Abstract
Inosine-5′-monophosphate dehydrogenase (IMPDH), a key regulatory enzyme in purine nucleotide biosynthesis, dynamically assembles filaments in response to changes in metabolic demand. Humans have two isoforms: IMPDH2 filaments reduce sensitivity to feedback inhibition, while IMPDH1 assembly remains uncharacterized. IMPDH1 plays a unique role in retinal metabolism, and point mutants cause blindness. Here, in a series of cryogenic-electron microscopy structures we show that human IMPDH1 assembles polymorphic filaments with different assembly interfaces in extended and compressed states. Retina-specific splice variants introduce structural elements that reduce sensitivity to GTP inhibition, including stabilization of the extended filament form. Finally, we show that IMPDH1 disease mutations fall into two classes: one disrupts GTP regulation and the other has no effect on GTP regulation or filament assembly. These findings provide a foundation for understanding the role of IMPDH1 in retinal function and disease and demonstrate the diverse mechanisms by which metabolic enzyme filaments are allosterically regulated.
Cells have evolved many ways to maintain precise and balanced pools of purine nucleotides. Purines are essential components of RNA and DNA, provide energy and are cofactors for many enzymatic reactions. Maintaining balanced purine pools is necessary to cell survival. In most tissues, complex and highly regulated interplay between salvage and de novo biosynthesis pathways maintains optimal nucleotide concentrations. Under high purine demands, as in proliferating cells, both pathways are upregulated1.
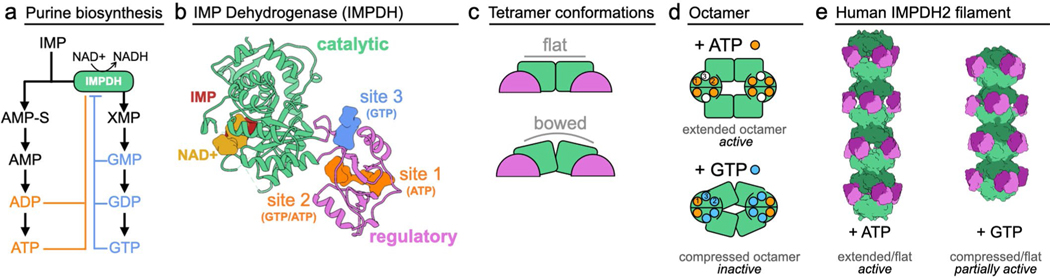
Inosine-5′-monophosphate dehydrogenase (IMPDH) is a highly conserved enzyme that catalyzes the first committed step in GTP synthesis. IMPDH sits at a branch point between adenine and guanine nucleotide synthesis, where its regulation is critical for balancing flux through the two pathways (Extended Data Fig. 1a). In vertebrate cells, IMPDH forms filamentous ultrastructures in response to high demand for guanine nucleotides2–4. Humans express two isoforms of IMPDH with 84% identity. Much of the research on human IMPDH has been focused on IMPDH2 because it plays a critical role in the immune response and is upregulated in proliferating cells5–7; for example, activation of T cells drives assembly of IMPDH2 into filaments8,9. In vitro studies have shown that assembly of IMPDH2 filaments prevents full inhibition, providing an additional layer of regulation that decreases GTP-feedback inhibition10. IMPDH1 also assembles filaments11, but whether polymerization plays a role in regulation of this isoform has been unclear. Understanding the mechanisms of IMPDH1 regulation is important, as point mutations in humans lead to retinal degeneration of varying severity, pointing to a key role of IMPDH1 in retinal metabolism12.
IMPDH quaternary structure is linked to activity and regulation by adenine and guanine nucleotides. The monomer is composed of a catalytic and a regulatory cystathionine β-synthase domain, and constitutively assembles into tetramers through interactions of the catalytic domains13 (Extended Data Fig. 1b,c). IMP is converted to xanthosine monophosphate in an NAD+ dependent reaction in the catalytic domain that has been extensively characterized14. The regulatory domain has three allosteric nucleotide binding sites for adenine and guanine nucleotides11,15,16 (Extended Data Fig. 1b). Site 1 has a preference for ATP/ADP, site 2 binds ATP/ADP and GTP/GDP competitively and site 3 exclusively binds GTP/GDP. Binding of nucleotides in sites 1 and 2 drives dimerization of the regulatory domains and formation of octamers, and the balance between ATP and GTP binding dictates whether octamers adopt an extended/active conformation or a compressed/inactive conformation (Extended Data Fig. 1d)11,17. GTP binding at both competitive site 2 and the GTP-only site 3 induces two conformational changes in IMPDH2: compression of the octamer and flexing of the catalytic domains from an active ‘flat’ to an partially inactive ‘bowed’ conformation (Extended Data Fig. 1c,d). These changes inactivate IMPDH2 by preventing essential loop movements in the core of the enzyme10,15,18. Nucleotide-dependent assembly of IMPDH2 into filaments stabilizes the flat tetramer conformation (Extended Data Fig. 1c), but does not prevent compression, allowing filamentous IMPDH2 to remain partially active even at high GTP concentrations, consistent with the role of IMPDH2 in expanding guanine nucleotide pools during proliferation10,17 (Extended Data Fig. 1e).
Mutations in IMPDH1 lead to autosomal dominant retinal degeneration in humans, a condition known as retinitis pigmentosa or Leber congenital amaurosis, but the molecular mechanism of disease remains unknown12,19–22. The activity of the mutant enzymes is normal, but some mutations reduce sensitivity to GDP inhibition11, change filament assembly parameters23,24, affect phosphorylation25 and disrupt association with nucleic acids22,26,27 yet not all mutations have the same effect. These observations have led to proposed mechanisms of disease including defective ability to bind single stranded DNA28, protein aggregation29 or disruption of phosphoregulation25 but there is not yet a cohesive theory for the molecular mechanism of disease.
Despite expression of IMPDH1 in almost all tissues6,30 and the ubiquitous need to maintain balanced purine pools, the retina is the only affected tissue. This may be due to the specific purine demands in the retina31. Photoreceptors have an unusually high demand for ATP, particularly in the dark32, and cyclic guanosine monophosphate (cGMP) is the key signaling molecule in the phototransduction cascade33–35. Furthermore, the retina is uniquely dependent on IMPDH1, because expression of both IMPDH2 and the major purine salvage enzyme HPRT are very low in the tissue29,30. The complex dependence of photoreceptor function on balanced purine production may lead to severe consequences36,37.
In the retina, IMPDH1 is expressed as two splice variants that have additions to each terminus. In the IMPDH1(546) variant, five residues at the C terminus of the canonical enzyme are replaced by 37 new residues, most of which are predicted to be unstructured. The IMPDH1(595) variant contains the same C-terminal extension, plus 49 residues at the N terminus. Most of the N-terminal addition is predicted to be unstructured, except a short, predicted helix near the canonical N terminus30,38,39 (Extended Data Fig. 2). Mouse retinal splice variants have reduced GTP inhibition compared to the canonical variant, but there is no structural explanation for the change40,41. IMPDH1(546) is the most abundant isoform in human retinas, followed by IMPDH1(595), although the opposite relative abundance has been reported in the retinas of other animals30,42.
Here, we show that canonical human IMPDH1 assembles into filaments with different structures depending on its activity state. Active IMPDH1 filaments closely resemble active IMPDH2 filaments. The GTP-bound IMPDH1 filament, however, uses completely different assembly contacts, which accommodate a bent tetramer conformation that allows for more complete inhibition. Thus, unlike IMPDH2, canonical IMPDH1 does not experience reduced feedback inhibition in the filament. The retinal variants, however, do have reduced sensitivity to GTP feedback, with independent mechanisms for the N- and C-terminal extensions in influencing IMPDH1 conformation. The N-terminal extension introduces specific structural changes in filament architecture that stabilize the partially active flat, compressed enzyme conformation, while the C-terminal extension likely reduces the stability of the compressed conformation. Finally, we characterize IMPDH1 retinopathy mutations in the retinal splice variants, revealing two distinct functional classes: class I are not inhibited by GTP and do not adopt the compressed conformation, while class II are identical to wildtype (WT) in biochemistry and filament assembly behavior.
Results
IMPDH1 assembles into a filament in response to ATP or GTP.
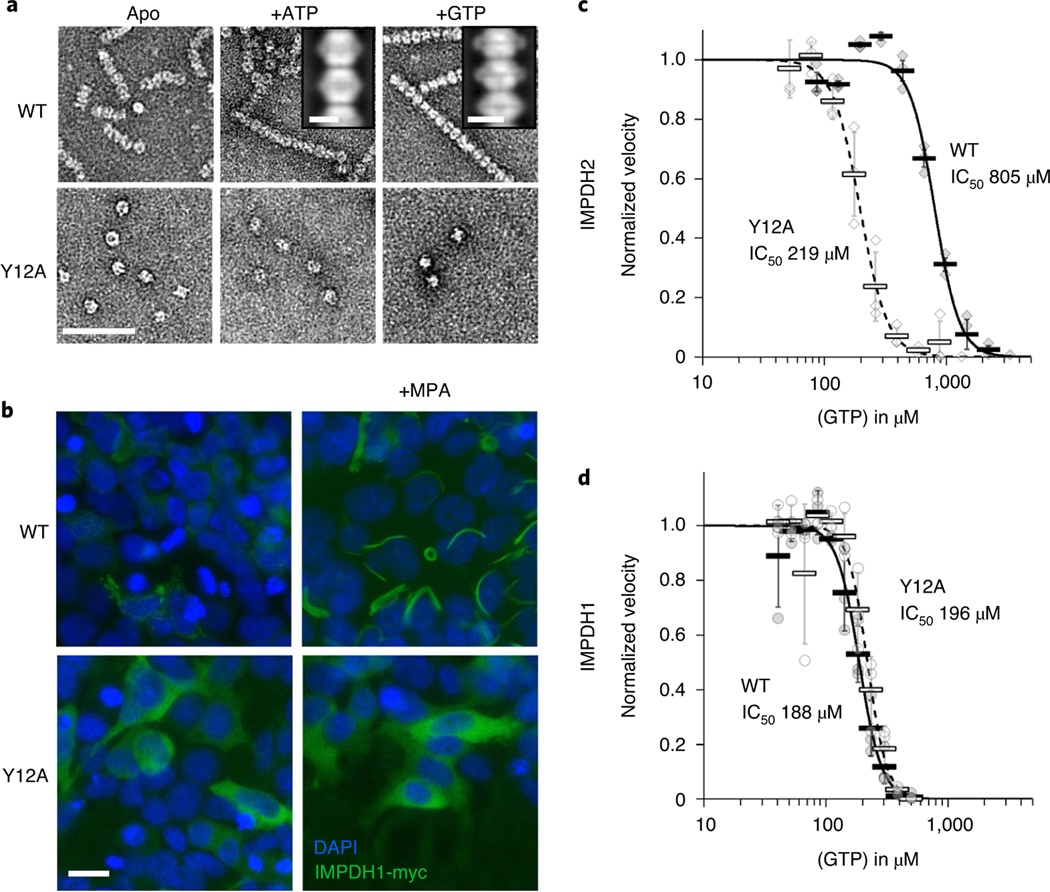
We tested the effects of the allosteric regulators ATP and GTP on IMPDH1 filament assembly and found broad similarities between IMPDH1 and IMPDH2. Using negative stain electron microscopy (EM), we found that, like IMPDH2, IMPDH1 assembles into filaments of stacked octamers in the presence of ATP or GTP (Fig. 1a). The ATP-bound IMPDH1 filaments have a 110 Å rise and the GTP-bound IMPDH1 filaments have a 95 Å rise, consistent with extended/active and compressed/inactive octamers seen in IMPDH2 filaments17. We previously engineered a separation of function point mutation in IMPDH2 at the filament assembly interface, Tyr12Ala, which disrupts polymerization but does not affect enzyme activity10,17. The Y12A mutation also inhibits polymerization of IMPDH1, suggesting both isoforms assemble with similar interfaces (Fig. 1a).
Fig. 1 |. IMPDH1 assembles filaments and is sensitive to gtP inhibition.
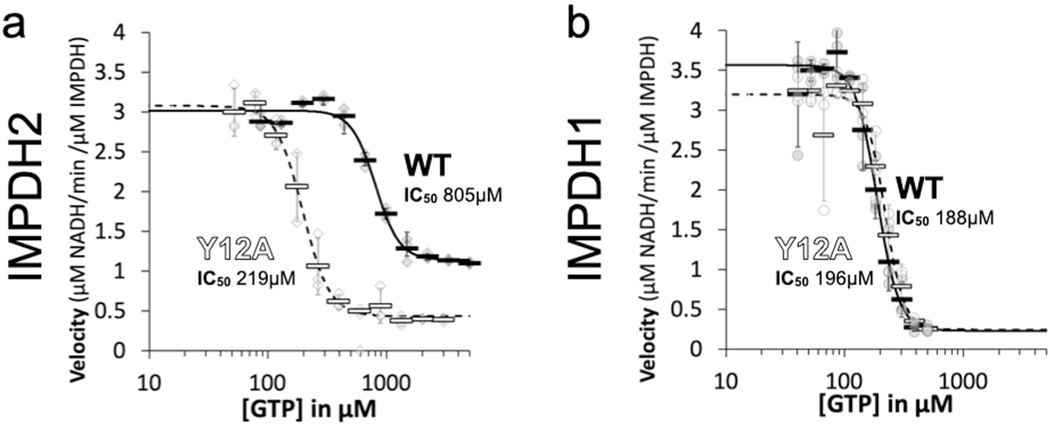
a, negative stain EM of purified human IMPDH1. Addition of ATP or GTP promotes filament assembly of WT. nonassembly mutation Y12A breaks both ATP- and GTP-dependent assembly. Insets are negative stain 2D class averages. Scale bar, 100 nm. b, Anti-myc immunofluorescence of HEK293 cells transfected with IMPDH1-myc constructs (green). DAPI staining in blue. Cells were either left untreated or treated with 10 μM MPA to induce IMPDH2 filament assembly. Scale bar, 20 μm. c,d, GTP inhibition curves of IMPDH2 (c) or IMPDH1-WT (d) (solid line) and the respective nonassembly Y12A protein (dashed line). Individual data points are shown as diamonds (IMPDH2) or circles (IMPDH1), where filled are WT and empty Y12A. reactions were performed in triplicate and data are presented as mean values for each concentration is shown as a bold rectangle (filled is WT, empty is Y12A) ± standard deviation for n = 3 error bars. reactions performed with 1 μM protein, 1 mM ATP, 1 mM IMP, 300 μM NAD+ and varying GTP.
Previous studies have shown that IMPDH1 forms ultrastructures in cells, but the question remains whether the cellular ultrastructures assemble by the same mechanism as filaments in vitro43. To test this, we transiently transfected human embryonic kidney 293 (HEK293) cells with IMPDH1-myc, and induced filament assembly by treatment with the IMPDH inhibitor mycophenolic acid (MPA), a standard assay for cellular filament assembly2,24,44. In cells, IMPDH filaments appear to bundle together to assemble large ultrastructures several micrometers in length45. Staining with an anti-myc antibody shows strong induction of ultrastructure assembly with WT IMPDH1, but not with IMPDH1-Y12A, suggesting that the ultrastructures observed in cells are composed of filaments with an architecture similar to those we observe in vitro (Fig. 1b).
IMPDH1 is more sensitive to GTP-feedback inhibition than IMPDH2.
We characterized the substrate kinetics of IMPDH1 and IMPDH2 and found them to be nearly identical (Supplementary Table 1). However, we found a large difference in sensitivity to GTP inhibition, with the GTP half-maximum inhibitory concentration (IC50) fourfold lower for IMPDH1 than for IMPDH2 (Fig. 1c,d)11. Consistent with our previous results10, the nonassembly mutant IMPDH2-Y12A has a lower IC50 than the WT enzyme, but the same mutation has no effect on GTP affinity in IMPDH1 (Fig. 1c,d).
It is notable that IMPDH1-WT and Y12A mutants in both isoforms have similar GTP sensitivity and are mostly inhibited at high GTP concentrations, while IMPDH2-WT retains basal activity even at GTP concentrations six times higher than the IC50 (Extended Data Fig. 3). This suggests conservation of the intrinsic allosteric regulation, with the difference being that polymerization reduces the inhibitory effect of GTP in IMPDH2 but appears to have no role in tuning the response of IMPDH1. This was surprising given the similarity of GTP-bound IMPDH1 and IMPDH2 filaments in our low-resolution negative stain imaging (Fig. 1a), so we next turned to higher resolution cryogenic-EM (cryo-EM) of IMPDH1 filaments to provide insight into differences in inhibition behavior.
IMPDH1 assembles polymorphic filaments.
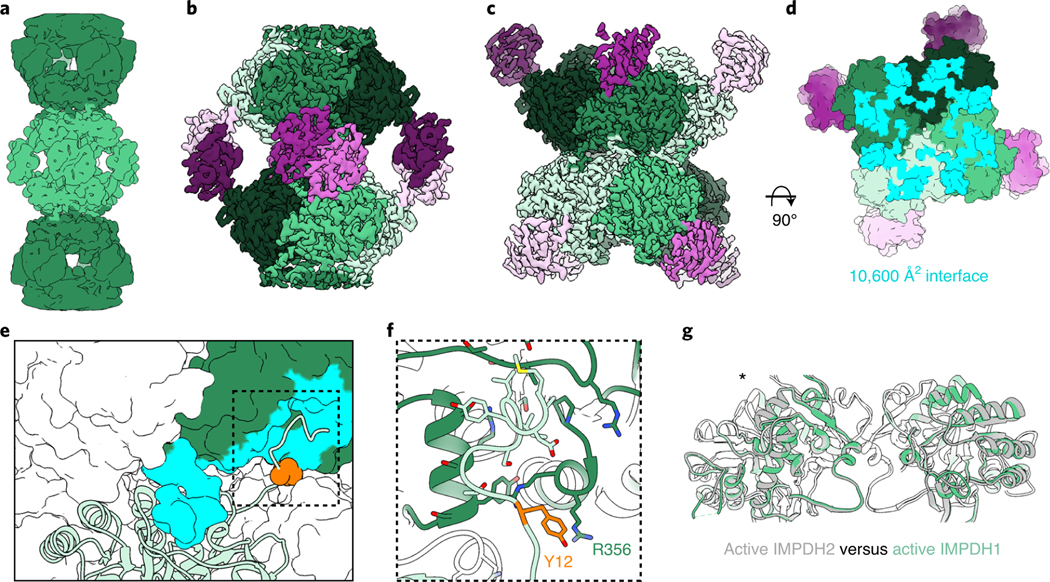
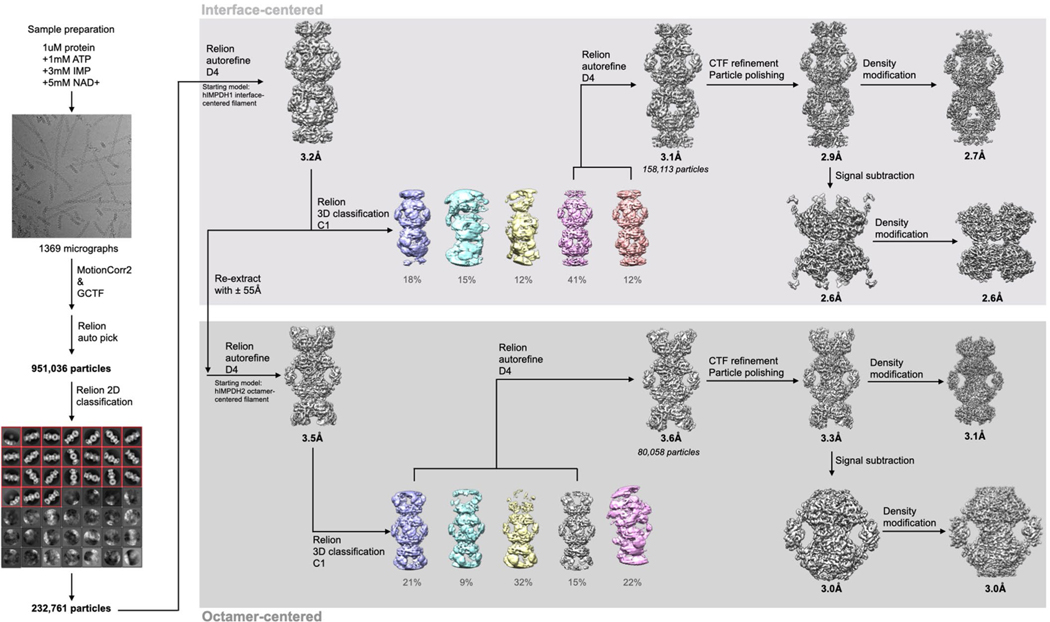
We determined the structure of the IMPDH1 extended filament bound to ATP, IMP and NAD+ by cryo-EM, using the approach we developed to solve structures of IMPDH2 filaments, which have a rigid assembly interface between octamers but are more flexible within the octamer10. This single-particle approach allows for high resolution structures of different regions of the filament by combining density subtraction, focused classification and focused refinement of two different regions of IMPDH1 filaments: full octamers and the interface between stacked octamers (Extended Data Fig. 4) that together represent the necessary information for the entire IMPDH1 filament. Because all substrates and cofactors are present in this reconstruction, the enzyme was actively turning over when prepared for cryo-EM analysis, so the active site likely contains a mixture of bound substrates and products; nonetheless, the active site was well-resolved in the final cryo-EM map (Extended Data Fig. 5). This approach yielded octamer- and interface-focused reconstructions at 3.1 and 2.6 Å resolution, respectively (Fig. 2a–c and Table 1) for extended IMPDH1 filaments.
Fig. 2 |. Structure of extended IMPDH1 filaments (AtP/IMP/NAD+ bound).
a, Low-pass filtered cryo-EM reconstruction colored by octamer. b, Octamer-centered single-particle reconstruction of the filament at 3.1 Å, with catalytic domains in different shades of green and regulatory domains in shades of pink. c, Interface-centered single-particle reconstruction of the filament at 2.6 Å. d, View of the top of an octamer from inside the filament. Surface representation of the atomic model at the assembly interface, with buried residues in cyan. e, Surface representation of the filament interface with one monomer in ribbon (light green). Tyr12 is shown in orange spheres. The monomer it contacts across the octamer interface is green with residues forming the interface in cyan. f, Close-up ribbon view of the interface where Tyr12 in orange contacts Arg356 in the opposing monomer. g, Comparison of the catalytic tetramers of active IMPDH2 ATP/IMP/nAD+ (PDB 6u8s) (gray) to active IMPDH1 ATP/IMP/nAD+ (green). Aligned on monomers with an asterisk.
Table 1.
Cryo-EM data collection, refinement and validation statistics
| No. 1 IMPDH1(514) extended filament (interface) (EMDB 24437) (PDB 7RER) | No. 2 IMPDH1(514) extended filament (octamer) (EMDB 24438) (PDB 7RES) | No. 3 IMPDH1(514) compressed filament (interface) (EMDB 24439) (PDB 7RFE) | No. 4 IMPDH1(514) compressed filament (octamer) (EMDB 24441) (PDB 7RFG) | No. 5 IMPDH1(546) extended filament (interface) (EMDB 24451) (PDB 7RGL) | No. 6 IMPDH1(546) extended filament (octamer) (EMDB 24452) (PDB 7RGM) | No. 7 IMPDH1(546) compressed filament (interface) (EMDB 24450) (PDB 7RGI) | No. 8 IMPDH1(546) compressed filament (octamer) (EMDB 24454) (PDB 7RGQ) | No. 9 IMPDH1(595) extended filament (interface) (EMDB 24440) (PDB 7RFF) | No. 10 IMPDH1(595) extended filament (octamer) (EMDB 24442) (PDB 7RFH) | No. 11 IMPDH1(595) compressed filament (interface) (EMDB 24443) (PDB 7RFI) | No. 12 IMPDH1(595) compressed filament (octamer) (EMDB 24448) (PDB 7RGD) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Data collection and processing | ||||||||||||
| Microscope | Titan Krios G3 | Titan Krios G3 | Titan Krios G3 | Glacios | Titan Krios G3 | Titan Krios G3 | ||||||
| Camera | Gatan K2 | Gatan K2 | Gatan K3 | Gatan K2 | Gatan K2 | Gatan K3 | ||||||
| Magnification | 130,000 | 130,000 | 130,000 | 130,000 | 130,000 | 130,000 | ||||||
| Voltage (kV) | 300 | 300 | 300 | 200 | 300 | 300 | ||||||
| Electron exposure (e−/Å2) | 90 | 90 | 90 | 90 | 90 | 90 | ||||||
| Defocus range (μm) | −1.5–−3.5 | −0.5–−1.8 | −0.6–−1.8 | −0.5–−3.2 | −0.6–−2.3 | −0.5–−2.5 | ||||||
| Pixel size (Å) | 0.525 | 0.525 | 0.4215 | 1.16 | 0.525 | 0.4215 | ||||||
| Camera mode | super-resolution | super-resolution | super-resolution | counting | super-resolution | super-resolution | ||||||
| Symmetry imposed | D4 | D4 | D4 | D4 | D4 | D4 | D4 | D4 | D4 | D4 | D4 | D4 |
| Initial particle images (no.) | 232,000 | 217,600 | 103,100 | 56,900 | 64,355 | 66,807 | 73,000 | 113,000 | 81,200 | 80,200 | 330,000 | 310,000 |
| Final particle images (no.) | 103,809 | 57,965 | 42,500 | 49,800 | 25,366 | 43,406 | 70,031 | 97,369 | 60,919 | 11,476 | 71,792 | 102,543 |
| Map resolution (Å) | 2.6 | 3.1 | 2.6 | 3.1 | 2.4 | 2.9 | 4.0 | 3.9 | 2.7 | 3.7 | 2.8 | 3.0 |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 |
| Map resolution range (Å) | 2.4–3.4 | 2.8–4.4 | 2.3–3.8 | 2.5–4.5 | 2.4–4.2 | 2.9–5.0 | 3.5–4.6 | 3.5–4.8 | 2.5–4,5 | 3.2–6.0 | 2.6–4.0 | 2.9–4.5 |
| Refinement | ||||||||||||
| Initial model used (PDB code) | 6U8E | 6U8N | 7RER | 7RES | 7RFE | 7RGL | 7RER | 7RES | ||||
| Model resolution (Å) | 3.1 | 2.5 | 2.2 | 2.1 | 1.9 | 2.4 | 3.0 | 3.2 | 2.1 | 3.0 | 2.1 | 2.3 |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 |
| Model resolution range (Å) | inf-3.1 | inf-3.1 | inf-2.6 | inf-3.1 | inf-2.4 | inf-2.9 | inf-4.0 | inf-3.9 | inf-2.7 | inf-3.7 | inf-2.8 | inf-3.0 |
| Map sharpening B factor (Å2) | −87 | −102 | −55 | −52 | −38 | −63 | −137 | −133 | −113 | −93 | −57 | −62 |
| Model composition | ||||||||||||
| Nonhydrogen atoms | 23,504 | 29,056 | 28,568 | 30,584 | 25,528 | 30,192 | 21,960 | 30,288 | 20,400 | 27,538 | 26,496 | 32,584 |
| Protein residues | 3,112 | 3,856 | 3,800 | 3,928 | 3,328 | 3,872 | 2,856 | 3,848 | 2,712 | 3,595 | 3,376 | 4,152 |
| Ligands | IMP (8), NAD+ (8) | IMP (8), NAD+ (8), ATP (16) | IMP (8), ATP (8), GTP (16) | IMP (8), ATP (8), GTP (16) | IMP (8), NAD+ (8) | IMP (8), NAD+ (8), ATP (16) | IMP (8), NAD+ (8) | IMP (8), NAD+ (8), ATP (8) | ATP (16) | IMP (8), NAD+ (8), GTP (16) | IMP (8), NAD+ (8), ATP (8), GTP (16) | |
| B factors (Å2) | ||||||||||||
| Protein | 28.95 | 62.88 | 42.20 | 37.49 | 50.18 | 49.58 | 48.86 | 41.67 | 85.89 | 51.89 | 34.42 | 44.08 |
| Ligand | 31.90 | 67.43 | 58.64 | 45.54 | 48.50 | 55.20 | 55.61 | 54.18 | - | 69.51 | 54.61 | 50.78 |
| R.m.s. deviations | ||||||||||||
| Bond lengths (Å) | 0.58 | 0.011 | 0.019 | 0.018 | 0.009 | 0.019 | 0.015 | 0.020 | 0.011 | 0.016 | 0.021 | 0.020 |
| Bond angles (°) | 0.89 | 1.136 | 2.195 | 2.179 | 1.296 | 1.986 | 1.958 | 2.129 | 1.950 | 2.069 | 2.051 | 2.196 |
| Validation | ||||||||||||
| MolProbity score | 1.72 | 2.54 | 1.44 | 1.34 | 1.85 | 1.75 | 1.57 | 1.82 | 1.41 | 2.13 | 1.60 | 1.87 |
| Clashscore | 7.75 | 11.30 | 1.13 | 1.16 | 3.77 | 1.92 | 2.18 | 2.77 | 1.56 | 4.93 | 3.51 | 2.52 |
| Poor rotamers (%) | 0.7 | 4.87 | 2.97 | 1.91 | 3.66 | 3.63 | 1.62 | 2.39 | 1.97 | 2.73 | 1.61 | 3.22 |
| Ramachandran plot | ||||||||||||
| Favored (%) | 95.72 | 92.96 | 96.24 | 95.64 | 96.18 | 94.56 | 93.80 | 92.63 | 95.90 | 90.17 | 95.63 | 93.08 |
| Allowed (%) | 4.28 | 6.99 | 3.54 | 4.18 | 3.66 | 5.18 | 6.02 | 6.71 | 3.80 | 8.75 | 3.97 | 6.60 |
| Disallowed (%) | 0 | 0.05 | 0.21 | 0.18 | 0.15 | 0.26 | 0.18 | 0.66 | 0.30 | 1.09 | 0.39 | 0.32 |
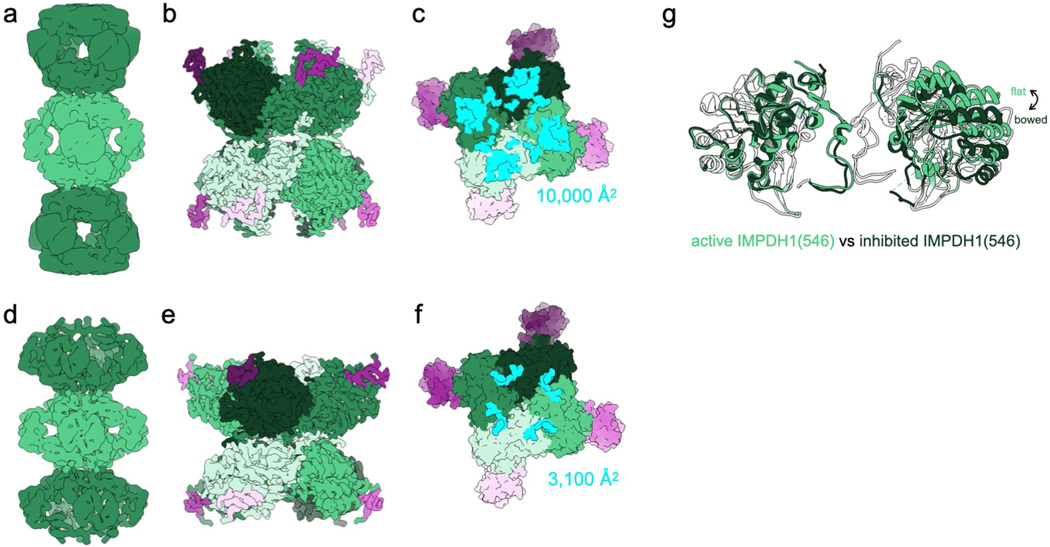
The extended filament structure is highly conserved between IMPDH1 and IMPDH2. Like IMPDH2, the IMPDH1 extended filament is composed of D4 symmetric stacked octamers with filament assembly contacts made between catalytic domains of opposing octamers. Each octamer has a rise of 113 Å and right-handed helical rotation of 30° between octamers. The interface buries a total of 10,600 Å2 (1,320 Å2 per monomer) (Fig. 2d) and is formed by residues 2–12 from the N terminus of one monomer that sit in a groove between two helices in the catalytic domain of the opposing monomer (Fig. 2e,f). Tyr12, which breaks filament assembly when mutated to alanine, packs against Arg356 on the opposing monomer (Fig. 2f). This filament architecture results in a tetramer in the flat conformation (Fig. 2g) similar to IMPDH2 (0.964 Å root mean-squared deviation (r.m.s.d.) for alpha carbons over the catalytic domains of the tetramer) (Supplementary Table 2). Overall, the filament interface and protomer conformations of IMPDH1 filaments are nearly identical to the IMPDH2 filament10.
The only major difference between isoforms in the extended filament appears to be the degree of flexibility in the octamer subunit. Active IMPDH2 shows extreme heterogeneity with mixed partially extended and partially compressed conformations in the same filament, due to flexibility between the catalytic and regulatory domains10. The interface-centered reconstruction reached a higher resolution than the octamer-focused reconstruction, likely due to some limited flexibility between domains. However, after extensive classification in the octamer-centered reconstruction we found that almost all protomers in the IMPDH1 filament structure are in the fully extended conformation and relatively homogenous despite the enzyme actively producing product. This may reflect a higher degree of cooperativity in the conformational state of IMPDH1 relative to IMPDH2.
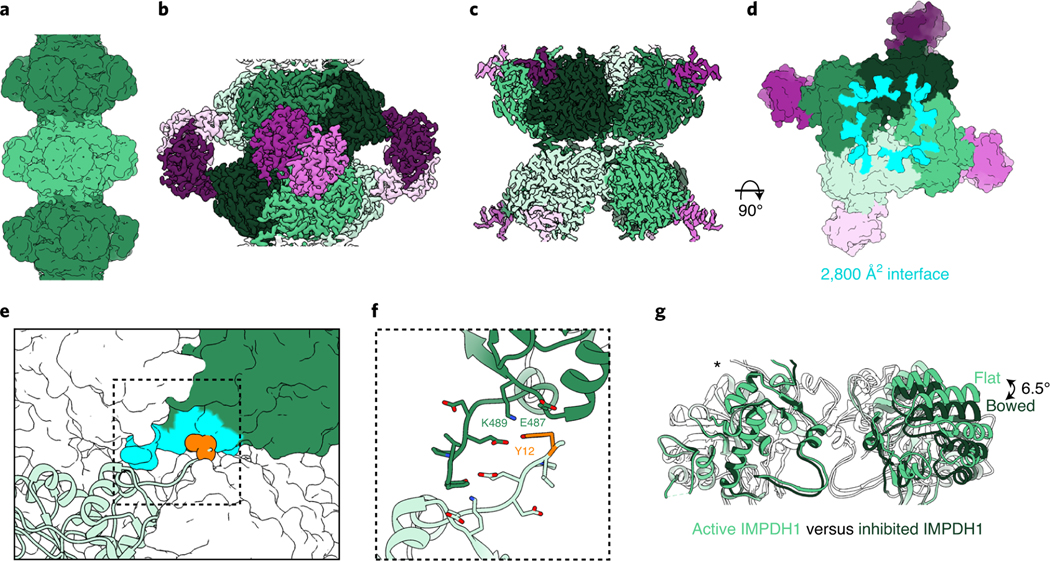
We next solved a cryo-EM structure of the IMPDH1 compressed filament bound to GTP, ATP and IMP, and found it assembles with a completely different architecture, with different interface contacts that lead to different helical symmetry (Fig. 3a–d, Table 1, Supplementary Video 1 and Supplementary Fig. 3b). The filament is still made up of D4 symmetric octamers but the N-terminal residues of one monomer now contact a different groove in the catalytic domain of the monomer in the opposing octamer (Fig. 3e). This change in interaction leads to a large shift in helical geometry where the rotation between stacked octamers increases from 30° to 74°. The major contacts are between Tyr12 and Glu15 on the N terminus and Glu487 and Lys489 on the opposing monomer (Fig. 3e,f). The involvement of Tyr12 in this interface explains why the Tyr12Ala point mutation also prevents assembly of this filament (Fig. 1a). The surface area buried by the interface is 75% smaller: 2,800 Å2 buried at the octamer–octamer interface (350 Å2 per monomer) for the compressed IMPDH1 filament compared to 10,600 Å2 for the extended IMPDH1 filament interface (Figs. 2d and 3d). We refer to this new interface as the ‘small interface’ and the previously characterized interface in all IMPDH2 filaments and extended IMPDH1 as the ‘large interface’ (Supplementary Video 1).
Fig. 3 |. Inhibited IMPDH1 assembles with an alternative filament architecture (gtP/AtP/IMP bound).
a, Low-pass filtered cryo-EM reconstruction colored by octamer. b, Octamer-centered single-particle reconstruction at 2.6 Å of the filament, with catalytic domains in different shades of green and regulatory domains in shades of pink. c, Interface-centered single-particle reconstruction of the filament at 2.6 Å. d, View of the top of an octamer from inside the filament. Surface representation of the atomic model at the assembly interface, with buried residues in cyan. e, Surface representation of filament interface with one monomer in ribbon (light green). Tyr12 is shown in orange spheres. The monomer it contacts across the octamer interface is green with residues forming the interface in aqua. f, Close-up ribbon view of the interface where Tyr12 in orange contacts Glu487 and Lys489 in the opposing monomer. g, Comparison of the catalytic tetramers of active IMPDH1 (light green) to mostly inhibited IMPDH1 (dark green), show that the GTP-bound structure is in the bowed conformation. Aligned on monomers with an asterisk.
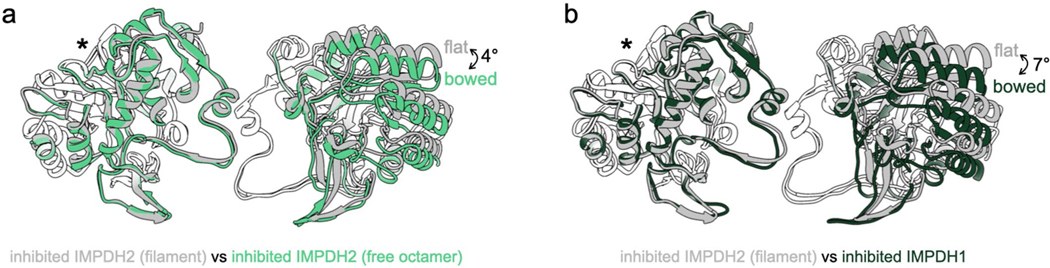
The GTP-bound IMPDH1 filament accommodates a bowed tetramer that is mostly inhibited (Fig. 3g). In contrast, GTP binding causes compression of IMPDH2, but filament contacts constrain the catalytic tetramers in a flat conformation, yielding a partially active flat, compressed state (Extended Data Figs. 3a and 6a). A free IMPDH2 octamer is not restrained and able to adopt a bent, compressed state that is completely inhibited (Extended Data Figs. 3 and 6a). GTP-bound IMPDH1 in filaments is nearly identical to the IMPDH2 free octamer bowed tetramer (Extended Data Fig. 6a,b) that is also mostly inhibited (Extended Data Fig. 3a). The conformational change can best be visualized by looking at protomers arranged diagonally to each other in the catalytic tetramer (chains A and C). Relative to the IMPDH1 extended octamer flat conformation, there is a rotation of 6.5° between these two protomers in the compressed bowed conformation, resulting in an r.m.s.d. of 3.8 Å (Supplementary Table 2). This finding that the IMPDH1 compressed filament accommodates a bowed tetramer provides an explanation for why IMPDH1 filaments do not appear to directly affect regulation by GTP. For IMPDH1, GTP binding causes compression, but shifting filament assembly contacts also accommodates the bowed tetramer conformation, yielding a bowed, compressed state that is mostly inhibited.
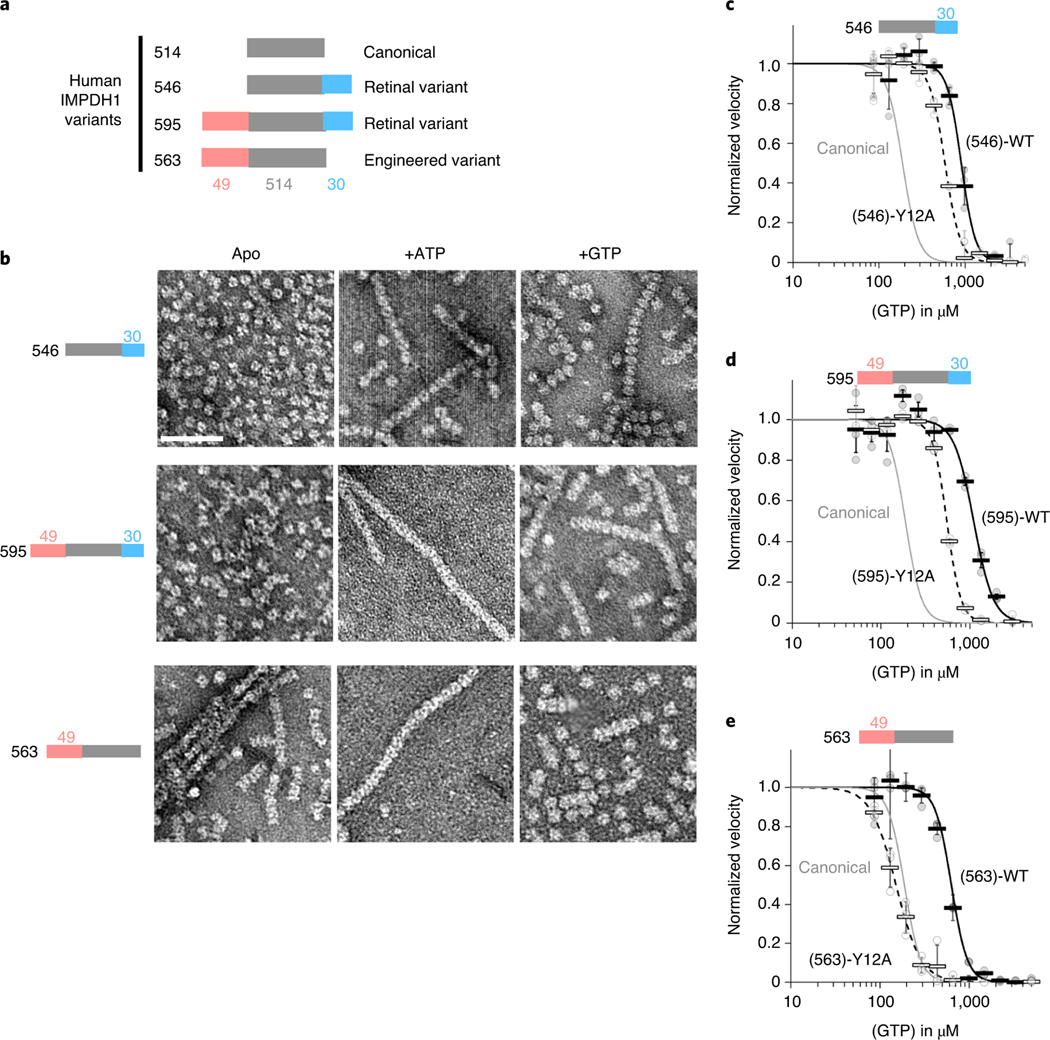
IMPDH1 retinal variants form filaments with ATP or GTP.
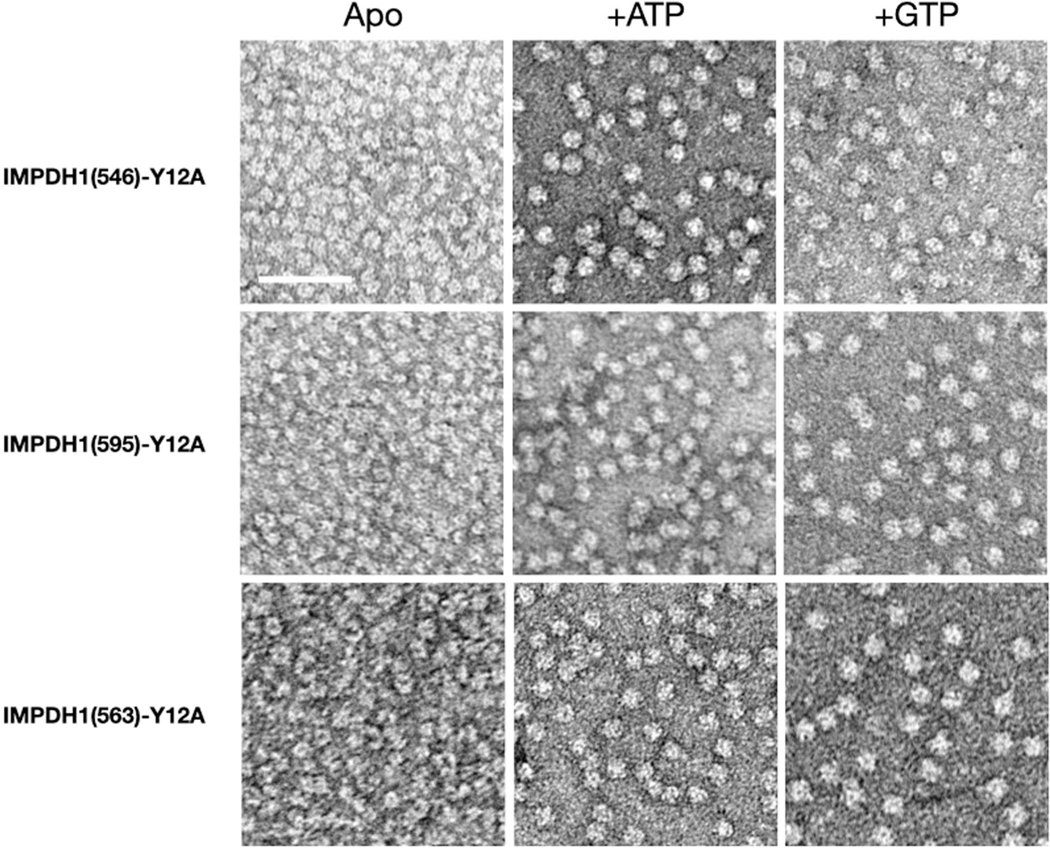
We tested the effects of the allosteric regulators ATP and GTP on filament assembly of both IMPDH1 retinal variants (Fig. 4a) and found the response to be similar to canonical IMPDH1. In addition, we engineered a variant IMPDH1(563) that only has the N-terminal extension, to specifically test its effect on filament assembly. Both retinal variants assemble filaments of stacked octamers in the presence of ATP or GTP, and IMPDH1(563) has a propensity to spontaneously assemble in the absence of ligands (Fig. 4b). ATP-bound IMPDH1 retinal variant filaments have a roughly 110 Å rise and GTP-bound a roughly 95 Å rise, which is consistent with the IMPDH1 canonical filaments, and it behaves like the retinal variants in negative stain EM. The mutation Tyr12Ala disrupts filament assembly in all the variants (Extended Data Fig. 7).
Fig. 4 |. IMPDH1 retinal variants assemble filaments that resist gtP inhibition.
a, representation of IMPDH1 variant sequences. b, negative stain EM of purified IMPDH1 variants in apo and nucleotide-bound states. Scale bar, 100 nm. c–e, GTP inhibition curves of IMPDH1 variants (black solid line) and the respective nonassembly Y12A protein (black dashed line) compared to IMPDH1 canonical (solid gray line). c, retinal variant IMPDH1(546). d, retinal variant IMPDH1(595). e, Engineered variant IMPDH1(563). Individual data points are shown as circles, where filled are WT and empty y12A. reactions were performed in triplicate and the average for each concentration is shown as a bold rectangle (filled WT, empty Y12A). Error bars show the standard deviation for n = 3. reactions performed with 1 μM protein, 1 mM ATP, 1 mM IMP, 300 μM NAD+ and varying GTP.
IMPDH1 retinal variants are less sensitive to GTP inhibition.
The K0.5 values for IMP and NAD+ for the IMPDH1 retinal splice variants are very similar to each other and to the canonical variant46 (Supplementary Table 2). However, the retinal variants have higher IC50s for inhibition by GTP (Fig. 4c–e). IMPDH1(563) is three times less sensitive to GTP inhibition, IMPDH1(546) is almost five times less sensitive and IMPDH1(595) six times less sensitive when compared to the canonical variant. These findings are consistent with previous results from the mouse IMPDH1 retinal splice variants40.
The sensitivity of IMPDH1 retinal variants and IMPDH2 to GTP is similar (Supplementary Table 3); given the role of filament assembly in tuning GTP sensitivity in IMPDH2, we tested the effect of the Y12A nonassembly mutant on inhibition of IMPDH1 variants. All variants had increased sensitivity to GTP inhibition when the Y12A mutation was introduced (Fig. 4c–e). However, for IMPDH1(563), which only has the N-terminal extension, the GTP IC50 dropped to canonical levels, indicating that the effect of the N-terminal extension on IC50 is dependent on the ability to assemble filaments. Thus, the retina-specific N- and C-terminal additions appear to have distinct mechanisms that function independently to increase the GTP IC50.
IMPDH1 retinal variants alter filament architecture.
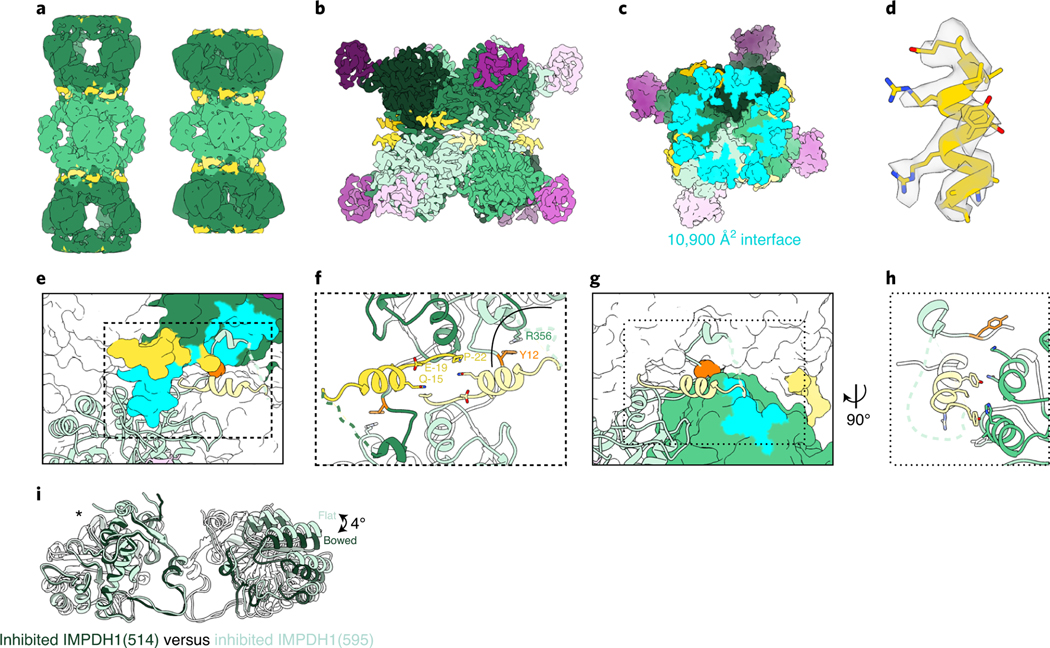
To gain insight into the mechanisms by which the splice variants alter GTP sensitivity, we determined structures of IMPDH1(595) and IMPDH1(546) in multiple ligand states by cryo-EM (Fig. 5a–c and Extended Data Figs. 5c–f and 8). The IMPDH1(546) extended structure is bound to the same ligand combination as the IMPDH1(514) extended structure (IMP, NAD+ and ATP) while IMPDH1(595) was solved in the presence of ATP alone to determine whether substrates affect conformational homogeneity in the filament. In the extended state, both variants closely resemble extended canonical filaments, with the enzyme in the fully extended, flat tetramer conformation and the large assembly interface between octamers (Fig. 5a). In both extended structures, most of the retinal C-terminal extension could not be resolved. Extended canonical IMPDH1 can be resolved to residue 514, while extended IMPDH1(595) could only be modeled to residue 504 and IMPDH1(546) to residue 515. In both IMPDH1(595) structures, there was clear density for a short alpha helix near the filament interface, composed of residues −22 to −4 (Fig. 5d). This helix packs between the catalytic core of the enzyme and the canonical N-terminal 15 residues that form the large interface (Fig. 5e,f). In this position it also makes contacts with the neighboring protomer in the same tetramer with the major contact between residues Tyr-5 and His288 of the neighbor (Fig. 5g,h). The N-terminal end of the variant helix also contacts its symmetry mate on the monomer across the interface with Gln-21 or Gln-15 (Fig. 5f). The helix appears to be positioned to stabilize the residues involved in the large interface filament interactions, and to stabilize the flat conformation of the catalytic tetramer.
Fig. 5 |. IMPDH1 retinal variant (595) constrains filament architecture.
a, Low-pass filtered cryo-EM reconstruction of the IMPDH1(595) extended filament (bound to ATP) and compressed filament (bound to GTP, ATP, IMP, NAD+). b–k, Compressed IMPDH1(595) filament (bound to GTP, ATP, IMP, NAD+). b, Interface-focused cryo-EM filament reconstruction. Eight monomers are colored with catalytic domain (green), regulatory domain (pink) and variant helix (yellow). c, View of the top of an octamer from inside the filament. Surface representation of the atomic model at the assembly interface, with buried residues in cyan. d, Additional n-terminal helix residues −18 to −7 in yellow in density. e, Surface representation of interface with one monomer in ribbon (light green). Additional n-terminal helix in yellow. The monomer it contacts across the octamer interface is colored dark green/pink with residues the ribbon monomer contacts in aqua. Tyr12 is shown in orange spheres. f, Close-up view of the interface contacts. g, Surface representation of interface with one monomer in ribbon (light green). Additional n-terminal helix in yellow. The neighbor monomer in the tetramer is colored green with residues the ribbon monomer contacts in aqua. Y12 is shown in orange spheres. h, Close-up view of the new n-terminal helix contacts with the adjacent monomer in the tetramer. i, Comparison of the catalytic tetramers of inhibited IMPDH1(514) GTP/IMP/NAD+ (dark green) to inhibited IMPDH1(595) GTP/ATP/IMP/NAD+ (light green). Aligned on monomers with an asterisk.
We next solved structures of IMPDH1(595) in a compressed filament at 3.7 Å resolution by cryo-EM. Unlike the canonical enzyme, IMPDH1(595) is maintained in the large interface and flat tetramer conformation in the compressed, GTP-bound state. The overall interface and position of the helix from the N-terminal extension are very similar to the extended filament described above (1.5 Å r.m.s.d. among Cαs of all eight catalytic domains at the interface when aligned on a single chain). Thus, the role of the N-terminal extension appears to be stabilizing the large interface, and through that stabilizing the flat enzyme conformation that is less sensitive to GTP inhibition. We would predict that the additional interactions at the interface would increase affinity for assembly and decrease the critical concentration for filament assembly. This explains why the effect of the N-terminal extension is dependent on filament assembly (Fig. 4e).
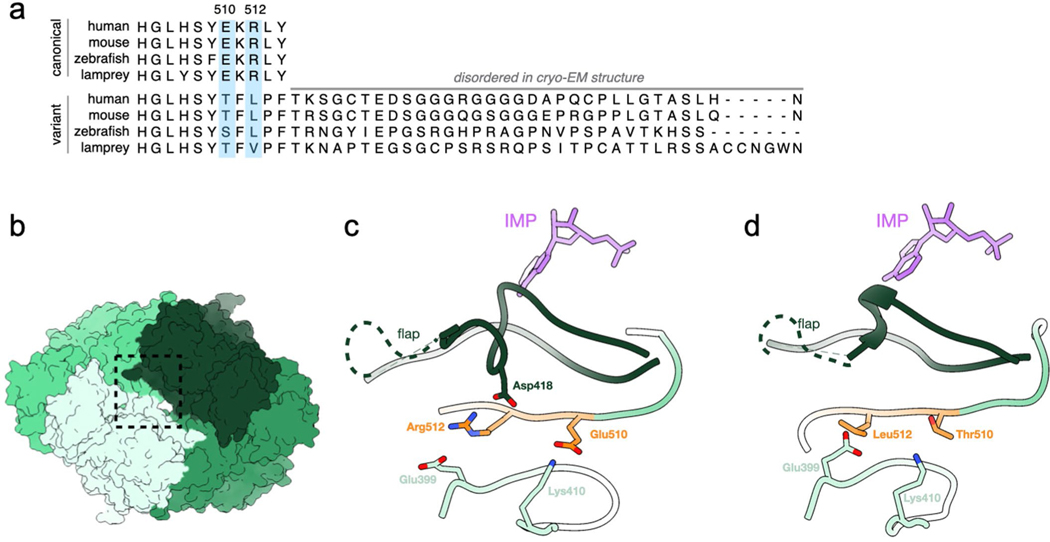
We then wondered how the C-terminal extension contributes to decreased GTP sensitivity. So, we solved a 3.6 Å cryo-EM structure of the compressed IMPDH1(546) filament, which only has the C-terminal extension (Extended Data Fig. 8a–f). In the compressed octamer conformation, bound to GTP/ATP/IMP/NAD+, IMPDH1(546) assembles with the small interface and in a bowed conformation, very similar to canonical IMPDH1 (Extended Data Fig. 8g). For both retinal compressed filament structures, we included all ligands (GTP/ATP/IMP/NAD+) when we had previously left out NAD+ for IMPDH1(514) with the prediction it might lead to a more rigid overall structure but it resulted in very little effect. In all other structures of compressed IMPDH, the conformation of the tetramer in the flat conformation, partially inhibited or bowed conformation or mostly inhibited has explained whether it is more or less sensitive to GTP inhibition. Even though IMPDH1(546) is less sensitive to GTP inhibition than the canonical variant (Fig. 4c), its tetramer is still in the bent conformation, which we had associated with mostly complete inhibition (Extended Data Fig. 6g). Therefore, we suspect the C-terminal extension prevents full octamer compression to resist GTP inhibition. In canonical IMPDH1 bound to GTP, Arg512 and Glu510 near the C terminus make ionic interactions with residues in protomers on the opposite face of the octamer, stabilizing the compressed conformation. In IMPDH1(546) these residues change to Thr510 and Leu512 (Extended Data Fig. 9). Disrupting these interactions likely disfavors octamer compression, potentially explaining why IMPDH1(546) resists GTP inhibition. A second possibility is that the remaining 31 residues that are not resolved in any of our IMPDH1(546) are highly flexible, and their presence near the core of the enzyme may sterically hinder compression.
Retinopathy mutations fall into two functional classes.
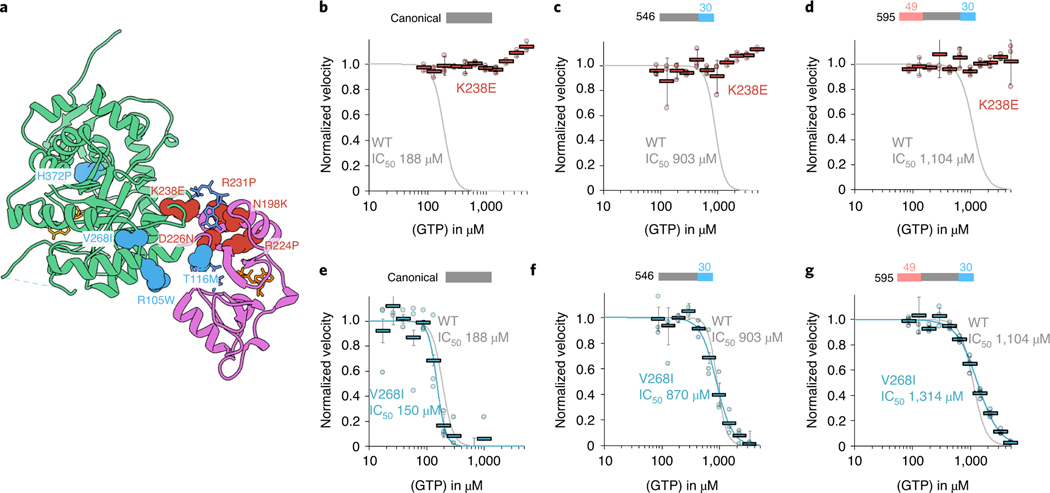
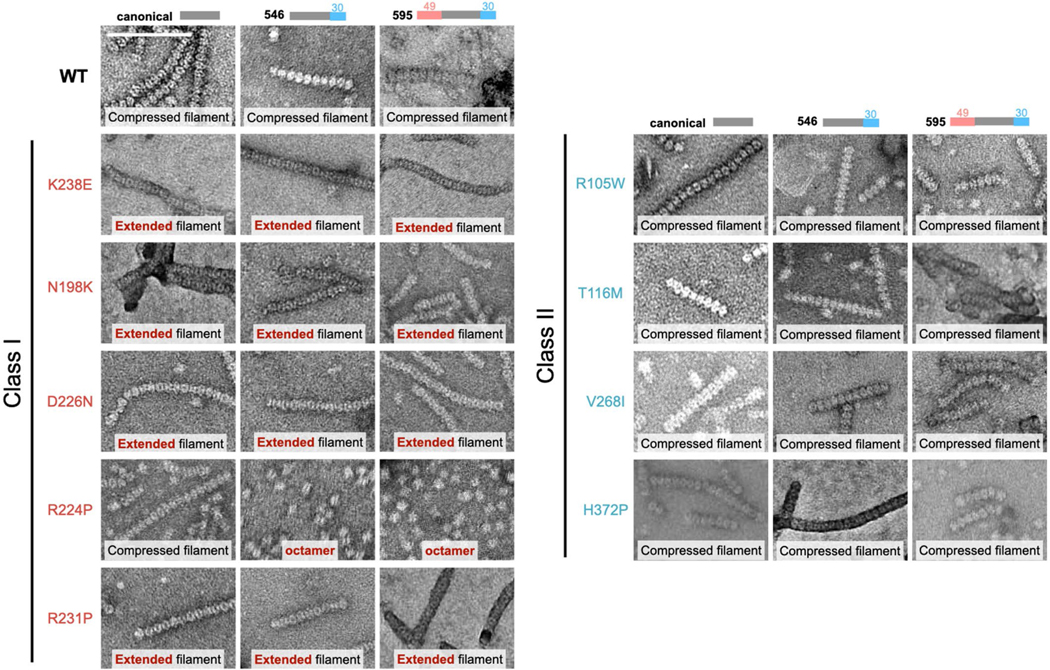
Previous studies have looked at the effect of retinitis pigmentosa mutations in canonical IMPDH1 and disease mutation D226 in both retinal variants and found there is no effect on substrate kinetics29,46, but a subset of mutations disrupt GTP regulation11. Since disease only occurs in the retina, where there is no expression of canonical IMPDH120, and given the large differences in GTP regulation we observed with splice variants, we wondered whether the mutations had specific effects in the retinal splice variants. We tested the mutant forms of canonical and retinal IMPDH1 variants for nine known retinitis pigmentosa mutations. Our findings for canonical IMPDH1 confirm the previous results11. We repeated these experiments with the retinal variants and found the effects of each mutation were similar in the variants and canonical IMPDH1, with several mutations affecting GTP regulation. Thus, in IMPDH1 splice variants, we can describe two classes of disease mutants. Mutations that are insensitive to GTP inhibition we describe as class I, which consists of five mutations around the third allosteric site that is specific for GTP (Fig. 6a–d and Supplementary Table 4). Class I mutations are N198K, R231P, R224P, D226N and K238E. Class II mutations are located at four positions more distal to the allosteric sites and had a GTP inhibition response nearly identical to WT (Fig. 6e–g and Supplementary Table 4). Class II mutations include R105W, T116M, V268I and H372P. The similar effects of disease mutants in all variants of IMPDH1 suggests that they likely have the same effect at the enzyme level across all tissues expressing IMPDH1.
Fig. 6 |. IMPDH1 retinopathy mutations fall into two classes.
a, IMPDH1 with catalytic domain (green), regulatory domain (pink), NAD+ (gold), GTP (blue) and ATP (orange). Class I (defective in GTP regulation) residues colored coral. Class II residues (normal GTP regulation) colored teal. b–g, GTP inhibition curves of IMPDH1 retinopathy mutant variants (colored lines) compared to IMPDH1-WT variants (gray lines). reactions performed with 1 μM protein, 1 mM ATP, 1 mM IMP, 300 μM NAD+ and varying GTP. Individual data points for mutants are shown as circles. reactions were performed in triplicate and the average for each concentration is shown as a bold rectangle (gray WT, red K238E, teal V268I). Error bars show the standard deviation for n = 3. Individual data points and averages for WT are in Fig. 1d. b, Canonical IMPDH1(514)-K238E. c, retinal variant IMPDH1(546)-K238E. d, retinal variant IMPDH1(595)-K238E. e, Canonical IMPDH1(514)-V268I. f, retinal variant IMPDH1(546)-V268I. g, retinal variant IMPDH1(595)-V268I.
Class II mutations do not have an effect on biochemical activity, so we wondered whether the reason they cause disease might be instead due to an effect on filament assembly.
We performed negative stain EM in the presence of ATP, substrates and inhibitory concentrations of GTP (Extended Data Fig. 10). Under this condition, all three WT variants assemble filaments of compressed octamers. Four of the five class I mutants form filaments in all variants, but are made up of extended/flexible octamers, which agrees with their inability to be inhibited by GTP. The only class I mutation that is different is R224P. IMPDH1(514)-R224P forms compressed filaments similar to WT, whereas R224P in both retinal variants does not assemble into filaments (Extended Data Fig. 10), which agrees with previous findings that IMPDH1(514)-R224P has a reduced propensity for forming ultrastructures in cells24. In this condition, the class II mutations form filaments that are indistinguishable from WT in negative stain EM. The lack of obvious in vitro biochemical or structural phenotypes for class II mutations suggests they may lead to misregulation in vivo that is dependent on other cellular factors.
Discussion
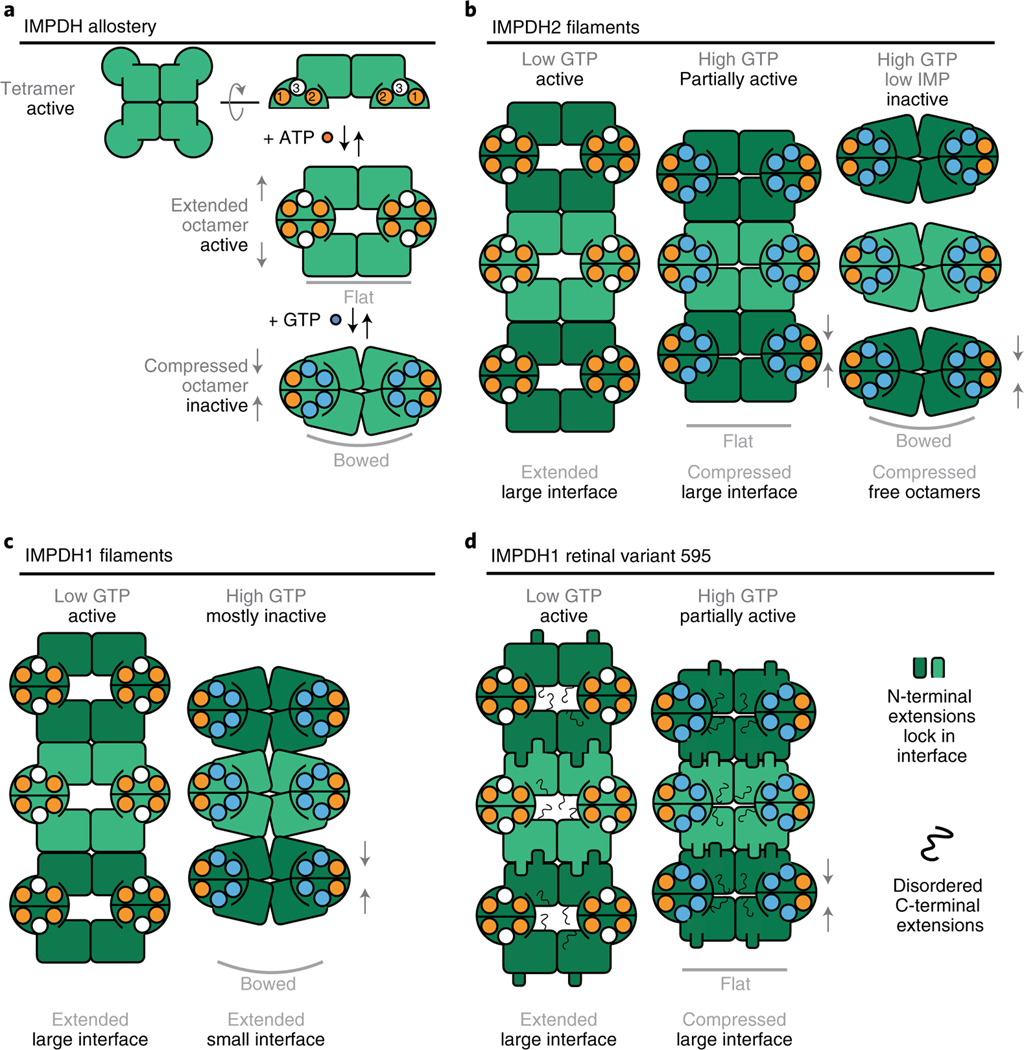
IMPDH ultrastructure assembly has been observed in many cell types and in vitro assembly of IMPDH2 has been thoroughly characterized2,10,24,44. Here, we showed that canonical IMPDH1 assembles filaments in both extended and compressed conformations (Figs. 1–3 and 7c). Other well-characterized filament-forming metabolic enzymes such as IMPDH2 and CTPS2 also switch enzyme conformations in the polymer, and constraints imposed by fixed assembly contacts give rise to filament-dependent changes in allosteric regulation10,47 (Fig. 7b). IMPDH1, on the other hand, changes both enzyme conformation and the nature of assembly contacts in transitioning between activity states. Thus, filament assembly of canonical IMPDH1 does not impose conformational constraints, explaining why we did not observe any differences in allosteric regulation in the filament compared to free enzyme (Fig. 1d).
Fig. 7 |. Model of IMPDH1 assembly and filament role in regulation.
a, ATP binding to sites 1 and 2 promotes formation of an extended octamer where the tetramer is in the flat conformation. GTP binding to sites 2 and 3 promotes formation of a compressed octamer that prefers the bowed tetramer conformation. b, In the presence of ATP, IMPDH2 assembles extended filaments. Binding of GTP leads to the assembly of partially inhibited filaments where the tetramer is in a flat conformation. In the presence of very high GTP, the tetramer is completely inhibited and enters a bowed conformation that promotes disassembly of filament into free octamers. c, For canonical IMPDH1, binding of ATP drives assembly of a filament composed of extended octamers. In the presence of GTP, canonical IMPDH1 assembles into a filament with the small interface made of fully compressed octamers that are mostly inhibited and have the bowed tetramer conformation. d, For IMPDH1 retinal variant 595, the binding of ATP drives assembly of a filament composed of extended octamers. Binding of GTP drives assembly of a filament composed of compressed octamers that are partially inhibited and have the strained tetramer confirmation. In both filaments, the N-terminal extension adds buried surface area to the large interface and the C-terminal extension is disordered.
This raises the question about the physiological function of canonical IMPDH1 filaments. Although we show that IMPDH1 assembles filaments in cells in response to inhibitors (Fig. 1b), how cellular filaments might influence metabolic flux is unclear. One possibility is that IMPDH1 filaments play some role other than direct tuning of enzyme activity, such as signaling or scaffolding of other enzymes48,49. Another possibility is that cellular factors shift the balance between the extended and compressed filament conformations, for example through posttranslational modifications or protein–protein interactions. There is precedent for the latter in the enzyme acetyl-CoA carboxylase, which assembles active filaments of one architecture, but binding to the regulator breast cancer type 1 susceptibility protein (BRCA1) stabilizes a different architecture with inactive enzyme50. Similarly, interactions with regulatory proteins may preferentially stabilize one IMPDH1 filament states. Indeed, in immunofluorescence experiments using antibodies that recognize both isoforms, IMPDH filaments colocalize with other enzymes in purine biosynthesis49, with the regulatory protein Ankyrin Repeat Domain 9 (ANKRD9)51 and with cytidine triphosphate synthase (CTPS)48. Future studies to identify interactors and probe the functional consequences of those interactions will provide valuable insight into the role of canonical IMPDH1 filament assembly in a physiological context.
This work also sheds light on the role of IMPDH1 filaments in regulating enzyme activity in the retina. The retina has extremely high purine nucleotide demands: maintenance of ion gradients across photoreceptor membranes consumes up to 108 molecules of ATP per second per cell and cGMP is essential for phototransduction33–35. IMPDH1 is the only isoform expressed in photoreceptors30, where it plays a critical role in balancing purine nucleotide pools. Two splice variants that add residues to the N and C termini of IMPDH1 are predominant in the retina30, and we found that both splice variants are less sensitive to GTP inhibition40 (Fig. 4c,d).
The N- and C-terminal retinal splice variant extensions independently increase the IC50 for GTP, consistent with the need for IMPDH1 to meet high guanine nucleotide demand in the retina31. Two structural rearrangements occur in IMPDH inhibition: the tetramer is in a bowed conformation and the octamer is fully compressed. We find that each retinal variant extension prevents one of these. The N-terminal extension effect is completely dependent on the ability of the protein to form filaments (Fig. 4e), as it tunes filament assembly so the inhibited protein assembles into the large interface filament that can only accommodate the flat/partially active tetramer. This prevents transitioning to the small filament interface, so that at high GTP concentrations the filament remains in a compressed and flat conformation that is partially active (Fig. 7d). The C-terminal extension, on the other hand, appears to destabilize the compressed conformation, either through changes at the compressed interface or through steric interference by the disordered region (Extended Data Fig. 9). The overall effect of the splice variants is to increase IMPDH1 activity under high GTP concentrations. Meeting this demand with IMPDH1 splice variants instead of IMPDH2 may provide unique regulatory advantages, through protein–protein interactions with, or posttranslational modifications of the flexible N- and C-terminal extensions.
Future studies of IMPDH1 retinal variants will be necessary to understand the layers of IMPDH1 regulation in the retina. One complication is the simultaneous presence of multiple splice variants30,42. The tetramerization and filament assembly contacts are identical among the variants so it is likely they coassemble into heterogeneous tetramers, octamers or filaments. A second complication is the interplay between three known phosphorylation sites25, assembly state and metabolic output. Phosphorylation site Ser477 is situated at the assembly interface of the large interface filament observed in the compressed retinal variant IMPDH1(595), bolstering previous speculation25 that phosphorylation at Ser477 may disrupt filament assembly, increasing GTP-feedback inhibition (Fig. 5d). Phosphomimetics at two phosphorylation sites, Thr159 and Ser150, decreased sensitivity to GTP inhibition in canonical IMPDH1 (ref. 25), but functional effects have yet to be characterized in the splice variants. Our structural and in vitro characterization of the IMPDH1 retinal variants has laid the groundwork for these future studies.
Some retinopathy mutations in IMPDH1 disrupt feedback inhibition11. Why these mutations lead to tissue-specific disease remains unclear, although because the retina only expresses IMPDH1 (refs. 29,30) it may be particularly sensitive to perturbations in the enzyme. In characterizing GTP-feedback inhibition, we found that disease-associated IMPDH1 mutants fall into two classes: class I are clustered near GTP allosteric sites and completely disrupt GTP inhibition, while class II are distal from the allosteric sites and have no effect on GTP inhibition. It seems likely that class I mutations lead to imbalanced nucleotide pools, which has been shown to lead to photoreceptor death52. Our in vitro characterization of class II mutations showed that most of them are indistinguishable from WT in terms of substrate kinetics and GTP regulation. This finding highlights the need to study these mutations in the complex photoreceptor environment.
Studies of IMPDH1 retinopathy mutants in the physiological context of the retina will be necessary to determine the molecular mechanisms of disease. Mutations are heterozygous and autosomal dominant12,19–22, so assembly of WT and mutant enzymes into heterogeneous structures made of different splice variants expressed at different levels is likely to further complicate effects on metabolic output. For example, coassembly of class I mutants and WT enzyme would likely prevent GTP-induced inhibitory conformational changes for all the copies in mixed oligomers. Another intriguing avenue of investigation is if class I and II mutations influence phosphorylation either directly or its allosteric effect. Plana-Bonamaisó et al.25 demonstrated that the disease mutations N198K and R224P in IMPDH1(546) reduced phosphorylation at 159/160 while H372P increased phosphorylation at the same site. Our characterization here of the functional differences among homogeneous assemblies of different splice variants, and of disease mutants in those assemblies, provides a basis for understanding the role of IMPDH1 in regulating nucleotide biosynthesis and mechanisms of retinopathies in the complex physiological context.
Many metabolic regulatory enzymes self-assemble into filamentous polymers53,54. In most cases filament assembly serves as an additional layer of allosteric regulation, taking advantage of existing allostery but imposing constraints on accessibility of different conformations53,54. For example, assembly of human CTPS1 into filaments stabilizes a conformation with higher specific activity to increase flux55, while CTPS2 assembles filaments that couple structural transitions to increase cooperativity of enzyme regulation47. Yeast glucokinase 1 was recently shown to assemble filaments that inactivate the enzyme, providing a mechanism to reduce overinvestment in early steps of glycolysis on sudden transition to nutrient rich environments56. Acetyl-CoA carboxylase filaments regulate activity by locking the enzyme into an active or inactive assembly57. Here, we have shown that another well-established means of metabolic regulation—tissue-specific splice variants—can add an additional layer of allosteric regulation on top of filament assembly to finely tune complex enzyme regulation (Fig. 7).
online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41594-021-00706-2.
Methods
Recombinant IMPDH expression and purification.
Purified IMPDH protein was prepared as described previously10,17. BL21 (DE3) Escherichia coli were transformed with a pSMT3-Kan vector expressing N-terminal 6xHis-SMT3/ SUMO-tagged IMPDH. Cells were cultured in Luria-Bertani medium at 37 °C until reaching an optical density (OD600) of 0.9 then expression induced with addition of isopropyl-β-D-thiogalactoside to 1 mM for 4 h at 30 °C and pelleted. The remainder of the purification was performed at 4 °C. Pellets were resuspended in lysis buffer (50 mM KPO4, 300 mM KCl, 10 mM imidazole, 800 mM urea, pH 8) and lysed with an Emulsiflex-05 homogenizer. Lysate was cleared by centrifugation and SUMO-tagged IMPDH chromatographically purified with HisTrap FF columns (GE Healthcare Life Sciences) and an Äkta Start chromatography system. After on-column washing with lysis buffer and elution (50 mM KPO4, 300 mM KCl, 500 mM imidazole, pH 8), peak fractions were treated with 1 mg of ULP1 protease58 per 100 mg IMPDH for 1 h at 4 °C, followed by the addition of 1 mM dithiothreitol (DTT) and 800 mM urea. Protein was then concentrated using a 30,000 molecular weight cutoff Amicon filter and subjected to size-exclusion chromatography using Äkta Pure system and a Superose 6 column preequilibrated in filtration buffer (20 mM HEPES, 100 mM KCl, 800 mM urea, 1 mM DTT, pH 8). Peak fractions were concentrated using a 10,000 molecular weight cutoff Amicon filter, then flash-frozen in liquid nitrogen and stored at −80 °C.
IMPDH activity assays.
Protein aliquots were diluted in activity buffer (20 mM HEPES, 100 mM KCl, 1 mM DTT, pH 7.0) and pretreated with varying concentrations of ATP, GTP and IMP for 30 min at 20 °C in 96-well ultraviolet transparent plates (Corning model 3635). Reactions (100 μl total) were initiated by addition of varying concentrations of NAD+. NADH production was measured by optical absorbance (340 nm) in real time using a Varioskan Lux microplate reader (Thermo Scientific) at 25 °C, 1 measurement per min, for 15 min; absorbance was correlated with NADH concentration using a standard curve.
Specific activity was calculated by linear interpretation of the reaction slope for a 4-min window beginning 1 min after reaction initiation. All data points reported are an average of three measurements from the same protein preparation. Error bars are standard deviation. Fits for activity assays were calculated using the Hill– Langmuir equation V = Vmax × [S]n/((K0.5)n + [S]n) and IC50 was calculated using a modified Hill equation V = Vmin × (Vmax − Vmin)/(1 + (I/IC50)n59. Where V = reaction velocity, Vmax = maximum reaction velocity, Vmin = minimum reaction velocity, [S] = substrate concentration, K0.5 = substrate concentration that gives half velocity of Vmax, n = Hill coefficient, I = inhibitor concentration, IC50 = half-maximum inhibitory concentration.
Negatively stained EM.
Protein samples were applied to glow-discharged continuous carbon EM grids and negatively stained with 2% uranyl formate as previously described10. Grids were imaged by transmission EM using an FEI Morgagni at 100 kV acceleration voltage and a Gatan Orius CCD. Micrographs were collected at a nominal ×22,000 magnification (pixel size 3.9 Å).
Cryo-EM sample preparation and data collection.
Protein preparations were applied to glow-discharged C-flat holey carbon EM grids (Protochips), blotted and plunge-frozen in liquid ethane using a Vitrobot plunging apparatus (FEI) at 4 °C, 100% relative humidity as previously described10. High-throughput data collection was performed using an FEI Titan Krios transmission electron microscope operating at 300 kV (equipped with a Gatan image filter and postGatan image filter Gatan K2 or K3 Summit direct electron detector) and an FEI Glacios (equipped with a Gatan K2 Summit direct electron detector) both using the Leginon software package60.
Cryo-EM image processing.
Videos were collected in super-resolution mode, then aligned and corrected for beam-induced motion using Motioncor2, with 2× Fourier binning and dose compensation applied during motion correction60,61. Contrast transfer function (CTF) was estimated using GCTF62. Relion v.3.1 was used for all subsequent image processing63,64. Each dataset was individually processed but using approximately the same previously published pipeline10, with some variations from dataset to dataset.
First, for most datasets, autopicking templates and initial three-dimensional (3D) references maps were prepared by manually picking and extracting boxed particles from a small subset of micrographs and classifying/refining in two and three dimensions. For a few datasets, Cryosparc Live65 was used for initial particle selection and two-dimensional (2D) classification. These particle coordinates were imported into Relion for 3D refinement. For these initial 3D refinements, a featureless, soft-edged cylinder was used as a refinement template of filaments. Because IMPDH filament segments possess D4 point-group symmetry, two different locations along filaments may be used as symmetry origins: the centers of canonical octamer segments, or the centers of the assembly interface between segments. For the filament datasets, we prepared and used autopicking templates centered on the filament assembly interface. Due to the expected flexibility of filaments, helical segments were processed as single particles, and at no point was helical symmetry applied during image processing. After template-based autopicking of each complete dataset, picked particles were boxed and extracted from micrographs, and subjected to hierarchical 2D classification to select the best-resolved classes. These selected particles were then auto-refined in 3D as a single class with D4 symmetry applied.
To improve resolution, partial signal subtraction was performed at this stage using a mask that left only the central eight catalytic domains of the filament assembly interface, and then subtracting the poorly resolved Bateman domains and neighboring segments, which often improved resolution after subsequent auto-refinement. Per-particle defocus and per-micrograph astigmatism were then optimized using CTF refinement followed by particle polishing, which generally improved resolution further (Table 1).
Model building and refinement.
Initial templates for model building were prepared from hIMPDH2 extended (Protein Data Bank (PDB) 6U8N) and compressed octamers (PDB 6U9O) with amino acid mutations to hIMPDH1 sequence made in Coot66. Where the N terminus (−22–12) location differed from IMPDH2, it was modeled by hand. After rigid-body fitting of templates into the cryo-EM densities using UCSF Chimera, repeated cycles of manual fitting with Coot, semiautomated or manual fitting with ISOLDE67, and automated fitting with phenix.real_space_ refine (using rigid-body refinement, NCS constraints, gradient-driven minimization and simulated annealing)66,68,69. Data collection parameters and refinement statistics are summarized in Table 1. Figures were prepared with UCSF Chimera68.
The sizes of interacting surfaces between IMPDH protomers were calculated using the PDB ePISA server70.
Cell culture and transfection.
HEK293 cells were grown in DMEM/10% fetal bovine serum (FBS)/1% L-glutamine on six-well dishes with coverslips in each dish to 50% confluency. They were transfected with 40 μl of Lipofectamine L-2000 (ThermoFisher catalog no. 11668030) and 16 μg of pcDNA3.1 plasmid with either IMPDH1-WT or IMPDH1-Y12A for 6 h and then the media was changed to either new DMEM/10%FBS/1% L-glutamine or DMEM/10%FBS/1% L-glutamine with 10 μM MPA. Twenty-four hours later, the cells were fixed with 4% formaldehyde/ PBS for 20 min and immunofluorescence was performed on the coverslips.
Immunofluorescence.
The coverslips with the transfected HEK293 cells were blocked with 2% bovine serum albumin (BSA)/PBS and stained with anti-myc antibody 9e10 DSHB (deposited in the Developmental Studies Hybridoma Bank by J.M. Bishop) diluted 1:50 in 2% BSA/PBS for 1 h at room temperature. Then, cells were incubated with Alexa 488 secondary goat anti-mouse (Invitrogen catalog no. A-11001) diluted 1:200 in 2%BSA/PBS for 30 min at room temperature. Finally, nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI) for 10 min at room temperature and coverslips were mounted with Vectashield mounting medium (Vector Laboratories). The cells were imaged on a Nikon Eclipse TE2000-U with a ×40 Nikon plan fluorescence objective and pictures were taken with Ocular QImaging software v.1.1 using a QImaging Retiga R1 CCD camera.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1 |. IMPDH structure and function.
a, Purine biosynthesis pathway. b, IMPDH monomer (6u9o) has a catalytic domain (green) that binds IMP and nAD+ in the active site, and a regulatory domain (pink) with three allosteric nucleotide binding sites. c, IMPDH is a tetramer in solution and can adopt a flat or bowed conformation. Side view of tetramers are depicted, so that only two monomers are visible. d, ATP (sites 1&2) or GTP (sites 2&3) binding promotes octamer assembly. e, IMPDH2 octamers can assemble into filaments of stacked octamers.
Extended Data Fig. 2 |. IMPDH1 Sequence Alignment.
Evolutionary conservation of the helix in the n-terminus of the longer retinal splice variant (blue) and the first 12 canonical residues particularly tyrosine 12 (pink).
Extended Data Fig. 3 |. IMPDH2-Wt filament resists gtP inhibition.
a,b, GTP inhibition curves of IMPDH2 or IMPDH1-WT (solid line) and the respective non-assembly y12A protein (dashed line). Individual data points are shown as diamonds (IMPDH2) or circles (IMPDH1), where filled are WT and empty y12A. reactions were performed in triplicate and the average for each concentration is shown as a bold rectangle (filled is WT, empty is y12A). Error bars are standard deviation calculated from n = 3. reactions performed with 1 μM protein, 1 mM ATP, 1 mM IMP, 300 μM nAD+, and varying GTP.
Extended Data Fig. 4 |. Cryo-eM workflow.
Flow chart summarizing data processing strategy for IMPDH1+ ATP/IMP/nAD+.
Extended Data Fig. 5 |. IMPDH1 active site map and model.
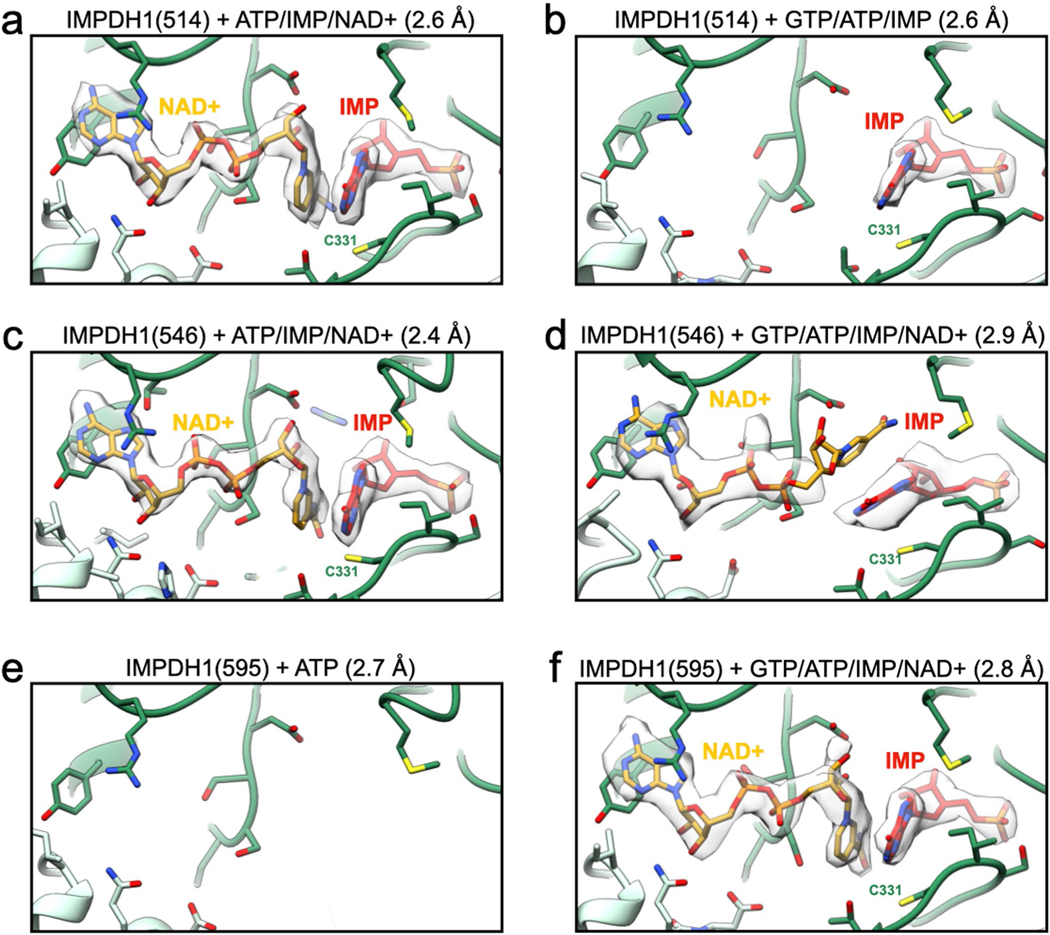
a-f, Cartoon representation of the active site. Side chains around the active site are shown as sticks. Chain A is dark green while the neighboring chain is light green. nAD+ is yellow and IMP red. Density for the ligand(s) is shown as a surface. a, IMPDH1(514) bound to ATP/IMP/nAD+. b, IMPDH1(514) bound to GTP/ATP/IMP. c, IMPDH1(546) bound to ATP/IMP/nAD+. d, IMPDH1(546) bound to GTP/ATP/IMP/nAD+. e, IMPDH1(595) bound to ATP. f, IMPDH1(595) bound to GTP/ATP/IMP/nAD+.
Extended Data Fig. 6 |. Inhibited IMPDH1-Wt tetramer is in a bowed conformation.
a, Comparison of the catalytic tetramers of inhibited IMPDH2 filament (gray; 6u8s) to inhibited IMPDH2 free octamer (6uaj). Aligned on monomers with asterisk, other monomer pair has an alpha carbon rMSD of 2.1 Å. b, Comparison of the catalytic tetramers of inhibited IMPDH2 filament (gray; 6u8s) to inhibited IMPDH1 filament. Aligned on monomers with asterisk, other monomer pair has an alpha carbon rMSD of 3.7 Å.
Extended Data Fig. 7 |. y12A non-assembly mutations prevents assembly in IMPDH1 variants.
negative stain EM of purified human IMPDH1. non-assembly mutation y12A breaks both ATP- and GTP-dependent assembly. Scale bar 100 nm. reactions performed with 1 μM protein, 1 mM ATP if used, 1 mM GTP if used.
Extended Data Fig. 8 |. IMPDH1 retinal variant (546) is similar to canonical IMPDH1.
a-c, Active IMPDH1(546) filament bound to ATP, IMP, nAD+. a, Low-pass filtered cryo-EM reconstruction b, Interface-focused cryo-EM reconstruction. 8 monomers are colored by catalytic domain (green) and regulatory domain (pink). c, View of the top of an octamer from inside the filament. The surface area buried by the octamer interface is in aqua with the indicated total buried surface area. (Surface representation of the atomic model at the assembly interface, with buried residues in cyan). d-f, Inhibited IMPDH1(546) filament bound to GTP, ATP, IMP, nAD+. d, Low-pass filtered cryo-EM reconstruction e, Interface-focused cryo-EM reconstruction. 8 monomers are colored by catalytic domain (green) and regulatory domain (pink). f, View of the top of an octamer from inside the filament. The surface area buried by the octamer interface is in aqua with the indicated total buried surface area. (Surface representation of the atomic model at the assembly interface, with buried residues in cyan).
Extended Data Fig. 9 |. IMPDH1 Retinal Variant C-term disrupts interactions.
a, Evolutionary conservation of the C-terminus in canonical IMPDH1 and both retinal splice variants. b, Surface representation of octamer side view. Dotted box indicates the region shown in c-d. c-d, Each chain is a different color green, C-term residues 510–512 in orange, and IMP in purple. c, inhibited canonical IMPDH1. d, Inhibited retinal variant IMPDH1(546).
Extended Data Fig. 10 |. IMPDH1 disease mutants have a variety of assembly phenotypes.
negative stain EM of purified human IMPDH1. Scale bar 100 nm. reactions performed with 1 μM protein, 1 mM ATP, 5 mM GTP, 3 mM IMP, 5 mM nAD+.
Supplementary Material
Acknowledgements
We thank the Arnold and Mabel Beckman Cryo-EM Center at the University of Washington for electron microscope use. We thank J. Calise for valuable feedback on the manuscript. This work was supported by the US National Institutes of Health (grant nos. R01GM118396 and R21EY031546 and S10OD032290 to J.M.K., R01GM-083025 to J.R.P., T32 CA-009035 to J.C.S. and F31EY030732 and T32GM008268 to A.L.B.), Universidad de Salamanca (Junta de Castilla y León fellowship to D.F.-J.), and Ministerio de Economía y Competitividad (grant no. BFU2016-79237-P to R.M.B.).
Footnotes
Competing interests
The authors declare no competing interests.
Data availability
The coordinates are deposited in the PDB with accession codes PDB 7RER (interface-centered extended IMPDH1(514)), PDB 7RES (octamer-centered active IMPDH1(514)), PDB 7RFE (interface-centered compressed IMPDH1(514)), PDB 7RFG (octamer-centered compressed IMPDH1(514)), PDB 7RGL (interface- centered extended IMPDH1(546)), PDB 7RGM (octamer-centered extended IMPDH1(546)), PDB 7RGI (interface-centered compressed IMPDH1(546)), PDB 7RGQ (octamer-centered compressed IMPDH1(546)), PDB 7RFF (interface- centered extended IMPDH1(595)), PDB 7RFH (octamer-centered extended IMPDH1(595)), PDB 7RFI (interface-centered compressed IMPDH1(595)), PDB 7RGD (octamer-centered compressed IMPDH1(595). The cryo-EM maps are deposited in the Electron Microscopy Data Bank (EMDB) with accession codes EMD-24437 (interface-centered extended IMPDH1(514)), EMD-24438 (octamer-centered extended IMPDH1(514)), EMD-24439 (interface-centered compressed IMPDH1(514)), EMD-24441 (octamer-centered compressed IMPDH1(514)), EMD-24451 (interface-centered extended IMPDH1(546)), EMD- 24452 (octamer-centered extended IMPDH1(546)), EMD-24450 (interface- centered compressed IMPDH1(546)), EMD-24454 (octamer-centered compressed IMPDH1(546)), EMD-24440 (interface-centered extended IMPDH1(595)), EMD24442 (octamer-centered extended IMPDH1(595)), EMD-24443 (interface- centered compressed IMPDH1(595)) and EMD-24448 (octamer-centered compressed IMPDH1(595). Source data are provided with this paper.
Additional information
Extended data are available for this paper at https://doi.org/10.1038/s41594-021-00706-2.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41594-021-00706-2.
Peer review information Nature Structural and Molecular Biology thanks Carsten Sachse and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Florian Ullrich was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team. Peer reviewer reports are available.
Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Kennan A. et al. On the role of IMPDH1 in retinal degeneration. Adv. Exp. Med. Biol 533, 13–18 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ji Y, Gu J, Makhov AM, Griffith JD & Mitchell BS Regulation of the interaction of inosine monophosphate dehydrogenase with mycophenolic acid by GTP. J. Biol. Chem 281, 206–212 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Carcamo WC, Calise SJ, von Mühlen CA, Satoh M. & Chan EKL Molecular cell biology and immunobiology of mammalian rod/ring structures. Int. Rev. Cell Mol. Biol 308, 35–74 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Calise SJ et al. Glutamine deprivation initiates reversible assembly of mammalian rods and rings. Cell. Mol. Life Sci 71, 2963–2973 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson RC, Weber G. & Morris HP IMP dehydrogenase, an enzyme linked with proliferation and malignancy. Nature 256, 331–333 (1975). [DOI] [PubMed] [Google Scholar]
- 6.Senda M. & Natsumeda Y. Tissue-differential expression of two distinct genes for human IMP dehydrogenase (E.C.1.1.1.205). Life Sci. 54, 1917–1926 (1994). [DOI] [PubMed] [Google Scholar]
- 7.Carr SF, Papp E, Wu JC & Natsumeda Y. Characterization of human type I and type II IMP dehydrogenases. J. Biol. Chem 268, 27286–27290 (1993). [PubMed] [Google Scholar]
- 8.Calise SJ, Abboud G, Kasahara H, Morel L. & Chan EKL Immune response-dependent assembly of IMP dehydrogenase filaments. Front. Immunol 9, 2789 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duong-Ly KC et al. T cell activation triggers reversible inosine5′-monophosphate dehydrogenase assembly. J. Cell Sci 10.1242/jcs.223289 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson MC & Kollman JM Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation. eLife 9, e53243 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernández-Justel D. et al. A nucleotide-dependent conformational switch controls the polymerization of Human IMP dehydrogenases to modulate their catalytic activity. J. Mol. Biol 431, 956–969 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Hedstrom L. IMP dehydrogenase-linked retinitis pigmentosa. Nucleosides Nucleotides Nucleic Acids 27, 839–849 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Zhang R. et al. Characteristics and crystal structure of bacterial inosine-5’-monophosphate dehydrogenase. Biochemistry 38, 4691–4700 (1999). [DOI] [PubMed] [Google Scholar]
- 14.Hedstrom L. IMP dehydrogenase: structure, mechanism, and inhibition. Chem. Rev 109, 2903–2928 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buey RM et al. Guanine nucleotide binding to the Bateman domain mediates the allosteric inhibition of eukaryotic IMP dehydrogenases. Nat. Commun 6, 8923 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernández-Justel D, Peláez R, Revuelta JL & Buey RM The Bateman domain of IMP dehydrogenase is a binding target for dinucleoside polyphosphates. J. Biol. Chem 294, 14768–14775 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anthony SA et al. Reconstituted IMPDH polymers accommodate both catalytically active and inactive conformations. Mol. Biol. Cell 10.1091/mbc.E17-04-0263 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buey RM et al. A nucleotide-controlled conformational switch modulates the activity of eukaryotic IMP dehydrogenases. Sci. Rep 7, 2648 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kennan A, Aherne A. & Palfi A. Identification of an IMPDH1 mutation in autosomal dominant retinitis pigmentosa (RP10) revealed following comparative microarray analysis of transcripts derived from retinas of wild-type and Rho(−/−) mice. Hum. Mol. Genet 11, 547–557 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Bowne SJ et al. Mutations in the inosine monophosphate dehydrogenase 1 gene (IMPDH1) cause the RP10 form of autosomal dominant retinitis pigmentosa. Hum. Mol. Genet 11, 559–568 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grover S, Fishman GA & Stone EM A novel IMPDH1 mutation (Arg231Pro) in a family with a severe form of autosomal dominant retinitis pigmentosa. Ophthalmology 111, 1910–1916 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Bowne SJ et al. Spectrum and frequency of mutations in IMPDH1 associated with autosomal dominant retinitis pigmentosa and Leber congenital amaurosis. Invest. Ophthalmol. Vis. Sci 47, 34–42 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Labesse G. et al. MgATP regulates allostery and fiber formation in IMPDHs. Structure 21, 975–985 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Thomas EC et al. Different characteristics and nucleotide binding properties of inosine monophosphate dehydrogenase (IMPDH) isoforms. PLoS ONE 7, e51096 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plana-Bonamaisó A. et al. Post-translational regulation of retinal IMPDH1 in vivo to adjust GTP synthesis to illumination conditions. eLife 9, e56418 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mortimer SE et al. IMP dehydrogenase type 1 associates with polyribosomes translating rhodopsin mRNA. J. Biol. Chem 283, 36354–36360 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sullivan LS et al. Prevalence of disease-causing mutations in families with autosomal dominant retinitis pigmentosa: a screen of known genes in 200 families. Invest. Ophthalmol. Vis. Sci 47, 3052–3064 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mortimer SE & Hedstrom L. Autosomal dominant retinitis pigmentosa mutations in inosine 5′-monophosphate dehydrogenase type I disrupt nucleic acid binding. Biochem. J 390, 41–47 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aherne A. et al. On the molecular pathology of neurodegeneration in IMPDH1-based retinitis pigmentosa. Hum. Mol. Genet 13, 641–650 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Bowne SJ et al. Why do mutations in the ubiquitously expressed housekeeping gene IMPDH1 cause retina-specific photoreceptor degeneration? Invest. Ophthalmol. Vis. Sci 47, 3754–3765 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong-Riley MTT Energy metabolism of the visual system. Eye Brain 2, 99–116 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Country MW Retinal metabolism: a comparative look at energetics in the retina. Brain Res. 1672, 50–57 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Arshavsky VY & Burns ME Photoreceptor signaling: supporting vision across a wide range of light intensities. J. Biol. Chem 287, 1620–1626 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo D-G, Xue T. & Yau K-W How vision begins: an odyssey. Proc. Natl Acad. Sci. USA 105, 9855–9862 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palczewski K. Chemistry and biology of the initial steps in vision: the Friedenwald lecture. Invest. Ophthalmol. Vis. Sci 55, 6651–6672 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du J, An J, Linton JD, Wang Y. & Hurley JB How excessive cGMP impacts metabolic proteins in retinas at the onset of degeneration. Adv. Exp. Med. Biol 1074, 289–295 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Charish J. cAMP and photoreceptor cell death in retinal degeneration. Adv. Exp. Med. Biol 1185, 301–304 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Spellicy CJ et al. Characterization of retinal inosine monophosphate dehydrogenase 1 in several mammalian species. Mol. Vis 13, 1866–1872 (2007). [PubMed] [Google Scholar]
- 39.Gunter JH et al. Characterisation of inosine monophosphate dehydrogenase expression during retinal development: differences between variants and isoforms. Int. J. Biochem. Cell Biol 40, 1716–1728 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Andashti B, Yazdanparast R, Barzegari E. & Galehdari H. The functional impact of the C/N-terminal extensions of the mouse retinal IMPDH1 isoforms: a kinetic evaluation. Mol. Cell. Biochem 465, 155–164 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Andashti B, Yazdanparast R, Motahar M, Barzegari E. & Galehdari H. Terminal peptide extensions augment the retinal IMPDH1 catalytic activity and attenuate the ATP-induced fibrillation events. Cell Biochem. Biophys 79, 221–229 (2021). [DOI] [PubMed] [Google Scholar]
- 42.Spellicy CJ et al. Characterization of retinal inosine monophosphate dehydrogenase 1 in several mammalian species. Mol. Vis 13, 1866–1872 (2007). [PubMed] [Google Scholar]
- 43.Keppeke GD et al. IMP/GTP balance modulates cytoophidium assembly and IMPDH activity. Cell Div. 13, 5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carcamo WC et al. Induction of cytoplasmic rods and rings structures by inhibition of the CTP and GTP synthetic pathway in mammalian cells. PLoS ONE 6, e29690 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Juda P, Smigová J, Kováčik L, Bártová E. & Raška I. Ultrastructure of cytoplasmic and nuclear inosine-5’-monophosphate dehydrogenase 2 ‘rods and rings’ inclusions. J. Histochem. Cytochem 62, 739–750 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Xu D. et al. Retinal isoforms of inosine 5′-monophosphate dehydrogenase type 1 are poor nucleic acid binding proteins. Arch. Biochem. Biophys 472, 100–104 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lynch EM & Kollman JM Coupled structural transitions enable highly cooperative regulation of human CTPS2 filaments. Nat. Struct. Mol. Biol 27, 42–48 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang C, Keppeke GD, Sung L. & Liu J. Interfilament interaction between IMPDH and CTPS cytoophidia. FEBS J. 285, 3753–3768 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao H. et al. Quantitative analysis of purine nucleotides indicates that purinosomes increase de novo purine biosynthesis. J. Biol. Chem 290, 6705–6713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hunkeler M. et al. Structural basis for regulation of human acetyl-CoA carboxylase. Nature 558, 470–474 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Hayward D. et al. ANKRD9 is a metabolically-controlled regulator of IMPDH2 abundance and macro-assembly. J. Biol. Chem 294, 14454–14466 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sharma AK & Rohrer B. Sustained elevation of intracellular cGMP causes oxidative stress triggering calpain-mediated apoptosis in photoreceptor degeneration. Curr. Eye Res 32, 259–269 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Simonet JC, Burrell AL, Kollman JM & Peterson JR Freedom of assembly: metabolic enzymes come together. Mol. Biol. Cell 31, 1201–1205 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lynch EM, Kollman JM & Webb BA Filament formation by metabolic enzymes—a new twist on regulation. Curr. Opin. Cell Biol 66, 28–33 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lynch EM et al. Human CTP synthase filament structure reveals the active enzyme conformation. Nat. Struct. Mol. Biol 24, 507–514 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stoddard PR et al. Polymerization in the actin ATPase clan regulates hexokinase activity in yeast. Science 367, 1039–1042 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hunkeler M. et al. Citrate-induced acetyl-CoA carboxylase (ACC-Cit) filament at 5.4 A resolution (Protein Data Bank, 2018); 10.2210/pdb6g2d/pdb [DOI] [Google Scholar]
- 58.Mossessova E. & Lima CD Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell 5, 865–876 (2000). [DOI] [PubMed] [Google Scholar]
- 59.Volpe DA, Hamed SS & Zhang LK Use of different parameters and equations for calculation of IC50 values in efflux assays: potential sources of variability in IC50 determination. AAPS J 16, 172–180 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suloway C. et al. Fully automated, sequential tilt-series acquisition with Leginon. J. Struct. Biol 167, 11–18 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang K. GCTF: real-time CTF determination and correction. J. Struct. Biol 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zivanov J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scheres SHW RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Punjani A. Algorithmic advances in single particle Cryo-EM data processing using CryoSPARC. Microsc. Microanal 26, 2322–2323 (2020). [Google Scholar]
- 66.Emsley P, Lohkamp B, Scott WG & Cowtan K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Croll TI ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr D. Struct. Biol 74, 519–530 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pettersen EF et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 69.Adams PD et al. in International Tables for Crystallography Vol. F, 539–547 (Wiley, 2012). [Google Scholar]
- 70.Krissinel E. & Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.