Abstract
The emergence of antibiotic resistance in mycobacteria involves the selection of mutant variants within a susceptible bacterial population. However, it is unclear whether antimycobacterial drugs act just as selective agents or can influence the rate of appearance of resistant mutants. The present study was initiated to address this issue by monitoring the effects of antimicrobial agents on the appearance and growth of clarithromycin (CLR)-resistant (CLRr) bacilli in broth cultures of Mycobacterium avium. Preexposure of M. avium to CLR had a significant dose effect on the emergence of resistance, with concentrations of 4 to 8 μg/ml resulting in a maximal (∼104-fold) increase in the number of CLRr bacilli after a 4-day incubation. In addition, a dose effect was found with azithromycin. The use of combinations of CLR with either ethambutol (EMB) or rifabutin (RFB) resulted in fewer resistant bacilli compared to the use of CLR alone. The lowest active concentration of EMB (4 μg/ml) was equivalent to the EMB MIC (4 to 8 μg/ml) for the parental CLRs strain and the emergent CLRr variants, and thus, the antiresistance effect was probably the result of the bacteriostatic effect of EMB on CLRr bacilli. However, RFB was an order of magnitude more active (0.05 μg/ml) at reducing resistance than suggested by the MIC of this agent (0.5 to 1 μg/ml). These results indicate that the emergence of resistance was not simply the selection of a preexisting subpopulation of resistant bacilli. Further analysis suggested that early events in the emergence of resistance involved organisms (progenitors) that acquired a resistance phenotype. In addition, the progenitors appeared to be in a transient state, able to develop into a stable resistant lineage in the presence of CLR, or able to revert to the wild type in nonselective conditions.
Drug resistance is a difficult and not uncommon problem in the treatment of mycobacterial diseases, especially tuberculosis. To reduce the likelihood of resistance emergence, patients with mycobacterioses are treated with multiple antimycobacterial agents. Yet, despite the use of combination therapy, new cases of secondary (i.e., emergent) drug resistance in Mycobacterium tuberculosis are continually arising (P. M. Simone and S. W. Dooley, http://www.cdc.gov/nchstp/tb/pubs/mdrtb/mdrtb.htm). Patient-associated determinants (e.g., compliance and immune state) are important risk factors in the emergence of resistance (Simone and Dooley, http://www.cdc.gov/nchstp/tb/pubs/mdrtb/mdrtb.htm); however, it is the genetic and metabolic states of the infecting microbial populations that ultimately determine if resistance will appear.
The focus of antimycobacterial treatment agents has been based, at least initially, on susceptibility studies with wild-type and isolated organisms expressing clinically significant levels of resistance. The choice of agents used in drug combinations is prioritized further largely on the basis of therapeutic efficacy. However, little is understood of the processes involved in the acquisition of resistance, and there may be agents that are relatively inactive as therapeutic agents but that may have significant activity at preventing the emergence of resistance. There is a precedent for this view, in that treatment with the combination of clarithromycin (CLR) and ethambutol (EMB) is effective at reducing the incidence of macrolide resistance during the treatment of disseminated Mycobacterium avium complex (MAC) disease (1). However, the addition of EMB has no significant effect on the reduction of total bacterial numbers.
Recently, Martinez and Baquero (10) reviewed the factors that are known to influence the appearance of mutations associated with increased levels of resistance to antimicrobial agents. Many of the environmental factors that affect the appearance of resistant mutants are clinically relevant, e.g., limiting nutrients, microbial competition, and antibiotic stress. The last factor is particularly interesting in that antibiotics can act as stressors (increasing mutation rates [10]) as well as be the selectors of any resulting resistant mutants. Furthermore, the concentration of antibiotic appears to influence the rate of drug resistance mutation emergence (10).
The present study was initiated to investigate the effect of antibiotic concentration and of antibiotic combinations on the emergence of drug-resistant mutants in susceptible mycobacterial populations. Furthermore, the study also questions whether the emergence of drug resistance derives from a drug-resistant subpopulation that existed prior to the addition of the antibiotic. To achieve these objectives, a simple in vitro model system was used to monitor the emergence of macrolide resistance in wild-type strains of MAC. Acquisition of macrolide resistance in MAC is a useful phenotype for study as it is based on a small number of possible mutations within the peptidyltransferase region of the 23S rRNA gene (12, 13, 14, 15). Furthermore, a single base substitution in the peptidyltransferase region is sufficient to confer high-level macrolide resistance to mycobacteria (18) and the drug resistance phenotype of the mutants is independent of the specific mutation involved.
MATERIALS AND METHODS
Antimicrobial agents.
CLR (Abbott Laboratories, Abbott Park, Ill.) was dissolved in methanol to give a maximum concentration of 6 mg/ml, which was then immediately diluted 1:2 with sterile 0.1 M phosphate buffer (pH 6.8). Stock solutions of 10 mg of azithromycin (AZM; Pfizer Inc., Groton, Conn.) and rifabutin (RFB; Pharmacia & Upjohn, Milano, Italy) per ml were prepared in methanol and ethanol, respectively. EMB (Sigma Chemical Co., St. Louis, Mo.) was dissolved in double-distilled water to 1 mg/ml and filter sterilized (pore size, 0.2 μm).
Media.
The media used for the culture of MAC were Middlebrook 7H11 agar (Hardy Diagnostics, Santa Maria, Calif.) and a modified Middlebrook 7H9 broth (7HSF broth) supplemented with 10% oleic acid-albumin-dextrose-catalase (OADC; BBL, Becton Dickinson, Cockeysville, Md.) and 1 g of Trypticase casein digest (BBL). Antibiotic-containing agar plates were prepared with 7H11 agar base (Difco, Detroit, Mich.) supplemented with 10% OADC and either 64 μg of CLR per ml (7H11-CLR), 256 μg of AZM per ml (7H11-AZM), or 8 μg of RFB per ml (7H11-RFB). All cultures were incubated at 37°C in room air.
Mycobacteria.
CLR-susceptible (CLRs) MAC strains 101 and 504 and CLR-resistant (CLRr) MAC strains 101R, 511, 512, and JJL004.2 are described elsewhere (14, 15). Strains 101R and 511 are variants of strains 101 and 504 (14). Five CLRr variants of strain 101 (strains 101ER-1 through 101ER-5) and five CLRr variants of strain 504 (strains 504ER1 through 504ER5) were selected on 7H11-CLR. The strain 504 variants, 504c5 and 504c8, express an intermediate-level CLR resistance (CLRi; CLR MIC, 16 to 32 μg/ml). These variants were isolated from cultures of strain 504 grown on 7H11 agar containing 16 μg of CLR per ml.
MIC determination and mutation analysis.
Susceptibility to antimicrobial agents was assessed by a broth microdilution assay based on previously described protocols (6). Briefly, drug dilution series (at a 1.5× concentration) were prepared in 7HSF, and 0.1 ml was dispensed into the wells of 96-well, U-bottom microtiter plates. Each well was inoculated with 7.5 × 104 CFU of MAC in 0.05 ml of 7HSF broth. Bacterial growth was assessed visually after the plates had been incubated for 5 days at 37°C. Previous analysis confirmed that the microdilution-based assay was consistent with a standard BACTEC radiometric, broth macrodilution assay (7). Macrolide resistance in MAC is defined as a CLR MIC of >32 μg/ml (although for CLRr MAC strains the CLR MIC is >512 μg/ml) or an AZM MIC of >256 μg/ml. Previous studies have confirmed that all CLRr MAC strains with 23S rRNA gene mutations are also resistant to AZM (13, 14, 15).
The presence of the 23S rRNA gene mutation was detected by using a modification of the Mismatch Detect II kit (Ambion, Inc., Austin, Tex.) as described elsewhere (15).
In vitro model of the emergence of macrolide resistance.
Seed cultures of MAC strains were cultured in 7HSF broth until the early stationary growth phase was reached (approximately 3 × 108 to 6 × 108 CFU/ml). Preliminary experiments showed that the emergence of resistance in vitro was more reproducible when seed cultures in the stationary growth phase were used. The seed cultures were diluted 5- to 10-fold with fresh 7HSF broth with or without the addition of antimicrobial agents (singly or in combination), and the cultures were incubated for 4 days at 37°C. During this period, samples (0.5 to 25 ml) were taken from the cultures. The samples were sonicated for 10 min in a bath sonicator (Gen-Probe, San Diego, Calif.) and then plated in triplicate onto 7H11 agar and 7H11-CLR to determine the total numbers of CFU per milliliter and the numbers of CLRr CFU per milliliter. Microscopic examination of the cultures showed that sonication thoroughly dispersed bacterial clumps. In selected experiments, the number of macrolide-resistant CFU was determined by plating on 7H11-AZM as well as 7H11-CLR.
For time course studies, 100- to 200-ml culture volumes were used; however, in selected experiments, triplicate 40-ml culture volumes were used in order to provide a sufficient sample size for statistical analysis (Student's t test). Each experiment was performed at least twice.
To characterize the heritable state of the emerging, resistant organisms, seed cultures were diluted 10-fold and preincubated in the absence of CLR for up to 10 h. Then CLR was added to the cultures at a final concentration of 4 μg/ml and the incubation was continued for a total of 4 days. Total numbers of CFU per milliliter and the numbers of CLRr CFU per milliliter were determined as described above.
RESULTS
Does the concentration of CLR affect the emergence of macrolide resistance?
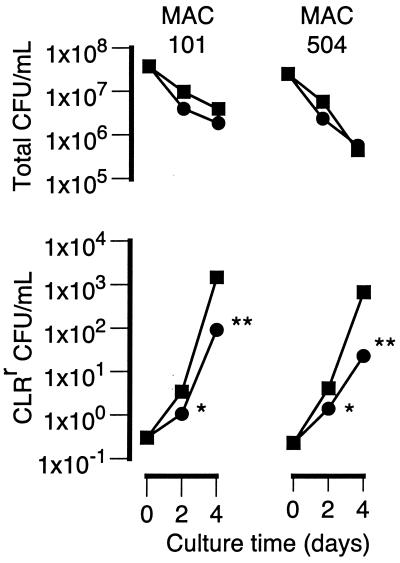
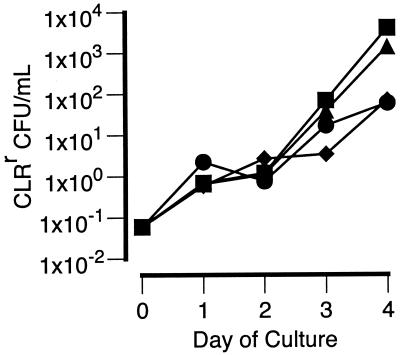
Figure 1 shows the effect of CLR concentrations of 8 and 32 μg/ml on the total numbers of CFU per milliliter and the numbers of CLRr CFU per milliliter over time in cultures of MAC strains 101 and 504. The number of resistant bacilli (numbers of CLRr CFU per milliliter; CLR MIC, >64 μg/ml) increased by more than 3 orders of magnitude after 4 days of culture with 8 μg of CLR per ml. However, the numbers of resistant bacilli in cultures containing 32 μg of CLR per ml were 15- to 30-fold less than the numbers in cultures containing 8 μg/ml. Furthermore, the difference between the numbers of resistant bacilli in cultures with 8 and 32 μg/ml was significant (P < 0.05) after only 2 days of incubation. Thus, the increase in the numbers of resistant bacilli was dependent on the concentration of CLR.
FIG. 1.
Effect of CLR (■, 8 μg/ml; ●, 32 μg/ml) on the total numbers of CFU per milliliter and the numbers of CLRr CFU per milliliter in cultures of MAC strains 101 and 504 over a 4-day incubation. Conditions that resulted in significantly lower numbers of CLRr organisms compared to the numbers in cultures containing 8 μg of CLR per ml are indicated (∗, P < 0.05; ∗∗, P < 0.001). For clarity, error bars are not shown, as they are all within the datum point symbols.
Clumping is often a confounding factor in relating the numbers of CFU to the number of mycobacterial bacilli. However, the results shown in Fig. 1 are unlikely to be a consequence of clumping, as sonication of the cultures prior to plating thoroughly dispersed bacterial aggregates. Furthermore, CLR had no effect on bacterial aggregates in cultures of CLRr MAC strains. Thus, clumping of any preexisting CLRr bacilli would be expected to be the same in the cultures containing CLR at either 8 or 32 μg/ml.
A decrease in the numbers of total viable organisms was expected, as the CLR concentrations were above the MICs for both strains (Table 1). In contrast to the numbers of CLRr CFU, however, the changes in the total numbers of CFU were relatively independent of the CLR concentration.
TABLE 1.
Susceptibilities of MAC strains to CLR, EMB, and RFB
| Strain | MIC (μg/ml)
|
||
|---|---|---|---|
| CLR | EMB | RFB | |
| 101 | 4 | 8 | 1 |
| 101R | >512 | 8 | 1 |
| 101ER-1 | >512 | 8 | 1 |
| 101ER-2 | >512 | 8 | 1 |
| 101ER-3 | >512 | 8 | 1 |
| 101ER-4 | >512 | 4 | 1 |
| 101ER-5 | >512 | 8 | 1 |
| 504 | 4 | 8 | 1 |
| 511 | >512 | 4 | 1 |
| 504ER-1 | >512 | 4 | 1 |
| 504ER-2 | >512 | 4 | 0.5 |
| 504ER-3 | >512 | 8 | 1 |
| 504ER-4 | >512 | 8 | 1 |
| 504ER-5 | >512 | 8 | 1 |
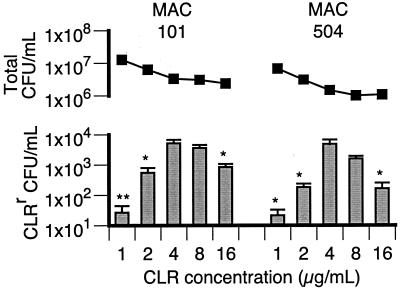
In order to expand on the results shown in Fig. 1, the effect of a wider range of CLR concentrations on the emergence of resistance was investigated (Fig. 2). As before, there was a dose effect of CLR on the numbers of resistant organisms present after 4 days of culture. Incubation in 4 μg of CLR per ml resulted in the greatest increase (approximately 10,000-fold) in the number of CLRr organisms, with significantly fewer resistant organisms emerging at CLR concentrations of ≤2 and 16 μg/ml. For MAC strain 101, 4 and 8 μg of CLR per ml tended to result in equivalent numbers of CLRr organisms. Interestingly, the peak CLR concentration for the emergence of resistance is the same as the CLR MICs for MAC strains 101 and 504 and at least 2 orders of magnitude below the CLR MICs for CLRr MAC strains (Table 1). The lower number of CLRr variants in the cultures with 1 μg of CLR per ml was not a result of out-competition by the more abundant CLRs organisms, since the total population density remained relatively low (<2 × 107 CFU/ml) during the 4-day incubation.
FIG. 2.
Total numbers of CFU per milliliter (■) and numbers of CLRr CFU per milliliter (bars) after 4 days of culture in the presence of CLR concentrations of 1 to 16 μg/ml. For each strain, conditions that resulted in significantly lower numbers of CLRr organisms compared to the numbers in cultures containing 4 μg of CLR per ml are indicated (∗, P < 0.05; ∗∗, P < 0.01).
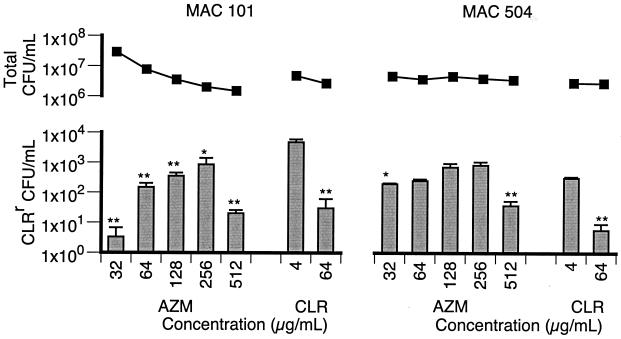
Similar to the results obtained with CLR, AZM showed a dose-dependent effect on the numbers of macrolide-resistant organisms (Fig. 3). Furthermore, the number of resistant organisms obtained by plating on 7H11-AZM was not significantly changed compared to the number obtained by plating on 7H11-CLR, confirming that CLR and AZM were selecting the same population of resistant organisms. The peak concentrations of AZM used were 256 μg/ml for strain 101 and 128 to 256 μg/ml for strain 504. However, unlike CLR, these concentrations were higher than the AZM MIC, i.e., 32 μg/ml for strain 101 and 16 μg/ml for strain 504. The phenotype of slightly greater susceptibility for strain 504 may explain the less distinct peak in the number of resistant bacilli and the greater bactericidal activity of AZM against this strain.
FIG. 3.
Total numbers of CFU per milliliter (■) and numbers of CLRr CFU per milliliter (bars) after 4 days of culture in the presence of AZM (32 to 512 μg/ml) and CLR (4 and 64 μg/ml). For each strain, conditions that resulted in significantly lower numbers of CLRr organisms compared to the numbers in cultures containing 4 μg of CLR per ml are indicated (∗, P < 0.05; ∗∗, P < 0.01).
Although the experiments represented by Fig. 1 to 3 were reproducible, the results were dependent on harvesting of the seed cultures in the stationary phase. Seed cultures that were in the early to the mid-exponential phase generated few resistant bacilli by day 4 in the experimental system. Furthermore, the resistant organisms that were present at day 4 tended to be unevenly distributed between experimental cultures, leading to so-called jackpot cultures.
Characterizing the emergent CLRr bacilli.
In order to confirm the mutational basis of the macrolide resistance, the DNAs isolated from CLRr variants were analyzed for 23S rRNA gene mutations. For each MAC strain, this analysis was applied to five CLRr variants derived from the seed cultures at time zero and 25 CLRr variants isolated from broth cultures on day 4. All 60 variants were found to have mutations in the peptidyltransferase region of the 23S rRNA gene, consistent with the macrolide resistance phenotype (data not shown). Although the specific mutations were not confirmed by DNA sequencing, the banding patterns generated by the mismatch assay did not indicate a shift in the base substitutions between the two time points.
Despite the mutation uniformity, there was a clear difference in the rate of colony appearance on the experimental 7H11-CLR plates inoculated with the seed cultures compared to that on the plates inoculated with the broth cultures from day 4. The agar plates inoculated with seed cultures required an incubation of ∼10 days before the first colonies appeared. Furthermore, the numbers of visible colonies increased over the following 4 to 5 days. In contrast, colonies were visible after an incubation of only 5 to 6 days for the agar plates inoculated with the experimental broth cultures from day 4, and all the colonies appeared concurrently. The 5- to 6-day incubation period was consistent with the growth of CLRr MAC strains (e.g., strains 101R and 511) plated on 7H11-CLR.
These observations suggest that the source organisms for the macrolide-resistant colonies derived from the seed cultures differed from the source organisms derived from the CLR-exposed broth cultures.
Modeling a preexisting macrolide-resistant subpopulation.
The low optimal concentration of CLR (4 μg/ml) for the emergence of resistance suggests that resistance does not derive from a highly resistant preexisting subpopulation. To confirm this, the emergence of resistance from a highly resistant preexisting subpopulation was modeled by using a collection of CLRr MAC strains (Table 2). Four of the strains emerged in vivo during treatment of disseminated disease with macrolide therapy (strains 101R, JJL004.2, 511, and 512), and the remaining two strains were selected in vitro (strains 504ER1 and 504ER2). The 23S rRNA genes of these strains were analyzed by either DNA sequencing (14) or mismatch assay (strains 504ER1 and 504ER2), and all strains were confirmed to have a mutation consistent a macrolide resistance phenotype. Furthermore, all strains expressed the resistance phenotype constitutively; i.e., resistance was not affected by preincubation in macrolide (data not shown).
TABLE 2.
Effect of CLR on the generation times of MAC strains
| Strain | Phenotypea | Generation time (h [mean ± SE]) with the following CLR concn (μg/ml):
|
||
|---|---|---|---|---|
| 0 | 4 | 64 | ||
| 101R | CLRr | 15.0 ± 2.2 | 12.4 ± 0.4 | 14.5 ± 2.4 |
| JJL004.2 | CLRr | 15.7 ± 0.5 | 15.2 ± 0.3 | 15.8 ± 0.4 |
| 511 | CLRr | 13.3 ± 0.3 | 13.1 ± 0.4 | 14.7 ± 0.5 |
| 512 | CLRr | 11.4 ± 0.9 | 11.3 ± 0.3 | 12.0 ± 1.2 |
| 504ER-1 | CLRr | 12.2 ± 0.7 | 13.4 ± 1.2 | 14.7 ± 1.1 |
| 504ER-2 | CLRr | 11.2 ± 0.3 | 11.0 ± 0.1 | 11.3 ± 0.3 |
| 511/504b | CLRr/CLRs | 13.5 ± 0.1 | 13.2 ± 0.6 | 13.3 ± 0.3 |
| 101 | CLRs | 8.3 ± 0.1 | —c | — |
| 504 | CLRs | 9.8 ± 0.5 | — | — |
For CLRr strains the MIC is ≥64 μg/ml; for CLRs strains the MIC is ≤4 μg/ml. All strains carry a 23S rRNA gene mutation, assessed either by mismatch assay (strains 504ER-1 and 504ER-2) or by DNA sequencing (14).
Mixture of strain 511 and strain 504 in an initial CFU ratio of 1:106. Generation times are for strain 511.
—, CLR concentrations of ≥4 μg/ml were bactericidal for MAC strains 101 and 504.
Table 2 shows the effect of CLR on the generation times of the six CLRr strains. Thus, growth was not significantly affected (P value range, 0.09 to 0.96) by the presence of either 4 or 64 μg of CLR per ml, irrespective of the origins of the resistant strains. Similarly, AZM (tested up to 256 μg/ml) had no significant effect on the growth rates of macrolide-resistant bacilli (data not shown). The growth rates of all the CLRr strains were significantly lower than those of CLRs strains 101 (P < 0.002) and 504 (P < 0.05).
In order to determine if the growth of CLRr bacilli was affected by an excess of CLRs bacilli, isogenous strains 504 (CLRs) and 511 (CLRr) were mixed at a ratio of 106:1. As before, the growth rate of the CLRr strain was unaffected by the presence of CLR (Table 2).
Thus, if the emergence of resistance from a wild-type bacterial population was simply the result of clonal expansion of preexisting CLRr bacilli, then the concentration of CLR should have had little effect on the increase in numbers of resistant organisms.
Drug combinations and the emergence of resistance.
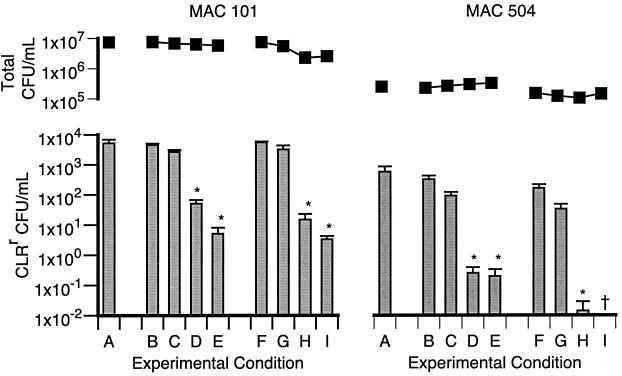
The results described above indicate that the emergence of resistance is affected by the concentration of macrolides in a way not predicted by the susceptibility profiles of either the susceptible or the resistant organisms (Table 1). This raises the possibility that susceptibility profiles may not predict how drug combinations affect the emergence of macrolide resistance. To investigate this, the emergence of macrolide resistance was compared between cultures containing CLR alone (4 μg/ml) and cultures containing CLR in combination with either EMB or RFB (Fig. 4).
FIG. 4.
Total numbers of CFU per milliliter (■) and numbers of CLRr CFU per milliliter (bars) after 4 days of culture in the presence of CLR (4 μg/ml) alone or CLR (4 μg/ml) in combination with either EMB (B, 1 μg/ml; C, 2 μg/ml; D, 4 μg/ml; E, 8 μg/ml) or RBT (F, 0.0125 μg/ml; G, 0.025 μg/ml; H, 0.05 μg/ml; I, 0.1 μg/ml). Conditions that resulted in significantly lower numbers of CLRr organisms compared to the numbers in cultures containing CLR alone are indicated (∗, P < 0.05).
The addition of EMB at concentrations ≥4 μg/ml resulted in significantly fewer resistant bacilli for both strains. Thus, the lowest concentration of EMB that significantly reduced the emergence of resistance was equivalent to the MIC of this agent for strains 101 and 504 and their derived CLRr variants (Table 1). This indicates that the effect of EMB was probably the result of bacteriostasis of the CLRr (and CLRs) bacilli.
The use of combinations of CLR with ≥0.05 μg of RFB per ml showed a significantly reduced rate of emergence of CLR resistance compared to that from the use of CLR alone (Fig. 4). In contrast to the effect of EMB, 0.05 μg of RFB per ml was 10- to 20-fold lower than the RFB MIC determined for the parental strains, 101 and 504, and the CLRr variants (Table 1). In fact, the level of growth of resistant organisms in 0.05 μg of RFB per ml, with or without 4 μg of CLR per ml, was not discernibly different from the level of growth in medium alone (data not shown). This indicates that the effect of RFB on the emergence of CLR resistance was not the result of inhibition of the growth of the CLRr bacilli by RFB.
Despite the significant effect on the emergence of resistance, the addition of EMB (up to 8 μg/ml) or RFB (up to 0.1 μg/ml) only marginally affected the bactericidal activity of CLR (Fig. 4).
Clonal expansion versus “nascent” resistance.
If the emergence of resistance does not derive from a preexisting resistant population, then there must be, at least initially, a period of nascent resistance (i.e., a period when organisms are acquiring a resistance phenotype). Furthermore, it must be this acquisition period that is dependent on the CLR concentration. Once high-level phenotypic resistance has been acquired, the numbers of resistant bacilli will increase by cell division (i.e., clonal expansion of the CLRr bacilli), which is unaffected by the CLR concentration (Table 2).
The time point when the emergence of resistance is predominantly the result of clonal expansion of CLRr bacilli can be assessed by determining when changes in number of CLRr bacilli are refractory to high concentrations of CLR (64 μg/ml). To determine this point, replicate 100-ml cultures of MAC 101 (7.6 × 107 CFU/ml) were set up with an initial CLR concentration of 8 μg/ml. At time zero and daily thereafter, a different replicate culture was supplemented with CLR to a final concentration of 64 μg/ml. In addition, samples were taken daily from all cultures for determination of the total numbers of CFU per milliliter and the numbers of CLRr CFU per milliliter.
The number of CLRr bacilli was equivalent in all cultures on day 1 and day 2 (Fig. 5). By day 4, however, the cultures maintained with 8 μg of CLR per ml for at least 3 days had significantly (P < 0.01) more CLRr bacilli than the other cultures, i.e., 2,375 ± 650 versus 55 ± 21 CFU/ml, respectively. The total numbers of CFU per milliliter were equivalent in all the cultures (data not shown). Strain 504 was analyzed in a similar manner, using a shift in the CLR concentration from 4 to 64 μg/ml. Cultures maintained with 4 μg CLR per ml for at least 2 days resulted in significantly (P < 0.01) higher numbers of CLRr CFU than cultures in which the CLR concentration was increased prior to day 2, i.e., 977 ± 294 versus 30 ± 14 CFU/ml, respectively.
FIG. 5.
Effects of increasing concentrations of CLR on the numbers of CLRr organisms in cultures of MAC strain 101. Replicate cultures were initiated with 8 μg of CLR per ml; and on day 0 (⧫), day 2 (●), day 3 (▴), or day 4 (■) different cultures were supplemented with CLR to final concentration of 64 μg/ml. For clarity, the results for the culture supplemented on day 1 are not shown. Error bars are not shown, as they are all within the datum point symbols.
Thus, clonal expansion appeared to become the predominant cause of the increase in the CLRr population after a 2- to 3-day exposure to concentrations of CLR optimal for the emergence of resistance.
Hypermutable organisms, intermediate-level resistance, and the emergence of resistance.
If the acquisition of resistance is a consequence of a stable hypermutable (mutator) state, then the probability of mutation should be higher for the emergent CLRr variants than for the parental strains. To investigate this, strain 101, 4 variants selected in vitro, and a pool representing 1,000 variants selected in vitro were each plated on 7H11 and 7H11-RFB, and the frequency of RFB-resistant (RFBr) variants was determined. The frequencies of RFBr bacilli of strain 101 and the mean frequencies for the individual CLRr variants and the variant pool were 6.2 × 10−9 ± 1.5 × 10−9, 4.7 × 10−9 ± 2.5 × 10−9, and 3.4 × 10−9 ± 2.7 × 10−9 respectively. Therefore, the probability of acquiring resistance to RFB for the CLRr variants was not greater than that for the parental strain.
An alternative explanation for the effect of CLR on the emergence of resistance is that CLR causes a transient increase in the general mutation rate in a dose-dependent manner. CLR could then select the CLRr organisms from the increased pool of variants. To explore this possibility, the frequency of resistance to RFB was determined in MAC cultures exposed to CLR (4 μg/ml) for 2 days. This period was long enough for a detectable increase in CLRr variants, without too severely depleting the total bacterial population (Fig. 1). In the experiment the numbers of viable organisms decreased from 7.45 × 107 to 6.09 × 107 CFU/ml, whereas the numbers of CLRr organisms increased from 0.7 to 6.8 CFU/ml; this represents a frequency shift of 9.4 × 10−9 to 1.1 × 10−7.
The frequency of RFBr bacilli in the strain 101 seed cultures was 7.9 × 10−9 ± 1.9 × 10−9, whereas it was 3.1 × 10−9 ± 1.3 × 10−9 for cultures incubated in 4 μg of CLR per ml for 2 days. Thus, incubation of MAC with CLR was not associated with an increase in the appearance of other (i.e., non-CLRr) variants. These results suggest that the progenitors of CLR resistance were drug specific.
It is possible that the appearance of CLRr bacilli is a multistep process, with organisms initially gaining an intermediate-level resistance phenotype (CLRi). If an intermediate phenotype is a step in the emergence of CLRr, then CLRi bacilli (CLR MIC, 16 to 32 μg/ml) should be more likely than wild-type bacilli to mutate to high-level resistance. To investigate this, the relative mutability of wild-type strain 504 and the CLRi derivatives, 504c5 and 504c8, was assessed by plating samples of each strain (1010 CFU) on 7H11-CLR. The frequencies of the derived numbers of CLRr CFU (relative to the total numbers of CFU plated) for strains 504, 504c5, and 504c8 were 7.0 × 10−9 ± 2.6 × 10−9, 4.4 × 10−9 ± 0.6 × 10−9, and 8.1 × 10−9 ± 0.8 × 10−9. Thus, strains with the wild-type and CLRi phenotypes were equally likely to mutate to high-level resistance. This suggests that an CLRi phenotype per se is not a required step in the acquisition of high-level resistance.
Characterization of the CLRr progenitors.
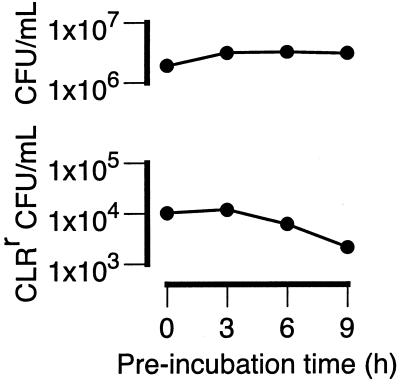
If the initial period of resistance acquisition is the consequence of phenotypic lag, then the mutated state (i.e., base substitution within the 23S rRNA gene) of the source organisms should be heritable. To investigate this, the heritable state of the resistance progenitors was determined by preincubation of the cultures at relatively low bacterial density (∼5 × 107 CFU/ml) for up to 10 h before the addition of CLR. This preincubation step placed the seed organisms (in the early-stationary-growth phase) in conditions suitable for exponential growth. The results of a representative experiment are shown in Fig. 6.
FIG. 6.
Effects of delaying the addition of CLR (4 μg/ml) on the emergence of CLR resistance in MAC strain 101. Seed cultures were diluted 10-fold with fresh medium and incubated from 0, 3, 6, and 9 h before the addition of CLR (4 μg/ml). The total numbers of CFU per milliliter and the numbers of CLRr CFU per milliliter were determined after a total of 4 days in culture. For clarity, error bars are not shown, as they are all within the datum point symbols.
In repeated experiments, preincubation of the cultures without CLR for approximately one generation (∼9 h) reduced the numbers of CLRr organisms by 5- to 10-fold, although in one case the reduction was 40-fold (data not shown). Delay of the addition of CLR resulted in a decrease in the numbers of emergent CLR bacilli. Thus, the phenotype of the resistant progenitors was not committed or heritable. Furthermore, the evidence suggests that the progenitors existed prior to the addition of CLR.
DISCUSSION
Study of the emergence of resistance to macrolides in mycobacteria has several advantages over the study of other resistance phenotypes. First, relatively few mutations are associated with macrolide resistance in mycobacteria, and all are in the peptidyltransferase region of the 23S rRNA gene (12, 13, 14, 15). Second, all resistant mutants have the same phenotype; i.e., the CLR MIC is high (>64 μg/ml [14, 15]) and the growth rates are refractory to the presence of macrolides (up to at least 64 μg/ml for CLR). However, the acquisition of a 23S rRNA gene mutation does confer a fitness disadvantage in the absence of macrolide, shown by the reduced rates of growth of macrolide-resistant M. avium relative to those for the wild type (Table 2). This effect has been reported previously for both mycobacteria (K. A. Nash, Program 37th Intersci. Conf. Antimicrob. Agents Chemother. abstr. S-84, 1987) and some clarithromycin-resistant Helicobacter pylori mutants (3, 22).
The effect of the drug concentration on the emergence of other types of resistance, such as quinolone resistance (4), is complicated by the fact that the phenotype (i.e., the level of resistance) can vary with the underlying mutation. This variability results in the selection of decreasing numbers of mutants as the concentration of antibiotic increases (4). This association between antibiotic concentration and the selection of different mutant populations has been modeled by Martinez and Baquero (10). However, the results presented here suggest that the emergence of macrolide resistance is more complex than just the selection of resistant populations.
Macrolides have a dose-dependent effect on the emergence of specific resistance in M. avium. The maximal increase in the numbers of resistant bacilli was at 4 μg of CLR per ml, which is the same as the CLR MIC for the susceptible parental M. avium strains. The use of combinations of CLR and either EMB or RFB resulted in fewer resistant organisms than the use of CLR alone. For EMB, this effect could be explained by its bacteriostatic effect on the resistant bacilli. However, the effect of RFB is not explained by a direct effect of RFB on resistant organisms. Thus, the emergence of macrolide-resistant M. avium is inconsistent with the hypothesis that drug resistance emerges predominantly from a preexisting resistant subpopulation.
The results presented here do not rule out the existence of preexisting resistant subpopulations, just that they have a minor role in the initial stages of the emergence of CLR resistance. In addition, the results of the present study are not contradictory to the role of random mutation, in the absence of selective pressure, as an important process in bacterial evolution. However, CLR-resistant M. avium appears to derive from a specific population of organisms that act as the progenitors of resistance. Furthermore, these progenitors appear to be in an unstable or transient state, i.e., the bacilli do not appear to be committed to the acquisition of resistance, although they are specific for the acquisition of CLR resistance. The progenitors appear to be more abundant in stationary-phase cultures. This is consistent with the increased mutability of other bacterial species, including Mycobacterium smegmatis, when they are in the stationary phase or under starvation conditions (8, 10, 21).
An important step in the progression of the progenitor to a resistant bacillus is the addition of the antimicrobial agent. Consequently, the antimicrobial agent probably has two effects, first, as an environmental stressor that affects the mutability of the progenitors and, second, as a selective agent for the bacilli that acquire a mutation that confers resistance. Since the only organisms that will be released from the inhibition by the antimicrobial agent are those that acquire the appropriate mutation, the combination of the two effects may explain the apparent drug specificity of the progenitors. Furthermore, since the mutant bacilli need to replace ∼50% of their ribosomes to acquire a high-level macrolide resistance phenotype (19), phenotypic lag probably accounts for a large proportion of the delay between the addition of CLR and the appearance of highly resistant bacilli.
Superficially, the results of the present study appear to support the directed mutation hypothesis (2). However, this is misleading since the most important step in the generation of the progenitors (i.e., acquisition of the uncommitted state) happens in the absence of CLR, and thus, CLR does not truly direct the evolution of the resistant variants. In addition, evidence presented by others has largely discounted the directed mutation hypothesis (5, 9, 16, 21). Previous reports proposed that hypermutable variants or mutators aid bacterial evolution (17, 20). The evidence presented here suggests that stable mutators do not play a significant role in the emergence of CLR resistance in MAC. However, the results are consistent with the presence of a population of organisms in a transient hypermutable state (17). In Escherichia coli, transient hypermutable (or adaptive mutation) is dependent on expression of the homologous recombination proteins RecA and RecBCD (17) and is regulated by the SOS response (11).
Understanding the mechanisms of the emergence of drug resistance and the factors that affect it will aid in the design of improved preventive strategies. This will occur by several means. Experimental data will help refine the mathematical models of the emergence of resistance and also provide a basis for predicting the probability of the development of resistance to new agents (10). Furthermore, the results of the drug combination experiments suggest that susceptibility results may not be a good indicator of the utility of antimicrobials for prevention of the emergence of resistance to other agents. Thus, drugs that are not considered highly active may still have a role in reducing or preventing resistance. The screening of antibiotics for the ability to reduce the level of resistance has been proposed by others (4, 10), and the results presented here lend support to that approach. However, the problem may be how to assess the emergence of resistance in a straightforward, reproducible, and clinically relevant manner (10).
ACKNOWLEDGMENTS
I thank Clark B. Inderlied for the use of laboratory resources and for comments in preparation of the manuscript and Priscilla A. Aralar for technical assistance.
This work was funded in part by NIH contract NO1-AI-25140.
REFERENCES
- 1.Bermudez L E, Nash K A, Petrofsky M, Young L S, Inderlied C B. Effect of ethambutol on emergence of clarithromycin-resistant Mycobacterium avium complex in the beige mouse model. J Infect Dis. 1996;174:1218–1222. doi: 10.1093/infdis/174.6.1218. [DOI] [PubMed] [Google Scholar]
- 2.Cairns J. Directed mutation. Science. 1993;260:1221–1224. doi: 10.1126/science.8493560. [DOI] [PubMed] [Google Scholar]
- 3.Debets-Ossenkopp Y J, Brinkman A B, Kuipers E J, Vandenbroucke-Grauls C M, Kusters J G. Explaining the bias in the 23S rRNA gene mutations associated with clarithromycin resistance in clinical isolates of Helicobacter pylori. Antimicrob Agents Chemother. 1998;42:2749–2751. doi: 10.1128/aac.42.10.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong Y, Zhao X, Domagala J, Drlica K. Effect of fluoroquinolone concentration on selection of resistant mutants of Mycobacterium bovis BCG and Staphylococcus aureus. Antimicrob Agents Chemother. 1999;43:1756–1758. doi: 10.1128/aac.43.7.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foster P L. Nonadaptive mutations occur on the F′ episome during adaptive mutation conditions in Escherichia coli. J Bacteriol. 1997;179:1550–1554. doi: 10.1128/jb.179.5.1550-1554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hindler J. Antimicrobial susceptibility testing. In: Isenberg H D, editor. Clinical microbiology procedures handbook. Vol. 1. Washington, D.C.: American Society for Microbiology; 1992. [Google Scholar]
- 7.Inderlied C B, Salfinger M. Antimicrobial agents and susceptibility tests: mycobacteria. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C.: American Society for Microbiology; 1995. pp. 1385–1404. [Google Scholar]
- 8.Karunakaran P, Davies J. Genetic antagonism and hypermutability in Mycobacterium smegmatis. J Bacteriol. 2000;182:3331–3335. doi: 10.1128/jb.182.12.3331-3335.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacPhee D G, Ambrose M. Spontaneous mutations in bacteria: chance or necessity? Genetica. 1996;97:87–101. doi: 10.1007/BF00132585. [DOI] [PubMed] [Google Scholar]
- 10.Martinez J L, Baquero F. Mutation frequencies and antibiotic resistance. Antimicrob Agents Chemother. 2000;44:1771–1777. doi: 10.1128/aac.44.7.1771-1777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKenzie G J, Harris R S, Lee P L, Rosenberg S M. The SOS response regulates adaptive mutation. Proc Natl Acad Sci USA. 2000;97:6646–6651. doi: 10.1073/pnas.120161797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meier A, Heifets L, Wallace R J, Jr, Zhang Y, Brown B A, Sander P, Böttger E C. Molecular mechanisms of clarithromycin resistance in Mycobacterium avium: observation of multiple 23S rDNA mutations in a clonal population. J Infect Dis. 1996;174:354–360. doi: 10.1093/infdis/174.2.354. [DOI] [PubMed] [Google Scholar]
- 13.Meier A, Kirschner P, Springer B, Steingrube V A, Brown B A, Wallace R J, Jr, Böttger E C. Identification of mutations in 23S rRNA gene of clarithromycin-resistant Mycobacterium intracellulare. Antimicrob Agents Chemother. 1994;38:381–384. doi: 10.1128/aac.38.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nash K A, Inderlied C B. Genetic basis of macrolide resistance in Mycobacterium avium isolated from patients with disseminated disease. Antimicrob Agents Chemother. 1995;39:2625–2630. doi: 10.1128/aac.39.12.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nash K A, Inderlied C B. Rapid detection of mutations associated with macrolide resistance in Mycobacterium avium complex. Antimicrob Agents Chemother. 1996;40:1748–1750. doi: 10.1128/aac.40.7.1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prival M J, Cebula T A. Adaptive mutation and slow-growing revertants of an Escherichia coli lacZ amber mutant. Genetics. 1996;144:1337–1341. doi: 10.1093/genetics/144.4.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenberg S M, Thulin C, Harris R S. Transient and heritable mutators in adaptive evolution in the lab and in nature. Genetics. 1998;148:1559–1566. doi: 10.1093/genetics/148.4.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sander P, Prammananan T, Meier A, Frischkorn K, Böttger E C. The role of ribosomal RNAs in macrolide resistance. Mol Microbiol. 1997;26:469–480. doi: 10.1046/j.1365-2958.1997.5811946.x. [DOI] [PubMed] [Google Scholar]
- 19.Sigmund C D, Ettayebi M, Borden A, Morgan E A. Antibiotic resistance mutations in ribosomal RNA genes of Escherichia coli. Methods Enzymol. 1988;164:673–690. doi: 10.1016/s0076-6879(88)64077-8. [DOI] [PubMed] [Google Scholar]
- 20.Sniegowski P D, Gerrish P J, Lenski R E. Evolution of high mutation rates in experimental populations of E. coli. Nature. 1997;387:703–705. doi: 10.1038/42701. [DOI] [PubMed] [Google Scholar]
- 21.Torkelson J, Harris R S, Lombardo M J, Nagendran J, Thulin C, Rosenberg S M. Genome-wide hypermutation in a subpopulation of stationary-phase cells underlies recombination-dependent adaptive mutation. EMBO J. 1997;16:3303–3311. doi: 10.1093/emboj/16.11.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang G, Rahman M S, Humayun M Z, Taylor D E. Multiplex sequence analysis demonstrates the competitive growth advantage of the A-to-G mutants of clarithromycin-resistant Helicobacter pylori. Antimicrob Agents Chemother. 1999;43:683–685. doi: 10.1128/aac.43.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]