Abstract
Novel antimicrobials for effective treatment of uncomplicated gonorrhea are essential, and the first-in-class, oral spiropyrimidinetrione DNA gyrase B inhibitor zoliflodacin appears promising. Using our newly developed Hollow Fiber Infection Model (HFIM), the pharmacodynamics of zoliflodacin was examined. A clinical zoliflodacin-susceptible N. gonorrhoeae strain, SE600/18 (harbouring a GyrB S467N amino acid substitution; MIC = 0.25 mg/L), and SE600/18-D429N (zoliflodacin-resistant mutant with a second GyrB substitution, D429N, selected in the HFIM experiments; zoliflodacin MIC = 2 mg/L), were examined. Dose-range experiments, simulating zoliflodacin single oral dose regimens of 0.5, 1, 2, 3, and 4 g, were performed for SE600/18. For SE600/18-D429N, dose-range experiments, simulating zoliflodacin single oral 2, 3, 4, and 6 g doses, and zoliflodacin oral dose-fractionation experiments with 4, 6, and 8 g administered as q12 h were performed. Both strains grew well in the untreated HFIM growth control arms and mostly maintained growth at 1010–1011 CFU/ml for 7 days. Zoliflodacin 3 and 4 g single dose oral regimens successfully eradicated SE600/18 and no growth was recovered during the 7-days experiments. However, the single oral 0.5, 1, and 2 g doses failed to eradicate SE600/18, and zoliflodacin-resistant populations with a GyrB D429N substitution were selected with all these doses. The zoliflodacin-resistant SE600/18-D429N mutant was not eradicated with any examined treatment regimen. However, this in vitro-selected zoliflodacin-resistant mutant was substantially less fit compared to the zoliflodacin-susceptible SE600/18 parent strain. In conclusion, the rare clinical gonococcal strains with GyrB S467N substitution are predisposed to develop zoliflodacin resistance and may require treatment with zoliflodacin ≥3 g. Future development may need to consider the inclusion of diagnostics directed at identifying strains resistant or predisposed to resistance development at a population level and to strengthen surveillance (phenotypically and genetically), and possibly also at the patient level to guide treatment.
Keywords: Neisseria gonorrhoeae, hollow fiber infection model, zoliflodacin, antimicrobial treatment, pharmacodynamics, pharmacokinetics, gyrB, mutant
Introduction
The high and increasing levels of antimicrobial resistance (AMR) in Neisseria gonorrhoeae globally (Wi et al., 2017; Day et al., 2018; Unemo et al., 2019; Unemo et al., 2021) are seriously threatening the management and control of gonorrhea. The WHO Global Action Plan to Control the Spread and Impact of Antimicrobial Resistance in N. gonorrhoeae (WHO, 2012) and WHO Global Action Plan on Antimicrobial Resistance (WHO, 2015) stress that new antimicrobials for treatment of urogenital and extragenital gonorrhea are imperative. Currently, solely two new antimicrobials, zoliflodacin (Jacobsson et al., 2014; Alm et al., 2015; Basarab et al., 2015; Foerster et al., 2015; Unemo et al., 2015; Taylor et al., 2018; Foerster et al., 2019; O’Donnell et al., 2019; Unemo et al., 2019; Bradford et al., 2020; Jacobsson et al., 2021) and gepotidacin (Jacobsson et al., 2018; Scangarella-Oman et al., 2018; Taylor et al., 2018), are in later stages of clinical development for treatment of uncomplicated gonorrhea.
Zoliflodacin is the first-in-class spiropyrimidinetrione and a type II topoisomerase inhibitor with unique target (GyrB) and mode of bactericidal action (Basarab et al., 2015; Kern et al., 2015). Zoliflodacin has a high in vitro activity against N. gonorrhoeae, including multi-drug-resistant clinical strains (Jacobsson et al., 2014; Unemo et al., 2015; Unemo et al., 2019; Bradford et al., 2020). No clinical N. gonorrhoeae isolates with zoliflodacin resistance have been reported when international gonococcal populations from the last decade have been examined (Jacobsson et al., 2014; Unemo et al., 2015; Unemo et al., 2019; Bradford et al., 2020; Le et al., 2021). However, in static in vitro laboratory experiments zoliflodacin-resistant mutants have been selected; all containing substitutions of amino acids D429 or K450 of GyrB (Alm et al., 2015; Foerster et al., 2015; Foerster et al., 2019; Jacobsson et al., 2021). No clinical isolate with amino acid substitution in GyrB K450 and only one single clinical isolate with a GyrB D429V substitution has been found (Jacobsson et al., 2014; Alm et al., 2015; Unemo et al., 2015; Unemo et al., 2019; Bradford et al., 2020; Le et al., 2021; Adamson et al., 2021). Additionally, an isolate with a GyrB S467N substitution was selected previously in static in vitro experiments (Alm et al., 2015). This substitution did not alone cause zoliflodacin resistance as a first step mutation (zoliflodacin MIC of 0.25 mg/L), however it further increased the MIC of zoliflodacin as a second step mutation (Alm et al., 2015). Rare clinical gonococcal isolates with a GyrB S467N substitution (https://pathogen.watch/collections/all?organismId=485), e.g., 1 of 143 isolates with zoliflodacin MICs of 0.125–0.25 mg/L in Nanjing, China (Le et al., 2021), and wild-type (susceptible) MICs of zoliflodacin have been found. However, the international prevalence of these strains is basically unknown. Overexpression of the MtrC-MtrD-MtrE efflux pump can also further increase the MICs of zoliflodacin (Foerster et al., 2015).
A phase 2 randomised controlled clinical trial (RCT) to evaluate the efficacy and safety of zoliflodacin 2 and 3 g single oral dose for treatment of uncomplicated gonorrhea was recently performed (Taylor et al., 2018). The zoliflodacin 3 g single oral dose was the most effective dose, providing microbiological cure rates of 100% (47/47), 100% (6/6), and 78% (7/9) for urogenital, rectal, and pharyngeal gonorrhea, respectively, in the per protocol analyses. This can be compared to microbiological cure rates of 98% (48/49), 100% (4/4), and 67% (4/6) for urogenital, rectal, and pharyngeal gonorrhea with zoliflodacin 2 g single oral dose. No N. gonorrhoeae isolates with in vitro resistance to zoliflodacin were found (Taylor et al., 2018). To provide further understanding of the findings of the zoliflodacin phase 2 RCT (Taylor et al., 2018), we developed, optimized and quality assured a dynamic in vitro hollow fiber infection model (HFIM) to simulate gonococcal infections and the pharmacokinetic (PK)/pharmacodynamic (PD) of antimicrobials acting against N. gonorrhoeae infections (Jacobsson et al., 2021), using geographically, phenotypically and genomically diverse WHO N. gonorrhoeae reference strains (Unemo et al., 2015). This HFIM for N. gonorrhoeae was used to study the PK/PD of zoliflodacin treatment against N. gonorrhoeae strains with full susceptibility to zoliflodacin and no gyrB mutations (Jacobsson et al., 2021). Further understanding of the microbiological cures and failures of the treatments in the zoliflodacin phase 2 RCT (Taylor et al., 2018), the zoliflodacin concentration-dependent killing of N. gonorrhoeae, and importance of also examining suppression of resistance emergence was provided (Jacobsson et al., 2021). According to the HFIM, for both effective killing and resistance suppression of zoliflodacin wild-type N. gonorrhoeae strains, zoliflodacin should be administered at ≥2 g as a single oral dose. However, it was also stated as essential to examine treatment of gonococcal strains with different gyrB mutations (Jacobsson et al., 2021).
The main aim of the present study was to examine the pharmacodynamics of zoliflodacin treatment against one clinical zoliflodacin-susceptible N. gonorrhoeae strain (SE600/18), with a zoliflodacin-target GyrB S467N substitution, and SE600/18-D429N (zoliflodacin-resistant mutant with an additional in vitro selected GyrB D429N substitution) in our dynamic HFIM for N. gonorrhoeae (Jacobsson et al., 2021). The biofitness of the zoliflodacin-resistant SE600/18-D429N mutant compared to the zoliflodacin-susceptible clinical SE600/18 strain was also evaluated in the HFIM.
Materials and Methods
Bacterial Strains
The clinical zoliflodacin-susceptible N. gonorrhoeae strain with GyrB S467N (SE600/18) was cultured in Sweden in 2018. Additionally, the zoliflodacin-resistant mutant of SE600/18 with an additional GyrB D429N substitution selected in the HFIM (SE600/18-D429N) was examined.
Antimicrobial Susceptibility Testing
For determination of zoliflodacin MICs (mg/L), agar dilution on GCVIT agar plates (Foerster et al., 2019) and microbroth dilution [in triplicates in the HFIM medium, i.e., modified Fastidious Broth (mFB)] were performed, as previously described (Jacobsson et al., 2021). Etest was used to determine MICs (mg/L) of ceftriaxone, cefixime, ciprofloxacin, and azithromycin, in accordance with the manufacturer’s instructions (bioMérieux, Marcy-l’Etoile, France).
Hollow Fiber Infection Model
For simulation of a gonococcal infection and the PK/PD of current and new antimicrobials, such as zoliflodacin, against N. gonorrhoeae, we recently developed and optimized a dynamic HFIM using cellulosic cartridges (FiberCell Systems Inc., Frederick, MD, United States) (Jacobsson et al., 2021). In brief, our HFIM is a two-compartment model system, in which N. gonorrhoeae cells grow in the extracapillary space of a cellulosic cartridge containing a bundle of microfibers (FiberCell Systems Inc., Frederick, MD, United States). A syringe pump administered zoliflodacin into the HFIM and peristaltic pumps isovolumetrically replaced zoliflodacin-containing liquid growth medium with zoliflodacin-free liquid growth medium to simulate the half-life of zoliflodacin and non-protein bound (free) zoliflodacin concentration-time profiles reported in human plasma throughout 7 days. N. gonorrhoeae quantitative cultures (colony forming units (CFUs)/mL) for total N. gonorrhoeae burden and zoliflodacin-resistant N. gonorrhoeae population and determination of zoliflodacin concentrations in the HFIM were performed over 7 days (Drusano, 2017).
Briefly, on the first day 0.5 ml of N. gonorrhoeae cultures (18–24 h) from GCAGP agar plates (Foerster et al., 2019) were inoculated in 49.5 ml of mFB and incubated at 36°C in a humidified 5% CO2-enriched atmosphere to mid-log phase. 10 ml (∼106 CFU/ml) of the N. gonorrhoeae suspension were then inoculated into each HFIM cartridge to mimic a clinically relevant N. gonorrhoeae cell concentration (Bissessor et al., 2011; Chow et al., 2016; Priest et al., 2017; Van Der Veer et al., 2020). Zoliflodacin was administrated to mimic an adult human PK concentration-time profile following a single oral dose of zoliflodacin [PK parameters for zoliflodacin 3 g oral dose were used (linearly adjusted for other doses): 17% fraction of zoliflodacin free (protein-unbound) in plasma, 6.47 h half-life (t1/2), and a 3 h infusion time] (O’Donnell et al., 2019), as previously described (Jacobsson et al., 2021). One HFIM cartridge per examined strain and experiment was used as an untreated growth control.
Dose-range experiments simulated zoliflodacin single dose oral regimens of 0.5, 1, 2, 3, and 4 g against the clinical SE600/18 strain, and single dose oral regimens of 2, 3, 4, and 6 g against SE600/18-D429N. Dose-fractionation experiments simulated zoliflodacin oral dose therapy with 4, 6, and 8 g administered as one half of the total dose given at 0 h and at 12 h (q12 h) against SE600/18-D429N. All experiments were followed for 7 days.
Quantification of Viable Bacterial Populations
To determine the N. gonorrhoeae total population and zoliflodacin-resistant subpopulations, bacterial solution (1 ml) was sampled from each HFIM cartridge at time points 3, 6.5, 24, 48, 72, 96, 120, 144, and 168 h for the dose-range experiments, and at 3, 6.5, 12, 15, 18.5, 24, 48, 72, 96, 120, 144, and 168 h for the q12 h dose-fractionation experiments. Samples were serially diluted in mFB and quantitatively plated on GCAGP agar plates (Foerster et al., 2019) and GCAGP agar plates (Foerster et al., 2019) containing 2 × MIC of zoliflodacin, resulting in a detection limit of ≥100 CFUs per HFIM cartridge, as previously described (Jacobsson et al., 2021). Colony counts (log10 CFU/ml) were quantified after incubation for up to 72 h at 36°C in a humid 5% CO2-enriched atmosphere using an automated colony counter (Scan 4000, Interscience, Saint-Nom-la-Bretèche, France).
Biofitness Experiments
To evaluate the biofitness of the zoliflodacin-resistant mutant selected in the HFIM (SE600/18-D429N) compared to the zoliflodacin-susceptible clinical SE600/18 parent strain, competition experiments using coculture were performed in the HFIM. Briefly, bacteria were harvested from GCAGP agar plates (Foerster et al., 2019) and suspended in mFB to a quantity of ∼106 CFU/ml. Equal volumes (5 ml/strain) of the suspensions of each strain were inoculated into the same HFIM cartridge. Aliquots (1 ml) were sampled at 24, 48, 72, 96, 120, 144, and 168 h, serially diluted in mFB and quantitatively plated on GCAGP agar plates (Foerster et al., 2019), as previously described (Jacobsson et al., 2021). Colony counts (log10 CFU/ml) were quantified after incubation for up to 72 h at 36°C in a humid 5% CO2-enriched atmosphere using an automated colony counter (Scan 4000, Interscience, Saint-Nom-la-Bretèche, France). The competitive index (CI) was determined by dividing the ratio of the SE600/18-D429N mutant to wild-type SE600/18 at each time point with the ratio of the SE600/18-D429N mutant to wild-type SE600/18 in the initial inoculum (Vincent et al., 2018).
Zoliflodacin Concentration Determination
To confirm that the predicted zoliflodacin PK profiles were observed in the HFIM, broth samples (500 µl) were collected at time points 1, 2, 3, 6.5, 18.5, 24, 48, 72, 96, 120, 144, and 168 h for the dose-range experiments, and at 1, 2, 3, 6.5, 12, 15, 18.5, 24, 48, 72, 96, 120, 144, and 168 h for the q12 h dose-fractionation experiments. All zoliflodacin concentrations were determined from 100 μl sample aliquots using liquid chromatography-tandem mass spectrometry (LC-MS/MS), as previously described (Jacobsson et al., 2021).
Population Pharmacokinetic/Pharmacodynamic Mathematical Modeling
We simultaneously modeled 3 system outputs for the analysis of the experimental data. The system outputs were: concentration of zoliflodacin, total N. gonorrhoeae burden, and burden of N. gonorrhoeae with lower susceptibility/resistance to zoliflodacin (containing MIC-increasing gyrB mutation selected during treatment). Population modeling was performed employing the Non-Parametric Adaptive Grid (NPAG) program of Leary et al. (2001) and Neely et al. (2012). Modeling choices (weighting, etc.) and goodness of fit evaluations were as previously published (Brown et al., 2015). Simulation was performed with the ADAPT V Program of D’Argenio et al. (2009) using Bayesian posterior parameter estimates.
Comparative Genomic Analysis
Whole genome sequencing (WGS) was performed, as previously described (Jacobsson et al., 2016; Golparian et al., 2020a), on selected colonies that grew on the zoliflodacin-containing plates and that also had increased MICs of zoliflodacin by agar dilution. The WGS was primarily performed to identify zoliflodacin resistance-associated gyrB mutations, i.e., the previously identified gyrB mutations that were verified to cause the increased MICs of zoliflodacin in in vitro selected zoliflodacin-resistant mutants (Alm et al., 2015; Foerster et al., 2015; Foerster et al., 2019; Jacobsson et al., 2021) or novel gyrB mutations. However, the whole genome sequences of the zoliflodacin-resistant mutants were examined to identify also any other zoliflodacin resistance-associated mutations selected in the HFIM. All reads were quality controlled and trimmed accordingly using our previously described CLC Genomics Workbench v20.0.4 workflow (Golparian et al., 2020b), and all quality-controlled reads were mapped to the gyrB reference obtained from Genbank (Genbank: AE004969.1) using local alignment with CLC Genomics Workbench with match score 1, mismatch cost of 2 and linear gap cost of 3, the variants across the gene were called with a minimum coverage of 10x and a minimum frequency of 35%. WGS reads of SE600/18 with a pre-existing GyrB S467N mutation and the zoliflodacin-resistant mutant of SE600/18 with an additional GyrB D429N substitution (SE600/18-D429N) are available through the European Nucleotide Archive (ENA) accession number PRJEB50904.
The main experiments of the zoliflodacin-susceptible N. gonorrhoeae SE600/18 parent strain (with a GyrB S467N mutation) and the zoliflodacin-resistant N. gonorrhoeae SE600/18-D429N mutant [with GyrB S467N plus GyrB D429N selected in the Hollow Fiber Infection Model (HFIM)] have been summarized in Supplementary Figure 1.
Results
Phenotypic and Genetic Characteristics of Examined N. gonorrhoeae Strains
The MICs of zoliflodacin determined using agar dilution and microbroth dilution methods, GyrB substitutions, and additional relevant characteristics of the two examined strains are summarised in Table 1.
TABLE 1.
Relevant phenotypic and genetic characteristics of N. gonorrhoeae strains. Differences between the clinical SE600/18 isolate and the SE600/18-D429N mutant selected in the HFIM are in bold letters.
| Strain characteristics | SE600/18 | SE600/18-D429N |
|---|---|---|
| Zoliflodacin MIC (microbroth MIC) a | 0.25 (0.5) | 2 (4) |
| Ceftriaxone MIC a | 0.032 | 0.032 |
| Cefixime MIC a | 0.125 | 0.125 |
| Ciprofloxacin MIC a | 0.5 | 0.5 |
| Azithromycin MIC a | 0.125 | 0.125 |
| Relevant GyrB mutations | S467N | S467N, D429N |
| GyrA codon S91, D95 | S91F, D95N | S91F, D95N |
| mtrR promoter region 13 bp inverted repeat | WT | WT |
| mtrR coding region | WT | WT |
| Mosaic mtrRCDE | — | — |
| PorB1b codon G120, A121 | A121D | A121D |
| NG-MAST | ST20643 | ST20643 |
| NG-STAR | ST3537 | ST3537 |
| MLST | ST7363 | ST7363 |
MIC, minimum inhibitory concentration; WT, wild type; NA, not applicable; NG-MAST, N. gonorrhoeae multiantigen sequence typing; ST, sequence type; NG-STAR, N. gonorrhoeae sequence typing antimicrobial resistance; MLST, multi-locus sequence typing.
MIC (mg/L) was determined using agar dilution and microbroth methods for zoliflodacin, and Etest (bioMérieux, Marcy-l’Etoile, France) for ceftriaxone, cefixime, ciprofloxacin, and azithromycin.
Briefly, the clinical zoliflodacin-susceptible N. gonorrhoeae SE600/18 strain, containing a GyrB S467N substitution, and the zoliflodacin-resistant SE600/18-D429N mutant (GyrB S467N plus an in vitro selected GyrB D429N substitution) were examined in the HFIM. The zoliflodacin MICs of both strains were one MIC dilution higher using microbroth dilution compared with agar dilution. Notably, SE600/18 belonged to MLST ST7363, which has been a common gonococcal ST internationally during latest decades and it has also been associated with multi-drug resistance, including decreased susceptibility and resistance to extended-spectrum cephalosporins such as ceftriaxone and particularly cefixime (Shimuta et al., 2015; Unemo, 2015; Harris et al., 2018; Sánchez-Busó et al., 2021) (Table 1).
Hollow Fiber Infection Model results
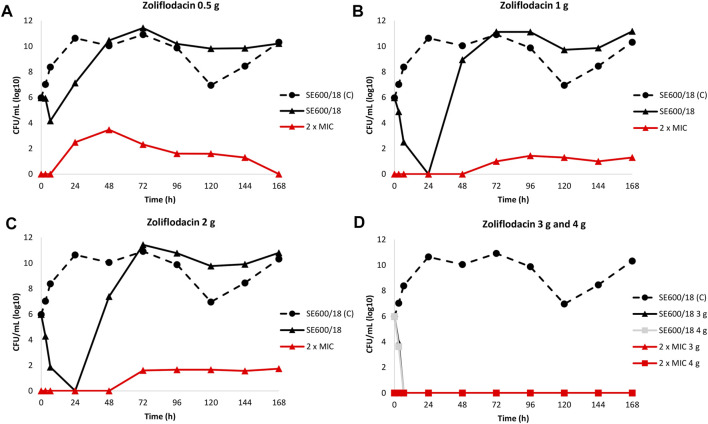
The results of the zoliflodacin dose-range studies for the zoliflodacin-susceptible clinical SE600/18 strain are summarised in Figure 1.
FIGURE 1.
Growth curves of the total population of the clinical zoliflodacin-susceptible N. gonorrhoeae strain SE600/18, containing a GyrB S467N amino acid substitution, in the dose-range Hollow Fiber Infection Model experiment simulating zoliflodacin single oral dose of 0.5 (A), 1 (B), 2 (C), 3, and 4 g (D) and followed for 7 days are shown (black solid lines). The total growth of zoliflodacin-resistant populations (red lines) on the zoliflodacin-containing plates (2 × MIC) and total growth of the untreated control (C; black dashed line) are also shown for each treatment.
Briefly, the SE600/18 strain grew well in the untreated growth control arms and reached a bacterial density of 1010–1011 CFU/ml at 24 h (Figure 1). All untreated controls also maintained growth at approximately 109–1011 CFU/ml throughout the 7-days experiments (Figure 1). A rapid bacterial kill was documented during the first 6.5 h for all doses. However, after zoliflodacin 0.5 g single dose therapy the SE600/18 strain had recovered growth at 24 h (approximately 107 CFU/ml) and at 48 h the bacterial density was as high as in the untreated control (>1010 CFU/ml) (Figure 1A). Using both the zoliflodacin 1 and 2 g treatment (Figures 1B,C, respectively), SE600/18 was rapidly killed and at 24 h no growth was detected. However, at 48 h SE600/18 had recovered growth at bacterial density of approximately 109 CFU/ml (Figure 1B) and 107 CFU/ml (Figure 1C), respectively. Using the zoliflodacin 3 and 4 g treatments, SE600/18 was eradicated at 6.5 h time point and the strain did not recover any growth during the 7-days experiments (Figure 1D).
Zoliflodacin-resistant mutants grew on the zoliflodacin-containing plates for all treatments arms where growth was recovered, i.e., in the 0.5, 1 and 2 g treatment arms (Figures 1A–C). These zoliflodacin-resistant populations emerged after 6.5 h (0.5 g treatment; Figure 1A) or 48 h (1 and 2 g treatments; Figures 1B,C). Notably, these zoliflodacin-resistant populations were maintained at low concentrations of approximately <102–104 CFU/ml during the 7-days experiment. Accordingly, further amplification of the zoliflodacin-resistant populations was not observed, and these zoliflodacin-resistant mutant populations appeared to grow slower and in smaller colonies compared to the SE600/18 strain on agar plates, which all may indicate a suboptimal biofitness. These selected zoliflodacin-resistant mutants had zoliflodacin MICs of 2 mg/L (agar dilution) and contained the GyrB D429N substitution in addition to the pre-existing GyrB S467N substitution (referred to as SE600/18-D429N hereafter). No other selected mutations that appeared to be associated with zoliflodacin susceptibility were found.
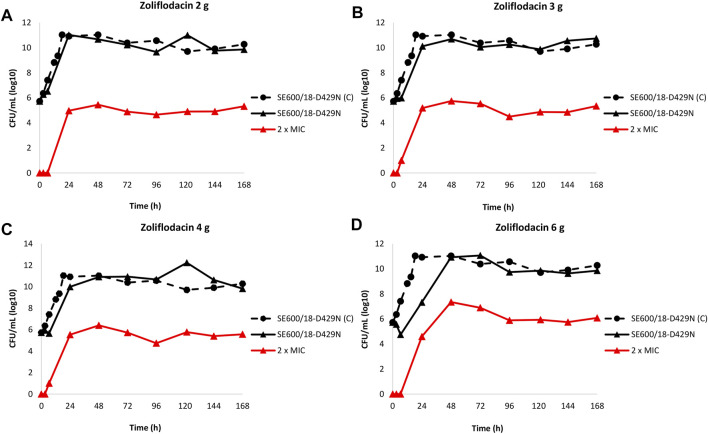
The results of the zoliflodacin dose-range studies for the zoliflodacin-resistant SE600/18-D429N mutant are summarised in Figure 2.
FIGURE 2.
Growth curves of the total population of the zoliflodacin-resistant Neisseria gonorrhoeae SE600/18-D429N mutant (containing GyrB S467N amino acid substitution plus an in vitro selected D429N substitution), in the dose-range Hollow Fiber Infection Model experiment simulating zoliflodacin single oral dose of 2 (A), 3 (B), 4 (C), and 6 g (D) and followed for 7 days are shown (black solid lines). The total growth of population with increased resistance (red lines) on the zoliflodacin-containing plates (2 × MIC) and total growth of the untreated control (C; black dashed line) are also shown for each treatment.
In brief, the SE600/18-D429N mutant grew well in the untreated growth control arms and reached a bacterial density of 1011 CFU/ml at 24 h (Figure 2). All untreated controls maintained growth at around 1010–1011 CFU/ml throughout the 7-day experiments (Figure 2). However, the zoliflodacin 2, 3, and 4 g single dose oral therapy did not result in any kill of SE600/18-D429N (Figures 2A–C) and the zoliflodacin 6 g single dose regimen only caused bacterial kill the first 6.5 h (Figure 2D). Growth was recovered in all treatment arms and ranged from approximately 107 CFU/ml (6 g arm) to 1011 CFU/ml (2 g arm) at 24 h and at the bacterial density of the untreated control at 48 h (1011 CFU/ml), which was maintained during the remaining 5 days of the experiments (Figure 2).
Furthermore, zoliflodacin-resistant gonococcal populations grew on the zoliflodacin-containing plates for all treatments arms (Figures 2A–D). These zoliflodacin-resistant populations grew at approximately 105 CFU/ml after 24 h in all treatment arms. Notably, these zoliflodacin-resistant populations fluctuated at 105–107 CFU/ml during the whole 7-days experiment (Figure 2). Accordingly, further amplification of these zoliflodacin-resistant populations was not observed, and these mutant populations appeared to grow slower and in smaller colonies on agar plates, which all may indicate a suboptimal biofitness. These zoliflodacin-resistant populations had zoliflodacin MICs of 2–8 mg/L (agar dilution) and contained the GyrB D429N substitution in addition to the pre-existing GyrB S467N substitution.
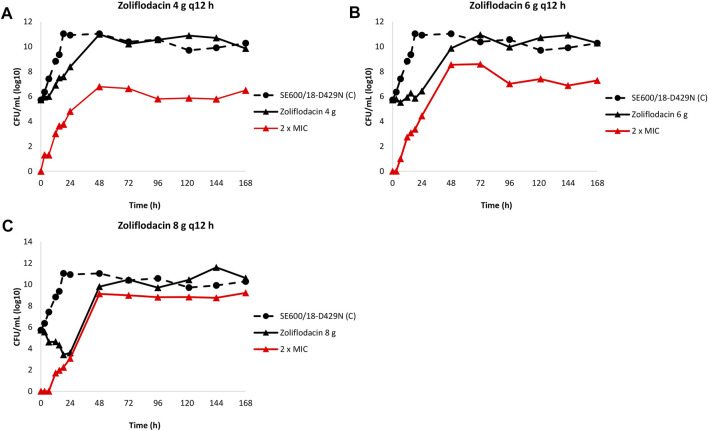
The results of the zoliflodacin dose-fractionation experiments for the zoliflodacin-resistant SE600/18-D429N mutant are summarised in Figure 3.
FIGURE 3.
Growth curves of the total population of the zoliflodacin-resistant Neisseria gonorrhoeae SE600/18-D429N mutant (containing the GyrB S467N amino acid substitution plus an in vitro selected D429N substitution), in the dose-range Hollow Fiber Infection Model experiment simulating a zoliflodacin oral fractionated dose of 4 g (2 g given at 0 and 12 h) (A), 6 g (3 g given at 0 and 12 h) (B), and 8 g (4 g given at 0 and 12 h) (C) over 24 h and followed for 7 days are shown (black solid lines). The total growth of population with increased resistance (red lines) on the zoliflodacin-containing plates (2 × MIC) and total growth of the untreated control (C; black dashed line) are also shown for each treatment.
Briefly, the SE600/18-D429N mutant grew well in the untreated growth control arms and reached a bacterial density of 1011 CFU/ml at 24 h (Figure 3). All untreated controls maintained growth at around 1010–1011 CFU/ml throughout the 7-day experiments (Figure 3). The equivalent zoliflodacin oral therapy of 4 and 6 g administered as equally divided doses at q12 h did not result in any substantial kill of SE600/18-D429N (Figures 3A,B) and the zoliflodacin 8 g q12 h regimen only caused bacterial kill the first 24 h (Figure 3C). Accordingly, growth was recovered in all treatment arms and ranged from approximately 103 CFU/ml (8 g q12 h arm) to 108 CFU/ml (4 g q12 h arm) at 24 h and the bacterial density at 48 h was at approximately 1010–1011 CFU/ml, which was maintained during the remaining 5 days of the experiments (Figure 3).
Zoliflodacin-resistant populations grew on the zoliflodacin-containing plates for all treatments arms (Figures 3A–C). These zoliflodacin-resistant populations grew at approximately 103–104 CFU/ml after 24 h and at >106–109 CFU/ml at 48 h in all treatment arms (Figure 3). The zoliflodacin-resistant population selected in the zoliflodacin 8 g q12 h treatment arm appeared to recover growth at nearly the same bacterial density as the untreated control (Figure 3C), however, also this mutant population appeared to grow slower and in smaller colonies compared to the SE600/18 strain on agar plates, which may indicate a suboptimal biofitness. These zoliflodacin-resistant populations had zoliflodacin MICs of 2–8 mg/L (agar dilution) and contained the GyrB D429N substitution in addition to the pre-existing GyrB S467N substitution.
Population Pharmacokinetic/Pharmacodynamic Modeling
The three output PK/PD model was fit to all the data for SE600/18. The mean and median values for SE600/18 are displayed in Table 2.
TABLE 2.
Mean, median and standard deviation of the parameter values for the Hollow Fiber Infection Model study with the N. gonorrhoeae clinical SE600/18 strain with a pre-existing GyrB S467N amino acid substitution.
| Parameter | Mean | Median | Standard deviation |
|---|---|---|---|
| Vc (L) | 1073 | 99.03 | 1050 |
| CL (L/hr) | 116.3 | 13.11 | 107.2 |
| Kg-s (hr−1) | 0.68 | 0.117 | 0.591 |
| Kg-r (hr−1) | 0.088 | 0.066 | 0.050 |
| Kkill-s (hr−1) | 9.087 | 5.218 | 4.065 |
| Kkill-r (hr−1) | 1.820 | 0.520 | 1.759 |
| C50-s (mg/L) | 0.724 | 0.134 | 0.670 |
| C50-r (mg/L) | 2.527 | 1.274 | 1.802 |
| Hs (—) | 4.202 | 3.191 | 2.350 |
| Hr (—) | 13.86 | 5.663 | 12.78 |
| POPMAX (CFU/ml) | 0.261 × 1011 | 0.109 × 1011 | 0.337 × 1011 |
| IC2 (CFU/ml) | 8.789 × 105 | 1.066 × 105 | 9.209 × 105 |
| IC3 (CFU/ml) | 5.012 | 3.059 | 3.769 |
Vc, apparent volume of the central compartment; CL, clearance; Kg-s and Kg-r, rate constants of growth for the susceptible and resistant population, respectively; Kkill-s and Kkill-r, rate constants of kill for the susceptible and resistant population, respectively; C50-s and C50-r, concentrations of zoliflodacin at which the kill rate is half maximal for the susceptible and resistant population, respectively; Hs and Hr, Hill’s constants for the susceptible and resistant populations, respectively (unitless); POPMAX, maximal population size; CFU, colony forming units; IC2 and IC3, sizes of the total and resistant populations, respectively, at therapy initiation.
The fit of the model to the data was acceptable, with exception of the pre-Bayesian (population) analysis for the zoliflodacin-resistant mutants (SE600/18-D429N). The predicted-observed regressions for the analysis are displayed in Supplementary Figure 2. The reason for the poor model fit for the zoliflodacin-resistant mutants is likely because of poor biofitness of these zoliflodacin-resistant mutants. This hypothesis is also supported by their very small Kg-r (Table 2) and, even when zoliflodacin was essentially gone, the zoliflodacin-resistant mutants did not amplify (Supplementary Figure 2; Figure 1).
Regarding the zoliflodacin PK profiles, the agreement between observed and predicted zoliflodacin concentrations in the HFIM during the experiments were high (Supplementary Figures 2A,D).
For the HFIM dose-ranging study for the clinical N. gonorrhoeae strain SE600/18, we performed simulation with the identified parameter values to obtain an approximate exposure of zoliflodacin that would suppress amplification of selected mutants with increased zoliflodacin MIC and gyrB resistance mutations. That exposure was a zoliflodacin 2.7 g single dose oral treatment in the HFIM.
The growth rate constant for the susceptible SE600/18 population (Table 2) was 58–60% of the growth rate constants previously measured for WHO F and WHO X (Jacobsson et al., 2021). However, the growth rate constant for the zoliflodacin-resistant SE600/18-D429N population (Table 2) was only 7–16% of the corresponding growth rate constants for zoliflodacin-resistant populations of WHO F and WHO X (Jacobsson et al., 2021). This suboptimal growth rate of the zoliflodacin-resistant SE600/18-D429N population shows that its growth is impaired and accordingly biofitness decreased, which was further supported by the kill rate constant for the zoliflodacin-resistant SE600/18-D429N population that was more than 20 times higher than the growth rate constant for the mutant (Table 2).
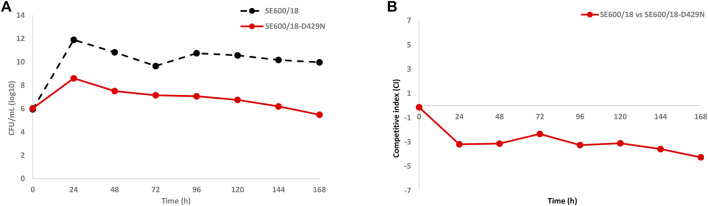
Competition Biofitness Experiments Using Coculture in the HFIM
To confirm if the in vitro-selected zoliflodacin resistance in the clinical zoliflodacin-susceptible SE600/18 strain impaired bacterial growth and accordingly decreased the biofitness, the zoliflodacin-susceptible parent SE600/18 and the zoliflodacin-resistant SE600/18-D429N mutant were cocultured in the same HFIM cartridge for 7 days (Figure 4A) and the competitive index calculated (Figure 4B). The growth of the zoliflodacin-susceptible clinical SE600/18 strain was maintained at ∼1010–1012 CFU/ml during the 7 days experiments, which was at a similar level as in the SE600/18 monocultures. However, the growth of the zoliflodacin-resistant SE600/18-D429N mutant was substantially lower particularly during the first 24 h and it peaked at ∼108 CFU/ml at 24 h and then decreased for each day resulting in a bacterial density of ∼106 CFU/ml after 7 days (Figure 4A). The clinical zoliflodacin-susceptible SE600/18 strain appeared to outcompete the zoliflodacin-resistant SE600/18-D429N mutant, which was even more clearly shown when plotting the competitive index over the 7 days experiments (Figure 4B). Accordingly, the in vitro-selected zoliflodacin-resistant mutant SE600/18-D429N was substantially less fit compared to the zoliflodacin-susceptible clinical SE600/18 parent strain (Figure 4).
FIGURE 4.
(A) Growth curves of the total population of the zoliflodacin-susceptible clinical Neisseria gonorrhoeae SE600/18 strain (black dashed line) and the zoliflodacin-resistant N. gonorrhoeae SE600/18-D429N mutant (containing the pre-existing GyrB S467N amino acid substitution plus an in vitro selected D429N substitution; red solid line), when cocultured in the same Hollow Fiber Infection Model (HFIM) cartridge and followed for 7 days. (B) Competitive indexes for the zoliflodacin-susceptible clinical Neisseria gonorrhoeae SE600/18 strain and the outcompeted zoliflodacin-resistant N. gonorrhoeae SE600/18-D429N mutant (red solid line), when cocultured in the same HFIM cartridge and followed for 7 days.
Discussion
The high levels of AMR in N. gonorrhoeae globally (Wi et al., 2017; Day et al., 2018; Unemo et al., 2019; Unemo et al., 2021) is seriously threatening the management and control of gonorrhea, and novel antimicrobials for effective treatment of urogenital and extragenital gonorrhea are urgently needed. The novel spiropyrimidinetrione zoliflodacin has been shown to be effective in treating gonococcal urogenital and rectal infections (Taylor et al., 2018). A single oral zoliflodacin 3 g dose was shown to cure all anogenital gonococcal infections and most (78%, 7/9) of the included pharyngeal infections, and no isolates with zoliflodacin resistance were found (Taylor et al., 2018). Recently, using our newly developed dynamic HFIM for N. gonorrhoeae, we verified that zoliflodacin administered as single oral doses ≥2 g is sufficient to eradicate infections caused by zoliflodacin-susceptible N. gonorrhoeae strains (wild-type gyrB gene and zoliflodacin MICs (0.064–0.125 mg/L)), which were effectively killed while also supressing resistance to zoliflodacin (Jacobsson et al., 2021). However, it was additionally stated as essential to examine treatment of N. gonorrhoeae strains with relevant gyrB mutations (Jacobsson et al., 2021).
In the present study, we examined the clinical zoliflodacin-susceptible N. gonorrhoeae strain SE600/18 with a pre-existing GyrB S467N substitution (zoliflodacin MIC = 0.25 mg/L in agar dilution). When treating SE600/18 with zoliflodacin single oral dose of 0.5–4 g in the HFIM, the SE600/18 strain was initially rapidly killed, however, with zoliflodacin 0.5, 1, and 2 g single oral doses the strain had recovered growth at 24 h (zoliflodacin 0.5 g) or at 48 h (zoliflodacin 1 and 2 g). Furthermore, zoliflodacin-resistant populations started to amplify after 6.5 h (zoliflodacin 0.5 g) or after 48 h (zoliflodacin 1 and 2 g). The zoliflodacin-resistant mutants of SE600/18 all contained an additional GyrB substitution (SE600/18-D429N; resulting in zoliflodacin MIC = 2 mg/L in agar dilution) (Alm et al., 2015; Foerster et al., 2015; Foerster et al., 2019; Jacobsson et al., 2021). Accordingly, zoliflodacin 2 g single oral dose failed to eradicate SE600/18 in the HFIM, which further strengthens previous evidence that single oral dose of zoliflodacin >2 g can be required for effective treatment of rare gonococcal strains (Taylor et al., 2018; Jacobsson et al., 2021), e.g., strains with the GyrB S467N substitution (require ≥2.7 g according to our PK/PD modeling). Our gonorrhea treatment simulations in the HFIM also showed that zoliflodacin-resistant mutants with GyrB S467N plus D429N substitution, if selected by suboptimal zoliflodacin exposures, may not be effectively treated with zoliflodacin single oral dose of 2–6 g or 4–8 g q12 h. Additionally, our HFIM results suggest that zoliflodacin-resistant mutants are selected at a higher frequency with zoliflodacin doses ≤2 g when a strain has the pre-existing GyrB S467N substitution, i.e., compared to the previously examined zoliflodacin-susceptible N. gonorrhoeae reference strains WHO F and WHO X with wild type gyrB gene (Jacobsson et al., 2021). Thus, the GyrB S467N substitution appears to predispose to emergence of zoliflodacin resistance, despite not conferring resistance to zoliflodacin on its own. Fortunately, N. gonorrhoeae strains with GyrB S467N substitution appear to be very rare internationally (https://pathogen.watch/collections/all?organismId=485; Le et al., 2021). Furthermore, the zoliflodacin-resistant SE600/18-D429N mutant suffered from a biofitness disadvantage and was outcompeted by the zoliflodacin-susceptible parent SE600/18 strain, which suggests that these zoliflodacin-resistant strains will be less effective at amplifying and spreading after emergence.
It is important to continue to survey phenotypic zoliflodacin susceptibility, but our data demonstrate the need to also consider the surveillance of known gyrB resistance mutations (in amino acid codons for D429 and K450), the GyrB S467N substitution and other mutations in gyrB or other genes that potentially cause resistance to zoliflodacin or could predispose for zoliflodacin resistance emergence. The failure to eradicate a N. gonorrhoeae GyrB S467N strain with up to zoliflodacin 2 g single oral dose (≥2.7 g required according to our PK/PD modeling) in this study, the higher selection rate of GyrB D429N resistance mutations in the GyrB S467N strains, and the underlying human inter-population PK variance could suggest that 100% of gonococcal infections caused by the sporadic gonococcal GyrB S467N strains may not be eradicated with a single oral dose of 3 g zoliflodacin, at all body sites.
The limitations of this study include the absence of zoliflodacin PK data from the infection sites for gonorrhea, such as the anogenital tract and the oropharynx. Consequently, the HFIM gonorrhea treatment simulations had to be based on concentrations of free zoliflodacin in human plasma which may not ideally reflect the urogenital and extragenital infection sites. Nevertheless, human plasma antimicrobial concentrations are commonly used as surrogates for concentrations of the antimicrobials at the infection sites for many bacterial infections (due to the lack of measured infection site concentrations), and mostly, these surrogates are sufficient to link drug exposure to effect (Drusano, 2004). It would be exceedingly valuable if appropriate studies could provide zoliflodacin PK data for the urogenital and extragenital infection sites, particularly in the pharynx. In fact, such PK data is lacking not only for potentially novel therapeutics but also for antimicrobials currently used for the treatment of gonorrhea, highlighting the urgent need in generating this type of PK data (Kong et al., 2019). Ideally, PK studies should be included in all RCTs for treatment of gonorrhea and other STIs, however, this may not be feasible and/or cost-effective in many studies. Furthermore, significantly enhanced understanding of pharyngeal gonorrhea and where and how to measure the relevant PK parameters of therapeutic antimicrobials in gonorrhea infection sites, especially in the pharynx, is urgently needed (Kong et al., 2019). Finally, inter-patient variance in PK parameters for zoliflodacin and other gonorrhea therapeutic antimicrobials from population modeling and employing these data in Monte Carlo simulations for target attainment is additionally imperative (Drusano et al., 2001; Drusano, 2004).
The ongoing international phase 3 RCT (ClinicalTrials.gov identifier NCT03959527) is comparing a zoliflodacin 3 g single oral dose to ceftriaxone plus azithromycin dual therapy for treatment of uncomplicated gonorrhea. This study will provide evidence on whether this single dose of oral zoliflodacin, is non-inferior to the globally recognised dual comparator but is not designed to address the suitability of the dose for infections caused by strains with emerging zoliflodacin resistance regardless of body site.
In conclusion, by examining the pharmacodynamics of zoliflodacin against one clinical zoliflodacin-susceptible N. gonorrhoeae strain with the pre-existing zoliflodacin-target GyrB S467N substitution in our dynamic HFIM for gonorrhea, we demonstrated that the rare N. gonorrhoeae clinical strains with a GyrB S467N substitution are predisposed to develop zoliflodacin resistance and require treatment with zoliflodacin ≥3 g. In the HFIM, zoliflodacin-resistant mutants (with an additional GyrB substitution, i.e., D429N) were selected using zoliflodacin single oral doses of 0.5–2 g. These selected zoliflodacin-resistant mutants (containing GyrB S467N and D429N substitutions) were not eradicated at any of the single- or multiple-dose regimens of zoliflodacin studied in the HFIM. A rapid point-of-care test simultaneously detecting N. gonorrhoeae and gyrB mutations causing or predisposing to zoliflodacin resistance may be valuable, i.e., for antimicrobial stewardship to avoid zoliflodacin treatment of patients without N. gonorrhoeae or with N. gonorrhoeae having relevant gyrB mutations. However, for this a clinical study evaluating this type of approach would be needed. It is additionally imperative to continue to survey zoliflodacin susceptibility phenotypically as well as genetically, i.e., with emphasis on mutations in gyrB or other genes that are verified to cause or predispose to zoliflodacin resistance. Finally, pharmacokinetic data for zoliflodacin (and other gonorrhea therapeutic antimicrobials) in urogenital and extragenital human infection sites, particularly in the pharynx, would be valuable.
Future use of zoliflodacin will require an additional evidence-base to support interventions such as dose adjustments, dual antimicrobial therapy to potentially enhance the bacterial eradication, prevent the emergence and or spread of resistance, and possibly also cure additional STIs.
Acknowledgments
We want to thank Entasis Therapeutics (Waltham, MA, United States), particularly John Mueller and John O’Donnell, for providing zoliflodacin, valuable PK/PD information, and critical review of the manuscript.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: National Center for Biotechnology Information (NCBI) BioProject database under accession numbers PRJEB50904 and ERP135503.
Author Contributions
SJ, FF, GD and MU designed, initiated and coordinated the study. SJ, DG and JO performed all the laboratory analyses. SJ, GD and MU analyzed and interpreted all the data, and wrote a first draft of the paper. All authors read, commented on and approved the final manuscript.
Funding
The present work was supported by grants from the Global Antibiotic Research and Development Partnership (GARDP), Geneva, Switzerland (2019); the Örebro County Council Research Committee, Örebro, Sweden; and the Foundation for Medical Research at Örebro University Hospital, Örebro, Sweden (2020).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2022.874176/full#supplementary-material
Schematic flow chart summarizing the main experiments of the zoliflodacin-susceptible Neisseria gonorrhoeae SE600/18 parent strain (with a GyrB S467N mutation) and the zoliflodacin-resistant N. gonorrhoeae SE600/18-D429N mutant (with GyrB S467N plus GyrB D429N selected in the Hollow Fiber Infection Model (HFIM)).
Predicted-Observed regressions for zoliflodacin concentrations, total Neisseria burden and resistant bacterial burden, respectively for the pre-Bayesian regression (A–C) and for the Bayesian regressions (D–F) for the clinical zoliflodacin-susceptible Neisseria gonorrhoeae SE600/18 strain with a GyrB S467N amino acid substitution.
References
- Adamson P. C., Lin E. Y., Ha S. M., Klausner J. D. (2021). Using a Public Database of Neisseria Gonorrhoeae Genomes to Detect Mutations Associated with Zoliflodacin Resistance. J. Antimicrob. Chemother. 76, 2847–2849. 10.1093/jac/dkab262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alm R. A., Lahiri S. D., Kutschke A., Otterson L. G., McLaughlin R. E., Whiteaker J. D., et al. (2015). Characterization of the Novel DNA Gyrase Inhibitor AZD0914: Low Resistance Potential and Lack of Cross-Resistance in Neisseria Gonorrhoeae . Antimicrob. Agents Chemother. 59, 1478–1486. 10.1128/AAC.04456-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basarab G. S., Kern G. H., McNulty J., Mueller J. P., Lawrence K., Vishwanathan K., et al. (2015). Responding to the Challenge of Untreatable Gonorrhea: ETX0914, a First-In-Class Agent with a Distinct Mechanism-Of-Action against Bacterial Type II Topoisomerases. Sci. Rep. 5, 11827. 10.1038/srep11827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissessor M., Tabrizi S. N., Fairley C. K., Danielewski J., Whitton B., Bird S., et al. (2011). Differing Neisseria Gonorrhoeae Bacterial Loads in the Pharynx and Rectum in Men Who Have Sex with Men: Implications for Gonococcal Detection, Transmission, and Control. J. Clin. Microbiol. 49, 4304–4306. 10.1128/JCM.05341-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford P. A., Miller A. A., O'Donnell J., Mueller J. P. (2020). Zoliflodacin: An Oral Spiropyrimidinetrione Antibiotic for the Treatment of Neisseria Gonorrheae, Including Multi-Drug-Resistant Isolates. ACS Infect. Dis. 6, 1332–1345. 10.1021/acsinfecdis.0c00021 [DOI] [PubMed] [Google Scholar]
- Brown A. N., Drusano G. L., Adams J. R., Rodriquez J. L., Jambunathan K., Baluya D. L., et al. (2015). Preclinical Evaluations to Identify Optimal Linezolid Regimens for Tuberculosis Therapy. mBio 6, e01741–1515. 10.1128/mBio.01741-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow E. P., Tabrizi S. N., Phillips S., Lee D., Bradshaw C. S., Chen M. Y., et al. (2016). Neisseria Gonorrhoeae Bacterial DNA Load in the Pharynges and Saliva of Men Who Have Sex with Men. J. Clin. Microbiol. 54, 2485–2490. 10.1128/JCM.01186-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Argenio D. Z., Schumitzky A., Wang X. (2009). ADAPT 5 User’s Guide: Pharmacokinetic/Pharmacodynamic Systems Analysis Software. Los Angeles: Biomedical Simulations Resource. [Google Scholar]
- Day M. J., Spiteri G., Jacobsson S., Woodford N., Amato-Gauci A. J., Cole M. J., et al. (2018). Stably High Azithromycin Resistance and Decreasing Ceftriaxone Susceptibility in Neisseria Gonorrhoeae in 25 European Countries, 2016. BMC. Infect. Dis. 18, 609. 10.1186/s12879-018-3528-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drusano G. L. (2004). Antimicrobial Pharmacodynamics: Critical Interactions of 'bug and Drug'. Nat. Rev. Microbiol. 2, 289–300. 10.1038/nrmicro862 [DOI] [PubMed] [Google Scholar]
- Drusano G. L. (2017). Pre-clinical In Vitro Infection Models. Curr. Opin. Pharmacol. 36, 100–106. 10.1016/j.coph.2017.09.011 [DOI] [PubMed] [Google Scholar]
- Drusano G. L., Preston S. L., Hardalo C., Hare R., Banfield C., Andes D., et al. (2001). Use of Preclinical Data for Selection of a Phase II/III Dose for Evernimicin and Identification of a Preclinical MIC Breakpoint. Antimicrob. Agents Chemother. 45, 13–22. 10.1128/aac.45.1.13-22.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foerster S., Drusano G., Golparian D., Neely M., Piddock L. J. V., Alirol E., et al. (2019). In Vitro antimicrobial Combination Testing of and Evolution of Resistance to the First-In-Class Spiropyrimidinetrione Zoliflodacin Combined with Six Therapeutically Relevant Antimicrobials for Neisseria Gonorrhoeae . J. Antimicrob. Chemother. 74, 3521–3529. 10.1093/jac/dkz376 [DOI] [PubMed] [Google Scholar]
- Foerster S., Golparian D., Jacobsson S., Hathaway L. J., Low N., Shafer W. M., et al. (2015). Genetic Resistance Determinants, In Vitro Time-Kill Curve Analysis and Pharmacodynamic Functions for the Novel Topoisomerase II Inhibitor ETX0914 (AZD0914) in Neisseria Gonorrhoeae . Front. Microbiol. 6, 1377. 10.3389/fmicb.2015.01377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golparian D., Bazzo M. L., Golfetto L., Gaspar P. C., Schörner M. A., Schwartz Benzaken A., et al. (2020a). Genomic Epidemiology of Neisseria Gonorrhoeae Elucidating the Gonococcal Antimicrobial Resistance and Lineages/sublineages across Brazil, 2015-16. J. Antimicrob. Chemother. 75, 3163–3172. 10.1093/jac/dkaa318 [DOI] [PubMed] [Google Scholar]
- Golparian D., Harris S. R., Sánchez-Busó L., Hoffmann S., Shafer W. M., Bentley S. D., et al. (2020b). Genomic Evolution of Neisseria Gonorrhoeae since the Preantibiotic Era (1928-2013): Antimicrobial Use/misuse Selects for Resistance and Drives Evolution. BMC. Genomics 21, 116. 10.1186/s12864-020-6511-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris S. R., Cole M. J., Spiteri G., Sánchez-Busó L., Golparian D., Jacobsson S., et al. (2018). Public Health Surveillance of Multidrug-Resistant Clones of Neisseria Gonorrhoeae in Europe: a Genomic Survey. Lancet Infect. Dis. 18, 758–768. 10.1016/S1473-3099(18)30225-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsson S., Golparian D., Alm R. A., Huband M., Mueller J., Jensen J. S., et al. (2014). High In Vitro Activity of the Novel Spiropyrimidinetrione AZD0914, a DNA Gyrase Inhibitor, against Multidrug-Resistant Neisseria Gonorrhoeae Isolates Suggests a New Effective Option for Oral Treatment of Gonorrhea. Antimicrob. Agents Chemother. 58, 5585–5588. 10.1128/AAC.03090-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsson S., Golparian D., Cole M., Spiteri G., Martin I., Bergheim T., et al. (2016). WGS Analysis and Molecular Resistance Mechanisms of Azithromycin-Resistant (MIC >2 mg/L) Neisseria Gonorrhoeae Isolates in Europe from 2009 to 2014. J. Antimicrob. Chemother. 71, 3109–3116. 10.1093/jac/dkw279 [DOI] [PubMed] [Google Scholar]
- Jacobsson S., Golparian D., Oxelbark J., Alirol E., Franceschi F., Gustafsson T. N., et al. (2021). Pharmacodynamic Evaluation of Dosing, Bacterial Kill, and Resistance Suppression for Zoliflodacin against Neisseria Gonorrhoeae in a Dynamic Hollow Fiber Infection Model. Front. Pharmacol. 12, 682135. 10.3389/fphar.2021.682135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsson S., Golparian D., Scangarella-Oman N., Unemo M. (2018). In Vitro activity of the Novel Triazaacenaphthylene Gepotidacin (GSK2140944) against MDR Neisseria Gonorrhoeae . J. Antimicrob. Chemother. 73, 2072–2077. 10.1093/jac/dky162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern G., Palmer T., Ehmann D. E., Shapiro A. B., Andrews B., Basarab G. S., et al. (2015). Inhibition of Neisseria Gonorrhoeae Type II Topoisomerases by the Novel Spiropyrimidinetrione AZD0914. J. Biol. Chem. 290, 20984–20994. 10.1074/jbc.M115.663534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong F. Y. S., Horner P., Unemo M., Hocking J. S. (2019). Pharmacokinetic Considerations Regarding the Treatment of Bacterial Sexually Transmitted Infections with Azithromycin: a Review. J. Antimicrob. Chemother. 74, 1157–1166. 10.1093/jac/dky548 [DOI] [PubMed] [Google Scholar]
- Le W., Su X., Lou X., Li X., Gong X., Wang B., et al. (2021). Susceptibility Trends of Zoliflodacin against Multidrug-Resistant Neisseria Gonorrhoeae Clinical Isolates in Nanjing, China, 2014 to 2018. Antimicrob. Agents Chemother. 65, e00863–20. 10.1128/AAC.00863-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary R., Jelliffe R., Schumitzky A., Van Guilder M. (2001). Proceedings of the 14th IEEE Symposium on Computer-Based Medical Systems. Bethesda, MD: IEEE Computer Society, 389–394.An Adaptive Grid Non-parametric Approach to Pharmacokinetic and Dynamic (PK/PD) Models [Google Scholar]
- Neely M. N., van Guilder M. G., Yamada W. M., Schumitzky A., Jelliffe R. W. (2012). Accurate Detection of Outliers and Subpopulations with Pmetrics, a Nonparametric and Parametric Pharmacometric Modeling and Simulation Package for R. Ther. Drug Monit. 34, 467–476. 10.1097/FTD.0b013e31825c4ba6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell J., Lawrence K., Vishwanathan K., Hosagrahara V., Mueller J. P. (2019). Single-Dose Pharmacokinetics, Excretion, and Metabolism of Zoliflodacin, a Novel Spiropyrimidinetrione Antibiotic, in Healthy VolunteersHealthy Volunteershealthy Volunteers. Antimicrob. Agents Chemother. 63, e01808–18. 10.1128/AAC.01808-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priest D., Ong J. J., Chow E. P. F., Tabrizi S., Phillips S., Bissessor M., et al. (2017). Neisseria Gonorrhoeae DNA Bacterial Load in Men with Symptomatic and Asymptomatic Gonococcal Urethritis. Sex. Transm. Infect. 93, 478–481. 10.1136/sextrans-2016-052950 [DOI] [PubMed] [Google Scholar]
- Sánchez-Busó L., Yeats C. A., Taylor B., Goater R. J., Underwood A., Abudahab K., et al. (2021). A Community-Driven Resource for Genomic Epidemiology and Antimicrobial Resistance Prediction of Neisseria Gonorrhoeae at Pathogenwatch. Genome Med. 13, 61. 10.1186/s13073-021-00858-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scangarella-Oman N. E., Hossain M., Dixon P. B., Ingraham K., Min S., Tiffany C. A., et al. (2018). Microbiological Analysis from a Phase 2 Randomized Study in Adults Evaluating Single Oral Doses of Gepotidacin in the Treatment of Uncomplicated Urogenital Gonorrhea Caused by Neisseria Gonorrhoeae . Antimicrob. Agents Chemother. 62. 10.1128/AAC.01221-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimuta K., Watanabe Y., Nakayama S., Morita-Ishihara T., Kuroki T., Unemo M., et al. (2015). Emergence and Evolution of Internationally Disseminated Cephalosporin-Resistant Neisseria Gonorrhoeae Clones from 1995 to 2005 in Japan. BMC. Infect. Dis. 15, 378. 10.1186/s12879-015-1110-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S. N., Marrazzo J., Batteiger B. E., Hook E. W., Seña A. C., Long J., et al. (2018). Single-dose Zoliflodacin (ETX0914) for Treatment of Urogenital Gonorrhea. N. Engl. J. Med. 379, 1835–1845. 10.1056/NEJMoa1706988 [DOI] [PubMed] [Google Scholar]
- Taylor S. N., Morris D. H., Avery A. K., Workowski K. A., Batteiger B. E., Tiffany C. A., et al. (2018). Gepotidacin for the Treatment of Uncomplicated Urogenital Gonorrhea: A Phase 2, Randomized, Dose-Ranging, Single-Oral Dose Evaluation. Clin. Infect. Dis. 67, 504–512. 10.1093/cid/ciy145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unemo M., Ahlstrand J., Sánchez-Busó L., Day M., Aanensen D., Golparian D., et al. (2021). High Susceptibility to Zoliflodacin and Conserved Target (GyrB) for Zoliflodacin Among 1209 Consecutive Clinical Neisseria Gonorrhoeae Isolates from 25 European Countries, 2018. J. Antimicrob. Chemother. 76, 1221–1228. 10.1093/jac/dkab024 [DOI] [PubMed] [Google Scholar]
- Unemo M. (2015). Current and Future Antimicrobial Treatment of Gonorrhoea - the Rapidly Evolving Neisseria Gonorrhoeae Continues to Challenge. BMC. Infect. Dis. 15, 364. 10.1186/s12879-015-1029-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unemo M., Lahra M. M., Cole M., Galarza P., Ndowa F., Martin I., et al. (2019). World Health Organization Global Gonococcal Antimicrobial Surveillance Program (WHO GASP): Review of New Data and Evidence to Inform International Collaborative Actions and Research Efforts. Sex. Health 16, 412–425. 10.1071/SH19023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unemo M., Ringlander J., Wiggins C., Fredlund H., Jacobsson S., Cole M. (2015). High In Vitro Susceptibility to the Novel Spiropyrimidinetrione ETX0914 (AZD0914) Among 873 Contemporary Clinical Neisseria Gonorrhoeae Isolates from 21 European Countries from 2012 to 2014. Antimicrob. Agents Chemother. 59, 5220–5225. 10.1128/AAC.00786-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unemo M., Lahra M. M., Escher M., Eremin S., Cole M. J., Galarza P., et al. (2021). WHO Global Antimicrobial Resistance Surveillance for Neisseria Gonorrhoeae 2017-18: a Retrospective Observational Study. Lancet Microbe. 10.1016/S2666-5247(21)00171-3 [DOI] [PubMed] [Google Scholar]
- van der Veer B. M. J. W., Hoebe C. J. P. A., Dukers-Muijrers N. H. T. M., van Alphen L. B., Wolffs P. F. G. (2020). Men and Women Have Similar Neisseria Gonorrhoeae Bacterial Loads: a Comparison of Three Anatomical Sites. J. Clin. Microbiol. 58, e01171–20. 10.1128/JCM.01171-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent L. R., Kerr S. R., Tan Y., Tomberg J., Raterman E. L., Dunning Hotopp J. C., et al. (2018). In Vivo-Selected Compensatory Mutations Restore the Fitness Cost of Mosaic penA Alleles that Confer Ceftriaxone Resistance in Neisseria Gonorrhoeae. mBio 9. 10.1128/mBio.01905-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wi T., Lahra M. M., Ndowa F., Bala M., Dillon J. R., Ramon-Pardo P., et al. (2017). Antimicrobial Resistance in Neisseria Gonorrhoeae: Global Surveillance and a Call for International Collaborative Action. Plos. Med. 14, e1002344. 10.1371/journal.pmed.1002344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2015). Global Action Plan on Antimicrobial Resistance. Available at: http://www.who.int/antimicrobial-resistance/publications/global-action-plan/en/ . [DOI] [PubMed]
- World Health Organization (2012). Global Action Plan to Control the Spread and Impact of Antimicrobial Resistance in Neisseria Gonorrhoeae. Available at: http://www.who.int/reproductivehealth/publications/rtis/9789241503501/en/ .
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic flow chart summarizing the main experiments of the zoliflodacin-susceptible Neisseria gonorrhoeae SE600/18 parent strain (with a GyrB S467N mutation) and the zoliflodacin-resistant N. gonorrhoeae SE600/18-D429N mutant (with GyrB S467N plus GyrB D429N selected in the Hollow Fiber Infection Model (HFIM)).
Predicted-Observed regressions for zoliflodacin concentrations, total Neisseria burden and resistant bacterial burden, respectively for the pre-Bayesian regression (A–C) and for the Bayesian regressions (D–F) for the clinical zoliflodacin-susceptible Neisseria gonorrhoeae SE600/18 strain with a GyrB S467N amino acid substitution.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: National Center for Biotechnology Information (NCBI) BioProject database under accession numbers PRJEB50904 and ERP135503.