Abstract
One of the most important health concerns in society is the development of pathogen-causing nosocomial infections. Since the first discovery of antibiotics, bacterial infections have been highly treatable. However, with evolution and the nondiscretionary usage of antibiotics, pathogens have also found new ways to survive the onslaught of antibiotics by surviving intracellularly or through the formation of obstinate biofilms, and through these, the outcomes of regular antibiotic treatments may now be unsatisfactory. Lipid-coated hybrid nanoparticles (LCHNPs) are the next-generation core–shell structured nanodelivery system, where an inorganic or organic core, loaded with antimicrobials, is enveloped by lipid layers. This core–shell structure, with multifarious decorations, not only improves the loading capabilities of therapeutics but also has the potential to improve therapeutic delivery, especially for targeting biofilm-based and intracellular bacterial infections. Although there has been significant interest in the development of LCHNPs, they have yet to be widely exploited for bacterial infections. In this review, we will provide an overview on the latest development of LCHNPs and the various approaches in synthesizing this nano-delivery system. In addition, a discussion on future perspectives of LCHNPs, in combination with other novel anti-bacterial technologies, will be provided towards the end of this review.
Lipid-coated hybrid nanoparticles are next-generation core–shell structured nanodelivery systems, which improve the loading capabilities of therapeutics and can improve therapeutic delivery, especially for targeting biofilm-based and intracellular bacterial infections.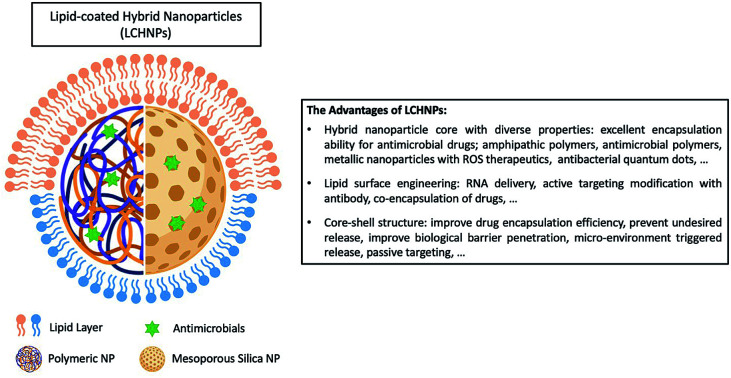
1. Introduction
Microbial infection is one of the biggest threats to global public health today due to the rapid evolution of antibiotic resistant strains.1–3 Increasingly, multidrug resistant pathogenic strains of bacteria are now routinely isolated from all parts of the world.4–7 Pathogenic microorganisms, such as Enterococcus, Staphylococcus and Pseudomonas, are closely related species that are responsible for a wide variety of common bacterial infections. However, the widespread non-discretionary usage of antibiotics over the last 70 years has driven the emergence and prevalence of bacteria that exhibit drug resistant characteristics. This makes infectious diseases, which were once easily treatable, fatal again. In the United States alone, antibiotic resistance has led to more than two million infections and about 23 000 deaths each year, resulting in $20 million of excess medical spending and $35 billion in lost productivity annually.8–10 In fact, it was recently projected that antibiotic resistant pathogens would cause more than 10 million deaths worldwide per year by 2050.11 For instance, the eradication of certain bacterial infections, such as methicillin-resistant Staphylococcus aureus (MRSA), still remains a challenge. Often, this is attributed not only to the complex pathogenic mechanisms in subverting the host's immune system, but also the cellular barriers and the formation of a biofilm that prevents antibiotics from reaching the foci of infection.12,13 Without doubt, addressing drug resistant and tolerant microbes is now a major global health challenge, and this calls for an urgent need to revisit approaches that can effectively control bacterial infections, either through novel formulations, delivery systems or through the discovery of more effective next generation antibiotics.
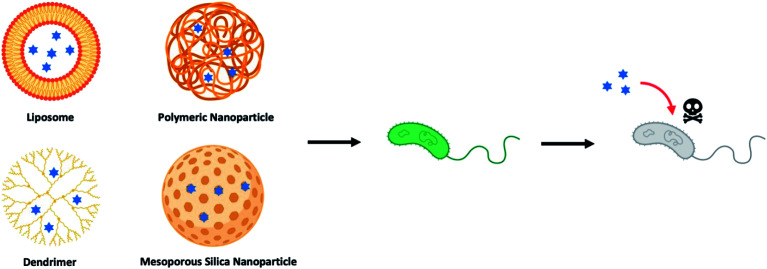
Over the last few decades, drug-loaded nanoparticles, as therapeutic delivery platforms, have been extensively developed and evaluated to overcome the inherent shortcomings of current antibiotic treatments.14–19 By protecting and delivering the antibiotic to the intended site of action, such delivery platforms are hypothesized to enhance effectiveness of the antibiotics, while potentially mitigating the emergence of drug-tolerant microbes. Of the myriad nano-delivery systems that have been reported for this purpose, most efforts have revolved around the use of liposomes,20–22 polymeric nanoparticles,23–25 dendrimers,26–28 and inorganic nanoparticles (Fig. 1).29–31 These studies have demonstrated that antimicrobial molecules loaded into such delivery nanoparticles, either through physical encapsulation or chemical conjugation, exhibited an improved pharmacokinetic profile and therapeutic index, as compared to their free drug counterparts. Nevertheless, there are also drawbacks with delivery systems, especially those that are composed of a single material (non-hybrid), such as low drug loading efficiency, premature drug release, and rapid clearance of these nanoparticles mediated by the mononuclear phagocyte system (MPS).32,33 To overcome these challenges, hybrid nanoparticles that tap on the advantages of two or more material systems are therefore preferred. For instance, a hybrid nanoparticle may consist of a core polymer that encapsulates the drug, while its surface may be coated or functionalized with another material to avoid the MPS. Such hybrid delivery system, through a coating layer, could also suppress any initial drug release, while allowing for targeting of the infection site.
Fig. 1. Illustration of different types of nanoparticle-based drug delivery system to against bacterial strains (blue hexagrams represent antibiotics and other therapeutic antimicrobials).
One novel progeny of a hybrid delivery system, that has been gaining much attention within the scientific community in the recent years,34–36 is the lipid-coated hybrid nanoparticle (LCHNP). LCHNPs can be described as composing of an organic or inorganic nanoparticulate core that is coated with single or multiple layers of lipids – constituting of simple lipids (i.e. DOTAP), compound lipids (i.e. phospholipids), or derived lipids (i.e. cholesterol). Possessing a core–shell architecture, the LCHNP exhibit promising attributes as a versatile and robust therapeutic, such as a stable structural integrity with good biocompatibility, holding the ability to encapsulate lipophilic and hydrophilic therapeutic agents, can be designed for controlled release, and flexibility for functionalization to suit a specific application.
Earlier scientific reviews on lipid-polymer hybrid nanoparticles chiefly focus on the preparation of LCHNPs, either organic or inorganic cores, and their applications in drug and gene delivery specifically for cancer therapy.37–41 As a contrast, this review aims to provide an up-to-date overview on the fabrication approaches of LCHNPs, before embarking on exploring how LCHNPs potentially can be exploited, as a therapeutic platform, for both intracellular and biofilm-mediated microbial infections. As a conclusion, this review will highlight the challenges and future perspectives of LCHNPs against infectious diseases.
2. Strategies to synthesize LCHNPs
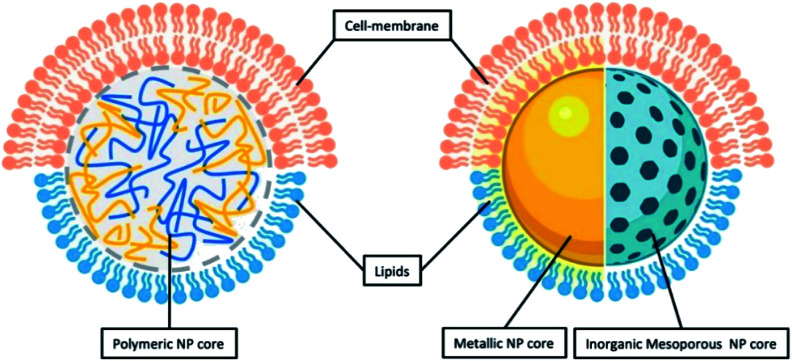
As inferred from its name, LCHNPs consist of two or more material systems as building blocks (Fig. 2), whereby the core is an organic or inorganic nanoparticle that is enveloped by a lipid layer. In some hybrid nanoparticulate systems, a polymeric/inorganic mesoporous core is chosen because of their excellent capabilities in encapsulating antibiotics. In other hybrid systems, a metallic core is preferred because of its other antimicrobial capabilities that are non-drug-related, i.e. ROS-generation, hyperthermia, etc. Payloads (i.e. antibiotics, antimicrobial peptides, RNA, etc.) can be encapsulated or associated in either the core or lipid layer, or, in some cases, both, depending on the construct of the hybrid nanoparticle. The lipid layer behaves as a molecular barrier that mitigates the loss of the entrapped agents (i.e. drugs) during fabrication while protecting the core from degradation in any physiological environment.42 Given the unique structure of this hybrid system, this review shall commence by highlighting some key strategies in the preparation of LCHNPs. In general, fabrication of LCHNPs can be achieved through two distinctive approaches: (1) a multi-step process whereby the nanoparticle core and lipid shell are prepared separately;43–46 or (2) a single-step process.47–50 The latter is often obtained through a one-pot emulsion–solvent–evaporation (ESE) or through a nanoprecipitation method. In this review, only the most widely employed approaches will be discussed, along with several recent representative examples of each fabrication method.
Fig. 2. General illustration of core–shell structure of LCHNP demonstrating either polymeric NP or inorganic NP could be coated with either lipids or cell-membrane.
2.1. Multi-step fabrication approaches
2.1.1. Lipid-coated nanoparticles
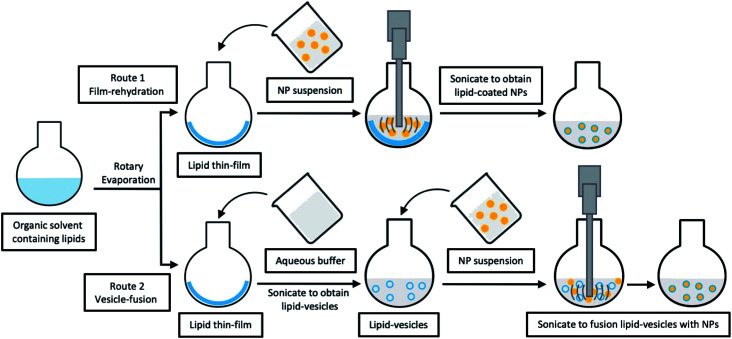
Typically, simple lipid-coated LCNHPs with either organic or inorganic core could be fabricated at a laboratory scale by using a lipid thin-film hydration method. Various studies on LCHNPs using this method are listed in Table 1. For LCHNPs with organic nanoparticulate cores, the two main components are first prepared separately. More specifically, the polymeric core is usually prepared through ESE or the nanoprecipitation methods, while the lipid thin-film is usually prepared by solvent evaporation of a lipid-containing organic solvent in a rotary evaporator. The core–shell structure is then obtained through direct hydration of the lipid thin film within the nanoparticle dispersion (i.e. film-rehydration method). Alternatively, it can also be prepared by fusing the preformed nanoparticles with lipid vesicles, followed by vortexing or sonication to achieve self-assembly of the lipids onto the nanoparticle surface through electrostatic or van der Waals interactions (vesicle-fusion method) (Fig. 3). The final LCHNPs could subsequently be harvested from any excessive, non-adsorbed lipids by centrifugation. For LCHNPs with inorganic cores, coating with lipids could be achieved similarly by using the vesicle-fusion or film-rehydration method to coat bare inorganic nanoparticles, so as to generate a symmetrical lipid bilayer on the inorganic nanoparticle surface.
Fabrication of lipid-coated NPs using two-step method.
| Core | Shell | Loaded agents | Remarks |
|---|---|---|---|
| Fusion of NPs and liposomal vesicles | |||
| PLGA NP | DOTAP | pDNA | The delivery system increased transfection efficiencies compared to free DNA. It is the first time to present NP carrying IL-12 to be efficient in gene delivery to liver cancer cells in the presence of a very high concentration of serum51 |
| CHOL-grafted PAMAM NP | DOTAP, DOPE, CHOL | siRNA | The core–shell nanoparticle carrying siRNA exhibited stronger downregulation of EGFR protein expression level in MCF-7 cells in vitro and the greatest inhibition on tumour growth in vivo44 |
| PLGA NP | DOTAP, CHOL | BSA | The lipid shell not only promoted in vitro cellular uptake of hybrid NPs, improved the stability and protected the integrity of the hybrid structure during long- term storage52 |
| PLGA NP | DOTAP, CHOL | HspX/EsxS protein | This delivery system has a good potential to enhance immune responses against HspX EsxS antigen after subcutaneous immunization of BALB/c mice53 |
| Silver NP | POPS | — | The coating presented excellent inhibition of Gram-positive and Gram-negative bacteria growth and reduced inflammatory response from bone marrow derived macrophages54 |
| Gold NPs | PC, PS | — | LCHNP-based methods allow for highly sensitive detection of protein-induced membrane apposition under conditions that minimize large-scale aggregation55 |
| MgP NP | DOTAP, CHOL | CAT | This LCHNP carrying CAT protected MCF-7 cells from lethal level of exogenous H2O2 and lowered the ROS levels in EA.hy 926 cells56 |
| PS NP | DOTAP, CHOL, DOPC, and DSPE-PEG-2000 | — | The lipid envelopes enable the diffusion of LCHNPs into the mucus layer and promote the interaction of LCHNPs with a bacterial biofilm57 |
| Re-hydration from lipid thin-film | |||
| CS-TPP NP | Egg PL, CHOL and DPPE | Moxifloxacin | The mean residence time, area under the curve, apparent permeability coefficient of LCHNPs were up to 6.74-fold, 4.29-fold, and 3.29-fold higher than commercials45 |
| CS NP | Egg PL, RHL, CHOL and DSPE-PEG-2000 | CLR | The CLR-loaded LCHNP exhibited excellent eradicating ability to H. pylori biofilm58 |
| PLGA NP | DSPC, DSPE-PEG-Mal and CHOL | ATRA | It firstly reported that LCHNP conjugated with CD133 aptamers showed a sustained release of ATRA, presented efficiently and specifically ATRA delivery to osteosarcoma initiating cells, and achieved superior therapeutic efficacy59 |
| MSN | PL and CHOL | HCPT | This HCPT-LCHNP had stronger inhibitory effects on hepatocellular carcinoma cells compared with free HCPT, providing an effective strategy for delivering poorly-soluble anticancer drugs60 |
| MSN | Soybean PL, CHOL and PEG-2000 | PTX, CUR | The co-delivery LCHNP system enabled the intravenous administration of hydrophobic drugs, and sustained released trend showed strong cytotoxic effect against breast cancer cells61 |
Fig. 3. Schematic illustration of typical multi-step fabrications for LCHNPs. Route 1, film-rehydration method, where the core–shell structure is obtained through direct hydration of the lipid thin film within the NP dispersion; route 2, vesicle-fusion method, where the core–shell structure is obtained by fusing the preformed NP into lipid-vesicles.
2.1.2. Cell-membrane coated nanoparticles
Consistent with the idea of fusing nanoparticles with lipid vesicles to form a core–shell structure, another category of LCHNPs using cellular membranes as the lipid coating, has recently also generated interest.62–66 Lipids from cellular membranes constitute 50% of the mass, with phospholipids being the most abundant.67 Membrane lipids typically form bilayers, complexed with membrane proteins and other components. Membrane lipids have thus been exploited to produce cell-membrane coated nanoparticles (i.e. cell-membrane coated LCHNPs). More specifically, these LCHNPs, with the same composition and functional moieties as cells, could easily penetrate biological barriers (i.e. blood–brain barrier), while promoting longer blood circulation half-lives.68,69 More importantly, the use of cellular membrane as a lipid coating, voids the need of subsequent surface functionalization of the nanoparticles, e.g. conjugation of ligands, as cell membranes already possess functional moieties from the original cell.62,70
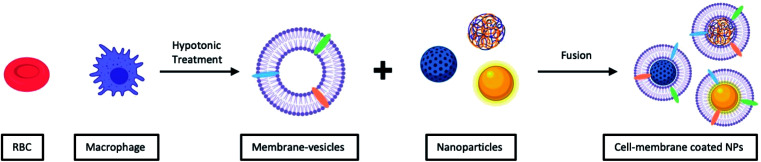
Unlike vesicle-fusion and film-rehydration methods, the preparation of cell-membrane coated nanoparticles involves an additional step of extracting membrane vesicles from cells (Fig. 4).71 Another point for consideration is that cell membrane extraction procedures are highly dependent on the cell source.72 For instance, to obtain bioactive cell membrane from nucleus-free cells (e.g. Red Blood Cells), the cells have to be first isolated from whole blood, which are then lysed with either a hypotonic treatment or a repeated freeze-thaw process, before extruding through polycarbonate membranes using nanosized pores to obtain purified vesicles. It is noted that the extraction and purification of membrane vesicles from eukaryotes (e.g. leukocytes), compared with nucleus-free cells, are more complicated. The harvested cells are then lysed through hypotonic treatment combined with mechanical membrane destruction, followed by the removal of the intracellular components (e.g. nucleus) by discontinuous sucrose gradient centrifugations. The final ideal vesicles are finally obtained by washing and extrusion. A list of different nanoparticulate cores coated with cell-membrane from various source cells is shown in Table 2. From this list, this recently developed strategy has demonstrated to be effective for surface functionalization of nanoparticles.
Fig. 4. Typical preparation of cell-membrane coated NPs. Cell-membrane-derived vesicles are collected from either nucleus-free cells or eukaryotes and then coated onto various types of NPs.
Fabrication of cell-membrane coated NPs.
| Core | Source cell | Loaded agents | Remarks |
|---|---|---|---|
| PLGA NP | Red blood cell | DOX | The LCHNP efficiently deliver DOX to solid tumour sites for significantly increased tumour growth inhibition compared with conventional free drug treatment73 |
| PLGA NP | Platelets | RAP | The plates-coated NP displayed 4.98-fold greater radiant efficiency than control in atherosclerotic arterial trees, indicating its effective homing to atherosclerotic plaques in vivo. NP loading RAP significantly attenuated the progression of atherosclerosis and stabilized atherosclerotic plaques74 |
| PLGA NP | Gastric epithelial cell (AGS cell) | CLR | The AGS-NPs preferentially accumulate on the H. pylori surfaces in vitro. The CLR-loaded AGS-NPs demonstrate superior therapeutic efficacy in a mouse model of H. pylori infection than free CLR75 |
| PCL and Pluronic F68 NP | 4T1 Breast cancer cell | PTX | The LCHNP achieved specific targeting of homotypic tumours or metastatic nodules in the lung. It further demonstrated a remarkable anti-tumour and metastasis efficacy, not only in the orthotopic transplantation tumour models but also in the advanced metastasis mice models76 |
| PPC8, PPiP NP | Mouse peritoneal macrophage | PTX | The designed macrophage-membrane-coated nanoparticle showed enhanced penetration efficiency on tumour cells, the loaded PTX was quickly released from the nanoparticles in response to the endosome pH. It exhibited an enhanced therapeutic effect inherited from both membrane-derived tumour homing and step-by-step controlled drug release77 |
| PCPDTBT and PEG-b-PPG-b-PEG NP | Activated fibroblast cell | — | The cell-membrane coated NP specifically target cancer-associated fibroblasts, leading to enhanced tumour accumulation. It generates enhanced cytotoxic heat and single oxygen to exert combinational photo-thermal and photo-dynamic therapy78 |
| Gold NP | Escherichia coli | — | The bacterial membrane-coated NPs induced rapid activation and maturation of dendritic cells in the lymph nodes of vaccinated mice. It generated strong Th1 and Th17 biased cell responses against the source bacteria79 |
| Gold nanocage | 4T1 Breast cancer cell | DOX | The LCHNP exhibit a stimuli-release of DOX under the hyperthermia and a high cell-specific targeting of the 4T1 cells in vitro. The therapy with about 98.9% and 98.5% inhibiting rates of the tumour volume and metastatic nodules is observed in the 4T1 orthotopic mammary tumour models80 |
| Gold–silver nanocage | Macrophage | — | The membrane-coated nano-system targets bacteria more efficiently. It retained at the infection site and showed greatly prolonged circulation time and excellent biocompatibility81 |
| MSNC | Macrophage | DOX | The MSNC greatly increase the loading capacity for DOX. The macrophage-coated MSNC not only offer camouflage function, but also provide active targeting ability due to surface proteins82 |
2.2. Singe-step fabrication approaches
Multi-step approaches are limited by the separate preparation of the core nanoparticles and lipid vesicles/cell membrane vesicles, and are likely to be less cost effective. In addition, there is always the possibility of drug leakages, especially for hydrophilic agents, during the assembly process. To improve on these drawbacks, single-step approaches have been improvised to synthesize LCHNPs, of which several of such approaches will be highlighted here.
2.2.1. Emulsification-solvent-evaporation method
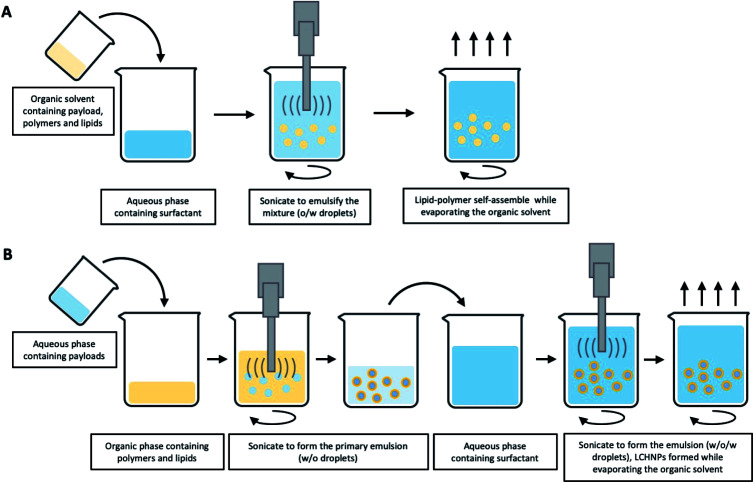
The ESE method, as the most common fabrication method of polymeric nanoparticles, is based on the emulsification of a water-immiscible organic solution in an aqueous phase.83 This process is typically carried out in the presence of surfactants and under high-shear forces. After the formation of an emulsion, evaporation of the organic solvent results in precipitation of the polymer and thus nanoparticle formation. This method can be further sub-classified into single and double emulsification methods,84–86 as illustrated in Fig. 4. An oil-in-water (o/w) single ESE method is suitable for loading hydrophobic agents.87 In this method, the o/w emulsion is formed when the organic phase containing the polymer, the lipid and the payload is mixed with an aqueous phase under ultra-sonication or constant stirring. Depending on their amphiphilicity, the lipids could be either dissolved in the organic or aqueous phase. During evaporation of the organic solvent, the polymer core is formed with the lipids assembling around the polymer core concomitantly (Fig. 5A). For example, in the formulation developed by Bose et al.,88 PLGA and lipids were dissolved in DCM, before adding slowly into an aqueous PVA solution under continuous stirring. The final core–shell structured nanoparticles are then formed with the removal, i.e. evaporation, of the organic solvent. Recently, our group developed a series of LCHNPs loaded with hydrophobic or amphiphilic antibiotics using the single o/w emulsion via ESE method, in which the PLGA nanoparticle was coated with a cationic DOTAP lipid shell.89 On the other hand, water-in-oil-in-water (w/o/w) double emulsions have mostly been used for the entrapment of hydrophilic small molecules, nucleic acids and antibiotics.90 In this case, the aqueous solution of hydrophilic agents was first emulsified with an organic solvent containing polymer and lipid to form a primary w/o emulsion. Next, a w/o/w double emulsion is generated when the primary emulsion is emulsified again with an aqueous phase, followed by subsequent organic solvent evaporation to yield hydrophilic drug-loaded LCHNPs (Fig. 5B).
Fig. 5. Schematic illustration of typical emulsification-solvent-evaporation method for LCHNPs. (A) An o/w single emulsion formulation is suitable for loading hydrophobic agents, where the emulsion is formed when the organic phase (polymer, lipid and payload) is emulsified with an aqueous phase (surfactant). (B) A w/o/w double emulsion is used for the entrapment of the hydrophilic payloads, where the aqueous solution (hydrophilic agents) is firstly emulsified with an organic solvent (polymer and lipid) to form a primary w/o emulsion, then a w/o/w double emulsion is generated when the primary emulsion is emulsified again with an aqueous phase (surfactant).
2.2.2. Nanoprecipitation method
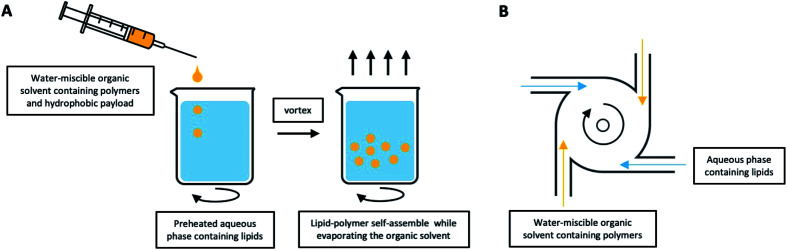
A typical nanoprecipitation method usually requires that the hydrophobic payloads and polymer are dissolved together in a water-miscible organic solvent, with the lipid dissolved in an aqueous phase.91,92 Compared with the aforementioned ESE method, the biggest difference is the need to heat the lipid-containing aqueous solution beyond its gel-to-liquid transition temperature in order to achieve a homogeneously dispersed liquid crystalline phase.41 After dropwise addition of the polymer into the lipid under continuous stirring, the lipids will self-assemble onto the polymer nanoparticles via hydrophobic interactions. The hydrophobic tails of the lipids are attached to the polymer core, while the hydrophilic head is affiliated to the external aqueous surrounding, thus leading to the formation and stabilization of LCHNPs (Fig. 6A). Ahmaditabar et al. synthesized N-Acetyl Cysteine (NAC) loaded LCHNPs using this nanoprecipitation method.93 Here, an acetonitrile solvent containing PLGA and NAC was added dropwise to a preheated 4% ethanol aqueous solution containing lecithin under gentle stirring. Next, the mixture was vortexed vigorously, followed by continuous stirring to evaporate organic solvent and finally obtaining the resulting LCHNPs by centrifugal filtration. As a further development of the nanoprecipitation method in advanced LCHNPs fabrication, macro-scale devices, called Multi-Inlet Vortex Reactors (MIVRs), can be used.94–96 The device can be made with either two or four radially symmetric inlets that lead into a circular reaction chamber (Fig. 6B). Rapid nanoprecipitation, with high production and improved size homogeneity, can be achieved to form polymeric particles with an organic phase containing the polymer, and an aqueous phase acting as the anti-solvent is added into separate inlets. Using this device, Fang et al. demonstrated large-scale reproducible LCHNPs production rate of 10 g per hour.97
Fig. 6. LCHNPs preparation using nanoprecipitation. (A) Schematic illustration of typical nanoprecipitation. The polymer-contained organic solution is dropwise added into the preheated lipid-contained aqueous phase under continuous stirring, the lipids will self-assemble onto the NPs. (B) An improved nanoprecipitation method using MIVRs for large-scale production of LCHNPs.
2.2.3. Chemical conjugation method
For certain lipid-coated inorganic nanoparticles, a chemical synthesis approach is available to introduce the lipid, based on the hydrophobic interaction with a pre-formed coating layer of various compositions to produce a hybrid lipid bilayer. Luchini et al. successfully achieved the reduction of aqueous metallic precursor in the presence of cationic lipids to fabricate lysophosphocholine (LPC)-coated SPIONs.98 Briefly, a cyclohexane dispersion of SPIONs pre-coated with oleic acid and oleylamine molecules was added to an aqueous solution containing LPC molecules, thus producing a biphasic system. Following sonication, cyclohexane was evaporated and the SPIONs were transferred into the aqueous phase where they are stabilized by the LPC coating. Hence, a hybrid lipid bilayer composed of oleic acid and oleylamine inner leaflet and LPC outer leaflet was formed on the SPIONs surface. This approach was further improved to produce stable LPC-coated AuNPs.99 Additionally, using nanoparticle-bound alkanethiols to anchor outer leaflet phospholipids based on the interactions between hydrophobic chains has been another popular route for encapsulating inorganic nanoparticles within lipid layers. It can be exemplified by sequential addition of POPC : POPS (70 : 30) liposomes and propanethiol to freshly synthesize AuNP, to produce a hybrid lipid bilayer in a single step. The thiol groups of the propanethiol molecules form a first coating layer on the AuNP surface, followed by the self-assembling of POPC and POPS phospholipids to form a second amphiphilic layer.55
In summary, the multi-step methods in preparing LCHNPs are more widely reported and are generally employed in lab-scale research. However, the drawbacks in preparing lipids/cell-membrane vesicles and NPs separately, renders this method far from efficient in terms of time and resources and this could possibly limit the clinical translation of these promising LCHNPs for treating infectious diseases. Although the one-step method for preparing LCHNPs overcomes the shortcomings of the multi-step processes, it is still unable to meet the large-scale production requirements for clinical usage.
3. LCHNPs as therapeutic delivery system against bacteria-derived diseases
Bacteria can become resistant or highly tolerant to antibiotics through different mechanisms.100 While antibiotic resistance is typically caused by inherited mutations stemming from diverse molecular mechanisms, non-inherited antibiotic tolerance is induced by phenotypic switching (e.g. reduced growth rate/metabolism) of a subpopulation of bacteria surviving in biofilms and/or within the host cells.101,102 Exploiting host cells as a shield against antibiotic appears to be one of the most effective evasion strategy. In fact, many severe, chronic and recurrent infections are mediated by biofilm bacteria or pathogens that persist intracellularly.103,104 The persistence of intracellular pathogens is not solely due to metabolic dormancy, but that intracellular lifestyle shield the pathogens from antibiotics owing to poor drug penetration and retention, as well as decreased intracellular drug activity.105 Such a tactic is commonly employed by many pathogens responsible for healthcare-associated and community-acquired infections, such as Mycobacterium tuberculosis, Escherichia coli, Listeria monocytogenes, Salmonella enterica, Enterococcus faecalis, Staphylococcus aureus, etc., in both wound and surgical-related infections.103,106,107 This mechanism is particularly relevant since most antibiotics are precluded from entering or are poorly retained by the host cells. Even if compounds do penetrate the host cells, they will either be rapidly degraded by the lysosomal acids or enzymes within the endolysosomes, or their concentration does not reach therapeutic levels for sufficient duration.108,109 When residing in the host cells, intracellular bacteria may also reduce their growth rate to lower their susceptibility to antibiotics.103,110 All these make eradication of intracellular pathogens extremely challenging. Of greater clinically significance is that intracellular pathogens can propagate inside phagocytic (i.e. macrophages, neutrophils, etc.) and non-phagocytic cells (i.e. fibroblasts, enterocytes, keratinocytes, hepatocytes, etc.),108 leading to the spread of infection even during antibiotic therapy.103,111 As such, the shielding effect conferred by the host cells has not only led to numerous chronic and recurrent infections, such as tuberculosis, urinary tract infections, pulmonary infections, endocarditis, atopic dermatitis, etc., but also contributed to the enrichment of antibiotic resistant strains during prolonged antibiotic treatment.104
3.1. Intracellular bacterial infections
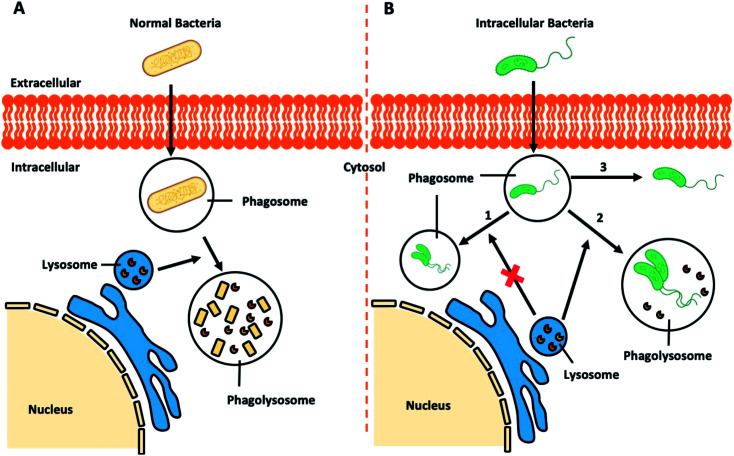
Regular pathogenic bacteria usually would be recognized and engulfed by professional phagocytes (i.e. macrophages), kept in a bubble-like structure – phagosome within the cytoplasm. This bacteria-harbouring phagosome then fuses with a lysosome, becoming a “phagolysosome”, where the lysosome provides enzymes and drastically lowers the internal pH to break down the contents (Fig. 7A).112 However, certain intracellular bacterial pathogens, such as Mycobacterium tuberculosis,113,114Salmonella enterica,115 and Klebsiella pneumoniae,116 develop various defence mechanisms to escape the phagocytosis pathways or remain viable in the phagolysosomes, which is the case for infectious diseases, such as tuberculosis,117 listeriosis,118 and salmonellosis.119
Fig. 7. Schematic illustration of (A) bacteria-involved phagocytosis and (B) resistance mechanisms developed by certain intracellular pathogens.
Recent research demonstrated that Klebsiella pneumoniae can promote the activation of Akt to arrest phagosome maturation, avoiding fusion into lysosomes, thus creating a Klebsiella-containing vacuole to survival intracellularly (Fig. 7B, route 1).120 Similar mechanisms are also observed in the phagocytosis pathways of Salmonella typhimurium and Mycobacterium tuberculosis.121 Previously considered as an extracellular bacterium, Staphylococcus aureus has also been shown to invade and survive in either professional or non-professional phagocytes, including keratinocytes, endothelial cells, epithelial cells, fibroblast and osteoblasts.122–125 The adhesion of Staphylococcus aureus to the host cell surface could lead to cytoskeletal rearrangement, allowing Staphylococcus aureus to move into cells and even replicate within the acidic phagolysosome (Fig. 7B, route 2).126 Other studies report that other bacteria could escape from the phagocytic vacuole into the cytosol.127 In the case of intracellular Listeria monocytogenes infection, pores can be formed in the vacuole membrane to release phospholipases to disrupt the vacuole membrane (Fig. 7B route 3). Once in the cytosol, Listeria monocytogenes will induce the polymerization of host actin filaments and the force generated by actin polymerization allow them to move into neighbouring cells resulting in a productive infection.128 Similar routes to infection is adopted by Shigella flexneri,129,130Salmonella enterica,131 and Mycobacterium tuberculosis.132
Thus, while professional phagocytes not only serve to eradicate intracellular pathogens, ironically it also provides a reservoir of latent infection representing a significant barrier to antibiotic treatment. Phagocytes harbouring such pathogens, acting as “Trojan Horses”, allow those intracellular pathogens to establish secondary infection foci, thus causing recurrent systemic infections.133,134 In such a situation, a delivery system, e.g. LCHNPs, that allows antibiotics to target intracellular infections would serve to be extremely beneficial. Theoretically, LCHNPs can either passively accumulate at the infection foci, or actively target the pathogen and macrophages. More importantly, upon macrophage phagocytosis, the nanoparticle shares the same pathway as pathogens, thus allowing the payloads to be delivered into the infected cells, thereby enhancing the penetration and permitting the release of therapeutics within these cells. To achieve this, the lipid coating on drug-loaded nanoparticles has emerged as an important step to not only promote colloidal stability, and to also provide a means for intracellular delivery of the payload. Diverse LCHNPs have been developed and employed recently for the delivery of therapeutics to intracellular pathogens.
3.1.1. Using lipid-coated inorganic core LCHNPs
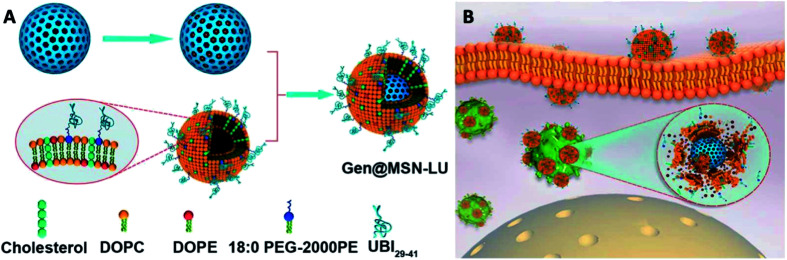
In recent years, mesoporous silica nanoparticle (MSN), due to its high surface area and large pore volume and its versatility to achieve stimuli-responsible drug release, have attracted wide public attention as a drug delivery system. However, the therapeutic efficacy of antibiotic-loaded MSNs is limited by its non-specific targeting and poor cellular permeability. To improve the efficiency of antibiotic delivery and targeting specificity, Yang et al. reported a unique gentamicin-loaded MSNs coated with bacteria toxin-responsive lipid bilayers that is decorated with the bacteria-targeting peptide, UBI29–41.135 Briefly, in their work, MSNs were pre-synthesized by co-condensation using TEOS as silica source, which were then modified with AEPTMS to obtain amine function group. The MSN-NH2 was further mixed with gentamicin by infusing the drug into the pores to yield Gen@MSNs. The liposome was separately prepared with DOPC, DOPE, CHOL and 18 : 0 PEG-2000 PE, using a lipid film-rehydration method and further modified with UBI29–41 (LU). The Gen@MSN-LU was assembled by vesicle-fusion method of these two components (Fig. 8A). The core–shell structure would prevent premature release of gentamicin and its shell, once at the infection foci, will be degraded by bacterial toxins to achieve rapid release of gentamicin (Fig. 8B). In their subsequent antimicrobial studies, the Gen@MSN-LU efficiently targeted Staphylococcus aureus from in vitro studies, and effectively inhibited bacteria growth in an intracellular-infected in vivo mouse model.
Fig. 8. (A) Fabrication route of Gen@MSN-LU. (B) Schematics of possible antimicrobial mechanism in phagocytic cells, where the liposomes outer layer can be degraded by the bacterium-secreted toxins, leading to the gentamicin release (adapted with permission from Yang et al., 2018.135 Copyright © 2018, American Chemical Society).
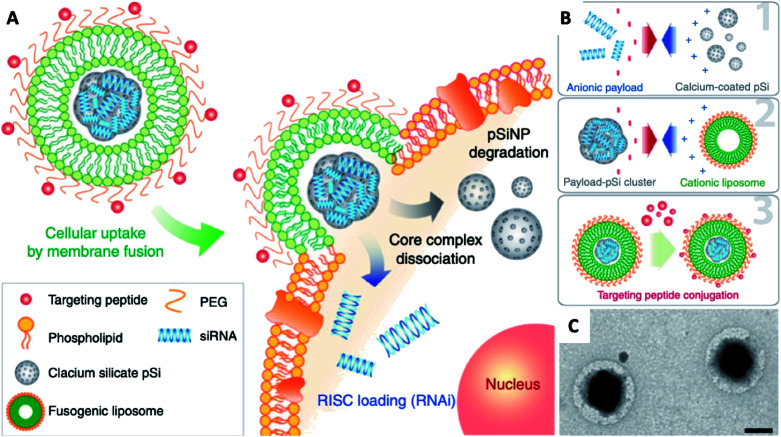
Naked gene therapeutics, such as small interfering RNA (siRNA), are proving to be highly promising but are limited by their short half-lives, and they need to be localized sub-cellularly to be functional. In view of this, a variety of nanoparticle delivery vehicles are explored to protect these oligonucleotides for intracellular delivery. Despite the use of delivery systems, it was reported more than 70% of siRNA payload would still be excreted extracellularly by cells, with most degraded in lysosomes; and typically, only less than 2% of the administered siRNA could escape the early endosomal pathway to potentially undergo RNA interference. In order to increase the quantity of RNA delivered, Kim et al. presented fusogenic liposomal-coated porous silicon nanoparticles, functionalized with a targeting peptide, for siRNA delivery (Fig. 9).136 Prepared with DMPC, DSPE-PEG, and DOTAP, the fusogenic thin-film was rehydrated within an aqueous calcium chloride solution containing pre-synthesized siRNA-loaded porous silicon nanoparticles. The final fusogenic liposomal siRNA-pSiNPs were obtained by subsequent co-extrusion through polycarbonate membrane. The fusogenic liposome, through membrane fusion mechanism, made it possible for hydrophilic payloads siRNA to be released directly from the nanoparticulate core into the cell cytoplasm. This dramatically increased the probability that siRNAs could reach the perinuclear region to undergo RNA interference, by entirely avoiding endocytosis. Their study demonstrated in vivo gene silencing immunotherapy for intracellular infection using a LCHNP delivery system. This macrophage-targeting fusogenic liposomal siRNA-pSiNPs provided a significantly high siRNA knockdown efficiency in vitro, which subsequently showed a strong therapeutic efficacy in terms of full recovery from a lethal dosage against intracellular Staphylococcus aureus infection in mice.
Fig. 9. Fusogenic pSi NP system. (A) Schematic showing mode of action of the fusogenic pSiNP. (B) Schematic showing nanoparticle synthesis, including (1) siRNA encapsulating into MSNs; (2) coating with cationic liposome; and (3) conjugation of targeting peptides. (C) TEM image of final LCHNP constructs (reprinted with permission from Kim et al., 2018.136 Copyright © 2018, Springer Nature).
3.1.2. Using cell membrane-coated organic core LCHNPs
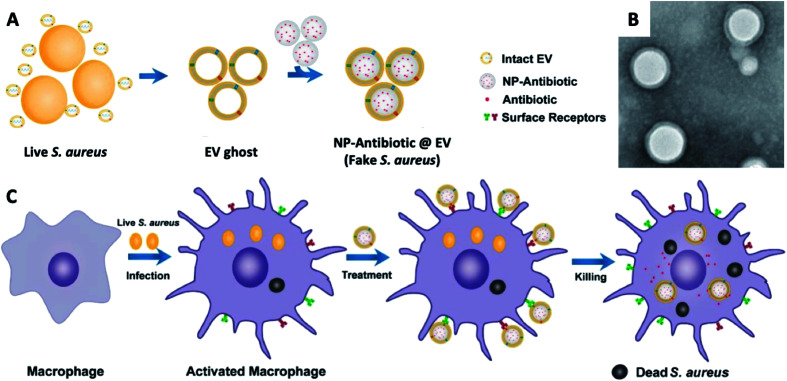
Notably, Gao et al. reported an active-targeting delivery platform established by using cell membrane-coated nanoparticles, which demonstrated promising prospects on intracellular pathogen-associated infections.137 More specifically, it is based on a phagocytosis mechanism of phagocytes that a rapid detection and clearance will be activated when a phagocyte is re-exposed to the same pathogen type. By fusing the membrane vesicles derived from Staphylococcus aureus secreted extracellular vesicles (EVs) with antibiotic preloaded PLGA nanoparticles, they developed membrane-coated polymeric nanoparticles, followed by the utilisation of these LCHNPs as fake Staphylococcus aureus to eliminate intracellular Staphylococcus aureus (Fig. 10). Hence, it can be found that an outer coating with Staphylococcus aureus EVs showed an active targeting property towards Staphylococcus aureus, which was confirmed by an in vitro study. Furthermore, these LCHNPs, when administrated intravenously into Staphylococcus aureus-harbouring mouse models, actively targets infected organs and significantly improved efficacy of the cargo in alleviating Staphylococcus aureus burdens in these organs. Interestingly, when switching the outer coating to Escherichia coli secreted membrane vesicles, these LCHNPs acquired active targeting capabilities to macrophages infected with Escherichia coli, indicating that these membrane-camouflaged nanoparticles were readily adaptable, and can be designed and tuned for specific intracellular pathogens.
Fig. 10. (A) Schematic illustration on the preparation of NP@EV, by fusing the derived membrane vesicle of the extracellular vesicles secreted by Staphylococcus aureus over the surface of NPs. (B) TEM of Staphylococcus aureus – EV coated NPs. (C) Exposure of a macrophage to a Staphylococcus aureus leads to phagocytosis and consequent antigen presentation, and re-exposure of the phagocyte to the same pathogen type (Staphylococcus aureus – EV coated NPs) leads to rapid pathogen detection and clearance (adapted with permission from Gao et al., 2019.137 Copyright © 2019, American Chemical Society).
3.2. Bacterial biofilms
Biofilms refer to a sessile community of bacteria embedded within a matrix of secreted extracellular polymeric substances (EPS), such as polysaccharides, proteins, enzymes and DNA, which are attached on surfaces and the compact EPS layer can protect the bacteria residing within it.138 Furthermore, it not only acts as a barrier to retard the penetration of antibiotics, but also allow pathogens to resist the actions of the host immune system. The failure of existing strategies makes it essential to develop novel treatments to treat biofilm infections. Thus, improving antibiotic delivery is a cornerstone for preventing biofilm-mediated infections. Among the various types of nano-delivery systems, LCHNPs possess huge advantages as the lipid layer presents a charged surface, and protects the payload from either being inactivated by the acidic microenvironment or degraded by extracellular enzymes.
3.2.1. Using lipid-coated organic core LCHNPs
Previously, our group have developed a highly programmable and reproducible LCHNP based on PLGA nanoparticulate core and lipids shell composited with DOTAP and DSPE-PEG5K by using the single emulsion solvent–evaporation method. These LCHNPs, presenting an ideal biofilm penetrating dimension of 100–130 nm, showed a superior affinity to be bound with a diverse bacterial species, including both Gram-positive and -negative pathogens in either planktonic or biofilm state. These LCHNPs were capable of encapsulating various hydrophobic antibiotics with relatively high loading efficiencies, demonstrating a remarkable decrease in MICs as well as a significant increase in biofilm inhibition, thus highlighting the potential of LCHNPs for antimicrobials delivery against bacterial biofilms.89
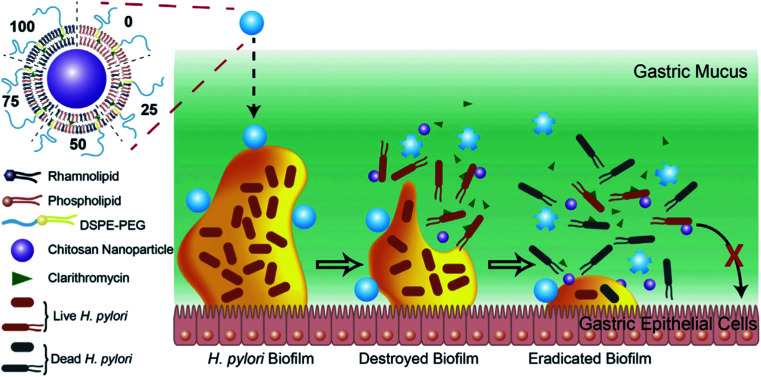
More recently, Li et al. developed a novel LCHNP to overcome both biofilm and mucus layer obstruction (Fig. 11).58 To be more specific, in their work, chitosan nanoparticle, employed as the core, was coated with a mixed lipid layer containing rhamnolipids and clarithromycin as the shell. The core–shell structure was obtained by using a film-rehydration method, followed by further modification with DSPE-PEG2000 to improve hydrophilicity. Combined with rhamnolipids and clarithromycin chitosan, these LCHNPs could disrupt biofilm architecture, remove EPS, prevent reformation of biofilm by effectively inhibiting bacterial adhesion and eliminate pathogens, thus exhibiting an excellent eradicating ability to Helicobacter pylori biofilm.
Fig. 11. Schematic illustration of the structure of LCHNPs and the process of eradicating bacterial Helicobacter pylori biofilm by LCHNPs (reprinted from Li et al., 2019,58 Copyright (2019), with permission from Elsevier).
3.2.2. Using lipid-coated inorganic core LCHNPs
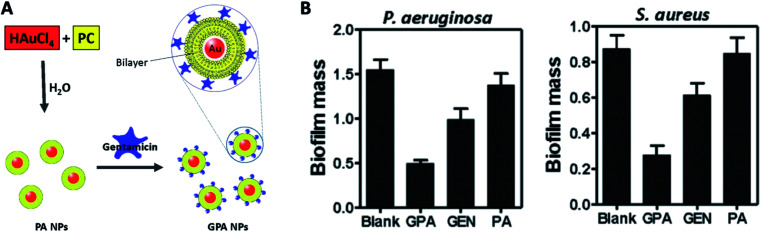
Mu et al. developed a facile approach to prepare phosphatidylcholine-decorated Au nanoparticles loaded with gentamicin.139 In their work, the phosphatidylcholine (PA) NPs were firstly acquired through PA thin-film rehydration with an aqueous solution of HAuCl4. The preformed PA NPs were subsequently loaded with gentamicin (GPA NPs) via mixing with gentamicin sulphate solution (Fig. 12A). These GPA NPs not only presented their antimicrobial effects against planktonic bacteria strains, but demonstrated effective destructive power towards established biofilms and ability to inhibit biofilm formation of both Gram-positive and negative bacteria strains (Fig. 12B). Their results suggested GPA NPs might be a promising antibacterial agent for effective treatment of chronic infections due to microbial biofilm.
Fig. 12. (A) Schematic of the procedure for preparing GPA NPs. (B) Crystal violet assay for biofilm biomass to assess the anti-biofilm activity of GPA against Pseudomonas aeruginosa and Staphylococcus aureus (adapted with permission from Mu et al., 2016.139 Copyright © 2016, Springer Nature).
4. Challenges and perspectives
4.1. Challenges for current nanoparticle formulations
Many of these nano-delivery systems, notably LCHNPs, have demonstrated significantly enhanced bactericidal activity compared to free drugs, particularly against intracellular bacterial strains and biofilms. In addition to delivering existing antibiotics, LCHNPs with inorganic and metallic materials also possess potent antimicrobial properties. However, despite of their outstanding potential, there has not been any clinical translations arising from this technology, not only because of their complex constituents but also the many barriers faced by this nano-delivery system. Some issues that need to be addressed include understanding the interactions of nanoparticles with biological systems, solving the issues of nanoparticle aggregation in physiological fluids, and characterizing these nanoparticles under physiological conditions.
Scalable manufacturing remains as one of the biggest challenges faced by nanoparticle therapies. The scale-up production of single-material systems or formulations, such as liposomes and polymeric nanoparticles loaded with an active pharmaceutical ingredient, have already been successfully achieved and are widely utilized by the pharmaceutical industry. The rapid development of new biomaterials, complex nanostructures and novel synthesis approaches will continue to pose new challenges to the scalable production of these technologies. In recent years, Particle Replication In Non-wetting Templates (PRINT), an advanced unique soft lithography particle moulding process, has been gaining interest.140–143 The PRINT process enables the specific design and large-scale synthesis of well-defined micro- and nanoparticles with uniformed size and shape. In addition, a double usage of spray-drying technique serves as a promising large-scale method to assemble lipids onto nanoparticle surfaces. Accompanied with these advanced techniques, the large-scale production of LCHNPs with acceptable batch-to-batch variation that is free of contamination could be achieved.
Given the usage of LCHNPs in the biological environment, it is therefore of great importance to evaluate its performance under relevant biological conditions. Usually, given limited research resources, in vitro evaluation of delivery systems on cells/bacterial cells is reported. However, in vitro studies by introducing nanoparticles onto planktonic bacteria often lack the complexities of a true biological environment.144 For clinical translation, the use of appropriate animal models to assess in vivo performances, such as tolerability, biodistribution, toxicity, and potential side effects, are often mandatory.145 In studies on bacterial infection, such models are highly diverse, usually employing either patient- or laboratory-derived bacteria strains infected zebrafish, Galleria mellonella larvae and mouse models. However, even these in vivo models do not fully replicate the complexity of a human infection, whereby the reticuloendothelial system (RES) would play a prominent role in the biodistribution of nanoparticles.69,146 For instance, to prolong blood circulation half-lives of nanoparticles to target extracellular pathogens, or to target intracellular bacteria, proper strategies must be adopted in designing ‘stealth’ nanoparticles to be less susceptible to the efficient RES and thus improving targeting capabilities.
4.2. Perspectives of LCHNPs
4.2.1. Active targeting delivery
There are two kinds of targeted drug delivery, passive and active targeting delivery. In passive targeting, the success of drug delivery system is directly related to circulation time of nanoparticles.147 Many studies have proven that by cloaking the nanoparticle surface with PEG, the water molecules have the ability to bind with the oxygen molecules on PEG via hydrogen interaction, resulting in a hydration film surrounding the nanoparticles.148–151 This endows nanoparticles with ‘stealth’ properties, thus anti-phagocytic to be recognized by the MPS. To achieve active targeting, surface modification with antibodies, lipid-coated magnetic nanoparticles, and bacterial membrane camouflaged nanoparticles may be considered for this purpose.
Surface functionalisation of LCHNPs with targeting ligands (i.e. antibodies) has the potential to significantly enhance delivery efficiency.152–154 A key concept in this active targeting strategy for infections is that the pathogens and the LCHNPs tend to accumulate in the same phagocytes, which could be exemplified by the current interest in antibody nanoparticle conjugates (ANCs).155,156 ANCs represent a relatively new approach that builds on the success and potential of both antibody antibiotic conjugates (AACs) and nano-technological delivery systems. AACs, consisting of monoclonal antibodies specifically targeted to pathogens with potent antibiotics, have been successfully developed for the treatment of Staphylococcus aureus infection.111 Conceptually ANCs are similar to AACs in that the antibodies can be used to specifically target pathogens or cells, which could significantly improve the possibilities to deliver encapsulated therapeutic cargoes to the right places.
The diagnostic application of magnetic nanoparticles (MNPs) has now been examined in a range of inflammatory pathologies. For example, Hoerr et al. labelled Staphylococcus aureus with iron oxide nanoparticles before in vivo inoculation, allowing real-time visualisation of the morphology of the infected organs and the resultant host inflammatory response.157 Interestingly, such MNPs with their superior superparamagnetic qualities hold the potential to be manipulated by an external magnetic field and be developed into active delivery systems.158,159 However, their application is strongly limited by the high cytotoxicity which is often associated with MNPs. Lipids are biocompatible molecules with an amphiphilic structure, emerging as perfect candidates to produce nano-biointerfaces by coating the MNPs surface, which improves the MNPs stability toward aggregation and reduces the related cytotoxic effects. More importantly, the lipid outer layers are the best candidates to encapsulate or load amphiphilic antibiotics as a protection to avoid undesired release. Thus, there is strong potential in combining diagnostic function, active targeting, and therapeutic delivery ability together in an antibiotic-loaded lipid-coated MNP formulation, which could be further exploited for pathogen imaging and antimicrobial therapy, known as antimicrobial nano-theranostics. Such proposed nano-theranostic anti-infection strategy is an interesting direction holding the potential to be personalized nanomedicines for pathogenic infections in the future.
4.2.2. Stimuli-responsive LCHNPs
Triggered release of the payloads from nano-formulations is another approach to increase the local transport efficiency of payloads. The feasibility of nanoparticle contributes such stimuli-responsive systems that recognize the micro-environment and react in a dynamic way. In general, there are several stimuli, such as electromagnetic radiation, heat, enzymes, virulence factors or pH changes.160,161 However, this approach is complex, which requires the use of “smart” materials. For now, only a few studies have focused on developing such an antimicrobial nano-formulation. For example, DPPC:CHOL liposomes could be triggered to release amikacin in the presence of rhamnolipids in Pseudomonas aeruginosa biofilms and CF sputum.162 Lipase-sensitive nanogels could be responsive to Staphylococcus aureus for triggered release of vancomycin to significantly inhibit pathogen growth and effectively eliminate intracellular bacteria.163 With LCHNPs, possibilities of developing responsive LCHNPs is to choose material systems that could respond to temperature-, pH-, bacterial toxin-responsive lipids or polymers that can be assembled in shell–core architectures in designing a stimuli-responsive delivery system.
4.2.3. CRISPR-Cas system using LCHNPs
Given the rapid evolution of antibiotic resistance and the lengthy development period for a novel antibiotic, the phenomenon of eliminating microbes using new biotechnologies, such as synthetic antimicrobial peptides and engineered bacteriophages, is gradually emerging. The CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats), a family of DNA sequences derived from bacteria and archaea, represents a unique opportunity to combat antibiotic resistant pathogens.164 More specifically, the CRISPR-Cas (CRISPR-associated) system has been recently applied to design antimicrobials working by targeting specific DNA sequences related to antibiotic resistance, biofilm formation and virulence.165–167 These CRISPR-Cas antimicrobials function by destroying the DNA through chromosomal cleavage by using the RNA-guided nuclease Cas9, selectively killing the target DNA-harbouring bacteria. Although recent studies have proven the effectiveness of CRISPRs, the lack of an efficient delivery vehicle remains to be one of the major barriers for translating CRISPR-Cas system into a suitable antimicrobial agent for human medicine.168 In terms of delivering this payload, a nano-delivery system can be considered. However, CRISPR delivery using nanoparticles is still in its infancy. It requires in-depth researches on the development of safety and efficiency for CRISPR-Cas delivery. By direct conjugation of Cas9 protein to a cationic polymer bPEI, Kang et al. successfully obtained polymer-derivatived Cas9 and subsequently complexed with single-guide RNA (sgRNA) to form a CRISPR nanoparticle, which transported the CRISPR successfully to methicillin-resistant Staphylococcus aureus and maintained the specific activity of Cas9 endonuclease to achieve gene-editing effect,169 thus demonstrating a much higher efficiency compared with using unmodified Cas9/sgRNA complex. In this aspect, the efficiency could be further enhanced with the use of LCHNPs instead of using conventional polymers. We postulate that by employing specific lipids as coatings, it is possible to deliver the Cas9 protein directly to the infection site, owing to the affinity that these lipids may have for the bacteria. This would result in a highly specific therapeutic system for combating multidrug-resistant bacterial infections.
4.2.4. Interference of biofilm-associated signalling pathways using LCHNPs
Disrupting signalling pathways within biofilms is a strategy rapidly gaining traction for the development of anti-biofilm strategies.170 Such signalling pathways include: (i) homoserine-lactone (HSL) quorum-sensing (QS) signalling;170 (ii) cyclic dimeric guanosine monophosphate (c-Di-GMP);171 and (iii) tetra- and penta-phosphate guanosines signalling network ((p)ppGpp).172 Interference with HSL signalling, used in combination with antibiotics, has been shown to successfully treat Pseudomonas aeruginosa biofilms in vivo. For instance, Christensen et al. demonstrated the synergistic effects of tobramycin and QS inhibitors on Pseudomonas aeruginosa biofilm infection mouse model.173 This combination was able to achieve significant reduction in the number of colony forming units (CFUs) as compared to using monotherapies of just tobramycin or QS inhibitors. The authors postulated that in their system, the QS inhibitors compromise the biofilm structure, resulting in enhanced susceptibility to antibiotics.
To successfully develop LCHNPs as anti-biofilm therapeutics, it is essential to understand the roles that each signalling pathway plays in the biofilm environment. The c-Di-GMP and (p)ppGPP signalling pathways are hypothesized to play an important role in promoting biofilm formation, virulence, antibiotic and stress resistances.170–173 However, there remain more to be uncovered on the exact mechanisms of these signalling pathways and how to disrupt these pathways. Moreover, different pathogenic strains may differ in signalling pathways that are dominant in regulating biofilm formation and antibiotic resistance. In this aspect, LCHNPs have the potential to act as carriers to deliver anti-biofilm compounds, such as the QS inhibitors mentioned earlier, to the biofilm infection site.
From these perspectives, we can definitely predict numerous opportunities for LCHNPs in antimicrobial therapies, and hopefully these new approaches will bring the LCHNPs closer to the clinic. In conclusion, challenges faced by this novel delivery system remain; but advances in manufacturing and a more detailed understanding of their antimicrobial properties will promote LCHNPs to be a feasible proposition for the treatment of seemingly untreatable pathogen-causing infections in the near future.
List of abbreviations
- Akt
Protein kinase B
- ATRA
All-trans retinoic acid
- BSA
Bovine serum albumin
- CAT
Catalase
- CFU
Colony formed units
- CHOL
Cholesterol
- CLR
Clarithromycin
- CRISPR
Clustered regularly interspaced short palindromic repeats
- CS
Chitosan
- CUR
Curcumin
- DMPC
14:0 PC; 1,2-dimyristoyl-sn-glycero-3-phosphocholine
- DOPC
18:1 PC; 1,2-dioleoyl-sn-glycero-3-phosphocholine
- DOPE
18:1 PE; 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine
- DOTAP
18:1 TAP; 1,2-dioleoyl-3-trimethylammonium-propane
- DOX
Doxorubicin
- DSPC
18:0 PC; 1,2-distearoyl-sn-glycero-3-phosphocholine
- DSPE
18:0 PE; 1,2-distearoyl-sn-glycero-3-phosphorylethanolamine
- EPR
Enhanced permeation and retention
- ESE
Emulsion-solvent-evaporation
- EVs
Extracellular vesicles
- HCPT
Hydroxycamptothecin
- LCHNPs
Lipid-coated hybrid nanoparticles
- LPC
Lysophosphocholine
- MgP
Magnesium phosphate
- MIVRs
Multi-inlet vortex reactors
- MNPs
Magnetic nanoparticles
- MPS
Mononuclear phagocyte system
- MRSA
Methicillin-resistant Staphylococcus aureus
- MSN
Mesoporous silica nanoparticle
- MSNC
Mesoporous silica nanocapsules
- NAC
N-Acetyl cysteine
- NP
Nanoparticle
- PA
Phosphatidylcholine
- PAMAM
Poly(amidoamine)
- PCL
Poly(caprolactone)
- PCPDTBT
Poly(cyclopentadithiophenealt-benzothiadiazole)
- PE
Phosphatidylethanolamine
- PEG
Polyethylene glycol
- PPG
Poly(propylene glycol)
- PEI
Polyethylenimine
- PL
Phospholipid
- PLGA
Poly(d,l-lactide-co-glycolide)
- POPC
16:0-18:1 PC; 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPS
16:0-18:1 PS; 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine
Particle replication in non-wetting templates
- PS
Polystyrene
- PTX
Paclitaxel
- RAP
Rapamycin
- RES
Reticuloendothelial system
- RHL
Rhamnolipid
- TPP
Tripolyphosphate sodium
Conflicts of interest
The authors declare no competing interests.
Supplementary Material
Acknowledgments
The authors would like to acknowledge the financial support from the Singapore Centre for Environmental Life Sciences Engineering (SCELSE) (MOE/RCE: M4330019.C70), Ministry of Education AcRF-Tier 1 grant (RG19/18), Agri-Food & Veterinary Authority of Singapore (APF LCK102), Biomedical Research Council (BMRC) – Therapeutics Development Review (TDR-G-004-001), NTU-HSPH grant (NTU-HSPH 17002), and the Bill and Melinda Gates Foundation (OPP1199116).
References
- Prestinaci F. Pezzotti P. Pantosti A. Antimicrobial resistance: a global multifaceted phenomenon. Pathog. Glob Health. 2015;109(7):309–318. doi: 10.1179/2047773215Y.0000000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventola C. L. The antibiotic resistance crisis: part 1: causes and threats. P & T. 2015;40(4):277–283. [PMC free article] [PubMed] [Google Scholar]
- Podolsky S. H. The evolving response to antibiotic resistance (1945–2018) Palgrave Commun. 2018;4(1):124. doi: 10.1057/s41599-018-0181-x. [DOI] [Google Scholar]
- Baker S. Thomson N. Weill F. X. Holt K. E. Genomic insights into the emergence and spread of antimicrobial-resistant bacterial pathogens. Science. 2018;360(6390):733–738. doi: 10.1126/science.aar3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. Antibiotic resistance breakers: can repurposed drugs fill the antibiotic discovery void? Nat. Rev. Drug Discovery. 2015;14(12):821–832. doi: 10.1038/nrd4675. [DOI] [PubMed] [Google Scholar]
- Exner M. Bhattacharya S. Christiansen B. Gebel J. Goroncy-Bermes P. Hartemann P. Heeg P. Ilschner C. Kramer A. Larson E. Merkens W. Mielke M. Oltmanns P. Ross B. Rotter M. Schmithausen R. M. Sonntag H.-G. Trautmann M. Antibiotic resistance: What is so special about multidrug-resistant Gram-negative bacteria? GMS Hyg. Infect. Control. 2017;12:Doc05. doi: 10.3205/dgkh000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhondoro M. Ndlovu N. Bangure D. Juru T. Gombe N. T. Shambira G. Nsubuga P. Tshimanga M. Trends in antimicrobial resistance of bacterial pathogens in Harare, Zimbabwe, 2012–2017: a secondary dataset analysis. BMC Infect. Dis. 2019;19(1):746. doi: 10.1186/s12879-019-4295-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B. Webster T. J. Bacteria antibiotic resistance: New challenges and opportunities for implant-associated orthopedic infections. J. Orthop. Res. 2018;36(1):22–32. doi: 10.1002/jor.23656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevention C. f. D. C. a., Antibiotic Resistance Threats in the United States, 2019. Services, U. S. D. o. H. a. H., Atlanta, 2019 [Google Scholar]
- Nelson D. W. Moore J. E. Rao J. R. Antimicrobial resistance (AMR): significance to food quality and safety. Food Qual. Saf. 2019;3(1):15–22. doi: 10.1093/fqsafe/fyz003. [DOI] [Google Scholar]
- O'Neill J., Review on Antimicrobial Resistance Antimicrobial Resistance: Tackling a crisis for the health and wealth of nations, https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf
- Gebreyohannes G. Nyerere A. Bii C. Sbhatu D. B. Challenges of intervention, treatment, and antibiotic resistance of biofilm-forming microorganisms. Heliyon. 2019;5(8):e02192. doi: 10.1016/j.heliyon.2019.e02192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S. Singh S. K. Chowdhury I. Singh R. Understanding the Mechanism of Bacterial Biofilms Resistance to Antimicrobial Agents. Open Microbiol. J. 2017;11:53–62. doi: 10.2174/1874285801711010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S. Joo J. Kang J. Kim B. Braun G. B. She Z. G. Kim D. Mann A. P. Molder T. Teesalu T. Carnazza S. Guglielmino S. Sailor M. J. Ruoslahti E. Antibiotic-loaded nanoparticles targeted to the site of infection enhance antibacterial efficacy. Nat. Biomed. Eng. 2018;2(2):95–103. doi: 10.1038/s41551-017-0187-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M. Curtis A. Hoskins C. Application of Nanoparticle Technologies in the Combat against Anti-Microbial Resistance. Pharmaceutics. 2018;10(1):11. doi: 10.3390/pharmaceutics10010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra J. K. Das G. Fraceto L. F. Campos E. V. R. Rodriguez-Torres M. d. P. Acosta-Torres L. S. Diaz-Torres L. A. Grillo R. Swamy M. K. Sharma S. Habtemariam S. Shin H.-S. Nano based drug delivery systems: recent developments and future prospects. J. Nanobiotechnol. 2018;16(1):71. doi: 10.1186/s12951-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista P. V. McCusker M. P. Carvalho A. Ferreira D. A. Mohan N. M. Martins M. Fernandes A. R. Nano-Strategies to Fight Multidrug Resistant Bacteria-“A Battle of the Titans”. Front. Microbiol. 2018;9:1441. doi: 10.3389/fmicb.2018.01441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemeg H. A. Nanomaterials for alternative antibacterial therapy. Int. J. Nanomed. 2017;12:8211–8225. doi: 10.2147/IJN.S132163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. Hu C. Shao L. The antimicrobial activity of nanoparticles: present situation and prospects for the future. Int. J. Nanomed. 2017;12:1227–1249. doi: 10.2147/IJN.S121956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catania R. Maguire O. Moore C. Falcone F. H. Chan W. Mantovani G. Stolnik S. Huett A. Functionalised liposomal formulations for delivery of antibiotic agents. Access Microbiology. 2019;1(1A) doi: 10.1099/acmi.ac2019.po0507. doi: 10.1099/acmi.ac2019.po0507. [DOI] [Google Scholar]
- Alhariri M. Azghani A. Omri A. Liposomal antibiotics for the treatment of infectious diseases. Expert Opin. Drug Delivery. 2013;10(11):1515–1532. doi: 10.1517/17425247.2013.822860. [DOI] [PubMed] [Google Scholar]
- Drulis-Kawa Z. Dorotkiewicz-Jach A. Liposomes as delivery systems for antibiotics. Int. J. Pharm. 2010;387(1–2):187–198. doi: 10.1016/j.ijpharm.2009.11.033. [DOI] [PubMed] [Google Scholar]
- Gao W. Chen Y. Zhang Y. Zhang Q. Zhang L. Nanoparticle-based local antimicrobial drug delivery. Adv. Drug Delivery Rev. 2018;127:46–57. doi: 10.1016/j.addr.2017.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radovic-Moreno A. F. Lu T. K. Puscasu V. A. Yoon C. J. Langer R. Farokhzad O. C. Surface Charge-Switching Polymeric Nanoparticles for Bacterial Cell Wall-Targeted Delivery of Antibiotics. ACS Nano. 2012;6(5):4279–4287. doi: 10.1021/nn3008383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheow W. S. and Hadinoto K., Antibiotic Polymeric Nanoparticles for Biofilm-Associated Infection Therapy, in Microbial Biofilms: Methods and Protocols, ed. G. Donelli, Springer New York, New York, NY, 2014, pp. 227–238 [DOI] [PubMed] [Google Scholar]
- Kalomiraki M. Thermos K. Chaniotakis N. A. Dendrimers as tunable vectors of drug delivery systems and biomedical and ocular applications. Int. J. Nanomed. 2016;11:1–12. doi: 10.2217/nnm.15.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintzer M. A. Dane E. L. O'Toole G. A. Grinstaff M. W. Exploiting dendrimer multivalency to combat emerging and re-emerging infectious diseases. Mol. Pharmaceutics. 2012;9(3):342–354. doi: 10.1021/mp2005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omolo C. A. Kalhapure R. S. Agrawal N. Jadhav M. Rambharose S. Mocktar C. Govender T. A hybrid of mPEG-b-PCL and G1-PEA dendrimer for enhancing delivery of antibiotics. J. Controlled Release. 2018;290:112–128. doi: 10.1016/j.jconrel.2018.10.005. [DOI] [PubMed] [Google Scholar]
- Cheng C. A. Deng T. Lin F. C. Cai Y. Zink J. I. Supramolecular Nanomachines as Stimuli-Responsive Gatekeepers on Mesoporous Silica Nanoparticles for Antibiotic and Cancer Drug Delivery. Theranostics. 2019;9(11):3341–3364. doi: 10.7150/thno.34576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. Clemens D. L. Lee B. Y. Dillon B. J. Horwitz M. A. Zink J. I. Mesoporous Silica Nanoparticles with pH-Sensitive Nanovalves for Delivery of Moxifloxacin Provide Improved Treatment of Lethal Pneumonic Tularemia. ACS Nano. 2015;9(11):10778–10789. doi: 10.1021/acsnano.5b04306. [DOI] [PubMed] [Google Scholar]
- Mebert A. M. Aimé C. Alvarez G. S. Shi Y. Flor S. A. Lucangioli S. E. Desimone M. F. Coradin T. Silica core–shell particles for the dual delivery of gentamicin and rifamycin antibiotics. J. Mater. Chem. B. 2016;4(18):3135–3144. doi: 10.1039/C6TB00281A. [DOI] [PubMed] [Google Scholar]
- Gustafson H. H. Holt-Casper D. Grainger D. W. Ghandehari H. Nanoparticle Uptake: The Phagocyte Problem. Nano Today. 2015;10(4):487–510. doi: 10.1016/j.nantod.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. Wang J. Wang Y. Gao H. Wei G. Huang Y. Yu H. Gan Y. Wang Y. Mei L. Chen H. Hu H. Zhang Z. Jin Y. Recent progress in drug delivery. Acta Pharm. Sin. B. 2019;9(6):1145–1162. doi: 10.1016/j.apsb.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas K. M. Shon Y. S. Hybrid lipid-nanoparticle complexes for biomedical applications. J. Mater. Chem. B. 2019;7(5):695–708. doi: 10.1039/C8TB03084G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy S. Vaiyapuri R. Zhang L. Chan J. M. Lipid-coated polymeric nanoparticles for cancer drug delivery. Biomater. Sci. 2015;3(7):923–936. doi: 10.1039/C4BM00427B. [DOI] [PubMed] [Google Scholar]
- Tahir N., Haseeb M. T., Madni M. A., Parveen F., Khan M., Khan S., Jan N. and Khan A., Lipid Polymer Hybrid Nanoparticles: A Novel Approach for Drug Delivery, Role of Novel Drug Delivery Vehicles in Nanobiomedicine, 2019 [Google Scholar]
- Bose R. J. C. Ravikumar R. Karuppagounder V. Bennet D. Rangasamy S. Thandavarayan R. A. Lipid–polymer hybrid nanoparticle-mediated therapeutics delivery: advances and challenges. Drug Discovery Today. 2017;22(8):1258–1265. doi: 10.1016/j.drudis.2017.05.015. [DOI] [PubMed] [Google Scholar]
- Forier K. Raemdonck K. De Smedt S. C. Demeester J. Coenye T. Braeckmans K. Lipid and polymer nanoparticles for drug delivery to bacterial biofilms. J. Controlled Release. 2014;190:607–623. doi: 10.1016/j.jconrel.2014.03.055. [DOI] [PubMed] [Google Scholar]
- Hadinoto K. Sundaresan A. Cheow W. S. Lipid–polymer hybrid nanoparticles as a new generation therapeutic delivery platform: a review. Eur. J. Pharm. Biopharm. 2013;85(3):427–443. doi: 10.1016/j.ejpb.2013.07.002. [DOI] [PubMed] [Google Scholar]
- Mandal B. Bhattacharjee H. Mittal N. Sah H. Balabathula P. Thoma L. A. Wood G. C. Core-shell-type lipid-polymer hybrid nanoparticles as a drug delivery platform. Nanomedicine. 2013;9(4):474–491. doi: 10.1016/j.nano.2012.11.010. [DOI] [PubMed] [Google Scholar]
- Mukherjee A. Waters A. K. Kalyan P. Achrol A. S. Kesari S. Yenugonda V. M. Lipid-polymer hybrid nanoparticles as a next-generation drug delivery platform: state of the art, emerging technologies, and perspectives. Int. J. Nanomed. 2019;14:1937–1952. doi: 10.2147/IJN.S198353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Toni A. M. Habila M. A. Labis J. P. Alothman Z. A. Alhoshan M. Elzatahry A. A. Zhang F. Design, synthesis and applications of core–shell, hollow core, and nanorattle multifunctional nanostructures. Nanoscale. 2016;8(5):2510–2531. doi: 10.1039/C5NR07004J. [DOI] [PubMed] [Google Scholar]
- Chan J. M. Zhang L. Yuet K. P. Liao G. Rhee J. W. Langer R. Farokhzad O. C. PLGA-lecithin-PEG core-shell nanoparticles for controlled drug delivery. Biomaterials. 2009;30(8):1627–1634. doi: 10.1016/j.biomaterials.2008.12.013. [DOI] [PubMed] [Google Scholar]
- Gao L.-Y. Liu X.-Y. Chen C.-J. Wang J.-C. Feng Q. Yu M.-Z. Ma X.-F. Pei X.-W. Niu Y.-J. Qiu C. Pang W.-H. Zhang Q. Core-Shell type lipid/rPAA-Chol polymer hybrid nanoparticles for in vivo siRNA delivery. Biomaterials. 2014;35(6):2066–2078. doi: 10.1016/j.biomaterials.2013.11.046. [DOI] [PubMed] [Google Scholar]
- Liu D. Lian Y. Fang Q. Liu L. Zhang J. Li J. Hyaluronic-acid-modified lipid-polymer hybrid nanoparticles as an efficient ocular delivery platform for moxifloxacin hydrochloride. Int. J. Biol. Macromol. 2018;116:1026–1036. doi: 10.1016/j.ijbiomac.2018.05.113. [DOI] [PubMed] [Google Scholar]
- Mandal B. Bhattacharjee H. Mittal N. Sah H. Balabathula P. Thoma L. A. Wood G. C. Core–shell-type lipid–polymer hybrid nanoparticles as a drug delivery platform. Nanomedicine. 2013;9(4):474–491. doi: 10.1016/j.nano.2012.11.010. [DOI] [PubMed] [Google Scholar]
- Dave V. Yadav R. B. Kushwaha K. Yadav S. Sharma S. Agrawal U. Lipid-polymer hybrid nanoparticles: Development & statistical optimization of norfloxacin for topical drug delivery system. Bioact. Mater. 2017;2(4):269–280. doi: 10.1016/j.bioactmat.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. Su L. Wu C. Wu J. Zhu C. Yuan G. RGD peptide targeted lipid-coated nanoparticles for combinatorial delivery of sorafenib and quercetin against hepatocellular carcinoma. Drug Dev. Ind. Pharm. 2016;42(12):1938–1944. doi: 10.1080/03639045.2016.1185435. [DOI] [PubMed] [Google Scholar]
- Yang X.-Z. Dou S. Wang Y.-C. Long H.-Y. Xiong M.-H. Mao C.-Q. Yao Y.-D. Wang J. Single-Step Assembly of Cationic Lipid–Polymer Hybrid Nanoparticles for Systemic Delivery of siRNA. ACS Nano. 2012;6(6):4955–4965. doi: 10.1021/nn300500u. [DOI] [PubMed] [Google Scholar]
- Zhang M. He J. Zhang W. Liu J. Fabrication of TPGS-Stabilized Liposome-PLGA Hybrid Nanoparticle Via a New Modified Nanoprecipitation Approach: In Vitro and In Vivo Evaluation. Pharm. Res. 2018;35(11):199. doi: 10.1007/s11095-018-2485-3. [DOI] [PubMed] [Google Scholar]
- Díez S. Miguéliz I. Tros de Ilarduya C. Targeted cationic poly(D,L-lactic-co-glycolic acid) nanoparticles for gene delivery to cultured cells. Cell. Mol. Biol. Lett. 2009;14(2):347. doi: 10.2478/s11658-009-0003-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y. Hoerle R. Ehrich M. Zhang C. Engineering the lipid layer of lipid-PLGA hybrid nanoparticles for enhanced in vitro cellular uptake and improved stability. Acta Biomater. 2015;28:149–159. doi: 10.1016/j.actbio.2015.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khademi F. Sahebkar A. Fasihi-Ramandi M. Taheri R. A. Induction of strong immune response against a multicomponent antigen of Mycobacterium tuberculosis in BALB/c mice using PLGA and DOTAP adjuvant. APMIS. 2018;126(6):509–514. doi: 10.1111/apm.12851. [DOI] [PubMed] [Google Scholar]
- Taheri S. Cavallaro A. Christo S. N. Majewski P. Barton M. Hayball J. D. Vasilev K. Antibacterial Plasma Polymer Films Conjugated with Phospholipid Encapsulated Silver Nanoparticles. ACS Biomater. Sci. Eng. 2015;1(12):1278–1286. doi: 10.1021/acsbiomaterials.5b00338. [DOI] [PubMed] [Google Scholar]
- Hamilton D. J. Coffman M. D. Knight J. D. Reed S. M. Lipid-Coated Gold Nanoparticles and FRET Allow Sensitive Monitoring of Liposome Clustering Mediated by the Synaptotagmin-7 C2A Domain. Langmuir. 2017;33(36):9222–9230. doi: 10.1021/acs.langmuir.7b01397. [DOI] [PubMed] [Google Scholar]
- Fang Y. Vadlamudi M. Huang Y. Guo X. Lipid-Coated, pH-Sensitive Magnesium Phosphate Particles for Intracellular Protein Delivery. Pharm. Res. 2019;36(6):81. doi: 10.1007/s11095-019-2607-6. [DOI] [PubMed] [Google Scholar]
- Wan F. Nylander T. Klodzinska S. N. Foged C. Yang M. Baldursdottir S. G. Nielsen H. M. Lipid Shell-Enveloped Polymeric Nanoparticles with High Integrity of Lipid Shells Improve Mucus Penetration and Interaction with Cystic Fibrosis-Related Bacterial Biofilms. ACS Appl. Mater. Interfaces. 2018;10(13):10678–10687. doi: 10.1021/acsami.7b19762. [DOI] [PubMed] [Google Scholar]
- Li P. Chen X. Shen Y. Li H. Zou Y. Yuan G. Hu P. Hu H. Mucus penetration enhanced lipid polymer nanoparticles improve the eradication rate of Helicobacter pylori biofilm. J. Controlled Release. 2019;300:52–63. doi: 10.1016/j.jconrel.2019.02.039. [DOI] [PubMed] [Google Scholar]
- Gui K. Zhang X. Chen F. Ge Z. Zhang S. Qi X. Sun J. Yu Z. Lipid-polymer nanoparticles with CD133 aptamers for targeted delivery of all-trans retinoic acid to osteosarcoma initiating cells. Biomed. Pharmacother. 2019;111:751–764. doi: 10.1016/j.biopha.2018.11.118. [DOI] [PubMed] [Google Scholar]
- Liu X. Li M. Yuan W. Liu Y. Wang Y. Wang Y. Lipid-coated mesoporous silica nanoparticles of hydroxycamptothecin for sustained release and cancer therapy. Pharmazie. 2018;73(8):447–453. [Google Scholar]
- Lin J. Cai Q. Tang Y. Xu Y. Wang Q. Li T. Xu H. Wang S. Fan K. Liu Z. Jin Y. Lin D. PEGylated Lipid bilayer coated mesoporous silica nanoparticles for co-delivery of paclitaxel and curcumin: Design, characterization and its cytotoxic effect. Int. J. Pharm. 2018;536(1):272–282. doi: 10.1016/j.ijpharm.2017.10.043. [DOI] [PubMed] [Google Scholar]
- Fang R. H. Kroll A. V. Gao W. Zhang L. Cell Membrane Coating Nanotechnology. Adv. Mater. 2018;30(23):e1706759. doi: 10.1002/adma.201706759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Z. Hu X. Lu W. Cell membrane-coated nanoparticles for tumor-targeted drug delivery. Sci. China Mater. 2017;60(6):504–510. doi: 10.1007/s40843-016-5163-4. [DOI] [Google Scholar]
- Sherwood J. Sowell J. Beyer N. Irvin J. Stephen C. Antone A. J. Bao Y. Ciesla L. M. Cell-membrane coated iron oxide nanoparticles for isolation and specific identification of drug leads from complex matrices. Nanoscale. 2019;11(13):6352–6359. doi: 10.1039/C9NR01292C. [DOI] [PubMed] [Google Scholar]
- Gao W. Zhang L. Coating nanoparticles with cell membranes for targeted drug delivery. J. Drug Targeting. 2015;23(7–8):619–626. doi: 10.3109/1061186X.2015.1052074. [DOI] [PubMed] [Google Scholar]
- Jin J. Krishnamachary B. Barnett J. D. Chatterjee S. Chang D. Mironchik Y. Wildes F. Jaffee E. M. Nimmagadda S. Bhujwalla Z. M. Human Cancer Cell Membrane-Coated Biomimetic Nanoparticles Reduce Fibroblast-Mediated Invasion and Metastasis and Induce T-Cells. ACS Appl. Mater. Interfaces. 2019;11(8):7850–7861. doi: 10.1021/acsami.8b22309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper G. M., The cell: a molecular approach, ASM Press, Washington, D.C, 2nd edn, 2000 [Google Scholar]
- Cox A. Andreozzi P. Dal Magro R. Fiordaliso F. Corbelli A. Talamini L. Chinello C. Raimondo F. Magni F. Tringali M. Krol S. Jacob Silva P. Stellacci F. Masserini M. Re F. Evolution of Nanoparticle Protein Corona across the Blood–Brain Barrier. ACS Nano. 2018;12(7):7292–7300. doi: 10.1021/acsnano.8b03500. [DOI] [PubMed] [Google Scholar]
- Blanco E. Shen H. Ferrari M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat. Biotechnol. 2015;33(9):941–951. doi: 10.1038/nbt.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk B. T. Zhang L. Cell membrane-camouflaged nanoparticles for drug delivery. J. Controlled Release. 2015;220(Pt B):600–607. doi: 10.1016/j.jconrel.2015.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M. Le W. Mei T. Wang Y. Chen B. Liu Z. Xue C. Cell membrane camouflaged nanoparticles: a new biomimetic platform for cancer photothermal therapy. Int. J. Nanomed. 2019;14:4431–4448. doi: 10.2147/IJN.S200284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y. Su J. Ran W. Zhang P. Yin Q. Zhang Z. Yu H. Li Y. Preparation and Application of Cell Membrane-Camouflaged Nanoparticles for Cancer Therapy. Theranostics. 2017;7(10):2575–2592. doi: 10.7150/thno.20118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk B. T. Fang R. H. Hu C.-M. J. Copp J. A. Thamphiwatana S. Dehaini D. Gao W. Zhang K. Li S. Zhang L. Safe and Immunocompatible Nanocarriers Cloaked in RBC Membranes for Drug Delivery to Treat Solid Tumors. Theranostics. 2016;6(7):1004–1011. doi: 10.7150/thno.14471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y. Huang Z. Liu X. Pang Z. Chen J. Yang H. Zhang N. Cao Z. Liu M. Cao J. Li C. Yang X. Gong H. Qian J. Ge J. Platelet membrane-coated nanoparticle-mediated targeting delivery of Rapamycin blocks atherosclerotic plaque development and stabilizes plaque in apolipoprotein E-deficient (ApoE(-/-)) mice. Nanomedicine. 2019;15(1):13–24. doi: 10.1016/j.nano.2018.08.002. [DOI] [PubMed] [Google Scholar]
- Angsantikul P. Thamphiwatana S. Zhang Q. Spiekermann K. Zhuang J. Fang R. H. Gao W. Obonyo M. Zhang L. Coating Nanoparticles with Gastric Epithelial Cell Membrane for Targeted Antibiotic Delivery against Helicobacter pylori Infection. Adv. Ther. 2018;1(2):1800016. doi: 10.1002/adtp.201800016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H. Su J. Meng Q. Yin Q. Chen L. Gu W. Zhang P. Zhang Z. Yu H. Wang S. Li Y. Cancer-Cell-Biomimetic Nanoparticles for Targeted Therapy of Homotypic Tumors. Adv. Mater. 2016;28(43):9581–9588. doi: 10.1002/adma.201602173. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Cai K. Li C. Guo Q. Chen Q. He X. Liu L. Zhang Y. Lu Y. Chen X. Sun T. Huang Y. Cheng J. Jiang C. Macrophage-Membrane-Coated Nanoparticles for Tumor-Targeted Chemotherapy. Nano Lett. 2018;18(3):1908–1915. doi: 10.1021/acs.nanolett.7b05263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. Zhen X. Lyu Y. Jiang Y. Huang J. Pu K. Cell Membrane Coated Semiconducting Polymer Nanoparticles for Enhanced Multimodal Cancer Phototheranostics. ACS Nano. 2018;12(8):8520–8530. doi: 10.1021/acsnano.8b04066. [DOI] [PubMed] [Google Scholar]
- Gao W. Fang R. H. Thamphiwatana S. Luk B. T. Li J. Angsantikul P. Zhang Q. Hu C.-M. J. Zhang L. Modulating Antibacterial Immunity via Bacterial Membrane-Coated Nanoparticles. Nano Lett. 2015;15(2):1403–1409. doi: 10.1021/nl504798g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H. Su J. Meng Q. Yin Q. Chen L. Gu W. Zhang Z. Yu H. Zhang P. Wang S. Li Y. Cancer Cell Membrane-Coated Gold Nanocages with Hyperthermia-Triggered Drug Release and Homotypic Target Inhibit Growth and Metastasis of Breast Cancer. Adv. Funct. Mater. 2017;27(3):1604300. doi: 10.1002/adfm.201604300. [DOI] [Google Scholar]
- Wang C. Wang Y. Zhang L. Miron R. J. Liang J. Shi M. Mo W. Zheng S. Zhao Y. Zhang Y. Pretreated Macrophage-Membrane-Coated Gold Nanocages for Precise Drug Delivery for Treatment of Bacterial Infections. Adv. Mater. 2018;30(46):1804023. doi: 10.1002/adma.201804023. [DOI] [PubMed] [Google Scholar]
- Xuan M. Shao J. Dai L. He Q. Li J. Macrophage Cell Membrane Camouflaged Mesoporous Silica Nanocapsules for In Vivo Cancer Therapy. Adv. Healthcare Mater. 2015;4(11):1645–1652. doi: 10.1002/adhm.201500129. [DOI] [PubMed] [Google Scholar]
- Wang Y. Li P. Truong-Dinh Tran T. Zhang J. Kong L. Manufacturing Techniques and Surface Engineering of Polymer Based Nanoparticles for Targeted Drug Delivery to Cancer. Nanomaterials. 2016;6(2):26. doi: 10.3390/nano6020026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhand C. Dwivedi N. Loh X. J. Jie Ying A. N. Verma N. K. Beuerman R. W. Lakshminarayanan R. Ramakrishna S. Methods and strategies for the synthesis of diverse nanoparticles and their applications: a comprehensive overview. RSC Adv. 2015;5(127):105003–105037. doi: 10.1039/C5RA19388E. [DOI] [Google Scholar]
- McCall R. L. Sirianni R. W. PLGA nanoparticles formed by single- or double-emulsion with vitamin E-TPGS. J. Visualized Exp. 2013;(82):51015. doi: 10.3791/51015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sah E. Sah H. Recent Trends in Preparation of Poly(lactide-co-glycolide) Nanoparticles by Mixing Polymeric Organic Solution with Antisolvent. J. Nanomater. 2015;2015:22. doi: 10.1155/2015/794601. [DOI] [Google Scholar]
- Kumar M. Bishnoi R. S. Shukla A. K. Jain C. P. Techniques for Formulation of Nanoemulsion Drug Delivery System: A Review. Prev. Nutr. Food Sci. 2019;24(3):225–234. doi: 10.3746/pnf.2019.24.3.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jc Bose R. Lee S. Park H. Lipid polymer hybrid nanospheres encapsulating antiproliferative agents for stent applications. J. Ind. Eng. Chem. 2016 [Google Scholar]
- Baek J.-S. Tan C. H. Ng N. K. J. Yeo Y. P. Rice S. A. Loo S. C. J. A programmable lipid-polymer hybrid nanoparticle system for localized, sustained antibiotic delivery to Gram-positive and Gram-negative bacterial biofilms. Nanoscale Horiz. 2018;3(3):305–311. doi: 10.1039/C7NH00167C. [DOI] [PubMed] [Google Scholar]
- Dima C. Dima S. Water-in-oil-in-water double emulsions loaded with chlorogenic acid: release mechanisms and oxidative stability. J. Microencapsulation. 2018;35(6):584–599. doi: 10.1080/02652048.2018.1559246. [DOI] [PubMed] [Google Scholar]
- Bilati U. Allémann E. Doelker E. Development of a nanoprecipitation method intended for the entrapment of hydrophilic drugs into nanoparticles. Eur. J. Pharm. Sci. 2005;24(1):67–75. doi: 10.1016/j.ejps.2004.09.011. [DOI] [PubMed] [Google Scholar]
- Zhuang J. Fang R. H. Zhang L. Preparation of particulate polymeric therapeutics for medical applications. Small Methods. 2017;1(9) doi: 10.1002/smtd.201700147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmaditabar P. Momtazi-Borojeni A. A. Rezayan A. H. Mahmoodi M. Sahebkar A. Mellat M. Enhanced Entrapment and Improved in Vitro Controlled Release of N-Acetyl Cysteine in Hybrid PLGA/Lecithin Nanoparticles Prepared Using a Nanoprecipitation/Self-Assembly Method. J. Cell. Biochem. 2017;118(12):4203–4209. doi: 10.1002/jcb.26070. [DOI] [PubMed] [Google Scholar]
- Liu Z. Ramezani M. Fox R. O. Hill J. C. Olsen M. G. Flow Characteristics in a Scaled-up Multi-inlet Vortex Nanoprecipitation Reactor. Ind. Eng. Chem. Res. 2015;54(16):4512–4525. doi: 10.1021/ie5041836. [DOI] [Google Scholar]
- Bokare A. Takami A. Kim J. H. Dong A. Chen A. Valerio R. Gunn S. Erogbogbo F. Herringbone-Patterned 3D-Printed Devices as Alternatives to Microfluidics for Reproducible Production of Lipid Polymer Hybrid Nanoparticles. ACS Omega. 2019;4(3):4650–4657. doi: 10.1021/acsomega.9b00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. Cheng J. C. Fox R. O. Olsen M. G. Measurements of turbulence in a microscale multi-inlet vortex nanoprecipitation reactor. J. Micromech. Microeng. 2013;23(7):075005. doi: 10.1088/0960-1317/23/7/075005. [DOI] [Google Scholar]
- Fang R. H. Chen K. N. H. Aryal S. Hu C.-M. J. Zhang K. Zhang L. Large-Scale Synthesis of Lipid–Polymer Hybrid Nanoparticles Using a Multi-Inlet Vortex Reactor. Langmuir. 2012;28(39):13824–13829. doi: 10.1021/la303012x. [DOI] [PubMed] [Google Scholar]
- Luchini A. Irace C. Santamaria R. Montesarchio D. Heenan R. K. Szekely N. Flori A. Menichetti L. Paduano L. Phosphocholine-decorated superparamagnetic iron oxide nanoparticles: defining the structure and probing in vivo applications. Nanoscale. 2016;8(19):10078–10086. doi: 10.1039/C5NR08486E. [DOI] [PubMed] [Google Scholar]
- Luchini A. D'Errico G. Leone S. Vaezi Z. Bortolotti A. Stella L. Vitiello G. Paduano L. Structural organization of lipid-functionalized-Au nanoparticles. Colloids Surf., B. 2018;168:2–9. doi: 10.1016/j.colsurfb.2018.04.044. [DOI] [PubMed] [Google Scholar]
- Blair J. M. Webber M. A. Baylay A. J. Ogbolu D. O. Piddock L. J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015;13(1):42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- Balaban N. Q. Merrin J. Chait R. Kowalik L. Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305(5690):1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- Brauner A. Fridman O. Gefen O. Balaban N. Q. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 2016;14(5):320–330. doi: 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- Rollin G. Tan X. Tros F. Dupuis M. Nassif X. Charbit A. Coureuil M. Intracellular Survival of Staphylococcus aureus in Endothelial Cells: A Matter of Growth or Persistence. Front. Microbiol. 2017;8:1354. doi: 10.3389/fmicb.2017.01354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon B. P. Staphylococcus aureus chronic and relapsing infections: Evidence of a role for persister cells: An investigation of persister cells, their formation and their role in S. aureus disease. BioEssays. 2014;36(10):991–996. doi: 10.1002/bies.201400080. [DOI] [PubMed] [Google Scholar]
- Xie S. Tao Y. Pan Y. Qu W. Cheng G. Huang L. Chen D. Wang X. Liu Z. Yuan Z. Biodegradable nanoparticles for intracellular delivery of antimicrobial agents. J. Controlled Release. 2014;187:101–117. doi: 10.1016/j.jconrel.2014.05.034. [DOI] [PubMed] [Google Scholar]
- Pei Y. Yeo Y. Drug delivery to macrophages: Challenges and opportunities. J. Controlled Release. 2016;240:202–211. doi: 10.1016/j.jconrel.2015.12.014. [DOI] [PubMed] [Google Scholar]
- Flores-Mireles A. L. Walker J. N. Caparon M. Hultgren S. J. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015;13(5):269–284. doi: 10.1038/nrmicro3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zazo H. Colino C. I. Lanao J. M. Current applications of nanoparticles in infectious diseases. J. Controlled Release. 2016;224:86–102. doi: 10.1016/j.jconrel.2016.01.008. [DOI] [PubMed] [Google Scholar]
- Lebeaux D. Chauhan A. Letoffe S. Fischer F. de Reuse H. Beloin C. Ghigo J. M. pH-mediated potentiation of aminoglycosides kills bacterial persisters and eradicates in vivo biofilms. J. Infect. Dis. 2014;210(9):1357–1366. doi: 10.1093/infdis/jiu286. [DOI] [PubMed] [Google Scholar]
- Helaine S. Thompson J. A. Watson K. G. Liu M. Boyle C. Holden D. W. Dynamics of intracellular bacterial replication at the single cell level. Proc. Natl. Acad. Sci. 2010;107(8):3746. doi: 10.1073/pnas.1000041107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehar S. M. Pillow T. Xu M. Staben L. Kajihara K. K. Vandlen R. DePalatis L. Raab H. Hazenbos W. L. Morisaki J. H. Kim J. Park S. Darwish M. Lee B. C. Hernandez H. Loyet K. M. Lupardus P. Fong R. Yan D. Chalouni C. Luis E. Khalfin Y. Plise E. Cheong J. Lyssikatos J. P. Strandh M. Koefoed K. Andersen P. S. Flygare J. A. Wah Tan M. Brown E. J. Mariathasan S. Novel antibody-antibiotic conjugate eliminates intracellular S. aureus. Nature. 2015;527(7578):323–328. doi: 10.1038/nature16057. [DOI] [PubMed] [Google Scholar]
- Flannagan R. S. Cosio G. Grinstein S. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat. Rev. Microbiol. 2009;7(5):355–366. doi: 10.1038/nrmicro2128. [DOI] [PubMed] [Google Scholar]
- Zhai W. Wu F. Zhang Y. Fu Y. Liu Z. The Immune Escape Mechanisms of Mycobacterium Tuberculosis. Int. J. Mol. Sci. 2019;20(2):340. doi: 10.3390/ijms20020340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahamed D. Boulle M. Ganga Y. Mc Arthur C. Skroch S. Oom L. Catinas O. Pillay K. Naicker M. Rampersad S. Mathonsi C. Hunter J. Wong E. B. Suleman M. Sreejit G. Pym A. S. Lustig G. Sigal A. Intracellular growth of Mycobacterium tuberculosis after macrophage cell death leads to serial killing of host cells. eLife. 2017;6 doi: 10.7554/eLife.22028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diacovich L. Lorenzi L. Tomassetti M. Meresse S. Gramajo H. The infectious intracellular lifestyle of Salmonella enterica relies on the adaptation to nutritional conditions within the Salmonella-containing vacuole. Virulence. 2017;8(6):975–992. doi: 10.1080/21505594.2016.1270493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengoechea J. A. Sa Pessoa J. Klebsiella pneumoniae infection biology: living to counteract host defences. FEMS Microbiol. Rev. 2018;43(2):123–144. doi: 10.1093/femsre/fuy043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussi C. Gutierrez M. G. Mycobacterium tuberculosis infection of host cells in space and time. FEMS Microbiol. Rev. 2019;43(4):341–361. doi: 10.1093/femsre/fuz006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoy D. A. Auerbuch V. Glomski I. J. The cell biology of Listeria monocytogenes infection: the intersection of bacterial pathogenesis and cell-mediated immunity. J. Cell Biol. 2002;158(3):409–414. doi: 10.1083/jcb.200205009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucciarelli M. G. García-Del Portillo F. Salmonella Intracellular Lifestyles and Their Impact on Host-to-Host Transmission. Microbiol. Spectrum. 2017;5(4) doi: 10.1128/microbiolspec.mtbp-0009-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano V. March C. Insua J. L. Aguiló N. Llobet E. Moranta D. Regueiro V. Brennan G. P. Millán-Lou M. I. Martín C. Garmendia J. Bengoechea J. A. Klebsiella pneumoniae survives within macrophages by avoiding delivery to lysosomes. Cell. Microbiol. 2015;17(11):1537–1560. doi: 10.1111/cmi.12466. [DOI] [PubMed] [Google Scholar]
- Weiss G. Schaible U. E. Macrophage defense mechanisms against intracellular bacteria. Immunol. Rev. 2015;264(1):182–203. doi: 10.1111/imr.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanses F. Kopp A. Bala M. Buechler C. Falk W. Salzberger B. Schäffler A. Intracellular Survival of Staphylococcus aureus in Adipocyte-Like Differentiated 3T3-L1 Cells Is Glucose Dependent and Alters Cytokine, Chemokine, and Adipokine Secretion. Endocrinology. 2011;152:4148–4157. doi: 10.1210/en.2011-0103. [DOI] [PubMed] [Google Scholar]
- Lacoma A. Cano V. Moranta D. Regueiro V. Dominguez-Villanueva D. Laabei M. Gonzalez-Nicolau M. Ausina V. Prat C. Bengoechea J. A. Investigating intracellular persistence of Staphylococcus aureus within a murine alveolar macrophage cell line. Virulence. 2017;8(8):1761–1775. doi: 10.1080/21505594.2017.1361089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzoni C. Kelley W. L. Return of the Trojan horse: intracellular phenotype switching and immune evasion by Staphylococcus aureus. EMBO Mol. Med. 2011;3(3):115–117. doi: 10.1002/emmm.201100123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reott Jr M. A. Ritchie-Miller S. L. Anguita J. Hudson M. C. TRAIL expression is induced in both osteoblasts containing intracellular Staphylococcus aureus and uninfected osteoblasts in infected cultures. FEMS Microbiol. Lett. 2008;278(2):185–192. doi: 10.1111/j.1574-6968.2007.00988.x. [DOI] [PubMed] [Google Scholar]
- Fraunholz M. Sinha B. Intracellular Staphylococcus aureus: live-in and let die. Front. Cell. Infect. Microbiol. 2012;2:43. doi: 10.3389/fcimb.2012.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamason R. L. Welch M. D. Actin-based motility and cell-to-cell spread of bacterial pathogens. Curr. Opin. Microbiol. 2017;35:48–57. doi: 10.1016/j.mib.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabiri G. A. Sanger J. M. Portnoy D. A. Southwick F. S. Listeria monocytogenes moves rapidly through the host-cell cytoplasm by inducing directional actin assembly. Proc. Natl. Acad. Sci. U. S. A. 1990;87(16):6068–6072. doi: 10.1073/pnas.87.16.6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellouk N. Enninga J. Cytosolic Access of Intracellular Bacterial Pathogens: The Shigella Paradigm. Front. Cell. Infect. Microbiol. 2016;6:35. doi: 10.3389/fcimb.2016.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agaisse H. Molecular and Cellular Mechanisms of Shigella flexneri Dissemination. Front. Cell. Infect. Microbiol. 2016;6(29) doi: 10.3389/fcimb.2016.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumell J. H. Tang P. Zaharik M. L. Finlay B. B. Disruption of the Salmonella-containing vacuole leads to increased replication of Salmonella enterica serovar typhimurium in the cytosol of epithelial cells. Infect. Immun. 2002;70(6):3264–3270. doi: 10.1128/IAI.70.6.3264-3270.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamwal S. V. Mehrotra P. Singh A. Siddiqui Z. Basu A. Rao K. V. Mycobacterial escape from macrophage phagosomes to the cytoplasm represents an alternate adaptation mechanism. Sci. Rep. 2016;6:23089. doi: 10.1038/srep23089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Tirado F. H. Doering T. L. False friends: Phagocytes as Trojan horses in microbial brain infections. PLoS Pathog. 2017;13(12):e1006680. doi: 10.1371/journal.ppat.1006680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotkin A. Chilmonczyk S. Eyngor M. Hurvitz A. Ghittino C. Eldar A. Trojan Horse Effect: Phagocyte-Mediated Streptococcus iniae Infection of Fish. Infect. Immun. 2003;71(5):2318. doi: 10.1128/IAI.71.5.2318-2325.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. Han X. Yang Y. Qiao H. Yu Z. Liu Y. Wang J. Tang T. Bacteria-Targeting Nanoparticles with Microenvironment-Responsive Antibiotic Release To Eliminate Intracellular Staphylococcus aureus and Associated Infection. ACS Appl. Mater. Interfaces. 2018;10(17):14299–14311. doi: 10.1021/acsami.7b15678. [DOI] [PubMed] [Google Scholar]
- Kim B. Pang H.-B. Kang J. Park J.-H. Ruoslahti E. Sailor M. J. Immunogene therapy with fusogenic nanoparticles modulates macrophage response to Staphylococcus aureus. Nat. Commun. 2018;9(1):1969. doi: 10.1038/s41467-018-04390-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F. Xu L. Yang B. Fan F. Yang L. Kill the Real with the Fake: Eliminate Intracellular Staphylococcus aureus Using Nanoparticle Coated with Its Extracellular Vesicle Membrane as Active-Targeting Drug Carrier. ACS Infect. Dis. 2019;5(2):218–227. doi: 10.1021/acsinfecdis.8b00212. [DOI] [PubMed] [Google Scholar]
- Nazir R., Zaffar M. R. and Amin I., Bacterial biofilms: the remarkable heterogeneous biological communities and nitrogen fixing microorganisms in lakes, in Freshwater Microbiology, ed. S. A. Bandh, S. Shafi and N. Shameem, Academic Press, 2019, ch. 8, pp. 307–340 [Google Scholar]
- Mu H. Tang J. Liu Q. Sun C. Wang T. Duan J. Potent Antibacterial Nanoparticles against Biofilm and Intracellular Bacteria. Sci. Rep. 2016;6:18877. doi: 10.1038/srep18877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan W. Chu K. Gullapalli A. Dunn S. S. Enlow E. M. Luft J. C. Tian S. Napier M. E. Pohlhaus P. D. Rolland J. P. DeSimone J. M. Delivery of Multiple siRNAs Using Lipid-Coated PLGA Nanoparticles for Treatment of Prostate Cancer. Nano Lett. 2012;12(1):287–292. doi: 10.1021/nl2035354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beletskii A. Galloway A. Rele S. Stone M. Malinoski F. Engineered PRINT® nanoparticles for controlled delivery of antigens and immunostimulants. Hum. Vaccines Immunother. 2014;10(7):1908–1913. doi: 10.4161/hv.28817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J. Wong D. H. C. Byrne J. D. Chen K. Bowerman C. DeSimone J. M. Future of the Particle Replication in Nonwetting Templates (PRINT) Technology. Angew. Chem., Int. Ed. 2013;52(26):6580–6589. doi: 10.1002/anie.201209145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X. Cai J. Zhang X. Li W.-D. Ge H. Hu Y. Top-down fabrication of shape-controlled, monodisperse nanoparticles for biomedical applications. Adv. Drug Delivery Rev. 2018;132:169–187. doi: 10.1016/j.addr.2018.07.006. [DOI] [PubMed] [Google Scholar]
- Hegde K. Brar S. K. Verma M. Surampalli R. Y. Current understandings of toxicity, risks and regulations of engineered nanoparticles with respect to environmental microorganisms. Nanotechnol. Environ. Eng. 2016;1(1):5. doi: 10.1007/s41204-016-0005-4. [DOI] [Google Scholar]
- Hua S. de Matos M. B. C. Metselaar J. M. Storm G. Current Trends and Challenges in the Clinical Translation of Nanoparticulate Nanomedicines: Pathways for Translational Development and Commercialization. Front. Pharmacol. 2018;9:790. doi: 10.3389/fphar.2018.00790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.-D. Huang L. Nanoparticles evading the reticuloendothelial system: role of the supported bilayer. Biochim. Biophys. Acta. 2009;1788(10):2259–2266. doi: 10.1016/j.bbamem.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. Lillard Jr J. W. Nanoparticle-based targeted drug delivery. Exp. Mol. Pathol. 2009;86(3):215–223. doi: 10.1016/j.yexmp.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J. Y. Kim H. S. Palanikumar L. Go E. M. Jana B. Park S. A. Kim H. Y. Kim K. Seo J. K. Kwak S. K. Kim C. Kang S. Ryu J.-H. Cloaking nanoparticles with protein corona shield for targeted drug delivery. Nat. Commun. 2018;9(1):4548. doi: 10.1038/s41467-018-06979-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokerst J. V. Lobovkina T. Zare R. N. Gambhir S. S. Nanoparticle PEGylation for imaging and therapy. Nanomedicine. 2011;6(4):715–728. doi: 10.2217/nnm.11.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandara S. R. Molley T. G. Kim H. Bharath P. A. Kilian K. A. Leal C. The structural fate of lipid nanoparticles in the extracellular matrix. Mater. Horiz. 2020 doi: 10.1039/C9MH00835G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S. W. Kim W. S. Kim J. H. Surface Modification of Functional Nanoparticles for Controlled Drug Delivery. J. Dispersion Sci. Technol. 2003;24(3–4):475–487. doi: 10.1081/DIS-120021803. [DOI] [Google Scholar]
- Sun D. Chen J. Wang Y. Ji H. Peng R. Jin L. Wu W. Advances in refunctionalization of erythrocyte-based nanomedicine for enhancing cancer-targeted drug delivery. Theranostics. 2019;9(23):6885–6900. doi: 10.7150/thno.36510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Din F. U. Aman W. Ullah I. Qureshi O. S. Mustapha O. Shafique S. Zeb A. Effective use of nanocarriers as drug delivery systems for the treatment of selected tumors. Int. J. Nanomed. 2017;12:7291–7309. doi: 10.2147/IJN.S146315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene M. K. Richards D. A. Nogueira J. C. F. Campbell K. Smyth P. Fernández M. Scott C. J. Chudasama V. Forming next-generation antibody–nanoparticle conjugates through the oriented installation of non-engineered antibody fragments. Chem. Sci. 2018;9(1):79–87. doi: 10.1039/C7SC02747H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M. C. Scott C. J. Antibody conjugated nanoparticles as a novel form of antibody drug conjugate chemotherapy. Drug Discovery Today: Technol. 2018;30:63–69. doi: 10.1016/j.ddtec.2018.10.003. [DOI] [PubMed] [Google Scholar]
- Billingsley M. M. Riley R. S. Day E. S. Antibody-nanoparticle conjugates to enhance the sensitivity of ELISA-based detection methods. PLoS One. 2017;12(5):e0177592. doi: 10.1371/journal.pone.0177592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoerr V. Tuchscherr L. Hüve J. Nippe N. Loser K. Glyvuk N. Tsytsyura Y. Holtkamp M. Sunderkötter C. Karst U. Klingauf J. Peters G. Löffler B. Faber C. Bacteria tracking by in vivo magnetic resonance imaging. BMC Biol. 2013;11(1):63. doi: 10.1186/1741-7007-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D. McMillan J. M. Kabanov A. V. Sokolsky-Papkov M. Gendelman H. E. Bench-to-bedside translation of magnetic nanoparticles. Nanomedicine. 2014;9(4):501–516. doi: 10.2217/nnm.14.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K. Su D. Liu J. Saha R. Wang J. P. Magnetic nanoparticles in nanomedicine: a review of recent advances. Nanotechnology. 2019;30(50):502003. doi: 10.1088/1361-6528/ab4241. [DOI] [PubMed] [Google Scholar]
- Karimi M. Ghasemi A. Sahandi Zangabad P. Rahighi R. Moosavi Basri S. M. Mirshekari H. Amiri M. Shafaei Pishabad Z. Aslani A. Bozorgomid M. Ghosh D. Beyzavi A. Vaseghi A. Aref A. R. Haghani L. Bahrami S. Hamblin M. R. Smart micro/nanoparticles in stimulus-responsive drug/gene delivery systems. Chem. Soc. Rev. 2016;45(5):1457–1501. doi: 10.1039/C5CS00798D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros S. F. Santos A. M. Fessi H. Elaissari A. Stimuli-responsive magnetic particles for biomedical applications. Int. J. Pharm. 2011;403(1):139–161. doi: 10.1016/j.ijpharm.2010.10.011. [DOI] [PubMed] [Google Scholar]
- Meers P. Neville M. Malinin V. Scotto A. W. Sardaryan G. Kurumunda R. Mackinson C. James G. Fisher S. Perkins W. R. Biofilm penetration, triggered release and in vivo activity of inhaled liposomal amikacin in chronic Pseudomonas aeruginosa lung infections. J. Antimicrob. Chemother. 2008;61(4):859–868. doi: 10.1093/jac/dkn059. [DOI] [PubMed] [Google Scholar]
- Xiong M.-H. Bao Y. Yang X.-Z. Wang Y.-C. Sun B. Wang J. Lipase-Sensitive Polymeric Triple-Layered Nanogel for “On-Demand” Drug Delivery. J. Am. Chem. Soc. 2012;134(9):4355–4362. doi: 10.1021/ja211279u. [DOI] [PubMed] [Google Scholar]
- Shabbir M. A. B. Shabbir M. Z. Wu Q. Mahmood S. Sajid A. Maan M. K. Ahmed S. Naveed U. Hao H. Yuan Z. CRISPR-cas system: biological function in microbes and its use to treat antimicrobial resistant pathogens. Ann. Clin. Microbiol. Antimicrob. 2019;18(1):21. doi: 10.1186/s12941-019-0317-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strich J. R. Chertow D. S. CRISPR-Cas Biology and Its Application to Infectious Diseases. J. Clin. Microbiol. 2019;57(4):e01307. doi: 10.1128/JCM.01307-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath D. Amlinger L. Rath A. Lundgren M. The CRISPR-Cas immune system: biology, mechanisms and applications. Biochimie. 2015;117:119–128. doi: 10.1016/j.biochi.2015.03.025. [DOI] [PubMed] [Google Scholar]
- Hille F. Charpentier E. CRISPR-Cas: biology, mechanisms and relevance. Philos. Trans. R. Soc., B. 2016;371(1707):20150496. doi: 10.1098/rstb.2015.0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lino C. A. Harper J. C. Carney J. P. Timlin J. A. Delivering CRISPR: a review of the challenges and approaches. Drug Delivery. 2018;25(1):1234–1257. doi: 10.1080/10717544.2018.1474964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y. K. Kwon K. Ryu J. S. Lee H. N. Park C. Chung H. J. Nonviral Genome Editing Based on a Polymer-Derivatized CRISPR Nanocomplex for Targeting Bacterial Pathogens and Antibiotic Resistance. Bioconjugate Chem. 2017;28(4):957–967. doi: 10.1021/acs.bioconjchem.6b00676. [DOI] [PubMed] [Google Scholar]
- Romling U. Balsalobre C. Biofilm infections, their resilience to therapy and innovative treatment strategies. J. Intern. Med. 2012;272(6):541–561. doi: 10.1111/joim.12004. [DOI] [PubMed] [Google Scholar]
- Itoh Y. Wang X. Hinnebusch B. J. Preston 3rd J. F. Romeo T. Depolymerization of beta-1,6-N-acetyl-D-glucosamine disrupts the integrity of diverse bacterial biofilms. J. Bacteriol. 2005;187(1):382–387. doi: 10.1128/JB.187.1.382-387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan R. M. Abbott J. C. Burhenne H. Kaever V. Gründling A. c-di-AMP Is a New Second Messenger in Staphylococcus aureus with a Role in Controlling Cell Size and Envelope Stress. PLoS Pathog. 2011;7(9):e1002217. doi: 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen L. D. van Gennip M. Jakobsen T. H. Alhede M. Hougen H. P. Hoiby N. Bjarnsholt T. Givskov M. Synergistic antibacterial efficacy of early combination treatment with tobramycin and quorum-sensing inhibitors against Pseudomonas aeruginosa in an intraperitoneal foreign-body infection mouse model. J. Antimicrob. Chemother. 2012;67(5):1198–1206. doi: 10.1093/jac/dks002. [DOI] [PubMed] [Google Scholar]