Summary
SUMOylation is involved in various aspects of plant biology, including drought stress. However, the relationship between SUMOylation and drought stress tolerance is complex; whether SUMOylation has a crosstalk with ubiquitination in response to drought stress remains largely unclear. In this study, we found that both increased and decreased SUMOylation led to increased survival of apple (Malus × domestica) under drought stress: both transgenic MdSUMO2A overexpressing (OE) plants and MdSUMO2 RNAi plants exhibited enhanced drought tolerance. We further confirmed that MdDREB2A is one of the MdSUMO2 targets. Both transgenic MdDREB2A OE and MdDREB2AK192R OE plants (which lacked the key site of SUMOylation by MdSUMO2A) were more drought tolerant than wild‐type plants. However, MdDREB2AK192R OE plants had a much higher survival rate than MdDREB2A OE plants. We further showed SUMOylated MdDREB2A was conjugated with ubiquitin by MdRNF4 under drought stress, thereby triggering its protein degradation. In addition, MdRNF4 RNAi plants were more tolerant to drought stress. These results revealed the molecular mechanisms that underlie the relationship of SUMOylation with drought tolerance and provided evidence for the tight control of MdDREB2A accumulation under drought stress mediated by SUMOylation and ubiquitination.
Keywords: Apple, Drought, SUMOylation, Ubiquitination, MdSUMO2, MdDREB2A, MdRNF4
Introduction
Drought stress is one of the main abiotic constraints that limit agricultural development and productivity (Geng et al., 2020; Li et al., 2019). As the global climate warms in the twenty‐first century, the frequency of severe drought conditions is increasing (Dai, 2013). Water shortage impairs plant growth and development, limiting plant production and reducing the performance of crop plants and fruit trees (Basu et al., 2016). In fruit trees, water deficit inhibits flower bud differentiation and tree vegetative growth, thereby causing flowers and fruits to drop (Niu et al., 2019; Virlet et al., 2015). To cope with drought stress, plants respond at both the morphological and molecular levels, exhibiting changes in photosynthesis, stomatal movement, hormone content, leaf development, stem extension, root proliferation, hydraulic conductivity and gene expression (Basu et al., 2016; Geng et al., 2020; Li et al., 2020; Liao et al., 2016; Seiler et al., 2011; Sun et al., 2018; Yordanov et al., 2000). Therefore, decoding the molecular mechanisms that underlie drought responses is critical to the development of new cultivars for future agriculture (Geng et al., 2018; Li et al., 2020; Liao et al., 2016; Sun et al., 2013b, 2018).
Small ubiquitin‐like modifier (SUMO) is a ~100‐amino‐acid polypeptide that is structurally related to ubiquitin (Vierstra, 2012). Similar to ubiquitin, SUMOs are encoded as precursor proteins. To attain their mature form, precursor SUMOs require SUMO protease to cleave a C‐terminal peptide and expose two consecutive glycine residues that are essential for conjugation to substrates. The biochemical pathway of SUMOylation is also analogous to that of ubiquitination. The first step is SUMO activation, an ATP‐dependent reaction that is catalysed by the heterodimeric E1‐activating enzyme, SAE1/SAE2. In the second step, activated SUMO is transferred from SAE to the SUMO‐conjugating enzyme (SCE). Finally, the conjugation of SUMO to its substrates is catalysed by SCE (Colby et al., 2006). The consensus sequence ψKXE/D (where ψ is a hydrophobic aliphatic residue; X can be any residue; K, E and D are standard one‐letter symbols for amino acids; and K is the attachment site for SUMO) is considered to be the canonical SUMO attachment site (Elrouby et al., 2014; Novatchkova et al., 2004), although other sites may exist. SUMO proteases de‐SUMOylate the SUMOylated substrates to recycle SUMO. In addition to its covalent attachment to target proteins, SUMO can also establish noncovalent interaction with cellular proteins (Galanty et al., 2012).
In addition to the regulation of development and cellular homeostasis under normal growth conditions, SUMOylation is also involved in various biotic and abiotic stress responses, including the response to drought stress (Augustine and Vierstra, 2018; Benlloch and Lois, 2018; Castro et al., 2012; Staudinger, 2017). OsbZIP23 is a SUMOylation substrate that is targeted by the SUMO protease OTS1. SUMOylation of OsbZIP23 causes the transcriptional activation of drought protection genes and improves drought tolerance (Srivastava et al., 2017). Overexpression of SUMO E2‐conjugating enzyme (SCE1) in rice (Oryza sativa) impairs drought tolerance by reducing the accumulated proline content relative to the wild type (Nurdiani et al., 2018). However, overexpression of SaSce9 from the halophyte grass Spartina alterniflora enhances salinity and drought stress tolerance in Arabidopsis (Karan and Subudhi, 2012), indicating that SCE plays various roles in different plants. The SUMO E3 ligase MMS21 negatively influences Arabidopsis drought response through an ABA‐dependent pathway (Zhang et al., 2013). Likewise, the rice SUMO protease OsOTS1 also plays a negative role in drought stress response through an ABA‐dependent pathway (Srivastava et al., 2017). Another SUMO E3 ligase, SIZ1, plays more complicated roles in drought stress response in different plant species. Rice OsSIZ1 confers drought tolerance in transgenic bentgrass and cotton (Mishra et al., 2017). Transgenic tobacco plants ectopically expressing tomato SlSIZ1 are more tolerant to drought stress (Zhang et al., 2017b), as are Arabidopsis plants overexpressing SIZ1. However, SIZ1 has opposite roles in same species such as Arabidopsis (Catala et al., 2007; Kim et al., 2017; Miura et al., 2013). Reports on Arabidopsis SIZ1 overexpressing (OE) plants and siz1 mutants indicate that either increased or decreased SUMOylation levels can improve drought resistance. However, the physiological and molecular basis for this effect is unclear. In addition, despite the identification of drought‐related SUMO targets, the biological function of their SUMOylation modification is largely unknown.
The SUMO‐interacting proteins (SIPs) play a crucial role in the regulation of SUMOylated proteins; they usually interact with SUMO through SUMO‐interacting motifs (SIMs). Proteins with SIMs include a group of RING‐type ubiquitin E3 ligases, DNA methyltransferases or demethylases, and histone methyltransferases or demethylases (Elrouby et al., 2014; Kumar et al., 2017). The best‐studied SIPs are the RING‐type ubiquitin E3 ligases that target SUMOylated proteins for degradation by the proteasome pathway. RING finger protein 4 (RNF4, also known as SUMO‐targeted ubiquitin E3 ligase or STUbL) ubiquitinates promyelocytic leukaemia protein (PML) or the nuclear receptor NR4A1 that has been SUMOylated by SUMO2/3 in mammals (Geoffroy and Hay, 2009; Lallemand‐Breitenbach et al., 2008; Zhang et al., 2017a). RNF4 also ubiquitinates SUMOylated proteins in the fission yeast Schizosaccharomyces pombe (Sun et al., 2007) and promotes the ubiquitination of activated MEK1 in a RING‐finger‐dependent manner in Dictyostelium (Sobko et al., 2002). SUMOylation of PML recruits RNF4, triggering its Lys 48‐linked polyubiquitination and degradation (Lallemand‐Breitenbach et al., 2008; Tatham et al., 2008). NR4A1 is SUMOylated by SUMO2/3 and targeted by RNF4 for polyubiquitination and subsequent degradation to control macrophage cell death (Zhang et al., 2017a). Although several studies have reported the important and conserved role of RNF4 in multicellular eukaryotes, only one study has investigated the role of AT‐STUbL4 in the floral transition in plants (Elrouby et al., 2014). At‐stubl4 mutant plants flowered later than the wild type, whereas AT‐STUbL4 OE plants flowered earlier (Elrouby et al., 2014). To date, it remains unclear whether RNF4 can recognize and ubiquitinate SUMOylated proteins in plants, especially during the response to drought stress.
Dehydration‐responsive element‐binding factor (DREB2A) is a transcription factor that binds specifically to the DRE/CRT cis‐element and is rapidly induced by dehydration (Li et al., 2019; Liu et al., 1998). DREB2A is a key factor in plant drought stress tolerance. Overexpression of full‐length DREB2A in apple, Pennisetum glaucum, Zea mays and O. sativa enhances tolerance to drought stress (Agarwal et al., 2007; Cui et al., 2011; Liao et al., 2016; Qin et al., 2007). In Arabidopsis, DREB2A is unstable under control conditions owing to its negative regulatory domain (NRD) (Mizoi et al., 2013; Morimoto et al., 2017; Qin et al., 2008; Sadhukhan et al., 2014; Sakuma et al., 2006a). The overexpression of DREB2A‐CA (a constitutively active form of DREB2A with the NRD domain deleted) increases drought tolerance in Arabidopsis (Sakuma et al., 2006a). Various post‐translational modifications of DREB2A, including SUMOylation and ubiquitination, are tightly associated with its stability and transcriptional activity in Arabidopsis (Qin et al., 2008; Wang et al., 2020). Two types of ubiquitin E3 ligase, BPMs and DRIPs, mediate the degradation of DREB2A in Arabidopsis (Morimoto et al., 2017; Qin et al., 2008). However, SUMOylation of DREB2A by SCE1 can repress the interaction between DREB2A and BPM2, thereby increasing DREB2A protein stability under high temperature (Wang et al., 2020). Whether SUMOylated DREB2A can be targeted by ubiquitin E3 ligases for degradation remains unclear.
Apple DREB2A does not contain the NRD domain that is targeted for degradation in Arabidopsis. Unlike Arabidopsis DREB2A, MdDREB2A is consistently stable under normal conditions (Li et al., 2019). Overexpression of MsDREB6.2 (a MdDREB2A homolog) or MpDREB2A enhanced drought tolerance of apple or Arabidopsis (Li et al., 2019; Liao et al., 2016). Given the lack of an NRD domain in MdDREB2A, its post‐translational modifications and the molecular mechanisms of its protein stability under stress require clarification. In this study, we found that both increased and decreased SUMOylation levels improved apple drought tolerance. We further identified MdDREB2A as one of the SUMOylation target proteins and demonstrated that SUMOylation of MdDREB2A was critical for protein stability and drought tolerance. In addition, we further provided evidence that SUMOylated MdDREB2A could be recognized and ubiquitinated by MdRNF4 under drought stress, leading to the degradation of MdDREB2A. Our results highlight the roles of SUMOylation in apple drought tolerance and provide insight into the RNF4‐mediated ubiquitination of SUMOylated MdDREB2A in response to drought.
Results
Expression patterns and localization of SUMO2s in apple
The apple genome contains six SUMO2 genes (Figure 1a). Due to genome duplication, each pair of genes on different chromosomes has almost identical coding sequences, and we therefore named the three pairs MdSUMO2A, MdSUMO2B and MdSUMO2C. Protein alignment revealed a protein sequence similarity of 76%–88% among these three MdSUMO2 proteins (Figure 1a), indicating the high conservation among MdSUMO2 isoforms. To characterize the function of apple SUMO2 proteins in response to drought stress, we first examined their expression patterns under drought. We found that the MdSUMO2s had similar expression patterns in response to drought (Figure 1b), suggesting that they may have similar functions under drought stress. Apple SUMO2A and SUMO2B were more abundant in all tissues, whereas SUMO2C was less abundant in all tissues examined (Figure S1A and Appendix S1). When MdSUMO2A::GUS was ectopically expressed in Arabidopsis, similar results were observed, and GUS signal was detected in all tissues (Figure S1B–I).
Figure 1.
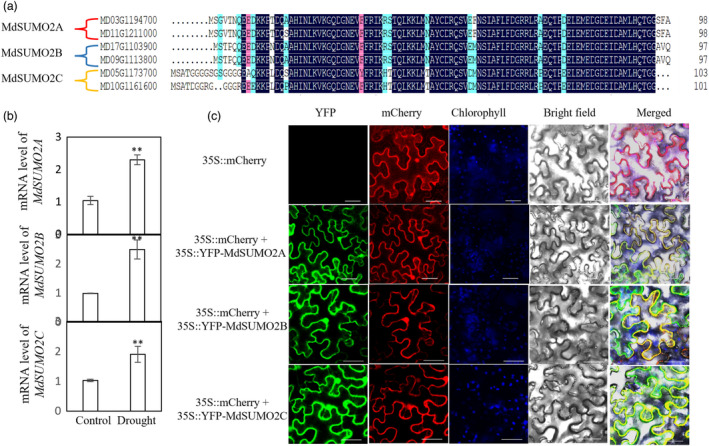
Sequences, responses to drought stress and localization of MdSUMO2s. (a) Comparison of amino acid sequences of MdSUMO2A, MdSUMO2B, and MdSUMO2C in apple. MD03G1194700 and MD11G1211000 were named MdSUMO2A; MD17G1103900 and MD09G1113800 were named MdSUMO2B; MD05G1173700 and MD10G1161600 were named MdSUMO2C. (b) MdSUMO2 expression in response to drought in 2‐month‐old GL‐3 plants, which were exposed to drought for 0 and 6 days. (c) Subcellular localization of MdSUMO2s. YFP‐MdSUMO2A, YFP‐MdSUMO2B or YFP‐MdSUMO2C was transformed into 5‐week‐old tobacco (Nicotiana benthamiana) leaves for 3 days, and YFP and mCherry fluorescent signals were then observed. Bars = 40 μm. Error bars indicate standard error (n = 3). Asterisks indicate significant differences based on one‐way ANOVA and Tukey test (**, P < 0.01).
We aligned SUMO2A proteins from different plant species and found that their sequences were highly conserved throughout the plant kingdom (Figure S2A). MdSUMO2A was highly similar to SUMO2A from Prunus mume (Figure S2B). We then cloned SUMO2A, SUMO2B and SUMO2C from the apple genome. Co‐localization with mCherry suggested that apple SUMO2A, SUMO2B and SUMO2C are localized in the nucleus, plasma membrane and cytoplasm (Figure 1c).
Knocking down MdSUMO2s or knocking in one MdSUMO2 gene leads to drought stress tolerance
To understand the biological function of the MdSUMO2s, we generated a series of transgenic plants: MdSUMO2A OE (over expression) with a higher MdSUMO2A expression level; MdSUMO2A RNAi with reduced expression of MdSUMO2A only; and MdSUMO2 RNAi with reduced expression of MdSUMO2A, MdSUMO2B and MdSUMO2C (Figure 2).
Figure 2.
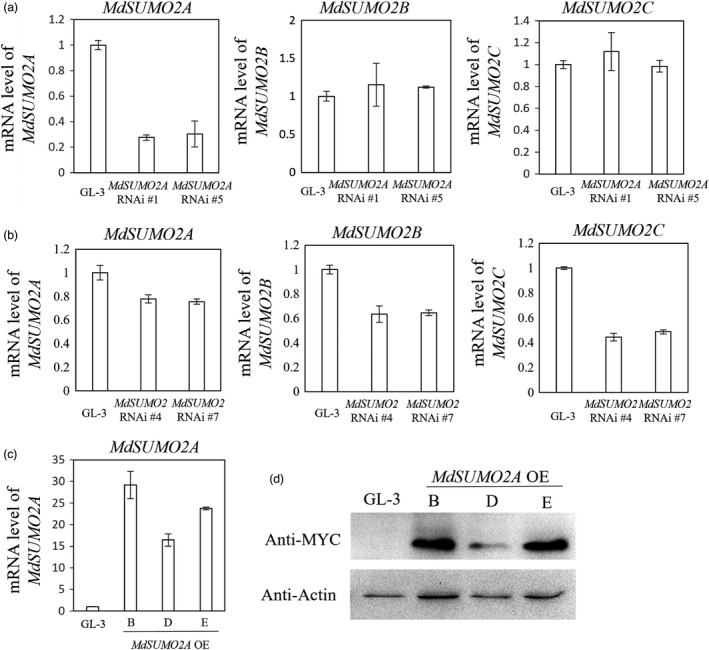
Expression of MdSUMO2s in MdSUMO2A RNAi plants (a), MdSUMO2 RNAi plants (b) and MdSUMO2A OE plants (c). (d) Protein level of MdSUMO2A in Gl‐3 and MdSUMO2A OE plants.
After transplant, the transgenic plants and nontransgenic plants (GL‐3) were exposed to prolonged drought stress by maintaining soil volumetric water content of 18%–23% for 3 months. As shown in Figure 3a,b, long‐term moderate drought stress reduced the growth of all plants. However, compared with GL‐3 plants, MdSUMO2A OE plants were taller, and MdSUMO2 RNAi plants were shorter (Figure 3a,b, Figure S3). In addition to differences in plant height, MdSUMO2A OE plants had longer internodes than GL‐3 plants under drought stress, whereas MdSUMO2 RNAi plants had shorter internodes (Figure S4). Moreover, MdSUMO2A OE plants had greater shoot dry weights under control and drought conditions, whereas MdSUMO2 RNAi plants had lower aboveground biomass (Figure S5). These results indicate that MdSUMO2A OE plants grew more vigorously under long‐term drought, whereas MdSUMO2 RNAi plants grew more slowly.
Figure 3.
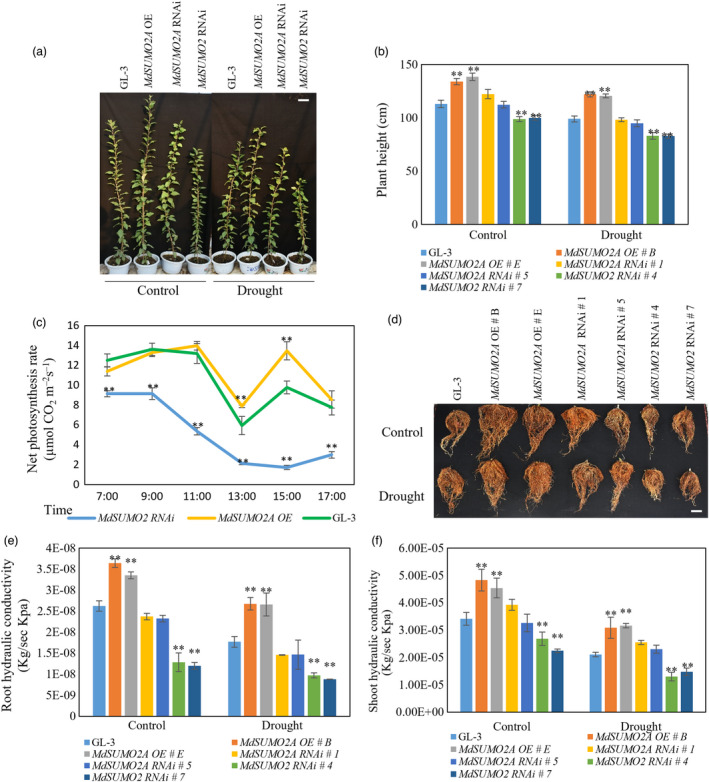
MdSUMO2A OE plants show increased tolerance to drought stress. (a) Morphology of MdSUMO2 transgenic plants under control and long‐term drought stress. Bar = 10 cm. (b–f) Plant height (b), net photosynthesis under long‐term drought stress condition (c), root morphology (d), bar = 5 cm, root hydraulic conductivity (e) and shoot hydraulic conductivity (f) of GL‐3 and MdSUMO2 transgenic plants under control and long‐term drought stress. Plants were exposed to drought for up to 3 months. During treatment, 43%–48% soil volumetric water content (VWC) was maintained as control and 18%–23% of VWC was maintained as drought treatment. Error bars indicate standard error [n = 12 in (b), 7 in (c), 5 in (e) and (f)]. Asterisks indicate significant differences based on one‐way ANOVA and Tukey test (*, P < 0.05; **, P < 0.01). OE, overexpression.
Drought can adversely affect crop photosynthetic capacity, water use efficiency and yield (Mao et al., 2015; Sun et al., 2013a; Xu et al., 2008), and drought stress reduced the photosynthetic capacity of all plants in the current experiment (Figure S6). However, MdSUMO2A OE plants had a greater photosynthetic capacity than GL‐3 plants under drought stress, whereas that of MdSUMO2 RNAi plants was lower (Figure S6A). Under drought stress, stomatal conductance and transpiration rate were also higher in MdSUMO2A OE plants than in GL‐3 plants, and both parameters were lower in MdSUMO2 RNAi plants (Figure S6B,C). We also measured the photosynthetic capacity of GL‐3 and transgenic plants under drought during the daytime from 7:00 AM to 5:00 PM. Similar results were observed. That is, MdSUMO2A OE plants maintained the highest photosynthetic rate under drought stress and exhibited a higher transpiration rate and stomatal conductance after noon, whereas MdSUMO2 RNAi plants had the lowest values for these parameters (Figure 3c and Figure S7).
The root system plays an important role in plant drought resistance (Geng et al., 2018; Hu et al., 2018; Liao et al., 2016). After long‐term drought, the root systems of MdSUMO2A OE plants were much more extensive (Figure 3d), as indicated by root dry weight in Figure S8. However, the root systems of MdSUMO2 RNAi plants were much smaller than those of GL‐3 under control and drought conditions (Figure 3d and Figure S8). Consistent with their strong root systems and greater shoot growth, MdSUMO2A OE plants had higher hydraulic conductivity of roots and shoots (Figure 3e,f), whereas those of MdSUMO2 RNAi plants were lower. These results suggest that MdSUMO2A OE plants performed better under drought, exhibiting vigorous shoot and root growth, as well as higher hydraulic conductivity and photosynthetic capacity.
Leaf morphology is important for drought tolerance (Anyia and Herzog, 2004; Sun et al., 2013a; Wu et al., 2014). MdSUMO2 RNAi leaves were smaller than those of GL‐3 and MdSUMO2A OE plants under control and drought conditions (Figure 4a), as indicated by leaf area measurements in Figure 4b. MdSUMO2 RNAi leaves were much thicker than GL‐3 and MdSUMO2A OE leaves under control and drought conditions (Figure 4c,d). By contrast, the leaf area, thickness and water holding capacity of MdSUMO2A OE leaves were comparable to those of GL‐3 leaves under control and drought conditions (Figure 4a–e). As a high‐throughput and cost‐efficient method to assess WUE (water use efficiency), δ13C, has been reported to has significantly positive correlation with apple WUE. The results showed that MdSUMO2 RNAi plants maintained a higher WUE than GL‐3 and MdSUMO2 OE plants under control and drought conditions (Figure 4f). Plants accumulate the phytohormone abscisic acid (ABA) after drought stimulus (Zhu, 2016). After drought stress, the ABA content of MdSUMO2A OE plants was lower than that of GL‐3 plants, whereas that of MdSUMO2 RNAi plants was higher (Figure S9). These results suggest that MdSUMO2 RNAi plants resist drought by adjusting their leaf morphology, increasing their WUE, and accumulating more ABA.
Figure 4.
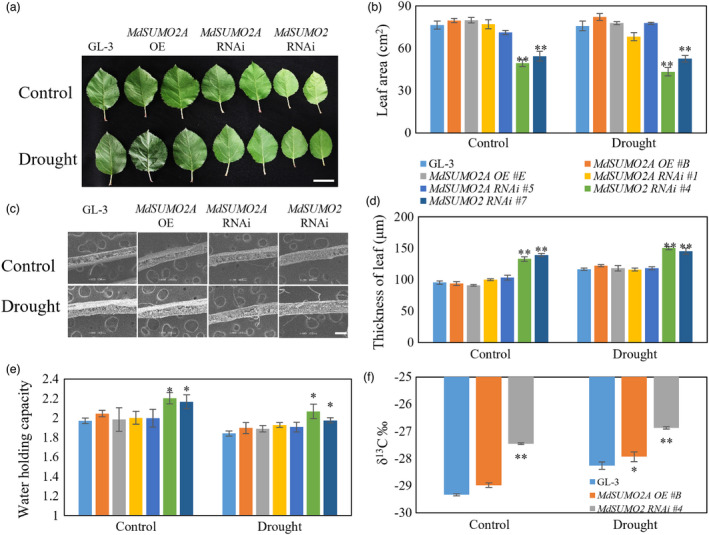
MdSUMO2 RNAi plants display increased tolerance to drought stress. (a–e) Leaf morphology (a, bar = 5 cm), leaf area (b), leaf thickness (c and d, bar = 200 μm), water holding capacity (e) and water use efficiency (f) of GL‐3 and MdSUMO2 transgenic plants under control and long‐term drought stress. Leaf thickness was observed using tungsten filament scanning electron microscope (TEM); water holding capacity = (leaf saturated weight − dry weight)/dry weight; water use efficiency was detected by carbon isotope (δ13C) composition. Plants were exposed to drought for up to 3 months. During treatment, 43%–48% soil volumetric water content (VWC) was maintained as control and 18%–23% of VWC was maintained as drought treatment. Error bars indicate standard error [n = 9 in (b), 16 in (d), 6 in (e), 3 in (f)]. Asterisks indicate significant differences based on one‐way ANOVA and Tukey test (*, P < 0.05; **, P < 0.01). OE, overexpression.
There were no significant differences in the parameters mentioned above between MdSUMO2A RNAi and GL‐3 plants under drought stress (Figures 1, 2, 3, 4, 5, Figures S3–S9), suggesting that the MdSUMO2s have redundant functions in response to drought.
Figure 5.
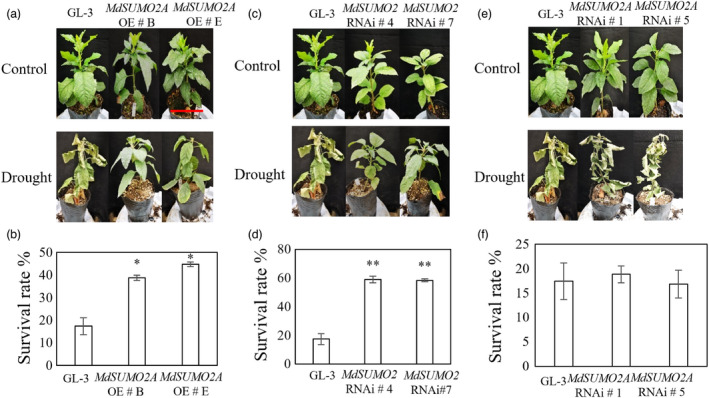
Tolerance of MdSUMO2 transgenic plants and GL‐3 in response to short‐term drought stress. (a) Tolerance of MdSUMO2A OE and GL‐3 plants under short‐term drought. Bar = 5 cm. (b) Survival rate of plants shown in (a). (c) Tolerance of MdSUMO2 RNAi and GL‐3 plants under short‐term drought. (d) Survival rate of plants shown in (c). (e) Tolerance of MdSUMO2A RNAi and GL‐3 plants under short‐term drought. (f) Survival rate of plants shown in (e). Error bars indicate standard error (n = 3). Asterisks indicate significant differences based on one‐way ANOVA and Tukey test (*, P < 0.05; **, P < 0.01). OE, overexpression.
To further support the notion that both MdSUMO2A OE and MdSUMO2 RNAi plants were tolerant to drought stress, we treated all plants with a shorter‐term drought stress. After 3 weeks of drought treatment, 83% of the GL‐3 plants had wilted, whereas 40% of the MdSUMO2A OE plants and 58% of the MdSUMO2 RNAi plants were still alive (Figure 5a–d), suggesting that the MdSUMO2 RNAi plants had a higher survival capacity than the MdSUMO2A OE plants. By contrast, MdSUMO2A RNAi plants did not differ in survival rate from GL‐3 plants under drought stress (Figure 5e,f). We also performed an extreme drought treatment after the long‐term drought treatment by withholding water for 10 days. Both the MdSUMO2 RNAi and MdSUMO2A OE plants performed better than the GL‐3 plants under drought, and the MdSUMO2 RNAi plants were more drought tolerant than the MdSUMO2A OE plants (Figure S3B). All these data suggest that MdSUMO2A OE and MdSUMO2 RNAi plants were more drought tolerant than GL‐3 plants and that MdSUMO2 RNAi plants had higher survival ability than MdSUMO2A OE plants.
In addition, we examined the SUMOylation of GL‐3 and MdSUMO2 transgenic plants under control and prolonged drought stress conditions. As shown in Figure S10, MdSUMO2A OE plants had a slightly higher SUMOylation level than GL‐3 plants under control and drought conditions, whereas the SUMOylation level of MdSUMO2 RNAi plants was lower.
Identification of MdSUMO2 targets reveals SUMOylation of MdDREB2A by MdSUMO2s
To obtain reliable MdSUMO2 targets, we generated transgenic apple calli carrying 6His‐H89R‐MdSUMO2A and performed affinity purification with three steps as described previously (Miller et al., 2010; Rytz et al., 2016), including two Ni‐NTA affinity chromatography steps and one anti‐SUMO1 antibody affinity chromatography step. Since Arabidopsis SUMO1 has high sequence similarity with MdSUMO2 (Figure S2A), we used the anti‐SUMO1 antibody to recognize three MdSUMO2s. Using this stringent approach, we identified 988 and 1110 potential targets of MdSUMO2A under control and simulated drought condition, respectively (Data S1), including MdDREB2A, MdALI, MdAQP2, MdHSP90, MdWRKY, MdH2B, MdCAT1 and MdbZIP (Figure S11). Using a SUMOylation reconstitution assay in Escherichia coli, in which MdSUMO2 and the candidate substrates were expressed (Elrouby and Coupland, 2010), we verified the SUMOylation of MdDREB2A, MdAQP2 and MdALI by the MdSUMO2s (Figure 6a–g). Three and one lysine sites are potential SUMO conjugation sites in MdDREB2A and MdAQP2 respectively. To determine the actual SUMOylation sites, each candidate lysine (K) was replaced by arginine (R) singly or in combinations. SUMOylation assays using the E. coli system showed that K192 was the main mapped SUMOylation site for the MdSUMO2A‐mediated SUMOylation of MdDREB2A (Figure 6a and Figure S12A,B). For MdSUMO2B‐mediated SUMOylation of MdDREB2A, mutation in either K192, K217 or K369 effectively blocks isopeptide linkage of SUMO to MdDREB2A, although the reduction by K369R was slight (Figure 6b). In addition, when K192 of MdDREB2A was mutated, MdDREB2A SUMOylation by MdSUMO2C was dramatically reduced (Figure 6c). The E.coli system also showed that MdSUMO2A, instead of MdSUMO2B or MdSUMO2C, could SUMOylate MdAQP2 (Figure 6d). Moreover, K272 was required for MdSUMO2A‐mediated MdAQP2 SUMOylation (Figure 6e and Figure S12C). For MdALI, there are five lysine sites and one SIM for potential SUMO conjugation. Deleting the SIM or mutating each lysine to R could not abolish the SUMOylation of MdALI by MdSUMO2A (Figure S12D). However, mutation of all five lysine sites to R or mutation of four lysine sites to R and in combination with SIM deletion could almost completely abolish the SUMO conjugation by MdSUMO2A, indicating that these five lysine sites and the SIM were all required for SUMOylation of MdALI by MdSUMO2A (Figure 6f,g and Figure S12E).
Figure 6.
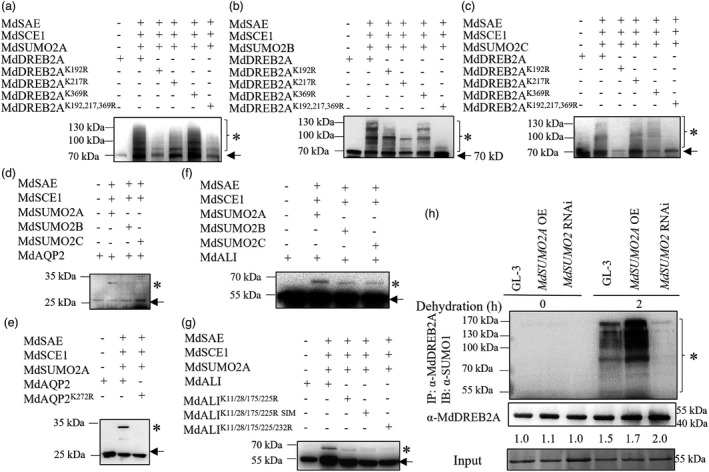
SUMOylation of MdDREB2A, MdAQP2 and MdALI using E. coli system. (a–c) MdDREB2A was SUMOylated by MdSUMO2A, MdSUMO2B and MdSUMO2C. Putative SUMOylation sites (K) of MdDREB2A were mutated to arginine (R). (d) and (e) SUMOylation of AQP2 by MdSUMO2A. Putative SUMOylation site (K272) of MdAQP2 was mutated to arginine (R). (f) and (g) SUMOylation of MdALI by MdSUMO2A, MdSUMO2B and MdSUMO2C. Putative SUMOylation sites (K) or SIM (V) of MdALI were mutated to arginine (R) or alanine (A). (h) SUMOylation of MdDREB2A and MdDREB2A protein in GL‐3, MdSUMO2 RNAi and MdSUMO2A OE plants under control or dehydration conditions. * Indicates SUMOylated substrates; arrows indicate substrates. OE, overexpression.
Because it is an important factor in plant drought stress response (Chen et al., 2007; Qin et al., 2007; Reis et al., 2014; Sakuma et al., 2006a), we next focused on MdDREB2A. Since MdDREB2A could be SUMOylated in the E. coli system, and MdDREB2A did not contain the SIM, we tested the interaction of MdDREB2A and MdSCE1, the SUMO E2‐conjugating enzyme. MST and CO‐IP analysis revealed that MdSCE1 interacts with MdDREB2A in vitro and vivo (Figure S13). SUMOylation can affect target protein localization, protein–protein interaction and protein stability. We co‐localized MdSUMO2A with MdDREB2A and found that SUMOylation of MdDREB2A did not affect its subcellular localization (Figure S14). We also examined the effect of SUMOylation on the stability of MdDREB2A. As shown in Figure 6h, MdDREB2A protein level was significantly increased in the MdSUMO2 RNAi plants under drought conditions but also slightly higher in the MdSUMO2A OE plants.
In addition to the in vitro SUMOylation of MdDREB2A, we also examined the in vivo SUMOylation of MdDREB2A by MdSUMO2 under control and drought conditions. After immunoprecipitation using anti‐MdDREB2A antibody, SUMOylation of MdDREB2A was detected in GL‐3 plants under drought stress, but much less SUMOylation was observed in MdSUMO2 RNAi plants (Figure 6h).
SUMOylation of MdDREB2A is critical for drought stress tolerance and is coupled with ubiquitination during drought
DREB2A is a positive regulator of plant drought and heat stress tolerance (Cui et al., 2011; Kim et al., 2011; Li et al., 2019). Arabidopsis wild‐type plants overexpressing DREB2AK163R (in which K was mutated to R) exhibited decreased thermotolerance (Wang et al., 2020). We therefore examined whether SUMOylation of MdDREB2A affected apple drought stress resistance. We transformed 35S::MdDREB2A (MdDREB2A OE) and 35S::MdDREB2AK192R (MdDREB2AK192R OE, in which K192 was mutated to arginine) into wild‐type GL‐3 apple plants. Both transgenic plants had better survival ability under drought stress compared with the wild type (Figure 7a,b). However, MdDREB2AK192R OE plants had a higher survival rate than MdDREB2A OE plants (Figure 7a,b). In addition, after drought stress, MdDREB2A OE plants had higher photosynthetic capacity than MdDREB2AK192R OE plants (Figure 7c). These data suggest that SUMOylation of MdDREB2A tightly controls plant drought tolerance.
Figure 7.
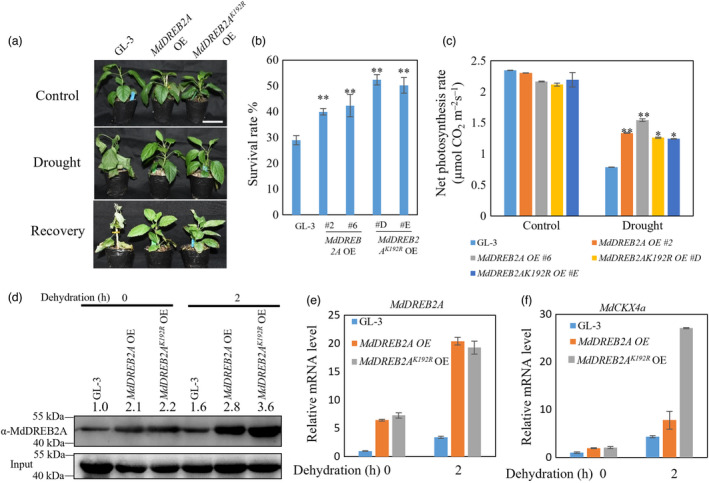
SUMOylation of MdDREB2A is critical for drought stress tolerance. (a) Morphology of MdDREB2A OE and MdDREB2AK192R OE transgenic plants under drought treatment for 3 weeks. Bar = 5 cm. (b) Survival rate of the plants shown in (a). (c) Net photosynthesis rate of MdDREB2A OE and MdDREB2AK192R OE transgenic plants under drought treatment. (d) MdDREB2A accumulation in MdDREB2AK192R OE and MdDREB2A OE plants under dehydration treatment. (e) and (f) mRNA level of MdDREB2A and MdCKX4a in MdDREB2A OE and MdDREB2AK192R OE transgenic plants under dehydration treatment. Error bars indicate standard error [n = 4 in (b), 13 in (c), 3 in (e) and (f)]. Asterisks indicate significant differences based on one‐way ANOVA and Tukey test (*, P < 0.05; **, P < 0.01). OE, overexpression.
Because SUMOylation can affect protein stability, we then examined MdDREB2A protein levels in both transgenic plants under control and drought conditions. As shown in Figure 7d, both transgenic plants had more MdDREB2A than GL‐3 plants under control conditions. Under drought conditions, MdDREB2A OE plants accumulated more MdDREB2A protein than GL‐3 plants, but less than transgenic plants carrying 35S::MdDREB2AK192R (Figure 7d). In addition, the transcripts of MdDREB2A were comparable between two transgenic plants (Figure 7e). We also transformed 35S::MdDREB2A and 35S::MdDREB2AK192R into apple calli and found that transgenic calli carrying either constructs were more tolerant to simulated drought treatment than wild‐type calli. Furthermore, calli carrying 35S::MdDREB2AK192R were more tolerant to PEG than calli carrying 35S::MdDREB2A (Figure S15). In apple, MdDREB2A targets MdCKX4a to modulate drought tolerance (Liao et al., 2016). We next evaluated the MdCKX4a expression in transgenic plants and GL‐3. As shown in Figure 7f, MdCKX4a expression was higher in transgenic apple plants under normal and drought conditions and much higher in plants expressing 35S::MdDREB2AK192R than in plants carrying 35S::MdDREB2A. These results indicate that SUMOylation of MdDREB2A is important for its stability and activity.
The above phenomena prompted us to investigate whether other protein modifications were involved. Indeed, we found that MdDREB2A accumulation was similar in both genotypes of transgenic plants under drought stress when they were treated with MG132, a 26S proteasome inhibitor (Figure 8a). The 26S proteasome is essential for the degradation of ubiquitin‐modified proteins (Smalle and Vierstra, 2004). We then examined SUMOylation and ubiquitination in transgenic plants. As shown in Figure 8b, both transgenic plants had higher levels of SUMOylation and ubiquitination after drought stress. Compared with that of MdDREB2A OE plants, the SUMOylation level of MdDREB2AK192R OE plants was slightly lower. However, their ubiquitination level was also lower (Figure 8b), suggesting that SUMOylated MdDREB2A may undergo ubiquitination in response to drought in MdDREB2A OE plants.
Figure 8.
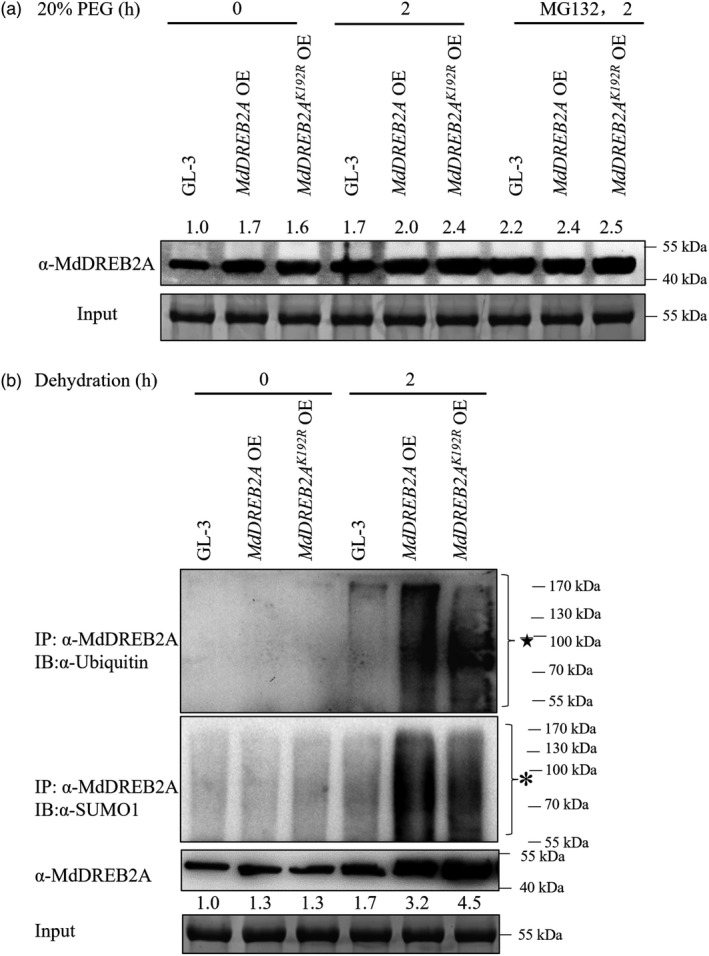
SUMOylation of MdDREB2A couples with ubiquitination mediated by 26S proteasome pathway under drought stress. (a) MdDREB2A accumulation in MdDREB2A OE and MdDREB2AK192R OE transgenic plants under simulated drought stress with or without 50 μm MG132 treatment. (b) Ubiquitination and SUMOylation of MdDREB2A in MdDREB2A OE and MdDREB2AK192R OE plants in response to dehydration. * Indicates SUMOylated substrates; ★ indicates ubiquitinated substrates. OE, overexpression.
MdRNF4 mediates ubiquitination of SUMOylated MdDREB2A
To identify the proteins responsible for the ubiquitination of SUMOylated MdDREB2A, we performed affinity purified mass spectrometry (AP‐MASS) analysis of MdDREB2A under control and drought stress conditions. We identified 1414 and 1472 proteins that may associate with MdDREB2A in planta under control and drought conditions respectively (Data S3). One of the potential MdDREB2A interacting proteins under drought stress was MdRNF4, which encodes an E3 ubiquitin ligase. Homologs of MdRNF4 in mammalian cells and yeast target SUMOylated proteins for degradation by the proteasome pathway (Kumar et al., 2017; Sun et al., 2007; Tatham et al., 2008). We verified the in vivo association of MdDREB2A with MdRNF4 using co‐immunoprecipitation (Co‐IP) analysis (Figure S16). MdRNF4 contains two SUMO‐interacting motifs (SIMs) (Figure S17A). To investigate whether SUMO could be bound to the SIMs of MdRNF4, we performed an Y2H analysis and found that MdRNF4 could interact with MdSUMO2A. When both SIMs were deleted, no interaction was detected. However, deletions of only one SIM did not impair the interaction, indicating that both SIMs are required for the interaction of MdSUMO2A with MdRNF4 (Figure S17B,C). A microscale thermophoresis (MST) approach and Co‐IP assay further verified the interaction between MdSUMO2A and MdRNF4 (Figure S17D,E).
RNF4 is a SUMO‐targeted ubiquitin E3 ligase that is required for degradation of SUMOylated substrates in mammals (Geoffroy and Hay, 2009; Lallemand‐Breitenbach et al., 2008; Zhang et al., 2017a) and the fission yeast Schizosaccharomyces pombe (Sun et al., 2007). We hypothesized that this protein is responsible for the ubiquitination of SUMOylated MdDREB2A. To test our hypothesis, we extracted total proteins from GL‐3 and MdDREB2A OE plants under drought stress and then added purified MdRNF4 to the protein extracts for specific durations. The addition of MdRNF4 for 2 h increased the ubiquitination level of MdDREB2A. However, greater MdDREB2A ubiquitination was observed in MdDREB2A OE plants that had higher MdDREB2A SUMOylation levels (Figure 9a). To further confirm the requirement of MdRNF4 for degradation of SUMOylated MdDREB2A, we generated transgenic plants with a reduced level of MdRNF4 (Figure S18). After immunoprecipitation with anti‐MdDREB2A antibody, MdDREB2A ubiquitination decreased in MdRNF4 RNAi plants under drought conditions (Figure 9b), further suggesting that MdRNF4 mediates the ubiquitination of MdDREB2A.
Figure 9.
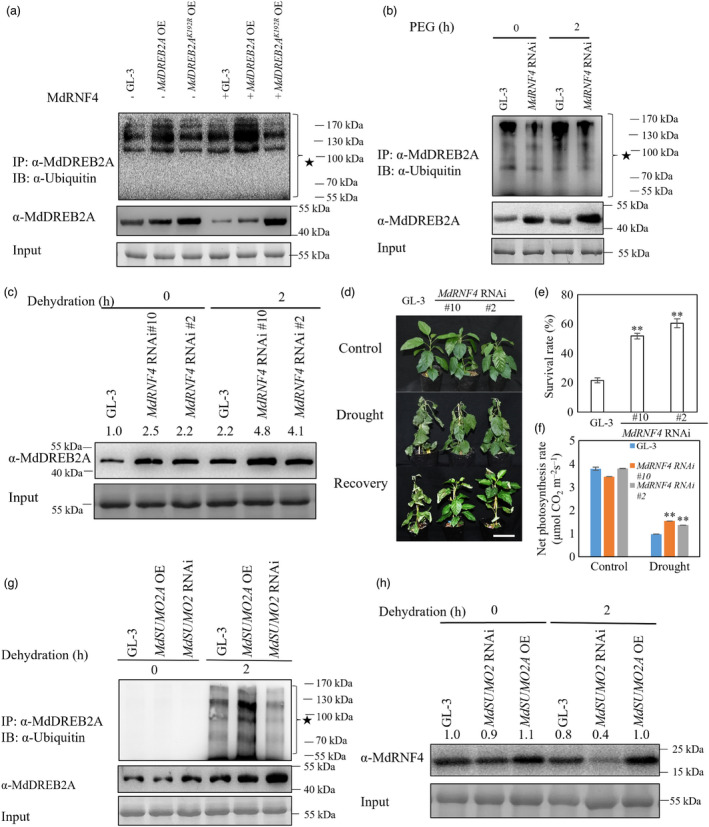
MdRNF4 mediates degradation of SUMOylated MdDREB2A under dehydration conditions. (a) Effects of recombinant MdRNF4 on ubiquitination of MdDREB2A under simulated drought stress. Proteins were extracted from PEG‐treated GL‐3 and MdDREB2A OE plants, and recombinant MdRNF4 was added. (b) Ubiquitination of MdDREB2A in MdRNF4 RNAi plants in response to dehydration. (c) MdDREB2A accumulation in MdRNF4 RNAi plants in response to dehydration. (d) Morphology of GL‐3 and MdRNF4 RNAi plants under control and drought stress conditions. Bar = 5 cm. (e) and (f) Survival rate (e) and photosynthetic capacity (f) of the plants shown in (d). (g) Ubiquitination of MdDREB2A in MdSUMO2 RNAi or MdSUMO2A OE plants in response to dehydration. (h) MdRNF4 level in MdSUMO2 RNAi or MdSUMO2A OE plants in response to dehydration. Error bars indicate standard error [n = 3 in (e) and 20 in (f)]. Asterisks indicate significant differences based on one‐way ANOVA and Tukey test (*, P < 0.05; **, P < 0.01). OE, overexpression. ★ Indicates ubiquitinated substrates. OE, overexpression.
To further analyse the modulation of MdDREB2A stability by MdRNF4, we performed immunoblot analysis of plants under control and drought conditions. MdDREB2A protein levels were higher in MdRNF4 RNAi plants than in GL‐3 plants, either under control or drought conditions; although drought stress induced MdDREB2A accumulation (Figure 9c). In addition, the MdRNF4 RNAi plants were more tolerant to drought stress, consistent with the increased tolerance of MdRNF4 RNAi calli to simulated drought (Figure 9d–f; Figure S19A–C).
Because SUMO2s affect MdDREB2A SUMOylation and stability (Figure 6h), we asked whether this effect was related to MdRNF4. We examined the ubiquitination of MdDREB2A in MdSUMO2 transgenic plants under control and drought conditions. After drought stress, MdSUMO2A OE plants had higher levels of MdDREB2A ubiquitination, and MdSUMO2 RNAi plants had lower levels (Figure 9g). In addition, less MdRNF4 accumulated in MdSUMO2 RNAi plants under drought stress, while more in MdSUMO2A OE plants (Figure 9h), implying the involvement of ubiquitination mediated by MdRNF4 in the MdDREB2A SUMOylation and stability.
Discussion
Drought stress is one of the major environmental fluctuations that affect plant productivity and survival (Geng et al., 2018; Li et al., 2019). During evolution, plants have acquired divergent strategies to respond to water deficiency, including shortening their life cycles to complete vegetative growth and reproduction before soil water is depleted, evolving unique morphologies and root systems to avoid drought stress, and developing the ability to withstand low tissue water content under drought stress. The latter ability may involve processes such as osmotic adjustment, cellular elasticity, and epicuticular wax formation (Polania et al., 2016; Wei et al., 2016; Yıldırım and Kaya, 2017). As a perennial woody plant, apple trees evolved divergent strategies to respond to water deficiency, including establishing unique morphologies and root systems to avoid drought stress, and developing the ability to withstand low tissue water content under drought stress. In our study, both MdSUMO2A OE plants and MdSUMO2 RNAi plants were more drought tolerant than the wild type. However, both transgenic plants applied different approaches of drought resistance. The MdSUMO2A OE plants exhibited greater root system, more vigorous growth and higher photosynthetic capacity and hydraulic conductivity (Figures 3, 4, 5; Figures S3–S9). The MdSUMO2 RNAi transgenic plants had smaller but thicker leaves, much lower stomatal conductance and higher water use efficiency (Figures 3 and 4; Figures S3–S9). In addition, when treated with shorter‐term extreme drought stress condition, both MdSUMO2 RNAi plants and MdSUMO2A OE plants were more tolerant than GL‐3; with MdSUMO2 RNAi plants had higher survival rate than MdSUMO2A OE plants under drought (Figure 5). Tukey's HSD analysis also showed that drought stress had a significant effect on all the traits we tested, and a significant interaction between different genotypes and treatments (Table S1). Taken together, our data suggest that both increased and decreased SUMOylation levels can increase plant drought tolerance.
SUMO is a crucial post‐translational modifier in plants that is covalently conjugated with target substrates to maintain chromatin integrity, transduce signals, stabilize proteins and change cell locations (Dohmen, 2004; Elrouby, 2015; Rytz et al., 2016). Previous studies identified a large number of SUMO substrates in Arabidopsis under heat and oxidative stress, including TPL (TOPLESS), ARF, JAZ, ABF and NAC proteins (Miller et al., 2010; Rytz et al., 2016, 2018). Here, we identified 988 and 1110 potential targets of MdSUMO2A in apple under control and simulated drought condition, respectively, according to the previously described methods in Arabidopsis (Miller et al., 2010; Rytz et al., 2016) (Data S1). Since we used the wild‐type apple calli as the background of gene transformation, it is possible that some potential targets might be missing in our data due to the potential competition between endogenous SUMO2A and transgene of MdSUMO2A. We found that some MdSUMO2A target proteins were homologous to proteins in Arabidopsis, whereas the majority was unique proteins in the apple genome (Data S1). The reconstituted Arabidopsis SUMOylation cascade in E. coli is a rapid and effective method for evaluating the SUMOylation of potential SUMO target proteins (Okada et al., 2009; Saitoh et al., 2009). We used the apple SUMOylation cascade in E. coli as a powerful tool to elucidate the SUMOylation level of targets and confirmed that MdDREB2A, MdALI and MdAQP2 were MdSUMO2 substrates in apple (Figure 6), highlighting the power and reliability of this system.
DREB2A encodes a transcription factor that binds to the dehydration‐responsive element (DRE) (Liu et al., 1998; Yamaguchi‐shinozaki and Shinozaki, 1994). Numerous studies have reported the positive role of DREB2A in response to drought stress in various plants, including apple, Arabidopsis, rice, maize and Pennisetum glaucum. However, DREB2A sequences from these species did not show high similarity outside of the conserved DNA binding domain in the N‐terminal region that may function as a nuclear localization signal (Agarwal et al., 2007; Cui et al., 2011; Liao et al., 2016; Qin et al., 2007, 2008). The N‐terminal region of DREB2A that contains the DNA binding and NRD domains is responsible for its protein stability. The DREB2A NRD domain has been shown to interact with DRIP and BPM ubiquitin E3 ligases, leading to ubiquitination and degradation of DREB2A (Morimoto et al., 2017; Qin et al., 2008). DREB2A can be transformed into a stable and constitutively active form (DREB2A‐CA) by deleting its NRD domain, thereby facilitating plant drought and heat stress tolerance (Sakuma et al., 2006b). In addition, SUMOylation of DREB2A can increase its protein stability under heat stress by suppressing its interaction with BPM2 (Wang et al., 2020). However, apple MdDREB2A does not contain the NRD domain (Figure S20). Whether apple MdDREB2A undergoes any protein modifications was previously unknown. In this study, we found that MdDREB2A was a SUMOylation target of the MdSUMO2s (Figure 6a–c). The critical SUMOylation site of MdDREB2A by MdSUMO2A was the K192 (Figure 6a). Similar to DREB2As in other plant species, MdDRBE2A was a positive regulator of apple drought stress resistance (Figure 7a–c). SUMOylation of targets often increases their stability, as well as overall environmental stress resistance (Miura et al., 2007; Wang et al., 2020; Zhou et al., 2017). To our surprise, we found that the mutation of K192 to R caused MdDREB2A protein levels to be more stable in MdDREB2AK192R OE plants (Figure 7d). In addition, transgenic plants carrying MdDREB2AK192R had a higher survival rate than MdDREB2A OE plants, implying that DREB2A SUMOylation may serve different functions and proceed by different mechanisms in different plant species in response to stress.
In addition to its covalent attachment to target substrates, SUMO can also interact noncovalently with proteins that contain SIMs (Elrouby et al., 2014; Kumar et al., 2017; Sun et al., 2007). SIPs in mammals and Arabidopsis include ubiquitin E3 ligases, DNA methyltransferases or demethylases, and histone methyltransferases or demethylases (Elrouby et al., 2014; Kumar et al., 2017). We also identified SIPs in the apple genome and obtained similar results (Data S4). Among the SIPs, we identified a RING finger protein 4, MdRNF4, that appeared with the highest frequency in the Y2H screen. Similar RING‐type ubiquitin E3 ligases (RNF4s) have been reported to interact with SUMO and ubiquitinate SUMOylated substrates via the 26S proteasome in mammals and yeast (Geoffroy and Hay, 2009; Lallemand‐Breitenbach et al., 2008; Sun et al., 2007; Zhang et al., 2017a). Our study found that MdRNF4‐mediated ubiquitination of SUMOylated MdDREB2A by a 26S proteasome pathway, resulting in the degradation of SUMOylated MdDREB2A (Figure 9). These results suggest a widely conserved function for RNF4 in ubiquitination among eukaryotes.
In summary, we investigated the relationship between SUMOylation and drought stress tolerance in perennial apple trees. Using MdSUMO2A OE and MdSUMO2 RNAi plants, we observed that both decreased and increased SUMOylation can increase plant drought tolerance, although decreased SUMOylation was associated with relatively higher survival rates. We also showed that increased SUMOylation of MdDREB2A in MdSUMO2A OE and MdDREB2A OE plants was associated with MdRNF4‐mediated greater ubiquitination under drought stress, thereby relatively decreasing MdDREB2A accumulation in MdSUMO2A OE and MdDREB2A OE plants compared with MdSUMO2 RNAi and MdDREB2AK192R OE plants.
Methods
Plant materials and growth conditions
The experiments were conducted at Northwest A&F University, Yangling, China (34°20′N, 108°24′E). The transgenic lines and GL‐3 plants after rooting on MS were transplanted to soil and grown for 3 months at 25 °C under a long‐day photoperiod (14 h : 10 h, light : dark). The general management was conducted as described by Xie et al. (2018).
The leaves of apple ‘Golden delicious’ (Malus x domestica) were used for gene cloning. A line isolated from ‘Royal Gala’ (Malus x domestica) named GL‐3 (Dai et al., 2013), which has high regeneration capacity, was used for genetic transformation. GL‐3 tissue‐cultured plants were subcultured every 4 weeks. They were grown on MS medium (4.43 g/L MS salts, 30 g/L sucrose, 0.2 mg/L 6‐BA, 0.2 mg/L IAA and 7.5 g/L agar, pH 5.8) under long‐day conditions (14 h : 10 h, light : dark) at 25 °C.
Generation of transgenic apple plants and calli
201‐bp (3′ UTR region), 121‐bp (conserved CDS of MdSUMO2s) or 74‐bp fragments of MdSUMO2A, MdSUMO2 or MdRNF4 were individually cloned into the pDONR222 vector by multisite Gateway recombination, as described by Karimi and Hilson (2007) and subsequently transferred to RNA silencing vector pK7GWIWG2, a destination vector containing an N‐terminal GFP tag by LR recombination. To overexpress genes, the coding sequences of MdSUMO2A, MdDREB2A or MdDREB2AK192R were constructed to pCambia 2300 with N‐myc tag or pGWB418. All the constructed vectors were transformed into Agrobacterium strain EHA105. Agrobacterium‐mediated transformation of apple was carried out as described, using GL‐3 as the genetic background (Dai et al., 2013; Holefors et al., 1998).
To generate transgenic apple calli, ‘Orin’ (Malus× domestica) calli grown on MS media (1.5 mg/L 2,4‐Dichlorophenoxyacetic acid (2,4‐D) and 0.4 mg/L 6‐BA at the dark environment) were used as the wild type. The coding sequence of MdDREB2A, MdDREB2AK192R or MdRNF4 was cloned into plant binary vector pGWB418. A 300‐bp sequence of MdRNF4 was cloned into pK7GWIWG2 to knock down MdRNF4 expression. The resulting plasmids were transformed into Agrobacterium strain EHA105 and then transformed to ‘Orin’ calli according to previous methods (An et al., 2019, 2020) The primers used for constructing these vectors are shown in Data S5.
Stress treatment
For long‐term drought treatment, 3‐month‐old GL‐3 and transgenic apple plants were transplanted to a greenhouse at the beginning of April 2019. The drought treatment was performed 2 months later in June. The plants were grown in plastic pots (15 cm × 20 cm, ~1.3 L) filled with a mixture of sand and substrate (Pindstrup Mosebrug, Fabriksvej 2, Denmark) (1:1, v/v). The measurement of soil volumetric water content (VWC) was conducted by TDR (FS6430, Spectrum Technologies, Aurora, USA). At the beginning of drought treatment, uniform trees of each line (30 trees for each line) were divided into two groups for the following treatments (15 trees for each treatment for each line): (1) control, well‐watered, irrigated daily to maintain 43%–48% of VWC and (2) moderate drought, irrigated daily to maintain 18%–23% of VWC. The treatment was lasted for 3 months. The photosynthetic capacity was determined with LI‐Cor 6400 portable photosynthesis system (LI‐COR, Huntington Beach, CA). Hydraulic conductivity of roots and shoots was conducted by an HPFM (Dynamax, Houston, TX) as described previously (Geng et al., 2018). The thickness of leaves was measured by using tungsten filament scanning electron microscope (JSM‐6360LV, JEOL, Tokyo, Japan) according to the methods described by Liao (Liao et al., 2016) with modifications. For the detection of leaves δ13C ‰, mature leaves were collected. Leaves were oven‐dried at 105 °C for 0.5 h and then 70 °C for 3 days to dry completely. Dried leaves were ground and filtered through a sieve (80 holes per cm2). The δ13C ‰ of leaves was determined with an elementary analysis‐isotope ratio mass spectrometer (Flash EA 1112 HT‐Delta V Advantages, Thermo Fisher Scientific, Waltham, MA, USA) as described previously (Wang et al., 2018).
For short‐term drought treatment, 3‐month‐old uniform trees of GL‐3 and transgenic apples were used. Before treatment, plants were irrigated to maintain saturation of soil water content. Then plants were withheld with water until VWC reached 0, and survival rate was calculated after rewatering for 1 week. The soil VWC was measured by TDR (FS6430).
RNA extraction and quantitative real‐time RT‐PCR
Total RNA from apple leaves was extracted by a CTAB method. DNase I (Fermentas, Thermo Fisher Scientific, Waltham, MA, USA) was used to remove residual genomic DNA. We used total RNA to generate cDNA according to the manufacturer's instructions by using the RevertAid™ First Strand cDNA synthesis kit (K1622, Thermo Fisher Scientific, Waltham, MA, USA). The qRT‐PCR was performed in a reaction containing GoTaq® qPCR Master Mix (Promega, Madison, WI, USA), cDNA and primers (described in Data S5) on an CFX96 real‐time PCR detection systems (Bio‐Rad, Hercules, Ca, USA). MdMDH (malate dehydrogenases) was used as the reference gene.
Subcellular localization
To generate the constructs for subcellular localization assay, coding region of MdSUMO2A, MdSUMO2B or MdSUMO2C was amplified and cloned into pEarleyGate104 vector by BP and LR reactions (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA), and were then transformed into Agrobacterium strain C58C1. The empty vector pGWB455, which carries 35S::mCherry, was also transformed into Agrobacterium strain C58C1. The C58C1 carrying the resulting plasmid, 35S::mCherry and 35S:p19 (p19 is an RNA silencing repressor protein from Tomato bushy stunt virus) was coinfiltrated into tobacco leaves (Nicotiana benthamiana). Three days later, the leaf epidermal cells were observed by Nikon A1R/A1 confocal microscope system (Nikon, Tokyo, Japan) for yellow fluorescence observation.
For colocalization of MdSUMO2A with MdDREB2A or MdDREB2AK192R, the full‐length sequence of MdDREB2A or MdDREB2AK192R was cloned into pGWB455 and then transformed into C58C1. Mature fragments of MdSUMO2A, MdSUMO2B and MdSUMO2C (MdSUMO2s with exposed GG) were amplified and individually cloned into pEarleyGate104 vector by BP and LR reactions (Invitrogen), and were then transformed into Agrobacterium strain C58C1. The C58C1 carrying mCherry‐MdDREB2A, 35S:p19, and YFP‐MdSUMO2A, YFP‐MdSUMO2B or YFP‐MdSUMO2C were resuspended in the buffer containing 10 mm MgCl2, 10 mm MES–KOH and 180 μm acetosyringone and then coinfiltrated into the tobacco leaves for 3 d to detect signals with confocal microscope. The primers used are listed in Data S5.
Histochemical and fluorometric assays for GUS activity
For the promoter‐GUS reporter assay, an ~1000‐bp DNA fragment upstream of the MdSUMO2A was cloned into pMDC164 and then transformed into Agrobacterium strain GV3101. The resulting plasmid was introduced into Col‐0 using the floral‐dipping method (Clough and Bent, 1998) for stable transformation in Arabidopsis. GUS activity was observed after staining with 0.5 mg/mL 5‐bromo‐4‐chloro‐3‐indolyl‐b‐Dglucoronide as described previously (Guan et al., 2013). The primers used are listed in Data S5.
Endogenous ABA determination
After 3 months of moderate drought treatment, the mature leaves were collected from GL‐3 and MdSUMO2 transgenic lines to determine ABA content. Leaves were weighed and immediately frozen in liquid nitrogen. Frozen leaves were then pulverized, and ABA was extracted as described previously (Chen et al., 2012; Xie et al., 2021). Quantitative determination of endogenous ABA was performed on a UPLC–MS/MS system (QTRAP™ 5500 LC/MS/MS, SCIEX, Framingham, MA, USA) and a Shimadzu LC‐30AD UPLC system (Tokyo, Japan).
SUMOylation assay in E. coli
SUMOylation assays in E. coli were conducted as described previously (Elrouby and Coupland, 2010). The coding region of MdSAE1 or MdSAE2 was amplified and cloned into binary expression vector pCDFDuet‐1, and mature MdSUMO2s or MdSCE1 was cloned into pACYCDuet‐1. Prokaryotic expression vector PGEX‐4T‐1 was used to express GST‐MdDREB2A, MdALI and MdAQP2 protein. Subsequently, the resulting plasmids in certain combination were introduced into Escherichia coli stain BL21 (DE3). After incubation at 37 °C until OD600 reached 0.6, 1 mm IPTG (Isopropyl β‐D‐1‐thiogalactopyranoside) was added to induce protein expression. Eight hours later, the bacterium was harvested and denaturized for Western blot analysis with GST antibody (M20007, Abmart, Shanghai, China). The primers used are listed in Data S5.
Identification of MdSUMO2 targets
To obtain reliable MdSUMO2 targets, we generated transgenic apple calli carrying 6His‐H89R‐MdSUMO2A and performed affinity purification according to the previously described methods (Miller et al., 2010; Rytz et al., 2016): 1. Ni‐NTA affinity chromatography; 2. immunoprecipitation with anti‐SUMO1 antibody; 3. Ni‐NTA affinity chromatography. The purified SUMOylated proteins were subjected to a MASS spectrometry analysis (Rytz et al., 2018). This experiment was performed with two biological replicates; only proteins displayed in both replicates were considered. The wild‐type apple calli were used as the negative control, and the detected proteins are listed in Data S2.
Immunoblot analysis
The proteins of transgenic apple plants and GL‐3 were extracted with protein extraction buffer [50 mm Tris‐HCl, pH 8.0, 150 mm NaCl, 2 mm EDTA, 1 mm DTT, 10% glycerol, 1% Triton X‐100, 1 mm phenylmethylsulfonyl fluoride (PMSF) and 1× Halt protease inhibitor cocktail (78427, Thermo Fisher Scientific, Shanghai, China)] and centrifuged at 14 000 g at 4 °C for 10 min. The extracted proteins were used for Western blot analysis with polyclonal MdDREB2A antibody against rabbit, or anti‐SUMO (ab5316, Abcam, Waltham, MA, USA), anti‐Ubiquitin (P4D1, Cell Signaling Technology®, Shanghai, China), anti‐MdRNF4 (rabbit polyclonal antibody, ABclonal Technology, Wuhan, China) or anti‐Actin (AC009, ABclonal Technology, Wuhan, China).
In vivo SUMOylation and ubiquitination analysis
Total proteins extracted from transgenic plants (MdSUMO2A OE, MdSUMO2 RNAi, MdDREB2A OE, MdDREB2AK192R OE, MdRNF4 RNAi and GL‐3) were immunoprecipitated with anti‐MdDREB2A and immunoblotted with anti‐SUMO (ab5316, Abcam) or anti‐Ubiquitin (P4D1, Cell Signaling Technology®) antibodies. To examine the ubiquitination and SUMOylation of MdDREB2A under drought stress conditions, plants were dehydrated for 2 h.
To detect the effects of recombinant MdRNF4 on ubiquitination and SUMOylation of MdDREB2A under simulated drought stress, proteins were extracted from PEG‐treated GL‐3 and MdDREB2A OE plants, and recombinant MdRNF4 was added for 2 h (An et al., 2019, 2020). Total proteins were extracted and immunoprecipitated with anti‐MdDREB2A and immunoblotted with anti‐SUMO (ab5316, Abcam) or anti‐Ubiquitin (P4D1, Cell Signaling Technology®) antibodies.
Yeast two‐hybrid assay
To identify MdSUMO2 interacting proteins, 1‐95 aa of MdSUMO2A (mature MdSUMO2A with exposed GG) was amplified and cloned into pGBKT7 vector to generate bait plasmid. Y2H screen was performed to screen the apple library according to the user manual of MatchmakerTM Gold Yeast Two Hybrid System (Clontech, Shiga, Japan) by using Saccharomyces cerevisiae strain Y2H Gold.
To perform the point‐to‐point Y2H, mature MdSUMO2A with exposed GG was cloned into pGBKT7, resulting in MdSUMO2A‐pGBKT7. Full‐length or truncated MdRNF4 with SIM deletion was constructed to pGADT7 vector. MdSUMO2A‐pGBKT7 and MdRNF4‐pGADT7 or truncated MdRNF4‐pGADT7 was co‐transformed into yeast strain Y2H Gold. The positive clones were selected on SD‐Leu‐Trp and then on SD‐Leu‐Trp‐His‐Ade + x‐α‐gal plates for growth observation and the x‐α‐gal assay. The primers used are listed in Data S5.
Co‐IP assay
For Co‐IP analysis, the leaves of GL‐3 and MdSUMO2A OE (35S::myc‐MdSUMO2A) transgenic plants were dehydrated for 2 h. Total proteins were extracted from leaf samples with extraction buffer [50 mm Tris‐HCl, pH 8.0, 150 mm NaCl, 2 mm EDTA, 1 mm DTT, 10% glycerol, 1% Triton X‐100, 1 mm phenylmethylsulfonyl fluoride (PMSF) and 1× Halt protease inhibitor cocktail]. The protein extracts were incubated overnight with polyclonal MdDREB2A antibody. The immunocomplexes were collected by adding protein A/G agarose beads (78610, Thermo Fisher Scientific, Shanghai, China) and were washed with immunoprecipitation buffer [50 mm Tris‐HCl, pH 8.0, 150 mm NaCl, 2 mm EDTA, 1 mm DTT, 10% glycerol, 0.15% Triton X‐100, 1 mm PMSF and 1× Halt protease inhibitor cocktail]. The pellet (immunocomplexes with beads) was resuspended in 1× SDS‐PAGE loading buffer. Eluted proteins were analysed by immunoblotting using anti‐MdRNF4 antibody or anti‐MdDREB2A antibody. Chemiluminescence signals were detected by autoradiography.
AP‐MASS assay
To identify the interacting proteins of MdDREB2A in vivo, AP‐MASS assay was performed as described previously (Maio et al., 2020) with modifications. Total proteins were extracted in leaves of GL‐3 plants with or without 2‐h dehydration treatments using extraction buffer [50 mm Tris‐HCl, pH 8.0, 150 mm NaCl, 2 mm EDTA, 1 mm DTT, 10% glycerol, 1% Triton X‐100, 1 mm phenylmethylsulfonyl fluoride (PMSF) and 1× Halt protease inhibitor cocktail]. The protein extracts were incubated overnight with polyclonal MdDREB2A antibody and then added protein A/G agarose beads to incubate at 4 °C for additional 4–5 h. After incubation, the beads were captured with a magnetic rack and washed three times in 0.5 mL of washing buffer (10 mm Tris‐HCl pH 7.5, 150 mm NaCl, 0.5 mm EDTA, 1 mm PMSF protease inhibitor). The pellet (immunocomplexes with beads) was resuspended in 1× SDS‐PAGE loading buffer and subjected to mass spectrometry analysis (Applied Protein Technology, Shanghai, China).
Microscale thermophoresis (MST) assay
Full‐length MdSUMO2A and MdDREB2A were cloned into pET‐32a. MdSCE1 or MdRNF4 was cloned into pGEX‐4T and pMAL‐c5X respectively. The resulting plasmids were expressed in E. coli BL21. Recombinant protein MdSUMO2A‐HIS and MdDREB2A were purified by HIS Sepharose beads (GE Healthcare, Fairfield, CT), GST‐MdSCE1 was purified by Pierce™ Glutathione Spin Columns (16105, Thermo Scientific™), and MBP‐MdRNF4 was purified by MBP TRAP HP (GE Healthcare). MST was conducted according the manufacturer's manual (NanoTemper, Munich, Germany). The primers used are listed in Data S5.
Statistical analysis
Statistical analysis was performed with one‐way ANOVA and Tukey test according to the SPSS 20.0 (IBM, Armonk, New York, USA). Differences were considered statistically significant when *P < 0.05 and **P < 0.01. The transgenic lines were compared with GL‐3 under control or drought conditions.
Accession numbers
The accession numbers in GDR are as follows: MdSUMO2B (MD17G1103900, MD09G1113800), MdSUMO2A (MD03G1194700, MD11G1211000) and MdSUMO2C (MD05G1173700, MD10G1161600), and in NCBI under the following: MdDREB2A (NP_001280947.1), MdSAE1 (XP_028948277.1), MdSAE2 (XP_008382303.1), MdSCE1 (XP_008338336.1), MdRNF4 (XP_008346210.1), MdALI (XP_008341016.1) and MdAQP2 (XP_008363507.1).
Conflict of interest
The authors declare that they have no conflicts of interest.
Author contributions
We thank Dr. Zhihong Zhang from Shenyang Agricultural University for providing tissue‐cultured GL‐3 plants. Q.G. and F.M. designed the project. X.L., S.Z., L.L., H.D., Z.L., P. C., Z.M., S.Z. and B.C. performed the experiments. Q.G., X.L., H.D., B.C., Z.L. and L.L. analysed the data. Q.G., X.L. and F.M. wrote the manuscript.
Supporting information
Figure S1. Expression patterns of apple SUMO2s.
Figure S2. Protein alignment (A) and phylogenetic analysis (B) of SUMO2A from different species.
Figure S3. Phenotypes of MdSUMO2 transgenic plants and GL‐3 under prolonged drought stress.
Figure S4. Internode length of GL‐3, MdSUMO2A OE plants, MdSUMO2A RNAi, and MdSUMO2 RNAi plants under long‐term drought conditions.
Figure S5. Dry weight of shoots of MdSUMO2 transgenic plants and GL‐3 under prolonged drought stress.
Figure S6. Photosynthesis related parameters of GL‐3, MdSUMO2A overexpression plants, MdSUMO2A RNAi, and MdSUMO2 RNAi plants under long‐term drought conditions.
Figure S7. Transpiration rate (A) and stomatal conductance (B) of MdSUMO2 transgenic plants and GL‐3 under long‐term drought stress conditions.
Figure S8. Dry weight of roots of MdSUMO2A transgenic plants and GL‐3 under control and long‐term drought stress condition.
Figure S9. The ABA content of MdSUMO2A transgenic plants and GL‐3 under control and long‐term drought stress conditions.
Figure S10. SUMOylation of MdSUMO2 transgenic plants and GL‐3 under control and long‐term drought stress conditions.
Figure S11. The putative SUMOylation lysine residues in MdDREB2A (A), MdAQP2 (B), and MdALI (C) proteins.
Figure S12. SUMOylation of MdDREB2A, MdAQP2, and MdALI using E. coli system.
Figure S13. MdDREB2A interacts with MdCE1 in vitro and vivo.
Figure S14. Effects of MdSUMO2A on the subcellular localization of MdDREB2A.
Figure S15. SUMOylation of MdDREB2A is critical for drought stress tolerance.
Figure S16. MdDREB2A interacts with MdRNF4 in vivo.
Figure S17. MdRNF4 interacts with MdSUMO2A by SIMs.
Figure S18. Expression of MdRNF4 in MdRNF4 RNAi transgenic plants.
Figure S19. Tolerance of MdRNF4 RNAi calli in response to simulated drought stress.
Figure S20. Protein alignment of DREB2A from different species.
Table S1. P‐values of Turkey's HSD test to compare plant traits among GL‐3 and transgenic plants exposed to control and long‐term drought stress conditions.
Data S1. Potential targets of MdSUMO2.
Data S2. Proteins identified in the MS/MS analysis of the wild‐type apple calli control.
Data S3. Potential interacted proteins of MdDREB2A.
Data S4. Y2H screening of MdSUMO2A in apple.
Data S5. Primers used in this study.
Appendix S1. Various tissues of Malus fusca.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2019YFD1000100), the Key S&T Special Projects of Shaanxi Province (2020zdzx03‐01‐02), the China Postdoctoral Science Foundation (2021M692661), the Chinese Universities Scientific Fund (2452020216) and National Natural Science Foundation of China (31872080).
Li, X. , Zhou, S. , Liu, Z. , Lu, L. , Dang, H. , Li, H. , Chu, B. , Chen, P. , Ma, Z. , Zhao, S. , Li, Z. , van Nocker, S. , Ma, F. and Guan, Q. (2022) Fine‐tuning of SUMOylation modulates drought tolerance of apple. Plant Biotechnol. J., 10.1111/pbi.13772
Contributor Information
Fengwang Ma, Email: fwm64@nwafu.edu.cn.
Qingmei Guan, Email: qguan@nwafu.edu.cn.
References
- Agarwal, P. , Agarwal, P.K. , Nair, S. , Sopory, S.K. and Reddy, M.K. (2007) Stress‐inducible DREB2A transcription factor from Pennisetum glaucum is a phosphoprotein and its phosphorylation negatively regulates its DNA‐binding activity. Mol. Genet. Genomics, 277, 189. [DOI] [PubMed] [Google Scholar]
- An, J.P. , Wang, X.F. , Zhang, X.W. , Bi, S.Q. , You, C.X. and Hao, Y.J. (2019) MdBBX22 regulates UV‐B‐induced anthocyanin biosynthesis through regulating the function of MdHY5 and is targeted by MdBT2 for 26S proteasome‐mediated degradation. Plant Biotechnol. J. 17, 2231–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, J.P. , Wang, X.F. , Espley, R.V. , Lin‐Wang, K. , Bi, S.Q. , You, C.X. and Hao, Y.J. (2020) An apple B‐Box protein MdBBX37 modulates anthocyanin biosynthesis and hypocotyl elongation synergistically with MdMYBs and MdHY5. Plant Cell Physiol. 61, 130–143. [DOI] [PubMed] [Google Scholar]
- Anyia, A.O. and Herzog, H. (2004) Water‐use efficiency, leaf area and leaf gas exchange of cowpeas under mid‐season drought. Eur. J. Agron. 20, 327–339. [Google Scholar]
- Augustine, R.C. and Vierstra, R.D. (2018) SUMOylation: re‐wiring the plant nucleus during stress and development. Curr. Opin. Plant Biol. 45, 143–154. [DOI] [PubMed] [Google Scholar]
- Basu, S. , Ramegowda, V. , Kumar, A. and Pereira, A. (2016) Plant adaptation to drought stress. F1000Research, 5, 1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benlloch, R. and Lois, L.M. (2018) Sumoylation in plants: mechanistic insights and its role in drought stress. J. Exp. Bot. 69, 4539–4554. [DOI] [PubMed] [Google Scholar]
- Castro, P.H. , Tavares, R.M. , Bejarano, E.R. and Azevedo, H. (2012) SUMO, a heavyweight player in plant abiotic stress responses. Cell Mol. Life Sci. 69, 3269–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala, R. , Ouyang, J. , Abreu, I.A. , Hu, Y. , Seo, H. , Zhang, X. and Chua, N.H. (2007) The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell, 19, 2952–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M.L. , Fu, X.M. , Liu, J.Q. , Ye, T.T. , Hou, S.Y. , Huang, Y.Q. , Yuan, B.F. et al. (2012) Highly sensitive and quantitative profiling of acidic phytohormones using derivatization approach coupled with nano‐LC‐ESI‐Q‐TOF‐MS analysis. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 905, 67–74. [DOI] [PubMed] [Google Scholar]
- Chen, M. , Wang, Q. , Cheng, X. , Xu, Z. , Li, L. , Ye, X. , Xia, L. et al. (2007) GmDREB2, a soybean DRE‐binding transcription factor, conferred drought and high‐salt tolerance in transgenic plants. Biochem. Biophys. Res. Commun. 353, 299–305. [DOI] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Colby, T. , Matthai, A. , Boeckelmann, A. and Stuible, H.P. (2006) SUMO‐conjugating and SUMO‐deconjugating enzymes from Arabidopsis. Plant Physiol. 142, 318–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, M. , Zhang, W. , Zhang, Q. , Xu, Z. , Zhu, Z. , Duan, F. and Wu, R. (2011) Induced over‐expression of the transcription factor OsDREB2A improves drought tolerance in rice. Plant Physiol. Biochem. 49, 1384–1391. [DOI] [PubMed] [Google Scholar]
- Dai, A.G. (2013) Increasing drought under global warming in observations and models. Nat. Clim. Change, 3, 52–58. [Google Scholar]
- Dai, H.Y. , Li, W.R. , Han, G.F. , Yang, Y. , Ma, Y. , Li, H. and Zhang, Z.H. (2013) Development of a seedling clone with high regeneration capacity and susceptibility to Agrobacterium in apple. Sci. Hortic. 164, 202–208. [Google Scholar]
- Dohmen, R.J. (2004) SUMO protein modification. Biochim. Biophys. Acta, 1695, 113–131. [DOI] [PubMed] [Google Scholar]
- Elrouby, N. (2015) Analysis of Small Ubiquitin‐Like Modifier (SUMO) targets reflects the essential nature of protein SUMOylation and provides insight to elucidate the role of SUMO in plant development. Plant Physiol. 169, 1006–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrouby, N. , Bonequi, M.V. , Porri, A. and Coupland, G. (2014) Identification of Arabidopsis SUMO‐interacting proteins that regulate chromatin activity and developmental transitions. Proc. Natl. Acad. Sci. USA, 110, 19956–19961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrouby, N. and Coupland, G. (2010) Proteome‐wide screens for small ubiquitin‐like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes. Proc. Natl. Acad. Sci. USA, 107, 17415–17420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanty, Y. , Belotserkovskaya, R. , Coates, J. and Jackson, S.P. (2012) RNF4, a SUMO‐targeted ubiquitin E3 ligase, promotes DNA double‐strand break repair. Genes Dev. 26, 1179–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng, D.L. , Chen, P.X. , Shen, X.X. , Zhang, Y. , Li, X.W. , Jiang, L.J. , Xie, Y.P. et al. (2018) MdMYB88 and MdMYB124 enhance drought tolerance by modulating root vessels and cell walls in apple. Plant Physiol. 178, 1296–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng, D.L. , Shen, X.X. , Xie, Y.P. , Yang, Y.S. , Bian, R.L. , Gao, Y.Q. , Li, P.M. et al. (2020) Regulation of phenylpropanoid biosynthesis by MdMYB88 and MdMYB124 contributes to pathogen and drought resistance in apple. Hortic. Res. 7, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoffroy, M.‐C. and Hay, R.T. (2009) An additional role for SUMO in ubiquitin‐mediated proteolysis. Nat. Rev. Mol. Cell Biol. 10, 564. [DOI] [PubMed] [Google Scholar]
- Guan, Q. , Lu, X. , Zeng, H. , Zhang, Y. and Zhu, J. (2013) Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J. 74, 840–851. [DOI] [PubMed] [Google Scholar]
- Holefors, A. , Xue, Z.T. and Welander, M. (1998) Transformation of the apple rootstock M26 with the rolA gene and its influence on growth. J. Plant Physiol. 136, 69–78. [Google Scholar]
- Hu, L. , Xie, Y. , Fan, S. , Wang, Z. , Wang, F. , Zhang, B. , Li, H. et al. (2018) Comparative analysis of root transcriptome profiles between drought‐tolerant and susceptible wheat genotypes in response to water stress. Plant Sci. 272, 276. [DOI] [PubMed] [Google Scholar]
- Karan, R. and Subudhi, P.K. (2012) A stress inducible SUMO conjugating enzyme gene (SaSce9) from a grass halophyte Spartina alterniflora enhances salinity and drought stress tolerance in Arabidopsis. BMC Plant Biol. 12, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi, M. , Depicker, A. , Hilson, P. (2007) Recombinational cloning with plant gateway vectors. Plant Physiol, 145(4), 1144–1154. 10.1104/pp.107.106989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.S. , Mizoi, J. , Yoshida, T. , Fujita, Y. , Nakajima, J. , Ohori, T. , Todaka, D. et al. (2011) An ABRE promoter sequence is involved in osmotic stress‐responsive expression of the DREB2A gene, which encodes a transcription factor regulating drought‐inducible genes in Arabidopsis. Plant Cell Physiol. 52, 2136–2146. [DOI] [PubMed] [Google Scholar]
- Kim, J. , Song, J. and Seo, H. (2017) Post‐translational modifications of Arabidopsis E3 SUMO ligase AtSIZ1 are controlled by environmental conditions. FEBS Open Bio. 7, 1622–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, R. , Gonzalez‐Prieto, R. , Xiao, Z. , Verlaan‐de Vries, M. and Vertegaal, A.C.O. (2017) The STUbL RNF4 regulates protein group SUMOylation by targeting the SUMO conjugation machinery. Nat. Commun. 8, 1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand‐Breitenbach, V. , Jeanne, M. , Benhenda, S. , Rihab, N. , Lei, M. , Peres, L. , Zhou, J. et al. (2008) Arsenic degrades PML or PML‐RARalpha through a SUMO‐triggered RNF4/ubiquitin‐mediated pathway. Nat. Cell Biol. 10, 547–555. [DOI] [PubMed] [Google Scholar]
- Li, X.W. , Chen, P.X. , Xie, Y.P. , Yan, Y. , Wang, L.P. , Dang, H. , Zhang, J. et al. (2020) Apple SERRATE negatively mediates drought resistance by regulating MdMYB88 and MdMYB124 and microRNA biogenesis. Hortic. Res. 7, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X.W. , Xie, Y.P. , Lu, L.Y. , Yan, M.J. , Fang, N. , Xu, J.D. , Wang, L.P. et al. (2019) Contribution of methylation regulation of MpDREB2A promoter to drought resistance of Mauls prunifolia. Plant Soil, 441, 15–32. [Google Scholar]
- Liao, X. , Guo, X. , Wang, Q. , Wang, Y.T. , Zhao, D. , Yao, L.P. , Wang, S. et al. (2016) Overexpression of MsDREB6. 2 results in cytokinin‐deficient developmental phenotypes and enhances drought tolerance in transgenic apple plants. Plant J. 89, 510–526. [DOI] [PubMed] [Google Scholar]
- Liu, Q. , Kasuga, M. , Sakuma, Y. , Abe, H. , Miura, S. , Yamaguchi‐Shinozaki, K. and Shinozaki, K. (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought‐and low‐temperature‐responsive gene expression, respectively, in Arabidopsis. Plant Cell, 10, 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maio, F. , Helderman, T.A. , Arroyo‐Mateos, M. , van der Wolf, M. , Boeren, S. , Prins, M. and van den Burg, H.A. (2020) Identification of tomato proteins that interact with replication initiator protein (Rep) of the geminivirus TYLCV. Front. Plant Sci. 11, 1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, H. , Wang, H. , Liu, S. , Li, Z. , Yang, X. , Yan, J. , Li, J. et al. (2015) A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat. Commun. 6, 8326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, M.J. , Barrett‐Wilt, G.A. , Hua, Z. and Vierstra, R.D. (2010) Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin‐like modifier conjugation in Arabidopsis. Proc. Natl. Acad. Sci. USA, 107, 16512–16517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra, N. , Sun, L. , Zhu, X. , Smith, J. , Prakash Srivastava, A. , Yang, X. , Pehlivan, N. et al. (2017) Overexpression of the rice SUMO E3 ligase gene OsSIZ1 in cotton enhances drought and heat tolerance, and substantially improves fiber yields in the field under reduced irrigation and rainfed conditions. Plant Cell Physiol. 58, 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura, K. , Jin, J.B. , Lee, J. , Yoo, C.Y. , Stirm, V. , Miura, T. , Ashworth, E.N. et al. (2007) SIZ1‐mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell, 19, 1403–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura, K. , Okamoto, H. , Okuma, E. , Shiba, H. , Kamada, H. , Hasegawa, P. and Murata, Y. (2013) SIZ1 deficiency causes reduced stomatal aperture and enhanced drought tolerance via controlling salicylic acid‐induced accumulation of reactive oxygen species in Arabidopsis. Plant J. 73, 91–104. [DOI] [PubMed] [Google Scholar]
- Mizoi, J. , Ohori, T. , Moriwaki, T. , Kidokoro, S. , Todaka, D. , Maruyama, K. , Kusakabe, K. et al. (2013) GmDREB2A;2, a canonical DEHYDRATION‐RESPONSIVE ELEMENT‐BINDING PROTEIN2‐type transcription factor in soybean, is posttranslationally regulated and mediates dehydration‐responsive element‐dependent gene expression. Plant Physiol. 161, 346–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto, K. , Ohama, N. , Kidokoro, S. , Mizoi, J. , Takahashi, F. , Todaka, D. , Mogami, J. et al. (2017) BPM‐CUL3 E3 ligase modulates thermotolerance by facilitating negative regulatory domain‐mediated degradation of DREB2A in Arabidopsis. Proc. Natl. Acad. Sci. USA, 114, E8528–E8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu, C.D. , Li, H.Y. , Jiang, L.J. , Yan, M.K. , Li, C.Y. , Geng, D.L. , Xie, Y.P. et al. (2019) Genome‐wide identification of drought‐responsive microRNAs in two sets of Malus from interspecific hybrid progenies. Hortic. Res. 6, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novatchkova, M. , Budhiraja, R. , Coupland, G. , Eisenhaber, F. and Bachmair, A. (2004) SUMO conjugation in plants. Planta, 220, 1–8. [DOI] [PubMed] [Google Scholar]
- Nurdiani, D. , Widyajayantie, D. and Nugroho, S. (2018) OsSCE1 encoding SUMO E2‐conjugating enzyme involves in drought stress response of Oryza sativa. Rice Sci. 25, 73–81. [Google Scholar]
- Okada, S. , Nagabuchi, M. , Takamura, Y. , Nakagawa, T. , Shinmyozu, K. , Nakayama, J.‐I. and Tanaka, K. (2009) Reconstitution of Arabidopsis thaliana SUMO pathways in E. coli: Functional evaluation of SUMO machinery proteins and mapping of SUMOylation sites by mass spectrometry. Plant Cell Physiol. 50, 1049–1061. [DOI] [PubMed] [Google Scholar]
- Polania, J.A. , Poschenrieder, C. , Beebe, S. and Rao, I.M. (2016) Effective use of water and increased dry matter partitioned to grain contribute to yield of common bean improved for drought resistance. Front. Plant Sci. 7, 660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, F. , Kakimoto, M. , Sakuma, Y. , Maruyama, K. , Osakabe, Y. , Tran, L. , Shinozaki, K. et al. (2007) Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. Plant J. 50, 54–69. [DOI] [PubMed] [Google Scholar]
- Qin, F. , Sakuma, Y. , Tran, L.S. , Maruyama, K. , Kidokoro, S. , Fujita, Y. , Fujita, M. et al. (2008) Arabidopsis DREB2A‐interacting proteins function as RING E3 ligases and negatively regulate plant drought stress‐responsive gene expression. Plant Cell, 20, 1693–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis, R.R. , Brito, D. , da Cunha, B.A. , Martins, P.K. , Bazzo Martins, M.T. , Alekcevetch, J.C. , Chalfun‐Junior, A. et al. (2014) Induced over‐expression of ArDREB2A CA improves drought tolerance in sugarcane. Plant Sci. 221, 59–68. [DOI] [PubMed] [Google Scholar]
- Rytz, T.C. , Miller, M.J. and Vierstra, R.D. (2016) Purification of SUMO conjugates from Arabidopsis for mass spectrometry analysis. Methods Mol. Biol. 1475, 257–281. [DOI] [PubMed] [Google Scholar]
- Rytz, T.C. , Miller, M.J. , McLoughlin, F. , Augustine, R.C. , Marshall, R.S. , Juan, Y.T. , Charng, Y.Y. et al. (2018) SUMOylome profiling reveals a diverse array of nuclear targets modified by the SUMO ligase SIZ1 during heat stress. Plant Cell, 30, 1077–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadhukhan, A. , Panda, S.K. and Sahoo, L. (2014) The cowpea RING ubiquitin ligase VuDRIP interacts with transcription factor VuDREB2A for regulating abiotic stress responses. Plant Physiol. Biochem. 83, 51–56. [DOI] [PubMed] [Google Scholar]
- Saitoh, H. , Uwada, J. and Azusa, K. (2009) Strategies for the expression of SUMO‐modified target proteins in Escherichia coli . Methods Mol. Biol. 497, 211–221. [DOI] [PubMed] [Google Scholar]
- Sakuma, Y. , Maruyama, K. , Osakabe, Y. , Qin, F. , Seki, M. , Shinozaki, K. and Yamaguchi‐Shinozaki, K. (2006a) Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought‐responsive gene expression. Plant Cell, 18, 1292–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma, Y. , Maruyama, K. , Qin, F. , Osakabe, Y. , Shinozaki, K. and Yamaguchi‐Shinozaki, K. (2006b) Dual function of an Arabidopsis transcription factor DREB2A in water‐stress‐responsive and heat‐stress‐responsive gene expression. Proc. Natl. Acad. Sci. USA, 103, 18822–18827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler, C. , Harshavardhan, V.T. , Rajesh, K. , Reddy, P.S. , Strickert, M. , Rolletschek, H. , Scholz, U. et al. (2011) ABA biosynthesis and degradation contributing to ABA homeostasis during barley seed development under control and terminal drought‐stress conditions. J. Exp. Bot. 62, 2615–2632. [DOI] [PubMed] [Google Scholar]
- Smalle, J. and Vierstra, R.D. (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55, 555–590. [DOI] [PubMed] [Google Scholar]
- Sobko, A. , Ma, H. and Firtel, R.A. (2002) Regulated SUMOylation and ubiquitination of DdMEK1 Is required for proper chemotaxis. Dev. Cell, 2, 745–756. [DOI] [PubMed] [Google Scholar]
- Srivastava, A. , Zhang, C. , Caine, R. , Gray, J. and Sadanandom, A. (2017) Rice SUMO protease Overly Tolerant to Salt 1 targets the transcription factor, OsbZIP23 to promote drought tolerance in rice. Plant J. 92, 1031–1043. [DOI] [PubMed] [Google Scholar]
- Staudinger, J.L. (2017) The molecular Interface between the SUMO and ubiquitin systems. Adv. Exp. Med. Biol. 963, 99–110. [DOI] [PubMed] [Google Scholar]
- Sun, H. , Leverson, J.D. and Hunter, T. (2007) Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 26, 4102–4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, J. , Gu, J. , Zeng, J. , Han, S. , Song, A.P. , Chen, F.D. , Fang, W.M. et al. (2013a) Changes in leaf morphology, antioxidant activity and photosynthesis capacity in two different drought‐tolerant cultivars of chrysanthemum during and after water stress. Sci. Hortic. 161, 249–258. [Google Scholar]
- Sun, X. , Wang, P. , Jia, X. , Huo, L.Q. , Che, R.M. and Ma, F.W. (2018) Improvement of drought tolerance by overexpressing MdATG18a is mediated by modified antioxidant system and activated autophagy in transgenic apple. Plant Biotechnol. J. 16, 545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X.P. , Yan, H.L. , Kang, X.Y. and Ma, F.W. (2013b) Growth, gas exchange, and water‐use efficiency response of two young apple cultivars to drought stress in two scion‐one rootstock grafting system. Photosynthetica, 51, 404–410. [Google Scholar]
- Tatham, M.H. , Geoffroy, M.C. , Shen, L.N. , Anna, P. , Hattersley, N. , Jaffray, E.G. , Palvimo, J.J. et al. (2008) RNF4 is a poly‐SUMO‐specific E3 ubiquitin ligase required for arsenic‐induced PML degradation. Nat. Cell Biol. 10, 538–546. [DOI] [PubMed] [Google Scholar]
- Vierstra, R.D. (2012) The expanding universe of ubiquitin and ubiquitin‐like modifiers. Plant Physiol. 160, 2–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virlet, N. , Costes, E. , Martinez, S. , Kelner, J.J. and Regnard, J.L. (2015) Multispectral airborne imagery in the field reveals genetic determinisms of morphological and transpiration traits of an apple tree hybrid population in response to water deficit. J. Exp. Bot. 66, 5453–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, F.G. , Liu, Y.Y. , Shi, Y.Q. , Han, D.L. , Wu, Y.Y. , Ye, W.X. , Yang, H.L. et al. (2020) SUMOylation stabilizes the transcription factor DREB2A to improve plant thermotolerance. Plant Physiol. 183, 41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H.B. , Zhao, S. , Mao, K. , Dong, Q.L. , Liang, B.W. , Li, C. , Wei, Z.W. et al. (2018) Mapping QTLs for water‐use efficiency reveals the potential candidate genes involved in regulating the trait in apple under drought stress. BMC Plant Biol. 18, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, W. , Hu, Y. , Han, Y.T. , Zhang, K. , Zhao, F.L. and Feng, J.Y. (2016) The WRKY transcription factors in the diploid woodland strawberry Fragaria vesca: Identification and expression analysis under biotic and abiotic stresses. Plant Physiol. Biochem. 105, 129–144. [DOI] [PubMed] [Google Scholar]
- Wu, S. , Liang, D. and Ma, F.W. (2014) Leaf micromorphology and sugar may contribute to differences in drought tolerance for two apple cultivars. Plant Physiol. Biochem. 80, 249–258. [DOI] [PubMed] [Google Scholar]
- Xie, Y.P. , Chen, P.X. , Yan, Y. , Bao, C.N. , Li, X.W. , Wang, L.P. , Shen, X.X. et al (2018) An atypical R2R3 MYB transcription factor increases cold hardiness by CBF‐dependent and CBF‐independent pathways in apple. New Phytol, 218(1), 201–218. 10.1111/nph.14952 [DOI] [PubMed] [Google Scholar]
- Xie, Y.P. , Bao, C.N. , Chen, P.X. , Cao, F.G. , Liu, X.F. , Geng, D.L. , Li, Z.X. et al. (2021) Abscisic acid homeostasis is mediated by feedback regulation of MdMYB88 and MdMYB124. J. Exp. Bot. 72, 592–607. [DOI] [PubMed] [Google Scholar]
- Xu, G. , Duan, B. and Li, C. (2008) Different adaptive responses of leaf physiological and biochemical aspects to drought in two contrasting populations of seabuckthorn. Can. J. For. Res. 38, 584–591. [Google Scholar]
- Yamaguchi‐shinozaki, K. and Shinozaki, K. (1994) A novel cis‐acting element in an Arabidopsis gene is involved in responsiveness to drought, low‐temperature, or high‐salt stress. Plant Cell, 6, 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yıldırım, K. and Kaya, Z. (2017) Gene regulation network behind drought escape, avoidance and tolerance strategies in black poplar (Populus nigra L.). Plant Physiol. Biochem. 115, 183–199. [DOI] [PubMed] [Google Scholar]
- Yordanov, I. , Velikova, V. and Tsonev, T. (2000) Plant responses to drought, acclimation, and stress tolerance. Photosynthetica, 38, 171–186. [Google Scholar]
- Zhang, L. , Xie, F. , Zhang, J. , Dijke, P.T. and Zhou, F.F. (2017a) SUMO‐triggered ubiquitination of NR4A1 controls macrophage cell death. Cell Death Differ. 24, 1530–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S.C. , Qi, Y.L. , Liu, M. and Yang, C.W. (2013) SUMO E3 ligase AtMMS21 regulates drought tolerance in Arabidopsis thaliana . J. Integr. Plant Biol. 55, 83–95. [DOI] [PubMed] [Google Scholar]
- Zhang, S. , Zhuang, K. , Wang, S. , Lv, J. , Ma, N. and Meng, Q. (2017b) A novel tomato SUMO E3 ligase, SlSIZ1, confers drought tolerance in transgenic tobacco. J. Integr. Plant Biol. 59, 102–117. [DOI] [PubMed] [Google Scholar]
- Zhou, L.J. , Li, Y.Y. , Zhang, R.F. , Zhang, C.L. , Xie, X.B. , Zhao, C. and Hao, Y.J. (2017) The small ubiquitin‐like modifier E3 ligase MdSIZ1 promotes anthocyanin accumulation by sumoylating MdMYB1 under low‐temperature conditions in apple. Plant Cell Environ. 40, 2068–2080. [DOI] [PubMed] [Google Scholar]
- Zhu, J.K. (2016) Abiotic stress signaling and responses in plants. Cell, 167, 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Expression patterns of apple SUMO2s.
Figure S2. Protein alignment (A) and phylogenetic analysis (B) of SUMO2A from different species.
Figure S3. Phenotypes of MdSUMO2 transgenic plants and GL‐3 under prolonged drought stress.
Figure S4. Internode length of GL‐3, MdSUMO2A OE plants, MdSUMO2A RNAi, and MdSUMO2 RNAi plants under long‐term drought conditions.
Figure S5. Dry weight of shoots of MdSUMO2 transgenic plants and GL‐3 under prolonged drought stress.
Figure S6. Photosynthesis related parameters of GL‐3, MdSUMO2A overexpression plants, MdSUMO2A RNAi, and MdSUMO2 RNAi plants under long‐term drought conditions.
Figure S7. Transpiration rate (A) and stomatal conductance (B) of MdSUMO2 transgenic plants and GL‐3 under long‐term drought stress conditions.
Figure S8. Dry weight of roots of MdSUMO2A transgenic plants and GL‐3 under control and long‐term drought stress condition.
Figure S9. The ABA content of MdSUMO2A transgenic plants and GL‐3 under control and long‐term drought stress conditions.
Figure S10. SUMOylation of MdSUMO2 transgenic plants and GL‐3 under control and long‐term drought stress conditions.
Figure S11. The putative SUMOylation lysine residues in MdDREB2A (A), MdAQP2 (B), and MdALI (C) proteins.
Figure S12. SUMOylation of MdDREB2A, MdAQP2, and MdALI using E. coli system.
Figure S13. MdDREB2A interacts with MdCE1 in vitro and vivo.
Figure S14. Effects of MdSUMO2A on the subcellular localization of MdDREB2A.
Figure S15. SUMOylation of MdDREB2A is critical for drought stress tolerance.
Figure S16. MdDREB2A interacts with MdRNF4 in vivo.
Figure S17. MdRNF4 interacts with MdSUMO2A by SIMs.
Figure S18. Expression of MdRNF4 in MdRNF4 RNAi transgenic plants.
Figure S19. Tolerance of MdRNF4 RNAi calli in response to simulated drought stress.
Figure S20. Protein alignment of DREB2A from different species.
Table S1. P‐values of Turkey's HSD test to compare plant traits among GL‐3 and transgenic plants exposed to control and long‐term drought stress conditions.
Data S1. Potential targets of MdSUMO2.
Data S2. Proteins identified in the MS/MS analysis of the wild‐type apple calli control.
Data S3. Potential interacted proteins of MdDREB2A.
Data S4. Y2H screening of MdSUMO2A in apple.
Data S5. Primers used in this study.
Appendix S1. Various tissues of Malus fusca.


