ABSTRACT
The HIV Env glycoprotein is the surface glycoprotein responsible for viral entry into CD4+ immune cells. During infection, Env also serves as a primary target for antibody responses, which are robust but unable to control virus replication. Immune evasion by HIV-1 Env appears to employ complex mechanisms to regulate what antigenic states are presented to the immune system. Immunodominant features appear to be distinct from epitopes that interfere with Env functions in mediating infection. Further, cell-cell transmission studies indicate that vulnerable conformational states are additionally hidden from recognition on infected cells, even though the presence of Env at the cell surface is required for viral infection through the virological synapse. Cell-cell infection studies support that Env on infected cells is presented in distinct conformations from that on virus particles. Here we review data regarding the regulation of conformational states of Env and assess how regulated sorting of Env within the infected cell may underlie mechanisms to distinguish Env on the surface of virus particles versus Env on the surface of infected cells. These mechanisms may allow infected cells to avoid opsonization, providing cell-to-cell infection by HIV with a selective advantage during evolution within an infected individual. Understanding how distinct Env conformations are presented on cells versus viruses may be essential to designing effective vaccine approaches and therapeutic strategies to clear infected cell reservoirs.
KEYWORDS: HIV, envelope, conformation, endocytosis, virological synapse, Env, neutralizing antibodies, protein trafficking
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) is a lentivirus that infects CD4 receptor-expressing (CD4+) immune cells. Untreated, HIV-1 causes a chronic infection that leads to AIDS, characterized by CD4+ T cell depletion that leaves patients vulnerable to opportunistic infections and malignancies (1). The use of effective anti-retroviral therapy (ART) suppresses viral replication to prevent HIV-1 transmission and progression to AIDS (2, 3). However, there remain numerous obstacles to overcome, including developing a vaccine and cure for HIV-1.
The HIV-1 envelope (Env) glycoprotein is particularly important in HIV-1 prevention and treatment efforts because it mediates viral entry into host cells (4). The mature Env consists of three exterior gp120 subunits that bind the target cell receptor (CD4) and coreceptor (CCR5 or CXCR4), as well as three non-covalently associated transmembrane gp41 subunits that mediate membrane fusion (5). Env is derived from a gp160 precursor synthesized, folded, trimerized, and glycosylated in the host cell rough endoplasmic reticulum (ER) and subsequently cleaved by host furin-like proteases in the Golgi apparatus (6, 7). The mature trimers transit to the cell surface and are incorporated into budding virions via the endosomal recycling compartment (8). This mature Env is capable of mediating both cell-free infection and cell-cell infection: in the former, virions released from infected host cells infect non-adjacent, uninfected target cells; in the latter, direct cell-cell connections called virological synapses (VS) mediate direct HIV-1 transmission from an infected host cell to an adjacent, uninfected target cell (9, 10).
In addition to mediating viral entry, Env is the only virus-specific antigen exposed on the surface of virions and infected cells (4). Consequently, it is the primary target of host humoral responses, including neutralizing antibodies and Fc-dependent cell-mediated mechanisms such as antibody-dependent cellular cytotoxicity (ADCC), antibody-dependent cellular phagocytosis (ADCP), and complement activation (11–14). The HIV-1 Env has evolved multiple mechanisms to successfully evade these immune responses, including extensive glycan shielding, sequence-variable loops, and conformational flexibility (11–13, 15). Recent single-molecule fluorescence resonance energy transfer (FRET) imaging studies of HIV-1 Env on virus particles reveal that the mature, unliganded Env also samples multiple conformational states to conceal conserved, immunodominant antigens (16).
Here, we review current data surrounding the HIV-1 conformational states of Env on cell-free virus versus the infected cell surface and examine the influence of endosomal recycling, sorting, and post-translational processing of Env in influencing the states that Env can assume in different sites. We consider evidence that differential regulation of the Env conformational equilibrium on virus particles verses the cell surface is regulated by the host cell biosynthetic pathway that recycles heterogeneous Env forms from the cell surface and specifically sorts mature Env to budding virus particles. A clear understanding of how the Env conformational states present on infected cells and virions may be essential in the future development of HIV-1 vaccines and therapies.
ENV BIOSYNTHESIS AND THE PATHWAY TO THE VIRUS PARTICLE
During infection, Env is synthesized in the late stage of the virus life cycle on the rough endoplasmic reticulum as a polyprotein precursor from a singly-spliced viral mRNA. The nascent Env contains a signal sequence at its N-terminus that targets the cotranslational insertion of the protein into the ER and is removed by a signal peptidase resulting in a type I membrane topology with the N-terminus inserted in the lumen of the ER. Variations in the signal peptide (SP) can influence the glycosylation of Env and its ability to bind to antibodies (17). It is proposed that the SP remains attached to the nascent gp160 in the ER and is cleaved before delivery to the Golgi apparatus (18, 19). Interestingly, the substitution of SP processed with greater efficiency can result in under-processed glycans and altered structure and antigenicity (20).
The Env glycoprotein is heavily glycosylated with around 30 N-linked glycosylation sites modified within the ER-Golgi system by many cellular enzymes. Glycan represents up to half of the mass of Env and occupies 50% to 70% of the surface of Env and thus has a major effect on the antigenicity of the protein. The proper folding of Env and trimerization requires that correct disulfide bonds be made, and interactions with chaperones may facilitate this process. The dense glycan shield contributes greatly to the heterogeneity of the recognized structures on the cell and virion surface. The dense network of high mannose glycans at the surface is often referred to as a glycan shield, decreasing the overall affinity of antibodies that target Env (21–23). Many broadly neutralizing antibodies are glycan dependent, and the loss or gain of N-linked glycan sites is thought to be a common immune escape mechanism during chronic infection (12, 24).
In the Golgi, gp160 precursor is proteolytically cleaved by furin-like proteases into gp120/gp41, an event that is required for the activation of Env (25–30). Uncleaved Env cannot mediate viral membrane fusion and has been described as assuming a non-native state with a higher degree of processed glycans than seen on cleaved gp120 (31). Not all Env is cleaved before trafficking to the cell surface, and a large fraction of Env that is in the cell is uncleaved (32). However, a selective sorting mechanism supports preferential packaging of Env into virus particles which occurs at the plasma membrane where virus particles are formed (33).
The cell surface density of Env is maintained at very low levels by clathrin-mediated endocytic pathways (34). When at the cell surface, a membrane-proximal tyrosine motif, YXXL, is recognized by adapter protein 2 (AP-2) to recruit the clathrin machinery that mediates rapid endocytosis of Env from the surface (35–37). Perturbations of this endosomal internalization signal can alter the efficiency with which individual broadly neutralizing antibodies can block cell-to-cell transmission of HIV (38).
ENV CONFORMATIONAL STATES
Historically, one of the most significant challenges in structural biology has been developing a three-dimensional structural model for the HIV-1 Env glycoprotein, as its membrane association, conformational heterogeneity, and structural instability initially resisted crystallographic approaches (5, 39). Biophysical studies have suggested that the Env machinery is metastable, transitioning through different conformational states to enable receptor-driven membrane fusion (5). Env binding to target cell CD4 receptor triggers dramatic restructuring of Env from a “closed” to an “open” conformation that exposes the gp120 coreceptor binding site (CoRBS) as well as critical gp41 elements such as the heptad repeat 1 (HR1) coiled-coil and the fusion peptide (FP). Subsequent binding of the target cell coreceptor (CCR5 or CXCR4) to Env promotes FP insertion into the target cell membrane, as well as the association of HR1 and HR2 to form the highly stable six-helix bundle (6HB) that is believed to mediate viral-cell membrane fusion (40–42).
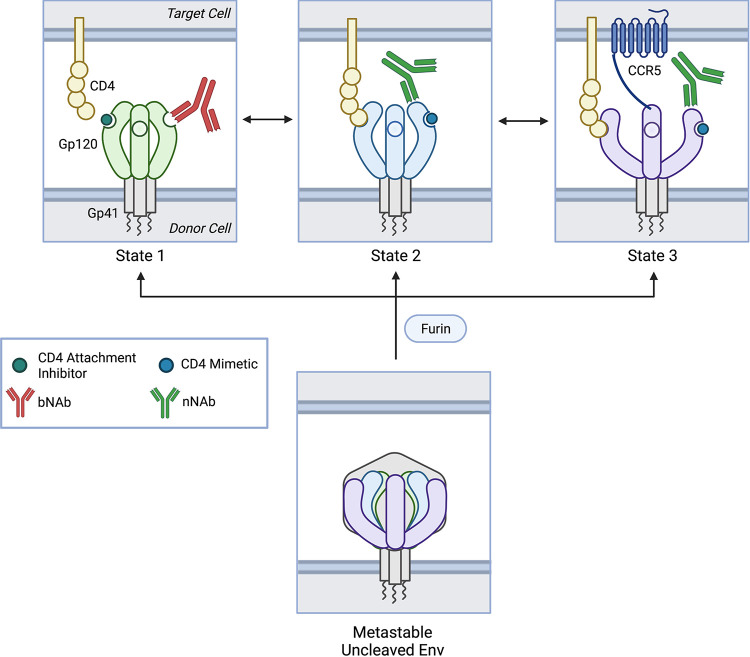
Recent single-molecule fluorescence resonance energy transfer (smFRET) studies have demonstrated that the native, mature, membrane-associated Env trimer is, in fact, inherently dynamic. In its unliganded form, Env spontaneously transitions between three distinct, intrinsic prefusion conformations as depicted in Fig. 1: the “closed” State 1, the obligate intermediate State 2, and the “open” State 3 (43, 44). The relative free energies of these conformational states dictate their occupancy in Env’s dynamic conformational equilibrium (44).
FIG 1.
Model of cleaved and uncleaved HIV-1 Env conformational states. The native, cleaved, membrane-associated HIV-1 Env trimer is metastable, occupying an equilibrium of three conformational states that are modulated by CD4 receptor and conformation-sensitive ligands. State 1 or the “closed” conformation is predominant. CD4 receptor binding triggers State 2, an obligate intermediate. The fully “open” conformation, State 3 or the three-CD4-bound conformation, is characterized by coreceptor binding site (CoRBS) formation. State 1 is bound and stabilized by attachment inhibitors and broadly neutralizing antibodies (bNAbs) whereas States 2 and 3 are favored by CD4 mimetic compounds (CD4mc) and non-neutralizing antibodies (nNAbs). The uncleaved Env trimer is similar metastable but favors States 2 and 3. Created with Biorender.com.
State 1 represents the predominant conformational state of the native, mature, unliganded membrane-associated Env trimer on the virus particle (43) (Fig. 1). In this conformation, the gp120 variable loops 1 and 2 (V1/V2) obscures most of the V3 loop at the trimer apex (43) while crucial structural elements such as the CoRBS and the HR1 coiled-coil are concealed.
State 2 represents an obligate intermediate conformation between States 1 and 3 (43) (Fig. 1). It likely encompasses a set of related conformations that reside in a local energy well in the Env trimer conformational landscape and are therefore accessed during the structural changes between States 1 and 3 (44). CD4 receptor binding stabilizes the downstream, lower energy states, leading to time-dependent progression from State 1, through State 2, to State 3 (45).
State 3 represents a more open three-CD4-bound Env trimer conformational state (43, 45–47) (Fig. 1). CD4 binding induces several structural changes such as movement of the V1/V2 loops from the trimer apex to the periphery (43, 46–52), reordering of the gp120 beta sheet bridging domain (46, 50, 52–54), CoRBS formation (46, 47, 55–58), and gp41 HR1 coiled-coil assembly at the trimer axis (46, 47, 52, 59–61). HIV-1 Env trimers that favor State 3 can mediate infection of target cells with low or no CD4 expression (44, 53, 62–70).
Notably, uncleaved Env (71) and Env with gp41 C-terminal tail truncation (ΔCT) (51, 54, 68, 72, 73) tend to sample States 2 and 3 more readily than mature, cleaved, membrane-associated Env. Similarly, all of the current high-resolution HIV-1 Env structures including BG505 SOSIP.664 gp140 complexed with various bNAbs and/or soluble CD4 (46, 47, 51, 54), JR-FL ΔCT complexed with bNAb PGT151 (73), and aldrithiol-2 (AT-2)-treated BaL (74) appear to predominantly occupy States 2 and 3 (45, 74). Therefore, the structure of State 1 remains unknown.
Env may also access off-pathway conformational states such as State 2A where membrane-associated CD4 interacts with Env to induce exposure of cluster A epitopes in the gp120 inner domain C1 to C2 region. These epitopes are targeted by anti-cluster A antibodies that mediate the majority of ADCC (75). Envs that favor States 2 and 3 (e.g., uncleaved Env) also tend to sample State 2A more readily (76).
The class of antiretrovirals that act as entry inhibitors has expanded recently to include conformation-sensitive attachment inhibitors (also known as conformational blockers), CD4 mimetic compounds (CD4mc). Other Env targeted antagonists include broadly neutralizing antibodies (bNAbs), and poorly or non-neutralizing antibodies (nNAbs), which can also be conformationally sensitive. Conformation-sensitive ligands are unique in that they bind Env and alter the relative free energies and/or activation barriers of the conformational equilibrium in order to stabilize certain states along the entry pathway (43, 44). They not only interfere with the first step of the viral life cycle, but also have the potential to stabilize Env in conformational states that may theoretically present a simplified antigenic target for evolving host immune responses.
Attachment inhibitors are small molecules that inhibit CD4-induced conformational changes at lower concentrations and CD4 binding at higher concentrations (50, 77–83). They reversibly bind to a gp120 pocket comprised of the α1-helix and the β20-21 hairpin adjacent to the CD4 binding loop (77, 84). The prototypical attachment inhibitor is fostemsavir (also known as BMS-663068), which was approved by the United States Food & Drug Administration (FDA) in July 2020 for the treatment of adults with multidrug-resistant HIV-1 infection (85). Temsavir is the active metabolite of fostemsavir, BMS-525629 and other molecules BMS-378806, 484, and 18a are described as having related properties (53, 79, 86, 87). Attachment inhibitors stabilize State 1 (Fig. 1) and, therefore, potently neutralize Env that predominantly occupies State 1, such as mature, cleaved, membrane-associated Env (43–45, 53, 87, 88). In contrast, Envs that favor States 2 and 3 tend to resist neutralization by attachment inhibitors (44, 53).
CD4mc are small molecules the bind the well-conserved gp120 Phe 43 cavity near the CD4 binding site (52, 63, 89, 90). They inhibit Env-mediated fusion and entry through multiple mechanisms, including competitively inhibiting CD4 binding (91), increasing gp120 shedding (65), and triggering a transient, premature, activated conformational state similar to that induced by CD4 that spontaneously and irreversibly inactivates (50, 62, 64, 65). Examples include NBD-556, YYA-021, the DMJ compounds, and BNM-III-170 (100-103). Because CD4mc stabilizes State 2 and 3 (Fig. 2), Env that predominantly occupies State 1 is relatively resistant to neutralization by CD4mc (88, 92). Conversely, Envs that favor States 2 and 3 are highly susceptible to neutralization by CD4mc (43, 44, 53, 65, 70, 87, 90). CD4mc can also stabilize State 2A in combination with certain nNAbs or HIV-positive human sera (76, 93–98).
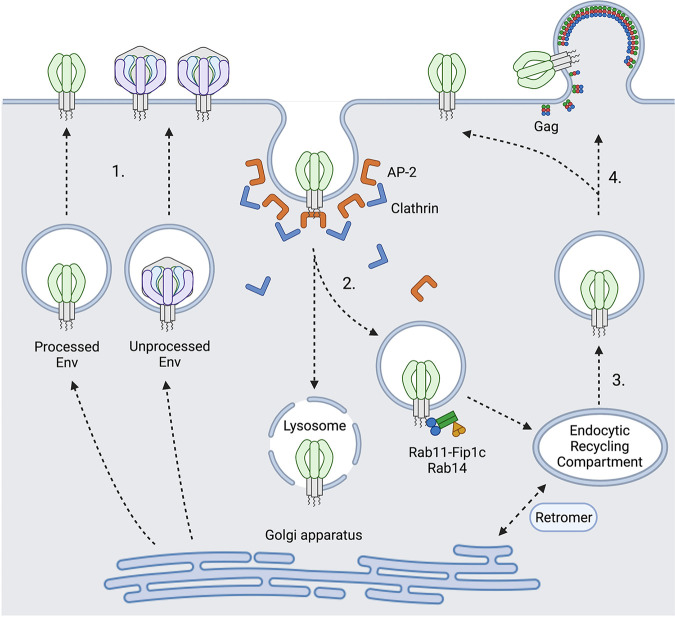
FIG 2.
HIV-1 Env trafficking, recycling, and egress. (1) Env trafficking to plasma membrane after synthesis and transit through endoplasmic reticulum and Golgi apparatus. Both fully processed and unprocessed forms of Env can transit to the cell surface. (2) Env endocytosis is initiated by AP2-dependent recruitment of the clathrin endocytosis pathway at the plasma membrane. Early endosomes traffic to recycling endosomes mediated by an interaction of the gp41 cytoplasmic tail with Rab11-FIP1C and Rab14. Alternative pathways for lysosomal degradation of a fraction of endocytosed Env are proposed, but not well documented. (3) Endocytosed Env sorted for outward trafficking or hypothetically could travel to retrograde transport pathways to Golgi apparatus for further proteolytic processing and/or glycosylation. (4) Virus particles are enriched for fully processed Env, which is targeted to the viral assembly site at the plasma membrane. Figure created with Biorender.com.
BNAbs can bind heavily shielded, conserved epitopes on the surface of the “closed” State 1 conformation and can neutralize a wide range of HIV-1 strains (11, 99–101). They rarely occur in natural HIV infection, usually require several years to develop (102–104), and have proven quite challenging to elicit through vaccination (105). Common epitopes include the CD4 binding site (CD4bs), the gp120-gp41 interface, the membrane-proximal external region (MPER), and quaternary and/or glycan-dependent epitopes in the V1V2 and V3 domains (106). Envs that predominantly occupy State 1 are highly susceptible to neutralization by bNAbs (32, 43–45) (Fig. 1), while Envs that favor States 2 and 3 are relatively resistant (44, 66, 68, 70, 107).
NNAbs preferentially recognize non-State 1 conformational states with narrow to no ability to neutralize HIV-1 strains. They appear readily elicited during natural HIV-1 infection and by animal or human Env protein immunization (11, 32, 100, 101, 108–119). Common epitopes include the CD4-induced (CD4i) or CoRBS and V3 epitopes (100, 101, 109). While nNAbs poorly neutralize Envs that predominantly occupy State 1 (50, 77, 80, 82, 88), they are able to potently neutralize Envs favoring States 2 and 3 (43, 44, 50, 53, 66–70, 87, 90, 107, 110, 120–126) (Fig. 1). Anti-cluster A and CoRBS nNAbs can stabilize State 2A in combination with membrane-associated CD4 or CD4mc (76, 93–98). NNAbs also can mediate ADCC, ADCP, and complement activation if they can detect relevant forms of Env displayed on the cell surface. Non-neutralizing responses can also impart significant selective pressure on viral evolution and have been associated with protection in several animal models and one human vaccine clinical trial (127–129).
Overall, the HIV-1 Env trimer is far more dynamic and heterogeneous than initially anticipated, occupying an equilibrium of conformational states dictated by each Env’s intrinsic conformational landscape. This energy landscape is in turn modulated by receptor and coreceptor binding, as well as conformation-sensitive ligands. The ability of a ligand to neutralize an Env trimer depends on its binding affinity as well as the conformational state equilibrium of the specific Env (130). Conversely, the conformational stability or instability of Env can influence its resistance to neutralization (43).
HIV-1 cell-to-cell infection.
HIV-1 infection can be initiated efficiently by cell-free virus particles or by infected cells through virological synapses (VS), which mediate efficient cell-to-cell infection in vitro (131–133). For the purposes of this review, we focus on the role of T cell-to-T cell infection in comparison with cell-free viral infection, to better understand how antibody responses may recognize these modes of infection with variable efficiency. HIV-1 cell-to-cell transmission between T cells and macrophage and trans-infection between dendritic cells and T cells are reviewed elsewhere (134, 135).
The VS is an adhesive structure formed at the cell-cell contact region. It is initiated by interaction between Env on the surface of HIV-infected T cells and CD4 on adjacent uninfected CD4 T cells (9, 131), and is further stabilized by intercellular adhesion molecules including intercellular adhesion molecule 1 (ICAM1), ICAM3 and lymphocyte function-associated antigen 1 (LFA-1) (136). These molecules are enriched at the VS and may enhance VS formation to facilitate cell-to-cell infection (136). Activated leukocyte cell adhesion molecule (ALCAM), which is critical for HIV-1 infected monocyte transmigration through the blood brain barrier in the CNS (137), promotes cell-cell aggregation and VS formation. The loss of ALCAM can diminish cell-to-cell transmission, which can be rescued by restoring cell-cell aggregation through other means (138). A minor allele of ALCAM is reported to be associated with accelerated disease progression (139). VS promotes cell-to-cell infection through dynamic recruitment of viral proteins to the cell-cell contact region (133) with the involvement of cytoskeletal rearrangement (131, 140–142). In addition to supporting transmission between infected and uninfected T cells, cell-to-cell transmission can also occur from infected macrophages to uninfected CD4 T cells (143).
Compared with cell-free infection, cell-to-cell infection demonstrates greater efficiency in vitro (9, 144, 145) and evidence for cell-cell infection is supported by in vivo small animal models (146, 147). In one model system, Murooka et al. determined that migration of HIV infected T cells from local draining lymph nodes to systemic circulation is required for systemic spread of HIV infection, implying that the dissemination of the infection requires cells as vehicles of dissemination (146). In another humanized mouse study, Law et al. observed that infections that were seeded with HIV-infected cells that were co-infected with two viral genotypes gave rise to newly infected co-infected cells, indicating that the genotypes inherited by newly infected cells is determined by the genotypes present in the input cells (147). In lymphoid tissues where susceptible CD4+ T cells are densely packed, immune surveillance creates continual opportunities for cell-cell interactions between infected and uninfected T cells (146, 147). HIV-infected donor cells can interact with multiple adjacent target cells and form polysynapses. The virus transmits through polysynapses at multiple membrane regions simultaneously to facilitate exponential viral growth, potentially contributing to evasion from immune responses (132). Another key feature of HIV-1 cell-to-cell transmission is that multiple copies of viral genomes may be transferred from donor to target cell, resulting in productively infected cells that carry multiple proviruses (147, 148). Multicopy infection may contribute to the virus’s genetic diversity by enabling co-transmission of genetic variants with non-mutant viruses and increasing the opportunity for mutational RT events that occur during each round of infection (149–152). Several studies from Sigal group find that the high copy number of HIV that occurs from cell-to-cell infection can contribute to enhanced resistance to antiretroviral drugs, facilitate persistence in the presence of antiretroviral drugs, and increase the speed of viral gene expression and cell death in infected cells (153–155). Seemingly in contrast to the evidence for multicopy infection seen in experimental models, studies of cells from infected patients have found that the HIV copy number in individual cells from people with HIV have observed that a large majority of cells >90% carry a single copy (156, 157). This may indicate that of those cells that survive or persist with HIV genomes in them, a majority are single copy. Further studies are needed to determine if these single copy infected cells are overrepresented in the cells that survive infection, while cells infected with multiple copies, may produce greater levels of virus experience greater rates of cell death (134, 154).
During chronic HIV-1 infection, circulating virus strains evolve to resist antibody neutralization where mutations allow viral isolates to evade contemporaneous antibody responses (12, 158, 159). Individuals that generate abundant high titer neutralizing antibodies against HIV Env have been identified from screens of chronically infected patients (104, 160–163). Because antibodies that can neutralize a broad range of viruses in vitro were not isolated from patients in whom viral replication was spontaneously controlled, it can be inferred that the virus replicating in these patients is resistant to the levels of bNAb that are circulating in these individuals (164, 165). The virus has evolved diverse mechanisms to evade and adapt rapidly to immune responses during each infection, including error-prone RNA pol, reverse transcriptase, and innate cellular cytosine deaminases. Cell-to-cell transmission is generally considered as another mechanism by which HIV-1 can evade antibody neutralization (149).
Cell-to-cell infection is more resistant to bNAb neutralization than cell-free transmission.
The development of single-cell antibody cloning technology led to the discovery of dozens of new generation bNAbs with extraordinary neutralizing breadth and potency (166). These bNAbs have been shown to provide protective immunity against challenges with chimeric simian-human immunodeficiency viruses in macaques and HIV-1 in humanized mice (161, 167–174). Early-stage clinical trials have tested passive infusions of new generation bNAbs, including VRC01, 3BNC117, and 10–1074, and have demonstrated safety and some efficacy (175–178), invigorating interest in antibody therapy for established infections. Evidence of pre-existing resistance to individual bNabs makes it likely that these approaches will require careful consideration of sensitivity and the use of combinations to prevent rapid escape.
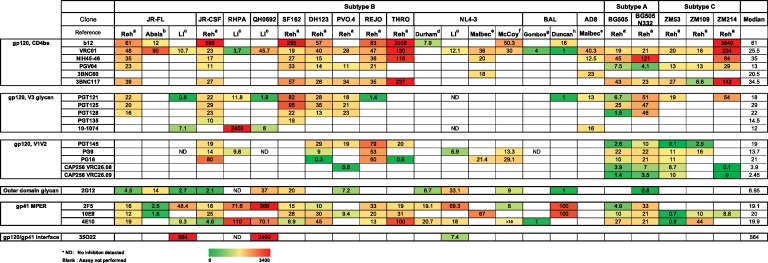
In studies specifically focused on cell-to-cell transmission, VS-mediated infection is more resistant to inhibition by antiretroviral agents, HIV-1 patient sera, and new generation neutralizing antibodies compared to the same virus during cell-free infection in vitro (38, 155, 167, 179–182) Table 1. This has been shown in the case for both lab-adapted viral strains and viruses with transmitted founder Env (9, 38, 167, 179, 180, 183–186). The differences in IC50 between cell-free and cell-to-cell infection are dependent upon viral strain, and the epitopes targeted (38, 183). Discrepancies are observed among different studies regarding the magnitude of differences in IC50 of cell-free and cell-to-cell transmission. For example, Abela et al. found CD4bs antibody VRC01 displayed dramatic potency loss while MPER antibody such as 2F5 remained potent against cell-to-cell transmission relative to cell-free infection (184). However, in the study from Duncan et al., MPER antibody 2F5 was much less effective inhibiting cell-to-cell infection over cell-free infection, while CD4bs antibody VRC01 remained equally potent (181). These differences are likely attributable to the use of different viral strains, cell lines, and/or experimental designs (9, 38, 167, 179, 180, 183, 184, 187–192). Despite what may be strain- or cell-specific differences in the study of neutralization of cell-to-cell transmission, in nearly all cases, cell-to-cell infection is more difficult to neutralize than cell-free infection, which supports that it may play an important mechanism for immune evasion. The largest differences in neutralization sensitivity are 2,000- to 3,500-fold less sensitive to neutralization of cell-cell infection relative to infection by cell-free virus. bNabs showing these large differences include 35O22, the gp120/gp41 interface antibody, 10-1074, a V3 glycan targeted antibody, and b12, a CD4 bs antibody (Table 1). The mechanisms that mediate these stark differences in inhibitory potency are still unclear.
TABLE 1.
Fold change in half maximal inhibitory concentration (IC50) of neutralizing antibodies comparing cell-free versus cell-to-cell infection
Relative resistance of cell-to-cell neutralization over cell-free infection is reflected as increased IC50, and reduced efficacy. A previous study with transmitted founder (T/F) Env carrying constructs in the T cell line system observed incomplete neutralization (38). In many cases, we found an inability of bNAbs to completely neutralize the virus at high antibody concentrations unique to cell-to-cell infection and not seen with infection by cell-free virus. In a severe case, we observed incomplete inhibition where the peak neutralization blocked only 36% of the infection. When a significant fraction of the virus bypasses antibody neutralization through cell-to-cell transmission (38) this is indicative of incomplete neutralization or a decreased efficacy. The Burton group reported an incomplete neutralization phenotype with a very high concentration of highly potent bNAbs (193). They have postulated that the observed incomplete neutralization is partly attributable to the heterogeneity in glycosylation of HIV-1 Env (193). These studies underscore the importance of characterizing the in vitro efficacy of bNAbs for their clinical use and vaccine designs, as resistant viral populations would result in failed viral control in patients. The precise mechanisms of how cell-to-cell infection promotes neutralization resistance remain unclear, although one clue reported by Li et al., is that the mutation of a recycling motif in Env, a membrane proximal tyrosine motif YXXL, can have strong effects on neutralization of cell-cell infection, in both positive or negative ways (38). The sensitivity of cell-to-cell transmission mediated by T/F Env QH0692 to b12 (CD4bs Ab) increased in the context of the YXXL mutation, whereas the sensitivity to 35O22 (gp120/gp41 interface Ab) decreased. These results may implicate endocytic recycling in sorting different antigenic forms of Env, or alternatively suggests that the engagement of Env recycling machinery on the intracytoplasmic domain can directly affect Env conformation (38).
Role of the cytoplasmic tail in modulating Env conformation on cells versus cell-free virus particles.
Recent studies support the hypothesis that cell-associated Env presents a distinct conformation from that presented on cell-free particles, showing different neutralization sensitivities (38, 179, 194). Dale et al. proposed a two-step entry model of HIV-1 cell-to-cell infection where cell-associated virus maturation occurred after viral transfer through the VS, and where viral maturation is required for viral fusion to be activated (194). Based on this model, cell-associated Env is held as a pre-fusogenic conformation during the process of Env-CD4 engagement and the virus material transfer step and is thus likely not well recognized by bNAbs as mature Envs, contributing to neutralization resistance. The study revealed that in VS-mediated, cell-to-cell infection, viral fusion did not occur until cell-associated viral particles were transferred into a trypsin-resistant endocytic compartment of the recipient CD4+ cell. It is postulated that Env conformational transitions occur concomitantly with viral maturation from within the endocytic compartment through regulation by the cytoplasmic tail (CT) (194).
In agreement with the notion that the Env CT can control epitope exposure on the cell surface, it is reported that truncation of the cytoplasmic tail of gp41 enhanced neutralization sensitivity in cell-to-cell infection while not in cell-free infection (179). Studies of cell-free viral infection have found that the maturation status of the virus particle, i.e., the proteolytic cleavage of Gag, through interaction with cytoplasmic tail, can control the fusogenic activity of the Env on the particle (72, 195, 196). Cytoplasmic tail truncation mutants exhibit altered neutralization sensitivity compared with wild-type counterparts. These studies support an allosteric model whereby the cytoplasmic tail can regulate the fusogenic potential and the ability of Env to make conformational transitions in the ectodomain of gp120. In support of conformational regulation by the Env CT, both bNAbs and non-neutralizing antibodies are reported to bind to WT cell-surface Envs differently from CT truncation mutants (72).
Inefficient Env processing and recycling.
While Env that is packaged onto virus particles is typically enriched for fully processed forms, significant levels of uncleaved gp160 Env are present at the cell surface (33, 197) and may present a means by which heterogeneous Env states may be presented on the surface of infected cells. We consider here evidence that a heterogeneous processing state of Env at the cell surface may affect antibody recognition and neutralization.
Proteolytic cleavage affects Env conformations.
Env trimers once synthesized, glycosylated and proteolytically processed can then travel to the plasma membrane via the secretory pathway and are rapidly recycled from the cell surface into the endocytic compartment to avoid immune detection (Fig. 2) (198–202). Early studies observed that only 5% to 15% of intracellular gp160 was processed into mature gp120, which is then transported to the cell surface and incorporated into virus particles (203). Cleavage deficient gp160 was also found to be directed to the surface of infected T cells (197). Proteolytic processing significantly affects epitope presentation of cell surface Env, alters neutralizing and non-neutralizing antibody recognition, and restrains conformational flexibility (130, 204–206). Zhang et al. observed that the uncleaved gp160 is heterogeneous in its extent of modification by complex carbohydrates (33). A larger population of uncleaved Env without complex glycans cross-links into diverse oligomeric forms, and can be recognized by poorly neutralizing antibodies. smFRET studies show that uncleaved precursor gp160 was the most extensively characterized by smFRET (207) and can sample all three conformational states, displaying a high level of conformational flexibility (45, 71), and shifting more toward state 2 and 3 conformations, resulting in increased binding to non-neutralizing antibodies (71, 204–206). Considering its presence on the surface of infected cells, uncleaved gp160 could distract the immune system so that only non-neutralizing epitopes are present, protecting functional pre-triggered state 1 conformation from being recognized (33).
Uncleaved Env and mature cleaved Env have different antibody binding profiles; most bNAbs preferentially bind to cleaved Env (204), while the uncleaved Env shows increased binding to non-neutralizing antibodies (71, 204–206). Assuming a conformation not recognized by bNAbs, the uncleaved Env at the cell surface may limit recognition, providing a mechanism for resistance of antibody neutralization during HIV-1 cell-to-cell transmission. The heterogeneous characteristics of gp160 on the infected cell surface could also contribute to the incomplete neutralization phenotype during cell-to-cell infection, in which a large fraction of cell-associated virus was not inhibited by bNAbs at saturating concentrations (38).
Env endocytosis and outward sorting pathway.
HIV-1 infected cells maintain a low abundance of Env on their surface (34). HIV-1 Env is either incorporated into virions or rapidly internalized through Clathrin/AP2 mediated endocytosis (35, 37, 200, 202, 208–210). This is mediated by the tyrosine-based, membrane-proximal sorting motif Y712XXL (35, 37, 200–202, 211) and a dileucine motif in the cytoplasmic tail of gp41 (212). The Y712XXL motif is highly conserved across HIV-1, HIV-2, chimpanzee simian immunodeficiency virus (SIVcpz) and sooty mangabey SIV (SIVsmm) (211, 213). In addition to constituting a strong endocytic (35, 37, 200, 202) and basolateral sorting signal (214, 215), Y712XXL is also required for virion infectivity, viral entry, and can affect Env incorporation in HIV-1 (216). Mutant Env with Y712A packages about 50% of gp120 into virus particles relative to the wild-type counterpart, which correlates with decreased infectivity (216). The virions with Y712A were less fusogenic, as demonstrated by reduced syncytia formation (216). The Y712XXL motif also contributes to the polarized budding of the virus. Mutants with the Y712A mutation displayed reduced cell-to-cell transmission efficiency (214). Compared with wild-type Env, the Y712A mutant expressed not only a higher level of Env on the surface of infected cells but also showed differential antibody binding and neutralization sensitivity by bNAbs (38). Studies of the role of the Y712XXL motif in SIV suggested that this motif is dispensable for in vitro viral replication but is critical in vivo. Loss of the Y712XXL motif resulted in significant reduction of peak viral load and delayed disease progression in rhesus macaques (217). SIVmac239 with a disrupted YXXL motif by a ΔGY mutation did not deplete mucosal CD4 T cells when infecting rhesus macaques, but was still able to induce immune activation and progression to AIDS (218). Interestingly, pig-tail macaques were able to control replication of SIVmac239 with the ΔGY mutation and preserve mucosal CD4 as well (219), suggesting that the YXXL motif can alter pathogenesis of SIVmac239 in a species-specific manner. A recent NMR structure of the transmembrane segment of gp41 placed the Y712XXL motif within the hydrophilic core and demonstrated interaction of Y712 and P714 on one protomer with L704 and V708 on an adjacent protomer respectively, contributing to Env trimer stability (220).
Studies from the Spearman group showed that the outward trafficking of Env from endocytic recycling compartment (ERC) to the virus assembly area is mediated by host factors Rab11-FIP1C and Rab14 (Fig. 2). A tyrosine-based motif Y795W in the cytoplasmic tail of gp41 (210, 221) was identified to directly interact with the Rab14-FIP1c complex. A key finding from these studies was a pulse-chase experiment that demonstrated that Env’s internalization from the plasma membrane via Y712XXL and Clathrin/AP2 into the endocytic recycling compartment is a prerequisite step for Env incorporation via the Rab11-FIP1c/Rab14 pathway. (8, 210, 221). Recent imaging results from Wang et al. find that a surface labeled Env traffics first to an internal compartment before focally accumulating at a VS when cocultured with CD4+ uninfected target cells (222).
The mechanism whereby uncleaved Env traffics to the plasma membrane is not completely understood. A recent study from Zhang et al. confirmed that there is abundant uncleaved gp160 present on the surface of HIV-1 infected cells (33). A dual pathway model was proposed in which cleaved and uncleaved Env adopt different conformations and are transported to the cell surface via different routes (Fig. 2). The uncleaved Env fraction bypassed the Golgi apparatus, traveled to the cell surface without being proteolytically processed or fully glycosylated, and was selectively excluded from virus particles. Mature cleaved Env, with less abundance on the cell surface in many cases, is better represented in cell-free virus particles, can be recognized by bNAbs, and presumably comprises the functional Envs (33).
Jolly et al. proposed the retrograde trafficking model in which the C-terminal 100 amino acids of gp41 directly interact with Vps35 and Vps26 of the mammalian retromer complex and modulate retrieval of recycled Env from the endosomal recycling compartment to the Golgi apparatus following endocytosis (223) (Fig. 2). A potential corollary of this model is that uncleaved Env recycled from the plasma membrane could regain access to the trans-Golgi network Env processing that is thought to occur. Additional studies are needed to determine if Env can be additionally modified when trafficking through recycling pathways.
During VS-mediated cell-to-cell HIV-1 infection, Env accumulates at the VS; however, the physical movements of Env that support the enrichment of Env at the VS are not well characterized. The extent to which Env diffuses laterally during recruitment by the VS from surface pools or is concentrated by a secretory pathway that targets the VS warrants further investigation. Based on the model described above (33, 223) cleaved and uncleaved Envs can both be transported to the plasma membrane and form adhesive structures by transient interaction with CD4, triggering VS formation. Cell surface Env protein is recycled through Y712XXL-Clathrin-AP2 into ERC. From there, uncleaved gp160 could also retrograde traffic to the trans Golgi network (TGN). Through unknown cell sorting mechanisms cleaved Env is then selectively packaged onto virus particles. A recent report from the Finzi group suggests that conformation specific antibodies are differentially endocytosed, which raises the possibility that different conformational states are trafficked differently (224). They reported that bNAbs targeting “closed” conformation of Env induces its internalization from surface of cells infected with both lab-adapted and T/F Envs in a dynamin dependent manner. Interestingly, Envs bound to non-neutralizing antibodies are displayed on cell surface for prolonged period of times.
Additional considerations that may affect the neutralization of cell-to-cell transmission by bNAbs.
(i) High avidity of Env at VSs. A plausible explanation for the neutralization resistance of cell-to-cell HIV infection is that high concentrations of cellular entry receptors and cell-associated Env form a high avidity interaction that requires higher concentrations of antibodies to inhibit. However, a previous study (179) showed that cytoplasmic tail truncation mutants of NL4-3 Env that expressed much higher levels of surface Env expression showed enhanced (rather than diminished) cell-to-cell neutralization sensitivity without changing neutralization sensitivity in cell-free infection.
(ii) Steric exclusion. Some studies have examined if the synaptic space at the virologic synapses is structurally restrictive, hiding cell-associated Env epitopes in a privileged space that limits the access of neutralizing antibodies. Abela et al. examined pre-CD4 attachment and post-CD4 attachment steps during cell-to-cell transmission and observed that gp120 directed antibodies, CD4bs antibodies in particular, showed significantly decreased efficiency. In contrast, MPER antibodies and fusion inhibitors still maintained potency against cell-to-cell transmission. They suggested that this may be due to a limited time window when neutralizing epitopes were accessible to antibodies during the formation of virological synapses (184). Duncan et al. examined cell-to-cell transmission from infected macrophages to uninfected CD4+ T cells and hypothesized steric restriction as a mechanism for the reduced activity of MPER antibody in cell-to-cell neutralization. They reported that macrophage-to-CD4 T cell transmission was susceptible to CD4bs antibodies and glycan or glycan-dependent epitopes such as those targeted by monoclonal antibody PGT121. Interestingly, they observed that MPER Fab antibodies showed enhanced neutralization efficiency to cell-to-cell infection than the full-length counterpart, suggesting that a smaller Fab domain can better inhibit cell-to-cell infection and that the MPER epitope may be particularly susceptible to steric restriction (181).
Other studies provide evidence against this hypothesis. A study from the Jolly group showed that recombinant llama antibody Fc fusion against CD4bs inhibited cell-to-cell infection even better than the original (smaller) 80kD Ilama antibody J3, indicating that it is not limited by the antibody size (191). Malbec et al. observed that bNAbs that effectively inhibited cell-to-cell infection interfere with the formation of donor-target conjugates and cell-to-cell transfer by accumulating at the VS (167). One important observation was that virus particles captured by target cells in cell-to-cell neutralization assays were mostly coated with bNAbs (167). In another study, anti-gp41 MPER antibodies failed to block virological synapse formation, but could still inhibit cell-to-cell infection (190). These findings together support that epitopes presented by cell-associated viruses are generally accessible by antibodies (191).
In this review we describe potential mechanisms by which differential Env sorting, proteolytic processing, glycosylation, interactions with host factors or newly formed Gag lattice, or trafficking through endocytic pathways may collectively modulate the antigenic state(s) of cell-surface Env distinguishing it from virion-associated Env. Further study of how the trafficking of Env may impact its conformation on its way to assembling virus particles at the VS, and how Env is selected for packaging will provide a better understanding for how HIV-1 infected cells evade antibody neutralization by bNAbs and limit antibody-mediated clearance of infected cells. The results may explain how HIV evades immune responses with such high efficiency.
ACKNOWLEDGMENTS
We thank Svenja Weiss, Susan Zolla Pazner, and Dania Figueroa for helpful comments and suggestions during the preparation of this review.
Contributor Information
Benjamin K. Chen, Email: ben.chen@mssm.edu.
Li Wu, Carver College of Medicine, University of Iowa.
Vinayaka R. Prasad, Albert Einstein College of Medicine
REFERENCES
- 1.Fauci AS. 1996. Host factors and the pathogenesis of HIV-induced disease. Nature 384:529–534. doi: 10.1038/384529a0. [DOI] [PubMed] [Google Scholar]
- 2.Cohen MS, Chen YQ, McCauley M, Gamble T, Hosseinipour MC, Kumarasamy N, Hakim JG, Kumwenda J, Grinsztejn B, Pilotto JHS, Godbole SV, Mehendale S, Chariyalertsak S, Santos BR, Mayer KH, Hoffman IF, Eshleman SH, Piwowar-Manning E, Wang L, Makhema J, Mills LA, de Bruyn G, Sanne I, Eron J, Gallant J, Havlir D, Swindells S, Ribaudo H, Elharrar V, Burns D, Taha ET, Nielsen-Saines K, Celentano D, Essex M, Fleming TR, Team HS. 2011. Prevention of HIV-1 Infection with early antiretroviral therapy. N Engl J Med 365:493–505. doi: 10.1056/NEJMoa1105243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palella FJ, Delaney KM, Moorman AC, Loveless MO, Fuhrer J, Satten GA, Aschman DJ, Holmberg SD, Investigators H. 1998. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N Engl J Med 338:853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- 4.Wyatt R, Sodroski J. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 5.Chen B. 2019. Molecular mechanism of HIV-1 entry. Trends Microbiol 27:878–891. doi: 10.1016/j.tim.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Checkley MA, Luttge BG, Freed EO. 2011. HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation. J Mol Biol 410:582–608. doi: 10.1016/j.jmb.2011.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCune JM, Rabin LB, Feinberg MB, Lieberman M, Kosek JC, Reyes GR, Weissman IL. 1988. Endoproteolytic cleavage of gp160 is required for the activation of human immunodeficiency virus. Cell 53:55–67. doi: 10.1016/0092-8674(88)90487-4. [DOI] [PubMed] [Google Scholar]
- 8.Kirschman J, Qi ML, Ding LM, Hammonds J, Dienger-Stambaugh K, Wang JJ, Lapierre LA, Goldenring JR, Spearman P. 2018. HIV-1 envelope glycoprotein trafficking through the endosomal recycling compartment is required for particle incorporation. J Virol 92. doi: 10.1128/JVI.01893-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen P, Hubner W, Spinelli MA, Chen BK. 2007. Predominant mode of human immunodeficiency virus transfer between T cells is mediated by sustained Env-dependent neutralization-resistant virological synapses. J Virol 81:12582–12595. doi: 10.1128/JVI.00381-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agosto LM, Uchil PD, Mothes W. 2015. HIV cell-to-cell transmission: effects on pathogenesis and antiretroviral therapy. Trends Microbiol 23:289–295. doi: 10.1016/j.tim.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, Katinger H, Parren P, Robinson J, Van Ryk D, Wang LP, Burton DR, Freire E, Wyatt R, Sodroski J, Hendrickson WA, Arthos J. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 12.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 13.Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, Sodroski JG. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 14.Ahmad A, Menezes J. 1996. Antibody-dependent cellular cytotoxicity in HIV infections. FASEB J 10:258–266. doi: 10.1096/fasebj.10.2.8641559. [DOI] [PubMed] [Google Scholar]
- 15.Chung AW, Isitman G, Navis M, Kramski M, Center RJ, Kent SJ, Stratov I. 2011. Immune escape from HIV-specific antibody-dependent cellular cytotoxicity (ADCC) pressure. Proc Natl Acad Sci USA 108:7505–7510. doi: 10.1073/pnas.1016048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Q, Finzi A, Sodroski J. 2020. The conformational states of the HIV-1 envelope glycoproteins. Trends Microbiol 28:655–667. doi: 10.1016/j.tim.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lambert GS, Upadhyay C. 2021. HIV-1 envelope glycosylation and the signal peptide. Vaccines (Basel) 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Luo L, Thomas DY, Kang CY. 2000. The HIV-1 Env protein signal sequence retards its cleavage and down-regulates the glycoprotein folding. Virology 272:417–428. doi: 10.1006/viro.2000.0357. [DOI] [PubMed] [Google Scholar]
- 19.Snapp EL, McCaul N, Quandte M, Cabartova Z, Bontjer I, Kallgren C, Nilsson I, Land A, von Heijne G, Sanders RW, Braakman I. 2017. Structure and topology around the cleavage site regulate post-translational cleavage of the HIV-1 gp160 signal peptide. Elife 6. doi: 10.7554/eLife.26067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Upadhyay C, Feyznezhad R, Cao L, Chan KW, Liu K, Yang W, Zhang H, Yolitz J, Arthos J, Nadas A, Kong XP, Zolla-Pazner S, Hioe CE. 2020. Signal peptide of HIV-1 envelope modulates glycosylation impacting exposure of V1V2 and other epitopes. PLoS Pathog 16:e1009185. doi: 10.1371/journal.ppat.1009185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Behrens AJ, Crispin M. 2017. Structural principles controlling HIV envelope glycosylation. Curr Opin Struct Biol 44:125–133. doi: 10.1016/j.sbi.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daniels CN, Saunders KO. 2019. Antibody responses to the HIV-1 envelope high mannose patch. Adv Immunol 143:11–73. doi: 10.1016/bs.ai.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klasse PJ, Ozorowski G, Sanders RW, Moore JP. 2020. Env exceptionalism: why are HIV-1 Env glycoproteins atypical immunogens? Cell Host Microbe 27:507–518. doi: 10.1016/j.chom.2020.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crispin M, Ward AB, Wilson IA. 2018. Structure and immune recognition of the HIV glycan shield. Annu Rev Biophys 47:499–523. doi: 10.1146/annurev-biophys-060414-034156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Decroly E, Vandenbranden M, Ruysschaert JM, Cogniaux J, Jacob GS, Howard SC, Marshall G, Kompelli A, Basak A, Jean F. 1994. The convertases furin and PC1 can both cleave the human immunodeficiency virus (HIV)-1 envelope glycoprotein gp160 into gp120 (HIV-1 SU) and gp41 (HIV-I TM). J Biol Chem 269:12240–12247. doi: 10.1016/S0021-9258(17)32707-2. [DOI] [PubMed] [Google Scholar]
- 26.Decroly E, Wouters S, Di Bello C, Lazure C, Ruysschaert JM, Seidah NG. 1996. Identification of the paired basic convertases implicated in HIV gp160 processing based on in vitro assays and expression in CD4(+) cell lines. J Biol Chem 271:30442–30450. doi: 10.1074/jbc.271.48.30442. [DOI] [PubMed] [Google Scholar]
- 27.Franzusoff A, Volpe AM, Josse D, Pichuantes S, Wolf JR. 1995. Biochemical and genetic definition of the cellular protease required for HIV-1 gp160 processing. J Biol Chem 270:3154–3159. doi: 10.1074/jbc.270.7.3154. [DOI] [PubMed] [Google Scholar]
- 28.Moulard M, Decroly E. 2000. Maturation of HIV envelope glycoprotein precursors by cellular endoproteases. Biochim Biophys Acta 1469:121–132. doi: 10.1016/s0304-4157(00)00014-9. [DOI] [PubMed] [Google Scholar]
- 29.Vollenweider F, Benjannet S, Decroly E, Savaria D, Lazure C, Thomas G, Chretien M, Seidah NG. 1996. Comparative cellular processing of the human immunodeficiency virus (HIV-1) envelope glycoprotein gp160 by the mammalian subtilisin/kexin-like convertases. Biochem J 314:521–532. doi: 10.1042/bj3140521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hallenberger S, Bosch V, Angliker H, Shaw E, Klenk HD, Garten W. 1992. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature 360:358–361. doi: 10.1038/360358a0. [DOI] [PubMed] [Google Scholar]
- 31.Pritchard LK, Vasiljevic S, Ozorowski G, Seabright GE, Cupo A, Ringe R, Kim HJ, Sanders RW, Doores KJ, Burton DR, Wilson IA, Ward AB, Moore JP, Crispin M. 2015. Structural constraints determine the glycosylation of HIV-1 envelope trimers. Cell Rep 11:1604–1613. doi: 10.1016/j.celrep.2015.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zou ST, Zhang SJ, Gaffney A, Ding HT, Lu ML, Grover JR, Farrell M, Nguyen HT, Zhao ON, Anang S, Zhao MQ, Mohammadi M, Blanchard SC, Abrams C, Madani N, Mothes W, Kappes JC, Smith AB, Sodroski J. 2020. Long-acting BMS-378806 analogues stabilize the state-1 conformation of the human immunodeficiency virus type 1 envelope glycoproteins. J Virol 94. doi: 10.1128/JVI.00148-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang S, Nguyen HT, Ding H, Wang J, Zou S, Liu L, Guha D, Gabuzda D, Ho DD, Kappes JC, Sodroski J. 2021. Dual pathways of human immunodeficiency virus (HIV-1) envelope glycoprotein trafficking modulate the selective exclusion of uncleaved oligomers from virions. J Virol 95. doi: 10.1128/JVI.01369-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Egan MA, Carruth LM, Rowell JF, Yu X, Siliciano RF. 1996. Human immunodeficiency virus type 1 envelope protein endocytosis mediated by a highly conserved intrinsic internalization signal in the cytoplasmic domain of gp41 is suppressed in the presence of the Pr55gag precursor protein. J Virol 70:6547–6556. doi: 10.1128/JVI.70.10.6547-6556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boge M, Wyss S, Bonifacino JS, Thali M. 1998. A membrane-proximal tyrosine-based signal mediates internalization of the HIV-1 envelope glycoprotein via interaction with the AP-2 clathrin adaptor. J Biol Chem 273:15773–15778. doi: 10.1074/jbc.273.25.15773. [DOI] [PubMed] [Google Scholar]
- 36.Noble B, Abada P, Nunez-Iglesias J, Cannon PM. 2006. Recruitment of the adaptor protein 2 complex by the human immunodeficiency virus type 2 envelope protein is necessary for high levels of virus release. J Virol 80:2924–2932. doi: 10.1128/JVI.80.6.2924-2932.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohno H, Aguilar RC, Fournier MC, Hennecke S, Cosson P, Bonifacino JS. 1997. Interaction of endocytic signals from the HIV-1 envelope glycoprotein complex with members of the adaptor medium chain family. Virology 238:305–315. doi: 10.1006/viro.1997.8839. [DOI] [PubMed] [Google Scholar]
- 38.Li H, Zony C, Chen P, Chen BK. 2017. Reduced potency and incomplete neutralization of broadly neutralizing antibodies against cell-to-cell transmission of HIV-1 with transmitted founder Envs. J Virol 91. doi: 10.1128/JVI.02425-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ward AB, Wilson IA. 2015. Insights into the trimeric HIV-1 envelope glycoprotein structure. Trends Biochem Sci 40:101–107. doi: 10.1016/j.tibs.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chan DC, Fass D, Berger JM, Kim PS. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263–273. doi: 10.1016/S0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 41.Melikyan GB, Markosyan RM, Hemmati H, Delmedico MK, Lambert DM, Cohen FS. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J Cell Biol 151:413–423. doi: 10.1083/jcb.151.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 43.Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, Burton DR, Koff WC, Courter JR, Smith AB, 3rd, Kwong PD, Blanchard SC, Mothes W. 2014. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 346:759–763. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herschhorn A, Ma X, Gu C, Ventura JD, Castillo-Menendez L, Melillo B, Terry DS, Smith AB, 3rd, Blanchard SC, Munro JB, Mothes W, Finzi A, Sodroski J. 2016. Release of gp120 restraints leads to an entry-competent intermediate state of the HIV-1 envelope glycoproteins. mBio 7. doi: 10.1128/mBio.01598-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu M, Ma X, Castillo-Menendez LR, Gorman J, Alsahafi N, Ermel U, Terry DS, Chambers M, Peng D, Zhang B, Zhou T, Reichard N, Wang K, Grover JR, Carman BP, Gardner MR, Nikic-Spiegel I, Sugawara A, Arthos J, Lemke EA, Smith AB, 3rd, Farzan M, Abrams C, Munro JB, McDermott AB, Finzi A, Kwong PD, Blanchard SC, Sodroski JG, Mothes W. 2019. Associating HIV-1 envelope glycoprotein structures with states on the virus observed by smFRET. Nature 568:415–419. doi: 10.1038/s41586-019-1101-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang H, Barnes CO, Yang Z, Nussenzweig MC, Bjorkman PJ. 2018. Partially open HIV-1 envelope structures exhibit conformational changes relevant for coreceptor binding and fusion. Cell Host Microbe 24:579–592. doi: 10.1016/j.chom.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang H, Cohen AA, Galimidi RP, Gristick HB, Jensen GJ, Bjorkman PJ. 2016. Cryo-EM structure of a CD4-bound open HIV-1 envelope trimer reveals structural rearrangements of the gp120 V1V2 loop. Proc Natl Acad Sci USA 113:E7151–E7158. doi: 10.1073/pnas.1615939113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109–U76. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.White TA, Bartesaghi A, Borgnia MJ, Meyerson JR, de la Cruz MJV, Bess JW, Nandwani R, Hoxie JA, Lifson JD, Milne JLS, Subramaniam S. 2010. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: strain-dependent variation in quaternary structure. PLoS Pathog 6:e1001249. doi: 10.1371/journal.ppat.1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guttman M, Garcia NK, Cupo A, Matsui T, Julien JP, Sanders RW, Wilson IA, Moore JP, Lee KK. 2014. CD4-induced activation in a soluble HIV-1 Env trimer. Structure 22:974–984. doi: 10.1016/j.str.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB. 2013. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jette CA, Barnes CO, Kirk SM, Melillo B, Smith AB, Bjorkman PJ. 2021. Cryo-EM structures of HIV-1 trimer bound to CD4-mimetics BNM-III-170 and M48U1 adopt a CD4-bound open conformation. Nat Commun 12. doi: 10.1038/s41467-021-21816-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Herschhorn A, Gu C, Moraca F, Ma XC, Farrell M, Smith AB, Pancera M, Kwong PD, Schon A, Freire E, Abrams C, Blanchard SC, Mothes W, Sodroski JG. 2017. The beta 20-beta 21 of gp120 is a regulatory switch for HIV-1 Env conformational transitions. Nat Commun 8. doi: 10.1038/s41467-017-01119-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. 2013. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rizzuto CD, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong PD, Hendrickson WA, Sodroski J. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 57.Wu LJ, Gerard NP, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso AA, Desjardin E, Newman W, Gerard C, Sodroski J. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 58.Trkola A, Dragic T, Arthos J, Binley JM, Olson WC, Allaway GP, Cheng-Mayer C, Robinson J, Maddon PJ, Moore JP. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 59.Furuta RA, Wild CT, Weng YK, Weiss CD. 1998. Capture of an early fusion-active conformation of HIV-1 gp41. Nat Struct Biol 5:276–279. doi: 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- 60.Koshiba T, Chan DC. 2003. The prefusogenic intermediate of HIV-1 gp41 contains exposed C-peptide regions. J Biol Chem 278:7573–7579. doi: 10.1074/jbc.M211154200. [DOI] [PubMed] [Google Scholar]
- 61.He Y, Vassell R, Zaitseva M, Nguyen N, Yang ZN, Weng YK, Weiss CD. 2003. Peptides trap the human immunodeficiency virus type 1 envelope glycoprotein fusion intermediate at two sites. J Virol 77:1666–1671. doi: 10.1128/jvi.77.3.1666-1671.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Haim H, Si ZH, Madani N, Wang LP, Courter JR, Princiotto A, Kassa A, DeGrace M, McGee-Estrada K, Mefford M, Gabuzda D, Smith AB, Sodroski J. 2009. Soluble CD4 and CD4-mimetic compounds inhibit HIV-1 infection by induction of a short-lived activated state. PLoS Pathog 5:e1000360. doi: 10.1371/journal.ppat.1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Madani N, Schon A, Princiotto AM, LaLonde JM, Courter JR, Soeta T, Ng D, Wang LP, Brower ET, Xiang SH, Do Kwon Y, Huang CC, Wyatt R, Kwong PD, Freire E, Smith AB, Sodroski J. 2008. Small-molecule CD4 mimics interact with a highly conserved pocket on HIV-1 gp120. Structure 16:1689–1701. doi: 10.1016/j.str.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schon A, Madani N, Klein JC, Hubicki A, Ng D, Yang X, Smith AB, Sodroski J, Freire E. 2006. Thermodynamics of binding of a low-molecular-weight CD4 mimetic to HIV-1 gp120. Biochemistry 45:10973–10980. doi: 10.1021/bi061193r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Madani N, Princiotto AM, Zhao C, Jahanbakhshsefidi F, Mertens M, Herschhorn A, Melillo B, Smith AB, Sodroski J. 2017. Activation and inactivation of primary human immunodeficiency virus envelope glycoprotein trimers by CD4-mimetic compounds. J Virol 91. doi: 10.1128/JVI.01880-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kolchinsky P, Kiprilov E, Sodroski J. 2001. Increased neutralization sensitivity of CD4-independent human immunodeficiency virus variants. J Virol 75:2041–2050. doi: 10.1128/JVI.75.5.2041-2050.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Taylor BM, Foulke JS, Flinko R, Heredia A, DeVico A, Reitz M. 2008. An alteration of human immunodeficiency virus gp41 leads to reduced CCR5 dependence and CD4 independence. J Virol 82:5460–5471. doi: 10.1128/JVI.01049-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Edwards TG, Wyss S, Reeves JD, Zolla-Pazner S, Hoxie JA, Doms RW, Baribaud F. 2002. Truncation of the cytoplasmic domain induces exposure of conserved regions in the ectodomain of human immunodeficiency virus type 1 envelope protein. J Virol 76:2683–2691. doi: 10.1128/jvi.76.6.2683-2691.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang PF, Bouma P, Park EJ, Margolick JB, Robinson JE, Zolla-Pazner S, Flora MN, Quinnan GV. 2002. A variable region 3 (V3) mutation determines a global neutralization phenotype and CD4-independent infectivity of a human immunodeficiency virus type 1 envelope associated with a broadly cross-reactive, primary virus-neutralizing antibody response. J Virol 76:644–655. doi: 10.1128/jvi.76.2.644-655.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haim H, Strack B, Kassa A, Madani N, Wang LP, Courter JR, Princiotto A, McGee K, Pacheco B, Seaman MS, Smith AB, Sodroski J. 2011. Contribution of intrinsic reactivity of the HIV-1 envelope glycoproteins to CD4-independent infection and global inhibitor sensitivity. PLoS Pathog 7:e1002101. doi: 10.1371/journal.ppat.1002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Haim H, Salas I, Sodroski J. 2013. Proteolytic processing of the human immunodeficiency virus envelope glycoprotein precursor decreases conformational flexibility. J Virol 87:1884–1889. doi: 10.1128/JVI.02765-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen J, Kovacs JM, Peng HQ, Rits-Volloch S, Lu JM, Park D, Zablowsky E, Seaman MS, Chen B. 2015. HIV-1 envelope. Effect of the cytoplasmic domain on antigenic characteristics of HIV-1 envelope glycoprotein. Science 349:191–195. doi: 10.1126/science.aaa9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee JH, Ozorowski G, Ward AB. 2016. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science 351:1043–1048. doi: 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li Z, Li W, Lu M, Bess J, Jr, Chao CW, Gorman J, Terry DS, Zhang B, Zhou T, Blanchard SC, Kwong PD, Lifson JD, Mothes W, Liu J. 2020. Subnanometer structures of HIV-1 envelope trimers on aldrithiol-2-inactivated virus particles. Nat Struct Mol Biol 27:726–734. doi: 10.1038/s41594-020-0452-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Veillette M, Desormeaux A, Medjahed H, Gharsallah NE, Coutu M, Baalwa J, Guan YJ, Lewis G, Ferrari G, Hahn BH, Haynes BF, Robinson JE, Kaufmann DE, Bonsignori M, Sodroski J, Finzi A. 2014. Interaction with cellular CD4 exposes HIV-1 envelope epitopes targeted by antibody-dependent cell-mediated cytotoxicity. J Virol 88:2633–2644. doi: 10.1128/JVI.03230-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alsahafi N, Bakouche N, Kazemi M, Richard J, Ding SL, Bhattacharyya S, Das D, Anand SP, Prevost J, Tolbert WD, Lu H, Medjahed H, Gendron-Lepage G, Delgado GGO, Kirk S, Melillo B, Mothes W, Sodroski J, Smith AB, Kaufmann DE, Wu XL, Pazgier M, Rouiller I, Finzi A, Munro JB. 2019. An asymmetric opening of HIV-1 envelope mediates antibody-dependent cellular cytotoxicity. Cell Host Microbe 25:578–587.e5. doi: 10.1016/j.chom.2019.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pancera M, Lai YT, Bylund T, Druz A, Narpala S, O'Dell S, Schon A, Bailer RT, Chuang GY, Geng H, Louder MK, Rawi R, Soumana DI, Finzi A, Herschhorn A, Madani N, Sodroski J, Freire E, Langley DR, Mascola JR, McDermott AB, Kwong PD. 2017. Crystal structures of trimeric HIV envelope with entry inhibitors BMS-378806 and BMS-626529. Nat Chem Biol 13:1115–1122. doi: 10.1038/nchembio.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lin PF, Blair W, Wang T, Spicer T, Guo Q, Zhou NN, Gong YF, Wang HGH, Rose R, Yamanaka G, Robinson B, Li CB, Fridell R, Deminie C, Demers G, Yang Z, Zadjura L, Meanwell N, Colonno R. 2003. A small molecule HIV-1 inhibitor that targets the HIV-1 envelope and inhibits CD4 receptor binding. Proc Natl Acad Sci USA 100:11013–11018. doi: 10.1073/pnas.1832214100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang T, Zhang Z, Wallace OB, Deshpande M, Fang H, Yang Z, Zadjura LM, Tweedie DL, Huang S, Zhao F, Ranadive S, Robinson BS, Gong Y-F, Ricarrdi K, Spicer TP, Deminie C, Rose R, Wang H-GH, Blair WS, Shi P-Y, Lin P-F, Colonno RJ, Meanwell NA. 2003. Discovery of 4-benzoyl-1- (4-methoxy-1H-pyrrolo 2,3-b lpyridin-3-yl)oxoacetyl -2-(R)- methylpiperazine (BMS-378806): A novel HIV-1 attachment inhibitor that interferes with CD4-gp120 interactions. J Med Chem 46:4236–4239. doi: 10.1021/jm034082o. [DOI] [PubMed] [Google Scholar]
- 80.Ho HT, Fan L, Nowicka-Sans B, McAuliffe B, Li CB, Yamanaka G, Zhou NN, Fang H, Dicker I, Dalterio R, Gong YF, Wang T, Yin ZW, Ueda Y, Matiskella J, Kadow J, Clapham P, Robinson J, Colonno R, Lin PF. 2006. Envelope conformational changes induced by human immunodeficiency virus type 1 attachment inhibitors prevent CD4 binding and downstream entry events. J Virol 80:4017–4025. doi: 10.1128/JVI.80.8.4017-4025.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guo Q, Ho HT, Dicker I, Fan L, Zhou NN, Friborg J, Wang T, McAuliffe BV, Wang HGH, Rose RE, Fang H, Scarnati HT, Langley DR, Meanwell NA, Abraham R, Colonno RJ, Lin PF. 2003. Biochemical and genetic characterizations of a novel human immunodeficiency virus type 1 inhibitor that blocks gp120-CD4 interactions. J Virol 77:10528–10536. doi: 10.1128/jvi.77.19.10528-10536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Si ZH, Madani N, Cox JM, Chruma JJ, Klein JC, Schon A, Phan N, Wang L, Biorn AC, Cocklin S, Chaiken I, Freire E, Smith AB, Sodroski JG. 2004. Small-molecule inhibitors of HIV-1 entry block receptor-induced conformational changes in the viral envelope glycoproteins. Proc Natl Acad Sci USA 101:5036–5041. doi: 10.1073/pnas.0307953101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Madani N, Perdigoto AL, Srinivasan K, Cox JM, Chruma JJ, LaLonde J, Head M, Smith AB, Sodroski JG. 2004. Localized changes in the gp120 envelope glycoprotein confer resistance to human immunodeficiency virus entry inhibitors BMS-806 and #155. J Virol 78:3742–3752. doi: 10.1128/jvi.78.7.3742-3752.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Langley DR, Kimura SR, Sivaprakasam P, Zhou NN, Dicker I, McAuliffe B, Wang T, Kadow JF, Meanwell NA, Krystal M. 2015. Homology models of the HIV-1 attachment inhibitor BMS-626529 bound to gp120 suggest a unique mechanism of action. Proteins-Structure Function and Bioinformatics 83:331–350. doi: 10.1002/prot.24726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kozal M, Aberg J, Pialoux G, Cahn P, Thompson M, Molina JM, Grinsztejn B, Diaz R, Castagna A, Kumar P, Latiff G, DeJesus E, Gummel M, Gartland M, Pierce A, Ackerman P, Llamoso C, Lataillade M, Team BT. 2020. Fostemsavir in adults with multidrug-resistant HIV-1 infection. N Engl J Med 382:1232–1243. doi: 10.1056/NEJMoa1902493. [DOI] [PubMed] [Google Scholar]
- 86.Wang T, Ueda Y, Zhang ZX, Yin ZW, Matiskella J, Pearce BC, Yang Z, Zheng M, Parker DD, Yamanaka GA, Gong YF, Ho HT, Colonno RJ, Langley DR, Lin PF, Meanwell NA, Kadow JF. 2018. Discovery of the human immunodeficiency virus type 1 (HIV-1) attachment inhibitor temsavir and its phosphonooxymethyl prodrug fostemsavir. J Med Chem 61:6308–6327. doi: 10.1021/acs.jmedchem.8b00759. [DOI] [PubMed] [Google Scholar]
- 87.Herschhorn A, Gu C, Espy N, Richard J, Finzi A, Sodroski JG. 2014. A broad HIV-1 inhibitor blocks envelope glycoprotein transitions critical for entry. Nat Chem Biol 10:845–852. +. doi: 10.1038/nchembio.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pacheco B, Alsahafi N, Debbeche O, Prevost J, Ding SL, Chapleau JP, Herschhorn A, Madani N, Princiotto A, Melillo B, Gu C, Zeng X, Mao YD, Smith AB, Sodroski J, Finzi A. 2017. Residues in the gp41 ectodomain regulate HIV-1 envelope glycoprotein conformational transitions induced by gp120-directed inhibitors. J Virol 91. doi: 10.1128/JVI.02219-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kwon YD, LaLonde JM, Yang YP, Elban MA, Sugawara A, Courter JR, Jones DM, Smith AB, Debnath AK, Kwong PD. 2014. Crystal structures of HIV-1 gp120 envelope glycoprotein in complex with NBD analogues that target the CD4-binding site. PLoS One 9:e85940. doi: 10.1371/journal.pone.0085940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Do Kwon Y, Finzi A, Wu XL, Dogo-Isonagie C, Lee LK, Moore LR, Schmidt SD, Stuckey J, Yang YP, Zhou TQ, Zhu J, Vicic DA, Debnath AK, Shapiro L, Bewley CA, Mascola JR, Sodroski JG, Kwong PD. 2012. Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc Natl Acad Sci USA 109:5663–5668. doi: 10.1073/pnas.1112391109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhao Q, Ma LY, Jiang SB, Lu H, Liu SW, He YX, Strick N, Neamati N, Debnath AK. 2005. N-phenyl-N'-(2,2,6,6-tetramethyl-piperidin-4-yl)-oxalamides as a new class of HIV-1 entry inhibitors that prevent gp120 binding to CD4. Virology 339:213–225. doi: 10.1016/j.virol.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 92.Kassa A, Madani N, Schon A, Haim H, Finzi A, Xiang SH, Wang LP, Princiotto A, Pancera M, Courter J, Smith AB, Freire E, Kwong PD, Sodroski J. 2009. Transitions to and from the CD4-bound conformation are modulated by a single-residue change in the human immunodeficiency virus type 1 gp120 inner domain. J Virol 83:8364–8378. doi: 10.1128/JVI.00594-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ding SL, Grenier MC, Tolbert WD, Vezina D, Sherburn R, Richard J, Prevost J, Chapleau JP, Gendron-Lepage G, Medjahed H, Abrams C, Sodroski J, Pazgier M, Smith AB, Finzi A. 2019. A new family of small-molecule CD4-mimetic compounds contacts highly conserved aspartic acid 368 of HIV-1 gp120 and mediates antibody-dependent cellular cytotoxicity. J Virol 93. doi: 10.1128/JVI.01325-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ding SL, Veillette M, Coutu M, Prevost J, Scharf L, Bjorkman PJ, Ferrari G, Robinson JE, Sturzel C, Hahn BH, Sauter D, Kirchhoff F, Lewis GK, Pazgier M, Finzi A. 2016. A highly conserved residue of the HIV-1 gp120 inner domain is important for antibody-dependent cellular cytotoxicity responses mediated by anti-cluster A antibodies. J Virol 90:2127–2134. doi: 10.1128/JVI.02779-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Richard J, Pacheco B, Gohain N, Veillette M, Ding S, Alsahafi N, Tolbert WD, Prévost J, Chapleau J-P, Coutu M, Jia M, Brassard N, Park J, Courter JR, Melillo B, Martin L, Tremblay C, Hahn BH, Kaufmann DE, Wu X, Smith AB, Sodroski J, Pazgier M, Finzi A. 2016. Co-receptor Binding Site Antibodies Enable CD4-Mimetics to Expose Conserved Anti-cluster A ADCC Epitopes on HIV-1 Envelope Glycoproteins. Ebiomedicine 12:208–218. doi: 10.1016/j.ebiom.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Richard J, Veillette M, Brassard N, Iyer SS, Roger M, Martin L, Pazgier M, Schön A, Freire E, Routy J-P, Smith AB, Park J, Jones DM, Courter JR, Melillo BN, Kaufmann DE, Hahn BH, Permar SR, Haynes BF, Madani N, Sodroski JG, Finzi A. 2015. CD4 mimetics sensitize HIV-1-infected cells to ADCC. Proc Natl Acad Sci USA 112:E2687–E2694. doi: 10.1073/pnas.1506755112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vezina D, Gong SY, Tolbert WD, Ding SL, Nguyen D, Richard J, Gendron-Lepage G, Melillo B, Smith AB, Pazgier M, Finzi A. 2021. Stabilizing the HIV-1 Envelope Glycoprotein State 2A Conformation. J Virol 95. doi: 10.1128/JVI.01620-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Anand SP, Prevost J, Baril S, Richard J, Medjahed H, Chapleau JP, Tolbert WD, Kirk S, Smith AB, Wines BD, Kent SJ, Hogarth PM, Parsons MS, Pazgier M, Finzi A. 2019. Two Families of Env Antibodies Efficiently Engage Fc-Gamma Receptors and Eliminate HIV-1-Infected Cells. J Virol 93. doi: 10.1128/JVI.01823-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Walker LM, Simek MD, Priddy F, Gach JS, Wagner D, Zwick MB, Phogat SK, Poignard P, Burton DR. 2010. A Limited Number of Antibody Specificities Mediate Broad and Potent Serum Neutralization in Selected HIV-1 Infected Individuals. PLoS Pathog 6:e1001028. doi: 10.1371/journal.ppat.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Guttman M, Cupo A, Julien JP, Sanders RW, Wilson IA, Moore JP, Lee KK. 2015. Antibody potency relates to the ability to recognize the closed, pre-fusion form of HIV Env. Nat Commun 6. doi: 10.1038/ncomms7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yuan W, Bazick J, Sodroski J. 2006. Characterization of the multiple conformational states of free monomeric and trimeric human immunodeficiency virus envelope glycoproteins after fixation by cross-linker. J Virol 80:6725–6737. doi: 10.1128/JVI.00118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gray ES, Madiga MC, Hermanus T, Moore PL, Wibmer CK, Tumba NL, Werner L, Mlisana K, Sibeko S, Williamson C, Karim SSA, Morris L, Team CAIS, CAPRISA002 Study Team . 2011. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4(+) t cell decline and high viral load during acute infection. J Virol 85:4828–4840. doi: 10.1128/JVI.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wibmer CK, Bhiman JN, Gray ES, Tumba N, Karim SSA, Williamson C, Morris L, Moore PL. 2013. Viral escape from HIV-1 neutralizing antibodies drives increased plasma neutralization breadth through sequential recognition of multiple epitopes and immunotypes. PLoS Pathog 9:e1003738. doi: 10.1371/journal.ppat.1003738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sather DN, Armann J, Ching LK, Mavrantoni A, Sellhorn G, Caldwell Z, Yu XS, Wood B, Self S, Kalams S, Stamatatos L. 2009. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J Virol 83:757–769. doi: 10.1128/JVI.02036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sliepen K, Sanders RW. 2016. HIV-1 envelope glycoprotein immunogens to induce broadly neutralizing antibodies. Expert Rev Vaccines 15:349–365. doi: 10.1586/14760584.2016.1129905. [DOI] [PubMed] [Google Scholar]
- 106.Karuna ST, Corey L. 2020. Broadly neutralizing antibodies for HIV prevention. Annu Rev Med 71:329–346. doi: 10.1146/annurev-med-110118-045506. [DOI] [PubMed] [Google Scholar]
- 107.Xiang SH, Kwong PD, Gupta R, Rizzuto CD, Casper DJ, Wyatt R, Wang LP, Hendrickson WA, Doyle ML, Sodroski J. 2002. Mutagenic stabilization and/or disruption of a CD4-bound state reveals distinct conformations of the human immunodeficiency virus type 1 gp120 envelope glycoprotein. J Virol 76:9888–9899. doi: 10.1128/jvi.76.19.9888-9899.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Labrijn AF, Poignard P, Raja A, Zwick MB, Delgado K, Franti M, Binley J, Vivona V, Grundner C, Huang CC, Venturi M, Petropoulos CJ, Wrin T, Dimitrov DS, Robinson J, Kwong PD, Wyatt RT, Sodroski J, Burton DR. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gpl120 is sterically restricted on primary human immunodeficiency virus type 1. J Virol 77:10557–10565. doi: 10.1128/jvi.77.19.10557-10565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Davis KL, Gray ES, Moore PL, Decker JM, Salomon A, Montefiori DC, Graham BS, Keefer MC, Pinter A, Morris L, Hahn BH, Shaw GM. 2009. High titer HIV-1 V3-specific antibodies with broad reactivity but low neutralizing potency in acute infection and following vaccination. Virology 387:414–426. doi: 10.1016/j.virol.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Decker JM, Bibollet-Ruche F, Wei XP, Wang SY, Levy DN, Wang WQ, Delaporte E, Peeters M, Derdeyn CA, Allen S, Hunter E, Saag MS, Hoxie JA, Hahn BH, Kwong PD, Robinson JE, Shaw GM. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J Exp Med 201:1407–1419. doi: 10.1084/jem.20042510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Davis KL, Bibollet-Ruche F, Li H, Decker JM, Kutsch O, Morris L, Salomon A, Pinter A, Hoxie JA, Hahn BH, Kwong PD, Shaw GM. 2009. Human immunodeficiency virus type 2 (HIV-2)/HIV-1 envelope chimeras detect high titers of broadly reactive HIV-1 V3-specific antibodies in human plasma. J Virol 83:1240–1259. doi: 10.1128/JVI.01743-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Forsell MNE, McKee K, Feng Y, Mascola JR, Wyatt RT. 2014. HIV-1 envelope glycoprotein trimer immunogenicity elicited in the presence of human CD4 alters the neutralization profile. Aids Res Hum Retroviruses 30:1089–1098. doi: 10.1089/AID.2014.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Forsell MNE, Dey B, Morner A, Svehla K, O’Dell S, Hogerkorp CM, Voss G, Thorstensson R, Shaw GM, Mascola JR, Hedestam GBK, Wyatt RT. 2008. B cell recognition of the conserved HIV-1 Co-receptor binding site is altered by endogenous primate CD4. PLoS Pathog 4:e1000171. doi: 10.1371/journal.ppat.1000171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Douagi I, Forsell MNE, Sundling C, O'Dell S, Feng Y, Dosenovic P, Li YX, Seder R, Lore K, Mascola JR, Wyatt RT, Hedestam GBK. 2010. Influence of novel CD4 binding-defective HIV-1 envelope glycoprotein immunogens on neutralizing antibody and T-Cell responses in nonhuman primates. J Virol 84:1683–1695. doi: 10.1128/JVI.01896-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dey B, Pancera M, Svehla K, Shu Y, Xiang SH, Vainshtein J, Li YX, Sodroski J, Kwong PD, Mascola JR, Wyatt R. 2007. Characterization of human immunodeficiency virus type 1 monomeric and trimeric gp120 glycoproteins stabilized in the CD4-bound state: Antigenicity, biophysics, and immunogenicity. J Virol 81:5579–5593. doi: 10.1128/JVI.02500-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dey B, Svehla K, Xu L, Wycuff D, Zhou TQ, Voss G, Phogat A, Chakrabarti BK, Li YX, Shaw G, Kwong PD, Nabel GJ, Mascola JR, Wyatt RT. 2009. Structure-based stabilization of HIV-1 gp120 enhances humoral immune responses to the induced co-receptor binding site. PLoS Pathog 5:e1000445. doi: 10.1371/journal.ppat.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sullivan N, Sun Y, Sattentau Q, Thali M, Wu D, Denisova G, Gershoni J, Robinson J, Moore J, Sodroski J. 1998. CD4-induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J Virol 72:4694–4703. doi: 10.1128/JVI.72.6.4694-4703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kassa A, Dey AK, Sarkar P, Labranche C, Go EP, Clark DF, Sun YD, Nandi A, Hartog K, Desaire H, Montefiori D, Carfi A, Srivastava IK, Barnett SW. 2013. Stabilizing exposure of conserved epitopes by structure guided insertion of disulfide bond in HIV-1 envelope glycoprotein. PLoS One 8:e76139. doi: 10.1371/journal.pone.0076139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Moore JP, Sodroski J. 1996. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J Virol 70:1863–1872. doi: 10.1128/JVI.70.3.1863-1872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Otsuki H, Hishiki T, Miura T, Hashimoto C, Narumi T, Tamamura H, Yoshimura K, Matsushita S, Igarashi T. 2013. Generation of a replication-competent simian-human immunodeficiency virus, the neutralization sensitivity of which can be enhanced in the presence of a small-molecule CD4 mimic. J Gen Virol 94:2710–2716. doi: 10.1099/vir.0.055590-0. [DOI] [PubMed] [Google Scholar]
- 121.Madani N, Princiotto AM, Easterhoff D, Bradley T, Luo K, Williams WB, Liao HX, Moody MA, Phad GE, Bernat NV, Melillo B, Santra S, Smith AB, Hedestam GBK, Haynes B, Sodroski J. 2016. Antibodies elicited by multiple envelope glycoprotein immunogens in primates neutralize primary human immunodeficiency viruses (HIV-1) sensitized by CD4-mimetic compounds. J Virol 90:5031–5046. doi: 10.1128/JVI.03211-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Madani N, Princiotto AM, Schon A, LaLonde J, Feng Y, Freire E, Park J, Courter JR, Jones DM, Robinson J, Liao HX, Moody MA, Permar S, Haynes B, Smith AB, Wyatt R, Sodroski J. 2014. CD4-mimetic small molecules sensitize human immunodeficiency virus to vaccine-elicited antibodies. J Virol 88:6542–6555. doi: 10.1128/JVI.00540-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lusso P, Earl PL, Sironi F, Santoro F, Ripamonti C, Scarlatti G, Longhi R, Berger EA, Burastero SE. 2005. Cryptic nature of a conserved, CD4-inducible V3 loop neutralization epitope in the native envelope glycoprotein oligomer of CCR5-restricted, but not CXCR4-using, primary human immunodeficiency virus type 1 strains. J Virol 79:6957–6968. doi: 10.1128/JVI.79.11.6957-6968.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. 1993. Characterization of conserved human-immunodeficiency-virus type-1 GP120 neutralization epitopes exposed upon GP120-CD4 binding. J Virol 67:3978–3988. doi: 10.1128/JVI.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yoshimura K, Harada S, Shibata J, Hatada M, Yamada Y, Ochiai C, Tamamura H, Matsushita S. 2010. Enhanced exposure of human immunodeficiency virus type 1 primary isolate neutralization epitopes through binding of CD4 mimetic compounds. J Virol 84:7558–7568. doi: 10.1128/JVI.00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.O’Rourke SM, Schweighardt B, Phung P, Fonseca D, Terry K, Wrin T, Sinangil F, Berman PW. 2010. Mutation at a single position in the V2 domain of the HIV-1 envelope protein confers neutralization sensitivity to a highly neutralization-resistant virus. J Virol 84:11200–11209. doi: 10.1128/JVI.00790-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Horwitz JA, Bar-On Y, Lu CL, Fera D, Lockhart AAK, Lorenzi JCC, Nogueira L, Golijanin J, Scheid JF, Seaman MS, Gazumyan A, Zolla-Pazner S, Nussenzweig MC. 2017. Non-neutralizing antibodies alter the course of HIV-1 infection in vivo. Cell 170:637–648 e10. doi: 10.1016/j.cell.2017.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mielke D, Bandawe G, Zheng J, Jones J, Abrahams MR, Bekker V, Ochsenbauer C, Garrett N, Abdool Karim S, Moore PL, Morris L, Montefiori D, Anthony C, Ferrari G, Williamson C. 2021. ADCC-mediating non-neutralizing antibodies can exert immune pressure in early HIV-1 infection. PLoS Pathog 17:e1010046. doi: 10.1371/journal.ppat.1010046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kim JH, Excler JL, Michael NL. 2015. Lessons from the RV144 Thai phase III HIV-1 vaccine trial and the search for correlates of protection. Annu Rev Med 66:423–437. doi: 10.1146/annurev-med-052912-123749. [DOI] [PubMed] [Google Scholar]
- 130.Haim H, Salas I, McGee K, Eichelberger N, Winter E, Pacheco B, Sodroski J. 2013. Modeling virus- and antibody-specific factors to predict human immunodeficiency virus neutralization efficiency. Cell Host Microbe 14:547–558. doi: 10.1016/j.chom.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jolly C, Kashefi K, Hollinshead M, Sattentau QJ. 2004. HIV-1 cell to cell transfer across an Env-induced, actin-dependent synapse. J Exp Med 199:283–293. doi: 10.1084/jem.20030648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rudnicka D, Feldmann J, Porrot F, Wietgrefe S, Guadagnini S, Prevost MC, Estaquier J, Haase AT, Sol-Foulon N, Schwartz O. 2009. Simultaneous cell-to-cell transmission of human immunodeficiency virus to multiple targets through polysynapses. J Virol 83:6234–6246. doi: 10.1128/JVI.00282-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hubner W, McNerney GP, Chen P, Dale BM, Gordon RE, Chuang FY, Li XD, Asmuth DM, Huser T, Chen BK. 2009. Quantitative 3D video microscopy of HIV transfer across T cell virological synapses. Science 323:1743–1747. doi: 10.1126/science.1167525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bracq L, Xie M, Benichou S, Bouchet J. 2018. Mechanisms for cell-to-cell transmission of HIV-1. Front Immunol 9:260. doi: 10.3389/fimmu.2018.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Piguet V, Sattentau Q. 2004. Dangerous liaisons at the virological synapse. J Clin Invest 114:605–610. doi: 10.1172/JCI22812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jolly C, Mitar I, Sattentau QJ. 2007. Adhesion molecule interactions facilitate human immunodeficiency virus type 1-induced virological synapse formation between T cells. J Virol 81:13916–13921. doi: 10.1128/JVI.01585-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Veenstra M, Leon-Rivera R, Li M, Gama L, Clements JE, Berman JW. 2017. Mechanisms of CNS viral seeding by HIV(+) CD14(+) CD16(+) monocytes: establishment and reseeding of viral reservoirs contributing to HIV-associated neurocognitive disorders. mBio 8. doi: 10.1128/mBio.01280-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Park RJ, Wang T, Koundakjian D, Hultquist JF, Lamothe-Molina P, Monel B, Schumann K, Yu H, Krupzcak KM, Garcia-Beltran W, Piechocka-Trocha A, Krogan NJ, Marson A, Sabatini DM, Lander ES, Hacohen N, Walker BD. 2017. A genome-wide CRISPR screen identifies a restricted set of HIV host dependency factors. Nat Genet 49:193–203. doi: 10.1038/ng.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kruize Z, Maurer I, Boeser-Nunnink BDM, Booiman T, Kootstra NA. 2020. Genetic variations in the host dependency factors ALCAM and TPST2 impact HIV-1 disease progression. AIDS 34:1303–1312. doi: 10.1097/QAD.0000000000002540. [DOI] [PubMed] [Google Scholar]
- 140.Fais S, Capobianchi MR, Abbate I, Castilletti C, Gentile M, Cordiali Fei P, Ameglio F, Dianzani F. 1995. Unidirectional budding of HIV-1 at the site of cell-to-cell contact is associated with co-polarization of intercellular adhesion molecules and HIV-1 viral matrix protein. AIDS 9:329–335. doi: 10.1097/00002030-199504000-00003. [DOI] [PubMed] [Google Scholar]
- 141.Jolly C, Mitar I, Sattentau QJ. 2007. Requirement for an intact T-cell actin and tubulin cytoskeleton for efficient assembly and spread of human immunodeficiency virus type 1. J Virol 81:5547–5560. doi: 10.1128/JVI.01469-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jolly C, Sattentau QJ. 2004. Retroviral spread by induction of virological synapses. Traffic 5:643–650. doi: 10.1111/j.1600-0854.2004.00209.x. [DOI] [PubMed] [Google Scholar]
- 143.Groot F, Welsch S, Sattentau QJ. 2008. Efficient HIV-1 transmission from macrophages to T cells across transient virological synapses. Blood 111:4660–4663. doi: 10.1182/blood-2007-12-130070. [DOI] [PubMed] [Google Scholar]
- 144.Sourisseau M, Sol-Foulon N, Porrot F, Blanchet F, Schwartz O. 2007. Inefficient human immunodeficiency virus replication in mobile lymphocytes. J Virol 81:1000–1012. doi: 10.1128/JVI.01629-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Dimitrov DS, Willey RL, Sato H, Chang LJ, Blumenthal R, Martin MA. 1993. Quantitation of human immunodeficiency virus type 1 infection kinetics. J Virol 67:2182–2190. doi: 10.1128/JVI.67.4.2182-2190.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Murooka TT, Deruaz M, Marangoni F, Vrbanac VD, Seung E, von Andrian UH, Tager AM, Luster AD, Mempel TR. 2012. HIV-infected T cells are migratory vehicles for viral dissemination. Nature 490:283–287. doi: 10.1038/nature11398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Law KM, Komarova NL, Yewdall AW, Lee RK, Herrera OL, Wodarz D, Chen BK. 2016. In vivo HIV-1 cell-to-cell transmission promotes multicopy micro-compartmentalized infection. Cell Rep 15:2771–2783. doi: 10.1016/j.celrep.2016.05.059. [DOI] [PubMed] [Google Scholar]
- 148.Del Portillo A, Tripodi J, Najfeld V, Wodarz D, Levy DN, Chen BK. 2011. Multiploid inheritance of HIV-1 during cell-to-cell infection. J Virol 85:7169–7176. doi: 10.1128/JVI.00231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kreger J, Garcia J, Zhang H, Komarova NL, Wodarz D, Levy DN. 2021. Quantifying the dynamics of viral recombination during free virus and cell-to-cell transmission in HIV-1 infection. Virus Evol 7:veab026. doi: 10.1093/ve/veab026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kreger J, Komarova NL, Wodarz D. 2020. Effect of synaptic cell-to-cell transmission and recombination on the evolution of double mutants in HIV. J R Soc Interface 17:20190832. doi: 10.1098/rsif.2019.0832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wodarz D, Levy DN. 2011. Effect of different modes of viral spread on the dynamics of multiply infected cells in human immunodeficiency virus infection. J R Soc Interface 8:289–300. doi: 10.1098/rsif.2010.0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wodarz D, Levy DN, Komarova NL. 2019. Multiple infection of cells changes the dynamics of basic viral evolutionary processes. Evol Lett 3:104–115. doi: 10.1002/evl3.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Boulle M, Muller TG, Dahling S, Ganga Y, Jackson L, Mahamed D, Oom L, Lustig G, Neher RA, Sigal A. 2016. HIV cell-to-cell spread results in earlier onset of viral gene expression by multiple infections per cell. PLoS Pathog 12:e1005964. doi: 10.1371/journal.ppat.1005964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Jackson L, Hunter J, Cele S, Ferreira IM, Young AC, Karim F, Madansein R, Dullabh KJ, Chen CY, Buckels NJ, Ganga Y, Khan K, Boulle M, Lustig G, Neher RA, Sigal A. 2018. Incomplete inhibition of HIV infection results in more HIV infected lymph node cells by reducing cell death. Elife 7. doi: 10.7554/eLife.30134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sigal A, Kim JT, Balazs AB, Dekel E, Mayo A, Milo R, Baltimore D. 2011. Cell-to-cell spread of HIV permits ongoing replication despite antiretroviral therapy. Nature 477:95–98. doi: 10.1038/nature10347. [DOI] [PubMed] [Google Scholar]
- 156.Josefsson L, King MS, Makitalo B, Brannstrom J, Shao W, Maldarelli F, Kearney MF, Hu WS, Chen J, Gaines H, Mellors JW, Albert J, Coffin JM, Palmer SE. 2011. Majority of CD4+ T cells from peripheral blood of HIV-1-infected individuals contain only one HIV DNA molecule. Proc Natl Acad Sci USA 108:11199–11204. doi: 10.1073/pnas.1107729108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Josefsson L, Palmer S, Faria NR, Lemey P, Casazza J, Ambrozak D, Kearney M, Shao W, Kottilil S, Sneller M, Mellors J, Coffin JM, Maldarelli F. 2013. Single cell analysis of lymph node tissue from HIV-1 infected patients reveals that the majority of CD4+ T-cells contain one HIV-1 DNA molecule. PLoS Pathog 9:e1003432. doi: 10.1371/journal.ppat.1003432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Frost SD, Liu Y, Pond SL, Chappey C, Wrin T, Petropoulos CJ, Little SJ, Richman DD. 2005. Characterization of human immunodeficiency virus type 1 (HIV-1) envelope variation and neutralizing antibody responses during transmission of HIV-1 subtype B. J Virol 79:6523–6527. doi: 10.1128/JVI.79.10.6523-6527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Richman DD, Wrin T, Little SJ, Petropoulos CJ. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci USA 100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Simek MD, Rida W, Priddy FH, Pung P, Carrow E, Laufer DS, Lehrman JK, Boaz M, Tarragona-Fiol T, Miiro G, Birungi J, Pozniak A, McPhee DA, Manigart O, Karita E, Inwoley A, Jaoko W, Dehovitz J, Bekker LG, Pitisuttithum P, Paris R, Walker LM, Poignard P, Wrin T, Fast PE, Burton DR, Koff WC. 2009. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J Virol 83:7337–7348. doi: 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.McCoy LE, Weiss RA. 2013. Neutralizing antibodies to HIV-1 induced by immunization. J Exp Med 210:209–223. doi: 10.1084/jem.20121827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Stamatatos L, Morris L, Burton DR, Mascola JR. 2009. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nat Med 15:866–870. doi: 10.1038/nm.1949. [DOI] [PubMed] [Google Scholar]
- 163.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Koff WC, Wilson IA, Burton DR, Poignard P, Protocol G Principal Investigators . 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Mascola JR, Haynes BF. 2013. HIV-1 neutralizing antibodies: understanding nature's pathways. Immunol Rev 254:225–244. doi: 10.1111/imr.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Moog C, Fleury HJ, Pellegrin I, Kirn A, Aubertin AM. 1997. Autologous and heterologous neutralizing antibody responses following initial seroconversion in human immunodeficiency virus type 1-infected individuals. J Virol 71:3734–3741. doi: 10.1128/JVI.71.5.3734-3741.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, Ott RG, Anthony RM, Zebroski H, Hurley A, Phogat A, Chakrabarti B, Li Y, Connors M, Pereyra F, Walker BD, Wardemann H, Ho D, Wyatt RT, Mascola JR, Ravetch JV, Nussenzweig MC. 2009. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 167.Malbec M, Porrot F, Rua R, Horwitz J, Klein F, Halper-Stromberg A, Scheid JF, Eden C, Mouquet H, Nussenzweig MC, Schwartz O. 2013. Broadly neutralizing antibodies that inhibit HIV-1 cell to cell transmission. J Exp Med 210:2813–2821. doi: 10.1084/jem.20131244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hessell AJ, Poignard P, Hunter M, Hangartner L, Tehrani DM, Bleeker WK, Parren PW, Marx PA, Burton DR. 2009. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat Med 15:951–954. doi: 10.1038/nm.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hessell AJ, Rakasz EG, Poignard P, Hangartner L, Landucci G, Forthal DN, Koff WC, Watkins DI, Burton DR. 2009. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog 5:e1000433. doi: 10.1371/journal.ppat.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hessell AJ, Rakasz EG, Tehrani DM, Huber M, Weisgrau KL, Landucci G, Forthal DN, Koff WC, Poignard P, Watkins DI, Burton DR. 2010. Broadly neutralizing monoclonal antibodies 2F5 and 4E10 directed against the human immunodeficiency virus type 1 gp41 membrane-proximal external region protect against mucosal challenge by simian-human immunodeficiency virus SHIVBa-L. J Virol 84:1302–1313. doi: 10.1128/JVI.01272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Pietzsch J, Gruell H, Bournazos S, Donovan BM, Klein F, Diskin R, Seaman MS, Bjorkman PJ, Ravetch JV, Ploss A, Nussenzweig MC. 2012. A mouse model for HIV-1 entry. Proc Natl Acad Sci USA 109:15859–15864. doi: 10.1073/pnas.1213409109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Barouch DH, Whitney JB, Moldt B, Klein F, Oliveira TY, Liu J, Stephenson KE, Chang HW, Shekhar K, Gupta S, Nkolola JP, Seaman MS, Smith KM, Borducchi EN, Cabral C, Smith JY, Blackmore S, Sanisetty S, Perry JR, Beck M, Lewis MG, Rinaldi W, Chakraborty AK, Poignard P, Nussenzweig MC, Burton DR. 2013. Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature 503:224–228. doi: 10.1038/nature12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Shingai M, Nishimura Y, Klein F, Mouquet H, Donau OK, Plishka R, Buckler-White A, Seaman M, Piatak M, Jr, Lifson JD, Dimitrov DS, Nussenzweig MC, Martin MA. 2013. Antibody-mediated immunotherapy of macaques chronically infected with SHIV suppresses viraemia. Nature 503:277–280. doi: 10.1038/nature12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Klein F, Halper-Stromberg A, Horwitz JA, Gruell H, Scheid JF, Bournazos S, Mouquet H, Spatz LA, Diskin R, Abadir A, Zang T, Dorner M, Billerbeck E, Labitt RN, Gaebler C, Marcovecchio PM, Incesu RB, Eisenreich TR, Bieniasz PD, Seaman MS, Bjorkman PJ, Ravetch JV, Ploss A, Nussenzweig MC. 2012. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature 492:118–122. doi: 10.1038/nature11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Caskey M, Klein F, Lorenzi JC, Seaman MS, West AP, Jr, Buckley N, Kremer G, Nogueira L, Braunschweig M, Scheid JF, Horwitz JA, Shimeliovich I, Ben-Avraham S, Witmer-Pack M, Platten M, Lehmann C, Burke LA, Hawthorne T, Gorelick RJ, Walker BD, Keler T, Gulick RM, Fatkenheuer G, Schlesinger SJ, Nussenzweig MC. 2015. Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature 522:487–491. doi: 10.1038/nature14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Caskey M, Schoofs T, Gruell H, Settler A, Karagounis T, Kreider EF, Murrell B, Pfeifer N, Nogueira L, Oliveira TY, Learn GH, Cohen YZ, Lehmann C, Gillor D, Shimeliovich I, Unson-O'Brien C, Weiland D, Robles A, Kummerle T, Wyen C, Levin R, Witmer-Pack M, Eren K, Ignacio C, Kiss S, West AP, Jr, Mouquet H, Zingman BS, Gulick RM, Keler T, Bjorkman PJ, Seaman MS, Hahn BH, Fatkenheuer G, Schlesinger SJ, Nussenzweig MC, Klein F. 2017. Antibody 10–1074 suppresses viremia in HIV-1-infected individuals. Nat Med 23:185–191. doi: 10.1038/nm.4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lynch RM, Boritz E, Coates EE, DeZure A, Madden P, Costner P, Enama ME, Plummer S, Holman L, Hendel CS, Gordon I, Casazza J, Conan-Cibotti M, Migueles SA, Tressler R, Bailer RT, McDermott A, Narpala S, O'Dell S, Wolf G, Lifson JD, Freemire BA, Gorelick RJ, Pandey JP, Mohan S, Chomont N, Fromentin R, Chun TW, Fauci AS, Schwartz RM, Koup RA, Douek DC, Hu Z, Capparelli E, Graham BS, Mascola JR, Ledgerwood JE, VRCS 601 Study Team . 2015. Virologic effects of broadly neutralizing antibody VRC01 administration during chronic HIV-1 infection. Sci Transl Med 7:319ra206. [DOI] [PubMed] [Google Scholar]
- 178.Ledgerwood JE, Coates EE, Yamshchikov G, Saunders JG, Holman L, Enama ME, DeZure A, Lynch RM, Gordon I, Plummer S, Hendel CS, Pegu A, Conan-Cibotti M, Sitar S, Bailer RT, Narpala S, McDermott A, Louder M, O'Dell S, Mohan S, Pandey JP, Schwartz RM, Hu Z, Koup RA, Capparelli E, Mascola JR, Graham BS, Team VRCS . 2015. Safety, pharmacokinetics and neutralization of the broadly neutralizing HIV-1 human monoclonal antibody VRC01 in healthy adults. Clin Exp Immunol 182:289–301. doi: 10.1111/cei.12692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Durham ND, Yewdall AW, Chen P, Lee R, Zony C, Robinson JE, Chen BK. 2012. Neutralization resistance of virological synapse-mediated HIV-1 Infection is regulated by the gp41 cytoplasmic tail. J Virol 86:7484–7495. doi: 10.1128/JVI.00230-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Gombos RB, Kolodkin-Gal D, Eslamizar L, Owuor JO, Mazzola E, Gonzalez AM, Korioth-Schmitz B, Gelman RS, Montefiori DC, Haynes BF, Schmitz JE. 2015. Inhibitory effect of individual or combinations of broadly neutralizing antibodies and antiviral reagents against cell-free and cell-to-cell HIV-1 transmission. J Virol 89:7813–7828. doi: 10.1128/JVI.00783-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Duncan CJ, Williams JP, Schiffner T, Gartner K, Ochsenbauer C, Kappes J, Russell RA, Frater J, Sattentau QJ. 2014. High-multiplicity HIV-1 infection and neutralizing antibody evasion mediated by the macrophage-T cell virological synapse. J Virol 88:2025–2034. doi: 10.1128/JVI.03245-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Titanji BK, Aasa-Chapman M, Pillay D, Jolly C. 2013. Protease inhibitors effectively block cell-to-cell spread of HIV-1 between T cells. Retrovirology 10:161. doi: 10.1186/1742-4690-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Reh L, Magnus C, Schanz M, Weber J, Uhr T, Rusert P, Trkola A. 2015. Capacity of broadly neutralizing antibodies to inhibit HIV-1 cell-cell transmission is strain- and epitope-dependent. PLoS Pathog 11:e1004966. doi: 10.1371/journal.ppat.1004966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Abela IA, Berlinger L, Schanz M, Reynell L, Gunthard HF, Rusert P, Trkola A. 2012. Cell-cell transmission enables HIV-1 to evade inhibition by potent CD4bs directed antibodies. PLoS Pathog 8:e1002634. doi: 10.1371/journal.ppat.1002634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Keele BF. 2010. Identifying and characterizing recently transmitted viruses. Curr Opin HIV AIDS 5:327–334. doi: 10.1097/COH.0b013e32833a0b9b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, Li H, Decker JM, Wang S, Baalwa J, Kraus MH, Parrish NF, Shaw KS, Guffey MB, Bar KJ, Davis KL, Ochsenbauer-Jambor C, Kappes JC, Saag MS, Cohen MS, Mulenga J, Derdeyn CA, Allen S, Hunter E, Markowitz M, Hraber P, Perelson AS, Bhattacharya T, Haynes BF, Korber BT, Hahn BH, Shaw GM. 2009. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J Exp Med 206:1273–1289. doi: 10.1084/jem.20090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Gupta P, Balachandran R, Ho M, Enrico A, Rinaldo C. 1989. Cell-to-cell transmission of human immunodeficiency virus type 1 in the presence of azidothymidine and neutralizing antibody. J Virol 63:2361–2365. doi: 10.1128/JVI.63.5.2361-2365.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ganesh L, Leung K, Lore K, Levin R, Panet A, Schwartz O, Koup RA, Nabel GJ. 2004. Infection of specific dendritic cells by CCR5-tropic human immunodeficiency virus type 1 promotes cell-mediated transmission of virus resistant to broadly neutralizing antibodies. J Virol 78:11980–11987. doi: 10.1128/JVI.78.21.11980-11987.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.van Montfort T, Nabatov AA, Geijtenbeek TB, Pollakis G, Paxton WA. 2007. Efficient capture of antibody neutralized HIV-1 by cells expressing DC-SIGN and transfer to CD4+ T lymphocytes. J Immunol 178:3177–3185. doi: 10.4049/jimmunol.178.5.3177. [DOI] [PubMed] [Google Scholar]
- 190.Massanella M, Puigdomenech I, Cabrera C, Fernandez-Figueras MT, Aucher A, Gaibelet G, Hudrisier D, Garcia E, Bofill M, Clotet B, Blanco J. 2009. Antigp41 antibodies fail to block early events of virological synapses but inhibit HIV spread between T cells. AIDS 23:183–188. doi: 10.1097/QAD.0b013e32831ef1a3. [DOI] [PubMed] [Google Scholar]
- 191.McCoy LE, Groppelli E, Blanchetot C, de Haard H, Verrips T, Rutten L, Weiss RA, Jolly C. 2014. Neutralisation of HIV-1 cell-cell spread by human and llama antibodies. Retrovirology 11:83. doi: 10.1186/s12977-014-0083-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Martin N, Welsch S, Jolly C, Briggs JA, Vaux D, Sattentau QJ. 2010. Virological synapse-mediated spread of human immunodeficiency virus type 1 between T cells is sensitive to entry inhibition. J Virol 84:3516–3527. doi: 10.1128/JVI.02651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.McCoy LE, Falkowska E, Doores KJ, Le K, Sok D, van Gils MJ, Euler Z, Burger JA, Seaman MS, Sanders RW, Schuitemaker H, Poignard P, Wrin T, Burton DR. 2015. Incomplete neutralization and deviation from sigmoidal neutralization curves for HIV broadly neutralizing monoclonal antibodies. PLoS Pathog 11:e1005110. doi: 10.1371/journal.ppat.1005110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Dale BM, McNerney GP, Thompson DL, Hubner W, de Los Reyes K, Chuang FY, Huser T, Chen BK. 2011. Cell-to-cell transfer of HIV-1 via virological synapses leads to endosomal virion maturation that activates viral membrane fusion. Cell Host Microbe 10:551–562. doi: 10.1016/j.chom.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Murakami T, Ablan S, Freed EO, Tanaka Y. 2004. Regulation of human immunodeficiency virus type 1 env-mediated membrane fusion by viral protease activity. J Virol 78:1026–1031. doi: 10.1128/jvi.78.2.1026-1031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Jiang J, Aiken C. 2007. Maturation-dependent human immunodeficiency virus type 1 particle fusion requires a carboxyl-terminal region of the gp41 cytoplasmic tail. J Virol 81:9999–10008. doi: 10.1128/JVI.00592-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Moulard M, Chaloin L, Canarelli S, Mabrouk K, Darbon H, Challoin L. 1998. Retroviral envelope glycoprotein processing: structural investigation of the cleavage site. Biochemistry 37:4510–4517. doi: 10.1021/bi972662f. [DOI] [PubMed] [Google Scholar]
- 198.Miranda LR, Schaefer BC, Kupfer A, Hu Z, Franzusoff A. 2002. Cell surface expression of the HIV-1 envelope glycoproteins is directed from intracellular CTLA-4-containing regulated secretory granules. Proc Natl Acad Sci USA 99:8031–8036. doi: 10.1073/pnas.122696599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Byland R, Vance PJ, Hoxie JA, Marsh M. 2007. A conserved dileucine motif mediates clathrin and AP-2-dependent endocytosis of the HIV-1 envelope protein. Mol Biol Cell 18:414–425. doi: 10.1091/mbc.e06-06-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Rowell JF, Stanhope PE, Siliciano RF. 1995. Endocytosis of endogenously synthesized HIV-1 envelope protein. Mechanism and role in processing for association with class II MHC. J Immunol 155:473–488. [PubMed] [Google Scholar]
- 201.Wyss S, Berlioz-Torrent C, Boge M, Blot G, Honing S, Benarous R, Thali M. 2001. The highly conserved C-terminal dileucine motif in the cytosolic domain of the human immunodeficiency virus type 1 envelope glycoprotein is critical for its association with the AP-1 clathrin adaptor [correction of adapter]. J Virol 75:2982–2992. doi: 10.1128/JVI.75.6.2982-2992.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Sauter MM, Pelchen-Matthews A, Bron R, Marsh M, LaBranche CC, Vance PJ, Romano J, Haggarty BS, Hart TK, Lee WM, Hoxie JA. 1996. An internalization signal in the simian immunodeficiency virus transmembrane protein cytoplasmic domain modulates expression of envelope glycoproteins on the cell surface. J Cell Biol 132:795–811. doi: 10.1083/jcb.132.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Willey RL, Bonifacino JS, Potts BJ, Martin MA, Klausner RD. 1988. Biosynthesis, cleavage, and degradation of the human immunodeficiency virus 1 envelope glycoprotein gp160. Proc Natl Acad Sci USA 85:9580–9584. doi: 10.1073/pnas.85.24.9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Castillo-Menendez LR, Witt K, Espy N, Princiotto A, Madani N, Pacheco B, Finzi A, Sodroski J. 2018. Comparison of uncleaved and mature human immunodeficiency virus membrane envelope glycoprotein trimers. J Virol 92. doi: 10.1128/JVI.00277-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Chakrabarti BK, Pancera M, Phogat S, O’Dell S, McKee K, Guenaga J, Robinson J, Mascola J, Wyatt RT. 2011. HIV type 1 Env precursor cleavage state affects recognition by both neutralizing and nonneutralizing gp41 antibodies. AIDS Res Hum Retroviruses 27:877–887. doi: 10.1089/AID.2010.0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Pancera M, Wyatt R. 2005. Selective recognition of oligomeric HIV-1 primary isolate envelope glycoproteins by potently neutralizing ligands requires efficient precursor cleavage. Virology 332:145–156. doi: 10.1016/j.virol.2004.10.042. [DOI] [PubMed] [Google Scholar]
- 207.Lu M, Ma X, Reichard N, Terry DS, Arthos J, Smith AB, 3rd, Sodroski JG, Blanchard SC, Mothes W. 2020. Shedding-resistant HIV-1 envelope glycoproteins adopt downstream conformations that remain responsive to conformation-preferring ligands. J Virol 94. doi: 10.1128/JVI.00597-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Wyss S, Dimitrov AS, Baribaud F, Edwards TG, Blumenthal R, Hoxie JA. 2005. Regulation of human immunodeficiency virus type 1 envelope glycoprotein fusion by a membrane-interactive domain in the gp41 cytoplasmic tail. J Virol 79:12231–12241. doi: 10.1128/JVI.79.19.12231-12241.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Berlioz-Torrent C, Shacklett BL, Erdtmann L, Delamarre L, Bouchaert I, Sonigo P, Dokhelar MC, Benarous R. 1999. Interactions of the cytoplasmic domains of human and simian retroviral transmembrane proteins with components of the clathrin adaptor complexes modulate intracellular and cell surface expression of envelope glycoproteins. J Virol 73:1350–1361. doi: 10.1128/JVI.73.2.1350-1361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Qi M, Williams JA, Chu H, Chen X, Wang JJ, Ding L, Akhirome E, Wen X, Lapierre LA, Goldenring JR, Spearman P. 2013. Rab11-FIP1C and Rab14 direct plasma membrane sorting and particle incorporation of the HIV-1 envelope glycoprotein complex. PLoS Pathog 9:e1003278. doi: 10.1371/journal.ppat.1003278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.LaBranche CC, Sauter MM, Haggarty BS, Vance PJ, Romano J, Hart TK, Bugelski PJ, Marsh M, Hoxie JA. 1995. A single amino acid change in the cytoplasmic domain of the simian immunodeficiency virus transmembrane molecule increases envelope glycoprotein expression on infected cells. J Virol 69:5217–5227. doi: 10.1128/JVI.69.9.5217-5227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Bhakta SJ, Shang L, Prince JL, Claiborne DT, Hunter E. 2011. Mutagenesis of tyrosine and di-leucine motifs in the HIV-1 envelope cytoplasmic domain results in a loss of Env-mediated fusion and infectivity. Retrovirology 8. doi: 10.1186/1742-4690-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Marsh M, Pelchen-Matthews A, Hoxie JA. 1997. Roles for endocytosis in lentiviral replication. Trends Cell Biol 7:1–4. doi: 10.1016/S0962-8924(97)20038-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Deschambeault J, Lalonde JP, Cervantes-Acosta G, Lodge R, Cohen EA, Lemay G. 1999. Polarized human immunodeficiency virus budding in lymphocytes involves a tyrosine-based signal and favors cell-to-cell viral transmission. J Virol 73:5010–5017. doi: 10.1128/JVI.73.6.5010-5017.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Lodge R, Lalonde JP, Lemay G, Cohen EA. 1997. The membrane-proximal intracytoplasmic tyrosine residue of HIV-1 envelope glycoprotein is critical for basolateral targeting of viral budding in MDCK cells. EMBO J 16:695–705. doi: 10.1093/emboj/16.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Day JR, Munk C, Guatelli JC. 2004. The membrane-proximal tyrosine-based sorting signal of human immunodeficiency virus type 1 gp41 is required for optimal viral infectivity. J Virol 78:1069–1079. doi: 10.1128/jvi.78.3.1069-1079.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Fultz PN, Vance PJ, Endres MJ, Tao B, Dvorin JD, Davis IC, Lifson JD, Montefiori DC, Marsh M, Malim MH, Hoxie JA. 2001. In vivo attenuation of simian immunodeficiency virus by disruption of a tyrosine-dependent sorting signal in the envelope glycoprotein cytoplasmic tail. J Virol 75:278–291. doi: 10.1128/JVI.75.1.278-291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Breed MW, Jordan AP, Aye PP, Lichtveld CF, Midkiff CC, Schiro FR, Haggarty BS, Sugimoto C, Alvarez X, Sandler NG, Douek DC, Kuroda MJ, Pahar B, Piatak M, Jr, Lifson JD, Keele BF, Hoxie JA, Lackner AA. 2013. Loss of a tyrosine-dependent trafficking motif in the simian immunodeficiency virus envelope cytoplasmic tail spares mucosal CD4 cells but does not prevent disease progression. J Virol 87:1528–1543. doi: 10.1128/JVI.01928-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Breed MW, Elser SE, Torben W, Jordan AP, Aye PP, Midkiff C, Schiro F, Sugimoto C, Alvarez-Hernandez X, Blair RV, Somasunderam A, Utay NS, Kuroda MJ, Pahar B, Wiseman RW, O'Connor DH, LaBranche CC, Montefiori DC, Marsh M, Li Y, Piatak M, Jr, Lifson JD, Keele BF, Fultz PN, Lackner AA, Hoxie JA. 2015. Elite control, gut CD4 T cell sparing, and enhanced mucosal T Cell responses in Macaca nemestrina Infected by a Simian immunodeficiency virus lacking a gp41 trafficking motif. J Virol 89:10156–10175. doi: 10.1128/JVI.01134-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Dev J, Park D, Fu Q, Chen J, Ha HJ, Ghantous F, Herrmann T, Chang W, Liu Z, Frey G, Seaman MS, Chen B, Chou JJ. 2016. Structural basis for membrane anchoring of HIV-1 envelope spike. Science 353:172–175. doi: 10.1126/science.aaf7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Qi M, Chu H, Chen X, Choi J, Wen X, Hammonds J, Ding L, Hunter E, Spearman P. 2015. A tyrosine-based motif in the HIV-1 envelope glycoprotein tail mediates cell-type- and Rab11-FIP1C-dependent incorporation into virions. Proc Natl Acad Sci USA 112:7575–7580. doi: 10.1073/pnas.1504174112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Wang L, Sandmeyer A, Hubner W, Li H, Huser T, Chen BK. 2021. A replication-competent HIV clone carrying GFP-Env reveals rapid Env recycling at the HIV-1 T cell virological synapse. Viruses 14:38. doi: 10.3390/v14010038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Groppelli E, Len AC, Granger LA, Jolly C. 2014. Retromer regulates HIV-1 envelope glycoprotein trafficking and incorporation into virions. PLoS Pathog 10:e1004518. doi: 10.1371/journal.ppat.1004518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Anand SP, Grover JR, Tolbert WD, Prevost J, Richard J, Ding S, Baril S, Medjahed H, Evans DT, Pazgier M, Mothes W, Finzi A. 2019. Antibody-induced internalization of HIV-1 Env proteins limits surface expression of the closed conformation of Env. J Virol 93. doi: 10.1128/JVI.00293-19. [DOI] [PMC free article] [PubMed] [Google Scholar]