SUMMARY
To elucidate mechanisms by which T-cells eliminate leukemia, we study donor lymphocyte infusion (DLI), an established immunotherapy for relapsed leukemia. We model T-cell dynamics by integrating longitudinal, multimodal data from 94,517 bone-marrow-derived single T-cell transcriptomes in addition to chromatin accessibility and single T-cell receptor sequencing from patients undergoing DLI. We find that responsive tumors are defined by enrichment of late-differentiated T-cells before DLI, and rapid, durable expansion of early-differentiated T-cells after treatment, highly similar to ‘terminal’ and ‘precursor’ exhausted subsets, respectively. Resistance, by contrast, is defined by heterogeneous T-cell dysfunction. Surprisingly, early-differentiated T-cells in responders mainly originate from pre-existing and novel clonotypes recruited to the leukemic microenvironment, rather than the infusion. Our work provides a paradigm for analyzing longitudinal single-cell profiling of scenarios beyond adoptive cell therapy, and introduces Symphony, a Bayesian approach to infer regulatory circuitry underlying T-cell subsets, with broad relevance to exhaustion antagonists across cancers.
Keywords: immunotherapy, donor lymphocyte infusion, leukemia, scRNA-seq, probabilistic models, statistical machine learning, exhaustion, ATAC-seq, gene regulatory networks, allogeneic hematopoietic stem cell transplant
INTRODUCTION
Despite the potency of cancer immunotherapy for a subset of patients with cancer, the variability in responses and efficacy suggests that the fundamental mechanisms, cell types and pathways driving clinical outcomes remain elusive (Yofe et al., 2020). Single-cell transcriptomic profiling is a powerful technology that can characterize the full range of immune cell states and gene programs in the tumor microenvironment (TME) in a comprehensive and unbiased manner. Studying the evolution of the TME at single-cell resolution before and after therapy can thus reveal how heterogeneous cell states evolve in relation to distinct clinical outcomes and illuminate the molecular and cellular determinants of immunotherapeutic response or resistance (Lesterhuis et al., 2017; Yofe et al., 2020). However, working with highly variable patient material presents unique confounding factors and logistical challenges that can hinder high-resolution studies of such temporal dynamics.
Here, we develop strategies to overcome these limitations and apply them to a single-cell study of the human TME undergoing immunotherapy, using serial biopsies from the same patients before and after treatment. As an instructive demonstration, we focus on donor lymphocyte infusion (DLI), a widely used adoptive cellular immunotherapy for relapsed leukemia after allogeneic stem cell transplant. The clear, binary outcomes of response or resistance; the clinical samples collected over a multi-year time-span; and the lack of confounding chemotherapy or immunomodulators have made DLI an attractive immunotherapeutic setting to study the essential ‘search and destroy’ functions of donor-derived T cell responses that underlie the therapeutic graft-versus-leukemia (GvL) effect of allo-SCT (Bachireddy and Wu, 2014; Jenq and van den Brink, 2010). Over the last 30 years, DLI has directly demonstrated the potency of GvL by inducing durable molecular remissions in ~75% of patients with relapsed chronic myelogenous leukemia (CML) following allo-SCT (Collins et al., 1997; Kolb et al., 1995). These experiences have provided the foundation for the current development of newer generations of effective adoptive cellular therapies (Schmid et al., 2021).
Response to DLI modified by CD8-depletion has been associated with decreased toxicity (Alyea et al., 1998; Champlin et al., 1991; Giralt et al., 1995; Soiffer et al., 2002), increased T cell receptor (TCR) repertoire diversity (Claret et al., 1997), expansion of endogenous, tumor-specific, marrow resident CD8+ T cells (Zhang et al., 2010), and reversal of T cell exhaustion (Bachireddy et al., 2014). Similar observations in acute myelogenous leukemia (Liu et al., 2018) suggest that the study of DLI in CML can reveal insights that are broadly relevant across hematologic malignancies. Yet despite the long-established use of DLI for the treatment of relapsed disease following allo-SCT (Collins et al., 1997; Porter et al., 1994; Schmid et al., 2021), the mechanistic basis for its effectiveness remains incompletely understood. While allo-SCT is no longer a first-line therapy for CML, we hypothesized that studying the biological basis for its increased DLI sensitivity would elucidate the pathways driving GvL clinical outcomes and inform therapeutic strategies to prevent or treat relapse following allo-SCT for which DLI remains a standard of care therapy.
To identify the T cell subsets mediating DLI resistance, response and exhaustion after DLI therapy, we analyze single-cell T cell transcriptomes, bulk chromatin accessibility profiles, and single T cell clonality data from bone marrow biopsies of a longitudinal cohort of patients with relapsed CML after allo-SCT treated with DLI (Alyea et al., 1998). We introduce computational models to integrate data across multiple timepoints and modalities and use this framework to detect and characterize the intratumoral T cells whose divergent dynamics and regulatory circuitries define immunotherapeutic response. Our findings link the hierarchy of ‘terminal’ and ‘precursor’ exhausted T cell subsets directly to immunotherapeutic responses in human leukemia, extend their relevance beyond checkpoint blockade to adoptive cellular therapies, and nominate this cellular program as a potent effector of the graft-versus-leukemia effect. Finally, we present a general computational framework for modeling the temporal dynamics of therapy response, applicable also to other cancer types and therapeutic scenarios beyond oncology.
RESULTS
A high-resolution map of T cell states in the leukemic microenvironment
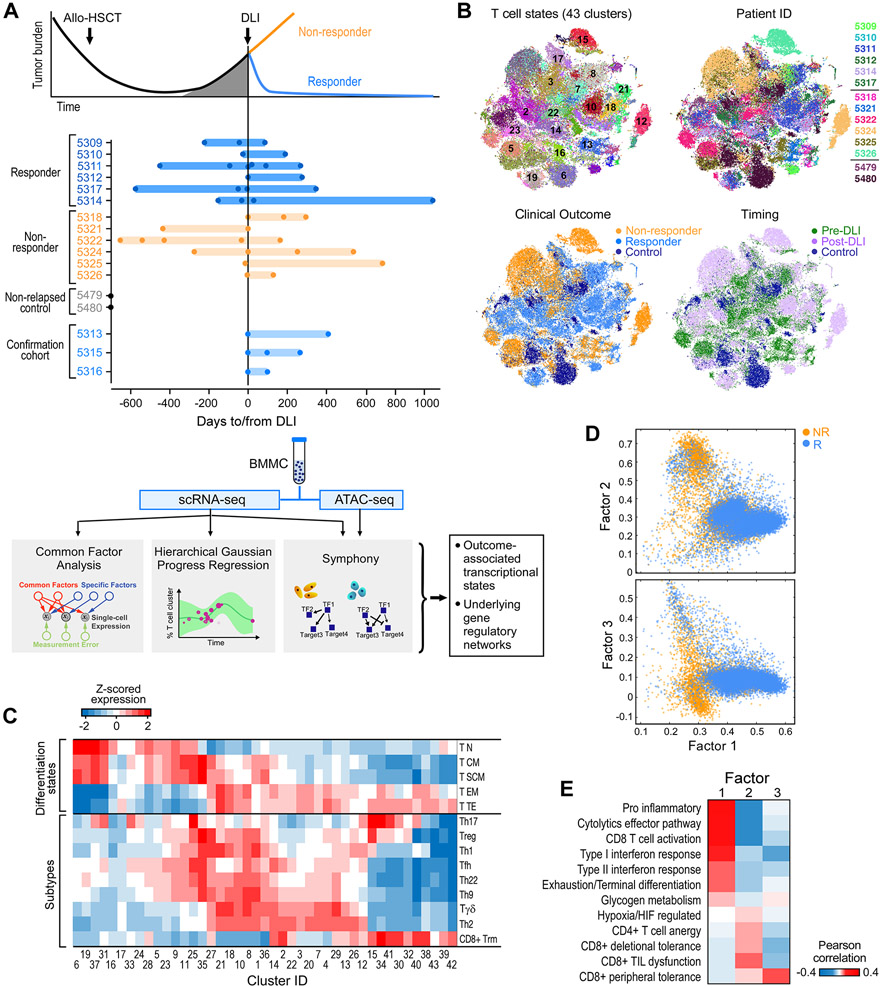
To delineate the evolving landscape of cellular phenotypic states for marrow-infiltrating T cells in relation to DLI therapy, we assembled a cohort of 12 patients treated with CD8-depleted DLI for relapsed CML (Alyea et al., 1998). Six patients were long-term DLI responders (“Rs”), defined as having achieved molecular remission (i.e. RT-PCR negative for the BCR-ABL transcript) after DLI, and 6 were nonresponders (“NRs”), who did not achieve measurable tumor reduction following DLI. None of the patients developed acute graft-versus-host disease (GvHD) after DLI (Table S1). Serial bone marrow (BM) biopsies were collected before and after DLI treatment at a median of 3 timepoints per patient (STAR Methods). The cohorts had comparable timing between allo-SCT and DLI therapy (median 702 (R) and 1064 (NR) days), and between pre- and post-DLI sampling (Figure S1A; Table S1). As reference, we also analyzed post-transplant BM biopsies from two patients with CML who never relapsed after allo-SCT; as an extension cohort, we assembled an independent set of 3 long-term DLI responders. From each of the 46 total BM samples, we obtained scRNA-seq on viable mononuclear cells and, for 41 samples, chromatin accessibility profiles (using ATAC-seq) on isolated CD45RA+ and CD45RA−, CD4+ and CD8+ T cells (Figure 1A, STAR Methods).
Figure 1. Experimental design and global map of T cell states.
(A) Clinical cohorts, and flow chart of experimental and analysis schema. (B) t-SNE projection of normalized scRNA-seq data for all T cells from 41 samples from discovery cohort. Each dot represents a cell colored by cluster, patient ID, clinical outcome and timing respectively (also expanded in Figure S6B). (C) Mean expression for a curated set of transcriptomic signatures representing T cell subtypes and differentiation states for each T cell cluster; expression values are z-scored relative to all T cell clusters. Additional signatures appear in Figure S1D. (D) Common Factor Analysis of T cells identifying 3 common latent factors distinguishing T cells between responders (R) and non-responders (NR). Each dot represents a cell colored by patient outcome and axes show factor loadings. (E) Pearson correlation between common latent factors and mean expression of curated signatures. See also Figures S1, S2, S6 and Tables S1, S2.
In total, from the discovery cohort, we identified 381,462 cells that passed our quality metrics, with a median of 8735 cells/sample (Table S2). We used Phenograph (Levine et al., 2015) to cluster the data into 62 distinct cell states, including subtypes of T, B, NK, monocytes, progenitor cells and CD34+ stem cells (STAR Methods). Given the established critical role of T cells in the anti-leukemic potency of DLI (Bachireddy and Wu, 2014), we normalized and clustered the 87,939 T cells in our data, using Biscuit (Azizi et al., 2018; Prabhakaran et al., 2016) which robustly accounts for artifacts such as batch effects and library size variation (STAR Methods). This analysis yielded 43 distinct T cell states spanning combinations of subtypes and functional or differentiation states with variably expressed gene programs related to environmental stimuli (Figure 1B,C; Figure S1B-D). For example, clusters 6, 19, 37 and 31 exhibited similar differentiation states and subtypes, for which we observed differential enrichment of pathways involving adenosine suppression, glucose deprivation, and anergy. Thus our global T cell map reveals substantial diversity corresponding to established T cell subtypes and states, marked by known and previously unexplored markers, that are shared across groups of patients.
DLI resistance comprises multiple states of T cell dysfunction
While most T cell clusters were shared across patients, they were variably distributed across clinical features such as timing relative to DLI and clinical outcome (R vs NR) (Figure S1E, Figure 1B), motivating us to identify the gene expression programs that might underlie these clinical variables. We tested standard techniques used to decompose single-cell data to identify trends underlying its variance (Figure S2A), but no principal or diffusion component was associated with R or NR status. Instead, the unsupervised approach of common factor analysis (Zientek, 2008), selected for its potential to uncover latent factors that explain shared variance across T cells while ignoring the portion of variance unique to cells and hence de-emphasizing patient-specific variation, was informative (Figure S2B, STAR Methods). We identified 3 factors that explained 67% of the variation in our data which segregated R and NR T cells (Figure 1D). Co-variation in R T cells was defined by Factor 1, which correlated with profiles associated with T cell activation (i.e. cytolytic effectors, interferon response, glycogen metabolism, CD8+ T cell activation, T cell exhaustion; Figure 1E). We further confirmed enrichment of T cell exhaustion pre-DLI in R compared to NR, as previously observed (Bachireddy et al., 2014) (P<10−6; Figure S2C). In contrast, Factors 2 and 3, which defined the NR T cells, correlated with non-overlapping signatures related to multiple, distinct T cell dysfunctional states (i.e. hypoxia, anergy, peripheral and deletional tolerance, tumor-infiltrating lymphocyte dysfunction; Figure 1E, Figure S2D, STAR Methods), suggesting that DLI resistance may be driven by not one, but multiple types of T cell dysfunction.
DLI response is heralded by pre-treatment enrichment of activated and cytotoxic T cells
Given the substantial diversity of T cell subsets and gene programs in the leukemic microenvironment, we aimed to quantify this heterogeneity and study its change with outcome. T cell states are known to reside on continuous trajectories, which explain the majority of their variation (Azizi et al., 2018; Li et al., 2019a; Singer et al., 2017). We thus quantified their diversity across all clusters using phenotypic volume (Azizi et al., 2018), defined as the pseudo-determinant of covariance between genes. Phenotypic volume serves as a measure of the diversity of co-expressed transcriptional programs, which increases with the number and degree of independence of gene programs (STAR Methods). We found substantially higher phenotypic diversity in pre-DLI Rs compared to pre-DLI NRs (Figure 2A, log fold change=104.6, P<10−6), suggesting that diverse T cell phenotypes pre-DLI could be essential for response.
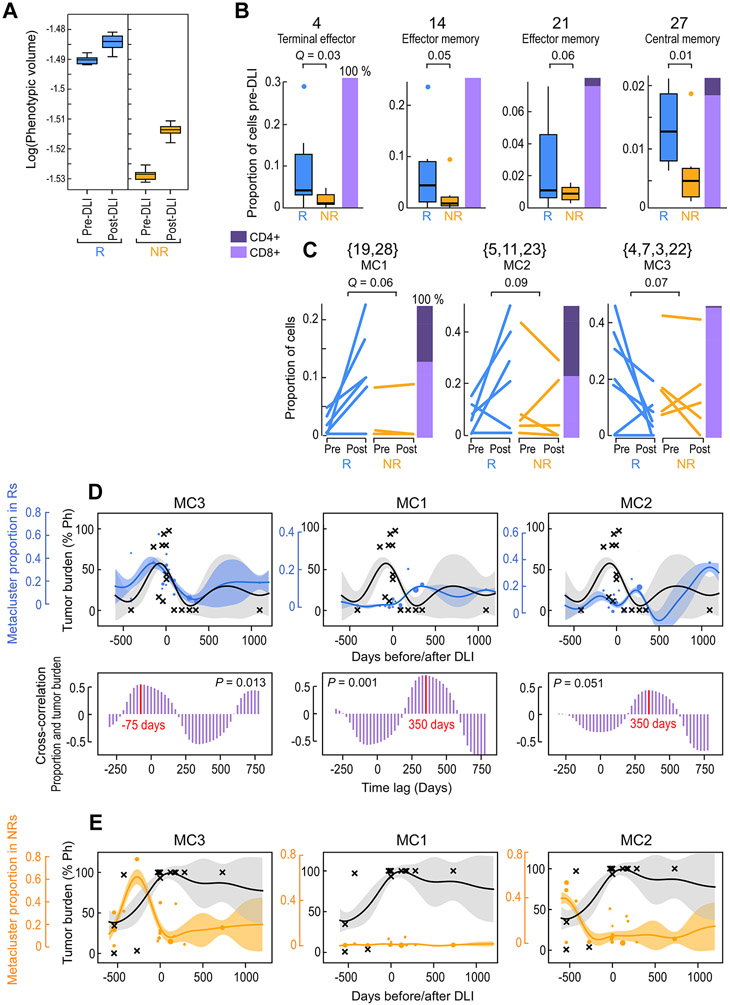
Figure. 2. Identification of outcome-associated transcriptional states and their temporal dynamics.
(A) Phenotypic volume in log-scale (metric of transcriptional diversity (Azizi et al., 2018)) of T cells before and after DLI in n=6 responders (R) and n=5 non-responders (NR). Box plot elements display the center line as median; box limits as first and third quartiles; (B-C) Proportion of T cells for pre-DLI only (B) or paired pre-/post-DLI (C) samples from n=6 Rs and n=5 NRs assigned to the indicated cluster or meta-cluster. Q-values determined from weighted t-test and empirical FDR estimation. Box plot elements display the center line as median; box limits as first and third quartiles; whiskers extend to maximum/minimum data points (A) or 1.5x interquartile range with points as outliers (B). Each line in (C) indicates one patient. Stacked bars on the right indicate the proportion of CD4+ and CD8+ T cells. (D-E) Hierarchical GP regression models (STAR Methods; Figure S3D,E; Figure S6E,F) for both the proportion of the indicated meta-cluster (in blue dots for Rs and in orange dots for NRs) and the percentage of tumor burden (in grey crosses) (indicated by percent positivity of the Philadelphia chromosome) per longitudinal samples from n=6 Rs (D) or n=6 NRs (E). Each dot is one sample and dot size is proportional to sample size (total cells); inferred model mean is shown with lines and shaded area shows +/−1 standard deviation (SD). Cross-correlation plots (D; purple) indicate the time shift between the models for meta-cluster proportion and tumor percentage, showing in-sync dynamics for MC3 and tumor (left) and a lag between MC1/MC2 and tumor (middle, right). See also Figures S3 and S6.
In addition to finding increased overall phenotypic diversity in pre-DLI Rs, we sought to identify distinct transcriptional states associated with clinical outcome. We tested each cluster for enrichment in baseline pre-DLI samples from Rs compared to NRs (Table S2). No cluster was consistently enriched in NRs, attesting to the notion of multiple pathways to DLI resistance rather than a common resistance mechanism shared across NRs. In contrast, within Rs, we identified four individual clusters (4, 14, 21, 27) that were consistently enriched pre-DLI (Figure 2B, FDR<0.1), comprised predominantly CD8+ T cells, and shared expression of genes involved in T cell activation (CD160, HAVCR2, CD38) and cytotoxicity (CRTAM, GNLY, GZMK, GZMB) (Figure S3A). Nevertheless, their distinct differentiation states (4, 14, 21: TEM/TTE; 27: TCM), subtypes (21: Tγδ), and varied expression of chemokine receptors (14: XCL2, CXCR4; 21: CXCR1, CXCR2), tissue residency (14: ITGA1, RGS1; high score for “CD8+ TRM”) and cell cycle (27: CDKN2A, TAF5, RRM2) programs indicated the baseline diversity of these T cell states (Figure 1C, Figure S3A). The majority of these T cell states (i.e. 4, 14, 21) implicate a ‘late differentiated’ program that is enriched in Rs pre-DLI.
We observed a marked increase in the number of T cell clusters in post-DLI samples compared to matched pre-DLI samples (mean 41 [range: 35-46] versus mean 38 [range: 34-41], P<0.001; STAR Methods), and correspondingly, increases in phenotypic volume following DLI for both R and NR cases (P<10−6) (Figure 2A). Rs displayed higher phenotypic volume than NRs at both pre- and post-DLI timepoints (P<10−5), whereas NRs displayed a far greater increase in phenotypic volume after DLI than Rs (P<10−6). Thus, despite an absent clinical response, NRs undergo marked T cell phenotypic remodeling. Of note, the phenotypic volumes of the non-relapsed reference samples were lower than samples from the study cohort, (P<10−6; Figure S3B). These results implicate more transcriptionally diverse local microenvironments within the leukemic bed that may persist even after leukemia remission following DLI.
Distinct temporal dynamics of T cell expression clusters define DLI response
To identify T cell clusters that expand after DLI, we compared the cluster proportions in baseline pre-DLI samples to those from the remission timepoint following DLI. To increase our statistical power for detecting changes induced by DLI, we grouped transcriptionally similar clusters into meta-clusters (Figure S3C, STAR Methods). In this fashion, we identified two meta-clusters which consistently expanded (MC1:{19,28}, MC2:{5,11,23}) and one that consistently contracted (MC3:{4,7,3,22}) after DLI therapy, only in Rs (Figure 2C). The T cell states that expanded in response to DLI comprised both CD4+ and CD8+ T cells; enriched for TN (19, 28, and 5), TCM (11), or both (23) states; and expressed corresponding gene programs for proliferation (CDK20, CDK14, CDKL3), lymph node homing (SELL, CCR7), and survival/self-renewal (TCF7, IL7R, SATB1) (Figure S3A). Overall, these programs identify a set of ‘early-differentiated’ T cell states that expand in response to DLI. Analogous to the clusters enriched in pre-DLI R samples, the T cell states contracting in response to DLI comprised mostly CD8+ T cells, enriched similarly for TEM and TTE states, and expressed similar gene programs of cytotoxicity and activation. In contrast, no clusters or meta-clusters consistently changed in NRs.
Having identified response-associated T cell meta-clusters with diverging patterns after DLI (expanding MC1 and MC2, and contracting MC3), we then characterized their evolution over time by merging samples across all timepoints for each clinical outcome and thereafter modelling their temporal dynamics over the 4.5 year time period. To account for variability in timing, total cell number, and meta-cluster size on a per-sample basis, we constructed a hierarchical Gaussian Process (GP) regression model to capture dependencies between all pairs of time points per clinical group (R,NR) (Figure S3D,E; S6E,F; STAR Methods). We quantified these dynamics through correlation between model fit to cluster proportion and tumor burden. Indeed, the MC3 meta-cluster tracked with leukemic growth in Rs and sharply contracted during DLI response (p=0.013, Figure 2D-left) whereas both MC1 and MC2 meta-clusters robustly expanded as early as 3 weeks and endured even 3 years after DLI (Figure 2D-middle, right). No association was detected between these meta-clusters and leukemic burden in NRs (Figure 2E).
Transcriptional and immunophenotypic properties implicate exhausted T cell subsets in DLI response
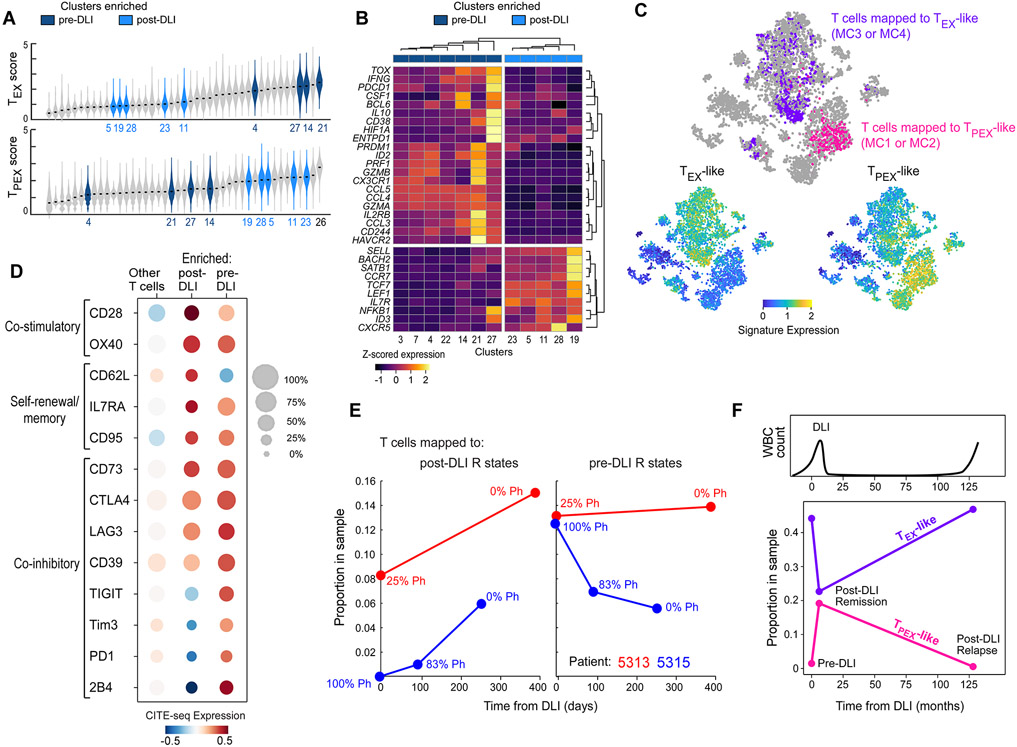
Recent studies in murine models of chronic viral infection and cancer have delineated two major subsets of exhausted T cells distinguishable on the basis of gene expression signatures: terminal exhausted (TEX) cells that possess relatively greater cytotoxicity but shorter lifespan compared to precursor exhausted (TPEX) cells which have greater polyfunctionality, expand following PD-1 blockade, and exert tumor control (Kallies et al., 2020; Miller et al., 2019). We hypothesized that the human CD8+ ‘late differentiated’ T cell clusters enriched pre-DLI and the rapidly expanding ‘early differentiated’ T cell clusters enriched post-DLI might be phenotypically similar to these murine subsets. Indeed, by scoring all clusters for TEX- or TPEX-defining signatures derived from a viral murine model of exhaustion (Im et al., 2016) (Table S3), we found that clusters enriched in pre-DLI Rs (4, 14, 21, 27) scored highest for TEX expression profiles whereas clusters consistently expanded post-DLI in Rs (MC1, MC2) scored highest for TPEX expression profiles (Figure 3A). Cluster 26 was the highest TPEX scoring cluster and expanded only in R patient 5309 but did not meet the threshold for significance due to its small size and patient-dominant variation. Because patient 5309 was the only R without expansion in either of the two meta-clusters, MC1 or MC2 (Figure S3F), the expansion of cluster 26 suggests that all six Rs, in fact, demonstrated post-DLI expansion of TPEX-enriched clusters. TEX- or TPEX-defining signatures from an alternate, tumor murine model of exhaustion (Miller et al., 2019) also segregated pre- and post-DLI enriched clusters in an unsupervised analysis (Figure 3B). While pre-DLI enriched clusters expressed transcription factors (TOX, ID2, PRDM1), co-inhibitory receptors (HAVCR2, PDCD1, ENTPD1, CD160, CD244), chemokines and associated receptors (CCL3, CCL4, CCL5, CX3CR1), and effector molecules (PRF1, GZMA, GZMB) classically associated with TEX cells, post-DLI enriched clusters expressed transcription factors (TCF7, ID3, LEF1), surface receptors (CXCR5, IL7R), and chromatin regulators (SATB1) consistent with TPEX cells (Alfei et al., 2019; Brummelman et al., 2018; Im et al., 2016; Kallies et al., 2020; Khan et al., 2019; Leong et al., 2016; Scott et al., 2019; Wu et al., 2016) (Figure 3B). Finally, unlike many studies that used antigen-specific models of CD8+ T cell responses (Im et al., 2016), we found a mixture of both CD4+ and CD8+ T cells to constitute these expanding, early differentiated clusters. Within the MC1 and MC2 meta-clusters, both subtypes exhibited global transcriptional similarity, with similar TPEX scores and similar expression of key TFs such as TCF7, indicating the importance of both CD4+ and CD8+ subtypes to DLI response (Figure S3G).
Figure 3. T cell states defining DLI response correspond to exhausted subsets.
(A) Violin plots showing density of viral-specific TEX (top) or TPEX (bottom) signature scores (Im et al., 2016) across T cells grouped by cluster. Clusters are ordered by median score. Colored violins refer to clusters enriched in pre-DLI Rs (dark blue, Figure 2B) or expanding in post-DLI Rs (light blue, Figure 2C). Full labels provided in Figure S6D. (B) Unsupervised hierarchical clustering based on tumor infiltrating TPEX or TEX genes (Miller et al., 2019) segregates dark/light blue clusters. (C) t-SNE projection of all T cells from patients 5313 and 5315, with cells colored by mapped metacluster (top), expression of TPEX gene set (bottom left) or TEX gene set (bottom right). (D) Normalized CITE-seq expression data for defined T cell phenotypes for enriched pre-DLI, enriched post-DLI and all other T cells from patients 5313 and 5315. (E) Post-DLI kinetics of enriched post-DLI (left) and enriched pre-DLI (right) T cells measured by their sample proportion with indicated percentage of tumor burden (%Ph+) for patients 5313 (red) and 5315 (blue). (F) Kinetics of TPEX-like and TEX-like cells in a patient with CLL in post-DLI remission as well as long-term relapse (after 11 years). See also Figures S3, S6, S7 and Table S3.
To further investigate the exhausted immunophenotypes of these DLI response-associated clusters, we generated combined single cell transcriptome and barcoded antibody (CITE-seq) measurements from matched longitudinal bone marrow samples (n=5) collected from 2 additional long-term DLI responders (Figure 1A). We first mapped the scRNA-seq profile of each T cell in the confirmation cohort to a cluster identified in the discovery cohort, finding clear separation between cells mapping to pre-DLI enriched metaclusters (“pre-DLI T cells”) with late differentiated programs, and cells mapping to post-DLI enriched metaclusters (“post-DLI T cells”) with early differentiated programs (Figure 3C; STAR Methods). We analyzed the paired CITE-seq protein expression for each group, revealing classic co-expression of multiple co-inhibitory receptors (CTLA4, LAG3, TIGIT, TIM3, PD1, and 2B4) on pre-DLI T cells, especially in relation to post-DLI T cells and all other T cells in our dataset (Figure 3D). Similarly, post-DLI T cells demonstrated co-expression of a few exhaustion markers (i.e. co-inhibitory receptors CTLA4 and LAG3) as well as ectonucleotidase enzymes CD39 and CD73 (Chen et al., 2019a; Sade-Feldman et al., 2019), indicating their exhausted lineage, though clearly not to the extent, by either magnitude of expression or number of expressed receptors, seen on pre-DLI T cells. Key co-stimulatory receptors (OX40 and CD28) shown to be critical for efficacy of exhaustion resolution (Kamphorst et al., 2017) and known self-renewal/memory markers (CD62L, IL7RA, CD95) were also maximally expressed on post-DLI T cells indicating the same “late versus early” differentiation distinction seen in our discovery cohort. The post-DLI kinetics of expanding early and contracting late differentiated T cells mirrored those observed in the discovery cohort, confirming that the mapping strategy selected appropriate counterpart cells (Figure 3E). Of note, we observed similar post-DLI kinetics of expanding and contracting TPEX/TEX-like cells after DLI response in a patient with chronic lymphocytic leukemia (CLL) (Figure 3F; Figure S7D, STAR Methods). Analysis of the CLL recurrence 11 years after DLI therapy revealed reversion back to the pre-DLI states. Overall, this index case supports the notion that these T cell subsets define graft-versus-leukemia responses following DLI, beyond CML.
Thus, late and early differentiated T cells enriched pre- or post-DLI in responding patients exhibit transcriptional, dynamic and immunophenotypic profiles of TEX and TPEX cells, respectively; in addition, we confirm these properties in an independent cohort of DLI responders, both for CML and CLL. Taken together, our data shows that resolution of T cell exhaustion is driven not by changes in gene expression, but rather by shifts in cell type composition – specifically, the expansion of early differentiated, TPEX-like populations and contraction of late differentiated, TEX-like subsets.
Cell-state specific gene regulatory networks affirm exhausted subset identities
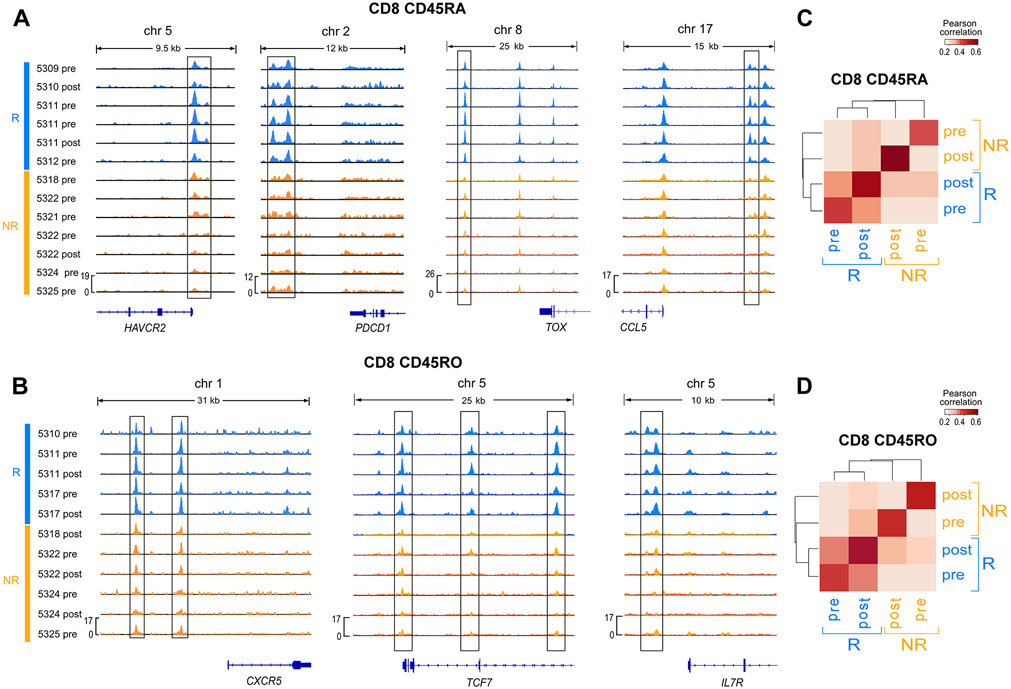
While recent work has described epigenetic (i.e. changes in gene expression not due to alterations in the DNA sequence) T cell states that drive dedifferentiation (Youngblood et al., 2017), effector “poising” (Akondy et al., 2017) and exhaustion (Pauken et al., 2016; Sen et al., 2016), their relevance to clinical immunotherapeutic outcomes is unclear. To investigate the regulatory circuitry underlying the T cell transcriptional states associated with DLI outcome, we compared chromatin accessibility profiles between Rs and NRs (STAR Methods). Consistent with our scRNA-seq analysis, we found increased chromatin accessibility in Rs in regions near TEX- and TPEX-associated genes in CD8+CD45RA+ and CD8+CD45RO+ cells, respectively, further supporting the association of these exhausted subsets with DLI response (Figure 4A-B, Figure S4A). Notably, we found similar accessibility for these genes among R samples, regardless of timing relative to DLI. In fact, we observed that the genome-wide accessibility landscape of T cells is more similar between pre- and post-DLI timepoints of Rs, than between Rs and NRs (Figure 4C-D), suggesting that DLI response does not involve the global rewiring of epigenetic landscapes. This potentially inflexible global landscape in response to DLI is similar to observations made in murine models of response to PD-1 blockade (Pauken et al., 2016; Sen et al., 2016).
Figure 4. Epigenetic landscape of T cell subsets.
(A-B) Chromatin accessibility signal from ATAC-seq data for CD8+ CD45RA+ (A) and CD8+ CD45RO+ (B) T cells indicating differential accessibility (p<0.05 indicated with boxes) between R and NR in regions near exhaustion marker genes. (C-D) Average pairwise Pearson correlation between normalized ATAC-seq peak heights for CD8+ CD45RA+ (C) and CD8+ CD45RO+ (D) T cells from different clinical groups. See also Figure S4.
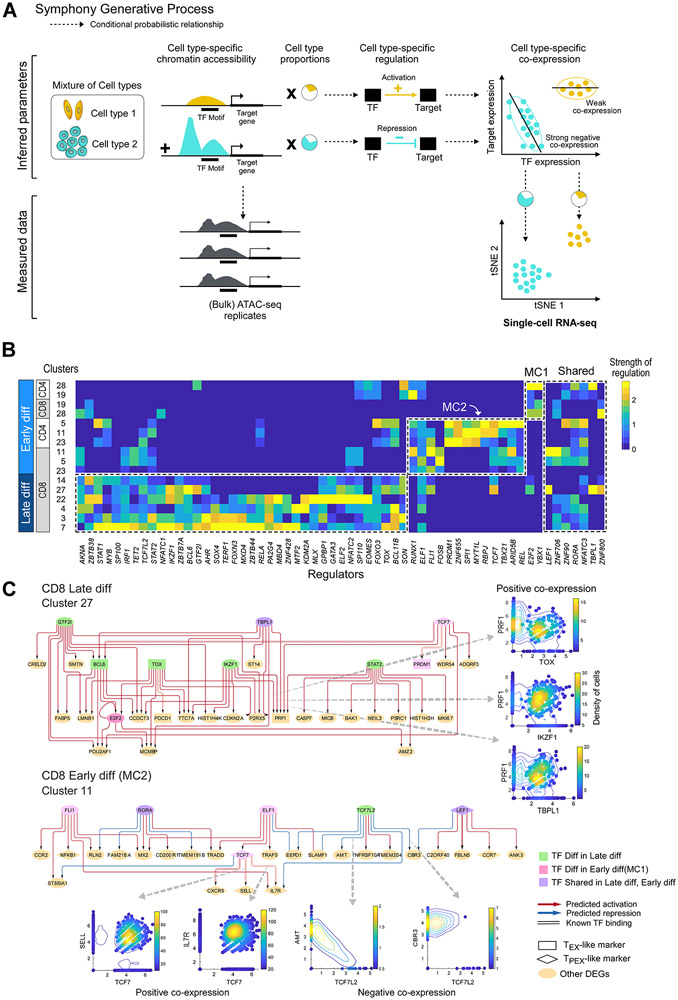
To further study the circuitry underlying the distinct expanding and contracting subsets, we developed Symphony (C. Burdziak, E. Azizi, S. Prabhakaran, D. Pe’er, 2019), a probabilistic multi-view model to infer gene regulation in each exhausted cluster (Figure 5A; Figure S7A,B). Symphony uses co-expression patterns between transcription factors (TF) and targets as evidence suggesting a potential regulatory impact. However, since co-expression between genes could be a by-product of indirect regulation or co-regulation, Symphony integrates scRNA-seq data with chromatin accessibility data from ATAC-seq, together with TF motif information to resolve direct links between genes. We first evaluated the performance of Symphony on data from well-characterized PBMCs (STAR Methods; Figure S7C) and then confirmed the robustness of predicted links in our cohort with leave one (patient) out analysis (Figure S4B; STAR Methods).
Figure 5. Regulatory circuitry underlying exhausted T cell subsets.
(A) Generative process for Symphony, a probabilistic model that infers regulation from integration of ATAC-seq and scRNA-seq data. Dotted line arrows indicate conditional probabilistic relationship. (B) Heatmap showing scaled values of predicted regulatory strength of TFs (i.e. magnitude of regulation independent of sign) from Symphony (STAR Methods; Figure S4B, M10-12), averaged across differentially expressed genes characterizing each cluster. Master regulators that are differential (t-test p<0.05) or shared between late and early differentiated (“diff”) T cell subsets are shown in dotted lines. (C) Predicted regulatory circuitry for two example clusters; arrows between nodes indicate regulatory impact of a TF on a target gene. Master regulators that are differentially enriched in late differentiated, TEX-like and early differentiated, TPEX-like subsets are shown in green or pink nodes, respectively. Circuitry for other enriched clusters are shown in Figure S5. See also Figures S5 and S7.
To determine the strongest regulators underlying the differences in gene expression across the clusters, we summarized predicted gene regulatory networks (GRNs) in each cluster and defined master-regulators as TFs with strong average regulatory impact (either activation or repression) on the differentially expressed genes (DEGs) characterizing each cluster. Strikingly, the inferred master regulators organized into distinct groups associated with early or late differentiated subsets (Figure 5B). From our unsupervised analysis, we predicted many TFs previously known to associate with exhaustion in general (e.g. EOMES, TBX21) (Paley et al., 2012; Utzschneider et al., 2016) or regulate TEX (e.g. MYB, NFATC1, TOX) (Chen et al., 2019b) and TPEX subsets (e.g. TCF7, PRDM1, LEF1) (Utzschneider et al., 2016) in particular. Two of the identified TFs, MTF2 and GATA3, were recently defined as mediators of intratumoral CD8+ T cell dysfunction in murine models (Singer et al., 2017). While master regulators identified by TEX-associated DEGs were largely shared among disparate late differentiated clusters, the two early differentiated meta-clusters were well-discriminated by two distinct sets of master regulators. We also observed a smaller group of master regulators including LEF1 and RORA that were shared across early and late differentiated subsets (Figure 5B), suggesting a core shared regulatory program. Finally, we confirmed the differential expression of the predicted master regulators in early and late differentiated subsets in our confirmation cohort (Figure S4C).
Despite shared master regulators even within highly related transcriptional late or early differentiated states (dotted line boxes in Figure 5B), Symphony revealed a distinct regulatory network architecture for each cluster (Figure 5C, Figure S5) suggesting differences in wiring and target genes influenced by these regulators. Importantly, these cluster-specific regulatory networks imply that master regulators (shown in green, Figure 5C e.g. TOX) for pre-DLI enriched clusters appear to be directly linked to known TEX markers; similarly, master regulators (shown in pink) for post-DLI enriched meta-clusters directly regulate known TPEX markers. For example, in pre-DLI enriched cluster 27, PDCD1 is inferred to be activated by TOX, while the effector molecule PRF1 is predicted to be combinatorially activated by TOX, IKZF1, TBPL1 and STAT2 which are all up-regulated in this subset. Similarly, in post-DLI enriched cluster 11, TCF7 acts as a hub, predicted to be regulated by ELF1 and activating known TPEX markers IL7R, SELL and CXCR5 as expected. These connections, between regulators found from our unbiased approach and known markers of exhaustion, support the central role of these TFs in defining the identities of potentially exhausted T cell clusters. Furthermore, their regulatory function, inferred with Symphony, is supported by evidence in TF and target gene co-expression (Figure 5C) and/or chromatin accessibility (STAR Methods). Thus, in addition to identifying known, exhaustion-related regulators driving these DLI response-associated T cell clusters, Symphony provides a roadmap for future investigation on the role of previously unexplored regulators.
TCR sequencing reveals sources of expanding early differentiated T cells
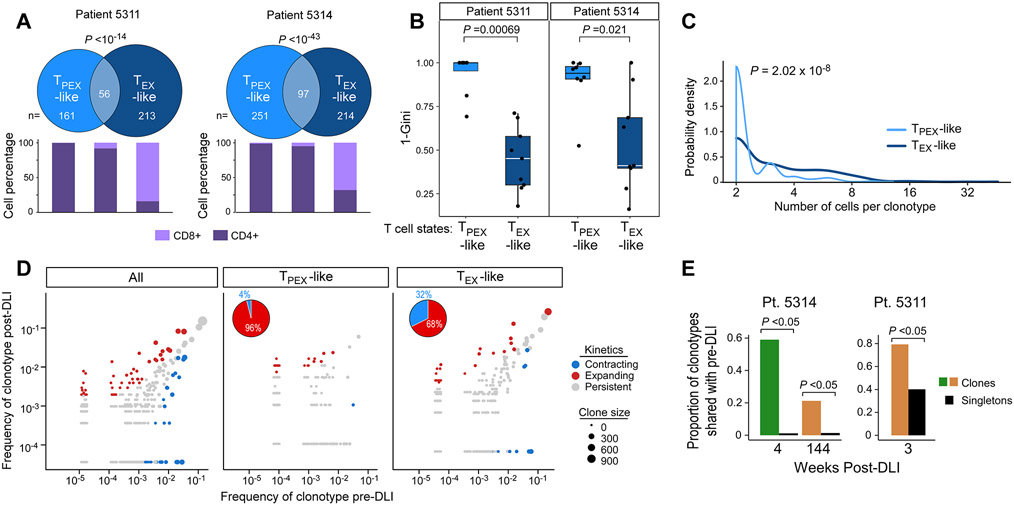
In murine models, TPEX and TEX subsets have been reported to share a lineage relationship in which the former self-renews and gives rise to the latter (Kallies et al., 2020). For two Rs (5311, 5314) with multiple timepoints, we used paired single-cell TCR- and RNA-seq to compare TCR clonotype sequences of TPEX-like and TEX-like clones (defined as >1 cell sharing the same TCR and enriched for TEX/TPEX gene scores). We observed that 27% of TPEX-like clones overlapped with TEX-like clones (p<10−14 for both patients), confirming their common ancestry (Figure 6A; STAR Methods, Table S9). The clones with TPEX-like phenotype were predominantly CD4+ T cells (81%) and clones with TEX-like phenotype were predominantly CD8+ T cells (99%) as were TEX/TPEX-like overlapping clones (93%). Clonotype diversity was higher in cells with a TPEX-like phenotype than in those with a TEX-like phenotype (P<0.05) for both patients (Figure 6B), consistent with previous reports in murine and human studies (Miller et al., 2019); and TEX-like clonotypes resided in larger clones than TPEX-like clonotypes (Figure 6C).
Figure 6. Clonal properties, distributions and evolution during response to DLI.
(A) Venn diagrams showing clonotype overlap between TPEX-like and TEX-like cells from two R patients (5311 and 5314), and stacked bars indicating percentage of CD8+ and CD4+ T cells in TPEX-like, TEX-like and overlap categories. P value calculated from hypergeometric test. (B) TPEX-like clusters show increased TCR diversity (quantified with Gini coefficient) compared to TEX-like clusters, Wilcoxon. Box plot elements display center line as median; box limits as first and third quartiles; whiskers extend to 1.5x interquartile range with points as outliers. TPEX-like clusters: 5311, n=6; 5314, n=8; TEX-like clusters: 5311,5314 n=9. (C) Probability densities of clone sizes for all TEX-like and TPEX-like cells from samples derived from R patients 5314 and 5311. (D) Clonotype frequencies 1 month before and 1 month after DLI from both R patients. Each dot represents a clonotype with dot size proportional to size of clone for each cell subset. Expanding/contracting clonotypes determined with Fisher’s exact test (P<0.05). Left, clonotypes from all cells colored by one of three dynamic patterns: contracting, expanding, persistent. Middle, right dynamic clonotypes from TPEX-like clusters are less likely to be contracting compared to clonotypes from TEX-like clusters (pie charts). (E) Barplots of proportions of clonal versus singleton TCRs shared with pre-DLI samples from each post-DLI timepoint from 5314 (n=2 post-DLI samples) and 5311 (n=1 post-DLI sample).
To study the dynamics of how clonal populations initially shifted in response to DLI in these two patients, we evaluated their TCR repertoire within one month before and after DLI and identified significantly expanding and contracting clonotypes (Figure 6D, left). Consistent with our observation of expanding TPEX-like states following DLI, dynamic clonotypes from TPEX-like clusters were more likely to expand than contract compared to those from TEX-like clusters (Figure 6D, middle and right). Thus, the evolution of TCRs mirrors that of TEX/TPEX-like transcriptional states after DLI.
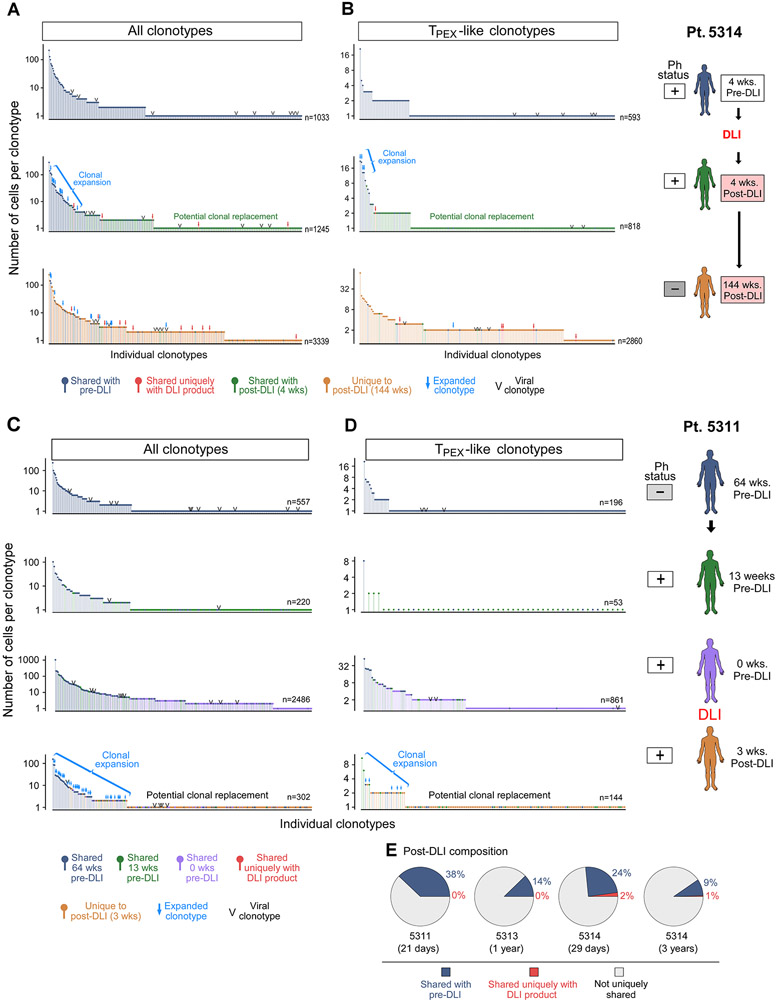
We noted that clonal TCRs following DLI were more likely to be shared with pre-DLI timepoints than were singletons, and many of these shared clones persisted even 3 years after DLI (Figure 6E, 7A-D; P<10−15, 4 wks and 144 wks post-DLI). Because the post-DLI expansion of TPEX-like cells was tightly linked to DLI response, we sought to determine its source by also profiling the DLI infusion products. We found that only 1.4% of TPEX-like cells from all post-DLI timepoints share clonotypes exclusively with the infusion product (Figure 7B, D pie charts). Although viral reactivity can be common in the post-transplant period (Link et al., 2016), we found scant evidence for viral antigen recognition among the post-DLI clonotypes (<1.5% across the 2 patients), suggesting it did not explain the expansion or durability of TPEX-like cells (STAR Methods). Thus, for these two patients, the vast majority of post-DLI expanding TPEX-like cells either shared clonotypes with pre-DLI samples or exhibited clonotypes specific to that timepoint. Single cell TCR analysis of a marrow specimen from an independent R patient (5313) again demonstrated a higher proportion of their post-DLI clonotypes to be shared only with the pre-DLI sample rather than with the DLI product (Figure 7E; STAR Methods). For another independent R patient (5316), we performed bulk TCR sequencing due to lower cell viability, and only the post-DLI specimen and the infusion product were of sufficient quality for analysis. Comparison of the clonotypes between these two compartments reveal a modest overlap (14%), suggesting that a minority of clonotypes may be contributed by the DLI product though their specificity to the DLI product could not be determined given the low quality of the pre-DLI sample (Figure S7E; STAR Methods). Altogether, these results demonstrate that the DLI product may not directly introduce the clonotypes that constitute the post-DLI TPEX-like expansion in Rs (Figure 7B, D); instead, it may predominantly drive expansion of pre-existing clonotypes as well as the recruitment of new T cell clones.
Figure 7. Source of expanding early differentiated T cells.
A-B) Frequency distribution of all (A) or TPEX-like (B) clonotypes per timepoint for patient 5314. Blue arrows indicate clonotype expansion from pre-DLI (P<0.05, Fisher’s exact test). (C-D) Frequency distribution of all (C) or TPEX-like (D) clonotypes per timepoint for patient 5311. Post-DLI clonotypes marked in red indicate match exclusive to DLI product in all (A,C) or TPEX-like (B,D) post-DLI clonotypes. Arrows indicate P<0.05, Exact Fisher’s Test, for clonotype expansion from pre-DLI. (E) Pie charts displaying the percentage of overlap between expanded clonotypes in post-DLI sample with those in pre-DLI (blue) or DLI product (red) using scTCR data from three patients; expanded clonotypes are defined as clonotypes detected in at least 2 cells. See also Figure S7.
DISCUSSION
In 1878, Leo Tolstoy published his masterpiece Anna Karenina and its eponymous principle that “all happy families are alike; each unhappy family is unhappy in its own way.” Likewise, our analysis of the evolution of T cell states following DLI unveiled common, shared pathways defining DLI response whereas multiple dysfunctional T cell states shaped DLI resistance, evoking a clinical outcome paradigm characteristic of other therapeutic scenarios where a limited set of targetable alterations predicts response in contrast to a diversified set of resistance mechanisms (Goetz and Garraway, 2012; Ricordel et al., 2019).
To enable such clear insights from a limited patient cohort, we leveraged two critical features: samples collected from an informative clinical setting and innovative computational tools. Specifically, we exploited a scenario with unambiguous, binary clinical outcomes (response or resistance) in the absence of any toxicities; longitudinal sample collection; and uniform patient treatment with CD8-depleted DLI for relapsed CML in the absence of any confounding chemotherapy or immunomodulators. Furthermore, we consistently sampled a single leukemic microenvironment (i.e. bone marrow) for all patient-timepoints as opposed to varied sites of metastases.
To overcome limitations of experimental design inherent to clinical studies such as variable timing of sample collection, patient heterogeneity, sample quality, measurement uncertainty, and challenges in hypothesis testing on key populations, we adapted statistical techniques and developed longitudinal and integrative probabilistic models. These models, in turn, allowed us to detect and define intratumoral T-cell dynamics in relation to immunotherapeutic outcome in humans. Importantly, these computational approaches for dissecting global heterogeneity, identifying immune states related to dynamics of tumor burden, and integrative gene regulatory network inference are readily generalizable to other longitudinal, clinical settings. Indeed, with the increasing number of clinical correlative studies using longitudinal tumor biopsies (Olson et al., 2011; TRACERx Renal consortium, 2017), we anticipate a growing need for such analytic frameworks.
Through direct interrogation of the human bone marrow microenvironment, we readily identified T cell states enriched pre- and post-DLI in Rs who followed late and early differentiation programs, respectively. Intriguingly, their dynamic, transcriptional, immunophenotypic, epigenetic and clonal properties mirror those of TEX and TPEX exhaustion subsets, previously identified from murine models of chronic viral infections (Kallies et al., 2020; Leong et al., 2016; Miller et al., 2019; Pauken et al., 2016). Our results now implicate the hierarchy of both TEX- and TPEX-like states for immunotherapeutic responses in leukemia, extending the scope of their relevance to adoptive cellular therapies and nominating this cellular program as a potent effector of GvL. Furthermore, these data indicate that resolution of T cell exhaustion may be driven not by changes in gene expression, but rather by shifts in cell type composition – namely, expansion of TPEX-like populations and contraction of TEX-like subsets. Because such distinctions cannot be delineated by bulk measurements, our findings highlight the advantages of single cell transcriptomics for discriminating between these possibilities. Future studies that demonstrate the hypofunctionality of these T cell subsets ex vivo or in vitro will be important in confirming their exhausted status.
Remarkably, the rapid expansion of TPEX-like states after DLI dovetail with similar observations in murine models of response to PD-1 pathway blockade (He et al., 2016; Im et al., 2016; Miller et al., 2019; Siddiqui et al., 2019; Utzschneider et al., 2016). In conjunction with recent studies indicating a role for TPEX cells during outcomes to checkpoint blockade in advanced melanoma (Miller et al., 2019; Sade-Feldman et al., 2019), these data now suggest similar mechanisms of action between PD-1 blockade and DLI. Our data moreover offer mechanistic insight into DLI efficacy. Our scTCR analysis not only confirmed the common ancestry shared between TEX- and TPEX-like states but now also explains that previous independent observations of increased TCR diversity detected in the setting of DLI response (Claret et al., 1997) are a consequence of TPEX-like subset expansion. Provocatively, this expansion of TPEX-like cells during DLI response did not primarily arise directly from the DLI product. Instead, we observed both marked recruitment of previously undetected clonotypes (potential clonal replacement (Yost et al., 2019)) and expansion of pre-existing ones (clonal expansion), suggesting that immunologic ‘help’ from DLI, rather than direct transfer of anti-leukemic T cells, may drive leukemic remission. Similar results have been observed in murine models of exhaustion reversal after adoptive transfer of CD4+ T cells (Aubert et al., 2011; Zander et al., 2019). These data suggest that TEX/TPEX-like subsets serve as both marker and mechanism for DLI response. Our findings motivate future clinical trial designs to test the status of TEX cells as a biomarker for predicting DLI response and to evaluate therapeutic strategies that enhance TPEX recruitment and expansion. Pursuing such approaches offers the possibility of enhancing the GvL effect during relapse after allo-SCT. In addition, recent observations that chimeric antigen receptor (CAR)-T cells also activate endogenous, non-CAR T cells (Chen et al., 2020), affirm the relevance of our findings to newer generations of ACT and warrant study of exhausted-like cells in these contexts as well.
Functional interrogation of the regulatory networks proposed by our joint analysis of scRNA- and bulk ATAC-seq datasets through Symphony should accelerate these efforts with identification of potential targets for therapeutic drug development. Future studies should also address the mechanism of DLI-induced TPEX-like expansion and whether molecular therapies can recapitulate this effect. The critical roles likely played by leukemia cells and alloreactivity should also be better understood given their known influence on GvL escape (Bachireddy et al., 2020). In addition, while these T cell exhausted subsets have now been observed in multiple clinical settings, which aspects of their underlying molecular machinery and distinct regulatory circuits remain specific to the leukemic or GvL setting and which generally extend to other cancers and human diseases should be explored. Finally, our analytic approaches serve as a template for future studies that seek to harness such multidimensional data sets for clinical and therapeutic relevance within oncology and beyond.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contacts, Catherine J Wu (cwu@partners.org).
Materials Availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
Single cell transcriptome and TCR as well as chromatin accessibility data will be submitted to NCBI’s Database of Genotypes and Phenotype (dbGaP; https://www.ncbi.nlm.nih.gov/gap) under study number phs001998.v3 and will be made publicly available as of the date of publication. Accession numbers are listed in the key resources table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE |
SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Human CD14, FITC | BD Biosciences | Cat#555397; AB_395798 |
| Anti-Human CD19, FITC | BD Biosciences | Cat#555412; AB_395812 |
| Anti-Human CD3, PE | BD Biosciences | Cat#555339; AB_395745 |
| Anti-Human CD4, BUV395 | BD Biosciences | Cat#563550; AB_2738273 |
| Anti-Human CD8, APC-Vio770 | Miltenyi Biotec | Cat#130-113-155; AB_2725983 |
| Anti-Human CD45RA, BV510 | BD Biosciences | Cat#563031; AB_2722499 |
| Human BD Fc Block | BD Biosciences | Cat#564219; AB_2728082 |
| DAPI solution | BD Biosciences | Cat#564907; AB_2869624 |
| Anti-Human CD11c, FITC | BD Biosciences | Cat#561355; AB_10611872 |
| Anti-Human CD14, FITC | BD Biosciences | Cat#555397; AB_395798 |
| Anti-Human CD36, FITC | BD Biosciences | Cat#555454; AB_2291112 |
| Anti-Human CD33, FITC | BD Biosciences | Cat#555626; AB_395992 |
| Anti-Human CD16, FITC | BD Biosciences | Cat#555406; AB_395806 |
| Anti-Human CD11b, FITC | BD Biosciences | Cat#562793; AB_2737798 |
| Anti-Human CD15, FITC | BD Biosciences | Cat#555401; AB_395801 |
| Anti-Human CD34, FITC | BD Biosciences | Cat#348053; AB_2228982 |
| Anti-Human CD56, FITC | BD Biosciences | Cat#562794; AB_2737799 |
| Anti-Human CD123, FITC | BD Biosciences | Cat#558663; AB_1645485 |
| Anti-Human CD235a, FITC | BD Biosciences | Cat#559943; AB_397386 |
| IgG1 isotype | Biolegend | Cat#400187; AB_2888921 |
| IgG2a isotype | Biolegend | Cat#400293; AB_2888922 |
| IgG2b isotype | Biolegend | Cat#400381; AB_2888923 |
| Anti-Human B2M | Biolegend | Cat#316323; AB_2800837 |
| Anti-Human B7H4 | Biolegend | Cat#358116; AB_2800986 |
| Anti-Human CD10 | Biolegend | Cat#312233; AB_2800817 |
| Anti-Human CD117 | Biolegend | Cat#313243; AB_2810474 |
| Anti-Human CD11a | Biolegend | Cat# 350617; AB_2800935 |
| Anti-Human CD11b | Biolegend | Cat# 301359; AB_2800732 |
| Anti-Human CD11c | Biolegend | Cat# 371521; AB_2801018 |
| Anti-Human CD127 | Biolegend | Cat# 351356; AB_2800937 |
| Anti-Human CD134 | Biolegend | Cat# 350035; AB_2800932 |
| Anti-Human CD137 | Biolegend | Cat# 309839; AB_2800807 |
| Anti-Human CD138 | Biolegend | Cat# 356539; AB_2810567 |
| Anti-Human CD14 | Biolegend | Cat# 301859; AB_2800736 |
| Anti-Human CD15 | Biolegend | Cat# 323053; AB_2800847 |
| Anti-Human CD152 | Biolegend | Cat# 369621; AB_2801015 |
| Anti-Human CD16 | Biolegend | Cat# 302065; AB_2800738 |
| Anti-Human CD163 | Biolegend | Cat# 333637; AB_2810510 |
| Anti-Human CD18 | Biolegend | Cat# 302129; AB_2800739 |
| Anti-Human CD183 | Biolegend | Cat# 353747; AB_2800949 |
| Anti-Human CD184 | Biolegend | Cat# 306533; AB_2800791 |
| Anti-Human CD19 | Biolegend | Cat# 302265; AB_2800741 |
| Anti-Human CD194 | Biolegend | Cat# 359425; AB_2800988 |
| Anti-Human CD197 | Biolegend | Cat# 353251; AB_2800943 |
| Anti-Human CD1c | Biolegend | Cat# 331547; AB_2800871 |
| Anti-Human CD1d | Biolegend | Cat# 350319; AB_2800934 |
| Anti-Human CD20 | Biolegend | Cat# 302363; AB_2800743 |
| Anti-Human CD223 | Biolegend | Cat# 369335; AB_2814327 |
| Anti-Human CD226 | Biolegend | Cat# 338337; AB_2800899 |
| Anti-Human CD244 | Biolegend | Cat# 329529; AB_2800857 |
| Anti-Human CD25 | Biolegend | Cat# 302649; AB_2800745 |
| Anti-Human CD27 | Biolegend | Cat# 302853; AB_2800747 |
| Anti-Human CD274 | Biolegend | Cat# 329751; AB_2800860 |
| Anti-Human CD278 | Biolegend | Cat# 313553; AB_2800823 |
| Anti-Human CD279 | Biolegend | Cat# 329963; AB_2800862 |
| Anti-Human CD28 | Biolegend | Cat# 302963; AB_2800751 |
| Anti-Human CD3 | Biolegend | Cat# 300479; AB_2800723 |
| Anti-Human CD31 | Biolegend | Cat# 303139; AB_2800757 |
| Anti-Human CD314 | Biolegend | Cat# 320837; AB_2800844 |
| Anti-Human CD33 | Biolegend | Cat# 366633; AB_2801008 |
| Anti-Human CD335 | Biolegend | Cat# 331941; AB_2800874 |
| Anti-Human CD34 | Biolegend | Cat# 343537; AB_2749972 |
| Anti-Human CD38 | Biolegend | Cat# 303543; AB_2800758 |
| Anti-Human CD39 | Biolegend | Cat# 328237; AB_2800853 |
| Anti-Human CD4 | Biolegend | Cat# 300567; AB_2800725 |
| Anti-Human CD40 | Biolegend | Cat# 334348; AB_2800886 |
| Anti-Human CD44 | Biolegend | Cat# 338827; AB_2800900 |
| Anti-Human CD45 | Biolegend | Cat# 304068; AB_2800762 |
| Anti-Human CD45RA | Biolegend | Cat# 304163; AB_2800764 |
| Anti-Human CD45RO | Biolegend | Cat# 304259; AB_2800766 |
| Anti-Human CD49f | Biolegend | Cat# 313635; AB_2800825 |
| Anti-Human CD5 | Biolegend | Cat# 300637; AB_2800726 |
| Anti-Human CD56 | Biolegend | Cat# 392425; AB_2801024 |
| Anti-Human CD57 | Biolegend | Cat# 393321; AB_2801030 |
| Anti-Human CD62L | Biolegend | Cat# 304851; AB_2800770 |
| Anti-Human CD69 | Biolegend | Cat# 310951; AB_2800810 |
| Anti-Human CD70 | Biolegend | Cat# 355119; AB_2800955 |
| Anti-Human CD73 | Biolegend | Cat# 344031; AB_2800916 |
| Anti-Human CD80 | Biolegend | Cat# 305243; AB_2800783 |
| Anti-Human CD86 | Biolegend | Cat# 305447; AB_2800786 |
| Anti-Human CD8a | Biolegend | Cat# 301071; AB_2800730 |
| Anti-Human CD95 | Biolegend | Cat# 305651; AB_2800787 |
| Anti-Human HLA-DR | Biolegend | Cat# 307663; AB_2800795 |
| Anti-Human KLRG1 | Biolegend | Cat# 138433; AB_2800649 |
| Anti-Human TCRab | Biolegend | Cat# 306743; AB_2800793 |
| Anti-Human TCRgd | Biolegend | Cat# 331231; AB_2814199 |
| Anti-Human TIGIT | Biolegend | Cat# 372729; AB_2801021 |
| Anti-Human Tim3 | Biolegend | Cat# 345049; AB_2800925 |
| Biological Samples | ||
| Cryopreserved bone marrow mononuclear cells | Dana-Farber Cancer Institute | Pasquarello Tissue Bank in Hematologic Malignancies |
| Cryopreserved donor lymphocyte infusion products | Dana-Farber Cancer Institute | Pasquarello Tissue Bank in Hematologic Malignancies |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DNase I | StemCell Technologies | Cat#07900 |
| Digitonin | Promega | Cat#G9441 |
| AMPure XP beads | Beckman Coulter | A63881 |
| Critical Commercial Assays | ||
| MACS Dead Cell Removal Kit | Miltenyi Biotec | Cat#130-090-101 |
| Pan T Cell Isolation Kit, human | Miltenyi Biotec | Cat#130-096-535 |
| MACS CD19 MicroBeads | Miltenyi Biotec | Cat#130-050-301 |
| 10X Chromium Single Cell 3′ Library & Gel Bead Kit (v2) | 10x Genomics | Cat#PN-120237 |
| Bioanalyzer High Sensitivity DNA Kit | Agilent | Cat#5067-4626 |
| 10x Chromium Single Cell 5’ Library & Gel Bead Kit | 10x Genomics | PN-1000006 |
| 10x Chromium Single Cell V(D)J Enrichment Kit, Human T Cell | 10x Genomics | PN-1000005 |
| 5' Feature Barcode Kit | 10x Genomics | PN-1000256 |
| 10x Chromium Next GEM Single Cell 5' Kit v2 | 10x Genomics | PN-1000263 |
| 10x Chromium Single Cell Human TCR Amplification Kit | 10x Genomics | PN-1000252 |
| Nextera DNA Library Prep Kit | Illumina | FC-121-1030 |
| NEBNext High Fidelity PCR Mix | New England Biolabs | M0541S |
| MinElute Reaction Cleanup kit | Qiagen | 28206 |
| Deposited Data | ||
| 10x scRNA-seq | dbGaP | phs001998.v3 |
| 10x scTCR-seq | dbGaP | phs001998.v3 |
| 10x CITE-seq | dbGaP | phs001998.v3 |
| ATAC-seq | dbGaP | phs001998.v3 |
| Symphony | This paper | DOI: https://zenodo.org/record/5498358 |
| Gaussian process regression models | This paper | DOI: doi.org/10.5281/zenodo.5498361 |
| Oligonucleotides | ||
| Primers for rhTCR-seq | Translational Immunogenomics Laboratory, Dana-Farber Cancer Institute | (Li et al., 2019) |
| Software and Algorithms | ||
| Symphony | This paper | DOI: https://zenodo.org/record/5498358 |
| Gaussian process regression models | This paper | DOI: doi.org/10.5281/zenodo.5498361 |
| SEQC | (Azizi et al., 2018) | https://github.com/dpeerlab/seqc |
| Cell Ranger 5.0.1 | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest |
| Cell Ranger V(D)J 2.1.0 | 10x Genomics | https://support.10xgenomics.com/single-cell-vdj/software/downloads/latest? |
| scanpy 1.8.0 | (Wolf et al., 2018) | https://github.com/theislab/Scanpy |
| t-SNE | (Maaten and Hinton, 2008) | https://lvdmaaten.github.io/software/ |
| Biscuit | (Azizi et al., 2018) | https://github.com/dpeerlab/BISCUIT_SingleCell_IMM_ICML_2016 |
| PhenoGraph | (Levine et al., 2015) | https://github.com/dpeerlab/phenograph |
| Pyro | (Bingham et al., 2019) | https://pyro.ai/ |
| ATAC-seq pipeline | ENCODE consortium | https://doi.org/10.5281/zenodo.156534; https://github.com/ENCODE-DCC/atac-seq-pipeline |
| MACS2 2.2.7.1 | (Zhang et al., 2008) | https://pypi.org/project/MACS2/ |
Code availability:
The hierarchical Gaussian Process model is implemented using the probabilistic programming language pyro (Bingham, Eli and Chen, Jonathan P and Jankowiak, Martin and Obermeyer, Fritz and Pradhan, Neeraj and Karaletsos, Theofanis and Singh, Rohit and Szerlip, Paul and Horsfall, Paul and Goodman, Noah D, 2019) available at: https://github.com/dpeerlab/dli_gpr. The integrative model Symphony is implemented using the probabilistic language Edward (Tran et al., 2016) with code available at: https://github.com/dpeerlab/Symphony. All original code has been deposited at [repository] and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human Subjects
Bone marrow (BM) biopsies were obtained pre- and post-DLI after relapse following allo-SCT (or during remission following allo-SCT) from patients enrolled in Dana-Farber Cancer Institute (DFCI) clinical trials (94-009, 95-011, 96-372, 96-022, and 96-277) between 1994-2001 that were approved by the DFCI Human Subjects Protection Committee. The sex for each patient is reported in Table S1. These studies were conducted in accordance with the Declaration of Helsinki; informed consent was obtained from the patients. Bone marrow mononuclear cells (BMMCs) were isolated via Ficoll-Hypaque density gradient centrifugation, cryopreserved with 10% dimethyl sulfoxide, and stored in vapor-phase liquid nitrogen until the time of sample processing.
Cohort sample characteristics
All 17 patients had CML that was treated with CD6-T cell depleted allo-SCT. Of these, 15 patients had CML relapse after allo-SCT that was treated with CD8-depleted DLI, and 2 patients never had CML relapse and served as non-relapse controls (Table S2). DLIs were infused at weekly intervals until the target cell dose was reached. No significant differences were observed between the number of infusions or total cells infused between responders and nonresponders. No post-DLI samples were collected in between the infusions (which, if more than one session were required, occurred 1 week apart). None of the patients received imatinib or any other TKIs after transplant or during the course of relapse treatment. All study samples reported in this manuscript were obtained between 1994-1998, before imatinib was FDA-approved. The presence of acute and chronic GVHD was graded by standard criteria (Przepiorka et al., 1995); grades 0 and I acute GvHD were considered clinically equivalent (Gratwohl et al., 1995). The median age of all samples was 23 years, ranging from 20-25 years. In the discovery cohort, a median of 3 timepoints was available for each R and NR patient (range: 2-6), and there were no significant differences between R and NR cohorts regarding time from allo-SCT to DLI (R: median 702, range 362-2371 days; NR: median 1064, range 422-1787 days; P=0.6) (Figure S1A), time from allo-SCT to pre-DLI sample (R: median 583, range 138-2344 days; NR: median 809, range 147-1783 days; P=0.2), nor time from allo-SCT to post-DLI sample (R: median 925, range 447-2561 days; NR: median 1512, range 674-1916 days; P=0.2). The times from transplant to sample for the two non-relapsed control samples fell within this range of times (1113 and 1817 days). Time from allo-SCT to sample for the non-relapsed controls was 1817 days for 5379 and 1113 days for 5380. Sample characteristics are listed in Table S2. Given the size of the cohort, no association of sex with the results of the study could be detected.
The single patient with CLL (5283) was also treated with CD6-T cell depleted allo-SCT. His subsequent relapse initially responded to CD8-depleted DLI until a repeat relapse 11 years after DLI infusion (Table S2).
Cytogenetic and molecular information on CML tumor burden
The percent positivity of the Philadelphia (Ph) chromosome for each BM sample was extracted from the clinical record where available (as described previously (Alyea et al., 1998)). Molecular remission was defined as achievement of molecular response (defined as the absence of BCR-ABL transcripts by RT-PCR). This data is shown in grey crosses in Figure 2D,E.
METHOD DETAILS
Sample processing
Cryopreserved primary bone marrow mononuclear cells (BMMCs) were thawed on the day of sequencing at 37°C and dispensed drop-wise into a warmed solution of 10% FBS, 10% DNaseI (StemCell Technologies, cat. No. 07900) in PBS. The cell suspension was centrifuged at 200g for 10 minutes at room temperature. Viable cells were negatively selected using MACS Dead Cell Removal Kit (Miltenyi Biotec, cat. No. 130-090-101), running on MS columns to prevent sample loss. Collected live cells were resuspended in 0.04% BSA in PBS and diluted to a concentration of 1000 cells/uL. These cells were then divided into portions taken immediately for scRNA-seq (samples B1-B46) or for FACS isolation (described below) for subsequent ATAC-seq. For paired scTCR- and scRNA-seq on samples D1-D7, E1-E3, and E6-E7 (Table S9), BMMCs were processed as described here and then samples D1-D7 were taken for FACS enrichment of T cells described below while confirmation cohort samples E1-E3 and E6-E7 were taken directly for scRNA/TCR-seq.
For cryopreserved PBMCs of DLI products (D8, D9, E9; Table S9), cells were thawed as described above, T cells were enriched using the human Pan T Cell Isolation Kit (Miltenyi Biotec), and then processed with the MACS Dead Cell Removal Kit (Miltenyi Biotec) before scRNA- and TCR-seq.
For cryopreserved BMMCs from patient 5283 with CLL, samples were processed as previously described for subsequent inDrop sequencing (Bachireddy et al., 2020). Briefly, dead cells were removed via an OptiPrep selection protocol, and viable cells were then subjected to immunomagnetic selection (MACS CD19 MicroBeads; Miltenyi Biotec) using MS columns to isolate C19+ and CD19−, which were then mixed at a 1:1 ratio at a total concentration of 1.15 x 105 cells/mL in a 15% OptiPrep solution in PBS and kept on ice until time of encapsulation.
Fluorescence activated cell sorting (FACS)
For downstream ATAC-seq which was performed on samples B1-B46 (Table S8), viable BMMC single-cell suspensions (prepared as above) were stained using antibody cocktails in the dark at 4oC, washed and run on a 5-laser FACSAria II (BD Biosciences) cell sorter. Cells then underwent FACS for the following CD14−CD19−CD3+ T cell populations: CD45RA+CD4+, CD45RA−CD4+, CD45RA+CD8+, and CD45RA−CD8+. The following fluorochrome-conjugated antibodies were used: CD14-FITC (M5E2, BD Biosciences); CD19-FITC (HIB19, BD Biosciences); CD3-PE (HIT3A, BD Biosciences); CD4-BUV395 (SK3, BD Biosciences); CD8-APC Vio770 (BW135/80, Miltenyi Biotec); CD45RA-BV510 (HI100, BD Biosciences) (Figure S6A).
In order to perform paired scRNA- and scTCR-seq on samples D1-D7 (Table S9), BMMCs were thawed as above without dead cell removal, stained with human Fc block (BD Pharmingen) for 10 minutes in the dark at 4oC, stained with antibody cocktail, washed and run on a 4-laser, FACSAria II (BD Biosciences) cell sorter. DAPI (BD Pharmingen) was used to exclude dead cells, and the following fluorochrome-conjugated antibodies were used to negatively select for T cells (to avoid stimulation of gene expression by anti-CD3 antibodies): Lineage 1: CD11c-FITC (B-ly6, BD Biosciences); CD14-FITC (M5E2, BD Biosciences); CD36-FITC (CB38, BD Biosciences); CD33-FITC (HIM3-4, BD Biosciences); CD16-FITC (3G8, BD Biosciences)
Lineage 2: CD11b-PE (ICRF44, BD Biosciences); CD15-PE (HI98, BD Biosciences); CD34-PE (8G12, BD Biosciences); CD56-PE (B159, BD Biosciences); CD123-PE (7G3, BD Biosciences); CD235a-PE (GA-R2, BD Biosciences).
Library preparation for scRNA- , scTCR-seq, and scCITE-seq
For BMMC samples B1-B46 (Table S2), approximately 17,000 BMMCs (after dead cell removal) were loaded across 2 lanes onto a 10x Genomics Chromium™ instrument (10x Genomics) according to the manufacturer’s instructions. The scRNAseq libraries were processed using Chromium Single Cell 3’ Library & Gel Bead v2 Kit (10x Genomics). Quality control for amplified cDNA libraries and final sequencing libraries were performed using Bioanalyzer High Sensitivity DNA Kit (Agilent). scRNAseq libraries were normalized to 4nM concentration and pooled before loading onto Illumina sequencer. The pooled libraries were sequenced on the Illumina HiSeq X or NovaSeq S4 platform. The sequencing data were demultiplexed and processed as described below.
For BMMC samples processed for CITE-seq (E1-E3 and E6-E7; Table S9), 500,000 cells were labeled with a pool of 70 CITE-seq antibodies selected for identification and characterization of key immune cell populations. For all BMMC samples processed for scRNA- and sc-TCRseq (Table S9), approximately 17,000 cells were loaded across two lanes onto a 10x Genomics Chromium™ instrument (10x Genomics) according to the manufacturer’s instructions. The scRNAseq libraries were processed using Chromium™ single cell 5’ library & gel bead kit, coupled scTCRseq libraries were obtained using Chromium™ single cell V(D)J enrichment kit (human T cell) (10x Genomics), and coupled scCITE-seq libraries were obtained using Chromium™ single cell Feature Barcode kit. Quality control for amplified cDNA libraries and final sequencing libraries were performed using Bioanalyzer High Sensitivity DNA Kit (Agilent). Both scRNAseq, scTCRseq, and CITE-seq libraries were normalized to 4nM concentration and pooled in a volume ratio of 4:1. The pooled libraries were sequenced on an Illumina NovaSeq S4 platform. The sequencing parameters were: Read 1 of 150bp, Read 2 of 150bp and Index 1 of 8bp. The scRNA-, sxTCR-seq, and CITE-seq data were processed as described in Suppl. Text.
For BMMC samples from patient 5283, cell encapsulation and subsequent library preparation were performed as previously described (Bachireddy et al., 2020). Briefly, cells were encapsulated with RT/lysis mix and barcoded hydrogen beads (BHBs; from 1CellBio) and maintained at 4oC in their respective syringes throughout using refrigerated copper coiling. Similar working flow rates as previously described were used to obtain similar encapsulation times and calculated cell doublet percentages. Libraries were prepared using overnight in vitro transcription (16 hours at 37oC), followed by fragmentation of amplified RNA, and PCR amplification. Sequencing was performed on NextSeq Illumina Sequencer.
Library preparation for ATAC-seq
After FACS isolation of CD45RA+CD4+, CD45RA−CD4+, CD45RA+CD8+, and CD45RA−CD8+ T cell populations, the Fast-ATAC protocol was then performed as previously described(Corces et al., 2016). Briefly, fifty microliters of transposase mixture (25 μl of 2× TD buffer, 2.5 μl of TDE1, 0.5 μl of 1% digitonin, and 22 μl of nuclease-free water) (FC-121-1030, Illumina; G9441, Promega) was added to a cell pellet consisting of 10000-50000 cells and incubated at 37°C for 30 minutes. Transposed DNA was purified using a MinElute Reaction Cleanup kit (Qiagen), and purified DNA was eluted in 10 μl of elution buffer (10 mM Tris-HCl, pH 8). Libraries were barcoded (Nextera Index Kit, Illumina), amplified with NEBNext High Fidelity PCR Mix (New England Biolabs), and cleaned using a 1x volume of AMPure XP beads. Libraries were quantified using Agilent BioAnalyzer and sequenced on the HiSeq High Output and NovaSeq Illumina Sequencers (25 bp, paired-end).
Bulk TCR-seq
Because we were unable to isolate sufficient numbers of viable BMMCs for samples from patient 5316, we were not able to collect single-cell TCR data and, instead, followed the rhTCRseq protocol for bulk TCR sequencing and repertoire analysis as described previously (Li et al., 2019b). Total RNA samples were extracted from DLI products, pre-infusion, and post-infusion BMMCs. From the RNA samples, we generated cDNA libraries with a reverse transcriptase reaction that appends a Unique Molecular Identifier (UMI) to each cDNA molecule to facilitate frequency calculations in later steps. We ensure TCRs are specifically amplified by performing RNase H-dependent PCR (rhPCR)–which uses 3’ blocked primers that incorporate a single ribo residue. The blocked ends are cleaved, and the proceeding sequence is amplified if, and only if, the primer is hybridized to the appropriate target. After rhPCR, we performed a second PCR on the pooled samples to create a sequencing library, followed by sequencing on the Miseq platform.
QUANTIFICATION AND STATISTICAL ANALYSIS
Preprocessing single-cell RNA-seq data
FASTQ files were preprocessed using the Sequence Quality Control (SEQC) bioinformatics pipeline (Azizi et al., 2018) with aligning reads to the hg38 genome and turning off the mitochondrial filter (using the option --no-filter-mitochondrial-rna). Empty droplets were identified using SEQC default parameters followed by further filtering of cell barcodes per sample. Specifically, if the histogram of log10 of library size (i.e. sum of counts per cell) was bimodal, the lower mode was removed. Characteristics of samples and quality control (QC) metrics are provided in Table S2. In total, 381,462 total cells including 87,939 T cells (identified in the next section) from the combination of 41 bone marrow (BM) samples passed SEQC QC metrics, with a median of 2548 UMIs/cell and 8735 cells/sample.
Constructing global single cell map of T cells
Identifying T cells.
To select T cells, we first normalized all n=381K BM cells to median library size and computed the log of normalized expression as for each cell j = (1,…,n) where contains the normalized expression of genes in cell j. To identify major cell types, we filtered genes expressed in less than 2% of cells (resulting in 9767 genes) and performed PCA on the log-transformed normalized expression. The number of PCs was selected based on the knee-point (defined as minimum curvature radius) of eigenvalues. Then cells were clustered by applying Phenograph (Levine et al., 2015) with the number of nearest neighbors set to 30, on the first 24 principal components (PCs), resulting in 94 clusters.
The normalized expression of {CD3D, CD3E} gene markers were averaged across cells in each Phenograph cluster and clusters with a high average expression of CD3 (right tail of distribution across all clusters) were selected as T cells, which consisted of 97,355 cells. A lower threshold of CD3 expression selected clusters with high expression of markers of other major cell types (myeloid, B or NK cells).
Biscuit normalizing and clustering.
To construct a more refined map of T cells, we performed simultaneous clustering and cluster-dependent normalization on raw counts for n = 97,355 T cells using Biscuit (Azizi et al., 2018; Prabhakaran et al., 2016) . Using a hierarchical Dirichlet process mixture model, Biscuit performs a cell-type dependent normalization on the count matrix where each column contains the expression (number of unique mRNA molecules) of d genes in cell j, while simultaneously inferring robust subsets of cells with zj denoting assignment of cell j to cluster k. Biscuit assumes that the log of counts follow a multivariate Normal distribution: where are the mean and covariance, respectively, of the k-th mixture component (cluster), and scalars αj,βj are cell-dependent scaling factors used for normalization. We have previously shown that this cluster-dependent normalization removes batch effects while retaining biological signal (Azizi et al., 2018). In particular, Biscuit helps retain biological processes that are entwined with library size. For example in the case of immune cell activation, activated cells have a higher number of transcripts (Blackinton and Keene, 2016; Cheadle, 2005; Marrack et al., 2000; Singer et al., 2016) leading to higher total counts captured, hence variation due to real immune activation can be partially removed with methods that normalize cells by library size, whereas Biscuit performs a more careful normalization of cells conditioned on the cell state (captured by cluster assignment).
For faster inference, we used the implementation described in (Azizi et al., 2018) (from https://github.com/sandhya212/BISCUIT_SingleCell_IMM_ICML_2016) which deploys a conjugate prior for the multivariate Gaussian, namely the Normal-inverse Wishart distribution for joint inference of cluster means and covariances.
After fitting the model, we transform the data from to in which the expression is corrected for cell-specific factors αjβj using a linear transformation with , such that imputed expression for cell j follows N(μk, Σk) and hence all cells assigned to the same cluster follow the same distribution after correction.
Using Biscuit with 500 iterations; gene batch size set to 50, and alpha (dispersion parameter) set to 200, we identified 65 unique clusters. This choice of parameters led to both relatively good mixing of samples (Figure 1B and Figure S1E), and distinct sets of differentially-expressed genes (Figure S1C). Only 3 clusters were found to be exclusive to one single patient (all 3 in NR 5326), who was the only patient with CML in blast crisis (Figure S1E, Table S1).
Figure S1E shows the distribution of each cluster across clinical groups of R/NR and pre/post-DLI. Prior to computing the distribution, the number of cells in each cluster was first normalized by the total number of cells in each clinical group to account for imbalanced cell/sample numbers. The size of bubbles in each cluster is proportional to the distribution of normalized values and each cluster (column) sums to 100%.
Importantly, the interpretability of Biscuit enables the use of inferred parameters in downstream characterization of clusters: The inferred cluster mean μk and its conjugate prior are used for estimating differentially expressed genes as detailed in the Cluster Annotation section below. To ensure each cluster is a legitimate cell population, we then scanned the clusters for doublets as explained below.
Removing doublets.
Doublet cells were identified by applying DoubletDetection (https://github.com/dpeerlab/DoubletDetection), using the Biscuit derived clusters, with 50 iterations and p_thresh=1e-6, voter_thresh=0.8 followed by inspection of the co-occurrence of contradictory markers (including T cell and B cell markers; T cell and myeloid markers, T cell and erythroid markers etc). With this approach, 8.4% of cells were marked as doublets, which matches expectations given our cell loading (described in Method Details). This resulted in 87,939 cells in 43 T cell clusters that were not flagged as doublets and retained for the remainder of the analysis.
Visualization.
The Biscuit-normalized data for the 87,939 cells are projected to 2D in Figure 1B and also expanded in Figure S6B using tSNE (Maaten and Hinton, 2008),(Amir et al., 2013) on the first 18 PCs (identified based on knee-point of eigenvalues - defined as min curvature radius).
Cluster annotation.
T cell clusters were annotated through: (1) identifying cell type signatures enriched in each cluster (listed in Table S7) by computing the expression of each signature (defined as average expression across all genes in a signature) per cluster and comparing to all other clusters using a t-test with p<0.1. The list of signatures compiled from literature are provided in Table S7. The expression of enriched cell type signatures are shown in Figure 1C and Figure S1D; (2) differentially expressed genes (DEGs) (Figure S1C, Figure S3A) were computed with t-test (p<0.01) comparing inferred mean expression of a gene in each cluster (listed in Table S5) to its prior mean μ′ which represents expression across the entire population of cells. Since Biscuit fits a multivariate Gaussian mixture model to log-transformed data, the assumptions for a t-test are satisfied. Figure S1C shows the specificity of most DEGs to clusters as a block diagonal structure. The DEGs are listed in Table S6.
The genesets derived from murine models of chronic viral infection (Im et al., 2016) were used for characterizing exhausted T cell subsets (Figure 3A) listed in Table S3. The TEX and TPEX score per cell was defined as normalized expression averaged across all genes in the geneset. Cell scores are aggregated by cluster in Figure 3A.
For signatures related to T cell differentiation states (Figure 1C, top), we used genesets from Gattinoni et al. (Gattinoni et al., 2017) To consider both up-regulated and down-regulated genes, we defined the expression of these signatures as a weighted sum of expression of genes in the geneset, with the weights being +1 or −1 for up-regulated and down-regulated genes respectively. We replaced CD45RO with the gene HNRNPLL gene which has been shown to regulate alternative splicing of CD45 (Oberdoerffer et al., 2008).
Quantifying Diversity of T cell states
We evaluated if response to DLI was associated with a change in the number of distinct T cell transcriptional states. We found a marked increase in the number of T cell clusters in post-DLI samples compared to matched pre-DLI samples after controlling for cell number (t-test p-value <0.001). For this test, we corrected for differences in the number of cells. We downsampled each clinical group (R/NR, pre-/post-DLI, control) to 5000 cells by uniformly sampling with replacement from each group and clustering using Phenograph (using 20 PCs, K=30). This process was repeated 20 times and the number of clusters were compared with a t-test (Figure S6C).
However, because T cell states are known to reside on continuous trajectories explaining the majority of variation (Azizi et al., 2018; Li et al., 2019a; Singer et al., 2016) we used the Phenotypic Volume metric devised in (Azizi et al., 2018) to compare the global transcriptional diversity between clinical groups and before/after DLI.
Phenotypic volume (V) for a subpopulation of cells is defined as the determinant of the gene expression covariance matrix for that subpopulation, which considers covariance between all gene pairs in addition to their variance. The covariance matrix can be written as Σ d x d and its pseudo-determinant det (Σ) is equal to the volume of a parallelepiped spanned by vectors of the covariance matrix (Tao and Vu, 2005) and can be computed as the product of nonzero eigenvalues of the covariance matrix. To improve sensitivity to noise and avoid multiplication of small nonzero eigenvalues, we compute the log of phenotypic volume which is the sum of log of non-zero eigenvalues:
for λe > ε representing the e-th non-zero eigenvalue (ε is set to a small value instead of zero to improve stability of the metric).
To correct for differences in number of cells, we downsampled each clinical group (R/NR, pre-/post-DLI, control) to 5000 cells by uniformly sampling with replacement from each group and computing the phenotypic volume. Only time points immediately pre-DLI and at remission post DLI (in Rs) were considered in this analysis. Patient 5321 was excluded in this analysis, as it did not have any post-DLI samples. Below is a list of samples used in this analysis.
List of scRNA-seq sample IDs from baseline pre-DLI and the remission timepoint following DLI:
| Patient ID | Outcome | Time | scRNA-seq Sample ID |
|---|---|---|---|
| 5309 | Responder | Pre | B05 |
| 5309 | Responder | Post | B06 |
| 5310 | Responder | Pre | B01 |
| 5310 | Responder | Post | B02 |
| 5311 | Responder | Pre | B09 |
| 5311 | Responder | Post | B12 |
| 5312 | Responder | Pre | B21 |
| 5312 | Responder | Post | B22 |
| 5314 | Responder | Pre | B25 |
| 5314 | Responder | Post | B26 |
| 5317 | Responder | Pre | B23 |
| 5317 | Responder | Post | B24 |
| 5318 | Non-Responder | Pre | B27 |
| 5318 | Non-Responder | Post | B28 |
| 5322 | Non-Responder | Pre | B03 |
| 5322 | Non-Responder | Post | B04 |
| 5324 | Non-Responder | Pre | B07 |
| 5324 | Non-Responder | Post | B08 |
| 5325 | Non-Responder | Pre | B17 |
| 5325 | Non-Responder | Post | B18 |
| 5326 | Non-Responder | Pre | B19 |
| 5326 | Non-Responder | Post | B20 |
This process was repeated 50 times to achieve a range summarized in boxplots in Figure 2A, Figure S3B showing statistically significant expansion of volume after DLI in both Rs and NRs. Importantly, the phenotypic volume is higher in Rs compared to NRs in particular in baseline (pre-DLI). Both R and NR cases exhibited increases in phenotypic volume induced by DLI (log fold change=104.6, p<10−6). At both pre- and post-DLI timepoints, phenotypic volumes in R cases were higher than that of NR cases, (mean R-pre vs mean NR-pre, log-fold change = 199.1, p<10−6; mean R-post vs mean NR-post, log-fold change = 49.3, p=1.5x10−6), but a far greater increase in phenotypic volume was observed within NRs than within R’s (log-fold change [NR-post vs pre] = 203.8 vs log fold change [R-post vs pre) = 54.1; p<10−6].
Comparing the pre-DLI volume to that in non-relapse control samples in Figure S3B reveals greater diversity of T cells in the leukemic microenvironment (in R/NR pre-DLI samples) than in non-relapse control samples which are leukemia-free.
Common Factor Analysis
We aimed to decompose the T cells to uncover components potentially corresponding to response/resistance. The samples in from baseline pre-DLI and the remission timepoint following DLI were used in this analysis. To correct for differences in numbers of cells across samples, we first downsampled T cells from each sample to 1000 cells, resulting in a total of 20,682 cells.
Applying PCA or diffusion component analysis (Coifman et al., 2010; Setty et al., 2016) showed that the top linear/nonlinear components explaining most of the variance across T cells are not highly correlated with response (Figure S2A). Instead, we used Common Factor Analysis (CFA), a method that assumes there are underlying latent (unknown) factors that explain shared variance between cells, and thus explains co-variation of cells Figure S2B illustrates an example where cells are varying along two trajectories that could be related to different gene programs, e.g. T cell activation and exhaustion. If these trajectories are correlated but not colinear, dimensionality reductions methods that maximize explained variance will capture the two trajectories. CFA however will seek underlying (latent) factors that explain the shared variance between the two trajectories, ignoring the portion of variance unique to cells. Our assumption is that response or resistance might involve underlying latent factors associated with multiple distinct processes that might co-vary across the cells. Thus, common factors identified through CFA could potentially be related to response or resistance mechanisms affecting the majority of cells through multiple pathways (Figure S2B). A brief description of CFA follows:
Shared factors are denoted as f1, f2,…,fm for expression of n = 20,682 cells denoted with x1,…,xn:
CFA assumes that cov(fi, fj) and cov(ϵi, ϵj) = 0 for i ≠ j and cov(ϵi, fj) = 0.
Common factors were extracted using factanal function in R (https://www.rdocumentation.org/packages/FAiR/versions/0.2-0/topics/Factanal) with the method of maximum likelihood and “varimax” rotation. Setting the number of factors to two, a chi-square test rejected the hypothesis of model fit (p<0.05). Hence, we increased the number of factors to three which indicated that the hypothesis of perfect fit cannot be rejected. The first three common factors (Figure 1D.) explain 67% of variance (29%, 20%, 18% of variance by each of factor 1 to 3 respectively) and separate groups of T cells enriched in Rs or NRs. To annotate the factors, we correlated the loadings of cells on each factor with expression of gene signatures. Figure 1E shows gene signatures with the highest correlations with factors 1-3. Figure S2D shows that the signatures enriched for factors 2 and 3 are mostly non-overlapping, thus suggesting the involvement of different T cell dysfunction mechanisms in DLI resistance. Increasing the number of common factors to 4 and 5, we did not find any gene signatures highly correlated with the additional factors and factor 4 showed weak correlation with Hypoxia. We repeated this analysis on multiple downsampled sets and achieved the same conclusions with regard to signatures most correlated with factors.
We also performed permutation tests with creating 500 randomly selected genesets of the same size of each manually curated geneset shown in Figure 1E and computing a null distribution for correlation with factors. We confirmed that the correlations observed between factors and manually curated genesets are indeed statistically significant compared to the null distribution (p<0.05).
Identifying T cell clusters enriched pre-therapy
We aimed to find any pre-DLI T cell states that are differentially enriched between Rs and NRs, that could potentially be predictive of response or resistance. Since different samples had differences in the total number of cells collected, this impacted our resolution of detecting a T cell state (cluster) in a patient. We therefore accounted for this uncertainty using a weighted one-sided t-test (using statsmodels.stats.weightstats.ttest_ind in Python). Within each clinical group (Rs or NRs), the weight of the i-thpatient was given by
with ni denoting total number of T cells in patient i pre-DLI and P = 6 being the total number of patients in that group (R or NR).
We also corrected the p-values for the size of clusters using a bootstrapping technique: For each cluster k with size uk, we randomly select uk number of cells from the pool of all (R or NR) samples, and compute the p-value using the above test. Repeating this for 2000 iterations, we achieve a null hypothesis for p-values. The actual p-value for the cluster is then compared to the null, resulting in an empirical FDR (q-value) calculation. Applying this to pre-DLI samples, we found clusters 4, 14, 21, and 27 were differentially enriched consistently across R patients compared to NRs (FDR<0.1) as shown in Figure 2B. These clusters are enriched for TEX gene signatures shown in Figure 3A,B.
Aligned with our global observation with common factor analysis, we did not find any clusters to be differentially enriched consistently across NR patients compared to Rs, and we rather found multiple clusters each mostly present in one NR patient (Figure S1E) suggesting that NR patients might be driven by different resistance mechanisms (Figure 1E, Figure S2D).
Identifying T cell dynamics associated with therapy outcome
We used a weighted t-test similar to the previous section to compare the change in proportion of each cluster from pre-DLI to post-DLI. We performed a weighted one-sided t-test, summing the total cells in the pre- and post-DLI samples (from P = 6 R patients and P = 5 NR patients who had both pre and post therapy samples). to determine the weights. Specifically, the expression we used for weights was:
Where ni,Pre represents total number of cells in the pre-DLI sample of i-th patient and ni,Post represents the total number of cells in the post-DLI sample of i-th patient. Similarly, nj,Pre represents total number of cells in the pre-DLI sample of j-th patient and nj,Post represents the total number of cells in the post-DLI sample of j-th patient.
Compared to the test in the previous section which was performed on cluster proportions at one time-point (pre-DLI), this test involves computing the change in proportion from pre-DLI to post-DLI. Hence, the variance in the variable being tested is higher while the sample size (in this case number of patients) remains the same, meaning we have lower statistical power. In fact, across paired, pre- to post-DLI timepoints, we found no single cluster to consistently expand or contract over time in Rs or NRs using the above weighted t-test. Thus, to improve our statistical power in detecting consistent changes in clusters over time, we combined clusters that are transcriptionally most similar as described below.
Defining meta-clusters.
We computed the pairwise distance between each pair of clusters by comparing the distribution of expression of each gene across all cells in one cluster (from Biscuit normalized data) and comparing it to the distribution in another cluster using the Bhattacharyya distance metric (Bhattacharyya, 1990), which is effective in pairwise comparisons of distributions. The advantage of computing cluster distances based on distribution is that we go beyond cluster means and also account for within-cluster variability, e.g. two clusters can have a similar mean expression but different variance. The total distance is then summarized across all genes, resulting in the distance matrix in Figure S3C. We then merged clusters that were most similar, resulting in 8 meta-clusters shown with white boxes.
Identifying expanding or contracting meta-clusters.
By applying the weighted t-test above, we identified two metaclusters consistently expanded and one consistently contracted after DLI therapy (weighted t-test p<0.1), only in Rs, shown in Figure 2C. The two expanding meta-clusters (MC1 consisting of clusters {19,28} and MC2 consisting of clusters {5,11,23}) are enriched for the Precursor Exhausted T cell gene signature TPEX shown in violin plots in Figure 3A and Figure S6D.
Interestingly, one expanding cluster (19 in MC1) is also enriched in the non-relapse control samples (Figure S1E), suggesting a transformation to normal T cell states after DLI in Rs. It should be noted that no meta-clusters or clusters consistently changed (expanding or contracting) in NRs, mirroring the Anna Karenina principle (Ahmed et al., 2019).
MC3 consisting of clusters {3,4,7,22} and MC4 consisting of clusters {2,14} (Figure S3C) are enriched for the Terminally Exhausted T cell gene signature TTEX.
Hierarchical Gaussian Process regression model
To study the dynamics of meta-clusters and tumor burden over time, we used a Gaussian Process (GP) model. The advantages of a GP model are (1) it is nonparametric, hence we do not assume a functional form over time and rather learn a distribution over all functions that explain temporal dynamics; (2) we account for dependencies between all pairs of time points which tackles the problem of non-uniform distribution of time-points in our cohort (Figure 1A), for example in patient 5311, we have time-points within 19 days of each other, whereas in patient 5314 we have time-points 2.8 years (1059-29 days) apart from each other post-DLI and including them in the study can elucidate long-term sustainability of T cell states; (3) the probabilistic framework is flexible and we can therefore add priors representing uncertainty in measurements as explained below.
Tumor burden dynamics.
We fit two GP regression models (, ), each with a Radial Basis Function (RBF) kernel(Vert), to model the temporal changes in tumor burden in each outcome group (R or NR) separately in response to DLI therapy:
where biR is tumor burden (see definition in section “Cytogenetic and molecular information on CML tumor burden” in Materials and Methods) in sample i in Rs and tiR is time relative to DLI therapy in sample i of Rs. Similarly for NR samples:
We optimized σs with the gradient-based algorithm Adam to maximize the log likelihood of our observed data. We set σϵ2 = 10 and λ = 285 (which is the median distance between pairs of points). Results were robust to the choice of these parameters as shown in the next section.
Prior to regression, the mean tumor burden in each clinical group was subtracted so that our target variable bi would have zero mean, consistent with the distribution over fbR/NR. This resulted in one model inferred for tumor burden in Rs (fbR) and one model for tumor burden in NRs (fbNR) shown in grey lines (mean) and shaded grey area (+/−1 standard deviation) in Figure 2D,E. The data points for tumor burden are shown in grey crosses.
Temporal dynamics of T cell clusters.
Similarly, we aimed to use a GP regression model to track the temporal dynamics of proportions of T cell meta-clusters in each outcome group. In other words, we learn two models fpkR, fpkNR on the proportion of each meta-cluster k over time separately in Rs and in NRs respectively. The proportion of a meta-cluster k in a sample i is defined as νi,k = mi,k/ni with mi,k being the number of cells in meta-cluster k in sample i and ni defined as sample size, i.e. total number of T cells in sample i.
Since there were significant differences in the size of samples and meta-clusters, we aimed to account for the uncertainty in detecting a metacluster in each sample (Figure S3C). For example, if metacluster k is not observed in two samples i1 and i2 such that: νi1,k = νi2,k, = 0 and sample i1 contains ni1 = 10000 total cells compared to ni2 = 1000 cells in sample i2, we have more certainty about the absence of metacluster k (representing a T cell state) in sample i1 than in sample i2 and the true value for νi2,k could be missing or underestimated due to lack of statistical power.
To build this uncertainty into the probabilistic framework, we use a Gaussian process regression model that accounts for heteroscedastic noise. The measurement precision (βi) has a conjugate Gamma prior, whose mean is inversely proportional to the number of T cells measured in a given sample. Specifically we set the shape parameter of the prior distribution for βi as r = 1, and use the inverse of the number of cells collected for sample i as the rate parameter θ. This places more confidence on samples with larger sizes. For this model we use the RBF kernel K, with entries Kij = k(ti, tj) and scale parameter σs set to the empirical variance of the response variable.
The full generative model is as follows:
where:
As with standard GP regression, after we fit our model to data t and ν, we use the following joint marginal distribution to estimate the expected μ* for an input t*. Specifically, let k* = k(ti, t*) be a vector representing the kernel function computed between each input time point ti in our training data, and our out of sample point t*, and let c* = k(ν*, ν*) be the kernel function computed on the out-of-sample time point. The joint distribution between our training data ν and the new point ν* is then as follows:
Because this is a multivariate normal distribution, we can use this distribution to compute the conditional distribution over fp* given our training data and t*:
where the predicted mean and covariance are defined as follows:
The plate model for this hierarchical GP model is shown in Figure S3F. We implemented this model in the probabilistic programming language pyro(Bingham, Eli and Chen, Jonathan P and Jankowiak, Martin and Obermeyer, Fritz and Pradhan, Neeraj and Karaletsos, Theofanis and Singh, Rohit and Szerlip, Paul and Horsfall, Paul and Goodman, Noah D, 2019) (https://pyro.ai/) and inferred the weights and temporal function with Stochastic Variational Inference, which computes an efficient approximation to the posterior by taking stochastic gradient steps to maximize the evidence lower bound (ELBO) (Blei et al., 2017). The code for our hierarchical GP model is available at: https://github.com/dpeerlab/dli_gpr.
We first benchmarked this model on data simulated from a sinusoidal process y = 5sin(x) (shown as a grey line below) with two different noise variances representing levels of uncertainty in measurement: y1 = 5sin(x1)+ϵ1 with ϵ1 ~ N(0,1) (data points shown in blue) and y2 = 5sin(x2) + ϵ2 with ϵ2 ~ N (0,10) (data points shown in red) in Figure S6E. Please note the y notation here is not to be confused with expression in the Biscuit or Symphony models.
We combined these two datasets and fit the above hierarchical GPR model and compared it to the fit of a standard GPR (without prior) showing that the hierarchical model performs better in reconstructing the underlying sinusoidal function while a standard GPR model can overfit the noisy portion of data as shown in Figure S6E.
For quantitative comparison of the two models, we computed the log likelihood of unobserved noiseless simulated data along with the R2 score of the noiseless data vs. mean of the conditional distribution. The performance of hierarchical GP on simulated data compared to standard GP regression is listed below.
| Model | Negative log likelihood | R2 score |
|---|---|---|
| Hierarchical GPR | 193.24 | 0.801 |
| Standard GPR | 336.68 | 0.412 |
We then applied the hierarchical GPR mode to all meta-clusters in both Rs and NRs (Figure 2D,E) and use (TEX-like) metacluster MC3 in Rs as an illustration. As reference, we compared the fit of the hierarchical model to a standard (vanilla) GP model (Figure S3G). The blue dots show the actual data points with the size of dots proportional to sample size ni. The blue line and shaded area shows mean and standard deviation of fpTE*.
Interestingly, the inferred hierarchical GP model shows that the TEX meta-cluster tracks the tumor burden dynamics. The strong similarity between the inferred fpMC3 and fb in Rs is quantified by correlation, i.e. cross correlation at zero lag. MC1 and MC2 (TPEX-like) meta-clusters did not show a correlation with tumor burden. Below is the similarity between inferred GP model for metacluster proportion and model for tumor burden in Rs:
| Metacluster | Correlation (Pearson R) at lag = 0 for Rs |
|---|---|
| MC1 | −0.7436 |
| MC2 | −0.2633 |
| MC3 | 0.9852 |
We found that the dynamics of MC3 do not follow tumor burden in NRs. Below is the similarity between inferred GP model for metacluster proportion and model for tumor burden in NRs
| Metacluster | Correlation (Pearson R) at lag = 0 for NR |
|---|---|
| MC1 | 0.6907 |
| MC2 | −0.7549 |
| MC3 | −0.7009 |
Additionally, the expansion of early differentiated, TPEX-like clusters post-DLI is durable in Rs and nonexistent in NRs. Results were robust to choice of of σϵ and λ. As shown in Figure S6F, similar fit is achieved on a range of values for λ from 150-300 compared to 285, which is the median distance between pairs of points, and the value used to generate Figure 2D,E.
This example shows tumor burden and proportion of late differentiated, TEX-like metacluster MC3 in non-responders. To quantify the relative timing of TEX-like and TPEX-like meta-clusters, we computed the cross-correlation between fp* and fb shown as purple bars in Figure 2D (second row). The max cross-correlation between MC3 and tumor burden in Rs (max{fpTE* ⋆ fbR} with ⋆ indicating cross-correlation) is at 75 days which is 1/4 of median time interval between samples (marked with a red line in Figure 2D left bottom; t-statistic=8.58, p=0) indicating they are almost in sync, whereas for the TPEX meta-clusters, max{fpPE* ⋆ fbR} occurs at 703 days (MC1: t-statistic=2.05, p=0.02; MC2: t-statistic=0.72, p=0.23) indicating a significant lag compared to the tumor burden.
Preprocessing ATAC-seq data
Bulk ATAC-seq data for each sorted subset of T cells from each bone marrow sample was processed using the automated end-to-end quality control (QC) and processing pipeline (https://github.com/kundajelab/atac_dnase_pipelines) from the ENCODE consortium with configuration SPECIES=hg38. Alignment is performed using Bowtie2(Langmead and Salzberg, 2012) and peak calling and normalization is done with MACS2 (Zhang et al., 2008). MACS2 normalization involves comparing ATAC signal to local background noise using a Poisson test (Zhang et al., 2008)(Reske et al., 2020). The full list of samples and QC metrics for ATAC-seq data are provided in Table S8.
Correlation between accessibility profiles
We first aimed to study the potential impact of DLI in the global epigenetic landscape of T cells. We thus compared ATAC-seq samples with ID listed below. To compare chromatin accessibility between pairs of samples, we first created a consensus peak set similar to (Corces et al., 2016) as follows: Peak summits were extended to 150bp windows and a set of maximally non-overlapping peaks was generated across all samples, resulting in 133,968 peaks for CD8+ CD45RO+ and 169,740 peaks for CD8+ CD45RA+ samples. Then Pearson correlation was computed between all pairs of 14 samples in each subset, and then correlations were averaged by pairs of clinical groups (Figure 4C,D).
List of ATAC-seq samples from IDs from baseline pre-DLI and the remission timepoint following DLI; n/a denotes low sample quality or excluded based on data preprocessing QC:
| Patient | Outcome | Time | ATAC-seq sample ID for CD8 CD45RA sorted T cells |
ATAC-seq sample ID for CD8 CD45RA sorted T cells |
|---|---|---|---|---|
| 5309 | Responder | Pre | C44 | C45 |
| 5309 | Responder | Post | n/a | n/a |
| 5310 | Responder | Pre | C31 | C32 |
| 5310 | Responder | Post | C35 | C36 |
| 5311 | Responder | Pre | C63 | C66 |
| 5311 | Responder | Post | C79 | C81 |
| 5312 | Responder | Pre | C105, C106 | n/a |
| 5312 | Responder | Post | n/a | n/a |
| 5314 | Responder | Pre | n/a | n/a |
| 5314 | Responder | Post | n/a | C130 |
| 5317 | Responder | Pre | n/a | C114 |
| 5317 | Responder | Post | C118 | C119 |
| 5318 | Non-Responder | Pre | C133, C134 | n/a |
| 5318 | Non-Responder | Post | n/a | C156 |
| 5322 | Non-Responder | Pre | C38 | C39 |
| 5322 | Non-Responder | Post | C41 | C42 |
| 5324 | Non-Responder | Pre | C48 | C49 |
| 5324 | Non-Responder | Post | C54 | C56 |
| 5325 | Non-Responder | Pre | C91 | C92 |
| 5325 | Non-Responder | Post | C95 | n/a |
| 5326 | Non-Responder | Pre | n/a | n/a |
| 5326 | Non-Responder | Post | n/a | n/a |
Symphony model for cell type-specific gene regulatory networks
To study the underlying circuitry of distinct clusters, we developed an integrative model named Symphony (C. Burdziak, E. Azizi, S. Prabhakaran, D. Pe’er, 2019), for inferring gene regulatory networks (GRNs) specific to subsets of cells.
Gene regulatory networks (GRNs) are directed weighted networks between genes depicting the extent to which a regulator gene influences (activation or repression) the expression of each of its downstream target genes. Symphony estimates these networks in each subset by extracting co-expression patterns between TFs and target genes from scRNA-seq and combining them with the presence of TF motifs within regions of chromatin accessibility in the vicinity of targets as derived from ATAC-seq. This is accomplished in Symphony by constructing a generative model that mimics transcriptional regulation illustrated in Figure 5A.
Since the ATAC-seq data in this study measures accessibility summarized across all cells in a sorted compartment (e.g. CD8+CD45RO+) each consisting of multiple TTEX or TPEX clusters, we also leveraged the deconvolution capability of Symphony: bulk epigenetic data is deconvolved into cluster-specific epigenetic profiles. The deconvolved profiles are then used to explain gene co-expression patterns through GRNs, and thus resolve direct links from indirect links in the network (Figure 5A).
Symphony (C. Burdziak, E. Azizi, S. Prabhakaran, D. Pe’er, 2019) is an extension of the Biscuit (Azizi et al., 2018; Prabhakaran et al., 2016) model which clusters cells while simultaneously distinguishing biological heterogeneity from technical noise in single-cell gene expression data. Symphony extends this model by replacing the hyperparameter for gene co-expression in Biscuit with a generative process exclusively driven by epigenetic data (collected from the same sample or a sample with similar composition of cell types). Thus, Symphony models the biological mechanism responsible for the observed gene co-expressions per cell type.
The model also simultaneously deconvolves the bulk epigenetic profiles (which denote accessible DNA) into cell-type (cluster)-specific accessible regions (Figure 5A) within a unified statistical framework. Within these regions, the binding of transcription factors (TF associated with open regions based on known DNA binding motifs) impacts the expression of nearby genes, such that accessible regions may help explain gene-gene interactions.
Given the observed bulk chromatin accessibility profiles and single-cell RNA-seq count matrix, the model finds a deconvolution of the bulk accessibility data into cluster-specific accessibility profiles that are best able to explain the gene-gene relationships observed in scRNA-seq. We note that Symphony can infer whether a TF impacts a target gene without requiring epigenetic evidence as well, which facilitates inferring the regulatory influence of the many TFs (e.g. TOX) for which a binding motif is unknown.
Symphony input, output and model specification are provided below:
Input data to Symphony.
The observed paired datasets are:
(1) Epigenetic data measured with ATAC-seq (Buenrostro et al., 2015), denoted as Cw×r = [c1,…,ct,…,cr] where ct ∈ Rw is epigenetic data for one patient (as replicate), containing accessibility (quantified as peak height) in genomic regions m = [1,…,w] (identified from MACS2(Zhang et al., 2008)).
(2) Single-cell RNA-seq data where denotes log-transformed normalized single-cell expression data for cell j with d genes.
Symphony output.
The main latent variables being estimated (Figure 5A) are:
(1) Epigenetic profile for each cluster k represented as pk ∈ R+w which contains estimated genome accessibility in w genomic regions.
(2) Gene Regulatory Network (GRN) represented as Rk for each cluster k. Rkd×d is an asymmetric matrix with nonzero entries Rka,b ≠ 0 if gene b is predicted to be regulated by gene a. Positive and negative values for Rka,b suggest activation and repression respectively.
Model details.
These latent parameters are estimated simultaneously in an integrative model with three components explained below:
Epigenetic model.
Bulk epigenetic profiles (ct) are assumed to be represented as a weighted sum of cluster-specific epigenetic profiles (pk) such that:
where the weights πk represent the proportion of clusters in the sample. This assumption is validated in (C. Burdziak, E. Azizi, S. Prabhakaran, D. Pe’er, 2019) using data on PBMCs with ground truth deconvolved profiles.
We set a Gamma prior for accessibility: pk ~ Gamma(η, Λ) to ensure a positive domain.
GRN model.
We assume a regulatory link is dependent on genome accessibility as well as motif information within an accessible region. Specifically, a genomic region m in C is mapped to an interaction between genes a, b in Y with a predefined function g(a, b) = m. We also define Md×d based on prior knowledge: Ma,b = 1 if the motif sequence for gene a exists in region m in the vicinity of gene b, suggesting a potential regulatory interaction from gene a to gene b. Motifs were scanned using FIMO (Grant et al., 2011) in this study.
We thus model Rka,b as:
Where Sis a sign indicator representing activation or repression set according to the sign of empirical covariance:
Σ′a,b is an empirical prior set to the covariance between genes a, b across all cells in the scRNA-seq data. The variance λallows for Rka,b to have non-zero value, even when Ma,b = 0.
Expression model.
Similar to Biscuit(Azizi et al., 2018; Prabhakaran et al., 2016), Symphony assumes that log-transformed normalized single-cell expression data follows a multivariate Normal distribution:
where zj denotes the assignment of cell j to cluster k modeled as:
Since the single cell expression data was already normalized and clustered with Biscuit as explained in section “Constructing global single cell map of T cells”, we did not use the clustering feature of Symphony and instead fixed the assignments (zj) of cells to clusters as assigned by Biscuit; the proportions πk are thus also fixed. The full normalized expression matrix (output from Biscuit) is thus used as the second input to Symphony in this case. However, as a more general tool Symphony is also able to successfully cluster de-novo as demonstrated in simulated data (C. Burdziak, E. Azizi, S. Prabhakaran, D. Pe’er, 2019).
The parameters are the mean and covariance, respectively, of the k-th cluster. We define the prior for in Symphony as follows:
where μ′ is set to the empirical mean expression across all cells and Σ′ was set to I (identity) in this study.
Importantly, the covariance in observed gene expression is related to a graph power of the regulatory network, capturing the propagated impact of regulation in the network (indirect regulation) as depicted in Figure S7A. Specifically, co-expressed in each cluster is modeled as:
While using a Wishart instead of Inverse Wishart is not conjugate, this is valid as both distributions satisfy the positive semi-definite requirements for priors on the covariance matrix. The plate model for Symphony used in this study is shown in Figure S7B.
Inference, approximations and scalable implementation.
An EM-VI inference procedure was presented for Symphony in (C. Burdziak, E. Azizi, S. Prabhakaran, D. Pe’er, 2019). We also showed the performance of Symphony on well-characterized peripheral blood mononuclear cells (PBMCs), and significant improvement over other deconvolution methods (C. Burdziak, E. Azizi, S. Prabhakaran, D. Pe’er, 2019). In this study, given the complexity of the model and size of data, we used a scalable implementation of Symphony (C. Burdziak, E. Azizi, S. Prabhakaran, D. Pe’er, 2019) in the probabilistic programming language Edward (Tran et al., 2016). This implementation in Edward is provided in https://github.com/dpeerlab/Symphony with example input data for group 1.
The use of variational algorithms in Edward (Tran et al., 2016) allows for fast approximations of the posterior for large gene-by-gene matrices including GRNs and covariances per cluster, and scales well to additional cells and ATAC-seq replicates. Setting constraints on covariance matrices of a multivariate normal distribution are difficult to enforce in the optimization setting of variational inference. Thus, to avoid non-singularity issues during optimization, we define the Wishart distribution in Edward using the Bartlett Decomposition, rather than the built-in Wishart function of tensorflow, which allows us to more easily define variational parameters.
Specifically, we replace the sampling of covariance matrices Σk∣Rk ~ Wishart with a generative model constructed from univariate chi-squared distributions and normal distributions, which can be shown to produce a valid sample from the Wishart distribution (Kshirsagar, 1959). Given Lk as a cholesky factor of the prior (Rk + RkT)2, we sample the cluster-specific covariance as follows:
where Ak is a lower triangular matrix whose diagonal elements are composed of χ2 random variables with γ – i + 1 degrees of freedom, where i indexes the rows of Ak, and the off-diagonal elements in the lower triangle are independent normal distributions. Hence each Σk is a positive semi-definite matrix centered at LkLkT or equivalently (Rk + RkT)2. In this setting, we define variational distributions corresponding to the dummy variables h ~ chi squared and v ~ Normal, as opposed to defining a matrix variate distribution which, during the course of optimization, must fit all the constraints of valid covariance matrices.
Still, in the Edward implementation, we observed that the Barlett product often produced matrices which are not positive semi-definite due to numerical instability, and hence did not generate a valid covariance matrix. As such, we approximated the mean of Σk with the highly-related (unitarily similar) matrix LkTLk, which we ensured produced a posterior in covariance which is highly correlated with its mean derived from the posterior GRNs (minimum correlation r=0.745 across all groups in this paper). For additional speed, the cholesky factor was computed from Gram matrix (Rk + RkT)2 using the QR decomposition of (Rk + RkT) where (Rk′ + LkT) given Qk′Rk′ = QR(Rk + RkT).
In addition to the use of the Bartlett Decomposition, the Edward version of Symphony replaces the standard Wishart with a scaled Wishart for added flexibility of the model in the variational inference case. The scaled Wishart necessitates addition of a latent parameter per cluster δk, such that Σk′ ~ Wishart and δk ~ Normal and Σk = ΔkΣk′Δk where diag(Δk) = δk.
Addition of the normal distribution above to the generative process infuses flexibility to the Wishart, whose variance is usually defined by a single parameter (degrees of freedom (Alvarez et al., 2014)). In addition, the resulting matrix will have a diagonal scaled by δki2, hence allowing better fit to the empirical per-gene variances which are not captured directly by the regulatory model driving the prior for covariances. Off-diagonal elements are scaled by δkiδkj, a transformation which decouples the correlation structure embedded in the off-diagonal elements from the scaling of the diagonal. Specifically, correlations between genes in the original matrix Σk′ are encoded as Σ′k,ij/σ′iσ′j. After scaling, δ’s in the numerator and denominator cancel, hence allowing the overall structure to be maintained under any arbitrary scaling of per-gene variances to fit the empirical data per cluster.
We note that with the above approximations, the constraint on the sign of Rk is not always enforced to be the same as Σ′. Thus, we have more confidence in the inferred strength of regulation (magnitude of Rk). The estimated regulatory strength is used to identify master regulators in Figure 5B (as explained in section “master regulators” below). We also show the robustness of inferred regulatory strength in the section “robustness analysis” below.
Guide for choice of parameters.
The variational inference implementation of Symphony requires choice of several hyperparameters. By default, priors on cluster mean expression are set with empirical means across the cells in that cluster as explained above, and shape and rate parameters for the Gamma prior on peak heights are set as 4.5 and 1 respectively for a relatively uninformative prior. Other parameters, particularly those controlling the variance of distributions in the generative model, are user-defined and should be tuned to each dataset.
As Symphony is designed to manage a trade-off between fitting to expression covariance and chromatin accessibility in the posterior distribution over GRNs, the choice of variance parameter on the prior distribution for each Rk denoted by λ, as well as the degrees of freedom in Wishart linking Rk to Σk denoted by γ, can be chosen to prioritize fit to each type of data. To inform the choice of these parameters, we recommend setting these parameters with small values and checking the empirical fit of the posterior to both data types. For example, the parameter settings used in this study (λ = 0.005, γ = d + 1 where d is the number of genes) ensured strong correlation of posterior GRNs with both the inferred peak heights, which in turn associated strongly with the bulk accessibility data, and further with the posterior covariance which itself associated with the empirical covariance. We also track these correlations over inference to ensure they increase over iterations. Details can be found in https://github.com/dpeerlab/Symphony.
Performance of Symphony on PBMC Data.
Prior to utilizing Symphony for discovery of regulators in our DLI cohort, we evaluated the performance of Symphony on an independent dataset from the well-studied PBMC system. We obtained publicly available PBMC scRNA-seq data from (Zheng et al., 2017) and chose a subset of 6825 cells expressing markers for either B, T, NK cells, or Monocytes. For proper normalization of these subsets, we corrected data using the Biscuit algorithm (Azizi et al., 2018; Prabhakaran et al., 2016) prior to applying Symphony, and selected a subset of 500 highly variable genes. Clusters from Biscuit were manually merged to obtain a distinct cluster defining each of 5 major celltypes: B cells, CD8 T cells, CD4 T cells, NK cells, and Monocytes (Figure S7C). These assignments were used as input to Symphony to fix the GRN and peak accessibility inference to these specific phenotypic categories. For epigenetic data, we obtained processed bulk ATAC-seq on PBMCs from (Kukurba et al., 2016) for two samples. Peaks were lifted over from hg19 to hg38 genome builds using the UCSC genome browser (Kent et al., 2002) liftover tool. To derive Symphony inputs, motifs were identified in peak regions using FIMO (Grant et al., 2011). The ATAC-seq peak counts reported in the original study were used as inputs for bulk chromatin accessibility measurements.
Figure S7C shows the results of Symphony applied to the PBMC data. First, we evaluated the extent to which key cell type regulators are identified through the cell type -specific GRN posteriors. To do so, we considered the absolute value (magnitude) of edge weights emanating from each of 8 regulators per cell type, which would indicate the degree of predicted activity for each TF in particular clusters (Figure S7C). As expected, we observed key regulators CEBPA, CEBPB, and CEBPD are substantially more active in Monocytes. A recent study (Jaitin et al., 2016) has shown that knock-outs of CEBPB block monocyte differentiation. We observe similarly strong regulatory edges in the GRNs for CD8 T cells between TFs RUNX3, EOMES and their target genes, and indeed these TFs are known to be associated with CD8 T cell development (Woolf et al., 2003) and activation of cytotoxic T cells (Pearce et al., 2003) respectively. We find that GATA3, a well-known master regulator of Th2 T cell subsets (Zheng and Flavell, 1997; Zhu et al., 2004), has the strongest (highest mean) regulatory interactions in CD4 T cells, followed by CD8 T cells where it has also been shown to be functionally active (Tai et al., 2013). Finally, PAX5 has several strong links exclusively within B cell subsets, as would be expected for its role as a regulator of B cell maintenance and function (Cobaleda et al., 2007; Schebesta et al., 2007). Thus, the GRNs output from Symphony successfully recovered many well-known master regulators in PBMC subsets.
To then evaluate the peak deconvolution function of Symphony, we inspected the posterior peak heights across each of the 5 cell subsets. Figure S7C also shows a heatmap of the posterior peak accessibilities z-scored to display differences across the cell types, with peaks (rows) ordered by a kmeans clustering of normalized peak heights to highlight accessibility modules. We observe several modules which are clearly differential across the cell subsets, suggesting that the posterior of our model peak heights can capture cell type specific regulatory activities. To interpret these modules, we annotated peaks’ target genes for several cell type markers. This revealed that cell type markers tend to fall in modules which are more highly accessible in their respective cell types. For instance, we find peaks associated with B cell marker CD79A fall within a B cell -specific module, and likewise for Monocyte marker CD14 and NK cell marker GNLY. This suggests that the deconvolution is identifying biologically-meaningful differences in peak accessibility. We also confirmed that the deconvolution correlates highly with the original bulk data (0.39<Spearman r<.51 for pairwise correlations of each cell type specific peak height vs. each bulk replicate) to ensure that the deconvolution is not over-fitting to covariances, but rather fits strongly to the input ATAC-seq data as well.
ATAC-seq samples used in Symphony.
Prior to running Symphony, TEX-like and TPEX-like clusters that fell in the same sort compartment of CD4 or CD8, CD45RA or CD45RO were grouped together as listed below. Figure 4A,B and Figure S4A show ATAC-seq accessibility profiles for these samples (full list of samples and QC metrics are provided in Table S8). Bigwig files were loaded to IGV(Robinson et al., 2011) to visualize normalized accessibility signal with differential accessibility identified with DESeq2(Love et al., 2014).
Groups of ATAC-seq samples used for deconvolution of accessibility profiles in Symphony:
| Symphony deconvolution group |
Enriched exhaustion state |
Enriched exhausted clusters |
cell type | ATAC-seq sample ID |
|---|---|---|---|---|
| 1 | T_EX-like | 14,27 | CD8 CD45RO | C149 |
| 1 | T_EX-like | 14,27 | CD8 CD45RO | C156 |
| 1 | T_EX-like | 14,27 | CD8 CD45RO | C45 |
| 1 | T_EX-like | 14,27 | CD8 CD45RO | C66 |
| 1 | T_EX-like | 14,27 | CD8 CD45RO | C75 |
| 2 | T_EX-like | MC3 (4,7,3,22) | CD8 CD45RA | C70 |
| 2 | T_EX-like | MC3 (4,7,3,22) | CD8 CD45RA | C171 |
| 2 | T_EX-like | MC3 (4,7,3,22) | CD8 CD45RA | C167 |
| 2 | T_EX-like | MC3 (4,7,3,22) | CD8 CD45RA | C144 |
| 2 | T_EX-like | MC3 (4,7,3,22) | CD8 CD45RA | C139 |
| 2 | T_EX-like | MC3 (4,7,3,22) | CD8 CD45RA | C106 |
| 3 | P_EX-like | MC2 (5,11,23) | CD8 CD45RO | C39 |
| 3 | P_EX-like | MC2 (5,11,23) | CD8 CD45RO | C36 |
| 3 | P_EX-like | MC2 (5,11,23) | CD8 CD45RO | C156 |
| 3 | P_EX-like | MC2 (5,11,23) | CD8 CD45RO | C164 |
| 3 | P_EX-like | MC2 (5,11,23) | CD8 CD45RO | C168 |
| 4 | P_EX-like | MC2 (5,11,23) | CD4 CD45RO | C37 |
| 4 | P_EX-like | MC2 (5,11,23) | CD4 CD45RO | C34 |
| 4 | P_EX-like | MC2 (5,11,23) | CD4 CD45RO | C127 |
| 4 | P_EX-like | MC2 (5,11,23) | CD4 CD45RO | C158 |
| 4 | P_EX-like | MC2 (5,11,23) | CD4 CD45RO | C162 |
| 5 | P_EX-like | MC1 (19,28) | CD8 CD45RA | C41 |
| 5 | P_EX-like | MC1 (19,28) | CD8 CD45RA | C79 |
| 5 | P_EX-like | MC1 (19,28) | CD8 CD45RA | C118 |
| 5 | P_EX-like | MC1 (19,28) | CD8 CD45RA | C35 |
| 6 | P_EX-like | MC1 (19,28) | CD4 CD45RA | C76 |
| 6 | P_EX-like | MC1 (19,28) | CD4 CD45RA | C33 |
| 6 | P_EX-like | MC1 (19,28) | CD4 CD45RA | C116 |
In each group 1-6, scRNA-seq data and ATAC-seq data from the same samples are used as input to Symphony. Bulk ATAC-seq samples from different patients are assumed as biological replicates, and deconvolved using Symphony to achieve accessibility profiles for each cluster. Combined with scRNA-seq data for the clusters, Symphony infers a GRN for each cluster shown in Figure 5C and Figure S5. We limited target genes to the pool of differentially expressed markers (Table S6) across clusters. We filtered inferred regulatory links (entries of Rk) that had a magnitude less than two (∣Rk∣<2 selected based on knee-point of distribution, ∣CV∣>0.5).
Runtime.
With this implementation, the runtime for Symphony was 1h 52m on group 4 containing 2593 cells and 1305 pooled DEGs and 5h 54m on group 1 with 7181 cells and 1459 DEGs, on a local machine with 64GB of RAM and 12 CPU cores (2.7 GHz processors). This runtime is at least 40 times faster than MCMC inference used in Biscuit which has a similar model structure.
Robustness analysis.
To test the robustness of GRN inference, we performed a leave one (patient) out analysis in the TEX CD8 and TPEX CD4 groups. Specifically, we fit Symphony to scRNA-seq and ATAC-seq data for each group and excluded ATAC-seq data from one patient at a time. We then compared the coefficient of variation (CV) of predicted regulatory links across the leave-one-out iterations to the inferred regulation from the entire data. As shown in Figure S4B, CV is lower for stronger regulatory links and the majority of links have CV<1.
Master regulators.
We used the output GRNs from Symphony to identify master regulators of each cluster as follows: For cluster k, we averaged the inferred impact of each TF a, across all targets b that are differentially expressed genes (DEGs) in the cluster (listed in Table S7): Σb ∈ DEGk∣Rka,b∣/Dk with Dk being the number of DEGs for cluster k. The resulting average regulatory strength of each TF in each cluster is shown in Figure 5B. We performed a one-sided t-test between late differentiated, TEX-like clusters and all other exhausted clusters to find “differential regulators” of TEX-like clusters shown with dotted line box in Figure 5B, and green nodes in Figure 5C and Figure S5. Similarly, we identified differential regulators of early differentiated, TPEX-like MC1 and MC2 subsets (Figure 5B) shown as pink nodes in Figure 5C and Figure S5.
Regulatory network.
To elucidate the target genes impacted most by these master regulators, we filtered the GRNs by centrality or out-degree of regulators (defined as number of target genes predicted to be regulated by the TF) as well as regulatory strength (∣Rk∣>2). Figure 5C and Figure S5 show subnetworks containing individual known (Man et al., 2017)(Wu et al., 2016) and previously unexplored links. The circuitry for exhausted clusters reveals similarity and differences in network architecture across clusters. We identified mediating regulators such as BCL6 connecting two other regulators (TBPL1 and E2F2) differentially regulating cluster 27. The network link predictions are supported by co-expression and/or accessibility (Figure 5C). Other predicted repressors such as TCF7L2 are supported by mutually exclusive (negative) co-expression patterns with DEGs.
Analysis of paired single-cell TCR and RNA-seq data
Single cell 5’ RNA-seq reads were processed with the Cell Ranger pipeline available from 10x Genomics. QC metrics for this data is provided in Table S9. A total of 23K total T cells were identified based on {CD3D, CD3E} expression (similar to section “Constructing global single cell map of T cells”) and normalized and clustered using Biscuit with the same parameters mentioned before. The 29 newly identified clusters were scored for the same TPEX and TEX signatures (listed in Table S3), and the clusters with the highest scores were identified as TPEX-like and TEX-like clusters.
Preprocessing and analysis of TCR clonotypes.
Single cell TCR-seq reads were aligned to the GRCh38 reference genome and consensus TCR annotation was performed using Cell Ranger V(D)J (10x Genomics, version 2.1.0.). QC metrics are provided in Table S9.
Clonotypes mapping to TRB loci were used to annotate each cell, similar to others(Yost et al., 2019). Overlap between clonotypes from TEX-like cells and TPEX-like cells (Figure 6A) was measured by counting the number of cells from each group per clonotype and performing a hypergeometric test using the phyper function with R. Venn diagrams were drawn using the eulerr package.
TCR diversity (Figure 6B) was calculated between all RNA clusters on a per patient basis via Gini coefficient(Dixon et al., 1987) using the ineq() function within the ineq package.
To determine the kinetics of TEX-like and TPEX-like clonotypes after DLI (Figure 6D, Figure 7A-D), the proportion of pre- and post-treatment cells were calculated for both patients together. Clonotypes were defined as expanding if they significantly enriched pre-DLI (p<0.05 according to Fisher’s exact test), contracting if they were enriched post-DLI (p<0.05 by Fisher’s exact test), and persistent otherwise. Viral-specific clonotypes were identified via VDJdb(Bagaev et al., 2020) and marked (V). Statistical analysis was performed in R version 3.5.3. Plots were generated using the ggplot package.
For analysis of bulk TCRseq, following sequencing we analyzed the resulting fastq files for unique TCR clonotypes using an in-house rhTCRseq analysis pipeline. The pipeline first separates reads by alignment to either TRA or TRB locus using BLAST. After separating the reads by locus of origin, the pipeline assembles component V, D, and J regions into TCR clonotypes with MiXCR and collapses the assemblies by CDR3 similarity into a list of unique clonotypes and associated frequencies. Figure S7E shows the percentage of TCR clonotypes (based on mapping to TRB loci) in the post-DLI sample (total, n=471) shared with the DLI product (total, n=68).
Analysis of Validation Cohort.
CITE-seq data was processed using Cellranger 5.0.1 count function and setting Gene Expression and Antibody Capture library types in the libraries argument. The chemistry argument was set to SC5P-R2. Transcriptomic data for T cells was normalized with Biscuit similar to the discovery cohort analysis explained above (Figure 3C). CITE protein expression data was normalized by median of total counts across all proteins per cell (Figure 3D, Figure S4C). The proportion of T cells assigned to TEX-like or TPEX-like groups in each sample is shown in Figure 3E.
We mapped the 5-prime transcriptomic data for each individual T cell in the validation cohort to a cluster identified from the 3-prime discovery cohort (Figure 1B) by computing the likelihood of its assignment to the cluster according to mean and covariance model parameters inferred for the cluster. We limited this computation to 2517 genes that were differentially expressed in at least one 3-prime cluster. We identified T cells with maximum likelihood of mapping to either MC1 or MC2 metacusters and labeled them as TPEX-like and similarly T cells mapped to either MC3 or MC4 metaclusters as TEX-like.
Analysis of CLL inDrop.
Raw count data were initially processed using the dropEst software suite as previously described (Bachireddy et al., 2020; Petukhov et al., 2018). Count matrices from bone marrow inDrop datasets for the patient with CLL were first filtered by library size, with a total of 12,254 T cells (3112, 5487, 3655 T cells from F1, F2, F3 samples respectively) remaining cells over 3 timepoints, which included 0 (pre-DLI), and 6 and 128 months post-DLI (Figure 3F; Figure S7D). Normalization and UMAP projection were performed using scanpy. T cells were then isolated by clusters with the highest expression of CD3E and CD3D. T-cells were renormalized by median of total counts across all genes per cell and then clustered using PhenoGraph clustering and k=15. TEX-like and TPEX-like clusters were identified using gene expression markers in Figure 3B. Top TEX-like and TPEX-like cluster groupings were identified as clusters [11,2,1,15,13,6] and [5,16] respectively. Line plots were generated for each TEX-like and TPEX-like group by summing the number of cells belonging to each cluster grouping at each time point and then dividing by the total number of cells in that time point. Heatmap was generated by z-scoring expression across each gene of interest for all clusters.
ADDITIONAL RESOURCES
Because the clinical trials from which these samples were obtained (94-009, 95-011, 96-372, 96-022, and 96-277) were conducted before Clinicaltrials.gov was made available to the public in 2000, these trials are not associated with a clinical registry number.
An earlier version of this manuscript was published at https://www.biorxiv.org/content/10.1101/2020.07.08.194332v1
Supplementary Material
ACKNOWLEDGMENTS
We thank John Ray, Jaeyoung Chun, Manu Setty, and Daniel Kim for their valuable discussions on ATAC and scRNA-seq profiling and analysis, and Sandhya Prabhakaran for discussions on Symphony. We also thank all members of the Wu and Pe’er laboratories for helpful discussions and Brian Miller, Tal Nawy, Eva Petschnigg and Mandar Muzumdar for valuable feedback on the manuscript. We appreciate the assistance of Nikolaos Barkas and Peter Kharchenko for preprocessing of inDrop data. Finally, we are grateful for the study nurses and clinical staff that obtained samples, the Pasquarello Tissue Bank in Hematologic Malignancies for processing and banking samples, and the patients who generously consented for the research use of these samples. This work was supported in part by the NCI grants 1R01CA155010 (C.J.W.), P01CA206978 (C.J.W.), U10CA180861 (C.J.W.), U24CA224331 (C.J.W.), P01CA229092 (R.J.S, J.R, C.J.W), R50CA251956 (S.L.) and U54CA209975 (D.P.); Ludwig Cancer Research (D.P.); The G. Harold and Leila Y. Mathers Foundation (C.J.W) and the Parker Institute for Cancer Immunotherapy (C.J.W, D.P.). P.B. was supported by a Physician-Scientist Training Award from the Damon Runyon Cancer Research Foundation, an Amy Strelzer Manasevit Scholar Award from the Be The Match Foundation, an American Society of Hematology Fellow Scholar Award, and NCI grant 1K08CA248458-01. E.A. was supported by NCI grant K99CA230195 and an American Cancer Society Postdoctoral Fellowship (PF-17-243-01-RMC). C.B. was supported by NCI grant F31CA246901. D.N. was supported by a grant from the NIH (5P30 CA006516). C.J.W. is a Scholar of the Leukemia and Lymphoma Society.
Footnotes
DECLARATION OF INTERESTS
C.J.W. is an equity holder of BioNTech. P.B. reports equity in Agenus, Amgen, Breakbio Corp., Johnson & Johnson, Exelixis, and BioNTech. C.J.W. and D.N. receive research funding from Pharmacyclics. D.N. reports stock ownership in Madrigal Pharmaceuticals. V.N.N. is an employee of Bluebird Bio. D.B.K. has previously advised Neon Therapeutics and has received consulting fees from Neon Therapeutics, and owns equity in Aduro Biotech, Agenus, Armata Pharmaceuticals, Breakbio, BioMarin Pharmaceutical, Bristol-Myers Squibb, Celldex Therapeutics, Editas Medicine, Exelixis, Gilead Sciences, IMV, Lexicon Pharmaceuticals, Moderna and Regeneron Pharmaceuticals. BeiGene, a Chinese biotech company, supports unrelated research at the DFCI Translational Immunogenomics Laboratory (TIGL). J.R. receives research funding from Amgen, Equillium, Novartis and Kite/Gilead and serves on Data Safety Monitoring Committees for AvroBio and Scientific Advisory Boards for Akron Biotech, Clade Therapeutics, Garuda Therapeutics, Immunitas Therapeutics, LifeVault Bio, Novartis, Rheos Medicines, Talaris Therapeutics and TScan Therapeutics. R.J.S. serves on the Board of Directors for Kiadis and Be The Match/National Marrow Donor Program; provided consulting for Gilead, Rheos Therapeutics, Cugene, Precision Bioscience, Mana Therapeutics, VOR Biopharma, and Novartis; and Data Safety Monitoring Board for Juno/Celgene. D.P serves on the Board of Insitro. The remaining authors declare no competing financial interests.
References
- Ahmed HI, Herrera M, Liew YJ, and Aranda M (2019). Long-Term Temperature Stress in the Coral Model Aiptasia Supports the “Anna Karenina Principle” for Bacterial Microbiomes. Frontiers in Microbiology 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akondy RS, Fitch M, Edupuganti S, Yang S, Kissick HT, Li KW, Youngblood BA, Abdelsamed HA, McGuire DJ, Cohen KW, et al. (2017). Origin and differentiation of human memory CD8 T cells after vaccination. Nature 552, 362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfei F, Kanev K, Hofmann M, Wu M, Ghoneim HE, Roelli P, Utzschneider DT, von Hoesslin M, Cullen JG, Fan Y, et al. (2019). TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature 571, 265–269. [DOI] [PubMed] [Google Scholar]
- Alvarez I, Niemi J, and Simpson M (2014). BAYESIAN INFERENCE FOR A COVARIANCE MATRIX. Conference on Applied Statistics in Agriculture. [Google Scholar]
- Alyea EP, Soiffer RJ, Canning C, Neuberg D, Schlossman R, Pickett C, Collins H, Wang Y, Anderson KC, and Ritz J (1998). Toxicity and efficacy of defined doses of CD4(+) donor lymphocytes for treatment of relapse after allogeneic bone marrow transplant. Blood 91, 3671–3680. [PubMed] [Google Scholar]
- Amir E-AD, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, Shenfeld DK, Krishnaswamy S, Nolan GP, and Pe’er D (2013). viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol 31, 545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert RD, Kamphorst AO, Sarkar S, Vezys V, Ha S-J, Barber DL, Ye L, Sharpe AH, Freeman GJ, and Ahmed R (2011). Antigen-specific CD4 T-cell help rescues exhausted CD8 T cells during chronic viral infection. Proc. Natl. Acad. Sci. U. S. A 108, 21182–21187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azizi E, Carr AJ, Plitas G, Cornish AE, Konopacki C, Prabhakaran S, Nainys J, Wu K, Kiseliovas V, Setty M, et al. (2018). Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell 174, 1293–1308.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachireddy P, and Wu CJ (2014). Understanding anti-leukemia responses to donor lymphocyte infusion. Oncoimmunology 3, e28187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachireddy P, Hainz U, Rooney M, Pozdnyakova O, Aldridge J, Zhang W, Liao X, Hodi FS, O’Connell K, Haining WN, et al. (2014). Reversal of in situ T-cell exhaustion during effective human antileukemia responses to donor lymphocyte infusion. Blood 123, 1412–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachireddy P, Ennis C, Nguyen VN, Gohil SH, Clement K, Shukla SA, Forman J, Barkas N, Freeman S, Bavli N, et al. (2020). Distinct evolutionary paths in chronic lymphocytic leukemia during resistance to the graft-versus-leukemia effect. Sci. Transl. Med 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagaev DV, Vroomans RMA, Samir J, Stervbo U, Rius C, Dolton G, Greenshields-Watson A, Attaf M, Egorov ES, Zvyagin IV, et al. (2020). VDJdb in 2019: database extension, new analysis infrastructure and a T-cell receptor motif compendium. Nucleic Acids Research 48, D1057–D1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya A (1990). On a Geometrical Representation of Probability Distributions and its use in Statistical Inference. Calcutta Statistical Association Bulletin 40, 23–49. [Google Scholar]
- Bingham Eli and Chen Jonathan P and Jankowiak Martin and Obermeyer Fritz and Pradhan Neeraj and Karaletsos Theofanis and Singh Rohit and Szerlip Paul and Horsfall Paul and Goodman Noah D (2019). Pyro: deep universal probabilistic programming. The Journal of Machine Learning Research 20, 973–978. [Google Scholar]
- Blackinton JG, and Keene JD (2016). Functional coordination and HuR-mediated regulation of mRNA stability during T cell activation. Nucleic Acids Res. 44, 426–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blei DM, Kucukelbir A, and McAuliffe JD (2017). Variational Inference: A Review for Statisticians. Journal of the American Statistical Association 112, 859–877. [Google Scholar]
- Brummelman J, Mazza EMC, Alvisi G, Colombo FS, Grilli A, Mikulak J, Mavilio D, Alloisio M, Ferrari F, Lopci E, et al. (2018). High-dimensional single cell analysis identifies stem-like cytotoxic CD8 T cells infiltrating human tumors. J. Exp. Med 215, 2520–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, and Greenleaf WJ (2015). ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr. Protoc. Mol. Biol 109, 21.29.1–21.29.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdziak C, Azizi E, Prabhakaran S, Pe’er D (2019). A Nonparametric Multi-view Model for Estimating Cell Type-Specific Gene Regulatory Networks. [Google Scholar]
- Champlin R, Jansen J, Ho W, Gajewski J, Nimer S, Lee K, Territo M, Winston D, Tricot G, and Reichert T (1991). Retention of graft-versus-leukemia using selective depletion of CD8-positive T lymphocytes for prevention of graft-versus-host disease following bone marrow transplantation for chronic myelogenous leukemia. Transplant. Proc 23, 1695–1696. [PubMed] [Google Scholar]
- Cheadle C (2005). Stability Regulation of mRNA and the Control of Gene Expression. Ann. N. Y. Acad. Sci 1058, 196–204. [DOI] [PubMed] [Google Scholar]
- Chen P-H, Lipschitz M, Weirather JL, Jacobson C, Armand P, Wright K, Hodi FS, Roberts ZJ, Sievers SA, Rossi J, et al. (2020). Activation of CAR and non-CAR T cells within the tumor microenvironment following CAR T cell therapy. JCI Insight 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Fan J, Zhang M, Qin L, Dominguez D, Long A, Wang G, Ma R, Li H, Zhang Y, et al. (2019a). CD73 expression on effector T cells sustained by TGF-β facilitates tumor resistance to anti-4-1BB/CD137 therapy. Nat. Commun 10, 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Ji Z, Ngiow SF, Manne S, Cai Z, Huang AC, Johnson J, Staupe RP, Bengsch B, Xu C, et al. (2019b). TCF-1-Centered Transcriptional Network Drives an Effector versus Exhausted CD8 T Cell-Fate Decision. Immunity 51, 840–855.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claret EJ, Alyea EP, Orsini E, Pickett CC, Collins H, Wang Y, Neuberg D, Soiffer RJ, and Ritz J (1997). Characterization of T cell repertoire in patients with graft-versus-leukemia after donor lymphocyte infusion. J. Clin. Invest 100, 855–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coifman R, Coppi A, Hirn M, and Warner F (2010). Diffusion Geometry Based Nonlinear Methods for Hyperspectral Change Detection. [Google Scholar]
- Collins RH Jr, Shpilberg O, Drobyski WR, Porter DL, Giralt S, Champlin R, Goodman SA, Wolff SN, Hu W, Verfaillie C, et al. (1997). Donor leukocyte infusions in 140 patients with relapsed malignancy after allogeneic bone marrow transplantation. J. Clin. Oncol 15, 433–444. [DOI] [PubMed] [Google Scholar]
- Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, Snyder MP, Pritchard JK, Kundaje A, Greenleaf WJ, et al. (2016). Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet 48, 1193–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon PM, Weiner J, Mitchell-Olds T, and Woodley R (1987). Bootstrapping the Gini Coefficient of Inequality. Ecology 68, 1548–1551. [Google Scholar]
- Gattinoni L, Speiser DE, Lichterfeld M, and Bonini C (2017). T memory stem cells in health and disease. Nature Medicine 23, 18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giralt S, Hester J, Huh Y, Hirsch-Ginsberg C, Rondón G, Seong D, Lee M, Gajewski J, Van Besien K, Khouri I, et al. (1995). CD8-depleted donor lymphocyte infusion as treatment for relapsed chronic myelogenous leukemia after allogeneic bone marrow transplantation. Blood 86, 4337–4343. [PubMed] [Google Scholar]
- Goetz EM, and Garraway LA (2012). Mechanisms of Resistance to Mitogen-Activated Protein Kinase Pathway Inhibition in BRAF-Mutant Melanoma. American Society of Clinical Oncology Educational Book 680–684. [DOI] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, and Noble WS (2011). FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratwohl A, Hermans J, Apperley J, Arcese W, Bacigalupo A, Bandini G, di Bartolomeo P, Boogaerts M, Bosi A, and Carreras E (1995). Acute graft-versus-host disease: grade and outcome in patients with chronic myelogenous leukemia. Working Party Chronic Leukemia of the European Group for Blood and Marrow Transplantation. Blood 86, 813–818. [PubMed] [Google Scholar]
- He R, Hou S, Liu C, Zhang A, Bai Q, Han M, Yang Y, Wei G, Shen T, Yang X, et al. (2016). Follicular CXCR5-expressing CD8 T cells curtail chronic viral infection. Nature 537, 412–416. [DOI] [PubMed] [Google Scholar]
- Im SJ, Hashimoto M, Gerner MY, Lee J, Kissick HT, Burger MC, Shan Q, Hale JS, Lee J, Nasti TH, et al. (2016). Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenq RR, and van den Brink MRM (2010). Allogeneic haematopoietic stem cell transplantation: individualized stem cell and immune therapy of cancer. Nat. Rev. Cancer 10, 213–221. [DOI] [PubMed] [Google Scholar]
- Kallies A, Zehn D, and Utzschneider DT (2020). Precursor exhausted T cells: key to successful immunotherapy? Nat. Rev. Immunol 20, 128–136. [DOI] [PubMed] [Google Scholar]
- Kamphorst AO, Wieland A, Nasti T, Yang S, Zhang R, Barber DL, Konieczny BT, Daugherty CZ, Koenig L, Yu K, et al. (2017). Rescue of exhausted CD8 T cells by PD-1-targeted therapies is CD28-dependent. Science 355, 1423–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, and Haussler D (2002). The human genome browser at UCSC. Genome Res. 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan O, Giles JR, McDonald S, Manne S, Ngiow SF, Patel KP, Werner MT, Huang AC, Alexander KA, Wu JE, et al. (2019). TOX transcriptionally and epigenetically programs CD8 T cell exhaustion. Nature 571, 211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb HJ, Schattenberg A, Goldman JM, Hertenstein B, Jacobsen N, Arcese W, Ljungman P, Ferrant A, Verdonck L, Niederwieser D, et al. (1995). Graft-versus-leukemia effect of donor lymphocyte transfusions in marrow grafted patients. Blood 86, 2041–2050. [PubMed] [Google Scholar]
- Kshirsagar AM (1959). Bartlett Decomposition and Wishart Distribution. The Annals of Mathematical Statistics 30, 239–241. [Google Scholar]
- Kukurba KR, Parsana P, Balliu B, Smith KS, Zappala Z, Knowles DA, Favé M-J, Davis JR, Li X, Zhu X, et al. (2016). Impact of the X Chromosome and sex on regulatory variation. Genome Res. 26, 768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong YA, Chen Y, Ong HS, Wu D, Man K, Deleage C, Minnich M, Meckiff BJ, Wei Y, Hou Z, et al. (2016). CXCR5(+) follicular cytotoxic T cells control viral infection in B cell follicles. Nat. Immunol 17, 1187–1196. [DOI] [PubMed] [Google Scholar]
- Lesterhuis WJ, Joost Lesterhuis W, Bosco A, Millward MJ, Small M, Nowak AK, and Lake RA (2017). Dynamic versus static biomarkers in cancer immune checkpoint blockade: unravelling complexity. Nature Reviews Drug Discovery 16, 264–272. [DOI] [PubMed] [Google Scholar]
- Levine JH, Simonds EF, Bendall SC, Davis KL, Amir E-AD, Tadmor MD, Litvin O, Fienberg HG, Jager A, Zunder ER, et al. (2015). Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell 162, 184–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, van der Leun AM, Yofe I, Lubling Y, Gelbard-Solodkin D, van Akkooi ACJ, van den Braber M, Rozeman EA, John BA, Blank CU, et al. (2019a). Dysfunctional CD8 T Cells Form a Proliferative, Dynamically Regulated Compartment within Human Melanoma. Cell 176, 775–789.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Sun J, Allesøe R, Datta K, Bao Y, Oliveira G, Forman J, Jin R, Olsen LR, Keskin DB, et al. (2019b). RNase H-dependent PCR-enabled T-cell receptor sequencing for highly specific and efficient targeted sequencing of T-cell receptor mRNA for single-cell and repertoire analysis. Nat. Protoc 14, 2571–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link CS, Eugster A, Heidenreich F, Rücker-Braun E, Schmiedgen M, Oelschlägel U, Kühn D, Dietz S, Fuchs Y, Dahl A, et al. (2016). Abundant cytomegalovirus (CMV) reactive clonotypes in the CD8(+) T cell receptor alpha repertoire following allogeneic transplantation. Clin. Exp. Immunol 184, 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Chang Y-J, Xu L-P, Zhang X-H, Wang Y, Liu K-Y, and Huang X-J (2018). Reversal of T Cell Exhaustion by the First Donor Lymphocyte Infusion Is Associated with the Persistently Effective Antileukemic Responses in Patients with Relapsed AML after Allo-HSCT. Biol. Blood Marrow Transplant 24, 1350–1359. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten L, and Hinton G (2008). Visualizing Data using t-SNE. J. Mach. Learn. Res 9, 2579–2605. [Google Scholar]
- Man K, Gabriel SS, Liao Y, Gloury R, Preston S, Henstridge DC, Pellegrini M, Zehn D, Berberich-Siebelt F, Febbraio MA, et al. (2017). Transcription Factor IRF4 Promotes CD8 T Cell Exhaustion and Limits the Development of Memory-like T Cells during Chronic Infection. Immunity 47, 1129–1141.e5. [DOI] [PubMed] [Google Scholar]
- Marrack P, Bender J, Hildeman D, Jordan M, Mitchell T, Murakami M, Sakamoto A, Schaefer BC, Swanson B, and Kappler J (2000). Homeostasis of αβ TCR T cells. Nat. Immunol 1, 107–111. [DOI] [PubMed] [Google Scholar]
- Miller BC, Sen DR, Al Abosy R, Bi K, Virkud YV, LaFleur MW, Yates KB, Lako A, Felt K, Naik GS, et al. (2019). Subsets of exhausted CD8 T cells differentially mediate tumor control and respond to checkpoint blockade. Nat. Immunol 20, 326–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdoerffer S, Moita LF, Neems D, Freitas RP, Hacohen N, and Rao A (2008). Regulation of CD45 alternative splicing by heterogeneous ribonucleoprotein, hnRNPLL. Science 321, 686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EM, Lin NU, Krop IE, and Winer EP (2011). The ethical use of mandatory research biopsies. Nat. Rev. Clin. Oncol 8, 620–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paley MA, Kroy DC, Odorizzi PM, Johnnidis JB, Dolfi DV, Barnett BE, Bikoff EK, Robertson EJ, Lauer GM, Reiner SL, et al. (2012). Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science 338, 1220–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauken KE, Sammons MA, Odorizzi PM, Manne S, Godec J, Khan O, Drake AM, Chen Z, Sen DR, Kurachi M, et al. (2016). Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 354, 1160–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petukhov V, Guo J, Baryawno N, Severe N, Scadden DT, Samsonova MG, and Kharchenko PV (2018). dropEst: pipeline for accurate estimation of molecular counts in droplet-based single-cell RNA-seq experiments. Genome Biol. 19, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter DL, Roth MS, McGarigle C, Ferrara J, and Antin JH (1994). Induction of Graft-versus-Host Disease as Immunotherapy for Relapsed Chronic Myeloid Leukemia. New England Journal of Medicine 330, 100–106. [DOI] [PubMed] [Google Scholar]
- Prabhakaran S, Azizi E, Carr A, and Pe’er D (2016). Dirichlet Process Mixture Model for Correcting Technical Variation in Single-Cell Gene Expression Data. In International Conference on Machine Learning, pp. 1070–1079. [PMC free article] [PubMed] [Google Scholar]
- Przepiorka D, Weisdorf D, Martin P, Klingemann HG, Beatty P, Hows J, and Thomas ED (1995). 1994 Consensus Conference on Acute GVHD Grading. Bone Marrow Transplant. 15, 825–828. [PubMed] [Google Scholar]
- Reske JJ, Wilson MR, and Chandler RL (2020). ATAC-seq normalization method can significantly affect differential accessibility analysis and interpretation. Epigenetics Chromatin 13, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricordel C, Friboulet L, Facchinetti F, and Soria J-C (2019). Molecular mechanisms of acquired resistance to third-generation EGFR-TKIs in EGFR T790M-mutant lung cancer. Ann. Oncol 30, 858. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011). Integrative genomics viewer. Nat. Biotechnol 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sade-Feldman M, Yizhak K, Bjorgaard SL, Ray JP, de Boer CG, Jenkins RW, Lieb DJ, Chen JH, Frederick DT, Barzily-Rokni M, et al. (2019). Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell 176, 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid C, Kuball J, and Bug G (2021). Defining the Role of Donor Lymphocyte Infusion in High-Risk Hematologic Malignancies. J. Clin. Oncol 39, 397–418. [DOI] [PubMed] [Google Scholar]
- Scott AC, Dündar F, Zumbo P, Chandran SS, Klebanoff CA, Shakiba M, Trivedi P, Menocal L, Appleby H, Camara S, et al. (2019). TOX is a critical regulator of tumour-specific T cell differentiation. Nature 571, 270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen DR, Kaminski J, Barnitz RA, Kurachi M, Gerdemann U, Yates KB, Tsao H-W, Godec J, LaFleur MW, Brown FD, et al. (2016). The epigenetic landscape of T cell exhaustion. Science 354, 1165–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty M, Tadmor MD, Reich-Zeliger S, Angel O, Salame TM, Kathail P, Choi K, Bendall S, Friedman N, and Pe’er D (2016). Wishbone identifies bifurcating developmental trajectories from single-cell data. Nat. Biotechnol 34, 637–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui I, Schaeuble K, Chennupati V, Fuertes Marraco SA, Calderon-Copete S, Pais Ferreira D, Carmona SJ, Scarpellino L, Gfeller D, Pradervand S, et al. (2019). Intratumoral Tcf1PD-1CD8 T Cells with Stem-like Properties Promote Tumor Control in Response to Vaccination and Checkpoint Blockade Immunotherapy. Immunity 50, 195–211.e10. [DOI] [PubMed] [Google Scholar]
- Singer M, Wang C, Cong L, Marjanovic ND, Kowalczyk MS, Zhang H, Nyman J, Sakuishi K, Kurtulus S, Gennert D, et al. (2016). A Distinct Gene Module for Dysfunction Uncoupled from Activation in Tumor-Infiltrating T Cells. Cell 166, 1500–1511.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M, Wang C, Cong L, Marjanovic ND, Kowalczyk MS, Zhang H, Nyman J, Sakuishi K, Kurtulus S, Gennert D, et al. (2017). A Distinct Gene Module for Dysfunction Uncoupled from Activation in Tumor-Infiltrating T Cells. Cell 171, 1221–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soiffer RJ, Alyea EP, Hochberg E, Wu C, Canning C, Parikh B, Zahrieh D, Webb I, Antin J, and Ritz J (2002). Randomized trial of CD8 T-cell depletion in the prevention of graft-versus-host disease associated with donor lymphocyte infusion. Biology of Blood and Marrow Transplantation 8, 625–632. [DOI] [PubMed] [Google Scholar]
- Tao T, and Vu V (2005). On random ±1 matrices: Singularity and determinant. Random Struct. Algorithms 28, 1–23. [Google Scholar]
- TRACERx Renal consortium (2017). TRACERx Renal: tracking renal cancer evolution through therapy. Nat. Rev. Urol 14, 575–576. [DOI] [PubMed] [Google Scholar]
- Tran D, Kucukelbir A, Dieng AB, Rudolph M, Liang D, and Blei DM (2016). Edward: A library for probabilistic modeling, inference, and criticism. [Google Scholar]
- Utzschneider DT, Charmoy M, Chennupati V, Pousse L, Ferreira DP, Calderon-Copete S, Danilo M, Alfei F, Hofmann M, Wieland D, et al. (2016). T Cell Factor 1-Expressing Memory-like CD8(+) T Cells Sustain the Immune Response to Chronic Viral Infections. Immunity 45, 415–427. [DOI] [PubMed] [Google Scholar]
- Vert J-P Kernel Methods in Genomics and Computational Biology. Kernel Methods in Bioengineering, Signal and Image Processing; 42–63. [Google Scholar]
- Wherry EJ, John Wherry E, Ha S-J, Kaech SM, Nicholas Haining W, Sarkar S, Kalia V, Subramaniam S, Blattman JN, Barber DL, et al. (2007). Molecular Signature of CD8 T Cell Exhaustion during Chronic Viral Infection. Immunity 27, 824. [DOI] [PubMed] [Google Scholar]
- Wu T, Ji Y, Moseman EA, Xu HC, Manglani M, Kirby M, Anderson SM, Handon R, Kenyon E, Elkahloun A, et al. (2016). The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci Immunol 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yofe I, Dahan R, and Amit I (2020). Single-cell genomic approaches for developing the next generation of immunotherapies. Nat. Med 26, 171–177. [DOI] [PubMed] [Google Scholar]
- Yost KE, Satpathy AT, Wells DK, Qi Y, Wang C, Kageyama R, McNamara KL, Granja JM, Sarin KY, Brown RA, et al. (2019). Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat. Med 25, 1251–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngblood B, Hale JS, Kissick HT, Ahn E, Xu X, Wieland A, Araki K, West EE, Ghoneim HE, Fan Y, et al. (2017). Effector CD8 T cells dedifferentiate into long-lived memory cells. Nature 552, 404–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander R, Schauder D, Xin G, Nguyen C, Wu X, Zajac A, and Cui W (2019). CD4 T Cell Help Is Required for the Formation of a Cytolytic CD8 T Cell Subset that Protects against Chronic Infection and Cancer. Immunity 51, 1028–1042.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Choi J, Zeng W, Rogers SA, Alyea EP, Rheinwald JG, Canning CM, Brusic V, Sasada T, Reinherz EL, et al. (2010). Graft-versus-leukemia antigen CML66 elicits coordinated B-cell and T-cell immunity after donor lymphocyte infusion. Clin. Cancer Res 16, 2729–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. (2017). Massively parallel digital transcriptional profiling of single cells. Nat. Commun 8, 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zientek LR (2008). Exploratory and Confirmatory Factor Analysis: Understanding Concepts and Applications. Struct. Equ. Modeling 15, 729–734. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single cell transcriptome and TCR as well as chromatin accessibility data will be submitted to NCBI’s Database of Genotypes and Phenotype (dbGaP; https://www.ncbi.nlm.nih.gov/gap) under study number phs001998.v3 and will be made publicly available as of the date of publication. Accession numbers are listed in the key resources table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE |
SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Human CD14, FITC | BD Biosciences | Cat#555397; AB_395798 |
| Anti-Human CD19, FITC | BD Biosciences | Cat#555412; AB_395812 |
| Anti-Human CD3, PE | BD Biosciences | Cat#555339; AB_395745 |
| Anti-Human CD4, BUV395 | BD Biosciences | Cat#563550; AB_2738273 |
| Anti-Human CD8, APC-Vio770 | Miltenyi Biotec | Cat#130-113-155; AB_2725983 |
| Anti-Human CD45RA, BV510 | BD Biosciences | Cat#563031; AB_2722499 |
| Human BD Fc Block | BD Biosciences | Cat#564219; AB_2728082 |
| DAPI solution | BD Biosciences | Cat#564907; AB_2869624 |
| Anti-Human CD11c, FITC | BD Biosciences | Cat#561355; AB_10611872 |
| Anti-Human CD14, FITC | BD Biosciences | Cat#555397; AB_395798 |
| Anti-Human CD36, FITC | BD Biosciences | Cat#555454; AB_2291112 |
| Anti-Human CD33, FITC | BD Biosciences | Cat#555626; AB_395992 |
| Anti-Human CD16, FITC | BD Biosciences | Cat#555406; AB_395806 |
| Anti-Human CD11b, FITC | BD Biosciences | Cat#562793; AB_2737798 |
| Anti-Human CD15, FITC | BD Biosciences | Cat#555401; AB_395801 |
| Anti-Human CD34, FITC | BD Biosciences | Cat#348053; AB_2228982 |
| Anti-Human CD56, FITC | BD Biosciences | Cat#562794; AB_2737799 |
| Anti-Human CD123, FITC | BD Biosciences | Cat#558663; AB_1645485 |
| Anti-Human CD235a, FITC | BD Biosciences | Cat#559943; AB_397386 |
| IgG1 isotype | Biolegend | Cat#400187; AB_2888921 |
| IgG2a isotype | Biolegend | Cat#400293; AB_2888922 |
| IgG2b isotype | Biolegend | Cat#400381; AB_2888923 |
| Anti-Human B2M | Biolegend | Cat#316323; AB_2800837 |
| Anti-Human B7H4 | Biolegend | Cat#358116; AB_2800986 |
| Anti-Human CD10 | Biolegend | Cat#312233; AB_2800817 |
| Anti-Human CD117 | Biolegend | Cat#313243; AB_2810474 |
| Anti-Human CD11a | Biolegend | Cat# 350617; AB_2800935 |
| Anti-Human CD11b | Biolegend | Cat# 301359; AB_2800732 |
| Anti-Human CD11c | Biolegend | Cat# 371521; AB_2801018 |
| Anti-Human CD127 | Biolegend | Cat# 351356; AB_2800937 |
| Anti-Human CD134 | Biolegend | Cat# 350035; AB_2800932 |
| Anti-Human CD137 | Biolegend | Cat# 309839; AB_2800807 |
| Anti-Human CD138 | Biolegend | Cat# 356539; AB_2810567 |
| Anti-Human CD14 | Biolegend | Cat# 301859; AB_2800736 |
| Anti-Human CD15 | Biolegend | Cat# 323053; AB_2800847 |
| Anti-Human CD152 | Biolegend | Cat# 369621; AB_2801015 |
| Anti-Human CD16 | Biolegend | Cat# 302065; AB_2800738 |
| Anti-Human CD163 | Biolegend | Cat# 333637; AB_2810510 |
| Anti-Human CD18 | Biolegend | Cat# 302129; AB_2800739 |
| Anti-Human CD183 | Biolegend | Cat# 353747; AB_2800949 |
| Anti-Human CD184 | Biolegend | Cat# 306533; AB_2800791 |
| Anti-Human CD19 | Biolegend | Cat# 302265; AB_2800741 |
| Anti-Human CD194 | Biolegend | Cat# 359425; AB_2800988 |
| Anti-Human CD197 | Biolegend | Cat# 353251; AB_2800943 |
| Anti-Human CD1c | Biolegend | Cat# 331547; AB_2800871 |
| Anti-Human CD1d | Biolegend | Cat# 350319; AB_2800934 |
| Anti-Human CD20 | Biolegend | Cat# 302363; AB_2800743 |
| Anti-Human CD223 | Biolegend | Cat# 369335; AB_2814327 |
| Anti-Human CD226 | Biolegend | Cat# 338337; AB_2800899 |
| Anti-Human CD244 | Biolegend | Cat# 329529; AB_2800857 |
| Anti-Human CD25 | Biolegend | Cat# 302649; AB_2800745 |
| Anti-Human CD27 | Biolegend | Cat# 302853; AB_2800747 |
| Anti-Human CD274 | Biolegend | Cat# 329751; AB_2800860 |
| Anti-Human CD278 | Biolegend | Cat# 313553; AB_2800823 |
| Anti-Human CD279 | Biolegend | Cat# 329963; AB_2800862 |
| Anti-Human CD28 | Biolegend | Cat# 302963; AB_2800751 |
| Anti-Human CD3 | Biolegend | Cat# 300479; AB_2800723 |
| Anti-Human CD31 | Biolegend | Cat# 303139; AB_2800757 |
| Anti-Human CD314 | Biolegend | Cat# 320837; AB_2800844 |
| Anti-Human CD33 | Biolegend | Cat# 366633; AB_2801008 |
| Anti-Human CD335 | Biolegend | Cat# 331941; AB_2800874 |
| Anti-Human CD34 | Biolegend | Cat# 343537; AB_2749972 |
| Anti-Human CD38 | Biolegend | Cat# 303543; AB_2800758 |
| Anti-Human CD39 | Biolegend | Cat# 328237; AB_2800853 |
| Anti-Human CD4 | Biolegend | Cat# 300567; AB_2800725 |
| Anti-Human CD40 | Biolegend | Cat# 334348; AB_2800886 |
| Anti-Human CD44 | Biolegend | Cat# 338827; AB_2800900 |
| Anti-Human CD45 | Biolegend | Cat# 304068; AB_2800762 |
| Anti-Human CD45RA | Biolegend | Cat# 304163; AB_2800764 |
| Anti-Human CD45RO | Biolegend | Cat# 304259; AB_2800766 |
| Anti-Human CD49f | Biolegend | Cat# 313635; AB_2800825 |
| Anti-Human CD5 | Biolegend | Cat# 300637; AB_2800726 |
| Anti-Human CD56 | Biolegend | Cat# 392425; AB_2801024 |
| Anti-Human CD57 | Biolegend | Cat# 393321; AB_2801030 |
| Anti-Human CD62L | Biolegend | Cat# 304851; AB_2800770 |
| Anti-Human CD69 | Biolegend | Cat# 310951; AB_2800810 |
| Anti-Human CD70 | Biolegend | Cat# 355119; AB_2800955 |
| Anti-Human CD73 | Biolegend | Cat# 344031; AB_2800916 |
| Anti-Human CD80 | Biolegend | Cat# 305243; AB_2800783 |
| Anti-Human CD86 | Biolegend | Cat# 305447; AB_2800786 |
| Anti-Human CD8a | Biolegend | Cat# 301071; AB_2800730 |
| Anti-Human CD95 | Biolegend | Cat# 305651; AB_2800787 |
| Anti-Human HLA-DR | Biolegend | Cat# 307663; AB_2800795 |
| Anti-Human KLRG1 | Biolegend | Cat# 138433; AB_2800649 |
| Anti-Human TCRab | Biolegend | Cat# 306743; AB_2800793 |
| Anti-Human TCRgd | Biolegend | Cat# 331231; AB_2814199 |
| Anti-Human TIGIT | Biolegend | Cat# 372729; AB_2801021 |
| Anti-Human Tim3 | Biolegend | Cat# 345049; AB_2800925 |
| Biological Samples | ||
| Cryopreserved bone marrow mononuclear cells | Dana-Farber Cancer Institute | Pasquarello Tissue Bank in Hematologic Malignancies |
| Cryopreserved donor lymphocyte infusion products | Dana-Farber Cancer Institute | Pasquarello Tissue Bank in Hematologic Malignancies |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DNase I | StemCell Technologies | Cat#07900 |
| Digitonin | Promega | Cat#G9441 |
| AMPure XP beads | Beckman Coulter | A63881 |
| Critical Commercial Assays | ||
| MACS Dead Cell Removal Kit | Miltenyi Biotec | Cat#130-090-101 |
| Pan T Cell Isolation Kit, human | Miltenyi Biotec | Cat#130-096-535 |
| MACS CD19 MicroBeads | Miltenyi Biotec | Cat#130-050-301 |
| 10X Chromium Single Cell 3′ Library & Gel Bead Kit (v2) | 10x Genomics | Cat#PN-120237 |
| Bioanalyzer High Sensitivity DNA Kit | Agilent | Cat#5067-4626 |
| 10x Chromium Single Cell 5’ Library & Gel Bead Kit | 10x Genomics | PN-1000006 |
| 10x Chromium Single Cell V(D)J Enrichment Kit, Human T Cell | 10x Genomics | PN-1000005 |
| 5' Feature Barcode Kit | 10x Genomics | PN-1000256 |
| 10x Chromium Next GEM Single Cell 5' Kit v2 | 10x Genomics | PN-1000263 |
| 10x Chromium Single Cell Human TCR Amplification Kit | 10x Genomics | PN-1000252 |
| Nextera DNA Library Prep Kit | Illumina | FC-121-1030 |
| NEBNext High Fidelity PCR Mix | New England Biolabs | M0541S |
| MinElute Reaction Cleanup kit | Qiagen | 28206 |
| Deposited Data | ||
| 10x scRNA-seq | dbGaP | phs001998.v3 |
| 10x scTCR-seq | dbGaP | phs001998.v3 |
| 10x CITE-seq | dbGaP | phs001998.v3 |
| ATAC-seq | dbGaP | phs001998.v3 |
| Symphony | This paper | DOI: https://zenodo.org/record/5498358 |
| Gaussian process regression models | This paper | DOI: doi.org/10.5281/zenodo.5498361 |
| Oligonucleotides | ||
| Primers for rhTCR-seq | Translational Immunogenomics Laboratory, Dana-Farber Cancer Institute | (Li et al., 2019) |
| Software and Algorithms | ||
| Symphony | This paper | DOI: https://zenodo.org/record/5498358 |
| Gaussian process regression models | This paper | DOI: doi.org/10.5281/zenodo.5498361 |
| SEQC | (Azizi et al., 2018) | https://github.com/dpeerlab/seqc |
| Cell Ranger 5.0.1 | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest |
| Cell Ranger V(D)J 2.1.0 | 10x Genomics | https://support.10xgenomics.com/single-cell-vdj/software/downloads/latest? |
| scanpy 1.8.0 | (Wolf et al., 2018) | https://github.com/theislab/Scanpy |
| t-SNE | (Maaten and Hinton, 2008) | https://lvdmaaten.github.io/software/ |
| Biscuit | (Azizi et al., 2018) | https://github.com/dpeerlab/BISCUIT_SingleCell_IMM_ICML_2016 |
| PhenoGraph | (Levine et al., 2015) | https://github.com/dpeerlab/phenograph |
| Pyro | (Bingham et al., 2019) | https://pyro.ai/ |
| ATAC-seq pipeline | ENCODE consortium | https://doi.org/10.5281/zenodo.156534; https://github.com/ENCODE-DCC/atac-seq-pipeline |
| MACS2 2.2.7.1 | (Zhang et al., 2008) | https://pypi.org/project/MACS2/ |
Code availability:
The hierarchical Gaussian Process model is implemented using the probabilistic programming language pyro (Bingham, Eli and Chen, Jonathan P and Jankowiak, Martin and Obermeyer, Fritz and Pradhan, Neeraj and Karaletsos, Theofanis and Singh, Rohit and Szerlip, Paul and Horsfall, Paul and Goodman, Noah D, 2019) available at: https://github.com/dpeerlab/dli_gpr. The integrative model Symphony is implemented using the probabilistic language Edward (Tran et al., 2016) with code available at: https://github.com/dpeerlab/Symphony. All original code has been deposited at [repository] and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.