ABSTRACT
Biomaterials offer unique properties that make them irreplaceable for next-generation applications. Fibrous proteins, such as various caterpillar silks and especially spider silk, have strength and toughness not found in human-made materials. In early studies, proteins containing long tandem repeats, such as major ampullate spidroin 1 (MaSp1) and flagelliform silk protein (FSLP), were produced using a large DNA template composed of many tandem repeats. The hierarchical DNA assembly of the DNA template is very time-consuming and labor-intensive, which makes the fibrous proteins difficult to study and engineer. In this study, we designed a circularized mRNA (cmRNA) employing the RNA cyclase ribozyme mechanism. cmRNAs encoding spider silk protein MaSp1 and FSLP were designed based on only one unit of the template sequence but provide ribosomes with a circular and infinite translation template for production of long peptides containing tandem repeats. Using this technique, cmRNAs of MaSp1 and FSLP were successfully generated with circularization efficiencies of 8.5% and 36.7%, respectively, which supported the production of recombinant MaSp1 and FSLP larger than 110 and 88 kDa, containing tens of repeat units. Western blot analysis and mass spectrometry confirmed the authenticity of MaSp1 and FSLP, which were produced at titers of 22.1 and 81.5 mg · liter−1, respectively.
IMPORTANCE Spider silk is a biomaterial with superior properties. However, its heterologous expression template is hard to construct. The cmRNA technique simplifies the construction and expression strategy by proving the ribosome a circular translation template for expression of long peptides containing tandem repeats. This revolutionary technique will allow researchers to easily build, study, and experiment with any fiber proteins with sequences either from natural genes or artificial designs. We expect a significantly accelerated development of fibrous protein-based biomaterials with the cmRNA technique.
KEYWORDS: spider silk, cmRNA, polypeptide
INTRODUCTION
Biomaterials represent unique properties and irreplaceable applications. As unique biomaterials, fibrous proteins such as various caterpillar and spider silk proteins have strength and toughness not found in human-made materials (1, 2). Spider dragline silk is one of the strongest fibers in nature. Due to its excellent mechanical properties, good biocompatibility, and slow degradation, spider dragline has a wide spectrum of potential applications as a biomaterial (3–8). The dragline is secreted by the main ampullate gland of spiders as the main skeleton of the spider’s web and is composed of major ampullate spidroin 1 (MaSp1) and major ampullate spidroin 2 (MaSp2) (9, 10). MaSp1 contains around 3,500 amino acid residues in repeat regions, along with 100 nonrepeating residues on both sides of the protein repeats. The molecular weight of natural MaSp1 protein is about 250 to 320 kDa (11, 12). The amino acid sequence repeat regions are rich in alanine and glycine as well as (GGX)n motifs (X is tyrosine, leucine, or glutamine) (10, 13). The alanine-rich areas provide the high tensile strength of the spider silk fibers by forming β-sheets (14, 15).
In contrast with dragline silk, flagelliform silk (FLAG) does not have high tensile strength, but it has excellent extensibility that enables it to form the capture spiral of an orb-web (16). The mRNA of FLAG is 15.5 kb in Nephila clavipes (17, 18). FLAG has a molecular mass of 500 kDa and is rich in proline and valine, where the alanine content is lower than that in MaSp1 (19, 20). FLAG is comprised of (GGX)n and GPGXX blocks, which fold into 31 helices and β-turn spirals, respectively, leading to the high elasticity and flexibility (21, 22). We chose several characteristic sequences and designed an artificial silk sequence named flagelliform silk-like protein (FSLP) to produce FSLP polypeptides (18).
A major aim of spider silk research is the development of possible large-scale production techniques. However, since spiders are generally aggressive, territorial, and even cannibalistic, it is practically impossible to culture wild spiders to produce spider silk in analogy to silkworm sericulture (18). With the development of synthetic biology techniques, researchers attempted to use microbial cell factories to produce recombinant spider silk proteins (23, 24). The core expression template of MaSp1 is a long and highly repetitive DNA sequence. DNA cassettes with different numbers of tandem repeats were assembled using the isocaudamer enzyme ligation method, and the resulting sequences were cloned into vectors for expression in Escherichia coli BL21 (25).
Several groups successfully obtained MaSp1 protein in various engineered hosts (26, 27). However, this method has intrinsic limitations that have not been resolved. First, there are too many repetitive sequences in the DNA template, which leads to DNA recombination, gene fragment loss, and premature termination of protein translation (28). The repetitive genes carried on plasmids are unstable in the bacterial host (29). The expression of spidroin 1 analog genes in E. coli is a big problem due to the termination error rate being 4 to 12 times higher than that of a natural gene, depending on the precise DNA sequence used to encode the protein (30). This also makes it very difficult to express high-molecular-weight recombinant spider silk proteins. Second, the hierarchical DNA assembly method used to construct sequences with multiple tandem fragments is very time-consuming and labor-intensive, which makes it difficult to study and experiment with different silk protein sequences. These drawbacks greatly limit the development of spider silk protein production techniques. Circular mRNA was designed to express green fluorescent protein (GFP) polypeptide, which is a feasible method to express highly repetitive silk protein (31).
In this work, we developed an alternative approach by designing a circularized mRNA (cmRNA) employing the RNA cyclase ribozyme for autocatalytic splicing, which possesses an internal sequence circularization mechanism (32). We designed a cmRNA carrying only one spider silk protein open reading frame (ORF). Due to its circular structure, theoretically the cmRNA provides ribosomes with an infinite translation template, and long repeat peptides will be produced before ribosomes disengage from the circular mRNA template. Since subtle changes in amino acid sequence can achieve broad functionality of protein, it is much easier to translate cmRNA to produce novel spider silk combination protein than mRNA from a DNA template (29). In this work, we report the production of recombinant MaSp1 protein of up to ∼110 kDa and recombinant FSLP of up to ∼88.2 kDa.
RESULTS AND DISCUSSION
Design of MaSp1 and FSLP cmRNAs for expression in E. coli.
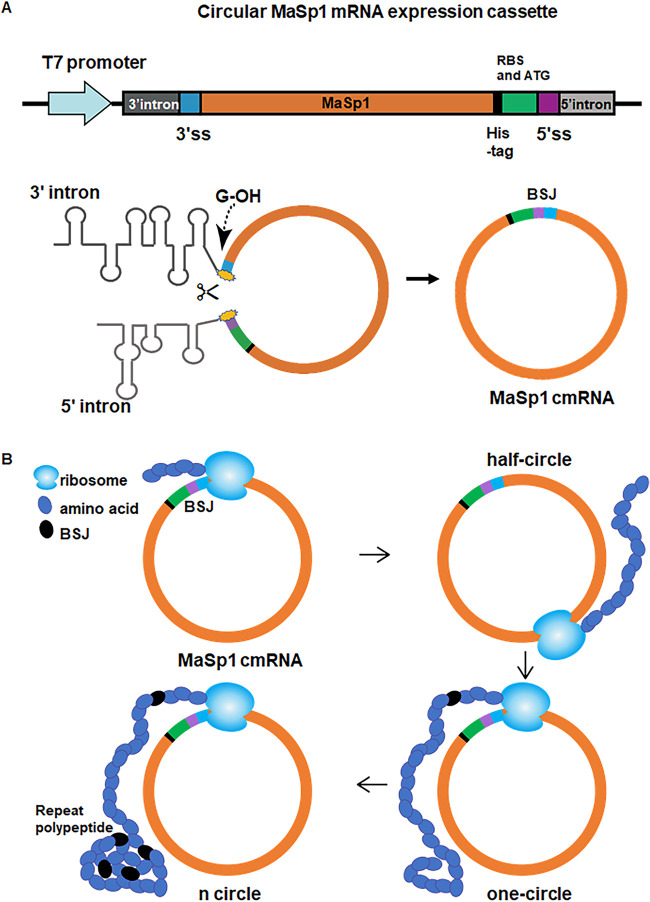
To generate a cmRNA encoding spider silk protein, we utilized the td intron, an intron of the td gene from T4 phage belonging to group IE, which circularizes the exon to form a back-splice junction (BSJ) in a reaction catalyzed by guanosine (33–35). To ensure that the ribosomes do not translate the ORF of the spider silk protein from unprocessed linear mRNA, the ribosome binding sequence (RBS) and translation initiation codon ATG were placed downstream of the MaSp1 or FSLP coding sequence (Fig. 1A). Consequently, the regulatory sequences were located upstream of the coding sequence only after circularization of the mRNA. To purify the resulting MaSp1 or FSLP polypeptides, a His tag was incorporated into the ORF (Fig. 1A). If the mRNA is circularized, the ribosome could circle the cmRNA, producing a long repeating polypeptide (Fig. 1B).
FIG 1.
Design of a circularized spider silk mRNA based on td flanking introns. (A) Schematic of the MaSp1 mRNA circularization mechanism. The MaSp1 coding sequence was cloned between the upstream and downstream td introns, and the whole sequence was transcribed using a T7 promoter. 3′ss, 3′ splice site; 5′ss, 5′ splice site. Upon transcription, the introns form secondary structures, and the 3′ hydroxyl group of guanosine initiates the generation of MaSp1 cmRNA. BSJ, back-splice junction. (B) The ribosome continuously translates spider silk polypeptide by traveling along the round cmRNA.
Analysis of the proportion of circularized mRNA.
Following transcription and exon circularization, the RBS and initial codon were fused with the MaSp1 or FSLP coding sequence to generate the cmRNA. To determine whether the MaSp1 and FSLP mRNAs were circularized in vivo, we extracted the total RNA from E. coli for quantitative PCR (qPCR) amplification. We first used the method of Costello et al. to measure the circularization efficiency of mRNA of MaSp1 and FSLP (36). However, by primer-amplifying BSJ of cDNA produced by reverse transcription (RT) of cmRNA according to Chen et al. (37), larger PCR products were presented, suggesting the presence of long DNA template generated from reverse transcription of more than 1 cycle (Fig. S1). These results indicated that the normal quantification method generates biased data.
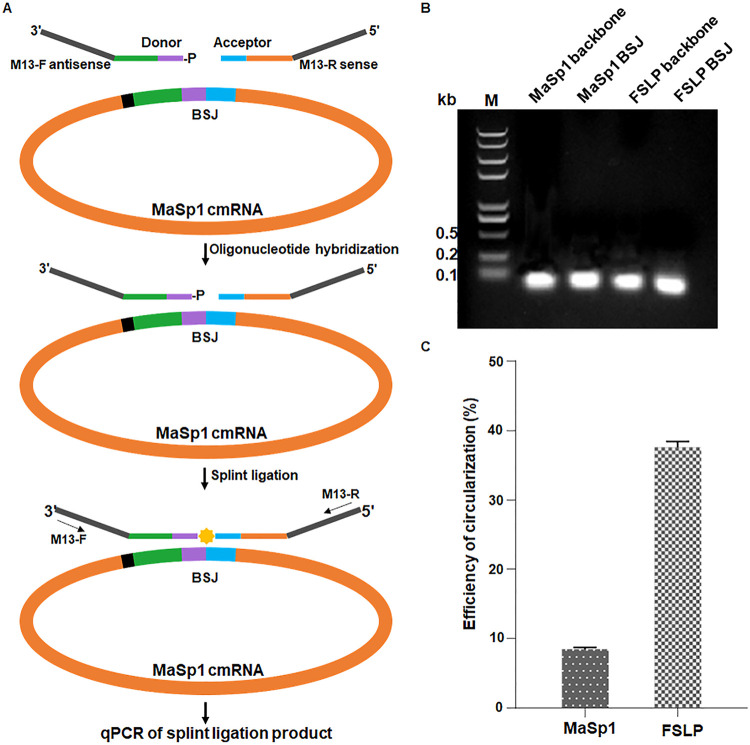
Therefore, we employed a SplintQuant method described by Conn and Conn (38) to quantify the copy number of mRNA containing the BSJ, which represent the cmRNA, along with a region without BSJ, representing the total mRNA of MaSp1 or FSLP. DNA oligonucleotides flanking the MaSp1 BSJ were designed with M13 primer sequences incorporated flanking the cmRNA complementary oligonucleotides, permitting amplification and quantification of cmRNA transcripts by qPCR with the ligated DNA oligonucleotides as the template (Fig. 2A). Accordingly, DNA oligonucleotides flanking the backbone of MaSp1 transcripts were used to quantify the total mRNA. The DNA oligonucleotides used to quantify total transcripts and cmRNA of FSLP were designed on the same principle as MaSp1. This method eliminated the overly reverse transcribed problem caused by the circular RNA template.
FIG 2.
SplintQuant for quantification of cmRNA relative to the total mRNA. (A) Schematic of SplintQuant method mechanism. (B) Circularization efficiency was evaluated by M13 primer sets for all transcripts and cmRNA (MaSp1 and FSLP), respectively. MaSp1 backbone, the PCR product of common area of cDNA obtained from MaSp1 linear mRNA and MaSp1 cmRNA. MaSp1 BSJ, the PCR product of BSJ sequence of cDNA obtained from MaSp1 cmRNA. FSLP backbone, the PCR product of common area of cDNA obtained from FSLP linear mRNA and FSLP cmRNA. FSLP BSJ, the PCR product of cDNA obtained from FSLP cmRNA. (C) The circularization efficiency of the MaSp1 and FSLP transcripts was determined by RT-qPCR.
Following splint reaction, ligated product of all transcripts and cmRNA (MaSp1 and FSLP) were identified by M13 primer pair amplification (Fig. 2B), quantified spectrophotometrically, and serially diluted to generate standard samples at different concentrations. After the Cq (cycle quantification) values of samples from both standard sets were determined by RT-PCR, the logarithmic value of the copy number of the standard sample was taken as the abscissa, and the measured Cq value was taken as the ordinate to draw a standard curve (see Fig. S2A and B in the supplemental material). Thus, the copy number of a given sample could be determined from the standard curve according to its Cq value. The quantification of the Cq value indicated that the ratio of MaSp1 and FSLP transcript circularization was 8.5% and 36.7%, respectively (Fig. 2C).
It was reported that the optimal length of the td exon between the 5′ and 3′ intron splice sites is 300 to 500 nucleotides (39). One unit of FSLP cmRNA contains 327 nucleotides, while that of MaSp1 cmRNA has only 156 nucleotides, which is less than the optimal size of the exons. Thus, the size of MaSp1 mRNA might cause the lower circularization efficiency. The result suggested that we need to design and express fibrous protein within the optimal length to obtain a relatively high cyclization efficiency.
Production of large spider silk peptides.
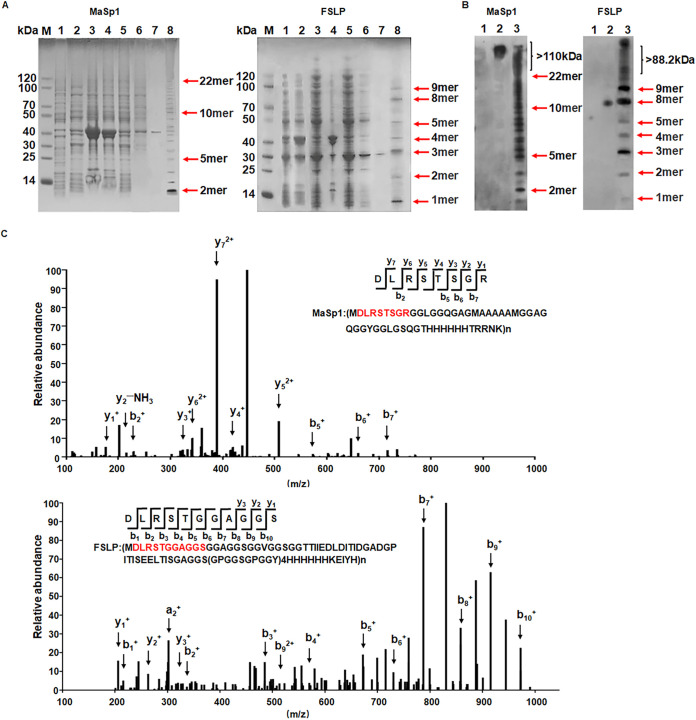
E. coli BL21 was individually transformed with the plasmids pcirMaSp1 and pcirFSLP to produce MaSp1 and FSLP protein, respectively. MaSp1 was expressed and collected after being cultured for 18 h at 30°C. After nickel nitrilotriacetic acid (Ni-NTA) column purification, the protein sample was subjected to SDS-PAGE and visualized with Coomassie brilliant blue. The MaSp1 sample was a mixture of proteins with various sizes that formed a ladder on the gel. According to the protein marker, the sizes of the proteins ranged from about 10 to 110 kDa (Fig. 3A). The MaSp1 polypeptide was composed of repeating units with a size of 5.1 kDa, which was formed by the ribosome traveling one round along the circularized MaSp1 mRNA. The smallest band was found to be around 10 kDa, which was most possibly a dimeric MaSp1 peptide, and the largest differentiable band on the gel was around 110 kDa, indicating that the ribosome could travel along the MaSp1 cmRNA at least 22 rounds (Fig. 1B and 3A). Although there were still larger bands above the 22-mer band on the gel, they formed a smear instead of a differentiable ladder. These large proteins were later confirmed to be MaSp1 proteins larger than 110 kDa by Western blotting. This suggested that ribosomes could bind quite stably to the MaSp1 cmRNA, which provided a circular translation template for ribosomes to travel many rounds and produce large proteins containing more than 22 repeating units.
FIG 3.
Identification of MaSp1 and FSLP. (A) Coomassie brilliant blue staining of purified spider silk protein. (Left) SDS-PAGE gel of MaSp1. (Right) SDS-PAGE gel of FSLP. M, marker; lane 1, uninduced cell lysate; lanes 2 to 7, induced cell sample; lane 2, whole-cell lysate; lane 3, precipitate; lane 4, supernatant; lane 5, sample washed with lysis buffer; lane 6, sample washed with lysis buffer with 10 mM imidazole; lane 7, sample washed with lysis buffer with 50 mm imidazole; lane 8, freeze-dried sample. (B) Western blot of purified spider silk protein. (Left) Western image of MaSp1 protein. (Right) Western blot of FSLP protein. Lane 1, uninduced cell lysate; lane 2, positive control; lane 3, freeze-dried sample. (C) Mass spectrum of purified spider silk protein. (Upper) Mass spectrogram of MaSp1 polypeptide. y1, R; y2, RG; y3, RGS; y4, RGST; y5, RGSTS; y6, RGSTSR; y7, RGSTSRL; b2, DL; b5, DLRST; b6, DLRSTS; b7, DLRSTSG. (Lower) Mass spectrogram of FSLP polypeptide. y1, S; y2, SG; y3, SGG; b1, D; b2, DL; b3, DLR; b4, DLRS; b5, DLRST; b6, DLRSTG; b7, DLRSTGG; b8, DLRSTGGA; b9, DLRSTGGAG; b10, DLRSTGGAGS. The characteristic peptide sequence of spider silk protein is highlighted in red. The peaks representing digested signature oligopeptides are indicated with arrows. y and b variables represent the digestion products from the C and N terminus, respectively.
FSLP was expressed, collected, purified, and characterized in the same way. The FSLP sample also formed a ladder on the SDS-PAGE gel, and the size of the protein bands spanned from ∼10 to 90 kDa (Fig. 3A). A single unit of the repeat FSLP polypeptide had a molecular weight of 9.8 kDa, indicating that the smallest band on the gel represented the single FSLP, which was produced by the ribosome travelling only one round along the FSLP cmRNA. Similar to MaSp1, the purified FSLP was a mixture of FSLPs with various numbers of repeat units.
In addition, according to the SDS gels and Western images, there was almost no smear of the purified proteins. Instead, both spider silk proteins were presented as the ladder form. This indicated that in the translation process of cmRNA of the spider proteins, the ribosome stopped at a relatively specific site instead of at random stops. Since rare codons might cause stop and detachment of ribosomes (40), we checked the cmRNA sequence to see if there was a rare codon that might cause the specific stop. The gene sequences of MaSp1 and FSLP were optimized according to the codon preference of E. coli BL21, which did not contain rare codons. However, we found there was a rare codon, ATA, in the RBS region (41), which might cause the detachment of ribosomes from cmRNA. In the future, we might be able to improve the translation persistence of spider silk cmRNA by eliminating rare codons.
Identification of MaSp1 and FSLP polypeptides.
The purified MaSp1 and FSLP samples were separated by SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane for Western blot analysis. An antibody targeting the His tag was used to detect the expressed target protein. The distribution of MaSp1 polypeptides on the Western blot membrane was the same as that of the Coomassie blue-stained SDS-PAGE bands, indicating that the bands observed on SDS page were indeed MaSp1. Due to the higher sensitivity of Western blotting, the MaSp1 polypeptides larger than 110 kDa were also visible, which was more obvious than that on the Coomassie blue gel and further confirmed the authenticity of MaSp1 proteins larger than 110 kDa (Fig. 3B). The distribution of FSLP polypeptides again was similar to that of MaSp1, as indicated in Fig. 3B.
The MaSp1 and FSLP samples were subsequently analyzed by liquid chromatography mass spectrometry (LC-MS). The protein sample was in-gel digested with proteinase K and subjected to LC-MS identification. A characteristic peptide sequence of MaSp1 was successfully identified in the mass spectrum. As indicated in Fig. 3C, a series of peaks representing digested signature oligopeptides was found in the mass spectrum, including the following: y1, R; y2, RG; y3, RGS; y4, RGST; y5, RGSTS; y6, RGSTSR; and y7, RGSTSRL, the digestion products by proteinase K from the C terminus. Digestion products from the N terminus were also present in the spectrum, including the following: b2, DL; b5, DLRST; b6, DLRSTS; and b7, DLRSTSG. Similarly, FSLP was successfully identified with LC-MS. The following signature digestion products were found in the mass spectrum: y1, S; y2, SG; y3, SGG; b1, D; b2, DL; b3, DLR; b4, DLRS; b5, DLRST; b6, DLRSTG; b7, DLRSTGG; b8, DLRSTGGA; b9, DLRSTGGAG; and b10, DLRSTGGAGS (Fig. 3C).
The results of Western blot analysis and LC-MS further confirmed the successful expression of recombinant MaSp1 and FSLP proteins.
Product titer of MaSp1 and FSLP polypeptides.
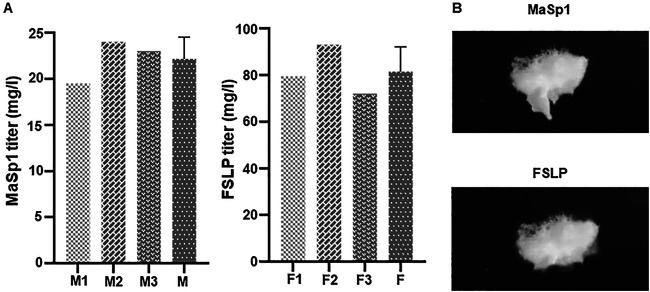
The protein samples were prepared from 2-liter shake-flask cultures of E. coli and subjected to Ni-NTA resin column purification. Subsequently, the eluted protein samples were dialyzed against water and the protein concentration was measured by the Bradford method. Although a loss of target protein during the purification process is unavoidable, we were able to calculate the product titer for both proteins, 22.1 mg · liter−1 for MaSp1 and 81.5 mg · liter−1 for FSLP. The cell dry weight yields of MaSp1 and FSLP were 4.4 g/kg and 16.5 g/kg, respectively (Fig. 4A). We also obtained protein samples of MaSp1 and FSLP by freeze-drying (Fig. 4B).
FIG 4.
Product titer of MaSp1 protein and FSLP protein. (A) Concentration of purified spider silk protein was measured by the Bradford method. (Left) The product titer of purified MaSp1 protein. M1, M2, and M3 represent the weight of three independently produced samples; M is the average of M1, M2, and M3. (Right) The product titer of purified FSLP. F1, F2, and F3 represent the weights of three independently produced samples; F is the average of F1, F2, and F3. (B) The dialyzed silk samples were freeze-dried. (Upper) Freeze-dried sample of MaSp1. (Lower) Freeze-dried sample of FSLP.
As we mentioned previously, the mRNA circularization efficiency of MaSp1 and FSLP was 8.5% and 36.7%, respectively (Fig. 2C). We hypothesized that the 4-fold difference between the titer of FSLP and MaSp1 might be related to the mRNA circularization efficiency. The size of MaSp1 mRNA might cause the low circularization efficiency, resulting in a low working cmRNA concentration and decreased product titer, so that the length of the repeat unit might be a key factor to decide whether the cmRNA is an optimal strategy to express fibrous proteins.
Compared with previous work (Table 1), this work provides a simple innovative strategy to express recombinant spidroins. For intensively studied MaSp1, our production titer was relatively low compared with production based on long repeat DNA template. However, for the production of larger and new FSLP, the cmRNA expression strategy provides a much higher yield than that of the traditional method, although different hosts were employed (42, 43). The comparison suggested the largest advantage of this technique was the time, investment, and labor to construct the DNA to express various fibrous proteins, which allows research on many more forms of fibrous protein than before.
TABLE 1.
Different host system performances in the production of recombinant spidroins
| Expression host | Spider silk protein | Size (kDa) | Reported maximum yield | Reference or source |
|---|---|---|---|---|
| E. coli | MaSp1 | 100.7 | 2,700 mg/liter | 25 |
| 284.9 | 500 mg/liter | |||
| E. coli | MaSp1 | 285 | 2,000 mg/liter | 47 |
| 556 | 1,240 mg/liter | |||
| P. pastoris | MaSp1 | 65 | 663 mg/liter | 28 |
| S. cerevisiae | MaSp1 | 94 | 450 mg/liter | 48 |
| E. coli | MaSp1 | 10–110 | 22.1 mg/liter | This work |
| Transgenic mice | MaSp1 | 40 | 11.7 mg/liter | 49 |
| N. tobacum (leaf) | Flagelliform | >72 to >250 | 36 mg/kg | 42 |
| N. tobacum (seed) | Flagelliform | >72 to >460 | 190 mg/kg | 43 |
| E. coli | FSLP | 10–90 | 16.5 g/kg | This work |
Conclusions.
In this work, we designed and established the cmRNA technique for convenient heterologous expression of large proteins containing tandem repeats, such as MaSp1 and FSLP, which belong to a group of important biomaterials with superior performance. The cmRNA provides ribosomes with an infinite translation template to travel along, and long repeat peptides were produced. With this technique, recombinant MaSp1 and FSLPs larger than 110 and 90 kDa, respectively, containing tens of repeat units, were produced. This technique makes the artificial construction of various silk proteins and proteins with similar structure very convenient with minimal cost and labor intensity. Such a revolutionary technique will allow researchers to easily build, study, and experiment with a large number of silk-like proteins with sequences derived from either natural genes or artificial designs. We expect a significantly accelerated development of fibrous protein-based biomaterials with the cmRNA technique.
MATERIALS AND METHODS
Plasmid construction.
The sequences encoding MaSp1 protein monomer, FSLP protein monomer, and group I self-splicing intron were ordered as fully synthetic DNA (GenScript, China) and cloned into the pET21b vector by Golden Gate assembly (44). All the DNA templates were amplified by PCR using Phusion DNA polymerase (NEB, USA). PCR products were gel purified, digested with DpnI (NEB, USA) to remove the methylated template, and assembled via Golden Gate assembly (45). The primers and plasmids are listed in Table 2 and Table S1 in the supplemental material, respectively.
TABLE 2.
Oligonucleotides used for this study
| Primer | Sequence | Note |
|---|---|---|
| cirMaSp1 BK-F | CCAGGTCTCACACCACCACCACCACCAC | pcirMaSp1 construction |
| cirMaSp1 BK-R | CCAGGTCTCAGGTAGCATTATGTTCAGATAAGGTCGTTAATCT | |
| cirMaSp1-F | CCAGGTCTCATACCAGCGGTCGCGGCGGTCTGGG | |
| cirMaSp1-R | CCAGGTCTCAGGTGAGTACCCTGAGAACCCAGGCCGC | |
| cirFSLP-F | TACCGGGGGAGCAGGCGGATCAGG | pcirFSLP construction |
| cirFSLP-R | GGTGGTAGCCACCCGGACCGCTGC | |
| MaSp1 linear donor | TTGCAGCCGCAGCCTGGCCGTCGTTTTAC | Hybridization with non-BSJ sequence of MaSp1 transcripts |
| MaSp1 linear acceptor | CAGGAAACAGCTATGACCCAGCACCGCCCA | |
| MaSp1 circular donor | AACGCAGATCCATCTGGCCGTCGTTTTAC | Hybridization with BSJ sequence of MaSp1 cmRNA |
| MaSp1 circular acceptor | CAGGAAACAGCTATGACCGACCGCTGGTAG | |
| FSLP linear donor | GCGCCGTCGATCGCTGGCCGTCGTTTTAC | Hybridization with non-BSJ sequence of FSLP transcripts |
| FSLP linear acceptor | CAGGAAACAGCTATGACGATCGGACCATCT | |
| FSLP circular donor | AACGCAGATCCATCTGGCCGTCGTTTTAC | Hybridization with BSJ sequence of FSLP cmRNA |
| FSLP circular acceptor | CAGGAAACAGCTATGACGCTCCCCCGGTAG | |
| M13-F | GTAAAACGACGGCCAG | Amplification of ligated products |
| M13-R | CAGGAAACAGCTATGAC |
Silk protein expression in shake flasks.
For MaSp1 protein expression, E. coli BL21 cells were incubated in a 2-liter flask containing 500 mL of TB (Terrific broth) medium at 30°C, with shaking at 220 rpm. FSLP was expressed at 25°C under the same conditions. The culture was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) when the optical density (OD) reached 0.4, and cells were collected after 18 h of expression.
SplintQuant method.
We used the RNAprep pure cell/bacterium kit (Tiangen) to extract RNA of strains expressing MaSp1 and FSLP. DNase (Promega) was used to digest the genome and plasmid. The SplintQuant method was adapted from Conn and Conn (38) to quantify the copy number of mRNA containing the BSJ and a region without BSJ. Two custom DNA oligonucleotides (donor and acceptor) were designed to be complementary to all isoforms of the MaSp1 and FSLP transcripts, while another two custom DNA oligonucleotides were used to be complementary to the regions containing the BSJ, which only exists in the circularized mRNA form (all oligonucleotides are listed in Table 1). The DNA/RNA hybrid-forming system was the following: 5× oligonucleotide annealing buffer (2 μL), 10 μM donor oligonucleotide (1 μL), 10 μM acceptor oligonucleotide (1.1 μL), total RNA (100 ng), and nuclease-free water added to 10 μL. The no template control was a system without the RNA. The reaction condition was 95°C to 85°C for 2 min and then cooled to 25°C at a rate of −0.1°C s−1. SplintR ligase (NEB) then was used to ligate the DNA oligomers on the splint.
We performed qPCR of ligated product using the M13 primer pair. Absolute expression abundance was calculated using standard curves. The standard substances of mRNA and cmRNA (MaSp1 and FSLP) were amplified by M13 primer pair, quantified spectrophotometrically, and serially diluted to produce standard samples at different concentrations. After the Cq values of samples from both standard sets were determined by RT-PCR, the logarithm of the copy number of the standard sample was taken as the abscissa, and the measured threshold cycle (CT) value was taken as the ordinate to draw a standard curve. RT-PCR was performed on a LightCycler 96 instrument (Roche) using the Fast SYBR green master mix (Toyobo). Each experiment was performed in biological triplicates. The primer sequences and standard curves are shown in Table 2 and Fig. S2, respectively.
Protein purification.
Cells were resuspended in ice-cold lysis buffer (50 mM NaCl, Tris-HCl, 10 mM imidazole, pH 8.0) and passed through a homogenizer (JNBIO) 3 times at 1,000 lb/in2. Cell debris was pelleted by centrifugation at 18,000 rpm and 4°C for 30 min. The resulting supernatant was mixed with 1 mL of Ni-NTA resin (GE Healthcare), which was preequilibrated with lysis buffer. The protein-bound resin was subsequently loaded onto a column at 4°C and then washed three times with 20 mL wash buffer (150 mM NaCl, 50 mM imidazole, 50 mM Tris-HCl, pH 8.0). Finally, 5 mL of elution buffer (250 mM imidazole, 50 mM NaCl, 50 mM Tris-HCl, pH 8.0) was used to elute the protein. The obtained MaSp1 protein solution was dialyzed against acetate buffer, pH 4.0, for 24 h and freeze-dried. FSLP was purified using the same method.
Western blot analysis.
The identity of the target protein was confirmed by Western blotting after protein purification. Proteins were separated on 5 to 20% SDS-PAGE gels and transferred to a polyvinylidene difluoride (PVDF) membrane. The membranes were blocked with 5% bovine serum albumin in Tris-buffered saline Tween (TBST) buffer for 1 h at room temperature and incubated with an antibody against the His tag overnight at 4°C. The membrane then was incubated with a goat anti-mouse secondary antibody for 1 h at room temperature, and the bands were developed using enhanced chemiluminescence (ECL) reagent, which was used to expose X-ray photographic films in a darkroom. Finally, the bands were recorded using Quantity One software.
In-gel protein digestion.
Following Coomassie brilliant blue staining, protein bands were excised from the SDS-PAGE gel, and in-gel digestion was performed by following a previously reported protocol (46). We used proteinase K (50 μg/mL; Sigma, USA) to digest the proteins for 3 h at 56°C; 100 μL acetonitrile (60%, vol/vol) with formic acid (5%, vol/vol) then was added to stop the digestion, and the supernatant was pipetted out and collected. This step was repeated several times, and the collected supernatants were combined and dried under vacuum.
Mass spectrometry.
About 2 μg total peptides was loaded onto a C18 nanocolumn (25 cm) using the EASY-nLC system (Thermo Fisher Scientific) and eluted for 120 min by using a 4 to 90% acetonitrile fraction-optimized nonlinear gradient with 0.1% formic acid. The MS analysis was conducted on an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific). The difference multiple was >1.2 (Beijing ZD-Health BioMedical Technology Company Limited).
Data availability.
We have provided supporting and necessary data for publication of the article. All supporting data are present in the article and the supplemental material. The strains and plasmid associated with this work will be made physically available by the authors upon reasonable request.
ACKNOWLEDGMENTS
We thank the reviewer for pointing out the problem of the original quantification method for calculation of circulation efficiency of cmRNAs so that we could find a better method for that.
A provisional patent has been submitted in part entailing the reported approach.
This research was financially supported by the National Key Research and Development Program of China (2018YFA0903700), the National Natural Science Foundation of China (32171449 and 31770105), and a Tianjin Synthetic Biotechnology Innovation Capacity Improvement Project (TSBICIP-KJGG-017).
X.Z. and C.B. designed the study, analyzed data, and wrote the manuscript. L.L. designed the study, performed experiments, analyzed the data, and wrote the manuscript. P.W. designed the study and analyzed the data. D.Z. designed the study. L.Z., J.T., W.L., J.S., and Y.L. performed experiments.
Footnotes
Supplemental material is available online only.
Contributor Information
Changhao Bi, Email: bi_ch@tib.cas.cn.
Xueli Zhang, Email: zhang_xl@tib.cas.cn.
Haruyuki Atomi, Kyoto University.
REFERENCES
- 1.Gu L, Jiang Y, Hu J. 2019. Scalable spider-silk-like supertough fibers using a pseudoprotein polymer. Adv Mater 31:e1904311. 10.1002/adma.201904311. [DOI] [PubMed] [Google Scholar]
- 2.Rockwood DN, Preda RC, Yücel T, Wang X, Lovett ML, Kaplan DL. 2011. Materials fabrication from Bombyx mori silk fibroin. Nat Protoc 6:1612–1631. 10.1038/nprot.2011.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ricki L. 1996. Unraveling the weave of spider silk. Bioscience 46:636–638. [Google Scholar]
- 4.Newman J, Newman C. 1995. Oh what a tangled web: the medicinal uses of spider silk. Int J Dermatol 34:290–292. 10.1111/j.1365-4362.1995.tb01600.x. [DOI] [PubMed] [Google Scholar]
- 5.Bai H, Ju J, Zheng Y, Jiang L. 2012. Functional fibers with unique wettability inspired by spider silks. Adv Mater 24:2786–2791. 10.1002/adma.201200289. [DOI] [PubMed] [Google Scholar]
- 6.Gustafsson L, Jansson R, Hedhammar M, van der Wijngaart W. 2018. Structuring of functional spider silk wires, coatings, and sheets by self-assembly on superhydrophobic pillar surfaces. Adv Mater. 10.1002/adma.201704325. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Zhou Z, Sun L, Liu Z, Xia X, Tao TH. 2018. Genetically engineered biofunctional triboelectric nanogenerators using recombinant spider silk. Adv Mater 30:e1805722. 10.1002/adma.201805722. [DOI] [PubMed] [Google Scholar]
- 8.Liu X, Shi L, Wan X, Dai B, Yang M, Gu Z, Shi X, Jiang L, Wang S. 2021. A spider-silk-inspired wet adhesive with supercold tolerance. Adv Mater 33:e2007301. 10.1002/adma.202007301. [DOI] [PubMed] [Google Scholar]
- 9.Xu M, Lewis RV. 1990. Structure of a protein superfiber: spider dragline silk. Proc Natl Acad Sci USA 87:7120–7124. 10.1073/pnas.87.18.7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinman MB, Lewis RV. 1992. Isolation of a clone encoding a second dragline silk fibroin. Nephila clavipes dragline silk is a two-protein fiber. J Biol Chem 267:19320–19324. 10.1016/S0021-9258(18)41777-2. [DOI] [PubMed] [Google Scholar]
- 11.Ayoub NA, Garb JE, Tinghitella RM, Collin MA, Hayashi CY. 2007. Blueprint for a high-performance biomaterial: full-length spider dragline silk genes. PLoS One 2:e514. 10.1371/journal.pone.0000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sponner A, Schlott B, Vollrath F, Unger E, Grosse F, Weisshart K. 2005. Characterization of the protein components of Nephila clavipes dragline silk. Biochemistry 44:4727–4736. 10.1021/bi047671k. [DOI] [PubMed] [Google Scholar]
- 13.Winkler S, Kaplan DL. 2000. Molecular biology of spider silk. J Biotechnol 74:85–93. 10.1016/S1389-0352(00)00005-2. [DOI] [PubMed] [Google Scholar]
- 14.Thiel BL, Viney C. 1996. Beta sheets and spider silk. Science 273:1480–1481. 10.1126/science.273.5281.1480. [DOI] [PubMed] [Google Scholar]
- 15.Simmons AH, Michal CA, Jelinski LW. 1996. Molecular orientation and two-component nature of the crystalline fraction of spider dragline silk. Science 271:84–87. 10.1126/science.271.5245.84. [DOI] [PubMed] [Google Scholar]
- 16.Gosline JM, DeMont ME, Denny MW. 1986. The structure and properties of spider silk. Endeavour 10:37–43. 10.1016/0160-9327(86)90049-9. [DOI] [Google Scholar]
- 17.Hayashi CY, Lewis RV. 1998. Evidence from flagelliform silk cDNA for the structural basis of elasticity and modular nature of spider silks. J Mol Biol 275:773–784. 10.1006/jmbi.1997.1478. [DOI] [PubMed] [Google Scholar]
- 18.Hayashi CY, Lewis RV. 2000. Molecular architecture and evolution of a modular spider silk protein gene. Science 287:1477–1479. 10.1126/science.287.5457.1477. [DOI] [PubMed] [Google Scholar]
- 19.Andersen SO. 1970. Amino acid composition of spider silks. Comp Biochem Physiol Physiol 35:705–711. 10.1016/0010-406X(70)90988-6. [DOI] [Google Scholar]
- 20.Babb PL, Lahens NF, Correa-Garhwal SM, Nicholson DN, Kim EJ, Hogenesch JB, Kuntner M, Higgins L, Hayashi CY, Agnarsson I, Voight BF. 2017. The Nephila clavipes genome highlights the diversity of spider silk genes and their complex expression. Nat Genet 49:895–903. 10.1038/ng.3852. [DOI] [PubMed] [Google Scholar]
- 21.Brooks AE, Steinkraus HB, Nelson SR, Lewis RV. 2005. An investigation of the divergence of major ampullate silk fibers from Nephila clavipes and Argiope aurantia. Biomacromolecules 6:3095–3099. 10.1021/bm050421e. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, Sponner A, Porter D, Vollrath F. 2008. Proline and processing of spider silks. Biomacromolecules 9:116–121. 10.1021/bm700877g. [DOI] [PubMed] [Google Scholar]
- 23.Fahnestock SR, Yao Z, Bedzyk LA. 2000. Microbial production of spider silk proteins. J Biotechnol 74:105–119. 10.1016/s1389-0352(00)00008-8. [DOI] [PubMed] [Google Scholar]
- 24.Widmaier DM, Tullman-Ercek D, Mirsky EA, Hill R, Govindarajan S, Minshull J, Voigt CA. 2009. Engineering the Salmonella type III secretion system to export spider silk monomers. Mol Syst Biol 5:309. 10.1038/msb.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia XX, Qian ZG, Ki CS, Park YH, Kaplan DL, Lee SY. 2010. Native-sized recombinant spider silk protein produced in metabolically engineered Escherichia coli results in a strong fiber. Proc Natl Acad Sci USA 107:14059–14063. 10.1073/pnas.1003366107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vendrely C, Scheibel T. 2007. Biotechnological production of spider-silk proteins enables new applications. Macromol Biosci 7:401–409. 10.1002/mabi.200600255. [DOI] [PubMed] [Google Scholar]
- 27.Heidebrecht A, Scheibel T. 2013. Recombinant production of spider silk proteins. Adv Appl Microbiol 82:115–153. 10.1016/B978-0-12-407679-2.00004-1. [DOI] [PubMed] [Google Scholar]
- 28.Fahnestock SR, Bedzyk LA. 1997. Production of synthetic spider dragline silk protein in Pichia pastoris. Appl Microbiol Biotechnol 47:33–39. 10.1007/s002530050884. [DOI] [PubMed] [Google Scholar]
- 29.O’Brien JP, Fahnestock SR, Termonia Y, Gardner KH. 1998. Nylons from nature: synthetic analogs to spider silk. Adv Mater 10:1185–1195. . [DOI] [Google Scholar]
- 30.Fahnestock SR, Irwin SL. 1997. Synthetic spider dragline silk proteins and their production in Escherichia coli. Appl Microbiol Biotechnol 47:23–32. 10.1007/s002530050883. [DOI] [PubMed] [Google Scholar]
- 31.Perriman R, Ares M, Jr.. 1998. Circular mRNA can direct translation of extremely long repeating-sequence proteins in vivo. RNA 4:1047–1054. 10.1017/s135583829898061x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ford E, Ares M, Jr.. 1994. Synthesis of circular RNA in bacteria and yeast using RNA cyclase ribozymes derived from a group I intron of phage T4. Proc Natl Acad Sci USA 91:3117–3121. 10.1073/pnas.91.8.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puttaraju M, Been MD. 1992. Group I permuted intron-exon (PIE) sequences self-splice to produce circular exons. Nucleic Acids Res 20:5357–5364. 10.1093/nar/20.20.5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hedberg A, Johansen SD. 2013. Nuclear group I introns in self-splicing and beyond. Mob DNA 4:17. 10.1186/1759-8753-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eger N, Schoppe L, Schuster S, Laufs U, Boeckel JN. 2018. Circular RNA splicing. Adv Exp Med Biol 1087:41–52. 10.1007/978-981-13-1426-1_4. [DOI] [PubMed] [Google Scholar]
- 36.Costello A, Lao NT, Barron N, Clynes M. 2019. Continuous translation of circularized mRNA improves recombinant protein titer. Metab Eng 52:284–292. 10.1016/j.ymben.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 37.Chen D-F, Zhang L-J, Tan K, Jing Q. 2018. Application of droplet digital PCR in quantitative detection of the cell-free circulating circRNAs. Biotechnol Biotec Eq 32:116–123. 10.1080/13102818.2017.1398596. [DOI] [Google Scholar]
- 38.Conn V, Conn SJ. 2019. SplintQuant: a method for accurately quantifying circular RNA transcript abundance without reverse transcription bias. RNA 25:1202–1210. 10.1261/rna.070953.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quentin V, Paukstelis PJ, Eric W, Lambowitz AM, Cech TR. 2008. Toward predicting self-splicing and protein-facilitated splicing of group I introns. RNA 14:2013–2029. 10.1261/rna.1027208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dittmar KA, Sørensen MA, Elf J, Ehrenberg M, Pan T. 2005. Selective charging of tRNA isoacceptors induced by amino-acid starvation. EMBO Rep 6:151–157. 10.1038/sj.embor.7400341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim S, Lee SB. 2006. Rare codon clusters at 5'-end influence heterologous expression of archaeal gene in Escherichia coli. Protein Expr Purif 50:49–57. 10.1016/j.pep.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 42.Hauptmann V, Weichert N, Menzel M, Knoch D, Paege N, Scheller J, Spohn U, Conrad U, Gils M. 2013. Native-sized spider silk proteins synthesized in planta via intein-based multimerization. Transgenic Res 22:369–377. 10.1007/s11248-012-9655-6. [DOI] [PubMed] [Google Scholar]
- 43.Weichert N, Hauptmann V, Helmold C, Conrad U. 2016. Seed-specific expression of spider silk protein multimers causes long-term stability. Front Plant Sci 7:6. 10.3389/fpls.2016.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Engler C, Kandzia R, Marillonnet S. 2008. A one pot, one step, precision cloning method with high throughput capability. PLoS One 3:e3647. 10.1371/journal.pone.0003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hillson NJ, Rosengarten RD, Keasling JD. 2012. j5 DNA assembly design automation software. ACS Synth Biol 1:14–21. 10.1021/sb2000116. [DOI] [PubMed] [Google Scholar]
- 46.Shevchenko A, Wilm M, Vorm O, Mann M. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68:850–858. 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 47.Bowen CH, Dai B, Sargent CJ, Bai W, Ladiwala P, Feng H, Huang W, Kaplan DL, Galazka JM, Zhang F. 2018. Recombinant spidroins fully replicate primary mechanical properties of natural spider silk. Biomacromolecules 19:3853–3860. 10.1021/acs.biomac.8b00980. [DOI] [PubMed] [Google Scholar]
- 48.Sidoruk KV, Davydova LI, Kozlov DG, Gubaidullin DG, Glazunov AV, Bogush VG, Debabov VG. 2015. Fermentation optimization of a Saccharomyces cerevisiae strain producing 1F9 recombinant spidroin. Appl Biochem Microbiol 51:766–773. 10.1134/S0003683815070066. [DOI] [Google Scholar]
- 49.Xu HT, Fan BL, Yu SY, Huang YH, Zhao ZH, Lian ZX, Dai YP, Wang LL, Liu ZL, Fei J, Li N. 2007. Construct synthetic gene encoding artificial spider dragline silk protein and its expression in milk of transgenic mice. Anim Biotechnol 18:1–12. 10.1080/10495390601091024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1, Fig. S1 and S2, and DNA sequences for vector components. Download aem.00028-22-s0001.pdf, PDF file, 0.1 MB (153.5KB, pdf)
Data Availability Statement
We have provided supporting and necessary data for publication of the article. All supporting data are present in the article and the supplemental material. The strains and plasmid associated with this work will be made physically available by the authors upon reasonable request.