Abstract
Colorectal cancer (CRC) is one of the most malignant cancers resulting from abnormal metabolism alterations. As one of the essential amino acids, tryptophan has a variety of physiological functions, closely related to regulation of immune system, central nervous system, gastrointestinal nervous system and intestinal microflora. Colorectal cancer, a type of high-grade malignancy disease, stems from a variety of factors and often accompanies inflammatory reactions, dysbacteriosis, and metabolic disorders. Colorectal cancer accompanies inflammation and imbalance of intestinal microbiota and affects tryptophan metabolism. It is known that metabolites, rate-limiting enzymes, and ARH in tryptophan metabolism are associated with the development of CRC. Specifically, IDO1 may be a potential therapeutic target in colorectal cancer treatment. Furthermore, the reduction of tryptophan amount is proportional to the poor quality of life for colorectal cancer patients. This paper aims to discuss the role of tryptophan metabolism in a normal organism and investigate the relationship between this amino acid and colorectal cancer. This study is expected to provide theoretical support for research related to targeted therapy for colorectal cancer. Furthermore, strategies that modify tryptophan metabolism, effectively inhibiting tumor progression, may be more effective for CRC treatment.
Colorectal cancer (CRC) is one of the most malignant cancers resulting from abnormal metabolism alterations.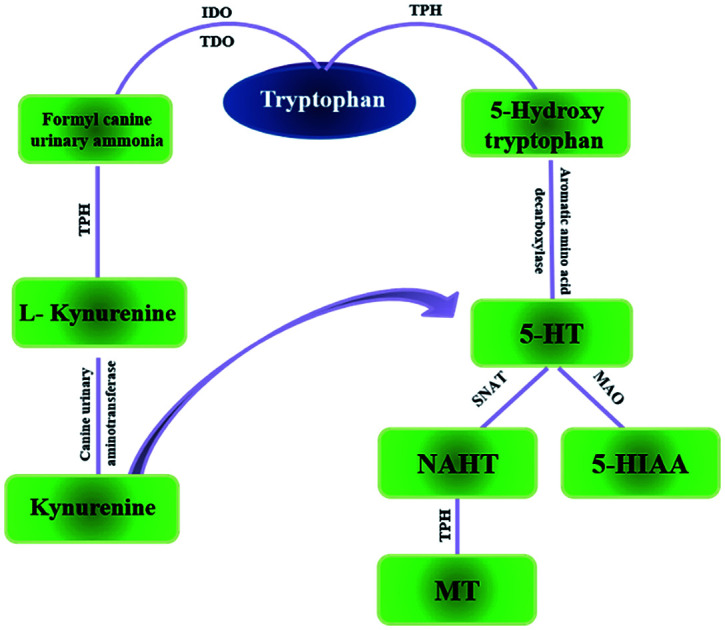
1. Introduction
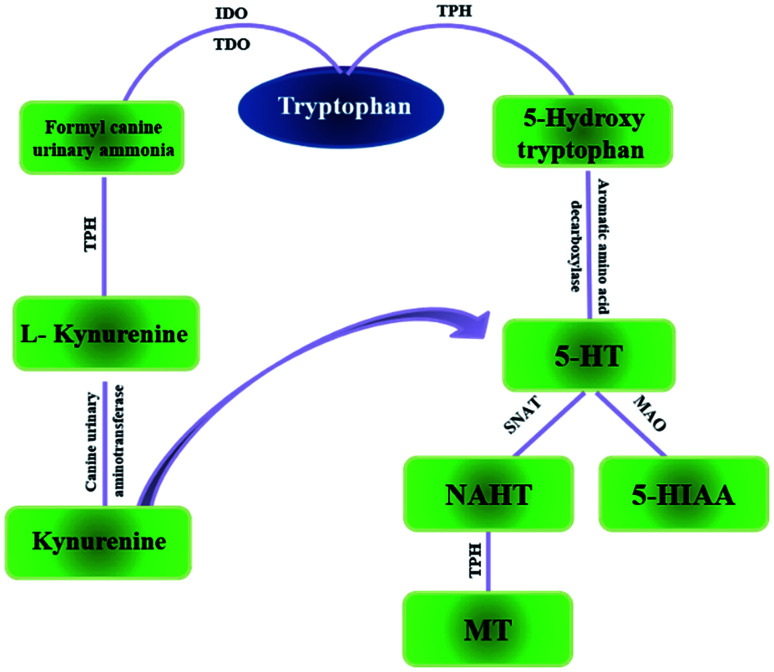
Tryptophan (TRP) is one of the essential amino acids for humans. It is an important functional amino acid with a variety of nutritional and physiological functions.1 It is indicated that many diseases are associated with TRP metabolism disorders, such Huntington's disease,3 and gastrointestinal diseases4 to name a few, also, miscarriage.2 In recent years, a large number of studies have shown that tumor growth is closely related to TRP metabolism.5 TRP is mainly a part of two metabolic pathways in vivo: serotonin metabolism and kynurenine metabolism (Fig. 1). Kynurenine metabolism is the main metabolic pathway of TRP. Under normal circumstances, serotonin metabolism is closely related to the nervous system and provides an important neurotransmitter 5-HT. Relatively speaking, the kynurenine pathway also plays an important role which is associated with the function of the immune system.6 Generally, TRP is an important regulator of mucosal homeostasis. The endogenous TRP metabolites play an important role in intestinal immune homeostasis in mammals.7,8
Fig. 1. The two main pathways of tryptophan metabolism in vivo. IDO: indoleamine-2,3-dioxygenase; TDO: toluene dioxygenase; TPH: tryptophan hydroxylase MAO: monoamine oxidase; SNAT: N-acetyltransferase.
Colorectal cancer (CRC), one of the most common worldwide malignancies, always starts with an inner tumor or tissue in the rectum or colon, grows abnormally after becoming a polyp, and eventually transform into cancer. The incidence of CRC is always occult, and many advanced-stage cancer patients, so the mortality rate is high. In 2016, CRC became the third most deadly cancer in the United States9 and the number of patients is on the rise. Gene mutations, inflammatory stimuli, and immune disorders can lead to precancerous lesions. In addition, many studies have confirmed that improper diet and unhealthy lifestyle play a role in the development of CRC.10 In addition, various metabolic abnormalities affect the development of the disease.11 Among them, TRP metabolic disorder is one of the metabolic pathways closely associated with CRC. The article summarizes the role of TRP metabolism in the development of CRC and aims to guide related scientific research.
2. The status of CRC related studies
CRC is divided into several fields by Traditional Chinese Medicine (TCM) as follows: “intestinal accumulation”, “accumulation”, “diagnosis”, “intestinal fistula”, “intestinal wind”, “dirty poison” and “snoring” as well as “anal fistula”. The hypostatic cause of malignancy is caused by healthy energy deficiency and evil excess, dyspeptic convulsion with internal injury and self-mood disorders. The viscera deficiency with deficiency of Qi and blood is the core reason for CRC.12 Modern studies have shown that the occurrence of CRC is related to various factors, such as inflammation and gene mutations. It was reported earlier that the inflammatory cytokine interleukin-6 (IL-6), isolated from mesenchymal stem cells, promotes the progression of colorectal cancer cells via IL-6/JAK2/STAT3 signaling.13 The classical “adenomas-carcinoma” doctrine describes CRC as a multi-gene and multi-step complex process. Cancer progresses slowly from normal epithelium to dysplasia, adenoma, carcinogenesis, and cancer metastasis in turn. Some of the mutated genes associated with CRC lesions include APC,14TP53,15,16KRAS,17 which are mainly involved Wnt, TGF-β, and p53 pathways. Mutations in the gene APC initiate CRC tumor progression, resulting in 50% of all CRC cases.18,19 At the same time, at the genomic level, the study of the gene mutation collaboration effect on CRC development has also received attention.20 A series of gene mutations, mismatches, activation of oncogenes, and inactivation of tumor suppressor genes have constituted a process of malignant disease. In addition, diet, lifestyle and other factors affect the development and prognosis of CRC.21 For example, obesity can accelerate the progression of CRC.22 The negative habits need to ameliorate in time, such as high-fat diet, smoking, and drinking.23–27 In other words, the occurrence of CRC is mainly caused by the combination of genetic and environmental factors. According to epidemiological statistics data, there are more than 1 million new cases of enlarged bowel cancer each year.28 Therefore, healthy lifestyle, active prevention, and accurate diagnosis and treatment programs are effective measures to curb CRC.
At present, fecal occult blood tests29 and colonoscopy30 are the main screening technologies for CRC. However, fecal occult blood tests are not always accurate and colonoscopy is an invasive procedure. Therefore, a non-invasive fecal exfoliated cell technology and serum markers such as CEA, CEA199, CEA242, CA742, and CA125 are often used in clinical practice.31,32 Drugs, nutritional supplements, diet, and exercise can effectively prevent the occurrence of CRC. In addition to medication, clinicians often use colonoscopy and other methods to treat CRC. However, the results are not satisfactory, owing to the hidden incidence of CRC. In addition, surgery removes only the local lesions and cannot intrinsically treat the disease. It is noteworthy that immunotherapy has brought new hopes for malignant disease.33–35 The common immunosuppressive sites are programmed death 1(PD-1),36 cytotoxic T-lymphocyte-associated antigen (CTLA-4),37,38etc. However, most of the current immunotherapy is to achieve immune activators, such as T cell activation,39 and the realization of therapy depends on microbiota in a certain extent. In summary, the urgent need for diagnosis and treatment of CRC requires more sensitive screening and effective therapy.
Intestinal microorganisms are one of the important microenvironmental factors associated with occurance of intestinal tumors. At present, various studies have confirmed the role of intestinal microbiota in carcinomatosis, especially in colorectal cancer. Intestinal microbiota affects inflammatory response, chemotherapy resistance, and prognosis, as well as therapeutic target research.40–43 For example, Ackermania can induce remodeling of intestinal flora44 and Bifidobacteria and Lactobacillus are associated with immune response. In addition, intestinal microbes impact the treatment of cancer tumor suppressors.45 Notably, the role of metabolites of the gut microflora on host diseases cannot be ignored, such as indoles, short-chain fatty acids (SCFAs) and fatty acids etc.,46,47 which are closely related to host immunity, cancer progression, and participate in immune regulation and metabolism.48 Metabolomics is an effective way to study small molecule metabolites,49–68 and it has shown that CRC precancerous lesions are highly correlated with TRP metabolism, energy metabolism, polyamine metabolism, and composition of the intestinal flora. Abnormal metabolism in the pathogenesis of CRC is very important for clinical diagnosis, treatment, and prognosis evaluation, especially for screening of early adenomas.69 It is worth noting that TRP metabolism affects multiple systems, such as central nervous system, gastrointestinal tract, and immunity, which is closely related to intestinal flora. These factors are closely related to the occurrence, development, and prognosis of CRC. For example, indoles is an important class of bacterial-host co-metabolites, which is the metabolite in the TRP metabolism pathway.70 Therefore, TRP metabolism and CRC are inextricably linked.
3. The physiological function of TRP metabolism
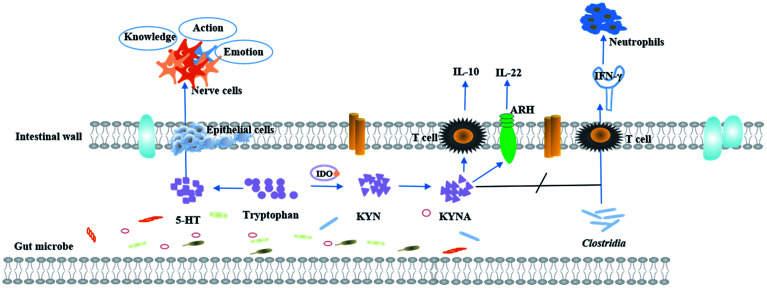
TRP is mainly obtained from food and has a variety of physiological functions. In vivo, TRP metabolism mainly occurs in the small intestine and central nervous system. The main metabolic pathways and metabolites of TRP are shown in Fig. 2.
Fig. 2. A schematic diagram of the physiological function of tryptophan metabolism.
3.1. TRP metabolism on the nervous system
TRP metabolic pathways are associated with sleep and wakefulness, mental and emotional activity, and endocrine regulation. The lacking TRP in diet can cause inflammation of the central nervous system in mice.70,71 TRP is an important precursor of the neurotransmitter serotonin. In the central nervous system, 5-HT is an important neurotransmitter.72 95% of 5-HT production occurs in the gastrointestinal tract, and 90% of 5-HT is stored in the intestinal chromaffin cell, and serotonin can use intestinal chromaffin cells as chemoreceptors to transmit perceived chemical information to the nervous system.73 Recently, serotonin is considered as a type of neurotransmitter that transmits intestinal signals to the brain quickly.74 Therefore, local changes in the gastrointestinal tract are affected by the concentration of serotonin, signals are transmitted along the stent along the intestine axis to affect central neurotransmission.72,75 During the process, the symbiotic bacteria are involved in the biosynthesis of 5-HT.76 In the central nervous system, 5-HT participates in a series of emotional, behavioral and cognitive functions and is the target of many psychotic drugs. TRP is converted to 5-hydroxy tryptophan by the action of enzymes, further decarboxylation to 5-HT, and finally metabolized to 5-hydroxy indole acetic acid (5-HIAA) in vivo. Reduced levels of TRP can decrease the production of 5-HT, which causes the emotional disturbances, depression, and cognitive impairment. 90% of TRP is metabolized into the kynurenine (KYNA) metabolic pathway, which is one of the activation pathways of the intestinal brain axis. The imbalance of metabolites is associated with multiple neurodegenerative and psychiatric diseases, such as the alzheimer's syndrome, depression, etc.77
3.2. Immunoregulatory effects of TRP metabolites
TRP is a key regulator of inflammation and immunity. Its metabolites are thought to be endogenous ligands of the aryl hydrocarbon receptor (AHR),78,79 which regulates intestinal immune function80,81 and plays an important role in mammalian intestinal immune homeostasis. Animal experiments have indicated that the lack of TRP affects the intestinal immunity in mice. For example, CD4+CD8αα+ double positive intraepithelial T cells (DP IELs) in the small intestine have immunoregulatory functions and are derived from CD4+ T cells. Activation of DP IELs depends on Lactobacillus reuteri metabolizing the product of TRP indole-3-lactic acid. Experimental results suggest that high TRP intake from the diet and Lactobacillus reuteri can increase the number of immune T cells.82 Predictive experiments of host–microbe interactions have shown that TNF-α and IFN-γ are associated with specific microbial metabolic pathways and palmitic acid and TRP degrade alcohols.83 Intestinal diseases are usually accompanied by inflammatory reactions.84 The indole acrylic acid, a metabolite of TRP, improves the intestinal epithelial barrier function, reduces the inflammatory response, and has a therapeutic effect.69 Furthermore, experiments have shown that TRP and its metabolites (such as kynurenine) have inhibitory effects on inflammation. A certain concentration of TRP can significantly inhibit gene expression of inflammatory factors, thereby, curbing the disease progression and playing an active role in human health. Metabolites 3-hydroxy ethyl amino benzoic acid (3-HAA) and quinolinic acid, produced by the TRP-kynurenine pathway, can induce apoptosis of murine lymphocytes (mainly Th1 cells) in vitro.85 Studies have shown that swollen cells can consume a large amount of TRP to produce a tumor microenvironment with immunosuppressive effects, which facilitates their own proliferation.
4. The relationship between TRP metabolism and CRC
4.1. The function of callback immune system
Under normal conditions, the immune system is its own line of defense against disease, which can effectively regulate the balance between the organism and the disease. However, while the tumor occurs in vivo, its microenvironment can produce immune tolerance against tumor antigens. Studies have shown that tumor cells can consume a large amount of TRP to produce a tumor microenvironment with immunosuppressive effects that facilitate their proliferation. Microorganisms can regulate the metabolism of tryptophan and, thus, regulate intestinal immunity.81 In addition, the development of CRC is accompanied by various inflammatory processes, and inflammation has been one of the hallmarks of cancer.86 It means that anti-inflammatory agents should be effective in preventing and treating CRC.
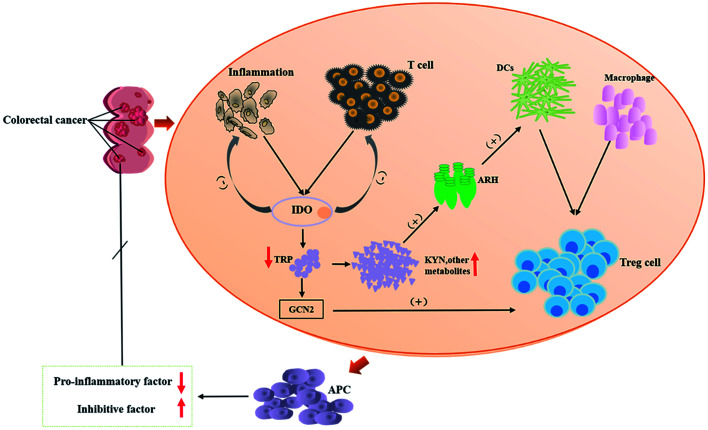
CRC is the fourth most fatal malignancy after lung cancer, liver cancer, and gastric cancer.87 The survival rates of CRC are related to the extent of the spread of cancer cells from primary tumors to regional lymph nodes.88,89 Surveys have revealed that the quality of life of CRC patients is positively correlated with the serum TRP levels. Studies have shown that KYNA reduces PI3K/Akt and p38, ERK1/2 cellular kinases in the HT-29 cell line, and inhibits the Wnt pathway.90 Tryptophan is mainly degraded by IDO (indoleamine 2,3-dioxygenase (IDO1)) in tumor cells.91,92 The KYNA pathway is closely related to the tumor immune escape mechanisms93 within gastrointestinal anti-inflammatory properties94 and inhibit the proliferation of colon cancer cells, which would be the way to immunotherapy for CRC.95 It is known that the risk of developing CRC is associated with Clostridia which is an intrinsic inflammatory bacterium.96 KYNA inhibits Clostridium-mediated IFN-γ-induced neutrophils and, thereby, counteracts the infection.97 The prerequisite for this process is that KYNA is synthesized by IDO1. Inhibitors of the TRP metabolic enzyme IDO1 may accelerate the infection by Clostridium.98 There have been reports of an increased expression of IDO1 in many types of cancer99 and processes related to tumor immune tolerance, which has implications for targeted immunotherapy.100 For example, experiments confirmed that Salmonella can achieve anti-tumor effects by inhibiting IDO1 expression and eliciting T cell immune responses in vitro.92 It means that IDO1 plays a role in regulating immunity in the TRP metabolism, and blocking or eliminating IDO1 causes a significant increase in inflammation.101,102 IDO can cause anti-regulation to inhibit T cell activity, which leads to inhibition of the immune system's response to tumors and promotes tumor progression. Therefore, IDO is an important point of immunosuppression in CRC immunotherapy.103 Tryptophan receptor ARH, mentioned above, is associated with maintaining mucosal homeostasis.104 It has been reported that ARH has a dual role in the regulation of tumors. While ARH is activated, it will have an effect on tumor progression. ARH has been used as an anti-tumor target for anti-tumor therapy.105 Based on the data mentioned above, the KYNA-IDO1-ARH pathway becomes a new therapeutic target for CRC. The immunomodulation process of TRP metabolism in the process of CRC development is presented in Fig. 3.
Fig. 3. Schematic representation of tryptophan metabolism in the process of CRC development. TRP: tryptophan; DC: dendritic cells; IDO: indoleamine-2,3-dioxygenase, IDO mainly is IDO1 in this paper; ARH: aromatic hydrocarbon receptor; GCN2: stress response kinase GCN2; APC: antigen-presenting cell.
4.2. Repair of the intestinal barrier structure
Normally, intestinal epithelial cells are barriers which can block the external environment and the internal environment.106 Dysfunction of the intestinal barrier may lead to increased intestinal permeability and bacterial translocation of the intestinal mucosa, which promote local and whole immune activation. TRP metabolites have an inhibitory effect on activated T cells and mediate immune tolerance and response. Compared with the healthy group, if inflammatory factors are detected in the normal mucosa of patients with colonic adenomas, the synthesis of 5-HT in the intestine increases at the same time. Beside mediating anti-inflammatory factors and fighting inflammation associated with cancer, TRP can increase the length of the intestinal wall and glandular villous, and crypt depth. It means that TRP can repair the intestinal wall and delay the progression of cancer.107 In the disease state, it is necessary to ingest more TRP to provide raw materials for TRP metabolism to meet the need of the body for TRP, and it may also help to elevate the patient's mood.
4.3. Interaction between TRP metabolism and intestinal microecology
TRP is a complex amino acid, which is a biosynthetic precursor to a great many complex microbial natural products.108 Intestinal ecosystem is the largest micro-ecological system in the human body which plays an important role in shaping host immune and metabolic activities.109 As a part of the intestinal microflora metabolism, TRP produces indole-3-acetic acid, indophenol-3-sulfate, indole-3-propionic acid, indole-3-acetaldehyde and other derivatives.80 Under the action of Lactobacillus, activation of ARH receptors mediates transcription of IL-22,110 protecting the mucosa from inflammation.111 The intestinal flora regulates kynurenine metabolism via numerous mechanisms, which affect the nervous system and gastrointestinal function.112 For example, both spore producing bacteria (SP) and short-chain fatty acids (SCFA) affect the synthesis of 5-HT. Specifically, SP promotes the expression of IDO1 to increase 5-HT production.97 Meanwhile, it was reported that the tryptophan metabolites, produced by the colonic bacteria, will have some adverse effects.113 Under normal physiological conditions, though the intestinal structure of adults varies, the type and the total amount of bacterial flora are basically unchanged, mainly containing Bacteroidetes, Firmicutes, actinomycetes, and Verrucomicrobia.112 With the presence of the digestive tract tumor, the intestinal wall barrier is damaged and the intestinal wall is prone to congestion and edema, which induces a change in colonization environment of the gut microbiota. Meanwhile, inflammation can induce the synthesis of genotoxic gut microbiota and abnormal composition of the intestinal flora, which promotes tumorigenesis.114 Dysregulated intestinal microflora can affect the TRP metabolism, disturb the intestinal immune balance, and worsen the disease.
TRP, an important precursor of neurotransmitter and metabolic regulator, plays an important role in the regulation of nutrient metabolism. Compared with mice in the control group, the villus height and villus height/crypt depth are increased in the TRP supplemented group, which suggests that TRP uptake positively affects the intestinal structure.4 In addition, the intake of TRP has an impact on bacterial diversity. For example, healthy pig larvae with Escherichia coli F4 susceptibility need to consume more TRP to increase intestinal flora diversity.115 TRP protects against CD4 T cell-mediated death of Mycobacteria in an emergency state.116 More and more experiments have confirmed the role of intestinal flora and TRP metabolites in the central nervous system health, and the relationship between the metabolites of the two pathways of 5-HT and KYNA mentioned above and neurological diseases. In conclusion, dietary intake of TRP exerts a physiological effect on metabolism of the intestinal microflora and affects the intestinal microecology.117 The process is in a dynamic equilibrium state. Once this relationship is broken, it will tremendous influence an organism.
4.4. The role of prevention
Because of the inhibition of TRP in potential carcinogenic diseases of colorectal cancer, it is usually used for the prevention of CRC. Tryptophan immunomodulators play a key role in the intestinal barrier maintenance and the integrity of its functions. In addition, CRC patients have reduced levels of TRP that impair quality of life.118 Therefore, nutritional supplements, such as probiotics, regulate tryptophan metabolism and can also be used for prevention of CRC.119 It has been reported that indoleamine 2,3-dioxygenase (IDO) activity is the first line of defense against invading cells. In the first stage of infection, IDO-mediated TRP consumption is mainly antimicrobial. At a further stage, it is an inhibitor of cell growth.120
5. Conclusions
TRP is an essential amino acid in animals. It not only participates in regulation of physiological processes, such as pain and sleep, but also takes part in relieving stress. Tryptophan is a precursor of biologically active substances 5-HT and KYNA and plays an important role in regulating immune function. During the development of CRC, an organism produces substances that cause inflammatory reactions, and the intestinal microenvironment simultaneously changes. The intestinal barrier is damaged and its permeability increases, leading to an outflow of endotoxin-like toxic substances which affect other sites. Composition of the intestinal microflora changes and the proportion of harmful bacteria, such as Enterococci and Clostridium, is increased to postpone the development of CRC. Metabolites of TRP in vivo not only inhibit inflammation, but also repair the intestinal wall structure and interact with beneficial bacteria in the intestinal tract, which can delay the progression of CRC. TRP metabolism plays an active role in inhibiting the development of CRC, and clarifying the relationship between the TRP metabolism and CRC is beneficial to subsequent studies.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was supported by grants from the Key Program of Natural Science Foundation of State (Grant No. 81830110, 81430093, 81373930, 81673586, 81703685, 81302905, 81503386), National Key Subject of Drug Innovation (Grant No. 2015ZX09101043-005, 2015ZX09101043-011), TCM State Administration Subject of Public Welfare (Grant No. 2015468004), Major Projects of Application Technology Research and Development Plan in Heilongjiang Province (GX16C003), TCM State Administration Subject of Public Welfare (Grant No. 2015468004), Specialized Research Fund for the Doctoral Program of Higher Education (20132327130001, 20122327120006), University Nursing Program for Young Scholars with Creative Talents in Heilongjiang Province (UNPYSCT-2015118, UNPYSCT-2016213), Young Talent Lift Engineering Project of China Association of Traditional Chinese Medicine (QNRC2-B06), Outstanding Talents Foundation of Heilongjiang University of Chinese Medicine (2018jc01), Doctoral Innovation Fund of Heilongjiang University of Chinese Medicine (2018bs02), University Nursing Program for Young Scholars with Creative Talents in Heilongjiang Province (UNPYSCT-2015118, UNPYSCT-2016213, UNPYSCT-2016212), Application Technology and Development of Youth Talents Project in Harbin (2014RFQXJ116), Chinese Postdoctoral Science Foundation (2017M621319b), Returned Overseas Scholars Program of Heilongjiang Province (2017QD0025), and Natural Science Foundation of Heilongjiang Province of China (QC2018117).
References
- Bai M. Liu H. Xu K. et al., A review of the immunomodulatory role of dietary tryptophan in livestock and poultry. Amino Acids. 2016;49(1):1–8. doi: 10.1007/s00726-016-2351-8. [DOI] [PubMed] [Google Scholar]
- Yang H. L. Zhou W. J. Li D. J. et al., Pleiotropic roles of melatonin in endometriosis, recurrent spontaneous abortion, and polycystic ovary syndrome. Am. J. Reprod. Immunol. 2018;80(1):e12839. doi: 10.1111/aji.12839. [DOI] [PubMed] [Google Scholar]
- Breda C. Sathyasaikumar K. V. Idrissi S. S. et al., L18 Tryptophan-2,3-dioxygenase (TDO) inhibition ameliorates neurodegeneration by modulation of kynurenine pathway metabolites. Proc. Natl. Acad. Sci. U. S. A. 2016;113(19):201604453. doi: 10.1073/pnas.1604453113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamas B. Richard M. L. Leducq V. et al., CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 2016;22(6):598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotti S. D'Angelo E. Bedin C. et al., Tryptophan metabolism along the kynurenine and serotonin pathways reveals substantial differences in colon and rectal cancer. Metabolomics. 2017;13(12):148. doi: 10.1007/s11306-017-1288-6. [DOI] [Google Scholar]
- Wettersten H. I. Aboud O. A. Lara Jr P. N. L. Metabolic reprogramming in clear cell renal cell carcinoma. Nat. Rev. Nephrol. 2017;13(7):410–419. doi: 10.1038/nrneph.2017.59. [DOI] [PubMed] [Google Scholar]
- Mondanelli G. Bianchi R. Pallotta M. T. et al., A Relay Pathway between Arginine and Tryptophan Metabolism Confers Immunosuppressive Properties on Dendritic Cells. Immunity. 2017;46(2):233. doi: 10.1016/j.immuni.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelante T. Iannitti R. G. Cunha C. et al., Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39(2):372. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Keum N. N. and Giovannucci E. L., Epidemiology of Colorectal Cancer, Pathology and Epidemiology of Cancer, Springer International Publishing, 2017 [Google Scholar]
- Lee J. Jeon J. Meyerhardt J. A. Diet and Lifestyle in Colorectal Cancer Survivors. Hematol. Oncol. Clin. North Am. 2015;29(1):1. doi: 10.1016/j.hoc.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A. Sun H. Yan G. Wang P. Han Y. Wang X. Metabolomics in diagnosis and biomarker discovery of colorectal cancer. Cancer Lett. 2014;345(1):17–20. doi: 10.1016/j.canlet.2013.11.011. [DOI] [PubMed] [Google Scholar]
- Liu Q. L., Master's thesis, Effect of anti-cancer solution on expression of P53, VEGF and EGFR in human CRC nude mice transplanted with tumor, Henan University of Chinese Medicine, 2014, pp. 32–33 [Google Scholar]
- Zhang X. Hu F. Li G. et al., Human colorectal cancer-derived mesenchymal stem cells promote colorectal cancer progression through IL-6/JAK2/STAT3 signaling. Cell Death Dis. 2018;9(2):25. doi: 10.1038/s41419-017-0176-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell M. J. Yang M. Teer J. K. et al., A multigene mutation classification of 468 colorectal cancers reveals a prognostic role for APC. Nat. Commun. 2016;7:11743. doi: 10.1038/ncomms11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. Zhang X. Han C. et al., TP53 loss creates therapeutic vulnerability in colorectal cancer. Nature. 2015;520(7549):697–701. doi: 10.1038/nature14418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. Zheng W. Guo Z. et al., A novel TP53 pathway influences theHGS-mediated exosome formation in colorectal cancer. Sci. Rep. 2016;6:28083. doi: 10.1038/srep28083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimbert J. Tachon G. Junca A. et al., Association between clinicopathological characteristics and RAS mutation in colorectal cancer. Mod. Pathol. 2017;31(3):517–526. doi: 10.1038/modpathol.2017.119. [DOI] [PubMed] [Google Scholar]
- Gregorieff A. Liu Y. Inanlou M. R. et al., Yap-dependent reprogramming of Lgr5+ stem cells drive intestinal regeneration and cancer. Nature. 2015;526(7575):715. doi: 10.1038/nature15382. [DOI] [PubMed] [Google Scholar]
- Kang D. H. Lee D. J. Lee S. et al., Interaction of tankyrase and peroxiredoxin II is indispensable for the survival of colorectal cancer cells. Nat. Commun. 2017;8(1):40. doi: 10.1038/s41467-017-00054-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlston R. S. Cogent M. O. COGENT (COlorectal cancer GENeTics) revisited. Mutagenesis. 2012;27(2):143–151. doi: 10.1093/mutage/ger059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belcheva A. Irrazabal T. Robertson S. J. et al., Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell. 2014;158(2):288–299. doi: 10.1016/j.cell.2014.04.051. [DOI] [PubMed] [Google Scholar]
- Wunderlich C. M. Ackermann P. J. Ostermann A. L. et al., Obesity exacerbates colitis-associated cancer via IL-6-regulated macrophage polarisation and CCL-20/CCR-6-mediated lymphocyte recruitment. Nat. Commun. 2018;9:1646. doi: 10.1038/s41467-018-03773-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J. S. Bradbury K. E. Cross A. J. et al., Physical activity, sedentary behaviour and colorectal cancer risk in the UK Biobank. Br. J. Cancer. 2018;118:920–929. doi: 10.1038/bjc.2017.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross A. J. Boca S. Freedman N. D. et al., Metabolites of tobacco smoking and colorectal cancer risk. Carcinogenesis. 2014;35(7):1516–1522. doi: 10.1093/carcin/bgu071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oluyemi A. O. Welch A. R. Yoo L. J. et al., Colorectal cancer screening in high-risk groups is increasing, although current smokers fall behind. Cancer. 2014;120(14):2106–2113. doi: 10.1002/cncr.28707. [DOI] [PubMed] [Google Scholar]
- Song M. Garrett W. S. Chan A. T. Nutrients, Foods, and Colorectal Cancer Prevention. Gastroenterology. 2015;148(6):1244–1260. doi: 10.1053/j.gastro.2014.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira A. R. Abar L. Chan D. et al., Foods and beverages and colorectal cancer risk: a systematic review and meta-analysis of cohort studies, an update of the evidence of the WCRF-AICR continuous update project. Ann. Oncol. 2017;28(8):1788. doi: 10.1093/annonc/mdx171. [DOI] [PubMed] [Google Scholar]
- Wang M. Gu D. Du M. et al., Common genetic variation in ETV6 is associated with colorectal cancer susceptibility. Nat. Commun. 2016;7:11478. doi: 10.1038/ncomms11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski M. F. Wieszczy P. Rupinski M. et al., Increased Rate of Adenoma Detection Associates With Reduced Risk of Colorectal Cancer and Death. Gastroenterology. 2017;153(1):98. doi: 10.1053/j.gastro.2017.04.006. [DOI] [PubMed] [Google Scholar]
- Gao Y. Wang J. Zhou Y. et al., Evaluation of Serum CEA, CA19-9, CA72-4, CA125 and Ferritin as Diagnostic Markers and Factors of Clinical Parameters for Colorectal Cancer. Sci. Rep. 2018;8(1):2732. doi: 10.1038/s41598-018-21048-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. Dai W. Li Y. et al., Nomograms for predicting the prognostic value of serological tumor biomarkers in colorectal cancer patients after radical resection. Sci. Rep. 2017;7:46345. doi: 10.1038/srep46345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkin W. Wooldrage K. Brenner A. et al., Adenoma surveillance and colorectal cancer incidence: a retrospective, multicentre, cohort study. Lancet Oncol. 2017;18(6):823. doi: 10.1016/S1470-2045(17)30187-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcgrail D. J. Federico L. Li Y. et al., Multi-omics analysis reveals neoantigen-independent immune cell infiltration in copy-number driven cancers. Nat. Commun. 2018;9(1):1317. doi: 10.1038/s41467-018-03730-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson V. Fessler J. Bao R. et al., The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359(6371):104. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson V. Fessler J. Bao R. et al., The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359(6371):104. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand F. Montfort A. Marcheteau E. et al., TNFα blockade overcomes resistance to anti-PD-1 in experimental melanoma. Nat. Commun. 2017;8(1):2256. doi: 10.1038/s41467-017-02358-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ock C. Y. Hwang J. E. Keam B. et al., Genomic landscape associated with potential response to anti-CTLA-4 treatment in cancers. Nat. Commun. 2017;8(1):1050. doi: 10.1038/s41467-017-01018-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauriello D. V. F. Palomoponce S. Stork D. et al., TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature. 2018;554(7693):538–543. doi: 10.1038/nature25492. [DOI] [PubMed] [Google Scholar]
- Lee P. Gujar S. Potentiating prostate cancer immunotherapy with oncolytic viruses. Nat. Rev. Neurol. 2018;15:235–250. doi: 10.1038/nrurol.2018.10. [DOI] [PubMed] [Google Scholar]
- Rosshart S. P. Vassallo B. G. Angeletti D. et al., Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell. 2017;171:1015–1028. doi: 10.1016/j.cell.2017.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudnovskiy A. Mortha A. Kana V. et al., Host-Protozoan Interactions Protect from Mucosal Infections through Activation of the Inflammasome. Cell. 2016;167(2):444–456. doi: 10.1016/j.cell.2016.08.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos Y. A. Hemann M. T. Drugs, Bugs, and Cancer: Fusobacterium nucleatum Promotes Chemoresistance in Colorectal Cancer. Cell. 2017;170(3):411–413. doi: 10.1016/j.cell.2017.07.018. [DOI] [PubMed] [Google Scholar]
- García-González A. P. Ritter A. D. Shrestha S. et al., Bacterial Metabolism Affects the C. elegans Response to Cancer Chemotherapeutics. Cell. 2017;169(3):431–441. doi: 10.1016/j.cell.2017.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänninen A. Toivonen R. Pöysti S. et al., Akkermansia muciniphila induces gut microbiota remodelling and controls islet autoimmunity in NOD mice. Gut. 2017;67:1373–1374. doi: 10.1136/gutjnl-2017-314508. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan V. Spencer C. N. Nezi L. et al., Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359(6371):97. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan G. V. Choi K. Klemashevich C. et al., Prediction and quantification of bioactive microbiota metabolites in the mouse gut. Nat. Commun. 2014;5:5492. doi: 10.1038/ncomms6492. [DOI] [PubMed] [Google Scholar]
- Zhao L. Ni Y. Su M. et al., High Throughput and Quantitative Measurement of Microbial Metabolome by Gas Chromatography/Mass Spectrometry Using Automated Alkyl Chloroformate Derivatization. Anal. Chem. 2017;89(10):5565–5577. doi: 10.1021/acs.analchem.7b00660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belcheva A. Irrazabal T. Martin A. Gut microbial metabolism and colon cancer: Can manipulations of the microbiota be useful in the management of gastrointestinal health? BioEssays. 2015;37(4):403–412. doi: 10.1002/bies.201400204. [DOI] [PubMed] [Google Scholar]
- Sun H. Zhang H. L. Zhang A. H. Zhou X. H. Wang X. Q. Han Y. Yan G. L. Liu L. Wang X. J. Network pharmacology combined with functional metabolomics discover bile acid metabolism as a promising target for mirabilite against colorectal cancer. RSC Adv. 2018;8:30061–30070. doi: 10.1039/C8RA04886J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H. L. Zhang A. H. Yu J. B. Sun H. Kong L. Wang X. Q. Yan G. L. Liu L. Wang X. J. High-throughput lipidomics characterize key lipid molecules as potential therapeutic targets of Kaixinsan protects against Alzheimer's disease in APP/PS1 transgenic mice. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2018;1092:286–295. doi: 10.1016/j.jchromb.2018.06.032. [DOI] [PubMed] [Google Scholar]
- Zhang A. H. Yu J. B. Sun H. et al., Identifying quality-markers from Shengmai San protects against transgenic mouse model of Alzheimer's disease using chinmedomics approach. Phytomedicine. 2018;45:84–92. doi: 10.1016/j.phymed.2018.04.004. [DOI] [PubMed] [Google Scholar]
- Liu C. Zong W. J. H Zhang A. Zhang H. M. Luan Y. H. Sun H. Cao H. X. Wang X. J. Lipidomic characterisation discovery for coronary heart disease diagnosis based on high-throughput ultra-performance liquid chromatography and mass spectrometry. RSC Adv. 2018;8:647–654. doi: 10.1039/C7RA09353E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. Kong L. Zhang A. H. Han Y. Liu Z. Sun H. Liu L. Wang X. J. High-throughput metabolomics for discovering potential metabolite biomarkers and metabolic mechanism from APPswe/PS1dE9 transgenic model of Alzheimer's disease. J. Proteome Res. 2017;16(9):3219–3228. doi: 10.1021/acs.jproteome.7b00206. [DOI] [PubMed] [Google Scholar]
- Zhao Q. Zhang A. H. Zong W. J. An N. Zhang H. M. Luan Y. H. Sun H. Wang X. J. Cao H. X. Exploring potential biomarkers and determining the metabolic mechanism of type 2 diabetes mellitus using liquid chromatography coupled to high-resolution mass spectrometry. RSC Adv. 2017;7:44186–44198. doi: 10.1039/C7RA05722A. [DOI] [Google Scholar]
- Zhang A. H. Sun H. Wang X. J. Emerging role and recent applications of metabolomics biomarkers in obesity diseases research. RSC Adv. 2017;7:14966–14973. doi: 10.1039/C6RA28715H. [DOI] [Google Scholar]
- Zhang A. H. Yan G. Zhou X. Wang Y. Han Y. Guan Y. Sun H. Wang X. J. High resolution metabolomics technology reveals widespread pathway changes of alcoholic liver disease. Mol. BioSyst. 2016;12(1):262–273. doi: 10.1039/C5MB00603A. [DOI] [PubMed] [Google Scholar]
- Cao H. Zhang A. H. Sun H. Zhou X. Y. Guan Y. Liu Q. Kong L. Wang X. J. Metabolomics-proteomics profiles delineate metabolic changes in kidney fibrosis disease. Proteomics. 2015;15(21):3699–3710. doi: 10.1002/pmic.201500062. [DOI] [PubMed] [Google Scholar]
- Sun H. Zhang S. Zhang A. H. Yan G. L. Wu X. Han Y. Wang X. J. Metabolomic analysis of diet-induced type 2 diabetes using UPLC/MS integrated with pattern recognition approach. PLoS One. 2014;9(3):e93384. doi: 10.1371/journal.pone.0093384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A. H. Yan G. L. Han Y. Wang X. J. Metabolomics approaches and applications in prostate cancer research. Appl. Biochem. Biotechnol. 2014;174(1):6–12. doi: 10.1007/s12010-014-0955-6. [DOI] [PubMed] [Google Scholar]
- Zhang A. H. Sun H. Qiu S. Wang X. J. Metabolomics insights into pathophysiological mechanisms of nephrology. Appl. Biochem. Biotechnol. 2014;46(5):1025–1030. doi: 10.1007/s11255-013-0600-2. [DOI] [PubMed] [Google Scholar]
- Zhang A. H. Sun H. Yan G. L. Han Y. Wang X. J. Metabolomics analysis of marker metabolites for patients with pancreatic cancer. Med. Chem. 2014;4:6. [Google Scholar]
- Wang X. J. Zhang A. H. Sun H. Power of metabolomics in diagnosis and biomarker discovery of hepatocellular carcinoma. Hepatology. 2013;57(5):2072–2077. doi: 10.1002/hep.26130. [DOI] [PubMed] [Google Scholar]
- Sun H. Zhang A. H. Yan G. L. Piao C. Li W. Sun C. Wu X. Li X. Chen Y. Wang X. J. Metabolomic analysis of key regulatory metabolites in hepatitis C virus-infected tree shrews. Mol. Cell. Proteomics. 2013;12(3):710–719. doi: 10.1074/mcp.M112.019141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A. H. Sun H. Wang X. J. Power of metabolomics in biomarker discovery and mining mechanisms of obesity. Obes. Rev. 2013;14(4):344–349. doi: 10.1111/obr.12011. [DOI] [PubMed] [Google Scholar]
- Zhang A. H. Sun H. Han Y. Yan G. Wang X. J. Urinary metabolic biomarker and pathway study of hepatitis B virus infected patients based on UPLC-MS system. PLoS One. 2013;8(5):e64381. doi: 10.1371/journal.pone.0064381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Zhang A. H. Yan G. L. Cheng W. Sun H. Meng X. C. Liu L. Xie N. Wang X. J. High-throughput metabolomic approach revealed the acupuncture exerting intervention effects by perturbed signatures and pathways. Mol. BioSyst. 2014;10(1):65–73. doi: 10.1039/C3MB70352E. [DOI] [PubMed] [Google Scholar]
- Zhang A. H. Wang P. Sun H. Yan G. L. Han Y. Wang X. J. High-throughput ultra-performance liquid chromatography-mass spectrometry characterization of metabolites guided by a bioinformatics program. Mol. BioSyst. 2013;9(9):2259–2265. doi: 10.1039/C3MB70171A. [DOI] [PubMed] [Google Scholar]
- Zhang A. H. Sun H. Qiu S. Wang X. J. Metabolomics in noninvasive breast cancer. Clin. Chim. Acta. 2013;424:3–7. doi: 10.1016/j.cca.2013.05.003. [DOI] [PubMed] [Google Scholar]
- Wlodarska M. Luo C. Kolde R. et al., Indoleacrylic Acid Produced by Commensal Peptostreptococcus Species Suppresses Inflammation. Cell Host Microbe. 2017;22(1):25–37. doi: 10.1016/j.chom.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kałużna-Czaplińska J. Gątarek P. Chartrand M. S. et al., Is there a relationship between intestinal microbiota, dietary compounds, and obesity? Trends Food Sci. Technol. 2017;70:105–113. doi: 10.1016/j.tifs.2017.10.010. [DOI] [Google Scholar]
- Jenkins T. A. Nguyen J. C. D. Polglaze K. E. Bertrand P. P. Influence of Tryptophan and Serotonin on Mood and Cognition with a Possible Role of the Gut-Brain Axis. Nutrients. 2016;8:56. doi: 10.3390/nu8010056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Mahony S. M. Clarke G. Borre Y. E. et al., Serotonin, tryptophan metabolism and the brain-gut-microbiome axis. Behav. Brain Res. 2015;277:32–48. doi: 10.1016/j.bbr.2014.07.027. [DOI] [PubMed] [Google Scholar]
- Bellono N. W. Bayrer J. R. Leitch D. B. et al., Enterochromaffin Cells Are Gut Chemosensors that Couple to Sensory Neural Pathways. Cell. 2017;170(1):185–198. doi: 10.1016/j.cell.2017.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maya Kaelberer M. Buchanan K. L. Marguerita E. et al., A gut-brain neural circuit for nutrient sensory transduction. Science. 2018;361:6408. doi: 10.1126/science.aat5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reigstad C. S. Salmonson C. E. Szurszewski J. H. et al., Gut microbes promote colonic serotonin production through an effect of short-chain fatty acids on enterochromaffin cells. FASEB J. 2015;29(4):1395. doi: 10.1096/fj.14-259598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano J. M. Yu K. Donaldson G. P. et al., Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161(2):264–276. doi: 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux G. A. Cunningham K. A. Lin L. et al., Kynurenic acid is a nutritional cue that enables behavioral plasticity. Cell. 2015;160(1–2):119–131. doi: 10.1016/j.cell.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jie C. Li W. Bo K. et al., Tryptophan derivatives regulate the transcription of Oct4 in stem-like cancer cells. Nat. Commun. 2015;6:7209. doi: 10.1038/ncomms8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam J. Sato S. Watanabe K. et al., Dietary tryptophan alleviates dextran sodium sulfate-induced colitis through aryl hydrocarbon receptor in mice. J. Nutr. Biochem. 2017;42:43. doi: 10.1016/j.jnutbio.2016.12.019. [DOI] [PubMed] [Google Scholar]
- Marsland B. J. Regulating inflammation with microbial metabolites. Nat. Med. 2016;22(6):581. doi: 10.1038/nm.4117. [DOI] [PubMed] [Google Scholar]
- Gao J. Xu K. Liu H. et al., Impact of the Gut Microbiota on Intestinal Immunity Mediated by Tryptophan Metabolism. Front. Cell. Infect. Microbiol. 2018;8:13. doi: 10.3389/fcimb.2018.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Barragan L. Chai J. N. Tianero M. D. et al., Lactobacillus reuteri induces gut intraepithelial CD4 (+) CD8αα (+) T cells. Science. 2017;357(6353):806. doi: 10.1126/science.aah5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer M. Smeekens S. Vlamakis H. et al., Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell. 2016;167(4):1125. doi: 10.1016/j.cell.2016.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing W. Yun X. Lu X. et al., GFI1 downregulation promotes inflammation-linked metastasis of colorectal cancer. Cell Death Differ. 2017;24(5):929–943. doi: 10.1038/cdd.2017.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. Kwak J. H. Pyo S. Inhibition of LPS-induced inflammatory mediators by 3-hydroxyanthranilic acid in macrophages through suppression of PI3K/NF-κB signaling pathways. Food Funct. 2016;7(7):3073–3082. doi: 10.1039/C6FO00187D. [DOI] [PubMed] [Google Scholar]
- Lasry A. Zinger A. Benneriah Y. Inflammatory networks underlying colorectal cancer. Nat. Immunol. 2016;17(3):230. doi: 10.1038/ni.3384. [DOI] [PubMed] [Google Scholar]
- Ferlay J. Soerjomataram I. Dikshit R. et al., Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- Naxerova K. Reiter J. G. Brachtel E. et al., Origins of lymphatic and distant metastases in human colorectal cancer. Science. 2017;357(6346):55. doi: 10.1126/science.aai8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothhammer V. Mascanfroni I. D. Bunse L. et al., Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and CNS inflammation via the aryl hydrocarbon receptor. Nat. Med. 2016;22(6):586–597. doi: 10.1038/nm.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freewan M. Rees M. D. Plaza T. S. Glaros E. Lim Y. J. Wang X. S. et al., Human Indoleamine 2,3-Dioxygenase is a Catalyst of Physiological Heme Peroxidase Reactions-Implications for the Inhibition of Dioxygenase Activity by Hydrogen Peroxide. J. Biol. Chem. 2013;288:1548–1567. doi: 10.1074/jbc.M112.410993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaker A. I. Rao M. S. Bishnupuri K. S. et al., IDO1 metabolites activate β-catenin signaling to promote cancer cell proliferation and colon tumorigenesis in mice. Gastroenterology. 2013;145(2):1–4. doi: 10.1053/j.gastro.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn D. H. Mellor A. L. IDO in the Tumor Microenvironment: Inflammation, Counter-Regulation, and Tolerance. Trends Immunol. 2016;37(3):193. doi: 10.1016/j.it.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puccetti P. Fallarino F. Italiano A. et al., Accumulation of an endogenous tryptophan-derived metabolite in colorectal and breast cancers. PLoS One. 2015;10(4):e0122046. doi: 10.1371/journal.pone.0122046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak K. Turski W. A. Rajtar G. Kynurenic acid inhibits colon cancer proliferation in vitro: effects on signaling pathways. Amino Acids. 2014;46(10):2393–2401. doi: 10.1007/s00726-014-1790-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santhanam S. Alvarado D. M. Ciorba M. A. Therapeutic targeting of inflammation and tryptophan metabolism in colon and gastrointestinal cancer. Transl. Res. 2016;167(1):67–79. doi: 10.1016/j.trsl.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J. Sinha R. Pei Z. et al., Human gut microbiome and risk for colorectal cancer. J. Natl. Cancer Inst. 2013;105(24):1907. doi: 10.1093/jnci/djt300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K. Tanoue T. Oshima K. et al., Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500(7461):232. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- Xue Y. Xiao H. Guo S. et al., Indoleamine 2,3-dioxygenase expression regulates the survival and proliferation of Fusobacterium nucleatum in THP-1-derived macrophages. Cell Death Dis. 2018;9(3):355. doi: 10.1038/s41419-018-0389-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai L. Ladomersky E. Lenzen A. et al., IDO1 in cancer: a Gemini of immune checkpoints. Cell. Mol. Immunol. 2018;15:447–457. doi: 10.1038/cmi.2017.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai L. Spranger S. Binder D. C. et al., Molecular Pathways: Targeting IDO1 and Other Tryptophan Dioxygenases for Cancer Immunotherapy. Clin. Cancer Res. 2015;21(24):5427. doi: 10.1158/1078-0432.CCR-15-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y. Giver C. R. Sharma A. et al., IFN-γ and indoleamine 2,3-dioxygenase signaling between donor dendritic cells and T cells regulates graft versus host and graft versus leukemia activity. Blood. 2012;119(4):1075. doi: 10.1182/blood-2010-12-322891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engin A. and Engin A. B., Indoleamine 2,3-Dioxygenase-Competent Regulatory Dendritic Cells and Their Role in Alloimmune Regulation and Transplant Immune Tolerance, Tryptophan Metabolism: Implications for Biological Processes, Health and Disease, Springer International Publishing, 2015, pp. 335–360 [Google Scholar]
- Zhang Y. Sun Z. Mao X. et al., Impact of mismatch-repair deficiency on the colorectal cancer immune microenvironment. Oncotarget. 2017;8(49):85526–85536. doi: 10.18632/oncotarget.20241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiering C. Wincent E. Metidji A. et al., Feedback control of AHR signalling regulates intestinal immunity. Nature. 2017;542(7640):242. doi: 10.1038/nature21080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. Liang X. Dong W. et al., Tumor-Repopulating Cells Induce PD-1 Expression in CD8+ T Cells by Transferring Kynurenine and AhR Activation. Cancer Cell. 2018;33(3):480. doi: 10.1016/j.ccell.2018.02.005. [DOI] [PubMed] [Google Scholar]
- Luissint L. A. C. Parkos C. A. Nusrat A. Inflammation and the Intestinal Barrier: Leukocyte–Epithelial Cell Interactions, Cell Junction Remodeling, and Mucosal Repair. Gastroenterology. 2016;151(4):616–632. doi: 10.1053/j.gastro.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade M. E. Araújo R. S. de Barros P. A. et al., The role of immunomodulators on intestinal barrier homeostasis in experimental models. Clin. Nutr. 2015;34(6):1080–1087. doi: 10.1016/j.clnu.2015.01.012. [DOI] [PubMed] [Google Scholar]
- Alkhalaf L. M. Ryan K. S. et al., Biosynthetic Manipulation of Tryptophan in Bacteria: Pathways and Mechanisms. Chem. Biol. 2015;22(3):317–318. doi: 10.1016/j.chembiol.2015.02.005. [DOI] [PubMed] [Google Scholar]
- Romani L. Zelante T. Luca A. D. et al., Microbiota control of a tryptophan-AhR pathway in disease tolerance to fungi. Eur. J. Immunol. 2014;44(11):3192. doi: 10.1002/eji.201344406. [DOI] [PubMed] [Google Scholar]
- Zelante T. Iannitti R. G. Cunha C. et al., Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39(2):372. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Ruan Z. Yang Y. Wen Y. et al., Metabolomic analysis of amino acid and fat metabolism in rats with L-tryptophan supplementation. Amino Acids. 2014;46(12):2681. doi: 10.1007/s00726-014-1823-y. [DOI] [PubMed] [Google Scholar]
- Kennedy P. J. Cryan J. F. Dinan T. G. et al., Kynurenine pathway metabolism and the microbiota-gut-brain axis. Neuropharmacology. 2017;112:399. doi: 10.1016/j.neuropharm.2016.07.002. [DOI] [PubMed] [Google Scholar]
- Fernstrom J. D. A Perspective on the Safety of Supplemental Tryptophan Based on Its Metabolic Fates. J. Nutr. 2016;146(12):2601S. doi: 10.3945/jn.115.228643. [DOI] [PubMed] [Google Scholar]
- Arthur J. C. Perezchanona E. Mühlbauer M. et al., Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338(6103):120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messori S. Trevisi P. Simongiovanni A. et al., Effect of susceptibility to enterotoxigenic Escherichia coli F4 and of dietary tryptophan on gut microbiota diversity observed in healthy young pigs. Vet. Microbiol. 2013;162(1):173–179. doi: 10.1016/j.vetmic.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Zhang Y. J. Reddy M. C. Ioerger T. R. et al., Tryptophan biosynthesis protects mycobacteria from CD4 T cell-mediated killing. Cell. 2013;155(6):1296–1308. doi: 10.1016/j.cell.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervenka I. Agudelo L. Z. Ruas J. L. Kynurenines: Tryptophan's metabolites in exercise, inflammation, and mental health. Science. 2017;357(6349):9794. doi: 10.1126/science.aaf9794. [DOI] [PubMed] [Google Scholar]
- Huang A., Fuchst D. and Widnert B., et al., Tryptophan and Quality of Life in Colorectal Cancer, Developments in Tryptophan and Serotonin Metabolism, Springer US, 2003, pp. 353–358 [DOI] [PubMed] [Google Scholar]
- Strasser B. Geiger D. Schauer M. et al., Probiotic Supplements Beneficially Affect Tryptophan–Kynurenine Metabolism and Reduce the Incidence of Upper Respiratory Tract Infections in Trained Athletes: A Randomized, Double-Blinded, Placebo-Controlled Trial. Nutrients. 2016;8(11):752. doi: 10.3390/nu8110752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engin A. B. Karahalil B. Karakaya A. E. Engin A. Helicobacter pylori and serum kynurenine-tryptophan ratio in patients with colorectal cancer. World J. Gastroenterol. 2015;21(12):3636–3643. doi: 10.3748/wjg.v21.i12.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]