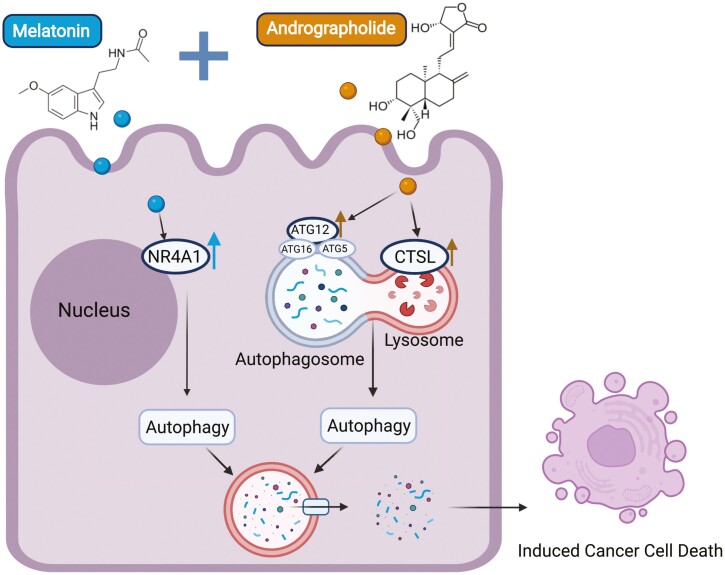
Abstract
Colorectal cancer (CRC) is one of the most frequent malignancies worldwide and remains one of the leading causes of cancer-related deaths in the USA. The high degree of morbidity and mortality associated with this disease is largely due to the inadequate efficacy of current treatments as well the development of chemoresistance. In recent years, several pharmaceutical agents screened from natural products have shown the promise to offer a safe, inexpensive and synergistically multi-targeted treatment option in various cancers. Given the growing evidence of anti-carcinogenic properties of two natural compounds, melatonin (MLT) and andrographis (Andro), we aimed to evaluate their synergistic anticancer effects in CRC. We demonstrate that indeed these two compounds possessed a synergistic anticancer effect in terms of their ability to inhibit cell viability, suppression of colony-formation and induction of apoptosis (P < 0.05). In line with our in vitro findings, we were able to validate this combinatorial anticancer activity in xenograft animal models (P < 0.001) as well as tumor-derived 3D organoids (P < 0.01). RNA-sequencing analysis revealed candidate pathways and genes that mediated antitumor efficacy of MLT and Andro in CRC, among which autophagy pathway and related genes, including NR4A1, CTSL and Atg12, were found to be primarily responsible for the increased anticancer effect by the two natural products. In conclusion, our data reveal a potent and synergistic therapeutic effect of MLT and Andro in the treatment of CRC and provides a rationale for suppressing autophagy in cancer cells as a potential therapeutic strategy for CRC.
We systematically used multiple in vitroand in vivomodels to demonstrate melatonin and andrographis exhibit pronounced synergistic activity in CRC inhibition, mediated in part by autophagy-associated pathways induced apoptotic activation, highlighting the combinatorial therapeutic potential of these two natural products in patients with CRC.
Graphical Abstract
Introduction
Colorectal cancer (CRC) ranks as the third most common malignancy and the second leading cause of cancer-related deaths worldwide (1). It is estimated that by the end of 2030, the global burden of CRC will increase with an estimated 2.2 million new cases and over 1.1 million CRC-related deaths (2). Currently, surgical resection followed by postoperative chemotherapy remains the most common treatment modality for the therapeutic management of patients with stage II and III CRC (3–5). For patients with advanced metastatic CRC that cannot be cured with surgery, chemotherapy is used mainly with a palliative intent (6).
However, the efficacy of these treatments, particularly chemotherapy, is somewhat limited due to the acquisition of secondary chemoresistance leading to an overall poor prognosis (5,7). The use of combinational chemotherapy regimens is becoming a more frequently used approach that also aims to help overcome acquired chemoresistance and facilitate improved survival of patients with cancer (8,9). Unfortunately, however, such treatments are often accompanied by adverse cytotoxic adverse effects which lead to the killing of normal cells along with rapidly dividing tumor cells (9). These data highlight that instead of the clinical implementation of multiple cytotoxic drugs, treatment strategies that exploit synergistic drug combinations that are safer yet more effective are critically needed for the management of patients with CRC.
Several pharmaceutical agents identified from natural plants and microorganisms have been shown to play a pivotal role in the chemotherapeutic and prophylactic treatment of various malignant diseases (10–25). Melatonin (MLT) is one such naturally occurring amine hormone synthesized and secreted in the pineal gland or epiphysis of mammals and humans (26). It is well known that the primary function of MLT is to participate in the regulation of biological rhythms and endocrine function (27). However, in recent years, accumulating evidence has also demonstrated that it also possesses potent cytotoxic activity in several types of malignant cells, including CRC cells (15,28–30). A previous study from our group demonstrated that MLT functioned as an anticancer agent against CRC cells via down-regulation of thymidylate synthase (15). Due to its low to no toxicity for normal cells and high antitumor efficacy, MLT is emerging as a promising compound of choice in the field of anticancer drug development (28–32). Emerging evidence also indicates that associations between multiple drugs may produce an overtly enhanced synergistic protective effect, allowing the use of significantly lower concentrations of drugs in combination vs. when used individually—a scenario that potentially leads to reduced adverse events (33,34). However, there is lack of an understanding on the potential use of MLT as a potential synergistic treatment of choice for an improved management of patients with CRC.
Numerous studies have now confirmed that the active constituent within the plant Andrographis paniculate, andrographolide, has a potential anticancer efficacy in various malignancies (16,35–39). In addition, it has been demonstrated that Andrographis (Andro) may serve as an effective adjuvant therapeutic option in CRC by virtue of its synergistic activity with the conventional chemotherapeutic drugs such as 5-fluorouracil (5-FU) in CRC (16,35,38,39). Although the antitumor effects of Andro and MLT in CRC have been studied individually, whether their combination has any synergistic efficacy in this malignancy remains unclear. Accordingly, in this study, we aimed to interrogate whether MLT and Andro have any synergistic activity in CRC and if so, understand the underlying mechanisms responsible for their antitumor activity.
We undertook a series of systematic experiments in CRC cells to reveal that Andro synergistically enhanced the anticancer activity of MLT. Subsequently, we followed up these findings in preclinical animal models and patient-derived 3D tumor organoids, wherein the combination of MLT and Andro also exhibited a significantly pronounced synergistic activity versus the antitumor activity of these compounds individually. A whole-genome transcriptomic profiling analysis further revealed that the combined treatment with MLT and Andro and the observed synergism was in part mediated through activation of apoptosis induced by autophagy-associated pathways. In summary, to the best of our knowledge, ours is the first study to demonstrate the synergistic anticancer effects of MLT and Andro in CRC, highlighting their combinatorial therapeutic potential in this malignancy.
Materials and methods
Cell culture
For in vitro experiments, the human CRC cell lines HCT116, LoVo, DLD1, SW480, HT29 and SW620 were purchased from the American Type Culture Collection (ATCC, Manassas, VA). These cells were cultured, respectively, in Dulbecco’s modified Eagle medium (Gibco, Carlsbad, CA) or Roswell Park Memorial Institute (RPMI)-1640 (Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum (Gibco, Waltham, MA), penicillin (100 U/ml) and streptomycin (100 μg/ml). All cells were maintained in a 37°C incubator with a 5% humidified CO2 atmosphere. All the human CRC cell lines were authenticated by a panel of genetic and epigenetic markers and tested periodically for mycoplasma.
Reagents
MLT (Sigma-Aldrich, St Louis, MO) and Andro (standardized to 20% andrographolide content; EuroPharma USA, Green Bay, WI) were both dissolved in dimethyl sulfoxide (DMSO, Sigma-Aldrich). The final concentration of DMSO in the experimental wells did not exceed 1% for in vitro culture assays. The stock solution of MLT and Andro was stored at −20°C in light protection and diluted with complete medium to appropriate experimental concentrations prior to use.
Cell viability assay
Cell viability was measured by cell counting kit-8 (CCK-8) (Dojindo, Kumamoto, Japan). For the determination of 50% cell growth inhibition (IC50) of MLT in CRC cell lines, cells were plated at a density of 3 × 104 cells per well (HCT116 and RKO) or 4 × 104 cells per well (LoVo, DLD1, SW480, HT29 and SW620) in 96-well plates and treated with gradient concentrations of the MLT for 48 h. Subsequently, 10 μl CCK-8 solution (10 μg/ml) was added to each well and incubated at 37°C for 2 h. Then the absorbance was measured at 450 nm wavelength (OD450) by an enzymatic plate analyzer (Molecular Devices, San Jose, CA). Three independent experiments in triplicate ensured repeatability of results. For the growth curve of CRC cell lines after MLT treatment, HCT116 and SW480 cells were cultured in the 96-well plates at the density of 3 × 104 cells per well and 4 × 104 cells per well, respectively. Cells were treated with MLT (2 mM) for 0, 12, 24, 36, 48, 60 and 72 h. At the indicated time points, CCK-8 assays were performed to assess cell viability as described above.
Colony formation assay
The colony-forming capacity of CRC cells was measured by colony-formation assay. Specifically, HCT116 and SW480 cells were counted and seeded into six-well plates at a density of 800 cells per well and maintained in complete medium for 1 week. Subsequently, the cells were treated with MLT (2 mM), Andro (15 μg/ml) and their combination for another week. The colonies were fixed with methanol for 10 min and stained with 0.1% crystal violet (Acros Organics, Gujarat, India) for 10 min. Stained colonies were then counted using Image J software (National Institute of Health [NIH], Bethesda, MD). The fresh medium was replaced every 3 days during the assay. The experiments were performed in triplicate and repeated three times.
Caspase-3/7 activity analysis
The activation of caspase-3 and caspase-7 orchestrates cellular demolition largely through cleavage of specific substrates and influences the terminal events of the biochemical apoptotic pathway (40). The number and percentage of each stage of apoptosis were determined by the activity of caspase-3/7 in combination with a dead cell dye (7-aminoactinomycin D [7-AAD]) via Muse™ Caspase-3/7 Kit (Merck Millipore, Guyancourt, France). Briefly, HCT116 and SW480 cells were seeded in six-well plates at a density of 5 × 104 cells per well and treated with MLT (1 or 2 mM), Andro (15 μg/ml) and their combination for 48 h. A total of 1 × 105cells were collected by centrifugation (3000 r.p.m., 5 min) and resuspended in 50 µl phosphate-buffered saline (PBS). Then, 5 µl Muse™ Caspase-3/7 working solution was added into a cell suspension and then incubated at 37°C for 30 min protected from light. After incubation, 150 μl of Muse™ Caspase 7-AAD working solution (7-AAD dye) was added to each tube and mixed thoroughly by pipetting up and down. Assay results were obtained using the Muse™ Cell Analyzer (Merck Millipore). The quantitative measurements of apoptotic status were based on the four cell populations: live (caspase−/7-AAD−), dead (caspase−/7-AAD+), early apoptosis (caspase+/7-AAD−) and late apoptosis/dead (caspase+/7-AAD+). All treatments were performed in triplicate and repeated three times.
Preclinical xenograft model
All experiments with xenografted mice were conducted according to the ‘Guide for the Care and Use of Laboratory Animals’ published by the NIH, and approved by the Institutional Animal Care and Use Committee at City of Hope. For tumor induction, 5-week-old male athymic nude mice (Envigo, Houston, TX) were subcutaneously grafted into the left flank with 5 × 106 HCT116 cells (n = 40 mice) in 100 μl PBS. After a week of injection of cells, mice were randomly assigned into four groups: MLT (25 mg/kg body weight), Andro (125 mg/kg body weight), their combination and control group (equal amount of normal saline), n = 10 per group, which were given intraperitoneally doses of each compound every other day for up to 15 days. Tumor size and body weight were measured every 2 days. The tumor volume was calculated using the following formula: V = 0.5 × length × width × width. After euthanasia, each tumor was dissected, weighed and frozen in liquid nitrogen for subsequent total RNA and protein extraction.
Patient-derived 3D organoids
The isolation, culture and passage of tumor organoids derived from patients with CRC were carried out as previously described (38). All experiments have been approved by the Institutional Review Board and written informed consents were obtained from all enrolled patients in accordance with the Declaration of Helsinki. For treatments, organoids were randomly divided into four groups, and appropriate concentrations of MLT (2 mM), Andro (15 μg/ml), as well as their combination, were prepared with the IntestiCult Organoid Growth Medium (STEMCELL Technologies, Vancouver, British Columbia, Canada) of each group and then cultured for 10 days. The control group was treated with a very low concentration of DMSO, which was equivalent to the solvent dissolution of MLT. After treatment, the organoids were observed and counted under a microscope and collected by Gentle Cell Dissociation Reagent (STEMCELL Technologies) followed by ice-cold PBS washes (290 × g, 4°C, 5 min). Harvested organoids were aliquoted and frozen in liquid nitrogen for subsequent total RNA and protein extraction.
RNA-sequencing and data analysis
Total RNA was isolated from 2 × 106 HCT116 and SW480 cells treated with MLT (2 mM), Andro (15 μg/ml) and their combination for 48 h by miRNeasy kit (Qiagen, Hilden, Germany). For RNA-sequencing (RNA-seq) library preparation, 1 μg qualified RNA for each sample was used as input material for the library preparation via the TruSeq RNA Sample Prep Kit (Illumina, San Diego, CA) according to the manufacturer’s instruction. Then the purified library products were validated by Bioanalyzer DNA High Sensitivity Kit (Agilent Technologies, Santa Clara, CA) to determine their size distribution and concentration. Paired-end sequencing (150 bp each end) of the validated library products were then analyzed at the HiSeq X-Ten system (Illumina).
For data analysis, clean reads were obtained by removing reads containing adapter and with low quality, which were then mapped onto reference genome independently. Gene expression was estimated by fragments per kilobase of transcript per million mapped reads. Differentially expressed gene (DEG) analyses were performed using the ‘edgeR’ package (version 4.0.2; R Foundation). DEGs were selected by a threshold of false discovery rate <0.05 and absolute fold change >2.0. Biological functions and canonical pathways were analyzed by Ingenuity Pathway Analysis (IPA, Qiagen).
RNA extraction and quantitative RT-PCR
Total RNA of CRC cells, dissociated patient-derived tumor organoids, and minced xenograft fragments (~1 mm3 in size) were extracted with TRIzol (Invitrogen, Carlsbad, CA). The purity and concentration were determined by a NanoDrop-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA). One microgram of total RNA was reverse-transcribed to cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). Subsequently, 5 ng of the cDNA was subjected for real-time quantitative PCR (qPCR) analyses using SensiFAS SYBR Lo-ROX Kit (Bioline, London, UK) according to the manufacturer’s instruction: briefly, the cDNA samples were mixed with 0.5 µl (10 µM) target gene-specific forward and reverse primers, 5 µl SYBR Lo-ROX mix (2×), and final volume was made up with nuclease-free water. The expression for target genes was analyzed by the 2−ΔΔCT method normalized against the housekeeping β-actin gene. The primer sequences used for qPCR were listed as follows: β-actin: forward 5ʹ-CACCATTGGCAATGAGCGGTTC-3ʹ and β-actin: reverse 5ʹ-AGGTCTTTGCGGATGTCCACGT-3ʹ; ATG12: forward 5ʹ-GGGAAGGACTTACGGATGTCTC-3ʹ and ATG12 reverse 5ʹ- AGGAGTGTCTCCCACAGCCTTT-3ʹ; CTSL: forward 5ʹ- GAAAGGCTACGTGACTCCTGTG-3ʹ and CTSL: reverse 5ʹ- CCAGATTCTGCTCACTCAGTGAG-3ʹ; NR4A1: forward 5ʹ- GGACAACGCTTCATGCCAGCAT-3ʹ and NR4A1: reverse 5ʹ- CCTTGTTAGCCAGGCAGATGTAC-3ʹ.
Protein extraction and western immunoblotting
For protein extraction, CRC cells, dissociated organoids as well as chopped xenograft fragments (~1 mm3 in size) were incubated with a mixture of RIPA lysate buffer (Thermo Fisher Scientific) and protease inhibitors (Thermo Fisher Scientific) on ice for 30 min followed by centrifugation (12 000 × g, 4°C, 15 min). The protein samples were then diluted in Laemmli’s buffer (Bio-Rad, Hercules, CA) containing β-mercaptoethanol (Bio-Rad) and boiled for 10 min. Prepared protein samples were aliquoted and stored at −20°C for subsequent western blot analysis.
For western blot analysis, the protein samples were separated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis, then transferred to polyvinylidene fluoride membranes (Cytiva, Marlborough, MA). After blocked non-specific binding in 5% bovine serum albumin (Sigma-Aldrich) for 1 h, the membranes were incubated with the primary antibodies overnight at 4°C, which are listed as follows: anti-β-actin (#A2228, Sigma-Aldrich), anti-LC3B (#3868, Cell Signaling Technology), anti-NR4A1 (#ab109180, Abcam), anti-CTSL (#71298, Cell Signaling Technology) and anti-Atg12 (#4180, Cell Signaling Technology). After being washed by PBS with 1% Tween-20 three times, the membranes were incubated with horseradish-peroxidase-conjugated secondary antibodies (#7074 or #7076, CST) at room temperature for 1 h. ECL chemiluminescence kit (Thermo Fisher Scientific) was used to detect the signals of protein strip in Gel Imaging Systems (Bio-Rad). Image J software was used to evaluate the relative expression level of each target protein normalized to β-actin.
Statistical analysis
The statistical analyses were performed using GraphPad Prism 8.0 (GraphPad Software, Inc., La Jolla, CA). Chou–Talalay combination index (CI) values were calculated using the CompuSyn software (CompuSyn Inc., Paramus, NJ). All results were presented as mean ± SD of three independent experiments, each of them performed in triplicate. Comparisons between groups were performed by Student t-test and multiple comparisons were analyzed by one-way analysis of variance test. P-value <0.05 was considered statistically significant.
Results
MLT inhibits cell viability and induces cell apoptosis in a dose-dependent manner in CRC cells
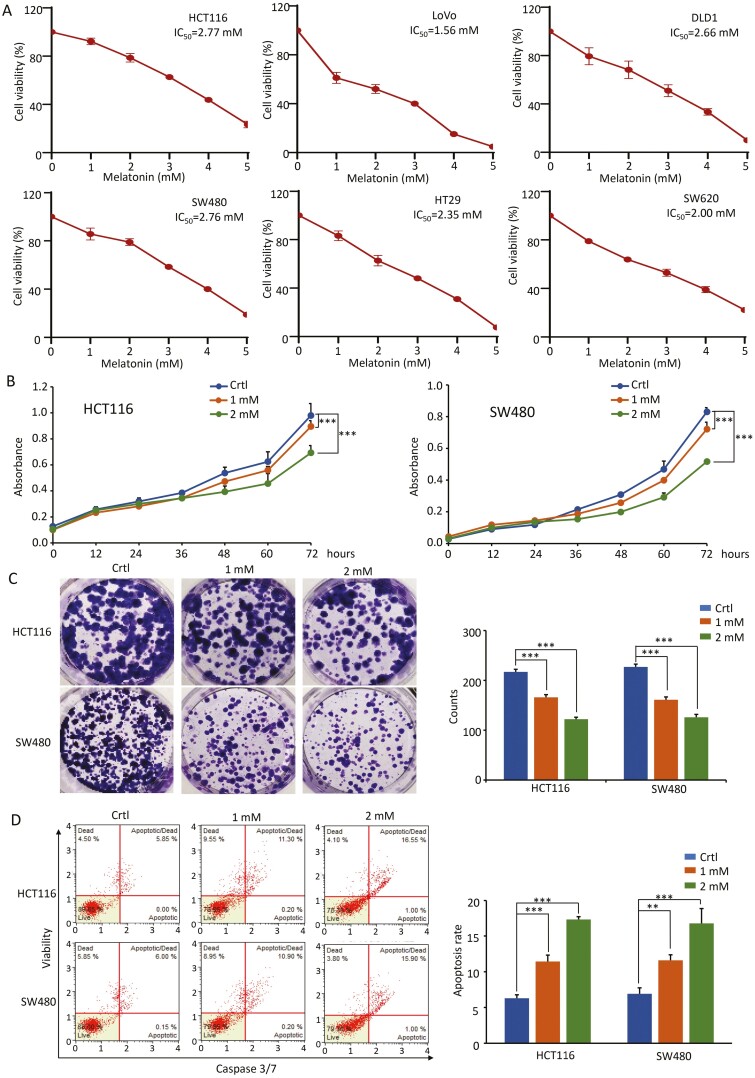
To explore the anticancer effects of MLT on CRC cells, the present study first investigated its effect on cell viability in six human CRC cell lines (HCT116, LoVo, DLD1, SW480, HT29, RKO and SW620). The CCK-8 viability assay was performed to measure IC50 values in different CRC cells following cell culture in the presence of MLT (0–5 mM) for 48 h. The results showed that treatment with MLT resulted in a dose-dependent inhibition of cell viability in all six CRC cell lines, with similar IC50 values ranging from 1.56 to 2.77 mM (Figure 1A). Representative cell lines including HCT116 (with microsatellite instability) and SW480 (with microsatellite stable phenotype) were selected for subsequent investigation for functional studies. To confirm the anti-proliferative effects of MLT, an independent CCK-8 assay was performed to evaluate the growth curves in HCT116 and SW480, using even lower concentrations of MLT. The obtained results revealed that MLT even at relatively low concentrations inhibited the proliferation of HCT116 and SW480 cells in a time-dependent manner (P < 0.001; Figure 1B).
Figure 1.
MLT inhibits the growth of CRC cells in a dose-dependent manner. (A) The cell viability of six CRC cell lines treated with different concentrations of MLT (1–5 mM) for 48 h was determined by CCK-8 assay. (B) Results of CCK-8 assay showing the effect of different MLT concentrations (1 mM, 2 mM) on CRC cell viability. Complete medium containing MLT was added at 24 h and the absorbance was measured at 450 nm after cell culture for 0, 12, 24, 36, 48, 60 and 72 h. (C) Representative results of colony formation assay in CRC cells after exposure to MLT (1 mM, 2 mM) for 7 days. Colony formation count analysis is shown in the right panel. (D) Representative results of FACS analysis using the Muse Caspase-3/7 Kit after treatment on CRC cells by MLT (1 mM, 2 mM) for 48 h. Apoptosis rate analysis is shown in the right panel. Statistical significance: ∗∗P < 0.01, ∗∗∗P < 0.001.
To assess the kinetics of cell survival during long-term MLT treatment, colony formation assays were performed. It was noted that MLT inhibited the capacity of HCT116 and SW480 cells to form colonies, even at low concentrations (P < 0.001; Figure 1C). Furthermore, caspase-3/7-based cellular apoptosis was analyzed using flow cytometric Muse™ Cell Analyzer to determine whether MLT-mediated growth inhibition involved cell apoptosis. MLT was found to induce cellular apoptosis in a dose-dependent manner in both HCT116 and SW480 cells (P < 0.01; Figure 1D). Taken together, these data suggest the potent anticancer effects of MLT on CRC cells even at low concentrations.
A combined treatment with MLT and Andro results in synergistic anticancer effects in CRC cells
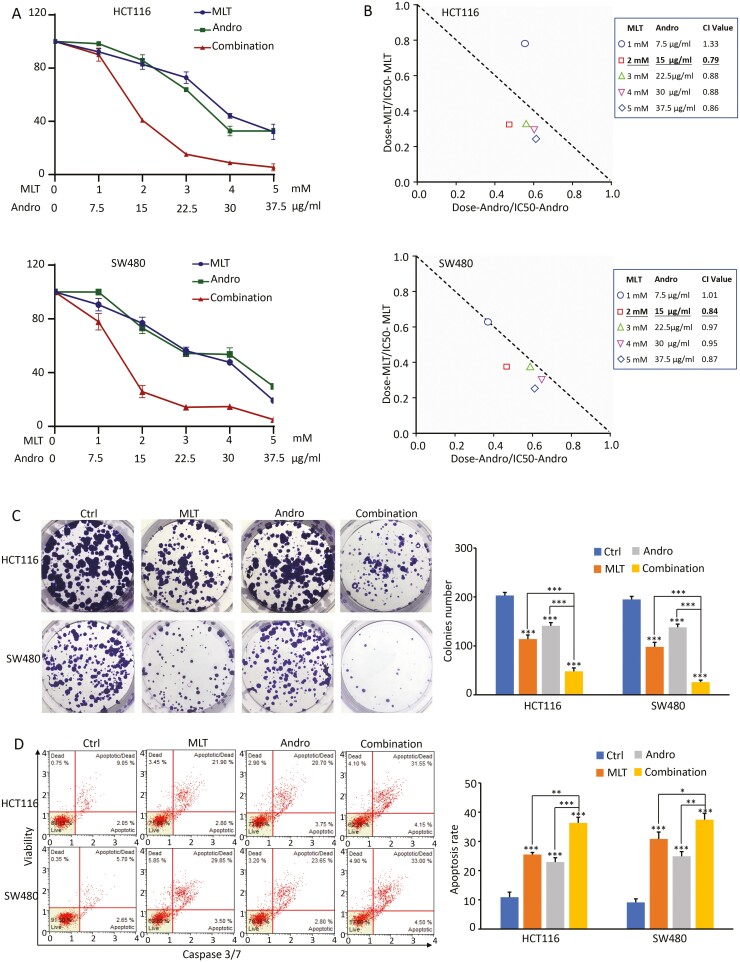
Next, to investigate the combinatorial effect of MLT together with Andro on CRC cell viability, HCT116 and SW480 cells were treated with different concentrations of MLT (1, 2, 3, 4 and 5 mM) and Andro (7.5, 15, 22.5, 30 and 37.5 µg/ml) independently, or in combination for 48 h. As shown in Figure 2A, both MLT and Andro exhibited a dose-dependent cytotoxic effect in CRC cells. To test synergies, the CI values were calculated using the method of Chou and Talalay, which has become a standard approach for analyzing interactions between biological agents, where CI <1, =1 and >1 indicates synergistic, additive and antagonistic effects, respectively (41). The calculations results showed a synergistic effect (CI < 1) between MLT and Andro in both HCT116 and SW480 cells, indicating that Andro sensitized MLT-induced anticancer effects, especially when the concentrations of MLT and Andro were 2 mM and 15 µg/ml, respectively (Figure 2B).
Figure 2.
The combination of MLT and Andro results in synergistic anticancer effects in CRC cells. (A) Results of CCK-8 assay comparing cell viability following treatment with MLT, Andro and their combination for 48 h in CRC cell lines. (B) Isobologram analysis of CI values based on the results of CCK-8 assay to determine synergistic effects of MLT and Andro in CRC cell lines. (C) Representative results of colony formation assay following treatment with MLT, Andro and their combination for 48 h in CRC cell lines. Colony count analysis is shown in the right panel. (D) Representative results of FACS analysis using the Muse Caspase-3/7 Kit after treatment on CRC cells by MLT, Andro and their combination for 48 h. Apoptosis rate analysis is shown in the right panel. Statistical significance: ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
Next, based on the above concentration’s exploration, to validate the above-noticed synergistic effect, we investigated the combinational anticancer effects of MLT (2 mM) and Andro (15 µg/ml) using a colony formation assay and caspase-3/7-based apoptosis assays. Both HCT116 and SW480 cells treated with the MLT or Andro alone drastically inhibited the colony formation capacity (P < 0.001) and exhibited increased apoptosis (P < 0.001) compared with the corresponding control groups. It’s worth noting that the CRC cells treated with the combination of MLT and Andro showed a more effective reduction in colony formation capacity and increase in apoptosis compared with either treatment alone (P < 0.05; Figure 2C and D), indicating a superiority in the anticancer properties of the combination of MLT and Andro versus either compound individually.
Genomewide transcriptomic profiling identifies autophagy-related pathway to be significantly abrogated in response to the combination treatment of MLT and Andro in CRC cells
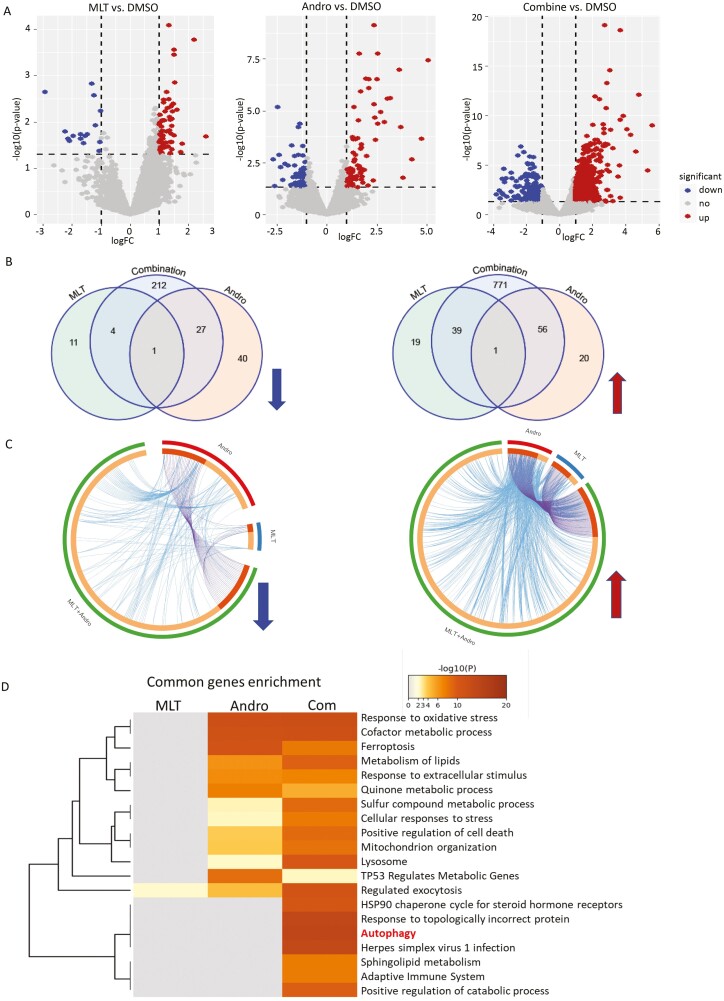
In order to further decipher the potential mechanisms involved in the synergistic antitumor effects of MLT and Andro, we next performed RNA-seq to analyze the transcriptomic profiling in HCT116 and SW480 cells treated with MLT (2 mM), Andro (15 μg/ml) and their combination (Figure 3A). The transcriptomic data revealed that single-agent treatment with MLT and Andro caused significant gene expression changes in 145 genes (68 down-regulated, 77 up-regulated) and 74 (15 down-regulated, 59 up-regulated) genes, in both cell lines, respectively. However, these changes were significantly more pronounced when the cells were treated with the combination of MLT and Andro, wherein we observed that 1111 transcripts (244 down-regulated, 867 up-regulated) were differentially expressed relative to the controls (Figure 3B).
Figure 3.
Genomewide transcriptomic profiling changes in CRC cells following treatment with MLT, Andro and their combination. (A) The volcano plot obtained from genomewide transcriptomic analyses in HCT116 and SW480 cells in each treatment group (MLT, Andro and their combination) relative to vehicle-treated controls. (B) The Venn diagrams representing the overlap of DEGs among various treatment groups. The up- and down-regulated genes were analyzed and represented separately. (C) The circos diagram depicting the gene correlations among different treatment groups in the up- and down-regulated DEGs using Metascape software. The outermost ring depicts different treatment groups of MLT, Andro and their combination. The inner ring represents the genes that are common in multiple groups (darker orange) and that are unique to each group (lighter orange). (D) Comparative heatmap representing the convergence and difference among different treatment groups by biological functions and pathway enrichment using Metascape software.
Venn diagrams and circus plots were thereafter used to describe the numeric and functional association of these transcriptomic changes in different treatment groups. The circus plots depicted the functional connection and overlap between the up-regulated and down-regulated genes in each treatment group (Figure 3C). To further identify these functional interactions based on the above DEGs caused by MLT, Andro and combined treatment, we next performed comparative pathway enrichment analyses using the Kyoto Encyclopedia of Genes and Genomes (KEGG) databases from Metascape (https://metascape.org). If the combination therapy targets a differential pathway than MLT or Andro, the different pathway may be due to drug combination synergy. Comparative pathway enrichment analyses showed that the combination therapy most significantly altered the expression profiles enriched in the autophagy pathway, which was not significantly affected by either compound alone (Figure 3D), hence providing a putative mechanistic based for the synergistic efficacy of the combined treatment with these two compounds together in CRC cells.
Andro sensitizes MLT-induced anticancer effects by promoting autophagy in CRC cells
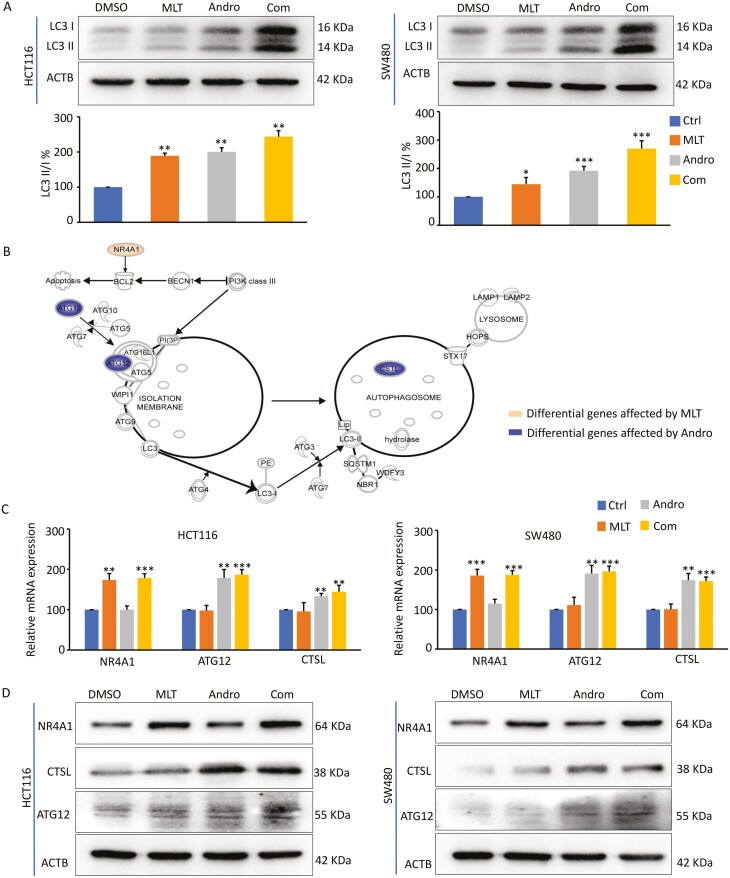
To verify the results of KEGG analyses based on genomewide transcriptomic profiling, we next investigated whether the expression of specific autophagy-associated genes was altered by MLT, Andro or their combination in HCT116 and SW480 cells. In this regard, LC3, an important protein marker of autophagy, was selected for the measurement of autophagy. When autophagy occurs, LC3-I is converted to LC3-II through ubiquitin-like modification; thus, the conversion of LC3-I to LC3-II reflects the progression of autophagy (42). The western blot analyses revealed that while MLT and Andro induced autophagy in CRC cells to some degree, their combined treatment significantly promoted autophagic activity vis-à-vis individual compounds (Figure 4A). In order to explore the potential mechanism(s) of the enhanced autophagy induced by the combination treatment of MLT with Andro, we focused on the expression levels of autophagy-related genes based on transcriptome profiling. Interestingly, according to IPA (Qiagen Inc.), MLT promoted autophagy by up-regulating the expression of NR4A1, while Andro elicited an autophagic response by regulating CTSL and Atg12 (Figure 4B). Both of these compounds promote autophagy by influencing the expression of different genes, respectively, which helps explain, at least in part, their synergistic activity through the autophagy signaling cascade. To further confirm these alterations, subsequently, qPCR and western blot assays were carried out to determine the degree of these changes at both transcript and protein expression levels. Here, we observed that consistent with the results of IPA analysis, both mRNA and protein expression levels of NR4A1, CTSL and Atg12 were significantly increased in CRC cells after the combination treatment with MLT and Andro (Figure 4C and D), supporting the hypothesis that enhanced autophagy is likely one of the molecular mechanisms for the observed synergism for the antitumor activity of MLT and Andro in CRC cells.
Figure 4.
Andro sensitizes MLT-induced anticancer effects by promoting autophagy in CRC cells. (A) Representative images of LC3B expression in the extracts of CRC cells in each treatment group by western blot analysis. The ratios of LC3-II to LC3-I based on the quantification of the bands in the immunoblot were shown in the lower panel. (B) Pattern diagram representing the significantly DEGs in the autophagy pathway after MLT and ANDRO treatment, respectively, by IPA analysis. (C) Bar graph showing relative mRNA expression of NR4A1, ATG12 and CTSL in each treatment group by qPCR analysis. (D) Representative images of NR4A1, ATG12 and CTSL expression in the extracts of CRC cells in each treatment group by western blot analysis. Statistical significance: ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
A combination of MLT and Andro synergistically inhibit tumor growth in a xenograft animal model
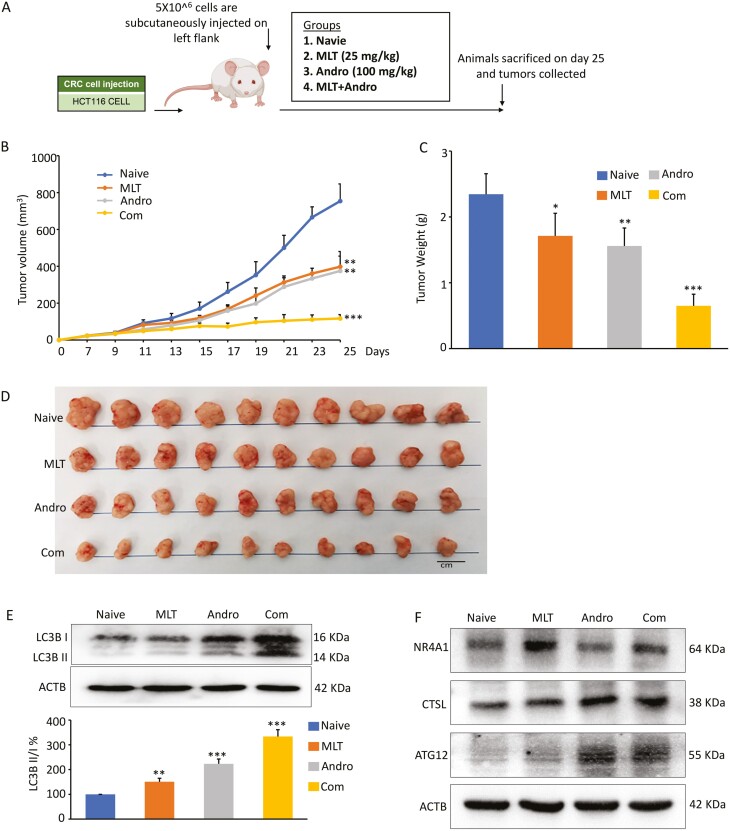
Based on the synergistic anticancer efficacy of MLT and Andro in CRC cells, tumor xenografts transplanted with the HCT-116 cells were used to further evaluate whether similar inhibitory effects of tumor growth could also be observed in a preclinical animal model system. As shown in the study design (Figure 5A), HCT116 xenografts were generated by subcutaneous injection of 5 × 106 cells. Once xenografts could be measured, nude mice carrying xenograft tumors were given appropriate concentrations of MLT, Andro and their combination, and tumor volumes were measured and calculated every other day for a 2-week treatment duration. Finally, nude mice were sacrificed approximately 25 days after subcutaneous injections. The measured results of tumor volumes (Figure 5B) and tumor weight (Figure 5C) revealed that both MLT and Andro treatments significantly inhibited tumor growth compared to the controls, as time progressed, while the combination treatment exhibited the most pronounced antitumor effects. These results once again confirmed that Andro had a dramatically synergistic antitumor effect with MLT even in the xenograft animal model (Figure 5D).
Figure 5.
The combination of MLT and Andro synergistically inhibit xenograft growth in an animal model. (A) Schematic diagram of HCT116 cell-derived xenograft model in nude mice and the treatment schedule of MLT, Andro and their combination. (B) Tumor volume alterations in each treatment group were measured at different time points after inoculation. (C) The measurement of tumor weights within each treatment group 25 days post-inoculation. (D) Representative images of harvested tumors surgically removed from nude mice 25 days post-inoculation. (E) Representative images of LC3B expression in the extracts of xenografts in each treatment group by western blot analysis. The ratios of LC3-II to LC3-I based on the quantification of the bands in the immunoblot were shown in the lower panel. (F) Representative images of NR4A1, ATG12 and CTSL expression in the extracts of xenografts in each treatment group by western blot analysis. Statistical significance: ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
For determining the autophagy levels in vivo, the conversion of LC3-I to LC3-II in the tumor xenografts extracts was also examined. In line with our in vitro findings, we observed that while MLT and Andro increased autophagy, the combination treatment with these two compounds had a much more potent autophagic effect in the animal model as well (Figure 5E). Thereafter, we assessed the expression levels of NR4A1, CTSL and ATG12 in these xenografts and found that MLT and Andro up-regulated the expression of NR4A1, CTSL, and ATG12, respectively, while combination of MLT and Andro exerted a significant increase in the expression of these three autophagy-related genes (Figure 5F). These results were consistent with our cell culture studies and indicate that MLT-induced tumor autophagy was enhanced in combination with Andro in CRC.
Patient-derived tumor organoid model further demonstrated the synergistic anticancer effects of CRC
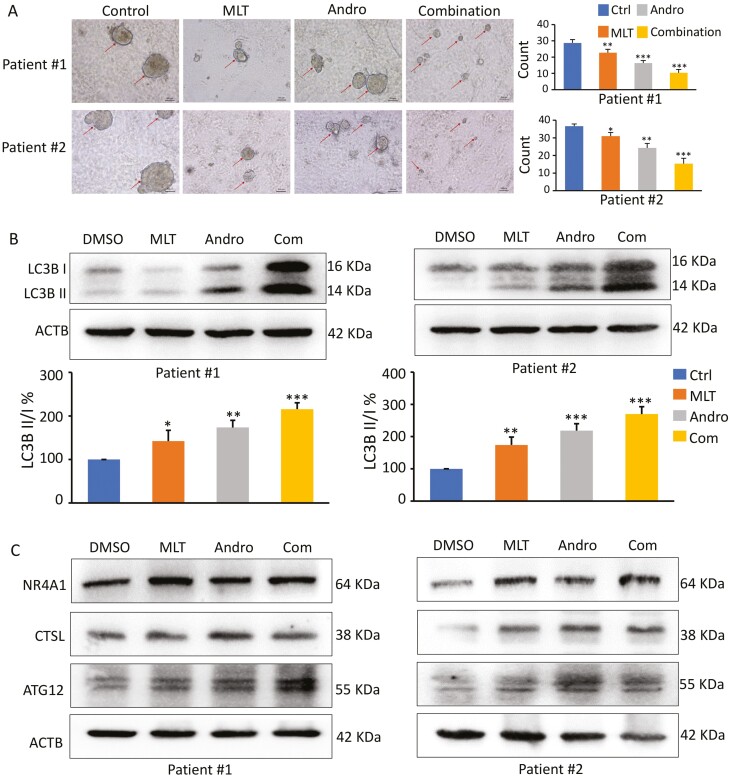
Given that the 3D tumor-derived organoids provide a physiological microenvironment that is closer to humans for determining cellular interactions and responses, it is considered superior to the traditional 2D monolayer cells for evaluating the activity of various anticancer drugs. To further confirm our in vitro and in vivo observations, we generated tumor organoids from CRC patients and examined the synergistic anticancer effect of MLT and Andro. Organoid cultures derived from human CRC specimens were treated with MLT, Andro and their combination for a week. Consistent with the results in cell lines and mice xenografts, the MLT and Andro combination significantly decreased the number and size of patient-derived tumor organoids compared to treatment with these compounds individually (Figure 6A). Furthermore, the expression of a protein marker of autophagy, LC3, was analyzed. Western blot analysis revealed that both MLT and Andro promoted autophagy; however, the combination group exhibited a significantly higher degree of autophagy compared to treatments with individual compounds (Figure 6B). In addition, we also examined the expression of autophagy-related genes, where we observed that expression of NR4A1, CTSL and Atg12 has drastically increased in the tumor organoids with MLT and Andro combination treatment compared to controls (Figure 6C)—once again validating the findings that we observed in cultured cells and the animal model for the combinatorial anticancer activity of these compounds in CRC.
Figure 6.
A combination of MLT and Andro synergistically inhibit patient-derived tumor organoid growth. (A) Microscopic images of patient-derived tumor organoids derived from two patients cultured in each treatment group (20× magnification); the organoid count analysis are shown in the right panel. (B) Representative images of LC3B expression in the extracts of organoids in each treatment group by western blot analysis. The ratios of LC3-II to LC3-I based on the quantification of the bands in the immunoblot were shown in the lower panel. (C) Representative images of NR4A1, ATG12 and CTSL expression in the extracts of organoids in each treatment group by western blot analysis. Statistical significance: ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
Discussion
Although the incidence of CRC in the United States has declined in recent decades, mortality associated with this malignancy still ranks as the second-leading cause of cancer-related deaths with a 5-year survival rate of only ~60% (43,44). For the treatment of this malignancy, surgery and chemotherapy have long been the mainstay as the first choice of treatment (45,46). Currently, the first-line chemotherapy strategies for CRC are considered to be more intensive FOLFOX, FOLFIRI, CAPEOX, and FOLFOXIRI regimens, which comprise of 5-FU, leucovorin calcium, capecitabine plus oxaliplatin/irinotecan (47). Unfortunately, however, their clinical efficacy is somewhat limited due to their toxicity and other adverse effects. Some evidence indicated that a combination of various chemotherapeutic agents (e.g. methotrexate and trimethoprim) may improve treatment outcomes and reduce various side effects, however, the potential for the emergence of a multi-drug resistance phenotype with such agents is a serious concern (48,49). In this regard, encouraging evidence suggests that certain naturally-occurring compounds might offer a safe and cost-effective strategy to target cancer cells and reduce adverse side effects (37,50–52). As reported, 49% of the 175 small-molecule antitumor agents approved by the US Food and Drugs Administration from the 1940s to 2014, were actually either natural products or analogs directly derived from various natural botanicals (53). In this context, natural compounds that target multiple genes and show a synergistic inhibitory effect on tumor growth are more likely to be suitable for combination therapy in CRC. Herein, our present study is another confirmation of this hypothesis that a combination of two natural products, MLT and Andro, exhibited significantly superior anticancer effects in CRC cells, a xenograft animal model, and patient-derived tumor organoids vs. the individual compounds.
Being a natural endogenous body hormone, MLT’s property for synchronizing the circadian rhythms is suggested to improve the sleep quality in cancer patients with insomnia, which is one of several side effects of chemotherapy (54). Moreover, MLT has been of interest for years due to its anticancer efficacies and the underlying molecular mechanisms of action responsible for these effects (15). MLT is known to block multiple epithelial–mesenchymal transition-related pathways for inhibiting migration and invasion in lipopolysaccharide-stimulated and -unstimulated prostate cancer cells (55). MLT has also been reported to sensitize oxidative stress-mediated cancer cell death by inhibiting the SIRT3/SOD2-Akt pathway, which is an established important regulator of mitochondrial redox balance (56). Furthermore, the antitumor activity of MLT, either alone or in combination with other chemotherapy regimens, has been demonstrated. MLT enhances the cytotoxicity and apoptosis of a variety of chemotherapy-induced tumor cells and may be used as a potent synergist in cancer therapy. For example, MLT has been shown to act as an adjuvant agent with rapamycin to improve the efficacy and minimize its side effects for patients with head and neck squamous cell carcinoma (57). In the present study, we demonstrate that a combination of MLT and Andro had a synergistic inhibitory effect on CRC. Furthermore, our data revealed that these two compounds could increase the level of intracellular autophagy by influencing different autophagy-related genes, respectively, so as to achieve synergistic up-regulation of autophagy level when used in combination.
Autophagy is a well-evolved and conserved cell degradation system that plays a critical role in maintaining intracellular homeostasis (58). The function of autophagy is considered a double-edged sword since it varies significantly in distinct cellular contexts. Normally, cells utilize basal levels of autophagy to eliminate or mitigate harmful stimuli to help maintain biological functions and protect cells from damage. Yet, on the contrary, persistent or excessive autophagy can induce autophagy-related cell death to exert a tumor-suppressive effect during tumor therapy (59). The roles of autophagy in the pathobiology of CRC are complex and contradictory, and the underlying mechanism(s) require further investigation. Our present study for the first-time reports that MLT promoted autophagy in CRC cells via up-regulation of NR4A1, and the same phenomena have been further confirmed in a xenograft model as well as patient-derived tumor organoids. NR4A1, a nuclear receptor, was reported to interact with the tumor suppressor p53, in which protein interactions further mediate autophagy-related cell death (60). In addition, we also observed increased autophagic characteristics in CRC cells in response to treatment with these compounds. Furthermore, RNA-seq indicated that Andro triggered autophagy by targeting CTSL and Atg12, both of which have been proven to be autophagy-related regulatory factors. Up-regulation of CTSL has been demonstrated to promote lysosomal membrane permeability leading to autophagy-related cell death (61). Atg12 is a major autophagy gene required for autophagosome synthesis. The human ATG12~ATG5 conjugate is required for LC3 lipidation during autophagy, whereas non-canonical LC3 lipidation has been reported to be positively correlated with human tumor cell death (62,63). Autophagy is an extremely complex process in which multiple genes have to participate in a temporally and spatially coordinated manner. Importantly, we discerned that an enhanced anti-CRC effect based on Andro synergizing MLT-induced autophagy by regulating their respective autophagy-related genes.
In summary, our data reveal a potent and synergistic therapeutic effect of MLT and Andro in the treatment of CRC. The dual activation of autophagy by these two anticancer compounds can effectively trigger CRC cell death and arrest tumor development. These findings provide ample evidence for the combinational targeting of autophagy-related genes as a potential therapeutic strategy in CRC, either on its own or in conjunction with conventional chemotherapy.
Acknowledgements
A.G. and C.W. conceived and designed the whole project. Y.Z. performed the experiments, analyzed the data and wrote the manuscript. Geeta G. Sharma and Souvick Roy performed the RNA-Seq library preparation and sequencing. All authors have read the final manuscript and agreed to its publication.
Conflict of interest statement: None declared.
Glossary
Abbreviations
- 5-FU
5-fluorouracil
- 7-AAD
7-aminoactinomycin D
- Andro
andrographis
- ATCC
American Type Culture Collection
- CCK-8
cell counting kit-8;
- CI
combination index
- CRC
colorectal cancer
- DEGs
differentially expressed genes
- DMSO
dimethyl sulfoxide
- IC50
50% cell growth inhibition
- IPA
Ingenuity Pathway Analysis
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MLT
melatonin
- RNA-seq
RNA-sequencing
- PBS
phosphate-buffered saline
- RPMI
Roswell Park Memorial Institute
Contributor Information
Yinghui Zhao, Department of Molecular Diagnostics and Experimental Therapeutics, Beckman Research Institute of City of Hope, Biomedical Research Center, Monrovia, CA, USA; Department of Clinical Laboratory, The Second Hospital, Cheeloo College of Medicine, Shandong University, Jinan, China.
Chuanxin Wang, Department of Clinical Laboratory, The Second Hospital, Cheeloo College of Medicine, Shandong University, Jinan, China; Shandong Engineering & Technology Research Center for Tumor Marker Detection, Jinan, China; Shandong Provincial Clinical Medicine Research Center for Clinical Laboratory, Jinan, China.
Ajay Goel, Department of Molecular Diagnostics and Experimental Therapeutics, Beckman Research Institute of City of Hope, Biomedical Research Center, Monrovia, CA, USA; City of Hope Comprehensive Cancer Center, Duarte, CA, USA.
Funding
This work was supported by CA184792, CA187956, CA227602, CA072851 and CA202797 grants from the National Cancer Institute, National Institutes of Health.
References
- 1. Siegel, R.L. et al. (2021) Cancer Statistics, 2021. CA. Cancer J. Clin., 71, 7–33. [DOI] [PubMed] [Google Scholar]
- 2. Arnold, M. et al. (2017) Global patterns and trends in colorectal cancer incidence and mortality. Gut, 66, 683–691. [DOI] [PubMed] [Google Scholar]
- 3. Mlecnik, B. et al. (2020) Multicenter International Society for Immunotherapy of Cancer Study of the Consensus Immunoscore for the Prediction of Survival and Response to Chemotherapy in Stage III Colon Cancer. J. Clin. Oncol., 38, 3638–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Modest, D.P. et al. (2018) Surgical treatment options following chemotherapy plus cetuximab or bevacizumab in metastatic colorectal cancer—central evaluation of FIRE-3. Eur. J. Cancer, 88, 77–86. [DOI] [PubMed] [Google Scholar]
- 5. Xie, Y.H. et al. (2020) Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct. Target. Ther., 5, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chiorean, E.G. et al. (2020) Treatment of patients with late-stage colorectal cancer: ASCO Resource-Stratified Guideline. JCO Glob. Oncol., 6, 414–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vodenkova, S. et al. (2020) 5-Fluorouracil and other fluoropyrimidines in colorectal cancer: past, present and future. Pharmacol. Ther., 206, 107447. [DOI] [PubMed] [Google Scholar]
- 8. Madani Tonekaboni, S.A. et al. (2018) Predictive approaches for drug combination discovery in cancer. Brief. Bioinform., 19, 263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nurgali, K. et al. (2018) Editorial: adverse effects of cancer chemotherapy: anything new to improve tolerance and reduce sequelae? Front. Pharmacol., 9, 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhao, Y. et al. (2021) Andrographis overcomes 5-fluorouracil-associated chemoresistance through inhibition of DKK1 in colorectal cancer. Carcinogenesis, 42, 814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang, X.M. et al. (2019) Natural products for treating colorectal cancer: a mechanistic review. Biomed. Pharmacother., 117, 109142. [DOI] [PubMed] [Google Scholar]
- 12. Catalani, E. et al. (2016) Natural products from aquatic eukaryotic microorganisms for cancer therapy: perspectives on anti-tumour properties of ciliate bioactive molecules. Pharmacol. Res., 113(Pt A), 409–420. [DOI] [PubMed] [Google Scholar]
- 13. Ravindranathan, P. et al. (2018) Mechanistic insights into anticancer properties of oligomeric proanthocyanidins from grape seeds in colorectal cancer. Carcinogenesis, 39, 767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ravindranathan, P. et al. (2019) Oligomeric proanthocyanidins (OPCs) from grape seed extract suppress the activity of ABC transporters in overcoming chemoresistance in colorectal cancer cells. Carcinogenesis, 40, 412–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sakatani, A. et al. (2019) Melatonin-mediated downregulation of thymidylate synthase as a novel mechanism for overcoming 5-fluorouracil associated chemoresistance in colorectal cancer cells. Carcinogenesis, 40, 422–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sharma, P. et al. (2020) Andrographis-mediated chemosensitization through activation of ferroptosis and suppression of β-catenin/Wnt-signaling pathways in colorectal cancer. Carcinogenesis, 41, 1385–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Weng, W. et al. (2020) Curcumin and colorectal cancer: an update and current perspective on this natural medicine. Semin. Cancer Biol. doi: 10.1016/j.semcancer.2020.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ma, R. et al. (2021) Antitumor effects of Andrographis via ferroptosis-associated genes in gastric cancer. Oncol. Lett., 22, 523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ravindranathan, P. et al. (2018) A combination of curcumin and oligomeric proanthocyanidins offer superior anti-tumorigenic properties in colorectal cancer. Sci. Rep., 8, 13869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Toden, S. et al. (2018) Oligomeric proanthocyanidins (OPCs) target cancer stem-like cells and suppress tumor organoid formation in colorectal cancer. Sci. Rep., 8, 3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yoshida, K. et al. (2017) Curcumin sensitizes pancreatic cancer cells to gemcitabine by attenuating PRC2 subunit EZH2, and the lncRNA PVT1 expression. Carcinogenesis, 38, 1036–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Toden, S. et al. (2015) Novel evidence for curcumin and Boswellic acid-induced chemoprevention through regulation of miR-34a and miR-27a in colorectal cancer. Cancer Prev. Res. (Phila)., 8, 431–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chandran, B. et al. (2012) A randomized, pilot study to assess the efficacy and safety of curcumin in patients with active rheumatoid arthritis. Phytother. Res., 26, 1719–1725. [DOI] [PubMed] [Google Scholar]
- 24. Goel, A. et al. (2010) Curcumin, the golden spice from Indian saffron, is a chemosensitizer and radiosensitizer for tumors and chemoprotector and radioprotector for normal organs. Nutr. Cancer, 62, 919–930. [DOI] [PubMed] [Google Scholar]
- 25. Goel, A. et al. (2008) Multi-targeted therapy by curcumin: how spicy is it? Mol. Nutr. Food Res., 52, 1010–1030. [DOI] [PubMed] [Google Scholar]
- 26. Buonfiglio, D. et al. (2020) Angelman syndrome and melatonin: what can they teach us about sleep regulation. J. Pineal Res., 69, e12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pham, L. et al. (2021) The interplay between mast cells, pineal gland, and circadian rhythm: links between histamine, melatonin, and inflammatory mediators. J. Pineal Res., 70, e12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lin, P.H. et al. (2020) Melatonin activates cell death programs for the suppression of uterine leiomyoma cell proliferation. J. Pineal Res., 68, e12620. [DOI] [PubMed] [Google Scholar]
- 29. Lee, J.H. et al. (2018) Melatonin and 5-fluorouracil co-suppress colon cancer stem cells by regulating cellular prion protein-Oct4 axis. J. Pineal Res., 65, e12519. [DOI] [PubMed] [Google Scholar]
- 30. Wei, J.Y. et al. (2015) Melatonin induces apoptosis of colorectal cancer cells through HDAC4 nuclear import mediated by CaMKII inactivation. J. Pineal Res., 58, 429–438. [DOI] [PubMed] [Google Scholar]
- 31. Hasan, M. et al. (2020) Novel melatonin, estrogen, and progesterone hormone therapy demonstrates anti-cancer actions in MCF-7 and MDA-MB-231 breast cancer cells. Breast Cancer (Auckl)., 14, 1178223420924634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gurunathan, S. et al. (2020) Melatonin enhances palladium-nanoparticle-induced cytotoxicity and apoptosis in human lung epithelial adenocarcinoma cells A549 and H1229. Antioxidants (Basel), 9, 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tallarida, R.J. (2011) Quantitative methods for assessing drug synergism. Genes Cancer, 2, 1003–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Malyutina, A. et al. (2019) Drug combination sensitivity scoring facilitates the discovery of synergistic and efficacious drug combinations in cancer. PLoS Comput. Biol., 15, e1006752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhou, J. et al. (2012) Andrographolide sensitizes cisplatin-induced apoptosis via suppression of autophagosome-lysosome fusion in human cancer cells. Autophagy, 8, 338–349. [DOI] [PubMed] [Google Scholar]
- 36. Peng, Y. et al. (2018) Andrographolide inhibits breast cancer through suppressing COX-2 expression and angiogenesis via inactivation of p300 signaling and VEGF pathway. J. Exp. Clin. Cancer Res., 37, 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shimura, T. et al. (2021) Enhanced anti-cancer activity of andrographis with oligomeric proanthocyanidins through activation of metabolic and ferroptosis pathways in colorectal cancer. Sci. Rep., 11, 7548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhao, Y. et al. (2021) Andrographis overcomes 5-fluorouracil associated chemoresistance through inhibition of DKK1 in colorectal cancer. Carcinogenesis, 42, 814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang, W. et al. (2016) Andrographolide reversed 5-FU resistance in human colorectal cancer by elevating BAX expression. Biochem. Pharmacol., 121, 8–17. [DOI] [PubMed] [Google Scholar]
- 40. D’Amelio, M. et al. (2010) Neuronal caspase-3 signaling: not only cell death. Cell Death Differ., 17, 1104–1114. [DOI] [PubMed] [Google Scholar]
- 41. Chou, T.C. (2006) Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev., 58, 621–681. [DOI] [PubMed] [Google Scholar]
- 42. Barth, S. et al. (2010) Autophagy: assays and artifacts. J. Pathol., 221, 117–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Siegel, R.L. et al. (2020) Colorectal cancer statistics, 2020. CA Cancer J. Clin., 70, 145–164. [DOI] [PubMed] [Google Scholar]
- 44. Jung, G. et al. (2020) Epigenetics of colorectal cancer: biomarker and therapeutic potential. Nat. Rev. Gastroenterol. Hepatol., 17, 111–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rentsch, M. et al. (2016) Surgery for colorectal cancer—trends, developments, and future perspectives. Visc. Med., 32, 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ruiz-Banobre, J. et al. (2019) Predictive biomarkers in metastatic colorectal cancer: a systematic review. JCO Precis. Oncol., 3, 1–17. doi: 10.1200/PO.18.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Benson, A.B. et al. (2021) Colon cancer, version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J. Natl Compr. Canc. Netw., 19, 329–359. [DOI] [PubMed] [Google Scholar]
- 48. Dik, V.K. et al. (2016) Frequent use of antibiotics is associated with colorectal cancer risk: results of a nested case-control study. Dig. Dis. Sci., 61, 255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jo, Y.K. et al. (2017) Enhancement of the antitumor effect of methotrexate on colorectal cancer cells via lactate calcium salt targeting methionine metabolism. Nutr. Cancer, 69, 663–673. [DOI] [PubMed] [Google Scholar]
- 50. Reuter, S. et al. (2011) Epigenetic changes induced by curcumin and other natural compounds. Genes Nutr., 6, 93–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Goel, A. et al. (2008) Curcumin as ‘Curecumin’: from kitchen to clinic. Biochem. Pharmacol., 75, 787–809. [DOI] [PubMed] [Google Scholar]
- 52. Weng, W. et al. (2015) An update on miRNAs as biological and clinical determinants in colorectal cancer: a bench-to-bedside approach. Future Oncol., 11, 1791–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Newman, D.J. et al. (2016) Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod., 79, 629–661. [DOI] [PubMed] [Google Scholar]
- 54. Kurdi, M.S. et al. (2016) The efficacy of oral melatonin in improving sleep in cancer patients with insomnia: a randomized double-blind placebo-controlled study. Indian J. Palliat. Care, 22, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tian, Q.X. et al. (2021) Melatonin inhibits migration and invasion in LPS-stimulated and -unstimulated prostate cancer cells through blocking multiple EMT-relative pathways. J. Inflamm. Res., 14, 2253–2265. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 56. Li, M. et al. (2020) Melatonin sensitises shikonin-induced cancer cell death mediated by oxidative stress via inhibition of the SIRT3/SOD2-AKT pathway. Redox Biol., 36, 101632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shen, Y.Q. et al. (2018) Combination of melatonin and rapamycin for head and neck cancer therapy: suppression of AKT/mTOR pathway activation, and activation of mitophagy and apoptosis via mitochondrial function regulation. J. Pineal. Res., 64, E12461. doi: 10.1111/jpi.12461. [DOI] [PubMed] [Google Scholar]
- 58. Su, T. et al. (2020) Autophagy: an intracellular degradation pathway regulating plant survival and stress response. Front. Plant Sci., 11, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. White, E. et al. (2009) The double-edged sword of autophagy modulation in cancer. Clin. Cancer Res., 15, 5308–5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bouzas-Rodríguez, J. et al. (2012) The nuclear receptor NR4A1 induces a form of cell death dependent on autophagy in mammalian cells. PLoS One, 7, e46422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Thirusangu, P. et al. (2021) Quinacrine-induced autophagy in ovarian cancer triggers cathepsin-L mediated lysosomal/mitochondrial membrane permeabilization and cell death. Cancers (Basel), 13, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Otomo, C. et al. (2013) Structure of the human ATG12~ATG5 conjugate required for LC3 lipidation in autophagy. Nat. Struct. Mol. Biol., 20, 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gao, Y. et al. (2016) Golgi-associated LC3 lipidation requires V-ATPase in noncanonical autophagy. Cell Death Dis., 7, e2330. [DOI] [PMC free article] [PubMed] [Google Scholar]