Abstract
Background
Mutations in PIGN, resulting in a glycosylphosphatidylinositol (GPI) anchor deficiency, typically leads to multiple congenital anomalies-hypotonia-seizures syndrome. However, the link between PIGN and epilepsy or paroxysmal non-kinesigenic dyskinesia (PNKD) is not well-described. This study reported a patient with PIGN mutation leading to developmental and epileptic encephalopathy and PNKD, to expand upon the genotype–phenotype correlation of PIGN.
Case presentation
During the first 10 days of life, a girl exhibited paroxysmal staring episodes with durations that ranged from several minutes to hours. These episodes occurred 2–5 times daily and always occurred during wakefulness. Ictal electroencephalography revealed no abnormalities, and PNKD was diagnosed. The patient also exhibited severely delayed psychomotor development and generalized seizures at the age of 4 months. Results of brain magnetic resonance imaging and metabolic screenings were normal, but trio-based whole-exome sequencing identified two novel compound heterozygous PIGN mutations (NM_176787; c.163C > T [p.R55 > X] and c.283C > T [p.R95W]). Flow cytometry analysis of the patient’s granulocytes revealed dramatically reduced expression of GPI-anchored proteins. This indicated that the mutations compromised GPI functions. The patient got seizure-free for 1 year, and her dyskinesia episodes reduced significantly (1–2 times/month) after treatment with levetiracetam (600 mg/day) and clonazepam (1.5 mg/day). No progress was observed with respect to psychomotor development; however, no craniofacial dysmorphic features, cleft lip/palate, brachytelephalangy with nail hypoplasia, and internal malformations have been observed until now (6 years of age).
Conclusion
This is the first study to document developmental and epileptic encephalopathy with PNKD in a human with PIGN mutations. This report expanded our understanding of the genotype–phenotype correlation of PIGN, and PIGN may be considered a potentially relevant gene when investigating cases of epilepsy or PNKD.
Keywords: Multiple congenital anomalies-hypotonia-seizures syndrome, Paroxysmal nonkinesigenic dyskinesia, PIGN gene; seizure
Background
The genetic etiologies of many patients with developmental and epileptic encephalopathy (DEE) are still unknown, highlighting the urgent need to identify more DEE-related genes [1]. The PIGN gene (OMIM* 606,097) encodes an enzyme involved in the biosynthesis of glycosylphosphatidylinositol (GPI), which anchors various proteins to the cell surface [2, 3]. Mutations in PIGN, resulting in a GPI anchor deficiency, typically causes multiple congenital anomalies-hypotonia-seizures syndrome (OMIM# 614,080)) and Fryns syndrome (OMIM# 229,850) [2, 3]. However, some patients with PIGN mutation do not fully meet the diagnostic criteria of these syndromes [4, 5]. Therefore, the phenotypic understanding of PIGN-related spectrum disorders needs to be improved. No cases of PIGN-related epilepsy with paroxysmal non-kinesigenic dyskinesia (PNKD) have been reported in humans thus far. Aiming to expand upon the genotype-phenotypic correlation of PIGN, we described a patient with PIGN mutation with DEE and PNKD without dysmorphic features.
Case presentation
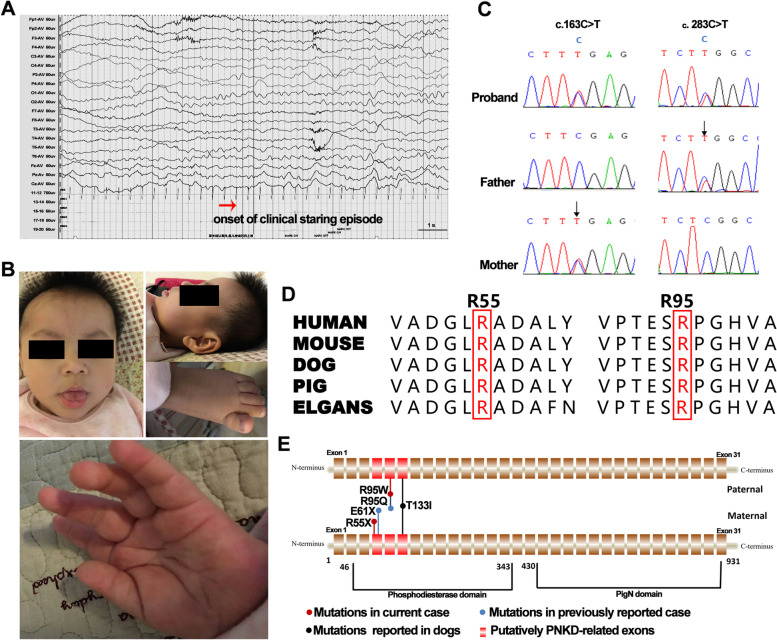
We report the case of a girl born to healthy parents, both of non-consanguineous Asian descent. An uncomplicated pregnancy was followed by a 39-week gestation period. She was delivered by cesarean section without complications. Her birth weight was 3.5 kg (75th–90th percentile). During the first 10 days of life, the girl exhibited paroxysmal staring episodes with durations that ranged from several minutes to hours. These episodes occurred 2–5 times daily and always occurred during wakefulness. Ictal electroencephalography revealed no abnormalities (Fig. 1A). Based on the durations and frequency of her dyskinesia episodes and the apparent lack of precipitating factors, a diagnosis of PNKD was established. Developmental delay was observed at the age of 3 months, as the girl had not yet developed a social smile or control over her own head. She began experiencing generalized seizures (generalized tonic–clonic seizures) 1–2 times daily since the age of 4 months. During the initial neurodiagnostic evaluation at the age of 4 months, her weight was 6.5 kg (25th–50th percentile), height was 65.5 cm (75th–90th percentile), and head circumference was 42.0 cm (25th–50th percentile). No dysmorphic features were observed at age of 1 year (Fig. 1B), and the results of metabolic screenings were normal. Brain magnetic resonance imaging revealed no abnormalities.
Fig. 1.
A Ictal electroencephalograms showing no abnormalities. B Photographs showing no obvious dysmorphic features at the age of 1 year. C The PIGN mutations. D Phylogenetic conservation of the R55 and R95 (highlighted in red). E Schematic of the PIGN gene showing the putatively PNKD-related exons. Abbreviation: PNKD, paroxysmal nonkinesigenic dyskinesia
Considering that the patient's early-onset epilepsy, with developmental delay, PNKD, without dysmorphic features, and hereditary causes other than chromosomal copy number abnormalities were considered, trio-based whole exome sequencing (WES) was selected for etiological exploration. The study protocol was approved by the Ethics Committee of the Affiliated Hospital of Zunyi Medical University, and the parents gave their written informed consents. The candidate variants were evaluated using the American College of Medical Genetics and Genomics guidelines [6]. The changes in protein stability were assessed using the free energy stability change (DDG, Kcal/mol) value (http://gpcr.biocomp.unibo.it/cgi/predictors/I-Mutant3.0/I-Mutant3.0.cgi). Effects of variants were predicted by Grantham scores [7] and multiple in silico tools. The conservation of mutated residues was evaluated using sequence alignment of phylogenetic species. Sanger sequencing was performed to identify potential clinically significant variants. Whole-exome sequencing identified two novel compound heterozygous PIGN mutations (NM_176787; c.163C > T [p.R55X] and c.283C > T [p.R95W]) (Fig. 1C): a maternally inherited nonsense mutation in exon 4 and a paternally inherited missense mutation in exon 5. Notably, the amino acid residues of the two variants are highly conserved among various species (Fig. 1D). The two variants were suggested to be damaging by at least six in silico tools (Table 1). The missense variant was predicted to have severe effects according to the Grantham scores (101), and was predicted to decrease the stability of PIGN with a DDG value of -0.33. Variant R55X was estimated as a pathogenic variant (PVS1 + PM2 + PP3), and variant R95W was estimated as a likely pathogenic variant (PM2 + PM3 + PP3 + PP4) based on the American College of Medical Genetics and Genomics guidelines. Flow cytometry analysis of the girl’s granulocytes revealed dramatically reduced expression of GPI-anchored proteins (34% of the expression corresponding to the control condition). This indicated that the mutations compromised GPI functions. She had no any dysmorphic features until she reached the age of 6 years. Although, she had global severe psychomotor retardation, hyperkinetic movements, non-verbal, occasionally displayed social smiles, but still couldn't control her head. Her weight was 16 kg (3rd–10th percentile), height was 112 cm (10th–25th percentile), and head circumference was 49 cm (50th–75th percentile). She was seizure-free for 1 year, and her dyskinesia episodes reduced significantly (1–2 times/month) after treatment with levetiracetam (600 mg/day) and clonazepam (1.5 mg/day).
Table 1.
Genetic features of the individual with PIGN mutations
| Position | cDNA change (NM_176787) |
Protein change | MAF | MAF-EAS | Mutation taster | SIFT | CADD | Polyphen2 | FATHMM | GERP + + | PhastCons | ClinPred |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| chr18:59,828,424 | 163C > T | R55X | 4.88e-05 | 3.0e-04 | DC (1.00) | - | D (36) | - | D (0.92) | C (2.83) | C (1.00) | - |
| chr18:59,824,980 | 283C > T | R95W | 0 | 0 | DC (1.00) | D (0) | D (34) | PD (1.00) | D (0.94) | C (4.51) | C (1.00) | P (0.99) |
Abbreviations: C Conserved, CADD Combined annotation dependent depletion, D Damaging, DC Disease causing, Chr Chromosome, MAF Minor allele frequency from Genome Aggregation Database, MAF-EAS Minor allele frequency from East Asia population in Genome Aggregation Database, NA Not applicable, P Pathogenic, PD Probably damaging
Discussion and conclusions
PIGN mapping to 18q21.33 encodes a protein with 931 amino-acids that successively contains a phosphodiesterase domain in the N-terminal and pigN domain in the C-terminal (Fig. 1E), and this protein is involved in GPI-anchor biosynthesis [3]. The GPI-anchor is a glycolipid found on various blood cells and serves to anchor proteins to the cell surface [8]. Biallelic PIGN variants often cause various clinical manifestations, including visceral abnormalities or dysmorphic features (> 90%), psychomotor development delay (100%), hypotonia (100%), and seizures (93%) [3, 8]. Such variants may even be lethal in the fetal or neonatal period, and can lead to multiple congenital anomalies-hypotonia-seizures syndrome and Fryns syndrome [2, 9]. However, many cases cannot be classified as either of the syndromes, as the genotype–phenotype associations of PIGN are not fully understood. Our patient only manifested DEE and PNKD, and no visceral abnormalities or dysmorphic features were observed until the age of 6 years. A novel term, namely developmental and epileptic-dyskinetic encephalopathy (DEDE), has been used to describe a subset of children with DEE and a hyperkinetic movement disorder including choreoathetosis, dyskinesia, dystonia, and non-epileptic myoclonus [10, 11]. DEDE is characterized by multiple types of refractory epilepsy, profound intellectual disability, and hyperkinetic movement disorder which often appears after epilepsy [10, 11]. Nevertheless, in this study, PNKD was the first presentation, which occurred independently before developmental delay and epilepsy; thus, PNKD is retained after epilepsy is controlled, indicating that PIGN mutations appear to be panethnic and may comprise an underappreciated cause of epilepsy and PNKD.
The PIGN gene is highly expressed in the brain (cerebral cortex, cerebellum, and basal ganglia) (www.proteinatlas.org/search/PIGN) and is ubiquitously expressed across the whole lifespan, with the highest expression being in the infancy stage (GD&P DataBase [gdap.org.cn]). The expressional stage of PIGN could explain the patient’s symptoms that gradually aggravate after birth and tend to stabilize with age. Paroxysmal dyskinesias can be attributed to abnormal basal ganglia and/or cerebellar activity [12]. Both non-acquired epilepsy and PNKD may be associated to gene mutations that affect the function of neuron membranes and lead to transient dysfunction of neuronal networks. It is speculated that these gene mutations may mainly affect the cortex-basal ganglia connection and cerebellum function, resulting in corresponding episodic presentations [12]. The high expression of PIGN in the cerebral cortex, cerebellum, and basal ganglia may help explain the combination of epilepsy and PNKD observed in this patient. This is the first study to document DEE with PNKD in a human with PIGN mutations.
The present patient carried a maternally inherited exon 4 mutation and a paternally inherited exon 5 mutation. Interestingly, a previous report described a patient with involuntary movements without dysmorphic features who harbored the biallelic PIGN variants involving exons 4 and 5, but the type of dyskinesia is not described in detail in this article [4]. Furthermore, the locus of the homozygous PIGN mutation (c.398C > T; p.T131I) previously reported in 25 affected dogs with PNKD corresponds to a locus in exon 6 in the human gene [13]. Given these findings, we can reasonably speculate that biallelic PIGN mutations affecting exons 4–6 may be more predisposed to cause PNKD (Fig. 1E); however, detailed molecular and phenotypic analyses of cases of PIGN-related disorders will be necessary to test this hypothesis.
Paroxysmal dyskinesias are characterized by recurrent episodes of involuntary dystonia, chorea, athetosis, or their combination with sudden onset and lasting a variable duration [12, 14, 15]. They are classified into three forms, namely, paroxysmal kinesigenic dyskinesia, PNKD, and exercise induced dyskinesia, based on their difference of triggers. Attacks in some patients with PNKD are triggered by unspecified factors, including emotional stress, fatigue, or consumption of alcohol or caffeine. Duration of PNKD is usually longer (up to several hours), and symptoms of onset tend to occur at a younger age compared to paroxysmal dyskinesia [14]. The MR-1 and KCNMA1 are reportedly related to PNKD [15–18]. The MR-1 mutation causes PNKD without epilepsy, whereas the KCNMA1 mutation was first reported in 2005 in a family with PNKD and epilepsy [17–19]. It was once again described in our reports of three unrelated children with PNKD and developmental delay without epilepsy [18]. Compared with MR-1 and KCNMA1 mutations, our patient’s PIGN mutations caused PNKD onset at an earlier age, including more severe DEE. Furthermore, the known pathogenetic MR-1 and KCNMA1 mutations are dominant, whereas our patient’s PIGN mutations are recessive. The clinical and genetic characteristics of patients with PIGN or KCNMA1 mutations are summarized in Table 2.
Table 2.
The clinical and genetic characteristic of patients with PNKD
| Genes | PIGN | KCNMA1 | |||
|---|---|---|---|---|---|
| Current case | Case 1 [18] | Case 2 [18] | Case 3a | Family cases [17] | |
| Sex | Female | Male | Male | Male | 10 male, 6 female |
| Perinatal | Unremarkable | Unremarkable | Unremarkable | Unremarkable | Unremarkable |
| Family | Negative | Negative | Negative | Negative | Inherited |
| Age of onset | 10 days (PNKD) | 20 days (PNKD) | 7 months (PNKD) | 13 months (PNKD) | 6 months -15 years |
| Triggers | No | No | No | No | Undefined (Alcohol?) |
| Development milestones | Severe delay | Severe delay | Mild delay | Mild delay | NA |
| Diagnosis | DEE + PNKD | PNKD (till 3.5 year of age) | PNKD (till 7 year of age) | No (till 5.7 year of age) | Epilepsy and/or PNKD |
| Effective medicine | Levetiracetam and clonazepam | No response to oxcarbazepine, valproate and levetiracetam | Clonazepam | Clonazepam | NA |
| Genetic data | |||||
| cDNA | 163C > T; 283C > T (NM_176787) | 2650G > A (NM_1161352) | 3158A > G (NM_1161352) | 3158A > G (NM_1161352) | 1301A > G (NM_1161352) |
| Protein | R55X; R95W | E884K | N1053S | N1053S | D434G |
| Inheritance manner | Autosomal recessive (inherited) | Autosomal dominant (de novo) | Autosomal dominant (de novo) | Autosomal dominant (de novo) | Autosomal dominant (inherited) |
aThis case was published in Chinese by our team
DEE Developmental epileptic encephalopathy, PNKD Paroxysmal nonkinesigenic dyskinesia, NA Not available
Patients with PNKD showed good response to clonazepam or diazepam [15, 18, 19]. Our patient also showed a good response to clonazepam, indicated by the reduction in the frequency of attacks from several times a day to 1–2 times per month. The patient’s epilepsy started early, but the progress of psychomotor development was not observed even the epilepsy was controlled, suggesting that PIGN-related DEDE was a more severe phenotype of DEE.
Our findings indicate that altered GPI anchor functions can be associated with epilepsy and PNKD, and DEDE may be one of the phenotypes of PIGN. This information enriches our understanding of the clinical and genetic spectrum of epilepsy and PNKD, and of the phenotypic consequences of PIGN mutations. We therefore conclude that PIGN should be considered as a candidate gene for screening in patients with early-onset epileptic encephalopathy and/or PNKD.
Acknowledgements
We thank the patient and her parents for consenting to the publication of this case report and cooperating with clinical assessments.
Abbreviations
- DEDE
Developmental and epileptic-dyskinetic encephalopathy
- DEE
Developmental and epileptic encephalopathy
- PNKD
Paroxysmal nonkinesigenic dyskinesia
Authors’ contributions
MT and JC collected the data from patients and wrote the paper; JL and HP analyzed genetic pathogenicity; WL analyzed EEG recordings and neuro-imaging data; XS designed and supervised the study. All authors have read and approved the manuscript.
Funding
All phases (collection, analysis, and interpretation of data and in writing the manuscript) of this study were supported by grants from grants from the National Natural Science Foundation of China (No. 81660219) and Basic Research Program of Guizhou Province: Guizhou Science and Technology Foundation (No. ZK [2021] General 418).
Availability of data and materials
The data that support the findings of this study will be available from supplementary materials and the corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
The study protocol was approved by the Ethics Committee of the Affiliated Hospital of Zunyi Medical University. The parents gave their informed consents for this report.
Consent for publication
The parents gave their informed consents for their child’s personal or clinical details along with any identifying images to be published in this study. A copy of the signed, written informed consent for publication form is available for review by the editor.
Competing interests
All authors have no financial relationships relevant to this article to disclose.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Maoqiang Tian and Jing Chen contributed equally to this work.
References
- 1.Saffari A, Schroter J, Garbade SF, et al. Quantitative retrospective natural history modeling of WDR45-related developmental and epileptic encephalopathy - a systematic cross-sectional analysis of 160 published cases. Autophagy. 2021:1–13. 10.1080/15548627.2021.1990671. [DOI] [PMC free article] [PubMed]
- 2.McInerney-Leo AM, Harris JE, Gattas M, et al. Fryns syndrome associated with recessive mutations in PIGN in two separate families. Hum Mutat. 2016;37:695–702. doi: 10.1002/humu.22994. [DOI] [PubMed] [Google Scholar]
- 3.Fleming L, Lemmon M, Beck N, et al. Genotype-phenotype correlation of congenital anomalies in multiple congenital anomalies hypotonia seizures syndrome (MCAHS1)/PIGN-related epilepsy. Am J Med Genet A. 2016;170A:77–86. doi: 10.1002/ajmg.a.37369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thiffault I, Zuccarelli B, Welsh H, et al. Hypotonia and intellectual disability without dysmorphic features in a patient with PIGN-related disease. BMC Med Genet. 2017;18:124. doi: 10.1186/s12881-017-0481-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jezela-Stanek A, Mierzewska H, Szczepanik E. Vertical nystagmus as a feature of PIGN-related glycosylphosphatidylinositol biosynthesis defects. Clin Neurol Neurosurg. 2020;196:106033. doi: 10.1016/j.clineuro.2020.106033. [DOI] [PubMed] [Google Scholar]
- 6.Rehder C, Bean LJH, Bick D, et al. Next-generation sequencing for constitutional variants in the clinical laboratory, 2021 revision: a technical standard of the American College of Medical Genetics and Genomics. Genet Med: ACMG); 2021. [DOI] [PubMed] [Google Scholar]
- 7.Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 8.Xiao SQ, Li MH, Meng YL, et al. Case report: compound heterozygous phosphatidylinositol-glycan biosynthesis class N (PIGN) mutations in a Chinese fetus with hypotonia-seizures syndrome 1. Front Genet. 2020;11:594078. doi: 10.3389/fgene.2020.594078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun L, Yang X, Xu Y, et al. Prenatal diagnosis of familial recessive PIGN mutation associated with multiple anomalies: a case report. Taiwan J Obstet Gynecol. 2021;60:530–533. doi: 10.1016/j.tjog.2021.03.026. [DOI] [PubMed] [Google Scholar]
- 10.Gorman KM, Peters CH, Lynch B, et al. Persistent sodium currents in SCN1A developmental and degenerative epileptic dyskinetic encephalopathy. Brain Commun. 2021;3:fcab235. doi: 10.1093/braincomms/fcab235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carvill GL, Helbig KL, Myers CT, et al. Damaging de novo missense variants in EEF1A2 lead to a developmental and degenerative epileptic-dyskinetic encephalopathy. Hum Mutat. 2020;41:1263–1279. doi: 10.1002/humu.24015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Gusmao CM, Garcia L, Mikati MA, et al. Paroxysmal genetic movement disorders and epilepsy. Front Neurol. 2021;12:648031. doi: 10.3389/fneur.2021.648031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolicheski AL, Johnson GS, Mhlanga-Mutangadura T, et al. A homozygous PIGN missense mutation in soft-coated wheaten terriers with a canine paroxysmal dyskinesia. Neurogenetics. 2017;18:39–47. doi: 10.1007/s10048-016-0502-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erro R, Sheerin UM, Bhatia KP. Paroxysmal dyskinesias revisited: a review of 500 genetically proven cases and a new classification. Mov Disord. 2014;29:1108–1116. doi: 10.1002/mds.25933. [DOI] [PubMed] [Google Scholar]
- 15.McGuire S, Chanchani S, Khurana DS. Paroxysmal Dyskinesias. Semin Pediatr Neurol. 2018;25:75–81. doi: 10.1016/j.spen.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 16.Yeh TH, Lin JJ, Lai SC, et al. Familial paroxysmal nonkinesigenic dyskinesia: clinical and genetic analysis of a Taiwanese family. J Neurol Sci. 2012;323:80–84. doi: 10.1016/j.jns.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 17.Du W, Bautista JF, Yang H, et al. Calcium-sensitive potassium channelopathy in human epilepsy and paroxysmal movement disorder. Nat Genet. 2005;37:733–738. doi: 10.1038/ng1585. [DOI] [PubMed] [Google Scholar]
- 18.Zhang ZB, Tian MQ, Gao K, et al. De novo KCNMA1 mutations in children with early-onset paroxysmal dyskinesia and developmental delay. Mov Disord. 2015;30:1290–1292. doi: 10.1002/mds.26216. [DOI] [PubMed] [Google Scholar]
- 19.Kim SY, Lee JS, Kim WJ, et al. Paroxysmal dyskinesia in children: from genes to the clinic. J Clin Neurol. 2018;14:492–497. doi: 10.3988/jcn.2018.14.4.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study will be available from supplementary materials and the corresponding author upon reasonable request.