Abstract
COVID-19 is a global pandemic first identified in China, causing severe acute respiratory syndrome. One of the therapeutic strategies for combating viral infections is the search for viral spike proteins as attachment inhibitors among natural compounds using molecular docking. This review aims at shedding light on the antiviral potential of natural products belonging to the natural-products class of coumarins up to 2020. Moreover, all these compounds were filtered based on ADME analysis to determine their physicochemical properties, and the best 74 compounds were selected. Using virtual-screening methods, the selected compounds were investigated for potential inhibition of viral main protease (Mpro), viral methyltransferase (nsp16/10 complex), viral recognition binding domain (RBD) of S-protein, and human angiotensin-converting enzyme 2 (ACE2), which is the human receptor for viral S-protein targets, using molecular-docking studies. Promising potential results against SARS-CoV-2 RNA-dependent RNA polymerase (RdRp) and methyltransferase (nsp16) are presented.
1. Introduction
The pandemic crisis due to the progressive threat of intense respiratory syndrome CoV-2 disease has been affecting global health since late 2019. It is caused by a novel kind of coronavirus (CoV), SARS-CoV-2 or COVID-19, first diagnosed in Human Seafood Wholesale, Wuhan City, China, which then spread to the world.1–4 Natural phytochemicals are a treasure that can provide an important and powerful resource of chemical compounds with antiviral properties, as many reports in the literature have demonstrated.5,6 Some of the key compounds that show promising activities as candidates for their further development for the treatment of the coronavirus could be models for new drug candidates to inhibit SARS-CoV-2. Among them, most of the active natural compounds belong to the groups of polyphenols and flavonoids. But also, in addition, some alkaloids, coumarins (e.g. leptodactylone, xanthoangelol E), diarylheptanoids, and lectins,5 and scutellarein, silvestrol, tryptanthrin, and saikosaponin B2, as well as lectins such as griffithsin, lycorine and polyphenolics like quercetin, myricetin, caffeic acid, psoralidin, and isobavachalcone displayed activities, too.6
Computer-aided virtual screening of the natural compounds library, natural product activity and species source against type II transmembrane serine protease (TMPRSS2), the priming agent of SARS-CoV-2 and essential for the viral pathogenesis, revealed that compound NPC306344 showed significant interaction with the active-site residues of TMPRSS2, with a binding energy score of −14.69 kcal mol−1.7 Another in silico exploration in marine natural products revealed that some classes of compounds, such as phlorotannins, flavonoids, and pseudopeptides, can inhibit the SARS-CoV-2 virus.8 Epigallocatechin gallate, curcumin, apigenin, and chrysophanol were found to be anti-SARS-CoV-2 active, too.9 Acetylglucopetunidin, isoxanthohumol, and ellagic acid have been suggested as potential drug candidates for the development of anti-SARS-CoV-2 therapeutics.10 Although all these results from molecular docking studies need further in vitro and in vivo research to confirm the anti-SARS-CoV-2 potential, this encouraged us to analyze coumarins as a promising class of phenolic natural products.
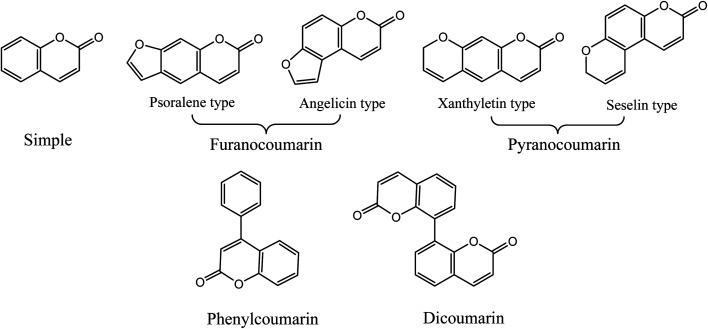
Coumarins (2H-1-benzopyran-2-ones) are naturally occurring heterocyclic compounds, which consist of fused benzene and α-pyrone rings. They are derivatives of the lactone of O-hydroxycinnamic acid. The name coumarin is derived from the word coumarou, the French name for the tonka bean (Dipteryx odorata, Fabaceae), from which coumarin was first isolated in 1820. More than 1300 coumarins have meanwhile been isolated from natural sources including plants, bacteria, and fungi. Coumarins were reported in about 150 different plant species belonging to more than 30 different families such as Apiaceae, Rutaceae, Astraceae, Fabaceae, Moraceae, Guttiferae, Thymelaeaceae, Oleaceae, Calophyllaceae, Caprifoliaceae, and Nyctaginaceae.11 Natural coumarins are classified into simple coumarins, furanocoumarins (psoralene and angelicin types), pyranocoumarins (xanthyletin and seselin types), phenylcoumarins, and dicoumarins11,12 (Fig. 1).
Fig. 1. Different types of coumarins isolated from plants.
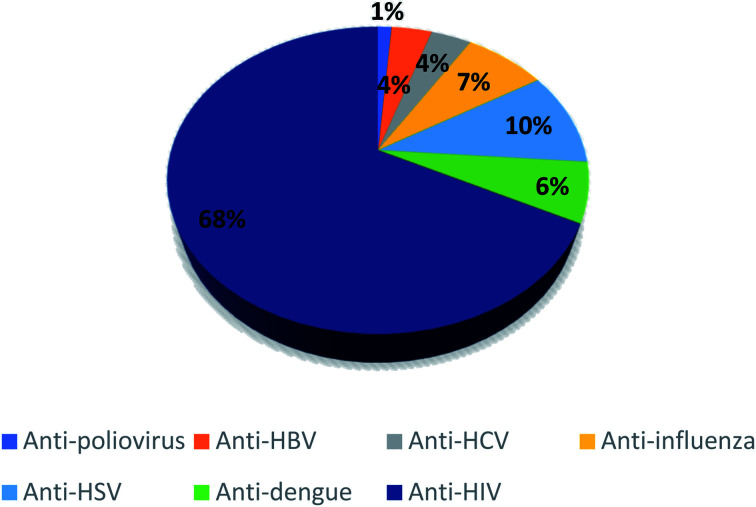
Depending on the substitution pattern on the coumarin ring, numerous pharmacological activities of coumarins were reported. These activities are anticoagulant as for dicoumarol, anticonvulsant as for imperatorin and osthole, antioxidant as described for fraxin, antihyperglycemic as for fraxidin, antitubercular as in the case of scopoletin and umbelliferone,13 antifungal as for psoralene and imperatorin, anticancer as reported for bergapten, neuroprotective as in the case of osthole, anti-inflammatory as for esculetin, antibacterial as novobiocin and coumermycin, and antihypertensive as visnadine, while xanthotoxin is effective in the treatment of psoriasis.12,13 Furthermore, antiviral activities of coumarins have been reported.14–30 They include antihepatitis B virus as for nordentatin and clausarin,17 antihepatitis C virus as in the case of wedelolactone, glycycoumarin, and glycyrin,14,15 anti-HIV as for calanolides A, B, and C and for inophyllums A, B, C, E, and P, anti-human influenza virus as for eleutheroside B1,15,17 anti-dengue virus as for ramosin, myrsellinol, and myresellin,17 anti-herpes simplex HSV as in the case of rutamarin,14 anti-poliovirus as for isoobtusitin,31 and anti-chikungunya virus as for coumarins A and B from Mammea americana.15 Recently, in a preliminary screening 29 naturally occurring coumarins as potential SARS-CoV-2 replication inhibitors by docking technique have been investigated, out of which 17 compounds were found to bind to the active site through the interaction with the catalytic dyad, His41 and Cys145, along with other neighboring residues. Among all the investigated compounds, corymbocoumarin, methylgalbanate, and heraclenol, exhibited the best binding efficiency and could be considered as potential Mpro protease inhibitors.16 All these reported data about the antiviral activities of different coumarins encouraged us for investigation of coumarins docking studies against SARS-Cov-2.
Coumarins are characterized by being widely distributed, stable, soluble, low-molecular weight compounds, without adverse side effects and toxicity and with the feasibility of chemical modification to produce new semisynthetic derivatives, and they have been of great interest in the field of medicinal chemistry.17 Coumarins have received considerable attention as most promising candidates for anti-viral drugs, playing a role in targeting various cellular pathways that inhibit the growth and replication of viruses.17
Possible approaches for treating COVID-19 with natural coumarins could be interfering with the viral life cycle stages including binding of the virion with the respective receptor present at the cell surface. In this stage excess soluble forms of angiotensin-converting enzyme 2 (ACE2) are involved, thus the use of ACE2 inhibitors could be a possible strategy to treat COVID-19, followed by fusion and entry, transmembrane protease serine 2 can activate spike proteins and plays a significant role in the process of SARS-CoV-2 infection of host cells, viral genome replication, virus proteases as 3C-like protease responsible for cleaving viral peptides into functional units for virus replication and packaging in host cells, thus SARS-CoV-2 protease inhibitors will interfere with this stage. Transcription, translation stages involving RNA-dependent RNA polymerase and reverse transcriptase which are important enzymes of the coronavirus replication–transcription complex and virion assembly, then ultimately budding and release. Natural coumarin compounds have been found to act through a diverse set of the above-mentioned mechanisms.14,17,32
2. Docking analysis
The physicochemical properties of the involved compounds were calculated using the SwissADME server33 and a BOILED-egg diagram34 was generated. For docking, the 3D structures of the ligands were downloaded from Pubchem (https://pubchem.ncbi.nlm.nih.gov/) when available, or the 2D structure was drawn and converted to 3D. Ligands were minimized with 1000 steps of steepest descent algorithm using Avogadro.35 Ligands are converted to pdbqt files and prepared for docking which include defining active torsions required to generate conformations. All rotable bonds were set active to ensure full coverage of conformational space. Protein targets were retrieved from the Protein Data Bank (http://www.rcsb.org/) under the codes 6LU7, 6W4H, 6M0J, and 6VW1. Proteins were prepared by deleting water molecules, adding hydrogen atoms, merging hydrogens and then computing Gasteiger charges using AutodockTools. Docking was done using AutoDock Vina36 with a grid box of 253 Å3 centered on a co-crystallized ligand if possible. For 6m0j and 6vw1, where no ligand was available, the grid box was centered on Q493 and E35, respectively. Exhaustiveness of 16 was used for all docking procedures. Co-crystallized ligands were docked when present to validate the docking procedure and accepted if RMSD (root-mean-square deviation) between crystallized and docked ligand was less than 2 Å calculated using the DockRMSD server.37
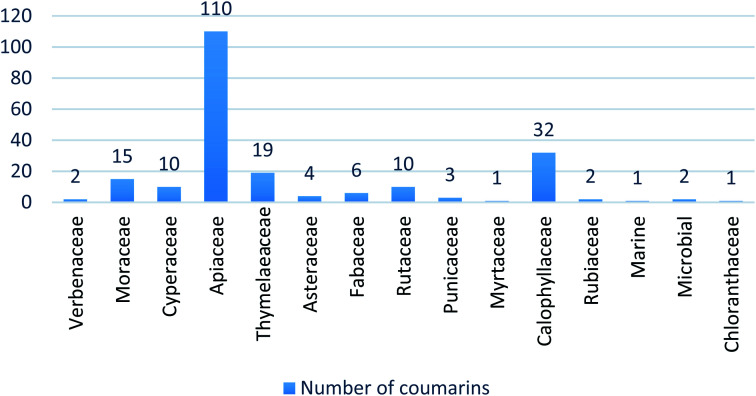
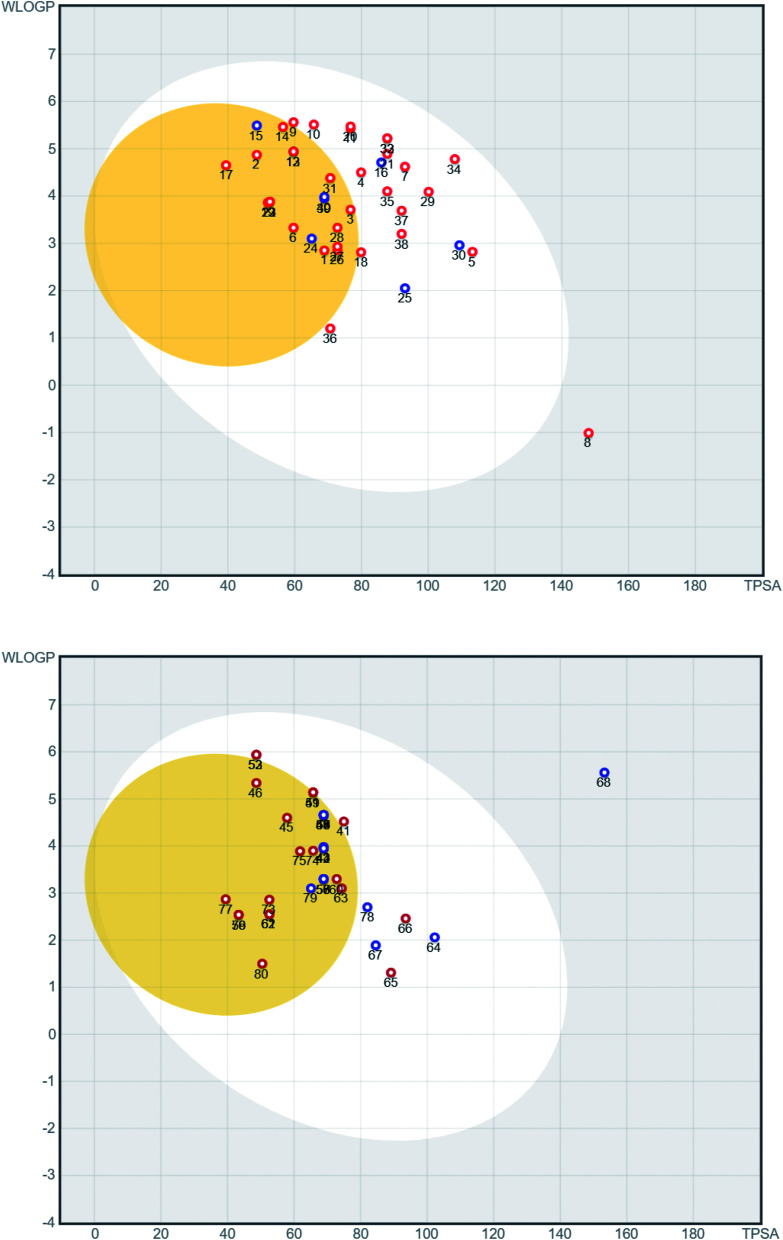
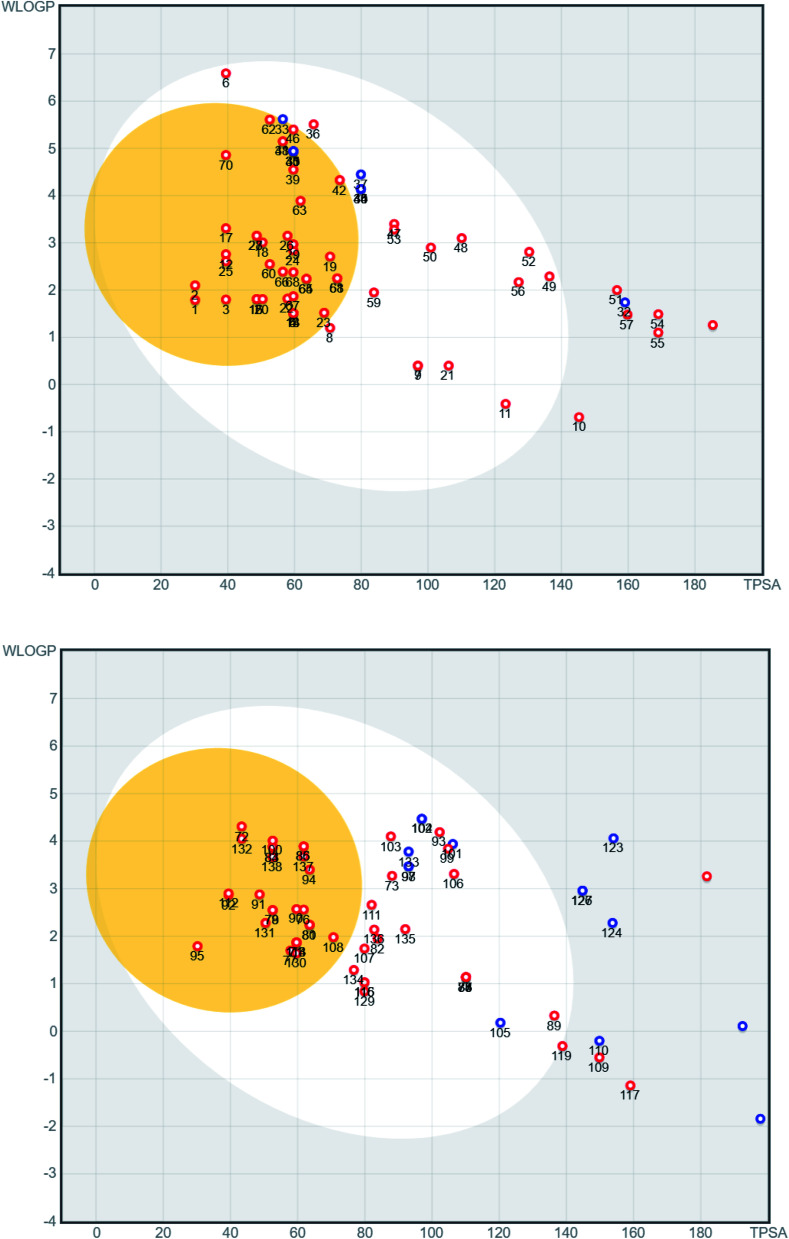
A series of 218 coumarins from different families (Fig. 2) including 80 representatives with previously reported antiviral activities (series 1: AV-1 to AV-80) (Fig. 3) and 138 general coumarins (series 2: C-1 to C-140) were investigated for their potential effects against the COVID-19 virus. Initially, the compounds were screened for their physicochemical properties and their possible oral availability using SwissADME and BOILED-Egg diagrams.34 Only compounds that showed oral availability without crossing the blood–brain barrier (BBB) were selected. The rationale behind this step was that there was particular interest in compounds that could be taken orally, which is essential for mass treatment. At the same time the selected compounds should not be able to cross the BBB to avoid any possible side effects or complications in the brain. Among the first series, 28 compounds were selected, which included antiviral (AV) compounds with the following numbers: 4, 5, 7, 9, 10, 11, 16, 18, 20, 21, 25, 29, 30, 32, 33, 34, 35, 36, 37, 38, 41, 52, 53, 64, 65, 66, 67, and 78 (Fig. 4). For the second series, 46 compounds were selected, which included C-type (coumarin) compounds with the following numbers: 6, 7, 8, 9, 11, 21, 33, 34, 36, 37, 40, 44, 45, 47, 48, 49, 50, 52, 53, 56, 59, 73, 74, 75, 82, 87, 88, 89, 93, 97, 98, 99, 101, 102, 103, 104, 105, 107, 111, 115, 116, 129, 133, 134, 135, and 136 (Fig. 5). For the structures, see the ESI.†
Fig. 2. Distribution of the origin of investigated 218 coumarins from different plant families.
Fig. 3. Different antiviral activities of the investigated 80 coumarins.
Fig. 4. BOILED-Egg diagram for the antiviral compounds AV-1 to AV-80 (top AV-1 to AV-40, bottom AV-41 to AV-80). Blue dots represent molecules that are predicted to be substrates of P-glycoprotein, while red dots stand for molecules that are not anticipated to be active.
Fig. 5. BOILED-Egg diagram of general coumarin compounds C-1 to C-138 (top C-1 to C-70, bottom C-71 to C-138). Blue dots stand for molecules that are predicted to be substrates of P-glycoprotein while red dots represent molecules that are not expected to be substrates.
The next step was the investigation of potential effects of these compounds against COVID-19-related targets. Using docking studies, four SARS-CoV-2 targets were investigated. These targets included Mpro, viral methyltransferase (nsp16/10 complex), RBD of S-protein and human angiotensin-converting enzyme 2 ACE2, which is the human receptor for viral S-protein. The Protein Data Bank (PDB) codes for these proteins are 6LU7, 6W4H, 6M0J, and 6VW1, respectively. After the preparation of protein targets and ligands, the compounds were docked in the active sites of the four targets. Initially, the co-crystallized ligand, if available, was redocked in the active site to validate the docking procedure and then the RMSD between the docked and the co-crystallized ligand was calculated using the DockRMSD server.24 Docking of test compounds was done after RMSD was found to reach an accepted value (<2 Å). The results of the test compounds are shown in Table 1. All results better than −8.0 kcal mol−1 were shown in bold and the top-ranked hits are shown in italics.
Results of compounds docking in SARS-CoV-targets. Compounds with better docking scores than the co-crystallized ligand are shown in bold. The best compounds are shown in italic and presented in the images.
| # | 6LU7 | 6W4H, nsp16 | 6VW1 | 6M0J |
|---|---|---|---|---|
| No. | Mpro | Methyl-transferase | Human ACE2 | RBD-COV-2-S protein |
| AV-4 | −6.9 | −7.4 | −5.9 | −6.2 |
| AV-5 | −8.4 | −7.6 | −6.5 | −6.7 |
| AV-7 | −7.5 | −8.2 | −7.0 | −6.7 |
| AV-9 | −6.6 | −8.1 | −5.9 | −6.4 |
| AV-10 | −6.8 | −7.9 | −5.9 | −6.3 |
| AV-11 | −7.2 | −8.1 | −7.2 | −6.6 |
| AV-16 | −7.5 | −8.8 | −6.1 | −6.9 |
| AV-18 | −7.6 | −7.4 | −6.7 | −5.6 |
| AV-20 | −7.5 | −7.8 | −6.1 | −6.6 |
| AV-21 | −7.7 | −7.1 | −5.9 | −6.3 |
| AV-25 | −7.1 | −7.8 | −6.1 | −6.0 |
| AV-29 | −7.5 | −9.2 | −6.4 | −6.7 |
| AV-30 | −7.6 | −7.8 | −6.9 | −6.8 |
| AV-32 | −8.2 | −7.2 | −5.7 | −6.6 |
| AV-33 | −7.9 | −7.2 | −5.7 | −6.9 |
| AV-34 | −8.0 | −7.1 | −5.6 | −6.9 |
| AV-35 | −7.0 | −7.6 | −8.0 | −6.2 |
| AV-36 | −6.2 | −6.4 | −5.7 | −5.9 |
| AV-37 | −8.2 | −7.3 | −5.4 | −7.3 |
| AV-38 | −7.8 | −7.2 | −5.2 | −6.5 |
| AV-41 | −7.6 | −7.8 | −5.9 | −6.7 |
| AV-52 | −8.5 | −8.8 | −6.6 | −7.2 |
| AV-53 | −8.8 | −7.9 | −6.8 | −7.3 |
| AV-64 | −6.8 | −7.8 | −6.1 | −6.2 |
| AV-65 | −6.7 | −7.3 | −6.4 | −5.9 |
| AV-66 | −7.6 | −8.3 | −6.7 | −6.7 |
| AV-67 | −7.9 | −8.3 | −6.4 | −6.8 |
| AV-78 | −7.0 | −7.5 | −5.7 | −6.1 |
| C-6 | −6.4 | −8.0 | −5.8 | −5.8 |
| C-7 | −7.7 | −7.6 | −6.2 | −6.4 |
| C-8 | −5.9 | −6.6 | −5.8 | −5.4 |
| C-9 | −7.4 | −7.6 | −5.7 | −6.1 |
| C-11 | −7.9 | −7.8 | −6.2 | −6.5 |
| C-21 | −7.1 | −7.6 | −5.7 | −6.6 |
| C-33 | −8.1 | −9.1 | −6.6 | −7.2 |
| C-34 | −7.5 | −8.8 | −6.2 | −6.7 |
| C-36 | −7.6 | −8.9 | −6.5 | −7.0 |
| C-37 | −7.4 | −8.4 | −7.4 | −6.5 |
| C-40 | −8.1 | −9.0 | −6.8 | −7.1 |
| C-44 | −7.8 | −8.8 | −6.5 | −6.9 |
| C-45 | −7.4 | −9.1 | −6.8 | −7.1 |
| C-47 | −7.7 | −9.0 | −7.7 | −6.9 |
| C-48 | −7.8 | −9.1 | −7.2 | −7.0 |
| C-49 | −8.4 | −9.8 | −6.6 | −8.2 |
| C-50 | −7.3 | −8.0 | −7.9 | −6.5 |
| C-52 | −8.5 | −9.1 | −6.7 | −6.9 |
| C-53 | −7.8 | −8.5 | −8.0 | −6.8 |
| C-56 | −8.4 | −8.2 | −5.5 | −7.1 |
| C-59 | −6.5 | −6.9 | −5.8 | −5.9 |
| C-73 | −6.9 | −7.5 | −5.4 | −6.0 |
| C-74 | −8.4 | −8.2 | −5.6 | −6.4 |
| C-75 | −8.3 | −8.2 | −5.2 | −6.2 |
| C-82 | −7.0 | −7.1 | −5.9 | −5.8 |
| C-87 | −8.2 | −7.9 | −6.4 | −6.6 |
| C-88 | −8.7 | −8.1 | −6.2 | −6.6 |
| C-89 | −8.2 | −8.2 | −5.1 | −6.2 |
| C-93 | −8.2 | −8.5 | −6.2 | −7.1 |
| C-97 | −8.7 | −9.1 | −6.9 | −6.8 |
| C-98 | −8.0 | −8.5 | −6.8 | −6.8 |
| C-99 | −7.4 | −7.7 | −5.9 | −6.6 |
| C-101 | −8.4 | −8.7 | −6.2 | −6.8 |
| C-102 | −8.4 | −8.0 | −6.6 | −7.5 |
| C-103 | −7.5 | −8.2 | −7.3 | −6.8 |
| C-104 | −8.3 | −8.5 | −6.3 | −6.8 |
| C-105 | −7.6 | −7.6 | −6.1 | −6.2 |
| C-107 | −6.5 | −6.4 | −6.3 | −5.6 |
| C-111 | −6.4 | −7.2 | −6.9 | −6.1 |
| C-115 | −6.8 | −7.5 | −6.9 | −6.2 |
| C-116 | −6.7 | −7.5 | −7.0 | −6.1 |
| C-129 | −6.8 | −7.2 | −6.7 | −6.0 |
| C-133 | −7.3 | −8.2 | −7.3 | −6.3 |
| C-134 | −6.1 | −6.5 | −5.9 | −5.7 |
| C-135 | −6.7 | −7.2 | −5.2 | −5.5 |
| C-136 | −6.5 | −7.0 | −6.5 | −5.7 |
| −7.9 (N3) | −8.2 (SAM) | — | — |
In general, the best docking scores (reaching −9.8 kcal mol−1) were seen with SARS-CoV-2 methyltransferase. Several of the compounds investigated here displayed better docking scores than the co-crystallized ligand for both the viral main protease and the methyltransferase. The other two targets, on the contrary, showed a lower number of hits and also lower docking scores.
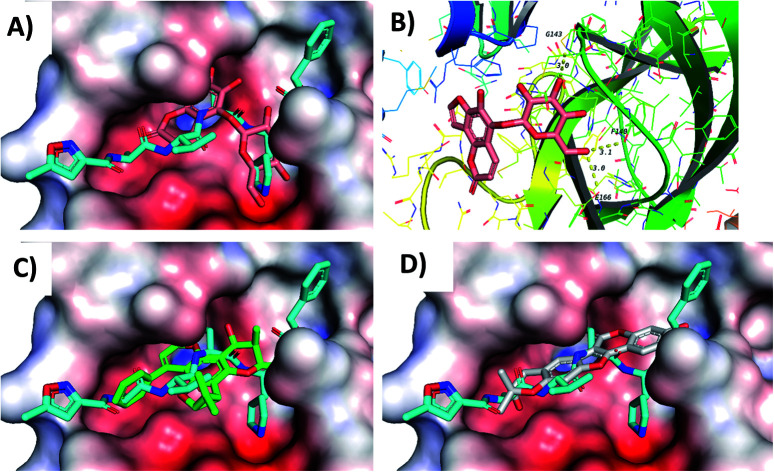
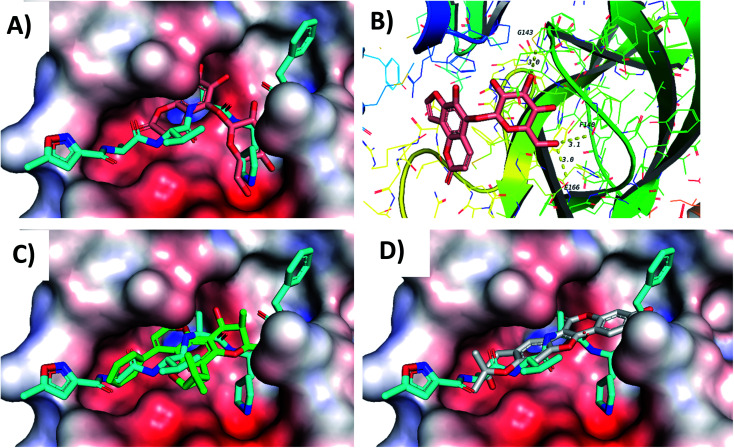
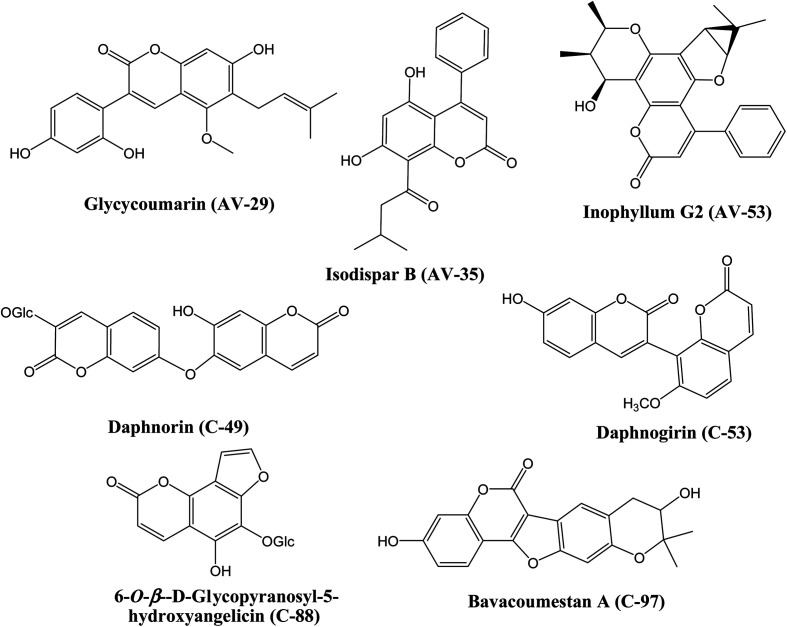
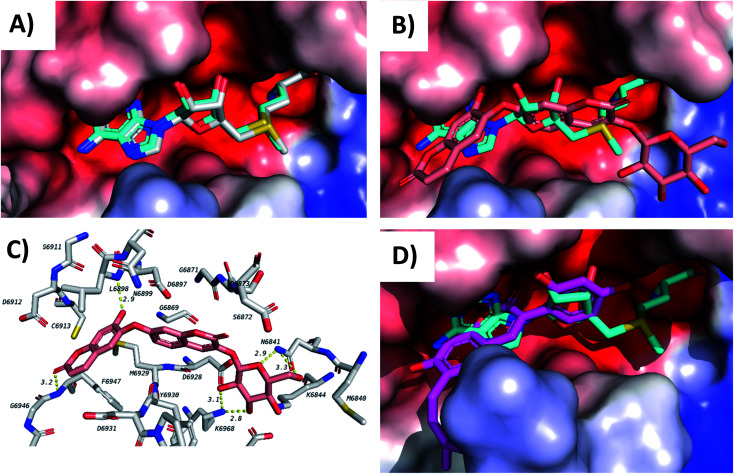
Screening of the compounds against the viral main protease picked 22 structures with docking scores better than that of the co-crystallized ligand N3 (Table 1). Validation of the docking procedure for Mpro was reported earlier38,39. Among these compounds, the top three representatives were 6-O-β-d-glucopyranosyl-5-hydroxyangelicin (C-88), inophyllum G2 (AV-53) and bavacoumestan A (C-97) (Fig. 7), which showed docking scores between −8.7 and −8.8 kcal mol−1. All the three compounds were able to fit in the same active site as N3 (Fig. 7A, C, and D). The top-ranked compound, 6-O-β-d-glucopyranosyl-5-hydroxyangelicin (C-88), was picked as a representative example; its specific interactions with the viral main protease are shown in Fig. 7B.
Fig. 7. Docking results of compounds in Mpro (6LU7). (A) C-88 (salmon) overlapped with N3 (blue, co-crystallized ligand) in the active site of the main protease. (B) Interactions of C-88. (C) AV-53 (green) overlapped with N3 (blue). (D) C-97 (gray) overlapped with N3 (blue).
Docking of the compounds in the active site of the viral methyltransferase showed the highest docking scores among all the tested targets (Table 1). Here the top compounds were daphnorin (C-49) and glycycoumarin (AV-29) (Fig. 6), which had docking scores of −9.8 and −9.2 kcal mol−1, respectively. Redocking of the internal ligand (S-adenosyl-l-methionine, SAM) was able to predict the binding pose with a very high accuracy (Fig. 8A). Daphnorin (C-49) was docked in the same position as SAM. The terminal coumarin ring overlapped with the purine ring of SAM, while the central coumarin ring overlapped with the SAM sugar moiety (Fig. 8B). In addition, daphnorin (C-49) was able to form several hydrogen bonds similar to those formed by SAM, along with extra hydrogen bonds. The interactions of daphnorin (C-49) with amino acids in the active site of the viral methyltransferase (6W4H) are shown in Fig. 8C. Another example of compounds with high affinity is glycycoumarin (AV-29), which is displayed in Fig. 8D, where the coumarin ring is also overlapped with the purine ring of SAM.
Fig. 6. The structures of natural coumarin compounds with top docking results in SARS-CoV-2 targets. This review presents structures as shown in the primary literature, even if sometimes without stereochemical details (like, e.g., for compound C-97).
Fig. 8. Docking results in the viral methyltransferase. (A) Validation (docked gray, crystallized blue). (B) C-49 (salmon) overlapped with SAM (blue) (C) C-49 interactions. (D) AV-29 (pink) overlapped with SAM (blue).
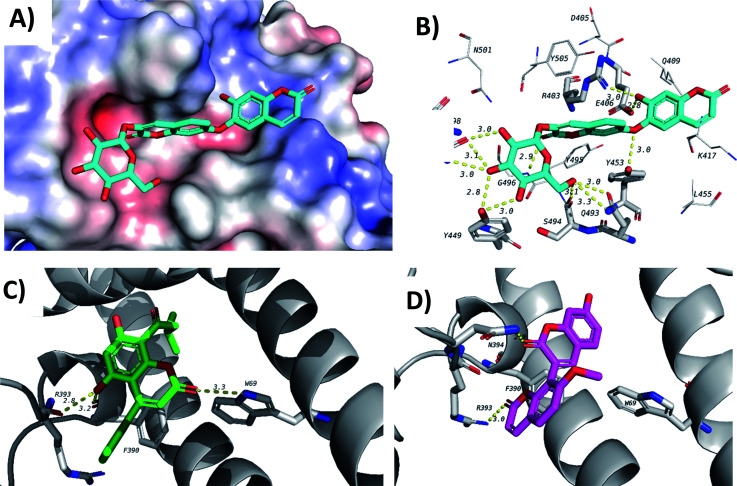
Finally, docking in the active site of viral S-protein and human ACE2 was more selective. Several compounds have shown low docking scores and only limited number of compounds have displayed docking scores above −8.0 kcal mol−1. With respect to RBD of S-protein (6M0J), daphnorin (C-49) was found to be the best-docked compound with a docking score of −8.2 kcal mol−1. The compound was found to bind at the surface that should interact with human ACE2 (Fig. 9A). Its sugar part forms two hydrogen bonds with Q493, which is known to be important in the interaction with ACE2,40 suggesting its potential role in preventing the recognition between S-protein and ACE2 (Fig. 9B). RBD of S-protein bound to ACE2 (6M0J) was found to align well with SARS-CoV-2 S trimer (7DK3) with RMSD of 1.47 suggesting similar interactions. Docking in human ACE2 provided similar results with only two compounds with a score of −8.0 kcal mol−1, which include isodispar B (AV-35) and daphnogirin (C-53). Both molecules were found to bind internally away from the interacting surface between S-protein and ACE2. Binding interaction of both compounds are shown in Fig. 9A and D.
Fig. 9. Docking results in the active sites of viral S-protein and human ACE2. C-49 (blue) in the active site of S-protein represented as a surface (A) and its interactions with amino acids in the active site (B). (C) Interactions of AV-35 (green) in the active site of human ACE. (D) Interactions of C-53 (pink) in the active site of human ACE.
It worth to mention that docking in compounds was proceeded by preparation steps which include stripping of water molecules. This step is essential to evacuate the active site for the docking step. Effect of water molecules on binding could be further investigated using other techniques such as molecular dynamics which is beyond the scope of this article.
3. Conclusions
COVID-19 is the causative agent of a global pandemic that challenges the economic, medical, and public health worldwide. Finding out known natural drugs that act as inhibitors of COVID-19 is a challenging task. In this study, we have screened several coumarins with and without reported antiviral activity against four different SARS-CoV-2 targets. 6-O-β-d-Glucopyranosyl-5-hydroxyangelicin (C-88) and bavacoumestan A (C-97) were among the compounds with the best docking score in the active site of main protease. The compounds were also tested against the viral methyl transferase, where glycycoumarin (AV-29) and daphnorin (C-49) showed the best results. Daphnorin also displayed good results when docked in the active site of RBD-COV-2-S protein when compared to other tested compounds. Finally, inophyllum G2 (AV-35) was the best coumarin against human ACE2. Further in vitro and in vivo testing of the natural coumarin candidates retrieved from this study is highly recommended as a promising starting point for the rapid development of drug leads against the newly emerged viral pathogen COVID-19.
Abbreviations
- ACE2
Angiotensin-converting enzyme 2
- ADME
Absorption, distribution, metabolism, and excretion
- BBB
Blood–brain barrier
- BOILED-Egg
Brain Or IntestinaL EstimateD permeation method
- Mpro
SARS-CoV-2 main protease
- Nsp
Non-structural protein
- PDB
Protein Data Bank
- RBD
Recognition binding domain
- RdRp
RNA-dependent RNA polymerase
- RMSD
Root-mean-square deviation
- SAM
S-Adenosyl-l-methionine
- TMPRSS2
Type II transmembrane serine protease
Funding
This publication has received no external funding.
Author contributions
Conceptualization, U. R. A., S. F. F., and G. B.; methodology, A. B. and B. S. A.; data curation, A. B., B. S. A., S. A. L. B., I. S. A. K., and L. G. M.; original draft preparation, A. B., B. S. A., S. A. L. B., I. S. A. K., and L. G. M.; writing, review, and editing, all authors. All authors have read and agreed to the published version of the manuscript.
Conflicts of interest
The authors declare no conflict of interest.
Supplementary Material
Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra01989a
References
- Acter T. Uddin N. Das J. Akhter A. Choudhury T. R. Kim S. Evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as coronavirus disease 2019 (COVID-19) pandemic: a global health emergency. Sci. Total Environ. 2020;730:138996. doi: 10.1016/j.scitotenv.2020.138996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Núñez-Delgado A. What do we know about the SARS-CoV-2 coronavirus in the environment? Sci. Total Environ. 2020;727:138647. doi: 10.1016/j.scitotenv.2020.138647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellingiri B. Jayaramayya K. Iyer M. Narayanasamy A. Govindasamy V. Giridharan B. Ganesan S. Venugopal A. Venkatesan D. Ganesan H. Rajagopalan K. Rahman P. K. S. M. Cho S. Kumar N. S. Subramaniam M. D. COVID-19: a promising cure for the global panic. Sci. Total Environ. 2020;725:138277. doi: 10.1016/j.scitotenv.2020.138277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarei A. Fardood S. T. Moradnia F. Ramazani A. A review on coronavirus family persistency and considerations of novel type, COVID-19 features. Eurasian Chem. Commun. 2020;2:798–811. doi: 10.33945/SAMI/ECC.2020.7.7. [DOI] [Google Scholar]
- Orhan I. E. Deniz F. S. Natural products as potential leads against coronaviruses: could they be encouraging structural models against SARS-CoV-2? Nat. Prod. Bioprospect. 2020;10:171–186. doi: 10.1007/s13659-020-00250-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani J. S. Johnson J. B. Steel J. C. Broszczak D. A. Neilsen P. M. Walsh K. B. Naiker M. Natural product-derived phytochemicals as potential agents against coronaviruses: a review. Virus Res. 2020;284:197989. doi: 10.1016/j.virusres.2020.197989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman N. Basharat Z. Yousuf M. Castaldo G. Rastrelli L. Khan H. Virtual screening of natural products against type II transmembrane serine protease (TMPRSS2), the priming agent of coronavirus 2 (SARS-CoV-2) Molecules. 2020;25:2271–2283. doi: 10.3390/molecules25102271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile D. Patamia V. Scala A. Sciortino M. T. Piperno A. Rescifina A. Putative inhibitors of SARS-CoV-2 main protease from a library of marine natural products: a virtual screening and molecular modeling study. Mar. Drugs. 2020;18:225. doi: 10.3390/md18040225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaiyan A. Ravichandran K. Singh S. V. Sankar M. Thomas P. Dhama K. Malik Y. S. Singh R. K. Chaudhuri P. In silico molecular docking analysis targeting SARS-CoV-2 spike protein and selected herbal constituents. J. Pure Appl. Microbiol. 2020;14:989–998. [Google Scholar]
- Sayed A. M. Khattab A. K. AboulMagd A. M. Hassan H. M. Rateb M. E. Zaidg H. Abdelmohsen U. R. Nature as a treasure trove of potential anti-SARS-CoV drug leads: a structural/mechanistic rationale. RSC Adv. 2020;10:19790. doi: 10.1039/D0RA04199H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barot K. P. Jain S. V. Kremer L. Singh S. Manjunath D. Ghate M. D. Recent advances and therapeutic journey of coumarins: current status and perspectives. Med. Chem. Res. 2015;24:2771–2798. doi: 10.1007/s00044-015-1350-8. [DOI] [Google Scholar]
- Kubrak T. Podgórski R. Stompor M. Natural and synthetic coumarins and their pharmacological activity. Eur. J. Clin. Exp. Med. 2017;15:169–175. doi: 10.15584/ejcem.2017.2.12. [DOI] [Google Scholar]
- Venugopala K. N. Rashmi V. Odhav B. Review on natural coumarin lead compounds for their pharmacological activity. BioMed Res. Int. 2013:963248. doi: 10.1155/2013/963248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan M. Z. Osman H. Ali M. A. Ahsan M. J. Therapeutic potential of coumarins as antiviral agents. Eur. J. Med. Chem. 2016;123:236–255. doi: 10.1016/j.ejmech.2016.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S. K. Pharmacological Potentiality of Coumarins as Anti-Viral Agent. Int. J. Res. Anal. Rev. 2018;5:281–291. [Google Scholar]
- Lyndem S. Sarmah S. Das S. Roy A. S. In silico screening of naturally occurring coumarin derivatives for the inhibition of the main protease of SARS-CoV-2. ChemRxiv. 2020 [Google Scholar]
- Mishra S. Pandey A. Manvat S. Coumarin: an emerging antiviral agent. Heliyon. 2020;6:e03217. doi: 10.1016/j.heliyon.2020.e03217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissanayake K. G. C. Perera W. P. R. T. Medicinal importance of Ferula asafetida oligogum resins against infective diseases. J. Med. Plants Stud. 2020;8:135–139. [Google Scholar]
- Venugopala K. N. Rashmi V. Odhav B. Review on natural coumarin lead compounds for their pharmacological activity. BioMed Res. Int. 2013:963248. doi: 10.1155/2013/963248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokoohinia Y. Sajjadi S.-E. Gholamzadeh S. Fattahi A. Behbahani M. Antiviral and cytotoxic evaluation of coumarins from Prangos ferulacea. Pharm. Biol. 2014;52:1543–1549. doi: 10.3109/13880209.2014.907322. [DOI] [PubMed] [Google Scholar]
- Bedoya L. M. Beltran M. Sancho R. Olmedo D. A. Sanchez-Palomino S. Olmo D. E. Lopez-Perez J. L. Munoz E. San A. F. Alcami J. 4-Phenylcoumarins as HIV transcription inhibitors. Bioorg. Med. Chem. Lett. 2005;15:4447–4450. doi: 10.1016/j.bmcl.2005.07.041. [DOI] [PubMed] [Google Scholar]
- Nahar L. Talukdar A. D. Nath D. Nath S. Mehan A. Ismail F. M. D. Sarker S. D. Naturally occurring calanolides: occurrence, biosynthesis, and pharmacological properties including therapeutic potential. Molecules. 2020;25:4983–5006. doi: 10.3390/molecules25214983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostova I. Mojzis J. Biologically active coumarins as inhibitor of HIV-1. Future HIV Ther. 2007;1:315–329. doi: 10.2217/17469600.1.3.315. [DOI] [Google Scholar]
- Patil A. D. Freyer A. J. Eggleston D. S. Haltiwanger R. C. Bean M. F. Taylor P. B. Caranfa M. J. Breen A. L. Bartus H. R. Johnson R. K. Hertzberg R. P. Westley J. W. The inophyllums, novel inhibitors of HIV-1 reverse transcriptase isolated from the Malaysian tree, Calophyllum inophyllum Linn. J. Med. Chem. 1993;36:4131–4138. doi: 10.1021/jm00078a001. [DOI] [PubMed] [Google Scholar]
- Silveira C. S. Martins F. O. Romanos M. T. V. Costa C. S. Kaplan M. A. C. Menezes F. S. In vitro cytotoxic, antioxidant and antiviral effects of Pterocaulon alopecuroides and Bidens segetum extracts. Rev. Bras. Farmacogn. 2009;19:343–348. doi: 10.1590/S0102-695X2009000300001. [DOI] [Google Scholar]
- Houghton P. J. Woldemariam T. Z. Khan A. I. Burke A. Mahmood N. A. Antiviral activity of natural and semi-synthetic chromone alkaloids. Antiviral Res. 1994;25:235–244. doi: 10.1016/0166-3542(94)90006-X. [DOI] [PubMed] [Google Scholar]
- Nonaka G. Nishioka I. Nishizawa M. Yamagishi T. Kashiwada Y. Dutschman G. E. Bodner A. J. Kilkuskie R. E. Cheng Y. Lee K. Anti-AIDS agents, 2. Inhibitory effects of tannins on HIV reverse transcriptase and HIV replication in H9 lymphocyte cells. J. Nat. Prod. 1990;53:587–595. doi: 10.1021/np50069a008. [DOI] [PubMed] [Google Scholar]
- Reusser F., Bannister B., Tarpley W. G., Althaus I. W. and Zapotocky B., Rubradirin derivatives for treatment of HIV infection, WO1988008707A3, 1988
- Barkat M. A. Rizwanullah M. Naim J. Pottoo F. H. Kumar R. Phytoconstituents as potential anti-HIV agents: a systematic review. Int. J. Biomed. Res. 2014;5:299–313. [Google Scholar]
- Rajtar B. Skalicka-Woźniak K. Świątek Ł. Stec A. Boguszewska A. Polz-Dacewicz M. Antiviral effect of compounds derived from Angelica archangelica L. on Herpes simplex virus-1 and Coxsackievirus B3 infections. Food Chem. Toxicol. 2017;109:1026–1031. doi: 10.1016/j.fct.2017.05.011. [DOI] [PubMed] [Google Scholar]
- Fortin H. Tomsai S. Jaccard P. Robin V. Boustie J. A. Prenyloxycoumarin from Psiadia dentate. Chem. Pharm. Bull. 2001;49:619–621. doi: 10.1248/cpb.49.619. [DOI] [PubMed] [Google Scholar]
- Barnard D. L. Xu Z.-Q. Stowell V. D. Yuan H. Smee D. F. Samy R. Sidwell R. W. Nielsen M. K. Sun L. Cao H. Li A. Quint C. Deignan J. Crabb J. Flavin M. T. Coumarins and pyranocoumarins, potential novel pharmacophores for inhibition of measles virus replication. Antiviral Chem. Chemother. 2002;13:39–59. doi: 10.1177/095632020201300104. [DOI] [PubMed] [Google Scholar]
- Daina A. Michielin O. Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina A. Zoete V. A BOILED-Egg to predict gastrointestinal absorption and brain penetration of small molecules. ChemMedChem. 2016;11:1117–1121. doi: 10.1002/cmdc.201600182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanwell M. D. Curtis D. E. Lonie D. C. Vandermeersch T. Zurek E. Hutchison G. R. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J. Cheminf. 2012;4:17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O. Olson A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell E. W. Zhang Y. DockRMSD: an open-source tool for atom mapping and RMSD calculation of symmetric molecules through graph isomorphism. J. Cheminf. 2019;11:40. doi: 10.1186/s13321-019-0362-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahran E. M. Albohy A. Khalil A. Ibrahim A. H. Ahmed H. A. El-Hossary E. M. Bringmann G. Abdelmohsen U. R. Bioactivity Potential of Marine Natural Products from Scleractinia-Associated Microbes and In Silico Anti-SARS-COV-2 Evaluation. Mar. Drugs. 2020;18:645. doi: 10.3390/md18120645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamad S. A. Zahran E. M. Abdel Fadeel M. R. Albohy A. Safwat M. A. New Acaciin-Loaded Self-Assembled Nanofibers as MPro Inhibitors Against BCV as a Surrogate Model for SARS-CoV-2. Int. J. Nanomed. 2021;16:1789–1804. doi: 10.2147/IJN.S298900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J. Ye G. Shi K. Wan Y. Luo C. Aihara H. Geng Q. Auerbach A. Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.