Summary
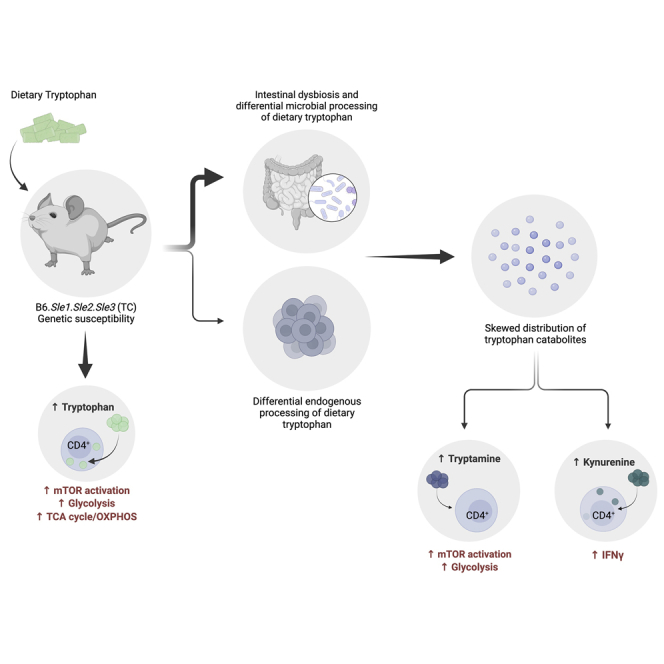
A skewed tryptophan metabolism has been reported in patients with lupus. Here, we investigated the mechanisms by which it occurs in lupus-susceptible mice, and how tryptophan metabolites exacerbate T cell activation. Metabolomic analyses demonstrated that tryptophan is differentially catabolized in lupus mice compared to controls and that the microbiota played a role in this skewing. There was no evidence for differential expression of tryptophan catabolic enzymes in lupus mice, further supporting a major contribution of the microbiota to skewing. However, isolated lupus T cells processed tryptophan differently, suggesting a contribution of T cell intrinsic factors. Functionally, tryptophan and its microbial product tryptamine increased T cell metabolism and mTOR activation, while kynurenine promoted interferon gamma production, all of which have been associated with lupus. These results showed that a combination of microbial and T cell intrinsic factors promotes the production of tryptophan metabolites that enhance inflammatory phenotypes in lupus T cells.
Subject areas: Biological sciences, Human metabolism, Immunology, Cell biology
Graphical abstract
Highlights
-
•
Intestinal dysbiosis skews tryptophan catabolism in lupus-prone mice
-
•
Murine lupus CD4+ T cells have an intrinsically different processing of tryptophan
-
•
Tryptophan and tryptamine increase mTOR activation and metabolism in CD4+ T cells
-
•
Kynurenine promotes IFNγ production in CD4+ T cells from lupus-prone mice
Biological sciences; Human metabolism; Immunology; Cell biology
Introduction
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease of complex etiology characterized by multi-organ damage and heterogeneous clinical manifestations, driven by an interplay between genetic susceptibility and environmental triggers. The impact of the microbiota on host immune reactivity (Belkaid and Harrison, 2017; Blander et al., 2017) has been a topic of great interest in the field of autoimmunity. Mounting evidence suggest that the microbiota plays a role in lupus, with intestinal dysbiosis reported in patients with SLE (Azzouz et al., 2019; He et al., 2016; Hevia et al., 2014; Zhang et al., 2014) and several mouse models of the disease (Choi et al., 2020; Luo et al., 2018; Manfredo Vieira et al., 2018; Zegarra-Ruiz et al., 2019; Zhang et al., 2014). We found perturbed gut microbial communities in the lupus-prone B6.Sle1.Sle2.Sle3 triple congenic (TC) mouse model, whereby it amplifies disease, at least in part, through microbial utilization of tryptophan and an imbalanced distribution of its catabolites (Choi et al., 2020). Therefore, specific microbial tryptophan metabolites may play an important role in disease pathogenesis in the TC model.
Tryptophan is an essential amino acid that is the precursor for the synthesis of numerous metabolites derived from both endogenous (i.e. host) and microbial enzymes. Microbes produce bioactive catabolites from dietary tryptophan (Agus et al., 2018; Gao et al., 2018; Roager and Licht, 2018), such as indole via the tryptophanase enzyme (TnaA), and various indole derivatives through separate pathways (Agus et al., 2018; Roager and Licht, 2018). Furthermore, some bacteria use tryptophan decarboxylase to produce tryptamine (Williams et al., 2014), which regulates intestinal transit by binding to serotonin receptors (Bhattarai et al., 2020 2018). Kynurenine, indoles, and tryptamine are AhR ligands (Cheng et al., 2015; Zelante et al., 2013) and thus have an immunomodulatory capacity in the context of autoimmunity (Rothhammer et al., 2016, 2018). We have previously reported that high dietary tryptophan exacerbated autoimmune phenotypes in TC mice, whereas low dietary tryptophan conferred protection (Choi et al., 2020). Although the mechanism by which it occurs remained elusive, our data suggested that this modulation was, at least to some extent, mediated by the gut microbiota (Choi et al., 2020). B6 and TC mice presented a differential distribution of fecal tryptophan metabolites, which was normalized by cohousing (Choi et al., 2020), suggesting that the TC microbiota has an altered capacity for tryptophan catabolism. In support of this hypothesis, TC mice responded with acute weight loss to dietary tryptophan deficiency compared to B6 controls (Choi et al., 2020), suggesting a higher abundance of microbes that catabolize tryptophan in the gut of TC mice. Indeed, some species of Prevotella, Paraprevotella, and Lactobacillus, which can catabolize tryptophan (Roager and Licht, 2018; Sasaki-Imamura et al., 2011; Zelante et al., 2013), were enriched in the TC gut microbiota. Moreover, fecal microbiota transfers from TC mice fed high tryptophan were more immunogenic in germ-free (GF) B6 recipients than transfers from TC mice fed low tryptophan (Choi et al., 2020).
Skewed tryptophan metabolism toward the kynurenine pathway (KP) has been observed in patients with SLE (Lood et al., 2015; Pertovaara et al., 2007; Widner et al., 1999, 2000; Åkesson et al., 2018), and its positive correlation with disease activity (Pertovaara et al., 2007; Åkesson et al., 2018) suggests that it plays a role in lupus pathogenesis. Kynurenine was one of the most prominently increased metabolites in the peripheral blood lymphocytes of patients with SLE, and its abundance was predictive of the clinical response to a treatment with N-acetylcysteine (Perl et al., 2015). Although kynurenine is primarily considered an immunosuppressive metabolite, its physiological role in lupus is not well understood. Exogenous kynurenine enhanced Th1 polarization in B6 and TC CD4+ T cells, and reduced Treg cell polarization in TC T cells (Choi et al., 2020), suggesting that kynurenine may promote pro-inflammatory T cell phenotypes, especially in a lupus-prone setting. Moreover, kynurenine promoted mTOR activation in human T cells (Perl et al., 2015), suggesting that it may contribute to the high level of mTOR activation that characterizes CD4+ T cells of patients with SLE (Perl, 2016) and lupus-prone mice (Choi et al., 2018; Yin et al., 2015).
It has been proposed that the increased kynurenine production in patients with SLE results from the upregulation of IDO by type 1 interferon (IFN), which plays a major role in lupus pathogenesis (Lood et al., 2015). However, the numerous enzymes and downstream catabolites in this pathway, including those of microbial origin, have not been assessed. Additional catabolites derived from endogenous or microbial processing of tryptophan may also exacerbate pro-inflammatory immune cell phenotypes. In the present study, we investigated the source of tryptophan metabolite skewing and the mechanisms by which tryptophan metabolites exacerbate CD4+ T cell phenotypes in lupus-susceptible TC mice. We demonstrated that tryptophan is differentially metabolized in TC mice and their CD4+ T cells, as compared to non-autoimmune B6 controls. We did not find evidence for differential expression of tryptophan catabolic enzymes in TC mice, suggesting that tryptophan metabolite skewing in this model is largely mediated by the intestinal microbiota. The differential effect of tryptophan on isolated TC T cells also suggests the existence of intrinsic factors. Furthermore, we showed that high tryptophan and tryptamine, but not kynurenine, activated mTOR in TC T cells. Finally, we showed that dietary tryptophan as well as exogenous tryptamine promoted glycolysis in TC T cells. These results suggest that tryptophan metabolites may modulate CD4+ T cell functions in lupus.
Results
Dietary tryptophan is differentially metabolized in lupus-prone TC mice
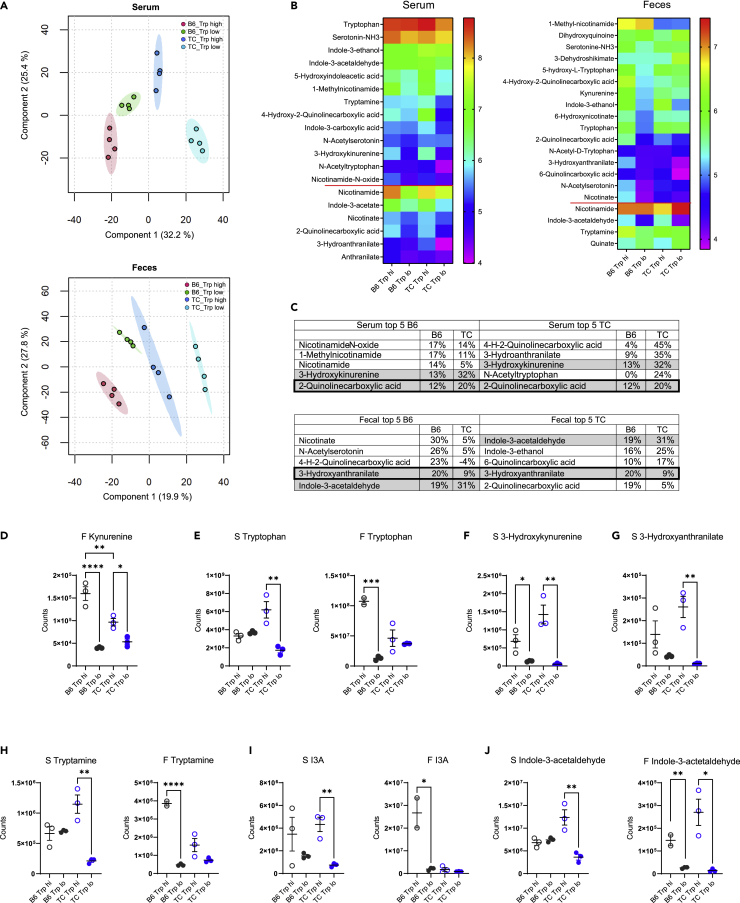
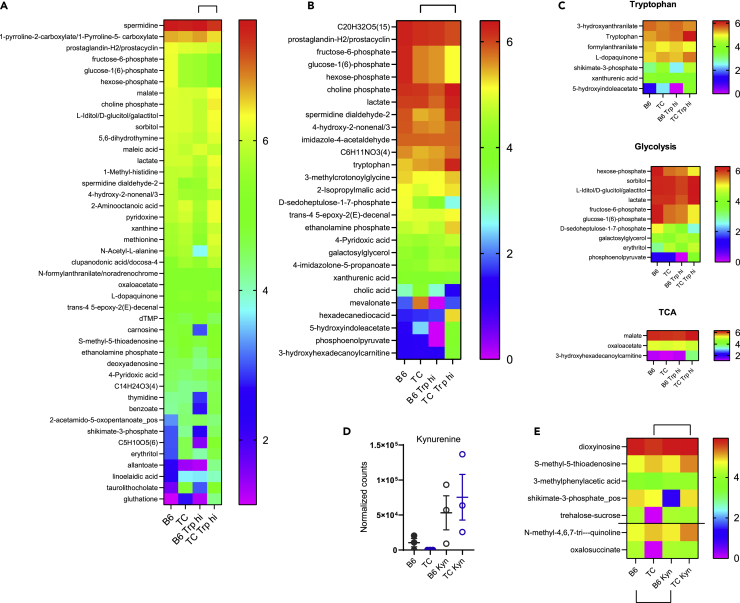
To understand how dietary tryptophan is metabolized in TC and B6 mice, we assessed serum and fecal metabolites from B6 and TC mice consuming high or low tryptophan for one month beginning at 3 months of age, before the onset of autoantibody production. This age was selected to exclude potential secondary effects of autoimmune activation. Global differences in metabolite profiles were observed between tryptophan diets and between strains for both serum and feces (Figures 1A and S1, and Tables S1 and S2). Among these, the abundance of 19 serum and 20 fecal tryptophan metabolites differed between high and low dietary tryptophan, the majority of which were shared between serum and feces (Figure 1B). Some metabolites, such as nicotinamide, responded similarly to tryptophan variations in both strains. Most metabolites, however, responded differently between strains. Three out of the top 5 tryptophan serum and fecal metabolites that were the most changed by dietary tryptophan differed between the two strains (Figure 1C). Fecal kynurenine was increased in mice fed with high tryptophan in both strains, but to a greater extent in B6 than TC mice (Figure 1D). Many metabolites showed a more exaggerated response in TC mice. Only TC mice fed high tryptophan showed increased tryptophan in the serum, while only B6 mice fed with high tryptophan showed increased tryptophan in feces (Figure 1E). In addition, metabolites of endogenous origin, such as 3-hydroxykynurenine (Figure 1F) and 3-hydroxyanthranilate (Figure 1G), were increased by dietary tryptophan to a greater extent in TC mice. Tryptophan metabolites of microbial origin were also differentially affected by dietary tryptophan between the two strains, including increased tryptamine (Figure 1H) and indole-3-acetate (I3A, Figure 1I) in TC sera and increased serum and fecal indole-3-acetaldehyde in TC mice (Figure 1J). No differences were observed for tryptophan, tryptamine, or I3A in TC feces, while high tryptophan increased their abundance in B6 feces (Figures 1E and 1H–1I, p < 0.05 in B6 for all 3 metabolites). Collectively, these data suggest that dietary tryptophan is differentially metabolized in TC mice compared to non-autoimmune B6 controls.
Figure 1.
Dietary tryptophan is processed differently in lupus-prone and control mice
(A) PLS-DA plots of serum and fecal metabolites from B6 and TC mice fed high or low tryptophan for 1 month (N = 4 per group). For simplicity, only the positive mode is shown.
(B) Heatmaps of tryptophan metabolites differently represented in serum and feces among the 4 groups (p < 0.05 analyzed by ANOVA). Data are shown as Log10 transformed mean for each group ordered from the highest to lowest in the B6 Trp low group. Metabolites below the red horizontal lines were changed by dietary tryptophan to a similar extent in both strains. Metabolites above the red lines were affected differently by tryptophan between the two strains, i.e. there was a significant difference between the Trp high groups with no difference between Trp low groups, or there was a difference between Trp low and high in only one of the strains.
(C) Top 5 serum and fecal tryptophan metabolites ranked separately for B6 and TC as the most changed by high dietary tryptophan. The values show the difference between high and low tryptophan as a percentage of the low tryptophan value, all Log10-transformed, as shown in the heatmaps.
(D–G) Values for tryptophan and endogenous metabolites.
(H–J) Values for microbial metabolites. C–J: Graphs show individual mice with mean ± SEM t tests, ∗p < 0.05, ∗∗p < 0.01.
Intestinal microbes contribute to tryptophan metabolite skewing in lupus-prone TC mice
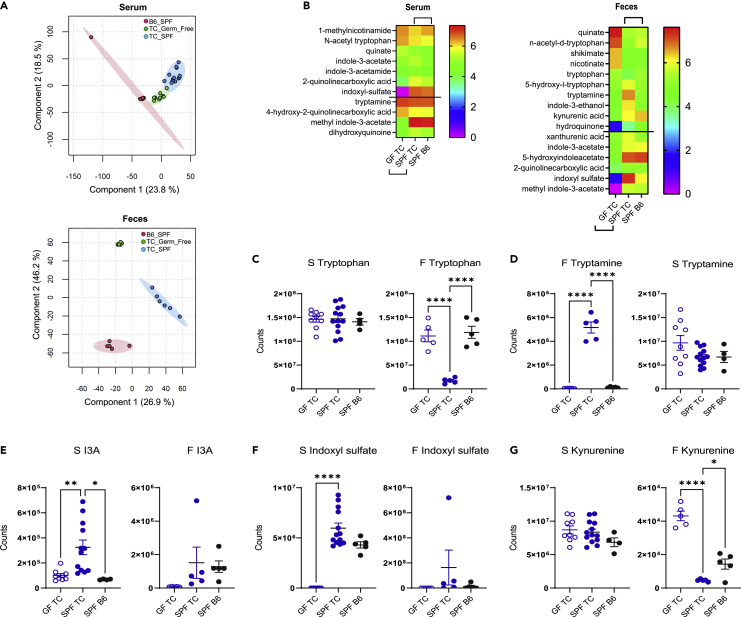
To elucidate whether the intestinal microbiota contributes to skewed tryptophan metabolites in TC mice, we compared serum and fecal metabolites between TC mice raised under GF and specific pathogen-free (SPF) conditions, with SPF B6 samples as controls, all obtained from the same institution (UC). As previously reported at the UF site (Choi et al., 2020), serum and fecal metabolites differ between TC and B6 SPF mice at the UC site, and, as expected, large differences were observed between GF and SPF TC samples (Figure 2A and S2 and Tables S4 and S5). More interestingly, the abundance of several tryptophan catabolites of microbial and endogenous origin were different in the serum and feces between groups (Figure 2B). Many catabolites that differed between SPF TC and B6 serum and fecal samples were also significantly altered in GF TC mice, indicating a major microbial contribution.
Figure 2.
Microbial contribution to serum and feces metabolites in TC mice
(A) PLS-DA plots of serum and fecal metabolite profiles in GF TC, SPF TC, and SPF B6 mice. For simplicity, only the positive mode is shown.
(B) Tryptophan metabolites in the serum and feces of GF and SPF TC as well as SPF B6 mice. All metabolites represented were significantly different between GF and SPF TC samples (microbiome effect shown by the lower brackets). Above the horizontal lines, samples were also different between SPF TC and SPF B6 samples (strain effect shown by the upper brackets). Data show Log10 transformed means for each group ordered from the highest to lowest in GF TC group (serum: N = 9 GF TC, 13 SPF TC, and 5 SPF B6; feces: N = 5 per group).
(C–G) Serum and fecal values in GF TC, SPF TC, and SPF B6 mice for tryptophan (C), tryptamine (D), indole-3-acetate (I3A) (E), indoxyl sulfate (F), and kynurenine (G). B–G: Graphs show individual mice with mean ± SEM 1-way ANOVA with T3 Dunnett’s multiple comparisons tests. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
The level of fecal tryptophan was dramatically reduced in SPF TC mice as compared to GF TC mice, which had a tryptophan concentration comparable to SPF B6 mice (Figure 2C), suggesting that the TC microbiota metabolizes more tryptophan than B6 intestinal microbes. On the other hand, fecal tryptamine (Figure 2D) and serum I3A (Figure 2E) were increased in SPF TC mice compared to GF TC mice and SPF B6 controls, suggesting that the TC microbiota produces more microbial-derived tryptophan metabolites than the B6 microbiota. Interestingly, tryptamine and I3A were also increased in the serum of TC mice fed high tryptophan (Figure 1). Indoxyl sulfate was also increased in the serum of SPF TC mice compared to GF TC and SPF B6 controls (Figure 2F), suggesting that more indole is produced by the TC microbiota, since indoxyl sulfate is an endogenous metabolite synthesized from bacterial indole in the liver. In addition to microbial tryptophan metabolites, we confirmed an increased level of serum kynurenine in SPF TC as compared to B6 (Choi et al., 2020), but contrary to results previously reported, fecal kynurenine was higher in SPF B6 than in SPF TC mice (Figure 2G). These differences may reflect environmental factors between the two institutions (UF and UC) that strongly impact fecal kynurenine levels. Serum kynurenine was unchanged in TC mice under GF relative to SPF conditions, but the fecal kynurenine concentration was decreased in SPF relative to GF TC mice (Figure 2G). This may reflect the expansion of bacteria in TC gut that can metabolize kynurenine into hydroxyanthranilic acid (Agus et al., 2018). Overall, these results strongly suggest that the intestinal microbiota contributes to the alteration of tryptophan metabolism in TC mice but not to their accumulation of kynurenine.
TC mice express normal levels of endogenous tryptophan metabolism enzymes
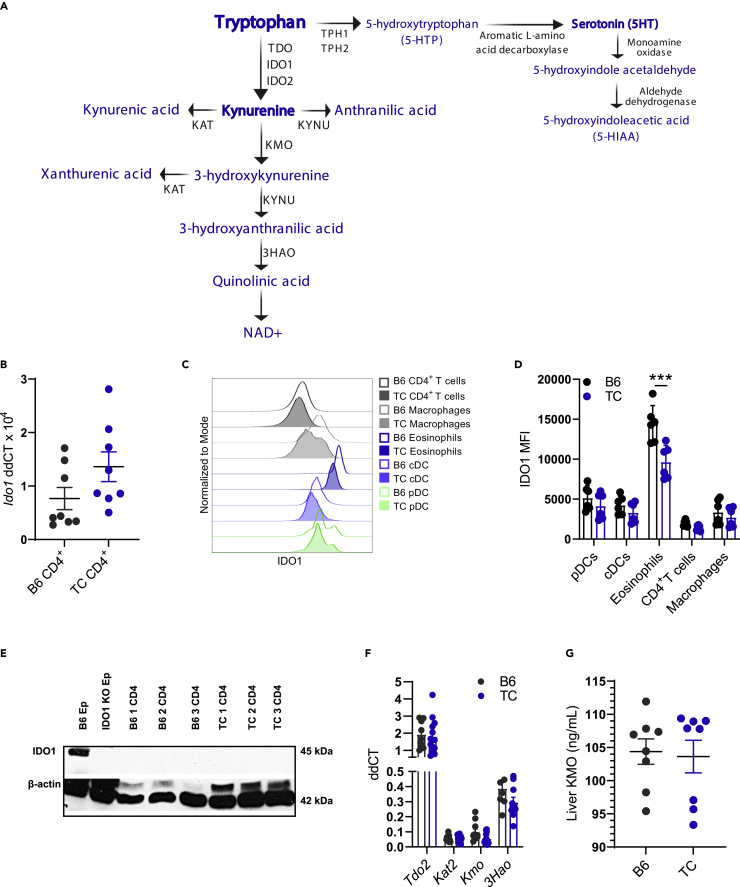
Endogenous enzymes in the tryptophan catabolism pathway (Figure 3A) could also contribute to the skewing of tryptophan metabolites in TC mice if their expression is modified by the inflammatory milieu, such as the high levels of type I and II IFN. Additionally, the genes for two of these enzymes, Kmo and Tph1, are located in the Sle1 and Sle3 loci, respectively, and genetic polymorphisms could lead to differential expression. To investigate whether endogenous enzymes contributed to the alterations in tryptophan metabolism in TC mice, we first focused on IDO1, which is expressed in numerous tissues, including immune cells, most notably dendritic cells (DCs) (Fallarino et al., 2002), and in the gut epithelium. We have previously reported a decreased Ido1 expression in DCs and no difference in the gut of TC compared to B6 mice (Choi et al., 2020). Here, we show a similar Ido1 expression in B6 and TC CD4+ T cells (Figure 3B). We confirmed by flow cytometry that IDO1 protein levels were similar between TC and B6 plasmacytoid DCs (pDCs), classical DCs (cDCs), and CD4+ T cells. However, eosinophils, a cell type that constitutively expresses IDO1 (Odemuyiwa et al., 2004), showed reduced levels of this enzyme in TC mice (Figures 3C–3D and S3A for gating strategy). The previously reported absence of IDO1 expression in T cells by Western blot analysis (Merlo et al., 2020) was confirmed in TC CD4+ T cells (Figure 3E). In addition, we found no difference in Ido2 expression between B6 and TC CD4+ T cells (Figure S3B), and IDO2 protein was not detected in CD4+ T cells from either strain (Figure S3C), which also aligns with a previous report (Merlo et al., 2020).
Figure 3.
Endogenous enzymes are not responsible for elevated kynurenine in TC mice
(A) Depiction of the endogenous enzymes and metabolites (blue) in the kynurenine and serotonin pathways.
(B) Ido1 gene expression in B6 and TC CD4+ T cells.
(C) Representative flow cytometry histogram overlays of IDO1 mean fluorescence intensity (MFI) in immune cell subsets from B6 and TC mice.
(D) Quantitation of (C), N = 6.
(E) Western blot analysis of B6 and TC CD4+ T cells (N = 3) for IDO1 and β-actin. Epididymis (Ep) from B6 and B6.Ido1−/− mice were used as positive and negative controls, respectively.
(F) Expression of KP genes in B6 and TC livers relative to Ppia, N = 8–13.
(G) Concentration of KMO protein in B6 and TC liver, N = 8. B, D–G: C–J: Graphs show individual mice with mean ± SEM t tests, ∗∗∗p < 0.001.
Hepatocytes express another major kynurenine synthesizing enzyme, TDO (encoded by Tdo2, Figure 3A), and accounts for a large part of tryptophan catabolism through the KP (Cervenka et al., 2017). We observed no difference in gene expression of any KP enzymes between TC and B6 livers (Figure 3F). We observed a trend for reduced Kmo expression in TC livers compared to B6 (Figure 3F), but KMO protein levels were similar between strains (Figure 3G). This indicates the buildup of kynurenine in TC mice is not due to its reduced catabolism by KMO. Overall, these results suggest that conventional endogenous enzymes are not responsible for the skewing of tryptophan metabolism in TC mice.
Tryptophan directly modifies the metabolism of CD4+ T cells
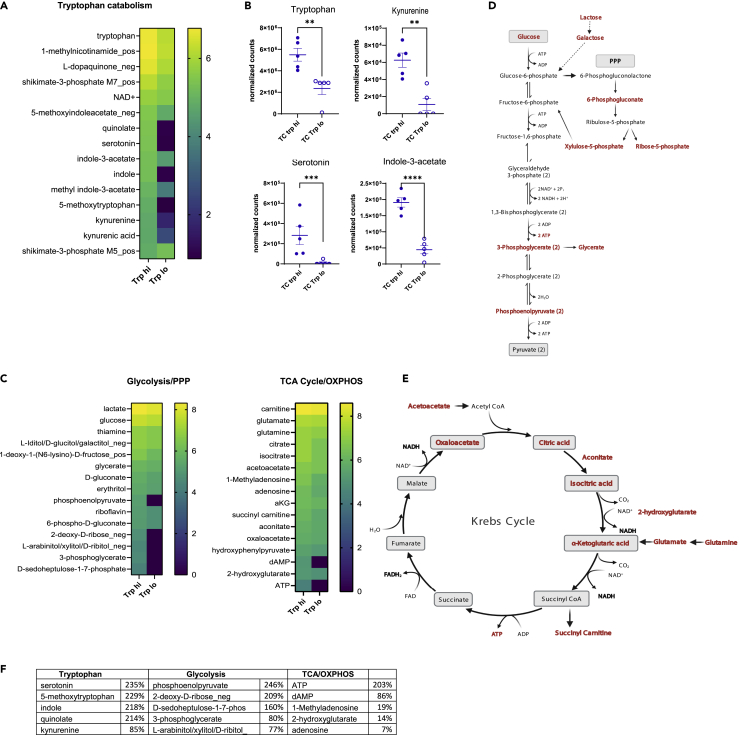
Because variations in dietary tryptophan altered the phenotypes of CD4+ cells from lupus-prone TC mice (Choi et al., 2020), we then assessed whether these phenotypes corresponded to changes in the metabolome of these cells. The abundance of 201 named metabolites differed between the CD4+ T cells from TC mice fed high or low tryptophan for 1 month (Table S3). We focused on the tryptophan, glycolysis, and tricarboxylic acid (TCA)/oxidative phosphorylation (OXPHOS) pathways because of relevance to T cell activation and previous results obtained in this model. The variation in dietary tryptophan resulted in differences in 15 tryptophan catabolites from both microbial and endogenous origins (Figure 4A). Increased amounts of tryptophan, kynurenine, serotonin, and I3A were found in CD4+ T cells from TC mice fed high tryptophan compared to those fed low tryptophan (Figure 4B). In these cells, we also observed a profound increase in metabolites associated with glycolysis and the pentose phosphate pathway (PPP) as well as the Krebs cycle (Figure 4C), which are depicted within their respective pathways (Figures 4D and 4E). Figure 4F summarizes these findings by showing the top 5 most differentially represented metabolites between low and high dietary tryptophan in TC CD4+ T cells for each of these three metabolic pathways. These results indicate that the distribution of tryptophan metabolites within TC CD4+ T cells is altered by high dietary tryptophan, which increased their glycolytic and mitochondria metabolism.
Figure 4.
Dietary tryptophan alters the metabolism of TC CD4+ T cells
(A) Heatmap of tryptophan metabolites in CD4+ T cells from TC mice fed with high or low tryptophan for 1 month (N = 5 per group).
(B) Values from (A) shown for selected metabolites. Graphs show individual mice with mean ± SEM t tests, ∗∗p < 0.01, ∗∗p < 0.001, ∗∗∗p < 0.0001.
(C) Heatmaps of metabolites in the glycolysis/PPP and TCA cycle/OXPHOS pathways showing Log10 transformed means for each group ordered from the highest to lowest in the Trp high group (N = 5).
(D) Representation of the glycolysis/PPP pathway.
(E) Representation of the TCA cycle. In (D) and (E), the metabolites present in the heatmaps in (C) are shown in red.
(F) Top 5 CD4+ T cells most changed by high dietary tryptophan in TC mice for the tryptophan, glycolysis, and TC/OXPHOS pathways. The values show the difference between high and low tryptophan as a percentage of the low tryptophan value, all Log10-transformed, as shown in the heatmaps.
Because we observed that high dietary tryptophan promotes increased glycolysis and mitochondrial metabolism in T cells from TC mice, we then sought to determine if exogenous tryptophan and kynurenine had a direct effect on CD4+ T cells. We compared the metabolome of CD4+ T cells purified from B6 and TC mice on a standard diet and activated in vitro in the presence of exogenous tryptophan or kynurenine for 24 h. At baseline in medium alone (15 uM tryptophan, no kynurenine), B6 and TC CD4+ T cells differed by 16 metabolites that belong to a variety of pathways (Table S6). Only one tryptophan metabolite, 3-hydroxyanthranilate, was found at a higher level in B6 T cells. The addition of tryptophan increased the number of metabolites that differed between strains to 48 (Figure 5A), which was higher than the number at baseline (Χ2 = 8.53, p = 0.0035). TC CD4+ T cells differed between baseline and high tryptophan at 32 metabolites, 27 of which were not affected by tryptophan in B6 CD4+ T cells (Figure 5B). Comparatively, only 10 metabolites differed between B6 CD4+ T cells at baseline and high tryptophan (Χ2 = 6.04, p = 0.012), 4 of which were unique to B6 (data not shown). Only 5 metabolites differed between strains at both baseline and high tryptophan, including 3-hydroxy anthranilate (Figure 5C). High tryptophan increased lactate levels and decreased the production of most glycolytic intermediates in TC but not in B6 T cells (Figure 5C). An increase in TCA cycle activity was also observed in TC T cells exposed to high tryptophan, with high levels of 3-hydroxyhexadecanoylcarnitine indicating an increased β-oxidation. High levels of malate and decreased levels of oxaloacetate suggest an active malate-aspartate shuttle, which is critical for CD4+ T cell activation, especially IFNγ production (Bailis et al., 2019). Overall, these results show that tryptophan is metabolized differently between B6 and TC CD4+ T cells in an intrinsic manner, and that, high levels of tryptophan increase glycolytic and TCA metabolites in TC CD4+ T cells.
Figure 5.
Exogenous tryptophan enhanced glycolysis and TCA cycle activity in TC CD4+ T cells in vitro
(A) Heatmaps of metabolites differentially represented between B6 and TC CD4+ T cells after 24 h activation with the addition of 50 uM tryptophan (Trp hi). Metabolites that differed between strains only at basal level of tryptophan were excluded.
(B) Heatmaps of metabolites differently represented between TC CD4+ T cells at baseline and with 50 uM tryptophan.
(C) Subsets of metabolites from A and B in the tryptophan, glycolysis, and TCA pathways.
(D) Kynurenine levels in B6 and TC CD4+ T cells after 24 h activation without or with the addition 50 uM kynurenine. Graphs show individual mice with mean ± SEM.
(E) Heatmap showing metabolites differently represented in TC and B6 T cells above and below the horizontal line respectively, with the addition of kynurenine. Data show Log10 transformed means for each group ordered from the highest to lowest in the B6 control group (N = 3 per group).
Kynurenine was not detected in CD4+ T cells from either strain in control medium but it was found in CD4+ T cells activated in the presence of kynurenine at similar levels between the two strains (Figure 5D). This confirmed that CD4+ T cells import kynurenine (Sinclair et al., 2018). Exogenous kynurenine altered the amounts of 15 and 28 metabolites in B6 and TC T cells, respectively, but most of them showed the same trends in both strains (Table S6). A small number of metabolites were uniquely changed by kynurenine in either TC or B6 cells (Figure 5E), but, contrary to tryptophan, none of them formed a coherent signature. These results suggest that kynurenine does not alter the metabolism of CD4+ T cells to the same extent as tryptophan, and that there was no specific metabolic enhancement of TC CD4+ T cells in response to kynurenine as observed for tryptophan.
High dietary tryptophan increases mTOR activation in lupus-prone CD4+ T cells
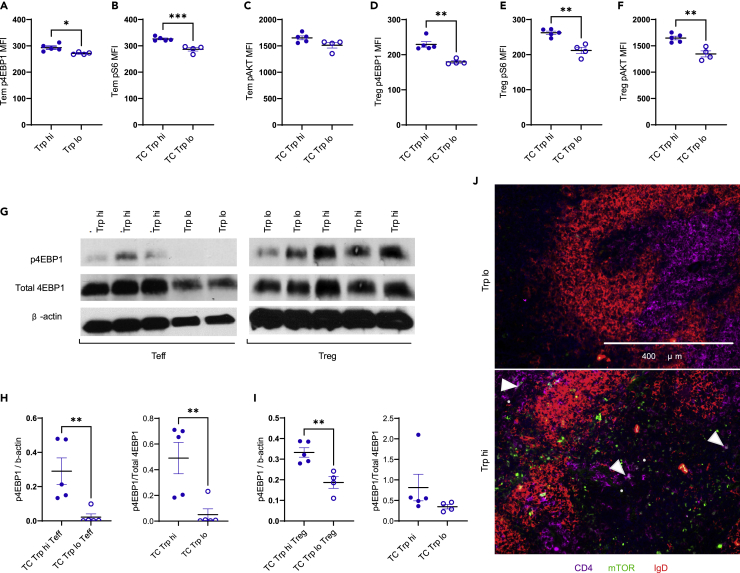
Kynurenine has been shown to activate mTOR in human T cells (Perl et al., 2015). mTOR activation regulates T cell metabolism by supporting glycolysis and mitochondrial respiration (Linke et al., 2017). Furthermore, mTOR activation as well as increased glycolysis and OXPHOS play a significant role in the expansion of pathogenic T cells in lupus (Yin et al., 2015). We hypothesized that the pathogenic effect of high dietary tryptophan in TC mice may be mediated at least in part through mTOR activation in T cells.
High dietary tryptophan increased mTORC1 activation in CD4+CD44+CD62L− effector memory (Tem) T cells and CD4+Foxp3+ regulatory (Treg) T cells from TC mice (Figure S4A), as demonstrated by increased p4EBP1 (Figures 6A and 6D) and pS6 (Figures 6B and 6E) expression. pAKTSer473 expression was unchanged in Tem cells (Figure 6C) but increased in Treg cells from TC mice fed high tryptophan (Figure 6F). Because AKT is phosphorylated by mTORC2 at the Ser473 site (Linke et al., 2017), this suggests that high dietary tryptophan activates mTORC1 in TC Tem cells, but that it promotes activation of both mTORC1 and mTORC2 in TC Treg cells. A Western blot analysis confirmed that p4EBP1 expression was increased in sorted CD4+CD25−CD44+ effector T (Teff) cells and CD4+CD25+ Treg cells from TC mice consuming high tryptophan (Figures 6G–6I and S4B–S4G). Finally, a high tryptophan diet increased the expression of mTOR in the spleen of TC mice, some of which co-localized to CD4+ T cells (Figure 6J). High dietary tryptophan did not promote mTOR activation in B6 CD4+T cells (Figures S4H–S4M), suggesting that the effect of high tryptophan on mTOR does not occur in the absence of autoimmune genetic predisposition. Therefore, our results show that dietary tryptophan increases mTOR activation in CD4+ T cells in TC mice.
Figure 6.
High dietary tryptophan promotes mTOR activation in CD4+ T cells
(A–F) MFI of p4EBP1, pS6, and pAKTSer473 in Tem (A–C) and Treg (D–F) cells from TC mice fed low or high tryptophan for 1 month, N = 4.
(G) Representative Western blot analysis of TC Teff and Treg cells for p4EBP1 and β-actin. For Teff samples, high tryptophan group is on the left. For Treg samples, high tryptophan group is on the right.
(H–I) Quantification of Western blot results for p4EBP1 relative to β-actin, N = 5.
(J) Representative immunofluorescence staining for CD4, mTOR, and IgD in spleen sections from TC mice fed high or low tryptophan for 2 months. Arrow heads indicate mTOR+ CD4+ T cells. Graphs show individual mice with mean ± SEM t tests, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
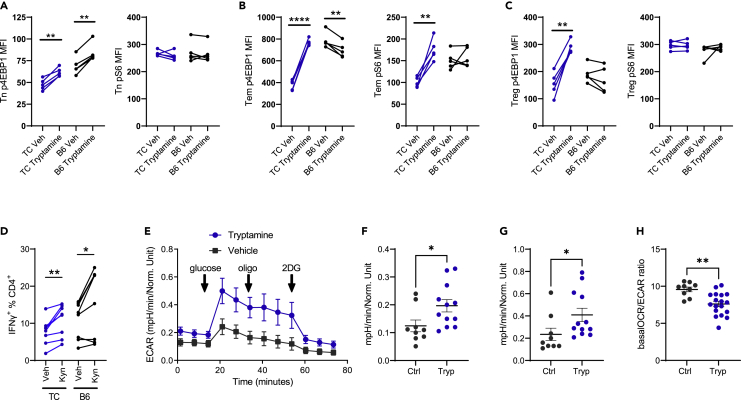
Tryptamine promotes mTOR activation and glycolysis in TC CD4+ T cells
To explore the effects of specific tryptophan metabolites on lupus CD4+ T cells, we tested the ability of kynurenine, tryptamine, and I3A to activate mTOR in CD4+ T cells in vitro. Tryptamine increased p4EBP1 expression in TC naive CD4+CD44− (Tn), Tem, and Treg cells (Figures 7A–7C), but only in Tn cells from B6 mice (Figure 7A). Tryptamine also increased pS6 expression in TC Tem cells (Figure 7B) but had no effect on pS6 in TC Tn and Treg cells (Figures 7A and 7C). In contrast, kynurenine reduced p4EBP1 expression in all TC T cell subsets, and it had no effect on pS6 expression (Figures S5A–S5C). However, kynurenine (Figure 7D), but not tryptamine (Figure S5D), increased IFNɣ production in activated B6 and TC CD4+ T cells. Therefore, tryptamine promoted mTORC1 activation in vitro across all TC CD4+ T cell subsets examined, whereas kynurenine promotes IFNɣ production from CD4+ T cells, both of which are pathogenic in lupus. The addition of I3A showed no effect on IFNɣ production and no effect (B6) or a decreased (TC) mTOR activation in CD4+ T cells (Figure S6). This suggests that individual tryptophan metabolites exert differential effects on the immune system, overall resulting in a more inflammatory setting.
Figure 7.
Tryptamine promotes mTOR activation in vitro and glycolysis in vivo in TC CD4+ T cells
(A–C) p4EBP1 and pS6 expression (MFI) in Tn (A), Tem (B), and Treg (C) cells gated from TC and B6 total CD4+ T cells incubated for 24 h with and without tryptamine.
(D) Frequency of IFNγ-producing CD4+ T cells after 96 h activation with and without kynurenine.
(E–H) Glycolysis stress assay on total CD4+ T cells from vehicle or tryptamine-treated TC mice.
(E) Complete assay with indications of the glucose, oligomycin, and 2DG injections.
(F) Basal ECAR before glucose injection. (F) Glycolysis: mean 3 values after glucose minus before glucose injections.
(H) Ratio of basal OCR and ECAR. (A–D) Graphs show paired t-tests between treated and control cells for each strain and subset (n = 5 for A-C, n = 8 for D).
(F–H) t tests (n = 9–12 from 2 cohorts). Graphs show individual mice with mean ± SEM ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001.
To further understand the effects of tryptamine on T cell phenotypes, we administered tryptamine to pre-autoimmune TC mice for one month. Contrary to direct exposure on isolated CD4+ T cells, tryptamine treatment did not increase mTOR activation in CD4+ T cells in vivo (Figures S5E–S5G), which may indicate either a dose effect or the interference of other cell types. However, the tryptamine treatment increased glucose metabolism in TC CD4+ T cells measured by a glycolysis stress test (Figure 7E), including basal extracellular acidification rate (ECAR) and glycolysis (Figures 7F and 7G). Furthermore, the basal oxygen consumption rate (OCR) to ECAR ratio was reduced in the CD4+ T cells of tryptamine-treated mice (Figure 7H). Taken together, these results show that tryptamine increases glycolysis in CD4+ T cells from TC mice.
Discussion
It has been proposed that microbial dysbiosis contributes to lupus pathogenesis through molecular mimicry between bacterial and auto-antigens (Azzouz et al., 2019; Greiling et al., 2018), or through bacterial translocation across a disrupted intestinal barrier to promote immune activation (Azzouz et al., 2019; Dehner et al., 2019; Manfredo Vieira et al., 2018; Zegarra-Ruiz et al., 2019). The lack of evidence for “leaky gut” and bacterial translocation in TC mice (Choi et al., 2020) suggested that microbial dysbiosis may contribute to autoimmune activation through metabolites. Indeed, TC mice present an altered tryptophan metabolism that was associated with their intestinal microbiota and that impacted autoimmune pathogenesis (Choi et al., 2020). This corresponded to numerous reports of skewed tryptophan catabolites favoring the kynurenine pathway in correlation with disease activity in patients with SLE (Brown et al., 2020). Although evidence suggests that type 1 IFN-mediated IDO1 upregulation increases kynurenine production in patients with SLE (Lood et al., 2015; Pertovaara et al., 2007), the TC lupus-prone mouse provided a model to perform a comprehensive evaluation of the microbial and endogenous contributions to the skewing of tryptophan catabolism in lupus. In addition, this model allowed the evaluation of the role of individual tryptophan metabolites to autoimmune activation.
The response of TC mice to variations in dietary tryptophan suggested that they metabolize this amino acid differently than controls (Choi et al., 2020). This hypothesis was validated by the finding that increased tryptophan intake resulted in different serum and fecal metabolite profiles between B6 and TC mice. These results demonstrated the expression of lupus-susceptible genes present in TC mice modulates tryptophan metabolism. Importantly, specific microbial tryptophan metabolites such as tryptamine and some indole compounds were increased by high dietary tryptophan only in TC mice. Therefore, these metabolites may promote pathogenesis in TC mice. High dietary tryptophan also induced a distinct profile of tryptophan metabolites in TC CD4+ T cells, including an increased amount of tryptophan, kynurenine, indole, and I3A, demonstrating that CD4+ T cells import some microbial tryptophan metabolites. Tryptophan also induced specific changes in isolated TC CD4+ T cells relative to B6 CD4+ T cells, further supporting altered tryptophan metabolism in that strain. Interestingly, we also observed an increased amount of tryptophan metabolites in the serum and CD4+ T cells from TC mice, while B6 feces presented an increased amount of tryptophan metabolites. These differences may reflect differences in tryptophan catabolism integrated with metabolite consumption by the host and its microbiota. The significance of the differential distribution of these metabolites in the general circulation (serum), in cells, or in association with the microbiota in feces is unknown. Tracer studies to compare the processing of dietary tryptophan between the two strains in various anatomic locations can provide some answers about the metabolic processes that are involved. The functional significance of these differences in term of impact on the immune system will be however more difficult to define.
We showed here that, as in B6 mice (Merlo et al., 2020), CD4+ T cells from TC mice do not express IDO1 or IDO2, even in presence of high amounts of tryptophan. We confirmed however that isolated CD4+ T cells import kynurenine (Sinclair et al., 2018). We also found no evidence of elevated IDO1 protein or mRNA expression in other immune cells of TC mice that could account for elevated kynurenine levels. In fact, we found a reduced IDO1 expression in TC eosinophils, and trends for decreased IDO1 in the remaining cell subsets examined. This observation is surprising considering the high type 1 and 2 IFN activity in TC mice (Choi et al., 2018; Sang et al., 2014), which should upregulate Ido1 expression (Lood et al., 2015; Pertovaara et al., 2007).
The TC microbiota synthesizes more indole compounds and tryptamine than the B6 microbiota in response to dietary tryptophan. The comparison of GF and SPF TC metabolites and the lack of differential expression of endogenous enzymes collectively suggest that tryptophan metabolite skewing in TC mice results largely from microbial enzymes. Therefore, as in a mouse model of multiple sclerosis (Sonner et al., 2019), dietary tryptophan potentiates pathogenic T cell responses largely through the intestinal microbiota in lupus-prone TC mice. Although some bacteria synthesize kynurenine from tryptophan using analogs of mammalian KP enzymes (Vujkovic-Cvijin et al., 2013), there was no difference in serum kynurenine between GF and SPF TC mice, suggesting that gut microbes do not directly contribute to elevated kynurenine in TC mice. The observation that fecal kynurenine was higher in GF TC mice relative to SPF TC mice also supports this conclusion. In addition, other endogenous KP metabolites, 3-hydroxykynurenine and 3-hydroxyanthranilate, were elevated only in TC mice consuming high dietary tryptophan, suggesting that there are other enzymes or cell types responsible for endogenous metabolite skewing. Finally, the different metabolite profiles between TC and B6 T cells directly exposed to tryptophan however indicate that tryptophan-catabolizing enzymes other than the classical KP enzymes must be differentially expressed in TC T cells. Therefore, in addition to a microbial origin, endogenous enzymes yet to be identified contribute to the increased KP activity in TC mice.
Besides the origin of skewed tryptophan metabolites, we investigated how they contribute to T cell activation. We showed using several approaches that mTOR activation is increased in Tn, Tem, and Treg cells from TC mice consuming high dietary tryptophan compared to those fed low tryptophan, while dietary tryptophan had a much weaker effect on mTOR in B6 T cells. This suggests that mTOR activation is a mechanism by which increased dietary tryptophan exacerbates disease in the TC model. Tryptamine activated mTOR in TC effector T cells and Treg cells in vitro, suggesting that tryptophan may activate mTOR through this metabolite directly. The observation that, contrary to human T cells, kynurenine did not activate mTOR in TC CD4+ T cells may be due to the specific mouse strain, or it may indicate that mouse T cells are less sensitive to kynurenine. However, we confirmed that kynurenine promotes IFNγ production in TC T cells (Choi et al., 2020), and it therefore also plays a pro-inflammatory role. mTOR activation by tryptamine in TC effector T cells was not sufficient to induce IFNγ production, while kynurenine promoted IFNγ production without mTOR activation, suggesting that these pathways are uncoupled in response to these metabolites.
High tryptophan either supplied systemically through diet or directly to CD4+ T cells from TC mice induced a pronounced increase in metabolites involved in glycolysis and the TCA cycle. We have shown that these two metabolic pathways were tightly linked with pathogenic T cells in lupus (Yin et al., 2015). Furthermore, we showed that exogenous tryptamine increased glycolysis in TC CD4+ T cells. These energy-producing metabolic pathways may be linked to the increased mTOR activation, and they may correspond to the mechanisms by which tryptophan increased T cell pathogenicity in TC mice. Tryptamine was not detected in CD4+ T cells from TC mice fed high tryptophan, which suggests that this metabolite is not imported but binds to a cell surface receptor to exert its effects on mTOR activation and glycolysis. Tryptamine binds to serotonin receptors (Bhattarai et al., 2020), including on T cells (Wu et al., 2020). Furthermore, serotonin activates T cells through some of these receptors (León-Ponte et al., 2007; Nordlind et al., 1992), and promotes IFNɣ production by natural killer cells (Hellstrand et al., 1993). Thus, it is possible that tryptamine activates TC CD4+ T cells by binding to serotonin receptors, or to a yet-to-be identified receptor.
It is unknown whether microbial tryptophan metabolites are increased in patients with SLE, or if these metabolites would have similar effects on CD4+ T cells in human subjects. Only a small number of bacteria have been specifically associated with lupus (Azzouz et al., 2019; Greiling et al., 2018; Manfredo Vieira et al., 2018; Zegarra-Ruiz et al., 2019). Among them, Ruminococcus gnavus, which is increased in patients with lupus nephritis (Azzouz et al., 2019), expresses tryptophan decarboxylase, the enzyme that synthesizes tryptamine from tryptophan. Therefore, it is possible that R. gnavus may contribute to elevated tryptamine levels, including in TC mice.
In summary, we showed that tryptophan metabolite skewing in murine lupus is affected by intestinal microbes but also by yet-to-be identified endogenous enzymes. We found that tryptophan increases the pathogenicity of TC CD4+ T cells directly and through the downstream metabolites kynurenine and tryptamine with non-overlapping functions. There is a growing interest to target the tryptophan pathway in multiple diseases, including cancer, with an increasing number of specific inhibitors for many enzymes in this pathway (Modoux et al., 2021). Thus, there is a need to better define how tryptophan availability may regulate autoreactive pathogenic CD4+ T cells in a variety of autoimmune settings.
Limitations of the study
We investigated the source of skewed tryptophan metabolites in the lupus-prone TC model and identified mechanisms by which some of these metabolites contribute to CD4+ T cell activation. One limitation to this study was that it did not identify specific cell types and enzymes that are responsible for the increased kynurenine found in TC mice, since neither the gut microbes, which skew the production of other tryptophan metabolites in TC mice, nor endogenous enzymes directly alter the production of kynurenine. Future studies should include a tracer approach to understand the production and utilization of kynurenine as part of the global metabolic fate of dietary tryptophan in lupus mice. The differential distribution of numerous tryptophan catabolites in isolated TC CD4+ T cells stimulated in presence of tryptophan suggests that they process tryptophan differently through enzyme that have not been yet identified. It is also possible that these metabolites are produced by non-T cells carried over in the culture, and then imported by T cells. In addition to the specific metabolites that were identified and tested in this study, it is possible that other tryptophan metabolites could individually or additively influence the functions of T cells or other immune cell types contributing to lupus pathogenesis.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| p4E-BP1 | Cell Signaling | RRID:AB_560835 |
| 4E-BP1 | Cell Signaling | RRID:AB_2097841 |
| p70S6K | Cell Signaling | RRID:AB_2265913 |
| IDO1 | Sigma | RRID:AB_1846221 |
| IDO2 | Sant Cruz | RRID:AB_374159 |
| beta-Actin | Cell Signaling | RRID:AB_2242334 |
| IDO1 | eBioscience | RRID:AB_831071 |
| CD4 | eBioscience | RRID:AB_10718983 |
| CD8 | Biolegend | RRID:AB_962672 |
| B220/CD45R | BD Bioscience | RRID:AB_394619 |
| CD11c | Biolegend | RRID:AB_959596 |
| CD11b | BD Bioscience | RRID:AB_1727421 |
| PDCA-1 | eBioscience | RRID:AB_763417 |
| CD45.2 | Biolegend | RRID:AB_492872 |
| Foxp3 | eBioscience | RRID:AB_465936 |
| p4EBP1 | Cell Signaling | RRID:AB_2097838 |
| pS6 | Cell Signaling | RRID:AB_2797646 |
| pAKT | eBioscience | RRID:AB_2574125 |
| IFNγ | Biolegend | RRID:AB_893526 |
| CD28 | BD Bioscience | RRID: AB_2737780 |
| CD3e | BD Bioscience | RRID:AB_394728 |
| mTOR | Invitrogen | RRID:AB_11155001 |
| IgDb | BD Bioscience | RRID:AB_394894 |
| AF594-donkey anti-rabbit IgG secondary | ThermoFisher | RRID:AB_141359 |
| HRP-anti-rabbit IgG secondary | Cell Signaling | RRID:AB_2099233 |
| HRP-anti-mouse IgG secondary | Cell Signaling | RRID:AB_330924 |
| hFAB™ Rhodamine Anti-GAPDH | Bio-Rad | Cat# 12004168 |
| Chemicals, peptides, and recombinant proteins | ||
| L-Tryptophan | Sigma | T0254 |
| L-Kynurenine | Sigma | K8625 |
| Indole-3-acetate | Sigma | I5148-2G |
| 5-Thio-D-Glucose | Sigma | 88635 |
| Tryptamine | Sigma | 193747 |
| Signal Fire Luminol Reagent | Cell Signaling | 6883 |
| HyGLO™ Chemiluminescent HRP Detection Reagent | Denville Scientific | E2500 |
| PVDF Stripping Buffer | ThermoFisher | 46430 |
| dsDNA | Sigma | D8515 |
| p-nitrophenol phosphate (PNPP) substrate | ThermoFisher | 34045 |
| McCoy's 5A media | ThermoFisher | 16600082 |
| Tryptophan synthetic chows | Research Diets | A11022501-03 |
| CD4 microbeads (mouse) | Miltenyi Biotech | 130-104-454 |
| CD11c microbeads (mouse) | Miltenyi Biotech | 130-125-835 |
| Rneasy mini kit | Qiagen | 74104 |
| Critical commercial assays | ||
| Xbridge Amide 3.5 μm × 2.1 mm x 100 mm chromatography column | Waters | 186004860 |
| Mouse KMO ELISA kit | Aviva Systems Biology | OKEH05543 |
| Experimental models: Organisms/strains | ||
| Specific Pathogen Free Adult C57BL/6J | UF Rodent Colony | N/A |
| Specific Pathogen Free Adult B6.Sle1.Sle2.Sle3 | UF Rodent Colony | N/A |
| Germ Free Adult C57BL/6J | UC Rodent Colony | N/A |
| Germ Free Adult B6.Sle1.Sle2.Sle3 | UC Rodent Colony | N/A |
| Oligonucleotides | ||
| Ido1 (F: CCCACACTGAGCACGGACGG; R: TTGCGGGGCAGCACCTTTCG) | Metz et al. (2014) | N/A |
| Tdo2 (F: CATGGCTGGAAAGAACAC; R: GGAGTGCACGGTATGAC) | Pilotte et al. (2012). | N/A |
| Ppia (F: GCTGTTTGCAGACAAAGTTCCA; R: CGTGTAAAGTCACCACCCTGG) | This Paper | N/A |
| Kmo (F: ATGGCATCGTCTGATACTCAGG; R: CCCTAGCTTCGTACACATCAACT) | Wang et al. (2018) | N/A |
| Kat2 (F: ATGAATTACTCACGGTTCCTCAC; R: AACATGCTCGGGTTTGGAGAT) | Wang et al. (2018) | N/A |
| 3Hao (F: GAACGCCGTGTGAGAGTGAA; R: CCAACGAACATGATTTTGAGCTG) | Wang et al. (2018) | N/A |
| Software and algorithms | ||
| FlowJo V10 | Tree Star | https://www.flowjo.com/solutions/flowjo/downloads |
| Prism 9.0 | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
Resource availability
Lead contact
Further information and reasonable requests for resources and reagents should be directed to the lead contact: morel@ufl.edu.
Materials availability
This study did not generate any unique materials.
Experimental model and subject details
Mice and treatments
C57BL/6J (B6) mice were purchased from the Jackson Laboratory and maintained at the University of Florida (UF). The TC, B6.Ido1-/- and B6.Ido2−/− strains have been described previously (Merlo et al., 2020; Morel et al., 2000). Only female mice were used, and they were housed in the same SPF conditions at UF unless indicated otherwise. For metabolite measurements in GF TC mice, SPF TC mice, and SPF B6 controls, serum and fecal samples were obtained from mice housed at the University of Chicago in either GF or SPF conditions. Mice were kept under SPF and GF conditions at the University of Chicago (UC) Animal Resource Center. GF status was monitored by aerobic and anaerobic fecal cultures and PCR amplification of bacterial 16S rRNA genes from fecal DNA as previously described (Kane et al., 2011).
All groups within an experiment were age-matched and tested at the same time. Mice were provided with autoclaved drinking water and fed with either standard chow (Envigo 7912) or with synthetic chows that differ only by their tryptophan concentration (A11022501-03 Research Diets, Inc.) for the indicated amount of time. Tryptophan high chow corresponded to 1.19% tryptophan, while the tryptophan low regimen corresponded to a tryptophan-deficient chow on week days and recharge with 0.19% tryptophan on week-end days, averaging 0.08% tryptophan as previously described (Choi et al., 2020). For in vivo assessment of exogenous tryptamine, mice were injected intraperitoneally with 0.5 mg of tryptamine three times per week for 1 month. All experiments were conducted according to approved protocols by the IACUC at UF and UC.
Method details
Gene expression
Total CD4+ T cells and CD11c+ DCs were purified from spleens with microbeads using negative and positive selection, respectively. Proximal colon and duodenum were collected after flushing contents with PBS and were homogenized separately in lysis buffer along with liver. RNA was isolated with RNeasy Mini Kit and cDNA was synthesized using ImProm II Reverse Transcriptase. Gene expression was quantified using SYBR Green Dye on the BioRad CFX Connect, with qRT-PCR primers listed in the STAR Methods table. The thermo-cycling protocol consisted of 30 s at 95°C, 30 s at 60°C, and 30 s at 72°C repeated for 40 cycles. Relative expression was calculated using the ΔΔCq method with difference in Cq values taken between the Ppia gene against the gene of interest.
Protein analyses
Lysates were prepared from either purified total CD4+ T cells, or from sorted Tem cells (CD4+CD44+CD25−) and Treg cells (CD4+ CD25+) from TC mice fed tryptophan high or tryptophan low chow for 1 month. Cells in lysis buffer and protease inhibitor cocktail were sonicated for 30 s. Total protein was quantified in supernatants using the Bradford Coomassie assay against a standard curve. Lysates were normalized to 30–40 ug/ul using a cocktail of DTT/dye solution at a ratio of 1:10 before heat denaturation. Protein lysates were separated by SDS-PAGE and transferred onto a PVDF membrane using the BioRad transblot mini-gel system. After blocking with 5% milk proteins in TBS-T at room temperature, the membrane was incubated serially with primary antibodies (1:1000) against p4E-BP1, 4E-BP1, p70S6K, IDO1, IDO2, β-Actin and GAPDH at 4cC. HRP-conjugated anti-rabbit or anti-mouse IgG were used as secondary antibodies (1:2000 in TBS-T with 5% milk proteins). Signal was detected using Signal-Fire, Signal-Fire-Elite or HYGLO Quickspray chemiluminescent HRP reagents. For repeated measurements of different proteins on the same membrane, PVDF stripping buffer was applied for 15 min and the membrane was re-blocked before re-probing. Band intensities were quantified using ImageJ software. Mouse hepatic KMO was quantified in liver lysates using an ELISA kit.
Flow cytometry
Single cell suspensions were prepared from spleens using standard procedures. Red blood cells were lysed, followed by fluorochrome-conjugated antibody staining in FACS buffer (2.5% FBS, 0.05% sodium azide in PBS). Dead cells were excluded with fixable viability dye. For intracellular staining, cells were fixed and permeabilized using the FOXP3/Transcription Factor Staining Buffer Set. Staining for IFNγ was preceded by a 4h incubation with Leukocyte Activation Cocktail. Data were acquired on an LSRFortessa and analyzed with FlowJo V10.
Cellular assays
Purified B6 and TC CD4+ T cells were activated with 2 μg/mL plate-bound anti-CD3 and 1 μg/mL soluble anti-CD28 antibody for 24 h. The complete McCoy’s 5A media (10% FBS, 1000× penicillin/streptomycin, 0.1 mM sodium pyruvate, 1 mM Hepes) containing 15 μM tryptophan was supplemented or not with tryptophan, L-kynurenine, I3A, or tryptamine (50 μM each). Intracellular IFNγ, p4EBP1, pS6 were assessed by flow cytometry.
Extracellular flux assays
CD4+ T cells were purified by negative selection with magnetic beads. Glycolysis assessed as Extracellular Acidification Rate (ECAR) and the Oxygen Consumption Rate (OCR) were measured using an XF96 Extracellular Flux Analyzer (Agilent) in a standard mitochondrial stress assay with non-buffered RPMI medium supplemented with 2.5 μM dextrose, 2 mM glutamine and 1 μM sodium pyruvate. Samples were assayed at least in triplicates for 3 successive 8 min time intervals between inhibitor injections. Baseline ECAR and OCR values were averaged between replicates for the first 3 time points before the oligomycin injection.
Immunofluorescence staining
Immunofluorescence staining of frozen spleen sections was performed as previously described (Choi et al., 2018) using an mTOR polyclonal antibody followed by AF594-conjugated donkey anti-rabbit IgG secondary antibody, anti-IgDb-PE and anti-CD4-APC. Images were acquired on an EVOS fluorescent microscope.
C-MS metabolomics
Metabolomic analysis of T cell lysates
Three million cells were washed in 1 mL of PBS, pelleted at 1500 rpm in an Eppendorf centrifuge at 4°C, and resuspended in 100 μL of 80% methanol (−80°C) and 10 μL of 0.3 mM 5-thio-glucose, used as internal standard to allow correction for sample recovery. After freezing at −80°C and thawing once, the sample was centrifuged at 13,000 × g for 30 min at 4°C, and 100μL of supernatant was saved. A 2nd 100 μL of 80% methanol (−80°C) was added to the pellet, the sample was vortexed, centrifuged at 13,000 × g for 30 min at 4°C, and the 2nd 100 μL of supernatant was saved. The two 100-μL supernatants were combined, dried and stored −80°C until analysis. Each sample was resuspended in 20 μL of LC/MS grade water, and 10 μL per sample was injected into a ThermoFisher Q-Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer attached to a Vanquish Horizon UHPLC System, using a Waters Xbridge Amide chromatography column and quantitative polar metabolomics profiling platform with selected reaction monitoring (SRM) that covers all major metabolic pathways. The platform uses hydrophilic interaction liquid chromatography (HILIC) method with positive/negative ion switching to analyze ∼516 metabolites from a single 25-min acquisition run with a 3-ms dwell time and a 1.55-s duty cycle time. The HESI probe parameters were the following in both polarity modes: sheath gas flow rate 30, aux gas flow rate 10, sweep gas flow rate 0, spray voltage 3.60 kV, aux gas heater temp 120°C, S-lens RF level 55, ion transfer capillary temp 320°C. In positive mode, the instrument acquired Full Scan spectra with a m/z range of 61–915, while in negative mode, the Full Scan mass range was m/z 70–920. The resolution of the scans was 70,000, the AGC target was 3e6, while maximum IT was 200 ms. Mobile phase component A was 10 mM ammonium acetate and 7.5 mM ammonium hydroxide in water with 3% (v/v) acetonitrile (pH 9.0) while mobile phase component B was 100% acetonitrile. The 25-minute-long gradient was as follows: 0 min, 85% B; 1.5 min, 85% B; 5.5 min, 35% B; 14.5 min, 35% B; 15.0 min, 85% B; 25.0 min, 85% B. The mobile phase flow rate was the following: 0 min, 0.150 mL/min; 10.0 min, 0.150 mL/min; 10.5 min, 0.300 mL/min; 14.5 min, 0.300 mL/min; 15.0 min, 0.150 mL/min, 25.0 min, 0.150 mL/min. Metabolites were identified with retention time and m/z value to a reference standard, such as kynurenine.
Metabolomic analysis of sera and feces
All samples were thawed on ice to preserve sample quality. Serum samples were aliquoted at 25 μL into Eppendorf tubes for extraction without pre-normalization. Fecal homogenates in 5 mM ammonium formate were pre-normalized to lowest protein concentration (47 μg/mL) and aliquoted at 150 μL into clean Eppendorf tubes for extraction. All samples were spiked with internal standards (IS) solution that consisted of creatine-D3 (4 μg/mL), leucine-D10 (4 μg/mL), L-tryptophan-2,3,3D3 (40 μg/mL), L-tyrosine Ring-13C6 (4 μg/mL), L-leucine-13C6 (4 μg/mL), L-phenylalanine 13C6 (4 μg/mL), N-BOC-L-tert-leucine (4 μg/mL), N-BOC-L-aspartic acid (4 μg/mL), succinic acid-2,2,3,3D4 (4 μg/mL), salicylic acid D6 (4 μg/mL), and caffeine-D3 (4 μg/mL). Metabolites were extracted by protein precipitation with a solution of 8/1/1 (v/v/v) Acetonitrile/Methanol/Acetone. IS spiked serum and fecal samples were precipitated with 200 and 800 μL; respectively. Further protein precipitation was allowed by incubating the samples at 4°C for 20 min. Samples were centrifuged at 20,000 × g for 10 min at 4°C to pellet the protein. Supernatants were transferred into clean Eppendorf tube and dried under a gentle stream of nitrogen at 30°C. The dried extracts were resuspended with 25 μL reconstitution solution consisting of 10 ug/mL injection standards (BOC-L-Tyrosine, BOC-L-Tryptophan and BOC-D-Phenylalanine). Resuspension was allowed at 4°C for 10–15 min then samples were centrifuged at 20,000 × g for 10 min at 4°C. Supernatants were collected into clean LC-vials for LC-MS analysis. Global metabolomics was performed using high resolution mass spectrometry coupled with ultra-high performance liquid chromatography (UHPLC) as previously described (Yazd et al., 2021). Briefly, samples were analyzed in positive and negative heated electrospray ionization with the following source parameters: spray voltage = 3500 V (+mode) and 3000 V (−mode), aux gas = 10, sheath gas = 50, capillary temperature = 325°C, spray gas = 1, and S-lens RF level = 30. Separation was achieved on an ACE 18-PFP 100 × 2.1 mm, 2 μm column (Mac-Mod Analytical, Chadds Ford, PA) using a gradient with mobile phase A as 0.1% formic acid in water and mobile phase B as acetonitrile with a column temperature of 25°C. Gradient elution was ramped from 0% B to 80% B over 13.0 min at 350 μL/min, which increased to 600 μL/min between 16.80 and 17.50 min for column flush and re-equilibration. The runtime was 20.50 min and full scan at 35 000 mass resolution was acquired from 2 μL injection in positive and 4 μL injection in negative ion mode. Metabolites from this method level 1 identified as they are matched with retention time and m/z value to a reference standard.
Quantification and statistical analysis
Metabolomics analysis
Metabolite identification of named metabolites was performed through reference to metabolite library of 1,000 compounds, curated by running each standard at the Southeast Center for Integrated Metabolomics (www.secim.ufl.edu). Feature alignment and curation were performed by MZmine through an automated routine developed in-house. From the same type of global metabolite profiling from serum and fecal samples, we quantified six tryptophan metabolites (tryptophan, serotonin, kynurenine, kynurenic acid, xanthurenic acid, and anthranilic acid), which were referenced to an external calibration curve covering the expected concentration range. The metabolomics data were first log2-transformed and then centered. They were further examined to ensure that the normality assumption was satisfied. All groups were first analyzed using principal components analysis, to obtain a global viewpoint of the data and the groups. Subsequently, individual tryptophan metabolites were compared between groups by ANOVA.
Statistical analysis
Statistical analyses were performed using the GraphPad Prism 9.0 software. Unless indicated, data were normally distributed, and graphs show means and standard deviations of the mean (SEM) for each group. Unless indicated, results were compared with 1-way ANOVA with correction for multiple tests comparisons or with 2-tailed t tests with a minimal level of significance set at p < 0.05.
Acknowledgments
We thank Morel lab members and Laura Mandik-Nayak for technical assistance and discussion. This work was supported by a grant from the NIH R01 AI143313 to LM.
Author contributions
J.B. and L.M. conceived the study, designed the experiments, and wrote the paper; J.B., G.A., S.C.C., Longhuan M., J.L., N.K., L.Z.-S., L.M., J.C., W.P., and T.F. performed experiments; J.B., J.C., T.G., A.P., and L.M. analyzed data. M.M., L.M.N., and A.C. provided samples and contributed to data interpretation.
Declaration of interests
The authors declare that they have no conflict of interest.
Inclusion and diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
Published: May 20, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104241.
Supplemental information
Data and code availability
This study did not generate any new codes. Metabolomics data have been included as Table S1. Raw data from metabolomic analyses of serum and feces from B6 and TC mice fed high or low tryptophan for 1 month, related to Figure 1, Table S2. Serum and feces metabolites from B6 and TC mice fed high or low tryptophan for 1 month with ANOVA p < 0.05, related to Figure 1, Table S3. Raw data from metabolomic analysis of CD4+ T cells from TC mice fed with high or low tryptophan for 1 month (N = 5 per group), related to Figure 2, Table S4. Raw data from metabolomic analysis of serum and feces from GF TC, SPF TC, and SPF B6 mice, related to Figure 3, Table S5. Serum and feces metabolites from GF TC, SPF TC, and SPF B6 mice with ANOVA p < 0.05, related to Figure 3, Table S6. Raw data from metabolomic analysis of CD4+ T cells from B6 and TC mice activated in vitro in medium alone (Ctrl), or with either 50 uM tryptophan or kynurenine (N = 3 per group), related to Figure 5. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Agus A., Planchais J., Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe. 2018;23:716–724. doi: 10.1016/j.chom.2018.05.003. [DOI] [PubMed] [Google Scholar]
- Åkesson K., Pettersson S., Ståhl S., Surowiec I., Hedenström M., Eketjäll S., Trygg J., Jakobsson P.J., Gunnarsson I., Svenungsson E., Idborg H. Kynurenine pathway is altered in patients with SLE and associated with severe fatigue. Lupus Sci. Med. 2018;5:e000254. doi: 10.1136/lupus-2017-000254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzouz D., Omarbekova A., Heguy A., Schwudke D., Gisch N., Rovin B.H., Caricchio R., Buyon J.P., Alekseyenko A.V., Silverman G.J. Lupus nephritis is linked to disease-activity associated expansions and immunity to a gut commensal. Ann. Rheum. Dis. 2019;78:947–956. doi: 10.1136/annrheumdis-2018-214856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailis W., Shyer J.A., Zhao J., Canaveras J.C.G., Al Khazal F.J., Qu R., Steach H.R., Bielecki P., Khan O., Jackson R., et al. Distinct modes of mitochondrial metabolism uncouple T cell differentiation and function. Nature. 2019;571:403–407. doi: 10.1038/s41586-019-1311-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y., Harrison O.J. Homeostatic immunity and the microbiota. Immunity. 2017;46:562–576. doi: 10.1016/j.immuni.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai Y., Jie S., Linden D.R., Ghatak S., Mars R.A.T., Williams B.B., Pu M., Sonnenburg J.L., Fischbach M.A., Farrugia G., et al. Bacterially derived tryptamine increases mucus release by activating a host receptor in a mouse model of inflammatory bowel disease. iScience. 2020;23:101798. doi: 10.1016/j.isci.2020.101798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander J.M., Longman R.S., Iliev I.D., Sonnenberg G.F., Artis D. Regulation of inflammation by microbiota interactions with the host. Nat. Immunol. 2017;18:851–860. doi: 10.1038/ni.3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J., Robutso B., Morel L. Intestinal dysbiosis and tryptophan metabolism in autoimmunity. Front. Immunol. 2020;11:1741. doi: 10.3389/fimmu.2020.01741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervenka I., Agudelo L.Z., Ruas J.L. Kynurenines: tryptophan's metabolites in exercise, inflammation, and mental health. Science. 2017;357:eaaf9794. doi: 10.1126/science.aaf9794. [DOI] [PubMed] [Google Scholar]
- Cheng Y., Jin U.H., Allred C.D., Jayaraman A., Chapkin R.S., Safe S. Aryl hydrocarbon receptor activity of tryptophan metabolites in young adult mouse colonocytes. Drug Metab. Dispos. 2015;43:1536–1543. doi: 10.1124/dmd.115.063677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S., Brown J., Gong M., Ge Y., Zadeh M., Li W., Croker B., Michailidis G., Garrett T., Mohamadzadeh M., Morel L. Gut microbiota dysbiosis and altered tryptophan catabolism contribute to autoimmunity in lupus-susceptible mice. Sci. Transl. Med. 2020;12:eaax2220. doi: 10.1126/scitranslmed.aax2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S.C., Titov A.A., Abboud G., Seay H.R., Brusko T.M., Roopenian D.C., Salek-Ardakani S., Morel L. Inhibition of glucose metabolism selectively targets autoreactive follicular helper T cells. Nat. Commun. 2018;9:4369. doi: 10.1038/s41467-018-06686-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehner C., Fine R., Kriegel M.A. The microbiome in systemic autoimmune disease: mechanistic insights from recent studies. Curr. Opin. Rheumatol. 2019;31:201–207. doi: 10.1097/BOR.0000000000000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallarino F., Vacca C., Orabona C., Belladonna M.L., Bianchi R., Marshall B., Keskin D.B., Mellor A.L., Fioretti M.C., Grohmann U., Puccetti P. Functional expression of indoleamine 2,3-dioxygenase by murine CD8 alpha(+) dendritic cells. Int. Immunol. 2002;14:65–68. doi: 10.1093/intimm/14.1.65. [DOI] [PubMed] [Google Scholar]
- Gao J., Xu K., Liu H., Liu G., Bai M., Peng C., Li T., Yin Y. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front. Cell Infect. Microbiol. 2018;8:13. doi: 10.3389/fcimb.2018.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiling T.M., Dehner C., Chen X., Hughes K., Iñiguez A.J., Boccitto M., Ruiz D.Z., Renfroe S.C., Vieira S.M., Ruff W.E., et al. Commensal orthologs of the human autoantigen Ro60 as triggers of autoimmunity in lupus. Sci. Transl. Med. 2018;10:eaan2306. doi: 10.1126/scitranslmed.aan2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z., Shao T., Li H., Xie Z., Wen C. Alterations of the gut microbiome in Chinese patients with systemic lupus erythematosus. Gut. Pathog. 2016;8:64. doi: 10.1186/s13099-016-0146-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellstrand K., Czerkinsky C., Ricksten A., Jansson B., Asea A., Kylefjord H., Hermodsson S. Role of serotonin in the regulation of interferon-gamma production by human natural killer cells. J. Interferon. Res. 1993;13:33–38. doi: 10.1089/jir.1993.13.33. [DOI] [PubMed] [Google Scholar]
- Hevia A., Milani C., López P., Cuervo A., Arboleya S., Duranti S., Turroni F., González S., Suárez A., Gueimonde M., et al. Intestinal dysbiosis associated with systemic lupus erythematosus. MBio. 2014;5:e01548-14. doi: 10.1128/mBio.01548-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane M., Case L.K., Kopaskie K., Kozlova A., MacDearmid C., Chervonsky A.V., Golovkina T.V. Successful transmission of a retrovirus depends on the commensal microbiota. Science. 2011;334:245–249. doi: 10.1126/science.1210718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- León-Ponte M., Ahern G.P., O'Connell P.J. Serotonin provides an accessory signal to enhance T-cell activation by signaling through the 5-HT7 receptor. Blood. 2007;109:3139–3146. doi: 10.1182/blood-2006-10-052787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke M., Fritsch S.D., Sukhbaatar N., Hengstschläger M., Weichhart T. mTORC1 and mTORC2 as regulators of cell metabolism in immunity. FEBS Lett. 2017;591:3089–3103. doi: 10.1002/1873-3468.12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lood C., Tydén H., Gullstrand B., Klint C., Wenglén C., Nielsen C.T., Heegaard N.H., Jönsen A., Kahn R., Bengtsson A.A. Type I interferon-mediated skewing of the serotonin synthesis is associated with severe disease in systemic lupus erythematosus. PLoS One. 2015;10:e0125109. doi: 10.1371/journal.pone.0125109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X.M., Edwards M.R., Mu Q., Yu Y., Vieson M.D., Reilly C.M., Ahmed S.A., Bankole A.A. Gut microbiota in human systemic lupus erythematosus and a mouse model of lupus. Appl. Environ. Microbiol. 2018;84:e02288-17. doi: 10.1128/AEM.02288-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredo Vieira S., Hiltensperger M., Kumar V., Zegarra-Ruiz D., Dehner C., Khan N., Costa F.R.C., Tiniakou E., Greiling T., Ruff W., et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science. 2018;359:1156–1161. doi: 10.1126/science.aar7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo L.M.F., DuHadaway J.B., Montgomery J.D., Peng W.D., Murray P.J., Prendergast G.C., Caton A.J., Muller A.J., Mandik-Nayak L. Differential roles of Ido1 and Ido2 in T and B cell inflammatory immune responses. Front. Immunol. 2020;11:1861. doi: 10.3389/fimmu.2020.01861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz R., Smith C., DuHadaway J.B., Chandler P., Baban B., Merlo L.M., Pigott E., Keough M.P., Rust S., Mellor A.L., et al. Ido2 is critical for Ido1-mediated T-cell regulation and exerts a non-redundant function in inflammation. Internatl. Immunol. 2014;26:357–367. doi: 10.1093/intimm/dxt073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modoux M., Rolhion N., Mani S., Sokol H. Tryptophan metabolism as a pharmacological target. Trends. Pharmacol. Sci. 2021;42:60–73. doi: 10.1016/j.tips.2020.11.006. [DOI] [PubMed] [Google Scholar]
- Morel L., Croker B.P., Blenman K.R., Mohan C., Huang G., Gilkeson G., Wakeland E.K. Genetic reconstitution of systemic lupus erythematosus immunopathology with polycongenic murine strains. Proc. Natl. Acad. Sci. U S A. 2000;97:6670–6675. doi: 10.1073/pnas.97.12.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordlind K., Sundström E., Bondesson L. Inhibiting effects of serotonin antagonists on the proliferation of mercuric chloride stimulated human peripheral blood T lymphocytes. Int. Arch. Allergy Immunol. 1992;97:105–108. doi: 10.1159/000236104. [DOI] [PubMed] [Google Scholar]
- Odemuyiwa S.O., Ghahary A., Li Y., Puttagunta L., Lee J.E., Musat-Marcu S., Moqbel R. Cutting edge: human eosinophils regulate T cell subset selection through indoleamine 2,3-dioxygenase. J. Immunol. 2004;173:5909–5913. doi: 10.4049/jimmunol.173.10.5909. [DOI] [PubMed] [Google Scholar]
- Perl A. Activation of mTOR (mechanistic target of rapamycin) in rheumatic diseases. Nat. Rev. Rheumatol. 2016;12:169–182. doi: 10.1038/nrrheum.2015.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perl A., Hanczko R., Lai Z.W., Oaks Z., Kelly R., Borsuk R., Asara J.M., Phillips P.E. Comprehensive metabolome analyses reveal. Metabolomics. 2015;11:1157–1174. doi: 10.1007/s11306-015-0772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertovaara M., Hasan T., Raitala A., Oja S.S., Yli-Kerttula U., Korpela M., Hurme M. Indoleamine 2,3-dioxygenase activity is increased in patients with systemic lupus erythematosus and predicts disease activation in the sunny season. Clin. Exp. Immunol. 2007;150:274–278. doi: 10.1111/j.1365-2249.2007.03480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilotte L., Larrieu P., Stroobant V., Colau D., Dolusic E., Frédérick R., De Plaen E., Uyttenhove C., Wouters J., Masereel B., Van den Eynde B.J. Reversal of tumoral immune resistance by inhibition of tryptophan 2,3-dioxygenase. Proc. Natl. Acad. Sci. U S A. 2012;109:2497–2502. doi: 10.1073/pnas.1113873109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roager H.M., Licht T.R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018;9:3294. doi: 10.1038/s41467-018-05470-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothhammer V., Borucki D.M., Tjon E.C., Takenaka M.C., Chao C.C., Ardura-Fabregat A., de Lima K.A., Gutiérrez-Vázquez C., Hewson P., Staszewski O., et al. Microglial control of astrocytes in response to microbial metabolites. Nature. 2018;557:724–728. doi: 10.1038/s41586-018-0119-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothhammer V., Mascanfroni I.D., Bunse L., Takenaka M.C., Kenison J.E., Mayo L., Chao C.C., Patel B., Yan R., Blain M., et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 2016;22:586–597. doi: 10.1038/nm.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang A., Zheng Y.Y., Yin Y., Dozmorov I., Li H., Hsu H.C., Mountz J.D., Morel L. Dysregulated cytokine production by dendritic cells modulates B cell responses in the NZM2410 mouse model of lupus. PLoS One. 2014;9:e102151. doi: 10.1371/journal.pone.0102151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki-Imamura T., Yoshida Y., Suwabe K., Yoshimura F., Kato H. Molecular basis of indole production catalyzed by tryptophanase in the genus Prevotella. FEMS Microbiol. Lett. 2011;322:51–59. doi: 10.1111/j.1574-6968.2011.02329.x. [DOI] [PubMed] [Google Scholar]
- Sinclair L.V., Neyens D., Ramsay G., Taylor P.M., Cantrell D.A. Single cell analysis of kynurenine and System L amino acid transport in T cells. Nat. Commun. 2018;9:1981. doi: 10.1038/s41467-018-04366-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonner J.K., Keil M., Falk-Paulsen M., Mishra N., Rehman A., Kramer M., Deumelandt K., Röwe J., Sanghvi K., Wolf L., et al. Dietary tryptophan links encephalogenicity of autoreactive T cells with gut microbial ecology. Nat. Commun. 2019;10:4877. doi: 10.1038/s41467-019-12776-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vujkovic-Cvijin I., Dunham R.M., Iwai S., Maher M.C., Albright R.G., Broadhurst M.J., Hernandez R.D., Lederman M.M., Huang Y., Somsouk M., et al. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci. Transl. Med. 2013;5:193ra191. doi: 10.1126/scitranslmed.3006438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Lian Y.J., Su W.J., Peng W., Dong X., Liu L.L., Gong H., Zhang T., Jiang C.L., Wang Y.X. HMGB1 mediates depressive behavior induced by chronic stress through activating the kynurenine pathway. Brain Behav. Immun. 2018;72:51–60. doi: 10.1016/j.bbi.2017.11.017. [DOI] [PubMed] [Google Scholar]
- Widner B., Sepp N., Kowald E., Kind S., Schmuth M., Fuchs D. Degradation of tryptophan in patients with systemic lupus erythematosus. Adv. Exp. Med. Biol. 1999;467:571–577. doi: 10.1007/978-1-4615-4709-9_71. [DOI] [PubMed] [Google Scholar]
- Widner B., Sepp N., Kowald E., Ortner U., Wirleitner B., Fritsch P., Baier-Bitterlich G., Fuchs D. Enhanced tryptophan degradation in systemic lupus erythematosus. Immunobiology. 2000;201:621–630. doi: 10.1016/S0171-2985(00)80079-0. [DOI] [PubMed] [Google Scholar]
- Williams B.B., Van Benschoten A.H., Cimermancic P., Donia M.S., Zimmermann M., Taketani M., Ishihara A., Kashyap P.C., Fraser J.S., Fischbach M.A. Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. Cell Host Microbe. 2014;16:495–503. doi: 10.1016/j.chom.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Herr D., MacIver N.J., Rathmell J.C., Gerriets V.A. CD4 T cells differentially express cellular machinery for serotonin signaling, synthesis, and metabolism. Int. Immunopharmacol. 2020;88:106922. doi: 10.1016/j.intimp.2020.106922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazd H.S., Rubio V.Y., Chamberlain C.A., Yost R.A., Garrett T.J. Metabolomic and lipidomic characterization of an X-chromosome deletion disorder in neural progenitor cells by UHPLC-HRMS. J. Mass. Spectrom. Adv. Clin. Lab. 2021;20:11–24. doi: 10.1016/j.jmsacl.2021.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Choi S.C., Xu Z., Perry D.J., Seay H., Croker B.P., Sobel E.S., Brusko T.M., Morel L. Normalization of CD4+ T cell metabolism reverses lupus. Sci. Transl. Med. 2015;7:274ra218. doi: 10.1126/scitranslmed.aaa0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegarra-Ruiz D.F., El Beidaq A., Iñiguez A.J., Lubrano Di Ricco M., Manfredo Vieira S., Ruff W.E., Mubiru D., Fine R.L., Sterpka J., Greiling T.M., et al. A Diet-sensitive commensal lactobacillus strain mediates TLR7-dependent systemic autoimmunity. Cell Host Microbe. 2019;25:113–127.e6. doi: 10.1016/j.chom.2018.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelante T., Iannitti R.G., Cunha C., De Luca A., Giovannini G., Pieraccini G., Zecchi R., D'Angelo C., Massi-Benedetti C., Fallarino F., et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Zhang H., Liao X., Sparks J.B., Luo X.M. Dynamics of gut microbiota in autoimmune. Lupus. Appl. Environ. Microbiol. 2014;80:7551–7560. doi: 10.1128/AEM.02676-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any new codes. Metabolomics data have been included as Table S1. Raw data from metabolomic analyses of serum and feces from B6 and TC mice fed high or low tryptophan for 1 month, related to Figure 1, Table S2. Serum and feces metabolites from B6 and TC mice fed high or low tryptophan for 1 month with ANOVA p < 0.05, related to Figure 1, Table S3. Raw data from metabolomic analysis of CD4+ T cells from TC mice fed with high or low tryptophan for 1 month (N = 5 per group), related to Figure 2, Table S4. Raw data from metabolomic analysis of serum and feces from GF TC, SPF TC, and SPF B6 mice, related to Figure 3, Table S5. Serum and feces metabolites from GF TC, SPF TC, and SPF B6 mice with ANOVA p < 0.05, related to Figure 3, Table S6. Raw data from metabolomic analysis of CD4+ T cells from B6 and TC mice activated in vitro in medium alone (Ctrl), or with either 50 uM tryptophan or kynurenine (N = 3 per group), related to Figure 5. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.