Abstract
Hematopoietic stem cells (HSCs) have been studied extensively since their initial functional description in 1961 when Dr. James Till and Dr. Ernest McCulloch developed the first in vivo clonal strategy, termed the spleen colony-forming unit (CFU-S) assay, to assess the functional capacity of bone marrow–derived hematopoietic progenitors at the single-cell level. Through transplantation of bone marrow cells and analysis of the resulting cellular nodules in the spleen, the CFU-S assay revealed both the self-renewal and clonal differentiation capacity of hematopoietic progenitors. Further development and use of this assay have identified highly proliferative, self-renewing, and differentiating HSCs that possess clonal, multilineage differentiation. The CFU-S strategy has also been adapted to interrogating single purified hematopoietic stem and progenitor cell populations, advancing our knowledge of the hematopoietic hierarchy. In this review, we explore the major discoveries made with the CFU-S assay, consider its modern use and recent improvements, and compare it with commonly used long-term transplantation assays to determine the continued value of the CFU-S assay for understanding HSC biology and hematopoiesis.
Graphical Abstract
Hematopoietic stem cells (HSCs) are the only cells within the hematopoietic system that possess the combined ability to differentiate into all lineages of functional blood cells and self-renew indefinitely to sustain hematopoiesis throughout life. HSCs were originally hypothesized to exist after the discovery that transplantation of healthy bone marrow (BM) cells could rescue irradiated recipient animals and replenish their hematopoietic cells through a tremendously dynamic process [1]. The hypothesis for the existence of HSCs was reinforced when cells with multilineage capacity were discovered in 1961 by Dr. James Till and Dr. Ernest McCulloch [2], who developed the first in vivo functional assay for quantification of the clonal and differentiation potential of hematopoietic progenitor cells. Since then, HSCs have remained one of the best-characterized tissue-specific stem cells, both from a basic biology perspective and for their use in regenerative medicine, with particular emphasis on clonal function [3–13].
In their seminal studies, Till and McCulloch developed the first in vivo assay to assess the proliferative and differentiation capacity of primitive hematopoietic cells in mouse BM [2,14,15]. In these early experiments, the identity of cells capable of forming multilineage spleen colonies was still uncertain, and these cells were appropriately and carefully termed “colony forming units” (CFUs) based on their functional capacity. This initial demonstration of a BM cell population capable of multilineage blood cell reconstitution resulted in a paradigm shift in the field of hematopoiesis and opened several questions that this elegant assay is uniquely positioned to answer. Here, we explore the major utilities of the spleen colony-forming unit (CFU-S) assay, consider its modern use and recent improvements, and discuss its utility in current hematopoiesis research.
ONE FROM ALL OR ALL FROM ONE: ARE SPLEEN COLONIES CLONAL?
Spleen colony formation is rare, with transplantation of approximately 10,000 BM cells into conditioned recipients required to yield one spleen colony [2]. Initially, the linear relationship between the number of hematopoietic nucleated BM cells transplanted and the number of spleen colonies formed indicated that single cells may be able to give rise to individual spleen colonies [2]. The CFU-S assay used by Till, McCulloch, and their colleagues consisted of intravenously injecting BM cells from the femora of healthy donor mice into recipients conditioned through lethal irradiation for host cell ablation. In a subsequent version of this assay, the donor cells were irradiated prior to transplantation to induce unique, random chromosomal breaks that distinguish them from host cells and distinguish single donor cells from one another. After 10–11 days, recipient mice presented macroscopic nodules of cellular expansion in the spleen that were formed by rapidly proliferating hematopoietic cells and composed of undifferentiated (stem and progenitor) cells along with erythroblasts, granulocytes, and megakaryocytes (Figure 1A) [14,16,17]. If more than one differentiated cell type within a spleen colony, as readily scored by cell morphology under a microscope, contained the same unique chromosome aberration, these cells must have a shared cellular origin. As this was indeed the outcome, these studies definitively demonstrated that individual cells with multilineage capacity exist within mouse BM. Additionally, the vast majority of scored cells from a single colony harbored the same unique chromosomal aberration, indicating a single, shared progenitor cell. Inducing chromosomal breaks via irradiation to establish clonality was a particularly clever strategy when more modern tools, such as flow cytometry (late 1960s [18]), polymerase chain reaction (PCR) (1983 [19]), and monoclonal antibodies (1975 [20]), had not yet been established. Thus, this CFU-S strategy provided direct cytological evidence indicating that most, if not all, cells within a single colony arose from a single, highly proliferative, multipotent CFU-S cell.
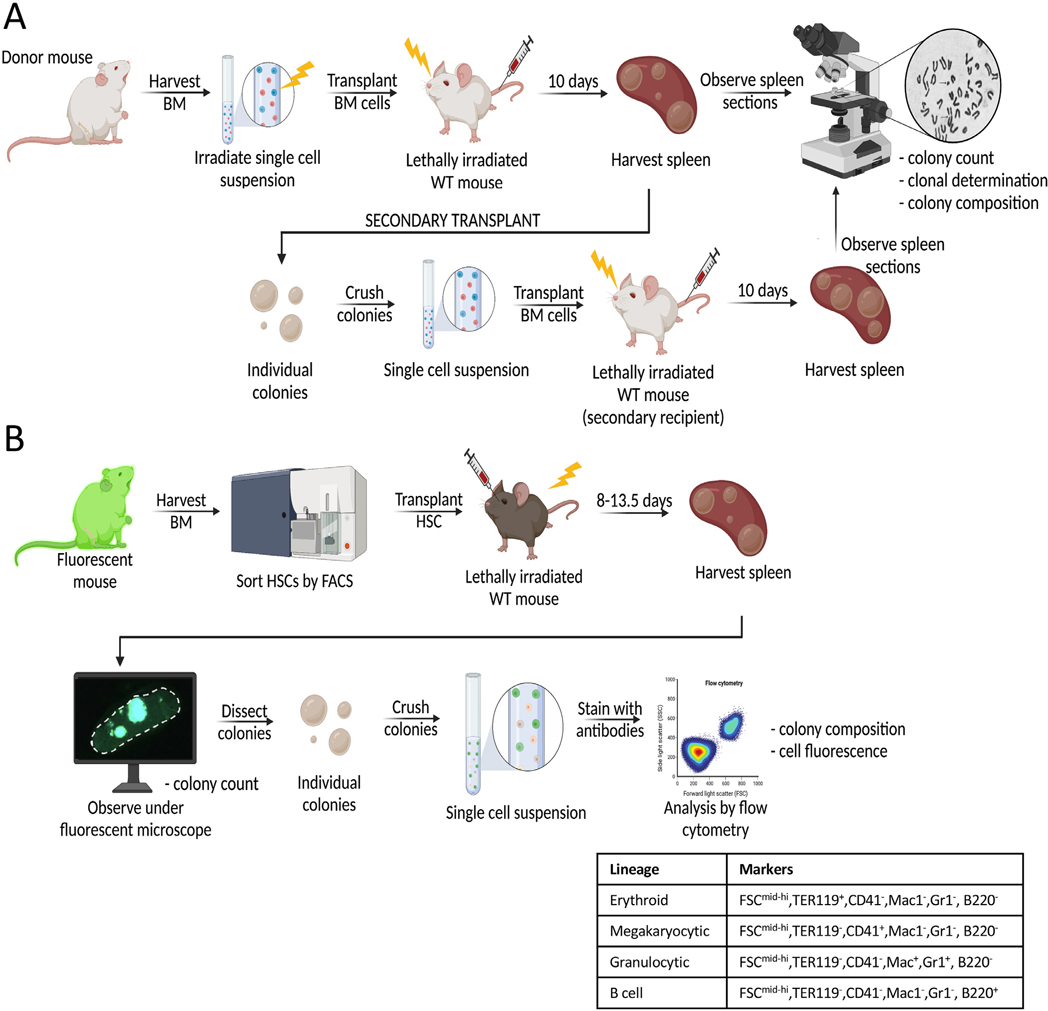
Figure 1.
Schematic of CFU-S assays. (A) In the original CFU-S assay developed by Till and McCulloch in 1961 [2], bone marrow cells were harvested from donor mice and irradiated prior to intravenous injection into irradiated recipient mice. Ten days post transplant, spleens were counted, harvested, and sectioned for histologic analysis. For serial transplantations to determine self-renewal of CFU-S cells within spleen colonies [15], individual colonies were dissected and transplanted into secondary recipients as single-cell suspensions. Spleen colonies formed after secondary transplantation were analyzed similarly to those from primary transplantations [15]. (B) In the updated CFU-S assay with high throughput, quantitative analysis, HSCs (or other hematopoietic progenitors) were FACS-purified and transplanted into irradiated recipient mice. At 13.5 days post transplan, individual spleen colonies were counted, harvested, and dissected under a fluorescent microscope [22]. Single-cell suspensions from each colony were then analyzed by flow cytometry to assess colony composition and cell fluorescence. Four lineages (erythroid, megakaryocytic, granulocytic, and B cell) were identified using the markers listed in the table [22,41]. BM=bone marrow; CFU-S=spleen colony-forming unit; HSCs=hematopoietic stem cells.
One caveat of the early CFU-S strategy was that radiation-induced chromosomal aberration was only obvious in a small fraction of the colonies obtained and that only intact cells in metaphase could be scored based on their karyotype, leaving the possibility that the unscored cells were derived from one or more additional cells. More recent data from our group and others support the evidence that each colony consists of progeny from a single cell. In one of our studies, transplantation of as few as 10 HSCs directly into the spleen resulted in several colonies, with some mice approaching a 1:1 HSC-to-colony ratio [21]. In a second study using fluorescence microscopy and flow cytometry, we observed only single-color splenic colonies when an equal mixture of green fluorescent protein (GFP) and Tomato fluorescent cells were transplanted into the same recipient [22]. Similarly, Fehse’s group assessed clonality using a red-green–blue cell tracking methodology that resulted in mostly homogenously colored spleen colonies upon transplantation [23]. In their study, the very few CFU-S colonies that contained more than one color were potentially due to two colonies initiating in close proximity and fusing together on growth. Definitive evidence for clonality could be obtained via the relatively recent clonal tracking strategies, such as single-cell barcoding [24–27]. Though not entirely unequivocal, the collective evidence uniformly supports the original conclusion by Till and McCulloch that spleen colonies are clonal.
PERSISTENCE MATTERS: DO CFU-S SELF-RENEW?
One hallmark of a true HSC is the ability to self-renew: to give rise to additional cells with the same properties. Self-renewal is often demonstrated by the cell’s ability to maintain multilineage reconstitution on secondary transplantation. In one of the first direct demonstrations of in vivo self-renewal, injection of day-10 spleen colony content into secondary irradiated recipient mice revealed that colony-forming cells include cells with the regenerative capacity expected of stem cells (Figure 1A) [15]. Supporting the conclusion of CFU-S self-renewal was the observation that the number of colony forming cells rapidly increased between days 10 and 14 after transplant, thus revealing that CFU-S cells can give rise to more cells with CFU-S capacity [15]. Additional studies from Schofield et al. [28] calculated the probability of CFU-S self-renewal and estimated that, on average, 68.3% of cells will produce more CFU-S while 31.7% will differentiate. Thus, CFU-S cells were defined as colony-forming hematopoietic progenitor cells that are highly proliferative and capable of differentiation and self-renewal.
CFU-S KINETICS: WHAT DO EARLY AND LATE SPLEEN COLONY FORMATION TELL US?
On transplantation of BM cells, a fraction of CFU-S cells (containing both stem and progenitor populations) will home to the spleen of the recipient mouse and give rise to colonies of heterogeneous composition. Given that transplanted, heterogeneous BM contains various hematopoietic progenitor populations along all stages of differentiation, the cellular output kinetics could inform relative contributions by unique CFU-S subpopulations. Indeed, an initial study found that colonies present at days 7–8 post-BM transplan consisted primarily of erythroblasts and were formed by unilineage, non–self-renewing mature erythroid precursors [29]. Importantly, cell purification technology progressed in parallel with CFU-S assays, allowing more purified populations, rather than whole BM, to be transplanted. Thus, although a small proportion of megakaryocyte/erythrocyte progenitor (MEP)–derived erythroid colonies persisted through day 12 [29–32], transplantation of MEPs purified via fluorescence-activated cell sorting (FACS) confirmed that day 8 erythroid-only colonies originated primarily from the MEP, and not the HSC, population [22,31,33].
Conversely, cells higher in the hematopoietic hierarchy form colonies with slower kinetics. On transplantation of purified HSCs or multipotent progenitors (MPPs), no spleen colonies formed at day 8, potentially because their differentiation kinetics cause a delay in the production of effector cells [22,31,33]. Instead, HSCs and MPPs formed colonies primarily at day 12 when transplanted together [17] or at days 11–12 (MPPs) or 12–14 (HSCs) when transplanted separately, with HSCs having a much higher CFU-S frequency than MPPs [22,33–35]. Moreover, purified long-term (LT) HSCs and short-term (ST) HSCs exhibited similar day 11–12 CFU-S activity and frequency, but ST-HSCs were able to form visible spleen colonies at day 8, which is consistent with their increased radioprotective capacity compared with LT-HSCs [22,33–38]. The composition of both HSC- and MPP-derived spleen colonies was heterogeneous and multilineage, with most colonies containing erythrocytes, granulocytes, and some megakaryocytes [22]. Previously, all day 12 colonies had been found to contain CFU-S cells of varying self-renewal and differentiation potential [15,17,30,39]. Thus, the splenic colonies appearing at this time point contain self-renewing primitive stem cells that are multipotent at the single-cell level [30,39,40]. HSC-Derived colonies arise between days 12 and 14, and only colonies with this timing contain multipotent HSCs. Of note, beyond 14 days, satellite colonies start forming, confounding identification and analysis of primary colonies. Taken together, multiple progenitor cells can form spleen colonies with different kinetics and cell output, and the CFU-S assay can resolve these temporal and cellular features.
TECHNOLOGY BOOSTS THE RESOLUTION AND THROUGHPUT OF CFU-S ASSAYS
Historically, determination of the composition of splenic colonies relied solely on histological analysis of dissected spleens. New technologies, most importantly the development of monoclonal antibodies and flow cytometry, have enabled strategies with increased resolution and higher throughput. To improve the measurement of CFU-S cell output, we recently published an updated, flow cytometry-based version of the original CFU-S assay (Figure 1B) [22,41]. This consists of transplanting specific FACS-purified hematopoietic populations, scoring colony frequency, then dissecting and individually analyzing the resulting splenic colonies qualitatively and quantitatively by flow cytometry for erythroid, megakaryocyte, granulocyte, and B-cell lineages (Figure 1B). The detection of B cells (Ter119−,CD41, Mac1−, Gr1−, B220+ cells) is an important addition as previous histology-based analyses were unable to assess B-cell production, and lymphoid output would reinforce both the identity and multilineage potential of the input cells. Importantly, by first excluding erythroid (Ter119), megakaryocyte (CD41), myeloid (Mac1/CD11b), and granulocytic (Gr1, Ly6C/Ly6G) cells, B220 expression is highly selective for B-lineage cells. As expected based on the slower kinetics for B-cell development, especially from HSCs [22,33], the number of B cells within colonies was low relative to that of erythroid and other cell types. We note that T-cell output cannot be assessed as more time and thymic involvement are required to produce these cells. The enhanced sensitivity flow cytometry provides also allows for the quantitative determination of rare stem and progenitor populations among individual colonies. Using this updated method, we found that day 13.5 colonies formed by purified HSCs, and day 11.5 colonies from Flk2-positive MPPs, contained cells from the erythroid, myeloid, megakaryocytic, and B-cell lineages, with erythroid cells constituting the most cells within a colony. Collectively, these data illustrate that this updated CFU-S assay can determine the multilineage potential of HSCs beyond what histological analyses provide, coupled with magnitudes higher throughput and sensitivity. Additionally, as indicated earlier, combining this improvement with cellular barcoding and intrasplenic transplantation may provide even more robust experimental CFU-S determination.
The CFU-S assay can also be used to determine the clonal differentiation capacity of many hematopoietic progenitors, similar to what was initially done to determine that MEPs were the cells responsible for producing most day 8 splenic colonies [22,31]. Transplantation of FACS-purified common myeloid progenitors (CMPs), which are classically placed upstream of both myeloid and erythroid/megakaryocytic lineages, gave rise to colonies containing primarily erythroid cells at day 9.5; the scarcity of nonerythroid cells is possibly due to their low burst size [22,31]. Transplantation of megakaryocyte progenitors (MkPs) did not produce colonies visible to the eye between days 8 and 12, however, histological analysis revealed microscopic foci of megakaryocytes in recipient mice [42]. Finally, common lymphoid progenitors (CLPs) [43] and granulocyte/monocyte progenitors (GMPs) [31,33] do not possess day 8–12 CFU-S capacity. This could be due either to inefficient homing to the spleen, their low burst size, and/or differentiation kinetics outside the optimal 8- to 14-day window. Lack of CLP- or GMP-derived CFU-S colonies also reinforces the notion that erythroid potential is a hallmark of CFU-S. Further research into the dynamics of specific progenitor populations in the CFU-S assay may reveal additional and/or differential functional capacity, further informing hematopoietic progenitor biology.
COMBINING CFU-S ASSAYS WITH MODERN GENETICS
Although not currently as common as other in vivo analyses, CFU-S assays remain valuable as a qualitative and quantitative method to assess the properties and function of various stem and progenitor cell populations. Genetic manipulation of hematopoietic cells and/or their environment is increasingly common, and the CFU-S assay is uniquely positioned to, quickly and accurately, provide functional insights into subsequent effects on hematopoietic stem and progenitor cell (HSPC) differentiation, expansion, and homing. For example, Kruse et al. (2009) observed that transplantation of whole BM from double heterozygous mutants for Fli-1 and Erg, two Ets proteins known to play roles in hematopoiesis and leukemia, respectively, formed significantly fewer and smaller day 11 spleen colonies compared with wild-type or single heterozygous mice for either gene [44]. This suggested that the genetic interaction between Fli-1 and Erg is critical for normal HSC and progenitor function. Similarly, Summers et al. [45] observed that loss of histone deacetylase 3 (HDAC3) yielded no colonies at either day 8 or 12 following BM transplantation, confirming HDAC3’s role in supporting the proliferation of HSC and progenitor cells.
The CFU-S assay is also used to study extramedullary hematopoiesis in the splenic environment. For example, Mehatre et al. [46] investigated the role of periostin (POSTN)–integrin-αv signaling in splenic HSC function by transplanting healthy BM cells into wild-type or Postn knockout (KO) mice and compared the number of spleen colonies at day 12. They observed a significant decrease in the number of spleen colonies in Postn KO mice, suggesting that the POSTN-deficient splenic microenvironment may not be able to support either the homing and/or growth of hematopoietic progenitors. In another study by our group, Rajendiran et al. [41] transplanted wild-type HSCs into control and CXCL12-overexpressing mice, which exhibited no differences in size, number, or composition of splenic colonies. This suggested that overexpression of CXCL12, which is essential for HSC trafficking, does not affect homing of HSCs to the spleen. Relevantly, a modified CFU-S assay can be used to study homing itself. On transplantation of BM cells or more purified populations, only a fraction of the colony-forming cells will home to the spleen while the rest will migrate elsewhere. To address this homing issue, we previously altered the CFU-S assay protocol to inject donor cells directly into the spleen (intrasplenically [IS]). By comparison, the CFU-S frequency of MPPs injected IS was comparable to the CFU-S frequency of HSCs injected retroorbitally, suggesting that homing efficiency affects, but does not entirely account for, the differential efficiency of colony formation [21]. Given the importance of homing, the traditional application of this assay underestimated the CFU-S frequency potential of the cell population of interest. Collectively, the historical importance combined with recent advancements underscores the power of the CFU-S assay and supports its continued use for assessing the functional capacity of hematopoietic stem and progenitor cells.
THE HEAVYWEIGHT CHAMPIONSHIP: CFU-S VERSUS LONG-TERM RECONSTITUTION ASSAYS
The most widely accepted methods for investigating in vivo HSPC self-renewal, differentiation, and expansion capacity are the CFU-S assay and the more recently developed long-term repopulation assay (LTRA) [47]. Similar to CFU-S assays, LTRA requires transplantation of donor BM or purified hematopoietic stem/progenitor cells into (usually) preconditioned hosts. Blood is then monitored at different time points to assess long-term reconstitution of hematopoietic lineages [21,22,41,48–51]. A transplanted cell population is considered to have long-term multilineage reconstitution (LTMR) potential, a key HSC property, if it continues to self-renew and differentiate in primary recipients beyond 16 weeks posttransplantation and on secondary transplantation [52,53]. LTRAs are performed to verify that one or more bona fide HSCs are present among the transplanted population of interest; persistence and secondary reconstitution distinguish HSCs from hematopoietic progenitors, as the latter will not support hematopoiesis beyond a few weeks post-transplantation.
The CFU-S and LTRA assays readily complement each other (Table 1). For example, Forsberg et al. (2006) transplanted HSCs, ST-HSCs, MPPs, and CMP/MEPs for both CFU-S and LTRA analyses [33]. The CFU-S assay indicated the short-term kinetics of erythropoietic output potential and revealed that erythroid cell generation is a clonal feature of all these populations (but not of GMPs), whereas LTRA allowed investigation of their long-term kinetics of peripheral blood reconstitution. Transplantation of bulk cell populations into a recipient can be used to determine self-renewal and LTMR at the population level; however, it cannot determine whether the donor cells are homogeneous or heterogeneous. To determine the clonal capacity of each transplanted cell, single-cell in vivo clonal analyses are required. These include CFU-S assays and single-cell transplantation [36]. It is important to note that CFU-S assays cannot, alone, be used to assess long-term multilineage reconstitution and self-renewal because spleen colonies get resorbed before LTMR can be determined. Importantly, however, relative to the resource-intensive and technically challenging single-cell transplantation, CFU-S assays are fast and straightforward, and can be used to answer similar questions. For example, CFU-S assays, such single-cell transplantation [22,54], illustrated that a substantial fraction of HSCs and MPPs are multipotent at the single-cell level and can differentiate into both erythromyeloid and lymphoid lineages [22].
Table 1.
Comparison of similarities and differences between CFU-S and LTRA
| CFU-S assay | LTRA | |
|---|---|---|
|
| ||
| Assay details and considerations | ||
| Input cell population | Heterogeneous or sorted populations | Heterogeneous or sorted populations |
| Cell purification | Bulk | Bulk or single cell |
| Assay speed | Fast (8–14 days) | Slow (16+ weeks) |
| Assay type | Terminal | Allows serial sampling from same recipient |
| Assessment of progeny | Progeny developing in the spleen within 8–14 days; does not assess T cell production | Cells that may take longer to be generated across all tissues; in spleen: T cells, tissue |
| resident cells | ||
| Relative assay cost | $ | $$$ |
| Can perform secondary transplantation? | Yes | Yes |
| Questions that can be addressed | ||
| Evaluate stem cell clonality? | Yes, inferred | Only if single-cell transplant |
| Determine stem cell self-renewal and LTMR? | Only if secondary transplants are performed | LTMR yes; HSC self-renewal only if secondary transplants are performed |
| Evaluate blood cell reconstitution capacity? | Yes, limited repertoire | Yes |
| Reconstitution kinetics? | Yes, only between 8–14 days | Yes, between 6 days to >16 weeks |
| Evaluate extramedullary hematopoiesis and the splenic microenvironment? | Optimal | Possible |
| Evaluate mature cell production in genetic models or following treatments? | Yes, this rapid assay can allow many conditions to be screened | Yes; however, feasibility of rapid screens is potentially lower than with CFU-S |
CFU-S= spleen colony-forming unit; LTRA=long-term repopulation assay.
CONCLUSION: CFU-S STRATEGIES STANDING STRONG
The CFU-S assay revolutionized hematopoiesis and stem cell biology at a time when rare hematopoietic cells had not yet been identified based on immunophenotypic markers. It provided the first direct in vivo evidence of stem cells and led to both transformational strategies and pioneering discoveries that we continue to build on today. This includes the first “draft” of the hematopoietic hierarchy that places the multipotent CFU-S at the top, followed by more committed progenitors that give rise to mature myeloid and lymphoid effector cells [15,55–57]. To this day, CFU-S are an important complement to in vivo assays and in vitro clonal assays with the advantage of being able to address questions of stem cell clonality under a spectrum of physiological, disease, and experimental conditions.
HIGHLIGHTS.
A perspective on the first in vivo clonal stem cell assay is provided.
The self-renewal and differentiation capacity of spleen colony-forming cells is discussed
Technology boosts the resolution and throughput of CFU-S assays.
The current day value of CFU-S assessment is determined.
Acknowledgments
We thank Connor Mattingly for pre-reviewing the article. This work was supported by NIH NIDDK and NIA awards (R01DK100917 and R01 AG062879) to ECF; by a Tobacco-Related Disease Research Program (TRDRP) Predoctoral Fellowship (T31DT1690) to ARyB; and by NIGMS IRACDA Postdoctoral Training Grant (K12GM139185) to BAM. Figure 1 was created with BioRender.com, Agreement Nos. YL22WB7VLT and NV22WB7PTB. ECF is the 2021 recipient of the McCulloch & Till Award, bestowed by the International Society for Experimental Hematology.
REFERENCES
- 1.Ford CE, Hamerton JL, Barnes DW, Loutit JF. Cytological identification of radiation-chimaeras. Nature 1956;177:452–4. [DOI] [PubMed] [Google Scholar]
- 2.Till JE, McCulloch EA. A direct measurement of the radiation sensitivity of normal mouse bone marrow cells. Radiat Res 1961;178:Av3–7. [DOI] [PubMed] [Google Scholar]
- 3.Ayachi S, Buscarlet M, Busque L. 60 Years of clonal hematopoiesis research: from X-chromosome inactivation studies to the identification of driver mutations. Exp Hematol 2020;83:2–11. [DOI] [PubMed] [Google Scholar]
- 4.Fujino T, Kitamura T. ASXL1 mutation in clonal hematopoiesis. Exp Hematol 2020;83:74–84. [DOI] [PubMed] [Google Scholar]
- 5.Cook EK, Luo M, Rauh MJ. Clonal hematopoiesis and inflammation: partners in leukemogenesis and comorbidity. Exp Hematol 2020;83:85–94. [DOI] [PubMed] [Google Scholar]
- 6.Steensma DP, Ebert BL. Clonal hematopoiesis as a model for premalignant changes during aging. Exp Hematol 2020;83:48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park SJ, Bejar R. Clonal hematopoiesis in cancer. Exp Hematol 2020;83:105–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swierczek S, Prchal JT. Clonal hematopoiesis in hematological disorders: three different scenarios. Exp Hematol 2020;83:57–65. [DOI] [PubMed] [Google Scholar]
- 9.Pardali E, Dimmeler S, Zeiher AM, Rieger MA. Clonal hematopoiesis, aging, and cardiovascular diseases. Exp Hematol 2020;83:95–104. [DOI] [PubMed] [Google Scholar]
- 10.Wang K, Yan Z, Zhang S, Bartholdy B, Eaves CJ, Bouhassira EE. Clonal origin in normal adults of all blood lineages and circulating hematopoietic stem cells. Exp Hematol 2020;83:25–34.e22. [DOI] [PubMed] [Google Scholar]
- 11.Ganuza M, Hall T, Obeng EA, McKinney-Freeman S. Clones assemble! The clonal complexity of blood during ontogeny and disease. Exp Hematol 2020;83:35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King KY, Huang Y, Nakada D, Goodell MA. Environmental influences on clonal hematopoiesis. Exp Hematol 2020;83:66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee-Six H, Kent DG. Tracking hematopoietic stem cells and their progeny using whole-genome sequencing. Exp Hematol 2020;83:12–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Becker AJ, Mc CE, Till JE. Cytological demonstration of the clonal nature of spleen colonies derived from transplanted mouse marrow cells. Nature 1963;197:452–4. [DOI] [PubMed] [Google Scholar]
- 15.Siminovitch L, McCulloch EA, Till JE. The distribution of colony-forming cells among spleen colonies. J Cell Comp Physiol 1963;62:327–36. [DOI] [PubMed] [Google Scholar]
- 16.Wu AM, Till JE, Siminovitch L, McCulloch EA. A cytological study of the capacity for differentiation of normal hemopoietic colony-forming cells. J Cell Physiol 1967;69:177–84. [DOI] [PubMed] [Google Scholar]
- 17.Fowler JH, Wu AM, Till JE, McCulloch EA. Siminovitch L. The cellular composition of hemopoietic spleen colonies. J Cell Physiol 1967;69:65–71. [Google Scholar]
- 18.Herzenberg LA, Parks D, Sahaf B, Perez O, Roederer M, Herzenberg LA. The history and future of the fluorescence activated cell sorter and flow cytometry: a view from Stanford. Clin Chem 2002;48:1819–27. [PubMed] [Google Scholar]
- 19.Mullis K, Faloona F, Scharf S, Saiki R, Horn G, Erlich H. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb Symp Quant Biol 1986;51(Pt 1):263–73. [DOI] [PubMed] [Google Scholar]
- 20.Kohler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 1975;256:495–7. [DOI] [PubMed] [Google Scholar]
- 21.Beaudin AE, Boyer SW, Forsberg EC. Flk2/Flt3 promotes both myeloid and lymphoid development by expanding non-self-renewing multipotent hematopoietic progenitor cells. Exp Hematol 2014;42:218–229.e214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyer SW, Rajendiran S, Beaudin AE, et al. Clonal and quantitative in vivo assessment of hematopoietic stem cell differentiation reveals strong erythroid potential of multipotent cells. Stem Cell Reports 2019;12:801–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weber K, Thomaschewski M, Warlich M, et al. RGB marking facilitates multicolor clonal cell tracking. Nat Med 2011;17:504–9. [DOI] [PubMed] [Google Scholar]
- 24.Lu R, Neff NF, Quake SR. Weissman IL. Tracking single hematopoietic stem cells in vivo using high-throughput sequencing in conjunction with viral genetic barcoding. Nat Biotechnol 2011;29:928–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naik SH, Perié L, Swart E, et al. Diverse and heritable lineage imprinting of early haematopoietic progenitors. Nature 2013;496:229–32. [DOI] [PubMed] [Google Scholar]
- 26.Verovskaya E, Broekhuis MJ, Zwart E, et al. Heterogeneity of young and aged murine hematopoietic stem cells revealed by quantitative clonal analysis using cellular barcoding. Blood 2013;122:523–32. [DOI] [PubMed] [Google Scholar]
- 27.Grosselin J, Sii-Felice K, Payen E, Chretien S, Tronik-Le D Roux, Leboulch P. Arrayed lentiviral barcoding for quantification analysis of hematopoietic dynamics. Stem Cells 2013;31:2162–71. [DOI] [PubMed] [Google Scholar]
- 28.Schofield R, Lord BI, Kyffin S, Gilbert CW. Self-maintenance capacity of CFU-S. J Cell Physiol 1980;103:355–62. [DOI] [PubMed] [Google Scholar]
- 29.Magli MC, Iscove NN, Odartchenko N. Transient nature of early haematopoietic spleen colonies. Nature 1982;295:527–9. [DOI] [PubMed] [Google Scholar]
- 30.Spangrude GJ, Heimfeld S, Weissman IL. Purification and characterization of mouse hematopoietic stem cells. Science 1988;241:58–62. [DOI] [PubMed] [Google Scholar]
- 31.Na Nakorn T, Traver D, Weissman IL, Akashi K. Myeloerythroid-restricted progenitors are sufficient to confer radioprotection and provide the majority of day 8 CFU-S. J Clin Invest 2002;109:1579–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolf NS, Priestley GV. Kinetics of early and late spleen colony development. Exp Hematol 1986;14:676–82. [PubMed] [Google Scholar]
- 33.Forsberg EC, Serwold T, Kogan S, Weissman IL, Passegué E. New evidence supporting megakaryocyte–erythrocyte potential of flk2/flt3+ multipotent hematopoietic progenitors. Cell 2006;126:415–26. [DOI] [PubMed] [Google Scholar]
- 34.Yang L, Bryder D, Adolfsson J, et al. Identification of Lin(−)Sca1(+)kit(+)CD34(+)Flt3− short-term hematopoietic stem cells capable of rapidly reconstituting and rescuing myeloablated transplant recipients. Blood 2005;105:2717–23. [DOI] [PubMed] [Google Scholar]
- 35.Pietras EM, Reynaud D, Kang YA, et al. Functionally distinct subsets of lineage-biased multipotent progenitors control blood production in normal and regenerative conditions. Cell Stem Cell 2015;17:35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science 1996;273:242–5. [DOI] [PubMed] [Google Scholar]
- 37.Nakauchi H, Takano H, Ema H, Osawa M. Further characterization of CD34-low/negative mouse hematopoietic stem cells. Ann NY Acad Sci 1999;872:57–66.; discussion 66–70. [DOI] [PubMed] [Google Scholar]
- 38.Zhao Y, Lin Y, Zhan Y, et al. Murine hematopoietic stem cell characterization and its regulation in BM transplantation. Blood 2000;96:3016–22. [PubMed] [Google Scholar]
- 39.Harris RA, Hogarth PM, Wadeson LJ, Collins P, McKenzie IF, Penington DG. An antigenic difference between cells forming early and late haematopoietic spleen colonies (CFU-S). Nature 1984;307:638–41. [DOI] [PubMed] [Google Scholar]
- 40.Morrison SJ, Weissman IL. The long-term repopulating subset of hematopoietic stem cells is deterministic and isolatable by phenotype. Immunity 1994;1:661–73. [DOI] [PubMed] [Google Scholar]
- 41.Rajendiran S, Smith-Berdan S, Kunz L, et al. Ubiquitous overexpression of CXCL12 confers radiation protection and enhances mobilization of hematopoietic stem and progenitor cells. Stem Cells 2020;38:1159–74. [DOI] [PubMed] [Google Scholar]
- 42.Nakorn TN, Miyamoto T, Weissman IL. Characterization of mouse clonogenic megakaryocyte progenitors. Proc Natl Acad Sci USA 2003;100:205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell 1997;91:661–72. [DOI] [PubMed] [Google Scholar]
- 44.Kruse EA, Loughran SJ, Baldwin TM, et al. Dual requirement for the ETS transcription factors Fli-1 and Erg in hematopoietic stem cells and the megakaryocyte lineage. Proc Natl Acad Sci USA. 2009;106:13814–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Summers AR, Fischer MA, Stengel KR, et al. HDAC3 is essential for DNA replication in hematopoietic progenitor cells. J Clin Invest 2013;123:3112–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mehatre SH, Roy IM, Biswas A, et al. Niche-mediated integrin signaling supports steady-state hematopoiesis in the spleen. J Immunol 2021;206:1549–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perry JM, Li L. Functional assays for hematopoietic stem cell self-renewal. Methods Mol Biol 2010;636:45–54. [DOI] [PubMed] [Google Scholar]
- 48.Cool T, Worthington A, Poscablo D, Hussaini A, Forsberg EC. Interleukin 7 receptor is required for myeloid cell homeostasis and reconstitution by hematopoietic stem cells. Exp Hematol 2020;90:39–45.e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leung GA, Cool T, Valencia CH, Worthington A, Beaudin AE, Forsberg EC. The lymphoid-associated interleukin 7 receptor (IL7R) regulates tissue-resident macrophage development. Development 2019;146:dev176180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poscablo DM, Worthington AK, Smith-Berdan S, Forsberg EC. Megakaryocyte progenitor cell function is enhanced upon aging despite the functional decline of aged hematopoietic stem cells. Stem Cell Rep 2021;16:1598–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith-Berdan S, Bercasio A, Kramer L, Petkus B, Hinck L, Forsberg EC. Acute and endothelial-specific Robo4 deletion affect hematopoietic stem cell trafficking independent of VCAM1. PLoS One 2021;16:e0255606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Szilvassy SJ, Humphries RK, Lansdorp PM, Eaves AC, Eaves CJ. Quantitative assay for totipotent reconstituting hematopoietic stem cells by a competitive repopulation strategy. Proc Natl Acad Sci USA 1990;87:8736–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Domen J, Weissman IL. Self-renewal, differentiation or death: regulation and manipulation of hematopoietic stem cell fate. Mol Med Today 1999;5:201–8. [DOI] [PubMed] [Google Scholar]
- 54.Yamamoto R, Morita Y, Ooehara J, et al. Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell 2013;154:1112–26. [DOI] [PubMed] [Google Scholar]
- 55.Till JE, McCulloch EA, Siminovitch L. A stochastic model of stem cell proliferation, based on the growth of spleen colony-forming cells. Proc Natl Acad Sci USA 1964;51:29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Worton RG, McCulloch EA, Till JE. Physical separation of hemopoietic stem cells differing in their capacity for self-renewal. J Exp Med 1969;130:91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iscove NN, Till JE, McCulloch EA. The proliferative states of mouse granulopoietic progenitor cells. Proc Soc Exp Biol Med 1970;134:33–6. [DOI] [PubMed] [Google Scholar]