Abstract
Genomic medicine aims to improve health using the individual genomic data of people to inform care. While clinical utility of genomic medicine in many monogenic, Mendelian disorders is amply demonstrated, clinical utility is less evident in polygenic traits, e.g., coronary artery disease or breast cancer. Polygenic risk scores (PRS) are subsets of individual genotypes designed to capture heritability of common traits, and hence to allow the stratification of risk of the trait in a population. We systematically reviewed the PubMed database for unequivocal evidence of clinical utility of polygenic risk scores, using stringent inclusion and exclusion criteria. While we identified studies demonstrating clinical validity in conditions where medical intervention based on a PRS is likely to benefit patient outcome, we did not identify a single study demonstrating unequivocally such a benefit, i.e. clinical utility. We conclude that while the routine use of PRSs hold great promise, translational research is still needed before they should enter mainstream clinical practice.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00439-022-02452-x.
Introduction
Genomic medicine aims to improve health using an individual’s genomic information, e.g. a SNP genotype or DNA sequence, to inform care. Genomic medicine is defined by the National Human Genome Research Institute as a rapidly growing field involving the application of genomic information in clinical care (NHGRI website 2021). While many successful examples of genomic medicine involve implementation of programs to identify and manage monogenic disease, i.e. disease with Mendelian inheritance, it is not clear to what extent genomic medicine is being successful regarding disease with complex inheritance. Examples of such diseases are coronary artery disease, type 2 diabetes, and cancer. Complexity in such genetic traits with intricate inheritance is twofold. Firstly, the role of the environment in disease expression is usually significant, decreasing the contribution of the genome typically to around 50% (Polderman et al. 2015), which ultimately limits the predictability of the trait based on genome analysis alone. Secondly, many complex traits result from the interaction of several independent loci. Thus, complex traits can be seen as polygenic predispositions from multiple quantitative trait loci, that eventually produced disease under the influence of a particular environmental or epigenetic modifier. Progress in Genome-wide association studies (GWAS) have identified many such quantitative trait loci, but more remain to be discovered. GWAS are research methods utilized to detect the association between genetic variants and traits in population samples. These studies are designed to improve the understanding of the biology of disease, under the assumption that a better understanding will lead to better prevention or better treatment. The GWAS data generated from human studies proved to be useful in creating genetic predictors for complex traits by estimating the effect size at multiple loci in a discovery sample and using those estimated SNP effects in independent samples to generate a polygenic risk score (PRS). (Visscher et al. 2017). Different PRS methods model the polygenic associations to the phenotype or traits in different ways, and often make distinct or similar modeling assumptions on the effect size distribution. These assumptions can frequently help in the understanding of the performance of PRS methods across phenotype with distinct genetic architectures. While most PRSs have been developed from defined populations, e.g., FinnGen (Mars et al. 2020), they seem at least partially valid in other populations as well (Dikilitas et al. 2020; Ho et al. 2020). Nonetheless, genetically diverse studies are mandatory to cover different world populations to ensure equitable clinical utilization of PRSs (Martin et al. 2019).
The generation of PRSs is a relatively novel statistical method that associates the collectively weighted risk alleles at many of a person’s SNP loci to a trait. Thus, PRS is a quantifiable genetic risk score, determined by the cumulative impact of genome-wide variants, aimed to improve risk prediction for common chronic diseases such as coronary artery disease. (Khera et al. 2018).
With empirical improvements over time, PRSs have been widely applied in many research studies of common chronic diseases, confirming their ability to predict disease risk or status, i.e., demonstrating clinical validity. According to the CDC ACCE model (Analytical validity, Clinical validity, Clinical utility and Ethical, legal & Social implication) refers to the power of a test to predict a particular clinical outcome or phenotype (CDC website: https://www.cdc.gov/genomics/gtesting/acce/index.htm). Clinical utility, on the other hand, is focused on the effect of the use of a given test on patient health outcomes. (Haddow and Palomaki 2003). The ability to predict disease occurrence using a PRS should eventually translate into clinical utility if these are to be implemented in clinical care. PRSs for some diseases were able to identify subgroups of patients with high relative risks, and absolute relative risks that approach risk values conveyed by highly penetrant, single-gene mutations (Khera et al. 2018), considered clinically actionable. PRSs have been shown to provide additional risk stratification when combined with single-gene mutation testing for monogenic disorders with incomplete penetrance, e.g., hereditary breast and ovarian cancer or familial hypercholesterolemia (Fahed et al. 2020). Stratifying the risks of common cancers, or of coronary artery disease, should presumably help tailor screening intervals, or drug regimens, respectively, and hence mitigate the disease associated with the genetic risks. Evidence for such clinical utility has however been lagging. This is because the complexity of genetic architecture and multidimensionality of genetic and environmental contributions to disease phenotypes continue to pose significant challenges for the clinical utility as well as broad-scale use of PRSs.
The aim of this study was to perform a systematic review of the existing evidence of clinical utility of PRS for genomic medicine applications. We focused our search on studies which demonstrated a benefit on patient clinical outcome, be it process outcome, intermediate outcome, or health outcome. In the case of hypercholesterolemia-related vascular disease, these would correspond for example to: the effective adoption of a healthy diet; a lowered blood LDL-cholesterol; and a decreased rate of myocardial infarction, respectively.
Methods
Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guidelines were followed (Moher et al. 2009; Parums 2021). This review was not registered in a systematic review register. Search terms were selected by convening a group of expert researchers in the field to consider PubMed’s Medical Subject Headings (MeSH) terms associated with the inclusion criteria. Searches combined sets of terms for genomic medicine, clinical utility and multifactorial inheritance. Terms were iteratively refined through review of results for relevance by the expert panel until consensus on these terms was reached. The full search strategy is shown in Table 1. The literature search was conducted in PubMed on articles published on or before December 16, 2020. The search was limited to publications in English. Articles retrieved were downloaded into an Excel spreadsheet, where duplicates were removed. Title and abstract screening were undertaken by five pairs of researchers (JK, LF, AB, TW, TK, CC, MT, EM, GE, HM) who each worked independently on 20% of the retrieved articles to determine if the article should be included or excluded. An identical search was conducted in PubMed on November 03, 2021 by JK, MA, FA, to ensure any recent publications were included in this review.
Table 1.
Search terms
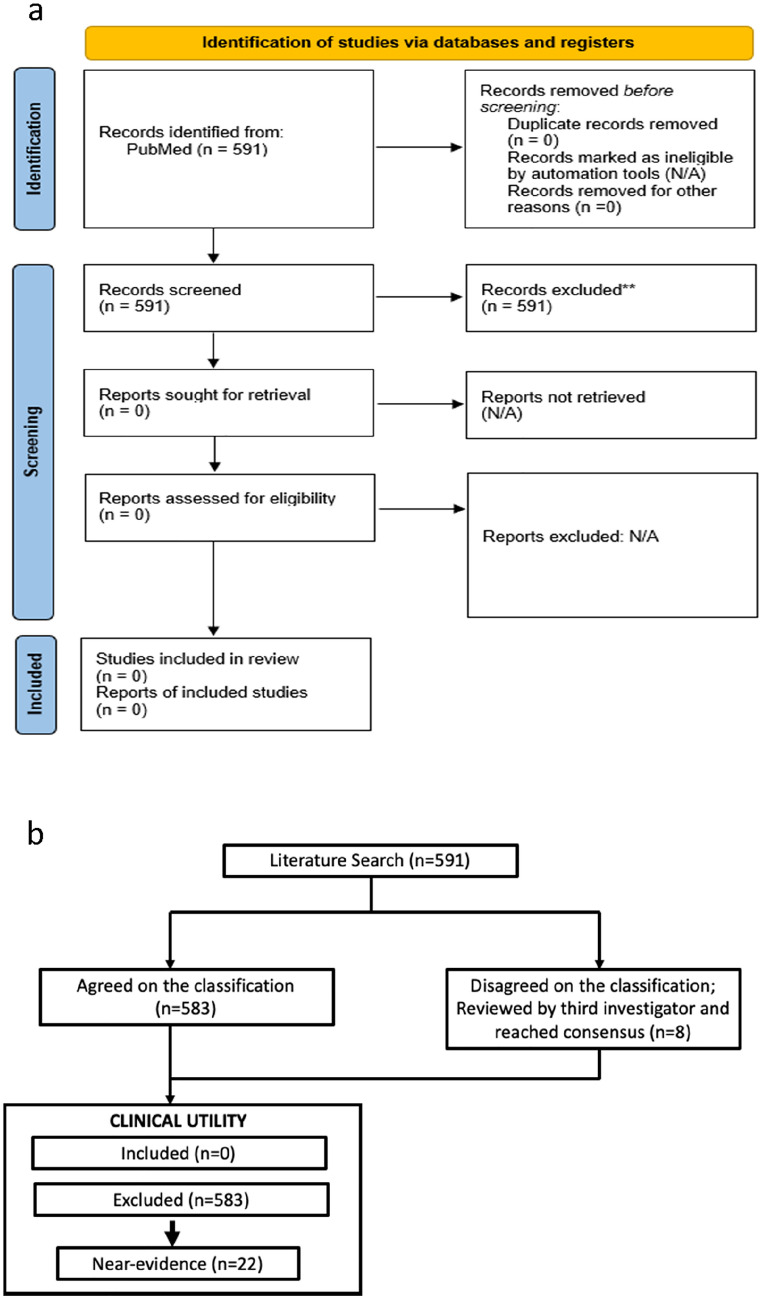
Included articles presented evidence of clinical utility of genomic medicine for conditions stemming from a polygenic risk where PRSs were used to inform intervention. Articles were excluded if the research findings were specific to monogenic disease, pharmacogenomics, microbial/metagenomics, expression profiling, somatic genome or methodology only. Articles were also excluded if they did not contain genomic data or health outcomes or if the articles were reviews, or association/observation studies. Studies that fulfilled all inclusion criteria and passed all exclusion criteria but failed to unequivocally demonstrate an effect on patient health outcome were additionally assigned the label of “near evidence.” These articles included evidence of clinical validity and were suggestive of utility but lacked clinical outcome data. Upon completion of the independent review of the articles, researchers compared screening results and resolved discordance with a third researcher. The final set of articles were agreed on by the entire research team (Fig. 1a). Inclusion/exclusion criteria are shown in Table 2 and additionally assigned a label of “near evidence” (see Table S1).
Fig. 1.
Study design and results. a Overview of the literature review process. b Outcome of the systematic review process of peer-reviewed literature following PRISMA guidelines
Table 2.
Exclusion criteria
|
● Monogenic disease ● NOT genomic data ● NOT clinical utility, no health outcome ● Pharmacogenomics ● Cancer studies—tumor profiling ● Microbial/metagenomics ● Expression profiling ● Association or observation study ● Methodology only ● Review |
| ● Other: meta-analysis, case report, interview, educational article |
Results
The initial PubMed query run on December 16, 2020, retrieved 530 articles. Ten investigators manually curated 105 articles each, and each article was curated in duplicate. Duplicate curations agreed on classifying 522 and disagreed on 8. These 8 articles were reviewed by a third investigator and discussed with the two initial investigators to reach a final consensus. This process was repeated on a second PubMed query run on November 03, 2021, and retrieved 61 additional items. The same paired review process was followed, and the results were 100% in agreement with having no clinical utility (Fig. 1b).
No study was found that showed unequivocal demonstration of clinical utility of any PRS. The study team therefore excluded the studies that were categorized as “Near-Evidence” in analysis (Fig. 1b).
22/591 studies showed robust evidence of clinical validity, i.e. some PRSs accurately stratified individual disease susceptibility, e.g., breast cancer (Mavaddat et al. 2019) or atrial fibrillation (Mars 2020). One example was PRS for breast cancer, where enhanced screens (mammograms) were likely, but not proven, to benefit women with highest risk scores, by analogy with BRCA1&2 (Kramer et al. 2020).
Discussion
We followed PRISMA guidelines to systematically review the PubMed database for published evidence of clinical utility of using a PRS for improving patient health, and manually curated the retrieved items to systematically remove studies dealing with monogenic disease, pharmacogenomics, microbial/metagenomics, expression profiling, somatic genome or methodology only. Our screen did not identify a single study demonstrating evidence of clinical utility of a PRS, as of November 3rd, 2021. This suggests that PRSs are not ready to be implemented in the clinic without further research. We did find studies that demonstrated clinical validity of PRSs in clinical conditions where medical action based on the PRS is likely to produce a benefit to patient outcome, which we referred to as ‘near evidence’ of clinical utility. For example, Kramer et al. (2020) demonstrated that a PRS was clinically valid in women with breast cancer for stratifying the risk of contralateral breast cancer, and concluded that this PRS “can be incorporated into contralateral breast cancer risk prediction models to help improve stratification and optimize surveillance and treatment strategies”. However, further studies are needed to demonstrate the utility of PRS prospectively does improve morbidity and mortality.
Our study has several limitations. We screened the PubMed database only, because it is a large repository of regularly updated peer-reviewed medical articles that captures a very large portion of medical knowledge. We chose not to include “gray” literature in our search, to minimize the chance of reporting false positive results, i.e. PRSs with no demonstrated clinical utility.
Our search of the PubMed literature was designed to minimize alpha and beta-type errors, but was not perfect. An additional, non-systematic approach identified a study that did demonstrate clinical utility on an intermediate outcome (blood level of LDL-cholesterol), albeit not on health outcome per se (myocardial infarction) (Kullo et al. 2016). It remains possible that more studies were not included despite demonstrating evidence of clinical utility, but we consider this possibility unlikely because our manual curation of the PubMed screen was performed in duplicate and reviewed by a third expert in case of disagreement.
We believe the main limitation of our review arises from the pragmatic decisions made to cope with the massive volume of literature on this topic. We searched for a single, albeit comprehensive source but did not attempt to identify unpublished studies or gray literature, and we did not intend to generate pooled estimates. With the huge number of studies being produced and published every year, it is very difficult to synthesize the evidence with traditional systematic review methods. One alternative is to apply artificial intelligence and other technologies that automate or semi-automate the different steps of the systematic review process. However, it is important to be very specific about what artificial intelligence can provide and where its use might be inappropriate. Exemplary reviews combining artificial intelligence with rigorous systematic review methods have been produced in the context of COVID-19 (Boutron et al. 2020; Pierre et al. 2021; Siemieniuk et al. 2020).
In spite of the current absence of unequivocal evidence of clinical utility using stringent criteria, the routine use of PRSs hold great promise. They can be assessed at low cost (< $30) at any point in time. Further research should now aim at comparing the current standard of care with and without the use of PRSs in cohorts of patients with complex traits to demonstrate a benefit for patient health. Randomized controlled studies are the best approach. An example of such a study design might consist of implementing the use of a PRS in making decisions regarding apparently benign breast tumors identified on routine mammogram screens in asymptomatic women, versus non-use of PRS in a random control cohort, and assess outcome in terms of invasive breast cancer after a defined interval, e.g., one year. It should be noted however that randomized controlled studies will be much harder to achieve where the relevant health outcomes take many years or decades to manifest (e.g. myocardial infarction). Furthermore, they must cover the diverse ethnicities of patients. Hence, some PRSs are likely to be implemented empirically for clinical decision-making, by analogy with monogenic testing, in extreme strata (both tails) of polygenic risk, e.g., for shortening screening intervals in women with high breast cancer PRS, with a posteriori, retrospective evaluation of clinical outcome. Another line of research should continue to improve the technical ability of PRSs to capture trait heritability. It is essential that future studies of PRSs for both clinical validity and utility engage diverse populations to improve their relevance to all population groups and avoid exacerbation of health inequities.
In conclusion, although our search could not identify published evidence of unequivocal clinical utility of a PRS, we found numerous examples of near evidence of clinical utility and ample demonstration of clinical validity. As PRS continue to improve in their ability to capture heritability of polygenic traits, we can expect demonstration of clinical utility by appropriate clinical trials in the coming years in a variety of disorders like coronary artery disease or common cancers, ushering a new era of genomic medicine.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
JK is in part funded by the National Institutes of Health Common Fund H3ABioNet grant, U24HG006941. The G2MC evidence investigators are Alassane B Maiga: Faculty of Medicine and Odontostomatology, University of Sciences, techniques and Technologies of Bamako, Mali; Elisa J.F. Houwink: Department of Public Health and Primary Care (PHEG), Leiden University Medical Center, Leiden, The Netherlands; George P. Patrinos: University of Patras, School of Health Sciences, Department of Pharmacy, Laboratory of Pharmacogenomics and Individualized Therapy, University Campus, Rion, GR-26504, Patras, Greece, United Arab Emirates University, College of Medicine and Health Sciences, Department of Pathology, Al-Ain, Abu Dhabi, UAE, United Arab Emirates University, Zayed Center for Health Sciences, Al-Ain, Abu Dhabi, UAE, Isabelle Vandernoot: Center of Human Genetics, Hôpital Erasme, Université Libre de Bruxelles, Brussels, Belgium Jeffrey Braithwaite: Australian Institute of Health Innovation, Macquarie University, Sydney, New South Wales, Australia, Saumya Shekhar Jamuar: Genetics Service, KK Women’s and Children’s Hospital, SingHealth Duke-NUS Genomic Medicine Centre, Singapore, Singapore, Sheri Schully: National Human Genome Research Institute, National Institutes of Health, Bethesda, Maryland, United States. schullys@mail.nih.gov, Yiping Shen: Division of Genetics and Genomics, Boston Children's Hospital, Harvard Medical School, Boston USA, Woman's Hospital Zhejiang University School of Medicine, Hangzhou Zhejiang, China. Fahd Al-Mulla and Veeramani Marimuthu: are funded by Kuwait Foundation for the Advancement of Sciences, Kuwait.
Author contributions
MA and FAM conceived the study and third-reviewed the discordant paired reviews; BZ implemented the PRISMA method and performed the PubMed searches; JK, AB, CC, GEK, LF, TK, EBM, HM, MKT, TW performed the paired reviews; HK Implemented the PRISMA method; GR commented on artificial intelligence; JK, BZ, MSW and MA wrote the manuscript; All authors had meetings to adjust the MESH search terms, participated in the discussion of the results, and reviewed the manuscript.
Funding
JK is in part funded by the National Institutes of Health Common Fund H3ABioNet grant, U24HG006941.
Declarations
Conflict of interest
GPP is Full Member and National Representative at the European Medicines Agency, Committee for Human Medicinal Products (CHMP)—Pharmacogenomics Working Party, Amsterdam, the Netherlands.
Footnotes
The G2MC Evidence investigators are given in the Acknowledgements section.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Judit Kumuthini, Email: jkumuthini@gmail.com.
Brittany Zick, Email: brittany@g2mc.org.
Angeliki Balasopoulou, Email: abalaso@eie.gr.
Constantina Chalikiopoulou, Email: cchalikio@eie.gr.
Collet Dandara, Email: collet.dandara@uct.ac.za.
Ghada El-Kamah, Email: ghadaelkamah@hotmail.com.
Laura Findley, Email: laurafindley5@gmail.com.
Theodora Katsila, Email: thkatsila@eie.gr.
Rongling Li, Email: lir2@mail.nih.gov.
Ebner Bon Maceda, Email: egmaceda@up.edu.ph.
Henrietta Monye, Email: henrietta.monye@gmail.com.
Gabriel Rada, Email: radagabriel@gmail.com.
Meow-Keong Thong, Email: thongmk@um.edu.my.
Thilina Wanigasekera, Email: thilinaw71@gmail.com.
Hannah Kennel, Email: hannah@g2mc.org.
Veeramani Marimuthu, Email: veeramani.marimuthu@dasmaninstitute.org.
Marc S. Williams, Email: Mswilliams1@geisinger.edu
Fahd Al-Mulla, Email: fahd@al-mulla.org.
Marc Abramowicz, Email: marc.abramowicz@unige.ch.
References
- Boutron I, Chaimani A, Meerpohl JJ, Hróbjartsson A, Devane D, Rada G, Tovey D, Grasselli G, Ravaud P, COVID-NMA Consortium The COVID-NMA Project: building an evidence ecosystem for the COVID-19 pandemic. Ann Intern Med. 2020;173(12):1015–1017. doi: 10.7326/M20-5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC website: https://www.cdc.gov/genomics/gtesting/acce/index.htm. Accessed 15 Mar 2022
- Dikilitas O, Schaid DJ, Kosel ML, Carroll RJ, Chute CG, Denny JA, Fedotov A, Feng Q, Hakonarson H, Jarvik GP, Lee M, Pacheco JA, Rowley R, Sleiman PM, Stein CM, Sturm AC, Wei WQ, Wiesner GL, Williams MS, Zhang Y, Kullo IJ. Predictive utility of polygenic risk scores for coronary heart disease in three major racial and ethnic groups. Am J Hum Genet. 2020;106(5):707–716. doi: 10.1016/j.ajhg.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahed AC, Wang M, Homburger JR, Patel AP, Bick AG, Neben CL, Lai C, Brockman D, Philippakis A, Ellinor PT, Cassa CA, Lebo M, Ng K, Lander ES, Zhou AY, Kathiresan S, Khera AV. Polygenic background modifies penetrance of monogenic variants for tier 1 genomic conditions. Nat Commun. 2020;11(1):3635. doi: 10.1038/s41467-020-17374-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddow JE, Palomaki GE (2003) ACCE: a model process for evaluating data on emerging genetic tests. In: Khoury M, Little J, Burke W (eds) Human genome epidemiology: a scientific foundation for using genetic information to improve health and prevent disease. Oxford University Press, pp 217–233
- Ho WK, Tan MM, Mavaddat N, Tai MC, Mariapun S, Li J, Ho PJ, Dennis J, Tyrer JP, Bolla MK, Michailidou K, Wang Q, Kang D, Choi JY, Jamaris S, Shu XO, Yoon SY, Park SK, Kim SW, Shen CY, Antoniou AC. European polygenic risk score for prediction of breast cancer shows similar performance in Asian women. Nat Commun. 2020;11(1):3833. doi: 10.1038/s41467-020-17680-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT, Kathiresan S. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50(9):1219–1224. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopans DB. The wisdom trial is based on faulty reasoning and has major design and execution problems. Breast Cancer Res Treat. 2021;185(3):549–556. doi: 10.1007/s10549-020-06020-7. [DOI] [PubMed] [Google Scholar]
- Kramer I, Hooning MJ, Mavaddat N, Hauptmann M, Keeman R, Steyerberg EW, Giardiello D, Antoniou AC, Pharoah P, Canisius S, Abu-Ful Z, Andrulis IL, Anton-Culver H, Aronson KJ, Augustinsson A, Becher H, Beckmann MW, Behrens S, Benitez J, Bermisheva M, Schmidt MK. Breast cancer polygenic risk score and contralateral breast cancer risk. Am J Hum Genet. 2020;107(5):837–848. doi: 10.1016/j.ajhg.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullo IJ, Jouni H, Austin EE, Brown SA, Kruisselbrink TM, Isseh IN, Haddad RA, Marroush TS, Shameer K, Olson JE, Broeckel U, Green RC, Schaid DJ, Montori VM, Bailey KR. Incorporating a genetic risk score into coronary heart disease risk estimates: effect on low-density lipoprotein cholesterol levels (the MI-GENES Clinical Trial) Circulation. 2016;133(12):1181–1188. doi: 10.1161/CIRCULATIONAHA.115.020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mars N, Koskela JT, Ripatti P, Kiiskinen T, Havulinna AS, Lindbohm JV, Ahola-Olli A, Kurki M, Karjalainen J, Palta P, FinnGen, Neale BM, Daly M, Salomaa V, Palotie A, Widén E, Ripatti S (2020). Polygenic and clinical risk scores and their impact on age at onset and prediction of cardiometabolic diseases and common cancers. Nat Med 26(4):549–557. 10.1038/s41591-020-0800-0 [DOI] [PubMed]
- Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019;51(4):584–591. doi: 10.1038/s41588-019-0379-xPMID30926966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavaddat N, Michailidou K, Dennis J, Lush M, Fachal L, Lee A, Tyrer JP, Chen TH, Wang Q, Bolla MK, Yang X, Adank MA, Ahearn T, Aittomäki K, Allen J, Andrulis IL, Anton-Culver H, Antonenkova NN, Arndt V, Aronson KJ, Easton DF. Polygenic risk scores for prediction of breast cancer and breast cancer subtypes. Am J Hum Genet. 2019;104(1):21–34. doi: 10.1016/j.ajhg.2018.11.002PMID30554720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ (clin Res Ed) 2009;339:b2535. doi: 10.1136/bmj.b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NHGRI website (2021). https://www.genome.gov/health/Genomics-and-Medicine. Accessed 30 Nov 2021
- Parums DV. Editorial: review articles, systematic reviews, meta-analysis, and the updated preferred reporting items for systematic reviews and meta-analyses (PRISMA) 2020 guidelines. Me Science Moni. 2021;27:e934475. doi: 10.12659/MSM.934475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre O, Riveros C, Charpy S, Boutron I. Secondary electronic sources demonstrated very good sensitivity for identifying studies evaluating interventions for COVID-19. J Clin Epidemiol. 2021;141:46–53. doi: 10.1016/j.jclinepi.2021.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polderman TJ, Benyamin B, de Leeuw CA, et al. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet. 2015;47(7):702–709. doi: 10.1038/ng.3285. [DOI] [PubMed] [Google Scholar]
- Siemieniuk RA, Bartoszko JJ, Ge L, Zeraatkar D, Izcovich A, Kum E, Pardo-Hernandez H, Qasim A, Martinez J, Rochwerg B, Lamontagne F, Han MA, Liu Q, Agarwal A, Agoritsas T, Chu DK, Couban R, Cusano E, Darzi A, Devji T, Brignardello-Petersen R. Drug treatments for covid-19: living systematic review and network meta-analysis. BMJ (clinical Research Ed) 2020;370:m2980. doi: 10.1136/bmj.m2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, Yang J. 10 years of GWAS discovery: biology, function, and translation. Am J Hum Genet. 2017;101(1):5–22. doi: 10.1016/j.ajhg.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.