Abstract
Growing evidence indicates an important link between gut microbiota, obesity, and metabolic syndrome. Alterations in exocrine pancreatic function are also widely present in patients with diabetes and obesity. To examine this interaction, C57BL/6J mice were fed a chow diet, a high-fat diet (HFD), or an HFD plus oral vancomycin or metronidazole to modify the gut microbiome. HFD alone leads to a 40% increase in pancreas weight, decreased glucagon-like peptide 1 and peptide YY levels, and increased glucose-dependent insulinotropic peptide in the plasma. Quantitative proteomics identified 138 host proteins in fecal samples of these mice, of which 32 were significantly changed by the HFD. The most significant of these were the pancreatic enzymes. These changes in amylase and elastase were reversed by antibiotic treatment. These alterations could be reproduced by transferring gut microbiota from donor C57BL/6J mice to germ-free mice. By contrast, antibiotics had no effect on pancreatic size or exocrine function in C57BL/6J mice fed the chow diet. Further, 1 week vancomycin administration significantly increased amylase and elastase levels in obese men with prediabetes. Thus, the alterations in gut microbiota in obesity can alter pancreatic growth, exocrine function, and gut endocrine function and may contribute to the alterations observed in patients with obesity and diabetes.
Introduction
Commensal bacteria of the gastrointestinal tract have an important role in host physiological processes, including aiding in digestion and metabolism of nutrients and drugs, and development of the intestinal tract and immune systems (1–4). Direct evidence that changes in gut microbiota play a role in disease pathogenesis comes from studies showing that oral treatment with nonabsorbable antibiotics (5–7) can reverse some disease phenotypes as well as by studies in which transfer of the microbiome from a diseased mouse or human to germ-free (GF) mouse results in the transfer of some aspects of the phenotype of the donor (8,9).
Using three inbred mice strains, we previously showed that in addition to diet, genetic background plays an important role in shaping the gut microbiome (10). This interaction between diet and genetic background on host metabolism can be further modulated by treatment with oral antibiotics, such as vancomycin, which targets Gram-positive bacteria, or metronidazole, which targets anaerobic bacteria. These effects of diet and antibiotics are mediated in part through changes in bile acid metabolism, inflammatory signaling, and alterations in insulin signaling in liver and muscle (5). Importantly, transfer of gut microbiota from mice on a high-fat diet (HFD) or HFD plus antibiotics to GF recipient mice can transfer many of these features of the insulin-resistance syndrome, indicating the role of gut microbiota in these metabolic pathways (5,11). Some of these changes also appear to be mediated by changes in plasma metabolites, a number of which strongly correlate positively or negatively with host insulin resistance (6). Similar effects on insulin resistance have also been shown with vancomycin and bacitracin treatment (7).
In the current study, we explored the effects of the gut microbiota on function of the exocrine pancreas and the gut endocrine system. Here, we show that gut microbiota affect pancreas size, function, and the secretion of gut hormones in mice and exocrine function in humans, expanding the importance of the gut microbiome in host physiology.
Research Design and Methods
Human Fecal Samples
The human fecal samples (n = 12 per group) used in this study for Western blot analysis were from individuals recruited for a previous study (12). In this randomized, placebo-controlled, double-blind study, researchers investigated the effects of 7-day administration of amoxicillin, vancomycin, or placebo on host metabolism using samples obtained from 57 obese men with prediabetes (n = 19). The median age of these male subjects was 60.9 ± 1.7, and the median BMI was 31.0 ± 0.5 kg/m2. The subjects were then randomized and received oral vancomycin, 1.5 g/day, or placebo for 7 consecutive days, followed by a 2-day washout period to ensure complete systemic and gastrointestinal clearance of antibiotics. All subjects gave written informed consent for participation in this study, which was reviewed and approved by the local Medical Ethical Committee of Maastricht University Medical Center. All procedures were according to the Declaration of Helsinki (revised version, October 2008). The fecal samples were collected before and after the procedure and stored at −80°C until used for extraction.
Animals
Male C57BL/6J mice were purchased from The Jackson Laboratory. After an acclimation period, 6-week-old male mice (n = 4–6) were randomized into different experimental groups to give the same average initial starting weight for each of the treatment groups. Mice were then maintained for 6 weeks on a normal chow diet containing 22% of calories from fat, 23% from protein, and 55% from carbohydrates (Mouse diet 9F 5020; PharmaServ) or an HFD (Open Source Diet, D12492; Research Diets) containing 60% of calories from fat, 20% from protein, and 20% from carbohydrates. During the last 2 weeks, some of the HFD- and chow-fed mice were treated with vancomycin or metronidazole (1 g/L in the drinking water). In addition, for some studies there was a long-term cohort in which mice on the HFD from age 7 weeks to age 18 weeks, with different cohorts receiving placebo, vancomycin (1 g/L), or metronidazole (1 g/L) in the drinking water from age 6 to 18 weeks. For GF experiments, cecal contents from donor mice from the shorter-term protocol described above were collected immediately after euthanasia, suspended in PBS, and filtered through a 40-μm cell strainer. Bacterial transfer was performed by gastric gavage with 200 μL of diluted cecal contents into 6-week-old male GF C57BL/6J mice (obtained from the gnotobiotic core facility of Brigham and Women’s Hospital). For the chow diet donors, the GF mice were on the chow diet, while for the HFD donors, the GF mice were on the HFD (double-irradiated D1249ii, Research Diets) from 4 weeks of age. The colonized mice were continued on chow or the HFD for 2 additional weeks. All mice were housed at 22°C on a 12 h light/dark cycle. All animal studies were approved by the Institutional Animal Care and Use Committee of Joslin Diabetes Center (no. 97-05) and Harvard Medical School (no. 05131) and Boston College (no. 2019-004-01) and were in accordance with National Institutes of Health guidelines.
Fecal Sample Collection, Protein Extraction, and Analysis
Two pellets of feces were collected daily (3 p.m.–5 p.m.) during the week before the sacrifice of the mice, chilled immediately on ice, and stored at −80 C until used for extraction. Fecal samples were lysed in ice-cold 1× PBS supplemented with 1× protease and phosphatase inhibitors (Biotool Chemicals). Homogenates were then filtered with a 0.22-μm filter, and fecal proteins were concentrated with a 3-kDa filter. To extract proteins, frozen pancreas tissues were lysed in ice-cold lysis buffer (radioimmunoprecipitation assay buffer complemented with 0.1% SDS and 1× protease and phosphatase inhibitor cocktail [Biotool Chemicals]). Protein concentrations were determined using the Pierce bicinchoninic acid protein assay reagent (Thermo Fisher Scientific). Lysates (15–20 μg) were separated on NuPage 4–12% polyacrylamide gels (Thermo Fisher Scientific) and transferred to nitrocellulose membranes (Thermo Fisher Scientific). Antibodies used were as follows: β-actin (1:10 000l 1616-horseradish peroxidase [HRP], Santa Cruz Biotechnology), amylase (1:1000; ab21156, Abcam), elastase (elastase-3 (V-14), 1:1000; 167740, Santa Cruz Biotechnology), pancreatic lipase (1:1000; 374612, Santa Cruz Biotechnology), PLA2G1B (D1T4C) rabbit monoclonal antibody (no. 4635; CST), and vinculin (1:1000; MAB3574, EMD Millipore). Appropriate secondary antibodies conjugated to HRP (1:7000) were from Santa Cruz Biotechnology. Membranes were visualized with Immobilon Western HRP Substrate (Millipore, Billerica, MA). Quantification was performed using ImageJ software (National Institutes of Health, Bethesda, MD; https://rsb.info.nih.gov/ij/).
Metabolic Studies
For the short-term studies, after 2 weeks of antibiotic treatment, mice that had been fasted for 6 h were assessed by both glucose tolerance test (GTT) and insulin tolerance test (ITT). For the GTT, mice were injected intraperitoneally with 2 g/kg glucose, and blood glucose was measured through the lateral tail vein at 0, 15, 30, 60, 90, and 120 min. For the ITT, mice were injected intraperitoneally with 0.5 IU/kg insulin (Humulin), and blood glucose was measured as for the GTT. Fasting plasma insulin levels were measured after the 6-h food restriction period using the murine ultrasensitive insulin ELISA kit (no. ALPCO).
Proteomics Analysis
Each sample was submitted for a single liquid chromatography tandem mass spectrometry (MS/MS) experiment, performed on a LTQ Orbitrap Elite (Thermo Fisher Scientific) equipped with Waters (Milford, MA) nanoACQUITY high-performance liquid chromatography pump for the electrostatic repulsion-hydrophilic interaction chromatography (ERLIC) analysis. Peptides were initially separated onto a 100-µm inner diameter microcapillary trapping column packed first with 5 cm of C18 5 µm, 100 Å Reprosil resin (Dr. Maisch GmbH, Tübingen, Germany), followed by an analytical 20-cm column of 1.8 µm, 200 Å Reprosil resin (Dr. Maisch GmbH). Separation was achieved using a gradient of 5–27% acetonitrile in 0.1% formic acid over 90 min at 200 nL/min.
The LTQ Orbitrap Elite was operated in data-dependent mode. The MS survey scan was performed in the Orbitrap in the range of 395 to 1,800 m/z at a resolution of 6 × 104, followed by the selection of the 20 most intense ions (TOP20) for collision-induced dissociation-tandem MS (MS2) fragmentation in the ion trap; the same TOP20 ions were subjected to higher energy collisional dissociation MS2 event in the Orbitrap part of the instrument. Raw data were submitted for analysis in Proteome Discoverer 2.1.0.81 (Thermo Fisher Scientific) software. Sequest HT searches were performed using a 20 ppm precursor ion tolerance and requiring each peptides N-/C termini to adhere with trypsin protease specificity, while allowing up to two missed cleavages. Tenplex tandem mass tags (TMT) tags on peptide N termini and lysine residues (+229.162932 Da) were set as static modifications, while methionine oxidation (+15.99492 Da) was set as the variable modification. A MS2 spectra assignment false discovery rate (FDR) of 0.25% on protein level was achieved by applying the target-decoy database search. Filtering was performed using Percolator (64-bit version) (13). Reporter ion intensities were exported in a result file of Proteome Discoverer 2.1 search engine as Excel tables. Total signal intensity across all peptides quantified was summed for each TMT channel, and all intensity values were adjusted to account for potentially uneven TMT labeling and/or sample handling variance for each labeled channel.
Bioinformatics Analysis
For all proteins appearing in the final analysis, only proteins that were identified in all 10 samples were included. All values were normalized to make the samples have the same total intensity. The statistical significance of peptides was assessed with empirical Bayesian linear modeling using the limma package. P values were corrected using the Benjamini-Hochberg FDR, and FDR <0.25 and P < 0.05 were considered statistically significant. Although we had small samples sizes for the antibiotic groups in the proteomics analysis, our moderated differential expression testing with the limma R package had statistical power, because it estimated pooled variances per protein over all groups and applied empirical Bayesian modeling of the variance, increasing its degrees of freedom (14). Gene sets based on canonical pathways and gene ontology from the molecular signatures database were tested using the Fry function of the limma R package. Heat maps were plotted with the gplots R package. Principal component analysis and volcano plots were plotted with the ggplot2 R package. The codes for the bioinformatics analysis are publicly available and can be found in this link: https://github.com/jdreyf/antibiotic-stool-proteomics/blob/master/R/analyze_emrah_stoolproteomics.r.
Immunostaining and BrdU Labeling for Pancreas Proliferation
BrdU was injected intraperitoneally three times per mouse (100 mg/kg of body wt; MP Biomedicals, LLC) daily, the last injection being 6 h before time of sacrifice. Formalin-fixed pancreata were processed through dehydration, clearing, and paraffin embedding at Harvard Rodent Histology Core, as described previously (15). Pancreas sections were cut to 5-μm thickness on positively charged tissue slides. Sections were cleared and rehydrated, blocked (5% normal donkey serum in PBS), and immunostained with anti-insulin (guinea pig, 1:400; Abcam, ab7842), anti-BrdU (mouse, 1:50; Dako, cat no. m0744,), anti–phospho-histone H3 (rabbit, 1:250; Millipore, cat no. 06-570,), anti-Ki67 (mouse, 1:50; BD, cat no. 556003), and anti-amylase (rabbit, 1:100; CST, cat no. 3796) diluted in antibody diluent (Dako, S0809) overnight at 4°C. Sections were then washed and subsequently stained with secondary antibodies (donkey anti-mouse Alexa 488, donkey anti-guinea pig Alexa 594, and donkey anti-rabbit Alexa 488, 1:400; Jackson Immunoresearch) for 1 h at room temperature. Nuclei were stained with DAPI. Sections were mounted with Fluorescence Mounting Medium (Dako, cat no. s3023) and imaged on a Zeiss Axio Imager A2 fluorescence microscope. For counting Ki67-positive cells, we used ImageJ software to manually count the number of Ki67-positive cells among at least 6,000 acinar cells per mouse. The same protocol was used for counting BrdU-positive cells.
β-Cell Mass
Mouse pancreas tissue was fixed by formaldehyde and embedded in paraffin. Sections of pancreas were cut to 5 μm thickness. Immunofluorescent staining for insulin was done by using guinea pig anti-insulin antibody (Abcam), followed by the secondary antibody conjugated with Alexa 594. Images were captured using Zeiss Axio Imager A2 upright fluorescence microscope and then analyzed using ImageJ software. The β-cell mass was calculated using the formula (β-cell area/pancreas area) × the wet weight of the pancreas.
Acinar Cell Counting
Sections were stained with rabbit anti-amylase antibody (Cell Signaling), followed by the secondary antibody conjugated with Alexa 488. Costaining with mouse anti-BrdU antibody or anti-Ki67 antibody (BD) was performed sequentially, followed by Alexa 595 conjugated secondary antibody. ImageJ software was used to manually count the number of Ki67-positive cells from at least 6,000 amylase-positive cells per mouse.
Gut Hormones
The level of circulating gut hormones were determined using a cholecystokinin (CCK) ELISA assay (RayBiotech) and a mouse metabolic hormone panel for eight different metabolic hormones, including ghrelin, gastric inhibitory polypeptide (GIP), glucagon-like peptide 1 (GLP-1), amylin, insulin, leptin, pancreatic polypeptide, and pancreatic peptide YY (PYY) (MILLIPLEX, Millipore, MMHMAG-44K) according to vendors’ protocols.
Statistical Analyses for GTT, ITT, and Western Blot Quantification
All data are presented as mean ± SEM. Data were analyzed by unpaired t test for GTT, ITT, and Western blot quantification of the mouse samples. We used the paired t test to analyze Western blot quantification of the human fecal samples (n indicates the number of samples per group). A P value <0.05 was considered statistically significant.
Data and Resource Availability
The proteomics data sets generated during the current study (Supplementary Table 1) and the raw data (Supplementary Table 2) are available in the Supplementary Tables. No applicable resources were generated or analyzed during the current study.
Results
Diet and Gut Microbiota Alter Intestinal Host Proteome in HFD-Fed Mice
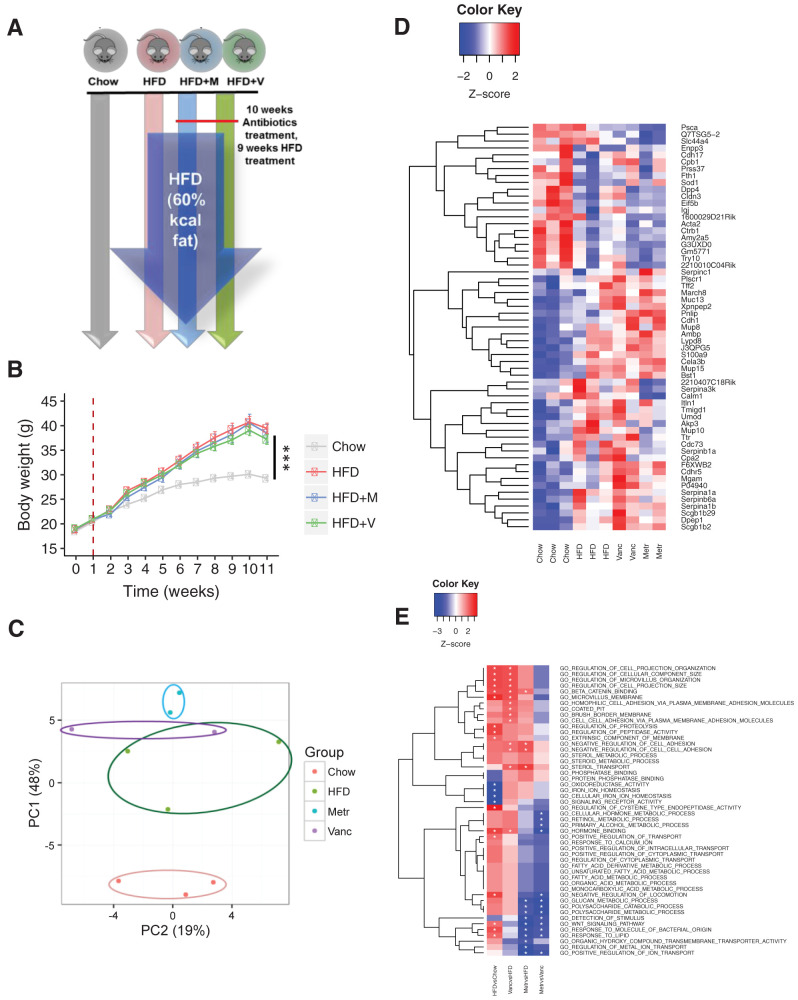
To determine the impact of diet and changing gut microbiome on the host gastrointestinal tract, 6-week-old male C57BL/6J mice were fed a chow diet (22% fat), HFD (60% fat), or HFD plus oral vancomycin or metronidazole to further modify the gut microbiome for 10 weeks (Fig. 1A). As expected, the HFD led to an ∼40% increase in weight gain over this 10-week period compared with the chow diet, and treatment with antibiotics did not significantly change this (Fig. 1B). The fecal proteins were extracted at the end of the experiment and subjected to a quantitative proteomics analysis using TMT labeling (16,17). In total, 138 mouse proteins were identified (Supplementary Table 1A). PCA of the fecal host proteome showed that the HFD induced significant changes (Fig. 1C). Thus, the relative abundance of 15 proteins significantly decreased, and 17 proteins significantly increased in the fecal samples of the HFD-fed mice compared with chow-fed mice (Fig. 1D and Supplementary Table 2).
Figure 1.
Gut microbiota alters host intestinal proteome. A: Schematic representation of the experimental design. B: Body weight gain of mice on chow or HFD with or without antibiotics (Abx) treatment with metronidazole (M) or vancomycin (V) (n = 8 per group). Time 0 is the beginning of the HFD, and the dashed line represents the beginning of Abx treatment. ***P < 0.001. C: Principal component (PC) analysis of intestinal proteomes of mice fed chow, HFD, HFD + vancomycin (Vanc), or HFD + metronidazole (Metr). Each dot represents one mouse sample. D: Heat map shows the top 60 proteins affected by Abx and diet. E: Heat map shows the top functional classes affected by Abx treatment.
Although it is difficult to reach a conclusion about the effects of the individual antibiotic treatment, because of the small sample size in the 10-plex quantitative analysis, proteomics screening indicated that both antibiotics further modified the host fecal proteome during exposure to the HFD. For example, among the 32 proteins altered by the HFD, 2 (Cela3B and Mup10) showed a trend of further alteration in metronidazole-treated HFD-fed mice (Fig. 1D and Supplementary Table 1C). In addition, four proteins (Sh3d21, Psca, Calm1, and SLC44A4) that were not altered by the HFD challenge tended to decrease, and two proteins (Pnlip and March8) tended to increase by metronidazole treatment (Fig. 1D, Supplementary Table 1C, and Supplementary Fig. 1).
Further analysis revealed that diet and antibiotics significantly changed the abundance of different functional classes of proteins (Fig. 1E). For example, proteolysis-related proteins, such as elastase and dipeptidase 1, were increased by the HFD, whereas proteins involved in iron homeostasis, such as ferritin heavy chain, were reduced with the HFD. Notably, serine protease inhibitors (serpin) proteins, which have been previously linked to metabolic diseases, were one of the main groups affected by diet and gut microbiota (Supplementary Fig. 2A and Supplementary Table 1D). Thus, Serpina1a (α1-antitrypsin 1-1), Serpina1b (α1-antitrypsin 1-2), and Serpina6a were increased with the HFD challenge (Supplementary Table 1B).
The third group of proteins that were altered in response to HFD challenge were those involved in inflammatory pathways and protection of gut integrity. HFD is known to promote intestinal inflammation through its action on the gut microbiome (18). Our analysis indicates that an HFD can modulate the host proteome in a way that would be protective to the host from the proinflammatory effects of changing gut microbiota. For example, two of the top three proteins most upregulated by the HFD challenge were related to immune system function (Supplementary Fig. 1B and Supplementary Table 1D). One of these was Lypd8 (LY6/PLAUR domain containing 8), a recently characterized antimicrobial protein that targets the flagella of Gram-negative bacteria and thus protects the colonic epithelium (19) (Fig. 2A). The other increased by the HFD was mucin (Muc) 13, a protein with a protective role in the colonic epithelium (20) (Fig. 2A).
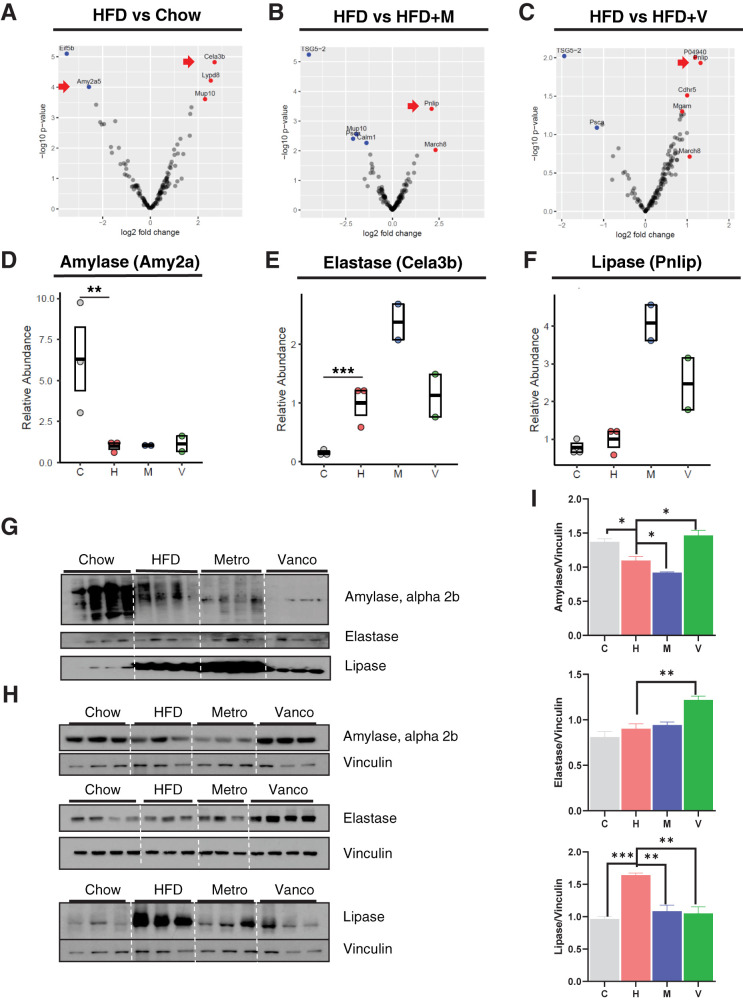
Figure 2.
Gut microbiota alters exocrine pancreas function. Volcano plot shows the distribution of differentially identified proteins by HFD (A), metronidazole (M) (B), and vancomycin (V) (C) in a log10 scale. C, chow; H, HFD. Dot/box plots show the relative abundance of amylase (D), elastase (E), and lipase (F). Data represent mean ± SEM (n = 2–3 per group). *P ≤ 0.05, **P ≤ 0.01. G: Western blots of fecal protein extracts (9 weeks on the HFD; 10 weeks on antibiotics). Metro, metronidazole; Vanco, vancomycin. Western blots for protein extracts of pancreas (H) and quantification of amylase, elastase, and lipase levels normalized to vinculin (I). Data represent mean ± SEM (n = 4 per group). *P ≤ 0.05, **P ≤ 0.01.
HFD and Gut Microbiota Modulate Function of the Exocrine Pancreas
The most significantly regulated proteins in response to the HFD and antibiotic treatment in the fecal proteome were exocrine pancreatic enzymes, including amylase, pancreatic lipase, and elastase (Fig. 2A–C). The HFD and antibiotic treatment differentially regulated the levels of each of these enzymes found in the feces. Amylase was significantly decreased by HFD treatment and not changed by antibiotics (Fig. 2D), whereas elastase was increased by the HFD and showed a further twofold increase by metronidazole treatment (Fig. 2E). Lipase protein levels in fecal samples did not change with the HFD and further increased after treatment with both antibiotics, but especially with metronidazole (Fig. 2F). The changes in exocrine pancreatic enzymes in fecal samples identified by proteomics were confirmed by Western blotting of fecal samples from a separate cohort of HFD and antibiotic-treated mice (Fig. 2G). Consistent with the lower levels in fecal samples, amylase levels were decreased in the pancreas by the HFD. Interestingly, however, these changes at the level of the pancreas were totally reversed by vancomycin treatment, while metronidazole treatment further decreased the amylase levels (Fig. 2H and I). Elastase, on the other hand, showed a trend of increase in the pancreas of the HFD and metronidazole-treated groups and significantly increased with vancomycin treatment in the pancreas. Lipase in pancreatic samples was also significantly increased with HFD feeding but normalized by both antibiotics (Fig. 2H and I). Overall, these results reveal that both HFD feeding and antibiotic-induced modifications in gut microbiota have a significant impact on the regulation of exocrine pancreas function as reflected by the abundance of pancreatic enzyme levels.
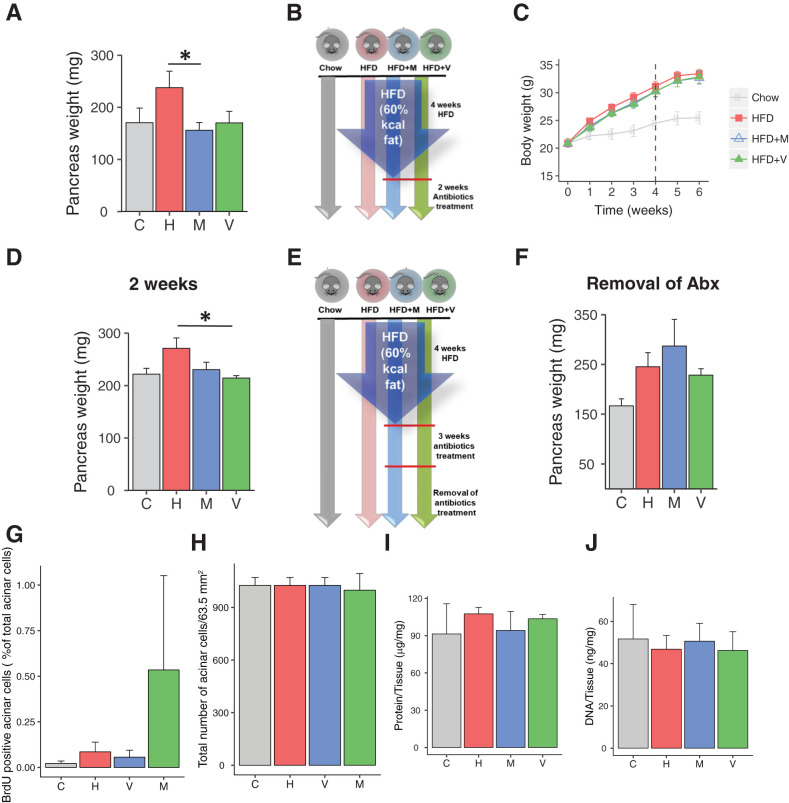
Diet and Gut Microbiota Regulate Pancreatic Growth in HFD-Fed Mice
To better understand these changes in exocrine function of the pancreas, we assessed pancreas weight in a new cohort of mice subjected to a similar protocol, but for 12 weeks. Again, pancreas weight was increased by an average of 40% in mice on the HFD (Fig. 3A). Furthermore, although antibiotic treatment had no effect on weight gain in the HFD-fed mice, antibiotic treatment completely blocked the increase in pancreatic weight of the HFD-fed mice (Fig. 3A). Similar effects on pancreas weight were also observed with only 6 weeks of the HFD and were reversed in these mice with as little as 2 weeks of antibiotic treatment (Fig. 3B–D). Thus, the effect of diet on pancreatic enlargement is rapid and can persist over multiple weeks in the mouse. Changes in gut microbiota due to antibiotic treatment are usually reversed following discontinuation of the antibiotics (21). When we removed antibiotics from the drinking water and continued to feed mice the HFD for 4 additional weeks, pancreatic weights of the groups that had been previously treated with antibiotics were as high as the HFD group (Fig. 3E and F). Thus, the reduction of pancreatic mass in HFD-fed mice requires continued remodeling of the microbiome by antibiotic treatment and parallels the changes in gut microbiota when antibiotics are discontinued.
Figure 3.
Gut microbiota alters pancreas mass. A: Pancreas weight of mice on chow, HFD, HFD + vancomycin (V), or HFD + metronidazole (M) (n = 7–8 per group) at the end of 10 weeks of antibiotics (Abx) treatment. B: Schematic representation of the modified short-term experimental design. C: Body weight gain of mice on chow or HFD with or without Abx treatment. Time 0 is the beginning of the HFD, and the dashed line represents the beginning of Abx treatment (n = 5 per group). D: Pancreas weight of mice on chow, HFD, HFD + vancomycin, or HFD + metronidazole (n = 5 per group) at the end of 2 weeks of Abx treatment. E: Experimental design in which mice were treated for 4 weeks with chow or HFD, then 3 weeks with water or Abx in addition to the diet, and then all mice on Abx were placed on normal water (Abx were removed). F: Pancreas weight of mice 3 weeks after the Abx removal (n = 4 per group). G: Quantification of amylase+ BrdU+ cells. Data represent mean ± SEM (n = 4–5 per group). **P ≤ 0.01. H: Total number of amylase+, acinar cells for a constant area (63.5 mm2). Data represent mean ± SEM. Protein content (I) and DNA content (J) of 1 mg pancreas tissue (n = 3; 6 images/pancreata). Data represent mean ± SEM.
Pancreatic Growth With HFD Is Caused by Acinar Cell Hyperplasia in HFD-Fed Mice
We observed an increase in both β-cell mass and β-cell area in all three mouse groups on HFD treatment for 10 weeks, although with this small cohort, this did not quite reach statistical significance. In contrast to the change in total pancreas weight, however, this was not blocked by antibiotic treatment, and indeed, there was a trend of a further increase in β-cell area in response to metronidazole treatment (Supplementary Fig. 3A and B). Furthermore, since islets only constitute 1–2% of pancreatic mass (22) and acinar cells constitute more than 95% of the pancreatic mass (23), the large increase in pancreatic weight in HFD-treated mice must represent a change in the exocrine pancreas. To better understand this change, we counted the number of acinar cells per high-power field (63.5 mm2) and observed no change in acinar cell number among groups (Fig. 3H and Supplementary Fig. 4). Furthermore, neither protein nor DNA levels per milligram of pancreas tissue differed among groups, suggesting that the size of the acinar cells in all groups was comparable (Fig. 3I and J and Supplementary Fig. 4).
To directly assess proliferation of pancreatic acinar cells, mice fed the HFD for 12 weeks were labeled with BrdU, and pancreatic sections were taken and costained with anti-amylase antibodies. This revealed no significant difference in the number of proliferating acinar cells (Fig. 3G and Supplementary Fig. 4A–D). Likewise, Ki67+ staining of the pancreas sections showed no differences in acinar cell proliferation among the four groups (Supplementary Fig. 5A–D). Thus, the pancreatic enlargement in HFD-fed mice must be due to an increase in the number of acinar cells (i.e., hyperplasia), rather than a change in acinar cell size (i.e., hypertrophy), and this proliferation must occur early after the HFD challenge.
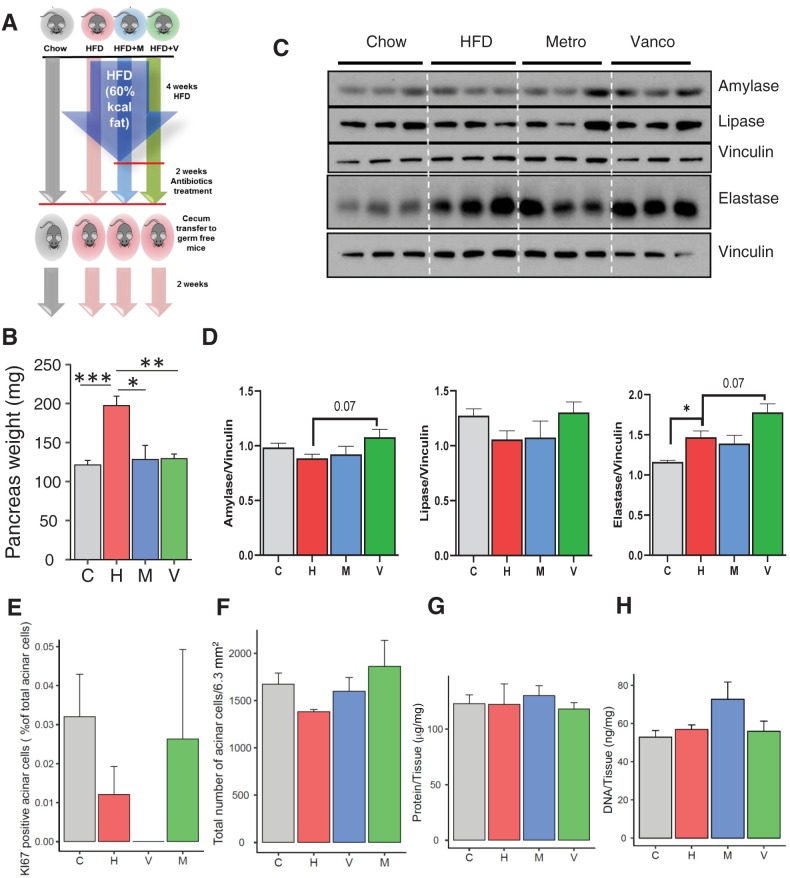
Transplantation of Gut Microbiota Transfers the Effects on Pancreatic Weight and Function to HFD-Fed GF Mice
To more directly assess the role of gut microbiota on pancreas growth and function, we performed cecal bacterial transfer from mice on chow or the HFD, with and without antibiotic treatment, into GF mice. The GF mice were maintained on irradiated normal chow or, for GF mice receiving microbiota from any of the three HFD groups, started on irradiated HFD 4 weeks prior to the microbiome transfer and continued on these diets for 2 weeks after the transfer (Fig. 4A). Consistent with the findings observed in the donor mice, pancreas weight of GF mice receiving the HFD and transplanted with the microbiome of HFD-challenged mice were ∼50% higher (P < 0.01) than those of chow-fed GF mice receiving gut microbiota of the chow-fed group (Fig. 4B). By contrast, the pancreata of GF mice transplanted with gut microbiota of mice on the HFD that had received antibiotic treatment were no different than those of mice on the chow diet (Fig. 4B). Gut microbiome transfer also modulated the expression of the exocrine pancreatic enzymes. Thus, as in the donor mice (Fig. 2H and I), HFD-fed GF mice tended to have lower levels of amylase and higher levels of lipase than GF mice on the chow diet (Fig. 4C and D), and transfer of cecal contents from vancomycin-treated mice showed a trend of reversing the levels of amylase (P = 0.07) and lipase to those observed in normal chow-fed mice, while further increasing elastase levels. Also, consistent with our results for the donor mice, at the time of sacrifice, we did not identify any increase in markers of acinar cell proliferation between groups (Fig. 4E and Supplementary Fig. 6A–D). Likewise, the number of acinar cells (Fig. 4F) and the protein and DNA levels per milligram of pancreas (Fig. 4G and H) were not different.
Figure 4.
Alteration of pancreas mass and pancreatic function can be transferred to GF mice. A: Schematic representation of the experimental design of bacterial transfer from treated donor mice to GF mice. B: Pancreas weight of GF mice colonized with bacteria from mice on chow (C), HFD (H), HFD + vancomycin (V), or HFD + metronidazole (M), weighed 2 weeks after transfer (n = 6 colonized mice). Results are shown as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001. C: Western blots for pancreatic enzymes in the pancreas of chow- or HFD-fed GF mice that received bacterial transfer from chow + placebo, HFD + placebo, HFD + vancomycin-, or HFD + metronidazole-treated mice and quantitation of amylase, lipase, and elastase proteins normalized to vinculin (n = 3 per group; 2 weeks after colonization). D: Quantification of Ki67+ amylase+ cells. Data represent mean ± SEM (n = 3 per group). **P ≤ 0.01. E: Total number of amylase+ acinar cells for a constant area (63.5 mm2, n = 5). Data represent mean ± SEM. Protein content (F) and DNA content (G) of 1 mg pancreas tissue (n = 5, 6 images for pancreata). Data represent mean ± SEM.
Diet and Gut Microbiota Regulate Gastrointestinal Hormone Secretion in HFD-Fed Mice
To determine the effects of gut microbiota on the secretion of enteroendocrine hormones (24), we measured plasma levels of nine hormones. GLP-1 is known to stimulate insulin secretion. GLP-1 can also act via the GLP-1 receptor on acinar cells and induce amylase secretion (25). In the donor mice, the HFD caused a decrease in circulating GLP-1 levels, and this was reversed with vancomycin treatment (Fig. 5A). Transferring microbiota from antibiotic-treated mice to GF mice was sufficient to transfer significant changes in GLP-1 secretion (Fig. 5B). In contrast to GLP-1, GIP tended to be increased in response to the HFD, and this small increase was reversed with antibiotic treatment (Fig. 5C). Interestingly, in recipient mice, this phenotype was exaggerated, with more marked increased in GIP in the HFD group and reversed in the antibiotics groups (Fig. 5D).
Figure 5.
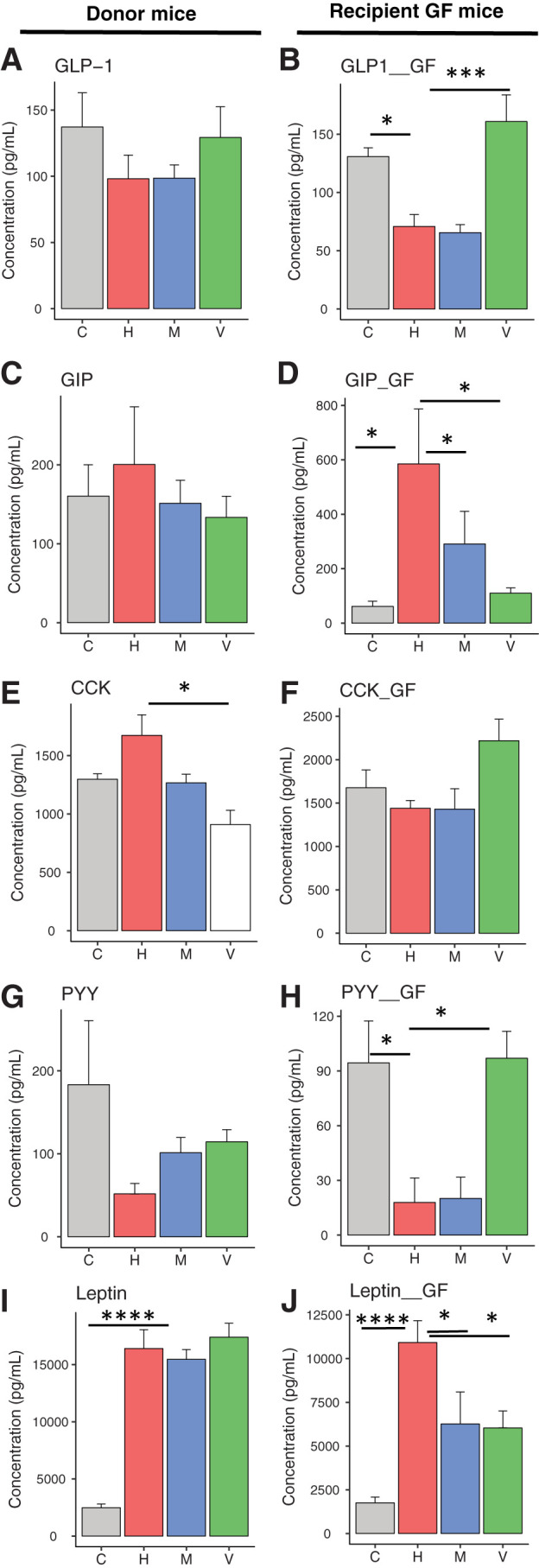
Gastrointestinal hormone secretion is altered by gut microbiota. Plasma levels of various intestinal hormones of mice fed chow (C) or HFD (H), HFD + metronidazole (M) or HFD + vancomycin (V) (left side) and of GF recipient mice, 2 weeks after the bacterial transfer (right side). Plasma levels of GLP-1 (A and B), GIP (C and D), CCK (E and F), PYY (G and H), and leptin (I and J) were measured. Data represent mean ± SEM (n = 6–8 for donor mice and n = 6 per mice colonized with cecal bacteria from antibiotic-treated mice). *P ≤ 0.05, **P ≤ 0.01, *** P ≤ 0.001.
CCK stimulates acinar cell growth and secretion (26). Because GIP can potentiate CCK-stimulated pancreatic enzyme secretion (27), it is possible that altered GIP secretion can modulate CCK secretion. Thus, CCK secretion was increased in the serum of donor mice on the HFD, and this was reversed with vancomycin treatment (Fig. 5E). This effect, however, was not transferred to the GF mice (Fig. 5F). PYY has a role in body weight regulation. Earlier studies suggested that PYY has an inhibitory role on acinar cell secretion (28). PYY was decreased in mice on the HFD, and this also tended to be reversed with vancomycin treatment (Fig. 5G). Significant changes were observed in PYY levels of GF mice after gut microbiota transfer (Fig. 5H). Finally, plasma levels of the adipocyte-secreted hormone leptin, which regulates food intake and energy expenditure, were increased in the HFD group, and this was not modified by antibiotic treatment (Fig. 5I). Leptin levels were also elevated in GF mice on the HFD. Interestingly, these were significantly decreased when the GF mice received microbiota from HFD mice treated with either metronidazole or vancomycin (Fig. 5J), suggesting some cross talk between the gut microbiome and the adipocytes.
Consistent with our previous studies (5), circulating insulin levels were increased with the HFD, and metronidazole treatment lowered it (Supplementary Fig. 7A). Although this did not reach statistical significance, there was a trend for similar changes after microbiota transplantation (Supplementary Fig. 7B). We did not observe a significant change in amylin, ghrelin, or pancreatic polypeptide levels in either donor or GF recipient mice (Supplementary Fig. 7C–F).
Effects of Antibiotic Treatment on Metabolism and Pancreas Function in Chow-Fed Mice
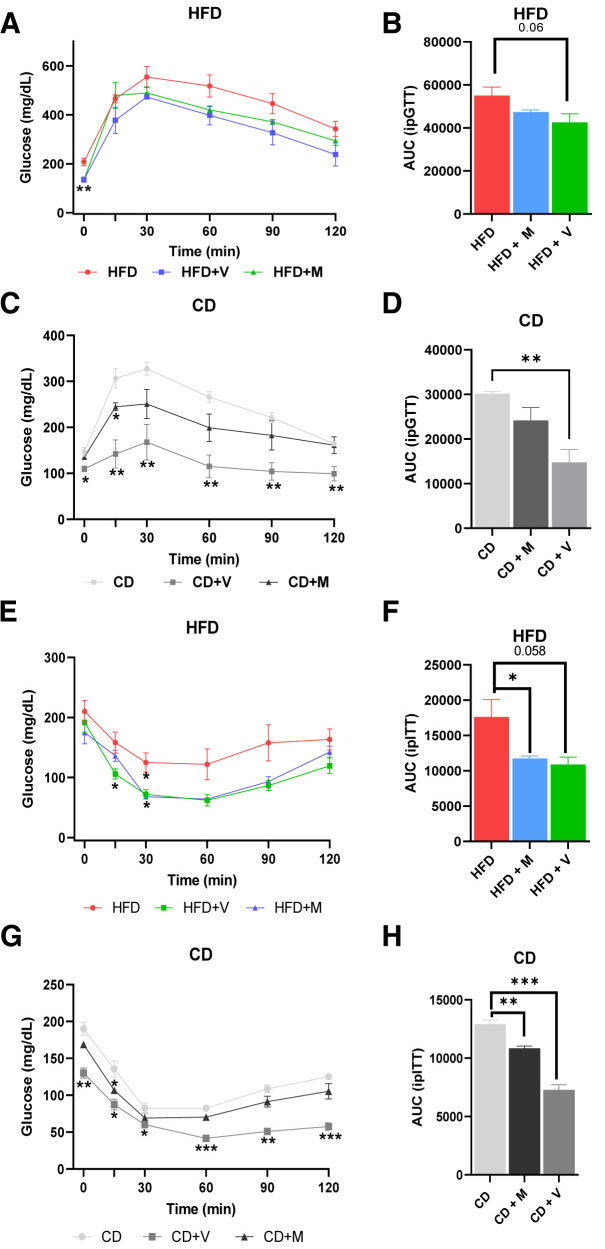
To determine whether the alterations produced by antibiotics are dependent on the presence of an “HFD microbiome,” C57BL/6J mice were fed chow for 6 weeks or chow plus oral vancomycin or metronidazole in the last 2 weeks of the experiment. We also repeated the HFD experiments using the short-term protocol (Fig. 3B). Consistent with the HFD-fed mice body weight data (Supplementary Fig. 8A), antibiotic treatment did not change the body weight in the chow-fed mice (Supplementary Fig. 8B). Vancomycin treatment significantly reduced fasted blood glucose levels in both HFD-fed (Fig. 6A) and chow-fed (Fig. 6B) mice. Intraperitoneal GTT of the HFD-fed mice revealed a trend toward improved glucose tolerance (P = 0.06) (Fig. 6A), consistent with our previous study (11). In the chow-fed mice, vancomycin treatment significantly increased glucose tolerance, and a similar trend was observed for metronidazole treatment (Fig. 6B). As previously observed (5,11), both antibiotics significantly increased insulin sensitivity in HFD-challenged mice as assessed by the ITT (Fig. 6C), while in the chow-fed mice, vancomycin treatment, but not metronidazole, improved insulin sensitivity (Fig. 6D). Consistent with increased insulin sensitivity, antibiotic treatment also significantly decreased insulin levels in chow-fed mice (Fig. 7A).
Figure 6.
Effects of antibiotic treatment on insulin sensitivity and glucose tolerance. A: Intraperitoneal GTTs (ipGTT) on 12-week old C57Bl/6J fed the HFD for 2 weeks (n = 4 per group). M, metronidazole; V, vancomycin. B: Area under curve (AUC) of ipGTT curve of HFD-fed mice. C: ipGTT on same age and genotype of chow diet (CD)-fed mice (n = 4 per group). D: Area under curve of ipGTT curve of CD-fed mice. E: ipITT on HFD-fed mice (n = 3–4 per group). F: Area under curve of ipITT curve of HFD-fed mice. G: ipITT on CD-fed mice (n = 3–4 per group). H: Area under curve of ipITT curve of CD-fed mice. Data are shown as mean ± SEM. Statistical analyses were performed by two-tailed, unpaired Student t test. *P ≤ 0.05, **P ≤ 0.01, *** P ≤ 0.001.
Figure 7.
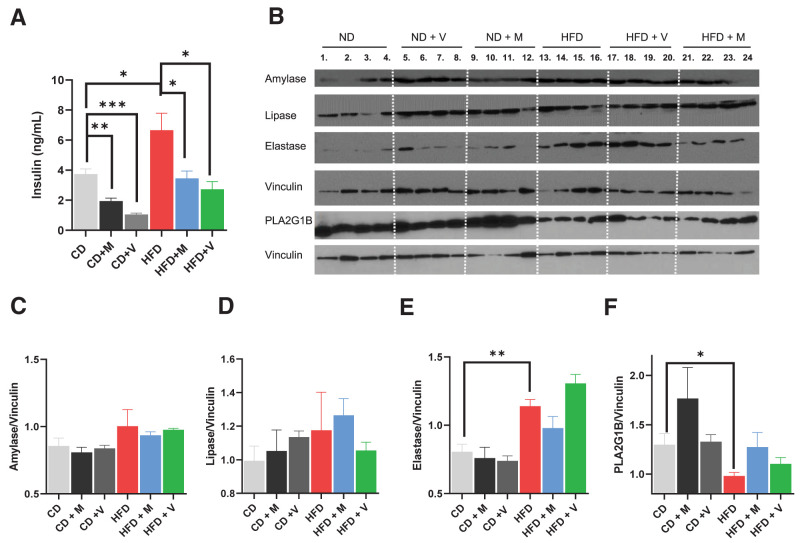
Antibiotic treatment alters fasting plasma insulin levels but not exocrine enzymes in chow-fed mice. A: Plasma insulin levels were measured on 12-week-old C57Bl/6J after mice were starved for 6 h (n = 4 per group). CD, chow diet; M, metronidazole; V, vancomycin. B: Western blot analysis of amylase, lipase, elastase, and PLA2G1B protein from pancreatic lysate (n = 4 per group). Relative intensity of amylase (C), lipase (D), elastase (E), and PLA2G1B (F). Vinculin was used as the loading control and for normalization in the quantification. Data are shown as mean ± SEM. Chow-fed and HFD-fed mice treated with antibiotics were compared with chow-fed control. HFD-fed mice treated with antibiotics were compared with HFD-fed control. Statistical analyses were performed by two-tailed, unpaired Student t test. **P < 0.01, ***P < 0.001.
In contrast to the effect of antibiotics to reduce pancreatic size in HFD-fed mice (Fig. 3D), antibiotic treatment did not affect pancreas size in chow-fed mice (Supplementary Fig. 8C). Western blot analysis (Fig. 7B) revealed that antibiotic treatment did not alter amylase (Fig. 7C) or pancreatic lipase (Fig. 7D) in pancreata of chow-fed or short-term HFD-fed mice. On the other hand, the HFD significantly increased elastase levels (Fig. 7E). Pancreatic phospholipase A2 (PLA2), one of the best measure of secretory function of the pancreas (29), on the other hand, was significantly decreased in the pancreata of mice on the HFD and tended to return to normal with antibiotic treatment (Fig. 7F). We previously observed that the cecum weight was reduced by ∼50% in HFD-fed mice, and this was restored with antibiotic treatment (5). Similar to the effect with the HFD, we now find that both vancomycin and metronidazole treatment significantly increased cecum weight in chow-fed mice (Supplementary Fig. 8D). Overall, these results reveal that antibiotic treatment has a significant impact on the regulation of metabolism in both chow-fed and HFD-fed mice, while the effects on exocrine pancreas are specific to the “HFD microbiome.”
Vancomycin Treatment Alters Exocrine Function in Obese Men With Prediabetes
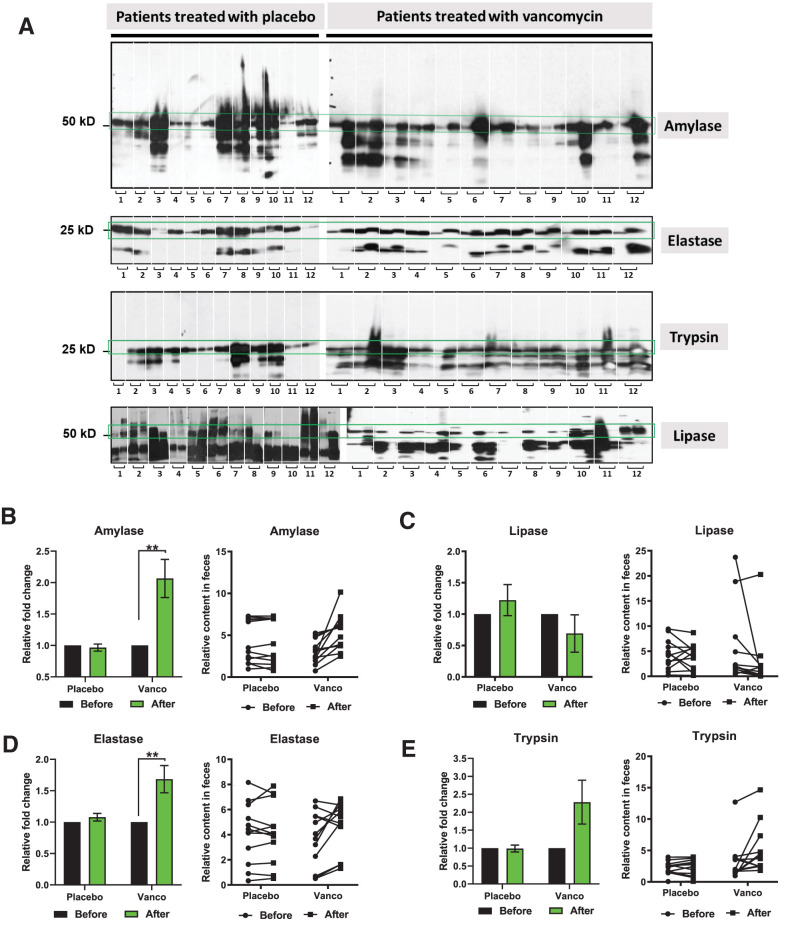
To determine whether similar effects of gut microbiota on pancreatic function could be identified in humans, we analyzed fecal samples obtained from 24 obese men with prediabetes who were treated with placebo or oral vancomycin for 7 days. Reijnders et al. (12) previously reported that 1 week of oral vancomycin treatment in humans changed the microbiota composition and short-chain fatty acid and bile acid concentrations, although it was not sufficient to improve insulin sensitivity or metabolic health in these individuals. Western blot analysis of these human fecal samples showed that vancomycin treatment caused a significant increase in the levels of two pancreatic enzymes (Fig. 8A). In particular, fecal amylase increased twofold (Fig. 8B), elastase increased 1.6-fold (Fig. 8D), and trypsin increased twofold (P = 0.06) (Fig. 8E), while fecal lipase levels did not change (Fig. 8C). These changes were not observed in the placebo group (Fig. 8). These results are consistent with our observations in mice and indicate that even short-term vancomycin treatment can alter pancreatic exocrine function in humans.
Figure 8.
Gut microbiota alters exocrine pancreas function in obese and insulin-resistant men. A: Western blots of amylase, elastase, trypsin, and lipase in the fecal samples of men who were treated with placebo (left) or vancomycin (Vanco) (right) for 1 week. The two adjacent columns correspond for each patient to before and after treatment. Individuals are separated with white dashes (n = 12). Quantification of amylase (B), lipase (C), elastase (D), and trypsin (E) levels in placebo and vancomycin groups. Molecular weight: amylase, 57 kDa; lipase, 51 kDa; elastase, 28 kDa; and trypsin, 27 kDa. Data represent mean ± SEM. * P < 0.05, ** P < 0.01 by paired t test.
Discussion
We and others have previously shown that diet and the gut microbiota interact with host genetics in the pathophysiology and development of the metabolic syndrome (5,10,30). Obesity and metabolic syndrome can also disrupt the enteroendocrine axis and pancreatic exocrine physiology, including increasing the risk of pancreatitis (31,32). In this study, we demonstrate that alterations in diet and gut microbiota can produce striking changes in pancreas mass. This is associated with changes in the levels of exocrine pancreatic enzymes, including amylase, elastase, and lipase found in the fecal proteome. We also observe that the levels of multiple gastrointestinal hormones, including GLP-1, GIP, PYY, and CCK, are altered by these changes in the gut microbiota. While previous reports have shown that an HFD challenge can increase pancreatic weight (33), the extent to which this is occurring in response to dietary content versus changes in gut microbiota was unclear. Here we show that transplantation of gut microbiota of high-fat–fed and antibiotic-treated mice to GF mice can transfer most of these phenotypes, including altered pancreas weight, pancreatic exocrine function, and gut endocrine function.
In addition to our findings in the mouse, analyses of fecal samples of obese individuals with prediabetes reveal that these observations can be translated to humans. Indeed, vancomycin treatment causes an increase in levels of amylase and elastase in these individuals. While further studies will be necessary to define the specific microbiota and molecular mechanism(s) underlying these changes, our findings have the potential to help us better understand the dysfunction of pancreas in obesity and diabetes. Insulin and insulin resistance may also play a role in the changes in the exocrine pancreas, since acinar cells do express insulin receptors (34). Indeed, pancreatic weight, which reflects acinar mass, is lower in patients with type 1 diabetes and higher in individuals with obesity-related insulin resistance (35), the latter mirroring the findings in mice. In addition, pancreatic exocrine insufficiency is prevalent in patients with type 1 diabetes (25–74%) and also those with type 2 diabetes (28–54%) (36,37).
In this study, we also show that vancomycin and metronidazole treatment can both affect insulin sensitivity and glucose tolerance in both HFD-fed and chow-fed mice. The effects of vancomycin are significantly stronger than metronidazole, suggesting that clearance of Gram-positive bacteria has a specific role in increased insulin sensitivity and glucose tolerance in these mice. Our results are consistent with a previous study showing the effects of antibiotics treatment on metabolic homeostasis in chow-fed mice (38). However, that study used a cocktail of antibiotics and antifungals and did not provide any data related to the exocrine pancreas function. While more studies are needed, fecal transfer from lean individuals to patients with metabolic syndrome has shown some potential to improve insulin sensitivity (39).
Our data indicate that the gut microbiome and its modification by diet and antibiotics is a significant determinant of pancreas function, not only in mice but also in humans. The change in pancreatic size is associated with changes in exocrine pancreas function and is related to an increase in acinar cell number (i.e., hyperplasia), rather than a change in acinar cell size. We do think that the lack of increase in BrdU labeling at 10 weeks suggests that most of the pancreatic growth occurs earlier in the HFD challenge. CCK stimulates acinar cell growth and secretion (26) and might be one of the factors altering the pancreas weight and function, since CCK levels tend to be increased by HFD feeding and are significantly decreased by vancomycin treatment. In addition, an insulin-acinar axis could contribute to the growth phenotype, since insulin has also been shown to act as a paracrine regulator of the growth and function of the exocrine pancreas (40). In our study, insulin levels correlate with increased pancreatic size, both in mice fed the HFD or GF mice receiving gut microbiota of HFD-fed mice, which have higher insulin levels, or in mice fed the HFD plus antibiotics or GF mice receiving their gut microbiota, which have lower insulin levels and decreased pancreatic size.
With regard to which microbiota drive these changes in the exocrine pancreas, we find that treatment with either vancomycin or metronidazole can reverse the enlargement of the pancreas caused by an HFD challenge in mice and that this effect can be reproduced by microbiota transfer to GF animals. On the other hand, only transfer of microbiota from vancomycin-treated mice to GF mice tended to reverse the effects of the HFD on pancreatic amylase and lipase secretion (P = 0.07), suggesting a role for specific types of bacteria in the regulation of pancreatic function versus pancreatic growth. Using this protocol, we previously showed that in B6 mice, an HFD decreases the level of Bacteroidetes and increases the level of Firmicutes (74%) and Verrucomicrobia and that antibiotic treatment further decreased bacterial diversity and increased in the relative of abundance of Proteobacteria (5,6,11). Vancomycin treatment reduces the abundance of Firmicutes to 37% and increases the relative abundance of Proteobacteria (5). In addition to the effects of these changes that might contribute to pancreatic growth, we previously identified multiple differences in cecal and plasma metabolites reflecting the effects of diet and antibiotics on the gut microbiome (6). To what extent these changes in the metabolome contribute to the changes in pancreatic exocrine and endocrine function remains to be determined.
Our study also suggests the importance of microbiota in the regulation of host enteroendocrine function. The microbiota transfer experiments show that GLP-1, GIP, PYY, and even leptin levels in circulation are significantly altered by the composition of gut microbiota. Indeed, transplant of microbiota obtained from vancomycin-treated mice is able to change the levels of all four hormones significantly in GF mice, whereas transfer of microbiota from metronidazole-treated mice does not alter the hormone levels except leptin, indicating roles of specific vancomycin-sensitive organisms in this response. Interestingly, while direct antibiotic treatment was able to produce only modest changes in the levels of these hormones in the donor mice, all of these changes reached statistical significance in the GF mice receiving the microbiota from the antibiotic-treated mice, indicating the strength of the microbiome transfer approach to define a direct role of the gut microbiome in the changes in these enteroendocrine hormones. This adds to the growing evidence for cross talk between gut microbiota and the host endocrine system (7,41–43).
Taken together, our findings demonstrate the importance of the gut microbiome in control of pancreas growth, as well as pancreas exocrine and endocrine function. These novel insights provide new target opportunities for development of therapeutics or preventive tools for pancreatic disease through modification of the gut microbiome or its products.
Article Information
Acknowledgments. The authors want to thank Nandita Srayoshi and Dr. Venkata Sabbisetti (Brigham and Women’s Hospital) for their help with gut hormone analysis. The authors also would like to acknowledge Renee Robinson and Dr. Bogdan Budnik for their proteomics service (Mass Spectrometry and Proteomics Resource Laboratory of Harvard University). The authors thank Hui Pan and Jonathan Dreyfuss (Joslin Diabetes Center) for the bioinformatics and statistical analysis and acknowledge Boston College Biology Department undergraduate students David Kim, Typhania Zanou, and Yena Sung for their help with the animal experiments. Thanks to Nancy McGilloway and Todd Gaines for their support in the Boston College Animal Care Facility.
Funding. This work was supported by National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases grant 1K01DK117967-01 and G. Harold & Leila Y. Mathers Foundation grant MF-1905-00311 to E.A. and the Joslin Diabetes Research Center grant P30 DK036836. R.N.K is supported by grant R01DK067536 and C.R.K. is supported by grants R01DK031026 and R01DK033201.
Duality of Interest. The human antibiotics study is funded by TI Food and Nutrition, a public-private partnership on precompetitive research in food and nutrition to E.E.B. No other potential conflicts of interest relevant to this article were reported.
Author Contributions. K.G. assisted with data analysis, Western blot analysis, and animal experiments with HFD- and chow-fed mice. M.S. assisted with data analysis and conducting the animal experiments with the HFD. Q.H. and B.S. assisted with immunoblotting of the human samples. L.O. assisted with immunoblotting and animal experiments. C.C., J.H., E.D., and R.N.K. assisted with the imaging of pancreatic sections, and quantification, analysis, and the interpretation of pancreatic proliferation data. M.F. and A.R. assisted with immunoblotting of the mouse samples and maintaining the mice. E.E.C., G.H.G., and E.E.B. designed the human antibiotics study and collected human fecal samples. S.F. assisted with the protocol and first animal experiments. C.R.K. and E.A. designed research. All authors helped with the analysis of the data that they contributed to produce. E.A. wrote the paper and assisted with all experiments. E.A. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. This study was presented as abstract at the 79th Scientific Sessions of the American Diabetes Association, San Francisco, CA, 7–11 June 2019 (https://diabetesjournals.org/diabetes/article/68/Supplement_1/187-OR/59851/187-OR-Gut-Microbiota-Regulate-Pancreatic-Growth).
Footnotes
This article contains supplementary material online at https://doi.org/10.2337/figshare.19193105.
References
- 1. Schroeder BO, Bäckhed F. Signals from the gut microbiota to distant organs in physiology and disease. Nat Med, 2016;22:1079–1089 [DOI] [PubMed] [Google Scholar]
- 2. Roy S, Trinchieri G. Microbiota: a key orchestrator of cancer therapy. Nat Rev Cancer 2017;17:271–285 [DOI] [PubMed] [Google Scholar]
- 3. Yurkovetskiy LA, Pickard JM, Chervonsky AV. Microbiota and autoimmunity: exploring new avenues. Cell Host Microbe 2015;17:548–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dedrick S, Sundaresh B, Huang Q, et al., The role of gut microbiota and environmental factors in type 1 diabetes pathogenesis. Front Endocrinol (Lausanne) 2020;11:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fujisaka S, Ussar S, Clish C, et al., Antibiotic effects on gut microbiota and metabolism are host dependent. J Clin Invest 2016;126:4430–4443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fujisaka S, Avila-Pacheco J, Soto M, et al., Diet, genetics, and the gut microbiome drive dynamic changes in plasma metabolites. Cell Rep 2018;22:3072–3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hwang I, Park YJ, Kim YR, et al., Alteration of gut microbiota by vancomycin and bacitracin improves insulin resistance via glucagon-like peptide 1 in diet-induced obesity. FASEB J 2015;29:2397–2411 [DOI] [PubMed] [Google Scholar]
- 8. Ridaura VK, Faith JJ, Rey FE, et al., Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 2013;341:1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Routy B, Le Chatelier E, Derosa L, et al., Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018;359:91–97 [DOI] [PubMed] [Google Scholar]
- 10. Ussar S, Griffin NW, Bezy O, et al., Interactions between gut microbiota, host genetics and diet modulate the predisposition to obesity and metabolic syndrome. Cell Metab 2015;22:516–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Soto M, Herzog C, Pacheco JA, et al., Gut microbiota modulate neurobehavior through changes in brain insulin sensitivity and metabolism. Mol Psychiatry 2018;23:2287–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reijnders D, Goossens GH, Hermes GD, et al., Effects of gut microbiota manipulation by antibiotics on host metabolism in obese humans: a randomized double-blind placebo-controlled trial. Cell Metab 2016;24:63–74 [DOI] [PubMed] [Google Scholar]
- 13. Käll L, Storey JD, Noble WS. Non-parametric estimation of posterior error probabilities associated with peptides identified by tandem mass spectrometry. Bioinformatics 2008;24:i42–i48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ritchie ME, Phipson B, Wu D, et al., limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kulkarni RN, Winnay JN, Daniels M, et al., Altered function of insulin receptor substrate-1-deficient mouse islets and cultured β-cell lines. J Clin Invest 1999;104:R69–R75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kolmeder CA, de Been M, Nikkilä J, et al., Comparative metaproteomics and diversity analysis of human intestinal microbiota testifies for its temporal stability and expression of core functions. PLoS One 2012;7:e29913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rauniyar N, Yates JR, 3rd. Isobaric labeling-based relative quantification in shotgun proteomics. J Proteome Res 2014;13:5293–5309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ding S, Chi MM, Scull BP, et al., High-fat diet: bacteria interactions promote intestinal inflammation which precedes and correlates with obesity and insulin resistance in mouse. PLoS One 2010;5:e12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Okumura R, Kurakawa T, Nakano T, et al., Lypd8 promotes the segregation of flagellated microbiota and colonic epithelia. Nature 2016;532:117–121 [DOI] [PubMed] [Google Scholar]
- 20. Sheng YH, Lourie R, Lindén SK, et al., The MUC13 cell-surface mucin protects against intestinal inflammation by inhibiting epithelial cell apoptosis. Gut 2011;60:1661–1670 [DOI] [PubMed] [Google Scholar]
- 21. Palleja A, Mikkelsen KH, Forslund SK, et al., Recovery of gut microbiota of healthy adults following antibiotic exposure. Nat Microbiol 2018;3:1255–1265 [DOI] [PubMed] [Google Scholar]
- 22. Sweet IR, Cook DL, Lernmark A, Greenbaum CJ, Krohn KA. Non-invasive imaging of beta cell mass: a quantitative analysis. Diabetes Technol Ther 2004;6:652–659 [DOI] [PubMed] [Google Scholar]
- 23. Mfopou JK, Baeyens L, Bouwens L. Hedgehog signals inhibit postnatal beta cell neogenesis from adult rat exocrine pancreas in vitro. Diabetologia 2012;55:1024–1034 [DOI] [PubMed] [Google Scholar]
- 24. Murphy KG, Bloom SR. Gut hormones and the regulation of energy homeostasis. Nature 2006;444:854–859 [DOI] [PubMed] [Google Scholar]
- 25. Hou Y, Ernst SA, Heidenreich K, Williams JA. Glucagon-like peptide-1 receptor is present in pancreatic acinar cells and regulates amylase secretion through cAMP. Am J Physiol Gastrointest Liver Physiol 2016;310:G26–G33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Murphy JA, Criddle DN, Sherwood M, et al., Direct activation of cytosolic Ca2+ signaling and enzyme secretion by cholecystokinin in human pancreatic acinar cells. Gastroenterology 2008;135:632–641 [DOI] [PubMed] [Google Scholar]
- 27. Mueller MK, Scheck T, Dreesmann V, Miodonski A, Goebell H. GIP potentiates CCK stimulated pancreatic enzyme secretion: correlation of anatomical structures with the effects of GIP and CCK on amylase secretion. Pancreas 1987;2:106–113 [DOI] [PubMed] [Google Scholar]
- 28. Huang SC, Tsai MF. Receptors for peptide YY and neuropeptide Y on guinea pig pancreatic acini. Peptides 1994;15:405–410 [DOI] [PubMed] [Google Scholar]
- 29. Levink IJM, Nesteruk K, Visser DI, et al., Optimization of pancreatic juice collection: a first step toward biomarker discovery and early detection of pancreatic cancer. Am J Gastroenterol 2020;115:2103–2108 [DOI] [PubMed] [Google Scholar]
- 30. Ussar S, Fujisaka S, Kahn CR. Interactions between host genetics and gut microbiome in diabetes and metabolic syndrome. Mol Metab 2016;5:795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Abu Hilal M, Armstrong T. The impact of obesity on the course and outcome of acute pancreatitis. Obes Surg 2008;18:326–328 [DOI] [PubMed] [Google Scholar]
- 32. Chen SM, Xiong GS, Wu SM. Is obesity an indicator of complications and mortality in acute pancreatitis? An updated meta-analysis. J Dig Dis 2012;13:244–251 [DOI] [PubMed] [Google Scholar]
- 33. Baldassano S, Amato A, Caldara GF, Mulè F. Glucagon-like peptide-2 treatment improves glucose dysmetabolism in mice fed a high-fat diet. Endocrine 2016;54:648–656 [DOI] [PubMed] [Google Scholar]
- 34. Korc M, Sankaran H, Wong KY, Williams JA, Goldfine ID. Insulin receptors in isolated mouse pancreatic acini. Biochem Biophys Res Commun 1978;84:293–299 [DOI] [PubMed] [Google Scholar]
- 35. Saisho Y. Pancreas volume and fat deposition in diabetes and normal physiology: consideration of the interplay between endocrine and exocrine pancreas. Rev Diabet Stud 2016;13:132–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Piciucchi M, Capurso G, Archibugi L, Delle Fave MM, Capasso M, Delle Fave G. Exocrine pancreatic insufficiency in diabetic patients: prevalence, mechanisms, and treatment. Int J Endocrinol 2015;2015:595649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Atkinson MA, Campbell-Thompson M, Kusmartseva I, Kaestner KH. Organisation of the human pancreas in health and in diabetes. Diabetologia 2020;63:1966–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zarrinpar A, Chaix A, Xu ZZ, et al., Antibiotic-induced microbiome depletion alters metabolic homeostasis by affecting gut signaling and colonic metabolism. Nat Commun 2018;9:2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kootte RS, Levin E, Salojärvi J, et al., Improvement of insulin sensitivity after lean donor feces in metabolic syndrome is driven by baseline intestinal microbiota composition. Cell Metab 2017;26:611–619.e6 [DOI] [PubMed] [Google Scholar]
- 40. Williams JA, Goldfine ID. The insulin-pancreatic acinar axis. Diabetes 1985;34:980–986 [DOI] [PubMed] [Google Scholar]
- 41. Neuman H, Debelius JW, Knight R, Koren O. Microbial endocrinology: the interplay between the microbiota and the endocrine system. FEMS Microbiol Rev 2015;39:509–521 [DOI] [PubMed] [Google Scholar]
- 42. Chen H, Nwe PK, Yang Y, et al., A forward chemical genetic screen reveals gut microbiota metabolites that modulate host physiology. Cell 2019;177:1217–1231.e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Perry RJ, Peng L, Barry NA, et al., Acetate mediates a microbiome-brain-β-cell axis to promote metabolic syndrome. Nature 2016;534:213–217 [DOI] [PMC free article] [PubMed] [Google Scholar]