Abstract
Polymorphisms in microRNA (miRNA) genes could influence the expression of miRNAs that regulate the PI3K/Akt signalling pathway and play crucial roles in cancer susceptibility. To investigate the association of single nucleotide polymorphisms (SNPs) in miRNA genes of PI3K/Akt with cervical intraepithelial neoplasia (CIN) and cervical cancer (CC), nine SNPs located in miRNA genes were selected for genotyping, and the association of these SNPs with CIN and CC risk was evaluated. A total of 1,402 participants were enrolled in the current study, including 698 healthy individuals in the control group, 431 patients with CC, and 273 patients with CIN. Nine SNPs in miRNA genes (rs107822 in miR-219a, rs10877887 in let-7i, rs2292832 in miR-149, rs353293 in miR-143, rs3746444 in miR-499, rs3803808 in miR-132, rs4078756 in miR-10b, rs629367 in let-7a, and rs7372209 in miR-26a) were genotyped using MassArray, and the association of these SNPs with CIN and CC were analysed. The results showed that the frequencies of rs107822 in miR-219a and rs2292832 in miR-149 were significantly different between the control and CC groups (p < 0.005). The C allele of rs107822 in miR-219a was associated with an increased risk of CC (OR = 1.29, 95%CI:1.09–1.54) whereas the C allele of rs2292832 in miR-149 was associated with a decreased risk of CC (OR = 0.77, 95%CI:0.64–0.92). The results of inheritance model analysis showed that the best-fit inheritance models for rs107822 and rs2292832 were log-additive. The 2CC + CT genotype of rs107822 could be a risk factor for CC when compared with the TT genotype (OR = 1.28, 95%CI:1.08–1.51). The 2CC + CT genotype of rs2292832 could be a protective factor against CC when compared with the TT genotype (OR = 0.76, 95%CI:0.64–0.92). However, no association of these SNPs with CIN was found in the current study. Our results suggest that rs107822 in the promoter region of miR-219a and rs2292832 in pre-miR-149 region are associated with the risk of CC.
Keywords: MicroRNAs, Phosphatidylinositol 3 kinase, Signalling pathway, Single nucleotide polymorphisms, Association, Cervical cancer, Chinese population
Introduction
Cervical cancer (CC) is the fourth most common malignancy and the second most common gynaecological malignancy in women worldwide (Torre et al., 2015). It is predominantly caused by the persistent infection of high-risk human papilloma virus (HR-HPV) (Burd, 2003; zur Hausen, 2009). Malignant progression involves two main stages: cervical intraepithelial neoplasia (CIN) and CC, and occurs over a long period of time (more than 10 years) after HPV infection (Sasagawa et al., 2012).
The initiation and development of CC is also accompanied by aberrant regulation of host signalling pathways involving in essential cellular mechanisms (proliferation, invasion, survival, inflammation, and immunity), such as PI3K/Akt (Bossler et al., 2019). The PI3K/Akt signalling cascade regulates various fundamental aspects of cellular biology by promoting cell survival, growth, proliferation, migration, and energy metabolism (Morgensztern and McLeod, 2005; Sadeghi and Gerber, 2012; LoRusso, 2016; Aoki and Fujishita, 2017). The aberrant activation of PI3K/Akt signalling pathway has been found to be involved in various human cancers (Sharma et al., 2017; Ediriweera et al., 2019; Liu et al., 2020). In 2006, Bertelsen et al. reported PIK3CA amplification and increased Akt activation in cervical neoplasia (Bertelsen et al., 2006). In 2019, Zhang et al. found that PI3k/Akt/mTOR gene and protein levels increased in the CC tissues compared with the corresponding adjacent tissues (Zhang et al., 2019). Moreover, many studies have revealed by inhibiting or promoting PI3K signalling pathway, that genes could inhibit or promote the CC cells (Fu et al., 2020; Shi et al., 2020; Bai et al., 2021), these indicated the important roles of PI3K signalling pathway in CC.
Dysregulation of microRNAs (miRNAs) in human cancers highlights the important roles of these small single-stranded non-coding RNAs in human cancers (Garzon et al., 2006; Di Leva et al., 2014; Acunzo et al., 2015). They negatively regulate the expression of their target genes through the direct cleavage of mRNA or inhibition of mRNA translation, depending on the degree of complementarity between the seed sequence of miRNAs and their target UTR regions (Lai, 2002; Bartel, 2004; de Moor et al., 2005). Many studies have reported that miRNAs regulate components of the PI3K/Akt signalling pathway (Rahmani et al., 2020a; Rahmani et al., 2020b), and abnormal expression of these miRNAs might induce an out-of-control expression of their targets, which leads to disorders of the corresponding signalling pathway (Akbarzadeh et al., 2021). Studies have observed the abnormal expression of miR-219a (Xu et al., 2020), let-7i (Chhabra, 2018), miR-149 (Zhou and Xu, 2021), miR-143 (Tang et al., 2021), miR-132 (Zhang et al., 2021), miR-10b (Zou et al., 2016), let-7a (Wu et al., 2016) and miR-26a (Dong et al., 2014) in CC or other human cancers, which indicated the important roles of these miRNAs in human cancers. Single nucleotide polymorphisms (SNPs) in miRNA genes can modify the expression of mature miRNAs (Slezak-Prochazka et al., 2010; Króliczewski et al., 2018). Thus, SNPs in miRNA genes are associated with susceptibility to human cancers (Du et al., 2014; Wu et al., 2015; Wang et al., 2018). Previously, we found that rs4636297 in pri-miR-126 and rs11614913 in mature miR-196a2 were associated with CC risk (Yan et al., 2019), which indicates that SNPs in miRNAs might be associated with the development of CC.
In the current study, we first predicted potential targets of candidate miRNAs and enriched them in cancer signalling pathways. Next, miRNAs involved in the PI3K/Akt signalling pathway were screened. Finally, nine SNPs related to nine miRNA genes of PI3K/Akt (rs107822 in miR-219a, rs10877887 in let-7i, rs2292832 in miR-149, rs353293 in miR-143, rs3746444 in miR-499, rs3803808 in miR-132, rs4078756 in miR-10b, rs629367 in let-7a, and rs7372209 in miR-26a) were selected, and the association of SNPs with CIN and CC was evaluated in a Chinese population.
Materials and Methods
Subjects
A total of 273 patients with CIN and 431 with CC were recruited. The patients were diagnosed with CIN and CC at the Third Affiliated Hospital of Kunming Medical University from 2017 to 2019 according to “Diagnosis and Treatment: Obstetrics and Gynaecology” and the International Federation of Gynaecology and Obstetrics (FIGO 2009). The exclusion criteria for the study were as follows: 1) a prior history of primary cancer other than CC, 2) malignant tumours other than CC, 3) currently receiving radiotherapy or chemotherapy, and 4) an unclear diagnosis. According to the cervical pathological diagnostic criteria, CIN was classified into CIN I, II and III. CIN I is characterized as having slight atypical hyperplasia; CIN II as medium atypical hyperplasia; and CIN III as severe atypical hyperplasia (Schiffman et al., 2007). During the same period, 698 healthy women who underwent physical examinations at the same hospital were recruited as the control group. The genomic DNA of the samples was obtained from EDTA anti-coagulated whole blood of the subjects using QIAamp Blood Mini Kit (Qiagen NV, Venlo, Netherlands).
Target Prediction and Signal Pathway Enrichment
The target genes of the miRNAs were predicted using the TargetScan Human 8.0 database (http://www.targetscan.org/vert_80/) (McGeary et al., 2019). Target enrichment was performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) v6.8 (Huang et al., 2009).
SNP Selection and Genotyping
First, the miRNAs involved in the regulation of the PI3K/Akt pathway were chosen through target prediction and enrichment. Then, SNPs which were located in the primary sequences, precursor sequences, or transcriptional regulatory regions of these miRNAs were selected. In addition, the MAF (minor allele frequency) of the SNPs was used as the selection criteria that only the SNPs with MAF over 0.05, were selected. As a result, nine SNPs (rs107822 in promotor region of miR-219a, rs10877887 in promotor region of let-7i, rs2292832 in pre-miRNA sequence of miR-149, rs353293 in promotor region of miR-143, rs3746444 in mature sequence of miR-499, rs3803808 in primary sequence region of miR-132, rs4078756 in promotor region of miR-10b, rs629367 in primary sequence region of let-7a, and rs7372209 in promotor region of miR-26a) were used. Information regarding the miRNA-SNPs selected in this study is presented in Table 1. Genotypes of the nine SNPs were determined using the Agena MassArray system. The PCR primers were designed using AssayDesigner 3.1 (Sequenom Inc., San Diego, CA, United States) (Supplementary Table S1). The PCR conditions and program have been described in our previous study (Li et al., 2020). A MALDI-TOF mass spectrometer (Agena, Inc, San Diego, CA, United States) was used to read SpectroCHIP, and the raw genotyping data was obtained using TYPER4.0 software. Samples were selected for sequencing to confirm the genotyping results for each SNP.
TABLE 1.
The information of the nine SNPs selected in the current study.
| SNPs | Genes | Function Consequence | Location | Alleles | MAF in EAS |
|---|---|---|---|---|---|
| rs107822 | MIR219A | promotor region | Chr 6:33207798 | T > C | 0.396 |
| rs10877887 | MIRLET7I | promotor region | Chr 12:62603400 | T > C | 0.343 |
| rs2292832 | MIR149 | pre-miRNA sequence | Chr 2:240456086 | T > C | 0.363 |
| rs353293 | MIR143 | promotor region | Chr 5:149427663 | C > T | 0.156 |
| rs3746444 | MIR499 | mature miR-499-5p | Chr20:34990448 | A > G | 0.145 |
| rs3803808 | MIR132 | 500bp Downstream | Chr17:2049683 | A > G | 0.455 |
| rs4078756 | MIR10B | promotor region | Chr2:176139387 | T > C | 0.271 |
| rs629367 | MIRLET7A | 500bp Downstream | Chr11:122146306 | A > C | 0.219 |
| rs7372209 | MIR26A | promotor region | Chr3:37969217 | C > T | 0.272 |
Statistical Analysis
Microsoft Excel software and the SPSS 19.0 statistical package were used for statistical analysis in the current study. The Hardy-Weinberg equilibrium (HWE) for each SNP in each group was evaluated. One-way analysis of variance (ANOVA) was used to compare the differences in age among the CIN, CC, and control groups. The differences in allele distributions of these SNPs in the CIN, CC, and control groups were analysed using Fisher’s chi-square test, and the odds ratios (ORs) with associated 95% confidence intervals (CIs) were calculated. Differences in the genotype distribution of these SNPs in the three groups were evaluated by inheritance model analysis using SNPstats software (Solé et al., 2006). The statistical power of the SNPs was calculated using “Power and sample size” software (Dupont and Plummer, 1990; Dupont and Plummer, 1998). The Bonferroni correction was performed for multiple comparisons, and the significance threshold was set at p < 0.005 (0.05/n, n = 9).
Results
Characteristics of the Subjects
A total of 1,402 participants were enrolled in this study. The general clinical characteristics of the participants are presented in Table 2. The average ages for the CIN, CC, and control groups were 46.80 ± 10.01, 47.74 ± 9.78, and 47.91 ± 7.18, respectively. No significant differences in age were found among the CIN, CC, and control groups (Table 2).
TABLE 2.
The clinical characteristics of the subjects enrolled in the current study.
| CC | CIN | Control | F | p value | ||
|---|---|---|---|---|---|---|
| N | 431 | 273 | 698 | |||
| Ages (year) | 47.74 ± 9.78 | 46.80 ± 10.01 | 47.91 ± 7.18 | 1.662 | 0.190 | |
| Pathologic types | SCC (n) | 359 | ||||
| AC(n) | 53 | |||||
| Others (n) | 19 | |||||
| Stages of CC | I (n) | 244 | ||||
| II (n) | 157 | |||||
| III and IV (n) | 30 | |||||
| Stages of CIN | I (n) | 71 | ||||
| II (n) | 57 | |||||
| III (n) | 145 |
Signal Pathway Enrichment of the miRNAs
Potential target genes of the miRNAs were predicted using TargetScan Human 8.0. The potential target genes were then submitted to DAVID to convert a gene list for enrichment. The enrichment results showed that nine miRNAs were involved in the PI3K/Akt signalling pathway (Figure 1).
FIGURE 1.
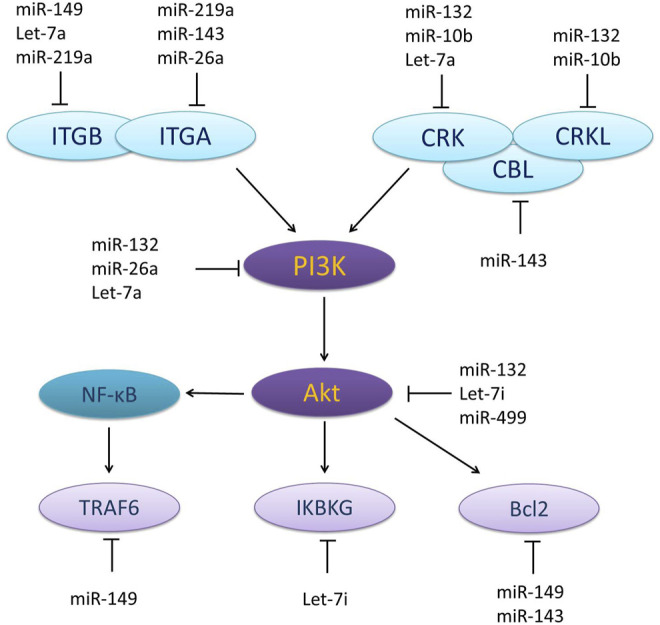
Nine miRNAs involving in PI3K/Akt signalling pathway.
Association of the Alleles of the Nine SNPs With CIN and CC
The nine SNPs were all in HWE in the CIN, CC, and control groups (p > 0.05) (Table 3). The allelic distributions of the nine SNPs are presented in Table 4. The results showed that the allelic distribution of rs107822 in miR-219a and rs2292832 in miR-149 was significantly different between the CC and control groups (p = 0.004 and 0.004, respectively). The C allele of rs107822 in miR-219a was associated with an increased risk of CC (OR = 1.29, 95%CI:1.09–1.54). The C allele of rs2292832 in miR-149 was associated with a decreased risk of CC (OR = 0.77, 95%CI:0.64–0.92). No significant difference in the allelic distribution of the other SNPs was observed among the three groups (p > 0.005). And no association of all nine SNPs with CIN was found (p > 0.005).
TABLE 3.
The Hardy–Weinberg equilibrium significance tests of the nine miRNA SNPs selected in the current study.
| SNPs | Genotypes n (%) | HWE (p-value) | ||
|---|---|---|---|---|
| rs107822 | T/T | T/C | C/C | |
| Control | 275 (39.4) | 314 (45.0) | 109 (15.6) | 0.221 |
| CIN | 101 (37.0) | 124 (45.4) | 48 (17.6) | 0.355 |
| CC | 139 (32.3) | 202 (46.9) | 90 (20.9) | 0.296 |
| rs10877887 | T/T | T/C | C/C | |
| Control | 286 (41.0) | 335 (48.0) | 77 (11.0) | 0.150 |
| CIN | 126 (46.2) | 123 (45.1) | 24 (8.8) | 0.435 |
| CC | 185 (42.9) | 200 (46.4) | 46 (10.7) | 0.457 |
| rs2292832 | T/T | T/C | C/C | |
| Control | 293 (42.0) | 316 (45.3) | 89 (12.8) | 0.792 |
| CIN | 114 (41.8) | 132 (48.4) | 27 (9.9) | 0.207 |
| CC | 209 (48.5) | 189 (43.9) | 33 (7.7) | 0.275 |
| rs353293 | C/C | C/T | T/T | |
| Control | 499 (71.5) | 180 (25.8) | 19 (2.7) | 0.570 |
| CIN | 181 (66.3) | 84 (30.8) | 8 (2.9) | 0.640 |
| CC | 322 (74.7) | 100 (23.2) | 9 (2.1) | 0.707 |
| rs3746444 | A/A | A/G | G/G | |
| Control | 490 (70.2) | 183 (26.2) | 25 (3.6) | 0.130 |
| CIN | 184 (67.4) | 75 (27.5) | 14 (5.1) | 0.090 |
| CC | 292 (67.7) | 119 (27.6) | 20 (4.6) | 0.088 |
| rs3803808 | A/A | A/G | G/G | |
| Control | 246 (35.2) | 340 (48.7) | 112 (16.0) | 0.761 |
| CIN | 83 (30.4) | 134 (49.1) | 56 (20.5) | 0.887 |
| CC | 137 (31.8) | 215 (49.9) | 79 (18.3) | 0.739 |
| rs4078756 | T/T | T/C | C/C | |
| Control | 384 (55.0) | 277 (39.7) | 37 (5.3) | 0.152 |
| CIN | 152 (55.7) | 105 (38.5) | 16 (5.9) | 0.702 |
| CC | 250 (58.0) | 155 (36.0) | 26 (6.0) | 0.762 |
| rs629367 | A/A | A/C | C/C | |
| Control | 407 (58.3) | 250 (35.8) | 41 (5.9) | 0.751 |
| CIN | 176 (64.5) | 82 (30.0) | 15 (5.5) | 0.192 |
| CC | 255 (59.2) | 145 (33.6) | 31 (7.2) | 0.105 |
| rs7372209 | C/C | C/T | T/T | |
| Control | 323 (46.3) | 294 (42.1) | 81 (11.6) | 0.262 |
| CIN | 117 (42.9) | 126 (46.2) | 30 (11.0) | 0.651 |
| CC | 208 (48.3) | 189 (43.9) | 34 (7.9) | 0.321 |
TABLE 4.
The Allele distribution of the nine SNPs in control, CIN and CC groups.
| SNPs | Alleles | Control n (%) | CIN n (%) | CC n (%) | CIN vs. control | CC vs. control | |||
|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) | p value | OR (95%CI] | p value | ||||||
| rs107822 | T | 864 (61.9) | 326 (59.7) | 480 (55.7) | 1.10 (0.90–1.34) | 0.374 | 1.29 (1.09–1.54) | 0.004 | |
| C | 532 (38.1) | 220 (40.3) | 382 (44.3) | ||||||
| rs10877887 | T | 907 (65.0) | 375 (68.7) | 570 (66.1) | 0.85 (0.68–1.05) | 0.121 | 0.95 (0.80–1.14) | 0.575 | |
| C | 489 (35.0) | 171 (31.3) | 292 (33.9) | ||||||
| rs2292832 | T | 902 (64.6) | 360 (65.9) | 607 (70.4) | 0.94 (0.77–1.16) | 0.583 | 0.77 (0.64–0.92) | 0.004 | |
| C | 494 (35.4) | 186 (34.1) | 255 (29.6) | ||||||
| rs353293 | C | 1,178 (84.4) | 446 (81.7) | 744 (86.3) | 0.83 (0.64–1.07) | 0.148 | 0.86 (0.67–1.09) | 0.2241 | |
| T | 218 (15.6) | 100 (18.3) | 118 (13.7) | ||||||
| rs3746444 | A | 1,163 (83.3) | 443 (81.1) | 703 (81.6) | 0.86 (0.67–1.11) | 0.255 | 0.89 (0.71–1.11) | 0.285 | |
| G | 233 (16.7) | 103 (18.9) | 159 (18.4) | ||||||
| rs3803808 | A | 832 (59.6) | 300 (54.9) | 489 (56.7) | 0.83 (0.68–1.01) | 0.062 | 0.89 (0.75–1.06) | 0.179 | |
| G | 564 (40.4) | 246 (45.1) | 373 (43.3) | ||||||
| rs4078756 | T | 1,045 (74.9) | 409 (74.9) | 655 (76.0) | 1.00 (0.79–1.25) | 0.981 | 0.94 (0.77–1.15) | 0.546 | |
| C | 351 (25.1) | 137 (25.1) | 207 (24.0) | ||||||
| rs629367 | A | 1,064 (76.2) | 434 (79.5) | 655 (76.0) | 1.21 (0.95–1.54) | 0.123 | 0.99 (0.81–1.21) | 0.900 | |
| C | 332 (23.8) | 112 (20.5) | 207 (24.0) | ||||||
| rs7372209 | C | 940 (67.3) | 360 (65.9) | 605 (70.2) | 0.94 (0.76–1.16) | 0.555 | 1.14 (0.95–1.38) | 0.157 | |
| T | 456 (32.7) | 186 (34.1) | 257 (29.8) | ||||||
Inheritance Model Analysis of the Nine SNPs With CIN and CC
Five inheritance models (codominant, dominant, recessive, overdominant, and log-additive) were analysed. Akaike Information Criterion (AIC) and Bayesian Information Criterion (BIC) values were used to determine the best-fit model, of which the AIC and BIC values were the lowest for each SNP (Solé et al., 2006). The association of the genotypes of the nine SNPs with CIN and CC was evaluated using inheritance model analysis (Tables 5, 6). The results showed that the genotypes of rs107822 and rs2292832 were significantly different between the CC and control groups (p = 4.6 × 10−3 and 0.004). The best-fit inheritance models for rs107822 and rs2292832 were log-additive. In this model, the 2CC + CT genotype of rs107822 was a risk factor for CC compared to the TT genotype (OR = 1.28, 95%CI:1.08–1.51). For rs2292832, the 2CC + CT genotype was a protective factor against CC compared with the TT genotype in this model (OR = 0.76, 95%CI:0.64–0.92). However, the results showed no association between the other SNPs and CIN or CC (p > 0.005).
TABLE 5.
The inheritance model analysis of these SNPs between CIN and control groups.
| SNPs | Model | Genotypes | Control (n%) | CIN (n%) | OR (95%CI) | p value | AIC | BIC |
|---|---|---|---|---|---|---|---|---|
| rs107822 | Codominant | T/T | 275 (39.4) | 101 (37.0) | 1.00 | 0.680 | 1,157.1 | 1,176.6 |
| C/T | 314 (45.0) | 124 (45.4) | 1.07 (0.79–1.46) | |||||
| C/C | 109 (15.6) | 48 (17.6) | 1.20 (0.80–1.81) | |||||
| Dominant | T/T | 275 (39.4) | 101 (37.0) | 1.00 | 0.500 | 1,155.4 | 1,170.1 | |
| C/T-C/C | 423 (60.6) | 172 (63.0) | 1.10 (0.83–1.48) | |||||
| Recessive | T/T-C/T | 589 (84.4) | 225 (82.4) | 1.00 | 0.440 | 1,155.3 | 1,169.9 | |
| C/C | 109 (15.6) | 48 (17.6) | 1.16 (0.80–1.68) | |||||
| Overdominant | T/T-C/C | 384 (55.0) | 149 (54.6) | 1.00 | 0.930 | 1,155.9 | 1,170.5 | |
| C/T | 314 (45.0) | 124 (45.4) | 1.01 (0.76–1.34) | |||||
| Log-additive | --- | --- | --- | 1.09 (0.90–1.33) | 0.380 | 1,155.1 | 1,169.8 | |
| rs10877887 | Codominant | T/T | 286 (41.0) | 126 (46.2) | 1.00 | 0.320 | 1,155.6 | 1,175.1 |
| C/T | 335 (48.0) | 123 (45.0) | 0.84 (0.62–1.12) | |||||
| C/C | 77 (11.0) | 24 (8.8) | 0.73 (0.44–1.21) | |||||
| Dominant | T/T | 286 (41.0) | 126 (46.2) | 1.00 | 0.160 | 1,153.9 | 1,168.5 | |
| C/T-C/C | 412 (59.0) | 147 (53.9) | 0.82 (0.62–1.08) | |||||
| Recessive | T/T-C/T | 621 (89.0) | 249 (91.2) | 1.00 | 0.360 | 1,155.1 | 1,169.7 | |
| C/C | 77 (11.0) | 24 (8.8) | 0.80 (0.49–1.30) | |||||
| Overdominant | T/T-C/C | 363 (52.0) | 150 (55.0) | 1.00 | 0.400 | 1,155.2 | 1,169.8 | |
| C/T | 335 (48.0) | 123 (45.0) | 0.89 (0.67–1.17) | |||||
| Log-additive | --- | --- | --- | 0.85 (0.68–1.05) | 0.130 | 1,153.6 | 1,168.3 | |
| rs2292832 | Codominant | T/T | 293 (42.0) | 114 (41.8) | 1.00 | 0.390 | 1,156.0 | 1,175.5 |
| T/C | 316 (45.3) | 132 (48.4) | 1.07 (0.80–1.45) | |||||
| C/C | 89 (12.8) | 27 (9.9) | 0.78 (0.48–1.26) | |||||
| Dominant | T/T | 293 (42.0) | 114 (41.8) | 1.00 | 0.960 | 1,155.9 | 1,170.5 | |
| T/C-C/C | 405 (58.0) | 159 (58.2) | 1.01 (0.76–1.34) | |||||
| Recessive | T/T-T/C | 609 (87.2) | 246 (90.1) | 1.00 | 0.200 | 1,154.2 | 1,168.9 | |
| C/C | 89 (12.8) | 27 (9.9) | 0.75 (0.47–1.18) | |||||
| Overdominant | T/T-C/C | 382 (54.7) | 141 (51.6) | 1.00 | 0.380 | 1,155.1 | 1,169.8 | |
| T/C | 316 (45.3) | 132 (48.4) | 1.13 (0.86–1.50) | |||||
| Log-additive | --- | --- | --- | 0.94 (0.76–1.16) | 0.570 | 1,155.6 | 1,170.2 | |
| rs353293 | Codominant | C/C | 499 (71.5) | 181 (66.3) | 1.00 | 0.280 | 1,155.3 | 1,174.9 |
| C/T | 180 (25.8) | 84 (30.8) | 1.29 (0.94–1.76) | |||||
| T/T | 19 (2.7) | 8 (2.9) | 1.18 (0.51–2.75) | |||||
| Dominant | C/C | 499 (71.5) | 181 (66.3) | 1.00 | 0.110 | 1,153.4 | 1,168.0 | |
| C/T-T/T | 199 (28.5) | 92 (33.7) | 1.28 (0.95–1.72) | |||||
| Recessive | C/C-C/T | 679 (97.3) | 265 (97.1) | 1.00 | 0.830 | 1,155.8 | 1,170.5 | |
| T/T | 19 (2.7) | 8 (2.9) | 1.10 (0.47–2.54) | |||||
| Overdominant | C/C-T/T | 518 (74.2) | 189 (69.2) | 1.00 | 0.120 | 1,153.5 | 1,168.1 | |
| C/T | 180 (25.8) | 84 (30.8) | 1.28 (0.94–1.74) | |||||
| Log-additive | --- | --- | --- | 1.21 (0.94–1.58) | 0.150 | 1,153.8 | 1,168.4 | |
| rs3746444 | Codominant | A/A | 490 (70.2) | 184 (67.4) | 1.00 | 0.460 | 1,156.4 | 1,175.9 |
| A/G | 183 (26.2) | 75 (27.5) | 1.09 (0.79–1.50) | |||||
| G/G | 25 (3.6) | 14 (5.1) | 1.51 (0.77–2.98) | |||||
| Dominant | A/A | 490 (70.2) | 184 (67.4) | 1.00 | 0.390 | 1,155.1 | 1,169.8 | |
| A/G-G/G | 208 (29.8) | 89 (32.6) | 1.14 (0.85–1.54) | |||||
| Recessive | A/A-A/G | 673 (96.4) | 259 (94.9) | 1.00 | 0.270 | 1,154.7 | 1,169.3 | |
| G/G | 25 (3.6) | 14 (5.1) | 1.47 (0.75–2.88) | |||||
| Overdominant | A/A-G/G | 515 (73.8) | 198 (72.5) | 1.00 | 0.690 | 1,155.7 | 1,170.4 | |
| A/G | 183 (26.2) | 75 (27.5) | 1.07 (0.78–1.46) | |||||
| Log-additive | --- | --- | --- | 1.15 (0.90–1.48) | 0.260 | 1,154.6 | 1,169.3 | |
| rs3803808 | Codominant | A/A | 246 (35.2) | 83 (30.4) | 1.00 | 0.180 | 1,154.4 | 1,173.9 |
| G/A | 340 (48.7) | 134 (49.1) | 1.18 (0.85–1.62) | |||||
| G/G | 112 (16.1) | 56 (20.5) | 1.47 (0.98–2.21) | |||||
| Dominant | A/A | 246 (35.2) | 83 (30.4) | 1.00 | 0.140 | 1,153.8 | 1,168.4 | |
| G/A-G/G | 452 (64.8) | 190 (69.6) | 1.25 (0.92–1.69) | |||||
| Recessive | A/A-G/A | 586 (83.9) | 217 (79.5) | 1.00 | 0.110 | 1,153.4 | 1,168 | |
| G/G | 112 (16.1) | 56 (20.5) | 1.34 (0.94–1.91) | |||||
| Overdominant | A/A-G/G | 358 (51.3) | 139 (50.9) | 1.00 | 0.870 | 1,155.9 | 1,170.5 | |
| G/A | 340 (48.7) | 134 (49.1) | 1.02 (0.77–1.36) | |||||
| Log-additive | --- | --- | --- | 1.21 (0.99–1.48) | 0.064 | 1,152.5 | 1,167.1 | |
| rs4078756 | Codominant | T/T | 384 (55.0) | 152 (55.7) | 1.00 | 0.930 | 1,157.8 | 1,177.3 |
| C/T | 277 (39.7) | 105 (38.5) | 0.97 (0.72–1.29) | |||||
| C/C | 37 (5.3) | 16 (5.9) | 1.08 (0.58–1.99) | |||||
| Dominant | T/T | 384 (55.0) | 152 (55.7) | 1.00 | 0.880 | 1,155.9 | 1,170.5 | |
| C/T-C/C | 314 (45.0) | 121 (44.3) | 0.98 (0.74–1.30) | |||||
| Recessive | T/T-C/T | 661 (94.7) | 257 (94.1) | 1.00 | 0.780 | 1,155.8 | 1,170.4 | |
| C/C | 37 (5.3) | 16 (5.9) | 1.09 (0.60–2.00) | |||||
| Overdominant | T/T-C/C | 421 (60.3) | 168 (61.5) | 1.00 | 0.780 | 1,155.8 | 1,170.4 | |
| C/T | 277 (39.7) | 105 (38.5) | 0.96 (0.72–1.28) | |||||
| Log-additive | --- | --- | --- | 1.00 (0.79–1.26) | 0.990 | 1,155.9 | 1,170.5 | |
| rs629367 | Codominant | A/A | 407 (58.3) | 176 (64.5) | 1.00 | 0.210 | 1,154.8 | 1,174.3 |
| C/A | 250 (35.8) | 82 (30.0) | 0.76 (0.56–1.03) | |||||
| C/C | 41 (5.9) | 15 (5.5) | 0.86 (0.46–1.60) | |||||
| Dominant | A/A | 407 (58.3) | 176 (64.5) | 1.00 | 0.084 | 1,152.9 | 1,167.5 | |
| C/A-C/C | 291 (41.7) | 97 (35.5) | 0.78 (0.58–1.04) | |||||
| Recessive | A/A-C/A | 657 (94.1) | 258 (94.5) | 1.00 | 0.870 | 1,155.9 | 1,170.5 | |
| C/C | 41 (5.9) | 15 (5.5) | 0.95 (0.52–1.75) | |||||
| Overdominant | A/A-C/C | 448 (64.2) | 191 (70.0) | 1.00 | 0.088 | 1,153.0 | 1,167.6 | |
| C/A | 250 (35.8) | 82 (30.0) | 0.77 (0.57–1.04) | |||||
| Log-additive | --- | --- | --- | 0.84 (0.66–1.06) | 0.140 | 1,153.7 | 1,168.3 | |
| rs7372209 | Codominant | C/C | 323 (46.3) | 117 (42.9) | 1.00 | 0.540 | 1,156.7 | 1,176.2 |
| T/C | 294 (42.1) | 126 (46.1) | 1.18 (0.88–1.59) | |||||
| T/T | 81 (11.6) | 30 (11.0) | 1.03 (0.65–1.66) | |||||
| Dominant | C/C | 323 (46.3) | 117 (42.9) | 1.00 | 0.340 | 1,155.0 | 1,169.6 | |
| T/C-T/T | 375 (53.7) | 156 (57.1) | 1.15 (0.86–1.52) | |||||
| Recessive | C/C-T/C | 617 (88.4) | 243 (89.0) | 1.00 | 0.830 | 1,155.8 | 1,170.5 | |
| T/T | 81 (11.6) | 30 (11.0) | 0.95 (0.61–1.49) | |||||
| Overdominant | C/C-T/T | 404 (57.9) | 147 (53.9) | 1.00 | 0.270 | 1,154.7 | 1,169.3 | |
| T/C | 294 (42.1) | 126 (46.1) | 1.17 (0.88–1.55) | |||||
| Log-additive | --- | --- | --- | 1.07 (0.87–1.31) | 0.540 | 1,155.5 | 1,170.2 |
TABLE 6.
The inheritance model analysis of these SNPs between CC and control groups.
| SNPs | Models | Genotypes | Control n (%) | CC n (%) | OR (95%CI) | p value | AIC | BIC |
|---|---|---|---|---|---|---|---|---|
| rs107822 | Codominant | T/T | 275 (39.4) | 139 (32.2) | 1.00 | 0.018 | 1,501.2 | 1,521.4 |
| C/T | 314 (45.0) | 202 (46.9) | 1.27 (0.97–1.67) | |||||
| C/C | 109 (15.6) | 90 (20.9) | 1.63 (1.15–2.31) | |||||
| Dominant | T/T | 275 (39.4) | 139 (32.2) | 1.00 | 0.015 | 1,501.4 | 1,516.5 | |
| C/T-C/C | 423 (60.6) | 292 (67.8) | 1.36 (1.06–1.76) | |||||
| Recessive | T/T-C/T | 589 (84.4) | 341 (79.1) | 1.00 | 0.026 | 1,502.3 | 1,517.4 | |
| C/C | 109 (15.6) | 90 (20.9) | 1.42 (1.05–1.94) | |||||
| Overdominant | T/T-C/C | 384 (55.0) | 229 (53.1) | 1.00 | 0.540 | 1,506.9 | 1,522.0 | |
| C/T | 314 (45.0) | 202 (46.9) | 1.08 (0.85–1.37) | |||||
| Log-additive | --- | --- | --- | 1.28 (1.08–1.51) | 4.6 x 10−3 | 1,499.0 | 1,514.0 | |
| rs10877887 | Codominant | T/T | 286 (41.0) | 185 (42.9) | 1.00 | 0.810 | 1,508.9 | 1,529.0 |
| T/C | 335 (48.0) | 200 (46.4) | 0.92 (0.72–1.19) | |||||
| C/C | 77 (11.0) | 46 (10.7) | 0.93 (0.61–1.40) | |||||
| Dominant | T/T | 286 (41.0) | 185 (42.9) | 1.00 | 0.520 | 1,506.9 | 1,522.0 | |
| T/C-C/C | 412 (59.0) | 246 (57.1) | 0.92 (0.72–1.18) | |||||
| Recessive | T/T-T/C | 621 (89.0) | 385 (89.3) | 1.00 | 0.860 | 1,507.2 | 1,522.3 | |
| C/C | 77 (11.0) | 46 (10.7) | 0.97 (0.66–1.42) | |||||
| Overdominant | T/T-C/C | 363 (52.0) | 231 (53.6) | 1.00 | 0.600 | 1,507.0 | 1,522.1 | |
| T/C | 335 (48.0) | 200 (46.4) | 0.94 (0.74–1.19) | |||||
| Log-additive | --- | --- | --- | 0.95 (0.79–1.14) | 0.570 | 1,507.0 | 1,522.0 | |
| rs2292832 | Codominant | T/T | 293 (42.0) | 209 (48.5) | 1.00 | 0.009 | 1,499.9 | 1,520.0 |
| T/C | 316 (45.3) | 189 (43.9) | 0.84 (0.65–1.08) | |||||
| C/C | 89 (12.8) | 33 (7.7) | 0.52 (0.34–0.80) | |||||
| Dominant | T/T | 293 (42.0) | 209 (48.5) | 1.00 | 0.033 | 1,502.7 | 1,517.8 | |
| T/C-C/C | 405 (58.0) | 222 (51.5) | 0.77 (0.60–0.98) | |||||
| Recessive | T/T-T/C | 609 (87.2) | 398 (92.3) | 1.00 | 0.006 | 1,499.8 | 1,514.9 | |
| C/C | 89 (12.8) | 33 (7.7) | 0.57 (0.37–0.86) | |||||
| Overdominant | T/T-C/C | 382 (54.7) | 242 (56.1) | 1.00 | 0.640 | 1,507.1 | 1,522.2 | |
| T/C | 316 (45.3) | 189 (43.9) | 0.94 (0.74–1.20) | |||||
| Log-additive | --- | --- | --- | 0.76 (0.64–0.92) | 0.004 | 1,499.0 | 1,514.0 | |
| rs353293 | Codominant | C/C | 499 (71.5) | 322 (74.7) | 1.00 | 0.460 | 1,507.7 | 1,527.8 |
| C/T | 180 (25.8) | 100 (23.2) | 0.86 (0.65–1.14) | |||||
| T/T | 19 (2.7) | 9 (2.1) | 0.73 (0.33–1.64) | |||||
| Dominant | C/C | 499 (71.5) | 322 (74.7) | 1.00 | 0.230 | 1,505.9 | 1,520.9 | |
| C/T-T/T | 199 (28.5) | 109 (25.3) | 0.85 (0.65–1.11) | |||||
| Recessive | C/C-C/T | 679 (97.3) | 422 (97.9) | 1.00 | 0.500 | 1,506.8 | 1,521.9 | |
| T/T | 19 (2.7) | 9 (2.1) | 0.76 (0.34–1.70) | |||||
| Overdominant | C/C-T/T | 518 (74.2) | 331 (76.8) | 1.00 | 0.320 | 1,506.3 | 1,521.4 | |
| C/T | 180 (25.8) | 100 (23.2) | 0.87 (0.66–1.15) | |||||
| Log-additive | --- | --- | --- | 0.86 (0.68–1.09) | 0.210 | 1,505.7 | 1,520.8 | |
| rs3746444 | Codominant | A/A | 490 (70.2) | 292 (67.8) | 1.00 | 0.560 | 1,508.1 | 1,528.3 |
| A/G | 183 (26.2) | 119 (27.6) | 1.09 (0.83–1.43) | |||||
| G/G | 25 (3.6) | 20 (4.6) | 1.34 (0.73–2.46) | |||||
| Dominant | A/A | 490 (70.2) | 292 (67.8) | 1.00 | 0.390 | 1,506.5 | 1,521.6 | |
| A/G-G/G | 208 (29.8) | 139 (32.2) | 1.12 (0.86–1.45) | |||||
| Recessive | A/A-A/G | 673 (96.4) | 411 (95.4) | 1.00 | 0.380 | 1,506.5 | 1,521.6 | |
| G/G | 25 (3.6) | 20 (4.6) | 1.31 (0.72–2.38) | |||||
| Overdominant | A/A-G/G | 515 (73.8) | 312 (72.4) | 1.00 | 0.610 | 1,507.0 | 1,522.1 | |
| A/G | 183 (26.2) | 119 (27.6) | 1.07 (0.82–1.41) | |||||
| Log-additive | --- | --- | --- | 1.12 (0.90–1.39) | 0.310 | 1,506.2 | 1,521.3 | |
| rs3803808 | Codominant | A/A | 246 (35.2) | 137 (31.8) | 1.00 | 0.400 | 1,507.4 | 1,527.5 |
| G/A | 340 (48.7) | 215 (49.9) | 1.14 (0.87–1.49) | |||||
| G/G | 112 (16.1) | 79 (18.3) | 1.27 (0.89–1.81) | |||||
| Dominant | A/A | 246 (35.2) | 137 (31.8) | 1.00 | 0.230 | 1,505.8 | 1,520.9 | |
| G/A-G/G | 452 (64.8) | 294 (68.2) | 1.17 (0.91–1.51) | |||||
| Recessive | A/A-G/A | 586 (83.9) | 352 (81.7) | 1.00 | 0.320 | 1,506.3 | 1,521.4 | |
| G/G | 112 (16.1) | 79 (18.3) | 1.17 (0.86–1.61) | |||||
| Overdominant | A/A-G/G | 358 (51.3) | 216 (50.1) | 1.00 | 0.690 | 1,507.1 | 1,522.2 | |
| G/A | 340 (48.7) | 215 (49.9) | 1.05 (0.83–1.33) | |||||
| Log-additive | --- | --- | --- | 1.13 (0.95–1.34) | 0.170 | 1,505.4 | 1,520.5 | |
| rs4078756 | Codominant | T/T | 384 (55.0) | 250 (58.0) | 1.00 | 0.450 | 1,507.7 | 1,527.8 |
| C/T | 277 (39.7) | 155 (36.0) | 0.86 (0.67–1.11) | |||||
| C/C | 37 (5.3) | 26 (6.0) | 1.08 (0.64–1.82) | |||||
| Dominant | T/T | 384 (55.0) | 250 (58.0) | 1.00 | 0.330 | 1,506.3 | 1,521.4 | |
| C/T-C/C | 314 (45.0) | 181 (42.0) | 0.89 (0.70–1.13) | |||||
| Recessive | T/T-C/T | 661 (94.7) | 405 (94.0) | 1.00 | 0.610 | 1,507.0 | 1,522.1 | |
| C/C | 37 (5.3) | 26 (6.0) | 1.14 (0.68–1.92) | |||||
| Overdominant | T/T-C/C | 421 (60.3) | 276 (64.0) | 1.00 | 0.210 | 1,505.7 | 1,520.8 | |
| C/T | 277 (39.7) | 155 (36.0) | 0.85 (0.67–1.10) | |||||
| Log-additive | --- | --- | --- | 0.94 (0.77–1.15) | 0.540 | 1,506.9 | 1,522.0 | |
| .rs629367 | Codominant | A/A | 407 (58.3) | 255 (59.2) | 1.00 | 0.580 | 1,508.2 | 1,528.3 |
| C/A | 250 (35.8) | 145 (33.6) | 0.93 (0.72–1.20) | |||||
| C/C | 41 (5.9) | 31 (7.2) | 1.21 (0.74–1.97) | |||||
| Dominant | A/A | 407 (58.3) | 255 (59.2) | 1.00 | 0.780 | 1,507.2 | 1,522.3 | |
| C/A-C/C | 291 (41.7) | 176 (40.8) | 0.97 (0.76–1.23) | |||||
| Recessive | A/A-C/A | 657 (94.1) | 400 (92.8) | 1.00 | 0.380 | 1,506.5 | 1,521.6 | |
| C/C | 41 (5.9) | 31 (7.2) | 1.24 (0.77–2.01) | |||||
| Overdominant | A/A-C/C | 448 (64.2) | 286 (66.4) | 1.00 | 0.460 | 1,506.7 | 1,521.8 | |
| C/A | 250 (35.8) | 145 (33.6) | 0.91 (0.71–1.17) | |||||
| Log-additive | --- | --- | --- | 1.01 (0.83–1.23) | 0.900 | 1,507.3 | 1,522.3 | |
| rs7372209 | Codominant | C/C | 323 (46.3) | 208 (48.3) | 1.00 | 0.130 | 1,505.2 | 1,525.3 |
| T/C | 294 (42.1) | 189 (43.9) | 1.00 (0.78–1.29) | |||||
| T/T | 81 (11.6) | 34 (7.9) | 0.65 (0.42–1.01) | |||||
| Dominant | C/C | 323 (46.3) | 208 (48.3) | 1.00 | 0.520 | 1,506.9 | 1,522.0 | |
| T/C-T/T | 375 (53.7) | 223 (51.7) | 0.92 (0.73–1.18) | |||||
| Recessive | C/C-T/C | 617 (88.4) | 397 (92.1) | 1.00 | 0.043 | 1,503.2 | 1,518.3 | |
| T/T | 81 (11.6) | 34 (7.9) | 0.65 (0.43–0.99) | |||||
| Overdominant | C/C-T/T | 404 (57.9) | 242 (56.1) | 1.00 | 0.570 | 1,506.9 | 1,522.0 | |
| T/C | 294 (42.1) | 189 (43.9) | 1.07 (0.84–1.37) | |||||
| Log-additive | --- | --- | --- | 0.88 (0.73–1.05) | 0.160 | 1,505.3 | 1,520.0 |
Association Analysis of Nine SNPs With Different Pathological Types of CC
To investigate the association of the nine SNPs with the pathological types of CC, we analysed the distribution characteristics of the nine SNPs in different pathological types of CC. However, there were no significant differences in these SNPs between AC and SCC after Bonferroni correction (p > 0.005) (Supplementary Table S2).
Association Analysis of Nine SNPs With Different Stages of CIN and CC
To investigate the association of the nine SNPs with different stages of CIN and CC, the CIN group was divided into CIN I + II and CIN III, and the CC group was divided into stages I and II + III + IV. No significant associations of these SNPs were observed between CIN I + II and CIN III and between CC stage I and stage II + III + IV after Bonferroni correction (p > 0.005) (Supplementary Tables S3, S4).
Discussion
Alterations in the PI3K/Akt signalling pathway have been found in human cancers (Vara et al., 2004). These alterations might be a consequence of aberrant miRNA expression (Peng et al., 2019; Ichikawa et al., 2020). To explore the role of SNPs in miRNA genes involved in the PI3K/Akt pathway in CC susceptibility, the association of nine SNPs located in the miRNA genes involved in the PI3K/Akt pathway with CIN and CC was investigated. Results showed that the frequencies of rs107822 in miR-219a and rs2292832 in miR-149 were significantly different between the control and CC groups (p < 0.005).
To date, many studies have revealed that miR-219a functions as a tumour suppressor in different cancers, such as ovarian and breast cancer (Long et al., 2017; Xing et al., 2018; Wang et al., 2020; Ye et al., 2021). In the current study, we predicted that miR-219a could target integrins (ITGA and ITGB) which can participate in the activation of the PI3K/Akt signalling pathway. Moreover, our results showed that the rs107822C allele and CC genotype were risk factors for CC. Similarly, rs107822 has been reported to be associated with lung cancer (Zheng et al., 2017) and oesophageal squamous cell carcinoma (Song et al., 2015), and the C allele was associated with an increased risk of cancer. These results are consistent with those found in CC in the current study. Rs107822 is located at the 2 Kb upstream of miR-219a, which may be the transcriptional regulatory region of miR-219a. In 2012, Greliche et al. found that rs107822 in miR-219a could affect HLA-DPB1 expression in monocytes through interaction with rs1042448 in the 3′-UTR of HLA-DPB1 (Greliche et al., 2012). The distance between rs107822 in miR-219a and rs1042448 in the 3′UTR of HLA-DPB1 is approximately 100 kb on chromosome 6, and these two SNPs show modest linkage disequilibrium (Greliche et al., 2012). Genome-wide association studies have revealed that loci susceptible for CC are located in the HLA-DP region (Chen et al., 2013; Shi et al., 2013), which indicates the important role of HLA-DP in CC. Thus, rs107822 may be associated with CC by affecting the expression of HLA-DPB1 through interaction with rs1042448 in the 3′UTR of HLA-DPB1. The interaction between rs107822 and the 3′UTR SNP (rs1042448) may be affected by miRNA expression (miRSNP) and miRNA binding specificity (3′UTR SNP) (Greliche et al., 2012). However, no association of this SNP with CIN was found in the current study, which was not consistent with the results of CC. As we known, the precancerous lesions and the carcinogenesis are different stages during the development of cervical cancer. Thus, one of the reasons of the discrepancy between CIN and CC could be miR-219a might play different roles in these two stages.
In 2020, Shao et al. reported that miR-149 functions as a tumour suppressor in CC by negatively regulating AURKA (Shao et al., 2020). Similarly, Zhou et al. found that miR-149 inhibits CC by targeting POU2F2 (Zhou and Xu, 2021). These results indicate a suppressive role of miR-149 in CC. In the current study, the results showed that rs2292832 was associated with CC susceptibility, and the C allele was associated with a decreased risk of CC. Our results are consistent with those of another study on CC by Wang et al. (2019). Similarly, the rs2292832 has been documented to be associated with various types of human cancer, such as gastric (Zhang et al., 2018), hepatocellular (Wang et al., 2014) and breast cancers (He et al., 2015). However, other studies have reported no such association (Dai et al., 2015; Li et al., 2016; Cîmpeanu et al., 2017; Yu et al., 2017). One of the reasons for the discrepancy between different studies is that rs2292832 may play different roles in different cancers. The other reason could be the different genetic background populations enrolled in the different studies. The third reason could be the different sample sizes in different studies which might affect the reliability of the association studies. Rs2292832 is located at the lower stream of the stem-loop structure of precursor miR-149, which might be related to the cleavage of pri-miRNA by DROSHA (Han et al., 2006; Auyeung et al., 2013). Thus, rs2292832 might be associated with CC through modulation of the maturation process of miR-149, subsequently affecting the expression of its target genes (ITGB and TRAF6) in the PI3K/Akt signalling pathway. Similar to rs107822, rs2292832 only exhibit an association with CC, not CIN, which might due to that miR-149 plays different roles between the precancerous lesions and the carcinogenesis stages in the cervical cancer development.
One limitation in the current study could be the lack of HPV status for our every patient, which makes it difficult to perform combined analyses of HPV status and gene SNPs interaction. Therefore, the roles of the interactions of HPV and host gene SNPs in the CC development should be investigated in the future.
Conclusion
In summary, nine miRNAs involved in the PI3K/Akt signalling pathway were selected, and nine SNPs located in regions related to miRNA transcription or processing were chosen to investigate their association with CC. Our results showed that rs107822 of miR-219a and rs2292832 of miR-149 were associated with CC risk. The statistical power in the comparison between CC and control groups for rs107822 and rs2292832 were 0.818 and 0.803 respectively. Thus, the function of these two SNPs in the CC development should be investigated and verified in the future.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics Statement
The studies involving human participants were reviewed and approved by the Institutional Review Board of the Third Affiliated Hospital of Kunming Medical University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This work was supported by grant from the National Science Foundation of China (82103190); CAMS Innovation Fund for Medical Sciences (2021-I2M-1-004); Fundamental Research Funds for the Central Universities (3332019111); Yunnan Province Clinical Research Center for Gynecological and Obstetric Disease (2022YJZX-FC06); Applied Basic Research Projects of Yunnan province (202101AS070205, 202101AU070191), Special Funds for High-level Healthy Talents of Yunnan Province (L-201615, H-2018014, and D-2018037). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.856505/full#supplementary-material
References
- Acunzo M., Romano G., Wernicke D., Croce C. M. (2015). MicroRNA and Cancer - A Brief Overview. Adv. Biol. Regul. 57, 1–9. 10.1016/j.jbior.2014.09.013 [DOI] [PubMed] [Google Scholar]
- Akbarzadeh M., Mihanfar A., Akbarzadeh S., Yousefi B., Majidinia M. (2021). Crosstalk between miRNA and PI3K/AKT/mTOR Signaling Pathway in Cancer. Life Sci. 285, 119984. 10.1016/j.lfs.2021.119984 [DOI] [PubMed] [Google Scholar]
- Aoki M., Fujishita T. (2017). Oncogenic Roles of the PI3K/AKT/mTOR Axis. Curr. Top. Microbiol. Immunol. 407, 153–189. 10.1007/82_2017_6 [DOI] [PubMed] [Google Scholar]
- Auyeung V. C., Ulitsky I., McGeary S. E., Bartel D. P. (2013). Beyond Secondary Structure: Primary-Sequence Determinants License Pri-miRNA Hairpins for Processing. Cell 152 (4), 844–858. 10.1016/j.cell.2013.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Li H., Lv R. (2021). Interleukin-17 Activates JAK2/STAT3, PI3K/Akt and Nuclear factor κB Signaling Pathway to Promote the Tumorigenesis of Cervical Cancer. Exp. Ther. Med. 22 (5), 1291. 10.3892/etm.2021.10726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P. (2004). MicroRNAs. Cell 116 (2), 281–297. 10.1016/s0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Bertelsen B. I., Steine S. J., Sandvei R., Molven A., Laerum O. D. (2006). Molecular Analysis of the PI3K-AKT Pathway in Uterine Cervical Neoplasia: FrequentPIK3CAamplification and AKT Phosphorylation. Int. J. Cancer 118 (8), 1877–1883. 10.1002/ijc.21461 [DOI] [PubMed] [Google Scholar]
- Bossler F., Hoppe-Seyler K., Hoppe-Seyler F. (2019). PI3K/AKT/mTOR Signaling Regulates the Virus/Host Cell Crosstalk in HPV-Positive Cervical Cancer Cells. Int. J. Mol. Sci. 20 (9), 2188. 10.3390/ijms20092188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd E. M. (2003). Human Papillomavirus and Cervical Cancer. Clin. Microbiol. Rev. 16 (1), 1–17. 10.1128/cmr.16.1.1-17.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Juko-Pecirep I., Hammer J., Ivansson E., Enroth S., Gustavsson I., et al. (2013). Genome-wide Association Study of Susceptibility Loci for Cervical Cancer. J. Natl. Cancer Inst. 105 (9), 624–633. 10.1093/jnci/djt051 [DOI] [PubMed] [Google Scholar]
- Chhabra R. (2018). Let-7i-5p, miR-181a-2-3p and EGF/PI3K/SOX2 axis Coordinate to Maintain Cancer Stem Cell Population in Cervical Cancer. Sci. Rep. 8 (1), 7840. 10.1038/s41598-018-26292-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cîmpeanu R. A., Popescu D. M., Burada F., Cucu M. G., Gheonea D. I., Ioana M., et al. (2017). miR-149 Rs2292832 C>T Polymorphism and Risk of Gastric Cancer. Rom. J. Morphol. Embryol. 58 (1), 125–129. [PubMed] [Google Scholar]
- Dai Z.-J., Shao Y.-P., Wang X.-J., Xu D., Kang H.-F., Ren H.-T., et al. (2015). Five Common Functional Polymorphisms in microRNAs (Rs2910164, Rs2292832, Rs11614913, Rs3746444, Rs895819) and the Susceptibility to Breast Cancer: Evidence from 8361 Cancer Cases and 8504 Controls. Curr. Pharm. Des. 21 (11), 1455–1463. 10.2174/1381612821666141208143533 [DOI] [PubMed] [Google Scholar]
- de Moor C. H., Meijer H., Lissenden S. (2005). Mechanisms of Translational Control by the 3′ UTR in Development and Differentiation. Semin. Cel Develop. Biol. 16 (1), 49–58. 10.1016/j.semcdb.2004.11.007 [DOI] [PubMed] [Google Scholar]
- Di Leva G., Garofalo M., Croce C. M. (2014). MicroRNAs in Cancer. Annu. Rev. Pathol. Mech. Dis. 9, 287–314. 10.1146/annurev-pathol-012513-104715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Sui L., Wang Q., Chen M., Sun H. (2014). MicroRNA-26a Inhibits Cell Proliferation and Invasion of Cervical Cancer Cells by Targeting Protein Tyrosine Phosphatase Type IVA 1. Mol. Med. Rep. 10 (3), 1426–1432. 10.3892/mmr.2014.2335 [DOI] [PubMed] [Google Scholar]
- Du M., Lu D., Wang Q., Chu H., Tong N., Pan X., et al. (2014). Genetic Variations in microRNAs and the Risk and Survival of Renal Cell Cancer. Carcinogenesis 35 (7), 1629–1635. 10.1093/carcin/bgu082 [DOI] [PubMed] [Google Scholar]
- Dupont W. D., Plummer W. D., Jr. (1990). Power and Sample Size Calculations. Controlled Clin. Trials 11 (2), 116–128. 10.1016/0197-2456(90)90005-m [DOI] [PubMed] [Google Scholar]
- Dupont W. D., Plummer W. D., Jr. (1998). Power and Sample Size Calculations for Studies Involving Linear Regression. Controlled Clin. Trials 19 (6), 589–601. 10.1016/s0197-2456(98)00037-3 [DOI] [PubMed] [Google Scholar]
- Ediriweera M. K., Tennekoon K. H., Samarakoon S. R. (2019). Role of the PI3K/AKT/mTOR Signaling Pathway in Ovarian Cancer: Biological and Therapeutic Significance. Semin. Cancer Biol. 59, 147–160. 10.1016/j.semcancer.2019.05.012 [DOI] [PubMed] [Google Scholar]
- Fu K., Zhang L., Liu R., Shi Q., Li X., Wang M. (2020). MiR-125 Inhibited Cervical Cancer Progression by Regulating VEGF and PI3K/AKT Signaling Pathway. World J. Surg. Onc. 18 (1), 115. 10.1186/s12957-020-01881-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R., Fabbri M., Cimmino A., Calin G. A., Croce C. M. (2006). MicroRNA Expression and Function in Cancer. Trends Mol. Med. 12 (12), 580–587. 10.1016/j.molmed.2006.10.006 [DOI] [PubMed] [Google Scholar]
- Greliche N., Zeller T., Wild P. S., Rotival M., Schillert A., Ziegler A., et al. (2012). Comprehensive Exploration of the Effects of miRNA SNPs on Monocyte Gene Expression. PLoS One 7 (9), e45863. 10.1371/journal.pone.0045863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Lee Y., Yeom K.-H., Nam J.-W., Heo I., Rhee J.-K., et al. (2006). Molecular Basis for the Recognition of Primary microRNAs by the Drosha-DGCR8 Complex. Cell 125 (5), 887–901. 10.1016/j.cell.2006.03.043 [DOI] [PubMed] [Google Scholar]
- He B., Pan Y., Xu Y., Deng Q., Sun H., Gao T., et al. (2015). Associations of Polymorphisms in microRNAs with Female Breast Cancer Risk in Chinese Population. Tumor Biol. 36 (6), 4575–4582. 10.1007/s13277-015-3102-2 [DOI] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Lempicki R. A. (2009). Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat. Protoc. 4 (1), 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Ichikawa R., Kawasaki R., Iwata A., Otani S., Nishio E., Nomura H., et al. (2020). MicroRNA-126 3p Suppresses HeLa Cell Proliferation, Migration and Invasion, and Increases Apoptosis via the PI3K/PDK1/AKT Pathway. Oncol. Rep. 43 (4), 1300–1308. 10.3892/or.2020.7512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Króliczewski J., Sobolewska A., Lejnowski D., Collawn J. F., Bartoszewski R. (2018). microRNA Single Polynucleotide Polymorphism Influences on microRNA Biogenesis and mRNA Target Specificity. Gene 640, 66–72. 10.1016/j.gene.2017.10.021 [DOI] [PubMed] [Google Scholar]
- Lai E. C. (2002). Micro RNAs Are Complementary to 3′ UTR Sequence Motifs that Mediate Negative post-transcriptional Regulation. Nat. Genet. 30 (4), 363–364. 10.1038/ng865 [DOI] [PubMed] [Google Scholar]
- Li H., Ren Y., Xia L., Qu R., Kong L., Yin Z., et al. (2016). Association of MicroRNA-149 Polymorphism with Lung Cancer Risk in Chinese Non-Smoking Female: A Case-Control Study. PLoS One 11 (9), e0163626. 10.1371/journal.pone.0163626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Dai S., Yan Z., Zhang X., Liu S., Wang X., et al. (2020). Genetic Polymorphisms of Proteasome Subunit Genes of the MHC-I Antigen-Presenting System Are Associated with Cervical Cancer in a Chinese Han Population. Hum. Immunol. 81 (8), 445–451. 10.1016/j.humimm.2020.07.002 [DOI] [PubMed] [Google Scholar]
- Liu Z., Hong Z., Ma H., Yu D., Qu P. (2020). Key Factors Mediated by PI3K Signaling Pathway and Related Genes in Endometrial Carcinoma. J. Bioenerg. Biomembr. 52 (6), 465–473. 10.1007/s10863-020-09854-4 [DOI] [PubMed] [Google Scholar]
- Long J., Menggen Q., Wuren Q., Shi Q., Pi X. (2017). MiR-219-5p Inhibits the Growth and Metastasis of Malignant Melanoma by Targeting BCL-2. Biomed. Res. Int. 2017, 1–7. 10.1155/2017/9032502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoRusso P. M. (2016). Inhibition of the PI3K/AKT/mTOR Pathway in Solid Tumors. J. Clin. Oncol. 34 (31), 3803–3815. 10.1200/jco.2014.59.0018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeary S. E., Lin K. S., Shi C. Y., Pham T. M., Bisaria N., Kelley G. M., et al. (2019). The Biochemical Basis of microRNA Targeting Efficacy. Science 366 (6472), eaav1741. 10.1126/science.aav1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgensztern D., McLeod H. L. (2005). PI3K/Akt/mTOR Pathway as a Target for Cancer Therapy. Anticancer Drugs 16 (8), 797–803. 10.1097/01.cad.0000173476.67239.3b [DOI] [PubMed] [Google Scholar]
- Peng L.-n., Shi W.-t., Feng H.-r., Wei C.-y., Yin Q.-n. (2019). Effect of miR-301a/PTEN Pathway on the Proliferation and Apoptosis of Cervical Cancer. Innate Immun. 25 (4), 217–223. 10.1177/1753425919840702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmani F., Ferns G. A., Talebian S., Nourbakhsh M., Avan A., Shahidsales S. (2020a). Role of Regulatory miRNAs of the PI3K/AKT Signaling Pathway in the Pathogenesis of Breast Cancer. Gene 737, 144459. 10.1016/j.gene.2020.144459 [DOI] [PubMed] [Google Scholar]
- Rahmani F., Ziaeemehr A., Shahidsales S., Gharib M., Khazaei M., Ferns G. A., et al. (2020b). Role of Regulatory miRNAs of the PI3K/AKT/mTOR Signaling in the Pathogenesis of Hepatocellular Carcinoma. J. Cel Physiol. 235 (5), 4146–4152. 10.1002/jcp.29333 [DOI] [PubMed] [Google Scholar]
- Sadeghi N., Gerber D. E. (2012). Targeting the PI3K Pathway for Cancer Therapy. Future Med. Chem. 4 (9), 1153–1169. 10.4155/fmc.12.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasagawa T., Takagi H., Makinoda S. (2012). Immune Responses against Human Papillomavirus (HPV) Infection and Evasion of Host Defense in Cervical Cancer. J. Infect. Chemother. 18 (6), 807–815. 10.1007/s10156-012-0485-5 [DOI] [PubMed] [Google Scholar]
- Schiffman M., Castle P. E., Jeronimo J., Rodriguez A. C., Wacholder S. (2007). Human Papillomavirus and Cervical Cancer. Lancet 370 (9590), 890–907. 10.1016/s0140-6736(07)61416-0 [DOI] [PubMed] [Google Scholar]
- Shao S., Wang C., Wang S., Zhang H., Zhang Y. (2020). Hsa_circ_0075341 is Up-Regulated and Exerts Oncogenic Properties by Sponging miR-149-5p in Cervical Cancer. Biomed. Pharmacother. 121, 109582. 10.1016/j.biopha.2019.109582 [DOI] [PubMed] [Google Scholar]
- Sharma V. R., Gupta G. K., Sharma A. K., Batra N., Sharma D. K., Joshi A., et al. (2017). PI3K/Akt/mTOR Intracellular Pathway and Breast Cancer: Factors, Mechanism and Regulation. Curr. Pharm. Des. 23 (11), 1633–1638. 10.2174/1381612823666161116125218 [DOI] [PubMed] [Google Scholar]
- Shi Y., Li L., Hu Z., Li S., Wang S., Liu J., et al. (2013). A Genome-wide Association Study Identifies Two New Cervical Cancer Susceptibility Loci at 4q12 and 17q12. Nat. Genet. 45 (8), 918–922. 10.1038/ng.2687 [DOI] [PubMed] [Google Scholar]
- Shi W.-J., Liu H., Ge Y.-F., Wu D., Tan Y.-J., Shen Y.-C., et al. (2020). LINC00673 Exerts Oncogenic Function in Cervical Cancer by Negatively Regulating miR-126-5p Expression and Activates PTEN/PI3K/AKT Signaling Pathway. Cytokine 136, 155286. 10.1016/j.cyto.2020.155286 [DOI] [PubMed] [Google Scholar]
- Slezak-Prochazka I., Durmus S., Kroesen B.-J., van den Berg A. (2010). MicroRNAs, Macrocontrol: Regulation of miRNA Processing. RNA 16 (6), 1087–1095. 10.1261/rna.1804410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solé X., Guinó E., Valls J., Iniesta R., Moreno V. (2006). SNPStats: a Web Tool for the Analysis of Association Studies. Bioinformatics 22 (15), 1928–1929. 10.1093/bioinformatics/btl268 [DOI] [PubMed] [Google Scholar]
- Song X., You W., Zhu J., Cui X., Hu J., Chen Y., et al. (2015). A Genetic Variant in miRNA-219-1 Is Associated with Risk of Esophageal Squamous Cell Carcinoma in Chinese Kazakhs. Dis. Markers 2015, 1–10. 10.1155/2015/541531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Pan H., Wang W., Qi C., Gu C., Shang A., et al. (2021). MiR-495-3p and miR-143-3p Co-target CDK1 to Inhibit the Development of Cervical Cancer. Clin. Transl. Oncol. 23 (11), 2323–2334. 10.1007/s12094-021-02687-6 [DOI] [PubMed] [Google Scholar]
- Torre L. A., Bray F., Siegel R. L., Ferlay J., Lortet-Tieulent J., Jemal A. (2015). Global Cancer Statistics, 2012. CA Cancer J. Clin. 65 (2), 87–108. 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- Vara J. Á. F., Casado E., de Castro J., Cejas P., Belda-Iniesta C., González-Barón M. (2004). PI3K/Akt Signalling Pathway and Cancer. Cancer Treat. Rev. 30 (2), 193–204. 10.1016/j.ctrv.2003.07.007 [DOI] [PubMed] [Google Scholar]
- Wang R., Zhang J., Ma Y., Chen L., Guo S., Zhang X., et al. (2014). Association Study of miR-149 Rs2292832 and miR-608 Rs4919510 and the Risk of Hepatocellular Carcinoma in a Large-Scale Population. Mol. Med. Rep. 10 (5), 2736–2744. 10.3892/mmr.2014.2536 [DOI] [PubMed] [Google Scholar]
- Wang B.-G., Jiang L.-Y., Xu Q. (2018). A Comprehensive Evaluation for Polymorphisms in Let-7 Family in Cancer Risk and Prognosis: a System Review and Meta-Analysis. Biosci. Rep. 38 (4), BSR20180273. 10.1042/bsr20180273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Zhu H., Ding B., Feng X., Zhao W., Cui M., et al. (2019). Genetic Variants in microRNAs Are Associated with Cervical Cancer Risk. Mutagenesis 34 (2), 127–133. 10.1093/mutage/gez005 [DOI] [PubMed] [Google Scholar]
- Wang W., Hou Z., Wen C., Ge L., Ge L. (2020). Long Non-coding RNA Colon Cancer-Associated Transcript-1 Promotes Migration, Invasion, and Epithelial Mesenchymal Transition of Lung Adenocarcinoma by Suppressing miR-219-1. Front. Genet. 11, 929. 10.3389/fgene.2020.00929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Hao X., Feng Z., Liu Y. (2015). Genetic Polymorphisms in miRNAs and Susceptibility to Colorectal Cancer. Cell Biochem. Biophys. 71 (1), 271–278. 10.1007/s12013-014-0195-y [DOI] [PubMed] [Google Scholar]
- Wu T., Chen X., Peng R., Liu H., Yin P., Peng H., et al. (2016). Let-7a Suppresses Cell Proliferation via the TGF-β/SMAD Signaling Pathway in Cervical Cancer. Oncol. Rep. 36 (6), 3275–3282. 10.3892/or.2016.5160 [DOI] [PubMed] [Google Scholar]
- Xing F., Song Z., He Y. (2018). MiR-219-5p Inhibits Growth and Metastasis of Ovarian Cancer Cells by Targeting HMGA2. Biol. Res. 51 (1), 50. 10.1186/s40659-018-0199-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Shi J., Mo D., Yang Y., Fu Q., Luo Y. (2020). miR-219a-1 Inhibits colon Cancer Cells Proliferation and Invasion by Targeting MEMO1. Cancer Biol. Ther. 21 (12), 1163–1170. 10.1080/15384047.2020.1843897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z., Zhou Z., Li C., Yang X., Yang L., Dai S., et al. (2019). Polymorphisms in miRNA Genes Play Roles in the Initiation and Development of Cervical Cancer. J. Cancer 10 (20), 4747–4753. 10.7150/jca.33486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q., Wang X., Yuan M., Cui S., Chen Y., Hu Z., et al. (2021). miR-219-5p Targets TBXT and Inhibits Breast Cancer Cell EMT and Cell Migration and Invasion. Biosci. Rep. 41 (8), BSR20210318. 10.1042/bsr20210318 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yu J.-Y., Hu F., Du W., Ma X.-L., Yuan K. (2017). Study of the Association between Five Polymorphisms and Risk of Hepatocellular Carcinoma: A Meta-Analysis. J. Chin. Med. Assoc. 80 (4), 191–203. 10.1016/j.jcma.2016.09.009 [DOI] [PubMed] [Google Scholar]
- Zhang L., Liu Q., Wang F. (2018). Association Between miR-149 Gene Rs2292832 Polymorphism and Risk of Gastric Cancer. Arch. Med. Res. 49 (4), 270–277. 10.1016/j.arcmed.2018.09.012 [DOI] [PubMed] [Google Scholar]
- Zhang W., Zhou Q., Wei Y., Da M., Zhang C., Zhong J., et al. (2019). The Exosome-Mediated PI3k/Akt/mTOR Signaling Pathway in Cervical Cancer. Int. J. Clin. Exp. Pathol. 12 (7), 2474–2484. [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Wu X., Li Y., Teng X., Zou L., Yu B. (2021). LncRNA SNHG5 Promotes Cervical Cancer Progression by Regulating the miR-132/SOX4 Pathway. Autoimmunity 54 (2), 88–96. 10.1080/08916934.2020.1864731 [DOI] [PubMed] [Google Scholar]
- Zheng C., Li X., Xia L., Fang X., Quan X., Yin Z., et al. (2017). Polymorphisms of Pri-miR-219-1 Are Associated with the Susceptibility and Prognosis of Non-small Cell Lung Cancer in a Northeast Chinese Population. Oncotarget 8 (34), 56533–56541. 10.18632/oncotarget.17035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Xu X.-l. (2021). Long Non-Coding RNA ARAP1-AS1 Facilitates the Progression of Cervical Cancer by Regulating miR-149-3p and POU2F2. Pathobiology 88 (4), 301–312. 10.1159/000507830 [DOI] [PubMed] [Google Scholar]
- Zou D., Zhou Q., Wang D., Guan L., Yuan L., Li S. (2016). The Downregulation of MicroRNA-10b and its Role in Cervical Cancer. Oncol. Res. 24 (2), 99–108. 10.3727/096504016x14611963142173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Hausen H. (2009). Papillomaviruses in the Causation of Human Cancers - a Brief Historical Account. Virology 384 (2), 260–265. 10.1016/j.virol.2008.11.046 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.


