Abstract
The objective of this study was to build a machine learning model that can predict healing of diabetes-related foot ulcers, using both clinical attributes extracted from electronic health records (EHR) and image features extracted from photographs. The clinical information and photographs were collected at an academic podiatry wound clinic over a three-year period. Both hand-crafted color and texture features and deep learning-based features from the global average pooling layer of ResNet-50 were extracted from the wound photographs. Random Forest (RF) and Support Vector Machine (SVM) models were then trained for prediction. For prediction of eventual wound healing, the models built with hand-crafted imaging features alone outperformed models built with clinical or deep-learning features alone. Models trained with all features performed comparatively against models trained with hand-crafted imaging features. Utilization of smartphone and tablet photographs taken outside of research settings hold promise for predicting prognosis of diabetes-related foot ulcers.
Keywords: Diabetic foot ulcers, Machine learning, Image processing, Photographs, Clinical prediction model, Electronic health records
1. Introduction
Diabetes-related foot ulcers (DFU) remain a significant cause of increased morbidity in patients with diabetes. Recent studies suggest that a diabetic patient’s lifetime incidence of developing a DFU may be as high as 25% [1], and greater than 85% of amputations in diabetic patients are preceded by a foot ulcer [2]. The current prognostication of the DFUs are mainly assessed by the clinicians based on various clinical characteristics of the patients, including the infection status of the wounds, comorbidities, and information obtained from imaging modalities, which requires a great deal of mental and manual labor of clinicians. To reduce such burden, several technologies have been developed for management of DFUs. For example, a few smartphone applications were created for image capturing and analysis, such as wound area determination and healing score evaluation [3,4]. Deep learning approaches, given its widespread success in medical image segmentation [5,6], have been utilized in several analyses of diabetic foot ulcer images as well: Gamage et al. [7] applied low-rank matrix factorization method on features extracted by convolutional neural network (CNN)-based architecture to predict six different severity stages of the DFUs, achieving accuracy of over 96%. Wound area segmentation from photographs using CNN is another well-explored area for DFU studies [8,9]. Incorporation of such tools and techniques into a telemedicine system is another area that’s actively pursued in management of DFUs [10,11].
In this study, we built machine learning models with the imaging biomarkers extracted from the wound photographs and the clinical attributes extracted from the electronic health records (EHR) to predict eventual healing of the DFUs, introducing additional potential machine learning tool in management and prognostication of DFUs. Identification of the wounds that would heal upon appropriate treatments is important in wound healing prognostication as it can save a patient from potential limb loss. While many studies have utilized machine learning techniques with wound photographs for diagnosis and staging, the prognosticative prediction of eventual wound healing has been limited to usage of clinical and laboratory markers [12,13].
DFU’s have demonstrated clinical features of the wound that can predict healing or infection; for example, prior work has demonstrated that the rate of wound surface area changes over time is highly predictive of healing [14]. A study by Valenzueula-Silva et al. found that wound base granulation tissue >50% at 2 weeks, >75% at the end of treatment, and 16% reduction in area had >70% positive predictive value for healing [15]. Such features of the wounds, as well as the advancement of image analysis techniques discussed earlier, hold promise for smartphone and tablet camera technologies to advance telemedicine and clinical prognostication of the DFU. The aim of this study is to build prediction models for wound healing using hand-crafted and deep learning-based imaging biomarkers extracted from the ulcer photographs as well as clinical attributes obtained during the clinic visits.
2. Research design and methods
2.1. Clinical setting
The electronic health records (EHR) of 2291 visits for 381 ulcers from 155 patients who visited the Michigan Medicine Podiatry and Wound clinic for diabetic foot ulcers and/or complications thereof from November 2014 to July 2017 were collected from its EHR system, along with smartphone or tablet photographs of the ulcers taken by the medical staff during each visit. Each ulcer was followed further down the EHR to assess for healing.
2.2. Clinical data
A total of 48 clinical attributes were extracted from the EHR. The entire list of clinical features is available in Supplementary Table 1 along with the % of the missing data that had to be imputed. The missing clinical parameters were imputed as the Euclidean distance-weighted mean of the three most similar data points (n = 3) using the k-nearest neighbor (k-NN) algorithm [16]. All of the non-numeric features were discretized – for example, categorical features, such as total contact cast or offload use, were binarized with one-hot encoding, and the University of Texas San Antonio (UTSA) foot ulcer grading system was separated into the score and grade. The estimated glomerular filtration rate (eGFR) was discretized into chronic kidney disease (CKD) stages, with a score of 0 given to CKD stage 1 and 2 because eGFR greater than 60 were right-censored in the dataset. However, the magnetic resonance imaging (MRI) signs of infection, Technetium-99 scan signs of infection, toe systolic pressure, and ankle systolic pressure were binarized into whether they were obtained at all or not (rather than imputing the unobtained values), since they were not measured in greater than 75% of the visits and the fact that they were obtained suggests its clinical significance to the treating clinician.
2.3. Image processing
The foot ulcers were manually segmented from the photograph by a podiatry fellow and a trained medical student using either Microsoft Paint or Adobe Photoshop (Fig. 1). The images were then processed using MATLAB R2019a to extract color and texture features. The radiomic texture texture features were extracted using a toolkit provided in Ref. [17]. The entire list of hand-crafted image-related features is presented in Supplementary Table 2. In addition, 2048 deep learning-based features were extracted from the global average pooling (GAP) layer of ResNet50 [18]: The network was initially loaded with the pre-trained weights from ImageNet. The manually segmented wound region was resized to (224, 224) pixels, which ResNet50 can take in as an input. Then each value output by the 2048 nodes in the GAP layer upon the input of each wound image was used as a feature.
Fig. 1.
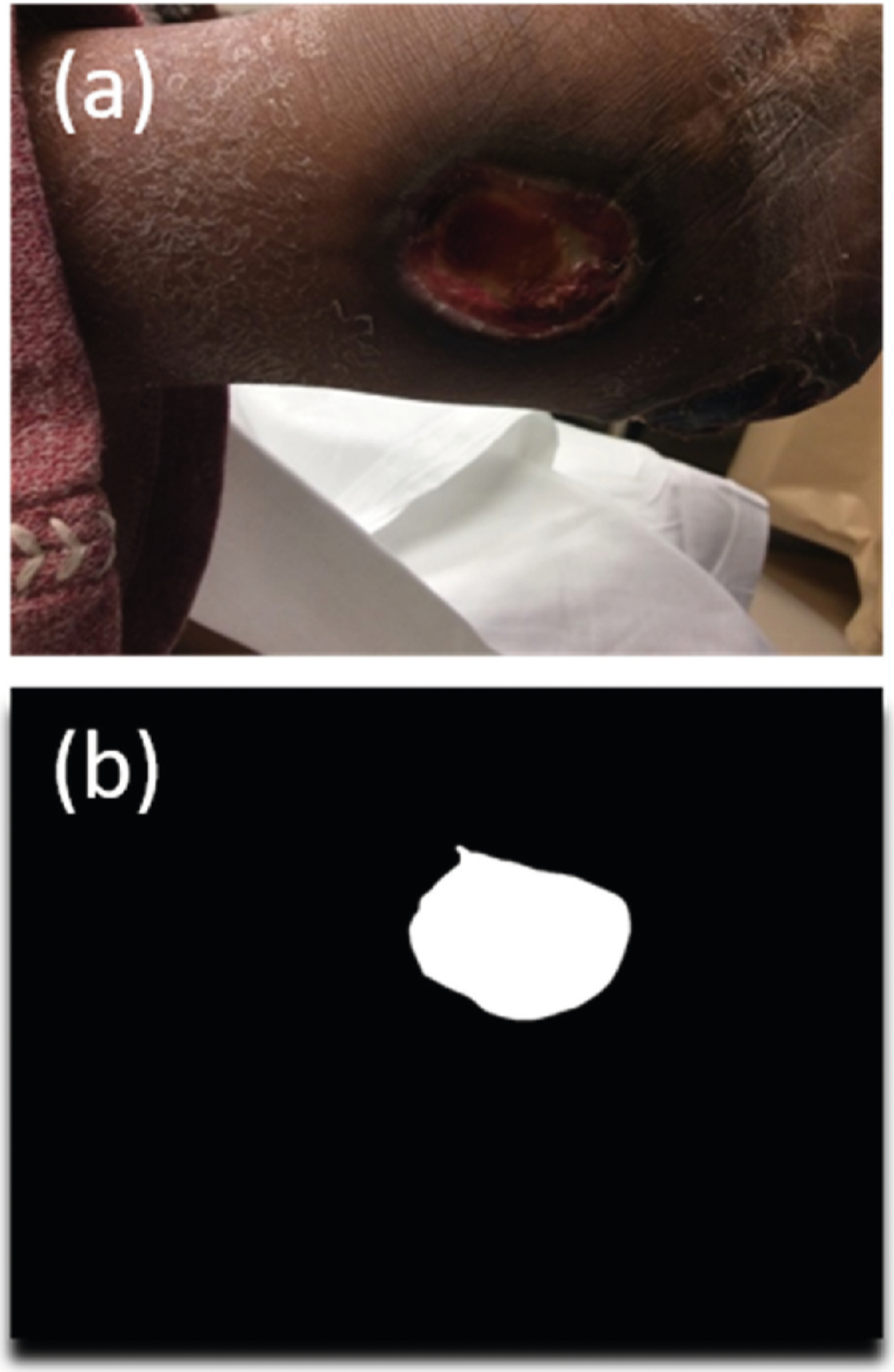
Example of manual wound segmentation. (a) Original wound image. (b) Segmented wound.
2.4. Prediction models
Two kinds of machine learning models, namely random forests (RF) [19] and support vector machine (SVM) [20] with an RBF kernel, were built to predict whether or not the wound would eventually heal. Outcomes including outpatient infection leading to hospitalization and/or amputation were considered “Not healed,” and complete epithelization was considered “Healed” – However, if a previously infected ulcer did eventually result in complete epithelization, then it was excluded from our study. In addition, those lost to follow up were entirely excluded from the study. If an ulcer previously marked as healed re-ulcerated at the same location, the recurrence event was excluded from the study. Any ulcer without an image at the initial visit was also excluded. This resulted in 208 wounds from 113 patients. Among these wounds, 25% were held out as the final test set and 3-fold cross validation was performed on the remaining 75% of the data as the training set for hyperparameter selection. Wounds from the same patient remained in the same fold throughout the entire analysis pipeline to ensure no data leakage between the folds and between the training and test sets.
Randomized grid-search was performed on the training set with 3-fold cross validation to find the model with the best area under the receiver operating characteristic (AUROC) curve, and the hyperparameter combination that produced the best AUROC during the validation was used to predict the outcomes in the test set. For the RF model, 2000 combinations of hyperparameters were included, including the use of bootstrapping, tree selection criteria (Gini impurity or entropy), the maximum depth, the minimum number of samples in a leaf, the minimum number of samples required to split a node, and the number of principal components in PCA. For the SVM model, 2028 combinations of hyperparameters were included, including C, the penalty parameter of the error term; gamma, the kernel coefficient; and the number of principal components in PCA. To accelerate the training process, the best hyperparameter combination was searched for among 300 randomly selected hyperparameter combinations for each model. The entire hyperparameter sets are available in Supplementary Table 3.
Furthermore, a separate RF model trained from clinical and hand-crafted image features alone without the PCA (since the deep learning features cannot be interpreted in semantic terms) from the training set to obtain feature importance values and shed light on which features had most impact on prediction.
3. Results
Table 1 shows the patient characteristics of the two groups based on whether or not the wound healed eventually. Of the 48 clinical features, 12 features were found to have a statistically significant difference (P < 0.05) as determined by an unpaired Student’s t-test.
Table 1.
Patient Characteristics. The values in the Healed and Not Healed Column indicates the mean value of the data points. The P-value was determined by Student’s unpaired t-test with unequal variance.
| Feature Name | Healed (n = 164) | Not Healed (n = 44) | P-value |
|---|---|---|---|
| Wound length | 0.788 | 0.823 | 0.817 |
| Wound width | 1.063 | 1.712 | 0.004 |
| Wound depth | 0.306 | 0.434 | 0.053 |
| Foot | 0.556 | 0.666 | 0.129 |
| Age | 60.458 | 58.594 | 0.387 |
| Gender | 0.347 | 0.219 | 0.052 |
| Ethnicity_Hispanic | 0.007 | 0.016 | 0.613 |
| CCI | 7.139 | 6.438 | 0.216 |
| DR | 0.174 | 0.234 | 0.330 |
| ALC | 1.69E+09 | 1.71E+09 | 0.822 |
| HbA1c | 8.370 | 8.569 | 0.537 |
| Albumin | 3.971 | 3.890 | 0.180 |
| Pre-Albumin | 22.534 | 25.698 | 0.010 |
| CKD Stage | 0.713 | 1.171 | 0.022 |
| CRP | 3.081 | 3.984 | 0.205 |
| ESR | 41.873 | 51.001 | 0.047 |
| Infection | 0.288 | 0.516 | 0.002 |
| PTB | 0.039 | 0.208 | 0.001 |
| X-ray | 0.102 | 0.150 | 0.296 |
| MRI | 0.083 | 0.219 | 0.020 |
| tc99 | 0.007 | 0.031 | 0.294 |
| DP_measured | 0.850 | 0.830 | 0.690 |
| PT_measured | 0.837 | 0.808 | 0.579 |
| AnkSys_measured | 0.104 | 0.266 | 0.010 |
| ToeSys_measured | 0.125 | 0.266 | 0.026 |
| TcPO2 | 0.000 | 0.063 | 0.045 |
| BSA | 2.843 | 3.398 | 0.491 |
| BMI | 35.175 | 33.909 | 0.297 |
| TCC | 0.033 | 0.005 | 0.058 |
| Offload | 0.395 | 0.352 | 0.513 |
| Immunosuppressants | 0.049 | 0.078 | 0.443 |
| Oral Steroids | 0.368 | 0.250 | 0.084 |
| Antihypertensives | 0.722 | 0.828 | 0.082 |
| Oral Hypoglycemics | 0.431 | 0.375 | 0.453 |
| Canagliflozin | 0.014 | 0.000 | 0.158 |
| Insulin | 0.694 | 0.750 | 0.407 |
| Heparin | 0.056 | 0.094 | 0.359 |
| Allopurinol | 0.076 | 0.063 | 0.713 |
| NSAIDs | 0.563 | 0.594 | 0.676 |
| ASA | 0.528 | 0.531 | 0.963 |
| Coumadin | 0.146 | 0.078 | 0.133 |
| Xa inhibitors | 0.000 | 0.000 | Undefined |
| Race_African | 0.111 | 0.234 | 0.041 |
| Race_Asian | 0.007 | 0.047 | 0.151 |
| Race_Caucasian | 0.875 | 0.703 | 0.008 |
| Race_Other | 0.007 | 0.016 | 0.613 |
| UTSA Stage | 1.155 | 1.274 | 0.069 |
| ischemia | 0.343 | 0.316 | 0.545 |
CCI: Charlson comorbidity index. DR: Diabetic retinopathy. ALC: Absolute lymphocyte count. CKD: Chronic Kidney Disease. CRP: C-reactive protein. ESR: Erythrocyte sedimentation rate. PTB: Probe to bone test. Tc99: Technetium 99 bone scan. DP: Dorsalis pedis pulse. PT: Posterior tibial pulse. TcPO2: Transcutaneous oxygen pressure. BSA: Body surface area. BMI: Body mass index. TCC: Total contact cast use. NSAID: Nonsteroidal anti-inflammatory drug. ASA: Aspirin.
Table 2 shows the precision, recall, F1 score, accuracy and AUROC on the test set for the SVM and RF models chosen based on the best AUROC obtained on the training set. The P-values for the significant difference between the AUROCs were obtained by using Nadeau-Bengio corrected t-test [21,22] on 500 random subsamples of the test set (Table 3). Each random subsample was created by randomly choosing, with replacement, 90% of the entries in the test set.
Table 2.
Performance of each model, with varying usage of features. The values for validation sets are Mean ± Standard deviation from the 3 cross validations.
| Models built with all available features | |||||
|---|---|---|---|---|---|
| Metric | AUROC | Accuracy | Precision | Recall | F1 |
| RF_val | 0.691 ± 0.075 | 0.671 ± 0.064 | 0.743 ± 0.042 | 0.823 ± 0.096 | 0.777 ± 0.043 |
| RF_test | 0.734 | 0.811 | 0.828 | 0.923 | 0.873 |
| SVM_val | 0.735 ± 0.083 | 0.726 ± 0.066 | 0.780 ± 0.061 | 0.854 ± 0.053 | 0.814 ± 0.044 |
| SVM_test | 0.734 | 0.811 | 0.828 | 0.923 | 0.873 |
| Models built only with hand-crafted image features | |||||
| RF_val | 0.683 ± 0.042 | 0.646 ± 0.033 | 0.795 ± 0.033 | 0.671 ± 0.033 | 0.727 ± 0.017 |
| RF_test | 0.760 | 0.811 | 0.852 | 0.885 | 0.868 |
| SVM_val | 0.691 ± 0.046 | 0.561 ± 0.037 | 0.840 ± 0.074 | 0.471 ± 0.032 | 0.600 ± 0.015 |
| SVM_test | 0.794 | 0.784 | 0.909 | 0.769 | 0.833 |
| Models built only with clinical features | |||||
| RF_val | 0.693 ± 0.056 | 0.707 ± 0.025 | 0.757 ± 0.042 | 0.875 ± 0.104 | 0.805 ± 0.026 |
| RF_test | 0.636 | 0.784 | 0.765 | 1 | 0.867 |
| SVM_val | 0.701 ± 0.086 | 0.591 ± 0.084 | 0.810 ± 0.119 | 0.567 ± 0.136 | 0.652 ± 0.094 |
| SVM_test | 0.657 | 0.703 | 0.800 | 0.769 | 0.784 |
| Models built only with deep learning features | |||||
| RF_val | 0.692 ± 0.051 | 0.720 ± 0.055 | 0.764 ± 0.070 | 0.880 ± 0.025 | 0.815 ± 0.033 |
| RF_test | 0.670 | 0.757 | 0.793 | 0.885 | 0.836 |
| SVM_val | 0.726 ± 0.052 | 0.707 ± 0.068 | 0.737 ± 0.069 | 0.912 ± 0.033 | 0.813 ± 0.044 |
| SVM_test | 0.670 | 0.757 | 0.793 | 0.885 | 0.836 |
Table 3.
P-values obtained by Nadeau-Bengio corrected t-test [11,12] on 500 random subsamples of the test set between different models.
| Model type | Features used | Features used | P-value |
|---|---|---|---|
| RF | All | Hand-crafted image | 0.515 |
| RF | All | Deep learning | 0.047 |
| RF | All | Clinical | 0.085 |
| RF | Deep learning | Hand-crafted image | 0.013 |
| RF | Clinical | Hand-crafted image | 0.562 |
| RF | Deep learning | Clinical | 0.562 |
| SVM | All | Hand-crafted image | 0.280 |
| SVM | All | Deep learning | 0.110 |
| SVM | All | Clinical | 0.086 |
| SVM | Deep learning | Hand-crafted image | 0.013 |
| SVM | Clinical | Hand-crafted image | 0.031 |
| SVM | Deep learning | Clinical | 0.780 |
The feature importance chart obtained from the RF model without the PCA and deep learning-based features is presented in Table 4.
Table 4.
The 30 most important features as determined by a RF model without PCA, using only hand-crafted image and clinical features (i.e. without deep learning features). The sum of importances of all features equals 1.
| Feature name | Feature importance value |
|---|---|
| img_LAB_Amean | 0.034231 |
| img_LAB_Lstd | 0.033047 |
| img_RGB_Rstd | 0.032891 |
| Pre_Alb | 0.027744 |
| img_HSV_Vstd | 0.023306 |
| img_RGB_Gmean | 0.021559 |
| img_HSV_Sstd | 0.018536 |
| img_HSV_Smean | 0.017932 |
| img_RGB_Gstd | 0.017794 |
| img_glrlm_SRHGE | 0.017419 |
| PTB | 0.017041 |
| Infection | 0.017015 |
| img_LAB_Bstd | 0.016309 |
| img_RGB_Bmean | 0.015348 |
| BSA | 0.014811 |
| img_glrlm_LGRE | 0.012993 |
| img_gabor_amp_std_4 | 0.012843 |
| WndWdth | 0.012599 |
| img_glrlm_RLV | 0.01215 |
| img_ngtdm_Complexity | 0.011454 |
| img_glszm_GLV | 0.011221 |
| img_HSV_Hmean | 0.011206 |
| img_glszm_LGZE | 0.01048 |
| img_glcm_Correlation | 0.010403 |
| Age | 0.010116 |
| img_gabor_amp_mean_4 | 0.010031 |
| img_RGB_Bstd | 0.009844 |
| img_glcm_Variance | 0.00958 |
| UTSAStage | 0.009379 |
| img_glrlm_LRLGE | 0.009364 |
Std: Standard deviation. SRHGE: Short-run high gray-level run emphasis Amp: Amplitude. Std: Standard deviation. LAB: CIELAB color space. RGB: RGB color space. HSV: HSV color space. GLRLM: Gray-Level Run Length Matrix. SRHGE: Short-run high gray-level run emphasis. PTB: Probe to bone, BSA: Body surface area. LGRE: Low gray-level run emphasis. GLV: Gray-level variance. RLV: Run level variance. NGTDM: Neighborhood Gray Tone Difference Matrix. GLSZM: Gray-level size-zone matrix. LGZE: Low gray-level zone emphasis. LRE: Long run emphasis. LZE: Level zone emphasis. LGRE: Low gray-level run emphasis. GLCM: Gray-level co-occurrence matrix. ZSV: Zone-size variance. ZP: Zone percentage. SZHGE: Small zone high gray-level emphasis. LRLGE: Long-run low gray-level run emphasis.
4. Discussion & conclusions
Both the RF and SVM models trained with hand-crafted imaging features alone outperformed models trained with deep learning-based or clinical features alone (P < 0.05; Table 3). They also slightly outperformed the models trained with all features, although they did not reach a statistical significance. Models trained with deep learning or clinical features alone performed comparatively against each other. Models trained with all of the available features outperformed the models trained with deep learning or clinical features alone, but they did not attain statistical significance.
This finding is interesting, as hand-crafted color and texture features, which require less computations than extraction of deep learning features, may actually suggest more about the prognostication of the wound healing outcomes. The color features, especially, consist of the simple mean and standard deviation of color intensity values in different color spaces and can therefore be intuitive to the clinicians; for example, the feature importance chart (Table 4) suggests that the mean of green color is one of the most important features, and one would clinically expect that the non-red colors in ulcers are concerning for more serious or gangrened infections. Deep learning architectures are known to outperform hand-crafted images for wound image segmentation, but as long as the wound is already segmented by any means, the hand-crafted features may still be useful in clinical prognostication.
The feature importance chart also demonstrates that most important features are predominantly hand-crafted imaging features when the PCA is not performed. Only 7 features out of 30 were clinical features. Among the clinical features, the patient’s nutritional status (pre-albumin) and the size of the ulcer are ranked among the 30 most important features. The nutritional status is known to affect any kind of wound healing [23,24] and has been found to be independently associated with prognosis of diabetic foot ulcers [25]. While our study did not include the size change at a certain time point, the wound surface area change at 4 weeks has been found to be a robust predictor of ulcer healing [14].
Although there has been no other study that combined hand-crafted image features, deep learning features and clinical features to build a prediction model for wound healing like our study, there have been studies that have tested clinical or molecular biomarkers’ predictive values in the healing of DFU. Fife et al. [12] created a multivariable logistic regression model using multiple clinical indices and demonstrated AUROCs between 0.648 and 0.668 for both the whole course model and the first encounter model. Margolis et al. [26] also used several clinical prognostic factors to create various multivariable logistic regression models and demonstrated the AUROCs of 0.66–0.70. Another study, which used the ratio of serum matrix metalloproteinase-9 (MMP-9) to tissue inhibitor of MMPs (TIMP) as a predictor for wound healing, demonstrated an AUROC of 0.658 [27]. Compared to these models, our models demonstrated higher AUROC value when using either all features (0.734) or hand-crafted imaging features alone (0.760–0.794) and comparatively when only deep learning features (0.670) or clinical features (0.636–0.657) were used in the models.
A few limitations exist for this study. First of all, Although the initial dataset contains almost 2300 visit records including the follow-up visits, only 208 ulcers were used because of the lack of images, those lost to follow-up, and sometimes questionable segmentations of ulcers. Using information from each follow-up visit may be used to refine the models, and a further verification of this methodology and models will be necessary with a larger patient population and dataset. In addition, some of the clinical parameters had to be imputed because the EHR do not contain every single feature used in this study. This allows data points with more features to have more impact than those with fewer features in building the models, since the missing features are inferred from those visits that have those features recorded. Also, because the EHR are entered by different clinicians, there may be slight inconsistencies in clinical attributes that are manually charted, such as the ulcer size or UTSA grade. However, all EHR are internally audited within the health system for the billing practice and this problem is thereby mitigated.
The tools that can originate from and the findings included in this study hold promise for the telemedicine system in the management of the DFU’s. First of all, all of the images utilized in this study were taken with a smartphone or tablet and did not require high-tech imaging only available in research settings. The use of hand-crafted image features and raw clinical attributes in the prediction algorithm more easily provides insights to both clinicians and patients on what attributes may be more important in positive outcomes in wound healing. Also, the fact that hand-crafted image features were of the most outperforming of all features means that a tool incorporating such pipeline may not have to require graphics processing units (GPU) necessary for deep learning-based analyses.
Supplementary Material
Acknowledgements
JSW, AM, JG, RBK and KN conceived of the presented study. All authors contributed to collection and preprocessing of the data. RBK designed and developed the experimental pipeline and performed the computations with support from SMRS, JG, JSW and KN. RBK wrote the manuscript with support from all other authors. All authors have discussed the results and contributed to the final manuscript. RBK is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. The authors declare no conflicts of interest.
This work was supported by NIH DiaComp Collaborative Funding Program project, 17AU3752, NIDDK Diabetic Complications Consortium (DiaComp, www.diacomp.org), grant DK076169 and NIH NIDDK U01, DFU Clinical Research Unit, 1U01DK119083-01. This work was approved by IRB HUM00128252. The authors would like to thank Tianyi Bao and for collection of the data at the initial phase of this study.
Footnotes
Declaration of competing interest
None declared.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2020.104042.
References
- [1].Singh N, Armstrong DG, Lipsky BA, Preventing foot ulcers in patients with diabetes, J. Am. Med. Assoc 293 (2) (2005. Jan 12) 217. [DOI] [PubMed] [Google Scholar]
- [2].Boulton AJM, The diabetic foot: grand overview, epidemiology and pathogenesis, in: Diabetes/Metabolism Research and Reviews, John Wiley & Sons, Ltd, 2008. S3–6. [DOI] [PubMed] [Google Scholar]
- [3].Wang L, Pedersen PC, Strong DM, Tulu B, Agu E, Ignotz R, Smartphone-based wound assessment system for patients with diabetes, IEEE Trans. Biomed. Eng 62 (2) (2015. Feb 1) 477–488. [DOI] [PubMed] [Google Scholar]
- [4].Wang L, Pedersen PC, Strong DM, Tulu B, Agu E, Ignotz R, et al. , An automatic assessment system of diabetic foot ulcers based on wound area determination, color segmentation, and healing score evaluation, J. Diabetes Sci. Technol 10 (2) (2016. Mar 1) 421–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ronneberger O, Fischer P, Brox T, U-net: convolutional networks for biomedical image segmentation, Springer, Cham, 2015. [cited 2019 Jul 12]. p. 234–41. Available from, http://link.springer.com/10.1007/978-3-319-24574-4_28. [Google Scholar]
- [6].Wang G, Li W, Zuluaga MA, Pratt R, Patel PA, Aertsen M, et al. , Interactive medical image segmentation using deep learning with image-specific fine tuning, IEEE Trans. Med. Imag 37 (7) (2018) 1562–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gamage C, Wijesinghe I, Perera I, Automatic scoring of diabetic foot ulcers through deep CNN based feature extraction with low rank matrix factorization, in: 2019 IEEE 19th International Conference on Bioinformatics and Bioengineering, (BIBE), 2019, pp. 352–356. [Google Scholar]
- [8].Cui C, Thurnhofer-hemsi K, Soroushmehr R, Mishra A, Gryak J, Dom E, et al. , Diabetic wound segmentation using convolutional neural networks, 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) (2019) 1002–1005, 10.1109/EMBC.2019.8856665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gamage H, Wijesinghe W, Perera I, Instance-based segmentation for boundary detection of neuropathic ulcers through Mask-RCNN, in: International Conference on Artificial Neural Networks, 2019, pp. 511–522. [Google Scholar]
- [10].Wijesinghe I, Gamage C, Perera I, Chitraranjan C, A Smart telemedicine system with deep learning to manage diabetic retinopathy and foot ulcers, in: 2019 Moratuwa Engineering Research Conference, MERCon, 2019, pp. 686–691. [Google Scholar]
- [11].Hazenberg CEVB, de Stegge WB, Van Baal SG, Moll FL, Bus SA, Telehealth and telemedicine applications for the diabetic foot: a systematic review, Diabetes Metab. Res. Rev 36 (3) (2020), e3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Fife CE, Horn SD, Smout RJ, Barrett RS, Thomson B, A predictive model for diabetic foot ulcer outcome: the Wound Healing Index, Adv. Wound Care 5 (7) (2016) 279–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Forsythe RO, Apelqvist J, Boyko EJ, Fitridge R, Hong JP, Katsanos K, et al. , Performance of prognostic markers in the prediction of wound healing or amputation among patients with foot ulcers in diabetes: a systematic review, Diabetes Metab. Res. Rev 36 (2020), e3278. [DOI] [PubMed] [Google Scholar]
- [14].Sheehan P, Jones P, Caselli A, Giurini JM, Veves A, Percent change in wound area of diabetic foot ulcers over a 4-week period is a robust predictor of complete healing in a 12-week prospective trial, Diabetes Care 26 (6) (2003. Jun 1) 1879–1882. [DOI] [PubMed] [Google Scholar]
- [15].Valenzuela-Silva CM, Tuero-Iglesias AD, Garcia-Iglesias E, Gonzalez-Diaz O, Del Rio-Martin A, Alos IBY, et al. , Granulation response and partial wound closure predict healing in clinical trials on advanced diabetes foot ulcers treated with recombinant human epidermal growth factor, Diabetes Care 36 (2) (2013. Feb) 210–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Stekhoven DJ, Bühlmann P, Missforest-Non-parametric missing value imputation for mixed-type data, Bioinformatics 28 (1) (2012. Jan 1) 112–118. [DOI] [PubMed] [Google Scholar]
- [17].Vallières M, Freeman CR, Skamene SR, El Naqa I, A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities, Phys. Med. Biol 60 (14) (2015. Jul 7) 5471–5496. [DOI] [PubMed] [Google Scholar]
- [18].He K, Zhang X, Ren S, Sun J, Deep residual learning for image recognition, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2016, pp. 770–778. [Google Scholar]
- [19].Breiman L, Leo, Random forests, Mach. Learn 45 (1) (2001) 5–32. [Google Scholar]
- [20].Cortes C, Vapnik V, Support-vector networks, Mach. Learn 20 (3) (1995) 273–297. [Google Scholar]
- [21].Nadeau C, Bengio Y, Inference for the generalization error, in: Advances in Neural Information Processing Systems, 2000, pp. 307–313. [Google Scholar]
- [22].Bouckaert RR, Frank E, Evaluating the replicability of significance tests for comparing learning algorithms, in: Pacific-asia Conference on Knowledge Discovery and Data Mining, 2004, pp. 3–12. [Google Scholar]
- [23].Dickhaut SC, DeLee JC, Page CP, Nutritional status: importance in predicting wound-healing after amputation, J. Bone Joint Surg. Am 66 (1) (1984. Jan) 71–75. [PubMed] [Google Scholar]
- [24].Stechmiller JK, Understanding the role of nutrition and wound healing, Nutr. Clin. Pract 25 (1) (2010. Feb 3) 61–68. [DOI] [PubMed] [Google Scholar]
- [25].Zhang S-S, Tang Z-Y, Fang P, Qian H-J, Xu L, Ning G, Nutritional status deteriorates as the severity of diabetic foot ulcers increases and independently associates with prognosis, Exp. Ther. Med 5 (1) (2013) 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Margolis DJ, Allen-Taylor L, Hoffstad O, Berlin JA, Diabetic neuropathic foot ulcers: predicting which ones will not heal, Am. J. Med 115 (8) (2003) 627–631. [DOI] [PubMed] [Google Scholar]
- [27].Li Z, Guo S, Yao F, Zhang Y, Li T, Increased ratio of serum matrix metalloproteinase-9 against TIMP-1 predicts poor wound healing in diabetic foot ulcers, J. Diabet. Complicat 27 (4) (2013) 380–382. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


