Abstract
Background
Inhibition of the sonic hedgehog (SHH) pathway with Smoothened (SMO) inhibitors is a promising treatment strategy in SHH-activated medulloblastoma, especially in adult patients. However, the problem is that tumors frequently acquire resistance to the treatment. To understand the underlying resistance mechanisms and to find ways to overcome the resistance, preclinical models that became resistant to SMO inhibition are needed.
Methods
To induce SMO inhibitor resistant tumors, we have treated a patient-derived xenograft (PDX) model of SHH medulloblastoma, sensitive to SMO inhibition, with 20 mg/kg Sonidegib using an intermitted treatment schedule. Vehicle-treated and resistant models were subjected to whole-genome and RNA sequencing for molecular characterization and target engagement. In vitro drug screens (76 drugs) were performed using Sonidegib-sensitive and -resistant lines to find other drugs to target the resistant lines. One of the top hits was then validated in vivo.
Results
Nine independent Sonidegib-resistant PDX lines were generated. Molecular characterization of the resistant models showed that eight models developed missense mutations in SMO and one gained an inactivating point mutation in MEGF8, which acts downstream of SMO as a repressor in the SHH pathway. The in vitro drug screen with Sonidegib-sensitive and -resistant lines identified good efficacy for Selinexor in the resistant line. Indeed, in vivo treatment with Selinexor revealed that it is more effective in resistant than in sensitive models.
Conclusions
We report the first human SMO inhibitor resistant medulloblastoma PDX models, which can be used for further preclinical experiments to develop the best strategies to overcome the resistance to SMO inhibitors in patients.
Keywords: resistance, Selinexor, SHH Medulloblastoma, SMO inhibitor, Sonidegib
Key Points.
Intermittent treatment with Sonidegib induces resistance in human SHH medulloblastoma PDX model.
Acquired resistance mainly due to missense mutations in SMO.
Detected mutations overlapping with mutations known of patients’ samples.
Importance of the Study.
Medulloblastoma and basal cell carcinoma with upstream activation of the SHH pathway can be treated with SMO inhibitors. Initial clinical responses are promising, but tumors often develop resistance to the treatment. To overcome resistance and evaluate new treatment strategies appropriate preclinical models reflecting the patients’ situation are needed. Previously, genetically engineered mouse models with Ptch1/Tp53 mutations were used to model resistance. However, TP53 mutations almost never co-occur with PTCH1 mutations in human medulloblastoma. Here we report about an orthotopic PTCH1-mutated PDX model of medulloblastoma that was treated in cycles with Sonidegib to develop resistant lines. Genomic analyses of the resistant sub-lines showed re-activation of the SHH pathway by mutations that have also been found in patients’ tumors. The models generated here are a good resource for translational research to improve treatment strategies for medulloblastoma patients that are resistant to SMO inhibitors.
Introduction
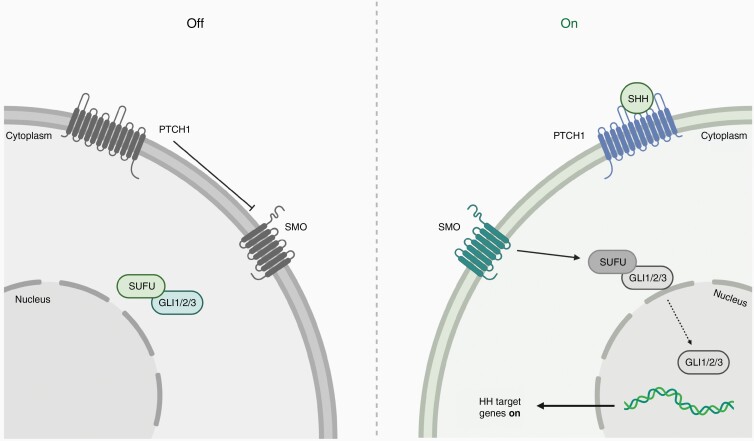
The hedgehog (HH) signaling pathway is a highly conserved pathway that was first identified in fruit flies.1 It transmits signals from the cell membrane to the nucleus and plays an important role during normal embryonic development but is mostly inactive in adult tissues.2 Nevertheless, it is involved in maintenance of somatic stem cells and pluripotent cells and plays a crucial role for tissue repair of different cell types including epithelial cells.3,4 In other tissues, the HH pathway is active in primary cilia, which receive mechanical, chemical, and thermal signals.5 The HH genes Sonic HH (SHH), Indian HH (IHH), and Desert HH (DHH) are ligands of the pathway and are relevant for polarity of organisms.6 In the inactive state of the pathway, patched 1 (PTCH1), a transmembrane protein, inhibits the seven-pass transmembrane protein Smoothened (SMO) and Suppressor of fused (SUFU) is bound to glioma-associated oncogene (GLI) in the cytoplasm (Figure 1). If one of the ligands, such as SHH, binds to PTCH1, the transmembrane protein undergoes internalization and degradation, and SMO gets activated and is released to enter the primary cilia. This promotes the dissociation of the SUFU-GLI complex and detached GLI is translocated to the nucleus where it acts as a transcription factor activating the expression of HH target genes such as GLI1/2 (in a feed-forward loop) and MYCN as well as PTCH1 that acts as a negative feedback loop regulator.7
Figure 1.
Schematic overview of the SHH signaling pathway. In inactivated status PTCH1 inhibits SMO and SUFU is bound to GLI1, GLI2 or GLI3 in cytoplasm (left panel); if SHH (ligand) binds to PTCH1 transmembranal SMO gets activated; GLI1/2/3 is released and translocted to nucleus where it functions as transcription factor (right panel); Adapted from “Hedgehog Signaling Pathway”, by BioRender.com (2021). Retrieved from https://app.biorender.com/biorender-templates.
Dysfunction or aberrant activation of the HH pathway can lead to developmental problems such as in patients with Gorlin syndrome who have germline mutations in HH pathway members PTCH1 or SUFU.8,9 HH signaling can also be activated in many different adult tumor types, including basal cell carcinoma (BCC) and brain tumors.10–13 In children, tumor entities that are well known for activated HH signaling are rhabdomyosarcoma and medulloblastoma (MB).7,14–16 Especially in MB, where activated SHH signaling is characteristic for the SHH subgroup, representing ~30% of all MB patients, the pathway has been intensively studied and many mutations affecting different pathway members have been identified in large genomic studies.17–19
Since aberrant activation of the HH pathway is the driving force in many different tumor types, inhibiting the pathway for therapy is a promising strategy and many different drugs have been developed.20 Most of the drugs that are on the market and approved target SMO.21 Early preclinical and clinical experiences have shown that these SMO inhibitors were highly effective in BCC and SHH MB tumors with aberrant activation of the HH pathway upstream of SMO, e.g. harboring a mutation in PTCH1.17,22,23 Tumors with mutations downstream of SMO, for instance in SUFU or with amplification of MYCN/GLI2 were intrinsically resistant to the treatment.17,24,25 Moreover, in contrast to the assumption stated in Kool et al. 2014, tumors harboring a mutation in SMO are also primarily resistant to the treatment with SMO inhibitors as the inhibitors cannot bind anymore.17,26
Even though SMO inhibitors were highly effective in preclinical and clinical studies, side effects such as premature closure of growth plates in young children are a major concern, thereby restricting the use of these inhibitors to post-pubertal and adult patients only.24 The other concern is that tumors frequently become resistant against these inhibitors. For instance, metastatic MB patients treated with Vismodegib rapidly developed resistance caused by mutations in SMO.27 Also BCC tumors treated with Vismodegib showed frequent resistance to SMO inhibition with 50% of resistant cases harboring mutations in SMO but also other mutations downstream of SMO in the SHH pathway were observed.26
Based on clinical data from patients treated with SMO inhibitors and the resistance they developed against these inhibitors there is a high need to either prevent the development of resistance against SMO inhibitors or to overcome resistance and find new treatment strategies for resistant tumors. To develop such new strategies, preclinical models mimicking the patients’ situation are urgently needed. Initial studies used mouse MB models harboring Ptch1 and Tp53 mutations to study the resistance mechanisms against SMO inhibitors. Mouse MB models were treated with either Sonidegib or Vismodegib and resistant tumors developed quickly depending on the applied treatment schedules.28,29 Molecular characterization of the resistant tumors revealed Gli2-amplifications in 50% of the models. Only 7/135 resistant tumors harbored missense point mutations in Smo. However, acquired mutations in SMO is the prevalent resistance mechanism seen in patients.26,30
Even though these mouse MB models provided first insights into the mechanisms that may explain the resistance these tumors developed against inhibition of SMO, they do not fully mimic the human situation. The main difference are the Tp53 mutations in these mouse models as mutations in TP53 almost never co-occur with PTCH1 mutations in human SHH MB patients.18TP53 mutations that are found in SHH MB patients are most prevalent in older children and almost always occur in the context of the Li-Fraumeni syndrome where the first hit in TP53 is already present in the germline. However, in human MB they almost always co-occur with MYCN and/or GLI2 amplifications and these tumors are resistant to SMO inhibitors.17,25 Moreover, the presence of Tp53 mutations in the mouse tumors makes them also more genomically unstable, which may facilitate the development of drug resistance through genomic rearrangements or amplifications. To study resistance mechanisms against inhibition of SMO, a better model mimicking the human situation is needed. Here, we describe the first Sonidegib-resistant SHH MB PDX models.
Material and Methods
Animal Studies
All experiments were conducted in accordance with legal and ethical regulations and approved by the regional council (Regierungspräsidium Karlsruhe, Germany; G259/14, G228/19). Mice were monitored daily for the presence of tumor-related symptoms. Animals were euthanized when termination criteria described in the protocol were observed. All in vivo work was performed with immune-compromised NSG mice (Nod-scid IL2Rgammanull) obtained from the laboratory breeding colony (German Cancer Research Center).
Intracranial injections (analgesia: 5 mg/kg Carprofen or 200 mg/kg Metamizol; depending on used protocol) and bioluminescence imaging were performed as published previously.31 The PDX models need to be propagated in vivo by re-injection and cannot be cultured indefinitely in vitro.
Extracted tumors were split and stored either as viable cryostocks with dissociated cells in medium (NeuroCult + 10% Proliferation Supplement) with 10% DMSO, as snap-frozen samples, or fixed in 10% formalin-solution for embedding in paraffin using standard procedures.
For treatments, animals were randomized into vehicle (0.5% methyl cellulose/0.5% Tween-80) or Sonidegib group (20 mg/kg; p.o.; MedChem Express, HY-16582), or into vehicle (0.6% Plasdone PVP K-29/32 + 0.6% Poloxamer Pluronic F-68 in water) and Selinexor group (5 mg/kg; Karyopharm Therapeutics) respectively, and treated in a five days on/2 days off schedule. The chosen dosings ensure clinically relevant drug exposures as well as good toleration of the mice.32
Histological Stainings
After antigen retrieval with citrate buffer, FFPE sections were blocked (1hr) with 10% normal donkey serum in 0.1% Tween20/PBS. Primary antibody (anti-cleaved Capsase-3, 1:500, #9661, Cell Signaling, USA) was incubated o/n at room temperature. Secondary antibody (anti-rabbit-biotin-SP-conjugated, 1:400, Jackson ImmunoResearch, USA) and ABC-staining (Vectastain Elite ABC Kit, Vector Laboratories, USA) were added and sections were stained with DAB (DAB-2V, Nichirei Bioscience). Staining for Ki67 (clone MIB-1, Dako Agilent, USA) was done on a Ventana BenchMark ULTRA Immunostainer using the OptiView DAB IHC Detection Kit for Ki67 (Ventana Medical Systems, USA). Hematoxylin and Eosin (H&E) staining was performed for 1.5 min and 5 min, respectively.
Methylation and Sequencing Analysis
DNA and RNA isolation, as well as whole-genome, whole-exome, and RNA sequencing, were performed as published previously.31 DNA methylation analysis using the EPIC (850k) BeadChip (Illumina, San Diego, USA) was performed as published previously.33
In Vitro Drug Screen With a Library of 76 Compounds
Cryopreserved cells were taken in short-term culture in TSM complete 34 and seeded in drug-preprinted 384-well round bottom ultra-low attachment spheroid microplates (Corning, #3830; 1000 cells/well; 25 µl TSM complete/well). The drug library consisted of 76 drugs of which most were approved and some were investigative compounds and the plate production was performed as previously published.35 All concentrations were tested in duplicates, readout (after 72 hrs) was done on a PHERAstar FS microplate reader (BMG Labtech) using CellTiterGlo 2.0 (Promega) according to the manufacturer’s protocol. Drug sensitivity analyses were performed using the web-based drug analysis pipeline BREEZE (https://breeze.fimm.fi), developed at the Institute for Molecular Medicine Finnland (FIMM).36
Results
Intermitted Dosing of Sonidegib Induces Resistance in SHH MB PDX
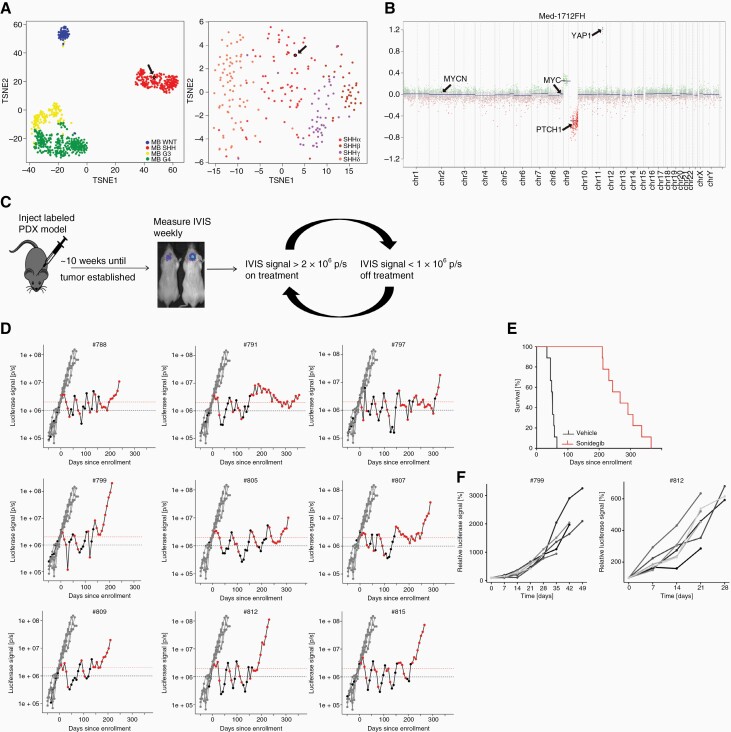
To study SMO inhibitor resistance mechanisms in human MB, we used the patient-derived orthotopic xenograft (PDX) model Med-1712FH.31 Methylation-based TSNE clustering of a reference cohort published in Cavalli et al. 2017 showed that the model Med-1712FH clusters within the SHH subgroup and can be further subtyped as MB-SHH-3 (previously annotated as SHH-α) (Figure 2A).37 Other molecular characteristics of the primary tumor and the model are missense mutations in PTCH1, ELP1, and CREBBP as well as heterozygous loss of chromosome 9q and amplification of YAP1 (Figure 2B). To generate resistant clones of the model we used luciferase-labeled PDX cells of the model intracranially injected in mice. Tumor volume was determined weekly using bioluminescence and mice were randomized into treatment (SMO inhibitor Sonidegib) or vehicle group after a threshold of luciferase signal of 2 x 106 p/s was reached (Figure 2C).
Figure 2.
Intermittent dosing of Sonidegib induces resistant SHH MB sub-lines. (A) Clustering of MB reference cohort published by Cavalli et al. 2017 showing the different subgroups of MB (left; n = 764) and further division of the SHH MB sugroup into four different subtypes (right; n = 224)37; the used model Med-1712FH clusters with the SHH MB group and in more detail as SHH-3 (highlighted by black arrow); (B) Copy number profile of the model Med-1712FH showing loss of chromosome 9q and amplification of 11q including YAP1; (C) PDX cells were injected intracranially and mice were randomized based on IVIS signal; treatment was started when threshold of 2 x 106 p/s was reached and stopped when IVIS signal was below 1 x 106 p/s; (D) Plots showing IVIS signal for time of treatment; each plot shows IVIS signal of vehicle-treated mice in gray (n = 6) and IVIS signals of one mouse in Sonidegib-treatment group; IVIS signals during time on treatment are shown in red, values measured during treatment breaks are shown in black; (E) Kaplan-Meier plot showing survival of vehicle- (n = 6) and Sonidegib-treated (n = 9) group; (F) two generated resistant models (#799, #812) were re-injected after cryopreservation and treated with Sonidegib as soon as IVIS signal reached 1 x 106 p/s; all mice (n = 10) show constant tumor growth indicating resistance to Sonidegib.
To induce resistance over time we went for an intermittent dosing schedule based on tumor burden measured by luciferase intensity. Treatment was stopped when luciferase intensity dropped below 1 x 106 p/s, tumors were allowed to re-grow and treatment was reinitiated when the IVIS signal reached 2 x 106 p/s again (Figure 2C). The treatment in cycles was conducted until we reached a stage that IVIS signals increased under treatment indicating that the tumors were not responding to treatment anymore and mice had to be euthanized because of tumor symptoms. The mice developed resistance to Sonidegib after 3–6 treatment cycles and the intermittent treatment was performed for 30–51 weeks (Figure 2D). The vehicle animals had to be euthanized 4–10 weeks after treatment start and treatment with Sonidegib led to significant survival benefit (Figure 2E). All nine resistant tumors were resected, parts were reserved for molecular analysis and other parts were cryopreserved as single cell suspension to allow re-passaging of the PDX for future preclinical testing.
To confirm that the resistant models can indeed be cryopreserved and used for further experiments, frozen cells of two generated resistant lines Med-1712FH_#812 and Med-1712FH_#799 were thawed and re-injected into 10 NSG mice each. This time treatment was started when a threshold of 1x106 p/s in IVIS was reached. All mice showed constant tumor growth and no effect of Sonidegib could be observed indicating that the tumors were resistant to Sonidegib (Figure 2F).
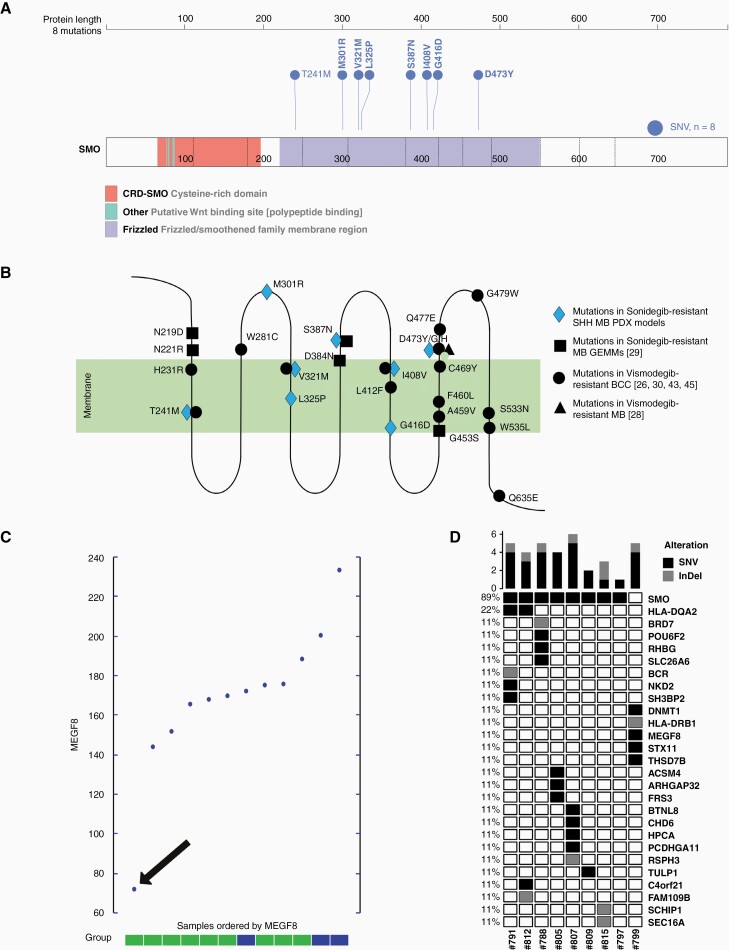
Molecular characterization of resistant models reveals eight different mutations in SMO and one mutation in MEGF8
To determine the mechanism(s) how resistant lines became resistant to Sonidegib treatment all nine lines were subjected to whole-genome sequencing (WGS) and DNA methylation analysis. Cluster analyses of methylation data showed that all nine resistant and the three vehicle-treated lines cluster closely with the original med-1712FH model (Supplementary Figure S1A+B) and they are all classified as MB-SHH-3 (Supplementary Figure S1C). The models form a separate cluster in proximity to the SHH and the SHH-3 clusters, respectively, since the methylation profiles of the original and resistant lines are highly similar. Histological analyses of the original, treatment-naïve, and the resistant models did not show any differences for HE-staining, proliferation (Ki67-staining), and apoptosis (cleaved Caspase-3-staining) (Supplementary Figure S1D). Sequencing data showed that 8/9 resistant lines acquired missense mutations in SMO as compared to the original Med-1712FH_#140 model used as starting material for injections (Figure 3 A+B). All mutations occurred in the proximity of the frizzled heptahelical membrane region of the protein that spans from amino acid 221 to amino acid 558 38 and partly overlapped with previously published mutations, one with a mutation of a GEMM model and four with mutations detected in Vismodegib-resistant MB or BCC patients (Figure 3B). Only one of the resistant lines did not have a mutation in SMO but instead had an inactivating mutation (S1358X) in MEGF8 (multiple epidermal growth factor-like domains protein 8). Expression of MEGF8 is also significantly lower in the MEGF8-mutated model compared to the other models (Figure 3C). MEGF8 is a single-pass type I transmembrane protein that acts as a negative regulator of HH signaling.39 The protein is involved in degradation of SMO and thereby involved in regulating HH signaling. If MEGF8 is mutated, SMO accumulates and HH signaling is elevated.40 Besides the mutations in SMO or MEGF8 each resistant model gained only few additional SNVs (1–5) and InDels (1–2) (Figure 3D). Copy number profiles of the original and the resistant models did not show any differences (data not shown).
Figure 3.
Genomic analysis of generated resistant samples reveals mutations in SMO and mutation in MEGF8. A) protein sequence showing different regions of SMO and eight different mutations discovered in the resistant SHH MB PDX models (visualized by using ProteinPaint; https://proteinpaint.stjude.org/; 46); B) schematic visualization of SMO with cytoplasmic, transmembrane and extracellular areas as well as localizations of the eight mutations detected in the resistant clones indicated in blue; mutations detected in previously published studies are shown in black; C) expression levels of MEGF8 for the nine resistant samples (green) and three vehicle samples (blue); sample with MEGF8-mutation is highlighted with a black arrow; D) Oncoplot showing all SNVs and InDels analyzed in the generated resistant models when compared to the original model used for injection.
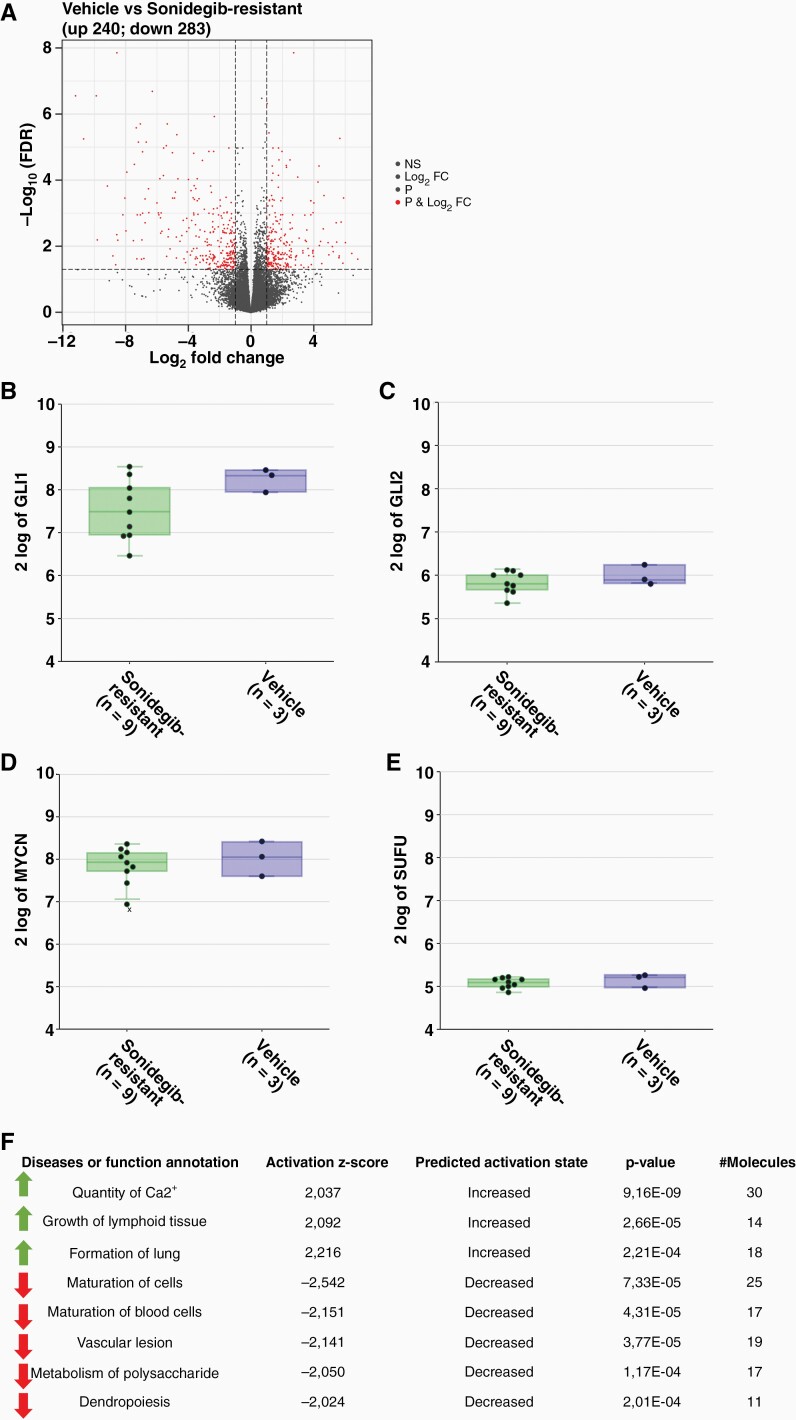
To investigate transcriptional differences between vehicle and resistant lines RNA sequencing of three vehicle samples and nine resistant lines was performed. Analysis revealed that 240 genes were significantly (P-value < .05) up- and 283 genes downregulated in the resistant samples (Figure 4A). The data also showed that GLI1/2 was still expressed and not significantly downregulated in the resistant lines indicating that the HH pathway was still active (Figure 4B+C). This is also depicted by expression of MYCN and SUFU (Figure 4D+E). Ingenuity Pathway Analysis (IPA) of resistant models compared to vehicle samples revealed three increased (activation z-score > 2) and five decreased (activation z-score < –2) pathways (Figure 4F). However, none of these annotated pathways seem to be related to HH signaling, growth of (cancer) cells, or tumorigenesis.
Figure 4.
Differential expression of genes of in resistant models shows re-activation of SHH pathway in resistant samples. A) RNA-seq analysis of vehicle and resistant samples showing up- (240) and downregulated (283) genes; expression levels of GLI1 (B), GLI2 (C), MYCN (D) and SUFU (E) of vehicle-treated (n = 3) and resistant models (n = 9) do not show differences; F) IPA analysis of pathways increased and decreased in resistant samples compared to vehicle samples does not reveal pathways related to (cancer) cell proliferation or tumor growth.
Short-Time Treatment of Models with Sonidegib Reveals Intermediate Response in Gene Expression
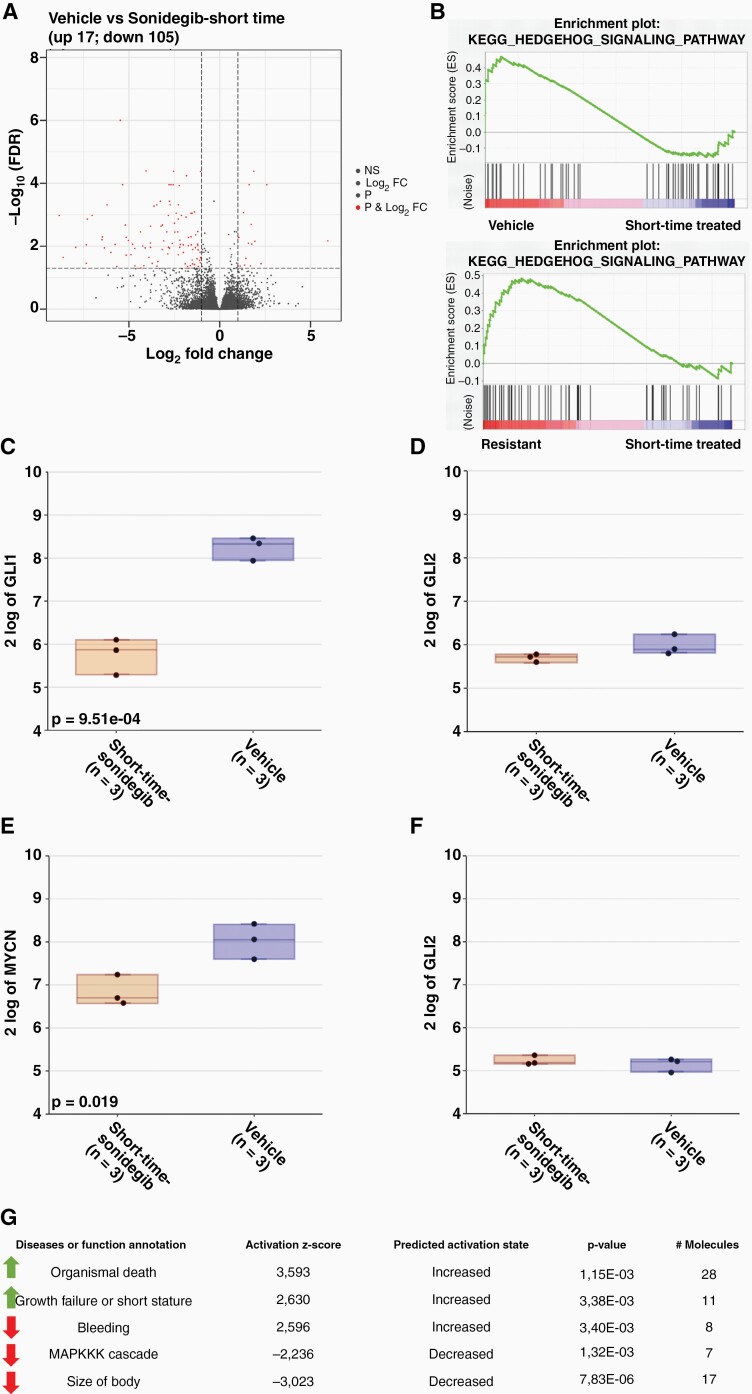
To better understand the initial response of the SHH PDX model to Sonidegib treatment, three vehicle-treated animals were treated with Sonidegib (20 mg/kg) on two consecutive days and euthanized four hours after the second dosing. Tumor cell pellets were subsequently used for RNA-sequencing identifying 105 down- and 17 upregulated genes in the treated samples (P-value < .05) (Figure 5A). GLI1 and MYCN expression were significantly downregulated in short-time treated samples compared to vehicle samples (Figure 5C+E), but expression of SUFU and GLI2 was not altered (Figure 5D+F). Gene set enrichment analysis (GSEA) of the short-time treated samples compared to vehicle but also to resistant samples showed that the KEGG HH signaling pathway was upregulated in vehicle and resistant samples but not in short-time treated samples (Figure 5B), indicating that the HH signaling pathway was inhibited in the short-time treated samples. Pathway analysis with IPA showed that pathways or functions related to organismal death and growth failure were increased (activation z-score > 2) in short-time treated samples. In contrast, pathways related to the MAPKKK cascade and body size were decreased (activation z-score < –2) in short-time treated samples, indicating that cell proliferation and cell division was downregulated (Figure 5G).
Figure 5.
RNA-seq analysis of short-time treated samples confirms target engagement of Sonidegib. A) RNA-seq analysis of Sonidegib (short time) treated samples showing 17 up- and 105 downregulated genes compared to vehicle-treated samples; B) GSEA shows that HH signaling pathway is upregulated in vehicle and resistant samples compared to short-time treated samples; expression levels of GLI1 (C), GLI2 (D), MYCN (E), SUFU (F) show downregulation in short-time treated samples compared to vehicle samples; G) IPA analysis of pathways increased and decreased in short-time treated samples compared to vehicle samples does show upregulation of pathways related to apoptosis and downregulation of the MAPKKK pathway in short-time treated samples.
Selinexor Treatment Prolongs Survival of Resistant Lines
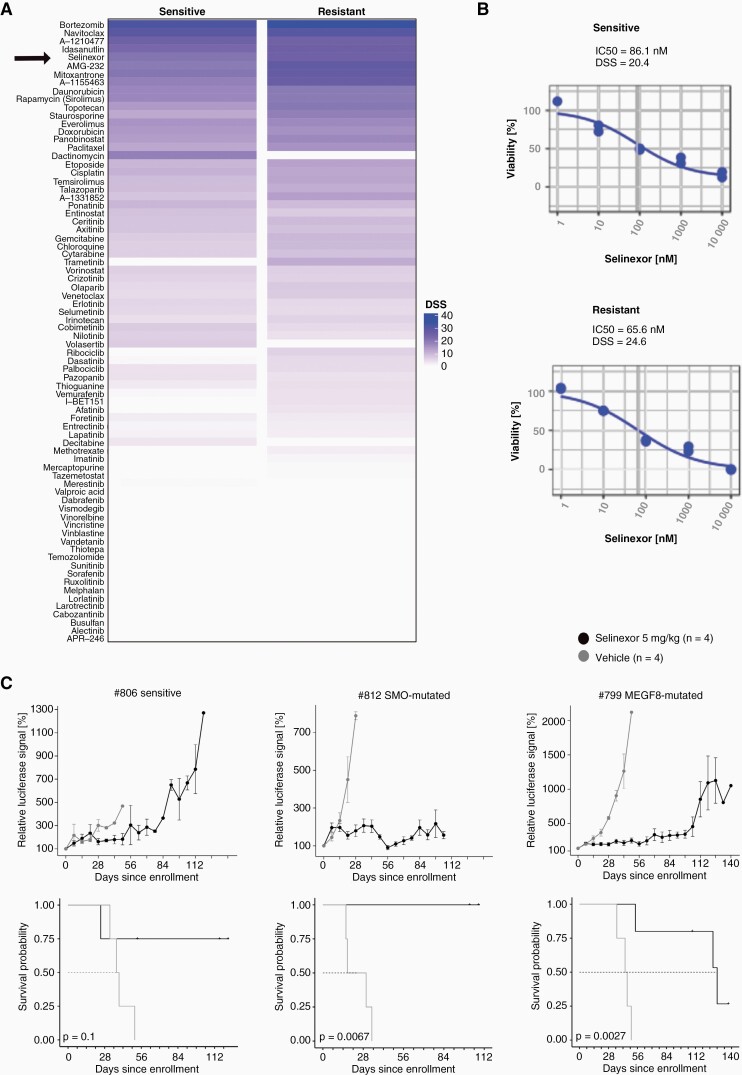
To evaluate new treatment strategies a drug screen with 76 drugs was performed using the Sonidegib-sensitive, treatment-naïve Med-1712FH model, and a resistant model with a SMO mutation (#791). One of the top hits, being effective in both models was Selinexor (Figure 6A), a blood-brain-barrier penetrant XPO1-inhibitor that is approved and in clinical use for multiple myeloma. It inhibits nuclear export of many tumor suppressor proteins as for example TP53 and was already shown to be effective for a MB model with SUFU mutation.41,42 For the in vitro used treatment-naïve sample the IC50 of Selinexor was 86.1 nM, for the resistant model it was 65.6 nM (Figure 6B). To verify efficacy of the XPO1-inhibitor in vivo, cells of one vehicle-treated model (#806) and of two resistant models (#812, SMO-mutated and #799, MEGF8-mutated) were injected intracranially into NSG mice. After bioluminescence signal reached 1x106 p/s, mice were randomized into vehicle or Selinexor group and treated with solvent control or 5 mg/kg Selinexor (p.o.) respectively in a 5 days on/2 days off schedule. Comparison of tumor volumes showed that for the sensitive model treatment with Selinexor did not induce huge differences in tumor volumes between the two groups, however, mice in the treatment group survived 12 weeks longer (Figure 6C). Treatment of both mutated models revealed a significant difference in tumor volumes between the vehicle-treated and Selinexor-treated group and the mice treated with Selinexor also lived significantly longer (Figure 6C). For the SMO-mutated model (#812), tumors treated with Selinexor did grow during the first week of treatment but then stopped growing and disease was stable for five weeks. Between weeks six and eight mice showed tumor regression with slow growing tumors afterwards. Mice with the MEGF8-mutated model (#799) showed stable disease for 13 weeks under treatment and growth of tumor after this time point.
Figure 6.
In vitro and in vivo treatment with Selinexor of sensitive and resistant med-1712FH-models does show higher efficacy in resistant cells. A) Drug sensitivity scores (DSS) of the in vitro drug screen with 76 drugs using sensitive (treatment-naïve) and resistant (#791) Med-1712FH cells reveals Selinexor as one of the top hits; B) IC50-values for both sensitive (86.1 nM) and resistant (65.6 nM) cells were low and treatment showed good dose-dependency; C) In vivo treatment of one sensitive (#806) and two resistant (#812, SMO-mutated; #799, MEGF8-mutated) models does lead to tumor growth inhibition in resistant but less in the sensitive models; vehicle group is shown in gray (n = 4), Selinexor group (5 mg/kg; n = 5; n = 4 for #806 sensitive) in black; tumor volume was measured once per week and is displayed relative to starting signal.
Discussion
Treatment of pediatric cancer patients is mostly a combination of surgery, radiation, and/or chemotherapy. Although this treatment strategy can lead to stable disease, patients often suffer from long-term side effects of the aggressive treatment, and more precise and less toxic treatments are thus needed. With increasing use of sequencing methods, tumor samples can be better molecularly characterized and tumor-specific driving alterations can be identified. Treatment strategies can become more personalized and precise when targeting those alterations in tumors with specific inhibitors. However, an arising problem of using specific inhibitors is that tumors often become quickly resistant to these monotherapies. Understanding the primary and acquired resistant mechanisms and how to overcome or prevent them with other drugs or combination therapies is thus important to improve treatment efficacy.
SMO inhibitors have been shown to be very effective in MB and BCC tumors with aberrant activation of the SHH pathway.26–28 However, not all MB and BCC tumors with SHH activation will respond and tumors that harbor mutations in SMO or downstream of SMO will be resistant.17,24,25 Moreover, also responding tumors may rapidly become resistant due to aquired mutations in SMO or other genes downstream of SMO.26–28 To overcome resistance, either with monotherapy with another drug or with a combination therapy, more research using model systems that mimic the patient’s situation is necessary. Here we generated for the first time a human SMO inhibitor resistant MB PDX model.
The MB SHH PDX model with a PTCH1-mutation utilized here responded well to the treatment with 20 mg/kg Sonidegib. With an intermitted dosing schedule based on bioluminescence signals of the tumors it was possible to generate tumors resistant to Sonidegib. Molecular characterization of the nine resistant models revealed that 89% (8 of 9) developed a tumor with a single, newly acquired SMO mutation, which leads to constitutive activation of the SHH pathway. Only one treated tumor developed a mutation downstream of SMO, in MEGF8, a negative regulator for HH signaling.39 More than half of the detected mutations in SMO (5/8 mutations) are overlapping with mutations found before in BCC tumors treated with Vismodegib or MB mouse allografts treated with Sonidegib.26,29,30,43 The remaining mutations in SMO were not detected before in a resistant model. Interestingly, one of the generated resistant PDX models harbors a mutation at position D473, which was also found mutated in a patient’s biopsy taken after developing resistance to treatment with Vismodegib.27 This overlap shows that the generated resistant PDX models mimic the patient’s situation and can be used for further development of treatment strategies to overcome the resistance.
Besides the different mutations in SMO, only one generated model harbored a mutation downstream of SMO in MEGF8. In contrast to previous published studies about models with aquired resistance to SMO inhibitors, none of the models generated here harbored mutations in SUFU or showed amplification of GLI1/2. Analyzed BCC tumor patient samples that were resistant to Vismodegib harbored mutations in SMO but also mutations or gene amplifications downstream of SMO.26 Published Sonidegib-resistant MB mouse models showed only few mutations in Smo and 50% of generated resistant models harbored amplification of Gli2.29 However, the resistant allograft models became resistant already after 16 days of constant treatment compared to more than 100 days of intermitted dosing in our study. This significant difference in dosing time is probably due to the homozygous loss of tp53 in these allograft mouse models.
More in-depth analysis of our generated resistant models by RNA-sequencing revealed no changes in expression levels of GLI1/2 and MYCN in resistant and vehicle lines indicating that in both conditions the HH signaling pathway is active. Compared to this, expression of the genes is decreased in short-time treated samples and confirms target engagement of the applied drug leading to suppression of the HH pathway. GSEA analysis of vehicle, short-time treated and resistant samples gave evidence that the HH signaling pathway is upregulated in vehicle and resistant samples when compared to the short-time treated models. IPA analysis revealed up- or downregulation of pathways related to organismal death or proliferation respectively for the short-time treated samples. These findings indicate that even the short treatment of only two days sensitizes the cells to apoptosis. In contrast, IPA analysis of the resistant samples does not reveal any affected pathways related to growth of (cancer) cells suggesting that the resistant models do not have alterations in their tumor-related metabolism.
The generated resistant models can be used to evaluate futher treatment strategies to overcome resistance. With an in vitro drug screen Selinexor was detected as one of the top hits being effective in both models, a treatment-naïve and a resistant, and even slightly more effective in the resistant model. The XPO1-inhibitor Selinexor was already used in a previous study with a SUFU-mutated MB model, where it showed good efficacy.42 The mutation in SUFU leads to aberrant activation of the SHH pathway and likely primary resistance to treatment with SMO inhibitors. To further evaluate efficacy of the drug in our resistant models we used one sensitive and two resistant models in vivo and treated them with Selinexor. Even though we observed a survival benefit of all the Sonidegib-treatment groups, weekly bioluminescence measurements showed that tumor growth was significantly inhibited for the two mutated samples. We were able to show that the treatment with Selinexor prolonged the survival for both groups with a resistant tumor significantly (P = .0027). For the sensitive group the effect seemed to be still present but smaller, though this could not be proven here in either direction.
In the study presented here we were able to generate, for the first time, an SMO inhibitor resistant patient-derived tumor in an orthotopic setting. With these resistant models, additional experiments to develop new treatment strategies can be performed. On the one hand, resistant tumors can be treated with inhibitors targeting proteins downstream of SMO as for example GLI. Unfortunately, until now, no specific GLI-inhibitors are on the market or in clinical trial. On the other hand, and more likely, combinatorial approaches as for example described in Hau et al. 2021 to prevent resistance can be applied and studied in vivo.44
Supplementary Material
Acknowledgments
We thank the DKFZ Genomics and Proteomics Core Facility for sequencing our samples. And we thank Apurva Gopisetty and Lukas Madenach for helping with data access and normalization. In addition we thank the High-Throughput Biomedicine Unit (Institute for Molecular Medicine Finland, Helsinki, Finland) for the provision of ready-to-go drug plates. Moreover, we thank Karyopharm Therapeutics for providing Selinexor.
Funding
S.K., S.B., and F.S.-H. were funded by the DKFZ International PhD Program. Additionally S.M.P. and M.K. have received funding from the Deutsche Krebshilfe (111537) and the Innovative Medicines Initiative2 Joint Undertaking under grant agreement No. 116064. This Joint Undertaking receives support from the European Union’s Horizon 2020 research and innovation program and EFPIA.
Conflict of interest statement. None.
Authorship statement. Coordination of the project and experiments: S.K., S.B.; In vivo experiments: N.L.M.; GSEA, IPA and clustering analysis: F.S-H.; Drug and sample preparation: B.S.; In vitro drug screen: H.P., A.M., I.O.; Analysis of sequencing data; N.J.; Statistic analysis: T.H-L.; Histological analysis: L.S., A.K.; Concept and Design of the paper: O.W., S.M.P., M.K.
References
- 1. Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287(5785):795–801. [DOI] [PubMed] [Google Scholar]
- 2. Skoda AM, Simovic D, Karin V, et al. The role of the Hedgehog signaling pathway in cancer: a comprehensive review. Bosn J Basic Med Sci. 2018;18(1):8–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lowry W, Richter L, Yachechko R, et al. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci USA. 2008;105(8):2883–2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature. 2004;432(7015):324–331. [DOI] [PubMed] [Google Scholar]
- 5. Plotnikova OV, Golemis EA, Pugacheva EN. Cell cycle–dependent ciliogenesis and cancer. Cancer Res. 2008;68(7):2058–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Varjosalo M, Taipale J. Hedgehog: functions and mechanisms. Genes Dev. 2008;22(18):2454–2472. [DOI] [PubMed] [Google Scholar]
- 7. MacDonald TJ. Hedgehog pathway in pediatric cancers: they’re not just for brain tumors anymore. Am Soc Clin Oncol Educat Book. 2012;32(1):605–609. [DOI] [PubMed] [Google Scholar]
- 8. Booms P, Harth M, Sader R, Ghanaati S. Vismodegib hedgehog-signaling inhibition and treatment of basal cell carcinomas as well as keratocystic odontogenic tumors in Gorlin syndrome. Ann Maxillofac Surg. 2015;5(1):14–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Onodera S, Nakamura Y, Azuma T. Gorlin syndrome: recent advances in genetic testing and molecular and cellular biological research. Int J Mol Sci . 2020;21(20):7559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bailey JM, Mohr AM, Hollingsworth MA. Sonic hedgehog paracrine signaling regulates metastasis and lymphangiogenesis in pancreatic cancer. Oncogene. 2009;28(40):3513–3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ma X, Sheng T, Zhang Y, et al. Hedgehog signaling is activated in subsets of esophageal cancers. Int J Cancer. 2006;118(1):139–148. [DOI] [PubMed] [Google Scholar]
- 12. Karhadkar SS, Bova GS, Abdallah N, et al. Hedgehog signalling in prostate regeneration, neoplasia and metastasis. Nature. 2004;431(7009):707–712. [DOI] [PubMed] [Google Scholar]
- 13. Sheng T, Li C, Zhang X, et al. Activation of the hedgehog pathway in advanced prostate cancer. Mol Cancer. 2004;3(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Roma J, Almazán-Moga A, Sánchez de Toledo J, Gallego S. Notch, wnt, and hedgehog pathways in rhabdomyosarcoma: from single pathways to an integrated network. Sarcoma. 2012;2012:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Almazán-Moga A, Zarzosa P, Molist C, et al. Ligand-dependent Hedgehog pathway activation in Rhabdomyosarcoma: the oncogenic role of the ligands. Br J Cancer. 2017;117(9):1314–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang SY, Yang J-Y. Targeting the hedgehog pathway in pediatric medulloblastoma. Cancers 2015;7(4):2110–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kool M, Jones DT, Jäger N, et al. Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer cell. 2014;25(3):393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Northcott PA, Buchhalter I, Morrissy AS, et al. The whole-genome landscape of medulloblastoma subtypes. Nature. 2017;547(7663):311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Skowron P, Farooq H, Cavalli FM, et al. The transcriptional landscape of Shh medulloblastoma. Nat Commun. 2021;12(1):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Amakye D, Jagani Z, Dorsch M. Unraveling the therapeutic potential of the Hedgehog pathway in cancer. Nat Med. 2013;19(11):1410–1422. [DOI] [PubMed] [Google Scholar]
- 21. Girardi D, Barrichello A, Fernandes G, Pereira A. Targeting the Hedgehog pathway in cancer: current evidence and future perspectives. Cells. 2019;8(2):153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Romer JT, Kimura H, Magdaleno S, et al. Suppression of the Shh pathway using a small molecule inhibitor eliminates medulloblastoma in Ptc1+/− p53−/− mice. Cancer cell. 2004;6(3):229–240. [DOI] [PubMed] [Google Scholar]
- 23. Atwood SX, Whitson RJ, Oro AE. Advanced treatment for basal cell carcinomas. Cold Spring Harbor perspectives in medicine 2014;4(7):a013581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Robinson GW, Orr BA, Wu G, et al. Vismodegib exerts targeted efficacy against recurrent sonic hedgehog–subgroup medulloblastoma: results from phase II pediatric brain tumor consortium studies PBTC-025B and PBTC-032. J Clin Oncol. 2015;33(24):2646–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee Y, Kawagoe R, Sasai K, et al. Loss of suppressor-of-fused function promotes tumorigenesis. Oncogene. 2007;26(44):6442–6447. [DOI] [PubMed] [Google Scholar]
- 26. Atwood SX, Sarin KY, Whitson RJ, et al. Smoothened variants explain the majority of drug resistance in basal cell carcinoma. Cancer Cell. 2015;27(3):342–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rudin CM, Hann CL, Laterra J, et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N Engl J Med. 2009;361(12):1173–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yauch RL, Dijkgraaf GJ, Alicke B, et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science. 2009;326(5952):572–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Buonamici S, Williams J, Morrissey M, et al. Interfering with resistance to smoothened antagonists by inhibition of the PI3K pathway in medulloblastoma. Sci Transl Med. 2010;2(51):51ra70–51ra70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sharpe HJ, Pau G, Dijkgraaf GJ, et al. Genomic analysis of smoothened inhibitor resistance in basal cell carcinoma. Cancer Cell. 2015;27(3):327–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brabetz S, Leary SE, Gröbner SN, et al. A biobank of patient-derived pediatric brain tumor models. Nat Med. 2018;24(11):1752–1761. [DOI] [PubMed] [Google Scholar]
- 32. Pan S, Wu X, Jiang J, et al. Discovery of NVP-LDE225, a potent and selective smoothened antagonist. ACS Med Chem Lett. 2010;1(3):130–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Capper D, Jones DT, Sill M, et al. DNA methylation-based classification of central nervous system tumours. Nature 2018;555(7697):469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin GL, Monje M. A protocol for rapid post-mortem cell culture of diffuse intrinsic pontine glioma (DIPG). J Visual Exp. 2017;(121):55360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. ElHarouni D, Berker Y, Peterziel H, et al. iTReX: Interactive exploration of mono-and combination therapy dose response profiling data. Pharmacol Res. 2021;175:105996. [DOI] [PubMed] [Google Scholar]
- 36. Potdar S, Ianevski A, Mpindi J-P, et al. Breeze: an integrated quality control and data analysis application for high-throughput drug screening. Bioinformatics 2020;36(11):3602–3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cavalli FM, Remke M, Rampasek L, et al. Intertumoral heterogeneity within medulloblastoma subgroups. Cancer cell. 2017;31(6):737–754.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang C, Wu H, Katritch V, et al. Structure of the human smoothened receptor bound to an antitumour agent. Nature 2013;497(7449):338–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pusapati GV, Kong JH, Patel BB, et al. CRISPR screens uncover genes that regulate target cell sensitivity to the morphogen sonic hedgehog. Dev Cell. 2018;44(1):113–129.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kong JH, Young CB, Pusapati GV, et al. A membrane-tethered ubiquitination pathway regulates Hedgehog signaling and heart development. Dev Cell. 2020;55(4):432–449.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bader JC, Razak ARA, Shacham S, Xu H. Pharmacokinetics of selinexor: the first-in-class selective inhibitor of nuclear export. Clin Pharmacokinet. 2021;60(8):957–969. [DOI] [PubMed] [Google Scholar]
- 42. Smith MA, Lock R, Carol H, et al. Pharmacodynamic and genomic markers associated with response to the XPO1/CRM1 inhibitor selinexor (KPT-330): a report from the Pediatric Preclinical Testing Program: AACR; 2015https://www.karyopharm.com/wp-content/uploads/2019/03/6_Malcolm_Smith_Sarcoma_poster_AACR_2015-1.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pricl S, Cortelazzi B, Dal Col V, et al. Smoothened (SMO) receptor mutations dictate resistance to vismodegib in basal cell carcinoma. Mol Oncol. 2015;9(2):389–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hau P, Frappaz D, Hovey E, et al. Development of randomized trials in adults with medulloblastoma—the example of eortc 1634-btg/noa-23. Cancers 2021;13(14):3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Atwood SX, Sarin KY, Li JR, et al. Rolling the genetic dice: neutral and deleterious Smoothened mutations in drug-resistant basal cell carcinoma. J Invest Dermatol. 2015;135(8):2138–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhou X, Edmonson MN, Wilkinson MR, et al. Exploring genomic alteration in pediatric cancer using ProteinPaint. Nat Genet. 2016;48(1):4–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.