Abstract
Head and neck cancer is the seventh most common cancer in the world, and most cases manifest as head and neck squamous cell carcinoma. Despite the prominent role of fucosylated carbohydrate antigens in tumor cell adhesion and metastasis, little is known about the functional role of fucose-modified glycoproteins in head and neck cancer pathobiology. Inactivating polymorphisms of the fut2 gene, encoding for the α1,2-fucosyltransferase FUT2, are associated with an increased incidence of head and neck cancer among tobacco users. Moreover, the presence of the α1,2-fucosylated Lewis Y epitope, with both α1,2- and α1,3-linked fucose, has been observed in head and neck cancer tumors while invasive regions lose expression, suggesting a potential role for α1,2-fucosylation in the regulation of aggressive tumor cell characteristics. Here, we report an association between fut2 expression and head and neck cancer survival, document differential surface expression of α1,2-fucosylated epitopes in a panel of normal, dysplastic, and head and neck cancer cell lines, identify a set of potentially α1,2-fucosylated signaling and adhesion molecules including the epidermal growth factor receptor (EGFR), CD44 and integrins via tandem mass spectrometry, and finally, present evidence that EGFR is among the α1,2-fucosylated and LeY-displaying proteins in head and neck cancer. This knowledge will serve as the foundation for future studies to interrogate the role of LeY-modified and α1,2-fucosylated glycoproteins in head and neck cancer pathogenesis. Data are available via ProteomeXchange with identifier PXD029420.
Keywords: α1, 2-fucosylation, EGFR, epidermal growth factor receptor, FUT2, head and neck squamous cell carcinoma
Introduction
Head and neck squamous cell carcinoma (HNSCC) is a debilitating disease with an estimated incidence of >650,000 cases and an annual toll of >350,000 deaths worldwide (Vigneswaran and Williams 2014; Siegel et al. 2019). In the United States, HNSCC accounts for over 60,000 cases and 13,000 deaths annually (Siegel et al. 2019). HNSCCs can develop at multiple subsites within the head and neck region, but most commonly arise in the oral cavity and oropharynx (Marur and Forastiere 2016). Major risk factors that contribute to the development of this cancer include tobacco use, alcohol use and infection with human papillomavirus (HPV) (Jethwa and Khariwala 2017). Current treatment for HNSCC includes surgical resection of the tumor followed by radiotherapy that may be combined with chemotherapy, but this form of cancer is aggressive and invasive, with a poor survival rate. Worldwide, up to 60% of patients with HNSCC are seen in the late stage of clinical progress, marked by large tumors with local invasion or evidence of metastases, and the 5-year survival rate is proximately 50%. In addition, the current metastatic rate is >65% in patients with HNSCC (Marur and Forastiere 2016; Chow 2020).
Dysregulation of Wnt/β-catenin (Molinolo et al. 2009; Varelas and Kukuruzinska 2014; Kartha et al. 2018), NOTCH1 (Agrawal et al. 2011; Stransky et al. 2011), PI3K (Lui et al. 2013), IL-6/JAK/STAT3 (Johnson et al. 2018) and epidermal growth factor receptor (EGFR) signaling (Ribeiro et al. 2014; Xu et al. 2017; Tian et al. 2018) contribute to HNSCC pathogenesis, and TP53 mutations occur with high frequency in tobacco-related HNSCC (Stransky et al. 2011; Cancer Genome Atlas Network 2015). In HPV-associated HNSCC, the PD-1:PD-L1 pathway also contributes to disease progression via suppression of the adaptive immune response (Lyford-Pike et al. 2013). However, despite these advances in the understanding of HNSCC development and progression, cetuximab and pembrolizumab/nivolumab, monoclonal antibodies targeting EGFR and PD-1, respectively, are the only FDA-approved targeted therapies for HNSCC, and persistent high mortality continues to be driven by recurrent, metastatic and treatment-resistant disease (Wen and Grandis 2015; Johnson et al. 2020). Due to the poor prognosis associated with HNSCC, it is imperative to delve further into the molecular mechanisms of tumorigenesis in search of additional targets for therapy and treatment.
A largely unexplored aspect of HNSCC pathobiology is the role of aberrant protein glycosylation. Glycosylation is a common co- and post-translational modification, and changes in glycosylation are known to alter cell–cell adhesion and metastasis (Chen et al. 2013). Changes in cell surface glycosylation have long been associated with the transformation of normal epithelia into malignant cells, and tumor-associated carbohydrate antigens have been extensively documented (Lin et al. 2014; Mehta et al. 2020), and there is a growing understanding of the functional role of glycosylation in tumor biology. For example, N-glycosylation regulates the function of receptor tyrosine kinases (RTKs) involved in cell growth and proliferation, and glycan modifications mediate the interaction of adhesion molecules that contribute to tumor metastasis (Seales et al. 2003; Seales et al. 2005; Pinho and Reis 2015; Munkley and Elliott 2016; Chandler et al. 2017; Chandler et al. 2019; Chandler et al. 2020). Dysregulation of multiple enzymes in the N-glycosylation pathway endows tumor cells with aggressive traits by promoting epithelial-to-mesenchymal transition (Britain et al. 2021), survival (Holdbrooks et al. 2018), stem cell maintenance (Swindall et al. 2013; Schultz et al. 2016), tumor angiogenesis (Croci et al. 2014; Chung et al. 2017; Chandler et al. 2019), drug resistance (Schultz et al. 2013; Very et al. 2018) and metastasis (Schultz et al. 2012; Engle et al. 2019). Changes in the protein N-glycosylation pathway also promote HNSCC pathogenesis (Resto et al. 2008; Nita-Lazar et al. 2009; Sengupta et al. 2010; Jamal et al. 2012; Lin et al. 2014; Mehta et al. 2020). Although fucosylated glycan epitopes including Lewis x and sialyl-Lewis x are known to play a role in many types of cancer (Jacobs and Sackstein 2011; Shan et al. 2019), little is known regarding the functional role of fucosylated carbohydrate epitopes in HNSCC.
The fut2 (secretor or Se) gene is one of two genes in the human genome that encode for α1,2-fucosyltransferases, with the second being fut1. The fut2 gene is primarily responsible for expression of α1,2-fucosylated epitopes in endoderm-derived epithelial tissue including the digestive tract and salivary glands, while fut1 is expressed in cells of erythroid lineage and in vascular endothelial cells (Watkins 1980). Approximately, 80% of individuals harbor an active Se gene (i.e. display the Secretor phenotype). High levels of the α1,2-fucosylated Lewis Y (LeY) carbohydrate epitope (α-Fuc-(1 → 2)-β-Gal-(1 → 4)-(α-Fuc-[1 → 3])-GlcNAc) have been observed in HNSCC, though invasive regions lose expression (Hotta et al. 2013), suggesting that fucosylated glycans could be involved in the suppression of cell growth and invasion. In addition, inactivating polymorphisms of the fut2 gene, which encodes for the FUT2 enzyme, are associated with an increased incidence of HNSCC among tobacco users (Campi et al. 2012; Su et al. 2016). LeY expression reportedly downregulates EGF signaling via the EGF receptor (Hotta et al. 2021), although there is no direct evidence that the receptor itself displays LeY epitopes in HNSCC, or whether LeY expressed on EGFR-interacting molecules may influence EGFR signaling. There is a critical need to identify α1,2-fucosylated and LeY-modified proteins in HNSCC, to advance our understanding of the role that FUT2-mediated α1,2-fucosylation plays in dysregulated signaling and adhesion in HNSCC.
Here, we examine the association between HNSCC survival and α1,2-fucosyltransferase gene (fut1, fut2) expression, measure surface expression of α1,2-fucosylated epitopes in a panel of normal, dysplastic and HNSCC cell lines, identify potentially α1,2-fucosylated glycoproteins via lectin enrichment to inform the role of FUT2 in HNSCC pathobiology and finally, present evidence that EGFR is among the α1,2-fucosylated and LeY epitope-displaying proteins in HNSCC. Our results suggest that FUT2-modified signaling and adhesion molecules play essential roles in HNSCC pathobiology and lay the groundwork to pursue a functional understanding of the role of α1,2-fucosylation in HNSCC.
Results
High expression of the α1,2-fucosyltransferase FUT2 correlates with improved overall survival in head and neck cancer
Given that α1,2-fucosylated epitopes alter signaling in HNSCC (Hotta et al. 2021), we sought to examine the association between survivorship and mRNA transcript levels of fut1 or fut2, in HNSCC. Kaplan–Meier and Cox proportional-hazards regression analyses were performed to examine HNSCC primary tumor patient data (n = 499) from The Cancer Genome Atlas (TCGA) (Grossman et al. 2016; Campbell et al. 2018). Patient characteristics, including history of alcohol and tobacco use, HPV status and primary tumor site, can be found in Table I. Categorization of patients into high and low fut2 expression groups was performed using expression cut-offs from the Human Protein Atlas (Uhlen et al. 2015). Based on the Kaplan–Meier survival curve and log-rank test (Figure 1, Supplemental File 1, 1–1 to 1–3), high fut2 expression is statistically significantly associated with improved overall survival (OS) (P = 6.18 × 10−4). Within the patient population analyzed, the median survival of the high fut2 expression group was 4.7 years, while the median survival of patients with low fut2 expression was 2.2 years. Moreover, the group with high fut2 expression demonstrated a 5-year survival rate of 49% while low fut2 expressors demonstrated a 5-year survival rate of 38%. When adjusted for age, sex, ethnicity, HPV status, alcohol usage, tobacco usage, Cox proportional-hazards regression demonstrated that high fut2 expression was found to be a significant predictor of increased survival (hazard ratio [HR] = 0.601, 95% CI: 0.452–0.799, P = 4.5 × 10−4). As the HR is less than one, this indicates that high fut2 confers a survival advantage. Other factors associated with OS were age in years (HR = 1.02, 95% CI: 1.01–1.04, P = 0.001) and current smoking status (HR = 1.56, 95% CI: 1.03–2.35). Alcohol usage, reformed smoker status, gender, ethnicity and primary tumor site were not independently associated with survival in the TCGA data set. HPV positive patients accounted for a small percentage of individuals in the TCGA data set, and therefore conclusions about the impact of HPV status on survival were not addressed in this study. The results of the multivariate analysis for fut2 transcript levels are shown in Supplementary Table I. In contrast, no association was found between FUT1 expression and survival in the TCGA data set (Supplementary Figure 1, Supplementary Table II, Supplemental File 1, 1–4 to 1–6).
Table I.
Head and neck cancer patient characteristics
| Characteristic | Value |
|---|---|
| Age (years): Mean (±SD) | 61.1 (11.9) |
| Sex: N (%) | |
| Male | 366 (73.3%) |
| Female | 133 (26.7%) |
| Ethnicity: N (%) | |
| Not Hispanic | 441 (88.4%) |
| Hispanic | 24 (4.8%) |
| Not reported | 34 (6.8%) |
| Alcohol Exposure History: N (%) | |
| No | 157 (31.5%) |
| Yes | 331 (66.3%) |
| Not reported | 11 (2.2%) |
| Tobacco Smoking History: N (%) | |
| Non-smoker | 111 (22.2%) |
| Current smoker | 169 (33.9%) |
| Reformed smoker | 209 (41.9%) |
| Not reported | 10 (2.0%) |
| HPV Status: N (%) | |
| Negative | 72 (14.4%) |
| Positive | 30 (6.0%) |
| Not reported | 397 (79.6%) |
| Primary Site: N (%) | |
| Oral cavity | 255 (51.1%) |
| Larynx | 111 (22.2%) |
| Other | 133 (26.7%) |
Head and neck cancer patient (n = 499) primary tumor tissue data from TCGA HNSCC genomic data commons data set version 18.0 was utilized for this study (Grossman et al. 2016; Campbell et al. 2018). Alcohol and tobacco exposure and HPV infection, which are known risk factors for HNSCC, are shown. Head and neck cancer occurs at diverse subsites, and major subsites represented in the population are also shown. TCGA clinical data were accessed and extracted for analysis via UCSC Xena browser (Goldman et al. 2020). Frequency (N) and percent (%) are indicated.
Fig. 1.
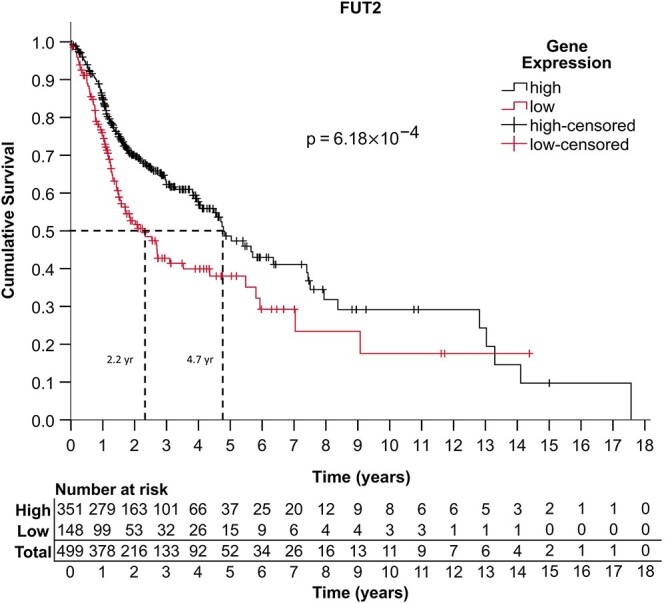
Kaplan–Meier and survival analyses indicate that FUT2 is a positive prognostic indicator in head and neck cancer. A Kaplan–Meier analysis was performed to evaluate the association between fut2 mRNA expression and OS in HNSCC. The study group consisted of all patient primary tumor tissue data from the head and neck cancer dataset found on TCGA genomic data commons database version 18.0 (n = 499) (Grossman et al. 2016). For statistical purposes, the exposure for this study was considered to be high expression of fut2 in primary cancer tissue samples, based on previously established cutoff values from the human protein atlas (Uhlen et al. 2015). Survivorship is measured by 5-year survival percentage and median survival time. Expression of fut2 correlated with improved OS (P = 6.18 × 10−4), based on a univariate log-rank test. P values of <0.05 were considered to be statistically significant. As further evidence, multivariate Cox regression analysis of FUT2 demonstrates that high fut2 expression is a significant predictor of increased survival (HR 0.601, 95% CI 0.452–0.799, P = 4.5 × 10−4) independent of the covariates considered in this analysis (Supplementary Table I). Analyses were conducted using IBM SPSS (IBM Corp., Armonk, NY) version 26.0.
Surface expression of α1,2-fucosylated epitopes varies dramatically between head and neck cancer cell lines and aggressive cells display low levels of α1,2-fucosylated epitopes
Following the identification of fut2 expression as a potential positive prognostic indicator of OS in head and neck cancer, we next sought to determine if α1,2-fucosylated epitopes could be detected in normal, dysplastic and HNSCC tumor cell lines. Given that fut2 is expressed primarily in the digestive tract and salivary glands (Watkins 1980) and that its expression in HNSCC correlates with improved OS, we hypothesized that α1,2-fucosylated epitopes would be higher in indolent tumor tissue, and low in aggressive tumors. To investigate the surface expression of α1,2-fucosylated epitopes in HNSCC, we used flow cytometry to probe for the surface expression of these epitopes in a panel of head and neck tumor cell lines and control cell lines derived from normal and dysplastic oral epithelium. A panel of head and neck cancer cell lines representing multiple subsites within the head and neck region, in addition to normal oral keratinocytes (NOK) and dysplastic oral keratinocytes (DOK), were analyzed via flow cytometry to detect cell surface levels of α1,2-fucosylated epitopes, including H-antigen, Lewis Y (LeY), and Lewis B (LeB) epitopes (Figures 2 and 3A–C, Supplementary Figures 2–4).
Fig. 2.
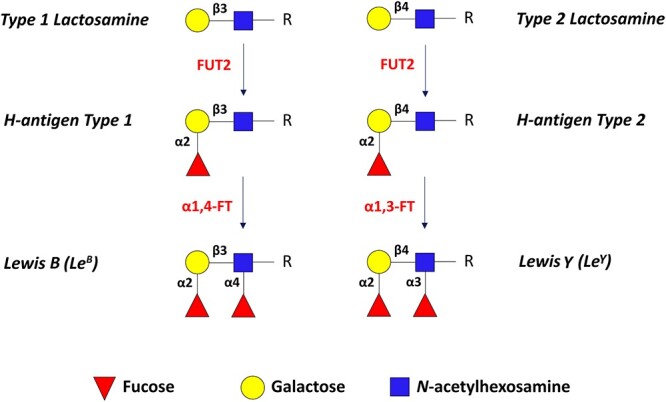
Schematic of α1,2-fucosylated epitopes: H-antigen, Lewis Y (LeY) and Lewis B (LeB). FUT2 catalyzes the addition of fucose to galactose residues in type 1 or type 2 lactosamine motifs (top) via α1,2-linkage to produce H-antigen types 1 and 2. Subsequent α1,4-fucosylation of N-acetylglucosamine in H-antigen type 1 or α1,3-fucosylation of N-acetylglucosamine in H-antigen type 2, leads to the production of Lewis B (LeB) or Lewis Y (LeY) epitopes, respectively.
Fig. 3.
Flow cytometry analysis of α1,2-fucosylated epitopes in HNSCC panel. A panel of cell lines including two control cell lines, NOK and DOK, and eight HNSCC cell lines spanning multiple HNSCC subsites, including A-253 salivary gland carcinoma cells, FaDu and Detroit-562 (Det-562) pharyngeal squamous carcinoma cells, and CAL-27, SCC-9, SCC-15, SCC-25 and HSC-3 tongue squamous carcinoma cells, was analyzed via flow cytometry for three α1,2-fucosylated epitopes, including (A) H antigen, (B), Lewis Y (LeY) and (C) Lewis B (LeB). For all flow cytometry experiments, unstained controls and isotype controls were performed. Mean fluorescence intensity is reported in relative fluorescence units (technical replicates, n = 3).
NOK, used as a control, expressed the highest level of H-antigen, high levels of LeY, and lower levels of LeB (Figure 3A–C) compared to other cell lines in the panel. DOK also expressed high levels of H-antigen, but expressed lower levels of LeY, and intermediate levels of LeB compared to the tumor cell lines. Next, we examined tumor cell lines from multiple subsites within the head and neck region. H-antigen was low in A-253 salivary gland carcinoma cells and pharyngeal squamous cell carcinoma cell lines FaDu and Detriot-562 (Det-562), while the tongue squamous carcinoma cell lines displayed a wide range of H-antigen expression (Figure 3A). Similarly, LeY expression varied widely in tongue squamous carcinoma cell lines, and pharyngeal SCC cell lines FaDu and Det-562 displayed intermediate levels. CAL-27 tongue squamous carcinoma cells displayed the highest levels of LeY followed by NOK cells, while A-253, SCC-15 and HSC-3 cells were among the lowest expressors of the LeY epitope and displayed similar levels as DOK cells (Figure 3B). Finally, CAL-27 and Det-562 cells displayed the highest levels of LeB epitopes, while A-253, SCC-9 and HSC-3 cells displayed the lowest levels of LeB epitopes, like NOK cells (Figure 3C). The wide range of α1,2-fucosylated epitope expression within the cell line panel parallels the range of mRNA expression levels observed in TCGA Head and Neck Cancer patient data. Notably, CAL-27 cells expressed the highest levels of all three α1,2-fucosylated epitopes, while HSC-3 cells expressed the lowest H-antigen levels, and among the lowest LeB and LeY levels within the cell line panel.
Identification of potentially α1,2-fucosylated proteins in HNSCC via lectin enrichment and mass spectrometry
Following identification of HNSCC cell lines with high and low expression of α1,2-fucosylated epitopes via flow cytometry, we next sought to identify glycoproteins bearing α1,2-fucosylated epitopes. CAL27 and HSC-3 cell lines, displaying high and low levels of α1,2-fucosylated epitopes, respectively, were selected for proteomic analyses. Based on RT-qPCR analyses, CAL27 cells express >10-fold higher levels of fut2 mRNA compared to HSC-3 cells (Figure 4A, right), consistent with flow cytometry results. Expression of fut1, a second α1,2-fucosyltransferase, was also evaluated (Figure 4A, left). Based on mRNA expression, FUT2 appears to be the dominant α1,2-fucosyltransferase in CAL27 cells. FUT2 protein levels were higher in CAL27 compared to HSC-3 cells based on western blot analysis of whole cell lysates (WCL; Figure 4B), and western blot analyses of the α1,2-fucosylated Lewis Y epitope was detected exclusively in proteins from CAL27 WCL (Figure 4C). Next, proteomic analyses were performed to build knowledge of the protein space and therefore inform subsequent glycoproteomic analyses. WCL digests from CAL27 and HSC-3 tongue squamous carcinoma cell lines were analyzed via nLC–MS/MS, and 3121 proteins were identified in CAL27 cell lysates and 2924 proteins in HSC-3 cell lysates (1% FDR cutoff), with 2490 proteins common to both cell lines (Figure 4D, and Supplemental File 2). Membrane proteins and adhesion molecules were well-represented (Supplemental File 3). Glycoproteins with a known role in HNSCC pathobiology, including EGFR, CD44, ICAM-1 (CD54), E-cadherin, Galectin-3 and the integrin subunits integrin beta-4 (ITGB4; CD104) and integrin alpha-6 (ITGA6; CD49f), were among the proteins identified in both cell lines. However, only 41 glycoproteins with at least one high confidence glycopeptide (score ≥ 200) were detected and of these only nine high confidence fucosylated glycopeptides were assigned in CAL27 and HSC3 cell lysate digests, emphasizing the need for deeper characterization of glycoproteins.
Fig. 4.
Analyses of tongue squamous carcinoma cell line lysates reveals the presence of multiply-fucosylated glycopeptides consistent with LeY epitope expression. (A) Comparison of fut1 and fut2 expression (mRNA) between CAL27 and HSC-3 cells (n = 3). (B) Western blot of WCL from CAL27 and HSC-3 cells, to assess levels of FUT2. GAPDH is shown as a loading control. (C) Western blot of WCL from CAL27 (lane 1) and HSC-3 (lane 2) cells, with an anti-Lewis Y (LeY) antibody. (D) Venn diagram of human proteins identified in CAL27 and HSC-3 WCL digests following nLC-MS/MS analyses (FDR ≤ 1%, unique peptides ≥1). (E) EICs of HexNAc1Hex1NeuAc1 oxonium ion (m/z 512.1974 ± 5 ppm) from representative nLC-MS/MS runs from CAL27 WCL, CAL27 AAL-enriched glycopeptides, HSC-3 WCL and HSC-3 AAL-enriched glycopeptides (from top to bottom). The relative abundance is normalized to the most intense EIC (HSC3-AAL). (F) Venn diagram of AAL-enriched glycoproteins with ≥1 high-confidence glycopeptide assigned, from CAL27 and HSC-3 lysate digests. (G) EThcD mass spectrum of CD59 site N43 glycopeptide, [M + 4H]4+ m/z 966.3972, demonstrating evidence of fucosylation. The presence of the HexNAc1Hex1dHex1 oxonium ion at m/z 512.1964 and the HexNAc1Hex1dHex2 oxonium ion at m/z 658.2533, in combination with the loss of HexNAc1Hex1dHex1 or HexNAc1Hex1dHex2 from the charge-reduced species [M + 3H]3+ ∙ provide evidence of a double-fucosylated lactosamine motif on the N-glycan antennae consistent with the presence of an α1,2-fucosylated LeY epitope. A deoxyhexose shift associated with fragments that contain only the trimannosyl chitobiose core (pN2H3), suggests that an additional fucose residue is located at the N-glycan core. p = peptide, N = HexNAc, H = hexose, F = dHex. # indicates unassigned peaks from potentially co-isolated species.
Next, fucosylated glycopeptides were enriched from CAL27 and HSC-3 WCL digests using Aleuria aurantia Lectin (AAL), with a broad affinity for fucosylated epitopes including α1,2-fucosylated glycopeptides (Baldus et al. 1996; Matsumura et al. 2007; Zhou et al. 2017), to generate a list of candidate α1,2-fucosylated glycoproteins. Glycoproteins detected in this study were considered candidate α1,2-fucosylated glycoproteins if (i) MS spectra indicate the presence of fucose, and (ii) the glycoprotein was either uniquely detected in high fut2 CAL27 cells but not in low fut2 HSC-3 cells, or the protein was detected in both cell lines but displayed glycans with higher numbers of fucose residues in CAL27 cells compared to HSC-3 cells. We reasoned that fucosylated glycoproteins identified in CAL27 cells could be considered among the candidate α1,2-fucosylated glycoproteins, and that fucosylated glycoproteins identified in low-fut2 expressing HSC-3 cells could help determine a set of α1,3- and/or α1,4-fucosylated HNSCC glycoproteins to be excluded from consideration as uniquely α1,2-fucosylated substrates in CAL27 cells. Moreover, given that fut2 encodes for an α1,2-fucosyltransferase that catalyzes a unique glycan linkage (i.e. introduces an additional potential site of fucosylation on galactose residues) compared to more frequently studied enzymes with α1,3- and/or α1,4-fucosyltransferase activity, we might expect to find higher numbers of terminal fucose residues in FUT2-modified glycoproteins from CAL27 cells.
AAL-enriched glycopeptides were analyzed using nLC-MS/MS with higher energy collisional dissociation (HCD) followed by oxonium ion-triggered electron-transfer dissociation with supplemental activation (EThcD), to facilitate assignment of outer arm and core fucosylated configurations of N-glycopeptides (Acs et al. 2018; Yuan et al. 2019; Chandler et al. 2020). Extracted ion chromatograms (EICs) of m/z 512.1974 (HexNAc-Hex-Fuc), an oxonium ion marker indicative of nonreducing terminal (antennary) fucosylation, from nLC-MS/MS runs demonstrate that AAL-enriched fractions from CAL27 and HSC-3 lysates are enriched for fucosylated epitopes compared to WCL digests (Figure 4E). AAL enrichment and nLC-MS/MS analysis led to the successful assignment of fucosylated N- and O-glycoproteins, including 212 unique high confidence glycopeptides (score ≥ 200) in CAL27 lysates representing 73 N- and O-glycosylated proteins, and 52% (111) of the unique CAL27 glycopeptides were fucosylated (Figure 4F, Supplemental File 4). In HSC-3 cell lysates, 169 unique high confidence glycopeptides were assigned representing 57 glycoproteins, including 31 fucosylated glycoproteins, and 95 unique fucosylated glycopeptides were assigned (Figure 4C, Supplemental File 4).
Fourteen fucosylated glycoproteins were identified in common between CAL27 and HSC-3 AAL-enriched fractions, while 24 unique fucosylated glycoproteins were assigned in CAL27 lysates and 17 unique fucosylated glycoproteins were assigned in HSC-3 lysates (Supplemental File 4, 4–6). In all fucosylated glycopeptide MS2 spectra, the common fucose-diagnostic oxonium ion HexNAc1Hex1Fuc1 at m/z 512.1964 can be observed (Figure 4D). Many of the MS2 spectra of multiply-fucosylated glycopeptides from CAL27 cells additionally contain the less frequently observed HexNAc1Hex1Fuc2 (m/z 658.2533) oxonium ion, which is present in the MS2 spectrum of the CD59 N-glycopeptide TAVNCSSDFDACLITK + HexNAc4Hex5Fuc3 (m/z 966.3972, 4+) (Figure 4G). This second ion provides evidence that that the N-acetylhexosamine (likely N-acetylglucosamine) and the hexose (likely galactose) each have an attached fucose residue, consistent with but not confirmatory of the presence of LeY epitopes on CAL27-derived glycoproteins.
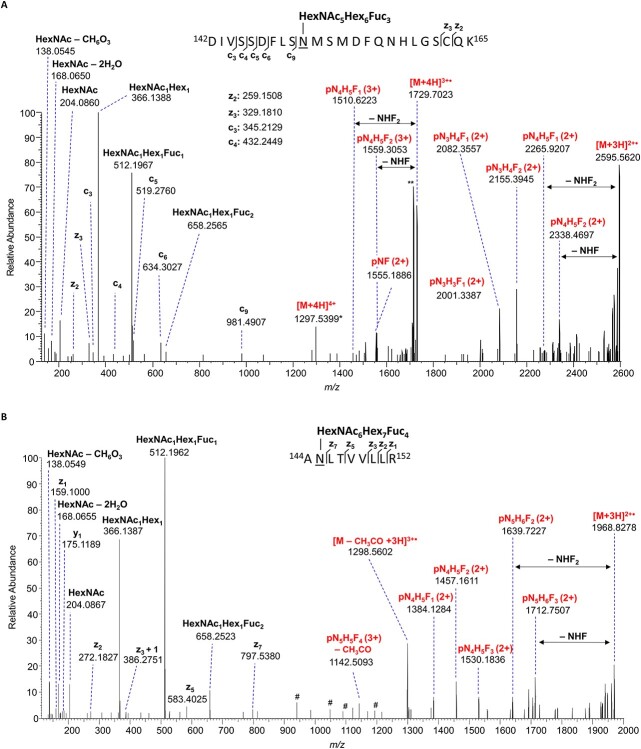
The EGFR, ICAM1 (CD54), CD166, LAMP1 and Galectin-3-binding protein (LG3BP), all glycoproteins with critical roles in tumor cell signaling and adhesion, are among the AAL-enriched fucosylated glycoproteins in common between the two cell lines (Supplemental File 4, 4–7). Of particular interest is EGFR, a current target of HNSCC therapy and known to play a critical signaling role in HNSCC (Wen and Grandis 2015; Xu et al. 2017). EGFR glycopeptides displaying up to three fucose residues were assigned in AAL-enriched fractions from CAL27 cells, while EGFR from HSC-3 cells contained a maximum of two fucose residues (Supplemental File 4, 4–8). Moreover, tri-fucosylated glycopeptides from CAL27-derived EGFR contained both m/z 512.1967 and m/z 658.2565 oxonium ions (Figure 5A), suggesting that LeY may be present on the glycan antennae, while fucosylated glycopeptides from HSC-3-derived EGFR contained only the m/z 512 ion but lacked the m/z 658 ion (Supplementary Figure 6), suggesting a paucity of LeY epitopes. The cell adhesion molecule ICAM1 (CD54) derived from CAL27 cells contained up to four fucose residues on a single glycan and its composition and MS2 spectra are consistent with the presence of LeY epitopes on the N-glycan antennae (Figure 5B). In contrast, ICAM1 from HSC-3 cells contained a maximum of two fucose residues.
Fig. 5.
EGFR and intercellular adhesion molecule 1 (ICAM-1, CD54) are highly fucosylated in HNSCC cells. (A) EThcD mass spectrum of a triple-fucosylated EGFR site N151 glycopeptide, [M + 4H]4+ m/z 1297.2791, demonstrating evidence of fucosylation. The presence of the HexNAc1Hex1dHex1 (m/z 512.1967) and HexNAc1Hex1dHex2 (m/z 658.2565) oxonium ions, and the loss of HexNAc1Hex1dHex1 or HexNAc1Hex1dHex2 from the charge-reduced species [M + 3H]3+ ∙ provide evidence of a double-fucosylated lactosamine motif on the N-glycan antennae consistent with the presence of the α1,2-fucosylated LeY epitope. A deoxyhexose (fucose) associated with fragments that contain only the core N-acetylhexosamine (pNF), suggests that an additional fucose residue is located at the N-glycan core. (B) EThcD mass spectrum of an ICAM-1 site N145 N-linked glycan with four fucose residues, [M + 4H]4+ m/z 984.6834, demonstrating evidence of fucosylation. As with the EGFR spectrum, the presence of the HexNAc1Hex1dHex1 (m/z 512.1962) and HexNAc1Hex1dHex2 (m/z 658.2523) oxonium ions, and the loss of HexNAc1Hex1dHex1 or HexNAc1Hex1dHex2 from the charge-reduced species [M + 3H]3+ ∙ is consistent with the presence of the α1,2-fucosylated LeY epitope. P = peptide, N = HexNAc, H = hexose, F = dHex. # indicates unassigned peaks from potentially CO-isolated species. * indicates that the monoisotopic peak was not detected. ** indicates the loss of CH3CO.
Among the fucosylated glycoproteins unique to indolent CAL27 cells, adhesion molecules including basal cell-adhesion molecule (BCAM) and CD47, and the tumor-associated calcium signal transducer-2 (TACD2) were identified (Supplemental File 4, 4–9). BCAM is known to promote tumor cell migration by competing with integrins to bind laminin α5, a major component of the basement membrane (Kikkawa et al. 2013), while CD47 is known to inhibit NK cell-mediated cytotoxicity (Kim et al. 2008). Tumor-associated calcium signal transducer 2 (TACD2 or TROP-2) is a surface protein with adhesive properties and may also mediate calcium-dependent signal transduction (Fornaro et al. 1995; Ripani et al. 1998). Fucosylated glycoprotein assignments unique to the HSC-3 lysate include multiple integrin α- and β-subunits (ITGB4, ITGB1, ITGA3, ITGA2, ITGA5) and Laminin subunit alpha-3. Surprisingly, while CD44 was detected in CAL27 cells, no fucosylated glycopeptides were detected. Notably, fucosylated CD44 glycopeptides were identified in HSC-3 cells. However, glycopeptide spectra suggest that the fucose resides at the core position of the N-linked glycan (α1,6-linked), eliminating the possibility that the fucose on HSC-3-derived CD44 is α1,2-linked.
Anti-Lewis Y antibody enrichment and analysis of HNSCC glycoproteins
We sought additional evidence that the potentially α1,2-fucosylated HNSCC glycoproteins identified in AAL-enriched fractions display the α1,2-fucosylated LeY epitope. An anti-LeY (F3) antibody was used to enrich proteins from CAL27 WCL, and LeY-enriched glycoproteins were analyzed via nUPLC-MS/MS. Glycoproteins with known involvement in HNSCC pathogenesis, including EGFR, CD44 and integrins, were identified (Figure 6A, Supplementary Figure 1, 1–5). Several of these proteins including EGFR, LAMP1 and cathepsin D, overlap with those identified in AAL-enriched fractions. In addition, LeY-enriched proteins were subject to SDS-PAGE followed by western blotting with an anti-EGFR antibody, and the results offer strong evidence that the LeY epitope is present on EGFR derived from HNSCC cells (Figure 6B). Notably, EGFR was not detected in the control. Anti-LeY antibody-enriched fractions from HSC-3 cells (from an aggressive, metastatic tongue squamous carcinoma) contained very few proteins (data not shown), consistent with flow cytometry results that demonstrated low cell surface levels of LeY on HSC-3 cells compared to other cell lines.
Fig. 6.

The EGFR displays the LeY epitope in head and neck squamous carcinoma cells. (A) An anti-Lewis Y (F3) antibody was covalently linked to protein A/G agarose beads, and then the anti-LeY or control beads were incubated with CAL27 WCL to enrich for LeY-displaying glycoproteins. Protein digests were analyzed via nLC-MS/MS in triplicate (n = 3). Proteins (FDR ≤ 1%, unique peptides ≥1) that appeared in at least 2 out of 3 nLC-MS/MS runs, and that were not present in the control, were considered. Selected proteins are shown. The full set of proteins can be accessed in Supplementary Figure 1, 1–5. (B) Anti-LeY (F3)-enriched proteins were resolved via SDS-PAGE under reducing conditions, and a western blot analysis was performed using an anti-EGFR antibody for detection to probe for EGFR. (C) EThcD mass spectrum of a triple-fucosylated EGFR site N579 glycopeptide, [M + 4H]4+ m/z 1051.4380, demonstrating evidence of fucosylation. The presence of the HexNAc1Hex1dHex1 (m/z 512.1959) and HexNAc1Hex1dHex2 (m/z 658.2532) oxonium ions, and the loss of HexNAc1Hex1dHex1 or HexNAc1Hex1dHex2 from the charge-reduced species [M + 3H]3+ ∙ provide evidence of a double-fucosylated lactosamine motif on the N-glycan antennae consistent with the presence of the α1,2-fucosylated LeY epitope.
Immunoprecipitation of EGFR, followed by tandem mass spectrometry analysis of EGFR glycopeptides using HCD and EThcD was performed with the goal of identifying EGFR fucosylated glycopeptides in a glycosylation-site specific manner. Multiple fucosylated EGFR glycopeptides were assigned, representing single-, double- and triple-fucosylated N-glycopeptides. Critically, the spectra provide strong evidence that sites N175 and N579 display triple-fucosylated N-glycopeptides with the composition HexNAc5Hex6Fuc3 (Figure 6C). Moreover, site N175 and N579 glycopeptide spectra are consistent with the presence of a Lewis Y structure in which two fucose residues are located on the same lactosamine subunit at the N-glycan nonreducing terminus, including oxonium ions at m/z 512.1959 (HexNAc1Hex1Fuc1) and m/z 658.2532 (HexNAc1Hex1Fuc2), and the loss of HexNAc1Hex1Fuc1 and HexNAc1Hex1Fuc2 from the precursor ion (m/z 1847.2649, 2+ and m/z 1773.7294, 2+, respectively). The MS2 spectrum of the EGFR glycopeptide TCPAGVMGENNTLVWK + HexNAc5Hex6Fuc3 (1051.4380, 4+) also suggests that a third fucose residue is present at the N-glycan core based on the presence of a fragment ion at m/z 1509.6399, assigned as the intact peptide (p) with the covalently linked glycan composition HexNAc3Hex3Fuc1. Together, the fragment ions demonstrating loss of nonreducing terminus fucose residues, and the presence of the noted oxonium ions, strongly suggest the presence of the LeY epitope is located on EGFR sites N175 and N579 and may also be displayed on additional EGFR N-glycosylation sites.
Discussion
Despite the prominent role of fucosylated carbohydrate antigens in tumor cell adhesion and metastasis (Jacobs and Sackstein 2011; Shan et al. 2019), little is known about the functional role of fucose-modified glycoproteins in head and neck cancer pathobiology. Here, we report an association between HNSCC survivorship and the expression of the FUT2 α1,2-fucosyltransferase, document differential surface expression of α1,2-fucosylated epitopes in a panel of normal, dysplastic, and HNSCC cell lines, identify a set of potentially α1,2-fucosylated signaling and adhesion molecules via nLC-MS/MS, and finally, present evidence that EGFR is among the α1,2-fucosylated and LeY-displaying proteins in HNSCC. It has been previously reported that inactivating polymorphisms of fut2 are associated with an increased incidence of HNSCC among tobacco users (Campi et al. 2012; Su et al. 2016). Furthermore, high levels of the α1,2-fucosylated Lewis Y epitope have been observed in HNSCC tumors while invasive regions lose expression (Hotta et al. 2013), suggesting a specific role for α1,2-fucosylation in the regulation of aggressive tumor cell characteristics. Our initial observation of an association between high fut2 expression and improved OS based on univariate (Log-rank test) and multivariate (Cox proportional-hazards model) analyses of the HNSCC patient data set from TCGA (Uhlen et al. 2015; Campbell et al. 2018), reinforces the notion that α1,2-fucosylation may play a prominent role in HNSCC pathobiology. This led us to undertake additional experiments to explore the expression of α1,2-fucosylated epitopes in HNSCC tumor cell lines and identify potential FUT2 glycoprotein substrates.
Analysis of α1,2-fucosylated epitope expression in a panel of head and neck cancer cell lines demonstrated a range of epitope expression. Cell lines derived from normal oral epithelium and from DOK demonstrated high expression of α1,2-fucosylated epitopes, while tumor cell lines exhibited a broad range of α1,2-fucosylated epitope expression. For a given cell line, the expression of H-antigen, Lewis Y and Lewis B epitopes generally trended together, which might be expected given that the epitopes are related and depend in part on the expression of an α1,2-fucosyltransferase such as FUT2. Among the HNSCC subsites represented in the cell line panel, a subsite-specific pattern of α1,2-fucosylated epitope expression was difficult to determine, in part due to the underrepresentation of certain subsites in the cell panel. However, we did succeed in identifying HNSCC cell lines with low- and high- α1,2-fucosylated epitope expression. Low-fut2-expressing HSC-3 cells are derived from an aggressive metastatic tumor, and high-fut2-expressing CAL27 cells derived from a primary tumor with low metastatic potential (i.e. it does not show the ability to metastasize in mouse orthotopic tumor xenograft models of HNSCC), which might be expected based on the association between high fut2 expression, OS, and less aggressive disease.
Comparison of fucosylated glycoproteins in CAL27 (high fut2) and HSC-3 (low fut2) cell lines via mass spectrometry, led to the identification of a set of candidate α1,2-fucosylated glycoproteins in HNSCC that are unique to high fut2-expressing CAL27 cells. Signaling molecules, including the EGFR, as well as adhesion molecules involved in both innate and adaptive immune responses and tumor metastasis, are among the candidate α1,2-fucosylated glycoproteins. Of particular interest among this set of α1,2-fucosylated glycoproteins is EGFR. Changes in EGFR glycosylation, including via core (α1,6) fucosylation (Wang et al. 2006), GALNT2 (Lin et al. 2014) and α1,3-fucosyltransferases (Liu et al. 2011) reportedly modify EGFR dimerization and signaling, and sialyltransferases impart EGFR resistance to tyrosine kinase inhibitors (Yen et al. 2015). The expression of the α1,2-fucosylated LeY epitope in HNSCC reportedly downregulates EGF signaling via the EGF receptor (Lin et al. 2015; Hotta et al. 2021), though to date there has been little direct evidence that the receptor itself displays LeY epitopes in HNSCC. Determining if LeY is present on EGFR is important, as EGFR interacts with other membrane-resident glycoproteins and glycolipids, and LeY expression on EGFR-interacting molecules rather than EGFR might itself influence EGFR-mediated signaling. Here, we report that the EGFR is α1,2-fucosylated, and displays LeY epitopes on the antennae of N-glycans at sites N175 and N579 of the receptor in HNSCC cells (Figures 5A and 6D). As glycosylation is known to influence EGFR conformation (Fernandes et al. 2001; Kaszuba et al. 2015), the presence of LeY on EGFR at sites N175 and N579 may alter receptor conformation and signaling. Given that α1,3-fucosylation of EGFR N-linked glycans alters EGFR dimerization, it may be that FUT2 contributes to this effect by the additional contribution of an α1,2-linked fucose to galactose residues within lactosamine motifs frequently displayed on the antennae of N-linked glycans to create LeY motifs. To what degree the addition of an α1,2-linked fucose amplifies or otherwise alters this effect is not known. However, glycosylation at N579 reportedly has an outsized impact on EGFR function (Whitson et al. 2005). Finally, among fucose modifications, α1,2-linked fucosylation has the unique ability to prevent addition of terminal sialic acid residues (Zerfaoui et al. 2000). Therefore, the α1,2-fucosylation of EGFR may alter its function and activity by blocking the addition of sialic acid residues, known to play a role in development of resistance to tyrosine kinase inhibitors (Yen et al. 2015). Considering this new evidence, we intend to investigate the impact of FUT2-mediated α1,2-fucosylation on EGFR-mediated signaling in HNSCC.
Adhesion molecules involved in innate and adaptive immune responses and tumor metastasis, are among the putative α1,2-fucosylated glycoproteins with high confidence glycopeptide spectra and evidence of multiply-fucosylated N-glycan antennae consistent with the presence of LeY epitopes. Putative α1,2-fucosylated glycoproteins reported here include CD166, an adhesion molecule and a marker of stem-like cells in HNSCC (Xiao et al. 2017), LAMP1, known to mediate tumor cell metastasis via presentation of sialofucosylated ligands to selectins (Sawada et al. 1993), and Galectin-3-binding protein (LG3BP), which mediates adhesive interactions with integrins and is known to be immunostimulatory when presenting galectin-3 ligands (Ullrich et al. 1994; Sasaki et al. 1998). ICAM1 from high-fut2 expressing CAL27 cells demonstrate high levels of fucosylation with up to four fucose residues on a single N-linked glycan and MS2-level evidence consistent with the presence of LeY on N-glycan antennae (Figure 5B), while ICAM1 from low-fut2 expressing HSC-3 cells was less fucosylated with a maximum of two fucose residues on N-linked glycans and lacked evidence of LeY epitope presence. ICAM1 (CD54) is an adhesion molecule frequently expressed on endothelial cells and immune cells, and also in tumor cells (Roland et al. 2007). LeY-modified ICAM family proteins are known to serve as ligands of the C-type lectin DC-SIGN, which recognizes high-mannose and LeY carbohydrate epitopes and mediates adhesion of dendritic cells (Geijtenbeek et al. 2000; van Liempt et al. 2006; García-Vallejo et al. 2008), which play a role in antitumor immunity (Veglia and Gabrilovich 2017). It is tempting to speculate that differences in FUT2-mediated α1,2-fucosylation could alter the adhesive interactions of these cells, and that reduced fut2 expression might therefore lead to an immunosuppressive effect in HNSCC. This will be a target of future investigations into the role of FUT2 in HNSCC.
We also detected highly fucosylated CD59 glycopeptides with evidence of LeY modification in our analyses. CD59 is a glycosylphosphatidylinositol (GPI)-anchored protein and complement inhibitor that serves as an integral component of the innate immune system and protects cells by inhibiting the formation of the complement membrane attack complex (MAC). CD59 is regulated by the tumor microenvironment in head and neck cancer, enabling cells to escape complement-mediated cell lysis (Kesselring et al. 2014) and that EGFR modulates this process in head and neck cancer (Abu-Humaidan et al. 2020). The presence of the HexNAc1Hex1dHex1 oxonium ion at m/z 512.1964 and the HexNAc1Hex1dHex2 oxonium ion at m/z 658.2533, in combination with the loss of HexNAc1Hex1dHex1 or HexNAc1Hex1dHex2 from the charge-reduced species [M + 3H]3+ ∙ provide evidence of a double-fucosylated lactosamine motif on the N-glycan antennae consistent with the presence of an α1,2-fucosylated LeY epitope on CD59 (Figure 4D). Whether α1,2-fucosylation regulates the function of CD59, including its ability to inhibit formation of the MAC complex, remains to be investigated.
There is a critical need to understand the role that FUT2-mediated α1,2-fucosylation plays in dysregulated signaling, adhesion, and immune evasion in HNSCC. Here, we document an association between high fut2 expression and improved OS in patients diagnosed with head and neck cancer, compare levels of α1,2-fucosylated epitopes in a panel of head and neck cancer and control cells, and report set of putative α1,2-fucosylated and LeY-modified signaling and adhesion molecules with known involvement in HNSCC pathogenesis. Finally, we document evidence that the EGFR displays the LeY epitope on N-glycans occupying sites N175 and N579, the latter with a known role in the modulation of EGFR conformation, dimerization, and signaling. This knowledge will serve as the foundation for future studies to interrogate how the reduction or loss of fut2 expression alters the function of these putative LeY-modified and α1,2-fucosylated glycoproteins and thereby contributes to HNSCC pathogenesis.
Materials and methods
Kaplan–Meier and Cox regression analyses of TCGA data
A retrospective cohort study of 499 tissue samples from patients with head and neck cancer was conducted using the Head and Neck Cancer dataset from TCGA Genomic Data Commons version 18.0, most recently updated in 2019 (Grossman et al. 2016; Campbell et al. 2018). The data were extracted for analysis utilizing the University of California Santa Cruz (UCSC) Xena Browser exploration tool, which allow for open access analysis and visualization of large-scale cancer data sets, including TCGA (Goldman et al. 2020). Samples in the database were deidentified prior to establishment of the database. Individuals considered for the study (n = 499) had a diagnosis of head and neck cancer with a tissue sample removed from the primary tumor site and analyzed via RNA-Seq to assess mRNA expression, supplied in the database as fragments per kilobase of transcript per million (FPKM) mapped reads. This value is then transformed in Xena Browser in log2(FPKM + 1) scale. To explore the link between fut2 mRNA gene expression and survivorship, survival and mRNA gene expression data were extracted from Xena Browser using the Head and Neck Cancer-TCGA data set. High and low levels of fut2 expression were established using an expression cut-off determined from the Human Protein Atlas (Uhlen et al. 2015). A Kaplan–Meier survival curve was generated using the time to event (OS) data grouped by fut2 mRNA expression-level. The significance of the difference between the survivorship of the low- and high-expression groups was first calculated by conducting a univariate log-rank test. P values of <0.05 were deemed to be statistically significant. Survival was assessed by the 5-year survival rate and median survival time (in years). A multivariate Cox proportional hazards regression analysis was performed to determine the HRs and their 95% confidence intervals (CIs) for high mRNA fut2 expression when adjusted for age (years), gender, ethnicity, HPV status, primary tumor location, alcohol usage history and tobacco usage history. HR values of >1.0 signify that the variable is an independent predictor of decreased survival, and those <1.0 signify the variable is protective. A P-value threshold of <0.05 was used for Cox analysis. All statistical analyses were conducted through IBM SPSS (IBM Corp., Armonk, NY) version 26.0.
Cell lines and reagents
NOK, derived from gingival tissues, were a gift from Dr Karl Munger (Tufts University, Medford, MA) (Piboonniyom et al. 2003). DOK, originally isolated from the dorsal tongue, were purchased from the European Collection of Authenticated Cell Cultures (ECACC) via Sigma (St. Louis, MO). A-253 salivary gland carcinoma cells, FaDu pharyngeal squamous carcinoma cells (SCC), Detroit 562 pharyngeal SCC (ATCC CCL-138), and CAL-27, SCC9, SCC15, and SCC25 tongue SCC, were purchased from the American Type Culture Collection (ATCC, Manassas, VA). HSC-3 tongue SCC, originally isolated from a lymph node metastasis, was purchased from the Japanese Collection of Research Bioresources (JCRB) Cell Bank via Sekisui XenoTech LLC (Kansas City, KS). Anti-Lewis b antibody 2-25LE was purchased from Novus Biologicals (Littleton, CO). Anti-Lewis Y antibody (F3) was purchased from ThermoFisher Scientific (Rockford, IL). Anti-Blood Group H ab antigen antibody [87-N] was purchased from Abcam (Cambridge, United Kingdom). All three antibodies were labeled using the Lightning-Link APC Conjugation Kit (Abcam), according to the manufacturer’s instructions. APC mouse IgM, K (MM-30) and APC mouse IgG1, K isotype control antibodies were purchased from BioLegend (San Diego, CA). Additional reagents: TRIzol (Invitrogen), Direct-zol™ RNA Miniprep Plus (Zymo Research). Glycoprotein Eluting Solution (Fucose/Arabinose), Aleuria Aurantia Lectin (AAL) Agarose Bound and Ulex Europaeus Agglutinin I (UEA I) Agarose Bound (Vector Laboratories, Burlingame, CA). MS grade trypsin (tosyl phenylalanine chloromethyl ketone treated) was purchased from ThermoFisher Scientific (Waltham, MA). Pierce C18 Tips (100 mL), dithiothreitol (DTT), ammonium bicarbonate (NH4HCO3) and iodoacetamide were purchased from Sigma Aldrich (St. Louis, MO). Small (10 mL) C18 ZipTips were purchased from Millipore (Billerica, MA).
Flow cytometry of HNSCC cell lines
To prepare adherent cells for flow cytometry, cells were washed once in phosphate buffered saline (PBS) followed by treatment with 5 mM EDTA in PBS for 10 min at 37°C. Cells were washed twice with 5 mL PBS, resuspended, and an automated cell counter was used to determine the number of live cells. Cells were blocked by washing twice in 2% FBS in PBS, and 2 × 105 cells were labeled with each antibody by adding 98 uL of 2% FBS in PBS and 2 uL of 0.1 mg/mL antibody (APC-labeled anti-Blood Group H, anti-Lewis Y, or anti-Lewis B). For each cell line, an unstained control and an antibody isotype control were processed in an identical manner. Cells were incubated with the antibodies for 30 min at 4°C, then washed twice with PBS and resuspended in PBS for flow cytometry analyses. All analyses were performed on a BD FACSCelesta™ Cell Analyzer (BD Biosciences, Franklin Lakes, NJ). Prior to analyses, the FSC and SSC gating voltages were optimized using unstained cells, and voltages were maintained (constant) for all analyses. Results were processed using FlowJo v10.6.1 (BD Biosciences), and populations of live, single cells were utilized for final analyses. All analyses were performed in triplicate.
RT-qPCR
CAL-27 and HSC-3 cells were plated into 6-well tissue culture plates and grown to 60–70% confluence, treated with TRIzol, and stored at −80°C. RNA was extracted using the Direct-zol™ RNA Miniprep Plus kit according to the manufacturer’s instructions. RNA and cDNA concentrations were determined via UV spectroscopy on a Cytation 5 Cell Imaging Multi-Mode Reader (BioTek, Winooski, VT). Synthesis of cDNA was achieved using SuperScript™ VILO™ Master Mix (ThermoFisher Scientific), according to the manufacturer’s instructions. For each qPCR reaction, 10 ng of cDNA, 10 uL PowerUp SYBR Green Master Mix (ThermoFisher Scientific), 2 uL each of 10 μM forward and reverse primers (see sequence below), and an appropriate volume of water were combined. Each reaction was performed in triplicate. qPCR analyses were performed on a QuantStudio™ 7 Flex Real-Time PCR System, with initial enzyme activation (95°C for 2 min), followed by 40 cycles at 95°C for 15 s (denaturing), 62°C for 30 s (annealing/extension), for 40 cycles.
Human fut2 Forward Primer: 5′-CCG ACT ACA TCA CCG AGA AGC T-3′.
Human fut2 Reverse Primer: 5′-CTA CCA CCT GAA CGA CTG GAT G-3′.
Proteolysis and lectin enrichment
CAL27 and HSC-3 WCLs were prepared as follows: four flask (175 cm2) of adherent CAL27 and HSC-3 cells (60–70% confluent) were washed four times with PBS, followed by addition of 500 μL ice cold lysis buffer (8 M urea, 75 mM NaCl, 50 mM TEAB, 1 mM EDTA, and MS-SAFE Protease Inhibitor Cocktail), and cells were scraped, then transferred to new tubes. Lysate was placed on ice for 20 min and mixed every 5 min, then centrifuged at 16,100 RCF for 15 min. The supernatant was transferred to a new tube. Proteins were reduced (5 mM DTT, 37°C, 1 h) and alkylated (15 mM iodoacetamide, room temperature, 30 min), followed by quenching (30 mM DTT, 1 h). Protein was precipitated by addition of ice-cold ethanol/acetone/0.1% acetic acid and storage overnight at −20°C. Following precipitation, samples were centrifuged at 8000 RCF and 4°C for 10 min, decanted, and pellets containing proteins were retained and washed with 500 μL ethanol/acetone/0.1% acetic acid. Pellets were then resuspended in 100 mM TEAB, and trypsin was added at a ratio of 1:100 and digested overnight at 37°C with periodic mixing. Peptides were dried, resuspended, and desalted with Oasis HLB 1 cc (30 mg) extraction cartridges. Peptides were eluted in 80% acetonitrile/20% water with 0.1% formic acid, and dried under vacuum, then quantified via BCA. AAL enrichment of fucosylated glycopeptides was performed according to Zhou et al. (2017). Briefly, 150 μg of agarose-bound AAL was used for each 150 μg peptide sample. AAL agarose was washed three times with TBS, followed by addition of 150 μg peptides in 500 uL TBS with Halt protease inhibitors, and incubated overnight on a rocker at 4°C. The samples were then centrifuged for 1 min at 2500 RCF and the supernatant removed, and washed four times with 500 μL TBS followed by vortexing for 10 s. AAL-bound peptides were eluted by addition of 400 μL Glycoprotein Eluting Solution (Fucose/Arabinose) from Vector Laboratories, followed by vortexing for 30 s. This was repeated once, and eluates were combined. Eluate was passed through activated SepPak C18 1 cc 100 mg cartridges twice, washed 3 × 500 uL with water, and eluted with 50% acetonitrile/50% water with 0.1% formic acid, and dried under vacuum.
nanoLC-MS/MS analyses
Nano-liquid chromatography tandem mass spectrometry analyses were performed on an Orbitrap Eclipse Tribrid Mass Spectrometer with an online EASY nLC 1200 system (Thermo Fisher Scientific, Waltham, MA). An Acclaim PepMap 100 (75 μm, 2 cm) trapping column and a PepMap RSLC C18 analytical column (2 μm, 100 Å, 75 μm × 15 cm) were employed for chromatographic separation. Peptides were separated according to the following gradient: starting conditions 2% B, 2–6% B from 0 to 5 min, 6–35% B from 5 to 75 min, 35–60% B from 75 to 80 min, 60–95% B for 30 s, and 95% B for 9.5 min (solvents A and B consisted of 1% acetonitrile/99% water +0.1% formic acid and 80% acetonitrile/20% water +0.1% formic acid, respectively). All MS analyses were performed in positive mode and spectra were acquired using the orbitrap. For proteomic analyses, MS1 scans were acquired using the following parameters: RF lens 30%; resolution 120,000; m/z range 375–2000; cycle time 3 s; 50 ms injection time; AGC target 4 × 105; 1 μscan. For MS2 scans, peptides with charge states 2–6 were selected; min. Intensity 2 × 104; and dynamic exclusion of 1 min. An isolation window of 1.2 was used. HCD at 30% collision energy, and a maximum injection time of 45 ms, and first mass at m/z 130 were used. MS spectra were recorded as profile spectra, and MS2 as centroided spectra. For AAL-enriched samples and additional glycopeptide analyses, MS1 scans were acquired as above. For MS2, charge states 2–7 were considered, min intensity 2 × 104 and an exclusion time of 10 s were used. An HCD spectrum (35% collision energy, 15,000 resolution, 35 ms, 5 × 104 AGC target) was acquired, and detection of ≥3 glycan oxonium ion peaks (m/z 138.055, 168.066, 186.076, 204.087, 274.092, 292.103, 366.139; with 25 ppm mass tolerance) in the HCD spectrum triggered a second EThcD MS2 scan (ETD with 20% supplemental activation, 30,000 resolution, 1 μscan, 54 ms max. Injection time, 200% AGC, AGC target 1 × 105). MS and MS2 spectra were recorded as profile spectra. The mass spectrometry data have been deposited to the ProteomeXchange Consortium (Deutsch et al. 2017) via the PRIDE (Perez-Riverol et al. 2019) partner repository with the dataset identifier PXD029420.
Data availability statement
The data underlying this article are available in the article and in its online supplementary material. The mass spectrometry proteomics and glycoproteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al. 2019) partner repository with the dataset identifier PXD029420.
Author contributions
K.B.C. conceived the study, and K.B.C., B.M., N.H., G.D.Y., J.M.H. and R.S. contributed to study design. Material preparation, data collection and analysis were performed by K.B.C., B.M. and A.S. The first draft of the manuscript was written by K.B.C. and B.M., and all authors commented on versions of the manuscript. All authors read and approved the final manuscript.
Abbreviations
- AAL
Aleuria aurantia Lectin
- DOK
dysplastic oral keratinocytes
- EGFR
epidermal growth factor receptor
- EThcD
electron transfer dissociation with supplemental activation
- Fuc
fucose (F)
- HCD
higher-energy collisional dissociation
- Hex
hexose (H)
- HexNAc
N-acetylhexosamine (N)
- HNSCC
head and neck squamous cell carcinoma
- HPV
human papillomavirus
- LeB
Lewis B
- LeY
Lewis Y
- NOK
normal oral keratinocytes
- NeuAc
N-acetylneuraminic acid (S)
Supplementary Material
Acknowledgements
NOK were a gift from Dr Karl Munger at Tufts University (Medford, MA, USA).
Contributor Information
Brittany Montesino, Department of Translational Medicine, Herbert Wertheim College of Medicine, Translational Glycobiology Institute, Florida International University, 11200 SW 8th St., Miami, FL 33199, USA.
Agata Steenackers, Department of Translational Medicine, Herbert Wertheim College of Medicine, Translational Glycobiology Institute, Florida International University, 11200 SW 8th St., Miami, FL 33199, USA.
Juan M Lozano, Division of Medical and Population Health Science Education and Research, Herbert Wertheim College of Medicine, Florida International University, 11200 SW 8th St., Miami, FL 33199, USA.
Geoffrey D Young, Miami Cancer Institute, 8900 N Kendall Dr, Miami, FL 33176, USA; Department of Surgery, Herbert Wertheim College of Medicine, Florida International University, 11200 SW 8th St., Miami, FL 33199, USA.
Nan Hu, Department of Biostatistics, Robert Stempel College of Public Health and Social Work, Florida International University, 11200 SW 8th St., Miami, FL 33199, USA.
Robert Sackstein, Department of Translational Medicine, Herbert Wertheim College of Medicine, Translational Glycobiology Institute, Florida International University, 11200 SW 8th St., Miami, FL 33199, USA.
Kevin Brown Chandler, Department of Translational Medicine, Herbert Wertheim College of Medicine, Translational Glycobiology Institute, Florida International University, 11200 SW 8th St., Miami, FL 33199, USA; Biomolecular Sciences Institute, Florida International University, 11200 SW 8th St., Miami, FL 33199, USA.
Funding
K12 Harvard Career Development Award in Translational Glycobiology (ProTG): “Bridging Glycoscience and Clinical Medicine” from the National Heart, Lung, and Blood Institute, 5K12HL141953–02 (to K.B.C., awarded to R.S.); the Florida International University Herbert Wertheim College of Medicine Summer Research Fellowship Program (to B.M.).
Conflict of interest statement
None declared.
References
- Abu-Humaidan AHA, Ekblad L, Wennerberg J, Sørensen OE. 2020. EGFR modulates complement activation in head and neck squamous cell carcinoma. BMC Cancer. 20(1):121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acs A, Ozohanics O, Vekey K, Drahos L, Turiak L. 2018. Distinguishing core and antenna fucosylated glycopeptides based on low-energy tandem mass spectra. Anal Chem. 90(21):12776–12782. [DOI] [PubMed] [Google Scholar]
- Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J et al. 2011. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 333(6046):1154–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldus SE, Thiele J, Park YO, Hanisch FG, Bara J, Fischer R. 1996. Characterization of the binding specificity of Anguilla anguilla agglutinin (AAA) in comparison to Ulex europaeus agglutinin I (UEA-I). Glycoconj J. 13(4):585–590. [DOI] [PubMed] [Google Scholar]
- Britain CM, Bhalerao N, Silva AD, Chakraborty A, Buchsbaum DJ, Crowley MR, Crossman DK, Edwards YJK, Bellis SL. 2021. Glycosyltransferase ST6Gal-I promotes the epithelial to mesenchymal transition in pancreatic cancer cells. J Biol Chem. 296:100034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JD, Yau C, Bowlby R, Liu Y, Brennan K, Fan H, Taylor AM, Wang C, Walter V, Akbani R et al. 2018. Genomic, pathway network, and immunologic features distinguishing squamous carcinomas. Cell Rep. 23(1):194–212.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campi C, Escovich L, Moreno A, Racca L, Racca A, Cotorruelo C, Biondi C. 2012. Expression of the gene encoding secretor type galactoside 2 alpha fucosyltransferase (FUT2) and ABH antigens in patients with oral lesions. Med Oral Patol Oral Cir Bucal. 17(1):e63–e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network . 2015. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 517(7536):576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler KB, Leon DR, Meyer RD, Rahimi N, Costello CE. 2017. Site-specific N-glycosylation of endothelial cell receptor tyrosine kinase VEGFR-2. J Proteome Res. 16(2):677–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler KB, Leon DR, Kuang J, Meyer RD, Rahimi N, Costello CE. 2019. N-glycosylation regulates ligand-dependent activation and signaling of vascular endothelial growth factor receptor 2 (VEGFR2). J Biol Chem. 294(35):13117–13130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler KB, Alamoud KA, Stahl VL, Nguyen BC, Kartha VK, Bais MV, Nomoto K, Owa T, Monti S, Kukuruzinska MA et al. 2020. β-Catenin/CBP inhibition alters epidermal growth factor receptor fucosylation status in oral squamous cell carcinoma. Mol Omics. 16(3):195–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YT, Chong YM, Cheng CW, Ho CL, Tsai HW, Kasten FH, Chen YL, Chang CF. 2013. Identification of novel tumor markers for oral squamous cell carcinoma using glycoproteomic analysis. Clin Chim Acta. 420:45–53. [DOI] [PubMed] [Google Scholar]
- Chow LQM. 2020. Head and neck cancer. N Engl J Med. 382(1):60–72. [DOI] [PubMed] [Google Scholar]
- Chung TW, Kim EY, Kim SJ, Choi HJ, Jang SB, Kim KJ, Ha SH, Abekura F, Kwak CH, Kim CH et al. 2017. Sialyllactose suppresses angiogenesis by inhibiting VEGFR-2 activation, and tumor progression. Oncotarget. 8(35):58152–58162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croci DO, Cerliani JP, Dalotto-Moreno T, Mendez-Huergo SP, Mascanfroni ID, Dergan-Dylon S, Toscano MA, Caramelo JJ, Garcia-Vallejo JJ, Ouyang J et al. 2014. Glycosylation-dependent lectin-receptor interactions preserve angiogenesis in anti-VEGF refractory tumors. Cell. 156(4):744–758. [DOI] [PubMed] [Google Scholar]
- Deutsch EW, Csordas A, Sun Z, Jarnuczak A, Perez-Riverol Y, Ternent T, Campbell DS, Bernal-Llinares M, Okuda S, Kawano S et al. 2017. The ProteomeXchange consortium in 2017: Supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 45(D1):D1100–d1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engle DD, Tiriac H, Rivera KD, Pommier A, Whalen S, Oni TE, Alagesan B, Lee EJ, Yao MA, Lucito MS et al. 2019. The glycan CA19-9 promotes pancreatitis and pancreatic cancer in mice. Science. 364(6446):1156–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes H, Cohen S, Bishayee S. 2001. Glycosylation-induced conformational modification positively regulates receptor-receptor association: A study with an aberrant epidermal growth factor receptor (EGFRvIII/DeltaEGFR) expressed in cancer cells. J Biol Chem. 276(7):5375–5383. [DOI] [PubMed] [Google Scholar]
- Fornaro M, Dell'Arciprete R, Stella M, Bucci C, Nutini M, Capri MG, Alberti S. 1995. Cloning of the gene encoding Trop-2, a cell-surface glycoprotein expressed by human carcinomas. Int J Cancer. 62(5):610–618. [DOI] [PubMed] [Google Scholar]
- García-Vallejo JJ, van Liempt E, da Costa MP, Beckers C, van het Hof B, Gringhuis SI, Zwaginga JJ, van Dijk W, Geijtenbeek TB, van Kooyk Y et al. 2008. DC-SIGN mediates adhesion and rolling of dendritic cells on primary human umbilical vein endothelial cells through LewisY antigen expressed on ICAM-2. Mol Immunol. 45(8):2359–2369. [DOI] [PubMed] [Google Scholar]
- Geijtenbeek TB, Torensma R, van Vliet SJ, van Duijnhoven GC, Adema GJ, van Kooyk Y, Figdor CG. 2000. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell. 100(5):575–585. [DOI] [PubMed] [Google Scholar]
- Goldman MJ, Craft B, Hastie M, Repečka K, McDade F, Kamath A, Banerjee A, Luo Y, Rogers D, Brooks AN et al. 2020. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 38(6):675–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman RL, Heath AP, Ferretti V, Varmus HE, Lowy DR, Kibbe WA, Staudt LM. 2016. Toward a shared vision for cancer genomic data. N Engl J Med. 375(12):1109–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdbrooks AT, Britain CM, Bellis SL. 2018. ST6Gal-I sialyltransferase promotes tumor necrosis factor (TNF)-mediated cancer cell survival via sialylation of the TNF receptor 1 (TNFR1) death receptor. J Biol Chem. 293(5):1610–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta H, Hamamura K, Yamashita K, Shibuya H, Tokuda N, Hashimoto N, Furukawa K, Yamamoto N, Hattori H, Toyokuni S et al. 2013. Lewis y antigen is expressed in oral squamous cell carcinoma cell lines and tissues, but disappears in the invasive regions leading to the enhanced malignant properties irrespective of sialyl-Lewis x. Glycoconj J. 30(6):585–597. [DOI] [PubMed] [Google Scholar]
- Hotta H, Hamamura K, Shibuya H, Ohmi Y, Furukawa K, Furukawa K. 2021. Lewis y expressed in oral squamous cell carcinoma attenuates malignant properties via down-regulation of EGF Signaling. Anticancer Res. 41(4):1821–1830. [DOI] [PubMed] [Google Scholar]
- Jacobs PP, Sackstein R. 2011. CD44 and HCELL: Preventing hematogenous metastasis at step 1. FEBS Lett. 585(20):3148–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamal B, Sengupta PK, Gao ZN, Nita-Lazar M, Amin B, Jalisi S, Bouchie MP, Kukuruzinska MA. 2012. Aberrant amplification of the crosstalk between canonical Wnt signaling and N-glycosylation gene DPAGT1 promotes oral cancer. Oral Oncol. 48(6):523–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jethwa AR, Khariwala SS. 2017. Tobacco-related carcinogenesis in head and neck cancer. Cancer Metastasis Rev. 36(3):411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DE, O'Keefe RA, Grandis JR. 2018. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol. 15(4):234–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DE, Burtness B, Leemans CR, Lui VWY, Bauman JE, Grandis JR. 2020. Head and neck squamous cell carcinoma. Nat Rev Dis Primers. 6(1):92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartha VK, Alamoud KA, Sadykov K, Nguyen BC, Laroche F, Feng H, Lee J, Pai SI, Varelas X, Egloff AM et al. 2018. Functional and genomic analyses reveal therapeutic potential of targeting β-catenin/CBP activity in head and neck cancer. Genome Med. 10(1):54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaszuba K, Grzybek M, Orlowski A, Danne R, Rog T, Simons K, Coskun U, Vattulainen I. 2015. N-glycosylation as determinant of epidermal growth factor receptor conformation in membranes. Proc Natl Acad Sci U S A. 112(14):4334–4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesselring R, Thiel A, Pries R, Fichtner-Feigl S, Brunner S, Seidel P, Bruchhage KL, Wollenberg B. 2014. The complement receptors CD46, CD55 and CD59 are regulated by the tumour microenvironment of head and neck cancer to facilitate escape of complement attack. Eur J Cancer. 50(12):2152–2161. [DOI] [PubMed] [Google Scholar]
- Kikkawa Y, Ogawa T, Sudo R, Yamada Y, Katagiri F, Hozumi K, Nomizu M, Miner JH. 2013. The lutheran/basal cell adhesion molecule promotes tumor cell migration by modulating integrin-mediated cell attachment to laminin-511 protein. J Biol Chem. 288(43):30990–31001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Lee JC, Lee JJ, Kim S, Lee SG, Park SW, Sung MW, Heo DS. 2008. Association of CD47 with natural killer cell-mediated cytotoxicity of head-and-neck squamous cell carcinoma lines. Tumour Biol. 29(1):28–34. [DOI] [PubMed] [Google Scholar]
- van Liempt E, Bank CM, Mehta P, Garciá-Vallejo JJ, Kawar ZS, Geyer R, Alvarez RA, Cummings RD, Kooyk Y, van Die I. 2006. Specificity of DC-SIGN for mannose- and fucose-containing glycans. FEBS Lett. 580(26):6123–6131. [DOI] [PubMed] [Google Scholar]
- Lin MC, Huang MJ, Liu CH, Yang TL, Huang MC. 2014. GALNT2 enhances migration and invasion of oral squamous cell carcinoma by regulating EGFR glycosylation and activity. Oral Oncol. 50(5):478–484. [DOI] [PubMed] [Google Scholar]
- Lin WL, Lin YS, Shi GY, Chang CF, Wu HL. 2015. Lewisy promotes migration of oral cancer cells by glycosylation of epidermal growth factor receptor. PLoS One. 10(3):e0120162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YC, Yen HY, Chen CY, Chen CH, Cheng PF, Juan YH, Chen CH, Khoo KH, Yu CJ, Yang PC et al. 2011. Sialylation and fucosylation of epidermal growth factor receptor suppress its dimerization and activation in lung cancer cells. Proc Natl Acad Sci U S A. 108(28):11332–11337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui VW, Hedberg ML, Li H, Vangara BS, Pendleton K, Zeng Y, Lu Y, Zhang Q, Du Y, Gilbert BR et al. 2013. Frequent mutation of the PI3K pathway in head and neck cancer defines predictive biomarkers. Cancer Discov. 3(7):761–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyford-Pike S, Peng S, Young GD, Taube JM, Westra WH, Akpeng B, Bruno TC, Richmon JD, Wang H, Bishop JA et al. 2013. Evidence for a role of the PD-1:PD-L1 pathway in immune resistance of HPV-associated head and neck squamous cell carcinoma. Cancer Res. 73(6):1733–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marur S, Forastiere AA. 2016. Head and neck squamous cell carcinoma: Update on epidemiology, diagnosis, and treatment. Mayo Clin Proc. 91(3):386–396. [DOI] [PubMed] [Google Scholar]
- Matsumura K, Higashida K, Ishida H, Hata Y, Yamamoto K, Shigeta M, Mizuno-Horikawa Y, Wang X, Miyoshi E, Gu J et al. 2007. Carbohydrate binding specificity of a fucose-specific lectin from aspergillus oryzae: A novel probe for core fucose. J Biol Chem. 282(21):15700–15708. [DOI] [PubMed] [Google Scholar]
- Mehta KA, Patel KA, Pandya SJ, Patel PS. 2020. Aberrant sialylation plays a significant role in oral squamous cell carcinoma progression. J Oral Pathol Med. 49(3):253–259. [DOI] [PubMed] [Google Scholar]
- Molinolo AA, Amornphimoltham P, Squarize CH, Castilho RM, Patel V, Gutkind JS. 2009. Dysregulated molecular networks in head and neck carcinogenesis. Oral Oncol. 45(4–5):324–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkley J, Elliott DJ. 2016. Hallmarks of glycosylation in cancer. Oncotarget. 7(23):35478–35489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nita-Lazar M, Noonan V, Rebustini I, Walker J, Menko AS, Kukuruzinska MA. 2009. Overexpression of DPAGT1 leads to aberrant N-glycosylation of E-cadherin and cellular discohesion in oral cancer. Cancer Res. 69(14):5673–5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M et al. 2019. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47(D1):D442–d450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piboonniyom SO, Duensing S, Swilling NW, Hasskarl J, Hinds PW, Münger K. 2003. Abrogation of the retinoblastoma tumor suppressor checkpoint during keratinocyte immortalization is not sufficient for induction of centrosome-mediated genomic instability. Cancer Res. 63(2):476–483. [PubMed] [Google Scholar]
- Pinho SS, Reis CA. 2015. Glycosylation in cancer: Mechanisms and clinical implications. Nat Rev Cancer. 15(9):540–555. [DOI] [PubMed] [Google Scholar]
- Resto VA, Burdick MM, Dagia NM, McCammon SD, Fennewald SM, Sackstein R. 2008. L-selectin-mediated lymphocyte-cancer cell interactions under low fluid shear conditions. J Biol Chem. 283(23):15816–15824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro FA, Noguti J, Oshima CT, Ribeiro DA. 2014. Effective targeting of the epidermal growth factor receptor (EGFR) for treating oral cancer: A promising approach. Anticancer Res. 34(4):1547–1552. [PubMed] [Google Scholar]
- Ripani E, Sacchetti A, Corda D, Alberti S. 1998. Human Trop-2 is a tumor-associated calcium signal transducer. Int J Cancer. 76(5):671–676. [DOI] [PubMed] [Google Scholar]
- Roland CL, Harken AH, Sarr MG, Barnett CC Jr. 2007. ICAM-1 expression determines malignant potential of cancer. Surgery. 141(6):705–707. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Brakebusch C, Engel J, Timpl R. 1998. Mac-2 binding protein is a cell-adhesive protein of the extracellular matrix which self-assembles into ring-like structures and binds beta1 integrins, collagens and fibronectin. EMBO J. 17(6):1606–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada R, Lowe JB, Fukuda M. 1993. E-selectin-dependent adhesion efficiency of colonic carcinoma cells is increased by genetic manipulation of their cell surface lysosomal membrane glycoprotein-1 expression levels. J Biol Chem. 268(17):12675–12681. [PubMed] [Google Scholar]
- Schultz MJ, Swindall AF, Bellis SL. 2012. Regulation of the metastatic cell phenotype by sialylated glycans. Cancer Metastasis Rev. 31(3–4):501–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz MJ, Swindall AF, Wright JW, Sztul ES, Landen CN, Bellis SL. 2013. ST6Gal-I sialyltransferase confers cisplatin resistance in ovarian tumor cells. J Ovarian Res. 6(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz MJ, Holdbrooks AT, Chakraborty A, Grizzle WE, Landen CN, Buchsbaum DJ, Conner MG, Arend RC, Yoon KJ, Klug CA et al. 2016. The tumor-associated glycosyltransferase ST6Gal-I regulates stem cell transcription factors and confers a cancer stem cell phenotype. Cancer Res. 76(13):3978–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seales EC, Jurado GA, Singhal A, Bellis SL. 2003. Ras oncogene directs expression of a differentially sialylated, functionally altered beta1 integrin. Oncogene. 22(46):7137–7145. [DOI] [PubMed] [Google Scholar]
- Seales EC, Jurado GA, Brunson BA, Wakefield JK, Frost AR, Bellis SL. 2005. Hypersialylation of beta1 integrins, observed in colon adenocarcinoma, may contribute to cancer progression by up-regulating cell motility. Cancer Res. 65(11):4645–4652. [DOI] [PubMed] [Google Scholar]
- Sengupta PK, Bouchie MP, Kukuruzinska MA. 2010. N-glycosylation gene DPAGT1 is a target of the Wnt/beta-catenin signaling pathway. J Biol Chem. 285(41):31164–31173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan M, Yang D, Dou H, Zhang L. 2019. Fucosylation in cancer biology and its clinical applications. Prog Mol Biol Transl Sci. 162:93–119. [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. 2019. Cancer statistics, 2019. CA Cancer J Clin. 69(1):7–34. [DOI] [PubMed] [Google Scholar]
- Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A et al. 2011. The mutational landscape of head and neck squamous cell carcinoma. Science. 333(6046):1157–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su KJ, Ho CC, Lin CW, Chen MK, Su SC, Yu YL, Yang SF. 2016. Combinations of FUT2 gene polymorphisms and environmental factors are associated with oral cancer risk. Tumour Biol. 37(5):6647–6652. [DOI] [PubMed] [Google Scholar]
- Swindall AF, Londoño-Joshi AI, Schultz MJ, Fineberg N, Buchsbaum DJ, Bellis SL. 2013. ST6Gal-I protein expression is upregulated in human epithelial tumors and correlates with stem cell markers in normal tissues and colon cancer cell lines. Cancer Res. 73(7):2368–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Lin J, Tian Y, Zhang G, Zeng X, Zheng R, Zhang W, Yuan Y. 2018. Efficacy and safety of anti-EGFR agents administered concurrently with standard therapies for patients with head and neck squamous cell carcinoma: A systematic review and meta-analysis of randomized controlled trials. Int J Cancer. 142(11):2198–2206. [DOI] [PubMed] [Google Scholar]
- Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A et al. 2015. Proteomics. Tissue-based map of the human proteome. Science. 347(6220):1260419. [DOI] [PubMed] [Google Scholar]
- Ullrich A, Sures I, D'Egidio M, Jallal B, Powell TJ, Herbst R, Dreps A, Azam M, Rubinstein M, Natoli C et al. 1994. The secreted tumor-associated antigen 90K is a potent immune stimulator. J Biol Chem. 269(28):18401–18407. [PubMed] [Google Scholar]
- Varelas X, Kukuruzinska MA. 2014. Head and neck cancer: From research to therapy and cure. Ann N Y Acad Sci. 1333(1):1–32. [DOI] [PubMed] [Google Scholar]
- Veglia F, Gabrilovich DI. 2017. Dendritic cells in cancer: The role revisited. Curr Opin Immunol. 45:43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Very N, Lefebvre T, El Yazidi-Belkoura I. 2018. Drug resistance related to aberrant glycosylation in colorectal cancer. Oncotarget. 9(1):1380–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneswaran N, Williams MD. 2014. Epidemiologic trends in head and neck cancer and aids in diagnosis. Oral Maxillofac Surg Clin North Am. 26(2):123–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Gu J, Ihara H, Miyoshi E, Honke K, Taniguchi N. 2006. Core fucosylation regulates epidermal growth factor receptor-mediated intracellular signaling. J Biol Chem. 281(5):2572–2577. [DOI] [PubMed] [Google Scholar]
- Watkins WM. 1980. Biochemistry and genetics of the ABO, Lewis, and P blood group systems. Adv Hum Genet. 10:1–136 379-85. [DOI] [PubMed] [Google Scholar]
- Wen Y, Grandis JR. 2015. Emerging drugs for head and neck cancer. Expert Opin Emerg Drugs. 20(2):313–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitson KB, Whitson SR, Red-Brewer ML, McCoy AJ, Vitali AA, Walker F, Johns TG, Beth AH, Staros JV. 2005. Functional effects of glycosylation at Asn-579 of the epidermal growth factor receptor. Biochemistry. 44(45):14920–14931. [DOI] [PubMed] [Google Scholar]
- Xiao M, Yan M, Zhang J, Xu Q, Qi S, Wang X, Chen W. 2017. Cancer stem-like cell related protein CD166 degrades through E3 ubiquitin ligase CHIP in head and neck cancer. Exp Cell Res. 353(1):46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu MJ, Johnson DE, Grandis JR. 2017. EGFR-targeted therapies in the post-genomic era. Cancer Metastasis Rev. 36(3):463–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen HY, Liu YC, Chen NY, Tsai CF, Wang YT, Chen YJ, Hsu TL, Yang PC, Wong CH. 2015. Effect of sialylation on EGFR phosphorylation and resistance to tyrosine kinase inhibition. Proc Natl Acad Sci U S A. 112(22):6955–6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Wei R, Goldman R, Sanda M. 2019. Optimized fragmentation for quantitative analysis of Fucosylated N-glycoproteins by LC-MS-MRM. Anal Chem. 91(14):9206–9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerfaoui M, Fukuda M, Sbarra V, Lombardo D, El-Battari A. 2000. Alpha(1,2)-fucosylation prevents sialyl Lewis x expression and E-selectin-mediated adhesion of fucosyltransferase VII-transfected cells. Eur J Biochem. 267(1):53–61. [DOI] [PubMed] [Google Scholar]
- Zhou J, Yang W, Hu Y, Hoti N, Liu Y, Shah P, Sun S, Clark D, Thomas S, Zhang H. 2017. Site-specific fucosylation analysis identifying glycoproteins associated with aggressive prostate cancer cell lines using tandem affinity enrichments of intact glycopeptides followed by mass spectrometry. Anal Chem. 89(14):7623–7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material. The mass spectrometry proteomics and glycoproteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al. 2019) partner repository with the dataset identifier PXD029420.