Abstract
Although plant pathogens are traditionally controlled using synthetic agrochemicals, the availability of commercial bactericides is still limited. One potential control strategy could be the use of plant growth‐promoting bacteria (PGPB) to suppress pathogens via resource competition or the production of antimicrobial compounds. This study aimed to conduct in vitro and in vivo screening of eight Pseudomonas strains against Ralstonia solanacearum (the causative agent of bacterial wilt) and to investigate underlying mechanisms of potential pathogen suppression. We found that inhibitory effects were Pseudomonas strain‐specific, with strain CHA0 showing the highest pathogen suppression. Genomic screening identified 2,4‐diacetylphloroglucinol, pyoluteorin, and orfamides A and B secondary metabolite clusters in the genomes of the most inhibitory strains, which were investigated further. Although all these compounds suppressed R. solanacearum growth, only orfamide A was produced in the growth media based on mass spectrometry. Moreover, orfamide variants extracted from Pseudomonas cultures showed high pathogen suppression. Using the “Micro‐Tom” tomato cultivar, it was found that CHA0 could reduce bacterial wilt disease incidence with one of the two tested pathogen strains. Together, these findings suggest that a better understanding of Pseudomonas–Ralstonia interactions in the rhizosphere is required to successfully translate in vitro findings into agricultural applications.
Keywords: environmental microbiology, microbial ecology, microbial interactions and pathogenesis, plant–microbe interactions
This study aimed to conduct in vitro and in vivo screening of eight Pseudomonas strains against Ralstonia solanacearum (the causative agent of bacterial wilt) and to investigate underlying mechanisms of potential pathogen suppression. Genomic screening identified 2,4‐diacetylphloroglucinol, pyoluteorin, and orfamides A and B secondary metabolite clusters in the genomes of the most inhibitory strains, which all suppressed R. solanacearum growth. Although Pseudomonas could reduce bacterial wilt disease incidence with tomato, the effect depended on the R. solanacearum strain, highlighting the need to better understand Pseudomonas–Ralstonia interactions in the rhizosphere.
1. INTRODUCTION
The use of chemical pesticides has declined in recent years due to elevated costs, environmental toxicity, and stricter legislation (Chen et al., 2016). As a result, new methods and approaches are required to control plant pathogens and to ensure future food security in the face of expanding human population (Hayward, 1991; Kaczmarek et al., 2014; Wang et al., 2019). One alternative to traditional agrochemicals is biocontrol, which depends on using natural microbial competitors against a variety of pathogens to restrict pathogens' growth and survival. Several biocontrol agents have been identified and found to show a broad range of activity against bacterial, fungal, viral, and nematode pathogens (Hansen & Keinath, 2013; Nagachandrabose, 2020; Sofrata et al., 2011; Wang et al., 2019). Plant growth‐promoting bacteria (PGPB) are one example of potentially highly effective biocontrol agents, with a broad‐range biocontrol activity (Pierson et al., 1998). PGPB biocontrol outcomes are, however, often variable in field conditions, despite clear inhibition observed under laboratory conditions (Compant et al., 2005). One reason for this is that lab conditions fail to replicate actual field conditions in terms of nutrient availability, competition with rhizospheric bacteria, degradation, and adsorption of secreted antimicrobials, and the likelihood of successful colonization in natural environments, these can all affect the efficacy of disease suppression (Weller, 2007). In addition, even though biocontrol agents can show a broad range of effects against different types of pathogens, they might not be able to inhibit different genotypes of one given pathogen species (Xue et al., 2013). Considering pathogen‐biocontrol agent interactions at the genotype level, and the mechanisms by which inhibition is mediated in more realistic environmental conditions, are therefore required to develop functionally robust biocontrol methods.
Plant pathogenic bacteria belonging to the Ralstonia solanacearum species complex (RSSC) cause bacterial wilt disease in many wild and cultivated plants. It is ranked the second most important bacterial plant pathogen globally and possesses numerous virulence mechanisms that allow it to infect more than 250 plant species from 54 plant families (Genin & Denny, 2012; Hayward, 1991; Mansfield et al., 2012; Nion & Toyota, 2015). RSSC has spread globally across the world and has a quarantine status in many countries meaning it is monitored regularly to prevent further spread (EPPO, 2021). In addition to the trade of infected plant materials, RSSC global distribution is likely explained by its high genetic variability and ability to rapidly adapt to surrounding environmental conditions (Genin, 2010; Genin & Denny, 2012). Between‐strain differences include variation in pathogenicity‐related genes, such as the presence of Type III secretion effectors and genes involved in chemotaxis, adherence, secondary metabolic pathways, biosynthesis of phytohormones, and the detoxification of various antimicrobial compounds which are important in determining R. solanacearum host range (Genin, 2010; Genin & Denny, 2012). This variation can be generated via accumulation of mutations (Gopalan‐Nair et al., 2021; Guidot et al., 2014) or horizontal gene transfer between different R. solanacearum strains, which can result in an exchange of virulence traits between different genotypes (Guidot et al., 2009). Although R. solanacearum strains in Europe are considered to be clonal, belonging to the phylotype IIB group (Castillo & Greenberg, 2007; Clarke et al., 2015), some strain variation exists at the local scale within countries (Caruso et al., 2017; Parkinson et al., 2013). To what extent this variation affects the pathogen response to biocontrol PGPBs is, however, currently unknown.
Traditionally, the development of a biocontrol method begins in vitro in the lab by identifying bacterial strains with potential biocontrol activity (Fravel, 2005). Biocontrol efficiency within Pseudomonas plant growth‐promoting species is often mediated by a variety of secreted secondary metabolites, including several antimicrobials and iron‐scavenging siderophores (Becker et al., 2012). Some biocontrol characteristics are more common amongst these strains, including the ability to produce 2,4‐diacetylphloroglucinol (DAPG) and hydrogen cyanide—which are effective antimicrobial against bacterial, fungal, and nematode plant pathogens (Almario et al., 2017; Compant et al., 2005; Cronin et al., 1997; Haas & Défago, 2005; Haas & Keel, 2003; Humair et al., 2009). In contrast, other biocontrol properties are strain‐specific, such as the production of cyclic orfamide lipopeptides by certain subgroups of Pseudomonas fluorescens such as Pseudomonas protegens strains: CHA0 and Pf‐5 (Ma et al., 2016). Methods for screening bacterial biocontrol agents against R. solanacearum include exposing pathogenic bacterial strains to biocontrol bacteria directly in co‐cultures and measuring pathogen growth or survival as a function of time (Fravel, 2005). Supernatant assays are also often used to test if secretions in the media suppress the growth of the pathogens (Kaur et al., 2019; C. Yang et al., 2019). However, the successful identification of biocontrol strains in vitro does not always translate to successful biocontrol outcomes in vivo and Pseudomonas biocontrol outcomes are often variable in field conditions despite clear inhibition observed in laboratory conditions. For example, Pseudomonas strains have been shown to unsuccessfully replicate in vitro protection against R. solanacearum during in vivo experiments with Eucalyptus trees, and it was suspected this may have been due to low‐level expression of genes responsible for biocontrol activity (Ran et al., 2005). The success of biocontrol is further influenced by competition with native bacteria in the rhizosphere, and it has also been shown that the effectiveness of their secondary metabolites, such as DAPG, can be influenced by the age and species of the host plant is has been selected to protect (Notz et al., 2001; Siddiqui & Shaukat, 2003). Soil properties such as moisture, temperature, clay content, and pH can also influence the success of biocontrol (van der Putten et al., 2006), while common bacterial feeders, such as protozoa and nematodes, could also predate the biocontrol bacteria (Pedersen et al., 2009). Therefore, it is vital that in vitro screening is coupled with in vivo validation of biocontrol effectiveness with plants to develop successful biocontrol applications with translational potential.
Here we used such an approach to screen and identify efficient Pseudomonas biocontrol bacterial strains against six UK and one Polish R. solanacearum isolates that belong to an economically important potato‐specific RSSC lineage (Phylotype IIB sequevar 1; formerly known as race3 biovar2 strains). We first tested direct and indirect interactions between R. solanacearum strains and eight plant growth‐promoting Pseudomonas strains in vitro, which were chosen for their previously characterized antimicrobial activity against a Chinese R. solanacearum isolate belonging to Phylotype I (Hu et al., 2016). We then conducted genomic screening of Pseudomonas strains to identify potential antimicrobial secondary metabolite clusters and directly tested whether these compounds were produced and if they inhibited R. solanacearum under laboratory conditions. Finally, the biocontrol potential of the P. protegens CHA0 strain was tested against two R. solanacearum strains in the tomato rhizosphere. Results revealed that antimicrobial effects of Pseudomonas were strain‐specific, and the strongest inhibitory effects were mediated by P. protegens strain CHA0. Of the tested compounds, the pyoluteorin antibiotic had the strongest inhibitory effect followed by DAPG and orfamides A and B. While CHA0 also showed biocontrol efficiency with tomato, this was clear only with one of the tested UK R. solanacearum isolates. Together, these results show that while in vitro screening can be used to identify potential Pseudomonas biocontrol strains against R. solanacearum, more work is needed to understand their biocontrol activity in vivo in the plant rhizosphere.
2. METHODS
2.1. Culturing and maintenance of bacterial strains
Eight fluorescent Pseudomonad strains (CHA0, Pf‐5, Q2‐87, Q8R1‐96, 1M1‐96, MVP1‐4, F113, and Ph11C2) which have been studied extensively previously in biocontrol (Hu et al., 2016), and seven Ralstonia solanacearum strains (#1–#7), which were isolated as a part of annual river sampling survey in England and Wales by Fera Science Ltd., were used in the experiments (listed in Tables A1 and A2). All bacteria were stored in 20% glycerol stocks at −80°C. Before experiments, bacterial starting cultures were prepared as follows: frozen samples were inoculated in 5 ml of LB, NB, or CPG broth (media recipes described in Table A3) and incubated with shaking at 200 rpm at 28°C for 24 h. Rich media preparations were used throughout all experiments to ensure the efficient growth of both bacterial genera. Bacterial cultures were prepared similarly throughout all experiments unless stated otherwise.
2.2. Measuring R. solanacearum inhibition by Pseudomonas strains in direct contact
Soft agar overlay assays were used to test direct inhibition of R. solanacearum by Pseudomonas strains. Pseudomonas strains were grown for 24 h in 5 ml of 100% LB broth at 28°C with shaking at 200 rpm and R. solanacearum strains were grown for 30 h under the same conditions. Soft agar overlay plates were prepared by first filling sterile 90‐mm Petri dishes with a layer of hard LB which was left to solidify. Two hundred microliters of each R. solanacearum culture were then mixed with 20 ml of cooled (below 55°C) liquid soft agar and poured on top of the hard agar layer. As R. solanacearum grows more slowly than Pseudomonas, plates were left to incubate for ∼8 h before spotting the Pseudomonas cultures on top of the soft agar overlay as follows. Each plate was divided evenly into quarters and 2 μl of a Pseudomonas strain (∼1.0 × 106 CFU/ml) was spotted on the center of each quarter of the plate (example shown in Figure A1). Plates were then incubated upside down at 28°C and zones of inhibition were recorded after 96 h. The distance of the inhibition zones was measured from the outer edge of the Pseudomonas spot to the R. solanacearum lawn (in millimeters with a ruler). Each strain combination treatment was carried out in triplicate.
2.3. Measuring R. solanacearum inhibition by Pseudomonas strains indirectly using supernatant assays
To investigate R. solanacearum inhibition by Pseudomonas in the absence of direct contact, we exposed R. solanacearum strains to supernatants of each Pseudomonas species in pairwise supernatant cultures. The Pseudomonas supernatants included all secondary metabolites excreted when Pseudomonas strains were grown alone in LB media and were prepared as follows. All Pseudomonas strains were first grown individually in 20 ml of LB broth (Table A3) for 24 h with shaking at 200 rpm. The supernatant was then prepared by centrifuging cultures for 10 min at 4000g before separating bacterial cells and fragments from soluble material including secondary metabolites using 0.2‐μm filters. The inhibition was measured using flat bottomed 96‐well plates in 50:50 Pseudomonas supernatant to LB mixtures. As a negative control R. solanacearum was grown in 50:50 LB in the sterile water mixture. At the start of the experiment, every supernatant mixture was inoculated with 2 μL of each R. solanacearum strain (∼1.0 × 106 CFU/ml), and microplates were then incubated at 28°C for 3 days and their bacterial densities were recorded as optical density at 24 and 72 h (OD 600 nm; Tecan Infinite spectrophotometer). All pairwise combinations were replicated four times and control treatments three times.
2.4. Genome sequencing of Pseudomonas strains
Single colonies of each Pseudomonas strain were inoculated in NB broth and grown for 12 h at 30°C with shaking at 170 rpm. Genomic DNA was extracted using the QIAGEN blood and cell culture DNA kit (catalog No. QIAGEN Genomic‐tip 100/G, Midi 13343) following the manufacturer's protocol, and DNA quality was tested using nanodrop. All strains were sequenced using the Miseq platform (2 × 300 bp) at the Utrecht Sequencing Facilities. To obtain more accurate genome assemblies, reads were preprocessed as follows. We first removed adapter sequences, read shorter than 50 bp, and low‐quality nucleotides using a Phred quality score threshold of <20. Genome assemblies were carried out in two steps. First, we used SOAPdenovo v2.04 to assemble reads into contigs and scaffolds based on K‐mer size (available at https://soap.genomics.org.cn/). Second, GapCloser v1.12 was used to close gaps emerging during the scaffolding process by SOAPdenovo (Li et al., 2010).
2.5. Investigating Pseudomonas secondary metabolite biosynthesis gene clusters using antiSmash5.0
The de novo assembled draft genomes of the eight Pseudomonas strains were analyzed to identify potential secondary metabolic clusters linked with antibiosis using the antiSMASH 5.0 pipeline (https://antismash.secondarymetabolites.org; Blin et al., 2019, 2017), which allows rapid genome‐wide identification and in‐depth analysis of secondary metabolite biosynthesis gene clusters based on several open‐source databases. Fasta files were uploaded to antiSMASH5.0 bacterial version with detection strictness set to “relaxed” and all search features included to maximize potential metabolic clusters identified. Based on this analysis, we identified four antimicrobial compounds which could potentially be produced by our Pseudomonas strains. One of these compounds, DAPG, is a common antimicrobial produced by all eight fluorescent Pseudomonas strains, while three specific gene clusters for pyoluteorin (an antimicrobial with unknown mechanisms of action; Kidarsa et al., 2011) and orfamides A and B (cyclic lipopeptides that can cause membrane pore formation; Ma et al., 2016) were found only in CHA0 and Pf‐5 Pseudomonas genomes.
2.6. Determining the effects of identified Pseudomonas metabolites on R. solanacearum growth in single‐compound and multicompound mixtures
Pure compounds of orfamide A (Santa Cruz Biotechnology), orfamide B (Santa Cruz Biotechnology), Pyoluteorin (Santa Cruz Biotechnology), and DAPG (Santa Cruz Biotechnology) were purchased to test their efficacy against R. solanacearum strains. Ten millimolar stocks were made in 100% dimethyl sulfoxide (DMSO) and stored at −20°C, except for DAPG, for which a 100 mM stock was prepared in 100% methanol and stored at −20°C. The effect of DAPG was measured by inoculating 2 μl of each R. solanacearum strain (∼1.0 × 106 CFU/ml) to 100% LB broth with DAPG at the following concentrations: 1000, 500, 100, 50, and 0 μM (negative control). Due to the relatively high costs of chemicals, only two R. solanacearum strains (#1 and #7) were tested for their susceptibility to pyoluteorin (100 μM concentration only). Similarly, only Strain #1 susceptibility was tested to orfamides A and B (100 μM concentration only) following the same methods as with DAPG. Bacterial densities were recorded as optical density at 0, 24, 48, and 72 h after inoculation with a spectrophotometer (OD 600 nm; Tecan Sunrise spectrophotometer).
2.7. Confirmation of the production of secondary metabolites in the Pseudomonas supernatant using mass spectrometry
Mass spectrometry was used to identify and quantify any secondary metabolites produced by CHA0 and Pf‐5 in the growth conditions they were exposed to in vitro (compounds were identified against chemical standards). We also used untargeted analysis to identify any novel antimicrobials produced by comparing peaks against existing databases.
The strains were grown in triplicate in LB broth with 200 rpm shaking at 28°C for 24 h. Following incubation, bacterial densities were normalized to an optical density of 0.1 (OD 600 nm), and the cultures were centrifuged for 10 min at 4000g. The supernatant was then filtered using 0.2‐μm filters to separate bacterial cells and fragments from secondary metabolites present in the supernatants. A total of six samples were produced (two Pseudomonas strains in one growth condition in triplicate). Two hundred microliters of each sample were provided to the Centre of Excellence in Mass Spectrometry at the University of York for mass spectrometry analysis. The LC separation was performed by an Acquity UPLC I class system (Waters), on a BEH C18 100 × 2.1, 1.7 U column (Waters). The sample injection volume was 7.5 µl. The MS end was a Synapt G2S‐Si QTOF (Waters) mass spectrometer which was operated in positive ESI, resolution mode, using the HDMSE acquisition technique (alternating scans of MS and MS2 acquisitions [fragmentation by CID] alongside traveling wave ion mobility separation). Voltages in the low energy function (MS) were 4 V both in the trap and transfer cell; and in the high energy function (MS2), the voltage was 4 V in the trap and a 20–100 V ramp in the transfer cell. The source was operated with the capillary set to 3 kV, source temperature 150°C, desolvation temperature 450°C, cone gas flow 20 L/h, desolvation gas flow 450 L/h, and nebulizer 6.5 bar. Trap gas flow was 2 ml/min, helium cell flow 180 ml/min, and IMS gas flow 80 ml/min. Mass range was 100–1000 m/z, scan time 0.2 s, lock mass was leucine enkephalin at m/z 556.2771.
Data was collected using MassLynx Software Version V4.2 SNC983 (Waters) and analyzed using UNIFI 1.9 (Waters) for semi‐quantitative analysis of target analytes (orfamide A, orfamide B, pyoluteorin, and DAPG). Calibration curves were constructed for target analytes at 10, 50, 100, 300, and 500 nM in LB media; the MS response for all analytes was linear in this range. Analytes were quantified using the calibration curve in the appropriate medium. Blanks, standards, and samples were injected in technical triplicates from the same vial. Progenesis QI v.2.0 (Waters) was used for the untargeted analysis of metabolomics data: putative compound identification was based on a score comprising MS match of exact mass of the compound precursor (ChemSpider database search with 10 ppm mass tolerance) and a match to in silico MS2 fragmentation pattern. Further data processing and downstream analysis were performed using Bioconductor package XCMS in R, combined with a database (LipidMaps) search.
Untargeted analyses were also conducted by comparing the collected dataset with the Knapsack database (https://www.knapsackfamily.com/KNApSAcK/). Briefly, Waters.raw files were converted to zlib compressed 32‐bit precision.mzML files using ProteoWizard MSConvert [1] version 3.0.19172, using the combineIonMobilitySpectra filter, and spectra centroided using the qtofpeakpicker with resolution set to 20,000 and threshold 1. Using custom scripts in R 3.0.0 operating in a Linux 64‐bit environment.mzML files were further processed using the mzR package [1] to lockmass correct spectra against leucine enkephalin ([M + H]+ = 556.27568, [M − H]− = 554.26202). Lockmasses were identified within a ±0.1 Da m/z window, and the running mean of lockmasses in a 60 s moving window was used to adjust the m/z values for every scan. Across files, feature detection was achieved using the xcmsSet() function from the xcms package [2], using the following parameters: method = ‘centWaveWithPredictedIsotopeROIs’, ppm = 10, snthresh = 10, peakwidth = c(3, 20), prefilter = c(3, 1000), integrate = 2, mzdiff = −0.1, firstBaselineCheck = FALSE. Custom scripts were used to retain Gaussian features, which were then aligned and missing values recalculated across samples using the xcms group() and fillPeaks() functions, respectively. Feature areas were adjusted by subtracting the mean + 3 × the standard deviation of blank samples, with adjusted values <0 being set to 0. The feature list was then filtered to retain only the most intense monoisotope belonging to a single compound as identified by CAMERA [3], with areas above 0 in at least 90% of blank‐subtracted samples. These filtered features were reported as masstags (unique m/z and retention time pairs) and annotated where possible against authentic standards or putative compounds from literature or databases based on exact mass. The Progenesis QI v.2.0 (Waters) was also used to search for compounds by their monoisotopic mass which has been reported to be present in Pseudomonas genomes from the literature. The potential metabolites were identified if their observed monoisotopic mass matched with the accurate known ppm within a range of ±10 ppm. However, without a chemically verified standard for definitive identity matches, this can only be considered speculative.
2.8. Extraction and isolation of Pseudomonas orfamide antimicrobials and testing their effects on R. solanacearum growth
A protocol was adapted from Ma et al. (2016) to extract cyclic lipopeptide orfamides from the CHA0 Pseudomonas strain. Briefly, a starting culture of CHA0 was grown in a 250 ml flask containing 50 ml of King's Broth growth medium at 28°C with shaking at 200 rpm. This was then inoculated into a 2 L flask containing 500 ml of liquid media and shaken at 150 rpm for 48 h. Pseudomonas supernatant was collected via centrifugation at 10,000g for 10 min (J25 XP series centrifuge). The supernatant was then acidified to a pH of 2.0 using 8 M HCl and stored overnight at 4°C. The precipitate was collected after centrifugation at 10,000g for 20 min and extracted with 1 ml of 100% methanol. At this stage in the centrifuge tube, there were two distinguishable precipitates, one at the bottom of the tube and one close to the top. It was decided to collect these separately and call them “A” and “B” as it was unknown if they would have different or similar properties. The organic phase was collected by spinning samples until dry in a vacuum concentrator (Savant svc SpeedVac 100 h concentrator) and samples were then dissolved in 300 μl of 100% DMSO to make orfamide variant stocks. Due to a limited amount of these orfamide variant stocks, only R. solanacearum strains #1 and #7 were used for inhibition testing. Inoculant cultures were grown in 5 ml of LB broth overnight and 2 μl of each R. solanacearum strain was inoculated with 1% orfamide variants in LB broth (200 μl final volume). Bacterial densities were recorded at 24, 48, and 72 h (OD 600 nm; Tecan Infinite spectrophotometer), and each treatment was carried out in triplicate.
2.9. Testing Pseudomonas biocontrol efficiency in vivo using a tomato plant model
To explore the translational potential of Pseudomonas CHA0 strain in vivo, we tested if any of the inhibitory effects observed in vitro could protect plants from R. solanacearum infections using the Micro‐Tom tomato cultivar. This cultivar of tomato was selected as it is well‐established and often used when studying R. solanacearum infections (Gu et al., 2016). The infectivity of two R. solanacearum UK strains (#1 and #7) was tested in the absence and presence of CHA0 (a negative water control treatment was included). Tomato (Solanum lycopersicum “Micro‐Tom”) plants were grown in an incubated light chamber at 28°C with 16:8 h light:dark conditions with regular watering. Seeds were sown in 7 cm seedling trays in 35 g of autoclaved compost (John Innes #2) where they remained for the entire experiment. Experiments were conducted between July and September 2019 in temperature‐controlled plant growth chambers (Sanyo MLR‐352) at the University of York. To test the biocontrol efficacy of CHA0, the Pseudomonas culture was prepared by inoculating 100 ml of CPG broth with 100 μl of frozen stock bacterial culture in 300 ml glass culture flasks and the strain was grown for 24 h at 28°C with shaking at 200 rpm. Bacterial culture was then washed of nutrient media by centrifuging at 4000g for 10 min and resuspending in sterile dH2O. Seeds were germinated and sown in 7 cm seedling trays with 35 g of autoclaved compost and regularly watered using sterile water (for growth conditions see earlier section). Pseudomonas strain was inoculated onto the soil of 4‐week old seedlings 1 week before R. solanacearum strains to allow time for the biocontrol bacteria to effectively colonize the soil and roots. Before inoculation, Pseudomonas cell densities were adjusted to an OD 600 nm of 0.25 (∼1.0 × 09 CFU/ml) and 6 ml was poured into each pot, soaking through the soil and roots, and any excess culture remained in the saucer below. Roots of tomato seedlings were cut before R. solanacearum inoculation using a sterile scalpel to mimic natural pathogen entry points in the field. R. solanacearum strains #1 and #7 were cultured in 30 ml of CPG broth (Table A3) for 48 h with shaking at 200 rpm at 28°C and adjusted to an OD 600 nm of 0.25. A total of 1 ml of each R. solanacearum strain was inoculated at the base of the visible stems 1 week after Pseudomonas inoculation. Three replicates were used for each treatment and each replicate consisted of nine individual plants. All plants were watered 1 h before monitoring infection to be certain that wilting was not due to dehydration. Bacterial wilting symptoms were recorded daily by scoring the plants as “uninfected” or “infected” when plants showed clear wilting symptoms. The experiment was terminated 3 weeks after pathogen inoculation and above‐ground dry weight was recorded for each replicate plant.
2.10. Statistical analyses
We used repeated‐measures analysis of variance to analyze mean differences between pathogen densities for all experiments including temporal data such as R. solanacearum exposure to different secondary metabolites. Two‐way ANOVA was used to analyze the mean differences between treatments when only a one‐time point was used for analysis, followed by pairwise post hoc Tukey tests with 95% confidence levels and Bonferroni‐corrected p values. These analyses were used when interpreting the direct and indirect soft agar and bacterial supernatant interactions between R. solanacearum and Pseudomonas. Poisson glm models with χ 2 tests were used to analyze the binomial in vivo tomato infection data. All statistical analyses and graphs were produced using R (R Foundation for Statistical Computing, R Studio Version [3. 4. 4], Packages: ggplot, tidyverse, nlme, rcompanion).
3. RESULTS
3.1. Measuring the direct inhibition of R. solanacearum by Pseudomonas strains using soft agar assays
Clear inhibition zones were detected in all pathogen strain treatments indicating that each Pseudomonas strain could inhibit the growth of R. solanacearum when in direct contact. However, inhibition zone sizes differed depending on the identities of interacting Pseudomonas and R. solanacearum strains (Ralstonia × Pseudomonas: F 42, 112 = 3.877, p < 0.0001, Figure 1a). This variation appeared to be mainly driven by Pseudomonas strains, as post hoc analyses revealed only a small and nonsignificant variation between different R. solanacearum strains (Ralstonia: F 6, 161 = 0.6364, p = 0.7009, Figure 1a). In contrast, much higher variation was observed between the inhibitory activity of Pseudomonas strains (Pseudomonas: F 7, 160 = 60.81, p = 0.0001, Figure 1a), and post hoc analyses revealed that CHA0 was the most inhibitory strain, followed by Q8R1‐96 and MVP1‐4, which showed modest levels of inhibition (Tukey: p < 0.05). The strains 1M1‐96 and Ph11C2 caused the smallest zones of inhibition on R. solanacearum lawns overall (Tukey: p < 0.05). These results suggest that all Pseudomonas strains inhibited the growth of R. solanacearum in direct contact, which varied between different Pseudomonas strains.
Figure 1.
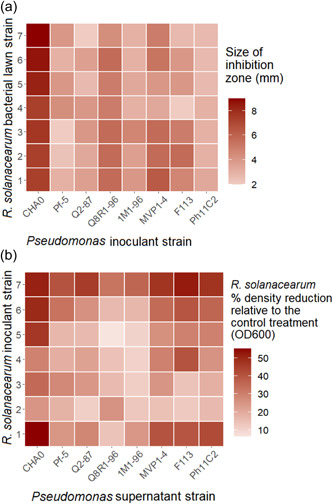
Direct and indirect inhibition of Ralstonia solanacearum by Pseudomonas strains. (a) Direct inhibition on soft agar assays, where each interaction represents a different Pseudomonas strain and the average diameter of the inhibition zones (mm) against different R. solanacearum strains after 96 h growth on soft agar lawns (N = 3). (b) Indirect inhibition of R. solanacearum by Pseudomonas strains' supernatant‐LB mixes (set up in 96‐well plates containing a 50:50 ratio of SN and 100% LB broth). The shade of red represents the growth reduction in R. solanacearum densities compared to the control (100% LB broth) treatment after 72 h of growth (N = 3)
3.2. Measuring the indirect inhibition of R. solanacearum by Pseudomonas strains using supernatant assays
Almost all Pseudomonas supernatants suppressed the growth of R. solanacearum compared to the no‐supernatant LB control treatments (Supernatant treatment: F 8, 236 = 3.697, p = 0.01567, Figure 1b). In contrast to agar plate assays, R. solanacearum strain variation was more evident and the growth reduction of Strains #2 and #3 was significantly lower compared to other strains (Ralstonia: F 6, 217 = 19.1102, p < 0.001, Figure 1b). The growth of Strains #1, #6, and #7 was reduced most by the Pseudomonas supernatants overall. In line with direct inhibition assays, the Pseudomonas CHA0 strain caused the greatest reduction in R. solanacearum growth, followed by strains Ph11C2 and MVP1‐4, which showed intermediate growth reduction (Pseudomonas strain: F 7, 216 = 11.3595, p < 0.001, Figure 1b). Pseudomonas strains 1M1‐96 and Q8R1‐96 showed relatively small growth reduction. Together, these findings are qualitatively similar to the direct inhibition assay (Figure 1a), highlighting the high efficacy of the Pseudomonas CHA0 strain in suppressing the growth of R. solanacearum.
3.3. Comparative genomic analysis reveals variation in the presence of secondary metabolic clusters between different Pseudomonas strains
Comparative genomic analysis of secondary metabolite clusters based on the antiSMASH output identified between 11 and 17 metabolic clusters in each of the eight Pseudomonas genomes (Tables A4, A5, A6, A7, A8, A9, A10, A11). The type and diversity of recognized clusters are shown in Table 1. Nonribosomal peptide synthetases (NRPS) were the most abundant secondary metabolite clusters, which was expected as these account for several common antimicrobials produced by fluorescent Pseudomonas strains (Girard et al., 2020; Hesse et al., 2018). Similarly, DAPG metabolite (belonging to the T3PKS cluster), as well as the pyoverdine siderophore (NRPS cluster) metabolite clusters, were found in all strains. Overall, the highest number of clusters were detected in CHA0 and Pf‐5 strains (17). These strains also harbored some unique metabolite clusters such as the T1PKS metabolic cluster, which encodes pyoluteorin antimicrobial, and the CDPS cluster, which encodes unknown metabolites. CHA0 and Pf‐5 also had the greatest number of NRPS clusters and were the only two strains capable of producing the cyclic lipopeptides known as orfamides. Other Pseudomonas strains also possessed some unique clusters such as the ectoine metabolic cluster found in the Q2‐87 strain. A more in‐depth insight into the clusters and the percentage similarity with various Pseudomonas and other bacterial genomes based on the antiSMASH database is shown in Tables A4, A5, A6, A7, A8, A9, A10, A11. The all clusters which were investigated further had 95%–100% similarity with already characterized secondary metabolite clusters, increasing the certainty of their predicted identity and functioning.
Table 1.
antiSMASH5.0 cluster types recognized in eight Pseudomonas genomes used in this study including a brief definition of their function
| Pseudomonas strain | Total no. of clusters | Cluster types | Description of metabolic clusters and examples of metabolites |
|---|---|---|---|
| CHA0 | 17 | NRPS (8), Bacteriocin (2), CDPS (1), T1PKS (1), T3PKS (1), NAGGN (1), Arylpolyene (1), Butyrolactone (1), Other (1) |
–NRPS (nonribosomal peptide synthetase cluster) e.g., orfamide, lipopeptide, viscosin, rhizomide, pyoverdine, enantiopyochelin –NRPS‐like (NRPS‐like fragment) e.g., mangotoxin, lankacidin –Bacteriocin (unspecified ribosomally synthesized and posttranslationally modified peptide product [RiPP] cluster) e.g., unknown –CDPS (tRNA‐dependent cyclodipeptide synthases) e.g., unknown –T1PKS (type I polyketide synthase) e.g., pyoluteorin –T3PKS (type III polyketide synthase) e.g., unknown –NAGGN (N‐acetylglutaminylglutamine amide) e.g., unknown –Arylpolyene (arylpolyene cluster) e.g., APE Vf –Butyrolactone (butyrolactone cluster) e.g., unknown –Betalactone (beta‐lactone containing protease inhibitor) –Ectoine (ectoine cluster) e.g., unknown –Lanthipeptide (lanthipeptide cluster) e.g., putative class II –LAP (linear azol(in)e‐containing peptides e.g., unknown –Other (unknown) e.g., pyrrolnitrin |
| Pf‐5 | 17 | NRPS (7), NRPS‐like (1), Bacteriocin (2), CDPS (1), T1PKS (1), T3PKS (1), NAGGN (1), Arylpolyene (1), Betalactone (1), Other (1) | |
| Q2‐87 | 15 | NRPS (4), NRPS‐like (1), Bacteriocin (3), T3PKS (1), NAGGN (1), Arylpolyene (1), Butyrolactone (1), Betalactone (1), Ectoine (1), Lanthipeptide (1) | |
| Q8R1‐96 | 13 | NRPS (6), NRPS‐like (1), Bacteriocin (1), T3PKS (1), NAGGN (1), Arylpolyene (1), Butyrolactone (1), Betalactone (1) | |
| 1M1‐96 | 12 | NRPS (3), NRPS‐like (1), Bacteriocin (1), T3PKS (1), NAGGN (1), Arylpolyene (1), Butyrolactone (1), Betalactone (1), Lanthipeptide (2) | |
| MVP1‐4 | 13 | NRPS (4), NRPS‐like (1), Bacteriocin (1), T3PKS (1), NAGGN (1), Arylpolyene (1), Butyrolactone (1), Betalactone (1), Lanthipeptide (2), LAP (1) | |
| F113 | 11 | NRPS (2), NRPS‐like (2), Bacteriocin (1), T3PKS (1), NAGGN (1), Arylpolyene (1), Butyrolactone (1), Betalactone (1), Lanthipeptide (1) | |
| Ph11C2 | 14 | NRPS (4), NRPS‐like (1), Bacteriocin (3), T3PKS (1), NAGGN (1), Arylpolyene (1), Butyrolactone (1), Betalactone (1), Lanthipeptide (1) |
3.4. Confirming the production of Pseudomonas secondary metabolites using mass spectrometry
Our genome screening results suggest that Pseudomonas inhibitory activity could have been due to the presence of DAPG or certain, less common, secondary metabolism clusters encoding pyoluteorin and Orfamides that were identified only in CHA0 and Pf‐5 strains. Due to the presence of these two unique clusters, CHA0 and Pf‐5 strains were chosen for a more detailed study. To verify the production of these compounds in LB media, the supernatants of CHA0 and Pf‐5 were analyzed using mass spectrometry against chemical standards for DAPG, pyoluteorin, orfamide A, and orfamide B. Based on matching monoisotopic masses to a database of Pseudomonas metabolites, a list of putative metabolites present in the Pseudomonas supernatants of CHA0 and Pf‐5 was determined using Progenesis QI v.2.0 (Waters) analysis (Table A12). These included compounds such as enantiopyochelin, rhizoxin, and indole‐3‐acetic acid which have different metabolic functions that can enhance bacterial survival (Table A12). For example, pyochelin is a siderophore that can aid iron acquisition when Pseudomonas may be in an iron‐limited environment to improve its survival chances and outcompete other bacterial competitors (Duffy & Defago, 1999). Based on a comparison with four available standards, only orfamide A production by CHA0 strain was detected in LB supernatant with an average concentration of 7.5 mg/ml (Figure A2). In contrast, none of the other four candidate compounds were detected in Pf‐5 supernatant in LB media. These results suggest that only orfamide A was produced by CHA0 in the conditions used in the supernatant inhibition assays. However, approximately 70 unidentified metabolites were detected in the samples, which could also have contributed to R. solanacearum inhibition.
3.5. Testing the inhibitory effects of identified Pseudomonas secondary metabolites on R. solanacearum growth
The inhibitory effects of DAPG, pyoluteorin, and orfamides A and B were tested individually against R. solanacearum strains using commercially available chemical standards. The effect of DAPG was tested against all R. solanacearum strains, while the effect of orfamides and pyoluteorin were tested against a subset of strains due to the high costs of chemical standards (Ralstonia strain #1 for orfamides, and R. solanacearum strains #1 and #7 for pyoluteorin). It was found that DAPG suppressed every R. solanacearum strain in a concentration‐dependent manner (Concentration: F 4, 200 = 238.2599, p < 0.001, Figure 2a), and all tested strains were unable to grow at the two highest concentrations (500 and 1000 μM). When all concentrations were included in the analysis, the effect of DAPG was independent of the R. solanacearum strain identity (Ralstonia: F 6, 98 = 0.724, p = 0.6305, Figure 2a). However, excluding the two highest concentrations, where all growth was inhibited, some R. solanacearum strain differences were revealed (Ralstonia: F 6, 56 = 3.355, p = 0.0068, Figure 2a): Strain #4 was the least susceptible to DAPG, while strains #3, #5, and #7 were relatively more susceptible. We also considered growth reduction by DAPG with growth in the absence of DAPG at the final time point for all R. solanacearum strains. Growth reduction was greater in 100 μM DAPG than 50 μM (Concentration: F 1, 124 = 46.8, p < 0.001, Figure 2a) and between strain variation was evident at both concentrations (Ralstonia at 100 μM: F 6, 56 = 5.671, p < 0.001, Figure 2a; Ralstonia at 50 μM: F 6, 56 = 3.856, p = 0.00273, Figure 2a). Post hoc analyses revealed that Strains #5 and #6 were the least susceptible to DAPG, while Strains #2 and #3 were relatively more susceptible. Both orfamides A and B reduced the growth of R. solanacearum strain #1 and this effect became clearer over time (Metabolite: F 2, 6 = 460.7, p < 0.001; Metabolite × Time: F 4, 12 = 45.806, p = 0.0359, Figure 2b). However, no difference was found between orfamides A and B (p = 0.11). No visible growth was observed when either R. solanacearum strains #1 or #7 were exposed to pyoluteorin (Ralstonia: F 2, 16 = 48.596, p < 0.001, Figure 2c), suggesting that both strains were highly susceptible to this compound.
Figure 2.
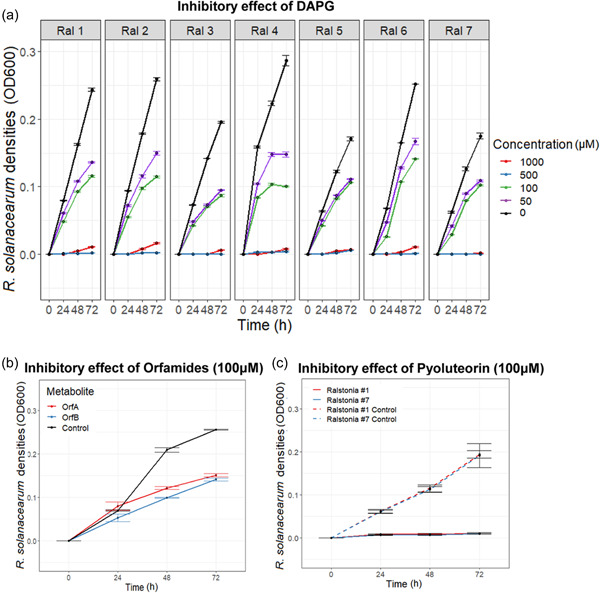
Testing the inhibitory effects of identified Pseudomonas secondary metabolites on Ralstonia solanacearum growth. Panel (a) shows the growth of seven R. solanacearum strains (in different panels) in various DAPG concentrations over time (1000 μM—red, 500 μM—blue, 100 μM—green, 50 μM—purple, and 0 μM LB broth [0 μM—control]—black). Panel (b) shows R. solanacearum strain #1 growth in the absence and presence of 100 μM of orfamides A and B. Panel (c) shows the growth of R. solanacearum strains #1 and #7 in the absence and presence of 100 μM of pyoluteorin. In all panels, bars show the standard error of the mean (±1 SEM) based on three replicates
3.6. Testing the inhibitory effects of orfamide variants isolated from Pseudomonas CHA0 on R. solanacearum growth
Orfamide variants “A” and “B” were isolated from the Pseudomonas CHA0 strain and their effects were tested only against R solanacearum strains #1 and #7 due to limited quantities of extracted compounds (Figure 3). The growth of both Ralstonia strains was constrained by both orfamide variants (Ralstonia × Treatment: F 2, 12 = 0.0476, p = 0.9537, Figure 3) and the growth suppression by the orfamide variants became visible only after 48 h during the assays (Treatment × Time: F 4, 30 = 75.652, p < 0.0001, Figure 3). At the final time point, there were clear differences between orfamide variants on both strain #1 (Treatment: F 2, 10 = 681.3, p < 0.001, Figure 3) and strain #7 (Treatment: F 4, 10 = 65.77, p < 0.001, Figure 3), with orfamide variant “B” showing a slightly higher pathogen growth suppression. Together these results suggest that the Pseudomonas CHA0 strain produces pathogen‐suppressing orfamide variants in vitro.
Figure 3.
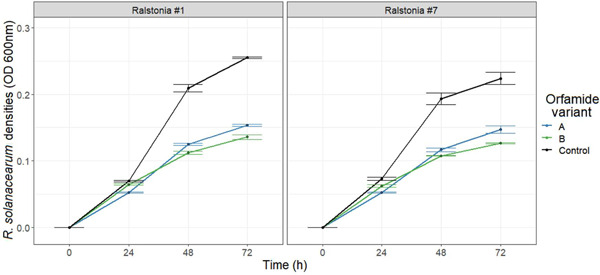
Testing the inhibitory effects of orfamide variants isolated from Pseudomonas CHA0 strain on Ralstonia solanacearum growth. The black lines denote R. solanacearum density in the absence of the orfamide variants over time (control), while colored lines show the growth in the presence of orfamide variants isolated from CHA0. Orfamide variants “A” and “B” refer to two different fractions isolated separately during the extraction process. All error bars show the standard error of the mean (±1 SEM) based on 3 replicates.
3.7. Testing Pseudomonas CHA0 biocontrol efficacy in vivo in a tomato system
The biocontrol potential of CHA0 was tested using the Micro‐Tom tomato cultivar infected by the UK R. solanacearum strains #1 and #7. In the absence of CHA0, the levels of disease incidence were higher when tomatoes were infected with Strain #7 compared to strain #1 (Ralstonia: F 1, 34 = 48.788, p = 0.315, Figure 4a). The CHA0 treatment did not affect wilting incidence compared to the water control treatment in the case of Ralstonia strain #1 (Pseudomonas: F 1, 16 = 24.731, p = 1, Figure 4a). However, CHA0 reduced wilting incidence of R. solanacearum strain #7 (Pseudomonas: F 1, 16 = 17.736, p = 0.01114, Figure 4a). The presence of CHA0 also increased the plant dry weight overall (Pseudomonas: F 1, 34 = 4.623, p = 0.0387, Figure 4b) and this effect was the same for both R. solanacearum strains (Ralstonia: F 1, 34 = 1.668, p = 0.205, Figure 4b). Together, these results suggest that CHA0 biocontrol efficacy was dependent on the R. solanacearum strain.
Figure 4.
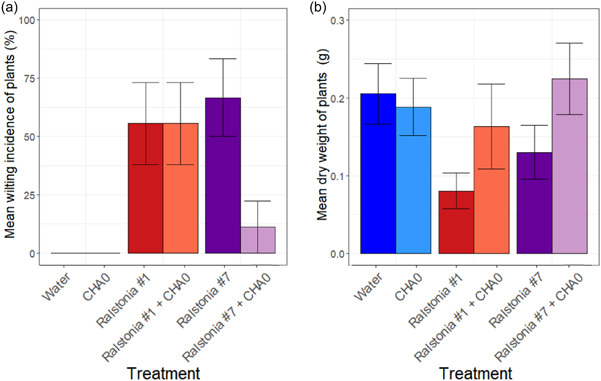
Tomato plant infections by Ralstonia solanacearum strains #1 and #7 in the absence and presence of Pseudomonas strains. Panel (a) displays the percentage of wilted plants (mean wilting incidence) and panel (b) the mean aboveground dry weight of individual plants at the end of the experiment for all treatments. All error bars show the standard error of the mean (±1 SEM) based on three replicates (every replicate consisting of nine plants)
4. DISCUSSION
This study aimed to screen and identify effective Pseudomonas bacterial biocontrol strains against Ralstonia solanacearum species complex, understand potential underlying mechanisms of inhibition, and validate their efficacy in vivo in the tomato rhizosphere. Pseudomonas strain CHA0 was the most suppressive strain in both direct and indirect R. solanacearum inhibition assays. The comparative genomic analysis highlighted that metabolite clusters encoding DAPG, pyoluteorin, and orfamides A and B were unique for the most suppressive CHA0 Pseudomonas strain, and while only orfamide A production was detected through mass spectrometry, extracted orfamide variants showed high inhibitory activity against R. solanacearum. In vivo tests revealed that CHA0 was effective at reducing bacterial wilt incidence. Interestingly, plant protection depended on the R. solanacearum strain identity and was only observed with one of the two tested strains. Despite the successful identification of potential Pseudomonas biocontrol species, more work is needed to harness their biocontrol activity in the plant rhizosphere.
When screening for the most effective Pseudomonas strains against R. solanacearum through direct and indirect assays, P. protegens strain CHA0 showed the most suppressive activity against all the tested R. solanacearum strains. The strong inhibition effects of CHA0 were observed through both direct and indirect assays. This result is in line with previous studies demonstrating the high biocontrol activity of CHA0 against various plant pathogens (Hu et al., 2016). Although multiple Pseudomonas strains have been described in the literature with biocontrol abilities, CHA0 is one of the most well‐established, studied, and successful biocontrol agents against plant‐parasitic nematodes, fungal, and bacterial pathogens (Flury et al., 2017; Humair et al., 2009; Jamali et al., 2009; Jousset et al., 2006; Neidig et al., 2011; Siddiqui & Shaukat, 2003, 2004). Interestingly, some less studied strains, such as MVP1‐4 and Ph11C2, showed also good biocontrol potential and warrant more study in the future. Overall, all Pseudomonas strains suppressed all tested R. solanacearum strains to some extent in vitro. The likely explanation for this is that European R. solanacearum strains are likely to be very similar as they belong to a clonal lineage of Phylotype 2b strains that are adapted to grow in cold climates (Hayward, 1991). In contrast, biocontrol strains belonged to different P. fluorescens subgroups (e.g., P. protegens and Pseudomonas corrugata) and originated from different countries across Europe and America. Hence, Pseudomonas strains were likely genetically more dissimilar compared to R. solanacearum strains. A more detailed analysis of European R. solanacearum strains is however required to link observed phenotypic similarities (equal susceptibility) and differences (infectivity in vivo) with underlying genetic differences.
Comparative genomic analyses revealed that Pseudomonas strains CHA0 and Pf‐5 (which belong to the P. protegens subgroup of P. fluorescens) had the greatest number of secondary metabolite clusters (17), while the other six Pseudomonas strains (the majority of which belong to the P. corrugata subgroup of P. fluorescens) harbored between 11 and 15 metabolic clusters. Based on previous literature, CHA0 and Pf‐5 can produce a similar range of secondary metabolites such as pyoluteorin and cyclic lipopeptides, which could potentially explain why CHA0 exhibited the greatest inhibitory effects (Haas & Keel, 2003; Loper & Gross, 2007; Ma et al., 2016). When testing these candidate metabolites on R. solanacearum growth, pyoluteorin stood out as the most suppressive compound, leading to the poorest R. solanacearum growth. Only Pseudomonas strains CHA0 and Pf‐5 were found to harbor the T1PKS metabolic cluster, which encodes pyoluteorin production. However, no pyoluteorin was produced in vitro by either of the strains in the growth conditions used. Pyoluteorin inactivity in our experiments was potentially due to the use of a rich growth medium (Heidari‐Tajabadi et al., 2011; Jamali et al., 2009; T. Yang et al., 2018). Pyoluteorin is a chlorinated polyketide antibiotic and its production is regulated by the DAPG precursor, monoacetylphloroglucinol (Kidarsa et al., 2011). It is thus possible that efficient production of pyoluteorin limits the production of DAPG, and that these compounds are not produced simultaneously. While CHA0 and Pf‐5 strains harbored the highest amount of NRPS clusters (8 and 7, respectively), they were also associated with a wide variety of other putative metabolites that could have been linked with pathogen suppression (e.g., enantiopyochelin, rhizomide, and pyoverdine). Furthermore, we also isolated uncharacterized orfamide variants from the CHA0 supernatant and observed highly inhibitory effects against the tested R. solanacearum strain. This suggests that orfamides at least partly explain the inhibitory capacity of the CHA0 strain in lab conditions. A wealth of other potential antimicrobial compounds was also identified in the CHA0 supernatant using nontargeted analysis. In the future, these secondary metabolites could be isolated and tested in more detailed studies to identify potentially novel antimicrobials (Geudens & Martins, 2018). For example, Rose et al. (2021) have shown that Pf‐5 has notable antialgal properties when interacting with microalga Chlamydomonas reinhardtii through the production of rhizoxins and they were also able to detect the production of DAPG, pyrrolnitrin, pyoluteorin, and orfamide A in their experiments. A key difference between ours and their study is that Pf‐5 was grown in a TAP medium under continuous illumination which could have changed the Pf‐5 metabolism and antimicrobial production (Rose et al., 2021). It is thus possible that the use of rich culture media constrained the production of certain secondary metabolites in our assays. In the future, it will be important to characterize the secondary metabolite gene expression and metabolite production across different environments and to identify metabolite production potential in more realistic biocontrol conditions (Deveau et al., 2016; Köhl et al., 2019).
To validate Pseudomonas efficacy in vivo, we first tested that both selected R. solanacearum strains could infect tomato plants and found that they both caused around 50% bacterial wilt disease incidence. Such variation in disease incidence is typical for R. solanacearum as its virulence is determined by a combination of host immunity and environmental conditions that can vary considerably even in controlled greenhouse experiments (Hu et al., 2016; Wei et al., 2019). We found that bacterial wilt incidence was clearly reduced in the presence of CHA0 but only in the case of R. solanacearum strain #7. These results contrast with the in vitro results as R. solanacearum strains #1 and #7 did not show any differences in their susceptibility to DAPG, pyoluteorin, or orfamide variants produced by CHA0. There are multiple potential biological explanations for this result. For example, it is possible that Ralstonia strain #1, was less able to colonize tomato roots, failed to express genes required for protection against Pseudomonas inhibition (e.g., efflux pumps or other deactivation of antimicrobials), or was not able to activate virulence gene expression (Ran et al., 2005) in the presence of CHA0. Alternatively, the conditions used in tomato experiments may not have been optimal for R. solanacearum survival. Moreover, autoclaving of the soil could have influenced soil properties and microbial metabolism as heating soil over 120°C can result in increased levels of ammonium and nitrogen (Serrasolsas & Khanna, 1995). Together, these findings demonstrate the challenges of translating in vitro results to in vivo applications, highlighting the importance of studying biocontrol effects in more realistic in vivo conditions.
In conclusion, here we show that a combination of in silico, in vitro, and in planta approaches can be used to identify and validate effective biocontrol agents against R. solanacearum. By using simple microbiological assays based on direct and indirect interactions with the pathogen we were able to identify P. protegens CHA0 as the most inhibitory biocontrol strain. Comparative genomics was used to identify potential secondary metabolite clusters responsible for the inhibition and efficacy of identified compounds and their production was validated in additional experiments. In addition to identified clusters, orfamide variants were also found to be highly inhibitory which could be characterized in the future (Keel et al., 1992; Yasmin et al., 2017). Based on these results, screening for pyoluteorin and orfamide secondary clusters could be used as a rapid way to identify effective biocontrol strains in the future. Such screening methods have been previously used to identify siderophore‐producing biocontrol agents against rice fungal pathogens (Chaiharn et al., 2009). Despite the ability to produce a repertoire of secondary metabolites, CHA0 was able to reduce disease incidence with only one of the two tested R. solanacearum strains in vivo. Biocontrol effects should thus be studied in more realistic in vivo conditions in the future to test which mechanisms are expressed and active in the plant rhizosphere and if the natural plant microbiota is also affected by or affects the activity of identified biocontrol bacterial strains. Together, our results suggest that while the developed low‐cost in vitro screening process can be used to identify effective biocontrol agents based on their secondary metabolism, further optimization is needed to predict the function of different strains in the plant rhizosphere.
AUTHOR CONTRIBUTIONS
Sophie E. Clough: Conceptualization (equal); data curation (equal); formal analysis (equal); investigation (equal); methodology (equal); validation (equal); visualization (equal); writing–original draft (equal); writing–review and editing (equal). Alexandre Jousset: Data curation (equal); investigation (equal); methodology (equal); writing–review and editing (equal). John G. Elphinstone: Investigation (equal); supervision (equal); writing–review and editing (equal). Ville‐Petri Friman: Conceptualization (equal); formal analysis (equal); funding acquisition (equal); investigation (equal); project administration (equal); supervision (equal); writing–review and editing (equal).
CONFLICTS OF INTEREST
None required.
ETHICS STATEMENT
None required.
ACKNOWLEDGMENTS
The authors thank Tony Larson and Swen Wagner of the Centre of Excellence in Mass Spectrometry group for performing Pseudomonas metabolite identification and semi‐quantification of select target compounds by ultra‐performance liquid chromatography‐mass spectrometry (UPLC‐MS) analysis. Sophie E. Clough was funded by the White Rose PhD Studentship Network and N8 AgriFood pump‐priming grant at the University of Leeds. Ville‐Petri Friman is funded by the Royal Society (grant nos. RSG\R1\180213 and CHL\R1\180031) and jointly by a grant from UKRI, Defra, and the Scottish Government, under the Strategic Priorities Fund Plant Bacterial Diseases program (BB/T010606/1).
1.
See Figure A1, Figure A2, Tables A1 and A2, Table A3, and Tables A4, A5, A6, A7, A8, A9, A10, A11, Table A12.
Figure A1.
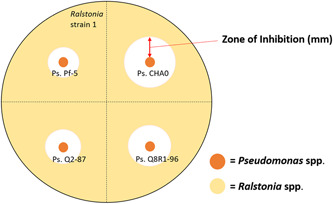
Experimental design quantifying direct inhibition of Ralstonia solanacearum strain #1 by four types of Pseudomonas strains using soft agar overlay assay
Figure A2.
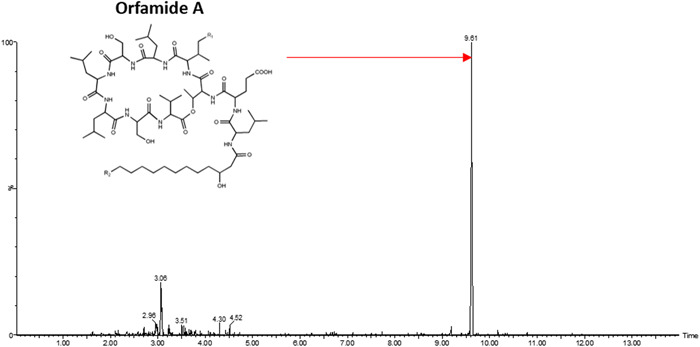
Mass spectrometry analysis indicating the presence of Pseudomonas metabolite orfamide A in the supernatant of strain CHA0. Orfamide A is identified with a monoisotopic mass (m/z) of 1317.826 [M + Na]+ and a retention time (t R) of 9.61 s at 15 ppm. This spectrum represents the replicate with the strongest peak out of the three replicates
Table A1.
Pseudomonas strains used in these experiments, and their geographical origin
| Pseudomonas strains | Pseudomonas fluorescens subgroup | Pseudomonas strains' origin |
|---|---|---|
| CHA0 | (Pseudomonas protegens) | Tobacco, Switzerland (Natsch et al., 1994) |
| Pf‐5 | (P. protegens) | Cotton, USA (Howell & Stipanovic, 1979) |
| Q2‐87 | (Pseudomonas corrugata) | Wheat, USA (Bangera & Thomashow, 1999) |
| Q8R1‐96 | (P. corrugata) | Wheat, USA (Raaijmakers & Weller, 1998) |
| 1M1‐96 (P. fluorescens) | Not reported | Wheat, USA (Raaijmakers & Weller, 2001) |
| MVP1‐4 (P. fluorescens) | Not reported | Pea, USA (Landa et al., 2002) |
| F113 | (P. corrugata) | Sugar beet, Ireland (Shanahan et al., 1992) |
| Ph11C2 (P. fluorescens) | Not reported | Tomato, France (Govaerts et al., 2007) |
Table A2.
Ralstonia solanacearum strains used in these experiments, and their geographical origin
| Ralstonia strains | York sample collection number | Ralstonia strains' origin |
|---|---|---|
| #1 | YO352 | River water, UK (2014) |
| #2 | YO354 | River water, UK (2015) |
| #3 | YO355 | River water, UK (2015) |
| #4 | YO356 | River water, UK (2013) |
| #5 | YO162 | Commercial culture, Poland (used for verification tests) (2014) |
| #6 | YO351 | River water, UK (2013) |
| #7 | YO353 | River water, UK (2014) |
Table A3.
Media used for bacterial cultures
| Media recipes | |
|---|---|
| LB broth (1 L) | 10 g Tryptone (pancreatic digest of casein) |
| 5 g Yeast extract | |
| 5 g NaCl | |
| (+15 g agar for solid media) | |
| CPG broth (1 L) | 1 g Casamino acids (casein hydrolysate) |
| 10 g Peptone | |
| 5 g Glucose | |
| (+17 g agar for solid media) | |
| NB broth (1 L) | 10 g Glucose |
| 5 g Tryptone | |
| 3 g Beef extract | |
| 0.5 g Yeast extract | |
| Kings Medium B broth (1 L) | 20 g Proteose peptone |
| 1.5 g K2HPO4 | |
| 1.5 g MgSO4·7H2O | |
| 10 ml Glycerol | |
| Cryomedia for freezing bacterial stocks | 50% vol/vol glycerol |
Table A4.
antiSMASH5.0 cluster types recognized in Pseudomonas CHA0 genome including the types of clusters and % hit in the database with the most similar known cluster
| Cluster | Type | Most similar known cluster |
|---|---|---|
| 1.1 | Bacteriocin | |
| 1.2 | CDPS | |
| 1.3 | NRPS | Orfamide (94%) |
| 2.1 | other | Pyrrolnitrin (100%) |
| 2.2 | NRPS | Pyochelin (100%) |
| 2.3 | T1PKS | Pyoluteorin (100%) |
| 3.1 | NRPS | Lipopeptide (6%) |
| 3.2 | NAGGN | |
| 3.3 | NRPS | Pyoverdine (16%) |
| 4.1 | T3PKS | DAPG (100%) |
| 4.2 | Bacteriocin | |
| 5.1 | Arylpolyene | APE Vf (40%) |
| 6.1 | NRPS | Viscosin (31%) Pyoverdine (14%) |
| 6.2 | Betalactone | |
| 8.1 | NRPS | Pyoverdine (19%) |
| 10.1 | NRPS | Pyoverdine (3%) |
| 13.1 | NRPS |
Table A5.
antiSMASH5.0 cluster types recognized in Pseudomonas Pf‐5 genome including the types of clusters and % hit in the database with the most similar known cluster
| Cluster | Type | Most similar known cluster |
|---|---|---|
| 1.1 | NRPS | Pyoverdine (14%) |
| 1.2 | Betalactone | |
| 1.3 | Other | Pyrrolnitrin (100%) |
| 1.4 | NRPS | Pyochelin 100%) |
| 1.5 | NRPS‐like | Rhizoxin (100%) |
| 1.6 | T1PKS | Pyoluteorin (100%) |
| 2.1 | Bacteriocin | |
| 2.2 | CDPS | |
| 2.3 | NRPS | Orfamide (94%) |
| 3.1 | NRPS | Pyoverdine (21%) |
| 3.2 | NAGGN | |
| 3.3 | NRPS | Lipopeptide (6%) |
| 4.1 | T3PKS | DAPG (100%) |
| 4.2 | Bacteriocin | APE Vf (40%) |
| 5.1 | Arylpolyene | |
| 7.1 | NRPS | Pyoverdine (19%) |
| 9.1 | NRPS | Rhizomide (100%) |
Table A6.
antiSMASH5.0 cluster types recognized in Pseudomonas Q2‐87 genome including the types of clusters and % hit in the database with the most similar known cluster
| Cluster | Type | Most similar known cluster |
|---|---|---|
| 1.1 | NAGGN | |
| 1.2 | NRPS | Pyoverdine (10%) |
| 1.3 | Betalactone | |
| 2.1 | Butyrolactone | |
| 4.1 | T3pks | DAPG (100%) |
| 4.2 | Bacteriocin | |
| 5.1 | Bacteriocin | |
| 6.1 | NRPS | Cupriachelin (35%) |
| 7.1 | Arylpolyene | APE vf (40%) |
| 8.1 | NRPS | Ishigamide (11%) |
| 9.1 | NRPS | Pyoverdine (19%) |
| 9.2 | Ectoine | |
| 15.1 | Bacteriocin | |
| 19.1 | Lanthipeptide |
Table A7.
antiSMASH5.0 cluster types recognized in Pseudomonas Q8R196 genome including the types of clusters and % hit in the database with the most similar known cluster
| Cluster | Type | Most similar known cluster |
|---|---|---|
| 1.1 | NRPS | Pyoverdine (10%) |
| 1.2 | Betalactone | |
| 1.3 | T3PKS | DAPG (100%) |
| 1.4 | NRPS | Cupriachelin (11%) |
| 1.5 | Butyrolactone | |
| 4.1 | Arylpolyene | APE Vf (40%) |
| 6.1 | NAGGN | Taiwachelin (11%) |
| 9.1 | NRPS | Viscosin (31%) Pyoverdine (10%) |
| 12.1 | Bacteriocin | |
| 20.1 | NRPS‐Like | Mangotoxin (71%) |
| 23.1 | NRPS | Pyoverdine (3%) |
| 24.1 | NRPS | Taiwachelin (11%) Pyoverdine (3%) |
| 25.1 | NRPS |
Table A8.
antiSMASH5.0 cluster types recognized in Pseudomonas 1M1‐96 genome including the types of clusters and % hit in the database with the most similar known cluster
| Cluster | Type | Most similar known cluster |
|---|---|---|
| 2.1 | T3PKS | DAPG (100%) |
| 2.2 | NRPS | Cupriachelin (11%) |
| 2.3 | Butyrolactone | |
| 3.1 | NAGGN | |
| 4.1 | NRPS | Serobactin (23%) Pyoverdine (10%) |
| 8.1 | Betalactone | |
| 9.1 | Bacteriocin | |
| 17.1 | Arylpolene | APE Vf (40%) |
| 32.1 | Lanthipeptide | Putative class II |
| 44.1 | NRPS‐like | Mangotoxin (71%) |
| 49.1 | Lanthipeptide | Putative class II |
| 62.1 | NRPS | Viscosin (25%) Pyoverdine (6%) |
Table A9.
antiSMASH5.0 cluster types recognized in Pseudomonas MVP1‐4 genome including the types of clusters and % hit in the database with the most similar known cluster
| Cluster | Type | Most similar known cluster |
|---|---|---|
| 2.1 | Betalactone | |
| 2.2 | LAP | |
| 4.1 | NRPS | Cupriachelin (11%) |
| 4.2 | T3PKS | DAPG (100%) |
| 6.1 | Arylpolyene | APE Vf (40%) |
| 9.1 | NRPS‐like | Mangotoxin (71%) |
| 10.1 | Bacteriocin | |
| 13.1 | NAGGN | |
| 24.1 | Lanthipeptide | Putative class II |
| 30.1 | Butyrolactone | |
| 32.1 | NRPS | Viscosin (31%) Pyoverdine (10%) |
| 35.1 | NRPS | Pyoverdine (10%) |
| 36.1 | NRPS | Pyoverdine (8%), Serobactin (15%), Taiwachelin (11%) |
Table A10.
antiSMASH5.0 cluster types recognized in Pseudomonas F113 genome including the types of clusters and % hit in the database with the most similar known cluster
| Cluster | Type | Most similar known cluster |
|---|---|---|
| 2.1 | Arylpolyene | APE Vf (40%) |
| 4.1 | NAGGN | |
| 4.2 | NRPS | Pyoverdine (10%), Serobactins (23%) |
| 5.1 | NRPS‐like | Lankacidin (26%) |
| 6.1 | Bacteriocin | |
| 11.1 | Betalactone | |
| 12.1 | T3PKS | DAPG (100%) |
| 16.1 | NRPS | Viscosin (31%), Pyoverdine (11%) |
| 19.1 | Butyrolactone | |
| 20.1 | Lantipeptide |
Table A11.
antiSMASH5.0 cluster types recognized in Pseudomonas Ph11C2 genome including the types of clusters and % hit in the database with the most similar known cluster
| Cluster | Type | Most similar known cluster |
|---|---|---|
| 1.1 | NAGGN | |
| 1.2 | NRPS | Serobactins (23%) Pyoverdine (10%) |
| 1.3 | Betalactone | |
| 2.1 | NRPS | |
| 2.2 | Bacteriocin | |
| 2.3 | Butyrolactone | |
| 4.1 | Bacteriocin | |
| 5.1 | NRPS‐like | Mangotoxin (71%) |
| 5.2 | Bacteriocin | Pyoverdine (1%) |
| 5.3 | Arylpolyene | APE Vf (40%) |
| 6.1 | NRPS | WLIP (28%), Cupriachelin (11%), Pyoverdine (2%) |
| 6.2 | T3PKS | DAPG (100%) |
| 7.1 | NRPS | Viscosin (31%), Taiwachelin (27%), Pyoverdine (19%) |
| 12.1 | Lanthipeptide | Putative class II |
Table A12.
Putative Pseudomonas metabolites produced in LB media identified using Progenesis software
| Candidate compound | Metabolite function | Present in CHA0 supernatant | Present in Pf‐5 supernatant |
|---|---|---|---|
| Indole‐3‐acetic acid | Auxin | Yes | Yes |
| Pyochelin | Siderophore | Yes | |
| Rhizoxin | Interferes with mitosis by binding to Beta‐tubulin affecting microtubule dynamics | Yes | Yes |
Clough, S. E. , Jousset, A. , Elphinstone, J. G. , & Friman, V.‐P. (2022). Combining in vitro and in vivo screening to identify efficient Pseudomonas biocontrol strains against the phytopathogenic bacterium Ralstonia solanacearum . MicrobiologyOpen, 11, e1283. 10.1002/mbo3.1283
DATA AVAILABILITY STATEMENT
Raw reads for all Pseudomonas strains are available at the NCBI Sequence Read Archive under BioProject accession number: PRJNA784243: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA784243.
REFERENCES
- Almario, J. , Bruto, M. , Vacheron, J. , Prigent‐Combaret, C. , Moënne‐Loccoz, Y. , & Muller, D. (2017). Distribution of 2,4‐diacetylphloroglucinol biosynthetic genes among the Pseudomonas spp. reveals unexpected polyphyletism. Frontiers in Microbiology, 8, 1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangera, M. G. , & Thomashow, L. S. (1999). Identification and characterization of a gene cluster for synthesis of the polyketide antibiotic 2,4‐diacetylphloroglucinol from Pseudomonas fluorescens Q2‐87. Journal of Bacteriology, 181(10), 3155–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, J. , Eisenhauer, N. , Scheu, S. , & Jousset, A. (2012). Increasing antagonistic interactions cause bacterial communities to collapse at high diversity. Ecology Letters, 15(5), 468–474. 10.1111/j.1461-0248.2012.01759 [DOI] [PubMed] [Google Scholar]
- Blin, K. , Shaw, S. , Steinke, K. , Villebro, R. , Ziemert, N. , Lee, S. Y. , Medema, M. H. , & Weber, T. (2019). antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Research, 47(W1), W81–W87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin, K. , Wolf, T. , Chevrette, M. G. , Lu, X. , Schwalen, C. J. , Kautsar, S. A. , Suarez Duran, H. G. , de Los Santos, E. L. , Kim, H. U. , Nave, M. , & Dickschat, J. S. (2017). antiSMASH 4.0—Improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Research, 45(W1), W36–W41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso, P. , Biosca, E. G. , Bertolini, E. , Marco‐Noales, E. , Gorris, M. T. , Licciardello, C. , & López, M. M. (2017). Genetic diversity reflects geographical origin of Ralstonia solanacearum strains isolated from plant and water sources in Spain. International Microbiology, 20(4), 155–164. [DOI] [PubMed] [Google Scholar]
- Castillo, J. A. , & Greenberg, J. T. (2007). Evolutionary dynamics of Ralstonia solanacearum . Applied and Environmental Microbiology, 73(4), 1225–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaiharn, M. , Chunhaleuchanon, S. , & Lumyong, S. (2009). Screening siderophore producing bacteria as potential biological control agent for fungal rice pathogens in Thailand. World Journal of Microbiology and Biotechnology, 25(11), 1919–1928. [Google Scholar]
- Chen, J. , Yu, Y. , Li, S. , & Ding, W. (2016). Resveratrol and coumarin: Novel agricultural antibacterial agent against Ralstonia solanacearum in vitro and in vivo. Molecules, 21(11), 1501. 10.3390/molecules21111501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, C. R. , Studholme, D. J. , Hayes, B. , Runde, B. , Weisberg, A. , Cai, R. , Wroblewski, T. , Daunay, M. C. , Wicker, E. , Castillo, J. A. , & Vinatzer, B. A. (2015). Genome‐enabled phylogeographic investigation of the quarantine pathogen Ralstonia solanacearum race 3 biovar 2 and screening for sources of resistance against its core effectors. Phytopathology, 105(5), 597–607. [DOI] [PubMed] [Google Scholar]
- Compant, S. , Duffy, B. , Nowak, J. , Clément, C. , & Barka, E. A. (2005). Use of plant growth‐promoting bacteria for biocontrol of plant diseases: Principles, mechanisms of action, and future prospects. Applied and Environmental Microbiology, 71(9), 4951–4959. 10.1128/AEM.71.9.4951-4959.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin, D. , Moenne‐Loccoz, Y. , Fenton, A. , Dunne, C. , Dowling, D. N. , & O'gara, F. (1997). Role of 2,4‐diacetylphloroglucinol in the interactions of the biocontrol Pseudomonas strain F113 with the potato cyst nematode Globodera rostochiensis . Applied and Environmental Microbiology, 63(4), 1357–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau, A. , Gross, H. , Palin, B. , Mehnaz, S. , Schnepf, M. , Leblond, P. , Dorrestein, P. C. , & Aigle, B. (2016). Role of secondary metabolites in the interaction between Pseudomonas fluorescens and soil microorganisms under iron‐limited conditions. FEMS Microbiology Ecology, 92(8), fiw107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy, B. K. , & Défago, G. (1999). Environmental factors modulating antibiotic and siderophore biosynthesis by Pseudomonas fluorescens biocontrol strains. Applied and Environmental Microbiology, 65(6), 2429–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EPPO . (2021). Ralstonia solanacearum species complex. EPPO datasheets on pests recommended for regulation. https://gd.eppo.int
- Flury, P. , Vesga, P. , Péchy‐Tarr, M. , Aellen, N. , Dennert, F. , Hofer, N. , Kupferschmied, K. P. , Kupferschmied, P. , Metla, Z. , Ma, Z. , & Siegfried, S. (2017). Antimicrobial and insecticidal: Cyclic lipopeptides and hydrogen cyanide produced by plant‐beneficial Pseudomonas strains CHA0, CMR12a, and PCL1391 contribute to insect killing. Frontiers in Microbiology, 8, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fravel, D. R. (2005). Commercialization and implementation of biocontrol. Annual Review of Phytopathology, 43, 337–359. [DOI] [PubMed] [Google Scholar]
- Genin, S. (2010). Molecular traits controlling host range and adaptation to plants in Ralstonia solanacearum . New Phytologist, 187(4), 920–928. 10.1111/j.1469-8137.2010.03397.x [DOI] [PubMed] [Google Scholar]
- Genin, S. , & Denny, T. P. (2012). Pathogenomics of the Ralstonia solanacearum species complex. Annual Review of Phytopathology, 50, 67–89. 10.1146/annurev-phyto-081211-173000 [DOI] [PubMed] [Google Scholar]
- Geudens, N. , & Martins, J. C. (2018). Cyclic lipodepsipeptides from Pseudomonas spp.—Biological Swiss‐army knives. Frontiers in Microbiology, 9, 1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard, L. , Höfte, M. , & Mot, R. D. (2020). Lipopeptide families at the interface between pathogenic and beneficial Pseudomonas‐plant interactions. Critical Reviews in Microbiology, 46(4), 397–419. [DOI] [PubMed] [Google Scholar]
- Gopalan‐Nair, R. , Jardinaud, M. F. , Legrand, L. , Landry, D. , Barlet, X. , Lopez‐Roques, C. , Vandecasteele, C. , Bouchez, O. , Genin, S. , & Guidot, A. (2021). Convergent rewiring of the virulence regulatory network promotes adaptation of Ralstonia solanacearum on resistant tomato. Molecular Biology and Evolution, 38(5), 1792–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govaerts, B. , Fuentes, M. , Mezzalama, M. , Nicol, J. M. , Deckers, J. , Etchevers, J. D. , Figueroa‐Sandoval, B. , & Sayre, K. D. (2007). Infiltration, soil moisture, root rot and nematode populations after 12 years of different tillage, residue and crop rotation managements. Soil and Tillage Research, 94(1), 209–219. 10.1016/j.still.2006.07.013 [DOI] [Google Scholar]
- Gu, Y. , Wei, Z. , Wang, X. , Friman, V.‐P. , Huang, J. , Wang, X. , Mei, X. , Xu, Y. , Shen, Q. , & Jousset, A. (2016). Pathogen invasion indirectly changes the composition of soil microbiome via shifts in root exudation profile. Biology and Fertility of Soils, 52(7), 997–1005. [Google Scholar]
- Guidot, A. , Coupat, B. , Fall, S. , Prior, P. , & Bertolla, F. (2009). Horizontal gene transfer between Ralstonia solanacearum strains detected by comparative genomic hybridization on microarrays. The ISME Journal, 3(5), 549–562. [DOI] [PubMed] [Google Scholar]
- Guidot, A. , Jiang, W. , Ferdy, J. B. , Thébaud, C. , Barberis, P. , Gouzy, J. , & Genin, S. (2014). Multihost experimental evolution of the pathogen Ralstonia solanacearum unveils genes involved in adaptation to plants. Molecular Biology and Evolution, 31(11), 2913–2928. [DOI] [PubMed] [Google Scholar]
- Haas, D. , & Défago, G. (2005). Biological control of soil‐borne pathogens by fluorescent pseudomonads. Nature Reviews Microbiology, 3, 307–319. 10.1038/nrmicro1129 [DOI] [PubMed] [Google Scholar]
- Haas, D. , & Keel, C. (2003). Regulation of antiobiotic production in root‐colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annual Review of Phytopathology, 41, 117–153. 10.1146/annurev.phyto.41.052002.095656 [DOI] [PubMed] [Google Scholar]
- Hansen, Z. R. , & Keinath, A. P. (2013). Increased pepper yields following incorporation of biofumigation cover crops and the effects on soilborne pathogen populations and pepper diseases. Applied Soil Ecology, 63, 67–77. [Google Scholar]
- Hayward, A. C. (1991). Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum . Annual Review of Phytopathology, 29, 65–87. [DOI] [PubMed] [Google Scholar]
- Heidari‐Tajabadi, F. , Ahmadzadeh, M. , Moinzadeh, A. , & Khezri, M. (2011). Influence of some culture media on antifungal activity of Pseudomonas fluorescens UTPF61 against the Sclerotinia sclerotiorum . African Journal of Agricultural Research, 6(30), 6340–6347. [Google Scholar]
- Hesse, C. , Schulz, F. , Bull, C. T. , Shaffer, B. T. , Yan, Q. , Shapiro, N. , Hassan, K. A. , Varghese, N. , Elbourne, L. D. , Paulsen, I. T. , & Kyrpides, N. (2018). Genome‐based evolutionary history of Pseudomonas spp. Environmental Microbiology, 20(6), 2142–2159. [DOI] [PubMed] [Google Scholar]
- Howell, C. R. , & Stipanovic, R. D. (1979). Control of Rhizoctonia solani on cotton seedlings with Pseudomonas fluorescens and with an antibiotic produced by the bacterium. Phytopathology, 69(5), 480–482. [Google Scholar]
- Hu, J. , Wei, Z. , Friman, V.‐P. , Gu, S. H. , Wang, X. F. , Eisenhauer, N. , Yang, T. J. , Ma, J. , Shen, Q. R. , Xu, Y. C. , & Jousset, A. (2016). Probiotic diversity enhances rhizosphere microbiome function and plant disease suppression. mBio, 7(6):e01790‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humair, B. , González, N. , Mossialos, D. , Reimmann, C. , & Haas, D. (2009). Temperature‐responsive sensing regulates biocontrol factor expression in Pseudomonas fluorescens CHA0. The ISME Journal, 3(8), 955–965. [DOI] [PubMed] [Google Scholar]
- Jamali, F. , Sharifi‐Tehrani, A. , Lutz, M. P. , & Maurhofer, M. (2009). Influence of host plant genotype, presence of a pathogen, and coinoculation with Pseudomonas fluorescens strains on the rhizosphere expression of hydrogen cyanide‐ and 2,4‐diacetylphloroglucinol biosynthetic genes in P. fluorescens biocontrol strain CHA0. Microbial Ecology, 57(2), 267–275. [DOI] [PubMed] [Google Scholar]
- Jousset, A. , Lara, E. , Wall, L. G. , & Valverde, C. (2006). Secondary metabolites help biocontrol strain Pseudomonas fluorescens CHA0 to escape protozoan grazing. Applied and Environmental Microbiology, 72(11), 7083–7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczmarek, A. , Mackenzie, K. , Kettle, H. , & Blok, V. C. (2014). Influence of soil temperature on Globodera rostochiensis and Globodera pallida . Phytopathologia Mediterranea, 396–405. [Google Scholar]
- Kaur, T. , Rani, R. , & Manhas, R. K. (2019). Biocontrol and plant growth promoting potential of phylogenetically new Streptomyces sp. MR14 of rhizospheric origin. AMB Express, 9, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keel, C. , Schnider, U. , Maurhofer, M. , Voisard, C. , Laville, J. , Burger, U. , Wirthner, P. J. , Haas, D. , & Défago, G. (1992). Suppression of root diseases by Pseudomonas fluorescens CHA0: Importance of the bacterial secondary metabolite 2,4‐diacetylphloroglucinol. Molecular Plant‐Microbe Interactions, 5(1), 4–13. [Google Scholar]
- Kidarsa, T. A. , Goebel, N. C. , Zabriskie, T. M. , & Loper, J. E. (2011). Phloroglucinol mediates cross‐talk between the pyoluteorin and 2,4‐diacetylphloroglucinol biosynthetic pathways in Pseudomonas fluorescens Pf‐5. Molecular Microbiology, 81(2), 395–414. [DOI] [PubMed] [Google Scholar]
- Köhl, J. , Kolnaar, R. , & Ravensberg, W. J. (2019). Mode of action of microbial biological control agents against plant diseases: Relevance beyond efficacy. Frontiers in Plant Science, 10, 845. 10.3389/fpls.2019.00845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landa, B. B. , Mavrodi, O. V. , Raaijmakers, J. M. , Gardener, B. B. M. , Thomashow, L. S. , & Weller, D. M. (2002). Differential ability of genotypes of 2,4‐diacetylphloroglucinol‐producing Pseudomonas fluorescens strains to colonize the roots of pea plants. Applied and Environmental Microbiology, 68(7), 3226–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Hu, Y. , Bolund, L. , & Wang, J. (2010). State of the art de novo assembly of human genomes from massively parallel sequencing data. Human Genomics, 4(4), 271–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loper, J. E. , & Gross, H. (2007). Genomic analysis of antifungal metabolite production by Pseudomonas fluorescens Pf‐5. European Journal of Plant Pathology, 119(3), 265–278. 10.1007/s10658-007-9179-8 [DOI] [Google Scholar]
- Ma, Z. , Geudens, N. , Kieu, N. P. , Sinnaeve, D. , Ongena, M. , Martins, J. C. , & Höfte, M. (2016). Biosynthesis, chemical structure, and structure‐activity relationship of orfamide lipopeptides produced by Pseudomonas protegens and related species. Frontiers in Microbiology, 7, 382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield, J. , Genin, S. , Magori, S. , Citovsky, V. , Sriariyanum, M. , Ronald, P. , Dow, M. A. X. , Verdier, V. , Beer, S. V. , Machado, M. A. , & Toth, I. A. N. (2012). Top 10 plant pathogenic bacteria in molecular plant pathology. Molecular Plant Pathology, 13(6), 614–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagachandrabose, S. (2020). Management of potato cyst nematodes using liquid bioformulations of Pseudomonas fluorescens, Purpureocillium lilacinum and Trichoderma viride . Potato Research, 63, 479–496. [Google Scholar]
- Natsch, A. , Keel, C. , Pfirter, H. A. , Haas, D. , & Défago, G. (1994). Contribution of the global regulator gene gacA to persistence and dissemination of Pseudomonas fluorescens biocontrol strain CHA0 introduced into soil microcosms. Applied and Environmental Microbiology, 60(7), 2553–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidig, N. , Paul, R. J. , Scheu, S. , & Jousset, A. (2011). Secondary metabolites of Pseudomonas fluorescens CHA0 drive complex non‐trophic interactions with bacterivorous nematodes. Microbial Ecology, 61(4), 853–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nion, Y. A. , & Toyota, K. (2015). Recent trends in control methods for bacterial wilt diseases caused by Ralstonia solanacearum . Microbes and Environments, 30, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notz, R. , Maurhofer, M. , Schnider‐Keel, U. , Duffy, B. , Haas, D. , & Défago, G. (2001). Biotic factors affecting expression of the 2,4‐diacetylphloroglucinol biosynthesis gene phlA in Pseudomonas fluorescens biocontrol strain CHA0 in the rhizosphere. Phytopathology, 91(9), 873–881. [DOI] [PubMed] [Google Scholar]
- Parkinson, N. , Bryant, R. , Bew, J. , Conyers, C. , Stones, R. , Alcock, M. , & Elphinstone, J. G. (2013). Application of variable‐number tandem‐repeat typing to discriminate Ralstonia solanacearum strains associated with English watercourses and disease outbreaks. Applied and Environmental Microbiology, 79(19), 6016–6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen, A. L. , Nybroe, O. , Winding, A. , Ekelund, F. , & Bjørnlund, L. (2009). Bacterial feeders, the nematode Caenorhabditis elegans and the flagellate Cercomonas longicauda, have different effects on outcome of competition among the Pseudomonas biocontrol strains CHA0 and DSS73. Microbial Ecology, 57(3), 501–509. [DOI] [PubMed] [Google Scholar]
- Pierson, L. S., III , Wood, D. W. , Pierson, E. A. , & Chancey, S. T. (1998). N‐acyl‐homoserine lactone‐mediated gene regulation in biological control by fluorescent pseudomonads: Current knowledge and future work. European Journal of Plant Pathology, 104, 1–9. [Google Scholar]
- Raaijmakers, J. M. , & Weller, D. M. (1998). Natural plant protection by 2,4‐diacetylphloroglucinol‐producing Pseudomonas spp. in take‐all decline soils. Molecular Plant‐Microbe Interactions, 11(2), 144–152. [Google Scholar]
- Raaijmakers, J. M. , & Weller, D. M. (2001). Exploiting genotypic diversity of 2,4‐diacetylphloroglucinol‐producing Pseudomonas spp.: Characterization of superior root‐colonizing P. fluorescens strain Q8r1‐96. Applied and Environmental Microbiology, 67(6), 2545–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran, L. X. , Liu, C. Y. , Wu, G. J. , van Loon, L. C. , & Bakker, P. A. H. M. (2005). Suppression of bacterial wilt in Eucalyptus urophylla by fluorescent Pseudomonas spp. in China. Biological Control, 32(1), 111–120. [Google Scholar]
- Rose, M. M. , Scheer, D. , Hou, Y. , Hotter, V. S. , Komor, A. J. , Aiyar, P. , Scherlach, K. , Vergara, F. , Yan, Q. , Loper, J. E. , & Jakob, T. (2021). The bacterium Pseudomonas protegens antagonizes the microalga Chlamydomonas reinhardtii using a blend of toxins. Environmental Microbiology, 23(9), 5525–5540. [DOI] [PubMed] [Google Scholar]
- Serrasolsas, I. , & Khanna, P. K. (1995). Changes in heated and autoclaved forest soils of SE Australia. I. Carbon and nitrogen. Biogeochemistry, 29(1), 3–24. [Google Scholar]
- Shanahan, P. , O'Sullivan, D. J. , Simpson, P. , Glennon, J. D. , & O'Gara, F. (1992). Isolation of 2,4‐diacetylphloroglucinol from a fluorescent pseudomonad and investigation of physiological parameters influencing its production. Applied and Environmental Microbiology, 58(1), 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui, I. A. , & Shaukat, S. S. (2003). Plant species, host age and host genotype effects on Meloidogyne incognita biocontrol by Pseudomonas fluorescens strain CHA0 and its genetically‐modified derivatives. Journal of Phytopathology, 151(4), 231–238. 10.1046/j.1439-0434.2003.00716.x. [DOI] [Google Scholar]
- Siddiqui, I. A. , & Shaukat, S. S. (2004). Systemic resistance in tomato induced by biocontrol bacteria against the root‐knot nematode, Meloidogyne javanica is independent of salicylic acid production. Journal of Phytopathology, 152(1), 48–54. 10.1046/j.1439-0434.2003.00800.x [DOI] [Google Scholar]
- Sofrata, A. , Santangelo, E. M. , Azeem, M. , Borg‐Karlson, A. K. , Gustafsson, A. , & Pütsep, K. (2011). Benzyl isothiocyanate, a major component from the roots of Salvadora persica is highly active against Gram‐negative bacteria. PLoS ONE, 6(8), e23045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Putten, W. H. , Cook, R. , Costa, S. , Davies, K. G. , Fargette, M. , Freitas, H. , Hol, W. G. , Kerry, B. R. , Maher, N. , Mateille, T. , & Moens, M. (2006). Nematode interactions in nature: Models for sustainable control of nematode pests of crop plants. Advances in Agronomy, 89, 227–260. [Google Scholar]
- Wang, X. , Wei, Z. , Yang, K. , Wang, J. , Jousset, A. , Xu, Y. , Shen, Q. , & Friman, V.‐P. (2019). Phage combination therapies for bacterial wilt disease in tomato. Nature Biotechnology, 37(12), 1513–1520. [DOI] [PubMed] [Google Scholar]
- Wei, Z. , Gu, Y. , Friman, V.‐P. , Kowalchuk, G. A. , Xu, Y. , Shen, Q. , & Jousset, A. (2019). Initial soil microbiome composition and functioning predetermine future plant health. Science Advances, 5(9):eaaw0759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller, D. M. (2007). Pseudomonas biocontrol agents of soilborne pathogens: Looking back over 30 years. Phytopathology, 97(2), 250–256. [DOI] [PubMed] [Google Scholar]
- Xue, Q. Y. , Ding, G. C. , Li, S. M. , Yang, Y. , Lan, C. Z. , Guo, J. H. , & Smalla, K. (2013). Rhizocompetence and antagonistic activity towards genetically diverse Ralstonia solanacearum strains—An improved strategy for selecting biocontrol agents. Applied Microbiology and Biotechnology, 97(3), 1361–1371. [DOI] [PubMed] [Google Scholar]
- Yang, C. , Dong, Y. , Friman, V.‐P. , Jousset, A. , Wei, Z. , Xu, Y. , & Shen, Q. (2019). Carbon resource richness shapes bacterial competitive interactions by alleviating growth‐antibiosis trade‐off. Functional Ecology, 33(5), 868–875. [Google Scholar]
- Yang, T. , Han, G. , Yang, Q. , Friman, V.‐P. , Gu, S. , Wei, Z. , Kowalchuk, G. A. , Xu, Y. , Shen, Q. , & Jousset, A. (2018). Resource stoichiometry shapes community invasion resistance via productivity‐mediated species identity effects. Proceedings of the Royal Society B, 285(1893), 20182035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasmin, S. , Hafeez, F. Y. , Mirza, M. S. , Rasul, M. , Arshad, H. M. , Zubair, M. , & Iqbal, M. (2017). Biocontrol of bacterial leaf blight of rice and profiling of secondary metabolites produced by rhizospheric Pseudomonas aeruginosa BRp3. Frontiers in Microbiology, 8, 1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw reads for all Pseudomonas strains are available at the NCBI Sequence Read Archive under BioProject accession number: PRJNA784243: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA784243.


