Abstract
Plant pathogenic bacteria have developed effectors to manipulate host cell functions to facilitate infection. A certain number of effectors use the conserved ubiquitin–proteasome system in eukaryotic to proteolyze targets. The proteasome utilization mechanism is mainly mediated by ubiquitin interaction with target proteins destined for degradation. Phyllogens are a family of protein effectors produced by pathogenic phytoplasmas that transform flowers into leaves in diverse plants. Here, we present a noncanonical mechanism for phyllogen action that involves the proteasome and is ubiquitin-independent. Phyllogens induce proteasomal degradation of floral MADS-box transcription factors (MTFs) in the presence of RADIATION-SENSITIVE23 (RAD23) shuttle proteins, which recruit ubiquitinated proteins to the proteasome. Intracellular localization analysis revealed that phyllogen induced colocalization of MTF with RAD23. The MTF/phyllogen/RAD23 ternary protein complex was detected not only in planta but also in vitro in the absence of ubiquitin, showing that phyllogen directly mediates interaction between MTF and RAD23. A Lys-less nonubiquitinated phyllogen mutant induced degradation of MTF or a Lys-less mutant of MTF. Furthermore, the method of sequential formation of the MTF/phyllogen/RAD23 protein complex was elucidated, first by MTF/phyllogen interaction and then RAD23 recruitment. Phyllogen recognized both the evolutionarily conserved tetramerization region of MTF and the ubiquitin-associated domain of RAD23. Our findings indicate that phyllogen functionally mimics ubiquitin as a mediator between MTF and RAD23.
Phyllogen, a bacterial effector produced by pathogenic phytoplasma, targets host proteins for proteasomal degradation by mediating their interaction with proteasome shuttle proteins instead of ubiquitin.
IN A NUTSHELL.
Background: Phyllogens are the protein family conserved among phytoplasmas, a group of plant pathogenic bacteria. Phyllogens transform flowers into leaves by interacting with and inducing degradation of host proteins required for flower development, such as MADS-domain transcription factors (MTFs). Phyllogen utilizes the proteasome, a protein degradation system of host plants that usually degrades proteins in two steps: first, ubiquitin is conjugated to proteins as a degradation tag. Then, the ubiquitinated proteins interact with the proteasome, either directly or with the help of shuttle proteins that transport ubiquitinated proteins to the proteasome. Several bacterial proteins utilize the proteasome by conjugating ubiquitin to their target proteins. On the contrary, phyllogen binds to the shuttle protein RADIATION SENSITIVE23 (RAD23) directly. Here, we examined the interaction between phyllogen, MTF, and RAD23 simultaneously in Nicotiana benthamiana leaves.
Questions: The main question is: what is the precise molecular mechanism of phyllogen-mediated MTF degradation? Also, what is the importance of ubiquitin in the phyllogen-mediated MTF degradation?
Findings: Our localization and coimmunoprecipitation (co-IP) analyses with phyllogen, MTF, and RAD23 showed that phyllogen mediated the interaction between MTF and RAD23. Ubiquitination of phyllogen, but not MTF, was observed in the co-IP. We also confirmed that phyllogen could work independently of ubiquitination. Moreover, co-IP assays using purified phyllogen, MTF, and RAD23 showed that phyllogen mediated the interaction between MTF and RAD23 without requiring ubiquitin or other proteins. Phyllogen acted as a mediator between targets and shuttle proteins instead of ubiquitin. Thus, we concluded that phyllogen induced proteasome-dependent but ubiquitin-independent degradation of MTFs by functionally mimicking ubiquitin, demonstrating a unique mechanism for bacterial utilization of the host proteasome.
Next steps: Unlike most proteasome-utilizing bacterial proteins, phyllogen and another recently reported phytoplasma protein (SAP05) function independently of ubiquitin. It is fascinating to consider that this novel proteasome utilization may be a common feature of phytoplasma proteins.
Introduction
Bacterial diseases are a serious threat to crop production (Sundin et al., 2016). To facilitate infection, bacterial pathogens can secrete virulence factors to control plant–microbe interactions. A notable example is effector proteins, which are injected into host cells by bacterial secretion systems such as the type III secretion system (Portaliou et al., 2016). Bacterial pathogens have developed diverse effectors that manipulate different host–cell pathways, leading to defense suppression, efficient pathogen colonization, and disease development (He et al., 2004; Tseng et al., 2009). Understanding the function(s) of these effectors is critical for understanding the molecular mechanisms underlying plant–microbe interactions and disease development.
Targets for these effectors include host proteolytic systems such as the 26S proteasome, a multisubunit proteolytic complex conserved among eukaryotes (Tomko and Hochstrasser, 2013). The 26S proteasome plays an integral role in maintaining eukaryotic cell homeostasis by modulating the levels of numerous cell regulators and degrading abnormal proteins. It consists of a 20S core particle, capped at one or both ends by 19S regulatory particles (Tomko and Hochstrasser, 2013; Livneh et al., 2016). The 20S core particle is composed of two outer α-rings and two inner β-rings with proteolytic activity. Canonically, the 26S proteasome degrades substrates in two steps (ubiquitination and degradation) that are collectively referred to as the ubiquitin-proteasome system (UPS; Ravid and Hochstrasser, 2008; Vierstra, 2009; Yu and Matouschek, 2017). In the first step, a ubiquitin tag (usually polyubiquitin) is conjugated to substrates by E1–E3 enzymes. Then, the ubiquitinated substrates interact directly or indirectly with the 19S regulatory particle, followed by ubiquitin clearing, unfolding, and entry into the 20S core particle for degradation. The direct interaction involves only the ubiquitinated substrates and the 19S regulatory particle, whereas the indirect interaction is mediated by shuttle proteins, which contain a ubiquitin-like (UbL) domain and ubiquitin-associated (UBA) domains for binding to the proteasome and the ubiquitinated substrates, respectively. In yeast, the interactions between the ubiquitinated substrates and the proteasome are largely dependent on shuttle proteins (Tsuchiya et al., 2017). RADIATION SENSITIVE23 (RAD23), which is one of the most studied shuttle proteins conserved among eukaryotes (Dantuma et al., 2009; Saeki, 2017), contains a UbL domain, two UBA (UBA1 and UBA2) domains, and RAD4 domains. In Arabidopsis thaliana (Arabidopsis), RAD23 functions as a shuttle protein essential for plant development (Farmer et al., 2010).
Many pathogenic bacteria secrete effectors that degrade target proteins, such as transcription factors and immune response proteins, via the host proteasome (Kim et al., 2014; Banfield, 2015). The effectors usually have E3 ligase activity and induce the ubiquitination of target proteins (Ashida et al., 2014; Lin et al., 2017). For example, AvrPtoB from Pseudomonas syringae contains a U-box E3 ligase domain and mediates the ubiquitination of several host immune-related kinases such as FLS2, followed by proteasomal degradation (Rosebrock et al., 2007; Göhre et al., 2008; Gimenez-Ibanez et al., 2009). XopL from Xanthomonas campestris does not share homology with known eukaryotic E3 domains, but it has E3 ligase activity (Singer et al., 2013; Erickson et al., 2018) and a XopL homolog from X. oryzae was recently reported to catalyze the ubiquitination and proteasomal degradation of Nicotiana benthamiana ferredoxin (Ma et al., 2020). In addition to these E3 effectors, a recent study suggested that P. syringae HopBB1 induces proteasomal degradation of the target transcription factor TCP14 by conjugating it to a plant hormone signaling repressor JAZ3, which is naturally degraded by the UPS (Yang et al., 2017). These studies indicate that, similar to the canonical UPS, the proteasome-utilizing effectors usually exploit ubiquitin as a key mediator of proteasomal degradation of targets. However, for many effectors that induce proteasomal degradation of targets, the mechanisms remain to be elucidated.
The phytoplasmas are a group of phloem-limited plant pathogenic bacteria that lack a cell wall and can infect more than 1,000 plant species including a number of major crops (Marcone, 2014). In addition to their devastating effect on plants, they often induce unique symptoms and developmental abnormalities, exemplified by phyllody, the transformation of floral organs into leaf-like structures (Namba, 2019). Recently, some phytoplasma effectors have been suggested to utilize the host proteasome to aid infection (Wang et al., 2018; Strohmayer et al., 2019). The best studied examples are phyllogens, a family of effectors that is conserved in phytoplasma and induces phyllody in diverse plants (MacLean et al., 2011; Maejima et al., 2014; Kitazawa et al., 2017; Iwabuchi et al., 2020). Phyllogens target floral MADS-box transcription factors (MTFs), which are key regulators of the flower development of angiosperms. These MTFs are functionally classified into classes A–E based on their requirement for the formation of floral organs. They form unique tetramers that determine the identity of the different floral organs (the floral quartet model; Honma and Goto, 2001; Theißen et al., 2016). To induce phyllody, phyllogens induce proteasomal degradation of A- and E-class MTFs of angiosperms through recognition of their K domains (MacLean et al., 2014; Maejima et al., 2014, 2015; Kitazawa et al., 2017), which are involved in tetramer formation (Yang and Jack, 2004; Lai et al., 2019). Recent structural analyses showed that phyllogens contain two α helices (Iwabuchi et al., 2019; Liao et al., 2019), similar to the K domain of MTFs (Puranik et al., 2014). This suggests that the interaction between phyllogens and the MTFs may be mediated by a mechanism similar to the one between MTFs. The previous studies have suggested the relevance of UPS-related factors in phyllogen-mediated MTF degradation processes. Maejima et al. (2014) reported ubiquitination of SEP3 or its interactor in the presence of phyllogen. MacLean et al. (2014) identified Arabidopsis RAD23 proteins as another interactor of phyllogen that is required for induction of phyllody. However, the precise molecular mechanism by which phyllogen targets MTFs for degradation to induce phyllody symptoms remains unknown.
In this study, we analyzed the relationships and functions of phyllogen, MTF, RAD23, and ubiquitin in the phyllody induction process. We found that phyllogen serves as an intermediator between MTF and RAD23 to initiate the shuttling of MTF to the proteasome. Unlike known canonical ubiquitin-dependent effectors, phyllogen acts independently of ubiquitin, suggesting that it functionally mimics ubiquitin, allowing it to utilize the host UPS.
Results
PHYLOY induces in planta colocalization of SEP3 with RAD23C and a proteasomal subunit
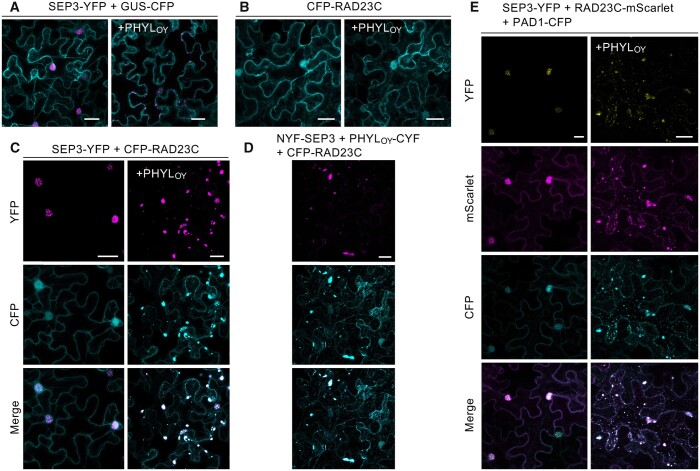
To explore whether phyllogen forms a protein complex with Arabidopsis MTF and RAD23 during the MTF degradation process, we used PHYLOY, a phyllogen of ‘Candidatus Phytoplasma asteris' onion yellows strain (Maejima et al., 2014). Note that the gene name of phyllogens has been described as PHYL1, but for simplicity based on their subgroup names of PHYL-A to PHYL-D (Iwabuchi et al., 2020), it will be referred to as PHYL hereafter. We determined the intracellular localizations of both SEP3 (Arabidopsis E-class MTF) and RAD23C (one of four Arabidopsis RAD23 homologs, RAD23A–D) with and without PHYLOY. First, YFP-fused SEP3 (SEP3-YFP) was transiently coexpressed with PHYLOY in N. benthamiana epidermal cells by agroinfiltration (Figure 1A). GUS-CFP was coexpressed to visualize the introduced cells. It was previously reported that overexpression of PHYLOY drastically reduces the fluorescence of SEP3-YFP such that fluorescence was barely observed (Kitazawa et al., 2017). Therefore, for observation of SEP3-YFP under the PHYLOY-mediated degradation process, the Agrobacterium tumefaciens cells for PHYLOY expression were infiltrated at a concentration lower than that of the previous study to induce a moderate level of PHYLOY-mediated SEP3-YFP degradation (Supplemental Figure S1A). Whereas SEP3-YFP was localized in the nucleus without PHYLOY, it showed scattered punctate localization in the cytoplasm when coexpressed with PHYLOY (Figure 1A). In contrast, the localization of CFP-RAD23C that was observed in both the cytoplasm and nucleus was not affected by the presence of PHYLOY (Figure 1B). When SEP3-YFP and CFP-RAD23C were coexpressed, their localizations were both closely associated with the nucleus, but clearly distinct from each other (Figure 1C). However, in the presence of PHYLOY, SEP3-YFP, and CFP-RAD23C were clearly colocalized and showed punctate structures in the cytoplasm (Figure 1C). These results indicate that PHYLOY affects RAD23C localization only when SEP3 exists and redistributes them into the same punctate structures.
Figure 1.
SEP3 colocalizes with RAD23C and a proteasomal subunit under PHYLOY expression in N. benthamiana epidermal cells. A–C, Confocal imaging of cells expressing SEP3-YFP (A), CFP-RAD23C (B), and both SEP3-YFP and CFP-RAD23C (C) without (left) and with (right) PHYLOY. GUS-CFP was coexpressed in (A) to visualize the cells. D, Localization of CFP-RAD23C and the BiFC signal of NYF-SEP3 and PHYLOY-CYF. In these experiments, Agrobacterium cultures (OD600 = 1.0) for the expression of SEP3, PHYLOY, RAD23C, or GUS, and P19 were mixed at a ratio of 4:2:3:1 before infiltration. E, Confocal imaging of cells expressing SEP3-YFP, RAD23C-mScarlet, and PAD1-CFP without (left) and with (right) PHYLOY. Agrobacterium cultures (OD600 = 1.0) for the expression of SEP3, PHYLOY, RAD23C, PAD1, and P19 were mixed at a ratio of 3:2:2:2:1. Nicotiana benthamiana cells were observed 40 h after infiltration. Fluorescence of YFP (A–D) or mScarlet (E) was pseudo-colored in magenta. Bars = 25 μm.
The localization of PHYLOY-CFP was uniform in both cytoplasm and nucleus, and it was not affected by the presence of SEP3 and/or RAD23C (Supplemental Figure S1B). In bimolecular fluorescence complementation (BiFC) analyses, however, the punctate YFP signals were observed when using SEP3 fused with an N-terminal fragment of YFP (NYF-SEP3) and PHYLOY fused with a C-terminal fragment of YFP (PHYLOY-CYF) (Supplemental Figure S1C). Induction of degradation of NYF-SEP3 by PHYLOY-CYF was confirmed as in the case of PHYLOY (Supplemental Figure S1A). The signals were not observed when using NYF-fused APETALA3 (AP3; B-class MTF), a nontarget MTF of phyllogen (MacLean et al., 2014). These results suggest that although the majority of overexpressed PHYLOY molecules show a uniform subcellular localization due to the lack of an interaction partner, PHYLOY interacting with SEP3 localizes in the cytoplasmic punctate structures. The BiFC signals of NYF-SEP3 and PHYLOY-CYF colocalized with fluorescence of CFP-RAD23C (Figure 1D), indicating that the interaction of SEP3 and PHYLOY induces colocalization of SEP3, PHYLOY, and RAD23C. Considering these results, it seems that SEP3 interacts with RAD23C via interaction with PHYLOY, resulting in the formation of these punctate structures, which contain at least of the three proteins SEP3, PHYLOY, and RAD23C.
Next, to examine the spatial relationship between SEP3 and the proteasome, SEP3-YFP was coexpressed with RAD23C-mScarlet (Bindels et al., 2017), and CFP-tagged PAD1, a proteasome subunit (Fu et al., 1998), that was used to visualize the cellular distribution of proteasomes in budding yeast (Enenkel et al., 1998; Laporte et al., 2008) and mammals (Bingol and Schuman, 2006). In the absence of PHYLOY, PAD1-CFP localized in both the cytoplasm and nucleus (Figure 1E). However, when PHYLOY was coexpressed with PAD1-CFP, the localization of PAD1-CFP was drastically altered, as was the localization of SEP3-YFP and RAD23C-mScarlet, which coexisted as punctate structures in cytoplasm. These data strongly suggest that PHYLOY recruits SEP3 to the proteasome via the formation of the SEP3/PHYLOY/RAD23C ternary protein complex.
SEP3 interacts with RAD23C via PHYLOY and enhances the interaction between PHYLOY and RAD23C
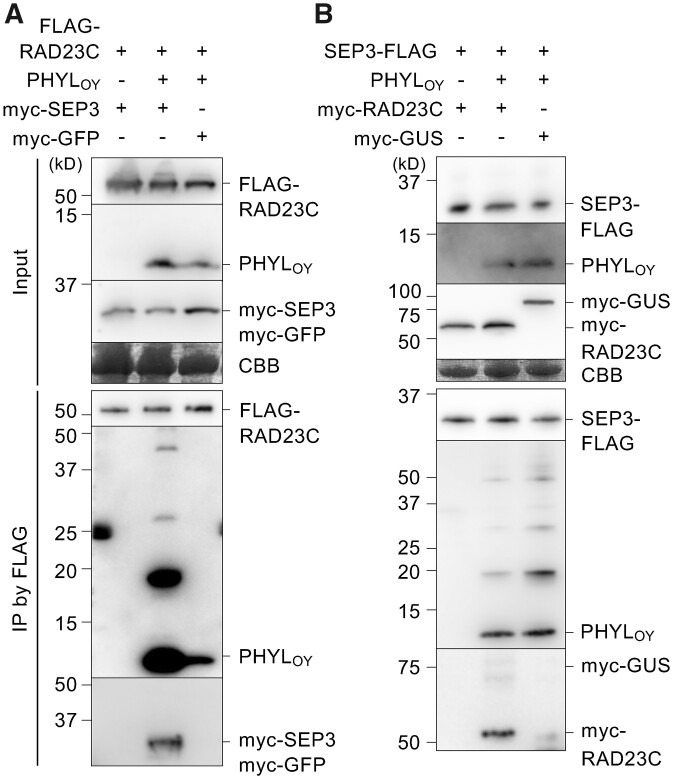
To demonstrate the formation of the SEP3/PHYLOY/RAD23C protein complex during the PHYLOY-mediated SEP3-degradation process, in planta coimmunoprecipitation (co-IP) assays were performed. The Agrobacterium cells for PHYLOY expression were infiltrated at a low concentration to reduce the effect of PHYLOY on the amount of myc-SEP3, with the goal being efficient observation of protein complexes formed during degradation of myc-SEP3. First, FLAG-RAD23C or FLAG-GUS was coexpressed with PHYLOY in N. benthamiana leaves and immunoprecipitated using an anti-FLAG antibody. The results showed that PHYLOY was coimmunoprecipitated with FLAG-RAD23C but not with FLAG-GUS (Supplemental Figure S2), indicating an in planta interaction between PHYLOY and RAD23C. Next, FLAG-RAD23C and myc-SEP3 were coexpressed with or without PHYLOY. Co-IP using an anti-FLAG antibody detected myc-SEP3 in the immunoprecipitates (IP) with FLAG-RAD23C only when coexpressed with PHYLOY (Figure 2A). The myc-GFP-negative control was not detected in the IP under the same conditions (Figure 2A). These results indicate that PHYLOY induces the interaction between SEP3 and RAD23C via the formation of the SEP3/PHYLOY/RAD23C ternary complex. Furthermore, additional ladder-like bands (at 6- to 10-kDa intervals) with sizes larger than the expected size of PHYLOY (10.6 kDa) were detected in the IP with myc-SEP3 (Figure 2A, middle lane). Multiple bands at <10-kDa intervals were also observed when PHYLOY-CYF (21.5 kDa) was used instead of PHYLOY (Supplemental Figure S3A), indicating that the band shifts were not due to multimerization of PHYLOY but to some posttranslational modification similar to ubiquitination (the mass of ubiquitin is 8.5 kDa). Such additional ladder-like bands were not observed above the expected size of myc-SEP3 in IP (Figure 2A, middle lane).
Figure 2.
PHYLOY induces interaction of SEP3 and RAD23C in planta. A, co-IP assays using FLAG-RAD23C, PHYLOY, and myc-SEP3. Myc-GFP was used as a negative control for myc-SEP3 (right lane). Agrobacterium cultures for the expression of FLAG-RAD23C (OD600 = 0.5), PHYLOY (OD600 = 0.1), either myc-SEP3 or myc-GFP (OD600 = 1.0), and P19 (OD600 = 1.0) were mixed at a ratio of 10:10:10:1 and infiltrated into N. benthamiana leaves. IP was performed using an anti-FLAG antibody 36-h after infiltration. The input and immunoprecipitated (IP) proteins were analyzed by immunoblot using anti-FLAG, anti-PHYLOY, and anti-myc antibodies. Coomassie brilliant blue (CBB) staining is shown as a loading control. B, co-IP assay using SEP3-FLAG, PHYLOY, and myc-RAD23C. Co-IP and detection were performed as in (A). Agrobacterium cultures for the expression of SEP3-FLAG (OD600 = 1.0), PHYLOY (OD600 = 0.1), either myc-RAD23C or myc-GUS (OD600 = 1.0), and P19 (OD600 = 1.0) were mixed at a ratio of 10:10:10:1. In both experiments, accumulation of SEP3 (input) was not obviously affected by PHYLOY, possibly because PHYLOY was more weakly expressed than in the previous experiments.
Interestingly, the amount of PHYLOY that coimmunoprecipitated with FLAG-RAD23C was highly increased by myc-SEP3 expression in comparison with myc-GFP expression (Figure 2A, middle and right lanes, respectively). This indicates that RAD23 interacts more strongly with PHYLOY in the presence of SEP3. This result was consistent with the fluorescence observation, showing that CFP-RAD23C localization changed when both PHYLOY and SEP3 were expressed (Figure 1C) but not when only PHYLOY was expressed (Figure 1B). Then, to test whether RAD23C affects PHYLOY/SEP3 interaction, SEP3-FLAG, PHYLOY, and myc-RAD23C were expressed in planta and coimmunoprecipitated using an anti-FLAG antibody. Myc-RAD23C was coimmunoprecipitated with SEP3-FLAG only when PHYLOY was coexpressed (Figure 2B), indicating the formation of SEP3/PHYLOY/RAD23C. However, the signal of PHYLOY in IP was not increased by coexpression of myc-RAD23C, suggesting that RAD23C does not affect PHYLOY/SEP3 interaction. These data indicate that RAD23 interacts with MTF/phyllogen complex rather than phyllogen, whereas MTF interacts with phyllogen independently of RAD23. The additional ladder-like bands larger in size than PHYLOY in IP were also detected independently of myc-RAD23C expression, suggesting that transient RAD23C expression does not modify PHYLOY. When PAD1-CFP was coexpressed with SEP3-FLAG and myc-RAD23C, PAD1-CFP was also coimmunoprecipitated with SEP3-FLAG in the presence of PHYLOY (Supplemental Figure S3B), indicating that SEP3 is recruited to the 26S proteasome in the presence of PHYLOY.
In planta ubiquitination of PHYLOY and SEP3 is not required for SEP3 degradation
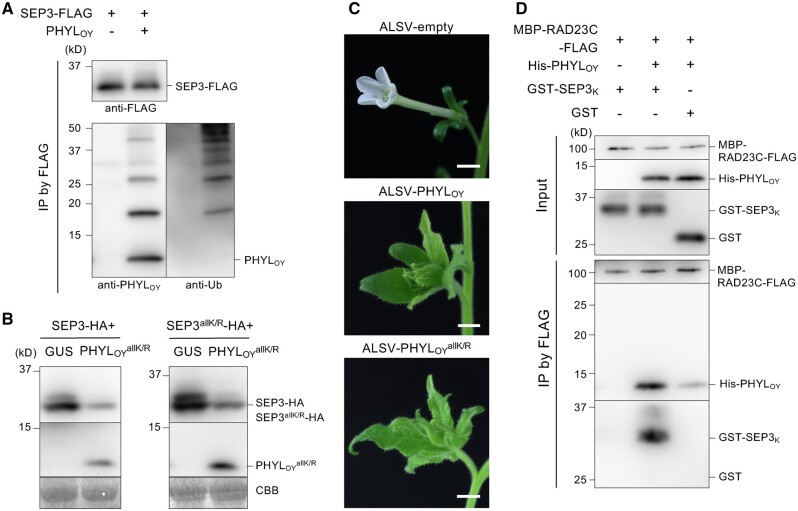
Next, we tested whether the band shifts of PHYLOY observed in co-IP assay were due to ubiquitination. PHYLOY was coimmunoprecipitated with SEP3-FLAG using the anti-FLAG antibody, and the ubiquitin signal in IP was analyzed using an anti-ubiquitin antibody (Figure 3A). Specific ladder-like bands larger in size than PHYLOY were detected at the same intervals as those detected by the anti-PHYLOY antibody (as shown in Figure 2, A and B). This indicated that both MTF- and RAD23-associated PHYLOY are ubiquitinated. Previously, MacLean et al. (2014) reported that ubiquitinated phyllogen (SAP54) was not detected in immunoprecipitants when phyllogen was used as bait. Taken together, the results indicate that most cellular phyllogen molecules are not normally ubiquitinated but become so only during the MTF degradation process.
Figure 3.
Ubiquitination of PHYLOY is not essential for interaction with SEP3. A, Detection of ubiquitinated PHYLOY. Co-IP and protein detection were performed as in Figure 2 using anti-FLAG, anti-PHYLOY, and anti-ubiquitin antibodies. Agrobacterium cultures for the expression of FLAG-SEP3, PHYLOY, and P19 (OD600 = 1.0) were mixed at a ratio of 10:1:1. B–C, Functional analyses of , a Lys-less PHYLOY mutant. B, induces SEP3 degradation in planta. Agrobacterium cultures (OD600 = 1.0) for the expression of HA-fused proteins, either GUS or PHYLOY, and P19 were mixed at a ratio of 10:10:1 and infiltrated into N. benthamiana leaves. Immunoblot analyses were performed 36 h after infiltration using anti-HA and anti-PHYLOY antibodies. CBB staining is shown as a loading control. C, Flower malformation-inducing activity of . expressed apple latent spherical virus (ALSV-) induced phyllody-like flower malformations in N. benthamiana, similar to ALSV-PHYLOY. Bars = 1 cm. D, PHYLOY directly mediates interaction between SEP3 and RAD23C in vitro. Co-IP was performed using purified MBP-RAD23C-FLAG, His-PHYLOY, and the GST-fused K domain of SEP3 (GST-SEP3K). These proteins were mixed (input), immunoprecipitated using an anti-FLAG antibody, and detected by anti-FLAG, anti-PHYLOY, and anti-GST antibodies.
Because ubiquitin is usually attached to the Lys residues of substrate proteins (Ye and Rape, 2009; Kwon and Ciechanover, 2017), a Lys-less PHYLOY mutant, , in which all Lys residues were replaced by Arg was expressed with SEP3-FLAG and immunoprecipitated using an anti-FLAG antibody. The bands from ubiquitinated PHYLOY were detected in IP (black arrows) but those from were not, indicating that was not ubiquitinated (Supplemental Figure S4). When PHYLOY and were expressed alone, immunoblotting analysis showed that the accumulation of was not enhanced compared to that of PHYLOY (Supplemental Figure S5), suggesting that degradation of phyllogen due to ubiquitination of Lys residues does not occur. Rather, it indicates that some Lys residues are involved in the accumulation of phyllogen. These results are consistent with the previous suggestion that phyllogen is not degraded by the proteasome (MacLean et al., 2014). Then, we examined the role of ubiquitin in phyllogen-mediated MTF degradation by analyzing the MTF degradation and phyllody induction activity of . When was expressed with HA tag-fused SEP3 (SEP3-HA) in planta, immunoblotting analysis showed that decreased the amount of SEP3-HA, as in the case of PHYLOY (Figure 3B). Additionally, the amount of a Lys-less SEP3 mutant (SEP3allK/R) was also diminished by . Furthermore, induced phyllody-like flower malformation (Figure 3C), when it was expressed in N. benthamiana using the apple latent spherical virus (ALSV) vector system for foreign protein expression (Li et al., 2004). No clear difference was observed in phyllody induction activity between PHYLOY and .
Finally, to examine whether the SEP3/PHYLOY/RAD23C complex is formed independently of ubiquitination, the in vitro co-IP assay was performed using each protein expressed in and purified from Escherichia coli. Due to the insolubility of full-length SEP3, its K domain (SEP3K; Supplemental Figure S6A), which is targeted by phyllogens (MacLean et al., 2014; Liao et al., 2019; Supplemental Figure S7A), was used in the study. Purified MBP-RAD23C-FLAG, His-PHYLOY, and GST-SEP3K were mixed and immunoprecipitated using an anti-FLAG antibody (Figure 3D). This result was similar to the results of the in planta assay: GST-SEP3K was immunoprecipitated only in the presence of His-PHYLOY, and His-PHYLOY was immunoprecipitated more strongly when mixed with GST-SEP3K than GST. No ubiquitinated PHYLOY bands were observed in the experiment. These results indicate that the SEP3/PHYLOY/RAD23C complex is formed without ubiquitin because PHYLOY mediates the interaction between SEP3 and RAD23C directly.
Identification of PHYLOY-targeted regions in MTF and RAD23
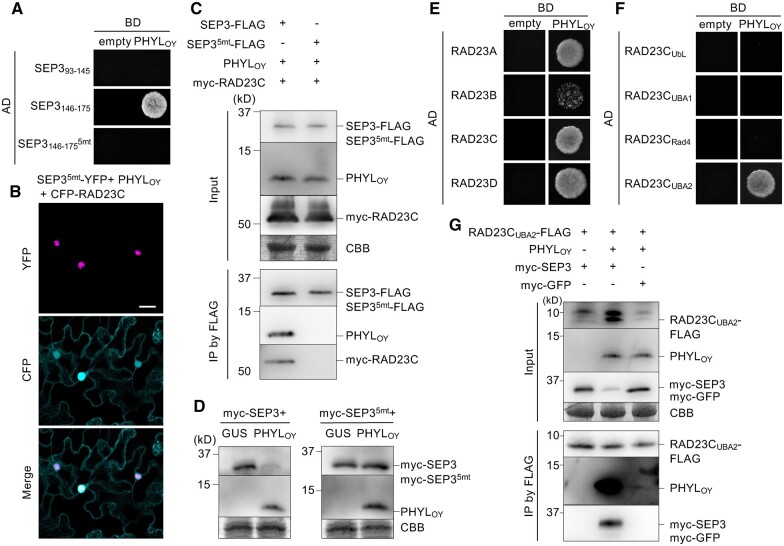
To explore the mechanisms underlying the formation of the ternary protein complex, we focused on the region in MTF or RAD23 that is recognized by PHYLOY. As shown for other phyllogens (MacLean et al., 2014; Liao et al., 2019), PHYLOY interacted with A- (APETALA1; AP1) and E-class MTFs in a K domain-dependent manner (Supplemental Figure S7, A and B). Previous structural analysis had revealed that the K domain forms two helices containing residues related to homodimerization or tetramerization (Puranik et al., 2014; Supplemental Figure S6A). Our yeast two-hybrid (Y2H) assay using SEP394–145 and SEP3146–175 (comprised of residues 94–145 and 146–175 in the SEP3 K domain, respectively) showed that PHYLOY interacted with SEP3146–175 including the residues involved in tetramerization residues (Figure 4A), as suggested by previous Y2H screening (MacLean et al., 2014). PHYLOY did not interact with SEP, an Ala substitution mutant of five hydrophobic residues (Met150, Leu154, Leu157, Leu164, and Leu171) that are reported to be involved in tetramerization of the SEP3 K domain (Puranik et al., 2014; Supplemental Figure S6A). In N. benthamiana cells, YFP-fused SEP35mt, a full-length SEP3 mutant with Ala substitutions for the hydrophobic residues described above, localized in the nucleus irrespective of PHYLOY coexpression (Figure 4B). The in planta co-IP assay showed that PHYLOY and myc-RAD23C were not immunoprecipitated with SEP35mt-FLAG (Figure 4C). When myc-SEP35mt was expressed with PHYLOY in planta, the immunoblotting assay showed that PHYLOY did not induce degradation of myc-SEP35mt (Figure 4D). These results indicate that PHYLOY interacts with the region of the SEP3 K domain that is involved in tetramerization.
Figure 4.
PHYLOY recognizes the region of SEP3 involved in tetramerization and the UBA2 domain of RAD23C. A, Y2H assay of PHYLOY and specific regions of the SEP3 K domain. Yeast cells harboring the AD and BD vectors were adjusted to an OD600 of 0.1. Aliquots (10 μL) of these cells were spotted on a SD medium lacking Leu/Trp/His and containing 5 mM 3-amino-1,2,4-triazole (–LWH+3AT). The plates were incubated for 4 days at 30°C. B, Subcellular localization of SEP35mt-YFP under PHYLOY and RAD23C expression. Protein expression and fluorescence observation were performed as described in Figure 1. Bar = 25 μm. C, In planta co-IP using SEP35mt-FLAG with PHYLOY and myc-RAD23C. The experiment was performed as described in Figure 2B. D, PHYLOY does not induces in planta degradation of SEP35mt. Agrobacterium cultures (OD600 = 1.0) for the expression of myc-fused proteins, either GUS or PHYLOY, and P19 were mixed at a ratio of 10:10:1 and infiltrated into N. benthamiana leaves. Immunoblot analyses were performed as described in Figure 3B using anti-myc and anti-PHYLOY antibodies. E–F, Y2H assay using PHYLOY and A. thaliana RAD23 homologs (E), and the domains of RAD23C (F). Both Y2H assays were performed as described in (A). G, In planta co-IP using RAD23CUBA2-FLAG, PHYLOY, and myc-SEP3. The experiment was performed as described in Figure 2A.
Next, interactions between PHYLOY and RAD23 homologs were investigated. RAD23 homologs in angiosperms are divided into two clades: one clade in Arabidopsis contains RAD23A/B and another contains RAD23C/D (Farmer et al., 2010). Previously, it was suggested that phyllogen uses the RAD23C/D pair because SAP54 recognizes only RAD23C/D and does not induce phyllody in the Arabidopsis rad23C/D mutant (MacLean et al., 2014). Our Y2H assays showed that PHYLOY bound not only to RAD23C/D but also to RAD23A/B (Figure 4E), suggesting that phyllogen can use a wider spectrum of RAD23 homologs than previously thought. To determine the domain of RAD23 responsible for the interaction with PHYLOY, we performed Y2H using PHYLOY and each of four domains of RAD23C (Supplemental Figure S6B). PHYLOY interacted only with the UBA2 domain (RAD23CUBA2; 370–412 amino acids; Figure 4F). PHYLOY also interacted with the UBA2 domain of other RAD23 homologs (Supplemental Figure S7C), indicating that PHYLOY recognizes RAD23 proteins through the UBA2 domain. In an in planta FLAG-tag co-IP assay using FLAG tag-fused RAD23CUBA2 (RAD23CUBA2-FLAG; 370–418 amino acids, including six additional C-terminal residues), myc-SEP3, and PHYLOY, we found that that RAD23CUBA2-FLAG interacted with myc-SEP3 only in the presence of PHYLOY (Figure 4G), as with RAD23C-FLAG. Moreover, PHYLOY interacted well with RAD23CUBA2-FLAG in the presence of myc-SEP3. One slight difference with the co-IP assay using full-length RAD23 was that the amount of myc-SEP3 (input) tended to decrease in the presence of PHYLOY. These data indicate that only the UBA2 domain of RAD23 is necessary for its PHYLOY-mediated interaction with SEP3.
The interactional relationships of the MTF/phyllogen/RAD23 protein complex are conserved when using different phyllogen or host proteins
Considering the diversity of phyllogen among phytoplasma strains (Maejima et al., 2014; Yang et al., 2015; Iwabuchi et al., 2020) and the wide host range of phytoplasmas (Marcone, 2014), we investigated whether the interaction relationships of phyllogen and host proteins are conserved among phytoplasmas or plants. First, in planta co-IP and Y2H assays were performed using PHYLPnWB from peanut witches’ broom phytoplasma (Chung et al., 2013) instead of PHYLOY. Although PHYLPnWB shows only 50% amino acid identity with the secreted region of PHYLOY, it induces degradation of SEP3 and flower malformations in plants (Yang et al., 2015; Kitazawa et al., 2017). Results from the in planta FLAG-tag co-IP assay using FLAG-RAD23C, myc-SEP3, and PHYLPnWB showed that FLAG–RAD23C/myc-SEP3 interaction occurred via PHYLPnWB (Supplemental Figure S8). In the presence of SEP3, the FLAG–RAD23C/PHYLPnWB interaction was enhanced and ladder-like band shifts larger than PHYLPnWB in IP were detected (Supplemental Figure S8A). The Y2H assay showed that PHYLPnWB bound to SEP3146–175 and RAD23CUBA2 but not to SEP (Supplemental Figure S8, B and C), indicating that phyllogens recognized the same region in the host factors. Furthermore, PHYLPnWB bound to RAD23A as to RAD23C and RAD23D (Iwabuchi et al., 2020), although PHYLPnWB/RAD23B interaction was not detected (Supplemental Figure S8C). These results indicate that PHYLPnWB induced MTF/RAD23 interaction in a manner quite similar to PHYLOY.
Next, we examined the in planta interaction of PHYLOY with the host factors of rice (Oryza sativa, a monocot) instead of those of Arabidopsis (a dicot); OsMADS8 (E-class MTF; Cui et al., 2010) and OsRAD23 (RAD23 homolog related to RAD23D; Schultz and Quatrano, 1997). In planta, degradation of OsMADS8 by PHYLOY has been reported (Kitazawa et al., 2017). The Y2H assay showed that OsRAD23 and its UBA2 domain interacted with PHYLOY (Supplemental Figure S8D). The in planta IP assay showed that OsMADS8 and OsRAD23 interacted only in the presence of PHYLOY and that OsRAD23 interacted with PHYLOY more strongly in the presence of OsMADS8 (Supplemental Figure S8E). These results indicate that formation of the MTF/phyllogen/RAD23 complex occurs similarly in rice and Arabidopsis.
Discussion
In the UPS, ubiquitination of substrates by E1–E3 ligases induces interactions of substrates with proteasomes either directly or indirectly via shuttle proteins. Similarly, many bacterial effectors induce proteasomal degradation of target proteins by recognizing ubiquitin as the tag for degradation (Ashida et al., 2014; Banfield, 2015; Lin and Machner, 2017). On the other hand, although ubiquitinated PHYLOY was detected during MTF degradation (Figure 3A), a Lys-less PHYLOY mutant () induced degradation of both SEP3 and its Lys-less mutant (Figure 3B), and caused phyllody phenotypes of flowers as with PHYLOY (Figure 3C). It cannot be ruled out the possibility that the mutant proteins are ubiquitinated on other residues, because some proteins are known to be ubiquitinated on residues other than Lys (McClellan et al., 2019). However, the ubiquitination of and SEP3 was not detected in our study (Figure 2A; Supplemental Figure S4). Moreover, in vitro co-IP assays showed that PHYLOY mediates the interaction between SEP3 and RAD23C independently of ubiquitin (Figure 3D). As RAD23 is thought to be one of major factors connecting ubiquitinated substrates with the 26S proteasome (Tsuchiya et al., 2017), the results strongly indicate that phyllogen induces ubiquitin-independent proteasomal degradation of target MTFs by recruiting them to RAD23. Our current model of phyllogen-mediated MTF degradation is shown in Figure 5. In eukaryotic cell homeostasis, RAD23 recognizes ubiquitinated proteins (Target/Ub) via two UBA domains and forms a ternary complex (Target/Ub/RAD23) to initiate proteasomal degradation of targets. Phyllogen induces the ubiquitin-independent formation of the MTF/phyllogen/RAD23 ternary complex by recognizing the K domain of MTFs and the UBA2 domain of RAD23. This indicates that phyllogen induces the ubiquitin-independent degradation of MTF by functionally mimicking ubiquitin, in that it acts as a mediator between the target and the proteasomal shuttle protein and, thereafter, the proteasome.
Figure 5.
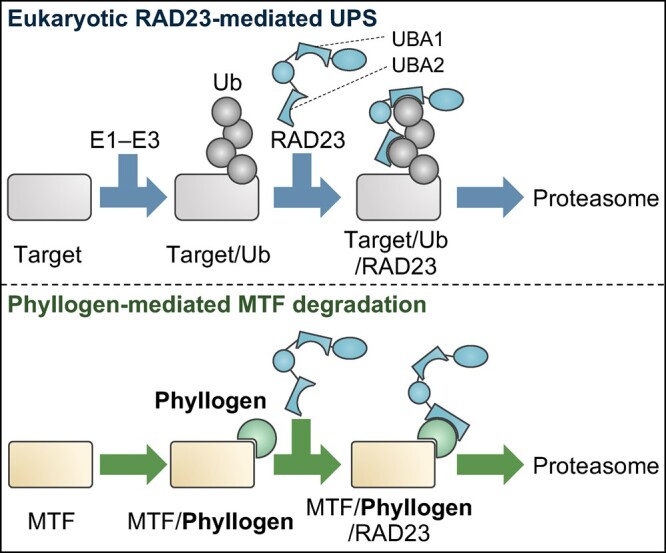
Model of the interaction between phyllogen and host factors. In the eukaryotic RAD23-mediated UPS (upper), target proteins are ubiquitinated by E1–E3 enzymes (Target/Ub), recognized by RAD23 (Target/Ub/RAD23) and sent to the proteasome. In phyllogen-mediated MTF degradation (lower), phyllogen mediates the ubiquitin-independent interaction between MTF and RAD23, causing proteasomal degradation of MTF. Target MTFs interact first with phyllogen instead of ubiquitin (MTF/phyllogen), then with RAD23 via phyllogen (MTF/phyllogen/RAD23), and are then sent to the proteasome.
Interestingly, many MTFs regulate the development of organs other than flowers (Theißen et al., 2016), and phyllogen interacts with diverse nonfloral MTFs (MacLean et al., 2014). Together with the present study, the findings suggest that phyllogen can induce the degradation of the nonfloral MTFs by mediating interactions between the MTFs and RAD23, resulting in changes in the gene expression and development of diverse plant organs. Actually, phyllogen contributes to insect colonization in Arabidopsis in a RAD23-dependent but phyllody-independent manner (Orlovskis and Hogenhout, 2016). Both these studies and our study suggest that phyllogen has multiple roles in promoting phytoplasma infection by acting a ubiquitin-mimicking tag for proteasomal degradation of multiple MTFs. It will be important to examine whether and how phyllogen induces degradation of nonfloral MTFs to fully understand the function of the effector family.
To the best of our knowledge, effectors that induce ubiquitin-independent proteasomal degradation of targets have not been reported in pathogenic bacteria except for phytoplasmas. Phyllogen appears to function in diverse plants, because it requires conserved RAD23 but not E1–E3 ligases for target degradation. Actually, our co-IP assay showed that PHYLOY connects rice MTF to rice RAD23 (Supplemental Figure S8), as in the case of the Arabidopsis homologs. Furthermore, skipping ubiquitination, the initial step in the UPS, might facilitate faster degradation of targets. Interestingly, SAP05, another effector of phytoplasmas, was reported during the revision of this manuscript (Huang et al., 2021). It induces ubiquitin-independent proteasomal degradation of plant developmental regulators by intermediating interaction between them and RPN10 (a subunit of the 19S regulatory particle). It is quite fascinating that the noncanonical ubiquitin-independent mechanisms utilizing the proteasome are common to two different phytoplasma effectors. The studies of phyllogen and SAP05 suggest that phytoplasmas have acquired a consistent host manipulation strategy of inducing ubiquitin-independent proteasomal degradation of plant-conserved developmental regulators by their effectors. Because at least one other phytoplasma effector, SWP1, was suggested to utilize proteasome (Wang et al., 2018), it is will be interesting to determine whether such phytoplasma effectors utilizing proteasome work independently of ubiquitination.
In this work, phyllogen interacted with the UBA2 domain of RAD23, as did ubiquitin. However, it was difficult to determine how phyllogen interacts with the domain because, in contrast to its functional similarity, phyllogen does not show sequential homology or structural similarity with ubiquitin. The UBA2 domain preferentially interacts with the K48 polyubiquitin chain by recognizing a hydrophobic patch in the β-sheet of each ubiquitin molecule (Dantuma et al., 2009; Dikic et al., 2009). However, phyllogen contains no β-sheet (Iwabuchi et al., 2019; Liao et al., 2019; Aurin et al., 2020) and may not function as a multimer (Iwabuchi et al., 2019; Liao et al., 2019). These data strongly suggest that phyllogen interacts with the UBA2 domain in a manner different from ubiquitin.
Exploring how phyllogen utilizes the domain will reveal the molecular mechanisms of the interaction between phyllogen and RAD23. The Y2H assay showed that the interaction of phyllogen with RAD23B was relatively weak (PHYLOY; Figure 4E) or not detectable (PHYLPnWB and SAP54; Supplemental Figure S8 and MacLean et al., 2014, respectively) despite the high sequence similarity with the UBA2 domain of other RAD23s (86% for RAD23A, and 72% for RAD23C and D). A previous report suggested that RAD23A–D have largely redundant functions, but that RAD23B also has a nonoverlapping role because rad23B mutants have pleiotropic developmental defects (Farmer et al., 2010). Phyllogen may not inhibit the unique function of RAD23B because it targets the other RAD23s. Mutation analysis of these domains based on sequence differences will be helpful for understanding the mechanisms underlying the molecular interaction between phyllogen and RAD23. Interestingly, a previous report showed that hydrophobic residues in the helices of phyllogen are well conserved (Iwabuchi et al., 2019), suggesting that such residues play a role in phyllogen/UBA2 interaction instead of the hydrophobic patch in the β-sheet of ubiquitin.
This study revealed that phyllogen recognized the region of the SEP3 K domain involved in tetramerization (Figure 4, A–D). Tetramerization of this domain primarily occurred through interactions of hydrophobic surfaces in the region formed by the five residues needed for tetramerization, and mutations in the residues disrupt the tetramerization (Puranik et al., 2014). Interestingly, Ala substitutions of the five residues also disrupted the SEP3/phyllogen interaction (Figure 4, A–D; Supplemental Figure S8B), suggesting that the surface of SEP3 involved in tetramerization is also recognized by phyllogen. This supports the idea that the MTF/phyllogen interaction is mediated in a manner similar to the tetrameric interaction of MTF, which was discussed previously based on the structural similarity of the two proteins (Rümpler et al., 2015; Iwabuchi et al., 2019). Furthermore, these five residues are highly conserved among not only SEP proteins from diverse flowering plants (Rümpler et al., 2018) and even hypothetical ancestral SEP proteins (Ruelens et al., 2017). Rümpler et al. (2018) showed that these positions in AP1-subfamily proteins are also occupied by hydrophobic amino acids, although with a lower frequency of Leu. Based on these reports, our data indicate that phyllogen recognizes the evolutionarily conserved hydrophobic interface in MTFs, consistent with the notion that phyllogen functions in diverse plants.
The in planta co-IP assays showed that the basal interaction between RAD23C and phyllogen was very slight, whereas the interaction is significantly enhanced in the presence of SEP3 (Figure 2A; Supplemental Figure S8A). On the other hand, the interaction of PHYLOY with SEP3 was not affected by transient expression of RAD23C (Figure 2B). This strongly suggested that phyllogen-mediated MTF degradation is mostly through the sequential interaction of the proteins: the target MTFs is first recognized by phyllogen, and then RAD23 interacts with MTF/phyllogen complex resulting in the initiation to the proteasome (Figure 5). It is of interest how and why such sequential interaction occurs. Recent research indicated that the in vitro interaction of SEP3K and phyllogen follows a conformational change of the SEP3K/phyllogen complex (Liao et al., 2019). Such conformational change of the proteins may induce enhancement of the interaction with RAD23. The simplest hypothesis is that the conformational change of the phyllogen structure can result in efficient binding to RAD23. Such structure-based changes in affinity have been recognized in several bacterial effectors (Popa et al., 2016). Further study for exploring the structure and stoichiometry of the ternary complex will be important for understanding the interaction mode.
Regarding the question of why phyllogen interacts well with RAD23 in the presence of MTF, one possible reason is that it minimizes the phyllogen/RAD23 interaction in the absence of the targets during phytoplasma infection. Because phytoplasmas are phloem-limited bacteria (Namba, 2019), phyllogen should be secreted in phloem cells (Sugio et al., 2011) remote from floral meristems and organs in which the target MTFs are expressed and function (Alvarez-Buylla et al., 2010; Kaufmann et al., 2010; Liu and Mara, 2010). Thus, phyllogen may have to move from phloem to cells in which target MTFs are expressed, as is the case with other phytoplasma effectors (Bai et al., 2009; Hoshi et al., 2009). On the other hand, RAD23 is expressed in the entire plant (Ishikawa et al., 2004; Farmer et al., 2010; Wang et al., 2017), and proteasome is even detected in phloem sap (Ostendorp et al., 2017). Based on these reports, we hypothesize the following spatiotemporal relationship of phyllogen with host proteins in phytoplasma infection. After secretion from phytoplasma, phyllogen spreads throughout plant cells where target MTFs do not exist. By maintaining the weak interaction with RAD23 in such cells, phyllogen might minimize being trapped by RAD23 and pass through the cells efficiently. Conversely, when phyllogen reaches target cells expressing MTFs, MTF recognition might cause phyllogen to interact strongly with RAD23 and form the MTF/phyllogen/RAD23 complex, to in turn initiate the interaction of MTF with the proteasome. Furthermore, in target cells, the A- and E-class MTFs mainly localize in nuclei (Immink et al., 2009; Smaczniak et al., 2012), whereas RAD23 localizes to both nuclei and cytoplasm (Katiyar and Lennarz, 2005; Farmer et al., 2010), suggesting that the interaction with RAD23 causes phyllogen to appear in different cellular compartments than the MTFs. Considering such differences between the intercellular and intracellular localization of MTF and RAD23, the function of phyllogen might be less perturbed by minimizing the interaction with RAD23. Although the model should be further evaluated, it seems to explain the function of phyllogen according to the different localizations of the pathogen and target cells. Future studies of the in planta dynamics of phyllogen during phytoplasma infection will elucidate the spatial relationship between phyllogen and host factors.
Materials and methods
Plant material
Nicotiana benthamiana was grown from seeds in soil under natural light at 25°C before use, and maintained in a growth chamber with 16-h light (about 7,000 lx) with a fluorescent lamp (FL40SW; Mitsubishi Electric)/8-h dark conditions at 25°C during experiments.
Materials
Previously cloned sequences (optimized for plant codon usage) that code for putative secreted regions of two phyllogens, PHYLOY (PHYLOYopt2; Iwabuchi et al., 2019) and PHYLPnWB (Kitazawa et al., 2017), were used in this study. Sequences coding for a Lys-less PHYLOYopt2 (; Supplemental Figure S9A), in which all Lys residues are replaced by Arg, were synthesized by Thermo Fisher Scientific and cloned into pENTA vector (Himeno et al., 2010), digested with SalI-HF and EcoRV-HF (NEB, Ipswich, MA, USA), using the GeneArt Seamless Cloning and Assembly Kit (Invitrogen Waltham, MA, USA). Sequences for a Lys-less SEP3 mutant (SEP3allK/R; Supplemental Figure S9B) were also synthesized and cloned into pENTA digested with SalI-HF and EcoRV-HF. Sequences for residues 146–175 in SEP3, in which five residues (M150, L154, L157, L164, and L171) were replaced by Ala (SEP3146–1755mt), were synthesized and cloned into pGADT7 (Clontech, Mountain View, CA, USA) digested with NdeI and EcoRI-HF (NEB), after amplification by polymerase chain reaction (PCR) using pAD_SEP3_5mt_F/R (Supplemental Table S1). To introduce the Ala mutations into full-length SEP3 (SEP35mt), the fragments were also amplified using 30aa5mt_to_SEP3_F/R and ligated with SEP3-containing pENTA fragments amplified by SEP3_inverse_for30aacloning_F and SEP3_inverse_R. The other constructions used in the study are described below.
Y2H assays
The Matchmaker GAL4 Two-Hybrid System 3 kit (Clontech) was used for Y2H assays as described previously (Yamaji et al., 2006). Briefly, lithium acetate-treated yeast cells (strain AH109) were cotransformed with appropriate pairs of pGADT7 and pGBKT7 vectors. The cotransformants were plated on synthetically defined (SD) leucine/tryptophan/histidine-lacking medium containing 5-mM 3-amino-1,2,4-triazole (Sigma-Aldrich, St. Louis, MO, USA) (SD/–LWH + 3AT). The plates were incubated for 4 days at 30°C. Some of constructions used in the experiments were created in previous reports (Maejima et al., 2014; Kitazawa et al., 2017; Iwabuchi et al., 2019, 2020). For further constructions, RAD23A and RAD23B were amplified by PCR from Arabidopsis (col-0) cDNA using the primers listed in Supplemental Table S1 and cloned into pGADT7 digested with NdeI and EcoRI-HF (NEB), using the GeneArt Seamless Cloning and Assembly Kit. In the same way, domains of the Arabidopsis MTF and RAD23 used in the experiments were also amplified from Col-0 using the appropriate pair of primers (Supplemental Table S1) and cloned into NdeI/EcoRI-HF-digested pGADT7. OsRAD23 or its UBA2 domain was amplified from rice (cv. Koshihikari) cDNA and cloned in the same way. SEP394–145 or SEP3146–175 was also amplified using the primer pair of SEP3-AD277F/pAD_SEP3_145R, or pAD_SEP3_146F/SEP3-AD525R, respectively, and cloned in the same way.
Protein expression in planta
Transient protein expression in N. benthamiana leaves was achieved via agroinfiltration, as described previously (Maejima et al., 2014) using A. tumefaciens strain EHA105 (Hood et al., 1993). Ratios of A. tumefaciens cells used for expressing each protein are described in the figure legends. Some constructions for transient protein expression were created in previous reports: NYF-SEP3 (Maejima et al., 2014), OsMADS8-YFP and PHYLPnWB (Kitazawa et al., 2017), myc-SEP3 (Iwabuchi et al., 2019), YFP (Ishikawa et al., 2013), myc-GFP (Hashimoto et al., 2015), FLAG-GUS (Okano et al., 2014), and P19 (Okano et al., 2020). The others were created as follows. Prior to construction, AP3 and PAD1 were amplified by PCR from the col-0 cDNA using the primers listed in Supplemental Table S1 and cloned into pENTA, as described above. OsRAD23 from rice (cv. Koshihikari) cDNA was cloned in the same way. Then, pENTA-cloned PAD1, OsRAD23, SEP3 (Maejima et al., 2014), RAD23C (Iwabuchi et al., 2019), GUS (Hashimoto et al., 2015), and PHYLOYopt2 (Iwabuchi et al., 2019) were subcloned using the Gateway Technology (Thermo Fisher Scientific, Waltham, MA, USA) into appropriate vectors: pFASTG02 (Shimada et al., 2010) for PHYLOY, pEarleyGate 100 (Earley et al., 2006) for SEP3, GUS and RAD23C, 102 for PHYLOY-CFP, PAD1-CFP and GUS-CFP, 202 for FLAG-RAD23C and FLAG-OsRAD23, 203 for myc-RAD23C and myc-GUS, pEarleyGate 104-CFP (YFP was replaced by CFP) for CFP-RAD23C, pEarleyGate 101-mScarlet (YFP was replaced by plant codon-optimized mScarlet; Supplemental Figure S9C) for RAD23C-mScarlet, pEarleyGate-NYF (Maejima et al., 2014) for NYF-AP3, and pEarleyGate-CBiFC-C (Maejima et al., 2014) for PHYLOY-CYF. If necessary, the stop codon of PHYLOY, or RAD23C was deleted by the GeneArt Site-Directed Mutagenesis System (Thermo Fisher Scientific). For expression of HA-fused PHYLOY (PHYLOY-HA), the gene was amplified by PCR using PHYL_to_pENTA_F and PHYL_G8_HA_R, ligated with pENTA fragments amplified by G7-HA-pENTA_F and pENTA_R, and subcloned into pEarleyGate 100, as described above. For expression of FLAG-fused UBA2 of RAD23C (RAD23CUBA2-FLAG), the region was amplified by PCR using UBA2_to_pENTA_F and Rad23C_G8_R, ligated with a pENTA fragment amplified by G8_FLAG_to_pENTA_F and pENTA_R, and subcloned into pEarleyGate 100, as described above. Constructs for HA- and FLAG-fused SEP3 were created as in the case of PHYLOY-HA and RAD23CUBA2-FLAG, respectively. Constructs for SEP3 and PHYLOY mutants (SEP35mt, SEP3allK/R, and ) were created in the same way.
Fluorescent microscopy observations
N. benthamiana epidermal cells were observed ∼40 h after agroinfiltration using a Leica TCS SP5 scanning confocal microscope (Leica Microsystems, Wetzlar, Germany) with a 40× oil immersion objective. CFP was excited by the 458-nm argon laser line, and the emission was visualized at 460–495 nm; the 514-nm argon laser line was used for YFP, and the emission was visualized at 525–550 nm; the 543-nm helium/neon laser line was used for mScarlet, and the emission was visualized at 585–660 nm. To observe double (YFP and CFP) and triple (YFP, CFP, and mScarlet) expression, images were scanned and captured by z-stack acquisition in the line-switching and frame-switching modes, respectively. Experiments were performed at least twice with two individual plants, and at least eight randomly chosen regions of infiltrated leaves were observed.
Protein extraction, co-IP, and immunoblotting
For protein extraction from N. benthamiana, leaves were collected 36 h after infiltration and pulverized in liquid nitrogen. Proteins were extracted with SDS sample buffer (62.5-mM Tris–HCl [pH 6.8], 2% sodium dodecyl sulfate, 5% sucrose, 0.005% bromophenol blue) containing 1% 3-mercapto-1,2-propanediol. The extract was centrifuged at 21,600g for 10 min at 4°C. The supernatant (100 µL) was precipitated by acetone, dissolved in SDS sample buffer, and incubated for 5 min at 95°C.
For in planta anti-FLAG co-IPs, the inoculated leaves were homogenized in RIPA buffer (Yamaji et al., 2006) containing complete mini EDTA-free protease inhibitors (Roche, Basel, Switzerland) and 0.1% 3-mercapto-1,2-propanediol. The homogenate was centrifuged at 13,800g for 10 min at 4°C. The supernatant (60 µL) was denatured by heating in SDS sample buffer after acetone precipitation, as described above (inputs). An aliquot of 1 mL of the supernatant was incubated for 90 min at 4°C with 30 µL of EZview Red ANTI-FLAG M2 Affinity Gel (Sigma-Aldrich). The resin was collected by centrifugation at 1,000g for 30 s and washed 6 times with 1 mL of RIPA buffer. Proteins were eluted from the resin complex by boiling with 60 µL of SDS sample buffer (IP). Experiments were performed at least twice, using two individual plants in each experiment.
Proteins were separated by SDS–PAGE and detected by immunoblotting, as described previously (Maejima et al., 2014). The commercial antibodies used in this study were Anti-FLAG antibody (F1804; Sigma-Aldrich), anti-myc antibody (4A6; Merck Millipore Burlington, MA, USA), anti-GST antibody (#2622; Cell Signaling Technology, Danvers, MA, USA), anti-HA antibody (#561; MBL, Aichi, Japan), monoclonal anti-GFP antibody for PAD1-CFP and SEP3-YFP (7.1 and 13.1; Roche), polyclonal anti-GFP antibody for NYF-SEP3, NYF-AP3 and GUS-CFP (A11122; Thermo Fisher Scientific), and anti-ubiquitin antibody (anti-UBQ11; Agrisera, Vännäs, Sweden). Polyclonal rabbit anti-PHYLOY or anti-PHYLPnWB antibody was generated by Eurofins Genomics KK using purified His-PHYLOY or His-PHYLPnWB, obtained as below and purified by NHS-activated Sepharose 4 Fast Flow (GE Healthcare Chicago, IL, USA). These antibodies were used at 1:1,000 dilution.
In vitro co-IP
For expression of phyllogens fused with a His tag, PHYLOYopt (Kitazawa et al., 2017) and PHYLPnWB genes were amplified by PCR and cloned into pCold I vector (Takara, Shiga, Japan) digested by NdeI and XhoI (NEB). Similarly, the GST-fused K domain of SEP3 (GST-SEP3K) was cloned into pGEX-4T-1 (GE Healthcare) digested by BamHI (NEB) and XhoI. For expression of MBP-RAD23C-FLAG, RAD23C was amplified by PCR using 23C_to_pMAL_F2 and Rad23C_G8_R, and then ligated with a pMAL-c5X vector (NEB) fragment amplified by G8_Flag_pMALX_F and pMALX_R. Each vector was transformed into E. coli strain BL21(DE3). Each protein was overexpressed in the transformed E. coli cells as described previously (Miura et al., 2015). For purification of His-fused proteins, the cells were resuspended in TBS (20-mM Tris/HCl [pH 7.9], 500-mM NaCl) with 5-mM imidazole, sonicated, and then centrifuged at 21,600g for 15 min at 4°C. The proteins were purified from the supernatants using Ni-NTA agarose (Qiagen, Hilden, Germany) under native conditions and eluted as described previously (Kakizawa et al., 2001). For purification of GST-SEP3K, the cells were resuspended in GST extraction buffer (20-mM Tris/HCl (pH 7.5), 150-mM NaCl, 1-mM EDTA (pH 8.0), 0.5% Triton X-100), sonicated, and centrifuged at 21,600g for 15 min at 4°C. The supernatant was incubated with Glutathione Sepharose 4B (GE Healthcare) for 1 h at 4°C. The resin was washed 6 times with GST extraction buffer containing 1.5% Triton X-100, and then washed 3 times with GST buffer containing 5-mM glutathione. GST-SEP3K was eluted with GST elution buffer (100-mM Tris/HCl [pH 9.5], 200-mM NaCl, 15-mM glutathione). MBP-RAD23C-FLAG was purified using the pMAL Protein Fusion and Purification System (NEB) according to the manufacturer’s protocol.
For in vitro co-IP, 100 pmol of MBP-RAD23C-FLAG, 50 pmol of His-PHYLOY, and 200 pmol of GST-SEP3K were mixed in 600-µL RIPA buffer and incubated for 2 h at 4°C. Then, 60 µL of the mixture was denatured at 95°C in 4× SDS sample buffer (inputs). FLAG-fused proteins were immunoprecipitated from a 500-µL aliquot of the mixture, and proteins in IP were detected as described above. Experiments were repeated 3 times.
ALSV inoculations
DNA fragments harboring the cauliflower mosaic virus 35S promoter, viral cDNA of pEALSR1 or pEALSR2L5R5, and the nopaline synthase terminator were amplified from pEALSR1 and pEALSR2L5R5 (Li et al., 2004) with the M13M3 and M13RV universal primers, respectively. pCAMBIA1301 (Accession number AF234297), an Agrobacterium binary vector, was partially amplified with M13R-pCAM2338F and M13F-pCAM8718 primers (Supplemental Table S1) and then assembled with each of the DNA fragments containing ALSV cDNAs (designated pCAM-ALSR1 and pCAM-ALSR2L5R5II, respectively) using the GeneArt Seamless Cloning and Assembly Kit (Thermo Fisher Scientific). PHYLOYopt2 or , amplified with the appropriate primers (Supplemental Table S1), was inserted into the pCAM-ALSR2L5R5II digested by XhoI and SmaI, as described previously (Kitazawa et al., 2017), and each construction was transformed into A. tumefaciens strain EHA105. Each virus was inoculated into 3-week-old N. benthamiana, as described previously (Kitazawa et al., 2017).
Accession numbers
Sequence data of genes used in this article can be found by the following accession numbers in the parentheses: SEP3 (AT1G24260), AP1 (AT1G69120), AP3 (AT3G54340), AG (AT4G18960), RAD23A (AT1G16190), RAD23B (AT1G79650), RAD23C (AT3G02540), RAD23D (AT5G38470), PAD1 (AT3G51260), PHYLOY (AB812838), and PHYLPnWB (PNWB_v1c0190 in AMWZ01000001) in the EMBL/GenBank/DDBJ data libraries; OsMADS8 (LOC_Os09g32948), OsRAD23 (LOC_Os09g24200) in the MSU Rice Genome Annotation Project Database (http://rice.plantbiology.msu.edu).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Supporting information for fluorescent microscopy analysis.
Supplemental Figure S2. In planta coimmunoprecipitation assay using FLAG-RAD23C and PHYLOY.
Supplemental Figure S3. In planta coimmunoprecipitation using (A) FLAG-RAD23C, PHYLOY-CYF, and myc-SEP3 or (B) SEP3-FLAG, PHYLOY, myc-RAD23C, and PAD1-CFP.
Supplemental Figure S4. Comparison of ubiquitination between PHYLOY and .
Supplemental Figure S5. Comparison of in planta protein accumulation between PHYLOY and .
Supplemental Figure S6. Protein structure of SEP3 and RAD23C.
Supplemental Figure S7. Y2H assay using PHYLOY and domains in A. thaliana MTF or RAD23.
Supplemental Figure S8. Interactions of the MTF/phyllogen/RAD23 protein complex using different phyllogen or host proteins.
Supplemental Figure S9. Modified nucleotide sequences cloned in the study.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr Nobuyuki Yoshikawa (Faculty of Agriculture, Iwate University, Iwate, Japan) for the ALSV vector.
Funding
This research was supported by funds from the Japan Society for the Promotion of Science (JSPS) (nos. 25221201, 19K15840, 20H02991, 20K22562, 21H04722, 21K14847, and 21K14853).
Conflict of interest statement. None declared.
Contributor Information
Yugo Kitazawa, Department of Agricultural and Environmental Biology, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo 113-8657, Japan.
Nozomu Iwabuchi, Department of Agricultural and Environmental Biology, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo 113-8657, Japan.
Kensaku Maejima, Department of Agricultural and Environmental Biology, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo 113-8657, Japan.
Momoka Sasano, Department of Agricultural and Environmental Biology, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo 113-8657, Japan.
Oki Matsumoto, Department of Agricultural and Environmental Biology, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo 113-8657, Japan.
Hiroaki Koinuma, Department of Agricultural and Environmental Biology, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo 113-8657, Japan.
Ryosuke Tokuda, Department of Agricultural and Environmental Biology, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo 113-8657, Japan.
Masato Suzuki, Department of Agricultural and Environmental Biology, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo 113-8657, Japan.
Kenro Oshima, Faculty of Bioscience and Applied Chemistry, Hosei University, Tokyo 184-8584, Japan.
Shigetou Namba, Department of Agricultural and Environmental Biology, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo 113-8657, Japan.
Yasuyuki Yamaji, Department of Agricultural and Environmental Biology, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo 113-8657, Japan.
Y.K., N.I., and K.M. designed the research. Y.K., and M.Sa. performed the experiments and analyzed data. Y.K. wrote the initial draft of the manuscript. N.I., K.M., K.H., O.K., S.N., and Y.Y. critically revised and edited the manuscript. All authors discussed the results and contributed to writing.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is Kensaku Maejima (amaejima@mail.ecc.u-tokyo.ac.jp).
References
- Alvarez-Buylla ER, Benítez M, Corvera-Poiré A, Chaos Cador A, de Folter S, Gamboa de Buen A, Garay-Arroyo A, García-Ponce B, Jaimes-Miranda F, Pérez-Ruiz RV, et al. (2010) Flower development. Arabidopsis Book 8: e0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashida H, Kim M, Sasakawa C (2014) Exploitation of the host ubiquitin system by human bacterial pathogens. Nat Rev Microbiol 12: 399–413 [DOI] [PubMed] [Google Scholar]
- Aurin MB, Haupt M, Görlach M, Rümpler F, Theißen G (2020) Structural requirements of the phytoplasma effector protein SAP54 for causing homeotic transformation of floral organs. Mol Plant Microbe Interact 33: 1129–1141 [DOI] [PubMed] [Google Scholar]
- Bai X, Correa VR, Toruño TY, Ammar el D, Kamoun S, Hogenhout SA (2009) AY-WB phytoplasma secretes a protein that targets plant cell nuclei. Mol Plant Microbe Interact 22: 18–30 [DOI] [PubMed] [Google Scholar]
- Banfield MJ (2015) Perturbation of host ubiquitin systems by plant pathogen/pest effector proteins. Cell Microbiol 17: 18–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindels DS, Haarbosch L, van Weeren L, Postma M, Wiese KE, Mastop M, Aumonier S, Gotthard G, Royant A, Hink MA, et al. (2017) mScarlet: a bright monomeric red fluorescent protein for cellular imaging. Nat Methods 14: 53–56 [DOI] [PubMed] [Google Scholar]
- Bingol B, Schuman EM (2006) Activity-dependent dynamics and sequestration of proteasomes in dendritic spines. Nature 441: 1144–1148 [DOI] [PubMed] [Google Scholar]
- Chung WC, Chen LL, Lo WS, Lin CP, Kuo CH (2013) Comparative analysis of the peanut witches'-broom phytoplasma genome reveals horizontal transfer of potential mobile units and effectors. PLoS One 8: e62770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui R, Han J, Zhao S, Su K, Wu F, Du X, Xu Q, Chong K, Theissen G, Meng Z (2010) Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa). Plant J 61: 767–781 [DOI] [PubMed] [Google Scholar]
- Dantuma NP, Heinen C, Hoogstraten D (2009) The ubiquitin receptor Rad23: at the crossroads of nucleotide excision repair and proteasomal degradation. DNA Repair 8: 449–460 [DOI] [PubMed] [Google Scholar]
- Dikic I, Wakatsuki S, Walters KJ (2009) Ubiquitin-binding domains—from structures to functions. Nat Rev Mol Cell Biol 10: 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway‐compatible vectors for plant functional genomics and proteomics. Plant J 45: 616. [DOI] [PubMed] [Google Scholar]
- Enenkel C, Lehmann A, Kloetzel PM (1998) Subcellular distribution of proteasomes implicates a major location of protein degradation in the nuclear envelope–ER network in yeast. EMBO J 17: 6144–6154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson JL, Adlung N, Lampe C, Bonas U, Schattat MH (2018) The Xanthomonas effector XopL uncovers the role of microtubules in stromule extension and dynamics in Nicotiana benthamiana. Plant J 93: 856–870 [DOI] [PubMed] [Google Scholar]
- Farmer LM, Book AJ, Lee KH, Lin YL, Fu H, Vierstra RD (2010) The RAD23 family provides an essential connection between the 26S proteasome and ubiquitylated proteins in Arabidopsis. Plant Cell 22: 124–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Doelling JH, Arendt CS, Hochstrasser M, Vierstra RD (1998) Molecular organization of the 20S proteasome gene family from Arabidopsis thaliana. Genetics 149: 677–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Hann DR, Ntoukakis V, Petutschnig E, Lipka V, Rathjen JP (2009) AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr Biol 19: 423–429 [DOI] [PubMed] [Google Scholar]
- Göhre V, Spallek T, Häweker H, Mersmann S, Mentzel T, Boller T, de Torres M, Mansfield JW, Robatzek S (2008) Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr Biol 18: 1824–1832 [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Komatsu K, Iwai R, Keima T, Maejima K, Shiraishi T, Ishikawa K, Yoshida T, Kitazawa Y, Okano Y, et al. (2015) Cell death triggered by a putative amphipathic helix of Radish mosaic virus helicase protein is tightly correlated with host membrane modification. Mol Plant Microbe Interact 28: 675–688 [DOI] [PubMed] [Google Scholar]
- He SY, Nomura K, Whittam TS (2004) Type III protein secretion mechanism in mammalian and plant pathogens. BBA-Mol Cell Res 1694: 181–206 [DOI] [PubMed] [Google Scholar]
- Himeno M, Maejima K, Komatsu K, Ozeki J, Hashimoto M, Kagiwada S, Yamaji Y, Namba S (2010) Significantly low level of small RNA accumulation derived from an encapsidated mycovirus with dsRNA genome. Virology 396: 69–75 [DOI] [PubMed] [Google Scholar]
- Honma T, Goto K (2001) Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409: 525–529 [DOI] [PubMed] [Google Scholar]
- Hood EE, Gelvin SB, Melchers LS, Hoekema A (1993) New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res 2: 208–218 [Google Scholar]
- Hoshi A, Oshima K, Kakizawa S, Ishii Y, Ozeki J, Hashimoto M, Komatsu K, Kagiwada S, Yamaji Y, Namba S (2009) A unique virulence factor for proliferation and dwarfism in plants identified from a phytopathogenic bacterium. Proc Natl Acad Sci USA 106: 6416–6421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, MacLean AM, Sugio A, Maqbool A, Busscher M, Cho ST, Kamoun S, Kuo CH, Immink RGH, Hogenhout SA (2021) Parasitic modulation of host development by ubiquitin-independent protein degradation. Cell 184: 5201–5214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immink RGH, Tonaco IAN, de Folter S, Shchennikova A, van Dijk ADJ, Busscher-Lange J, Borst JW, Angenent GC (2009) SEPALLATA3: the ‘glue’ for MADS box transcription factor complex formation. Genome Biol 10: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa K, Maejima K, Komatsu K, Netsu O, Keima T, Shiraishi T, Okano Y, Hashimoto M, Yamaji Y, Namba S (2013) Fig mosaic emaravirus p4 protein is involved in cell-to-cell movement. J Gen Viol 94: 682–686 [DOI] [PubMed] [Google Scholar]
- Ishikawa Y, Endo M, Abe K, Osakabe K, Nakajima N, Saji H, Ito Y, Ichikawa H, Kameya T, Toki S (2004) Isolation of four RAD23 genes from Arabidopsis thaliana and detection of alternative splicing variants. Plant Biotechnol 21: 65–71 [Google Scholar]
- Iwabuchi N, Kitazawa Y, Maejima K, Koinuma H, Miyazaki A, Matsumoto O, Suzuki T, Nijo T, Oshima K, Namba S, et al. (2020) Functional variation in phyllogen, a phyllody-inducing phytoplasma effector family, attributable to a single amino acid polymorphism. Mol Plant Pathol 21: 1322–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwabuchi N, Maejima K, Kitazawa Y, Miyatake H, Nishikawa M, Tokuda R, Koinuma H, Miyazaki A, Nijo T, Oshima K, et al. (2019) Crystal structure of phyllogen, a phyllody-inducing effector protein of phytoplasma. Biochem Biophys Res Commun 513: 952–957 [DOI] [PubMed] [Google Scholar]
- Kakizawa S, Oshima K, Kuboyama T, Nishigawa H, Jung H, Sawayanagi T, Tsuchizaki T, Miyata S, Ugaki M, Namba S (2001) Cloning and expression analysis of Phytoplasma protein translocation genes. Mol Plant Microbe Interact 14: 1043–1050 [DOI] [PubMed] [Google Scholar]
- Katiyar S, Lennarz WJ (2005) Studies on the intracellular localization of hHR23B. Biochem Biophys Res Commun 337: 1296–1300 [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Pajoro A, Angenent GC (2010) Regulation of transcription in plants: mechanisms controlling developmental switches. Nat Rev Genet 11: 830–842 [DOI] [PubMed] [Google Scholar]
- Kim M, Otsubo R, Morikawa H, Nishide A, Takagi K, Sasakawa C, Mizushima T (2014) Bacterial effectors and their functions in the ubiquitin-proteasome system: insight from the modes of substrate recognition. Cells 3: 848–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazawa Y, Iwabuchi N, Himeno M, Sasano M, Koinuma H, Nijo T, Tomomitsu T, Yoshida T, Okano Y, Yoshikawa N, Maejima K, Oshima K, Namba S (2017) Phytoplasma-conserved phyllogen proteins induce phyllody across the Plantae by degrading floral MADS domain proteins. J Exp Bot 68: 2799–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YT, Ciechanover A (2017) The ubiquitin code in the ubiquitin-proteasome system and autophagy. Trends Biochem Sci 42: 873–886 [DOI] [PubMed] [Google Scholar]
- Lai X, Daher H, Galien A, Hugouvieux V, Zubieta C (2019) Structural basis for plant MADS transcription factor oligomerization. Comput Struct Biotechnol J 17: 946–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D, Salin B, Daignan-Fornier B, Sagot I (2008) Reversible cytoplasmic localization of the proteasome in quiescent yeast cells. J Cell Biol 181: 737–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Sasaki N, Isogai M, Yoshikawa N (2004) Stable expression of foreign proteins in herbaceous and apple plants using Apple latent spherical virus RNA2 vectors. Arch Virol 149: 1541–1558 [DOI] [PubMed] [Google Scholar]
- Liao YT, Lin SS, Lin SJ, Sun WT, Shen BN, Cheng HP, Lin CP, Ko TP, Chen YF, Wang HC (2019) Structural insights into the interaction between phytoplasmal effector causing phyllody 1 and MADS transcription factors. Plant J 100: 706–719 [DOI] [PubMed] [Google Scholar]
- Lin YH, Machner MP (2017) Exploitation of the host cell ubiquitin machinery by microbial effector proteins. J Cell Sci 130: 1985–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Mara C (2010) Regulatory mechanisms for floral homeotic gene expression. Semin Cell Dev Biol 21: 80–86 [DOI] [PubMed] [Google Scholar]
- Livneh I, Cohen-Kaplan V, Cohen-Rosenzweig C, Avni N, Ciechanover A (2016) The life cycle of the 26S proteasome: from birth, through regulation and function, and onto its death. Cell Res 26: 869–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Xu X, Cai L, Cao Y, Haq F, Alfano JR, Zhu B, Zou L, Chen G (2020) A Xanthomonas oryzae type III effector XopL causes cell death through mediating ferredoxin degradation in Nicotiana benthamiana. Phytopathol Res 2: 16 [Google Scholar]
- MacLean AM, Orlovskis Z, Kowitwanich K, Zdziarska AM, Angenent GC, Immink RGH, Hogenhout SA (2014) Phytoplasma effector SAP54 hijacks plant reproduction by degrading MADS-box proteins and promotes insect colonization in a RAD23-dependent manner. PLoS Biol 12: e1001835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean AM, Sugio A, Makarova OV, Findlay KC, Grieve VM, Tóth R, Nicolaisen M, Hogenhout SA (2011) Phytoplasma effector SAP54 induces indeterminate leaf-like flower development in Arabidopsis plants. Plant Physiol 157: 831–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maejima K, Iwai R, Himeno M, Komatsu K, Kitazawa Y, Fujita N, Ishikawa K, Fukuoka M, Minato N, Yamaji Y, et al. (2014) Recognition of floral homeotic MADS domain transcription factors by a phytoplasmal effector, phyllogen, induces phyllody. Plant J 78: 541–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maejima K, Kitazawa Y, Tomomitsu T, Yusa A, Neriya Y, Himeno M, Yamaji Y, Oshima K, Namba S (2015) Degradation of class E MADS-domain transcription factors in Arabidopsis by a phytoplasmal effector, phyllogen. Plant Signal Behav 10: e1042635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcone C (2014) Molecular biology and pathogenicity of phytoplasmas. Ann Appl Biol 165: 199–221 [Google Scholar]
- McClellan AJ, Laugesen SH, Ellgaard L (2019) Cellular functions and molecular mechanisms of non-lysine ubiquitination. Open Biol 9: 190147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura C, Komatsu K, Maejima K, Nijo T, Kitazawa Y, Tomomitsu T, Yusa A, Himeno M, Oshima K, Namba S (2015) Functional characterization of the principal sigma factor RpoD of phytoplasmas via an in vitro transcription assay. Sci Rep 5: 11893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namba S (2019) Molecular and biological properties of phytoplasmas. P Jpn Acad B-Phys 95: 401–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano Y, Maejima K, Yoshida T, Nishida S, Tokuda R, Nishikawa M, Namba S, Yamaji Y (2020) Interfamily transfer of Arabidopsis lectin‐mediated antiviral gene confers resistance to pepino mosaic virus in tomato. J Gen Plant Pathol 86: 274 [Google Scholar]
- Okano Y, Senshu H, Hashimoto M, Neriya Y, Netsu O, Minato N, Yoshida T, Maejima K, Oshima K, Komatsu K, Yamaji Y, et al. (2014) In planta recognition of a double-stranded RNA synthesis protein complex by a potexviral RNA silencing suppressor. Plant Cell 26: 2168–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlovskis Z, Hogenhout SA (2016) A bacterial parasite effector mediates insect vector attraction in host plants independently of developmental changes. Front Plant Sci 7: 885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostendorp A, Pahlow S, Krüßel L, Hanhart P, Garbe MY, Deke J, Giavalisco P, Kehr J (2017) Functional analysis of Brassica napus phloem protein and ribonucleoprotein complexes. New Phytol 214: 1188–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popa CM, Tabuchi M, Valls M (2016) Modification of bacterial effector proteins inside eukaryotic host cells. Front Cell Infect Microbiol 6: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portaliou AG, Tsolis KC, Loos MS, Zorzini V, Economou A (2016) Type III Secretion: building and operating a remarkable nanomachine. Trends Biochem Sci 41: 175–189 [DOI] [PubMed] [Google Scholar]
- Puranik S, Acajjaoui S, Conn S, Costa L, Conn V, Vial A, Marcellin R, Melzer R, Brown E, Hart D, et al. (2014) Structural basis for the oligomerization of the MADS domain transcription factor SEPALLATA3 in Arabidopsis. Plant Cell 26: 3603–3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid T, Hochstrasser M (2008) Diversity of degradation signals in the ubiquitin–proteasome system. Nat Rev Mol Cell Biol 9: 679–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosebrock TR, Zeng L, Brady JJ, Abramovitch RB, Xiao F, Martin GB (2007) A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature 448: 370–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruelens P, Zhang Z, van Mourik H, Maere S, Kaufmann K, Geuten K (2017) The origin of floral organ identity quartets. Plant Cell 29: 229–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rümpler F, Gramzow L, Theißen G, Melzer R (2015) Did convergent protein evolution enable phytoplasmas to generate ‘zombie plants’? Trends Plant Sci 20: 798–806 [DOI] [PubMed] [Google Scholar]
- Rümpler F, Theißen G, Melzer R (2018) A conserved leucine zipper-like motif accounts for strong tetramerization capabilities of SEPALLATA-like MADS-domain transcription factors. J Exp Bot 69: 1943–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki Y (2017) Ubiquitin recognition by the proteasome. J Biochem 161: 113–124 [DOI] [PubMed] [Google Scholar]
- Schultz TF, Quatrano RS (1997) Characterization and expression of a rice RAD23 gene. Plant Mol Biol 34: 557–562 [DOI] [PubMed] [Google Scholar]
- Shimada TL, Shimada T, Hara-Nishimura I (2010) A rapid and non‐destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J 61: 519–528 [DOI] [PubMed] [Google Scholar]
- Singer AU, Schulze S, Skarina T, Xu X, Cui H, Eschen-Lippold L, Egler M, Srikumar T, Raught B, Lee J, et al. (2013) A pathogen type III effector with a novel E3 ubiquitin ligase architecture. PLoS Pathog 9: e1003121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaczniak C, Immink RGH, Muiño JM, Blanvillain R, Busscher M, Busscher-Lange J, Dinh QDP, Liu S, Westphal AH, Boeren S, et al. (2012) Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc Natl Acad Sci USA 109: 1560–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohmayer A, Moser M, Si-Ammour A, Krczal G, Boonrod K (2019) ‘Candidatus Phytoplasma mali’ genome encodes a protein that functions as an E3 ubiquitin ligase and could inhibit plant basal defense. Mol Plant Microbe Interact 32: 1487–1495 [DOI] [PubMed] [Google Scholar]
- Sugio A, MacLean AM, Kingdom HN, Grieve VM, Manimekalai R, Hogenhout SA (2011) Diverse targets of phytoplasma effectors: from plant development to defense against insects. Annu Rev Phytopathol 49: 175–195 [DOI] [PubMed] [Google Scholar]
- Sundin GW, Castiblanco LF, Yuan X, Zeng Q, Yang CH (2016) Bacterial disease management: challenges, experience, innovationand future prospects. Mol Plant Pathol 17: 1506–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theißen G, Melzer R, Rümpler F (2016) MADS-domain transcription factors and the floral quartet model of flower development: linking plant development and evolution. Development 143: 3259–3271 [DOI] [PubMed] [Google Scholar]
- Tomko RJ Jr, Hochstrasser M (2013) Molecular architecture and assembly of the eukaryotic proteasome. Annu Rev Biochem 82: 415–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng TT, Tyler BM, Setubal JC (2009) Protein secretion systems in bacterial-host associations, and their description in the Gene Ontology. BMC Microbiol 9: S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya H, Ohtake F, Arai N, Kaiho A, Yasuda S, Tanaka K, Saeki Y (2017) In vivo ubiquitin linkage-type analysis reveals that the Cdc48-Rad23/Dsk2 axis contributes to K48-linked chain specificity of the proteasome. Mol Cell 66: 488–502 [DOI] [PubMed] [Google Scholar]
- Vierstra RD (2009) The ubiquitin–26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Wang N, Gong XQ, Ma FW (2017) Genome-wide identification of the radiation sensitivity protein-23 (RAD23) family members in apple (Malus×domestica Borkh.) and expression analysis of their stress responsiveness. J Integr Agric 16: 820–827 [Google Scholar]
- Wang N, Yang H, Yin Z, Liu W, Sun L, Wu Y (2018) Phytoplasma effector SWP1 induces witches’ broom symptom by destabilizing the TCP transcription factor BRANCHED1. Mol Plant Pathol 19: 2623–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji Y, Kobayashi T, Hamada K, Sakurai K, Yoshii A, Suzuki M, Namba S, Hibi T (2006) In vivo interaction between Tobacco mosaic virus RNA-dependent RNA polymerase and host translation elongation factor 1A. Virology 347: 100–108 [DOI] [PubMed] [Google Scholar]
- Yang CY, Huang YH, Lin CP, Lin YY, Hsu HC, Wang CN, Liu LYD, Shen BN, Lin SS (2015) MicroRNA396-targeted SHORT VEGETATIVE PHASE is required to repress flowering and is related to the development of abnormal flower symptoms by the phyllody symptoms1 effector. Plant Physiol 168: 1702–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Teixeira PJPL, Biswas S, Finkel OM, He Y, Salas-Gonzalez I, English ME, Epple P, Mieczkowski P, Dangl JL (2017) Pseudomonas syringae type III effector HopBB1 promotes host transcriptional repressor degradation to regulate phytohormone responses and virulence. Cell Host Microbe 21: 156–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Jack T (2004) Defining subdomains of the K domain important for protein-protein interactions of plant MADS proteins. Plant Mol Biol 55: 45–59 [DOI] [PubMed] [Google Scholar]
- Ye Y, Rape M (2009) Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol 10: 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Matouschek A (2017) Recognition of client proteins by the proteasome. Ann Rev Biophys 46: 149–173 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.