Abstract
The genetic basis for fluoroquinolone resistance was examined in 30 high-level fluoroquinolone-resistant Escherichia coli clinical isolates from Beijing, China. Each strain also demonstrated resistance to a variety of other antibiotics. PCR sequence analysis of the quinolone resistance-determining region of the topoisomerase genes (gyrA/B, parC) revealed three to five mutations known to be associated with fluoroquinolone resistance. Western blot analysis failed to demonstrate overexpression of MarA, and Northern blot analysis did not detect overexpression of soxS RNA in any of the clinical strains. The AcrA protein of the AcrAB multidrug efflux pump was overexpressed in 19 of 30 strains of E. coli tested, and all 19 strains were tolerant to organic solvents. PCR amplification of the complete acrR (regulator/repressor) gene of eight isolates revealed amino acid changes in four isolates, a 9-bp deletion in another, and a 22-bp duplication in a sixth strain. Complementation with a plasmid-borne wild-type acrR gene reduced the level of AcrA in the mutants and partially restored antibiotic susceptibility 1.5- to 6-fold. This study shows that mutations in acrR are an additional genetic basis for fluoroquinolone resistance.
Fluoroquinolones are powerful broad-spectrum antimicrobial agents used for the treatment of a wide variety of community-acquired and nosocomial infections (35, 45). However, resistance to fluoroquinolones has increased markedly since their introduction in the late 1980s (1, 7, 26, 32, 39, 44, 49). In Beijing from 1997 to 1999, approximately 60% of Escherichia coli strains isolated from hospital-acquired infections and 50% of the E. coli strains isolated from the community were resistant to ciprofloxacin. Of those fluoroquinolone-resistant strains, 80% exhibited ciprofloxacin MICs of >32 μg/ml (references 54 and 61 and unpublished data). These findings contrast with much lower frequencies in other parts of the world.
Mechanisms of fluoroquinolone resistance fall into two principal categories: alterations in drug targets (e.g., DNA gyrase or topoisomerase IV) (12, 19, 34, 52) and decreased cellular accumulation of quinolones involving the major multidrug efflux pump, AcrAB (23, 37). Mutations causing quinolone resistance occur primarily in a highly conserved region (the quinolone resistance-determining region [QRDR]) of DNA gyrase and topoisomerase IV (9, 19, 25, 37, 52, 55, 59, 60). Other secondary mechanisms, such as those that affect the regulatory gene marA (multiple antibiotic resistance) (9, 10) or soxS (superoxide) (2), generally cause decreased expression of the OmpF porin (11) and overexpression of the AcrAB efflux pump (40). These porin and pump changes lead to resistance not only to the quinolones but also to a number of structurally unrelated compounds (2, 10, 40).
In this study, we sought to determine whether the regulatory genes (marA, soxS, acrR), in addition to the structural genes (gyrA/B, parC), contained mutations which contribute to the high-level fluoroquinolone resistance of clinical E. coli isolates from China.
MATERIALS AND METHODS
Bacterial strains.
Thirty clinical strains of E. coli with high levels of ciprofloxacin resistance (MIC > 32 μg/ml) were isolated from different patients in different wards from the 1,000-bed Peking Union Medical College Hospital in Beijing, China, from May to August 1999. These strains were selected for study because of their resistance to fluoroquinolones (Table 1). Additional strains used in this study were plasmid-free E. coli K-12 derivatives. Their properties are described in Table 2. Plasmid pHRP315, containing a spectinomycin cassette, was used for cloning (42), since all of the clinical strains were susceptible to spectinomycin. All isolates were grown in Luria-Bertani (LB) medium (Difco Laboratories, Detroit, Mich.) at 35°C, unless otherwise noted. Stock cultures were stored at −80°C in 30% glycerol or were lyophilized on dry disks until tested.
TABLE 1.
Sources and antibiotic phenotypes of 30 strains of ciprofloxacin-resistant clinical E. coli isolates (May to August 1999)
| Strain | Specimen | MIC (μg/ml)a
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Tetb | Chlb | Genc | Ampb | Timc | Ptac | Ftxc | Norbd | ||
| CH1 | Urine | 72 | 87 | 0.5 | 218 | 4 | 1.5 | 0.06 | 64 |
| CH2 | Urine | 1.3 | 125 | 3 | 76 | 256 | 12 | 0.2 | 250 |
| CH3 | Drainage | >400 | 6 | 24 | >400 | 256 | 3 | 256 | 167 |
| CH4 | Blood | 153 | 10 | 24 | 191 | 24 | 4 | 0.4 | 92 |
| CH5 | Urine | 83 | 250 | 1.5 | 258 | 16 | 4 | 0.3 | 194 |
| CH6 | Sputum | >400 | 16 | 24 | 262 | 32 | 12 | 4 | 250 |
| CH7 | Pus | >400 | 259 | 32 | 147 | 6 | 3 | 0.1 | 106 |
| CH8 | Pus | 138 | 250 | 24 | >400 | 128 | 1.5 | 0.3 | 69 |
| CH9 | Urine | 302 | 7 | 0.8 | >400 | 24 | 4 | 0.3 | 117 |
| CH10 | Blood | 258 | 258 | 64 | 191 | 24 | 4 | 0.4 | 250 |
| CH11 | Drainage | >400 | 3 | 64 | 182 | 8 | 4 | 0.4 | 158 |
| CH12 | Pus | 189 | 4 | 64 | >400 | 32 | 4 | 0.4 | 97 |
| CH13 | Vaginal swab | 50 | 316 | 192 | >400 | >256 | 2 | 0.06 | 25 |
| CH14 | Blood | 156 | 46 | >256 | >400 | 32 | 2 | 0.09 | 67 |
| CH15 | Abscess | 97 | 16 | 24 | 76 | 64 | 8 | 2 | 75 |
| CH16 | Blood | 128 | 147 | 48 | 142 | 96 | 6 | 0.5 | 28 |
| CH17 | Blood | 142 | 249 | 64 | 142 | 256 | 16 | 8 | 28 |
| CH18 | Urine | 111 | 250 | 64 | 173 | 16 | 4 | 0.2 | 21 |
| CH19 | Vaginal swab | 1.4 | 4 | 0.8 | >400 | 256 | 4 | >256 | 51 |
| CH20 | Urine | 267 | 236 | 64 | >400 | 256 | 3 | >256 | 156 |
| CH21 | Sputum | 160 | 187 | 1 | 129 | 32 | 3 | 0.2 | 58 |
| CH22 | Sputum | 167 | 236 | 256 | 267 | 64 | 24 | 6 | 69 |
| CH23 | Vaginal swab | 139 | 213 | >256 | >400 | >256 | 64 | >256 | 125 |
| CH24 | Throat | 169 | 259 | 192 | 142 | 192 | 16 | 6 | 24 |
| CH25 | Sputum | 236 | 6 | 4 | 209 | 64 | 3 | 0.4 | 28 |
| CH26 | Urine | 230 | 14 | 32 | 316 | 16 | 4 | 0.4 | 117 |
| CH27 | Urine | 44 | 8 | 32 | 244 | 96 | 4 | 0.5 | 236 |
| CH28 | Drainage | 191 | 187 | 48 | 351 | 48 | 2 | 0.4 | 33 |
| CH29 | Blood | 72 | 258 | 96 | 98 | 16 | 4 | 0.4 | 117 |
| CH30 | Urine | >400 | 19 | 32 | 222 | 48 | 3 | 0.3 | 158 |
Tet, tetracycline; Chl, chloramphenicol; Gen, gentamicin; Amp, ampicillin; Tim, ticarcillin-clavulanate; Pta, piperacillin-tazobactam; Ftx, cefotaxime; Nor, norfloxacin.
By gradient plate.
By E-test.
All strains showed ciprofloxacin MICs of >32 μg/ml. The susceptible E. coli K-12 strain AG100 showed a ciprofloxacin MIC of 0.015 μg/ml (by E-test) and a norfloxacin MIC of 0.08 μg/ml (by gradient plate).
TABLE 2.
Laboratory strains and plasmids
| Strain or plasmid | Description | Reference |
|---|---|---|
| Strains | ||
| AG100 | Wild-type E. coli K-12 | 16 |
| AG112 | marR mutant of AG100; 5-bp deletion (Δ1481–1485) | 37 |
| AG100A | AG100; ΔacrAB | 40 |
| AG100B | AG100; acrR::kan mutant | 40 |
| DJ901 | GC4468, soxRSΔ 901::Tn10Kan | 18 |
| JTG1078 | GC4468, soxR105 zjc-2204::Tn10Kan | 17 |
| Plasmids | ||
| pHRP315 | Apr, pUCMB20 with Ω Smr/Spr cassette in BamHI site | 42 |
| pHRP/acrR | acrR cloned into pHRP315 | This study |
Antimicrobial susceptibility testing.
Initial MIC profiles were screened by the disk diffusion method (M2-A6; National Committee for Clinical Laboratory Standards) and verified by E-test (AB Biodisk, Solna, Sweden) or gradient plate methodology (14). The MICs of ciprofloxacin, cefotaxime, imipenem, gentamicin, ticarcillin-clavulanic acid, and piperacillin-tazobactam were evaluated by E-test (a gift from AB Biodisk); in addition, ampicillin (400 μg/ml), tetracycline (250 μg/ml), chloramphenicol (300 μg/ml), and norfloxacin (250 μg/ml) (Sigma Chemical Co., St. Louis, Mo.) susceptibilities were tested on LB agar using the gradient plate method. The results were recorded after 24 h of incubation at 35°C. E. coli strains ATCC 25922 and AG100 were included as controls. Each assay was performed three times on separate occasions.
Antimicrobial susceptibility of clinical strains bearing pHRPacrR was determined using gradient plates.
Organic solvent tolerance.
Mid-logarithmic-phase cultures (A530, 0.4 to 0.5) grown in LB broth were diluted in phosphate-buffered saline to approximately 106 to 107 cells/ml. Five microliters was spotted onto LB agar and allowed to dry. The surface of the agar was overlaid with either hexane (99%; Sigma-Aldrich Chemical Co., Milwaukee, Wis.), cyclohexane (Fisher Scientific), or a mixture of hexane and cyclohexane (3:1, 1:1, or 1:3 [vol/vol]) to a depth of ∼2 to 3 mm. The plates were incubated at 30°C in a closed container to prevent evaporation of the solvent. After 24 to 36 h, the spots were scored for confluent growth, which demonstrated tolerance to the solvent(s) tested. Tests were run in duplicate, three times.
PCR amplification and DNA sequencing of gyrA, gyrB, parC, and acrR.
Mutations in the gyrA, gyrB, parC, and acrR genes of the E. coli isolates were identified by DNA sequencing of their PCR products. PCR amplification of the QRDRs of gyrA (nucleotides 100 to 368), gyrB (nucleotides 1223 to 1425), and parC (nucleotides 138 to 401) was performed with the following oligonucleotide primer pairs: the gyrA gene was amplified with 5′-12020TGCCAGATGTCCGAGAT12004-3′ and 5′-11753GTATAACGCATTGCCGC11769-3′ (AE000312; annealing temperature [Tm], 58°C), and parC was amplified with 5′-4664TATGCGATGTCTGAACTGGG4645-3′ and 5′-4401GCTCAATAGCAGCTCGGAAT4420-3′ (AE000384; Tm, 54°C). Likewise, amplification of the gyrB gene was with primers 5′-1910CAGACTGCCAGGAACGCGAT1891-3′ and 5′-1707AGCCAAGCGCGGTGATAAGC1726-3′ (AE000447; Tm, 60°C). Wild-type acrR (DNA from AG100) was amplified in its entirety from bases 9175 to 9822 (AE000152; Tm, 58°C), including the promoter-operator region, with the oligonucleotide primer pairs 5′-GCTCTAGA 8900ACTGTTACTACGCCAACG8918-3′ and 5′-AAACTGCAG 9934CTGAACCTGAAGAACGACCTG9913 - 3′. The underline denotes the XbaI and PstI sites, respectively, introduced into each primer for cloning into pHRP315.
A single colony of each bacterial isolate was used as the template for PCR amplification. The PCRs were performed using high-fidelity platinum Taq DNA polymerase (Gibco BRL) in a GeneAmp PCR System 9700 (PE Applied Biosystems). The PCR products were purified using a Qiaquick PCR purification kit (Qiagen, Inc.). Direct cycle sequencing in both directions was performed with the same primers, using an automatic 377A DNA Sequencer (Applied Biosystems) at the Tufts University Core Facility.
Computer analyses of the sequences were performed using the ClustalW (version 1.8) multiple sequence alignment program (51).
Cloning of acrR gene.
The 1.2-kb acrR PCR fragment was digested by XbaI and PstI, gel purified (QiaexII Gel Extraction Kit; Qiagen, Inc.), and ligated into the XbaI- and PstI-digested pHRP315. The resulting recombinant plasmid (pHRP/acrR) was first isolated in DH5α cells by selection with spectinomycin (30 μg/ml). It was then transformed by electroporation into clinical strains of E. coli (CH5, CH10, CH19, CH27, and CH29), with selection on LB agar plates containing 200 μg of spectinomycin/ml. CH15 was unable to be transformed using electroporation or CaCl2 heat shock, perhaps because it contained many endogenous plasmids.
Protein electrophoresis and Western blot analysis.
Freshly grown E. coli cells were harvested by centrifugation (794 × g, 10 min at 4°C), washed twice in ice-cold phosphate-buffered saline, resuspended in lysis buffer [20 mM Tris-HCl (pH 8), 100 mM NaCl, 30% glycerol, 2 mM 4-(2-aminoethyl) benzenesulfonyl fluoride, 1 mM EDTA (pH 8), 1 mM dithiothreitol], and sonicated on ice. After centrifugation (4,900 × g, 10 min at 4°C), the concentration of total whole-cell protein in the supernatant was assessed (Bio-Rad Protein Assay) using bovine serum albumin as a standard.
Whole-cell proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in a 15% separating gel (10% gel was used for detecting AcrA) by standard methods (28). Twenty micrograms of total protein was loaded for detection of MarA, while 15 μg was loaded for the detection of AcrA. The proteins from the gel were transferred electrophoretically for 30 min at 25 V (Trans-Blot SD Semi-Dry Transfer Cell; Bio-Rad) to a polyvinylidene difluoride membrane (Immobilon-P; Millipore, Bedford, Mass.), according to the manufacturer's protocol. The membrane was blocked overnight at room temperature in Tris-buffered saline (TBS; 0.15% NaCl, 10 mM Tris-HCl, pH 7.4) containing 5% dried milk, washed three times in wash buffer (0.05% Tween 20 in TBS), and hybridized at room temperature with anti-MarA polyclonal antibody (1:5,000) (31) or anti-AcrA polyclonal antibody (1:15,000; gift from H. Nikaido of the University of California, Berkeley) diluted in buffer A (0.5% Triton X-100, 0.2% SDS, 0.5% dry milk [wt/vol] in TBS) for 1 h with shaking. After three washes in wash buffer, the membrane was incubated at room temperature for 1 h with horseradish peroxidase conjugated to anti-rabbit immunoglobulin G (1:2,000; Life Technologies) diluted in wash buffer. Finally, after three additional washes, the blots were developed using the Renaissance Western Blot Chemiluminescence Reagent Plus (NEN Life Science Products, Inc., Boston, Mass.). AG100 and AG112 were used as negative and positive controls, respectively, for the detection of MarA. For the detection of AcrA, the following controls were used: AG100B (acrR mutant, AcrA overexpressed) was used to determine the location of overexpressed AcrA, and AG100A (acrAB deleted, no acrA expression) and AG100 (wild-type expression of AcrA) were employed to assess the relative amount of AcrA above wild-type levels. All Western blotting was run at least twice on separate occasions.
RNA extraction and Northern blot analysis.
soxS expression in the clinical E. coli strains was detected by Northern blot analysis. Briefly, total RNA was extracted from bacterial cultures, which were grown at 35°C to an A530 of 0.4 to 0.5, by using a modified hot acidic phenol extraction method (Sigma-Genosys Biotechnologies, Inc., The Woodlands, Tex.). The concentration of total RNA was determined spectrophotometrically at 260 nm. Samples of total RNA (5 μg/lane) were fractionated by electrophoresis on a 1.2% formaldehyde agarose gel and transferred to a nylon membrane (Hybond-N; Amersham Life Sciences, Inc.) overnight (48).
A 344-bp PCR fragment containing the complete soxS coding sequence was amplified from AG100 chromosomal DNA with the SoxS primer pair (Sigma-Genosys) according to the manufacturer's instructions. The PCR-amplified fragment was gel purified, radiolabeled with [32P]dCTP (New England Nuclear), and hybridized to the membrane-bound RNA overnight at 65°C, essentially as described by Barbosa and Levy (6). RNA from the strain with soxRS deleted (DJ901) and from the soxS-overexpressing strain (JTG1078) served as negative and positive controls, respectively (17, 18).
Effect of cloned acrR on AcrA expression.
Bacterial strains containing pHRP/acrR bearing the wild-type acrR gene or the strain alone were grown to an A600 of 0.6 to 0.8, cells were harvested, and total proteins were fractionated by SDS-PAGE, Western blotted, and hybridized with anti-AcrA antibody (see above). Densitometric analysis of the blot was assessed using the National Institutes of Health Image program (http://rsb.info.nih.gov/hih-image/manual/tech.html#analyze). Negative (AG100A) and positive (AG100B) control strains for acrA were included.
RESULTS
Antibiotic and organic solvent susceptibility.
Thirty fluoroquinolone-resistant E. coli isolates were obtained from a variety of clinical specimens from 13 different wards in one hospital, including samples of urine (nine samples), tissue drainage and abscess (seven samples), blood (six samples), and vaginal swabs (three samples). Each E. coli strain demonstrated high-level multidrug resistance (resistance to three or more structurally unrelated antibiotics) (Table 1). All 30 strains were highly resistant to ampicillin, and a large majority of the strains were resistant to tetracycline (28 of 30), chloramphenicol (23 of 30), gentamicin (23 of 30), ticarcillin-clavulanic acid (27 of 30), and piperacillin-tazobactam (24 of 30). Most strains were susceptible to cefotaxime, although four strains were highly resistant. All strains were resistant to ciprofloxacin (MIC, >32 μg/ml) and norfloxacin (MIC, ≥21 μg/ml) (Table 1).
All strains grew on LB agar overlaid with 99% hexane (log Pow, 3.9). However, only 19 of the 30 strains were able to grow in a mixture of 3:1 hexane:cyclohexane (log Pow, 3.4), and of these, only two strains (CH5 and CH29) could tolerate a 1:1 hexane:cyclohexane mixture (Table 3). These 19 strains were designated organic solvent tolerant (OST). None of the strains was able to grow in >50% cyclohexane.
TABLE 3.
Characterization of fluoroquinolone-resistant E. coli strainsa
| Strain | Topoisomerase mutations
|
AcrA overexpression | OSTb | ||
|---|---|---|---|---|---|
| GyrA | ParC | GyrBc | |||
| CH1 | S83L D87N | S80I | ND | − | − |
| CH2 | S83L D87N | S80I E84G | WT | − | − |
| CH3 | S83L D87N A93T | S80I | ND | + | + |
| CH4 | S83L D87N | S80I A108V | ND | − | − |
| CH5 | S83L D87N | S80I | ND | + | + |
| CH6 | S83L D87N | S80I | WT | + | + |
| CH7 | S83L D87N | S80I A108V | ND | + | + |
| CH8 | S83L D87N | S80I | ND | − | − |
| CH9 | S83L D87N A93T | E84K | ND | + | + |
| CH10 | S83L D87N | S80I E84G | WT | + | + |
| CH11 | S83L D87N A93T | S80I | ND | + | + |
| CH12 | S83L D87N | E84K | ND | + | + |
| CH13 | S83L D87N | S80I | ND | − | − |
| CH14 | S83L D87Y A93T | S80I | ND | − | − |
| CH15 | S83L D87N | S80I | ND | + | + |
| CH16 | S83L D87Y A93S | S80I | ND | − | − |
| CH17 | S83L D87N | S80I | ND | + | + |
| CH18 | S83L D87N | S80I | WT | − | − |
| CH19 | S83L D87N | E84K | ND | + | + |
| CH20 | S83L D87Y | S80I | ND | + | + |
| CH21 | S83L D87N A93T | S80I A108V | ND | + | + |
| CH22 | S83L D87N | S80I | ND | − | − |
| CH23 | S83L D87N | E84K | ND | + | + |
| CH24 | S83L D87N | S80I | WT | + | + |
| CH25 | S83L D87N | S80I | ND | − | + |
| CH26 | S83L D87N | E84K | ND | + | + |
| CH27 | S83L D87N | S80I | WT | + | + |
| CH28 | S83L D87N | S80I E84G | ND | − | − |
| CH29 | S83L D87N | S80I | ND | + | + |
| CH30 | S83L D87N | S80I | ND | + | + |
None of the strains showed overexpression of MarA or SoxS.
OST in hexane:cyclohexane (3:1).
WT, wild-type; ND, not determined.
Identification of mutations in DNA gyrA, gyrB, and parC.
DNA sequencing of the 268-bp PCR product covering the entire QRDR of gyrA demonstrated the presence of mutations at codons 83 and 87 in all 30 of the isolates when compared to wild-type E. coli K-12. A third mutation in gyrA was also noted in six isolates (Table 3). In every case, the mutation at codon 83 was a C→T transversion in the codon TCG, resulting in the substitution of leucine for serine. For 27 strains, a mutation at codon 87 (G→A transversion of codon GAC) resulted in an asparagine substitution for an aspartate. In the remaining three strains, Asp-87 was replaced by Tyr (G→T transversion of codon GAC). Isolates CH3, CH9, CH11, CH14, and CH21 contained a third mutation, Ala-93 Thr substitution due to a G→A transversion at codon GCG, whereas one strain (CH16) substituted a Ser for Ala at position 93 (GCG→TCG). Of note, 23 of 30 isolates had the same nucleotide changes at positions 255 (CAG→CAA), 273 (GCG→GCA), 300 (TAT→TAC), and 333 (AGA→AGG), none of which resulted in amino acid substitutions.
Analogous to the Ser-83 Leu substitution in gyrA, 83% of the isolates (25 of 30) contained a mutation (G→T) at codon 80 in the QRDR of parC, resulting in the substitution of isoleucine for serine (Table 3). The remaining five isolates demonstrated a Glu-84 Lys replacement (analogous to Asp-87 in gyrA). Among the 25 strains with a Ser-80 Ile substitution, six isolates had additional mutations: three had an Ala-108 Val substitution, and three had a Glu-84 Gly substitution.
The QRDR of the gyrB gene (204-bp fragment) was amplified from four strains with norfloxacin MICs of >200 μg/ml (CH2, CH6, CH10, CH27), as well as from one strain for which the MIC of norfloxacin was 19.4 μg/ml (CH24). No mutations were found (Table 3).
Expression of marA and soxS.
Overexpression of MarA protein was not observed in any of the 30 strains of E. coli by Western blot analysis, while MarA overexpression was clearly identified in the control Mar mutant strain, AG112 (data not shown). Likewise, Northern blot analysis was unable to detect the overexpression of soxS RNA in any of the clinical strains, while the 400-bp soxS-hybridizing band was easily detected in the control strain, JTG1078 (data not shown).
Genetic analysis of acrR.
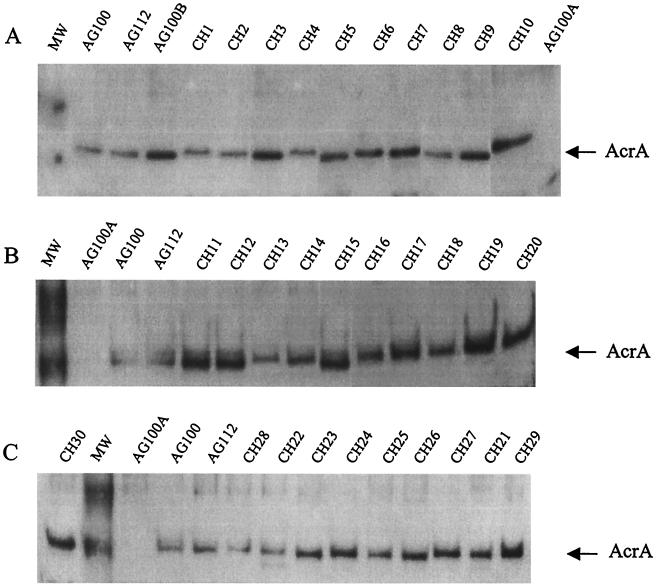
As measured by Western blot analysis, AcrA protein was overexpressed in 19 of the 30 strains of E. coli (Table 3 and Fig. 1), compared to control (AG100) levels and the Mar mutant AG112. All 19 of these strains were tolerant to organic solvents and, with the exception of five strains (CH15, CH17, CH19, CH21, and CH24), all demonstrated norfloxacin MICs of ≥100 μg/ml (Table 1 and 2). In contrast, none of the remaining 11 strains lacking AcrA overexpression could grow in the presence of organic solvents.
FIG. 1.
Western blot analysis of AcrA prepared from clinical E. coli strains, separated by SDS-PAGE, blotted, and developed by chemiluminescence. AG100B (AcrR mutant, AcrA overexpressed) was used to determine the location of AcrA; AG100A (acrAB deleted, no AcrA expression) was a negative control; and AG100 (wild-type expression of AcrA) was used to assess the relative amount of AcrA above wild-type levels. AG112 (a MarR mutant) was included to examine AcrA levels in a constitutive MarA background. The arrow points to the 50-kDa AcrA protein. MW = molecular mass markers of 62 and 51 kDa. Western analysis was performed at least twice for each series.
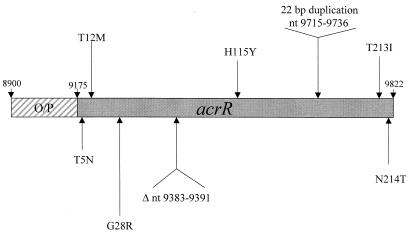
Strains demonstrating overexpression of AcrA (but not MarA or soxS) were selected for evaluation of acrR (the repressor/regulator for acrAB, the genes encoding the major multidrug efflux pump). PCR amplification of the complete acrR gene was performed on eight clinical strains overproducing AcrA. DNA sequencing revealed that four isolates contained point mutations (CH5, CH10, CH15, and CH29) (Table 4). In addition, CH19 had a 9-bp deletion and CH27 had a 22-bp duplication. The mutations were random throughout the repressor and were not localized to one region (Fig. 2 and Table 4). CH24 had three silent mutations. Silent mutations were also noted in all but two strains tested. Strain CH21, in which AcrA was elevated, had no mutations in the repressor or the operator-promoter region for acrR (Fig. 2).
TABLE 4.
Mutations in acrR
| Isolate | AcrA expression | DNA position | Codon substitution | No. of silent mutationsa |
|---|---|---|---|---|
| CH5 | + | 9188 (ACC→AAC) | T5N | 3 |
| CH10 | + | 9209 (ACG→ATG) | T12M | 4 |
| 9812 (ACT→ATT) | T213I | |||
| 9815 (AAC→ACC) | N214T | |||
| CH15 | + | 9516 (CCA→CTA) | H115Y | 2 |
| CH19 | + | Δ 9 nt (9383–9391)b | IGEL (70–73)→Y | |
| CH21 | + | None | 0 | |
| CH24 | + | None | 3 | |
| CH27 | + | 22-nt duplication (9715–9736) | FAPQSFDL (78–86) repeated, creating a frameshift | 1 |
| CH29 | + | 9256 (GGG→AGG) | G28R | 0 |
Changes in nucleotide sequence, with no change in amino acid.
nt, nucleotide.
FIG. 2.
Mutations within acrR, the repressor of the acrAB locus. The operator-promoter region is indicated by a hatched box, and nucleotide locations are indicated by the arrows.
Complementation studies were undertaken in these eight strains to determine if wild-type AcrR protein would restore antibiotic susceptibility and, in addition, lower the expression of AcrA. Only five of the eight strains accepted the complementing plasmid. Sensitivity to the tested antibiotic was partially restored when the clinical strain was complemented with the wild-type acrR gene on pHRP/acrR (Table 5). Resistance to chloramphenicol decreased by 2- to 3.5-fold, and tetracycline resistance decreased 1.3- to 3-fold. Norfloxacin resistance decreased 1.5- to 6-fold. Kanamycin and gentamicin resistances were not affected by the AcrAB efflux pump, as expected (47).
TABLE 5.
Restoration of sensitivity to select antibiotics in clinical E. coli strains complemented with wild-type acrR
| Strain | MIC (μg/ml)a
|
||
|---|---|---|---|
| Tetracycline | Chloramphenicol | Norfloxacin | |
| CH5 | 83 | 253 | 194 |
| CH5(pHRPacrR) | 27 | 73 | 138 |
| CH10 | 258 | 258 | 250 |
| CH10(pHRPacrR) | 81 | 103 | 78 |
| CH19 | ND | ND | 51 |
| CH19(pHRPacrR) | ND | ND | 18 |
| CH27 | 44 | 8 | 236 |
| CH27(pHRPacrR) | 20 | 2 | 146 |
| CH29 | 75 | 275 | 117 |
| CH29(pHRPacrR) | 56 | 80 | 21 |
All susceptibilities were determined using gradient plates. ND, not done, since these strains were already relatively sensitive to tetracycline and chloramphenicol.
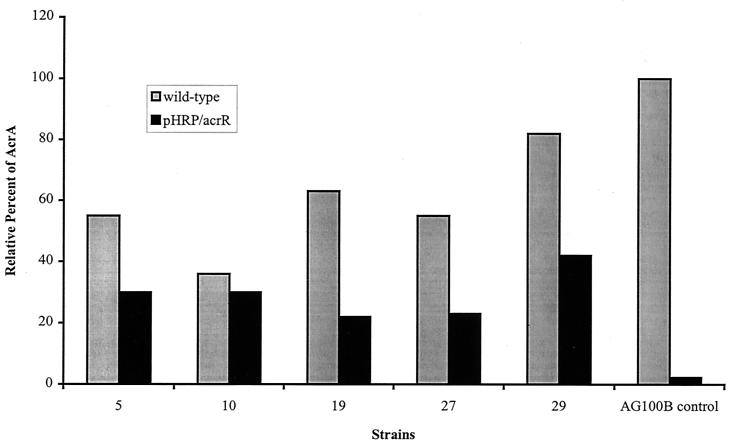
AcrA protein expression was examined in the complemented strains by Western blot analysis and compared to its isogenic clinical strain by densitometry. In every case, AcrA was reduced in the complemented strain compared to its isogenic parent (Fig. 3), showing that the wild-type AcrR supplanted the host mutant AcrR and that the region regulating acrAB in the host was susceptible to wild-type AcrR.
FIG. 3.
Effect of wild-type AcrR on the expression of AcrA protein. Hatched bars indicate AcrA expression in the clinical E. coli strains relative to that in AG100B (mutant in AcrR), the positive control. Solid bars denote AcrA expression in each isogenic strain when complemented with the wild-type acrR on pHRP/acrR. AG100B (acrR mutant) served as the positive control.
DISCUSSION
High-level fluoroquinolone resistance in E. coli has become a major problem in China and other countries. In the present study, 30 clinical E. coli isolates from a hospital in China showed multiple chromosomal mutations, including mutations in the topoisomerase genes, gyrA and parC, a finding in accord with other studies of fluoroquinolone-resistant E. coli (8, 19–21, 46, 50). While silent mutations in gyrA suggest some strains may be related, the variation of silent mutations in acrR and the different phenotypes and mutant genotypes indicate that more than one clone is involved.
Point mutations in GyrA of Ser-83 to either Tyr or Leu (both of which convert the polar amino acid serine to a nonpolar amino acid [13, 22, 41, 58, 60]), or a double mutation of Ser-83 Leu and Asp-87 Gly (53), are the most frequent bases for fluoroquinolone resistance in clinical E. coli isolates. In addition, mutations in ParC at Ser-80, Gly-78, and Glu-84 (corresponding to Ser-83, Gly-81, and Asp-87 of GyrA, respectively) have also been noted (27). Higher levels of quinolone resistance (MICs, 8 to 64 μg/ml) in E. coli result from double mutations in gyrA (Ser-83 Leu and Asp-87 Gly) with a single parC mutation (Ser-80 or Glu-84) (5, 15, 18, 19, 23, 24, 52). The highest level of resistance generally results from four mutations: two in gyrA and two in parC (52). Such findings are supported by studies of sequential mutations of gyrA in laboratory strains (4, 24), which correlate higher levels of resistance with increasing numbers of mutations.
In the present study, all 30 clinical strains of E. coli had at least three mutations in the target genes; all shared the common mutations at Ser83 and Asp87 in GyrA. Most notably, 11 strains had more than three mutations (10 strains with four mutations, and five mutations in 1 strain) (Table 3). This number of multiple mutations from individual clinical strains has not been reported before and undoubtedly contributes to the high resistance to fluoroquinolones observed (Table 1). Importantly, mutations affecting Ala-93 in gyrA and Ala-108 in parC have not been described previously.
Although less common, mutations in the QRDR of gyrB have been found by others (34, 46, 57, 58), but no mutations were found in gyrB in the isolates studied here. A study by Everett et al. (15) examining high-level fluoroquinolone resistance in E. coli isolates from humans and animals was also unable to detect mutations in either gyrB or parE.
Besides target gene mutations (gyrAB, parC/E), studies have shown that high-level fluoroquinolone resistance can be influenced, at least in part, by mutations in one or more of the known global regulator loci (24), such as marA (9, 10, 16), soxS (2, 56), and robA (3, 33). Mutations in the repressors of these loci lead to overexpression of the transcriptional activator marA or soxS (36). When overexpressed, MarA or SoxS decreases the synthesis of OmpF porin (responsible for outer membrane permeability of low-molecular-weight hydrophilic molecules, including many antibiotics) (11) and increases the expression of the multidrug efflux pump, AcrAB/TolC (40), ultimately resulting in increased resistance to fluoroquinolones and other structurally unrelated antibiotics. In the present study, we were unable to detect overexpression of either MarA protein or soxS RNA, although 63% of the strains (19 of 30) were resistant to organic solvents and overexpressed AcrA (Table 2). Therefore, another mechanism for AcrA overexpression was involved. The AcrAB efflux pump has been associated with increased OST (55). Studies by Kern et al. (24) and Oethinger et al. (36) found that while half of high-level fluoroquinolone-resistant E. coli clinical strains were OST, only a proportion of the strains overexpressed marA or soxS genes. The acrAB locus was not examined.
We looked for possible mutations in acrR, the regulator of acrAB, that might be the basis for increased AcrA expression. Of eight strains tested, six demonstrated different amino acid substitutions, deletions, or duplications in the AcrR repressor. When the six were complemented with a wild-type repressor, AcrA levels decreased, in most cases, by half. Of interest were the two strains (CH21 and CH24) where no mutations could be found in the acrR gene or the acrAB promoter/operator region, yet they overexpressed AcrA, were OST, and were highly resistant to multiple antimicrobials, including the fluoroquinolones. That other unidentified genes may be involved in the up-regulation of AcrA has been reported in an earlier study in which general stress signals were able to regulate the acrAB operon lacking a functional repressor (29). In that study, deletion of marRAB or soxRS had little effect on the transcription of acrAB, demonstrating that up-regulation of acrAB expression was not mediated by the known global regulators, MarA and SoxS. By utilizing gel mobility shift assays, Ma and colleagues suggested that a factor other than AcrR was able to bind to the promoter region in response to global stress conditions (29). This unknown factor, or another, may be operational in CH21 and CH24. While our work was under review, another group (30) reported the overexpression of AcrA in clinical E. coli isolates, but the genetic basis, e.g., mutations in acrR or in a regulatory locus, was not described. This present study demonstrates that acrR gene mutations can now be included with other known chromosomal mutations to explain high-level fluoroquinolone resistance in clinical strains of E. coli.
ACKNOWLEDGMENTS
We thank Vincent Perreten and Teresa Barbosa for their advice and intellectual input into this study and Laura McMurry for suggestions and critical reading. We also thank Xie Xiuli and Xu Yingchun and their group for collecting and identifying the clinical E. coli strains.
This work was supported in part by a grant from the U.S. National Institutes of Health (GM 51661) and a travel grant from the American Society for Microbiology (H.W.).
REFERENCES
- 1.Aguiar J M, Chacon J, Canton R, Baquero F. The emergence of highly fluoroquinolone-resistant Escherichia coli in community-acquired urinary tract infections. J Antimicrob Chemother. 1992;29:349–350. doi: 10.1093/jac/29.3.349. [DOI] [PubMed] [Google Scholar]
- 2.Amábile-Cuevas C F, Demple B. Molecular characterization of the soxRS genes of Escherichia coli: two genes control a superoxide stress regulon. Nucleic Acids Res. 1991;19:4479–4484. doi: 10.1093/nar/19.16.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ariza R R, Li Z, Ringstad N, Demple B. Activation of multiple antibiotic resistance and binding of stress-inducible promoters by Escherichia coli Rob protein. J Bacteriol. 1995;177:1655–1661. doi: 10.1128/jb.177.7.1655-1661.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachoual R, Tankovic J, Soussy C J. Analysis of mutations involved in fluoroquinolone resistance of in vivo and in vitro mutants of Escherichia coli. Microb Drug Resist. 1998;4:271–276. doi: 10.1089/mdr.1998.4.271. [DOI] [PubMed] [Google Scholar]
- 5.Bagel S, Hüllen V, Wiedemann B, Heisig P. Impact of gyrA and parC mutations on quinolone resistance, doubling time, and supercoiling degree of Escherichia coli. Antimicrob Agents Chemother. 1999;40:868–875. doi: 10.1128/aac.43.4.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbosa T M, Levy S B. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J Bacteriol. 2000;182:3467–3474. doi: 10.1128/jb.182.12.3467-3474.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blumberg H M, Rimland D, Carroll D J, Terry P, Wachsmuth I K. Rapid development of ciprofloxacin resistance in methicillin-susceptible and -resistant Staphylococcus aureus. J Infect Dis. 1991;163:1279–1285. doi: 10.1093/infdis/163.6.1279. [DOI] [PubMed] [Google Scholar]
- 8.Cambau E, Gutmann L. Mechanisms of resistance to quinolones. Drugs. 1993;45(Suppl. 3):5–23. doi: 10.2165/00003495-199300453-00005. [DOI] [PubMed] [Google Scholar]
- 9.Cohen S P, Hächler H, Levy S B. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J Bacteriol. 1993;175:1484–1492. doi: 10.1128/jb.175.5.1484-1492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen S P, McMurry L M, Hooper D C, Wolfson J S, Levy S B. Cross-resistance to fluoroquinolone in multiple-antibiotic-resistant (Mar) Escherichia coli selected by tetracycline or chloramphenicol: decreased drug accumulation associated with membrane changes in addition to OmpF reduction. Antimicrob Agents Chemother. 1989;33:1318–1325. doi: 10.1128/aac.33.8.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen S P, McMurry L M, Levy S B. marA locus causes decreased expression of OmpF porin in multiple-antibiotic-resistant (Mar) mutants of Escherichia coli. J Bacteriol. 1988;170:5416–5422. doi: 10.1128/jb.170.12.5416-5422.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conrad S, Oethinger M, Kaifel K, Klotz G, Marre R, Kern W V. gyrA mutations in high-level fluoroquinolone-resistant clinical isolates of Escherichia coli. J Antimicrob Chemother. 1996;38:443–455. doi: 10.1093/jac/38.3.443. [DOI] [PubMed] [Google Scholar]
- 13.Cullen M E, Wyke A W, Kuroda R, Fisher L M. Cloning and characterization of a DNA gyrase A gene from Escherichia coli that confers clinical resistance to 4-quinolones. Antimicrob Agents Chemother. 1989;33:886–894. doi: 10.1128/aac.33.6.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Curiale M S, Levy S B. Two complementation groups mediate tetracycline resistance determined by Tn10. J Bacteriol. 1982;151:209–215. doi: 10.1128/jb.151.1.209-215.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Everett M J, Jin Y F, Ricci V, Piddock L J V. Contributions of individual mechanisms to fluoroquinolone resistance in 36 strains of Escherichia coli isolated from humans and animals. Antimicrob Agents Chemother. 1996;40:2380–2386. doi: 10.1128/aac.40.10.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.George A M, Levy S B. Amplifiable resistance to tetracycline, chloramphenicol, and other antibiotics in Escherichia coli: involvement of a non-plasmid-determined efflux of tetracycline. J Bacteriol. 1983;155:531–540. doi: 10.1128/jb.155.2.531-540.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenberg J T, Chou J H, Monach P A, Chou J H, Demple B. Activation of oxidative stress genes by mutations at soxQ/cfxB/marA locus in Escherichia coli. J Bacteriol. 1991;173:4433–4439. doi: 10.1128/jb.173.14.4433-4439.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenberg J T, Monach P A, Chou J H, Josephy P D, Demple B. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:6181–6185. doi: 10.1073/pnas.87.16.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heisig P. Genetic evidence for a role of parC mutation in the development of high-level fluoroquinolone resistance in Escherichia coli. Antimicrob Agents Chemother. 1996;40:879–885. doi: 10.1128/aac.40.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heisig P, Schedletzky H, Falkenstein-Paul H. Mutations in the gyrA gene of a highly fluoroquinolone-resistant clinical isolate of Escherichia coli. Antimicrob Agents Chemother. 1993;37:696–701. doi: 10.1128/aac.37.4.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heisig P, Tschorny R. Characterization of fluoroquinolone-resistant mutants of Escherichia coli selected in vitro. Antimicrob Agents Chemother. 1994;38:1284–1291. doi: 10.1128/aac.38.6.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hooper D C. Quinolone mode of action—new aspects. Drugs. 1993;45(Suppl. 3):8–14. doi: 10.2165/00003495-199300453-00004. [DOI] [PubMed] [Google Scholar]
- 23.Hooper D C. Mechanisms of fluoroquinolone resistance. Drug Resist Updates. 1999;2:38–55. doi: 10.1054/drup.1998.0068. [DOI] [PubMed] [Google Scholar]
- 24.Kern W V, Oethinger M, Jellen-Ritter A S, Levy S B. Non-target gene mutations in the development of fluoroquinolone resistance in Escherichia coli. Antimicrob Agents Chemother. 2000;44:814–820. doi: 10.1128/aac.44.4.814-820.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khodursky A B, Zechiedrich E L, Cozzarelli N R. Topoisomerase IV is a target of quinolones in Escherichia coli. Proc Natl Acad Sci USA. 1995;92:11801–11805. doi: 10.1073/pnas.92.25.11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kresken M, Wiedemann B. Development of resistance to nalidixic acid and the fluoroquinolones after the introduction of norfloxacin and ofloxacin. Antimicrob Agents Chemother. 1988;32:1285–1288. doi: 10.1128/aac.32.8.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumagai Y, Kato J, Hoshino K, Akasaka T, Sato K, Ikeda H. Mutants of Escherichia coli DNA topoisomerase IV parC gene. Antimicrob Agents Chemother. 1996;40:710–714. doi: 10.1128/aac.40.3.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Ma D, Alberti M, Lynch C, Nikaido H, Hearst J E. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol Microbiol. 1996;19:101–112. doi: 10.1046/j.1365-2958.1996.357881.x. [DOI] [PubMed] [Google Scholar]
- 30.Mazzariol A, Tokue Y, Kanegawa T M, Cornaglia G, Nakaido H. High-level fluoroquinolone-resistant clinical isolates of Escherichia coli overproduce multidrug efflux protein AcrA. Antimicrob Agents Chemother. 2000;44:3441–3443. doi: 10.1128/aac.44.12.3441-3443.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDermott P F, White D G, Podglajen I, Alekshun M N, Levy S B. Multidrug resistance following expression of the Escherichia coli marA gene in Mycobacterium smegmatis. J Bacteriol. 1998;180:2995–2998. doi: 10.1128/jb.180.11.2995-2998.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muder R R, Brennen C, Goetz A M, Wagener M M, Rihs J D. Association with prior fluoroquinolone therapy of widespread ciprofloxacin resistance among gram-negative isolates in a Veterans Affairs medical center. Antimicrob Agents Chemother. 1991;35:256–258. doi: 10.1128/aac.35.2.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakajima H, Kobayashi K, Kobayashi M, Asako H, Aono R. Overexpression of the robA gene increases organic solvent tolerance and multiple antibiotic and heavy metal ion resistance in Escherichia coli. Appl Environ Microbiol. 1995;61:2302–2307. doi: 10.1128/aem.61.6.2302-2307.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakamura S, Nakmura M, Kojima T, Yoshida H. gyrA and gyrB mutations in quinolone-resistant clinical isolates of Escherichia coli. Antimicrob Agents Chemother. 1989;33:254–255. doi: 10.1128/aac.33.2.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oethinger M, Conrad S, Kaifel K, Cometta A, Belle J, Klotz G, Glauser M P, Marre R, Kern W V The International Antimicrobial Therapy Cooperative Group of the European Organization for Research and Treatment of Cancer. Molecular epidemiology of fluoroquinolone-resistant Escherichia coli bloodstream isolates from patients admitted to European cancer centers. Antimicrob Agents Chemother. 1996;40:387–392. doi: 10.1128/aac.40.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oethinger M, Kern W V, Goldman J D, Levy S B. Association of organic solvent tolerance and fluoroquinolone resistance in clinical isolates of Escherichia coli. J Antimicrob Chemother. 1998;41:111–114. doi: 10.1093/jac/41.1.111. [DOI] [PubMed] [Google Scholar]
- 37.Oethinger M, Kern W V, Jellen-Ritter A S, McMurry L M, Levy S B. Ineffectiveness of topoisomerase mutations in mediating clinically significant fluoroquinolone resistance in Escherichia coli in the absence of the AcrAB efflux pump. Antimicrob Agents Chemother. 2000;44:10–13. doi: 10.1128/aac.44.1.10-13.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oethinger M, Podglajen I, Kern W V, Levy S B. Overexpression of the marA or soxS regulatory gene in clinical topoisomerase mutants of Escherichia coli. Antimicrob Agents Chemother. 1998;42:2089–2094. doi: 10.1128/aac.42.8.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogle J W, Reller L B, Vasil M L. Development of resistance in Pseudomonas aeruginosa to imipenem, norfloxacin, and ciprofloxacin during therapy: proof provided by typing with a DNA probe. J Infect Dis. 1988;157:743–748. doi: 10.1093/infdis/157.4.743. [DOI] [PubMed] [Google Scholar]
- 40.Okusu H, Ma D, Nikaido H. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J Bacteriol. 1996;178:306–308. doi: 10.1128/jb.178.1.306-308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oram M, Fisher L M. 4-Quinolone resistance mutations in the DNA gyrase of Escherichia coli clinical isolates identified by using the polymerase chain reaction. Antimicrob Agents Chemother. 1991;35:387–389. doi: 10.1128/aac.35.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parales R E, Harwood C S. Construction and use of a new broad-host-range lacZ transcriptional fusion vector, pHRP309, for Gram-negative bacteria. Gene. 1993;133:23–30. doi: 10.1016/0378-1119(93)90220-w. [DOI] [PubMed] [Google Scholar]
- 43.Park Y H, Yoo J H, Huh D H, Cho Y K, Choi J H, Shin W S. Molecular analysis of fluoroquinolone-resistance in Escherichia coli. On the aspect of gyrase and multiple antibiotic resistance (mar) genes. Yonsie Med J. 1998;39:534–540. doi: 10.3349/ymj.1998.39.6.534. [DOI] [PubMed] [Google Scholar]
- 44.Pea C, Albareda J M, Pallares R, Pujol M, Tubau F, Ariza J. Relationship between quinolone use and emergence of ciprofloxacin-resistant Escherichia coli in bloodstream infections. Antimicrob Agents Chemother. 1995;39:520–524. doi: 10.1128/aac.39.2.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peterson L R. Quinolone resistance in clinical practice: occurrence and importance. In: Hooper D C, Wolfson J S, editors. Quinolone antimicrobial agents. 2nd ed. Washington, D.C.: American Society for Microbiology; 1993. pp. 119–137. [Google Scholar]
- 46.Quabdesselam S, Hooper D C, Tankovic J, Soussy C J. Detection of gyrA and gyrB mutations in quinolone-resistant clinical isolates of Escherichia coli by single-strand conformational polymorphism analysis and determination of levels of resistance conferred by two different single gyrA mutations. Antimicrob Agents Chemother. 1995;39:1667–1670. doi: 10.1128/aac.39.8.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosenberg E Y, Ma D, Nikaido H. AcrD of Escherichia coli is an aminoglycoside efflux pump. J Bacteriol. 2000;182:1754–1756. doi: 10.1128/jb.182.6.1754-1756.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 49.Tanaka M, Nakayama H, Haraoka M, Saika T, Kobayashi I, Naito S. Antimicrobial resistance of Neisseria gonorrhoeae and high prevalence of ciprofloxacin-resistant isolates in Japan, 1993 to 1998. J Clin Microbiol. 2000;38:521–525. doi: 10.1128/jcm.38.2.521-525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tavío M del M, Vila J, Ruiz J, Martín-Sánchez A M, de Anta M T J. Mechanisms involved in the development of resistance to fluoroquinolone in Escherichia coli. J Antimicrob Chemother. 1999;44:735–742. doi: 10.1093/jac/44.6.735. [DOI] [PubMed] [Google Scholar]
- 51.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vila J, Ruiz J, Goñi P, de Anta M T J. Detection of mutations in parC in quinolone-resistant clinical isolates of Escherichia coli. Antimicrob Agents Chemother. 1996;40:491–493. doi: 10.1128/aac.40.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vila J, Ruiz J, Marco F, Barcelo A, Goín P, Giralt E, de Anta T J. Association between double mutations in gyrA gene of ciprofloxacin-resistant clinical isolates of Escherichia coli and MICs. Antimicrob Agents Chemother. 1994;38:2477–2479. doi: 10.1128/aac.38.10.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang H, Xie X, Xu Y, Zhang X, Ren B, Chen C. Resistant rates of Gram-negative bacilli to 12 antibiotics by using Etest. Chin J Med Lab Sci. 1997;20:265–267. [Google Scholar]
- 55.White D G, Goldman J D, Demple B, Levy S B. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J Bacteriol. 1997;179:6122–6126. doi: 10.1128/jb.179.19.6122-6126.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu J, Weiss B. Two divergently transcribed genes, soxR and soxS, control a superoxide response regulon of Escherichia coli. J Bacteriol. 1991;173:2864–2871. doi: 10.1128/jb.173.9.2864-2871.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamagishi J, Yoshida H, Yamayoshi M, Nakamura S. Nalidixic acid-resistant mutations of the gyrB gene of Escherichia coli. Mol Gen Genet. 1986;204:367–373. doi: 10.1007/BF00331012. [DOI] [PubMed] [Google Scholar]
- 58.Yoshida H, Bogaki M, Nakamura M, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob Agents Chemother. 1990;34:1271–1272. doi: 10.1128/aac.34.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoshida H, Bogaki M, Nakamura M, Yamanaka L M, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrB gene of Escherichia coli. Antimicrob Agents Chemother. 1991;35:1647–1650. doi: 10.1128/aac.35.8.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoshida H, Kojima T, Yamagishi J, Nakamura S. Quinolone resistant mutations of the gyrase gyrA gene of Escherichia coli. Mol Gen Genet. 1988;211:1–7. doi: 10.1007/BF00338386. [DOI] [PubMed] [Google Scholar]
- 61.Zhang X, Wang H, Hu Y, Xie X, Xu Y, Chen M, Zhou G, Wu J, Xu S, Zhang S, Wang Q, Du X. Susceptibility of 5374 isolates to 18 antibiotics from 5 hospitals in Beijing. Natl Med J China. 1997;77:543–544. [Google Scholar]