Abstract
Plant defense responses against insect herbivores are induced through wound-induced signaling and the specific perception of herbivore-associated molecular patterns (HAMPs). In addition, herbivores can deliver effectors that suppress plant immunity. Here we review plant immune recognition of HAMPs and effectors, and argue that these initial molecular interactions upon a plant–herbivore encounter mediate and structure effective resistance. While the number of distinct HAMPs and effectors from both chewing and piercing–sucking herbivores has expanded rapidly with omics-enabled approaches, paired receptors and targets in the host are still not well characterized. Herbivore-derived effectors may also be recognized as HAMPs depending on the host plant species, potentially through the evolution of novel immune receptor functions. We compile examples of HAMPs and effectors where natural variation between species may inform evolutionary patterns and mechanisms of plant–herbivore interactions. Finally, we discuss the combined effects of wounding and HAMP recognition, and review potential signaling hubs, which may integrate both sensing functions. Understanding the precise mechanisms for plant sensing of herbivores will be critical for engineering resistance in agriculture.
This review describes the mechanisms by which plants perceive chewing and piercing–sucking herbivores, as well as the downstream signaling pathways that mediate specific responses.
Introduction
Global biomass is dominated by plant matter (Bar-On et al., 2018), partially due to the plants’ ability to resist parasitic attack. While herbivores can devastate plants in specific natural and agricultural contexts (Savary et al., 2019), the more frequent outcome of a specific plant–herbivore interaction is host tolerance or resistance. Why are these a typical outcome, and what explains exceptional pest outbreaks? The answers are not only fundamental for understanding the ecology and evolution of plant–herbivore interactions, but also inform strategies for crop protection in agriculture.
A half century of work primarily in the fields of chemical ecology and biochemistry has provided compelling mechanisms to explain plant resistance to herbivory (Fraenkel, 1959; Green and Ryan, 1972). Plants present physical barriers to attackers, as well as constitutive chemical defenses (Mithöfer and Boland, 2012). In addition, inducible responses to herbivores can further enhance plant resistance or tolerance through: (1) direct defenses to inhibit growth and survival; (2) indirect defenses via predator or parasitoid attraction; or (3) induced resource reallocation (Painter, 1951; Howe and Jander, 2008; Stenberg and Muola, 2017; Turlings and Erb, 2018; Erb and Reymond, 2019). Signaling mechanisms, including calcium influx, reactive oxygen species (ROS) production, propagation of electrical signals, and synthesis of defense hormones such as jasmonic acid (JA) are critical for both local induced responses and propagation to systemic tissues (Howe et al., 2018; Farmer et al., 2020). A strong theoretical grounding has developed around these empirical findings, helping to explain the diversity and structure of plant inducible chemistry in nature (Schuman and Baldwin, 2016).
In contrast to a high degree of mechanistic understanding of plant defense outputs, far less is known regarding mechanisms for initial recognition of herbivores. Here we review evidence that plant immune recognition of herbivory, and in contrast its evasion by successful pests, is also key to mediating and structuring plant resistance. We describe herbivore-associated molecular patterns (HAMPs), effectors, and receptors mediating induced plant defenses in plant–herbivore interactions. We also discuss the evolution of recognition and suppression functions and potential causes of specificity. Finally, we review HAMP recognition in the context of concomitant wound responses. We limit our discussion to plant-eating arthropods, but note that similar themes apply to plant–nematode interactions (Eves-van den Akker, 2021).
Pattern recognition by the plant immune system
Inducible responses to herbivory fit within a broad conceptual framework of innate immunity, and the plant immune system in particular (Medzhitov and Janeway, 2002; Jones and Dangl, 2006). A successful immune response requires two stages: (1) Recognition, that is, the perception of nonself or modified self-stimuli and (2) effective response, consisting of defensive outputs tailored to the attacker. Molecular interactions with the biotic attacker occur at both stages and control the outcome of an encounter. Predictably, strong selective pressures during biotic attack can lead to arms race dynamics over evolutionary time, in which immune recognition is evaded or re-established (Upson et al., 2018).
The first step in immune recognition is now well understood for plant resistance to many bacterial, fungal, and oomycete pathogens. Immune receptors, termed Pattern Recognition Receptors (PRRs), monitor the presence of pathogen-associated molecular patterns (PAMPs) via extracellular ligand-binding domains (Couto and Zipfel, 2016; Albert et al., 2020). Successful pathogens avoid recognition through several strategies; for example, shielding or modifying their PAMPs to avoid receptor binding, or interference with early signaling steps through secretion of effector proteins into the apoplast or plant cell (Toruño et al., 2016; Buscaill and van der Hoorn, 2021). However, effectors themselves are reliable signals of invasion and can betray the presence of a pathogen (Jones and Dangl, 2006; Cook et al., 2015). For example, many pathogenic effectors are recognized by intracellular NOD-like receptors (NLRs; Bentham et al., 2020). In summary, depending on the PAMPs, effectors, and corresponding immune receptors involved in a given plant–pathogen interaction, immune recognition either occurs or is instead evaded or suppressed.
Natural variation in PAMPs, effectors, and their cognate receptors can structure diverse plant–pathogen interactions. Analysis of pathogen genomes has revealed that presence–absence and sequence variation are pervasive in both PAMPs and effectors (Baltrus et al., 2011; Colaianni et al., 2021; Parys et al., 2021). Host immune receptors in plant genomes are also highly variable (Steinbrenner, 2020; Pruitt et al., 2021). For example, among 64 Arabidopsis thaliana accessions, only 53% of “core” NLRs were present in most accessions, while many NLRs (13%) were present in 12 or fewer accessions (Van de Weyer et al., 2019).
Although plant attack by herbivores and bacterial pathogens results in very distinct damage and damage patterns, several lessons inform the study of plant–herbivore interactions at the recognition level. First, immune receptor variation can determine recognition specificity and thus the outcome of a biotic interaction. Second, pathogen molecules can function as effectors and PAMPs, depending on the presence/absence of a recognizing immune receptor in the host genotype. Finally, immune recognition is an active molecular battleground driving the evolution of plants and pathogens. The same lessons almost certainly apply to plant sensing of herbivory, but demonstrated examples are still rare, mainly due to a lack of defined HAMP–receptor pairs. Below, we describe well-characterized and recently described molecular interactions between plants and herbivores in a plant immune system framework.
HAMPs and effectors in plant–herbivore interactions
A variety of herbivore-derived small molecules and proteins can elicit plant defense responses (Supplemental Data Set 1). In addition, endogenous, modified self-signals from damage, termed damage-associated molecular patterns (DAMPs), are involved in plant defense responses. The nature of both HAMPs and DAMPs has been extensively reviewed (Acevedo et al., 2015; Stahl et al., 2018; Tanaka and Heil, 2021). Precise characterization of HAMPs is required to understand recognition by host plants. We therefore focus here on pipelines for HAMP identification while giving special attention to recent studies and novel identification strategies.
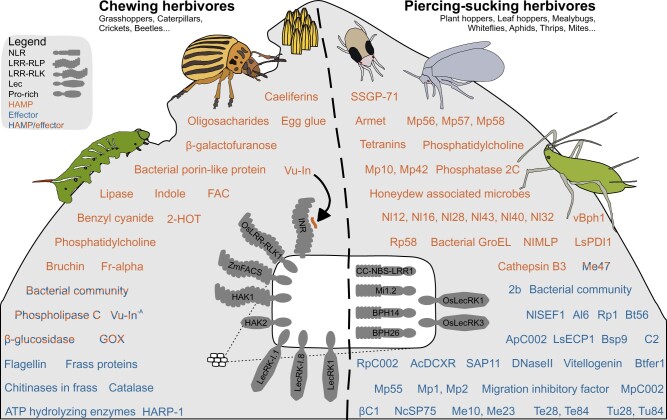
HAMPs were first characterized as elicitors of host volatile production purified as bioactive components from oral secretions (OS) (Supplemental Data Set 1). OS contain a mixture of salivary secretions, glandular secretions, and regurgitant deposited on the wounded leaf on feeding. Volicitin, 17-OH-C18:3-Gln, was the first purified HAMP; 300 pmol applied to wounded plants is sufficient to elicit volatile production (Alborn et al., 1997). Volicitin was first identified from fall armyworm (Spodoptera frugiperda), but related fatty acid-amino acid conjugates (FACs) are abundant HAMPs identified across larval OS of a subgroup of Lepidoptera, the Apoditrysia (Yoshinaga et al., 2010). For example, the related FAC molecule linoleic acid-Glu (C18:3-Glu) is abundant in OS of Manduca sp. and activates a variety of responses on coyote tobacco, Nicotiana attenuata (Halitschke et al., 2001). A similar HAMP discovery approach led to the biochemical purification of inceptin peptides, which are major HAMPs on specific legumes (Schmelz et al., 2006). The predominant inceptin when S. frugiperda feeds on cowpea (Vigna unguiculata), Vu-In, is an 11-amino acid peptide released upon proteolysis of chloroplastic ATP synthase gamma subunit (cATPC) in the Lepidopteran foregut. Inceptins derived from alternative host plant cATPC sequences are also bioactive (Schmelz et al., 2006; Steinbrenner et al., 2020). FACs and inceptins are thus ubiquitously present in OS during leaf herbivory (Yoshinaga et al., 2008).
Bioactive HAMPs/effectors from herbivores have now been identified in diverse sources and life history stages beyond larval OS. Phosphatidylcholines with C16–C18 acyl chains were recently shown to be an active fraction of crushed Pieris brassicae eggs (Stahl et al., 2020). Microgram quantities of a volatile sex pheromone component, namely E,S-conophthorin spiroacetal from goldenrod gall fly (Eurosta solidaginis), were sufficient to prime inducible JA production and to reduce herbivory on tall goldenrod (Solidago altissima) in a dose-dependent manner (Helms et al., 2017). Frass-associated, host plant-derived endochitinases can elicit specific maize (Zea mays) defenses (Ray et al., 2015, 2016). The aphid endosymbiont-derived protein GroEL elicits A. thaliana immune defenses (Chaudhary et al., 2014). Finally, host-associated microbes in OS and honeydew may confer immune elicitation, depending on the host plant species and the specific community acquired through herbivory (Acevedo et al., 2017; Wang et al., 2017b; Wari et al., 2019; Yamasaki et al., 2021).
In contrast to immune elicitation, herbivore-derived molecules can also act as effectors to suppress early steps in immune recognition. Similar to the HAMP discovery process, effectors have been characterized based on their suppression of various plant immune outputs (Supplemental Data Set 1). As a major salivary component of OS, glucose oxidase (GOX) was the first characterized effector associated with herbivory. Musser et al. (2002) described suppression of nicotine production in tobacco (Nicotiana tabacum) dependent on GOX enzymatic activity. Subsequent studies have confirmed broad suppressive effects on various host species, likely mediated by modulation of extracellular ROS (Louis et al., 2013; Lin et al., 2020).
Established and emerging methods for identification
Discovery of novel HAMPs/effectors increasingly leverages omics-enabled approaches rather than biochemical purification. For example, leveraging genomic resources for cotton bollworm (Helicoverpa armigera) allowed proteomic identification of OS components induced upon herbivory. One of these proteins, HARP1, was shown to interact with JAZ repressors preventing COI1-mediated, JA-induced degradation, thereby suppressing plant immune responses (Chen et al., 2019). Interestingly, HARP1 can be visualized via immunohistochemistry in cells adjacent to herbivorous wounding, and in vasculature upon exogenous application (Chen et al., 2019), and mechanisms of host uptake may be of great importance. Proteomic analysis of highly abundant molecules also enabled the discovery of deposited host chitinases, maize Pr4 and Endochitinase A, as frass-associated effectors able to suppress defense-related transcripts in maize (Ray et al., 2016). Other abundant or highly stable proteins identified in separate analyses of OS and frass are excellent candidates for novel effector functions (Chen et al., 2007; Acevedo et al., 2018; Rivera-Vega et al., 2018).
Secretome prediction has been an efficient strategy to identify HAMPs/effectors associated with piercing–sucking herbivores (Nalam et al., 2019; Favery et al., 2020; Naalden et al., 2021). The presence of salivary glands and associated endosymbionts has allowed for focused transcriptomic analyses; pioneering examples of this approach were used to identify effectors from aphids, whitefly, and galling Hessian fly species (Bos et al., 2010; Su et al., 2012; Zhao et al., 2015). In a recent example, tetranins were identified as predicted salivary gland proteins from the two-spotted spider mite (Tetranychus urticae) genome. When heterologously expressed in leaf tissue, two candidate genes Tet1 and Tet2 increased mite mortality and were sufficient to induce defensive marker transcripts, hormones, membrane depolarization, and/or ROS production (Iida et al., 2019). Similar transcriptomic characterization of planthopper salivary glands identified the planthopper HAMPs mucin-like protein NlMLP (Nilaparvata lugens) and disulfide isomerase LsPDI1 (Laodelphax striatellus), which elicit cell death and defense responses in both host cells and the nonhost Nicotiana benthamiana (Shangguan et al., 2018; Fu et al., 2020). Effectors that alter host plant morphology to promote transmission via insect vectors have also been identified in phytoplasma genomes (Sugio et al., 2011; MacLean et al., 2014). Finally, two effectors originating from begomoviruses were shown to suppress plant immunity against or increase performance and fecundity of a known transmitting vector, Bemisia tabaci (Li et al., 2014, 2019b).
From a comprehensive search of the literature, which identified 119 reports of HAMP or effector characterization, several broad conclusions can be drawn. From 1997 to 2009, volatile or hormonal markers coupled with biochemical purification was the primary method of HAMP identification (Figure 1; Supplemental Data Set 1). Since 2010, insect genomes have greatly enabled discovery: approximately 25 studies have used functional genomic approaches, primarily to identify HAMPs/effectors from piercing–sucking herbivores. Secretome prediction, transcriptomics across HAMP/effector-generating organs, and proteomics of diverse herbivore chemical fractions are among many exciting genome-enabled approaches.
Figure 1.
HAMPs and effectors with characterized effects on the plant immune response; HAMPs and effectors of chewing and piercing-sucking herbivores are shown respectively on the left and right side of the leaf separated by a dotted line (Supplemental Data Set 1). HAMPs are shown in orange, effectors are shown in blue, and molecules where both HAMP and effector activities have been observed are shown in orange-blue. Transmembrane and intracellular receptors involved in defense against herbivores are shown in an enlarged representative cell. Lec, Lectin domain, Pro-rich, Proline-rich sequence.
Genomic resources also facilitate forward genetic and comparative approaches to identify new HAMPs and effectors. Notably, effectors from Hessian fly have been successfully identified through a combination of genomics and traditional genetic mapping (Aggarwal et al., 2014; Zhao et al., 2015, 2016). Sequence data alone may also be sufficient to predict specific relevant biomolecules. For example, genomic screens based on positive selection have been employed to predict bacterial PAMPs as well as insect genes highly relevant for interaction with plant volatiles (Mott et al., 2016; Matsunaga et al., 2021). Such approaches may predict key HAMPs or effectors subject to co-evolution with host plants. In contrast to piercing–sucking herbivores, few functional predictions have been reported for effector repertoires of chewing herbivores (Supplemental Data Set 1), presenting an opportunity to discover additional factors (see Rivera-Vega et al., 2018; Acevedo et al., 2018). Additionally, biochemical approaches could be productive ways to find additional HAMPs/effectors of piercing–sucking herbivores, especially small molecule fractions (Jonckheere et al., 2016).
Mechanisms for immune recognition of herbivory
Physicochemical and DAMP-mediated effects
Upon herbivory, plants are exposed to a combination of mechanical damage, HAMPs, and effectors. Wounding alone is sufficient for complex host responses mediated by both physical properties of wounded cells and the active release of DAMPs (Hander et al., 2019; Farmer et al., 2020; Li et al., 2020; Vega-Muñoz et al., 2020; Tanaka and Heil, 2021). Proteins within OS may also actively alter membrane properties. For example, OS-derived pore-forming proteins may drive measurable current across an artificial bilayer; however, neither the in vivo effects nor specific gene products encoding porin activities have been characterized (Guo et al., 2013). The physicochemical properties of HAMPs (e.g. amphiphilic properties of FACs) may also contribute to induced responses; however, the high degree of compound and host specificity of most HAMP-elicited responses is inconsistent with general effects (Schmelz et al., 2009; Grissett et al., 2020).
Beyond physical effects of wounding, DAMPs are perceived by host cell surface receptors to activate defense responses. DAMPs are either constitutively or inducibly released upon wounding. Constitutive DAMPs include purine nucleotides, amino acids such as extracellular glutamate, and cell wall-derived oligosaccharides. These are perceived by receptor kinases (RKs) with either lectin (LecRK) or epidermal growth factor (EGF)-like ectodomains (Brutus et al., 2010; Choi et al., 2014; Wang et al., 2017a, 2019). Besides potential recognition by RKs, glutamate may also function as a HAMP through direct regulation of calcium channels (Toyota et al., 2018), although its in vivo regulation of ion channel activity has not yet been demonstrated (see “Do HAMPs interact with wound-induced cellular signaling”).
A second, inducible layer of DAMP recognition involves processing of induced signaling molecules, termed phytocytokines, followed by amplification of damage-induced effects through highly specialized recognition systems. Wound-inducible propeptide genes encode precursor proteins, which can be processed to release bioactive small peptides. DAMPs with defined receptors include systemin and the Pep and SCOOP families of related peptides; these are detected by RKs with leucine-rich repeat (LRR) ectodomains, namely Pep 1 Receptor (PEPR), Systemin Receptor 1 (SYR1), and MIK1 (Yamaguchi et al., 2006; Lori et al., 2015; Wang et al., 2018; Rhodes et al., 2021). Herbivore-induced volatiles can also prime or induce responses through as yet undefined perception mechanisms (Erb, 2018; Meents et al., 2019; Meents and Mithöfer, 2020).
Receptors mediating HAMP recognition
In contrast to knowledge regarding specific DAMPs, there are comparatively few HAMP receptors (Reymond, 2021). We discuss examples of defined and putative HAMP receptors below, including methods and criteria for further exploring receptor functions.
The peptide HAMP Vu-In was recently shown to be recognized by Inceptin Receptor (INR), a member of the LRR receptor-like protein (RLP) family of PRRs (Steinbrenner et al., 2020). RLPs are distinguished from RKs by lack of a kinase domain (KD), but function through related protein complexes with signaling competent co-RKs (Ma et al., 2016; DeFalco and Zipfel, 2021). When expressed in tobacco, INR confers a set of Vu-In-inducible responses consistent with a genuine receptor role: (1) binding of Vu-In, measured as retention of labeled Vu-In to crude membrane preparations; (2) rapid Vu-In induced immune signaling, measured as peptide-inducible ROS burst within 10 min of treatment; and (3) increased resistance to herbivory by Spodoptera exigua. INR is solely present in cowpea, common bean (Phaseolus vulgaris), and mung bean (Vigna radiata) genomes, but not in soybean (Glycine max) and barrel medic (Medicago truncatula). This is consistent with restricted bioactivity of inceptin peptides within the Phaseolinae. INR is the first PRR with demonstrated binding, signaling, and defensive functions in response to a defined HAMP derived from a chewing herbivore (Steinbrenner et al., 2020).
Other cell surface receptors families besides LRR-RLPs can also mediate plant defense against herbivory. Uemura et al. used Fr∝, a bioactive fraction of Spodoptera littoralis OS, to conduct a reverse genetic analysis of LRR-RKs involved in Fr∝ response in soybean. The Herbivore Danger Signal-ASSOCIATED (HAK) genes GmHAK1 and GmHAK2 could enhance Fr∝ response when overexpressed in soybean, and the A. thaliana homolog AtHAK1 is required for Fr∝ response (Uemura et al., 2020). A similar reverse genetic analysis of a highly upregulated receptor gene identified a role for the PSKR homolog OsLRR-RLK1 (Os06g47650) in resistance to the striped stem borer (Hu et al., 2018). Importantly, silencing OsLRR-RLK1 almost completely abolished striped stem borer-induced phosphorylation of Mitogen-Activated Protein Kinases (MAPKs) within 15 min of infestation, indicating an early role in DAMP and/or HAMP perception (Hu et al., 2018). A related maize RLK, ZmFACS, was recently shown to mediate specific sensitivity to FACs, but direct receptor association with FAC HAMPs is not yet demonstrated (Poretsky et al., 2021). Identified originally as an egg extract-induced gene, A. thaliana LecRK-I.8 was shown to be required for full-strength PR1 transcript upregulation and cell death induced by egg extract (Gouhier-Darimont et al., 2019, 2013). Interestingly, natural variation in egg-induced cell death was associated with a related gene family member, LecRK-I.1. Knockout of either LecRK-I.1 or LecRK-I.8 reduces cell death symptoms, consistent with redundant roles in signaling downstream of egg recognition (Groux et al., 2021).
Other receptors involved in herbivore resistance have also not been paired with known ligands. Mi1.2 is an NLR that mediates resistance to aphids and diverse piercing–sucking herbivores (Rossi et al., 1998; Casteel et al., 2006). Resistance genes for brown planthopper (N. lugens) include those encoding the NLRs Bph14 and Bph26, and the Bph3 locus encoding a set of three LecRKs (Du et al., 2009; Tamura et al., 2014; Liu et al., 2015). NaLecRK1 is transcriptionally induced by Manduca sexta OSs, and functions as a negative regulator of many herbivore-inducible outputs (Gilardoni et al., 2011). Despite the role of these receptors in resistance to herbivores, it is not yet clear if any of the RKs, NLRs, or LecRKs described directly recognize a HAMP or effector, and each may instead act downstream of wounding and/or immune recognition.
With many HAMPs and effectors now identified (Supplemental Data Set 1), additional examples of recognition mechanisms are needed to fully understand diverse plant–herbivore interactions. To identify candidate receptors, the field has traditionally relied on highly HAMP-induced receptor transcripts, but this approach may simply identify abundant or DAMP-responsive genes, rather than “true” HAMP-binding PRRs (Domazakis et al., 2020). An alternative strategy leverages the high degree of natural variation in immune receptors within and between species to identify germplasm for further comparison, an approach used to successfully identify INR (Steinbrenner et al., 2020). Forward genetics using responsive and unresponsive germplasm is the classical basis for mapping resistance genes for pathogens, and increasingly powerful comparative genomic approaches such as genome-wide association studies can leverage phenotypic response data without the need for a parental cross (Schultink and Steinbrenner, 2021). Variation in HAMP recognition (see “Recognition specificity of HAMPs/effectors”) is thus a powerful tool for generating candidate genes involved in recognition. In summary, a combination of approaches will be needed to pair HAMPs with PRRs, but omics-enabled approaches will facilitate this work in diverse plant models.
Recognition specificity of HAMPs/effectors
Plant sensitivity to a specific HAMP varies across populations and species, consistent with selective pressure imposed by diverse biotic environments as well as the highly variable gene families that likely mediate most HAMP recognition (Jamieson et al., 2018; Van de Weyer et al., 2019; Steinbrenner, 2020). Early HAMP characterization efforts noted that responses could be idiosyncratic (Spiteller et al., 2001; Schmelz et al., 2006). Studies that test multiple plant species are still rare, although they can be a powerful approach combined with species phylogenetics to discover new plant immune receptors. In an analysis of tested plant species for HAMP/effector characterization, the majority of studies used at most a few related plant species (Supplemental Data Set 1). Below we discuss FAC, GOX, and Vu-In, for which broad phylogenetic sampling of host responses has been reported. For other listed HAMPs/effectors in Supplemental Data Set 1, it is possible that (1) recognition is species-specific or (2) broad testing for multiple responses in diverse plant species is still needed.
HAMP/effector presence across chewing herbivores and the spectra of plants responding to them
GOX has been found to be an ubiquitous effector in chewing herbivore OS, but can also act as a HAMP on select members of the Solanaceae. Eichenseer et al. (2010) examined the labial gland GOX activity in 88 species of Lepidoptera and Hymenoptera and revealed that highly polyphagous species displayed high levels of GOX activity relative to species with a limited host range, possibly as an adaptation to broad host. GOX is an effector of plant immunity in N. attenuata, N. tabacum, and M. truncatula (Musser et al., 2002; Bede et al., 2006; Diezel et al., 2009), but instead elicits plant defenses in other species such as tomato (Solanum lycopersicum) and habanero pepper (Capsicum chinense), consistent with effector-triggered immunity (Tian et al., 2012; Lin et al., 2020). Solanum and Capsicum are highly related genera and may share a mechanism for recognition of GOX; further sampling within the Solanaceae is needed to pinpoint a potential gain of GOX recognition. The majority of GOX studies focused on Solanaceae species (Lin et al., 2020). An exception is Louis et al. (2013) who did not observe differential expression of a marker gene maize protease inhibitor (MPI) in maize upon application of GOX. Hence, the generality of suppression and elicitation of the plant immune defense by GOX is still unknown.
In contrast to GOX, FACs are bioactive on diverse plants, but chemical distinctions among the lipid conjugates affect host plant responses. Yoshinaga et al. (2010) detected FACs in 19 of 29 tested lepidopteran species. Within Lepidoptera, glutamine and glutamic acid conjugates can be synthesized by early diverging lineages, while mixtures including hydroxylated fatty acids were solely found in a monophyletic subgroup of the Macroheterocera (Mitter et al., 2017). The FAC volicitin (17OH-C18:3-Gln) triggers plant immune responses in a relatively wide range of plants including maize, soybean, and eggplant (Solanum melongena; Schmelz et al., 2009). An alternatively hydroxylated analog of volicitin (18OH-C18:3-Gln) also elicited a wide range of plant species including maize, soybean, and N. tabacum Yoshinaga et al. (2014). Another study focused instead on six closely related Nicotiana species (Xu et al., 2015), and observed differential responses to N-linolenoyl-glutamic acid (C18:3-Glu). Differential responses to another FAC N-linolenoyl-l-glutamine (C18:3-Gln) were also observed between five Nicotiana species by Grissett et al. (2020). Additionally, they reported C18:3-Glu and C18:3-Gln responses for bell pepper (Capsicum annuum), S. melongena, and petunia (Petunia hybrida), while potato (Solanum tuberosum), S. lycopersicum, and six wild tomato accessions were unable to respond. Intriguingly, C. annuum responded to C18:3-Gln and C18:3-Glu but not to volicitin (Grissett et al., 2020). In conclusion, FAC response is most consistent with a dynamic ancestral trait present in both monocots and eudicots, which most likely was lost repeatedly during the evolution of the Solanaceae and other plant families (e.g. Fabaceae; Schmelz et al., 2009). Differential responses to individual FACs depending on hydroxylation, amino acid identity, and fatty acid identity are also apparent (Alborn et al., 2003; Yoshinaga et al., 2014; Block et al., 2018; Grissett et al., 2020), strengthening the hypothesis that plants might have evolved receptors that detect specific FAC compounds (Block et al., 2018).
As a final example of a HAMP tested on multiple hosts, Vu-In is a specific HAMP of the Phaseolinae (subtribe of Fabaceae) likely due to the evolution of a cognate host receptor, INR. Inceptin related peptides including Vu-In were observed in all tested Lepidoptera (nine species; Schmelz et al., 2012). Vu-In activates plant defenses in the Phaseolinae; V. unguiculata, V. radiata, and P. vulgaris, but not in soybean (Schmelz et al., 2009; Steinbrenner et al., 2020). The Vu-In receptor INR was discovered in V. unguiculata and INR orthologs with >90% amino acid similarity were identified and validated for V. radiata and P. vulgaris. In contrast, soybean receptor homologs only share 73%–76% amino acid similarity with INR and show no response to Vu-In. The discovery of the Vu-In receptor INR will allow analysis of a defined HAMP response in the comparative genomic context of a cognate HAMP receptor. For example, it will be interesting to analyze the genomic origin of INR homologs in the genomes of early-branching Fabaceae. The INR case study also suggests a general strategy for receptor identification, using a HAMP-responsive clade of plants to reveal candidate receptors through comparative genomic analysis. Such an approach will require careful phylogenetic analysis of complex PRR and NLR gene families (Shiu and Bleecker, 2001; Jamieson et al., 2018; Prigozhin and Krasileva, 2021).
Most recently, Griese et al. (2021) studied hypersensitive response (HR)-like necrosis upon exposure to egg wash of nine different Pieridae species in an attempt to identify the phylogenetic distribution of this egg-killing trait within the order of the Brassicales (31 species). Their findings show a strong clade-specific induction mainly in species of the Brassiceae tribe elicited by Pierinae butterflies specialized on the Brassicaceae family. This suggests that the host ability to recognize egg wash to activate HR-like may be part of an arms-race between Pierinae eggs and Brassicaceae species. However, egg wash is potentially a complex mixture with multiple HAMPs, and the perception mechanisms involved in HR-like necrosis are unknown.
HAMP/effector presence across piercing-sucking herbivores and the spectra of plants responding to them
HAMPs/effectors from piercing-sucking herbivores have come to outnumber those of chewing herbivores in the past decade. The majority of the HAMPs/effectors were identified in aphids and planthoppers, but more recently several whitefly effectors were also identified (Supplemental Data Set 1). The piercing-sucking herbivore with the most identified HAMPs/effectors is the important polyphagous agricultural and model pest, the green peach aphid Myzus persicae (14 effectors). Due to the polyphagous nature of many piercing-sucking herbivores, candidate elicitors can more often be characterized in strong genetic model systems. For example, M. persicae accepts the model plant N. benthamiana as a host, facilitating genetic studies of factors controlling herbivore performance and fecundity.
Omics approaches have not only strongly facilitated the identification of candidate HAMPs/effectors in piercing–sucking herbivores (see “HAMPs and effectors in plant–herbivore interactions”), but also the comparison of effector repertoires between species (Pitino and Hogenhout, 2013; Chaudhary et al., 2015; Thorpe et al., 2016; Drurey et al., 2019; Huang et al., 2020). These analyses revealed a core set of conserved effector genes as well as more sequence-diverse and potentially species-specific effectors. For example, Drurey et al. (2019) identified Mp10 homologs in diverse plant-sucking insect species and earlier diverged species, and showed that five homologs were capable of suppressing an flg22 (PAMP) triggered ROS-burst. Combined with a species phylogeny, their data suggests that an ancestral Mp10-like sequence, which suppresses ROS-bursts, was acquired before the divergence of plant-sucking insect species. In addition to Mp10, other sets of orthologous effectors from piercing–sucking herbivores have been functionally validated: Armet, MpC002-ApC002-RpC002, Mp58-Me10-Rp58, Mp1-Rp1, Tu28-Te28, and Tu84-Te84 (Supplemental Data Set 1).
Most studies of piercing–sucking HAMPs/effectors only test a single host plant species. Response has been described in three species only for a single HAMP/effector, Me47 (Kettles and Kaloshian, 2016). In contrast to other studies using only the N. benthamiana system to demonstrate increased M. persicae performance, they also delivered Me47 to a natural host species of M. euphorbiae (S. lycopersicum), and observed enhanced M. euphorbiae fecundity. In contrast, when Me47 was delivered to A. thaliana, this resulted in a reduced M. persicae performance.
Effectors can be HAMPs on specific host plants
A given molecule can either suppress or elicit defenses depending on the host plant, and thus the entire category of effectors (i.e. immune suppressors) can be considered as candidate HAMPs on a different host variety or species. Besides GOX (discussed above; Tian et al., 2012; Lin et al., 2020), other molecules associated with chewing herbivores, such as phospholipase C and frass-associated chitinases, can either elicit or suppress specific defensive outputs (Ray et al., 2015; Acevedo et al., 2018), but equivalent early signaling responses should be analyzed across species to draw strong conclusions (see Box 1). Molecules delivered by several piercing–sucking herbivores can also have a host-dependent eliciting or suppressing function. The aphid effector Me47 induces defensive transcripts and aphid resistance in S. lycopersicum but not in A. thaliana (Kettles and Kaloshian, 2016). Similarly, although Me10 increased M. persicae fecundity on N. benthamiana (Atamian et al., 2013), the Me10 homologs Mp58 and Rp58 decreased M. persicae fecundity on N. benthamiana (Elzinga et al., 2014; Escudero-Martinez et al., 2020). Finally, Atamian et al. (2013) observed differential response of the M. euphorbiae effector Me23, which increased fecundity on N. benthamiana, but did not significantly change fecundity on the natural host S. lycopersicum. Hence, these studies highlight that a candidate HAMP/effector should ideally be tested on the natural host species, as the resulting response can be influenced by the choice of plant species used for validation.
Inceptin peptides provide another example of a molecule displaying host-dependent toggling between elicitation and suppression. The legume specialist Anticarsia gemmatalis rapidly truncates bioactive 11–13 amino acid length inceptin peptides to 10-amino acid, inactive Vu-In-A via proteolysis of the C-terminal alanine (Schmelz et al., 2012). Vu-In-A suppresses responses on the cowpea variety CB5, but instead weakly elicits ethylene on select lines of cowpea as revealed by screening a diversity panel of 364 varieties (Steinbrenner et al., 2020). Weak HAMP activity was associated with a single amino acid substitution in INR, which may sensitize the receptor against diverse inceptin ligands. Notably, both Vu-In and Vu-In-A bind cowpea INR (INR-Vu). It is possible that species besides cowpea have evolved to recognize Vu-In-A as a potent HAMP, possibly as an indicator of a specialist threat.
Ultimately, determining whether plant responses to any HAMP/effector are variable or fixed will answer whether forward or comparative genetics may lead toward specific mechanisms. Screening diverse germplasm within and between species should be a research priority. Thorough characterization of the plant response spectra of understudied HAMPs/effectors could inform us if a certain HAMP/effector perception or manipulation mechanism is species-specific or well conserved throughout the plant kingdom. Response variation within a few accessions could motivate further genetic analysis to identify candidate receptors. Once a mechanism is known, response data of closely related species in combination with genomic data can deepen our understanding of the evolution of plant immune receptors and their ecological functions.
Herbivore modulation of the wound response
In plant–bacterial pathosystems, cell damage and wound responses play a key role in mediating local immune response to PAMPs (Zhou et al., 2020; Manes et al., 2021). In plant–herbivore interactions, damage and the associated release of DAMPs also coincides with the presence of HAMPs at the feeding site; however, we do not know the extent to which these are required to mount strong herbivore-specific immune responses. The discovery of defined DAMP and HAMP receptors has provided a toolbox to address their relative contributions to the defensive output during a plant–herbivore interaction. Below we summarize roles of DAMPs in plant–herbivore interactions, and current methods to address the role of mechanical damage in the antiherbivore response.
Specific DAMPs are required for antiherbivore defenses
Only a few key DAMPs have been studied in the context of herbivory, mainly using tomato as a model for wound-inducible DAMP production. Tomato systemin was the first species-specific endogenous signal discovered to be released upon damage to systemically induce the wound and herbivory-related protease inhibitor (PIN) transcripts (Green and Ryan, 1972; Pearce et al., 1991; Coppola et al., 2019). Silencing of the prosystemin gene leads to increased susceptibility to M. sexta caterpillars, associated with lower PIN accumulation (Orozco-Cardenas et al., 1993). Similarly, tomato introgression lines lacking a functional SYR1 and SYR2 have lower PIN1 expression, and S. littoralis caterpillars feeding on them gain more weight than those feeding on WT plants (Wang et al., 2018). A variety of DAMP pathways associated with the antiherbivore response have been described in addition to systemin. For instance, hydroxyproline-rich systemins (HypSys) are found in tomato, other Solanaceae, and sweet potato (Ipomoea batatas). Like systemin, HypSys are induced by wounding and methyl jasmonate (MeJA), trigger media alkalinization and systemically induce PINs and sporamins (Pearce and Ryan, 2003; Chen et al., 2008; Pearce et al., 2009). CAPE1, another tomato-specific signal, was identified in a peptidomics assay with plants treated by wounding or combined wounding and MeJA application. PIN1 was also induced upon CAPE1 treatment, and S. litura larvae fed on leaves pretreated with CAPE1 were smaller than caterpillars fed on water-treated plants (Chen et al., 2014).
Unlike systemin, the PEP–PEPR system appears to be a hallmark of the wound response conserved throughout Angiosperms. Orthologs of the PROPEP gene family have been identified in several members of the Brassicaceae, Solanaceae, Fabaceae, and Poaceae (Huffaker et al., 2006; Lori et al., 2015; Poretsky et al., 2020). In A. thaliana, direct application of S. littoralis OS and feeding by S. littoralis, P. brassicae, Phaedon cochlearieae, and Thrips tabaci strongly activated the PEPR1 and PROPEP3 promoters compared to wounding alone (Klauser et al., 2015). Furthermore, caterpillars that were fed pepr1pepr2 plants gained more weight than those fed WT plants. In rice (Oryza sativa) (Shinya et al., 2018), application of OsPEP3 together with OS from the maize caterpillar (Mythimna loreyi) on cultured cells enhanced the production of ROS, p-coumaroylputrescine and momilactone compared to OS or peptide alone. In maize (Huffaker et al., 2013), treatment with low concentrations of ZmPEP3 produced higher amounts of VOCs, JA, and ethylene and attracted more parasitoids than the water control, and clustered regularly interspaced short palindromic repeats (CRISPR)-based mutants of ZmPEPR1 compromised ZmPep3-induced reduction in S. exigua larval weight gain (Huffaker et al., 2013, Poretsky et al., 2020). While this evidence collectively supports a requirement for DAMP signaling in antiherbivore responses, it is unknown if specific HAMPs induce the genes encoding precursor proteins as mechanisms to amplify the wound response, or if the precursor proteins can be targets of herbivore-derived effectors for downregulation.
A role for constitutive DAMPs and their receptors in plant–herbivore interaction is also an open question. For instance, eATP is released upon mechanical damage or changes in mechanical pressure to trigger JA and ethylene-dependent gene expression (Tripathi et al., 2018). Furthermore, high concentrations of eATP on common bean leaves induced the production of extrafloral nectar (Duran-Flores and Heil, 2014), a known indirect defense against herbivores, possibly mediated by known purinoreceptor gene families (Choi et al., 2014). However, there is a lack of evidence that directly links eATP wound-associated signaling to resistance against herbivores.
Combined effects of wounding and herbivore-derived molecules
While it is likely that a mix of other characterized DAMPs is released during herbivore feeding, it is unknown how they interplay with HAMPs/effectors and their signaling cascades. A hypothesis is that DAMPs establish the baseline of a general wound response, which is enhanced or suppressed by HAMPs and effectors. To address the combined effects of wounding and HAMPs/effectors, studies have relied on ablation, genetics, and/or robotic approaches. Future studies that combine receptor mutants to simultaneously remove DAMP and/or HAMP perception will also illuminate combined effects.
Ablation of salivary glands and other secretory structures outside of the mouth can effectively be used to measure herbivore damage (Musser et al., 2002, 2006, 2012). Helicoverpa zea caterpillars that fed on tobacco plants previously attacked by caterpillars with cauterized spinnerets and ablated labial salivary glands had a lower survival rate than those fed on plants previously attacked by intact caterpillars. This indicated that saliva has a role in counteracting immunity triggered by the damage or HAMPs. This hypothesis was further supported by a higher production of nicotine and H2O2 on tobacco and tomato leaves, respectively. The increased responses were partially attributed to the absence of GOX in accordance with its suppressing activity.
Genetic silencing or knockout of HAMP/effector components is an emerging method to understand plant responses in the absence of a specific HAMP or effector. Genetic evidence for the role of GOX has been provided by Lin et al. (2021). A gox H. zea knockout mutant line was generated with the ALT-R Cas9-HF and crRNA system and subsequently used to determine the effect of GOX on VOC emission in tomato and soybean. The authors found that different VOCs can be induced or suppressed by GOX, emphasizing that herbivore-derived molecules can have dual roles (see “HAMPs and effectors in plant–herbivore interactions” and “Recognition specificity of HAMPs/effectors”). Furthermore, Consales et al. (2012) demonstrated that the suppression of herbivore-related defenses by OS from S. littoralis and P. brassicae is independent of FACs and GOX. Briefly, caterpillars from both species were fed on A. thaliana fatty-acid desaturase triple mutant fad3 fad7 fad8 plants, which lack C18:3 fatty acids. This rendered FAC-depleted OS, which downregulated the expression of the transcription factor EFR/AP2, indicating that the suppression of defenses was not FAC-dependent. This downregulation was also partially independent of GOX activity, as it was detected for S. littoralis OS but not P. brassicae.
Robotics applications can also recapitulate herbivore chewing patterns and allow for controlled release of OS and HAMPs. MecWorm (Bricchi et al., 2010) and SpitWorm (Li et al., 2019a) can be used to simulate herbivory in the lab and compare to mechanical damage alone. For example, herbivore damage caused by S. littoralis on P. lunatus leaves led to greater membrane depolarization and intracellular calcium accumulation than mechanical damage caused by MecWorm or a pattern wheel. Conversely, equal accumulation of H2O2 and NO was detected on leaves exposed to herbivory and to MecWorm, but not to damage with the pattern wheel. This suggests that repeated mechanical damage is responsible to some extent for the herbivore-specific response. Further exploration of herbivore-specific responses has been addressed using SpitWorm, a device optimized to deliver OS quantities equivalent to S. littoralis caterpillars feeding. Relative amounts of VOCs from P. lunatus were quantified upon treatment with SpitWorm, MecWorm, or S. littoralis caterpillars. Only 4 out of 38 compounds differed when comparing SpitWorm and larvae. In contrast, 23 compounds were more abundant in MecWorm treated samples compared to S. littoralis or SpitWorm. These two studies emphasize the uniqueness of early signaling steps in response to herbivory and the suppression of the wound response by herbivore-derived molecules as a mechanism to overcome immunity. While robotic simulation of herbivory best mimics live herbivory, other models involving repeated wounding (Lin and Felton, 2020) may also be sufficient to recapitulate herbivore-induced responses.
Finally, transcriptomic comparisons of wounding to live herbivory, OS, or HAMPs also support a role for herbivore modulation of the wound response (Reymond et al., 2000; De Vos et al., 2005; Walley et al., 2007; Gilardoni et al., 2011; Appel et al., 2014; Zhou et al., 2016; Steinbrenner et al., 2021). Comparisons of wounding versus wounding + OS generally identify hundreds of differentially expressed genes. Studies that have narrowed these effects by using specific HAMPs include an analysis of FAC response in tobacco species (Zhou et al., 2016) and Vu-In response in cowpea (Steinbrenner et al., 2021). FAC response specifically leads to the upregulation of a co-expressed module termed M4, which contains genes related to JA biosynthesis and signaling. Vu-In treatment leads to similar upregulation of these gene families at either early (1 h) or late (6 h) timepoints. Interestingly, inceptin was also shown to partially reverse the effects of wounding for distinct sets of genes. These two studies provide a global overview of the modulation of the wound response by specific HAMPs, and are an excellent resource to identify and compare HAMP-specific defensive outputs.
Do HAMPs interact with wound-induced cellular signaling?
At the cellular level, many signaling factors are genetically required for effective defense against various herbivores (Wu et al., 2007; Yang et al., 2012; Lei et al., 2014). A list of specific genes where knockout, knockdown, or overexpression affects herbivore performance was recently reviewed (Erb and Reymond, 2019). However, it is often unclear if these factors mediate HAMP responses themselves, or instead facilitate general wound responses. Responses to HAMPs are also thought to operate through modulation of wound-induced responses (Erb and Reymond, 2019); however, molecular mechanisms by which defined HAMPs interact with the wound response are barely explored. Thus, a key challenge for the field is to link initial HAMP recognition to specific signaling targets in the local leaf. In this section, we review early steps in wound- and herbivore-induced signaling cascades. We also refer to excellent reviews with further detail on downstream responses, including induced JA biosynthesis, transcriptional reprogramming, and systemic propagation of signals (Howe et al., 2018; Wasternack and Feussner, 2018; Farmer et al., 2020). Specific signaling factors described below may serve as hubs to integrate HAMP recognition into the wound response (Figure 2).
Figure 2.
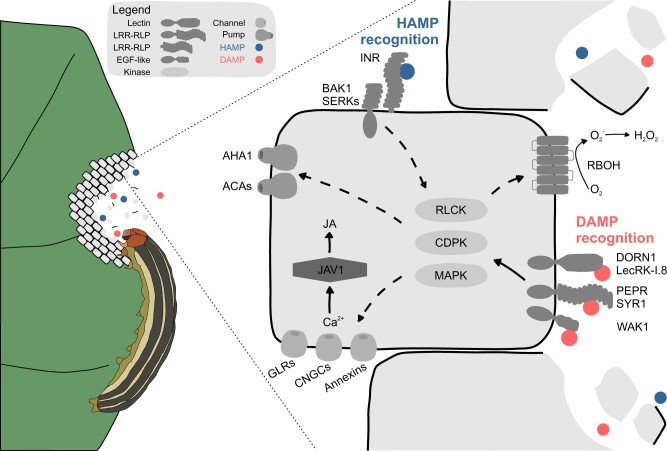
Potential points of signaling integration between DAMP and HAMP responses. Chewing herbivores lead to cellular damage and HAMP exposure to plant cells. HAMPs can be perceived by PRRs such as the INR LRR-RLP (top side of cell) and likely other receptor families, while DAMPs are perceived by defined receptors encoding various ectodomains (right side of cell). Known factors that are genetically linked to defense against herbivory are connected with arrows indicating potential signaling cascades. Other pictured signaling factors affect defense against herbivory but have not been specifically linked to recognition of HAMPs. Solid arrows indicate known activation of signaling factors. Hypothesized points of regulation are shown as dotted lines. See text for details of known receptor and signaling factors. Ultimately, calcium influx and jasmonate biosynthesis are modulated by the combined effect of wounding and HAMP recognition. WAK1, Wall-Associated Kinase 1; DORN1, Does Not Respond to Nucleotides 1; JAV1, Jasmonate Associated VQ motif.
Several studies have linked signaling machinery for PRRs to responses activated by HAMPs. Immediately downstream of recognition by putative LRR-containing HAMP receptors are co-RKs in the Somatic Embryogenesis RK family including BRI1-Associated RK 1 (BAK1). In N. attenuata, silencing the homolog NaBAK1 partially reduces OS-induced responses, strongly suggesting that LRR–RK/RLP receptors mediate responses to OS components such as FACs (Yang et al., 2011). Recently, it was shown that Vu-In induces its receptor INR to associate with BAK1 and other SERKs (Steinbrenner et al., 2020). Similarly, BAK1 is required for plant responses to M. persicae extract and the endosymbiont-derived GroEL elicitor (Chaudhary et al., 2014; Prince et al., 2014). Downstream of receptor activation, kinase cascades have also been linked to specific HAMP responses. For example, responses to S. littoralis OS fraction Fr∝ require the kinase PBL27, a member of the receptor-like cytoplasmic kinase family (Uemura et al., 2020). The N. attenuata kinases calcium-dependent cytoplasmic kinase 4 (CDPK4) and CDPK5 mediate responses to M. sexta OS (Yang et al., 2012). FAC treatment rapidly induces MAPK phosphorylation in N. attenuata and related Solanaceae, and silencing MAPK genes reduces responses to total OS (Wu et al., 2007; Grissett et al., 2020). The lipase-like protein PHYTOALEXIN DEFICIENT4, a well-known component of both PRR and NLR signaling (Pruitt et al., 2021), is required for responses to aphids (Louis et al., 2012). Finally, targets of aphid and whitefly effectors include TFs, 14-3-3 proteins, and other defined signaling factors important for piercing–sucking herbivore fecundity (Rodriguez et al., 2017; Chaudhary et al., 2019; Xu et al., 2019; Wang et al., 2021). In summary, HAMP recognition is likely mediated by typical signaling cascades downstream of PRRs and NLR-type receptors. Directly linking receptors to specific signaling targets is a research priority.
HAMP-induced signaling may also occur through modulation of factors in the wound response pathway. Spatial propagation of ion flux is a defining feature of the plant wound response; within seconds of mechanical wounding, membrane depolarization of 30–80 mV occurs alongside cytosolic Ca2+ influx (Demidchik et al., 2018; Farmer et al., 2020). Factors involved in these events have been elucidated using genetic approaches. For example, Mousavi et al. (2013) used a reverse genetic approach to identify a critical role for glutamate-receptor-like channels (GLRs) 3.3 and 3.6, which function redundantly for systemic propagation of electrical signals. Follow-up work has specifically isolated additional channel proteins involved in the propagation of wound responses, including MSL10, Cyclic Nucleotide Gated Channel 19 (CNGC19), ANNEXIN1, and TPC1 (Bonaventure et al., 2007; Kiep et al., 2015; Meena et al., 2019; Malabarba et al., 2021; Moe-Lange et al., 2021). The proton pump Autoinhibited H+-ATPase 1 (AHA1) is involved in membrane repolarization following wounding, and thus functions as a negative regulator of systemic signaling (Kumari et al., 2019). Channel family members have varying tissue-specific expression with respect to the vasculature, and can be regulated by calmodulins (Vadassery et al., 2012; Scholz et al., 2014; Nguyen et al., 2018). Both cell-specific expression and regulation likely determine which specific channel-encoding genes are critical for local propagation of the wound response. For example, CNGC19 is required for wound- or DAMP-induced signal propagation within the local leaf, but local responses are unaffected in tpc1-2, msl10, or glr3.3/glr3.6 mutants (Salvador-Recatalà et al., 2014; Kiep et al., 2015; Meena et al., 2019; Moe-Lange et al., 2021). Autoinhibited Ca2+-ATPase 8 (ACA8) also contributes to calcium homeostasis within the locally wounded leaf (Costa et al., 2017). A role for channels and pumps in HAMP response has not yet been studied, but channels and their associated regulators represent potential targets for HAMP- or effector-induced modulation (Figure 2).
Answers to several outstanding questions will help connect HAMPs to defined wound-induced signaling.
Which cells are HAMP responsive? Spatial connections between wound and HAMP perception are not well defined but are a potential point of interaction. For example, HAMP recognition in leaf mesophyll cells may breach a threshold for expanding a wound response throughout the local leaf. Single-cell datasets will also help to correlate expression of receptor-mediated signaling pathways with potential channel targets (Zhang et al., 2021). Feedback between adjacent cell types, for example through receptor upregulation upon damage (Zhou et al., 2020), may also play a role in full strength responses in the leaf. In general, model systems are needed in which both precise wounding and HAMP exposure in leaf cell populations can be manipulated, analogous to precise single-cell models for root damage facilitating recent breakthroughs in DAMP recognition mechanisms (Hander et al., 2019; Marhavý et al., 2019).
Are wound-responsive factors activated or modulated by HAMPs? As an example of a potential mechanism for HAMP action, ion flux may be directly regulated by DAMP treatment. Treatment of Pep1 DAMP induces dephosphorylation of tomato AHA1 (Haj Ahmad et al., 2019). While OS have been shown to interfere with specific channels or wound-induced signal propagation (Kiep et al., 2015; Camoni et al., 2018), specific effector molecules responsible for suppression have not been identified. Channel activators and ion substrate specificities are not well understood in planta; this knowledge will help to understand precise events following herbivory. Importantly, most mechanistic work to study wounding is performed in A. thaliana, a system where well-defined small molecule HAMPs and their receptors are currently lacking. Model plants that combine a strong reverse genetic toolkit with defined HAMP responses and receptors are badly needed to make further progress.
Do HAMPs act downstream of wound-induced ion fluxes? HAMPs may also interact with wounding via downstream signaling factors. For example, the NADPH oxidase respiratory burst oxidase homolog D (RBOHD) is a well-characterized target of pathogen-induced RK/RLP signaling, and its rapid phosphorylation is required for induced ROS production (DeFalco and Zipfel, 2021). Interestingly, RBOHD has contrasting functions as either a positive or negative regulator of defense against herbivores in different host plant species (Wu et al., 2013; Block et al., 2018), and it will be interesting to investigate signaling tradeoffs between herbivores and pathogens mediated by ROS homeostasis. Signal integration through other wound response regulators, such as the VQ-motif containing JAV proteins (Yan et al., 2018), could also potentially modulate sensitivity after HAMP recognition (Figure 2).
Conclusions
Induced responses to herbivory are now an accepted facet of the plant immune system (Karban and Baldwin, 2007; Howe and Jander, 2008). Just as deep mechanistic understanding of plant–microbe interactions has informed the use of immune receptors as traits (van Esse et al., 2020), we expect mechanisms for sensing insect herbivores to be equally valuable. Importantly, insect resistance traits can complement fragile biotechnological solutions based on transgenic plants expressing insecticidal proteins (Ortega et al., 2016; Tabashnik and Carrière, 2017). Identification of HAMP recognition mechanisms in diverse plant species will reveal principles for their effective deployment, complementing existing agricultural applications of chemical ecology (Turlings and Erb, 2018). While significant research questions remain, genome-enabled comparative approaches are beginning to illuminate the interface between herbivores and the plant immune system.
Box 1: Markers of HAMP/effector response.
Evolutionary analysis of HAMP/effector responses requires measuring plant immune responses in multiple species/accessions. Ideally, a marker/assay should: (1) be conserved in diverse plant accessions and species and (2) result in a rapid response upon treatment. Markers for herbivore activation/manipulation of plant immunity that fulfill these criteria are scarce. Here we analyze commonly measured defense outputs in HAMP/effector characterization (Supplemental Data Set 1).
Genes encoding conserved anti-herbivore defense responses are commonly used, but are themselves highly variable across species. For example, MPI in maize (Zm00001d011080) and the proteinase inhibitor 2 (PIN2) in tomato (Solyc11g020960), are commonly measured, but the markers are not orthologous but rather part of large gene families, and may respond differently to various treatments. Thus, marker genes are better suited for studies using a single species.
Measuring volatiles is another prevalent method to measure plant response (22 studies). Complex volatiles such as terpenoids have been used, but simple measurement of the gaseous hormone ethylene is more established in recent studies. Ethylene is a highly conserved marker that shows rapid elicitation (von Dahl and Baldwin, 2007; Schmelz et al., 2009). In contrast, more complex volatiles will be produced in a time- and species-dependent fashion. Similarly, the phytohormone JA is a conserved herbivore marker that shows rapid elicitation across multiple species (Schmelz et al., 2009). Due to these properties, and straightforward GC–MS quantification, JA is also a frequent marker in plant immunity (19 studies). Additionally, with the growing understanding of the key role of H2O2 in the regulation of multiple stress responses (Baxter et al., 2014), staining and quantification of H2O2 are more frequently used in the later years as a marker for HAMP/effector responses (six studies since 2017).
Alternatively, one can quantify the resulting defense compounds or phenotypes. However, most of these markers tend to have a restriction. For example, the measurement of trichome induction as a marker has been limited to tomato, and nicotine quantification is restricted to N. tabacum. In contrast, callose deposition has been observed as a resulting phenotype in multiple plant species such as Brassica spp., tomato, A. thaliana, rice, and Nicotiana spp., but its use has been restricted to piercing–sucking herbivores (seven studies). Similarly, trypsin protease inhibitors have been used as a plant immunity maker in multiple diverse host plants upon chewing herbivory (tomato, maize, Nicotiana spp., M. truncatula, C. chinense, and Cynodon dactylon) (six studies).
The ultimate proof of a functional immune response is a quantifiable effect on the herbivore itself. Herbivore performance (e.g. weight gain/loss) is often measured (10 studies), as well as herbivore fecundity for piercing–sucking herbivores (10 studies) and even the attraction of egg parasitoids. Nevertheless, herbivore assays can be challenging due to a plethora of reasons such as complex experimental setups and host range of available herbivores. These issues are more serious for studies that involve multiple plant species.
In summary, the choice of assay for HAMP/effector recognition can greatly affect interpretations. Researchers comparing HAMP/effector responses should focus on a unified set of assays that measure early and conserved signaling outputs such as MAPK activation, ROS production, and ethylene burst. Since these are broadly conserved across plant families as defense outputs, their routine use will facilitate phylogenetic analyses of HAMP/effector response. Preferably, pure HAMPs/effectors are used to test these responses. Finally, they should be aware of potential response variation between closely related species, accessions within the same species, and even growth conditions (Schmelz, 2009). Hence, claiming that a species “does not respond” requires rigorous testing.
Supplemental data
The following material is available in the online version of this article.
Supplemental Data Set 1 . Overview of known HAMPs and effectors from chewing and piercing–sucking herbivores.
Acknowledgement and Funding
We thank members of the Huffaker, Schmelz, Imaizumi, and Nemhauser labs for conversation and feedback. S.S., N.G.P., and A.D.S. are supported by start-up funding from the University of Washington. N.G.P. is partially supported by the UW Royalty Research Fund grant #A161929 and the Hereensperger and Walter and Margaret Sargent Awards. S.S. is supported as a Belgian American Educational Foundation postdoctoral fellow. A.D.S. is a Distinguished Investigator of the Washington Research Foundation.
Conflict of interest statement. None declared.
Supplementary Material
Contributor Information
Simon Snoeck, Department of Biology, University of Washington, Seattle, Washington, USA.
Natalia Guayazán-Palacios, Department of Biology, University of Washington, Seattle, Washington, USA.
Adam D Steinbrenner, Department of Biology, University of Washington, Seattle, Washington, USA.
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Adam D. Steinbrenner (astein10@uw.edu).
References
- Acevedo FE, Peiffer M, Ray S, Meagher R, Luthe DS, Felton GW (2018) Intraspecific differences in plant defense induction by fall armyworm strains. New Phytol 218: 310–321 [DOI] [PubMed] [Google Scholar]
- Acevedo FE, Peiffer M, Tan CW, Stanley BA, Stanley A, Wang J, Jones AG, Hoover K, Rosa C, Luthe D, et al. (2017) Fall armyworm-associated gut bacteria modulate plant defense responses. Mol Plant Microbe Interact 30: 127–137 [DOI] [PubMed] [Google Scholar]
- Acevedo FE, Rivera-Vega LJ, Chung SH, Ray S, Felton GW (2015) Cues from chewing insects - the intersection of DAMPs, HAMPs, MAMPs and effectors. Curr Opin Plant Biol 26: 80–86 [DOI] [PubMed] [Google Scholar]
- Aggarwal R, Subramanyam S, Zhao C, Chen MS, Harris MO, Stuart JJ (2014) Avirulence effector discovery in a plant galling and plant parasitic arthropod, the Hessian fly (Mayetiola destructor). PLoS One 9: e100958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert I, Hua C, Nürnberger T, Pruitt RN, Zhang L (2020) Surface sensor systems in plant immunity. Plant Physiol 182: 1582–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alborn HT, Brennan MM, Tumlinson JH (2003) Differential activity and degradation of plant volatile elicitors in regurgitant of tobacco hornworm (Manduca sexta) larvae. J Chem Ecol 29: 1357–1372 [DOI] [PubMed] [Google Scholar]
- Alborn HT, Turlings TCJ, Jones TH, Stenhagen G, Loughrin JH, Tumlinson JH (1997) An elicitor of plant volatiles from beet armyworm oral secretion. Science 276: 945–949 [Google Scholar]
- Appel HM, Fescemyer H, Ehlting J, Weston D, Rehrig E, Joshi T, Xu D, Bohlmann J, Schultz J (2014) Transcriptional responses of Arabidopsis thaliana to chewing and sucking insect herbivores. Front Plant Sci 5: 565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atamian HS, Chaudhary R, Cin VD, Bao E, Girke T, Kaloshian I (2013) In planta expression or delivery of potato aphid Macrosiphum euphorbiae effectors Me10 and Me23 enhances aphid fecundity. Mol Plant Microbe Interact 26: 67–74 [DOI] [PubMed] [Google Scholar]
- Baltrus DA, Nishimura MT, Romanchuk A, Chang JH, Mukhtar MS, Cherkis K, Roach J, Grant SR, Jones CD, Dangl JL (2011) Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathog 7: e1002132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-On YM, Phillips R, Milo R (2018) The biomass distribution on Earth. Proc Natl Acad Sci USA 115: 6506–6511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter A, Mittler R, Suzuki N (2014) ROS as key players in plant stress signalling. J Exp Bot 65: 1229–1240 [DOI] [PubMed] [Google Scholar]
- Bede JC, Musser RO, Felton GW, Korth KL (2006) Caterpillar herbivory and salivary enzymes decrease transcript levels of Medicago truncatula genes encoding early enzymes in terpenoid biosynthesis. Plant Mol Biol 60: 519–531 [DOI] [PubMed] [Google Scholar]
- Bentham AR, De la Concepcion JC, Mukhi N, Zdrzałek R, Draeger M, Gorenkin D, Hughes RK, Banfield MJ (2020) A molecular roadmap to the plant immune system. J Biol Chem 295: 14916–14935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block A, Christensen SA, Hunter CT, Alborn HT (2018) Herbivore-derived fatty-acid amides elicit reactive oxygen species burst in plants. J Exp Bot 69: 1235–1245 [DOI] [PubMed] [Google Scholar]
- Bonaventure G, Gfeller A, Proebsting WM, Hörtensteiner S, Chételat A, Martinoia E, Farmer EE (2007) A gain-of-function allele of TPC1 activates oxylipin biogenesis after leaf wounding in Arabidopsis. Plant J 49: 889–898 [DOI] [PubMed] [Google Scholar]
- Bos JIB, Prince D, Pitino M, Maffei ME, Win J, Hogenhout SA (2010) A functional genomics approach identifies candidate effectors from the aphid species Myzus persicae (green peach aphid). PLoS Genet 6: e1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricchi I, Leitner M, Foti M, Mithöfer A, Boland W, Maffei ME (2010) Robotic mechanical wounding (MecWorm) versus herbivore-induced responses: early signaling and volatile emission in Lima bean (Phaseolus lunatus L.). Planta 232: 719–729 [DOI] [PubMed] [Google Scholar]
- Brutus A, Sicilia F, Macone A, Cervone F, De Lorenzo G (2010) A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc Natl Acad Sci USA 107: 9452–9457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscaill P, van der Hoorn RAL (2021) Defeated by the nines: nine extracellular strategies to avoid MAMP recognition in plants. Plant Cell 33: 2116–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camoni L, Barbero F, Aducci P, Maffei ME (2018) Spodoptera littoralis oral secretions inhibit the activity of Phaseolus lunatus plasma membrane H+-ATPase. PLoS One 13: e0202142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casteel CL, Walling LL, Paine TD (2006) Behavior and biology of the tomato psyllid, Bactericerca cockerelli, in response to the Mi-1.2 gene. Entomol Exp Appl 121: 67–72 [Google Scholar]
- Chaudhary R, Atamian HS, Shen Z, Briggs SP, Kaloshian I (2014) GroEL from the endosymbiont Buchnera aphidicola betrays the aphid by triggering plant defense. Proc Natl Acad Sci USA 111: 8919–8924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary R, Atamian HS, Shen Z, Briggs SP, Kaloshian I (2015) Potato aphid salivary proteome: enhanced salivation using resorcinol and identification of aphid phosphoproteins. J Proteome Res 14: 1762–1778 [DOI] [PubMed] [Google Scholar]
- Chaudhary R, Peng HC, He J, MacWilliams J, Teixeira M, Tsuchiya T, Chesnais Q, Mudgett MB, Kaloshian I (2019) Aphid effector Me10 interacts with tomato TFT7, a 14-3-3 isoform involved in aphid resistance. New Phytol 221: 1518–1528 [DOI] [PubMed] [Google Scholar]
- Chen CY, Liu YQ, Song WM, Chen DY, Chen FY, Chen XY, Chen ZW, Ge SX, Wang CZ, Zhan S, et al. (2019) An effector from cotton bollworm oral secretion impairs host plant defense signaling. Proc Natl Acad Sci USA 116: 14331–14338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Gonzales-Vigil E, Wilkerson CG, Howe GA (2007) Stability of plant defense proteins in the gut of insect herbivores. Plant Physiol 143: 1954–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Siems WF, Pearce G, Ryan CA (2008) Six peptide wound signals derived from a single precursor protein in Ipomoea batatas leaves activate the expression of the defense gene sporamin. J Biol Chem 283: 11469–11476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YL, Lee CY, Cheng KT, Chang WH, Huang RN, Nam HG, Chen YR (2014) Quantitative peptidomics study reveals that a wound-induced peptide from PR-1 regulates immune signaling in tomato. Plant Cell 26: 4135–4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Tanaka K, Cao Y, Qi Y, Qiu J, Liang Y, Lee SY, Stacey G (2014) Identification of a plant receptor for extracellular ATP. Science 343: 290–294 [DOI] [PubMed] [Google Scholar]
- Colaianni NR, Parys K, Lee HS, Conway JM, Kim NH, Edelbacher N, Mucyn TS, Madalinski M, Law TF, Jones CD, et al. (2021) A complex immune response to flagellin epitope variation in commensal communities. Cell Host Microbe 29: 635–649.e9 [DOI] [PubMed] [Google Scholar]
- Consales F, Schweizer F, Erb M, Gouhier-Darimont C, Bodenhausen N, Bruessow F, Sobhy I, Reymond P (2012) Insect oral secretions suppress wound-induced responses in Arabidopsis. J Exp Bot 63: 727–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DE, Mesarich CH, Thomma BPHJ (2015) Understanding plant immunity as a surveillance system to detect invasion. Annu Rev Phytopathol 53: 541–563 [DOI] [PubMed] [Google Scholar]
- Coppola M, Di Lelio I, Romanelli A, Gualtieri L, Molisso D, Ruocco M, Avitabile C, Natale R, Cascone P, Guerrieri E, et al. (2019) Tomato plants treated with systemin peptide show enhanced levels of direct and indirect defense associated with increased expression of defense-related genes. Plants 8: 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Luoni L, Marrano CA, Hashimoto K, Köster P, Giacometti S, De Michelis MI, Kudla J, Bonza MC (2017) Ca2+-dependent phosphoregulation of the plasma membrane Ca2+-ATPase ACA8 modulates stimulus-induced calcium signatures. J Exp Bot 68: 3215–3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto D, Zipfel C (2016) Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol 16: 537–552 [DOI] [PubMed] [Google Scholar]
- von Dahl CC, Baldwin IT (2007) Deciphering the role of ethylene in plant–herbivore interactions. J Plant Growth Regul 26: 201–209 [Google Scholar]
- DeFalco TA, Zipfel C (2021) Molecular mechanisms of early plant pattern-triggered immune signaling. Mol Cell 81: 3449–3467 [DOI] [PubMed] [Google Scholar]
- Demidchik V, Shabala S, Isayenkov S, Cuin TA, Pottosin I (2018) Calcium transport across plant membranes: mechanisms and functions. New Phytol 220: 49–69 [DOI] [PubMed] [Google Scholar]
- De Vos M, Van Oosten VR, Van Poecke RMP, Van Pelt JA, Pozo MJ, Mueller MJ, Buchala AJ, Métraux JP, Van Loon LC, Dicke M, et al. (2005) Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol Plant Microbe Interact 18: 923–937 [DOI] [PubMed] [Google Scholar]
- Diezel C, von Dahl CC, Gaquerel E, Baldwin IT (2009) Different lepidopteran elicitors account for cross-talk in herbivory-induced phytohormone signaling. Plant Physiol 150: 1576–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazakis E, Wouters D, Lochman J, Visser R, Matthieu HA, Vivianne GA (2020) ELR is a true pattern recognition receptor that associates with elicitins from diverse Phytophthora species. biorxiv, https://doi.org/10.1101/2020.09.21.305813
- Drurey C, Mathers TC, Prince DC, Wilson C, Caceres-Moreno C, Mugford ST, Hogenhout SA (2019) Chemosensory proteins in the CSP4 clade evolved as plant immunity suppressors before two suborders of plant-feeding hemipteran insects diverged. bioRxiv, https://doi.org/10.1101/173278
- Du B, Zhang W, Liu B, Hu J, Wei Z, Shi Z, He R, Zhu L, Chen R, Han B, He G (2009) Identification and characterization of Bph14, a gene conferring resistance to brown planthopper in rice. Proc Natl Acad Sci USA 106: 22163–22168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran-Flores D, Heil M (2014) Damaged-self recognition in common bean (Phaseolus vulgaris) shows taxonomic specificity and triggers signaling via reactive oxygen species (ROS). Front Plant Sci 5: 585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenseer H, Mathews MC, Powell JS, Felton GW (2010) Survey of a salivary effector in caterpillars: glucose oxidase variation and correlation with host range. J Chem Ecol 36: 885–897 [DOI] [PubMed] [Google Scholar]
- Elzinga DA, De Vos M, Jander G (2014) Suppression of plant defenses by a Myzus persicae (green peach aphid) salivary effector protein. Mol Plant Microbe Interact 27: 747–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb M (2018) Volatiles as inducers and suppressors of plant defense and immunity-origins, specificity, perception and signaling. Curr Opin Plant Biol 44: 117–121 [DOI] [PubMed] [Google Scholar]
- Erb M, Reymond P (2019) Molecular interactions between plants and insect herbivores. Annu Rev Plant Biol 70: 527–557 [DOI] [PubMed] [Google Scholar]
- Escudero-Martinez C, Rodriguez PA, Liu S, Santos PA, Stephens J, Bos JIB (2020) An aphid effector promotes barley susceptibility through suppression of defence gene expression. J Exp Bot 71: 2796–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Esse HP, Reuber TL, van der Does D (2020) Genetic modification to improve disease resistance in crops. New Phytol 225: 70–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eves-van den Akker S (2021) Plant-nematode interactions. Curr Opin Plant Biol 62: 102035. [DOI] [PubMed] [Google Scholar]
- Farmer EE, Gao YQ, Lenzoni G, Wolfender JL, Wu Q (2020) Wound- and mechano-stimulated electrical signals control hormone responses. New Phytol 227: 1037–1050 [DOI] [PubMed] [Google Scholar]
- Favery B, Dubreuil G, Chen MS, Giron D, Abad P (2020) Gall-inducing parasites: convergent and conserved strategies of plant manipulation by insects and nematodes. Annu Rev Phytopathol 58: 1–22 [DOI] [PubMed] [Google Scholar]
- Fraenkel GS (1959) The raison d’ĕtre of secondary plant substances; these odd chemicals arose as a means of protecting plants from insects and now guide insects to food. Science 129: 1466–1470 [DOI] [PubMed] [Google Scholar]
- Fu J, Shi Y, Wang L, Zhang H, Li J, Fang J, Ji R (2020) Planthopper-secreted salivary disulfide isomerase activates immune responses in plants. Front Plant Sci 11: 622513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilardoni PA, Hettenhausen C, Baldwin IT, Bonaventure G (2011) Nicotiana attenuata LECTIN RECEPTOR KINASE1 suppresses the insect-mediated inhibition of induced defense responses during Manduca sexta herbivory. Plant Cell 23: 3512–3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouhier-Darimont C, Schmiesing A, Bonnet C, Lassueur S, Reymond P (2013) Signalling of Arabidopsis thaliana response to Pieris brassicae eggs shares similarities with PAMP-triggered immunity. J Exp Bot 64: 665–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouhier-Darimont C, Stahl E, Glauser G, Reymond P (2019) The Arabidopsis lectin receptor kinase LecRK-I.8 is involved in insect egg perception. Front Plant Sci 10: 623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green TR, Ryan CA (1972) Wound-induced proteinase inhibitor in plant leaves: a possible defense mechanism against insects. Science 175: 776–777 [DOI] [PubMed] [Google Scholar]
- Griese E, Caarls L, Bassetti N, Mohammadin S, Verbaarschot P, Bukovinszkine’Kiss G, Poelman EH, Gols R, Schranz ME, Fatouros NE (2021) Insect egg-killing: a new front on the evolutionary arms-race between brassicaceous plants and pierid butterflies. New Phytol 230: 341–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissett L, Ali A, Coble AM, Logan K, Washington B, Mateson A, McGee K, Nkrumah Y, Jacobus L, Abraham E, et al. (2020) Survey of sensitivity to fatty acid-amino acid conjugates in the Solanaceae. J Chem Ecol 46: 330–343 [DOI] [PubMed] [Google Scholar]
- Groux R, Stahl E, Gouhier-Darimont C, Kerdaffrec E, Jimenez-Sandoval P, Santiago J, Reymond P (2021) Arabidopsis natural variation in insect egg-induced cell death reveals a role for LECTIN RECEPTOR KINASE-I.1. Plant Physiol 185: 240–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Wielsch N, Hafke JB, Svatoš A, Mithöfer A, Boland W (2013) A porin-like protein from oral secretions of Spodoptera littoralis larvae induces defense-related early events in plant leaves. Insect Biochem Mol Biol 43: 849–858 [DOI] [PubMed] [Google Scholar]
- Haj Ahmad F, Wu XN, Stintzi A, Schaller A, Schulze WX (2019) The systemin signaling cascade as derived from time course analyses of the systemin-responsive phosphoproteome. Mol Cell Proteomics 18: 1526–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halitschke R, Schittko U, Pohnert G, Boland W, Baldwin IT (2001) Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. III. Fatty acid-amino acid conjugates in herbivore oral secretions are necessary and sufficient for herbivore-specific plant responses. Plant Physiol 125: 711–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hander T, Fernández-Fernández AD, Kumpf RP, Willems P, Schatowitz H, Rombaut D, Staes A, Nolf J, Pottie R, Yao P, et al. (2019) Damage on plants activates Ca2+-dependent metacaspases for release of immunomodulatory peptides. Science 363: 1–10 [DOI] [PubMed] [Google Scholar]
- Helms AM, De Moraes CM, Tröger A, Alborn HT, Francke W, Tooker JF, Mescher MC (2017) Identification of an insect-produced olfactory cue that primes plant defenses. Nat Commun 8: 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe GA, Jander G (2008) Plant immunity to insect herbivores. Annu Rev Plant Biol 59: 41–66 [DOI] [PubMed] [Google Scholar]
- Howe GA, Major IT, Koo AJ (2018) Modularity in jasmonate signaling for multistress resilience. Annu Rev Plant Biol 69: 387–415 [DOI] [PubMed] [Google Scholar]
- Huang HJ, Ye ZX, Lu G, Zhang CX, Chen JP, Li JM (2020) Identification of salivary proteins in the whitefly Bemisia tabaci by transcriptomic and LC-MS/MS analyses. Insect Sci 28: 1369–1381 [DOI] [PubMed] [Google Scholar]
- Huffaker A, Pearce G, Ryan CA (2006) An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc Natl Acad Sci USA 103: 10098–10103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A, Pearce G, Veyrat N, Erb M, Turlings TCJ, Sartor R, Shen Z, Briggs SP, Vaughan MM, Alborn HT, et al. (2013) Plant elicitor peptides are conserved signals regulating direct and indirect antiherbivore defense. Proc Natl Acad Sci USA 110: 5707–5712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Ye M, Kuai P, Ye M, Erb M, Lou Y (2018) OsLRR-RLK1, an early responsive leucine-rich repeat receptor-like kinase, initiates rice defense responses against a chewing herbivore. New Phytol 219: 1097–1111 [DOI] [PubMed] [Google Scholar]
- Iida J, Desaki Y, Hata K, Uemura T, Yasuno A, Islam M, Maffei ME, Ozawa R, Nakajima T, Galis I, et al. (2019) Tetranins: new putative spider mite elicitors of host plant defense. New Phytol 224: 875–885 [DOI] [PubMed] [Google Scholar]
- Jamieson PA, Shan L, He P (2018) Plant cell surface molecular cypher: receptor-like proteins and their roles in immunity and development. Plant Sci 274: 242–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonckheere W, Dermauw W, Zhurov V, Wybouw N, Van den Bulcke J, Villarroel CA, Greenhalgh R, Grbić M, Schuurink RC, Tirry L, et al. (2016) The salivary protein repertoire of the polyphagous spider mite Tetranychus urticae: a quest for effectors. Mol Cell Proteomics 15: 3594–3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Karban R, Baldwin IT (2007) Induced Responses to Herbivory, University of Chicago Press, Chicago, IL [Google Scholar]
- Kettles GJ, Kaloshian I (2016) The potato aphid salivary effector Me47 is a glutathione-s-transferase involved in modifying plant responses to aphid infestation. Front Plant Sci 7: 1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiep V, Vadassery J, Lattke J, Maaß JP, Boland W, Peiter E, Mithöfer A (2015) Systemic cytosolic Ca2+ elevation is activated upon wounding and herbivory in Arabidopsis. New Phytol 207: 996–1004 [DOI] [PubMed] [Google Scholar]
- Klauser D, Desurmont GA, Glauser G, Vallat A, Flury P, Boller T, Turlings TCJ, Bartels S (2015) The Arabidopsis Pep-PEPR system is induced by herbivore feeding and contributes to JA-mediated plant defence against herbivory. J Exp Bot 66: 5327–5336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari A, Chételat A, Nguyen CT, Farmer EE (2019) Arabidopsis H+-ATPase AHA1 controls slow wave potential duration and wound-response jasmonate pathway activation. Proc Natl Acad Sci USA 116: 20226–20231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J, A Finlayson S, Salzman RA, Shan L, Zhu-Salzman K (2014) BOTRYTIS-INDUCED KINASE1 modulates Arabidopsis resistance to green peach aphids via PHYTOALEXIN DEFICIENT4. Plant Physiol 165: 1657–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Bartram S, Guo H, Mithöfer A, Kunert M, Boland W (2019a) SpitWorm, a herbivorous robot: mechanical leaf wounding with simultaneous application of salivary components. Plants 8: 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Liu C, Deng WH, Yao DM, Pan LL, Li YQ, Liu YQ, Liang Y, Zhou XP, Wang XW (2019b) Plant begomoviruses subvert ubiquitination to suppress plant defenses against insect vectors. PLoS Pathog 15: e1007607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Wang C, Mou Z (2020) Perception of damaged self in plants. Plant Physiol 182: 1545–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Weldegergis BT, Li J, Jung C, Qu J, Sun Y, Qian H, Tee C, van Loon JJA, Dicke M, et al. (2014) Virulence factors of geminivirus interact with MYC2 to subvert plant resistance and promote vector performance. Plant Cell 26: 4991–5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PA, Chen Y, Chaverra-Rodriguez D, Heu CC, Zainuddin NB, Sidhu JS, Peiffer M, Tan CW, Helms A, Kim D, et al. (2021) Silencing the alarm: an insect salivary enzyme closes plant stomata and inhibits volatile release. New Phytol 230: 793–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PA, Felton GW (2020) Oral cues are not enough: induction of defensive proteins in Nicotiana tabacum upon feeding by caterpillars. Planta 251: 89. [DOI] [PubMed] [Google Scholar]
- Lin PA, Peiffer M, Felton GW (2020) Induction of defensive proteins in Solanaceae by salivary glucose oxidase of Helicoverpa zea caterpillars and consequences for larval performance. Arthropod Plant Interact 14: 317–325 [Google Scholar]
- Liu Y, Wu H, Chen H, Liu Y, He J, Kang H, Sun Z, Pan G, Wang Q, Hu J, et al. (2015) A gene cluster encoding lectin receptor kinases confers broad-spectrum and durable insect resistance in rice. Nat Biotechnol 33: 301–305 [DOI] [PubMed] [Google Scholar]
- Lori M, van Verk MC, Hander T, Schatowitz H, Klauser D, Flury P, Gehring CA, Boller T, Bartels S (2015) Evolutionary divergence of the plant elicitor peptides (Peps) and their receptors: interfamily incompatibility of perception but compatibility of downstream signalling. J Exp Bot 66: 5315–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis J, Gobbato E, Mondal HA, Feys BJ, Parker JE, Shah J (2012) Discrimination of Arabidopsis PAD4 activities in defense against green peach aphid and pathogens. Plant Physiol 158: 1860–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis J, Peiffer M, Ray S, Luthe DS, Felton GW (2013) Host-specific salivary elicitor(s) of European corn borer induce defenses in tomato and maize. New Phytol 199: 66–73 [DOI] [PubMed] [Google Scholar]
- MacLean AM, Orlovskis Z, Kowitwanich K, Zdziarska AM, Angenent GC, Immink RGH, Hogenhout SA (2014) Phytoplasma effector SAP54 hijacks plant reproduction by degrading MADS-box proteins and promotes insect colonization in a RAD23-dependent manner. PLoS Biol 12: e1001835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malabarba J, Meents AK, Reichelt M, Scholz SS, Peiter E, Rachowka J, Konopka-Postupolska D, Wilkins KA, Davies JM, Oelmüller R, et al. (2021) ANNEXIN1 mediates calcium-dependent systemic defense in Arabidopsis plants upon herbivory and wounding. New Phytol 231: 243–254 [DOI] [PubMed] [Google Scholar]
- Manes N, Brauer EK, Hepworth S, Subramaniam R (2021) MAMP and DAMP signalling contributes resistance to Fusarium graminearum in Arabidopsis. J Exp Bot erab285: 1–12 [DOI] [PubMed] [Google Scholar]
- Marhavý P, Kurenda A, Siddique S, Dénervaud Tendon V, Zhou F, Holbein J, Hasan MS, Grundler FM, Farmer EE, Geldner N (2019) Single-cell damage elicits regional, nematode-restricting ethylene responses in roots. EMBO J 38: e100972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga T, Reisenman CE, Goldman-Huertas B, Brand P, Miao K, Suzuki HC, Verster KI, Ramírez SR, Whiteman NK (2021) Evolution of olfactory receptors tuned to mustard oils in herbivorous Drosophilidae. Mol Biol Evol https://doi.org/10.1093/molbev/msab362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Xu G, He P, Shan L (2016) SERKing coreceptors for receptors. Trends Plant Sci 21: 1017–1033 [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Janeway CA Jr. (2002) Decoding the patterns of self and nonself by the innate immune system. Science 296: 298–300 [DOI] [PubMed] [Google Scholar]
- Meena MK, Prajapati R, Krishna D, Divakaran K, Pandey Y, Reichelt M, Mathew MK, Boland W, Mithöfer A, Vadassery J (2019) The Ca2+ channel CNGC19 regulates Arabidopsis defense against Spodoptera herbivory. Plant Cell 31: 1539–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meents AK, Chen SP, Reichelt M, Lu HH, Bartram S, Yeh KW, Mithöfer A (2019) Volatile DMNT systemically induces jasmonate-independent direct anti-herbivore defense in leaves of sweet potato (Ipomoea batatas) plants. Sci Rep 9: 17431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meents AK, Mithöfer A (2020) Plant-plant communication: is there a tole for volatile damage-associated molecular patterns? Front Plant Sci 11: 583275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mithöfer A, Boland W (2012) Plant defense against herbivores: chemical aspects. Annu Rev Plant Biol 63: 431–450 [DOI] [PubMed] [Google Scholar]
- Mitter C, Davis DR, Cummings MP (2017) Phylogeny and evolution of Lepidoptera. Annu Rev Entomol 62: 265–283 [DOI] [PubMed] [Google Scholar]
- Moe-Lange J, Gappel NM, Machado M, Wudick MM, Sies CSA, Schott-Verdugo SN, Bonus M, Mishra S, Hartwig T, Bezrutczyk M, et al. (2021) Interdependence of a mechanosensitive anion channel and glutamate receptors in distal wound signaling. Sci Adv 7: eabg4298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott GA, Thakur S, Smakowska E, Wang PW, Belkhadir Y, Desveaux D, Guttman DS (2016) Genomic screens identify a new phytobacterial microbe-associated molecular pattern and the cognate Arabidopsis receptor-like kinase that mediates its immune elicitation. Genome Biol 17: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi SAR, Chauvin A, Pascaud F, Kellenberger S, Farmer EE (2013) GLUTAMATE RECEPTOR-LIKE genes mediate leaf-to-leaf wound signalling. Nature 500: 422–426 [DOI] [PubMed] [Google Scholar]
- Musser RO, Farmer E, Peiffer M, Williams SA, Felton GW (2006) Ablation of caterpillar labial salivary glands: technique for determining the role of saliva in insect-plant interactions. J Chem Ecol 32: 981–992 [DOI] [PubMed] [Google Scholar]
- Musser RO, Hum-Musser SM, Eichenseer H, Peiffer M, Ervin G, Murphy JB, Felton GW (2002) Herbivory: caterpillar saliva beats plant defences. Nature 416: 599–600 [DOI] [PubMed] [Google Scholar]
- Musser RO, Hum-Musser SM, Lee HK, DesRochers BL, Williams SA, Vogel H (2012) Caterpillar labial saliva alters tomato plant gene expression. J Chem Ecol 38: 1387–1401 [DOI] [PubMed] [Google Scholar]
- Naalden D, van Kleeff PJM, Dangol S, Mastop M, Corkill R, Hogenhout SA, Kant MR, Schuurink RC (2021) Spotlight on the roles of whitefly effectors in insect-plant interactions. Front Plant Sci 12: 661141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalam V, Louis J, Shah J (2019) Plant defense against aphids, the pest extraordinaire. Plant Sci 279: 96–107 [DOI] [PubMed] [Google Scholar]
- Nguyen CT, Kurenda A, Stolz S, Chételat A, Farmer EE (2018) Identification of cell populations necessary for leaf-to-leaf electrical signaling in a wounded plant. Proc Natl Acad Sci USA 115: 10178–10183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco-Cardenas M, McGurl B, Ryan CA (1993) Expression of an antisense prosystemin gene in tomato plants reduces resistance toward Manduca sexta larvae. Proc Natl Acad Sci USA 90: 8273–8276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega MA, All JN, Boerma HR, Parrott WA (2016) Pyramids of QTLs enhance host-plant resistance and Bt-mediated resistance to leaf-chewing insects in soybean. Theor Appl Genet 129: 703–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter RH (1951) Insect resistance in crop plants. Macmillan Co., New York, https://doi.org/10.1097/00010694-195112000-00015
- Parys K, Colaianni NR, Lee HS, Hohmann U, Edelbacher N, Trgovcevic A, Blahovska Z, Lee D, Mechtler A, Muhari-Portik Z, et al. (2021) Signatures of antagonistic pleiotropy in a bacterial flagellin epitope. Cell Host Microbe 29: 620–634.e9 [DOI] [PubMed] [Google Scholar]
- Pearce G, Bhattacharya R, Chen YC, Barona G, Yamaguchi Y, Ryan CA (2009) Isolation and characterization of hydroxyproline-rich glycopeptide signals in black nightshade leaves. Plant Physiol 150: 1422–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce G, Ryan CA (2003) Systemic signaling in tomato plants for defense against herbivores. Isolation and characterization of three novel defense-signaling glycopeptide hormones coded in a single precursor gene. J Biol Chem 278: 30044–30050 [DOI] [PubMed] [Google Scholar]
- Pearce G, Strydom D, Johnson S, Ryan CA (1991) A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science 253: 895–898 [DOI] [PubMed] [Google Scholar]
- Pitino M, Hogenhout SA (2013) Aphid protein effectors promote aphid colonization in a plant species-specific manner. Mol Plant Microbe Interact 26: 130–139 [DOI] [PubMed] [Google Scholar]
- Poretsky E, Dressano K, Weckwerth P, Ruiz M, Char SN, Da S, Abagyan R, Yang B, Huffaker A (2020) Differential activities of maize plant elicitor peptides as mediators of immune signaling and herbivore resistance. Plant J 104: 1582–1602 [DOI] [PubMed] [Google Scholar]
- Poretsky E, Ruiz M, Ahmadian N, Steinbrenner AD, Dressano K, Schmelz EA, Huffaker A (2021) Comparative analyses of responses to exogenous and endogenous antiherbivore elicitors enable a forward genetics approach to identify maize gene candidates mediating sensitivity to herbivore-associated molecular patterns. Plant J 108: 1295–1316 [DOI] [PubMed] [Google Scholar]
- Prigozhin DM, Krasileva KV (2021) Analysis of intraspecies diversity reveals a subset of highly variable plant immune receptors and predicts their binding sites. Plant Cell 33: 998–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince DC, Drurey C, Zipfel C, Hogenhout SA (2014) The leucine-rich repeat receptor-like kinase BRASSINOSTEROID INSENSITIVE1-ASSOCIATED KINASE1 and the cytochrome P450 PHYTOALEXIN DEFICIENT3 contribute to innate immunity to aphids in Arabidopsis. Plant Physiol 164: 2207–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt RN, Locci F, Wanke F, Zhang L, Saile SC, Joe A, Karelina D, Hua C, Wan WL, Hu M, et al. (2021) The EDS1–PAD4–ADR1 node mediates Arabidopsis pattern-triggered immunity. Nature 598: 495–499 [DOI] [PubMed] [Google Scholar]
- Ray S, Alves PCMS, Ahmad I, Gaffoor I, Acevedo FE, Peiffer M, Jin S, Han Y, Shakeel S, Felton GW, et al. (2016) Turnabout is fair play: herbivory-induced plant chitinases excreted in gall armyworm frass suppress herbivore defenses in maize. Plant Physiol 171: 694–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Gaffor I, Acevedo FE, Helms A, Chuang WP, Tooker J, Felton GW, Luthe DS (2015) Maize plants recognize herbivore-associated cues from caterpillar frass. J Chem Ecol 41: 781–792 [DOI] [PubMed] [Google Scholar]
- Reymond P (2021) Receptor kinases in plant responses to herbivory. Curr Opin Biotechnol 70: 143–150 [DOI] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE (2000) Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12: 707–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes J, Yang H, Moussu S, Boutrot F, Santiago J, Zipfel C (2021) Perception of a divergent family of phytocytokines by the Arabidopsis receptor kinase MIK2. Nat Commun 12: 705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Vega LJ, Stanley BA, Stanley A, Felton GW (2018) Proteomic analysis of labial saliva of the generalist cabbage looper (Trichoplusia ni) and its role in interactions with host plants. J Insect Physiol 107: 97–103 [DOI] [PubMed] [Google Scholar]
- Rodriguez PA, Escudero-Martinez C, Bos JIB (2017) An aphid effector targets trafficking protein VPS52 in a host-specific manner to promote virulence. Plant Physiol 173: 1892–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi M, Goggin FL, Milligan SB, Kaloshian I, Ullman DE, Williamson VM (1998) The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc Natl Acad Sci USA 95: 9750–9754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador-Recatalà V, Tjallingii WF, Farmer EE (2014) Real-time, in vivo intracellular recordings of caterpillar-induced depolarization waves in sieve elements using aphid electrodes. New Phytol 203: 674–684 [DOI] [PubMed] [Google Scholar]
- Savary S, Willocquet L, Pethybridge SJ, Esker P, McRoberts N, Nelson A (2019) The global burden of pathogens and pests on major food crops. Nat Ecol Evol 3: 430–439 [DOI] [PubMed] [Google Scholar]
- Schmelz EA, Carroll MJ, LeClere S, Phipps SM, Meredith J, Chourey PS, Alborn HT, Teal PEA (2006) Fragments of ATP synthase mediate plant perception of insect attack. Proc Natl Acad Sci USA 103: 8894–8899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz EA, Engelberth J, Alborn HT, Tumlinson JH 3rd, Teal PEA (2009) Phytohormone-based activity mapping of insect herbivore-produced elicitors. Proc Natl Acad Sci USA 106: 653–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz EA, Huffaker A, Carroll MJ, Alborn HT, Ali JG, Teal PEA (2012) An amino acid substitution inhibits specialist herbivore production of an antagonist effector and recovers insect-induced plant defenses. Plant Physiol 160: 1468–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz SS, Vadassery J, Heyer M, Reichelt M, Bender KW, Snedden WA, Boland W, Mithöfer A (2014) Mutation of the Arabidopsis calmodulin-like protein CML37 deregulates the jasmonate pathway and enhances susceptibility to herbivory. Mol Plant 7: 1712–1726 [DOI] [PubMed] [Google Scholar]
- Schultink A, Steinbrenner AD (2021) A playbook for developing disease-resistant crops through immune receptor identification and transfer. Curr Opin Plant Biol 62: 102089. [DOI] [PubMed] [Google Scholar]
- Schuman MC, Baldwin IT (2016) The layers of plant responses to insect herbivores. Annu Rev Entomol 61: 373–394 [DOI] [PubMed] [Google Scholar]
- Shangguan X, Zhang J, Liu B, Zhao Y, Wang H, Wang Z, Guo J, Rao W, Jing S, Guan W, et al. (2018) A mucin-like protein of planthopper is required for feeding and induces immunity response in plants. Plant Physiol 176: 552–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinya T, Yasuda S, Hyodo K, Tani R, Hojo Y, Fujiwara Y, Hiruma K, Ishizaki T, Fujita Y, Saijo Y, et al. (2018) Integration of danger peptide signals with herbivore-associated molecular pattern signaling amplifies anti-herbivore defense responses in rice. Plant J 94: 626–637 [DOI] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB (2001) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA 98: 10763–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiteller D, Pohnert G, Boland W (2001) Absolute configuration of volicitin, an elicitor of plant volatile biosynthesis from lepidopteran larvae. Tetrahedron Lett 42: 1483–1485 [Google Scholar]
- Stahl E, Brillatz T, Ferreira Queiroz E, Marcourt L, Schmiesing A, Hilfiker O, Riezman I, Riezman H, Wolfender JL, Reymond P (2020) Phosphatidylcholines from Pieris brassicae eggs activate an immune response in Arabidopsis. Elife 9: e60293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl E, Hilfiker O, Reymond P (2018) Plant-arthropod interactions: who is the winner? Plant J 93: 703–728 [DOI] [PubMed] [Google Scholar]
- Steinbrenner AD, Muñoz-Amatriaín M, Chaparro AF, Aguilar-Venegas JM, Lo S, Okuda S, Glauser G, Dongiovanni J, Shi D, Hall M, et al. (2020) A receptor-like protein mediates plant immune responses to herbivore-associated molecular patterns. Proc Natl Acad Sci USA 117: 31510–31518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbrenner AD (2020) The evolving landscape of cell surface pattern recognition across plant immune networks. Curr Opin Plant Biol 56: 135–146 [DOI] [PubMed] [Google Scholar]
- Steinbrenner AD, Saldivar E, Hodges N, Chaparro AF, Schmelz EA (2021) Signatures of plant defense response specificity mediated by herbivore-associated molecular patterns in legumes. bioRxiv. https://doi.org/10.1101/2021.09.02.458788 [DOI] [PubMed]
- Stenberg JA, Muola A (2017) How should plant resistance to herbivores be measured? Front. Plant Sci 8: 663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio A, Kingdom HN, MacLean AM, Grieve VM, Hogenhout SA (2011) Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. Proc Natl Acad Sci USA 108: E1254–E1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YL, Li JM, Li M, Luan JB, Ye XD, Wang XW, Liu SS (2012) Transcriptomic analysis of the salivary glands of an invasive whitefly. PLoS One 7: e39303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabashnik BE, Carrière Y (2017) Surge in insect resistance to transgenic crops and prospects for sustainability. Nat Biotechnol 35: 926–935 [DOI] [PubMed] [Google Scholar]
- Tamura Y, Hattori M, Yoshioka H, Yoshioka M, Takahashi A, Wu J, Sentoku N, Yasui H (2014) Map-based cloning and characterization of a brown planthopper resistance gene BPH26 from Oryza sativa L. ssp. indica cultivar ADR52. Sci Rep 4: 5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Heil M (2021) Damage-associated molecular patterns (DAMPs) in plant innate immunity: applying the danger model and evolutionary perspectives. Annu Rev Phytopathol 59: 53–75 [DOI] [PubMed] [Google Scholar]
- Thorpe P, Cock PJA, Bos J (2016) Comparative transcriptomics and proteomics of three different aphid species identifies core and diverse effector sets. BMC Genomics 17: 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Peiffer M, Shoemaker E, Tooker J, Haubruge E, Francis F, Luthe DS, Felton GW (2012) Salivary glucose oxidase from caterpillars mediates the induction of rapid and delayed-induced defenses in the tomato plant. PLoS One 7: e36168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toruño TY, Stergiopoulos I, Coaker G (2016) Plant-pathogen effectors: cellular probes interfering with plant defenses in spatial and temporal manners. Annu Rev Phytopathol 54: 419–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota M, Spencer D, Sawai-Toyota S, Jiaqi W, Zhang T, Koo AJ, Howe GA, Gilroy S (2018) Glutamate triggers long-distance, calcium-based plant defense signaling. Science 361: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Tripathi D, Zhang T, Koo AJ, Stacey G, Tanaka K (2018) Extracellular ATP acts on jasmonate signaling to reinforce plant defense. Plant Physiol 176: 511–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turlings TCJ, Erb M (2018) Tritrophic interactions mediated by herbivore-induced plant volatiles: mechanisms, ecological relevance, and application potential. Annu Rev Entomol 63: 433–452 [DOI] [PubMed] [Google Scholar]
- Uemura T, Hachisu M, Desaki Y, Ito A, Hoshino R, Sano Y, Nozawa A, Mujiono K, Galis I, Yoshida A, Nemoto K, et al. (2020) Soy and Arabidopsis receptor-like kinases respond to polysaccharide signals from Spodoptera species and mediate herbivore resistance. Commun Biol 3: 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upson JL, Zess EK, Białas A, Wu CH, Kamoun S (2018) The coming of age of EvoMPMI: evolutionary molecular plant-microbe interactions across multiple timescales. Curr Opin Plant Biol 44: 108–116 [DOI] [PubMed] [Google Scholar]
- Vadassery J, Reichelt M, Hause B, Gershenzon J, Boland W, Mithöfer A (2012) CML42-mediated calcium signaling coordinates responses to Spodoptera herbivory and abiotic stresses in Arabidopsis. Plant Physiol 159: 1159–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Weyer AL, Monteiro F, Furzer OJ, Nishimura MT, Cevik V, Witek K, Jones JDG, Dangl JL, Weigel D, Bemm F (2019) A species-wide inventory of NLR genes and alleles in Arabidopsis thaliana. Cell 178: 1260–1272.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Muñoz I, Duran-Flores D, Fernández-Fernández ÁD, Heyman J, Ritter A, Stael S (2020) Breaking bad news: dynamic molecular mechanisms of wound response in plants. Front Plant Sci 11: 1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walley JW, Coughlan S, Hudson ME, Covington MF, Kaspi R, Banu G, Harmer SL, Dehesh K (2007) Mechanical stress induces biotic and abiotic stress responses via a novel cis-element. PLoS Genet 3: 1800–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Huang X, Li Q, Zhang Y, Li JL, Mou Z (2019) Extracellular pyridine nucleotides trigger plant systemic immunity through a lectin receptor kinase/BAK1 complex. Nat Commun 10: 4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Zhou M, Zhang X, Yao J, Zhang Y, Mou Z (2017a) A lectin receptor kinase as a potential sensor for extracellular nicotinamide adenine dinucleotide in Arabidopsis thaliana. Elife 6: e25474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Peiffer M, Hoover K, Rosa C, Zeng R, Felton GW (2017b) Helicoverpa zea gut-associated bacteria indirectly induce defenses in tomato by triggering a salivary elicitor(s). New Phytol 214: 1294–1306 [DOI] [PubMed] [Google Scholar]
- Wang L, Einig E, Almeida-Trapp M, Albert M, Fliegmann J, Mithöfer A, Kalbacher H, Felix G (2018) The systemin receptor SYR1 enhances resistance of tomato against herbivorous insects. Nat Plants 4: 152–156 [DOI] [PubMed] [Google Scholar]
- Wang Z, Lü Q, Zhang L, Zhang M, Chen L, Zou S, Zhang C, Dong H (2021) Aphid salivary protein Mp1 facilitates infestation by binding phloem protein 2-A1 in Arabidopsis. Biochem Biophys Res Commun 572: 105–111 [DOI] [PubMed] [Google Scholar]
- Wari D, Kabir MA, Mujiono K, Hojo Y, Shinya T, Tani A, Nakatani H, Galis I (2019) Honeydew-associated microbes elicit defense responses against brown planthopper in rice. J Exp Bot 70: 1683–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C, Feussner I (2018) The oxylipin pathways: biochemistry and function. Annu Rev Plant Biol 69: 363–386 [DOI] [PubMed] [Google Scholar]
- Wu J, Hettenhausen C, Meldau S, Baldwin IT (2007) Herbivory rapidly activates MAPK signaling in attacked and unattacked leaf regions but not between leaves of Nicotiana attenuata. Plant Cell 19: 1096–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Wang L, Wünsche H, Baldwin IT (2013) Narboh D, a respiratory burst oxidase homolog in Nicotiana attenuata, is required for late defense responses after herbivore attack. J Integr Plant Biol 55: 187–198 [DOI] [PubMed] [Google Scholar]
- Xu HX, Qian LX, Wang XW, Shao RX, Hong Y, Liu SS, Wang XW (2019) A salivary effector enables whitefly to feed on host plants by eliciting salicylic acid-signaling pathway. Proc Natl Acad Sci USA 116: 490–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Zhou W, Pottinger S, Baldwin IT (2015) Herbivore associated elicitor-induced defences are highly specific among closely related Nicotiana species. BMC Plant Biol 15: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Pearce G, Ryan CA (2006) The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proc Natl Acad Sci USA 103: 10104–10109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki Y, Sumioka H, Takiguchi M, Uemura T, Kihara Y, Shinya T, Galis I, Arimura GI (2021) Phytohormone-dependent plant defense signaling orchestrated by oral bacteria of the herbivore Spodoptera litura. New Phytol 231: 2029–2038 [DOI] [PubMed] [Google Scholar]
- Yan C, Fan M, Yang M, Zhao J, Zhang W, Su Y, Xiao L, Deng H, Xie D (2018) Injury activates Ca2+/calmodulin-dependent phosphorylation of JAV1-JAZ8-WRKY51 complex for jasmonate biosynthesis. Mol Cell 70: 136–149.e7 [DOI] [PubMed] [Google Scholar]
- Yang DH, Hettenhausen C, Baldwin IT, Wu J (2011) BAK1 regulates the accumulation of jasmonic acid and the levels of trypsin proteinase inhibitors in Nicotiana attenuata’s responses to herbivory. J Exp Bot 62: 641–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DH, Hettenhausen C, Baldwin IT, Wu J (2012) Silencing Nicotiana attenuata calcium-dependent protein kinases, CDPK4 and CDPK5, strongly up-regulates wound- and herbivory-induced jasmonic acid accumulations. Plant Physiol 159: 1591–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga N, Aboshi T, Abe H, Nishida R, Alborn HT, Tumlinson JH, Mori N (2008) Active role of fatty acid amino acid conjugates in nitrogen metabolism in Spodoptera litura larvae. Proc Natl Acad Sci USA 105: 18058–18063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga N, Alborn HT, Nakanishi T, Suckling DM, Nishida R, Tumlinson JH, Mori N (2010) Fatty acid-amino acid conjugates diversification in lepidopteran caterpillars. J Chem Ecol 36: 319–325 [DOI] [PubMed] [Google Scholar]
- Yoshinaga N, Ishikawa C, Seidl-Adams I, Bosak E, Aboshi T, Tumlinson JH, Mori N (2014) N-(18-hydroxylinolenoyl)-L-glutamine: a newly discovered analog of volicitin in Manduca sexta and its elicitor activity in plants. J Chem Ecol 40: 484–490 [DOI] [PubMed] [Google Scholar]
- Zhang TQ, Chen Y, Wang JW (2021) A single-cell analysis of the Arabidopsis vegetative shoot apex. Dev Cell 56: 1056–1074.e8 [DOI] [PubMed] [Google Scholar]
- Zhao C, Navarro L, Chen H, Benatti T, Qu J, Chellapilla S, Waterhouse RM, Wheeler D, Bao R, Batterton M, et al. (2015) A massive expansion of effector genes underlies gall-formation in the wheat pest Mayetiola destructor. Curr Biol 25: 613–620 [DOI] [PubMed] [Google Scholar]
- Zhao C, Shukle R, Navarro-Escalante L, Chen M, Richards S, Stuart JJ (2016) Avirulence gene mapping in the Hessian fly (Mayetiola destructor) reveals a protein phosphatase 2C effector gene family. J Insect Physiol 84: 22–31 [DOI] [PubMed] [Google Scholar]
- Zhou F, Emonet A, Dénervaud Tendon V, Marhavy P, Wu D, Lahaye T, Geldner N (2020) Co-incidence of damage and microbial patterns controls localized immune responses in roots. Cell 180: 440–453.e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Brockmöller T, Ling Z, Omdahl A, Baldwin IT, Xu S (2016) Evolution of herbivore-induced early defense signaling was shaped by genome-wide duplications in Nicotiana. Elife 5: e19531. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.