Abstract
Determining the transmissibility, prevalence and patterns of movement of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections is central to our understanding of the impact of the pandemic and to the design of effective control strategies. Phylogenies (evolutionary trees) have provided key insights into the international spread of SARS-CoV-2 and enabled investigation of individual outbreaks and transmission chains in specific settings. Phylodynamic approaches combine evolutionary, demographic and epidemiological concepts and have helped track virus genetic changes, identify emerging variants and inform public health strategy. Here, we review and synthesize studies that illustrate how phylogenetic and phylodynamic techniques were applied during the first year of the pandemic, and summarize their contributions to our understanding of SARS-CoV-2 transmission and control.
Subject terms: Evolutionary genetics, Phylogenetics, Viral genetics, SARS-CoV-2
In this Review, the authors describe how phylogenetic and phylodynamic methods provide insight into viral evolution, focusing on the SARS-CoV-2 pandemic. The approaches reveal routes and timings of transmission events, and they can assess the effectiveness of various intervention measures aimed at controlling the virus.
Introduction
The coronavirus disease 2019 (COVID-19) pandemic has triggered an unprecedented global response in pathogen genome sequencing, and nearly 400,000 full or partial severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genomes were generated and shared publicly within its first year. Although phylogenetic tools have become increasingly relevant to the public health management of a range of viral epidemics1–4, the COVID-19 crisis is the first global health emergency during which large-scale, real-time genomic sequencing and analysis have underpinned public health decisions. The first 12 months of the pandemic were characterized by continual change in the global epidemiological and virological situation, and the analysis of genome sequences was essential in tracking the changing situation. Phylogenetic and phylodynamic approaches (Box 1) can unlock information contained in sampled genomes and are often analysed in conjunction with other data sources5. Such analyses have been used to quantify international virus spread, identify outbreaks and transmission chains in specific settings, estimate growth rates and reproduction numbers, account for surveillance gaps and lags, identify and track mutations of interest, discover and analyse variants of concern, and investigate intra-host virus evolution.
This Review focuses on how SARS-CoV-2 transmission, epidemiology and spatial dispersal have been measured and investigated through phylogenetic and phylodynamic analyses of SARS-CoV-2 genomes (Fig. 1). It is intended to be a retrospective overview that uses examples from the first year of the pandemic to demonstrate the contributions of phylogenetics in the context of different phases of pandemic response. We examine how such analyses have informed global efforts to understand, control and predict the pandemic, and outline arising new challenges and how they are being addressed. We do not review events that precede the widespread emergence of SARS-CoV-2 (such as the evolutionary origins of the pandemic in non-human host species) or its functional genomics (that is, how virus mutations contribute to phenotypes such as transmissibility). Given the scale of the field and the size of the literature on SARS-CoV-2 genomic epidemiology, we do not attempt to provide a systematic review. Instead we focus on studies that represent the first year of the pandemic, which saw evolutionary approaches applied to a wide variety of public health interventions worldwide, often in an ad hoc or pragmatic manner. We further highlight research that was influential in contributing to epidemiological understanding and public health decision making. The pandemic’s first year also best illustrates the potential of these methods for urgent risk assessment, prediction and control of future emerging viruses. We mostly refer to the genetic diversity of SARS-CoV-2 using the Pango dynamic nomenclature6 (Box 1), but also sometimes use the WHO ‘Greek letter’ nomenclature scheme for particular variants of concern (VOCs) and variants of interest (VOIs).
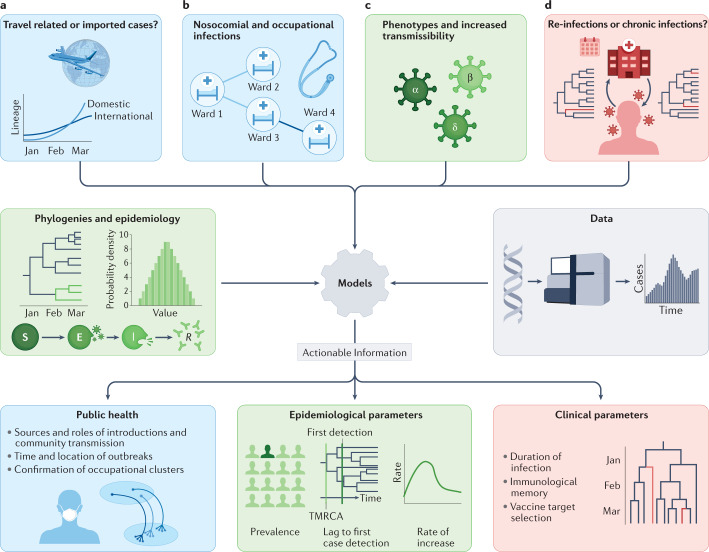
Fig. 1. Phylodynamic approaches to the investigation of SARS-CoV-2 transmission.
Relevant clinical and public health questions are defined (top row), phylodynamic and epidemiological data and models are then combined (middle row), and used in combined or joint analyses to provide actionable insight into virus transmission (bottom row). a | Phylogenetic approaches estimate the rate of international lineage introductions and distinguish introductions from community transmission. b | Genome sequences and phylogenetics support outbreak analyses by identifying or refuting links between local cases; this can lead to identification of outbreak sources and drivers or assessment of nosocomial transmission. c | Phylodynamic techniques using epidemiological demographic models, such as the susceptible–exposed–infected–recovered (SEIR) model, allow us to compare transmission rates between lineages bearing different key genotypes (for example, variants of concern (VOCs) and pre-existing lineages). d | Relative timing of variant and lineage emergence from the global (or regional) phylogeny, and scattering of case genomes across clades can distinguish persistent from repeat infections in some scenarios. Phylogenetics is also useful in studies of lineage turnover and interactions within the host. Panel colours indicate related themes: blue, public health; green, epidemiological parameters; red, clinical parameters. TMRCA, time to the most recent common ancestor.
Box 1 Phylogenetic terminology and concepts.
Phylogenetics
Phylogenetics provides a method for the generation of hypotheses about ancestor–descendant relationships using character-state data. The resulting phylogeny attempts to explain the observed character states in the sequences that we have sampled as having evolved from a single common ancestor in the past, via a sequence of usually unobserved (unsampled or extinct) hypothesized intermediate ancestors represented by internal nodes or branch points on a bifurcating tree (see the figure). Phylogenetic methods typically search for the solution with the minimum number of evolutionary steps (parsimony) or the one that maximizes the likelihood of the data given the tree. A third alternative is a Bayesian approach, which applies Bayes theorem to estimate a probability distribution for population parameters of interest. The ability to incorporate prior information (priors) for the events (for example, a prior distribution for outbreak onset time) gives the approach an advantage over maximum likelihood estimation.
Phylogenies have also formed the basis of a system for the identification, definition and monitoring of outbreak clusters and variants of concern (VOCs). Although nomenclatures such as that currently adopted by the WHO assign names to definitive constellations of substitutions that commonly occur together (for example, VOC delta), most other current nomenclatures are lineage based (for example, Pango and Nextstrain). In the case of the Pango nomenclature, lineages correspond either loosely or exactly to clades estimated on a reference phylogeny. A clade is a monophyletic subtree on a phylogeny; such subtrees include all descendants of their most recent common ancestor represented by the node joining them to the global phylogeny and no others (see lineage A in the figure, which forms a clade). Nevertheless, Pango lineages can include any fairly cohesive and exclusive (or nearly so) clustering of sequences on the global severe acute respiratory coronavirus 2 (SARS-CoV-2) phylogeny, particularly where that cluster associates with an outbreak, epidemiologically significant phenotype (for example, greater transmissibility) or any noteworthy characteristic, whether proven or awaiting investigation.
Phylodynamics
Phylodynamics focuses on the estimation of population dynamic parameters from genetic sequences and molecular phylogenies, such as epidemic growth rates, generation times and reproductive numbers. Phylodynamic analysis relies on other statistical models such as nucleotide substitution models and molecular clock models. Phylodynamics has been used to incorporate epidemiological data in phylogenetic studies of the pandemic. Such models allow estimation of demographic or epidemic parameters over time; these often include changes in relative population size (including reproductive number and growth rate) and selection coefficients. Phylodynamics can help date the first cases in a region and can provide public health officials with an estimate of the lag between importation and first-case detection by estimating the time to the most recent common ancestor (TMRCA) of a clade. Phylogeography has been used during the pandemic to estimate rates of virus (lineage) movement between regions and may be considered as a form of phylodynamics that uses phylogenetic methods to understand the spatial dissemination of lineages.
The coalescent model is central to a large class of phylodynamic methods. The coalescent model considers mutation drift (that is, evolution without selection) backwards in time, with pairs of lineages coalescing rather than diverging. The model can be visualized as a genealogy, is computationally efficient, and deviations from the expected distribution of coalescence intervals (in time) can be used to infer processes such as selection and migration. Phylodynamic applications of the coalescent model often involve the use of ‘skyline plots’ or related methods to estimate historical changes in population size (for example, virus demographics); these are estimates of effective population size (Ne) change through time. Various skyline methods exist, and these generally differ in the parameterization and smoothing of the population size changes. The coalescent model can also be modified to allow for expected population structure (structured coalescent). Similarly, epidemiological models, such as the susceptible–exposed–infected–recovered (SEIR) model, are incorporated into the phylodynamic framework as compartmental models to model disease transmission and prevalence. Compartmental models involve the partitioning of the individuals (for example, hosts) within a population into mutually exclusive groups according to their properties, with their progression between the groups permitted according to the rules that underlie the model. Owing to computational advances, more mathematically complex birth–death (BD) models, including multitype BD models (Table 1), are also being used in phylodynamic analysis5 in addition to coalescent approaches. These BD models explicitly represent lineage diversification events and can estimate growth rates for distinct ‘types’ or sub-populations.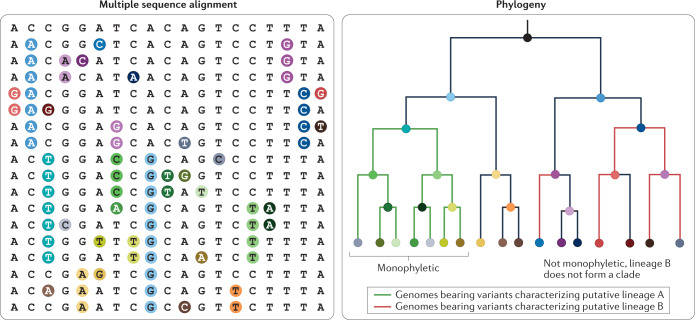
Tracking the global pandemic
Revealing how SARS-CoV-2 spread globally in early 2020 was important in informing public health strategies. Phylodynamic, particularly phylogeographic, methods can be used to estimate the timing and location of ancestral nodes within a molecular phylogeny7–9, allowing inference of the route and rate of spread of pandemic lineages, from the site of its initial detection in Wuhan, China, to the location of each sampled patient from which a virus genome was obtained.
International travel restrictions
Phylogeographic studies have investigated the impact of international travel restrictions, quantifying the number of lineage introductions from abroad and the relative contribution of local transmission. For example, a global phylogeny of the pandemic showed that earlier lineages were highly cosmopolitan, whereas later lineages tended to be continent-specific, which may reflect the rapid declines in mobility as many countries concurrently imposed restrictions on international travel10, although early sampling in some countries may have been biased towards cases in international travellers.
At the national scale, studies have typically observed reduced numbers of introductions along international routes covered by travel restrictions; however, the overall effects of this on controlling national transmission depended on the extent to which lineages were already locally well established. During the global expansion of SARS-CoV-2, international exportations were driven initially by dispersal from China; however, the number of exports declined rapidly following the cessation of China’s international flights in January 2020 (ref.11). Endemic transmission began in Italy during mid-February 2020, with establishment in other European countries soon thereafter12. The shift in global dissemination towards greater intercontinental exportation from Europe was associated with the expansion of a lineage bearing the D614G spike mutation13. Virus lineage migrations from Europe to North America increased until the declaration by WHO of a pandemic on 11 March 2020, suggesting that air travel restrictions subsequently slowed international spread14. In South Africa, international introductions plummeted after travel restrictions began on 26 March 2020 (ref.15). Similar observations were made in other nationally focused studies, including those from Italy16, New Zealand, Australia, Iceland, Taiwan17 and the UK18.
The impact of international travel restrictions depended on the level of domestic transmission control and whether restrictions were implemented before full establishment of local transmission. A study of 427 genomes from Brazil applied a discrete asymmetric phylogeographic model and estimated at least 104 international introductions during March and April 2020; these fell into three monophyletic clusters (Box 1) of apparently European origin, and a molecular clock approach indicated that they arrived in late February 2020. Domestic transmission in Brazil was already well established by early March, suggesting that international restrictions implemented thereafter may have had little impact19. In the USA, an early study investigated the efficacy of international travel restrictions in Connecticut20. Seven of nine Connecticut genomes fell into a clade of mostly Washington State genomes, whereas two clustered with genomes from China and Europe. As the Connecticut genomes were derived from people with no history of recent travel, their phylogenetic placement in a cluster of genetically similar genomes indicated community transmission of recently imported lineages; again, flight restrictions may have been more effective in reducing cases if they had been implemented earlier20. Similar patterns were observed in other countries, including Italy12 and the UK21.
Many countries strengthened travel restrictions later in 2020, aiming to slow the spread of variants associated with changes in transmissibility (see the section Tracking lineages of interest). In Brazil, a phylogeny of SARS-CoV-2 from cases detected in São Paulo in late December 2020 indicated two independent international introductions of lineage B.1.1.7 (the alpha VOC) from London, UK22. These introductions occurred despite the suspension of flights to and from the UK. Similarly, phylodynamics suggested multiple international introductions to the USA and hidden transmission of B.1.1.7 since November 2020, and that lineage B.1.1.7 expanded to 33 US states by January 2021 with a doubling time of 9.8 days23. Investigations have also considered the factors that drove the resurgence of transmission in Europe in late summer 2020. A recent study using a Bayesian time-scaled phylogeographic model (Box 1) found that by mid-August a large fraction of the lineages then circulating in European countries had been introduced after 15 June, the date when many countries in the Schengen area opened their borders24. The study also found that newly introduced lineages tended to expand more quickly when entering a region of low incidence, and that for most countries resurgence was driven by new introductions rather than persistence of lineages from the spring24.
Phylogeographic inference of SARS-CoV-2 migration patterns has typically used either a discrete trait analysis (DTA; for example, analyses with travel and/or mobility data)18,19,25 or structured event-based birth–death (BD) models12,17,26. The approaches differ in that DTA assigns discrete states (locations) to nodes on a phylogeny, whereas structured models explicitly model migration events and rates at a population level. The advantage of DTA is its relatively low computational demand26 and its ability to incorporate discrete metadata, such as travel histories (for example, ref.25), in a straightforward manner. However, DTA does not accommodate the interdependency of tree shape and migration rate or population size, and it is more difficult to interpret DTA model parameters26. Structured BD approaches are more computationally costly, but can model variable sampling between regions (DTA is less robust to sampling patterns27), and they infer parameters that can be more readily compared with those obtained from epidemiological or mobility data sets17.
Local transmission and interventions
Non-pharmaceutical interventions (NPIs) include travel restrictions, person-to-person distancing and mandatory mask wearing. Two phylogenetic approaches were typically adopted to investigate the impact of NPIs. First, the frequencies of lineage movement between regions within a country were assessed using phylogeographic analyses (as discussed above for international dissemination). Second, estimates of virus population size, epidemic doubling time and general reproduction number (Rt) were calculated from virus genome sequences using phylodynamic approaches.
Molecular clock dating of SARS-CoV-2 lineages indicated multiple introductions from Wuhan to Guangdong in early January 2020, with a fall in lineage diversity thereafter, suggesting that within-country travel restrictions combined with comprehensive tracing and isolation in Guangdong were effective in controlling transmission28. A phylogenetic study of transmission in Boston, USA, also reported a drop in importations to Boston from other domestic locations after national restrictions began14, and phylodynamic methods estimated a reduction in Rt in Israel of at least two-thirds, coincident with the imposition of quarantine measures29. By contrast, a study of NPIs in Italy16 suggested that domestic travel restrictions failed to prevent community transmission, although there was evidence that transmission was inhibited, and the relatively low genome sampling density means that NPIs could have greatly restricted the transmission of lower-frequency lineages. One global study of 29,000 SARS-CoV-2 genome sequences used a compartmental structured coalescent model to estimate the time of epidemic seeding in 57 different locations30. The authors found that locations with early implementation of strong NPIs experienced less severe morbidity and mortality during the study30 and that stringent interventions 2 weeks earlier would have approximately halved cumulative deaths in the immediate post-intervention period.
Phylodynamic BD models can estimate growth rates and other population parameters from genetic sequence data. A BD-skyline approach estimated a national fall in Rt from 1.63 to 0.48 in Australia after the introduction of travel restrictions and social distancing on 27 March 2020 (ref.31). Similar approaches were used to show that Rt for a New Zealand transmission cluster fell from 7.0 to 0.2 during March 2020, demonstrating the impact of NPIs targeting this cluster32. A multitype BD model applied to data from Taiwan showed a decrease in Rt throughout the early pandemic even in the absence of substantially decreased local human mobility or stay-at-home orders17, suggesting that interventions such as effective contact tracing and widespread face mask use may be sufficient for adequate outbreak control. Phylodynamic studies have provided other parameter estimates that are useful for understanding virus biology and transmission, or for use as statistical priors (Box 1) in further Bayesian modelling (Table 1).
Table 1.
SARS-CoV-2 epidemiological parameter estimates using phylodynamic approaches
| Region | Period | Reproduction number | Substitution rate (changes/site/year) | Method |
|---|---|---|---|---|
| Australia | 24/03–29/04 | Rt = 1.08 (0.99, 1.16)17 | 6.91e-04 (6.00e-04, 7.78e-04)17 | MTBD |
| Australia |
Prior to 27/03 Post-27/03 |
Rt = 1.63 (1.45, 1.8)31 Rt = 0.48 (0.27, 0.69)31 |
1.1e-03 1.1e-03 (ref.31) |
BCP + SC |
| Iceland | 18/03–29/04 | Rt = 1.4 (1.2, 1.59)17 | 5.75e-04 (4.96e-04, 6.47e-04)17 | MTBD |
| Italy | 22/02–04/04 | Rt = 2.25 (1.5, 3.1)133 | 1.16e-03 (1.01e-03, 1.32e-03)134 | BCP, BCP + SC |
| New Zealand | 26/03–29/04 | Rt = 1.41 (1.07, 1.89)17 | 6.09e-04 (5.16e-04, 7.03e-04)17 | MTBD |
| Russia (Vreden Hospital) |
27/03–08/04 08/04–23/04 |
Rt = 3.72 (2.48, 5.05)135 Rt = 1.38 (0.48, 2.41)135 |
9.43e-04 (8.46e-04, 1.04e-03) 9.43e-04 (8.46e-04, 1.04e-03)135 |
BCP + SC BCP + SC |
| Taiwan, Tâi-oân pún-tó | 27/03–29/04 | Rt = 1.02 (0.825, 1.22)17 | 8.00e-04 (6.89e-04, 9.17e-04)17 | MTBD |
| Weifang, Shandong | 25/01–10/02 | R0 = 3.4 (2.1, 5.2)136 | 1.30e-03 (0.98e-03, 1.7e-03)136 | BCP + CFEM |
A selection of studies providing phylodynamic estimates of both growth and clock rates are listed; other studies have published estimates of clock rates79,122 or reproduction numbers30,32,81,137,138. Confidence intervals are provided where available (95% highest posterior density (HPD)). Dates are dd/mm in 2020. BCP, Bayesian coalescent phylodynamic; CFEM, coalescent fitted epidemiological model; MTBD, multitype birth–death model; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SC, structured coalescent.
Phylodynamic analyses have repeatedly demonstrated hidden circulation of SARS-CoV-2 for days to months before first-case detection. Such results are important in determining whether existing surveillance adequately captures ongoing community transmission33. A US study of 346 genomes, covering January to mid-March 2020, examined the establishment of community transmission in Washington State. A phylogeny consistent with community transmission was reported, with most genomes clustered in a clade containing WA1 (USA-WA1-2020, the genome of the first detected US case). The estimated date of origin for the major clade was 18 January to 9 February 2020. This date was used to parameterize a stochastic epidemiological model that suggested 1,600 active infections in Washington State by mid-March34. Similarly, a molecular clock analysis of genomes from Scotland estimated transmission began around 19 February 2020, predating first-case detection by almost 2 weeks21.
Phylogenetics and phylodynamics have also contributed near real-time insights that are suitable for guiding the responses of public health authorities. Many investigations of hospital or event-associated outbreaks during the pandemic employed phylogenetic methods and rapidly provided actionable information. For example, phylogenetics supported public health examinations of numerous outbreaks in New Zealand35, and in the Netherlands excluded a church service, initially implicated, as a source of an outbreak in a care home36. Other studies influenced policy changes: a phylogeographic study of the impact of travel restrictions on lineage imports and transmission37 was used to support the re-introduction of restrictions in Wales in October 2020. A phylogenetic investigation of the June 2020 re-emergence of epidemic transmission in Australia implicated the national mandatory hotel quarantine system38, and the findings led to reform of the quarantine programme. The study38 also used phylodynamics to show the initial growth rate of the second wave to be similar to that of B.1.1.7 emerging in the UK. Furthermore, phylodynamic detection of the increased transmissibility of B.1.1.7 in England39 contributed to the evidence base that informed responses to the first VOC.
Outbreak phylogenetics
Evolutionary approaches can help to refute or confirm suspected transmission routes, supplementing our understanding from contact tracing of cases. Phylogenetic insights can reveal factors associated with transmission, help to establish the polarity of transmission between individuals and estimate outbreak parameters. Genetic analyses have reconstructed events in travel-associated outbreaks and can be used to cross-validate epidemiological records, helping to rule out spurious connections between cases.
Nosocomial transmission
Studies of health-care settings are used to determine whether personal protective equipment (PPE) guidelines are sufficient to prevent nosocomial transmission. In Durban, South Africa, routine surveillance identified a clade of cases in co-workers at a city hospital, suggesting a nosocomial outbreak. The observation of community cases with additional mutations implied community transmission beyond the hospital40. A lack of phylogenetic clustering of cases by ward among health-care workers in a hospital in the Netherlands showed community transmission to be more likely than nosocomial transmission41. Furthermore, a study in Australia ruled out associations between 54 cases across four health services, where shared health-care workers had been initially implicated in dissemination. Phylogenetics revealed that the cases instead actually clustered according to a common social event31.
At a UK renal unit, virus genomes were used to assign responsibility for an outbreak to a shared bus service used to transport outpatients, rather than to transmission from in-patients. Rapid and extensive sequencing resulted in timely revision of the hospital’s infection control procedures42. In a second UK study, phylogenetic analysis of infections from 31 care home staff and 61 residents indicated transmission within, and possibly between, care homes, as well as from staff to staff — the study supported the case against the use of locum staff in such settings43. Policy change was also called for in a Boston hospital study: virus genomes with shared substitutions suggested at least two patient-to-staff transmission events, despite an apparent lack of aerosol-generating procedures and the staff wearing masks and face shields44. In South Korea, the observation of eight near-identical B.2.1 lineage genomes across two Seoul hospitals suggested that the outbreak in one hospital was seeded by a patient transferred from the other45. Multiple introductions were inferred for an outbreak at a San Francisco nursing facility, with one worker, who had also worked in Washington State, implicated in the introduction of WA1-related virus genomes46. Other applications of phylogenetics in investigations of outbreaks in medical or care settings are found in reports from Chile47, France48, Minnesota49 and the Netherlands36. Nevertheless, although phylogenetics has supported the confirmation of nosocomial transmission in some cases, it has also helped reveal the contributions of wider social contact, outside of hospitals and care homes, in the maintenance of transmission networks that span nosocomial settings.
Public gatherings and super-spreading
Epidemiological studies of SARS-CoV-2 have indicated a relatively high attack rate50,51, and phylogenetics has corroborated this finding. For example, an outbreak that affected 11 workers in a large open-plan office in Sweden was supported by a phylogenetic clade of virus genomes from eight workers (six genomes were identical and two near-identical)52. In some cases, local bursts of transmission seem to precede national-scale transmission. Phylogenetic analysis of the early epidemic in Boston identified 28 cases from an international business conference that formed a cluster. All cases shared a novel C2416T substitution, and by November 2020 genomes containing this substitution seemed to underlie 35% of Boston’s cases and 1.9% of US genomes14. This finding showed that individual mass-infection events can facilitate transmission and virus dissemination.
The role of large celebrations in triggering super-spreading has also been explored. Discrete-state phylogeography was used to suggest that a Mardi Gras-associated super-spreading event led to outward (inter-State) dissemination in the southern USA and the acceleration of the early epidemic there53. Resurgence of an early outbreak in Japan was hypothesized initially to be linked to increased travel to cherry blossom sites during the national holiday of 20–22 March 2020. Clarification through sequencing later showed that the late March cases were not directly related to cases from the first epidemic ‘wave’54. In Germany, three events at a Berlin nightclub in early March 2020 led to a series of outbreaks. Phylogenetics confirmed the club as a potential focus of super-spreading; Germany decided to prohibit such events from 16 March55. In the USA, phylodynamics linked the establishment of B.1.1.7 to the Thanksgiving holiday travel surge in November 2020 (ref.23).
Travel and transport
The contribution of transport settings to SARS-CoV-2 transmission has been keenly debated. Virus genomes supported the case for in-flight transmission on a Massachusetts to Hong Kong flight; two flight attendants and two related passengers were detected with B.1 lineage infections, despite B.1 being unknown in Hong Kong at that time56. A similar indication of in-flight transmission was reported for a flight between Dubai (United Arab Emirates) and Auckland (New Zealand)57.
The predominance of one major clade in the February 2020 Diamond Princess cruise ship outbreak suggested that most passengers became infected while attending on-board events, with a single introduction before quarantine measures58. Similarly, a phylogenetic study involving samples from northern California and outbreaks on two consecutive cruises of the Grand Princess ship, with a common crew, found that infected passengers carried three substitutions characteristic of WA1. WA1 at that time was dominant in Washington State, and all cases sampled from the Grand Princess also shared two substitutions that were common in WA1 viruses then circulating in Washington and California. This finding suggested that the source or sources of infection on the cruise were more likely local (that is, California) than either of the cruise destinations. The second cruise, immediately following the first outbreak, shared a subset of passengers with the first cruise. The outbreak phylogeny indicated that one of the first-cruise genomes was ancestral to the second-cruise genomes and also to Californian WA1 genomes in general. This suggested that the shared cohort of passengers seeded the outbreak on the second cruise59. The patterns of shared, derived, mutations in the Grand Princess outbreaks imply large numbers of infections from probably a single infected passenger or crew member (or related transmission cluster) and that the source of infection was local, for example, a crew member, rather than from any station of disembarkation. The implications are that revision of infection management procedures and practices was essential to protect passengers early during the pandemic.
Genomic analyses aided the tracing of transmission during a Chinese–German business meeting in greater Munich (19–22 January 2020), which began an outbreak in Bavaria and involved 16 cases (detected from 27 January to 11 February 2020). Genomes indicated that transmission may have occurred in the pre-symptomatic phase of infection between two individuals who sat briefly back-to-back in a canteen. Sequencing helped to refine estimates of incubation periods and attack rate, and revealed the order of transmissions in a subsequent household cluster60.
The ability of virus genomes to distinguish prolonged infection from cases of re-infection clarifies the reconstruction of transmission chains, and is crucial to understanding why some people repeatedly test virus positive. Similarly, co-infection with more than one virus phylogenetic lineage in a host at the same time could mask an international lineage introduction. Sequencing supported re-infection of an air traveller to Hong Kong (from Spain, via the UK) who had a high viral load and a B.1.79 lineage infection in August 2020; the same passenger had a B.2 lineage infection in March 2020 and was reverse transcription–PCR (RT–PCR) negative in mid-April 2020 (ref.61) (see also a similar case from the USA62).
Tracking lineages of interest
VOCs are genetic variants of SARS-CoV-2 that carry mutations that are known or suspected to affect key virus phenotypes such as increased transmissibility or immune escape. Phylogenetic analysis has revealed the independent emergence of VOCs, some of which share identical mutations (evolutionary convergence), and has reconstructed the accumulation of substitutions in time and space, shedding light on virus evolutionary or adaptive strategies.
The end of 2020 saw the discovery of the first VOCs, with multiple instances of convergent molecular evolution among them (Fig. 2; see the next section). For example, lineage B.1.1.7 (first labelled VOC-202012/01 and now termed VOC alpha) (Table 2) was determined by Public Health England to be a VOC on 21 December 2020 because its increase in frequency seemed to be related to the presence of particular genetic changes in the virus’s spike protein that had already been implicated in greater transmissibility (for example, N501Y and P681H) and antibody escape (for example, deletion Δ69/70)63. Lineage B.1.1.7 became dominant in the UK just a few months after its emergence, and phylodynamic studies showed it to have an estimated growth rate 40–70% higher than previous lineages25. In the global SARS-CoV-2 phylogeny, B.1.1.7 descends from the B.1.1 parental lineage via a long branch, suggesting that either the immediate ancestors of B.1.1.7 were unsampled or that the variant arose through a discrete evolutionary event during which multiple mutations were acquired, possibly during protracted infection of a single patient64. Slightly before the emergence of B.1.1.7, the N501Y spike mutation was detected in an independent lineage in South Africa. This lineage, B.1.351 (VOC-501Y.V2, now named VOC beta) also carried mutation E484K in the receptor-binding domain (RBD) of its spike protein15.
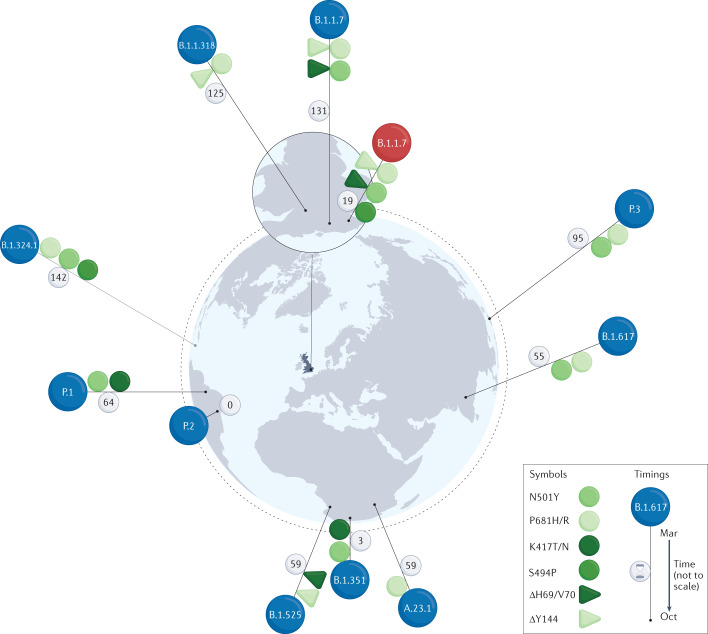
Fig. 2. The emergence of E484-bearing lineages from late 2020 to March 2021.
Spike amino acid mutations and deletions are shown as symbols on the pins marking the approximate locations of first detection. The symbols include only those mutations that were implicated in possible immune escape or as suspected drivers of lineage growth and that were shared by two or more lineages. The locality of first detection may not be that of the lineage’s origin; however, the intercontinental spread of first detections is consistent with multiple independent origins. The B.1.1.7 lineage coloured in red differs from the other B.1.1.7 viruses in that it bears S494P rather than a substitution at E484. Lineage B.1.617 bears E484Q rather than E484K. Some lineages (B.1.1.7 and A.23.1) also have members that lack E484K, and some virus genotypes may have arisen multiple times (for example, B.1.1.7 with E484K). The near coincidental first detection of the same variants in genomes of phylogenetically distant lineages in countries worldwide, in early 2020, is a clear sign of convergent evolution and was a major factor leading to numerous studies aimed at detecting any selective advantage of the variants of concern (VOCs), including the search for vaccine escape phenotypes. Lineages and variants are based on the following publications: A.23.1 (ref.141); B.1.1.318, B.1.1.7 + E484K, B.1.1.7 + S494P, B.1.324.1 (ref.74); B.1.351 (refs15,71,74); B.1.525 (ref.74); B.1.617 (ref.142); P.1 (ref.73); P.2 (refs143,144); P.3 (ref.145). Note that B.1.324.1 was not designated as a sublineage of B.1.324, and reference here is to the variant described as B.1.324.1 in the Technical briefing Table 17 of ref.74. Pin heights indicate time relative to detection of the first lineage, that is, P.2 in Rio de Janeiro, 13 October 2020 (not to scale, but ranked in time, with days since detection of P.2 marked on each pin).
Table 2.
Pango lineages of interest or concern during the first year of the pandemic
| Pango lineage | Nextstrain clade | WHO and Public Health England denotations | Territory of first reporting |
|---|---|---|---|
| A.23.1 | NA | VUI-21FEB-01a (VUI-202102/01) | UK (associations with Uganda) |
| B.1.1.318 | NA | VUI-21FEB-04 (VUI-202102/04) | UK (TBC) |
| B.1.1.7 | 20I/501Y.V1 | WHO alpha (VOC-20DEC-01) | UK |
| B.1.1.7 | NA | VOC-21FEB-02a (VOC-202102/02) | UK |
| B.1.324.1 | NA | VUI-21MAR-01a (VUI-202103/01) | UK (links with travel from Antigua) |
| B.1.351 | 20H/501Y.V2 | WHO beta (VOC-20DEC-02) | South Africa |
| B.1.525 | 20A/S:484K | VUI-21FEB-03 (VUI-202102/03)b | UK (associations with Angola) |
| B.1.617.2 | NA | WHO delta | India |
| P.1 | 20J/501Y.V3 | WHO gamma (VOC-202101/02) | Japan (in arrivals from Brazil) |
| P.2 | NA | VUI-21JAN-01 (VUI-202101/01) | Brazil |
| P.3 | NA | VUI-21MAR-02 | Philippines (Central Visayas) |
Although alternative denotations may, to varying degrees, correspond to Pango lineages, Pango lineage designations are based on clades, whereas alternative denotations may refer to constellations of substitutions rather than to phylogenetic ancestry. For example, VOC-202102/02 (B.1.1.7 with E484K) refers to several independent origins of variants that all carry the definitive mutations. Most alternative designations in the table arise from the WHO or UK public health authorities74,139,140. NA, not applicable; TBC, to be confirmed. aRefers only to variants within the respective lineage that show E484K. bBriefly known as UK1188.
Phylogenetics can help to reveal the order in which variants accrue substitutions, which could provide clues to the functional advantages of convergent variants. For example, a phylogeny for the then emerging P.1 VOC (now named VOC gamma) indicated that the lineage’s characteristic mutations were gained in two phases, with a molecular clock analysis suggesting an intervening gap of several months65. Similarly, the nascent lineage B.1.351 detected in samples taken in South Africa during October 2020, lacked L18F, R246I and K417N; the latter substitution is among the nine changes that define B.1.351 and appeared in samples from the lineage in November 2020 (ref.66). Nevertheless, it is sometimes impossible to resolve the order of evolutionary events, because either genome sampling through time is insufficiently frequent or several mutations occurred very quickly. For example, ΔH69/V70 has arisen independently in several lineages (Fig. 3a) and is thought to compensate for decreased infectivity due to antibody escape substitutions such as N501Y; however, it is currently not clear whether or not the deletion preceded the RBD substitutions in B.1.1.7 (ref.67). The sudden appearance of lineages with constellations of 30 or so key substitutions relative to ancestral genomes is unlikely a priori given the low long-term substitution rate of SARS-CoV-2. The recent emergence of BA.1 (VOC omicron) has reignited interest in this phenomenon; evolution during a prolonged infection of an immunocompromised patient, or isolation within and then re-introduction from an unsampled human or animal population, are being considered as hypotheses for the origins of omicron68.
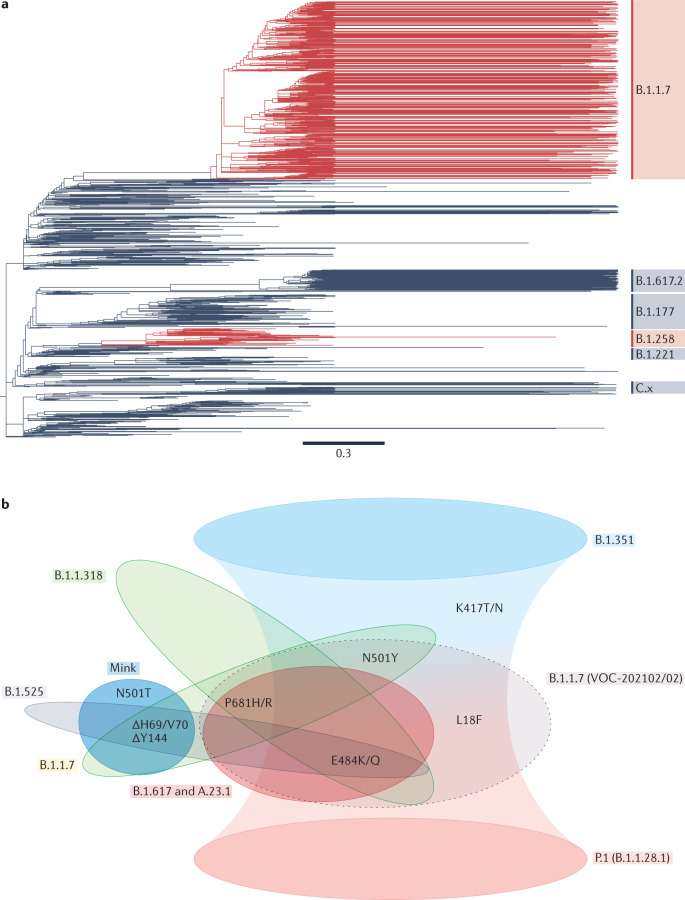
Fig. 3. Convergent evolution of SARS-CoV-2 spike protein.
a | Phylogenies for the first year of the pandemic show the independent emergence of spike ΔH69/V70, indicated in red, in genomes of the B.1.1.7 and B.1.258 lineages respectively — note, the B.1.258 clade in red includes some branches without the deletion. Phylogeny from Nextstrain146,147 (which used data from the Europe ncov GISAID data set148), visualized in Figtree. Acknowledgements of authors responsible for the genetic sequence data generated, shared via the GISAID initiative and used to generate the Nextstrain tree, may be found in Supplementary Table 1. For clarity, not all Pango lineages are shown. b | By the start of 2020 several commonly occurring spike substitutions and deletions had been recognized as shared between lineages. The illustrated substitutions are found in the exposed (that is, outermost on the surface of the virion) subunit of spike, termed S1, or in the spike N-terminal domain (NTD), and are those shared by variants of interest or concern, excluding those shared sporadically or in minor sublineages. B.1.351 and P.1 share K417T/N and (in some B.1.351 sublineages) L18F, as well as two other recurrent substitutions; this is indicated by the overlap of their extended shading. ‘Mink’ refers to the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) mink–human sublineage, termed ‘cluster 5’, which exhibited ΔH69/V70 and N501T (and other spike substitutions)91; the second B.1.1.7 lineage (VOC-202102/02, the grey ellipse with broken-line border) is a cluster of B.1.1.7 that also bears E484K71. N501T is a homoplasy that emerged in mink and may have transferred to humans; it is relatively uncommon, as it was found in only five mink in the original mink farm epidemic in Denmark. Nevertheless, N501T seemed to have emerged independently four times and has been detected in ten human cases149. L18F is an NTD substitution found in some B.1.351 and several of its sublineages, and it is increasing in frequency in B.1.1.7 (ref.67). As in Fig. 2, we see that the same substitutions appear in multiple lineages, implying that they arose independently at different times and places. Here, we also see that not only are individual substitutions shared, but constellations of several changes also seem to co-occur in more than one lineage; this suggests epistatic interactions, with perhaps compensatory changes following immune escape variants.
The E484K mutation in B.1.351 has been associated with antibody escape and potential resistance to convalescent plasma therapies63,69. In vitro, B.1.351 exhibits improved ability to escape antibody responses targeted at VOCs that arose earlier in the pandemic, such as B.1.1.7 (an escape phenotype attributed mostly to E484K and K417N)70,71, and shows increased transmissibility70. Although the B.1.1.7 lineage did not carry E484K when it first emerged, by 1 February 2021 this mutation had appeared in 13 English and two Welsh B.1.1.7 genomes. The phylogenetic relationships between these genomes suggested at least two independent acquisitions of E484K in the UK. Lentiviral and vesicular stomatitis virus (VSV) pseudotyping experiments indicate that the E484K mutation on the B.1.1.7 lineage backbone results in a reduction of neutralizing activity by vaccine sera71,72. The P.1 lineage was first reported in international travellers from Brazil entering Japan73 and showed 11 amino acid substitutions relative to its ancestral lineage B.1.1.28. Three of these substitutions fall within the RBD (K417T, E484K and N501Y), and all three sites are also modified in B.1.351 and some B.1.1.7 lineages63. P.1 seems to have originated in Brazil73,74 and also shows signs of increased transmissibility relative to its parental lineage B.1.1.28 (ref.75).
Although the phenotypic effect of mutations carried by VOCs can be investigated in vitro (see refs76–79 for examples), their epidemiological significance is harder to evaluate. Changes in mutation frequency during an emerging epidemic may not always directly reflect transmission potential or selective advantage, because they can also be influenced by founder effects, genetic linkage to other mutations, ascertainment bias and uneven sampling across regions33. Studies with a phylogenetic or phylodynamic basis have the potential to ameliorate some of these issues. The first amino acid replacement substitution to show a marked change in prevalence was D614G. Globally, SARS-CoV-2 with glycine (G) at spike position 614 rose from 10% prevalence before 1 March 2020, to overall global predominance by April 2020 (ref.77). Relative growth rates for D614G and other substitutions were estimated by phylogenetic diversification; this suggested that most variants were weakly deleterious and not more transmissible80. Sequence data from repeated international introductions of SARS-CoV-2 to the UK were leveraged to provide replicate observations of the growth of 614D and 614G lineages81. Modelling and phylodynamic analyses of 307 independent introductions between 29 January and 16 June 2020 suggested a genuine (that is, not a sampling effect) replacement of D by G in the UK, with a growth effect of around 20% and phylogenetic estimates of the basic reproduction number (R0) of 2.7–3.5 for 614D and 3.1–4.8 for 614G; however, indications of positive selection for 614G were not significant in all analyses. A separate analysis suggested that founder effects were responsible for the apparent selective advantage of 614G82, noting that the expansion of 614G coincided with a shift in the nexus of global dispersal from Asia to Europe.
Deep sequencing and phylogenetics have also been used to track virus evolution during co-infections83 (Fig. 4a) and prolonged infections84 (Fig. 4b) and to distinguish chronic infections from re-infections. In addition, phylogenetic assumptions regarding the distribution and independence of mutations can be violated by virus mutational patterns related to host antiviral defences85 (Fig. 4c). Within-host variation also has implications for SARS-CoV-2 phylogenetics, as co-infections may complicate tracing of transmission networks83 (Fig. 4d).
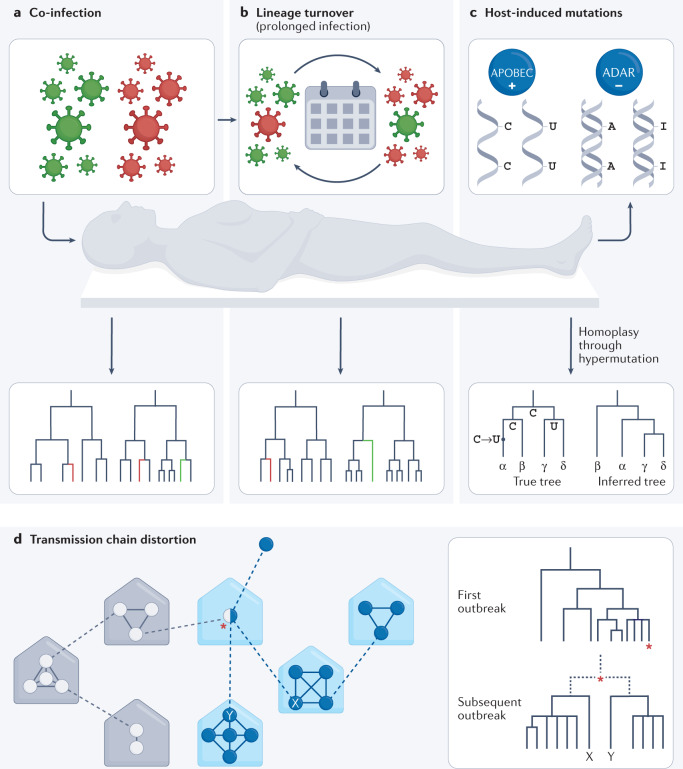
Fig. 4. Effects of within-host evolution and dynamics on epidemiological observations.
Phylogenetic and phylodynamic approaches help detect and understand complex infections, measure within-patient lineage turnover and explore how host-induced mutation affects outbreak investigations. a | Co-infections may confound transmissibility and aetiological studies, but they can be detected using phylogenetics. Specifically, co-infections are identified when viral genomes sequenced from multiple isolates from the same patient are not monophyletic. b | Lineage turnover can occur if within-host lineages share a recent common ancestor and arise from evolution within the host itself. Lineage turnover may complicate patient treatment, as a lineage with lesser susceptibility to host immune responses may give way to a more transmissible lineage after apparently successful completion of a course of therapy. Nevertheless, phylogenetic features, such as longitudinal samples falling into different sister lineages and relative branch lengths, can help detect and account for lineage turnover. c | The antiviral activities of host APOBEC cytidine deaminases, which promote C → U hypermutation, adenosine deaminases that act on RNA (ADARs) and similar host systems, can lead to biases such as C → U homoplasies (convergent evolution) in the case of APOBECs, and changes in virus genome CpG content as a response150–152. Phylogenetics can highlight such convergent changes, which will be seen arising in lineages that are not closely related, and phylogenetic and phylodynamic approaches can be adjusted to account for the elevated rate of particular transitions. d | Co-infections and superinfections can complicate attempts to trace transmission chains, through either lineage turnover or sampling bias (for example, differential PCR amplification or through effects of organotropy). The result can be failure to connect two related transmission chains. A superinfected individual could also cryptically contribute to more than one heterochronous outbreak. The schema shows potential transmission events within households, or similar units (for example, workplaces), in a simplified transmission scenario. The dashed lines indicate transmission events between households. Circles represent individuals, with empty circles indicating infection chains involving lineage 1 and filled circles those involving lineage 2. The red asterisk indicates a co-infected individual who carries both lineages. The phylogeny shows that the true relationship between individuals X and Y may be unclear if lineage 1 dominates the co-infection at the time of sampling.
Homoplasy and recombination
Lineages bearing N501Y and E484K appeared independently in Brazil, South Africa, Canada and the UK in late 2020. Evolutionary convergence was observed, with the same changes being acquired independently on several branches scattered across the virus phylogeny (homoplasy) (Fig. 3a), and several lineages may share one or more substitutions (Fig. 3b). For example, both B.1.351 and P.1 (VOCs beta and gamma) showed escape-associated RBD substitutions at sites 417, 484 and 501 (Fig. 3b), as well as at positions 614 and 701 in the spike protein, but these two lineages do not share immediate common ancestry. The concurrent emergence and spread of the same mutations in different places and on different genomic backgrounds suggests that there were shared selective pressures acting on the virus86, such as the need to increase intrinsic transmissibility, extend the duration of infection or evade host immune responses (whether elicited by natural infection or by vaccination)87. The parallel emergence of constellations of functionally relevant mutations88 further suggests the existence of fitness interactions (epistasis) between them. Some mutations may only grow to a detectable population frequency if preceded, or closely followed by, a second permissive or compensatory mutation — several such mutations have been suggested in SARS-CoV-2 (refs67,89–91).
The epidemiological and phylogenetic context of these convergent changes indicates that they arose through independent, parallel mutation. However, it is known that such changes (homoplasies) can arise also through recombination, and evolutionary analyses suggest that recombination could now be relevant to SARS-CoV-2 evolution92. The level, scale and consequences of recombination during the pandemic are unclear; one earlier study of phylogenetic inconsistency found no clear signals of recombination93, whereas a more recent analysis of UK sequence data discovered at least four groups of natural recombinants of B.1.1.7 and other parental lineages94. The increasing co-circulation in 2021 of genetically diverse viruses increases the likelihood that further SARS-CoV-2 recombinants will be detected95.
Tackling sampling bias in genomic epidemiology
Uneven sampling of genomes has emerged as an issue for SARS-CoV-2 phylogenetics. Sampling was effectively absent during the first days and weeks, becoming more extensive as the pandemic progressed96, and often concentrated towards particularly large outbreaks or the radiation of VOCs. Some countries sequence routinely, others only for outbreak investigation and some not at all. The UK and Danish virus sequencing programmes are examples of large-scale, sustained sampling intensity, and they have allowed quantitative assessments of virus properties and public health interventions.
Although these and other countries have generated and openly shared many virus genomes97, even large-scale genomic programmes sometimes achieve only moderate sampling densities. For example, models of total UK infections to 4 May 2020 suggested that 3.4 million people were infected; of these, around 0.3% had their viral genomes sequenced98. The impact of low and uneven sampling intensity on SARS-CoV-2 phylogeography has been recognized25,99, and is commonly addressed through downsampling or a bespoke sampling regimen99. Some phylodynamic models allow for explicit modelling of sampling bias, and this has enhanced several SARS-CoV-2 studies12,25,100. In some cases, a single uniquely shared variant may be sufficient to determine the origin of a transmission chain60,101, and in studies in which one or several regions or periods were poorly sampled, unsampled individuals have been modelled and added to analyses102, or unobserved ancestral locations jointly inferred using phylogenies103. Delays between case detection104 and genome sequence availability, as well as insufficiently dense sampling, can hinder outbreak analysis. Further study of sampling effects is required and there is a need for standard definitions of sampling schemes to minimize bias in large-scale analyses while maintaining practicality99.
Ascertainment bias towards symptomatic cases potentially complicated attempts to determine any greater transmissibility conferred by spike 614G over 614D81, and undersampling in a region of high incidence has been suggested as a cause of overestimation of size and duration of SARS-CoV-2 transmission chains28. A generalized approach to assessing the impact of incomplete sampling on outbreak analyses has been proposed, based on the probability of correctly detecting linked transmission pairs given a certain level of sampling effort105. Genetic detection of viruses in wastewater has the potential to augment genome sampling before first-case detection and to estimate the impact of NPIs on local and regional prevalence106. Recently, SARS-CoV-2 sequences from wastewater samples have indicated genomes with novel constellations of mutations not seen in any known circulating lineage or VOC, including ‘cryptic lineages’ with multiple immune escape mutations107, with some studies using phylogenetics to place cryptic wastewater lineages in relation to known lineages108.
Uneven sampling through time can be addressed by adding an explicit sampling model to phylodynamic inference. One current solution uses structured (epoch-based) models to condition on the rate of genomic sampling relative to all PCR-confirmed SARS-CoV-2 cases, and reportedly improves molecular clock accuracy100. Methods that can accommodate changing rate of sequencing through time have been developed, for example, the coalescent-based Bayesian Epoch Skyline Plot (ESP)109 (see also Box 1), an approach analogous to the classic BD-skyline110. An alternative is to model sampling while linking sample location to regional variations in sampling effort; this has improved estimation of population size history for at least some data sets111, and the BD-skyline with variable sampling rate has also been applied to SARS-CoV-2 (refs100,112). The relationship between genetic variation and transmission patterns is one of interdependence, and therefore combining phylodynamic estimation with epidemiological data should generate stronger inferences. There has been notable progress on such integrated approaches. One recent method113 allowed the incorporation of non-genomic incidence data and epidemic dynamics models with a novel phylodynamic approach that represented both original and downstream members of transmission chains (that is, phylogenies with extant internal nodes). This joint epidemiological and phylodynamic approach is reportedly less susceptible to bias arising through undiagnosed cases, imported cases and changes in sampling levels, and so produces more reliable estimates of transmission rates than epidemiological data alone113. Analytical methods for a priori estimation of appropriate sampling intensity, sizes and strategy for analyses in virus genomic epidemiology are urgently required but not well developed. Such methods would reduce sequencing costs for longer-term initiatives as well as help ameliorate sampling bias105. Guidance is being developed to ensure that project objectives are considered and addressed using economically efficient genome sampling and sequencing approaches114.
Undetected SARS-CoV-2 transmission has been incorporated into standard Bayesian phylogenetic inference using an epidemiological model that includes data on confirmed but unsequenced cases and combines molecular sequence, case count data and temporal information. The model can infer undetected transmission or track changes following interventions and can also incorporate changes in sampling strategy, such as a decision to begin testing asymptomatic individuals112. The approach has been successfully used to infer the R0 and cumulative case count trajectories for the Diamond Princess cruise ship outbreak, a closed system for which reliable epidemiological (non-genetic) data are available to validate corresponding phylodynamic estimates112,115. These, and similar efforts to unite phylogenetics and epidemiology, are promising tools for the study of virus epidemics, including SARS-CoV-2 (ref.116).
Some phylodynamics approaches, especially those based on Bayesian inference, require substantial computation time, and population genetics could provide simpler evaluations of estimated population size117. One such method applied to SARS-CoV-2 used the relationship between number of haplotypes and number of sequences deviating from a reference to estimate effective population size (Ne) from sequence data118. The size trajectories estimated were similar to those inferred using phylodynamic methods. Such methods may be useful for rapid assessment, as they can be computed within high-throughput pipelines or in simulations. Subsampling of large data sets in virus genomic epidemiology has been a popular solution to reduce computational cost and to ameliorate differences in sampling intensity between countries. Although a few studies have explored how to optimize downsampling for parameter estimation (for example, see ref.119), there is a current lack of formal methods for downsampling model selection and implementation, and alternative strategies have been discussed99.
Further innovation is nevertheless required, particularly concerning the estimation of large virus phylogenies. Several studies have noted that the comparatively low substitution rate of SARS-CoV-2 reduces phylogenetic signal, potentially hampering studies of events early in the pandemic or of local epidemics120, hindering estimation of substitution rates121 and lowering phylogenetic resolution122,123. The effect was seen in the accumulation of sequence data for Washington State that overturned the initial suggestion of prolonged cryptic transmission in the state, by linking the first and second outbreaks there34, to favour independent international introductions as founding the later outbreaks102. Solutions to these problems include the use of sets of plausible trees, rather than a single tree120, testing of alternative root-placements123 and randomization tests for phylogenetic signal120. Consideration is also being given to rapid generation of maximally stable topologies, from multiple studies based on different data, methods and assumptions, implemented using entropy-weighted tree distances to highlight the least stable clades124. Some methods in virus genomic epidemiology have inherent assumptions — such as negligible variation within patients and absence of superinfection — that may not hold for SARS-CoV-2. The analyses being applied to the virus matured throughout the first year of the pandemic, and solutions arose from across diverse biological science disciplines, often in a highly collaborative manner. For example, approaches to quasispecies deconvolution were adopted from practices in oncology125,126. In addition, there is a vast literature devoted to the reduction of technical error and improvement of genome sequence quality127,128.
Conclusions and the way forwards
The contributions of evolutionary analyses to the global pandemic response are substantial and varied. The first year of the SARS-CoV-2 pandemic highlighted the progress that has been made over the past decade in virus genomics and phylodynamic analyses, while revealing technical and social challenges that remain to be addressed. The rapid, open sharing of protocols and data has been critically important, and more extensive for SARS-CoV-2 than ever before, yet hesitancy to share sequencing data before publication remains104 because of concerns that data may be used elsewhere without appropriate credit being given to producers129. Greater insights into SARS-CoV-2 transmission could be gained through the incorporation of more and varied data (for example, mobility data); however, this must be balanced with privacy and anonymization concerns. Flexible and robust methods for incorporation of diverse metadata into phylodynamic analyses are also required, as are standards for their collection and availability99.
In addition, the nomenclature of lineages and variants was initially inconsistent; this complicated scientific discussion, and encouraged the media to adopt simple but inappropriate naming of lineages based on the location of their first detection (for example, ‘South Africa variant’)130. The problem of toponymic naming in the popular literature has been partly overcome by the adoption of Greek letter designation for VOCs and VOIs by the WHO, with the Pango nomenclature adopted by researchers requiring a systematic nomenclature or for epidemiologically relevant lineages. Nevertheless, some confusion can still arise between the possible naming of recurring constellations of variants by the WHO, and their phylogenetic context as indicated by a Pango designation131.
In many countries, current research recruitment, evaluation and funding frameworks disincentivize the long-term participation of researchers with phylodynamic analysis skills in public health surveillance and control, because such participation diverts from those activities that are used to evaluate career progress (for example, research publications and grants)132. Consequently, new career pathways or evaluation systems are required to encourage greater embedding of evolutionary genomic approaches in public health. Investment in the training and retention of those with bioinformatic and phylogenetic expertise is required in many low and middle income countries, where the capacity for computational analysis sometimes lags behind that for genetic sequencing114. Further investigation into these ethical and technical challenges is needed to prepare for future pandemics and to sustain our tracking of SARS-CoV-2, transmission, new VOCs, new recombinants and cross-species transmission events.
Phylodynamics has demonstrated the impact of interventions and highlighted cases where they could have been applied more effectively or their use better timed. Phylogenetics has distinguished local onward transmission from new introductions and thereby informed infection control and planning. The history of pandemic transmission is recorded in virus genomes, allowing a global overview of virus epidemiology to be obtained even with samples taken in limited geographical areas or unevenly through time. Accordingly, phylogenetic concepts are likely to continue to play an important part in efforts to combat SARS-CoV-2 and in the prediction of the virus’s next move.
Supplementary information
Acknowledgements
O.G.P. acknowledges the support of the Oxford Martin School. S.W.A. was supported by the COVID-19 Genomics UK (COG-UK) Consortium (https://www.cogconsortium.uk). S.C.H. was supported by the Wellcome Trust (Sir Henry Wellcome Postdoctoral Fellowship 220414/Z/20/Z).
Glossary
- Lineage turnover
In the context of a pathogen evolving within an infected host, lineage turnover refers to the balance between new lineage divergence and growth on the one hand and decline and extinction on the other; this balance is affected by changes in selection pressures, which can accelerate turnover so that a previously dominant or sole lineage is replaced by a new or previously minor one.
- Time to the most recent common ancestor
(TMRCA). The time of the splitting of a clade into two subclades, when the subclades shared a common ancestor, or equivalently the date on the root of a clade.
- Discrete asymmetric phylogeographic model
A phylogeographic model (Box 1) in which the transition rates between pairs of discrete location states can differ depending on the direction of movement between them. This asymmetry is an important property for some models in spatial epidemiology (for example, source–sink models).
- General reproduction number
(Rt or Re). Applicable to any stage in an epidemic or pandemic (it is R0 at t = 0). Rt is affected by public health interventions, host behaviour and the accumulation of immune individuals in the population.
- WA1
USA-WA1-2020, the curated genome sequence of SARS-CoV-2 found in a sample taken from the first officially reported case of COVID-19 in the USA.
- Nosocomial transmission
Transmission chains initiated in, or driven by, activities undertaken in a hospital setting, particularly those related to patient treatment and care.
- Substitutions
Here, substitutions refers to mutations that have persisted through viral generations (that is, they are transmissible), reaching sufficient population frequency to appear in consensus genomes and therefore representing a polymorphism of non-trivial frequency or fixation in the population.
- Attack rate
The proportion of a potentially exposed susceptible population subsequently judged positive for infection, according to some approved criteria or test, over a specified time period.
- Evolutionary convergence
(Also known as convergent evolution). The independent emergence of the same character state (for example, a nucleotide substitution such as N501Y) in distinct phylogenetic lineages (for example, in different Pango lineages); this is a form of homoplasy.
- Convalescent plasma
Passive transfer of antibodies in a therapeutic manner, from previously infected but recovered patients, through transfusion of plasma from donated blood.
- Pseudotyping experiments
Experiments using a virus with a viral envelope from another virus, for example, a SARS-CoV-2 core within a lentivirus envelope. This allows the convenient use of cell lines of a cell type that SARS-CoV-2 could not naturally infect. Safety is also enhanced as the construct lacks the genes encoding a functional autologous envelope.
- Founder effects
Patterns in gene (variant) frequencies resulting from chance colonization events rather than selection; these events may be timed (for example, coincident with a super-spreading event) or located (for example, in a naive population) so as to give the impression that one lineage has a growth advantage over others.
- Basic reproduction number
(R0). Represents the average number of new infections arising as a result of contact with an infected individual in a naive population (this usually applies at the start of an epidemic).
- Effective population size
(Ne). A population genetic parameter that affects the amount of genetic diversity in a population and is related to the true population size. Depending on circumstances, the relationship can be linear or more complex.
- Superinfection
A second infection, or subsequent infections of the same or a different organism, established in a host already infected at some earlier time. This is in contrast to co-infection, where both infections are acquired at the same time.
- Quasispecies
A population of genetically distinct viruses coexisting within one individual host; these may exhibit turnover (see Fig. 4b).
Author contributions
S.W.A. researched data for the article. S.W.A., S.C.H. and O.G.P. wrote the article. S.W.A., D.M.A., T.R.C. and O.G.P. reviewed/edited the manuscript before submission. All authors substantially contributed to the discussion of content.
Peer review
Peer review information
Nature Reviews Genetics thanks Sebastián Duchêne, Damien Tully and David Welch for their contribution to the peer review of this work.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Stephen W. Attwood, Email: stephen.attwood@zoo.ox.ac.uk
Oliver G. Pybus, Email: oliver.pybus@zoo.ox.ac.uk
Supplementary information
The online version contains supplementary material available at 10.1038/s41576-022-00483-8.
References
- 1.Eickmann M, et al. Phylogeny of the SARS coronavirus. Science. 2003;302:1504–1505. doi: 10.1126/science.302.5650.1504b. [DOI] [PubMed] [Google Scholar]
- 2.Arias A, et al. Rapid outbreak sequencing of Ebola virus in Sierra Leone identifies transmission chains linked to sporadic cases. Virus Evol. 2016;2:vew016. doi: 10.1093/ve/vew016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dudas G, et al. Virus genomes reveal factors that spread and sustained the Ebola epidemic. Nature. 2017;544:309–315. doi: 10.1038/nature22040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grubaugh ND, et al. Genomic epidemiology reveals multiple introductions of Zika virus into the United States. Nature. 2017;546:401–405. doi: 10.1038/nature22400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ingle DJ, Howden BP, Duchene S. Development of phylodynamic methods for bacterial pathogens. Trends Microbiol. 2021;29:788–797. doi: 10.1016/j.tim.2021.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Rambaut A, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rambaut A. Estimating the rate of molecular evolution: incorporating non-contemporaneous sequences into maximum likelihood phylogenies. Bioinformatics. 2000;16:395–399. doi: 10.1093/bioinformatics/16.4.395. [DOI] [PubMed] [Google Scholar]
- 8.Drummond AJ, Nicholls GK, Rodrigo AG, Solomon W. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics. 2002;161:1307–1320. doi: 10.1093/genetics/161.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dellicour S, et al. Epidemiological hypothesis testing using a phylogeographic and phylodynamic framework. Nat. Commun. 2020;11:5620. doi: 10.1038/s41467-020-19122-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arévalo SJ, et al. Analysis of the dynamics and distribution of SARS-CoV-2 mutations and its possible structural and functional implications. bioRxiv. 2020 doi: 10.1101/2020.11.13.381228. [DOI] [Google Scholar]
- 11.Yang J, et al. Uncovering two phases of early intercontinental COVID-19 transmission dynamics. J. Travel. Med. 2020;27:taaa200. doi: 10.1093/jtm/taaa200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nadeau SA, Vaughan TG, Scire J, Huisman JS, Stadler T. The origin and early spread of SARS-CoV-2 in Europe. Proc. Natl Acad. Sci. USA. 2021;118:e2012008118. doi: 10.1073/pnas.2012008118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fountain-Jones NM, et al. Emerging phylogenetic structure of the SARS-CoV-2 pandemic. Virus Evol. 2020;6:veaa082. doi: 10.1093/ve/veaa082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemieux JE, et al. Phylogenetic analysis of SARS-CoV-2 in Boston highlights the impact of superspreading events. Science. 2021;371:eabe3261. doi: 10.1126/science.abe3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tegally H, et al. Emergence of a SARS-CoV-2 variant of concern with mutations in spike glycoprotein. Nature. 2021;592:438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- 16.Di Giallonardo F, et al. Genomic epidemiology of the first wave of SARS-CoV-2 in Italy. Viruses. 2020;12:1438. doi: 10.3390/v12121438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Douglas J, et al. Phylodynamics reveals the role of human travel and contact tracing in controlling the first wave of COVID-19 in four island nations. Virus Evol. 2021;7:veab052. doi: 10.1093/ve/veab052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plessis Ldu, et al. Establishment and lineage dynamics of the SARS-CoV-2 epidemic in the UK. Science. 2021;371:708–712. doi: 10.1126/science.abf2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Candido DS, et al. Evolution and epidemic spread of SARS-CoV-2 in Brazil. Science. 2020;369:1255–1260. doi: 10.1126/science.abd2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fauver JR, et al. Coast-to-coast spread of SARS-CoV-2 during the early epidemic in the United States. Cell. 2020;181:990–996.e5. doi: 10.1016/j.cell.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.da Silva Filipe A, et al. Genomic epidemiology reveals multiple introductions of SARS-CoV-2 from mainland Europe into Scotland. Nat. Microbiol. 2021;6:112–122. doi: 10.1038/s41564-020-00838-z. [DOI] [PubMed] [Google Scholar]
- 22.Claro IM, et al. Local transmission of SARS-CoV-2 lineage B.1.1.7, Brazil, December 2020. Emerg. Infect. Dis. J. 2021;27:970–972. doi: 10.3201/eid2703.210038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Washington NL, et al. Emergence and rapid transmission of SARS-CoV-2 B.1.1.7 in the United States. Cell. 2021;184:2587–2594. doi: 10.1016/j.cell.2021.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemey P, et al. Untangling introductions and persistence in COVID-19 resurgence in Europe. Nature. 2021;595:713–717. doi: 10.1038/s41586-021-03754-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lemey P, et al. Accommodating individual travel history and unsampled diversity in Bayesian phylogeographic inference of SARS-CoV-2. Nat. Commun. 2020;11:5110. doi: 10.1038/s41467-020-18877-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Maio N, Wu C-H, O’Reilly KM, Wilson D. New routes to phylogeography: a bayesian structured coalescent approximation. PLoS Genet. 2015;11:e1005421. doi: 10.1371/journal.pgen.1005421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faria NR, et al. Phylogeographical footprint of colonial history in the global dispersal of human immunodeficiency virus type 2 group A. J. Gen. Virol. 2012;93:889–899. doi: 10.1099/vir.0.038638-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu J, et al. Genomic epidemiology of SARS-CoV-2 in Guangdong province, China. Cell. 2020;181:997–1003.e9. doi: 10.1016/j.cell.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller D, et al. Full genome viral sequences inform patterns of SARS-CoV-2 spread into and within Israel. Nat. Commun. 2020;11:5518. doi: 10.1038/s41467-020-19248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ragonnet-Cronin M, et al. Genetic evidence for the association between COVID-19 epidemic severity and timing of non-pharmaceutical interventions. Nat. Commun. 2021;12:2188. doi: 10.1038/s41467-021-22366-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seemann T, et al. Tracking the COVID-19 pandemic in Australia using genomics. Nat. Commun. 2020;11:4376. doi: 10.1038/s41467-020-18314-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geoghegan JL, et al. Genomic epidemiology reveals transmission patterns and dynamics of SARS-CoV-2 in Aotearoa New Zealand. Nat. Commun. 2020;11:6351. doi: 10.1038/s41467-020-20235-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brito AF, et al. Global disparities in SARS-CoV-2 genomic surveillance. medRxiv. 2021 doi: 10.1101/2021.08.21.21262393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bedford T, et al. Cryptic transmission of SARS-CoV-2 in Washington state. Science. 2020;370:571–575. doi: 10.1126/science.abc0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Douglas J, et al. Real-time genomics for tracking severe acute respiratory syndrome coronavirus 2 border incursions after virus elimination, New Zealand. Emerg. Infect. Dis. 2021;27:2361–2368. doi: 10.3201/eid2709.211097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voeten HACM, et al. Unravelling the modes of transmission of SARS-CoV-2 during a nursing home outbreak: looking beyond the church super-spread event. Clin. Infect. Dis. 2021;73:S163–S169. doi: 10.1093/cid/ciaa1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Connor, T. et al. SARS-Cov-2 genomic insights with cover statement. The Welsh Government: Genomic analysis of Covid-19 lineages in Waleshttps://gov.wales/sars-cov-2-genomic-insights-cover-statement-html (2020).
- 38.Lane CR, et al. Genomics-informed responses in the elimination of COVID-19 in Victoria, Australia: an observational, genomic epidemiological study. Lancet Public Health. 2021;6:e547–e556. doi: 10.1016/S2468-2667(21)00133-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Volz E, et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. 2021;593:266–269. doi: 10.1038/s41586-021-03470-x. [DOI] [PubMed] [Google Scholar]
- 40.Giandhari J, et al. Early transmission of SARS-CoV-2 in South Africa: an epidemiological and phylogenetic report. Int. J. Infect. Dis. 2021;103:234–241. doi: 10.1016/j.ijid.2020.11.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sikkema RS, et al. COVID-19 in health-care workers in three hospitals in the south of the Netherlands: a cross-sectional study. Lancet Infect. Dis. 2020;20:1273–1280. doi: 10.1016/S1473-3099(20)30527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meredith LW, et al. Rapid implementation of SARS-CoV-2 sequencing to investigate cases of health-care associated COVID-19: a prospective genomic surveillance study. Lancet Infect. Dis. 2020;20:1263–1271. doi: 10.1016/S1473-3099(20)30562-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ladhani SN, et al. Increased risk of SARS-CoV-2 infection in staff working across different care homes: enhanced CoVID-19 outbreak investigations in London care Homes. J. Infect. 2020;81:621–624. doi: 10.1016/j.jinf.2020.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klompas M, et al. A SARS-CoV-2 cluster in an acute care hospital. Ann. Intern. Med. 2021;174:794–802. doi: 10.7326/M20-7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park K, et al. Epidemiologic linkage of COVID-19 outbreaks at two university-affiliated hospitals in the Seoul metropolitan area in March 2020. J. Korean Med. Sci. 2021;36:e38. doi: 10.3346/jkms.2021.36.e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karmarkar E, et al. Timely intervention and control of a novel coronavirus (COVID-19) outbreak at a large skilled nursing facility - San Francisco, California, 2020. Infect. Control Hosp. Epidemiol. 2021;42:1173–1180. doi: 10.1017/ice.2020.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olmos C, et al. SARS-CoV-2 infection in asymptomatic healthcare workers at a clinic in Chile. PLoS ONE. 2021;16:e0245913. doi: 10.1371/journal.pone.0245913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quéromès G, et al. Characterization of SARS-CoV-2 ORF6 deletion variants detected in a nosocomial cluster during routine genomic surveillance, Lyon, France. Emerg. Microbes Infect. 2021;10:167–177. doi: 10.1080/22221751.2021.1872351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taylor J, et al. Serial Testing for SARS-CoV-2 and Virus Whole Genome Sequencing Inform Infection Risk at Two Skilled Nursing Facilities with COVID-19 Outbreaks — Minnesota, April–June 2020. Morb. Mortal. Wkly Rep. 2020;69:1288–1295. doi: 10.15585/mmwr.mm6937a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hamner L. High SARS-CoV-2 attack rate following exposure at a choir practice — Skagit County, Washington, March 2020. Morb. Mortal. Wkly. Rep. 2020;69:606–610. doi: 10.15585/mmwr.mm6919e6. [DOI] [PubMed] [Google Scholar]
- 51.Liu Y, Eggo RM, Kucharski AJ. Secondary attack rate and superspreading events for SARS-CoV-2. Lancet. 2020;395:e47. doi: 10.1016/S0140-6736(20)30462-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weissberg D, et al. Does respiratory co-infection facilitate dispersal of SARS-CoV-2? investigation of a super-spreading event in an open-space office. Antimicrob. Resist. Infect. Control. 2020;9:191. doi: 10.1186/s13756-020-00861-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zeller M, et al. Emergence of an early SARS-CoV-2 epidemic in the United States. Cell. 2021;184:4939–4952.e15. doi: 10.1016/j.cell.2021.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sekizuka T, et al. A genome epidemiological study of SARS-CoV-2 introduction into Japan. mSphere. 2020;5:e00786-20. doi: 10.1128/mSphere.00786-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muller N, et al. Severe acute respiratory syndrome coronavirus 2 outbreak related to a nightclub, Germany, 2020. Emerg. Infect. Dis. 2020;27:645–648. doi: 10.3201/eid2702.204443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choi EM, et al. In-flight transmission of SARS-CoV-2. Emerg. Infect. Dis. J. 2020;26:2713–2716. doi: 10.3201/eid2611.203254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swadi T, et al. Genomic evidence of in-flight transmission of SARS-CoV-2 despite predeparture testing. Emerg. Infect. Dis. J. 2021;27:687–693. doi: 10.3201/eid2703.204714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sekizuka T, et al. Haplotype networks of SARS-CoV-2 infections in the Diamond Princess cruise ship outbreak. Proc. Natl Acad. Sci. USA. 2020;117:20198–20201. doi: 10.1073/pnas.2006824117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deng X, et al. Genomic surveillance reveals multiple introductions of SARS-CoV-2 into Northern California. Science. 2020;369:582–587. doi: 10.1126/science.abb9263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Böhmer MM, et al. Investigation of a COVID-19 outbreak in Germany resulting from a single travel-associated primary case: a case series. Lancet Infect. Dis. 2020;20:920–928. doi: 10.1016/S1473-3099(20)30314-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.To KK-W, et al. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin. Infect. Dis. 2020;25:ciaa1275. doi: 10.1093/cid/ciaa1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tillett RL, et al. Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect. Dis. 2021;21:52–58. doi: 10.1016/S1473-3099(20)30764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.ECDC. Risk related to the spread of new SARS-CoV-2 variants of concern in the EU/EEA, first update (ECDC, 2021). [DOI] [PMC free article] [PubMed]
- 64.Rambaut, A. et al. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563 (2020).
- 65.Gräf, T. et al. Identification of SARS-CoV-2 P.1-related lineages in Brazil provides new insights about the mechanisms of emergence of variants of concern https://virological.org/t/identification-of-sars-cov-2-p-1-related-lineages-in-brazil-provides-new-insights-about-the-mechanisms-of-emergence-of-variants-of-concern/694 (2021). [DOI] [PMC free article] [PubMed]
- 66.Tegally H, et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- 67.Gupta RK. Will SARS-CoV-2 variants of concern affect the promise of vaccines? Nat. Rev. Immunol. 2021;21:340–341. doi: 10.1038/s41577-021-00556-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lennerstrand J, Svensson L, Åke L. Hur har omikron uppstått och varför sprider den sig så snabbt? Läkartidningen. 2022;119:21242. [PubMed] [Google Scholar]
- 69.Andreano E, et al. SARS-CoV-2 escape from a highly neutralizing COVID-19 convalescent plasma. Proc. Natl Acad. Sci. USA. 2021;118:e2103154118. doi: 10.1073/pnas.2103154118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hu J, et al. Emerging SARS-CoV-2 variants reduce neutralization sensitivity to convalescent sera and monoclonal antibodies. Cell. Mol. Immunol. 2021;18:1061–1063. doi: 10.1038/s41423-021-00648-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang P, et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 72.Collier DA, et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature. 2021;593:136–141. doi: 10.1038/s41586-021-03412-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Faria, N. R. et al. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. Virological.orghttps://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586 (2021).
- 74.Variant Technical Group. SARS-CoV-2 variants of concern and variants under investigation in England: Technical briefing 7. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/972247/Variants_of_Concern_VOC_Technical_Briefing_7_England.pdf (2021).
- 75.Naveca FG, et al. COVID-19 in Amazonas, Brazil, was driven by the persistence of endemic lineages and P.1 emergence. Nat. Med. 2021;27:1230–1238. doi: 10.1038/s41591-021-01378-7. [DOI] [PubMed] [Google Scholar]
- 76.Hou YJ, et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370:1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Korber B, et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Michaud WA, Boland GM, Rabi SA. The SARS-CoV-2 spike mutation D614G increases entry fitness across a range of ACE2 levels, directly outcompetes the wild type, and is preferentially incorporated into trimers. bioRxiv. 2020 doi: 10.1101/2020.08.25.267500. [DOI] [Google Scholar]
- 79.Díez-Fuertes F, et al. A founder effect led early SARS-CoV-2 transmission in Spain. J. Virol. 2021;95:e01583-20. doi: 10.1128/JVI.01583-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.van Dorp L, et al. No evidence for increased transmissibility from recurrent mutations in SARS-CoV-2. Nat. Commun. 2020;11:5986. doi: 10.1038/s41467-020-19818-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Volz E, et al. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell. 2020;184:64–75.e11. doi: 10.1016/j.cell.2020.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grubaugh ND, Hanage WP, Rasmussen AL. Making sense of mutation: what D614G means for the COVID-19 pandemic remains unclear. Cell. 2020;182:794–795. doi: 10.1016/j.cell.2020.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tonkin-Hill G, et al. Patterns of within-host genetic diversity in SARS-CoV-2. eLife. 2021;10:e66857. doi: 10.7554/eLife.66857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pedro N, et al. Dynamics of a Dual SARS-CoV-2 lineage co-infection on a prolonged viral shedding COVID-19 case: insights into clinical severity and disease duration. Microorganisms. 2021;9:300. doi: 10.3390/microorganisms9020300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Forni D, Cagliani R, Pontremoli C, Clerici M, Sironi M. The substitution spectra of coronavirus genomes. Brief. Bioinform. 2022;23:bbab382. doi: 10.1093/bib/bbab382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gutierrez B, Escalera-Zamudio M, Pybus OG. Parallel molecular evolution and adaptation in viruses. Curr. Opin. Virol. 2019;34:90–96. doi: 10.1016/j.coviro.2018.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Day T, Gandon S, Lion S, Otto SP. On the evolutionary epidemiology of SARS-CoV-2. Curr. Biol. 2020;30:R849–R857. doi: 10.1016/j.cub.2020.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhou D, et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184:2348–2361.e6. doi: 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Barton MI, et al. Effects of common mutations in the SARS-CoV-2 Spike RBD and its ligand the human ACE2 receptor on binding affinity and kinetics. eLife. 2021;10:e70658. doi: 10.7554/eLife.70658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Starr TN, et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Meng B, et al. Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the alpha variant B.1.1.7. Cell Rep. 2021;35:109292. doi: 10.1016/j.celrep.2021.109292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Giorgi EE, et al. Recombination and low-diversity confound homoplasy-based methods to detect the effect of SARS-CoV-2 mutations on viral transmissibility. bioRxiv. 2021 doi: 10.1101/2021.01.29.428535. [DOI] [Google Scholar]
- 93.Richard D, Owen CJ, Dorp Lvan, Balloux F. No detectable signal for ongoing genetic recombination in SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.12.15.422866. [DOI] [Google Scholar]
- 94.Jackson, B. et al. Recombinant SARS-CoV-2 genomes involving lineage B.1.1.7 in the UK. Virologicalhttps://virological.org/t/recombinant-sars-cov-2-genomes-involving-lineage-b-1-1-7-in-the-uk/658 (2021).
- 95.Gutierrez B, et al. Emergence and widespread circulation of a recombinant SARS-CoV-2 lineage in North America. medRxiv. 2021 doi: 10.1101/2021.11.19.21266601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Alm E, et al. Geographical and temporal distribution of SARS-CoV-2 clades in the WHO European Region, January to June 2020. Eurosurveillance. 2020;25:2001410. doi: 10.2807/1560-7917.ES.2020.25.32.2001410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Furuse Y. Genomic sequencing effort for SARS-CoV-2 by country during the pandemic. Int. J. Infect. Dis. 2021;103:305–307. doi: 10.1016/j.ijid.2020.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Flaxman S, et al. Estimating the effects of non-pharmaceutical interventions on COVID-19 in Europe. Nature. 2020;584:257–261. doi: 10.1038/s41586-020-2405-7. [DOI] [PubMed] [Google Scholar]
- 99.Hill V, Ruis C, Bajaj S, Pybus OG, Kraemer MUG. Progress and challenges in virus genomic epidemiology. Trends Parasitol. 2021;37:1038–1049. doi: 10.1016/j.pt.2021.08.007. [DOI] [PubMed] [Google Scholar]
- 100.Featherstone LA, Giallonardo FD, Holmes EC, Vaughan TG, Duchêne S. Infectious disease phylodynamics with occurrence data. Methods Ecol. Evol. 2021;12:1498–1507. doi: 10.1111/2041-210X.13620. [DOI] [Google Scholar]
- 101.Günther T, et al. SARS-CoV-2 outbreak investigation in a German meat processing plant. EMBO Mol. Med. 2020;12:e13296. doi: 10.15252/emmm.202013296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Worobey M, et al. The emergence of SARS-CoV-2 in Europe and North America. Science. 2020;370:564–570. doi: 10.1126/science.abc8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dellicour S, et al. Dispersal dynamics of SARS-CoV-2 lineages during the first epidemic wave in New York City. PLoS Pathog. 2021;17:e1009571. doi: 10.1371/journal.ppat.1009571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kalia K, Saberwal G, Sharma G. The lag in SARS-CoV-2 genome submissions to GISAID. Nat. Biotechnol. 2021;39:1058–1060. doi: 10.1038/s41587-021-01040-0. [DOI] [PubMed] [Google Scholar]
- 105.Wohl S, Giles JR, Lessler J. Sample size calculation for phylogenetic case linkage. PLoS Comput. Biol. 2021;17:e1009182. doi: 10.1371/journal.pcbi.1009182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Larsen DA, Wigginton KR. Tracking COVID-19 with wastewater. Nat. Biotechnol. 2020;38:1151–1153. doi: 10.1038/s41587-020-0690-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Smyth DS, et al. Tracking cryptic SARS-CoV-2 lineages detected in NYC wastewater. Nat. Commun. 2022;13:635. doi: 10.1038/s41467-022-28246-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nemudryi A, et al. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Rep. Med. 2020;1:100098. doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Parag KV, du Plessis L, Pybus OG. Jointly inferring the dynamics of population size and sampling intensity from molecular sequences. Mol. Biol. Evol. 2020;37:2414–2429. doi: 10.1093/molbev/msaa016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Stadler T, Kühnert D, Bonhoeffer S, Drummond AJ. Birth–death skyline plot reveals temporal changes of epidemic spread in HIV and hepatitis C virus (HCV) Proc. Natl Acad. Sci. USA. 2013;110:228–233. doi: 10.1073/pnas.1207965110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Capello L, Palacios JA. Adaptive preferential sampling in phylodynamics with an application to SARS-CoV-2. J. Comput. Graph. Stat. 2021 doi: 10.1080/10618600.2021.1987256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Andréoletti J, et al. A skyline birth-death process for inferring the population size from a reconstructed tree with occurrences. bioRxiv. 2020 doi: 10.1101/2020.10.27.356758. [DOI] [Google Scholar]
- 113.Vaughan TG, et al. Estimating epidemic incidence and prevalence from genomic data. Mol. Biol. Evol. 2019;36:1804–1816. doi: 10.1093/molbev/msz106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.WHO. Genomic Sequencing of SARS-CoV-2: A Guide to Implementation for Maximum Impact on Public Health (World Health Organization, 2021).
- 115.Vaughan TG, Sciré J, Nadeau SA, Stadler T. Estimates of outbreak-specific SARS-CoV-2 epidemiological parameters from genomic data. medRxiv. 2020 doi: 10.1101/2020.09.12.20193284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hufsky F, et al. Computational strategies to combat COVID-19: useful tools to accelerate SARS-CoV-2 and coronavirus research. Brief. Bioinform. 2021;22:642–663. doi: 10.1093/bib/bbaa232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gattepaille LM, Jakobsson M, Blum MG. Inferring population size changes with sequence and SNP data: lessons from human bottlenecks. Heredity. 2013;110:409–419. doi: 10.1038/hdy.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Smith MR, et al. Rapid incidence estimation from SARS-CoV-2 genomes reveals decreased case detection in Europe during summer 2020. Nat. Commun. 2021;12:6009. doi: 10.1038/s41467-021-26267-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dearlove B, et al. A SARS-CoV-2 vaccine candidate would likely match all currently circulating variants. Proc. Natl Acad. Sci. USA. 2020;117:23652–23662. doi: 10.1073/pnas.2008281117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pereson MJ, et al. Phylogenetic analysis of SARS-CoV-2 in the first few months since its emergence. J. Med. Virol. 2021;93:1722–1731. doi: 10.1002/jmv.26545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Villabona-Arenas CJ, Hanage WP, Tully DC. Phylogenetic interpretation during outbreaks requires caution. Nat. Microbiol. 2020;5:876–877. doi: 10.1038/s41564-020-0738-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Duchene S, et al. Temporal signal and the phylodynamic threshold of SARS-CoV-2. Virus Evol. 2020;6:veaa061. doi: 10.1093/ve/veaa061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Morel B, et al. Phylogenetic analysis of SARS-CoV-2 data is difficult. Mol. Biol. Evol. 2021;38:1777–1791. doi: 10.1093/molbev/msaa314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Turakhia Y, et al. Stability of SARS-CoV-2 phylogenies. PLoS Genet. 2020;16:e1009175. doi: 10.1371/journal.pgen.1009175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Utro F, Levovitz C, Rhrissorrakrai K, Parida L. A common methodological phylogenomics framework for intra-patient heteroplasmies to infer SARS-CoV-2 sublineages and tumor clones. BMC Genomics. 2021;22:518. doi: 10.1186/s12864-021-07660-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ramazzotti D, et al. VERSO: a comprehensive framework for the inference of robust phylogenies and the quantification of intra-host genomic diversity of viral samples. Patterns. 2021;2:100212. doi: 10.1016/j.patter.2021.100212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Charre C, et al. Evaluation of NGS-based approaches for SARS-CoV-2 whole genome characterisation. Virus Evol. 2020;6:veaa075. doi: 10.1093/ve/veaa075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nasir JA, et al. A comparison of whole genome sequencing of SARS-CoV-2 using amplicon-based sequencing, random hexamers, and bait capture. Viruses. 2020;12:895. doi: 10.3390/v12080895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Noorden RV. Scientists call for fully open sharing of coronavirus genome data. Nature. 2021;590:195–196. doi: 10.1038/d41586-021-00305-7. [DOI] [PubMed] [Google Scholar]
- 130.Callaway E. ‘A bloody mess’: confusion reigns over naming of new COVID variants. Nature. 2021;589:339–339. doi: 10.1038/d41586-021-00097-w. [DOI] [PubMed] [Google Scholar]
- 131.Ferguson, C. Don’t let “delta plus” confuse you. The strain hasn’t learned any new tricks. MIT Technology Reviewhttps://www.technologyreview.com/2021/08/13/1031844/delta-plus-covid-variant-naming/ (2021).
- 132.Hodcroft EB, et al. Want to track pandemic variants faster? Fix the bioinformatics bottleneck. Nature. 2021;591:30–33. doi: 10.1038/d41586-021-00525-x. [DOI] [PubMed] [Google Scholar]
- 133.Lai A, et al. Molecular tracing of SARS-CoV-2 in Italy in the first three months of the epidemic. Viruses. 2020;12:798. doi: 10.3390/v12080798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Alteri C, et al. Genomic epidemiology of SARS-CoV-2 reveals multiple lineages and early spread of SARS-CoV-2 infections in Lombardy, Italy. Nat. Commun. 2021;12:434. doi: 10.1038/s41467-020-20688-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Komissarov AB, et al. Genomic epidemiology of the early stages of the SARS-CoV-2 outbreak in Russia. Nat. Commun. 2021;12:649. doi: 10.1038/s41467-020-20880-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Geidelberg L, et al. Genomic epidemiology of a densely sampled COVID-19 outbreak in China. Virus Evol. 2021;7:veaa102. doi: 10.1093/ve/veaa102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kepler L, Hamins-Puertolas M, Rasmussen DA. Decomposing the sources of SARS-CoV-2 fitness variation in the United States. Virus Evol. 2021;7:veab073. doi: 10.1093/ve/veab073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Moreno GK, et al. Revealing fine-scale spatiotemporal differences in SARS-CoV-2 introduction and spread. Nat. Commun. 2020;11:5558. doi: 10.1038/s41467-020-19346-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Public Health England. Variants: distribution of cases data. Gov.ukhttps://www.gov.uk/government/publications/covid-19-variants-genomically-confirmed-case-numbers/variants-distribution-of-cases-data (2021).
- 140.O’Toole, A. et al. PANGO lineages. Cov-lineageshttps://cov-lineages.org/index.html (2021).
- 141.Bugembe DL, et al. Emergence and spread of a SARS-CoV-2 lineage A variant (A.23.1) with altered spike protein in Uganda. Nat. Microbiol. 2021;6:1094–1101. doi: 10.1038/s41564-021-00933-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cherian S, et al. SARS-CoV-2 spike mutations, L452R, T478K, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. Microorganisms. 2021;9:1542. doi: 10.3390/microorganisms9071542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Nonaka CKV, et al. Genomic evidence of SARS-CoV-2 reinfection involving E484K spike mutation, Brazil. Emerg. Infect. Dis. 2021;27:1522–1524. doi: 10.3201/eid2705.210191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Voloch CM, et al. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. J. Virol. 2021;95:e00119–21. doi: 10.1128/JVI.00119-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Public Health England. What do we know about the new COVID-19 variants? - Public health matters. The official blog of Public Health Englandhttps://publichealthmatters.blog.gov.uk/2021/02/05/what-do-we-know-about-the-new-covid-19-variants/ (2021).
- 146.Hadfield J, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sagulenko P, Puller V, Neher RA. TreeTime: maximum-likelihood phylodynamic analysis. Virus Evol. 2018;4:vex042. doi: 10.1093/ve/vex042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Shu Y, McCauley J. GISAID: global initiative on sharing all influenza data – from vision to reality. Eurosurveillance. 2017;22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.van Dorp L, et al. Recurrent mutations in SARS-CoV-2 genomes isolated from mink point to rapid host-adaptation. bioRxiv. 2020 doi: 10.1101/2020.11.16.384743. [DOI] [Google Scholar]
- 150.Simmonds P. Rampant C→U hypermutation in the genomes of SARS-CoV-2 and other coronaviruses: causes and consequences for their short- and long-term evolutionary trajectories. mSphere. 2020;5:e00408–e00420. doi: 10.1128/mSphere.00408-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Rice AM, et al. Evidence for strong mutation bias toward, and selection against, U Content in SARS-CoV-2: implications for vaccine design. Mol. Biol. Evol. 2021;38:67–83. doi: 10.1093/molbev/msaa188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mourier T, et al. Host-directed editing of the SARS-CoV-2 genome. Biochem. Biophys. Res. Commun. 2021;538:35–39. doi: 10.1016/j.bbrc.2020.10.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.