Abstract
Background and aims:
Normal gestation involves reprogramming of maternal gut microbiome (GM) that contributes to maternal metabolic changes by unclear mechanisms. This study aimed to understand the mechanistic underpinnings of GM – maternal metabolism interaction.
Methods:
The GM and plasma metabolome of CD1, NIH-Swiss and C57 mice were analyzed using 16S rRNA sequencing and untargeted LC-MS throughout gestation. Pharmacologic and genetic knockout mouse models were used to identify the role of indoleamine-2,3-dioxygenase (IDO1) in pregnancy-associated insulin resistance (IR). Involvement of gestational GM was studied using fecal microbial transplants (FMT).
Results:
Significant variation in gut microbial alpha diversity occurred throughout pregnancy. Enrichment in gut bacterial taxa was mouse strain and pregnancy time-point specific, with species enriched at gestation day 15/19 (G15/19), a point of heightened IR, distinct from those enriched pre- or post-pregnancy. Metabolomics revealed elevated plasma kynurenine at G15/19 in all three mouse strains. IDO1, the rate limiting enzyme for kynurenine production, had increased intestinal expression at G15, which was associated with mild systemic and gut inflammation. Pharmacologic and genetic inhibition of IDO1 inhibited kynurenine levels and reversed pregnancy-associated IR. FMT revealed that IDO1 induction and local kynurenine levels effects on IR derive from the GM in both mouse and human pregnancy.
Conclusions:
GM changes accompanying pregnancy shift IDO1-dependent tryptophan metabolism toward kynurenine production, intestinal inflammation and gestational IR, a phenotype reversed by genetic deletion or inhibition of IDO1.
Keywords: gut microbiome, IDO1, kynurenine, pregnancy
Lay summary:
This study demonstrates that the gestational gut microbiome mediates metabolic adaptations in pregnancy through effects on gut IDO1 activity and the production of kynurenine.
Introduction
Dynamic physiological and metabolic changes occur throughout pregnancy 1. Aberrations in these processes can result in pregnancies complicated by hyperglycemia and/or gestational diabetes mellitus, a harbinger of future diabetes in mothers and metabolic disease in the offspring. In early gestation, the maternal body is in an anabolic state with increased insulin sensitivity promoting adipose lipid stores. In contrast, late gestation is a catabolic state with reduced insulin sensitivity resulting in increased circulating free fatty acids and decreased fasting glucose levels 1. These changes are considered beneficial for growth of the fetus and preparing the maternal body for lactation 1.
Insulin resistance (IR) of normal pregnancies is multifactorial, with poorly understood underlying mechanisms 1. Recently, the composition of the gut microbiome (GM) was shown to be altered during pregnancy and associated with altered metabolic features in mothers 2, 3. Koren et al. 3 showed that the third trimester maternal GM resembles the dysbiotic microbiome of metabolic syndrome and promotes metabolic syndrome-like features in germ-free mice. Whether these changes are pre-requisite to the physiological adaptations during pregnancy is unclear, and there is lack of consensus on the changes in the microbiota of mothers 2–5.
Thus, highly controlled mouse studies can clarify the exact nature of GM changes during pregnancy and their influence on metabolism during pregnancy. Significant strain and vendor differences in the GM populations of mice 6 were leveraged in this study to capture changes in gut microbial and metabolomic features through pregnancy. This approach identified a metabolite, kynurenine, increased during pregnancy and further demonstrate its importance in pregnancy-associated metabolic physiology.
Results
Mouse genetic background influences the metabolic response to pregnancy.
Physiological and metabolic responses of the three mouse strains (C57, CD1, and NIH-Swiss) were, in general, similar during pregnancy1, with early increases in adiposity and circulating glucose, followed by IR and a concomitant reduction in adipose stores to fuel fetal growth and maternal lactation (Fig.S1). However, these data illustrate some strain-specific variations in the specific responses.
Strain specific gut microbial composition changes and phenotypic associations during pregnancy.
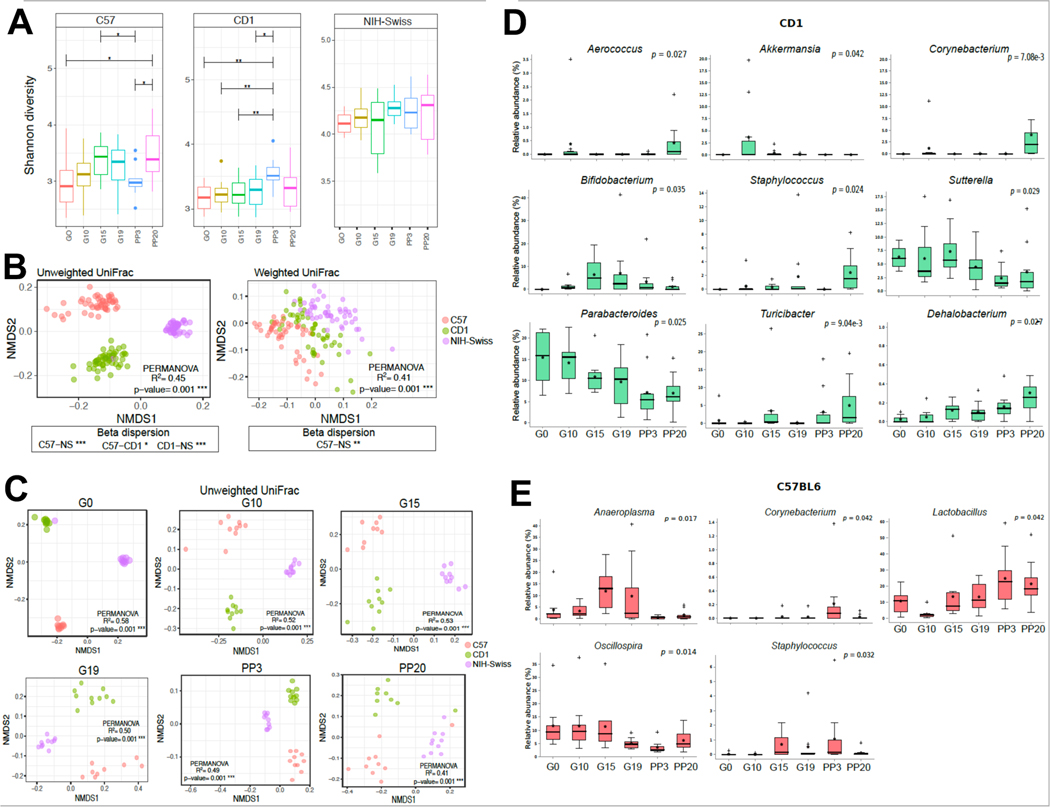
To systematically explore how pregnancy influences the GM, we investigated the fecal microbial communities of the three mouse strains (EMBL-ENA ID PRJEB45047) 5. Alpha diversity, calculated using Shannon index (accounts for richness and evenness), was significantly different between pregnancy stages and between mouse strains (PFDR<.05, non-parametric, Fig.1A) with NIH-Swiss mice being more microbially diverse than CD1 or C57 mice (PFDR<.05). Microbiota differences between mouse strains and across pregnancy stage were calculated by beta diversity using the unweighted and weighted UniFrac distance metrics (Fig. 1B-C, S2A). Arrayed using non-metric multidimensional scaling (NMDS) plots, these data demonstrated significantly differential clustering pattern by mouse strain (PFDR<.05) (Fig.1B), but not by pregnancy time-point (Fig.1C, S2A). As, unweighted UniFrac distance metrics, unlike weighted UniFrac, equally weighs both rare and dominant species, these results suggest that the three mouse strains do not exhibit commonalities in low-relative abundance taxa.
Figure 1. Pregnancy impacts fecal bacterial diversity and select bacterial taxa.
(A) Box plots of alpha diversity (Shannon index); (B-C) NMDS plots based on the weighted and unweighted UniFrac dissimilarity matrix between mouse strains (B) and across pregnancy stages (C); (D-E) multi-group non-parametric analyses of composition of microbiome (ANCOM) followed by Mann-Whitney U test indicating differentially abundant genera at various stages of pregnancy in CD1 (D) and C57 (E) mice. P-values (Benjamini-Hochberg FDR correction) by Kruskal-Wallis (A), permutational multivariate analysis of variance (PERMANOVA) (B-C).
To identify the compositional variations at both genus and Exact Amplicon Sequence Variant (ESV) level between pregnancy phases in each strain, we performed multi-group ANCOM analysis (Fig.1D-E; S2B). In CD1 mice, Aerococcus, Akkermansia, Bifidobacterium, Sutterella, Turicibacter, and Dehalobacterium were significantly enriched in pregnancy; Parabacteroides was most abundant at gestation day (G) 0 and reduced at G15 to postpartum day (PP) 20, and Corynebacterium and Staphylococcus were enriched at PP20 (for all PFDR<.05; Fig.1D). In C57, Lactobacillus increased significantly during gestation while Aneroplasma was significantly more abundant at G15 and G19 compared to other time-points (for both PFDR<.05; Fig.1E). In NIH-Swiss, Candidatus arthromitus peaked at G15 (PFDR<.05) (Fig.S2B). These ESVs when arranged in modules based on co-occurrence exhibited distinct correlations with above parameters (supplemental information, SI; Fig.S2C).
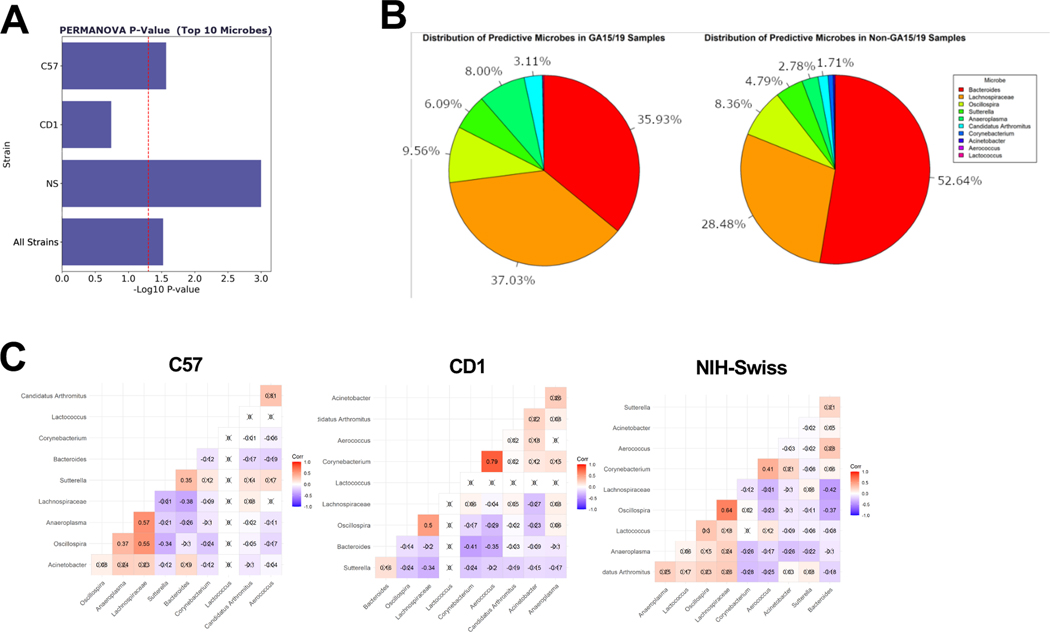
Using K clustering (Fig.S2D, SI), mice were grouped based on the gestational time (either as G15/G19 or non-G15/19 including G0, G10, PP3 and PP20). Meta-Signer, a tool that produces a ranked list of the taxa based on machine learning methods, was used to identify the most discriminative taxa between the two groups 7. Top 10 microbes identified by MetaSigner could discriminate between the G15/19 and non-G15/19 groups as supported by the PERMERNOVA P=.006 (Fig.2A) where Anaeroplasma, Candidatus Arthromitus, Sutterella, and family Lachnospiraceae showed higher average abundance at G15/G19; and Bacteroides, Corynebacterium, Acinetobacter, and Lactococcus showed a lower average abundance (Fig.2B). Additionally, these taxa were found with high level of co-occurrence based on Spearman’s rank correlation coefficients (Fig.2C, P<.05) that was consistent across mouse strains. Interestingly, the full microbe set could also significantly separate the G15/19 and non-G15/19 groups (P=.009) (Fig.S2E). These data corroborate earlier reports on distinctive shift in GM in later pregnancy in both humans and mice 3, 5.
Figure 2. Microbial taxa predict gestational age.
(A) Bar-chart of PERMANOVA P values using the top 10 microbes in each strain and all strains combined together; (B) distributions of the top 10 microbes within G15/19 and non-G15/19 samples; (C) Spearman correlation values between the top 10 microbes’ abundance values within strains.
Plasma metabolome is unique to mouse gestational age and strain
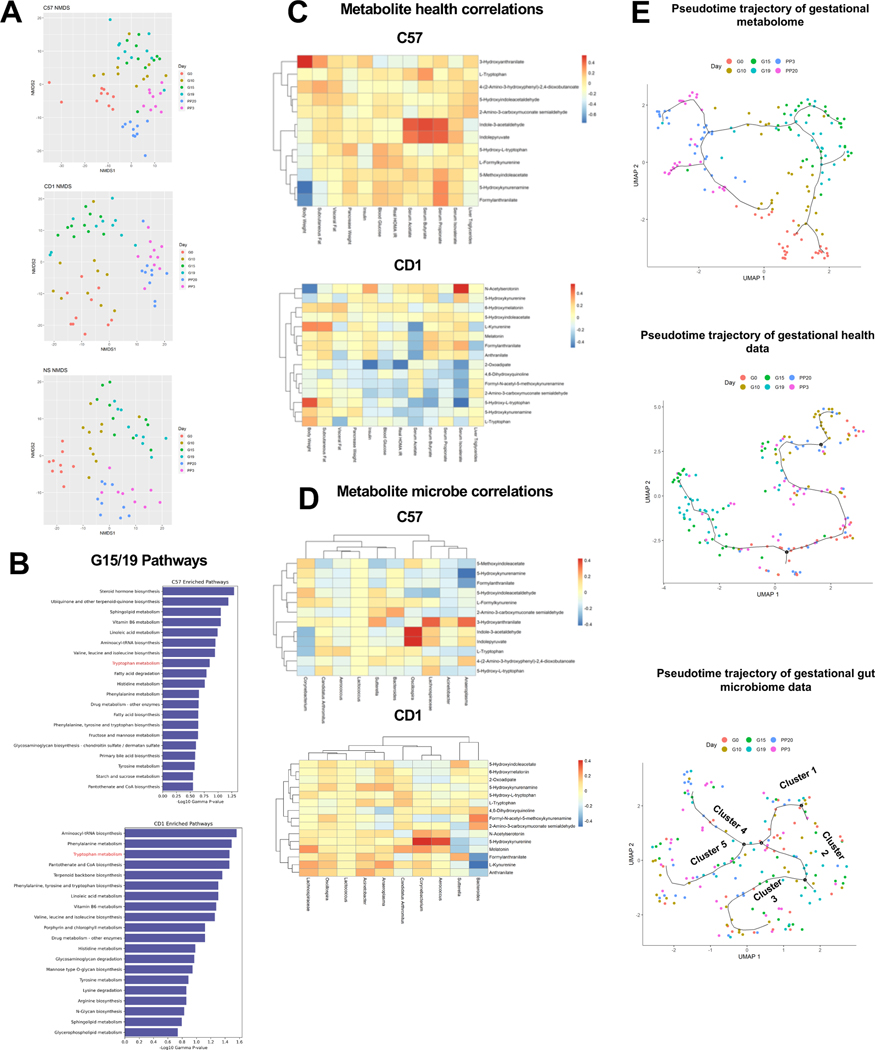
Metabolomic analyses by untargeted LC-MS revealed substantial changes in maternal metabolism in pregnancy, especially in later stages (MetaboLight ID MTBLS3598). Overall, there were 39,323 and 2,636 features in the positive and negative mode datasets, respectively, after filtering to remove features that were present in less than 40% of the samples. First, PERMANOVA analysis identified >700 significantly changed metabolites in NIH-Swiss and CD1 mice (P<.05) and 974 such metabolites in C57 mice (P<.05) during the course of gestation and postpartum (Fig.S3A). Further, even though the metabolome showed strong strain differences at each time-point (Fig.S3B), it was ordered similarly by the gestational state within each strain (Fig.3A). Using MetaboAnalyst 8 and Mummichog algorithm 9, we next identified top pathways significantly altered at G15/19 (Tables S1-3). As expected, several pathways of fatty acid metabolism, amino acid metabolism, steroid hormone biosynthesis and vitamin metabolism were enriched at G15/19. Among the amino acid metabolism pathways, tryptophan metabolism was significantly affected in all three strains (Fig.3B, S3C&F). As the GM and plasma metabolome could order the gestational time-points (Fig.2A; S2E; 3A; S3B), we next determined the association between the metabolomic, gut microbial and health profiles. We observed both common and strain specific associations in plasma metabolites with pregnancy metabolic parameters (Fig.3C; S3D). Tryptophan metabolites like L-kynurenine and its derivatives (e.g. 3-hydroxyanthranilate) correlated with body weight, adipose and HOMA-IR as observed in other human metabolic states 10. We also found previously unknown associations of 5-hydroxykynurenamine, a serotonin antagonist 11, and the melatonin precursor, N-acetylserotonin with pregnancy body weight. With known roles of melatonin and serotonin in energy metabolism and pregnancy 12, 13, these associations suggest additional metabolites involved in the regulation of pregnancy adaptations. Associations were also observed between the top 10 G15/19 predictive microbes and metabolites (Fig.3D, S3E), including between tryptophan metabolites and microbial features associated with obesity, diabetes and inflammation e.g. Lachnospiraceae and Sutterella.
Figure 3. Plasma metabolome strongly clusters by gestational age and mouse strain.
(A) NMDS plots of metabolomic data within each strain colored by gestational state; (B) the top 20 most significantly enriched pathways based on mummichog analyses on the differential metabolites for C57 and CD1; (C) Spearman correlation analyses between the top 10 most significant metabolites from the tryptophan metabolism pathway that were able to be mapped using mummichog candidates and both health parameters and (D) the top 10 microbes identified from Meta-Signer for C57 and CD1 strains; (E) gestational trajectories identified using Monocle3 using UMAP reduction and Louvain clustering.
To measure potential shifts of the metabolic profiles, microbiome, and plasma metabolome during the course of gestation and postpartum, Monocle3 14 was used to visualize potential trajectories based on their profiles over pseudo-time (Fig.3E). The G15/G19 mice were clustered based on the metabolic parameters and metabolomic data, with the latter giving an even tighter cluster, with the following temporal order: G0/G10, G15/G19 and PP3/PP20. The microbiome data, on the other hand, provided a mixed clustering pattern regarding the gestational stages.
The tryptophan metabolite, kynurenine, is elevated during pregnancy
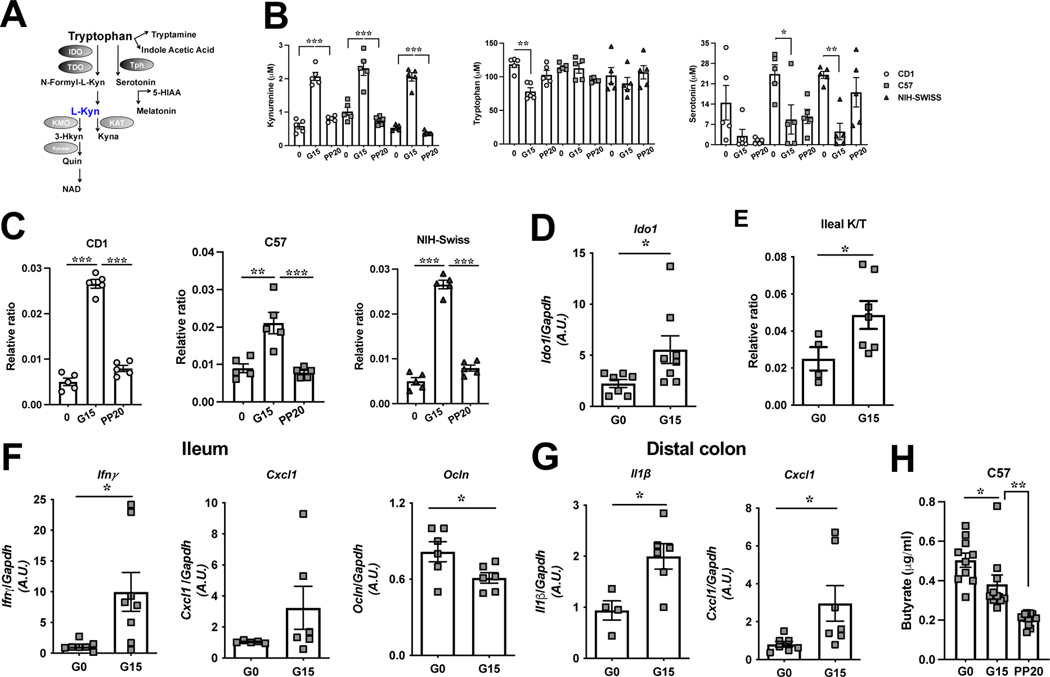
Our untargeted metabolomics data revealed enrichment of the tryptophan catabolic pathway in pregnancy in all three mouse strains. Since tryptophan metabolism is strongly associated with the GM, and to understand the potential metabolic role of pregnancy related tryptophan metabolism, we employed targeted metabolomics to assess this pathway (Fig.4A). Plasma kynurenine was elevated in each mouse strain during the IR phase of pregnancy and returned to pre-gestation levels postpartum (Fig.4B). Serotonin decreased during pregnancy in each mouse line, while tryptophan only decreased in CD1 mice (Fig.4B) and indoleacetic acid remained unchanged (Fig.S4A). With known roles of serotonin in pregnancy induced adaptation of beta cells 13, we assessed these metabolites in the pancreas, observing tryptophan, kynurenine and serotonin to be significantly increased although tryptophan levels remained unaltered in NIH-Swiss mice (Fig.S4B).
Figure 4. The tryptophan metabolite, kynurenine, is elevated during pregnancy.
(A) Schematic of the tryptophan metabolism pathways; (B) plasma kynurenine, tryptophan and serotonin levels and (C) the plasma kynurenine/tryptophan ratio (K/T) in C57, CD1 and NIH-Swiss mice at depicted time-points; (D-F) in the ileum of C57 mice at G0 and G15, Ido1 mRNA expression (D), K/T (E) and mRNA expression of Ifnγ, Cxcl1, and Ocln (F) and (G) mRNA expression of Il1β and Cxcl1 in DC of same mice; (H) plasma butyrate levels in C57 at depicted time-points (n=10 per time-point). Data are mean±SE, n=5–7 per time-point analyzed by one-way ANOVA with Tukey’s post-hoc analysis (B-C), two-tailed Student’s unpaired t-test (D-G), one-way ANOVA (Mixed effects analysis) (H). *P<.05, **P<.01, ***P<.0005. TDO-Tryptophan-2,3dioxygenase; Tph-Tryptophan hydroxylase; KAT-Kynurenine aminotransferase; KMO-Kynurenine-3-Monooxygenase; Kynase-Kynureninase; N-Formyl-L-Kyn-N-Formyl-LKynurenine; L-Kyn-Kynurenine; HIAA-5-Hydroxyindoleacetic acid; Quin-Quinolinic acid; Kyna-Kyneurenic acid; 3-Hkyn-3-Hydroxykynurenine.
Elevated kynurenine suggested increased tryptophan catabolism and flux through the kynurenine pathway; therefore, we computed kynurenine to tryptophan ratio (K/T) in the plasma and the pancreas and found it to be significantly higher in each mouse strain (Fig.4C, Fig.S4D). K/T is reflective of indoleamine-2,3-dioxygenase 1 (IDO1) activity, the rate limiting enzyme of tryptophan degradation in kynurenine pathway which is known to be expressed in immune, adipose, and intestinal cells 15. During pregnancy (G15), Ido1 mRNA expression increased in the ileum of C57 mice (Fig.4D), but not in colon, and trended higher in the ileum and was significantly higher in the colon of CD1 mice (Fig.S4E). Consequent to this increased Ido1 expression, K/T was significantly higher in C57 ileal tissue at G15 (Fig.4E). Subsequent experiments used C57 mice for simplicity and the availability of a IDO1 knockout model on the C57 background.
Low grade inflammation in pregnancy modulates gut IDO1 activity
Normal pregnancy is a state of low-grade inflammation, specifically at gut mucosal surfaces and adipose with a build-up of proinflammatory cytokines 3, 16 and inflammation upregulates IDO1 expression in the intestinal epithelium 17. Thus, we assessed mRNA expression of cytokine genes in G0 and G15 ileum and distal colon (DC) mucosa. At G15, ileal Ifnγ : expression, the main inducer of IDO1 15, was significantly elevated while expression of the pro-inflammatory chemokine, Cxcl1, trended high (Fig.4F). Expression of pro-inflammatory Il1β : and Cxcl1 increased significantly in the DC (Fig.4G). The tight junction proteins of the intestinal epithelium guard the entry across the epithelium and their expression is negatively impacted by gut inflammation 18. Accordingly, expression of occludin (Ocln), a tight junction protein, reduced significantly in G15 ileum (Fig.4F). Additionally, short chain fatty acids (SCFAs) modulate gut IDO1 expression, where butyrate can downregulate IDO1 expression in intestinal epithelial cells 19. Of note, plasma levels of total SCFAs and butyrate declined significantly from G0 to G15 (Fig.S4F; Fig.4H).
IDO1 activity affects pregnancy IR
As kynurenine was specifically elevated at the height of gestational IR, we assessed the role of kynurenine/IDO1 in pregnancy first by inhibiting IDO1 utilizing specific inhibitor L-1Methyltryptophan (L1MT), by supplementing the drinking water 20. L1MT treated mice showed no difference in plasma but reduction in ileal tissue K/T (Fig.5A-B), during pregnancy. No differences in pregnancy body weight gain and offspring number were observed between the control and L1MT treated mice. While L1MT treated mice displayed pre-pregnancy glucose intolerance (Fig.5C) that could reflect indirect effects of L1MT, these mice at G15 exhibited improved glucose tolerance (Fig.5C), reduced IR (Fig.5D) and corresponding increase in insulin signaling (phosphorylated-Akt) in the soleus muscle (Fig.5E, S5A).
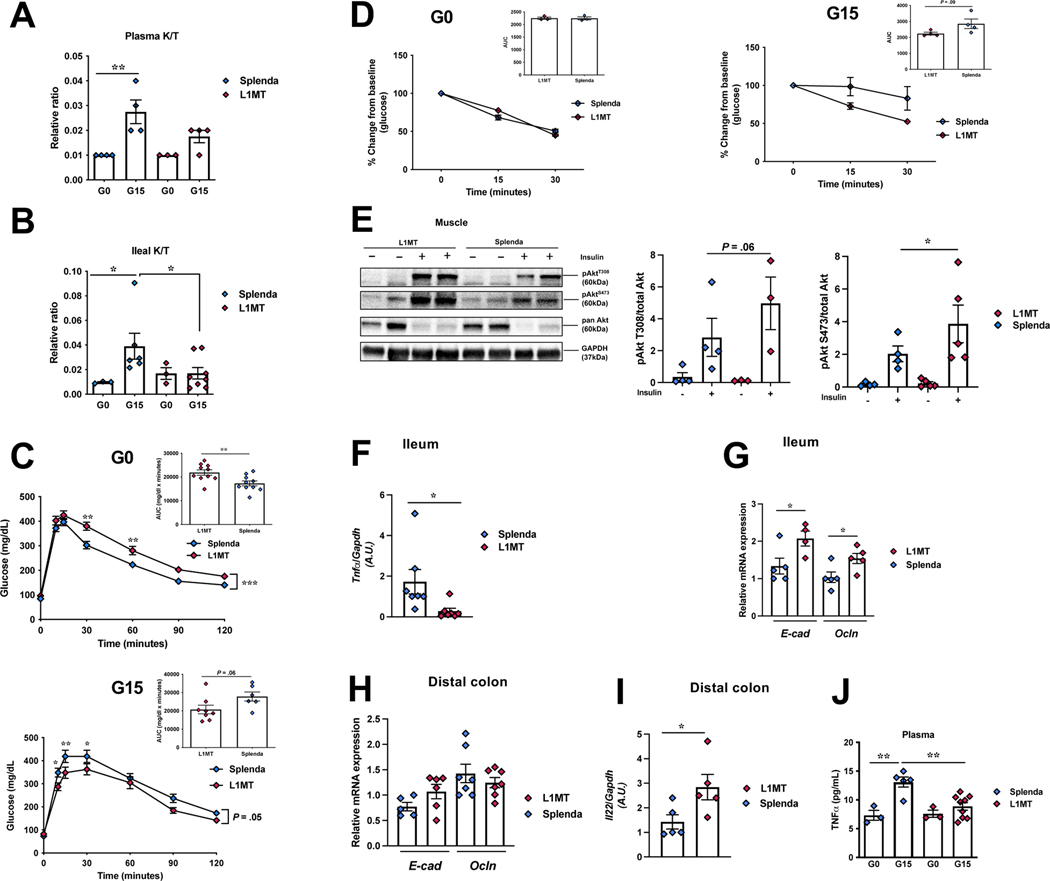
Figure 5. IDO1 activity affects pregnancy induced IR.
1-methyl-L-tryptophan (L1MT) treated and control mice were analyzed for kynurenine/tryptophan ratio (K/T) (A) in the plasma, and (B) in the ileum; (C-D) intraperitoneal glucose (C) and insulin tolerance tests (D), respectively, at G0 (n=3–10) and G15 (n=4–8) (inset, area under curve, AUC); (E) insulin signaling (p-Akt-T308 and p-Akt-S473) in soleus muscle with corresponding densitometric analysis (n=4–5); mRNA expression of (F) Tnfα, (G) E-cad, Ocln in the ileum and (H) E-cad, Ocln, (I) Il22 in the DC, at G15 (n=5–7); (J) plasma Tnfα levels at G0 (n=3) and G15 (n=5–7). Data are mean±SE analyzed by one-way ANOVA with Tukey’s post-hoc analysis (A-B), two-tailed Student’s unpaired t-test (insets C-D, E-H), two-way ANOVA with Tukey’s post-hoc analysis (C, D). *P<.05, **P<.01.
IDO1 activity inhibition partially protected against pregnancy induced gut inflammation, in the ileum, noted by significantly lower expression of pro-inflammatory Tnfα 17, and significant upregulation of barrier proteins, Ocln and E-cadherin (E-cad) and a modest increase in expression of claudin 4 (Cldn 4) in the ileum of L1MT treated mice (Fig.5F-G, S5B). The expression of other tested proinflammatory transcripts showed modest changes (Il1β and/or Il6) in L1MT treated mice (Fig.S5B). While in the DC, no differences were observed proinflammatory cytokines or gut barrier protein expression (Fig.5H; Fig.S5B), expression of the anti-inflammatory cytokine, Il22, was significantly upregulated in L1MT treated mice (Fig.5I). Interestingly, plasma levels of Tnfα, known to be increased in IR phase of pregnancy and obesity, 1 were significantly reduced in L1MT treated mice (Fig.5J). Thus, IDO1 inhibition prevented an increase in the K/T systemically and locally, moderately enhanced gut barrier and reduced IR. However, due to dysregulated glucose tolerance prior to pregnancy (Fig.5C), we used a more definitive model, mice with whole body genetic knockout of IDO1 (IDO-KO).
Genetic IDO1 deficiency protects against pregnancy induced IR
IDO-KO mice were characterized relative to control, C57 mice, prior to gestation and at G15. Contrasting controls, in IDO-KO mice, K/T ratios was markedly suppressed in plasma at G15 and in ileum at G0 and G15 (Fig.6A-B). Compared to controls, IDO-KO pregnant mice displayed significant protection against glucose intolerance (Fig.6C-D) and pregnancy induced IR (Fig.6E) with correspondingly higher insulin signaling (phosphorylated Akt) in adipose (Fig.6F; S5C).
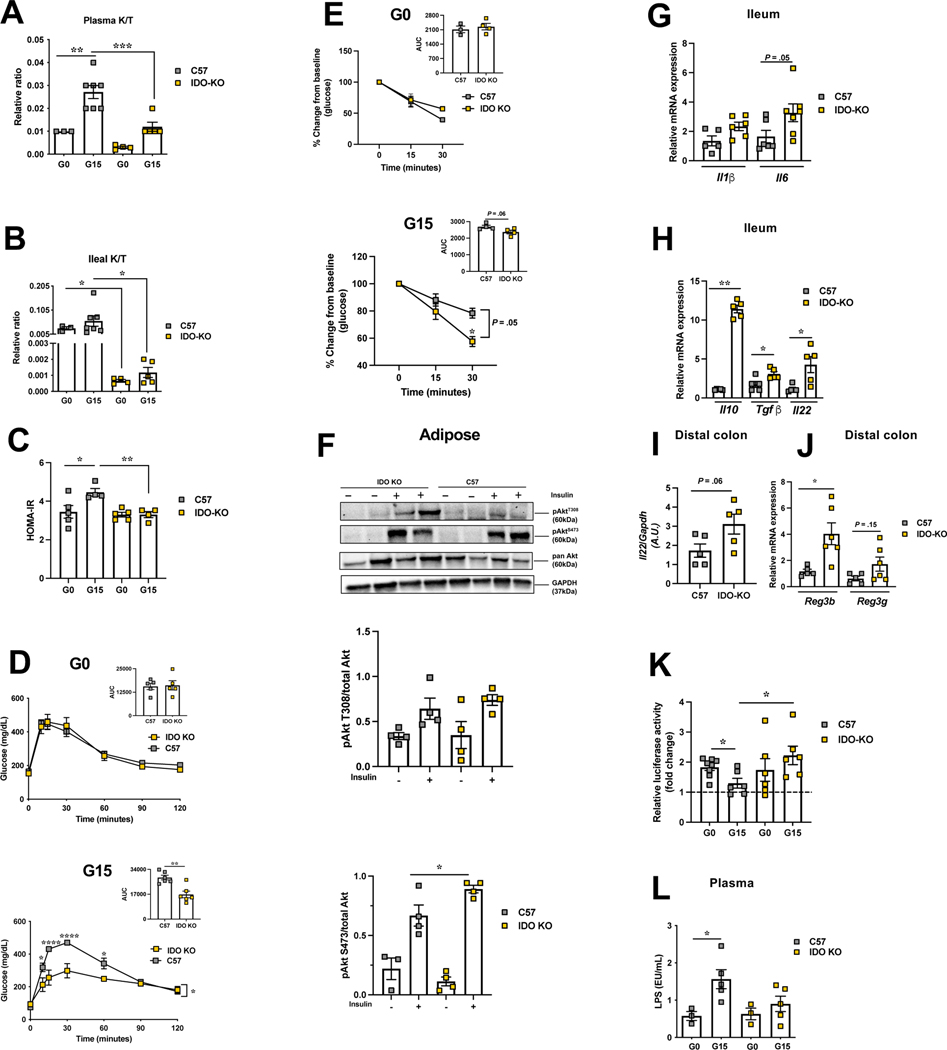
Figure 6. Genetic deficiency of IDO1 protects against pregnancy induced IR.
IDOKO and control mice were analyzed for kynurenine/tryptophan ratio (K/T) in plasma (A) and in ileum (B); (C) HOMA-IR, (D-E) intraperitoneal glucose (D) and insulin tolerance (E) tests, respectively, at G0 (n=4–7) and G15 (n=5–7) (inset, AUC); (F) insulin signaling (p-Akt-T308 and p-Akt-S473) at G15 in subcutaneous adipose with corresponding densitometric analysis (n=4–5); mRNA expression of (G) Il1β, Il6 and (H) Il10, Tgfβ, Il22 in the ileum and of (I) Il22 and (J) Reg3b and Reg3g in the DC at G15 (n= 5–7); (K) fecal AHR activity (n= 6–7); (L) plasma LPS levels at G0 (n=3) and G15 (n=5). Data are mean±SE analyzed by one-way ANOVA with Tukey’s post-hoc analysis (A-B, K), two-tailed Student’s unpaired t-test (insets D-E & F-J), and two-way ANOVA with Tukey’s post-hoc analysis (C-E, L). *P<.05, **P<.01, ****P<.0001.
As well known, IDO1 has immunoregulatory effects 15. No differences, however, were noted in expression of proinflammatory Cxcl1, Ifnγ, or Tnfα transcripts in ileum or DC (data not shown) except increased Il6 in the ileum of IDO-KO mice at G15 (Fig.6G). As the activity of IDO1 sustains an immunostimulatory state by inhibiting IL-10 production 21, we analyzed the expression of anti-inflammatory cytokine, Il10, observing in the ileum of G15-IDO-KO mice 11-fold enhanced expression of Il10 along with upregulation of another anti-inflammatory cytokine, Tgfβ 22 (Fig.6H). Additionally, expression of Il22, a cytokine promoting gut homeostasis and tissue regeneration20 was increased in both ileum and DC of G15-IDO-KO mice (Fig.6H-I) along with increase of Il22 target genes such as antimicrobial proteins, regenerating islet‐derived protein (Reg) 3g and Reg3b in DC of IDO-KO mice (Fig.6J). Il22 is induced by the arylhydrocarbon receptor (AHR) 23. Reduced kynurenine levels in IDO-KO mice and increased expression of AHR targets suggested shift in metabolism of tryptophan away from kynurenine to indole production. Using an AHR reporter system (Fig.6K, S7B) and targeted metabolomics (Fig.S7A), we detected significantly reduced AHR activity and agonist concentration in the feces of C57 mice at G15 than at G0. More importantly, at G15, IDO-KO mice feces had significantly higher AHR activity and agonist concentration than C57 mice (Fig.6K). As AHR agonists are derived from gut microbial activity 20, we analyzed gut microbiome of IDO-KO and control mice pre- and during pregnancy (Fig.S6, SI). At G15, besides beta-diversity differences (unweighted, PERMANOVA, P=.012), IDO-KO mice showed higher abundance of families S24–7, Rikenellaceae and Lactobacillaceae compared to C57 mice (Fig.S6). Thus, a higher fecal AHR activity and ligand concentration and gut gene expression are consistent with the GM composition in G15-IDO-KO mice with a higher abundance of features reported to metabolize tryptophan to AHR ligands 23, 24. Also, levels of beneficial gut microbial metabolite, butyrate, were maintained in pregnant the IDO-KO mice (Fig.S4G) but were lower in C57 G15 mice (Fig.4H). Consequent to the anti-inflammatory milieu in IDOKO gut, plasma lipopolysaccharide (LPS) levels were not elevated at G15 relative to G0, in contrast to control mice (Fig.6L).
Gut microbiome-IDO1 axis mediates pregnancy IR
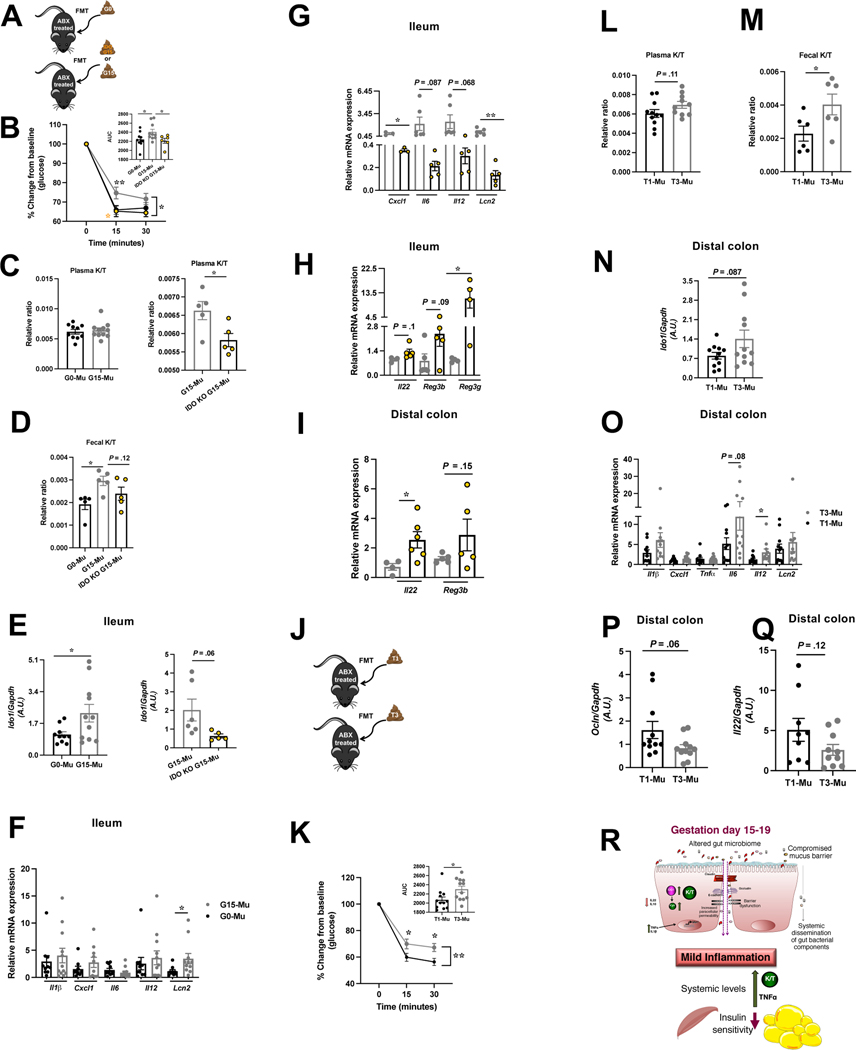
GM can influence kynurenine pathway either by modulating inflammation which impacts IDO1 expression or by affecting tryptophan availability 15. To test the involvement of the GM in IDO1 mediation of pregnancy IR, we explored whether fecal microbial transfer (FMT) from pre-gestation and G15 C57 or IDO-KO mice to antibiotic induced pseudo-germ free (GF) C57 mice (G0-Mu, G15-Mu, IDO-KO G15-Mu) could recapitulate the respective G0 and G15 pregnancy phenotypes (Fig.7A, S7C) where G15, unlike G0, is characterized by increased IR, K/T ratio, gut IDO1 expression and inflammation. While no weight difference emerged (Fig.S7D), G15 recipients, compared to G0 or IDO-KO-G15 recipients, exhibited higher IR, increased fecal K/T (with a trend toward increased ileal K/T) and higher mRNA expression of both Ido1, and the inflammation marker lipocalin 2 (Lcn2) (Fig.7B-F, S7E). Importantly, IDO-KOG15 recipients exhibited significantly lower plasma K/T (Fig.7C) and ileal Lcn2 and Cxcl1 mRNA expression (Fig.7G) but higher expression of Il22 (DC) and Reg3g (ileum) (Fig.7H-I). These data suggest that the G15 GM may be promoting tryptophan metabolism through IDO1 and leading to the IR phenotype whereas IDO-KO-G15 GM increase AHR ligand production. Indeed, G15 recipients showed reduced fecal AHR activity compared to G0 and IDO-KO-G15 recipients (Fig.S7F). Additionally, G15 recipients showed a significant decline in total agonists and the specific AHR agonist, indole (Fig.S7F).
Figure 7. Transfer of pregnancy associated gut microbiota impacts IR, IDO1 levels, and intestinal inflammation.
Schematic of fecal microbial transfer (FMT) to antibiotic (Abx) treated mice (A, J); insulin tolerance test (inset, AUC) (B, K); kynurenine/tryptophan ratio (K/T) in plasma (C, L) and feces (D, M); mRNA expression of ldo1 (E, N), of proinflammatory cytokines and anti-microbial peptide Lcn2 (F-G, O) in the ileum of G0 C57, G15 C57, and G15 IDO-KO and in DC of T1 and T3 recipients, respectively; mRNA expression of Il22 and Reg3b/3g in the ileum (H) and DC (I) of IDO-KO-G15 and of Ocln (P), Il22 (Q) in DC of T1 and T3 recipients. (R) summary: gut microbial change and mild inflammatory milieu at gut mucosal surfaces drives an increase in gut IDO1 expression/activity (K/T) and shift tryptophan metabolism to enhanced kynurenine (kyn) production in IR phase of pregnancy. Data are mean±SE (n=6–12) analyzed by two-tailed Student’s unpaired t-test (insets B and K, C-I, L-Q) and two-way ANOVA with Tukey’s post-hoc analysis (B, K). *P<.05, ** P<0.01.
To identify if a similar mechanism in human pregnancy occurs, we transferred first trimester (T1) and third trimester (T3) fecal microbiota samples to pseudo-GF mice (T1-Mu and T3-Mu) (Fig.7J, S7G). No weight differences emerged but T3 recipients displayed significantly increased IR and fecal K/T (Fig.S7H, 7K-M) and trend toward increased plasma and ileal K/T (Fig.7L, S7I). Analysis of DC of T3 recipients revealed a trend toward higher Ido1 mRNA expression (Fig.7N), and significant differences in gut inflammation evident from significantly elevated Il12 and trending high Lcn2, declined Ocln and Il22 transcripts (Fig.7O-Q). We further identified modulation of AHR axis in human pregnancy. T3 recipients compared to T1 recipients showed reduced fecal AHR activity with significant decline in tryptamine levels and total agonists trending low (Fig.S7J).
Discussion
The GM serves as crucial auxiliary in nutrition acquisition, maintaining gut homeostasis, and immune programming playing important role in host health and disease 25. During pregnancy, GM impacts maternal and fetal health 3, 26. However, GM modulation of maternal adaptations to pregnancy are poorly understood. Through our comprehensive approach, we have discovered a role for kynurenine in gestational metabolism. Our data describes: (1) GM changes, throughout the course of gestation and postpartum across three distinct mouse lines, possibly impacting gut inflammation; (2) these changes impact tryptophan metabolism via modulation of IDO1 expression and activity as indicated by higher K/T, (3) possibly modulates gut microbial AHR ligand production and activity; and (4) the mouse and human gestational GM mediates transferable aspects of these metabolic changes. Through these changes, kynurenine levels are impacted which in turn impact gestational metabolism, most notably IR.
Earlier studies on gestational GM changes in rodents have explored fewer pregnancy time-points 27, 28 and have additional GM modifying components, such as a high fat diet, in the study design, making extrapolation of the results to normal pregnancy less clear 27. Choice of 3 different mouse strains (from different vendors) each with distinct pre-pregnancy microbiomes 5, 6, phenotypic characteristics and propensities for metabolic disorders 29, 30, have allowed us to uniquely capture genotypic and phenotypic diversity of the microbial population cohorts 30. Our GM profiling results recapitulated aspects of previously reported pregnancy-specific GM features during the course of gestation and post-partum including changes in alpha diversity 3 and relatively stable beta diversity 31, with distinctive phyla, and genus level changes at each gestational time-point 3. With progression of pregnancy, a drift towards GM features associated with obesity, IR, gestational diabetes mellitus (GDM) and a proinflammatory states occurs 3, 16. G15/19 stages accompanied similar changes, specifically a rise in the family Lachnospiraceae 20, 30, 32 and the genera: Sutterella from phylum Proteobacteria 33, Staphylococcus from phylum Firmicutes 4, Aneroplasma from phylum Tenericutes, 34 and a decline in Bacteriodes 32. Corroborating earlier findings, transfer of G15 or T3 gut microbes to pseudo-GF mice elicited aspects of gut inflammation and mild IR 3. The gut microbial changes seen during pregnancy, thus, seem to impact maternal metabolic adaptation to pregnancy 4, 26.
The plasma metabolome was extensively remodeled by pregnancy. While expected enrichment of pregnancy signature features at G15/19, additional gut microbial signature metabolites were modulated, for example SCFAs and several tryptophan metabolites. These data indicated associations between plasma metabolome, pregnancy and the GM. We explored one such relationship, of the tryptophan metabolite kynurenine. A small percentage of tryptophan is metabolized directly by the GM for production of indole derivatives 20. Alternatively, the GM can indirectly impact the major arm of tryptophan metabolism catalyzed by IDO1 where gut dysbiosis affects gut IDO1 levels and/or activity, thereby influencing kynurenine levels. Importantly, our data suggest that while the shift toward greater kynurenine levels may impact gestational glucose homeostasis, it may have contributions from reduced AHR axis. Also, butyrate levels associate with and impact insulin sensitivity 25 and may activate AHR pathway and transcription of AHR dependent genes in intestine epithelial cells 24. In our data, a decline in butyrate levels support disturbed intestinal ecosystem at G15 and may represent another factor affecting pregnancy IR.
Current understanding of the contribution of different gut bacteria to plasma metabolites is incomplete. Studies assessing the effect of selective colonization of pregnant mice with defined bacterial populations on plasma features of tryptophan catabolic pathway and other significantly changing metabolites will help deconvolute complex interaction between maternal and gut bacterial metabolism. Nonetheless, our data reveals a crosstalk between gut environment and maternal systemic circulation that uniquely impacts maternal metabolism.
While IDO1 is a well-known immunoregulatory enzyme 15, its activity has only recently been implicated in metabolic disorders like obesity and diabetes 15, 20. The data presented here suggests a role of IDO1 in pregnancy IR (Fig.7R). In the presence of the pregnancy associated proinflammatory milieu, systemically and at gut mucosal surfaces (indicated by the presence of dysbiotic gut bacteria), there is an increase in IDO1 expression and activity that drives the formation of kynurenine, a molecule with multiple effects on peripheral tissues 17, 20, 35 including the development of IR. IDO1 deficiency, in contrast, reduces kynurenine production, promoting an anti-inflammatory gut environment and increased AHR ligand production. Lastly, the role of IDO1 in mediating pregnancy IR is associated, at least partially, with the GM in both mice and human pregnancy.
Future steps include the delineation of specific features of the human GM affecting IDO1 expression/activity and approaches by which those can be modified to minimize this gutinflammation-IDO1 activation, promoting AHR activation and thereby reducing its impact on gestational metabolism. Interestingly, human studies have emerged, although with unclear mechanisms, that probiotics/prebiotics may minimize the development of GDM 36.
Materials and Methods
Experimental animals.
Female C57 (Jackson Laboratory), CD-1 (Charles River Labs), and NIH-Swiss (Envigo) mice (age-10 weeks, n=10/strain/time-point) were individually housed in a temperature and humidity controlled specific pathogen free barrier facility with ad lib access to autoclaved food (Envigo-7912) and water. Following acclimation, pregnancy was induced using strain specific males. Upon pregnancy confirmation, the male was removed to avoid contamination. Feces, cecal material, blood, liver, pancreas, and adipose tissues were collected on gestation days: G0, G10, G15, G19 and post-partum days: PP3, PP20, and were flash-frozen in liquid N2 and stored at −80°C until processed. For separate set of experiments, female C57 mice (age 10–11 weeks) were given 2mg/ml IDO1 inhibitor (1-methyl-L-tryptophan, L1MT; Sigma-Aldrich) in drinking water supplemented with Splenda sweetener (2 sachets/liter) or Splenda alone as before 37 and female IDO1-KO mice (purchased from Jackson Laboratory and maintained in house) were followed during pregnancy.
To create pseudo germ free mice (GF; antibiotic treated) before FMT, C57 female mice (age 10–11 weeks) received penicillin (2000 units/ml) and streptomycin (2000μg/ml) in drinking water for 3 days. GF mice were next colonized (single 200μl gavage of inoculums prepared as before 3, 38) with either de-identified stool samples from T1 and T3 from 2 women (age 18–33 years, BMI 24–26.6), with G0 and G15 fecal samples from 4 C57 mice or G15 fecal samples from 4 IDO1-KO mice collected earlier and monitored for 2 weeks followed by an insulin tolerance test (day 14) and sacrificed at day 15. Human sample collection protocols and mouse studies were approved by the University of Illinois at Chicago (UIC) Institutional Review Board (IRB# 2014–0325) and the IACUC of the Jesse Brown VA Medical Center and UIC, respectively and performed in accordance with the Guide for the Care and Use of Laboratory Animals.
Metabolic assays and measurements
In vivo studies.
Mice were fasted 16h and 6h prior to intraperitoneal glucose (2g/kg) or insulin (1U/kg) injection, for glucose and insulin tolerance tests, respectively. 16h fasted mice received 2U/kg insulin or saline intraperitoneally euthanized 15min later. Protein lysates preparation and immunoblotting, cf. supplementary information.
Analyte measurements:
Plasma Tnfα was assayed by ELISA (Proteintech), LPS by pierce chromogenic endotoxin quant kit (Thermoscientific) and liver triglyceride by reagents from Wako Diagnostics (Richmond). Plasma or fecal samples at different pregnancy time points were assayed via LC-MS/MS using an untargeted approach as before 39 and via a targeted approach for SCFAs 40, tryptophan and indole metabolites. For the latter, homogenized ileal tissues and plasma were deproteinized with methanol. L-kynurenine sulfate was used as an internal standard. LC-MS/MS measurements were performed at the Mass Spectrometry Core facility of the UIC Research Resource Center.
Luciferase assay cf. supplementary information
RNA extraction and qPCR cf. supplementary information
16S rRNA gene sequencing and data analyses
Briefly, bacterial DNA extracted from feces was used for amplification of the V4 region of the 16S rRNA gene (515F-806R). Samples were sequenced on the Illumina MiSeq platform at the Argonne National Laboratory core sequencing facility according to EMP standard protocols (http://www.earthmicrobiome.org/emp-standard-protocols/its/). Sequence processing and analyses were conducted as before 41.
Metabolomics, clustering, metabolite-microbe/health correlations and Monocle3 analyses
Feature detection and basic quantitation of raw LC-MS data was performed using the OpenMS toolkit separately for positive and negative acquisition modes 42. Within each strain, mean imputation was used to replace missing values and for each feature, the mean value at G0 was subtracted from all values in order to normalize each strain to a baseline. The values were then normalized to 0 mean and unit variance within each strain. K-means clustering was used across all strains using 4 clusters. Microbes predictive of the G15/G19 gestational states were identified by Meta-Signer, which is an ensemble of four machine learning models to rank microbes based on their predictive power. The top 10 microbes from the rank list were selected for further analysis.
Before differential analysis of metabolites, values were clipped to a maximum value of 3 times the metabolite’s maximum within each strain to shrink the effect of outlier values. Metabolite values were then log-transformed and scaled to 0 mean and unit variance within each strain. Metabolites not appearing within 20% of any strain were removed. For each strain, PERMANOVA was then used to identify metabolites significantly different between GA15/19 samples and the rest with a P-value cut-off of .05 after Benjamini-Hochberg correction. Significantly enriched pathways for each strain were identified using the mummichog analysis in MetaboAnalyst. The candidate annotated metabolites were mapped back to the untargeted features, selecting the most significantly differential metabolites based on the original PERMANOVA analysis if a candidate mapped to multiple features. Spearman correlations were calculated using the top 10 microbes identified by Meta-Signer, the top 10 most significant annotated metabolites, and the set of health parameters. Benjamini-Hochberg corrected P-values were reported. Monocle3 was used for trajectory analysis for each data type, where the strain specific preprocessed data were grouped into a single set. Monocle3 was run using UMAP dimension reduction and Louvain clustering.
Statistical analyses
Microbial diversity (alpha diversity, based on Shannon and Inverse Simpson indices and beta diversity) was assessed for significance as before 41. The differences in alpha and beta diversity indices were then tested for significance using Kruskal-Wallis and permutational multivariate analysis of variance (PERMANOVA), respectively, with P-values corrected using Benjamini-Hochberg FDR correction. The analyses of composition of microbiome (ANCOM) followed by Mann-Whitney U test was used to identify differentially abundant bacterial ESVs between different groups, i.e. three strains and different pregnancy stages at the P-value cut-off of .05 and Benjamini-Hochberg FDR correction. Weighted correlation network analysis (WGCNA) package in R was used to identify clusters (modules) of significantly correlated ESVs and examine their associations with physiological variables as before 41.
All other data are expressed as means±SE and analyzed by Student’s two tailed unpaired t-tests, one-way ANOVA or two-way ANOVA with post hoc tests (GraphPad Software 9.0) as applicable. P<.05 was considered significant.
[Gestational gut microbiome-IDO1 axis mediates pregnancy insulin resistance. EMBLENA, 2021, https://www.ebi.ac.uk/ena/browser/view/PRJEB45047]
[EMBL-EBI, MetaboLight, www.ebi.ac.uk/metabolights/MTBLS3598, release date:2022–10-11]
Supplementary Material
What You Need to Know:
Background and Context:
Compositional shifts in the gut microbiome during pregnancy contribute to maternal metabolic adaptation though the underlying mechanisms are unclear.
New Findings:
Tryptophan metabolism shifts to enhanced kynurenine production during insulin resistance (IR) in pregnancy owing to increased gut IDO1 expression/activity in response to gut microbial changes and pro-inflammatory milieu at gut mucosal surfaces.
Limitations:
Delineation of immune versus epithelial role of IDO1 in the production of kynurenine in pregnancy is required.
A similar pathway may exist in human pregnancy as third trimester microbiota confer IDO1 dependent insulin resistant phenotype; however, translational human studies are required.
Impact:
Gut IDO1 activity is important for development of IR during pregnancy and might represent a therapeutic target in pregnancies complicated by new-onset diabetes.
Acknowledgments
Grant support: BTL-NIH R01DK104927-01A1, P30DK020595, VA merit 1I01BX003382-01-A1. YD-NVIDIA Corporation donation of Titan-Xp GPU. RKG-NIH R01DK098170. CRM-NIH F30-DK113703. BPB, JG, PMM-R03 HD095056. BPB-Arnold O. Beckman Postdoctoral Fellow Award, BIRCWH Training award (K12 HD101373). GHP-NIH ES028244.
Abbreviations:
- 16S rRNA
16S ribosomal RNA
- AHR
aryl hydrocarbon receptor
- Akt
protein kinase B
- ANCOM
analyses of composition of microbiome
- ANOVA
analysis of variance
- Cldn4
claudin 4
- Cxcl1
C-X-C Motif Chemokine Ligand 1
- DC
distal colon
- E-cad
E-cadherin
- ESV
Exact Amplicon Sequence Variant
- FDR
false discovery rate
- FMT
fecal microbial transfer
- G
gestation day
- GDM
gestational diabetes mellitus
- GM
gut microbiome
- IDO1
indoleamine-2,3-dioxygenase
- IDO-KO
IDO1 knockout
- Ifnγ
interferon γ
- Il
interleukin
- IR
insulin resistance
- K/T
kynurenine to tryptophan ratio
- L1MT
L-1Methyltryptophan
- LC-MS
Liquid chromatography–mass spectrometry
- Lcn2
lipocalin 2
- LPS
lipopolysaccharide
- NMDS
non-metric multidimensional scaling
- Ocln
occludin
- PERMANOVA
permutational multivariate analysis of variance
- PP
postpartum day
- GF
pseudo germ-free
- Reg
regenerating islet‐derived protein
- SCFA
short chain fatty acid
- SE
standard error
- T
trimester
- Tgfβ
transforming growth factor beta 1
- Tnfα
tumor necrosis factor α
Footnotes
Data transparency statement: Data, analytic methods, and study materials will be made available to other researchers upon request.
Conflict of interest: none
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Khan MW, Layden BT. Chapter 6 - Gestational Glucose Metabolism: Focus on the Role and Mechanisms of Insulin Resistance. In: Kovacs CS, Deal CL, eds. Maternal-Fetal and Neonatal Endocrinology: Academic Press, 2020:75–90. [Google Scholar]
- 2.DiGiulio DB, Callahan BJ, McMurdie PJ, et al. Temporal and spatial variation of the human microbiota during pregnancy. Proc Natl Acad Sci USA 2015;112:11060–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koren O, Goodrich JK, Cullender TC, et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell 2012;150:470–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collado MC, Isolauri E, Laitinen K, et al. Distinct composition of gut microbiota during pregnancy in overweight and normal-weight women. Am J Clin Nutr 2008;88:894–9. [DOI] [PubMed] [Google Scholar]
- 5.Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol 2011;9:279–90. [DOI] [PubMed] [Google Scholar]
- 6.Ericsson AC, Davis JW, Spollen W, et al. Effects of vendor and genetic background on the composition of the fecal microbiota of inbred mice. PLoS One 2015;10:e0116704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reiman D, Metwally AA, Sun J, et al. Meta-Signer: Metagenomic Signature Identifier based on Rank Aggregationof Features. bioRxiv 2020:2020.05.09.085993.
- 8.Chong J, Wishart DS, Xia J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr Protoc Bioinformatics 2019;68:e86. [DOI] [PubMed] [Google Scholar]
- 9.Li S, Park Y, Duraisingham S, et al. Predicting network activity from high throughput metabolomics. PLoS Comput Biol 2013;9:e1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christensen MHE, Fadnes DJ, Rost TH, et al. Inflammatory markers, the tryptophan-kynurenine pathway, and vitamin B status after bariatric surgery. PLoS One 2018;13:e0192169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okuma M, Tokuyama T, Senoh S, et al. Antagonism of 5-hydroxykynurenamine against serotonin action on platelet aggregation. Proc Natl Acad Sci USA 1976;73:643–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCarthy R, Jungheim ES, Fay JC, et al. Riding the Rhythm of Melatonin Through Pregnancy to Deliver on Time. Front Endocrinol (Lausanne) 2019;10:616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yabut JM, Crane JD, Green AE, et al. Emerging Roles for Serotonin in Regulating Metabolism: New Implications for an Ancient Molecule. Endocr Rev 2019;40:10921107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao J, Spielmann M, Qiu X, et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 2019;566:496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Platten M, Nollen EAA, Rohrig UF, et al. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat Rev Drug Discov 2019;18:379–401. [DOI] [PubMed] [Google Scholar]
- 16.Edwards SM, Cunningham SA, Dunlop AL, et al. The Maternal Gut Microbiome During Pregnancy. MCN Am J Matern Child Nurs 2017;42:310–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ciorba MA. Indoleamine 2,3 dioxygenase in intestinal disease. Curr Opin Gastroenterol 2013;29:146–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chelakkot C, Ghim J, Ryu SH. Mechanisms regulating intestinal barrier integrity and its pathological implications. Exp Mol Med 2018;50:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin-Gallausiaux C, Larraufie P, Jarry A, et al. Butyrate Produced by Commensal Bacteria Down-Regulates Indolamine 2,3-Dioxygenase 1 (IDO-1) Expression via a Dual Mechanism in Human Intestinal Epithelial Cells. Front Immunol 2018;9:2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laurans L, Venteclef N, Haddad Y, et al. Genetic deficiency of indoleamine 2,3dioxygenase promotes gut microbiota-mediated metabolic health. Nat Med 2018;24:1113–1120. [DOI] [PubMed] [Google Scholar]
- 21.Metghalchi S, Ponnuswamy P, Simon T, et al. Indoleamine 2,3-Dioxygenase FineTunes Immune Homeostasis in Atherosclerosis and Colitis through Repression of Interleukin-10 Production. Cell Metab 2015;22:460–71. [DOI] [PubMed] [Google Scholar]
- 22.Han G, Li F, Singh TP, et al. The pro-inflammatory role of TGFbeta1: a paradox? Int J Biol Sci 2012;8:228–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamas B, Natividad JM, Sokol H. Aryl hydrocarbon receptor and intestinal immunity. Mucosal Immunol 2018;11:1024–1038. [DOI] [PubMed] [Google Scholar]
- 24.Marinelli L, Martin-Gallausiaux C, Bourhis JM, et al. Identification of the novel role of butyrate as AhR ligand in human intestinal epithelial cells. Sci Rep 2019;9:643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arora T, Tremaroli V. Therapeutic Potential of Butyrate for Treatment of Type 2 Diabetes. Front Endocrinol (Lausanne) 2021;12:761834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang S, Ryan CA, Boyaval P, et al. Maternal Vertical Transmission Affecting Early-life Microbiota Development. Trends Microbiol 2020;28:28–45. [DOI] [PubMed] [Google Scholar]
- 27.Gohir W, Kennedy KM, Wallace JG, et al. High-fat diet intake modulates maternal intestinal adaptations to pregnancy and results in placental hypoxia, as well as altered fetal gut barrier proteins and immune markers. J Physiol 2019;597:30293051. [DOI] [PubMed] [Google Scholar]
- 28.Elderman M, Hugenholtz F, Belzer C, et al. Changes in intestinal gene expression and microbiota composition during late pregnancy are mouse strain dependent. Sci Rep 2018;8:10001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stockli J, Fisher-Wellman KH, Chaudhuri R, et al. Metabolomic analysis of insulin resistance across different mouse strains and diets. J Biol Chem 2017;292:1913519145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujisaka S, Avila-Pacheco J, Soto M, et al. Diet, Genetics, and the Gut Microbiome Drive Dynamic Changes in Plasma Metabolites. Cell Rep 2018;22:3072–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang H, Guo R, Li S, et al. Systematic analysis of gut microbiota in pregnant women and its correlations with individual heterogeneity. NPJ Biofilms Microbiomes 2020;6:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gomez-Arango LF, Barrett HL, McIntyre HD, et al. Connections Between the Gut Microbiome and Metabolic Hormones in Early Pregnancy in Overweight and Obese Women. Diabetes 2016;65:2214–23. [DOI] [PubMed] [Google Scholar]
- 33.Chen T, Zhang Y, Zhang Y, et al. Relationships between gut microbiota, plasma glucose and gestational diabetes mellitus. J Diabetes Investig 2021;12:641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Granado-Serrano AB, Martin-Gari M, Sanchez V, et al. Faecal bacterial and shortchain fatty acids signature in hypercholesterolemia. Sci Rep 2019;9:1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oxenkrug GF. Increased Plasma Levels of Xanthurenic and Kynurenic Acids in Type 2 Diabetes. Mol Neurobiol 2015;52:805–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Callaway LK, McIntyre HD, Barrett HL, et al. Probiotics for the Prevention of Gestational Diabetes Mellitus in Overweight and Obese Women: Findings From the SPRING Double-Blind Randomized Controlled Trial. Diabetes Care 2019;42:364–371. [DOI] [PubMed] [Google Scholar]
- 37.Polyzos KA, Ovchinnikova O, Berg M, et al. Inhibition of indoleamine 2,3dioxygenase promotes vascular inflammation and increases atherosclerosis in Apoe−/− mice. Cardiovasc Res 2015;106:295–302. [DOI] [PubMed] [Google Scholar]
- 38.Dugas LR, Bernabe BP, Priyadarshini M, et al. Decreased microbial cooccurrence network stability and SCFA receptor level correlates with obesity in African-origin women. Sci Rep 2018;8:17135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reutrakul S, Chen H, Chirakalwasan N, et al. Metabolomic profile associated with obstructive sleep apnoea severity in obese pregnant women with gestational diabetes mellitus: A pilot study. J Sleep Res 2021:e13327. [DOI] [PMC free article] [PubMed]
- 40.Barengolts E, Green SJ, Chlipala GE, et al. Predictors of Obesity among Gut Microbiota Biomarkers in African American Men with and without Diabetes. Microorganisms 2019;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Navarro G, Sharma A, Dugas LR, et al. Gut microbial features can predict host phenotype response to protein deficiency. Physiol Rep 2018;6:e13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rost HL, Sachsenberg T, Aiche S, et al. OpenMS: a flexible open-source software platform for mass spectrometry data analysis. Nat Methods 2016;13:741–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.