Abstract
Both katanosin B and plusbacin A3 are naturally occurring cyclic depsipeptide antibiotics containing a lactone linkage. They showed strong antibacterial activity against methicillin-resistant Staphylococcus aureus and VanA-type vancomycin-resistant enterococci, with MICs ranging from 0.39 to 3.13 μg/ml, as well as against other gram-positive bacteria. They inhibited the incorporation of N-acetylglucosamine, a precursor of cell wall synthesis, into peptidoglycan of S. aureus whole cells at concentrations close to their MICs. In vitro studies with a wall-membrane particulate fraction of S. aureus showed that katanosin B and plusbacin A3 inhibited the formation of lipid intermediates, with 50% inhibitory concentrations (IC50s) of 2.2 and 2.3 μg/ml, respectively, and inhibited the formation of nascent peptidoglycan, with IC50s of 0.8 and 0.4 μg/ml, respectively. Vancomycin, a well-known inhibitor of transglycosylation, did not inhibit the formation of lipid intermediates but did inhibit the formation of nascent peptidoglycan, with an IC50 of 4.1 μg/ml. Acetyl-Lys-d-Ala-d-Ala, an analog of the terminus of the lipid intermediates, effectively suppressed the inhibition of transglycosylation by vancomycin, but did not suppress those by katanosin B and plusbacin A3. These results indicate that the antibacterial activity of katanosin B and plusbacin A3 is due to blocking of transglycosylation and its foregoing steps of cell wall peptidoglycan synthesis via a mechanism differing from that of vancomycin.
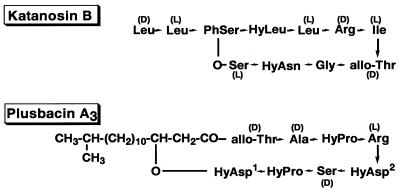
Katanosin B and plusbacin A3 were previously isolated from a strain related to the genus Cytophaga and a strain of Pseudomonas, respectively (35, 36). They exhibited in vitro activity against gram-positive bacteria and showed therapeutic effects in mice infected with Staphylococcus aureus by subcutaneous administration (35, 36). Structure analysis revealed them both to be cyclic depsipeptide antibiotics containing a lactone linkage, although their amino acid residues and sequences were very different from each other (17, 37) (Fig. 1). They were reported to be inhibitors of cell wall synthesis, judging from their inhibition of radio-labeled diaminopimelic acid incorporation into the cell wall peptidoglycan of a Bacillus strain.
FIG. 1.
Structures of katanosin B and plusbacin A3. PhSer, l-threo-β-phenylserine; HyLeu, l-threo-β-hydroxyleucine; HyAsn, l-threo-β-hydroxyasparagine; HyAsp1, l-threo-β-hydroxyaspartic acid; HyAsp2, d-threo-β-hydroxyaspartic acid; HyPro, l-trans-3-hydroxyproline.
Vancomycin has been used to treat gram-positive bacterial infection and is regarded as a last resort for the treatment of methicillin-resistant S. aureus infection. However, the prevalence of vancomycin-resistant enterococci has already become an important problem (21, 26), and even the emergence of S. aureus clinical isolates with reduced vancomycin susceptibility has been reported recently (6, 11, 12, 34, 38, 39). Thus, novel drugs to replace vancomycin are urgently required. Drugs with different modes of action from vancomycin should be promising candidates against vancomycin-resistant strains. In this study, the mechanism of action of katanosin B and plusbacin A3 was studied using target organisms with vancomycin for comparison.
MATERIALS AND METHODS
Bacteria and growth conditions.
The organisms used in this experiment are listed in Table 1. Unless otherwise stated, S. aureus strains were grown in tryptic soy broth (TSB; Difco Laboratories, Detroit, Mich.) at 37°C with aeration.
TABLE 1.
Properties of bacterial strains used in this study
| Strain | Relevant phenotypea | MIC (μg/ml)
|
Sourceb or reference | ||
|---|---|---|---|---|---|
| Katanosin B | Plusbacin A3 | Vancomycin | |||
| Staphylococcus aureus | |||||
| NCTC8325 | Mcs | 0.39 | 0.78 | 1.56 | 29 |
| Smith (diffuse) | Mcs | 0.39 | 0.78 | 0.78 | 13 |
| SRM133 | Mcr | 0.39 | 0.78 | 1.56 | Laboratory strain |
| SR3637 | Mcr | 0.39 | 1.56 | 1.56 | Clinical isolate |
| Mu50 | Mcr Vmi | 0.39 | 1.56 | 12.5 | 12 |
| Enterococcus faecalis | |||||
| SR1004 | Vms | 0.78 | 3.13 | 1.56 | Clinical isolate |
| SR7914 | Vmr (VanA) | 0.78 | 1.56 | >50 | Clinical isolate |
| Enterococcus faecium | |||||
| SR15941 | Vms | 0.78 | 3.13 | 1.56 | Clinical isolate |
| SR7917 | Vmr (VanA) | 0.78 | 3.13 | >50 | Clinical isolate |
Abbreviations: Mc, methicillin; Vm, vancomycin; s, susceptible; r, resistant; i, intermediate.
Clinical isolates were from Japan.
Drugs.
Katanosin B and plusbacin A3 were isolated as described previously (35, 36). Vancomycin is commercially available (Shionogi, Osaka, Japan).
Susceptibility test.
MICs were determined by using serial twofold microdilutions of antibiotic in cation-adjusted Mueller-Hinton broth (Difco Laboratories) (28). The overnight culture of bacteria was inoculated at 5 × 105 CFU/ml and incubated at 35°C for 20 h before the MIC was scored.
Incorporation of 14C-labeled N-acetylglucosamine into cell wall peptidoglycan.
S. aureus NCTC8325 was cultivated at 37°C overnight in CGPY broth (Na2HPO4, 6 g; NaCl, 3 g; MgCl2 · 6H2O, 0.1 g; NH4Cl, 2 g; Na2SO4, 0.15 g; KH2PO4, 3 g; Bactopeptone, 10 g; yeast extract, 0.1 g; and glucose, 5 g [per liter], pH 7.0) (19). The culture was diluted 100-fold with the same medium and further cultivated until an optical density at 660 nm (OD660) of 0.2 was reached by monitoring the density with a Spectronic 20A spectrophotometer (Shimazu, Kyoto, Japan). The cells were pelleted by centrifugation at 8,000 × g for 10 min and resuspended in modified cell wall synthesis medium (for [14C]GlcNAc incorporation experiment, KH2PO4 [6 g], K2HPO4 [6 g], NH4Cl [2 g], MgSO4 · 7H2O [5 mg], FeSO4 [5 mg], glucose [100 mg], uracil [40 mg], l-alanine [50 mg], l-glutamic acid [120 mg], l-lysine [50 mg], chloramphenicol [100 mg] [per liter]) to an OD660 of 0.1 (19, 23). One 1-ml portion of the cell suspension containing each concentration of the drug and 5 μM [14C]GlcNAc (1.85 GBq/mmol; Amersham Pharmacia Biotech UK Limited, Buckinghamshire, England) was incubated at 37°C with aeration. After 30 min of incubation, a 0.5-ml portion of the cell suspension was transferred to a microcentrifuge tube containing 0.5 ml of ice-cold 10% trichloroacetic acid (TCA). The mixture was incubated at 90°C for 15 min, placed on ice for 30 min, and filtered with a membrane filter (type HA; pore size, 0.45 μm; diameter, 25 mm; Millipore Corp., Bedford, Mass.) followed by 5% TCA washing. The membrane filter was then immersed in 5 ml of Pico-Fluor 40 (Packard Instrument Company, Meriden, Conn.), and the radioactivity was counted with a liquid scintillation analyzer, Tri-Carb 2000CA (Packard Instrument Company).
Preparation of wall-membrane particulate and supernatant fraction.
The overnight culture of S. aureus SRM133 was diluted 50-fold in warm TSB and further cultivated at 37°C with shaking to the logarithmic phase of growth. Next, cells were harvested and washed with cold 0.05 M Tris-HCl buffer (pH 8.0) containing 0.1 mM MgCl2 (buffer A). The cells were resuspended in buffer A and disrupted with glass beads in a mechanical cell homogenizer, HOM-MSK (B. Braun Biotech Inc., Allentown, Pa). A wall-membrane particulate fraction was prepared by differential centrifugation between 8,000 × g for 10 min and 100,000 × g for 30 min. The pellet was washed once with buffer A, resuspended in it, and stored at −80°C. For preparation of a supernatant fraction, S. aureus SRM133 cells grown overnight were harvested and disrupted as described above. The supernatant after differential centrifugation was collected and stored as supA at −80°C. The protein contents of a wall-membrane particulate fraction and supA were determined by using the Bio-Rad Dc protein assay reagent (Bio-Rad Laboratories, Hercules, Calif.) with bovine serum albumin as the standard.
Enzymatic reactions.
Enzymatic syntheses of lipid intermediates and nascent peptidoglycan were detected with labeled glycine. The reaction mixture contained 60 mM Tris-HCl (pH 8.5), 30 mM MgCl2, 1 mM 2-mercaptoethanol, 330 μM UDP-GlcNAc, 1.7 mM ATP, 0.5 mg of protein/ml of supA, 7.1 μM [14C]glycine (4.17 GBq/mmol; NEN Life Science Products, Inc. Boston, Mass.), and 0.5 mg of a wall-membrane particulate fraction per ml. If necessary, each concentration of the drugs was added to the reaction mixture. The reaction was performed at 30°C for 60 min. The reaction mixture was then spotted onto a cellulose thin-layer chromatography (TLC) plate (TLC plates, cellulose F precoated; Merck, Darmstadt, Germany) and developed with isobutyric acid–1 M ammonia (5:3) as the solvent for 17 h at room temperature. Radioactivity on the plate was detected with Bio-Imaging Analyzer BAS2000 (Fuji Photo Film Co., Ltd., Tokyo, Japan). For the suppression experiment, each concentration of acetyl-Lys-d-Ala-d-Ala (Sigma Chemical Co., St. Louis, Mo.) was added to the reaction mixture containing the respective drug (12.5 μg of vancomycin 1.56 μg of katanosin B, or 0.78 μg of plusbacin A3 per ml, at which concentrations the formation of nascent peptidoglycan was almost inhibited).
Suppression of growth inhibition.
The overnight culture of S. aureus NCTC8325 was streaked over a TSA (Difco) plate with a swab. Paper disks containing 5 μg of each antibiotic were placed on it, and then 50 μg of protein of a wall-membrane particulate was spotted near the each disk. The plate was incubated at 37°C overnight.
RESULTS
Antibacterial activity.
MICs of katanosin B, plusbacin A3, and vancomycin against representative staphylococci and enterococci are listed in Table 1. Katanosin B and plusbacin A3 showed strong activity irrespective of methicillin and vancomycin resistance.
Effect on incorporation of [14C]GlcNAc into peptidoglycan of S. aureus whole cells.
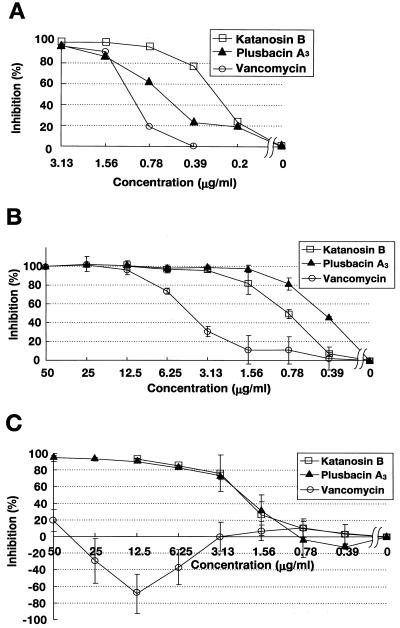
Both katanosin B and plusbacin A3 strongly inhibited the incorporation of [14C]GlcNAc into staphylococcal cell wall peptidoglycan (Fig. 2A). The degree of inhibition correlated with the concentration of the drug. The 50% inhibitory concentrations (IC50s) of katanosin B, plusbacin A3, and vancomycin were 0.28, 0.62, and 1.0 μg/ml, respectively, which were all close to their MICs.
FIG. 2.
(A) Inhibition of incorporation of [14C]GlcNAc into peptidoglycan of S. aureus whole cells. Assays were carried out as described in Materials and Methods. Radioactivity was measured and expressed as the inhibition rate by comparison with a control without antibiotic. (B and C) Inhibition of formation of (B) lipid intermediates and (C) nascent peptidoglycan. Radioactivity incorporated into each product shown in Fig. 4 was measured and expressed as the inhibition rate by comparison with a control without antibiotic. Results are means ± standard deviations.
Formation of lipid intermediates and nascent peptidoglycan in vitro.
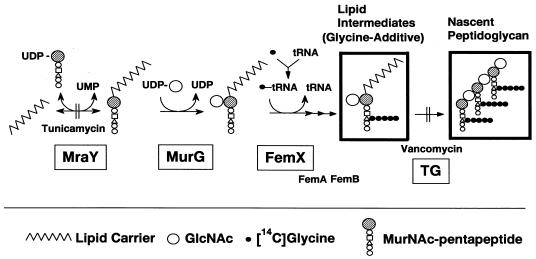
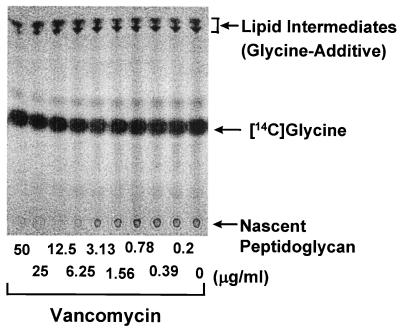
Newly formed lipid intermediates and nascent peptidoglycan were separated on TLC and detected by incorporation of [14C]glycine via successive reactions catalyzed by several enzymes, including MraY (phospho-N-acetylmuramoyl-pentapeptide translocase) (3, 31), MurG (N-acetylglucosaminyl transferase) (22, 24, 41), FemX (the postulated enzyme involved in attachment of the first glycine to the pentaglycine interpeptide, and fmhB was recently shown as the strong candidate for its gene) (18, 33, 42), FemA, FemB, and transglycosylase (Fig. 3 and 4). The Rf values of the signals indicated the positions of lipid intermediates and nascent peptidoglycan according to a previous report (30), whereas the signal corresponding to lipid intermediates was obviously not a single band. Although no information is available, the signal might represent a mixture of glycine additives with different numbers of glycine or might represent lipid I and lipid II. Tunicamycin, an MraY inhibitor, inhibited the formation of both lipid intermediates and nascent peptidoglycan simultaneously (data not shown), and vancomycin, a transglycosylase inhibitor, inhibited the formation of only nascent peptidoglycan (as shown later). Lipid intermediates and nascent peptidoglycan were hardly detected in the experiment without UDP-GlcNAc, which proved that we could detect these products through successive reactions from MraY to transglycosylation. Katanosin B and plusbacin A3 inhibited nascent peptidoglycan formation, with IC50s of 0.8 and 0.4 μg/ml, respectively (Fig. 2B), which were close to the MICs, as well as the case of [14C]GlcNAc incorporation into peptidoglycan of whole cell. Vancomycin inhibited nascent peptidoglycan formation with an IC50 of 4.1 μg/ml, which was several times higher than those of katanosin B and plusbacin A3. Katanosin B and plusbacin A3 also inhibited lipid intermediates formation, with IC50s of 2.2 and 2.3 μg/ml, respectively (Fig. 2C), which were a few times higher than those for nascent peptidoglycan formation. On the other hand, vancomycin did not inhibit the formation of lipid intermediates even at the highest concentration tested (50 μg/ml), as shown in the literature (20). Excessive accumulation of lipid intermediates was observed at concentrations of vancomycin ranging from 6.25 to 25 μg/ml, which seemed to be caused by inhibition of the following reaction, transglycosylation, with little effect on lipid intermediate formation. These results indicated that katanosin B and plusbacin A3 inhibited the steps preceding transglycosylation in the peptidoglycan synthesis pathway. Both katanosin B and plusbacin A3 showed IC50 differences between lipid intermediates and nascent peptidoglycan formation, which indicated that both katanosin B and plusbacin A3 also inhibited the transglycosylation step.
FIG. 3.
Membrane pathways of peptidoglycan synthesis in S. aureus. [14C]glycine is incorporated into lipid intermediates (glycine-additive) and nascent peptidoglycan, surrounded by bold rectangles. TG, transglycosylase. Enzymes essential for bacterial growth are boxed.
FIG. 4.
Detection of inhibition of peptidoglycan synthesis. The reaction products were separated by TLC as described in Materials and Methods. A representative autoradiogram indicating the effect of vancomycin on the formation of lipid intermediates (Rf ≈ 0.9) and nascent peptidoglycan (at the origin) is shown.
Suppression of inhibition by acetyl-Lys-d-Ala-d-Ala.
Vancomycin inhibits transglycosylation via binding to the acyl-d-alanyl-d-Alanine (d-Ala-d-Ala) terminus of the lipid intermediates (1, 32). Acetyl-Lys-d-Ala-d-Ala, an analog of the terminus, suppressed the inhibition of transglycosylation by vancomycin effectively, but did not suppress those by katanosin B and plusbacin A3 even at the highest concentration tested (800 μg/ml). MICs of katanosin B and plusbacin A3 against S. aureus SRM133 were not affected by addition of 50 μg of acetyl-Lys-d-Ala-d-Ala per ml, while the MIC of vancomycin increased drastically, from 1.56 to 50 μg/ml.
Antagonism of antibacterial activity by a wall-membrane particulate.
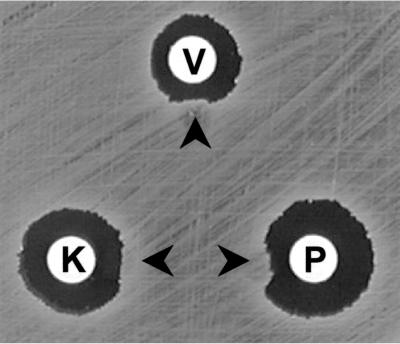
Inhibition zones surrounding disks containing katanosin B, plusbacin A3, and vancomycin were distorted by the presence of a wall-membrane particulate (Fig. 5). On the other hand, inhibition zones made by methicillin, fosfomycin, and erythromycin were not affected (data not shown).
FIG. 5.
Antagonism of antibacterial activity by a wall-membrane particulate. Wall membrane particulates were spotted at the positions shown by arrow heads. V, vancomycin; K, katanosin B; P, plusbacin A3.
DISCUSSION
This study showed that both katanosin B and plusbacin A3 can inhibit peptidoglycan synthesis. Their close MIC and IC50 values for [14C]GlcNAc incorporation and nascent peptidoglycan formation suggested that inhibition of peptidoglycan synthesis leads to the antimicrobial activity. Katanosin B and plusbacin A3 inhibited nascent peptidoglycan formation as well as vancomycin, whereas they also inhibited lipid intermediate formation, unlike vancomycin, although their IC50s for lipid intermediate formation were a few times higher than those for nascent peptidoglycan formation. Acetyl-Lys-d-Ala-d-Ala neither suppressed the inhibition of nascent peptidoglycan formation by katanosin B and plusbacin A3 nor reduced susceptibility to the two drugs, unlike its suppressive effects on vancomycin. These results indicated that katanosin B and plusbacin A3 are inhibitors of peptidoglycan synthesis with a mode distinct from that of vancomycin. The different mode of action would mean that mechanisms of resistance to vancomycin would be ineffective against katanosin B and plusbacin A3. In fact, the two drugs were active against vancomycin-resistant enterococci as well as against intermediate vancomycin-resistant S. aureus, whose resistance has been indicated to be due to increased vancomycin-binding ability (9, 10). The peptidoglycan synthesis pathway is an attractive target for antibacterial agents in terms of specificity for bacteria. Thus, katanosin B and plusbacin A3 are potential candidates for the development of new therapeutic drugs.
The mode of action of katanosin B and plusbacin A3 is not yet precisely understood. However, their inhibition seems to be the result of binding to lipid intermediates, substrates of several successive enzymes, rather than a direct effect on some enzyme, because both drugs inhibited at least two steps, the formation of lipid intermediates and transglycosylation. This speculation was supported by the antagonism of antibacterial activity by a wall-membrane particulate. The previous report showed that lysobactin, an analog of katanosin B, became bound to a cell wall preparation from S. aureus (2), and ramoplanin, an inhibitor of MurG, which is also a cyclic depsipeptide containing a lactone linkage, became bound to lipid intermediates (5, 40). The binding site of katanosin B and plusbacin A3 should be other than acyl-d-Ala-d-Ala, thus differing from vancomycin.
The MIC of vancomycin was lower than expected from the IC50 for nascent peptidoglycan formation, which might be due to the additional effect of inhibition of transpeptidation, another target of vancomycin following transglycosylation (32).
Direct assay for in vitro transglycosylase activity entailed laborious work with S. aureus enzyme because of the difficulty of purifying both the substrate and enzyme. While high-molecular-weight penicillin-binding proteins (PBPs) of Escherichia coli proved to have transglycosylase activity (15, 16, 27), none of the PBPs of S. aureus had such activity, although gene analysis revealed that staphylococcal PBP2 included a transglycosylase domain (25, 30). The staphylococcal major transglycosylase gene remains to be identified. The preceding reactions were also too cumbersome to detect because of the laborious preparation of the substrates and enzymes, although recently, MraY and MurG activities are being assayed with E. coli enzymes prepared by gene cloning (4, 7, 8, 14, 22). The staphylococcal murG gene has yet to be identified, while the staphylococcal mraY gene has already been identified and cloned, showing it to have the same function as that of E. coli (3). The fmhB gene was recently identified as being essential and specific for staphylococcal cells (33, 42). We could detect both staphylococcal transglycosylation and its foregoing reactions without particular purification of enzymes and substrates by taking advantage of staphylococcus-specific incorporation of glycine into peptidoglycan. Thus, this crude assay system may be useful for simple evaluation of inhibitors of staphylococcal peptidoglycan synthesis, especially MraY, MurG, FemX, and transglycosylase reactions, which are regarded as important targets for drug discovery.
ACKNOWLEDGMENTS
We are grateful to K. Hiramatsu, Juntendo University, for the generous gift of S. aureus Mu50 and to T. Kamigauchi, Shionogi & Co., Ltd., for the supply of katanosin B and plusbacin A3.
REFERENCES
- 1.Barna J C J, Williams D H. The structure and mode of action of glycopeptide antibiotics of the vancomycin group. Annu Rev Microbiol. 1984;38:339–357. doi: 10.1146/annurev.mi.38.100184.002011. [DOI] [PubMed] [Google Scholar]
- 2.Bonner D P, O'Sullivan J, Tanaka S K, Clark J M, Whitney R R. Lysobactin, a novel antibacterial agent produced by Lysobacter sp. II. Biological properties. J Antibiot. 1988;41:1745–1751. doi: 10.7164/antibiotics.41.1745. [DOI] [PubMed] [Google Scholar]
- 3.Bouhss A, Mengin-Lecreulx D, Beller D L, van Hijenoort J. Topological analysis of the MraY protein catalysing the first membrane step of peptidoglycan synthesis. Mol Microbiol. 1999;34:576–585. doi: 10.1046/j.1365-2958.1999.01623.x. [DOI] [PubMed] [Google Scholar]
- 4.Brandish P E, Burnham M K, Lonsdale J T, Southgate R, Inukai M, Bugg T D H. Slow binding inhibition of phospho-N-acetylmuramyl-pentapeptide-translocase (Escherichia coli) by mureidomycin A. J Biol Chem. 1996;271:7609–7614. doi: 10.1074/jbc.271.13.7609. [DOI] [PubMed] [Google Scholar]
- 5.Brötz H, Josten M, Wiedemann I, Schneider U, Götz F, Bierbaum G, Sahl H-G. Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol Microbiol. 1998;30:317–327. doi: 10.1046/j.1365-2958.1998.01065.x. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. Update: Staphylococcus aureus with reduced susceptibility to vancomycin—United States, 1997. Morb Mortal Wkly Rep. 1997;46:813–815. [PubMed] [Google Scholar]
- 7.Crouvoisier M, Mengin-Lecreulx D, van Heijenoort J. UDP-N-acetylglucosamine: N-acetylmuramoyl-(pentapetide) pyrophosphoryl undecaprenol N-acetylglucosamine transferase from Escherichia coli: overproduction, solubilization, and purification. FEBS Lett. 1999;449:289–292. doi: 10.1016/s0014-5793(99)00412-3. [DOI] [PubMed] [Google Scholar]
- 8.Ha S, Chang E, Lo M-C, Men H, Park P, Ge M, Walker S. The kinetic characterization of Escherichia coli MurG using synthetic substrate analogues. J Am Chem Soc. 1999;121:8415–8426. [Google Scholar]
- 9.Hanaki H, Kuwahara-Arai K, Boyle-Vavra S, Daum R S, Labischinski H, Hiramatsu K. Activated cell-wall synthesis is associated with vancomycin resistance in methicillin-resistant Staphylococcus aureus clinical strains Mu3 and Mu50. J Antimicrob Chemother. 1998;42:199–209. doi: 10.1093/jac/42.2.199. [DOI] [PubMed] [Google Scholar]
- 10.Hanaki H, Labischinski H, Inaba Y, Kondo N, Murakami H, Hiramatsu K. Increase in glutamine-non-amidated muropeptides in the peptidoglycan of vancomycin-resistant Staphylococcus aureus strain Mu50. J Antimicrob Chemother. 1998;42:315–320. doi: 10.1093/jac/42.3.315. [DOI] [PubMed] [Google Scholar]
- 11.Hiramatsu K, Aritaka N, Hanaki H, Kawasaki S, Hosoda Y, Hori S, Fukuchi Y, Kobayashi I. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet. 1997;350:1670–1673. doi: 10.1016/S0140-6736(97)07324-8. [DOI] [PubMed] [Google Scholar]
- 12.Hiramatsu K, Hanaki H, Ino T, Yabuta K, Oguri T, Tenover F C. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J Antimicrob Chemother. 1997;40:135–146. doi: 10.1093/jac/40.1.135. [DOI] [PubMed] [Google Scholar]
- 13.Hunt G A, Moses A J. Acute infection of mice with Smith strain of Staphylococcus aureus. Science. 1958;128:1574–1575. doi: 10.1126/science.128.3338.1574. [DOI] [PubMed] [Google Scholar]
- 14.Ikeda M, Wachi M, Jung H K, Ishino F, Matsuhashi M. The Escherichia coli mraY gene encoding UDP-N-acetylmuramoyl-pentapeptide: undecaprenyl-phosphate phospho-N-acetylmuramoyl-pentapeptide transferase. J Bacteriol. 1991;173:1021–1026. doi: 10.1128/jb.173.3.1021-1026.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishino F, Matsuhashi M. Peptidoglycan synthetic enzyme activities of highly purified penicillin-binding protein 3 in Escherichia coli: a septum-forming reaction sequence. Biochem Biophys Res Commun. 1981;101:905–911. doi: 10.1016/0006-291x(81)91835-0. [DOI] [PubMed] [Google Scholar]
- 16.Ishino F, Mitsui K, Tamaki S, Matsuhashi M. Dual enzyme activities of cell wall peptidoglycan synthesis, peptidoglycan transglycosylase and penicillin-sensitive transpeptidase, in purified preparations of Escherichia coli penicillin-binding protein 1A. Biochem Biophys Res Commun. 1980;97:287–293. doi: 10.1016/s0006-291x(80)80166-5. [DOI] [PubMed] [Google Scholar]
- 17.Kato T, Hinoo H, Terui Y, Kikuchi J, Shoji J. The structures of katanosins A and B. J Antibiot. 1988;41:719–725. doi: 10.7164/antibiotics.41.719. [DOI] [PubMed] [Google Scholar]
- 18.Kopp U, Roos M, Wecke J, Labischinski H. Staphylococcal peptidoglycan interpeptide bridge biosynthesis: a novel antistaphylococcal target? Microb Drug Resist. 1996;2:29–41. doi: 10.1089/mdr.1996.2.29. [DOI] [PubMed] [Google Scholar]
- 19.Lugtenberg E J J, de Haan P G. A simple method for following the fate of alanine-containing components in murein synthesis in Escherichia coli. Antonie van Leeuwenhoek J Microbiol Serol. 1971;37:537–552. doi: 10.1007/BF02218524. [DOI] [PubMed] [Google Scholar]
- 20.Matsuhashi M, Dietrich C P, Strominger J L. Incorporation of glycine into the cell wall glycopeptide in Staphylococcus aureus: role of sRNA and lipid intermediates. Proc Natl Acad Sci USA. 1965;54:587–594. doi: 10.1073/pnas.54.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDonald L C, Kuehnert M J, Tenover F C, Jarvis W R. Vancomycin-resistant enterococci outside the health-care setting: prevalence, sources, and public health implications. Emerg Infect Dis. 1997;3:311–317. doi: 10.3201/eid0303.970307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Men H, Park P, Ge M, Walker S. Substrate synthesis and activity assay for MurG. J Am Chem Soc. 1998;120:2484–2485. [Google Scholar]
- 23.Mengin-Lecreulx D, Allen N E, Hobbs J N, van Haijenoort J. Inhibition of peptidoglycan biosynthesis in Bacillus megaterium by daptomycin. FEMS Micobiol Lett. 1990;69:245–248. doi: 10.1016/0378-1097(90)90074-z. [DOI] [PubMed] [Google Scholar]
- 24.Mengin-Lecreulx D, Texier L, Rousseau M, van Heijenoort J. The murG gene of Escherichia coli codes for the UDP-N-acetylglucosamine:N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase involved in the membrane steps of peptidoglycan synthesis. J Bacteriol. 1991;173:4625–4636. doi: 10.1128/jb.173.15.4625-4636.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murakami K, Fujimura T, Doi M. Nucleotide sequence of the structural gene for the penicillin-binding protein 2 of Staphylococcus aureus and the presence of a homologous gene in other staphylococci. FEMS Microbiol Lett. 1994;117:131–136. doi: 10.1111/j.1574-6968.1994.tb06754.x. [DOI] [PubMed] [Google Scholar]
- 26.Murray B E. Vancomycin-resistant enterococcal infections. N Engl J Med. 2000;342:710–721. doi: 10.1056/NEJM200003093421007. [DOI] [PubMed] [Google Scholar]
- 27.Nakagawa J, Matsuhashi M. Molecular divergence of a major peptidoglycan synthetase with transglycosylase-transpeptidase activities in Escherichia coli—penicillin-binding protein 1Bs. Biochem Biophys Res Commun. 1982;105:1546–1553. doi: 10.1016/0006-291x(82)90964-0. [DOI] [PubMed] [Google Scholar]
- 28.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically—fourth edition; approved standard M7–A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 29.Novick R. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology. 1967;33:155–166. doi: 10.1016/0042-6822(67)90105-5. [DOI] [PubMed] [Google Scholar]
- 30.Park W, Matsuhashi M. Staphylococcus aureus and Micrococcus luteus peptidoglycan transglycosylases that are not penicillin-binding proteins. J Bacteriol. 1984;157:538–544. doi: 10.1128/jb.157.2.538-544.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pless D D, Neuhaus F C. Initial membrane reaction in peptidoglycan synthesis. J Biol Chem. 1973;248:1568–1576. [PubMed] [Google Scholar]
- 32.Reynolds P E. Structure, biochemistry and mechanism of action of glycopeptide antibiotics. Eur J Clin Microbiol Infect Dis. 1989;8:943–950. doi: 10.1007/BF01967563. [DOI] [PubMed] [Google Scholar]
- 33.Rohrer S, Ehlert K, Tschierske M, Labischinski H, Berger-Bächi B. The essential Staphylococcus aureus gene fmhB is involved in the first step of peptidoglycan pentaglycine interpeptide formation. Proc Natl Acad Sci USA. 1999;96:9351–9356. doi: 10.1073/pnas.96.16.9351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rotun S S, McMath V, Schoonmaker D J, Maupin P S, Tenover F C, Hill B C, Ackman D M. Staphylococcus aureus with reduced susceptibility to vancomycin isolated from a patient with fatal bacteria. Emerg Infect Dis. 1999;5:147–149. doi: 10.3201/eid0501.990118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shoji J, Hinoo H, Matsumoto K, Hattori T, Yoshida T, Matsuura S, Kondo E. Isolation and characterization of katanosins A and B. J Antibiot. 1988;41:713–718. doi: 10.7164/antibiotics.41.713. [DOI] [PubMed] [Google Scholar]
- 36.Shoji J, Hinoo H, Katayama T, Matsumoto K, Tanimoto T, Hattori T, Higashiyama I, Miwa H, Motokawa K, Yoshida T. Isolation and characterization of new peptide antibiotics, plusbacins A1 ∼ A4 and B1∼B4. J Antibiot. 1992;45:817–823. doi: 10.7164/antibiotics.45.817. [DOI] [PubMed] [Google Scholar]
- 37.Shoji, J., H. Hinoo, T. Katayama, Y. Nakagawa, Y. Ikenishi, K. Iwatani, and T. Yoshida. 1992. Structures of new peptide antibiotics, plusbacins A1∼A4 and B1∼B4. 45:824–831. [DOI] [PubMed]
- 38.Sieradzki K, Roberts R B, Haber S W, Tomasz A. The development of vancomycin resistance in a patient with methicillin-resistant Staphylococcus aureus infection. N Engl J Med. 1999;340:517–523. doi: 10.1056/NEJM199902183400704. [DOI] [PubMed] [Google Scholar]
- 39.Smith T L, Pearson M L, Wilcox K R, Cruz C, Lancaster M V, Robinson-Dunn B, Tenover F C, Zervos M J, Band J D, White E, Jarvis W R. Emergence of vancomycin resistance in Staphylococcus aureus. N Engl J Med. 1999;340:493–501. doi: 10.1056/NEJM199902183400701. [DOI] [PubMed] [Google Scholar]
- 40.Somner E A, Reynolds P E. Inhibition of peptidoglycan biosynthesis by ramoplanin. Antimicrob Agents Chemother. 1990;34:413–419. doi: 10.1128/aac.34.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strominger J L, Matsuhashi M, Anderson J S, Dietrich C P, Meadow P M, Katz W, Siewert G, Gilbert J M. Glycopeptide synthesis in Staphylococcus aureus and Micrococcus lysodeikticus. Methods Enzymol. 1966;8:473–486. [Google Scholar]
- 42.Tschierske M, Mori C, Rohrer S, Ehlert K, Shaw K J, Berger-Bächi B. Identification of three additional femAB-like open reading frames in Staphylococcus aureus. FEMS Microbiol Lett. 1999;171:97–102. doi: 10.1111/j.1574-6968.1999.tb13417.x. [DOI] [PubMed] [Google Scholar]