Abstract
Antimicrobial resistance (AMR) represents a critical obstacle to public health worldwide, due to the high incidence of strains resistant to available antibiotic therapies. In recent years, there has been a significant increase in the prevalence of resistant epidemic strains, associated with this, public health authorities have been alarmed about a possible scenario of uncontrolled dissemination of these microorganisms and the difficulty in interrupting their transmission, as nosocomial pathogens with resistance profiles previously considered sporadic. They become frequent bacteria in the community. In addition, therapy for infections caused by these pathogens is based on broad-spectrum antibiotic therapy, which favors an increase in the tolerance of remaining bacterial cells and is commonly associated with a poor prognosis. In this review, we present the current status of epidemic strains of methicillin-resistant Staphylococcus aureus (MRSA), Vancomycin-resistant Enterococcus (VRE), MDR Mycobacterium tuberculosis, extended-spectrum β-lactamase-producing Enterobacterales (ESBL), Klebsiella pneumoniae carbapenemase (KPC), and—New Delhi Metallo-beta-lactamase-producing Pseudomonas aeruginosa (NDM).
Introduction
About 90 years ago, one of the greatest medical advances in therapeutics was described, revolutionizing antimicrobial therapy. The discovery of penicillin by Alexander Fleming in 1928 promoted innovation in the management of infected patients. However, upon exposure to penicillin, the antimicrobial resistance (AMR) of bacteria was accelerated [1].
Between the 1980s and 1990s, the first outbreaks were caused by resistant bacterial clone strains that spread through hospitals in Europe, Latin America, and the United States reported [2]. The increase in the international flow of people and the food trade has also directly contributed to the spread and global expansion of clones of various pathogens in a hospital environment and different communities [3].
Since then, outbreaks of bacterial infections are evolving into complex phenomena, involving multiple species, and facilitated by natural selection or the ability to acquire resistance through horizontal gene transfer by mobile genetic elements. These genetic alterations are responsible for the higher incidence of resistance, through the establishment of regulatory mechanisms that ensure bacterial survival against different concentrations of the antimicrobial agent and other bacteria, favoring the multiplication, colonization, and advancement of the infectious process [3].
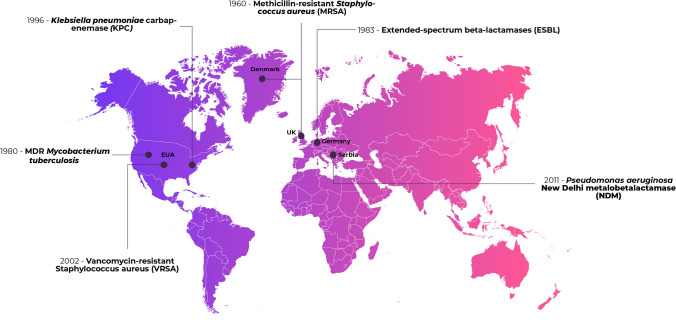
Over the decades, clinically important bacteria began to show resistance to more than one drug, even showing resistance to several classes of antimicrobials. The main microorganisms causing infections in the hospital environment are Escherichia coli, Klebsiella pneumoniae, Enterococcus spp., Staphylococcus aureus, Staphylococcus epidermidis, Mycobacterium tuberculosis, Pseudomonas aeruginosa, Acinetobacter baumannii, Salmonella spp., and Burkholderia spp. [4]. Figure 1 shows the place and year of the appearance of the first strains of methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant Staphylococcus aureus (VRSA), Vancomycin-resistant Enterococcus (VRE), Mycobacterium tuberculosis MDR, extended-spectrum β-lactamase-producing Enterobacterales (ESBL), Klebsiella pneumoniae carbapenemase (KPC), and New Delhi Metallo-beta-lactamase-producing Pseudomonas aeruginosa (NDM).
Fig. 1.
First report of resistant bacterial strains
The description of the multidrug-resistant (MDR) profile between Gram-negative and Gram-positive bacteria worldwide resulted in a scenario of uncertainty in the medical-scientific community [5]. Due to the variety of pathogens present in this group, a subclassification has been proposed in recent years for epidemiological purposes, including pathogens recognized as extensively drug-resistant (XDR), susceptible to only two or fewer categories between tested antimicrobials, and resistant to all tested antimicrobials (PDR), which pose a global threat to public health [6]. Thus, a bacterium present in the PDR subgroup is also classified as XDR and the XDR subgroup is also included in the MDR group (Fig. 2).
Fig. 2.
Schematic representation of the subclassification of strength profiles
Resistance to almost all classes of antibiotics has been reported, including aminoglycosides, cephalosporins, fluoroquinolones, β-lactams, and more recently colistin, an antimicrobial considered one of the last therapeutic options in the treatment of infections caused by MDR or XDR Gram-negative bacilli [7]. This scenario, which was previously limited to some regions of the country, started to gain a global proportion, crossing borders, affecting a greater number of people, leaving the world population in a state of alert [8].
About 90% of infections caused by MDR pathogens are based on empirical antibiotic treatment, associated with a prolonged hospital stay, generating a higher cost compared to infections caused by their antimicrobial-susceptible counterparts. Annually, the cost of treatment using MDR bacteria treatment (direct or indirectly) is estimated at US$ 45 billion and in Brazil could reach US$ 36 million annually, corresponding to 20% of federal expenditure on health [9].
The “post-antibiotic era” once such a distant idea may soon become a 21st-century reality. According to the WHO and the United Nations, AMR is configured as the critical issue of global public health. In which multisectoral and coordinated efforts are necessary, as no action today can result in no therapeutic option tomorrow [10]. Associated with this, the current situation that humanity has been going through, due to the pandemic caused by COVID-19, raises concerns about other possible epidemics/pandemics. It is estimated that the annual mortality rate related to resistance will reach 10 million by 2050. And the indiscriminate and excessive prescription of antibiotics in patients with COVID-19, in the face of possible co-infection or secondary bacterial infection, is expected to negatively impact this rate, accelerating this trend [11].
Thus, this review aims to describe some of the main epidemic pathogens, their resistance mechanisms, their therapeutic options and their current epidemic panorama, as a tool to assess the immediate challenges, and other possible epidemics/pandemics that may occur.
Antibiotic-Resistant Strains
Methicillin- and Vancomycin-Resistant Staphylococcus aureus (MRSA and VRSA)
Since its first report and after the introduction of β-lactams into clinical practice, Staphylococcus aureus remains one of the main global causes of nosocomial infections, which demonstrates its versatility in different epidemiological contexts [12].
In 1960 MRSA strains were detected in the United Kingdom and since then several strains have been spreading around the world. MRSA is associated with more complicated clinical outcomes when compared to that observed in methicillin-sensitive S. aureus (MSSA), as it can cause infective endocarditis and osteomyelitis, which can lead to sepsis and septic shock [13].
Although it has been considered a nosocomial pathogen (HA-MRSA) for years in the United States in the 1990s, MRSA infections were reported in the community (CA-MRSA), and in the early 2000s, MRSA strains were also identified associated with exposure to livestock (LA-MRSA) [14].
Vancomycin is one of the antimicrobials used for the treatment of MRSA; however, its administration has been limited due to the presence of vancomycin-intermediate Staphylococcus aureus (VISA) and vancomycin-resistant Staphylococcus aureus (VRSA) (Fig. 3). The first report of VISA in the literature is dated 1996, from a patient hospitalized in Japan. Later, VRSA was described in 2002 in the United States, in 2013 in Europe, and since then it has been expanding exponentially, making antibacterial therapy more difficult [15, 16].
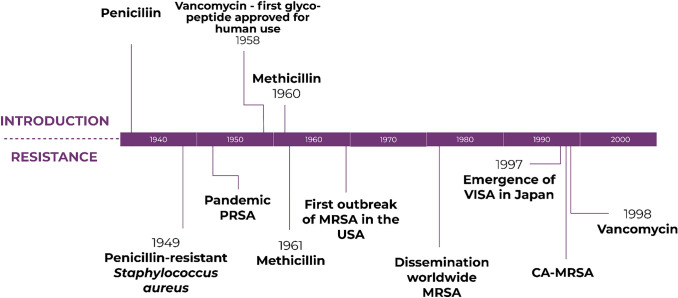
Fig. 3.
Timeline related to the introduction and emergence of antimicrobial resistance in Gram-positive bacteria
Infections caused by VRSA have high morbidity and mortality and are usually associated with co-infection between vancomycin-resistant Enterococci (VRE) and MRSA. It generally affects intensive care unit patients who are hospitalized for a long period and who have associated comorbidities, such as diabetes and gangrenous wounds [17].
Currently, MRSA and VRSA are the main Gram-positive pathogens involved in severe nosocomial infections with high rates of morbidity and mortality, resulting in a substantial economic burden, estimated at US$ 450 million in the last decade [15].
According to data from the Centers for Disease Control and Prevention (CDC), MRSA was responsible for 119.000 infections, and 20.000 deaths in 2019 in the US [14]. The epidemiological factors that contributed to the spread of some clones are still not well known. However, its high pathogenicity can be attributed to its genetic repertoire and diversity of virulence factors, which enabled its adaptation to different hostile environments [15].
Resistance Mechanism
Methicillin resistance occurs through the acquisition of the staphylococcal cassette mec (SCCmec) which encodes penicillin-binding protein 2a (PBP2a), a transpeptidase with low affinity for β-lactams responsible for inactivating the pharmacological activity of penicillin [17]. This is due to the acquisition of the mecA gene, responsible for encoding PBP2a. The regulation of methicillin resistance and the production of PBP2a are carried out by mecR1 and the repressor gene mecL [18].
A variant of mecA, called mecC, which encodes membrane proteins known as PBP2aLGA, was identified from milk isolates from herds in England in 2007 and carcasses in 2011 and 2012 in Belgium and France [19]. However, it was not only identified in animal products but also humans, as it showed compatibility with an isolate of 1975 from Denmark, thus suggesting that although it was later identified, it has probably been causing infections for more than 40 years [20]. In 2018, in Germany, a new variant of the mec gene, mecB, was reported, but the mechanism of resistance encoded has not yet been elucidated [18].
Resistance Rate
Regarding the prevalence rate, countries such as Switzerland, Canada, and the USA have an average of 18.6% of colonization by MRSA, Brazil has 35%, the UK 36%, and Africa and Portugal the rates of infections caused by MRSA are 49% [21, 22]. A similar panorama is seen in the Asian continent, where infection rates caused by MRSA are above 50% in countries such as Japan, China, Taiwan, and Singapore [23].
Based on the SCC elements, MRSA can be subdivided into 11 different types, but the types I-V are the main strains commonly isolated from patients. Infections by MRSA type I, II, and III are usually caused by HA-MRSA, whereas in CA-MRSA infections, types IV and V are involved [24, 25]. In China, the most identified type is SCCmec IV, found in 14.1% of MRSA isolates [26], whereas in Japan 68.4% is type IV, and 73.3% is type II [25].
In India, the most prevalent MRSA is SCCmec type III, corresponding to 58% of the isolates [20]. In Brazil, 65.4% of MRSA are type II, 37% type I, and 15.4% type IV [27, 28], and in the United States 29.9% are type II and 30.9% are type IV [29].
MRSA and VRSA Therapeutic Options
Therapeutic choices for MRSA infections take into account several factors, such as the resistance profile, risk factors, associated comorbidities, and response to previously used antibiotics. Thus, for most cases, vancomycin and daptomycin are the first choices; however, considering the VISA and VRSA strains, vancomycin has been little recommended [30].
Daptomycin, for example, is contraindicated for secondary infections of pneumonia, since pulmonary surfactants inactivate it, in addition, vancomycin has difficulty in penetrating the lung tissue, so clindamycin and linezolid are recommended for this clinical condition if the strain is susceptible [31]. Ceftaroline, in turn, is recommended for the treatment of uncomplicated infections located in sites such as the skin and skin structures, so for complicated cases, the recommendations are to use quinupristin/dalfopristin. As for cases of patients with valve prostheses, although there are no completed studies that condition their use, the use of the combination of gentamicin or rifampicin with daptomycin has been recommended [32].
Since identifications of VISA and VRSA strains are increasing, and taking into account that these causes significant public health impacts, the guidelines established by the Infectious Diseases Society of America (IDSA) indicate that other choices should be considered, such as combining daptomycin with another antibiotic, such as gentamicin, rifampicin, linezolid, or trimethoprim-sulfamethoxazole for VRSA [33]. For intermediate vancomycin resistance, combination or single use of quinupristin-dalfopristin, trimethoprim-sulfamethoxazole, linezolid, or telavancin is recommended [34].
Vancomycin-Resistant Enterococcus (VRE)
Enterococcus spp. are Gram-positive opportunistic microorganisms present in the human gastrointestinal tract, responsible for causing urinary tract infection, bacteremia, infective endocarditis, wound infections, neonatal sepsis, and meningitis [35]. Among Enterococcus spp., E. faecium and E. faecalis are the third leading cause of healthcare-associated infections, after S. aureus and P. aeruginosa, with considerable potential for healthcare-acquired outbreaks [35]. Its clinical relevance is due to its low intrinsic susceptibility to a wide range of antimicrobials such as aminoglycosides, lincosamides, streptogramins, sulfonamides, cephalosporins, and mainly due to the resistance to vancomycin, which causes great difficulties in clinical anti-infective therapy [36].
Vancomycin-resistant enterococcus (VRE) was first detected in the late 1980s in the UK and shortly after in France and USA [37]. Vancomycin-resistant occurs mainly in E. faecium (VREfm) isolates and they are associated with high mortality rates, especially in hospitalized individuals and longer hospital stays [37]. Since then, its reports have increased alarmingly, posing a threat to global public health, such that in some regions of the USA and Australia, about 50% or more of all blood culture isolates are VREfm [38]. In view of this, the World Health Organization (WHO) in 2017 classified VRE as a high-priority pathogen in its global list of antibiotic-resistant bacteria for which the discovery of new and effective therapeutic options is urgent.
Resistance Mechanism
Vancomycin-resistance in Enterococcus spp. involves a genetic change at the locus that harbors different vancomycin resistance genes (Van) [39]. These genes encode enzymes necessary for peptidoglycan synthesis, where instead of having D-Alanine-D-Alanine (D-Ala-D-Ala) in their terminal portion, they will have D-Alanine-D-Lactate (D- Ala-D-Lac) (A, B, D) and D-Ala-D-Serine (C, E, G) [40]. This alteration generates a reduction in the binding affinity of vancomycin to its target site (peptidoglycan), preventing its action [38].
There are six resistance phenotypes in Enterococcus (VanA, VanB, VanC, VanD, VanE, VanG), with vanA being the most prevalent type worldwide [41]. Most VRE outbreaks in human populations are attributed to the VanA and VanB resistance phenotypes, also already identified in animals and environmental samples [40]. Both are cited mainly in E. faecalis and E. faecium isolates, but the ability to transfer vanA genes to S. aureus and other Gram-positive organisms has already been proven in the laboratory [42] and VanB has already been identified in Streptococcus bovis [43].
The VanA phenotype, mediated by the vanA gene, confers a high degree of resistance to vancomycin and teicoplanin [41]. Whereas VanB (vanB gene) confers resistance to vancomycin but susceptibility to teicoplanin [43]. The VanC phenotype (vanC gene) has been described in E. casseliflavus and E. gallinarum and demonstrates a low or moderate level of vancomycin resistance and teicoplanin susceptibility [44]. Less common phenotypes include VanD, with moderate resistance to vancomycin and teicoplanin, and VanE and VanG, which have a low level of resistance to vancomycin alone [39, 43].
Resistance Rate
VRE was described more than 30 years ago and has since attracted special attention due to its increasing global prevalence. In the US and Europe, Enterococci spp. represent the main pathogens responsible for HAIs [38]. Despite its global distribution, its isolation rate is highest in North America. In the US, a CDC estimates about 100,000 cases of infection and VRE, leading to an average of 650 deaths per year, particularly in critically ill patients [44].
From 2009 to 2010, 35.5% of clinical strains were VREs according to the National Healthcare Safety Network (NHSN) [45]. Between 2011 and 2014, the same body reported that about 82% of E. faecium isolates from bloodstream infections in the US were vancomycin-resistant [46]. These results confirmed the concern raised by the CDC about threats to public health, which led to the inclusion of this microorganism among the pathogens known as ESKAPE (E. faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.), for which new effective therapies are urgent [47].
While colonization of hospitalized patients with VRE in the US was detected shortly after the first report of VRE, in Europe colonization rates only began to increase from the year 2000 onwards, and to this day VRE is much less prevalent than in the American continent [41, 45]. In Europe, VREs are very heterogeneously distributed, and only 4% VRE prevalence was reported by the European Antimicrobial Resistance Surveillance System [45]. Scandinavian countries such as the Netherlands, Luxembourg, and Belgium have less than 5% of their clinical isolates of Enterococcus spp. are VREs, while in Ireland, Latvia, Lithuania, and Romania, more than 30% of E. faecium are vancomycin-resistant [47]. Increasing numbers of VRE infections have already been reported in Switzerland, Australia, and Canada. And also in Asia, they have been reported in several countries such as Korea, Taiwan, India, and Nepal [43].
VRE Therapeutic Options
Due to its extensive intrinsic resistance to a variety of classes of antibiotics, Enterococcus ssp. has a certain advantage over other bacteria of the intestinal microbiota, which favors their dissemination in the most varied environments [47]. Treatment options for VREfm infections, especially vanA, are limited and antibiotics such as linezolid, daptomycin, tigecycline [38]. However, with the emergence and spread of VREs, reports of resistance have increased for the last-line agents used, such as linezolid and daptomycin. For infections caused by VREfm with a VanB and VanC phenotype, despite the emergence of resistance having been reported, teicoplanin remains a therapeutic option [38]. Tezidolide is another FDA-approved option for skin infections caused by E. faecali alone. A viable option for the treatment of VRE urinary tract infections is doxycycline and chloramphenicol, both of which act by inhibiting protein synthesis [45]. In the case of invasive E. faecalis infections, ampicillin and ceftriaxone combination therapy has been used successfully, as well as ampicillin plus gentamicin in the treatment of endocarditis. Another combination has been Quinupristine/Dalfopristine versus VREfm [47].
MDR Mycobacterium Tuberculosis
Mycobacterium tuberculosis is one of the microorganisms responsible for tuberculosis (TB) being an infectious disease that affects humans since ancient times. With the discovery of streptomycin in 1943, there was a revolution in treatment, leading to a significant reduction in the incidence of TB worldwide. With the expansion of the use of rifampicin, resistant M. tuberculosis strains emerged in 1980, making TB once again considered a public health concern [48, 49]. By showing simultaneous resistance to rifampicin and isoniazid, M. tuberculosis strains were defined as MDR, causing difficulties in TB control, especially in developing countries [48].
According to the WHO [50], in 2018, 10 million people were infected with TB in the world, being 5.7 million men, 3.2 million women, and 1.1 million children. Regarding the mortality rate, a total of 1.5 million people died from tuberculosis in 2018, is considered one of the 10 leading causes of death worldwide, as well as the leading cause of a single infectious agent, with a high rate of mortality above HIV/AIDS [50, 51].
Resistance Mechanism
The resistance mechanism of M. tuberculosis appears to be related to chromosomal mutation. Resistance-causing factors that affect the mutation rate are cellular mechanisms such as incompatible repair inefficiency, microsatellites, inappropriate translations, and error-prone DNA polymerases in addition to external factors that include lack of rapid diagnosis, inadequate prescription of anti-TB drugs, expression of environmental, genetic, and immunological determinants of the host, and exposure to smoke and/or pollution. Low patient adherence to TB treatment, as well as the high costs of achieving a complete cure for the disease, are the main factors contributing to drug resistance [51].
In about 96% of rifampicin-resistant M. tuberculosis strains were identified mutations in the 81 bp "hot spot" region, covering the 507–533 codons of the rpoB gene. While isoniazid resistance is related to mutations in several genes, such as katG, inhA, ahpC, kasA, and NDH. Mutations in the embB, rpsA, pncA, rpsL, and rrs genes have been identified as responsible for the resistance of M. tuberculosis to ethambutol, pyrazinamide, and streptomycin [51, 52].
Resistance Rate
Regarding the incidence rates of rifampicin-resistant TB cases in 2018, 484.000 cases were estimated, including about 378.000 cases of MDR TB and 214.000 deaths [53]. The highest levels of TB MDR are in the countries of Eastern Europe and Asia, with more than half of the global TB MDR load located in India (27%), China (14%), and Russia (9%). Regarding the drugs that this bacterium has already shown resistance to, in addition to rifampicin, they include ofloxacin, levofloxacin, and moxifloxacin in 20.8% of cases [54, 55].
About 3.4% of cases of resistance occur in patients who have never been treated for TB, while 18% of cases are from patients who have been previously treated, with the highest proportion of cases occurring in countries of the former Soviet Union [55]. In 2018, register a disease rate of 45 cases/100.000 inhabitants, with a TB-related mortality rate of 2.3 deaths/100.000 inhabitants [56].
MDR TB Treatment
The current WHO-recommended TB treatment is based on a 2-month program of isoniazid, rifampicin, pyrazinamide, and ethambutol, followed by 4 months of isoniazid and rifampicin [57]. Despite having a great bactericidal activity, some strains have developed resistance to some antibiotics of this therapeutic program. MDR TB strains are resistant to both rifampicin and isoniazid. Therefore, new classes of drugs have been evaluated for inclusion in the therapeutic regimen of MDR TB, as in the case of amoxicillin/clavulanate, carbapenem with clavulanic acid, clofazimine, fluoroquinolones, macrolides, among others [57].
According to Nahid et al. [58], some drugs that are strongly recommended include bedaquiline, moxifloxacin, and levofloxacin. Linezolid, clofazimine, cycloserine, amikacin, streptomycin, ethambutol, pyrazinamide, carbapenems with clavulanic acid, and delamanid may also be indicated. For short-term treatment (9 to 12 months) for MDR TB, the WHO recommends the use of kanamycin, as well as other drugs with documented resistance such as isoniazid, ethionamide, and pyrazinamide.
The Emergence of β-Lactamases
β-Lactamases represent a group of enzymes present in some bacterial species, responsible for hydrolyzing and inactivating the β-lactam ring of antibiotics. The detection of β-lactamases in bacteria dates to the beginning of the '40 s, among them the extended-spectrum β-lactamases (ESBL) and carbapenemases stand out. Infections caused by bacteria that contain these enzymes are considered an emerging public health problem [59].
Despite being present in both Gram-positive and Gram-negative bacteria, β-lactamases correspond to one of the main mechanisms of resistance to β-lactams in Gram-negative bacteria (Fig. 4) [59].
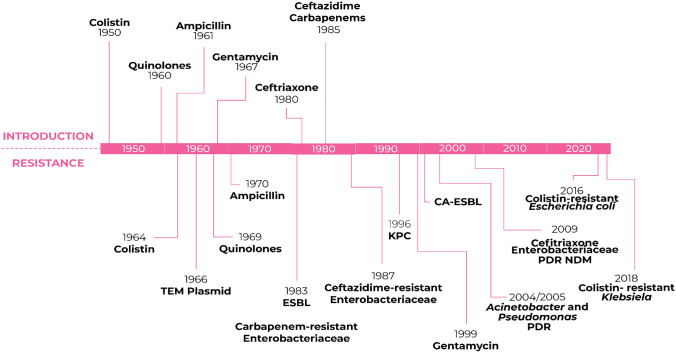
Fig. 4.
Timeline related to the introduction and emergence of antimicrobial resistance in Gram-negative bacteria
Extended-Spectrum β-Lactamase-Producing Enterobacterales (ESBL)
The first report of ESBL-producing Enterobacterales appeared in 1940, described by the production of cephalosporinase AmpC by Escherichia coli, and later in Europe, in 1983, in Klebsiella pneumoniae isolates, after the introduction of tigecycline in clinic experiments [1, 60]. Since then, ESBL-producing strains are currently reported on almost every continent [46]. According to the CDC, ESBLs are responsible for causing about 26,000 cases of HAI, and about 1700 deaths per year [60].
The nosocomial outbreak of Klebsiella pneumoniae isolates producing extended-spectrum of β-lactamases took place between 1980 and 1990, from genetic mutations in classical β-lactamases (TEM-1, TEM-2, and SHV-1), which later developed spread around the world. Nosocomial infections attributed to these microorganisms are associated with a high rate of morbidity and mortality. In 2017, in the United States alone, infections caused by ESBL strains were responsible for 9100 deaths [61].
ESBL-producing strains are described as a group of bacteria resistant to most antibiotics commonly used as a last line of treatment. In such cases, the remaining treatment option is carbapenems. Due to their broad spectrum of bactericidal action and their stability against most β-lactamases, carbapenems have been one of the drugs of choice in the therapy of MDR pathogens, until the appearance and spread of carbapenemase-producing strains [62].
Resistance Mechanism
ESBL consists of three groups of enzymes: TEM (Temorina Escherichia coli mutant), SHV (Sulfhydryl variant), and CTX-M (Cefotaximase-Munich), which are propagated by clonal expansion and dissemination of resistance genes (blaCTX-M, blaSHV, blaTEM, and its variations) via plasmids, soon spreading rapidly and causing outbreaks [59, 63].
TEM-type ESBLs are derived from TEM-1 and TEM-2 (non-ESBL), and found mostly in E. coli and K. pneumoniae, and can also be found in other bacteria of the Enterobacterales family and other Gram-negative bacteria. They have more than 130 types and are capable of hydrolyzing ampicillin, 1st, 2nd, and 3rd generation cephalosporins, as well as monobactams. Like the TEM-type ESBL, the SHV-type ESBL also have this hydrolyzing activity and are mainly present in Enterobacterales. SHV-type ESBLs have more than 50 types and are derived from SHV-1 (non-ESBL) [59, 64].
ESBLs of the CTX-M type are capable to hydrolyze cefotaxime, in addition to cephalosporins, penicillins, and monobactams. They have more than 40 types and are widely distributed worldwide, being found mainly in Enterobacterales. In addition, they are divided into 5 subgroups, according to similarities in the amino acid sequence: CTX-M-1, CTX-M-2, CTX-M-8, CTX-M-9, and CTX-M-25 [64].
Resistance Rate
In North America, ESBL-producing Enterobacterales have been more prevalent in the US, in which a study conducted between 2013 and 2017 with 1.112.312 enterobacterial isolates from 411 US hospitals found that about 12.05% were producers ESBL [65], with the CTX-M class being the most widespread in that country [97]. In Brazil, a study conducted with 435 isolates of enterobacteria from the southeast region of the country, showed that 48 isolates (11%) were ESBL producers, with the blaCTX-M-1 and blaCTX-M-8/25 genes being the most found in samples, as well as combinations of CTX-M, TEM, SHV and OXA [28].
In Europe, studies carried out between 2011 and 2013, showed that the rates of ESBL-producing strains were high, especially among the Klebsiella pneumoniae group, in which the rate was 45.6% (476). The most prevalent ESBL classes in European countries are CTX-M, followed by SHV [66]. In the countries of the Asia–Pacific region, a study carried out between 2008 and 2014 identified among 2.893 isolates of enterobacteria, 2.728 (94.3%) were ESBL producers, with the CTX-M-15 gene being the most found in total [67]. In Asia, CTX-M-14 was most prevalent in Hong Kong as well as Korea and Taiwan, CTX-M-27 in Japan and Vietnam, and CTX-M-15 in Kazakhstan, Malaysia, the Philippines, Singapore, and Thailand. In Oceania, CTX-M-15 was more prevalent in New Zealand and AmpC ACT and CTX-M-15 in Australia [67].
On the African continent, studies between 2009 and 2014 in Algeria, a rate of 16.4% [68] and 99% [69] of ESBL-producing strains were reported among the isolates identified clinicians.
ESBL-Producing Enterobacterales Therapeutic Approach
The rate of ESBL-producing Enterobacterales continues to rise, therefore limiting therapeutic options. Carbapenems are shown to be the most reliable antimicrobials to treat infections caused by ESBL-producing Enterobacterales, despite the increase of drug resistance by their overuse [70].
Hence, the Infectious Diseases Society of America (IDSA) recommends a guideline for treatment that includes mostly quinolones and carbapenems. When it is shown susceptibility, the prefered treatment for cystitis is with nitrofurantoin or trimethoprim-sulfamethoxazole, or with ciprofloxacin, levofloxacin, ertapenem, meropenem, imipenem-cilastatin, as an alternative treatment [71].
For pyelonephritis or complicated urinary tract infections, the preferred treatment is with ertapenem, meropenem, imipenem-cilastatin, ciprofloxacin, levofloxacin, or trimethoprim-sulfamethoxazole, while for other infections outside of the urinary tract, the preferred treatment is with meropenem, imipenem-cilastatin, or ertapenem [71].
Carbapenemases
Although Enterobacterales have developed several resistance mechanisms, the detection of carbapenemases is still a concern. The increasing number of reports of Carbapenem-resistant Enterobacterales (ERC) and the high mortality rate associated with these infections in the last decade poses a serious health threat [51].
Despite two decades of experience with KPC-producing bacteria, efforts to contain its global spread have failed. This critical data is worrying, given the scarcity of effective treatment options, due to the high rate of transmission of genetic material, and consequently of the resistance gene, that this microorganism presents [52, 53].
Carbapenemases are increasingly reported in Enterobacterales, mainly in the genera Klebsiella, Enterobacter, Escherichia, Serratia, Citrobacter, Salmonella, Proteus, and Morganela. The emergence and rapid spread of ERCs is one of the greatest challenges to global health [52].
Klebsiella Pneumoniae Carbapenemase (KPC)
Klebsiella pneumoniae carbapenemase (KPC) is the most isolated enzyme in the world and can hydrolyze a wide variety of β-lactams, carbapenems, cephalosporins, monobactams, conferring resistance to them [73].
Yigit et al. [72] described the presence of a carbapenemase enzyme in a clinical isolate of Klebsiella pneumoniae collected in 1996 in a hospital in the United States, which showed high levels of resistance to imipenem and meropenem. After analyzing the isolate, through phenotypic methods and molecular biology, the researchers confirmed the existence of a new β-lactamase enzyme. Serine-carbapenemase, described by Yigit et al. [72], showed no inhibition when exposed to EDTA and was weakly inhibited by clavulanic acid, thus it was classified in Ambler's group A and was named KPC-1. Subsequently, researchers described, in 2003, the enzyme KPC-2, which had a single amino acid variant about KPC-1, resulting in a point mutation [72, 73].
Currently, 24 variants of the KPC enzyme are described in Gram-negative bacteria, with endemic levels reported in several countries, including the United States, Greece, Italy, Israel, Colombia, and Brazil [74, 75]. The treatment of infections caused by microorganisms that have genes encoding carbapenemase enzymes in the plasmid is extremely difficult, due to the ability of these microorganisms to show resistance to multiple drugs [75].
Resistance Mechanism
The genes encoding class A serine-carbapenemase enzymes can be located on the chromosome, such as the genes of the SME, IMI, and NMC-A enzymes, or in mobile genetic elements (plasmids, transposons, and integrons), such as the genes of the KPC and GES. Until the early 1990s, carbapenemases were believed to be encoded only by chromosomal genes in some bacterial species, such as L1 in Stenotrophomonas maltophilia and BcII in Bacillus cereus. Subsequently, researchers identified IMP-1, oxacillinase-23 (OXA-23), and Klebsiella pneumoniae carbapenemase-1 enzymes encoded by mobile genetic elements in Pseudomonas aeruginosa, Acinetobacter baumannii, and Klebsiella pneumoniae strains, respectively. This finding aroused greater concern in health organizations due to the potential for dispersal among 23 species and clonal spread in the community and nosocomial infections [73, 76, 77].
Resistance Rate
The rapid spread of K. pneumoniae carrying blaKPC gene variants is mainly associated with the spread of a single clonal group (CG) designated CG258. This group is made up of 43 different “sequence types” (STs), in which the two predominant STs are ST258 and ST512. KPC-Kpn ST258 is associated with 90% of all infections caused by carbapenemase enzyme-producing K. pneumoniae in Israel and is related to 80% of endemic outbreaks caused by KPC-Kpn in the United States. Colombia was the first country in South America to report the presence of the KPC enzyme. In that country, the blaKPC gene was initially identified in K. pneumoniae isolates in 2005 [78]. Since then, the presence of this gene has been reported in other species of Gram-negative bacteria, such as P. aeruginosa. In Colombia, P. aeruginosa carrying the blaKPC gene is considered one of the main endemic bacterial strains.
Klebsiella Pneumoniae Carbapenemase (KPC) Therapeutic Options
KPC isolates are commonly resistant to fluoroquinolones, aminoglycosides, and most β-lactams; however, temocillin has some activity against some isolates, being seen as an option for lower UTIs by blaKPC-containing K. pneumoniae [79]. Thus, antibiotics such as colistin, polymyxin B, fosfomycin, tigecycline, and rifampicin have been seen as agents of last choice [123]. Other antimicrobials such as fosfomycin and nitrofurantoin, if active, may be used to a limited extent for lower UTIs [123].
Another recommendation to minimize bacterial resistance has been combined therapy. The ceftazidime/avibactam (CZA) combination was approved in 2015 for use by the FDA, consists of a cephalosporin and a new class A beta-lactamase inhibitor (KPC, CTX-M, SHV) and Ambler's C and some class D β-lactamases, which has been successful for the treatment of urinary tract infections, bacteremia and complicated intra-abdominal infections [80, 81]. Wang et al. [82] showed that combined administration of aztreonam and avibactam was effective in treating bacterial infections production of metallo-β-lactamases (MBL). However, given the emergence of ceftazidime/avibactam-resistant strains, the new meropenem/vaborbactam combination approved in 2017, could become an alternative for these conditions, including pyelonephritis by carbapenem-resistant Enterobacterales (CRE). Vaborbactam, in addition to being a class A (KPC, CTX-M, SHV) and class C β-lactamases inhibitor, also potentiates the action of meropenem against most Enterobacterales species [81]. Sulfamethoxazole/trimethoprim and colistin, even in the face of resistance reports, can still be considered as potential therapeutic options [83].
NDM Pseudomonas aeruginosa
Pseudomonas aeruginosa is an important and opportunistic pathogen responsible for several infections in the hospital environment, such as skin and soft tissue infections, urinary tract and lower respiratory tract infections [84]. Due to its intrinsic resistance and the ability to develop other resistance mechanisms through the acquisition of mobile genetic elements and mutations, an alarming increase in the number of reports of P. aeruginosa MDR strains has been observed [85].
Carbapenems are commonly used as antimicrobials of the last choice in infections caused by MDR P. aeruginosa; however, carbapenem-resistant strains have been reported around the world over the last decades with the production of MBL, enzymes that degrade carbapenems leading to resistance [86].
New Delhi MBL was reported in 2009 in New Delhi, India, in carbapenem-resistant Klebsiella pneumoniae strain from a Swiss patient who, while traveling to the Indian city, acquired a urinary tract infection. Since then, its variants have been reported worldwide [85, 87–89].
Resistance Mechanism
β-Lactamases are structurally classified according to the Ambler classification into class A (GES, KPC), class C (CMY, PDC), class D (OXA), all having the amino acid serine in their active site, and class B (VIM, IMP, SIM, GIM, SPM, and NDM), which comprises the β-lactamases that contain zinc in their active site, being called MBL. They are capable to hydrolyze all available β-lactams, except for aztreonam, a monobactam [87].
NDM consists of a subgroup of MBL currently formed by 31 variants, being the most prevalent blaNDM-1 among all species [55, 56]. The blaNDM-1 gene, which encodes NDM-1, is present in plasmids of different sizes that can undergo genetic rearrangements that provide rapid propagation and structural modifications that can increase its activity [54]. Unlike other MBL, this enzyme is a lipoprotein anchored in the inner layer of the outer membrane of Gram-negative bacteria. Some structural changes in NDM-1 do not occur in its active site and sometimes are not related to increased activity, but the stability of its structure in zinc-deprived environments, since one of the host's defense mechanisms is the production of metal-chelating proteins such as calprotectin present in neutrophils [90].
Resistance Rate
Since its discovery in 2009 in India, several countries in the Indian subcontinent (Bangladesh, Pakistan, and Sri Lanka), Europe, Asia, and the Americas have detected the presence of the blaNDM-1 gene diffused heterogeneously among members of the order Enterobacterales and the genus Pseudomonas. Studies suggest that air travel and migration allowed the transport of plasmids between countries and continents, being an explanation for the rapid spread of this gene [91]. One hundred and sixty-one strains of Pseudomonas aeruginosa have already been identified, carrying the variants blaNDM-1 (159/161) and blaNDM-4 (2/161). Among the strains carrying the blaNDM-1 gene, 45 were detected in Singapore, 19 strains in the United States, and 13 strains in India [91].
In Brazil, the NDM-1 variant was first detected in 2013 in an isolate of Providencia rettgeri from the culture of the amputated finger of the lower limb of a diabetic patient with peripheral vascular disease admitted to a public hospital in the Rio Grande do Sul, Brazil. However, the same author identified, in a retrospective study, that this variant was already present in the hospital since 2012 in isolates of Enterobacter hormaechei [92]. Since then, its presence in other Brazilian states has been reported among species of E. cloacae, K. pneumoniae, K. oxytoca, Acinetobacter baumannii complex, Proteus mirabilis, Citrobacter freundii, and Escherichia coli [93]. In 2018, during the investigation of the mechanism of resistance on 14 isolates of Pseudomonas aeruginosa resistant to β-lactams, aminoglycosides, and quinolones, the presence of the blaNDM-1 gene was detected in two strains from urine and tracheal aspirate samples from two patients hospitalized in a public hospital in Pernambuco, becoming the first cases of Pseudomonas aeruginosa carrying NDM-1 described in Brazil [94].
NDM Pseudomonas aeruginosa Therapeutic Options
The therapeutic options available for patients with infections caused by Pseudomonas aeruginosa carrying the blaNDM gene are scarce. Considered as antibiotics of last resort, Colistin and Polymyxin B, despite having in vitro antibacterial activity, have low clinical utility against these microorganisms, since toxicity and emergence of resistance during treatment are strictly related to the therapeutic dose range used [95].
Used as an alternative to monotherapy treatment, therapeutic combinations have been seen as a means of circumventing the enzymatic mechanisms of resistance to carbapenems; however, in NDM-producing isolates, the action of some of them may be insufficient [96]. Experimental studies show that the combination of Ceftazidime-Avibactam (CZA), a third-generation cephalosporin combined with a β-lactamases inhibitor, lacks antibacterial activity against P. aeruginosa NDM due to the inactivity of Avibactam against MBLs (NDM, VIM, IMP, EBV, and PER) resulting from the absence of a serine residue in its active site [97]. Mikhail et al. [98] observed a synergistic effect when using Aztreonam in combination with CZA against MDR isolates of P. aeruginosa; however, none of these isolates produced MBL.
Approved by the US Food and Drug Administration (FDA) in 2019 for the treatment of complicated urinary tract infections, nosocomial bacterial pneumonia, and ventilator-associated pneumonia, Cefiderocol, an injectable siderophore from the cephalosporin group, has been shown to be more stable to hydrolysis by β-lactamases, including ESBLs, KPCs, NDM, IMP, and VIM, when compared to Ceftazidime and Meropenem [99].
However, a UK study revealed an alarming rate of resistance between P. aeruginosa NDM and IMP-positive isolates with MICs greater than 128 mg/L [100]. Other study carried out in continental Europe did not report resistance to cyfederocol in P. aeruginosa NDM, VIM, IMP, and GES-positive isolates [99].
Conclusion
Bacterial resistance is a serious problem for the pharmaceutical industry, the scientific community, as well as the community at large, which will persist beyond the COVID-19 crisis. Epidemics caused by these strains represent one of the main problems that will be faced worldwide in the coming decades. In this review, we provide a perspective on the current scenario of important epidemic strains, describe their scenario of dissemination, discuss some resistance mechanisms, and finally, address some of the therapeutic options currently implemented.
It is difficult to predict the future situation of antimicrobial resistance, particularly in developing countries, but without raising awareness, and short-term and long-term global, regional and local measures in infection control, surveillance, and diagnosis, we will likely not be able to slow the spread of MDRs microorganisms. Implementation of interventions aimed at a more critical outpatient prescription of antibiotics should be encouraged, reducing risks to human, animal, and environmental health. Comparative genomic analysis of clinical pathogens before and after the current pandemic may elucidate different mechanisms related to the acquisition of circulating resistance genes.
The discovery of new antibiotics has become a necessity to mitigate antimicrobial resistance against existing antimicrobials. However, these microorganisms are highly adaptable and rapidly evolve in the face of different and new antimicrobial therapies. Thus, we cannot resort to the idea that new antibiotics are sufficient to control these pathogens. These data only reinforce the importance of continuous epidemiological surveillance aiming at caution in the clinical application of these antimicrobials in order to preserve their activity in the treatment of more serious infections. In addition, techniques for removing antibiotic residues and pathogens from the environment/hospital are also necessary.
Acknowledgements
J.V.d.O.S. thanks the Foundation for the Support of Science and Technology of the State of Pernambuco–FACEPE by the master’s scholarship (IBPG-1113-2.12/19).
Author Contributions
JVdeOS: participated in all stages from study design of the review to the final version of the article. SDdaCJ: participated in all stages from study design of the review to the final version of the article. SMdeFRdSM: participated in all stages from study design of the review to the final version of the article. IDLC: participated in the writing of the article and review of the final version. JBdeS: participated in the writing of the article and review of the final version. DLC: participated in the writing of the article and review of the final version. WRCdaS: participated in the writing of the article and review of the final version. MHMEA: participated in the writing of the article and review of the final version. IMFC: participated in all stages from study design of the review to the final version of the article.
Funding
The authors thank the National Council for Scientific and Technological Development–CNPq (426065/2018-2).
Data Availability
Not applicable.
Code Availability
Not applicable.
Declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Ethical Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lobanovska M, Pilla G. Focus: drug development: Penicillin’s discovery and antibiotic resistance: lessons for the future? Yale J Biol Med. 2017;90(1):135. [PMC free article] [PubMed] [Google Scholar]
- 2.Teixeira LA, et al. Geographic spread of epidemic multiresistant Staphylococcus aureus clone in Brazil. J Clin Microbiol. 1995;33(9):2400–2404. doi: 10.1128/jcm.33.9.2400-2404.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Partridge SR, et al. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev. 2018;31(4):e00088–e117. doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giedraitienė A, et al. Antibiotic resistance mechanisms of clinically important bacteria. Medicina. 2011;47(3):19. doi: 10.3390/medicina47030019. [DOI] [PubMed] [Google Scholar]
- 5.Fodor A, et al. Multidrug Resistance (MDR) and collateral sensitivity in bacteria, with special attention to genetic and evolutionary aspects and to the perspectives of antimicrobial peptides—a review. Pathogens. 2020;9(7):522. doi: 10.3390/pathogens9070522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Magiorakos AP, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 7.Perez F, et al. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2007;51(10):3471–3484. doi: 10.1128/AAC.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelly H. The classical definition of a pandemic is not elusive. Bull World Health Org. 2011;89(1):540–541. doi: 10.2471/BLT.11.088815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.World Health Organization (WHO). Global action plan on antimicrobial resistance, 2015. http://apps.who.int/gb/ebwha/pdf_files/WHA68/A68_ACONF1Rev1-en.pdf. Accessed 21 Apr 2020
- 10.Reardon S. Antibiotic resistance sweeping developing world. Nat News. 2021;509(7499):141. doi: 10.1038/509141a. [DOI] [PubMed] [Google Scholar]
- 11.Ghosh S, Bornman C, Zafer MM. Antimicrobial resistance threats in the emerging COVID-19 pandemic: where do we stand? J Infect Public Health. 2021;14:555–560. doi: 10.1016/j.jiph.2021.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nickol ME, et al. Characterization of host and bacterial contributions to lung barrier dysfunction following co-infection with 2009 pandemic influenza and methicillin resistant Staphylococcus aureus. Viruses. 2019;11(2):1–18. doi: 10.3390/v11020116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hassoun A, Linden PK, Friedman B. Incidence, prevalence, and management of MRSA bacteremia across patient populations—a review of recent developments in MRSA management and treatment. Crit Care. 2017;21(1):1–10. doi: 10.1186/s13054-017-1801-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kourtis AP, et al. Vital signs: epidemiology and recent trends in methicillin-resistant and in methicillin-susceptible Staphylococcus aureus bloodstream infections—United States. Morb Mortal Wkly Rep. 2019;68(9):214–219. doi: 10.15585/mmwr.mm6809e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGuinness WA, Malachowa N, Deleo FR. Focus: infectious diseases: vancomycin resistance in Staphylococcus aureus. Yale J Biol Med. 2017;90(2):269. [PMC free article] [PubMed] [Google Scholar]
- 16.Panthee S, et al. Genomic analysis of vancomycin-resistant Staphylococcus aureus VRS3b and its comparison with other VRSA isolates. Drug Discov Therapeut. 2017;11(1):1–6. doi: 10.5582/ddt.2017.01024. [DOI] [PubMed] [Google Scholar]
- 17.Cong Y, Yang S, Rao X. Vancomycin resistant Staphylococcus aureus infections: a review of case updating and clinical features. J Adv Res. 2020;21:169–176. doi: 10.1016/j.jare.2019.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becker K, et al. Plasmid-encoded transferable mecB-mediated methicillin resistance in Staphylococcus aureus. Emerg Infect Dis. 2018;24(2):242–252. doi: 10.3201/eid2402.171074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Duijkeren E, et al. Prevalence of methicillin-resistant Staphylococcus aureus carrying mecA or mecC in dairy cattle. Vet Microbiol. 2014;171(1):364–367. doi: 10.1016/j.vetmic.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 20.Lakhundi S, Zhang K. Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin Microbiol Rev. 2018;31(4):e00020–e118. doi: 10.1128/CMR.00020-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Junnila J, et al. Changing epidemiology of methicillin-resistant Staphylococcus aureus in a low endemicity area—new challenges for MRSA control. Eur J Clin Microbiol Infect Dis. 2020;2020(1):1–9. doi: 10.1007/s10096-020-03824-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Packer S, et al. Clonal expansion of community-associated meticillin-resistant Staphylococcus aureus (MRSA) in people who inject drugs (PWID): prevalence, risk factors and molecular epidemiology, Bristol, United Kingdom, 2012 to 2017. Eurosurveillance. 2019;24(13):1–10. doi: 10.2807/1560-7917.ES.2019.24.13.1800124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osaka S, et al. Genetic shifts in methicillin-resistant Staphylococcus aureus epidemic clones and toxin gene profiles in Japan: comparative analysis among pre-epidemic, epidemic and post-epidemic phases. J Med Microbiol. 2018;67(3):392–399. doi: 10.1099/jmm.0.000687. [DOI] [PubMed] [Google Scholar]
- 24.Dhawan B, et al. Dissemination of methicillin-resistant Staphylococcus aureus SCCmec type IV and SCCmec type V epidemic clones in a tertiary hospital: challenge to infection control. Epidemiol Infect. 2015;143(2):343–353. doi: 10.1017/S095026881400065X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harada D, et al. Change in genotype of methicillin-resistant Staphylococcus aureus (MRSA) affects the antibiogram of hospital-acquired MRSA. J Infect Chemother. 2018;24(7):563–569. doi: 10.1016/j.jiac.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 26.Zhong YM, et al. Emergence of methicillin-resistant Staphylococcus aureus SCCmec type IV/V epidemic clones in a large teaching hospital in China. J S Med Univ. 2017;37(7):861–865. doi: 10.3969/j.issn.1673-4254.2017.07.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carvalho SPD, et al. Molecular characteristics of methicillin-resistant Staphylococcus aureus isolates from hospital and community environments in northeastern Brazil. Braz J Infect Dis. 2019;23(2):134–138. doi: 10.1016/j.bjid.2019.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pereira JL, et al. Antimicrobial resistance and molecular characterization of extended-spectrum β-lactamases of Escherichia coli and Klebsiella spp. isolates from urinary tract infections in Southern Brazil. Microbial Drug Resist. 2019;25(2):173–181. doi: 10.1089/mdr.2018.0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tickler IA, et al. Continued expansion of USA300-like methicillin-resistant Staphylococcus aureus (MRSA) among hospitalized patients in the United States. Diagn Microbiol Infect Dis. 2017;88(4):342–347. doi: 10.1016/j.diagmicrobio.2017.04.016. [DOI] [PubMed] [Google Scholar]
- 30.Morrisette T, et al. The evolving reduction of vancomycin and daptomycin susceptibility in MRSA—salvaging the gold standards with combination therapy. Antibiotics. 2020;9(11):762–783. doi: 10.3390/antibiotics9110762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khan A, et al. Current and future treatment options for community-associated MRSA infection. Expert Opin Pharmacother. 2018;19(5):457–470. doi: 10.1080/14656566.2018.1442826. [DOI] [PubMed] [Google Scholar]
- 32.Ovchinnikov KV, et al. Successful development of bacteriocins into therapeutic formulation for treatment of MRSA skin infection in a murine model. Antimicrob Agents Chemother. 2020;64(12):829–920. doi: 10.1128/AAC.00829-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okada N, et al. A case report of postoperative VRSA enteritis: effective management of rifampicin for vancomycin resistant Staphylococcus aureus enteritis after esophagectomy and colon reconstruction. Int J Surg Case Rep. 2018;52:75–78. doi: 10.1016/j.ijscr.2018.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Godley MD, et al. A randomized trial of volunteer recovery support for adolescents (VRSA) following residential treatment discharge. J Subst Abuse Treatm. 2019;98:15–25. doi: 10.1016/j.jsat.2018.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Markwart R, et al. The rise in vancomycin-resistant Enterococcus faecium in Germany: data from the German Antimicrobial Resistance Surveillance (ARS) Antimicrob Resist Infect Control. 2019;8(1):1–11. doi: 10.1186/s13756-019-0594-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Driscoll T, Crank CW. Vancomycin-resistant enterococcal infections: epidemiology, clinical manifestations, and optimal management. Infect Drug Resist. 2015;8:217. doi: 10.2147/IDR.S54125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.López-Luis BA, et al. Risk factors and outcomes associated with vancomycin-resistant Enterococcus faecium and ampicillin-resistant Enterococcus faecalis bacteraemia: a 10-year study in a tertiary-care centre in Mexico City. J Glob Antimicrob Resist. 2021;24:198–204. doi: 10.1016/j.jgar.2020.12.005. [DOI] [PubMed] [Google Scholar]
- 38.Gorrie C, et al. Genomics of vancomycin-resistant Enterococcus faecium. Microb Genom. 2019;5:7. doi: 10.1099/mgen.0.000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faron ML, Ledeboer NA, Buchan BW. Resistance mechanisms, epidemiology, and approaches to screening for vancomycin-resistant Enterococcus in the health care setting. J Clin Microbiol. 2016;54(10):2436–2447. doi: 10.1128/JCM.00211-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahmed MO, Baptiste KE. Vancomycin-resistant enterococci: a review of antimicrobial resistance mechanisms and perspectives of human and animal health. Microb Drug Resist. 2018;24(5):590–606. doi: 10.1089/mdr.2017.0147. [DOI] [PubMed] [Google Scholar]
- 41.Kafil HS, Asgharzadeh M. Vancomycin-resistant enteroccus faecium and enterococcus faecalis isolated from education hospital of iran. Maedica. 2014;9(4):323. [PMC free article] [PubMed] [Google Scholar]
- 42.Poyart C, et al. Emergence of vancomycin resistance in the genus Streptococcus: characterization of a vanB transferable determinant in Streptococcus bovis. Antimicrob Agents Chemother. 1997;41(1):24–29. doi: 10.1128/AAC.41.1.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eliopoulos GM, Gold HS. Vancomycin-resistant enterococci: mechanisms and clinical observations. Clin Infect Dis. 2001;33(2):210–219. doi: 10.1086/321815. [DOI] [PubMed] [Google Scholar]
- 44.Cetinkaya Y, Falk P, Mayhall CG. Vancomycin-resistant enterococci. Clin Microbiol Rev. 2000;13(4):686–707. doi: 10.1128/CMR.13.4.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barber KE, et al. Therapeutic options for vancomycin-resistant enterococcal bacteremia. Expert Rev Anti Infect Ther. 2015;13(3):363–377. doi: 10.1586/14787210.2015.1001839. [DOI] [PubMed] [Google Scholar]
- 46.Weiner LM, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect Control Hosp Epidemiol. 2016;37(11):1288–1301. doi: 10.1017/ice.2016.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Riccardi N, et al. Therapeutic options for infections due to vanB genotype vancomycin-resistant Enterococci. Microb Drug Resist. 2021;27(4):536–545. doi: 10.1089/mdr.2020.0171. [DOI] [PubMed] [Google Scholar]
- 48.Marques M, et al. Antituberculosis-drug resistance in the border of Brazil with Paraguay and Bolivia/Resistencia as drogas antituberculose na fronteira do Brasil com Paraguai e Bolivia. Rev Panam Salud Publ. 2017;41(8):1. doi: 10.26633/RPSP.2017.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prasanna A, Niranjan V. Classification of Mycobacterium tuberculosis DR, MDR, XDR isolates and identification of signature mutationpattern of drug resistance. Bioinformation. 2019;15(4):261. doi: 10.6026/97320630015261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.WHO – World Health Organization. Tuberculose. 2020. https://www.who.int/news-room/fact-sheets/detail/tuberculosis. Accessed 13 May 2020.
- 51.Hameed HM, et al. Molecular targets related drug resistance mechanisms in MDR-, XDR-, and TDR-Mycobacterium tuberculosis Strains. Front Cell Infect Microbiol. 2018;8:114. doi: 10.3389/fcimb.2018.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Palomino JC, Martin A. Drug resistance mechanisms in Mycobacterium tuberculosis. Antibiotics. 2014;3(3):317–340. doi: 10.3390/antibiotics3030317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.WHO – World Health Organization. Global Tuberculosis Report 2019. World Health Organization: Geneva, Switzerland, 2019.
- 54.Kumar K, Abubakar I. Clinical implications of the global multidrug-resistant tuberculosis epidemic. Clin Med. 2015;15(6):s37–42. doi: 10.7861/clinmedicine.15-6-s37. [DOI] [PubMed] [Google Scholar]
- 55.Iacobino A, Fattorini L, Giannoni F. Drug-resistant tuberculosis 2020: where we stand. Appl Sci. 2020;10(6):2153. doi: 10.3390/app10062153. [DOI] [Google Scholar]
- 56.Silva DR, Mello FCQ, Migliori GB. Tuberculosis series 2020. J Bras Pneumol. 2020;46(2):e20200027. doi: 10.36416/1806-3756/e20200027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Silva DR, Mello FCQ, Migliori GB. Shortened tuberculosis treatment regimens: what is new? J Bras Pneumol. 2020;46(2):e20200009. doi: 10.36416/1806-3756/e20200009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nahid P, et al. Treatment of drug-resistant tuberculosis: An official ATS/CDC/ERS/IDSA clinical practice guideline. Am J Respir Crit Care Med. 2019;200(10):e93–e142. doi: 10.1164/rccm.201909-1874ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sah SK, Hemalatha S. Extended spectrum Beta lactamase (ESBL) Mechanism of antibiotic resistance and Epidemiology. Int J Pharm Res. 2015;7(2):303–309. [Google Scholar]
- 60.Tal Jasper R, et al. The complex epidemiology of extended-spectrum β-lactamase-producing Enterobacteriaceae. Future Microbiol. 2015;10(5):819–839. doi: 10.2217/fmb.15.16. [DOI] [PubMed] [Google Scholar]
- 61.CDC. Antibiotic Resistance Threats in the United States, 2019. Atlanta, GA: U.S. Department of Health and Human Services, CDC; 2019.
- 62.Yair Y, Gophna U. Escherichia coli, a Versatile Pathogen. Cham: Springer; 2018. Pandemic Bacteremic Escherichia coli Strains: evolution and emergence of drug-resistant pathogens; pp. 163–180. [DOI] [PubMed] [Google Scholar]
- 63.Brolund A. Overview of ESBL-producing Enterobacteriaceae from a Nordic perspective. Infect Ecol Epidemiol. 2014;4(1):24555. doi: 10.3402/iee.v4.24555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chong Y, Ito Y, Kamimura T. Genetic evolution and clinical impact in extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Infect Genet Evol. 2011;11(7):1499–1504. doi: 10.1016/j.meegid.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 65.Gupta V, et al. Trends in resistant Enterobacteriaceae and Acinetobacter species in hospitalized patients in the United States: 2013–2017. BMC Infect Dis. 2019;19(1):742. doi: 10.1186/s12879-019-4387-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lob SH, et al. Antimicrobial resistance and resistance mechanisms of Enterobacteriaceae in ICU and non-ICU wards in Europe and North America: SMART 2011–2013. J Glob Antimicrob Resist. 2015;3(3):190–197. doi: 10.1016/j.jgar.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 67.Jean SS, Hsueh PR. Distribution of ESBLs, AmpC β-lactamases and carbapenemases among Enterobacteriaceae isolates causing intra-abdominal and urinary tract infections in the Asia-Pacific region during 2008–14: results from the Study for Monitoring Antimicrobial Resistance Trends (SMART) J Antimicrob Chemother. 2016;72(1):166–171. doi: 10.1093/jac/dkw398. [DOI] [PubMed] [Google Scholar]
- 68.Iabadene H, et al. Prevalence of plasmid-mediated AmpC β-lactamases among Enterobacteriaceae in Algiers hospitals. Int J Antimicrob Agents. 2009;34(4):340–342. doi: 10.1016/j.ijantimicag.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 69.Naas T, et al. Outbreak of Salmonella enterica serotype Infantis producing ArmA 16S RNA methylase and CTX-M-15 extended-spectrum β-lactamase in a neonatology ward in Constantine, Algeria. Int J Antimicrob Agents. 2011;38(2):135–139. doi: 10.1016/j.ijantimicag.2011.04.012. [DOI] [PubMed] [Google Scholar]
- 70.Sheu C, et al. Management of infections caused by extended-spectrum β-lactamase-producing Enterobacteriaceae: current evidence and future prospects. Expert Rev Anti Infect Ther. 2018;16(3):205–218. doi: 10.1080/14787210.2018.1436966. [DOI] [PubMed] [Google Scholar]
- 71.Tamma PD et al. Infectious Diseases Society of America Guidance on the Treatment of AmpC β-lactamase-Producing Enterobacterales, Carbapenem-Resistant Acinetobacter baumannii, and Stenotrophomonas maltophilia Infections. Clinical Infectious Diseases 2021. [DOI] [PubMed]
- 72.Yigit H, et al. Novel carbapenem-hydrolyzing-lactamase, KPC-1, from a carbapenem resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45(4):1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Andrade LS, Darini ALC. Bacilos gram-negativos produtores de beta-lactamases: que bla bla bla é esse? J Infect Control. 2017;6(1):16–25. [Google Scholar]
- 74.Mathers AJ, et al. Risk factors for Klebsiella pneumoniae carbapenemase (KPC) gene acquisition and clinical outcomes across multiple bacterial species. J Hosp Infect. 2020;104(4):456–458. doi: 10.1016/j.jhin.2020.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Monteiro J, et al. First report of KPC-2-producing Klebsiella pneumoniae strains in Brazil. Antimicrob Agents Chemother. 2009;53(1):333–334. doi: 10.1128/AAC.00736-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Scaife W, et al. Transferable imipenem-resistance in Acinetobacter species from a clinical source. J Antimicrob Chemother. 1995;36(3):585–586. doi: 10.1093/jac/36.3.585. [DOI] [PubMed] [Google Scholar]
- 77.Watanabe M, et al. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1991;35(1):147–151. doi: 10.1128/AAC.35.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Villegas MV, et al. First detection of the plasmid-mediated class A carbapenemase KPC-2 in clinical isolates of Klebsiella pneumoniae from South America. Antimicrob Agents Chemother. 2006;50(8):2880–2882. doi: 10.1128/AAC.00186-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pitout JDD, Nordmann P, Poirel L. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother. 2015;59(10):5873–5884. doi: 10.1128/AAC.01019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang W, et al. In vitro and in vivo bactericidal activity of ceftazidime-avibactam against Carbapenemase-producing Klebsiella pneumoniae. Antimicrob Resist Infect Control. 2018;7(1):1–9. doi: 10.1186/s13756-018-0435-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Romanelli F, et al. Meropenem/vaborbactam activity in vitro: a new option for Klebsiella pneumoniae carbapenemase (KPC)-producing Klebsiella pneumoniae treatment. Future Microbiol. 2021;16(16):1261–1266. doi: 10.2217/fmb-2021-0007. [DOI] [PubMed] [Google Scholar]
- 82.Wang X, et al. In vitro activities of ceftazidime-avibactam and aztreonam-avibactam against 372 Gram-negative bacilli collected in 2011 and 2012 from 11 teaching hospitals in China. Antimicrob Agents Chemother. 2014;58(3):1774–1778. doi: 10.1128/AAC.02123-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bedenić B, et al. Klebsiella pneumoniae carbapenemase (KPC) in urinary infection isolates. Arch Microbiol. 2021;203(4):1825–1831. doi: 10.1007/s00203-020-02161-x. [DOI] [PubMed] [Google Scholar]
- 84.Shaaban M, et al. Molecular characterization of resistance mechanisms in Pseudomonas aeruginosa isolates resistant to carbapenems. J Infect Dev Count. 2018;11:935–943. doi: 10.3855/jidc.9501. [DOI] [PubMed] [Google Scholar]
- 85.Rahman M, et al. Prevalence and molecular characterization of new Delhi metallo-beta-lactamases in multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii from India. Microb Drug Resist. 2018;24(6):792–798. doi: 10.1089/mdr.2017.0078. [DOI] [PubMed] [Google Scholar]
- 86.Cornaglia G, Giamarellou H, Rossolini GM. Metallo-β-lactamases: a last frontier for β-lactams? Lancet Infect Dis. 2011;11(5):381–393. doi: 10.1016/S1473-3099(11)70056-1. [DOI] [PubMed] [Google Scholar]
- 87.Khan AU, Maryam L, Zarrilli R. Structure, genetics and worldwide spread of New Delhi metallo-β-lactamase (NDM): a threat to public health. BMC Microbiol. 2017;17(1):1–12. doi: 10.1186/s12866-017-1012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rasheed JK, et al. New Delhi metallo-β-lactamase–producing enterobacteriaceae, United States. Emerg Infect Dis. 2013;19(6):870. doi: 10.3201/eid1906.121515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Farhat N, Khan AU. Evolving trends of New Delhi Metallo-betalactamse (NDM) variants: a threat to antimicrobial resistance. Infect Genet Evol. 2020;2020:104588–104589. doi: 10.1016/j.meegid.2020.104588. [DOI] [PubMed] [Google Scholar]
- 90.Cheng Z, et al. Evolution of New Delhi metallo-β-lactamase (NDM) in the clinic: effects of NDM mutations on stability, zinc affinity, and mono-zinc activity. J Biol Chem. 2018;293(32):12606–12618. doi: 10.1074/jbc.RA118.003835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kumarasamy KK, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10(9):597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Carvalho-Assef APD, et al. Isolation of NDM-producing Providencia rettgeri in Brazil. J Antimicrob Chemother. 2013;68(12):2956–2957. doi: 10.1093/jac/dkt298. [DOI] [PubMed] [Google Scholar]
- 93.Silva IR, et al. Distribution of clinical NDM-1-producing Gram-negative bacteria in Brazil. Microb Drug Resist. 2019;25(3):394–399. doi: 10.1089/mdr.2018.0240. [DOI] [PubMed] [Google Scholar]
- 94.Scavuzzi AML, et al. Emergence of blaVIM-2, blaNDM-1, blaIMP-7 and blaGES-1 in blaKPC-2-harbouring Pseudomonas aeruginosa isolates in Brazil. J Glob Antimicrob Resist. 2019;19:181–182. doi: 10.1016/j.jgar.2019.09.009. [DOI] [PubMed] [Google Scholar]
- 95.Tsuji, et al. International Consensus Guidelines for the Optimal Use of the Polymyxins: Endorsed by the American College of Clinical Pharmacy (ACCP), European Society of Clinical Microbiology and Infectious Diseases (ESCMID), Infectious Diseases Society of America (IDSA), International Society for Anti-infective Pharmacology (ISAP), Society of Critical Care Medicine (SCCM), and Society of Infectious Diseases Pharmacists (SIDP) Pharmacotherapy. 2019;39(1):10–39. doi: 10.1002/phar.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Falcone M, Paterson D. Spotlight on ceftazidime/avibactam: a new option for MDR Gram-negative infections. J Antimicrob Chemother. 2016;71(10):2713–2722. doi: 10.1093/jac/dkw239. [DOI] [PubMed] [Google Scholar]
- 97.Khan, et al. Evaluation of susceptibility testing methods for aztreonam and ceftazidime-avibactam combination therapy on extensively drug-resistant gram-negative organisms. Antimicrob Agents Chemother. 2021;65(11):e00846–e921. doi: 10.1128/AAC.00846-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mikhail, et al. Evaluation of the synergy of ceftazidime-avibactam in combination with meropenem, amikacin, aztreonam, colistin, or fosfomycin against well-characterized multidrug-resistant Klebsiella pneumoniae and Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2019;63(8):e00779–e819. doi: 10.1128/AAC.00779-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kazmierczak, et al. In vitro activity of cefiderocol, a siderophore cephalosporin, against a recent collection of clinically relevant carbapenem-non-susceptible Gram-negative bacilli, including serine carbapenemase- and metallo-β-lactamase-producing isolates (SIDERO-WT-2014 study) Int J Antimicrob Agents. 2019;53(2):177–184. doi: 10.1016/j.ijantimicag.2018.10.007. [DOI] [PubMed] [Google Scholar]
- 100.Mushtaq, et al. In vitro activity of cefiderocol, a siderophore cephalosporin, against multidrug-resistant gram-negative bacteria. Antimicrob Agents Chemother. 2020;64(12):e01582–e1620. doi: 10.1128/AAC.01582-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.
Not applicable.