Abstract
The oxidation of protein-bound methionines to form methionine sulfoxides has a broad range of biological ramifications, making it important to delineate factors that influence methionine oxidation rates within a given protein. This is especially important for biopharmaceuticals, where oxidation can lead to deactivation and degradation. Previously, neighboring residue effects and solvent accessibility have been shown to impact the susceptibility of methionine residues to oxidation. In this study, we provide proteome-wide evidence that oxidation rates of buried methionine residues are also strongly influenced by the thermodynamic folding stability of proteins. We surveyed the Escherichia coli proteome using several proteomic methodologies and globally measured oxidation rates of methionine residues in the presence and absence of tertiary structure, as well as the folding stabilities of methionine-containing domains. These data indicated that buried methionines have a wide range of protection factors against oxidation that correlate strongly with folding stabilities. Consistent with this, we show that in comparison to E. coli, the proteome of the thermophile Thermus thermophilus is significantly more stable and thus more resistant to methionine oxidation. To demonstrate the utility of this correlation, we used native methionine oxidation rates to survey the folding stabilities of E. coli and T. thermophilus proteomes at various temperatures and propose a model that relates the temperature dependence of the folding stabilities of these two species to their optimal growth temperatures. Overall, these results indicate that oxidation rates of buried methionines from the native state of proteins can be used as a metric of folding stability.
Keywords: methionine, oxidation-reduction, protein stability, proteomics, mass spectrometry (MS)
Abbreviations: ACN, acetonitrile; AH, ammonium hydroxide; HDX, hydrogen-deuterium exchange; MS, mass spectrometry; PDB, protein data bank; PF, protection factor; ROS, reactive oxygen species; SA, solvent accessibility; SPROX, stability of proteins from rates of oxidation; TMT, tandem mass tag
Sidechains of methionine and cysteine residues contain sulfur atoms that are readily oxidizable by reactive oxygen species (ROS). While not as well-studied as cysteine oxidation, methionine oxidation is an important enzymatically reversible posttranslational modification that has been shown to play a role in a number of physiological processes and cellular pathways (1, 2, 3, 4, 5). Methionine oxidation converts a hydrophobic residue to a polar residue and as such can greatly alter the biochemical and structural properties of proteins (1, 2, 6, 7, 8, 9). Methionine oxidation can destabilize proteins and deactivate enzymes and thus is often considered a form of oxidative damage (10, 11, 12). This effect has been particularly well documented in the context of aging and neurological diseases where dysregulation of antioxidant pathways and loss of proteostasis can bring about elevated levels of methionine oxidation and other detrimental protein modifications (3, 13, 14). It has also been shown that methionine oxidation can exacerbate protein misfolding or aggregation and worsen the symptoms of age-related diseases (15).
More recent studies have indicated that methionine oxidation is not solely a form of spontaneous protein damage and in certain contexts can modify the normal biological function of proteins in a regulated manner (16, 17, 18, 19). For example, the oxidation of specific methionine residues in actin is catalyzed by the MICAL family of oxidoreductases that regulate cytoskeletal remodeling (5, 20). It has also been suggested that methionine oxidation can act as an antioxidant scavenger in cells (1, 2, 4). This theory posits that surface methionines can become reversibly oxidized by ROS to prevent more dangerous, irreversible oxidation elsewhere in the same protein or other macromolecules. Importantly, unlike some other oxidatively modified macromolecules, oxidized methionines can be enzymatically reduced by methionine sulfoxide reductases providing a regeneratable sink for the removal of damaging ROS (21).
Analysis of methionine oxidation is also of great significance to commercial production of protein therapeutics, where trace levels of peroxides during production or storage can cause degradation and loss of activity. Antibody-based therapeutics can be especially sensitive to methionine oxidation, making it important to understand and prevent this modification (22, 23). Because the levels of methionine oxidation can change significantly in different environments, it is important to monitor oxidation levels during development or long-term stability studies of biopharmaceuticals (24, 25).
It has been shown that, within the same protein, different methionines have widely different susceptibilities to oxidation. As just one example, among the 10 methionines in α1-antitrypsin, five are prone to oxidation and five are relatively protected from oxidation (26). The five oxidizable methionines in antitrypsin have widely different oxidation rates that are contingent on the conformation of the protein (27). Because of its importance to protein stability and function, it is important to understand the factors that influence the susceptibility of methionines to oxidation. There have been a number of published studies that have investigated factors that drive methionine oxidation (25, 28, 29, 30, 31, 32, 33). These studies have shown that solvent accessibility (SA) and neighboring sequence effects are two major determinants of methionine oxidation rates across the proteome. For example, a significant fraction of methionines are located near aromatic residues, which have been shown to reduce methionine oxidation rates in vivo (31, 34). The fact that SA is a strong global predictor of methionine oxidation is supported by a number of studies showing that exposed methionines are typically oxidized faster than buried methionines (27, 28, 35, 36). Indeed, this correlation has allowed methionine oxidation to be used as an experimental biochemical probe to monitor structural changes in proteins (37). However, a number of studies have suggested that SA and neighboring residue effects may not be sufficient to fully predict the propensity of methionines toward oxidation. Specifically, it has been argued that the oxidation of buried methionines may also be influenced by the dynamics of the native structure and conformational flexibility (29).
In this study, we considered the potential role of thermodynamic folding stabilities in determining the oxidation rates of buried methionines. Within a cell, most protein domains exist in a dynamic equilibrium between folded and unfolded conformations (38, 39, 40). The free energy difference between these two states (ΔGfolding or ‘thermodynamic folding stability’) establishes the fraction of its population that is in a folded conformation at equilibrium (41). For some buried methionines, transient protein unfolding may be necessary to expose the protein to oxidation. Thus, folding stability can modulate the folding equilibrium and establish the rate of methionine oxidation for buried methionines. To date, correlations between oxidation rates and protein stabilities have been reported for a few individual proteins (42, 43). However, the general correlation between folding stability and methionine oxidation rates has not been studied on a proteome-wide scale.
We used mass spectrometry (MS)-based proteomics to quantify oxidation rates of methionines within folded and unfolded proteins and calculated relative oxidation protection factors (PFs) for methionines on a global scale. We then measured folding stabilities for the detected proteins using SPROX (stability of proteins from rates of oxidation), a proteome-wide methodology for conducting denaturation experiments (37, 44). The resulting largescale datasets allowed us to explore the correlation between PFs and folding stabilities within the proteomes of the mesophile Escherichia coli and the thermophile Thermus thermophilus. The analysis globally quantified the influence of folding stabilities on methionine oxidation rates and provided insights into the relationship between proteome stabilities and optimal growth temperatures of these two bacterial organisms.
Results
Overview of experimental strategy
Oxidation of protein-bound methionines that are fully or partially solvent-exposed can potentially occur from the folded state of the protein without transient unfolding of the native structure. For these exposed methionines, SA and neighboring residue effects may strongly influence the rate of oxidation. However, we reasoned that for buried methionine residues, transient unfolding of the native structure is required to expose the sulfur atom to oxidation (Fig. 1A). In this scenario, transient protein unfolding may be the limiting step in the oxidation process. Accordingly, under conditions where the folding reaction is in a rapid pre-equilibrium relative to the oxidation reaction, the folding equilibrium constant will be directly correlated to the rate of oxidation (Fig. 1A and Discussion). This concept is analogous to the EX2 model commonly used to interpret hydrogen-deuterium exchange (HDX) experiments where the rate of exchange of hydrogen-bonded backbone protons are known to correlate with folding stabilities (45). Thus, primary sequence, SA, and folding stability can all potentially influence the rate of methionine oxidation, and the relative contribution of each parameter may depend on the conformational properties of the methionine sidechain. To quantify the contribution of each of these parameters to oxidation propensities of protein-bound methionines, we conducted three orthogonal proteomic experiments (Fig. 1B) to globally measure the following: (a) intrinsic rates of methionine oxidation within unstructured peptides, (b) rates of methionine oxidation within intact native proteins, and (c) folding stabilities of methionine-containing protein domains.
Figure 1.
Schematic representations of proposed model and methods.A, for buried methionines, protein unfolding precedes oxidation. Oxidation protection factors (PFs) of buried methionines are defined as the ratio of methionine oxidation rates from the unfolded state (kintrinsic) and the observed oxidation rate (kprotein). According to a two-state folding model, PFs are related to folding stability (ΔGfolding) and the midpoint of denaturation (C1/2) as indicated in the right side of the figure. B, the three proteomic methods employed in this study. Intrinsic oxidation rates (kintrinsic) were measured using trypsin-digested proteins. Oxidation rates in intact proteins (kprotein) were measured using native protein lysates. Folding stabilities (C1/2 values) were measured by SPROX. This method exposes native extracts to increasing concentrations chemical denaturants prior to oxidation. SPROX, stability of proteins from rates of oxidation.
Intrinsic methionine oxidation rates within unstructured peptides
To measure intrinsic rate constants for methionine oxidation in unstructured peptides (kintrinsic), E. coli protein extracts were digested with trypsin prior to treatment with H2O2 (Fig. 2A). Peptides were treated with varying concentrations of 18O-labeled H2O2 at 37 °C for 30 min and were subsequently blocked by the addition of excess levels of 16O-labeled H2O2. Fractional oxidation levels were determined with LC-MS/MS by quantifying relative intensities of 18O- and 16O-oxidized peptides using a bottom-up proteomics workflow. As previously described, this approach allows for accurate quantitation of methionine oxidation while minimizing the effects of spurious oxidation that could occur during sample processing and LC-MS/MS analyses (46). kintrinsic values were measured by analyzing the relationship between fractional oxidation and concentrations of 18O-labeled H2O2 treatments using a first-order kinetic model. We limited our analyses to peptides containing single methionines and no cysteine residues in order to minimize the confounding effects of multiple methionine oxidation events and cysteine oxidation. In all, we were able to measure kintrinsic values for approximately 3900 methionine-containing peptides (Fig. 2B). Measured kintrinsic values spanned approximately one order of magnitude with a median of 1.02 M−1 min−1, which is in line with previously measured oxidation rates for free methionine at this temperature (23, 43).
Figure 2.
kintrinsicmeasurements and neighboring residue effects in the Escherichia coli proteome.A, experimental method. Trypsin-digested peptide samples were exposed to varying concentrations of H2O2. Fractional oxidation levels were measured with LC-MS/MS and used to calculate pseudo-first order oxidation rate constants. In the schematic, closed ovals indicate methionines in the native state, open squares indicate exposed methionines in peptides and red squares indicate oxidized methionines. B, relative distribution of kintrinsic measurements in the E. coli proteome. The dotted line indicates the distribution median. C and D, effect of neighboring residues on kintrinsic of methionines. E, effect of position relative to the N-terminus on kintrinsic of methionines. For C–E, boxes indicate the interquartile range and whiskers show the entire range of values excluding outliers (>2 SD). ∗∗ indicates a p-value < 0.001 using a Mann-Whitney U test, corrected for family-wise error rate with a Holm–Šidák method (significance level = 0.001).
We then used our proteome-wide data to analyze the effects of primary sequence on kintrinsic values (Fig. 2, C–E). We observed that methionines located at the N-termini of peptides have significantly faster oxidation rates in comparison to methionines that are internal in the sequence (Fig. 2E). Other neighboring residue effects were comparatively less pronounced. For example, the presence of prolines at +1 positions and glycines at −1 positions of methionines correlated with slightly slower oxidation rates, whereas the presence of arginines at +1 positions correlated with slightly faster oxidation rates. It should be noted that because peptides were generated by trypsin digestion (which hydrolyzes proteins at the carboxyl side of lysines and arginines), certain neighboring residue combinations (e.g., arginines and lysines at -1 positions and methionines at C-termini) could not be observed in our analyses. Furthermore, methionines that are at -1 positions relative to lysines and arginines are necessarily positioned near the C-termini of peptides which may bias their oxidation rates. Nonetheless, our analysis indicates that regardless of primary sequence context, most protein-bound methionines in unstructured peptides are oxidized at relatively similar rates that are close to oxidation rates of free methionines.
Rates of methionine oxidation within intact proteins
To investigate the effects of higher order structure on methionine oxidation, we next measured methionine oxidation rates within intact proteins (kprotein) by exposing E. coli native extracts to H2O2 prior to digestion and LC-MS/MS analyses (Fig. 3A). To facilitate these experiments, two additional changes were made to the experimental protocols described above. First, oxidation reactions were carried out as a function of time rather than as a function of H2O2 concentration. Second, we took advantage of an isobaric tagging strategy (tandem mass tag, TMT) to multiplex the larger number of samples analyzed in this experiment (See Experimental procedures for additional details and rationale for these protocol modifications).
Figure 3.
kproteinmeasurements within native proteins and correlations with solvent accessibility (SA).A, experimental method. Native lysates were treated with H2O2 using a range of oxidation times. Fractional oxidation levels were measured with LC-MS/MS and used to calculate pseudo-first order oxidation rate constants. In the schematic, ovals indicate methionines in the native state, squares indicate exposed methionines in peptides, black indicates unoxidized methionines, and red indicates oxidized methionines. B, the relative log10 distribution of kprotein measurements. The dotted line indicates the distribution median. Gray and white bars indicate protected and unprotected methionines as defined in the text. C, the relative distribution of PFs in the E. coli proteome. D, the relationship between PF and SA measurements. Methionines were categorized into three groups (buried, partially buried, and exposed) based on their SA values. Boxes indicate the interquartile range and whiskers show the entire range of values excluding outliers (>2 SD). The total pairwise comparison between PFs and SAs had a significant level of correlation (Spearman rank test ρ <0.001). PFs, protection factors.
We were able to measure native oxidation rates (kprotein) for 1778 methionines mapped to 831 different proteins in the E. coli proteome. Among these methionines, ∼600 were oxidized at fast rates that were within the range of peptide kintrinsic measurements and free methionines (Fig. 3B). We refer to these residues as “unprotected” methionines. The remaining ∼1200 “protected” methionines are oxidized at significantly slower rates in comparison to unstructured peptides. The overall range of kprotein measurements spans at least three orders of magnitude, representing a significantly broader distribution than kintrinsic values. This observation indicates that for most protected methionines in proteins, higher order structure, and not primary sequence, is the dominant factor in establishing oxidation rates. The distribution of measured PFs for protected methionines (the ratio of kprotein and kintrinsic measurements for individual methionines) is plotted in Figure 3C. According to the simplified two-state unfolding model illustrated in Figure 1A and described in Experimental procedures, PF values are expected to be equal to the folding equilibrium constant and are logarithmically correlated with thermodynamic folding stabilities (ΔGfolding).
We next examined the relationship between methionine SAs and PFs. SA values for methionines were calculated as described previously (44) using either their experimentally determined structure in the Protein Data Bank (PDB) or as predicted by Alphafold2 (47). In total, we were able to calculate both PF and SA values for 1731 distinct methionines in E. coli. Overall, there is a significant statistical correlation between PF and SA (Spearman Rank Correlation ρ < 0.001). A closer examination of the relationship between SA and PF values indicated that methionine residues can be categorized into one of the three general categories based on their oxidation properties (Fig. 3D). Highly exposed methionines (SA ≳ 44 Å2) are oxidized rapidly at rates that are equivalent to free methionines (PF ≈ 1). Partially buried methionine residues (SA ≈ 3–44 Å2) have a range of PF values that are correlated to their SA. Buried methionine residues (SA ≲ 3 Å2) have high PF values that are independent of SA.
The above data suggest that oxidation rates of fully exposed methionines are likely dictated by their primary sequence context. For partially buried methionines, oxidation is correlated to SA established by the native structure of the protein. This suggests that for this class of methionines, oxidation likely occurs from the native state and does not require transient conformational unfolding of the protein structure. For fully buried methionines, oxidation rates are significantly slower and independent of SA. Yet, these methionines have a wide range of PFs spanning three orders of magnitude. We reasoned that for this class of methionines, oxidation occurs from the unfolded state of the protein and hence transient protein unfolding must precede oxidation. Hence, we hypothesized that for fully buried methionines, oxidation rates may be correlated to protein thermodynamic folding stabilities.
Thermodynamic folding stabilities of methionine-containing protein domains
We used the proteomic methodology SPROX to conduct denaturation experiments on E. coli extracts and quantify protein folding stabilities on a global scale (Fig. 4A). In SPROX, proteins are gradually unfolded by the addition of increasing concentrations of a chemical denaturant such as guanidinium chloride and subsequently pulse-oxidized by exposure to H2O2 (37, 44). Measurements of relative fractional oxidation as a function of denaturant concentration are used to generate denaturation curves and determine C1/2 values (the denaturant concentration that induces the unfolding of half of the protein population). Protein domains with higher C1/2 values have higher conformational stabilities and lower ΔGfolding values (See Experimental procedures for a detailed description and assumptions of the folding model).
Figure 4.
C1/2measurements within native proteins and correlations with PFs.A, experimental method (SPROX). Native lysates were treated with increasing concentrations of denaturant (GdmCl) prior to oxidation with H2O2 and subsequent digestion with trypsin. Fractional oxidation levels were measured with LC-MS/MS and used to measure C1/2 values. Symbols in the schematic are described in Figure 3. B, the relative distribution of C1/2 values in the Escherichia coli proteome. C and D, the relationship between PF and C1/2 values for all methionines (C) and highly protected methionines (D). Boxes indicate the interquartile range and whiskers show the entire range of values excluding outliers (>2 SD). The Spearman rank correlation values indicate the correlation for total pairwise comparisons. PFs, protection factors; SPROX, stability of proteins from rates of oxidation.
Using this method, we were able to measure C1/2 values for 434 protein domains (Fig. 4B). Overall, there was a significant positive correlation between methionines’ PF and C1/2 values (Fig. 4 C, Spearman Rank Correlation ρ = 1.8⋅10−10). As predicted, this correlation was particularly significant for well-buried methionine residues, where most stable protein domains generally contained the most highly protected methionine residues (Fig. 4D). These results demonstrate that thermodynamic folding stabilities can play a major role in establishing the propensity of protein-bound methionines toward oxidation.
Oxidation PFs of thermophilic bacteria
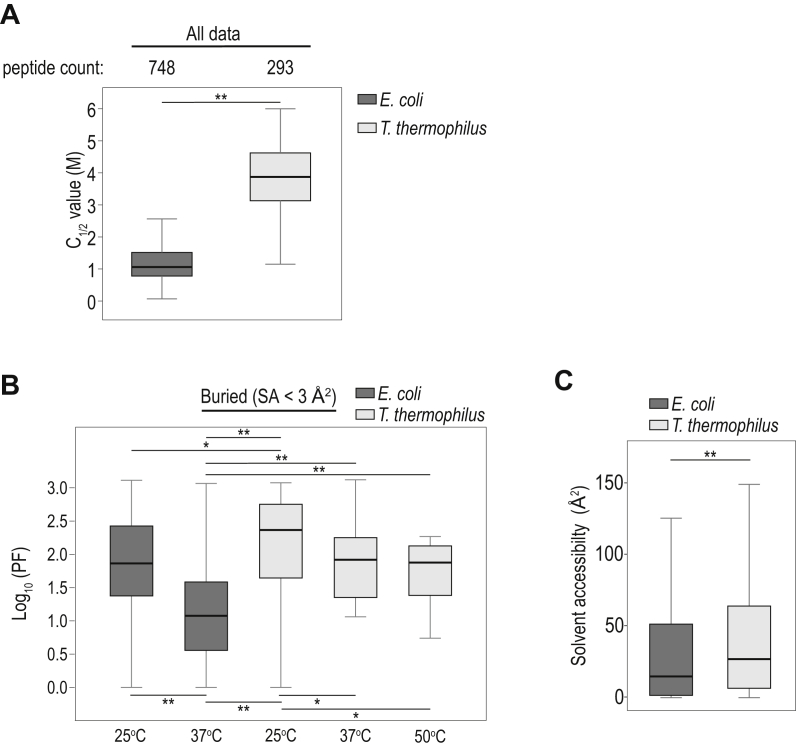
In order to verify that thermodynamic folding stabilities can strongly influence methionine oxidation rates, we analyzed the oxidation properties of the highly stable proteome of a model thermophile, T. thermophilus. By conducting SPROX experiments, we showed that, as expected, the proteome of T. thermophilus has significantly higher C1/2 values in comparison to E. coli (Fig. 5A). We subsequently quantified methionine PFs at multiple temperatures and observed that the T. thermophilus proteome is considerably more resistant to methionine oxidation than the E. coli proteome (Fig. 5B). It is important to note that overall, methionines within the T. thermophilus and E. coli proteomes measured in this study have similar SAs (Fig. 5C), with T. thermophilus being slightly higher. Hence, the high PF values of the T. thermophilus proteome cannot be explained by increased buriedness of methionines and are instead likely due to increased conformational stabilities.
Figure 5.
Comparison of C1/2and PF measurements between Escherichia coli and Thermus thermophilus proteomes.A, comparison of the distribution of C1/2 values for E. coli and T. thermophilus at 25 °C. B, comparison of the distributions of PF values for buried methionines within E. coli and T. thermophilus at varying temperatures. C, comparison of the distributions of SAs for methionines analyzed in (B). Boxes indicate the interquartile range and whiskers show the entire range of values excluding outliers (>2 SD). ∗ and ∗∗ indicate p-values greater than 0.05 and 0.001, respectively, using a Mann-Whitney U test. PF, protection factor; SA, solvent accessibility.
Our observations suggest that oxidation PFs of buried methionines from the native state can be leveraged as a quantitative metric of protein folding stabilities. Considering PFs as a proxy for folding stabilities, our data indicate that of the stabilities of the E. coli and T. thermophilus proteomes decrease as a function of temperature, and that, at both 25 °C and 37 °C, the T. thermophilus proteome is more stable than the E. coli proteome (Fig. 5B). The folding stabilities of the T. thermophilus proteome at higher temperatures (37 °C and 50 °C) are similar to the folding stabilities of the E. coli proteome at 25 °C. It is interesting to note that T. thermophilus is not viable at 25 °C, a temperature at which its proteome is highly stable. This observation is consistent with the idea that both destabilization and overstabilization of proteomes may be detrimental to viability. As has been previously suggested, optimal function may require proteomes to retain folded structures, but at marginal stabilities (48).
Discussion
Our results indicate that protein-folding stabilities can play a major role in limiting the oxidation of buried methionine residues. Accordingly, we showed that the proteome of the thermophile T. thermophilus is significantly more resistant to oxidation than the mesophile E. coli. Interestingly, T. thermophilus has the highest levels of oxidation protection at low temperatures where it is unviable. Natural proteins generally have marginal folding stabilities (10 kcal/mol or less). It is generally accepted that the lower limit of protein stability is established by the requirement of folded structures for most protein functions. Additionally, unstable proteins have larger unfolded populations that are nonfunctional and prone to degradation and aggregation. However, the reasons for upper limits of protein stability within proteomes are generally less understood and are typically attributed to decreased flexibility that may be required for optimal activity (49, 50). Our results suggest an additional explanation as to why overstabilization may be detrimental to function. Similar to oxidation of buried methionines observed in this study, important posttranslational modifications of buried residues may be limited by the folding stability of proteins. Hyperstable proteomes may be generally less conducive to covalent modifications required for posttranslational regulation. This effect may provide one explanation as to why thermophiles are not viable under conditions where their proteomes surpass a maximal stability threshold.
From an application standpoint, our results indicate that oxidation rates of buried methionines from the native state can be used as a convenient proxy for thermodynamic folding stabilities. To date, measurement of protein stabilities using MS-based proteomics has required methodologies that are based on the incremental unfolding of proteins using denaturants or temperature (37, 51, 52). However, these approaches are complicated by the fact that protein denaturation often results in irreversible aggregation. Furthermore, measurement of protein stability from denaturation curves typically involves making assumptions regarding the two-state nature of protein unfolding and model-dependent extrapolation from measurements under denaturing conditions to native conditions (40, 53). Here, we used principles previously outlined for native HDX (45) to show that oxidation rates under native conditions can be used to directly measure protein stabilities without extrapolation from denaturing conditions. This approach should significantly facilitate future proteomic analyses of folding stabilities.
The phenomenon of methionine oxidation is of particular practical interest for the development antibody-based therapeutics where methionine oxidation over long-term storage can lead to deactivation and degradation (22, 23). Understanding the factors that influence methionine oxidation will enable the identification of labile methionine residues and streamline the development of drug candidates (25, 54). The results of this study indicate that enhancing folding stabilities through genetic or chemical approaches may significantly mitigate the detrimental oxidation of protein therapeutics.
Experimental procedures
Culture growth and lysate preparations
E. coli K-12 W3110 was a generous gift from Dr Gloria Culver (University of Rochester) and was cultured in lab-made lysogeny broth at 37 °C. T. thermophilus strain HB8 (ATCC) was cultured at 70 °C in Thermus medium (ATCC medium 697) made in house. For both species, initial overnight growths were streaked onto agar plates, from which single colonies were picked for 200-ml batches, grown to late-log phase, and aliquoted into glycerol stocks at −80 °C. All further cultures were propagated from these stocks. For both species, 5- or 10-ml overnight cultures were diluted at a ratio of 1:100, grown to late-log phase, and collected on ice. Resulting cell pellets were immediately frozen at −80 °C until use. All cells were lysed at 4 °C using gentle sonication in a native lysis buffer (EDTA-free protease inhibitor mini tablets (Pierce) in filtered 20 mM sodium phosphate buffer (pH 7.4) with 50 mM NaCl), with 1 min resting periods on ice. Protein concentration was subsequently measured using a bicinchoninic acid assay and standardized to a final concentration of 2.5 mg/ml for all intact protein experiments. After standardization, lysates were aliquoted and flash-frozen in liquid N2 and kept at −80 °C until use.
Peptide oxidation
Proteomic peptide samples were prepared from native lysate, as above, which was immediately purified and digested with trypsin (see ‘Sample purification and TMT labeling’ section below). Oxidation was then performed on the purified peptides in a manner similar to that published previously (46). Peptides were brought to a final concentration of 1 mg/ml, and oxidative labeling was accomplished using a concentration-gradient of heavy-labeled H218O2 (Cambridge Isotope Laboratories). Specifically, samples were treated with 0, 0.02, 0.04, 0.08, or 0.16 M O-18 peroxide, for 30 min at 37 °C in a heat block. All samples were quenched using an excess of 1 M sodium sulfite and lyophilized. Quenching reagent was removed via desalting using lab-made C18 spin columns. Columns were first conditioned using elution buffer containing 50% acetonitrile (ACN) in 0.1% TFA (Thermo Scientific) and washed with sample buffer (0.1% TFA in water). Samples were resuspended in sample buffer, added to the columns, and washed. After elution, samples were lyophilized and then oxidized using a final concentration of 0.16 M H216O2 for 30 min at 37 °C to block any remaining methionines. Samples were lyophilized again to remove peroxide and resuspended in sample buffer for MS analysis.
Native oxidation
Samples were thawed on ice and then incubated for 10 min at RT with a final concentration of 100 μM sodium azide to prevent the degradation of peroxide by native catalases. Oxidation was achieved via a time course, as this was more amenable to measuring the slow rates expected for buried residues. Time courses were optimized for their given temperatures to account for the change in kinetics, and exact time points are listed in Table S1. For each set of samples, a native, unoxidized control and a fully denatured, oxidized (final concentration of 5% SDS heated to 90 °C for 10 min) control were also prepared. Samples were oxidized using a final concentration of 0.98 M peroxide for 25 °C and 37 °C, or 0.245 M for 50 °C, then purified as described in below in ‘Sample purification and TMT labeling’. An Amplex Red (Invitrogen) hydrogen peroxide assay kit was used to validate the steady state levels of peroxide in samples treated with 100 μM sodium azide under each condition (data not shown). To capture the large range of rates expected for these samples, it was advantageous to adopt a labeling method more amenable to high-throughput analysis. Thus, we opted for using Tandem Mass Tag (TMT)10plex (see TMT labeling section below) reagents to quantify the fraction of oxidation in multiplexed samples. This allowed us to maximize coverage by using peptide fractionation protocols while cutting down on the number of MS runs.
SPROX
SPROX oxidation, denaturation, and sample cleanup were performed as described previously (44). The guanidinium chloride concentrations used for each experiment are listed in Table S1. Stability measurements were performed using quality control filters as described previously (44).
Sample purification and TMT labeling
Native oxidation and peptide samples were purified using an S-Trap mini kit 2.1 (ProtiFi) using manufacturer’s protocol. Hundred millimolar iodoacetamide was used as the alkylating agent, and overnight trypsin digests were conducted at a trypsin-protein ratio of 1:25 at 37 °C. SPROX sample cleanup was performed as previously described (44) and digested overnight with a trypsin-protein ratio of 1:100 at 37 °C. After digestion, TMT labeling for SPROX and native oxidation samples was conducted with a TMT10plex mass tag labeling kit (Thermo Scientific), using 0.2 mg of isobaric label per 10 μg of lysate digest.
Peptide fractionation by spin column
To increase coverage, all peptide samples were fractionated using C18 spin columns prior to LC-MS/MS analysis. Columns were first conditioned using ACN and 100 mM ammonium hydroxide (AH). Samples were resuspended in 50 to 100 μl of AH and added to the columns. After washing the columns with ACN and AH, samples were eluted with a gradient of 16 concentrations of ACN ranging from 2 to 50%. Eluent fractions were then recombined into eight fractions in a staggered manner (#1 with #8, #2 with #9, etc.), dried down, and resuspended in 0.1% TFA at a concentration of 0.25 mg/ml for LC-MS/MS analysis.
LC-MS/MS
TMT-labeled peptides originating from the native oxidation and SPROX methods were injected onto an Easy nLC-1200 HPLC instrument (Thermo Fisher) using a 30 cm C18 column packed with 1.8 μm beads (Sepax), made in-house, for analysis via a Fusion Lumos Tribrid mass spectrometer (Thermo Fisher). Peptides were eluted from the column using a gradient of two solvents: A (0.1% formic acid in water) and B (0.1% formic acid in 80% ACN). The gradient started with 3% B for 2 min, increased to 10% B over 7 min, then to 38% B over 94 min, finally ramped up to 90% B over 5 min, and held for 3 min, before returning to starting conditions in 2 min and re-equilibrating for 7 min, for a total runtime of 120 min. Ionization was carried out using a Nanospray Flex source operating at 2 kV. The Fusion Lumos was operated in data-dependent mode, acquiring both MS1 and MS2 scans in the Orbitrap with a cycle time of 3 s. Monoisotopic Precursor Selection was set to Peptide. The scan range was 400 to 1500 m/z, with an AGC target of 4e5, a resolution of 120,000 at m/z of 200, and a maximum injection time of 50 ms. Only peptides with a charge state between 2 to 5 were picked for fragmentation. Precursor ions were fragmented by higher-energy collisional dissociation, using a collision energy of 38% and an isolation width of 1.0 m/z. MS2 scans were collected with a resolution of 50,000, a maximum injection time of 105 ms, and an AGC setting of 1e5. Dynamic exclusion was set to 45 s.
Analysis of samples generated by the peptide oxidation experiment was conducted as above with the following changes. The runtime was reduced to 90 min, while the cycle time was set to 1 s to ensure enough scans across the peak for accurate MS1 quantitation. Precursor ions were fragmented by collision-induced dissociation, using a collision energy of 30 and an isolation width of 1.1 m/z. MS2 scans were collected in the ion trap with the scan rate set to rapid, a maximum injection time of 35 ms, and an AGC setting of 1e4. Dynamic exclusion was set to 20 s.
Database searches and quantitation
For SPROX and native oxidation experiments, MS2 data were searched against the E. coli (4352 entries, downloaded 1/18/2019) or T. thermophilus (2227 entries, downloaded 1/18/2019) UniProt databases using the integrated Andromeda search engine with MaxQuant (version 1.6.5) software (55), with mass tolerances of 4.5 ppm for precursor ions and 0.003 Da for reporter ions, respectively. Searches included the TMT10plex labels as fixed modifications and methionine oxidation as a variable modification (Δmass = 15.99491) and allowed for up to two missed cleavages of trypsin protease (cutting at R and K) with maximum false discovery rate thresholds of 1%. Resulting ion intensities were then normalized using total channel intensity of non-Met peptides and median normalization. MS searching, quantification, and curve fitting to acquire intrinsic rate constants for proteomic E. coli peptides were performed as previously described (46). Coverage for all MS experiments is shown in Table S2.
SA measurements
SA measurements from PDB structures were computed for available E. coli and T. thermophilus proteins as previously described (44). Briefly, solvent accessible surface areas of methionine residues were calculated in Pymol, with dot density set to three. If a protein was represented by more than one PDB file, the value reported is the median value for all structures. A maximum of three PDB files were analyzed for each protein. Multimeric structures were analyzed with all subunits present. Additionally, SA values for E. coli were computed using structures available from the AlphaFold (AF) Protein Structure Database using AlphaFold2 (47). Structures for T. thermophilus were not available in the AF database. Therefore, they were predicted using ColabFold (56), utilizing the fast homology search of MMseqs2 with AlphaFold2, with the AlphaFold2_advanced option set to the default parameters for all proteins. This analysis was conducted using the computational resources of the University of Rochester’s Center for Integrated Research Computing Linux cluster. The top five ranked models for each protein were then used to calculate SAs as above, keeping only models with an average plDDT value >70. Where SA measurements for both PDB and AF structures were available, the two values were averaged. SA measurements are reported in Table S3.
Measurements of oxidation rate constants and PFs
Pseudo-first order observed rate constants for oxidation of peptides and proteins ( and , respectively) were measured by least-squares fitting of fractional oxidation (fox) of each methionine-containing peptide to the following equation:
| (1) |
Where t is the oxidation time in minutes. Given that concentration of H2O2 was in excess of methionines in all experiments, the above pseudo-first order rate constants can be converted to second-order oxidation rate constants for proteins and peptides (kprotein and kintrinsic, respectively) by dividing kobs values by the molar concentration of H2O2. For intact proteins, PFs were measured by taking the ratio of kprotein and kintrinsic:
| (2) |
Since measured peptide kintrinsic values closely matched the oxidation rate of free methionine (23), we used the latter as our reference kintrinsic value for measuring PFs [after correcting for temperature using the Arrhenius equation (43)]. For E. coli, where it was possible to calculate PFs for a portion of the data using kintrinsic values derived from peptide rate measurements, the measured PFs were very similar to those measured using kintrinsic values derived using free methionine. We chose to use the latter as it significantly increased the number of measured PFs (as there was limited overlap between proteomic coverages of peptide and protein oxidation experiments). Since our time course was optimized to survey slow, protected methionines, the maximum value of kprotein in each experiment was set to the respective value for kintrinsic. Rates for proteins and peptides were kept only for tryptic peptides containing a single Met, which also lacked Cys, and had a r2 (goodness-of-fit to Equation 1) ≥ 0.75. The filtered rate data and resulting PFs are listed in Table S3.
Model for relating PFs to folding stabilities
For protected methionines where protein unfolding is required for oxidation, the oxidation reaction can be modeled based on the following reaction:
| (3) |
Where kopen and kclose are the folding and unfolding rate constants, respectively, and can be related to the folding equilibrium constant:
| (4) |
Analogous to the model used for analysis of HDX experiments (45, 57), the following relationship is established between the observed rate constant for oxidation (kprotein) and the three rate constants employed in the model:
| (5) |
A survey of available protein-folding rates from the protein folding database indicated that the average protein has a folding rate roughly 15,000 times faster than the oxidation of free methionine (58). Therefore, this reaction is expected to be in an EX2 regime (45, 57) where kclose is significantly faster than kopen and kintrinsic, and therefore:
| (6) |
| (7) |
PFs can thus be related to folding stabilities (ΔGfolding) and midpoints of denaturation curves (C1/2):
| (8) |
Where R is the gas constant, T is the temperature (in Kelvin), and m is the slope of the linear relationship between ΔGfolding and denaturant concentration in accordance to a two-state folding model (44). Therefore, if the assumptions of the above model are valid, oxidation PFs are expected to be correlated with folding stabilities and C1/2 values.
Data availability
All raw and processed data are available in the included Supporting Information and at the ProteomeXchange Consortium via the PRIDE (59) partner repository (accession number PXD030245).
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank the members of the Ghaemmaghami lab at the University of Rochester for helpful discussions and suggestions.
Author contributions
E. J. W. and S. G. conceptualization; E. J. W., J. Q. B., K. A. W., J. R. H., A. M. M. V., M. R. O’C., and S. G. methodology; E. J. W., J. Q. B., A. M. M. V., and M. R. O’C. software; E. J. W., J. Q. B., A. M. M. V., M. R. O’C., and S. G. formal analysis; E. J. W., K. A. W., J. R. H., A. M. M. V., and M. R. O’C. investigation; E. J. W. and S. G. writing–original draft; S. G. supervision.
Funding and additional information
This work was supported by grants from the National Institutes of Health (R35 GM119502 and S10 OD025242 to S. G, and R35 GM133462 to M. R. O’C.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Biography

Ethan J. Walker (https://www.linkedin.com/in/ethan-j-walker) is a graduate researcher in the Department of Biochemistry at the University of Rochester. They study proteomics and mass spectrometry, with a special interest in method development for the purpose of interrogating protein folding and stability on a global scale. They are defending their PhD thesis in the summer of 2022 and looking forward to the possibility of applying their work in pharmaceutical research.
Edited by Ursula Jakob
Supporting information
(Supp_table1.xlsx) contains detailed descriptions of proteomic experiments and serves as the key for the raw MS data uploaded to PRIDE.
(Supp_table2.xlsx) contains descriptions and coverage of all MS experiments for each method.
(Supp_table3.xlsx) contains descriptions and lists of all the relevant measured parameters presented in this study that were gathered from our three methods and passed our filtering criteria (see text).
References
- 1.Levine R.L., Moskovitz J., Stadtman E.R. Oxidation of methionine in proteins: Roles in antioxidant defense and cellular regulation. IUBMB Life. 2000;50:301–307. doi: 10.1080/713803735. [DOI] [PubMed] [Google Scholar]
- 2.Levine R.L., Mosoni L., Berlett B.S., Stadtman E.R. Methionine residues as endogenous antioxidants in proteins. Proc. Natl. Acad. Sci. U. S. A. 1996;93:15036–15040. doi: 10.1073/pnas.93.26.15036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stadtman E.R., Van Remmen H., Richardson A., Wehr N.B., Levine R.L. Methionine oxidation and aging. Biochim. Biophys. Acta. 2005;1703:135–140. doi: 10.1016/j.bbapap.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 4.Moskovitz J. Roles of methionine suldfoxide reductases in antioxidant defense, protein regulation and survival. Curr. Pharm. Des. 2005;11:1451–1457. doi: 10.2174/1381612053507846. [DOI] [PubMed] [Google Scholar]
- 5.Fremont S., Romet-Lemonne G., Houdusse A., Echard A. Emerging roles of MICAL family proteins - from actin oxidation to membrane trafficking during cytokinesis. J. Cell Sci. 2017;130:1509–1517. doi: 10.1242/jcs.202028. [DOI] [PubMed] [Google Scholar]
- 6.Chao C.C., Ma Y.S., Stadtman E.R. Modification of protein surface hydrophobicity and methionine oxidation by oxidative systems. Proc. Natl. Acad. Sci. U. S. A. 1997;94:2969–2974. doi: 10.1073/pnas.94.7.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.von Eckardstein A., Walter M., Holz H., Benninghoven A., Assmann G. Site-specific methionine sulfoxide formation is the structural basis of chromatographic heterogeneity of apolipoproteins A-I, C-II, and C-III. J. Lipid Res. 1991;32:1465–1476. [PubMed] [Google Scholar]
- 8.Schoneich C., Zhao F., Wilson G.S., Borchardt R.T. Iron-thiolate induced oxidation of methionine to methionine sulfoxide in small model peptides. Intramolecular catalysis by histidine. Biochim. Biophys. Acta. 1993;1158:307–322. doi: 10.1016/0304-4165(93)90030-c. [DOI] [PubMed] [Google Scholar]
- 9.Dalle-Donne I., Rossi R., Giustarini D., Gagliano N., Di Simplicio P., Colombo R., Milzani A. Methionine oxidation as a major cause of the functional impairment of oxidized actin. Free Radic. Biol. Med. 2002;32:927–937. doi: 10.1016/s0891-5849(02)00799-2. [DOI] [PubMed] [Google Scholar]
- 10.Liu D., Ren D., Huang H., Dankberg J., Rosenfeld R., Cocco M.J., Li L., Brems D.N., Remmele R.L., Jr. Structure and stability changes of human IgG1 Fc as a consequence of methionine oxidation. Biochemistry. 2008;47:5088–5100. doi: 10.1021/bi702238b. [DOI] [PubMed] [Google Scholar]
- 11.Brot N., Weissbach H. Biochemistry and physiological role of methionine sulfoxide residues in proteins. Arch. Biochem. Biophys. 1983;223:271–281. doi: 10.1016/0003-9861(83)90592-1. [DOI] [PubMed] [Google Scholar]
- 12.Mulinacci F., Capelle M.A., Gurny R., Drake A.F., Arvinte T. Stability of human growth hormone: Influence of methionine oxidation on thermal folding. J. Pharm. Sci. 2011;100:451–463. doi: 10.1002/jps.22293. [DOI] [PubMed] [Google Scholar]
- 13.Smith C.D., Carney J.M., Starke-Reed P.E., Oliver C.N., Stadtman E.R., Floyd R.A., Markesbery W.R. Excess brain protein oxidation and enzyme dysfunction in normal aging and in Alzheimer disease. Proc. Natl. Acad. Sci. U. S. A. 1991;88:10540–10543. doi: 10.1073/pnas.88.23.10540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berlett B.S., Stadtman E.R. Protein oxidation in aging, disease, and oxidative stress. J. Biol. Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- 15.Younan N.D., Nadal R.C., Davies P., Brown D.R., Viles J.H. Methionine oxidation perturbs the structural core of the prion protein and suggests a generic misfolding pathway. J. Biol. Chem. 2012;287:28263–28275. doi: 10.1074/jbc.M112.354779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manta B., Gladyshev V.N. Regulated methionine oxidation by monooxygenases. Free Radic. Biol. Med. 2017;109:141–155. doi: 10.1016/j.freeradbiomed.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erickson J.R., Joiner M.L., Guan X., Kutschke W., Yang J., Oddis C.V., Bartlett R.K., Lowe J.S., O'Donnell S.E., Aykin-Burns N., Zimmerman M.C., Zimmerman K., Ham A.J., Weiss R.M., Spitz D.R., et al. A dynamic pathway for calcium-independent activation of CaMKII by methionine oxidation. Cell. 2008;133:462–474. doi: 10.1016/j.cell.2008.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoshi T., Heinemann S. Regulation of cell function by methionine oxidation and reduction. J. Physiol. 2001;531:1–11. doi: 10.1111/j.1469-7793.2001.0001j.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bigelow D.J., Squier T.C. Redox modulation of cellular signaling and metabolism through reversible oxidation of methionine sensors in calcium regulatory proteins. Biochim. Biophys. Acta. 2005;1703:121–134. doi: 10.1016/j.bbapap.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 20.Ortegon Salas C., Schneider K., Lillig C.H., Gellert M. Signal-regulated oxidation of proteins via MICAL. Biochem. Soc. Trans. 2020;48:613–620. doi: 10.1042/BST20190866. [DOI] [PubMed] [Google Scholar]
- 21.Moskovitz J. Methionine sulfoxide reductases: Ubiquitous enzymes involved in antioxidant defense, protein regulation, and prevention of aging-associated diseases. Biochim. Biophys. Acta. 2005;1703:213–219. doi: 10.1016/j.bbapap.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 22.Lam X.M., Yang J.Y., Cleland J.L. Antioxidants for prevention of methionine oxidation in recombinant monoclonal antibody HER2. J. Pharm. Sci. 1997;86:1250–1255. doi: 10.1021/js970143s. [DOI] [PubMed] [Google Scholar]
- 23.Yin J., Chu J.W., Ricci M.S., Brems D.N., Wang D.I., Trout B.L. Effects of antioxidants on the hydrogen peroxide-mediated oxidation of methionine residues in granulocyte colony-stimulating factor and human parathyroid hormone fragment 13-34. Pharm. Res. 2004;21:2377–2383. doi: 10.1007/s11095-004-7692-4. [DOI] [PubMed] [Google Scholar]
- 24.Zang L., Carlage T., Murphy D., Frenkel R., Bryngelson P., Madsen M., Lyubarskaya Y. Residual metals cause variability in methionine oxidation measurements in protein pharmaceuticals using LC-UV/MS peptide mapping. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2012;895-896:71–76. doi: 10.1016/j.jchromb.2012.03.016. [DOI] [PubMed] [Google Scholar]
- 25.Sankar K., Hoi K.H., Yin Y., Ramachandran P., Andersen N., Hilderbrand A., McDonald P., Spiess C., Zhang Q. Prediction of methionine oxidation risk in monoclonal antibodies using a machine learning method. MAbs. 2018;10:1281–1290. doi: 10.1080/19420862.2018.1518887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffiths S.W., Cooney C.L. Development of a peptide mapping procedure to identify and quantify methionine oxidation in recombinant human alpha1-antitrypsin. J. Chromatogr. A. 2002;942:133–143. doi: 10.1016/s0021-9673(01)01350-4. [DOI] [PubMed] [Google Scholar]
- 27.Griffiths S.W., Cooney C.L. Relationship between protein structure and methionine oxidation in recombinant human alpha 1-antitrypsin. Biochemistry. 2002;41:6245–6252. doi: 10.1021/bi025599p. [DOI] [PubMed] [Google Scholar]
- 28.Delmar J.A., Buehler E., Chetty A.K., Das A., Quesada G.M., Wang J., Chen X. Machine learning prediction of methionine and tryptophan photooxidation susceptibility. Mol. Ther. Methods Clin. Dev. 2021;21:466–477. doi: 10.1016/j.omtm.2021.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu K., Uversky V.N., Xue B. Local flexibility facilitates oxidization of buried methionine residues. Protein Pept. Lett. 2012;19:688–697. doi: 10.2174/092986612800494084. [DOI] [PubMed] [Google Scholar]
- 30.Chu J.W., Yin J., Wang D.I., Trout B.L. Molecular dynamics simulations and oxidation rates of methionine residues of granulocyte colony-stimulating factor at different pH values. Biochemistry. 2004;43:1019–1029. doi: 10.1021/bi0356000. [DOI] [PubMed] [Google Scholar]
- 31.Valley C.C., Cembran A., Perlmutter J.D., Lewis A.K., Labello N.P., Gao J., Sachs J.N. The methionine-aromatic motif plays a unique role in stabilizing protein structure. J. Biol. Chem. 2012;287:34979–34991. doi: 10.1074/jbc.M112.374504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharp J.S., Becker J.M., Hettich R.L. Analysis of protein solvent accessible surfaces by photochemical oxidation and mass spectrometry. Anal. Chem. 2004;76:672–683. doi: 10.1021/ac0302004. [DOI] [PubMed] [Google Scholar]
- 33.Veredas F.J., Canton F.R., Aledo J.C. Methionine residues around phosphorylation sites are preferentially oxidized in vivo under stress conditions. Sci. Rep. 2017;7:40403. doi: 10.1038/srep40403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghesquiere B., Jonckheere V., Colaert N., Van Durme J., Timmerman E., Goethals M., Schymkowitz J., Rousseau F., Vandekerckhove J., Gevaert K. Redox proteomics of protein-bound methionine oxidation. Mol. Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang R., Jain T., Lynaugh H., Nobrega R.P., Lu X., Boland T., Burnina I., Sun T., Caffry I., Brown M., Zhi X., Lilov A., Xu Y. Rapid assessment of oxidation via middle-down LCMS correlates with methionine side-chain solvent-accessible surface area for 121 clinical stage monoclonal antibodies. MAbs. 2017;9:646–653. doi: 10.1080/19420862.2017.1290753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aledo J.C., Canton F.R., Veredas F.J. A machine learning approach for predicting methionine oxidation sites. BMC Bioinformatics. 2017;18:430. doi: 10.1186/s12859-017-1848-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.West G.M., Tang L., Fitzgerald M.C. Thermodynamic analysis of protein stability and ligand binding using a chemical modification- and mass spectrometry-based strategy. Anal. Chem. 2008;80:4175–4185. doi: 10.1021/ac702610a. [DOI] [PubMed] [Google Scholar]
- 38.Henzler-Wildman K., Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 39.Bryngelson J.D., Onuchic J.N., Socci N.D., Wolynes P.G. Funnels, pathways, and the energy landscape of protein folding: A synthesis. Proteins. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 40.Pace C.N., Martin Scholtz J. Measuring the conformational stability of a protein. Protein Struct. A Pract. approach. 1997;2:299–321. [Google Scholar]
- 41.Schellman J.A. The thermodynamic stability of proteins. Annu. Rev. Biophys. Biophys. Chem. 1987;16:115–137. doi: 10.1146/annurev.bb.16.060187.000555. [DOI] [PubMed] [Google Scholar]
- 42.Thirumangalathu R., Krishnan S., Bondarenko P., Speed-Ricci M., Randolph T.W., Carpenter J.F., Brems D.N. Oxidation of methionine residues in recombinant human interleukin-1 receptor antagonist: Implications of conformational stability on protein oxidation kinetics. Biochemistry. 2007;46:6213–6224. doi: 10.1021/bi700321g. [DOI] [PubMed] [Google Scholar]
- 43.Pan B., Abel J., Ricci M.S., Brems D.N., Wang D.I., Trout B.L. Comparative oxidation studies of methionine residues reflect a structural effect on chemical kinetics in rhG-CSF. Biochemistry. 2006;45:15430–15443. doi: 10.1021/bi061855c. [DOI] [PubMed] [Google Scholar]
- 44.Walker E.J., Bettinger J.Q., Welle K.A., Hryhorenko J.R., Ghaemmaghami S. Global analysis of methionine oxidation provides a census of folding stabilities for the human proteome. Proc. Natl. Acad. Sci. U. S. A. 2019;116:6081–6090. doi: 10.1073/pnas.1819851116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huyghues-Despointes B.M., Pace C.N., Englander S.W., Scholtz J.M. Measuring the conformational stability of a protein by hydrogen exchange. Methods Mol. Biol. 2001;168:69–92. doi: 10.1385/1-59259-193-0:069. [DOI] [PubMed] [Google Scholar]
- 46.Bettinger J.Q., Welle K.A., Hryhorenko J.R., Ghaemmaghami S. Quantitative analysis of in Vivo methionine oxidation of the human proteome. J. Proteome Res. 2020;19:624–633. doi: 10.1021/acs.jproteome.9b00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zidek A., Potapenko A., Bridgland A., Meyer C., Kohl S.A.A., Ballard A.J., Cowie A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeldovich K.B., Chen P., Shakhnovich E.I. Protein stability imposes limits on organism complexity and speed of molecular evolution. Proc. Natl. Acad. Sci. U. S. A. 2007;104:16152–16157. doi: 10.1073/pnas.0705366104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Braselmann E., Chaney J.L., Clark P.L. Folding the proteome. Trends Biochem. Sci. 2013;38:337–344. doi: 10.1016/j.tibs.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karshikoff A., Nilsson L., Ladenstein R. Rigidity versus flexibility: The dilemma of understanding protein thermal stability. FEBS J. 2015;282:3899–3917. doi: 10.1111/febs.13343. [DOI] [PubMed] [Google Scholar]
- 51.Mateus A., Maatta T.A., Savitski M.M. Thermal proteome profiling: Unbiased assessment of protein state through heat-induced stability changes. Proteome Sci. 2016;15:13. doi: 10.1186/s12953-017-0122-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leuenberger P., Ganscha S., Kahraman A., Cappelletti V., Boersema P.J., von Mering C., Claassen M., Picotti P. Cell-wide analysis of protein thermal unfolding reveals determinants of thermostability. Science. 2017;355 doi: 10.1126/science.aai7825. [DOI] [PubMed] [Google Scholar]
- 53.Pace C.N. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- 54.Swann P.G., Tolnay M., Muthukkumar S., Shapiro M.A., Rellahan B.L., Clouse K.A. Considerations for the development of therapeutic monoclonal antibodies. Curr. Opin. Immunol. 2008;20:493–499. doi: 10.1016/j.coi.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 55.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 56.Mirdita M., Schütze K., Moriwaki Y., Heo L., Ovchinnikov S., Steinegger M. ColabFold - making protein folding accessible to all. bioRxiv. 2021 doi: 10.1101/2021.08.15.456425. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Konermann L., Tong X., Pan Y. Protein structure and dynamics studied by mass spectrometry: H/D exchange, hydroxyl radical labeling, and related approaches. J. Mass Spectrom. 2008;43:1021–1036. doi: 10.1002/jms.1435. [DOI] [PubMed] [Google Scholar]
- 58.Manavalan B., Kuwajima K., Lee J. Pfdb: A standardized protein folding database with temperature correction. Sci. Rep. 2019;9:1588. doi: 10.1038/s41598-018-36992-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., Perez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S., et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(Supp_table1.xlsx) contains detailed descriptions of proteomic experiments and serves as the key for the raw MS data uploaded to PRIDE.
(Supp_table2.xlsx) contains descriptions and coverage of all MS experiments for each method.
(Supp_table3.xlsx) contains descriptions and lists of all the relevant measured parameters presented in this study that were gathered from our three methods and passed our filtering criteria (see text).
Data Availability Statement
All raw and processed data are available in the included Supporting Information and at the ProteomeXchange Consortium via the PRIDE (59) partner repository (accession number PXD030245).