Abstract
As carriers of multiple human diseases, understanding the mechanisms behind mosquito reproduction may have implications for remediation strategies. Transfer RNA (tRNA) acts as the adapter molecule of amino acids and are key components in protein synthesis. A critical factor in the function of tRNAs is chemical modifications which contribute to codon-anticodon interactions. Here, we provide an assessment of tRNA modifications between sexes for three mosquito species and examine the correlation of transcript levels underlying key proteins involved in tRNA modification. Thirty-three tRNA modifications were detected among mosquito species and most of these modifications are higher in females compared to males for three mosquito species. Analysis of previous male and female RNA-seq datasets indicated a similar increase in transcript levels of tRNA modifying enzymes in females among six mosquito species, supporting our observed female enrichment of tRNA modifications. Tissues-specific expressional studies revealed higher transcript levels for tRNA modifying enzymes in the ovaries for Aedes aegypti, but not male reproductive tissues. These studies suggest that tRNA modifications may be critical to reproduction in mosquitoes, representing a potential novel target for control through suppression of fecundity.
Keywords: tRNA, chemical modifications, reproduction, sex-associated, mosquitoes, post-transcriptional modifications
Graphical Abstract
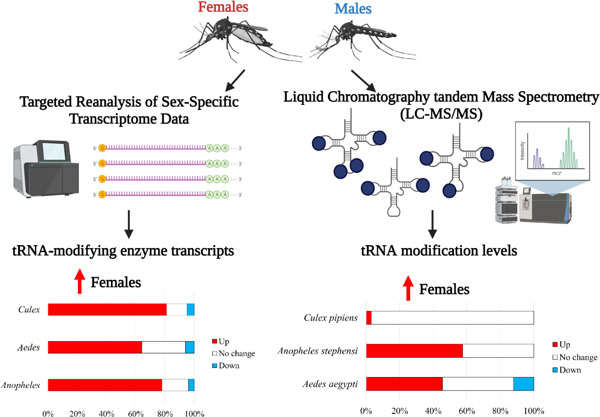
1. Introduction
Mosquitoes are vectors for human disease causing millions of deaths each year and compromising 17% of global infectious diseases (World Health Organization, 2020). As such, remediation techniques, such as vector population control are beneficial for preventing disease transmission. Current methods consist of pesticides, genetic modification, and other experimental techniques (Balatsos et al., 2021; Benelli et al., 2016). However, there are limitations to each of these methods and the fear of permanent environmental effects should also be considered (Benelli et al., 2016). Proteomics, transcriptomics, and small RNA sequencing have all been performed in mosquitoes (Camargo et al., 2020; Eng et al., 2018; Gamez et al., 2020; Khan et al., 2005; Shettima et al., 2021), which has proven critical to understanding mosquito biology. Based on these molecular techniques, males and females exhibit significant differences but there are still gaps in understanding RNA modifications in mosquitoes.
Transfer RNA (tRNA) is responsible for carrying the amino acid to the ribosome based on the codon in messenger RNA (mRNA). As such, the tRNA is critical in protein synthesis. tRNA requires chemical modifications throughout the molecule to ensure stability and alter codon-anticodon interactions (Agris et al., 2007; Edwards et al., 2020; Motorin and Helm, 2010). For example, methylations on the ribose group prevent nuclease cleavage and stabilize the structure of tRNAs to increase thermal tolerance (Hori, 2014). Chemical modifications to tRNA are diverse and range from methylations to more complex hypermodifications (Boccaletto et al., 2018). The hypermodifications are often located on the anticodon and require multi-step synthesis with several enzymes (Boccaletto et al., 2018; Klassen et al., 2016; Noma et al., 2006). Modifications on the anticodon loop may alter the codon preference for a given tRNA (Agris et al., 2007). Regardless of complexity, tRNA modifications have important roles in maintaining translational speed and accuracy.
tRNA modifications have central roles in translation and are potential targets for new classes of pesticides and other population control methods. In Aedes aegypti, cleavage of tRNA transcripts into tRNA fragments is sex-specific (Eng et al., 2018). As various tRNA modifications have been shown to impact nuclease cleavage of tRNAs (Kawai et al., 1991; Wang et al., 2018), it is possible tRNA modifications are different between the sexes as well. However, the tRNA modification status is unknown in mosquito species. In fact, there are few studies on tRNA modifications in insects, particularly in vectors for disease (Koh and Sarin, 2018). In fruit flies, the anticodon hypermodification N6-threonyladenosine (t6A) was found to be crucial for timely progression into developmental stages and loss of the modification led to smaller larvae likely due to codon-specific defects (Lin et al., 2015; Rojas-Benítez et al., 2017). Still, there has not been a characterization of tRNA modifications or a sex-associated comparison in mosquitoes or other insect species.
Here, we present data that female mosquitoes have a higher abundance of tRNA modifications than their male counterparts. This is supported by tRNA-modifying enzyme transcript levels which are higher in female mosquitoes compared to males for all mosquito species. Further, several tRNA modifications detected in Aedes and Anopheles were sex-specific. Likewise, the relative abundance of many tRNA modifications is higher in females than in males in Aedes and Anopheles. While Culex pipiens exhibited less dramatic differences based on sex, there was one modification elevated in females compared to males and in general, there were slightly higher levels in females. Altogether, female mosquitoes likely utilize chemical modifications to tRNA more abundantly than males, which could underlie factors associated with female reproduction.
2. Methods
2.1. Mosquitoes
tRNA modifications were examined between males and females for Aedes aegypti, Culex pipiens, and Anopheles stephensi. Larvae were maintained on groundfish food (Tetramin) supplemented with yeast extract (Fisher). All mosquitoes were maintained at 27°C and 80% RH (vapor pressure deficit = 0.71 kPa) under a 15h light: 9h dark cycle with access to water and 10% sucrose ad libitum, unless otherwise mentioned. Adult mosquitoes were 4–5 days old when sampled.
2.2. Identification of tRNA modifying enzymes
To identify possible tRNA modifying enzymes in the mosquito species, seventy protein sequences of Homo sapiens tRNA modifying enzymes were obtained from the UniProt database (UniProt Consortium, 2021). Mosquito proteome datasets were acquired from Vectorbase (Giraldo-Calderón et al., 2015). The reference sequences were used to BLAST against the proteome for each mosquito species. A protein was considered to be homologous if the E-value < 1E-29 and the corresponding gene used in RNA-seq analyses. The specific genome for each organism that was surveyed for tRNA modifying enzymes are listed in Table S1.
tRNA gene numbers were identified in the genomes of C. quinquefasciatus, A. stephensi, and A. aegypti using tRNAscan-SE 2.0 (Chan et al., 2021). Briefly, genomes were analyzed with tRNAscan-SE 2.0 with an initial minimum quality score of 50 to obtain high quality tRNAs. The output from these initial analyses were subsequently assessed through the Eukaryote Confidence Filter to discriminate tRNA-derived repetitive elements from canonical tRNA. The high confidence sets were considered for comparison. To estimate the composition of the tRNA pool, tRNA gene copy numbers were assumed to be reflective of active tRNAs as has been shown previously (Tuller et al., 2010). A proportional Z-test was used to evaluate if there was a difference in the proportion of tRNA isotypes (Ala, Asn, Asp, etc.) between species. Only one isotype, valine (Val), had more tRNA genes when A. aegypti was compared to A. stephensi. However, when evaluating the tRNA pool composition using isodecoder copy numbers, no differences were observed between species. For example, Val tRNAs with the anticodon AAC were in the same proportion in all three species. Likewise, the other Val tRNAs, Val-CAC and Val-TAC, were also in similar proportions across species. The proportion of tRNA genes for the isotypes and isodecoders are listed in Table S2 and Table S3, respectively. This indicates that differences in the number of tRNA types between species is unlikely to have major impacts on the tRNA modification abundances.
2.3. Total RNA and tRNA isolation from female and male mosquitoes
For the RNA isolation protocol, 15 mosquitos were placed in individual bead beater tubes containing 15 beads and 0.5 mL of TRI reagent (Sigma Aldrich). To isolate the total RNA, the samples were inverted and then stored at −80 ℃ until completely frozen. The samples were homogenized by bead beating for three cycles of 20 seconds of shaking and 10 seconds of still. Another 0.5 mL of TRI reagent was added and vortexed. Separation was performed by adding 300 μL of acid-phenol-chloroform (Invitrogen), the samples were then vortexed until homogenous and incubated at room temperature for 10 minutes. The samples were centrifuged for 15 minutes at 12,000 rpm at 4 °C and the aqueous phase was collected into a new tube and precipitated with 1.5X volume of isopropyl alcohol (Sigma Aldrich) overnight at −20°C. Total RNA was precipitated by centrifuging for 30 minutes at 12,000 rpm at 4°C. The supernatant was discarded and the RNA pellets were washed once with 75% ethanol (Thermo Fisher Scientific) and centrifuged again for 15 minutes at 12,000 rpm at 4°C. The RNA pellets were left to dry for 15 minutes and then resuspended in 100 μL of sterile water and then stored in the −80°C until tRNA isolation.
For the isolation of tRNAs, total RNA was separated using anion exchange chromatography with a Nucleobond AX100 column (Macherey-Nagel, Duren, Germany). 95 µg of total RNA was resuspended in 100 mM Tris acetate (Research Products International, Mt Prospect, IL) pH 6.3, and 15% v/v ethanol (Thermo Fisher Scientific) solution with pH 7. To equilibrate the column 10 mL of 200 mM NH4Cl (Sigma Aldrich), 100 mM Tris acetate pH 6.3, and 15% ethanol solution were added to the column. The total RNA sample was passed through the column three times. Small RNAs are separated in a buffer of 400 mM NH4Cl, 100 mM Tris acetate pH 6.3, and 15% ethanol solution. The tRNA fraction was eluted by adding 500 μL of 650 mM NH4Cl, 100 mM Tris acetate pH 6.3, and 15% ethanol solution until 12 fractions were collected. The tRNA was precipitated by adding 750 μL of isopropyl alcohol to each fraction, mixed, and stored at −20°C overnight. One biological replicate was defined as tRNA isolated from approximately 300 µg of total RNA and three biological replicates worth of tRNA were collected. Concentrations were assessed via NanoPhotometer (Nanodrop 2000C, Thermo Scientific) and a 1% agarose gel confirmed tRNA presence and purity (Figure S1, Figure S2, Figure S3).
2.4. Nucleoside analysis of tRNAs to identify modifications
Samples of tRNA were digested into nucleosides using previously reported conditions (Ross et al., 2017). Briefly, 2 µg of tRNA were denatured by incubating at 95°C for 10 min and immediately cooled for 10 min at - 20°C. Next, tRNA was incubated with 1/10 volume of 0.1 M ammonium acetate (Sigma Aldrich) and nuclease P1 (2 U/µg tRNA, Sigma Aldrich) at 45°C for 2 h. Then 1/10 volume of 1 M ammonium bicarbonate (Sigma Aldrich) was added with snake venom phosphodiesterase (1.2×10−4 U/µg tRNA, Worthington Biochemical) to catalyze the formation of individual nucleotides. Phosphates were removed by adding bacterial alkaline phosphatase (0.1 U/µg tRNA, Worthington Biochemical) and the mixture was incubated for an additional 2 h at 37°C before vacuum drying. An internal standard of 2-bromo-deoxycytidine (m/z = 306.0078) was spiked into each sample to ensure matrix effects were the same across samples.
For detection and relative quantification of modifications, nucleosides were resuspended in mobile phase A and separated via reversed-phase liquid chromatography (RP-LC) using a high-strength silica column (Acquity UPLC HSS T3, 1.8 μm, 1.0 mm× 100 mm, Waters) and an ultra-high-performance liquid chromatography (UHPLC) system (Vanquish Flex Quaternary, Thermo Fisher Scientific). Mobile phase A was composed of 5.3 mM ammonium acetate in water, pH 4.5, and mobile phase B was composed of a mixture of acetonitrile/water (40:60) with 5.3 mM ammonium acetate, pH 4.5. The flow rate was 100 μL min −1 and the column compartment temperature was set at 30°C. The following gradient was used: 0% B (from 0 to 7.6 min), 2% B at 15.7 min, 3% B at 19.2 min, 5% B at 25.7 min, 25% B at 29.5 min, 50% B at 32.3 min, 75% B at 36.4 min (hold for 0.2 min), 99% B at 39.6 min (hold for 7.2 min), then returning to 0% B at 46.9 min.
Next, an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific) with an H-ESI source (Thermo Fisher Scientific) was used for data acquisition as previously reported (Jora et al., 2018). The analyses were carried out in positive polarity and the settings to obtain the full scan data were a resolution of 120,000, a mass range of 220–900 m/z, automatic gain control 7.5 × 104, and injection time of 100 ms. MS/MS fragmentation was carried out with the following collision energy setting for CID to 42% and the setting for HCD 80 arbitrary units. Other instrumental settings consisted of the following: quadrupole isolation of 1.6 m/z; ion funnel radiofrequency level of 30%; sheath gas, auxiliary gas, and sweep gas of 30, 10, and 0 arbitrary units, respectively; ion transfer tube temperature of 299 °C; vaporizer temperature of 144 °C; and spray voltage of 3.5 kV.
Data processing was performed using Qual Browser in Xcalibur 3.0 (Thermo Fisher Scientific). Three characteristics were used to identify a given nucleoside: retention time (RT), molecular ion m/z, and fragment ion m/z. Molecular ions m/z and fragment ions m/z within a mass error of 5 ppm were considered for quantification. Relative abundance was determined by integration of the extracted ion chromatographic peak area. To account for potential differences in tRNA amounts, the modified nucleoside peak areas were normalized with the summation of integrated peak areas of the canonical nucleosides (A, G, C, and U). To determine the percentage of modifications differing between the sexes, the relative abundance for three biological replicates of females and males was compared using a Student’s t-test in which a p-value < 0.05 was significant.
2.5. RNA-seq analyses of sex and tissue-specific differences in tRNA modifying enzymes.
Sequence Read Archive (SRA) experiments for the six mosquito species used in this study are in Table S4 (Aryan et al., 2020; Grigoraki et al., 2020; Honnen et al., 2016; Jayaswal et al., 2021; Matthews et al., 2018; Xu et al., 2019). These male and female samples are at similar biological states as those collected for the tRNA modifications, where females were pre-vitellogenic before blood feeding and males are a few days of age. RNA-seq analyses were performed as previously published (Attardo et al., 2019; Scott et al., 2020). First, the adapter sequences were trimmed and assessed for quality using Trimmomatic (version 0.38.0) with default settings (Bolger et al., 2014). Reads were mapped with Sailfish (version 0.10.1.1) under the default settings and generated transcripts per million mapped (TPM) (Patro et al., 2014). Next, DEseq2 (version 2.11.40.6) was utilized to determine differential expression levels following an FDR at 0.05 (Love et al., 2014). To determine sex-associated trends, genes that were two-fold enriched or reduced were considered for pairwise comparisons. The genes enriched in the whole-body samples of one sex were considered to be sex-associated in A. aegypti, C. quinquefaciatus, and A. stephensi. Tissue analyses focused on expression profiles in relation to the whole-body transcriptome levels in A. aegypti.
2.6. Statistics
To assess the relationship between sex, species, tRNA modification, and transcript levels of tRNA-modifying enzymes, a linear regression model was applied to the data (lm function in the lme4 R package (Bates et al., 2015)). Specific modifications and transcripts were selected because the homologous enzyme gene that performs the respective modification has been identified in all three mosquito species. Therefore, twenty modifications and twenty-nine perspective genes were examined for interactions between sex, transcript levels, and modification abundance. Modification abundances and transcript levels were also subjected to permanova (Permutational Analysis of Variance, function “adonis” in R in “vegan” package, 999 permutations) to test differences in transcript levels and tRNA modification abundance between species and sex on Bray-Curtis abundance matrices.
3. Results
3.1. tRNA-modifying expression is higher in female mosquitoes
In general, the transcript levels of tRNA modifying enzymes is significantly higher in females than in males in mosquitoes (Figure 1A). Moreover, the general trends of expression seem to be held regardless of species which could be due to the similar proportion of tRNA gene numbers between species (Table S2, Table S3). For example, females typically have higher transcript levels for PUS3 and DUS3L that modify uridine to pseudouridine (Ψ) and dihydrouridine (D), respectively (Charette and Gray, 2000; Dai et al., 2021). Similarly, tRNA-modifying enzymes associated with anticodon modifications also demonstrated transcript enrichment in females. For example, TRIT1 and TrmO modify adenosine at the position adjacent to the anticodon to form N6-isopentenyladenosine (i6A) and N6-methyl-N6-threonylcarbamoyladenosine (m6t6A) (Edvardson et al., 2017; Spinola et al., 2005). Transcripts of other enzymes that modify the wobble position are also in higher abundance in female mosquitoes. With few exceptions, the vast majority of tRNA modifying enzyme transcripts are enriched in females.
Figure 1. Summary of tRNA modification identified for mosquitoes and expressional levels of tRNA modifying enzymes.
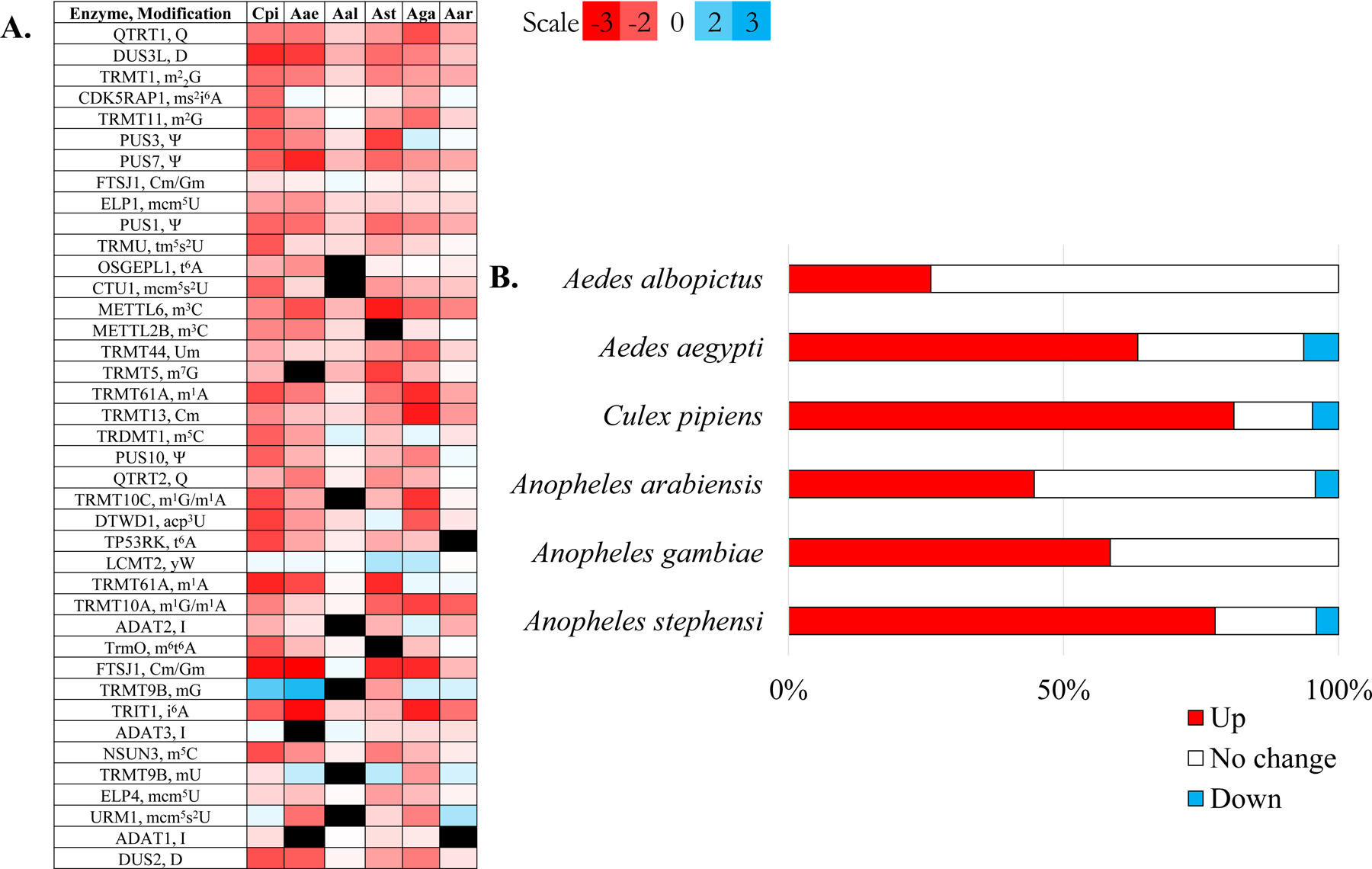
A. Homologous tRNA-modifying enzymes identified in A. albopictus, A. aegypti, A. stephensi, A. gambiae, A. arabiensis, and C. pipiens. Values are log2 fold change and a negative value (red) indicates the expression is higher in females while a positive value (blue) indicates higher expression in males. B. Bar graph of the percentage of differentially expressed tRNA-modifying enzymes when comparing males and females. The red bar represents the percentage of tRNA-modifying enzyme transcripts higher in females while the blue indicates the percentage higher in males. The white bar indicates the percentage of tRNA-modifying enzyme transcripts that exhibited the same expression in males and females.
There are a couple of tRNA-modifying enzymes that exhibit higher transcript levels in males. For example, LCMT2 is upregulated and is a component of the wybutosine (yW) biosynthesis pathway. However, there were not detectable levels of yW or yW-biosynthesis intermediates (i.e., imG) (Table 1). The modification yW is typically located at position 37 of the anticodon of primarily Phe-tRNA (Boccaletto et al., 2018; Guy and Phizicky, 2015). Thus, it is possible yW was not detected due to the low abundance of Phe-tRNA at the conditions the samples were collected. Higher transcripts of TRMT9B and LCMT2 suggest modification of Phe-tRNA is likely correlated with specific aspects of male mosquito biology. All in all, differential expression of the tRNA-modifying enzymes suggests there may be alternative modifications present in a sex-biased manner.
Table 1. List of 33 modifications detected in tRNA of mosquitoes.
tRNA modifications detected using LC-MS/MS. 33 modifications were detected in all female samples and are listed here. The abbreviation for the modification, the name, the molecular ion m/z ([MH+]), and retention time (RT) are listed here.
| Modification | Name | MH+ | RT (min) |
|---|---|---|---|
| Ψ | Pseudouridine | 245.0773 | 1.6 |
| D | Dihydrouridine | 247.0930 | 1.5 |
| m5C | 5-methylcytidine | 258.1090 | 4.8 |
| m3C | 3-methylcytidine | 258.1090 | 3.4 |
| Cm | 2’-O-methylcytidine | 258.1090 | 6.8 |
| Ψm | 2’-O-methylpseudouridine | 259.0930 | 5.1 |
| m3U | 3-methyluridine | 259.0930 | 13 |
| Um | 2’-O-methyluridine | 259.0930 | 13 |
| m5U | 5-methyluridine | 259.0930 | 8.5 |
| I | Inosine | 269.0886 | 8 |
| Am | 2’-O-methyladenosine | 282.1202 | 29.5 |
| m6A | N6-methyladenosine | 282.1202 | 31.2 |
| m1A | 1-methyladenosine | 282.1202 | 4.2 |
| m1I | 1-methylinosine | 283.1042 | 18.3 |
| ac4C | N4-acetylcytidine | 286.1039 | 20.3 |
| m62A | N6,N6-dimethyladenosine | 296.1359 | 33.3 |
| m2G | 2-methylguanosine | 298.1151 | 21.4 |
| Gm | 2’-O-methylguanosine | 298.1151 | 17.9 |
| m1G | 1-methylguanosine | 298.1151 | 18.8 |
| m7G | 7-methylguanosine | 298.1151 | 7.4 |
| m22G | N2,N2-dimethylguanosine | 312.1308 | 29.3 |
| mcm5U | 5-methylcarbonylmethyluridine | 317.0985 | 18.8 |
| mcm5s2U | 5-methoxycarbonylmethyl-2-thiouridine | 333.0756 | 30.3 |
| i6A | N6-isopentenyladenosine | 336.1672 | 36.5 |
| acp3U | 3-(3-amino-3-carboxypropyl)uridine | 346.1250 | 2.8 |
| acp3D | 3-(3-amino-3-carboxypropyl)-5,6-dihydrouridine | 348.1407 | 1.3 |
| ms2i6A | 2-methylthio-N6-isppentenyladenosine | 382.1549 | 41.7 |
| Q | Queuosine | 410.1675 | 18.3 |
| t6A | N6-threonylcarbamoyladenosine | 413.1421 | 31.1 |
| isomer oQ | Epoxyqueuosine | 426.1625 | 16.4 |
| m6t6A | N6-methyl-N6-threonylcarbamoyladenosine | 427.1577 | 32.3 |
| ms2t6A | 2-methylthio-N6-threonylcarbamoyladenosine | 459.1298 | 32.8 |
| manQ | Mannosyl-queuosine | 572.2204 | 19 |
Overall, the majority of tRNA-modifying enzyme transcripts are significantly higher in female mosquitoes. Regardless of species, at least 25% of the identified genes have enriched transcript levels in females (Figure 1B). Likewise, approximately 5% of tRNA-modifying enzyme transcripts are elevated in males. This trend was consistent across six species of mosquitoes but the extent of sex-associated transcript enrichment remains unique to each species.
3.2. Census of tRNA modifications in mosquitoes
The census of tRNA modifications detected in A. aegypti, A. stephensi, and C. pipiens is listed in Table 1. All four of the canonical nucleosides (A, C, G, U) were detected in each sample as well as Ψ and dihydrouridine (D). Ψ and D are important for the structure of all tRNAs since these modifications are central to the TΨC-loop and D-loop, respectively (Dalluge et al., 1996; Ge and Yu, 2013). Inosine (I) was also detected and is a common wobble modification used to expand decoding capacity. I34 is formed by tRNA-dependent adenosine deaminases (ADATs) and can base pair with C, A, and U on the third position of the codon (Nishikura, 2016; Wolf et al., 2002). As expected, many common ribose and base methylations (e.g., m5C, m3C, m3U, m5U, m6A, m1A, m1I, m2G, m1G, m7G, and m22 G) were detected. In mosquitoes, the 2’-O-methylations detected in tRNA are Ψm, Am, Gm, Cm, and Um. The addition of a methyl group to the canonical nucleoside is common in many kinds of RNA (Boccaletto et al., 2018). In tRNA, methylations are critical for structure and thermal tolerance through increasing the melting temperature of tRNA (Hori, 2014; Ishida et al., 2011). In addition, methylation on the 2’ position of the ribose occurs to prevent nuclease cleavage and provide structural support in tRNA (Kurth and Mochizuki, 2009). Overall, the tRNA modifications detected are commonly present in tRNA of eukaryotes and other domains.
One modification detected in these experiments is not reported to be present in tRNA and is likely from small contamination of other types of RNA. The modification N6,N6-dimethyladeonsine (m62A) has been previously reported in ribosomal RNA of bacteria and eukaryotes (Boccaletto et al., 2018). As a common rRNA modification, presence of m62A is likely the result of small ribosomal subunit RNA contamination or possible coelution of degraded rRNA with tRNA fractions. However, this modification was detected in low abundance, supporting that this is a minor component. Further, before digestion of the tRNA into nucleosides, samples were visualized on a 1% agarose to ensure there was no rRNA contamination (Figure S1, Figure S2, Figure S3). Thus, if contamination of rRNA or coelution of degraded rRNA is present, it is likely minimal.
As for anticodon modifications, several wobble position modifications were detected. Wobble modifications such as these are important in codon interactions and are altered in response to stress in microorganisms (Chan et al., 2012; Fernández-Vázquez et al., 2013). Two of the wobble modifications detected in mosquito tRNA were 5-methylcarbonylmethyluridine (mcm5U) and 5-methylcarbonylmethyl-2-thiouridine (mcm5s2U). The hypermodifications mcm5U and mcm5s2U are performed by the URM and ELP pathways (Rezgui et al., 2013). The type of U34 modification promotes the decoding of certain codons over others. For example, mcm5U improves the decoding of G-ending codons (Johansson et al., 2008). In contrast, mcm5s2U enhances the decoding of A- and G-ending codons (Johansson et al., 2008). In addition, queuosine (Q) and mannosyl-queuosine (manQ) are also wobble position modifications. Previously, manQ was thought to always be present with the isomer galactosyl-queuosine (galQ) in eukaryotes (Nishimura, 1983). Q replaces G at position 34 and has only been mapped to four types of tRNAs histidine, aspartic acid, asparagine, and tyrosine (Harada and Nishimura, 1972). The presence of Q on His tRNAs determined the preference of codon (Meier et al., 1985). Altogether, U34 modifications, Q, and its derivatives have established roles and likely contribute to codon selection in mosquitoes as well.
The anticodon modifications located at position 37 that were detected in mosquito tRNA were: t6A, i6A, 2-methylthio-N6-isopentenyladenosine (ms2i6A), m6t6A, and 2-methylthio-N6-threonylcarbamoyladenosine (ms2t6A). Position 37 is located adjacent to the anticodon and serves many functions in regulating translation. For example, t6A is universally present across domains of life and stabilizes anticodon interactions (Weissenbach and Grosjean, 1981). Notably, t6A is located at position 37 on tRNAs decoding ANN codons. As methionine (Met) tRNAs contain the anticodon CAU, they decode an ANN codon and contain t6A (Boccaletto et al., 2018). Met-CAU is also the tRNA that initiates translation by decoding the start codon, AUG. Thus, the presence of t6A has been proposed to be a regulator of the initiation of protein synthesis. Ultimately, there are numerous anticodon loop modifications that demonstrate sex-specific abundances in mosquitoes which highlight the importance of translational efficiency and accuracy.
3.3. tRNA modifications detected are sex-associated in mosquitoes
Detected modifications in tRNA of C. pipiens, A. aegypti, and A. stephensi have a few specific variations between sexes and species (Figure 2A). In C. pipiens, sex-biased tRNA modification was not observed as all 33 of the modifications were detected in both groups. However, A. aegpti and A. stephensi demonstrated sex-biased tRNA modifications. Two modifications were absent in A. aegypti males that were detected in females, oQ and mcm5s2U. Both of these modifications are typically present on the anticodon loop and mcm5s2U is a product of several enzymes which requires multistep synthesis (Björk et al., 2007; Boccaletto et al., 2018). oQ is commonly found in bacteria similar to its final product, Q (Boccaletto et al., 2018; Zallot et al., 2017). Bacteria-specific modifications that are typically in high abundance, such as 2-thiocytidine (s2C) and 4-thiouridine (s4U), were not detected in any of the samples. This suggests oQ and Q are present in mosquito tRNAs and not from co-isolated bacterial tRNAs from the microbiome. Collectively, the differences of anticodon modifications suggest sex-associated modification occurs at the anticodon loop in A. aegypti.
Figure 2. Comparison of detectable modifications across species and between sexes.
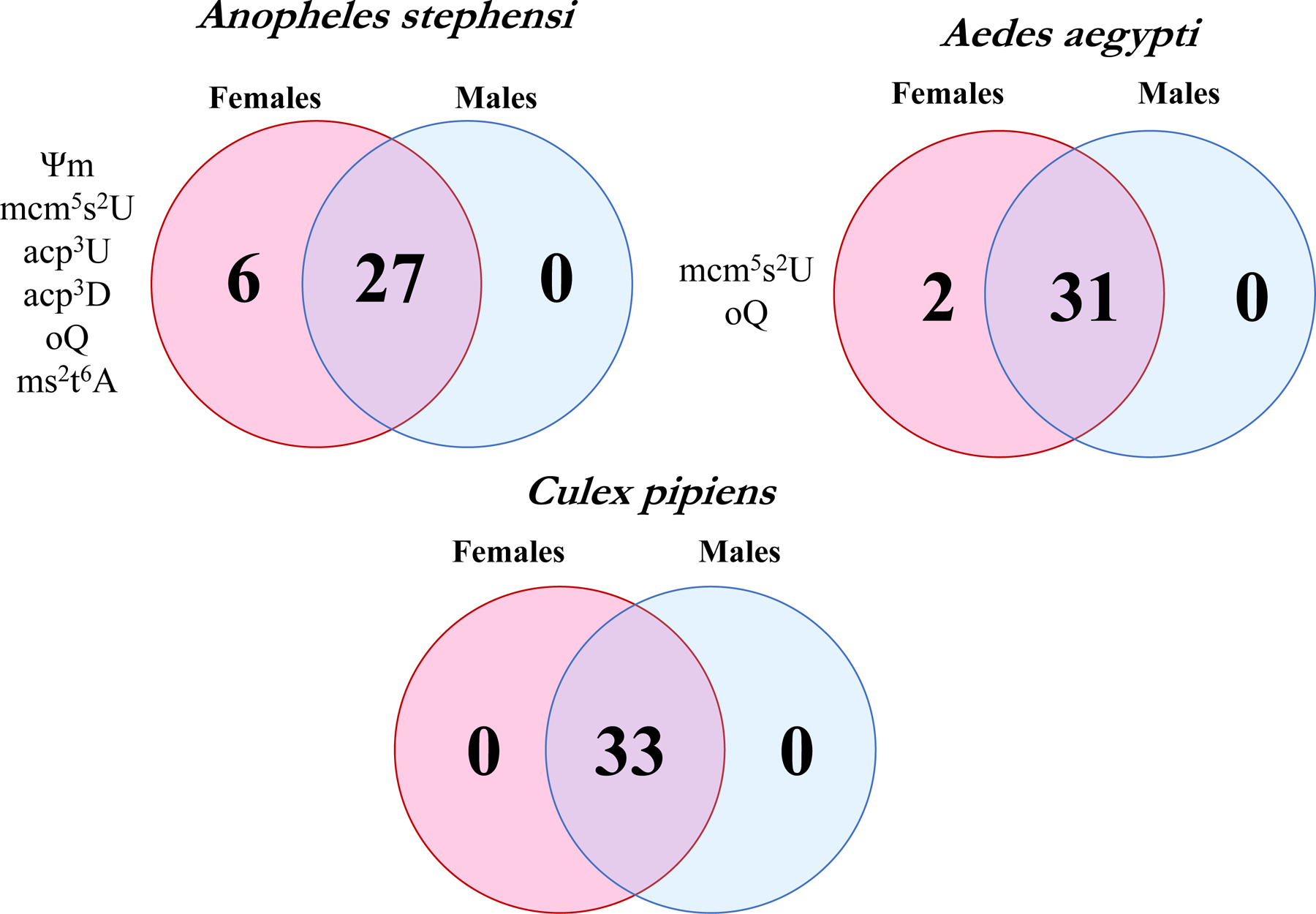
The Venn diagrams of tRNA modifications detected using LC-MS/MS in each species. There were some modifications detected only in females in A. aegypti and A. stephensi. However, all thirty-three modifications were detected in male and female C. pipiens.
Likewise, oQ and mcm5s2U were not at detectable levels in A. stephensi males. In addition, four other modifications were not detected in A. stephensi males: 2’-O-methylpseudouridine (Ψm), 3-(3-amino-3-carboxypropyl)-5,6-uridine (acp3U), 3-(3-amino-3-carboxypropyl)-5,6-dihydrouridine (acp3D), and ms2t6A. The modification acp3U has been mapped to the variable and D loop of tRNA and the enzymes required for this modification were recently documented in E. coli (Meyer et al., 2019; Takakura et al., 2019). While little is known on the function of acp3D, the modification acp3U increases thermal stability and loss of this modification impairs growth in mammals (Takakura et al., 2019). Expectedly, the modification t6A was detected across samples and is known to be present in all domains of life (Lorenz et al., 2017). Hypermodification of t6A forms ms2t6A or m6t6A which are also located at position 37 (Boccaletto et al., 2018). It is proposed that ms2t6A stabilizes the anticodon interaction with the codon through additional stacking interactions by the ms2 group (Durant et al., 2005). The other t6A derivative, m6t6A, improves the efficiency of the tRNA to read ACC codons in E. coli (Qian et al., 1998). Female A. stephensi possess both t6A derivatives, ms2t6A and m6t6A, which suggests stabilization in the anticodon loop may be more common in females. However, functional studies of these modifications are necessary to fully understand their sex-associated role in mosquitoes.
Ultimately, the majority of the detected tRNA modifications are shared regardless of sex and species. Twenty-six modifications were identified in all samples suggesting that the general census of tRNA modifications is relatively conserved in mosquitoes. Furthermore, the composition of the tRNA pool likely affects the ability to detect certain modifications. It is possible modifications present on a single tRNA or tRNA that are in low abundance would not be detected with current methods. Therefore, the modifications detected presently may only be representative of abundant tRNA levels and future studies may reveal additional lower abundance modifications.
3.4. tRNA modification abundances are higher in female mosquitoes
In each species, females had a higher abundance of at least one tRNA modification (Figure 3A). C. pipiens demonstrated the most similar abundances between the sexes with only one modification being higher in females, N6-methyladenosine (m6A). However, the trend of elevated modification abundance in females is common for A. aegypti and A. stephensi which have heightened levels of at least 13 modifications. Notably, several anticodon modifications were higher in females than in males. For example, m6t6A and ms2t6A were more abundant in females as well as Q and manQ. The levels of Q are significantly higher in female tRNA for A. aegypti and A. stephensi. However, Q abundance is not sex-specific in C. pipiens (Figure 3B).
Figure 3. Summary of tRNA modification abundance differences and correlation of specific enzyme transcript levels.
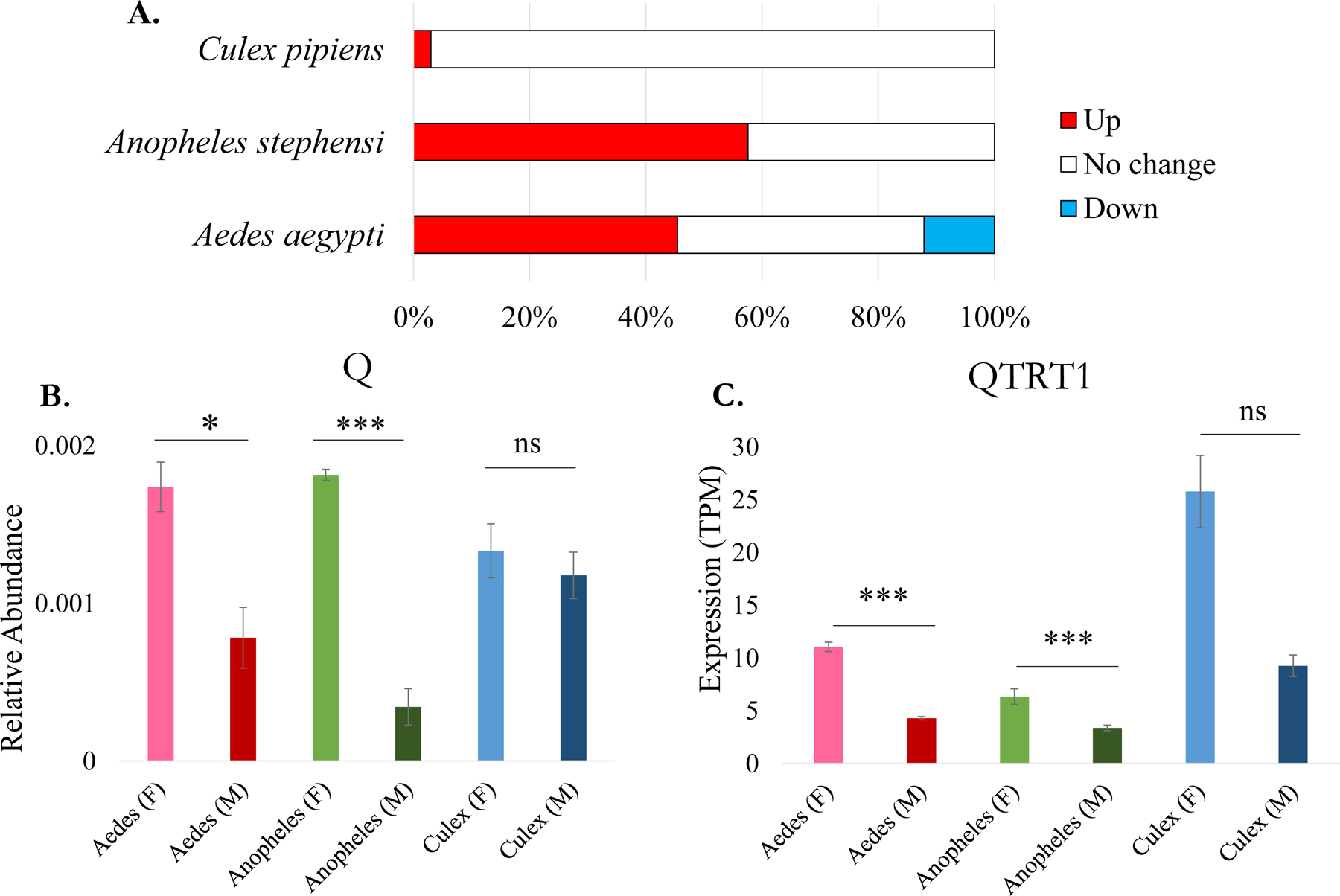
A. Percentage of tRNA modifications with sex-specific differences in abundance.The percentage of tRNA modifications exhibiting relative abundance changes as increases in females (pink), no change (gray), or decrease in females (blue). B. Relative abundance of the tRNA modification queuosine (QtRNA) in each of the groups. The peak area of QtRNA was normalized with the sum of the canonical peak areas. Significance was considered if p-value < 0.05 between the males and females. C. Expression values of the catalytic subunit of the tRNA-modifying enzyme Q transferase, QTRT1, in TPM.
Next, tRNA-modifying enzyme expression differences were compared to tRNA modification abundances. For A. aegypti and A. stephensi, the catalytic subunit of the enzyme that transfers Q onto tRNAs is Q transferase (QTRT1) follows the sex-associated trend observed in modifications (Figure 3C). There is not a significant difference in the expression of QTRT1 in C. pipiens which correlates with the unchanging modifications levels. We determined general trends in both sets of data to correlate tRNA modifying enzyme expression and modification abundances. To do this, twenty modifications were assessed for the tRNA-modifying enzyme expression in the different sexes. If the modification and enzyme expression is higher in females then the trend was considered to “match”. On the contrary, if the modification levels are not reflected in the enzyme expression, then they are considered not a match (Do not Match). In Figure 4, the percentage of tRNA modification abundances that follow the corresponding tRNA-modifying enzyme expression trend is shown for all three species. In A. stephensi, the majority of modifications (63%) had abundances that correlated with tRNA-modifying enzyme expression levels. A. aegypti tRNA modifications were slightly less in line with the tRNA-modifying enzyme expression with only 40% matching expression. Again, C. pipiens demonstrates discrepancy in tRNA modification abundance and tRNA-modifying enzyme expression. Only 20% of the tRNA modifications correlate with enzyme expression patterns. Altogether, these differences highlight there is a stronger correlation of transcript and modification levels in some mosquito species than in others.
Figure 4. Frequency of tRNA modification abundances correlating with tRNA-modifying enzyme transcript levels.
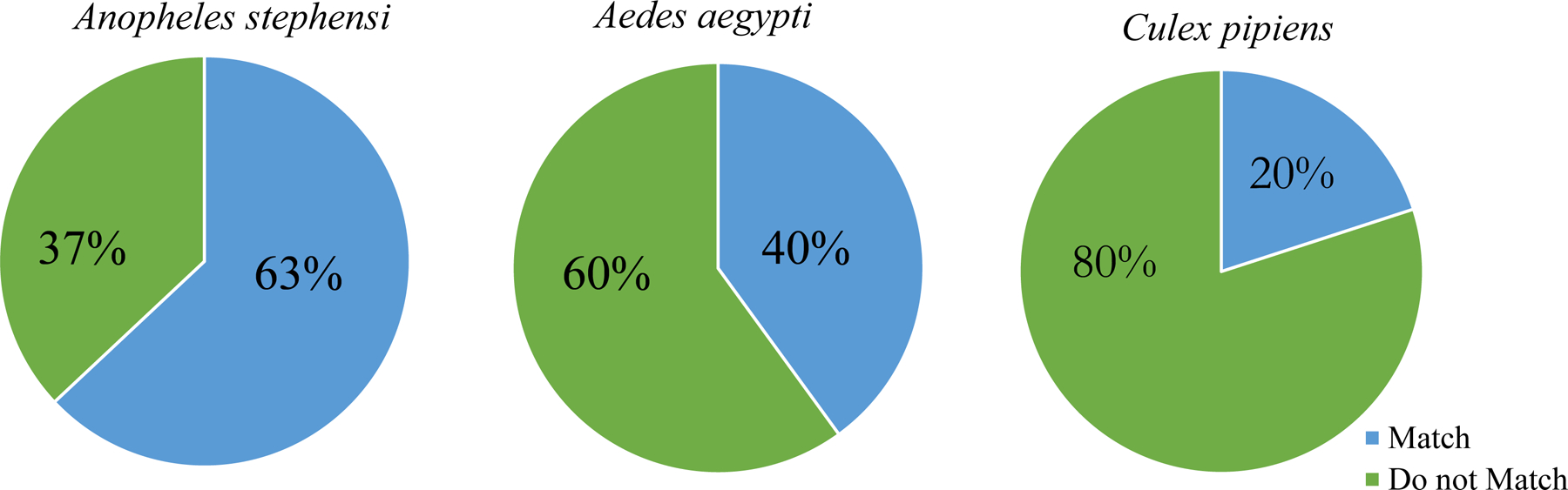
Percentage of tRNA modifications that correlate with tRNA-modifying enzyme transcript levels. Twenty modifications with known tRNA-modifying enzymes were compared to the corresponding transcript levels. A match was defined as a tRNA modification with an abundance that aligned with transcript differences between males and females (i.e, modification and transcript significantly higher in females). The tRNA modification was not considered a match when the tRNA modification abundance did not follow transcript levels (i.e., modification significantly higher but transcript levels were the same).
To increase confidence the correlation of modification and transcript levels were sex-associated, a linear regression model was applied to the relative abundance of modifications and the transcript levels of the respective genes. Table 2 lists the output of the linear regression model on the relationship between sex, modification, and trascript levels. Of the twenty-nine genes assessed, twenty exhibited high correlation (p-value < 0.05) with the modification of interest and sex across species in relation to the transcript abundance of the modifying enzyme. To increase confidence in this relationship, permanova demonstrated the transcript levels and tRNA modification abundance are significantly correlated to sex across species (P < 0.001). Therefore, there is statistical evidence that the trends in modifications and transcript levels for modifying enzymes are sex-associated in mosquitoes.
Table 2. There is a correlation between sex, tRNA modification abundances, and tRNA-modifying enzyme transcript levels.
Table of regression model values generated by the relationship between sex, tRNA modification abundance, and tRNA-modifying enzyme expression across species. There are 20 enzymes/modifications that are sex associated following linear model trends. The trend was considered sex-associated if the p-value < 0.05.
| Gene | Modification | Coefficient | Std. Error | t-value | p-value |
|---|---|---|---|---|---|
| PUS3 | Ψ | −8.4762 | 0.8001 | −10.594 | 9.34E-07 |
| PUS1 | Ψ | −29.307 | 4.199 | −6.98 | 3.81E-05 |
| DUS3L | D | −23.302 | 5.816 | −4.007 | 0.00249 |
| DUS2 | D | −6.042 | 2.592 | −2.331 | 0.04197 |
| TRMT9B | m3U | 0.37334 | 3.43793 | 0.109 | 0.91567 |
| TRMT13B | Am | −4.5451 | 0.6187 | −7.346 | 2.47E-05 |
| ADAT2 | I | −0.1946 | 2.2575 | 1.076 | 0.93301 |
| TRMT61A | m1A | −5.525 | 2.074 | −2.663 | 0.02376 |
| TRMT10A | m1A | −2.197 | 0.9163 | −2.398 | 0.03746 |
| TRIT1 | i6A | −6 | 2.6791 | −2.24 | 0.04904 |
| METTL6 | m3C | −18.2294 | 2.1163 | −8.614 | 6.13E-06 |
| NSUN3 | m5C | −3.9764 | 0.7025 | −5.661 | 0.00021 |
| TRDMT1 | m5C | −4.5451 | 0.6187 | −7.346 | 2.47E-05 |
| FTSJ1 | Cm | −29.278 | 7.934 | −3.69 | 0.00418 |
| TRMT13A | Cm | −1.9868 | 0.748 | −2.656 | 0.02406 |
| TRMT44 | Um | −0.2357 | 0.5395 | −0.437 | 0.67152 |
| TRMT11 | m2G | −2.861 | 1.825 | −1.568 | 0.14799 |
| FTSJ1 | Gm | −29.278 | 7.934 | −3.69 | 0.00418 |
| TRMT10C | m1G | −5.098 | 2.714 | −1.878 | 0.08975 |
| TRMT1 | mcm22G | −4.734 | 3.113 | −1.521 | 0.15933 |
| ELP1 | mcm5U | −1.848 | 1.514 | −1.221 | 0.25026 |
| ELP4 | mcm5U | −0.7833 | 1.3064 | −0.6 | 0.5621 |
| CTU1 | mcm5s2U | −1.978 | 1.712 | −1.156 | 0.27461 |
| URM1 | mcm5s2U | 9.667 | 2.145 | 4.507 | 0.00113 |
| DTWD1 | acp3U | −11.076 | 1.801 | −6.151 | 0.00011 |
| QTRT1 | Q | −6.747 | 1.977 | −3.413 | 0.00663 |
| QTRT2 | Q | −4.8298 | 0.7702 | −6.271 | 9.25E-05 |
| OSGEPL1 | t6A | −3.09186 | 0.80339 | −3.849 | 0.00322 |
| TP53RK | t6A | −13.535 | 5.534 | −2.446 | 0.03451 |
3.5. Reproductive tissues have higher expression of tRNA-modifying enzymes in A. aegypti
To determine if there is a relationship between tRNA-modifying enzyme expression and reproduction, the percentage of tRNA-modifying genes with different transcript levels were measured in the accessory glands, testes, and ovaries in A. aegypti in respect to levels within the entire body. Of the three, the ovaries demonstrated the most tRNA modifying enzyme transcript enrichment compared to the whole body. Approximately 55% of tRNA-modifying enzyme transcripts are elevated in the ovaries, suggesting this may be contributing to the overall expression patterns and higher tRNA modifications in females. Enriched transcript levels for specific tRNA modifying enzymes are also higher in testes (20%) and the accessory glands (5%), suggesting a potential role in male reproduction as well (Figure 5A). Ultimately, enrichment in reproductive tissues imply there may be a biological role of tRNA modifications in the reproduction of mosquitoes.
Figure 5. tRNA-modifying enzyme transcripts demonstrate elevation in reproductive tissues.
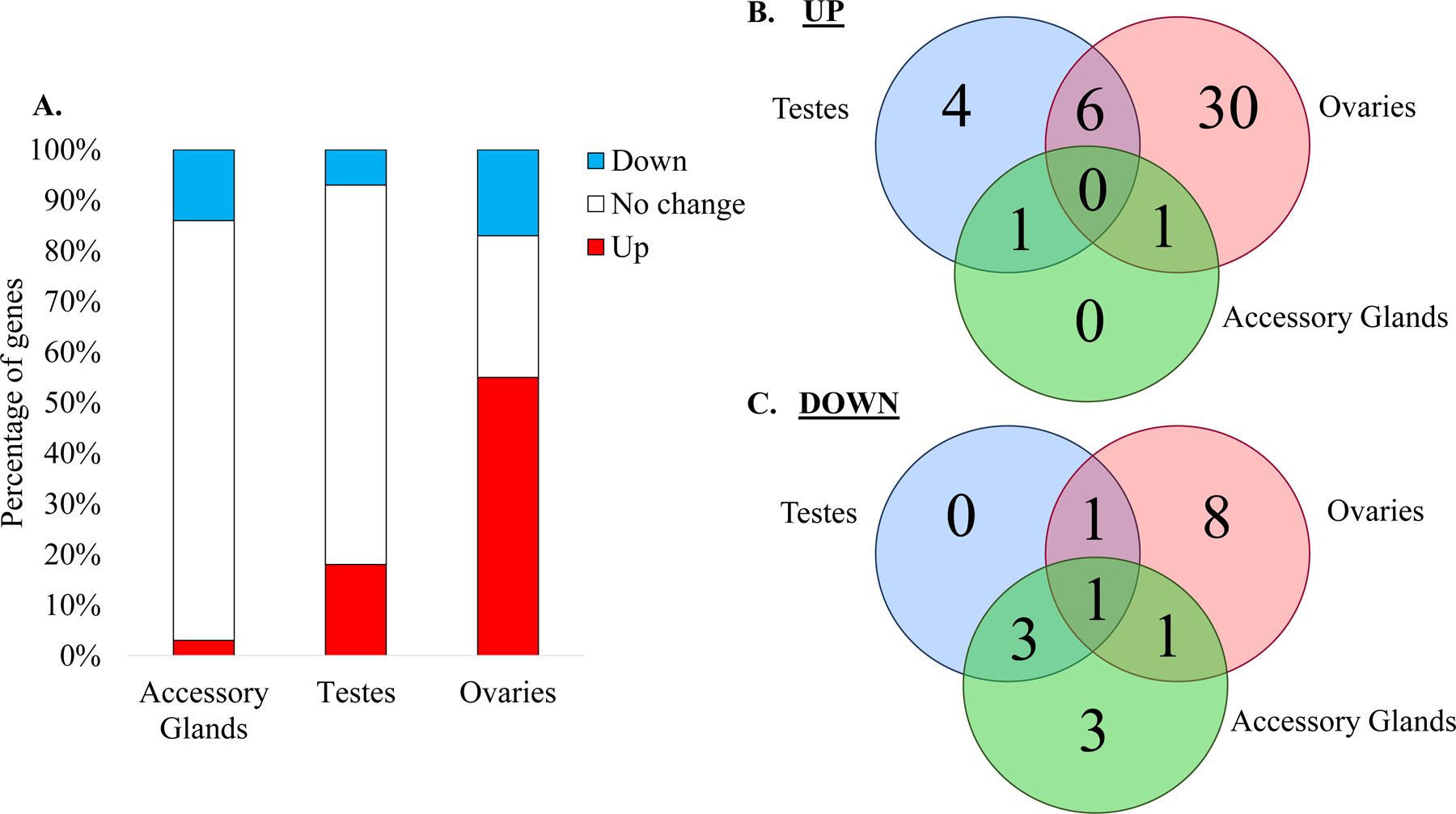
A. Percentage of tRNA-modifying enzyme gene expression trends in tissues of A. aegypti compared to whole body expression. The red bar indicates the percentage of tRNA-modifying enzymes upregulated in the tissue. The white bar represents the percentage of genes that do not exhibit differential expression between the whole body and tissue. The blue bar indicates the percentage of genes that are lower in the tissue than the whole body. B. Venn diagram of tRNA-modifying enzymes that have transcript levels higher in specific tissues compared to the whole body. C. Venn diagram of tRNA-modifying enzymes that have transcript levels lower in tissues.
There is little overlap in transcript abundance of tRNA-modifying enzymes in male and female reproductive tissues compared to the whole body levels. For example, two enzymes that form Ψ in tRNA, PUS3 and PUS10, are enriched in the testes and ovaries. Transcript levels for two anticodon modifications are also in higher abundance in the reproductive tissues: CTU2 and QTRT1. The enzyme CTU2 thiolates mcm5U to form mcm5s2U and transcripts are elevated in testes and ovaries in A. aegypti. Six of the sixty genes considered had higher transcriptional levels in reproductive tissues compared to the whole body (Figure 5B). Only one gene is consistently down in reproductive tissues (Figure 5C). Despite this observation, the ovaries demonstrated the most dramatic difference with over half of the genes having higher levels of transcripts. With such little overlap between the three tissues, it is likely that the tRNA modifications are tissue specific much like the expression trends, suggesting their is likely critical in the function of these organs that necessitates further study.
In surveying the specific genes enriched in the ovaries, many of the enzymes are for modifications found in every tRNA. For example, A. aegypti ovaries had a higher expression of tRNA-dihydrouridine synthase (i.e., DUS2L) which adds the typical D in the D-loop of tRNAs (Yu et al., 2011). Likewise, there was a higher expression of tRNA-pseudouridine synthase (i.e., PUS10) which adds Ψ to tRNAs (McCleverty et al., 2007). Enrichment of these enzymes along with many tRNA-specific methyltransferases potentially suggests tRNAs are modified differently in the ovaries than the whole body. However, tRNA modifications do not always reflect expression levels and therefore the tissue-specific tRNA modifications must be assessed for insight on trends.
Other enzymes upregulated in the ovaries were for anticodon loop modifications. Both Q transferase subunits (i.e., QTRT1 and QTRT2) had higher expression in the ovaries than the whole body. Q transferase catalyzes the exchange of guanosine with Q at the wobble position of GUN anticodons (Harada and Nishimura, 1972). Several enzymes that modify position 37 were also enriched in the ovaries. While the transcripts for the enzyme that catalyzes the formation of t6A were in lower abundance, the enzymes involved in the formation of t6A-hypermodifications, m6t6A and ms2t6A, exhibited higher expression in the ovaries. Therefore, several anticodon modifications also demonstrate differing transcript levels in reproductive organs which may indicate the tRNA modification demand is tissue-specific.
4. Discussion
Altogether, we present that female mosquitoes have a higher abundance of tRNA modifications than male mosquitoes. This is supported by higher transcript levels in females for genes associated with tRNA modifications evaluated in six mosquito species (Figure 1). This sex-associated impact is more substantial in some species over others, but even so, this species does show more enrichment. Tissue-specific analyses highlighted that there is high enrichment in the ovaries, suggesting that the increase in modifications and enzyme expression in females may be associated with reproduction. The substantial differences between sexes and transcript levels in reproductive organs suggest that tRNA modifications could have critical importance to mosquito biology.
The tRNA modifications detected in mosquito tRNA are largely shared between the three species (Figure 2). While A. stephensi males had the least detectable modifications, the females followed the other species and thirty-three modifications were present. Of the thirty-three modifications characterized (Table 1), at least nine are known to be located at the wobble position of tRNAs (Boccaletto et al., 2018). As for the other anticodon modifications, six nucleosides were identified that have been previously reported at the position adjacent to the anticodon (Boccaletto et al., 2018). Intriguingly the majority of differences between the males and females were modifications on the anticodon loop. As anticodon loop modifications affect the decoding of codons, the sex-specific differences suggest females have different codon usage than males. Oligonucleotide analysis to map the modifications would confirm the locations of tRNA modifications. However, this would be a major challenge since there are more than 300 tRNA genes in these mosquito species (Eng et al., 2018; Lawson et al., 2009). Such a large number of potential tRNAs, mapping the position of modifications would likely require isolation of individual tRNAs and may still be a challenge. As tRNA modifications are highly conserved, many modifications are found on certain tRNA types and in specific locations on the tRNA molecule. Due to this conserved nature, the inferences of modifications on types of tRNA are from reports in other organisms for which the data is available (Boccaletto et al., 2018). Overall, the census of tRNA modifications is largely the same with 79% of the modifications being shared in mosquito tRNA.
A blood meal is required for reproduction in all three of the mosquito species assessed (Attardo et al., 2005; Hansen et al., 2014). However, a blood meal also induces several stresses on the organism including thermal stress (Benoit et al., 2019, 2010; Benoit and Denlinger, 2017). Chemical modifications to tRNA contribute to thermal stability and thermophilic organisms often have higher levels of methylation in tRNAs (Hori et al., 2018; McCloskey et al., 2000). For example, in the thermophile Thermus thermophilus unmodified phenylalanine tRNAs had a lower melting temperature than those of modified transcripts (Tomikawa et al., 2010). Furthermore, 2’-O-methylations (Am, Gm, Cm, Um, Ψm) increased the melting temperature of tRNA by more than 20 oC in another thermophile (Noon et al., 2003). Thus, tRNA modifications are a strategy to combat elevated temperatures by reinforcing structure and stability (Hori, 2014). In female mosquitoes, four methylations are in higher abundance in tRNA of A. aegypti and A. stephensi. In addition, C. pipiens females had one modification more abundant in tRNA and it is the methyl modification, m6A. While additional functional studies are necessary, the higher incidence of methylations in tRNAs of female mosquitoes is potentially in preparation for the thermal stress that will occur during a blood meal.
Substantial transcriptome remodeling also occurs in females in the hours following a blood meal. For example, Culex quinquefasciatus and other mosquitoes upregulate the egg yolk protein vitellogenin post-blood meal (Isoe and Hagedorn, 2007; Reid et al., 2015). This is accompanied by many other enriched transcripts such as those for cytochrome p450, trypsins, and lipases (Reid et al., 2015). The remodeling of the transcriptome in response to a blood meal likely impacts codon usage as well. In turn, tRNA modifications in females may be necessary to account for the changing codon demand. Many modifications were in higher abundance in females compared to males (Figure 3). The linear regression model indicated that the majority of transcript levels and modification abundances are correlated with sex (Table 2). This relationship was further evaluated through permanova which also indicated sex is significant in relation to species for both modification abundances (P < 0.001) and transcript levels (P < 0.001). Two of the enriched enzymes catalyze the transfer of the modification Q onto G34 of tRNAs. Hypomodified histidine tRNAs with G34 exhibit preference toward the codon CAC while the presence of Q34 demonstrates no preference and can decode CAC or CAU (Meier et al., 1985). In the tobacco mosaic virus, a hypomodified tyrosine tRNA with G34 suppresses stop codons by failing to halt protein synthesis at the stop codon (Beier and Grimm, 2001; Bienz and Kubli, 1981). In both cases, the presence of Q on the wobble position affects the decoding of codons and subsequently improves translational efficiency. The abundance of Q correlates with the transcript levels of QTRT1. However, the levels of modification were not always in agreement with transcript abundance and are likely influenced by species (Figure 4).
As Q and other anticodon modifications are in higher abundance in females, the enrichment of these enzymes suggests anticodon modifications may be important for translational efficiency in mosquitoes. Anticodon modifications are also sex-specific in mosquitoes and the expression of enzymes involved in anticodon modification is higher in the ovaries than the whole body in A. aegypti. The majority of tRNA-modifying enzymes have higher transcript levels in the ovaries. In addition, there is little overlap with male-specific tissues such as the testes and accessory glands (Figure 5). This observation suggests tRNA modification is likely related to reproductive tissues which is in support of potential differences in codon usage between tissues.
Some limitations must be addressed to better understand the role of higher tRNA modification in females and their significance in reproduction. Primarily, the method of assessing modification levels is in relative abundance and not absolute abundance. Ideally, the absolute amount of each nucleoside would illustrate the differences between males and females. Additionally, the differences in tRNA modifications detected and their abundances may be caused by differences within the tRNA pool. Previous work in Anopheles gambiae notes tRNA transcript abundances are not tissue-specific (Bryant et al., 2020). While it is still possible the tRNA pool may influence relative abundance of modifications, tRNA gene numbers for each species is similar (Table S2 & Table S3). Since the tRNA gene numbers are assumed to correlate with the tRNA levels (Tuller et al., 2010), it is likely the differences in modification abundances are minimally impacted by tRNA pool composition. Furthermore, there are discrepancies in the enzyme transcript levels and tRNA modification abundance. In C. pipiens, nearly all of the tRNA modification abundances are the same regardless of sex. However, tRNA-modifying enzyme expression is consistently higher in females than in males. This demonstrates that the tRNA-modifying enzyme expression is not always reflective of the modification status. This complicated relationship between tRNA-modifying enzyme expression and tRNA modifications in mosquitoes suggests that this may be a fruitful and underexplored area of mosquito biology.
Altogether, we have shown that tRNA modifying enzyme expression and tRNA modification abundance is sex-associated in mosquitoes. The consistent elevation of tRNA modification and enzyme transcript levels suggests that female mosquitoes require more mechanisms for translational efficiency. Modifications can contribute to the thermal stability of tRNAs which may be required in females coping with a blood meal. Additionally, the higher levels of anticodon modification in females could suggest preparation for altered codon usage in transcripts necessary for blood meal response and egg production. However, future studies to evaluate the effects of a blood meal on tRNA modifications will provide more insight into their relevance concerning blood digestion and reproduction.
Supplementary Material
Figure S1. Confirmation of tRNA in Anopheles stephensi samples. 1% agarose gel on A. stephensi tRNA samples. From left to right samples are female-1, female-2, female-3, male-1, male-2, male-3, yeast tRNA.
Figure S2. Confirmation of tRNA in Aedes aegypti samples. 1% agarose gel on A. aegypti tRNA samples confirming presence of tRNA. From left to right the samples are male-1, male-2, male-3, female-1, female-2, female-3, yeast tRNA.
Figure S3. Confirmation of tRNA in Culex pipiens samples. 1% agarose gel on C. pipiens tRNA samples confirming presence of tRNA. From left to right the samples are female-1, female-2, female-3, male-1, male-2, and male-3.
Figure S4. The peak area of the internal standard spiked into each sample for LC-MS/MS. The internal standard of 5-bromo-2’-deoxycytidine (m/z = 306.0078) shares similar properties to the analytes of interest. The average peak area is shown for three replicates and error bars are standard deviation.
Highlights.
Transcript levels of tRNA-modifying enzymes are higher in female mosquitoes.
33 modifications are detected in tRNA of mosquitoes using LC-MS/MS.
Abundances of tRNA modifications are sex-associated in mosquitoes.
tRNA-modifying enzyme transcripts are higher in reproductive tissues in A. aegypti.
5. Acknowledgements
Research reported in this publication was partially supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number R01AI148551 (to J.B.B.) and the National Science Foundation (CHE 1507357 to P.A.L). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations
- RNA
ribonucleic acid
- tRNA
transfer RNA
- LC-MS/MS
liquid chromatography tandem mass spectrometry
- rRNA
ribosomal RNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest: none
References
- Agris PF, Vendeix FAP, Graham WD, 2007. tRNA’s wobble decoding of the genome: 40 years of modification. J. Mol. Biol 366, 1–13. [DOI] [PubMed] [Google Scholar]
- Aryan A, Anderson MAE, Biedler JK, Qi Y, Overcash JM, Naumenko AN, Sharakhova MV, Mao C, Adelman ZN, Tu Z, 2020. Nix alone is sufficient to convert female Aedes aegypti into fertile males and myo-sex is needed for male flight. Proc. Natl. Acad. Sci. U. S. A 117, 17702–17709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attardo GM, Abd-Alla AMM, Acosta-Serrano A, Allen JE, Bateta R, Benoit JB, Bourtzis K, Caers J, Caljon G, Christensen MB, Farrow DW, Friedrich M, Hua-Van A, Jennings EC, Larkin DM, Lawson D, Lehane MJ, Lenis VP, Lowy-Gallego E, Macharia RW, Malacrida AR, Marco HG, Masiga D, Maslen GL, Matetovici I, Meisel RP, Meki I, Michalkova V, Miller WJ, Minx P, Mireji PO, Ometto L, Parker AG, Rio R, Rose C, Rosendale AJ, Rota-Stabelli O, Savini G, Schoofs L, Scolari F, Swain MT, Takáč P, Tomlinson C, Tsiamis G, Van Den Abbeele J, Vigneron A, Wang J, Warren WC, Waterhouse RM, Weirauch MT, Weiss BL, Wilson RK, Zhao X, Aksoy S, 2019. Comparative genomic analysis of six Glossina genomes, vectors of African trypanosomes. Genome Biol 20, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attardo GM, Hansen IA, Raikhel AS, 2005. Nutritional regulation of vitellogenesis in mosquitoes: implications for anautogeny. Insect Biochem. Mol. Biol 35, 661–675. [DOI] [PubMed] [Google Scholar]
- Balatsos G, Puggioli A, Karras V, Lytra I, Mastronikolos G, Carrieri M, Papachristos DP, Malfacini M, Stefopoulou A, Ioannou CS, Balestrino F, Bouyer J, Petrić D, Pajović I, Kapranas A, Papadopoulos NT, Milonas PG, Bellini R, Michaelakis A, 2021. Reduction in egg fertility of Aedes albopictus mosquitoes in Greece following releases of imported sterile males. Insects 12. 10.3390/insects12020110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Bolker BM, Machler M, Walker SC, 2015. Fitting linear mixed-effects models using lme4. J of Stat Softw 10.18637/jss.v067.i01 [DOI]
- Beier H, Grimm M, 2001. Misreading of termination codons in eukaryotes by natural nonsense suppressor tRNAs. Nucleic Acids Res 29, 4767–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benelli G, Jeffries CL, Walker T, 2016. Biological control of mosquito vectors: past, present, and future. Insects 7. 10.3390/insects7040052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit JB, Denlinger DL, 2017. Bugs battle stress from hot blood. Elife 10.7554/eLife.33035 [DOI] [PMC free article] [PubMed]
- Benoit JB, Lazzari CR, Denlinger DL, Lahondère C, 2019. Thermoprotective adaptations are critical for arthropods feeding on warm-blooded hosts. Curr Opin Insect Sci 34, 7–11. [DOI] [PubMed] [Google Scholar]
- Benoit JB, Lopez-Martinez G, Phillips ZP, Patrick KR, Denlinger DL, 2010. Heat shock proteins contribute to mosquito dehydration tolerance. J. Insect Physiol 56, 151–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienz M, Kubli E, 1981. Wild-type tRNATyrG reads the TMV RNA stop codon, but Q base-modified tRNATyrQ does not. Nature 294, 188–190. [DOI] [PubMed] [Google Scholar]
- Björk GR, Huang B, Persson OP, Byström AS, 2007. A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA 13, 1245–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, de Crécy-Lagard V, Ross R, Limbach PA, Kotter A, Helm M, Bujnicki JM, 2018. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res 46, D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B, 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant WB, Ray S, Mills MK, 2020. Global analysis of small non-coding RNA Populations across tissues in the malaria vector, Anopheles gambiae. Insects 11. 10.3390/insects11070406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo C, Ahmed-Braimah YH, Alexandra Amaro I, Harrington LC, Wolfner MF, Avila FW, 2020. Mating and blood-feeding induce transcriptome changes in the spermathecae of the yellow fever mosquito Aedes aegypti. Sci. Rep 10, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CTY, Pang YLJ, Deng W, Ramesh Babu I, Dyavaiah M, Begley TJ, Dedon PC, 2012. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat. Commun 3, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PP, Lin BY, Mak AJ, Lowe TM, 2021. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res 49, 9077–9096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charette M, Gray MW, 2000. Pseudouridine in RNA: what, where, how, and why. IUBMB Life 49, 341–351. [DOI] [PubMed] [Google Scholar]
- Dai W, Li A, Yu NJ, Nguyen T, Leach RW, Wühr M, Kleiner RE, 2021. Activity-based RNA-modifying enzyme probing reveals DUS3L-mediated dihydrouridylation. Nat. Chem. Biol 17, 1178–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalluge JJ, Hashizume T, Sopchik AE, McCloskey JA, Davis DR, 1996. Conformational flexibility in RNA: the role of dihydrouridine. Nucleic Acids Res 24, 1073–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durant PC, Bajji AC, Sundaram M, Kumar RK, Davis DR, 2005. Structural effects of hypermodified nucleosides in the Escherichia coli and human tRNALys anticodon loop: the effect of nucleosides s2U, mcm5U, mcm5s2U, mnm5s2U, t6A, and ms2t6A. Biochemistry 44, 8078–8089. [DOI] [PubMed] [Google Scholar]
- Edvardson S, Prunetti L, Arraf A, Haas D, Bacusmo JM, Hu JF, Ta-Shma A, Dedon PC, de Crecy-Lagard V, Elpeleg O, 2017. tRNA N6-adenosine threonylcarbamoyltransferase defect due to KAE1/TCS3 (OSGEP) mutation manifest by neurodegeneration and renal tubulopathy. Eur. J. Hum. Genet 25, 545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AM, Addo MA, Dos Santos PC, 2020. Extracurricular functions of tRNA modifications in microorganisms. Genes 11. 10.3390/genes11080907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng MW, Clemons A, Hill C, Engel R, Severson DW, Behura SK, 2018. Multifaceted functional implications of an endogenously expressed tRNA fragment in the vector mosquito Aedes aegypti. PLoS Negl. Trop. Dis 12, e0006186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Vázquez J, Vargas-Pérez I, Sansó M, Buhne K, Carmona M, Paulo E, Hermand D, Rodríguez-Gabriel M, Ayté J, Leidel S, Hidalgo E, 2013. Modification of tRNA(Lys) UUU by elongator is essential for efficient translation of stress mRNAs. PLoS Genet 9, e1003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamez S, Antoshechkin I, Mendez-Sanchez SC, Akbari OS, 2020. The developmental transcriptome of Aedes albopictus, a major worldwide human disease vector. G3 Genes|Genomes|Genetics 10, 1051–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge J, Yu Y-T, 2013. RNA pseudouridylation: new insights into an old modification. Trends Biochem. Sci 38, 210–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo-Calderón GI, Emrich SJ, MacCallum RM, Maslen G, Dialynas E, Topalis P, Ho N, Gesing S, VectorBase Consortium, Madey G, Collins FH, Lawson D, 2015. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res 43, D707–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoraki L, Grau-Bové X, Carrington Yates H, Lycett GJ, Ranson H, 2020. Isolation and transcriptomic analysis of Anopheles gambiae oenocytes enables the delineation of hydrocarbon biosynthesis. Elife 9. 10.7554/eLife.58019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy MP, Phizicky EM, 2015. Conservation of an intricate circuit for crucial modifications of the tRNAPhe anticodon loop in eukaryotes. RNA 21, 61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen IA, Attardo GM, Rodriguez SD, Drake LL, 2014. Four-way regulation of mosquito yolk protein precursor genes by juvenile hormone-, ecdysone-, nutrient-, and insulin-like peptide signaling pathways. Front. Physiol 5, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada F, Nishimura S, 1972. Possible anticodon sequences of tRNAHis, tRNAAsn, and tRNAAsp from Escherichia coli. Universal presence of nucleoside O in the first position of the anticodons of these transfer ribonucleic acid. Biochemistry 11, 301–308. [DOI] [PubMed] [Google Scholar]
- Honnen A-C, Johnston PR, Monaghan MT, 2016. Sex-specific gene expression in the mosquito Culex pipiens f. molestus in response to artificial light at night. BMC Genomics 17, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori H, 2014. Methylated nucleosides in tRNA and tRNA methyltransferases. Front. Genet 5, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori H, Kawamura T, Awai T, Ochi A, Yamagami R, Tomikawa C, Hirata A, 2018. Transfer RNA modification enzymes from thermophiles and their modified nucleosides in tRNA. Microorganisms 6. 10.3390/microorganisms6040110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida K, Kunibayashi T, Tomikawa C, Ochi A, Kanai T, Hirata A, Iwashita C, Hori H, 2011. Pseudouridine at position 55 in tRNA controls the contents of other modified nucleotides for low-temperature adaptation in the extreme-thermophilic eubacterium Thermus thermophilus. Nucleic Acids Res 39, 2304–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isoe J, Hagedorn HH, 2007. Mosquito vitellogenin genes: comparative sequence analysis, gene duplication, and the role of rare synonymous codon usage in regulating expression. J. Insect Sci 7, 1–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaswal V, Ndo C, Ma H-C, Clifton BD, Pombi M, Cabrera K, Couhet A, Mouline K, Diabaté A, Dabiré R, Ayala D, Ranz JM, 2021. Intraspecific transcriptome variation and sex-biased expression in Anopheles arabiensis. Genome Biol. Evol 13. 10.1093/gbe/evab199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MJO, Esberg A, Huang B, Björk GR, Byström AS, 2008. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol. Cell. Biol 28, 3301–3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jora M, Burns AP, Ross RL, Lobue PA, Zhao R, Palumbo CM, Beal PA, Addepalli B, Limbach PA, 2018. Differentiating positional isomers of nucleoside modifications by higher-energy collisional dissociation mass spectrometry (HCD MS). J. Am. Soc. Mass Spectrom 29, 1745–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai G, Ue H, Yasuda M, Sakamoto K, Hashizume T, McCloskey JA, Miyazawa T, Yokoyama S, 1991. Relation between functions and conformational characteristics of modified nucleosides found in tRNAs. Nucleic Acids Symp. Ser 49–50. [PubMed]
- Khan SM, Franke-Fayard B, Mair GR, Lasonder E, Janse CJ, Mann M, Waters AP, 2005. Proteome analysis of separated male and female gametocytes reveals novel sex-specific Plasmodium biology. Cell 121, 675–687. [DOI] [PubMed] [Google Scholar]
- Klassen R, Ciftci A, Funk J, Bruch A, Butter F, Schaffrath R, 2016. tRNA anticodon loop modifications ensure protein homeostasis and cell morphogenesis in yeast. Nucleic Acids Res 44, 10946–10959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh CS, Sarin LP, 2018. Transfer RNA modification and infection - implications for pathogenicity and host responses. Biochim. Biophys. Acta Gene Regul. Mech 1861, 419–432. [DOI] [PubMed] [Google Scholar]
- Kurth HM, Mochizuki K, 2009. 2’-O-methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. RNA 15, 675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson D, Arensburger P, Atkinson P, Besansky NJ, Bruggner RV, Butler R, Campbell KS, Christophides GK, Christley S, Dialynas E, Hammond M, Hill CA, Konopinski N, Lobo NF, MacCallum RM, Madey G, Megy K, Meyer J, Redmond S, Severson DW, Stinson EO, Topalis P, Birney E, Gelbart WM, Kafatos FC, Louis C, Collins FH, 2009. VectorBase: a data resource for invertebrate vector genomics. Nucleic Acids Res 37, D583–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C-J, Smibert P, Zhao X, Hu JF, Ramroop J, Kellner SM, Benton MA, Govind S, Dedon PC, Sternglanz R, Lai EC, 2015. An extensive allelic series of Drosophila kae1 mutants reveals diverse and tissue-specific requirements for t6A biogenesis. RNA 21, 2103–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz C, Lünse CE, Mörl M, 2017. tRNA modifications: impact on structure and thermal adaptation. Biomolecules 7. 10.3390/biom7020035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S, 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BJ, Dudchenko O, Kingan SB, Koren S, Antoshechkin I, Crawford JE, Glassford WJ, Herre M, Redmond SN, Rose NH, Weedall GD, Wu Y, Batra SS, Brito-Sierra CA, Buckingham SD, Campbell CL, Chan S, Cox E, Evans BR, Fansiri T, Filipović I, Fontaine A, Gloria-Soria A, Hall R, Joardar VS, Jones AK, Kay RGG, Kodali VK, Lee J, Lycett GJ, Mitchell SN, Muehling J, Murphy MR, Omer AD, Partridge FA, Peluso P, Aiden AP, Ramasamy V, Rašić G, Roy S, Saavedra-Rodriguez K, Sharan S, Sharma A, Smith ML, Turner J, Weakley AM, Zhao Z, Akbari OS, Black WC 4th, Cao H, Darby AC, Hill CA, Johnston JS, Murphy TD, Raikhel AS, Sattelle DB, Sharakhov IV, White BJ, Zhao L, Aiden EL, Mann RS, Lambrechts L, Powell JR, Sharakhova MV, Tu Z, Robertson HM, McBride CS, Hastie AR, Korlach J, Neafsey DE, Phillippy AM, Vosshall LB, 2018. Improved reference genome of Aedes aegypti informs arbovirus vector control. Nature 563, 501–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleverty CJ, Hornsby M, Spraggon G, Kreusch A, 2007. Crystal structure of human Pus10, a novel pseudouridine synthase. J. Mol. Biol 373, 1243–1254. [DOI] [PubMed] [Google Scholar]
- McCloskey JA, Liu XH, Crain PF, Bruenger E, Guymon R, Hashizume T, Stetter KO, 2000. Posttranscriptional modification of transfer RNA in the submarine hyperthermophile Pyrolobus fumarii. Nucleic Acids Symp. Ser 267–268. [DOI] [PubMed]
- Meier F, Suter B, Grosjean H, Keith G, Kubli E, 1985. Queuosine modification of the wobble base in tRNAHis influences “in vivo” decoding properties. EMBO J 4, 823–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B, Immer C, Kaiser S, Sharma S, Yang J, Watzinger P, Weiß L, Kotter A, Helm M, Seitz H-M, Kötter P, Kellner S, Entian K-D, Wöhnert J, 2019. Identification of the 3-amino-3-carboxypropyl (acp) transferase enzyme responsible for acp3U formation at position 47 in Escherichia coli tRNAs. Nucleic Acids Res 48, 1435–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin Y, Helm M, 2010. tRNA stabilization by modified nucleotides. Biochemistry 49, 4934–4944. [DOI] [PubMed] [Google Scholar]
- Nishikura K, 2016. A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol 17, 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura S, 1983. Structure, Biosynthesis, and Function of Queuosine in Transfer RNA, in: Cohn WE (Ed.), Progress in Nucleic Acid Research and Molecular Biology Academic Press, pp. 49–73. [DOI] [PubMed] [Google Scholar]
- Noma A, Kirino Y, Ikeuchi Y, Suzuki T, 2006. Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J 25, 2142–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noon KR, Guymon R, Crain PF, McCloskey JA, Thomm M, Lim J, Cavicchioli R, 2003. Influence of temperature on tRNA modification in archaea: Methanococcoides burtonii (optimum growth temperature [Topt], 23°C) and Stetteria hydrogenophila (Topt, 95°C). J. Bacteriol 185, 5483–5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Mount SM, Kingsford C, 2014. Sailfish enables alignment-free isoform quantification from RNA-seq reads using lightweight algorithms. Nat. Biotechnol 32, 462–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Q, Curran JF, Björk GR, 1998. The methyl group of the N6-methyl-N6-threonylcarbamoyladenosine in tRNA of Escherichia coli modestly improves the efficiency of the tRNA. J. Bacteriol 180, 1808–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid WR, Zhang L, Liu N, 2015. Temporal gene expression profiles of pre blood-fed adult females immediately following eclosion in the southern house mosquito Culex quinquefasciatus. Int. J. Biol. Sci 11, 1306–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezgui VAN, Tyagi K, Ranjan N, Konevega AL, Mittelstaet J, Rodnina MV, Peter M, Pedrioli PGA, 2013. tRNA tKUUU, tQUUG, and tEUUC wobble position modifications fine-tune protein translation by promoting ribosome A-site binding. Proc. Natl. Acad. Sci. U. S. A 110, 12289–12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Benítez D, Eggers C, Glavic A, 2017. Modulation of the proteostasis machinery to overcome stress caused by diminished levels of t6A-modified tRNAs in Drosophila. Biomolecules 7. 10.3390/biom7010025 [DOI] [Google Scholar]
- Ross RL, Cao X, Limbach PA, 2017. Mapping post-transcriptional modifications onto transfer ribonucleic acid sequences by liquid chromatography tandem mass spectrometry. Biomolecules 7. 10.3390/biom7010021 [DOI] [Google Scholar]
- Scott MJ, Benoit JB, Davis RJ, Bailey ST, Varga V, Martinson EO, Hickner PV, Syed Z, Cardoso GA, Torres TT, Weirauch MT, Scholl EH, Phillippy AM, Sagel A, Vasquez M, Quintero G, Skoda SR, 2020. Genomic analyses of a livestock pest, the New World screwworm, find potential targets for genetic control programs. Commun Biol 3, 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shettima A, Joseph S, Ishak IH, Abdul Raiz SH, Abu Hasan H, Othman N, 2021. Evaluation of total female and male Aedes aegypti proteomes reveals significant predictive protein-protein interactions, functional ontologies, and differentially abundant proteins. Insects 12. 10.3390/insects12080752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinola M, Galvan A, Pignatiello C, Conti B, Pastorino U, Nicander B, Paroni R, Dragani TA, 2005. Identification and functional characterization of the candidate tumor suppressor gene TRIT1 in human lung cancer. Oncogene 24, 5502–5509. [DOI] [PubMed] [Google Scholar]
- Takakura M, Ishiguro K, Akichika S, Miyauchi K, Suzuki T, 2019. Biogenesis and functions of aminocarboxypropyluridine in tRNA. Nat. Commun 10, 5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomikawa C, Yokogawa T, Kanai T, Hori H, 2010. N7-Methylguanine at position 46 (m7G46) in tRNA from Thermus thermophilus is required for cell viability at high temperatures through a tRNA modification network. Nucleic Acids Res 38, 942–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, Zaborske J, Pan T, Dahan O, Furman I, Pilpel Y, 2010. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell 141, 344–354. [DOI] [PubMed] [Google Scholar]
- UniProt Consortium, 2021. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res 49, D480–D489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Matuszek Z, Huang Y, Parisien M, Dai Q, Clark W, Schwartz MH, Pan T, 2018. Queuosine modification protects cognate tRNAs against ribonuclease cleavage. RNA 24, 1305–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenbach J, Grosjean H, 1981. Effect of threonylcarbamoyl modification (t6A) in yeast tRNA Arg III on codon-anticodon and anticodon-anticodon interactions, a thermodynamic and kinetic evaluation. Eur. J. Biochem 116, 207–213. [DOI] [PubMed] [Google Scholar]
- Wolf J, Gerber AP, Keller W, 2002. tadA, an essential tRNA-specific adenosine deaminase from Escherichia coli. EMBO J 21, 3841–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization, 2020. Vector-borne disease WHO. [Google Scholar]
- Xu Y, Dong Y, Xu Y, Lai Z, Jin B, Hao Y, Gao Y, Sun Y, Chen X-G, Gu J, 2019. Differentiation of long non-coding RNA and mRNA expression profiles in male and female Aedes albopictus. Front. Genet 10, 975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Tanaka Y, Yamashita K, Suzuki T, Nakamura A, Hirano N, Suzuki T, Yao M, Tanaka I, 2011. Molecular basis of dihydrouridine formation on tRNA. Proc. Natl. Acad. Sci. U. S. A 108, 19593–19598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallot R, Ross R, Chen W-H, Bruner SD, Limbach PA, de Crécy-Lagard V, 2017. Identification of a novel epoxyqueuosine reductase family by comparative genomics. ACS Chem. Biol 12, 844–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Confirmation of tRNA in Anopheles stephensi samples. 1% agarose gel on A. stephensi tRNA samples. From left to right samples are female-1, female-2, female-3, male-1, male-2, male-3, yeast tRNA.
Figure S2. Confirmation of tRNA in Aedes aegypti samples. 1% agarose gel on A. aegypti tRNA samples confirming presence of tRNA. From left to right the samples are male-1, male-2, male-3, female-1, female-2, female-3, yeast tRNA.
Figure S3. Confirmation of tRNA in Culex pipiens samples. 1% agarose gel on C. pipiens tRNA samples confirming presence of tRNA. From left to right the samples are female-1, female-2, female-3, male-1, male-2, and male-3.
Figure S4. The peak area of the internal standard spiked into each sample for LC-MS/MS. The internal standard of 5-bromo-2’-deoxycytidine (m/z = 306.0078) shares similar properties to the analytes of interest. The average peak area is shown for three replicates and error bars are standard deviation.


