Abstract
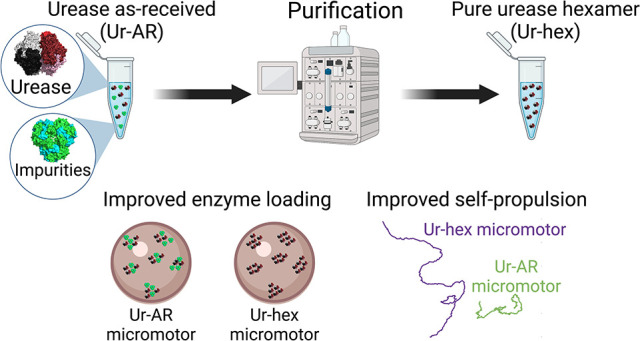
Enzyme-powered micro- and nanomotors make use of biocatalysis to self-propel in aqueous media and hold immense promise for active and targeted drug delivery. Most (if not all) of these micro- and nanomotors described to date are fabricated using a commercially available enzyme, despite claims that some commercial preparations may not have a sufficiently high degree of purity for downstream applications. In this study, the purity of a commercial urease, an enzyme frequently used to power the motion of micro- and nanomotors, was evaluated and found to be impure. After separating the hexameric urease from the protein impurities by size-exclusion chromatography, the hexameric urease was subsequently characterized and used to functionalize hollow silica microcapsules. Micromotors loaded with purified urease were found to be 2.5 times more motile than the same micromotors loaded with unpurified urease, reaching average speeds of 5.5 μm/s. After comparing a number of parameters, such as enzyme distribution, protein loading, and motor reusability, between micromotors functionalized with purified vs unpurified urease, it was concluded that protein purification was essential for optimal performance of the enzyme-powered micromotor.
Keywords: micromotors, enzyme, catalysis, self-propulsion, super-resolution microscopy, DLS
Enzyme-powered micro- and nanomotors (EMNMs) are particles that self-propel due to a chemical reaction catalyzed by an enzyme attached to the particle’s surface. Catalytic self-propulsion of micro–nanomotors has been demonstrated for a number of highly efficient enzymes, including catalase,1−7 urease,8−12 and lipase,13,14 as well as combinations of multiple enzymes, such as catalase and glucose oxidase.15−19 These microscopic sized motors have been intensely studied for their biomedical applications, where they show great potential to be used as active drug delivery vehicles for site-specific cancer therapies. Indeed, compared to their inorganic counterparts, enzyme-powered micro- and nanomotors are more biocompatible, as enzymes are highly specific to their bioavailable fuels, and thus produce fewer toxic byproducts.20−22 Although there has been demonstrative progress recently for enzyme-powered micro- and nanomotors reaching the clinic, there are still a number of biochemical and biophysical challenges associated with EMNM propulsion in biological fluids that have to be overcome in order for EMNM-based therapies to reach their full potential. Indeed, biological fluids tend to be highly viscous and have a high saline content, both of which affect EMNM self-propulsion.23−26
Overcoming these challenges is a high priority in the field of EMNMs. Fortunately, there has been a lot of incentive to understand the underlying fundamental aspects of EMNM motion in recent years, but improvements to the motors’ self-propulsion properties have mostly been focused on changing the shape and distribution of the enzyme on the surface of the particle. What is more, and to the best of our knowledge, all EMNMs reported in the literature have been fabricated using unprocessed, commercially supplied enzymes. As pointed out by Zhang and Hess,27 enzyme products bought commercially are often mixtures of isoenzymes with different molecular weights, which could have different kinetic properties. Seeing as most enzyme immobilization strategies for fabricating EMNMs are nonspecific (most of which rely on functionalizing the particles with a coupling agent that covalently bonds to surface lysines), more careful consideration should be given to analyzing the protein species that are present in the commercial enzyme product.
The study performed here seeks to investigate how enzyme purity affects the self-propulsion of EMNMs, using urease as an example. After determining that the urease reagent used by many groups to make EMNMs constitutes an impure mixture of proteins of different molecular weights, we have been able to isolate the pure urease hexamer using size-exclusion chromatography. This pure urease sample was then used to functionalize hollow silica microcapsules (HSMCs), and the resulting micromotors showed significantly enhanced self-propulsion compared to micromotors functionalized with unpurified urease. Micromotors functionalized with pure urease also demonstrated better reusability and propelled with less protein attached compared to their impure counterparts. The role of the impurities on self-propulsion was evaluated; we found that the impurities significantly hinder motor motility and speed. Finally, an asymmetric distribution of urease was found for both types of motors, ruling out the possibility that the enhanced propulsion could be due to differences in the enzyme distribution.
Results/Discussion
Urease Purification
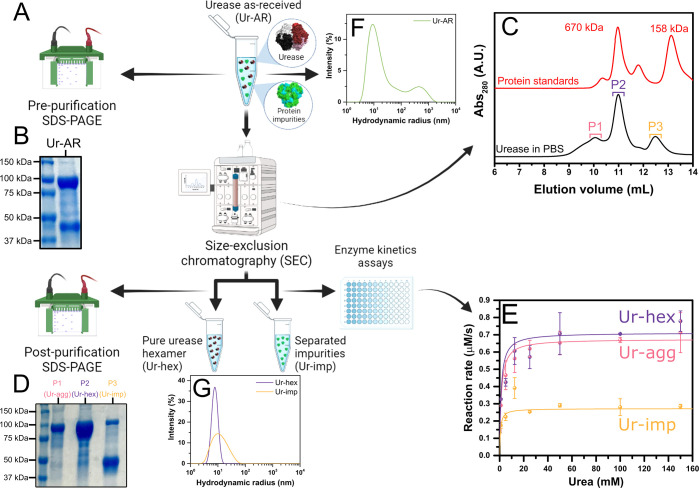
The commercial as-received urease type IX from Canavalia ensiformis (jack bean), supplied by the company Merck (previously Sigma-Aldrich), is reported to be composed of one major protein species with a molecular mass range of 440–480 kDa and two less abundant protein species with molecular mass ranges of 230–260 and 660–740 kDa.28,29 Although unspecified in the suppliers’ documentation, it is presumed that the species with a molecular mass of 440–480 kDa corresponds to the native hexameric conformation for jack bean urease, though it is widely reported to have a molecular weight of 540 kDa, equivalent to the combined sum of the six identical subunits of ∼90 kDa composing its structure.30 It is also not specified in the suppliers’ documentation whether there are any protein impurities found in the urease type IX preparation from Sigma-Aldrich nor whether the protein species of various molecular weights all possess enzymatic activity toward urea. To investigate this further, the urease type IX as-received (Ur-AR) in this study was analyzed by SDS-PAGE, and urease hexamer was subsequently purified by size-exclusion chromatography (Figure 1A).
Figure 1.
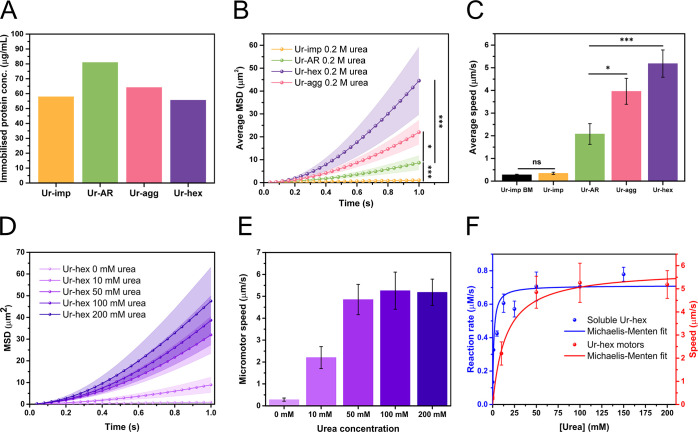
Purification of commercial urease type IX (Sigma-Aldrich) and characterization of postpurification products. (A) Scheme of the purification of urease as received (Ur-AR) by size-exclusion chromatography (SEC) and analysis of urease by SDS-PAGE before purification, as well as the SEC fractions (postpurification). (B) Reducing SDS-PAGE gel showing the protein composition of Ur-AR. (C) SEC chromatograms of the Ur-AR purification (with major peaks highlighted in blue, purple, and orange) and gel filtration protein standards (670 and 158 kDa protein peaks correspond to thyroglobulin and bovine γ-globulin, respectively). (D) Reducing SDS-PAGE gel showing the fractions corresponding to the three major SEC peaks found in C. (E) Michaelis–Menten fits of the recorded reaction rates for urease activity from the fractions corresponding to SEC peaks P1, P2, and P3. (F, G) Characterization by dynamic light scattering of urease samples before (F) and after (G) purification.
Under reducing conditions, two major proteins species were found represented by two distinct bands on the polyacrylamide gel: one broad band at the expected molecular weight for monomeric urease (∼91 kDa) and another band between 37 and 50 kDa, representing approximately 40% of the total protein mass (according to a quantification of the gel bands in ImageJ)31 and corresponding to an unknown protein impurity (Figure 1B). The SDS-PAGE thus confirmed that the Ur-AR from the commercial supplier was not pure. The size-exclusion chromatography (SEC) of Ur-AR yielded three observable peaks in the absorbance at 280 nm, confirming the presence of the three protein species of different molecular mass ranges disclosed in the suppliers’ documentation (Figure 1C). An additional SDS-PAGE of the fractions corresponding to the maxima of the three SEC peaks confirmed that the chromatography successfully separated the hexameric urease (Ur-hex), found mostly in peak 2, from the protein impurity (Ur-imp), which eluted in peak 3, and an aggregated urease form (Ur-agg) in peak 1 (Figure 1D).
To determine the identity of the protein impurity found in peak 3 of the SEC of Ur-AR, the corresponding band in the SDS-PAGE gel was excised, digested with trypsin, and analyzed by liquid chromatography tandem mass spectrometry (LC-MS/MS). The resulting peptides were searched against Arabidopsis thaliana Swissport release 2021 and the common contaminants databases; with a coverage of 75% of the protein, the database search concluded that the protein impurity was canavalin (UniProt accession number: P50477), a major storage protein in jack bean.32−34 The structure of canavalin is trimeric, and with each subunit possessing a molecular mass of 50.3 kDa (matching the band cut out of the SDS-PAGE gel in Figure 1B and D), this would equate to a molecular mass of ∼150 kDa for the trimer. It was unclear why there was a second band in the lane corresponding to SEC peak 3 on the SDS-PAGE gel (Figure 1D) at the expected molecular weight for urease monomer, but it was hypothesized that this could correspond to trimeric urease (expected molecular mass of ∼272 kDa), which might be interacting in some way with the canavalin to coelute in the same SEC peak. To investigate this further, P3 was further analyzed by SEC-MALS (Figure S1 and Table S1). The results of the SEC-MALS run on P3 confirmed that this sample was a composite of protein species: 51.41% of the mass fraction corresponded to the canavalin contaminant (measured molecular mass of 143 kDa), 27.49% corresponded to trimeric urease (measured molecular mass of 254.35 kDa), and 21.11% corresponded to hexameric urease (molecular mass of 532.55 kDa).
The catalytic activity of the three SEC peaks was characterized by means of a urease assay (Figure 1E). By fitting the change in reaction rate to increasing substrate (urea) concentration to a Michaelis–Menten equation and comparing the derived kinetic parameters, we observe that SEC P2 (corresponding to Ur-hex) has the highest turnover number (kcat = 1421 s–1, see Table S2). Although this is apparently lower compared to the turnover values cited in the literature for the same enzyme,35 we believe this is a more accurate determination of the urease activity of the native hexameric enzyme, as this was measured subsequently to its purification. Indeed, as evidenced by dynamic light scattering (DLS), the Ur-hex sample shows much more monodispersity in that a single peak is observed, compared to the Ur-AR and Ur-imp samples, with Ur-AR displaying a particularly high degree of polydispersity (Figure 1F and G). The measured hydrodynamic radius of urease in the Ur-hex sample is 7.9 nm, which correlates with the measured hydrodynamic radius of urease found in previous reports.36,37
It is thought that urease is capable of dissociating in the presence of its substrate, urea, at concentrations higher than the KM.35 Enzyme dissociation can be misinterpreted as enhanced diffusion, as the smaller enzyme oligomers diffuse faster in solution.27,38−40 While investigating this report of urease dissociation using DLS and SEC, we found no evidence for this phenomena, even at urea concentrations well above KM (Figure S2 and Figure S3).35 Indeed, in the presence of 0.2 M urea, the peak measured by DLS at a hydrodynamic radius corresponding to urease becomes slightly smaller (characteristic of enhanced diffusion), but does not split, as claimed by Jee etal.35 We do however observe peaks at a higher radius in the presence of urea, indicative of protein aggregation. We believe that this protein aggregation occurs as a result of the chaotropic nature of urea, which perturbs the hydrophobic effect maintaining the protein’s solubility and causes urease to denature and thus aggregate.41
Micromotor Fabrication and Characterization
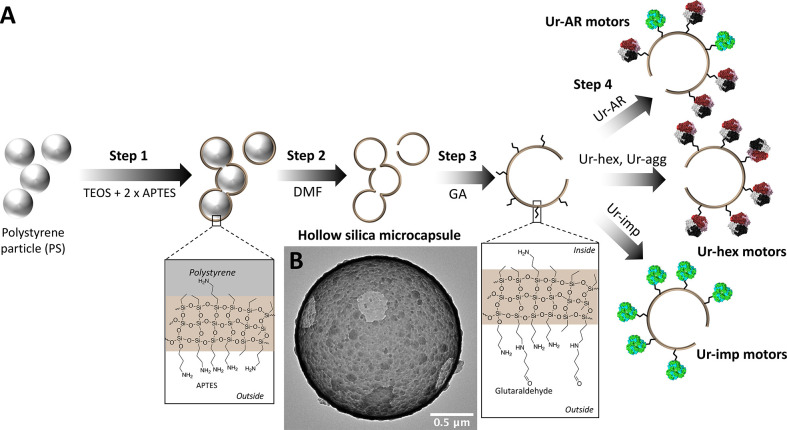
Non-Janus hollow silica microcapsules were chosen to demonstrate the enhanced self-propulsion properties of micromotors functionalized with purified urease, due to their ease of surface modification and their proven efficacy for exhibiting enzyme-powered motion in a propulsive regime.25,42,43 Silica particles are also biocompatible, making them suitable for downstream biomedical applications, and the surface chemistry is well understood.9,44−46 The HSMCs were synthesized according a procedure previously described,42 with the exception of an additional step of 3-aminopropyltriethanolamine (APTES) functionalization after creation of the silica layer on top of the polystyrene (PS) core (step 1 in Scheme 1). This additional step allowed for more APTES reactive groups to be present on the outer surface of the microparticle, in order to increase glutaraldehyde (GA)/urease loading on the outside of the particles in steps 3 and 4 of Scheme 1. The positive effect of glutaraldehyde cross-linking on protein stability, and in some cases on enzyme biocatalytic activity, is also well-documented (reviewed in ref (47)). The synthesized microparticle (after step 2) was analyzed by TEM (Scheme 1B) to verify that the extra APTES had no effect of compromising the structure of the particle, and the ζ-potential was recorded at each step to verify the success of the functionalization process (Figure S4). A total of four types of micromotors were synthesized: Ur-AR motors, HSMCs functionalized with the unpurified Ur-AR enzyme sample; Ur-hex motors, HSMCs functionalized with purified Ur-hex enzyme sample (P2 in Figure 1C); Ur-agg motors, HSMCs functionalized with the aggregated from of urease (P1 in Figure 1C); and Ur-imp motors, HSMCs functionalized with the impurities from the SEC purification of Ur-AR (P3 in Figure 1C).
Scheme 1. Fabrication of urease motors. (A) Synthesis steps for the three different urease motors described in this work. Ur-hex and Ur-agg are represented as the same motor because in both cases the urease is pure (see Figure 1D). (B) TEM image of the hollow silica microcapsules (HSMCs) before functionalization with urease.
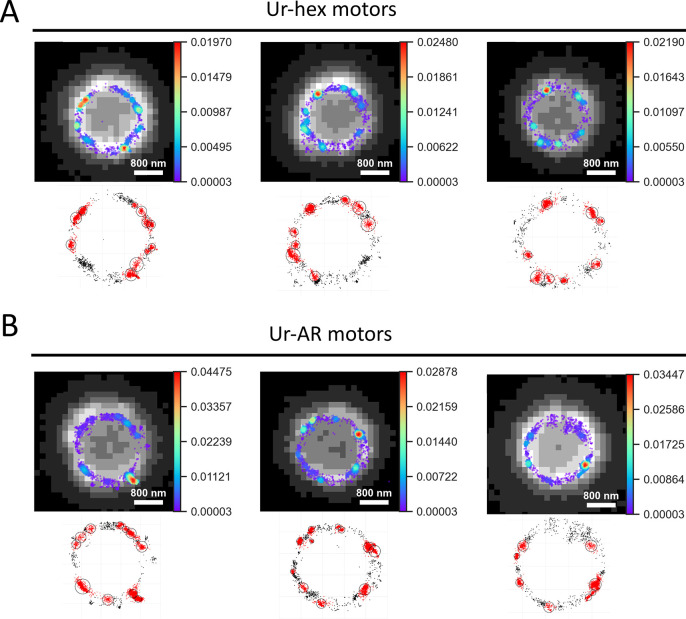
To study the enzyme coating of the micromotors, Ur-AR (which contained impurities) and purified Ur-hex enzyme samples were labeled with a Cy5 fluorescent dye and used to functionalize the microparticles, in a similar procedure to that previously described.48 Then, the resulting micromotors were imaged by STORM super-resolution microscopy (Figure 2). The 2D STORM images obtained were further analyzed using a Python script to generate heat maps of localization density (top panels of Figures 2A and 4B), as well as a clustering script written in Matlab to analyze the characteristics of the localization clusters found on the Ur-AR and Ur-hex motors, which included cluster size, diameter, density, and clusters per particle (Figure S5).
Figure 2.
Analysis of localization clusters on surface of urease micromotors. (A) Upper panels: Density maps of localizations detected by STORM superposed over low-resolution images of three representative Ur-hex motors, with color bars showing the relative density in probability values; lower panels: clusters of localizations found using the Matlab clustering algorithm. (B) Same as in panel A, but for three representative Ur-AR motors.
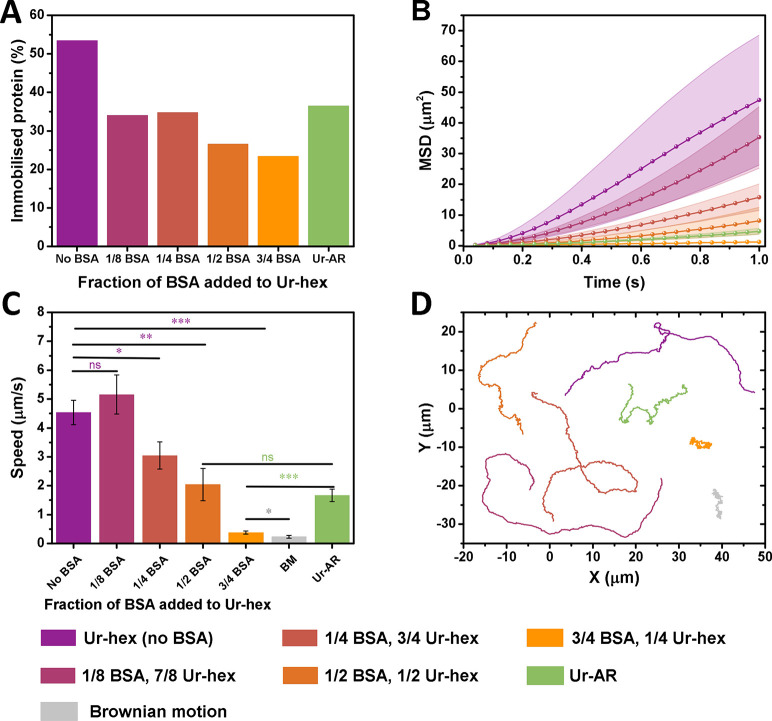
Figure 4.
Influence of protein impurities on mobility of urease micromotors. All plots follow the color legend at the bottom of the figure, and the urea concentration for all plots is 0.2 M. (A) Portion of immobilized protein from the 200 μg/mL used to functionalize the urease micromotors with different fractions of BSA to Ur-hex. (B) Average MSD, (C) average speeds, and (D) representative trajectories of the BSA/Ur-hex urease micromotors over 15 s. The p values determined from pairwise t tests are represented in panel C in the NEJM format: ***: p value ≤ 0.001; **: p value ≤ 0.01; *: p value ≤ 0.05; ns: nonsignificant. The p values in purple are being compared with the Ur-hex (no BSA) motors, p values in green are being compared with Ur-AR motors, and p values in gray are comparing the 3/4th BSA with the Brownian motion control.
Both Ur-AR and Ur-hex motors showed an asymmetric distribution on the microparticles, similar to the results obtained by Patiño etal. (2018).48 Moreover, the clusters on each type of Ur-AR and Ur-hex motors showed no significant difference in all of the parameters used to characterize them (Figure S5). The cluster size for both types of motors was ∼350 localizations per cluster, the density was ∼1.7 localizations per nm, the average cluster diameter was ∼220 nm, and ∼7.6 clusters were found per particle. It can be concluded from these data that the impurities only seem to affect enzyme distribution minimally on the surface of the particles, as a slightly higher number of clusters and cluster density could be found for the Ur-hex motors (Figure S5D and B, respectively).
The self-propulsion characteristics for the four types of micromotors were assessed by optical tracking of single particles in the microscope, and the motion of at least 15 particles (recorded for at least 15 s) for each type of micromotor was analyzed using a custom-made Python script, as previously described in the works of Samuel Sánchez and coauthors (see Methods/Experimental section).25,42,43,48 The mean-squared displacement (MSD) and average propulsive speed for each type of micromotor, along with their respective immobilized protein content, is plotted in Figure 3. Although the measured protein content on the Ur-AR motors is higher than that of the Ur-hex (Figure 3A), both the MSD and the average speed of the Ur-hex motors is significantly enhanced (p value ≤ 0.001) compared to the Ur-AR motors (Figure 3B and Figure 2C). Indeed, the Ur-hex motors could reach an average propulsive speed of up to 5.5 μm/s, which is in a similar range to the highest speed recorded for a non-Janus urease-powered micromotor (Video S1).43 Moreover, the MSD and speed of the Ur-hex motors increase with increasing concentration of the urea substrate (Figure 3D and E, respectively); this increase in speed could be fit to the Michaelis–Menten enzyme kinetics equation (Figure 3F), similarly to previously described urease micromotors.9,10,42,49
Figure 3.
Motion characterization of urease micromotors. (A) Measured protein content immobilized onto HSMCs for Ur-AR (green bar), Ur-hex (purple bar), Ur-agg (pink bar), and Ur-imp (orange bar) motors. (B) Average mean-squared displacement (MSD) of the Ur-AR (green), Ur-hex (purple), Ur-agg (pink), and Ur-imp (orange) motors with 0.2 M urea. (C) Average speed of the Ur-AR (green bar), Ur-hex (violet bar), Ur-agg (pink bar), and Ur-imp (orange bar) with 0.2 M urea and the Brownian motion (BM) (speed in the absence of fuel) of Ur-imp motors (black bar). The p values determined from pairwise t tests are represented in panels B and C in the New England Journal of Medicine (NEJM) format: ***: p value ≤ 0.001; **: p value ≤ 0.01; *: p value ≤ 0.05; ns: nonsignificant. (D) Average MSD of the Ur-hex motors at 0, 10, 50, 100, and 200 mM urea (light to dark purple). (E) Average speed of the Ur-hex motors at 0, 10, 50, 100, and 200 mM urea (light to dark purple). (F) Michaelis–Menten fit of the data for the activity assays for soluble Ur-hex (blue line and spheres, left y-axis, same data as shown in Figure 1E) compared to the Michaelis–Menten fit of the Ur-hex motor self-propulsion speeds at different concentrations of urea (red line and spheres, right y-axis, same data as shown in panel B).
The Ur-agg motors also had significantly higher MSD and speed compared to the Ur-AR (p value ≤ 0.05), demonstrating that purified urease enhances micromotor self-propulsion even in its aggregated form (Figure 3B and C). Nevertheless, the Ur-agg motors did not reach the same motility observed for Ur-hex motors, despite both soluble forms of Ur-hex and Ur-agg having very similar kinetic properties (Figure 1E and Table S2). It is likely that the urease active sites on the surface of the particles are less accessible when the purified urease is aggregated. Thus, the reaction rate, and by consequence the self-propulsion, is slower for the Ur-agg motors compared to the Ur-hex motors.
For the Ur-imp motors, there was no significant difference found when comparing the propulsive speed in the absence and presence of the 0.2 M urea fuel (Figure 3C). This result was to be expected given that the Ur-imp sample, which corresponds to the SEC P3 in Figure 1, is primarily composed of the catalytically inert protein impurity found in the Ur-AR commercial preparation.
Influence of Protein Impurities on Micromotor Motion and Enzyme Distribution
To study how protein impurities in the commercial sample might affect the self-propulsion of urease micromotors, a series of micromotors were functionalized with varying ratios of purified urease and bovine serum albumin (BSA). In this case, BSA was used as an inert protein to replace the canavalin, which we were unable to isolate from the Ur-AR preparation due to its coelution with urease during the SEC purification (Figure 1D). Indeed, BSA was chosen not only because of its wide use as a nonreactive protein but also owing to its similar pI and hydrophobicity to canavalin (Table S3). To this effect, the motion of the Ur-hex motors (no BSA) was compared to that of motors functionalized with a 1/8th to 7/8th, 1/4th to 3/4th, 1/2 to 1/2, and 3/4th to 1/4th BSA to Ur-hex ratio (Figure 4). These motors were also compared to Ur-AR motors, which contain canavalin.
Although the protein concentration used to functionalize all motors was adjusted to approximately 200 μg/mL, the determined amount of protein immobilized onto the HSMC particles varied (Figure 4A). These differences could be explained by taking into account the lower molecular mass of BSA (∼ 66 kDa compared to the 545 kDa of the urease hexamer).
When comparing the average MSDs and speeds of the six different micromotors, it is apparent that the motion of the urease micromotors is tolerant to lower amounts of BSA (1/8th), but that mobility drops substantially with increased BSA concentration (Figure 4B, C, and D and Video S2). Moreover, the speed of the Ur-AR seems to be most comparable to the motors functionalized with 1/2 BSA, themselves showing a 55% drop in speed compared to the Ur-hex motors with no BSA, evidencing the dramatic effect that impurities binding to the particle exert on its self-propulsion (Figure 4C and Video S2).
Enhanced Properties of Purified Urease Motors
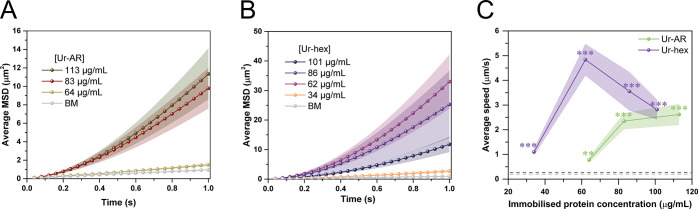
In order to highlight the clear advantages offered by the Ur-hex motors compared to the Ur-AR, the influence of immobilized enzyme quantity and reusability of both urease motors was studied. Regarding the influence of immobilized enzyme quantity, Ur-hex motors were found to have enhanced motion compared to the Ur-AR motors with less immobilized enzyme (Figure 5). Indeed, the MSD and speed of the Ur-AR motors for 64 and 84 mg/mL of immobilized urease (Figure 5A and C) were 22 times and 2.6 times lower, respectively, compared to the MSD of the Ur-hex motors at similar concentrations of immobilized urease (Figure 5B and C). Given that the proportion of canavalin in the Ur-AR sample is estimated to be approximately 40%, a motion enhancement of the micromotors of at least 2-fold was to be expected for the particles that are coated with pure urease. We also observe that the MSD of the Ur-AR motors drops suddenly below 80 mg/mL of attached urease, a similar result as has been described previously for the same micromotors (Figure 5A).48 This could be due to there not being enough urease to power the motion of Ur-AR motors below 80 mg/mL, with a significant portion of the attached protein being the canavalin impurity. In the case of Ur-hex, although a similar drop in the MSD to the of Ur-AR could be observed between 62 and 34 mg/mL, the MSD was lower when more urease (80 and 100 mg/mL) was attached to the motors (Figure 5B). This effect could be due to enzyme overcrowding on the particle surface, which can result in lower enzymatic activity due to steric hindrance.50,51 Although (to the best of our knowledge) this behavior has not been observed for urease micromotors before, Yang etal. do observe a similar trend for their MOFtors, where an increase in catalase loading resulted in less generation of the O2 product.4 Thus, the optimal concentration of immobilized urease for self-propulsion of Ur-hex motors is ∼60 mg/mL.
Figure 5.
Influence of immobilized enzyme quantity on self-propulsion of Ur-AR and Ur-hex motors. (A) Average MSD for motors functionalized with 113 μg/mL (olive), 83 μg/mL (maroon), and 64 μg/mL (beige) and Brownian motion (BM) of Ur-AR. (B) Average MSD for motors functionalized with 101 μg/mL (dark purple), 86 μg/mL (purple), 62 μg/mL (magenta-pink), and 34 μg/mL (orange) and Brownian motion of Ur-hex motors. The urea concentration for panels A and B is 0.2 M. (C) Average speeds for Ur-hex (purple) and Ur-AR (green) as a function of immobilized protein concentration. The p values determined from pairwise t tests comparing the speed of urease motors in the presence of 0.2 M urea with the no fuel control are represented in panel C in the NEJM format: ***: p value ≤ 0.001; **: p value ≤ 0.01; *: p value ≤ 0.05. The no-fuel (Brownian motion) baseline is represented as a green dashed line for Ur-AR and a purple dashed line for Ur-hex.
Motor reusability is a parameter routinely studied, especially in the context of environmental applications, as the micromotors need to be recuperated from reaction once it is over, which increases the micromotors’ cost-efficiency.52−55 Reusability has been demonstrated for Janus micromotors functionalized with urease, but only in the context of motion control.9 In the study in question, the motors were exposed to a Hg+ urease inhibitor to stop motion, and the authors demonstrated that motion could be regained following an injection of dithiothreitol (DTT), which removes the inhibitor, with little to no effect on the performance of the motor. In these “on–off” cycles, the urease-powered motion of the micromotors appears to be mostly unaffected, even after 8 cycles.
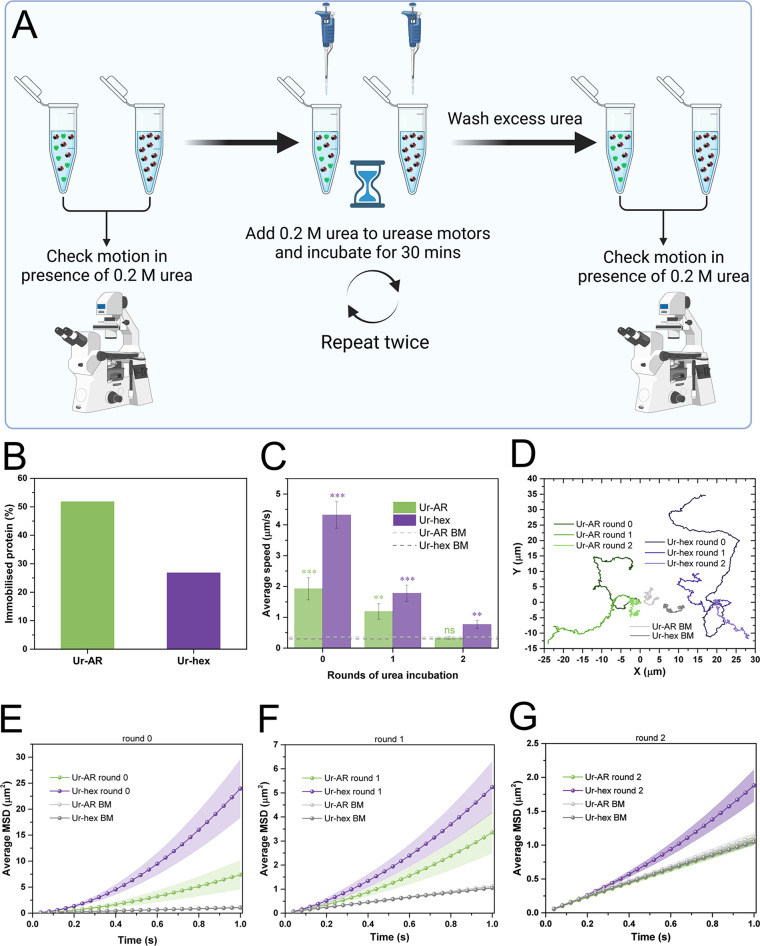
The reusability of the motors in this study was evaluated by exposing the urease on the motors to repetitive incubations with 0.2 M urea, with the intention of evaluating the effect of repetitive rounds of catalytic turnover on the motors’ self-propulsion. Unlike the study previously mentioned, nothing more was added to the motors apart from the urea substrate, as this experiment did not seek to study motion control, but rather evaluate the effect of substrate exposure on the motion of the micromotors. This experimental setup is represented in Figure 6A. Briefly, videos of the Ur-AR and Ur-hex motors were recorded using the same experimental setup described throughout this work before exposing the stock solution of the motors to urea. Then, 0.2 M urea was added to the stock solution of both motors and allowed to react with the urease for 30 min. After washing the particles thoroughly from any excess urea leftover from the reaction, another set of videos was recorded. This cycle was repeated twice, after which point the self-propulsion of both motors was significantly reduced. Nevertheless, the Ur-hex were the only motors to retain some motion after two rounds of incubation with the substrate (Figure 6B–G, Video S3). This is most likely a result of there being more initial active urease on the particles, because both motors follow the same downward trend in their average speeds (Figure 6B) and MSDs (Figure 6D–G). Therefore, we hypothesize that the reusability of the urease motors is affected by enzyme deactivation, which occurs as a result of repeated exposure to the substrate and consequently to the ammonia product, both of which have been reported to poison urease.56,57
Figure 6.
Reusability of urease motors. (A) Schematic diagram of reusability experiment. An aliquot of both motors was used to visualize the self-propulsion in the presence of 0.2 M urea. Then, 0.2 M urea is added to both motors and left to react for 30 min. Particles were washed from excess urea by centrifugation, and loss of motility was evaluated by taking an aliquot of the washed stock. This cycle was repeated twice. (B) Quantity of immobilized protein on the urease motors, expressed as a percentage of the initial quantity used to functionalized the particles (both approximately 200 μg/mL). (C) Average speeds for both types of urease motors before (0) and after the first and second incubation with urea. The p values from the pairwise t tests comparing the motion at 0.2 M urea with the no-fuel control (represented by green and purple dashed lines, for Ur-AR and Ur-hex, respectively) are represented as stars above the bars (same NEJM format as in previous figures). (D) Representative trajectories for the Ur-AR (shades of green) and Ur-hex (shades of purple) motors at 0.2 M urea and after 0, 1, and 2 rounds of incubation. The Brownian motion of the Ur-AR and Ur-hex motors is represented in two shades of gray. (E–G) Average MSDs of the Ur-AR (green) and Ur-hex (purple) motors before (E) and after first (F) and second (G) rounds of incubation with 0.2 M urea. The MSDs from the Brownian motion of Ur-AR and Ur-hex motors are also represented in all plots in shades of gray.
Conclusions
In this work, we have demonstrated that the urease sample used by many groups to functionalize micro/nanomotors contains protein impurities and that motors functionalized with purified urease exhibit improved self-propulsion compared to those functionalized with the impure commercial urease preparation. We have also demonstrated that the presence of inert impurities in the protein samples used to functionalize the micromotors results in less active urease binding to the particle and thus slower motors. Moreover, we find no differences in the enzyme distribution on the surface of the particles between the Ur-AR and Ur-hex motors, which both result in an asymmetric distribution. Finally, the critical concentration necessary to self-propel Ur-hex motors is lower than that of the Ur-AR, and Ur-hex motors can be reused more than the Ur-AR. Both are a consequence of higher active enzyme loading on the particles when using a purified urease sample.
This work illustrates the necessity to evaluate the purity of the enzyme samples used to fabricate enzyme-powered micro/nanomotors, especially when using a protein immobilization strategy involving a chemoligation between surface lysines and linker molecules with terminal NHS esters (e.g., EDC/NHS) or aldehydes (e.g., glutaraldehyde). Indeed, lysines can be found on the surface of virtually all proteins, although in varying prevalence. As an alternative to protein purification, a more specific immobilization strategy could be adopted; strategies that rely on a protein tag, such as biotin, which reacts with streptavidin linkers on the particle surface,5,10,58 avoid binding of undesirable protein impurities and could potentially improve micro/nanomotor self-propulsion.
Protein stability may also be lost by purifying out potentially stabilizing protein species found in commercial samples, which in turn might have a deleterious effect on motors’ lifetime. Future work on increasing protein stability on the surface of nano/micromotors should evaluate the suitability of less bulky chemical stabilizers, such as polyethylene glycol (PEG),25,59 that would not interfere with the chemoligation of the protein onto the surface of the particles.
Methods/Experimental
Purification of Urease from Canavalia ensiformis
Approximately 50 mg of jack bean urease from C. ensiformis (type IX, Sigma-Aldrich cat. no. U4002) was solubilized in PBS (pH = 7.4) and loaded onto an ENrich SEC 650 10 × 300 size-exclusion column (Biorad cat. no. 780-1650), pre-equilibrated in the same buffer. The column was mounted on an NGC Quest 10 chromatography system (Biorad). Two successive chromatography runs were performed, with fraction sizes of 0.5 mL, which were collected in the same tubes; thus the final volume of the fractions was 1 mL. The fractions corresponding to the major peaks in the chromatogram were concentrated using a Vivaspin 500 Centricon (MWCO: 30 kDa, Sartorius cat. no. VS0122), mixed with 4× SDS loading buffer (composition: 50 mM Tris-HCl pH 6.8, 2% sodium dodecyl sulfate, 10% glycerol, 1% β-mercaptoethanol, 12.5 mM EDTA, 0.02% bromophenol blue) and heated to 95 °C for 10 min. The denatured protein samples were loaded onto a 10% SDS Mini-Protean precast gel (Biorad cat. no. 4568036), along with a Precision Plus Protein all blue prestained protein standards ladder (Biorad cat. no. 1610393), in a Mini-Protean Tetra electrophoresis cell. SDS-PAGE gels were stained with InstantBlue (Expedeon cat. no. ISB01L).
Dynamic Light Scattering
The hydrodynamic radius of the urease samples was determined by DLS using a Wyatt Mobius light scattering instrument. The unlabeled enzyme samples were diluted to a concentration of 30 nM with 1× PBS that had been filtered through a 0.22 μm pore. Light scattering measurements were performed using disposable cuvettes to avoid sample contamination.
Urease Activity Assays
The assays used to characterize urease activity were based on the Berthelot method.60,61 Briefly, in a 96-well plate, reactions of 50 μL of urea solubilized in 1× PBS at a set concentration were mixed with 50 μL of soluble urease (diluted to a final concentration of 0.5 nM in 1× PBS) or particles and incubated for 2 min at RT. Wells containing 0–200 μM ammonium chloride solution (NH4Cl, Sigma-Aldrich, cat. no. 254134) were prepared alongside the assays in order to make a calibration curve to quantify ammonium production. To stop the ureolytic reactions, 80 μL of phenol nitroprusside solution (Sigma-Aldrich, cat. no. P6994) was added to each assay and calibration standard, followed by 40 μL of an alkaline hypochlorite solution (Sigma-Aldrich, cat. no. A1727). The plates were mixed thoroughly, then incubated for 30 min at 37 °C, before reading the absorbance of the plate at 630 nm. The rate of the urease reaction was determined using the NH4Cl calibration curve.
Synthesis of Hollow Silica Microcapsules
The HSMCs were synthesized by growing a layer of silica dioxide on top of 2 μm particles based on PS (Sigma-Aldrich cat. no. 78452) according (but with slight modifications) to a co-condensation method previously described.42,43,48 Briefly, 1 mL of ethanol 99% (PanReac AppliChem cat. no. 131086-1214) and 0.8 mL of ultrapure water were added to 500 μL of PS particles, followed by 25 μL of ammonium hydroxide (28–30%, Sigma-Aldrich cat. no. 221228). This mixture was left to magnetically stir for 5 min, after which point 5 μL of APTES (99%, Sigma-Aldrich cat. no. 440140) was added, and the reaction was left to proceed for 6 h. Then, 7.5 μL of tetraethylorthosilicate (TEOS, >99%, Sigma-Aldrich cat. no. 86578) was added to the particle mixture, and the reaction was left overnight at RT. The resulting particles were washed three times with ethanol by centrifuging them at 3500 rpm for 3.5 min. After the final wash, 50 μL of APTES was added to 950 μL of particles, and the reaction was mixed for 5 h. Unreacted APTES was removed with one wash in ethanol and three washes in dimethylformamide (DMF, 99.8%, Sigma-Aldrich cat. no. 319937), mixing for 15 min between washes, the later also serving to remove the PS. An additional three washes in ethanol were necessary to remove the excess DMF, and the synthesized HSMCs could be stored at 4 °C or further functionalized (vide infra). The size and morphology of the HSMCs were characterized by TEM.
Functionalization of HSMCs with Enzyme Samples
The HSMCs were functionalized with glutaraldehyde (GA) and urease following a protocol that has been previously described.42 Briefly, the HSMC particles were washed three times with ultrapure water, then once with 1× PBS pH 7.4, before 100 μL of 25% GA (Sigma-Aldrich cat. no. G6257) was added to the particles, previously resuspended in 1× PBS (GA final concentration = 2.5%). This mixture was left to mix for 2.5 h. The newly GA-functionalized HSMCs were subsequently washed three times with 1× PBS and resuspended again in one of three enzyme mixtures: (1) 3 mg/mL of urease type IX powder solubilized in 1× PBS to make the Ur-AR motors, (2) 100–200 μg/mL (depending on the yield of the purification, see Purification of Urease from Canavalia ensiformis section) of purified hexameric urease to make the Ur-hex motors, or (3) 100–200 μg/mL (also depending on the urease purification) of protein impurities found in the urease powder to make the Ur-imp motors. Regardless of the enzyme sample used to functionalize the particles, the solution was mixed overnight at RT, after which the particles were washed three times in 1× PBS, collecting the supernatants from each wash for total protein quantification. Either the resulting urease motors were stored at 4 °C, or an aliquot of 50 μL was diluted in ultrapure water and washed three more times in ultrapure water for optical tracking experiments.
Total Protein Quantification of Enzyme-Functionalized HSMC Particles
Total protein quantification was calculated following a similar procedure to that described by Arqué etal.42 Briefly, the protein concentrations of the Ur-AR, Ur-hex, and Ur-imp enzyme samples used to functionalize the particles, as well as the supernatants from the three postfunctionalization washes in 1× PBS, were determined using a Pierce BCA protein assay kit (Thermo Fisher cat. no. 23227). The protein detected in the supernatant served to establish the quantity of protein that had not reacted with the GA; thus the quantity of enzyme that had been successfully immobilized onto the particles could be determined by subtracting the protein found in the supernatants from the initial protein concentration used for particle functionalization.
Optical Video Recording
Videos of the urease micromotors were recorded using a Hammatsu C11440 digital camera mounted onto an inverted optical microscope (Leica DMi8), using a 63× water immersion objective. A 5 μL drop of the urease micromotor solution (prepared in water) was mixed with 5 μL of either water (to record samples in the absence of fuel) or urea (generally 0.4 M in water, which when mixed with the motors reaches a final concentration of 0.2 M). The glass slide was covered with a coverslip, and videos of at least 15 s at 25 frames/second were recorded for the first minutes after mixing. A total of at least 15 particles were recorded for each urease micromotor and/or for each specified condition studied.
Particle Tracking and Motion Analysis
The recorded videos were analyzed using a custom-made Python script, capable of tracking the trajectories of the particles and calculating the MSD using the following equation:
| 1 |
where t is the time and i = 2, for an analysis in 2D. The propulsive speed (ν) was derived from the fitting of the MSD to the following equation:
| 2 |
where Dt is the diffusion coefficient and v is the speed in a propulsive regime, when t ≪ τr, with τr being the rotational diffusion time and t the time of the MSD in question.62,63
Data Processing of Motion Analyses
Once all the videos had been analyzed, the resulting data were further processed using the Python-based Nanomicromotor Analysis Tool (NMAT) v. 0.5 (https://github.com/rafamestre/NMAT-nanomicromotor-analysis-tool). This script concatenates all the data for a single condition (15 particles) and creates .csv files to visualize a number of motion analysis parameters, including MSD, propulsive speed, and trajectories of single particles.64
Urease Labeling
To study the differences in enzyme coating between the Ur-AR and Ur-hex micromotors, both samples were labeled with Cy5 dye using slightly different protocols. The Ur-AR sample was labeled using a similar procedure to that described by Patiño etal.48 Briefly, the urease type IX from Sigma-Aldrich was solubilized in PBS buffered to pH 8.4 by addition of NaOH, to make up a solution of 1 μM Ur-AR, to which 1.5 molar equiv of Cy5-NHS ester reactive dye was added (Lumiprobe, cat. no. 13020). For the Ur-hex sample, the urease type IX was first purified by size-exclusion chromatography in PBS pH 8.4 (using the same procedure as described previously), the peak corresponding to the urease hexamer was collected, the protein concentration was determined to be similar to that of Ur-AR (∼1 μM), and the same 1.5 molar equiv of Cy5-NHS ester dye was mixed in. Both reactions of Ur-AR and Ur-hex with Cy5 reactive dye were left to react overnight on an orbital shaker. The urease–Cy5 conjugates were purified from the excess dye by gel filtration using a 5 mL HiTrap desalting column (Cytiva, cat. no. 17-1408-01) and PBS pH 7.4 buffer. The Cy5-labeled Ur-AR and Ur-hex samples were then mixed with unlabeled Ur-AR and Ur-hex, respectively, to make up a solution of 4% Cy5–urease. A 900 mL amount of this solution was then used to functionalize the HSMC particles, using the same protocol as previously described.
The protein concentration was ascertained by measuring the absorbance of the sample at 280 nm, assuming an extinction coefficient (e) of 325 365 M–1·cm–1 (determined using Protparam from the protein sequence of UniProt accession code P07374).65 The Cy5 to protein ratio was estimated by molar equivalence, using an e650 = 250 000 M–1·cm–1.
STORM Imaging
STORM images were acquired using a Nikon N-STORM system configured for total internal reflection fluorescence imaging. Cy5-labeled urease on motors was imaged by means of a 647 nm laser (160 mW). Fluorescence was collected by means of a Nikon 100×, 1.49 NA oil immersion objective and passed through a quadband pass dichroic filter (97335 Nikon). Images were acquired onto a 256 × 256 pixel region (pixel size 0.16 μm) of a Hamamatsu ORCAFlash 4.0 camera at 10 ms integration time. A total of 20 000 frames were acquired for the 647 channel, and the total time required to acquire one image was about 2 min. Bright field images were taken for assessing the number of motors per field. STORM images were analyzed with the STORM module of the NIS element Nikon software.
STORM Data Analysis
The data obtained from the STORM images were processed and analyzed using a custom-made Python script adapted from ref (66) and a Matlab script adapted from ref (67). Briefly, the Python script was designed to automatically detect the centroids of the micromotors based on the low-resolution images, and the density of the clusters of localizations could be visualized locally on each microparticle. The Matlab script also worked by detecting localization clusters from a coordinates output file from the NIS element Nikon software and by filtering for relevant clusters according to a set of parameters (including minimum number of localizations set to 100, maximum diameter of cluster set to 500, and minimum distance between clusters set to 75 nm). The output of the Matlab script is a cell array with locations for the selected clusters and an array for the number of localizations in each selected cluster (termed cluster size) and the diameter of each cluster.
Acknowledgments
The research leading to these results has received funding from the grant RTI2018-098164-B-I00 funded by MICIN/AEI/10.13039/5011000110333 and by “FEDER Una manera de hacer Europa” (BOTSinFluids project), the CERCA program by the Generalitat de Catalunya, and the “Centro de Excelencia Severo Ochoa”, funded by Agencia Estatal de Investigación (CEX2018-000789-S). This project has also received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement no. 866348; i-NanoSwarms). S.P. and L.A acknowledge the financial support by the Spanish Ministry of Science and Innovation (PID2019-109450RB-I00/AEI/10.13039/501100011033), European Research Council/Horizon 2020 (ERC-StG-757397), “la Caixa” Foundation (ID 100010434), and the Generalitat de Catalunya through the CERCA program. The authors would like to acknowledge Marta Vilaseca Casas and Mar Vilanova, who performed the mass spectrometry at the IRB Barcelona Mass Spectrometry and Proteomics Core Facility, which actively participated in the BMBS European COST Action BM 1403 and is a member of Proteored, PRB3-ISCII, supported by grant PRB3 (IPT17/0019 – ISCII-SGEFI/ERDF. The authors would also like to thank Laura Company from IBMB-CSIC (Barcelona, Spain) for her help in performing the SEC-MALS experiments and Joaquin Llacer-Wintle for his help in the STORM data processing.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.1c10520.
Experimental details, SEC-MALS analysis, DLS and SEC experiments of soluble urease in the presence of urea, and further characterization of micromotors (Figures S1–S5 and Tables S1–S3) (PDF)
Comparison of Ur-hex, Ur-AR, and Ur-imp motors (MP4)
Influence of impurities on micromotor motion (MP4)
Reusability of urease motors (MP4)
The authors declare no competing financial interest.
Supplementary Material
References
- Ma X.; Jannasch A.; Albrecht U.-R.; Hahn K.; Miguel-López A.; Schäffer E.; Sánchez S. Enzyme-Powered Hollow Mesoporous Janus Nanomotors. Nano Lett. 2015, 15 (10), 7043–7050. 10.1021/acs.nanolett.5b03100. [DOI] [PubMed] [Google Scholar]
- Ma X.; Sánchez S. Bio-Catalytic Mesoporous Janus Nano-Motors Powered by Catalase Enzyme. Tetrahedron 2017, 73 (33), 4883–4886. 10.1016/j.tet.2017.06.048. [DOI] [Google Scholar]
- Solovev A. A.; Sanchez S.; Pumera M.; Mei Y. F.; Schmidt O. G. Magnetic Control of Tubular Catalytic Microbots for the Transport, Assembly, and Delivery of Micro-Objects. Adv. Funct. Mater. 2010, 20 (15), 2430–2435. 10.1002/adfm.200902376. [DOI] [Google Scholar]
- Yang Y.; Arqué X.; Patiño T.; Guillerm V.; Blersch P.-R.; Pérez-Carvajal J.; Imaz I.; Maspoch D.; Sánchez S. Enzyme-Powered Porous Micromotors Built from a Hierarchical Micro- and Mesoporous UiO-Type Metal–Organic Framework. J. Am. Chem. Soc. 2020, 142 (50), 20962–20967. 10.1021/jacs.0c11061. [DOI] [PubMed] [Google Scholar]
- Dey K. K.; Zhao X.; Tansi B. M.; Méndez-Ortiz W. J.; Córdova-Figueroa U. M.; Golestanian R.; Sen A. Micromotors Powered by Enzyme Catalysis. Nano Lett. 2015, 15 (12), 8311–8315. 10.1021/acs.nanolett.5b03935. [DOI] [PubMed] [Google Scholar]
- Orozco J.; García-Gradilla V.; D’Agostino M.; Gao W.; Cortés A.; Wang J. Artificial Enzyme-Powered Microfish for Water-Quality Testing. ACS Nano 2013, 7 (1), 818–824. 10.1021/nn305372n. [DOI] [PubMed] [Google Scholar]
- Guo Z.; Liu J.; Li Y.; McDonald J. A.; Bin Zulkifli M. Y.; Khan S. J.; Xie L.; Gu Z.; Kong B.; Liang K. Biocatalytic Metal-Organic Framework Nanomotors for Active Water Decontamination. Chem. Commun. 2020, 56 (94), 14837–14840. 10.1039/D0CC06429G. [DOI] [PubMed] [Google Scholar]
- Ma X.; Hortelao A. C.; Miguel-López A.; Sánchez S. Bubble-Free Propulsion of Ultrasmall Tubular Nanojets Powered by Biocatalytic Reactions. J. Am. Chem. Soc. 2016, 138 (42), 13782–13785. 10.1021/jacs.6b06857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X.; Wang X.; Hahn K.; Sánchez S. Motion Control of Urea-Powered Biocompatible Hollow Microcapsules. ACS Nano 2016, 10 (3), 3597–3605. 10.1021/acsnano.5b08067. [DOI] [PubMed] [Google Scholar]
- Luo M.; Li S.; Wan J.; Yang C.; Chen B.; Guan J.. Enhanced Propulsion of Urease-Powered Micromotors by Multilayered Assembly of Ureases on Janus Magnetic Microparticles. Langmuir 2020. 10.1021/acs.langmuir.9b03315. [DOI] [PubMed] [Google Scholar]
- Tang S.; Zhang F.; Gong H.; Wei F.; Zhuang J.; Karshalev E.; De vila B. E. F.; Huang C.; Zhou Z.; Li Z.; Yin L.; Dong H.; Fang R. H.; Zhang X.; Zhang L.; Wang J. Enzyme-Powered Janus Platelet Cell Robots for Active and Targeted Drug Delivery. Sci. Robot. 2020, 5 (43), 41–43. 10.1126/scirobotics.aba6137. [DOI] [PubMed] [Google Scholar]
- Somasundar A.; Sen A. Chemically Propelled Nano and Micromotors in the Body: Quo Vadis?. Small 2021, 17 (5), 1–7. 10.1002/smll.202007102. [DOI] [PubMed] [Google Scholar]
- Wang L.; Hortelão A. C.; Huang X.; Sánchez S. Lipase-Powered Mesoporous Silica Nanomotors for Triglyceride Degradation. Angew. Chem. 2019, 131 (24), 8076–8080. 10.1002/ange.201900697. [DOI] [PubMed] [Google Scholar]
- Wang L.; Marciello M.; Estévez-Gay M.; Rodriguez P. E. D. S.; Morato Y. L.; Iglesias-Fernández J.; Huang X.; Osuna S.; Filice M.; Sanchez S.. Enzyme Conformation Influences the Performance of Lipase-powered Nanomotors. Angew. Chemie Int. Ed. 2020, 59, 21080. 10.1002/anie.202008339. [DOI] [PubMed] [Google Scholar]
- Toebes B. J.; Cao F.; Wilson D. A. Spatial Control over Catalyst Positioning on Biodegradable Polymeric Nanomotors. Nat. Commun. 2019, 10 (1), 5308. 10.1038/s41467-019-13288-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelmohsen L. K. E. A.; Nijemeisland M.; Pawar G. M.; Janssen G.-J. A.; Nolte R. J. M.; van Hest J. C. M.; Wilson D. A. Dynamic Loading and Unloading of Proteins in Polymeric Stomatocytes: Formation of an Enzyme-Loaded Supramolecular Nanomotor. ACS Nano 2016, 10 (2), 2652–2660. 10.1021/acsnano.5b07689. [DOI] [PubMed] [Google Scholar]
- Schattling P.; Thingholm B.; Städler B. Enhanced Diffusion of Glucose-Fueled Janus Particles. Chem. Mater. 2015, 27 (21), 7412–7418. 10.1021/acs.chemmater.5b03303. [DOI] [Google Scholar]
- Kutorglo E. M.; Elashnikov R.; Rimpelova S.; Ulbrich P.; ŘíhováAmbrožová J.; Svorcik V.; Lyutakov O. Polypyrrole-Based Nanorobots Powered by Light and Glucose for Pollutant Degradation in Water. ACS Appl. Mater. Interfaces 2021, 13 (14), 16173–16181. 10.1021/acsami.0c20055. [DOI] [PubMed] [Google Scholar]
- Fernández-Medina M.; Ramos-Docampo M. A.; Hovorka O.; Salgueiriño V.; Städler B. Recent Advances in Nano- and Micromotors. Adv. Funct. Mater. 2020, 30 (12), 1908283. 10.1002/adfm.201908283. [DOI] [Google Scholar]
- Yuan H.; Liu X.; Wang L.; Ma X. Fundamentals and Applications of Enzyme Powered Micro/Nano-Motors. Bioact. Mater. 2021, 6 (6), 1727–1749. 10.1016/j.bioactmat.2020.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathesh M.; Sun J.; Wilson D. A. Enzyme Catalysis Powered Micro/Nanomotors for Biomedical Applications. J. Mater. Chem. B 2020, 8 (33), 7319–7334. 10.1039/D0TB01245A. [DOI] [PubMed] [Google Scholar]
- Ma X.; Hortelão A. C.; Patiño T.; Sánchez S. Enzyme Catalysis To Power Micro/Nanomachines. ACS Nano 2016, 10 (10), 9111–9122. 10.1021/acsnano.6b04108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katuri J.; Ma X.; Stanton M. M.; Sánchez S. Designing Micro-and Nanoswimmers for Specific Applications. Acc. Chem. Res. 2017, 50 (1), 2–11. 10.1021/acs.accounts.6b00386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nsamela A.; Sharan P.; Garcia-Zintzun A.; Heckel S.; Chattopadhyay P.; Wang L.; Wittmann M.; Gemming T.; Saenz J.; Simmchen J. Effect of Viscosity on Microswimmers: A Comparative Study. ChemNanoMat 2021, 7, 1–10. 10.1002/cnma.202100119. [DOI] [Google Scholar]
- Arqué X.; Andrés X.; Mestre R.; Ciraulo B.; Ortega Arroyo J.; Quidant R.; Patiño T.; Sánchez S. Ionic Species Affect the Self-Propulsion of Urease-Powered Micromotors. Research 2020, 2020, 1–14. 10.34133/2020/2424972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X.; Wang J.; Xiong Z.; Zhang X.; Zhou Y.; Zheng J.; Chen J.; Feng S. P.; Tang J. Enhanced Ion Tolerance of Electrokinetic Locomotion in Polyelectrolyte-Coated Microswimmer. Nat. Commun. 2019, 10 (1), 1–9. 10.1038/s41467-019-11907-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Hess H. Enhanced Diffusion of Catalytically Active Enzymes. ACS Cent. Sci. 2019, 5 (6), 939–948. 10.1021/acscentsci.9b00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesareo S. D.; Langton S. R. Kinetic Properties of Helicobacter Pylori Urease Compared with Jack Bean Urease. FEMS Microbiol. Lett. 1992, 99 (1), 15–21. 10.1111/j.1574-6968.1992.tb05535.x. [DOI] [PubMed] [Google Scholar]
- Krajewska B.; Ciurli S. Jack Bean (Canavalia Ensiformis) Urease. Probing Acid-Base Groups of the Active Site by PH Variation. Plant Physiol. Biochem. 2005, 43 (7), 651–658. 10.1016/j.plaphy.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Kappaun K.; Piovesan A. R.; Carlini C. R.; Ligabue-Braun R. Ureases: Historical Aspects, Catalytic, and Non-Catalytic Properties – A Review. J. Adv. Res. 2018, 13, 3–17. 10.1016/j.jare.2018.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A.; Rasband W. S.; Eliceiri K. W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9 (7), 671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. C.; Johnson S.; Andrews J.; McPherson A. Biochemical Characterization of Canavalin, the Major Storage Protein of Jack Bean. Plant Physiol. 1982, 70 (4), 1199–1209. 10.1104/pp.70.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko T. P.; Ng J. D.; McPherson A. The Three-Dimensional Structure of Canavalin from Jack Bean (Canavalia Ensiformis). Plant Physiol. 1993, 101 (3), 729–744. 10.1104/pp.101.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng J. D.; Ko T. P.; McPherson A. Cloning, Expression, and Crystallization of Jack Bean (Canavalia Ensiformis) Canavalin. Plant Physiol. 1993, 101 (3), 713–728. 10.1104/pp.101.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jee A.-Y.; Chen K.; Tlusty T.; Zhao J.; Granick S. Enhanced Diffusion and Oligomeric Enzyme Dissociation. J. Am. Chem. Soc. 2019, 141 (51), 20062–20068. 10.1021/jacs.9b06949. [DOI] [PubMed] [Google Scholar]
- Follmer C.; Pereira F. V.; Da Silveira N. P.; Carlini C. R. Jack Bean Urease (EC 3.5.1.5) Aggregation Monitored by Dynamic and Static Light Scattering. Biophys. Chem. 2004, 111 (1), 79–87. 10.1016/j.bpc.2004.03.009. [DOI] [PubMed] [Google Scholar]
- Feng M.; Gilson M. K. A Thermodynamic Limit on the Role of Self-Propulsion in Enhanced Enzyme Diffusion. Biophys. J. 2019, 116 (10), 1898–1906. 10.1016/j.bpj.2019.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther J.-P.; Börsch M.; Fischer P. Diffusion Measurements of Swimming Enzymes with Fluorescence Correlation Spectroscopy. Acc. Chem. Res. 2018, 51 (9), 1911–1920. 10.1021/acs.accounts.8b00276. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Hess H.. Chemically-Powered Swimming and Diffusion in the Microscopic World. Nat. Rev. Chem. 2021, 5, 500. 10.1038/s41570-021-00281-6. [DOI] [PubMed] [Google Scholar]
- Feng M.; Gilson M. K. Enhanced Diffusion and Chemotaxis of Enzymes. Annu. Rev. Biophys. 2020, 49, 87–105. 10.1146/annurev-biophys-121219-081535. [DOI] [PubMed] [Google Scholar]
- Salvi G.; De Los Rios P.; Vendruscolo M. Effective Interactions between Chaotropic Agents and Proteins. Proteins Struct. Funct. Genet. 2005, 61 (3), 492–499. 10.1002/prot.20626. [DOI] [PubMed] [Google Scholar]
- Arqué X.; Romero-Rivera A.; Feixas F.; Patiño T.; Osuna S.; Sánchez S. Intrinsic Enzymatic Properties Modulate the Self-Propulsion of Micromotors. Nat. Commun. 2019, 10 (1), 2826. 10.1038/s41467-019-10726-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patino T.; Porchetta A.; Jannasch A.; Lladó A.; Stumpp T.; Schäffer E.; Ricci F.; Sánchez S. Self-Sensing Enzyme-Powered Micromotors Equipped with PH-Responsive DNA Nanoswitches. Nano Lett. 2019, 19 (6), 3440–3447. 10.1021/acs.nanolett.8b04794. [DOI] [PubMed] [Google Scholar]
- Li Z.; Barnes J. C.; Bosoy A.; Stoddart J. F.; Zink J. I. Mesoporous Silica Nanoparticles in Biomedical Applications. Chem. Soc. Rev. 2012, 41 (7), 2590. 10.1039/c1cs15246g. [DOI] [PubMed] [Google Scholar]
- Slowing I. I.; Vivero-Escoto J. L.; Wu C.-W.; Lin V. S.-Y. Mesoporous Silica Nanoparticles as Controlled Release Drug Delivery and Gene Transfection Carriers. Adv. Drug Delivery Rev. 2008, 60 (11), 1278–1288. 10.1016/j.addr.2008.03.012. [DOI] [PubMed] [Google Scholar]
- Trewyn B. G.; Slowing I. I.; Giri S.; Chen H. T.; Lin V. S. Y. Synthesis and Functionalization of a Mesoporous Silica Nanoparticle Based on the Sol-Gel Process and Applications in Controlled Release. Acc. Chem. Res. 2007, 40 (9), 846–853. 10.1021/ar600032u. [DOI] [PubMed] [Google Scholar]
- Barbosa O.; Ortiz C.; Berenguer-Murcia Á.; Torres R.; Rodrigues R. C.; Fernandez-Lafuente R. Glutaraldehyde in Bio-Catalysts Design: A Useful Crosslinker and a Versatile Tool in Enzyme Immobilization. RSC Adv. 2014, 4 (4), 1583–1600. 10.1039/C3RA45991H. [DOI] [Google Scholar]
- Patiño T.; Feiner-Gracia N.; Arqué X.; Miguel-López A.; Jannasch A.; Stumpp T.; Schäffer E.; Albertazzi L.; Sánchez S. Influence of Enzyme Quantity and Distribution on the Self-Propulsion of Non-Janus Urease-Powered Micromotors. J. Am. Chem. Soc. 2018, 140 (25), 7896–7903. 10.1021/jacs.8b03460. [DOI] [PubMed] [Google Scholar]
- Feng Y.; Yuan Y.; Wan J.; Yang C.; Hao X.; Gao Z.; Luo M.; Guan J.. Self-Adaptive Enzyme-Powered Micromotors with Switchable Propulsion Mechanism and Motion Directionality. Appl. Phys. Rev. 2021, 8 ( (1), ), 011406. 10.1063/5.0029060. [DOI] [Google Scholar]
- Zhang D. H.; Li Y. Q.; Peng L. J.; Chen N. Lipase Immobilization on Magnetic Microspheres via Spacer Arms: Effect of Steric Hindrance on the Activity. Biotechnol. Bioprocess Eng. 2014, 19 (5), 838–843. 10.1007/s12257-013-0495-x. [DOI] [Google Scholar]
- Zhang D. H.; Peng L. J.; Wang Y.; Li Y. Q. Lipase Immobilization on Epoxy-Activated Poly(Vinyl Acetate-Acrylamide) Microspheres. Colloids Surfaces B Biointerfaces 2015, 129, 206–210. 10.1016/j.colsurfb.2015.03.056. [DOI] [PubMed] [Google Scholar]
- Evli S.; Öndes̨ B.; Uygun M.; Uygun D. A. Asparaginase Immobilized, Magnetically Guided, and Bubble-Propelled Micromotors. Process Biochem. 2021, 108 (June), 103–109. 10.1016/j.procbio.2021.06.009. [DOI] [Google Scholar]
- Uygun M.; Asunción-Nadal V. de la; Evli S.; Uygun D. A.; Jurado-Sánchez B.; Escarpa A. Dye Removal by Laccase-Functionalized Micromotors. Appl. Mater. Today 2021, 23, 23. 10.1016/j.apmt.2021.101045. [DOI] [Google Scholar]
- Parmar J.; Vilela D.; Villa K.; Wang J.; Sánchez S. Micro- and Nanomotors as Active Environmental Microcleaners and Sensors. J. Am. Chem. Soc. 2018, 140 (30), 9317–9331. 10.1021/jacs.8b05762. [DOI] [PubMed] [Google Scholar]
- Maria-Hormigos R.; Jurado-Sánchez B.; Escarpa A. Surfactant-Free β-Galactosidase Micromotors for “On-The-Move” Lactose Hydrolysis. Adv. Funct. Mater. 2018, 28 (25), 1704256. 10.1002/adfm.201704256. [DOI] [Google Scholar]
- Vasdudevan P. T.; Gokarn Y. Deactivation of Jack Bean Urease in Urea Hydrolysis. Appl. Biochem. Biotechnol. 1996, 60 (1), 49–61. 10.1007/BF02788059. [DOI] [Google Scholar]
- Vasudevan P. T.; Ruggiano L.; Weiland R. H. Studies on the Deactivation of Immobilized Urease. Biotechnol. Bioeng. 1990, 35 (11), 1145–1149. 10.1002/bit.260351110. [DOI] [PubMed] [Google Scholar]
- Ghosh S.; Mohajerani F.; Son S.; Velegol D.; Butler P. J.; Sen A. Motility of Enzyme-Powered Vesicles. Nano Lett. 2019, 19 (9), 6019–6026. 10.1021/acs.nanolett.9b01830. [DOI] [PubMed] [Google Scholar]
- Hortelão A. C.; Carrascosa R.; Murillo-Cremaes N.; Patiño T.; Sánchez S. Targeting 3D Bladder Cancer Spheroids with Urease-Powered Nanomotors. ACS Nano 2019, 13 (1), 429–439. 10.1021/acsnano.8b06610. [DOI] [PubMed] [Google Scholar]
- Patton C. J.; Crouch S. R. Spectrophotometric and Kinetics Investigation of the Berthelot Reaction for the Determination of Ammonia. Anal. Chem. 1977, 49 (3), 464–469. 10.1021/ac50011a034. [DOI] [Google Scholar]
- Riedel C.; Gabizon R.; Wilson C. A. M.; Hamadani K.; Tsekouras K.; Marqusee S.; Pressé S.; Bustamante C. The Heat Released during Catalytic Turnover Enhances the Diffusion of an Enzyme. Nature 2015, 517 (7533), 227–230. 10.1038/nature14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunderdale G.; Ebbens S.; Fairclough P.; Howse J. Importance of Particle Tracking and Calculating the Mean-Squared Displacement in Distinguishing Nanopropulsion from Other Processes. Langmuir 2012, 28 (30), 10997–11006. 10.1021/la301370y. [DOI] [PubMed] [Google Scholar]
- Howse J. R.; Jones R. A. L.; Ryan A. J.; Gough T.; Vafabakhsh R.; Golestanian R. Self-Motile Colloidal Particles: From Directed Propulsion to Random Walk. Phys. Rev. Lett. 2007, 99 (4), 48102. 10.1103/PhysRevLett.99.048102. [DOI] [PubMed] [Google Scholar]
- Mestre R.; Palacios L. S.; Miguel-López A.; Arqué X.; Pagonabarraga I.; Sánchez S. Extraction of the Propulsive Speed of Catalytic Nano- and Micro-Motors under Different Motion Dynamics. ArXiv: 2007.15316v 2020, 1–18. 10.48550/arXiv.2007.15316. [DOI] [Google Scholar]
- Gasteiger E.; Hoogland C.; Gattiker A.; Duvaud S.; Wilkins M. R.; Appel R. D.; Bairoch A.. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker J. M., Ed.; Humana Press: Totowa, NJ, 2005; pp 571–607. 10.1385/1-59259-890-0:571. [DOI] [Google Scholar]
- Patiño T.; Llacer-Wintle J.; Pujals S.; Albertazzi L.; Sánchez S. Protein Corona Formation around Biocatalytic Nanomotors Unveiled by STORM. Chemrxiv 2021, 16–19. 10.33774/chemrxiv-2021-rhtps. [DOI] [PubMed] [Google Scholar]
- Delcanale P.; Albertazzi L. DNA-PAINT Super-Resolution Imaging Data of Surface Exposed Active Sites on Particles. Data Br. 2020, 30, 105468. 10.1016/j.dib.2020.105468. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.