ABSTRACT
The gastrointestinal ecosystem is formed from interactions between the host, indigenous gut microbiota, and external world. When colonizing the gut, bacteria must overcome barriers imposed by the intestinal environment, such as host immune responses and microbiota-mediated nutrient limitation. Thus, understanding bacterial colonization requires determining how the gut landscape interacts with microbes attempting to establish within the ecosystem. However, the complicated network of interactions between elements of the intestinal environment makes it challenging to uncover emergent properties of the system using only reductionist methods. A systems biology approach, which aims to investigate complex systems by examining the behavior and relationships of all elements of the system, may afford a more holistic perspective of the colonization process. Here, we examine the confluence between the gut landscape and bacterial colonization through the lens of systems biology. We offer an overview of the conceptual and methodological underpinnings of systems biology, followed by a discussion of key elements of the gut ecosystem as they pertain to bacterial establishment and growth. We conclude by reintegrating these elements to guide future comprehensive investigations of the ecosystem in the context of bacterial intestinal colonization.
KEYWORDS: gut microbiota, colonization resistance, systems biology
INTRODUCTION
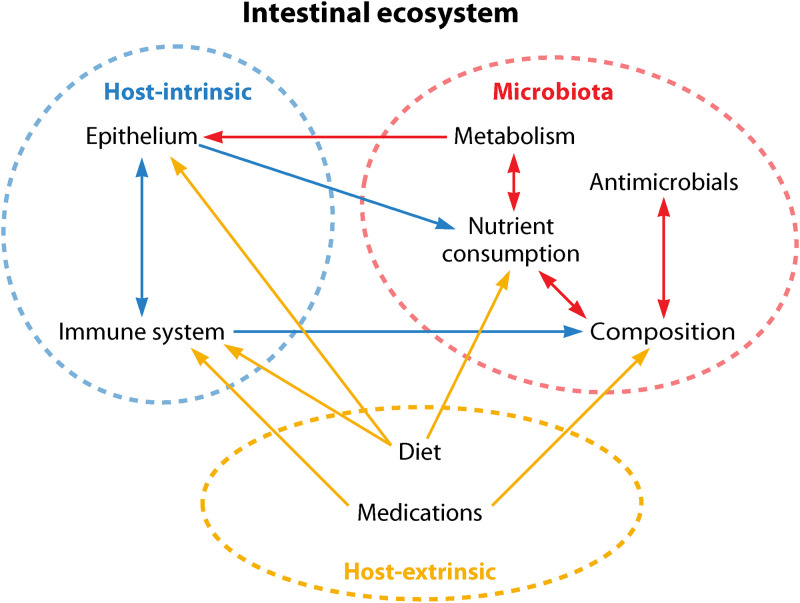
The mammalian gastrointestinal tract is a dynamic ecosystem shaped by interactions between the host, indigenous gut microbiota (the community of microorganisms inhabiting the gut), and external world (Fig. 1). Diverse bacteria continuously attempt to integrate within this ecosystem; while some are harmless and potentially beneficial, others pose a threat to host health. However, regardless of their effects on host well-being, to successfully colonize the gut bacteria must overcome challenges imposed by the intestinal environment, such as host-derived antimicrobial defenses and microbiota-mediated nutrient competition. Understanding bacterial intestinal colonization, therefore, requires uncovering mechanisms by which the gut ecosystem interfaces with microbes attempting to associate with the established community.
FIG 1.
The gut is a complex ecosystem. Factors inherent to the host, microbiota, and external environment interact with others within and between sectors to shape the dynamic intestinal landscape. While there are numerous elements that influence the gut ecosystem beyond those depicted here (e.g., age, pH, oxygen concentrations, lifestyle factors, etc.), we have chosen to highlight only those discussed in this review.
The myriad interactions between elements of the gastrointestinal ecosystem make it difficult to detect emergent properties of the system when studied via reductive methods. As such, a systems biology approach, which broadly seeks to understand complex biological systems by studying their components collectively (1), may be the best way to obtain an integrated perspective on the colonization process. However, the use of holistic experimental frameworks for deciphering the complexity of the gut is still in its infancy.
In this minireview, we explore the relationship between the intestinal environment and bacterial colonization from a systems biology standpoint. We first provide an overview of the theoretical and methodological foundations of systems biology. Then, we “break apart” the gut ecosystem to discuss key elements as they relate to bacterial intestinal colonization. Finally, we recombine these elements to highlight the complexity of the gut landscape and provide a basis for future integrative investigations of this landscape as it pertains to bacterial establishment and growth.
SYSTEMS BIOLOGY: AN OVERVIEW
Systems biology seeks to investigate complex systems, from the inner workings of a single cell to the multifaceted gut landscape, by examining the behavior and relationships of all elements of the system (1). Conceptually, the approach is rooted in the theory of emergence, which posits that the characteristics and functions of complex systems are not entirely deducible from their individual components (2, 3). To that end, emergent properties are those that cannot be assigned to single system elements but stem from the “togetherness” of those elements (3). Colonization resistance is a relevant example, whereby the interplay between the host and microbiota and their interactions with the external environment prevent integration of foreign microbes into the gut ecosystem (4, 5). As such, the permissiveness of the gut is a novel attribute of the system that varies depending on the nature of interactions between its composite parts.
Methodologically, systems biology sits at the crux of technology, computation, and biological experimentation (1, 6). “Omics” technologies have been foundational to the field by allowing researchers to broadly profile the genomic, metabolic, transcriptomic, and proteomic facets of systems in response to specific perturbations (e.g., genetic deletions, disease, introduction of a pathogen, etc.) in biological models (6). In the context of the gut, these models can range in complexity, from simple in vitro cell culture, through germfree animals, to vertebrates with replete microbial communities, including humans. The choice of model depends on the experimental goals and the type of data to be collected; the degree of model complexity depends on the components of the system to be analyzed (6). Thus, it can be beneficial to use models of various complexities to gain diverse insights into the functions of system elements.
Once generated via omics methods, high-throughput data are integrated using computational modeling to visualize and explore complex phenomena (6). While beyond the scope of this review, there are multiple discussions of the different modeling schemes for systems biology (6–9). Ultimately, constructing and refining a model is an iterative process, one which yields new hypotheses that can be experimentally tested and validated. The results of these experiments produce data that seed new questions to promote model refinement, hypothesis generation, biological experimentation, and subsequent data integration and modeling, thus highlighting the repetitious nature of systems biology (1).
Because of its emphasis on holistic, interactions-based research, systems biology is often viewed in direct opposition to reductionist approaches that have historically dominated biological research (1, 6). However, reductionist methodologies are not distinct from, but part of, holistic experimental frameworks. Indeed, prior to integration of a system, it is necessary to define its components and characterize the functions and responses of those components by methodically perturbing and monitoring the system, as outlined above (1). These steps lie at the heart of reductionist science, which emphasizes dissecting parts of a system to understand its workings as a whole; systems biology builds on this framework by elucidating relationships between those parts and characterizing properties resulting from their interactions (6, 10). This synergy of holistic and reductive methodologies under the systems biology umbrella is perhaps best summarized by Francis Crick, who stated that “while the whole [system] may not be the sum of the separate parts, its behavior can, at least in principle, be understood from the nature and behavior of its parts plus the knowledge of how all those parts interact” (11).
“BREAKING APART” THE GUT ECOSYSTEM
As discussed, systems biology is defined by iterative cycles of experimentation and modeling. The approach involves delineating a system’s components and then combining those components to describe the system at large. To the first point, a great deal of work has been done to understand specific gut environmental factors derived from the host, microbiota, and external world that modulate bacterial intestinal colonization. We highlight some of these factors below, focusing on those that are well established and backed by strong experimental evidence. Such knowledge provides a basis for holistic investigations of the ecosystem moving forward.
HOST-INTRINSIC FACTORS
The host is traditionally viewed as providing the structural and biochemical bases of the intestinal ecosystem; the epithelium and immune system are two such host-intrinsic pillars of the gut environment (Fig. 1). These factors directly and indirectly interface with microbes to promote or inhibit their growth. Essentially, they serve as filters that actively select for the integration of certain microbes into the gut ecosystem while excluding others.
The intestinal epithelium.
The intestinal epithelium is a single layer of diverse cell types lining the gastrointestinal tract that exhibit unique functions, such as mucus secretion and production of antimicrobial compound or hormones (12). The epithelium forms a dynamic barrier between the lumen and host circulation that allows selective passage of macromolecules across the intestinal wall (13). Epithelial secretion of antimicrobials, including antimicrobial peptides like defensins and cathelicidins, proteins like lysozyme and calprotectin, and C-type lectins such as RegIII-γ, prevent bacteria from traversing this barrier (14, 15). Moreover, these compounds influence which bacteria will survive and thrive within the gut (16, 17). For example, epithelial-derived antimicrobials inhibit intestinal colonization and survival of Salmonella enterica serovar Typhimurium and Listeria monocytogenes (18, 19). Lysozyme deficiency in mice promotes expansion of lysozyme-sensitive, mucus-degrading bacteria, such as Ruminococcus gnavus and Akkermansia muciniphila (20), indicating that lysozyme contributes to the host barrier against these species. Similarly, RegIII-γ-deficient mice have more bacteria directly associated with their small intestinal epithelium than wild-type animals, demonstrating that RegIII-γ promotes physical separation between luminal bacteria and the epithelial surface (15).
The epithelium also offers spatial and nutritional niches that support microbial growth. For instance, mucus secreted by the epithelium forms a scaffold for bacterial adherence (21), provides nutrients for microbial consumption, contributes to spatial organization of bacterial communities (e.g., by impacting transport of nutrients throughout the gut), and provides regulatory signals that influence bacterial behavior and survival within the gut landscape, such as biofilm formation (22). Thus, the epithelium serves as a framework on which bacterial populations are built and regulated.
The immune system.
Innate and adaptive immune cells reside below the epithelial surface in the lamina propria, as well as in Peyer’s patches and mesenteric lymph nodes embedded within the epithelium, where they sample and engage with intestinal bacteria (13, 23). Host secretion of cytokines, antibodies, and antimicrobial compounds determines how bacteriologically hostile the gut is, and which microbes are targeted, at a given time (24). For example, immunoglobulin A (IgA) secreted by plasma cells is abundant at mucosal surfaces and is integral in regulating the composition of gut bacterial populations (25). IgA can bind bacteria to limit motility and invasion, as well as aggregate microbes to promote their elimination from the gut (25, 26). On the other hand, IgA can promote colonization of bacteria, such as Bacteroides fragilis, by helping them anchor to the epithelial surface (27). Whether the immune system tolerates or inhibits bacteria depends on the microbe, and how the host distinguishes between friend and foe is not entirely understood. Indeed, a misguided immune attack on symbiotic organisms is a well-recognized feature of chronic intestinal diseases like inflammatory bowel disease (28).
Beyond direct interactions between bacteria and immune cells or their secretory products, immune responses alter the nutritional landscape of the gut to make it hospitable or hostile to specific microbes. For example, pathogens like S. Typhimurium, Vibrio cholerae, and Clostridioides difficile capitalize on nutrients liberated from host cells during infection-associated inflammation to occupy a niche that is absent under homeostatic conditions (29–32). However, intestinal inflammation can also limit concentrations of bacterially coveted micronutrients, such as zinc, thereby preventing colonization and infection (33). In addition to metabolic modifications, inflammation increases intestinal oxygen concentrations, which promote expansion of aerobic Enterobacteriaceae species while inhibiting growth of anaerobic bacteria (34–36).
MICROBIOTA-ASSOCIATED FACTORS
The intestinal environment is densely populated, with nearly 100 trillion microbes inhabiting the gut (37). To establish residence in the gut, bacteria must secure sufficient resources to survive within the intestinal ecosystem. The indigenous gut microbiota determines the relative hostility of the intestinal landscape to invading microbes by creating nutritional niches in the intestine, releasing metabolic by-products that facilitate or inhibit growth of other bacteria, producing signaling molecules that foster communication between microbiota members, and secreting compounds that target and kill microbial competitors. As a result, the ability of bacteria to engraft within the gut is largely regulated by the metabolic and antimicrobial defenses of the established bacterial community.
Microbial nutrient competition, cross-feeding, and production of metabolites.
By sequestering nutrients within the intestine, the indigenous microbiota constitutes a barrier to colonization and growth of adventitious bacteria. The importance of nutrient availability in shaping gut bacterial populations was first recognized by Rolf Freter, whose “nutrient niche” hypothesis posits that populations of gut bacteria are controlled by competition for distinct nutritional niches, and that each particular species is more efficient than others in utilizing one or a few specific substrates (38). The population of a given species, therefore, is regulated by the concentrations of these limiting substrates.
For instance, in one proof-of-concept study, Bacteroides ovatus was engineered to metabolize the marine polysaccharide porphyran, a substrate that is not endogenous to the murine diet, thus equipping this gut bacterium to stably engraft in the gut of porphyrin-fed mice in the presence of a complex microbiota (39). Without porphyran supplementation, the bacteria were excluded from the gut (39). Another study illustrated that three nonpathogenic strains of Escherichia coli collectively eliminated a pathogenic strain (EDL933) from the gut of streptomycin-treated mice, despite exhibiting variable individual success in reducing EDL933 colonization (40). These results suggest that indigenous E. coli strains saturate available niches for invading strains and, in light of Freter’s hypothesis, consume distinct nutrients to coexist in the gut (40, 41). However, such coexistence could also be explained by an alternative theory, known as the Restaurant hypothesis (42, 43), which theorizes that organisms with the same nutritional preferences can coexist within the gut if they reside in spatially distinct biofilms (42, 43). The hypothesis emerged from work postulating that E. coli occupies mixed mucosal biofilms (“restaurants”), where it consumes polysaccharides released by other, namely anaerobic, species (44, 45). The Restaurant hypothesis refines Freter’s theory by adding a spatial dynamic to microbial competition and coexistence within the gut. Both hypotheses, however, highlight the necessity of nutrient accessibility for bacterial intestinal colonization and growth.
In addition to nutrient utilization and competition, the microbiota produces thousands of metabolites that modulate intestinal microbial community composition and behavior. For example, short-chain fatty acids (SCFAs) generated via microbial fermentation of dietary fiber serve as a food source for microbes and support cross-feeding relationships (46–48). SCFAs can also be directly toxic to bacteria by dissociating within and acidifying the intracellular environment (49). As such, SCFAs prevent potentially pathogenic bacteria from colonizing the gut (50). Indeed, butyrate can inhibit growth of C. difficile, and a reduction in SCFA levels in murine models of C. difficile infection is associated with an altered microbiota and susceptibility to infection (51–53).
Bile acids represent another class of microbiota-associated metabolites with well-recognized roles in bacterial intestinal colonization. These compounds disrupt bacterial membrane integrity and induce DNA damage and oxidative stress, among other inhibitory effects (54). Primary bile acids are produced in the liver and conjugated to the amino acids taurine and glycine (54); upon passage into the large intestine, they are metabolized by members of the microbiota via deconjugation and 7α-dehydroxylation. The resulting secondary bile acids are particularly important for inhibiting growth of a range of bacteria, including members of the genera Lactobacillus and Bifidobacteria (55) and C. difficile (56, 57).
In addition to these inhibitory compounds, the microbiota also secretes signaling molecules (i.e., autoinducers) that, via quorum sensing, regulate the density and behavior of intestinal bacterial populations. For instance, Thompson and colleagues demonstrated that treatment with streptomycin alters the microbiota of mice, leading to an enrichment in members of the phylum Bacteroidetes and a decrease in Firmicutes species (58). However, when antibiotic-treated mice were colonized by a strain of E. coli capable of producing high levels of AI-2, an autoinducer that fosters cross-species communication, animals exhibited an increased abundance of Firmicutes and fewer Bacteroidetes species (58). This study points to bacterial communication as an important regulator of microbial cooperation and community structure within the gut ecosystem.
Microbiota production of antimicrobial compounds.
In addition to the aforementioned metabolites, bacteria secrete antimicrobial compounds that directly target and kill other bacteria, thus giving the secreting cell a competitive edge (59, 60). For example, bacterial cells produce peptides called bacteriocins, which come in various sizes and structures, and elicit their bactericidal activity in several ways, including forming pores in target cell membranes and inhibiting DNA, RNA, or protein synthesis (61–66). Bacteriocins are highly prevalent within the microbiota and are secreted by both Gram-positive and Gram-negative bacteria; lactic acid bacteria, including members of the genera Lactobacillus and Enterococcus, are some well-known producers (67). Importantly, bacteriocins can promote resistance to colonization by enteric invaders, like Yersinia enterocolitica, Salmonella enterica, and L. monocytogenes (60, 68, 69).
While some antimicrobial compounds are secreted into the extracellular milieu, others are directly injected into competitors via the type VI secretion system (T6SS), a mechanism employed by both pathogenic and nonpathogenic bacteria (69, 70). Effectors secreted by T6SS include cell wall-degrading enzymes, pore-forming toxins, and nucleases, and gut microbes employ T6SS to effectively colonize the gut. For example, B. fragilis uses a T6SS to deploy toxins that antagonize other Bacteroidales species and create a niche within the intestine (71).
HOST-EXTRINSIC FACTORS
The external world plays an important role in regulating conditions within the gut environment and the bacterial populations that reside there. Factors associated with host lifestyle, such as exercise and smoking, affect intestinal processes like gut transit time and gut microbiota composition and metabolism, which impact bacterial intestinal colonization and proliferation (72, 73). The bacteria present within a host’s surroundings determine which species contact the gut in the first place. For example, mode of delivery at birth (e.g., vaginal birth versus Cesarean section) dictates whether “seeding” gut bacterial populations are primarily maternal or environmental (74). Moreover, the composition of the microbiota varies with age, whereby the community rapidly expands and diversifies from birth through early childhood before reaching a relatively stable state characteristic of adulthood (75, 76). However, as is true for host-intrinsic and microbiota facets of the gut environment, there are several extrinsic factors with paramount roles in modulating which bacteria survive and thrive in the gut. Diet and medications are two examples (Fig. 1).
Diet.
Diet is one of the most important modulators of the intestinal environment; it influences host intestinal physiology and function and controls which bacteria will be “well fed” within the gut. Dietary constituents, such as fiber, protein, fat, vitamins, and sugars, impact host intestinal function in different ways, including by regulating immune responses and the integrity of the epithelial barrier (77). Moreover, diet affects host digestive processes and metabolism. For example, increased bile secretion in mice fed a high-fat diet promotes intestinal colonization by S. Typhimurium, which has higher bile resistance than other gut bacterial colonizers (78). Thus, food can select for bacteria that survive in the potentially hostile metabolic landscape of the intestine.
As discussed above, nutrient availability plays a major role in determining the composition of the gut microbiota. Notably, though microbiota composition is largely stable, daily changes in diet can transiently alter community structure (79, 80). Such changes are partially modulated by the community itself; the microbiota regulate host appetite (81) and may influence dietary choices (82, 83) via the gut-brain axis, highlighting the cross talk between host and microbiota in shaping the intestinal nutritional repertoire. Along these lines, gut microbiota structure fluctuates in response to host circadian rhythms and feeding patterns (84, 85), which likely reflects temporal variations in the nutritional landscape of the gut. As such, diet serves to shape the competitive pressures on the microbial community. For instance, low-fiber diets promote expansion of mucus-degrading bacteria and subsequent susceptibility to mucosal pathogens (86). In contrast, high-fiber diets create a nutritionally permissive environment for bacteria that degrade complex carbohydrates, like those of the genus Bacteroides (e.g., Bacteroides thetaiotaomicron) (87). Interestingly, a diet high in simple sugars (glucose and sucrose) inhibits B. thetaiotaomicron from colonizing the mouse gut (88). These sugars suppress expression of colonization factors needed for B. thetaiotaomicron to become established and persist within the gut, suggesting that dietary components effect bacterial physiology beyond acting as a food source (88).
Drugs: antibiotics and beyond.
Medications and other xenobiotics regulate the composition and function of gut microbial populations. Antibiotics can affect the structure of the intestinal bacterial community by obliterating large swaths of the gut microbiota, thus opening niches for bacteria that might normally be barred from the gut. Nonantibiotic medications also influence bacterial survival and proliferation within the intestine. Drugs, including proton pump inhibitors (PPIs; used to decrease stomach acid), metformin (an antidiabetic), laxatives, and antipsychotics, among others, are associated with structural and functional changes in the gut microbial community (89, 90). PPIs in particular are known for increasing risk for infection by enteric pathogens, like C. difficile (50). While the mechanisms are relatively unclear, it is likely that such drugs modulate the gut microbiota in both indirect and direct ways. For instance, PPI-induced reduction in stomach acid may inadvertently select for bacterial species that are normally suppressed (91). Similarly, drugs that target the host immune response, such as biologic therapies and immunosuppressants, influence the inflammatory (and thus antimicrobial) profile of the gut. Indeed, mice administered common immunosuppressants exhibit alterations in gut microbiota structure, as well as decreased expression of antimicrobial peptides within their small intestine (92). These changes are accompanied by an increase in the abundance of endogenous Enterobacteriaceae and increased susceptibility to colonization by pathogenic E. coli, suggesting that conditions are favorable for survival of these inflammation-associated microbes (92).
In terms of direct modulation, pantoprazole, a PPI, was recently shown to inhibit the in vitro growth of various members of a defined bacterial community isolated from human stool (93). On a broader scale, Maier and colleagues discovered that a range of antipsychotics, antimetabolites, and calcium channel blockers, among other drugs, inhibit the growth of representative gut bacterial colonizers in vitro (94). Moreover, therapeutic drugs can bioaccumulate within gut bacterial cells and alter their metabolism, ultimately leading to changes in community composition via formation of new cross-feeding opportunities (95). Together, these findings support the idea that drugs can alter bacterial community composition beyond their host-associated mechanisms of action.
REINTEGRATING THE GUT ECOSYSTEM
Investigations of host-intrinsic, microbiota, and host-extrinsic factors have yielded essential insight into the elements shaping the structural and functional foundations of the gut ecosystem and their role in bacterial colonization. However, while controlled experimental systems provide the opportunity to study specific aspects of physiology, they miss out on important interactions between system components. Therefore, the challenge in research going forward is how to study specific mechanisms in the context of these complex relationships.
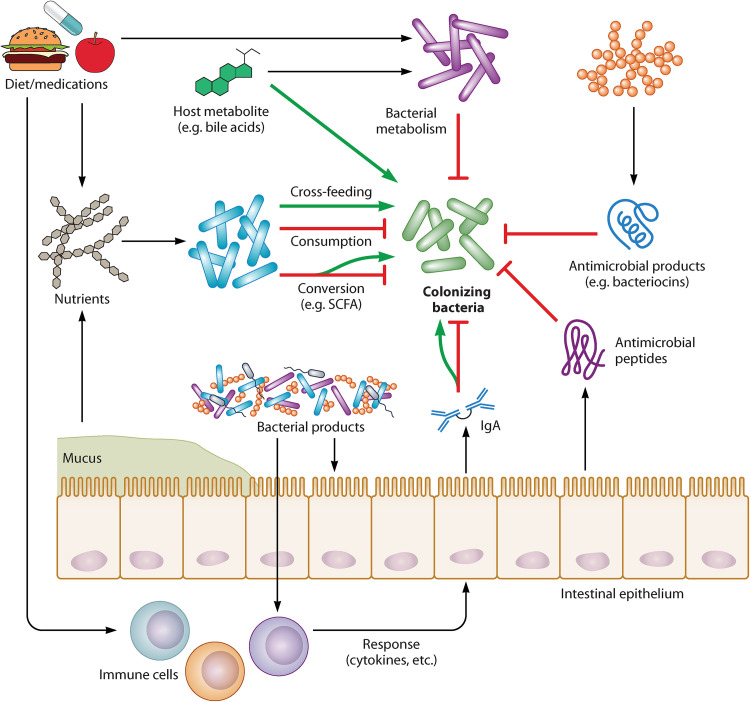
For example, the epithelial barrier continuously interacts with immune cells to modulate their response to intestinal microbes, essentially acting as a portal through which immune-microbe communications are initiated and regulated (96) (Fig. 2). In addition, the gut microbiota regulates epithelial and immune barrier functions, including secretion of mucus, IgA, and antimicrobial peptides, via production of metabolites and other products (97–99) (Fig. 2). The host, in turn, deploys these defenses to shape the composition and metabolic output of the microbiota, which determine the hostility of the metabolic environment for colonizing microbes (Fig. 2). Finally, factors like diet and medications modify host and microbial metabolic and antimicrobial defense mechanisms (97, 98) (Fig. 2); in fact, it is impossible to discuss these factors without accounting for their relationships with the host and microbiota. Furthermore, it is impossible to fully grasp emergent biological phenomena, like colonization resistance, without a holistic view of the gut landscape.
FIG 2.
Mechanisms by which the gut ecosystem modulates bacterial intestinal colonization. All facets of the gut landscape are directly or indirectly linked with the others. The gut microbiota influences the success of bacterial colonizers by regulating nutrient availability, producing metabolites that inhibit bacterial growth, and secreting antimicrobial molecules. Microbiota-associated processes depend on host-extrinsic factors (e.g., diet and medications). Moreover, the microbiota shapes the gut landscape via interactions with key host-intrinsic gut environmental factors, like immune cells and the intestinal epithelium, both of which secrete products like antibodies (e.g., IgA) and antimicrobial peptides and mucus, respectively, that further influence the hostility or permissiveness of the gut. The functions of these host-intrinsic elements are also shaped by interactions with one another, as well as other factors, like diet. Green arrows indicate functions that can support colonization, depending on context.
Due to the development of technologies that allow examination of the gut in all its complexity and nuance, investigators have begun adopting a systems biology approach to studying the intestinal ecosystem. Many studies have centered on using integrative methods to mine the microbiome for features, like specific microbial taxa and metabolites, that associate with particular host outcomes, including diseases like IBD, colorectal cancer, obesity, and type 2 diabetes (100–105). There are studies, however, that have employed such methods specifically to investigate bacterial intestinal colonization (106–112). Indeed, by integrating microbiota and metabolomics data with machine learning, researchers predicted microbial and metabolic features associated with susceptibility to intestinal colonization and persistence of C. difficile in antibiotic-treated mice (110). To this end, multiple studies have taken a modeling approach toward identifying bacterial taxa predictive of C. difficile infection (109, 113) and disentangling how indigenous microbes modulate infection by the pathogen (114). Likewise, Midani and colleagues integrated 16S rRNA sequencing data and computational modeling to predict individuals’ susceptibility to V. cholerae colonization after exposure to cholera patients, based on their microbiotas and other clinical and epidemiologic factors (108). Beyond colonization by individual microbes, investigators have also applied systems biological techniques to understand microbial succession in the intestine (112), as well as to determine factors governing successful engraftment of donor bacteria in the gut of fecal microbiota transplant recipients (111).
Nevertheless, while progress has been made, many studies thus far have been partially integrative, methodologically and in terms of how they view the gut landscape. From a methodological standpoint, omics have been extensively employed to profile elements of the gut landscape and their association with bacteria (e.g., the use of microbiota sequencing to identify taxa correlated with intestinal colonization by diverse bacterial species, such as C. difficile, Salmonella enterica, and Lactobacillus reuteri) (115, 116). However, a relatively small number of studies use these data to construct computational models that form the backbone of systems biology. Those that do, including the examples above, tend to be narrow in their scope of the intestinal landscape by focusing on one or a few facets of the environment (e.g., microbiota composition/metabolic output) while leaving others (e.g., host-intrinsic/host-extrinsic elements) out of the equation.
With this in mind, moving toward a comprehensive understanding of gut microbial colonization will require incorporating elements of the gut ecosystem stemming from the host, microbiota, and the external environment into analyses. Technically, it will require adopting a bona fide systems biology framework. This means going beyond omics to computationally integrate high-throughput data and develop models that take all system elements and their interactions into account. These in silico efforts will be informed by, and inform, experimentation in intestinal model systems, the number and diversity of which continue to increase as technology advances (e.g., development of organotypic intestinal cell culture, gut-on-a-chip technologies, etc.). Benchmarking biological and computational models will be necessary for delineating their strengths and weaknesses in diverse experimental contexts (105). Moreover, collaborations between bench and data scientists will be key for recognizing and meeting the challenges that come with such complex, multifactorial analyses.
CONCLUDING REMARKS
Understanding how bacteria integrate within the gut requires thorough, integrative investigations of the intestinal ecosystem. This can be achieved via a systems biology approach informed by new technologies; biological experimentation; advancements in methods for generating, integrating, and analyzing high-throughput data sets; and the increasingly collaborative nature of biological research. Given the demonstrated role of bacteria in host health, there is interest in using bacteria to bolster health and prevent disease. Ultimately, a holistic perspective of the intestinal environment could facilitate the rational design of strategies to promote colonization by symbiotic organisms and prevent colonization by potentially pathogenic ones.
ACKNOWLEDGMENTS
We thank Dr. Michele Swanson, Dr. Nicole Koropatkin, and Matthew Schnizlein for their critical review of the manuscript during the early stages of its development.
This work was supported by the National Institutes of Health cooperative agreement AI124255 to V.B.Y. and training award AI007528 to M.R.B.
V.B.Y. has served as a consultant to Vedanta Biosciences.
Biographies

Madeline R. Barron, B.S., is a doctoral candidate in the Department of Microbiology and Immunology at the University of Michigan. She is completing her training in the laboratory of Vincent B. Young, where she uses systems biology methods to study mechanisms by which the gastrointestinal environment modulates bacterial colonization of the gut. Outside her research, Madeline is passionate about science communication and plans to pursue a career where she can make scientific research accessible and digestible to diverse audiences.

Vincent B. Young, M.D., Ph.D., is the William Henry Fitzbutler Collegiate Professor in the Department of Internal Medicine/Infectious Diseases Division at the University of Michigan Medical School. Dr. Young has a long-standing interest in understanding the pathogenesis of bacterial infections of the gastrointestinal tract and the role of the indigenous microbiota in human health and disease. As part of the NIH Human Microbiome Project, Dr. Young led a team that studied the role of the microbiome in inflammatory bowel disease. Current research in the Young lab includes systems biology efforts to understand the pathogenesis of Clostridioides difficile infection by an integrated team science approach that combines clinical research, bacterial genomics, microbial ecology, and immunology/host response projects.
Contributor Information
Vincent B. Young, Email: youngvi@umich.edu.
Vanni Bucci, University of Massachusetts Medical School.
REFERENCES
- 1.Ideker T, Galitski T, Hood L. 2001. A new approach to decoding life: systems biology. Annu Rev Genomics Hum Genet 2:343–372. doi: 10.1146/annurev.genom.2.1.343. [DOI] [PubMed] [Google Scholar]
- 2.Casadevall A, Fang FC, Pirofski L-A. 2011. Microbial virulence as an emergent property: consequences and opportunities. PLoS Pathog 7:e1002136. doi: 10.1371/journal.ppat.1002136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ablowitz R. 1939. The theory of emergence. Philos Sci 6:1–16. doi: 10.1086/286529. [DOI] [Google Scholar]
- 4.Leshem A, Liwinski T, Elinav E. 2020. Immune-microbiota interplay and colonization resistance in infection. Mol Cell 78:597–613. doi: 10.1016/j.molcel.2020.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Lawley TD, Walker AW. 2013. Intestinal colonization resistance. Immunology 138:1–11. doi: 10.1111/j.1365-2567.2012.03616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eckhardt M, Hultquist JF, Kaake RM, Hüttenhain R, Krogan NJ. 2020. A systems approach to infectious disease. Nat Rev Genet 21:339–354. doi: 10.1038/s41576-020-0212-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartocci E, Lió P. 2016. Computational modeling, formal analysis, and tools for systems biology. PLoS Comput Biol 12:e1004591. doi: 10.1371/journal.pcbi.1004591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dahal S, Yurkovich JT, Xu H, Palsson BO, Yang L. 2020. Synthesizing systems biology knowledge from omics using genome-scale models. Proteomics 20:e1900282. doi: 10.1002/pmic.201900282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bauer E, Thiele I. 2018. From network analysis to functional metabolic modeling of the human gut microbiota. mSystems 3:e00209-17. doi: 10.1128/mSystems.00209-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Damper RI. 2000. Editorial for the special issue on 'emergent properties of complex systems': emergence and levels of abstraction. Int J Systems Sci 31:811–818. doi: 10.1080/002077200406543. [DOI] [Google Scholar]
- 11.Crick F. 1994. The astonishing hypothesis: the scientific search for the soul. Scribner, Maxwell Macmillan International, New York, NY. [Google Scholar]
- 12.Allaire JM, Crowley SM, Law HT, Chang S-Y, Ko H-J, Vallance BA. 2018. The intestinal epithelium: central coordinator of mucosal immunity. Trends Immunol 39:677–696. doi: 10.1016/j.it.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Caricilli AM, Castoldi A, Câmara NOS. 2014. Intestinal barrier: a gentlemen's agreement between microbiota and immunity. World J Gastrointest Pathophysiol 5:18–32. doi: 10.4291/wjgp.v5.i1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tollin M, Bergman P, Svenberg T, Jörnvall H, Gudmundsson GH, Agerberth B. 2003. Antimicrobial peptides in the first line defence of human colon mucosa. Peptides 24:523–530. doi: 10.1016/s0196-9781(03)00114-1. [DOI] [PubMed] [Google Scholar]
- 15.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. 2011. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science 334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ostaff MJ, Stange EF, Wehkamp J. 2013. Antimicrobial peptides and gut microbiota in homeostasis and pathology. EMBO Mol Med 5:1465–1483. doi: 10.1002/emmm.201201773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjöberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, Stoel M, Zhou Y, Sodergren E, Weinstock GM, Bevins CL, Williams CB, Bos NA. 2010. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol 11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salzman NH, Ghosh D, Huttner KM, Paterson Y, Bevins CL. 2003. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature 422:522–526. doi: 10.1038/nature01520. [DOI] [PubMed] [Google Scholar]
- 19.Brandl K, Plitas G, Schnabl B, DeMatteo RP, Pamer EG. 2007. MyD88-mediated signals induce the bactericidal lectin RegIIIγ and protect mice against intestinal Listeria monocytogenes infection. J Exp Med 204:1891–1900. doi: 10.1084/jem.20070563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu S, Balasubramanian I, Laubitz D, Tong K, Bandyopadhyay S, Lin X, Flores J, Singh R, Liu Y, Macazana C, Zhao Y, Béguet-Crespel F, Patil K, Midura-Kiela MT, Wang D, Yap GS, Ferraris RP, Wei Z, Bonder EM, Häggblom MM, Zhang L, Douard V, Verzi MP, Cadwell K, Kiela PR, Gao N. 2020. Paneth cell-derived lysozyme defines the composition of mucolytic microbiota and the inflammatory tone of the intestine. Immunity 53:398–416.E8. doi: 10.1016/j.immuni.2020.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sicard J-F, Le Bihan G, Vogeleer P, Jacques M, Harel J. 2017. Interactions of intestinal bacteria with components of the intestinal mucus. Front Cell Infect Microbiol 7:387. doi: 10.3389/fcimb.2017.00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang BX, Wu CM, Ribbeck K. 2021. Home, sweet home: how mucus accommodates our microbiota. FEBS J 288:1789–1799. doi: 10.1111/febs.15504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jung C, Hugot JP, Barreau F. 2010. Peyer's patches: the immune sensors of the intestine. Int J Inflam 2010:823710. doi: 10.4061/2010/823710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hooper LV, Littman DR, Macpherson AJ. 2012. Interactions between the microbiota and the immune system. Science 336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weis AM, Round JL. 2021. Microbiota-antibody interactions that regulate gut homeostasis. Cell Host Microbe 29:334–346. doi: 10.1016/j.chom.2021.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moor K, Diard M, Sellin ME, Felmy B, Wotzka SY, Toska A, Bakkeren E, Arnoldini M, Bansept F, Co AD, Völler T, Minola A, Fernandez-Rodriguez B, Agatic G, Barbieri S, Piccoli L, Casiraghi C, Corti D, Lanzavecchia A, Regoes RR, Loverdo C, Stocker R, Brumley DR, Hardt W-D, Slack E. 2017. High-avidity IgA protects the intestine by enchaining growing bacteria. Nature 544:498–502. doi: 10.1038/nature22058. [DOI] [PubMed] [Google Scholar]
- 27.Donaldson GP, Ladinsky MS, Yu KB, Sanders JG, Yoo BB, Chou W-C, Conner ME, Earl AM, Knight R, Bjorkman PJ, Mazmanian SK. 2018. Gut microbiota utilize immunoglobulin A for mucosal colonization. Science 360:795–800. doi: 10.1126/science.aaq0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sartor RB, Wu GD. 2017. Roles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approaches. Gastroenterology 152:327–339.E4. doi: 10.1053/j.gastro.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Bäumler AJ. 2011. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci USA 108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rivera-Chávez F, Mekalanos JJ. 2019. Cholera toxin promotes pathogen acquisition of host-derived nutrients. Nature 572:244–248. doi: 10.1038/s41586-019-1453-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fletcher JR, Pike CM, Parsons RJ, Rivera AJ, Foley MH, McLaren MR, Montgomery SA, Theriot CM. 2021. Clostridioides difficile exploits toxin-mediated inflammation to alter the host nutritional landscape and exclude competitors from the gut microbiota. Nat Commun 12:462–462. doi: 10.1038/s41467-020-20746-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Bäumler AJ. 2010. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zackular JP, Moore JL, Jordan AT, Juttukonda LJ, Noto MJ, Nicholson MR, Crews JD, Semler MW, Zhang Y, Ware LB, Washington MK, Chazin WJ, Caprioli RM, Skaar EP. 2016. Dietary zinc alters the microbiota and decreases resistance to Clostridium difficile infection. Nat Med 22:1330–1334. doi: 10.1038/nm.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeng MY, Inohara N, Nuñez G. 2017. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol 10:18–26. doi: 10.1038/mi.2016.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chanin RB, Winter MG, Spiga L, Hughes ER, Zhu W, Taylor SJ, Arenales A, Gillis CC, Büttner L, Jimenez AG, Smoot MP, Santos RL, Winter SE. 2020. Epithelial-derived reactive oxygen species enable AppBCX-mediated aerobic respiration of Escherichia coli during intestinal inflammation. Cell Host Microbe 28:780–788.E5. doi: 10.1016/j.chom.2020.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cevallos SA, Lee J-Y, Tiffany CR, Byndloss AJ, Johnston L, Byndloss MX, Bäumler AJ, Ehrt S, Sigal M, Nassif X. 2019. Increased epithelial oxygenation links colitis to an expansion of tumorigenic bacteria. mBio 10:e02244-19. doi: 10.1128/mBio.02244-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thursby E, Juge N. 2017. Introduction to the human gut microbiota. Biochem J 474:1823–1836. doi: 10.1042/BCJ20160510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Freter R, Brickner H, Botney M, Cleven D, Aranki A. 1983. Mechanisms that control bacterial populations in continuous-flow culture models of mouse large intestinal flora. Infect Immun 39:676–685. doi: 10.1128/iai.39.2.676-685.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shepherd ES, DeLoache WC, Pruss KM, Whitaker WR, Sonnenburg JL. 2018. An exclusive metabolic niche enables strain engraftment in the gut microbiota. Nature 557:434–438. doi: 10.1038/s41586-018-0092-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leatham MP, Banerjee S, Autieri SM, Mercado-Lubo R, Conway T, Cohen PS. 2009. Precolonized human commensal Escherichia coli strains serve as a barrier to E. coli O157:H7 growth in the streptomycin-treated mouse intestine. Infect Immun 77:2876–2886. doi: 10.1128/IAI.00059-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fabich AJ, Jones SA, Chowdhury FZ, Cernosek A, Anderson A, Smalley D, McHargue JW, Hightower GA, Smith JT, Autieri SM, Leatham MP, Lins JJ, Allen RL, Laux DC, Cohen PS, Conway T. 2008. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect Immun 76:1143–1152. doi: 10.1128/IAI.01386-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conway E, Cohen PS. 2015. Commensal and pathogenic Escherichia coli metabolism in the gut. Microbiol Spectr 3:MBP-0006-2014. doi: 10.1128/microbiolspec.MBP-0006-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pereira FC, Berry D. 2017. Microbial nutrient niches in the gut: microbial nutrient niches in the gut. Environ Microbiol 19:1366–1378. doi: 10.1111/1462-2920.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leatham-Jensen MP, Frimodt-Møller J, Adediran J, Mokszycki ME, Banner ME, Caughron JE, Krogfelt KA, Conway T, Cohen PS. 2012. The streptomycin-treated mouse intestine selects Escherichia coli envZ missense mutants that interact with dense and diverse intestinal microbiota. Infect Immun 80:1716–1727. doi: 10.1128/IAI.06193-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Conway T, Cohen PS. 2015. Applying the restaurant hypothesis to intestinal microbiota: anaerobes in mixed biofilms degrade polysaccharides, sharing locally prepared sugars with facultative anaerobes that also colonize the intestine. Microbe Magazine 10:324–328. doi: 10.1128/microbe.10.324.1. [DOI] [Google Scholar]
- 46.Besten G, Lange K, Havinga R, Dijk THv, Gerding A, Eunen K, Müller M, Groen AK, Hooiveld GJ, Bakker BM, Reijngoud D-J. 2013. Gut-derived short-chain fatty acids are vividly assimilated into host carbohydrates and lipids. Am J Physiol Gastrointest Liver Physiol 305:G900–G910. doi: 10.1152/ajpgi.00265.2013. [DOI] [PubMed] [Google Scholar]
- 47.Rios-Covian D, Gueimonde M, Duncan SH, Flint HJ, de los Reyes-Gavilan CG. 2015. Enhanced butyrate formation by cross-feeding between Faecalibacterium prausnitzii and Bifidobacterium adolescentis. FEMS Microbiol Lett 362:fnv176. doi: 10.1093/femsle/fnv176. [DOI] [PubMed] [Google Scholar]
- 48.Moens F, Verce M, De Vuyst L. 2017. Lactate- and acetate-based cross-feeding interactions between selected strains of lactobacilli, bifidobacteria and colon bacteria in the presence of inulin-type fructans. Int J Food Microbiol 241:225–236. doi: 10.1016/j.ijfoodmicro.2016.10.019. [DOI] [PubMed] [Google Scholar]
- 49.Sun Y, O'Riordan MXD. 2013. Regulation of bacterial pathogenesis by intestinal short-chain fatty acids. Adv Appl Microbiol 85:93–118. doi: 10.1016/B978-0-12-407672-3.00003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ducarmon QR, Zwittink RD, Hornung BVH, van Schaik W, Young VB, Kuijper EJ. 2019. Gut microbiota and colonization resistance against bacterial enteric infection. Microbiol Mol Biol Rev 83:e00007-19. doi: 10.1128/MMBR.00007-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ferreyra JA, Wu KJ, Hryckowian AJ, Bouley DM, Weimer BC, Sonnenburg JL. 2014. Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe 16:770–777. doi: 10.1016/j.chom.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koenigsknecht MJ, Theriot CM, Bergin IL, Schumacher CA, Schloss PD, Young VB. 2015. Dynamics and establishment of Clostridium difficile infection in the murine gastrointestinal tract. Infect Immun 83:934–941. doi: 10.1128/IAI.02768-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hryckowian AJ, Van Treuren W, Smits SA, Davis NM, Gardner JO, Bouley DM, Sonnenburg JL. 2018. Microbiota-accessible carbohydrates suppress Clostridium difficile infection in a murine model. Nat Microbiol 3:662–669. doi: 10.1038/s41564-018-0150-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Begley M, Gahan CG, Hill C. 2005. The interaction between bacteria and bile. FEMS Microbiol Rev 29:625–651. doi: 10.1016/j.femsre.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 55.Kurdi P, Kawanishi K, Mizutani K, Yokota A. 2006. Mechanism of growth inhibition by free bile acids in lactobacilli and bifidobacteria. J Bacteriol 188:1979–1986. doi: 10.1128/JB.188.5.1979-1986.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thanissery R, Winston JA, Theriot CM. 2017. Inhibition of spore germination, growth, and toxin activity of clinically relevant C. difficile strains by gut microbiota derived secondary bile acids. Anaerobe 45:86–100. doi: 10.1016/j.anaerobe.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink MRM, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, Pamer EG. 2015. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thompson JA, Oliveira RA, Djukovic A, Ubeda C, Xavier KB. 2015. Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota. Cell Rep 10:1861–1871. doi: 10.1016/j.celrep.2015.02.049. [DOI] [PubMed] [Google Scholar]
- 59.Cleusix V, Lacroix C, Vollenweider S, Duboux M, Le Blay G. 2007. Inhibitory activity spectrum of reuterin produced by Lactobacillus reuteri against intestinal bacteria. BMC Microbiol 7:101. doi: 10.1186/1471-2180-7-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sassone-Corsi M, Nuccio SP, Liu H, Hernandez D, Vu CT, Takahashi AA, Edwards RA, Raffatellu M. 2016. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature 540:280–283. doi: 10.1038/nature20557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cotter PD, Ross RP, Hill C. 2013. Bacteriocins—a viable alternative to antibiotics? Nat Rev Microbiol 11:95–105. doi: 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
- 62.Parks WM, Bottrill AR, Pierrat OA, Durrant MC, Maxwell A. 2007. The action of the bacterial toxin, microcin B17, on DNA gyrase. Biochimie 89:500–507. doi: 10.1016/j.biochi.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 63.Vincent PA, Morero RD. 2009. The structure and biological aspects of peptide antibiotic microcin J25. Curr Med Chem 16:538–549. doi: 10.2174/092986709787458461. [DOI] [PubMed] [Google Scholar]
- 64.Metlitskaya A, Kazakov T, Kommer A, Pavlova O, Praetorius-Ibba M, Ibba M, Krasheninnikov I, Kolb V, Khmel I, Severinov K. 2006. Aspartyl-tRNA synthetase is the target of peptide nucleotide antibiotic microcin C. J Biol Chem 281:18033–18042. doi: 10.1074/jbc.M513174200. [DOI] [PubMed] [Google Scholar]
- 65.Wiedemann I, Breukink E, van Kraaij C, Kuipers OP, Bierbaum G, de Kruijff B, Sahl H-G. 2001. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J Biol Chem 276:1772–1779. doi: 10.1074/jbc.M006770200. [DOI] [PubMed] [Google Scholar]
- 66.Scherer KM, Spille J-H, Sahl H-G, Grein F, Kubitscheck U. 2015. The lantibiotic nisin induces lipid II aggregation, causing membrane instability and vesicle budding. Biophys J 108:1114–1124. doi: 10.1016/j.bpj.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dobson A, Cotter PD, Ross RP, Hill C. 2012. Bacteriocin production: a probiotic trait? Appl Environ Microbiol 78:1–6. doi: 10.1128/AEM.05576-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Corr SC, Li Y, Riedel CU, O'Toole PW, Hill C, Gahan CG. 2007. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc Natl Acad Sci USA 104:7617–7621. doi: 10.1073/pnas.0700440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bosák J, Micenková L, Hrala M, Pomorská K, Kunova Bosakova M, Krejci P, Göpfert E, Faldyna M, Šmajs D. 2018. Colicin F(Y) inhibits pathogenic Yersinia enterocolitica in mice. Sci Rep 8:12242. doi: 10.1038/s41598-018-30729-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sana TG, Lugo KA, Monack DM. 2017. T6SS: The bacterial “fight club” in the host gut. PLoS Pathog 13:e1006325. doi: 10.1371/journal.ppat.1006325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chatzidaki-Livanis M, Geva-Zatorsky N, Comstock LE. 2016. Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc Natl Acad Sci USA 113:3627–3632. doi: 10.1073/pnas.1522510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Monda V, Villano I, Messina A, Valenzano A, Esposito T, Moscatelli F, Viggiano A, Cibelli G, Chieffi S, Monda M, Messina G. 2017. Exercise modifies the gut microbiota with positive health effects. Oxid Med Cell Longev 2017:3831972–3831972. doi: 10.1155/2017/3831972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gui X, Yang Z, Li MD. 2021. Effect of cigarette smoke on gut microbiota: state of knowledge. Front Physiol 12:673341. doi: 10.3389/fphys.2021.673341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, Belzer C, Delgado PS, Arboleya MS, Mancabelli L, Lugli GA, Rodriguez JM, Bode L, de Vos W, Gueimonde M, Margolles A, van Sinderen D, Ventura M. 2017. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev 81:e00036-17. doi: 10.1128/MMBR.00036-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kumbhare SV, Patangia DV, Patil RH, Shouche YS, Patil NP. 2019. Factors influencing the gut microbiome in children: from infancy to childhood. J Biosci 44:49. doi: 10.1007/s12038-019-9860-z. [DOI] [PubMed] [Google Scholar]
- 76.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. 2011. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA 108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zmora N, Suez J, Elinav E. 2019. You are what you eat: diet, health and the gut microbiota. Nat Rev Gastroenterol Hepatol 16:35–56. doi: 10.1038/s41575-018-0061-2. [DOI] [PubMed] [Google Scholar]
- 78.Wotzka SY, Kreuzer M, Maier L, Arnoldini M, Nguyen BD, Brachmann AO, Berthold DL, Zünd M, Hausmann A, Bakkeren E, Hoces D, Gül E, Beutler M, Dolowschiak T, Zimmermann M, Fuhrer T, Moor K, Sauer U, Typas A, Piel J, Diard M, Macpherson AJ, Stecher B, Sunagawa S, Slack E, Hardt WD. 2019. Escherichia coli limits Salmonella Typhimurium infections after diet shifts and fat-mediated microbiota perturbation in mice. Nat Microbiol 4:2164–2174. doi: 10.1038/s41564-019-0568-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.David LA, Materna AC, Friedman J, Campos-Baptista MI, Blackburn MC, Perrotta A, Erdman SE, Alm EJ. 2014. Host lifestyle affects human microbiota on daily timescales. Genome Biol 15:R89. doi: 10.1186/gb-2014-15-7-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Leeming ER, Johnson AJ, Spector TD, Le Roy CI. 2019. Effect of diet on the gut microbiota: rethinking intervention duration. Nutrients 11:2862. doi: 10.3390/nu11122862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Han H, Yi B, Zhong R, Wang M, Zhang S, Ma J, Yin Y, Yin J, Chen L, Zhang H. 2021. From gut microbiota to host appetite: gut microbiota-derived metabolites as key regulators. Microbiome 9:162. doi: 10.1186/s40168-021-01093-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Alcock J, Maley CC, Aktipis CA. 2014. Is eating behavior manipulated by the gastrointestinal microbiota? Evolutionary pressures and potential mechanisms. Bioessays 36:940–949. doi: 10.1002/bies.201400071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.García-Cabrerizo R, Carbia C, Or KJ, Schellekens H, Cryan JF. 2021. Microbiota-gut-brain axis as a regulator of reward processes. J Neurochem 157:1495–1524. doi: 10.1111/jnc.15284. [DOI] [PubMed] [Google Scholar]
- 84.Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, Abramson L, Katz MN, Korem T, Zmora N, Kuperman Y, Biton I, Gilad S, Harmelin A, Shapiro H, Halpern Z, Segal E, Elinav E. 2014. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell 159:514–529. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- 85.Zarrinpar A, Chaix A, Yooseph S, Panda S. 2014. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab 20:1006–1017. doi: 10.1016/j.cmet.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, Young VB, Henrissat B, Wilmes P, Stappenbeck TS, Núñez G, Martens EC. 2016. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 167:1339–1353.E21. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. 2012. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3:289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Townsend GE, Han W, Schwalm ND, Raghavan V, Barry NA, Goodman AL, Groisman EA. 2019. Dietary sugar silences a colonization factor in a mammalian gut symbiont. Proc Natl Acad Sci USA 116:233–238. doi: 10.1073/pnas.1813780115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vich Vila A, Collij V, Sanna S, Sinha T, Imhann F, Bourgonje AR, Mujagic Z, Jonkers DMAE, Masclee AAM, Fu J, Kurilshikov A, Wijmenga C, Zhernakova A, Weersma RK. 2020. Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nat Commun 11:362. doi: 10.1038/s41467-019-14177-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, Mujagic Z, Vila AV, Falony G, Vieira-Silva S, Wang J, Imhann F, Brandsma E, Jankipersadsing SA, Joossens M, Cenit MC, Deelen P, Swertz MA, Weersma RK, Feskens EJM, Netea MG, Gevers D, Jonkers D, Franke L, Aulchenko YS, Huttenhower C, Raes J, Hofker MH, Xavier RJ, Wijmenga C, Fu J, LifeLines Cohort Study. 2016. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 352:565–569. doi: 10.1126/science.aad3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jaynes M, Kumar AB. 2019. The risks of long-term use of proton pump inhibitors: a critical review. Ther Adv Drug Saf 10:2042098618809927. doi: 10.1177/2042098618809927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tourret J, Willing BP, Dion S, MacPherson J, Denamur E, Finlay BB. 2017. Immunosuppressive treatment alters secretion of ileal antimicrobial peptides and gut microbiota, and favors subsequent colonization by uropathogenic Escherichia coli. Transplantation 101:74–82. doi: 10.1097/TP.0000000000001492. [DOI] [PubMed] [Google Scholar]
- 93.Guzman Rodriguez M, Hyde R, Vanner S, Allen-Vercoe E, Sheth P, Petrof EO. 2019. Direct bactericidal effect of pantoprazole on bacterial isolates from the gut microbiome. J Can Assoc Gastroenterol 2:305–306. doi: 10.1093/jcag/gwz006.153. [DOI] [Google Scholar]
- 94.Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE, Brochado AR, Fernandez KC, Dose H, Mori H, Patil KR, Bork P, Typas A. 2018. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 555:623–628. doi: 10.1038/nature25979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Klünemann M, Andrejev S, Blasche S, Mateus A, Phapale P, Devendran S, Vappiani J, Simon B, Scott TA, Kafkia E, Konstantinidis D, Zirngibl K, Mastrorilli E, Banzhaf M, Mackmull M-T, Hövelmann F, Nesme L, Brochado AR, Maier L, Bock T, Periwal V, Kumar M, Kim Y, Tramontano M, Schultz C, Beck M, Hennig J, Zimmermann M, Sévin DC, Cabreiro F, Savitski MM, Bork P, Typas A, Patil KR. 2021. Bioaccumulation of therapeutic drugs by human gut bacteria. Nature 597:533–538. doi: 10.1038/s41586-021-03891-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Okumura R, Takeda K. 2017. Roles of intestinal epithelial cells in the maintenance of gut homeostasis. Exp Mol Med 49:e338. doi: 10.1038/emm.2017.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Belkaid Y, Hand TW. 2014. Role of the microbiota in immunity and inflammation. Cell 157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Soderholm AT, Pedicord VA. 2019. Intestinal epithelial cells: at the interface of the microbiota and mucosal immunity. Immunology 158:267–280. doi: 10.1111/imm.13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pabst O, Slack E. 2020. IgA and the intestinal microbiota: the importance of being specific. Mucosal Immunol 13:12–21. doi: 10.1038/s41385-019-0227-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang Q, Li L, Xu R. 2018. A systems biology approach to predict and characterize human gut microbial metabolites in colorectal cancer. Sci Rep 8:6225. doi: 10.1038/s41598-018-24315-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Greenblum S, Turnbaugh PJ, Borenstein E. 2012. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc Natl Acad Sci USA 109:594–599. doi: 10.1073/pnas.1116053109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, Andrews E, Ajami NJ, Bonham KS, Brislawn CJ, Casero D, Courtney H, Gonzalez A, Graeber TG, Hall AB, Lake K, Landers CJ, Mallick H, Plichta DR, Prasad M, Rahnavard G, Sauk J, Shungin D, Vázquez-Baeza Y, White RA, Bishai J, Bullock K, Deik A, Dennis C, Kaplan JL, Khalili H, McIver LJ, Moran CJ, Nguyen L, Pierce KA, Schwager R, Sirota-Madi A, Stevens BW, Tan W, ten Hoeve JJ, Weingart G, Wilson RG, Yajnik V, Braun J, Denson LA, Jansson JK, Knight R, Kugathasan S, McGovern DPB, Petrosino JF, IBDMDB Investigators. 2019. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569:655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhou W, Sailani MR, Contrepois K, Zhou Y, Ahadi S, Leopold SR, Zhang MJ, Rao V, Avina M, Mishra T, Johnson J, Lee-McMullen B, Chen S, Metwally AA, Tran TDB, Nguyen H, Zhou X, Albright B, Hong B-Y, Petersen L, Bautista E, Hanson B, Chen L, Spakowicz D, Bahmani A, Salins D, Leopold B, Ashland M, Dagan-Rosenfeld O, Rego S, Limcaoco P, Colbert E, Allister C, Perelman D, Craig C, Wei E, Chaib H, Hornburg D, Dunn J, Liang L, Rose S-F, Kukurba K, Piening B, Rost H, Tse D, McLaughlin T, Sodergren E, Weinstock GM, Snyder M. 2019. Longitudinal multi-omics of host–microbe dynamics in prediabetes. Nature 569:663–671. doi: 10.1038/s41586-019-1236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fernández-Edreira D, Liñares-Blanco J, Fernandez-Lozano C. 2021. Machine learning analysis of the human infant gut microbiome identifies influential species in type 1 diabetes. Expert Syst Appl 185:115648. doi: 10.1016/j.eswa.2021.115648. [DOI] [Google Scholar]
- 105.Topçuoğlu BD, Lesniak NA, Ruffin MT, Wiens J, Schloss PD, Blaser MJ. 2020. A framework for effective application of machine learning to microbiome-based classification problems. mBio 11:e00434-20. doi: 10.1128/mBio.00434-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kurkjian HM, Akbari MJ, Momeni B. 2021. The impact of interactions on invasion and colonization resistance in microbial communities. PLoS Comput Biol 17:e1008643. doi: 10.1371/journal.pcbi.1008643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schubert AM, Sinani H, Schloss PD, Fraser CM. 2015. Antibiotic-induced alterations of the murine gut microbiota and subsequent effects on colonization resistance against Clostridium difficile. mBio 6:e00974-15. doi: 10.1128/mBio.00974-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Midani FS, Weil AA, Chowdhury F, Begum YA, Khan AI, Debela MD, Durand HK, Reese AT, Nimmagadda SN, Silverman JD, Ellis CN, Ryan ET, Calderwood SB, Harris JB, Qadri F, David LA, LaRocque RC. 2018. Human Gut Microbiota Predicts Susceptibility to Vibrio cholerae Infection. J Infect Dis 218:645–653. doi: 10.1093/infdis/jiy192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Henson MA. 2021. Computational modeling of the gut microbiota reveals putative metabolic mechanisms of recurrent Clostridioides difficile infection. PLoS Comput Biol 17:e1008782. doi: 10.1371/journal.pcbi.1008782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jenior ML, Leslie JL, Young VB, Schloss PD. 2018. Clostridium difficile alters the structure and metabolism of distinct cecal microbiomes during initial infection to promote sustained colonization. mSphere 3:e00261-18. doi: 10.1128/mSphere.00261-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Smillie CS, Sauk J, Gevers D, Friedman J, Sung J, Youngster I, Hohmann EL, Staley C, Khoruts A, Sadowsky MJ, Allegretti JR, Smith MB, Xavier RJ, Alm EJ. 2018. Strain tracking reveals the determinants of bacterial engraftment in the human gut following fecal microbiota transplantation. Cell Host Microbe 23:229–240.E5. doi: 10.1016/j.chom.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Marino S, Baxter NT, Huffnagle GB, Petrosino JF, Schloss PD. 2014. Mathematical modeling of primary succession of murine intestinal microbiota. Proc Natl Acad Sci USA 111:439–444. doi: 10.1073/pnas.1311322111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Skraban J, Dzeroski S, Zenko B, Mongus D, Gangl S, Rupnik M. 2013. Gut microbiota patterns associated with colonization of different Clostridium difficile ribotypes. PLoS One 8:e58005. doi: 10.1371/journal.pone.0058005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Girinathan BP, DiBenedetto N, Worley JN, Peltier J, Arrieta-Ortiz ML, Immanuel SRC, Lavin R, Delaney ML, Cummins CK, Hoffman M, Luo Y, Gonzalez-Escalona N, Allard M, Onderdonk AB, Gerber GK, Sonenshein AL, Baliga NS, Dupuy B, Bry L. 2021. In vivo commensal control of Clostridioides difficile virulence. Cell Host Microbe 29:1693–1708.E7. doi: 10.1016/j.chom.2021.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Theriot CM, Koenigsknecht MJ, Carlson PE, Jr, Hatton GE, Nelson AM, Li B, Huffnagle GB, Li J, Young VB. 2014. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun 5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Stecher B, Chaffron S, Käppeli R, Hapfelmeier S, Freedrich S, Weber TC, Kirundi J, Suar M, McCoy KD, von Mering C, Macpherson AJ, Hardt W-D. 2010. Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog 6:e1000711. doi: 10.1371/journal.ppat.1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]