Abstract
In a recent article, Immel et al. (Immel A, Key FM, Szolek A, Barquera R, Robinson MK, Harrison GF, Palmer WH, Spyrou MA, Susat J, Krause-Kyora B, et al. 2021. Analysis of genomic DNA from medieval plague victims suggests long-term effect of Yersinia pestis on human immunity genes. Mol Biol Evol. 38:4059–4076) extracted DNA from 36 individuals dead from plague in Ellwangen, Southern Germany, during the 16th century. By comparing their human leukocyte antigen (HLA) genotypes with those of 50 present-day Ellwangen inhabitants, the authors reported a significant decrease of HLA-B*51:01 and HLA-C*06:02 and a significant increase of HLA-DRB1*13:01/13:02 frequencies from ancient to modern populations. After comparing these frequencies with a larger sample of 8,862 modern Germans and performing simulations of natural selection, they concluded that these changes had been driven by natural selection. In an attempt to provide more evidence on such stimulating results, we explored the HLA frequency patterns over all of Europe, we predicted binding affinities of HLA-B/C/DRB1 alleles to 106,515 Yersinia pestis-derived peptides, and we performed forward simulations of HLA genetic profiles under neutrality. Our analyses do not sustain the conclusions of HLA protection or susceptibility to plague based on ancient DNA.
Keywords: HLA, Yersinia pestis, peptide-binding predictions, computer simulations, ancient DNA, pathogen-driven selection
Following the discovery of their crucial role in adaptive immunity, the classical human leukocyte antigen (HLA) genes have long been considered as candidates for genetic resistance or susceptibility to infectious diseases (Trowsdale and Knight 2013; Sanchez-Mazas 2020). Consequently, it may also be expected to detect, on HLA genes, signals of natural selection due to their adaptation to pathogens. Among these, Yersinia pestis (Y. pestis), a bacterium transmitted from rodents to humans that causes plague, was the major agent of several most-devastating pandemics in human history. According to historical records, Europe may have suffered the most severe death toll from the plague. The most dramatic plague pandemic known as the “Black Death” during the 14th century caused more than 50 million deaths in Europe, which was about 30–60% of the European population. More local outbreaks continued until the 18th century. Yersinia pestis is practically eradicated from Europe nowadays, whereas plague epidemics still occur sporadically in Africa, Asia, and South America (WHO 2017).
In a recent paper, Immel et al. (2021) analyzed 488 immune-related genes, among which 8 HLA Class I (-A, -B, and -C) and Class II (-DPA1, -DPB1, -DQA1, -DQB1, and -DRB1) genes, on ancient DNA extracted from 36 individuals buried in a mass grave located in Ellwangen in Southern Germany and who apparently died from plague during the 16th century. By comparing these data with those of 50 modern-day native inhabitants of Ellwangen, the authors reported that the frequencies of two HLA alleles decreased significantly, that is, that of B*51:01 from 15.3% to 6.0% and that of C*06:02 from 13.9% to 5.0% from ancient to modern inhabitants, whereas the combined frequency of DRB1*13:01 and DRB1*13:02 increased significantly from 5.6% to 17.0%. Based on these results and simulations of natural selection, the authors concluded that the changes in frequencies of the aforementioned HLA alleles were driven by natural selection due to the 16th-century plague epidemics, that is, a susceptibility effect of B*51:01 and C*06:02 and a protective effect of DRB1*13:01/13:02.
Here, we first explored this hypothesis in relation to current knowledge on both frequency distributions of HLA alleles in worldwide populations and peptide-binding properties of the corresponding HLA molecules, two approaches that were not considered in the study carried out by Immel et al. (2021). In this perspective, we collected allele frequency data (defined at the 2nd-field-level of resolution, thereafter named “2nd-field alleles”) for 81 modern populations from 10 major geographic regions of the world chosen according to several criteria to ensure data quality (see supplementary table S1, Supplementary Material online). We mainly focused on the frequency distributions of alleles B*51:01, C*06:02, DRB1*13:01, and DRB1*13:02 as well as of the “DRB1*13” allele family, which includes all DRB1*13 alleles found in each population (supplementary table S2, Supplementary Material online). However, in order to give a reliable representation of these alleles of interest at the global level, we also predicted binding affinities for the HLA molecules encoded by all HLA alleles observed in the ancient and modern Ellwangen samples studied by Immel et al. (2021) against Y. pestis-derived peptides (i.e., 17 HLA-B, 14 HLA-C, and 17 HLA-DRB1 2nd-field alleles). The whole Y. pestis proteome was extracted from the UniProt database (Parkhill et al. 2001; UniProt 2021) and a total of 3,915 distinct proteins were investigated by applying netMHCpan 4.1 (Reynisson, Alvarez, et al. 2020) for HLA Class I (HLA-B and HLA-C) and netMHCIIpan 4.0 (Reynisson, Alvarez, et al. 2020; Reynisson, Barra, et al. 2020) for HLA Class II (HLA-DRB1) molecules. Special attention was paid to the peptides derived from five Y. pestis proteins (rstB, YPO2385, hmuR, flaA1, psaB) that were recently reported as highly antigenic and immunogenic (Haq et al. 2021). The lengths of the most common bound peptides, 9-mer and 15-mer, were set for the two HLA Classes, respectively, resulting in a set of 1,220,545 9-mer and 1,127,562 15-mer different Y. pestis-derived peptides. For each HLA molecule, a binding affinity score (BAscore) was estimated, and the percentile rank of the predicted binding score (%Rank) was also reported to determine whether the protein was expected to bind a peptide with upper thresholds of 2/0.5 (Class I) and 10/2 (Class II) for weak/strong binders, respectively (supplementary tables S3 and S4, Supplementary Material online). Finally, we also performed new forward simulations of HLA genetic evolution for the Ellwangen population with the demographic parameters (generation numbers, population sizes) considered by Immel et al. (2021) but without selection by using the program Selector (see supplementary table S5, Supplementary Material online for detailed information about Selector and the setting information), a program designed for studying HLA evolution in human populations (Currat et al. 2015).
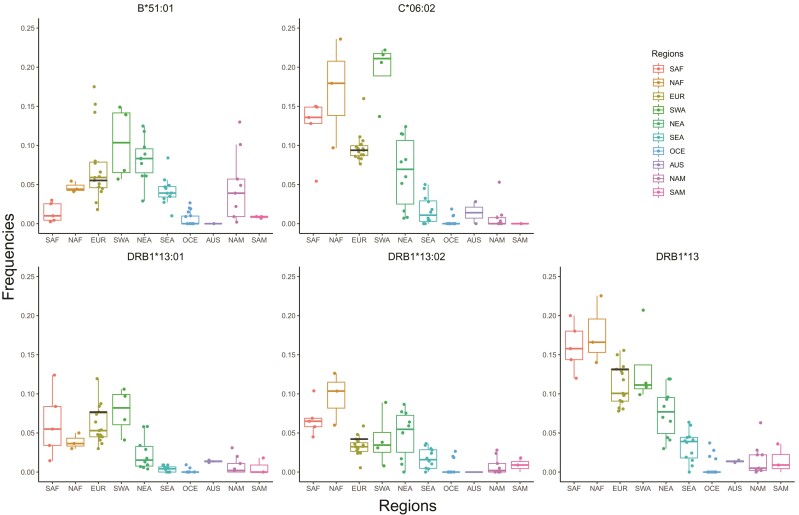
The frequency distributions of B*51:01, C*06:02, DRB1*13:01, DRB1*13:02, and DRB1*13 HLA alleles in the 10 worldwide geographic regions are shown in figure 1. As, according to Immel et al. (2021), B*51:01 and C*06:02 would have conferred susceptibility and DRB1*13 resistance to plague, respectively, we would expect reduced frequencies of B*51:01 and C*06:02, as a result of negative selection, and enhanced frequencies of DRB1*13 alleles (including DRB1*13:01 and DRB1*13:02 in Ellwangen and often a few additional alleles in other populations), as a result of positive selection in Europe, where the Black Death was particularly harmful. However, this is not the case. First, none of these 2nd-field alleles shows such tendencies compared with the other geographic regions (fig. 1): This is also true for the DRB1*13 allele family taken as a whole, which exhibits decreasing frequencies from Africa (14–23%) to Americas (<4%) at the global scale (fig. 1). Second, the frequencies of B*51:01, C*06:02, DRB1*13:01, and DRB1*13:02 vary, sometimes greatly, among populations across Europe (fig. 1 and supplementary table S2, Supplementary Material online). Although the Black Death unevenly affected this continent from a demographic point of view, devastating some regions and sparing others, as recently suggested on the basis of paleoecological data (Izdebski et al. 2022), the frequency distribution of these alleles does not match what would be expected in distinct European regions if they had been susceptibility or protective alleles (supplementary fig. S1, Supplementary Material online). For example, the putative susceptibility allele B*51:01, instead of being rare or absent in Greece where plague mortality had likely been very high, reaches its highest frequency (∼15%) in this region compared with the rest of Europe; by contrast, it is rare in Ireland (<3%) where plague likely had a negligible or no impact at all. Therefore, based on their present-day frequencies, there are no clear signatures of pathogen-driven selection for these HLA Class I and Class II alleles in Europe.
Fig. 1.
Box plots representing frequency distributions of four HLA alleles, B*51:01, C*06:02, DRB1*13:01, and DRB1*13:02, and one allele family, DRB1*13, in each of 10 geographic regions (SAF, Sub-Saharan Africa; NAF, North Africa; EUR, Europe; SWA, South-West Asia; NEA, North-East Asia; SEA, South-East Asia; OCE, Oceania; AUS, Australia; NAM, North America; SAM, South America). The frequencies in a German sample are marked by a black bar, whereas the dots represent the frequencies of these alleles observed in all other populations, as reported in supplementary table S2, Supplementary Material online.
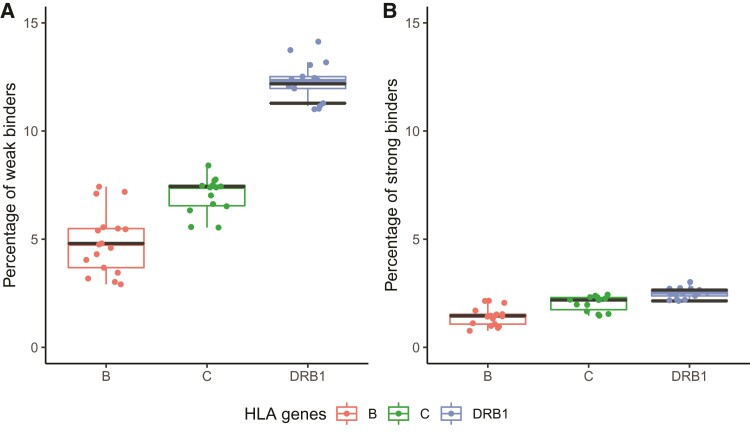
Although an efficient HLA response to infectious diseases may also depend upon other properties than peptide presentation, such as HLA gene expression levels (Apps et al. 2015), tapasin dependence (Bashirova et al. 2020), and/or coverage of specific epitopes, peptide-binding predictions between HLA molecules and pathogen-derived peptides were used in previous studies as indicators of HLA susceptibility or resistance to different pathogens (Arora et al. (2019) for HIV-1; Nguyen et al. (2020) and Barquera et al. (2020) for SARS-CoV-2, among others). This approach was shown to be relevant, for example, in a study by Krause-Kyora et al. (2018) on ancient DNA extracted from a sample of Danish medieval skeletons showing specific lesions of lepromatous leprosy; the authors found that HLA-DRB1*15:01 was a possible susceptible factor for this disease (as already suggested in modern populations, see Sanchez-Mazas (2020)), and this result was corroborated by the fact that the HLA-DRB1*15:01 molecule binds the second-smallest number of potential Mycobacterium leprae antigens. Here, we computed the percentage of Y. pestis-derived peptides predicted to bind either weakly or strongly each of the whole set of HLA-B, HLA-C, and HLA-DR molecules corresponding to the HLA alleles observed in Ellwangen. The results are visualized in figure 2, with values for B*51:01, C*06:02, DRB1*13:01, and DRB1*13:02 molecules highlighted with black bars. The putative susceptible B*51:01 and C*06:02 molecules do not show lower proportions of bound peptides than other HLA molecules, and the suggested protective DRB1*13:01 and/or DRB1*13:02 molecules do not show higher proportions of bound peptides than other ones, either. These results remain compatible when only peptides from the five strongly antigenic and immunogenic Y. pestis proteins rstB, YPO2385, hmuR, flaA1, and psaB identified by Haq et al. (2021) are considered (supplementary fig. S2, Supplementary Material online). Based on these results, we are not able to conclude to any particular susceptibility effect of B*51:01 and C*06:02 or protective effect of DRB1*13 molecules against Y. pestis in terms of peptide-binding properties.
Fig. 2.
Box plots representing the percentages of (A) weak binders and (B) strong binders, among all peptides derived from a total of 3,915 distinct proteins of Yersinia pestis proteome, predicted by netMHCpan and netMHCIIpan for HLA-B, HLA-C, and HLA-DRB1 molecules, with values for B*51:01, C*06:02, DRB1*13:01 (lower bar), and DRB1*13:02 (upper bar) alleles highlighted by black bars. The dots represent the percentages for the other HLA molecules, as reported in supplementary table S3, Supplementary Material online.
Although neither HLA frequency comparisons among populations nor peptide-binding predictions, which are two independent approaches, are consistent with possible associations to plague of the HLA-B, HLA-C, and HLA-DRB1 alleles suggested by Immel et al. (2021), this is not sufficient, of course, to reject their hypotheses, but more evidence is also necessary to retain them. Indeed, the small sample size of 36 plague victims used by these authors likely increases the chance of sampling errors, as the authors acknowledge in their discussion, and may lead to false-positives. Actually the P-values they obtained for B*51:01 and C*06:02 alleles are borderline (P = 0.044 and 0.041, respectively) and no longer significant after correction for multiple testing. A significant P-value (P = 0.026), here again not significant after correction for multiple testing, was obtained for the DRB1*13 allele family, which is actually a group of two alleles coding for two functionally distinct molecules. Taken individually, these two alleles do not yield significant results (P = 0.067 and P = 0.323 for DRB1*13:01 and DRB1*13:02, respectively). Although the authors argue that only one amino acid differs between DRB1*13:01 and DRB1*13:02 molecules, it is insufficient to consider that they assume identical functions. Our peptide-binding predictions indicate that their binding affinities are more different from each other than many other DRB1 molecule pairs (fig. 2 and supplementary fig. S2, Supplementary Material online), as they show important binding motif differences in their peptide-binding region (supplementary fig. S3, Supplementary Material online).
Regarding natural selection, the authors estimate selection coefficients s of −25% and −27% to produce the observed decrease in B*51:01 and C*06:02, respectively, and of 37% to produce the observed increase in DRB1*13:01 with simulations run since the 16th century. Although they argue that these values are “within the range of previously reported values of s acting on MHC (Radwan et al. 2020)”, previously estimated s values never exceed 5% for HLA in Homo sapiens (Satta et al. 1994; Currat et al. 2010; Yasukochi and Satta 2013; Di et al. 2015), except one unexpectedly high-reported value of 46.2% to explain HLA-A and HLA-B homozygotes deficiency in a Native American population (Havasupai), with no associations with particular alleles (Black and Hedrick 1997), and never confirmed by independent studies. Otherwise, s only reaches 4.3% at the DARC (Duffy) locus (McManus et al. 2017) and, depending on the method used, from 0.3% to 19% (2–2.5% based on ancient DNA; Mathieson 2020) at the LCT (lactase persistence) locus (Gerbault 2013), DARC and LCT being (among) the most strongly selected genomic regions in humans. Although the approaches to estimate s may differ and short-term estimates may lead to higher values than long-term estimates, s values of more than 20% for HLA, thus, seem unrealistic, mostly because this genomic region is predominantly submitted to balancing selection in the form of divergent allele advantage (Pierini and Lenz 2018), joint divergent asymmetric selection (Buhler et al. 2016; Di et al. 2021), and/or soft selective sweep (Sanchez-Mazas et al. 2017) rather than positive selection. Moreover, as the deadliest plague pandemic occurred in the 14th century in Europe (the Black Death decimating almost half of the European population during its 5-year period) and the same Y. pestis lineage was responsible for subsequent plague outbreaks in Europe (Spyrou et al. 2016), in case of such strong susceptibility or resistance effects, the frequencies of B*51:01, C*06:02, and DRB1*13:01 would have been affected by natural selection long (i.e., at least 200 years) before the 16th century. This conflicts with the fact that B*51:01 and C*06:02 frequencies are among the highest and that of DRB1*13:01 among the lowest of all allele frequencies observed at their respective locus in the 16th-century plague victims from Ellwangen (table 3 of Immel et al. (2021)). Immel et al. (2021) did not find any significant differences between the overall HLA frequency distributions estimated for Ellwangen plague victims and present-day Germans, which is also incompatible with such strong selective effects.
In this context, we further performed 10,000 simulations of the demographic history of Ellwangen population from the 16th to the 21st century (20 generations) using the computer program Selector, which was designed specifically for studying HLA evolution in human populations (Currat et al. 2015). We fixed a total duration of 120 generations for each simulation and 20 initial HLA alleles with random frequencies. These settings allowed us to create, after 100 generations of randomization (using a growth rate of 0.1), a population (p1) having a HLA allele frequency distribution comparable to that observed in the 16th-century Ellwangen sample (with effective population size equal to a third of the census size =5,000/3, and HLA allele number between 14 and 20 representing any of the HLA-B, HLA-C, and HLA-DRB1 loci). At this moment, the effective population size was reset to 25,000/3 and the growth rate to 0.085 (to be equivalent to the values used with a different model by Immel et al. (2021)), but without any type of natural selection (selection coefficient fixed at 0). The frequencies of HLA alleles were recorded in the p1 population and the final population (p2) after 20 additional generations (representing 500 years). Based on the empirical dataset of Immel et al. (2021), 36 “ancient” and 50 “modern” individuals were sampled from the p1 and p2 populations, respectively. None of the 10,000 simulations showed significant differences between the two samples, as measured by FST using Arlesumstat v3.5.2.2 program (Excoffier and Lischer 2010). For about 39% of the total simulations (3,891 among 10,000), a significant difference was detected by χ2 test (P < 0.05, without multiple testing correction) for at least one allele. These simulations show that the simplest scenario without selection can produce population samples that are not genetically distant and yet show at least one allele with significantly different frequencies without multiple testing correction, as found by Immel et al. (2021). More straightforwardly, our results indicate that it is not necessary to invoke selection to explain the observed differences in some allele frequencies between ancient and modern samples.
In conclusion, although the analysis of ancient DNA is one of the most promising approaches to unravel the evolution of human immunity genes in relation to past epidemics (such as the mechanisms of pathogen-driven selection that shaped the evolution of the HLA polymorphism), it needs to be put into context and, to our view, more robust evidence than that presented by Immel et al. (2021) through the study of medieval plague victims from Southern Germany is required to confirm the susceptibility or resistance effects of HLA alleles to Y. pestis.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by grants no. 310030_188820 and 31003A-144180 (to A.S.-M.) and no. 31003A_182577 (to M.C.) of the Swiss National Science Foundation. We sincerely thank two anonymous reviewers for their constructive comments allowing us to substantially improve a previous version of this manuscript.
Data Availability
All data used in this paper are available in Supplementary information, Supplementary Material online or on request to the authors.
References
- Apps R, Meng Z, Del Prete GQ, Lifson JD, Zhou M, Carrington M. 2015. Relative expression levels of the HLA class-I proteins in normal and HIV-infected cells. J Immunol. 194:3594–3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora J, McLaren PJ, Chaturvedi N, Carrington M, Fellay J, Lenz TL. 2019. HIV peptidome-wide association study reveals patient-specific epitope repertoires associated with HIV control. Proc Natl Acad Sci USA. 116:944–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barquera R, Collen E, Di D, Buhler S, Teixeira J, Llamas B, Nunes JM, Sanchez-Mazas A. 2020. Binding affinities of 438 HLA proteins to complete proteomes of seven pandemic viruses and distributions of strongest and weakest HLA peptide binders in populations worldwide. HLA 96:277–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashirova AA, Viard M, Naranbhai V, Grifoni A, Garcia-Beltran W, Akdag M, Yuki Y, Gao X, O’HUigin C, Raghavan M, et al. . 2020. HLA tapasin independence: broader peptide repertoire and HIV control. Proc Natl Acad Sci USA. 117:28232–28238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black FL, Hedrick PW. 1997. Strong balancing selection at HLA loci: evidence from segregation in South Amerindian families. Proc Natl Acad Sci USA. 94:12452–12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler S, Nunes JM, Sanchez-Mazas A. 2016. HLA class I molecular variation and peptide-binding properties suggest a model of joint divergent asymmetric selection. Immunogenetics 68:401–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currat M, Gerbault P, Di D, Nunes JM, Sanchez-Mazas A. 2015. Forward-in-time, spatially explicit modeling software to simulate genetic lineages under selection. Evol Bioinform Online 11:27–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currat M, Poloni ES, Sanchez-Mazas A. 2010. Human genetic differentiation across the Strait of Gibraltar. BMC Evol Biol. 10:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di D, Nunes JM, Jiang W, Sanchez-Mazas A. 2021. Like wings of a bird: functional divergence and complementarity between HLA-A and HLA-B molecules. Mol Biol Evol. 38:1580–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di D, Sanchez-Mazas A, Currat M. 2015. Computer simulation of human leukocyte antigen genes supports two main routes of colonization by human populations in East Asia. BMC Evol Biol. 15:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 10:564–567. [DOI] [PubMed] [Google Scholar]
- Gerbault P. 2013. The onset of lactase persistence in Europe. Hum Hered. 76:154–161. [DOI] [PubMed] [Google Scholar]
- Haq AU, Khan A, Khan J, Irum S, Waheed Y, Ahmad S, Nizam-Uddin N, Albutti A, Zaman N, Hussain Z, et al. . 2021. Annotation of potential vaccine targets and design of a multi-epitope subunit vaccine against Yersinia pestis through reverse vaccinology and validation through an agent-based modeling approach. Vaccines (Basel) 9:1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immel A, Key FM, Szolek A, Barquera R, Robinson MK, Harrison GF, Palmer WH, Spyrou MA, Susat J, Krause-Kyora B, et al. . 2021. Analysis of genomic DNA from medieval plague victims suggests long-term effect of Yersinia pestis on human immunity genes. Mol Biol Evol. 38:4059–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izdebski A, Guzowski P, Poniat R, Masci L, Palli J, Vignola C, Bauch M, Cocozza C, Fernandes R, Ljungqvist FC, et al. . 2022. Palaeoecological data indicates land-use changes across Europe linked to spatial heterogeneity in mortality during the Black Death pandemic. Nat Ecol Evol. 6:297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause-Kyora B, Nutsua M, Boehme L, Pierini F, Pedersen DD, Kornell SC, Drichel D, Bonazzi M, Möbus L, Tarp P, et al. . 2018. Ancient DNA study reveals HLA susceptibility locus for leprosy in medieval Europeans. Nat Commun. 9:1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieson I. 2020. Estimating time-varying selection coefficients from time series data of allele frequencies. bioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus KF, Taravella AM, Henn BM, Bustamante CD, Sikora M, Cornejo OE. 2017. Population genetic analysis of the DARC locus (Duffy) reveals adaptation from standing variation associated with malaria resistance in humans. PLoS Genet. 13:e1006560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A, David JK, Maden SK, Wood MA, Weeder BR, Nellore A, Thompson RF. 2020. Human leukocyte antigen susceptibility map for severe acute respiratory syndrome coronavirus 2. J Virol. 94:e00510-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J, Wren BW, Thomson NR, Titball RW, Holden MT, Prentice MB, Sebaihia M, James KD, Churcher C, Mungall KL, et al. . 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523–527. [DOI] [PubMed] [Google Scholar]
- Pierini F, Lenz TL. 2018. Divergent allele advantage at human MHC genes: signatures of past and ongoing selection. Mol Biol Evol. 35:2145–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radwan J, Babik W, Kaufman J, Lenz TL, Winternitz J. 2020. Advances in the evolutionary understanding of MHC polymorphism. Trends Genet. 36:298–311. [DOI] [PubMed] [Google Scholar]
- Reynisson B, Alvarez B, Paul S, Peters B, Nielsen M. 2020. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 48:W449–W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynisson B, Barra C, Kaabinejadian S, Hildebrand WH, Peters B, Nielsen M. 2020. Improved prediction of MHC II antigen presentation through integration and motif deconvolution of mass spectrometry MHC eluted ligand data. J Proteome Res. 19:2304–2315. [DOI] [PubMed] [Google Scholar]
- Sanchez-Mazas A. 2020. A review of HLA allele and SNP associations with highly prevalent infectious diseases in human populations. Swiss Med Wkly. 150:w20214. [DOI] [PubMed] [Google Scholar]
- Sanchez-Mazas A, Černý V, Di D, Buhler S, Podgorná E, Chevallier E, Brunet L, Weber S, Kervaire B, Testi M, et al. . 2017. The HLA-B landscape of Africa: signatures of pathogen-driven selection and molecular identification of candidate alleles to malaria protection. Mol Ecol. 26:6238–6252. [DOI] [PubMed] [Google Scholar]
- Satta Y, O’HUigin C, Takahata N, Klein J. 1994. Intensity of natural selection at the major histocompatibility complex loci. Proc Natl Acad Sci USA. 91:7184–7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spyrou MA, Tukhbatova RI, Feldman M, Drath J, Kacki S, de Heredia J B, Arnold S, Sitdikov AG, Castex D, Wahl J, et al. . 2016. Historical Y. pestis genomes reveal the European Black Death as the source of ancient and modern plague pandemics. Cell Host Microbe 19:874–881. [DOI] [PubMed] [Google Scholar]
- Trowsdale J, Knight JC. 2013. Major histocompatibility complex genomics and human disease. Annu Rev Genomics Hum Genet. 14:301–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium . 2021. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 49:D480–D489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2017. Plague key facts [Internet]. World Health Organization. [2021 Jun 20]. Available from: https://www.who.int/en/news-room/fact-sheets/detail/plague. [Google Scholar]
- Yasukochi Y, Satta Y. 2013. Current perspectives on the intensity of natural selection of MHC loci. Immunogenetics 65:479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this paper are available in Supplementary information, Supplementary Material online or on request to the authors.