Background:
Poststroke recovery depends on multiple factors and varies greatly across individuals. Using machine learning models, this study investigated the independent and complementary prognostic role of different patient-related factors in predicting response to language rehabilitation after a stroke.
Methods:
Fifty-five individuals with chronic poststroke aphasia underwent a battery of standardized assessments and structural and functional magnetic resonance imaging scans, and received 12 weeks of language treatment. Support vector machine and random forest models were constructed to predict responsiveness to treatment using pretreatment behavioral, demographic, and structural and functional neuroimaging data.
Results:
The best prediction performance was achieved by a support vector machine model trained on aphasia severity, demographics, measures of anatomic integrity and resting-state functional connectivity (F1=0.94). This model resulted in a significantly superior prediction performance compared with support vector machine models trained on all feature sets (F1=0.82, P<0.001) or a single feature set (F1 range=0.68–0.84, P<0.001). Across random forest models, training on resting-state functional magnetic resonance imaging connectivity data yielded the best F1 score (F1=0.87).
Conclusions:
While behavioral, multimodal neuroimaging data and demographic information carry complementary information in predicting response to rehabilitation in chronic poststroke aphasia, functional connectivity of the brain at rest after stroke is a particularly important predictor of responsiveness to treatment, both alone and combined with other patient-related factors.
Keywords: aphasia, language, machine learning, magnetic resonance imaging, neuroimaging, rehabilitation
Stroke is a leading cause of severe and complex long-term disability1 which affects various domains of social participation.2 Aphasia, one of the most devastating consequences of a stroke, affects approximately one-third of stroke survivors.3 A personalized prognosis on the evolution of aphasia not only helps patients and their relatives plan for the future but also provides guidance for clinicians to select the appropriate treatment. However, poststroke aphasia recovery varies widely across individuals4 and is influenced by multiple factors,5 which makes a prognosis difficult to determine for clinicians.
Previous studies have demonstrated that different factors can partially or independently explain the degree of spontaneous or treatment-related language recovery after stroke.5,6 Among the most consistent predictors found in the literature are initial aphasia severity7 and lesion size.8,9 Specific linguistic or nonlinguistic abilities at baseline assessment10,11 and demographic information5 may also play a role in the amount of language abilities recovered over time. However, the role of demographic data in therapy outcomes did not reach a consensus in the literature.12 Furthermore, better brain structural integrity9,13,14 and functional local activity and connectivity15–18 are positively related to the degree of spontaneous and treatment-related language recovery. Importantly, most of these studies investigated the value of individual variables or combined only a few of them. Therefore, it remains unclear (1) how each of the aforementioned factors comparatively predict natural language recovery and recovery after rehabilitation and (2) whether a combination of multiple factors is superior in prediction relative to a single type of factor. This question is important because clinicians need to know which types of data are necessary to provide an accurate prognosis to patients.
Recent advances in machine learning have allowed the application of multivariate analysis methods on multimodal neuroimaging data to predict language impairments at a single time point after brain damage19–22 or, in longitudinal studies, to predict natural language recovery over time after stroke.13,16,23,24 However, these studies present several limitations: the period of recovery investigated varied across participants,13,23 the amount of rehabilitation received by each individual was not controlled,13,16,23 and only one type of imaging data was included (ie, functional or structural).13,16,23,24
In this study, we sought to identify the independent and cumulative importance of behavioral, demographic, and multimodal structural and functional imaging data to predict treatment-related language recovery in chronic poststroke aphasia. Building on our pilot work,25 we investigate the efficacy of 2 different machine learning models, support vector machine (SVM) and random forest (RF), to predict the improvement in language ability after 12 weeks of rehabilitation. We hypothesized that model accuracy will be improved by the combination of behavioral, demographic, structural and functional imaging variables compared with single modality models.
Methods
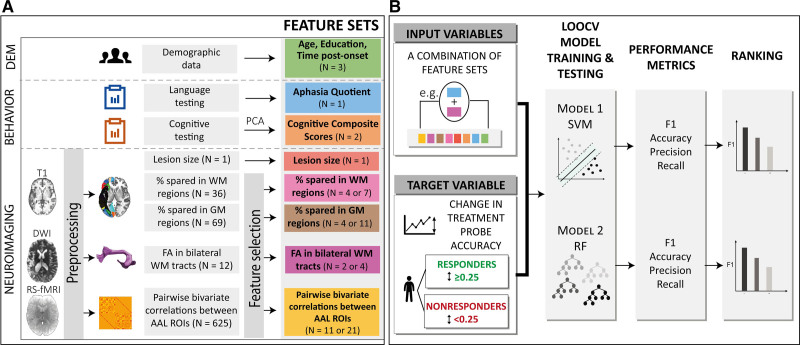
This study has been conducted in adherence to the TRIPOD guidelines (Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis; see the checklist in the Supplemental Material). Data can be shared upon request based on a formal data sharing agreement. Figure 1 presents the methodological framework of the study.
Figure 1.
Methodological framework of the study. A, Behavioral, demographic, and neuroimaging data were collected before the commencement of the treatment. Neuroimaging data were preprocessed and feature selection was performed on feature sets with a high number of variables. B, All combinations of feature sets (N=255) were tested as input to the support vector machine (SVM) and random forest (RF) models to classify participants into responders and nonresponders. F1 score was used to rank models’ performance. AAL indicates Automated Anatomical Labeling atlas; DEM, demographics; DWI, diffusion weighted-imaging; FA, fractional anisotropy; GM, gray matter; LOOCV, leave-one-out cross-validation; PCA, principal component analysis; ROI, region of interest; RS-fMRI, resting-state functional MRI; and WM, white matter.
Participants
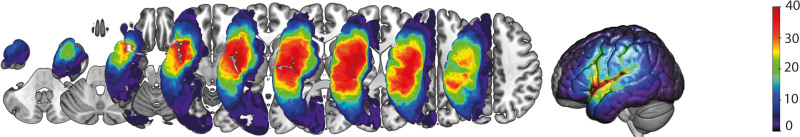
Participants were 55 individuals (37 male) with chronic poststroke aphasia due to a single left-hemisphere stroke (mean age =58.8 years, mean time poststroke onset =59.0 months, mean education =15.8 years). For individual demographic data and aphasia severity (Table S1). Participants were selected from a larger sample (n=81) who were enrolled in a multi-site study between 2015 and 2018 examining neurobiological features of aphasia recovery (https://cnlr.northwestern.edu/). Of the full sample, 55 participants were included based on data availability of all input features of interest for this investigation (see flowchart in Figure S1). The participants included in this study were recruited at Boston University (N=30), Johns Hopkins University (N=16), and Northwestern University (N=9). Figure 2 presents the lesion distribution of all participants. Exclusion criteria included premorbid neurological disease, history of multiple left-hemisphere strokes, and contraindications for magnetic resonance imaging (MRI). All participants provided written informed consent before study participation. The study protocol was approved by the institutional review boards at Boston University, Massachusetts General Hospital, Northwestern University, and Johns Hopkins University.
Figure 2.
Lesion overlay for all participants. Z coordinates: −40 −30 −20 −10 0 10 20 30 40.
Behavioral Assessment and Treatment
Participants completed a battery of standardized assessments at baseline. The Western Aphasia Battery—Revised26 was used to assess aphasia severity per the aphasia quotient (AQ). The Doors and People test,27 the Wechsler Adult Intelligence Scale Digit Span,28 the Raven’s Coloured Progressive Matrices,26 the Corsi block-tapping test,29 and the Serial Reaction Time Task30 were used to assess overall cognitive function. Participants at 3 sites received different types of language treatments (see Methods in the Supplemental Material). The treatment protocols and the successful results of these treatments have been reported elsewhere.11,15,31–34 For the purposes of this study, data for these patients are collapsed as we examine responsiveness to overall language rehabilitation (and not to any treatment type in particular).
MRI Data Acquisition
MRI was completed on a Siemens 3T Skyra with a 20-channel head/neck coil at the Martinos Center in Charlestown, MA, for Boston University; on a Siemens TIM Trio with a 32-channel head coil or a Siemens Prisma with a 64-channel head/neck coil at the Center for Translational Imaging in Chicago, IL, for Northwestern University; and on a Philips Intera with a 32-channel head coil at Johns Hopkins University. Imaging protocols were harmonized across sites to ensure similar quality and timing and these protocols have been reported in previous papers.35–37 Structural imaging included a T1-weighted sagittal sequence (voxel size=1×1×1 mm3), and a high-resolution whole-brain cardiac-gated diffusion-weighted imaging sequence (voxel size=1.983×1.983×2.000 mm3, 72 interleaved slices with 60 gradient directions and 10 nondiffusion weighted (b=0) volumes, b value=1500 s/mm2). Whole-brain functional images were collected using a gradient-echo T2*-weighted sequence (voxel size=1.72×1.72×3 mm3). Complete imaging sequence parameters are provided in the Supplemental Material.
MRI Data Preprocessing
Several brain structural and functional measures were calculated including (1) lesion volume extracted through in-house MATLAB scripts9 from lesion masks manually drawn using MRIcron software38 and normalized to MNI space, (2) the integrity of gray and white matter regions calculated by computing the percentage of spared tissue in 69 left-hemisphere gray matter regions of the Automated Anatomical Labeling atlas,39 and 36 white matter tracts of the BCBToolKit40 probabilistic atlas, respectively, (3) average fractional anisotropy (FA) in 12 tracts of interest, computed from diffusion tensor imaging data preprocessed using the Advanced Diffusion Preprocessing Pipeline41 and the Northwestern University Neuroimaging Data Archive,42 and converted to tracts using the Automated Fiber Quantification software (deterministic tractography),43 and (4) resting-state functional MRI (rs-fMRI) connectivity, involving the rs-fMRI data first preprocessed in fMRIPrep version 1.4.1, a Nipype-based tool,44 and then through the CONN toolbox45 to extract bivariate Fisher-transformed Pearson correlations between 50 bilateral anatomic regions of interest specified using the Automated Anatomical Labeling atlas 3,46 resulting in 625 pairwise correlations per individual.
Methodological details on the MRI preprocessing are available in the Supplemental Material.
Model Development and Evaluation
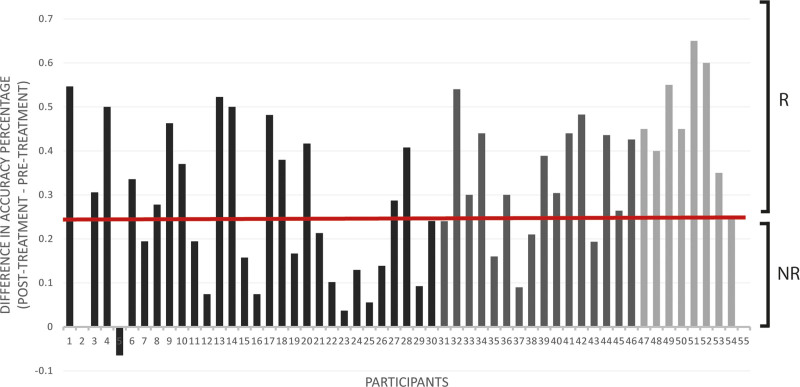
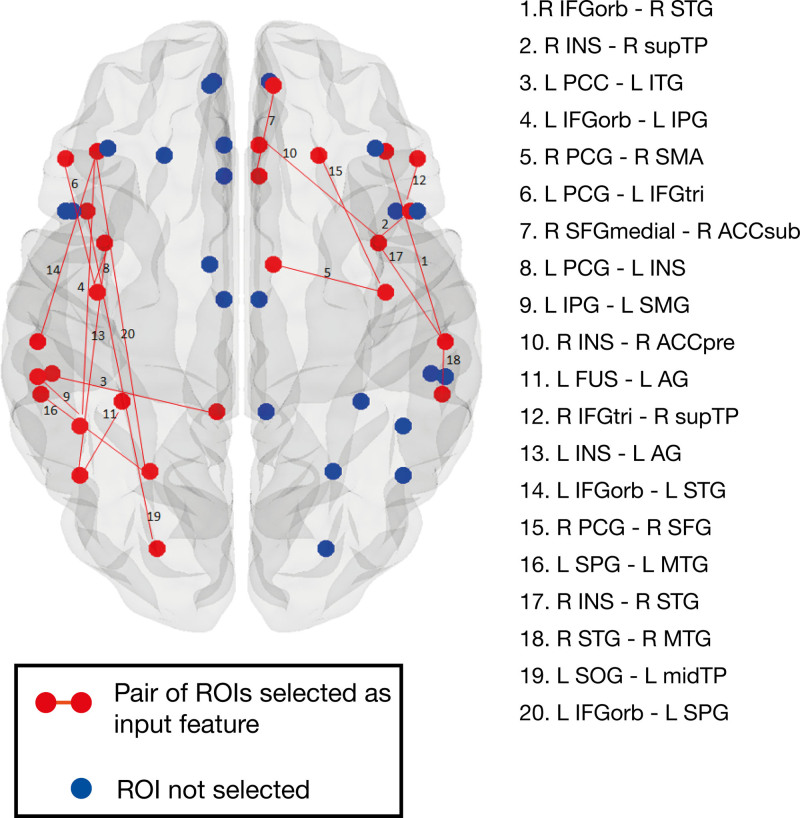
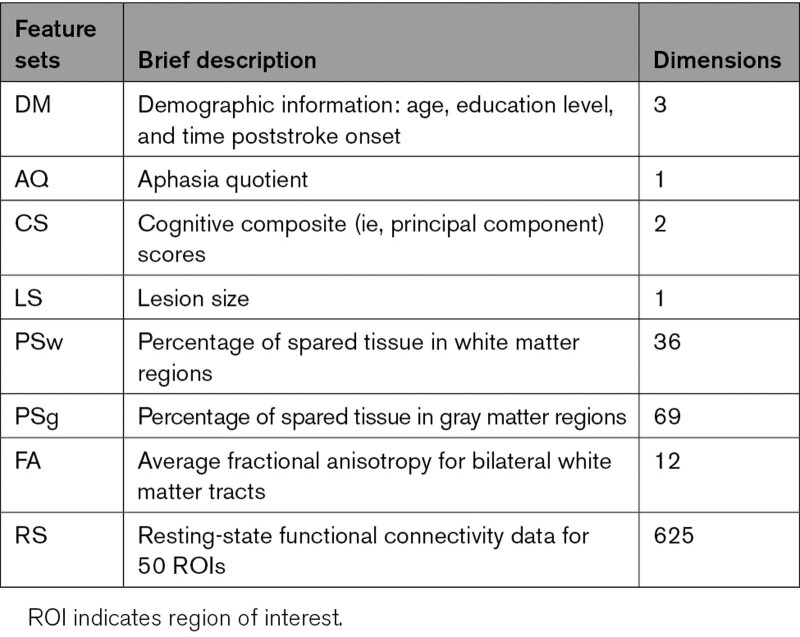
The construction and comparison of classification models predicting treatment response included several steps detailed in the Supplemental Material. First, responsiveness to rehabilitation was determined by calculating the change in accuracy percentage on the treatment probes. Participants were classified into responders (n=33) and nonresponders (n=22) based on a cutoff of 25 percentage points change in accuracy (Figure 3 and Supplemental Material). Second, given the large number of variables in the feature sets (Table), dimensionality reduction methods were used. The total scores of the 6 neuropsychological tests assessing overall cognitive function were entered in a varimax-rotated principal component analysis, performed in R, version 3.4.3.47 Two components (hereafter, cognitive composite scores), representing visuospatial processing and verbal working memory, were selected and explained 63% of the variance of the data (Table S2 and Supplemental Material). Additionally, some neuroimaging feature sets, namely percentage spared in white matter regions, percentage spared in gray matter regions, FA, and resting-state fMRI (RS) data, had dimensions much larger than any of the rest: 36, 69, 12, and 625, respectively. Therefore, supervised feature selection48 was performed on these variables and Pearson correlation coefficients between all selected features were computed (Figure 4 and Tables S3 and S4 and Figure S2).
Figure 3.
Change in accuracy on the treatment probes with accuracy measured as a percentage. Participants were classified into responders (R) and nonresponders (NR) to treatment based on a cutoff at 0.25. The different shades correspond to each participant’s site: Boston University (BU; dark), Johns Hopkins University (JHU; dark gray), and Northwestern University (NU; light gray).
Figure 4.
Resting-state functional connectivity features. Red dot and lines represent functional regions and connections selected after feature selection and included in the machine learning models. Blue dots represent regions excluded from the analyses after feature selection. ACC indicates anterior cingulate cortex; AG, angular gyrus; FUS, fusiform gyrus; IFG, inferior frontal gyrus; INS, insula; IPG, inferior parietal gyrus; ITG, inferior temporal gyrus; L, left; midTP, middle temporal pole; MTG, middle temporal gyrus; orb, pars orbitalis; PCC, posterior cingulate gyrus; PCG, precentral gyrus; pre, pregenual; R, right; ROI, region of interest; SFG, superior frontal gyrus; STG, superior temporal gyrus; supTP, superior temporal pole; SMA, supplementary motor area; sub, subgenual; SMG, supramarginal; SOG, superior occipital gyrus; SPG, superior parietal gyrus; and tri, pars triangularis.
Table.
List and Dimensions of Feature Sets (Before Feature Selection)
SVM and RF models were trained, tuned, and tested using a leave-one-out cross-validation procedure on all possible combinations of feature sets, including individual feature sets, to predict treatment response labels. Training and testing steps, hyperparameter values and details of fine-tuning are provided in the Supplemental Material and Tables S5 and S6. Prediction performance was evaluated using 4 metrics: accuracy, F1 score, precision, and recall, capturing different types of prediction errors (Supplemental Material). The F1 score was selected as the primary metric because it is less affected by an imbalanced class distribution (as is the case in this data set) than other metrics, and allows researchers to evaluate models based on a balance between precision and recall. Finally, distributions of F1 scores were computed for each of these models using a leave-one-out approach by iteratively removing one sample and computing the F1 scores based on performance on the 54 other samples. These scores were used in 2-tailed Wilcoxon signed-rank tests49 (function wilcox.test in R version 3.4.347) to compare the prediction performance of SVM/RF models based on a single feature set with (1) the optimal SVM/RF model, that is, the SVM/RF model trained on the feature set combination that resulted in the highest average F1 score and (2) the SVM/RF model trained on all feature sets combined. Statistical comparison was considered significant at an alpha level of 0.05.
Results
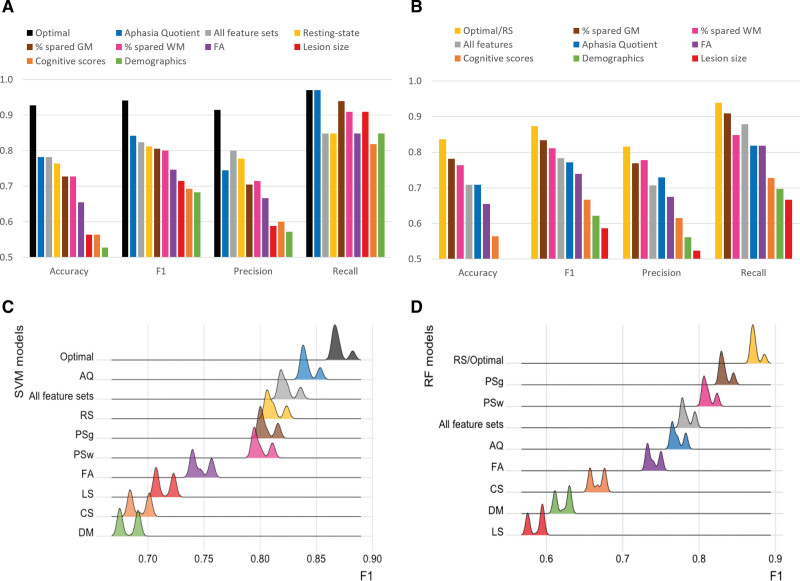
Figure 5A and 5B present all evaluation metrics for each individual-feature-set model, the all-feature-sets model and the optimal model (ie, best F1 score). Figure 5C and 5D show the distribution of F1 scores for each of these 3 model types. In addition, Tables S7 and S8 show evaluation metrics for (1) the top 20 models trained on the best combinations of feature sets (ranked by F1 score), (2) the model trained on all feature sets, and (3) individual-feature-set models, for both SVM and RF, respectively. Results of statistical comparisons between these 3 types of models are presented in Table S9.
Figure 5.
Predictive performance for support vector machine (SVM) and random forest (RF) models trained on a single feature set, all feature sets or the optimal combination of feature sets (aphasia quotient [AQ], demographics [DM], fractional anisotropy [FA], percentage spared in gray matter regions [PSg], resting-state [RS] for SVM and RS for RF). F1 scores of models trained on a single feature set were significantly lower than the models trained on the optimal combination of feature sets (P<0.001). A, Performance of SVM models on all 55 samples; (B) performance of RF models on all 55 samples; (C) distribution (kernel density estimation in R with automatic bandwidth selection) of SVM F1 scores computed from all 55 subsets of 54 samples each; and (D) distribution (kernel density estimation in R with automatic bandwidth selection) of RF F1 scores computed from all 55 subsets of 54 samples each.
Across all models and all evaluation metrics, responsiveness to rehabilitation at the chronic stage was best predicted by SVM models including multiple feature sets, with maximum accuracy (0.927), F1 (0.941), precision (0.914), and recall values (0.970). The optimal SVM model included aphasia severity, demographics, FA, percentage of spared tissue in gray matter regions and resting-state fMRI connectivity, and performed significantly better than any of the models trained on an individual feature set (P<0.001). This overall best performing model correctly classified 51/55 participants as responders and nonresponders to language treatment. All the top 10 SVM models (F1 scores ranging from 0.941 to 0.909 and accuracy values from 0.927 to 0.891) included combined information on the structural integrity of the brain (ie, percentage spared in gray matter regions, percentage of spared tissue in white matter regions, or LS) and the neural activity patterns at rest (ie, RS). Most of these combinations (8/10) also contained information on the behavioral performance (ie, AQ or cognitive composite scores).
Surprisingly, the SVM model using all feature sets (accuracy=0.782, F1=0.824, precision=0.800, and recall=0.848), did not perform better than aforementioned optimal SVM model (ie, AQ+demographics+FA+percentage spared in gray matter regions+RS; P<0.001). Notably, among SVM models trained on an individual feature set, aphasia severity (ie, AQ) and rs-fMRI connectivity (ie, RS) independently provided the best prediction performance scores, with an accuracy of 0.782 and 0.764 and an F1 score of 0.842 and 0.812, respectively.
Compared with SVM models, the top 10 RF models resulted in lower prediction performance. Three equally optimal RF models (accuracy=0.836, F1=0.873, precision=0.816, and recall=0.939) included RS alone, LS+RS and AQ+demographics+FA+RS. These top models correctly classified 46/55 participants as responders and nonresponders. Most of the top 10 RF models (9/10) included information on functional connectivity (ie, RS), and some models included information about the structural integrity of the brain (ie, LS, percentage spared in gray matter regions, percentage of spared tissue in white matter regions, or FA), the behavioral performance (ie, AQ or cognitive composite scores), and demographics. Similar to SVM models, the combination of all feature sets (accuracy=0.709, F1=0.784, precision=0.707, and recall=0.879) did not improve the prediction performance compared with the optimal models combining a subset of the feature sets (P<0.001).
Across the top combinations of feature sets ranked by F1 scores, the cumulative occurrence of each feature set shows that RS is predominant in both SVM and RF models, followed by percentage spared in gray matter regions in SVM models (Figure S3).
Discussion
Previous studies from our group on a subset of these data showed that brain function data,15,36 brain structure information,9,14 and language and cognitive performance11,36 independently predict treatment-related outcomes. This study is the first, to our knowledge, to investigate the cumulative importance of patient-related information using a comprehensive data set including behavioral, demographic, and multimodal neuroimaging information to predict rehabilitation-induced language recovery in individuals with chronic poststroke aphasia. Our results show that models that combine a subset of multimodal neuroimaging, behavioral, and demographic data outperform models trained on a single type of information and, more importantly, all the available types of information. Three types of patient-related data were consistently important in predicting responsiveness to language treatment: functional connectivity at rest (ie, rs-fMRI), the anatomical integrity, and the aphasia severity. Previous studies that have examined predictors of natural language recovery over time also demonstrated that combining information from language tests and structural MRI50,51 or task-based fMRI16 improved prognosis accuracy. In this study, we found an additional benefit of combining both functional and structural MRI information with behavioral abilities to predict responsiveness to language rehabilitation. Interestingly, the present study also demonstrates that combining all data types may not provide the best estimates for treatment outcomes. Instead, the results show that a combination of features that provide unique and salient information are sufficient in the optimal model.
Importantly, not all relevant behavioral, demographic and multimodal neuroimaging variables were equally important for the prognosis of language recovery after rehabilitation. In addition to confirming the importance of aphasia severity in predicting treatment-related language recovery,5 the present study showed that the status of neural connectivity at rest, even in the chronic stage, strongly predicts response to language treatment. Indeed, resting-state connectivity data contributed to all top performing models, regardless of the algorithm used. While structural neuroimaging data, such as lesion size or percentage of spared tissue in brain regions, informed the model on the extent and location of the initial damage, rs-fMRI connectivity between several undamaged left-hemisphere and a few right-hemisphere regions (Figure 4 and Table S4) provided information on the status of brain functional (re)organization and potential for relearning.52 Interestingly, all pairs of functional regions of interest selected in the models consisted of at least one region of interest from the language network, and all white matter tracts retained from the diffusion imaging data set corresponded to ventral or dorsal streams involved in language processing suggesting the importance of the left hemisphere in predicting recovery. In contrast, the regions where the degree of spared tissue was highly correlated with treatment response and selected as part of the final percentage of spared tissue feature sets, were not circumscribed to the left language network. Future studies are needed to draw specific conclusions on the relative importance of individual brain regions.
This study also shows that the choice of machine learning algorithm can influence the prediction performance. On our data set, SVM models seemed to better leverage complementary information from the patient-related data than RF models, resulting in higher prediction performance. These differences, however, may be related to not only the characteristics of each algorithm but also to the small sample size. Thus, differences between models should be interpreted with caution and replication studies are needed to generalize these findings.53
Importantly, this study demonstrates that language recovery after rehabilitation is multifactorial. In particular, language abilities at baseline, brain structural integrity and functional connectivity comprise unique and complementary information that can improve language treatment prognosis estimations. Machine learning models presented in this study are a first step towards a more personalized treatment approach for individuals with poststroke aphasia. By leveraging behavioral and multimodal neuroimaging data, future models trained on data from a larger sample size and different treatment types could assist clinicians with targeting the best treatment approach for individuals with poststroke aphasia. As a result of this advanced personalization, stronger evidence of the effectiveness of language treatment at the chronic stage could also be used to advocate for more insurance coverage beyond a few months after stroke.
Limitations
Although the sample size of this study was limited due to the availability of the multimodal MRI data, it was still representative of the heterogeneity among individuals with poststroke aphasia in terms of aphasia severity and treatment response (Table S1). Previous studies demonstrated that small sample sizes can lead to less accurate results when using machine learning models on neuroimaging data to predict behavior.54 Therefore, we performed feature selection on all eligible feature sets to improve learning accuracy and model stability. As part of this multi-site project, all participants received an impairment-based treatment of the same intensity, yet targeting a different language impairment at each site (ie, naming, syntax, and spelling). Importantly, the goal of this study was to investigate patient-specific factors that would predict recovery after language treatment and not to investigate treatment-related factors (eg, intensity, duration, and language target). Although differences in treatments may add noise to the results of this study, the accuracy-maximizing prediction based on site-information alone is to predict all participants as responders, irrespective of the site, as Figure 3 shows. This is so since at each site, the number of responders either equals or exceeds the number of nonresponders. Thus, site as a feature may have only a limited influence on performance.
Ideally, we should use k-fold cross-validation in all computer experiments, but we used leave-one-out cross-validation for both feature selection and model training and validation steps. Furthermore, the samples used in both steps should be different, but we used the same set of samples in both steps. Both decisions were motivated by the limited total number of samples in the data set. This approach is commonly used and is particularly appropriate for small sample sizes.55 Thus, testing models on a larger and independent data set will be needed in future studies to improve the generalizability of these results.
Article Information
Acknowledgments
We thank all individuals with aphasia who participated in this study and their families. We additionally express our gratitude to past and present members of the Boston University Aphasia Research Laboratory. We also acknowledge the work of our collaborators through the Center for the Neurobiology of Language Recovery.
Sources of Funding
This study was funded by the National Institutes of Health/National Institute on Deafness and Other Communication Disorders, Clinical Research Center Grant (P50DC012283), the Hariri Institute Artificial Intelligence Research Initiative and the Institute for Health System Innovation Policy at Boston University.
Disclosures
Dr Kiran is a scientific advisor for Constant Therapy Health, but there is no overlap between this role and the submitted investigation. The other authors report no conflicts.
Supplemental Material
Supplemental Materials and Methods
Tables S1–S10
Figures S1–S4
Supplementary Material
Nonstandard Abbreviations and Acronyms
- AQ
- aphasia quotient
- FA
- fractional anisotropy
- LS
- lesion size
- MRI
- magnetic resonance imaging
- RF
- random forest
- rs-fMRI or RS
- resting-state functional magnetic resonance imaging
- SVM
- support vector machine
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/STROKEAHA.121.036749.
For Sources of Funding and Disclosures, see page 1613.
Contributor Information
Sha Lai, Email: lais823@bu.edu.
Maria Varkanitsa, Email: mvarkan@bu.edu.
Emily J. Braun, Email: ejbraun@bu.edu.
Brenda Rapp, Email: brapp1@jhu.edu.
Todd B. Parrish, Email: toddp@northwestern.edu.
James Higgins, Email: james.higgins@northwestern.edu.
Ajay S. Kurani, Email: ajay.kurani@northwestern.edu.
David Caplan, Email: DCAPLAN@partners.org.
Cynthia K. Thompson, Email: ckthom@northwestern.edu.
Prakash Ishwar, Email: pi@bu.edu.
Margrit Betke, Email: betke@bu.edu.
Swathi Kiran, Email: kirans@bu.edu.
References
- 1.Adamson J, Beswick A, Ebrahim S. Is stroke the most common cause of disability? J Stroke Cerebrovasc Dis. 2004;13:171–177. doi: 10.1016/j.jstrokecerebrovasdis.2004.06.003 [DOI] [PubMed] [Google Scholar]
- 2.Daniel K, Wolfe CD, Busch MA, McKevitt C. What are the social consequences of stroke for working-aged adults? A systematic review. Stroke. 2009;40:e431–e440. doi: 10.1161/STROKEAHA.108.534487 [DOI] [PubMed] [Google Scholar]
- 3.Kauhanen ML, Korpelainen JT, Hiltunen P, Määttä R, Mononen H, Brusin E, Sotaniemi KA, Myllylä VV. Aphasia, depression, and non-verbal cognitive impairment in ischaemic stroke. Cerebrovasc Dis. 2000;10:455–461. doi: 10.1159/000016107 [DOI] [PubMed] [Google Scholar]
- 4.Wilson SM, Eriksson DK, Brandt TH, Schneck SM, Lucanie JM, Burchfield AS, Charney S, Quillen IA, de Riesthal M, Kirshner HS, et al. Patterns of recovery from aphasia in the first 2 weeks after stroke. J Speech Lang Hear Res. 2019;62:723–732. doi: 10.1044/2018_JSLHR-L-18-0254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watila MM, Balarabe SA. Factors predicting post-stroke aphasia recovery. J Neurol Sci. 2015;352:12–18. doi: 10.1016/j.jns.2015.03.020 [DOI] [PubMed] [Google Scholar]
- 6.Plowman E, Hentz B, Ellis C, Jr. Post-stroke aphasia prognosis: a review of patient-related and stroke-related factors. J Eval Clin Pract. 2012;18:689–694. doi: 10.1111/j.1365-2753.2011.01650.x [DOI] [PubMed] [Google Scholar]
- 7.Lazar RM, Speizer AE, Festa JR, Krakauer JW, Marshall RS. Variability in language recovery after first-time stroke. J Neurol Neurosurg Psychiatry. 2008;79:530–534. doi: 10.1136/jnnp.2007.122457 [DOI] [PubMed] [Google Scholar]
- 8.Benghanem S, Rosso C, Arbizu C, Moulton E, Dormont D, Leger A, Pires C, Samson Y. Aphasia outcome: the interactions between initial severity, lesion size and location. J Neurol. 2019;266:1303–1309. doi: 10.1007/s00415-019-09259-3 [DOI] [PubMed] [Google Scholar]
- 9.Meier EL, Johnson JP, Pan Y, Kiran S. The utility of lesion classification in predicting language and treatment outcomes in chronic stroke-induced aphasia. Brain Imaging Behav. 2019;13:1510–1525. doi: 10.1007/s11682-019-00118-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lambon Ralph MA, Snell C, Fillingham JK, Conroy P, Sage K. Predicting the outcome of anomia therapy for people with aphasia post CVA: both language and cognitive status are key predictors. Neuropsychol Rehabil. 2010;20:289–305. doi: 10.1080/09602010903237875 [DOI] [PubMed] [Google Scholar]
- 11.Gilmore N, Meier EL, Johnson JP, Kiran S. Nonlinguistic cognitive factors predict treatment-induced recovery in chronic poststroke aphasia. Arch Phys Med Rehabil. 2019;100:1251–1258. doi: 10.1016/j.apmr.2018.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leff AP, Nightingale S, Gooding B, Rutter J, Craven N, Peart M, Dunstan A, Sherman A, Paget A, Duncan M, et al. Clinical effectiveness of the queen square intensive comprehensive aphasia service for patients with poststroke aphasia. Stroke. 2021;52:e594–e598. doi: 10.1161/STROKEAHA.120.033837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hope TM, Seghier ML, Leff AP, Price CJ. Predicting outcome and recovery after stroke with lesions extracted from MRI images. Neuroimage Clin. 2013;2:424–433. doi: 10.1016/j.nicl.2013.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varkanitsa M, Peñaloza C, Charidimou A, Caplan D, Kiran S. White matter hyperintensities predict response to language treatment in poststroke aphasia. Neurorehabil Neural Repair. 2020;34:945–953. doi: 10.1177/1545968320952809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson JP, Meier EL, Pan Y, Kiran S. Pre-treatment graph measures of a functional semantic network are associated with naming therapy outcomes in chronic aphasia. Brain Lang. 2020;207:104809. doi: 10.1016/j.bandl.2020.104809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saur D, Ronneberger O, Kümmerer D, Mader I, Weiller C, Klöppel S. Early functional magnetic resonance imaging activations predict language outcome after stroke. Brain. 2010;133(Pt 4):1252–1264. doi: 10.1093/brain/awq021 [DOI] [PubMed] [Google Scholar]
- 17.Tao Y, Rapp B. The effects of lesion and treatment-related recovery on functional network modularity in post-stroke dysgraphia. Neuroimage Clin. 2019;23:101865. doi: 10.1016/j.nicl.2019.101865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purcell JJ, Wiley RW, Rapp B. Re-learning to be different: increased neural differentiation supports post-stroke language recovery. Neuroimage. 2019;202:116145. doi: 10.1016/j.neuroimage.2019.116145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pustina D, Branch Coslett H, Ungar L, Faseyitan OK, Medaglia JD, Avants B, Schwartz MF. Enhanced estimations of post‐stroke aphasia severity using stacked multimodal predictions. Hum Brain Mapp. 2017;38:5603–5615. doi: 10.1002/hbm.23752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halai AD, Woollams AM, Lambon Ralph MA. Investigating the effect of changing parameters when building prediction models for post-stroke aphasia. Nat Hum Behav. 2020;4:725–735. doi: 10.1038/s41562-020-0854-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hope TMH, Leff AP, Price CJ. Predicting language outcomes after stroke: is structural disconnection a useful predictor? Neuroimage Clin. 2018;19:22–29. doi: 10.1016/j.nicl.2018.03.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kristinsson S, Zhang W, Rorden C, Newman-Norlund R, Basilakos A, Bonilha L, Yourganov G, Xiao F, Hillis A, Fridriksson J. Machine learning-based multimodal prediction of language outcomes in chronic aphasia. Hum Brain Mapp. 2021;42:1682–1698. doi: 10.1002/hbm.25321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roohani YH, Sajid N, Madhyastha P, Price CJ, Hope TMH. Predicting Language Recovery after Stroke with Convolutional Networks on Stitched MRI. arXiv:1811.10520. 2018. http://arxiv.org/abs/1811.10520
- 24.Lahiri D, Dubey S, Ardila A, Sanyal D, Ray BK. Determinants of aphasia recovery: exploratory decision tree analysis. Lang Cogn Neurosci. 2020;0:1–8. [Google Scholar]
- 25.Lai S, Billot A, Varkanitsa M, Braun E, Rapp B, Parrish T, Kurani A, Higgins J, Caplan D, Thompson C, et al. An exploration of machine learning methods for predicting post-stroke aphasia recovery. In: The 14th PErvasive Technologies Related to Assistive Environments Conference. New York, NY, USA: Association for Computing Machinery; 2021. pp. 556–564. 10.1145/3453892.3461319 [DOI] [Google Scholar]
- 26.Kertesz A. WAB-R: Western Aphasia Battery-Revised. PsychCorp; 2007. [Google Scholar]
- 27.Baddeley AD, Emslie H, Nimmo-Smith I. Doors and People: A Test of Visual and Verbal Recall and Recognition. Harcourt Assessment; 2006. [Google Scholar]
- 28.Weschler D. Weschler adult intelligence scale. Archives of Clinical Neuropsychology. 1955. https://scholar.google.com/scholar?cluster=12516842848265392073&hl=fr&as_sdt=40000005&sciodt=0,22.
- 29.Kessels RP, van Zandvoort MJ, Postma A, Kappelle LJ, de Haan EH. The Corsi Block-Tapping Task: standardization and normative data. Appl Neuropsychol. 2000;7:252–258. doi: 10.1207/S15324826AN0704_8 [DOI] [PubMed] [Google Scholar]
- 30.Nissen MJ, Bullemer P. Attentional requirements of learning: evidence from performance measures. Cogn Psychol. 1987;19:1–32. [Google Scholar]
- 31.Gilmore N, Meier EL, Johnson JP, Kiran S. Typicality-based semantic treatment for anomia results in multiple levels of generalisation. Neuropsychol Rehabil. 2020;30:802–828. doi: 10.1080/09602011.2018.1499533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barbieri E, Mack J, Chiappetta B, Europa E, Thompson CK. Recovery of offline and online sentence processing in aphasia: Language and domain-general network neuroplasticity. Cortex. 2019;120:394–418. doi: 10.1016/j.cortex.2019.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shea J, Wiley R, Moss N, Rapp B. Pseudoword spelling ability predicts response to word spelling treatment in acquired dysgraphia. Neuropsychol Rehabil. 2020;1–37. doi: 10.1080/09602011.2020.1813596 [DOI] [PubMed] [Google Scholar]
- 34.Rapp B, Wiley RW. Re-learning and remembering in the lesioned brain. Neuropsychologia. 2019;132:107126. doi: 10.1016/j.neuropsychologia.2019.107126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Higgins J, Barbieri E, Wang X, Mack J, Caplan D, Kiran S, Rapp B, Thompson C, Zinbarg R, Parrish T. Reliability of BOLD signals in chronic stroke‐induced aphasia. Eur J Neurosci. 2020;52:3963–3978. doi: 10.1111/ejn.14739 [DOI] [PubMed] [Google Scholar]
- 36.Iorga M, Higgins J, Caplan D, Zinbarg R, Kiran S, Thompson CK, Rapp B, Parrish TB. Predicting language recovery in post-stroke aphasia using behavior and functional MRI. Sci Rep. 2021;11:8419. doi: 10.1038/s41598-021-88022-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lukic S, Thompson CK, Barbieri E, Chiappetta B, Bonakdarpour B, Kiran S, Rapp B, Parrish TB, Caplan D. Common and distinct neural substrates of sentence production and comprehension. Neuroimage. 2021;224:117374. doi: 10.1016/j.neuroimage.2020.117374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rorden C, Brett M. Stereotaxic display of brain lesions. Behav Neurol. 2000;12:191–200. doi: 10.1155/2000/421719 [DOI] [PubMed] [Google Scholar]
- 39.Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978 [DOI] [PubMed] [Google Scholar]
- 40.Foulon C, Cerliani L, Kinkingnéhun S, Levy R, Rosso C, Urbanski M, Volle E, Thiebaut de Schotten M. Advanced lesion symptom mapping analyses and implementation as BCBtoolkit. Gigascience. 2018;7:1–17. doi: 10.1093/gigascience/giy004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurani AI & Neuroimaging Laboratory. Advanced Diffusion Preprocessing Pipeline. 2020. [cited 2021 May 31]. https://www.kuranilab.fsm.northwestern.edu/software/adpp/
- 42.Alpert K, Kogan A, Parrish T, Marcus D, Wang L. The Northwestern University Neuroimaging Data Archive (NUNDA). Neuroimage. 2016;124(Pt B):1131–1136. doi: 10.1016/j.neuroimage.2015.05.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yeatman JD, Dougherty RF, Myall NJ, Wandell BA, Feldman HM. Tract profiles of white matter properties: automating fiber-tract quantification. PLoS One. 2012;7:e49790. doi: 10.1371/journal.pone.0049790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Esteban O, Markiewicz CJ, Blair RW, Moodie CA, Isik AI, Erramuzpe A, Kent JD, Goncalves M, DuPre E, Snyder M, et al. fMRIPrep: a robust preprocessing pipeline for functional MRI. Nat Methods. 2019;16:111–116. doi: 10.1038/s41592-018-0235-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whitfield-Gabrieli S, Nieto-Castanon A. Conn: a functional connectivity toolbox for correlated and anticorrelated brain networks. Brain Connect. 2012;2:125–141. doi: 10.1089/brain.2012.0073 [DOI] [PubMed] [Google Scholar]
- 46.Rolls ET, Huang CC, Lin CP, Feng J, Joliot M. Automated anatomical labelling atlas 3. Neuroimage. 2020;206:116189. doi: 10.1016/j.neuroimage.2019.116189 [DOI] [PubMed] [Google Scholar]
- 47.R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2018. https://www.R-project.org/. [Google Scholar]
- 48.Lai S, Billot A, Varkanitsa M, Braun EJ, Rapp B, Parrish TB, Higgins J, Caplan D, Thompson CK, Kiran S, et al. Predicting Post-stroke Aphasia Recovery: incorporating cognitive and brain imaging data. In: Proceedings of the 14th ACM International Conference on PErvasive Technologies Related to Assistive Environments. 2021. [Google Scholar]
- 49.Wilcoxon F. Individual comparisons by ranking methods. In: Breakthroughs in Statistics. Springer; 1992:196–202. [Google Scholar]
- 50.Osa García A, Brambati SM, Brisebois A, Désilets-Barnabé M, Houzé B, Bedetti C, Rochon E, Leonard C, Desautels A, Marcotte K. Predicting early post-stroke aphasia outcome from initial aphasia severity. Front Neurol. 2020;11:120. doi: 10.3389/fneur.2020.00120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tábuas-Pereira M, Beato-Coelho J, Ribeiro J, Nogueira AR, Cruz L, Silva F, Sargento-Freitas J, Cordeiro G, Santana I. Single word repetition predicts long-term outcome of aphasia caused by an Ischemic Stroke. J Stroke Cerebrovasc Dis. 2020;29:104566. doi: 10.1016/j.jstrokecerebrovasdis.2019.104566 [DOI] [PubMed] [Google Scholar]
- 52.Kiran S, Meier EL, Johnson JP. Neuroplasticity in aphasia: a proposed framework of language recovery. J Speech Lang Hear Res. 2019;62:3973–3985. doi: 10.1044/2019_JSLHR-L-RSNP-19-0054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davatzikos C. Machine learning in neuroimaging: progress and challenges. Neuroimage. 2019;197:652–656. doi: 10.1016/j.neuroimage.2018.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Varoquaux G. Cross-validation failure: small sample sizes lead to large error bars. Neuroimage. 2018;180(Pt A):68–77. doi: 10.1016/j.neuroimage.2017.06.061 [DOI] [PubMed] [Google Scholar]
- 55.Wong T-T. Performance evaluation of classification algorithms by k-fold and leave-one-out cross validation. Pattern Recognit. 2015;48:2839–2846. [Google Scholar]
- 56.Zhao Y, Halai AD, Lambon Ralph MA. Evaluating the granularity and statistical structure of lesions and behaviour in post-stroke aphasia. Brain Commun. 2020;2:fcaa062. doi: 10.1093/braincomms/fcaa062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rojkova K, Volle E, Urbanski M, Humbert F, Dell’Acqua F, Thiebaut de Schotten M. Atlasing the frontal lobe connections and their variability due to age and education: a spherical deconvolution tractography study. Brain Struct Funct. 2016;221:1751–1766. doi: 10.1007/s00429-015-1001-3 [DOI] [PubMed] [Google Scholar]
- 58.Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 1996;29:162–173. doi: 10.1006/cbmr.1996.0014 [DOI] [PubMed] [Google Scholar]
- 59.Cook PA, Bai Y, Hall MG, Nedjati-Gilani S, Seunarine KK, Alexander DC. Camino: Diffusion MRI reconstruction and processing. 2005;
- 60.Nachev P, Coulthard E, Jäger HR, Kennard C, Husain M. Enantiomorphic normalization of focally lesioned brains. Neuroimage. 2008;39:1215–1226. doi: 10.1016/j.neuroimage.2007.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rorden C, Bonilha L, Fridriksson J, Bender B, Karnath HO. Age-specific CT and MRI templates for spatial normalization. Neuroimage. 2012;61:957–965. doi: 10.1016/j.neuroimage.2012.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging. 2001;20:45–57. doi: 10.1109/42.906424 [DOI] [PubMed] [Google Scholar]
- 63.Lacey EH, Skipper-Kallal LM, Xing S, Fama ME, Turkeltaub PE. Mapping common aphasia assessments to underlying cognitive processes and their neural substrates. Neurorehabil Neural Repair. 2017;31:442–450. doi: 10.1177/1545968316688797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gorgolewski K, Burns CD, Madison C, Clark D, Halchenko YO, Waskom ML, Ghosh SS. Nipype: a flexible, lightweight and extensible neuroimaging data processing framework in python. Front Neuroinform. 2011;5:13. doi: 10.3389/fninf.2011.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging. 2010;29:1310–1320. doi: 10.1109/TMI.2010.2046908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal. 2008;12:26–41. doi: 10.1016/j.media.2007.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8 [DOI] [PubMed] [Google Scholar]
- 68.Greve DN, Fischl B. Accurate and robust brain image alignment using boundary-based registration. Neuroimage. 2009;48:63–72. doi: 10.1016/j.neuroimage.2009.06.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Behzadi Y, Restom K, Liau J, Liu TT. A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. Neuroimage. 2007;37:90–101. doi: 10.1016/j.neuroimage.2007.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Buckner RL, Andrews-Hanna JR, Schacter DL. The brain’s default network: anatomy, function, and relevance to disease. Ann NY Acad Sci. 2008;1124:1–38. doi: 10.1196/annals.1440.011 [DOI] [PubMed] [Google Scholar]
- 71.Walenski M, Europa E, Caplan D, Thompson CK. Neural networks for sentence comprehension and production: an ALE-based meta-analysis of neuroimaging studies. Hum Brain Mapp. 2019;40:2275–2304. doi: 10.1002/hbm.24523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Johnson JP, Meier EL, Pan Y, Kiran S. Treatment-related changes in neural activation vary according to treatment response and extent of spared tissue in patients with chronic aphasia. Cortex. 2019;121:147–168. doi: 10.1016/j.cortex.2019.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Purcell JJ, Turkeltaub PE, Eden GF, Rapp B. Examining the central and peripheral processes of written word production through meta-analysis. Front Psychol. 2011;2:239. doi: 10.3389/fpsyg.2011.00239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Seeley WW, Menon V, Schatzberg AF, Keller J, Glover GH, Kenna H, Reiss AL, Greicius MD. Dissociable intrinsic connectivity networks for salience processing and executive control. J Neurosci. 2007;27:2349–2356. doi: 10.1523/JNEUROSCI.5587-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.