GM, Hess HF, Jain V, Plaza SM. A connectome and analysis of the adult Drosophila central brain. eLife. 2020;9:e57443. doi: 10.7554/eLife.57443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nature Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel P, Bates AS, Stürner T, Jagannathan SR, Drummond N, Hsu J, Serratosa Capdevila L, Javier A, Marin EC, Barth-Maron A, Tamimi IF, Li F, Rubin GM, Plaza SM, Costa M, Jefferis GSXE. Information flow, cell types and stereotypy in a full olfactory connectome. eLife. 2021;10:e66018. doi: 10.7554/eLife.66018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K, Brady R, Cravchik A, Morozov P, Rzhetsky A, Zuker C, Axel R. A Chemosensory Gene Family Encoding Candidate Gustatory and Olfactory Receptors in Drosophila. Cell. 2001;104:661–673. doi: 10.1016/s0092-8674(01)00263-x. [DOI] [PubMed] [Google Scholar]
- Scott K. Gustatory Processing in Drosophila melanogaster. Annual Review of Entomology. 2018;63:15–30. doi: 10.1146/annurev-ento-020117-043331. [DOI] [PubMed] [Google Scholar]
- Seki Y, Dweck HKM, Rybak J, Wicher D, Sachse S, Hansson BS. Olfactory coding from the periphery to higher brain centers in the Drosophila brain. BMC Biology. 2017;15:0389-z. doi: 10.1186/s12915-017-0389-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearin HK, Macdonald IS, Spector LP, Stowers RS. Hexameric GFP and mCherry reporters for the Drosophila GAL4, Q, and LexA transcription systems. Genetics. 2014;196:951–960. doi: 10.1534/genetics.113.161141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi O. Neurogenetics of olfaction in Drosophila melanogaster. Trends in Genetics. 1987;3:137–142. doi: 10.1016/0168-9525(87)90204-6. [DOI] [Google Scholar]
- Siegal ML, Hartl DL. Transgene Coplacement and high efficiency site-specific recombination with the Cre/loxP system in Drosophila. Genetics. 1996;144:715–726. doi: 10.1093/genetics/144.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silbering AF, Rytz R, Grosjean Y, Abuin L, Ramdya P, Jefferis GSXE, Benton R. Complementary function and integrated wiring of the evolutionarily distinct Drosophila olfactory subsystems. The Journal of Neuroscience. 2011;31:13357–13375. doi: 10.1523/JNEUROSCI.2360-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smadja C, Shi P, Butlin RK, Robertson HM. Large Gene Family Expansions and Adaptive Evolution for Odorant and Gustatory Receptors in the Pea Aphid, Acyrthosiphon pisum. Molecular Biology and Evolution. 2009;26:2073–2086. doi: 10.1093/molbev/msp116. [DOI] [PubMed] [Google Scholar]
- Soffan A, Subandiyah S, Makino H, Watanabe T, Horiike T. Evolutionary Analysis of the Highly Conserved Insect Odorant Coreceptor (Orco) Revealed a Positive Selection Mode, Implying Functional Flexibility. Journal of Insect Science (Online) 2018;18:120. doi: 10.1093/jisesa/iey120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spletter ML, Liu J, Liu J, Su H, Giniger E, Komiyama T, Quake S, Luo L. Lola regulates Drosophila olfactory projection neuron identity and targeting specificity. Neural Development. 2007;2:14. doi: 10.1186/1749-8104-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stengl M. Pheromone transduction in moths. Frontiers in Cellular Neuroscience. 2010;4:133. doi: 10.3389/fncel.2010.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stensmyr MC, Dweck HKM, Farhan A, Ibba I, Strutz A, Mukunda L, Linz J, Grabe V, Steck K, Lavista-Llanos S, Wicher D, Sachse S, Knaden M, Becher PG, Seki Y, Hansson BS. A Conserved Dedicated Olfactory Circuit for Detecting Harmful Microbes in Drosophila. Cell. 2012;151:1345–1357. doi: 10.1016/j.cell.2012.09.046. [DOI] [PubMed] [Google Scholar]
- Stocker RF. The organization of the chemosensory system in Drosophila melanogaster: a review. Cell and Tissue Research. 1994;275:3–26. doi: 10.1007/BF00305372. [DOI] [PubMed] [Google Scholar]
- Stocker RF. Drosophila as a focus in olfactory research: mapping of olfactory sensilla by fine structure, odor specificity, odorant receptor expression, and central connectivity. Microscopy Research and Technique. 2001;55:284–296. doi: 10.1002/jemt.1178. [DOI] [PubMed] [Google Scholar]
- Su CY, Menuz K, Carlson JR. Olfactory perception: receptors, cells, and circuits. Cell. 2009;139:45–59. doi: 10.1016/j.cell.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Kumar S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Molecular Biology and Evolution. 2021;38:3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Task D, Lin CC, Vulpe A, Afify A, Ballou S, Brbić M, Schlegel P, Jefferis G, Li H, Menuz K, Potter CJ. Chemoreceptor Co-Expression in Drosophila Olfactory Neurons. bioRxiv. 2020 doi: 10.1101/2020.11.07.355651. [DOI] [PMC free article] [PubMed]
- Task D, Potter CJ. Rapid degeneration of Drosophila olfactory neurons in Orco mutant maxillary palps. MicroPublication Biology. 2021;14:000398. doi: 10.17912/micropub.biology.000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurmond J, Goodman JL, Strelets VB, Attrill H, Gramates LS, Marygold SJ, Matthews BB, Millburn G, Antonazzo G, Trovisco V, Kaufman TC, Calvi BR, FlyBase Consortium FlyBase 2.0: the next generation. Nucleic Acids Research. 2019;47:D759–D765. doi: 10.1093/nar/gky1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trible W, Olivos-Cisneros L, McKenzie SK, Saragosti J, Chang NC, Matthews BJ, Oxley PR, Kronauer DJC. Orco mutagenesis causes loss of antennal lobe glomeruli and impaired social behavior in ants. Cell. 2017;170:727–735. doi: 10.1016/j.cell.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Maaten L, Hinton G. Visualizing data using t-SNE. Journal of Machine Learning Research: JMLR. 2008;9:2579–2605. [Google Scholar]
- Venkatesh S, Naresh Singh R. Sensilla on the third antennal segment of Drosophila melanogaster meigen (Diptera: Drosophilidae) International Journal of Insect Morphology and Embryology. 1984;13:51–63. doi: 10.1016/0020-7322(84)90032-1. [DOI] [Google Scholar]
- Vilain S, Vanhauwaert R, Maes I, Schoovaerts N, Zhou L, Soukup S, da Cunha R, Lauwers E, Fiers M, Verstreken P. Fast and efficient Drosophila melanogaster gene knock-ins using MiMIC transposons. G3: Genes, Genomes, Genetics. 2014;4:2381–2387. doi: 10.1534/g3.114.014803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. A Spatial Map of Olfactory Receptor Expression in the Drosophila Antenna. Cell. 1999;96:725–736. doi: 10.1016/s0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Wong AM, Axel R. An Olfactory Sensory Map in the Fly Brain. Cell. 2000;102:147–159. doi: 10.1016/s0092-8674(00)00021-0. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Stocker RF. Molecular architecture of smell and taste in Drosophila. Annual Review of Neuroscience. 2007;30:505–533. doi: 10.1146/annurev.neuro.30.051606.094306. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Hansson BS. A unified nomenclature system for the insect olfactory coreceptor. Chemical Senses. 2011;36:497–498. doi: 10.1093/chemse/bjr022. [DOI] [PubMed] [Google Scholar]
- Vulpe A, Kim HS, Ballou S, Wu ST, Grabe V, Nava Gonzales C, Liang T, Sachse S, Jeanne JM, Su CY, Menuz K. An ammonium transporter is a non-canonical olfactory receptor for ammonia. Current Biology. 2021;31:3382–3390. doi: 10.1016/j.cub.2021.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulpe A, Menuz K. Ir76b is a Co-receptor for Amine Responses in Drosophila Olfactory Neurons. Frontiers in Cellular Neuroscience. 2021;15:759238. doi: 10.3389/fncel.2021.759238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RI. Early olfactory processing in Drosophila: mechanisms and principles. Annual Review of Neuroscience. 2013;36:217–241. doi: 10.1146/annurev-neuro-062111-150533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T, Ho MCW, Liu Q, Horiuchi W, Lin CC, Task D, Luan H, White BH, Potter CJ, Wu MN. A genetic toolkit for dissecting dopamine circuit function in Drosophila. Cell Reports. 2018;23:652–665. doi: 10.1016/j.celrep.2018.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z, Ren M, Wu M, Dai J, Rong YS, Gao G. Efficient gene knock-out and knock-in with transgenic Cas9 in Drosophila. G3: Genes, Genomes, Genetics. 2014;4:925–929. doi: 10.1534/g3.114.010496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Opachaloemphan C, Mancini G, Yang H, Gallitto M, Mlejnek J, Leibholz A, Haight K, Ghaninia M, Huo L, Perry M, Slone J, Zhou X, Traficante M, Penick CA, Dolezal K, Gokhale K, Stevens K, Fetter-Pruneda I, Bonasio R, Zwiebel LJ, Berger SL, Liebig J, Reinberg D, Desplan C. An Engineered orco Mutation Produces Aberrant Social Behavior and Defective Neural Development in Ants. Cell. 2017;170:736–747. doi: 10.1016/j.cell.2017.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Jafari S, Pask G, Zhou X, Reinberg D, Desplan C. Evolution, developmental expression and function of odorant receptors in insects. The Journal of Experimental Biology. 2020;223:jeb208215. doi: 10.1242/jeb.208215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao CA, Ignell R, Carlson JR. Chemosensory Coding by Neurons in the Coeloconic Sensilla of the Drosophila Antenna. The Journal of Neuroscience. 2005;25:8359–8367. doi: 10.1523/JNEUROSCI.2432-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z, Liu F, Sun H, Baker A, Zwiebel LJ. Discrete Roles of the Ir76b Ionotropic Co-Receptor Impact Olfaction, Blood Feeding, and Mating in the Malaria Vector Mosquito Anopheles Coluzzii. bioRxiv. 2021 doi: 10.1101/2021.07.05.451160. [DOI] [PMC free article] [PubMed]
- Younger MA, Herre M, Goldman OV, Lu TC, Caballero-Vidal G, Qi Y, Gilbert ZN, Gong Z, Morita T, Rahiel S, Ghaninia M, Ignell R, Matthews BJ, Li H, Vosshall LB. Non-Canonical Odor Coding in the Mosquito. bioRxiv. 2020 doi: 10.1101/2020.11.07.368720. [DOI] [PMC free article] [PubMed]
- Zhang YV, Ni J, Montell C. The molecular basis for attractive salt-taste coding in Drosophila. Science. 2013;340:1334–1338. doi: 10.1126/science.1234133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Lauritzen JS, Perlman E, Robinson CG, Nichols M, Milkie D, Torrens O, Price J, Fisher CB, Sharifi N, Calle-Schuler SA, Kmecova L, Ali IJ, Karsh B, Trautman ET, Bogovic JA, Hanslovsky P, Jefferis G, Kazhdan M, Khairy K, Saalfeld S, Fetter RD, Bock DD. A complete electron microscopy volume of the brain of adult Drosophila melanogaster. Cell. 2018;174:730–743. doi: 10.1016/j.cell.2018.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]