Abstract
Background
Predicting outcomes of patients with COVID-19 at an early stage is crucial for optimised clinical care and resource management, especially during a pandemic. Although multiple machine learning models have been proposed to address this issue, because of their requirements for extensive data preprocessing and feature engineering, they have not been validated or implemented outside of their original study site. Therefore, we aimed to develop accurate and transferrable predictive models of outcomes on hospital admission for patients with COVID-19.
Methods
In this study, we developed recurrent neural network-based models (CovRNN) to predict the outcomes of patients with COVID-19 by use of available electronic health record data on admission to hospital, without the need for specific feature selection or missing data imputation. CovRNN was designed to predict three outcomes: in-hospital mortality, need for mechanical ventilation, and prolonged hospital stay (>7 days). For in-hospital mortality and mechanical ventilation, CovRNN produced time-to-event risk scores (survival prediction; evaluated by the concordance index) and all-time risk scores (binary prediction; area under the receiver operating characteristic curve [AUROC] was the main metric); we only trained a binary classification model for prolonged hospital stay. For binary classification tasks, we compared CovRNN against traditional machine learning algorithms: logistic regression and light gradient boost machine. Our models were trained and validated on the heterogeneous, deidentified data of 247 960 patients with COVID-19 from 87 US health-care systems derived from the Cerner Real-World COVID-19 Q3 Dataset up to September 2020. We held out the data of 4175 patients from two hospitals for external validation. The remaining 243 785 patients from the 85 health systems were grouped into training (n=170 626), validation (n=24 378), and multi-hospital test (n=48 781) sets. Model performance was evaluated in the multi-hospital test set. The transferability of CovRNN was externally validated by use of deidentified data from 36 140 patients derived from the US-based Optum deidentified COVID-19 electronic health record dataset (version 1015; from January, 2007, to Oct 15, 2020). Exact dates of data extraction were masked by the databases to ensure patient data safety.
Findings
CovRNN binary models achieved AUROCs of 93·0% (95% CI 92·6–93·4) for the prediction of in-hospital mortality, 92·9% (92·6–93·2) for the prediction of mechanical ventilation, and 86·5% (86·2–86·9) for the prediction of a prolonged hospital stay, outperforming light gradient boost machine and logistic regression algorithms. External validation confirmed AUROCs in similar ranges (91·3–97·0% for in-hospital mortality prediction, 91·5–96·0% for the prediction of mechanical ventilation, and 81·0–88·3% for the prediction of prolonged hospital stay). For survival prediction, CovRNN achieved a concordance index of 86·0% (95% CI 85·1–86·9) for in-hospital mortality and 92·6% (92·2–93·0) for mechanical ventilation.
Interpretation
Trained on a large, heterogeneous, real-world dataset, our CovRNN models showed high prediction accuracy and transferability through consistently good performances on multiple external datasets. Our results show the feasibility of a COVID-19 predictive model that delivers high accuracy without the need for complex feature engineering.
Funding
Cancer Prevention and Research Institute of Texas.
Introduction
COVID-19 is an infectious disease caused by SARS-CoV-2, which emerged in December, 2019.1 By the end of 2021, there were more than 295 million confirmed SARS-CoV-2 infections worldwide and more than 825 000 deaths due to COVID-19 in the USA alone.2 Furthermore, there have been around 3·7 million COVID-19-related hospital admissions recorded since August 2020 in the USA.2 During the peaks of the pandemic waves, many US states reported near-capacity hospital and intensive care unit use. Accurate prediction of the future clinical trajectories of patients with COVID-19 at the time of admission is crucial for clinical decision making and enables the efficient allocation of resources. Indeed, several models for the prediction of COVID-19 outcomes have been developed. Wynants and colleagues3 reviewed 107 COVID-19 prognostic models published before July 1, 2020. The most common issue highlighted in this study was the high risk of bias associated with the reviewed models, which was caused by either a small, locally sourced training dataset and the subsequent high risk of model overfitting or the absence of model calibration or external validation.4, 5 Through an updated survey of the literature, as of Dec 31, 2021, we found that only four studies6, 7, 8, 9 involved training the proposed models of COVID-19 outcomes on data from more than 20 000 patients. Moreover, all four models are based on a small set of specific features and need a laborious data preprocessing and feature engineering process that limits the transferability, reliability, and sustainability of the models.
Research in context.
Evidence before this study
Although many methods for predicting COVID-19 outcomes have been developed, they have not been extensively externally validated due to their limited transferability. A key obstacle to the transferability of such methods is the need for laborious data preprocessing and feature engineering. A 2020 systematic review that critically assessed prediction models for diagnosing and prognosing COVID-19 revealed that the majority of 107 prognostic models published before July 1, 2020, have a high risk of overfitting bias. Using the Prediction model Risk Of Bias ASsessment Tool (known as PROBAST), the authors identified common reasons for biased results, including training the model on a small, locally sourced dataset, which leads to a high risk of model overfitting, and the absence of model calibration or external validation. To provide an updated survey of the literature, we searched Scopus and PubMed for articles published in English between July 1, 2020, and Dec 31, 2021 predicting COVID-19 outcomes using the keywords “COVID electronic health record (‘mortality’ or ‘ventilator’ or ‘length of stay’ or ‘real-time’) prediction”. The literature search retrieved a total of 466 unique articles, and, on review, we found 53 studies that describe the development and validation of machine learning predictive models for predicting prognosis for patients with COVID-19 after admission. Of the 53 studies, only four involved training and evaluating the models on a multi-sourced cohort of more than 20 000 patients with COVID-19. The proposed models in these studies, however, still require extensive data preprocessing and feature engineering, which limits the transferability, reliability, and sustainability of such models.
Added value of this study
We propose a machine learning model training framework that can flexibly adapt to the changing pandemic and requires minimal preprocessing. For convenience and practicality, our framework is designed to consume electronic health record data mapped to standard terminologies in common use without the need for specific feature selection or missing value imputation. Because they were trained and evaluated on large, heterogenous datasets collected from different health systems, our COVID-19 outcome prediction models (CovRNN) showed high accuracy in predicting three outcomes (in-hospital mortality, need for mechanical ventilation, and prolonged hospital stay), outperforming the prediction accuracy of state-of-the-art models in the literature, good calibration, and had a low risk of bias. In addition, our models can be fine-tuned on new data for continuous improvement, as recommended by the US Food and Drug Administration's Good Machine Learning Practice. Furthermore, our framework includes a utility for model predictions explanation to facilitate clinical judgment of the model predictions.
Implications of all the available evidence
While consuming structured, categorical data from electronic health records, deep learning-based models can achieve state-of-the-art prediction accuracy in their standard format without the need for features selection or missing value imputations, which implies that the trained models can be easily validated on new data sources. We validated our trained models across datasets from different sources, indicating the transferability of our models. Our model development framework can be further applied to train and evaluate predictive models for different types of clinical events. For clinicians who are fighting COVID-19 on the frontlines, there are two potentially actionable contributions of our work. Clinicians can (1) fine-tune our pretrained models on their local data (regardless of cohort size), establish utility, and then deploy the models and (2) use our comprehensive model development framework to train a predictive model using their own data.
In this study, we aimed to develop accurate and transferrable models of the outcomes on admission for patients with COVID-19. Our models, CovRNN, use a gated recurrent neural network architecture proven to be effective in modelling patients’ electronic health record data.10, 11, 12, 13, 14
Methods
Datasets and cohort description
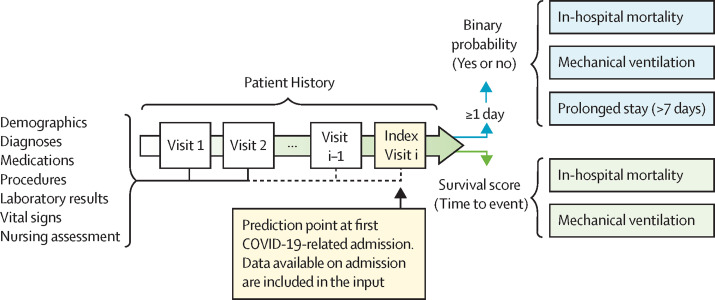
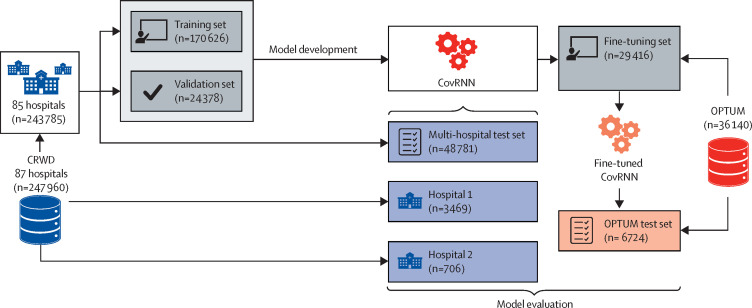
We extracted our main training set from the Cerner Real-World COVID-19 Q3 Dataset (CRWD) hosted on the Cerner HealtheDataLab, which is a cloud-based, large, heterogenous, deidentified dataset including clinical data from patients with COVID-19 from 87 US health systems up to the end of September, 2020 (appendix 1 p 1). The CRWD includes only patients who had a minimum of one emergency or inpatient encounter with a diagnosis code that could be associated with COVID-19 exposure or infection or a positive result from a COVID-19 laboratory test. The CRWD includes patients’ medical histories for up to 5 years before their first SARS-CoV-2 infection. In our study, we predefined our prediction point as the first day of COVID-19-related admission to an emergency, observation, or inpatient unit in hospital, and we refer to this point as the index date (figure 1 ). We thereby excluded all patients who had no recorded clinical information on or before the index date and patients who stayed in hospital for less than 1 day (appendix 1 p 3). We also excluded patients who had inconsistent dates, such as a discharge date before the hospitalisation date, and patients who were readmitted later and presented different outcomes. For further external validation outside of the CRWD, we extracted deidentified data from a US-based cohort derived from the Optum deidentified COVID-19 electronic health record dataset (version 1015; data from January, 2007, to Oct 15, 2020), which we refer to as the OPTUM cohort (figure 2 ). Further description of the Optum dataset, along with a discussion of the differences and commonalities between the CRWD and OPTUM cohorts, are available in appendix 1 (pp 1–2). Exact dates of data extraction were masked by the databases to ensure patient data safety. The Committee for the Protection of Human Subjects at the University of Texas Health Science Center in Houston, TX, USA, reviewed the Analysis of COVID-19 related data in Cerner's HealtheDataLab project (Institutional Review Board number HSC-SBMI-20-0836). The committee determined the project to qualify for exempt status according to 45 CFR 46.101(b).
Figure 1.
CovRNN prediction tasks
Visit i represents the index visit. Visit i–1 represents the visit before the index visit.
Figure 2.
Model development and external validation datasets
CRWD=Cerner Real-World COVID-19 Q3 Dataset. OPTUM=Optum deidentified COVID-19 electronic health record dataset.
Data preparation
We kept our data curation to a minimum to facilitate the transferability of our trained models to different datasets. We extracted all patient information on or before the date of their first hospital admission with COVID-19 (the index date), including demographics, diagnosis, medication, procedures, laboratory results, and observations (eg, nursing assessment and vital signs). To facilitate interoperability, we utilised standard terminologies or codes in common use: the ninth and tenth revisions of the International Classification of Diseases (ICD-9 and ICD-10) and the Systematized Nomenclature Of Medicine-Clinical Terms (SNOMED-CT) for diagnosis; Logical Observational Identifiers Names and Codes (known as LOINC) and SNOMED-CT for laboratory results and observations; Multum drug identifiers and categories for medications; and Current Procedural Terminology Fourth Edition (known as CPT-4), the Healthcare Common Procedure Coding System (known as HCPCS), and procedure codes in the ICD-9 and ICD-10 for procedures. Such standard terminologies are readily accesible in the majority of electronic health record systems. In cases for which Multum codes for medication were not used, we utilised pre-existing mapping tools15 to convert National Drug Codes to corresponding Multum information.
The majority of our features, such as diagnosis, medications, and procedures, were categorical. We converted numerical variables, such as laboratory results, to categorical variables as follows. For the CRWD, we converted numerical results into the “below normal low”, “normal”, or “above normal high” interpretation values that were provided in the CRWD; for OPTUM, we defined the result categories (“below normal low”, “normal”, or “above normal high”) on the basis of the corresponding normal result ranges. By doing so, we could further convert our input to either multi-hot or embedding matrices to feed to our models. On the basis of our previous study,16 we decided to use the clinical information in the coding standards with which it was recorded, as the normalisation of those codes to a more unified terminology provides minimal gain.16 Further details on our data curation are available in appendix 1 (p 3) and online.
Models
CovRNN models were based on a gated type of recurrent neural network, namely a gated recurrent unit, which is known for being an efficient sequential deep learning architecture for clinical event predictions.12, 17 The source code of our models is publicly available to enable its application and further evaluation by other researchers. For convenience and practicality, our models were designed to consume all demographics, diagnoses, medications, procedures, laboratory results, and other clinical event information readily available in electronic health records before or on the index date to predict patient outcomes without the need for specific feature selection or missing value imputation. CovRNN also consumes the time difference between visits for a better temporal representation of patient history, which is known to slightly improve the accuracy of predictions.18, 19 For binary classification tasks, we compared CovRNN against traditional machine learning algorithms: logistic regression20 and light gradient boost machine.21 For survival prediction, we used the DeepSurv22 architecture while replacing the multiple layer perceptron layers with gated recurrent unit layers for better sequential information modelling. We were unable to adequately compare against machine learning survival models, such as random survival forest or running proper factor analysis, due to computational resource restrictions on the Cerner HealtheDataLab, especially with the increased number of covariates and large training set size. Any version of the random survival forest model runnable on Cerner HealtheDataLab had a very small number of iterations or trees that led to poor and unreliable results; therefore, we decided not to report these results. Further implementation details are available in appendix 1 (p 5). We evaluated the prospective compliance of CovRNN against quality standards: the Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD) statement and the Prediction model Risk Of Bias ASsessment Tool (PROBAST).
Outcomes of prediction tasks
Our tasks comprised the prediction of patients’ in-hospital mortality, need for mechanical ventilation during the stay, and requirement for a prolonged hospital stay (>7 days) on admission. For defining in-hospital mortality, we relied on the preassigned mortality flags on the CRWD along with the “expired” encounter discharge disposition to confirm in-hospital mortality and identify the date of death. In-hospital mortality event definition was slightly different for the OPTUM data because there was no clear discharge disposition that indicated patient in-hospital death. We instead used the date of death and compared it against the hospitalisation discharge date to assign the proper label. For the need for mechanical ventilation outcome, we mainly used relevant mechanical ventilation procedure codes to define the outcome. In addition, with the CRWD, we used other relevant observations and recorded ventilator settings to identify the instance of the event and the earliest time of the event (appendix 1 p 4). For in-hospital mortality and mechanical ventilation prediction tasks, we trained survival-based and binary classification-based prediction models. CovRNN predicts a time-to-event risk score that can be interpreted as a binary prediction with a time horizon (survival prediction) and an all-time risk score (binary prediction). For the survival analysis, we defined the time to event as the number of days between our index date and the earliest date that indicated the occurrence of the event, either a laboratory result or a recorded procedure for mechanical ventilation or the discharge date for in-hospital mortality. We used the discharge date from hospital as the censoring time. We defined a prolonged hospital stay as a binary indicator for hospitalisations lasting longer than 7 days because the median length of stay in the CRWD was 3 days (IQR 1–6) and the median length of stay in the OPTUM cohort was 5 days (3–10); we only trained a binary classification model for the prolonged length of stay task.
On the basis of our literature review and recommendations from clinical collaborators, we used the area under the receiver operating characteristic curve (AUROC) as the main model-discriminative performance metric for the binary prediction models. We also report other clinically relevant metrics: specificity at 95% sensitivity, the area under the precision-recall curve (AUPRC), F1-score, and the sensitivity and specificity at the optimum threshold (the cutoff probability that leads to the highest sensitivity and specificity; appendix p 8) defined by use of the validation set. For the survival analysis, we report the concordance index23 as our main evaluation metric, and conducted a stratified analysis using the predicted survival scores to stratify patients into low (1–30th percentile), medium (31–80th percentile), or high (81–100th percentile) risk groups, comparing against patient survival plotted by Kaplan–Meier curves. In addition, we used the predicted survival score to calculate the AUROC at any time between day 1 and 128.
Experiments and calibration
The total CRWD cohort comprised data from 247 960 patients from 87 health systems, from which we removed (held out) the data of 4175 patients from two randomly selected hospitals (hospital 1 from the south region [n=3469] and hospital 2 from the west region [n=706]) for external validation, evaluating cross-hospital generalisability. The remaining 243 785 patients from the 85 health systems were grouped into training (n=170 626), validation (n=24 378), and multi-hospital test (n=48 781) sets in a ratio of 7:1:2 (figure 2). The validation set was used to determine the best model trained while controlling for overfitting. All of our reported CRWD results (model performance) are from the held-out multi-hospital test set.
For further external validation outside of the CRWD, we extracted deidentified data from the OPTUM cohort of 36 140 patients. We used the OPTUM cohort to evaluate the transferability of CovRNN models across different sources of electronic health records. Although the CovRNN models can be directly used and evaluated on the OPTUM cohort, it is recommended to fine-tune the transferred models on sample data from the destination for two reasons: (1) some clinical code distribution might vary or be newly presented at the destination data source, and, thus, during the models’ fine-tuning, these codes would get introduced to the models and become embedded closer to codes of similar meaning; and (2) the definition of the outcome variables can be slightly different, given the limitations of each data source (eg, need for mechanical ventilation was mainly defined by only the procedure codes in the OPTUM cohort but by procedure codes and additional clinical events in the CRWD). Therefore, to evaluate the value added by the models’ fine-tuning, we transferred the best models trained on the CRWD and evaluated them on the OPTUM test set before and after fine-tuning (figure 2). We also compared the performance of the fine-tuned models against new models that were trained only on the same OPTUM data used for fine-tuning (the fine-tuning set).
We evaluated the added value for each clinical data category, the models’ performances with single visit information (using either the information provided on the index visit or the last visit with clinical information) versus full patient history, and the impact of excluding intubated patients within the first 24 h (the restricted test set; n=2999) on the prediction of need for mechanical ventilation. We also did subgroup analyses in the CRWD multi-hospital test set to assess the performance of CovRNN on different populations based on different age groups, races, baseline comorbidities, and geographical US census regions. We constructed calibration plots for the binary classification models in the CRWD validation set, the CRWD multi-hospital test set, and the OPTUM test set.
Models interpretation
For the interpretation of CovRNN predictions, we used the integrated gradient technique24 to expose the factors that contribute to the personalised model predictions. We used the integrated gradient technique due to its good theoretical properties, such as implementation invariance and completeness, and its implementation simplicity; as opposed to methods like layer-wise relevance propagation or DeepLIFT, the integrated gradient technique does not require modification of the gradient backpropagation process and can be viewed as a deterministic and computationally efficient approximation of gradient Shapley additive explanations. For recurrent neural network-based models, we can achieve a more personalised explanation that shows the contribution scores for each code at each patient visit. This is unlike models based on logistic regression or light gradient boost machine algorithms, in which the existing interpretation utilities provide fixed feature-level importance by using either logistic regression coefficients or light gradient boost machine feature importance scores. For the preliminary evaluation, we randomly extracted 20 patients from the CRWD multi-hospital test set and internally shared their predicted risk scores and the contribution score assigned for each medical event and asked an infectious disease specialist (MD level expertise) to evaluate their relevance.
Role of the funding source
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
The prevalence of the outcome variables varied across the different data sources (table 1 ). The prevalences of in-hospital mortality and mechanical ventilation were higher in the OPTUM cohort than in the CRWD cohort, and the median length of hospital stay was longer in the OPTUM cohort than in the CRWD cohort. For further descriptive analyses for each subset, see appendix 1 (pp 6–7).
Table 1.
Descriptive statistics for CRWD and OPTUM extracted cohorts
| CRWD (n=247 960) | OPTUM (n=36 140) | ||
|---|---|---|---|
| Demographics | |||
| Median age at index visit, years | 57 (36–72) | 60 (44–72) | |
| Sex* | |||
| Female | 130 540 (52·6%) | 18 237 (50·5%) | |
| Male | 116 653 (47·0%) | 17 885 (49·5%) | |
| Race and ethnicity* | |||
| White | 168 606 (68·0%) | 19 704 (54·5%) | |
| African American | 36 762 (14·8%) | 7 836 (21·7%) | |
| Asian | 5494 (2·2%) | 930 (2·6%) | |
| American Indian or Alaska Native | 4285 (1·7%) | NA† | |
| Hispanic | 72 068 (29·1%) | 5782 (16·0%) | |
| Comorbidities | |||
| Hypertension | 114 387 (46·1%) | 22 035 (61·0%) | |
| Diabetes | 64 023 (25·8%) | 12 942 (35·8%) | |
| Congestive heart failure | 36 040 (14·5%) | 6568 (18·2%) | |
| Chronic kidney disease | 34 789 (14·0%) | 7517 (20·8%) | |
| Cancer | 19 145 (7·7%) | 5094 (14·1%) | |
| Outcomes | |||
| In-hospital mortality | 13 607 (5·5%) | 4831 (13·4%) | |
| Median time to event, days | 8 (4–16) | 5 (3–10) | |
| Mechanical ventilation | 33 505 (13·5%) | 9582 (26·5%) | |
| Intubated on first day (index date) | 17 811 (7·2%) | 4466 (12·4%) | |
| Median time to event, days | 2 (1–5) | 3 (2–7) | |
| Prolonged hospital stay | 46 421 (18·7%) | 12 457 (34·5%) | |
| Median length of stay, days | 3 (1–6) | 5 (3–10) | |
| Total number of unique features | 123 642 | 67 128 | |
| Number of health-care systems | 87 | 197 | |
Data are median (IQR) or n (%). CRWD=Cerner Real-World COVID-19 Q3 Dataset. NA=not applicable. OPTUM=Optum deidentified COVID-19 electronic health record dataset.
Data do not add up to N totals as some patients fell into the other or unknown category.
This race did not appear in OPTUM.
In the CRWD multi-hospital test set, CovRNN binary classification models achieved AUROCs of 93·0% (95% CI 92·6–93·4) for the prediction of in-hospital mortality, 92·9% (92·6–93·2) for the prediction of the need for mechanical ventilation, and 86·5% (86·2–86·9) for the prediction of a prolonged hospital stay, outperforming light gradient boost machine and logistic regression algorithms (table 2 ). External validation on held-out data from hospitals 1 and 2 showed that AUROCs ranged from 91·5% to 97·0% for CovRNN binary predictions of in-hospital mortality and mechanical ventilation (table 2). CovRNN had an AUROC of 87·2% for hospital 1 and an AUROC of 88·3% for hospital 2 for the prediction of prolonged hospital stay (table 2). External validation on the OPTUM cohort resulted in AUROCs of 91·3%, 91·5%, and 81·0% for in-hospital mortality, mechanical ventilation, and prolonged hospital stay, respectively, after fine-tuning (table 3 ). Additional metrics—specificity at 95% sensitivity, AUPRC, F1-score, and sensitivity and specificity at the optimum threshold—are presented in appendix 1 (p 8).
Table 2.
Model performance on different CRWD test sets
| n |
In-hospital mortality |
Mechanical ventilation |
Prolonged hospital stay (> 7 days) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Logistic regression | Light gradient boost machine | CovRNN binary prediction | CovRNN survival prediction* | Logistic regression | Light gradient boost machine | CovRNN binary prediction | CovRNN survival prediction* | Logistic regression | Light gradient boost machine | CovRNN binary prediction | ||
| Multi-hospital test set | 48 781 | 90·3% (89·8–90·8) | 91·5% (91·1–92·0) | 93·0% (92·6–93·4) | 86·0% (85·1–86·9) | 89·5% (89·1–89·9) | 91·2% (90·8–91·5) | 92·9% (92·6–93·2) | 92·6% (92·2–93·0) | 80·0% (79·5–80·4) | 81·7% (81·3–82·2) | 86·5% (86·2–86·9) |
| Hospital 1 | 3469 | 88·8% (86·9–90·5) | 91·0% (89·5–92·4) | 91·8% (90·3–93·2) | 86·0% (83·2–88·5) | 86·7% (85·1–88·4) | 88·4% (87·0–89·9) | 91·5% (90·2–92·8) | 90·8% (89·4–92·2) | 77·3% (75·5– 79·1) | 78·5% (76·7–80·2) | 87·2% (85·8–88·4) |
| Hospital 2 | 706 | 94·6% (91·9–96·9) | 95·1% (92·7–97·2) | 97·0% (95·2–98·6) | 91·6% (87·5–94·8) | 93·5% (90·7–95·8) | 95·6% (93·8–97·1) | 96·0% (94·2–97·7) | 93·8% (91·4–96·0) | 80·9% (76·9– 84·7) | 84·3% (80·5–87·7) | 88·3% (85·6–90·9) |
Data are area under the receiver operating characteristic curve (95% CI), unless otherwise indicated.
Unlike the binary outcomes used in the other models, CovRNN survival prediction uses time-to-event outcomes and the concordance index (95% CI) is shown. CRWD=Cerner Real-World COVID-19 Q3 Dataset.
Table 3.
Performance of CovRNN models on the OPTUM test set before and after fine-tuning
| CovRNN trained only on OPTUM fine-tuning set | CRWD-trained CovRNN before fine-tuning | CRWD-trained CovRNN after fine-tuning using OPTUM fine-tuning set | |
|---|---|---|---|
| In-hospital mortality binary prediction | 88·6% | 87·0% | 91·3% |
| Mechanical ventilation binary prediction | 90·4% | 72·5% | 91·5% |
| Prolonged hospital stay (>7 days) binary prediction | 78·1% | 68·0% | 81·0% |
| In-hospital mortality survival prediction* | 86·1% | 77·1% | 88·9% |
| Mechanical ventilation survival prediction* | 90·2% | 69·2% | 93·7% |
Data are area under the receiver operating characteristic curve, unless otherwise indicated. All data are based on evaluation in the OPTUM test set. CRWD=Cerner Real-World COVID-19 Q3 Dataset. OPTUM=Optum deidentified COVID-19 electronic health record dataset.
Unlike the binary classifications used in other models, values for the survival models represent the concordance index.
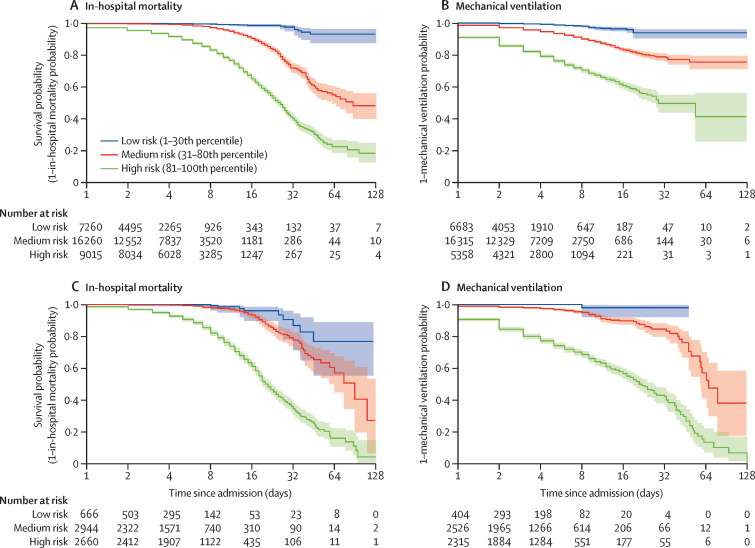
Evaluation of CovRNN survival models on the CRWD test set found a concordance index of 86·0% for the prediction of in-hospital mortality and 92·6% for the prediction of mechanical ventilation (table 2). Using the survival models to predict patient risk of developing the event at a certain timepoint between day 1 and day 60 since admission, AUROCs ranged from 88·8% to 93·6% for in-hospital mortality and from 91·4% to 95·5% for mechanical ventilation (appendix p 11). Similarly, the concordance index ranged from 86·0% to 93·8% for the prediction of in-hospital mortality or mechanical ventilation on the held-out hospital sets (table 2; figure 3A ) and the OPTUM test set after fine-tuning (table 3; figure 3B). For CovRNN binary classification and survival models, transferred models that were fine-tuned consistently achieved better performances than when new models were trained only on the OPTUM fine-tuning set (table 3). Similarly, transferred, fine-tuned survival models had a concordance index of 88·9% for the prediction of in-hospital mortality and 93·7% for the mechanical ventilation task, outperforming newly trained models (table 3).
Figure 3.
Kaplan-Meier curves in the stratified survival analysis
In-hospital mortality (A) and mechanical ventilation (B) in the multi-hospital test set of the Cerner Real-World COVID-19 Q3 Dataset. In-hospital mortality (C) and mechanical ventilation (D) in the test set of the Optum deidentified COVID-19 electronic health record dataset. Stratification of patients is according to their predicted survival score over time in days since admission. Shaded areas indicate 95% CIs calculated on the logarithmic scale from the SEs of the Kaplan–Meier estimator with the centre values corresponding to the Kaplan–Meier estimate.
We conducted three experiments. The first experiment was an ablation study that showed that each clinical data category contributed to an increase in the model prediction accuracy (appendix 1 p 9). The use of the patient's known comorbidities alone achieved an AUROC of 85·9% for in-hospital mortality and 83·7% for mechanical ventilation (appendix 1 p 9), which was expected as this category summarises the patient's health condition. The addition of medication history and then laboratory results increased model performance by around 3–4 percentage points each time, as these categories introduced useful information to the models that was not captured by the recorded diagnoses. A relatively small degree of optimisation (<1%) was obtained from adding procedures and other assessments, as this category mostly introduced redundant information already captured by the previously fed data. The second experiment showed that using full patient history continuously resulted in better model performance than did using information from the last (or index) visit only, especially for the prediction of prolonged hosptial stay (appendix 1 pp 9–10). In the final experiment, we found that CovRNN's performance in predicting mechanical ventilation remained unchanged on the restricted test set (ie, regardless of whether we kept or excluded patients who were intubated within the first 24 h from the training set; appendix 1 p 10).
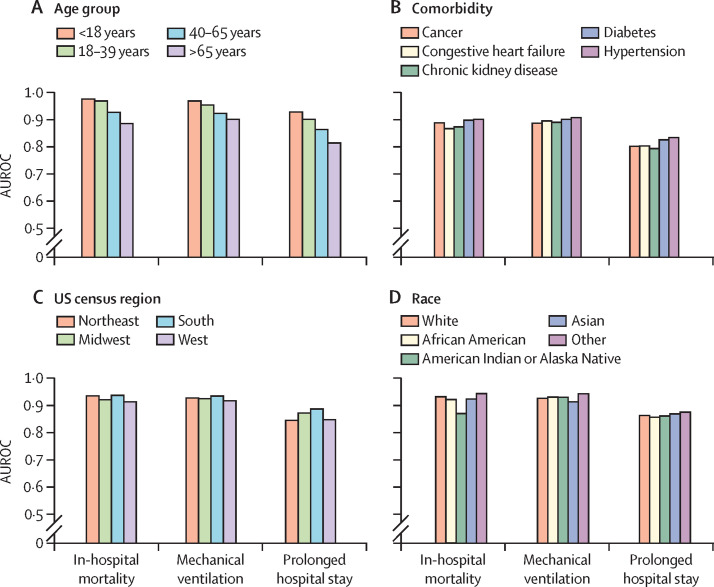
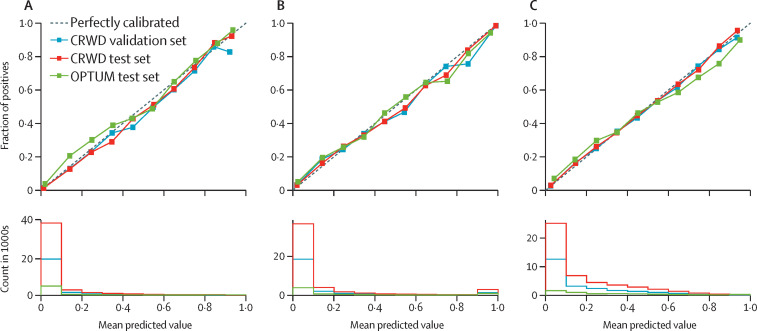
The subgroup analysis showed that CovRNN's prediction accuracy remained mainly consistent among people with different comorbidities, of different ages and races, and from different US census regions (figure 4 ). The most notable trend was that prediction accuracy was numerically better among the younger population than among the older population (figure 4). In addition, CovRNN binary classification models showed good calibration without sacrificing high prediction accuracy, as shown in the calibration plots (figure 5 ). In appendix 1 (p 12), we present a sample visualisation that shows the integrated gradient-based explanation of CovRNN models’ true positive prediction for a patient who had a prolonged hospital stay. The information in this visualisation, however, is based on a sample of the patient's data and not the full patient data; the data for 20 randomly selected patients for each prediction task was presented to one infectious disease specialist, who found it informative and relevant when he assessed the displayed contribution scores against his clinical knowledge. To show our efforts to abide by transparent reporting standards, we provide the PROBAST assessment in appendix 2 and the TRIPOD assessment in appendix 3.
Figure 4.
Subgroup analysis using the CRWD multi-hospital test set
(A) Age group. (B) Comorbidity. (C) US census region. (D) Race. AUROC=area under the receiver operating characteristic curve. CRWD=Cerner Real-World COVID-19 Q3 Dataset.
Figure 5.
Calibration plots for the CRWD validation set, CRWD multi-hospital test set, and OPTUM test set
(A) In-hospital mortality. (B) Mechanical ventilation. (C) Prolonged hospital stay. CRWD=Cerner Real-World COVID-19 Q3 Dataset. OPTUM=Optum deidentified COVID-19 electronic health record dataset.
Discussion
Our experiments showed that CovRNN models trained on a large heterogeneous dataset of approximately 200 000 patients with COVID-19 required minimal data curation to achieve high prediction accuracy (AUROC 86·0–97·0%) for different patient clinical outcomes, namely in-hospital mortality, mechanical ventilation, and prolonged hospital stay. CovRNN not only showed high prediction accuracy but also good transferability between two large deidentified electronic health record databases with different structures, good external validity, proper model calibration, and the utility of fine-tuning for continuous improvement. In addition, we used integrated gradients to expose the factors that contribute to the model-predicted scores.
CovRNN models consistently outperformed other methods (logistic regression and light gradient boost machine). Interestingly, we found that the maximum difference between the AUROC estimates made by logistic regression, light gradient boost machine, and CovRNN models was around 3% for in-hospital mortality and mechanical ventilation, whereas the difference exceeded 6% for the prediction of prolonged hospital stay. Similarly, we observed that the accuracy of predicting a prolonged hospital stay was highly affected by the inclusion of full patient history versus information from the last (index) visit only. Therefore, we believe that considering the sequence of events that occurred in the past is of higher importance for the prolonged hospital stay prediction task than for the in-hospital mortality and mechanical ventilation prediction tasks, for which we infer that the most recent events are of higher importance.
Although several studies have reported on predictive machine learning models for COVID-19 outcomes with prediction accuracies similar to those offered by our models,6, 9, 25, 26, 27 our models were trained and evaluated on larger, multicentre cohorts from two large, well known, deidentified electronic health record databases from the USA (a total of 284 100 patients). CovRNN outperforms other prediction models for COVID-19 outcomes that have been trained and evaluated on more than 50 000 patients with COVID-19 and commonly rely on boosting-based algorithms.7, 8 The N3C study9 included a similar number of patients with COVID-19 (160 000) in their training set; however, their reported prediction accuracy (AUROC) for in-hospital mortality and mechanical ventilation (combined as a severity indicator) was 87% (95% CI 86–88). In addition, the majority of published studies with machine learning models predict outcomes in a very short follow-up window, such as 1 h or 1 day from the index timepoint.6, 27 Furthermore, some studies did not specify the time window of prediction or used limited historical data.28 As window periods become shorter, the prediction task becomes easier, and, thus, accuracy increases; nevertheless, the results are less valuable as physicians can predict short-term clinical outcomes better without using models. We reported the results of our CovRNN survival models to show the flexibility of our approach. We believe, however, that predicting the probability of adverse events occurring within the hospital stay should be informative enough for clinicians to make appropriate decisions on admission and might not be limited to a specific time range. Therefore, we also focused on the evaluation and calibration of the binary classification models.
In our study, we included data available on or before the index admission to predict clinical outcomes throughout the hospital stay. Of note, our cohorts also included patients who stayed in observation units without hospitalisation. Many physicians often encounter considerable dilemmas when deciding on the patient's disposition, such as discharge or transfer to a higher level of care, in the observation unit or at the time of hospitalisation. Furthermore, patients with COVID-19 often progress rapidly, especially after about 7 days from the onset of symptoms, even when they initially present with mild symptoms.29 This characteristic clinical course in patients with COVID-19 makes it substantially difficult for clinicians to predict future outcomes on the first day of hospital encounters. Our models are particularly helpful in those clinical scenarios because they predicted the occurrence of in-hospital mortality with a specificity of 70·93% at 95% sensitivity. The threshold can be easily adjusted to prioritise sensitivity or specificity to meet clinicians’ needs. For example, in a situation in which our models predict the patient's death with high specificity, physicians could initiate an early discussion of poor outcomes with the patient and goals of care in appropriate cases. As the risk of further COVID-19 surges still cannot be ruled out and scenarios of health-care systems being overwhelmed with patients are still a distinct possibility, CovRNN can be a useful tool while triaging patients. The score provided by CovRNN can be used to risk-stratify large numbers of patients on the basis of their readily available data in a few seconds. The minimal need for data curation and reliance on the power of the deep learning model architecture for learning proper feature representations from large data are key advantages of our CovRNN models. We were able to transfer the models between two datasets that differ in several ways, such as in the distribution of clinical codes. With a simple model fine-tuning step on sample data from the destination dataset, the models consistently achieved high prediction accuracy. Although we focused on the outcomes of patients with COVID-19, this study is proof of concept that we could apply the same methodology to predict different clinical circumstances.
Our study has several limitations. First, our data analysis included only retrospective data. Despite our efforts to avoid potential bias by separating training, validation, and test datasets and conducting external validation on a different data source, potential biases are inevitable. A prospective validation study is warranted, ideally, in hospitals that did not participate in data sharing with the database that we used to secure the validation of transferability. Second, our models focused only on predicting clinical outcomes at the time of hospital admission. It is possible to use multiple timepoints during the hospital stay to update models to achieve real-time predictions. Because minimal data preprocessing is required, our models can be easily modified to use different datapoints to predict future clinical outcomes. Third, real-world structured data from electronic health records are not always associated with standard codes. For example, data from Cerner Millennium might not be codified at all in the source system or can only be associated with clients’ proprietary event codes. Hospitals commonly have access to utilities to map their structured electronic health record data into industry-standard codes, which we used in our models, to facilitate interoperability, Fast Healthcare Interoperability Resources (known as FHIR) queries, data sharing, billing, and public health reporting tasks. Such utilities are sometimes provided natively by their electronic health record vendors. Such mappings are required to get benefit from our pretrained CovRNN models; otherwise, we recommend using our CovRNN training framework to train compatible models utilising the system's proprietary event codes. However, these codes or representations are only meaningful in the context of the originating system, and they are not helpful to train transferable models.
Fourth, we only conducted a preliminary evaluation for the model predictions explanations, whereby we extracted data from 20 randomly selected patients and presented their predicted risk scores and the contribution scores assigned for each medical event and asked an infectious disease specialist to evaluate their relevance. Although we acknowledge that this evaluation method is not rigorous, it showed that our proposed models allow for model transparency and help to engage clinicians and facilitate their judgment on model predictions. Future work is warranted to systematically evaluate the models’ transparency. Further evaluation of the model explanation should take into consideration that the evaluation of such personalised explanations, whereby the same clinical code can have different contribution scores at each patient and visit level, given the different contexts, is laborious. Finally, the dynamics of COVID-19 management in hospitals and patient surges from pandemic waves have changed with time, which modifies patient outcomes over time. Thus, the accuracy of our models, for which we mostly used data from the first pandemic wave (up to September, 2020), might differ for future datasets. For example, patients who receive COVID-19 vaccines probably have different clinical outcomes than those who do not.30 Because our models are trained on historical data, they can be easily fine-tuned on more current data to improve prediction accuracy, which is one of the major advantages of deep learning models. Future work is warranted to fine-tune and evaluate our models on data from later pandemic waves.
Through benchmarking, we found that CovRNN can provide accurate and transferable predictive models for a wide range of outcomes and that we can continuously improve upon the models through periodic fine-tuning. Furthermore, our data preparation pipeline was kept to a minimum to facilitate the transferability of the models and facilitate further validation on new data sources. Our model development framework can be further applied to train and evaluate predictive models for different types of clinical events. For clinicians who are fighting COVID-19 on the frontlines, there are two potentially actionable contributions of our work. Clinicians can (1) fine-tune our pretrained models on their local data, regardless of cohort size, establish utility, and then deploy the models and (2) use our comprehensive model development framework to train a predictive model using their own data.
In conclusion, to the best of our knowledge, CovRNN models are the first COVID-19 outcome prediction models that can simultaneously accurately predict different outcomes on admission for patients with COVID-19 and use readily available structured data from electronic health records in their categorical format without the need for specific feature selection or missing value imputation. We also showed the value added by the fine-tuning utility of CovRNN and how it can be used to improve models’ prediction accuracy. Such utility can be further used to continually improve the models, as per Good Machine Learning Practice recommendations, to secure the models’ reliability and sustainability.
Data sharing
The data that support the findings of this study, the CRWD and the Optum dataset, are available for licensing at Cerner and Optum, respectively. Data access might require a data sharing agreement and might incur data access fees.
Declaration of interests
We declare no competing interests.
Acknowledgments
Acknowledgments
This work was supported by the Cancer Prevention and Research Institute of Texas (CPRIT; CPRIT grant number RP170668) and the University of Texas Health Science Center in Houston (UTHealth) Innovation for Cancer Prevention Research Training Program Pre-Doctoral Fellowship (CPRIT grant number RP160015 and CPRIT grant number RP210042). We would like to acknowledge the use of the CRWD and the support from the Cerner research team, especially Cheryl Akridge. We also acknowledge the assistance provided by the UTHealth School of Biomedical Informatics Data Service team, the UTHealth Center of Healthcare Data, and the Glassell Innovation Fund.
Contributors
LR and DZ conceived the idea for this study. LR led the design and implementation of experiments. LR, KP, MN, and BSK reviewed the evidence before the study. MN contributed to the discussion and the model explanation evaluation. ZX contributed to the model explanation. LR, BM, and KP ran the experiments on the OPTUM data. LR and YZ extracted the electronic health records data. WZ added the visualisations. LR led the manuscript writing. BSK, MN, HX, and DZ contributed to the writing. AR assessed the study against TRIPOD and PROBAST standards. HX and DZ supervised the project. LR and DZ finalised the manuscript. LR, BSK, and MN accessed and verified the CRWD data. LR, YZ, BM, and KP accessed and verified the OPTUM data. All coauthors reviewed and approved the manuscript. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Supplementary Materials
References
- 1.WHO Coronavirus disease (COVID-19) pandemic. https://www.who.int/emergencies/diseases/novel-coronavirus-2019
- 2.Centers for Disease Control and Prevention COVID data tracker. March 28, 2020. https://covid.cdc.gov/covid-data-tracker
- 3.Wynants L, Van Calster B, Collins GS, et al. Prediction models for diagnosis and prognosis of covid-19: systematic review and critical appraisal. BMJ. 2020;369 doi: 10.1136/bmj.m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sperrin M, Grant SW, Peek N. Prediction models for diagnosis and prognosis in Covid-19. BMJ. 2020;369 doi: 10.1136/bmj.m1464. [DOI] [PubMed] [Google Scholar]
- 5.Leeuwenberg AM, Schuit E. Prediction models for COVID-19 clinical decision making. Lancet Digit Health. 2020;2:e496–e497. doi: 10.1016/S2589-7500(20)30226-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwab P, Mehrjou A, Parbhoo S, et al. Real-time prediction of COVID-19 related mortality using electronic health records. Nat Commun. 2021;12 doi: 10.1038/s41467-020-20816-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He F, Page JH, Weinberg KR, Mishra A. The development and validation of simplified machine learning algorithms to predict prognosis of hospitalized patients with COVID-19: multicenter, retrospective study. J Med Internet Res. 2022;24 doi: 10.2196/31549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng A. Using electronic health records to accurately predict COVID-19 health outcomes through a novel machine learning pipeline. Proceedings of the 12th ACM Conference on Bioinformatics, Computational Biology, and Health Informatics; Aug 1, 2021 (abstr 61).
- 9.Bennett TD, Moffitt RA, Hajagos JG, et al. Clinical characterization and prediction of clinical severity of SARS-CoV-2 infection among US adults using data from the US National COVID Cohort Collaborative. JAMA Netw Open. 2021;4 doi: 10.1001/jamanetworkopen.2021.16901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rasmy L, Wu Y, Wang N, et al. A study of generalizability of recurrent neural network-based predictive models for heart failure onset risk using a large and heterogeneous EHR data set. J Biomed Inform. 2018;84:11–16. doi: 10.1016/j.jbi.2018.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiang Y, Ji H, Zhou Y, et al. Asthma exacerbation prediction and risk factor analysis based on a time-sensitive, attentive neural network: retrospective cohort study. J Med Internet Res. 2020;22 doi: 10.2196/16981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rasmy L, Zhu J, Li Z, et al. Simple recurrent neural networks is all we need for clinical events predictions using EHR data. arXiv. 2021 https://arxiv.org/abs/2110.00998 published online Oct 3. (preprint). [Google Scholar]
- 13.Wanyan T, Honarvar H, Jaladanki SK, et al. Contrastive learning improves critical event prediction in COVID-19 patients. arXiv. 2021 doi: 10.1016/j.patter.2021.100389. https://arxiv.org/abs/2101.04013 published online Jan 11. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajkomar A, Oren E, Chen K, et al. Scalable and accurate deep learning with electronic health records. NPJ Digit Med. 2018;1:1–10. doi: 10.1038/s41746-018-0029-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.National Institutes of Health. National Library of Medicine. Unified Medical Language System (UMLS) MMSL (multum)—synopsis. https://www.nlm.nih.gov/research/umls/sourcereleasedocs/current/MMSL/index.html
- 16.Rasmy L, Tiryaki F, Zhou Y, et al. Representation of EHR data for predictive modeling: a comparison between UMLS and other terminologies. J Am Med Inform Assoc. 2020;27:1593–1599. doi: 10.1093/jamia/ocaa180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi E, Schuetz A, Stewart WF, Sun J. Using recurrent neural network models for early detection of heart failure onset. J Am Med Inform Assoc. 2017;24:361–370. doi: 10.1093/jamia/ocw112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi E, Bahadori MT, Sun J, Kulas J, Schuetz A, Stewart W. RETAIN: an interpretable predictive model for healthcare using reverse time attention mechanism. 2016. https://proceedings.neurips.cc/paper/2016/file/231141b34c82aa95e48810a9d1b33a79-Paper.pdf
- 19.Wu S, Liu S, Sohn S, et al. Modeling asynchronous event sequences with RNNs. J Biomed Inform. 2018;83:167–177. doi: 10.1016/j.jbi.2018.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.scikit learn sklearn.linear_model.LogisticRegression—scikit-learn 0.24.2 documentation. https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.LogisticRegression.html
- 21.LightGBM Welcome to LightGBM's documentation! LightGBM 3.2.1.99 documentation. https://lightgbm.readthedocs.io/
- 22.Katzman JL, Shaham U, Cloninger A, Bates J, Jiang T, Kluger Y. DeepSurv: personalized treatment recommender system using a Cox proportional hazards deep neural network. BMC Med Res Methodol. 2018;18:24. doi: 10.1186/s12874-018-0482-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrell FE, Jr, Califf RM, Pryor DB, Lee KL, Rosati RA. Evaluating the yield of medical tests. JAMA. 1982;247:2543–2546. [PubMed] [Google Scholar]
- 24.Sundararajan M, Taly A, Yan Q. Axiomatic attribution for deep networks. 34th International Conference on Machine Learning; Aug 6–11, 2017; 70: 3319–28.
- 25.Villegas M, Gonzalez-Agirre A, Gutiérrez-Fandiño A, et al. Predicting the evolution of COVID-19 mortality risk: a recurrent neural network approach. medRxiv. 2021 doi: 10.1101/2020.12.22.20244061. published online Jan 11. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Razavian N, Major VJ, Sudarshan M, et al. A validated, real-time prediction model for favorable outcomes in hospitalized COVID-19 patients. NPJ Digit Med. 2020;3:130. doi: 10.1038/s41746-020-00343-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yadaw AS, Li Y-c, Bose S, Iyengar R, Bunyavanich S, Pandey G. Clinical features of COVID-19 mortality: development and validation of a clinical prediction model. Lancet Digit Health. 2020;2:e516–e525. doi: 10.1016/S2589-7500(20)30217-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Estiri H, Strasser ZH, Murphy SN. Individualized prediction of COVID-19 adverse outcomes with MLHO. Sci Rep. 2021;11 doi: 10.1038/s41598-021-84781-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Centers for Disease Control and Prevention Healthcare workers. Interim clinical guidance for management of patients with confirmed coronavirus disease (COVID-19) May 27, 2021. https://www.cdc.gov/coronavirus/2019-ncov/hcp/clinical-guidance-management-patients.html
- 30.CDC COVID-19 Vaccine Breakthrough Case Investigations Team COVID-19 vaccine breakthrough infections reported to CDC—United States, January 1–April 30, 2021. MMWR Morb Mortal Wkly Rep. 2021;70:792–793. doi: 10.15585/mmwr.mm7021e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study, the CRWD and the Optum dataset, are available for licensing at Cerner and Optum, respectively. Data access might require a data sharing agreement and might incur data access fees.