ABSTRACT
Binding to the receptor, CD4, drives the pretriggered, “closed” (state-1) conformation of the human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein (Env) trimer into more “open” conformations (states 2 and 3). Broadly neutralizing antibodies, which are elicited inefficiently, mostly recognize the state-1 Env conformation, whereas the more commonly elicited poorly neutralizing antibodies recognize states 2/3. HIV-1 Env metastability has created challenges for defining the state-1 structure and developing immunogens mimicking this labile conformation. The availability of functional state-1 Envs that can be efficiently cross-linked at lysine and/or acidic amino acid residues might assist these endeavors. To that end, we modified HIV-1AD8 Env, which exhibits an intermediate level of triggerability by CD4. We introduced lysine/acidic residues at positions that exhibit such polymorphisms in natural HIV-1 strains. Env changes that were tolerated with respect to gp120-gp41 processing, subunit association, and virus entry were further combined. Two common polymorphisms, Q114E and Q567K, as well as a known variant, A582T, additively rendered pseudoviruses resistant to cold, soluble CD4, and a CD4-mimetic compound, phenotypes indicative of stabilization of the pretriggered state-1 Env conformation. Combining these changes resulted in two lysine-rich HIV-1AD8 Env variants (E.2 and AE.2) with neutralization- and cold-resistant phenotypes comparable to those of natural, less triggerable tier 2/3 HIV-1 isolates. Compared with these and the parental Envs, the E.2 and AE.2 Envs were cleaved more efficiently and exhibited stronger gp120-trimer association in detergent lysates. These highly cross-linkable Envs enriched in a pretriggered conformation should assist characterization of the structure and immunogenicity of this labile state.
IMPORTANCE The development of an efficient vaccine is critical for combating HIV-1 infection worldwide. However, the instability of the pretriggered shape (state 1) of the viral envelope glycoprotein (Env) makes it difficult to raise neutralizing antibodies against HIV-1. Here, by introducing multiple changes in Env, we derived two HIV-1 Env variants that are enriched in state 1 and can be efficiently cross-linked to maintain this shape. These Env complexes are more stable in detergent, assisting their purification. Thus, our study provides a path to a better characterization of the native pretriggered Env, which should assist vaccine development.
KEYWORDS: human immunodeficiency virus, envelope, polymorphism, native conformation, state 1, stabilizing mutation, chemical cross-link
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) entry into target cells is mediated by the viral envelope glycoprotein (Env) trimer (1, 2). The Env trimer is composed of three gp120 exterior subunits and three gp41 transmembrane subunits (2). In infected cells, Env is first synthesized as an uncleaved precursor in the rough endoplasmic reticulum (ER), where signal peptide cleavage, folding, trimerization, and the addition of high-mannose glycans take place (3–6). Exiting the ER, the trimeric gp160 Env precursor follows two pathways to the cell surface (7). In the conventional secretory pathway, the Env precursor transits through the Golgi compartment, where it is cleaved into gp120 and gp41 subunits and is further modified by the addition of complex sugars. These mature Envs are transported to the cell surface and are incorporated into virions (8–11). In the second pathway, the gp160 precursor bypasses processing in the Golgi compartment and traffics directly to the cell surface; these Golgi compartment-bypassed gp160 Envs are excluded from virions (7).
Single-molecule fluorescence resonance energy transfer (smFRET) experiments indicate that, on virus particles, the trimer exists in three conformational states (states 1 to 3) (12). From its pretriggered conformation (state 1), the metastable Env trimer interacts with the receptors, CD4 and CCR5 or CXCR4, and undergoes transitions to lower-energy states (2, 12–23). Initially, the engagement with CD4 induces an asymmetric intermediate Env conformation, with the CD4-bound protomer in state 3 and the unliganded protomers in state 2 (24, 25). Binding of additional CD4 molecules to the Env trimer then induces the full CD4-bound, prehairpin intermediate conformation, with all three Env protomers in state 3 (24–31). An extended coiled coil consisting of the heptad repeat (HR1) region of gp41 is exposed in the prehairpin intermediate (23, 25, 29–31). State-3 Env protomers subsequently interact with the CCR5 or CXCR4 coreceptor to trigger the formation of a gp41 six-helix bundle, a process that results in fusion of the viral and target cell membranes (32–36).
Env strain variability, heavy glycosylation, and conformational flexibility contribute to HIV-1 persistence by avoiding the binding of potentially neutralizing antibodies. Mature Envs from primary HIV-1 strains largely reside in a state-1 conformation, which resists the binding of most antibodies elicited during natural infection (12, 23, 37–39); these high-titer, poorly neutralizing antibodies often recognize state-2/3 Env conformations (40–44). After years of infection, a small percentage of HIV-1-infected individuals generate broadly neutralizing antibodies (bNAbs), most of which recognize the state-1 Env conformation (12, 37, 38, 45–54). Passively administered monoclonal bNAbs have been shown to be protective in animal models of HIV-1 infection, suggesting that the elicitation of bNAbs is an important goal for vaccines (55–60). Unfortunately, bNAbs have not been efficiently and consistently elicited in animals immunized with current HIV-1 vaccine candidates, including stabilized soluble gp140 (sgp140) SOSIP.664 trimers (61–69). Compared with functional membrane Envs, differences in the antigenicity, glycosylation, and conformation of sgp140 SOSIP.664 trimers have been observed (70–77), potentially contributing to the inefficiency of bNAb elicitation. Single-molecule FRET (smFRET) analysis indicates that the sgp140 SOSIP.664 trimers assume a state-2-like conformation (78). These studies imply that the available structures of sgp140 SOSIP.664 and other detergent-solubilized Env trimer preparations (27, 28, 77, 79–91) differ from that of state-1 Env. The extent of the structural differences between the state-1 and state-2 Env conformations and their potential impact on Env immunogenicity are unknown. However, the importance of the state-1 Env as a likely target of vaccine-induced bNAbs provides a rationale for better characterization of this conformation.
HIV-1 is a polymorphic virus with a high mutation rate, allowing escape from host immune responses and antiretroviral drugs (92–99). Env polymorphisms that arise naturally or as a result of tissue culture adaptation can result in altered virus infectivity, receptor binding, or neutralization sensitivity (23, 40, 44–46, 100–119). Specifically, changes in “restraining residues” in gp120 have been shown to destabilize state 1, disrupt the closed pretriggered Env conformation, and lead to increased sampling of downstream conformations (45, 118, 119). These more “triggerable” Env mutants exhibit increased sensitivity to cold, soluble CD4 (sCD4), CD4-mimetic compounds, and poorly neutralizing antibodies (23, 37, 45, 118, 119). Less common Env alterations apparently decrease Env triggerability and stabilize a state-1 conformation (120–125).
Cross-linking of HIV-1 Env amino acid residues, in some cases combined with mass spectrometry, has been used to study Env conformations (37, 73, 126–131). Cross-linking protocols that target lysine or acidic amino acid residues on native proteins have been integrated with mass spectrometry to provide low-resolution structural information (132–135). Here, we introduced lysine and acidic amino acid residues into a primary HIV-1 Env, using natural polymorphisms as a guide. Env changes that were functionally tolerated were combined to create Envs that are potentially able to be conformationally fixed by treatment with specific cross-linking agents. In the process of generating these Env variants, we identified two common polymorphisms that increased virus resistance to cold, sCD4, and a CD4-mimetic compound, phenotypes associated with stabilization of a pretriggered (state-1) Env conformation (120–125). In combination, state-1-stabilizing changes resulted in additive Env phenotypes. Two lysine-rich variants with cold- and sCD4-resistant phenotypes were cleaved more efficiently and exhibited stronger gp120-trimer association in detergent lysates than the parental HIV-1 Env. Such highly cross-linkable and efficiently processed Envs enriched in a pretriggered conformation should assist characterization of state 1.
RESULTS
Env variants with common lysine and acidic residue polymorphisms.
We sought to create functional primary HIV-1 Env variants with an increased number of lysine/acidic residues that could be used to introduce stabilizing cross-links. To identify Env residues that might potentially tolerate such substitutions, we compared Env sequences from 193 representative group M, N, O, and P HIV-1 and SIVcpz strains (136). We identified Env residues where lysine or acidic substitutions occurred in at least 5% of these natural virus strains from more than one phylogenetic clade. The lysine polymorphisms were grouped by location in Env regions (gp41 and gp120 C terminus, gp120 trimer association domain, and gp120 inner domain) and by the number of substitutions in a set (sets 4 to 7 contain additional lysine substitutions compared with those in sets 1 to 3) (Fig. 1A). The ED2 set contains seven of the most common aspartic acid and glutamic acid polymorphisms in natural HIV-1/SIVcpz variants (Fig. 1A). None of these polymorphisms affect known sites of N- or O-linked Env glycosylation.
FIG 1.
HIV-1AD8 Env modification guided by natural polymorphisms. (A) Natural polymorphisms in HIV-1 Env were used to suggest amino acid residues that might tolerate replacement with a lysine residue or with acidic amino acid residues. The lysine substitutions are grouped according to the Env region in which the residues are located. Compared with sets 1 to 3, sets 4 to 7 contain an increased number of substitutions. (B) A schematic representation of the HIV-1AD8 Env glycoprotein is shown, with the gp120-gp41 cleavage site depicted as a black triangle. S, signal peptide; V1 to V5, gp120 major hypervariable regions; FP, fusion peptide; HR, heptad repeat region; TM, transmembrane region; CT, cytoplasmic tail. The amino acid changes associated with some of the key Env variants studied here are shown. Red vertical tick marks indicate changes in addition to those found in the 2–4 R Env.
We selected the primary clade B HIV-1AD8 as the source of the parental “wild-type” Env in this study. Primary HIV-1 Envs differ in triggerability by CD4, a property that influences virus resistance to sCD4, CD4-mimetic compounds, and some antibodies (23). The HIV-1AD8 Env is efficiently expressed and processed, is well characterized with respect to antibody binding and neutralization sensitivity (Tier 2), and, among primary HIV-1 Envs, exhibits an intermediate level of triggerability by CD4 (7, 23, 37, 73). Single, double, and triple sets of lysine substitutions were introduced into the wild-type HIV-1AD8 Env. For example, double sets included sets 1 + 2, 1 + 3, 2 + 3, 2 + 4, 3 + 5, etc.; triple sets included sets 1 + 2 + 3, 2 + 3 + 4, 2 + 3 + 6, 3 + 6 + 7, etc. (Fig. 1A). In a preliminary study, a total of 24 Env variants were analyzed for protein expression and processing, ability to support entry of a pseudotyped virus, and sensitivity of the viral pseudotype to neutralization by the 19b antibody. The 19b antibody is a poorly neutralizing antibody that recognizes the gp120 V3 loop and serves as a sensitive indicator of HIV-1 Env transitions to state-2/3 conformations (45, 71–74, 137). With a few exceptions, most of the lysine substitutions were well tolerated with respect to HIV-1AD8 Env processing, virus infectivity, and sensitivity to 19b neutralization (data not shown). However, Envs with set 3 + 7 and set 3 + 6 + 7 changes were poorly processed and inefficiently supported pseudovirus infection. Viruses with set 3 + 5 changes were more sensitive than the wild-type HIV-1AD8 to neutralization by the 19b antibody (data not shown). Thus, while most of the introduced lysine substitutions were well tolerated, some specific combinations apparently exert undesirable effects on HIV-1AD8 Env conformation and function.
Lysine-rich 2–4 R and 2–4 RED2 Envs.
Based on the results of our preliminary analysis, we selected the 2–4 R Env, which contains set 2 + 4 and R315K changes, for more detailed characterization. The ED2 set of acidic substitutions was also added to the 2–4 R Env to create the 2–4 RED2 Env (Fig. 1B). Both 2–4 R and 2–4 RED2 Envs mediated pseudovirus infection as efficiently as the wild-type HIV-1AD8 Env (data not shown). To evaluate Env expression, proteolytic processing, and gp120-trimer association, HOS cells were transfected with plasmids expressing the wild-type HIV-1AD8 Env and the 2–4 R and 2–4 RED2 Envs tagged at the C terminus with His6. Cell lysates were Western blotted directly (input) or were precipitated with nickel-nitrilotriacetic (Ni-NTA) beads in the presence of BMS-806, sCD4, or the dimethyl sulfoxide (DMSO) control. BMS-806 is a small-molecule HIV-1 entry inhibitor that binds gp120 and stabilizes a state-1-like Env conformation (12, 78, 138–140). The uncleaved gp160 Env precursor and mature gp120 and gp41 glycoproteins were detected in lysates of cells expressing the wild-type HIV-1AD8, 2–4 R, and 2–4 RED2 Envs (Fig. 2A, input lanes). Comparison of the gp120/gp160 ratio in the cell lysates indicates that the 2–4 R and 2–4 RED2 Envs are processed more efficiently than the wild-type HIV-1AD8 Env (Fig. 2A, input lanes). In the DMSO control sample, although wild-type HIV-1AD8 gp41 and gp160 were precipitated by the Ni-NTA beads, little gp120 was coprecipitated (Fig. 2A, Ni-NTA lanes). Apparently, under these conditions, gp120 dissociates from the wild-type HIV-1AD8 Env complex. BMS-806 increased the association of the wild-type HIV-1AD8 gp120 with the precipitated Env complex, as previously seen (138). In the presence of sCD4, no coprecipitated gp120 was detected, presumably as a result of CD4-induced gp120 shedding (141, 142). Compared with the wild-type HIV-1AD8 Env, the 2–4 R gp120 was precipitated more efficiently by the Ni-NTA beads in the DMSO control lysates. The coprecipitation of the 2–4 RED2 gp120 from the DMSO-treated cell lysates by the Ni-NTA beads was even more efficient. For both 2–4 R and 2–4 RED2 Envs, the association of gp120 with the Env complex was enhanced by BMS-806 and decreased by sCD4. Thus, the Env changes in 2–4 R and 2–4 RED2 can enhance Env processing and, in detergent lysates, strengthen the association of gp120 with solubilized Env trimers. Both phenotypes were more pronounced for the 2–4 RED2 Env than for the 2–4 R Env.
FIG 2.
Phenotypes of the 2–4 R and 2–4 RED2 Envs. (A) HOS cells were transfected transiently with plasmids expressing His6-tagged wild-type HIV-1AD8 Env or the 2–4 R or 2–4 RED2 Env variants. Forty-eight hours later, cells were lysed; the cell lysates were incubated with Ni-NTA beads for 1.5 h at 4°C in the presence of the DMSO control, 10 μM BMS-806, or 10 μg/mL sCD4. Total cell lysates (input) and proteins bound to the Ni-NTA beads were Western blotted with a goat anti-gp120 antibody (upper panels) or the 4E10 anti-gp41 antibody (lower panels). (B) 293T cells were transfected with plasmids encoding the indicated Envs, HIV-1 packaging proteins, and Tat and a luciferase-expressing HIV-1 vector. Forty-eight hours later, cell supernatants were filtered (0.45 μm) and incubated with different antibodies for 1 h at 37°C before the mixture was added to Cf2Th target cells expressing CD4 and CCR5. Forty-eight hours after infection, the target cells were lysed, and the luciferase activity was measured. The concentration of antibody required to inhibit 50% of virus infection (IC50) was calculated using the GraphPad Prism program. (C) Filtered cell supernatants containing recombinant viruses were incubated with sCD4 or the CD4-mimetic compound BNM-III-170 for 1 h at 37°C. The mixture was then added to target cells as described above. In the cold sensitivity assay, viruses were incubated on ice for the indicated times, after which the virus infectivity was measured. The infectivity values were normalized to those obtained without virus incubation in the cold or in the absence of inhibitor. The results shown in panels A and C are representative of those obtained in at least two independent experiments. The means and standard deviations derived from two independent experiments or triplicate measurements are shown in panels B and C, respectively.
The sensitivity of viruses with the wild-type HIV-1AD8, 2–4 R, and 2–4 RED2 Envs to neutralization by broadly and poorly neutralizing antibodies was examined. The broadly neutralizing antibodies (bNAbs) in our panel included VRC01 and VRC03 against the CD4-binding site of gp120 (143, 144), PG16 against a quaternary V2 epitope (145), PGT121 against a V3-glycan epitope on gp120 (146), and 35O22 against the gp120-gp41 interface (147). The poorly neutralizing antibodies included 17b against a CD4-induced epitope (148), 19b against the gp120 V3 loop (137), 902090 against a V2 gp120 epitope (149), and F105 against the CD4-binding site of gp120 (150). The 2–4 R and 2–4 RED2 viruses were neutralized by bNAbs comparably to the wild-type HIV-1AD8; like the wild-type HIV-1AD8, the 2–4 R and 2–4 RED2 viruses were resistant to poorly neutralizing antibodies (Fig. 2B).
The sensitivity of HIV-1 to inactivation by exposure to cold, sCD4, or CD4-mimetic compounds can provide an indication of Env “triggerability,” the tendency to make transitions from state 1 (23, 37, 45, 118, 120–125). Compared with the wild-type HIV-1AD8, the 2–4 R virus displayed slight but reproducible resistance to cold, sCD4, and BNM-III-170, a CD4-mimetic compound (151) (Fig. 2C). The 2–4 RED2 virus exhibited an even higher level of resistance to cold, sCD4, and BNM-III-170 than either the wild-type or the 2–4 R virus. These phenotypes are consistent with the stability of the state-1 Env conformation exhibiting the following rank order in these variants: 2–4 RED2 > 2–4 R > wild-type HIV-1AD8.
Q114E and Q576K changes determine state-1-stabilizing phenotypes.
We wished to identify the changes in 2–4 RED2 and 2–4 R responsible for the above-mentioned phenotypes. Because differences among the wild-type HIV-1AD8, 2–4 R, and 2–4 RED2 Envs were most apparent in the Ni-NTA coprecipitation and virus sensitivity experiments, we used these assays to characterize HIV-1AD8 Env mutants with single-residue changes corresponding to those in the 2–4 R and 2–4 RED2 Envs. Among the acidic residue substitutions found in the ED2 set, a single change, Q114E, was sufficient to recapitulate the 2–4 RED2 Env phenotypes (Fig. 3A). Similarly, a single lysine substitution originally found in set 4, Q576K, was responsible for most of the 2–4 R Env phenotypes (Fig. 3B). Thus, Q114E or Q567K alone can enhance HIV-1AD8 Env processing, gp120-trimer association, and virus resistance to cold, sCD4, and a CD4-mimetic compound.
FIG 3.
Major contributions of the Q114E and Q567K changes to the respective 2–4 RED2 and 2–4 R phenotypes. (A) The effects of the Q114E change on gp120-trimer association (left panel) and virus sensitivity to cold, sCD4, and BNM-III-170 (right panels) were measured as described in the legend to Fig. 2. The sensitivities of viruses with the wild-type HIV-1AD8 Env and the 2–4 RED2 Env are shown for comparison. (B) The effects of the Q567K change on gp120-trimer association (left panel) and virus sensitivity to cold, sCD4, and BNM-III-170 (right panels) were measured. The sensitivities of viruses with the wild-type HIV-1AD8 Env and 2–4 R Env are shown for comparison. The results shown are typical of those obtained in at least two independent experiments. The right panels report the means and standard deviations derived from triplicate measurements.
Gln 114 is located in the gp120 α1 helix, part of the gp120 inner domain that faces the trimer axis and interacts with gp41 (79–82, 152–155). Gln 567 resides in the N-terminal segment of the gp41 heptad repeat 1 (HR1N) region, which participates in the formation of the gp41 coiled coil after CD4 binding (32–34). In the available Env trimer structures, which have been suggested to represent a state-2-like conformation (78), the HR1N region is disordered or structurally heterogeneous (79–89). Although structural information on Gln 114 and Gln 567 in the context of a state-1 Env is currently unavailable, based on their approximate location near the trimer axis and the charge complementarity of the substitutions yielding similar phenotypes, we tested their functional dependence. The phenotypes of a panel of 18 single and double Q114/Q567 Env variants were characterized (Table 1). Only acidic residue substitutions at position 114 resulted in an improvement of the constellation of state-1-associated phenotypes. At position 567, lysine substitution yielded the strongest state-1-associated phenotypes, while arginine substitution exerted a more modest effect. Analysis of the double mutants yielded two insights. First, the phenotypes of the Q114E mutant were not significantly affected by changing Gln 567 to an alanine residue. Likewise, the phenotypes of the Q567K mutant were similar to those of the Q567K Q114A double mutant. Therefore, the state-1-associated phenotypes of the Q114E and Q567K mutants are not dependent on the formation of hydrogen bonds between the side chains of residues 114 and 567. Second, the phenotypic effects of the changes in residues 114 and 567 were additive. Combination of the strongest individual changes yielded the Q114E Q567K variant, with the most pronounced phenotype. Both changes are found in the 2–4 RED2 Env. In summary, the Q114E and Q567K changes independently impart their individual effects on Env function and these effects are additive.
TABLE 1.
Phenotypes of HIV-1AD8 Env variants with changes in Gln 114 and Gln 567a
| Env | Env processing | Infectivity (%) | Resistance/sensitivity compared to wild type |
gp120-trimer association | ||
|---|---|---|---|---|---|---|
| Cold | sCD4 | BNM-III-170 | ||||
| Wild type | ● | ● | ● | ● | ● | ● |
| Q114A | ● | ● | ● | ● | ● | + |
| Q114D | + | ● | RR | RR | RR | ++ |
| Q114E | + | + | RR | RR | RR | ++ |
| Q114K | None | ND | ND | ND | ND | NA |
| Q114N | ● | ND | ND | ND | ND | ● |
| Q114S | ● | ND | ND | ND | ND | ● |
| Q567A | ● | + | ● | ● | ● | ● |
| Q567E | ● | − − − | ● | ● | ● | ● |
| Q567K | + | ● | R | R | R | ++ |
| Q567R | + | − | Slight R | ● | ● | + |
| Q114A Q567K | ● | ● | Slight R | R | R | ++ |
| Q114A Q567R | ● | ● | ● | ● | ● | + |
| Q114D Q567R | + | ● | R | RR | RR | ++ |
| Q114E Q567A | + | + | RR | RR | RR | ++ |
| Q114E Q567K | + | ● | RRR | RRR | RRR | ++ |
| Q114E Q567E | + | − | R | ● | R | ++ |
| Q114E Q567R | + | ● | RR | RR | RR | ++ |
| Q114K Q567E | None | ND | ND | ND | ND | NA |
The phenotypes of the wild-type HIV-1AD8 Env and the indicated Gln 114 and Gln 567 variants were determined as described in the legend to Fig. 2. The values for Env processing efficiency, virus infectivity, and gp120-trimer association, relative to those observed for the wild-type HIV-1AD8 Env, are shown. The sensitivity or resistance of viruses with the Env variants to cold, sCD4, and BNM-III-170 is reported relative to that of the wild-type HIV-1AD8 virus. To ensure accurate comparison of the Env variant phenotypes across multiple assays, the wild-type HIV-1AD8 and key Env mutants (e.g., Q114E or Q567K) were included in all assays. Phenotypes are labeled as follows: •, wild-type level; +, increase; −, decrease; R, resistant; S, sensitive; ND, not determined; NA, not applicable. For virus infectivity: − − −, 0 to 25% of wild-type level; −, 51 to 75%; •, 76 to 125%; +, >125%. An increase in the number of R symbols indicates a greater level of resistancecompared to the wild-type level. The levels of gp120-trimer association exhibit the following order: ++ > + > •, where • indicates the wild-type level. The data shown are representative of results obtained in at least two independent experiments.
We extended our mutagenesis approach to evaluate the potential of other Env residues to influence the Q114E and Q567K phenotypes. A state-1 Env structure would be most relevant to the search for interacting partners but is currently not available. Therefore, we used the available structural models, many of which represent state-2-like Env conformations (78), to suggest candidate amino acid residues. In sgp140 SOSIP.664 trimers, the highly conserved His 72 is located ∼8 Å from Gln 114 (79–81). Replacing His 72 with lysine or glutamine residues resulted in increased sensitivity to sCD4 and BNM-III-170; these phenotypes were partially relieved when these His 72 changes were combined with Q114E (Table 2). Replacing His 72 with an alanine residue resulted in a virus with a neutralization sensitivity similar to that of the wild-type virus. Compared with the Q114E virus, the H72A Q114E virus was less resistant to cold and sCD4. Thus, some changes in His 72 result in an apparent increase in Env triggerability and can influence the Q114E phenotypes.
TABLE 2.
Effects of Env amino acid changes on the phenotypes of the Q114E and Q567K Env variantsa
| Env | Env processing | Infectivity (%) | Resistance/sensitivity compared to wild type |
gp120-trimer association | ||
|---|---|---|---|---|---|---|
| Cold | sCD4 | BNM-III-170 | ||||
| Wild type | ● | ● | ● | ● | ● | ● |
| Q114E | + | + | RR | RR | RR | ++ |
| Q567K | ● | ● | R | R | R | ++ |
| Q114E Q567K | + | ● | RRR | RRR | RRR | ++ |
| H72A | − − | − − − | R | ● | ● | NA |
| H72K | − − | − − − | ● | SS | SS | NA |
| H72Q | − − | − − − | ● | ND | SS | NA |
| H72A Q114E | − | − − | R | ● | RR | − |
| H72K Q114E | − | − − − | ● | S | SS | − |
| H72Q Q114E | − | − | R | S | ● | − |
| T202K | ● | − | SS | S | SS | ● |
| T202R | ● | − | SS | S | SS | ● |
| T202A | ● | − | SSS | SSS | SSS | − |
| T202Q | ● | ● | SSS | SSS | SSS | − |
| Q114E T202K | + | + | SS | S | SS | + |
| Q114E T202R | + | + | SS | S | SS | + |
| Q114E T202A | ● | ● | SSS | SS | SSS | ● |
| Q114E T202Q | ● | + | SSS | SS | SSS | ● |
| Q203A | ● | ● | SS | ND | SSS | − |
| Q114E Q203A | + | ● | ● | ND | ● | ● |
| K117A | ● | ● | RR | ND | RR | ● |
| K117Q | ND | − | R | ND | RR | ND |
| K121A | ● | − − | ● | ND | RR | − |
| K121Q | ND | − | R | ND | RR | ND |
| Q114E K117A | ● | ● | RR | ND | RR | ● |
| Q114E K121A | + | − | Slight R | ND | RR | + |
| H66N | ● | − | R | RRR | RRR | ● |
| A582T | ● | ● | RRR | RR | RR | + |
| L587A | − | ● | RR | R | RR | − |
| Q114E H66N | + | ● | RR | RRR | RRR | ++ |
| Q114E A582T | + | ● | RRR | RR | RRR | ++ |
| Q567K A582T | ● | ● | RRR | RR | RRR | + |
| Q114E L587A | + | ● | RR | RR | RR | ● |
| Q114E Q567K A582T | + | + | RRRR | RRRR | RRRR | ++ |
| Q114E Q567K L587A | + | − | RR | RR | RRR | + |
The phenotypes of the wild-type and mutant HIV-1AD8 Envs were determined as described in the legend to Fig. 2. The values, relative to those of the wild-type HIV-1AD8 Env, are reported as described in Table 1, footnote a. The phenotypes are labeled as follows: •, wild-type level; +, increase; −, decrease; R, resistant; S, sensitive; ND, not determined; NA, not applicable. Relative to the wild-type level (•), the levels exhibit the following order: ++ > + >•> − > − − > − − −. For virus infectivity: − − −, 0 to 25% of wild-type level; − −, 26 to 50%; −, 51 to 75%; •, 76 to 125%; +, >125%. An increase in the number of R or S symbols indicates a greater level of resistance or sensitivity, respectively, compared to the wild-type level. The data shown are representative of results obtained in at least two independent experiments.
In HIV-1/SIVcpz Envs, Thr/Lys polymorphism in residue 202 often exhibits covariance with Gln/Glu polymorphism in residue 114 (136). Compared with the wild-type HIV-1AD8, viruses with Thr 202 replaced by alanine, lysine, arginine, or glutamine residues were more sensitive to cold, BNM-III-170, and the 19b anti-V3 antibody (Table 2 and data not shown). These phenotypes, which are indicative of increased Env triggerability and state-1 destabilization, were minimally compensated by the addition of the Q114E change. Replacing the conserved Gln 203 residue with an alanine residue (Q203A in Table 2) also resulted in a state-1-destabilized phenotype, but in this case, the Q114E Q203A mutant exhibited phenotypes close to that of the wild-type HIV-1AD8. Thus, the Q114E change can compensate for some but not all state-1-destabilizing changes.
In the unliganded sgp140 SOSIP.664 and PGT151-bound EnvΔCT structures (PDB ID 4ZMJ and 5FUU, respectively) (82, 86), the side chains of Gln 114, Lys 117, and Lys 121 from each Env protomer point toward the trimer axis, stacking in three layers. Interprotomer Lys 117-Lys 117 and Lys 121-Lys 121 cross-links were formed in a cross-linking/mass spectrometry study of the sgp140 SOSIP.664 trimer, confirming the location of these residues in the trimer core in these Env structures (73). Substitution of Lys 117 or Lys 121 with an alanine or glutamine residue resulted in viruses that were more resistant to cold and BNM-III-170 than the wild-type virus (Table 2). No additive or synergistic effect was observed when the Q114E change was combined with the K117A or K121A changes. In fact, the double mutants exhibited less stable association of gp120 with solubilized Env trimers (Table 2). Thus, the effects of the Gln 114, Lys 117, and Lys 121 changes on the viral phenotypes are redundant, whereas in the detergent-solubilized Envs, the K117A and K121A changes nullify the trimer-stabilizing effects of the Q114E change. Similar phenotypic effects of the K117A and K121A changes were observed in the context of the E.2 and AE.2 HIV-1AD8 constructs discussed below (Table 3).
TABLE 3.
Effects of Env amino acid changes on the phenotypes of the E.2, AE.2, and 2–4 RM6 AE Env variantsa
| Env | Env processing | Infectivity (%) | Resistance/sensitivity compared to wild type |
gp120-trimer association | ||
|---|---|---|---|---|---|---|
| Cold | sCD4 | BNM-III-170 | ||||
| Wild type | ● | ● | ● | ● | ● | ● |
| E.2 | + | + | RRR | RRR | RRR | ++ |
| AE.2 | + | − | RRRR | RRRR | RRRR | ++ |
| E.2 K117A | + | ● | RRR | RRR | RRR | − |
| AE.2 K117A | + | − − | RRRR | RRRR | RRRR | + |
| K59A | ● | ● | R | R | RR | ● |
| V255I | ● | ● | RR | NA* | NA* | ● |
| E.2 K59A | + | − | RRR | RRR | RRR | ++ |
| AE.2 K59A | + | − − | RRRR | RRRR | RRRR | ++ |
| E.2 K59A V255I | + | − − − | RRRR | NA* | NA* | ++ |
| E.2 V255I | + | − | RRRR | NA* | NA* | ++ |
| AE.2 V255I | + | − − − | RRRR | NA* | NA* | ++ |
| AE.2 K59A V255I | + | − − − | RRRR | NA* | NA* | ++ |
| K59A Q114E V255I | + | − − | RRRR | NA* | NA* | ++ |
| R542A | − | − − − | S | SSS | SSS | − |
| I595F | − | − | R | SSS | SSS | − |
| L602H | − | − | S | SSS | SSS | − |
| 2−4 RM6 AE | + | − | RRR | RRR | RRR | ++ |
| 2−4 RM6 AE R542A | + | − − − | RR | RRR | RR | ++ |
| 2−4 RM6 AE I595F | + | − − − | R | ● | ● | ++ |
| 2−4 RM6 AE L602H | + | − − | RRR | RRR | RR | ++ |
The indicated amino acid changes were introduced into the HIV-1AD8 Env or into the E.2, AE.2, or 2–4 RM6 AE Envs. The phenotypes of these Env variants were determined as described in the legend to Fig. 2. The phenotypes are labeled as follows: •, wild-type level; +, increase; −, decrease; R, resistant; S, sensitive. Relative to the wild-type level (•), the levels exhibit the following order: ++ > + >•> − > − − > − − −. For virus infectivity: − − −, 0 to 25% of wild-type level; − −, 26 to 50%; -, 51 to 75%; •, 76 to 125%; +, >125%. An increase in the number of R or S symbols indicates a greater level of resistance or sensitivity, respectively, compared to the wild-type level. The values, relative to those of the wild-type HIV-1AD8 Env, are reported as described in Table 1, footnote a. The data shown are representative of results obtained in at least two independent experiments. *NA (not applicable); because Val 255 is near the binding site for sCD4 and the CD4-mimetic compounds, the V255I change may directly decrease the binding of these Env ligands. Therefore, the sensitivity of Env mutants containing the V255I change to sCD4 and BNM-III-170 does not allow conclusions about the conformational state of these Env variants.
As Gln 567 is disordered in most Env trimer structures, we used a low-resolution model of the uncleaved HIV-1JR-FL Env (156) to suggest potential interaction partners. However, alanine substitutions in these potentially interacting HIV-1AD8 residues (Glu 47, Glu 83, Glu 87, Glu 91, Asp230, Glu 492, and Glu 560) did not affect the phenotypes of the Q567K mutant virus (data not shown).
Q114E and Q567K synergize with other state-1-stabilizing Env changes.
Previous studies suggested that changes in His 66, Ala 582, and Leu 587 could enrich the state-1 HIV-1YU2 Env conformation through different proposed mechanisms: H66N destabilizes the CD4-bound conformation, A582T directly stabilizes the pretriggered conformation, and L587A destabilizes the gp41 3-helix bundle (121, 122, 125). We confirmed that individually these changes increased HIV-1AD8 resistance to cold, sCD4, and BNM-III-170 (Table 2). Of the three changes, only A582T enhanced gp120-trimer association in cell lysates. Both the H66N and A582T changes synergized with the Q114E and Q567K changes in producing viral phenotypes associated with state-1 stabilization (Table 2). A combination of three changes in the Q114E Q567K A582T Env resulted in the most robust phenotypes.
Cross-linkable E.2 and AE.2 Envs with enhanced state-1 stability.
To generate HIV-1 Envs enriched in a pretriggered conformation and containing multiple lysine residues for cross-linking, we added two lysine substitutions (R252K, A667K) and Q114E to the lysine-rich 2–4 R Env to create the E.2 Env construct (Fig. 1). The AE.2 Env contains, in addition, the A582T change. The A582T change was chosen because it not only resulted in viral phenotypes additive with those of Q114E and Q567K but also increased gp120 association with the detergent-solubilized Env, a property that K117A, K121A, H66N, and L587A lacked (Table 2). Both E.2 and AE.2 Env were cleaved more efficiently than the wild-type HIV-1AD8 Env and resisted gp120 dissociation from the solubilized Env trimer (Fig. 4A). By comparison, the wild-type Env from another primary strain, HIV-1JR-FL, was poorly processed and highly unstable in detergent.
FIG 4.
Phenotypes of the E.2 and AE.2 Envs. (A) HOS cells transiently expressing His6-tagged Envs (wild-type HIV-1AD8 Env, the E.2 Env, the AE.2 Env, or the HIV-1JR-FL Env) were lysed. Cell lysates were incubated with Ni-NTA beads for 1.5 h at 4°C in the presence of the DMSO control or 10 μM BMS-806. Total cell lysates (input) and Ni-NTA-bound proteins were Western blotted with a goat anti-gp120 antibody (upper panels) and the 4E10 anti-gp41 antibody (lower panels). (B) Recombinant luciferase-expressing viruses with the indicated Envs were incubated with antibodies for 1 h at 37°C, after which the mixture was added to Cf2Th-CD4/CCR5 target cells. Forty-eight hours later, the target cells were lysed and the luciferase activity was measured. The IC50 values were calculated using the GraphPad Prism program. (C) Recombinant luciferase-expressing viruses with the indicated Envs were incubated with sCD4 or BNM-III-170 for 1 h at 37°C before the mixture was added to Cf2Th-CD4/CCR5 target cells. Cold sensitivity was assessed by incubation of the viruses on ice for the indicated times, after which virus infectivity was measured on Cf2Th-CD4/CCR5 cells as described above. The results are representative of those obtained in at least two independent experiments. The values reported in panels B and C represent the means and standard deviations from at least two independent experiments or triplicate measurements, respectively.
To evaluate the functional E.2 and AE.2 Envs in more detail, we tested virus sensitivity to a panel of broadly and poorly neutralizing antibodies. In addition to the antibodies used in Fig. 2, we included two bNAbs, PGT151 against the gp120-gp41 interface (157) and PGT145 against a quaternary V2 epitope (158), and the poorly neutralizing F240 antibody against gp41 (159). Envs from the clade B HIV-1JR-FL and clade A HIV-1BG505 tier 2/3 strains were included for comparison. All Env variants resisted neutralization by poorly neutralizing antibodies, as expected (Fig. 4B). Compared to the wild-type HIV-1AD8, HIV-1JR-FL, and HIV-1BG505, the E.2 and AE.2 viruses were just as sensitive, and even more sensitive in some cases, to neutralization by broadly neutralizing antibodies.
The sensitivity of the viruses to cold inactivation, sCD4, and the CD4-mimetic compound BNM-III-170 is shown in Fig. 4C. Compared with the wild-type HIV-1AD8, the E.2 virus exhibited increased resistance to cold, sCD4, and BNM-III-170. Alteration of Glu 114 in the E.2 Env to glutamine largely reverted these phenotypes, suggesting that the Q114E change is a critical determinant of the stabilized pretriggered conformation in the E.2 Env (data not shown). The inclusion of the A582T change in the AE.2 Env further increased cold, sCD4, and BNM-III-170 resistance to the levels of the tier 2/3 HIV-1JR-FL and HIV-1BG505 strains. In addition, the E.2 and AE.2 viruses were more sensitive than the wild-type HIV-1AD8 to the state-1-preferring entry inhibitors, BMS-806 and 484 (45, 118); the AE.2 virus was more sensitive to these small-molecule inhibitors than the E.2 virus (data not shown).
In an attempt to improve the E.2 and AE.2 Envs further, we added the K59A and/or V255I changes. Lysine 59 is a highly conserved residue in the gp120 inner domain, within the disulfide loop (layer 1) that includes His 66, discussed above. Valine 255 packs against the critical Trp 112 and Trp 427 residues in the CD4-binding Phe 43 cavity of gp120 (152); the V255I change was associated with resistance to AAR029b, a cyclic peptide triazole inhibitor of CD4 binding (160). The K59A and V255I changes alone rendered HIV-1AD8 more cold resistant, and the K59A virus was also relatively resistant to sCD4 and BNM-III-170 (Table 3). However, the K59A and V255I changes had only modest effects in the E.2 and AE.2 background on state-1-associated phenotypes but led to significant reductions in infectivity (Table 3). These observations hint that further stabilization of state-1-associated phenotypes in the AE.2 context may be accompanied by decreases in Env function.
Effects of state-1-destabilizing changes in different Env contexts.
In the above-described studies, the Q114E change could revert the viral phenotypes associated with state-1 destabilization by the Q203A change but not by changes in the adjacent Thr 202 residue (Table 2). We evaluated whether an Env with multiple state-1-stabilizing changes, 2–4 RM6 AE, would better tolerate state-1 destabilization. The 2–4 RM6 AE and AE.2 Envs are identical except for the R252K change in the latter (Fig. 1). The 2–4 RM6 AE virus is resistant to cold, sCD4, and BNM-III-170 and exhibits strong gp120-trimer association in detergent (Table 3). We individually introduced the R542A, I595F, and L602H changes into the wild-type HIV-1AD8 Env or the 2–4 RM6 AE Env. These gp41 changes rendered HIV-1 more sensitive to the nonpeptidic inhibitory compound RPR103611, which suggested that they might destabilize the pretriggered (state-1) Env conformation (161). In agreement with this hypothesis, the R542A and L602H HIV-1AD8 variants exhibited increased sensitivity to cold, sCD4, and BNM-III-170 relative to HIV-1AD8 (Table 3). The I595F virus was sensitive to sCD4 and BNM-III-170 as well as to the 19b anti-V3 antibody but was slightly more resistant to cold inactivation than HIV-1AD8. Interestingly, increases in sensitivity to cold, sCD4, BNM-III-170, and 19b associated with these gp41 changes were not evident in the 2–4 RM6 AE background (Table 3). Thus, the state-1-stabilizing changes in 2–4 RM6 AE apparently resist the state-1-destabilizing effects of the R542A, I595F, and L602H changes in the gp41 ectodomain.
Correlations among key Env phenotypes.
To understand the relationships among key Env phenotypes and to visualize the effects of specific amino acid changes on the progression of successive generations of Env mutants, we plotted the relative levels of resistance to cold, BNM-III-170, and gp120-trimer dissociation for all characterized Env variants (Fig. 5). Virus resistance to cold inactivation reflects the stability of the functional Env trimer on virions and is independent of the binding of an Env ligand. Virus resistance to the CD4-mimetic compound generally correlates with resistance to sCD4 (122, 154, 162). Of interest, there exists a strong correlation between virus resistance to the CD4-mimetic compound and to cold (Fig. 5). Beginning with the wild-type HIV-1AD8 Env, Envs incorporating additive state-1-stabilizing changes displayed upward shifts toward highly resistant phenotypes, comparable to those of the HIV-1JR-FL and HIV-1BG505 Envs. Envs with state-1-destabilizing changes grouped together in the lower left quadrant.
FIG 5.
Correlations among key Env phenotypes. The plot shows the relative level of resistance to the CD4-mimetic compound BNM-III-170 versus the relative level of cold resistance for the HIV-1 Env variants tested in this study. The levels of resistance are scored as described in the footnotes to Tables 1 to 3: •, wild-type level; R, resistant; S, sensitive. Key Env variants are designated with stars. Envs are colored according to their relative gp120-trimer association level reported in Tables 1 to 3, as measured by Ni-NTA coprecipitation of gp120 with the His6-tagged gp41 glycoprotein: black, wild-type level; blue, not determined or not applicable; light green, +; green, ++; red, −. The V255I change is located near the binding site for CD4-mimetic compounds and may directly affect Env interaction with BNM-III-170. Note that the E.2 and AE.2 Envs exhibit resistance to cold and BNM-III-170 comparable to those of the HIV-1JR-FL and HIV-1BG505 Envs but also display significantly better gp160 processing. Relative to the HIV-1JR-FL Env, the E.2 and AE.2 Envs exhibit a tighter association of gp120 with the Env trimer solubilized in detergent.
Env variants that exhibited a higher level of gp120-trimer association in detergent, relative to that of the wild-type HIV-1AD8 Env, are colored green in Fig. 5. The skewed distribution of these Env variants in the upper right quadrant indicates that a tighter association of gp120 with the solubilized Env trimer is related to virus resistance to cold and BNM-III-170, phenotypes associated with state-1 stabilization. Note that several HIV-1AD8 Env variants and the natural HIV-1JR-FL Env achieve virus resistance to cold and BNM-III-170 without increasing gp120-trimer association in detergent-solubilized Envs (Fig. 5, black symbols in the upper right quadrant). Therefore, increasing gp120-trimer association is not the only means of achieving a more stable pretriggered (state-1) Env conformation.
Cross-linking efficiency of the wild-type AD8, E.2, and AE.2 Envs.
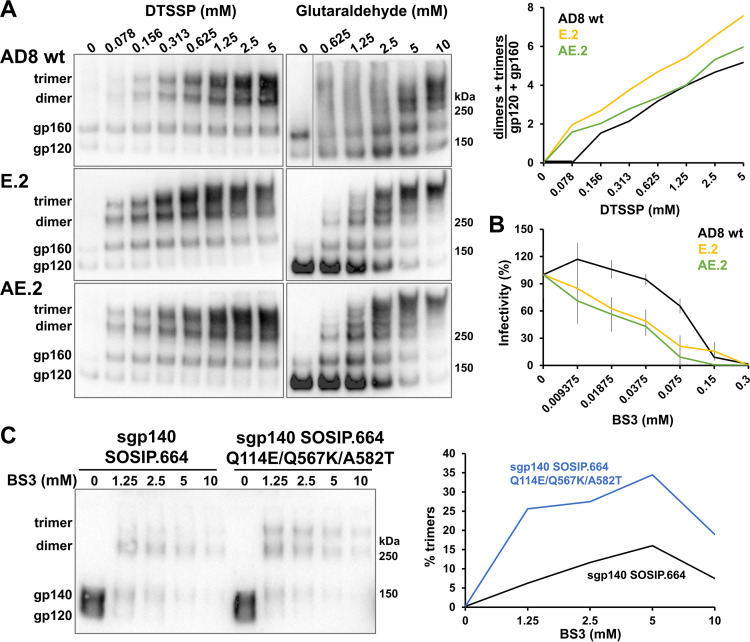
The lysine-rich E.2 and AE.2 Envs are expected to cross-link more efficiently than the wild-type HIV-1AD8 Env with lysine-reactive cross-linkers like 3,3′-dithiobis (sulfosuccinimidyl propionate) [DTSSP] and glutaraldehyde. DTSSP has a spacer arm of 12 Å, whereas, because of its tendency to polymerize, glutaraldehyde forms cross-links of more variable lengths (163). Both DTSSP and glutaraldehyde cross-linked the E.2 and AE.2 Envs more efficiently than the wild-type AD8 Env (Fig. 6A). For example, after treatment with 5 mM glutaraldehyde, the E.2 and AE.2 Envs cross-linked into gel-stable trimers, whereas the wild-type HIV-1AD8 Env mostly formed monomers and dimers. Apparently, a greater number of lysine residues accessible to the cross-linkers exist on the E.2 and AE.2 Env trimers than on the wild-type HIV-1AD8 Env.
FIG 6.
Cross-linking of membrane and soluble Envs with state-1-stabilizing changes. (A) VLPs consisting of the HIV-1 Gag-mCherry fusion protein and the wild-type HIV-1AD8 Env, the E.2 Env, or the AE.2 Env were incubated with different concentrations of the DTSSP or glutaraldehyde cross-linkers. After the reactions were quenched, VLPs were pelleted and lysed. The VLP proteins cross-linked with DTSSP or glutaraldehyde were analyzed by nonreducing or reducing PAGE, respectively, followed by Western blotting. The ratio of gel-stable (dimers + trimers) to (gp120 + gp160) provides an indication of interprotomer cross-linking by DTSSP (upper right panel). (B) Luciferase-expressing viruses pseudotyped with the wild-type HIV-1AD8, E.2, or AE.2 Envs were incubated with the BS3 cross-linker. After quenching the reaction, the viruses were added to Cf2Th-CD4/CCR5 cells. Luciferase activity in the target cells was measured 48 h later. (C) The HIV-1AD8 sgp140 SOSIP.664 and sgp140 SOSIP.664 Q114E/Q567K/A582T glycoproteins in the supernatants of transfected 293T cells were incubated with the indicated concentrations of the BS3 cross-linker and then analyzed by Western blotting. The percentage of the sgp140 SOSIP.664 glycoproteins that cross-link into trimers is plotted in the graph on the right. The results are representative of those obtained in at least two independent experiments. The values reported in panel B represent the means and standard deviations derived from triplicate experiments.
We also examined the relative sensitivity of the functional wild-type HIV-1AD8, E.2, and AE.2 Envs to bissulfosuccinimidyl suberate (BS3), another lysine-specific cross-linker with spacer arms of 12 Å. The infectivity of viruses pseudotyped with the E.2 and AE.2 Envs was inhibited by BS3 at 3- to 4-fold lower concentrations than those required for inhibition of viruses with the wild-type HIV-1AD8 Env (Fig. 6B). These results suggest that BS3 cross-links occur more efficiently on the E.2 and AE.2 Envs than on the wild-type HIV-1AD8 Env, leading to a loss of infectivity at lower BS3 concentrations.
We tested the potential effect of the three amino acid changes that stabilize state 1 in the HIV-1AD8 membrane Env on the integrity of a soluble Env trimer. Envs from different HIV-1 strains form sgp140 SOSIP.664 trimers with various levels of efficiency (63, 164) and could potentially benefit from additional stabilizing modifications. The effect of the three key state-1-stabilizing Env changes (Q114E/Q567K/A582T) on the oligomerization of the HIV-1AD8 sgp140 SOSIP.664 glycoprotein was examined by cross-linking with saturating levels of BS3. Compared with the HIV-1AD8 sgp140 SOSIP.664 glycoprotein, sgp140 SOSIP.664 with the Q114E/Q567K/A582T changes exhibited 2- to 4-fold more trimers (Fig. 6C). This result indicates that the Q114E, Q567K, and A582T changes can enhance the trimerization of the sgp140 SOSIP.664 glycoproteins.
DISCUSSION
Despite more than 3 decades of intense research, an effective HIV-1 vaccine remains elusive. The metastability and multiple conformational states of the HIV-1 Env create challenges for the generation of broadly neutralizing antibodies, either following vaccination or during natural HIV-1 infection. In Env-expressing cells, both uncleaved and cleaved (mature) Envs are present on the cell surface. A significant fraction of the uncleaved Env bypasses the conventional Golgi secretory pathway to traffic to the cell surface; these Envs differ from mature Envs in glycan processing, conformation, and recognition by antibodies (7). Uncleaved Envs may function as a decoy to the host immune system and divert antibody responses away from the mature Envs. The pretriggered (state-1) conformation of the mature virion Env of primary HIV-1 strains is the target for most broadly neutralizing antibodies (12, 37, 38, 45). This native conformation, however, is unstable and can transition into more open state-2/3 conformations that are able to be recognized by poorly neutralizing antibodies. Therefore, it is of significant interest to devise methods to lock Env in its native state-1 conformation by means that resist perturbation during Env purification, characterization, and immunization.
Here, we tackled the challenges posed by HIV-1 Env conformational flexibility in two ways. First, we used polymorphisms in naturally occurring HIV-1 strains to guide the introduction of extra lysine and acidic amino acid residues in the HIV-1AD8 Env. Chemical cross-linkers that couple lysine or acidic residues on proteins under physiological conditions are available (132–135). During the iterative process employed to identify HIV-1AD8 Envs that are potentially more susceptible to cross-linking, we required that the Env variants exhibit efficient processing, subunit association, and the ability to support virus entry. Some of the functional HIV-1AD8 Env variants developed by this approach contain up to 11 extra lysine residues (33 per Env trimer) and up to 7 extra acidic residues (21 per Env trimer). Using DTSSP or glutaraldehyde as cross-linking agents, two Env variants, E.2 and AE.2, were shown to form interprotomer cross-links more efficiently than the wild-type HIV-1AD8 Env. The infectivity of viruses with these Envs was inactivated more efficiently than that of viruses with the wild-type HIV-1AD8 Env by another lysine-specific cross-linker, BS3. These assays document the accessibility of some of the additional lysine residues introduced into the E.2 and AE.2 Envs. Chemical cross-linking can enrich the representation of otherwise labile native conformations in Env preparations for structural analysis or immunogenicity studies. Cross-linking/mass spectrometry can provide distance constraints between Env residues that can be used to validate available structural models or to derive new models (132–135). A previous study utilized cross-linking/mass spectrometry to detect differences between soluble and membrane-bound Envs (73). The inclusion of the 2–4 RED2, E.2, and AE.2 Envs in future cross-linking/mass spectrometry studies should increase the number of distance constraints and thereby improve our ability to discriminate among alternative structural models.
The second strategy employed in our approach was to screen the Env variants for function and viral phenotypes associated with stabilization of a state-1 Env conformation. For this purpose, we evaluated viral resistance to cold, sCD4, and the CD4-mimetic compound BNM-III-170. Cold inactivation reflects the resistance of the functional HIV-1 Env trimer to the detrimental effects of ice formation at near-freezing temperatures (165–167). The sensitivity of HIV-1 variants to cold inactivation is related to the intrinsic reactivity or triggerability of Env; Envs that more readily make the transition from state 1 to downstream conformations are invariably cold sensitive (23, 121, 122). HIV-1 sensitivity to sCD4 and BNM-III-170 inhibition is a function of Env triggerability (23, 45, 118, 119, 125); because we generally avoided changes to the highly conserved and well-defined BNM-III-170 binding site on gp120 (151, 162), most of the observed differences in virus sensitivity to this CD4-mimetic compound reflect changes in the ability of Env to negotiate transitions from a state-1 conformation. Our study documents the strong correlation between HIV-1 resistance to cold and resistance to BNM-III-170. This screening strategy identified two changes, Q114E in gp120 and Q567K in gp41, that individually increased the resistance of the HIV-1AD8 Env to inactivation by cold, sCD4, and BNM-III-170. These viral phenotypes were additively enhanced by combining the Q114E and Q567K changes in Env variants, such as the lysine-rich E.2 and AE.2 Envs. Cold, sCD4, and BNM-III-170 resistance were further increased by the inclusion in the AE.2 Env of the A582T gp41 change, which previously was shown to stabilize a pretriggered Env conformation (123, 125). Viruses with the E.2 and AE.2 Envs were inhibited efficiently by BMS-806, a small molecule that exhibits some preference for a state-1 Env conformation (12, 78, 138–140). The functional E.2 and AE.2 Envs exhibited an antigenic profile consistent with a state-1 conformation, conferring virus sensitivity to bNAbs that target quaternary epitopes (PG16, PGT145, PGT151, 35O22) and resistance to poorly neutralizing antibodies (17b, 19b, 902090, F105, F240). Of note, compared with the wild-type HIV-1AD8, viruses with the E.2 and AE.2 Envs were more sensitive to neutralization by the 10E8.v4 bNAb directed against the gp41 membrane-proximal external region (MPER) (168) (Fig. 4B). Although 10E8.v4 can recognize Env prior to receptor engagement (169), in contrast to most MPER-directed bNAbs (170), stabilization of the state-1 Env conformation would not necessarily be expected to increase virus sensitivity to 10E8.v4 neutralization. Indeed, upon further investigation, we found that an HIV-1AD8 mutant with the three main state-1-stabilizing changes (Q114E/Q567K/A582T) was inhibited by 10E8.v4 comparably to wild-type HIV-1AD8 (171). Therefore, the relatively increased sensitivity of the E.2 and AE.2 viruses to 10E8.v4 neutralization is apparently due to changes in these Envs other than those related to state-1 stabilization. We note that both E.2 and AE.2 Envs have lysine substitutions within and near the gp41 MPER that could potentially affect 10E8.v4 bNAb recognition.
Two unanticipated beneficial phenotypes associated with the E.2 and AE.2 Envs are more efficient Env processing and greater stability of solubilized Env trimers. HIV-1 Env cleavage has been suggested to contribute to the stability of the state-1 conformation (138, 139, 172–177). As uncleaved HIV-1 Envs sample multiple conformations, including those reactive with poorly neutralizing antibodies, achieving a high level of gp120-gp41 processing may be important for an effective vaccine immunogen. The E.2 and AE.2 Envs achieve levels of state-1-associated phenotypes comparable to those of the tier 2/3 HIV-1JR-FL and HIV-1BG505 but. notably, are processed much more efficiently. In addition, relative to the wild-type HIV-1AD8 and HIV-1JR-FL Envs, the E.2 and AE.2 Envs solubilized in detergent exhibit much greater gp120 association with the Env trimer. The Q114E, Q567K, and A582T changes individually strengthen the noncovalent association of gp120 with the solubilized Env trimers, a property that will assist purification and characterization. Of interest, the Q567K change was included in a combination of Env changes that were reported to stabilize HIV-1 Env trimers in different contexts (178–180). In our panel of HIV-1 Env variants, enhancement of Env trimer stability in detergent was strongly correlated with virus resistance to cold and BNM-III-170, state-1-associated phenotypes. We note that the binding of the state-1-preferring compound, BMS-806, also stabilizes gp120-trimer association (138). In future studies, the ability of Q114E, Q567K, A582T, and other state-1-stabilizing changes to enhance Env cleavage efficiency, strengthen gp120-trimer association, and increase the sampling of pretriggered Env conformations in other HIV-1 strains will be explored. That some of the individual Env changes identified in our study demonstrate similar phenotypes in the Envs of other HIV-1 strains (121, 122, 125, 178–180) is encouraging in this regard.
In addition to Q114E and Q567K, we identified other changes (K59A, K117A, K121A) that individually yielded Env phenotypes consistent with state-1 stabilization. These and previously identified state-1-stabilizing changes (H66N, L587A) (121, 122, 125) were tested in combination with the Q114E and/or Q567K changes in various Env backgrounds. In no case did we observe an additive improvement in viral phenotypes associated with state-1 stabilization, and several of these combinations resulted in attenuated virus replication or gp120-trimer dissociation in detergent. It is not surprising that as state-1 stability is increased, virus replication diminishes as the activation barriers governing state-1-to-state-2 transitions increase. However, the validation of state-1 stabilization would be less straightforward for replication-incompetent Envs, given current uncertainties about a state-1 Env structure. Therefore, we deferred investigation of these potentially state-1-stabilizing changes until better assays to characterize the conformations of nonfunctional Envs are established.
The amino acid changes identified in our study that stabilize a state-1 conformation could potentially be applied to membrane Env immunogens presented in proteoliposomes or virus-like particles, or expressed in an mRNA vaccine (181, 182). Soluble forms of HIV-1 Env trimers have attracted significant interest as possible vaccine immunogens (61–69). Although differences exist between the conformations of membrane and soluble Env trimers (70–78), we evaluated the reciprocity of trimer-stabilizing changes in these two contexts. The changes that stabilize state 1 in the membrane HIV-1AD8 Env (Q114E/Q567K/A582T) modestly increased the levels of trimerization of HIV-1AD8 sgp140 SOSIP.664 glycoproteins.
We also evaluated the effect of changes that stabilize soluble gp140 trimers on the native HIV-1AD8 Env. Changes in gp41 (I559P, L555P) that are intended to prevent the formation of the HR1 coiled coil have been used to stabilize sgp140 SOSIP.664 trimers (70, 88, 183). However, introduction of these changes in combination with the major state-1-stabilizing changes (Q114E/I559P, Q114E/Q567K/I559P, and Q114E/Q567K/L555P) resulted in membrane Envs that were not processed (data not shown). We also considered another gp41 change, Q658E, that has been shown to stabilize sgp140 SOSIP.664 trimers (184). Introduction of the Q658E change into the wild-type HIV-1AD8 Env resulted in increased virus sensitivity to cold, sCD4, BNM-III-170, and the 19b antibody (data not shown). These phenotypes are consistent with those reported in other HIV-1 strains (184), and as they suggest a lower occupancy of state 1, we did not evaluate the Q658E change in combination with the Q114E and Q567K changes. These results indicate that not all conformationally stabilizing changes identified in soluble Env trimers are applicable to membrane Env glycoprotein trimers.
Although a state-1 Env structure is currently unknown, mapping the Env residues identified in this study on available Env trimer models can provide some insights. Figure 7 uses a PGT151-bound HIV-1JR-FL EnvΔCT trimer structure (PDB ID 5FUU) (82) to show the locations of Env residues in which changes resulted in increases or decreases in state-1-associated phenotypes. The binding of the PGT151 antibody induces a state-2-like conformation that is asymmetric, with two antibody Fabs bound to the Env trimer (78, 82). We chose this structure because, unlike most HIV-1 trimer structures, the HR1N region containing Gln 567 is resolved; however, in keeping with the asymmetry of the PGT151-bound Env trimer, the positions of the Gln 567 residues differ among the three Env protomers. Gln 567, Gln 114, and Ala 582 are close to the trimer axis in the EnvΔCT structure (Fig. 7A). The Cα-Cα distances between Gln 114 and Gln 567 residues vary from 11.6 to 15.2 Å, and the side chains of these residues do not apparently interact in this Env conformation. Gln 114 is stacked above Lys 117 and Lys 121, the side chains of which project toward the Env trimer axis (Fig. 7B, right panel). Although a precise structural explanation for the observed state-1-stabilizing phenotypes will require more data, the implicated residues are positioned near intersubunit or interprotomer junctions and therefore could potentially modulate trimer opening. For example, electrostatic repulsion among Lys 117 residues that destabilizes the Env trimer could be mitigated by their conversion to alanine residues or by replacing Gln114 with acidic residues.
FIG 7.
Location of Env residues in a structural model of an HIV-1 Env trimer. Env residues studied herein are depicted as CPK spheres in a PGT151-bound HIV-1JR-FL Env ΔCT trimer (PDB ID 5FUU) (82). The binding of two PGT151 Fabs introduces asymmetry into the Env trimer. In this depiction, the PGT151 Fabs have been removed from the structure. The individual Env protomers are colored pink, light blue, and gray. In this orientation, the gp120 subunits are at the bottom and gp41 subunits are at the top of the figures. (A) Env residues (Gln 114 [magenta], Gln 567 [orange], and Ala 582 [blue]) associated with state-1-stabilizing changes are shown, with a closeup image in the panel on the right. The distances between the Cα atoms of Gln 114 and Gln 567 residues in this asymmetric trimer structure are 11.6, 13.1, and 15.2 Å. The HR1N regions of the three Env protomers differ in conformation. (B) Env residues (Lys 59, His 66, Gln 114, Gln 567, and Ala 582) associated with state-1-stabilizing changes are colored green. Env residues (His 72, Thr 202, Gln 203, Arg 542, Ile 595, Leu 602, and Gln 658) associated with state-1-destabilizing changes are colored red. Changes in the residues (Lys 117, Lys 121, and Leu 587) colored yellow resulted in Envs that were resistant to cold and a CD4-mimetic compound but were subject to gp120 dissociation from the Env trimer solubilized in detergent. The right panel shows the side chain stacking of residues Gln 114, Lys 117, and Lys 121 near the Env trimer axis.
The state-1-destabilizing changes identified in this study (red residues in Fig. 7B, left panel) are less localized than the state-1-stabilizing changes (green and yellow residues in Fig. 7B, left panel). This is consistent with the expectation that a metastable structure can be disrupted by a diverse set of changes, whereas a more limited and strategically placed set of changes is required to strengthen the structure. In this study, we provide an example of how state-1-stabilizing changes in Env can counter the phenotypic effects of state-1-destabilizing alterations, even when these changes involve amino acid residues very distant on current Env trimer structures.
In a related paper (171), we report the ability of the state-1-stabilizing changes identified herein to counter the phenotypic consequences of disruption of the gp41 MPER. Although further work will be required to understand fully the mechanisms underlying these observations, the ability of the Q114E, Q567K, and A582T changes to counteract the disruptive effects of distant changes suggests that they may have significant utility in preserving pretriggered Env conformations in multiple circumstances.
MATERIALS AND METHODS
Env glycoprotein constructs.
The HIV-1AD8 and HIV-1JR-FL Envs were coexpressed with the Rev protein in the pSVIIIenv expression vector, using the natural HIV-1 env and rev sequences (23). The Asp718 (Kpn I)-BamHI fragment of HIV-1AD8 env was cloned into the corresponding sites of the pSVIIIenv plasmid expressing the HIV-1HXBc2 Env and Rev. Thus, the “wild-type” HIV-1AD8 Env used in this study contains a signal peptide and part of the cytoplasmic tail from the HIV-1HXBc2 Env. Viruses with this chimeric Env exhibit a degree of CD4 dependence and an antibody neutralization profile consistent with those of a primary, tier 2 HIV-1 (23, 37). The initial single, double, and triple sets of lysine substitutions shown in Fig. 1A were introduced into the HIV-1AD8 Env lacking an epitope tag. A carboxy-terminal GGHHHHHH (His6) epitope tag was added to the Env variants shown in Fig. 1B and derivatives thereof. The mutations were introduced by site-directed PCR mutagenesis using Pfu Ultra II polymerase (Agilent Technologies), according to the manufacturer’s protocol. The plasmid expressing the HIV-1BG505 Env (BG505.W6M.ENV.C2) was obtained through the NIH HIV Reagent Program and was contributed by Julie Overbaugh.
To express sgp140 SOSIP.664 glycoproteins, the pSVIIIenv plasmid containing the wild-type HIV-1AD8 env gene was mutagenized using Q5 high-fidelity DNA polymerase (New England Biolabs) according to the manufacturer’s instructions. The following changes were introduced into the wild-type HIV-1AD8 Env: A501C, modification of the gp120-gp41 cleavage site (REKR) to RRRRRR, I559P, T605C, and a C-terminal GSGHHHHHH tag; the sgp140 SOSIP.664 glycoprotein is truncated due to a stop codon replacing the codon for Asp664 (73, 183). The Q114E, Q567K, and A582T changes were introduced into this sgp140 SOSIP.664 construct.
Cell lines.
293T cells (ATCC) and HOS cells (ATCC) were grown in Dulbecco’s modified Eagle’s medium/nutrient mixture F12 (DMEM-F12) supplemented with 10% fetal bovine serum (FBS) and 100 μg/mL of penicillin-streptomycin. A549 cells expressing HIV-1 Envs with Gag-mCherry fusion proteins were grown in DMEM-F12 medium supplemented with 10% FBS, 1× penicillin-streptomycin, 1× l-glutamine, and 0.2% amphotericin B. Cf2Th cells stably expressing the human CD4 and CCR5 coreceptors for HIV-1 were grown in the same medium supplemented with 0.4 mg/mL of G418 and 0.2 mg/mL of hygromycin. All cell culture reagents are from Life Technologies.
Env processing and gp120-trimer association in Ni-NTA precipitation assay.
HOS cells were cotransfected with a Rev/Env-encoding pSVIIIenv plasmid and a Tat-encoding plasmid at a 1:0.125 ratio using the Effectene transfection reagent (Qiagen). At 48 h after transfection, HOS cells were washed with 1× phosphate-buffered saline (PBS) and lysed in 100 mM (NH4)2SO4, 20 mM Tris-HCl, pH 8, 300 mM NaCl, and 1.5% Cymal-5 (Anatrace) containing DMSO, 10 μM BMS-806, or 10 μg/mL soluble CD4-Ig. Lysates were clarified, and aliquots were saved as the input samples. The remaining lysates were incubated with nickel-nitriloacetic acid (Ni-NTA) beads (Qiagen) for 1.5 h at 4°C. The beads were gently pelleted and washed 3 times with room temperature washing buffer [100 mM (NH4)2SO4, 20 mM Tris-HCl, pH 8, 1 M NaCl, and 0.5% Cymal-5]. The beads were then boiled in LDS sample buffer (Invitrogen), and the proteins were analyzed by Western blotting using 1:2,000 goat anti-gp120 polyclonal antibody (Thermo Fisher Scientific) and 1:2,000 horseradish peroxidase (HRP)-conjugated rabbit anti-goat IgG (Thermo Fisher Scientific) or 4E10 anti-gp41 antibody (Polymun) and 1:2,000 HRP-conjugated goat anti-human IgG (Santa Cruz).
Virus infectivity, neutralization, and cold sensitivity.
Single-round virus infection assays were used to measure the ability of the Env variants to support virus entry, as described previously (23). Briefly, 293T cells were cotransfected with the Rev/Env-encoding pSVIIIenv plasmid, a Tat-encoding plasmid, the pCMV HIV-1 Gag-Pol packaging construct, and a plasmid containing the luciferase-expressing HIV-1 vector at a weight ratio of 1:0.125:1:3 using a standard calcium phosphate transfection protocol. At 48 h after transfection, virus-containing supernatants were collected, filtered through a 0.45-μm membrane, and incubated with soluble CD4, BNM-III-170, or antibody for 1 h at 37°C. The mixture was then added to Cf2Th-CD4/CCR5 cells, which were cultured at 37°C and 5% CO2. To enhance infection by recombinant viruses with the HIV-1BG505 Env, virus-antibody mixtures were spinoculated with target cells at 1,800 rpm for 1 h at room temperature and then incubated for 1 more hour before additional medium was added. Luciferase activity in the Cf2Th-CD4/CCR5 target cells was measured 48 h later. To measure cold sensitivity, the viruses were incubated on ice for various lengths of time prior to measuring their infectivity. To measure the sensitivity of virus infectivity to cross-linking, the viruses were incubated with BS3 (Thermo Fisher Scientific) for 15 min at room temperature; the reaction was quenched with 15 mM Tris-HCl, pH 8.0, for 10 min, and the mixture was then added to the target cells.
Cross-linking of Envs on VLPs.
A549 cells inducibly expressing virus-like particles (VLPs) consisting of the HIV-1 Gag-mCherry fusion protein and the wild-type HIV-1AD8 Env have been previously described (7, 138). The D1253 A549-Gag/Env cell line expressing VLPs with wild-type HIV-1AD8 Env was selected by fluorescence-activated cell sorting (FACS) for Gag-positive and PGT145-positive cells. The D1555.042321.sort A549-Gag/E.2 Env cells and the D1553.042321.sort A549-Gag/AE.2 Env cells inducibly expressing VLPs with the E.2 and AE.2 Envs, respectively, were established similarly. After FACS sorting, these cells were >90% dual positive for Gag expression (KC567 antibody positive) and Env expression (PGT145 antibody positive).
Equivalent numbers of cells from the three cell lines described above were seeded, and the expression of Gag-mCherry/Env VLPs was induced with 2 μg/mL doxycycline. Forty-eight to 72 h later, supernatants containing VLPs were centrifuged at low speed to remove cell debris and then filtered (0.45 μm). Clarified supernatants were centrifuged at 100,000 × g for 1 h at 4°C. VLP pellets were resuspended in 1× PBS, aliquoted, and incubated with different concentrations of either DTSSP (Thermo Fisher Scientific) or glutaraldehyde cross-linkers. The cross-linking reaction with DTSSP was carried out for 30 min at room temperature, after which the reaction was quenched with 100 mM Tris-HCl, pH 8.0, for 10 min at room temperature. Glutaraldehyde cross-linking was carried out for 5 min at room temperature, after which the reaction was quenched with 50 mM glycine for 10 min at room temperature. VLPs were then pelleted at 20,000 × g for 30 min at 4°C. VLP pellets were resuspended in 1× PBS/LDS, boiled, and Western blotted with a goat anti-gp120 antibody, as described above. The intensity of the gp120, gp160, dimer, and trimer bands was quantified using the Bio-Rad Image Lab program.
Cross-linking of sgp140 SOSIP.664 glycoproteins.
293T cells were cotransfected with a 1:8 ratio of a Tat-expressing plasmid and a pSVIIIenv plasmid expressing either the HIV-1AD8 sgp140 SOSIP.664 glycoprotein or a sgp140 SOSIP.664 glycoprotein with Q114E/Q567K/A582T changes, using polyethyleneimine (PEI) according to the manufacturer’s protocol. Six hours later, the medium was replaced. Forty-eight hours after transfection, cell supernatants containing the sgp140 SOSIP.664 glycoproteins were collected, filtered (0.45 μm), and incubated with BS3 (Thermo Fisher Scientific) at different concentrations for 30 min at room temperature. The cross-linking reaction was quenched with 100 mM Tris-HCl, pH 8.0, for 10 min. LDS and dithiothreitol (DTT) were added to the samples, which were analyzed by reducing SDS-PAGE and Western blotting, as described above.
ACKNOWLEDGMENTS
We thank Elizabeth Carpelan for manuscript preparation. Antibodies against HIV-1 were kindly supplied by Dennis Burton (Scripps), Peter Kwong and John Mascola (Vaccine Research Center NIH), Barton Haynes (Duke University), Hermann Katinger (Polymun), James Robinson (Tulane University), and Marshall Posner (Mount Sinai Medical Center). We thank the NIH HIV Reagent Program for providing additional reagents.
This work was supported by grants from the National Institutes of Health (grant no. AI 145547, AI 124982, and AI 150471, including the Basic Science Core of the University of Alabama Center for AIDS Research [AI27767]) and by a gift from the late William F. McCarty-Cooper.
We declare no conflicts of interest.
Contributor Information
Hanh T. Nguyen, Email: hanht_nguyen@dfci.harvard.edu.
Joseph G. Sodroski, Email: joseph_sodroski@dfci.harvard.edu.
Viviana Simon, Icahn School of Medicine at Mount Sinai.
REFERENCES
- 1.Allan JS, Coligan JE, Barin F, McLane MF, Sodroski JG, Rosen CA, Haseltine WA, Lee TH, Essex M. 1985. Major glycoprotein antigens that induce antibodies in AIDS patients are encoded by HTLV-III. Science 228:1091–1094. 10.1126/science.2986290. [DOI] [PubMed] [Google Scholar]
- 2.Wyatt R, Sodroski J. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884–1888. 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 3.Fennie C, Lasky LA. 1989. Model for intracellular folding of the human immunodeficiency virus type 1 gp120. J Virol 63:639–646. 10.1128/JVI.63.2.639-646.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y, Luo L, Thomas DY, Kang CY. 2000. The HIV-1 Env protein signal sequence retards its cleavage and down-regulates the glycoprotein folding. Virology 272:417–428. 10.1006/viro.2000.0357. [DOI] [PubMed] [Google Scholar]
- 5.Willey RL, Bonifacino JS, Potts BJ, Martin MA, Klausner RD. 1988. Biosynthesis, cleavage, and degradation of the human immunodeficiency virus 1 envelope glycoprotein gp160. Proc Natl Acad Sci USA 85:9580–9584. 10.1073/pnas.85.24.9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Earl PL, Moss B, Doms RW. 1991. Folding, interaction with GRP78-BiP, assembly, and transport of the human immunodeficiency virus type 1 envelope protein. J Virol 65:2047–2055. 10.1128/JVI.65.4.2047-2055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang S, Nguyen HT, Ding H, Wang J, Zou S, Liu L, Guha D, Gabuzda D, Ho DD, Kappes JC, Sodroski J. 2021. Dual pathways of human immunodeficiency virus type 1 envelope glycoprotein trafficking modulate the selective exclusion of uncleaved oligomers from virions. J Virol 95:e01369-20. 10.1128/JVI.01369-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doores KJ, Bonomelli C, Harvey DJ, Vasiljevic S, Dwek RA, Burton DR, Crispin M, Scanlan CN. 2010. Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens. Proc Natl Acad Sci USA 107:13800–13805. 10.1073/pnas.1006498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Go EP, Herschhorn A, Gu C, Castillo-Menendez L, Zhang S, Mao Y, Chen H, Ding H, Wakefield JK, Hua D, Liao HX, Kappes JC, Sodroski J, Desaire H. 2015. Comparative analysis of the glycosylation profiles of membrane-anchored HIV-1 envelope glycoprotein trimers and soluble gp140. J Virol 89:8245–8257. 10.1128/JVI.00628-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merkle RK, Helland DE, Welles JL, Shilatifard A, Haseltine WA, Cummings RD. 1991. gp160 of HIV-I synthesized by persistently infected Molt-3 cells is terminally glycosylated: evidence that cleavage of gp160 occurs subsequent to oligosaccharide processing. Arch Biochem Biophys 290:248–257. 10.1016/0003-9861(91)90616-q. [DOI] [PubMed] [Google Scholar]
- 11.Brakch N, Dettin M, Scarinci C, Seidah NG, Di Bello C. 1995. Structural investigation and kinetic characterization of potential cleavage sites of HIV GP160 by human furin and PC1. Biochem Biophys Res Commun 213:356–361. 10.1006/bbrc.1995.2137. [DOI] [PubMed] [Google Scholar]
- 12.Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, Burton DR, Koff WC, Courter JR, Smith AB, III, Kwong PD, Blanchard SC, Mothes W. 2014. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 346:759–763. 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalgleish AG, Beverley PC, Clapham PR, Crawford DH, Greaves MF, Weiss RA. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763–767. 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 14.Cocchi F, DeVico AL, Garzino-Demo A, Arya SK, Gallo RC, Lusso P. 1995. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science 270:1811–1815. 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 15.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton RE, Hill CM, Davis CB, Peiper SC, Schall TJ, Littman DR, Landau NR. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661–666. 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 16.Feng Y, Broder CC, Kennedy PE, Berger EA. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872–877. 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 17.Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, Berger EA. 1996. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955–1958. 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 18.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, Wu L, Mackay CR, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135–1148. 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 19.Doranz BJ, Rucker J, Yi Y, Smyth RJ, Samson M, Peiper SC, Parmentier M, Collman RG, Doms RW. 1996. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 85:1149–1158. 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 20.Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA, Cayanan C, Maddon PJ, Koup RA, Moore JP, Paxton WA. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667–673. 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 21.Wu L, Gerard NP, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso AA, Desjardin E, Newman W, Gerard C, Sodroski J. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179–183. 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 22.Trkola A, Dragic T, Arthos J, Binley JM, Olson WC, Allaway GP, Cheng-Mayer C, Robinson J, Maddon PJ, Moore JP. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184–187. 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 23.Haim H, Strack B, Kassa A, Madani N, Wang L, Courter JR, Princiotto A, McGee K, Pacheco B, Seaman MS, Smith AB, III, Sodroski J. 2011. Contribution of intrinsic reactivity of the HIV-1 envelope glycoproteins to CD4-independent infection and global inhibitor sensitivity. PLoS Pathog 7:e1002101. 10.1371/journal.ppat.1002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma X, Lu M, Gorman J, Terry DS, Hong X, Zhou Z, Zhao H, Altman RB, Arthos J, Blanchard SC, Kwong PD, Munro JB, Mothes W. 2018. HIV-1 Env trimer opens through an asymmetric intermediate in which individual protomers adopt distinct conformations. Elife 7:e34271. 10.7554/eLife.34271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khasnis MD, Halkidis K, Bhardwaj A, Root MJ. 2016. Receptor activation of HIV-1 Env leads to asymmetric exposure of the gp41 trimer. PLoS Pathog 12:e1006098. 10.1371/journal.ppat.1006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109–113. 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H, Cohen AA, Galimidi RP, Gristick HB, Jensen GJ, Bjorkman PJ. 2016. Cryo-EM structure of a CD4-bound open HIV-1 envelope trimer reveals structural rearrangements of the gp120 V1V2 loop. Proc Natl Acad Sci USA 113:E7151–E7158. 10.1073/pnas.1615939113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ozorowski G, Pallesen J, de Val N, Lyumkis D, Cottrell CA, Torres JL, Copps J, Stanfield RL, Cupo A, Pugach P, Moore JP, Wilson IA, Ward AB. 2017. Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike. Nature 547:360–363. 10.1038/nature23010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Furuta RA, Wild CT, Weng Y, Weiss CD. 1998. Capture of an early fusion-active conformation of HIV-1 gp41. Nat Struct Biol 5:276–279. 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- 30.Koshiba T, Chan DC. 2003. The prefusogenic intermediate of HIV-1 gp41 contains exposed C-peptide regions. J Biol Chem 278:7573–7579. 10.1074/jbc.M211154200. [DOI] [PubMed] [Google Scholar]
- 31.He Y, Vassell R, Zaitseva M, Nguyen N, Yang Z, Weng Y, Weiss CD. 2003. Peptides trap the human immunodeficiency virus type 1 envelope glycoprotein fusion intermediate at two sites. J Virol 77:1666–1671. 10.1128/jvi.77.3.1666-1671.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan DC, Fass D, Berger JM, Kim PS. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263–273. 10.1016/S0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 33.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426–430. 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 34.Lu M, Blacklow SC, Kim PS. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat Struct Biol 2:1075–1082. 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- 35.Melikyan GB, Markosyan RM, Hemmati H, Delmedico MK, Lambert DM, Cohen FS. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J Cell Biol 151:413–423. 10.1083/jcb.151.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilen CB, Tilton JC, Doms RW. 2012. Molecular mechanisms of HIV entry. Adv Exp Med Biol 726:223–242. 10.1007/978-1-4614-0980-9_10. [DOI] [PubMed] [Google Scholar]
- 37.Haim H, Salas I, McGee K, Eichelberger N, Winter E, Pacheco B, Sodroski J. 2013. Modeling virus- and antibody-specific factors to predict human immunodeficiency virus neutralization efficiency. Cell Host Microbe 14:547–558. 10.1016/j.chom.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guttman M, Cupo A, Julien JP, Sanders RW, Wilson IA, Moore JP, Lee KK. 2015. Antibody potency relates to the ability to recognize the closed, pre-fusion form of HIV Env. Nat Commun 6:6144. 10.1038/ncomms7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, Katinger H, Parren PW, Robinson J, Van Ryk D, Wang L, Burton DR, Freire E, Wyatt R, Sodroski J, Hendrickson WA, Arthos J. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678–682. 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 40.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312. 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 41.Decker JM, Bibollet-Ruche F, Wei X, Wang S, Levy DN, Wang W, Delaporte E, Peeters M, Derdeyn CA, Allen S, Hunter E, Saag MS, Hoxie JA, Hahn BH, Kwong PD, Robinson JE, Shaw GM. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J Exp Med 201:1407–1419. 10.1084/jem.20042510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alsahafi N, Bakouche N, Kazemi M, Richard J, Ding S, Bhattacharyya S, Das D, Anand SP, Prevost J, Tolbert WD, Lu H, Medjahed H, Gendron-Lepage G, Ortega Delgado GG, Kirk S, Melillo B, Mothes W, Sodroski J, Smith AB, III, Kaufmann DE, Wu X, Pazgier M, Rouiller I, Finzi A, Munro JB. 2019. An asymmetric opening of HIV-1 envelope mediates antibody-dependent cellular cytotoxicity. Cell Host Microbe 25:578–587.e5. 10.1016/j.chom.2019.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Labrijn AF, Poignard P, Raja A, Zwick MB, Delgado K, Franti M, Binley J, Vivona V, Grundner C, Huang CC, Venturi M, Petropoulos CJ, Wrin T, Dimitrov DS, Robinson J, Kwong PD, Wyatt RT, Sodroski J, Burton DR. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J Virol 77:10557–10565. 10.1128/jvi.77.19.10557-10565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moore PL, Ranchobe N, Lambson BE, Gray ES, Cave E, Abrahams MR, Bandawe G, Mlisana K, Abdool Karim SS, Williamson C, Morris L, Study C, Immunology NCfHAV. 2009. Limited neutralizing antibody specificities drive neutralization escape in early HIV-1 subtype C infection. PLoS Pathog 5:e1000598. 10.1371/journal.ppat.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herschhorn A, Ma X, Gu C, Ventura JD, Castillo-Menendez L, Melillo B, Terry DS, Smith AB, III, Blanchard SC, Munro JB, Mothes W, Finzi A, Sodroski J. 2016. Release of gp120 restraints leads to an entry-competent intermediate state of the HIV-1 envelope glycoproteins. mBio 7:e01598-16. 10.1128/mBio.01598-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wibmer CK, Bhiman JN, Gray ES, Tumba N, Abdool Karim SS, Williamson C, Morris L, Moore PL. 2013. Viral escape from HIV-1 neutralizing antibodies drives increased plasma neutralization breadth through sequential recognition of multiple epitopes and immunotypes. PLoS Pathog 9:e1003738. 10.1371/journal.ppat.1003738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gray ES, Taylor N, Wycuff D, Moore PL, Tomaras GD, Wibmer CK, Puren A, DeCamp A, Gilbert PB, Wood B, Montefiori DC, Binley JM, Shaw GM, Haynes BF, Mascola JR, Morris L. 2009. Antibody specificities associated with neutralization breadth in plasma from human immunodeficiency virus type 1 subtype C-infected blood donors. J Virol 83:8925–8937. 10.1128/JVI.00758-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sather DN, Armann J, Ching LK, Mavrantoni A, Sellhorn G, Caldwell Z, Yu X, Wood B, Self S, Kalams S, Stamatatos L. 2009. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J Virol 83:757–769. 10.1128/JVI.02036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klein F, Diskin R, Scheid JF, Gaebler C, Mouquet H, Georgiev IS, Pancera M, Zhou T, Incesu RB, Fu BZ, Gnanapragasam PN, Oliveira TY, Seaman MS, Kwong PD, Bjorkman PJ, Nussenzweig MC. 2013. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell 153:126–138. 10.1016/j.cell.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walker LM, Simek MD, Priddy F, Gach JS, Wagner D, Zwick MB, Phogat SK, Poignard P, Burton DR. 2010. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog 6:e1001028. 10.1371/journal.ppat.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gray ES, Madiga MC, Hermanus T, Moore PL, Wibmer CK, Tumba NL, Werner L, Mlisana K, Sibeko S, Williamson C, Abdool Karim SS, Morris L, Team CS, CAPRISA002 Study Team. 2011. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J Virol 85:4828–4840. 10.1128/JVI.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Corti D, Langedijk JP, Hinz A, Seaman MS, Vanzetta F, Fernandez-Rodriguez BM, Silacci C, Pinna D, Jarrossay D, Balla-Jhagjhoorsingh S, Willems B, Zekveld MJ, Dreja H, O'Sullivan E, Pade C, Orkin C, Jeffs SA, Montefiori DC, Davis D, Weissenhorn W, McKnight A, Heeney JL, Sallusto F, Sattentau QJ, Weiss RA, Lanzavecchia A. 2010. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS One 5:e8805. 10.1371/journal.pone.0008805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, O'Dell S, Perfetto S, Schmidt SD, Shi W, Wu L, Yang Y, Yang ZY, Yang Z, Zhang Z, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Simek M, Burton DR, Koff WC, Doria-Rose NA, Connors M, Program NCS, Mullikin JC, Nabel GJ, Roederer M, Shapiro L, Kwong PD, Mascola JR, NISC Comparative Sequencing Program. 2011. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333:1593–1602. 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hraber P, Seaman MS, Bailer RT, Mascola JR, Montefiori DC, Korber BT. 2014. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS 28:163–169. 10.1097/QAD.0000000000000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hessell AJ, Poignard P, Hunter M, Hangartner L, Tehrani DM, Bleeker WK, Parren PW, Marx PA, Burton DR. 2009. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat Med 15:951–954. 10.1038/nm.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mascola JR, Lewis MG, Stiegler G, Harris D, VanCott TC, Hayes D, Louder MK, Brown CR, Sapan CV, Frankel SS, Lu Y, Robb ML, Katinger H, Birx DL. 1999. Protection of macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J Virol 73:4009–4018. 10.1128/JVI.73.5.4009-4018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mascola JR, Stiegler G, VanCott TC, Katinger H, Carpenter CB, Hanson CE, Beary H, Hayes D, Frankel SS, Birx DL, Lewis MG. 2000. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med 6:207–210. 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 58.Moldt B, Rakasz EG, Schultz N, Chan-Hui PY, Swiderek K, Weisgrau KL, Piaskowski SM, Bergman Z, Watkins DI, Poignard P, Burton DR. 2012. Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proc Natl Acad Sci USA 109:18921–18925. 10.1073/pnas.1214785109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parren PW, Marx PA, Hessell AJ, Luckay A, Harouse J, Cheng-Mayer C, Moore JP, Burton DR. 2001. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J Virol 75:8340–8347. 10.1128/jvi.75.17.8340-8347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pauthner MG, Nkolola JP, Havenar-Daughton C, Murrell B, Reiss SM, Bastidas R, Prevost J, Nedellec R, von Bredow B, Abbink P, Cottrell CA, Kulp DW, Tokatlian T, Nogal B, Bianchi M, Li H, Lee JH, Butera ST, Evans DT, Hangartner L, Finzi A, Wilson IA, Wyatt RT, Irvine DJ, Schief WR, Ward AB, Sanders RW, Crotty S, Shaw GM, Barouch DH, Burton DR. 2019. Vaccine-induced protection from homologous tier 2 SHIV challenge in nonhuman primates depends on serum-neutralizing antibody titers. Immunity 50:241–252.e6. 10.1016/j.immuni.2018.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pauthner M, Havenar-Daughton C, Sok D, Nkolola JP, Bastidas R, Boopathy AV, Carnathan DG, Chandrashekar A, Cirelli KM, Cottrell CA, Eroshkin AM, Guenaga J, Kaushik K, Kulp DW, Liu J, McCoy LE, Oom AL, Ozorowski G, Post KW, Sharma SK, Steichen JM, de Taeye SW, Tokatlian T, Torrents de la Pena A, Butera ST, LaBranche CC, Montefiori DC, Silvestri G, Wilson IA, Irvine DJ, Sanders RW, Schief WR, Ward AB, Wyatt RT, Barouch DH, Crotty S, Burton DR. 2017. Elicitation of robust tier 2 neutralizing antibody responses in nonhuman primates by HIV envelope trimer immunization using optimized approaches. Immunity 46:1073–1088.e6. 10.1016/j.immuni.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Torrents de la Pena A, de Taeye SW, Sliepen K, LaBranche CC, Burger JA, Schermer EE, Montefiori DC, Moore JP, Klasse PJ, Sanders RW. 2018. Immunogenicity in rabbits of HIV-1 SOSIP trimers from clades A, B, and C, given individually, sequentially, or in combination. J Virol 92:e01957-17. 10.1128/JVI.01957-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klasse PJ, LaBranche CC, Ketas TJ, Ozorowski G, Cupo A, Pugach P, Ringe RP, Golabek M, van Gils MJ, Guttman M, Lee KK, Wilson IA, Butera ST, Ward AB, Montefiori DC, Sanders RW, Moore JP. 2016. Sequential and simultaneous immunization of rabbits with HIV-1 envelope glycoprotein SOSIP.664 trimers from clades A, B and C. PLoS Pathog 12:e1005864. 10.1371/journal.ppat.1005864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu JK, Crampton JC, Cupo A, Ketas T, van Gils MJ, Sliepen K, de Taeye SW, Sok D, Ozorowski G, Deresa I, Stanfield R, Ward AB, Burton DR, Klasse PJ, Sanders RW, Moore JP, Crotty S. 2015. Murine antibody responses to cleaved soluble HIV-1 envelope trimers are highly restricted in specificity. J Virol 89:10383–10398. 10.1128/JVI.01653-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feng Y, Tran K, Bale S, Kumar S, Guenaga J, Wilson R, de Val N, Arendt H, DeStefano J, Ward AB, Wyatt RT. 2016. Thermostability of well-ordered HIV spikes correlates with the elicitation of autologous Tier 2 neutralizing antibodies. PLoS Pathog 12:e1005767. 10.1371/journal.ppat.1005767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dubrovskaya V, Tran K, Ozorowski G, Guenaga J, Wilson R, Bale S, Cottrell CA, Turner HL, Seabright G, O'Dell S, Torres JL, Yang L, Feng Y, Leaman DP, Vazquez Bernat N, Liban T, Louder M, McKee K, Bailer RT, Movsesyan A, Doria-Rose NA, Pancera M, Karlsson Hedestam GB, Zwick MB, Crispin M, Mascola JR, Ward AB, Wyatt RT. 2019. Vaccination with glycan-modified HIV NFL envelope trimer-liposomes elicits broadly neutralizing antibodies to multiple sites of vulnerability. Immunity 51:915–929.e7. 10.1016/j.immuni.2019.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu K, Acharya P, Kong R, Cheng C, Chuang GY, Liu K, Louder MK, O'Dell S, Rawi R, Sastry M, Shen CH, Zhang B, Zhou T, Asokan M, Bailer RT, Chambers M, Chen X, Choi CW, Dandey VP, Doria-Rose NA, Druz A, Eng ET, Farney SK, Foulds KE, Geng H, Georgiev IS, Gorman J, Hill KR, Jafari AJ, Kwon YD, Lai YT, Lemmin T, McKee K, Ohr TY, Ou L, Peng D, Rowshan AP, Sheng Z, Todd JP, Tsybovsky Y, Viox EG, Wang Y, Wei H, Yang Y, Zhou AF, Chen R, Yang L, Scorpio DG, McDermott AB, Shapiro L, et al. 2018. Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1. Nat Med 24:857–867. 10.1038/s41591-018-0042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, Arendt H, Kim HJ, Lee JH, Pugach P, Williams M, Debnath G, Moldt B, van Breemen MJ, Isik G, Medina-Ramirez M, Back JW, Koff WC, Julien JP, Rakasz EG, Seaman MS, Guttman M, Lee KK, Klasse PJ, LaBranche C, Schief WR, Wilson IA, Overbaugh J, Burton DR, Ward AB, Montefiori DC, Dean H, Moore JP. 2015. HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science 349:aac4223. 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.de Taeye SW, Ozorowski G, Torrents de la Pena A, Guttman M, Julien JP, van den Kerkhof TL, Burger JA, Pritchard LK, Pugach P, Yasmeen A, Crampton J, Hu J, Bontjer I, Torres JL, Arendt H, DeStefano J, Koff WC, Schuitemaker H, Eggink D, Berkhout B, Dean H, LaBranche C, Crotty S, Crispin M, Montefiori DC, Klasse PJ, Lee KK, Moore JP, Wilson IA, Ward AB, Sanders RW. 2015. Immunogenicity of stabilized HIV-1 envelope trimers with reduced exposure of non-neutralizing epitopes. Cell 163:1702–1715. 10.1016/j.cell.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, Golabek M, de Los Reyes K, Ketas TJ, van Gils MJ, King CR, Wilson IA, Ward AB, Klasse PJ, Moore JP. 2013. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog 9:e1003618. 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alsahafi N, Debbeche O, Sodroski J, Finzi A. 2015. Effects of the I559P gp41 change on the conformation and function of the human immunodeficiency virus (HIV-1) membrane envelope glycoprotein trimer. PLoS One 10:e0122111. 10.1371/journal.pone.0122111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alsahafi N, Anand SP, Castillo-Menendez L, Verly MM, Medjahed H, Prevost J, Herschhorn A, Richard J, Schon A, Melillo B, Freire E, Smith AB, III, Sodroski J, Finzi A. 2018. SOSIP changes affect human immunodeficiency virus type 1 envelope glycoprotein conformation and CD4 engagement. J Virol 92:e01080-18. 10.1128/JVI.01080-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Castillo-Menendez LR, Nguyen HT, Sodroski J. 2019. Conformational differences between functional human immunodeficiency virus envelope glycoprotein trimers and stabilized soluble trimers. J Virol 93:e01709-18. 10.1128/JVI.01709-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nguyen HT, Alsahafi N, Finzi A, Sodroski JG. 2019. Effects of the SOS (A501C/T605C) and DS (I201C/A433C) disulfide bonds on HIV-1 membrane envelope glycoprotein conformation and function. J Virol 93:e00304-19. 10.1128/JVI.00304-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Go EP, Ding H, Zhang S, Ringe RP, Nicely N, Hua D, Steinbock RT, Golabek M, Alin J, Alam SM, Cupo A, Haynes BF, Kappes JC, Moore JP, Sodroski JG, Desaire H. 2017. Glycosylation benchmark profile for HIV-1 envelope glycoprotein production based on eleven Env trimers. J Virol 91:e02428-16. 10.1128/JVI.02428-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cao L, Pauthner M, Andrabi R, Rantalainen K, Berndsen Z, Diedrich JK, Menis S, Sok D, Bastidas R, Park SR, Delahunty CM, He L, Guenaga J, Wyatt RT, Schief WR, Ward AB, Yates JR, 3rd, Burton DR, Paulson JC. 2018. Differential processing of HIV envelope glycans on the virus and soluble recombinant trimer. Nat Commun 9:3693. 10.1038/s41467-018-06121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Torrents de la Pena A, Rantalainen K, Cottrell CA, Allen JD, van Gils MJ, Torres JL, Crispin M, Sanders RW, Ward AB. 2019. Similarities and differences between native HIV-1 envelope glycoprotein trimers and stabilized soluble trimer mimetics. PLoS Pathog 15:e1007920. 10.1371/journal.ppat.1007920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lu M, Ma X, Castillo-Menendez LR, Gorman J, Alsahafi N, Ermel U, Terry DS, Chambers M, Peng D, Zhang B, Zhou T, Reichard N, Wang K, Grover JR, Carman BP, Gardner MR, Nikic-Spiegel I, Sugawara A, Arthos J, Lemke EA, Smith AB, III, Farzan M, Abrams C, Munro JB, McDermott AB, Finzi A, Kwong PD, Blanchard SC, Sodroski JG, Mothes W. 2019. Associating HIV-1 envelope glycoprotein structures with states on the virus observed by smFRET. Nature 568:415–419. 10.1038/s41586-019-1101-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. 2013. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342:1477–1483. 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB. 2013. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342:1484–1490. 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, Stewart-Jones GB, Stuckey J, Bailer RT, Joyce MG, Louder MK, Tumba N, Yang Y, Zhang B, Cohen MS, Haynes BF, Mascola JR, Morris L, Munro JB, Blanchard SC, Mothes W, Connors M, Kwong PD. 2014. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514:455–461. 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee JH, Ozorowski G, Ward AB. 2016. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science 351:1043–1048. 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bartesaghi A, Merk A, Borgnia MJ, Milne JL, Subramaniam S. 2013. Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Nat Struct Mol Biol 20:1352–1357. 10.1038/nsmb.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stewart-Jones GB, Soto C, Lemmin T, Chuang GY, Druz A, Kong R, Thomas PV, Wagh K, Zhou T, Behrens AJ, Bylund T, Choi CW, Davison JR, Georgiev IS, Joyce MG, Kwon YD, Pancera M, Taft J, Yang Y, Zhang B, Shivatare SS, Shivatare VS, Lee CC, Wu CY, Bewley CA, Burton DR, Koff WC, Connors M, Crispin M, Baxa U, Korber BT, Wong CH, Mascola JR, Kwong PD. 2016. Trimeric HIV-1-Env structures define glycan shields from clades A, B, and G. Cell 165:813–826. 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gristick HB, von Boehmer L, West AP, Jr, Schamber M, Gazumyan A, Golijanin J, Seaman MS, Fatkenheuer G, Klein F, Nussenzweig MC, Bjorkman PJ. 2016. Natively glycosylated HIV-1 Env structure reveals new mode for antibody recognition of the CD4-binding site. Nat Struct Mol Biol 23:906–915. 10.1038/nsmb.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kwon YD, Pancera M, Acharya P, Georgiev IS, Crooks ET, Gorman J, Joyce MG, Guttman M, Ma X, Narpala S, Soto C, Terry DS, Yang Y, Zhou T, Ahlsen G, Bailer RT, Chambers M, Chuang GY, Doria-Rose NA, Druz A, Hallen MA, Harned A, Kirys T, Louder MK, O'Dell S, Ofek G, Osawa K, Prabhakaran M, Sastry M, Stewart-Jones GB, Stuckey J, Thomas PV, Tittley T, Williams C, Zhang B, Zhao H, Zhou Z, Donald BR, Lee LK, Zolla-Pazner S, Baxa U, Schon A, Freire E, Shapiro L, Lee KK, Arthos J, Munro JB, Blanchard SC, Mothes W, Binley JM, et al. 2015. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat Struct Mol Biol 22:522–531. 10.1038/nsmb.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Guenaga J, de Val N, Tran K, Feng Y, Satchwell K, Ward AB, Wyatt RT. 2015. Well-ordered trimeric HIV-1 subtype B and C soluble spike mimetics generated by negative selection display native-like properties. PLoS Pathog 11:e1004570. 10.1371/journal.ppat.1004570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guenaga J, Dubrovskaya V, de Val N, Sharma SK, Carrette B, Ward AB, Wyatt RT. 2015. Structure-guided redesign increases the propensity of HIV Env to generate highly stable soluble trimers. J Virol 90:2806–2817. 10.1128/JVI.02652-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Georgiev IS, Joyce MG, Yang Y, Sastry M, Zhang B, Baxa U, Chen RE, Druz A, Lees CR, Narpala S, Schon A, Van Galen J, Chuang GY, Gorman J, Harned A, Pancera M, Stewart-Jones GB, Cheng C, Freire E, McDermott AB, Mascola JR, Kwong PD. 2015. Single-chain soluble BG505.SOSIP gp140 trimers as structural and antigenic mimics of mature closed HIV-1 Env. J Virol 89:5318–5329. 10.1128/JVI.03451-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang Z, Wang H, Liu AZ, Gristick HB, Bjorkman PJ. 2019. Asymmetric opening of HIV-1 Env bound to CD4 and a coreceptor-mimicking antibody. Nat Struct Mol Biol 26:1167–1175. 10.1038/s41594-019-0344-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pan J, Peng H, Chen B, Harrison SC. 2020. Cryo-EM structure of full-length HIV-1 Env bound with the Fab of antibody PG16. J Mol Biol 432:1158–1168. 10.1016/j.jmb.2019.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Roberts JD, Bebenek K, Kunkel TA. 1988. The accuracy of reverse transcriptase from HIV-1. Science 242:1171–1173. 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- 93.Smyth RP, Davenport MP, Mak J. 2012. The origin of genetic diversity in HIV-1. Virus Res 169:415–429. 10.1016/j.virusres.2012.06.015. [DOI] [PubMed] [Google Scholar]
- 94.Cuevas JM, Geller R, Garijo R, Lopez-Aldeguer J, Sanjuan R. 2015. Extremely high mutation rate of HIV-1 in vivo. PLoS Biol 13:e1002251. 10.1371/journal.pbio.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Monajemi M, Woodworth CF, Zipperlen K, Gallant M, Grant MD, Larijani M. 2014. Positioning of APOBEC3G/F mutational hotspots in the human immunodeficiency virus genome favors reduced recognition by CD8+ T cells. PLoS One 9:e93428. 10.1371/journal.pone.0093428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sadler HA, Stenglein MD, Harris RS, Mansky LM. 2010. APOBEC3G contributes to HIV-1 variation through sublethal mutagenesis. J Virol 84:7396–7404. 10.1128/JVI.00056-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mansky LM, Le Rouzic E, Benichou S, Gajary LC. 2003. Influence of reverse transcriptase variants, drugs, and Vpr on human immunodeficiency virus type 1 mutant frequencies. J Virol 77:2071–2080. 10.1128/jvi.77.3.2071-2080.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fraser C, Lythgoe K, Leventhal GE, Shirreff G, Hollingsworth TD, Alizon S, Bonhoeffer S. 2014. Virulence and pathogenesis of HIV-1 infection: an evolutionary perspective. Science 343:1243727. 10.1126/science.1243727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Davenport MP, Loh L, Petravic J, Kent SJ. 2008. Rates of HIV immune escape and reversion: implications for vaccination. Trends Microbiol 16:561–566. 10.1016/j.tim.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 100.Hraber P, Korber BT, Lapedes AS, Bailer RT, Seaman MS, Gao H, Greene KM, McCutchan F, Williamson C, Kim JH, Tovanabutra S, Hahn BH, Swanstrom R, Thomson MM, Gao F, Harris L, Giorgi E, Hengartner N, Bhattacharya T, Mascola JR, Montefiori DC. 2014. Impact of clade, geography, and age of the epidemic on HIV-1 neutralization by antibodies. J Virol 88:12623–12643. 10.1128/JVI.01705-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kolchinsky P, Mirzabekov T, Farzan M, Kiprilov E, Cayabyab M, Mooney LJ, Choe H, Sodroski J. 1999. Adaptation of a CCR5-using, primary human immunodeficiency virus type 1 isolate for CD4-independent replication. J Virol 73:8120–8126. 10.1128/JVI.73.10.8120-8126.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hoffman TL, LaBranche CC, Zhang W, Canziani G, Robinson J, Chaiken I, Hoxie JA, Doms RW. 1999. Stable exposure of the coreceptor-binding site in a CD4-independent HIV-1 envelope protein. Proc Natl Acad Sci USA 96:6359–6364. 10.1073/pnas.96.11.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dumonceaux J, Nisole S, Chanel C, Quivet L, Amara A, Baleux F, Briand P, Hazan U. 1998. Spontaneous mutations in the env gene of the human immunodeficiency virus type 1 NDK isolate are associated with a CD4-independent entry phenotype. J Virol 72:512–519. 10.1128/JVI.72.1.512-519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Taylor BM, Foulke JS, Flinko R, Heredia A, DeVico A, Reitz M. 2008. An alteration of human immunodeficiency virus gp41 leads to reduced CCR5 dependence and CD4 independence. J Virol 82:5460–5471. 10.1128/JVI.01049-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Beauparlant D, Rusert P, Magnus C, Kadelka C, Weber J, Uhr T, Zagordi O, Oberle C, Duenas-Decamp MJ, Clapham PR, Metzner KJ, Gunthard HF, Trkola A. 2017. Delineating CD4 dependency of HIV-1: adaptation to infect low level CD4 expressing target cells widens cellular tropism but severely impacts on envelope functionality. PLoS Pathog 13:e1006255. 10.1371/journal.ppat.1006255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zerhouni B, Nelson JA, Saha K. 2004. Isolation of CD4-independent primary human immunodeficiency virus type 1 isolates that are syncytium inducing and acutely cytopathic for CD8+ lymphocytes. J Virol 78:1243–1255. 10.1128/jvi.78.3.1243-1255.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bhattacharya J, Peters PJ, Clapham PR. 2003. CD4-independent infection of HIV and SIV: implications for envelope conformation and cell tropism in vivo. AIDS 17(Suppl 4):S35–S43. 10.1097/00002030-200317004-00004. [DOI] [PubMed] [Google Scholar]
- 108.LaBranche CC, Hoffman TL, Romano J, Haggarty BS, Edwards TG, Matthews TJ, Doms RW, Hoxie JA. 1999. Determinants of CD4 independence for a human immunodeficiency virus type 1 variant map outside regions required for coreceptor specificity. J Virol 73:10310–10319. 10.1128/JVI.73.12.10310-10319.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Edwards TG, Wyss S, Reeves JD, Zolla-Pazner S, Hoxie JA, Doms RW, Baribaud F. 2002. Truncation of the cytoplasmic domain induces exposure of conserved regions in the ectodomain of human immunodeficiency virus type 1 envelope protein. J Virol 76:2683–2691. 10.1128/jvi.76.6.2683-2691.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kolchinsky P, Kiprilov E, Sodroski J. 2001. Increased neutralization sensitivity of CD4-independent human immunodeficiency virus variants. J Virol 75:2041–2050. 10.1128/JVI.75.5.2041-2050.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shin Y, Yoon CH, Yang HJ, Lim H, Choi BS, Kim SS, Kang C. 2016. Functional characteristics of the natural polymorphisms of HIV-1 gp41 in HIV-1 isolates from enfuvirtide-naive Korean patients. Arch Virol 161:1547–1557. 10.1007/s00705-016-2807-x. [DOI] [PubMed] [Google Scholar]
- 112.Carmona R, Perez-Alvarez L, Munoz M, Casado G, Delgado E, Sierra M, Thomson M, Vega Y, Vazquez de Parga E, Contreras G, Medrano L, Najera R. 2005. Natural resistance-associated mutations to Enfuvirtide (T20) and polymorphisms in the gp41 region of different HIV-1 genetic forms from T20 naive patients. J Clin Virol 32:248–253. 10.1016/j.jcv.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 113.Moyo T, Ereno-Orbea J, Jacob RA, Pavillet CE, Kariuki SM, Tangie EN, Julien JP, Dorfman JR. 2018. Molecular basis of unusually high neutralization resistance in Tier 3 HIV-1 strain 253–11. J Virol 92:e02261-17. 10.1128/JVI.02261-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang PF, Bouma P, Park EJ, Margolick JB, Robinson JE, Zolla-Pazner S, Flora MN, Quinnan GV. Jr, 2002. A variable region 3 (V3) mutation determines a global neutralization phenotype and CD4-independent infectivity of a human immunodeficiency virus type 1 envelope associated with a broadly cross-reactive, primary virus-neutralizing antibody response. J Virol 76:644–655. 10.1128/jvi.76.2.644-655.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zolla-Pazner S, Cohen SS, Boyd D, Kong XP, Seaman M, Nussenzweig M, Klein F, Overbaugh J, Totrov M. 2016. Structure/function studies involving the V3 region of the HIV-1 envelope delineate multiple factors that affect neutralization sensitivity. J Virol 90:636–649. 10.1128/JVI.01645-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Powell RLR, Totrov M, Itri V, Liu X, Fox A, Zolla-Pazner S. 2017. Plasticity and epitope exposure of the HIV-1 envelope trimer. J Virol 91:e00410-17. 10.1128/JVI.00410-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bradley T, Trama A, Tumba N, Gray E, Lu X, Madani N, Jahanbakhsh F, Eaton A, Xia SM, Parks R, Lloyd KE, Sutherland LL, Scearce RM, Bowman CM, Barnett S, Abdool-Karim SS, Boyd SD, Melillo B, Smith AB, III, Sodroski J, Kepler TB, Alam SM, Gao F, Bonsignori M, Liao HX, Moody MA, Montefiori D, Santra S, Morris L, Haynes BF. 2016. Amino acid changes in the HIV-1 gp41 membrane proximal region control virus neutralization sensitivity. EBioMedicine 12:196–207. 10.1016/j.ebiom.2016.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Herschhorn A, Gu C, Moraca F, Ma X, Farrell M, Smith AB, III, Pancera M, Kwong PD, Schon A, Freire E, Abrams C, Blanchard SC, Mothes W, Sodroski JG. 2017. The beta20-beta21 of gp120 is a regulatory switch for HIV-1 Env conformational transitions. Nat Commun 8:1049. 10.1038/s41467-017-01119-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Guzzo C, Zhang P, Liu Q, Kwon AL, Uddin F, Wells AI, Schmeisser H, Cimbro R, Huang J, Doria-Rose N, Schmidt SD, Dolan MA, Connors M, Mascola JR, Lusso P. 2018. Structural constraints at the trimer apex stabilize the HIV-1 envelope in a closed, antibody-protected conformation. mBio 9:e00955-18. 10.1128/mBio.00955-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.McGee K, Haim H, Korioth-Schmitz B, Espy N, Javanbakht H, Letvin N, Sodroski J. 2014. The selection of low envelope glycoprotein reactivity to soluble CD4 and cold during simian-human immunodeficiency virus infection of rhesus macaques. J Virol 88:21–40. 10.1128/JVI.01558-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kassa A, Finzi A, Pancera M, Courter JR, Smith AB, III, Sodroski J. 2009. Identification of a human immunodeficiency virus type 1 envelope glycoprotein variant resistant to cold inactivation. J Virol 83:4476–4488. 10.1128/JVI.02110-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kassa A, Madani N, Schon A, Haim H, Finzi A, Xiang SH, Wang L, Princiotto A, Pancera M, Courter J, Smith AB, III, Freire E, Kwong PD, Sodroski J. 2009. Transitions to and from the CD4-bound conformation are modulated by a single-residue change in the human immunodeficiency virus type 1 gp120 inner domain. J Virol 83:8364–8378. 10.1128/JVI.00594-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Klasse PJ, McKeating JA, Schutten M, Reitz MS, Jr, Robert-Guroff M. 1993. An immune-selected point mutation in the transmembrane protein of human immunodeficiency virus type 1 (HXB2-Env:Ala 582(–>Thr)) decreases viral neutralization by monoclonal antibodies to the CD4-binding site. Virology 196:332–337. 10.1006/viro.1993.1484. [DOI] [PubMed] [Google Scholar]
- 124.Thali M, Charles M, Furman C, Cavacini L, Posner M, Robinson J, Sodroski J. 1994. Resistance to neutralization by broadly reactive antibodies to the human immunodeficiency virus type 1 gp120 glycoprotein conferred by a gp41 amino acid change. J Virol 68:674–680. 10.1128/JVI.68.2.674-680.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pacheco B, Alsahafi N, Debbeche O, Prevost J, Ding S, Chapleau JP, Herschhorn A, Madani N, Princiotto A, Melillo B, Gu C, Zeng X, Mao Y, Smith AB, III, Sodroski J, Finzi A. 2017. Residues in the gp41 ectodomain regulate HIV-1 envelope glycoprotein conformational transitions induced by gp120-directed inhibitors. J Virol 91:e02219-16. 10.1128/JVI.02219-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schiffner T, Pallesen J, Russell RA, Dodd J, de Val N, LaBranche CC, Montefiori D, Tomaras GD, Shen X, Harris SL, Moghaddam AE, Kalyuzhniy O, Sanders RW, McCoy LE, Moore JP, Ward AB, Sattentau QJ. 2018. Structural and immunologic correlates of chemically stabilized HIV-1 envelope glycoproteins. PLoS Pathog 14:e1006986. 10.1371/journal.ppat.1006986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shen X, Bogers WM, Yates NL, Ferrari G, Dey AK, Williams WT, Jaeger FH, Wiehe K, Sawant S, Alam SM, LaBranche CC, Montefiori DC, Martin L, Srivastava I, Heeney J, Barnett SW, Tomaras GD. 2017. Cross-linking of a CD4-mimetic miniprotein with HIV-1 Env gp140 alters kinetics and specificities of antibody responses against HIV-1 Env in macaques. J Virol 91:e00401-17. 10.1128/JVI.00401-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schiffner T, de Val N, Russell RA, de Taeye SW, de la Pena AT, Ozorowski G, Kim HJ, Nieusma T, Brod F, Cupo A, Sanders RW, Moore JP, Ward AB, Sattentau QJ. 2016. Chemical cross-linking stabilizes native-like HIV-1 envelope glycoprotein trimer antigens. J Virol 90:813–828. 10.1128/JVI.01942-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Schiffner T, Kong L, Duncan CJ, Back JW, Benschop JJ, Shen X, Huang PS, Stewart-Jones GB, DeStefano J, Seaman MS, Tomaras GD, Montefiori DC, Schief WR, Sattentau QJ. 2013. Immune focusing and enhanced neutralization induced by HIV-1 gp140 chemical cross-linking. J Virol 87:10163–10172. 10.1128/JVI.01161-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yuan W, Bazick J, Sodroski J. 2006. Characterization of the multiple conformational states of free monomeric and trimeric human immunodeficiency virus envelope glycoproteins after fixation by cross-linker. J Virol 80:6725–6737. 10.1128/JVI.00118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Witt KC, Castillo-Menendez L, Ding H, Espy N, Zhang S, Kappes JC, Sodroski J. 2017. Antigenic characterization of the human immunodeficiency virus (HIV-1) envelope glycoprotein precursor incorporated into nanodiscs. PLoS One 12:e0170672. 10.1371/journal.pone.0170672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Leitner A, Walzthoeni T, Aebersold R. 2014. Lysine-specific chemical cross-linking of protein complexes and identification of cross-linking sites using LC-MS/MS and the xQuest/xProphet software pipeline. Nat Protoc 9:120–137. 10.1038/nprot.2013.168. [DOI] [PubMed] [Google Scholar]
- 133.Merkley ED, Rysavy S, Kahraman A, Hafen RP, Daggett V, Adkins JN. 2014. Distance restraints from crosslinking mass spectrometry: mining a molecular dynamics simulation database to evaluate lysine-lysine distances. Protein Sci 23:747–759. 10.1002/pro.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Singh P, Panchaud A, Goodlett DR. 2010. Chemical cross-linking and mass spectrometry as a low-resolution protein structure determination technique. Anal Chem 82:2636–2642. 10.1021/ac1000724. [DOI] [PubMed] [Google Scholar]
- 135.Leitner A, Joachimiak LA, Unverdorben P, Walzthoeni T, Frydman J, Forster F, Aebersold R. 2014. Chemical cross-linking/mass spectrometry targeting acidic residues in proteins and protein complexes. Proc Natl Acad Sci USA 111:9455–9460. 10.1073/pnas.1320298111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Foley B, Leitner T, Apetrei C, Hahn B, Mizrachi I, Mullins J, Rambaut A, Wolinsky S, Korber B (ed). 2014. HIV sequence compendium 2014. Los Alamos National Laboratory, Los Alamos, NM. LA-UR-14-26717. [Google Scholar]
- 137.Moore JP, Trkola A, Korber B, Boots LJ, Kessler JA, 2nd, McCutchan FE, Mascola J, Ho DD, Robinson J, Conley AJ. 1995. A human monoclonal antibody to a complex epitope in the V3 region of gp120 of human immunodeficiency virus type 1 has broad reactivity within and outside clade B. J Virol 69:122–130. 10.1128/JVI.69.1.122-130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zou S, Zhang S, Gaffney A, Ding H, Lu M, Grover JR, Farrell M, Nguyen HT, Zhao C, Anang S, Zhao M, Mohammadi M, Blanchard SC, Abrams C, Madani N, Mothes W, Kappes JC, Smith AB, III, Sodroski J. 2020. Long-acting BMS-378806 analogues stabilize the state-1 conformation of the human immunodeficiency virus type 1 envelope glycoproteins. J Virol 94:e00148-20. 10.1128/JVI.00148-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lu M, Ma X, Reichard N, Terry DS, Arthos J, Smith AB, III, Sodroski JG, Blanchard SC, Mothes W. 2020. Shedding-resistant HIV-1 envelope glycoproteins adopt downstream conformations that remain responsive to conformation-preferring ligands. J Virol 94:e00597-20. 10.1128/JVI.00597-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Si Z, Madani N, Cox JM, Chruma JJ, Klein JC, Schon A, Phan N, Wang L, Biorn AC, Cocklin S, Chaiken I, Freire E, Smith AB, III, Sodroski JG. 2004. Small-molecule inhibitors of HIV-1 entry block receptor-induced conformational changes in the viral envelope glycoproteins. Proc Natl Acad Sci USA 101:5036–5041. 10.1073/pnas.0307953101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Moore JP, McKeating JA, Weiss RA, Sattentau QJ. 1990. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science 250:1139–1142. 10.1126/science.2251501. [DOI] [PubMed] [Google Scholar]
- 142.McKeating JA, McKnight A, Moore JP. 1991. Differential loss of envelope glycoprotein gp120 from virions of human immunodeficiency virus type 1 isolates: effects on infectivity and neutralization. J Virol 65:852–860. 10.1128/JVI.65.2.852-860.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Li Y, O'Dell S, Walker LM, Wu X, Guenaga J, Feng Y, Schmidt SD, McKee K, Louder MK, Ledgerwood JE, Graham BS, Haynes BF, Burton DR, Wyatt RT, Mascola JR. 2011. Mechanism of neutralization by the broadly neutralizing HIV-1 monoclonal antibody VRC01. J Virol 85:8954–8967. 10.1128/JVI.00754-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861. 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Pancera M, Shahzad-Ul-Hussan S, Doria-Rose NA, McLellan JS, Bailer RT, Dai K, Loesgen S, Louder MK, Staupe RP, Yang Y, Zhang B, Parks R, Eudailey J, Lloyd KE, Blinn J, Alam SM, Haynes BF, Amin MN, Wang LX, Burton DR, Koff WC, Nabel GJ, Mascola JR, Bewley CA, Kwong PD. 2013. Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1-V2-directed antibody PG16. Nat Struct Mol Biol 20:804–813. 10.1038/nsmb.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Julien JP, Sok D, Khayat R, Lee JH, Doores KJ, Walker LM, Ramos A, Diwanji DC, Pejchal R, Cupo A, Katpally U, Depetris RS, Stanfield RL, McBride R, Marozsan AJ, Paulson JC, Sanders RW, Moore JP, Burton DR, Poignard P, Ward AB, Wilson IA. 2013. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog 9:e1003342. 10.1371/journal.ppat.1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Huang J, Kang BH, Pancera M, Lee JH, Tong T, Feng Y, Imamichi H, Georgiev IS, Chuang GY, Druz A, Doria-Rose NA, Laub L, Sliepen K, van Gils MJ, de la Pena AT, Derking R, Klasse PJ, Migueles SA, Bailer RT, Alam M, Pugach P, Haynes BF, Wyatt RT, Sanders RW, Binley JM, Ward AB, Mascola JR, Kwong PD, Connors M. 2014. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature 515:138–142. 10.1038/nature13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol 67:3978–3988. 10.1128/JVI.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Madani N, Princiotto AM, Easterhoff D, Bradley T, Luo K, Williams WB, Liao HX, Moody MA, Phad GE, Vazquez Bernat N, Melillo B, Santra S, Smith AB, III, Karlsson Hedestam GB, Haynes B, Sodroski J. 2016. Antibodies elicited by multiple envelope glycoprotein immunogens in primates neutralize primary human immunodeficiency viruses (HIV-1) sensitized by CD4-mimetic compounds. J Virol 90:5031–5046. 10.1128/JVI.03211-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Posner MR, Hideshima T, Cannon T, Mukherjee M, Mayer KH, Byrn RA. 1991. An IgG human monoclonal antibody that reacts with HIV-1/GP120, inhibits virus binding to cells, and neutralizes infection. J Immunol 146:4325–4332. [PubMed] [Google Scholar]
- 151.Melillo B, Liang S, Park J, Schon A, Courter JR, LaLonde JM, Wendler DJ, Princiotto AM, Seaman MS, Freire E, Sodroski J, Madani N, Hendrickson WA, Smith AB, III.. 2016. Small-molecule CD4-mimics: structure-based optimization of HIV-1 entry inhibition. ACS Med Chem Lett 7:330–334. 10.1021/acsmedchemlett.5b00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659. 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, Sodroski JG. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705–711. 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 154.Finzi A, Xiang SH, Pacheco B, Wang L, Haight J, Kassa A, Danek B, Pancera M, Kwong PD, Sodroski J. 2010. Topological layers in the HIV-1 gp120 inner domain regulate gp41 interaction and CD4-triggered conformational transitions. Mol Cell 37:656–667. 10.1016/j.molcel.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Pancera M, Majeed S, Ban YE, Chen L, Huang CC, Kong L, Kwon YD, Stuckey J, Zhou T, Robinson JE, Schief WR, Sodroski J, Wyatt R, Kwong PD. 2010. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc Natl Acad Sci USA 107:1166–1171. 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mao Y, Wang L, Gu C, Herschhorn A, Desormeaux A, Finzi A, Xiang SH, Sodroski JG. 2013. Molecular architecture of the uncleaved HIV-1 envelope glycoprotein trimer. Proc Natl Acad Sci USA 110:12438–12443. 10.1073/pnas.1307382110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Blattner C, Lee JH, Sliepen K, Derking R, Falkowska E, de la Pena AT, Cupo A, Julien JP, van Gils M, Lee PS, Peng W, Paulson JC, Poignard P, Burton DR, Moore JP, Sanders RW, Wilson IA, Ward AB. 2014. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity 40:669–680. 10.1016/j.immuni.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Protocol GPI, Koff WC, Wilson IA, Burton DR, Poignard P, Protocol G Principal Investigators. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470. 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Cavacini LA, Emes CL, Wisnewski AV, Power J, Lewis G, Montefiori D, Posner MR. 1998. Functional and molecular characterization of human monoclonal antibody reactive with the immunodominant region of HIV type 1 glycoprotein 41. AIDS Res Hum Retroviruses 14:1271–1280. 10.1089/aid.1998.14.1271. [DOI] [PubMed] [Google Scholar]
- 160.Zhang S, Holmes AP, Dick A, Rashad AA, Enriquez RL, Canziani GA, Root MJ, Chaiken IM. 2021. Altered Env conformational dynamics as a mechanism of resistance to peptide-triazole HIV-1 inactivators. Retrovirology 18:31. 10.1186/s12977-021-00575-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Labrosse B, Treboute C, Alizon M. 2000. Sensitivity to a nonpeptidic compound (RPR103611) blocking human immunodeficiency virus type 1 Env-mediated fusion depends on sequence and accessibility of the gp41 loop region. J Virol 74:2142–2150. 10.1128/jvi.74.5.2142-2150.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Madani N, Schon A, Princiotto AM, Lalonde JM, Courter JR, Soeta T, Ng D, Wang L, Brower ET, Xiang SH, Kwon YD, Huang CC, Wyatt R, Kwong PD, Freire E, Smith AB, III, Sodroski J. 2008. Small-molecule CD4 mimics interact with a highly conserved pocket on HIV-1 gp120. Structure 16:1689–1701. 10.1016/j.str.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Migneault I, Dartiguenave C, Bertrand MJ, Waldron KC. 2004. Glutaraldehyde: behavior in aqueous solution, reaction with proteins, and application to enzyme crosslinking. Biotechniques 37:790–802. 10.2144/04375RV01. [DOI] [PubMed] [Google Scholar]
- 164.Joyce MG, Georgiev IS, Yang Y, Druz A, Geng H, Chuang GY, Kwon YD, Pancera M, Rawi R, Sastry M, Stewart-Jones GBE, Zheng A, Zhou T, Choe M, Van Galen JG, Chen RE, Lees CR, Narpala S, Chambers M, Tsybovsky Y, Baxa U, McDermott AB, Mascola JR, Kwong PD. 2017. Soluble prefusion closed DS-SOSIP.664-Env trimers of diverse HIV-1 strains. Cell Rep 21:2992–3002. 10.1016/j.celrep.2017.11.016. [DOI] [PubMed] [Google Scholar]
- 165.Privalov PL. 1990. Cold denaturation of proteins. Crit Rev Biochem Mol Biol 25:281–305. 10.3109/10409239009090612. [DOI] [PubMed] [Google Scholar]
- 166.Tsai CJ, Maizel JV, Jr, Nussinov R. 2002. The hydrophobic effect: a new insight from cold denaturation and a two-state water structure. Crit Rev Biochem Mol Biol 37:55–69. 10.1080/10409230290771456. [DOI] [PubMed] [Google Scholar]
- 167.Lopez CF, Darst RK, Rossky PJ. 2008. Mechanistic elements of protein cold denaturation. J Phys Chem B 112:5961–5967. 10.1021/jp075928t. [DOI] [PubMed] [Google Scholar]
- 168.Kwon YD, Georgiev IS, Ofek G, Zhang B, Asokan M, Bailer RT, Bao A, Caruso W, Chen X, Choe M, Druz A, Ko SY, Louder MK, McKee K, O'Dell S, Pegu A, Rudicell RS, Shi W, Wang K, Yang Y, Alger M, Bender MF, Carlton K, Cooper JW, Blinn J, Eudailey J, Lloyd K, Parks R, Alam SM, Haynes BF, Padte NN, Yu J, Ho DD, Huang J, Connors M, Schwartz RM, Mascola JR, Kwong PD. 2016. Optimization of the solubility of HIV-1-neutralizing antibody 10E8 through somatic variation and structure-based design. J Virol 90:5899–5914. 10.1128/JVI.03246-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, Alam SM, Wang T, Yang Y, Zhang B, Migueles SA, Wyatt R, Haynes BF, Kwong PD, Mascola JR, Connors M. 2012. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491:406–412. 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chakrabarti BK, Walker LM, Guenaga JF, Ghobbeh A, Poignard P, Burton DR, Wyatt RT. 2011. Direct antibody access to the HIV-1 membrane-proximal external region positively correlates with neutralization sensitivity. J Virol 85:8217–8226. 10.1128/JVI.00756-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wang Q, Esnault F, Zhao M, Chiu T-J, Smith AB, III, Nguyen HT, Sodroski J. 2021. Global increases in human immunodeficiency virus neutralization sensitivity due to alterations in the membrane-proximal external region of the envelope glycoprotein can be minimized by distant State 1-stabilizing changes. bioRxiv. 10.1101/2021.11.01.466860. [DOI] [PMC free article] [PubMed]
- 172.Herrera C, Klasse PJ, Michael E, Kake S, Barnes K, Kibler CW, Campbell-Gardener L, Si Z, Sodroski J, Moore JP, Beddows S. 2005. The impact of envelope glycoprotein cleavage on the antigenicity, infectivity, and neutralization sensitivity of Env-pseudotyped human immunodeficiency virus type 1 particles. Virology 338:154–172. 10.1016/j.virol.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 173.Pancera M, Wyatt R. 2005. Selective recognition of oligomeric HIV-1 primary isolate envelope glycoproteins by potently neutralizing ligands requires efficient precursor cleavage. Virology 332:145–156. 10.1016/j.virol.2004.10.042. [DOI] [PubMed] [Google Scholar]
- 174.Chakrabarti BK, Pancera M, Phogat S, O'Dell S, McKee K, Guenaga J, Robinson J, Mascola J, Wyatt RT. 2011. HIV type 1 Env precursor cleavage state affects recognition by both neutralizing and nonneutralizing gp41 antibodies. AIDS Res Hum Retroviruses 27:877–887. 10.1089/AID.2010.0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Li Y, O'Dell S, Wilson R, Wu X, Schmidt SD, Hogerkorp CM, Louder MK, Longo NS, Poulsen C, Guenaga J, Chakrabarti BK, Doria-Rose N, Roederer M, Connors M, Mascola JR, Wyatt RT. 2012. HIV-1 neutralizing antibodies display dual recognition of the primary and coreceptor binding sites and preferential binding to fully cleaved envelope glycoproteins. J Virol 86:11231–11241. 10.1128/JVI.01543-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Haim H, Salas I, Sodroski J. 2013. Proteolytic processing of the human immunodeficiency virus envelope glycoprotein precursor decreases conformational flexibility. J Virol 87:1884–1889. 10.1128/JVI.02765-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Castillo-Menendez LR, Witt K, Espy N, Princiotto A, Madani N, Pacheco B, Finzi A, Sodroski J. 2018. Comparison of uncleaved and mature human immunodeficiency virus membrane envelope glycoprotein trimers. J Virol 92:e00277-18. 10.1128/JVI.00277-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Gift SK, Leaman DP, Zhang L, Kim AS, Zwick MB. 2017. Functional stability of HIV-1 envelope trimer affects accessibility to broadly neutralizing antibodies at its apex. J Virol 91:e01216-17. 10.1128/JVI.01216-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Dey AK, David KB, Klasse PJ, Moore JP. 2007. Specific amino acids in the N-terminus of the gp41 ectodomain contribute to the stabilization of a soluble, cleaved gp140 envelope glycoprotein from human immunodeficiency virus type 1. Virology 360:199–208. 10.1016/j.virol.2006.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Dey AK, David KB, Ray N, Ketas TJ, Klasse PJ, Doms RW, Moore JP. 2008. N-terminal substitutions in HIV-1 gp41 reduce the expression of non-trimeric envelope glycoproteins on the virus. Virology 372:187–200. 10.1016/j.virol.2007.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Pardi N, LaBranche CC, Ferrari G, Cain DW, Tombacz I, Parks RJ, Muramatsu H, Mui BL, Tam YK, Kariko K, Polacino P, Barbosa CJ, Madden TD, Hope MJ, Haynes BF, Montefiori DC, Hu SL, Weissman D. 2019. Characterization of HIV-1 nucleoside-modified mRNA vaccines in rabbits and rhesus macaques. Mol Ther Nucleic Acids 15:36–47. 10.1016/j.omtn.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zhang P, Narayanan E, Liu Q, Tsybovsky Y, Boswell K, Ding S, Hu Z, Follmann D, Lin Y, Miao H, Schmeisser H, Rogers D, Falcone S, Elbashir SM, Presnyak V, Bahl K, Prabhakaran M, Chen X, Sarfo EK, Ambrozak DR, Gautam R, Martin MA, Swerczek J, Herbert R, Weiss D, Misamore J, Ciaramella G, Himansu S, Stewart-Jones G, McDermott A, Koup RA, Mascola JR, Finzi A, Carfi A, Fauci AS, Lusso P. 2021. A multiclade env-gag VLP mRNA vaccine elicits tier-2 HIV-1-neutralizing antibodies and reduces the risk of heterologous SHIV infection in macaques. Nat Med 27:2234–2245. 10.1038/s41591-021-01574-5. [DOI] [PubMed] [Google Scholar]
- 183.Sanders RW, Vesanen M, Schuelke N, Master A, Schiffner L, Kalyanaraman R, Paluch M, Berkhout B, Maddon PJ, Olson WC, Lu M, Moore JP. 2002. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol 76:8875–8889. 10.1128/jvi.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Wang Q, Liu L, Ren W, Gettie A, Wang H, Liang Q, Shi X, Montefiori DC, Zhou T, Zhang L. 2019. A single substitution in gp41 modulates the neutralization profile of SHIV during in vivo adaptation. Cell Rep 27:2593–2607.e5. 10.1016/j.celrep.2019.04.108. [DOI] [PubMed] [Google Scholar]