Summary
Background
African Americans (AA) remain underrepresented in Alzheimer's disease (AD) research, despite the prevalence of AD being double in AA compared to non-Hispanic whites. To address this disparity, our group has established the Florida Consortium for African American Alzheimer's Disease Studies (FCA3DS), focusing on the identification of genetic risk factors and novel plasma biomarkers.
Method
Utilizing FCA3DS whole exome sequence (WES) and plasma RNA samples from AD cases (n=151) and cognitively unimpaired (CU) elderly controls (n=269), we have performed differential gene expression (DGE) and expression quantitative trait locus (eQTL) analyses on 50 transcripts measured with a custom nanoString® panel. We designed this panel to measure, in plasma, cell-free mRNA (cf-mRNA) levels of AD-relevant genes.
Findings
Association with higher plasma CLU in CU vs. AD remained significant after Bonferroni correction. Study-wide significant eQTL associations were observed with 105 WES variants in cis with 22 genes, including variants in genes previously associated with AD risk in AA such as ABCA7 and AKAP9. Results from this plasma eQTL analysis identified AD-risk variants in ABCA7 and AKAP9 that are significantly associated with lower and higher plasma mRNA levels of these genes, respectively. Receiver operating characteristic analysis of age, sex APOE-ε4 dosage, CLU, APP, CD14, ABCA7, AKAP9 and APOE mRNA levels, and ABCA7 and AKAP9 eQTLs, achieved 77% area under the curve to discriminate AD vs. CU, an 8% improvement over a model that only included age, sex and APOE-ε4 dosage.
Interpretation
Incorporating plasma mRNA levels could contribute to improved predictive value of AD biomarker panels.
Funding
This work was supported by the National Institute on Aging [RF AG051504, U01 AG046139, R01 AG061796 to NET; P30 AG062677 to JAL and NGR]; Florida Health Ed and Ethel Moore Alzheimer's Disease grants [5AZ03 and 7AZ17 to NET; 7AZ07 to MMC; 8AZ08 to JAL].
Keywords: Alzheimer's disease, African Americans, Biomarkers, Plasma, cf-mRNA, eQTL, Gene expression, Whole exome sequencing
Research in context.
Evidence before this study
The identification of plasma biomarkers for Alzheimer's disease (AD) has become an important area of research given their potential to enhance accessibility to an accurate clinical diagnosis, which can in turn impact management and development of treatments. Several studies have shown promising results for the utility of plasma phosphorylated tau 217 (p-tau 217) and plasma phosphorylated tau 181 (p-tau 181) as potential diagnostic biomarkers of AD, both being able to distinguish AD from other dementias with higher accuracy than other plasma biomarkers tested thus far. In this study we focused on plasma cell-free mRNAs as potentially complementary biomarkers that may increase the diagnostic accuracy of AD and which may also provide insight into the pathobiological mechanisms underlying the disease. We conducted a PubMed search for articles published in any language up to August 15, 2021 that were related to the evaluation of plasma cell-free mRNA levels as biomarkers of AD, using search terms that included "Alzheimer's" AND “cell-free" AND "mRNA", OR "Alzheimer's" AND “plasma” AND “extracellular” AND “RNA”, OR "Alzheimer's" AND “plasma” AND “RNA levels”. We only found 6 publications that tested the association of plasma cell-free mRNA with AD diagnosis, and only 3 evaluated diagnostic performance.
Added value of this study
None of the prior studies that evaluated plasma transcripts as potential diagnostic biomarkers of AD were conducted in an African American cohort. Our study focuses on African Americans, a population that remains underrepresented in AD research despite having twice the risk of developing AD compared to non-Hispanic Whites. Our study identified a set of 6 plasma cf-mRNAs that along with sex, age and APOE-ε4 allelic dosage yield a receiver operating characteristic area under the curve of 0.77 to differentiate AD cases vs. cognitively unimpaired controls. Furthermore, unlike prior work, our study conducted a targeted screen of cf-mRNAs using a nanoString custom panel that measures mRNA transcript counts of genes implicated in AD, inflammation or the immune response, as these transcripts have the potential of being developed as theragnostic biomarkers. Therefore, the value of our study lies in the novelty of the targeted approach of testing specific plasma cf-mRNA levels as potential biomarkers of AD, its focus on an understudied and underserved population, and identification of a predictive set of plasma cf-mRNAs that may lead to improved biomarker panels of AD.
Implications of all available evidence
Collectively with prior plasma biomarkers studies, the findings of this study may contribute to the development of minimally invasive biomarker panels for improved accuracy of AD diagnosis which will enable more effective disease management. Further, our study provides an estimate of the AD predictive value for a plasma cf-mRNA panel in African Americans that will guide future studies of these novel biomarkers in this underrepresented and other populations.
Alt-text: Unlabelled box
Introduction
Alzheimer's disease (AD) is the most common form of dementia, currently affecting an estimated 6·2 million people in the United States.1 There is no cure yet for this devastating and deadly disease which robs patients from their memory and eventually renders them completely incapacitated. The clinical diagnosis of AD is challenging due to other dementias that share similar symptoms, and the heterogeneity of symptoms that AD patients present.2 A definite diagnosis of AD can only be made at autopsy upon confirmation of the presence of co-existing extracellular amyloid plaques and intraneuronal tau tangles, which are the neuropathological hallmarks of AD.3 Neuroimaging modalities such as amyloid PET and tau PET have been developed as diagnostic biomarkers that improve the accuracy of a clinical diagnosis of AD.3,4 Cerebrospinal fluid (CSF) levels of specific isotypes of amyloid ß (Aß) and tau protein, such as Aß42/Aß40 ratio and phosphorylated tau at residues 181 and 217 (p-tau 181 and p-tau 217) have been shown to have comparable diagnostic sensitivity and specificity as PET biomarkers.5,6 However, there is a need for biomarkers that are less costly and less invasive than PET and CSF biomarkers respectively, in order to make them more accessible to the general population. The development of plasma biomarkers for AD could achieve both of these goals. One of the most promising plasma biomarkers to date is plasma p-tau 217 which has been shown to discriminate AD from other dementias with accuracy equivalent to CSF p-tau 217 and tau PET (plasma p-tau 217 area under the curve [AUC] =0·96, CSF p-tau 217 AUC=0·99, tau PET AUC=0·98).7 Similar results have been obtained with plasma p-tau 181,8 although plasma p-tau 217 seems to outperform p-tau 181 (plasma p-tau 217 AUC=0·96, plasma p-tau 181 AUC=0·90)7 and both of these outperform plasma total tau.
Very few plasma AD biomarker studies have been performed in African Americans (AA), and only one of these, published by our group, analyzed tau levels in AD cases and cognitively unimpaired (CU) participants. In that study we found higher levels of plasma total tau in AD compared to CU controls, yet this marker alone was not sufficient to discriminate AD vs. CU participants (AUC=0·55).9 Despite the tremendous progress in AD biomarker research, there is still a need to develop additional plasma biomarkers that add to the predictive value of existing ones, and which assess the contribution of biological processes beyond Aß and tau that underlie the disease pathophysiology, such as inflammation, as this type of theragnostic biomarker could inform future treatment options.
Studies have explored the utility of cell free RNA (cf-RNA) as potential plasma AD biomarkers that inherently provide insight into the disease pathomechanism. Published work in this area has primarily focused on microRNAs,10 while few studies have profiled circulating, protein-coding messenger RNAs (cf-mRNA), but none of these studies have been performed in underrepresented populations such as AA. In the present study, we explore the utility of cf-mRNA as potential AD biomarkers in AA, a population that has twice the risk of developing AD as non-Hispanic whites (NHW)11 and which remains understudied in AD research.12 Given the wealth of evidence implicating the immune response and inflammation in the etiology of AD,13,14 in this study we evaluated the discriminatory potential of plasma cf-mRNA from genes that are involved in inflammation or that have been previously shown to associate with AD diagnosis. A total of 50 such transcripts detectable in plasma were prioritized to assess their potential as predictors of AD diagnosis in AA. Our results demonstrate that analyzing just 6 key cf-mRNAs (CLU, APP, CD14, ABCA7, AKAP9 and APOE) in conjunction with age, sex, allelic dosage of APOE-ε4 and allelic dosage of the most significant ABCA7 and AKAP9 plasma expression quantitative trait locus (eQTL) can achieve 77% AUC for the classification of AD cases vs. CU controls in AA.
Methods
Study population
A total of 530 self-reported AA study participants (242 AD cases and 288 CU controls) were recruited for this study at the Mayo Clinic in Jacksonville, Florida or at the Wien Center for Alzheimer's Disease and Memory Disorders, Mount Sinai, Miami, Florida (Table 1). All CU controls and AD cases included in this study consented to participate in Alzheimer's disease research as part of the Florida Consortium for African-American Alzheimer's Disease Studies (FCA3DS), and were diagnosed by a neurologist as having possible or probable AD according to the NINCDS-ADRDA criteria, or were CU elderly participants with a Clinical Dementia Rating scale score of 0 at their last examination, as previously described.15
Table 1.
Characteristics of case-control series. Summary characteristics are shown for FCA3DS AD and CU participants included in the DGE, eQTL and AD-risk association analyses. N represents sample size; Female (%) is the percentage of females in each group; Age is the mean and standard deviation of age at plasma draw, and APOE-ε4 (%) is the percentage of APOE-ε4 carriers in each group.
| Analysis | DGE |
eQTL |
AD-risk association |
|||
|---|---|---|---|---|---|---|
| Diagnosis | AD | CU | AD | CU | AD | CU |
| N | 151 | 269 | 139 | 225 | 230 | 244 |
| Females (%) | 69·5 | 76·6 | 67·6 | 74·7 | 70·0 | 74·2 |
| Age | 77·5 ± 9·2 | 80·1 ± 8·4 | 77·0 ± 9·3 | 79·8 ± 8·7 | 76·4 ± 9·5 | 79·6 ± 9·0 |
| APOE-ε4 (%) | 65·6 | 34·9 | 66·2 | 36·0 | 63·5 | 36·1 |
Plasma RNA sample preparation
A total of 10 ml of peripheral whole blood were collected from patients in EDTA-Vacutainer tubes and centrifuged at 3000 rpm (1408 rcf) for 10 min. Plasma was removed and stored as 0·5 ml aliquots at -80°C. For total RNA extraction from plasma a 0·5 ml frozen aliquot of plasma was thawed on ice and centrifuged at 4°C for 10 min at 3000 g to remove any potential cellular contamination. A plasma volume of 400 ul was carefully removed and processed using QIAGEN's miRNeasy Serum/Plasma Kit according to manufacturer's instructions with RWT buffer prepared using 45 ml isopropanol. The protocol included a column DNase treatment. RNA was eluted in 14ul RNase free water. The spectroscopic absorbance at 415 nm was recorded for each plasma sample using Thermo Scientific™ NanoDrop 2000. RNA was visualized on an Agilent 2100 Bioanalyzer using RNA 6000 HS Pico Chip, though concentrations could not be quantified accurately at these picogram concentrations. Low RNA integrity number (RIN) and absence of ribosomal 18s and 26s bands indicated lack of cellular RNA contamination. For RNA to cDNA conversion, 4 ul of RNA were reverse transcribed using SuperScript IV VILO cDNA Synthesis Kit (Life Technologies) following the workflow of the low input protocol (nCounter ®XT Assay User Manual).
Plasma RNAseq pilot study
To determine which transcripts could be detected in human plasma, we conducted RNA sequencing on 7 plasma RNA samples from cognitively unimpaired (CU) study participants. Total RNA was extracted from plasma as described in the main methods section and was utilized to prepare libraries for RNAseq with the Ovation® SoLo kit for ultra-low input RNA which produces rRNA depleted libraries. Between 7·1 and 14·9 million 50bp paired-end raw read pairs were obtained. The Mayo Clinic MAP-RSeq bioinformatic pipeline v2.1 was applied to map raw reads to the human reference genome build hg38.16 Gene read counts were calculated using Subread package v1.4.17 Between 2·3 and 5·7 million mapped reads were obtained, among which 1·8-4·9 million reads were mapped to known genes. To assess the robustness of our method to detect transcripts in plasma, we compared the read counts from our plasma samples to those from a public dataset GSE106804,18 which contains read counts from 15 plasma extracellular vesicle RNA samples of glioblastoma multiforme patients and healthy donor controls. In our samples, there are 1048 genes with median read counts ≥50, whereas in GSE106804, there are 396 such genes; 262 genes are shared, and these 262 genes have a Pearson correlation coefficient of 0·8 between our samples and GSE106804 samples (Figure S1).
Measurement of plasma transcript levels
A nanoString® 50-gene custom panel was designed for this study which targets mRNA from genes that are known to be involved in the immune response or inflammation and/or that harbor genetic variants known to influence AD risk (see Table S1), and which were detected in a plasma RNAseq pilot study. Additional evidence of the involvement of these genes in the immune response related to AD was obtained from our prior published work, in which AIF1, AOAH, BLNK, CD14, CSF1R, HCK, IL1B, ITGAL, ITGB2, LY86, LY96, LYZ, STAB1 were found to be part of a gene co-expression module in post-mortem temporal cortex tissue that was identified as being enriched for an immune gene ontology term and that has higher expression in temporal cortex of AD cases compared to non-AD.19 The RNAseq pilot study was performed as described in the section above called “Plasma RNAseq pilot study”. The 50 genes whose transcripts are targeted on this custom panel are listed on Table S1. The nanoString® custom panel included a single probe per gene. The nCounter™ CodeSet Design Report's Probe Design and Isoform Coverage are now shown in Tables S2 and S3. Six genes were included in the ROC analysis models: APP, CD14, CLU, ABCA7, AKAP9 and APOE, and the location of their probe in the context of their transcript sequence on the UCSC Genome Browser is shown in Figure S2. Given low levels of transcripts in plasma, a multiplex target enrichment (MTE) was performed prior to hybridization. MTE primers were pooled at a final concentration of 500 nM per oligo in TE Buffer (pH 7·5) (Integrated DNA Technologies). Using pooled primers designed for the 50 target genes, MTE reactions were performed on the cDNA using Taqman® PreAmp Master Mix (Applied Biosystems), for 10 cycles of amplification. Subsequently, 8 ul of the resulting MTE reaction was denatured for 2 min at 95°C and mixed with 8 ul of the nanoString® hybridization master mix consisting of hybridization buffer and Reporter CodeSet. Finally, 2 ul of the nanoString® Capture ProbeSet was added, mixed, and incubated for 18 h at 65°C. Following overnight hybridization, excess probes were removed using magnetic bead purification on the nanoString® nCounter Prep Station and levels of barcoded target molecules quantified using the nanoString® nCounter Digital Analyzer following the manufacturer's protocol. Of the 447 plasma samples tested, 430 passed the default sample quality control (QC) implemented in the NanoString® nSolver™ Analysis Software, which includes imaging, binding density, positive control, and limit of detection QC. Of the 50 genes tested in the custom panel, 41 passed QC (Table S1): 7 were excluded (CCL2, CR1, CRH, ICAM1, IL4, LY96 and MS4A6A) due to having mean transcript counts ≤ 50 among the samples that passed QC (50 counts is the lower end of the dynamic range for NanoString® technology regardless of input levels), and two genes were excluded due to a spillover effect of fluorescence signal from the most abundant transcript in plasma, ACTB and HDAC6. Given that there are no validated endogenous control genes in plasma that can be utilized to normalize gene counts across samples, inter-sample variability was adjusted in the statistical models by incorporating a unique identifier for each sample as a random effects variable into the statistical analyses. In addition, since cross-RLF calibration could not be implemented, RLF "batch" was incorporated as a covariate in the linear models (see statistical analyses section).
Validation of transcript level measurements
Genes whose transcript levels were significantly associated with disease status or with genetic variants were validated via relative quantification (qPCR) Taqman assays. The qPCR reactions were performed using 4 ul of plasma RNA from samples that based on nanoString® measurement had the highest and lowest levels of the transcripts of interest (15 highest and 15 lowest). The plasma RNA from these samples were reverse transcribed using SuperScript IV VILO cDNA and pre-amplified for 10 cycles using target gene probes Hs00156548_m1 (CLU), Hs00169098_m1 (APP), Hs02621496_s1 (CD14), Hs01105081_m1 (ABCA7), Hs00323978_m1 (AKAP9), Hs04931857_m1 (IL33), and HS0103996_m1 (STAT1). The resulting template was used for qPCR employing ABI Taqman® chemistry (Applied Biosystems) with the Taqman minor groove binder probes. A negative control lacking reverse transcription was included to confirm the specificity of the Taqman probes. IL33 and STAT1 were used as endogenous controls for normalization, as it was determined based on the nanoString® counts that these genes were most stably expressed across all plasma samples. Each sample was run in triplicate on a QuantStudio™ 7 Flex Real-Time PCR instrument. The analysis was performed using the QuantStudio™ Real-Time PCR Software v1.6.1 (Applied Biosystems).
Whole exome sequence (WES)
WES data was generated for 250 AD and 286 CU Florida Consortium for African American Alzheimer's Disease Studies (FCA3DS) study participants. Sample preparation and sequencing for a subset of these samples has been previously described.15 FastQ files were processed through Mayo Clinic's GenomeGPS pipeline. Reads were aligned to the GRCh38 human reference genome assembly using BWA-MEM20 and variant calling and joint genotyping was performed using Genome Analysis Tool Kit (GATK) v3.6 while implementing Best Practices Workflow.21 Samples underwent QC, which implemented the following criteria for inclusion: coverage of at least 90% at 10x and 40% at 40x, contamination VerifyBamID22 FREEMIX score less than 0.02, genotyping quality median GQ of 99, minimum call rate of 95%, transition to transversion (Ti/Tv) ratio of approximately 2·8 and a sex check in PLINK23 with inbreeding coefficient of the X-chromosome for males>0·7 and females <0·3. Subsequently, samples were evaluated for relatedness, and retained only one sample from each set of 1st, 2nd and 3rd degree relatives. Principal component analysis was performed on samples after resolving relatedness to evaluate population substructure and potential heterogeneity due to sequencing batch and flowcell. WES was performed in two batches and we performed comprehensive sample and variant QC including evaluating heterogeneity due to sequencing batches (Figure S3). We did not identify any significant variability due to sequencing batches and therefore did not include “batch” as a variable in the model. Bi-allelic variants passing VQSR filter, having a genotyping rate equal to or greater than 98%, a minor allele frequency of at least 2% and a Hardy-Weinberg p-value greater than 5e-08 in controls were retained. Variants in high variability regions of the genome were excluded. A total of 474 samples (230 AD cases and 244 CU controls) and 878,447 variants passed QC. Variants were annotated using ANNOVAR.24 Regulome scores provide an indication of the functional potential of a variant based on known and predicted regulatory elements and was obtained through https://regulomedb.org/.25
Statistics
Sample size, randomisation and blinding
We utilized every plasma sample that was available from AD cases and CU controls in the FCA3DS cohort to maximize statistical power. Samples were randomized based on age, sex and diagnosis in each batch for the quantification of plasma transcripts and WES. The generation of nanoString® and WES data were blinded to all grouping and outcome variables. Inclusion criteria was the availability of a plasma sample as well as demographic and clinical information required to perform the analyses.
Differential gene expression (DGE) analysis
Plasma transcript measures quantified using the custom nanoString® panel were log2-transformed and tested for differential expression between AD cases and CU controls using a linear mixed model adjusted for age, sex, optical density (OD), batch, inter-sample variability and cartridge using the ‘lmer()’ function in the ‘lme4’ package in R (v3.6.2). Given the low concentration of cf-RNA in plasma, RNA concentration and RIN cannot be reliably measured. Therefore, to adjust for all differences in sample quality, including concentration, RIN, potential differences due to banking time, and potential inter-individual differences such as fasting status or medications, we implemented a linear mixed model after obtaining random effect estimates for each sample. Cartridge was also encoded as random effects variable, while all other covariates, including plasma OD and batch were encoded as fixed effects in the model. Additionally, this analysis was repeated while adjusting for APOE-ε4 allelic dosage in the model. DGE was analyzed using the following model, as well as a similar model adjusted for APOE-ε4 allelic dosage: .
eQTL analysis
Utilizing plasma transcript measures and whole exome sequence (WES) genotypes from 139 FCA3DS AD cases and 225 FCA3DS CU controls (see Supplementary Methods), a cis-eQTL analysis was performed to test the association of exome variants with plasma transcript counts. Each variant within 1Mb of a targeted gene's Ensembl gene coordinates (GRCh38) was tested for association with log2-transformed transcript counts (Y) while accounting for diagnosis, age, sex, APOE-ε2 and APOE-ε4 allelic dosage, the first three principal components (PC), batch, OD, cartridge and a variable accounting for inter-sample variability, using the following linear mixed model implemented with the lme4 package in R (v3.6.2): . PCs to adjust for population substructure were derived from WES genotypes available for all samples in the FCA3DS AD case-control series as previously described.26 No significant variability due to sequencing batches was observed, therefore “batch” was not included as a covariate in the model. Analyses were repeated excluding APOE-ε2 and APOE-ε4 allelic dosage from the model. Denominator degrees of freedom for test statistic was obtained using Kenward-Roger restricted maximum likelihood approximation in the lmerTest package in R. False discovery rate (FDR) (Benjamini-Hochberg) adjusted q-values were calculated in R for all tested cis-eQTLs. All variants that were present in the WES data from this AA cohort that had a MAF ≥ 2% and that were in cis with genes of interest were included in our eQTL analyses, even if they have been observed in other populations besides AA. The rationale for this is that we expect that an eQTL may have biomarker potential in multiple populations. Therefore, we did not exclude variants based on their presence in other populations besides AA.
Association test of WES variants with AD-risk
The association of WES variants with AD-risk in the FCA3DS AD case-control series (230 AD cases and 244 CU controls) was tested using multivariable logistic regression in PLINK v1.9.23 Dosage of the minor allele was tested for association with AD while accounting for age, sex, APOE-ε2 and APOE-ε4 allelic dosage and the first three PCs to account for population substructure.
Receiver operating characteristic (ROC) analysis
The predictive value of variables to discern AD cases vs. CU controls was evaluated using the AUC ROC curves with the ‘pROC’ package in R v3.6.2. The base model (M1) evaluated age, sex and APOE-ε4 allelic dosage. Plasma cf-mRNA measures of significant differentially expressed genes (DEGs: CLU, APP and CD14) were added to the second model (M2). Allelic dosage of the variant that showed the most significant eQTL association at the two loci that also showed association with AD-risk in this dataset, ABCA7 and AKAP9 loci (rs3752232 and rs171764315, respectively), were added to the third model (M3). ABCA7 and AKAP9 plasma cf-mRNA measures were added to the fourth model (M4). While the levels of APOE plasma transcript only reached a suggestive level of significance in the differential expression analysis between AD cases and CU controls and no significant eQTLs were detected at the APOE locus, APOE is a known genetic risk factor for AD, therefore we evaluated the contribution of APOE plasma cf-mRNA levels in the fifth model (M5). The plasma cf-mRNA measures utilized for these analyses were the residuals of the log2-transformed counts of CLU, APP, CD14, AKAP9, ABCA7 and APOE, after adjustment for technical variables (OD, batch, cartridge, and inter-sample variability). A secondary ROC analysis was performed to evaluate plasma total tau levels that were available for a subset of the FCA3DS AD cases and CU controls (N=331) as part of a previously published study.9 In this subset of the samples, models M1-M5 were evaluated, and residuals of plasma total tau levels derived after adjusting for age of plasma, plate and batch were added to M5 generating a sixth model (M6). The relationship between plasma total tau levels and plasma levels of each of the six cf-mRNA measures utilized for the ROC analysis in M6 was assessed using Pearson correlation. Figure S4 shows the distribution of these measures in these 331 participants as well as the Pearson correlation coefficients. ROC curves comparing the AUC of the base model (M1) to all other models was generated using the ‘plot.roc()’ function in ‘pROC’ package in R.
Ethics
Approval for the study was provided by the Mayo Clinic Institutional Review Board (ID: 14-005465) and informed consent was obtained from all study participants.
Role of funders
This work was supported by the National Institute on Aging [RF AG051504, U01 AG046139, R01 AG061796 to NET; P30 AG062677 to JAL and NGR]; Florida Health Ed and Ethel Moore Alzheimer's Disease grants [5AZ03 and 7AZ17 to NET; 7AZ07 to MMC; 8AZ08 to JAL]. The funders of this study had no role in study design, data collection, data analysis, data interpretation, writing of the manuscript or the decision to submit it for publication.
Results
In this study, we utilized plasma cf-RNA from 420 FCA3DS participants (Table 1) to determine if plasma mRNA levels of key genes (Table S1) detectable in plasma (Fig. S1) that are known to be involved in AD, inflammation or the immune response differed between AD and CU study participants (plasma DGE analysis). We also evaluated the association between genetic variants identified through WES in this cohort and plasma transcript levels (plasma eQTL analysis), as well as the association of these variants with AD-risk. Lastly, we also examined the potential utility of plasma DEGs and eQTLs identified in this study as AD biomarkers that may aid in the classification of AD cases and CU controls in AA.
Plasma differential gene expression (DGE)
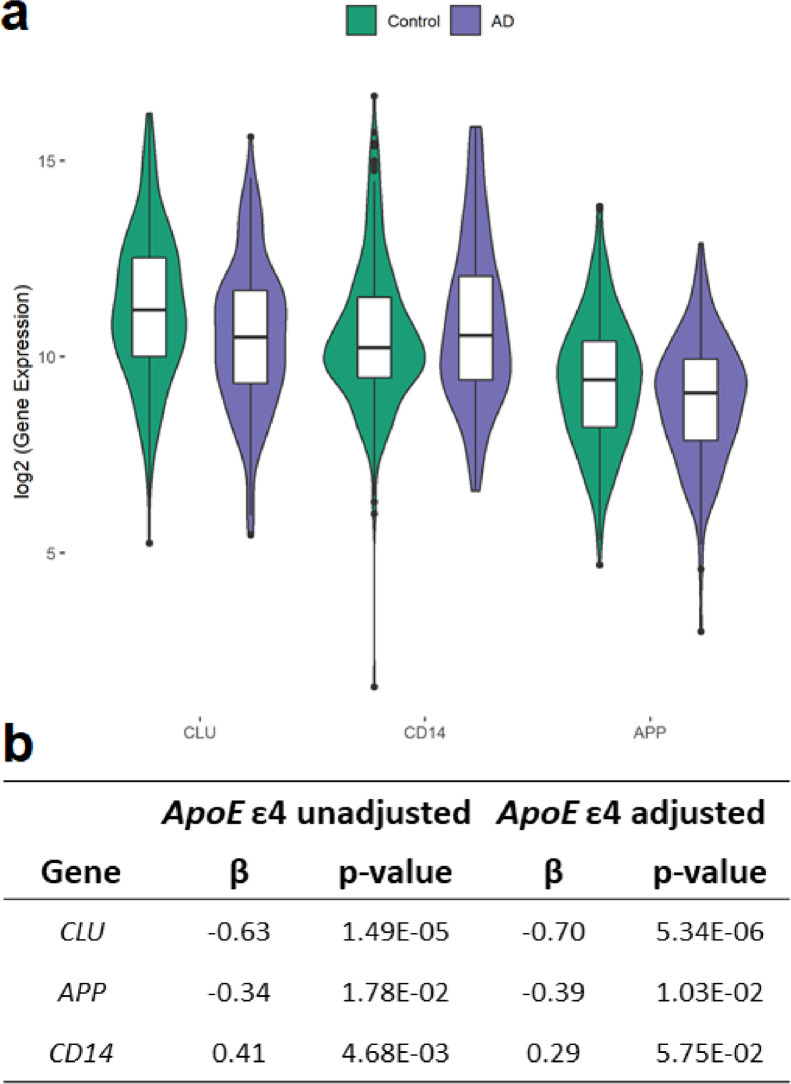
In a linear regression model that was adjusted for sex, age at plasma collection and technical covariates, we found a significant difference between AD cases and CU controls in plasma levels of 3 out of the 41 cf-mRNAs that passed quality control (QC): APP, CD14 and CLU (Figure 1, Table S4). The association observed with higher levels of APP and CLU transcripts in plasma from CU participants compared to AD cases remained significant after adjustment for APOE-ε4 allelic dosage. Furthermore, the association observed with plasma CLU levels survived Bonferroni correction in both linear regression models, adjusted for APOE-ε4 allelic dosage (AD vs. CU ß=-0·70, Bonferroni adjusted p=0·0002, t-test), and in the APOE-ε4 unadjusted model (AD vs. CU ß=-0·63, Bonferroni adjusted p=0·0006, t-test). These significant DGE results were validated by the observation that raw transcript counts, as measured with our nanoString® custom panel, correlated well with levels measured using relative quantification (qPCR) Taqman assays, with Spearman correlation r=0·72–0·86 (Figure S5).
Figure 1.
Log2-transformed plasma transcript counts for significant DEGs in the 420 FCA3DS AA AD vs. CU participants. (a) Log2-transformed counts are shown for the three genes that showed nominally significant DGE using a mixed linear regression model that includes relevant biological and technical covariates such as age, sex, batch, cartridge, optical density of the plasma sample, and includes sample ID as a random effect to adjust for inter-sample technical variation. Error bars represent 95% confidence intervals, the bottom and top of the box are the 25th and 75th percentiles, the line inside the box is the 50th percentile (median), and outliers are shown as dots. (b) Gene β-coefficient and p-value (T-test) from DGE analysis is shown. Only CLU and APP were nominally significant in the APOE-ε4 adjusted model. CLU was significant in both the APOE-ε4 adjusted and unadjusted models after Bonferroni correction for multiple testing.
Plasma eQTL analysis
In our plasma eQTL analysis, cf-mRNA levels from 22 of the 41 genes tested showed significant associations with 105 WES variants that are in cis with these genes. These eQTL associations achieved FDR q-values <0·05 (Table S5). All 105 eQTLs were also tested for association with AD-risk in the FCA3DS case-control series, which detected nominally significant associations with eQTLs at the ABCA7 and AKAP9 loci (Table S5). For each gene with a significant eQTL (Q<0.05), pairwise LD was calculated between the variant with the most significant p-value and all other variants tested at that locus using PLINK v1.9 utilizing WES genotypes from the 371 FCA3DS participants that were included in the eQTL analysis.
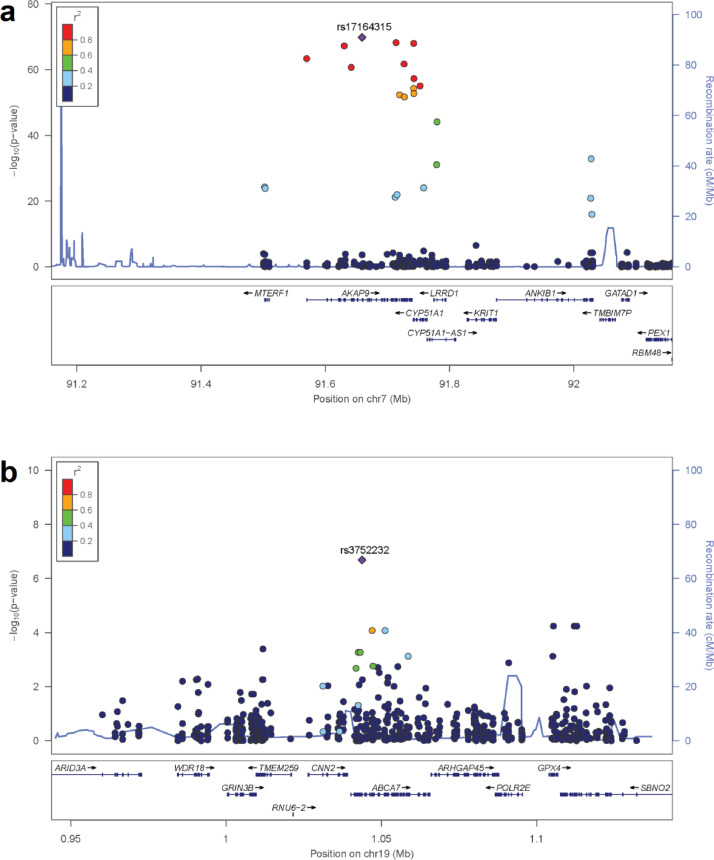
Two ABCA7 variants, rs3752232 and rs4147910, were significantly associated with lower plasma ABCA7 cf-mRNA levels (ß=-0·51, FDR q-value=2·74E-04; ß=-0·38, FDR q-value=4·01E-02, respectively), and with increased AD-risk in this FCA3DS AA case-control series (OR=1·65, unadjusted-p=1·13E-02; OR=1·52, unadjusted-p=2·99E-03, respectively) (Table 2)); however, these AD-risk associations do not remain significant after FDR adjustment (FDR q-value>0.05). These variants are in strong linkage disequilibrium (LD) with each other (r2=0·7) (Figure 2 and Table S5), and also reached nominal significance with increased AD-risk in a genome-wide association study (GWAS) conducted in a large NHW case-control series14 (Table 2). Annotation of significant eQTLs with their Regulome Score25 revealed that one of these two ABCA7 cf-mRNA eQTLs, rs3752232, had the strongest regulatory potential out of all 105 cf-mRNA eQTLs identified in this study, with a Regulome Score of 1f (Table S5). However, we note that regulatory potential indicated by Regulome scores may not be equivalent across all populations, and thus may not be applicable to AA.
Table 2.
Significant cis eQTLs that are also associated with AD-risk in the FCA3DS AD case-control series. WES variants within 1Mb of AKAP9 and ABCA7 that remained significant after FDR correction (cis eQTLs) and which had a nominal p-value for association with AD-risk in 420 FCA3DS AD cases and CU controls are shown. rsID: dbSNP ID of the eQTL; Chr: chromosome; Position: hg38; A1: tested allele; A2: reference allele; ß: coefficient of eQTL linear regression model; P: p-value (F-test); Q: FDR q-value. Also shown are the AD-risk association results, including AD-risk odds ratio (OR), p-value (T-test) and MAFs of variants in the 474 FCA3DS AD cases and controls, and ORs and p-values from the Kunkle et al. AD-risk GWAS in NHW.14 Variant annotation shows the gene proximal to the variant, its exonic function (syn: synonymous; nonsyn: nonsynonymous missense variant) and Regulome score25 obtained from ANNOVAR.24
| Plasma eQTL |
AD GWAS - FCA3DS |
AD GWAS - NHW |
Variant Annotation |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cf-mRNA | rs ID | Chr | Position | A1 | A2 | β | P | Q | MAF | OR | P | MAF | OR | P | Gene | Function | Regulome |
| AKAP9 | rs10228334 | 7 | 92084658 | C | T | 1·67 | 4·75E-69 | 1·18E-64 | 0·49 | 1·34 | 3·53E-02 | 0·49 | 1·03 | 5·83E-02 | AKAP9 | syn | 5 |
| rs57122389 | 7 | 92113034 | C | A | 1·69 | 8·45E-69 | 1·39E-64 | 0·48 | 1·42 | 1·36E-02 | 0·49 | 1·03 | 4·78E-02 | CYP51A1 | 3’UTR | 6 | |
| rs6964587 | 7 | 92001306 | G | T | 1·69 | 4·73E-68 | 5·85E-64 | 0·49 | 1·33 | 4·19E-02 | 0·49 | 1·03 | 5·58E-02 | AKAP9 | syn | 5 | |
| rs7793861 | 7 | 92113414 | G | C | -1·60 | 4·57E-58 | 2·83E-54 | 0·49 | 0·76 | 4·69E-02 | 0·48 | 0·97 | 4·70E-02 | CYP51A1 | 3’UTR | 6 | |
| rs11459 | 7 | 92112873 | G | A | -1·56 | 4·73E-55 | 2·34E-51 | 0·47 | 0·76 | 4·66E-02 | 0·46 | 0·97 | 4·18E-02 | CYP51A1 | 3’UTR | 6 | |
| rs6465348 | 7 | 92113288 | G | A | -1·55 | 1·55E-53 | 6·97E-50 | 0·47 | 0·74 | 2·92E-02 | 0·46 | 0·97 | 3·88E-02 | CYP51A1 | 3’UTR | 6 | |
| rs57433727 | 7 | 92400943 | C | T | 0·74 | 4·85E-05 | 2·90E-02 | 0·11 | 1·88 | 3·69E-03 | 0·12 | - | - | ANKIB1 | 3’UTR | 5 | |
| rs61740421 | 7 | 92397766 | T | C | 0·73 | 5·00E-05 | 2·91E-02 | 0·11 | 1·87 | 4·20E-03 | 0·12 | - | - | ANKIB1 | syn | 4 | |
| ABCA7 | rs3752232 | 19 | 1043749 | G | A | -0·51 | 2·10E-07 | 2·74E-04 | 0·25 | 1·65 | 2·99E-03 | 0·26 | 0·93 | 4·87E-02 | ABCA7 | nonsyn | 1f |
| rs4147910 | 19 | 1047079 | G | A | -0·38 | 8·24E-05 | 4·01E-02 | 0·25 | 1·52 | 1·13E-02 | 0·26 | 1·16 | 9·27E-09 | ABCA7 | intronic | 4 | |
Figure 2.
Regional association plots of cis eQTLs at the AKAP9 and ABCA7 loci. Locus zoom44 plots of all variants within 1Mb of the gene were tested for association with gene expression in 364 AD cases and CU controls are shown. The -log10(p-value) of the eQTL association is shown on the y-axis while the chromosomal position of each variant is show on the x-axis. The lead variant, relative to which LD was calculated for all other variants in cis, is shown as a purple diamond. LD values between the lead variant and all other variants is colour coded based on the r2 in a population of African ancestry in Southwest USA.44(a) Locus zoom plot for the AKAP9 locus. (b) Locus zoom plot for the ABCA7 locus.
At the AKAP9 locus, 31 variants showed significant association with plasma AKAP9 cf-mRNA, with FDR q-values<0·05 (Table S5). There were also nominally significant associations with AD-risk (p-values<0·05) in the FCA3DS AD case-control series with 8 of these AKAP9 cf-mRNA eQTLs (Table 2). We also observed nominal associations with AD-risk in the large NHW AD-risk GWAS for 11 of the AKAP9 cf-mRNA eQTLs (Table S5), 4 of which were also significant in FCA3DS (Table 2). These variants have a consistent direction of effect, such that variants that associate with higher risk of AD also show significant association with higher levels of AKAP9 cf-mRNA, and those with lower AD-risk association also significantly associate with lower levels of AKAP9 cf-mRNA. The LD between these eQTLs is indicative of at least two independent AD-risk association signals (Table S5 and Figure 2), one of which is detected only in FCA3DS with two variants, rs57433727 and rs6174042, whose frequencies are 12% in AA and 0·03% in NHW (Table S5).
ROC analysis
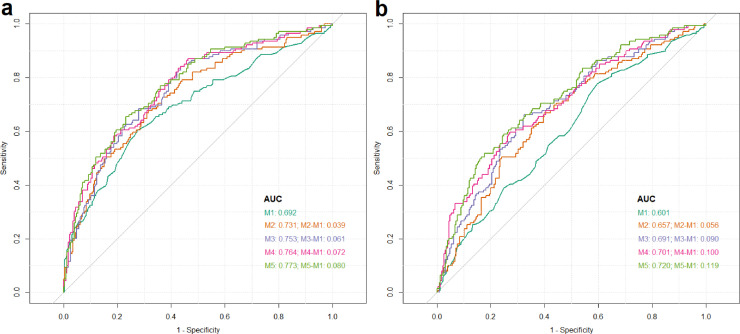
The utility of these plasma DEGs and eQTLs as potential biomarkers of AD was evaluated with the ROC analysis. Only the eQTL with the most significant AD-risk association at each locus was included in the ROC models. Figure 3. shows that the greatest AUC to differentiate AD cases vs. CU controls was achieved with a model that included age, sex APOE-ε4 allelic dosage, CLU, APP, CD14, ABCA7, AKAP9 and APOE cf-mRNA levels, and allelic dosage of the most significant ABCA7 and AKAP9 cf-mRNA eQTLs. This model showed 77% AUC to discriminate AD vs. CU (Figure 3a, model 5), an 8% improvement over the base model that only included age, sex and APOE-ε4 dosage (Figure 3a, model 1). Inclusion in this model of previously obtained plasma total tau levels9 only increased the AUC by an additional 0·06% (Figure S6). The effect of plasma total tau levels appears to be independent of the effect observed with the plasma levels of the six cf-mRNA that were included in the ROC AUC model (Figure S4).
Figure 3.
ROC curves for APOE-ε4 adjusted and unadjusted models. AUC for each model, along with the improvement of models M2 to M5 compared to the base model (M1) are shown. (a)APOE-ε4 adjusted ROC analysis wherein all models (M1 to M5) were adjusted for APOE-ε4 allelic dosage. (b)APOE-ε4 unadjusted models.
Discussion
Our study evaluates plasma transcript levels as potential novel biomarkers of AD in AA. We chose to measure gene expression specifically in plasma, as plasma samples are more readily available in the clinical setting. The feasibility of gene expression measurements in plasma had been established by others,27 but to our knowledge has not previously been studied as diagnostic biomarkers for AD in AA. Our study is of particular importance given its focus on AA, a population that remains vastly underrepresented in AD studies and whose risk of developing AD is twice as high as for NHW. In addition, the focus on genes functionally implicated in inflammation is especially relevant in this population whose high prevalence of diabetes and metabolic syndrome, conditions associated with systemic inflammation, may contribute to their increased risk of AD.28 Thus, levels of the targeted cf-mRNA transcripts could serve as biomarkers of the biological pathways underlying the disease and ultimately guide the selection of optimal therapies.
In this study we found that CLU transcript levels in plasma are significantly higher in CU compared to AD cases. This finding is in line with results from published studies that showed that higher baseline concentration of plasma clusterin was associated with slower rates of brain atrophy,29 and that the CLU rs11136000 AD-risk allele is associated with low clusterin plasma levels.30 Others have also shown a neuroprotective effect of clusterin,31 and that loss of CLU is associated with greater accumulation of pathological tau in a mouse model of tauopathy.32 However, other studies have observed contradicting results such as clusterin enhancing tau aggregate seeding in a cellular model.33 Although, the role of clusterin in the etiology of AD is not yet completely understood, our findings suggest that plasma CLU cf-mRNA levels may have the potential to serve as an AD biomarker in this population.
We also detected significant cf-mRNA eQTL associations with 105 variants in cis with genes targeted in our study. Importantly, significant ABCA7 and AKAP9 cf-mRNA eQTLs also show association with AD-risk in our FCA3DS AD case-control series. The direction of effect of the ABCA7 cf-mRNA eQTLs is concordant with published work in which ABCA7 variants that are associated with increased risk of AD have lower ABCA7 gene expression in brain tissue.34 This suggests that plasma transcript levels may reflect expression changes in disease relevant tissues. In addition, in a previous publication,15 we demonstrated that ABCA7 rs3752232 is associated with AD-risk in FCA3DS. In the present plasma eQTL study, this variant was the most significant plasma eQTL at the ABCA7 locus and had the strongest regulatory potential out of all 105 cf-mRNA eQTLs identified, with a Regulome Score of 1f (Table S5), suggesting that rs3752232 may be a functional variant underlying the association observed with AD risk in our FCA3DS case-control series. Interestingly, rs3752232 was previously shown to be correlated with worse cognitive scores using Rey Complex Figure Test copy score (β=-6·861, Pcorrected=0·013) in a Korean cohort.35 This variant is 3–10 times more frequent in AA (minor allele frequency, MAF=25%) than in other populations [Korean MAF=8%, European (non-Finnish) MAF=4%, Latino/Admixed American MAF=2%],36 further underscoring the need for population-specific studies.
AKAP9 rare variants were previously found to increase the risk of AD in AA.37,38 Although we did not detect an association in our study with those AKAP9 rare variants, likely due to their low frequency, we did observe significant plasma AKAP9 eQTL associations with 8 common variants (MAF>10% in FCA3DS participants), all of which showed consistent associations with higher risk of AD and higher AKAP9 cf-mRNA levels. Four of these AKAP9 eQTLs also have nominal associations with AD-risk in a NHW AD GWAS14 (Tables 2 and S5). Two other AKAP9 eQTLs, rs57433727 and rs6174042, that also associate with AD-risk in FCA3DS (OR=1·9, p=4E-3) had no reported associations in the NHW AD-risk GWAS, likely due to their very low frequency in NHW compared to AA (NHW MAF=0·03%, AA MAF=12%). To our knowledge this is the first study to test the AD-risk association of AKAP9 eQTLs, and to report an association with AKAP9 cf-mRNA eQTLs in any population. Further evaluation of these AKAP9 eQTLs is warranted as they could provide insight into the biological mechanism by which AKAP9 influences AD-risk.
ROC analysis of genes with significant plasma DGE and eQTLs revealed a predictive value that could contribute to improved AD biomarker panels, above and beyond the predictive value of age, sex and APOE-ε4 dosage. In our study, inclusion of the significant DGE and eQTLs in the predictive model yielded an 8·0% improvement over a model that only included age, sex and APOE-ε4 dosage. The addition of plasma total tau levels to this model only improved the AUC by 0·06% (Figure S5), underscoring the added value of the plasma cf-mRNA measures as potential biomarkers. Importantly, these transcript levels may also allow for discrimination of AD subtypes, such as those that may have a distinct inflammatory component. A few studies have evaluated in an AA cohort the use of plasma proteins or metabolites as a fluid biomarker for AD, but none of these have evaluated plasma cf-mRNAs.9,39, 40, 41, 42, 43 Future studies should systematically evaluate the biomarker potential of all transcripts detectable in plasma in diverse populations and incorporate these plasma cf-RNAs into a biomarker panel along with plasma levels of proteins and metabolites that have been shown to effectively discriminate AD from CU and other types of dementias, such as Aß42/Aß40 ratio and p-tau, in order to achieve greater discriminatory potential and add theragnostic value.
Despite the innovative aspects of our study including assessment of plasma cf-mRNA in AA AD and CU participants and evaluation of significant DGE and eQTL for their predictive potential, our study has several weaknesses. These include the relatively small sample size, clinically diagnosed AD without autopsy validation and lack of a replication cohort. Yet, a priori estimates of power indicated greater than 50% power to detect differential transcript levels at α=0.05 for ß=0.20 and a sample of 200 vs. 200, and 100% power for ß=0.60 for the same sample size. In this study consisting of 420 AD cases and CU controls, we were able to detect significant association with differential plasma cf-RNA levels of CD14, APP and CLU, with ß=0.29, -0.39, -0.70, respectively. Also, in this study we specifically focused on variants in our FCA3DS WES dataset that were significantly associated with plasma transcript levels. Therefore, other variants previously found to associate with AD-risk in AA that were not detected in our WES cohort, or which do not show association with plasma transcript levels were not evaluated. Additionally, the more recently established p-tau analytes were not available at the time of our study. This and sample limitations in our FCA3DS cohort precluded inclusion of these p-tau measures in our study. These weaknesses highlight the importance of increasing recruitment of underrepresented groups for AD research to achieve sample sizes as those for NHW. In summary, our study establishes the novel plasma cf-mRNA measures as potential future biomarkers that can inform on perturbed pathways beyond Aß and tau and improve diagnosis of AD in AA participants.
Contributors
Conceptualization: MMC, NET
Funding acquisition: NET, MMC, JL, NGR
Investigation: MMC, JSR, JJ, CCGH, XW, NET, have verified the underlying data.
Methodology: SJL, JJ, CCGH, JSR JEC, KGM, TN
Project administration: NET, MMC
Resources: NGR, JL, NET, MTG
Visualization: JSR, XW, NT
Writing – original draft: MMC, JSR, SJL
Writing – review & editing: all authors read and approved the final version of the manuscript.
Data sharing statement
Data may be made available after a reasonable and well-justified request to the principal investigator of FCA3DS, Dr. Nilufer Ertekin-Taner. Data cannot, however, be made freely available to the public, due to privacy regulations and informed consent. Codes and materials used in this study, may be made available for purposes of reproducing, or extending the analysis pending materials transfer agreements.
Declaration of interests
The authors declare that they have no competing interests.
Acknowledgements
We thank the participants and their families for their participation in research. Without them this work would not have been possible. We are also thankful to the Mayo Clinic Memory Disorders Center Coordinators including Michelle Fudge, Rita Fletcher, Francine Parfitt, Kelly Smith, and Sylvia Grant. We also thank the Mayo Clinic Center for Health Equity and Community Engagement Research for their support. This work was supported by the National Institute on Aging [RF AG051504, U01 AG046139, R01 AG061796 to NET; P30 AG062677 to JAL and NGR]; Florida Health Ed and Ethel Moore Alzheimer's Disease grants [5AZ03 and 7AZ17 to NET; 7AZ07 to MMC; 8AZ08 to JAL].
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2022.103929.
Appendix. Supplementary materials
References
- 1.2021 Alzheimer's disease facts and figures. Alzheimers Dement. 2021;17(3):327–406. doi: 10.1002/alz.12328. Epub 2021 Mar 23. PMID: 33756057. [DOI] [PubMed] [Google Scholar]
- 2.Petersen R.C. Clinical subtypes of Alzheimer's disease. Dement Geriatr Cogn Disord. 1998;9(3):16–24. doi: 10.1159/000051199. Suppl. [DOI] [PubMed] [Google Scholar]
- 3.Jack C.R., Bennett D.A., Blennow K., et al. NIA-AA research framework: toward a biological definition of Alzheimer's disease. Alzheimers Dement. 2018;14(4):535–562. doi: 10.1016/j.jalz.2018.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ossenkoppele R., Smith R., Mattsson-Carlgren N., et al. Accuracy of Tau positron emission tomography as a prognostic marker in preclinical and prodromal Alzheimer disease: a head-to-head comparison against amyloid positron emission tomography and magnetic resonance imaging. JAMA Neurol. 2021;78(8):961–971. doi: 10.1001/jamaneurol.2021.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blennow K., Mattsson N., Schöll M., Hansson O., Zetterberg H. Amyloid biomarkers in Alzheimer's disease. Trends Pharmacol Sci. 2015;36(5):297–309. doi: 10.1016/j.tips.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Janelidze S., Stomrud E., Smith R., et al. Cerebrospinal fluid p-tau217 performs better than p-tau181 as a biomarker of Alzheimer's disease. Nat Commun. 2020;11(1):1683. doi: 10.1038/s41467-020-15436-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palmqvist S., Janelidze S., Quiroz Y.T., et al. Discriminative accuracy of plasma phospho-tau217 for Alzheimer disease vs other neurodegenerative disorders. JAMA. 2020;324(8):772–781. doi: 10.1001/jama.2020.12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janelidze S., Mattsson N., Palmqvist S., et al. Plasma P-tau181 in Alzheimer's disease: relationship to other biomarkers, differential diagnosis, neuropathology and longitudinal progression to Alzheimer's dementia. Nat Med. 2020;26(3):379–386. doi: 10.1038/s41591-020-0755-1. [DOI] [PubMed] [Google Scholar]
- 9.Deniz K., Ho C.C.G., Malphrus K.G., et al. Plasma biomarkers of Alzheimer's disease in African Americans. J Alzheimers Dis. 2021;79(1):323–334. doi: 10.3233/JAD-200828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Y., Jaber V., Alexandrov P.N., et al. microRNA-based biomarkers in Alzheimer's disease (AD) Front Neurosci. 2020;14 doi: 10.3389/fnins.2020.585432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steenland K., Goldstein F.C., Levey A., Wharton W. A meta-analysis of Alzheimer's disease incidence and prevalence comparing African-Americans and Caucasians. J Alzheimers Dis. 2016;50(1):71–76. doi: 10.3233/JAD-150778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams M.M., Scharff D.P., Mathews K.J., et al. Barriers and facilitators of African American participation in Alzheimer disease biomarker research. Alzheimer Dis Assoc Disord. 2010;24(Suppl):S24–S29. doi: 10.1097/WAD.0b013e3181f14a14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kinney J.W., Bemiller S.M., Murtishaw A.S., Leisgang A.M., Salazar A.M., Lamb B.T. Inflammation as a central mechanism in Alzheimer's disease. Alzheimers Dement. 2018;4:575–590. doi: 10.1016/j.trci.2018.06.014. (N Y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kunkle B.W., Grenier-Boley B., Sims R., et al. Genetic meta-analysis of diagnosed Alzheimer's disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat Genet. 2019;51(3):414–430. doi: 10.1038/s41588-019-0358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.N'Songo A., Carrasquillo M.M., Wang X., et al. African American exome sequencing identifies potential risk variants at Alzheimer disease loci. Neurol Genet. 2017;3(2):e141. doi: 10.1212/NXG.0000000000000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalari K.R., Nair A.A., Bhavsar J.D., et al. MAP-RSeq: mayo analysis pipeline for RNA sequencing. BMC Bioinform. 2014;15(1):224. doi: 10.1186/1471-2105-15-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liao Y., Smyth G.K., Shi W. The subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41(10):e108. doi: 10.1093/nar/gkt214. -e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reátegui E., van der Vos K.E., Lai C.P., et al. Engineered nanointerfaces for microfluidic isolation and molecular profiling of tumor-specific extracellular vesicles. Nat Commun. 2018;9(1):175. doi: 10.1038/s41467-017-02261-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allen M., Wang X., Burgess J.D., et al. Conserved brain myelination networks are altered in Alzheimer's and other neurodegenerative diseases. Alzheimers Dement. 2018;14(3):352–366. doi: 10.1016/j.jalz.2017.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H. (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v1
- 21.Van der Auwera G.A., Carneiro M.O., Hartl C., et al. From FastQ data to high confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinform. 2013;43:11.10.1. doi: 10.1002/0471250953.bi1110s43. –11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jun G., Flickinger M., Hetrick K.N., et al. Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. Am J Hum Genet. 2012;91(5):839–848. doi: 10.1016/j.ajhg.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyle A.P., Hong E.L., Hariharan M., et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22(9):1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 27.Koh W., Pan W., Gawad C., et al. Noninvasive in vivo monitoring of tissue-specific global gene expression in humans. Proc Natl Acad Sci USA. 2014;111(20):7361–7366. doi: 10.1073/pnas.1405528111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yaffe K., Kanaya A., Lindquist K., et al. The metabolic syndrome, inflammation, and risk of cognitive decline. JAMA. 2004;292(18):2237–2242. doi: 10.1001/jama.292.18.2237. [DOI] [PubMed] [Google Scholar]
- 29.Thambisetty M., An Y., Kinsey A., et al. Plasma clusterin concentration is associated with longitudinal brain atrophy in mild cognitive impairment. Neuroimage. 2012;59(1):212–217. doi: 10.1016/j.neuroimage.2011.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schürmann B., Wiese B., Bickel H., et al. Association of the Alzheimer's disease clusterin risk allele with plasma clusterin concentration. J Alzheimers Dis. 2011;25(3):421–424. doi: 10.3233/JAD-2011-110251. [DOI] [PubMed] [Google Scholar]
- 31.Giannakopoulos P., Kövari E., French L.E., Viard I., Hof P.R., Bouras C. Possible neuroprotective role of clusterin in Alzheimer's disease: a quantitative immunocytochemical study. Acta Neuropathol. 1998;95(4):387–394. doi: 10.1007/s004010050815. [DOI] [PubMed] [Google Scholar]
- 32.Wojtas A.M., Carlomagno Y., Sens J.P., et al. Clusterin ameliorates tau pathology in vivo by inhibiting fibril formation. Acta Neuropathol Commun. 2020;8(1):210. doi: 10.1186/s40478-020-01079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuste-Checa P., Trinkaus V.A., Riera-Tur I., et al. The extracellular chaperone clusterin enhances Tau aggregate seeding in a cellular model. Nat Commun. 2021;12(1):4863. doi: 10.1038/s41467-021-25060-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allen M., Lincoln S.J., Corda M., et al. ABCA7 loss-of-function variants, expression, and neurologic disease risk. Neurol Genet. 2017;3(1):e126. doi: 10.1212/NXG.0000000000000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chung S.J., Kim M.J., Kim Y.J., et al. CR1, ABCA7, and APOE genes affect the features of cognitive impairment in Alzheimer's disease. J Neurol Sci. 2014;339(1-2):91–96. doi: 10.1016/j.jns.2014.01.029. [DOI] [PubMed] [Google Scholar]
- 36.Karczewski K.J., Francioli L.C., Tiao G., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Logue M.W., Schu M., Vardarajan B.N., et al. Two rare AKAP9 variants are associated with Alzheimer's disease in African Americans. Alzheimers Dement. 2014;10(6):609–618. doi: 10.1016/j.jalz.2014.06.010. .e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Logue M.W., Lancour D., Farrell J., et al. Targeted sequencing of Alzheimer disease genes in African Americans implicates novel risk variants. Front Neurosci. 2018;12:592. doi: 10.3389/fnins.2018.00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khan M.J., Desaire H., Lopez O.L., Kamboh M.I., Robinson R.A.S. Why inclusion matters for Alzheimer's disease biomarker discovery in plasma. J Alzheimers Dis. 2021;79(3):1327–1344. doi: 10.3233/JAD-201318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patel R.A., Wharton W., Bay A.A., Ni L., Barter J.D., Hackney M.E. Association between anti-inflammatory interleukin-10 and executive function in African American women at risk for Alzheimer's disease. J Clin Exp Neuropsychol. 2020;42(7):647–659. doi: 10.1080/13803395.2020.1798879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vardarajan B., Kalia V., Manly J., et al. Differences in plasma metabolites related to Alzheimer's disease, APOE epsilon4 status, and ethnicity. Alzheimers Dement. 2020;6(1):e12025. doi: 10.1002/trc2.12025. (N Y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grewal R., Haghighi M., Huang S., et al. Identifying biomarkers of dementia prevalent among amnestic mild cognitively impaired ethnic female patients. Alzheimers Res Ther. 2016;8(1):43. doi: 10.1186/s13195-016-0211-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tranah G.J., Yokoyama J.S., Katzman S.M., et al. Mitochondrial DNA sequence associations with dementia and amyloid-beta in elderly African Americans. Neurobiol Aging. 2014;35(2):442. doi: 10.1016/j.neurobiolaging.2013.05.023. e1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pruim R.J., Welch R.P., Sanna S., et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.