Abstract
Prostate‐specific membrane antigen (PSMA) is a validated target for molecular diagnostics and targeted radionuclide therapy. Our purpose was to evaluate PSMA expression in hepatocellular carcinoma (HCC), cholangiocarcinoma (CCA), and hepatic adenoma (HCA); investigate the genetic pathways in HCC associated with PSMA expression; and evaluate HCC detection rate with 68Ga‐PSMA‐11 positron emission tomography (PET). In phase 1, PSMA immunohistochemistry (IHC) on HCC (n = 148), CCA (n = 111), and HCA (n = 78) was scored. In a subset (n = 30), messenger RNA (mRNA) data from the Cancer Genome Atlas HCC RNA sequencing were correlated with PSMA expression. In phase 2, 68Ga‐PSMA‐11 PET was prospectively performed in patients with treatment‐naïve HCC on a digital PET scanner using cyclotron‐produced 68Ga. Uptake was graded qualitatively and semi‐quantitatively using standard metrics. On IHC, PSMA expression was significantly higher in HCC compared with CCA and HCA (P < 0.0001); 91% of HCCs (n = 134) expressed PSMA, which principally localized to tumor‐associated neovasculature. Higher tumor grade was associated with PSMA expression (P = 0.012) but there was no association with tumor size (P = 0.14), fibrosis (P = 0.35), cirrhosis (P = 0.74), hepatitis B virus (P = 0.31), or hepatitis C virus (P = 0.15). Overall survival tended to be longer in patients without versus with PSMA expression (median overall survival: 4.2 vs. 1.9 years; P = 0.273). FGF14 (fibroblast growth factor 14) mRNA expression correlated positively (rho = 0.70; P = 1.70 × 10‐5) and MAD1L1 (Mitotic spindle assembly checkpoint protein MAD1) correlated negatively with PSMA expression (rho = −0.753; P = 1.58 × 10‐6). Of the 190 patients who met the eligibility criteria, 31 patients with 39 HCC lesions completed PET; 64% (n = 25) lesions had pronounced 68Ga‐PSMA‐11 standardized uptake value: SUVmax (median [range] 9.2 [4.9‐28.4]), SUVmean 4.7 (2.4‐12.7), and tumor‐to‐liver background ratio 2 (1.1‐11). Conclusion: Ex vivo expression of PSMA in neovasculature of HCC translates to marked tumor avidity on 68Ga‐PSMA‐11 PET, which suggests that PSMA has the potential as a theranostic target in patients with HCC.
Abbreviations
- CCA
cholangiocarcinoma
- CT
computed tomography
- GSEA
gene‐set enrichment analysis
- HBV
hepatitis B virus
- HCA
hepatic adenoma
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- H&E
hematoxylin and eosin
- I.A.
intraarterial
- IHC
immunohistochemistry
- LI‐RADS
Liver Imaging and Reporting Data System
- MRI
magnetic resonance imaging
- mCRPC
metastatic castration‐resistant prostate cancer
- mRNA
messenger RNA
- PET
positron emission tomography
- PRRT
peptide receptor radionuclide therapy
- PSMA
prostate‐specific membrane antigen
- RNA‐seq
RNA sequencing
- SUV
standardized uptake value
- TACE
transarterial chemoembolization
- TBR
tumor‐to‐liver background ratio
- TCGA
The Cancer Genome Atlas
- TMA
tissue microarray
Hepatocellular carcinoma (HCC) is diagnosed by imaging using contrast‐enhanced computed tomography (CT) or magnetic resonance imaging (MRI) based on the Liver Imaging and Reporting Data System (LI‐RADS) criteria.( 1 ) However, anatomic imaging provides little insight into the biology of HCC, which is a neoplasm characterized by marked molecular and pathologic heterogeneity. Biopsy has limitations because of intratumor heterogeneity, sampling errors, and difficulty distinguishing between early‐stage HCC and dysplastic nodules.( 2 ) The ubiquitous positron emission tomography (PET) radiotracer, fluorodeoxyglucose (18F‐FDG), has suboptimal liver imaging kinetics and only a fraction of high‐grade HCCs tend to be FDG‐avid.( 3 ) Most importantly, there is a strong unmet need for precision therapeutics in HCC. Combination immunotherapy and anti‐angiogenic therapy (atezolizumab‐bevacizumab) is the new first‐line therapy in advanced HCC. However, grade 3 or 4 adverse events occur in more than 50% of patients, and median progression‐free survival is less than 7 months.( 4 ) Inevitably, the cancer progresses and patients are left with limited, if any, therapeutic options. Moreover, response to immunotherapy tends to be suboptimal in nonviral‐mediated HCC, such as nonalcoholic fatty liver–associated HCC, which is rapidly becoming a major driver of HCC incidence in Western countries.( 5 , 6 )
Prostate‐specific membrane antigen (PSMA), also called as glutamate carboxypeptidase II, is a zinc metalloenzyme that catalyzes the hydrolysis of N‐acetylaspartylglutamate to glutamate and N‐acetylaspartate.( 7 ) PSMA is overexpressed in prostate cancer cells and has been validated as a theranostic (i.e., a combined diagnostic and therapeutic) target. However, PSMA is not specific to prostate cancer but can be expressed in other solid tumors such as sarcomas, thyroid, and lung cancers.( 8 , 9 , 10 ) Recent immunohistochemistry (IHC) reports suggest that there is higher‐than‐average expression of PSMA in HCC.( 11 ) Anecdotal reports and retrospective studies( 12 , 13 , 14 ) suggest that expression of PSMA in HCC can be detected noninvasively on 68Ga‐PSMA‐11 PET. These data provide the rationale to prospectively investigate PSMA‐targeted PET imaging as a foundational step toward the prospect of PSMA‐targeted peptide receptor radionuclide therapy (PRRT) (e.g., 177Lu‐PSMA‐617) in HCC. PRRT, the therapeutic use of radiolabeled molecules that target specific receptors or proteins on tumor cells or in tumor microenvironment, represents a precision medicine paradigm because it tailors a minimally invasive treatment to the unique biological profile of a tumor. Only patients with tumors that sufficiently express those receptors or markers (as detected on targeted PET imaging) qualify for treatment, which provides a personalized selection strategy. Therefore, it is necessary to evaluate the detection rate of HCC and to define the thresholds of radiolabeled PSMA avidity on 68Ga‐PSMA‐11 PET adequate for PRRT consideration.
Other relevant knowledge gaps include the mechanistic basis for potential PSMA expression in HCC, and the expression of PSMA in other liver tumors such as cholangiocarcinoma (CCA) and hepatocellular adenomas (HCA), which can have overlapping imaging features with HCC. To address these knowledge gaps, the aims of our study were (1) to determine the protein expression and localization of PSMA in surgically resected HCC, CCA, and HCA; (2) to investigate the genes and/or pathways in HCC tumors that are associated with PSMA protein expression, based on RNA‐sequencing (RNA‐seq) data; and (3) to prospectively evaluate the detection rate of HCC with 68Ga‐PSMA‐11 PET/CT or PET/MRI using Food and Drug Administration–approved technology for cyclotron production of 68Ga.
Materials and Methods
This institutional review board–approved HIPAA‐compliant study was conducted in two phases (Fig. 1). Phase 1 involved assessment of PSMA protein expression in surgically resected HCC, CCA, and HCA and its association with genes and/or pathways in HCC. This phase was done through the use of tissue microarrays (TMAs), which had been previously created as part of our institutional Hepatobiliary Neoplasia Biorepository. Phase 2 was a prospective single‐arm evaluation of 68Ga‐PSMA‐11 PET using cyclotron‐produced 68Ga for detection of HCC in patients with localized HCC (blinded for review).
FIG. 1.
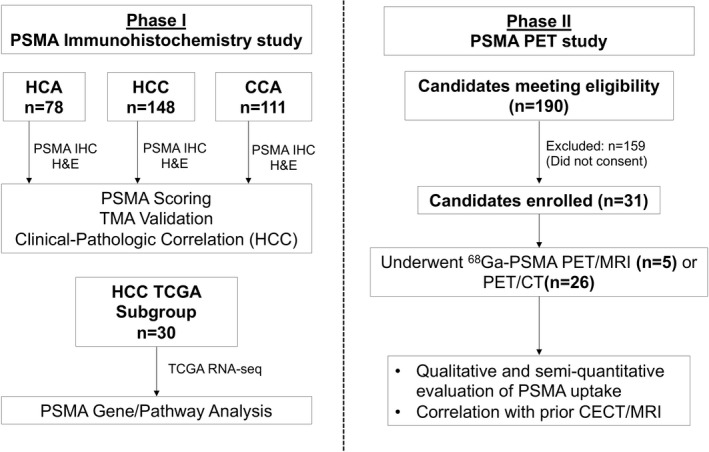
Study overview.
Phase I: PSMA IHC Study
TMAs previously created from surgically resected HCC (n = 148), CCA (n = 111), and HCA (n = 78) were used for this phase of the study.( 15 ) The TMAs were constructed using two tumor tissue samples per patient from paraffin blocks stored in the institutional pathology tissue archives. Histopathology features were analyzed by hematoxylin and eosin (H&E) and PSMA protein expression by IHC (additional details included as part of the Supporting Methods). Relevant clinical variables such as age at surgery, gender, race, height, weight, evidence of fibrosis or cirrhosis, hepatitis B virus (HBV), and hepatitis C virus (HCV) status, and tumor size were extracted from the electronic medical record. Additional information on the cohort of patients with HCC had been previously described.( 15 )
Pathology Review
All H&E and PSMA‐stained sections were evaluated in a blinded and random fashion by a hepatobiliary pathologist (M.S.T.) with more than 20 years of experience. HCC tumor grading, defined as the highest histologic grade component of the tumor, was performed from H&E‐stained sections using World Health Organization criteria: 0 = well differentiated, 1 = moderately differentiated, and 2 = poorly differentiated. PSMA‐stained sections were scored for percentage staining by area as follows: 0% staining = 0, <5% staining = 0.5, 5%‐30% staining = 1, 31%‐60% staining = 2, and 61%‐100% staining = 3. To evaluate the intrareader agreement in the PSMA IHC scoring system for the TMAs, the original HCC, CCA, and HCA tissue blocks were obtained, a 5‐mm‐thick tissue section was cut from the representative wax block, and PSMA IHC was repeated on the full tissue section. Intrarater agreement between PSMA‐IHC score from the TMA and the PSMA‐IHC score from the corresponding full section equal to or within one scoring level was determined by prevalence‐adjusted Cohen’s kappa.( 16 ) To evaluate interrater agreement in the PSMA‐IHC scoring system for HCC TMAs, a second hepatobiliary pathologist (Z.M.C.) with more than 14 years of experience evaluated the PSMA‐stained sections in a blinded and random fashion. Interrater agreement score equal to or within one scoring level was determined by prevalence‐adjusted Cohen’s kappa.( 16 )
HCC PSMA Protein Expression and HCC Gene/Pathway Analysis
A subset of the subjects in the HCC TMA were included in the Cancer Genome Atlas (TCGA) HCC project (n = 30).( 17 ) To investigate the genes and/or pathways in HCC that are associated with PSMA protein expression, mRNA expression data from TCGA‐HCC RNA‐seq were used to correlate with PSMA expression. Raw‐sequencing FASTQ files of 30 subjects overlapping with PSMA‐TMA study were downloaded from the Genomic Data Commons (https://gdc.cancer.gov/) and processed using RNA‐seq pipeline MAPRseq to reference genome HG38, resulting in mRNA expression–normalized values (reads per kilobase per million [RPKM]) of 19,518 protein‐coding genes.( 18 ) For each gene, mRNA RPKM values were correlated with HCC‐PSMA‐TMA scores (range 0‐3) using Spearman rank correlation in the same 30 tumors. For pathway enrichment analysis, genes were ranked based on correlation coefficient from highest to lowest, and gene‐set enrichment analysis (GSEA) was used to investigate statistical enrichment of canonical pathways defined in the Kyoto Encyclopedia of Genes and Genomes collections: Genes within a same pathway enriched toward the top of the ranked list were deemed as a positively associated pathway, and toward the bottom of the ranked list as a negatively associated pathway, with GSEA P < 0.05 as statistically significant.( 19 ) For GSEA, the raw P values were adjusted using the Benjamini‐Yekutieli method to control the false discovery rate.( 20 ) To assess for differences in HCC PSMA (FOLH1) mRNA expression by iCluster subtype from the entire TCGA cohort, normalized mRNA expression of PSMA (FOLH1) across the three iClusters within the TCGA HCC cohort were compared.( 17 )
Phase 2: 68Ga‐PSMA PET Study
The inclusion criteria for the prospective 68Ga‐PSMA PET study were treatment‐naïve patients with biopsy‐proven HCC or an image‐based diagnosis of HCC by CT or MRI (LI‐RADS 5; version 2018) who were able and willing to give written informed consent. Exclusion criteria were patients requiring emergent surgery for a ruptured or bleeding HCC or those with standard contraindications to PET. Enrolled patients underwent either 68Ga‐PSMA PET/MRI or, if ineligible for PET/MRI, 68Ga‐PSMA PET/CT, within 4 weeks of recruitment. Subsequent to the 68Ga‐PSMA PET, patients underwent standard‐of‐care treatment for HCC as determined by the care team.
Imaging Protocols
Synthesis of 68Ga‐PSMA‐HBED‐CC: Cyclotron‐produced 68Ga‐GaCl3 was used for automated synthesis of 68Ga‐PSMA‐HBED‐CC in full accordance with Good Manufacturing Practice requirements.
PET/MRI: All PSMA PET/MRI studies were performed on an integrated PET/MRI scanner equipped with Time‐of‐Flight technology (Signa; GE Healthcare). Patients received an injection of 5 ± 10% mCi of 68Ga‐PSMA‐HBED‐CC, with an approximately 90‐minute (±15 minutes) uptake period. PET/MRI protocol consisted of two main parts: (1) whole‐body survey PET/MRI and (2) focused abdominal PET/MRI. The acquisition protocol is summarized in the Supporting Methods and Supporting Fig. S1. Total imaging duration was approximately 60 minutes.
PET/CT: All PSMA PET/CT studies were performed on a state‐of‐the art digital PET/CT system (Siemens Biograph Vision 600; Siemens Healthineers) equipped with silicon photomultiplier PET detectors. Patients received an injection of 5 ± 10% mCi of 68Ga‐PSMA‐HBED‐CC, with an approximately 90‐minute (± 15 minutes) uptake period. A low‐dose, nongated, non‐contrast‐enhanced, free‐breathing CT was acquired from the orbits to upper thighs for attenuation correction (CTAC) and anatomic co‐localization. A whole‐body continuous bed motion PET scan from the orbits to upper thighs was then acquired. All PET data were reconstructed with fully 3D iterative reconstruction algorithms that included corrections for attenuation (using the CTAC scan), scatter, randoms, dead time, decay, and normalization.
Image Analyses
PET/MRI and PET/CTs were independently reviewed on a dedicated workstation (MIM Software Inc., Cleveland, OH) by an abdominal and nuclear radiologist (A.H.G.) with 11 years of postresidency experience who was blinded to all other imaging, clinical, and pathology information. Quality of images for the intended primary diagnostic task (i.e., PSMA‐avid hepatic lesion detection) was assessed with the use of a 4‐point image quality score: 4 = excellent, 3 = good, 2 = acceptable, and 1 = insufficient. This assessment considered image noise and sharpness, artifacts, registration accuracy of PET, and MRI or CT data.
PSMA uptake in the hepatic lesions was evaluated qualitatively and semi‐quantitatively on whole‐body survey PET/MRI or the PET/CT images. Qualitative evaluation was assessed for the intensity of PSMA uptake in hepatic lesions, which was graded as follows: grade 1: uptake <normal liver; grade 2: uptake = normal liver; grade 3: uptake > normal liver; and grade 4: uptake > spleen or kidneys. For statistical analyses and correlation with PSMA‐IHC scoring, grades 1 and 2 on PET/MRI were grouped as low PSMA expression. Conversely, grades 3 and 4 were grouped as high PSMA expression. Semi‐quantitative evaluation was performed on lesions showing high PSMA expression using a volume of interest incorporating the gross lesion volume and a previously validated gradient‐based method (PET Edge; MIM Software, Cleveland, OH).( 21 ) Maximum and mean standardized uptake value (SUVmax and SUVmean) of the lesion and the background liver were noted. Tumor‐to‐liver background ratio (TBR) of lesion SUVmax to liver SUVmax (TBRmax) and liver SUVmean (TBRmean), respectively, were calculated. In addition, whole‐body images were evaluated for lesions suggestive of metastases based on the morphology and intensity of PSMA uptake.
Available contrast‐enhanced CT or MRI scans done before PET/CT or PET/MRI were reviewed on a PACS workstation (Visage Imaging, Inc., San Diego, CA). Image quality was assessed on the 4‐point scale. HCC lesions were evaluated for morphology, signal intensity, and post‐contrast‐enhancement characteristics. Three patients who underwent surgical resection of their HCC following PSMA PET had PSMA IHC performed for comparison with PSMA uptake at PET.
Statistical Analyses
We compared the clinical characteristics and PSMA expression among HCC, CCA, and HCA groups. Categorical variables were compared using the chi‐square test or Fisher’s exact test. Continuous variables were compared using the one‐way analysis of variance test or Kruskal–Wallis test. Subgroup analysis of the HCC subjects was performed. Differences in HCC tumor grade, absence or presence of cirrhosis or fibrosis, HBV, or HCV were compared between absent versus positive PSMA expression using a chi‐square test. Differences in HCC tumor size were compared between absent versus positive PSMA expression using a Kruskal‐Wallis test. Univariate and multivariate logistic regression was performed to assess association between PSMA expression with absence or presence of cirrhosis, fibrosis, HBV, HCV, HCC tumor grade, or tumor size. Overall survival (OS) following surgical resection for HCC was estimated using Kaplan‐Meier analysis and compared between absent versus positive PSMA expression using the log‐rank test. Hazard ratios (HRs) and 95% confidence intervals (CIs) were estimated using multivariate Cox model adjusting for cirrhosis, fibrosis, HBV, HCV, fibrosis, tumor grade, and tumor size. Sensitivity analysis was conducted to associate clinical variables and survival outcomes with PSMA expression by considering PSMA expression as a continuous variable. Descriptive statistics were used for the phase 2 component of the study. Cross‐sectional (CT/MR) imaging findings were compared between lesions with low versus high PSMA expression on PET using chi‐square test.
Results
Phase I: PSMA IHC Study
HCC, CCA, and HCA PSMA Protein Expression and Localization
Clinical and PSMA scoring data by tumor type are summarized in Table 1. PSMA was expressed in all three tumor types, but PSMA expression was different across tumor types (P < 0.0001), with higher PSMA expression in HCC compared with CCA and HCA (Table 1, Fig. 2, and Supporting Figs. S2 and S3). Patients with HCC were more likely to have a PSMA expression higher than 30% (corresponding to a PSMA IHC score of 2 and above) compared with CCA or HCA (39.2% vs. 2.7% vs. 6.4%, respectively). Conversely, patients with HCA were more likely to have no PSMA expression compared with HCC and CCA (67.9% vs. 9.5% vs. 9.0%, respectively). Of note, while 40.5% of CCA had a PSMA score of 1 (5%‐30% expression), most had 10% or less expression. Among cases with high PSMA expression, the localization was to the tumor‐associated endothelial cells in all cases with rare concomitant intratumoral sinusoidal/canalicular PSMA expression in HCC (5.5%) and rare tumor cell PSMA expression in CCA (3.7%). Conversely, while only 32.1% of HCA cases demonstrated PSMA expression, among those positive HCA cases, intratumoral canalicular staining only was more common (20%). Finally, intrarater agreement for PSMA scoring between TMA and full section was very good for HCC (kappa = 0.94) and CCA (kappa = 0.86), and good for HCA (kappa = 0.68). Similarly, interrater agreement for PSMA scoring between experienced hepatobiliary pathologists for the HCC TMAs was very good (kappa = 0.96).
TABLE 1.
Demographic, Tumor, and PSMA IHC Data by Tumor Type
| HCC (n = 148) | CCA (n = 111) | HCA (n = 78) | Total (n = 337) | P Value | |
|---|---|---|---|---|---|
| Gender, n (%) | <0.0001* | ||||
| Female | 58 (39.2%) | 49 (44.1%) | 71 (91.0%) | 178 (52.8%) | |
| Male | 90 (60.8%) | 62 (55.9%) | 7 (9.0%) | 159 (47.2%) | |
| Race, n (%) | 0.022 † | ||||
| White | 131 (88.5%) | 107 (96.4%) | 75 (96.2%) | 313 (92.9%) | |
| Black or African American | 4 (2.7%) | 1 (0.9%) | 0 (0.0%) | 5 (1.5%) | |
| Asian | 9 (6.1%) | 2 (1.8%) | 0 (0.0%) | 11 (3.3%) | |
| American Indian/Alaskan Native | 1 (0.7%) | 1 (0.9%) | 0 (0.0%) | 2 (0.6%) | |
| Hispanic or Latino | 0 (0.0%) | 0 (0.0%) | 2 (2.6%) | 2 (0.6%) | |
| Unknown/decline | 3 (2.0%) | 0 (0.0%) | 1 (1.3%) | 4 (1.2%) | |
| BMI | 0.17 ‡ | ||||
| n | 138 | 103 | 66 | 307 | |
| Mean (SD) | 27.6 (5.51) | 28.9 (5.92) | 29.1 (7.15) | 28.3 (6.06) | |
| Median | 26.7 | 28.8 | 28.3 | 27.6 | |
| Range | 16.0, 50.7 | 18.9, 52.9 | 15.9, 46.6 | 15.9, 52.9 | |
| Tumor Size (cm) | 0.055 ‡ | ||||
| n | 148 | 110 | 74 | 332 | |
| Mean (SD) | 8.0 (5.78) | 6.2 (3.89) | 7.2 (4.08) | 7.2 (4.91) | |
| Median | 6.0 | 5.0 | 6.5 | 5.7 | |
| Range | 0.8, 33.0 | 0.8, 20.0 | 1.0, 18.0 | 0.8, 33.0 | |
| Age at Surgery (years) | <0.0001 ‡ | ||||
| N | 148 | 111 | 78 | 337 | |
| Mean (SD) | 63.5 (14.09) | 62.8 (11.49) | 38.6 (12.84) | 57.5 (16.59) | |
| Median | 66.0 | 64.0 | 39.0 | 61.0 | |
| Range | 20.0, 96.0 | 29.0, 82.0 | 5.0, 67.0 | 5.0, 96.0 | |
| PSMA Score, n (%) | <0.0001 † | ||||
| 0 (0%) | 14 (9.5%) | 10 (9.0%) | 53 (67.9%) | 77 (22.8%) | |
| 0.5 (<5%) | 21 (14.2%) | 53 (47.7%) | 13 (16.7%) | 87 (25.8%) | |
| 1 (5‐30%) | 55 (37.2%) | 45 (40.5%) | 7 (9.0%) | 107 (31.8%) | |
| 2 (31‐60%) | 47 (31.8%) | 3 (2.7%) | 5 (6.4%) | 55 (16.3%) | |
| 3 (61‐100%) | 11 (7.4%) | 0 (0.0%) | 0 (0.0%) | 11 (3.3%) |
Chi‐square P value.
Fisher exact P value.
Kruskal‐Wallis P value.
Abbreviation: BMI, body mass index.
FIG. 2.

PSMA expression in HCC. (A‐E) Tumor associated endothelial cell staining as a percentage of area: 0% (A) <5% (B), 5%‐30% (C), 31%‐60% (D), 61%‐100% (E). (F) Canalicular staining.
PSMA in HCC
Among HCCs (n = 148), 14 patients had no PSMA expression, and 134 patients had PSMA expression. Clinical and HCC tumor data stratified by PSMA expression are summarized in Supporting Table S1. Median tumor size was significantly larger among HCCs with PSMA expression than those without PSMA expression (6.0 vs. 4.2 cm, respectively; P = 0.019). A higher proportion of HCCs with moderate and poorly differentiated tumor grade were present among subjects with PSMA expression (85.5% vs. 50.0%, respectively; P = 0.003).
Higher tumor grade (well versus poorly differentiated: OR = 0.109, P = 0.008) and larger tumor size (OR = 1.196, P = 0.035) were predictors of positive PSMA expression in the univariate analysis. However, only higher tumor grade was associated with positive PSMA expression after adjusting for cirrhosis, HBV, HCV, fibrosis, tumor grade, and tumor size (Supporting Table S2). There was a trend for improved overall survival among HCCs without PSMA expression compared to those with PSMA expression (median OS: 4.2 vs. 1.9 years after surgery, HR = 1.36, 95% CI = 0.78‐2.38) but the difference was not significant (P = 0.273), which could potentially be due to small number of cases in the group without PSMA expression (n = 14) (Supporting Fig. S4 and Supporting Table S3). Consistent results were found when associating continuous PSMA score with OS, with adjustment for the clinical variables (Supporting Table S3).
HCC TCGA Pathway Analysis
Among 19,518 protein coding genes with correlation results among 30 subjects with HCC with both tumor RNA‐seq data from the TCGA‐HCC study and PSMA expression data from this study, 138 genes had a moderate positive correlation (coefficient ranging from 0.50 to 0.70), 98 genes had a moderate negative correlation (coefficient ranging from −0.50 to −0.70), and four genes had a strong negative correlation (coefficient greater than −0.70). The gene with the strongest positive correlation was fibroblast growth factor 14 (FGF14) with a correlation coefficient of 0.70 (P = 1.70 × 10‐5). The gene with the strongest negative correlation was Mitotic spindle assembly checkpoint protein MAD1 (MAD1L1) with a correlation coefficient of −0.753 (P = 1.58 × 10‐6) (Supporting Table S4). GSEA pathway analysis identified 45 of 280 pathways associated with PSMA expression (P < 0.05) with 30 enriched for positive correlations and 15 enriched for negative correlations (Supporting Table S5). Finally, comparison of the normalized PSMA (FOLH1) mRNA expression across the three iClusters with the TCGA HCC cohort demonstrated that PSMA separates iCluster 1 from iCluster 2 and 3 (7.48 ± 1.41 vs. 8.29 ± 1.89 vs. 8.37 ± 1.42, respectively; P value = 2.367 × 10‐3; q‐value = 7.561 × 10‐3).
Phase 2: PSMA PET Study
Between January 2019 and May 2021, 190 patients met the eligibility criteria and were evaluated for potential enrollment. Of these, 31 patients (16%) (23 men and 8 women; median age: 66 years, range 48‐80 years) provided written informed consent and completed imaging. In 24 patients, the diagnosis of HCC was based on LI‐RADS, while the remaining 7 patients had a histopathological confirmation as well. Five patients underwent 68Ga‐PSMA PET/MRI, and 26 patients underwent 68Ga‐PSMA PET/CT. There were no adverse events in any of the patients.
68Ga‐PSMA PET Findings: Image quality was scored as excellent in all studies. There were 39 total lesions: one HCC lesion each in 25 patients, two lesions each in 5 patients, and four lesions in 1 patient. On qualitative evaluation, 25 lesions (64%) had high PSMA uptake (grade 3 or 4) (Fig. 3, Supporting Fig. S5), while 14 (36%) of 39 lesions had low PSMA uptake (grade 1 or 2) (Fig. 4). Semi‐quantitative PET metrics (median [range]) in the lesions with high PSMA uptake were as follows: SUVmax 9.2 (4.9‐28.4), SUVmean 4.7 (2.4‐12.7), TBRmax 2 (1.1‐11), and TBRmean 2.8 (1.3‐10.1). The metrics for each lesion are summarized in Table 2. Of the 31 patients in our study, 11 (35%) had SUVmax of at least 10, and 3 (10%) had SUVmax > 20 in their HCC lesions. In 2 patients, the HCC‐associated tumor thrombus extending into portal vein also showed grade 4 PSMA uptake (Fig. 3). A PSMA‐avid, biopsy‐proven metastatic gastrohepatic lymph node was noted in 1 patient. There was no other PSMA‐avid metastasis in any other patient. Incidental grade 4 PSMA uptake was noted in a known concurrent intrahepatic CCA (SUVmax: 18.6, SUVmean: 7.7) in 1 patient. Incidental focal PSMA uptake was present in the prostate in 2 patients, 1 of whom had known prostate cancer.
FIG. 3.
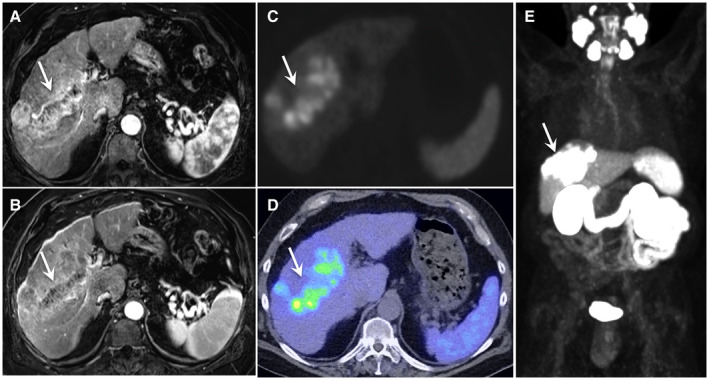
A 68‐year‐old male with alcohol‐associated liver disease–related cirrhosis and a LI‐RADS 5 observation. Axial dynamic contrast‐enhanced MRI images show a large, infiltrative lesion in right lobe of liver, showing nonrim arterial hyperenhancement (A) and washout in equilibrium phase (B) with tumor thrombus extending into right portal vein (arrows). Axial attenuation corrected gray‐scale PET image (C), fused PET/CT image (D), and maximum‐intensity projection image (E) from 68Ga‐PSMA PET showing intense PSMA uptake (grade 4) in the lesion as well as in the tumor thrombus.
FIG. 4.

A 60‐year‐old female with biopsy proven well‐differentiated HCC. Axial MRI images show a large exophytic lesion arising from left hepatic lobe with arterial hyperenhancement (A) and pseudocapsule (B) (arrows). Axial attenuation corrected gray‐scale PET image (C) and fused PET/MR image (D) from 68Ga‐PSMA PET show no uptake of PSMA in the lesion (grade 1).
TABLE 2.
Clinical, Laboratory and 68Ga‐PSMA PET Data in Patients Who Underwent 68Ga‐PSMA PET/CT or PET/MRI
| Patient | BMI | Background Liver Disease | AFP (ng/mL) | Number of Lesions | Lesion PSMA Expression | Lesion SUVmax | Lesion SUVmean | TBRmax | TBRmean | Treatment |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 32.6 | PBC | 1.8 | 2 | Both low | — | — | — | — | Transplant evaluation |
| 2 | 32.3 | NASH | 7.1 | 1 | High | 9.2 | 3.6 | 2.1 | 2.8 | Ablation |
| 3 | 41.2 | NASH | 6.5 | 1 | Low | — | — | — | — | TACE |
| 4 | 38.9 | ALD | 59 | 1 | High | 12 | 6.3 | 1.8 | 2.3 | Ablation |
| 5 | 39.4 | ALD | 3.1 | 1 | Low | — | — | — | — | Ablation |
| 6 | 20 | HCV | 3.1 | 1 | Low | — | — | — | — | Ablation |
| 7 | 33 | NASH status post‐ transplant | 3.3 | 1 | High | 23.9 | 5.2 | 8 | 10 | Ablation |
| 8 | 20.2 | None | 10 | 1 | Low | — | — | — | — | Surgery |
| 9 | 29.1 | None | 3.9 | 1 | Low | — | — | — | — | Surgery |
| 10 | 27.6 | HBV | 9.2 | 1 | Low | — | — | — | — | Transplant evaluation |
| 11 | 32.4 | HCV, ALD | 8.8 | 1 | High | 5.8 | 4.4 | 1.3 | 1.7 | Ablation |
| 12 | 28.9 | ALD | 4.6 | 2 | Both high | 7.3 and 8.2 | 5.1 and 4.7 | 1.3 and 1.1 | 1.7 and 1.4 | Ablation |
| 13 | 38.5 | None | 9.5 | 1 | High | 13.5 | 5.6 | 2.8 | 3.8 | Ablation |
| 14 | 29.5 | HCV | 28.6 | 1 | High | 19.3 | 12.7 | 3.5 | 4.8 | Ablation |
| 15 | 46.5 | NASH | 33 | 1 | High | 12.1 | 5 | 2 | 3.4 | Systemic chemotherapy |
| 16 | 28.8 | ALD | 5.5 | 2 | Both high | 8.4 and 4.9 | 3 and 2.6 | 1.9 and 1.1 | 2.8 and 1.6 | Ablation |
| 17 | 26.2 | HCV | 2.9 | 1 | Low | — | — | — | — | Ablation |
| 18 | 29 | NASH | 2.5 | 1 | High | 7.2 | 3.4 | 1.5 | 2.1 | Transplant evaluation |
| 19 | 37.9 | ALD | 2 | 1 | High | 26.5 | 11.2 | 4.3 | 7 | Systemic chemotherapy |
| 20 | 32.6 | HCV | 981 | 1 | High | 12.6 | 7.2 | 2 | 2.8 | Systemic chemotherapy |
| 21 | 24.6 | NASH | 2 | 1 | High | 6.1 | 3.8 | 11 | 1.3 | TARE |
| 22 | 31 | ALD | 3.6 | 1 | High | 8.2 | 4.4 | 1.5 | 2 | Transplant evaluation |
| 23 | 24 | HCV, ALD | 2.5 | 1 | Low | — | — | — | — | Transplant evaluation |
| 24 | 25 | Autoimmune hepatitis | 6.1 | 2 | Both high | 13.4 and 7.5 | 4.9 and 3 | 3.2 and 1.8 | 4.4 and 2.5 | Transplant evaluation, bridging resection |
| 25 | 30.2 | NASH | 4 | 4 | High in all lesions | 28.4, 5.7, 19.8, 5.3 | 8.7, 2.7, 6.3, 2.4 | 7.2, 1.4, 5, 1.4 | 10.1, 2, 7, 1.9 | Transplant evaluation |
| 26 | 32 | NASH | 4.5 | 1 | Low | — | — | — | — | HAE |
| 27 | 33.8 | NASH | 2.9 | 1 | Low | — | — | — | — | HAE |
| 28 | 28.1 | HCV, ALD | 4.6 | 1 | Low | — | — | — | — | Transplant evaluation |
| 29 | 23.4 | ALD | 3 | 2 | High and low | 5.2 | 3.5 | 1.3 | 1.7 | Transplant evaluation |
| 30 | 29.7 | HCV, ALD post‐ transplant | 2.9 | 1 | High | 12.2 | 4.7 | 2.2 | 3.3 | Ablation |
| 31 | 31.6 | Hemochromatosis | 3.8 | 1 | High | 18.6 | 7.2 | 3.4 | 4.5 | Ablation |
Abbreviations: ALD, alcohol‐associated liver disease; AFP, alpha‐fetoprotein; BMI, body mass index; HAE, hepatic artery embolization; NASH, nonalcoholic steatohepatitis; PBC, primary biliary cirrhosis; TARE, transarterial radioembolization.
CT and MRI Findings: Average time interval between CT (n = 9) or MRI (n = 22) and 68Ga‐PSMA PET was 18 days (range: 0‐92 days). There was no difference (P = 0.09) in the relative frequency of background cirrhotic liver in lesions with high PSMA uptake (24 of 25; 96%) versus low PSMA uptake (11 of 14; 79%), non‐rim arterial hyperenhancement (high PSMA uptake: 24 of 25, 96%; low PSMA uptake: 13 of 14, 93%; P = 0.3), washout (high PSMA uptake: 19 of 25, 76%; low PSMA uptake: 9 of 14, 64%; P = 0.5), pseudo‐capsule (high PSMA uptake: 13 of 25, 52%; low PSMA uptake: 5 of 14, 36%; P = 0.3), or restricted diffusion (high PSMA uptake: 12 of 15, 80%; low PSMA uptake: 6 of 10, 60%; P = 0.3). Finally, there was no difference in the size (mean ± SD) of lesions showing low versus high uptake (2.8 ± 2.2 cm vs. 2.2 ± 2.3 cm; P = 0.5).
PSMA Uptake at PET Versus PSMA IHC
Two patients with absent HCC PSMA uptake at PET (grade 1) had no HCC protein expression of PSMA confirmed by IHC (PSMA score = 0; true negative). One patient with positive HCC PSMA uptake at PET (grade 4) had positive HCC protein expression of PSMA confirmed by IHC (PSMA score = 3; true positive) (Supporting Fig. S6).
Discussion
In our IHC study, a high proportion of HCCs (about 90%) expressed PSMA, and about 40% of the HCC lesions had PSMA expression greater than 30%. The PSMA principally localized to the tumor‐associated endothelial cell membrane. The high intrarater agreement for PSMA scoring between the TMA core and the standard full tissue section also indicates generalizability of TMA findings to the full section. These findings in our U.S. cohort of surgically resected HCCs are concordant with the recent reports from Europe and Asia.( 11 , 22 , 23 ) Therefore, we posit that PSMA expression in HCC is generalizable across cohorts with diverse risk factors for development of HCC. Second, approximately 89% of CCAs also expressed PSMA on tumor‐associated endothelial cells. However, most of these demonstrated less than 10% PSMA expression. Of note, PSMA was also expressed in a small proportion of the CCA tumor cells (4%) themselves, while this was exceedingly rare in both HCC and HCA. PSMA was also expressed in some HCAs (32%), but most of these demonstrated expression closer to 5%. Although tumor endothelial cell staining was the most common site of PSMA localization even in HCAs, intratumoral canalicular PSMA staining was more common in HCA (20%) compared with HCC and CCA (4%‐6%). In summary, PSMA expression in tumor‐associated endothelial cells was present in a higher proportion of and with greater expression in HCC compared with CCA and HCA.
In multivariate analyses, higher tumor grade (moderate and poor differentiation) was associated with PSMA expression. There was a trend for longer overall survival in patients without PSMA expression (4.2 vs. 1.9 years), which is concordant with a recent study from Europe.( 11 ) However, higher HCC tumor grade is a known negative prognostic factor for survival in HCC. As such, there could be an interaction between tumor grade and PSMA expression. Given the principal localization of PSMA to the HCC tumor–associated endothelial cells and association with higher grade tumors, we speculate a role for PSMA in neo‐angiogenesis. PSMA has been shown to have a role in regulating angiogenesis through a complex mechanism involving matrix metalloprotease–mediated proteolysis of the extracellular matrix protein laminin with downstream activation of endothelial cell integrin signaling and angiogenesis.( 24 , 25 , 26 ) Moreover, PSMA has a vascular endothelial growth factor–independent role in retinal angiogenesis.( 27 ) As such, PSMA‐associated angiogenesis may represent a distinct mechanism of tumor‐associated angiogenesis.
To elucidate candidate molecular mechanisms of PSMA‐associated angiogenesis in HCC, we evaluated associations between HCC genes and/or pathways with PSMA expression. The gene with the strongest positive correlation with PSMA expression was FGF14. FGF signaling has been implicated in the pathogenesis of HCC, particularly with regulatory effects in the tumor microenvironment and angiogenesis.( 28 ) Moreover, FGF signaling is also being explored as a therapeutic target in HCC.( 29 ) However, currently there are no data on a potential role specifically for FGF14 in HCC. Conversely, the gene with the strongest negative correlation with PSMA expression was MAD1L1. MAD1 may play a role in cell‐cycle control and tumor suppression and studies have suggested a role for MAD1 in the pathogenesis of HCC.( 30 , 31 , 32 ) More specifically, the loss of MAD1 function in HCC results in loss of mitotic checkpoint control and is associated with recurrence following surgical resection and decreased overall survival.( 31 , 32 , 33 , 34 , 35 ) Several of the pathways positively associated with PSMA expression have roles in the tumor microenvironment, including cytokine–cytokine receptor interaction, extracellular matrix–receptor interaction, and phosphoinositide 3‐kinase–AKT signaling.( 36 ) These data suggest potential areas for future research into the mechanistic role of PSMA‐associated angiogenesis in HCC, which may include FGF14 and MAD1 genes as well as tumor microenvironmental and angiogenic signaling in the pathogenesis of HCC. However, such mechanistic research would need PSMA‐positive small animal HCC models, which currently do not exist.( 37 ) In summary, our data provide hypothesis‐generating insights regarding the potential mechanistic role of PSMA in HCC, which warrants further exploration in biologically relevant preclinical models.
PSMA expression on ex vivo pathology or IHC may not always translate to sufficient tumor avidity on PET imaging, as was recently observed in patients with metastatic colorectal cancer.( 38 ) Therefore, we prospectively investigated 68Ga‐PSMA‐11 PET in patients with HCC. The specific PSMA analog, 68Ga‐PSMA‐11, has minimal physiologic hepatic uptake and excretion, and sufficiently long half‐life (68 minutes), which makes it apt for liver imaging. 68Ga‐PSMA‐11 binds to PSMA through clathrin‐coated pits, resulting in endosome accumulation and the internalization of the PSMA‐drug complex. We used the technology of cyclotron production of 68GaCl3 to radiolabel 68Ga‐PSMA‐11. The 68Ga for labeling of PSMA is typically obtained from commercial 68Ge/68Ga generator. However, generators tend to be expensive, provide a maximum of two to three doses per elution, a maximum of three elutions per day, and only supply low doses of 68Ga‐PSMA (four to six mCi). Thus, the commercial supply for 68Ga from 68Ge/68Ga generators has not kept pace with the growing clinical demand, cannot scale production to meet increasing demand, and is not cost‐effective in the long run. On the other hand, cyclotron‐produced 68Ga to make 68Ga‐PSMA for the proposed study has the potential to provide a large supply of 68Ga in a cost‐effective way. In our study, 25 lesions (64%) had pronounced PSMA uptake (median SUVmax 9.2, range: 4.9‐28.4). This SUV range is comparable to the values reported by others, but direct comparison is limited due to differences in patient cohorts. For instance, our cohort included patients with newly diagnosed treatment‐naive HCC, which is in contrast to the cohort with advanced disease in the report by Kuyumcu et al.( 39 ) Others have also observed the potential of PSMA PET to identify unsuspected extrahepatic metastatic disease and to change management strategy in patients with liver‐limited disease on cross‐sectional imaging.( 40 , 41 ) The differences in patient‐selection strategy between our study versus those reports could be one reason for the absence of PSMA‐avid metastatic disease in our cohort with early HCC. Importantly, absent PSMA uptake at PET in 2 patients was confirmed by PSMA IHC, and positive PSMA uptake at PET in 1 patient was also confirmed by PSMA IHC, which supports the specificity of the radiotracer.
The most important reason to pursue PSMA‐targeted imaging in HCC is the prospect of PSMA‐targeted PRRT (e.g., 177Lu‐PSMA‐617) for patients with 68Ga‐PSMA‐11‐avid HCC. Toward that end, definition of thresholds of 68Ga‐PSMA‐11‐avidity adequate for PRRT consideration are needed.( 42 ) In the recent TheraP trial of metastatic castration‐resistant prostate cancer (mCRPC), the PSMA PET eligibility criteria for 177Lu‐PSMA PRRT were SUVmax of at least 20 at a site of disease and greater than 10 at all other measurable sites of metastatic disease.( 43 ) In that context, 10% of patients in our cohort met both criteria for PSMA‐avidity, and 35% of patients met the second criterion. In contrast, PSMA avidity in the recent LuPSMA phase 2 trial in patients with mCRPC was defined as SUVmax at dominant sites of tumor involvement to be at least 1.5 times the SUVmean of liver (i.e., TBRmean > 1.5).( 43 ) By that criterion, all but one of the lesions with high PSMA expression from our study met the criteria for PSMA avidity adequate for PSMA‐targeted PRRT. In contrast to the systemic distribution of metastatic prostate cancer, a key distinction is that the primary index hepatic lesion be the main target for therapy in HCC because it can serve as a biomarker of long‐term outcomes.( 44 ) In patients with PSMA‐avid HCC on 68Ga‐PSMA‐11 PET, intraarterial (I.A.) administration of 177Lu‐PSMA through the hepatic artery could further enhance tumor uptake while minimizing systemic toxicity due to non‐target systemic organ uptake. It is conceivable that PSMA localization to tumor‐associated microvasculature of HCC rather than to tumor cells per se could translate into a quicker washout of injected therapeutic 177Lu‐PSMA. However, recent dosimetry data of 177Lu‐PSMA treatment for glioblastoma multiforme, which is another tumor with angiogenesis‐associated PSMA expression, has shown otherwise.( 45 ) Thus, the SUVmax eligibility thresholds in the context of PRRT considerations for HCC may be lower than in mCRPC. A critical prerequisite, however, would be establishment of significant first‐pass effect of PSMA in HCC with direct I.A. versus systemic intravenous (I.V.) 68Ga‐PSMA administration. Therefore, there is a need to compare intra‐individual safety and pharmacokinetic tumor differences between I.A. versus I.V. 68Ga‐PSMA PET imaging in patients with PSMA‐avid HCC. In light of the limited therapeutic options in patients with advanced HCC, these investigations could lead to approaches for safe and selective delivery of targeted therapy, to reduce toxicity and improve outcomes of patients with HCC.
Our study has limitations. Our IHC data are from a U.S. cohort of patients with early‐stage disease. Potential generalization to patient cohorts with other stages of disease or different underlying risk factors will need further investigation. The correlative data between tissue HCC PSMA expression and TCGA genomic data are from a small cohort, and the definite mechanistic role of PSMA in HCC remains to be elucidated. We were not able to assess prognostic significance of 68Ga‐PSMA‐11 avidity in HCC in the phase 2 prospective study due to limited follow‐up currently. Finally, our inclusion criterion was treatment‐naïve patients with known HCC. Therefore, the diagnostic value of 68Ga‐PSMA PET in patients with suspected hepatobiliary malignancies or with equivocal lesions who do not meet LI‐RADS criteria for HCC remains to be evaluated.
In summary, PSMA has the potential as a theranostic target in patients with HCC, and its ex vivo expression on pathology specimens can be translated to sufficient tumor avidity on 68Ga‐PSMA‐11 PET. Compared with CCA and HCA, PSMA expression in tumor‐associated endothelial cells was more common and pronounced for HCC. FGF14 and MAD1 genes are potential candidates for further investigation into the mechanistic basis for PSMA expression in HCC, for which there is a need for PSMA‐positive small animal HCC models. Based on our experience, we propose that PSMA‐targeted PRRT be investigated as a therapeutic option in patients with PSMA‐avid HCC to complement existing therapeutic options. Comparison of intraindividual safety and pharmacokinetic tumor differences between intra‐arterial versus intravenous 68Ga‐PSMA‐11 PET could help determine a safe and selective delivery of targeted therapy to reduce toxicity and improve outcomes of HCC.
Supporting information
Fig S1
Fig S2
Fig S3
Fig S4
Fig S5
Fig S6
Supplementary Material
Supported by Department of Defense Peer‐Reviewed Cancer Research Program (CA190188), Mayo Clinic Office of Transform the Practice, Mayo Clinic Center for Cell Signaling in Gastroenterology (NIDDK P30DK084567), and Mayo Clinic Hepatobiliary SPORE (NCI P50CA210964).
Potential conflict of interest: Dr. Roberts advises and received grants from Bayer, Exact Sciences, and Gilead. He consults for AstraZeneca, pontifex, and Clinical Care Options. He advises GRAIL, Tavec, QED, Genentech, Envision, and Eisai. He received grants from Ariad, BTG/Boston Scientific, Fujifilm, TARGET, Glycotest, and RedHill. Dr. Johnson received grants from Novartis, Pfizer, and Clarity.
References
Author names in bold designate shared co‐firstauthorship.
- 1. Chernyak V, Fowler KJ, Kamaya A, Kielar AZ, Elsayes KM, Bashir MR, et al. Liver imaging reporting and data system (LI‐RADS) version 2018: imaging of hepatocellular carcinoma in at‐risk patients. Radiology 2018;289:816‐830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bruix J, Reig M, Sherman M. Evidence‐based diagnosis, staging, and treatment of patients with hepatocellular carcinoma. Gastroenterology 2016;150:835‐853. [DOI] [PubMed] [Google Scholar]
- 3. Ronot M, Clift AK, Vilgrain V, Frilling A. Functional imaging in liver tumours. J Hepatol 2016;65:1017‐1030. [DOI] [PubMed] [Google Scholar]
- 4. Finn RS, Qin S, Ikeda M, Galle PR, Ducreux M, Kim T‐Y, et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N Engl J Med 2020;382:1894‐1905. [DOI] [PubMed] [Google Scholar]
- 5. Pfister D, Núñez NG, Pinyol R, Govaere O, Pinter M, Szydlowska M, et al. NASH limits anti‐tumour surveillance in immunotherapy‐treated HCC. Nature 2021;592:450‐456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Haber PK, Puigvehi M, Castet F, Lourdusamy V, Montal R, Tabrizian P, et al. Evidence‐based management of HCC: systematic review and meta‐analysis of randomized controlled trials (2002*2020). Gastroenterology 2021;161(Suppl 1):879‐898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hofman MS, Hicks RJ, Maurer T, Eiber M. Prostate‐specific membrane antigen PET: clinical utility in prostate cancer, normal patterns, pearls, and pitfalls. Radiographics 2018;38:200‐217. [DOI] [PubMed] [Google Scholar]
- 8. Schmidt LH, Heitkötter B, Schulze AB, Schliemann C, Steinestel K, Trautmann M, et al. Prostate specific membrane antigen (PSMA) expression in non‐small cell lung cancer. PLoS One 2017;12:e0186280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bychkov A, Vutrapongwatana U, Tepmongkol S, Keelawat S. PSMA expression by microvasculature of thyroid tumors—potential implications for PSMA theranostics. Sci Rep 2017;7:5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Heitkötter B, Trautmann M, Grünewald I, Bögemann M, Rahbar K, Gevensleben H, et al. Expression of PSMA in tumor neovasculature of high grade sarcomas including synovial sarcoma, rhabdomyosarcoma, undifferentiated sarcoma and MPNST. Oncotarget 2017;8:4268‐4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jiao D, Li YU, Yang FA, Han D, Wu J, Shi S, et al. Expression of prostate‐specific membrane antigen in tumor‐associated vasculature predicts poor prognosis in hepatocellular carcinoma. Clin Transl Gastroenterol 2019;10:1‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sasikumar A, Joy A, Nanabala R, Pillai MR, Thomas B, Vikraman KR. (68)Ga‐PSMA PET/CT imaging in primary hepatocellular carcinoma. Eur J Nucl Med Mol Imaging 2016;43:795‐796. [DOI] [PubMed] [Google Scholar]
- 13. Taneja S, Taneja R, Kashyap V, Jha A, Jena A. 68Ga‐PSMA uptake in hepatocellular carcinoma. Clin Nucl Med 2017;42:e69‐e70. [DOI] [PubMed] [Google Scholar]
- 14. Hirmas N, Leyh C, Sraieb M, Barbato F, Schaarschmidt BM, Umutlu L, et al. [(68)Ga]Ga‐PSMA‐11 PET/CT improves tumor detection and impacts management in patients with hepatocellular carcinoma (HCC). J Nucl Med 2021;62:1235‐1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Blanc V, Riordan JD, Soleymanjahi S, Nadeau JH, Nalbantoglu I, Xie Y, et al. Apobec1 complementation factor overexpression promotes hepatic steatosis, fibrosis, and hepatocellular cancer. J Clin Invest 2021;131(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Byrt T, Bishop J, Carlin JB. Bias, prevalence and kappa. J Clin Epidemiol 1993;46:423‐429. [DOI] [PubMed] [Google Scholar]
- 17. Cancer Genome Atlas Research Network . Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 2017;169:1327‐1341.e1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kalari KR, Nair AA, Bhavsar JD, O’Brien DR, Davila JI, Bockol MA, et al. MAP‐RSeq: Mayo Analysis Pipeline for RNA sequencing. BMC Bioinformatics 2014;15:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge‐based approach for interpreting genome‐wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545‐15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat 2001;29:1165‐1188. [Google Scholar]
- 21. Werner‐Wasik M, Nelson AD, Choi W, Arai Y, Faulhaber PF, Kang P, et al. What is the best way to contour lung tumors on PET scans? Multiobserver validation of a gradient‐based method using a NSCLC digital PET phantom. Int J Radiat Oncol Biol Phys 2012;82:1164‐1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tolkach Y, Goltz D, Kremer A, Ahmadzadehfar H, Bergheim D, Essler M, et al. Prostate‐specific membrane antigen expression in hepatocellular carcinoma: potential use for prognosis and diagnostic imaging. Oncotarget 2019;10:4149‐4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen L‐X, Zou S‐J, Li D, Zhou J‐Y, Cheng Z‐T, Zhao J, et al. Prostate‐specific membrane antigen expression in hepatocellular carcinoma, cholangiocarcinoma, and liver cirrhosis. World J Gastroenterol 2020;26:7664‐7678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Conway RE, Petrovic N, Li Z, Heston W, Wu D, Shapiro LH. Prostate‐specific membrane antigen regulates angiogenesis by modulating integrin signal transduction. Mol Cell Biol 2006;26:5310‐5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Conway RE, Joiner K, Patterson A, Bourgeois D, Rampp R, Hannah BC, et al. Prostate specific membrane antigen produces pro‐angiogenic laminin peptides downstream of matrix metalloprotease‐2. Angiogenesis 2013;16:847‐860. [DOI] [PubMed] [Google Scholar]
- 26. Conway RE, Rojas C, Alt J, Nováková Z, Richardson SM, Rodrick TC, et al. Prostate‐specific membrane antigen (PSMA)‐mediated laminin proteolysis generates a pro‐angiogenic peptide. Angiogenesis 2016;19:487‐500. [DOI] [PubMed] [Google Scholar]
- 27. Grant CL, Caromile LA, Ho V, Durrani K, Rahman MM, Claffey KP, et al. Prostate specific membrane antigen (PSMA) regulates angiogenesis independently of VEGF during ocular neovascularization. PLoS One 2012;7:e41285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sandhu DS, Baichoo E, Roberts LR. Fibroblast growth factor signaling in liver carcinogenesis. Hepatology 2014;59:1166‐1173. [DOI] [PubMed] [Google Scholar]
- 29. Coleman SJ, Grose RP, Kocher HM. Fibroblast growth factor family as a potential target in the treatment of hepatocellular carcinoma. J Hepatocell Carcinoma 2014;1:43‐54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mcarthur GA, Laherty CD, Queva C, Hurlin PJ, Loo L, James L, et al. The Mad protein family links transcriptional repression to cell differentiation. Cold Spring Harb Symp Quant Biol 1998;63:423‐433. [DOI] [PubMed] [Google Scholar]
- 31. Sze KM, Ching YP, Jin DY, Ng IO. Role of a novel splice variant of mitotic arrest deficient 1 (MAD1), MAD1beta, in mitotic checkpoint control in liver cancer. Cancer Res 2008;68:9194‐9201. [DOI] [PubMed] [Google Scholar]
- 32. Sze KM, Chu GK, Mak QH, Lee JM, Ng IO. Proline‐rich acidic protein 1 (PRAP1) is a novel interacting partner of MAD1 and has a suppressive role in mitotic checkpoint signalling in hepatocellular carcinoma. J Pathol 2014;233:51‐60. [DOI] [PubMed] [Google Scholar]
- 33. Nam CW, Park NH, Park BR, Shin JW, Jung SW, Na YW, et al. Mitotic checkpoint gene MAD1 in hepatocellular carcinoma is associated with tumor recurrence after surgical resection. J Surg Oncol 2008;97:567‐571. [DOI] [PubMed] [Google Scholar]
- 34. Cui C, Lu Z, Yang L, Gao Y, Liu W, Gu L, et al. Genome‐wide identification of differential methylation between primary and recurrent hepatocellular carcinomas. Mol Carcinog 2016;55:1163‐1174. [DOI] [PubMed] [Google Scholar]
- 35. Liping X, Jia L, Qi C, Liang Y, Dongen L, Jianshuai J. Cell cycle genes are potential diagnostic and prognostic biomarkers in hepatocellular carcinoma. Biomed Res Int 2020;2020:6206157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rebouissou S, Nault JC. Advances in molecular classification and precision oncology in hepatocellular carcinoma. J Hepatol 2020;72:215‐229. [DOI] [PubMed] [Google Scholar]
- 37. Brown ZJ, Heinrich B, Greten TF. Mouse models of hepatocellular carcinoma: an overview and highlights for immunotherapy research. Nat Rev Gastroenterol Hepatol 2018;15:536‐554. [DOI] [PubMed] [Google Scholar]
- 38. Cuda TJ, Riddell AD, Liu C, Whitehall VL, Borowsky J, Wyld DK, et al. PET imaging quantifying (68)Ga‐PSMA‐11 uptake in metastatic colorectal cancer. J Nucl Med 2020;61:1576‐1579. [DOI] [PubMed] [Google Scholar]
- 39. Kuyumcu S, Has‐Simsek D, Iliaz R, Sanli Y, Buyukkaya F, Akyuz F, et al. Evidence of prostate‐specific membrane antigen expression in hepatocellular carcinoma using 68Ga‐PSMA PET/CT. Clin Nucl Med 2019;44:702‐706. [DOI] [PubMed] [Google Scholar]
- 40. Kunikowska J, Cieślak B, Gierej B, Patkowski W, Kraj L, Kotulski M, et al. [(68) Ga]Ga‐prostate‐specific membrane antigen PET/CT: a novel method for imaging patients with hepatocellular carcinoma. Eur J Nucl Med Mol Imaging 2021;48:883‐892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kesler M, Levine C, Hershkovitz D, Mishani E, Menachem Y, Lerman H, et al. (68)Ga‐PSMA is a novel PET‐CT tracer for imaging of hepatocellular carcinoma: a prospective pilot study. J Nucl Med 2019;60:185‐191. [DOI] [PubMed] [Google Scholar]
- 42. Kratochwil C, Fendler WP, Eiber M, Baum R, Bozkurt MF, Czernin J, et al. EANM procedure guidelines for radionuclide therapy with (177)Lu‐labelled PSMA‐ligands ((177)Lu‐PSMA‐RLT). Eur J Nucl Med Mol Imaging 2019;46:2536‐2544. [DOI] [PubMed] [Google Scholar]
- 43. Hofman MS, Violet J, Hicks RJ, Ferdinandus J, Thang SP, Akhurst T, et al. [(177)Lu]‐PSMA‐617 radionuclide treatment in patients with metastatic castration‐resistant prostate cancer (LuPSMA trial): a single‐centre, single‐arm, phase 2 study. Lancet Oncol 2018;19:825‐833. [DOI] [PubMed] [Google Scholar]
- 44. Riaz A, Miller FH, Kulik LM, Nikolaidis P, Yaghmai V, Lewandowski RJ, et al. Imaging response in the primary index lesion and clinical outcomes following transarterial locoregional therapy for hepatocellular carcinoma. JAMA 2010;303:1062‐1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kunikowska J, Charzynska I, Kulinski R, Pawlak D, Maurin M, Krolicki L. Tumor uptake in glioblastoma multiforme after IV injection of [(177)Lu]Lu‐PSMA‐617. Eur J Nucl Med Mol Imaging 2020;47:1605‐1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Fig S3
Fig S4
Fig S5
Fig S6
Supplementary Material


