Summary
Background
Atherosclerotic plaque quantification from coronary CT angiography (CCTA) enables accurate assessment of coronary artery disease burden and prognosis. We sought to develop and validate a deep learning system for CCTA-derived measures of plaque volume and stenosis severity.
Methods
This international, multicentre study included nine cohorts of patients undergoing CCTA at 11 sites, who were assigned into training and test sets. Data were retrospectively collected on patients with a wide range of clinical presentations of coronary artery disease who underwent CCTA between Nov 18, 2010, and Jan 25, 2019. A novel deep learning convolutional neural network was trained to segment coronary plaque in 921 patients (5045 lesions). The deep learning network was then applied to an independent test set, which included an external validation cohort of 175 patients (1081 lesions) and 50 patients (84 lesions) assessed by intravascular ultrasound within 1 month of CCTA. We evaluated the prognostic value of deep learning-based plaque measurements for fatal or non-fatal myocardial infarction (our primary outcome) in 1611 patients from the prospective SCOT-HEART trial, assessed as dichotomous variables using multivariable Cox regression analysis, with adjustment for the ASSIGN clinical risk score.
Findings
In the overall test set, there was excellent or good agreement, respectively, between deep learning and expert reader measurements of total plaque volume (intraclass correlation coefficient [ICC] 0·964) and percent diameter stenosis (ICC 0·879; both p<0·0001). When compared with intravascular ultrasound, there was excellent agreement for deep learning total plaque volume (ICC 0·949) and minimal luminal area (ICC 0·904). The mean per-patient deep learning plaque analysis time was 5·65 s (SD 1·87) versus 25·66 min (6·79) taken by experts. Over a median follow-up of 4·7 years (IQR 4·0–5·7), myocardial infarction occurred in 41 (2·5%) of 1611 patients from the SCOT-HEART trial. A deep learning-based total plaque volume of 238·5 mm3 or higher was associated with an increased risk of myocardial infarction (hazard ratio [HR] 5·36, 95% CI 1·70–16·86; p=0·0042) after adjustment for the presence of deep learning-based obstructive stenosis (HR 2·49, 1·07–5·50; p=0·0089) and the ASSIGN clinical risk score (HR 1·01, 0·99–1·04; p=0·35).
Interpretation
Our novel, externally validated deep learning system provides rapid measurements of plaque volume and stenosis severity from CCTA that agree closely with expert readers and intravascular ultrasound, and could have prognostic value for future myocardial infarction.
Introduction
Coronary CT angiography (CCTA) is a robust first-line test for the evaluation of coronary artery stenosis severity.1 When integrated into clinical decision making, CCTA guides the use of preventative therapies, improves eventfree survival, and enhances the diagnostic yield of invasive coronary angiography (ICA).2–4 Beyond assessment of stenosis severity, CCTA also enables non-invasive, wholeheart quantification of atherosclerosis. Advances in CT technology allow for semi-automated measurements of coronary atherosclerotic plaque with high accuracy when compared with intravascular ultrasound.5–8 CCTA-derived plaque volumes have shown prognostic value for lesion-specific and patient-level risk of acute coronary syndrome.9,10 More recently, low-attenuation plaque burden quantified from CCTA was shown to be the strongest independent predictor of myocardial infarction in patients from the landmark SCOT-HEART (Scottish Computed Tomography of the HEART) trial.11 However, semi-automated plaque quantification is time consuming and requires a high level of human expertise.
Artificial intelligence (AI) algorithms are increasingly being applied to CCTA to improve the efficiency and accuracy of image analysis, demonstrating high performance when compared with expert readers.12 Deep learning is a form of AI that uses artificial neural networks to generate automated predictions directly from image data.12
We aimed to develop and externally validate a novel deep learning system for CCTA-derived measurements of plaque volume and stenosis severity, and to evaluate the diagnostic performance of deep learning against that of expert readers, ICA, and intravascular ultrasound. We also aimed to assess the predictive value of deep learning-enabled atherosclerotic plaque quantification for risk of future myocardial infarction in a subanalysis of the SCOT-HEART trial.
Methods
Study population
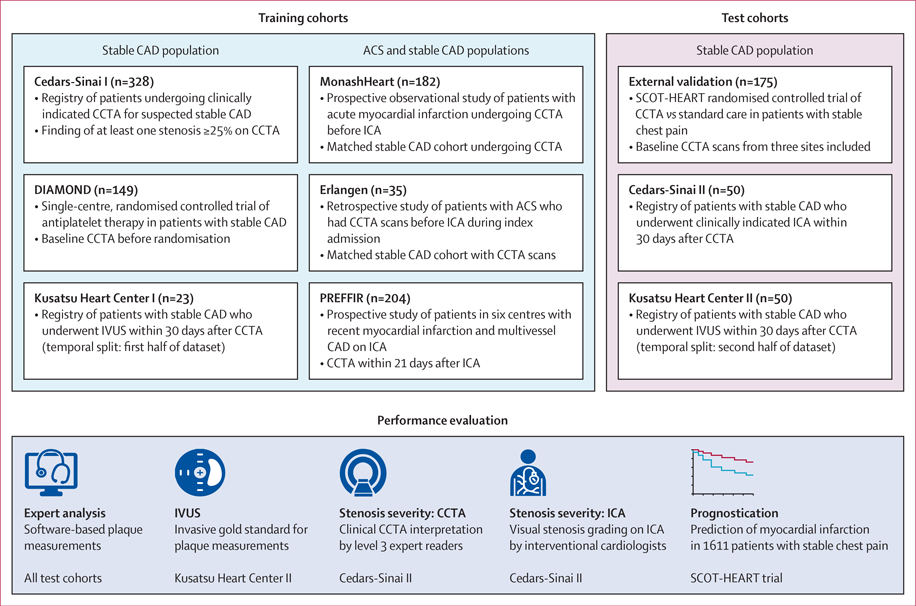
This international, multicentre study included nine cohorts (from 11 sites) split into training and test sets (figure 1). Sites were in the USA, UK, Germany, Australia, and Japan. Data were retrospectively collected on patients who underwent CCTA between Nov 18, 2010, and Jan 25, 2019. We excluded patients with CCTA scans of poor image quality that were deemed uninterpretable by expert readers. The training set consisted of six cohorts (921 patients, 5045 lesions), including a prospective observational study (MonashHeart),13 a randomised controlled trial (DIAMOND), and registries of patients with stable CAD (Cedars-Sinai I and Kusatsu Heart Center I).14 The clinical spectrum of coronary artery disease ranged from stable angina to acute myocardial infarction to the convalescent stage post-myocardial infarction. The test set, with unseen data, comprised an external validation cohort of 175 patients (1081 lesions) from the SCOT-HEART trial, and 100 patients with stable coronary artery disease (820 lesions) who had paired CCTA and invasive imaging data available in 30-day intervals (denoted as Cedars-Sinai II and Kusatsu Heart Center II). The SCOT-HEART trial was chosen for external validation because no sites from this trial were included in the training set. Figure 1 provides an overview of the training and test sets and the evaluation steps. Clinical characteristics of the whole study population are shown in the appendix (p 4).
Figure 1: Overview of the training and test cohorts and implemented evaluation steps.
The deep learning system was trained in 921 patients (5045 lesions) and applied to an independent test set of 275 patients (1901 lesions). The performance of deep learning for quantification of plaque volume and stenosis severity from CCTA was evaluated against expert readers and IVUS. The prognostic value of deep learning-based plaque measurements was evaluated in 1611 patients with stable chest pain from the SCOT-HEART trial. ACS=acute coronary syndrome. CAD=coronary artery disease. CCTA=coronary CT angiography. DIAMOND=Dual Antiplatelet Therapy to Reduce Myocardial Injury. ICA=invasive coronary angiography. IVUS=intravascular ultrasound. PREFFIR=Prediction of Recurrent Events with 18F-Fluorid. SCOT-HEART=Scottish Computed Tomography of the Heart.
This study was approved by the Cedars-Sinai Medical Center Institutional Review Board. The collection of patient data with written informed consent in each cohort was approved by the local ethics board at each institution.
Image acquisition
CCTA was performed at all sites using CT scanners with at least 64 detector rows, in accordance with the Society of Cardiovascular Computed Tomography (SCCT) guidelines.15 CT scan parameters for each site are given in the appendix (p 4). The Cedars-Sinai II cohort underwent clinically indicated ICA according to a standard protocol.16 In Kusatsu Heart Center II, intravascular ultrasound was performed before percutaneous coronary intervention in a standard fashion with a 40-MHz imaging catheter (Boston Scientific or Terumo). The imaging catheter was advanced beyond the distal portion of the target lesion for percutaneous coronary intervention, and automated pullback was performed at a speed of 0·5 mm/s.6
Image analysis
Clinical CCTA interpretation
In the Cedars-Sinai cohorts, two to three advanced imaging cardiologists who were SCCT-accredited level 3 readers performed joint clinical interpretation of CCTA images. The readers were aware of patients’ clinical data; however, no quantitative plaque analysis or invasive coronary imaging was done before CCTA interpretation. Each coronary segment that measured at least 1·5 mm was visually assessed for maximal diameter stenosis according to the 18-segment SCCT model of the coronary tree17 and Coronary Artery Disease Reporting and Data System (CAD-RADS) for categorisation of stenosis severity: 0 (0%), 1 (1–24%), 2 (25–49%), 3 (50–69%), 4 (70–99%), and 5 (100%).18 Per-vessel and per-patient CAD-RADS categories were assigned based on the highest-grade stenosis.
Expert plaque measurements from CCTA
Quantitative plaque analysis was performed using semiautomated research software (Autoplaque version 2.5; Cedars-Sinai Medical Center, Los Angeles, CA, USA)5,6 by expert readers with 3–8 years of experience who were unaware of clinical CCTA interpretation and invasive imaging findings (appendix p 2). Interobserver variability was assessed in a random sample of 100 patients (234 lesions) from the test set, between two experts (PM [3 years of experience] and HM [5 years of experience]) who independently performed plaque analysis and were unaware of each other’s measurements.
ICA stenosis interpretation
The SCCT 18-segment coronary tree17 and CAD-RADS categorisation of stenosis severity18 were applied to ICA to align with the same model used for CCTA. In the Cedars-Sinai II cohort, two interventional cardiologists (with 10–15 years of experience) who were unaware of CCTA findings performed visual assessment of maximal diameter stenosis in each coronary segment using two orthogonal views. An observer (AL) with 8 years of CCTA experience, who was unaware of the results of ICA and CCTA stenosis interpretation, performed co-registration of lesions between ICA and CCTA by comparing coronary segment coding and using vessel branch points as fiduciary landmarks. Per-vessel and per-patient CAD-RADS categories were assigned based on the highest-grade stenosis.
Intravascular ultrasound
Intravascular ultrasound images were analysed using computerised planimetry (EchoPlaque version 4.0.27; Indec Systems, Mountain View, CA, USA)6 by independent experienced observers with 3–5 years of experience who were unaware of CCTA findings (appendix p 2).
Deep learning plaque measurements from CCTA
We implemented our deep learning model with a novel architecture—the hierarchical convolutional long shortterm memory (ConvLSTM) network—using the PyTorch library to segment the coronary arteries. To enforce structural consistency, coronary segmentation was performed in a multitask approach for (1) the vessel wall and (2) lumen and plaque components (appendix p 10). The input to the network consisted of CCTA vessel cross-sections with a 0·3-mm slice thickness and 20-mm field of view around the predefined coronary centerline. The ground truth consisted of expert reader coronary segmentations reconstructed as straightened views and contained two labels (non-coronary structures and vessel wall) for task 1, and three labels (non-coronary structures, lumen, and plaque) for task 2. The hierarchical ConvLSTM network had two branches, each containing a feature extractor and segmentation head19 (appendix p 11). The first branch used a ConvLSTM to extract features from the current cross-section and five adjacent sections on either side. A ConvLSTM is a recurrent convolutional neural network that captures spatiotemporal information by learning about dependencies between sequential images;20 in this case, vessel cross-sections. The second branch used a DenseNet block to extract features from the current vessel cross-section. The segmentation head in both branches performed semantic segmentation using convolutional layers, batch normalisation, and a leaky rectified linear unit. The outputs of both branches were combined using an attention head to enable generation of a probability score (using a softmax function) which classified each voxel as belonging to a specific label.
To train the deep learning model, the training set was further randomly split into training (80%) and internal validation (20%) datasets (appendix p 2). The performance and computation speed of the deep learning model was evaluated using both a Nvidia GeForce RTX 3090 graphics processing unit and an Intel Core i7–6800K central processing unit. Additionally, we compared the performance and computation speed of our hierarchical ConvLSTM network against U-Net, a mainstream image segmentation method. Our network achieved a significantly higher Dice coefficient than U-Net for the vessel wall (0·94 vs 0·83; p<0·0001) and for lumen and plaque (0·90 vs 0·83; p<0·0001). The mean computation speed for our network was 0·1627 s with a graphics processing unit and 4·3 s with central processing units, compared with 0·4892 s and 14·4 s, respectively, for U-Net. The total cumulative memory consumption for our network was 19·0 gigabytes using the graphics processing unit, compared with a U-Net consumption of 21·6 gigabytes.
Given that coronary artery centreline extraction is a pre-processing step for our deep learning system, we assessed the reproducibility of deep learning-based plaque measurements in a random sample of 30 patients (148 lesions) from the test set with centrelines created by two independent experts (PM [3 years of experience] and AR [5 years of experience]).
Prognostic validation
Deep learning-based plaque measurements were performed in patients with stable chest pain from the CCTA group of the SCOT-HEART randomised controlled trial.3 The prognostic value of expert plaque measurements in this cohort has been previously published.11 Of 1778 patients who underwent CCTA, 1611 scans were available for deep learning plaque analysis. These included the 175 scans from the external validation cohort. Plaque volume and burden measurements for the entire coronary tree were summed on a per-patient level. Cardiovascular risk was calculated using the ASSIGN clinical risk score.21
The primary outcome for this subanalysis was the occurrence of fatal or non-fatal myocardial infarction. Outcome data were obtained electronically from Data Research and Innovation Services (National Health Service, Scotland) and were verified by review of electronic health records when required.3 Follow-up was administratively censored as of Jan 1, 2018, for patients without an event.
Statistical analysis
The performance of deep learning for per-lesion level plaque measurements was evaluated in the test set. Agreement between deep learning and expert readers or intravascular ultrasound was assessed using the intraclass correlation coefficient (ICC) and Bland-Altman analysis. Correlation was evaluated with Spearman’s rank correlation coefficient. Differences between deep learning and expert plaque measurements were assessed using the Wilcoxon rank-sum test. Cohen’s kappa coefficient (κ) was used to assess per-vessel and per-patient agreement in CAD-RADS categorisation between CCTA-based deep learning and expert interpretation of CCTA and ICA. The diagnostic performance of deep learning for detecting significant stenosis at two thresholds (maximal diameter stenosis ≥50% and ≥70%) was evaluated against these expert reference standards using accuracy, sensitivity, specificity, and positive and negative predictive values.
The predictive value of deep learning-based plaque measurements for fatal or non-fatal myocardial infarction in patients from the SCOT-HEART trial was assessed using Cox proportional hazards regression and presented graphically using Kaplan-Meier cumulative incidence curves. Univariable analysis was performed for the presence of deep learning-based obstructive stenosis (≥50%). To generate distinct clinical risk groups, we determined the optimum cutoff for total plaque volume to identify patients at increased risk of myocardial infarction using the Youden’s J statistic (sensitivity+specificity–1) on receiver operating characteristic (ROC) curve analysis. At this cutoff, we also calculated the area under the ROC curve (AUROC), positive predictive value, and negative predictive value for the discrimination of myocardial infarction. We tested the prognostic value of deep learning-based total plaque volume (as a dichotomous variable) using multivariable Cox regression analysis, with adjustment for the presence of deep learning-based obstructive stenosis and the ASSIGN clinical risk score. We performed ROC curve and univariable Cox regression analyses for deep learning-based low-attenuation plaque burden, given the strong prognostic value of this metric established in our previous work.11
We used Stata 14.0 (StataCorp, College Station, TX, USA) and Python 3.7.0, with the SciPy package, for analyses. A two-sided p value of <0·05 indicated statistical significance.
Role of the funding source
The funder had no role in the study design, data collection, data analysis, data interpretation, writing of the report, or the decision to submit for publication.
Results
The mean per-patient deep learning plaque analysis time was 5·65 s (SD 1·87) when computation was performed using a graphics processing unit and 3·82 min (0·77) with the use of a central processing unit. The mean analysis time taken by experts was 25·66 min (6·79) per patient.
In the overall test set, there was excellent agreement between deep learning and expert readers for volumes of total plaque (ICC 0·964, 95% CI 0·960–0·967), calcified plaque (0·945, 0·939–0·950), and noncalcified plaque (0·938, 0·932–0·944; all p<0·0001; table). Agreement was good for low-attenuation plaque volume (ICC 0·810, 0·786–0·831) and quantitative diameter stenosis (0·879, 0·863–0·895; both p<0·0001). Case examples of deep learning plaque segmentation are shown in figure 2.
Table:
Performance of deep learning versus expert plaque measurements in the test set (1901 lesions)
| ICC (95% CI) | Spearman correlation | |
|---|---|---|
|
| ||
| Total plaque volume | 0·964 (0·960—0·967) | 0·922 |
| Noncalcified plaque volume | 0·938 (0·932–0·944) | 0.906 |
| Calcified plaque volume | 0·945 (0·939–0·950) | 0·904 |
| Low-attenuation plaque volume | 0·810 (0·786–0·831) | 0.798 |
| Vessel volume | 0·992 (0·991–0·993) | 0·988 |
| Diameter stenosis | 0·879 (0·863–0·895) | 0·847 |
| Total plaque burden | 0·818 (0·796–0·838) | 0·788 |
| Noncalcified plaque burden | 0·813 (0·792–0·833) | 0·786 |
| Calcified plaque burden | 0·905 (0.895–0·914) | 0·857 |
| Low-attenuation plaque burden | 0·801 (0·781–0·837) | 0·772 |
ICC=intraclass correlation coefficient.
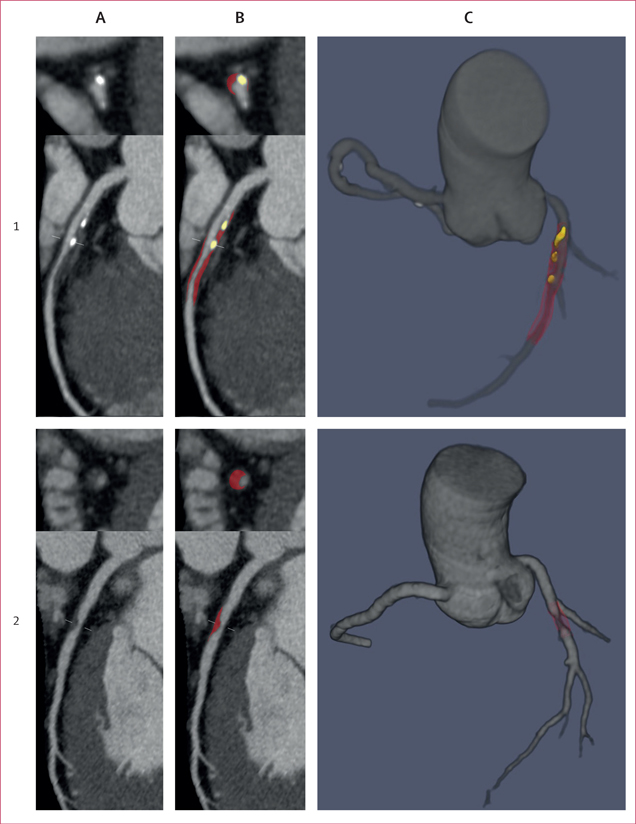
Figure 2. Case examples of deep learning plaque segmentation.
(A) Curved multiplanar reformation coronary CT angiography images showing lesions in the proximal-to-mid left anterior descending artery (1) and the mid left anterior descending artery (2). (B) Deep learning segmentation of calcified plaque (yellow) and noncalcified plaque (red). (C) Three-dimensional rendered view of the coronary tree showing deep learning plaque segmentation in the individual analysed segments. All lesions in each vessel were analysed by deep learning and measurements summed on a per-patient level.
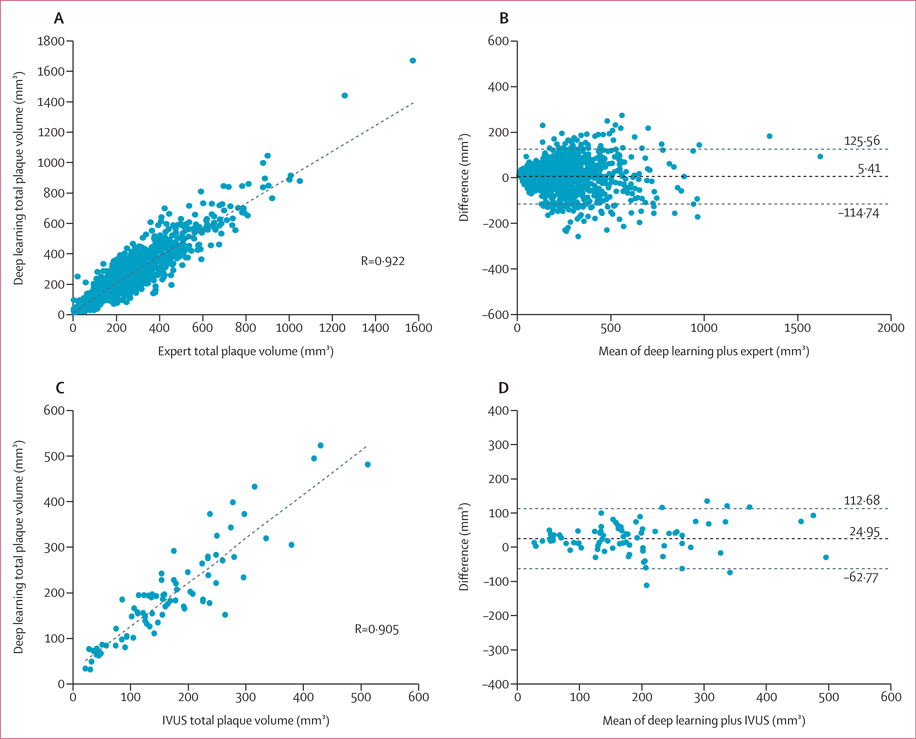
There was strong correlation between deep learning and expert measurements of total plaque volume in the overall test set (r=0·922, p<0·0001; figure 3A). On Bland-Altman analysis, the mean difference in total plaque volume between deep learning and experts was 5·41 mm3 (95% limits of agreement −114·74 to 125·56; figure 3B). Bland-Altman plots of noncalcified and calcified plaque volume measurements in the test set are shown in the appendix (p 12). Similar results were observed for deep learning versus expert measurements of total, noncalcified, and calcified plaque volume in the external validation cohort (appendix pp 6, 13).
Figure 3: Total plaque volume measured by deep learning versus expert readers and IVUS in the test set.
Correlation (A) and Bland-Altman (B) plots comparing total plaque volume measured by deep learning versus expert readers in 1901 lesions from the overall test set. Correlation (C) and Bland-Altman (D) plots of total plaque volume measured by deep learning versus IVUS in 84 lesions. Difference refers to deep learning – expert (B) and deep learning – IVUS (D). Horizontal lines refer to mean difference and 95% limits of agreement. IVUS=intravascular ultrasound.
In 100 patients from the test set, there was excellent agreement between two expert readers for volumes of all plaque components and quantitative diameter stenosis (ICC 0·915–0·973; all p<0·0001; appendix p 7). Bland-Altman analysis showed a mean difference in total plaque volume of 23·5 mm3 (95% limits of agreement −156·5 to 203·4).
There was excellent reproducibility of deep learning-based plaque analysis when applied to a subset of coronary artery centrelines derived from two different expert readers, with ICCs for plaque volumes and quantitative diameter stenosis ranging from 0·975 to 1 (all p<0·0001; appendix p 7).
Deep learning-based analysis had excellent agreement and correlation with intravascular ultrasound for measurements of total plaque volume (ICC 0·949, 95% CI 0·921–0·967, r=0·905; figure 3C) and minimal luminal area (ICC 0·904, 0·851–0·938, r=0·840; all p<0·0001). On Bland-Altman analysis, the mean difference in total plaque volume was 24·95 mm3 (95% limits of agreement −62·77 to 112·68; figure 3D).
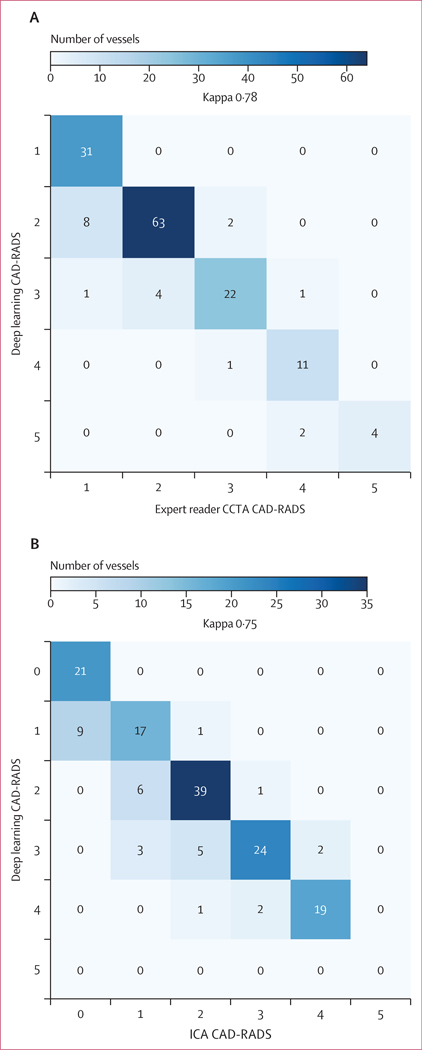
There was CAD-RADS categorical agreement between deep learning and expert CCTA interpretation in 131 (87%) of 150 vessels; agreement within one category (on either side of the correct category) was seen in 149 (99%) vessels (figure 4A). Cohen’s κ coefficient was 0·78. The most frequent disagreement occurred with deep learning CAD-RADS 2 and expert CAD-RADS 1 (eight vessels, 5%). At the patient level, CAD-RADS categorical agreement between deep learning and experts was strong (κ=0·81; appendix p 14), and there was 100% agreement within one CAD-RADS category. The diagnostic performance of deep learning versus expert CCTA interpretation for significant stenosis is shown in the appendix (p 9). Deep learning exhibited a per-patient sensitivity of 100·0% (95% CI 69·2–100·0) and specificity of 97·5% (86·8–99·9) for detecting stenosis of 70% or greater.
Figure 4: Per-vessel CAD-RADS categorisation by deep learning versus expert readers and invasive coronary angiography.
Confusion matrices of deep learning versus expert readers (A; 150 vessels) and ICA (B; 150 vessels) in the Cedars-Sinai cohorts for the categorisation of stenosis severity according to CAD-RADS. CAD-RADS=Coronary Artery Disease Reporting and Data System. CCTA=coronary CT angiography. ICA=invasive coronary angiography.
When comparing CCTA-based deep learning and ICA, there was CAD-RADS categorical agreement in 120 (80%) of 150 vessels and agreement within one category in 146 (97%) vessels (figure 4B); Cohen’s κ was 0·75. Per-patient CAD-RADS categorical agreement was strong (κ=0·80; appendix p 14). The diagnostic performance of CCTA-based deep learning versus ICA for significant stenosis is shown in the appendix (p 9). Deep learning had a sensitivity of 90·0% (95% CI 68·3–98·8) and negative predictive value of 93·6% (79·5–98·2) for detecting stenosis of 70% or greater at the patient level.
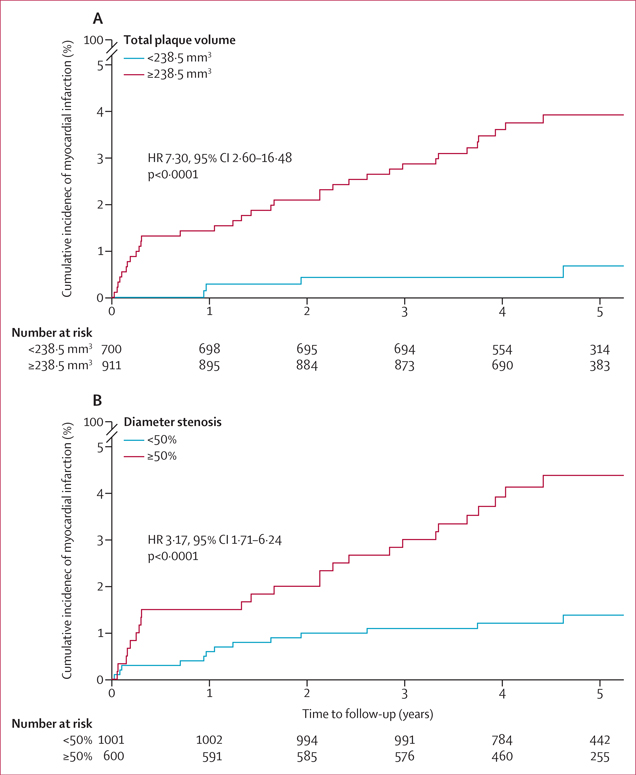
Over a median follow-up of 4·7 years (IQR 4·0–5·7), the primary outcome of fatal or non-fatal myocardial infarction occurred in 41 (2·5%) of 1611 patients from the SCOT-HEART trial. On ROC curve analysis, the optimum cutoff for total plaque volume was 238·5 mm3, which yielded an AUROC of 0·68 (95% CI 0·65–0·71), sensitivity of 90·2% (76·9–97·3), specificity of 44·3% (41·9–46·8), positive predictive value of 4·1% (3·7–4·5), and negative predictive value of 99·4% (98·6–99·8) for the discrimination of myocardial infarction. Patients with a total plaque volume of 238·5 mm3 or higher had 7-times greater risk of myocardial infarction than did patients with total plaque volume of less than 238·5 mm3 (unadjusted hazard ratio [HR] 7·30, 95% CI 2·60–16·48, p<0·0001; figure 5A). Deep learning-based obstructive stenosis (≥50%) exhibited an AUROC of 0·70 (95% CI 0·68–0·73), sensitivity of 65·9% (49·4–79·9), specificity of 63·5% (61·1–65·9), positive predictive value of 4·5% (3·6–5·6), and negative predictive value of 98·6% (97·9–99·1) for myocardial infarction discrimination. In univariable Cox regression analysis, deep learning-based obstructive stenosis was associated with an increased risk of myocardial infarction (unadjusted HR 3·17, 95% CI 1·71–6·24; p<0·0001; figure 5B). In multivariable Cox regression analysis, a total plaque volume of 238·5 mm3 or higher was associated with a greater risk of myocardial infarction (HR 5·36, 1·70–16·86; p=0·0042) after adjustment for deep learning-based obstructive stenosis (HR 2·49, 1·07–5·50; p=0·0089) and the ASSIGN clinical risk score (HR 1·01, 0·99–1·04; p=0·35). The optimum cutoff for low-attenuation plaque burden was 4%; patients with a low-attenuation plaque burden of 4% or higher had a 2·5-times increased risk of myocardial infarction compared with patients with total plaque volume of less than 238·5 mm3 (unadjusted HR 2·51, 1·38–4·73; p=0·0023; appendix p 15).
Figure 5: Prognostic value of deep learning-based total plaque volume and stenosis for myocardial infarction.
Kaplan-Meier cumulative incidence curves of fatal or non-fatal myocardial infarction in patients from the SCOT-HEART trial stratified by deep learning-based total plaque volume above or below 238·5 mm3, the optimum cutoff determined by receiver operating characteristic curve analysis (A) and by the presence of deep learning-based obstructive (≥50%) or non-obstructive (<50%) stenosis (B). HR=hazard ratio.
Discussion
In this international multicentre study, we developed and externally validated a novel deep learning system for rapid and automated plaque quantification from CCTA. To our knowledge, the present analysis is the first to show that: (1) deep learning has excellent agreement and correlation with expert readers and intravascular ultrasound for measurements of plaque volume and luminal stenosis; (2) CAD-RADS stenosis categorisation by deep learning agrees closely with expert CCTA interpretation and with ICA; and (3) deep learning-based plaque and stenosis measurements can predict risk of future myocardial infarction in a large cohort of patients with stable chest pain.
Several studies have described AI approaches for the automated detection and classification of coronary atherosclerotic lesions on CCTA. Kang and colleagues22 trained a supervised machine learning algorithm to detect diameter stenosis of 25% or greater with high sensitivity (93%) and specificity (95%) when compared with consensus reading by experts. Zreik and colleagues23 used a multi-task deep convolutional neural network to classify plaque composition as calcified, noncalcified, or mixed in 163 patients, achieving an accuracy of 80% when compared with experts. In 2021, Choi and colleagues24 used a series of convolutional neural networks to perform CAD-RADS stenosis categorisation in 232 patients, with close agreement seen with the consensus of expert readers (κ coefficients of 0·72 at the vessel level and 0·81 at the patient level). In previous work, we leveraged an M-Net-based convolutional neural network for automated measurements of minimal luminal area and percentage diameter stenosis in 156 patients, yielding excellent correlation (r=0·984 and r=0·957, respectively) with manual expert annotations.25 The present analysis extends these findings through a novel ConvLSTM-based neural network that is the first to quantify accurately coronary stenosis and volumes of all atherosclerotic plaque components. To our knowledge, this analysis is the largest study of AI-enabled CCTA plaque analysis to date, and included patients from five different countries who were scanned using multiple different CT scanners and protocols.
Our deep learning system demonstrated excellent agreement with expert CCTA interpretation for categorisation of stenosis severity by CAD-RADS, which is the current clinical standard endorsed by multiple medical societies.18 Although visual estimation of luminal stenosis remains the cornerstone of CCTA reporting, reader experience and invasive angiographic training are crucial for grading stenosis with acceptable accuracy. Furthermore, disagreement over the presence or absence of obstructive coronary artery disease is common even among experienced expert readers.26 In the present analysis, deep learning achieved 99% agreement within one CAD-RADS category at the vessel level and 100% agreement at the patient level referenced by level 3 expert readers, reflecting a reasonable estimation of stenosis severity compared with clinical practice. Moreover, the 93·6% negative predictive value of the deep learning approach for ICA stenosis of 70% or greater at the patient level suggests that it might be a useful tool for excluding severe obstructive disease and reducing unnecessary invasive angiography. A deep learning system that rapidly and accurately quantifies coronary stenosis has the potential for integration into routine CCTA workflow, where it could function as a second reader and clinical decision support tool. By providing automated and objective results, deep learning could reduce interobserver variability and interpretative error among physicians. The system could also be used to pre-screen CCTA scans, flagging patients with obstructive disease who should be prioritised for reporting.
The paradigm for CCTA imaging has evolved beyond assessment of luminal stenosis to include the characterisation and quantification of atherosclerosis.27 Measurements of plaque volume and composition from CCTA are now feasible using various semi-automated research software, with strong correlations with intravascular ultrasound.5–8 However, current quantitative plaque analysis is time-consuming and requires substantial manual input from an expert reader,7,8 thus limiting its implementation into clinical practice. Our deep learning approach automates the most complex steps in the plaque analysis workflow—vessel wall contouring, setting adaptive Hounsfield unit thresholds for the various plaque components, and differentiating coronary from non-coronary structures—to provide a graphical three-dimensional output of plaque segmentation and a comprehensive list of quantitative parameters. Using a standard desktop computer, this approach showed excellent agreement with expert reader measurements and the gold standard of intravascular ultrasound, at a fraction of the analysis time taken by experts (3·82 min vs 25·66 min). Although quantitative plaque analysis remains investigational, continued advancements in hardware (eg, graphics processing units) and software (eg, cloud-based computing services) will improve the time and computational efficiency of our deep learning system, thus increasing its potential for future clinical application.
CCTA-derived overall and noncalcified plaque volumes have demonstrated predictive value for incident acute coronary syndrome and cardiac death.9,10,28 In a recent subanalysis of the prospective SCOT-HEART trial, low-attenuation plaque burden (% plaque-to-vessel volume) measured by expert readers was the single greatest predictor of incident myocardial infarction, after adjustment for the ASSIGN clinical risk score and presence of obstructive stenosis.11 The current study showed total plaque volume in the coronary tree quantified by deep learning to have strong and independent prognostic value for myocardial infarction in the SCOT-HEART cohort. We identified an optimum deep learning cutoff (≥238·5 mm3) above which there was a steep increase in the risk of events. This value is close to the expert-derived total plaque volume threshold (>179 mm3) which was shown in our previous study28 to stratify stable patients into high-risk versus low-risk groups for future cardiac death. Finally, we used deep learning to corroborate the prognostic cutoff of 4% for low-attenuation plaque burden established in our previous work,11 reflecting the close agreement between deep learning and expert readers for this metric.
This study has several limitations. First, data on patient race or ethnicity were not uniformly available for all sites; however, our training dataset drew from diverse and geographically distinct populations. Second, a pre-processing step for our deep learning-based coronary segmentation is the extraction of coronary artery centrelines. This task can currently be performed rapidly by technologists using semi-automated software. Once coronary centreline extraction is fully automated by our deep learning system in the near future, we will have a true end-to-end solution for plaque quantification. Third, although we showed the robust performance of deep learning across several different CT vendors and scan parameters, we excluded CCTA studies of poor image quality that were deemed uninterpretable by expert readers. Fourth, validation of the deep learning system against intravascular ultrasound was done in a relatively small number of patients; thus larger studies are required. Lastly, in the SCOT-HEART trial, ICA images were not available for the identification of culprit and non-culprit lesions in patients with myocardial infarction, precluding a lesion-level prognostic analysis. In future work, we aim to apply our deep learning system to CCTA scans of patients in whom culprit lesion precursors have been identified. Despite these limitations, our study represents the first validation of a deep learning approach for atherosclerotic quantification from CCTA using invasive reference standards, and is the first demonstration of the predictive value of deep learning-based plaque measurements for risk of cardiac events.
In summary, a novel, externally validated deep learning system provides rapid measurements of plaque volume and stenosis severity from CCTA that agree closely with expert readers and intravascular ultrasound and carry prognostic value for future myocardial infarction. This system has the potential for implementation into routine CCTA workflow as a clinical decision support tool.
Supplementary Material
Research in context.
Evidence before this study
Deep learning has the potential to improve the speed and diagnostic performance of coronary CT angiography (CCTA) image analysis by providing automated and objective results. We searched PubMed and Google Scholar for papers published between database inception and Jan 31, 2021, using the search terms (“deep learning” OR “machine learning” OR “artificial intelligence”) AND (“coronary computed tomography angiography” OR “coronary CT angiography”) AND (“plaque” OR “stenosis”), with no language restrictions. We identified 26 articles exploring the use of deep learning to assess coronary lesions on CCTA, most of which were small proof-of-concept studies. To date, no studies had performed external validation of a deep learning model for CCTA or compared deep learning-based plaque measurements with the invasive reference standard of intravascular ultrasound. Notably, the prognostic value of deep learning from CCTA remains unknown.
Added value of this study
In this international multicentre study, we developed and externally validated a novel deep learning system for rapid and automated plaque quantification from CCTA. To our knowledge, the present analysis is the first to show that deep learning has excellent agreement and correlation with expert readers and intravascular ultrasound for measurements of plaque volume and luminal stenosis, and that deep learning agrees closely with expert readers in their clinical grading of stenosis severity. We also show that deep learning-based plaque and stenosis measurements predict the risk of future myocardial infarction in a large cohort of patients with stable chest pain from the landmark SCOT-HEART (Scottish Computed Tomography of the Heart) trial.
Implications of all the available evidence
A deep learning system that rapidly and accurately quantifies coronary artery stenosis has the potential for integration into routine CCTA workflow, where it could function as a second reader and clinical decision support tool. By providing automated and objective results, deep learning could reduce interobserver variability and interpretative error among physicians. Deep learning-based plaque volume measurements have independent prognostic value for future cardiac events, and could enhance risk stratification in patients with stable chest pain who are undergoing CCTA.
Acknowledgments
This study was supported by grants from the National Heart, Lung, and Blood Institute, USA (1R01HL148787–01A1) and the Miriam & Sheldon G Adelson Medical Research Foundation.
Footnotes
Declaration of interests
SC, PJS, DD, and DSB received software royalties from Cedars-Sinai Medical Center outside of the current work. DSB, PJS, and DD hold a single patent (US8885905B2/WO2011069120A1, Method and System for Plaque Characterisation). KP reports a non-financial advisory role with Kardiollytics. KG reports grants or contracts from the Foundation for Polish Science and Polish Society of Cardiology. ACK reports partial effort support from the Doris Duke Charitable Foundation Grant. KK reports grants or contracts from The Society of Nuclear Medicine and Molecular Imaging Wagner-Torizuka Fellowship and Nihon University School of Medicine Alumni Association. ET is supported by a grant from the British Heart Foundation. SC reports financial support from the Miriam & Sheldon G Adelson Medical Research Foundation Grant. SJN reports grants or contracts from AstraZeneca, New Amsterdam Pharma, Amgen, Anthera, Eli Lilly, Esperion, Novartis, Cerenis, The Medicines Company, Resverlogix, InfraReDx, Roche, Sanofi-Regeneron, and LipoScience; and consulting fees from AstraZeneca, Amarin, Akcea, Eli Lilly, Anthera, Omthera, Merck, Takeda, Resverlogix, Sanofi-Regeneron, CSL Behring, Esperion, and Boehringer Ingelheim. MDw reports consulting fees from Novartis, Jupiter Bioventures, and Silence Therapeutics; and payment or honoraria from Pfizer and Novartis. DEN reports a grant from the British Heart Foundation and Wellcome Trust. MCW reports a grant from the British Heart Foundation; declares payment or honoraria from Cannon Medical Systems; and a leadership or fiduciary roles with British Society of Cardiovascular Imaging, Society of Cardiovascular Computed Tomography, and European Society of Cardiovascular Radiology. All other authors declare no competing interests.
Data sharing
Deidentified final results supporting this study are available for research purposes upon reasonable written request to the corresponding author. Access to such data is available from the date of publication and requires a Data Access Agreement, which is examined and approved by the ethics committees who approved this research. Details of the deep learning model used in this study are further described in the appendix (pp 10–11). The source code can be shared using a Creative Commons NC-ND 4.0 international license upon reasonable written request to the corresponding author and requires a data use agreement.
Contributor Information
Andrew Lin, Biomedical Imaging Research Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA; Monash Cardiovascular Research Centre, Victorian Heart Institute, Monash University, Melbourne, VIC, Australia; MonashHeart, Monash Health, Melbourne, VIC, Australia.
Nipun Manral, Biomedical Imaging Research Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Priscilla McElhinney, Biomedical Imaging Research Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Aditya Killekar, Division of Artificial Intelligence, Department of Medicine, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Hidenari Matsumoto, Division of Cardiology, Showa University School of Medicine, Tokyo, Japan.
Jacek Kwiecinski, Division of Artificial Intelligence, Department of Medicine, Cedars-Sinai Medical Center, Los Angeles, CA, USA; British Heart Foundation Centre for Cardiovascular Science, University of Edinburgh, Edinburgh, UK; Department of Interventional Cardiology and Angiology, Institute of Cardiology, Warsaw, Poland.
Konrad Pieszko, Division of Artificial Intelligence, Department of Medicine, Cedars-Sinai Medical Center, Los Angeles, CA, USA; Department of Interventional Cardiology, Collegium Medicum, University of Zielona Góra, Poland.
Aryabod Razipour, Biomedical Imaging Research Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Kajetan Grodecki, Biomedical Imaging Research Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA; Department of Cardiology, Medical University of Warsaw, Warsaw, Poland.
Caroline Park, Biomedical Imaging Research Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Yuka Otaki, Department of Imaging, Cedars-Sinai Medical Center, Los Angeles, CA, USA; Smidt Heart Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Mhairi Doris, British Heart Foundation Centre for Cardiovascular Science, University of Edinburgh, Edinburgh, UK.
Alan C Kwan, Department of Imaging, Cedars-Sinai Medical Center, Los Angeles, CA, USA; Smidt Heart Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Donghee Han, Department of Imaging, Cedars-Sinai Medical Center, Los Angeles, CA, USA; Smidt Heart Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Keiichiro Kuronuma, Department of Imaging, Cedars-Sinai Medical Center, Los Angeles, CA, USA; Smidt Heart Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Guadalupe Flores Tomasino, Department of Imaging, Cedars-Sinai Medical Center, Los Angeles, CA, USA; Smidt Heart Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Evangelos Tzolos, Department of Imaging, Cedars-Sinai Medical Center, Los Angeles, CA, USA; Smidt Heart Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA; British Heart Foundation Centre for Cardiovascular Science, University of Edinburgh, Edinburgh, UK.
Aakash Shanbhag, Division of Artificial Intelligence, Department of Medicine, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Markus Goeller, Department of Cardiology, Friedrich-Alexander Universität Erlangen-Nürnberg, Erlangen, Germany.
Mohamed Marwan, Department of Cardiology, Friedrich-Alexander Universität Erlangen-Nürnberg, Erlangen, Germany.
Heidi Gransar, Department of Imaging, Cedars-Sinai Medical Center, Los Angeles, CA, USA; Smidt Heart Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Balaji K Tamarappoo, Department of Imaging, Cedars-Sinai Medical Center, Los Angeles, CA, USA; Smidt Heart Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Sebastien Cadet, Department of Imaging, Cedars-Sinai Medical Center, Los Angeles, CA, USA; Smidt Heart Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Stephan Achenbach, Department of Cardiology, Friedrich-Alexander Universität Erlangen-Nürnberg, Erlangen, Germany.
Stephen J Nicholls, Monash Cardiovascular Research Centre, Victorian Heart Institute, Monash University, Melbourne, VIC, Australia; MonashHeart, Monash Health, Melbourne, VIC, Australia.
Dennis T Wong, Monash Cardiovascular Research Centre, Victorian Heart Institute, Monash University, Melbourne, VIC, Australia; MonashHeart, Monash Health, Melbourne, VIC, Australia.
Daniel S Berman, Department of Imaging, Cedars-Sinai Medical Center, Los Angeles, CA, USA; Smidt Heart Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Marc Dweck, British Heart Foundation Centre for Cardiovascular Science, University of Edinburgh, Edinburgh, UK.
David E Newby, British Heart Foundation Centre for Cardiovascular Science, University of Edinburgh, Edinburgh, UK.
Michelle C Williams, British Heart Foundation Centre for Cardiovascular Science, University of Edinburgh, Edinburgh, UK.
Piotr J Slomka, Division of Artificial Intelligence, Department of Medicine, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Damini Dey, Biomedical Imaging Research Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
References
- 1.Narula J, Chandrashekhar Y, Ahmadi A, et al. SCCT 2021 expert consensus document on coronary computed tomographic angiography: a report of the Society of Cardiovascular Computed Tomography. J Cardiovasc Comput Tomogr 2021; 15: 192–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williams MC, Hunter A, Shah ASV, et al. Use of coronary computed tomographic angiography to guide management of patients with coronary disease. J Am Coll Cardiol 2016; 67: 1759–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Newby DE, Adamson PD, Berry C, et al. Coronary CT angiography and 5-year risk of myocardial infarction. N Engl J Med 2018; 379: 924–33. [DOI] [PubMed] [Google Scholar]
- 4.Chang HJ, Lin FY, Gebow D, et al. Selective referral using CCTA versus direct referral for individuals referred to invasive coronary angiography for suspected CAD: a randomized, controlled, openlabel trial. JACC Cardiovasc Imaging 2019; 12: 1303–12. [DOI] [PubMed] [Google Scholar]
- 5.Dey D, Schepis T, Marwan M, Slomka PJ, Berman DS, Achenbach S. Automated three-dimensional quantification of noncalcified coronary plaque from coronary CT angiography: comparison with intravascular US. Radiology 2010; 257: 516–22. [DOI] [PubMed] [Google Scholar]
- 6.Matsumoto H, Watanabe S, Kyo E, et al. Standardized volumetric plaque quantification and characterization from coronary CT angiography: a head-to-head comparison with invasive intravascular ultrasound. Eur Radiol 2019; 29: 6129–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voros S, Rinehart S, Qian Z, et al. Coronary atherosclerosis imaging by coronary CT angiography: current status, correlation with intravascular interrogation and meta-analysis. JACC Cardiovasc Imaging 2011; 4: 537–48. [DOI] [PubMed] [Google Scholar]
- 8.Nalcanishi R, Motoyama S, Leipsic J, Budoff MJ. How accurate is atherosclerosis imaging by coronary computed tomography angiography? J Cardiovasc Comput Tomogr 2019; 13: 254–60. [DOI] [PubMed] [Google Scholar]
- 9.Versteylen MO, Kietselaer BL, Dagnelie PC, et al. Additive value of semiautomated quantification of coronary artery disease using cardiac computed tomographic angiography to predict future acute coronary syndrome. J Am Coll Cardiol 2013; 61: 2296–305. [DOI] [PubMed] [Google Scholar]
- 10.Chang HJ, Lin FY, Lee SE, et al. Coronary atherosclerotic precursors of acute coronary syndromes. J Am Coll Cardiol 2018; 71: 2511–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams MC, Kwiecinski J, Doris M, et al. Low-attenuation noncalcified plaque on coronary computed tomography angiography predicts myocardial infarction: results from the multicenter SCOT-HEART trial (Scottish Computed Tomography of the HEART). Circulation 2020; 141: 1452–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dey D, Slomka PJ, Leeson P, et al. Artificial intelligence in cardiovascular imaging: JACC state-of-the-art review. J Am Coll Cardiol 2019; 73: 1317–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin A, Kolossváry M, Yuvaraj J, et al. Myocardial infarction associates with a distinct pericoronary adipose tissue radiomic phenotype: a prospective case-control study. JACC Cardiovasc Imaging 2020; 13: 2371–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doris MK, Meah MN, Moss AJ, et al. Coronary 18F-fluoride uptake and progression of coronary artery calcification. Circ Cardiovasc Imaging 2020; 13: e011438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abbara S, Blanke P, Maroules CD, et al. SCCT guidelines for the performance and acquisition of coronary computed tomographic angiography: a report of the society of Cardiovascular Computed Tomography Guidelines Committee: endorsed by the North American Society for Cardiovascular Imaging (NASCI). J Cardiovasc Comput Tomogr 2016; 10: 435–49. [DOI] [PubMed] [Google Scholar]
- 16.Scanlon PJ, Faxon DP, Audet AM, et al. ACC/AHA guidelines for coronary angiography. A report of the American College of Cardiology/American Heart Association Task Force on practice guidelines (Committee on Coronary Angiography). Developed in collaboration with the Society for Cardiac Angiography and Interventions. J Am Coll Cardiol 1999; 33: 1756–824. [DOI] [PubMed] [Google Scholar]
- 17.Leipsic J, Abbara S, Achenbach S, et al. SCCT guidelines for the interpretation and reporting of coronary CT angiography: a report of the Society of Cardiovascular Computed Tomography Guidelines Committee. J Cardiovasc Comput Tomogr 2014; 8: 342–58. [DOI] [PubMed] [Google Scholar]
- 18.Cury RC, Abbara S, Achenbach S, et al. Coronary artery disease - reporting and data system (cad-rads): an expert consensus document of SCCT, ACR and NASCI: endorsed by the ACC. JACC Cardiovasc Imaging 2016; 9: 1099–113. [DOI] [PubMed] [Google Scholar]
- 19.Tao A, Sapra K, Catanzaro B. Hierarchical multi-scale attention for semantic segmentation. arXiv 2020; published online May 20. https://arxiv.org/abs/2005.10821 (preprint). [Google Scholar]
- 20.Shi X, Chen Z, Wang H, Yeung D-Y, Wong W-K, Woo W-C. Convolutional LSTM network: a machine learning approach for precipitation nowcasting. In: NIPS’15: proceedings of the 28th International Conference on Neural Information Processing Systems. Cambridge: MIT Press, 2015: 802–10. [Google Scholar]
- 21.Woodward M, Brindle P, Tunstall-Pedoe H. Adding social deprivation and family history to cardiovascular risk assessment: the ASSIGN score from the Scottish Heart Health Extended Cohort (SHHEC). Heart 2007; 93: 172–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang D, Dey D, Slomka PJ, et al. Structured learning algorithm for detection of nonobstructive and obstructive coronary plaque lesions from computed tomography angiography. J Med Imaging (Bellingham) 2015; 2: 014003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zreik M, van Hamersvelt RW, Wolterink JM, Leiner T, Viergever MA, Isgum I. A Recurrent CNN for automatic detection and classification of coronary artery plaque and stenosis in coronary CT angiography. IEEE Trans Med Imaging 2019; 38: 1588–98. [DOI] [PubMed] [Google Scholar]
- 24.Choi A, Marques H, Kumar V, et al. CT evaluation by artificial intelligence for atherosclerosis, stenosis and vascular morphology (CLARIFY): a multi-center, international study. J Cardiovasc Comput Tomogr 2021; 15: 470–76. [DOI] [PubMed] [Google Scholar]
- 25.Hong Y, Commandeur F, Cadet S, et al. Deep learning-based stenosis quantification from coronary CT angiography. Proc SPIE Int Soc Opt Eng 2019; 10949: 109492I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arbab-Zadeh A, Hoe J. Quantification of coronary arterial stenoses by multidetector CT angiography in comparison with conventional angiography methods, caveats, and implications. JACC Cardiovasc Imaging 2011; 4: 191–202. [DOI] [PubMed] [Google Scholar]
- 27.Williams MC, Earls JP, Hecht H. Quantitative assessment of atherosclerotic plaque, recent progress and current limitations. J Cardiovasc Comput Tomogr 2021; published online July 16. 10.1016/j.jcct.2021.07.001. [DOI] [PubMed] [Google Scholar]
- 28.Hell MM, Motwani M, Otaki Y, et al. Quantitative global plaque characteristics from coronary CT angiography for the prediction of future cardiac mortality during long-term follow-up. Eur Heart J Cardiovasc Imaging 2017; 18: 1331–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.