ABSTRACT
The successful infection of a host plant by a phytopathogenic bacterium depends on a finely tuned molecular cross talk between the two partners. Thanks to transposon insertion sequencing techniques (Tn-seq), whole genomes can now be assessed to determine which genes are important for the fitness of several plant-associated bacteria in planta. Despite its agricultural relevance, the dynamic molecular interaction established between the foliar hemibiotrophic phytopathogen Xanthomonas hortorum pv. vitians and its host, lettuce (Lactuca sativa), remains completely unknown. To decipher the genes and functions mobilized by the pathogen throughout the infection process, we conducted a Tn-seq experiment in lettuce leaves to mimic the selective pressure occurring during natural infection. This genome-wide screening identified 170 genes whose disruption caused serious fitness defects in lettuce. A thorough examination of these genes using comparative genomics and gene set enrichment analyses highlighted that several functions and pathways were highly critical for the pathogen’s survival. Numerous genes involved in amino acid, nucleic acid, and exopolysaccharide biosynthesis were critical. The xps type II secretion system operon, a few TonB-dependent transporters involved in carbohydrate or siderophore scavenging, and multiple genes of the carbohydrate catabolism pathways were also critical, emphasizing the importance of nutrition systems in a nutrient-limited environment. Finally, several genes implied in camouflage from the plant immune system and resistance to immunity-induced oxidative stress were strongly involved in host colonization. As a whole, these results highlight some of the central metabolic pathways and cellular functions critical for Xanthomonas host adaptation and pathogenesis.
IMPORTANCE Xanthomonas hortorum was recently the subject of renewed interest, as several studies highlighted that its members were responsible for diseases in a wide range of plant species, including crops of agricultural relevance (e.g., tomato and carrot). Among X. hortorum variants, X. hortorum pv. vitians is a reemerging foliar hemibiotrophic phytopathogen responsible for severe outbreaks of bacterial leaf spot of lettuce all around the world. Despite recent findings, sustainable and practical means of disease control remain to be developed. Understanding the host-pathogen interaction from a molecular perspective is crucial to support these efforts. The genes and functions mobilized by X. hortorum pv. vitians during its interaction with lettuce had never been investigated. Our study sheds light on these processes by screening the whole pathogen genome for genes critical for its fitness during the infection process, using transposon insertion sequencing and comparative genomics.
KEYWORDS: Xanthomonas hortorum, phytopathogen, lettuce, Tn-seq, comparative genomics
INTRODUCTION
Most Gram-negative bacterial plant pathogens behave as hemibiotrophs when interacting with their host plant (1). Xanthomonas species are indeed mostly considered hemibiotrophic pathogens that cause necrosis after a first asymptomatic phase (2–4). The infection process of a host plant by a nonvascular Xanthomonas is sequential: the pathogen displays different abilities in the cross talk with its host and ensures its own survival through nutrient acquisition systems and transformation of its microniche (5). Xanthomonas species initially start as epiphytes in the phyllosphere, an environment generally considered stressful and oligotrophic (6). There, in addition to deploying strategies to adhere to the leaf surface and survive abiotic stresses (7, 8), they rely on high-affinity TonB-dependent transporters (TBDTs) to take up simple sugars like sucrose and xylose (9, 10) or exploit more complex carbon sources (11). Biofilm formation at this stage participates in creating a safer and stable microenvironment for the population to develop (12).
The pathogen penetrates the leaves through natural openings (i.e., stomata, hydathodes, and wounds) and establishes itself within the apoplast to initiate its endophytic biotroph phase (5). It has to withstand the early basal responses triggered by the recognition of microbial-associated molecular patterns (MAMPs) by the plant pattern recognition receptors (PRRs) (13), feed on available nutrients, and synthesize compounds that are not directly available within its habitat. Polysaccharides such as cell-surface lipopolysaccharides (LPS) and the secreted exopolysaccharide (EPS) xanthan help to dampen the host immune response (14, 15). The type III secretion system (T3SS) also plays a major role in this phase, as it allows Xanthomonas species to disrupt the immune response signaling cascades and remodel the host metabolism to their own benefit through the intensive secretion of type III effectors (T3Es) (16, 17). Finally, the pathogen evolves toward a necrotrophic lifestyle: other cell death-inducing T3Es and the secretion of cell-wall-degrading enzymes (CWDEs) by the type II secretion system (T2SS) allow it to feed on leaked intracellular compounds and degraded cell components (1, 16).
Xanthomonas hortorum pv. vitians is a foliar pathogen of cultivated lettuce (Lactuca sativa); it causes a disease called bacterial leaf spot of lettuce (BLSL) (18), characterized by the progressive formation of small necrotic spots a few days after the pathogen has been inoculated. Then, these spots coalesce to form large necrotic patches that alter the quality and yield of the harvest without killing the entire plants (19, 20). BLSL has become a significant threat for lettuce producers over the past decades, and X. hortorum pv. vitians is considered a reemerging pathogen all around the world (18, 20–23). Nowadays, very little is known about the biology of this pathogen during the infection process, and the molecular bases sustaining the host-pathogen interaction remain mostly unelucidated (24).
To investigate the molecular biology and describe the essential genome of X. hortorum pv. vitians in vitro, a transposon insertion sequencing (Tn-seq) approach was recently developed for our model strain LM16734 (25). Tn-seq techniques rely on the generation of a saturated mutant library through large-scale transposon mutagenesis coupled with high-throughput next-generation sequencing and statistical analysis to estimate the essentiality and/or fitness contribution of each genetic feature in a selective environment (26). Tn-seq applications to bacteria of agricultural interest recently improved our understanding of the molecular cross talk between symbiotic or pathogenic bacteria and their host plants (27). For the plant-pathogenic species Pantoea stewartii subsp. stewartii (28), Dickeya dadantii (29), Pseudomonas syringae (30, 31), Ralstonia solanacearum (32), and Agrobacterium tumefaciens (33), Tn-seq provided an overall picture of the cellular functions and processes mobilized during plant infection and identified novel virulence factors.
We used our Tn-seq mutant library to screen for genes implied in lettuce colonization by X. hortorum pv. vitians. We applied a combination of comparative genomics, gene set enrichment analysis, and metabolic and cellular pathway mapping to draw biological sense from the list of genes defined as conditionally essential in lettuce. This approach successfully identified critical genetic features involved in amino acid and nucleic acid biosynthesis, CWDE secretion by the T2SS, carbohydrate nutrition and iron uptake, cellular polysaccharide synthesis, and oxidative stress resistance systems.
RESULTS AND DISCUSSION
Validation of the Tn-seq experimental design and assessment of bottleneck effects.
To investigate the fitness contribution of each genetic feature of X. hortorum pv. vitians LM16734 to growth in rich medium broth and during lettuce leaf infection, we used the previously developed and published Mariner-based Tn-seq mutant library (25). This library consists of at least 60,715 chromosomal mutants and 1,218 plasmidic mutants with transposon insertions at individual TA dinucleotide sites, out of a total of 85,314 chromosomal and 1,361 plasmidic TA sites within the X. hortorum pv. vitians LM16734 complete genome (5.23-Mb circular chromosome and 57-kb native plasmid pLM16734). The insertion densities in the library were ca. 71% and 89% on the chromosome and the plasmid, respectively.
First, the infection bottleneck effect arising from our inoculation procedure in young lettuce plants was estimated. The central paradigm behind Tn-seq experiments states that the mean read count per genetic feature is correlated with the corresponding mutant fitness in the tested selective environment (26). However, sampling bottleneck effects—which are stochastic events of loss of genetic diversity within a population when its size is drastically reduced (e.g., during host penetration and infection by a pathogen)—are known to blur data interpretation if not carefully considered in the experimental design (34, 35). Moreover, the selective DNA library preparation steps and random nature of mass sequencing can result in another type of bottleneck by reducing the genetic diversity within the extracted genomic library DNA (36). The number of generations experienced by the mutant library is also a critical parameter to be considered, as it directly impacts the fitness score calculated for each genetic feature (34, 37). Conceptually, a Tn-seq experiment consists of thousands of competition assays conducted simultaneously (34). Thus, variations in a mutant’s abundance caused by selective pressure will be correlated with the number of generations experienced by the population.
A preliminary experiment was performed to estimate the number of bacterial cells spray-inoculated on lettuce leaves and the number of cells penetrating the leaves, as well as to monitor the population size over the course of infection. Approximately 2.5 × 104 CFU · cm−2 on average were deposited on the leaves during inoculation, and 5 × 103 CFU · cm−2 were measured inside the leaves after 24 h (Fig. 1a). Therefore, the theoretical successful infection ratio was 1:5, provided that no generation event occurred within this time interval. In reality, the founding population was likely to have been smaller than 5 × 103 CFU · cm−2, as a few generations probably occurred inside the leaves during the first 24 h. The generation time of X. hortorum pv. vitians is ca. 4.5 h in rich medium in vitro (data not shown). The total foliar surface for 30 plants was estimated to be approximately 6,000 cm2, meaning that a total of 1.5 × 108 CFU were inoculated on the leaves and that 3 × 107 CFU were inside the leaves after 24 h. Regarding these estimations, a theoretical total mutant library coverage of 2,500× was inoculated, and a rough estimate of 50× to 500× maximum successfully invaded the plant tissue. Finally, the highest population level was achieved 10 days postinoculation (dpi), with an average of 5.6 × 106 internal CFU · cm−2 (Fig. 1a). Theoretically, 10 bacterial generation events occurred between 1 and 10 dpi, without considering mortality events. In other comparable in planta Tn-seq experiments, similar numbers of generations proved to be adequate for efficient and unbiased fitness score assessment (30, 31, 38).
FIG 1.
Validation of the experimental design and bottleneck assessment. (a) Evolution of the X. hortorum pv. vitians LM16734 population during lettuce infection, expressed as log10-transformed CFU · cm−2 of inoculated leaf. Red triangles, measured epiphytic populations following spray inoculation; green triangles, measured endophytic populations at each time point; gray bars, standard errors calculated on triplicates. (b and c) Read count correlation among Tn-seq duplicates for the X. hortorum pv. vitians LM16734 chromosome in (b) the inoculum and (c) lettuce leaves. (d and e) Volcano plots of the resampling results in (d) the inoculum versus the in vitro library condition and (e) lettuce leaves versus the inoculum condition. Plots represent the log2 fold change of each genetic feature as a function of the negative log10 of the resampling test P value. Genetic features that passed the FDR adjustment test (q value of ≤0.05) are indicated in red if they are conditionally essential and in blue if they provide a growth advantage. Nonsignificant genetic features are in gray.
Read mapping statistics showed that 22 to 26 million reads mapped to unique TA sites in the inoculum duplicates (T1 and T2), and 26 to 30 million in the lettuce infection duplicates (P1 and P2) (Table 1). The insertion densities were 69.9 and 71.0% on the chromosome and 88.5 and 87.9% on pLM16734 under the inoculum condition and 61.9 and 64.5% on the chromosome and 71.9 and 77.3% on pLM16734 under the in planta condition. Almost all library mutants were still present in the inoculum, and ca. 90% chromosomal and 80% plasmidic mutants were recovered in lettuce after 10 days. Moreover, Pearson correlation coefficients of read counts at unique TA sites between duplicates were 0.98 and 0.97 for the chromosome and plasmid pLM16734 under the inoculum condition, respectively, and 0.77 and 0.84 under the lettuce condition (Fig. 1b and 1c). These sequencing statistics confirmed that our experimental design bypassed the experimental bottleneck and that the technical bottleneck resulting from the DNA library preparation steps and sequencing was negligible.
TABLE 1.
Transposon-insertion sequencing statistics before TTR normalization
| Replicate | Sequencing yield | No. of Tn end-containing reads | Replicon | Read count (no. of unique TA sites) | No. of TA hitsa | Insertion densityb | Median read count over non-zero TAc |
|---|---|---|---|---|---|---|---|
| T1 | 25,830,465 | 22,029,814 | Chromosome | 21,916,438 | 59,675 | 69.9 | 100.9 |
| pLM16734 | 1,262,371 | 1,205 | 88.5 | 62.6 | |||
| T2 | 39,102,386 | 26,462,594 | Chromosome | 26,330,348 | 60,579 | 71.0 | 99.6 |
| pLM16734 | 956,994 | 1,196 | 87.9 | 65.6 | |||
| P1 | 30,091,641 | 26,195,853 | Chromosome | 26,067,795 | 52,850 | 61.9 | 99.9 |
| pLM16734 | 1,227,568 | 979 | 71.9 | 77.3 | |||
| P2 | 34,333,964 | 30,135,609 | Chromosome | 29,982,890 | 55,013 | 64.5 | 93.8 |
| pLM16734 | 1,236,398 | 1,052 | 77.3 | 70.05 | |||
The X. hortorum pv. vitians LM16734 genome contains 85,314 chromosomal TA sites and 1,361 plasmidic TA sites.
Insertion density reflects the percentage of TA sites with at least one read mapped over the total number of TA sites on the replicon.
Mean read count per TA site containing at least one read.
Identification of 170 critical genes in planta and overrepresentation of anabolic processes.
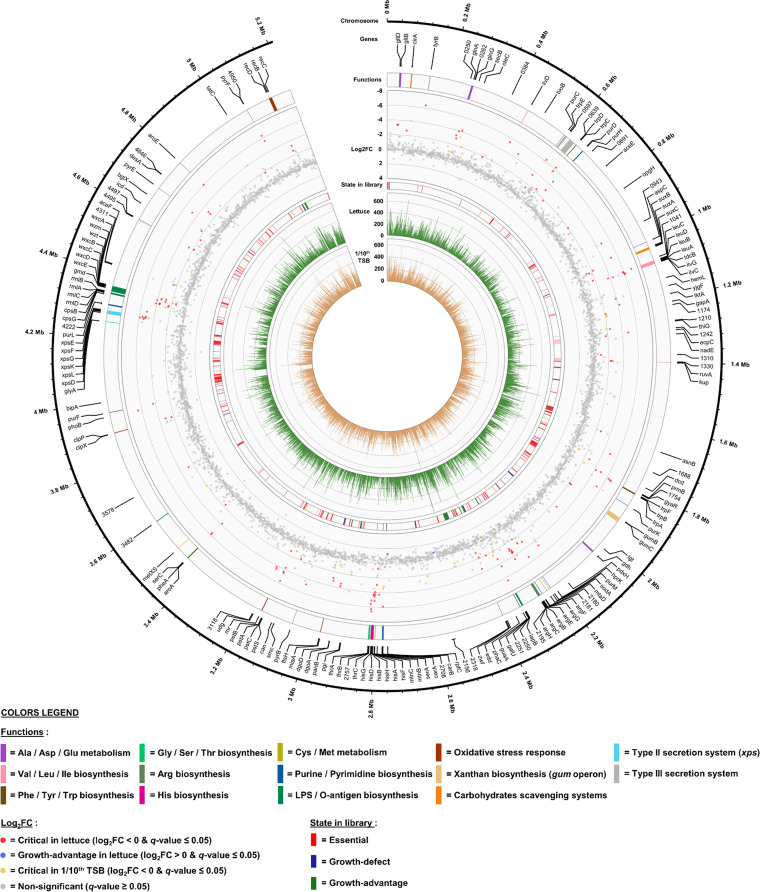
The resampling analyses conducted with TRANSIT identified 36 critical genes in 1/10th tryptic soy broth (TSB) medium (Fig. 1d) and 170 critical genes and two growth advantage genes in lettuce (Fig. 1e). The complete gene lists and associated resampling metrics are available in Tables S1 to S4 in the supplemental material. There was a partial overlap between the two critical gene sets, as 29 genes in lettuce leaf were already considered critical in 1/10th TSB. Additionally, we applied an extra filter by considering that mutants presenting a mean gene read count below 5 under the 1/10th TSB condition showed decreased fitness in vitro, and 8 more genes were classified as critical in 1/10th TSB. Examining the distribution of the critical genes in lettuce along the X. hortorum pv. vitians LM16734 chromosome revealed that a significant part seemed to be colocalized and organized in operons or gene clusters (Fig. 2).
FIG 2.
Genome atlas of the critical genetic features required for lettuce infection by X. hortorum pv. vitians LM16734. From the inner to outer track are the mean read count over 1 kb in 1/10th TSB and in lettuce, the gene state in the in vitro library, as described in reference 24, the log2 FC score in lettuce leaves compared to the inoculum, clustering of critical genes, pathways, functions and structural components in lettuce, gene names or locus tag identifiers, and chromosomal tracks.
Complete genome annotation and Tn-seq results in vitro and in planta. Download Table S1, XLSX file, 1.2 MB (1.2MB, xlsx) .
Copyright © 2022 Morinière et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To determine which biological processes, pathways, and functions were overrepresented in our lettuce critical gene set, Gene Ontology (GO) terms (39) and KEGG ortholog (KO) (40) identifiers were assigned to all X. hortorum pv. vitians LM16734 genes. The GO term enrichment analysis indicated that 43 GO terms were significantly enriched in our gene set (Table 2). Among the 35 biological process-related enriched GO terms, nucleotide, amino acid, polysaccharide, carbohydrate, nitrogen, and phosphorus metabolism-associated terms were most abundant. Overall, biosynthetic processes seemed to be more represented than degradation processes. For example, 67.7% of genes related to “Nucleobase biosynthetic process” (GO:0046112) versus 54.5% for “Nucleobase metabolic process” (GO:0009112) and 42.5% of genes associated with “Cellular amino acid biosynthetic process” (GO:0008652) versus 34.9% for “Cellular amino acid metabolic process” (GO:0006520) were identified. Interestingly, 5 out of 7 genes for “Phosphate ion transport” (GO:0006817) were also critical in planta.
TABLE 2.
Significantly enriched Gene Ontology terms in the critical genes in the lettuce leaf subset
| GO identifier | GO term | Total no. of genesa | No. of genes in subsetb | Enrichment scorec | q valued |
|---|---|---|---|---|---|
| Biological processes | |||||
| General processes | |||||
| GO:0006082 | Organic acid metabolic process | 298 | 64 | 2.686 | 0.0 |
| GO:0016053 | Organic acid biosynthetic process | 167 | 49 | 2.188 | 0.0 |
| GO:0044283 | Small molecule biosynthetic process | 226 | 56 | 1.965 | 0.0 |
| GO:1901362 | Organic cyclic compound biosynthetic process | 233 | 39 | 1.696 | 0.006043 |
| GO:0018130 | Heterocycle biosynthetic process | 223 | 37 | 1.7 | 0.008476 |
| GO:0019438 | Aromatic compound biosynthetic process | 199 | 38 | 1.984 | 0.000196 |
| GO:0046176 | Aldonic acid catabolic process | 4 | 3 | 5.8 | 0.033252 |
| GO:0040007 | Growth | 90 | 14 | 2.543 | 0.035375 |
| Nucleotide metabolism | |||||
| GO:0052803 | Imidazole-containing compound metabolic process | 12 | 6 | 5.887 | 0.011874 |
| GO:0034654 | Nucleobase-containing compound biosynthetic process | 138 | 22 | 2.025 | 0.013182 |
| GO:0055086 | Nucleobase-containing small molecule metabolic process | 124 | 26 | 1.901 | 0.015897 |
| GO:0042435 | Indole-containing compound biosynthetic process | 6 | 5 | 5.292 | 0.017839 |
| GO:0046112 | Nucleobase biosynthetic process | 9 | 6 | 4.451 | 0.022403 |
| GO:1901293 | Nucleoside phosphate biosynthetic process | 73 | 20 | 2.205 | 0.000229 |
| GO:0042430 | Indole-containing compound metabolic process | 11 | 7 | 7.289 | 0.000771 |
| GO:0009112 | Nucleobase metabolic process | 11 | 6 | 4.42 | 0.03223 |
| Amino acid metabolism | |||||
| GO:0006520 | Cellular amino acid metabolic process | 166 | 53 | 3.702 | 0.0 |
| GO:0008652 | Cellular amino acid biosynthetic process | 94 | 40 | 2.453 | 0.0 |
| GO:0009072 | Aromatic amino acid family metabolic process | 24 | 9 | 3.338 | 0.027736 |
| GO:0000105 | Histidine biosynthetic process | 8 | 6 | 3.642 | 0.035375 |
| Polysaccharide metabolism | |||||
| GO:0000271 | Polysaccharide biosynthetic process | 23 | 8 | 7.75 | 3.8e−05 |
| GO:0033692 | Cellular polysaccharide biosynthetic process | 19 | 7 | 8.4 | 7.7e−05 |
| GO:0044264 | Cellular polysaccharide metabolic process | 26 | 8 | 7.754 | 0.000214 |
| GO:0005976 | Polysaccharide metabolic process | 35 | 9 | 6.524 | 0.00034 |
| GO:0008653 | Lipopolysaccharide metabolic process | 12 | 6 | 5.546 | 0.003351 |
| Carbohydrate metabolism | |||||
| GO:1901135 | Carbohydrate derivative metabolic process | 178 | 29 | 1.95 | 0.008547 |
| GO:0046394 | Carboxylic acid biosynthetic process | 167 | 49 | 1.346 | 0.009947 |
| GO:0006757 | ATP generation from ADP | 15 | 4 | 4.938 | 0.028161 |
| Nitrogen and phosphate metabolism | |||||
| GO:0019637 | Organophosphate metabolic process | 147 | 25 | 2.12 | 0.008476 |
| GO:0044106 | Cellular amine metabolic process | 17 | 7 | 5.333 | 0.008547 |
| GO:0019627 | Urea metabolic process | 4 | 4 | 8.25 | 0.009169 |
| GO:0071941 | Nitrogen cycle metabolic process | 6 | 4 | 7.754 | 0.027141 |
| GO:0000050 | Urea cycle | 4 | 4 | 5.515 | 0.028161 |
| GO:0006793 | Phosphorus metabolic process | 244 | 34 | 1.602 | 0.042509 |
| GO:0006817 | Phosphate ion transport | 7 | 5 | 2.5 | 0.04326 |
| Molecular functions | |||||
| GO:0016829 | Lyase activity | 93 | 21 | 3.246 | 0.000101 |
| GO:0016614 | Oxidoreductase activity, acting on CH-OH group of donors | 49 | 11 | 3.6 | 0.002604 |
| GO:0016879 | Ligase activity, forming carbon-nitrogen bonds | 42 | 12 | 1.907 | 0.008476 |
| GO:0048037 | Cofactor binding | 251 | 28 | 1.86 | 0.015897 |
| GO:0005315 | Inorganic phosphate transmembrane transporter activity | 4 | 3 | 10.533 | 0.018301 |
| GO:0016638 | Oxidoreductase activity, acting on the CH-NH2 group of donors | 8 | 4 | 8.333 | 0.024953 |
| GO:0042802 | Identical protein binding | 144 | 19 | 1.661 | 0.033252 |
| GO:0070279 | Vitamin B6 binding | 32 | 7 | 4.238 | 0.034385 |
Total number of genes associated with the GO term in the X. hortorum pv. vitians LM16734 genome.
Number of genes associated with the GO term in the subset of genes conditionally essential in planta.
The simplified formula for the enrichment score (without pseudocounts) is log[(b/q)/(m/p)], where b is the number of genes with the GO term in the subset, q is the number of genes in the subset with a parent of the GO term, m is the total number of genes with the GO term in the genome, and p is the number of genes in the genome with a parent of the GO term. Thus, the enrichment score is the log-transformed ratio of the two relative abundances of genes with the GO term compared to those with a parent GO term (i) in the subset and (ii) in the complete genome. The calculation was derived from the Ontologizer method (114).
Adjusted P value using the Benjamini-Hochberg false-discovery rate (FDR) test. GO terms with a q value of <0.05 were considered significantly enriched.
The KEGG pathway analysis reached more precision in the identification of the most critical metabolic and cellular pathways in planta (Table 3). For instance, it highlighted that even though a total of 49.5% of the genes involved in the de novo biosynthesis of amino acids were critical in lettuce, the relative proportions of genes varied among the specific amino acid pathways. The most impacted biosynthetic pathways were those of arginine (92.3% of the genes), valine, leucine, and isoleucine (76.9%), and alanine, aspartate, and glutamate (64%) biosynthesis. The genes involved in these pathways were often colocalized, like the leu and ilv genes encoding part of leucine and isoleucine biosynthesis or the arg and his operons for arginine and histidine, respectively (Fig. 2). Similarly, 19.6% of the carbon metabolism genes were critical, but the prevalent pathways were those of the 2-oxocarboxylic acid metabolism (57.1%), the pentose phosphate pathway (36%), the starch and sucrose metabolism (31.6%), and glycolysis/gluconeogenesis (29%). The genes related to the polysaccharide metabolism were mostly implied in O-antigen nucleotide sugar biosynthesis (56.3%). This analysis also pinpointed that 22.9% of the genes associated with the bacterial secretion systems were critical, which was not detected by the GO term enrichment analysis.
TABLE 3.
Critical KEGG pathways in planta (nonexhaustive)
| Pathway code | KEGG pathway | No. of genes: |
% of genes in pathway | |
|---|---|---|---|---|
| Criticala | Totalb | |||
| General and diverse pathways | ||||
| xhr01110 | Biosynthesis of secondary metabolites | 84 | 301 | 27.9 |
| xhr01240 | Biosynthesis of cofactors | 19 | 128 | 14.8 |
| xhr00910 | Nitrogen metabolism | 9 | 14 | 64.3 |
| xhr03070 | Bacterial secretion system | 16 | 70 | 22.9 |
| Amino acid metabolism | ||||
| xhr01230 | Biosynthesis of amino acids | 54 | 109 | 49.5 |
| xhr00220 | Arginine biosynthesis | 12 | 13 | 92.3 |
| xhr00290 | Valine, leucine, and isoleucine biosynthesis | 10 | 13 | 76.9 |
| xhr00250 | Alanine, aspartate, and glutamate metabolism | 16 | 25 | 64 |
| xhr00400 | Phenylalanine, tyrosine, and tryptophan biosynthesis | 11 | 26 | 42.3 |
| xhr00260 | Glycine, serine, and threonine metabolism | 14 | 35 | 40 |
| xhr00340 | Histidine metabolism | 6 | 18 | 33.3 |
| xhr00270 | Cysteine and methionine metabolism | 11 | 37 | 29.7 |
| Carbon compound metabolism | ||||
| xhr01200 | Carbon metabolism | 18 | 92 | 19.6 |
| xhr01210 | 2-Oxocarboxylic acid metabolism | 12 | 21 | 57.1 |
| xhr00030 | Pentose phosphate pathway | 9 | 25 | 36 |
| xhr00500 | Starch and sucrose metabolism | 12 | 38 | 31.6 |
| xhr00010 | Glycolysis/gluconeogenesis | 9 | 31 | 29 |
| xhr00052 | Galactose metabolism | 5 | 20 | 25 |
| xhr00051 | Fructose and mannose metabolism | 5 | 21 | 23.8 |
| Nucleobase metabolism | ||||
| xhr00240 | Pyrimidine metabolism | 7 | 25 | 28 |
| xhr00230 | Purine metabolism | 11 | 54 | 20.4 |
| Polysaccharide metabolism | ||||
| xhr00541 | O-antigen nucleotide sugar biosynthesis | 9 | 16 | 56.3 |
Number of genes identified as conditionally essential in planta (q value of ≤0.05).
Total number of genes in model pathway in the X. hortorum-specific KEGG pathway database.
Overall, our results matched those of the other Tn-seq studies conducted on plant-pathogenic bacteria so far, in which many genes involved in the amino acid and nucleic acid metabolisms and in polysaccharide biosynthesis were critical in planta (28–30, 32). Moreover, random transposon mutant screening in other Xanthomonas species had already shown that mutating genes involved in these processes impaired growth and virulence in planta (41–44). For example, out of the 75 genes randomly screened as required for the virulence of Xanthomonas campestris pv. campestris in cabbage, 10 were involved in amino acid biosynthesis, 3 in de novo purine biosynthesis, 10 in lipopolysaccharide biosynthesis, and 3 in xanthan biosynthesis (41). This suggests that the central metabolic pathways required for plant colonization are equivalent across biological models and pathosystems, probably because plant-pathogenic bacteria face similar challenges in these selective environments: e.g., scarce nutrient or proteogenic amino acid availability.
Interestingly, no chemotaxis- or motility-related genes were screened as critical for the in planta lifestyle of X. hortorum pv. vitians, even though these features are usually considered important for pathogenicity and plant colonization (45). This could be explained by a combination of several factors. First, we hypothesized that during the spray-inoculation of our mutant library, some mutants may have been directly inoculated into the stomata and hydathodes. Thus, chemotaxis-driven motility was not required by these mutants to penetrate the leaves at the initial stages of plant infection. Second, motility might not be an indispensable feature for pathogenicity and in planta colonization by Xanthomonas species. A comparative genomic study of Xanthomonas fuscans subsp. fuscans suggested that the absence of motility could occur in field populations of several Xanthomonas species and that nonmotile variants would not undergo fitness defects in mixed populations, including flagellated strains (46). Furthermore, a new transcriptomic analysis of X. campestris pv. campestris conducted during cauliflower hydathode colonization revealed that the expression of most chemotaxis and motility genes was strongly repressed during the first days of infection (4). A similar observation was made during early rice infection by Xanthomonas oryzae pv. oryzicola (47). Such repression could even confer a stealth advantage to the bacterial population by reducing the production of microbial-associated molecular pattern (MAMP) molecules such as flagellar or pilus proteins (4, 46). Finally, chemotaxis and motility genes were not detected in the in planta RB-Tn-seq study of P. syringae, neither in the epiphytic screening nor in the apoplastic one (30). The in planta transcriptome analysis of Pseudomonas syringae infecting Arabidopsis thaliana also demonstrated that these genes were downregulated during plant infection (48), consistent with the fact that plant-pathogenic Xanthomonas and Pseudomonas species share similar overall infection strategies.
Nutrient acquisition and assimilation as key features of the in planta lifestyle.
Numerous mutants of the carbohydrate metabolism were also strongly impaired in their fitness in planta (Tables 2 and 3). Interestingly, many genes of the central carbohydrate catabolism pathways—glycolysis and the pentose phosphate and Entner-Doudoroff pathways—were critical in lettuce, while none had been found indispensable on a glucose-rich medium (25). This time, most of the genes in these three pathways were conditionally essential either in lettuce or in 1/10th TSB medium (see Fig. S1 in the supplemental material), and so were some key genes of the starch and sucrose degradation and fructose and mannose degradation pathways (Table 3). Unfortunately, no information is available about the chemical composition of the lettuce leaf apoplast or surface. Sucrose and malate are generally the most abundant carbohydrates in the apoplast of eudicots (49–52). In any case, this apparent metabolic versatility most probably contributes to the pathogen’s ability to adapt to the lettuce leaf environment, both outside and inside the leaf.
Critical genes in the carbon metabolism pathways of X. hortorum pv. vitians LM16734 under the different Tn-seq experimental conditions, created with KEGG Mapper. Carbon numbers in compounds are indicated in circles or by asterisks for cofactors. Download FIG S1, TIF file, 2.6 MB (2.6MB, tif) .
Copyright © 2022 Morinière et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
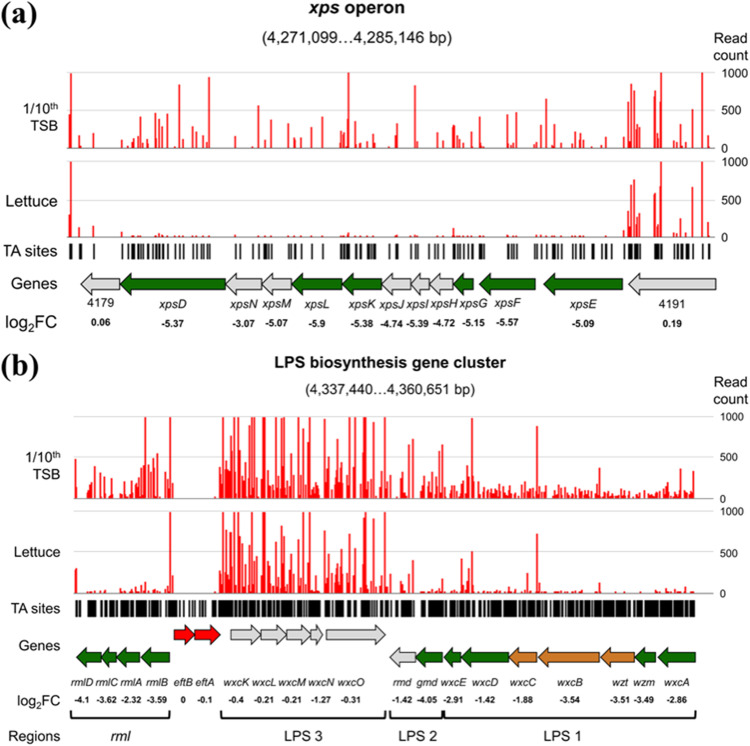
Consistent with numerous reports showing that xps-deficient Xanthomonas species mutants present reduced virulence in planta (41, 43, 44, 53), most of the xps T2SS mutants (XHV734_4180 to -4190) were positively screened as fitness deficient in our experiment (Fig. 3a). The xps genes that did not pass the significance threshold still displayed very low log2 fold change (FC) values (Table S1), suggesting that, despite their rejection by the statistical test, they were also critical in lettuce. Like the X. hortorum pv. gardneri genome (54), the X. hortorum pv. vitians LM16734 genome contained a large repertoire of genes encoding CWDEs, including various pectinolytic, cellulolytic, and hemicellulolytic enzymes. These genes were not critical in our analysis, either because of their high functional redundancy or because of potential complementation in trans by neighboring cells. Once the cell wall polymers have been degraded by the type II-secreted CWDEs, breakdown products and simple oligosaccharides have to be internalized by the pathogen to be assimilated. This role has been mainly attributed to TonB-dependent transporters (TBDTs) in Xanthomonas species (10). These TBDTs are primarily known for their role in iron-siderophore uptake and are often associated with carbohydrate sensor or degrading genes in so-called carbohydrate scavenging systems or carbohydrate utilization with TBDT (CUT) loci (10, 11). They allow for the scavenging of carbohydrates with high affinity and are thought to play an important role in bacterial adaptation to oligotrophic environments, such as plants for phytopathogens (9). Consequently, we searched for TBDT genes in our critical gene set and found three loci—XHV734_0066, -1016, and -1310—annotated as TonB-dependent receptors.
FIG 3.
Schematic representation of (a) the critical genes in lettuce within the xps T2SS operon and (b) the LPS biosynthesis gene cluster of X. hortorum pv. vitians LM16734. Numbers in brackets indicate the width of the viewing window on the LM16734 genome. Green arrows, critical genes in lettuce; gold arrows, critical genes in 1/10th TSB; red arrows, essential genes in the in vitro library; gray arrows, nonsignificant genes. Gene names, log2 FC values, and—for panel b—grouping in functional regions as described in reference 67 are displayed below. Black bars, localizations of TA sites; bar plots, read counts at each TA site under the control and experimental conditions. (Values of >1,000 are not displayed.)
First, XHV734_1016 was located in the middle of a cluster of 3 critical genes (XHV734_1015 to -1017) (Fig. 2) which displayed a very strong homology and synteny with the sux CUT locus of X. campestris pv. campestris, responsible for sucrose-specific uptake and degradation (10). The sux gene cluster consists of suxA (XHV734_1016), coding for a sucrose TBDT that transports sucrose into the periplasm; suxB (XHV734_1015), coding for an intracellular amylosucrase that cleaves sucrose into glucose-1-P and fructose (55); suxC (XHV734_1017), coding for an inner membrane sucrose transporter; and suxR (XHV734_1018), coding for a LacI/GalR-family transcriptional repressor of sucrose import, which was logically not critical in our screening. The phosphohexose mutase encoded by xanA (XHV734_4228), which allows glucose-1-P to enter the glycolysis pathway, was also screened as critical. The conditional essentiality of this sucrose utilization locus in lettuce correlates with a previous study showing that sucrose was the primary carbon source of X. campestris pv. campestris in low-molecular-weight cabbage leaf extract during the first hours of incubation (56). Sucrose can also stimulate production of the diffusible signal factor (DSF) (57)—the Xanthomonas quorum-sensing signaling molecule that plays a major role in virulence regulation and survival in planta (58). Thus, sucrose may well be the preferential carbon source of X. hortorum pv. vitians during the early biotrophic phase of lettuce leaf invasion.
Second, XHV734_0066 was nearly identical to the cirA gene of X. citri subsp. citri 306, an oligoxyloside-specific TBDT involved in the uptake and utilization of xylan breakdown products (59). This cirA ortholog was adjacent to XHV734_0067, a xylose repressor-like transcriptional regulator strongly homologous to xylR, which regulates xylan degradation (9). Xylan from the cell wall hemicelluloses is mainly degraded into xylooligosaccharides by β-1,4-endoxylanases (60). cirA has been hypothesized to transport methylglucuronoxylotriose (MeGX3)—a tetrasaccharide intermediate of xylan degradation—into the periplasm, where it can be converted into xylotriose by a membrane-bound α-glucuronidase (59). There, xylotriose can be either broken down into d-xylose or directly transported to the cytoplasm. Once in the cytoplasm, d-xylose is converted into d-xylulose-5-P by XylA1, XylA2, and XylB (XHV734_2842, -4908, and -2843), processed by the pentose phosphate pathway (PPP), and finally assimilated (9). Intriguingly, the tktA gene encoding the transketolase of the PPP that catalyzes the conversion of d-xylulose-5-P into glyceraldehyde-3-P was critical for fitness, but the xylA and xylB genes were not. Therefore, their function could be redundant or performed by other enzymes. Xylan exploitation is important for Xanthomonas nutrition during the early phyllosphere colonization stage (9), but xylan could also be an important nutrient during the late necrotrophic phase when host cells are actively degraded.
Finally, XHV734_1310 carries a homolog of the ferric enterobactin receptor bfeA of various Xanthomonas spp., which is flanked by two other similar copies (XHV734_1309 and -1312) and located upstream of the xanthoferrin siderophore biosynthesis and uptake gene cluster xss (XHV734_1298 to -1305) (61). The organization of these three bfeA-like genes upstream of xss is strikingly similar to what was observed in X. citri subsp. citri (62) and X. oryzae pv oryzae (42). Wang et al. (42) named the genes bfeA, bfeA2, and bfeA3 and reported that mutating bfeA alone ended up in a reduced-virulence phenotype in planta. Iron uptake using siderophores is indeed a well-known crucial feature of most bacterial phytopathogens necessary for successful host invasion (63). A recent proteomic analysis suggested that BfeA protein abundance could be directly correlated with virulence (64), which could partially explain why the gene is present in the form of three colocalized copies. However, further studies would have to elucidate why only one copy of bfeA is critical for fitness in planta.
Hiding from the plant immune system using cellular polysaccharides.
The Tn-seq screening in planta highlighted a few critical genetic factors allowing X. hortorum pv. vitians to evade and counteract the lettuce immune response triggered by pathogen recognition during leaf invasion. The type III secretion system and its effectors are probably the best-known factors used by Xanthomonas to disrupt the host immune response (17). However, the genes involved in the T3SS of X. hortorum pv. vitians LM16734 (XHV734_0611 to -0633) did not pass the statistical significance threshold in our Tn-seq analysis, even though almost all of them presented a moderate fitness defect (Table S1). Only hrpF (XHV734_0597) was considered critical; it encodes the translocon required for T3E translocation into host cells. In comparable in planta Tn-seq experiments conducted in D. dadantii (29) and R. solanacearum (32), genes of the T3SS apparatus were not critical either because of the variable efficiency of Tn-seq screens in detecting genes involved in production and secretion of “public goods” (65). On the other hand, the statistically significant screening of T3SS genes in P. syringae was attributed to low bacterial apoplastic densities that did not allow complementation in trans (30). Thus, it seems that the genetic diversity of the transposon mutants coexisting in lettuce apoplastic spaces was high enough to observe a partial complementation phenomenon of the T3SS roles.
Following the identification of the cellular polysaccharide metabolism as significantly enriched in the GO term enrichment analysis (Table 2), we searched for genes associated with these processes in our critical gene set. As a result, two genes of the gum gene cluster encoding the synthesis of the EPS xanthan and 19 genes involved in the biosynthesis and assembly of cell surface LPS, clustered in 3 distinct genomic regions, were identified (Fig. 2). We already discussed the lethal phenotypes observed in our Tn-seq library for the gumE, gumI, and gumJ mutants (25), but here only gumB (XHV734_1853) and gumC (XHV734_1854) were critical in lettuce leaves. These two genes, along with gumE, are responsible for polymerization of the lipid-linked pentasaccharide units and subsequent export of the final xanthan EPS out of the periplasm (66). Moreover, xanthan production by gumB and gumC mutants of X. campestris was completely abolished in vivo (67). Xanthan gum is required for full epiphytic survival and virulence in Xanthomonas species (12, 42, 68).
The remaining 19 genes associated with LPS biosynthesis and assembly emphasized the crucial importance of these surface components in the interaction with the host plant. Three main gene clusters were identified (Fig. 2), each encoding specific functions. The first region contained 4 genes: the conserved LPS assembly gene lapB (XHV734_2249) (69), a homolog of the rfb303 gene of X. citri subsp. citri (XHV734_2250) putatively associated with biosynthesis of the LPS core (70), a putative sugar epimerase gene (XHV734_2251), and the galU gene (XHV734_2252), which is involved in O-antigen nucleotide sugar biosynthesis (66, 71). Another small gene cluster consisting of XHV734_4495 and -4497 was related to LPS biosynthesis. XHV734_4497 is an O-antigen ligase-like enzyme putatively involved in ligating the O-antigen and the lipid A core.
The principal LPS gene cluster corresponded to the major rml/wxc cluster of LPS biosynthesis (72). Tn-seq screening distinguished which parts of this cluster were critical and consequently which particular functions of the LPS biosynthesis process were required for growth in lettuce (Fig. 3b). First, the rmlBACD critical genes (XHV734_4231 to -4233) encode a set of enzymes that convert glucose-1-phosphate into the nucleotide sugar dTDP-l-rhamnose (73), which is the direct precursor of the rhamnose moieties of the X. hortorum pv. vitians LPS core and O-antigen (74). Then, the three functional regions of the LPS biosynthesis gene cluster (72) showed different results (Fig. 3b). The LPS 1 region located between wxcA (XHV734_4249) and wxcE (XHV734_4243) was entirely critical in lettuce and is responsible for the biosynthesis of the water-soluble O-antigen (72). Genes wzt, wxcB, and wxcC (XHV734_4247, -4246, and -4245) were also important for bacterial growth in vitro, suggesting that the formation of a functional O-antigen is necessary even in rich medium. Diverse X. campestris pv. campestris and X. oryzae pv. oryzae mutants defective in O-antigen biosynthesis showed reduced or abolished virulence in planta (41, 42). The gmd gene of LPS region 2 (XHV734_4242) was also critical in lettuce. This region is implied in biosynthesis of the core oligosaccharide of the LPS (72). Finally, none of the LPS 3 region genes, ranging from wxcK (XHV734_4236) to wxcO (XHV734_4240), was critical. Genes within this last segment probably encode accessory synthesis and transfer of N-acetyl-3-amino hexose (72) and contribute to the side branching of the O-antigen (75). Mutating these genes resulted in the production of modified O-antigens in X. campestris pv. campestris (75).
Altogether, our findings indicate that certain genes directly implied in biosynthesis of the LPS core oligosaccharide and O-antigen are indispensable for successful bacterial invasion and survival inside lettuce leaves. Tn-seq also showed that some genes involved in biosynthesis of the LPS core and O-antigen were critical for P. syringae and R. solanacearum growth in planta (Table 4) (30, 32). Gram-negative bacterial LPS have a general protective role against stresses in diverse environments (76), including oxidative stress (70). They also participate in camouflaging pathogens from the host immune system, although they are also one of the major microbial-associated molecular patterns (MAMPs) recognized by host immunity receptors (77, 78). The long O-antigen of Xylella fastidiosa delays pathogen recognition and the basal immune response in grape (79). If a structurally intact LPS seems to be required for hiding from the lettuce immune system in X. hortorum pv. vitians, modifications of the O-antigen side branching does not alter this protective role.
TABLE 4.
X. hortorum pv. vitians critical genes in lettuce with at least one homolog in other in planta Tn-seq/RB-Tn-seq studies conducted on plant-pathogenic bacteria
| Locus | Gene | Product | Critical homologous gene(s) in: |
|||
|---|---|---|---|---|---|---|
| Pseudomonas syringae a | Ralstonia solanacearum b | Agrobacterium fabrum c | Dickeya dadantii d | |||
| XHV734_0038 | gltD | Glutamate synthase, 4Fe-4S protein, small subunit | ATU_RS17595 | |||
| XHV734_0039 | gltB | Glutamate synthase, large subunit | Psyr_0411 | ATU_RS17605 | ||
| XHV734_0130 | tyrB | Tyrosine aminotransferase, tyrosine-repressible, PLP-dependent | RS_RS05000 | |||
| XHV734_0259 | glnA | Glutamine synthetase | ATU_RS02965 | |||
| XHV734_0262 | Two-component system sensor protein | ATU_RS07120 | Dda3937_01117 | |||
| XHV734_0263 | glnG | Fused DNA-binding response regulator, sigma54 interaction protein | ATU_RS07125 | Dda3937_01116 | ||
| XHV734_0278 | secB | Protein export chaperone | RS_RS01755 | |||
| XHV734_0458 | ilvD | Dihydroxyacid dehydratase | Psyr_0469 | ATU_RS09365 ATU_RS18520 ATU_RS14860 |
||
| XHV734_0506 | bioB | Biotin synthase | Psyr_4687 | |||
| XHV734_0595 | trpE | Anthranilate synthase component 1 | Psyr_4609 | RS_RS14430 | ATU_RS11170 | |
| XHV734_0640 | trpD | Anthranilate phosphoribosyltransferase | Psyr_4580 | ATU_RS08260 | ||
| XHV734_0642 | trpC | Indole-3-glycerol phosphate synthase | ATU_RS08265 | |||
| XHV734_0670 | purD | Phosphoribosylglycinamide synthetase phosphoribosylamine-glycine ligase | ATU_RS03175 | |||
| XHV734_0671 | purH | Fused IMP cyclohydrolase; phosphoribosylaminoimidazolecarboxamide formyltransferase | ATU_RS13745 | Dda3937_00244 | ||
| XHV734_0831 | opgH | Glucans biosynthesis glucosyltransferase H | Psyr_0378 | RS_RS14570 | Dda3937_03563 | |
| XHV734_0943 | Transcriptional regulator of N-acetylglucosamine utilization, LacI family | ATU_RS21970 | ||||
| XHV734_0998 | aspC | Putative aspartate aminotransferase | ATU_RS10725 | |||
| XHV734_1041 | Type I secretion outer membrane protein, TolC precursor | Dda3937_00726 | ||||
| XHV734_1046 | leuC | 3-Isopropylmalate isomerase subunit, dehydratase component | Psyr_1983 | ATU_RS13195 | Dda3937_01352 | |
| XHV734_1047 | leuD | 3-Isopropylmalate isomerase subunit | ATU_RS13585 | |||
| XHV734_1049 | leuB | 3-Isopropylmalate dehydrogenase | Psyr_1985 | ATU_RS13590 | Dda3937_04404 | |
| XHV734_1050 | leuA | 2-Isopropylmalate synthase | Psyr_1257 | ATU_RS11045 | Dda3937_04301 | |
| XHV734_1051 | tdcB | l-Threonine dehydratase catabolic TdcB | ATU_RS05955 | |||
| XHV734_1053 | ilvG | Acetolactate synthase isozyme 2 large subunit | Psyr_0846 | ATU_RS09945 | ||
| XHV734_1054 | ilvC | Ketol-acid reductoisomerase (NADP(+)) | Psyr_0848 | ATU_RS09860 | ||
| XHV734_1096 | hemL | Glutamate-1-semialdehyde aminotransferase (aminomutase) | ATU_RS22545 | |||
| XHV734_1124 | yjgF | Ketoacid-binding protein | ||||
| XHV734_1145 | tktA | Transketolase 1, thiamin-binding | ATU_RS17355 | |||
| XHV734_1167 | gapA | Glyceraldehyde-3-phosphate dehydrogenase A | ATU_RS17360 | |||
| XHV734_1174 | Putative fructose-bisphosphate aldolase class 1 | ATU_RS17375 | ||||
| XHV734_1213 | thiG | Thiamine biosynthesis ThiGH complex subunit | Psyr_4740 | |||
| XHV734_1262 | nadE | Glutamine-dependent NAD(+) synthetase | RS_RS11780 | |||
| XHV734_1330 | Hypothetical protein | ATU_RS17310 | ||||
| XHV734_1332 | ruvA | Component of RuvABC resolvasome, regulatory subunit | ATU_RS17290 | |||
| XHV734_1688 | Nicotinate-nucleotide adenylyltransferase | RS_RS11005 | ||||
| XHV734_1761 | trpF | N-(5′-Phosphoribosyl)anthranilate isomerase | Psyr_1663 | RS_RS09970 | ATU_RS00085 | |
| XHV734_1763 | trpB | Tryptophan synthase, beta subunit | Psyr_0034 | RS_RS09965 | ATU_RS00090 | |
| XHV734_1765 | trpA | Tryptophan synthase alpha chain | Psyr_0033 | RS_RS09955 | ATU_RS00095 | |
| XHV734_1808 | purK | N5-carboxyaminoimidazole ribonucleotide synthase | ATU_RS17455 | Dda3937_01683 | ||
| XHV734_1945 | tgt | tRNA-guanine transglycosylase | RS_RS13575 | |||
| XHV734_1977 | gdh | NAD-specific glutamate dehydrogenase | ATU_RS13460 | |||
| XHV734_2148 | purM | Phosphoribosylaminoimidazole synthetase | ATU_RS05630 | Dda3937_02515 | ||
| XHV734_2154 | sodA | Superoxide dismutase, Mn | ATU_RS04315 | |||
| XHV734_2180 | Dihydroorotase | ATU_RS06435 | ||||
| XHV734_2250 | UDP-N-acetylmuramyl pentapeptide phosphotransferase/UDP-N-acetylglucosamine-1-phosphate transferase | ATU_RS22585 | ||||
| XHV734_2252 | galU | UTP-glucose-1-phosphate uridylyltransferase | Psyr_2980 | ATU_RS17570 | ||
| XHV734_2285 | phaC | Poly(3-hydroxyalkanoate) polymerase subunit PhaC | RS_RS08220 | |||
| XHV734_2316 | zwf | Glucose-6-phosphate dehydrogenase | ATU_RS02955 | |||
| XHV734_2707 | carB | Carbamoyl-phosphate synthase large subunit | Dda3937_01389 | |||
| XHV734_2709 | carA | Carbamoyl phosphate synthetase small subunit, glutamine amidotransferase | Dda3937_01390 | |||
| XHV734_2726 | serA | d-3-Phosphoglycerate dehydrogenase | Psyr_4852 | ATU_RS17200 | ||
| XHV734_2742 | hisF | Imidazole glycerol phosphate synthase, catalytic subunit with HisH | Psyr_4894 | ATU_RS00190 | ||
| XHV734_2743 | hisA | N-(5′-Phospho-l-ribosyl-formimino)-5-amino-1-(5′-phosphoribosyl)-4-imidazolecarboxamide isomerase | Psyr_4894 | ATU_RS00195 | ||
| XHV734_2744 | hisH | Imidazole glycerol phosphate synthase, glutamine amidotransferase subunit with HisF | Psyr_4896 | ATU_RS00200 | ||
| XHV734_2745 | hisB | Fused histidinol-phosphatase; imidazoleglycerol-phosphate dehydratase | Psyr_4897 | ATU_RS00210 | ||
| XHV734_2747 | hisD | Bifunctional histidinal dehydrogenase and histidinol dehydrogenase | Psyr_4133 | ATU_RS02645 | ||
| XHV734_2748 | hisG | ATP phosphoribosyltransferase | Psyr_4134 | ATU_RS03340 | ||
| XHV734_2829 | pgi | Glucose-6-phosphate isomerase | Psyr_0826 | ATU_RS01945 | ||
| XHV734_2887 | miaA | Delta(2)-isopentenylpyrophosphate tRNA-adenosine transferase | RS_RS12855 | ATU_RS09955 | ||
| XHV734_2989 | pyrB | Aspartate carbamoyltransferase | ATU_RS06440 | Dda3937_01284 | ||
| XHV734_3113 | udg | UDP-glucose 6-dehydrogenase | ATU_RS12570; | |||
| ATU_RS19395 | ||||||
| XHV734_3365 | aroA | 3-Phosphoshikimate 1-carboxyvinyltransferase | RS_RS04510 | |||
| XHV734_3366 | pheA | Chorismate mutase/prephenate dehydratase | ATU_RS00480 | |||
| XHV734_3367 | serC | 3-Phosphoserine/phosphohydroxythreonine aminotransferase | RS_RS04490 | |||
| XHV734_3398 | metXS | Homoserine O-succinyltransferase | Psyr_0474 | |||
| XHV734_3771 | clpX | ATPase and specificity subunit of ClpX-ClpP ATP-dependent serine protease | Psyr_1748 | RS_RS08650 | ||
| XHV734_3772 | clpP | Proteolytic subunit of ClpA-ClpP and ClpX-ClpP ATP-dependent serine protease | RS_RS08645 | |||
| XHV734_3851 | purF | Amidophosphoribosyltransferase | Psyr_1668 | ATU_RS05325 | Dda3937_02099 | |
| XHV734_4155 | glyA | Serine hydroxymethyltransferase | Psyr_4270 | RS_RS03670 | ||
| XHV734_4196 | purL | Phosphoribosylformyl-glycineamide synthetase | Psyr_1269 | Dda3937_03379 | ||
| XHV734_4228 | cpsG | Phosphohexose mutase | Psyr_0219 | |||
| XHV734_4229 | cpsB | Mannose-1-phosphate guanyltransferase | ATU_RS15505 | |||
| XHV734_4230 | rmlD | dTDP-4-dehydrorhamnose reductase subunit, NAD(P)-binding, of dTDP-l-rhamnose synthase | RS_RS03435 | ATU_RS21640 | ||
| XHV734_4231 | rmlC | dTDP-4-deoxyrhamnose-3,5-epimerase | ATU_RS22530 | |||
| XHV734_4232 | rmlA | Glucose-1-phosphate thymidylyltransferase | RS_RS03440 | ATU_RS21635 | ||
| XHV734_4233 | rmlB | dTDP-glucose 4,6-dehydratase | ATU_RS21645 | Dda3937_03924 | ||
| XHV734_4242 | gmd | GDP-d-mannose dehydratase, NAD(P)-binding | Psyr_0915 | |||
| XHV734_4247 | wzt | ATP binding component of ABC-transporter | Psyr_0918 | |||
| XHV734_4248 | wzm | Transport permease protein | Psyr_0917 | |||
| XHV734_4497 | Lipid A core-O-antigen ligase-like enzyme | RS_RS11060 | ||||
| XHV734_4610 | pyrE | Orotate phosphoribosyltransferase | ATU_RS01925 | Dda3937_03258 | ||
| XHV734_4646 | Flavodoxin reductases (ferredoxin-NADPH reductases) family 1 | ATU_RS03585 | ||||
| XHV734_4897 | tatC | TatABCE protein translocation system subunit | RS_RS14730 | |||
Genes required for P. syringae epiphytic and/or apoplastic fitness in bean leaves according to Helmann et al. (30).
Genes required for R. solanacearum fitness in tomato plants according to Su et al. (32).
Genes required for A. fabrum fitness under at least one condition (i.e., tomato tumors, tomato roots, maize roots, or poplar tumors) according to Torres et al. (33).
Genes required for D. dadantii fitness in chicory leaves according to Royet et al. (29).
Diversity of resistance mechanisms to the immunity-triggered oxidative burst.
The rapid and localized accumulation of reactive oxygen species (ROS) inside the plant tissues following MAMP recognition by the plant PRRs is referred to as the oxidative burst (13). In general, protection against ROS-induced damage, which includes lipid peroxidation, enzyme inactivation, and nucleic acid degradation, can be achieved in three different ways: (i) prevention of ROS generation, (ii) detoxification of radicals, and (iii) repair of damaged elements (80). We found a set of 10 critical genes in lettuce that we hypothesized to actively participate in protecting the pathogen against the oxidative burst by detoxifying ROS and repairing damaged macromolecules (Fig. 2).
The Mn-superoxide dismutase-encoding gene sodA (XHV734_2154) was critical in planta. SodA plays a pivotal role in the virulence of several plant-pathogenic bacteria (81). The expression of this gene is induced within the first 3 h of infection in X. campestris pv. campestris and D. dadantii (82, 83), proving that it acts as a very early protection against the oxidative burst. Interestingly, sodA is not directly induced by ROS in X. campestris pv. campestris, but by redox cycling agents like plumbagin, a plant quinone with antimicrobial properties (84). Another gene, XHV734_2318, encodes an YgfZ-family folate-dependent protein that might also be involved in plumbagin resistance (85). On the other hand, phaC encodes a polymerase producing granules of poly(3-hydroxyalkanoate) (PHA), a polyester that can both serve as a form of carbon storage and can enhance resistance to cold and oxidative stresses in pseudomonads (81).
A variety of genes were implied in the recycling of misfolded proteins and damaged nucleic acids. The clpX (XHV734_3771), clpP (XHV734_3772), and ftsH (XHV734_2889) genes encode the ClpXP serine protease and an ATP-dependent Zn-metalloprotease, respectively. The ClpXP protease recycles misfolded proteins in the cytoplasm (86), and its inactivation in X. campestris pv. campestris through clpX or clpP mutations leads to pleiotropic effects, including increased stress sensitivity (87, 88). The clpX and clpP genes were also critical in planta in an R. solanacearum Tn-seq study (Table 4) (32). Moreover, oxidative stress induces the expression of ftsH in Lactobacillus plantarum, and ΔftsH mutants are highly sensitive to various stresses (89). These two proteases are probably mobilized to buffer the damaging effects of the oxidative burst on the bacterial proteome.
Finally, the RNase R-encoding rnr gene (XHV734_3096) and the homologous genes of the recombination system recBCD (XHV734_5034, -5035, and -5033) were all critical in lettuce. They are probably involved in repairing the damage caused to nucleic acids by immunity-induced ROS production. RNase R is indeed required for the trans-translation pathway that directs deficient proteins and transcripts toward degradation while rescuing stalled ribosomes and is important for pathogenesis in many bacterial pathogens (90). This protein is overexpressed in X. citri subsp. citri during orange tree infection, supporting that it is important for Xanthomonas virulence (91). The RecBCD homologous recombination system has been proposed to be the predominant repair mechanism of oxidative stress-induced DNA damage (92–94), probably because it specifically repairs double-stranded breaks—the most deleterious DNA damage caused by oxidative agents (95). However, the recC mutant already showed a growth defect in the inoculum, and the recBCD genes all displayed low mean read count values in vitro. Intriguingly, RecA did not appear to play a role in oxidative stress resistance in Xanthomonas (96), which tends to be confirmed by its neutral status in our analysis.
Specific and common traits of the in planta lifestyle unveiled by the comparison with other plant-pathogenic bacteria.
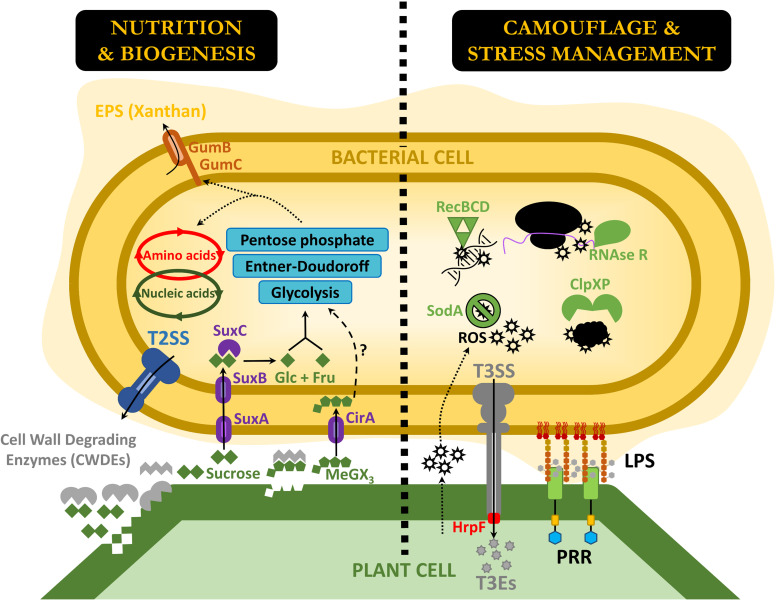
The present study presents a first overview of some of the molecular processes required for successful colonization and infection of lettuce leaves by the phytopathogenic bacterium X. hortorum pv. vitians (Fig. 4). Similar studies have been conducted on a handful of other plant-pathogenic bacteria in planta in the past 3 years, allowing us to compare the gene set we identified with those unveiled by other similar studies. We searched for homologs of the genes we identified in the data sets obtained for D. dadantii in chicory leaves (29), P. syringae in bean leaves (30), R. solanacearum in tomato roots (32), and A. fabrum under multiple conditions (tomato tumors and roots, maize roots, and poplar tumors) (33).
FIG 4.
Schematic overview of the critical genes and processes required for lettuce infection by X. hortorum pv. vitians LM16734 as discussed in this work. Gray objects are known important virulence factors not screened as critical but discussed in the text. Abbreviations: EPS, exopolysaccharide; T2SS, type II secretion system; Glc, glucose; Fru, fructose; MeGX3, methylglucuronoxylotriose; ROS, reactive oxygen species; T3SS, type III secretion system; T3Es, type III effectors; LPS, lipopolysaccharide; PRRs, pattern recognition receptors.
Eighty-three out of 170 critical genes of X. hortorum pv. vitians had at least one homolog in one of these other important plant-pathogenic bacterial genes in planta (Table 4). More precisely, 57 genes had homologs in A. fabrum, 33 in P. syringae, 21 in R. solanacearum, and 17 in D. dadantii, even though there was not a single gene found in all 5 studies. The importance of amino acid biosynthesis in planta was also true for A. fabrum and P. syringae, particularly in the Val/Leu/Ile, Ala/Asp/Glu, Phe/Tyr/Trp, Gly/Ser/Thr, and His biosynthesis pathways, where most of the key genes were critical. Only the genes in the Phe/Tyr/Trp and Gly/Ser/Thr biosynthesis pathways were also critical in R. solanacearum, and only a few genes involved in the Val/Leu/Ile biosynthesis pathway were critical in D. dadantii. Most of the genes of the PPP and dDTP-l-rhamnose (a rhamnolipid and LPS component in several bacteria) biosynthesis pathways (97) were critical in both X. hortorum pv. vitians and A. fabrum. Finally, some key genes of the purine and pyrimidine biosynthesis pathways were important too in A. fabrum and D. dadantii. Overall, it appears that despite a few similarities, each of these 5 pathogens has specific nutritional requirements during its life in planta, probably because of the differential nutrient availability in the respective host plants.
Xanthomonas hortorum pv. vitians seems to be more dependent than the other pathogens on de novo amino acid biosynthesis. Besides the above-cited pathways, the 6 genes argBCEFG of the Arg biosynthesis pathway, thrABC, gyaR, and XHV734_4311 of the Gly/Ser/Thr biosynthesis pathway, and mtaD and mtnBC of the Cys/Met biosynthesis pathway were also important for fitness in lettuce leaves. Exogenous amino acid availability might therefore be scarcer in the lettuce apoplast than in the tomato, bean, or poplar leaf apoplasts. Mutagenesis of the T2SS was detrimental for in planta fitness of X. hortorum pv. vitians, but not for fitness of R. solanacearum, P. syringae, and D. dadantii. Yet, the T2SS is an important virulence factor for these three pathogens (29, 98, 99). Even though an in trans complementation effect could account for this phenomenon in the D. dadantii Tn-seq study (29), our results suggest that the T2SS may play an unexpectedly important role in the virulence of X. hortorum pv. vitians. A thorough comparison with other plant-pathogenic bacterial LPSs would be difficult because each bacterium has a specific set of genes involved in LPS biosynthesis (100–102). Yet, the accurate identification of the LPS biosynthesis genomic regions critical for in planta fitness of X. hortorum pv. vitians is also a new finding that strengthens our understanding of the role of this major surface component in Xanthomonas pathogenesis. Finally, several genes putatively implied in the degradation and repair of damaged nucleic acids or misfolded proteins (recBCD, rnr, ftsH, and XHV734_2318) were critical only in X. hortorum pv. vitians, while the clpXP protease was also important for in planta fitness of P. syringae and R. solanacearum (Table 4). However, different proteases were important in planta for the other pathogens: e.g., the clpAP protease for A. fabrum and D. dadantii (29, 33). Although the detrimental consequences of the plant defense mechanisms are relatively similar in all these pathosystems, it seems that each pathogen displays a specific response based on different genes.
More in planta Tn-seq studies will undoubtedly be conducted on other plant-pathogenic bacteria in the years to come. The next step will consist of extending the comparison conducted here to see if the gene sets important for in planta fitness of plant pathogens belonging to a same genus overlap. A comparison between pathogens conventionally classified as biotrophs or necrotrophs, soilborne or not, or between pathogens infecting a same plant species would also refine our understanding of the complex genetic requirements sustaining these host-pathogen interactions.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and Tn-seq library generation.
Xanthomonas hortorum pv. vitians LM16734 (=CFBP 8638) was originally isolated from a diseased leaf sample of oakleaf lettuce (Lactuca sativa) collected in the Rhône-Alpes region (France) in 2016 (18). The bacterium was routinely cultured on 1/10th tryptic soy agar (TSA) plates and in 1/10th tryptic soy broth (TSB) at 28°C. The procedure for generating the Tn-seq mutant library was described previously (25). Briefly, mating the recipient X. hortorum pv. vitians LM16734 strain with a donor Escherichia coli MFDpir/pSamEc strain containing a Himar1-C9 transposase gene yielded approximately 250,000 individual mutant colonies. They were recovered with 1/10th TSB, pooled, and stored in 30% (vol/vol) glycerol vials at −80°C. The resulting mutant library population was estimated to 1010 CFU · mL−1.
Monitoring of the bacterial population in planta.
The bacterial population was estimated over the course of the infection process in leaf lettuce cv. Météore for 14 days. Plants were cultivated and inoculated as described previously (18). The population size was enumerated 0, 1, 2, 4, 7, 10, and 14 days postinoculation (dpi). At each time point, three infected leaves from three different plants were randomly collected and treated as independent technical replicates. Except for the initial sampling at t = 0 dpi, aimed at enumerating the inoculum deposited on the leaves, all other leaf samples were surface sterilized in a 70% ethanol bath for 20 s and rinsed in sterile deionized H2O (dH2O) to count only the bacteria that had penetrated the leaf tissue. In all cases, 4 disks of 3.14 cm2 each were cut with a sterilized 1-cm-diameter punch on either side of the leaf midrib and pooled in 10 mL of sterile dH2O. Leaf disks were crushed using a T25 IKA Ultra-turrax disperser (IKA, Staufen im Bresigau, Germany) at full speed. The resulting leaf homogenates were serially diluted and plated onto 1/10th TSA plates supplemented with cycloheximide at 50 μg · mL−1 with an easySpiral automated plater (Interscience, Saint-Nom-la-Bretèche, France). The plates were incubated at 28°C for 4 days and enumerated with a Scan 1200 automatic colony counter (Interscience). Bacterial population sizes were expressed in CFU · cm−2 of lettuce leaf.
Inoculation of lettuce with the transposon library.
Two transposon library aliquots were thawed at 4°C overnight and inoculated in two flasks containing 100 mL of 1/10th TSB supplemented with kanamycin at 25 μg · mL−1. The cultures were incubated overnight at 28°C under shaking at 120 rpm until they reached an optical density at 600 nm (OD600) of 1.4. Then, they were centrifuged at 3,344 × g at 4°C for 20 min, and the cell pellets were washed by being resuspended in 25 mL of cold sterile dH2O and repeating the centrifugation step. Each cleaned cell pellet was resuspended in sterile dH2O and spectrophotometrically adjusted to obtain 400 mL of inoculum at an OD600 of 0.2 supplemented with Tween 80 at 0.08% (vol/vol). Half of each inoculum was centrifuged, and the cell pellets were stored at −20°C for DNA extraction of the control condition. The remaining 200 mL of each inoculum was hand-sprayed on the leaves of 30 young lettuce cv. Météore plants per duplicate until runoff. All plants were immediately incubated in an Ineltec environmental chamber (Ineltec France, Vénissieux, France) at 25°C, with an 18-h photoperiod and 90% relative humidity (RH) for 48 h and then 70% RH for the rest of the experiment.
Recovery of bacterial cells from lettuce leaves.
Diseased lettuce leaves were collected at t = 10 dpi, when small water-soaked leaf spot lesions were abundant but not coalescent, and duplicates were treated separately. Leaves were crushed in 500 mL of sterile dH2O with a sterilized Moulinex Masterchef 58 electronic blender (SEB Moulinex G S M, Ecully, France) at middle speed for 2 to 3 min until <2-mm rough fragments were obtained. The resulting suspensions were sonicated in a sonication water bath 88155 (Fischer Bioblock Scientific, Illkirch, France) for 15 min to detach bacterial cells from leaf debris. To remove plant debris, the suspensions were successively filtered through 1-, 0.5-, 0.25-, and 0.05-mm-pore-diameter sieves, then twice through 20-μm-pore-diameter coffee filters, and finally vacuum filtered through 10-μm-pore ipPORE track-etched membrane filters (it4ip, Louvain-la-Neuve, Belgium). In the end, approximately 400 mL of filtered bacterial cell suspensions was recovered per duplicate and centrifuged at 10,647 × g for 20 min at 4°C, and the cell pellets were temporarily kept at −20°C.
DNA extraction and library preparation.
Genomic DNA library extraction and preparation were conducted mostly as described previously (25), with technical variations. Briefly, the cell pellets were thawed, and genomic DNA was extracted using a Promega Wizard genomic DNA purification kit (Promega, Madison, WI, USA) and resuspended in 200 μL of Tris-EDTA (pH 7). Fifty micrograms of DNA per sample was digested with the MmeI restriction enzyme (New England Biolabs, Ipswich, MA, USA), and digestion products were purified. Samples were deposited on a 1% (wt/vol) agarose gel, and the 1.3- to 1.8-kb DNA fragments were cut out from the gel and retrieved using a QIAquick gel extraction kit (Qiagen, Hilden, Germany). Then, 300 to 500 ng of each sample was ligated to double-stranded barcoded adapters with the T4 DNA ligase (New England Biolabs). The 20-bp genomic DNA regions flanking the transposon were amplified by 22 cycles of PCRs in 50-μL final volumes. The reaction mixtures contained 2 μL of ligated DNA matrix, 1 U of Q5 DNA polymerase (New England Biolabs), 1× Q5 buffer, 0.2 mM deoxynucleoside triphosphates (dNTPs), and 0.4 μM of each TruSeq primers (Illumina, Inc., San Diego, USA). The PCR products were deposited on a 2% (wt/vol) agarose gel, and the 125-bp bands were extracted using a QIAquick gel extraction kit (Qiagen). Library DNA samples were sent to the I2BC-sequencing platform (I2BC, Gif-sur-Yvette, France) to be sequenced in the “High Output Single Read” 75-bp mode on a NextSeq 500 instrument (Illumina, Inc.).
Pretreatment of the sequencing read data.
Raw reads were trimmed with CUTADAPT v1.15 (103) to filter those containing the Mariner inverted left repeat (i.e., ACAGGTTGGATGATAAGTCCCCGGTCTT) and remove the adapter sequences. Trimmed data sets were mapped without mismatches to unique TA sites with a modified version of the TPP script from the TRANSIT software suite v2.0.2 (104) on the complete genome sequence of X. hortorum pv. vitians LM16734 available from the NCBI GenBank database under accession no. GCA_014338485.1. The script generating the prot_table file used by TRANSIT was modified to integrate noncoding RNA (ncRNA) features. In vitro library sequencing reads available in the NCBI SRA database under accession no. SRR12067051 and SRR12067052 were retrieved and combined with the newly generated sequencing reads in a single .wig file, and subjected to the “Trimmed Total Reads” (TTR) normalization method in TRANSIT v3.2.1.
Assessment of the contribution of each gene to fitness in vitro and in planta.
The contribution of each genetic feature to bacterial fitness was determined both in 1/10th TSB (inoculum) and in lettuce by carrying out pairwise comparisons of (i) the inoculum condition with the previously generated in vitro library results (25) and (ii) the lettuce infection condition with the inoculum condition. Reads in the 5% N-terminal and 10% C-terminal portions of the genetic feature were discarded because they could lead to incorrect interpretations, and a LOESS (locally estimated scatterplot smoothing) correction for genome positional bias was applied. The “Resampling” method available in the TRANSIT v3.2.1 suite (105) was used to evaluate the fitness contribution and statistical significance of each genetic feature in the two comparisons, by (i) calculating the difference in the sum of read counts and log2 fold change (FC) between the control and experimental conditions, (ii) calculating P values using variation of the nonparametric permutation test, and (iii) correcting the P values by performing a Benjamini-Hochberg false-discovery rate (FDR) test (106), which yields q values expressing the probabilities of false-positive results. The genetic features with a q value of ≤0.05 were considered to present a significant fitness difference between the control and experimental conditions, and potential artifactual features were checked manually on the MicroScope platform (https://mage.genoscope.cns.fr) (107). Finally, the circular genome and data mapping were visualized with the R/Shiny application shinyCircos (108).
Gene annotation, Gene Ontology term enrichment, KEGG pathway mapping, and comparison with other Tn-seq or RB-Tnseq in planta experiments.
A mixed annotation strategy was applied to assign Gene Ontology (GO) terms (39) to X. hortorum pv. vitians LM16734 genes. Protein sequences were extracted from the genome using the cds_extractor script from the bac-genomic-scripts suite (109). Terms were assigned using the web-based tool eggNOG-Mapper v2 (110) against the eggNOG v5.0 database (111) and InterproScan v5.0.0 (112), available on the Galaxy for Genome Annotation (GGA) server (https://annotation.usegalaxy.eu/) (113). Finally, a GO term enrichment analysis was conducted with the “Pathway Enrichment Analysis” method available in TRANSIT, based on the Ontologizer method (114). GO terms with an FDR-adjusted q value of ≤0.05 were considered to be significantly enriched in the gene subset. EC numbers, eggNOG orthologs, and Cluster of Orthologous Groups (COG) categories were also assigned with eggNOG-Mapper v2. The Kyoto Encyclopedia of Genes and Genomes (KEGG) (https://www.kegg.jp/) database was used to assign KEGG Orthology (KO) terms to genes with BlastKOALA v2.2 (40). KO terms were mapped to the X. hortorum reference pathways (xhr) with KEGG Mapper (115). Synteny and gene homology assessment were performed on the MicroScope platform following the guidelines of the MicroScope team annotation (107). Our data were compared with the genes identified as important for in planta fitness of P. syringae (30), R. solanacearum (32), Agrobacterium fabrum (33), and D. dadantii (29) by searching our in planta critical gene set for homologs of these genes using BLASTP. Genes with more than 50% coverage, 30% protein identity and an E value of <10−5 were considered homologs as described previously (25), and gene annotations and functions were consistently checked to prevent potential inconsistencies.
Data availability.
The NCBI GenBank assembly accession number for the complete X. hortorum pv. vitians LM16734 genome is GCA_014338485.1. Transposon insertion sequencing raw reads have been deposited in the NCBI SRA database under accession no. SRR14385278 to SRR14385281. All data are associated with BioProject no. PRJNA530964 and PRJNA726806.
List of the 170 genes critical in lettuce leaves. Download Table S2, XLSX file, 0.06 MB (57.2KB, xlsx) .
Copyright © 2022 Morinière et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of the 36 genes critical in 1/10th TSB medium. Download Table S3, XLSX file, 0.02 MB (19.6KB, xlsx) .
Copyright © 2022 Morinière et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of the 2 growth-advantage genes in lettuce leaves. Download Table S4, XLSX file, 0.01 MB (10.1KB, xlsx) .
Copyright © 2022 Morinière et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplementary Material
ACKNOWLEDGMENTS
The authors are grateful to Élise Lacroix from the greenhouse platform at UCBL for her precious help in the maintenance of plants. The first author also thanks the EuroXanth COST Action CA16107 and the FNX network for the scientific meeting opportunities they provided. This work benefited from the facilities and expertise of the high-throughput sequencing core facility of I2BC (Centre de Recherche de Gif [http://www.i2bc.paris-saclay.fr/]). We also thank Annie Buchwalter for English proofreading and corrections.
The LABGeM (CEA/Genoscope and CNRS UMR8030) and the France Génomique and French Bioinformatics Institute national infrastructures (funded as part of Investissement d'Avenir program managed by Agence Nationale pour la Recherche, contracts ANR-10-INBS-09 and ANR-11-INBS-0013) are acknowledged for support within the MicroScope annotation platform.
Contributor Information
Franck Bertolla, Email: franck.bertolla@univ-lyon1.fr.
Christopher W. Schadt, Oak Ridge National Laboratory
Steven Lindow, University of California, Berkeley.
REFERENCES
- 1.Kraepiel Y, Barny M. 2016. Gram‐negative phytopathogenic bacteria, all hemibiotrophs after all? Mol Plant Pathol 17:313–316. doi: 10.1111/mpp.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pierella Karlusich JJ, Zurbriggen MD, Shahinnia F, Sonnewald S, Sonnewald U, Hosseini SA, Hajirezaei M-R, Carrillo N. 2017. chloroplast redox status modulates genome-wide plant responses during the non-host interaction of tobacco with the hemibiotrophic bacterium Xanthomonas campestris pv. vesicatoria. Front Plant Sci 8:1158. doi: 10.3389/fpls.2017.01158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Islam MT, Al Mamun M, Lee B-R, Van Hien L, Jung W-J, Bae D-W, Kim T-H. 2021. Role of salicylic acid signaling in the biotrophy-necrotrophy transition of Xanthomonas campestris pv. campestris infection in Brassica napus. Physiol Mol Plant Pathol 113:101578. doi: 10.1016/j.pmpp.2020.101578. [DOI] [Google Scholar]
- 4.Luneau JS, Cerutti A, Roux B, Carrère S, Jardinaud M-F, Gaillac A, Gris C, Lauber E, Berthomé R, Arlat M, Boulanger A, Noël LD. 2022. Xanthomonas transcriptome inside cauliflower hydathodes reveals bacterial virulence strategies and physiological adaptations at early infection stages. Mol Plant Pathol 23:159–174. doi: 10.1111/mpp.13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.An S-Q, Potnis N, Dow M, Vorhölter F-J, He Y-Q, Becker A, Teper D, Li Y, Wang N, Bleris L, Tang J-L. 2020. Mechanistic insights into host adaptation, virulence and epidemiology of the phytopathogen Xanthomonas. FEMS Microbiol Rev 44:1–32. doi: 10.1093/femsre/fuz024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vorholt JA. 2012. Microbial life in the phyllosphere. Nat Rev Microbiol 10:828–840. doi: 10.1038/nrmicro2910. [DOI] [PubMed] [Google Scholar]
- 7.Darsonval A, Darrasse A, Durand K, Bureau C, Cesbron S, Jacques M-A. 2009. Adhesion and fitness in the bean phyllosphere and transmission to seed of Xanthomonas fuscans subsp. fuscans. Mol Plant Microbe Interact 22:747–757. doi: 10.1094/MPMI-22-6-0747. [DOI] [PubMed] [Google Scholar]
- 8.Barcarolo MV, Gottig N, Ottado J, Garavaglia BS. 2020. Participation of two general stress response proteins from Xanthomonas citri subsp. citri in environmental stress adaptation and virulence. FEMS Microbiol Ecol 96:fiaa138. doi: 10.1093/femsec/fiaa138. [DOI] [PubMed] [Google Scholar]
- 9.Déjean G, Blanvillain‐Baufumé S, Boulanger A, Darrasse A, de Bernonville TD, Girard A-L, Carrére S, Jamet S, Zischek C, Lautier M, Solé M, Büttner D, Jacques M-A, Lauber E, Arlat M. 2013. The xylan utilization system of the plant pathogen Xanthomonas campestris pv campestris controls epiphytic life and reveals common features with oligotrophic bacteria and animal gut symbionts. New Phytol 198:899–915. doi: 10.1111/nph.12187. [DOI] [PubMed] [Google Scholar]
- 10.Blanvillain S, Meyer D, Boulanger A, Lautier M, Guynet C, Denancé N, Vasse J, Lauber E, Arlat M. 2007. Plant carbohydrate scavenging through TonB-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS One 2:e224. doi: 10.1371/journal.pone.0000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boulanger A, Zischek C, Lautier M, Jamet S, Rival P, Carrere S, Arlat M, Lauber E. 2014. The plant pathogen Xanthomonas campestris pv. campestris exploits N-acetylglucosamine during infection. mBio 5:e01527-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rigano LA, Siciliano F, Enrique R, Sendín L, Filippone P, Torres PS, Qüesta J, Dow JM, Castagnaro AP, Vojnov AA, Marano MR. 2007. Biofilm formation, epiphytic fitness, and canker development in Xanthomonas axonopodis pv. citri. Mol Plant Microbe Interact 20:1222–1230. doi: 10.1094/MPMI-20-10-1222. [DOI] [PubMed] [Google Scholar]
- 13.Boller T, Felix G. 2009. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 14.Aslam SN, Newman M-A, Erbs G, Morrissey KL, Chinchilla D, Boller T, Jensen TT, De Castro C, Ierano T, Molinaro A, Jackson RW, Knight MR, Cooper RM. 2008. Bacterial polysaccharides suppress induced innate immunity by calcium chelation. Curr Biol 18:1078–1083. doi: 10.1016/j.cub.2008.06.061. [DOI] [PubMed] [Google Scholar]
- 15.Silipo A, Erbs G, Shinya T, Dow JM, Parrilli M, Lanzetta R, Shibuya N, Newman M-A, Molinaro A. 2010. Glyco-conjugates as elicitors or suppressors of plant innate immunity. Glycobiology 20:406–419. doi: 10.1093/glycob/cwp201. [DOI] [PubMed] [Google Scholar]
- 16.Fatima U, Senthil-Kumar M. 2015. Plant and pathogen nutrient acquisition strategies. Front Plant Sci 6:750. doi: 10.3389/fpls.2015.00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Timilsina S, Potnis N, Newberry EA, Liyanapathiranage P, Iruegas-Bocardo F, White FF, Goss EM, Jones JB. 2020. Xanthomonas diversity, virulence and plant–pathogen interactions. Nat Rev Microbiol 18:415–427. doi: 10.1038/s41579-020-0361-8. [DOI] [PubMed] [Google Scholar]
- 18.Morinière L, Burlet A, Rosenthal ER, Nesme X, Portier P, Bull CT, Lavire C, Fischer-Le Saux M, Bertolla F. 2020. Clarifying the taxonomy of the causal agent of bacterial leaf spot of lettuce through a polyphasic approach reveals that Xanthomonas cynarae Trébaol et al. 2000 emend. Timilsina et al. 2019 is a later heterotypic synonym of Xanthomonas hortorum Vauterin et al. 1995. Syst Appl Microbiol 43:126087. doi: 10.1016/j.syapm.2020.126087. [DOI] [PubMed] [Google Scholar]
- 19.Bull CT, Koike ST. 2005. Evaluating the efficacy of commercial products for management of bacterial leaf spot on lettuce. Plant Health Prog doi: 10.1094/PHP-2005-1121-01-RS. [DOI] [Google Scholar]
- 20.Ozyilmaz U, Benlioglu K. 2018. Bacterial leaf spot of lettuce caused by Xanthomonas hortorum pv. vitians in the Aegean Region of Turkey. Australas Plant Dis Notes 13:37. doi: 10.1007/s13314-018-0325-2. [DOI] [Google Scholar]
- 21.Koike ST, Gilbertson RL. 2017. Detection of Xanthomonas campestris pv. vitians in lettuce seeds, p 173–178. In Fatmi M, Walcott RR, Schaad NW (ed), Detection of plant-pathogenic bacteria in seed and other planting material, 2nd ed. APS Press, St Paul, MN. [Google Scholar]
- 22.Sandoya GV, Maisonneuve B, Truco MJ, Bull CT, Simko I, Trent M, Hayes RJ, Michelmore RW. 2019. Genetic analysis of resistance to bacterial leaf spot in the heirloom lettuce cultivar Reine des Glaces. Mol Breed 39:160. doi: 10.1007/s11032-019-1072-6. [DOI] [Google Scholar]
- 23.Myung I-S, Moon SY, Jeong IH, Lee SW, Lee YH, Shim HS. 2010. Bacterial leaf spot of iceberg lettuce caused by Xanthomonas campestris pv. vitians type B, a new disease in South Korea. Plant Dis 94:790–790. doi: 10.1094/PDIS-94-6-0790B. [DOI] [PubMed] [Google Scholar]
- 24.Dia NC, Morinière L, Cottyn B, Bernal E, Jacobs JM, Koebnik R, Osdaghi E, Potnis N, Pothier JF. 2022. Xanthomonas hortorum—beyond gardens: current taxonomy, genomics, and virulence repertoires. Mol Plant Pathol doi: 10.1111/mpp.13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morinière L, Lecomte S, Gueguen E, Bertolla F. 2021. In vitro exploration of the Xanthomonas hortorum pv. vitians genome using transposon insertion sequencing and comparative genomics to discriminate between core and contextual essential genes. Microb Genom 7:e000546. doi: 10.1099/mgen.0.000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Opijnen T, Bodi KL, Camilli A. 2009. Tn-seq; high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods 6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fabian BK, Tetu SG, Paulsen IT. 2020. Application of transposon insertion sequencing to agricultural science. Front Plant Sci 11:291. doi: 10.3389/fpls.2020.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duong DA, Jensen RV, Stevens AM. 2018. Discovery of Pantoea stewartii ssp. stewartii genes important for survival in corn xylem through a Tn-Seq analysis. Mol Plant Pathol 19:1929–1941. doi: 10.1111/mpp.12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Royet K, Parisot N, Rodrigue A, Gueguen E, Condemine G. 2019. Identification by Tn-seq of Dickeya dadantii genes required for survival in chicory plants. Mol Plant Pathol 20:287–306. doi: 10.1111/mpp.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Helmann TC, Deutschbauer AM, Lindow SE. 2019. Genome-wide identification of Pseudomonas syringae genes required for fitness during colonization of the leaf surface and apoplast. Proc Natl Acad Sci USA 116:18900–18910. doi: 10.1073/pnas.1908858116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Helmann TC, Deutschbauer AM, Lindow SE. 2020. Distinctiveness of genes contributing to growth of Pseudomonas syringae in diverse host plant species. PLoS One 15:e0239998. doi: 10.1371/journal.pone.0239998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su Y, Xu Y, Liang H, Yuan G, Wu X, Zheng D. 2021. Genome-wide identification of Ralstonia solanacearum genes required for survival in tomato plants. mSystems 6:e00838-21. doi: 10.1128/mSystems.00838-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Torres M, Jiquel A, Jeanne E, Naquin D, Dessaux Y, Faure D. 2022. Agrobacterium tumefaciens fitness genes involved in the colonization of plant tumors and roots. New Phytol 233:905–918. doi: 10.1111/nph.17810. [DOI] [PubMed] [Google Scholar]
- 34.Chao MC, Abel S, Davis BM, Waldor MK. 2016. The design and analysis of transposon-insertion sequencing experiments. Nat Rev Microbiol 14:119–128. doi: 10.1038/nrmicro.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abel S, Abel Zur Wiesch P, Chang H-H, Davis BM, Lipsitch M, Waldor MK. 2015. Sequence tag-based analysis of microbial population dynamics. Nat Methods 12:223–226. doi: 10.1038/nmeth.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahmutovic A, Abel Zur Wiesch P, Abel S. 2020. Selection or drift: the population biology underlying transposon insertion sequencing experiments. Comput Struct Biotechnol J 18:791–804. doi: 10.1016/j.csbj.2020.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Opijnen T, Camilli A. 2013. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat Rev Microbiol 11:435–442. doi: 10.1038/nrmicro3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Z, Beskrovnaya P, Melnyk RA, Hossain SS, Khorasani S, O’Sullivan LR, Wiesmann CL, Bush J, Richard JD, Haney CH. 2018. A genome-wide screen identifies genes in rhizosphere-associated Pseudomonas required to evade plant defenses. mBio 9:e00433-18. doi: 10.1128/mBio.00433-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gene Ontology Consortium. 2021. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res 49:D325–D334. doi: 10.1093/nar/gkaa1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanehisa M, Sato Y, Morishima K. 2016. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 41.Qian W, Jia Y, Ren S-X, He Y-Q, Feng J-X, Lu L-F, Sun Q, Ying G, Tang D-J, Tang H, Wu W, Hao P, Wang L, Jiang B-L, Zeng S, Gu W-Y, Lu G, Rong L, Tian Y, Yao Z, Fu G, Chen B, Fang R, Qiang B, Chen Z, Zhao G-P, Tang J-L, He C. 2005. Comparative and functional genomic analyses of the pathogenicity of phytopathogen Xanthomonas campestris pv. campestris. Genome Res 15:757–767. doi: 10.1101/gr.3378705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang JC, So BH, Kim JH, Park YJ, Lee BM, Kang HW. 2008. Genome-wide identification of pathogenicity genes in Xanthomonas oryzae pv. oryzae by transposon mutagenesis. Plant Pathol 57:1136–1145. doi: 10.1111/j.1365-3059.2008.01884.x. [DOI] [Google Scholar]
- 43.Song X, Guo J, Ma W, Ji Z, Zou L, Chen G, Zou H. 2015. Identification of seven novel virulence genes from Xanthomonas citri subsp. citri by Tn5-based random mutagenesis. J Microbiol 53:330–336. doi: 10.1007/s12275-015-4589-3. [DOI] [PubMed] [Google Scholar]
- 44.Zou H-S, Yuan L, Guo W, Li Y-R, Che Y-Z, Zou L-F, Chen G-Y. 2011. Construction of a Tn5-tagged mutant library of Xanthomonas oryzae pv. oryzicola as an invaluable resource for functional genomics. Curr Microbiol 62:908–916. doi: 10.1007/s00284-010-9804-1. [DOI] [PubMed] [Google Scholar]
- 45.Matilla MA, Krell T. 2018. The effect of bacterial chemotaxis on host infection and pathogenicity. FEMS Microbiol Rev 42:fux052. doi: 10.1093/femsre/fux052. [DOI] [PubMed] [Google Scholar]
- 46.Darrasse A, Carrère S, Barbe V, Boureau T, Arrieta-Ortiz ML, Bonneau S, Briand M, Brin C, Cociancich S, Durand K, Fouteau S, Gagnevin L, Guérin F, Guy E, Indiana A, Koebnik R, Lauber E, Munoz A, Noël LD, Pieretti I, Poussier S, Pruvost O, Robène-Soustrade I, Rott P, Royer M, Serres-Giardi L, Szurek B, van Sluys M-A, Verdier V, Vernière C, Arlat M, Manceau C, Jacques M-A. 2013. Genome sequence of Xanthomonas fuscans subsp. fuscans strain 4834-R reveals that flagellar motility is not a general feature of xanthomonads. BMC Genomics 14:761. doi: 10.1186/1471-2164-14-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liao Z-X, Ni Z, Wei X-L, Chen L, Li J-Y, Yu Y-H, Jiang W, Jiang B-L, He Y-Q, Huang S. 2019. Dual RNA-seq of Xanthomonas oryzae pv. oryzicola infecting rice reveals novel insights into bacterial-plant interaction. PLoS One 14:e0215039. doi: 10.1371/journal.pone.0215039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nobori T, Velásquez AC, Wu J, Kvitko BH, Kremer JM, Wang Y, He SY, Tsuda K. 2018. Transcriptome landscape of a bacterial pathogen under plant immunity. Proc Natl Acad Sci USA 115:E3055–E3064. doi: 10.1073/pnas.1800529115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rico A, Preston GM. 2008. Pseudomonas syringae pv. tomato DC3000 uses constitutive and apoplast-induced nutrient assimilation pathways to catabolize nutrients that are abundant in the tomato apoplast. Mol Plant Microbe Interact 21:269–282. doi: 10.1094/MPMI-21-2-0269. [DOI] [PubMed] [Google Scholar]
- 50.Lohaus G, Winter H, Riens B, Heldt HW. 1995. Further studies of the phloem loading process in leaves of barley and spinach. The comparison of metabolite concentrations in the apoplastic compartment with those in the cytosolic compartment and in the sieve tubes. Bot Acta 108:270–275. doi: 10.1111/j.1438-8677.1995.tb00860.x. [DOI] [Google Scholar]
- 51.O'Leary BM, Neale HC, Geilfus C-M, Jackson RW, Arnold DL, Preston GM. 2016. Early changes in apoplast composition associated with defence and disease in interactions between Phaseolus vulgaris and the halo blight pathogen Pseudomonas syringae pv. phaseolicola. Plant Cell Environ 39:2172–2184. doi: 10.1111/pce.12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Joosten MHAJ, Hendrickx LJM, Wit PJGM. 1990. Carbohydrate composition of apoplastic fluids isolated from tomato leaves inoculated with virulent or avirulent races of Cladosporium fulvum (syn. Fulvia fulva). Neth J Plant Pathol 96:103–112. doi: 10.1007/BF02005134. [DOI] [Google Scholar]
- 53.Szczesny R, Jordan M, Schramm C, Schulz S, Cogez V, Bonas U, Büttner D. 2010. Functional characterization of the Xcs and Xps type II secretion systems from the plant pathogenic bacterium Xanthomonas campestris pv vesicatoria. New Phytol 187:983–1002. doi: 10.1111/j.1469-8137.2010.03312.x. [DOI] [PubMed] [Google Scholar]
- 54.Vandroemme J, Cottyn B, Baeyen S, De Vos P, Maes M. 2013. Draft genome sequence of Xanthomonas fragariae reveals reductive evolution and distinct virulence-related gene content. BMC Genomics 14:829. doi: 10.1186/1471-2164-14-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim H-S, Park H-J, Heu S, Jung J. 2004. Molecular and functional characterization of a unique sucrose hydrolase from Xanthomonas axonopodis pv. glycines. J Bacteriol 186:411–418. doi: 10.1128/JB.186.2.411-418.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watt TF, Vucur M, Baumgarth B, Watt SA, Niehaus K. 2009. Low molecular weight plant extract induces metabolic changes and the secretion of extracellular enzymes, but has a negative effect on the expression of the type-III secretion system in Xanthomonas campestris pv. campestris. J Biotechnol 140:59–67. doi: 10.1016/j.jbiotec.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 57.Zhang C, Lv M, Yin W, Dong T, Chang C, Miao Y, Jia Y, Deng Y. 2019. Xanthomonas campestris promotes diffusible signal factor biosynthesis and pathogenicity by utilizing glucose and sucrose from host plants. Mol Plant Microbe Interact 32:157–166. doi: 10.1094/MPMI-07-18-0187-R. [DOI] [PubMed] [Google Scholar]
- 58.He Y-W, Zhang L-H. 2008. Quorum sensing and virulence regulation in Xanthomonas campestris. FEMS Microbiol Rev 32:842–857. doi: 10.1111/j.1574-6976.2008.00120.x. [DOI] [PubMed] [Google Scholar]
- 59.Chow V, Shantharaj D, Guo Y, Nong G, Minsavage GV, Jones JB, Preston JF. 2015. Xylan utilization regulon in Xanthomonas citri pv. citri strain 306: gene expression and utilization of oligoxylosides. Appl Environ Microbiol 81:2163–2172. doi: 10.1128/AEM.03091-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Santos CR, Hoffmam ZB, de Matos Martins VP, Zanphorlin LM, de Paula Assis LH, Honorato RV, Lopes de Oliveira PS, Ruller R, Murakami MT. 2014. Molecular mechanisms associated with xylan degradation by Xanthomonas plant pathogens. J Biol Chem 289:32186–32200. doi: 10.1074/jbc.M114.605105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pandey SS, Patnana PK, Rai R, Chatterjee S. 2017. Xanthoferrin, the α-hydroxycarboxylate-type siderophore of Xanthomonas campestris pv. campestris, is required for optimum virulence and growth inside cabbage. Mol Plant Pathol 18:949–962. doi: 10.1111/mpp.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guerra GS, Balan A. 2020. Genetic and structural determinants on iron assimilation pathways in the plant pathogen Xanthomonas citri subsp. citri and Xanthomonas sp. Braz J Microbiol 51:1219–1231. doi: 10.1007/s42770-019-00207-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu Y, Kong D, Wu H-L, Ling H-Q. 2021. Iron in plant-pathogen interactions. J Exp Bot 72:2114–2124. doi: 10.1093/jxb/eraa516. [DOI] [PubMed] [Google Scholar]
- 64.Távora FTPK, Santos C, Maximiano MR, Murad AM, Oliveira‐Neto OB, Megias E, Junior FBR, Franco OL, Mehta A. 2019. Pan proteome of Xanthomonas campestris pv. campestris isolates contrasting in virulence. Proteomics 19:1900082. doi: 10.1002/pmic.201900082. [DOI] [PubMed] [Google Scholar]
- 65.Cain AK, Barquist L, Goodman AL, Paulsen IT, Parkhill J, van Opijnen T. 2020. A decade of advances in transposon-insertion sequencing. Nat Rev Genet 21:526–540. doi: 10.1038/s41576-020-0244-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vorhölter F-J, Schneiker S, Goesmann A, Krause L, Bekel T, Kaiser O, Linke B, Patschkowski T, Rückert C, Schmid J, Sidhu VK, Sieber V, Tauch A, Watt SA, Weisshaar B, Becker A, Niehaus K, Pühler A. 2008. The genome of Xanthomonas campestris pv. campestris B100 and its use for the reconstruction of metabolic pathways involved in xanthan biosynthesis. J Biotechnol 134:33–45. doi: 10.1016/j.jbiotec.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 67.Galván EM, Ielmini MV, Patel YN, Bianco MI, Franceschini EA, Schneider JC, Ielpi L. 2013. Xanthan chain length is modulated by increasing the availability of the polysaccharide copolymerase protein GumC and the outer membrane polysaccharide export protein GumB. Glycobiology 23:259–272. doi: 10.1093/glycob/cws146. [DOI] [PubMed] [Google Scholar]
- 68.Kim S-Y, Kim J-G, Lee B-M, Cho J-Y. 2009. Mutational analysis of the gum gene cluster required for xanthan biosynthesis in Xanthomonas oryzae pv oryzae. Biotechnol Lett 31:265–270. doi: 10.1007/s10529-008-9858-3. [DOI] [PubMed] [Google Scholar]
- 69.Klein G, Kobylak N, Lindner B, Stupak A, Raina S. 2014. Assembly of lipopolysaccharide in Escherichia coli requires the essential LapB heat shock protein. J Biol Chem 289:14829–14853. doi: 10.1074/jbc.M113.539494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Petrocelli S, Tondo ML, Daurelio LD, Orellano EG. 2012. Modifications of Xanthomonas axonopodis pv. citri lipopolysaccharide affect the basal response and the virulence process during citrus canker. PLoS One 7:e40051. doi: 10.1371/journal.pone.0040051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guo Y, Sagaram US, Kim J, Wang N. 2010. Requirement of the galU gene for polysaccharide production by and pathogenicity and growth in planta of Xanthomonas citri subsp. citri. Appl Environ Microbiol 76:2234–2242. doi: 10.1128/AEM.02897-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vorhölter F-J, Niehaus K, Pühler A. 2001. Lipopolysaccharide biosynthesis in Xanthomonas campestris pv. campestris: a cluster of 15 genes is involved in the biosynthesis of the LPS O-antigen and the LPS core. Mol Genet Genomics 266:79–95. doi: 10.1007/s004380100521. [DOI] [PubMed] [Google Scholar]
- 73.Köplin R, Wang G, Hötte B, Priefer UB, Pühler A. 1993. A 3.9-kb DNA region of Xanthomonas campestris pv. campestris that is necessary for lipopolysaccharide production encodes a set of enzymes involved in the synthesis of dTDP-rhamnose. J Bacteriol 175:7786–7792. doi: 10.1128/jb.175.24.7786-7792.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Molinaro A, Castro CD, Lanzetta R, Parrilli M, Petersen BO, Broberg A, Duus JØ. 2002. NMR and MS evidences for a random assembled O-specific chain structure in the LPS of the bacterium Xanthomonas campestris pv. vitians. Eur J Biochem 269:4185–4193. doi: 10.1046/j.1432-1033.2002.03138.x. [DOI] [PubMed] [Google Scholar]
- 75.Steffens T, Vorhölter F-J, Giampà M, Hublik G, Pühler A, Niehaus K. 2016. The influence of a modified lipopolysaccharide O-antigen on the biosynthesis of xanthan in Xanthomonas campestris pv. campestris B100. BMC Microbiol 16:93. doi: 10.1186/s12866-016-0710-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Raetz CRH, Whitfield C. 2002. Lipopolysaccharide endotoxins. Annu Rev Biochem 71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Matsuura M. 2013. Structural modifications of bacterial lipopolysaccharide that facilitate Gram-negative bacteria evasion of host innate immunity. Front Immunol 4:109. doi: 10.3389/fimmu.2013.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Newman M-A, Dow JM, Daniels MJ. 2001. Bacterial lipopolysaccharides and plant-pathogen interactions. Eur J Plant Pathol 107:95–102. doi: 10.1023/A:1008738817551. [DOI] [Google Scholar]
- 79.Rapicavoli JN, Blanco-Ulate B, Muszyński A, Figueroa-Balderas R, Morales-Cruz A, Azadi P, Dobruchowska JM, Castro C, Cantu D, Roper MC. 2018. Lipopolysaccharide O-antigen delays plant innate immune recognition of Xylella fastidiosa. Nat Commun 9:390. doi: 10.1038/s41467-018-02861-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lushchak VI. 2001. Oxidative stress and mechanisms of protection against it in bacteria. Biochem Mosc 66:476–489. doi: 10.1023/A:1010294415625. [DOI] [PubMed] [Google Scholar]
- 81.Fones H, Preston GM. 2012. Reactive oxygen and oxidative stress tolerance in plant pathogenic Pseudomonas. FEMS Microbiol Lett 327:1–8. doi: 10.1111/j.1574-6968.2011.02449.x. [DOI] [PubMed] [Google Scholar]
- 82.Santos R, Franza T, Laporte M-L, Sauvage C, Touati D, Expert D. 2001. Essential role of superoxide dismutase on the pathogenicity of Erwinia chrysanthemi strain 3937. Mol Plant Microbe Interact 14:758–767. doi: 10.1094/MPMI.2001.14.6.758. [DOI] [PubMed] [Google Scholar]
- 83.Hugouvieux V, Barber CE, Daniels MJ. 1998. Entry of Xanthomonas campestris pv. campestris into hydathodes of Arabidopsis thaliana leaves: a system for studying early infection events in bacterial pathogenesis. Mol Plant Microbe Interact 11:537–543. doi: 10.1094/MPMI.1998.11.6.537. [DOI] [PubMed] [Google Scholar]
- 84.Smith SG, Wilson TJ, Dow JM, Daniels MJ. 1996. A gene for superoxide dismutase from Xanthomonas campestris pv. campestris and its expression during bacterial-plant interactions. Mol Plant Microbe Interact 9:584–593. doi: 10.1094/mpmi-9-0584. [DOI] [PubMed] [Google Scholar]
- 85.Lin C-N, Syu W-J, Sun W-SW, Chen J-W, Chen T-H, Don M-J, Wang S-H. 2010. A role of ygfZ in the Escherichia coli response to plumbagin challenge. J Biomed Sci 17:84. doi: 10.1186/1423-0127-17-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Figaj D, Ambroziak P, Przepiora T, Skorko-Glonek J. 2019. The role of proteases in the virulence of plant pathogenic bacteria. Int J Mol Sci 20:672. doi: 10.3390/ijms20030672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li C-E, Liao C-T, Lo H-H, Hsiao Y-M. 2020. Functional characterization and transcriptional analysis of clpP of Xanthomonas campestris pv. campestris. Curr Microbiol 77:2876–2885. doi: 10.1007/s00284-020-02093-1. [DOI] [PubMed] [Google Scholar]
- 88.Lo H-H, Liao C-T, Li C-E, Chiang Y-C, Hsiao Y-M. 2020. The clpX gene plays an important role in bacterial attachment, stress tolerance, and virulence in Xanthomonas campestris pv. campestris. Arch Microbiol 202:597–607. doi: 10.1007/s00203-019-01772-3. [DOI] [PubMed] [Google Scholar]
- 89.Bove P, Capozzi V, Garofalo C, Rieu A, Spano G, Fiocco D. 2012. Inactivation of the ftsH gene of Lactobacillus plantarum WCFS1: effects on growth, stress tolerance, cell surface properties and biofilm formation. Microbiol Res 167:187–193. doi: 10.1016/j.micres.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 90.Matos RG, Bárria C, Moreira RN, Barahona S, Domingues S, Arraiano CM. 2014. The importance of proteins of the RNase II/RNB-family in pathogenic bacteria. Front Cell Infect Microbiol 4:68. doi: 10.3389/fcimb.2014.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Facincani AP, Moreira LM, Soares MR, Ferreira CB, Ferreira RM, Ferro MIT, Ferro JA, Gozzo FC, de Oliveira JCF. 2014. Comparative proteomic analysis reveals that T3SS, Tfp, and xanthan gum are key factors in initial stages of Citrus sinensis infection by Xanthomonas citri subsp. citri. Funct Integr Genomics 14:205–217. doi: 10.1007/s10142-013-0340-5. [DOI] [PubMed] [Google Scholar]
- 92.Imlay JA, Linn S. 1987. Mutagenesis and stress responses induced in Escherichia coli by hydrogen peroxide. J Bacteriol 169:2967–2976. doi: 10.1128/jb.169.7.2967-2976.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stohl EA, Seifert HS. 2006. Neisseria gonorrhoeae DNA recombination and repair enzymes protect against oxidative damage caused by hydrogen peroxide. J Bacteriol 188:7645–7651. doi: 10.1128/JB.00801-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cano DA, Pucciarelli MG, García-del Portillo F, Casadesús J. 2002. Role of the RecBCD recombination pathway in Salmonella virulence. J Bacteriol 184:592–595. doi: 10.1128/JB.184.2.592-595.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kuzminov A, Stahl FW. 1999. Double-strand end repair via the RecBC pathway in Escherichia coli primes DNA replication. Genes Dev 13:345–356. doi: 10.1101/gad.13.3.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mongkolsuk S, Rabibhadana S, Sukchavalit R, Vaughn G. 1998. Construction and physiological analysis of a Xanthomonas oryzae pv. oryzae recA mutant. FEMS Microbiol Lett 169:269–275. doi: 10.1111/j.1574-6968.1998.tb13328.x. [DOI] [PubMed] [Google Scholar]
- 97.Aguirre-Ramírez M, Medina G, González-Valdez A, Grosso-Becerra V, Soberón-Chávez G. 2012. The Pseudomonas aeruginosa rmlBDAC operon, encoding dTDP-l-rhamnose biosynthetic enzymes, is regulated by the quorum-sensing transcriptional regulator RhlR and the alternative sigma factor σS. Microbiology (Reading) 158:908–916. doi: 10.1099/mic.0.054726-0. [DOI] [PubMed] [Google Scholar]
- 98.Bronstein PA, Marrichi M, Cartinhour S, Schneider DJ, DeLisa MP. 2005. Identification of a twin-arginine translocation system in Pseudomonas syringae pv. tomato DC3000 and its contribution to pathogenicity and fitness. J Bacteriol 187:8450–8461. doi: 10.1128/JB.187.24.8450-8461.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Genin S, Denny TP. 2012. Pathogenomics of the Ralstonia solanacearum species complex. Annu Rev Phytopathol 50:67–89. doi: 10.1146/annurev-phyto-081211-173000. [DOI] [PubMed] [Google Scholar]
- 100.Ranjan M, Khokhani D, Nayaka S, Srivastava S, Keyser ZP, Ranjan A. 2021. Genomic diversity and organization of complex polysaccharide biosynthesis clusters in the genus Dickeya. PLoS One 16:e0245727. doi: 10.1371/journal.pone.0245727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jayaraman J, Jones WT, Harvey D, Hemara LM, McCann HC, Yoon M, Warring SL, Fineran PC, Mesarich CH, Templeton MD. 2020. Variation at the common polysaccharide antigen locus drives lipopolysaccharide diversity within the Pseudomonas syringae species complex. Environ Microbiol 22:5356–5372. doi: 10.1111/1462-2920.15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Georgoulis SJ, Shalvarjian KE, Helmann TC, Hamilton CD, Carlson HK, Deutschbauer AM, Lowe-Power TM. 2021. Genome-wide identification of tomato xylem sap fitness factors for three plant-pathogenic Ralstonia species. mSystems 6:e01229-21. doi: 10.1128/mSystems.01229-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetj 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 104.DeJesus MA, Ambadipudi C, Baker R, Sassetti C, Ioerger TR. 2015. TRANSIT—a software tool for Himar1 TnSeq analysis. PLoS Comput Biol 11:e1004401. doi: 10.1371/journal.pcbi.1004401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.DeJesus MA, Ioerger TR. 2016. Normalization of transposon-mutant library sequencing datasets to improve identification of conditionally essential genes. J Bioinform Comput Biol 14:1642004. doi: 10.1142/S021972001642004X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Benjamini Y, Yekutieli D. 2005. False discovery rate—adjusted multiple confidence intervals for selected parameters. J Am Stat Assoc 100:71–81. doi: 10.1198/016214504000001907. [DOI] [Google Scholar]
- 107.Vallenet D, Calteau A, Dubois M, Amours P, Bazin A, Beuvin M, Burlot L, Bussell X, Fouteau S, Gautreau G, Lajus A, Langlois J, Planel R, Roche D, Rollin J, Rouy Z, Sabatet V, Médigue C. 2020. MicroScope: an integrated platform for the annotation and exploration of microbial gene functions through genomic, pangenomic and metabolic comparative analysis. Nucleic Acids Res 48:D579–D589. doi: 10.1093/nar/gkz926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yu Y, Ouyang Y, Yao W. 2018. shinyCircos: an R/Shiny application for interactive creation of Circos plot. Bioinformatics 34:1229–1231. doi: 10.1093/bioinformatics/btx763. [DOI] [PubMed] [Google Scholar]
- 109.Leimbach A. 2016. bac-genomics-scripts: bovine E. coli mastitis comparative genomics edition. Zenodo 10.5281/zenodo.215824. Accessed 16 March 2021. [DOI]
- 110.Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, von Mering C, Bork P. 2017. Fast genome-wide functional annotation through orthology assignment by eggNOG-Mapper. Mol Biol Evol 34:2115–2122. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Huerta-Cepas J, Szklarczyk D, Heller D, Hernández-Plaza A, Forslund SK, Cook H, Mende DR, Letunic I, Rattei T, Jensen LJ, von Mering C, Bork P. 2019. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res 47:D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong S-Y, Lopez R, Hunter S. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rasche H, Grüning B, Dunn N, Bretaudeau A. 2018. GGA: Galaxy for genome annotation, teaching, and genomic databases. F1000Res 7:f1000research. doi: 10.7490/f1000research.1116181.1. [DOI] [Google Scholar]
- 114.Grossmann S, Bauer S, Robinson PN, Vingron M. 2007. Improved detection of overrepresentation of Gene-Ontology annotations with parent child analysis. Bioinformatics 23:3024–3031. doi: 10.1093/bioinformatics/btm440. [DOI] [PubMed] [Google Scholar]
- 115.Kanehisa M, Sato Y. 2020. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci 29:28–35. doi: 10.1002/pro.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complete genome annotation and Tn-seq results in vitro and in planta. Download Table S1, XLSX file, 1.2 MB (1.2MB, xlsx) .
Copyright © 2022 Morinière et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Critical genes in the carbon metabolism pathways of X. hortorum pv. vitians LM16734 under the different Tn-seq experimental conditions, created with KEGG Mapper. Carbon numbers in compounds are indicated in circles or by asterisks for cofactors. Download FIG S1, TIF file, 2.6 MB (2.6MB, tif) .
Copyright © 2022 Morinière et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of the 170 genes critical in lettuce leaves. Download Table S2, XLSX file, 0.06 MB (57.2KB, xlsx) .
Copyright © 2022 Morinière et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of the 36 genes critical in 1/10th TSB medium. Download Table S3, XLSX file, 0.02 MB (19.6KB, xlsx) .
Copyright © 2022 Morinière et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of the 2 growth-advantage genes in lettuce leaves. Download Table S4, XLSX file, 0.01 MB (10.1KB, xlsx) .
Copyright © 2022 Morinière et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The NCBI GenBank assembly accession number for the complete X. hortorum pv. vitians LM16734 genome is GCA_014338485.1. Transposon insertion sequencing raw reads have been deposited in the NCBI SRA database under accession no. SRR14385278 to SRR14385281. All data are associated with BioProject no. PRJNA530964 and PRJNA726806.
List of the 170 genes critical in lettuce leaves. Download Table S2, XLSX file, 0.06 MB (57.2KB, xlsx) .
Copyright © 2022 Morinière et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of the 36 genes critical in 1/10th TSB medium. Download Table S3, XLSX file, 0.02 MB (19.6KB, xlsx) .
Copyright © 2022 Morinière et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of the 2 growth-advantage genes in lettuce leaves. Download Table S4, XLSX file, 0.01 MB (10.1KB, xlsx) .
Copyright © 2022 Morinière et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.