Abstract
To date, identification of nonislet-specific transcriptional factors in the regulation of insulin gene expression has been little studied. Here, we report that the expression level of the transcription factor YY1 is increased dramatically in both human and mouse pancreatic β-cells after birth. Nevertheless, the physiological role of YY1 during β-cell development and its regulatory mechanism in β-cell function remain largely unknown. After β-cell ablation of Yy1, we observed rapid onset of hyperglycemia, impaired glucose tolerance, and reduced β-cell mass in neonatal and adult mice. These mice also had hypoinsulinemia with normal insulin sensitivity compared with their wild-type littermates, manifesting as a type 1 diabetic phenotype. Mechanistically, genome-wide RNA sequencing has defined dysregulated insulin signaling and defective glucose responsiveness in β-cells devoid of YY1. Integrative analyses coupled with chromatin immunoprecipitation assays targeting YY1, and histone modifications, including H3K4me1, H3K27ac, and H3K27me3, have further identified Ins1 and Ins2 as direct gene targets of YY1. Luciferase reporter assays and loss- and gain-of-function experiments also demonstrated that YY1 binds to the enhancer regions in exon 2 of Ins1 and Ins2, activating insulin transcription and, therefore, proinsulin and insulin production in pancreatic β-cells. YY1 also directly interacts with RNA polymerase II, potentially stabilizing the enhancer-promoter interaction in the multiprotein-DNA complex during transcription initiation. Taken together, our findings suggest a role for YY1 as a transcriptional activator of insulin gene expression, assisting β-cell maturation and function after birth. These analyses may advance our understanding of β-cell biology and provide clinically relevant insights targeting the pathophysiological origins of diabetes.
The primary regulator of glucose homeostasis in vertebrates is the pancreatic β-cell, which releases insulin (INS) to direct glucose uptake and glycogen conversion for energy storage. The mammalian Ins gene is exclusively expressed in pancreatic β-cells and is regulated by a group of transcription factors (TFs), including pancreas/duodenum homeobox protein 1 (Pdx1) (1,2), neurogenic differentiation 1 (NeuroD1, also known as Beta2) (3), and V-maf musculoaponeurotic fibrosarcoma oncogene homolog A (MafA) (4), that have been extensively studied. These islet-enriched TFs often bind to one of the three conserved promoter regions upstream of the Ins gene—known as A3 (−201 to −196 bp), RIPE3b (−126 to −101 bp), and E1 (−100 to −91 bp) elements—through protein–DNA and/or protein–protein interactions leading to the activation of Ins gene expression (5). Mutations in PDX1, NeuroD1, or MafA are also linked to the development of maturity-onset diabetes of the young or type 2 diabetes in humans (6–8). TFs regulating Ins gene expression are, therefore, key mediators of pancreatic β-cell development, maturation, and function. Identification and characterization of Ins gene activators are critical for understanding the pathophysiological origins of diabetes. Nevertheless, the functional role of nonislet-specific TFs in the regulation of Ins gene expression has been little studied.
Yin Yang 1 (YY1) is a DNA-binding zinc-finger TF that belongs to the GLI-Kruppel family. It is ubiquitously expressed in many cell types and is multifunctional, activating or repressing transcription of networks of genes required for essential cellular processes during organogenesis and development. Moreover, YY1 has been widely studied in tumorigenesis and metastasis of multiple cancers by virtue of its regulation in expression of genes involved in cell survival, replication, differentiation, metabolism, and inflammation (9). In the pancreas, YY1 has been reported to contribute to development of multiple pancreatic cancers, such as pancreatic ductal adenocarcinoma (10), insulinomas (11,12), and nonfunctional pancreatic neuroendocrine tumors (12). Nevertheless, understanding of the physiological role of YY1 during pancreatic β-cell development and its regulatory mechanism in β-cell function remains largely elusive.
In this study, we report that YY1 expression in both human and murine pancreas becomes more apparent after birth. Pancreatic β-cell ablation of Yy1 at the neonatal or adult stage of Pdx1CreERT/+;Yy1fl/fl mice results in glucose intolerance with a progressive loss of β-cell mass, ultimately leading to β-cell failure and diabetes. Mechanistically, integrative transcriptome-, genome-, and epigenome-wide analyses through RNA-sequencing (RNA-seq) and chromatin immunoprecipitation (ChIP-seq), respectively, demonstrate that the gene expression levels of Ins1 and Ins2 were downregulated in β-cells after Yy1 depletion, and YY1 directly bound to the enhancer regions in exon 2 of Ins1 and Ins2. Luciferase reporter assays further revealed that YY1 activates Ins transcription and, therefore, production of proinsulin and INS in β-cells. YY1 also interacts with RNA polymerase II (pol II) directly, potentially stabilizing the enhancer–promoter interaction by forming a large multiprotein-DNA complex for transcription initiation. Our findings show that YY1 has an unappreciated role as a transcriptional activator in the regulation of Ins gene expression.
Research Design and Methods
Integrative Transcriptomic, Genomic, and Epigenomic Data Analysis
RNA-seq Functional Annotation
The sequenced reads were aligned to the mouse reference genome (GRCm38) with an annotated gene model (vM23; GENCODE, www.gencodegenes.org/) using Spliced Transcripts Alignment to a Reference (STAR; version 2.5.3a; www.encodeproject.org/software/star/) with default parameters. The expression abundances of all genes were estimated by running RSEM (version 1.3.0; deweylab.github.io/RSEM/) across all of the samples. The differential expression analysis was performed using edgeR (version 3.28.1; bioconductor.org/packages/release/bioc/html/edgeR.html) with absolute logarithm of fold change (log2fc) cutoff of 1 and false discovery rate–adjusted P value <0.05. The identified differentially expressed genes isolated from the pancreatic islets of conditional knockout (cKO) and control mice were further annotated with gene ontology (GO) and the Kyoto encyclopedia of genes and genomes (KEGG), using the DAVID (Database for Annotation, Visualization and Integrated Discovery) Bioinformatics Tool (version 6.8; david.ncifcrf.gov/).
YY1 ChIP-seq Coupled With RNA-seq Analysis
The sequenced reads were aligned to the mouse reference genome (GRCm38) with the annotated gene model (vM23; GENCODE) using the Burrow-Wheeler Aligner (version 0.7.17) with the Maximum Exact Match algorithm. After filtering the GRCm38 ChIP-seq blacklisted regions and removal of duplicate reads, YY1 binding peaks were identified using the Model-Based Analysis of ChIP-Seq peak caller (MACS; version 2.1.3; pypi.org/project/MACS2/2.1.3/) with q value cutoff of 0.01. Annotation of peaks was performed with the Hypergeometric Optimization of Motif Enrichment (HOMER) software (version 4.10.3; homer.ucsd.edu/homer/) with the annotated gene model (vM23). With default settings, YY1 binding peaks were assigned to the nearest transcription start site (TSS) of genes, and further classified using the following features: TSS (−1 kb to +100 bp), transcription end site (−100 bp to +1kb), exons, introns, and intergenic regions. De novo motif discovery was performed with all the identified YY1 binding peaks by HOMER. Heat map visualization of YY1 ChIP-seq data was performed using deepTools (version 3.3.0; github.com/deeptools/deepTools). Selected genes with YY1 binding peaks were visualized using the UCSC (University of California, Santa Cruz) genome browser. YY1 target genes were determined by differential expression demonstrated in RNA-seq with reference to YY1 binding peaks that were further annotated with GO and KEGG by the DAVID Bioinformatics Tool.
Analysis of YY1 Target Gene Expression Through Histone ChIP-seq Analysis
Raw data of histone ChIP-seq targeting H3K4me1 (13), H3K27ac (14), and H3K27me3 (15) in pancreatic islets were downloaded from previous studies for subsequent analysis. The sequenced reads were aligned and potential YY1 gene targets with DNA regions enriched for histone modification marks were identified as described in the previous paragraph. The BEDtools intersect program was used to evaluate regions overlapped by YY1 and the histone modification marks. Enrichment profiles of selected genes were visualized using the UCSC genome browser. Additional bioinformatics analyses were performed to determine β-cell–specific superenhancers based on H3K27ac ChIP-seq signals using the Rank Ordering of Super-Enhancers (ROSE) algorithm as previously described (16). A 12.5-kb distance threshold was used to stitch enhancers together. All the stitched enhancers were ranked according to the ChIP-seq occupancy of H3K27ac signals, which revealed a geometric inflection point and established a cutoff that separated superenhancers from typical enhancers.
Statistics
All of the data were expressed as mean ± SEM with biological replicates (n = 6 unless otherwise specified) performed under the same conditions. Statistical analysis was performed using the two-sided, unpaired Student’s t test for comparing differences between two groups, whereas data from more than two groups were analyzed by ANOVA followed by Tukey’s method for multiple comparisons. Significance was accepted when P < 0.05. Other details of methodologies can be found in Supplementary Methods.
Data and Resource Availability
The data sets (RNA-Seq and ChIP-Seq) generated from the current study are deposited under NCBI Bioproject (ID: PRJNA806704).
Results
Expression Level of β-Cell YY1 Increases After Birth
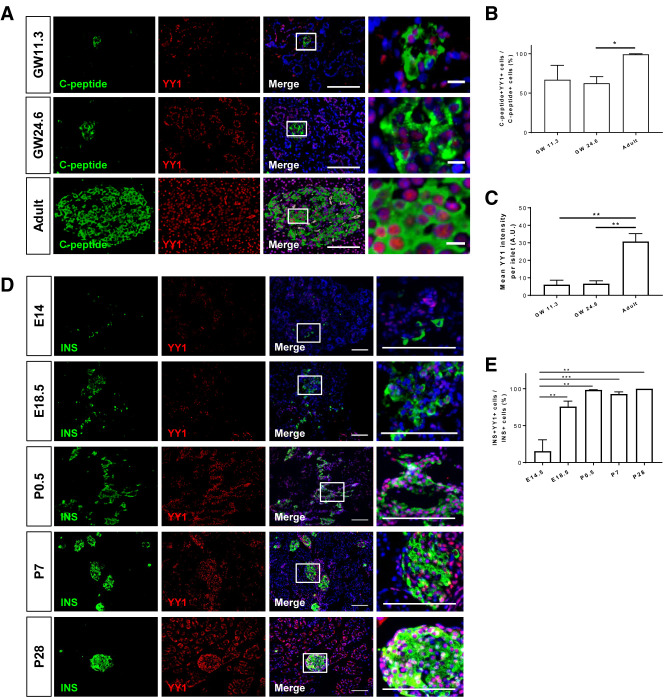
To investigate the functional role of YY1 during pancreatic β-cell development, we first studied its expression profile by collecting human fetal pancreata at gestational weeks 11.3 and 24.6 from aborted fetuses, and pancreatic tissue from adult patients. Immunostaining for C-peptide and YY1 on frozen pancreatic sections (Fig. 1A) demonstrated that the percentage (Fig. 1B) and expression level (Fig. 1C) of YY1 in C-peptide+ β-cells were significantly higher in the adult than in the fetal stage during human pregnancy. To better understand its expression profile during embryonic and neonatal development until adulthood, we collected pancreata from C57Bl/6 mice at embryonic day (E) 14.5 and E18.5, and at postnatal day (P) 0.5, P7, and P28. Immunostaining for INS and YY1 on frozen pancreatic sections revealed that β-cell–specific expression of YY1 became more apparent at E18.5, peaked at P0.5, and was maintained at high level in neonates and adults, with almost every INS+ cell was YY1+ (Fig. 1D and E). Given that the expression levels of YY1 in β-cells were significantly lower during prenatal development in both human and mouse, our findings might suggest that YY1 could be more important for β-cell function in the postnatal pancreas.
Figure 1.
Expression of YY1 in human and mouse pancreatic β-cells becomes more apparent after birth. (A) Immunostaining for C-peptide (green) and YY1 (red) with nuclear Hoechst 33342 counterstain (blue) in frozen sections collected from human pancreata at gestation week (GW) 11.3 and GW24.6 as well as at the adult stage. Scale bars: 100 μm. Squares denote magnified regions on the right (scale bars: 10 μm). (B and C) Quantification of (A) showing that (B) the number of YY1-expressing C-peptide+ cells and (C) the expression levels of YY1 in C-peptide+ cells are significantly higher in adult than fetal β-cells in humans (n = 3). (D) Immunostaining for INS (green) and YY1 (red) with nuclear Hoechst counterstain (blue) in frozen sections collected from C57Bl/6 pancreata at E14 and E18.5, as well as at P0.5, P7, and P28. Scale bars: 100 μm. Squares denote magnified regions on the right. Scale bars: 10 μm. (E) Quantification of (D) showing that the number of YY1-expressing INS+ cells is significantly increased shortly before birth and is maintained at a high level until the adulthood. (B, C, and E) Data are presented as mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001; n = 3–10 mice per group. AU, arbitrary unit.
Expression of YY1 in β-Cells Is Essential for the Maintenance of Glucose Homeostasis in Neonates and Adults
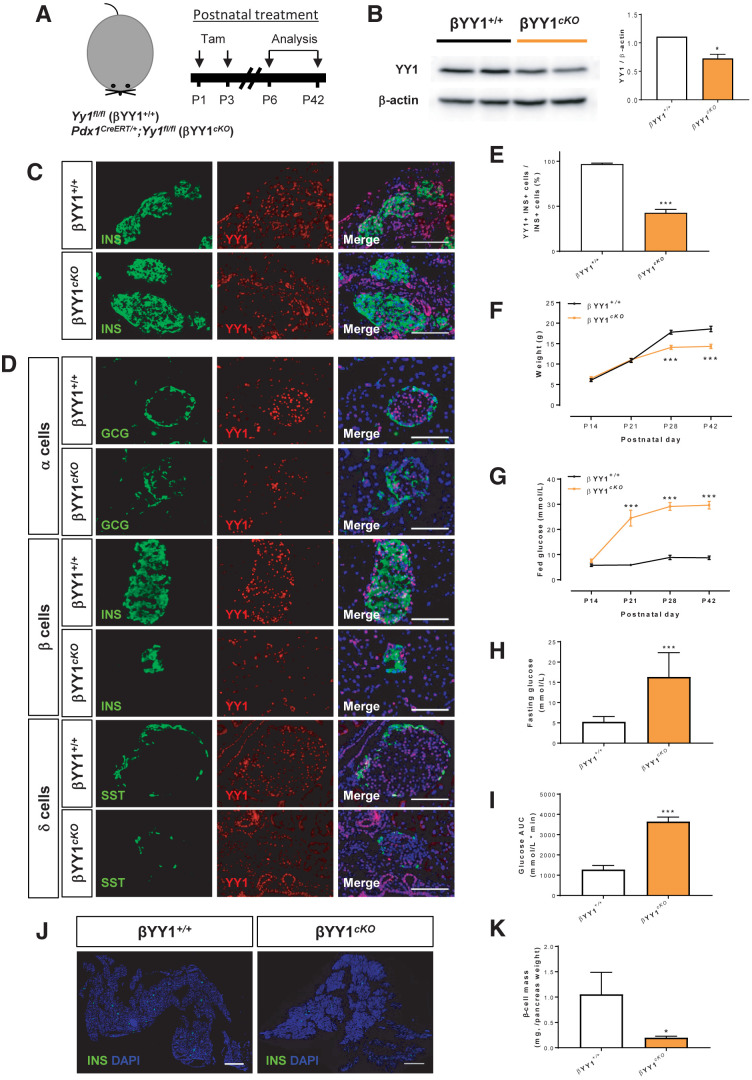
Next, we attempted to study β-cell function in the neonatal and adult pancreas. Pdx1 has been reported as a marker for tracing mature β-cells after birth (17–19). In particular, the Pdx1CreER mouse strain has been used to enable a more acute, tamoxifen (Tam)-induced manipulation of transgene expression in β-cells (20). Therefore, we generated a conditional knockout mouse (cKO) line, Pdx1CreERT/+;Yy1fl/fl, by crossing Pdx1CreERT/+ with Yy1fl/fl (control). In cKO mice, Yy1 expression in mature β-cells can be temporally ablated because Cre recombinase is only activated in β-cells to remove the Yy1 cassette via loxP recognition at specific time points after Tam induction.
We first treated cKO mice with Tam at P1 and P3 to deplete YY1 in neonatal β-cells, and we analyzed data from P6 to P42 (Fig. 2A). At 3 days after Tam, Western blotting revealed that the expression levels of YY1 were significantly reduced in pancreatic islets of cKO mice compared with their control littermates (Fig. 2B). Moreover, immunostaining of frozen pancreatic sections revealed reduced numbers of YY1+INS+ β-cells in the P6 cKO compared with control islets (Fig. 2C). At P42, immunostaining of frozen pancreatic sections for α-, β-, and δ- cell–specific markers, glucagon (GCG), INS, and somatostatin (SST), respectively, showed that ablation of YY1 was more elusive to INS+ β-cells of the cKO mice (Fig. 2D). Specifically, approximately 50% of INS+ β-cells were devoid of YY1 (Fig. 2E). We reasoned that the knockout efficiency was not very high likely due to replication of INS+YY1+ β-cells during neonatal pancreas development. Nevertheless, body weight was significantly reduced starting at P28 (Fig. 2F) and fed blood glucose levels significantly increased starting at P21 (Fig. 2G) in cKO compared with these measures in control littermates. Moreover, fasting blood glucose levels were significantly elevated in cKO mice (Fig. 2H), and intraperitoneal glucose tolerance testing also demonstrated significantly impaired glucose tolerance in cKO mice compared with control mice (Fig. 2I) at P42. The hyperglycemic phenotype appeared to become exacerbated with age, suggesting a progressive loss of β-cell function. Therefore, we quantified β-cell mass by immunostaining for INS, which revealed a significant reduction in β-cell mass in cKO mice relative to control mice at P42 (Fig. 2J and K).
Figure 2.
YY1 regulates glucose homeostasis in neonatal β-cells. (A) Schematic diagram showing the experimental strategy using neonatal Pdx1CreERT/+;Yy1fl/fl mice. Tam was administered at P1 and P3. (B) Western blotting showing a significant reduction in islet YY1 expression in Pdx1CreERT/+;Yy1fl/fl mice, compared with Yy1fl/fl (control) mice, at day 3 after Tam treatment. Data are presented as mean ± SEM; *P < 0.05; n = 9 mice per group; islets of three pancreata were pooled for each band. (C and D) Immunostaining of frozen pancreatic sections for GCG (green), INS (green), or SST (green), and YY1 (red) with nuclear Hoechst counterstain (blue) at (C) 3 days or (D) 6 weeks after Tam treatment, showing specific knockout of YY1 in INS+ β-cells. Scale bars: 100 μm. (E) Quantification of (D) showing at least 50% of INS+ cells were devoid of YY1 at 6 weeks after Tam treatment. (F–I) The physiological change in mice was recorded in terms of (F) body weight, (G) fed blood glucose levels, (H) fasting blood glucose levels, and (I) glucose tolerance test at 2–6 weeks after Tam treatment, showing the onset of hyperglycemia and impaired glucose tolerance after β-cell loss of YY1. Data are presented as mean ± SEM; ***P < 0.001 compared with that of the control group; n = 9 mice for control and n = 9–12 mice for the cKO group. (J) Immunostaining of frozen pancreatic sections for INS (green) with nuclear Hoechst counterstain (blue) at 6 weeks after Tam administration. Scale bars: 2,000 μm. (K) Quantification of (J) showing significantly reduced β-cell mass (INS+ area/total pancreas area × pancreas weight) at 6 weeks after Tam treatment. Data are presented as mean ± SEM; *P < 0.05; n = 3 mice per group.
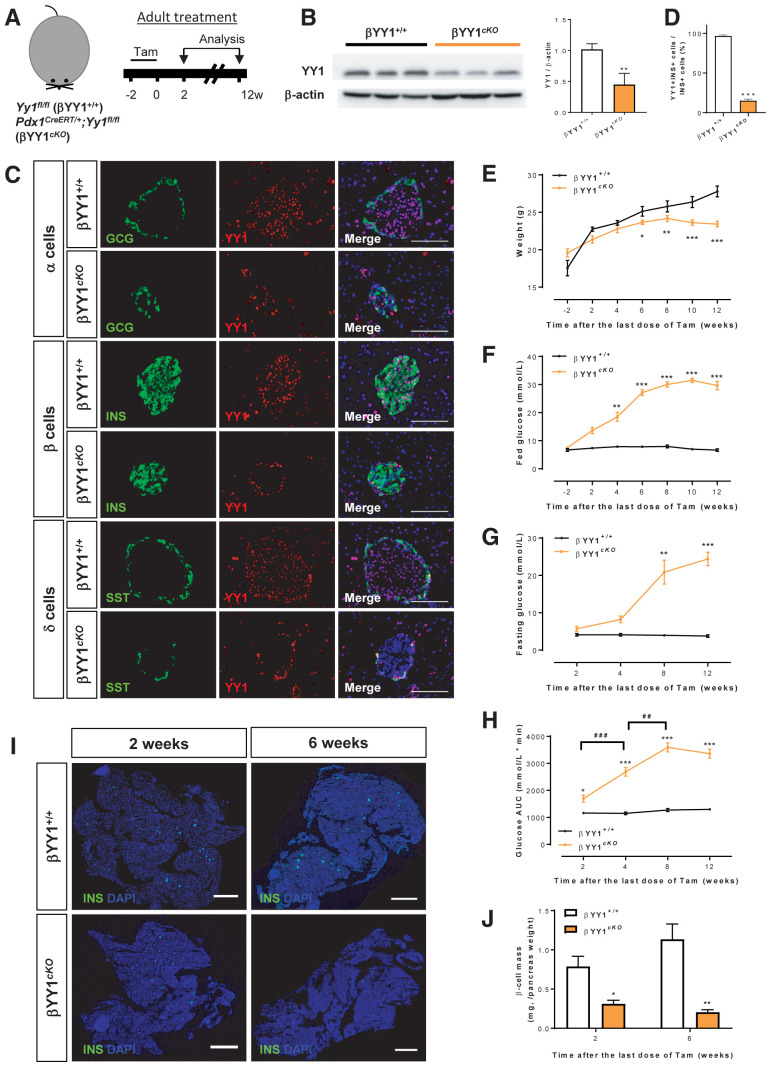
Furthermore, we treated 6–8 week-old cKO mice with Tam to specifically deplete YY1 in adult β-cells, and we analyzed data at 2 to 12 weeks after Tam administration (Fig. 3A). After the washout period at 2 weeks after Tam administration, Western blotting revealed significantly reduced expression levels of YY1 in pancreatic islets of cKO mice compared with control mice (Fig. 3B). Immunostaining of frozen pancreatic sections for GCG, INS, and SST showed that YY1 depletion was exclusive to INS+ β-cells of cKO mice at 2 weeks after administering Tam (Fig. 3C). Specifically, approximately 90% of INS+ β-cells were devoid of YY1 (Fig. 3D). Compared with the control littermates, the body weight of cKO mice was significantly reduced starting at 6 weeks after Tam treatment (Fig. 3E), fed blood glucose levels were significantly increased starting at 4 weeks after Tam treatment (Fig. 3F), and fasting blood glucose levels were significantly increased at 8 weeks after Tam (Fig. 3G). Intraperitoneal glucose tolerance testing also demonstrated significantly impaired glucose tolerance in cKO mice compared with control mice starting at 2 weeks after administering Tam (Fig. 3H). Quantification of β-cell mass by immunostaining for INS (Fig. 3I) revealed a rapid reduction by approximately 30% in cKO mice at 2 weeks after Tam administration, and at 6 weeks, that was significantly reduced by approximately 70%–80%, compared with the respective control mice (Fig. 3J). Altogether, our findings uncovered that β-cell loss of Yy1 resulted in an early onset of diabetes manifested as hyperglycemia, glucose intolerance, and progressive loss of β-cell mass in both neonates and adults.
Figure 3.
YY1 regulates glucose homeostasis in adult β-cells. (A) Schematic diagram showing the experimental strategy using adult Pdx1CreERT/+;Yy1fl/fl mice. Tam was administered within 2 weeks and analyses were performed at 2 weeks after Tam treatment; this was the washout period. (B) Western blotting showing a significant reduction in islet YY1 expression in Pdx1CreERT/+;Yy1fl/fl compared with Yy1fl/fl (control) mice at 2 weeks after Tam administration. Data are presented as mean ± SEM; **P < 0.01; n = 9 mice per group; islets of three pancreata were pooled for each band. (C) Immunostaining of frozen pancreatic sections for GCG (green), INS (green), or SST (green) and YY1 (red) with nuclear Hoechst counterstain (blue) at 2 weeks after Tam treatment showing specific knockout of YY1 in INS+ β-cells. Scale bars: 100 μm. (D) Quantification of (C) showing at least 90% of INS+ β-cells were YY1 deficient. Data are presented as mean ± SEM; ***P < 0.001; n = 9 mice per group. (E–H) The physiological change in mice was recorded in terms of (E) body weight, (F) fed blood glucose levels, (G) fasting blood glucose levels, and (H) glucose tolerance test at 2–12 weeks after Tam treatment, showing the onset of hyperglycemia and impaired glucose tolerance after β-cell loss of YY1. Data are presented as mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001 compared with the control group; ##P < 0.01, ###P < 0.001 compared with other time points; n = 7 mice in the control group and n = 7–13 mice in the cKO group. (I) Immunostaining of frozen pancreatic sections for INS (green) with nuclear Hoechst counterstain (blue) at 2 and 6 weeks after Tam administration, respectively. Scale bars: 2,000 μm. (J) Quantification of (I) showing significantly reduced β-cell mass at 2 and 12 weeks after Tam administration, respectively. Data are presented as mean ± SEM; *P < 0.05, **P < 0.01; n = 3–4 mice per group.
Genome-Wide Transcriptional Profiling Reveals Impaired INS Signaling and Glucose Responsiveness After β-Cell Loss of Yy1
We further evaluated the potential mechanisms by which YY1 regulates adult pancreatic β-cell function by performing genome-wide bulk RNA-seq with islets purified from cKO and control mice. The observed diabetic phenotype in cKO mice could be directly caused by the ablation of Yy1 or secondary to the loss of β-cell mass. To distinguish these possibilities, RNA-seq was performed at 2 weeks after Tam administration—the washout period—during which hyperglycemia was not detected. Although fewer (Supplementary Fig. 1A and C) and smaller (Supplementary Fig. 1E) INS+ β-cells were observed at this time point, the percentage of GCG+ α-cells (Supplementary Fig. 1A and B), the GCG+ α to INS+ β-cell ratio (Supplementary Fig. 1A and D), as well as the percentage of SST+ δ-cells (Supplementary Fig. 1A and F) in cKO islets were not different from that of the controls. Therefore, there was no difference in the gross cellular composition of cKO and control islets at 2 weeks after Tam treatment.
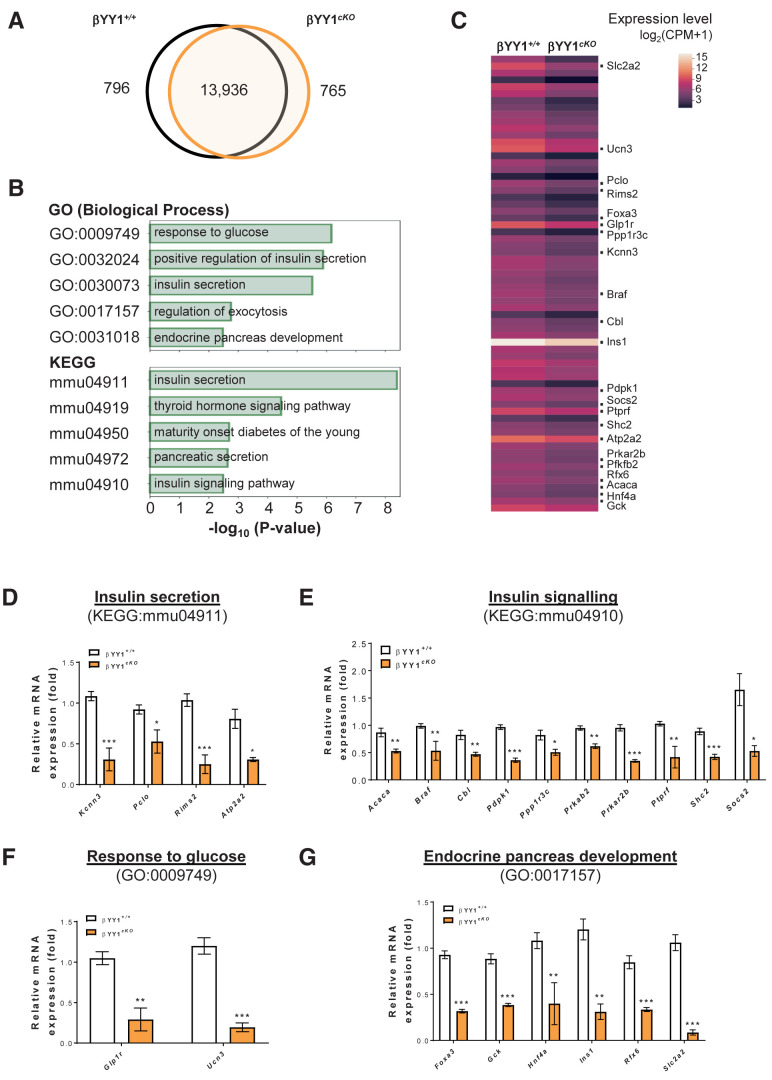
From the RNA-seq analyses, we found a total of 15,473 and 15,347 genes in cKO and control islets, respectively (Fig. 4A) and analyzed 1,561 differentially expressed genes in cKO islets compared with that in the control islets. Of those 1,561 genes, 796 were downregulated and functionally annotated by pathway analyses. Consistent with the observed β-cell dysfunction, our results showed that the top 5 pathways that were significantly downregulated in cKO islets, as demonstrated in terms of biological process by Gene Ontology (GO) enrichment analysis and by Kyoto encyclopedia of genes and genomes (KEGG) analysis, were associated with INS secretion, INS signaling, response to glucose, and endocrine cell development (Fig. 4B; Supplementary Tables 3 and 4). The differentially expressed genes of these pathways were further demonstrated by heat mapping (Fig. 4C).
Figure 4.
Genome-wide RNA-seq profiling reveals impaired glucose response and INS signaling after β-cell loss of Yy1. Tam was administered to adult Pdx1CreERT/+;Yy1fl/fl mice within 2 weeks of the start of the experiment, and analyses were performed at 2 weeks after Tam treatment. (A) Venn diagram illustrating transcripts that are significantly differentially expressed (log2fc ≥1 or log2fc ≤−1; false discovery rate–adjusted P value ≤0.05) in islets isolated from β-cell Yy1-deficient mice (i.e., cKO mice) compared with that of the Yy1fl/fl (control) mice. A total of 1,561 transcripts were altered in abundance in vivo, including 796 downregulated and 765 upregulated genes in cKO compared with control islets. (B) The top 5 most significant pathways by GO in terms of biological processes or KEGG pathways determined by downregulated genes expressed by the pancreatic islets of cKO compared with that of control mice. (C) Heat map of selected differentially expressed genes as determined by GO and KEGG functional clustering from (B). The red scale bar denotes the log2 expression level of genes in each group. (D–G) Quantitative RT-qPCR of the highlighted genes in terms of (D) INS secretion, (E) INS signaling, (F) response to glucose, and (G) endocrine pancreas development with reference to (C). Gene expression levels in cKO islets were compared with that of the control islets. Data are presented as mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001; n = 6 mice per group.
To validate our RNA-seq data, we purified islets from control and cKO mice at 2 weeks after Tam treatment and confirmed expression levels of the highlighted genes in Fig. 4C by quantitative RT-qPCR. In terms of INS secretion, the expression levels of Kcnn3, Pclo, Rims2, and Alp2a2 were significantly downregulated in cKO compared with control islets (Fig. 4D). In terms of INS signaling, the expression levels of Acaca, Braf, Cbl, Pdpk1, Ppp1r3c, Prkab2, Prkar2b, Ptprf, Shc2, and Socs2 were significantly reduced in cKO compared with control islets (Fig. 4E). In terms of response to glucose, the expression levels of Glp1r and Ucn3 were significantly lessened (Fig. 4F), and in terms of endocrine cell development, the expression levels of Foxa3, Gck, Hnf4a, Ins1, Rfx6, and Slc2a2 were significantly decreased in cKO compared with control islets (Fig. 4G). We also examined gene expression essential for β-cell identity, including Pdx1, Nkx6.1, and Ngn3, and β-cell maturation, such as Npy and Ucn3, by RT-qPCR and found that the expression levels of Nkx6.1, Npy, and Ucn3 were significantly reduced in cKO compared with control islets (Supplementary Fig. 1G).
From the RNA-seq data, we also found 765 genes that were upregulated in cKO islets and not in control islets, and these were further functionally annotated by pathway analyses. Compared with the controls, the top 3 significantly upregulated pathways in cKO islets, as demonstrated in terms of biological process by GO enrichment and KEGG analyses, were associated with immune signaling, inflammation, and cytokine response (Supplementary Fig. 2A, Supplementary Tables 5 and 6). To validate whether β-cell loss of Yy1 led to islet inflammation, we purified islets from control and cKO mice for RT-qPCR examination of cytokine expression, including pro-Il1b, pro-Il2, pro-Il6, pro-Il17, pro-Ifng and pro-Tnf-α; and anti-Il10 and anti-Tgf-β inflammatory cytokines that are often examined to indicate tissue inflammation (21,22). Although there was no significant difference in the expression levels of most of these genes, Il2 expression was significantly reduced, whereas Il17 expression was significantly increased in cKO islets relative to control islets (Supplementary Fig. 2B).
Because IL-17 is specifically produced by CD4+ T-helper 17 cells, we also performed immunostaining for CD3 and INS in frozen pancreatic sections and found no significant infiltration of CD3+ T cells in the cKO islets compared with control islets at 2 weeks and 12 weeks after Tam administration, respectively (Supplementary Fig. 2C). Moreover, flow cytometric analyses at 4 weeks after Tam administration (Supplementary Fig. 2D) further confirmed there was no significant increase in the percentage of CD3+CD4+ helper T cells (Supplementary Fig. 2E), CD3+CD8+ cytotoxic T cells (Supplementary Fig. 2F), or CD11b+F4/80+ macrophages (Supplementary Fig. 2G) among total CD45+ leukocytes in the cKO islets compared with control islets. Therefore, the early onset of diabetes observed after β-cell loss of Yy1 did not appear to be resulted directly from islet inflammation.
YY1 Ablation Does Not Directly Contribute to β-Cell Death
One of the acknowledged functions of YY1 is its regulation of cancer cell survival (9). To determine if β-cell dysfunction after YY1 ablation resulted from increased cell death, we performed immunostaining for INS and cleaved caspase 3 (cCASP3) in frozen pancreatic sections collected at 2 weeks after Tam treatment. INS+cCASP3+ cells were hardly detected in cKO and control islets (Supplementary Fig. 3A), and the number of cCAPS3+ cells per cKO and control islet was not significantly different (Supplementary Fig. 3B). To determine if β-cell dysfunction resulted from increased DNA damage, we also performed immunostaining for INS and phospho(p)-γH2AX, which marks chromatin areas flanking double-stranded DNA breaks (20). Our results revealed no staining of p-γH2AX in INS+ β-cells in either cKO or control islets (Supplementary Fig. 3C). We also performed Western blotting at 2 weeks after Tam administration to score the expression levels and overall activities of proteins involved in the PI3K/AKT/mTOR pathway that regulates cell survival (Supplementary Fig. 3D and E). Compared with the control islets, the expression levels of YY1 were significantly reduced in cKO islets (Supplementary Fig. 3F). However, the levels of PI3K expression (Supplementary Fig. 3G); AKT expression, phosphorylation at Ser 473, and overall activities (i.e., p-AKT/AKT; Supplementary Fig. 3H); mTOR expression, phosphorylation at Ser 2446, and overall activities (i.e., p-mTOR/mTOR; Supplementary Fig. 3I); as well as ribosomal protein S6 (rpS6) expression, phosphorylation at Ser 240/244, and overall activities (i.e., p-rpS6/rpS6; Supplementary Fig. 3J) were not significantly different between cKO and control islets. Altogether, our data suggested that the early onset of diabetes as a result of YY1 ablation in β-cells was not associated with apoptosis or DNA damage in β-cells.
YY1 Ablation Does Not Directly Contribute to Mitochondrial Dysfunction in β-Cells
A recent report documented that YY1 ablation in β-cells via a constitutively active Cre recombinase driven under the control of mouse (MipCre) or rat (RipCre) INS promoter leads to impaired INS secretion and mitochondrial dysfunction in β-cells (23). Therefore, we conducted a study to see if mitochondrial dysfunction is detected at 2 weeks after Tam treatment in Pdx1CreER;YY1fl/fl mice. Immunostaining for INS and MitoSOX Red did not reveal a significant difference in the accumulation of mitochondrial-specific reactive oxygen species in β-cells of cKO and control mice (Supplementary Fig. 4A). Tetramethylrhodamine, ethyl ester immunostaining (Supplementary Fig. 4B) or flow cytometry (Supplementary Fig. 4C), respectively, revealed no significant difference in the mitochondrial membrane potential of INS+ β-cells or isolated islets of cKO and control mice. RT-qPCR analyses also showed no significant difference in the expression levels of the mitochondrial gene cytochrome C (Cycs) in cKO and control islets (Supplementary Fig. 4D). In fact, both INS deficiency and resistance can alter mitochondrial function (24). Western blotting showed significantly reduced cytochrome C expression in the cKO islets compared with control islets at 6 weeks after Tam treatment (Supplementary Fig. 4E), indicating that mitochondrial damage was likely a secondary effect after prolonged β-cell dysfunction.
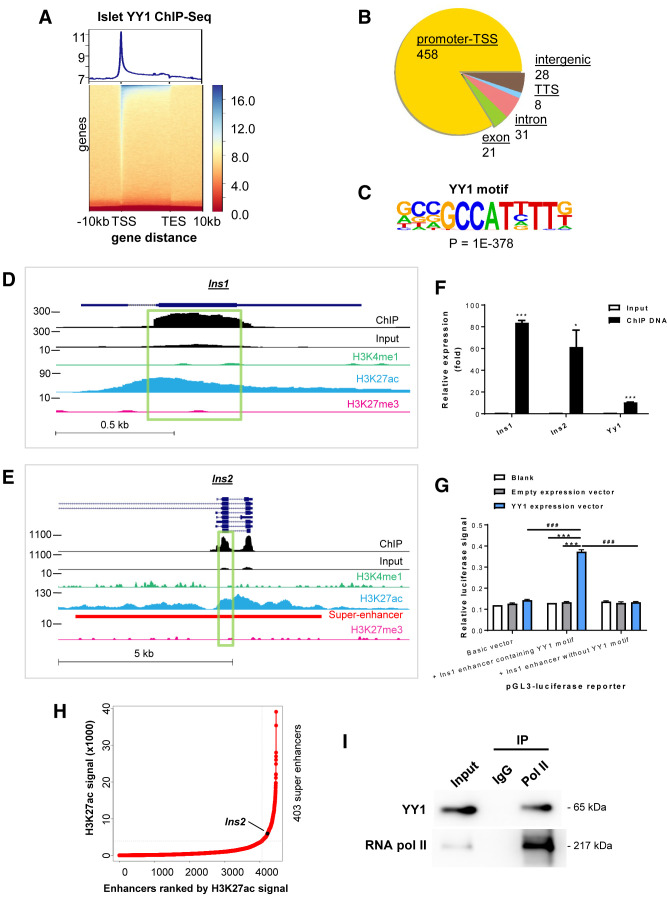
Integrative RNA-seq and ChIP-seq Analyses Reveal Ins1 and Ins2 as Direct Targets of YY1 in Pancreatic β-Cells
To further determine the direct targets of YY1 that regulate pancreatic β-cell function, we performed ChIP-seq analysis of mouse islets (Fig. 5A). We found that YY1 predominantly bound to the promoter regions or TSS of the genome (Fig. 5B). Consistent with the motif reported in muscle cells (25) and B lymphocytes (26), our de novo motif analysis also identified GCCAT as the core motif sequence in pancreatic β-cells (Fig. 5C) that was preferably bound by YY1 in 69.17% of peaks identified on its gene targets (P = 1 × 10−378) (Supplementary Table 7). There was a total of 546 peaks, 319 of which were from protein-coding transcripts and the remaining 227 were from noncoding elements. We focused on the protein-coding peaks that were found on 317 genes for subsequent analyses. To shortlist and identify potential target genes, we also performed integrative analyses using raw data from both YY1 ChIP-seq and RNA-seq of control and cKO islets isolated at 2 weeks after Tam administration to determine the regulatory networks of YY1 in maintaining β-cell function in vivo.
Figure 5.
Integrative transcriptomic, genetic, and epigenetic analyses demonstrate transcriptional activation of the INS genes by YY1 by stabilizing the enhancer–promoter interaction. (A) Heat map representing the normalized YY1 ChIP-seq intensities ± 10 kb over the gene body from TSS to transcription end site (TES) of YY1-bound DNA in pancreatic islets. The upper panel shows an enrichment plot representing the average distribution of YY1 intensities ± 10 kb over the gene body. (B) Typical peak annotation pie chart shows that the majority of the peaks fall into promoter/TSS regions. (C) De novo motif discovery using all the 546 peaks of YY1 ChIP-seq data. The motif of β-cells was compared with the available consensus and optimal motifs of other cell types in the indicated references. (D and E) Genome snapshots for YY1, H3K4me1, H3K27ac, and H3K27me3 ChIP-seq analyses performed in pancreatic islets at the (D) Ins1 and (E) Ins2 loci. The superenhancer region was also highlighted in Ins2. (F) ChIP-qPCR showing the enrichment of YY1 on exon 2 of Ins1 and Ins2. Chromatin of Ins-1 cells was immunoprecipitated using an anti-YY1 antibody. Input served as nonimmunoprecipitated controls. The relative enrichment values were normalized to β-actin because there is no YY1 binding site on this housekeeping gene. Data are presented as mean ± SEM; *P < 0.05, ***P < 0.001; n = 3 independent experiments. (G) Luciferase report assays showing that the luciferase signals driven under the control of a GCCAT-containing Ins1 enhancer were significantly increased compared with that driven under the Ins1 enhancer without the motif after forced expression of YY1 in Ins-1 cells. Data are presented as mean ± SEM; ***P < 0.001 compared with blank or empty expression vector; ###P < 0.001 compared with luciferase reporter containing Ins1 enhancer without YY1 binding motif; n = 3 independent experiments. (H) A 12.5 kb distance threshold was used to stitch islet-specific enhancers together, and the stitched enhancers based on H3K27ac ChIP-seq signals were ranked using the ROSE algorithm. Superenhancer of Ins is indicated. (I) Western blotting analyses of co-immunoprecipitation of YY1 and RNA pol II from nuclear extract of pancreatic islets using anti-RNA pol II antibody. IgG was used in control immunoprecipitation, and the amount of input loaded for immunoprecipitation was 2%.
We identified 37 differentially expressed protein-coding genes from our RNA-seq data that showed binding peaks of YY1, 36 of which displayed the canonical core motif sequence as determined by ChIP-seq. Among these genes, 7 were upregulated and 30 were downregulated in cKO islets, indicating that YY1 could act as both a repressor and an activator for regulating gene expression. With reference to the downregulated genes after YY1 depletion in β-cells that were involved in INS secretion, INS signaling, response to glucose, and endocrine pancreas development (Fig. 4D–G, and Supplementary Fig. 1G), ChIP-seq analyses showed that YY1 directly bound to the DNA regions of Rims2, Pdpk1, and Ins1 (Supplementary Fig. 5).
Next, we performed enrichment analyses with global histone modification profiling targeting H3K4me1 for primed enhancers, H3K27ac for active enhancers, and H3K27me3 for poised enhancers in pancreatic β-cells, as previously described (27). By combining the data from RNA-seq, YY1 ChIP-seq and histone H3 ChIP-seq analyses of pancreatic islets, we found that the Ins1 and Ins2 genes were potentially regulated by YY1, as evident by their reduced mRNA expression in islets after β-cell depletion of YY1, compared with that of the control, and that YY1 bound to an enhancer region on exon 2 of Ins1 (Fig. 5D) and on exons 2 and 3 of Ins2 (Fig. 5E). Of note, the exon 2 of Ins1 and Ins2 contained the core motif sequence, whereas exon 3 of Ins2 did not have the core motif sequence, suggesting a noncanonical binding site. Moreover, the YY1 binding peaks of these genes were enriched for the active H3K27ac but not the poised H3K27me3 marks (Fig. 5D and E).
Bioinformatics analysis showed that the YY1 binding enhancer regions of Ins1 (Supplementary Fig. 6A) and Ins2 (Supplementary Fig. 6B) were conserved across mouse and rat. Therefore, we performed YY1 ChIP-qPCR using a rat pancreatic β-cell line, Ins-1, to validate these bindings that respectively showed YY1 enrichment on exon 2 of the Ins1 and Ins2 genes and on the Yy1 gene, which is a known YY1 target gene (Fig. 5F). Therefore, our data showed that Ins1 and Ins2 were YY1-specific gene targets with β-cell–specific active enhancers. We also confirmed the function of YY1 in the regulation of Ins1 transcription in Ins-1 β-cells through luciferase reporter assays. The luciferase signals driven under the control of a GCCAT-containing Ins1 enhancer were significantly increased compared with that driven under the Ins1 enhancer without the core motif sequence after YY1 overexpression (Fig. 5G), suggesting a direct regulation of Ins1 expression in pancreatic β-cells by YY1. Additional bioinformatics analyses were performed to determine islet-specific superenhancers based on H3K27ac ChIP-seq signals within 12.5 kb genomic regions containing enhancers, as previously described (16). A total of 403 superenhancers were identified in pancreatic islets including YY1 binding sites on the superenhancer region of Ins2 (Fig. 5E and H).
To determine how YY1 might regulate gene transcription in β-cells, we performed co-immunoprecipitation assays that revealed direct binding of YY1 and RNA pol II in isolated pancreatic islets (Fig. 5I), suggesting the formation of a YY1–RNA pol II protein complex potentially facilitating the enhancer–promoter interaction during gene transcription. Altogether, our results indicated that YY1 could regulate INS biosynthesis by directly binding to the enhancer regions of its target genes and to RNA pol II, activating the transcription of the Ins1 and Ins2 genes in pancreatic β-cells.
YY1 Regulates Glucose Homeostasis by Controlling INS Production in β-Cells
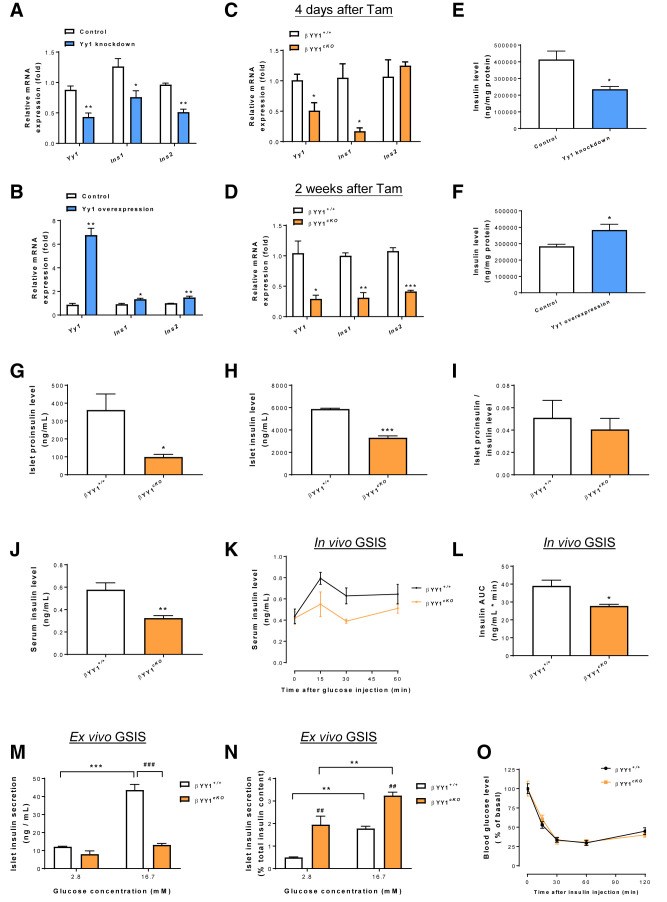
To further prove that YY1 regulates Ins1 and Ins2 gene transcription, we first knocked down and upregulated Yy1 expression in Ins-1 cells using siRNA and overexpression plasmid, respectively, and scored their gene expression levels by RT-qPCR. At 48 hours after treatment with siRNA and overexpression plasmids, the expression levels of Yy1, Ins1, and Ins2 were significantly reduced (Fig. 6A) and increased (Fig. 6B), respectively. We also validated these findings in vivo by purifying pancreatic islets from cKO and control mice at 4 days and 2 weeks after Tam treatment, respectively. RT-qPCR data showed that the gene expression levels of Ins1 were significantly reduced at both 4 days (Fig. 6C) and 2 weeks (Fig. 6D) after Tam treatment, and that of Ins2 were significantly downregulated at 2 weeks after Tam treatment (Fig. 6D) in cKO islets compared with control islets. These data further indicate that YY1 is a transcriptional activator of Ins1 and Ins2 in pancreatic β-cells both in vitro and in vivo.
Figure 6.
YY1 regulates glucose homeostasis by controlling INS production in β-cells. (A–D) Quantitative RT-qPCR analyses in (A and B) pancreatic β-cells in vitro or (C and D) pancreatic islets in vivo. (A and B) Compared with the control groups, significantly (A) reduced and (B) increased transcription levels of Ins1 and Ins2 were observed at 48 hours after treatment with Yy1 siRNA and overexpression vector, respectively, in pancreatic β-cells. (C and D) Compared with the Yy1fl/fl (control) groups, significantly reduced transcription levels of Ins1 and/or Ins2 were also found at (C) 4 days or (D) 2 weeks after Tam treatment in adult Pdx1CreERT/+;Yy1fl/fl (cKO) islets. (E and F) ELISA assays showing reduced and increased INS levels in pancreatic β-cells after stable knockdown of Yy1 for 96 hours and overexpression of Yy1 for 72 hours, respectively, compared with that of the control groups. (G–J) ELISA assays showing significantly (G) reduced islet proinsulin levels, (H) reduced islet INS levels in size-matched cKO islets, or (J) reduced serum INS levels in the circulating blood of cKO mice at 2 weeks after Tam administration compared with that of the control mice. (I) No significant difference in islet proinsulin/INS levels was found. (K and L) In vivo GSIS assays showing significantly impaired serum INS response to intraperitoneal (i.p.) glucose injection at 2 mg/g body weight in cKO compared with control mice at 2 weeks after Tam treatment. (A–L) Data are presented as mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001; n = 3 independent experiments in vitro or n = 4 mice per group in vivo. (M and N) Quantification of INS secretion from size-matched pancreatic islets in response to 2.8 and 16.7 mmol/L glucose showing (M) significantly impaired or (N) enhanced ex vivo GSIS in cKO islets at 2 weeks after Tam treatment without or with reference to total INS content, respectively. Data are presented as mean ± SEM; **P < 0.01, ***P < 0.001 compared within the same types of islets; ##P < 0.01, ###P < 0.001 compared between cKO and control islets; n = 4 mice per group per condition. (O) In vivo intention-to-treat assays showing no difference in INS sensitivity after i.p. INS injection at 0.5 units/kg body weight between the cKO and control groups at 2 weeks after Tam treatment. AUC, area under the curve.
To further validate the regulatory role of YY1 in INS biosynthesis, we measured INS levels by ELISA and found treatment with lentivirus-mediated Yy1 knockdown for 96 hours significantly reduced INS levels in β-cells in vitro (Fig. 6E), whereas treatment with Yy1 overexpression plasmid for 72 hours significantly increased INS levels (Fig. 6F). Moreover, we evaluated the proinsulin and INS levels in size-matched islets by ELISA and found significantly reduced proinsulin (Fig. 6G) and INS (Fig. 6H) levels in cKO islets compared with control islets at 2 weeks after Tam administration. There was no difference in the proinsulin to INS levels of cKO and control islets (Fig. 6I), indicating that reduced INS levels were not due to impaired INS processing. Moreover, we detected significantly reduced serum INS levels at 2 weeks after Tam administration in cKO mice compared with control mice (Fig. 6J).
To further evaluate whether YY1 regulates INS secretion in response to glucose challenge, we performed glucose-sensing INS secretion (GSIS) assays in cKO and control mice at 2 weeks after Tam treatment. In vivo GSIS assays demonstrated significantly reduced serum INS levels after glucose challenge in cKO mice compared with control mice (Fig. 6K and L). However, we could not use these data to distinguish between reduced INS production and reduced INS secretion in cKO mice. Therefore, we also performed ex vivo GSIS assays using size-matched islets isolated from cKO and control mice. Although the control islets significantly increased INS secretion at a high glucose concentration, the cKO islets did not demonstrate a significant difference from control islets in terms of in INS secretion at low and high glucose concentrations (Fig. 6M), consistent with the in vivo GSIS results. Nonetheless, if we measured INS secretion normalized to the total INS content, both the cKO and control islets responded to the increasing glucose concentrations by increasing INS secretion at a high glucose concentration (Fig. 6N), suggesting that there was no defect in INS secretion after glucose challenge to the cKO islets. In fact, the cKO islets secreted more INS than the control islets at both low and high concentrations (Fig. 6N), a characteristic of dysfunctional β-cells (28,29). These results suggested that the cKO islets were unable to produce sufficient INS to respond to the increasing glucose concentration.
Furthermore, we also performed an INS tolerance test that demonstrated no significant difference between the cKO and control groups at 2 weeks after Tam administration (Fig. 6O), indicating that the diabetic phenotype observed in cKO mice was not related to INS resistance. Taken together, our results indicated that YY1 controlled the biosynthesis rather than secretion or sensitivity of INS by regulating the transcription of Ins1 and Ins2 in β-cells.
Discussion
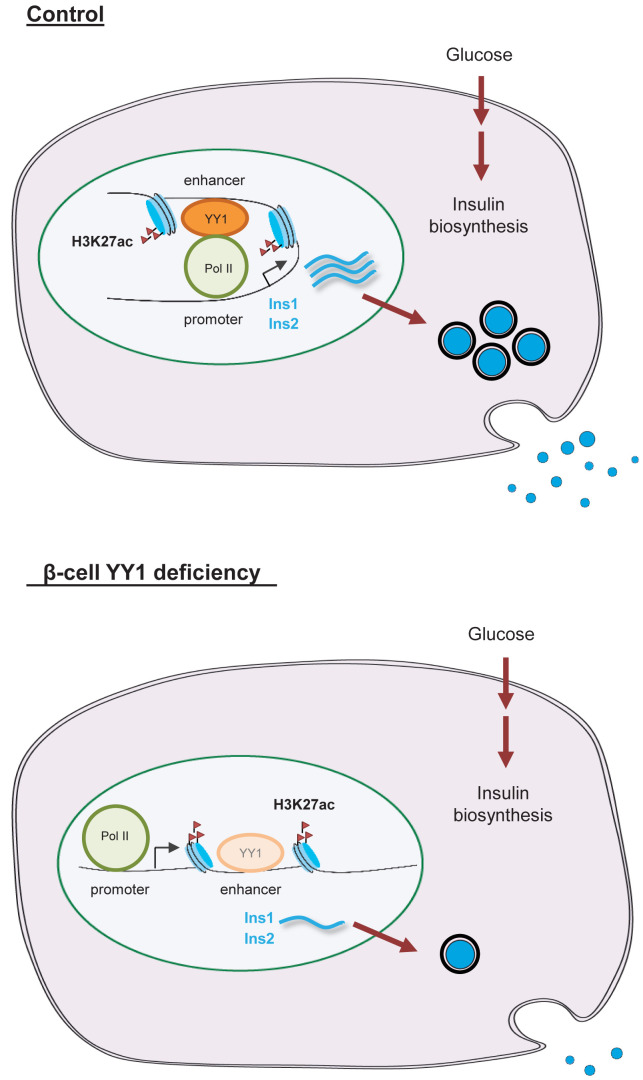
In this study, we found that YY1 was ubiquitously expressed in both human and mouse pancreatic β-cells after birth. Nevertheless, the physiological role of YY1 during β-cell development and the molecular mechanism by which YY1 regulates β-cell function remain largely unknown. By generating the Pdx1CreERT/+;Yy1fl/fl mice, we temporally deleted Yy1 in β-cells during neonatal and adult development. After Tam treatment, these mice developed hyperglycemia, impaired glucose tolerance, and reduced β-cell mass in both neonates and adults. Moreover, they also displayed hypoinsulinemia with normal INS sensitivity compared with the control mice, manifesting as a type 1 diabetic phenotype. Through integrative transcriptomic, genomic, and epigenomic data mining, we revealed an unappreciated role of YY1 as a transcriptional activator of the Ins gene in pancreatic β-cells (Fig. 7).
Figure 7.
A hypothetical model depicting the functional role of YY1 in regulating glucose metabolism in pancreatic β cells. Under homeostasis, YY1 can directly bind to the enhancer regions of Ins1 and Ins2 or to the superenhancer region of Ins2. YY1 can also directly bind to the RNA pol II, potentially stabilizing the enhancer–promoter interaction through the multiprotein-DNA complex formation during gene transcription in pancreatic β-cells after birth. On the other hand, β-cell loss of Yy1 leads to reduced transcription and production of INS. Therefore, cKO mice develop characteristics of type 1 diabetes manifested as hyperglycemia, hypoinsulinemia, and impaired glucose tolerance as a result of impaired INS production. Taken together, YY1 is a novel transcriptional activator of the INS genes in pancreatic β-cells.
Compared with the control islets, our genome-wide RNA-seq analyses showed an early defect in INS secretion, INS signaling, and glucose responsiveness of the cKO islets isolated at 2 weeks after Tam administration. Additional analyses through RT-qPCR, immunostaining, and flow cytometry indicated that impaired GSIS after β-cell loss of Yy1 displayed at this early time point did not directly result from islet inflammation, β-cell death, or mitochondrial dysfunction. In fact, TFs often regulate gene expression through protein–DNA interactions by binding to specific promoter–enhancer regions.
We then performed genome-wide ChIP-seq to identify target genes by identifying the promoter–enhancer regions of genes that showed the YY1 binding peaks. After integrative analyses using islet-specific RNA-seq, YY1 ChIP-seq, and histone ChIP-seq data, we then identified Ins1 and Ins2 as gene targets of YY1 with support from the following evidence: RNA-seq and RT-qPCR, respectively, showed significantly downregulated gene expression levels of Ins1 and Ins2 in cKO islets; ChIP-seq and ChIP-qPCR, respectively, demonstrated the canonical YY1 binding sites with core motif sequences on exon 2 of both Ins1 and Ins2; histone ChIP-seq revealed that the YY1 binding regions of these genes were enriched for the active H3K27ac but not the poised H3K27me3 marks; and H3K27ac enrichment analyses uncovered YY1 binding peaks on the superenhancer region of Ins2 in pancreatic β-cells. Although YY1 did not directly bind to the promoter regions of the INS genes, we further showed that it could directly bind to RNA pol II in pancreatic islets, suggesting the formation of YY1–RNA pol II protein complex, potentially stabilizing the enhancer–promoter interaction during gene transcription.
The novel regulation of YY1 in INS transcription was further supported by luciferase reporter assays that demonstrated significantly enhanced luciferase expression driven by the control of Ins1 enhancer containing the YY1 core motif sequence but not that without the YY1 binding site in rat Ins-1 β-cells after forced expression with YY1. Moreover, β-cells treated with Yy1 siRNA or lentivirus-mediated stable Yy1 knockdown had significantly downregulated gene expression of Ins1 and Ins2 and significantly reduced INS production, whereas β-cells treated with the Yy1 overexpression plasmid had significantly upregulated gene expression of Ins1 and Ins2 and significantly increased INS production. Ex vivo examination of purified pancreatic islets also demonstrated significantly reduced proinsulin and INS levels in cKO islets compared with control islets. Furthermore, results of in vivo and ex vivo GSIS and of in vivo ITT validated that β-cell failure observed in cKO mice after β-cell loss of Yy1 did not result from impaired INS secretion or INS resistance. Therefore, the reduced islet INS production and hypoinsulinemia of cKO mice were directly attributed to mitigated INS biosynthesis after the loss of the transcriptional activator YY1 in pancreatic β-cells.
We were aware of a recent publication that reported impaired INS secretion and β-cell mitochondrial dysfunction after YY1 ablation in MipCre;Yy1fl/fl or RipCre;Yy1fl/fl mice (23). In contrast, we used an ablation model driven by Pdx1CreER, which enables acute and temporal control of Yy1 expression at specific developmental stages, including neonatal and adult stages, coupled with a genome-wide screening approach at the mRNA, DNA, and histone levels that demonstrated YY1 is a transcriptional activator in controlling INS production rather than INS secretion by directly increasing Ins1 and Ins2 transcription in pancreatic β-cells. We believe the mitochondrial damage observed in β-cells devoid of YY1 reported in the previous study was likely a secondary effect following INS deficiency, because INS plays a pivotal role in maintaining mitochondrial biogenesis and function (24). Indeed, at 2 weeks after Tam treatment, when impaired INS synthesis was observed, we did not find a significant difference in the accumulation of mitochondrial reactive oxygen species, the mitochondrial membrane potential, or mitochondrial cytochrome C expression in β-cells of cKO and control islets. Significantly reduced cytochrome C expression was detected at 6 weeks after Tam administration, indicating reduced mitochondrial biogenesis after prolonged INS deficiency. Furthermore, there was no significant difference between the cKO and control islets in the number of cCASP3+ cells or in the expression levels or activities of PI3K/AKT/mTOR signaling that regulate β-cell survival (30,31), suggesting that the early onset of diabetes observed in Pdx1CreER;Yy1fl/fl mice did not directly result from β-cell death. We reasoned that the discrepancy in these findings could be due to the use of a constitutively active Cre recombinase in the Song et al. (23) study that limited examination at specific time points after Yy1 ablation and, therefore, the direct effects of YY1 in regulating β-cell function during development cannot be studied.
Recent lineage tracing experiments have demonstrated that reduced β-cell mass during the progression of diabetes, particularly in type 2 diabetes, can be attributed to β-cell dedifferentiation rather than β-cell death (32). In particular, characteristics of β-cell dedifferentiation include reduced Pdx1 expression, increased progenitor marker gene expression, and increased plasticity of dedifferentiated β-cells leading to conversion of β to other hormone-producing cells (32). In our model, we showed that the gene expression levels of Pdx1 and the endocrine progenitor marker Ngn3 were not significantly different in cKO and control islets at 2 weeks after Tam administration. Moreover, we did not observe increased GCG+ α-cells, SST+ δ-cells, or poised ratio of β-cells to α-cells in cKO than control islets at 2 weeks after Tam administration. Our findings suggested that YY1 might not regulate β-cell identity.
Taken together, in our study, we uncovered a novel transcriptional activator in the regulation of Ins gene expression essential for β-cell development, maturation, and function. In addition to PDX1, NeuroD1, and MafA, previously known as INS transcriptional regulators that bind to the upstream promoter regions of the Ins genes (33), YY1 could also govern glucose homeostasis by directly activating INS transcription through binding to the enhancer regions of the Ins genes and to RNA pol II for potentially stabilizing the enhancer–promoter interaction via the multiprotein-DNA complex during gene transcription. Hence, YY1 could promote INS production in pancreatic β-cells, assisting β-cell development and function after birth. Our findings could advance our understanding of β-cell biology with clinically relevant insights targeting disease-related INS biosynthesis, including diabetes or even cancer.
Article Information
Acknowledgments. We thank Prof. Douglas A. Melton (Harvard University) for the gift of Pdx1CreER.
Funding. This work was supported by the National Natural Science Foundation of China (grants 81922077 and 82070494), Research Grants Council of Hong Kong (grants 14100021, 14108420, C4026-17WF, and M-402-20), Croucher Foundation (Innovation Award), University Grants Committee Research Matching Grant Scheme (2019, 2020, and 2021), Research Committee Funding, Direct Grants, Faculty Innovation Award, Young Researcher Award, and postdoctoral fellowships (K.Y.Y., W.Y.) and a postgraduate studentship (D.L.) from the Chinese University of Hong Kong.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. D.L., K.Y.Y., V.W.C., and W.Y. performed experiments and analyzed the data; D.L., K.K.Y.C. and K.O.L. interpreted the data; C.C.N.C. and C.C.W. recruited human volunteers and provided clinical samples; H.W. and B.Z. provided reagents; and K.O.L. designed experiments, supervised the research, and wrote the manuscript. K.O.L. also is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
D.L. and K.Y.C. contributed equally to this work.
This article contains supplementary material online at https://doi.org/10.2337/figshare.19087874.
References
- 1. Iype T, Francis J, Garmey JC, et al. Mechanism of insulin gene regulation by the pancreatic transcription factor Pdx-1: application of pre-mRNA analysis and chromatin immunoprecipitation to assess formation of functional transcriptional complexes. J Biol Chem 2005;280:16798–16807 [DOI] [PubMed] [Google Scholar]
- 2. Qiu Y, Guo M, Huang S, Stein R. Insulin gene transcription is mediated by interactions between the p300 coactivator and PDX-1, BETA2, and E47. Mol Cell Biol 2002;22:412–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sharma A, Moore M, Marcora E, et al. The NeuroD1/BETA2 sequences essential for insulin gene transcription colocalize with those necessary for neurogenesis and p300/CREB binding protein binding. Mol Cell Biol 1999;19:704–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Olbrot M, Rud J, Moss LG, Sharma A. Identification of β-cell-specific insulin gene transcription factor RIPE3b1 as mammalian MafA. Proc Natl Acad Sci USA 2002;99:6737–6742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Harrington RH, Sharma A. Transcription factors recognizing overlapping C1-A2 binding sites positively regulate insulin gene expression. J Biol Chem 2001;276:104–113 [DOI] [PubMed] [Google Scholar]
- 6. Iacovazzo D, Flanagan SE, Walker E, et al. MAFA missense mutation causes familial insulinomatosis and diabetes mellitus. Proc Natl Acad Sci USA 2018;115:1027–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Oliver-Krasinski JM, Kasner MT, Yang J, et al. The diabetes gene Pdx1 regulates the transcriptional network of pancreatic endocrine progenitor cells in mice. J Clin Invest 2009;119:1888–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Malecki MT, Jhala US, Antonellis A, et al. Mutations in NEUROD1 are associated with the development of type 2 diabetes mellitus. Nat Genet 1999;23:323–328 [DOI] [PubMed] [Google Scholar]
- 9. Meliala ITS, Hosea R, Kasim V, Wu S. The biological implications of Yin Yang 1 in the hallmarks of cancer. Theranostics 2020;10:4183–4200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yuan P, He XH, Rong YF, et al. KRAS/NF-κB/YY1/miR-489 signaling axis controls pancreatic cancer metastasis. Cancer Res 2017;77:100–111 [DOI] [PubMed] [Google Scholar]
- 11. Cromer MK, Choi M, Nelson-Williams C, et al. Neomorphic effects of recurrent somatic mutations in Yin Yang 1 in insulin-producing adenomas. Proc Natl Acad Sci USA 2015;112:4062–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hong X, Qiao S, Li F, et al. Whole-genome sequencing reveals distinct genetic bases for insulinomas and non-functional pancreatic neuroendocrine tumours: leading to a new classification system. Gut 2020;69:877–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoffman BG, Robertson G, Zavaglia B, et al. Locus co-occupancy, nucleosome positioning, and H3K4me1 regulate the functionality of FOXA2-, HNF4A-, and PDX1-bound loci in islets and liver. Genome Res 2010;20:1037–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lu TT, Heyne S, Dror E, et al. The polycomb-dependent epigenome controls β cell dysfunction, dedifferentiation, and diabetes. Cell Metab 2018;27:1294–1308.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tennant BR, Robertson AG, Kramer M, et al. Identification and analysis of murine pancreatic islet enhancers. Diabetologia 2013;56:542–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Whyte WA, Orlando DA, Hnisz D, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013;153:307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gao T, McKenna B, Li C, et al. Pdx1 maintains β cell identity and function by repressing an α cell program. Cell Metab 2014;19:259–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ida S, Morino K, Sekine O, et al. Diverse metabolic effects of O-GlcNAcylation in the pancreas but limited effects in insulin-sensitive organs in mice. Diabetologia 2017;60:1761–1769 [DOI] [PubMed] [Google Scholar]
- 19. Gu G, Dubauskaite J, Melton DA. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development 2002;129:2447–2457 [DOI] [PubMed] [Google Scholar]
- 20. Tornovsky-Babeay S, Dadon D, Ziv O, et al. Type 2 diabetes and congenital hyperinsulinism cause DNA double-strand breaks and p53 activity in β cells. Cell Metab 2014;19:109–121 [DOI] [PubMed] [Google Scholar]
- 21. Leung OM, Li J, Li X, et al. Regulatory T cells promote apelin-mediated sprouting angiogenesis in type 2 diabetes. Cell Rep 2018;24:1610–1626 [DOI] [PubMed] [Google Scholar]
- 22. Liang C, Yang KY, Chan VW, et al. CD8+ T-cell plasticity regulates vascular regeneration in type-2 diabetes. Theranostics 2020;10:4217–4232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Song D, Yang Q, Jiang X, et al. YY1 deficiency in β-cells leads to mitochondrial dysfunction and diabetes in mice. Metabolism 2020;112:154353. [DOI] [PubMed] [Google Scholar]
- 24. Ruegsegger GN, Creo AL, Cortes TM, Dasari S, Nair KS. Altered mitochondrial function in insulin-deficient and insulin-resistant states. J Clin Invest 2018;128:3671–3681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou L, Sun K, Zhao Y, et al. Linc-YY1 promotes myogenic differentiation and muscle regeneration through an interaction with the transcription factor YY1. Nat Commun 2015;6:10026. [DOI] [PubMed] [Google Scholar]
- 26. Kleiman E, Jia H, Loguercio S, Su AI, Feeney AJ. YY1 plays an essential role at all stages of B-cell differentiation. Proc Natl Acad Sci USA 2016;113:E3911–E3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang A, Yue F, Li Y, et al. Epigenetic priming of enhancers predicts developmental competence of hESC-derived endodermal lineage intermediates. Cell Stem Cell 2015;16:386–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Blum B, Hrvatin S, Schuetz C, Bonal C, Rezania A, Melton DA. Functional beta-cell maturation is marked by an increased glucose threshold and by expression of urocortin 3. Nat Biotechnol 2012;30:261–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jermendy A, Toschi E, Aye T, et al. Rat neonatal beta cells lack the specialised metabolic phenotype of mature beta cells. Diabetologia 2011;54:594–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Blandino-Rosano M, Barbaresso R, Jimenez-Palomares M, et al. Loss of mTORC1 signalling impairs β-cell homeostasis and insulin processing. Nat Commun 2017;8:16014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang X, Liu G, Guo J, Su Z. The PI3K/AKT pathway in obesity and type 2 diabetes. Int J Biol Sci 2018;14:1483–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Talchai C, Xuan S, Lin HV, Sussel L, Accili D. Pancreatic β cell dedifferentiation as a mechanism of diabetic β cell failure. Cell 2012;150:1223–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhu Y, Liu Q, Zhou Z, Ikeda Y. PDX1, Neurogenin-3, and MAFA: critical transcription regulators for beta cell development and regeneration. Stem Cell Res Ther 2017;8:240. [DOI] [PMC free article] [PubMed] [Google Scholar]