Abstract
Oocyte developmental competence is defined as the capacity of the female gamete to be fertilized and sustain development to the blastocyst stage. Epigenetic reprogramming, a correct cell division pattern, and an efficient DNA damage response are all critical events that, before embryonic genome activation, are governed by maternally inherited factors such as maternal-effect gene (MEG) products. Although these molecules are stored inside the oocyte until ovulation and exert their main role during fertilization and preimplantation development, some of them are already functioning during folliculogenesis and oocyte meiosis resumption. This mini review summarizes the crucial roles played by MEGs during oocyte maturation, fertilization, and preimplantation development with a direct/indirect effect on the acquisition or maintenance of oocyte competence. Our aim is to inspire future research on a topic with potential clinical perspectives for the prediction and treatment of female infertility.
Keywords: Maternal effect genes, Folliculogenesis, Oocyte competence, Embryonic genome activation, Preimplantation development
Introduction
Oocyte developmental competence is the capacity of a mature female gamete to be fertilized and sustain the initial phases of embryonic development until the blastocyst stage [1–3].
During folliculogenesis, many factors contribute to the production of a good quality oocyte, and abnormalities in this process lead to infertility and recurrent failure of assisted reproductive technologies (ART). Infertility is a pathology affecting up to 15% of couples worldwide [4], with 40% of the causes that might be either maternal or paternal, and 20% attributed to both partners [5, 6]. Regardless of the main causes of infertility, adverse reproductive outcomes in humans are mainly due to advanced maternal age, because of its well-known double impact: the progressive depletion of the ovarian reserve and a heavier insult on oocyte quality. This impact mainly results in an exponential increase in the blastocyst aneuploidy rates especially beyond the age of 35 [7]. Nevertheless, oocyte quality is not defined only based on its euploid chromosomal constitution, but also by several other aspects that concur to its capacity to reach the blastocyst stage and implant. For instance, critical roles are covered by mechanisms ensuring correct (i) epigenetic reprogramming, (ii) cell division pattern, and (iii) DNA damage response. Before embryonic genome activation (EGA), all these events are governed by maternally inherited factors, which are carried by the oocyte and inevitably affect embryo competence downstream. These factors encompass several maternal-effect gene (MEG) products transcribed during folliculogenesis by the oocyte itself or its surrounding follicle cells through a tightly regulated bidirectional communication. Although MEGs are stored inside the oocyte during its growth [8] and exert their main role during fertilization and preimplantation development, some of them already interact with the autocrine, paracrine, and biomechanical pathways that orchestrate folliculogenesis and oocyte meiosis resumption. The aim of this review is to summarize the crucial roles played by MEGs during oocyte maturation, fertilization, and preimplantation development.
Material and methods
For each area discussed, a systematic bibliographical search was performed, without temporal limits, using PubMed Central, Web of Science, and Scopus search engines employing the keyword maternal-effect gene in combination with folliculogenesis; oocyte growth; meiosis; oocyte competence; fertilization; embryonic genome activation; and preimplantation development. A total of 1513 papers were yielded and further selected based on our focus. Following the manual elimination of duplicates, this selection process retrieved 116 papers that were cited in the manuscript.
Maternal-effect gene products that regulate early embryonic development
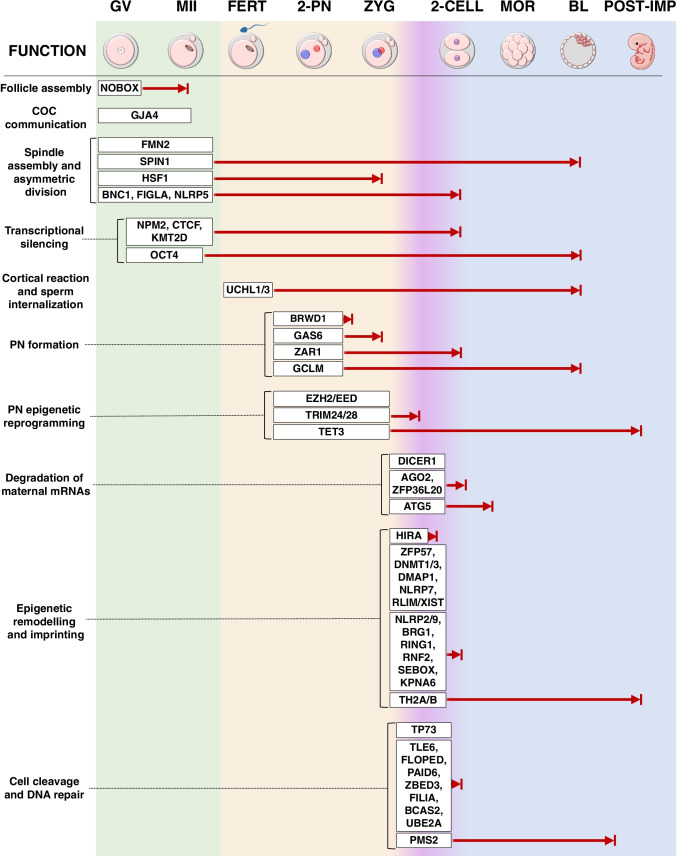
Preimplantation from the zygote to the blastocyst stage is a developmental window full of hurdles. These very early stages rely almost completely on the developmental capacity of the oocyte to regulate a sequence of key events that guide the zygote from a maternal to an embryonic control. Soon after fertilization, paternal chromatin decondensation [9–12], pronuclei (PN) DNA demethylation and fusion [9, 13], epigenetic reprogramming, EGA [14], and early embryo cell divisions are the main processes under the control of maternal factors. In the zygote, the large number of transcripts stored in the oocyte during folliculogenesis undergoes elimination or inactivation, except for a small group of MEG products that survives erasure and plays a critical role at the time of maternal-to-embryonic transition, when further development goes under the control of the embryo. In the next paragraphs and in Fig. 1, we will detail the main roles of MEGs involved in oocyte growth, meiosis resumption and progression to the metaphase II stage, fertilization, and preimplantation development.
Fig. 1.
Summary of 53 maternal-effect gene (MEG) products active during oocyte growth and meiosis resumption (green box), fertilization (orange box) and early preimplantation development (blue box). The purple-shaded area represents the moment when the embryonic genome activation occurs. MEGs are grouped according to their specific function and located depending on the site where they exert their main activity. For each MEG, the lethality stage observed in experimental models is indicated with a red arrow. All the experiments were performed in the mouse, except for Spind1, whose lethality was detected in the porcine model. The absence of the red arrow indicates that the lethality stage is not yet available. GV, germinal vesicle; MII, metaphase II; FERT, fertilization; PN, pronuclei; ZYG, zygote; MOR, morula; BL, blastocyst; POST-IMP, post-implantation; COC, cumulus-oocyte complex
MEGs with a role in oocyte growth, meiosis resumption, and progression to the metaphase II stage
Folliculogenesis is the process of growth and maturation from the primordial follicle, enclosing a primary oocyte arrested at the prophase meiosis I (MI), to the fully grown antral follicle, ready to ovulate a metaphase II (MII) oocyte. It entails timely morphological and functional changes mostly based on the bi-directional communication between the oocyte and the surrounding somatic cells [15]. While MEG products are well-known for their critical function at the time of maternal-to-embryonic transition in preimplantation embryos [14], here we highlight twelve of them that contribute also to follicle assembly and oocyte-cumulus cells communication, spindle assembly and asymmetric division, and transcriptional silencing during the GV to MII transition (Fig. 1, green area). Some interrelate with biomechanical pathways operating at the time of primordial follicle activation [16, 17], whereas others cooperate with autocrine and paracrine signalling cascades important for meiosis resumption, asymmetrical division, and oocyte epigenetic reprogramming.
By interacting with the Notch pathway, the newborn ovary homeobox-encoding gene (NOBOX) product is involved in the bi-directional communication between oocyte and granulosa cells, critical for follicle assembly [18]. This role becomes evident in Nobox knockout female mice lacking primary and secondary follicles [19]. Lastly, disruption of Nobox has been associated with non-syndromic ovarian failure in mice [19], as well as with premature ovarian insufficiency (POI) in women with specific mutations of this gene [20–22].
During folliculogenesis, the interaction between follicular cells and oocytes is facilitated by endocrine and paracrine signalling and intercellular gap junctional communication. Gap junction alpha-4 protein (CX37) is a gap junction connexin required in developing follicles [23]; the targeted deletion of its gene (Gja4) causes arrested folliculogenesis at the early antral stage, impaired oocyte maturation and meiotic competence, and premature follicles luteinization [24, 25]. During meiotic maturation, the prophase I-arrested oocyte undergoes nuclear envelope breakdown, chromosome recombination and condensation, and homolog segregation towards opposite poles thanks to the activity of the meiotic spindle [26, 27]. These events result in unequal cytokinesis and extrusion of the first polar body (PB-I), containing half genetic material. Then, meiotic maturation progresses with spindle reassembly, until the arrest at the MII oocyte ready for fertilization [27]. Some MEGs, including Fmn2, SPIN1, Hsf1, Bnc1, Figla, and Nlrp5, are involved in spindle assembly and asymmetric division. Formin-2 (FMN2) is the first actin nucleator during microtubules extension, migration, and spindle formation, a process regulated by the Rho-family GTPase RhoA ending with PB-I extrusion [28]. Female mice lacking FMN2 produce oocytes incapable of completing MI, resulting, when fertilized, in polyploid embryos with a consequently high incidence of pregnancy losses [29]. Spindlin-1 (SPIN1) is essential for chromosome stability and MII maintenance [30]. Its aberrant expression is associated with the extrusion of larger PB and a decreased blastocyst development [30]. Heat shock transcription factor 1 (HSF1) is known for its role in oogenesis and preimplantation development [31, 32], where its absence causes arrest at the zygote stage [33]. Interestingly, Hsf1 knockout mouse oocytes lack of Hsp90α expression and display delayed germinal vesicle breakdown (GVBD), altered G2/M transition, partial MI block, and defective asymmetrical division [34]. Basonuclin 1 (BNC1), a zinc-finger protein involved in the regulation of rRNAs transcription, is localized in the nucleolus of mouse oocytes and zygotes [35]. Experimental Bnc1 inactivation causes the formation of dark granules in the ooplasm [36], aberrant expression and activity of RNA polymerase I and II [37, 38], defective PB-I extrusion [36], and developmental arrest at the two-cell stage [36]. Folliculogenesis-specific basic helix-loop-helix α (FIGLA) is a transcription factor, whose deficiency arrests meiotic progression by dysregulating the transcription of Sycp3, Rad51, Cpeb1, and Ybx2 meiosis-related genes [37]. During follicle growth, FIGLA is also implicated in the interaction between oocytes and granulosa cells and in the coordination of zona pellucida (ZP) genes expression [39–41]. Interestingly, heterozygous mutations of Figla have been observed in patients with POI [42], suggesting its involvement in the pathogenesis of this condition [43]. Another MEG involved in asymmetric division is NACHT, LRR and PYD domains-containing protein 5 (Nlrp5, also known as Mater), whose transcripts and proteins are detected in oocytes of primary follicles and throughout folliculogenesis [44]. In growing oocytes, by interacting with the phosphoinositide 3-kinase pathway, NLRP5 participates in the F-actin polymerization during meiosis resumption [45]. Further evidence correlated its nucleolar localization during follicle growth with a specific type of chromatin organization [46] associated with oocyte developmental potential [47]. In particular, the great majority of Nlrp5-null fully grown antral oocytes displays a not-surrounded nucleolus (NSN) type of chromatin that is characteristic of developmentally incompetent female gametes, as opposed to SN oocytes that, instead, show a ring of chromatin around the nucleolus and are potentially developmentally competent [48, 49]. The compacted SN chromatin conformation is strictly associated with the global transcriptional silencing occurring in the final stages of oocyte growth that is essential to establish its developmental competence. Extensive chromatin modifications, including changes in histone methylation or acetylation, participate in this event, and transcription does not significantly resume until EGA [50–52]. Two additional factors associated with the oocyte’s chromatin organization and developmental competence are NPM2 and OCT4. Nucleoplasmin-2 (NPM2) is an oocyte-specific nuclear protein detected from the primary to the antral follicle and involved in chromatin remodelling at the time of fertilization and during early embryonic development [14, 53]. NPM2 is essential for the oocyte chromatin compaction during the final stages of maturation [54]. Indeed, Npm2−/− fully grown antral oocytes show an NSN type of chromatin [47, 55, 56]. These oocytes progress correctly through meiosis with no alterations in spindle formation and chromosomal segregation, although, after fertilization, they arrest their development at the two-cell stage [53]. As for Octamer-binding transcription factor 4 (Oct4), besides its role in the maintenance of pluripotency in primordial germ cells and in the blastocyst inner cell mass (ICM) [57], its protein is also present in fully grown antral developmentally competent SN and their derived MII oocytes [58, 59]. Although its role in oogenesis has not yet been detailed, OCT4 and a network of Oct4-regulated genes may represent a developmental link between the female gamete and the pluripotency of the preimplantation embryo [1, 60].
Among the best characterized epigenetic modifications, recent evidence supports that the accumulation of H3K4me3 in the late oocyte growth stages contributes to its transcriptional silencing [61, 62]. Lysine (K)-specific methyltransferase 2D (KMT2D) is a histone 3 lysine 4 (H3K4) methyltransferase, thought to be involved in most of the promoter-specific chromatin modifications during oogenesis and early embryo development. Conditional KMtd2 knockout female mice show increased serum gonadotropin hormones, decreased serum oestradiol, altered folliculogenesis, premature ovarian follicle loss, and developmental arrest between the one- and four-cell stage [50].
Alongside with its function in somatic epigenetic regulation and reprograming [63], CCCTC-binding factor (CTCF) has an important role in the establishment and maintenance of maternal imprinting during oocyte growth. Specifically, CTCF binding inhibits the methylation of the imprinting control region in female germ cells [64]. Using RNA interference (RNAi), the depletion of maternal Ctcf from growing mouse oocytes induced defects in maternal gene expression that, in turn, caused meiotic defects in eggs, mitotic defects in embryos, and apoptosis [65].
MEGs with a role during fertilization
Fertilization is a sequence of strictly controlled events involving the entry of the sperm inside the oocyte, the fusion of their PNs, and the establishment of the embryonic genome. In this paragraph, we will describe those MEGs acting at the time of cortical reaction and sperm internalization, PN formation, and subsequent epigenetic reprogramming of the parental genomes (Fig. 1, orange area).
The binding between sperm and oocyte ZP is the first process required for fertilization; it is regulated by several factors like ADAMs, Integrins, CRISP1, IZUMO, and JUNO. By triggering acrosomal exocytosis, this event allows the passage of the male gamete through the ZP and ends with sperm-egg fusion [66, 67]. Soon after this phase, sperm-derived factors, such as phospholipase C zeta 1, trigger the release of cortical granules, whose enzymes modify and harden the ZP, preventing polyspermy [68]. Concurrently, the second oocyte meiotic division and PB-II release take place. Within this context, two MEGs, ubiquitin carboxyl-terminal hydrolase isozyme L1 and L3 (Uchl1 and Uchl3), play an important role as, if inhibited in MII oocytes, they induce aberrant cortical granules release and, as a consequence, meiotic spindle defects and fertilization block [69]. Their key maternal effect role beyond fertilization and during preimplantation was shown in embryos from Uchl1 knockout females failing to undergo morula formation [69].
Immediately after internalization, the sperm nucleus undergoes a series of changes to form the male PN, which migrates towards and fuses with the female PN, resulting in the diploid zygote. During PNs formation and maturation, significant epigenetic reprogramming of the parental genomes occurs to allow the access and transcription of genes needed for the zygote further development [70]. First, active demethylation of the paternal genome takes place, whereas in the maternal genome, it mostly occurs passively [27, 71]. When demethylation is complete, the two PNs move closer together and fuse [27]. A group of four MEGs, Gas6, Zar1, Brwd1, and Gclm, are known for their role at the time of PNs formation. Growth arrest-specific protein 6 (Gas6) RNAi-treated oocytes arrested at MII stage, exhibited no exocytosis of cortical granules, and impaired PNs formation, suggesting that the decreased Gas6 expression influences sperm head decondensation [72]. Although zygote arrest protein 1 (Zar1) null mice have a normal oogenesis, more than 80% of their embryos arrest at the one-cell stage, with separated maternal and paternal PNs. The remaining embryos display marked reduction in the synthesis of the transcription-requiring complex, and none of them develops beyond the two-cell stage [73]. Bromodomain and WD repeat domain containing 1 (Brwd1) encodes a putative transcriptional regulator acting on chromatin through interactions with the Brg1-dependent SWI/SNF chromatin remodelling pathway [74]. Brwd1-mutant oocytes do not show any morphological alteration, but are defective in the oocyte–embryo transition, suggesting an altered transcription of genes involved in the developmental progression beyond the 2-PN stage [75]. Finally, mice lacking maternal glutamate cysteine ligase modifier subunit (Gclm) show a decreased glutathione concentration in ovulated oocytes, and although fertilization occurs, they have a reduced rate of male PN formation, decreased development to the blastocyst stage, reduced implantation, and smaller litter size [76].
During paternal PN maturation, methylcytosine dioxygenase TET3 (TET3) is particularly abundant and has recently been shown to contribute to the genome-wide loss of 5-methylcytosine, the main responsible of its epigenetic reprogramming [77]. Female mice depleted of Tet3 in the germline show severely reduced fertility and their heterozygous mutant offspring suffer from an increased incidence of developmental failures [9]. TRIM24 and TRIM28 are transcription intermediary factors that interact with numerous proteins involved in chromatin organization and zygotic genome activation in the PNs. In particular, TRIM24 enters the PNs to interact with sites enriched with chromatin remodellers BRG-1 and SNF2H. Zygotes lacking Trim24 proceed their development only until the two- to four-cell stage transition, because of mis-localization of RNA polymerase II and impaired expression of SNF2H-dependent genes [78]. In bovine oocytes, TRIM28 knockdown, instead, alters histone methylation at the two-cell stage [79]. Two other MEGs with histone methyltransferase activity are the histone-lysine N-methyltransferase EZH2 and the polycomb protein EED, part of a single multimeric complex (Ezh2/Eed). Depletion of the maternal Ezh2 allele affects the preferential localization of EED to the maternal PN in early zygotes, causing alterations to the parental histones’ methylation profile, and severe growth retardation in newborns [66].
MEGs with a role during preimplantation development
Shortly after fertilization, a second wave of transcriptional activation begins in mouse [80], involving the final transition from an oocyte to an embryonic control of development. This dynamic process, known as EGA, aims at restoring totipotency to the zygote. It takes place predominantly at the two-cell stage in mice and the eight-cell stage in humans. In this paragraph, we will describe MEGs that contribute to the degradation of maternal mRNAs and proteins, epigenetic remodelling, and imprinting, all essential for EGA. Then, we will highlight those MEGs involved in the processes of early cell cleavage and DNA repair [81] (Fig. 1, purple-blue area).
MEGs like Ago2, Dicer1, Atg5, and Zfp36l2 [82–85] participate in the timely degradation of subgroups of maternal mRNAs that facilitates the transition between embryonic developmental stages. Protein argonaute-2 (AGO2) and endoribonuclease Dicer (DICER1) are the best-known endonucleases of the RNA-induced silencing complex (RISC) [84, 85]. Specifically, Ago2 transcript is expressed in mouse oocytes and throughout preimplantation development and localizes in mRNA degradation P-bodies. RNAi of Ago2 leads to developmental arrest at the two-cell stage, by inactivating zygotic transcripts and stabilizing those maternal mRNAs that are normally degraded [86]. Another factor whose function is important at the time of mouse EGA is autophagy-related gene 5 (ATG5), which is a regulator of maternal transcripts degradation. In this case, though, when the maternal gene is experimentally knocked out, the paternal transcript can compensate its lack [82].
The embryonic development arrest in a transgenic mouse model also showed the involvement of ZFP36L2 in maternal mRNA turnover in normal embryogenesis, by binding mRNAs and destabilizing proteins [78].
In the rich sub-group of MEGs regulating epigenetic remodelling and imprinting, two histone variants, histone H2A and H2B types (Th2A and Th2B), are highly expressed in oocytes and contribute to the activation of the paternal genome after fertilization, by influencing its chromatin state [79]. Interestingly, the loss of TH2A/TH2B does not cause histological alterations to folliculogenesis, whereas the resulting oocytes display reduced developmental competence and altered paternal genome activation [87]. Zinc finger protein 57 (ZFP57), then, is a transcriptional repressor required for the post-fertilization maintenance of both maternal and paternal methylation imprints [88]. DNA methyltransferase 1 (DNMT1) and its associated protein DNMT1-associated protein 1 (DMAP1) are involved in the maintenance of embryonic stem cell pluripotency [89, 90] and preserve the correct methylation at the differentially methylated regions (DMRs) of all imprinted genes [82]. Homozygous Dmap1-/- embryos show a lethal phenotype [89]. Among the methyltransferase family, Dnmt3a and Dnmt3b are expressed in both male and female germlines and are required for de novo methylation of the DMRs and for the acquisition of functional imprints [83]. Specifically, embryos derived from Dnmt3a-, Dnmt3b-, and Dnmt3a⁄Dnmt3b-deficient oocytes show growth retardation and died at early post-implantation stages [84]. Three members of the Nlrp family are involved in the regulation of genome imprinting. Germline mutations in Nlrp2, whose knockdown in mouse oocytes leads to arrest at the 2-cell stage [85], result in a familial imprinting disorder (Beckwith–Wiedemann syndrome) in humans, suggesting that this gene might have a function in the establishment and/or maintenance of DNA methylation [86]. NLRP7, in association with FILIA, is required for the establishment or maintenance of the appropriate oocyte imprint. In fact, women affected from familial recurrent hydatidiform mole, due to mutations in both genes, show abnormal maternal imprinting patterns [91]. Recently, evidences of a NLRP9 function during preimplantation development are emerging: when mated with wild-type males, Nlrp9-deficient mice are fertile, but produce less and slower-growing blastocysts; on the contrary, when fertilized in vitro, development arrested at the two-cell stage after asymmetric cell divisions [92].
In early mammalian development, one of the two X chromosomes is randomly silenced in each female cell as a result of X chromosome inactivation. This choice is then transmitted to all daughter cells through mitosis, such that the adult female is a mosaic of two different cell lineages [93]. Maternal RING finger LIM domain-interacting protein (RLIM) is a ubiquitin ligase required for imprinted X chromosome inactivation by triggering the expression of the X-linked Xist [68]. A recent study demonstrated that knockdown of RLIM improved the developmental rate of cloned male pig embryos. This effect might be due to the suppression of Xist transcription and the consequent upregulation of several X‐linked and autosomal genes required for blastocyst development [69].
Another group of molecules that participate in epigenetic remodelling and regulate DNA transcription are HIRA, BRG1, RING1, and RNF2. HIRA, a chaperone for the histone variant H3.3, is a protein required for the extensive chromatin reprogramming that occurs during oogenesis, fertilization, and mouse development beyond the zygote stage. In fact, Hira-mutant oocytes show strongly reduced DNA replication and transcription levels, essential for the first cleavage [10, 70]. Transcription activator BRG1 encodes a catalytic subunit of the SWI/SNF-related chromatin remodelling complex which regulates transcription involved in zygotic gene expression reprogramming during the two-cell stage. In particular, BRG1-depleted mouse zygotes arrest at the two-cell stage displaying reduced gene expression and lower dimethyl-H3K4 levels [94]. RING1 and RNF2 are components of the polycomb-repressive complex 1 (PRC1), essential for proper EGA, replication, and cell cycle progression in early embryos. Genetic ablation of both genes in oocytes results in loss of chromatin-bound PRC1, induction of massive transcriptional mis-regulation during oocyte growth, and developmental arrest at the two-cell stage. These results support that PRC1 acts in the female germline to establish developmental competence by silencing differentiation-inducing genes and defining an appropriate chromatin organization [95]. Two other genes, Kpna6 and Sebox, whose mechanism of action is still unclear, have been highlighted experimentally for their role in regulating DNA transcription at the very beginning of preimplantation development. Oocytes lacking importin α7 (KPNA6) show correct fertilizability but arrest at the two-cell stage [96]. The homeobox protein SEBOX RNAi at the PN stage, instead, led to arrested embryo development between the two-cell (85% of cases) and the four-/eight-cell (15%) stage. Specifically, in Sebox-knockdown two-cell embryos, the embryonic transcriptional activity is reduced, with critical maternal mRNAs not fully degraded and several EGA markers under-expressed [97, 98].
During preimplantation development, soon after EGA, embryos go through several cell divisions and transit from totipotency to pluripotency, and their cells start their differentiation towards trophectoderm or ICM [99].
Early embryonic symmetrical cell division requires the action of maternally inherited complexes, such as the subcortical maternal complex (SCMC), the spindle assembly checkpoint (SAC), and strictly regulated mechanisms of DNA repair. The SCMC assembles underneath the oolemma and is essential to progress beyond the first embryonic cell division [100]. TLE6, NLRP5, PADI6, FLOPED, and FILIA are the main components of the SCMC; however, recently novel members, such as NLRP2, NLRP7, NLRP9, NLPR4, FILIA, and ZBED3, have been identified [92, 101, 102]. As described above, NLRP2, NLRP7, NLRP9, and FILIA have a role also in the establishment of genomic imprints and post-zygotic methylation maintenance [92, 101].
In the mouse, genetic ablation of individual components provides evidence that the SCMC is required for normal cleavage and development [103]. Transducin-like enhancer protein 6 (TLE6) is a transcriptional co-repressor [104, 105], and, together with FLOPED and NLRP5, its transcripts accumulate and reach the highest abundance in fully grown oocytes. The oocyte-expressed protein homolog (Floped) expression was detected in growing oocytes, and the protein is present in the subcortex of eggs where it overlaps with cortical F-actin but extends further into the cytoplasm. Although genetic ablation of either Floped and/or NLRP5, and the subsequent impairment of the SCMC, does not affect oocyte development up to fertilization, mutant embryos fail to reach the cleavage stage, thereby resulting in infertility [43]. Peptidylarginine deiminase 6 (PADI6) is a germ cell-specific enzyme mainly located in the cortex of eggs and preimplantation embryos. It is responsible for amino acid citrullination, a process crucial for the formation of the cytoplasmic lattices (CPLs), a fibrillar matrix composed of proteins and RNAs. Also, PADI6 has been identified as a member of the SCMC, because it co-localizes with its other components, and it is detectable from the oocyte to the blastocyst stage [106, 107]. Specifically, in mice oocytes, the absence of PADI6 protein results in impaired embryonic transcription, reduced ribosomal component levels, and dysregulated de novo protein synthesis, thus causing arrest at the zygote-to-embryo transition [107, 108].
To guarantee a correct segregation of genetic materials into daughter cells, eukaryotic organism developed the SAC complex to prevent premature metaphase-anaphase transition until all chromosomes successfully attach to the bipolar spindle. It has been reported that SAC is one of the main mechanisms essential for the regulation of mitotic cell cycle progression in cleavage stage embryos [109]. By regulating the activity of SAC, FILIA and TP73 ensure a correct spindle formation and chromosome alignment. Alterations to these processes are among the most common causes of embryonic aneuploidies [110]. FILIA (also known as KHDC3) enables SAC activity by guiding the interaction of AURKA, PLK1, and γ-tubulin to the microtubule-organizing center, and the attachment of MAD2 to kinetochores. Depletion of maternal Filia transcripts impairs mouse preimplantation development, and defects in the human homolog could play a similar role and cause recurrent miscarriages [111]. Tumor protein p73 (TP73) regulates SAC activity by interacting with its BUBR1 and BUB1 components; in fact, TP73-null mice are infertile and produce oocytes with spindle abnormalities [112].
In the early embryonic stages of development, to preserve and maintain the integrity of the genome, cells activate complex DNA repair mechanisms to prevent mutations caused by endogenous and exogenous factors. In response to a DNA damage, DNA repair proteins can stop cell cycle progression, allowing genome correction. If the damage cannot be repaired, a proapoptotic pathway is activated, resulting in cell death [113]. During early embryogenesis, DNA transcription is inactive; thus, the embryo’s ability to repair DNA is restricted to the function of maternally inherited DNA repair proteins, such as BCAS2, PMS2, and UBE2A [114–116]. Breast carcinoma amplified sequence 2 (BCAS2) plays an important role in the DNA damage response through the replication protein A (RPA) complex, a key regulator in the maintenance of genome integrity [117]. Maternal BCAS2 depletion leads to developmental arrest at the two-/four-cell stage [114]. Mouse zygotes deficient in the mismatch repair endonuclease PMS2 show unrepaired replication errors in early cell divisions, suggesting a role of maternal Pms2 in DNA mismatch repair [115].
Conclusions
With the beginning of preimplantation development, the major hurdle encountered by the embryo is the transition from a maternal to an embryonic control of development, a phase strictly regulated by maternal-effect factors like MEGs [118]. These molecules are the cargo that the oocyte inherits during folliculogenesis and oocyte maturation also via a mutual communication with its companion cumulus cells. MEGs therefore represent the molecular link between folliculogenesis and preimplantation development, thereby covering a key role in the acquisition and maintenance of oocytes developmental competence. Yet, the whole network of molecular regulations and all the mechanisms and pathways governed in this choreography still need to be unveiled.
Here we described the role of MEGs not only during preimplantation development, but also their contribution to the acquisition of oocytes developmental competence during folliculogenesis. Our view aims at inspiring future research on a topic with potential clinical perspectives for the prediction and treatment of female infertility.
Acknowledgements
This work was made possible thanks to support from the Italian Ministry of Education, University and Research (MIUR) Dipartimenti di Eccellenza Program (2018–2022) to the Department of Biology and Biotechnology ‘L. Spallanzani’, University of Pavia, and a grant from the University of Pavia (FRG 2020).
Author Contribution
FI, GF, and DS conducted the literature search. FI, GF, DC and MZ drafted the manuscript. FI, GF, DC, DS and MZ designed the figure. All other authors reviewed the manuscript and contributed to its summary and discussion.
Declarations
Conflict of Interest
The authors declare no competing interests.
Footnotes
Federica Innocenti and Giulia Fiorentino are co-first authors.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zuccotti M, Merico V, Cecconi S, Redi CA, Garagna S. What does it take to make a developmentally competent mammalian egg? Hum Reprod Update. 2011;17(4):525–540. doi: 10.1093/humupd/dmr009. [DOI] [PubMed] [Google Scholar]
- 2.Keefe D, Kumar M, Kalmbach K. Oocyte competency is the key to embryo potential. Fertil Steril. 2015;103(2):317–322. doi: 10.1016/j.fertnstert.2014.12.115. [DOI] [PubMed] [Google Scholar]
- 3.Conti M, Franciosi F. Acquisition of oocyte competence to develop as an embryo: integrated nuclear and cytoplasmic events. Hum Reprod Update. 2018;24(3):245–266. doi: 10.1093/humupd/dmx040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun H, Gong TT, Jiang YT, Zhang S, Zhao YH, Wu QJ. Global, regional, and national prevalence and disability-adjusted life-years for infertility in 195 countries and territories, 1990–2017: results from a global burden of disease study, 2017. Aging (Albany NY) 2019;11(23):10952–10991. doi: 10.18632/aging.102497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fidler AT, Bernstein J. Infertility: from a personal to a public health problem. Public Health Rep. 1999;114(6):494–511. doi: 10.1093/phr/114.6.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tabong PT, Adongo PB. Understanding the social meaning of infertility and childbearing: a qualitative study of the perception of childbearing and childlessness in Northern Ghana. PLoS ONE. 2013;8(1):e54429. doi: 10.1371/journal.pone.0054429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capalbo A, Hoffmann ER, Cimadomo D, Ubaldi FM, Rienzi L. Human female meiosis revised: new insights into the mechanisms of chromosome segregation and aneuploidies from advanced genomics and time-lapse imaging. Hum Reprod Update. 2017;23(6):706–722. doi: 10.1093/humupd/dmx026. [DOI] [PubMed] [Google Scholar]
- 8.Zhang K, Smith GW. Maternal control of early embryogenesis in mammals. Reprod Fertil Dev. 2015;27(6):880–896. doi: 10.1071/RD14441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477(7366):606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 10.Lin CJ, Koh FM, Wong P, Conti M, Ramalho-Santos M. Hira-mediated H3.3 incorporation is required for DNA replication and ribosomal RNA transcription in the mouse zygote. Dev Cell. 2014;30(3):268–79. doi: 10.1016/j.devcel.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McLay DW, Clarke HJ. Remodelling the paternal chromatin at fertilization in mammals. Reproduction. 2003;125(5):625–633. doi: 10.1530/rep.0.1250625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okada Y, Yamaguchi K. Epigenetic modifications and reprogramming in paternal pronucleus: sperm, preimplantation embryo, and beyond. Cell Mol Life Sci. 2017;74(11):1957–1967. doi: 10.1007/s00018-016-2447-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Payer B, Saitou M, Barton SC, Thresher R, Dixon JP, Zahn D, et al. Stella is a maternal effect gene required for normal early development in mice. Curr Biol. 2003;13(23):2110–2117. doi: 10.1016/j.cub.2003.11.026. [DOI] [PubMed] [Google Scholar]
- 14.Wu D, Dean J. Maternal factors regulating preimplantation development in mice. Curr Top Dev Biol. 2020;140:317–340. doi: 10.1016/bs.ctdb.2019.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gershon E, Dekel N. Newly identified regulators of ovarian folliculogenesis and ovulation. Int J Mol Sci. 2020;21(12). doi: 10.3390/ijms21124565. [DOI] [PMC free article] [PubMed]
- 16.Grosbois J, Demeestere I. Dynamics of PI3K and Hippo signaling pathways during in vitro human follicle activation. Hum Reprod. 2018;33(9):1705–1714. doi: 10.1093/humrep/dey250. [DOI] [PubMed] [Google Scholar]
- 17.Shah JS, Sabouni R, Cayton Vaught KC, Owen CM, Albertini DF, Segars JH. Biomechanics and mechanical signaling in the ovary: a systematic review. J Assist Reprod Genet. 2018;35(7):1135–1148. doi: 10.1007/s10815-018-1180-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vanorny DA, Mayo KE. The role of Notch signaling in the mammalian ovary. Reproduction. 2017;153(6):R187–R204. doi: 10.1530/REP-16-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rajkovic A, Pangas SA, Ballow D, Suzumori N, Matzuk MM. NOBOX deficiency disrupts early folliculogenesis and oocyte-specific gene expression. Science. 2004;305(5687):1157–1159. doi: 10.1126/science.1099755. [DOI] [PubMed] [Google Scholar]
- 20.Qin Y, Choi Y, Zhao H, Simpson JL, Chen ZJ, Rajkovic A. NOBOX homeobox mutation causes premature ovarian failure. Am J Hum Genet. 2007;81(3):576–581. doi: 10.1086/519496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferrari I, Bouilly J, Beau I, Guizzardi F, Ferlin A, Pollazzon M, et al. Impaired protein stability and nuclear localization of NOBOX variants associated with premature ovarian insufficiency. Hum Mol Genet. 2016;25(23):5223–5233. doi: 10.1093/hmg/ddw342. [DOI] [PubMed] [Google Scholar]
- 22.Li L, Wang B, Zhang W, Chen B, Luo M, Wang J, et al. A homozygous NOBOX truncating variant causes defective transcriptional activation and leads to primary ovarian insufficiency. Hum Reprod. 2017;32(1):248–255. doi: 10.1093/humrep/dew271. [DOI] [PubMed] [Google Scholar]
- 23.Harris AL. Emerging issues of connexin channels: biophysics fills the gap. Q Rev Biophys. 2001;34(3):325–472. doi: 10.1017/s0033583501003705. [DOI] [PubMed] [Google Scholar]
- 24.Denomme MM, White CR, Gillio-Meina C, Macdonald WA, Deroo BJ, Kidder GM, et al. Compromised fertility disrupts Peg1 but not Snrpn and Peg3 imprinted methylation acquisition in mouse oocytes. Front Genet. 2012;3:129. doi: 10.3389/fgene.2012.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carabatsos MJ, Sellitto C, Goodenough DA, Albertini DF. Oocyte-granulosa cell heterologous gap junctions are required for the coordination of nuclear and cytoplasmic meiotic competence. Dev Biol. 2000;226(2):167–179. doi: 10.1006/dbio.2000.9863. [DOI] [PubMed] [Google Scholar]
- 26.Eppig JJ. Coordination of nuclear and cytoplasmic oocyte maturation in eutherian mammals. Reprod Fertil Dev. 1996;8(4):485–489. doi: 10.1071/rd9960485. [DOI] [PubMed] [Google Scholar]
- 27.Swain JE, Pool TB. ART failure: oocyte contributions to unsuccessful fertilization. Hum Reprod Update. 2008;14(5):431–446. doi: 10.1093/humupd/dmn025. [DOI] [PubMed] [Google Scholar]
- 28.Namgoong S, Kim NH. Roles of actin binding proteins in mammalian oocyte maturation and beyond. Cell Cycle. 2016;15(14):1830–1843. doi: 10.1080/15384101.2016.1181239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leader B, Lim H, Carabatsos MJ, Harrington A, Ecsedy J, Pellman D, et al. Formin-2, polyploidy, hypofertility and positioning of the meiotic spindle in mouse oocytes. Nat Cell Biol. 2002;4(12):921–928. doi: 10.1038/ncb880. [DOI] [PubMed] [Google Scholar]
- 30.Choi JW, Zhao MH, Liang S, Guo J, Lin ZL, Li YH, et al. Spindlin 1 is essential for metaphase II stage maintenance and chromosomal stability in porcine oocytes. Mol Hum Reprod. 2017;23(3):166–176. doi: 10.1093/molehr/gax005. [DOI] [PubMed] [Google Scholar]
- 31.Mezger V, Renard JP, Christians E, Morange M. Detection of heat shock element-binding activities by gel shift assay during mouse preimplantation development. Dev Biol. 1994;165(2):627–638. doi: 10.1006/dbio.1994.1281. [DOI] [PubMed] [Google Scholar]
- 32.Christians E, Michel E, Adenot P, Mezger V, Rallu M, Morange M, et al. Evidence for the involvement of mouse heat shock factor 1 in the atypical expression of the HSP70.1 heat shock gene during mouse zygotic genome activation. Mol Cell Biol. 1997;17(2):778–88. doi: 10.1128/mcb.17.2.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christians E, Davis AA, Thomas SD, Benjamin IJ. Maternal effect of Hsf1 on reproductive success. Nature. 2000;407(6805):693–694. doi: 10.1038/35037669. [DOI] [PubMed] [Google Scholar]
- 34.Metchat A, Akerfelt M, Bierkamp C, Delsinne V, Sistonen L, Alexandre H, et al. Mammalian heat shock factor 1 is essential for oocyte meiosis and directly regulates Hsp90alpha expression. J Biol Chem. 2009;284(14):9521–9528. doi: 10.1074/jbc.M808819200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang D, Liu Y, Zhang Z, Lv P, Liu Y, Li J, et al. Basonuclin 1 deficiency is a cause of primary ovarian insufficiency. Hum Mol Genet. 2018;27(21):3787–3800. doi: 10.1093/hmg/ddy261. [DOI] [PubMed] [Google Scholar]
- 36.Ma J, Zeng F, Schultz RM, Tseng H. Basonuclin: a novel mammalian maternal-effect gene. Development. 2006;133(10):2053–2062. doi: 10.1242/dev.02371. [DOI] [PubMed] [Google Scholar]
- 37.Iuchi S, Green H. Basonuclin, a zinc finger protein of keratinocytes and reproductive germ cells, binds to the rRNA gene promoter. Proc Natl Acad Sci U S A. 1999;96(17):9628–9632. doi: 10.1073/pnas.96.17.9628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tseng H, Biegel JA, Brown RS. Basonuclin is associated with the ribosomal RNA genes on human keratinocyte mitotic chromosomes. J Cell Sci. 1999;112(Pt 18):3039–3047. doi: 10.1242/jcs.112.18.3039. [DOI] [PubMed] [Google Scholar]
- 39.Liang L, Soyal SM, Dean J. FIGalpha, a germ cell specific transcription factor involved in the coordinate expression of the zona pellucida genes. Development. 1997;124(24):4939–4947. doi: 10.1242/dev.124.24.4939. [DOI] [PubMed] [Google Scholar]
- 40.Soyal SM, Amleh A, Dean J. FIGalpha, a germ cell-specific transcription factor required for ovarian follicle formation. Development. 2000;127(21):4645–4654. doi: 10.1242/dev.127.21.4645. [DOI] [PubMed] [Google Scholar]
- 41.Joshi S, Davies H, Sims LP, Levy SE, Dean J. Ovarian gene expression in the absence of FIGLA, an oocyte-specific transcription factor. BMC Dev Biol. 2007;7:67. doi: 10.1186/1471-213X-7-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao H, Chen ZJ, Qin Y, Shi Y, Wang S, Choi Y, et al. Transcription factor FIGLA is mutated in patients with premature ovarian failure. Am J Hum Genet. 2008;82(6):1342–1348. doi: 10.1016/j.ajhg.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li L, Baibakov B, Dean J. A subcortical maternal complex essential for preimplantation mouse embryogenesis. Dev Cell. 2008;15(3):416–425. doi: 10.1016/j.devcel.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tong ZB, Gold L, De Pol A, Vanevski K, Dorward H, Sena P, et al. Developmental expression and subcellular localization of mouse MATER, an oocyte-specific protein essential for early development. Endocrinology. 2004;145(3):1427–1434. doi: 10.1210/en.2003-1160. [DOI] [PubMed] [Google Scholar]
- 45.Zheng P, Baibakov B, Wang XH, Dean J. PtdIns(3,4,5)P3 is constitutively synthesized and required for spindle translocation during meiosis in mouse oocytes. J Cell Sci. 2013;126(Pt 3):715–721. doi: 10.1242/jcs.118042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Monti M, Zanoni M, Calligaro A, Ko MS, Mauri P, Redi CA. Developmental arrest and mouse antral not-surrounded nucleolus oocytes. Biol Reprod. 2013;88(1):2. doi: 10.1095/biolreprod.112.103887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zuccotti M, Garagna S, Merico V, Monti M, Alberto RC. Chromatin organisation and nuclear architecture in growing mouse oocytes. Mol Cell Endocrinol. 2005;234(1–2):11–17. doi: 10.1016/j.mce.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 48.Zuccotti M, Giorgi Rossi P, Martinez A, Garagna S, Forabosco A, Redi CA. Meiotic and developmental competence of mouse antral oocytes. Biol Reprod. 1998;58(3):700–704. doi: 10.1095/biolreprod58.3.700. [DOI] [PubMed] [Google Scholar]
- 49.Zuccotti M, Ponce RH, Boiani M, Guizzardi S, Govoni P, Scandroglio R, et al. The analysis of chromatin organisation allows selection of mouse antral oocytes competent for development to blastocyst. Zygote. 2002;10(1):73–78. doi: 10.1017/s0967199402002101. [DOI] [PubMed] [Google Scholar]
- 50.Andreu-Vieyra CV, Chen R, Agno JE, Glaser S, Anastassiadis K, Stewart AF, et al. MLL2 is required in oocytes for bulk histone 3 lysine 4 trimethylation and transcriptional silencing. PLoS Biol. 2010;8(8). doi: 10.1371/journal.pbio.1000453. [DOI] [PMC free article] [PubMed]
- 51.De La Fuente R. Chromatin modifications in the germinal vesicle (GV) of mammalian oocytes. Dev Biol. 2006;292(1):1–12. doi: 10.1016/j.ydbio.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 52.Zuccotti M, Bellone M, Longo F, Redi CA, Garagna S. Fully-mature antral mouse oocytes are transcriptionally silent but their heterochromatin maintains a transcriptional permissive histone acetylation profile. J Assist Reprod Genet. 2011;28(12):1193–1196. doi: 10.1007/s10815-011-9562-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burns KH, Viveiros MM, Ren Y, Wang P, DeMayo FJ, Frail DE, et al. Roles of NPM2 in chromatin and nucleolar organization in oocytes and embryos. Science. 2003;300(5619):633–636. doi: 10.1126/science.1081813. [DOI] [PubMed] [Google Scholar]
- 54.Tamada H, Van Thuan N, Reed P, Nelson D, Katoku-Kikyo N, Wudel J, et al. Chromatin decondensation and nuclear reprogramming by nucleoplasmin. Mol Cell Biol. 2006;26(4):1259–1271. doi: 10.1128/MCB.26.4.1259-1271.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tan JH, Wang HL, Sun XS, Liu Y, Sui HS, Zhang J. Chromatin configurations in the germinal vesicle of mammalian oocytes. Mol Hum Reprod. 2009;15(1):1–9. doi: 10.1093/molehr/gan069. [DOI] [PubMed] [Google Scholar]
- 56.Bogolyubova I, Bogolyubov D. Heterochromatin morphodynamics in late oogenesis and early embryogenesis of mammals. Cells. 2020;9(6). doi: 10.3390/cells9061497. [DOI] [PMC free article] [PubMed]
- 57.Zernicka-Goetz M. Proclaiming fate in the early mouse embryo. Nat Cell Biol. 2011;13(2):112–114. doi: 10.1038/ncb0211-112. [DOI] [PubMed] [Google Scholar]
- 58.Zuccotti M, Merico V, Sacchi L, Bellone M, Brink TC, Stefanelli M, et al. Oct-4 regulates the expression of Stella and Foxj2 at the Nanog locus: implications for the developmental competence of mouse oocytes. Hum Reprod. 2009;24(9):2225–2237. doi: 10.1093/humrep/dep191. [DOI] [PubMed] [Google Scholar]
- 59.Zuccotti M, Merico V, Sacchi L, Bellone M, Brink TC, Bellazzi R, et al. Maternal Oct-4 is a potential key regulator of the developmental competence of mouse oocytes. BMC Dev Biol. 2008;8:97. doi: 10.1186/1471-213X-8-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zuccotti M, Merico V, Belli M, Mulas F, Sacchi L, Zupan B, et al. OCT4 and the acquisition of oocyte developmental competence during folliculogenesis. Int J Dev Biol. 2012;56(10–12):853–858. doi: 10.1387/ijdb.120174mz. [DOI] [PubMed] [Google Scholar]
- 61.Dahl JA, Jung I, Aanes H, Greggains GD, Manaf A, Lerdrup M, et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature. 2016;537(7621):548–552. doi: 10.1038/nature19360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang B, Zheng H, Huang B, Li W, Xiang Y, Peng X, et al. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature. 2016;537(7621):553–557. doi: 10.1038/nature19361. [DOI] [PubMed] [Google Scholar]
- 63.Wallace JA, Felsenfeld G. We gather together: insulators and genome organization. Curr Opin Genet Dev. 2007;17(5):400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Szabo PE, Tang SH, Silva FJ, Tsark WM, Mann JR. Role of CTCF binding sites in the Igf2/H19 imprinting control region. Mol Cell Biol. 2004;24(11):4791–4800. doi: 10.1128/MCB.24.11.4791-4800.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wan LB, Pan H, Hannenhalli S, Cheng Y, Ma J, Fedoriw A, et al. Maternal depletion of CTCF reveals multiple functions during oocyte and preimplantation embryo development. Development. 2008;135(16):2729–2738. doi: 10.1242/dev.024539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Evans JP. The molecular basis of sperm-oocyte membrane interactions during mammalian fertilization. Hum Reprod Update. 2002;8(4):297–311. doi: 10.1093/humupd/8.4.297. [DOI] [PubMed] [Google Scholar]
- 67.Georgadaki K, Khoury N, Spandidos DA, Zoumpourlis V. The molecular basis of fertilization (Review) Int J Mol Med. 2016;38(4):979–986. doi: 10.3892/ijmm.2016.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nomikos M, Swann K, Lai FA. Starting a new life: sperm PLC-zeta mobilizes the Ca2+ signal that induces egg activation and embryo development: an essential phospholipase C with implications for male infertility. BioEssays. 2012;34(2):126–134. doi: 10.1002/bies.201100127. [DOI] [PubMed] [Google Scholar]
- 69.Mtango NR, Sutovsky M, Susor A, Zhong Z, Latham KE, Sutovsky P. Essential role of maternal UCHL1 and UCHL3 in fertilization and preimplantation embryo development. J Cell Physiol. 2012;227(4):1592–1603. doi: 10.1002/jcp.22876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fraser R, Lin CJ. Epigenetic reprogramming of the zygote in mice and men: on your marks, get set, go! Reproduction. 2016;152(6):R211–R222. doi: 10.1530/REP-16-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guo F, Li X, Liang D, Li T, Zhu P, Guo H, et al. Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell. 2014;15(4):447–459. doi: 10.1016/j.stem.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 72.Kim KH, Kim EY, Kim Y, Kim E, Lee HS, Yoon SY, et al. Gas6 downregulation impaired cytoplasmic maturation and pronuclear formation independent to the MPF activity. PLoS ONE. 2011;6(8):e23304. doi: 10.1371/journal.pone.0023304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu X, Viveiros MM, Eppig JJ, Bai Y, Fitzpatrick SL, Matzuk MM. Zygote arrest 1 (Zar1) is a novel maternal-effect gene critical for the oocyte-to-embryo transition. Nat Genet. 2003;33(2):187–191. doi: 10.1038/ng1079. [DOI] [PubMed] [Google Scholar]
- 74.Huang H, Rambaldi I, Daniels E, Featherstone M. Expression of the Wdr9 gene and protein products during mouse development. Dev Dyn. 2003;227(4):608–614. doi: 10.1002/dvdy.10344. [DOI] [PubMed] [Google Scholar]
- 75.Philipps DL, Wigglesworth K, Hartford SA, Sun F, Pattabiraman S, Schimenti K, et al. The dual bromodomain and WD repeat-containing mouse protein BRWD1 is required for normal spermiogenesis and the oocyte-embryo transition. Dev Biol. 2008;317(1):72–82. doi: 10.1016/j.ydbio.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nakamura BN, Fielder TJ, Hoang YD, Lim J, McConnachie LA, Kavanagh TJ, et al. Lack of maternal glutamate cysteine ligase modifier subunit (Gclm) decreases oocyte glutathione concentrations and disrupts preimplantation development in mice. Endocrinology. 2011;152(7):2806–2815. doi: 10.1210/en.2011-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tsukada Y, Akiyama T, Nakayama KI. Maternal TET3 is dispensable for embryonic development but is required for neonatal growth. Sci Rep. 2015;5:15876. doi: 10.1038/srep15876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ramos SB, Stumpo DJ, Kennington EA, Phillips RS, Bock CB, Ribeiro-Neto F, et al. The CCCH tandem zinc-finger protein Zfp36l2 is crucial for female fertility and early embryonic development. Development. 2004;131(19):4883–4893. doi: 10.1242/dev.01336. [DOI] [PubMed] [Google Scholar]
- 79.Banaszynski LA, Allis CD, Lewis PW. Histone variants in metazoan development. Dev Cell. 2010;19(5):662–674. doi: 10.1016/j.devcel.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schultz RM. The molecular foundations of the maternal to zygotic transition in the preimplantation embryo. Hum Reprod Update. 2002;8(4):323–331. doi: 10.1093/humupd/8.4.323. [DOI] [PubMed] [Google Scholar]
- 81.Eckersley-Maslin MA, Alda-Catalinas C, Reik W. Dynamics of the epigenetic landscape during the maternal-to-zygotic transition. Nat Rev Mol Cell Biol. 2018;19(7):436–450. doi: 10.1038/s41580-018-0008-z. [DOI] [PubMed] [Google Scholar]
- 82.Hirasawa R, Chiba H, Kaneda M, Tajima S, Li E, Jaenisch R, et al. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev. 2008;22(12):1607–1616. doi: 10.1101/gad.1667008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Obata Y, Kono T. Maternal primary imprinting is established at a specific time for each gene throughout oocyte growth. J Biol Chem. 2002;277(7):5285–5289. doi: 10.1074/jbc.M108586200. [DOI] [PubMed] [Google Scholar]
- 84.Kaneda M, Hirasawa R, Chiba H, Okano M, Li E, Sasaki H. Genetic evidence for Dnmt3a-dependent imprinting during oocyte growth obtained by conditional knockout with Zp3-Cre and complete exclusion of Dnmt3b by chimera formation. Genes Cells. 2010;15(3):169–179. doi: 10.1111/j.1365-2443.2009.01374.x. [DOI] [PubMed] [Google Scholar]
- 85.Peng H, Chang B, Lu C, Su J, Wu Y, Lv P, et al. Nlrp2, a maternal effect gene required for early embryonic development in the mouse. PLoS ONE. 2012;7(1):e30344. doi: 10.1371/journal.pone.0030344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Meyer E, Lim D, Pasha S, Tee LJ, Rahman F, Yates JR, et al. Germline mutation in NLRP2 (NALP2) in a familial imprinting disorder (Beckwith-Wiedemann Syndrome) PLoS Genet. 2009;5(3):e1000423. doi: 10.1371/journal.pgen.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shinagawa T, Takagi T, Tsukamoto D, Tomaru C, Huynh LM, Sivaraman P, et al. Histone variants enriched in oocytes enhance reprogramming to induced pluripotent stem cells. Cell Stem Cell. 2014;14(2):217–227. doi: 10.1016/j.stem.2013.12.015. [DOI] [PubMed] [Google Scholar]
- 88.Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, et al. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell. 2008;15(4):547–557. doi: 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet. 2000;25(3):269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- 90.Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134(1):162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fallahian M, Sebire NJ, Savage PM, Seckl MJ, Fisher RA. Mutations in NLRP7 and KHDC3L confer a complete hydatidiform mole phenotype on digynic triploid conceptions. Hum Mutat. 2013;34(2):301–308. doi: 10.1002/humu.22228. [DOI] [PubMed] [Google Scholar]
- 92.Kanzaki S, Tamura S, Ito T, Wakabayashi M, Saito K, Kato S, et al. Involvement of Nlrp9a/b/c in mouse preimplantation development. Reproduction. 2020;160(2):181–191. doi: 10.1530/REP-19-0516. [DOI] [PubMed] [Google Scholar]
- 93.Disteche CM, Berletch JB. X-chromosome inactivation and escape. J Genet. 2015;94(4):591–599. doi: 10.1007/s12041-015-0574-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bultman SJ, Gebuhr TC, Pan H, Svoboda P, Schultz RM, Magnuson T. Maternal BRG1 regulates zygotic genome activation in the mouse. Genes Dev. 2006;20(13):1744–1754. doi: 10.1101/gad.1435106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Posfai E, Kunzmann R, Brochard V, Salvaing J, Cabuy E, Roloff TC, et al. Polycomb function during oogenesis is required for mouse embryonic development. Genes Dev. 2012;26(9):920–932. doi: 10.1101/gad.188094.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rother F, Shmidt T, Popova E, Krivokharchenko A, Hugel S, Vilianovich L, et al. Importin alpha7 is essential for zygotic genome activation and early mouse development. PLoS ONE. 2011;6(3):e18310. doi: 10.1371/journal.pone.0018310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kim KH, Kim EY, Lee KA. SEBOX is essential for early embryogenesis at the two-cell stage in the mouse. Biol Reprod. 2008;79(6):1192–1201. doi: 10.1095/biolreprod.108.068478. [DOI] [PubMed] [Google Scholar]
- 98.Park MW, Kim KH, Kim EY, Lee SY, Ko JJ, Lee KA. Associations among Sebox and other MEGs and its effects on early embryogenesis. PLoS ONE. 2015;10(2):e0115050. doi: 10.1371/journal.pone.0115050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rebuzzini P, Zuccotti M, Garagna S. Building pluripotency identity in the early embryo and derived stem cells. Cells. 2021;10(8). doi: 10.3390/cells10082049. [DOI] [PMC free article] [PubMed]
- 100.Bebbere D, Masala L, Albertini DF, Ledda S. The subcortical maternal complex: multiple functions for one biological structure? J Assist Reprod Genet. 2016;33(11):1431–1438. doi: 10.1007/s10815-016-0788-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Monk D, Sanchez-Delgado M, Fisher R. NLRPs, the subcortical maternal complex and genomic imprinting. Reproduction. 2017;154(6):R161–R170. doi: 10.1530/REP-17-0465. [DOI] [PubMed] [Google Scholar]
- 102.Gao Z, Zhang X, Yu X, Qin D, Xiao Y, Yu Y, et al. Zbed3 participates in the subcortical maternal complex and regulates the distribution of organelles. J Mol Cell Biol. 2018;10(1):74–88. doi: 10.1093/jmcb/mjx035. [DOI] [PubMed] [Google Scholar]
- 103.Yu XJ, Yi Z, Gao Z, Qin D, Zhai Y, Chen X, et al. The subcortical maternal complex controls symmetric division of mouse zygotes by regulating F-actin dynamics. Nat Commun. 2014;5:4887. doi: 10.1038/ncomms5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bajoghli B. Evolution of the Groucho/Tle gene family: gene organization and duplication events. Dev Genes Evol. 2007;217(8):613–618. doi: 10.1007/s00427-007-0167-y. [DOI] [PubMed] [Google Scholar]
- 105.Buscarlet M, Stifani S. The 'Marx' of Groucho on development and disease. Trends Cell Biol. 2007;17(7):353–361. doi: 10.1016/j.tcb.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 106.Jafari T, Faghihimani E, Feizi A, Iraj B, Javanmard SH, Esmaillzadeh A, et al. Effects of vitamin D-fortified low fat yogurt on glycemic status, anthropometric indexes, inflammation, and bone turnover in diabetic postmenopausal women: a randomised controlled clinical trial. Clin Nutr. 2016;35(1):67–76. doi: 10.1016/j.clnu.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 107.Yurttas P, Vitale AM, Fitzhenry RJ, Cohen-Gould L, Wu W, Gossen JA, et al. Role for PADI6 and the cytoplasmic lattices in ribosomal storage in oocytes and translational control in the early mouse embryo. Development. 2008;135(15):2627–2636. doi: 10.1242/dev.016329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Esposito G, Vitale AM, Leijten FP, Strik AM, Koonen-Reemst AM, Yurttas P, et al. Peptidylarginine deiminase (PAD) 6 is essential for oocyte cytoskeletal sheet formation and female fertility. Mol Cell Endocrinol. 2007;273(1–2):25–31. doi: 10.1016/j.mce.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 109.Wei Y, Multi S, Yang CR, Ma J, Zhang QH, Wang ZB, et al. Spindle assembly checkpoint regulates mitotic cell cycle progression during preimplantation embryo development. PLoS ONE. 2011;6(6):e21557. doi: 10.1371/journal.pone.0021557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bennabi I, Terret ME, Verlhac MH. Meiotic spindle assembly and chromosome segregation in oocytes. J Cell Biol. 2016;215(5):611–619. doi: 10.1083/jcb.201607062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zheng P, Dean J. Role of Filia, a maternal effect gene, in maintaining euploidy during cleavage-stage mouse embryogenesis. Proc Natl Acad Sci U S A. 2009;106(18):7473–7478. doi: 10.1073/pnas.0900519106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tomasini R, Tsuchihara K, Tsuda C, Lau SK, Wilhelm M, Rufini A, et al. TAp73 regulates the spindle assembly checkpoint by modulating BubR1 activity. Proc Natl Acad Sci U S A. 2009;106(3):797–802. doi: 10.1073/pnas.0812096106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Khokhlova EV, Fesenko ZS, Sopova JV, Leonova EI. Features of DNA Repair in the early stages of mammalian embryonic development. Genes (Basel). 2020;11(10). doi: 10.3390/genes11101138. [DOI] [PMC free article] [PubMed]
- 114.Xu Q, Wang F, Xiang Y, Zhang X, Zhao ZA, Gao Z, et al. Maternal BCAS2 protects genomic integrity in mouse early embryonic development. Development. 2015;142(22):3943–3953. doi: 10.1242/dev.129841. [DOI] [PubMed] [Google Scholar]
- 115.Gurtu VE, Verma S, Grossmann AH, Liskay RM, Skarnes WC, Baker SM. Maternal effect for DNA mismatch repair in the mouse. Genetics. 2002;160(1):271–277. doi: 10.1093/genetics/160.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Roest HP, Baarends WM, de Wit J, van Klaveren JW, Wassenaar E, Hoogerbrugge JW, et al. The ubiquitin-conjugating DNA repair enzyme HR6A is a maternal factor essential for early embryonic development in mice. Mol Cell Biol. 2004;24(12):5485–5495. doi: 10.1128/MCB.24.12.5485-5495.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zou Y, Liu Y, Wu X, Shell SM. Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses. J Cell Physiol. 2006;208(2):267–273. doi: 10.1002/jcp.20622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Braude P, Bolton V, Moore S. Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature. 1988;332(6163):459–461. doi: 10.1038/332459a0. [DOI] [PubMed] [Google Scholar]