Abstract
Though in vitro culture of primary intestinal organoids has gained significant momentum in recent years, little has been done to investigate the impact of microenvironmental cues provided by the encapsulating matrix on the growth and development of these fragile cultures. In this work, the impact of various in vitro culture parameters on primary adult murine organoid formation and growth are analyzed with a focus on matrix properties and geometric culture configuration. The air–liquid interface culture configuration was found to result in enhanced organoid formation relative to a traditional submerged configuration. Additionally, through use of a recombinantly engineered extracellular matrix (eECM), the effects of biochemical and biomechanical cues were independently studied. Decreasing mechanical stiffness and increasing cell adhesivity were found to increase organoid yield. Tuning of eECM properties was used to obtain organoid formation efficiency values identical to those observed in naturally harvested collagen I matrices but within a stiffer construct with improved ease of physical manipulation. Increased ability to remodel the surrounding matrix through mechanical or enzymatic means was also shown to enhance organoid formation. As the engineering and tunability of recombinant matrices is essentially limitless, continued property optimization may result in further improved matrix performance and may help to identify additional microenvironmental cues that directly impact organoid formation, development, differentiation, and functional behavior. Continued culture of primary organoids in recombinant matrices could therefore prove to be largely advantageous in the field of intestinal tissue engineering for applications in regenerative medicine and in vitro tissue mimics.
Introduction
Intestinal tissue engineering has emerged as a rapidly evolving field in which biopsies of intestinal tissue are cultured in vitro1–5 or propagated as in vivo cultures in living bioreactors.2,6–8 These cultures have wide-spread application for use as in vitro tissue mimics to study tissue development and maintenance,1,5,7–10 the progression of various intestinal diseases including cancer and pathogenic infections,2–4,10 and as platforms for drug screening.3,4,10–12 In addition, intestinal tissue engineering offers a potential regenerative medicine therapy for multiple, debilitating gastrointestinal diseases that often require resection of afflicted tissue. In extreme cases, an extensive length of tissue must be removed, thereby impacting the patient’s ability to digest food and absorb nutrients properly.13–17 Short Bowel Syndrome (SBS) is a life-threatening condition that leaves patients dependent upon intravenously administered nutrition until either lengthening procedures can be used to restore a functional intestinal length or the patient receives an intestinal transplant.7,13–20
To date, there is no ideal curative treatment for Short Bowel Syndrome. Bowel lengthening procedures require sufficient healthy tissue and are technically challenging, time and resource intensive, and are often reversed by the body’s natural adaptation and recovery processes.7,18–20 In addition to common concerns for organ transplants, such as host rejection and lifetime dependency on immunosuppressives, intestinal transplants suffer from markedly lower success rates than other whole organ procedures, likely due to the high degree of immune activity that occurs within this organ system.13,21–27 Chronic dependency on intravenously delivered nutrition imposes elevated risks for central line infection and sepsis, loss of venous access, and liver failure.21,22,24,27,28 As such, tissue engineering is a promising strategy that may one day enable the expansion of autologous tissue in vitro for subsequent implantation to restore healthy digestive functioning without the risk of rejection, the need for immunosuppressives, or the presence of chronically dwelling central lines to deliver nutrition.13,14
Primary intestinal epithelium has historically been difficult to maintain in vitro, though in 2009, two distinct culture protocols were published that enabled the long-term culture of primary neonatal intestinal epithelium harvested from mouse tissue as 3D organoids.29,30 In one method, isolated intestinal crypts are encapsulated within Matrigel and seeded onto the bottom of a culture dish and submerged in a heavily supplemented medium.30 In the alternative method, minced tissue sections containing both epithelial and supportive stromal cells are encapsulated within a collagen matrix and seeded onto a suspended porous transwell insert.29 The insert serves to maintain the organoids at a height that is equivalent to that of the surrounding air–liquid interface that results from the medium being placed around rather than inside of the insert (Fig. 1A). The top surface of the organoid-containing matrix is thus exposed to open-air conditions, and nutrient diffusion from the medium can only occur in a bottom-up fashion through the porous membrane.
Fig. 1.
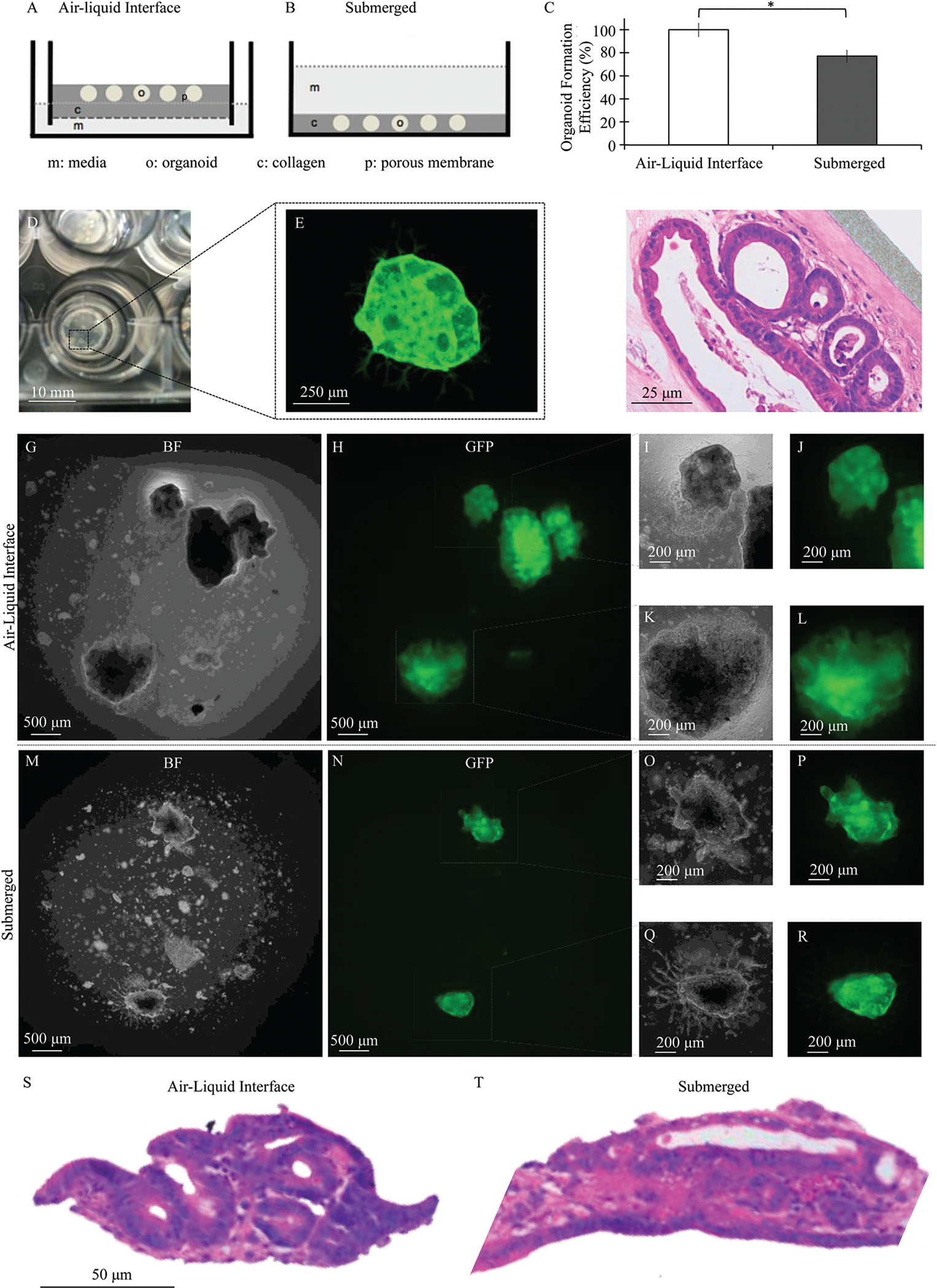
Schematic of (A) air–liquid interface and (B) submerged configurations used to culture primary adult murine intestinal organoids in collagen I matrices, with “c” indicating collagen I, “o” indicating organoids, “m” indicating cell culture media, and “p” indicating the porous membrane of a transwell insert. (C) Culture within the air–liquid interface configuration resulted in enhanced organoid formation efficiency compared to the submerged configuration, mean ± SEM compiled across two independent trials after 3 days of culture, * p < 0.05. (D) Image of primary adult murine intestinal organoids cultured under the air–liquid interface method. (E) Global expression of GFP enabled confocal imaging of intestinal organoids, demonstrating a large central body with mesenchymal myofibroblasts extending outward from the organoid surface. (F) Hematoxylin and eosin staining demonstrated organoid structures, each consisting of a sheet of epithelial cells enclosing a central lumen and surrounded by a network of stromal cells after 5 days of culture. Bright field and fluorescence images of GFP+ intestinal organoids cultured under both (G–L) air–liquid interface and (M–R) submerged configurations. (S and T) Hematoxylin and eosin staining demonstrated epithelial sheet formation under both configurations.
These primary intestinal organoid culture protocols have been further developed and studied since their initial introduction in 2009. Further studies have included the application of various genetic modifications to the primary cultures31,32 and the discovery of the role of various biochemical factors in promoting differentiation of specific epithelial lineages within primary organoids.33–35 To date, most investigation of the effects of the in vitro culture microenvironment on primary intestinal organoid behavior have focused on exogenous biochemical cues and soluble signaling molecules.29,30,33–36 Little work has been done to investigate the role of biomechanical and biochemical cues provided by the surrounding three-dimensional matrix on intestinal organoid formation and development, though we recently demonstrated that matrix mechanical and structural properties are critical in enabling coordinated contraction within primary neonatal intestinal organoid cultures.37 Furthermore, though both primary organoid culture techniques have been successfully demonstrated with adult murine tissue, most studies have focused on the use of neonatal tissue. Thus, little is known about how the in vitro microenvironment impacts the behavior of primary intestinal organoids derived from adult tissue, an inherently more fragile cell population.38
In this work, we investigate the impact of various microenvironmental properties on the formation and development of primary organoids derived from adult murine tissue, focused specifically on the role of cues provided by the surrounding 3D matrix material and culture configuration. We chose to focus on adult organoids containing both epithelial and mesenchymal cell types because we found these cultures to have the most direct relevance to the essential goal of tissue engineering efforts: the in vitro expansion of primary tissue to be used in the generation of tissue constructs that contain all predominant cell types found in healthy intestinal tissue.
Within this work, a recombinant engineered extracellular matrix (eECM) was used as a novel organoid culture matrix to enable the independent tuning and therefore investigation of the influences of matrix biomechanical and biochemical signals on primary intestinal organoids. This eECM contains peptide domains derived from naturally occurring extracellular matrix proteins.39–41 Within the polypeptide backbone, an extended RGD cell-binding domain derived from fibronectin has been linked with a structural domain derived from elastin.41 This pairing allows for the in vitro provision of cell-adhesive biochemical cues and elastomeric biomechanical cues that naturally exist within healthy intestinal tissue. Furthermore, by harnessing distinct bioactive and structural domains, cell-adhesive ligand concentration can be varied independently of crosslinking density and mechanical properties.41–43 This is a tremendous advantage over native matrix materials, such as collagen I and Matrigel, in which these properties are inherently coupled.44,45 Furthermore, several recombinant variants of elastin-like proteins have been reported for a variety of medical applications, and they have displayed excellent biocompatibility and scalability.46 Thus, our eECM allows for a level of microenvironment engineering previously unattained in the culture of primary intestinal organoids.
Results & discussion
Air–liquid interface configuration promotes enhanced organoid formation from primary adult murine intestinal tissue
Intestinal organoids containing both epithelial and mesenchymal cells were successfully isolated from primary adult murine tissue and encapsulated in a collagen I matrix according to established protocols.29 Organoids were cultured using a traditional, submerged configuration and within an air–liquid interface method through the use of transwell inserts (Fig. 1A and B).29,37 Organoids successfully formed under both conditions when encapsulated in collagen I, though air–liquid interface cultures resulted in higher yields than the submerged configuration. When normalized on a per harvest basis, the incidence of organoid formation in the submerged configuration was reduced to 77% of that found for the air–liquid configuration (Fig. 2C), with average maximum organoid counts ranging from 3 to 9 organoids per 25 μL of gel over various independent harvests (Fig. S1†).
Fig. 2.
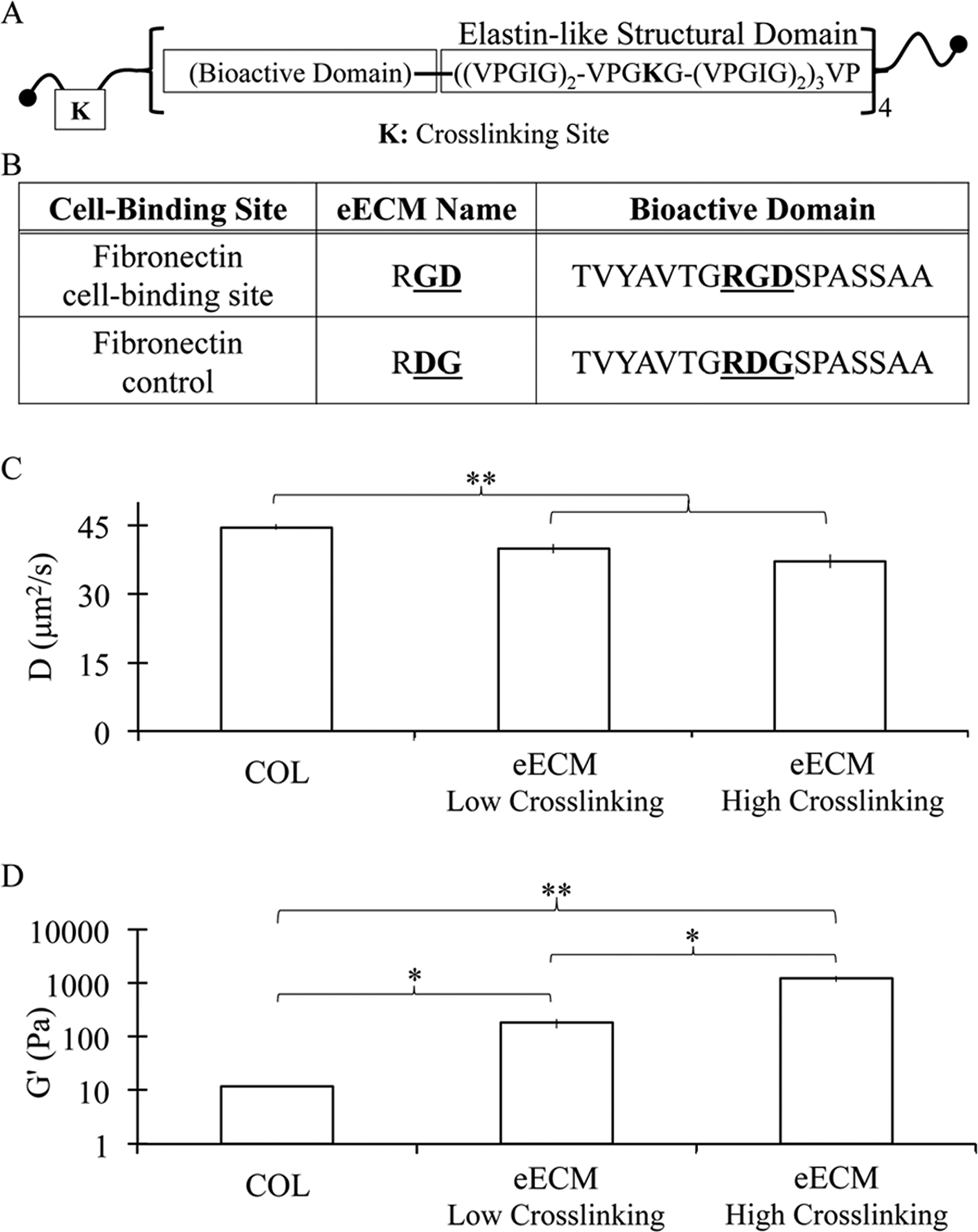
(A) Schematic of engineered extracellular matrix (eECM) polypeptide chain and (B) amino acid sequences within the cell-adhesive or non-adhesive versions of the extended RGD domain. (C) Diffusivity constants for fluorescently labeled dextran (150 kDa) demonstrate that eECM matrices have decreased transport rates relative to collagen I matrices. (D) Increased eECM crosslinking density results in increased mechanical stiffness, as indicated by storage moduli measured by oscillatory rheology. Collagen I matrices were significantly more compliant than both eECM formulations tested. All data is shown as mean ± SEM. * indicates statistical significance, Student’s t-test, *p < 0.05, **p < 0.01.
Organoids formed from adult tissue were similar to those previously observed from neonatal tissue and contained a sheet of epithelial cells that enclosed a central lumen and was surrounded by a network of stromal cells (Fig. 1D–F). Culture of neonatal organoids under the air–liquid interface configuration was previously shown to result in more spherically symmetric and larger organoids relative to the submerged configuration.37 Similar though less pronounced differences were observed for adult organoids (Fig. 1G–R). Organoid structures, each consisting of a central lumen and surrounded by stromal cells, were observed under both conditions (Fig. 1S and T).
The air–liquid interface culture configuration was also previously shown to be advantageous in the culture of primary neonatal intestinal organoids.37 For neonatal organoids, the air–liquid interface configuration resulted in enhanced contraction when compared to the submerged configuration. Air–liquid interface cultures were found to achieve higher equilibrium oxygen levels and more symmetric oxygen distributions throughout the culture microenvironment.37 These differences in oxygenation may contribute to the enhanced organoid formation and development observed for adult organoid cultures, although this hypothesis requires further study. Nevertheless, in considering these results altogether, continued use of the air–liquid interface culture configuration is recommended for the culture of both neonatal and adult primary murine intestinal organoids.
Development of an engineered extracellular matrix (eECM) for the culture of primary adult murine organoids
After observing the direct benefits of the air–liquid interface culture configuration, this set-up was utilized in the encapsulation and in vitro culture of primary adult organoids within recombinant matrices of an engineered extracellular matrix, eECM. Recombinant eECM contains a cell-adhesive, extended RGD amino acid sequence derived from fibronectin and an elastin-like structural backbone as previously reported (Fig. 2A and B).39–41 A non-adhesive, scrambled RDG domain-containing eECM was generated to serve as a negative control.41 Thus, hydrogels of 3 wt% eECM with RGD concentrations of 3.2 mM and 0 mM (i.e. 3.2 mM RDG) were generated. These matrices represent two extremes of ligand concentration. A chemical crosslinker, tetrakis(hydroxymethyl)phosphonium chloride, was utilized to specifically target lysine groups outside of the cell-adhesive ligand domain. This enables the decoupling and therefore independent tuning of cell-adhesive ligand concentration and substrate mechanical properties.39–41,44
The biomechanical properties of an encapsulating matrix have the potential to produce profound effects in the growth and development of encapsulated three-dimensional, multicellular structures. For example, low crosslinking density in elastin-like polypeptide hydrogels resulted in spontaneous differentiation of 100% of embryonic stem cell-derived embryoid bodies into cardiomyocytes, whereas spontaneous differentiation only occurred in 53% of hydrogels with high crosslinking density.39 Additionally, inclusion of specific cell-binding domains known to bind with integrin cell-surface receptors and propagate downstream cellular signaling can have a tremendous impact on cellular outgrowth and behavior. For example, inclusion of RGD cell-binding domains within elastin-like polypeptides enabled a two-fold increase in neurite outgrowth from encapsulated dorsal root ganglia.40
eECM matrices were found to have significantly different transport and mechanical properties relative to collagen I matrices (Fig. 2C and D). In general, diffusion was decreased within eECM relative to collagen I. The diffusion coefficient for 150 kDa dextran was found to be reduced from 44.6 ± 0.6 μm2 s−1 in collagen to 39.8 ± 1.0 μm2 s−1 and 37.1 ± 1.5 μm2 s−1 for eECM matrices of low and high crosslinking density, respectively. No statistically significant differences in transport were found between eECM matrices of varying crosslinking density, suggesting that both matrices had meshes sizes significantly larger than the hydrodynamic radius of 150 kDa dextran (~8.5 nm).47 As most nutrients and growth factors are smaller than this size, this suggests that transport should not be a limiting factor for these cultures. In contrast, altering the matrix crosslinking density resulted in significant differences in hydrogel stiffness, with storage moduli ranging from 180.8 ± 36.9 Pa to 1221.6 ± 158.7 Pa (Fig. 2D). This range of stiffness was previously shown to alter embryoid body behavior.39 Collagen I matrices were found to be significantly more compliant than both eECM matrices, with a measured storage modulus of 12.0 ± 0.4 Pa (Fig. 2D). As expected for this de-coupled eECM system, RGD concentration did not impact hydrogel mechanical properties (Fig. S2†).
Compliant eECM matrices with high concentrations of cell-adhesive binding domains enhanced organoid formation from primary adult murine intestinal tissue
Through recombinant engineering strategies, the cell-adhesive binding domain concentration and matrix stiffness of eECM matrices were varied independently, resulting in the formation of hydrogels with shear moduli of 180 Pa and 1220 Pa and RGD concentrations of 0 mM and 3.2 mM. Thus, four formulations of eECM were investigated for the culture of primary adult murine intestinal organoids. Interestingly, organoids were successfully formed in all four formulations of eECM, demonstrating that our recombinant eECM material is a suitable encapsulating matrix for the in vitro culture of primary adult intestinal organoids (Fig. 3A–H).
Fig. 3.
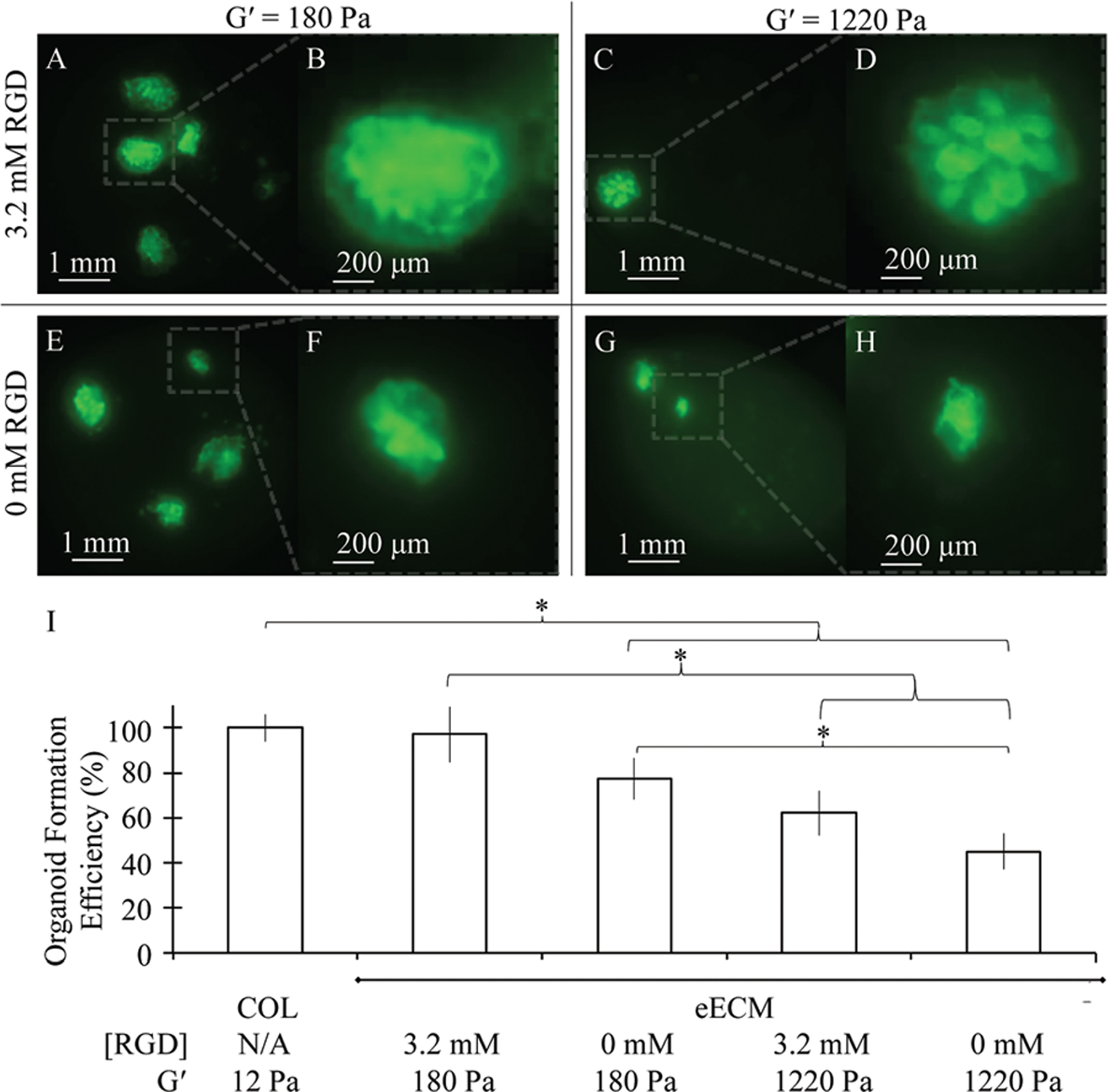
(A–H) Fluorescence images of GFP+ adult murine intestinal organoid cultured in eECM matrices of varying RGD concentration and mechanical stiffness under the air–liquid interface configuration for 3 days. (I) Increasing RGD concentration and decreasing mechanical stiffness of eECM matrices obtained organoid formation efficiency values nearly identical to those observed within collagen I matrices, whereas decreasing RGD concentration and increasing mechanical stiffness was found to negatively impact organoid formation. Organoid formation efficiency data shown is compiled across 3 independent trials. Data is shown as mean ± SEM. *indicates statistical significance (Student’s t-test, p < 0.05).
Enhanced organoid formation was observed in the most compliant matrices, and hydrogels with increased cell-adhesive domain concentration were found to further promote organoid formation (Fig. 2I). The eECM matrices with 3.2 mM RGD and storage modulus of 180 Pa resulted in the highest incidence of organoid formation and achieved nearly identical organoid yields as those observed in collagen I matrices. Decreasing RGD concentration to 0 mM reduced organoid formation efficiency to 77%, whereas increasing mechanical stiffness resulted in an even more pronounced reduction in organoid formation to 62% in 3.2 mM RGD and 45% in 0 mM RGD matrices relative to collagen I. This suggests that, if too high, matrix stiffness may actually prevent the re-organization of minced tissue explants into lumen-enclosing organoids. Similarly, though not critical in allowing organoids to form, cell–matrix interactions achieved through the inclusion of specific cell-binding domains have advantageous effects in facilitating elevated levels of organoid formation as well. One possible explanation for the mild dependence of organoid formation on the presence of cell-adhesive ligands is that the viable stromal cells that are present have supportive roles that enable the efficient formation of organoids in the absence of matrix ligands.
Intestinal organoids contain a variety of different cell types that arise from the Lgr5-positive intestinal stem cells.48 Quantitative real-time polymerase chain reaction (qRT-PCR) was employed to characterize the distribution of these intestinal cell types for organoids cultured in eECM (3.2 mM RGD, 180 Pa) or collagen I (Fig. S3†). The eECM culture resulted in enhanced expression of the enteroendocrine marker chromogranin A, while expression of the goblet cell marker mucin-2 as well as the myofibroblast marker vimentin were reduced. No statistical differences were observed in the stem cell marker Lgr5, the intestinal brush border cell marker amino peptidase, and the Paneth cell marker lysozyme.
It is worth noting that it is the recombinant nature of our eECM material that enabled the decoupling of biomechanical and biochemical cues to illuminate the relative importance of these two parameters on primary intestinal organoid cultures, something that cannot readily be accomplished using native, harvested materials such as collagen I. Furthermore, despite the fact that collagen I matrices were significantly more compliant than eECM matrices (Fig. 4C), an optimal eECM formulation that achieved the same level of organoid formation as that seen in collagen I was identified. Use of this eECM may be advantageous in the generation of a tissue engineered replacement construct due to the improved ease of physical manipulation that results from its increased mechanical stiffness relative to collagen I hydrogels. As physical manipulation will be necessary for the eventual clinical implantation of engineered tissue and the development of in vitro tissue arrays for high-throughput screening, this factor is an important practical consideration for future engineering efforts.
Fig. 4.
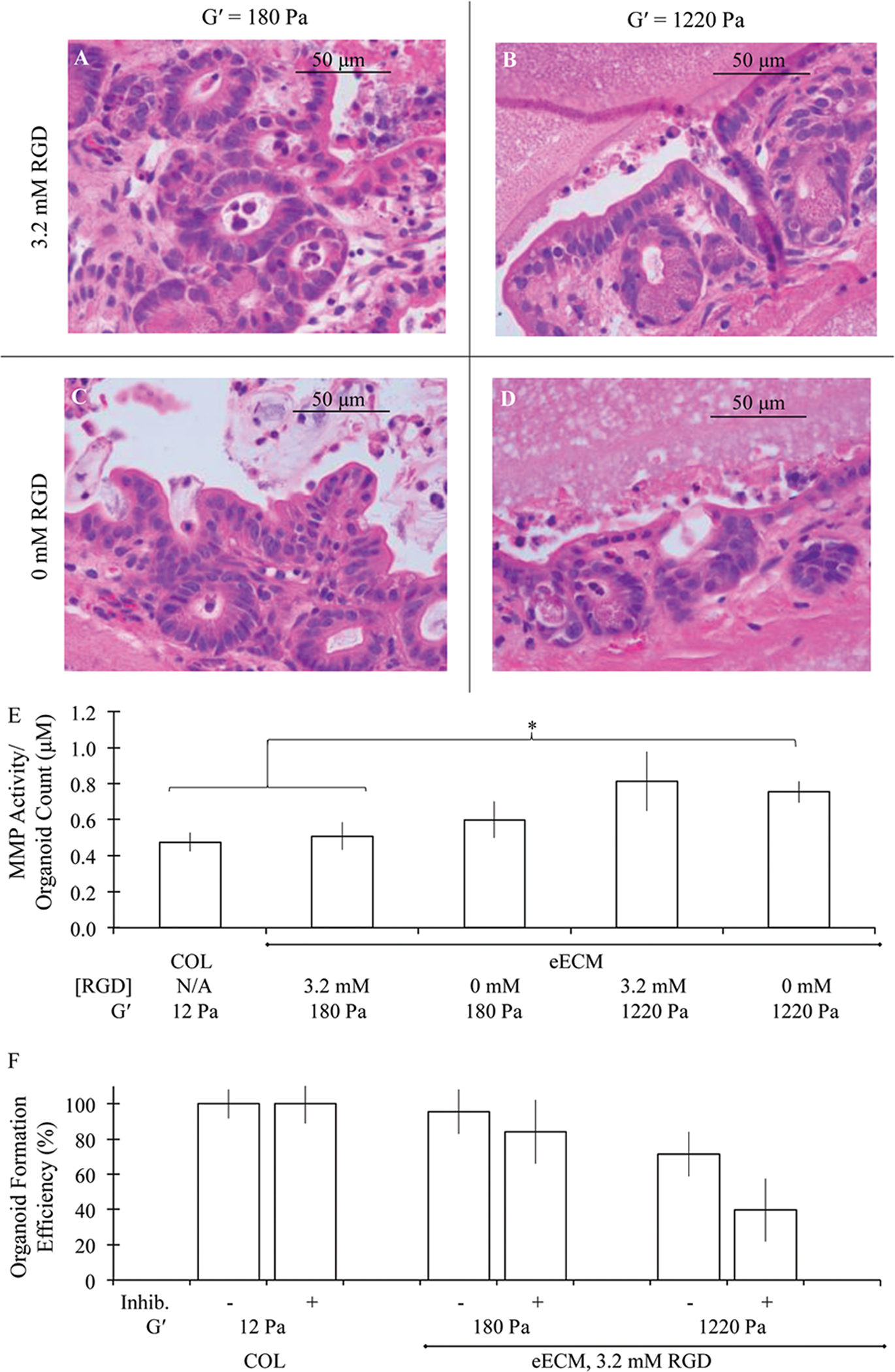
(A–D) Hematoxylin and eosin staining of adult murine intestinal organoids cultured in eECM matrices of varying RGD concentration and mechanical stiffness under the air–liquid interface configuration for 3 days. (E) MMP activity normalized to average organoid counts was found to increase with increasing matrix stiffness. (F) Inhibition of enzymatic degradation had an increasingly negative impact on organoid formation within stiffer matrices. All data is shown as mean ± SEM. *indicates statistical significance (Student’s t-test, p < 0.05).
Matrix degradation influences adult murine intestinal organoid formation
Organoids within eECM matrices were found to consist of epithelial sheets and surrounding stromal cell networks, as was previously shown for collagen I (Fig. 1F, Fig. 4A–D). As these multicellular structures continue to grow and expand into large structures in vitro, it is likely that a significant amount of mechanical disruption and chemical degradation of the surrounding matrix occurs. This notion is supported by the negative impact of increasing mechanical stiffness on organoid formation. Thus, the impact of matrix remodeling on organoid formation was investigated.
The activity level of organoid-produced matrix metalloproteinases (MMPs) was measured using a broad spectrum assay sensitive to range of enzyme activities including MMP-1, 2, 3, 7, 8, 9, 12, 13, and 14. Previous work has demonstrated that eECM is degraded by a collagenase, likely MMP-2 based upon the molecular weight band indicated in zymographic studies.39,49 To account for differences in yield across conditions, measured MMP activities were normalized to average organoid count values for each respective condition. Increased mechanical stiffness was found to result in enhanced MMP activity (Fig. 4E). The relative MMP activity measured in collagen I and compliant, highly cell-adhesive eECM matrices, 0.47 ± 0.05 μM and 0.51 ± 0.08 μM, respectively, was significantly increased to 0.81 ± 0.16 μM in stiffer, cell-adhesive eECM matrices (student’s T test, p < 0.05). MMP activity in non-adhesive eECM matrices was similar to that of cell-adhesive eECM matrices, with values of 0.60 ± 0.10 μM and 0.77 ± 0.06 μM measured in compliant and stiff formulations, respectively. This suggests that cells in stiffer matrices are likely responding to this environment by increasing their efforts to degrade the matrix, in essence working to remodel the matrix and potentially decrease the stiffness and increase the amount of free space to a more desirable state that is more conducive to organoid growth and development.
A broad MMP inhibitor, GM6001, was then added to culture medium from the time of initial encapsulation in collagen I and cell-adhesive eECM cultures. Addition of inhibitor had no effect on organoid formation in collagen I matrices. In the most compliant eECM matrices, organoid formation was slightly reduced due to MMP inhibition from 95.7% ± 12.7% to 84.1% ± 18.1% organoids per sample relative to collagen I. Organoid formation was most affected by the presence of MMP inhibitor in the stiffest eECM matrices, with a reduction in organoid formation from 71.5% ± 12.7% to 40.0% ± 17.9% (Fig. 4F). Therefore, MMP inhibition resulted in a modest 12% reduction in organoid formation in compliant eECM matrices, while in stiffer matrices the organoid formation was reduced 44%. These observations suggest that the low mechanical integrity of the more compliant materials may allow for organoid formation and growth without necessitating significant amounts of enzymatic degradation. Instead, it seems possible that the large multi-cellular organoid structures may be able to remodel a weak surrounding matrix through mechanical deformation rather than biochemical cleavage of peptide bonds. This mechanical deformation is much more difficult to achieve in stiffer matrices, thereby resulting in an enhanced reliance on enzymatic degradation to enable organoids to form and grow; therefore, organoid formation is decreased in stiffer matrices in the presence of a global MMP inhibitor, but this effect is diminished in more compliant systems.
These data suggest that even stiffer eECM scaffolds may be suitable for intestinal organoid culture if they are designed to enable sufficient matrix remodeling. Elastin-like protein matrices with a broad range of mechanical properties reaching into the 1000 kPa range have been reported.46 Remodeling within these recombinant matrices can be enhanced by encoding specific amino acid sequences into the polymer backbone that are targets for MMP cleavage.50 Furthermore, eECM scaffolds that employ adaptable crosslinks may enable matrix remodeling without the need for proteolytic degradation.51–53 The data provided here represent an encouraging starting point for further biomaterials engineering to produce intestinal organoids with scalable, reproducible matrices.
Conclusions
We have demonstrated successful culture of primary adult murine intestinal organoids within a recombinant, protein-engineered matrix. This engineered extracellular matrix (eECM) enabled the strategic decoupling of biochemical and biomechanical properties. Both matrix stiffness and cell-binding domain concentration were found to influence organoid formation. The highest yield of successful organoid formation was found to occur in compliant matrices with high cell-binding domain concentration. In fact, organoid formation within this eECM formulation was as efficient as that measured in collagen I matrices despite being an order of magnitude stiffer and hence more robust for physical manipulation. Matrix remodeling was also found to influence organoid formation, as inhibition of proteolytic matrix remodeling decreased organoid yield in stiffer matrices.
This first-pass optimization of our eECM enabled the identification of a recombinant matrix that proved to be just as successful as naturally derived collagen I in the formation of intestinal organoids from primary adult murine tissue. As the engineering and tunability of recombinant matrices is essentially limitless, further optimization, such as the addition of readily cleavable degradation sites, a further reduction in crosslinking density, or the inclusion of additional cell-binding ligands may result in improved matrix performance. This further tuning of matrix properties may also help to illuminate the microenvironmental cues that directly impact organoid formation, development, differentiation, and functional behavior. Thus, the use of recombinant matrices for in vitro culture of primary adult organoids has the potential to significantly advance the field of intestinal tissue engineering.
Materials & methods
Organoid isolation and culture
Actin-GFP C57BL/6 adult mice were used to generate intestinal organoids following previously described methods with modifications.29 Briefly, entire lengths of small intestine were isolated using aseptic techniques. The intestinal lumen was flushed with ice-cold phosphate buffered saline (PBS) containing 1% penicillin/streptomycin supplementation (Life Technologies, Carlsbad, CA) using a fine-gauge syringe needle to remove any residual contents. The tissue was then rinsed three times in sterile, ice-cold PBS and minced rapidly into tissue explants of <1 mm3 in size using iris scissors. Explants were then rinsed in cold PBS and centrifuged at 200g for 4 min. This rinsing and centrifugation step was repeated, after which excess supernatant was removed. The resulting finely minced tissue explants were immediately mixed into encapsulating protein matrices to constitute 5% of the total hydrogel volume. For samples encapsulated in collagen I, minced explants were mixed into bovine tendon type-I collagen (Cellmatrix I-A, Nitta Gelatin, Osaka, Japan). Collagen I matrices were formed according to the manufacturer’s protocol by mixing with 10% volume of 10× Ham’s F12 medium (Life Technologies, Carlsbad, CA) without sodium bicarbonate and 10% reconstitution buffer (2.2 g sodium bicarbonate (Sigma-Aldrich, St. Louis, MO) in 100 mL of 0.05 N NaOH and 200 mM HEPES, prepared according to the manufacturer’s instructions). Subsequently, tissue-containing collagen I gel mixtures were placed into either submerged or air–liquid interface culture configurations. For air–liquid interface cultures, tissue-containing gels were cast onto traditional 24-well or 6-well transwell inserts (hydrophilic polytetrafluoroethylene with 0.4 μm pores, EMD Millipore, Billerica, MA) depending on the end-point assay. For MMP activity analysis, 100 μL tissue-containing gels were analyzed from 24-well inserts. For imaging studies, four distinct 25 μL tissue-containing gel droplets were placed onto the acellular collagen surface of 30 mm, 6-well inserts. All organoids were cultured in medium containing 1% penicillin/streptomycin, 20% fetal bovine serum (Life Technologies, Carlsbad, CA), and 500 ng mL−1 of Chinese hamster ovary (CHO) cell-derived human recombinant R-spondin1 (R&D Systems, Minneapolis, MN). For air–liquid interface cultures, 300 μL or 1 mL of medium was added to the 24-well or 6-well plates, respectively, outside of the transwell insert.
For histological evaluation, samples were fixed in 10% buffered formalin (Fisher Scientific, Hampton, New Hampshire) overnight after 3 to 5 days of culture. Samples were then rinsed in PBS, removed from transwell inserts, and paraffin embedded prior to hematoxylin and eosin staining.
All animal experiments were performed in compliance with the Animal Welfare Act, the Public Health Service (PHS) Policy on Humane Care and Use of Laboratory Animals, and the Stanford University institutional guidelines. In particular, this work has been approved by the Stanford University Administrative Panel on Laboratory Animal Care (APLAC).
eECM synthesis
Recombinant eECM proteins were encoded within pET15b plasmids under control of the T7 promoter, expressed in BL21 (DE3) Escherichia coli, and purified through iterative thermal cycling as previously reported.39–41 Purified eECM was solubilized in phosphate-buffered saline (PBS) and mixed with isolated intestinal explants. Gels were covalently crosslinked using tetrakis (hydroxymethyl) phosphonium chloride42 (THPC, Sigma-Aldrich, St. Louis, MO) at a 0.5 : 1 or 1 : 1 reactive group stoichiometric ratio to form 3 wt% hydrogels of varying crosslinking density. As with the collagen I cultures, the volume of minced tissue explants encapsulated within the eECM scaffolds constituted 5% of the total hydrogel volume.
Image acquisition
Phase contrast and fluorescence images of intestinal organoids were obtained with a Zeiss Axiovert 200M fluorescence microscope with 2× and 10× objectives or a Leica TCS SPE confocal microscope with 10× objective. GFP+ organoids were manually counted per sample after 3 days of culture. Histological cross-sections were imaged with a Leica DM2000.
Rheological matrix characterization
To characterize the viscoelastic properties of the bovine tendon collagen I and eECM matrices, dynamic oscillatory shear measurements were performed using a 20 mm cone and plate geometry and a Discovery Hybrid Rheometer (TA Instruments, New Castle, DE). A gap height of 28 μm was used to test 40 μL of each sample. Samples were given 15 min at room temperature to crosslink, after which the lower plate temperature was increased to 37 °C for one min prior to testing. The storage modulus, G′, was taken as an average reading of values within the linear plateau region of a frequency sweep. For frequency sweeps ranging from 0.100 to 100 Hz with 1 Pa applied stress, a consistent plateau region from 1 to 10 Hz was found across all samples and conditions tested. A minimum of 3 samples was tested per condition.
Molecular diffusivity measurements
Diffusivities for 150 kDa FITC-dextran were measured by fluorescence recovery after photobleaching (FRAP).54 FITC-dextran was mixed with collagen I or eECM protein solution at 2 mg mL−1, after which gels were formed according to the previously described protocols. Distinct gel regions (110 μm × 110 μm) were bleached at 100% laser intensity for 30 s prior to capturing fluorescence recovery at 1.34 s imaging intervals for a duration of 1.5 min at 18% laser intensity. Open source MATLAB code (http://www.mathworks.com/matlabcentral/fileexchange/29388-frapanalysis) was used for image analysis and diffusivity calculations.
MMP activity measurements and inhibition
Samples were cultured in the presence of 25 μM of a global MMP inhibitor, GM6001 (EMD Millipore, Billerica, MA) from the time of initial seeding. Manual counts of GFP+ organoids were taken after 3 days of culture in the presence or absence of MMP inhibitor. MMP activities were quantified using a Senso-Lyte assay (AnaSpec, Freemont, CA) according to the manufacturer’s instructions. Enzymes were given 3 h to activate at 37 °C and incubated with substrate for 1 h prior to reading with a SpectraMax M2 fluorescence plate reader.
Supplementary Material
Acknowledgements
The authors acknowledge support from NIH Transformative R01 DK085720-01 (CK & SCH), 1DP2-OD006477-01 (SCH), 1R21-EB018407-01 (SCH), 1U19AI116484-01 (CK & SCH) and U01DK085527 (CK). RLD acknowledges support from the Stanford BioX Graduate Fellowship and the National Science Foundation Graduate Research Fellowship, DEG-1147470. The authors also acknowledge Pauline Chu, Department of Comparative Medicine, Histology Lab, Stanford University, for assistance with histology.
Footnotes
Electronic supplementary information (ESI) available. See DOI: 10.1039/c5bm00108k
References
- 1.Parris A and Williams MR, A Human Colonic Crypt Culture System to Study Regulation of Stem Cell-Driven Tissue Renewal and Physiological Function, Methods Mol. Biol, 2015, 1212, 141–161. [DOI] [PubMed] [Google Scholar]
- 2.Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, Watanabe T, Kanai T and Sato T, Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids, Nat. Med, 2015, 21(3), 256–262. [DOI] [PubMed] [Google Scholar]
- 3.Ohta Y and Sato T, Intestinal tumor in a dish, Front. Med, 2014, 1, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Es JH and Clevers H, Generation and analysis of mouse intestinal tumors and organoids harboring APC and K-Ras mutations, Methods Mol. Biol, 2015, 1267, 125–144. [DOI] [PubMed] [Google Scholar]
- 5.Bas T and Augenlicht LH, Real time analysis of metabolic profile in ex vivo mouse intestinal crypt organoid cultures, J. Visualized Exp, 2014, (93), e52026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levin DE, Sala FG, Barthel ER, Speer AL, Hou X, Torashima Y and Grikscheit TC, A “living bioreactor” for the production of tissue-engineered small intestine, Methods Mol. Biol, 2013, 1001, 299–309. [DOI] [PubMed] [Google Scholar]
- 7.Spurrier RG and Grikscheit TC, Tissue engineering the small intestine, Clin. Gastroenterol. Hepatol, 2013, 11(4), 354–358. [DOI] [PubMed] [Google Scholar]
- 8.Barthel ER, Speer AL, Levin DE, Sala FG, Hou X, Torashima Y, Wigfall CM and Grikscheit TC, Tissue engineering of the intestine in a murine model, J. Visualized Exp, 2012, (70), e4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grant CN, Garcia Mojica S, Sala FG, Hill JR, Levin DE, Speer AL, Barthel ER, Shimada H, Zachos NC and Grikscheit TC, Human and Mouse Tissue-Engineered Small Intestine Both Demonstrate Digestive And Absorptive Function, Am. J. Physiol.: Gastrointest. Liver Physiol, 2015, 308(8), G664–G677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lancaster MA and Knoblich JA, Organogenesis in a dish modeling development and disease using organoid technologies, Science, 2014, 345(6194), 1247125. [DOI] [PubMed] [Google Scholar]
- 11.Kuratnik A and Giardina C, Intestinal organoids as tissue surrogates for toxicological and pharmacological studies, Biochem. Pharmacol, 2013, 85(12), 1721–1726. [DOI] [PubMed] [Google Scholar]
- 12.Foulke-Abel J, In J, Kovbasnjuk O, Zachos NC, Ettayebi K, Blutt SE, Hyser JM, Zeng XL, Crawford SE, Broughman JR, Estes MK and Donowitz M, Human enteroids as an ex vivo model of host-pathogen interactions in the gastrointestinal tract, Exp. Biol. Med, 2014, 239(9), 1124–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howell JC and Wells JM, Generating intestinal tissue from stem cells: potential for research and therapy, Regen. Med, 2011, 6(6), 743–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grant CN and Grikscheit TC, Tissue engineering: a promising therapeutic approach to necrotizing enterocolitis, Semin. Pediatr. Surg, 2013, 22(2), 112–116. [DOI] [PubMed] [Google Scholar]
- 15.Yoshida A, Chitcholtan K, Evans JJ, Nock V and Beasley SW, In vitro tissue engineering of smooth muscle sheets with peristalsis using a murine induced pluripotent stem cell line, J. Pediatr. Surg, 2012, 47(2), 329–335. [DOI] [PubMed] [Google Scholar]
- 16.Sala FG, Kunisaki SM, Ochoa ER, Vacanti J and Grikscheit TC, Tissue-engineered small intestine and stomach form from autologous tissue in a preclinical large animal model, J. Surg. Res, 2009, 156(2), 205–212. [DOI] [PubMed] [Google Scholar]
- 17.Neu J and Mihatsch W, Recent developments in necrotizing enterocolitis, JPEN J. Parenter. Enteral. Nutr, 2012, 36(1 Suppl), 30S–35S. [DOI] [PubMed] [Google Scholar]
- 18.Jones BA, Hull MA and Kim HB, Autologous intestinal reconstruction surgery for intestinal failure management, Curr. Opin. Organ. Transplant, 2010, 15(3), 341–345. [DOI] [PubMed] [Google Scholar]
- 19.Sudan D and Rege A, Update on surgical therapies for intestinal failure, Curr. Opin. Organ. Transplant, 2014, 19(3), 267–275. [DOI] [PubMed] [Google Scholar]
- 20.Sulkowski JP and Minneci PC, Management of short bowel syndrome, Pathophysiology, 2014, 21(1), 111–118. [DOI] [PubMed] [Google Scholar]
- 21.Abu-Elmagd K, Reyes J, Bond G, Mazariegos G, Wu T, Murase N, Sindhi R, Martin D, Colangelo J, Zak M, Janson D, Ezzelarab M, Dvorchik I, Parizhskaya M, Deutsch M, Demetris A, Fung J and Starzl TE, Clinical intestinal transplantation: a decade of experience at a single center, Ann. Surg, 2001, 234(3), 404–416; discussion 16–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen MK and Beierle EA, Animal models for intestinal tissue engineering, Biomaterials, 2004, 25(9), 1675–1681. [DOI] [PubMed] [Google Scholar]
- 23.Grant D, Abu-Elmagd K, Reyes J, Tzakis A, Langnas A, Fishbein T, Goulet O and Farmer D, 2003 report of the intestine transplant registry: a new era has dawned, Ann. Surg, 2005, 241(4), 607–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato T, Nishida S, Mittal N, Levi D, Nery J, Madariaga J, Thompson J, Weppler D, Ruiz P and Tzakis A, Intestinal transplantation at the University of Miami, Transplant. Proc, 2002, 34(3), 868. [DOI] [PubMed] [Google Scholar]
- 25.Perez A, Grikscheit TC, Blumberg RS, Ashley SW, Vacanti JP and Whang EE, Tissue-engineered small intestine: ontogeny of the immune system, Transplantation, 2002, 74(5), 619–623. [DOI] [PubMed] [Google Scholar]
- 26.Reyes J, Bueno J, Kocoshis S, Green M, Abu-Elmagd K, Furukawa H, Barksdale EM, Strom S, Fung JJ, Todo S, Irish W and Starzl TE, Current status of intestinal transplantation in children, J. Pediatr. Surg, 1998, 33(2), 243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reyes J, Mazariegos GV, Bond GM, Green M, Dvorchik I, Kosmach-Park B and Abu-Elmagd K, Pediatric intestinal transplantation: historical notes, principles and controversies, Pediatr. Transplant, 2002, 6(3), 193–207. [DOI] [PubMed] [Google Scholar]
- 28.Scolapio JS, Fleming CR, Kelly DG, Wick DM and Zinsmeister AR, Survival of home parenteral nutrition-treated patients: 20 years of experience at the Mayo Clinic, Mayo Clin. Proc, 1999, 74(3), 217–222. [DOI] [PubMed] [Google Scholar]
- 29.Ootani A, Li X, Sangiorgi E, Ho QT, Ueno H, Toda S, Sugihara H, Fujimoto K, Weissman IL, Capecchi MR and Kuo CJ, Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche, Nat. Med, 2009, 15(6), 701–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ and Clevers H, Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche, Nature, 2009, 459(7244), 262–265. [DOI] [PubMed] [Google Scholar]
- 31.Koo BK, Stange DE, Sato T, Karthaus W, Farin HF, Huch M, van Es JH and Clevers H, Controlled gene expression in primary Lgr5 organoid cultures, Nat. Methods, 2012, 9(1), 81–83. [DOI] [PubMed] [Google Scholar]
- 32.Schwank G, Andersson-Rolf A, Koo BK, Sasaki N and Clevers H, Generation of BAC transgenic epithelial organoids, PLoS One, 2013, 8(10), e76871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan YY, Davidson LA, Callaway ES, Goldsby JS and Chapkin RS, Differential effects of 2- and 3-series E-prostaglandins on in vitro expansion of Lgr5+ colonic stem cells, Carcinogenesis, 2014, 35(3), 606–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petersen N, Reimann F, Bartfeld S, Farin HF, Ringnalda FC, Vries RG, van den Brink S, Clevers H, Gribble FM and de Koning EJ, Generation of L cells in mouse and human small intestine organoids, Diabetes, 2014, 63(2), 410–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yin X, Farin HF, van Es JH, Clevers H, Langer R and Karp JM, Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny, Nat. Methods, 2014, 11(1), 106–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD and Clevers H, Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium, Gastroenterology, 2011, 141(5), 1762–1772. [DOI] [PubMed] [Google Scholar]
- 37.DiMarco RL, Su J, Yan KS, Dewi R, Kuo CJ and Heilshorn SC, Engineering of three-dimensional microenvironments to promote contractile behavior in primary intestinal organoids, Integr. Biol, 2014, 6(2), 127–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scadden DT, The stem-cell niche as an entity of action, Nature, 2006, 441(7097), 1075–1079. [DOI] [PubMed] [Google Scholar]
- 39.Chung C, Pruitt BL and Heilshorn SC, Spontaneous cardiomyocyte differentiation of mouse embryoid bodies regulated by hydrogel crosslink density, Biomater. Sci, 2013, 1(10), 1082–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lampe KJ, Antaris AL and Heilshorn SC, Design of three-dimensional engineered protein hydrogels for tailored control of neurite growth, Acta Biomater, 2013, 9(3), 5590–5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Straley KS and Heilshorn SC, Independent tuning of multiple biomaterial properties using protein engineering, Soft Matter, 2009, 5(1), 114–124. [Google Scholar]
- 42.Chung C, Lampe KJ and Heilshorn SC, Tetrakis (hydroxylmethyl) phosphonium chloride as a covalent crosslinking agent for cell encapsulation within protein-based hydrogels, Biomacromolecules, 2012, 13(12), 3912–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Straley KS and Heilshorn SC, Design and Adsorption of Modular Engineered Proteins to Prepare Customized, Neuron-Compatible Coatings, Frontiers in Neuroengineering, 2009, 2, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DiMarco RL and Heilshorn SC, Multifunctional materials through modular protein engineering, Adv. Mater, 2012, 24(29), 3923–3940. [DOI] [PubMed] [Google Scholar]
- 45.Sengupta D and Heilshorn SC, Protein-engineered biomaterials: highly tunable tissue engineering scaffolds, Tissue Eng. Part B, Rev, 2010, 16(3), 285–293. [DOI] [PubMed] [Google Scholar]
- 46.Chilkoti A, Christensen T and MacKay JA, Stimulus responsive elastin biopolymers: Applications in medicine and biotechnology, Curr. Opin. Chem. Biol, 2006, 10(6), 652–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Granath KA, Solution properties of branched dextrans, J. Colloid Sci, 1958, 13(4), 308–328. [Google Scholar]
- 48.Yan KS, Chia LA, Li X, Ootani A, Su J, Lee JY, Su N, Luo Y, Heilshorn SC, Amieva MR, Sangiorgi E, Capecchi MR and Kuo CJ, The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations, Proc. Natl. Acad. Sci. U. S. A, 2012, 109(2), 466–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chung C, Anderson E, Pera RR, Pruitt BL and Heilshorn SC, Hydrogel crosslinking density regulates temporal contractility of human embryonic stem cell-derived cardiomyocytes in 3D cultures, Soft Matter, 2012, 8(39), 10141–10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gustafson JA, Price RA, Frandsen J, Henak CR, Cappello J and Ghandehari H, Synthesis and characterization of a matrix-metalloproteinase responsive silk-elastin-like protein polymer, Biomacromolecules, 2013, 14(3), 618–625. [DOI] [PubMed] [Google Scholar]
- 51.Liu Y, Liu B, Riesberg JJ and Shen W, In situ forming physical hydrogels for three-dimensional tissue morphogenesis, Macromol. Biosci, 2011, 11(10), 1325–1330. [DOI] [PubMed] [Google Scholar]
- 52.McKinnon DD, Domaille DW, Cha JN and Anseth KS, Biophysically defined and cytocompatible covalently adaptable networks as viscoelastic 3D cell culture systems, Adv. Mater, 2014, 26(6), 865–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang H and Heilshorn SC, Adaptable Hydrogel Networks with Reversible Linkages for Tissue Engineering, Adv. Mater, 2015, 27, 3717–3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jonsson P, Jonsson MP, Tegenfeldt JO and Hook F, A method improving the accuracy of fluorescence recovery after photobleaching analysis, Biophys. J, 2008, 95(11), 5334–5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


