Abstract
Natural language processing (NLP) is a set of automated methods to organize and evaluate the information contained in unstructured clinical notes, which are a rich source of real-world data from clinical care that may be used to improve outcomes and understanding of disease in cardiology. The purpose of this systematic review is to provide an understanding of NLP, review how it has been used to date within cardiology, and illustrate the opportunities that this approach provides for both research and clinical care. We systematically searched six scholarly databases (ACM Digital Library, Arxiv, Embase, IEEE Explore, PubMed, and Scopus) for studies published in 2015–2020 describing the development or application of NLP methods for clinical text focused on cardiac disease. Studies not published in English, lacking a description of NLP methods, non-cardiac focused, and duplicates were excluded. Two independent reviewers extracted general study information, clinical details, and NLP details, and appraised quality using a checklist of quality indicators for NLP studies. We identified 37 studies developing and applying NLP in heart failure, imaging, coronary artery disease, electrophysiology general cardiology and valvular heart disease. Most studies used NLP to identify patients with a specific diagnosis and extract disease severity using rule-based NLP methods. Some used NLP algorithms to predict clinical outcomes. A major limitation is the inability to aggregate findings across studies due to vastly different NLP methods, evaluation, and reporting. This review reveals numerous opportunities for future NLP work in cardiology with more diverse patient samples, cardiac diseases, datasets, methods, and applications.
Keywords: Cardiology, Natural Language Processing, Electronic Health Records
Introduction
A vast amount of data is collected during routine clinical care. Clinicians cognitively process this data, organizing it into contextual information which is then documented in clinical notes. Data has inherent structure, while the information contained in clinical notes is unstructured text. Structured data are managed as computable data elements (e.g., diagnosis codes, blood pressure reading, laboratory values), while unstructured text (clinical notes) lacks organization and standardized formatting, making it challenging to analyze at scale in its raw form. The ability to organize and evaluate the information contained in clinical notes at scale provides a rich source of real-world data from clinical care.1 Unfortunately, by some estimates, more than 80% of the information in electronic health records (EHRs) is in unstructured formats.2
Natural language processing (NLP) is a set of automated methods for interpreting different aspects of natural language, including syntax (the arrangement) and semantics (the meaning) of words and phrases (Figure 1). A spectrum of NLP approaches exists, ranging from identification of text strings to deep learning. Many NLP models can interpret the complex natural language contained in clinical text, including medical jargon, misspellings, and abbreviations, into accurate representations of clinical information.
Figure 1.
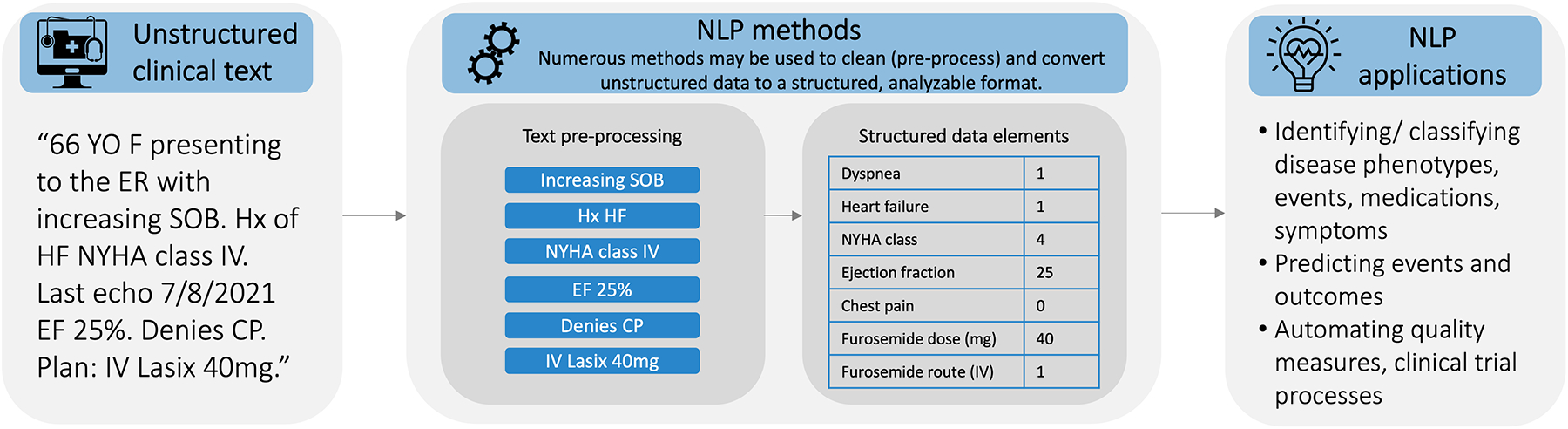
Basic steps of natural language processing (NLP) and NLP applications in cardiology
There is potential for researchers and clinicians to use NLP to extract information from unstructured clinical notes which may then be used in studies to improve outcomes and understanding of disease.3 The involvement of researchers and healthcare professionals in cardiology in the development and application of novel NLP methods is needed to ensure these methods are accurate and representative, and that envisioned use cases are relevant and feasible in cardiac contexts. Researchers are increasingly applying machine learning methods in cardiology,4 but NLP methods and its applications in clinical care have not been thoroughly described in systematic reviews.
The purpose of this systematic review is to provide investigators and clinicians in the field of cardiology with an understanding of NLP, to review how it has been used to date within cardiology, and to illustrate the opportunities that this approach provides for both research and clinical care. We synthesize and discuss current trends in clinical applications, applicability of test datasets, NLP methods, and primary findings of recent NLP research in cardiology, with the goal of increasing awareness of how these methods can be used to extract information from clinical text and encouraging future innovations and applications among researchers and healthcare professionals in cardiology.
Methods
This review follows the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines for reporting in systematic reviews.
Article retrieval
We searched the metadata of articles in six scholarly databases in medicine, science, and computer science: ACM Digital Library, Arxiv, Embase, IEEE Explore, PubMed, and Scopus. A combination of search terms relating to NLP and cardiology were selected based on the Medical Subject Headings vocabulary (U.S. National Library of Medicine) with additional terms identified from prior NLP-focused systematic reviews5, 6 through collaboration with a medical librarian (DW). We applied filters to only search publications in the English language and from 1/1/2015 through 12/31/2020 to ensure relevance given the rapid advances in NLP in recent years. The reference lists of included studies were reviewed to identify additional relevant studies for potential inclusion. Details of the search strategy are provided in Supplemental File 1.
Study selection
We used Covidence software (www.covidence.org) to organize and structure our review of retrieved studies. Studies considered eligible for inclusion were published in calendar years 2015–2020, described the development or application of NLP methods for clinical text (EHRs), and were clinically focused on a patient population with existing cardiac disease. Studies were excluded for the following reasons: (1) not published or available in English, (2) duplicate studies, (3) aspects of the same study published by the same research group in multiple publications, (4) lacking a description of NLP methods or applications, and (5) focus on patients without existing cardiac disease, such as patients with cardiac risk factors but no diagnosed disease; while these do represent additional areas of interest in NLP, they were felt to be beyond the scope of this work. Following this strategy, three reviewers (MRT, AV, DS) performed two rounds of study selection: title and abstract screening followed by full-text review. Each article was screened by two independent reviewers and disagreements were discussed among the three reviewers until consensus was achieved.
Data extraction and synthesis
Data from each included article was independently extracted by two of three reviewers (MRT, AV, DS). Extracted data included general study information (design, objectives), clinical details (cardiac focus, patient characteristics), and NLP details (NLP methods, evaluation metrics). To reduce complexity, evaluation metrics were reported as ranges when performance metrics for multiple cohorts or methods were reported separately. All reviewers worked from the same understanding of common NLP terms and methods, described in Table 1. Data for each article was extracted by two independent reviewers and discrepancies were resolved through discussion.
Table 1.
Glossary of NLP methods and metrics
| NLP methods | |
| Key term search | Identify and extract terms from pre-specified list of terms of interest. |
| Named entity recognition | Locate and translate terms, or named entities, into predefined categories of concepts, often using controlled medical vocabularies. |
| Rule-based methods | Detect concepts of interest based on an established set of rules or logic, often using regular expressions, which are sequences of characters that define a search pattern. |
| Convolutional neural network | A deep learning neural network approach identifying, weighting, and connecting “nodes” across multiple convolutional “layers” of nodes (including a convolutional layer) and applying filters between layers. |
| Conditional random fields | A classification approach that accounts for context in order to recognize patterns and make predictions. |
| Decision tree | Hierarchical trees of knowledge used to classify concepts of interest. |
| Logistic regression | A basic building block for neural networks; a classification approach used to discover links between concepts of interest. |
| Random forest | An “ensemble” of decision trees built using a combination of learning models and used to produce more accurate and stable predictions. |
| Recurrent neural network | A deep learning neural network approach designed to interpret temporal or sequential information and used to make predictions. |
| Evaluation Methods | |
| Manual annotation | The task of reading pre-selected texts and marking (i.e., annotating) linguistic components (paragraphs, sentences, phrases, or words) that represents concepts of interest. |
| Cross-validation; also called held out testing set | A technique to evaluate predictive models by partitioning the original sample into a training set to train the model, and a test set to evaluate it. |
| Performance metrics | |
| Positive predictive value (PPV); also called precision | The percentage of results that were actually relevant among all results that the system obtained. |
| Negative predictive value (NPV) | The percentage of results that were actually irrelevant among all results that the system did not obtain. |
| Sensitivity; also called recall | The percentage of results that were actually obtained by the system among all results that should have been obtained. |
| Specificity | The percentage of results that were actually not obtained by the system among all results that should not have been obtained. |
| F-score | A combination of PPV/precision and sensitivity/recall; can be weighted to give more significance to one measure. |
| Accuracy | The percentage of results that were actually relevant among all results that were and were not obtained. |
| Area under the curve (AUC) | Reflects the degree to which a model is capable of classifying or distinguishing between classes or events of interest. |
Quality appraisal
While relevant reporting standards for NLP research have not been established,7 we conducted a modified quality appraisal based on the approach described by Koleck and colleagues,6 who documented the presence of specific quality indicators in NLP articles. We included additional machine learning quality indicators described by Nascimento and colleagues.8 Each article was appraised by two of three reviewers (MRT, AV, DS) and disagreements were resolved through discussion.
Results
Article screening and included studies
After applying eligibility criteria, 37 articles were included in the review (Figure 2). We retrieved 653 studies from scholarly databases. Covidence automatically identified and excluded 261 studies as duplicates. During the title and abstract screening, the majority of studies were excluded for not having a cardiology focus (n=327) and not using NLP or providing a description of the NLP methods (n=200). During the full text screening, studies were mainly excluded for not having a cardiology focus (n=64) or not providing details about the NLP methods (n=51). The detailed exclusion cascade is provided in Supplemental File 1 and a complete list of screened articles and exclusion reason are provided in Supplemental File 2.
Figure 2.
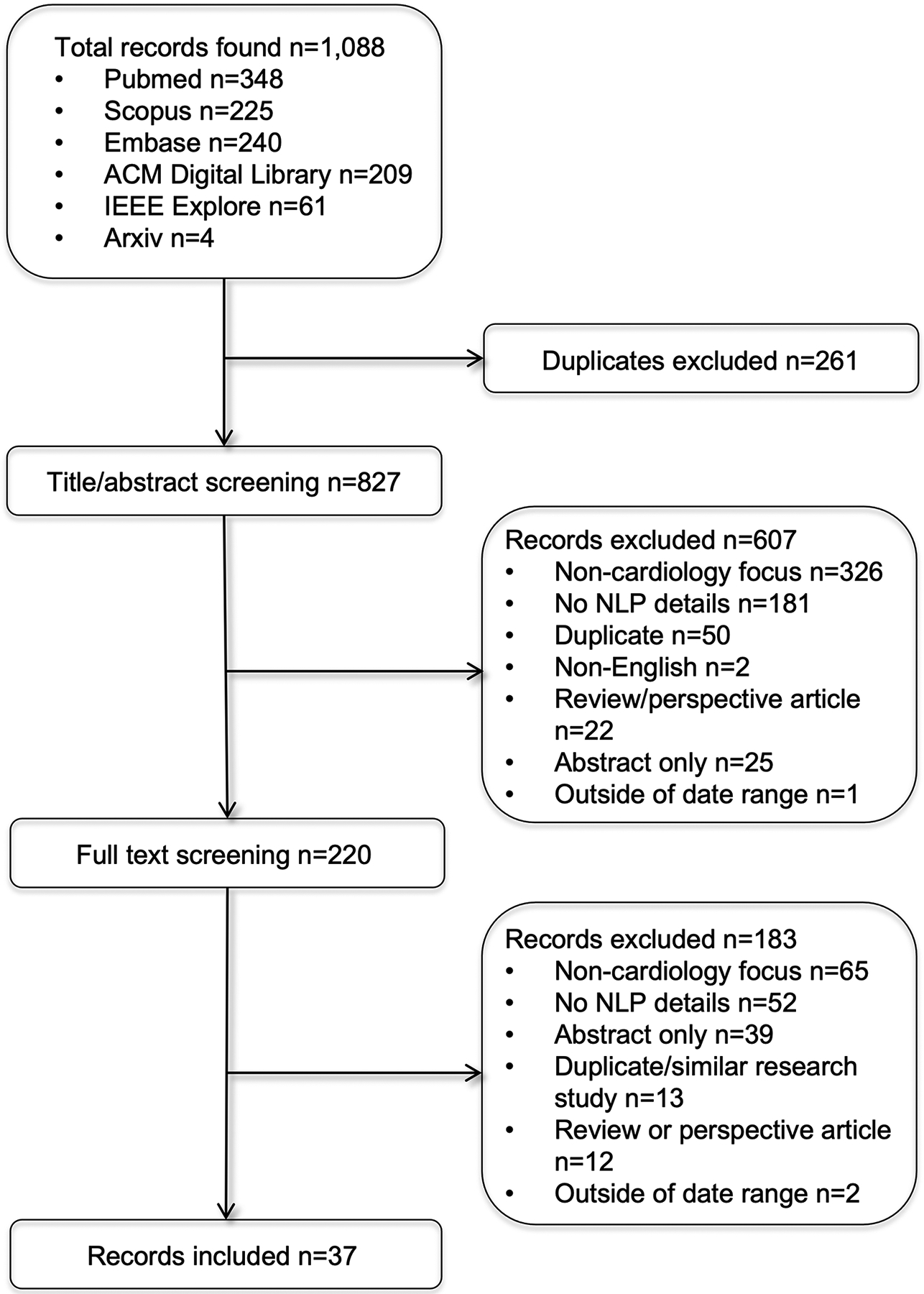
PRISMA flow diagram
Description of the included studies
Of the 37 included studies, 15 were published in biomedical informatics or engineering journals, 12 in cardiology journals, six in general biomedical research journals, and the remaining four in other disciplines including nursing, public health, and radiology. Table 2 reports on the patient populations, datasets, and NLP methods of included studies. The samples of cardiac patients included a mix of hospitalized and non-hospitalized patients with sample sizes ranging from 60 to over 621,000 patients. Among studies reporting demographic characteristics of patient samples (n=15), the mean age ranged from 56 to 90 years old, 45–99% were male, and 48–94% were Caucasian. Data sources included a single hospital (n=16), regional or national healthcare systems (n=10), and an existing corpus of notes or patient registry (n=11). The majority of studies (n=28) were conducted in the US. The number of documents analyzed ranged widely from 310 to over 2.1 million notes, and consisted primarily of inpatient progress notes, outpatient notes, and echocardiogram reports. Fifteen studies used rule-based methods (n=15), named entity recognition (n=13), key term search (n=11), and other methods (n=9) including convolutional neural networks, conditional random fields, decision trees, logistic regression, random forests, and recurrent neural networks. Several studies used previously developed tools, primarily Leo and MedTagger.
Table 2.
Quality appraisal results by cardiac disease focus
| Clearly defined purpose | Number of patients specified | Patient demographic information reported | Number of documents specified | Detailed description of NLP approach | Parameterization conducted | Inclusion of comparative evaluation | Detailed description of comparative evaluation design | Justification for evaluation design and metrics | Evaluation metrics reported | Statistical treatment of results (e.g., confidence tests) | Discussion of model costs (time, resources) and explainability | Availability of code and datasets for reproducibility | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Coronary Artery Disease | |||||||||||||
| Esteban et al, 20179 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Hu et al, 201613 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| Owlia et al, 201910 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||||
| Safarova et al, 201611 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||
| Shah et al, 201912 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Toerper et al, 201614 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||||||
| Electrophysiology | |||||||||||||
| Bean et al, 201916 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||
| Hu et al, 201921 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||||
| Moon et al, 201920 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Moon et al, 202019 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| Rosier et al, 201618 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||
| Shah et al, 2020a17 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||
| Shah et al, 2020b15 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| General cardiology | |||||||||||||
| Viani et al, 201922 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||
| Heart Failure | |||||||||||||
| Alnazzawi et al, 201623 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||||
| Bielinski et al, 201524 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Eggerth et al, 202036 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| Evans et al, 201630 | ✓ | ✓ | ✓ | ✓ | |||||||||
| Garvin et al, 201833 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| Jonnalagadda et al, 201734 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Kaspar et al, 201825 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||
| Leiter et al, 202037 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| Liu et al, 201931 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||
| Mahajan et al, 201932 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||||||
| Patel et al, 201826 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||||||
| Topaz et al, 201735 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| Wagholikar et al, 201827 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| Wang et al, 201528 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||
| Zhang et al, 201829 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||
| Imaging | |||||||||||||
| Adekkanattu et al, 201938 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||||||
| Nath et al, 201639 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| Patterson et al, 201742 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||
| Shi et al, 201540 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||
| Valtchinov et al, 202043 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||
| Xie et al, 201741 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| Zheng et al, 202044 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| Valvular Disease | |||||||||||||
| Galper et al, 201845 | ✓ | ✓ | ✓ | ✓ | ✓ | ||||||||
Study purposes and primary findings
By subspeciality, six studies focused on coronary artery disease (CAD), seven on electrophysiology (EP), 15 on heart failure (HF), seven on imaging, one on valvular heart disease, and one on general cardiology. Figure 3 presents a summary of the primary areas of application of the NLP methods. Supplemental File 1 reports on the purposes and study outcomes for each included study; below we describe these by subspecialty.
Figure 3.
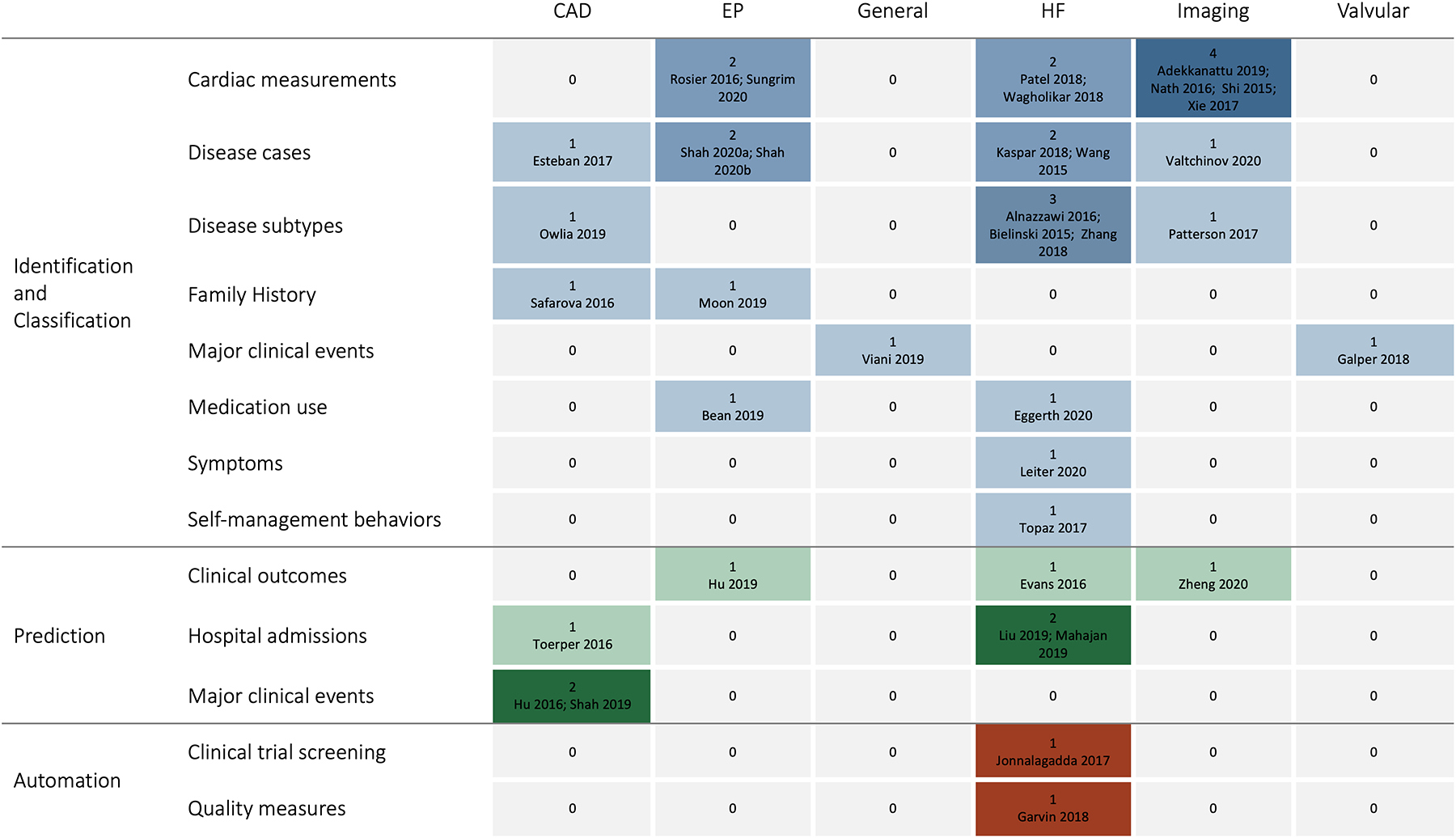
Summary of the number of included articles by area of NLP application and cardiac disease
Coronary Artery Disease
Within CAD, most studies focused on identification and classification of disease9–12 while two focused on prediction of major adverse cardiovascular events13 and inpatient admissions following cardiac catheterization.14 The studies demonstrated the ability to use NLP to identify CAD events and symptoms,9 Canadian Cardiovascular Society angina classification,10 symptoms and test results related to myocardial infarction,12 and patients with familial hypercholesterolemia11 with sensitivity, specificity, and positive and negative predictive values over 80% for most studies. The algorithm predicting major adverse cardiovascular events outperformed two widely used acute coronary syndrome risk score tools with an AUC of 72%.13 The algorithm predicting admissions following cardiac catheterization also reported an AUC of 72%, and identified age, gender, and past medical and surgical history-related factors associated with increased risk of admission.14
Electrophysiology
Study purposes within EP included identifying patients with atrial fibrillation15 and characterizing those most likely to receive guideline-directed thromboembolic prophylaxis at the time of hospital discharge,16, 17 an algorithm to evaluate the significance of atrial fibrillation alerts received from remote monitoring of cardiac implantable electronic devices based upon calculated CHA2DS2-VASc stroke risk score data obtained from the EHR,18, 19 extracting family history information,20 and predicting cardiac resynchronization therapy outcomes.21 One study was able to identify patient with atrial fibrillation with F-scores of 93–94%.15 Studies aiming to characterize anticoagulant use reported sensitivities of 90–97% and found that risk scores were more accurate for the CHA2DS2-VASc than HAS-BLED,16 and models including unstructured and structured data together were more accurate than unstructured data alone.17 Studies aiming to automatically detect heart rhythm reported an accuracy of 98%18 and F-scores of 92–98%;19 they also reported that unstructured data identified rhythm and therapy delivered from ICDs more accurately than structured data alone.19 One study aiming to extract family history information found a machine learning model that incorporated unstructured data had sensitivities of 91–95% and specificities of 90–98%.20 Another study predicted cardiac resynchronization therapy outcomes with a PPV of 79%, sensitivity of 26%, and AUC of 75%.21
General cardiology
One general study focused on extracting events from cardiology-focused notes in the Italian language, seeking to adapt existing NLP methods which have been primarily developed for the English language.22 The authors reported that models integrating recurrent neural networks with standard dictionary-lookup approaches performed better than recurrent neural networks alone.
Heart Failure
Within HF, most studies focused on either identification and classification of disease, including subtypes based on left ventricular ejection fraction and New York Heart Association (NYHA) heart failure class,23–29 and prediction of hospital readmissions and mortality.30–32 Other studies aimed to automate a HF quality measure,33 automate screening for a clinical trial,34 identify patients with ineffective self-management,35 evaluate medication adherence,36 and identify documented symptoms among patients undergoing cardiac resynchronization therapy.37 The studies aiming to identify and classify HF had sensitivities of 60–100%, specificities of 96–99%, PPV’s of 71–96%, NPV’s of 87–100%, F-scores of 74–94%, and accuracies of 77–100%.23–29 The studies aiming to predict hospital readmissions and mortality found NLP improved model performance over structured data alone30,31 and that deep learning models outperformed other machine learning models,31 but performance statistics varied widely (PPV 98%,30 F-score 73–76%,31 AUC 51–65%32). Other studies successfully used NLP methods to characterize HF patients for quality care metrics (PPV 89–99%, sensitivity 27–100%)33 and for clinical trials eligibility (PPV 86%, sensitivity 95%).34 The study aiming to use NLP to identify patients with ineffective self-management reported a PPV of 95% and sensitivity of 79%, and identified specific types of self-management deficits that were significantly associated with readmissions.35 Studies also demonstrated moderate success applying NLP to evaluate medication adherence (F-score 55–90%)36 and identify HF symptoms (F-score: 72%).37
Imaging
Most studies within imaging focused on extracting data elements from echocardiogram reports.38–42 One focused on identifying patients with implantable devices prior to MRIs,43 and one on interpreting exercise treadmill test results.44 The studies aiming to extract data elements from echocardiogram reports reported widely variable PPV’s (6–100%) and sensitivities (25–100%); some studies reported NLP was reliable across concepts of interest while others reported wide variability in performance metrics between concepts.38–42 One study found that expert-derived and ontology-derived NLP methods had similar accuracy (expert-derived: 83%, ontology-derived 91%) in identifying patients with implantable devices prior to MRIs.43 One study used NLP to extract relevant information from exercise treadmill test results with a sensitivity of 96% and specificity of 95%, and was able to associate test results with risk of severe 30-day outcomes (myocardial infarction, death).44
Valvular disease
The one article that was focused on valvular disease aimed to extract and analyze adverse events from transcatheter aortic valve replacement (TAVR) and MitraClip procedures.45 The study found that NLP found common events associated with TAVR and MitraClip procedures with high correlation to structured data alone (R2 0.86).45
Indicators of quality
Table 2 reports on indicators of quality6, 8 across the included studies. The following indicators were met by the fewest studies: discussion of model costs and other implementation considerations (n=9; 24%), availability of code and datasets for reproducibility (n=13; 35%), and patient demographic information reported (n=18; 49%).
Discussion
In this systematic review, we found the majority of NLP work in cardiology has been concentrated in a small number of clinical domains (primarily HF) and NLP methods (primarily rule-based). The most common applications were extracting information for disease identification and classification purposes; these studies reported fairly high accuracy, indicating that NLP algorithms are well developed towards this goal in cardiovascular disease.9–12, 16–20, 23–29, 38–45 Fewer studies used machine learning to predict outcomes and reported moderate predictive abilities (AUC 51–75%).13, 14, 30,21, 31 Below we describe how cardiology researchers and clinicians interested in engaging in NLP can leverage multiple opportunities to further explore patient and disease conditions, datasets, and novel NLP applications. These future research directions are summarized in Table 3.
Table 3.
Future research directions of NLP in cardiology
| 1. | Applying NLP to study a broader range of cardiovascular diseases (beyond heart failure) and more diverse, representative patient samples to reduce bias in trained models. |
| 2. | Developing and applying sophisticated NLP approaches, using machine learning, to accomplish complex tasks such as generating disease timelines, monitoring drug safety, and untangling symptom-physiology relationships. |
| 3. | Leveraging open-source, previously developed NLP tools to study portability and reliability of tools across health systems and use cases. |
| 4. | Exploring the value of other types of unstructured health data for cardiology beyond inpatient physician notes, such as nursing progress notes, primary care notes, patient-generated health data and social media content. |
| 5. | Conducting rigorous evaluations identifying strategies to improve explainability and address other challenges surrounding implementation of NLP algorithms in clinical practice (i.e., costs, clinical workflows, time burden). |
Our review showed that NLP research in cardiology has been concentrated on a few disease areas, potentially because author lists suggest very few research groups are working at the intersection of NLP and cardiology. In future work NLP may be used to address a broader range of cardiovascular diseases, especially those that are growing in prevalence such as atrial fibrillation, and study more diverse patient samples. In this review, patient samples were predominantly middle- to older-age, male, and Caucasian. This may be explained by the underlying patient populations seeking care. Nonetheless, increasing attention has been paid to the bias in machine learning; training models with unrepresentative patient samples causes them to work less effectively for, and potentially harm, underrepresented patient groups.46, 47 Future studies should consider the importance of diverse, representative samples when training, validating, and implementing NLP methods to ensure they work well for the diverse range of patients receiving cardiac care in the US and globally.
Another relevant issue is explainability of predictive models. Our review demonstrated that very few articles (24%) addressed implementation considerations such as explainability. Clinicians frequently mistrust machine learning-based predictive models because of challenges understanding how machine learning-based models generate outputs, which has slowed adoption of these models in clinical practice.48 Recognizing this problem, novel interpretability methods, such as SHAP (SHapley Additive exPlanations) values,49 have improved insight into predictions, but much work to improve explainability and usefulness of these models in clinical practice remains.
More than half of the included studies focused on formative methods development versus evaluation of previously developed tools, and only two studies described NLP methods being adopted in routine clinical care.28, 30 Most of the previously developed NLP tools employed by the included studies are now publicly available (Table 4). However, portability and reliability of previously developed tools are major concerns, as performance often differs between institutions and EHR systems with different document structures and linguistic expressions. Lack of portability remains a significant challenge in NLP research, and may explain why the majority of studies in this review developed novel NLP methods rather than reuse existing algorithms and tools. There are also opportunities to explore novel unstructured data sources, such as outpatient and primary care notes,12 nursing documentation, which has been used to predict mortality,50 and even non-clinical unstructured data sources, such as social media posts and patient-generated health data.
Table 4.
Portfolio of open-source NLP tools and datasets applied in cardiology contexts
| Name and Origin | Description | Accessibility |
|---|---|---|
| NLP tools | ||
| clinical Text Analysis and Knowledge Extraction System (cTakes); Mayo Clinic | A modular pipeline of components using both rule-based and machine learning methods to support information extraction; based on UIMA (Unstructured Information Management Architecture) standards. | Open-source at http://www.ohnlp.org |
| EchoExtractor; Veterans Affairs | An application which extracts Concept-Value pairs for metrics measured during an echocardiogram study. | Open-source at https://github.com/department-of-veterans-affairs/EchoExtractor |
| Leo; Veterans Affairs Informatics and Computing Infrastructure (VINCI) | A set of services and libraries that leverages UIMA standards to enable rapid creation and deployment of NLP analysis tools and incorporation of previously developed tools. | Open-source at https://department-of-veterans-affairs.github.io/Leo/userguide.html |
| MedTagger; Mayo Clinic | A set of tools developed for indexing based on dictionaries, information extraction based on patterns, and machine learning-based named entity recognition to support information extraction; based on UIMA standards. | Open-source at https://github.com/OHNLP/MedTagger |
| pyConText; University of Utah | A Python implementation of ConText, a simple text processing algorithm for identifying a large number of features and relationships between features. | Open-source at https://pypi.org/project/pyConTextNLP/0.6.0.5/ |
| semEHR; King’s College London, UK | A general-purpose search and analytics tool that processes heterogeneous data sources, covers a range of biomedical concepts, and captures context to support information extraction in study-specific or case-specific contexts. | Open-source at https://github.com/CogStack/CogStack-SemEHR |
| Datasets | ||
| The Medical Information Mart for Intensive Care III (MIMIC III), Massachusetts Institute of Technology | Deidentified, freely available, critical care database of over 60,000 intensive care unit admissions. | https://mimic.mit.edu/ |
| Electronic Medical Records and Genomics (eMERGE) network, National Human Genome Research Institute (NHGRI) | Combines DNA biorepositories with EHR data from several clinical sites nationally, and has been extensively used to develop phenotyping algorithms. | https://emerge-network.org/ |
| Integrating Biology and the Bedside (i2b2), Partners Healthcare | A dataset of deidentified patient discharge summaries made available for research purposes. | https://www.i2b2.org/ |
Finally, there are novel areas of NLP application that this review suggests are underexplored in cardiology. The few studies developing predictive algorithms demonstrated that unstructured data greatly improves algorithm performance,17, 19, 25, 26, 30, 32, 45 suggesting opportunities for greater use of NLP for prediction tasks in future work. Deep learning models incorporating NLP methods have been applied extensively in oncology to identify temporal events to generate clinical timelines, extract highly detailed tumor information, match patients to clinical trials, and conduct drug-safety surveillance.51 NLP has also been used in clinical decision support systems in other clinical contexts but was largely unexplored in the included articles. Finally, NLP may be applied to untangle symptom-physiology relationships, study symptom assessment and management practices, and support interventions to improve patient quality of life.6 In this review, few of the studies focused on symptoms, potentially because of the high degree of symptom overlap between conditions, which obfuscates a symptom’s etiology, and the lack of normalized symptom concepts in controlled vocabularies.6
Limitations of this review include the inability to aggregate findings across studies due to vastly different methods, evaluation, and reporting around NLP. Similarly, studies reported evaluation metrics for NLP with varying degrees of detail. We reported ranges of metrics for brevity, which removed detail necessary to understand nuanced differences between specific methods and concepts being extracted. Additionally, to maintain a focused scope of this review, we excluded some studies in interesting, related areas, such as prediction of cardiovascular disease among patients without existing disease, and cerebrovascular disease.
Finally, for conciseness, we identified several studies from the same author groups reporting on the same or highly similar research studies. In these cases, we included only the most recent study under the assumption that they represented the most evolved NLP methods from that project or body of work; however, this may have biased findings towards improved performance.
Conclusion
NLP is an underutilized method for unlocking information from unstructured notes in EHRs. This systematic review of the state of the science of NLP in cardiology identified several areas of success with NLP in cardiology, specifically the identification and classification of disease phenotypes and the augmentation of predictive outcome models through the inclusion of unstructured data. It also points to opportunities for future research and clinical opportunities, including novel patient and disease conditions, datasets, and applications.
Supplementary Material
Funding
This work was supported by a National Institute of Nursing Research career development award (K99NR019124; PI: Reading Turchioe).
Footnotes
Conflicts
MRT and JP are affiliated with Iris OB Health Inc., New York, a startup company focused on postpartum depression, and have equity ownership. MRT is a consultant for Boston Scientific Corporation.
Registration and study protocol/materials
This review was not registered. A protocol and other study materials will be provided upon reasonable request to the authors.
References
- 1.Sherman RE, Anderson SA, Dal Pan GJ, et al. Real-World Evidence — What Is It and What Can It Tell Us? New England Journal of Medicine. 2016;375(23):2293–2297. doi: 10.1056/NEJMsb1609216 [DOI] [PubMed] [Google Scholar]
- 2.Kong H-J. Managing Unstructured Big Data in Healthcare System. Healthc Inform Res. 2019;25(1):1–2. doi: 10.4258/hir.2019.25.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kilic A Artificial Intelligence and Machine Learning in Cardiovascular Healthcare. The Annals of thoracic surgery. May 2019;109(5):1323–1329. doi: 10.1016/j.athoracsur.2019.09.042 [DOI] [PubMed] [Google Scholar]
- 4.Thompson MP, Fanaroff AC, Parker JD, Vallabhajosyula S, Sterling MR. Focusing on the Future of Cardiovascular Outcomes Research: Highlights From the American Heart Association/American Stroke Association Quality of Care and Outcomes Research 2018 Scientific Sessions. Circ Cardiovasc Qual Outcomes. Jun 2018;11(6):e004871. doi: 10.1161/circoutcomes.118.004871 [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Wang L, Rastegar-Mojarad M, et al. Clinical information extraction applications: A literature review. Journal of Biomedical Informatics. 2018/January/01/ 2018;77:34–49. doi: 10.1016/j.jbi.2017.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koleck TA, Dreisbach C, Bourne PE, Bakken S. Natural language processing of symptoms documented in free-text narratives of electronic health records: a systematic review. Journal of the American Medical Informatics Association : JAMIA. Apr 1 2019;26(4):364–379. doi: 10.1093/jamia/ocy173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cunha W, Mangaravite V, Gomes C, et al. On the cost-effectiveness of neural and non-neural approaches and representations for text classification: A comprehensive comparative study. Information Processing & Management. 2021/May/01/ 2021;58(3):102481. doi: 10.1016/j.ipm.2020.102481 [DOI] [Google Scholar]
- 8.Borges do Nascimento IJ, Marcolino MS, Abdulazeem HM, et al. Impact of Big Data Analytics on People’s Health: Overview of Systematic Reviews and Recommendations for Future Studies. J Med Internet Res. Apr 13 2021;23(4):e27275. doi: 10.2196/27275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esteban S, Rodríguez Tablado M, Ricci RI, Terrasa S, Kopitowski K. A rule-based electronic phenotyping algorithm for detecting clinically relevant cardiovascular disease cases. BMC research notes. Jul 14 2017;10(1):281. doi: 10.1186/s13104-017-2600-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Owlia M, Dodson JA, King JB, et al. Angina Severity, Mortality, and Healthcare Utilization Among Veterans With Stable Angina. Journal of the American Heart Association. Aug 6 2019;8(15):e012811. doi: 10.1161/JAHA.119.012811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Safarova MS, Liu H, Kullo IJ. Rapid identification of familial hypercholesterolemia from electronic health records: The SEARCH study. Journal of clinical lipidology. Sep-Oct 2016;10(5):1230–9. doi: 10.1016/j.jacl.2016.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shah AD, Bailey E, Williams T, Denaxas S, Dobson R, Hemingway H. Natural language processing for disease phenotyping in UK primary care records for research: a pilot study in myocardial infarction and death. Journal of biomedical semantics. Nov 12 2019;10(Suppl 1):20. doi: 10.1186/s13326-019-0214-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu D, Huang Z, Chan TM, Dong W, Lu X, Duan H. Utilizing Chinese admission records for MACE prediction of acute coronary syndrome. International Journal of Environmental Research and Public Health. Sep 13 2016;13(9):912. doi: 10.3390/ijerph13090912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toerper MF, Flanagan E, Siddiqui S, Appelbaum J, Kasper EK, Levin S. Cardiac catheterization laboratory inpatient forecast tool: a prospective evaluation. Journal of the American Medical Informatics Association : JAMIA. Apr 2016;23(e1):e49–57. doi: 10.1093/jamia/ocv124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shah RU, Mutharasan RK, Ahmad FS, et al. Development of a Portable Tool to Identify Patients With Atrial Fibrillation Using Clinical Notes From the Electronic Medical Record. Circ Cardiovasc Qual Outcomes. Oct 2020;13(10):e006516. doi: 10.1161/circoutcomes.120.006516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bean DM, Teo J, Wu H, et al. Semantic computational analysis of anticoagulation use in atrial fibrillation from real world data. PloS one. 2019;14(11):e0225625. doi: 10.1371/journal.pone.0225625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shah RU, Mukherjee R, Zhang Y, et al. Impact of Different Electronic Cohort Definitions to Identify Patients With Atrial Fibrillation From the Electronic Medical Record. Journal of the American Heart Association. Mar 3 2020;9(5):e014527. doi: 10.1161/JAHA.119.014527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosier A, Mabo P, Temal L, et al. Personalized and automated remote monitoring of atrial fibrillation. Europace : European pacing, arrhythmias, and cardiac electrophysiology : journal of the working groups on cardiac pacing, arrhythmias, and cardiac cellular electrophysiology of the European Society of Cardiology. Mar 2016;18(3):347–52. doi: 10.1093/europace/euv234 [DOI] [PubMed] [Google Scholar]
- 19.Sungrim M, Andrew W, Christopher GS, et al. Real-World Data Analysis of Implantable Cardioverter Defibrillator (ICD) in Patients with Hypertrophic Cardiomyopathy (HCM). arXiv pre-print. 2020; [Google Scholar]
- 20.Moon S, Liu S, Scott CG, et al. Automated extraction of sudden cardiac death risk factors in hypertrophic cardiomyopathy patients by natural language processing. International journal of medical informatics. Aug 2019;128:32–38. doi: 10.1016/j.ijmedinf.2019.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu SY, Santus E, Forsyth AW, et al. Can machine learning improve patient selection for cardiac resynchronization therapy? PloS one. 2019;14(10):e0222397. doi: 10.1371/journal.pone.0222397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Viani N, Miller TA, Napolitano C, et al. Supervised methods to extract clinical events from cardiology reports in Italian. J Biomed Inform. Jul 2019;95:103219. doi: 10.1016/j.jbi.2019.103219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alnazzawi N, Thompson P, Ananiadou S. Mapping Phenotypic Information in Heterogeneous Textual Sources to a Domain-Specific Terminological Resource. PLoS One. 2016;11(9):e0162287. doi: 10.1371/journal.pone.0162287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bielinski SJ, Pathak J, Carrell DS, et al. A Robust e-Epidemiology Tool in Phenotyping Heart Failure with Differentiation for Preserved and Reduced Ejection Fraction: the Electronic Medical Records and Genomics (eMERGE) Network. Journal of cardiovascular translational research. Nov 2015;8(8):475–83. doi: 10.1007/s12265-015-9644-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaspar M, Fette G, Güder G, et al. Underestimated prevalence of heart failure in hospital inpatients: a comparison of ICD codes and discharge letter information. Clinical research in cardiology : official journal of the German Cardiac Society. Sep 2018;107(9):778–787. doi: 10.1007/s00392-018-1245-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel YR, Robbins JM, Kurgansky KE, et al. Development and validation of a heart failure with preserved ejection fraction cohort using electronic medical records. BMC cardiovascular disorders. Jun 28 2018;18(1):128. doi: 10.1186/s12872-018-0866-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagholikar KB, Fischer CM, Goodson A, et al. Extraction of Ejection Fraction from Echocardiography Notes for Constructing a Cohort of Patients having Heart Failure with reduced Ejection Fraction (HFrEF). Journal of medical systems. Sep 25 2018;42(11):209. doi: 10.1007/s10916-018-1066-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Luo J, Hao S, et al. NLP based congestive heart failure case finding: A prospective analysis on statewide electronic medical records. International journal of medical informatics. Dec 2015;84(12):1039–47. doi: 10.1016/j.ijmedinf.2015.06.007 [DOI] [PubMed] [Google Scholar]
- 29.Zhang R, Ma S, Shanahan L, Munroe J, Horn S, Speedie S. Discovering and identifying New York heart association classification from electronic health records. BMC medical informatics and decision making. Jul 23 2018;18(Suppl 2):48. doi: 10.1186/s12911-018-0625-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans RS, Benuzillo J, Horne BD, et al. Automated identification and predictive tools to help identify high-risk heart failure patients: pilot evaluation. Journal of the American Medical Informatics Association : JAMIA. Sep 2016;23(5):872–8. doi: 10.1093/jamia/ocv197 [DOI] [PubMed] [Google Scholar]
- 31.Liu X, Chen Y, Bae J, Li H, Johnston J, Sanger T. Predicting Heart Failure Readmission from Clinical Notes Using Deep Learning. 2019:2642–2648.
- 32.Mahajan SM, Ghani R. Combining Structured and Unstructured Data for Predicting Risk of Readmission for Heart Failure Patients. Studies in health technology and informatics. Aug 21 2019;264:238–242. doi: 10.3233/SHTI190219 [DOI] [PubMed] [Google Scholar]
- 33.Garvin JH, Kim Y, Gobbel GT, et al. Automating Quality Measures for Heart Failure Using Natural Language Processing: A Descriptive Study in the Department of Veterans Affairs. JMIR medical informatics. Jan 15 2018;6(1):e5. doi: 10.2196/medinform.9150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jonnalagadda SR, Adupa AK, Garg RP, Corona-Cox J, Shah SJ. Text Mining of the Electronic Health Record: An Information Extraction Approach for Automated Identification and Subphenotyping of HFpEF Patients for Clinical Trials. Journal of cardiovascular translational research. Jun 2017;10(3):313–321. doi: 10.1007/s12265-017-9752-2 [DOI] [PubMed] [Google Scholar]
- 35.Topaz M, Radhakrishnan K, Blackley S, Lei V, Lai K, Zhou L. Studying Associations Between Heart Failure Self-Management and Rehospitalizations Using Natural Language Processing. Western journal of nursing research. Jan 2017;39(1):147–165. doi: 10.1177/0193945916668493 [DOI] [PubMed] [Google Scholar]
- 36.Eggerth A, Kreiner K, Hayn D, et al. Natural Language Processing for Detecting Medication-Related Notes in Heart Failure Telehealth Patients. Stud Health Technol Inform. Jun 16 2020;270:761–765. doi: 10.3233/shti200263 [DOI] [PubMed] [Google Scholar]
- 37.Leiter RE, Santus E, Jin Z, et al. Deep Natural Language Processing to Identify Symptom Documentation in Clinical Notes for Patients With Heart Failure Undergoing Cardiac Resynchronization Therapy. J Pain Symptom Manage. Nov 2020;60(5):948–958.e3. doi: 10.1016/j.jpainsymman.2020.06.010 [DOI] [PubMed] [Google Scholar]
- 38.Prakash Adekkanattu a, Guoqian Jiang a, Yuan Luo a, et al. Evaluating the Portability of an {NLP} System for Processing Echocardiograms: A Retrospective, Multi-site Observational Study. arXiv pre-print. 2019; [PMC free article] [PubMed] [Google Scholar]
- 39.Nath C, Albaghdadi MS, Jonnalagadda SR. A Natural Language Processing Tool for Large-Scale Data Extraction from Echocardiography Reports. PLoS One. 2016;11(4):e0153749. doi: 10.1371/journal.pone.0153749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi Y, Li Z, Jia Z, et al. Automatic knowledge extraction and data mining from echo reports of pediatric heart disease: Application on clinical decision support. 2015. https://www.scopus.com/inward/record.uri?eid=2-s2.0-84952669071&doi=10.1007%2f978-3-319-25816-4_34&partnerID=40&md5=03a776ff8eef7cecdafe6b349e9557bf https://link.springer.com/chapter/10.1007%2F978-3-319-25816-4_34
- 41.Xie F, Zheng C, Yuh-Jer Shen A, Chen W. Extracting and analyzing ejection fraction values from electronic echocardiography reports in a large health maintenance organization. Health informatics journal. Dec 2017;23(4):319–328. doi: 10.1177/1460458216651917 [DOI] [PubMed] [Google Scholar]
- 42.Patterson OV, Freiberg MS, Skanderson M, S JF, Brandt CA, DuVall SL. Unlocking echocardiogram measurements for heart disease research through natural language processing. BMC cardiovascular disorders. Jun 12 2017;17(1):151. doi: 10.1186/s12872-017-0580-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Valtchinov VI, Lacson R, Wang A, Khorasani R. Comparing Artificial Intelligence Approaches to Retrieve Clinical Reports Documenting Implantable Devices Posing MRI Safety Risks. Journal of the American College of Radiology : JACR. Feb 2020;17(2):272–279. doi: 10.1016/j.jacr.2019.07.018 [DOI] [PubMed] [Google Scholar]
- 44.Zheng C, Sun BC, Wu YL, et al. Automated Identification and Extraction of Exercise Treadmill Test Results. Journal of the American Heart Association. Mar 3 2020;9(5):e014940. doi: 10.1161/JAHA.119.014940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Galper BZ, Beery DE, Leighton G, Englander LL. Comparison of adverse event and device problem rates for transcatheter aortic valve replacement and Mitraclip procedures as reported by the Transcatheter Valve Therapy Registry and the Food and Drug Administration postmarket surveillance data. American heart journal. Apr 2018;198:64–74. doi: 10.1016/j.ahj.2017.10.013 [DOI] [PubMed] [Google Scholar]
- 46.Mathur P, Srivastava S, Xu X, Mehta JL. Artificial Intelligence, Machine Learning, and Cardiovascular Disease. Clin Med Insights Cardiol. 2020;14:1179546820927404. doi: 10.1177/1179546820927404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang F, Preininger A. AI in Health: State of the Art, Challenges, and Future Directions. Yearb Med Inform. Aug 2019;28(1):16–26. doi: 10.1055/s-0039-1677908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diprose WK, Buist N, Hua N, Thurier Q, Shand G, Robinson R. Physician understanding, explainability, and trust in a hypothetical machine learning risk calculator. J Am Med Inform Assoc. Apr 1 2020;27(4):592–600. doi: 10.1093/jamia/ocz229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lundberg SM, Lee S-I. A unified approach to interpreting model predictions. 2017:4768–4777.
- 50.Collins SA, Cato K, Albers D, et al. Relationship between nursing documentation and patients’ mortality. Am J Crit Care. Jul 2013;22(4):306–13. doi: 10.4037/ajcc2013426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Savova GK, Danciu I, Alamudun F, et al. Use of Natural Language Processing to Extract Clinical Cancer Phenotypes from Electronic Medical Records. Cancer Res. Nov 1 2019;79(21):5463–5470. doi: 10.1158/0008-5472.Can-19-0579 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


