Summary:
Cytokines are powerful immune modulators that initiate signaling through receptor dimerization, but natural cytokines have structural limitations as therapeutics. We present a strategy to discover cytokine surrogate agonists, using modular ligands that exploit induced proximity and receptor dimer geometry as pharmacological metrics amenable to high-throughput screening. Using VHH and scFv to human Interleukin-2/15, Type I Interferon, and Interleukin-10 receptors, we generated combinatorial matrices of single-chain bispecific ligands that exhibited diverse spectrums of functional activities, including potent inhibition of SARS-CoV-2 by surrogate Interferons. Crystal structures of IL-2R:VHH complexes revealed that variation in receptor dimer geometries led to the functional diversification. This modular platform enabled engineering of surrogate ligands that compelled assembly of an IL-2R/IL-10R heterodimer, which does not exist in nature, that signaled through pSTAT5 on T and NK cells. This “cytokine med-chem” approach, rooted in principles of induced proximity, is generalizable for discovery of diversified agonists for many ligand-receptor systems.
Keywords: Cytokine engineering, Interleukin-2, Interferon, Interleukin-10, bispecific antibodies, cell signaling
Graphical Abstract
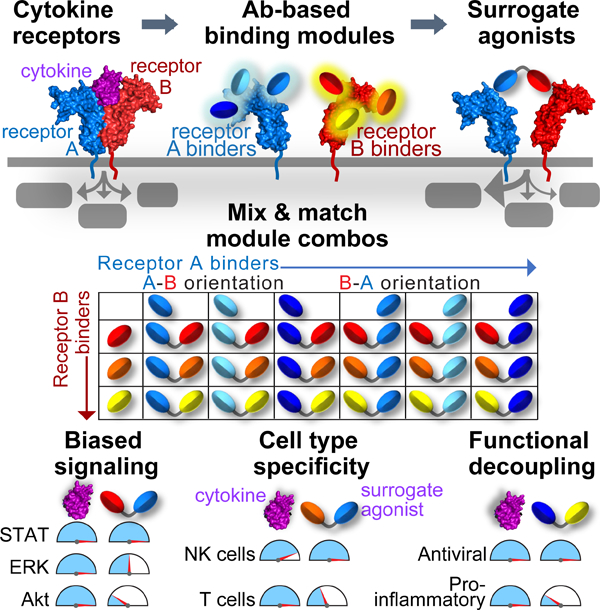
In brief:
A discovery platform for functionally diverse cytokine surrogates
Introduction:
Cytokines are garnering increasing interest as therapeutics. However, the process of therapeutic cytokine discovery is generally limited to exploring intrinsic biological properties of natural ligands, through modifications such as affinity maturation, half-life extension and/or tissue targeting (Berraondo et al., 2018; Mansurov et al., 2021; Overwijk et al., 2021). More recently, cytokine engineering strategies have succeeded in demonstrating that cytokine pleiotropy can be mitigated by selective structure-based engineering and protein design (Glassman et al., 2021b; Mendoza et al., 2019; Mitra et al., 2015; Saxton et al., 2021). However, unlike multi-pass transmembrane proteins such as GPCRs and ion channels that are responsive to small molecule agonists (Shoichet and Kobilka, 2012), cytokines are proteins that signal through Type I single-pass transmembrane receptors, which present immense challenges for small molecule agonist discovery. This is due to two principal reasons.
First, cytokines are globular proteins that function to dimerize their receptors via protein-protein interactions with their receptor extracellular domains (Spangler et al., 2015; Stroud and Wells, 2004). In contrast, small molecule agonists bind within pockets in GPCR and ion channel transmembrane helices to activate conformational changes. Thus, because of their structural properties, cytokine and growth factor Type I single-pass transmembrane receptors are generally unsuitable for small molecule library-based screening campaigns to discover agonists, with rare exceptions (Corman and Mohammad, 2010). Furthermore, cytokines themselves are single-domain four-helix bundle proteins that present structural limitations for ligand engineering (Silva et al., 2019), which is generally restricted to interface mutagenesis (Glassman et al., 2021a; Levin et al., 2012; Mitra et al., 2015).
Second, cytokine-mediated signaling is often assumed to be “on or off,” in contrast to graded, or rheostat-like GPCR (i.e. biased) signaling (Smith et al., 2018). However, recent studies have shown that strategic ligand affinity modulation, as well as the orientation and proximity of dimeric receptor assemblies can profoundly influence signaling output to exhibit rheostat-like behavior in a manner conceptually analogous to GPCRs (Glassman et al., 2021b; Mohan et al., 2019; Moraga et al., 2015; Saxton et al., 2021). Furthermore, antibodies to cytokine receptor ECDs can, in some instances, act as “surrogate” cytokine agonists by dimerizing the cytokine receptors into appropriate signaling geometries (Harris et al., 2021; Moraga et al., 2015; Zhang et al., 2013). Taken together, a class of surrogate ligands, with drug-like properties, that can induce varied Type I transmembrane receptor orientations and proximities, and is amenable to high-throughput screening, could bridge the gap between the power of medicinal chemistry and the limitations of cytokine engineering, which does not access the full scope of cytokine receptor signaling plasticity.
In the current study we utilized an induced proximity strategy on four pleiotropic immunoregulatory cytokines: Interleukin-2 and Interleukin-15, Type I Interferon, and Interleukin-10. Interleukin-2 (IL-2) is a stimulatory cytokine that directs proliferation and survival of T lymphocytes, natural killer (NK) cells, and B lymphocytes (Lin and Leonard, 2018). IL-2, like IL-15, signals through a receptor heterodimer composed of common gamma (γc) and IL-2Rβ which trigger signaling through JAK-STAT, MAP kinase/ERK, and PI3 kinase-Akt pathways (Leonard et al., 2019). IL-2 activates JAK1 and JAK3 kinases, which relay the signal primarily through STAT5 activity (Miyazaki et al., 1994; Russell et al., 1994; Xue et al., 2002). Type I Interferons (IFN) have a wide range of immunomodulatory, anti-viral, and anti-proliferative actions which are mediated by 16 different sub-types of IFN cytokines that dimerize IFNAR1/IFNAR2 to activate several STATs, principally STAT1 (Ng et al., 2016). Finally, IL-10 is an anti-inflammatory cytokine that dimerizes IL-10Rα and IL-10Rβ to elicit STAT1 and STAT3 activation (Ouyang and O’Garra, 2019).
We present an unbiased, structurally agonistic, and modular platform for surrogate cytokine discovery which samples the effects of varied receptor dimerization geometries as well as ligand-receptor affinities, on signaling and function of IL-2/15 receptors, Type I IFN receptors, and IL-10 receptors. This led to the discovery of a wide range of functionally diverse ligands, including 28 IL-2 “surrogate” agonists with differing patterns of STAT1/3/5, ERK, and PI3K signaling, preferential induction of memory T cell differentiation, and NK cell cytotoxicity relative to IL-2. IFN surrogate agonists showed biased induction of anti-viral genes and potent anti-viral activity against SARS-CoV-2. Finally, we demonstrate that surrogate ligands can compel heterodimerization of cytokine receptors for which a natural cytokine ligand does not exist. A surrogate bispecific ligand assembled a “non-natural” signaling entity between IL-2 and IL-10 receptors, resulting in a hybrid signaling entity on NK and CD8+ T cells. Overall, this approach exploits the intrinsic signaling and functional plasticity of cytokines using a scalable and modular strategy for agonist discovery that constitutes a form of “cytokine med-chem” applicable to many cellsurface receptor systems.
Results:
A platform for discovery of bispecific surrogate cytokine agonists
We devised a screening platform consisting of single-chain, bispecific ligands comprised of small antibody domains (VHH and/or scFv) that can be “mixed and matched” in modular fashion to create libraries of dimerizing ligands (Figures 1A and 1B). Using antibody-based binding domains offers the possibility of diverse epitope coverage of the target receptor ECDs, which will manifest as diverse dimerizing topologies in the context of bispecific ligands, in addition to their drug-like properties facilitating clinical translation.
Figure 1. Platform for generation and screening of bispecific IL-2Rβ/γC surrogate agonists.
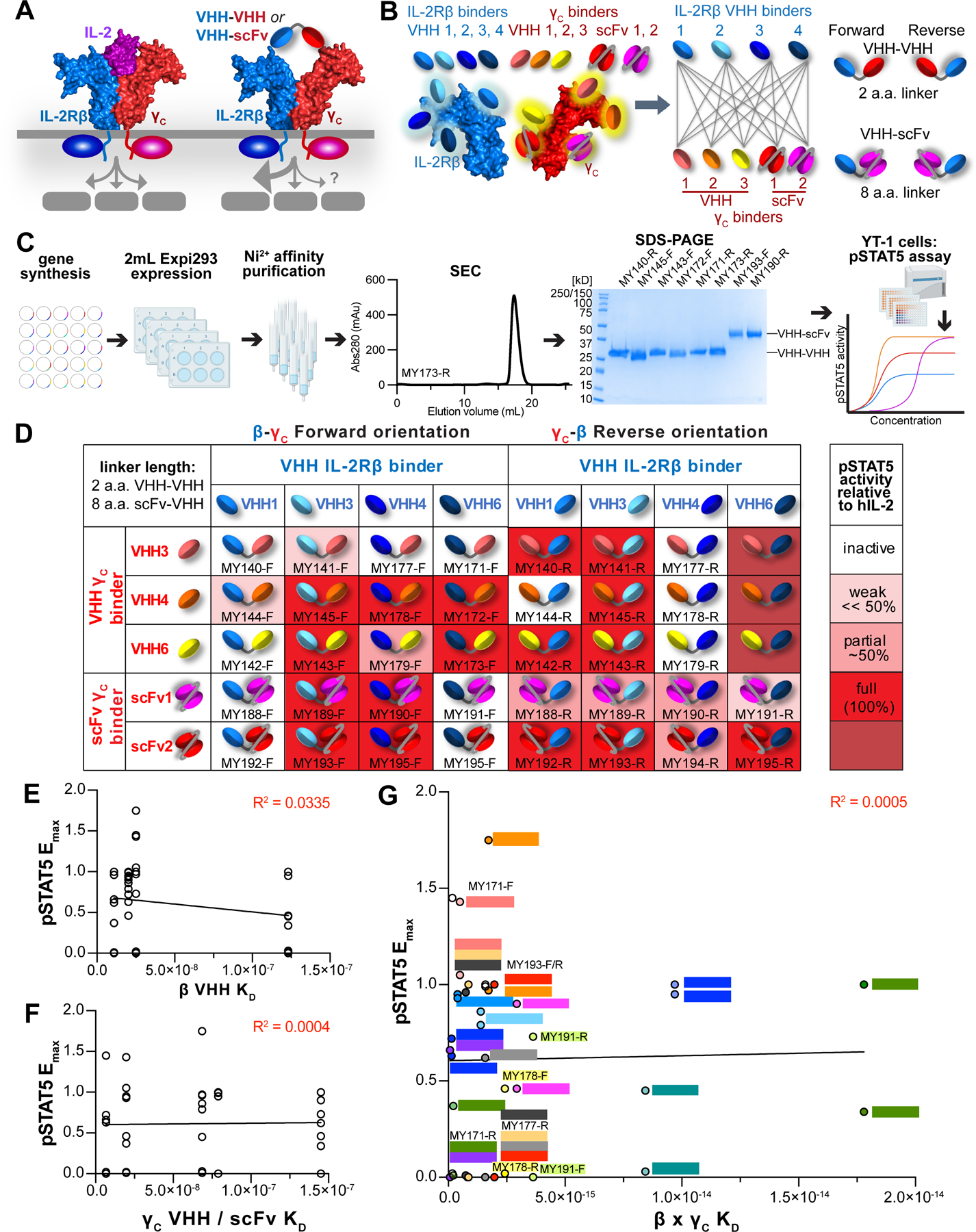
(A) Platform for discovery of surrogate IL-2 receptor agonists based on construction of bispecific VHH or scFv specific for IL-2Rβ and γC. (B) Schematic representation of VHH and scFv binding to diverse epitopes along the IL-2Rβ or γc extracellular domains (left), combinatorial matrix to generate a collection of β-γ dimerizing ligands (middle), and representation of VHH–VHH or VHH–scFv fusion constructs connected by short linkers in Forward or Reverse orientations (right). (C) Schematic pipeline for protein expression and activity screening of surrogate ligands. Bispecific VHH were produced by gene synthesis of VHH monomers and cloning, expressed at 2mL scale in Expi293 cells, and purified via their 6-His tags on Ni2+ affinity resin followed by size exclusion chromatography (SEC) and SDS-PAGE analysis. Protein activity was measured via a pSTAT5 phosphoflow assay on YT-1 cells. (D) Heatmap of pSTAT5 activity evoked by bispecific antibody pairings. YT-1 cells were stimulated with saturating ligand concentration for 20 min., fixed and permeabilized, then stained with α-STAT5(pY694)-AlexaFluor647 and analyzed via flow cytometry. (E) Affinity to IL-2Rβ does not predict STAT5 activity. Each circle represents a bispecific molecule, with pSTAT5 Emax (normalized to hIL-2) plotted against the affinity of its IL-2Rβspecific VHH. Data were fit by linear regression, with R2 = 0.0335. (F) Affinity to γC does not predict STAT5 activity. Each circle represents a bispecific molecule, with pSTAT5 Emax (normalized to hIL-2) plotted against the affinity of its γC-specific VHH or scFv. Data were fit by linear regression, with R2 = 0.0004. (G) Overall receptor binding affinity (IL-2Rβ × γC KD) is not predictive of STAT5 activity. Bispecific molecules with identical IL-2Rβ × γC antibody usage are depicted in the same color. Data were fit by linear regression, with R2 = 0.0005.
See also Figures S1 and S2A–S2B.
In the IL-2/15 system, we first generated a collection of small, single Ig-domain VHH binders against IL-2Rβ and γC selected from phage-displayed libraries of target-immunized Bactrian camels (Figures S1A, S1B, S1C, and S1D). ELISA-based screening of recombinantly expressed VHH clones identified 65 IL-2Rβ binders and 50 γC binders. Based on their CDR3 sequence diversity, 10 IL-2Rβ clones from the 4 VHH classes and 6 γC clones were selected for further evaluation. The binning into distinct VHH classes was to select for diverse epitope coverage on the receptor ECDs.
We assessed the ability of the isolated VHHs to bind to YT-1 cells, a human NK cell line which endogenously expresses IL-2Rβ and γC (Figures S1E and S1F). Four IL-2Rβspecific VHH clones (β-VHH1, 3, 4, and 6) were chosen for SPR analysis, and bound to IL2Rβ with steady-state affinities ranging from ~10–125 nM (Figure S2A). VHH against γC were also used for cell binding studies on YT-1 cells (Figure S1G), and SPR experiments yielded affinities ranging from ~7–70 nM (Figure S2B). We selected four IL-2Rβ binders (βVHH1, 3, 4, and 6), three γC clones (γC-VHH3, 4, and 6), along with two γC scFv clones (P1A3, P2B9) whose sequences we identified in a patent (Figure S1H) (Wang et al., 2016).
We generated bispecific molecules by fusing IL-2Rβ and γC binders through short, flexible Gly-Ser linkers (8 a.a. for VHH–scFv fusions and 2 a.a. for VHH–VHH fusions) in both Forward and Reverse orientations (Figure 1B). In total, the “all by all” matrix of 4 IL2Rβ binders × 5 γC binders × 2 orientations resulted in 40 molecules. These small protein constructs (~23kDa-40kDa) were rapidly produced by gene synthesis, expressed through transient transfection of approximately 2mL of Expi293 cells and purified via their 6-His tags using small Ni2+-agarose columns (Figure 1C).
The surrogate dimerizing ligands were then rapidly screened in parallel for induction of STAT5 phosphorylation in YT-1 cells (Figures 1D, S2C, and S2D). Of the 40 candidates, we found 28 agonists (~70% “hit rate”), spanning from minimally active (3 ligands), ~1/2 Emax relative to hIL-2 (5 ligands), full Emax (17 ligands), and supraphysiologic Emax (3 ligands) (Figures 1D and S2E). Notably, amongst the agonists, there was no discernable relationship between a ligand’s Emax and its affinity for individual IL-2Rβ or γC receptors (Figures 1E and 1F). This finding highlights the influence of VHH binding epitope on the receptor ECDs, which influences the overall geometry of the signaling dimer, in addition to ligand-receptor affinity, in determining signaling output. There was also a lack of correlation between Emax and the product of receptor binding affinities β × γC (Figure 1G). Even amongst ligand pairs with identical β and γC VHH, alternative (Forward or Reverse) orientations (β–γC vs. γC–β), elicited divergent activities in 15 of 20 total pairings (Figure 1G).
Profiling signaling properties of IL-2 surrogate ligands
We next examined the principal membrane-proximal outputs of IL-2 signaling: activation of pSTAT5, pERK, and PI3K/pAkt. For kinetics studies, YT-1 cells were stimulated with saturating concentration of hIL-2 or surrogate ligands for varying amounts of time (Figure 2A). For dose-response studies, we varied the concentration of ligand and measured phosphorylation at a fixed time point (3min.) corresponding to peak pERK and pAkt signal levels for hIL-2 (Figures S3A, S3C, and S3E). Surrogate agonists elicited a range of behaviors: some similar to IL-2, while others showed delayed activation and reduced peak responses (MY189-F, MY190-F), and impaired activation of some, but not all pathways (MY178-F, MY179-F) (Figures 2A–2B and S3B–S3E). Grouping ligands according to their relative strengths of pSTAT5, pERK, and pAkt signaling (relative to hIL-2) revealed distinct classes of signal patterns (Figure 2C). Across all ligands, pSTAT5 appeared to be preferentially activated, trailed by pERK and then pAkt: pSTAT5 activity ≥ pERK ≥ pAkt.
Figure 2. Profiling signaling properties of IL-2 surrogate ligands.
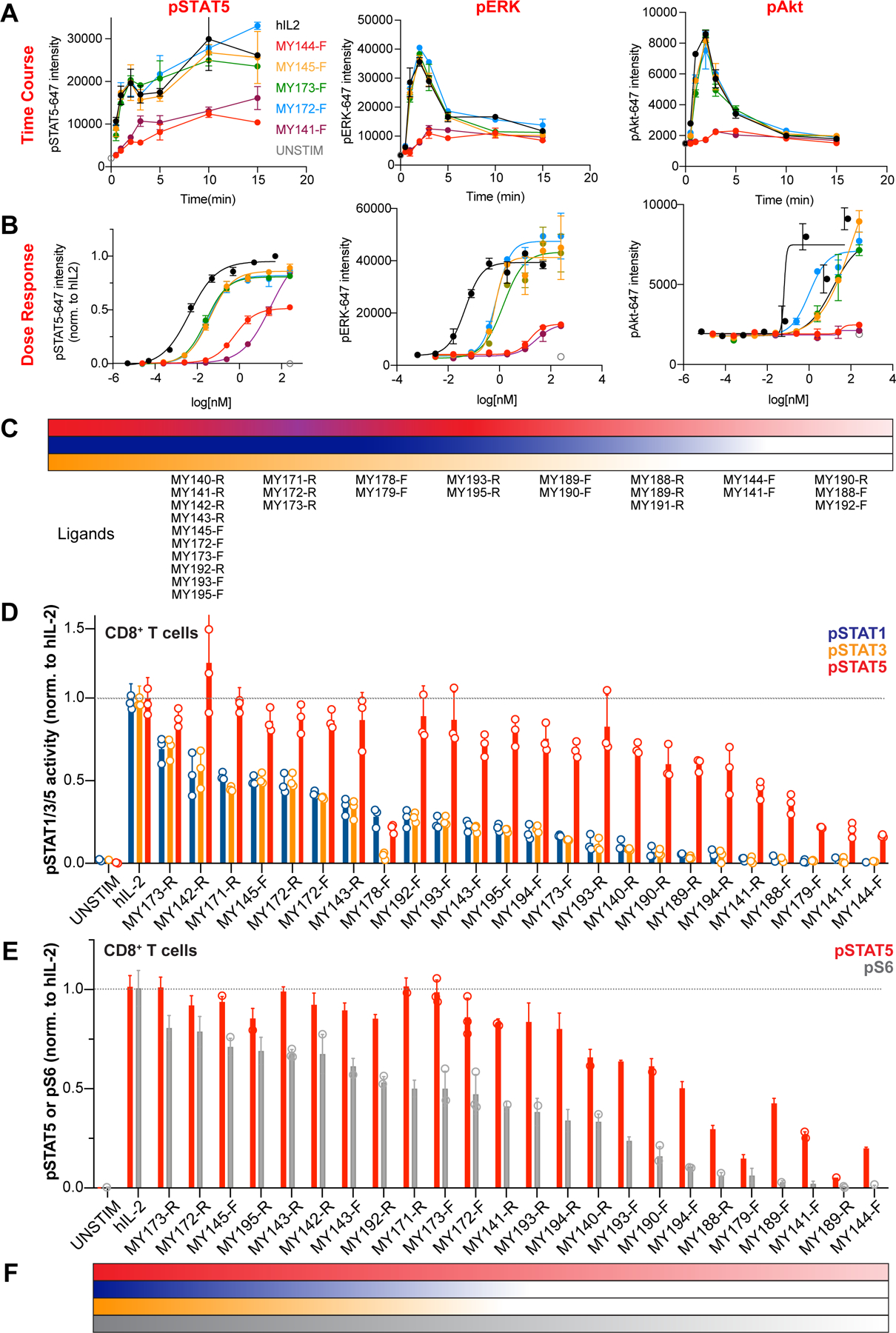
(A) Kinetics of pSTAT5, pERK, and pAkt signaling evoked by IL-2 or surrogate agonists. YT1 cells were serum-starved for 1–2hr., then stimulated with 50nM ligand for 0.5–15min. at 37°C, fixed and permeabilized, then stained with fluorescently-conjugated phospho-antibodies before reading on a flow cytometer. (B) Dose-response relationship of pSTAT5, pERK, and pAkt activity evoked by IL-2 or surrogate agonists. Serum-starved YT-1 cells were stimulated with varying concentration of ligand for 3min., then processed as in (A) for phosphoflow analysis. (C) Classification of signal strength for IL-2 surrogate agonists, with relative strength of activity encoded by colored gradients. (D) T cell blasts were stimulated with 50nM hIL-2 or surrogate agonist for 20min. at 37°C, fixed and permeabilized, then stained with fluorescently conjugated antibodies against pSTAT1, pSTAT3, or pSTAT5 and read on a flow cytometer. Raw fluorescence intensities were background subtracted against that of unstimulated cells, then normalized to hIL-2 values. (E) T cell blasts were stimulated with 50nM hIL-2 or surrogate agonist for 1hr. at 37°C. Cells were fixed and permeabilized, then stained with fluorescently-conjugated antibodies against pSTAT5 and pS6, and read on a flow cytometer. Data are baseline subtracted and normalized as in (D). (F) Summary of ligand signaling properties across pSTAT1/3/5 and pAkt pathways, with relative strength of activity encoded by colored gradients. Data were collected in triplicate, with graphs displaying the mean ± sem.
See also Figures S2 and S3.
IL-2 and IL-15 principally activate STAT5 but have also been shown to induce STAT1 and STAT3 activity (Delespine-Carmagnat et al., 2000; Ng and Cantrell, 1997), which are required for efficient maintenance of CD8+ memory T cells (Cui et al., 2011; Quigley et al., 2008; Siegel et al., 2011). We measured STAT1, 3, and 5 phosphorylation after stimulation by IL-2 analogs (Figures S3F and S3G). In pre-activated primary T and NK cells (Figures 2D, S3F, and S3G), ligand MY173-R was similar to IL-2 in its pSTAT1/3/5 balance. However, the remaining surrogate agonists favored dominant pSTAT5 signaling over pSTAT1 and pSTAT3. For example, in CD8+ T cells, MY193-R displayed ~90% pSTAT5 activity, but only ~25% pSTAT1 or pSTAT3 activity relative to hIL-2. We also found that in T cells, many surrogate ligands exhibited higher pSTAT5 activity relative to pS6, a substrate downstream of PI3K/Akt signaling (Ross and Cantrell, 2018) (Figure 2E). In general, the biased pSTAT5 vs. pS6 ratio was not as pronounced as in the pSTAT5 vs. pSTAT1/3 ratio. In addition to MY173-R (the only ligand with a balanced pSTAT1/3/5 ratio), MY172-R, MY145-F, and MY195-F stimulated balanced levels of pSTAT5 and pS6 phosphorylation in T cells, whereas the remaining ligands favored pSTAT5 activity over pS6.
Surrogate IL-2 ligands form alternative dimeric receptor geometries
To gain insight into the structural basis for signaling differences, we crystallized two different VHHs, one bound to IL-2Rβ and the other bound to γC. With structures of the individual receptor:VHH complexes in hand we generated approximate models of the complete dimeric receptor geometry since the short (2 a.a.) linker places constraints on the relative overall geometry of the two VHHs in the bispecific ligand. The IL-2Rβ:β-VHH6 complex was resolved at 1.9Å and revealed that the β-VHH6 binds to the D1 domain of the receptor, as opposed to binding at the “elbow” of the D1-D2 juncture like IL-2 (Wang et al., 2005) (Figures 3A and Table S1). We resolved the γC:γC-VHH6 complex to 2.6Å and found that the γC-VHH6 occupied a similar binding footprint on γC as IL-2 (Figures 3B and Table S1). We modeled the two (forward and reverse) orientations of ligands, β-VHH6—γC-VHH6 and γC-VHH6—β-VHH6. The two complexes predict significant differences from the IL-2 receptor heterodimer geometry in both distance and angular relationship between IL-2Rβ and γC (Figures 3C, 3D, and 3E). Surprisingly, we observed that in YT-1 cells γC-VHH6—βVHH6 (MY173-R) induced supraphysiologic STAT5 phosphorylation but normal ERK/Akt phosphorylation relative to hIL-2 (Figures 1D and 2C). The reverse orientation analog, βVHH6—γC-VHH6 (MY173-F), had a similar signaling profile to IL-2 in YT-1 cells. Whereas MY173-R had a signaling profile like IL-2 in CD8+ T and NK cells, MY173-F had high pSTAT5 activity (~70–100% of IL-2), and low pS6 and pSTAT1/3 activity (50% and ~14–24%, respectively). These structures support our hypothesis that differences in receptor dimerization geometry strongly influence proximal signaling.
Figure 3. Dimeric geometries from crystal structures of IL-2Rβ:VHH and γc:VHH complexes.
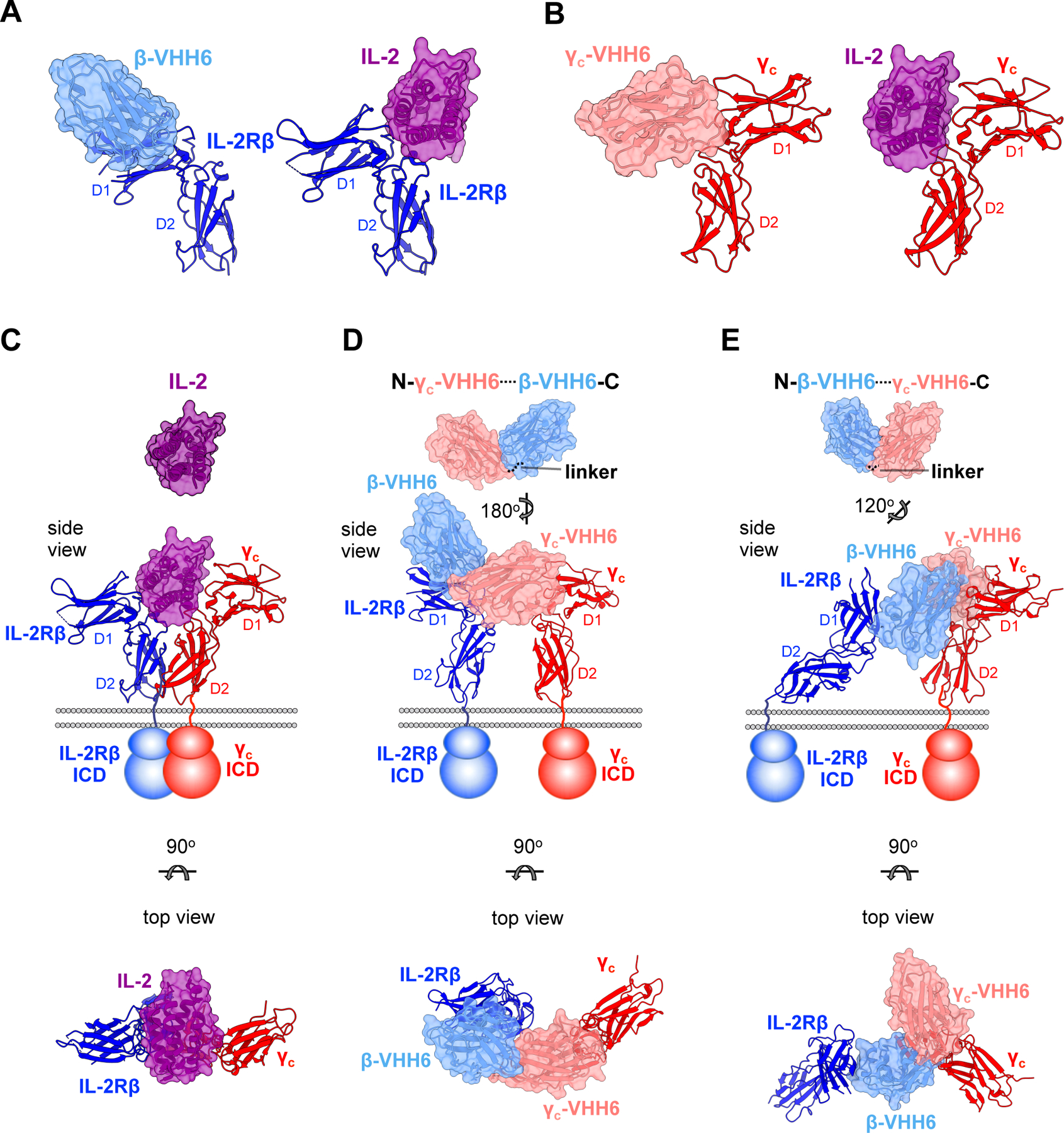
(A) Side view comparisons between the human IL-2:IL-2Rβ binary complex (PDB: 2B5I) (Wang et al., 2005) and β-VHH6:IL-2Rβ binary complex. Surface representations of IL-2 and β-VHH6 are colored in purple and light blue, respectively, while IL-2Rβ is shown in ribbon representation in navy. (B) Side view comparisons of IL-2:γC and γC-VHH6:γC receptor complexes. γC-VHH6 is shown in pink, with γC colored in red (C) Crystal structure of the human IL-2:IL-2Rβ:γC ternary complex (PDB: 2B5I) (Wang et al., 2005). Side view with membrane bilayer and schematic representation of receptor transmembrane and intracellular domains (ICD) is shown at middle. Top view (below) is related to the side view by a 90° rotation about the horizontal axis. (D) Model of γC-VHH6–β-VHH6 bound to its receptors. Structures of the γC-VHH6:γC and IL-2Rβ:β-VHH6 were determined separately. The γC-VHH6–β-VHH6 linker distance was modeled in and represented by a dotted line (top), with a side and top views of receptor-bound model shown underneath. (E) Model of β-VHH6–γC-VHH6 bound to its receptors. The VHH–VHH linker was modeled as in (D).
See also Table S1.
Transcriptional profiling of IL-2 surrogate agonists
We next performed mRNA sequencing to characterize the transcriptional profiles induced by different signaling classes of IL-2 surrogate ligands on T cells. Principal component analysis (PCA) indicated that the IL-2 analogs had a concerted effect on gene expression across the PC1 axis (Figure 4A). Within each donor, ligands MY173-R, MY143-R, MY145-F, and MY172-F clustered together with IL-2 and IL-15 (which are known to drive highly similar gene expression profiles (Ring et al., 2012)); MY141-F, MY188-R, and MY179-F clustered near the unstimulated sample, whereas MY190-R and MY193-R were close to IL-7, which is required for naïve and memory T cell homeostasis and opposes terminal effector T cell differentiation (Figure 4A) (Shourian et al., 2019). These relationships were also preserved on a group level, as seen in the distance calculation matrix (Figure 4B). Overall, a given ligand’s pSTAT5 activity (on CD8+ T cells) was linearly correlated with its potency at regulating gene expression (R2 = 0.82, Figure 4C). We identified 96 STAT5 target genes from a curated gene list (mSigDb) (Liberzon et al., 2015; Subramanian et al., 2005) whose expression was significantly altered by treatment with cytokines or IL-2 analogs (padj < 0.05), including targets known to be highly upregulated by IL-2 (SOCS1, CISH, BCL2, IL2RA). Overall, the IL-2 surrogates regulated STAT5 targets in a graded manner, with strong ligands (MY173-R, MY145-F, MY143-R, MY172-F) having a similar profile to IL-2 and IL-15, weak ligands (MY141-F, MY179-F, MY188-R) mildly upregulating robust STAT5 targets such as SOCS1, CISH, SELL, and having no effect on other targets, and IL-7/MY193-R/MY190-R having an intermediate effect on gene regulation (Figure 4D).
Figure 4. Transcriptional profiling of IL-2 surrogate agonists.
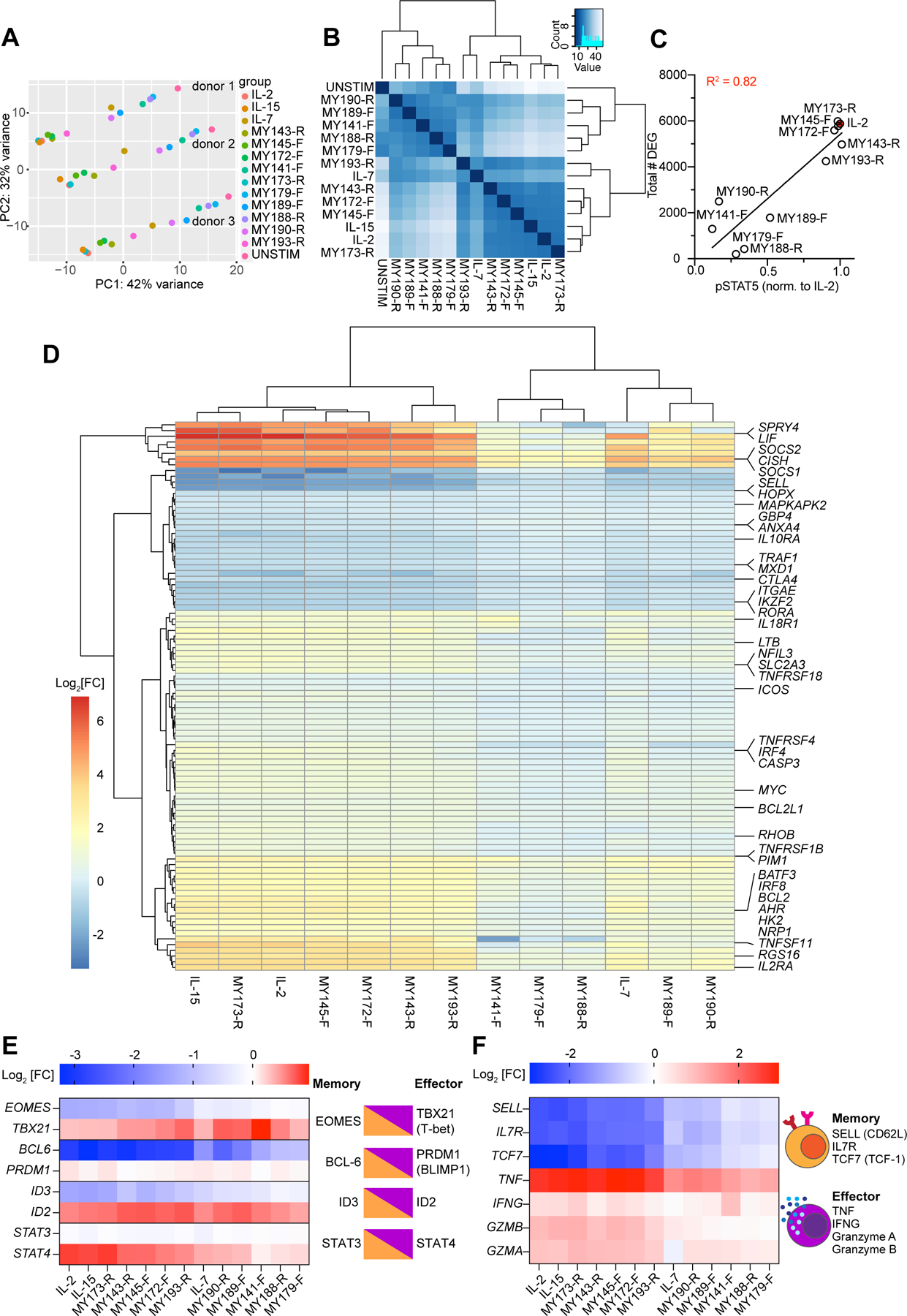
(A) Principal component analysis (PCA) of gene expression in CD8+ T cells from 3 donors stimulated with IL-2 or surrogate ligands for 24 hours. Samples from a given donor lie along a horizontal line, with unstimulated samples at the right and IL-2/IL-15 treated samples at the left. The effect of various ligand stimulations is largely described by PC1. (B) Euclidean distance matrix showing overall similarities in mRNA expression between treatment conditions. Samples within the same treatment condition were pooled together. Dendrograms depict the results of unsupervised hierarchical clustering between samples, with the branch length being proportional to the distance between samples. (C) Relationship between surrogate ligand pSTAT5 activity and the total number of differentially expressed genes (DEG) induced by ligand stimulation. STAT5 phosphorylation was normalized to that of hIL-2 stimulated cells. (D) Hierarchical clustering of “Hallmark” STAT5 targets (curated by mSigDB) (Liberzon et al., 2015; Subramanian et al., 2005) whose expression was significantly altered by treatment cytokine or surrogate agonist treatment (padj < 0.05). Differential gene expression is represented as the log2 fold-change (Log2[FC]) of normalized mRNA counts. Genes are arrayed by row and ligands by column. (E) Log2 fold expression change of transcription factors which play opposing roles in CD8+ memory vs. effector differentiation. Opposing transcription factor pairs are diagrammed (right) with the accompanying log2 fold changes induced by surrogate ligands (left). (F) Log2 fold expression change of selected markers of memory and effector T cells (left). Memory T cells express CD62L (encoded by SELL), IL7 receptor, and the transcription factor TCF1 (encoded by TCF7), whereas effector CD8+ T cells produce abundant amounts of cytokines TNFα and IFNγ and cytolytic molecules such as granzymes A and B (right).
An important function of IL-2 is to induce CD8+ T cell differentiation, so we examined expression of pairs of transcription factors that exert opposing effects on Tmemory vs. Teffector differentiation (Figure 4E) (Kaech and Cui, 2012). Four pairs of transcription factors, EOMES/TBX21, BCL-6/PRDM1, ID3/ID2, and STAT3/STAT4, regulate the balance of memory vs. effector potential based on their relative expression ratios and/or activities (Kaech and Cui, 2012). We found that along with IL-2/IL-15, ligands MY173-R, MY143-R, MY145-F, and MY172-F downregulated EOMES but not TBX21 and BCL6 and not PRDM1, while upregulating ID2 but not ID3, and STAT4 but not STAT3 expression. Taken together these four sets of ratios favor differentiation toward effector over memory cells (Kaech and Cui, 2012). Consistent with this, the same set of ligands downregulated expression of markers of naïve and central memory cells (such as SELL, IL7R, and TCF7) while upregulating expression of genes encoding the effector cytokines and cytolytic molecules TNFα, IFNγ, granzyme A, and granzyme B (Figure 4F) (Kaech and Cui, 2012).
IL-2 surrogate agonists support T and NK cell proliferation and cytolysis
One of the principal roles of IL-2 is to direct the differentiation of naïve CD8+ T cells into memory and cytotoxic effector cells, thus we probed the ability of our ligands to orchestrate development of naïve (Tn), central memory (TCM), effector memory (TEM), and more terminally differentiated effector memory CD45RA (TEMRA) T cells (Maecker et al., 2012) (Figures S4A and S4B). Ligands spanned a broad range of differentiation potential, ranging from IL-2-like, to central and effector memory biased, entirely CD8 selective, or nonfunctional despite triggering pSTAT5 signaling (Figures 5A, 5B, and 5D).
Figure 5. IL-2 surrogate agonists support T and NK cell proliferation and cytolysis.
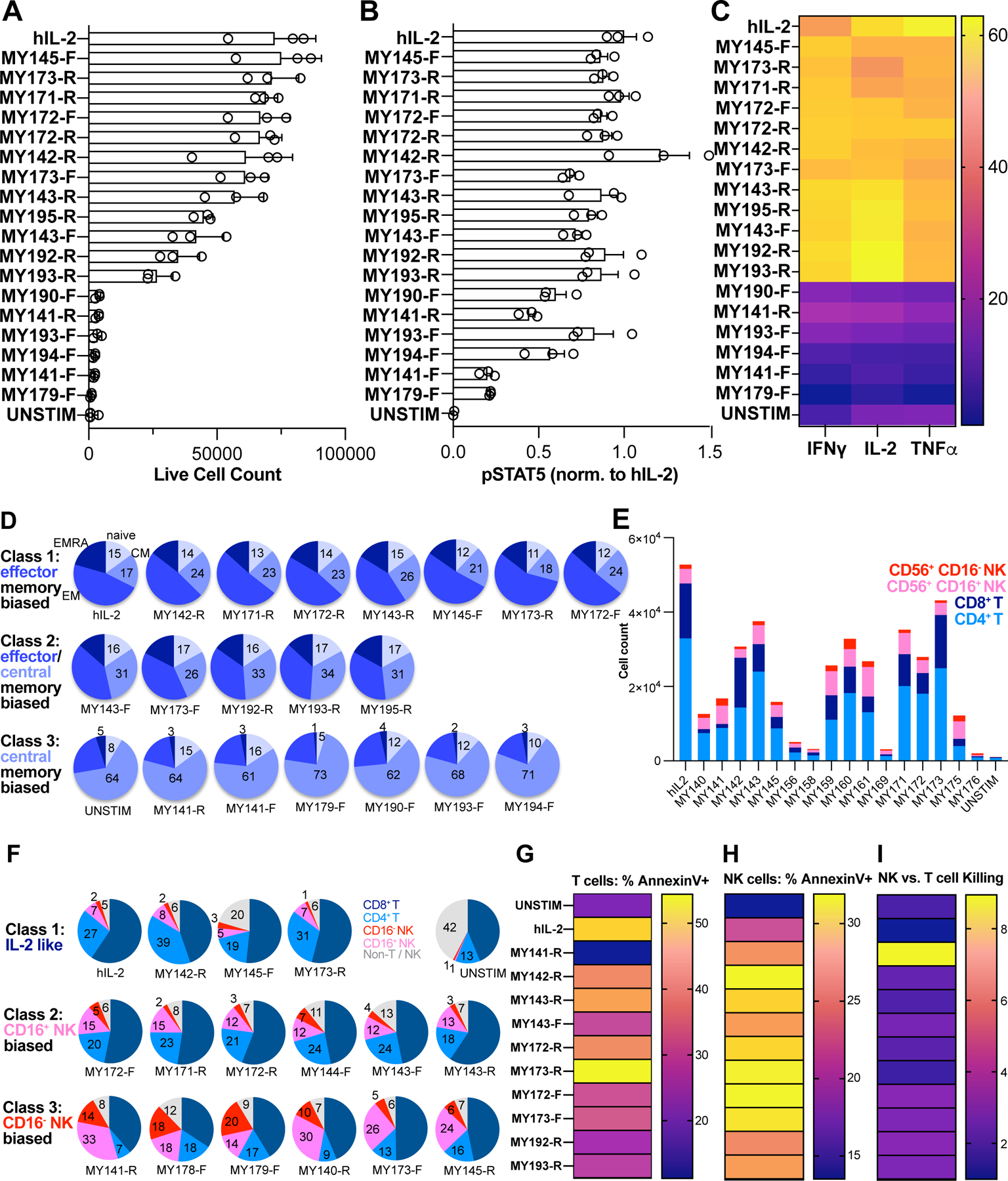
Naïve T cells were isolated from PBMC by negative magnetic selection, preactivated for 4d with surface-bound α-CD3 + soluble α-CD28, then cultured in the presence of 100nM hIL-2 or surrogate agonist for 8d. (A) Total T cell count after differentiation in the presence of indicated ligand. Wells were set up in triplicate. (B) pSTAT5 activity in preactivated T cells, normalized to that of hIL-2. (C) Cytokine profiling of CD8+ T cells was performed by stimulating cells with PMA + ionomycin in the presence of brefeldin A and monensin, followed by intracellular staining to assess IFNγ, IL-2, and TNFα production. Data represent an average of 3 replicate wells and are colored by heat map encoding the percentage of CD8+ cells expressing the indicated cytokine. (D) Cells were stained with surface antibodies against CD4, CD8, CCR7, and CD45RA to enumerate differentiation into T cell memory subtypes. The fraction of naïve, central memory, effector memory, and TEMRA cells are represented using pie charts. (E) PBMC were cultured for 2 weeks in the presence of 100nM hIL-2 or surrogate agonists, then stained with phenotyping markers for T and NK cells and enumerated using flow cytometry. The graph displays absolute live cell counts of CD8+ T, CD4+ T, CD16+ NK, and CD16−NK cells. (F) Pie charts of cell count data from (E) depict the fraction of T and NK cell types. (G) T cell cytolytic activity stimulated by culture with hIL-2 or surrogate agonists. Pre-activated human T cells were lentivirally transduced with A3A TCR and cultured for 10d in the presence of 100nM hIL-2 or IL-2 surrogate agonists to generate CTLs. Cytotoxicity was measured by mixing effector T cells with a fixed number of CTV-labeled A375 melanoma target cells for 4–6hr., then assessing apoptosis via annexin V staining. (H) NK cytolytic activity stimulated by culture with hIL-2 or surrogate agonists. Pre-activated NK cells were cultured for 4 weeks in the presence of 100nM hIL-2 or surrogate agonists and mixed with 25,000 CTV-labeled K562 target cells per well. Following 5hr. incubation, cells were stained with annexin V-PE, then analyzed for early apoptosis using flow cytometry. (I) Relative efficiency of NK vs. T cell cytolysis supported by surrogate IL-2 ligands. Annexin V positivity rates were normalized to hIL-2 in NK cells (H) or T cells (G) cultured with surrogate ligands, then ratioed and represented as a heat map.
See also Figures S4 and S5.
We also profiled expression of cytokines important for cytolytic function (Figures 5C, S4C, and S4D). Ligands that supported T cell expansion were tightly correlated with acquisition of proliferative and cytotoxic cytokine production (IL-2, TNFα, IFNγ). MY173-R supported equivalent T cell proliferation to IL-2, and the resultant cells had a CD8+ memory distribution phenotype (Figures 5A and 5D, top row). However, relative to hIL-2, a higher proportion of MY173-R-cultured cells produced IFNγ, with a lower proportion making IL-2 and TNFα (Figure 5C). Another differentiation phenotype is represented by MY173-F and MY193-R. These ligands evoked relatively high levels of pSTAT5 activity (~70–90% of IL-2) but had lower levels of pSTAT1/3 activity (<30% of hIL-2; Figure 2D) and promoted lower levels of proliferation as compared to hIL-2 (Figure 5D, middle row). Relative to IL-2 treatment, MY173-F and MY193-R drove higher proportions of central memory cells in addition to supporting effector memory differentiation, while inducing less TEMRA cells. A higher fraction of MY193-R treated cells produced IL-2 with a lower percentage of TNFα producers, consistent with a central memory phenotype (Figure 5C). A third category of ligands, which includes MY141-F, were strongly central memory dominant (Figure 5D, bottom row).
One clear IL-2 dependent functional readout is cytolysis of target cells. To test this we used preactivated human T cells transduced with the A3A T cell receptor (TCR), which recognizes the MAGE-3A peptide presented by HLA-A*01 on A375 melanoma cells (Cameron et al., 2013; Linette et al., 2013). The IL-2 surrogate ligands supported T cell cytotoxic function to varying degrees, largely matching their ability to support CD8+ T cell proliferation and generate effector cytokines (Figure 5G).
IL-2 and IL-15 are also known to support NK cell expansion and arm them with cytotoxic function (Wu et al., 2017). To assess proliferation, we expanded PBMCs with 100nM hIL-2 or IL-2 surrogates for 14d, then profiled T and NK cell types using surface antibody staining (Figures 5E and S5A). Culture with IL-2 supported the highest level of total cell expansion, while 12 of the surrogate agonists increased the proportion of NK cells in the population relative to IL-2-treated cells (Figure 5F), indicating an NK bias. Ligands which supported 24–50% of total cell number produced an expanded fraction of CD16+ NK cells (~3–5 fold increased relative to IL-2), which marks cytolytic NK cells (Cooper et al., 2001). Ligands MY141-R and MY178-F produced 7- and 9-fold expanded fractions of CD16−NK cells, which are thought to be specialized for cytokine production (Cooper et al., 2001).
We also directly measured the cytotoxic capacity of NK cells cultured with IL-2 or analogs. Surprisingly, a subset of IL-2 surrogate agonists including MY173-R and MY173-F supported annexin positivity rates that were ~23–45% higher than that produced by IL-2 or IL-15 (Figures 5H, S5B–S5D). The NK vs. T cell bias in cell expansion and survival (Figure 5F) suggested that the surrogate ligands might also preferentially drive the ability of NK cells to acquire cytotoxicity over that of T cells. To measure this, we normalized cytotoxicity values for surrogate ligands to that of hIL-2 in NK cells and in T cells, then plotted the normalized NK-to-T cell killing ratio (Figure 5I). Our IL-2 analogs all exhibited bias toward NK-mediated killing (ratio > 1), from slight NK bias (MY173-R, ratio = ~1.4) to moderate (most ligands, including MY173-F, ratio ~2–3) to highly NK selective (MY141-R, ratio ~9).
Type I Interferon surrogate agonists exhibit biased signaling and inhibit viral replication
We applied a similar strategy to create surrogate agonists in the Type I interferon system using a collection of VHH and scFv binders to human IFNAR1 and IFNAR2, fused via 2 a.a. or 5 a.a linkers (Figures 6A, S6A, and S6B). A subset of binders were selected for SPR analysis, and bound to their corresponding receptor with KD < 1nM (Figures S6A and S6B). Despite their high affinities, an initial 60-member screening matrix (10 IFNAR1 binders × 6 IFNAR2 binders) in the IFNAR1-IFNAR2 orientation produced only 12 active hits (20% hit rate, Figure 6A). These 12 hits were then expressed in the reverse (IFNAR2-IFNAR1) orientation and screened, and all of them were inactive. A subset of active molecules, which we term “Human Interferon Surrogates” (HIS) 1–7, were selected for further studies (Figure 6A). The HIS ligands induced dose-dependent pSTAT1 activation, the hallmark STAT activated by Type I IFNs, on YT-1 and A549 (lung epithelial) cell lines, as well as on human PBMCs, exhibiting partial agonist Emax relative to the natural cytokine human IFNω (Figures 6B, 6C, and 6D). Since Type I IFNs also activate additional STATs, we profiled STAT1-STAT6 phosphorylation. We observed reduced pSTAT1 activation relative to IFNω but equivalent pSTAT2 and pSTAT3 activation on both on the NK cell line YT-1 and A549 cells (Figures 6E–6F and S6C–S6D). Thus, the surrogate IFN ligands display signaling bias for pSTAT activation relative to human IFNω.
Figure 6. Type I Interferon surrogate agonists exhibit biased signaling and inhibit viral replication.
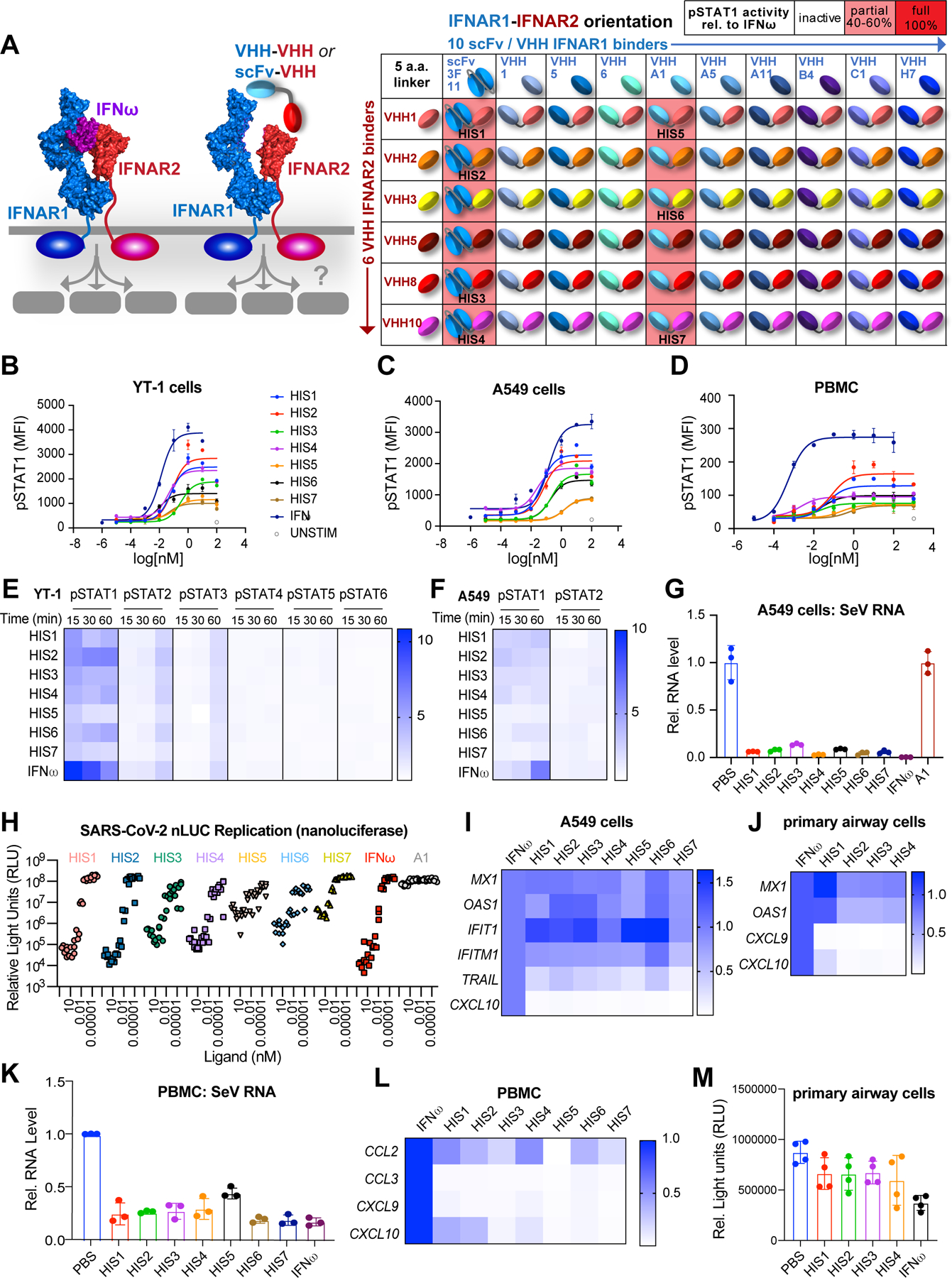
(A) Schematic representation of bispecific type I IFN surrogate ligands which heterodimerize IFNAR1 and IFNAR2 (left). A collection of 11 IFNAR1 binders (1 scFv, 10 VHH) were paired with 6 IFNAR2 binders (VHH), resulting in 66 combinations of IFNAR1-IFNAR2 fusion molecules connected via a 5 a.a. linker (right). Twelve of these molecules induced pSTAT1 activity on YT-1 cells (pink shading). The IFNAR2-specific scFv “3F11” was identified from the patent US7662381B2 (Cardarelli et al., 2010). Seven of the hits, “HIS1–7,” were selected for further analysis. (B-D) Dose-response relationship of STAT1 phosphorylation evoked by IFNω or surrogate agonists. YT-1 cells (B), A549 cells (C), or PBMCs (D) were stimulated with saturating ligand concentration for 20 min., fixed and permeabilized, then stained with α-STAT1(pY701)-AlexaFluor647 and analyzed via flow cytometry. (E) Heatmap representation of STAT1-STAT6 phosphorylation evoked by surrogate agonists in YT-1 cells at different time points and normalized to the activation induced by IFNω. (F) Heatmap representation of STAT1 and STAT2 phosphorylation evoked by HIS in A549 cells at varying time points, normalized to activation induced by IFNω. (G) qRT-PCR analysis of SeV RNA in A549 cells pre-treated with 10nM HIS or IFNω for 24hr. followed by SeV infection (MOI=0.1) for 24hr. (H) SARS-CoV-2 nLUC A549-hACE2 Antiviral Assay. A549-hACE2 cells were treated with varying concentration of HIS, IFNω, or negative control (monomer VHH “A1”) for 24hr. prior to infection with SARS-CoV2 nLUC. SARS-CoV-2 nLUC replication (relative light units) for triplicate wells per VHH dilution is shown. (I-J) Heatmap representation of selected ISGs induced by HIS in A549 cells (I) or human primary bronchial/tracheal epithelial cells (J). Gene expression is normalized to the level induced by IFNω. (K) qRT-PCR analysis of SeV RNA in PBMCs pre-treated with 10nM HIS or IFNω for 24hr. followed by SeV infection (MOI=0.5) for 24hr. (L) Heatmap representation of selected ISGs induced by HIS in PBMCs. (M) CellTiter-GLO assay of human primary bronchial/tracheal epithelial cells treated with 10nM HIS or IFNω for 72hr.
See also Figure S6.
Type I IFNs are a critical viral defense mechanism, so we asked whether the surrogate ligands exhibited antiviral activity on A549 cells infected with Sendai virus (SeV). All surrogate ligands showed similar inhibition of SeV replication as IFNω, despite their reduced pSTAT1 activation (Figure 6G). The surrogate ligands also inhibited SARS-CoV-2 replication in A549 cells expressing human ACE2 receptor, as measured with an antiviral assay using recombinant SARS-CoV-2 engineered to express nanoluciferase (Hou et al., 2020a) (Figure 6H). After 24hr. pretreatment, we observed a potent dose-dependent antiviral effect on SARS-CoV-2 replication. Interestingly, the antiviral potency of HIS agonists varied based on the identity of the IFNAR1 binder. Whereas all 4 ligands using the “3F11” scFv (HIS1–4) exhibited potent antiviral activity, 2/3 ligands with the “A1” VHH (HIS5–7) had poor activity (Figures 6A and 6H). Three HIS ligands were further examined for their ability to inhibit SARS-CoV-2 (Washington 1 strain) replication in primary human airway cells (Figure S6E) (Fulcher et al., 2005; Hou et al., 2020b). HIS2 significantly reduced SARS-CoV-2 titers at 100pM, and HIS7 reduced viral titers even at 0.01pM (Figure S6E), indicating that a subset of HIS ligands act as potent antiviral agents on primary cells.
Type I IFNs exhibit antiviral ability by inducing interferon stimulated genes (ISGs), and we observed biased induction of ISGs by the surrogate IFN ligands compared with IFNω. Specifically, HIS ligands maintained high levels of antiviral gene expression but induced lower levels of pro-inflammatory and pro-apoptotic gene expression (Figures 6I and S6F). In human primary airway epithelial cells, HIS agonists induced high levels of the antiviral genes MX1 and OAS1 with minimal induction of pro-inflammatory genes CXCL9 and CXCL10 (Figures 6J and S6G). Moreover, our agonists effectively inhibited SeV replication in PBMCs while barely inducing pro-inflammatory cytokine expression (Figures 6K, 6L, and S6H). Another functional property of type I IFNs is anti-proliferative activity, and we observed less pro-apoptotic gene induction by the HIS agonists. Consistent with this ISG bias, the HIS did not suppress cell proliferation as much as IFNω in primary airway epithelial cells (Figure 6M). Taken together, these ligands have biased ISG induction, resulting in preserved antiviral activity but restrained anti-proliferative and pro-inflammatory effects. Collectively, these data demonstrate that surrogate IFN agonists are exquisitely potent antiviral agents against SARS-CoV-2 and could be further explored as potential medical countermeasures for COVID-19, as well as for other viruses.
Induced proximity between IL-2Rβ and IL-10Rβ activates pSTAT5 signaling in T and NK cells
The availability of a large collection of VHH and scFv binders to cytokine receptor ECDs enabled us to reach beyond natural pairings of cytokine receptors that are driven by endogenously expressed cytokines, to explore induced proximity of “unnatural” cytokine receptor heterodimer pairs that might elicit non-canonical types of signals. Since IL-2Rβ and IL-10Rβ are co-expressed on T and NK cells, we designed a series of bispecific ligands to induce a “synthetic” IL-2Rβ/IL-10Rβ heterodimer on cells (Figures 7A, S7A, and S7B). In principle, this would create a heterodimeric entity on the cell surface with the potential for JAK1/TYK2 transphosphorylation and activation of pSTAT5 (via IL-2Rβ) and pSTAT3 (via IL-10Rβ). The IL-2Rβ/IL-10Rβ induced proximity approach yielded only 4 active agonist “hits” out of 30 attempted combinations (~13% hit rate, Figure 7B). These ligands showed partial agonism of pSTAT5 relative to hIL-2 (Figure 7B), but none stimulated measurable pSTAT3 activity. One of the most potent, 10Rβ1–2Rβ6, was selected for further optimization by linker length-modulation, which induced pSTAT5 Emax activity from very low (16 a.a. linker) to full agonism equivalent to IL-2 (0 a.a. linker) and demonstrated the “tunability” of the surrogate system (Figure 7C). We selected the most potent variant, 10Rβ1–2Rβ6 (0 a.a.) and tested the effect of C-terminal Fc-fusion on pSTAT5 signaling in human primary T cells (Figures 7D and 7E). The Fc fusion more than doubled the Emax relative to the monomeric 10Rβ1–2Rβ6 (Figures 7E and S7C), likely through avidity enhancement.
Figure 7. A surrogate agonist that enforces proximity between IL-2Rβ and IL-10Rβ activates pSTAT5 signaling in T and NK cells.
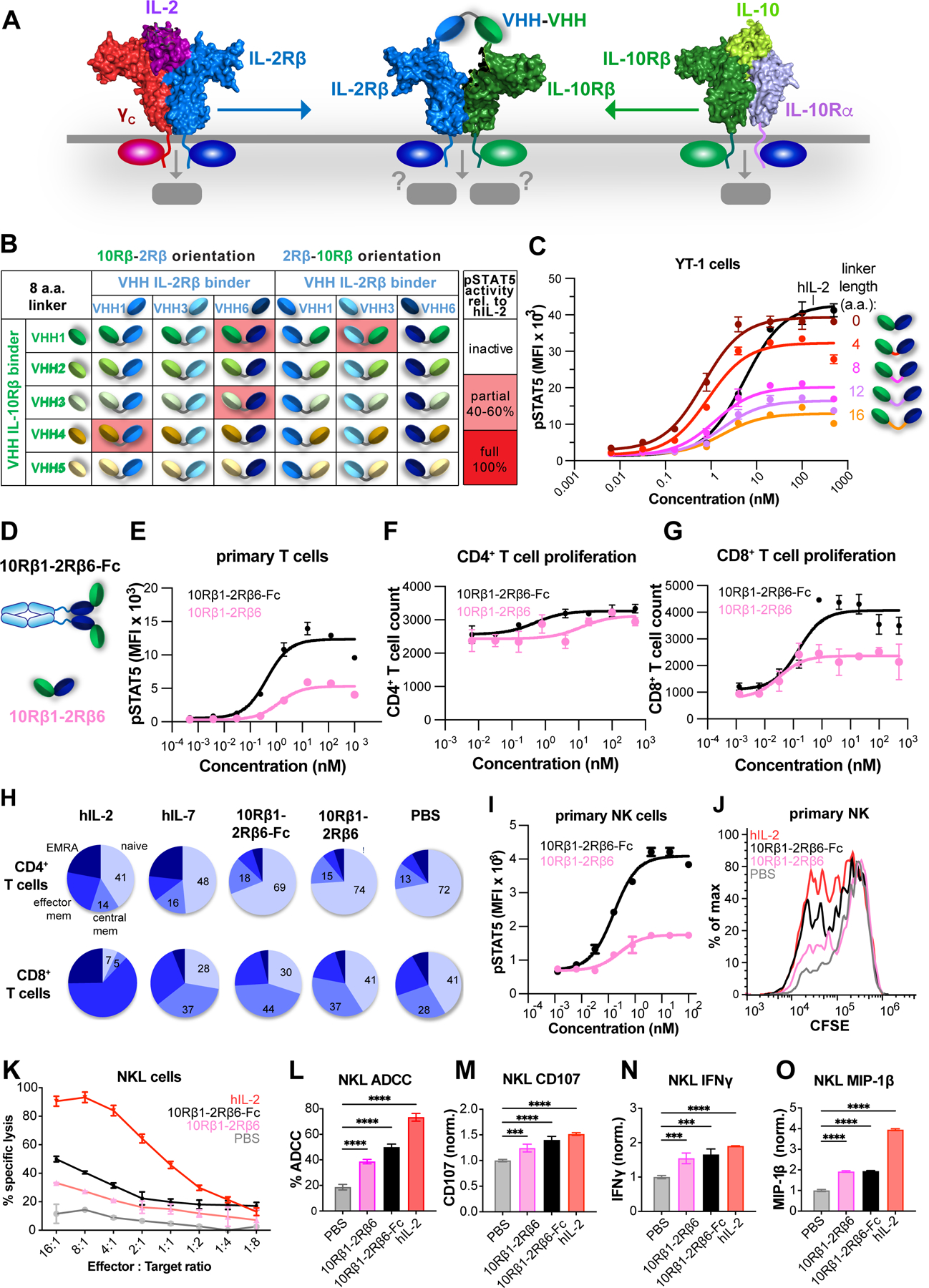
(A) Schematic showing non-natural receptor pairing of IL-10Rβ/IL2-Rβ to compel formation of a synthetic JAK1/TYK2 heterodimer. (B) Thirty IL-10β/IL-2Rβ VHH pairings (with 8 a.a. linkers) in forward and reverse orientations were expressed and assayed for pSTAT5 activity in YT-1 CD25−cells. Of the thirty combinations, four pairings had partial pSTAT5 activity (pink shading). (C) Modulation of ligand activity by linker length. 10Rβ12Rβ6 agonists with varying linker length between 0–16 a.a. were tested for pSTAT5 signaling in YT-1 cells. (D) Modulation of agonist activity by Fc-mediated dimerization. (E) Dimerization of the 10Rβ1–2Rβ6 ligand via Fc-fusion enhances pSTAT5 in primary human T cells. (F-G) CD8+ but not CD4+ T cell proliferation is driven by 10Rβ1–2Rβ6 and 10Rβ1– 2Rβ6-Fc. Pre-activated human T cells were cultured with varying concentrations of 10Rβ1– 2Rβ6 (pink) and 10Rβ1–2Rβ6-Fc agonists (black). Dose-response relationship of CD4+ (F) and CD8+ (G) T cells proliferation is indicated. (H) CD8+ but not CD4+ T cell differentiation is driven by 10Rβ1–2Rβ6 and 10Rβ1–2Rβ6-Fc. (I) Dose-response of 10Rβ1–2Rβ6 agonist (pink) and 10Rβ1–2Rβ6-Fc agonist (black) on pSTAT5 in primary effector NK cells. Data (mean ± SD) are from three independent replicates. (J) Effector NK cells were labelled with 5μM CFSE for 20min at 37°C. Histogram at 100nM ligand concentration displays proliferation of effector NK following 3d culture. (K) NKL killing of K562 tumor cells is enhanced by treatment with 10Rβ1–2Rβ6 (pink), 10Rβ1–2Rβ6-Fc (black) and hIL-2 (red). (L-O) Degranulation and activation of NKL cells in response to 10Rβ1–2Rβ6, 10Rβ1–2Rβ6-Fc, and hIL-2.
See also Figure S7.
Given the critical role of STAT5 activity in T cell proliferation, we asked whether the 10Rβ1–2Rβ6 dimerizers could drive primary T cells to proliferate. Monomeric and Fc-linked 10Rβ1–2Rβ6 induced expansion of CD8+ T cells but not CD4+ T cells (Figures 7F and 7G). Stimulation of naive T cells with these ligands in a differentiation assay resulted in a greater fraction of central memory T cells in CD8+ T cells than CD4+ T cells, which is more similar to the actions of IL-7 than to IL-2 (Figure 7H). The 10Rβ1–2Rβ6 and 10Rβ1–2Rβ6-Fc agonists also potentiated CD8+ T cell degranulation (Figure S7D), IFNγ production (Figure S7E), and activation in the A3A-MAGE 3A TCR:pMHC system (Figure S7F). On primary NK cells, 10Rβ1–2Rβ6 and 10Rβ1–2Rβ6-Fc induced different extents of STAT5 phosphorylation (Figure 7I) and proliferation (Figures 7J and S7G). 10Rβ1–2Rβ6 and 10Rβ1–2Rβ6-Fc promoted the lytic activity of NKL cells in a NK cytotoxicity assay against K562 tumor cells and an NK ADCC assay against rituximab-treated Raji tumor cells (Figures 7K and 7L). To assess the effect of these ligands on NK degranulation and activation, we co-cultured NK cells with K562 cells. Treatment with 10Rβ1–2Rβ6 and 10Rβ1–2Rβ6-Fc robustly enhanced CD107 (LAMP-1) surface expression and production of IFNγ and MIP-1β in both primary NK cells or NKL cells (Figures 7M–7O and S7H–S7J, respectively). Our results show that IL-10Rβ/IL-2Rβ agonists preferentially act on CD8+ T cells and NK cells versus IL-2, and that the IL-10Rβ/IL-2Rβ heterodimer signal more closely resembles an IL2 receptor partial agonist than an IL-10-mediated partial given its pSTAT5 bias.
Discussion:
We have exploited the principle of induced proximity of signaling receptors to create a class of modular, surrogate ligands with the capacity to dimerize cell surface receptors in ways that are structurally inaccessible to natural or engineered cytokines. These surrogate agonists have revealed an unexpectedly broad signaling plasticity and functional diversification downstream of these receptors, which can be exploited for drug discovery. The varied and unpredictable structure-activity relationships exhibited by these molecules highlight that the dimeric geometries and proximities of the receptor heterodimers, ligand affinity, and possibly steric effects, are key determinants that collectively modulate signaling in a complex and unpredictable interplay. This strategy is appropriate for dimeric receptor systems with limited or nonexistent structural knowledge, or for creating surrogate ligands when the native ligands present biochemical challenges (Janda et al., 2017). Since the binders can be utilized in a pairwise combinatorial manner, the approach can rapidly screen for diverse activities. Unlike combinatorial antibody library-based functional screens (Lerner et al., 2015), our smaller libraries are restricted to modules with biochemically validated binding to the target receptor ECDs so the attribution of agonist activity is unambiguous. In contrast to synthetic chimeric receptor screening systems (Engelowski et al., 2018), our ligands act on natural cell types not requiring genetic manipulation. Furthermore, while we fused the modules in a format to create single-chain bispecific ligands that are easily expressed, the approach is also amenable to a vast array of oligomerizing formats and stoichiometries beyond what we describe here. Our modular strategy could be applied towards other dimeric cell surface receptor pairs, such as Receptor Tyrosine Kinase (RTK) systems, BMP and TGF-β ligand-receptor systems, as well as trimeric death receptors where three, or more, VHHs could be fused. This overall strategy begins to position cytokine, and more broadly, protein-based agonist drug discovery as an induced proximity-based medicinal chemistry platform.
Prior work engineering IL-2 has focused on installing mutations on the natural cytokine scaffold in order to improve its affinity to IL-2Rβ and/or weaken its affinity to γC (Glassman et al., 2021a; Levin et al., 2012; Mitra et al., 2015). Here, our module-based approach allowed us to expand beyond the structural limitations imposed by the cytokine ligand. In comparison to previously reported bispecific antibody IL-2 agonist molecules (Harris et al., 2021), our single-chain platform enables rapid construction, expression and screening of large libraries and matrices of binders. The surrogate IL-2 analogs we identified enabled us to make several surprising observations about IL-2 signaling. First, we found that the principal signaling pathways downstream of IL-2R engagement – STAT5, ERK, and PI3K-Akt – were not necessarily coupled. ERK and Akt phosphorylation were extremely sensitive to structural perturbation. Whereas ~70% of molecules turned on pSTAT5 activity to some extent, pERK activity was sometimes much weaker or even nonexistent relative to the pSTAT5 amplitude, and pAkt activity was weaker still relative to pERK. These data indicate that signal strength across IL-2 pathways is not intrinsically linked like a rheostat that dials the amplitude in an isotropic manner across pathways, but that pSTAT5 activity may be decoupled from other signals. The differences in proximal signal patterning were manifested in proliferation, differentiation, cytokine secretion, and cytotoxicity. Some ligands exhibited biased abilities to differentiate naïve cells into central memory, effector memory, or “exhausted” T cells. Weaker ligands promoted growth only in CD8+ T cells, whereas robust ligands promoted strong central or effector memory phenotypes. Consistent with skewing to effector memory phenotype, these ligands also instructed the resultant T cells to produce cytolytic cytokines and conferred the ability to kill tumor cells.
The endogenous Type I IFN system represents a powerful example of “natural” protein engineering, with 16 cytokine sub-types that signal through a common IFNAR1/IFNAR2 heterodimer, yet elicit differentiated functional effects (Ng et al., 2016; Piehler et al., 2012). This is achieved through polymorphisms within IFN cytokine subtypes that occur at the ligand-receptor interfaces and that lead to different receptor-ligand affinities (Thomas et al., 2011). However, the overall dimeric geometry of the IFNAR1/IFNAR1 heterodimer is very similar among the different IFN sub-types (Thomas et al., 2011), thus the functional differentiation is largely based on differences in cytokinereceptor affinity and kinetics (Sandler et al., 2014; Sharma et al., 2016). The surrogate approach takes advantage of the epitope diversity of the VHH and scFv binders on the IFNAR1 and INFAR2 ECDs to achieve further Type I IFN functional diversification not seen with the natural IFNs. We find that surrogate IFN agonists do not elicit maximal levels of pSTAT1 compared to natural IFN, yet exhibit near equivalent anti-viral activity against several viruses, including SARS-CoV-2, but reduced expression of pro-inflammatory genes and anti-proliferative activities, two parameters that are thought to contribute to the doselimiting toxicity of Type I IFNs.
This platform offers additional flexibility to engineer ligands that compel formation of cytokine receptor heterodimers that are not found endogenously. In principle, if two cytokine receptors are expressed on the same cell, the relative promiscuity of JAK and TYK kinases could result in cross-talk and signaling if induced to form heterodimers by a synthetic ligand (Moraga et al., 2017). Here we have shown that an IL-2Rβ/IL-10Rβ heterodimer generates a pSTAT5 signal that is NK and CD8+ T cell biased and that more resembles an IL-7 signal in its functional effects compared to IL-2, although the investigation of this surrogate cytokine is at an early stage. The differentiated activity of this new signaling entity highlights an unexplored frontier beyond simply recapitulating natural cytokine receptor heterodimers that this platform can fully explore for drug discovery. Beyond JAK/STAT cytokine receptors, such unnatural pairings can be easily explored for other receptor systems using this modular “mix and match” approach.
In the future, this screening platform offers the possibility, with the advent of personalized medicine guided by immune profiling, to reimagine cytokine therapy tailored to bespoke cytokine analogs, chosen from a larger “toolkit” based in individual patient needs. In this sense, cytokine drug discovery will benefit from the indicia of choices that medicinal chemistry has afforded to many classes of small molecule drugs.
Limitations of the Study:
A limitation of our study is that, while our initial in vitro demonstration reveals the creation of a diverse set of cytokine agonists with granular signaling and functional differences, our ability to conduct in vivo studies in mouse models was severely limited due to lack of cross-species reactivity of our human receptor-specific VHH and scFv. Future studies will focus on exploring the in vivo properties of surrogate agonists either through development of mouse surrogate cytokines or the use of humanized mice.
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, K. Christopher Garcia (kcgarcia@stanford.edu).
Materials availability
Plasmids generated in this study will be provided by the lead contact upon completion of a Materials Transfer Agreement.
Data and code availability
RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the Key Resources Table.
Crystallography data have been deposited at RSCB PDB and are publicly available as of the date of publication. PDB IDs are listed in the Key Resources Table.
This paper does not report original code.
KEY RESOURCE TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER | |||
|---|---|---|---|---|---|
| Antibodies | |||||
| Anti-human CD3e | BioLegend | Cat#317326; clone OKT3 | |||
| Anti-human CD28 | BioLegend | Cat#302943; clone CD28.2 | |||
| Alexa Fluor 647 Mouse Anti-Stat5 (pY694) | BD Biosciences | Cat#612599; 47/Stat5(pY694) | |||
| Mouse Anti-Stat5 (pY694), PE | BD Biosciences | Cat#612567; 47/Stat5(pY694) | |||
| Mouse Anti-Stat3 (pY705), Alexa Fluor 647 | BD Biosciences | Cat#557815; clone 4/P-STAT3 | |||
| Phospho-Stat1 (Tyr701) Rabbit mAb, Alexa Fluor 647 Conjugate | Cell Signaling Technology | Cat#8009S; clone 58D6 | |||
| Phospho-Stat1 (Tyr701) Rabbit mAb, Alexa Fluor 488 Conjugate | Cell Signaling Technology | Cat#9174S; clone 58D6 | |||
| Phospho-S6 Ribosomal Protein (Ser240/244) XP Rabbit mAb, Alexa Fluor 488 Conjugate | Cell Signaling Technology | Cat#5018S; clone D68F8 | |||
| HA-Tag Mouse mAb, Alexa Fluor 647 Conjugate | Cell Signaling Technology | Cat#3444S; clone 6E2 | |||
| Ultra-LEAF Purified anti-human CD337 (NKp30) Antibody | BioLegend | Cat#325224; clone P30–15 | |||
| Anti-human CD107a (LAMP-1) Antibody, FITC | Biolegend | Cat#328606; clone H4A3 | |||
| BD GolgiPlug Protein Transport Inhibitor (containing Brefeldin A) | BD Biosciences | Cat#555029 | |||
| GolgiStop Protein Transport Inhibitor (Containing Monensin) | BD Biosciences | Cat#554724 | |||
| Fixation/Permeabilization Solution Kit | BD Biosciences | Cat#554714 | |||
| PE-Cy7 Mouse Anti-Human TNF | BD Biosciences | Cat#557647; clone Mab11 | |||
| Perm buffer III | BD Biosciences | Cat#558050 | |||
| Mouse Anti-Human CD8, PE | BD Biosciences | Cat#555635; clone HIT8a | |||
| Mouse Anti-Human MIP-1β, PE | BD Biosciences | Cat#550078; clone D21–1351 | |||
| Mouse Anti-Human IFN-γ, Alexa Fluor 647 | BD Biosciences | Cat#557729; clone B27 | |||
| Anti-human CD122 (IL-2Rβ) Antibody, APC | Biolegend | Cat#339008; clone TU27 | |||
| Anti-human CD3 Antibody, APC | Biolegend | Cat#300439; clone UCHT1 | |||
| Anti-human CD45RA Antibody, APC | Biolegend | Cat#304150; clone HI100 | |||
| Anti-human IFN-γ Antibody, APC | Biolegend | Cat#506510; clone B27 | |||
| Anti-human CD8 Antibody, APC/Cyanine7 | Biolegend | Cat#344714; clone SK1 | |||
| Anti-human CD56 (NCAM) Antibody, Brilliant Violet 605 | Biolegend | Cat#362538; clone 5.1H11 | |||
| Anti-human CD197 (CCR7) Antibody, Brilliant Violet 605 | Biolegend | Cat#353224; clone G043H7 | |||
| Anti-human CD8a Antibody, Brilliant Violet 605 | Biolegend | Cat#344742; clone SK1 | |||
| Anti-human CD4 Antibody, Brilliant Violet 785 | Biolegend | Cat#300554; clone RPA-T4 | |||
| Anti-human CD3 Antibody, FITC | Biolegend | Cat#300452; clone UCHT1 | |||
| Anti-human CD4 Antibody, FITC | Biolegend | Cat#300538; clone RPA-T4 | |||
| FITC anti-human CD69 Antibody | Biolegend | Cat#310904; Clone FN50 | |||
| Anti-human CD3 Antibody , Pacific Blue | Biolegend | Cat#300431; clone UCHT1 | |||
| Anti-human CD4 Antibody, Pacific Blue | Biolegend | Cat#300521; clone RPA-T4 | |||
| Anti-human IL-2 Antibody, Pacific Blue | Biolegend | Cat#500307; clone MQ117H12 | |||
| Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) Rabbit mAb, Alexa Fluor 647 Conjugate | Biolegend | Cat#13148s; clone 197G2 | |||
| Phospho-Akt (Ser473) XP Rabbit mAb, Alexa Fluor 647 Conjugate | Cell Signaling Technology | Cat#4075S; clone DE9 | |||
| Bacterial and Virus Strains | |||||
| Mix & Go Competent Cells - DH5α | Zymo Research | Cat#T3007 | |||
| Biological Samples | |||||
| Human peripheral mononuclear cells (PBMCs), isolated from leukocyte reduction shuttles | Stanford Blood Center | N/A | |||
| Chemicals, Peptides, and Recombinant Proteins | |||||
| SA sensor chip | Cytiva | Cat#BR-1005–31 | |||
| HBS-P | Cytiva | Cat#BR-1006–71 | |||
| EasySep Magnet | StemCell Technologies | Cat#18000 | |||
| Human CD8+ T cell Isolation Kit | Miltenyi | Cat#130–096-495 | |||
| LS magnetic selection column | Miltenyi | Cat#130–042-401 | |||
| EasySep Human Naïve Pan T Cell Isolation Kit | StemCell Technologies | Cat#17961 | |||
| ExpiFectamine 293 Transfection Kit | Gibco | Cat#A14525 | |||
| Sapphire Baculovirus DNA | Allele | Cat#ABP-BVD-10002 | |||
| RNeasy Plus Mini Kit | Qiagen | Cat#74134 | |||
| CellTrace Violet Proliferation Kit | Invitrogen | Cat#C34557 | |||
| Bio-Safe Coomassie G-250 Stain, 5L | Biorad | Cat#1610787 | |||
| UltraComp eBeads | eBiosciences | Cat#01–2222-42 | |||
| 16% Paraformaldehyde aqueous solution | Electron Microscopy Sciences | Cat#15710 | |||
| EndoH | Produced in house | N/A | |||
| Kifunensine | Toronto Research Chemicals | Cat#K450000 | |||
| BirA | Produced in house (Fairhead and Howarth, 2015) | N/A | |||
| Streptavidin | Sigma | Cat#189730 | |||
| Alexa Flour 647 C2 Maleimide | Invitrogen | Cat#A20347 | |||
| Human MSA-IL-2 | Produced in house (Glassman et al., 2021) | Produced in house | |||
| Human IFNω | Produced in house (Thomas et al., 2011) | Produced in house | |||
| Human IL-15 | R&D systems | Cat#247-ILB-025 | |||
| Human IL-7 | R&D systems | Cat#207-IL-010 | |||
| PE-AnnexinV | Biolegend | Cat#640947 | |||
| Ionomycin | Sigma | Cat#I9657–1MG | |||
| Phorbol 12-myristate 13-acetate (PMA) | Sigma | Cat#P8139–5MG | |||
| Cellfectin II | Gibco | Cat#10362100 | |||
| Carboxypeptidase A | Sigma | Cat#C9268 | |||
| Carboxypeptidase B | Sigma | Cat#217356 | |||
| Expi293 Expression Medium | Gibco | Cat#A1435101 | |||
| Sf-900 III Media | Invitrogen | Cat#12658019 | |||
| ESF 921 Insect Cell Culture Medium | Expression Systems | Cat#96–001-01 | |||
| MY140-F | This paper | N/A | |||
| MY140-R | This paper | N/A | |||
| MY141-F | This paper | N/A | |||
| MY141-R | This paper | N/A | |||
| MY177-F | This paper | N/A | |||
| MY177-R | This paper | N/A | |||
| MY171-F | This paper | N/A | |||
| MY171-R | This paper | N/A | |||
| MY144-F | This paper | N/A | |||
| MY144-R | This paper | N/A | |||
| MY145-F | This paper | N/A | |||
| MY145-R | This paper | N/A | |||
| MY178-F | This paper | N/A | |||
| MY178-R | This paper | N/A | |||
| MY172-F | This paper | N/A | |||
| MY172-R | This paper | N/A | |||
| MY142-F | This paper | N/A | |||
| MY142-R | This paper | N/A | |||
| MY143-F | This paper | N/A | |||
| MY143-R | This paper | N/A | |||
| MY179-F | This paper | N/A | |||
| MY179-R | This paper | N/A | |||
| MY173-F | This paper | N/A | |||
| MY173-R | This paper | N/A | |||
| MY188-F | This paper | N/A | |||
| MY188-R | This paper | N/A | |||
| MY189-F | This paper | N/A | |||
| MY189-R | This paper | N/A | |||
| MY190-F | This paper | N/A | |||
| MY190-R | This paper | N/A | |||
| MY191-F | This paper | N/A | |||
| MY191-R | This paper | N/A | |||
| MY192-F | This paper | N/A | |||
| MY192-R | This paper | N/A | |||
| MY193-F | This paper | N/A | |||
| MY193-R | This paper | N/A | |||
| MY194-F | This paper | N/A | |||
| MY194-R | This paper | N/A | |||
| MY195-F | This paper | N/A | |||
| MY195-R | This paper | N/A | |||
| IL2Rβ-Nb6 | This paper | N/A | |||
| γc-Nb6 | This paper | N/A | |||
| Human IL2Rβ ECD | This paper | N/A | |||
| Human γc ECD | This paper | N/A | |||
| HIS1 | This paper | N/A | |||
| HIS2 | This paper | N/A | |||
| HIS3 | This paper | N/A | |||
| HIS4 | This paper | N/A | |||
| HIS5 | This paper | N/A | |||
| HIS6 | This paper | N/A | |||
| HIS7 | This paper | N/A | |||
| A1 VHH monomer | This paper | N/A | |||
| 10Rβ1–2Rβ6 (8a.a. linker) | This paper | N/A | |||
| 10Rβ3–2Rβ6 (8a.a. linker) | This paper | N/A | |||
| 10Rβ4–2Rβ1 (8a.a. linker) | This paper | N/A | |||
| 2Rβ3–10Rβ1 (8a.a. linker) | This paper | N/A | |||
| 10Rβ1–2Rβ6 (0a.a. linker) | This paper | N/A | |||
| 10Rβ1–2Rβ6 (4a.a. linker) | This paper | N/A | |||
| 10Rβ1–2Rβ6 (12a.a. linker) | This paper | N/A | |||
| 10Rβ1–2Rβ6 (16a.a. linker) | This paper | N/A | |||
| 10Rβ1–2Rβ6-Fc | This paper | N/A | |||
| Critical Commercial Assays | |||||
| Superdex 200 Increase column 10/300 GL | Cytiva | Cat#28990944 | |||
| Superdex 75 Increase column 10/300 GL | Cytiva | Cat#29148721 | |||
| SA sensor chip | Cytiva | Cat#BR-1005–31 | |||
| HBS-P | Cytiva | Cat#BR-1006–71 | |||
| Yeast-Display Nanobody Library (NbLib) | Kerafast | Cat#EF0014-FP | |||
| Deposited Data | |||||
| γc:γc-Nb6 Complex Crystal Structure | RSCB | 7S2R | |||
| RNA-seq dataset | GEO | GSE183436 | |||
| IL2Rβ:β-Nb6 Complex Crystal Structure | RSCB | 7S2S | |||
| Experimental Models: Cell Lines | |||||
| Human: Expi293F | GIBCO | Cat#A14527 | |||
| Insect: Spodoptera frugiperda (Sf9) | ATCC | Cat#CRL-1711 | |||
| Insect: Trichoplusia ni (T. ni) | Expression Systems | Cat#94–002F | |||
| Human: YT-1 (CD25+) | N/A | ||||
| Human: K-562 | ATCC | Cat#CCL-243 | |||
| Human: A-375 | ATCC | Cat#CRL-1619 | |||
| Recombinant DNA | |||||
| DNA | |||||
| pD649 MY140-F | This paper | N/A | |||
| pD649 MY140-R | This paper | N/A | |||
| pD649 MY141-F | This paper | N/A | |||
| pD649 MY141-R | This paper | N/A | |||
| pD649 MY177-F | This paper | N/A | |||
| pD649 MY177-R | This paper | N/A | |||
| pD649 MY171-F | This paper | N/A | |||
| pD649 MY171-R | This paper | N/A | |||
| pD649 MY144-F | This paper | N/A | |||
| pD649 MY144-R | This paper | N/A | |||
| pD649 MY145-F | This paper | N/A | |||
| pD649 MY145-R | This paper | N/A | |||
| pD649 MY178-F | This paper | N/A | |||
| pD649 MY178-R | This paper | N/A | |||
| pD649 MY172-F | This paper | N/A | |||
| pD649 MY172-R | This paper | N/A | |||
| pD649 MY142-F | This paper | N/A | |||
| pD649 MY142-R | This paper | N/A | |||
| pD649 MY143-F | This paper | N/A | |||
| pD649 MY143-R | This paper | N/A | |||
| pD649 MY179-F | This paper | N/A | |||
| pD649 MY179-R | This paper | N/A | |||
| pD649 MY173-F | This paper | N/A | |||
| pD649 MY173-R | This paper | N/A | |||
| pD649 MY188-F | This paper | N/A | |||
| pD649 MY188-R | This paper | N/A | |||
| pD649 MY189-F | This paper | N/A | |||
| pD649 MY189-R | This paper | N/A | |||
| pD649 MY190-F | This paper | N/A | |||
| pD649 MY190-R | This paper | N/A | |||
| pD649 MY191-F | This paper | N/A | |||
| pD649 MY191-R | This paper | N/A | |||
| pD649 MY192-F | This paper | N/A | |||
| pD649 MY192-R | This paper | N/A | |||
| pD649 MY193-F | This paper | N/A | |||
| pD649 MY193-R | This paper | N/A | |||
| pD649 MY194-F | This paper | N/A | |||
| pD649 MY194-R | This paper | N/A | |||
| pD649 MY195-F | This paper | N/A | |||
| pD649 MY195-R | This paper | N/A | |||
| pD649 IL2Rβ-Nb6 | This paper | N/A | |||
| pD649 γc-Nb6 | This paper | N/A | |||
| pD649 | ATUM | Cat#PD649 | |||
| Software and Algorithms | |||||
| FlowJo v10.5 | Tree Star | RRID: SCR_008520 | |||
| GraphPad Prism 9.1.0 | GraphPad Software | RRID: SCR_002798 | |||
| BIAevaluation software | Cytiva | RRID: SCR_015936 | |||
| PHENIX | (Liebschner et al., 2019) | RRID:SCR_014224 | |||
| Coot | (Emsley et al., 2010) | RRID:SCR_014222 | |||
| SBGrid | (Morin et al., 2013) | RRID:SCR_003511 | |||
| DESEQ2 | (Love et al., 2014) | RRID:SCR_015687 | |||
| STAR | https://github.com/alexdobin/STAR/releases | RRID:SCR_004463 | |||
| UCSF ChimeraX | https://www.cgl.ucsf.edu/chimerax/ | RRID:SCR_015872 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines and culture conditions for functional studies
Human PBMC were isolated from LRS chambers (Stanford Blood Center) and cryopreserved until time of use. CD25+ YT-1 cells (Kuziel et al., 1993) and PBMC were maintained at 37°C in a 5% CO2 humidified chamber, and cultured in complete RPMI medium (RPMIc) containing 10% FBS and supplemented with 25mM HEPES, 2mM pyruvate, 4mM GlutaMAX, non-essential amino acids, and penicillin-streptomycin (all cell culture reagents were purchased from Gibco). Prior to stimulation for pERK and pAkt studies, cells were starved in serum-free RPMI for 1–2 hours. Primary cells were rested overnight without cytokine before measuring signaling.
Normal human primary bronchial/tracheal epithelial cells were purchased from ATCC (PCS-300–010) and grown in Airway Epithelial Cell Basal Media (ATCC PCS-300–030) supplemented with Bronchial/Tracheal Epithelial Cell Growth Kit components (ATCC PCS300–040) following manufacturer’s instructions. A549 cells were maintained in complete DMEM medium containing 10% FBS and supplemented with 25mM HEPES, 2mM sodium pyruvate, 4mM GlutaMAX, and penicillin-streptomycin. NKL cells were cultured in RPMIc containing 100IU human IL-2, with media and IL-2 changes every other day. All cell lines were maintained at 37°C in a 5% CO2 humidified incubator.
Cell lines and culture conditions for protein expression
Baculovirus was generated in Spodoptera frugiperda (Sf9) cells maintained in Sf-900 III SFM (Gibco) supplemented with 10% FBS (Sigma) and GlutaMAX (Gibco). For insect protein expression, Trichoplusia ni (High5, Expression Systems) cells grown in ESF 921 Insect Cell Culture Medium (Expression Systems) were infected with baculovirus. Insect cells were maintained at 27°C at ambient CO2 with shaking at 120rpm.
Expi293 suspension cells (Thermo Fisher) were maintained in Expi293 Expression Medium on a 120rpm shaking platform at 37°C in a 5% CO2 humidified incubator. Protein was produced by transient transfection using ExpiFectamine (Thermo Fisher).
METHOD DETAILS
Camel immunization
Human IL-2Rβ ECD (a.a. 27–240), human γC ECD (a.a. 23–262), human IL-10Rβ ECD (a.a. 20–220), human IFNAR1 ECD (a.a. 28–436), and IFNAR2 ECD (a.a. 27–243) were expressed as Fc fusions in HEK293F cells and purified by protein A affinity chromatography. Purified receptor ECDs were mixed with Freund’s adjuvant, then individually injected into healthy Bactrian camels (Camelus bactrianus). After the seventh immunization, antiserum titer reached 1.0×105 (indicating a strong immune response) and we collected 100 mL of peripheral blood for phage display library construction. All camel experiments were performed in compliance with ethics guidelines approved by Shanghai Science and Technology Committee (STCSM).
VHH library construction
Following isolation of peripheral blood lymphocytes (PBLs) from immunized camels, RNA was extracted, cDNAs were reverse transcribed, and VHHs were amplified by two-step nested PCR. Purified VHH fragments were subcloned into the phage-display phagemid pMECS and used to construct the phage display libraries. The quality of libraries was evaluated by size and insertion rate. Insertion rate was calculated by randomly screening 24 clones per library and determining insertion size by PCR amplification.
VHH library selection
VHHs specific for IL-2Rβ, γC, IL-10Rβ, IFNAR1 (VHH 1, 5, and 6), and IFNAR2 were selected from phage-display libraries using target proteins and enriched by three consecutive rounds of bio-panning with the infection of VCSM13 helper phages. Three hundred individual colonies were randomly selected from the enriched pool and positive clones were identified using periplasmic extract ELISA (PE-ELISA). VHH A1, A5, and A11 were isolated from a yeast-displayed VHH library using standard techniques (Kerafast).
Protein expression
VHH were fused using a 2–8 a.a. linker, and VHH–scFv were fused via an 8 a.a. Gly-Ser linker, unless otherwise indicated. VHH and scFv fusions were cloned into a pD649 mammalian expression vector (ATUM DNA 2.0), which carries an HA secretion signal peptide and a C-terminal 6-His tag. Proteins were expressed in Expi293F cells (Thermo Fisher) for 5–7 days according to manufacturer protocols, isolated using Ni2+ affinity chromatography, then further fractionated over a Superdex 200 increase column equilibrated with 20 mM HEPES (pH 7.4) and 150 mM NaCl. The 10Rβ1–2Rβ6 Fc-fusion was isolated using Protein G affinity chromatography (Thermo Fisher), but otherwise processed identically as the other ligands.
RNA-seq experiments
T cells were pre-activated for 4d with α-CD3/CD28, washed, and rested overnight without stimulation. The following day, CD8+ T cells were purified using MACS (CD8+ T cell isolation kit, Miltenyi Biotec), then stimulated with 100nM natural cytokine (hIL-2, hIL-7, hIL-15) or surrogate ligand for 24hr. at 37°C. Total RNA from 1–2 million cells per condition was extracted using an RNeasy micro kit (Qiagen). For each condition, we performed 3 biological replicates, representing samples from 3 independent donors. cDNA library preparation and RNA sequencing were performed by Novogene. cDNA libraries were loaded onto an Illumina NovaSeq 6000 sequencer, PE150 platform. Reference genome and gene model annotation files were downloaded from the genome website browser (NCBI/UCSC/Ensembl) directly. Paired-end clean reads were aligned to the reference genome using STAR software, and differential expression analysis was conducted using the DESeq2 R package (Love et al., 2014). Data (raw and processed) are deposited under GEO accession record GSE183436.
NK experiments
Primary NK cells (from a mixed human PBMC population) were pre-activated for 57d with 2µg/mL plate-bound α-NKp30 (Biolegend) with 9nM hIL-15 (R&D) in RPMIc media. Following activation, cells were rested for 1d in RPMIc without stimulation, then plated into 96-well microplates in the presence of hIL-2 or IL-2 surrogate ligands. Media and ligand were refreshed every 3–4d. Cells were analyzed at the indicated time points for cytokine profiling and cytotoxicity.
T cell proliferation
PBMC were pre-activated for 3–4d with 2.5μg/mL plate-bound α-CD3 (clone OKT3, Biolegend) and 5μg/mL soluble α-CD28 (Biolegend), then rested for 1d in RPMIc without stimulation. Cells were loaded with 5uM CellTrace Violet, then plated in 96-well format in media containing 100nM hIL-2 or surrogate ligands. Live CD4+ and CD8+ T cells were enumerated using cell surface antibodies and propidium iodide exclusion after 3–5d in culture.
T cell differentiation
Naïve pan-T cells were isolated to >90% purity from 5–10E+7 cryopreserved PBMCs using an EasySep Human Naïve Pan T Cell Isolation Kit (STEMCELL technologies). Cells were preactivated using 2µg/mL plate-bound α-CD3 (clone OKT3, Biolegend) and 1µg/mL soluble α-CD28 (Biolegend) in RPMIc for 4d. Prior to differentiation, cells were washed and rested in RPMIc without stimulation for 1d, then plated into 96-well microplates with 100nM hIL-2 or IL-2 surrogate ligands. Media and ligand were refreshed after 4d. Cells were analyzed for T memory surface markers or cytokine profiling at 8–10d post differentiation.
Crystallography
For IL-2R VHH crystallography, hIL-2Rβ extracellular domain (a.a. 27–233) and hγc extracellular domain (a.a. 55–254) were cloned into the pAcGP67a baculoviral vector carrying an N-terminal GP64 signal sequence and C-terminal 6xHis tag. Baculovirus was produced by transfection of Sf9 insect cells with Cellfectin II (Gibco) and Sapphire Baculovirus DNA (Allele) followed by viral amplification in Sf9 cells. Protein was expressed in T. ni cells infected for 48–72 h. For γc expression, cells were infected in the presence of the endoplasmic reticulum mannosidase I (ERM1) inhibitor, kifunensine (Toronto Research Chemicals). Protein was purified by Ni-NTA affinity chromatography followed by size exclusion chromatography (SEC) using a Superdex S75 increase column (Cytiva). For VHH expression, sequences were cloned into the pD649 vector with an N-terminal HA signal peptide and C-terminal AviTag and 6xHis tag. VHH were expressed by transient transfection in Expi293F cells (Gibco) using an ExpiFectamine 293 Transfection Kit (Gibco) according to manufacturer’s protocols. VHH were purified by Ni-NTA affinity chromatography and S75 SEC.
For γc:γc-VHH6 crystallography, γc and γc-VHH6 were complexed for 4 hours at 4°C in the presence of carboxypeptidase A (Sigma), carboxypeptidase B (Sigma), and endoglycosidase H (EndoH) in HBS, pH6.8. The complex was purified by S75 SEC and concentrated to 12.9mg/mL. Crystals were grown in a solution of 2M ammonium sulfate, 0.2M BIS-Tris pH5.5 and flash cooled in liquid nitrogen with the addition of 30% glycerol as cryoprotectant. Diffraction data were collected at Stanford Linear Accelerator SSRL beamline 12–1. Data were indexed, integrated, and scaled using the XDS package (Kabsch, 2010). The structure was solved by molecular replacement using PHASER with hγc (PDB: 2B5I) (Wang et al., 2005) and a VHH with loop deletions (PDB: 5LHR) (Kromann-Hansen et al., 2017). The final model was built by iterative rounds of model building in COOT (Emsley et al., 2010) and refinement in PHENIX (Liebschner et al., 2019). All crystallographic software was installed and configured by SBGrid (Morin et al., 2013).
For IL-2Rβ crystallography, IL-2Rβ and IL-2Rβ-VHH6 were methylated with borane dimethylamine complex (Sigma) and paraformaldehyde (Electron Microscopy Sciences) overnight at 4°C according to previously established protocols (Walter et al., 2006) in the presence of carboxypeptidase A and B (Sigma). The following morning, the reaction was quenched with 200mM Tris pH8.0 and purified by S75 SEC. The complex was concentrated to 11.7mg/mL and crystallized in a solution of 0.23M ammonium sulfate, 0.08M BisTris pH5.5 and 23% PEG3350. Crystals were cryoprotected with 20% PEG 400 and flash cooled in liquid nitrogen. Diffraction data were collected at Stanford Linear Accelerator (SSRL 122) and processed as described for the γc structure other than that molecular replacement was performed with IL-2Rβ (PDB: 2B5I) (Wang et al., 2005).
For both structures, data refinement and statistics can be found in Table S1 and were deposited in the RSCB protein databank with accession codes PDB: 7S2R (γc:γc-VHH6) and PDB: 7S2S (IL-2Rβ:β-VHH6).
qRT-PCR
RNA was extracted with an RNeasy Plus kit (QIAGEN), converted to cDNA by a RTPCR reaction (iSCRIPT reverse transcription kit, Bio-rad), and ISG induction relative to the untreated controls and normalized to GAPDH levels were measured by the PowerTrack SYBR green qPCR assay system (Thermo Fisher Scientific) on a StepOnePlus instrument (Thermo Fisher Scientific). Primer pairs used for transcript amplification are listed in Table S2.
Anti-proliferative activity assay
Human primary bronchial/tracheal epithelial cells were seeded at 1,000 cells/well in 96-well plates. The following day media was replaced with surrogate ligand or IFNω containing media. 3 days post IFN treatment cell density was measured using CellTiter-Glo (Promega) according to the manufacturer’s protocol.
A549-hACE2 SARS-CoV-2 antiviral assay
96-well plates were seeded with 20,000 A549-hACE2 cells/well. A549 is a human lung epithelial cell line stably expressing the SARS-CoV-2 receptor, hACE2, to facilitate efficient infection for antiviral assays (Hou et al., 2020a). Culture medium was removed 24 hr. post-seeding, and a 9-point agonist dose-response (top concentration 1000nM, 10-fold steps) was prepared in “infection medium” (DMEM (Gibco), 5% fetal bovine serum (Hyclone), 1x anti/anti (antibiotic, antimycotic, Gibco). Cells were transported to Biosafety Level 3 after 24hr. treatment with agonists, at which point cells were infected with recombinant SARS-CoV-2 engineered to express nanoluciferase at a multiplicity of infection of 0.25. After incubation for 1hr. at 37°C, input virus was removed, cells were washed once with infection medium and 100µL fresh infection medium was added. As a positive control, a similar dose-response of recombinant human IFNω was employed. As a negative control, the monomeric hIFNAR1-specific VHH “A1” was employed, which should not facilitate the dimerization of the type I interferon receptor subunits. After 48hr. of infection, levels of virus replication were measured by Promega NanoGlo assay measured on a Promega GloMax Luminometer. Similarly treated uninfected sister plates were generated in order to gauge potential cytotoxicity.
SARS-CoV-2 viral inhibition assay
Primary human airway epithelial cells were treated with varying concentrations of HIS ligands, human IFNω positive control, or “A1” VHH monomer negative control for 24hr. prior to infection with SARS-CoV-2 (Washington 1 strain) at a multiplicity of infection of 0.1. After 3d, apical washes were performed to measure viral titer.
NK and NKL functional assays
NK cells were stimulated with hIL-18 (100 ng/mL, R&D), hIL-15 (20 ng/mL, R&D), and hIL-12 (10 ng/mL, BioLegend) for 18hr, washed 3 times, then cultured in cRPMI for 2 days.
NKL cells were rested in cytokine-free media for 2 days. The rested NKL cells were preincubated with 100nM surrogate ligand or hIL-2 for 12h. K562 cells were labeled with 15μM Calcein-AM (BioLegend) for 30min at 37°C. The NKL cells were cocultured with 10,000 K562 cells at indicated effector:target ratios for 4h at 37°C in V bottom 96 well plate. The supernatants were transferred to a new 96 well plate and measured using a Spectramax Gemini dual-scanning microplate (excitation filter: 485 ± 9 nm; band-pass filter: 530 ± 9 nm).
For degranulation and activation of NKL or primary NK cells, cells were rested and pre-stimulated with surrogate agonists for 12h. K562 cells were labeled with 1uM CellTrace Violet (Thermo Fisher) for 20min at 37°C. NK cells were co-cultured with K562 cells for 4h in the presence of FITC-CD107 antibody (BioLegend), GolgiStop and GolgiPlug (BD). The cells were surface stained with NK markers and CD69 antibody for 30min. on ice. IFNγ staining was performed by following the intracellular staining protocol (Invitrogen). The samples were analyzed via flow cytometry.
QUANTIFICATION AND STATISTICAL ANALYSIS
SPR data was analyzed using BIAevaluation software (Cytiva). Flow cytometry data was analyzed using FlowJo software (BD). Statistical analyses were performed using Prism v9 (GraphPad Software). Statistics were determined using Prism v9.1.0 (GraphPad Software). For Figures 7 and S7, one-way ANOVA followed by Dunnett’s multiple comparisons tests was used to analyze experiments with more than two groups. For Figure S6E, statistics were calculated using two-way ANOVA followed by Tukey’s multiple comparisons tests. Significance is indicated as follows: *p < 0.05; **p < 0.01; ***p < 0.001, ****p<0.0001. Data are expressed as mean ± standard deviation of duplicate or triplicate wells, as indicated in the figure legends. Data are representative of two or more independent experiments, except for Figure S6E, which was performed once.
Supplementary Material
(A) SDS-PAGE of purified human IL-2Rβ (ECD)-Fc and γC (ECD)-Fc antigen used for camel immunization. (B) Phage library sizes were determined by counting the number of colonies after serial dilution onto plates containing selective antibiotics. (C) Insertion rate of the libraries measured by PCR of 24 randomly selected clones yielded correct insert rates of 95.8% and 100% for IL-2Rβ and γC libraries, respectively. (D) Enrichment following each round of bio-panning. (E) Sequence alignment of selected IL-2Rβ- and γC-VHH clones used for cell-based receptor binding assays. (F) Cell surface-based screening of IL-2Rβ-specific VHH candidates. YT-1 cells were incubated with varying concentration of HA-tagged VHH, washed, and stained with anti-HA-AlexaFluor488, then analyzed via flow cytometry. (G) Cell surface screening of γC-specific VHH candidates. YT-1 cells were incubated with biotinylated VHH, washed, then stained with Streptavidin-AlexaFluor647 followed by detection using flow cytometry. (H) Steady-state binding affinities between VHH/scFv and IL-2R, as determined by SPR. Values for scFv affinities are derived from patent US20160367664A1 (Wang et al., 2016).
(A) Biotinylated human IL-2Rβ ECD was immobilized on a streptavidin (SA) sensor chip, and varying concentrations of IL-2Rβ VHH were applied to determine binding parameters using SPR. Sensorgrams are shown on the left, and steady state binding is plotted on the right. Binding affinities (KD) were determined by fitting to steady state response values. (B) Biotinylated human γC ECD was immobilized on a SA chip, and varying concentrations of γC VHH were flowed over the chip to determine binding. Sensorgrams (left) and steady state binding (right) are displayed as in (A). (C) Dose-response relationship of pSTAT5 geometric mean flurorescence intensities (gMFI) of selected agonists. (D) Corresponding histograms showing STAT5 activity in YT-1 cells treated with human IL-2 vs. IL-2 surrogate agonists at saturating concentration. (E) Table shows Emax values of all 40 surrogate agonists. Emax was baseline subtracted from the unstimulated values and then normalized to that of hIL-2.
(A) Kinetics of pERK (left) and pAkt activation (right) in YT-1 cells stimulated with high dose (25nM) hIL-2. Geometric mean fluorescence intensity (gMFI) of replicate wells was averaged and plotted as a function of time (middle). Phosphorylation peaks by 3min. after stimulation. (B, D) Time course of pERK activation by hIL-2 (black) vs. IL-2 surrogate ligands (colored lines). (C, E) Dose-response of pERK activation at 3 minutes post-ligand addition. (F) Histograms for hIL-2 and selected agonists showing STAT5 (left), STAT1 (middle), and STAT3 (right) activity levels in pre-activated human T cells stimulated with 100nM ligand. (G) Pre-activated human T cells were stimulated as in (F), and the gMFI of STAT5, STAT1, or STAT3 antibody staining was plotted as indicated. To aid comparison, these raw data were baseline subtracted with the “unstimulated” values, then normalized to hIL-2 STAT levels in main Figure 2.
(A) Schematic outline of human T cell differentiation assay. (B) Gating scheme for distinguishing CD4+ and CD8+ memory cell subsets, and example T cell “input” after purification of naïve T cells (“post-enrichment”). (C) Gating scheme to distinguish CD4+ and CD8+ T cells for cytokine profiling. (D) Example intracellular cytokine staining histograms showing expression of effector cytokines induced by surrogate agonists, as described in (A).
(A) Gating scheme for enumerating T cell vs. NK cell types based on cell surface staining. (B) Histograms displaying NK cell intracellular cytokine staining for hIL-2 or surrogate agonist-cultured cells. (C) Gating scheme used to identify CTV-loaded target cells and quantify annexin V positivity. Top row shows a well containing target cells only, bottom row contains a mixture of NK effectors + target cells. (D) Histogram of target cell annexin V staining when mixed with NK cells cultured with 10nM hIL-2 vs. selected IL-2 analogs.
(A) SPR sensorgrams displaying dose-dependent binding of IFNAR2 VHHs to immobilized human IFNAR2 ECD. SPR experiments were performed using the same conditions as for IL2 specific VHHs (Figure S2). Binding constants were determined from kinetic fitting and summarized in (B). (C) Kinetics of STAT1-STAT6 phosphorylation induced by surrogate ligands or IFN junk in YT-1 cells. YT-1 cells were serum-starved for 1–2 hr., then stimulated with 10nM ligand for 15–60 min at 37°C, fixed and permeabilized, then stained with fluorescently conjugated phospho-antibodies before reading on a flow cytometer. (D) STAT1 and STAT2 phosphorylation induced by HIS or IFNω junk at different time points in A549 cells. (E) SARS-CoV-2 antiviral assay on primary human airway epithelial cells. Cells were pre-treated with HIS, IFNω, or VHH monomer “A1” for 24hr. prior to infection with SARS-CoV-2 infection (MOI = 0.1). Viral titers were quantified from apical washes collected at 72hr. post-infection. (F-H) qRT-PCR analysis of ISG transcripts induced by 10nM HIS or IFNω junk for 8hr. in (F) A549 cells, (G) primary human airway epithelial cells, or (H) human PBMCs.
(A) SPR sensorgrams displaying dose-dependent binding of IL-10Rβ VHHs to immobilized human IL-10Rβ ECD. SPR experiments were performed using the same conditions as for IL2 specific VHHs (Figure S2). Binding constants were determined from kinetic fitting and summarized in (B). (C) Proximal signaling of monomeric or Fc-linked 10Rβ1–2Rβ6 surrogate agonist vs. hIL-2, assessed by western blotting. YT-1 cells were treated with 100nM ligand for varying times, lysed, then examined for phosphorylation of IL-2Rβ, TYK2, JAK1 and STAT5. (D-F) Degranulation and activation of A3A-TCR+CD8+ T cells in response to monomeric or Fc-linked 10Rβ1–2Rβ6, hIL-7, and hIL-2. (G-J) Functional properties of monomeric or Fc-linked 10Rβ1–2Rβ6 on primary human NK cells. Human NK cells were treated with surrogate agonists, hIL-7, or hIL-2, then assayed for (G) proliferation or expression of (H) CD107, (I) IFNγ, or (J) MIP-1β.
Highlights.
A platform to expand and diversify cytokine biology with modular surrogate agonists
IL-2 surrogates reveal signaling plasticity and biased activities on T and NK cells
Type I IFN surrogates are potent antiviral agents with reduced cytotoxic properties
IL-2/10 surrogates drive non-natural receptor hetero-dimerization on T and NK cells
Acknowledgements:
We acknowledge Yakun Wan for sharing of VHH reagents. This work was supported by National Institute of Allergy and Infectious Diseases grant R37-AI051321 to K.C.G. and by the Mathers Charitable Foundation. K.C.G. is an investigator of the Howard Hughes Medical Institute. M.Y.’s training was supported by the Stanford Immunology Program Training Grant (5T32AI07290-33) and the Stanford Immunology and Rheumatology Training Grant (5T32AR050942-15). Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (P30GM133894). Portions of schematics in Figures 1C and S4A were generated using BioRender.com.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests:
K.C.G., M.Y., J.R., and Q.L. are co-inventors on a provisional patent 63/306,882 based upon the technology described in this manuscript. K.C.G. is the founder of Synthekine Therapeutics.
References:
- Berraondo P, Sanmamed MF, Ochoa MC, Etxeberria I, Aznar MA, Pérez-Gracia JL, Rodríguez-Ruiz ME, Ponz-Sarvise M, Castañón E, and Melero I (2018). Cytokines in clinical cancer immunotherapy. British Journal of Cancer 120, 6–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron BJ, Gerry AB, Dukes J, Harper JV, Kannan V, Bianchi FC, Grand F, Brewer JE, Gupta M, Plesa G, et al. (2013). Identification of a Titin-derived HLA-A1-presented peptide as a cross-reactive target for engineered MAGE A3-directed T cells. Sci Transl Med 5, 197ra103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardarelli JM, Witte A, and Srinivasan M (2010). Interferon Alpha Receptor 1 Antibodies and Their Uses. US Patent 7662381 B2, filed June 20, 2005, and granted February 16, 2010
- Cooper MA, Fehniger TA, and Caligiuri MA (2001). The biology of human natural killercell subsets. Trends Immunol 22, 633–640. [DOI] [PubMed] [Google Scholar]
- Corman SL, and Mohammad RA (2010). Eltrombopag: a novel oral thrombopoietin receptor agonist. Ann Pharmacother 44, 1072–1079. [DOI] [PubMed] [Google Scholar]
- Cui W, Liu Y, Weinstein JS, Craft J, and Kaech SM (2011). An interleukin-21interleukin-10-STAT3 pathway is critical for functional maturation of memory CD8+ T cells. Immunity 35, 792–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delespine-Carmagnat M, Bouvier G, and Bertoglio J (2000). Association of STAT1, STAT3 and STAT5 proteins with the IL-2 receptor involves different subdomains of the IL-2 receptor I chain. Eur. J. Immunol 30, 59–68. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelowski E, Schneider A, Franke M, Xu H, Clemen R, Lang A, Baran P, Binsch C, Knebel B, Al-Hasani H, et al. (2018). Synthetic cytokine receptors transmit biological signals using artificial ligands. Nat Commun 9, 2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairhead M, and Howarth M (2015). Site-Specific Protein Labeling: Methods and Protocols. In Methods in Molecular Biology, Gautier A and Hinner MJ, eds. (Springer Science+Business Media; ), pp. 171–184. [Google Scholar]
- Fulcher ML, Gabriel S, Burns KA, Yankaskas JR, and Randell SH (2005). Welldifferentiated human airway epithelial cell cultures. Methods Mol Med 107, 183–206. [DOI] [PubMed] [Google Scholar]
- Glassman CR, Su L, Majri-Morrison SS, Winkelmann H, Mo F, Li P, Perez-Cruz M, Ho PP, Koliesnik I, Nagy N, et al. (2021a). Calibration of cell-intrinsic interleukin-2 response thresholds guides design of a regulatory T cell biased agonist. Elife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glassman CR, Mathiharan YK, Jude KM, Su L, Panova O, Lupardus PJ, Spangler JB, Ely LK, Thomas C, Skiniotis G, et al. (2021b). Structural basis for IL-12 and IL-23 receptor sharing reveals a gateway for shaping actions on T versus NK cells. Cell 184, 983–999.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KE, Lorentsen KJ, Malik-Chaudhry HK, Loughlin K, Basappa HM, Hartstein S, Ahmil G, Allen NS, Avanzino BC, Balasubramani A, et al. (2021). A bispecific antibody agonist of the IL-2 heterodimeric receptor preferentially promotes in vivo expansion of CD8 and NK cells. Sci Rep 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou YJ, Chiba S, Halfmann P, Ehre C, Kuroda M, Dinnon KH, Leist SR, Schäfer A, Nakajima N, Takahashi K, et al. (2020a). SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 370, 1464–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou YJ, Okuda K, Edwards CE, Martinez DR, Asakura T, Dinnon KH, Kato T, Lee RE, Yount BL, Mascenik TM, et al. (2020b). SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell 182, 429–446.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda CY, Dang LT, You C, Chang J, de Lau W, Zhong ZA, Yan KS, Marecic O, Siepe D, Li X, et al. (2017). Surrogate Wnt agonists that phenocopy canonical Wnt and β-catenin signalling. Nature 545, 234–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (2010). XDS. Acta Crystallogr D Biol Crystallogr 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech SM, and Cui W (2012). Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol 12, 749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kromann-Hansen T, Louise Lange E, Peter Sørensen H, Hassanzadeh-Ghassabeh G, Huang M, Jensen JK, Muyldermans S, Declerck PJ, Komives EA, and Andreasen PA (2017). Discovery of a novel conformational equilibrium in urokinase-type plasminogen activator. Sci Rep 7, 3385–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuziel WA, Ju G, Grdina TA, and Greene WC (1993). Unexpected effects of the IL-2 receptor alpha subunit on high affinity IL-2 receptor assembly and function detected with a mutant IL-2 analog. J Immunol 150, 3357–3365. [PubMed] [Google Scholar]
- Leonard WJ, Lin J-X, and O’Shea JJ (2019). The γc Family of Cytokines: Basic Biology to Therapeutic Ramifications. Immunity 50, 832–850. [DOI] [PubMed] [Google Scholar]
- Lerner RA, Grover RK, Zhang H, Xie J, Han KH, Peng Y, and Yea K (2015). Antibodies from combinatorial libraries use functional receptor pleiotropism to regulate cell fates. Q Rev Biophys 48, 389–394. [DOI] [PubMed] [Google Scholar]
- Levin AM, Bates DL, Ring AM, Krieg C, Lin JT, Su L, Moraga I, Raeber ME, Bowman GR, Novick P, et al. (2012). Exploiting a natural conformational switch to engineer an interleukin-2 “superkine.” Nature 484, 529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, and Tamayo P (2015). The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1, 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner D, Afonine PV, Baker ML, Bunkóczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, et al. (2019). Macromolecular structure determination using Xrays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol 75, 861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J-X, and Leonard WJ (2018). The Common Cytokine Receptor γ Chain Family of Cytokines. Cold Spring Harb Perspect Biol 10, a028449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linette GP, Stadtmauer EA, Maus MV, Rapoport AP, Levine BL, Emery L, Litzky L, Bagg A, Carreno BM, Cimino PJ, et al. (2013). Cardiovascular toxicity and titin crossreactivity of affinity-enhanced T cells in myeloma and melanoma 122 6, 863–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maecker HT, McCoy JP, and Nussenblatt R (2012). Standardizing immunophenotyping for the Human Immunology Project. Nat. Rev. Immunol 12, 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansurov A, Lauterbach A, Budina E, Alpar AT, Hubbell JA, and Ishihara J (2021). Immunoengineering approaches for cytokine therapy. Am. J. Physiol., Cell Physiol 321, C369–C383. [DOI] [PubMed] [Google Scholar]
- Mendoza JL, Escalante NK, Jude KM, Bellon JS, Su L, Horton TM, Tsutsumi N, Berardinelli SJ, Haltiwanger RS, Piehler J, et al. (2019). Structure of the IFNγ receptor complex guides design of biased agonists. Nature 567, 56–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S, Ring AM, Amarnath S, Spangler JB, Li P, Ju W, Fischer S, Oh J, Spolski R, Weiskopf K, et al. (2015). Interleukin-2 Activity Can Be Fine Tuned with Engineered Receptor Signaling Clamps. Immunity 42, 826–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki T, Kawahara A, Fujii H, Nakagawa Y, Minami Y, Liu ZJ, Oishi I, Silvennoinen O, Witthuhn BA, Ihle JN, et al. (1994). Functional Activation of Jak1 and Jak3 by Selective Association with IL-2 Receptor Subunits. Science 266, 1045–1047. [DOI] [PubMed] [Google Scholar]
- Mohan K, Ueda G, Kim AR, Jude KM, Fallas JA, Guo Y, Hafer M, Miao Y, Saxton RA, Piehler J, et al. (2019). Topological control of cytokine receptor signaling induces differential effects in hematopoiesis. Science 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraga I, Spangler JB, Mendoza JL, Gakovic M, Wehrman TS, Krutzik P, and Garcia KC (2017). Synthekines are surrogate cytokine and growth factor agonists that compel signaling through non-natural receptor dimers. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraga I, Wernig G, Wilmes S, Gryshkova V, Richter CP, Hong W-J, Sinha R, Guo F, Fabionar H, Wehrman TS, et al. (2015). Tuning Cytokine Receptor Signaling by Reorienting Dimer Geometry with Surrogate Ligands. Cell 160, 1196–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin A, Eisenbraun B, Key J, Sanschagrin PC, Timony MA, Ottaviano M, and Sliz P (2013). Collaboration gets the most out of software. Elife 2, e01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng CT, Mendoza JL, Garcia KC, and Oldstone MB (2016). Alpha and Beta Type 1 Interferon Signaling: Passage for Diverse Biologic Outcomes. Cell 164, 349–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng J, and Cantrell D (1997). STAT3 is a serine kinase target in T lymphocytes. Interleukin 2 and T cell antigen receptor signals converge upon serine 727. J. Biol. Chem 272, 24542– 24549. [DOI] [PubMed] [Google Scholar]
- Ouyang W, and O’Garra A (2019). IL-10 Family Cytokines IL-10 and IL-22: from Basic Science to Clinical Translation. Immunity 50, 871–891. [DOI] [PubMed] [Google Scholar]
- Overwijk WW, Tagliaferri MA, and Zalevsky J (2021). Engineering IL-2 to Give New Life to T Cell Immunotherapy. Annu Rev Med 72, 281–311. [DOI] [PubMed] [Google Scholar]
- Piehler J, Thomas C, Garcia KC, and Schreiber G (2012). Structural and dynamic determinants of type I interferon receptor assembly and their functional interpretation. Immunol. Rev 250, 317–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley M, Huang X, and Yang Y (2008). STAT1 Signaling in CD8 T Cells Is Required for Their Clonal Expansion and Memory Formation Following Viral Infection In Vivo. J Immunol 180, 2158–2164. [DOI] [PubMed] [Google Scholar]
- Ring AM, Lin J-X, Feng D, Mitra S, Rickert M, Bowman GR, Pande VS, Li P, Moraga I, Spolski R, et al. (2012). Mechanistic and structural insight into the functional dichotomy between IL-2 and IL-15. Nat. Immunol 13, 1187–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross SH, and Cantrell DA (2018). Signaling and Function of Interleukin-2 in T Lymphocytes [DOI] [PMC free article] [PubMed]
- Russell SM, Johnston JA, Noguchi M, Kawamura M, Bacon CM, Friedmann M, Berg M, McVicar DW, Witthuhn BA, Silvennoinen O, et al. (1994). Interaction of IL-2Rb and gammac Chains with Jak1 and Jak3: Implication for XSCID and SCID. Science 266. [DOI] [PubMed] [Google Scholar]
- Sandler NG, Bosinger SE, Estes JD, Zhu RTR, Tharp GK, Boritz E, Levin D, Wijeyesinghe S, Makamdop KN, del Prete GQ, et al. (2014). Type I interferon responses in rhesus macaques prevent SIV infection and slow disease progression. Nature 511, 601– 605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton RA, Tsutsumi N, Su LL, Abhiraman GC, Mohan K, Henneberg LT, Aduri NG, Gati C, and Garcia KC (2021). Structure-based decoupling of the pro- and anti-inflammatory functions of interleukin-10. Science 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N, Longjam G, and Schreiber G (2016). Type I Interferon Signaling Is Decoupled from Specific Receptor Orientation through Lenient Requirements of the Transmembrane Domain. J. Biol. Chem 291, 3371–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoichet BK, and Kobilka BK (2012). Structure-based drug screening for G-proteincoupled receptors. Trends Pharmacol. Sci 33, 268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shourian M, Beltra J-C, Bourdin B, and Decaluwe H (2019). Common gamma chain cytokines and CD8 T cells in cancer. Semin. Immunol 42, 101307. [DOI] [PubMed] [Google Scholar]
- Siegel AM, Heimall J, Freeman AF, Hsu AP, Brittain E, Brenchley JM, Douek DC, Fahle GH, Cohen JI, Holland SM, et al. (2011). A critical role for STAT3 transcription factor signaling in the development and maintenance of human T cell memory. Immunity 35, 806–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva D-A, Yu S, Ulge UY, Spangler JB, Jude KM, Labão-Almeida C, Ali LR, QuijanoRubio A, Ruterbusch M, Leung I, et al. (2019). De novo design of potent and selective mimics of IL-2 and IL-15. Nature 565, 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Lefkowitz RJ, and Rajagopal S (2018). Biased signalling: from simple switches to allosteric microprocessors. Nature Reviews Drug Discovery 17, 243–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangler JB, Moraga I, Mendoza JL, and Garcia KC (2015). Insights into cytokinereceptor interactions from cytokine engineering. Annu. Rev. Immunol 33, 139–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud RM, and Wells JA (2004). Mechanistic diversity of cytokine receptor signaling across cell membranes. Sci STKE 2004, re7. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Moraga I, Levin D, Krutzik PO, Podoplelova Y, Trejo A, Lee C, Yarden G, Vleck SE, Glenn JS, et al. (2011). Structural Linkage between Ligand Discrimination and Receptor Activation by Type I Interferons. Cell 146, 621–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter TS, Meier C, Assenberg R, Au K-F, Ren J, Verma A, Nettleship JE, Owens RJ, Stuart DI, and Grimes JM (2006). Lysine methylation as a routine rescue strategy for protein crystallization. Structure 14, 1617–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C-I, Brauer P, Yeo SP, Tan HC, and Connelly JE (2016). IL2Rbeta/common gamma chain antibodies. U.S. Patent 20160367664 A1, filed August 5, 2016, and published December 22, 2016
- Wang X, Rickert M, and Garcia KC (2005). Structure of the quaternary complex of interleukin-2 with its alpha, beta, and gammac receptors. Science 310, 1159–1163. [DOI] [PubMed] [Google Scholar]
- Wu Y, Tian Z, and Wei H (2017). Developmental and Functional Control of Natural Killer Cells by Cytokines. Front Immunol 8, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue HH, Fink DWJ, Zhang X, Qin J, Turck CW, and Leonard WJ (2002). Serine phosphorylation of Stat5 proteins in lymphocytes stimulated with IL-2. International Immunology 14, 1263–1271. [DOI] [PubMed] [Google Scholar]
- Zhang H, Yea K, Xie J, Ruiz D, Wilson IA, and Lerner RA (2013). Selecting agonists from single cells infected with combinatorial antibody libraries. Chem. Biol 20, 734–741. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) SDS-PAGE of purified human IL-2Rβ (ECD)-Fc and γC (ECD)-Fc antigen used for camel immunization. (B) Phage library sizes were determined by counting the number of colonies after serial dilution onto plates containing selective antibiotics. (C) Insertion rate of the libraries measured by PCR of 24 randomly selected clones yielded correct insert rates of 95.8% and 100% for IL-2Rβ and γC libraries, respectively. (D) Enrichment following each round of bio-panning. (E) Sequence alignment of selected IL-2Rβ- and γC-VHH clones used for cell-based receptor binding assays. (F) Cell surface-based screening of IL-2Rβ-specific VHH candidates. YT-1 cells were incubated with varying concentration of HA-tagged VHH, washed, and stained with anti-HA-AlexaFluor488, then analyzed via flow cytometry. (G) Cell surface screening of γC-specific VHH candidates. YT-1 cells were incubated with biotinylated VHH, washed, then stained with Streptavidin-AlexaFluor647 followed by detection using flow cytometry. (H) Steady-state binding affinities between VHH/scFv and IL-2R, as determined by SPR. Values for scFv affinities are derived from patent US20160367664A1 (Wang et al., 2016).
(A) Biotinylated human IL-2Rβ ECD was immobilized on a streptavidin (SA) sensor chip, and varying concentrations of IL-2Rβ VHH were applied to determine binding parameters using SPR. Sensorgrams are shown on the left, and steady state binding is plotted on the right. Binding affinities (KD) were determined by fitting to steady state response values. (B) Biotinylated human γC ECD was immobilized on a SA chip, and varying concentrations of γC VHH were flowed over the chip to determine binding. Sensorgrams (left) and steady state binding (right) are displayed as in (A). (C) Dose-response relationship of pSTAT5 geometric mean flurorescence intensities (gMFI) of selected agonists. (D) Corresponding histograms showing STAT5 activity in YT-1 cells treated with human IL-2 vs. IL-2 surrogate agonists at saturating concentration. (E) Table shows Emax values of all 40 surrogate agonists. Emax was baseline subtracted from the unstimulated values and then normalized to that of hIL-2.
(A) Kinetics of pERK (left) and pAkt activation (right) in YT-1 cells stimulated with high dose (25nM) hIL-2. Geometric mean fluorescence intensity (gMFI) of replicate wells was averaged and plotted as a function of time (middle). Phosphorylation peaks by 3min. after stimulation. (B, D) Time course of pERK activation by hIL-2 (black) vs. IL-2 surrogate ligands (colored lines). (C, E) Dose-response of pERK activation at 3 minutes post-ligand addition. (F) Histograms for hIL-2 and selected agonists showing STAT5 (left), STAT1 (middle), and STAT3 (right) activity levels in pre-activated human T cells stimulated with 100nM ligand. (G) Pre-activated human T cells were stimulated as in (F), and the gMFI of STAT5, STAT1, or STAT3 antibody staining was plotted as indicated. To aid comparison, these raw data were baseline subtracted with the “unstimulated” values, then normalized to hIL-2 STAT levels in main Figure 2.
(A) Schematic outline of human T cell differentiation assay. (B) Gating scheme for distinguishing CD4+ and CD8+ memory cell subsets, and example T cell “input” after purification of naïve T cells (“post-enrichment”). (C) Gating scheme to distinguish CD4+ and CD8+ T cells for cytokine profiling. (D) Example intracellular cytokine staining histograms showing expression of effector cytokines induced by surrogate agonists, as described in (A).
(A) Gating scheme for enumerating T cell vs. NK cell types based on cell surface staining. (B) Histograms displaying NK cell intracellular cytokine staining for hIL-2 or surrogate agonist-cultured cells. (C) Gating scheme used to identify CTV-loaded target cells and quantify annexin V positivity. Top row shows a well containing target cells only, bottom row contains a mixture of NK effectors + target cells. (D) Histogram of target cell annexin V staining when mixed with NK cells cultured with 10nM hIL-2 vs. selected IL-2 analogs.
(A) SPR sensorgrams displaying dose-dependent binding of IFNAR2 VHHs to immobilized human IFNAR2 ECD. SPR experiments were performed using the same conditions as for IL2 specific VHHs (Figure S2). Binding constants were determined from kinetic fitting and summarized in (B). (C) Kinetics of STAT1-STAT6 phosphorylation induced by surrogate ligands or IFN junk in YT-1 cells. YT-1 cells were serum-starved for 1–2 hr., then stimulated with 10nM ligand for 15–60 min at 37°C, fixed and permeabilized, then stained with fluorescently conjugated phospho-antibodies before reading on a flow cytometer. (D) STAT1 and STAT2 phosphorylation induced by HIS or IFNω junk at different time points in A549 cells. (E) SARS-CoV-2 antiviral assay on primary human airway epithelial cells. Cells were pre-treated with HIS, IFNω, or VHH monomer “A1” for 24hr. prior to infection with SARS-CoV-2 infection (MOI = 0.1). Viral titers were quantified from apical washes collected at 72hr. post-infection. (F-H) qRT-PCR analysis of ISG transcripts induced by 10nM HIS or IFNω junk for 8hr. in (F) A549 cells, (G) primary human airway epithelial cells, or (H) human PBMCs.
(A) SPR sensorgrams displaying dose-dependent binding of IL-10Rβ VHHs to immobilized human IL-10Rβ ECD. SPR experiments were performed using the same conditions as for IL2 specific VHHs (Figure S2). Binding constants were determined from kinetic fitting and summarized in (B). (C) Proximal signaling of monomeric or Fc-linked 10Rβ1–2Rβ6 surrogate agonist vs. hIL-2, assessed by western blotting. YT-1 cells were treated with 100nM ligand for varying times, lysed, then examined for phosphorylation of IL-2Rβ, TYK2, JAK1 and STAT5. (D-F) Degranulation and activation of A3A-TCR+CD8+ T cells in response to monomeric or Fc-linked 10Rβ1–2Rβ6, hIL-7, and hIL-2. (G-J) Functional properties of monomeric or Fc-linked 10Rβ1–2Rβ6 on primary human NK cells. Human NK cells were treated with surrogate agonists, hIL-7, or hIL-2, then assayed for (G) proliferation or expression of (H) CD107, (I) IFNγ, or (J) MIP-1β.
Data Availability Statement
RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the Key Resources Table.
Crystallography data have been deposited at RSCB PDB and are publicly available as of the date of publication. PDB IDs are listed in the Key Resources Table.
This paper does not report original code.
KEY RESOURCE TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER | |||
|---|---|---|---|---|---|
| Antibodies | |||||
| Anti-human CD3e | BioLegend | Cat#317326; clone OKT3 | |||
| Anti-human CD28 | BioLegend | Cat#302943; clone CD28.2 | |||
| Alexa Fluor 647 Mouse Anti-Stat5 (pY694) | BD Biosciences | Cat#612599; 47/Stat5(pY694) | |||
| Mouse Anti-Stat5 (pY694), PE | BD Biosciences | Cat#612567; 47/Stat5(pY694) | |||
| Mouse Anti-Stat3 (pY705), Alexa Fluor 647 | BD Biosciences | Cat#557815; clone 4/P-STAT3 | |||
| Phospho-Stat1 (Tyr701) Rabbit mAb, Alexa Fluor 647 Conjugate | Cell Signaling Technology | Cat#8009S; clone 58D6 | |||
| Phospho-Stat1 (Tyr701) Rabbit mAb, Alexa Fluor 488 Conjugate | Cell Signaling Technology | Cat#9174S; clone 58D6 | |||
| Phospho-S6 Ribosomal Protein (Ser240/244) XP Rabbit mAb, Alexa Fluor 488 Conjugate | Cell Signaling Technology | Cat#5018S; clone D68F8 | |||
| HA-Tag Mouse mAb, Alexa Fluor 647 Conjugate | Cell Signaling Technology | Cat#3444S; clone 6E2 | |||
| Ultra-LEAF Purified anti-human CD337 (NKp30) Antibody | BioLegend | Cat#325224; clone P30–15 | |||
| Anti-human CD107a (LAMP-1) Antibody, FITC | Biolegend | Cat#328606; clone H4A3 | |||
| BD GolgiPlug Protein Transport Inhibitor (containing Brefeldin A) | BD Biosciences | Cat#555029 | |||
| GolgiStop Protein Transport Inhibitor (Containing Monensin) | BD Biosciences | Cat#554724 | |||
| Fixation/Permeabilization Solution Kit | BD Biosciences | Cat#554714 | |||
| PE-Cy7 Mouse Anti-Human TNF | BD Biosciences | Cat#557647; clone Mab11 | |||
| Perm buffer III | BD Biosciences | Cat#558050 | |||
| Mouse Anti-Human CD8, PE | BD Biosciences | Cat#555635; clone HIT8a | |||
| Mouse Anti-Human MIP-1β, PE | BD Biosciences | Cat#550078; clone D21–1351 | |||
| Mouse Anti-Human IFN-γ, Alexa Fluor 647 | BD Biosciences | Cat#557729; clone B27 | |||
| Anti-human CD122 (IL-2Rβ) Antibody, APC | Biolegend | Cat#339008; clone TU27 | |||
| Anti-human CD3 Antibody, APC | Biolegend | Cat#300439; clone UCHT1 | |||
| Anti-human CD45RA Antibody, APC | Biolegend | Cat#304150; clone HI100 | |||
| Anti-human IFN-γ Antibody, APC | Biolegend | Cat#506510; clone B27 | |||
| Anti-human CD8 Antibody, APC/Cyanine7 | Biolegend | Cat#344714; clone SK1 | |||
| Anti-human CD56 (NCAM) Antibody, Brilliant Violet 605 | Biolegend | Cat#362538; clone 5.1H11 | |||
| Anti-human CD197 (CCR7) Antibody, Brilliant Violet 605 | Biolegend | Cat#353224; clone G043H7 | |||
| Anti-human CD8a Antibody, Brilliant Violet 605 | Biolegend | Cat#344742; clone SK1 | |||
| Anti-human CD4 Antibody, Brilliant Violet 785 | Biolegend | Cat#300554; clone RPA-T4 | |||
| Anti-human CD3 Antibody, FITC | Biolegend | Cat#300452; clone UCHT1 | |||
| Anti-human CD4 Antibody, FITC | Biolegend | Cat#300538; clone RPA-T4 | |||
| FITC anti-human CD69 Antibody | Biolegend | Cat#310904; Clone FN50 | |||
| Anti-human CD3 Antibody , Pacific Blue | Biolegend | Cat#300431; clone UCHT1 | |||
| Anti-human CD4 Antibody, Pacific Blue | Biolegend | Cat#300521; clone RPA-T4 | |||
| Anti-human IL-2 Antibody, Pacific Blue | Biolegend | Cat#500307; clone MQ117H12 | |||
| Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) Rabbit mAb, Alexa Fluor 647 Conjugate | Biolegend | Cat#13148s; clone 197G2 | |||
| Phospho-Akt (Ser473) XP Rabbit mAb, Alexa Fluor 647 Conjugate | Cell Signaling Technology | Cat#4075S; clone DE9 | |||
| Bacterial and Virus Strains | |||||
| Mix & Go Competent Cells - DH5α | Zymo Research | Cat#T3007 | |||
| Biological Samples | |||||
| Human peripheral mononuclear cells (PBMCs), isolated from leukocyte reduction shuttles | Stanford Blood Center | N/A | |||
| Chemicals, Peptides, and Recombinant Proteins | |||||
| SA sensor chip | Cytiva | Cat#BR-1005–31 | |||
| HBS-P | Cytiva | Cat#BR-1006–71 | |||
| EasySep Magnet | StemCell Technologies | Cat#18000 | |||
| Human CD8+ T cell Isolation Kit | Miltenyi | Cat#130–096-495 | |||
| LS magnetic selection column | Miltenyi | Cat#130–042-401 | |||
| EasySep Human Naïve Pan T Cell Isolation Kit | StemCell Technologies | Cat#17961 | |||
| ExpiFectamine 293 Transfection Kit | Gibco | Cat#A14525 | |||
| Sapphire Baculovirus DNA | Allele | Cat#ABP-BVD-10002 | |||
| RNeasy Plus Mini Kit | Qiagen | Cat#74134 | |||
| CellTrace Violet Proliferation Kit | Invitrogen | Cat#C34557 | |||
| Bio-Safe Coomassie G-250 Stain, 5L | Biorad | Cat#1610787 | |||
| UltraComp eBeads | eBiosciences | Cat#01–2222-42 | |||
| 16% Paraformaldehyde aqueous solution | Electron Microscopy Sciences | Cat#15710 | |||
| EndoH | Produced in house | N/A | |||
| Kifunensine | Toronto Research Chemicals | Cat#K450000 | |||
| BirA | Produced in house (Fairhead and Howarth, 2015) | N/A | |||
| Streptavidin | Sigma | Cat#189730 | |||
| Alexa Flour 647 C2 Maleimide | Invitrogen | Cat#A20347 | |||
| Human MSA-IL-2 | Produced in house (Glassman et al., 2021) | Produced in house | |||
| Human IFNω | Produced in house (Thomas et al., 2011) | Produced in house | |||
| Human IL-15 | R&D systems | Cat#247-ILB-025 | |||
| Human IL-7 | R&D systems | Cat#207-IL-010 | |||
| PE-AnnexinV | Biolegend | Cat#640947 | |||
| Ionomycin | Sigma | Cat#I9657–1MG | |||
| Phorbol 12-myristate 13-acetate (PMA) | Sigma | Cat#P8139–5MG | |||
| Cellfectin II | Gibco | Cat#10362100 | |||
| Carboxypeptidase A | Sigma | Cat#C9268 | |||
| Carboxypeptidase B | Sigma | Cat#217356 | |||
| Expi293 Expression Medium | Gibco | Cat#A1435101 | |||
| Sf-900 III Media | Invitrogen | Cat#12658019 | |||
| ESF 921 Insect Cell Culture Medium | Expression Systems | Cat#96–001-01 | |||
| MY140-F | This paper | N/A | |||
| MY140-R | This paper | N/A | |||
| MY141-F | This paper | N/A | |||
| MY141-R | This paper | N/A | |||
| MY177-F | This paper | N/A | |||
| MY177-R | This paper | N/A | |||
| MY171-F | This paper | N/A | |||
| MY171-R | This paper | N/A | |||
| MY144-F | This paper | N/A | |||
| MY144-R | This paper | N/A | |||
| MY145-F | This paper | N/A | |||
| MY145-R | This paper | N/A | |||
| MY178-F | This paper | N/A | |||
| MY178-R | This paper | N/A | |||
| MY172-F | This paper | N/A | |||
| MY172-R | This paper | N/A | |||
| MY142-F | This paper | N/A | |||
| MY142-R | This paper | N/A | |||
| MY143-F | This paper | N/A | |||
| MY143-R | This paper | N/A | |||
| MY179-F | This paper | N/A | |||
| MY179-R | This paper | N/A | |||
| MY173-F | This paper | N/A | |||
| MY173-R | This paper | N/A | |||
| MY188-F | This paper | N/A | |||
| MY188-R | This paper | N/A | |||
| MY189-F | This paper | N/A | |||
| MY189-R | This paper | N/A | |||
| MY190-F | This paper | N/A | |||
| MY190-R | This paper | N/A | |||
| MY191-F | This paper | N/A | |||
| MY191-R | This paper | N/A | |||
| MY192-F | This paper | N/A | |||
| MY192-R | This paper | N/A | |||
| MY193-F | This paper | N/A | |||
| MY193-R | This paper | N/A | |||
| MY194-F | This paper | N/A | |||
| MY194-R | This paper | N/A | |||
| MY195-F | This paper | N/A | |||
| MY195-R | This paper | N/A | |||
| IL2Rβ-Nb6 | This paper | N/A | |||
| γc-Nb6 | This paper | N/A | |||
| Human IL2Rβ ECD | This paper | N/A | |||
| Human γc ECD | This paper | N/A | |||
| HIS1 | This paper | N/A | |||
| HIS2 | This paper | N/A | |||
| HIS3 | This paper | N/A | |||
| HIS4 | This paper | N/A | |||
| HIS5 | This paper | N/A | |||
| HIS6 | This paper | N/A | |||
| HIS7 | This paper | N/A | |||
| A1 VHH monomer | This paper | N/A | |||
| 10Rβ1–2Rβ6 (8a.a. linker) | This paper | N/A | |||
| 10Rβ3–2Rβ6 (8a.a. linker) | This paper | N/A | |||
| 10Rβ4–2Rβ1 (8a.a. linker) | This paper | N/A | |||
| 2Rβ3–10Rβ1 (8a.a. linker) | This paper | N/A | |||
| 10Rβ1–2Rβ6 (0a.a. linker) | This paper | N/A | |||
| 10Rβ1–2Rβ6 (4a.a. linker) | This paper | N/A | |||
| 10Rβ1–2Rβ6 (12a.a. linker) | This paper | N/A | |||
| 10Rβ1–2Rβ6 (16a.a. linker) | This paper | N/A | |||
| 10Rβ1–2Rβ6-Fc | This paper | N/A | |||
| Critical Commercial Assays | |||||
| Superdex 200 Increase column 10/300 GL | Cytiva | Cat#28990944 | |||
| Superdex 75 Increase column 10/300 GL | Cytiva | Cat#29148721 | |||
| SA sensor chip | Cytiva | Cat#BR-1005–31 | |||
| HBS-P | Cytiva | Cat#BR-1006–71 | |||
| Yeast-Display Nanobody Library (NbLib) | Kerafast | Cat#EF0014-FP | |||
| Deposited Data | |||||
| γc:γc-Nb6 Complex Crystal Structure | RSCB | 7S2R | |||
| RNA-seq dataset | GEO | GSE183436 | |||
| IL2Rβ:β-Nb6 Complex Crystal Structure | RSCB | 7S2S | |||
| Experimental Models: Cell Lines | |||||
| Human: Expi293F | GIBCO | Cat#A14527 | |||
| Insect: Spodoptera frugiperda (Sf9) | ATCC | Cat#CRL-1711 | |||
| Insect: Trichoplusia ni (T. ni) | Expression Systems | Cat#94–002F | |||
| Human: YT-1 (CD25+) | N/A | ||||
| Human: K-562 | ATCC | Cat#CCL-243 | |||
| Human: A-375 | ATCC | Cat#CRL-1619 | |||
| Recombinant DNA | |||||
| DNA | |||||
| pD649 MY140-F | This paper | N/A | |||
| pD649 MY140-R | This paper | N/A | |||
| pD649 MY141-F | This paper | N/A | |||
| pD649 MY141-R | This paper | N/A | |||
| pD649 MY177-F | This paper | N/A | |||
| pD649 MY177-R | This paper | N/A | |||
| pD649 MY171-F | This paper | N/A | |||
| pD649 MY171-R | This paper | N/A | |||
| pD649 MY144-F | This paper | N/A | |||
| pD649 MY144-R | This paper | N/A | |||
| pD649 MY145-F | This paper | N/A | |||
| pD649 MY145-R | This paper | N/A | |||
| pD649 MY178-F | This paper | N/A | |||
| pD649 MY178-R | This paper | N/A | |||
| pD649 MY172-F | This paper | N/A | |||
| pD649 MY172-R | This paper | N/A | |||
| pD649 MY142-F | This paper | N/A | |||
| pD649 MY142-R | This paper | N/A | |||
| pD649 MY143-F | This paper | N/A | |||
| pD649 MY143-R | This paper | N/A | |||
| pD649 MY179-F | This paper | N/A | |||
| pD649 MY179-R | This paper | N/A | |||
| pD649 MY173-F | This paper | N/A | |||
| pD649 MY173-R | This paper | N/A | |||
| pD649 MY188-F | This paper | N/A | |||
| pD649 MY188-R | This paper | N/A | |||
| pD649 MY189-F | This paper | N/A | |||
| pD649 MY189-R | This paper | N/A | |||
| pD649 MY190-F | This paper | N/A | |||
| pD649 MY190-R | This paper | N/A | |||
| pD649 MY191-F | This paper | N/A | |||
| pD649 MY191-R | This paper | N/A | |||
| pD649 MY192-F | This paper | N/A | |||
| pD649 MY192-R | This paper | N/A | |||
| pD649 MY193-F | This paper | N/A | |||
| pD649 MY193-R | This paper | N/A | |||
| pD649 MY194-F | This paper | N/A | |||
| pD649 MY194-R | This paper | N/A | |||
| pD649 MY195-F | This paper | N/A | |||
| pD649 MY195-R | This paper | N/A | |||
| pD649 IL2Rβ-Nb6 | This paper | N/A | |||
| pD649 γc-Nb6 | This paper | N/A | |||
| pD649 | ATUM | Cat#PD649 | |||
| Software and Algorithms | |||||
| FlowJo v10.5 | Tree Star | RRID: SCR_008520 | |||
| GraphPad Prism 9.1.0 | GraphPad Software | RRID: SCR_002798 | |||
| BIAevaluation software | Cytiva | RRID: SCR_015936 | |||
| PHENIX | (Liebschner et al., 2019) | RRID:SCR_014224 | |||
| Coot | (Emsley et al., 2010) | RRID:SCR_014222 | |||
| SBGrid | (Morin et al., 2013) | RRID:SCR_003511 | |||
| DESEQ2 | (Love et al., 2014) | RRID:SCR_015687 | |||
| STAR | https://github.com/alexdobin/STAR/releases | RRID:SCR_004463 | |||
| UCSF ChimeraX | https://www.cgl.ucsf.edu/chimerax/ | RRID:SCR_015872 |


