Abstract
Summary
CellWalkR is an R package that integrates single-cell open chromatin data with cell type labels and bulk epigenetic data to identify cell type-specific regulatory regions. A Graphics Processing Unit (GPU) implementation and downsampling strategies enable thousands of cells to be processed in seconds. CellWalkR’s user-friendly interface provides interactive analysis and visualization of cell labels and regulatory region mappings.
Availability and implementation
CellWalkR is freely available as an R package under a GNU GPL-2.0 License and can be accessed from https://github.com/PFPrzytycki/CellWalkR with an accompanying vignette.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Gene regulatory elements active in specific cell types can be identified using single-cell open chromatin (scATAC-seq). However, analysis is difficult due to extremely low coverage per region per cell (Chen et al., 2019). Bulk data have much higher resolution, enabling confident identification of regulatory regions, but it does not provide cell type-specific annotations. Single-cell RNA sequencing (scRNA-seq) is less sparse than scATAC-seq and hence can identify dozens of cell types in complex tissues, but it does not pinpoint regulatory elements (Chen et al., 2019).
We developed CellWalkR, an R package that combines the strengths of these different technologies to resolve regulatory elements to cell types (Supplementary Fig. S1). We extended a previously introduced network diffusion model (Przytycki and Pollard, 2021) by adding filters based on bulk epigenetic data, flexible scATAC-seq formats, statistical thresholds for cell labels and multiple label sets. Key features of the package include a GPU implementation using TensorFlow (Falbel et al., 2020) and an interactive R Shiny interface (Chang et al., 2020). A suite of visualizations is provided for cell labeling, determining label similarity, embedding cells into low-dimensional space and mapping regulatory regions to cell types.
2 Results
CellWalkR builds a network consisting of two types of nodes representing cells and labels (i.e. cell types or states). Building this network requires two types of input: labeling data and scATAC-seq input as a cell-by-region matrix (e.g. cell-by-peak or cell-by-bin) or as processed by SnapATAC (Fang et al., 2021), ArchR (Granja et al., 2021) or Cicero (Pliner et al., 2018). Cells are connected by edges weighted by similarity in the scATAC-seq experiment (Supplementary Fig. S2, edges between circle nodes). Labels are connected to cells by edges based on how open gene bodies, promoters or correlated distal peaks (Pliner et al., 2018) of marker genes are in each cell (Supplementary Fig. S2, edges from square nodes to circle nodes). Marker gene names can be user-supplied or generated from scRNA-seq data using Seurat (Stuart et al., 2019). This strategy can be used to build networks for an arbitrary number of label sets, representing cell types in different time points or disease states.
All label-cell and cell-cell edges are combined into a single network, with each set of labeling edges scaled by a parameter s. Once a complete network is built, a random walk with restarts model of diffusion is run to convergence (Przytycki and Pollard, 2021). CellWalkR can optionally run this step on a GPU using TensorFlow (Falbel et al., 2020) for a greater than 15-fold speedup (Supplementary Fig. S3). The output of this process is an influence matrix where each column represents the amount of information that flows from each node to each other node.
In addition to labeling data, CellWalkR can also take bulk epigenetic data as input. This data serves as a filter through which scATAC-seq peaks are passed before being considered for calculating cell-label edges. Each filter has a scaling parameter f for each label set that determines the strength of the filter. Filters are restrictive by default (remove scATAC-seq peaks overlapping filter regions) but can also be set to permissive (allow only overlapping peaks). They can apply to the entire associated gene or just overlapping peaks.
Setting the s parameter and the f parameters correctly for each label set affects how information is propagated through the network. CellWalkR has built-in functions that tune these parameters to optimize cell homogeneity, a measure of influence between cells of the same label versus between cells of different labels (Przytycki and Pollard, 2021). To accelerate optimization, CellWalkR can downsample cells, run these calculations in parallel or compute an approximate solution for the random walk (Supplementary Fig. S4).
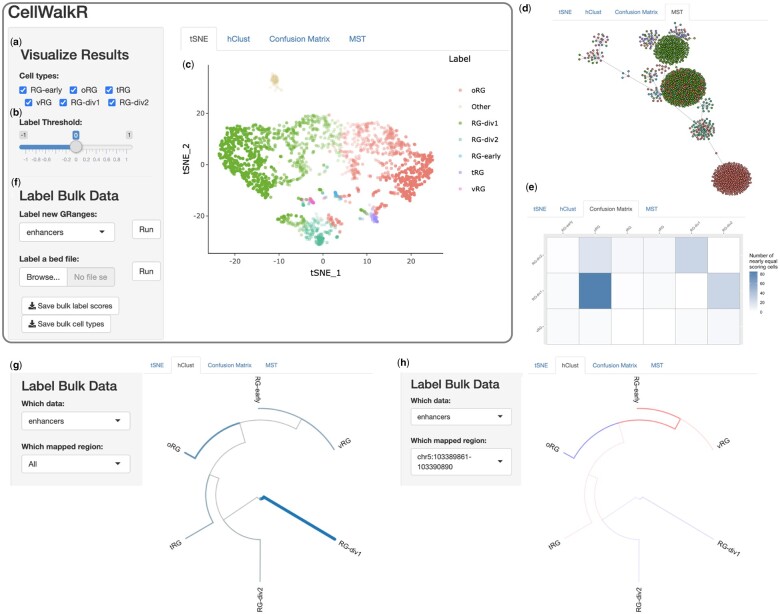
Once an influence matrix has been computed, the user has many options for downstream analysis. In addition to writing outputs to files or manipulating them with R code, one can launch CellWalkR’s Shiny interface (Fig. 1) to visualize and perform interactive analysis with different portions of the influence matrix. The label-to-cell influence portion is used to assign labels to cells, a subset of which can be excluded (Fig. 1a). These label estimates are fuzzy, meaning each cell has a distribution of scores from each label. Cells scoring below a chosen threshold can be determined to belong to no known type (Fig. 1b). Visualizing cells in a 2D space is a helpful strategy for understanding how cells relate to each other and to their labels. CellWalkR uses t-SNE (Krijthe and Van der Maaten, 2015) or Uniform Manifold Approximation and Projection (UMAP) (Melville et al., 2021) to embed cell-to-cell influence (Fig. 1c). By default, cells are colored by their highest scoring label. When only one cell type is selected, CellWalkR generates a t-SNE or UMAP plot showing the amount of influence a single label has on each cell (Supplementary Fig. S3a). If two labels are selected, it shows the difference in influence between two labels (Supplementary Fig. S5b), allowing the user to identify transition regions. Overall, embedding cell-to-cell influence rather than sparse, noisy cell accessibility profiles has clear advantages for separating cells of different types, especially rare cell types (Supplementary Fig. S6). To visualize relationships between cell types, cells can be drawn as a minimum spanning tree (MST) built by including edges of maximal influence (Fig. 1d). CellWalkR can determine how many cells receive nearly the same score for two different labels, which it summarizes in an uncertainty matrix (Fig. 1e).
Fig. 1.
Overview of CellWalkR interface. CellWalkR provides an interactive interface for downstream analysis and visualization of influence matrices. The user selects (a) their cell types of interest and (b) the label threshold, and CellWalkR dynamically generates (c) a t-SNE embedding and (d) an MST of cell-to-cell influence, and (e) a confusion matrix counting how many cells have very similar label scores. The user can then calculate cell type-specific labels for bulk data (f) either from their active session or from a BED format file. After bulk data are selected, new menu options are displayed for analyzing labels, allowing the user to (g) view enrichment for cell types across all bulk data in a hierarchical clustering of cell types, and (h) examine how a specific range is enriched or depleted in each cell type. Shown data are for cell type labels (Nowakowski et al., 2017) and scATAC-seq from a population of radial glia in the developing brain (Ziffra et al., 2021). RG, Radial Glia; oRG, outer RG; tRG, truncated RG and vRG, ventral RG
To resolve cell types/states for bulk-identified genomic regions (e.g. genetic variants, enhancers, protein binding sites and epigenetic segmentations) CellWalkR uses cell-to-label influence (Fig. 1f). Cell types for which labels are enriched across the set of regions are shown in a hierarchical clustering built using label-to-label influence (Fig. 1g). The user can then select a single region to see how strongly it maps to each cell type (Fig. 1h).
3 Conclusions
Many regulatory elements are only mapped in bulk data, and scATAC-seq data can be difficult to use due to its sparsity. CellWalkR is a flexible R package for scATAC-seq data that incorporates bulk epigenetic and expression data to multilabel cells. The package can resolve rare cell types or states and associate regulatory elements with those labels.
Supplementary Material
Acknowledgements
The authors thank to many people who tested CellWalkR including Sean Whalen, Amanda Everitt, Candace Chan, Maureen Pittman, Patrick Bradley and Laura Gunsalus.
Funding
This work was supported by research grants from NIH/NIMH [awards U01-MH116438, R01-MH109907 and R01-MH123178 to K.S.P.]. Funding bodies played no role in the design of the study and collection, analysis and interpretation of data or in writing the manuscript.
Conflict of Interest: none declared.
Contributor Information
Pawel F Przytycki, Gladstone Institutes, San Francisco, CA, USA.
Katherine S Pollard, Gladstone Institutes, San Francisco, CA, USA; Chan Zuckerberg Biohub, San Francisco, CA, USA; Department of Epidemiology and Biostatistics, Institute for Computational Health Sciences, University of California, San Francisco, CA, USA.
References
- Chang W. et al. (2020) shiny: web application framework for R. https://CRAN.R-project.org/package=shiny (6 July 2021, date last accessed).
- Chen H. et al. (2019) Assessment of computational methods for the analysis of single-cell ATAC-seq data. Genome Biol., 20, 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falbel D. et al. (2020) tensorflow: R interface to ‘TensorFlow’. https://CRAN.R-project.org/package=tensorflow (2 December 2021, date last accessed).
- Fang R. et al. (2021) Comprehensive analysis of single cell ATAC-seq data with SnapATAC. Nat. Commun., 12, 1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granja J.M. et al. (2021) ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat. Genet., 53, 403–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijthe J.H., Van der Maaten L. (2015) Rtsne: t-distributed stochastic neighbor embedding using Barnes-Hut implementation. https://github.com/jkrijthe/Rtsne.
- Melville J. et al. (2021) uwot: the Uniform Manifold Approximation and Projection (UMAP) method for dimensionality reduction. https://CRAN.R-project.org/package=uwot (24 December 2021, date last accessed).
- Nowakowski T.J. et al. (2017) Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science, 358, 1318–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pliner H.A. et al. (2018) Cicero predicts cis-regulatory DNA interactions from single-cell chromatin accessibility data. Mol. Cell, 71, 858–871.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przytycki P.F., Pollard K.S. (2021) CellWalker integrates single-cell and bulk data to resolve regulatory elements across cell types in complex tissues. Genome Biol., 22, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T. et al. (2019) Comprehensive integration of single-cell data. Cell, 177, 1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziffra R.S. et al. (2021) Single-cell epigenomics reveals mechanisms of human cortical development. Nature, 598, 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.