Abstract
Currently colorimetric paper lateral flow strips (PLFS) encounter two major limitations, that is, low sensitivity and severe interference from complex sample matrices such as blood. These shortcomings limit their application in detection of low-concentration analytes in complex samples. To solve these problems, a PLFS has been developed by utilizing surface-enhanced Raman scattering (SERS) for sensing signal transduction. In particular, a hierarchical three-dimensional nanostructure has been designed to create “hot spots”, which can significantly amplify the SERS sensing signal, leading to high sensitivity. As a result, this PLFS has demonstrated a limit of detection (LOD) of 5.0 pg mL−1 toward detection of S-100β, a traumatic brain injury (TBI) protein biomarker in blood plasma. The PLFS has been successfully used for rapid measurement of S-100β in clinical TBI patient samples taken in the emergency department. Availability of PLFS for blood testing would shift the paradigm of TBI patient management and clinical outcome in emergency departments. It is expected that this type of PLFS can be adapted for rapid detection of various human diseases due to its capability of measuring a low level of protein blood biomarkers in complex human fluids.
Keywords: traumatic brain injury, paper lateral flow assay, blood testing, biomarker, surface-enhanced Raman scattering, S-100β
1. Introduction
Traumatic brain injury (TBI) is sudden damage to brain due to an outside force, which causes a large number of mortality and permanent disability every year (CDC, 2020). About 27 million new cases of TBI were reported in the world in 2016 (Association, 2020). In U.S. alone, $48–56 billion are estimated for annual costs related to TBI directly and indirectly (Surgeons, 2020). About 2.87 million TBI-associated patients visited emergency departments in 2014 (CDC, 2020). A 15-point scale Glasgow Coma Score (GCS), which is based on eye opening, motor response and verbal response, is a common method for initial screening of TBI in emergency departments (Levin and Diaz-Arrastia, 2015). GCS is a subjective approach, which can cause errors in many cases. Confirmatory testing is performed on moderate or severe TBI patients with computed tomography (CT). Although magnetic resonance imaging (MRI) is capable of imaging more details than CT, MRI is not used in emergency departments due to its high cost and low efficacy in detecting bleeds and fractures. The current TBI assessment process is tedious and expensive. Blood testing based on biomarker measurement is an objective approach, which provides a TBI screening method alternative to GCS. For example, S-100β, neuron-specific enolase (NSE), myelin basic protein (MBP) and glial fibrillary acidic protein (GFAP) blood biomarkers are elevated in case of TBI (Agoston et al., 2017; Gordillo-Escobar et al., 2016; Wang et al., 2018). In particular, S-100β is a protein released into blood, which is mainly expressed from the central nervous system, mostly astroglial and neuronal cells (David et al., 2017; Thelin et al., 2017). It is considered to be a potential objective quantitative biomarker for screening of TBI and for prognosis of severe head injury (Biberthaler et al., 2006; Egea-Guerrero et al., 2012). Currently, proteins in blood are measured by enzyme-linked immunosorbent assay (ELISA) or Western Blot, which takes several hours to finish by professional staff in a central laboratory (the typical assay time is around three hours for ELISA, and about one day for Western Blot). This causes long delay in obtaining the blood test results for timely treatment of TBI patents. Therefore, there is a strong incentive to develop a portable device that can rapidly measure blood markers for TBI screening in emergency departments.
Paper-based lateral flow strips (PLFS) are characteristic of automation, miniaturization, low cost, easy operation and rapid detection of analytes (Gao et al., 2014a; Gao et al., 2016; Gao et al., 2014b; Gao et al., 2017). Pregnancy test strip is a successful example of PLFS. Currently the majority of PLFS are built on colorimetric transduction. There is no problem for them to detect a high level of analytes such as human chorionic gonadotrophin, the pregnancy biomarker in urine. However, when colorimetric PLFS are used to measure a low level of analyte in serum, plasma and whole blood samples, they suffer from poor sensitivity, large error and severe interference from sample matrices. For example, our previous study shows that colorimetric PLFS based on gold nanoparticles exhibited a limit of detection (LOD) of 0.5 ng mL−1 toward detection of IgG in a phosphate-buffered saline (PBS) solution (Gao et al., 2017). While it was used for IgG detection in a diluted blood plasma, its LOD dropped to 50 ng mL−1.
In order to solve this problem, we have utilized surface-enhanced Raman scattering (SERS) for sensing signal transduction in a PLFS (Gao et al., 2017). In SERS detection, the wavelength of laser (e.g., 785 nm) falls into the first biological transparency window (700–950 nm), minimizing auto-fluorescence, light absorption and scattering of blood matrix; and the SERS peaks possess a “fingerprint” feature. Therefore, SERS has a strong anti-interference resistance to non-specific molecules in complex sample matrices. Our previous study (Gao et al., 2017) has confirmed that the PLFS based on SERS transduction exhibited almost three orders of magnitude better than LOD of the PLFS based on colorimetric transduction. The SERS-PLFS achieved a LOD of 0.86 ng mL−1 toward detection of neuron-specific enolase (NSE) in blood plasma, which was below its cutoff value (12.4 ng mL−1) in TBI patients. As a result, the SERS-PLFS was successfully used for detection of NSE in blood plasma samples taken from clinical TBI patients in the emergency department. S-100β is another TBI protein biomarker with a cutoff value of 0.1 ng mL−1 in blood for acute TBI patients (Gordillo-Escobar et al., 2016). Because the cutoff value of S-100β is low, the SERS-PLFS developed previously (Gao et al., 2017) is incapable of blood plasma detection for clinical TBI patients. This demands development of ultrasensitive PLFS that can achieve a LOD at the pg mL−1 level.
To further improve LOD, herein a gold nano-pyramid array on a quartz substrate is incorporated into a PLFS to create “hot spots” between the SERS probes and the nano-pyramids, increasing the SERS enhancement factor. Coupling of SERS probes to the nano-pyramid array creates a hierarchical three-dimensional (3D) nanostructure to extend the SERS enhancement field in the 3D space, improving sensitivity. This can improve LOD of SERS-PLFS to the pg mL−1 level, which is five orders of magnitude better than that of colorimetric PLFS reported in the previous study (Gao et al., 2017). In short, the goal of this work is to develop an effective strategy to enable PLFS to detect protein biomarkers in blood plasma at the pg mL−1 level, which is comparable to the sensitivity of the state-of-the-art ultrasensitive ELISA. Also, the assay time for PLFS measurement is only 30 minutes, which will make blood testing an objective quantification method for rapid screening of TBI in emergency departments.
2. Material and Methods
Synthesis of the SERS probes (Figure S1) and fabrication of the gold nano-pyramid arrays were described in detail in Supporting Information. As shown in Figure 1a, the PLFS is composed of the following major components: a sample pad (17 mm long × 3 mm wide), a conjugate pad (8 mm × 3 mm), a wick/absorbent pad (17 mm × 3 mm), two nitrocellulose (NC) membranes (12.5 mm × 3 mm), and a gold nano-pyramid array on a quartz substrate (2 mm × 3 mm). Prior to assembly of the PLFS, the sample pad was saturated with a Tris-HCl buffer consisting of 0.05 M Tris-HCl, 0.15 M NaCl and 0.23% Triton X-100 at pH=8.0. The sample pad was heated in an oven at 37 °C for 2 h, and then stored in a desiccator at room temperature. The gold nano-pyramid array on the quartz substrate was functionalized with the capture antibody. All components were mounted on a common adhesive backing layer (typically an inert plastic, e.g., polyester). In particular, the quartz substrate was placed between the two nitrocellulose membranes (Figure 1). All other paper components were overlapped with 2 mm to ensure that liquid can flow uninterruptedly from the sample pad to the absorbent pad.
Figure 1.
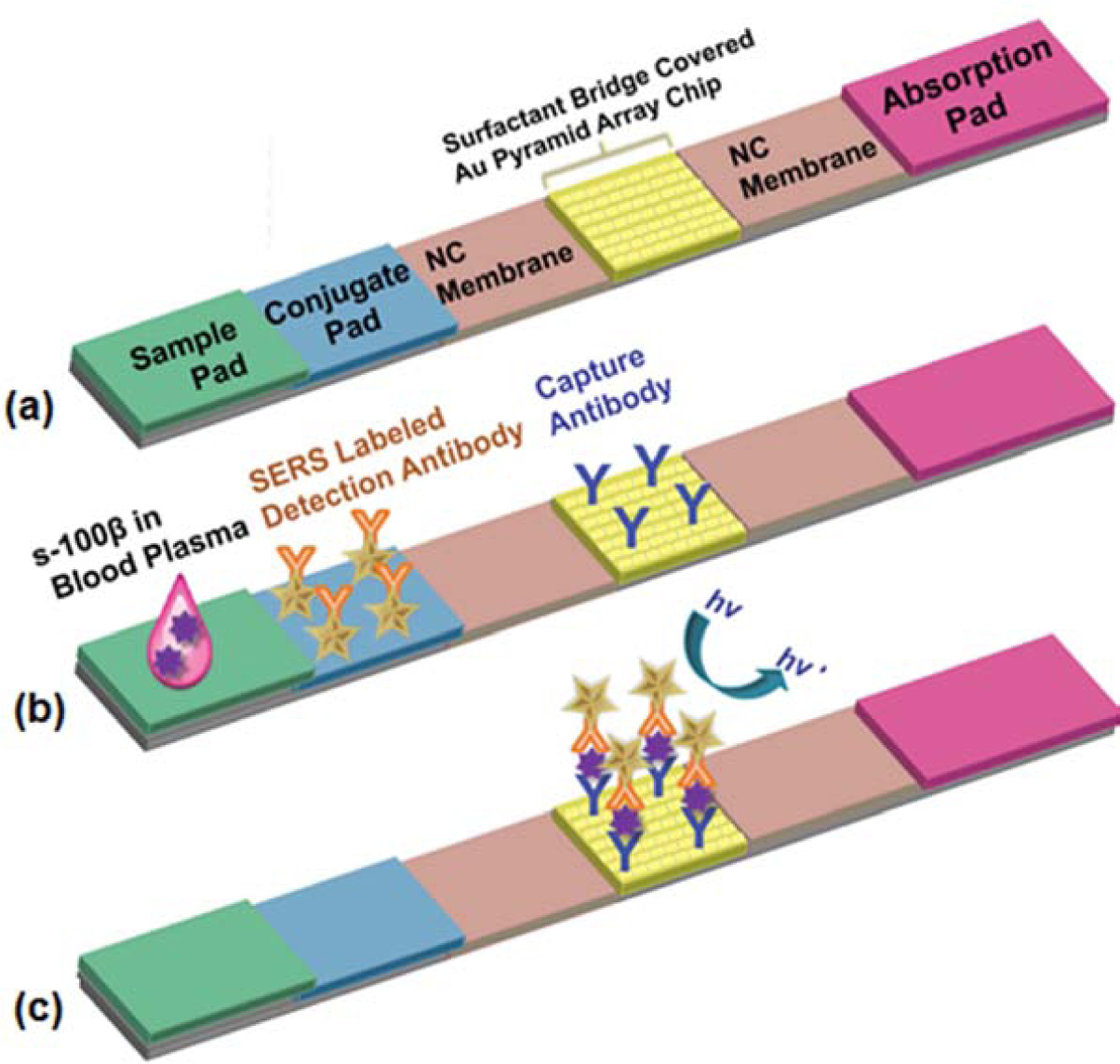
Schematic illustration of a paper-based test strip (PLFS). (a) PLFS structure, and (b) at the moment of sample loading, (c) the SERS signal readout
After the PLFS was assembled, 5 μL of solution containing the SERS probe-detection antibody complex was dropped onto the conjugate pad. The whole quartz substrate was then covered with a surfactant bridge, which consisted of Tween 20 and sucrose. Briefly, 1 μL of Tween 20 was dropped on and then spread evenly on the quartz substrate surface. 0.01 g of sucrose was then sprinkled on the Tween 20-covered surface. The surfactant bridge functionalized-substrate was dried under an ambient condition for 0.5 h. The resulting PLFS was stored at 4 °C for future use. Figure S2 and Supporting Information gave the details of preparation and functions of all components in the integrated PLFS.
When testing a sample, a small volume (100 μL) of liquid was loaded onto the sample pad, waiting for 30 min. Subsequently, Raman spectra were collected from the test zone by a portable spectrometer (B&W Tek i-Raman® Plus) under excitation by a 785 nm laser at 34 mW of power and 10 s of acquisition time. Such a low power and a short acquisition period were adopted to avoid any laser burning issue to the PLFS.
3. Results and Discussion
3.1. Operation of PLFS
Typically, 100 μL of liquid sample containing analytes was dropped onto the sample pad (Figure 1b). Liquid flowed through the conjugate pad by capillary forces. On the conjugate pad, the analyte (antigen) of interest was conjugated to the detection antibody linked to the SERS probes (Figure S1). The resulting “antigen/detection antibody-SERS probe” complex was delivered to the boundary between the nitrocellulose membrane and the quartz substrate. With assistance of the surfactant bridge, the liquid can migrate smoothly across the quartz substrate. The surfactant bridge did not block signal acquisition from the substrate surface because when liquid passed through the quartz substrate, the surfactant bridge was dissolved into liquid. Figure S3 illustrates the flowing behavior of the liquid sample when migrating along the PLFS. Owing to hydrophilicity of both the paper components and the surfactant bridge, the liquid sample flowed quickly and smoothly along the PLFS, passing through the quartz substrate covered by the surfactant bridge. As shown in Figure S3, after 15 s, liquid reached the absorption pad. After 1.5 min, the surfactant bridge was dissolved into liquid, and the capture antibody functionalized nanoarray pattern was exposed. This resulted in the formation of the “capture antibody/antigen/detection antibody-SERS probe” complex on the gold nano-pyramid array surface (Figure 1c), which was located on the test zone. Raman spectra were collected from the SERS probes captured on the test zone by a portable spectrometer. Under excitation by a 785 nm laser, the SERS signal was generated from the MBA molecules (Raman reporter) inside the SERS probes. The SERS signal was in direct proportion to the logarithmic concentration of antigen, allowing for quantification of the targeted antigen such as S-100β.
3.2. Optimization of PLFS
In order to optimize the PLFS, human IgG was chosen as model analyte. The SERS peak at 1084 cm−1 from MBA was measured in two different cases: (i) A PBS solution spiked with 10 ng mL−1 human IgG was loaded to the PLFS, and (ii) a blank PBS solution was loaded to the PLFS. The signal-to-noise (S/N) ratio was defined as the SERS peak intensity of Case (i) over Case (ii). The location of quartz substrate on the PLFS was optimized by adjusting the distance between the front edge of quartz substrate and the downstream edge of conjugation pad. Figure S4a reveals that the S/N ratio of SERS peak increased with an increase in the distance until the distance reached 11 mm. The S/N ratio dropped with further increasing the distance. This was because a longer distance gave more time for binding between the antigen and the detection antibody before the analyte reached the test zone. If the travelling distance was too long, the “antigen/detection antibody-SERS probe” complex had a higher probability in sediment in the nitrocellulose membrane before it arrived at the test zone. In addition, the amount of SERS probes on the conjugation pad was optimized by varying the volume of the “detection antibody-SERS probe” complex solution loaded. The optimal solution was determined to be 5 μL (Figure S4b). If the solution was less than 5 μL, the amount of the “detection antibody-SERS probe” complex was not enough for capture of a high level of antigens in the sample solution. The solution volume became saturated after it reached 5 μL. Moreover, the amount of the capture antibodies on the test zone was optimized by changing the concentration of capture antibody solution used for the gold nano-pyramid array functionalization, giving the optimal concertation of 1.5 mg mL−1 (Figure S4c). If capture antibody amount was not enough on the test zone, the analytes in the sample cannot be captured completely. If capture antibody amount was too high, excessive capture antibody molecules can lead to non-specific binding of the SERS probes on the test zone.
After the PLFS was optimized, the PLFS was used to measure various concentrations of S-100β spiked into the PBS solution. After 100 μL of sample solution was loaded on the sample pad, waited for 30 minutes, and the SERS signal was then recorded as shown in Figure 2a. The calibration curve was obtained by plotting the SERS peak intensity at 1084 cm−1 as a function of the logarithmic concentration of S-100β (Figure 2b). The regression equation was y = 184.3x + 570.5 with a correlation coefficient of 0.93. The LOD of PLFS was found to be 1.0 pg mL−1 in the running buffer solution, which was estimated based on three times the signal noise of the baseline.
Figure 2.
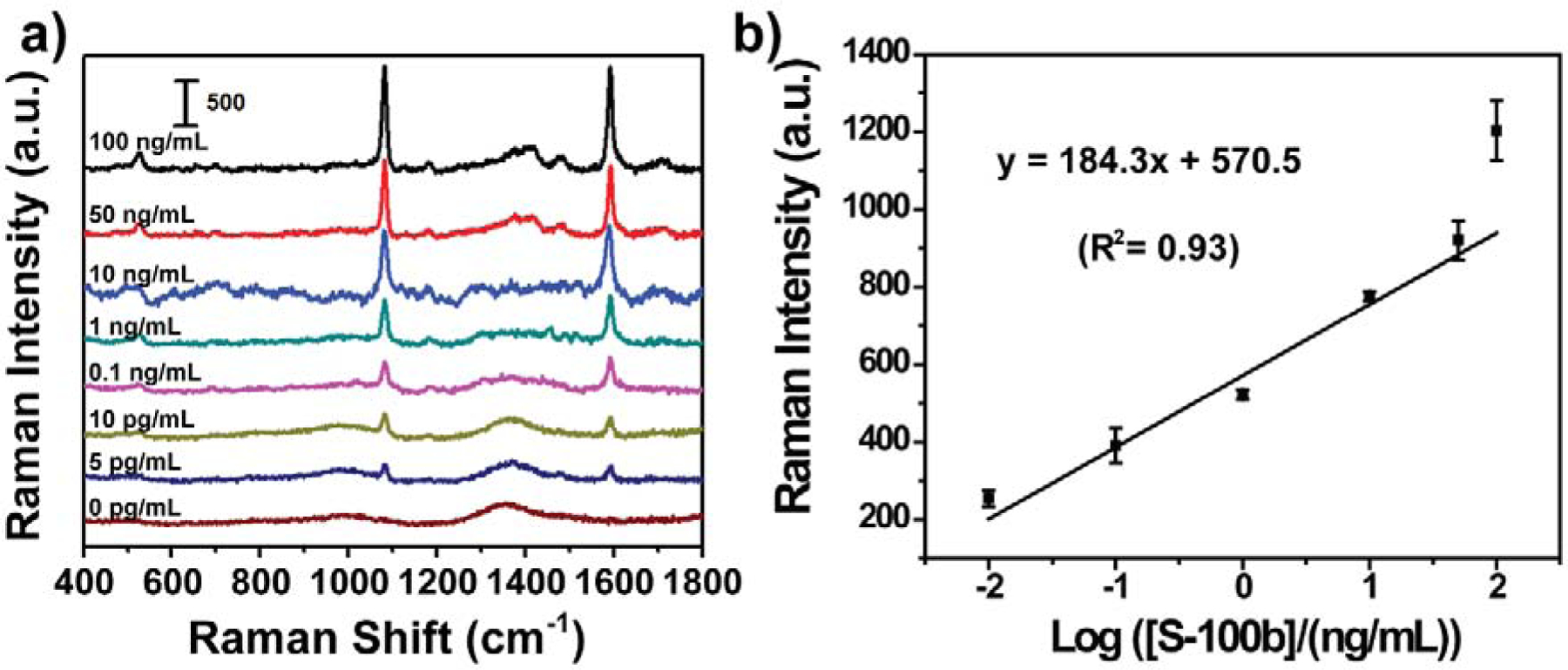
Calibration of PLFS in the buffer solution. (a) SERS spectra of PLFS acquired from the buffer solution containing various concentrations of S-100β, and (b) the calibration curve.
3.3. Measurement of S-100β in clinical blood plasma samples with PLFS
The PLFS was further calibrated with known concentrations of spiked S-100β in a mixture of 80% blood plasma and 20% PBS buffer. The blank sample was measured by an ELISA kit, and no any obvious signal was observed. Figure 3 shows the SERS spectra and the calibration curve. The SERS peak at 1084 cm−1 exhibited a linear correlation with the logarithmic concentration of S-100β in a range from 0.1 to 100 ng mL−1, which was fitted as y = 175.1x + 437.3 with the relative coefficient (R2) of 0.96. The LOD was estimated to be 5.0 pg mL−1 at the signal-to-noise ratio of 3. For normal adults, the upper limit of S-100β for intracranial damage detection is defined at 0.1 ng mL−1 based on the investigation of mild TBI (Gordillo-Escobar et al., 2016). Also, more than 0.13–0.20 ng mL−1 S-100β in plasma can be considered to be pathological. Hence the PLFS developed in this work is suitable for TBI screening.
Figure 3.
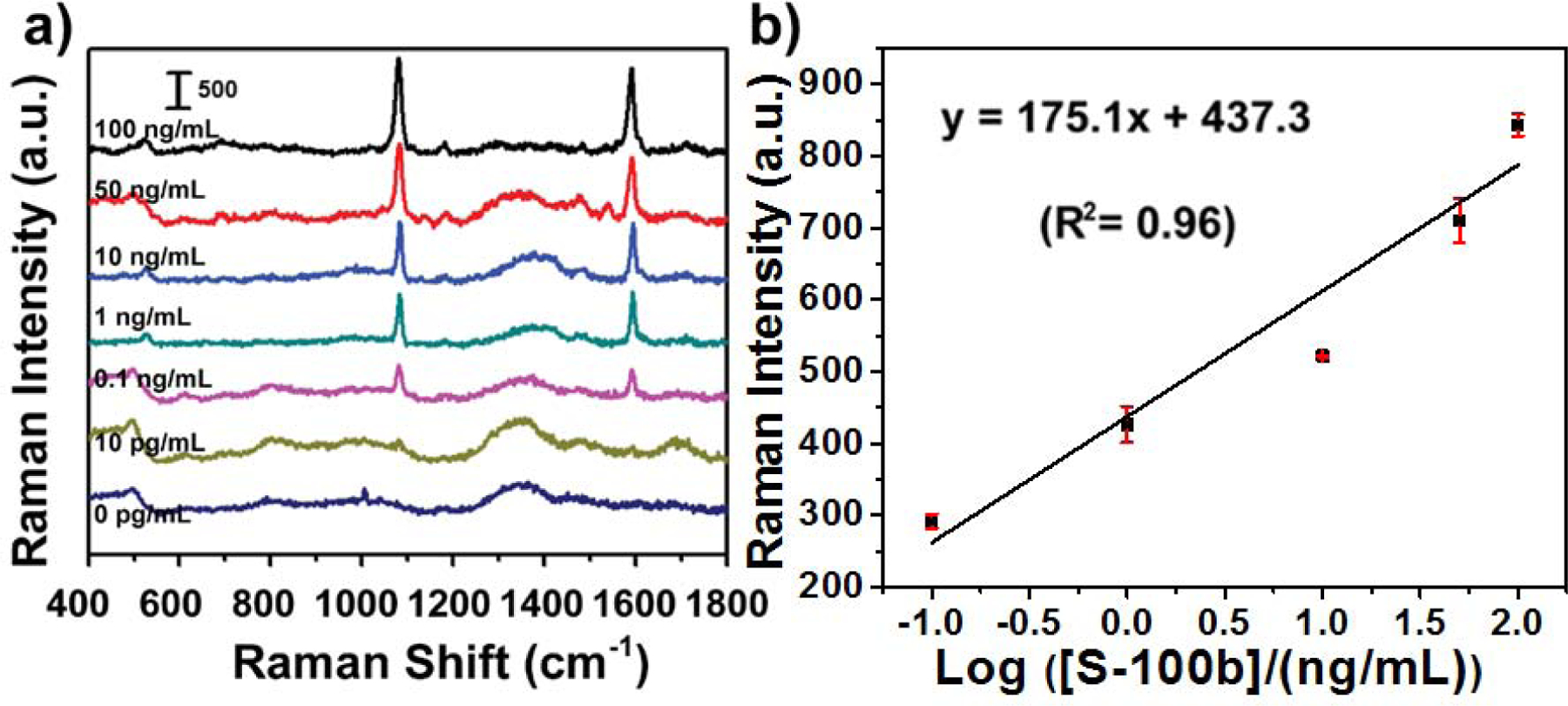
Calibration of PLFS in a mixture of 80% blood plasma and 20% PBS buffer. (a) SERS spectra of PLFS taken at various concentrations of S-100β, and (b) the calibration curve.
In addition, the selectivity tests were conducted on the PLFS in the solutions containing: (i) 1 ng mL−1 of target biomarker (S-100β); (ii) 100 ng mL−1 of interference protein biomolecules including carcinoembryonic antigen (CEA) and other traumatic brain injury biomarkers such as glial fibrillary acidic protein (GFAP) and neuron-specific enolase (NSE); and (iii) 0 ng mL−1 protein molecules. As shown in Figure S5, the obtained SERS signals from the solution containing the interference biomolecules remained as low as those obtained from the control sample, which indicated that the developed PLFS exhibited excellent selectivity toward the targeted S-100β. In addition, as shown in Figure 3a (the bottom spectrum in brown color in Figure 3a), the characteristic SERS peak at 1084 cm−1 from 4-MBA was absent in the spectrum in the absence of S-100β (0 pg mL−1) in plasma, which implied that the signals from the interference biomolecules in the complex plasma solution was negligible. The above results demonstrate that our developed PLFSs showed no cross-reactivity toward the interferencing biomolecules in the complex human fluid.
Table S1 gives comparison among the present 3D nanostructure integrated PLFS and those published previously. In the present work, the PLFS exhibited high sensitivity both in the buffer solution and in the blood plasma sample. Its LOD was reduced just a bit when the sample matrix switched from a buffer solution to blood plasma. One of the reasons is the use of gold nanostar@MBA@SiO2 sandwich nanoparticles as the SERS probes in the current PLFS (Figure 4a). In conventional colorimetric PLFS, bare gold nanospheres typically serve as the colorimetric probes. This type of colorimetric probes suffers from aggregation in sample matrices with high ionic strength, such as blood serum or plasma, which leads to low sensitivity and poor repeatability. As compared to the bare gold nanosphere-based colorimetric probes, the current SERS probes possess a thin silica outer shell (Figure S1 and 4a), which renders the SERS probes excellent water solubility and exceptional stability in blood plasma with high ionic strength, as confirmed in our previous study (Gao et al., 2017). On the other hand, bare gold nanospheres in the colorimetric PLFS, which are typically 10–30 nm in size, show the light absorption band (the localized surface plasmon resonance (LSPR)) that was overlapped with that of the blood sample matrix. In other words, the bare gold nanospheres and the blood samples exhibited the similar color, which reduces the signal-to-noise ratio. In the present work, the gold nanostar@MBA@SiO2 SERS probes show a LSPR peak at around 790 nm, and the laser excitation wavelength is 785 nm, which fall into the first biological transparency window. This minimizes auto-fluorescence and improves the signal-to-noise ratio. Furthermore, the Au nanostar acts as the SERS substrate, creating “hot spots” at the sharp tips. This achieves a SERS enhancement factor (|E/Eo|4) at a maximum of 109, as shown in the finite-difference time-domain (FDTD) simulation map in Figure 5a. Therefore, the SERS-PLFS shows higher sensitivity and better repeatability than the colorimetric PLFS when they are applied to the blood plasma samples.
Figure 4.
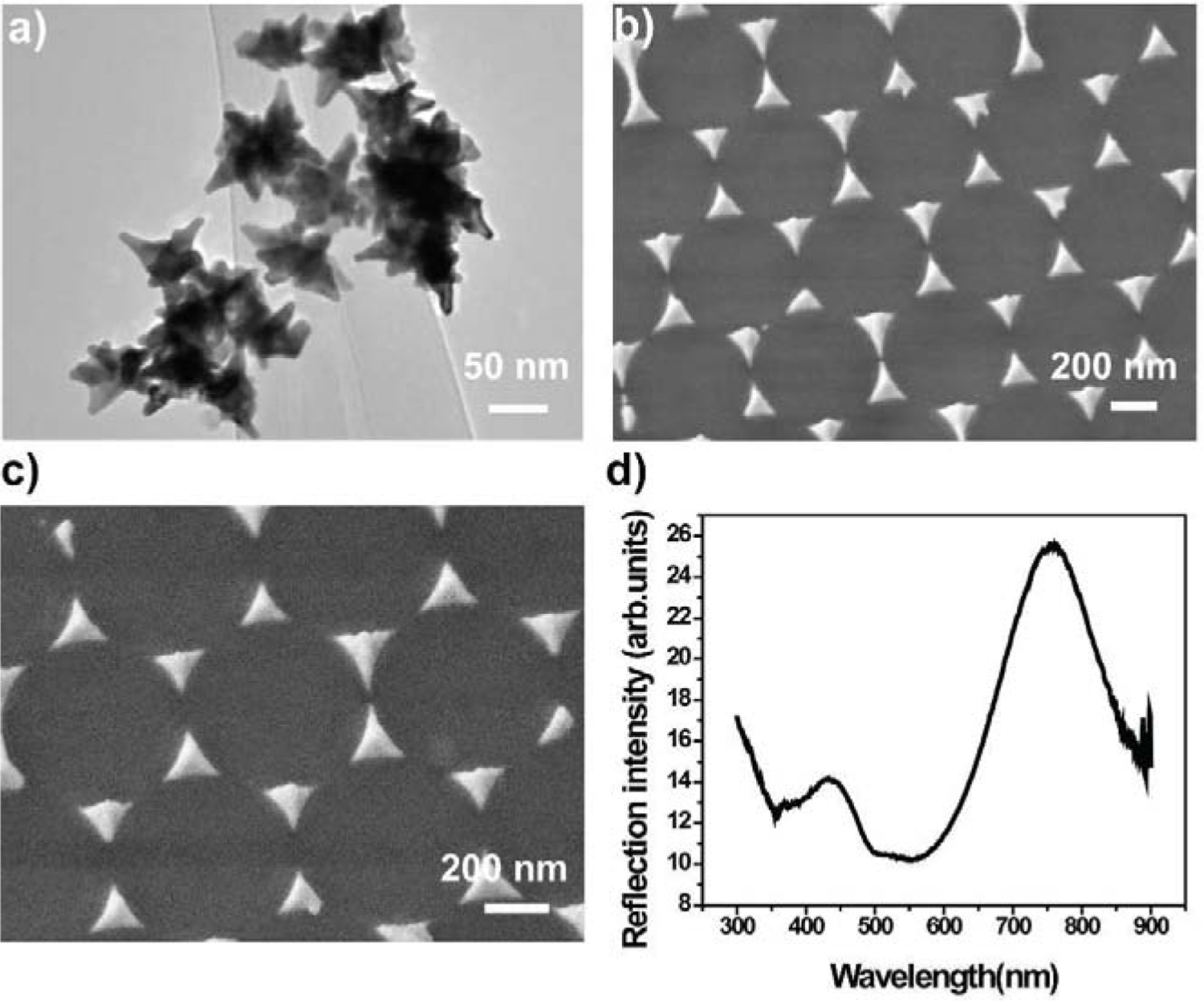
The SERS probes and the gold nano-pyramid array. (a) TEM images of the gold nanostar@MBA@SiO2 sandwich-structured SERS probes; (b) low-magnification, (c) high-magnification SEM images of the gold nano-pyramid array, and (d) UV-Visible spectrum of the gold nano-pyramid array.
Figure 5.
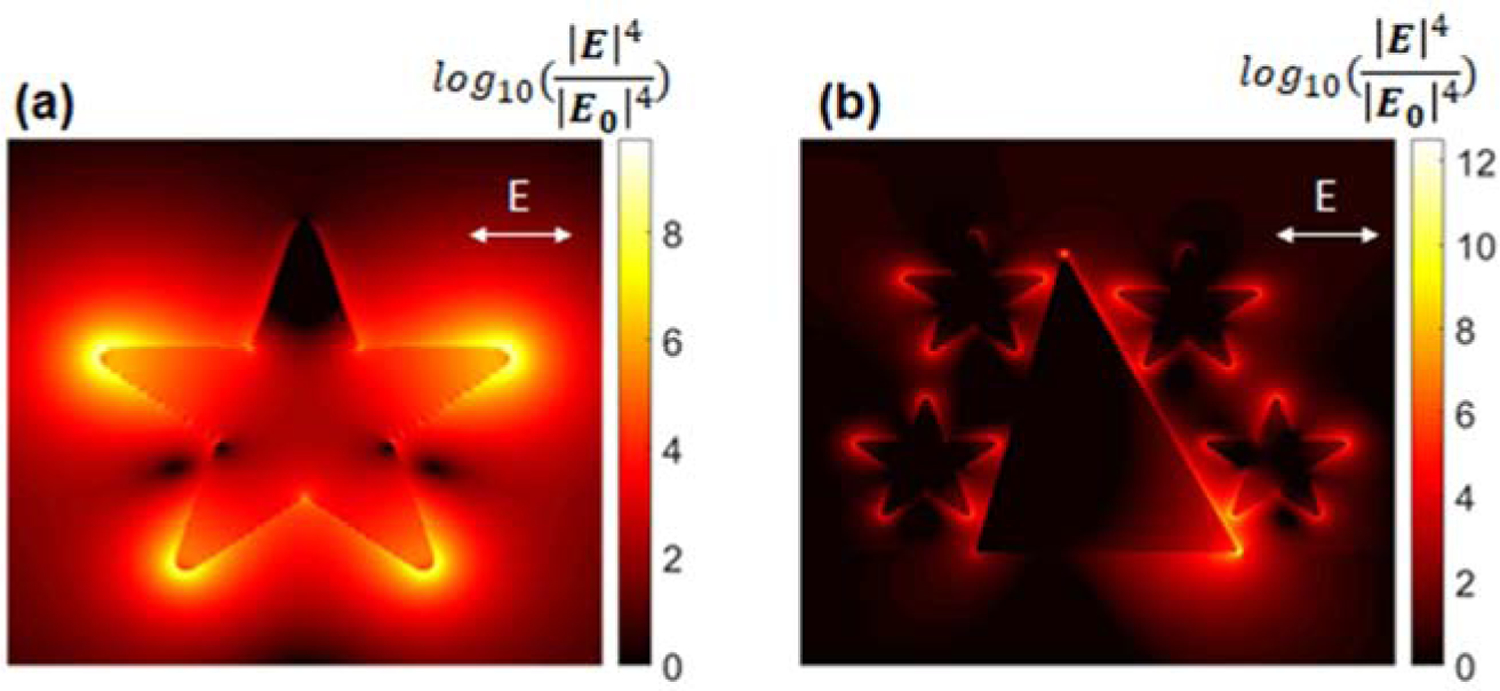
SERS enhancement field map. (a) The SERS probe alone, and (b) the SERS probes coupled to an individual nano-pyramid on the test zone of PLFS.
In addition, high sensitivity of the current PLFS is ascribed to the use of the gold nano-pyramid array pattern on the test zone (Figure 4b and 4c). When the antigen (analyte) is present on the test zone, the “capture antibody/antigen/detection antibody-SERS probe” complex is formed on the gold nano-pyramid array surface (Figure 1c), which brings one or multiple SERS probes into proximity with individual gold nano-pyramids, as shown in Figure 4b. The gap between a SERS probe and a nano-pyramid is estimated to be around 9 nm. The LSPR band of the gold nano-pyramid array centered at 773 nm (Figure 4d) is largely overlapped with that of the SERS probe centered at 790 nm. Hence strong plasmonic coupling takes place between the SERS probe and the nano-pyramid, which creates “hot spots” (Figure 5b), achieving a SERS enhancement factor (|E/Eo|4) at a maximum of 1012. Considering that there are thousands of nano-pyramids on the test zone, a hierarchical 3D nanostructure has been formed after the SERS probes are coupled to individual nano-pyramids. As a result, “hot spots” are created in this hierarchical 3D nanostructure in the extended space. This result demonstrates remarkable amplification of SERS signals, leading to high sensitivity of the PLFS.
In order to evaluate the applicability to real-world clinical sample, the developed PLFSs were employed to quantify the S-100β level in the blood plasma samples taken from clinical de-identified TBI patients who visited the emergency department (Figure 6). The commercial ELISA Kit was used to measure the same clinical samples for the sake of comparison. It can be seen from Figure 6 that the test results of PLFS are comparable to those of ELISA. Currently blood tests are performed with ELISA. The turnaround time for obtaining the blood test results requires several hours or even more than one day in emergency departments. This impedes fast screening of TBI for the suspended visitors in emergency departments. The PLFS developed in this work can quantify the TBI protein biomarker in a small volume of clinical blood plasma samples by nurses or other laypersons within 30 minutes. The use of the PLFS will enable rapid blood testing in emergency departments, allowing for fast screening of TBI. For the patients with the positive blood test results from PLFS, CT scan would be performed to confirm the results, which will provide useful information for medical treatment. In short, implementation of PLFS will shorten the blood testing time, reduce the unnecessary radiation exposure to CT scan, and save the costs for suspended TBI visitors in emergency departments.
Figure 6.
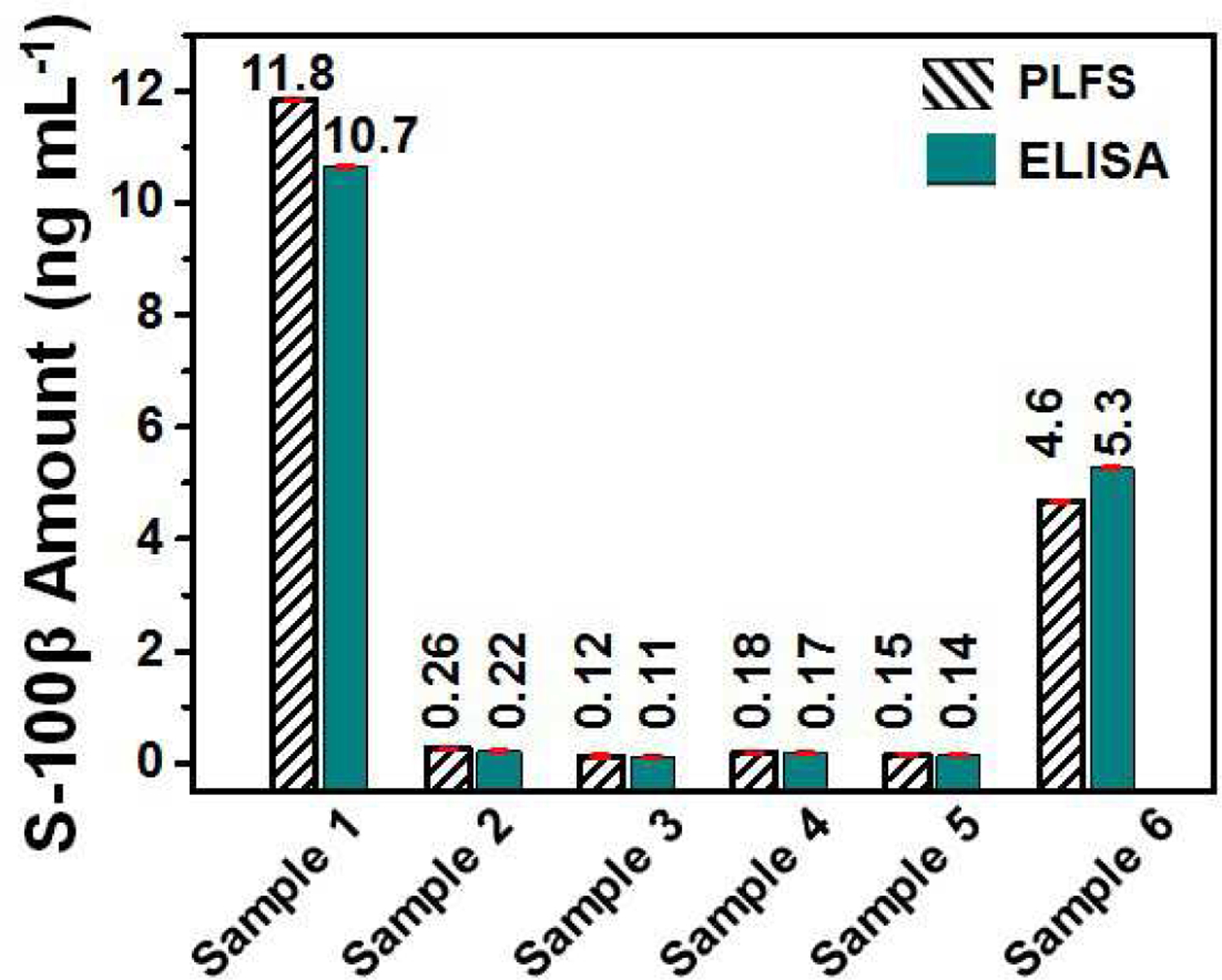
Comparative evaluation of PLFS with ELISA. The S-100β concentrations (in ng mL−1) in blood plasma samples taken from clinical TBI patients, which were measured by PLFS and ELISA, respectively.
Commercial colorimetric PLFS is inexpensive, which typically costs $0.8~$2 for each PLFS (Fabres-Klein et al., 2014; Rohrman et al., 2012). However, these PLFSs are unable to detect a low level of protein biomarkers at the pg mL−1 level in blood plasma, as discussed in the previous sections. To solve these problems, the sandwiched SERS probes, the plasmonic nanoarray pattern and the SERS spectrometer are used in the current portable detection system. This has improved the sensitivity dramatically. On the other hand, this will increase the cost to around $5 for each disposable PLFS base on our rough estimation if PLFSs are produced massively. However, it is still much more inexpensive than ELISA (typically $110–150 for each protein testing), the current gold stand method for protein measurement.
4. Conclusions
In summary, protein biomarkers play important roles in diagnosis and prognosis of various diseases. Many of protein biomarkers are at the low level in blood. This demands the development of portable devices for rapid and sensitive measurement of protein biomarkers. In this work, a hierarchical 3D nanostructure was developed on the test zone in the PLFS. When the targeted protein molecules were present on the test zone, the star-shaped SERS probes were brought close to the gold nano-pyramid array due to the antigen-antibody interaction, which created “hot spots” in the hierarchical 3D structure, leading to remarkable amplification of SERS signals. As a result, the PLFS exhibited a LOD of 5.0 pg mL−1 toward S-100β detection in blood plasma, which was comparable to the LOD of the start-of-the-art ultrasensitive ELISA kits. The PLFS was also successfully employed for rapid quantitative analysis of real-world clinical TBI patient samples, which showcased the advantages of the device. Implementation of the developed PLFS for blood testing would change the clinical TBI patient management in emergency departments. Because this PLFS can be used to quantify a low level of protein biomarker in blood by a layperson within 30 minutes, it can be adapted for early detection of various human diseases.
Supplementary Material
Highlights:
A paper biosensor for the detection of protein biomarker in blood plasma is proposed.
A hierarchical three-dimensional (3D) plasmonic nanostructure is developed and integrated into the paper biosensor to improve the detection sensitivity.
A two-order of magnitude improvement in detection sensitivity is obtained when the hierarchical three-dimensional (3D) plasmonic nanostructure is employed.
The proposed biosensor enables protein biomarker detection from clinical plasma samples and the test results are comparable to those by the standard ELISA.
The proposed paper biosensor provides guidance for rational design of highly sensitive and anti-interference point-of-care devices
Acknowledgments
This work was partially supported by the National Institute of Neurological Disorders and Stroke of the National Institutes of Health (R15NS087515). The views and opions in this aritcle presented in this article do not necessarily reflect those of NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
No competing financial interest was declared by the authors.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References:
- (CDC), C.f.D.C.a.P., 2020. Traumatic Brain Injury & Concussion.
- Agoston DV, Shutes-David A, Peskind ER, 2017. Brain Inj 31(9), 1195–1203. [DOI] [PubMed] [Google Scholar]
- Association AS-L-H, 2020. Traumatic Brain Injury in Adults.
- Biberthaler P, Linsenmeier U, Pfeifer K-J, Kroetz M, Mussack T, Kanz K-G, Hoecherl EF, Jonas F, Marzi I, Leucht P, 2006. Shock 25(5), 446–453. [DOI] [PubMed] [Google Scholar]
- David A, Mari C, Vignaud F, Masson D, Planche L, Bord E, Bourcier R, Frampas E, Batard E, Desal H, 2017. Diagn. Interv. Imaging 98(7–8), 551–556. [DOI] [PubMed] [Google Scholar]
- Egea-Guerrero J, Revuelto-Rey J, Murillo-Cabezas F, Munoz-Sanchez M, Vilches-Arenas A, Sanchez-Linares P, Dominguez-Roldan J, Leon-Carrion J, 2012. Brain Inj 26(1), 76–82. [DOI] [PubMed] [Google Scholar]
- Fabres-Klein MH, Aguilar AP, Silva MP, Silva DM, Ribon AO, 2014, Eur. J Clin. Microbiol. Infect Dis 33(12), 2095–104. [DOI] [PubMed] [Google Scholar]
- Gao X, Xu H, Baloda M, Gurung AS, Xu L-P, Wang T, Zhang X, Liu G, 2014a. Biosens. Bioelectron 54, 578–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Xu L-P, Wu T, Wen Y, Ma X, Zhang X, 2016. Talanta 146, 648–654. [DOI] [PubMed] [Google Scholar]
- Gao X, Xu L-P, Zhou S-F, Liu G, Zhang X, 2014b. Am. J. Biomed. Sci 6(1). [Google Scholar]
- Gao X, Zheng P, Kasani S, Wu S, Yang F, Lewis S, Nayeem S, Engler-Chiurazzi EB, Wigginton JG, Simpkins JW, 2017. Anal. Chem 89(18), 10104–10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordillo-Escobar E, Egea-Guerrero J, Rodríguez-Rodríguez A, Murillo-Cabezas F, 2016. Med. Intensive 40(2), 105–112. [DOI] [PubMed] [Google Scholar]
- Levin HS, Diaz-Arrastia RR, 2015. Lancet Neurol 14(5), 506–517. [DOI] [PubMed] [Google Scholar]
- Rohrman BA, Leautaud V, Molyneux E, Richards-Kortum RR, 2012, PLoS One, 7(9), e45611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surgeons A.A.o.N., 2020. Traumatic Brain Injury
- Thelin EP, Nelson DW, Bellander B-M, 2017. Acta Neurochir 159(2), 209–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KK, Yang Z, Zhu T, Shi Y, Rubenstein R, Tyndall JA, Manley GT, 2018. Expert Rev. Mol. Diagn 18(2), 165–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


