Abstract
Food intake profoundly affects systemic physiology. A large body of evidence has indicated a link between food intake and circadian rhythms, and ~24‐h cycles are deemed essential for adapting internal homeostasis to the external environment. Circadian rhythms are controlled by the biological clock, a molecular system remarkably conserved throughout evolution. The circadian clock controls the cyclic expression of numerous genes, a regulatory program common to all mammalian cells, which may lead to various metabolic and physiological disturbances if hindered. Although the circadian clock regulates multiple metabolic pathways, metabolic states also provide feedback on the molecular clock. Therefore, a remarkable feature is reprogramming by nutritional challenges, such as a high‐fat diet, fasting, ketogenic diet, and caloric restriction. In addition, various factors such as energy balance, histone modifications, and nuclear receptor activity are involved in the remodeling of the clock. Herein, we review the interaction of dietary components with the circadian system and illustrate the relationships linking the molecular clock to metabolism and critical roles in the remodeling process.
Keywords: circadian clock, energy metabolism, epigenetics, nutrition
Subject Categories: Chromatin, Transcription & Genomics; Metabolism
The intake of food and its composition affect the circadian clock. This review provides a comprehensive overview of the interconnections between the nutritional status, metabolic homeostasis and circadian rhythms.
Glossary
- ACC
acetyl‐CoA carboxylase
- AceCS1
acetyl‐CoA synthetase 1
- ACLY
ATP‐citrate lyase
- AMP
adenosine monophosphate
- AMPK
5' AMP‐activated protein kinase
- Angptl8
angiopoietin‐like 8
- ATP
adenosine triphosphate
- BMAL1
brain and muscle ARNT‐like 1
- cAMP
cyclic adenosine monophosphate
- CCGs
clock‐controlled genes
- CLOCK
circadian locomotor output cycles protein caput
- CREB
cAMP response element‐binding protein
- CRY
cryptochrome
- DBP
D site of albumin promoter binding protein
- DEC1
differentiated embryo chondrocyte 1
- E4bp4
E4 promotor‐binding protein 4
- E‐box
enhancer box
- EGCG
epigallocatechin gallate
- GCs
glucocorticoids
- GPC
sn‐glycero‐3‐phosphocholine
- GR
glucocorticoid receptor
- GSK3β
glycogen synthase kinase 3β
- HAT
histone acetyl transferase
- HDAC
histone deacetylases
- HFD
high‐fat diet
- HMGCS2
3‐hydroxy‐3‐methylglutaryl‐CoA synthase 2
- JMJD2B
jumonji C domain‐containing protein 2B
- KD
ketogenic diet
- LKB1
serine/threonine kinase liver kinase B1
- LXR
liver X receptor
- MAPK
mitogen‐activated protein kinase
- MLL1
mixed lineage leukemia 1
- mTOR
mammalian target of rapamycin
- NAD⁺
oxidized nicotinamide adenine dinucleotide
- NADH
reduced nicotinamide adenine dinucleotide
- NAM
nicotinamide mononucleotide
- NAMPT
nicotinamide phosphoribosyltransferase
- Npy
neuropeptide Y
- O‐GlcNAc
O‐linked N‐acetyl‐glucosamine
- O‐GlcNAcylation
O‐linked N‐acetylglucosamine modification
- OGT
O‐linked GlcNAc transferase
- PC
phosphatidylcholine
- Per
period
- PGC‐1α
peroxisome proliferator‐activated receptor‐gamma coactivator‐1α
- Pomc
pro‐opiomelanocortin
- PPAR
peroxisome proliferator‐activated receptor
- RBP4
retinol‐binding protein 4
- ROR
RAR‐related orphan receptor
- SAM
s‐adenosyl methionine
- SCN
suprachiasmatic nucleus
- SIRT
sirtuin
- SREBP‐1c
sterol‐regulatory element‐binding protein‐1c
- STAT5
signal transducer and activator of transcription 5
- TRF
time‐restricted feeding
- UDP‐GlcNAc
uridine diphosphate N‐acetylglucosamine
- β‐OHB
β‐hydroxybutyrate
Introduction
Every morning, after a night of sleep, we wake up, eat our regularly timed meals, go through our normal routines, sleep, and then repeat the same cycle. Various physiological functions, including sleep and being awake, body temperature, hormone secretion, locomotor activity, and appetite, are regulated by an autonomous, ~24‐h mechanism, termed the circadian clock (Sahar & Sassone‐Corsi, 2012). This endogenous timekeeper allows organisms to anticipate daily environmental fluctuations and time internal processes (Bass & Takahashi, 2010; Eckel‐Mahan & Sassone‐Corsi, 2013). Anatomically, the mammalian central clock or pacemaker is located in the suprachiasmatic nucleus (SCN) of the hypothalamus, with functions regulated by photic inputs from the retina in the form of light. Notably, light‐induced resetting of the SCN clock depends on wavelength. Blue light (380–500 nm) potentially exerts more robust effects on mammalian circadian rhythms than green and yellow wavelengths (Lockley et al, 2003). A transformative discovery around the turn of the century revealed that in addition to the brain, the circadian clock functions in peripheral organs, including the liver and muscle (Schibler & Sassone‐Corsi, 2002). These local or peripheral clocks are semi‐autonomous elements of a larger system and are synchronized by the SCN clock, functioning as an “orchestra director,” via neural, hormonal (e.g., glucocorticoids [GCs], insulin, and melatonin), and behavioral inputs (Saini et al, 2011).
The molecular machinery underlying the circadian clock consists of a transcriptional/translational feedback loop: the core transcription factors, brain and muscle Arnt‐like 1 (BMAL1) and circadian locomotor output cycles kaput (CLOCK), heterodimerize and drive the expression of core clock genes or output genes by binding to enhancer boxes (E‐boxes) on target promoters (Fig 1) (Crane & Young, 2014). As E‐boxes are among the most common promoter elements in the genome, the clock can transcriptionally control a large array of genes. In addition, CLOCK:BMAL1 directly activates the transcription of Period (Per1, Per2, and Per3) and Cryptochrome (Cry1 and Cry2) genes, known to encode transcriptional repressors that dimerize and generate a tightly regulated negative portion of the feedback loop (Gekakis et al, 1998; Kume et al, 1999; Shearman et al, 2000; Lee et al, 2001; Padmanabhan et al, 2012; Kim et al, 2014). An additional level of circadian regulation involves the nuclear receptors, i.e., RAR‐related orphan receptors (RORs) and REV‐ERBα (Nr1d1), which activate and repress Bmal1 transcription, respectively (Reppert & Weaver, 2002; Everett & Lazar, 2014; Partch et al, 2014). Furthermore, virtually all clock proteins are reportedly regulated by post‐translational modifications, including phosphorylation, acetylation, ubiquitination, and O‐linked N‐acetylglucosamine modification (O‐GlcNAcylation) (Asher & Schibler, 2011; Kaasik et al, 2013; Li et al, 2013).
Figure 1. The molecular organization of the circadian clock.
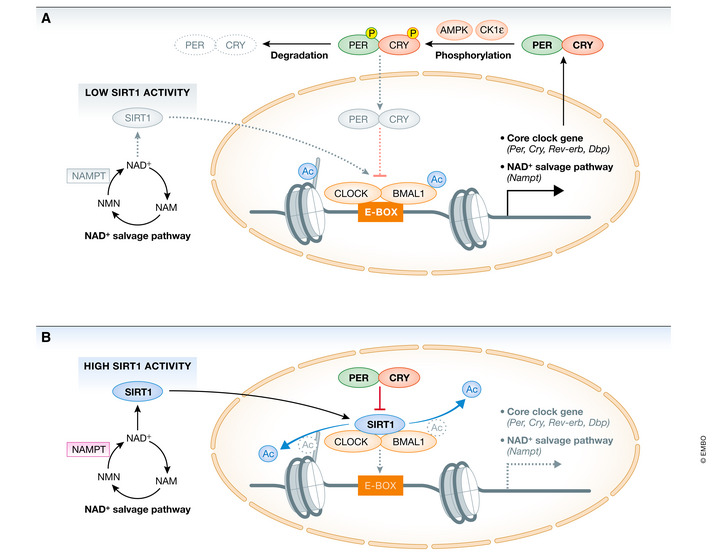
(A) Daytime: The core transcription factor BMAL1 heterodimerizes with CLOCK to form a CLOCK:BMAL1 complex. CLOCK acetylates BMAL1 and histone tails, leading to chromatin opening that promotes binding of CLOCK:BMAL1 to E‐box elements in promoter regions of the core‐clock genes and clock‐controlled genes (Per, Cry, Rev‐erv, Nampt). During the daytime AMPK and CK1ε contribute to phosphorylation and degradation of the negative regulators CRY and PER, respectively, thus relieving the negative feedback on CLOCK:BMAL1. The circadian activity of SIRT1, which regulates cyclic acetylation levels of BMAL1 and histones in nucleosomes associated with clock‐controlled genes, is controlled by the rhythmic cellular levels of its cofactor NAD⁺. Nampt gene expression and cellular NAD⁺ levels oscillate and peak at night, leading to lower SIRT1 activity during the daytime. (B) Nighttime: PER and CRY protein accumulate in the cytosol during the night, heterodimerize and translocate to the nucleus to repress CLOCK:BMAL1 transcriptional activity. SIRT1 deacetylase activity during the nighttime is high, deacetylating BMAL1 and histone tails. SIRT1‐mediated histone deacetylation induces packing of DNA (heterochromatin) and gene silencing. AMPK, 5′ AMP‐activated protein kinase; BMAL1, brain and muscle ARNT‐like 1; CK1ε, casein kinase 1 epsilon; CLOCK, circadian locomotor output cycles protein caput; CRY, cryptochrome; NAD⁺, oxidized nicotinamide adenine dinucleotide; Nampt, nicotinamide phosphoribosyltransferase; PER, period; SIRT1, sirtuin 1.
Numerous studies have highlighted how the clock system closely interacts with energy metabolism in peripheral organs (Bass, 2012; Eckel‐Mahan et al, 2013). Although the SCN clock contributes to circadian variations in glucose homeostasis via glucose uptake and insulin release, peripheral clocks can also constitute another layer of regulation of these processes. For example, melatonin secreted from the pineal gland in an SCN clock‐dependent manner directly regulates pancreatic insulin secretion (Peschke et al, 1997; Picinato et al, 2002). Pancreatic insulin secretion is regulated by multisynaptic projections from the SCN (Ueyama et al, 1999; Buijs et al, 2001). In addition to insulin‐mediated signaling, it has been suggested that the SCN may regulate glucose uptake in peripheral organs through the nervous system (La Fleur, 2003). Moreover, the clock machinery controls the expression of numerous metabolic output genes in peripheral tissues (Dibner et al, 2010; Maury et al, 2010). Accordingly, genetic disruption of mouse clock components can induce metabolic diseases, including obesity, by attenuating rhythmic changes in hormone concentration and metabolic gene expression (Dibner et al, 2010; Maury et al, 2010). In humans, disruption of circadian rhythms owing to jet lag, time shift work, and irregular meal timing has been linked to metabolic diseases (Kettner et al, 2015; Morris et al, 2016). These observations suggest that the circadian clock controls several signaling pathways encompassing major components of metabolic homeostasis.
Accumulated evidence has suggested that the quality and timing of meals markedly alter circadian metabolism (Eckel‐Mahan et al, 2013; Tognini et al, 2017). For example, a comparison of oscillating transcripts in different tissues revealed that approximately 15% of hepatic transcripts and 4% of muscle transcripts oscillate under ad libitum conditions of a normal chow diet (Hughes et al, 2010; Zhang et al, 2014b). A high‐fat diet (HFD), wherein energy from fat exceeds 40%, can disrupt normally oscillating genes and induce de novo oscillations (Eckel‐Mahan et al, 2013). Transcriptome analyses have revealed that a large fraction of the genome can be potentially controlled by the clock, and diet‐induced remodeling exerts tissue‐specific effects (Masri & Sassone‐Corsi, 2010). Moreover, varying the macronutrient composition and specific nutrients (nobiletin, resveratrol, and caffeine) have been reported to influence peripheral clock gene expression (Sherman et al, 2011; Sun et al, 2015; He et al, 2016).
In this review, we describe the relationship between the circadian clock and metabolic homeostasis from the perspective of energy balance and epigenome regulation. We focus on the effect of dietary composition on the molecular clock and discuss how nutritional approaches may contribute to the prevention of metabolic diseases.
Reciprocal regulation between the circadian clock and metabolites
According to several genome‐wide expression studies, genes involved in glucose metabolism, lipid metabolism, heme biosynthesis, and mitochondrial adenosine triphosphate (ATP) synthesis all exhibit a circadian pattern of expression (Ceriani et al, 2002; Panda et al, 2002; Oishi et al, 2003; Gachon et al, 2004). Importantly, several transcriptional factors central to regulating metabolic pathways have diurnal gene expression patterns and activity. Peroxisome proliferator‐activated receptors (PPAR) compose a group of nuclear receptor proteins that play essential roles in regulating glucose and lipid metabolism (Oishi et al, 2005; Canaple et al, 2006). Transcription of Pparα, a PPAR isotype, can be activated by CLOCK:BMAL1 via an intronic E‐box‐rich region. In contrast, it has been reported that Pparγ expression may be regulated by the products of two clock‐controlled genes, the D site albumin promoter binding protein (Dbp) and E4 promoter‐binding protein 4 (E4bp4). DBP and E4BP4 jointly induce the circadian expression of one Pparγ subtype by binding to D‐box sequences located in the first exon of the gene (Takahashi et al, 2010), thus indicating that Pparγ may be regulated by the circadian clock component. RORα plays a crucial role in the homeostasis of lipid metabolism in the liver by negatively regulating PPARγ signaling via histone deacetylase (HDAC) 3 recruitment to Pparγ target promoters (Kim et al, 2017). However, it remains unclear whether RORα regulates other PPAR isotypes.
REV‐ERBα directly regulates the transcription of genes controlling carbohydrate and lipid metabolism by binding to ROR‐responsive elements via its DNA‐binding domain (Zhang et al, 2015). Furthermore, REV‐ERBα exhibits a DNA‐binding domain‐independent function that modulates liver metabolism (Zhang et al, 2015). Based on experiments in cultured hepatocytes, downregulation of Rev‐erbα by siRNA increases the expression of glucose‐6‐phosphatase (G6Pase) and phosphoenolpyruvate carboxykinase (Pepck), which are rate‐limiting enzymes of gluconeogenesis (Yin et al, 2007). In addition, the administration of REV‐ERB agonists alters the rhythm and amplitude of metabolic genes in the liver, skeletal muscles, and adipose tissue (Solt et al, 2012).
Apart from direct transcriptional control, diverse studies have revealed that the clock control of metabolism is pervasive and multilayered. High‐throughput metabolomic investigations of human plasma and saliva samples have revealed that approximately 15% of all identified metabolites oscillate in a circadian manner (Minami et al, 2009; Dallmann et al, 2012). Notably, a high proportion of rhythmic metabolites in blood plasma are lipid metabolites, such as fatty acids and phospholipids. As outputs of the circadian system, some metabolites also provide feedback and operate as inputs to the circadian clock. Oxidized nicotinamide adenine dinucleotide (NAD⁺) is one such example. The circadian clock controls NAD⁺ levels by transcriptionally regulating the nicotinamide phosphoribosyltransferase (Nampt) gene, which encodes an enzyme that catalyzes the rate‐limiting step in the NAD⁺ salvage pathway (Fig 1B). The activator complex CLOCK:BMAL1 directly binds to the Nampt promoter to control its circadian expression, leading to the oscillation of NAD⁺ levels from recycled nicotinamide mononucleotide (NAM). Importantly, NAD⁺ operates as a coenzyme for Class III histone deacetylases and sirtuins (SIRTs), and its cyclic accumulation results in rhythmic deacetylation of SIRT targets, ultimately contributing to the circadian gene expression (Nakahata et al, 2008; Feng et al, 2011). We provide the detailed interaction mechanism between SIRT1 and the circadian clock system in the section on “Energy balance controls the clock and metabolic homeostasis”.
Several lipid metabolites, including phospholipids and free fatty acids, also contribute to circadian rhythmicity. PPARs are activated by dietary fatty acids and their metabolic derivatives, thus serving as lipid sensors that activate lipid metabolism, including lipogenesis and lipolysis. Liu et al (2013) reported that plasma levels of phosphatidylcholine (PC) 36:1 fluctuate diurnally and that PC production is regulated by hepatic PPARδ (Liu et al, 2013). Tandem mass spectrometry scanning identified PC 36:1 as PC (18:0/18:1), which acts as a ligand for PPARα in muscle, producing a rhythm of fatty acid uptake (Liu et al, 2013); this finding suggests that rhythmic oscillations of lipid metabolites derived from hepatic lipogenesis can integrate metabolic functions between the liver and muscle. PPARs are associated with sterol element‐binding protein‐1c (SREBP‐1c), a master regulator that controls hepatic de novo lipogenesis. Recent studies have shown that a sn‐glycero‐3‐phosphocholine (GPC) (16:0/18:1), presumed to be a ligand of PPARα, is produced in an SREBP‐1c‐dependent manner, establishing a circadian rhythm for lipid oxidation in the liver (Guan et al, 2018).
Collectively, these findings illustrate that NAD⁺ and lipid pathways are both outputs and inputs to the clock mechanism. The concept that specific metabolites influence clock function implies that consuming diets containing these nutrients, or their precursors, may enable genomic and metabolic reprogramming.
Energy balance controls the clock and metabolic homeostasis
The circadian clock system is essential for adapting to and anticipating the surrounding environment. Notably, it contains certain factors that act as sensors to recognize the nutritional state. The intracellular ratios of adenosine monophosphate and triphosphate (AMP/ATP) and NAD⁺/reduced nicotinamide adenine dinucleotide (NADH) reflect the nutritional and energetic states. Several studies have revealed that 5′ AMP‐activated protein kinase (AMPK) and SIRT1 act as nutritional sensors to relay metabolic information to the clock. For example, AMPK is activated under low‐energy conditions, as reflected by a high AMP/ATP ratio. Activated AMPK enhances catabolic processes to restore intracellular ATP levels. AMPK phosphorylation at Thr172 is required for AMPK activation, and serine/threonine kinase liver kinase B1 (LKB1) directly mediates this event (Hawley et al, 2003; Liang et al, 2007). AMPK activity was reportedly rhythmic in the mouse liver, hypothalamus, and fibroblasts (Um et al, 2011). Notably, AMPK activation can influence the circadian clock system by reducing the stability of the core clock repressors CRY1 and PER2, and thus, may contribute to the metabolic entrainment of clocks in peripheral tissues (Fig 1A). AMPK directly phosphorylates the serine (S)71 and S280 residues of CRY1 and enhances its degradation; mutation of either S71 or S280 to a non‐phosphorylatable amino acid (alanine) blocks this effect (Lamia et al, 2009). Phosphorylated CRY1 exhibits lower affinity for PER2 and instead increases binding to F‐box and Leu‐rich repeat protein 3 (FBXL3), a ubiquitin ligase that promotes CRY1 ubiquitination and degradation (Gatfield & Schibler, 2007). Furthermore, AMPK phosphorylates casein kinase 1ε (CK1ε), resulting in periodic phosphorylation and degradation of PER2 (Um et al, 2007). AMPK deletion leads to tissue‐specific alterations in circadian gene expression (Um et al, 2011). Basic helix‐loop‐helix transcription factor differentiated embryo chondrocyte 1 (Dec1) is a clock gene that acts on the negative limb by suppressing CLOCK and BMAL1 transcriptional activity. DEC1 protein expression can be inversely correlated with AMPK activity and negatively regulates AMPK activity via LKB1 (Sato et al, 2015). According to one report, the kinetics of metformin‐induced AMPK activation in the liver appeared remarkably dependent on circadian time (Henriksson et al, 2017), indicating that AMPK is clock‐controlled. AMPK regulates clock components, as well as directly phosphorylates cooperating transcription factors that play key roles in lipid metabolism. Activated AMPK phosphorylates SREBP‐1c, leading to SREBP‐1c protein degradation and suppression of de novo lipogenesis in the liver (Lee et al, 2015). Acetyl‐CoA carboxylase (ACC) is another AMPK target; ACC phosphorylation decreases malonyl‐CoA levels, thereby activating carnitine palmitoyltransferase 1 (CPT 1) and resulting in fatty acid oxidation (Fullerton et al, 2013).
SIRT1 is a sensor of energy balance that bridges circadian regulation and nutritional status. SIRT1 activity is modulated by the cellular redox state, which can be inferred from the NAD⁺/NADH ratio (Asher et al, 2008). SIRT1 regulates several metabolic processes, including gluconeogenesis, insulin sensitivity, and de novo lipogenesis via the deacetylation of various proteins and histones. Several SIRT1‐regulated transcription factors are known to be involved in nutrient flux, such as peroxisome proliferator‐activated receptor‐gamma coactivator‐1α (PGC‐1α), AMPK, and forkhead box O1 (FOXO1) (Schwer & Verdin, 2008). The deacetylase activity of SIRT1 is synchronized with the diurnal variation of NAD⁺, inducing rhythmic activity in target proteins. Recent studies have suggested that SIRT1 regulates the acetylation level of acetyl‐CoA synthetase 1 (AceCS1), which is localized in the cytoplasm and is required for cytosolic acetyl‐CoA metabolism, and may affect intracellular acetyl‐CoA levels and the total amount of acetylated protein (Sato et al, 2017). In addition, SIRT1 deacetylates and regulates several clock proteins and acts as a key modulator of the circadian clock machinery (Asher et al, 2008; Nakahata et al, 2008). SIRT1 interacts with the CLOCK:BMAL1 complex in a circadian fashion, inducing diurnal fluctuations in transcriptional activity (Fig 1B). Additionally, SIRT1‐mediated deacetylation of PER2 is critical for the stability of PER2 (Asher et al, 2008). A mutant SIRT1 was shown to enhance BMAL1 acetylation, concomitant with higher amplitude of circadian gene expression (Foteinou et al, 2018), and loss of SIRT1 reportedly led to a lower amplitude of the circadian rhythms in SIRT1 knockout mouse embryonic fibroblast cells (Asher et al, 2008). Recent computational and experimental approaches have supported PER2, not BMAL1, as the direct target of SIRT1 (Foteinou et al, 2018). Moreover, the authors suggested that SIRT1‐induced the deacetylation of PGC‐1α enhances ROR‐driven BMAL1 transcription via PGC‐1α coactivation.
The SIRT1 and AMPK signaling pathways are tightly coordinated (Ruderman et al, 2010). For example, AMPK increases the cellular NAD+/NADH ratio, which subsequently activates the deacetylase activity of SIRT1 (Canto et al, 2009). Conversely, SIRT1 controls AMPK activation via deacetylation of the AMPK‐activating kinase LKB1 (Hou et al, 2008; Lan et al, 2008). Therefore, AMPK and SIRT1 not only regulate each other, but their metabolic actions often converge, indicating that their close interrelationship might also play a role in circadian clock regulation (Ruderman et al, 2010).
AMPK and SIRT1 are sensors that detect energy deficiency, whereas the mammalian mechanistic target of rapamycin (mTOR) is a nutrient sensor that detects over‐nutrition. Notably, the mTOR signaling pathway is activated by growth factors (e.g., insulin and insulin‐like growth factor‐1) and amino acids (leucine and arginine) (Inoki et al, 2002; Nair & Short, 2005). In response to these extracellular signals, mTOR regulates various physiological processes, including cell growth, protein synthesis, and autophagy (Morita et al, 2013). mTOR protein expression and activity exhibit robust circadian rhythms (Cao et al, 2011). The expression of F‐box and WD repeat domain‐containing 7 (Fbxw7), a ubiquitin ligase that degrades mTOR protein, is transcriptionally regulated by DBP (Cao et al, 2011). The circadian clock system regulates mTOR, and conversely, mTOR directly affects the period and amplitude of the circadian clock at the cellular level. Ramanathan et al (2018) revealed that mTOR activation accelerates clock oscillations, whereas inhibition lengthens the circadian period in both the SCN and peripheral tissues (Ramanathan et al, 2018). Regulating BMAL1 can partly clarify the effect of mTOR on the CLOCK system. Reportedly, insulin‐mediated activation of mTOR‐ribosomal protein S6 kinase signaling can inhibit the nuclear localization of BMAL1 by phosphorylating S42 (Dang et al, 2016). Moreover, Cao et al reported that disruption of mTOR signaling attenuates the light‐induced phase delay of circadian locomotor activity (Cao et al, 2010). These findings suggest that mTOR may also function as a hub for peripheral and central clock regulation by external stimuli, such as insulin and light.
For intracellular proteins, O‐GlcNAcylation, a post‐translational modification incorporating O‐linked N‐acetyl‐glucosamine (O‐GlcNAc) into specific serine/threonine residues of proteins, is a key mediator of the metabolic response to nutrient availability (Hart, 2019). This widespread and dynamic glycosylation is mediated by O‐linked GlcNAc transferase (OGT) and O‐GlcNAcase (OGA), catalyzing sugar addition and removal, respectively (Yang & Qian, 2017; Hart, 2019). O‐GlcNAcylation depends on the concentration of uridine diphosphate N‐acetylglucosamine (UDP‐GlcNAc), the donor for O‐GlcNAcylation, produced via the hexosamine biosynthesis pathway and glucose flux. As the production of UDP‐GlcNAc requires glucose as a precursor, O‐GlcNAcylation is recognized as a cellular glucose sensor. OGT can modify numerous proteins, including key transcription factors of glucose and lipid metabolism, such as liver X receptor (LXR) and carbohydrate‐responsive element‐binding protein (ChREBP) (Anthonisen et al, 2010; Bindesboll et al, 2015). In addition, several core clock genes are known targets of O‐GlcNAcylation. O‐GlcNAcylation competitively inhibits the phosphorylation of PER and CRY, increasing their stabilization (Kim et al, 2012; Kaasik et al, 2013). O‐GlcNAcylation also stabilizes CLOCK:BMAL1, promoting target gene expression (Li et al, 2013). A recent study has revealed that REV‐ERBα, but not REV‐ERBβ, interacts with cytoplasmic and nuclear OGT. Moreover, the diurnal fluctuation of REV‐ERBα protein has been linked to rhythms of OGT protein stability and protein O‐GlcNAcylation (Berthier et al, 2018).
Nutritional control of epigenome modifications
Molecular signatures of epigenetic regulation and chromatin architecture are fundamental to genetically determined biological processes. Chromatin histone modifications, such as acetylation, methylation, and phosphorylation, regulate the chromatin structure and control gene expression (Fig 2). For example, histone H3 Lys27 (H3K27ac) acetylation indicates active gene transcription, whereas trimethylation of the same residue (H3K27me3) leads to gene silencing. Over the past decades, accumulating evidence has undoubtedly established that cellular metabolism exerts a profound and dynamic influence on histone modifications, as histone‐modifying enzymes utilize key metabolic intermediates.
Figure 2. Nutritional conditions affect the chromatin landscape.
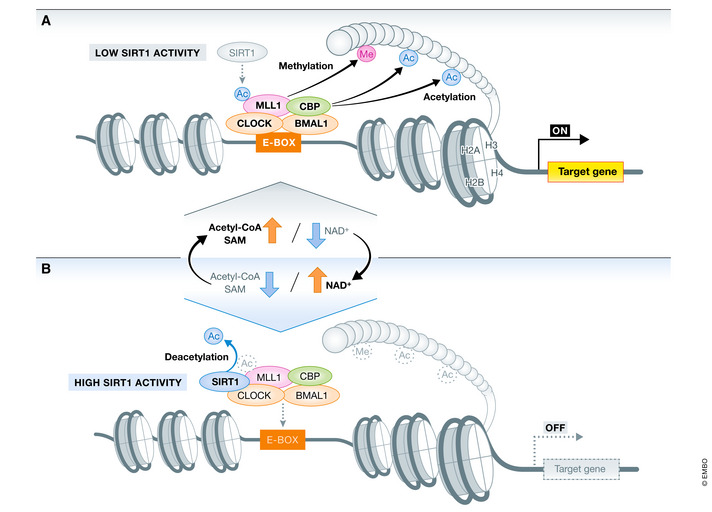
Most chromatin‐modifying enzymes use intermediary metabolites as cofactors or substrates, and their activity is regulated by the availability of these metabolites. Acetyl‐CoA, SAM, and NAD⁺ are substrates for histone acetylation, methylation, and deacetylation, respectively. Acetyl‐CoA is necessary for protein acetylation. SAM is an important methyl‐group donor metabolite in DNA and histone methylation. SIRT1 deacetylates histone lysine residues using NAD+ as a co‐substrate. (A) MLL1 physically interacts with CLOCK and contributes to CLOCK:BMAL1 recruitment to the E‐box regions. Together, MLL1 and CLOCK induce histone modifications (H3K4me4, H3K9Ac, and H3K14Ac) and enhance target gene expression. (B) Oscillating levels of NAD⁺ control SIRT1 deacetylase activity (histone and non‐histone proteins). Under high cellular NAD⁺ conditions, SIRT1 deacetylates MLL1, an event that reduces MLL1 enzymatic activity and leads to decreased H3K4me3 and H3K9/14ac levels. NAD⁺ thus controls MLL1 acetylation level and CLOCK:BMAL1 dependent transcription. BMAL1, brain and muscle ARNT‐like 1; CLOCK, circadian locomotor output cycles protein caput; MLL1, mixed‐lineage leukemia 1; NAD⁺, oxidized nicotinamide adenine dinucleotide; SAM, s‐adenosyl methionine; SIRT1, sirtuin 1.
As a precursor of anabolic reactions and acetyl group donor for acetylation of histone and non‐histone proteins, acetyl‐CoA is critical in cellular processes. The nuclear/cytosolic pool of acetyl‐CoA is maintained by two enzymes, AceCS1 and ATP‐citrate lyase (ACLY) (Albaugh et al, 2011). Acetate and citrate are metabolized to acetyl‐CoA by AceCS1 and ACLY, respectively. A decrease in acetyl‐CoA levels due to loss of both enzymes reportedly reduces global histone acetylation (Wellen et al, 2009; Ariyannur et al, 2010). Histone modification is reversible, and histone acetylation is removed by histone deacetylases (HDACs), which are components of transcriptional repressor complexes. SIRT1 catalyzes a deacetylation reaction that requires NAD⁺. NAD⁺ can be synthesized de novo from diverse dietary sources (e.g., tryptophan) or regenerated from NAM via the salvage pathway (Canto et al, 2015; Verdin, 2015). Transcript and protein levels of SIRT1 in peripheral tissues remain constant during day and night, but SIRT1 enzymatic activity is regulated by the availability of NAD⁺ (Fig 2) (Nakahata et al, 2008, 2009).
S‐adenosyl methionine (SAM) is a universal substrate for DNA methylation and methylation of arginine and lysine residues of histone and non‐histone proteins. SAM is synthesized from methionine and ATP by methionine adenosyltransferase (Grillo & Colombatto, 2008). After methyl group transfer by SAM‐dependent methylase, S‐adenosyl homocysteine, a byproduct of the reaction, acts as a potent methyltransferase inhibitor (Selhub & Miller, 1992). The importance of dietary methyl donors in epigenetic regulation has been examined in rodent models and humans, and the effects of maternal dietary intake have been the focus of recent studies (Donohoe & Bultman, 2012; Huang et al, 2014; Shorter et al, 2014).
The molecular clock hinges on epigenetic mechanisms. CLOCK itself has an intrinsic histone acetyltransferase (HAT) activity necessary for circadian function (Doi et al, 2006). The HAT activity of CLOCK is preferentially linked to targets H3K9 and H3K14. In addition, CLOCK acetylates its binding partner, BMAL1, a crucial event for circadian rhythmicity. Other proteins with HAT activity, such as CBP and P300, can associate with CLOCK:BMAL1 and participate in rhythmic histone acetylation (Curtis et al, 2004). SIRT1 is known to competitively regulate acetylation marks on CLOCK targets, such as H3K9 and H3K14, as well as K537 of BMAL1 (Nakahata et al, 2008).
According to Katada and Aguilar‐Arnala, a key event in circadian transcriptional activation is the interaction of CLOCK:BMAL1 with proteins associated with the Set1/COMPASS complex component mixed‐lineage leukemia 1 (MLL1), whose enzymatic activity results in the transcription‐activating histone mark H3K4me3 (Katada & Sassone‐Corsi, 2010; Aguilar‐Arnal et al, 2015). MLL1 and CLOCK physically interact at specific circadian times, paralleling the cyclic peaks of transcription. The H3K4me3 modification catalyzed by MLL1 favors the recruitment of CLOCK:BMAL1 to chromatin and subsequent H3K9/K14 acetylation (Fig 2).
The feeding‐fasting cycle remodels the clock and metabolic homeostasis
The circadian clock and metabolic homeostasis are regulated by the cellular environment, including energy and hormone balance. The energy balance and diurnal rhythmicity of blood hormone concentrations depend on feeding behavior and dietary composition. It has been established that feeding‐fasting cycles are potent timing cues for the clock in peripheral tissues. Mice primarily consume food during the night (active phase), consuming 20% of their daily caloric intake during the day (Bare, 1959; Koronowski et al, 2019). To experimentally clarify the feeding‐fasting cycle in mice, it is necessary to restrict food consumption by time‐restricted feeding (TRF). Disturbance of the feeding‐fasting cycle by restricted daytime feeding gradually inverts the phase of peripheral clocks (Damiola, 2000). Intriguingly, the impact of restricted daytime feeding on the SCN clock phase is considered negligible.
Fasting affects clock‐controlled genes (CCGs) and fasting‐sensitive transcription factors, and it suppresses CLOCK:BMAL1 recruitment to the E‐box region of Dbp and Rev‐erbα, consequently diminishing diurnal gene expression of these genes (Kinouchi et al, 2018). Under fasting conditions, the amount of cellular NAD⁺ is increased while acetylated BMAL1 protein is decreased, resulting in diminished transcriptional activity of CLOCK:BMAL1. Glucagon, a hormone secreted by pancreatic α cells under fasting conditions, induces the hepatic expression of Per1 and Per2 genes through the cyclic adenosine monophosphate (cAMP) response element‐binding protein (CREB) signaling pathway (Mukherji et al, 2015). In addition, Kinouchi et al reported that fasting activates transcription factors such as CREB and glucocorticoid receptor (GR) (Kinouchi et al, 2018). However, the authors revealed that Per2 expression was reduced under fasting conditions. This discrepancy is likely attributed to different fasting durations (12 h vs. 24 h) and possibly the timing of fasting initiation across studies. Moreover, fasting increases cellular levels of AMP, enhancing AMPK activation. Activated AMPK phosphorylates and destabilizes CRY1 protein. Collectively, these results suggest that fasting greatly remodels metabolic homeostasis by decreasing CLOCK:BMAL1 transcriptional activity and controlling metabolic gene expression by fasting‐induced transcription factors.
Feeding triggers an insulin surge that prompts the liver to switch to anabolic processes such as glycogenesis and de novo lipogenesis. Numerous studies have reported that insulin is a potent hormone regulating peripheral clock function, but not insulin‐independent tissues (Balsalobre et al, 2000; Yamajuku et al, 2012; Sato et al, 2014). Administration of insulin reportedly increased the hepatic expression of Per2 via the phosphatidylinositol 3‐kinase (PI3K) pathway and induced a phase shift of the clock (Yamajuku et al, 2012). In adipose tissues, insulin stimulated Per2 expression via the mitogen‐activated protein kinase (MAPK) pathway, resulting in a phase shift in the adipose clock similar to that observed in the liver (Sato et al, 2014). BMAL1 is also an essential regulator of insulin‐induced phase resetting and promotes lipid metabolism via the action of insulin (Zhang et al, 2014a). Bmal1 deficiency inhibited AKT phosphorylation by reducing the abundance of rapamycin‐insensitive companion of mammalian target of rapamycin (RICTOR), a key component of mTOR complex 2, and attenuated insulin signaling. In Bmal1‐depleted mice, refeeding failed to activate insulin signaling and increase gene expression related to the clock and de novo lipogenesis. Recently, it has been reported that in addition to insulin, oxyntomodulin, a peptide hormone released from the gastrointestinal tract after feeding, induces the expression of Per1 and Per2 in the liver (Landgraf et al, 2015). Based on these reports, insulin and oxyntomodulin appear to play important roles in the timing and output of peripheral clocks.
Glycogen synthase kinase 3β (GSK3β) has been proposed to play a critical role in insulin signaling (Wan et al, 2013). GSK3β is a ubiquitous kinase that regulates diverse cellular processes, including glucose homeostasis. Notably, GSK3β exhibits a circadian pattern of activity, and BMAL1 phosphorylation by this kinase controls its stability (Sahar et al, 2010). Moreover, GSK3β reportedly phosphorylates PER2, CRY2, and REV‐ERBα, leading to proteasomal degradation of CRY2 and stabilization of REV‐ERBα (Harada et al, 2005; Yin et al, 2006). O‐GlcNAc modification competitively inhibits the phosphorylation of PER and CRY proteins and GSK3β phosphorylates OGT. Thus, the circadian activity of GSK3β contributes to the development of a diurnal pattern of protein O‐GlcNAcylation (Kaasik et al, 2013; Li et al, 2013). Thus, GSK3β is involved in the tuning of the clock by post‐translational modification of its proteins.
Currently, the effects of hepatokines on the clock and metabolic homeostasis are gaining momentum (Ma et al, 2016; Chen et al, 2019). The liver is sensitive to food signals and responds via cytokine secretion. Chen et al (2019) reported that the secretion of angiopoietin‐like 8 (Angptl8) is a potential direct link between food intake, hepatic clock resetting, and metabolic gene transcription (Chen et al, 2019). Angptl8 induces the phosphorylation of P38 MAPK, nuclear factor‐κB (NF‐κB), and AKT, ultimately activating Per1 expression. In addition, retinol‐binding protein 4 (RBP4), a specific retinol carrier in the circulation (Quadro et al, 1999), is produced and released mainly by hepatocytes. RBP4 oscillates in a daily manner under Dbp control. Ma et al (2016) revealed that RBP4 acts as a hepatokine in the temporal regulation of glucose metabolism (Ma et al, 2016).
Clock remodeling by dietary composition
The rhythm of peripheral clocks can be maintained by normal chow TRF, independent of central clock synchronization (Chaix et al, 2018). In certain metabolic tissues, daytime‐restricted feeding results in 12‐h phase shifts of peripheral circadian clock gene expression (Vollmers et al, 2009; Bray et al, 2013). Thus, while meal timing is an important factor, dietary composition is also important for guiding the clock system. Several studies have demonstrated the molecular mechanisms of clock remodeling and circadian metabolism based on dietary composition. Herein, we review the effects of high fat and ketogenic diets (KD).
How does a high‐fat diet alter the circadian clock system and energy metabolism?
A typical rodent HFD contains 60% kcal of energy as lipids. Chronic HFD intake induces metabolic diseases, such as obesity, insulin resistance, and diabetes. Recent studies have revealed massive changes in circadian gene expression in diet‐induced obesity (Kohsaka et al, 2007; Eckel‐Mahan et al, 2013). How does HFD remodel the oscillation of peripheral gene expression? To address this question, Eckel‐Mahan et al (2013) analyzed the liver metabolome and transcriptome profile using high‐throughput “omics” analysis (Eckel‐Mahan et al, 2013). HFD induced the loss of oscillation in a large number of normally oscillating genes. Core‐clock genes are maintained even in HFD‐fed animals, indicating their high resistance to dietary composition. In contrast, HFD induced large‐scale de novo oscillating transcripts. HFD‐induced reprogramming is mediated by several mechanisms (Fig 3). Recruitment of CLOCK:BMAL1 to chromatin is impaired in genes that would normally be considered clock‐controlled. HFD decreased the transcriptional activity of CLOCK and BMAL1 without affecting their mRNA and protein expression (Fig 3A). Another study reported that HFD severely impaired the amplitude and rhythmicity of clock genes in the liver (Wang et al, 2018). Both experiments used the same HFD (60% fat, D12491 Research Diets, Inc.), but the administration period differed (10 weeks vs. 12 weeks). Recruitment of CLOCK:BMAL1 to E‐box regions of target genes decreased within three days of HFD administration. Further examination is required to reveal the complete course of acute and chronic effects of HFD on the clock.
Figure 3. High‐fat diet (HFD)‐induced reprogramming of the hepatic clock and metabolic homeostasis.
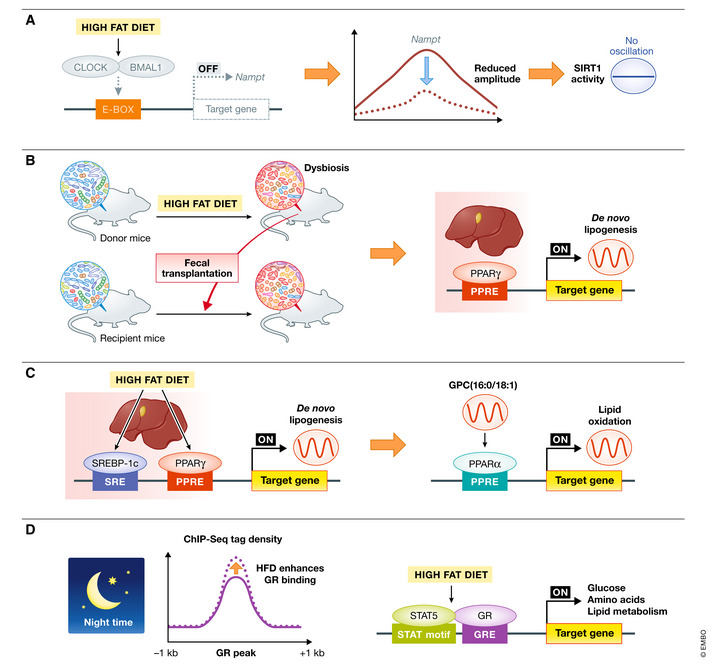
(A) HFD blunts CLOCK:BMAL1 chromatin recruitment to the promoter region of Nampt, dampening transcriptional oscillation and as a consequence the oscillation of SIRT1 activity. (B) HFD treatment induces dysbiosis. When chow‐diet fed mice receive fecal transplantation from HFD‐treated mice, the PPARγ pathway is activated in their liver in a ZT‐dependent manner, resulting in transcriptional reprogramming. (C) HFD leads to a remarkable and synchronous circadian oscillation of the anabolic transcriptional factors SREBP‐1c and PPARγ in the liver, leading to rhythmic de novo lipogenesis. SREBP‐1c elevates the levels of the PPARα ligand sn‐glycero‐3‐phosphocholine (GPC) (16:0/18:1). Rhythmic GPC (16:0/18:1) level induce circadian hepatic fatty acid oxidation. (D) HFD increases GR binding near promoters and enhances GR‐STAT5 co‐occupancy in target genes related to glucose, amino acid, and lipid metabolism. BMAL1, brain and muscle ARNT‐like 1; CBP, histone acetyltransferase CREB‐binding protein; CLOCK, circadian locomotor output cycles protein caput; Nampt, nicotinamide phosphoribosyltransferase; GR, glucocorticoid receptor; PPARα, peroxisome proliferator‐activated receptor‐alpha; PPARγ, peroxisome proliferator‐activated receptor‐gamma; SREBP‐1c, sterol‐regulatory element‐binding protein‐1c; STAT5, signal transducer and activator of transcription 5.
In addition, PPARγ is involved in HFD‐mediated metabolic remodeling. HFD enhanced nuclear PPARγ protein levels and rhythmic chromatin recruitment to target genes (Eckel‐Mahan et al, 2013). In liver‐specific PPARγ‐deficient mice, the expression of numerous genes involved in lipid uptake and lipid transport decreased remarkably, resulting in reduced hepatic steatosis (Moran‐Salvador et al, 2011). RORα specifically recruited HDAC3 to PPARγ target promoters and suppressed PPARγ transcriptional activity (Kim et al, 2017). HFD administration to liver‐specific RORα‐deficient mice induced severe steatosis and obesity by dysregulating PPARγ signaling (Kim et al, 2017). In addition, HFD regulated PPARγ gene expression through epigenetic modifications. The histone demethylase Jumonji C domain‐containing protein 2 B (JMJD2B) is involved in HFD‐induced PPARγ gene expression (Kim et al, 2018). Overexpression of JMJD2B increased the expression of PPARγ and caused hepatic steatosis. In addition, studies have demonstrated that histone H3K4 methyltransferase MLL4 is recruited to the peroxisome proliferator response elements of PPARγ and target genes in the liver, an event linked with steatosis (Kim et al, 2016). Moreover, it has been recently reported that HFD regulates the circadian clock in the liver by altering the gut microbiota. To investigate the direct effect of the microbiome on the hepatic clock, Murakami et al (2016) colonized control chow‐fed recipient mice with microbial communities harvested from HFD‐fed donors and analyzed the circadian clock machinery in the liver (Murakami et al, 2016). The authors demonstrated that fecal transplantation from HFD to chow mice induced rhythmic activation of hepatic PPARγ, which, in turn, leads to transcriptional reprogramming in the liver (Fig 3B). Additionally, in mice whose microbiome was ablated by antibiotic treatment, HFD disrupted the PPARγ rhythm and target gene expression. These results strongly indicate that PPARγ is involved in HFD‐induced steatosis and that hepatic lipid accumulation can be prevented by suppressing the PPARγ network. In contrast, Guan et al (2018) reported that SREBP‐1c is the primary factor for HFD‐induced metabolic remodeling (Fig 3C) (Guan et al, 2018). The authors reported that Pparγ deletion caused no significant changes in the amplitude or rhythmicity of hepatic de novo lipogenesis genes. HFD‐induced SREBP‐1c oscillations have been observed in other HFD studies as well. SIRT1 reportedly deacetylates SREBP‐1c and inhibits its activity (Wang et al, 2017). As described above, HFD inhibits Nampt transcription by attenuating CLOCK:BMAL1 transcriptional activity, which in turn lowers SIRT1 activity. This links HFD‐induced clock remodeling with the regulation of SREBP‐1c‐dependent de novo lipogenesis. HFD‐induced SREBP‐1c oscillations also leads to circadian production of GPC (16:0/18:1) and circadian fatty oxidation in the liver (Fig 3C).
In addition to PPARγ and SREBP‐1c, GR has recently been reported as a nuclear receptor responsible for HFD‐induced metabolic adaptations (Quagliarini et al, 2019). GR mediates most of the known biological effects of GCs and steroid hormones secreted by the zona fasciculata of the adrenal cortex. GC secretion exhibits a prominent circadian rhythm and peaks during feeding time, i.e., the early night in rodents and early morning in humans (Weitzman et al, 1971; Windle et al, 1998). GCs are GR ligands, and activated GRs are translocated to the nucleus to regulate target gene expression. The promoter region of Per contains a GR‐binding region, and Per gene expression rapidly increases after GR activation (Yamamoto et al, 2005; So et al, 2009). Subsequently, GR represses the transcriptional level of Rev‐erba (Torra et al, 2000). GR binding to target promoter regions exhibits a distinct daily pattern, closely mirroring serum corticosterone levels (Quagliarini et al, 2019). A 12‐week HFD administration maintained the typical circadian GR binding pattern while expanding the GR cistrome, especially during the feeding phase in the liver (Quagliarini et al, 2019). Simultaneously, the recruitment of signal transducer and activator of transcription 5 (STAT5), a transcriptional factor that genetically interacts with GR in liver homeostasis, is also increased, and the HFD‐mediated increase in GR occupancy is driven by STAT5 (Fig 3D). This finding suggests that GR is one of the factors inducing HFD‐mediated reprogramming. Other nuclear receptors, hepatocyte nuclear factor‐4α (HNF‐4α) and LXR, have also been reported to play a role in regulating the clock system (Noshiro et al, 2009; Qu et al, 2018), and concurrently, both nuclear receptors are involved in the homeostasis of carbohydrate and lipid metabolism (Joseph et al, 2002; Rhee et al, 2003; Gilardi et al, 2014). During HFD feeding, metabolic remodeling involves the interaction of numerous nuclear receptors.
HFD blunts feeding‐fasting cycles in mice, increases daily food consumption, and affects the circadian patterns of circulating hormones (Kohsaka et al, 2007; Hatori et al, 2012; Vieira et al, 2012). Leptin plays a major role in regulating appetite via mediobasal hypothalamic signaling (Begg & Woods, 2013). Reportedly, blood leptin levels exhibit circadian rhythmicity and are subjected to acute regulation by food intake. HFD increased blood leptin concentration (Sundaram & Yan, 2016). Hypothalamic neuropeptides linked to feeding are known to determine eating behavior. The saturated fatty acid palmitate, a largely abundant lipid, was shown to increase the expression of neuropeptide Y (Npy) and pro‐opiomelanocortin (Pomc) (Clemenzi et al, 2020). Other in vitro experiments have indicated that palmitate treatment enhanced Bmal1 and reduced Per2 in neuronal cells (Fick et al, 2011; Greco et al, 2014). BMAL1 rhythmically binds to Npy and Pomc promoters and increases Npy and Pomc gene expression (Fick et al, 2010; Loganathan et al, 2019). As palmitate treatment‐induced Npy and Pomc expression diminished in Bmal1 knockout neuronal cells, a palmitate‐BMAL1‐NPY axis in the hypothalamus may contribute to HFD‐altered feeding patterns (Clemenzi et al, 2020).
How does a ketogenic diet change the circadian clock system and energy metabolism?
In contrast to HFD, KD is a high‐fat diet with low carbohydrate and low protein content. KD has primarily been used for weight loss in obese individuals (Bueno et al, 2013; Nymo et al, 2017). Indeed, KD consumption can induce a switch to fatty acid oxidation and provide excess acetyl‐CoA, resulting in ketone body generation, including acetoacetate and β‐hydroxybutyrate (β‐OHB). KD mediates different effects on the liver and intestine circadian clocks (Fig 4) (Tognini et al, 2017). In the liver, administration of KD for 4 weeks enhanced the amplitude of genes with an E‐box sequence in the promoter region, such as Dbp, Nampt, and patatin‐like phospholipase domain‐containing 2 (Pnpla2) and increased the recruitment of BMAL1 to E‐boxes. Genome‐wide analysis showed that 2,339 genes displayed circadian expression in the liver when compared with 719 genes under normal dietary conditions. In contrast, oscillation of only 785 novel genes was observed in the intestine. Transcriptomic analysis revealed that the PPARα transcriptional network was enriched in KD‐fed mice. Intriguingly, although the PPARα pathway was induced both, in the liver and in the intestine during ketogenesis, the two tissues displayed different oscillation phases for PPARα nuclear accumulation and target gene expression. Serum and gut levels of β‐OHB, an endogenous HDAC I inhibitor (Shimazu et al, 2013) were increasingly correlating with increased histone H3 acetylation levels in the gut but not the liver, which might contribute to the oscillation of PPARα target genes in the gut. In addition, KD increased global protein acetylation levels in whole‐cell lysates and the mitochondrial fraction isolated from the liver (Newman et al, 2017). Recent evidence indicates a more direct epigenetic effect of β‐OHB via a novel histone modification: β‐hydroxybutyrylation of H3K9 (Xie et al, 2016). Further studies are required to clarify the effects of KD on histone modification.
Figure 4. Ketogenic diet (KD)‐induced reprogramming of peripheral clocks and metabolic homeostasis.
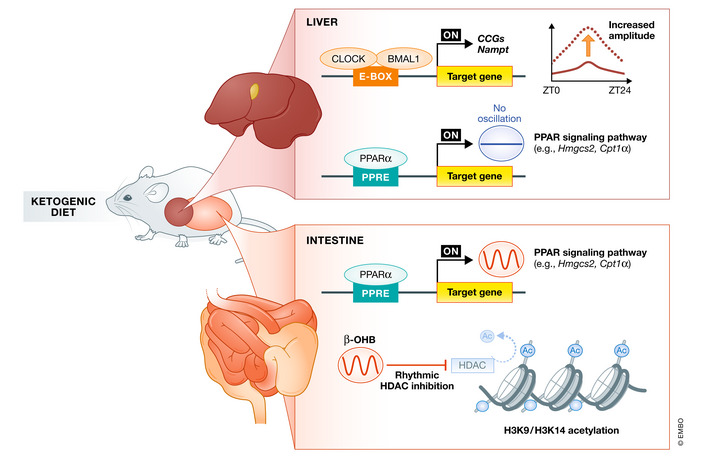
KD remodels rhythms differently in the liver and intestine. KD enhances BMAL1 recruitment to target genes in the liver, thereby increasing the amplitude of CCGs oscillations. KD activates PPARα signaling both in the liver and intestine. However, robust oscillations can be observed only in the intestine. KD‐induced ketogenesis increases serum β‐OHB concentration and circadian oscillations. The local intestinal concentration of β‐OHB mirrored the profile of serum β‐OHB, leading to time‐dependent HDAC activity and histone acetylation. BMAL1, brain and muscle ARNT‐like 1; β‐OHB, β‐hydroxybutyrate; CCGs, clock‐controlled genes; CPT‐1α, carnitine palmitoyltransferase 1α; HDAC, histone deacetylases; HMGCS2, 3‐hydroxy‐3‐methylglutaryl‐CoA synthase 2; PPARα, peroxisome proliferator‐activated receptor alpha.
Differences in metabolic reprogramming between HFD and KD
Both HFD and KD regimens contain a high‐fat component (HFD 60%, KD 90%) but differ in carbohydrate and protein content. KD contains < 1% carbohydrate and 10% protein, mimicking caloric restriction. Following the administration of KD and HFD, a transcriptional analysis identified the downregulation of multiple metabolic pathways such as insulin signaling and protein synthesis, common to KD and HFD (Newman et al, 2017). However, the effects on PPARα signaling differed between these two studies. Newman et al (2017) reported the upregulation of PPARα target genes unique to KD (Newman et al, 2017). However, Guan et al (2018) showed that HFD induced diurnal oscillation of PPARα (Guan et al, 2018). This discrepancy could be attributed to differences in diet composition and administration methods in each study. HFD and KD have different effects on the energy balance. Low glucose conditions, such as calorie restriction and fasting, enhanced AMPK activation (Zhu et al, 2005; Zhang et al, 2013). KD mimicked calorie restriction and resulted in the activation of AMPK. In contrast, HFD decreased hepatic (but not muscle) AMPK phosphorylation levels (Shiwa et al, 2015), which can be reversed by implementing a normal diet for three days (Shiwa et al, 2015). Another major difference between HFD and KD groups was the blood β‐OHB levels. After one week of dietary intervention, the blood β‐OHB concentration tended to be higher in the KD group than in the HFD group, and the value after 6 weeks was significantly higher in the KD group (Roberts et al, 2017). Mitochondrial 3‐hydroxy‐3‐methylglutaryl‐CoA synthase 2 (HMGCS2) is a key enzyme in ketogenesis (Puisac et al, 2012). It has been reported that insulin repressed the mRNA and protein expression of HMGCS2 (Nadal et al, 2002). As the blood insulin level under KD was markedly reduced when compared with that under HFD (Kennedy et al, 2007), HMGCS2 expression is expected to be higher, and the increase in β‐OHB concentration could also be attributed to differences in insulin concentration. As mentioned above, β‐OHB can inhibit HDAC activity. After one month of KD, total acetyl‐Lys levels were individually increased by 5‐ and 2.5‐fold in the liver and muscle of the KD‐treated group when compared with the HFD‐treated group (Roberts et al, 2017). Differences in comprehensive protein acetylation levels may therefore be one mechanism that induces differences in metabolic characteristics between the two diet groups and the difference in the carbohydrate content is one of the key factors resulting in this difference between HFD and KD.
How do carbohydrates affect the circadian clock system and energy metabolism?
Glucose is widely accepted as one of the factors that can influence cellular circadian rhythms (Hirota et al, 2002). However, most studies that examined the effect of carbohydrates on clock remodeling and metabolic homeostasis have been performed in vitro using cultured cells, and knowledge regarding its in vivo effects remain limited.
Hirota et al (2002) demonstrated that exchanging the culture medium induced circadian gene expression in cultured rat fibroblasts and that glucose is a key molecule that can directly reset the phase of cells derived from peripheral tissues (Hirota et al, 2002). Glucose reportedly increases expression of the transforming growth factor beta‐inducible early gene 1 (Tieg1) and transiently accumulates TIEG1 protein in the nucleus, which directly represses Bmal1expression, resulting in reduced Per1 and Per2 levels (Hirota et al, 2010).
Fructose and glucose are characterized by the same chemical formula; however, they differ in structure and metabolism. In hepatocytes, fructose led to disrupted Bmal1 mRNA expression and delayed the expression of Per1 mRNA (Chapnik et al, 2016). In myotubes, fructose induced a higher amplitude of Per1 and Bmal1 mRNA expression and delayed rhythms of Per1, Bmal1, and Clock (Chapnik et al, 2016). Based on these in vitro results, the influence of fructose on clock genes varies between organs and corresponding tissues.
AMPK activation is considered one of the factors responsible for differences in glucose‐ and fructose‐mediated effects on the molecular mechanism of the circadian clock. The phosphorylated AMPK/AMPK ratio in hepatocytes treated with fructose was approximately half that of glucose‐treated hepatocytes (Chapnik et al, 2016). On the other hand, fructose exposure increased the myocyte p‐AMPK/AMPK ratio by approximately 2.5‐fold when compared with that following glucose exposure (Chapnik et al, 2016). Thus, the effects of glucose and fructose on AMPK are tissue‐specific. As AMPK is involved in the degradation of CRY (Lamia et al, 2009), differences in the glucose‐ and fructose‐induced effects on clock molecules may be mediated by AMPK. In addition to the direct effects of carbohydrates on organs, it is necessary to consider the effects of hormones secreted following carbohydrate intake. Insulin is strongly involved in metabolic changes in peripheral organs after carbohydrate intake.
Glucose is a potent stimulant of insulin secretion; however, fructose intake either marginally or fails to increase the insulin concentration (Sato et al, 2019). Thus, to clarify the effects of carbohydrate intake on clock function in organisms, it is necessary to consider the effects of carbohydrates, as well as the associated effects of changes in hormone concentrations.
In contrast to the findings described above, four weeks of a high‐sucrose diet did not alter hepatic clock gene expression in rats (Sun et al, 2019a). Conversely, it modestly changed the amplitude and rhythm of gene expression related to fructolysis and de novo lipogenesis, leading to fatty liver and the development of hyperlipidemia. Hepatic phosphate levels were reduced under high‐sucrose diet, indicating lower ATP levels (Sun et al, 2019a). Since ATP is a putative mRNA stabilizer (Chen et al, 2007), these reports imply that variations in ATP‐dependent RNA stability contribute to the high‐sucrose‐induced amplitude enhancement of lipogenic enzyme mRNA in the liver.
In contrast to the liver, a four‐week high‐sucrose diet altered the expression of nutrient transporters and carbohydrate metabolism, as well as the oscillation patterns of circadian clock genes in the small intestine (Sun et al, 2019b). These results indicate that sucrose treatment has a greater impact on the intestinal clock and metabolism than the liver.
In mice experiments, each carbohydrate (glucose, fructose, and sucrose) causes distinct shifts in circadian timing in the liver (Hirao et al, 2009). Further studies in animal models are required to elucidate the precise mechanisms by which carbohydrates influence peripheral clock function in vivo.
Positive impact of clock remodeling on health and metabolism
It is widely established that circadian misalignment leads to obesity and several metabolic diseases, such as insulin resistance, dyslipidemia, and hyperglycemia (Shimba et al, 2011; Paschos et al, 2012; Shi et al, 2013). Conversely, these findings suggest that maintaining proper circadian rhythms can result in health benefits. Eating behavior is one of the most influential factors in maintaining a proper circadian clock. TRF, a feeding strategy that limits daily food intake, prolongs the temporary fasting time and forms a clear feeding/fasting cycle, enhancing the robustness or amplitude of rhythms. In mice, TRF during the dark phase can improve several detrimental metabolic consequences of HFD and a high‐fructose diet by correcting metabolic and physiological rhythms (Hatori et al, 2012; Sun et al, 2018). However, the effect of TRF on weight loss varies among diets and experimental protocols and needs to be further verified (Hatori et al, 2012; Chaix et al, 2014; Woodie et al, 2018). Based on findings from Drosophila studies, the TRF regimen can enhance the amplitude of oscillating transcripts in the peripheral tissue and head, leading to improved sleep, prevention of body weight gain, and deceleration of cardiac aging (Gill et al, 2015). In addition, the efficacy of TRF has been reported in human studies. Food consumption at an inappropriate circadian time has been associated with weight gain and metabolic disorders (Hibi et al, 2013). In contrast, an 8‐h TRF at the appropriate time (10:00 to 18:00 h) for 12 weeks reportedly reduced body weight and systolic blood pressure in obese subjects (Gabel et al, 2018). Additional studies in humans have shown that TRF in the early phase positively impacts human health, such as improving insulin sensitivity and blood pressure (Sutton et al, 2018). These reports suggest that circadian clock remodeling, especially enhancing the clock amplitude, affords positive health effects and maintains appropriate rhythms, critical for preventing obesity and metabolic disease.
Remodeling of the clock and metabolism by natural compounds
In addition to dietary composition, natural compounds can influence circadian and metabolic homeostasis. Several studies have identified single nutrients capable of driving or phase‐shifting circadian rhythms.
Flavonoids, polyphenolic compounds derived from plants and fungi, reportedly possess numerous biological and pharmacological activities, including anti‐inflammatory, antioxidant, and anticancer effects. Recent studies have revealed that flavonoids have great therapeutic potential for treating diet‐induced obesity and hepatic inflammation, given their ability to manipulate the clock system.
Epigallocatechin gallate (EGCG) is a major component of tea polyphenols. EGCG treatment of hepatocytes exposed to H2O2 demonstrated that EGCG had a protective effect against H2O2‐induced circadian misalignment (Qi et al, 2018). Another in vitro study reported that EGCG supplementation can reverse obesity‐induced insulin resistance and obesity by alleviating circadian desynchrony and metabolic misalignment (Mi et al, 2017).
Nobiletin, a polymethoxylated flavone isolated from citrus peels, affords potent protection against metabolic syndromes in a clock‐dependent manner (He et al, 2016). Nobiletin targets ROR, which in turn may stabilize and even enhance the transcriptional activity of the whole molecular oscillator. The anti‐obesity effect of nobiletin was diminished in ClockΔ19/Δ19 mutant mice, indicating that nobiletin exerts its metabolic effect through Clock. In the muscle, nobiletin reportedly promotes healthy aging by ROR activation under HFD treatment via a concerted optimization of mitochondrial respiration (Nohara et al, 2019). Similar effects were observed with tangeretin, a flavonoid present in citrus peels, using the PER2::LUC luciferase system (Shinozaki et al, 2017).
Resveratrol (3,4,5‐trihydroxy‐trans‐stilbene) is a natural polyphenolic compound found in several fruits and vegetables, such as grapes and peanuts. Several of the health benefits associated with resveratrol have been attributed to its ability to mimic the effects of calorie restriction. Resveratrol increases intracellular cAMP concentration by inhibiting cAMP phosphodiesterases, enhancing AMPK activity (Park et al, 2012). Moreover, resveratrol upregulates not only protein expression but also the enzyme activity of SIRT1 in multicellular animals (Howitz et al, 2003; Chai et al, 2017). Resveratrol intervention during HFD restored the rhythmic expression of Sirt1, concomitant with the restoration of clock genes (Sun et al, 2015). Numerous functional components and metabolites of food that impact the clock are likely to be discovered in future investigations.
Conclusions and future perspectives
During past decades, a large array of nutritional challenge studies have provided insights into the reciprocal relationship between the circadian system and metabolic homeostasis. In addition to dietary composition, macronutrients can influence circadian rhythmicity. However, numerous outstanding questions remain unresolved.
Over the last decade, several circadian transcriptome analyses have been conducted to characterize the circadian and metabolic genes expressed in different organs and under several dietary conditions. Interestingly, the effects of diet differ depending on the organ, which is influenced by tissue‐specific transcription factors. The interaction between organs via output metabolites has also been reported, although a comprehensive understanding is yet to be established. Future research should aim to determine the contribution of each peripheral clock to metabolic homeostasis in other organs.
Recent studies on various diets have revealed the effects of diet composition on clock genes and transcription factors. However, results across studies have been contradictory. As diet compositions vary substantially from study to study, a careful and comprehensive approach is required to elucidate the intricate mechanisms linking the clock and metabolism.
Most studies investigating clock changes and metabolic function changes attributed to dietary modifications have been performed in rodents. Available information regarding the impact of various dietary changes on the human clock system remains insufficient. Blood samples can be obtained using minimally invasive methods and are extremely valuable for extracting important biological information. For example, plasma levels of PC reflect the dietary carbohydrate–fat ratio in humans (Inoue et al, 2017). A recently published study has illustrated changes in metabolites in human serum in response to dietary changes over circadian time (Sato et al, 2018). Therefore, blood samples can be substantially useful for determining the effect of dietary composition on metabolic rhythms in humans. Further studies on chrono‐nutritional aspects of the circadian clock can provide potential therapeutics for treating metabolic diseases.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
In need of answers.
How does each organ’s clock interact with other peripheral clocks?
How do nutritional challenges impact tissue‐specific remodeling of the clock?
What is the precise relationship between diet composition‐period and remodeling of the clock?
How does β‐hydroxy butyrylation impact the clock at chromatin under a ketogenic diet?
Accurate methods for detecting the nutritional effect on the human circadian system are needed.
Acknowledgments
We thank all members of the Sassone‐Corsi laboratory for stimulating discussion and critically reading of the manuscript. T.S. was supported by a Japan Society for the Promotion of Science (JSPS) fellowship. Financial support for P.S.‐C. was provided by the National Institute of Health, INSERM, and a Novo Nordisk Challenge Grant.
EMBO reports (2022) 23: e52412.
See the Glossary for abbreviations used in this article.
References
- Aguilar‐Arnal L, Katada S, Orozco‐Solis R, Sassone‐Corsi P (2015) NAD(+)‐SIRT1 control of H3K4 trimethylation through circadian deacetylation of MLL1. Nat Struct Mol Biol 22: 312–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albaugh BN, Arnold KM, Denu JM (2011) KAT(ching) metabolism by the tail: insight into the links between lysine acetyltransferases and metabolism. ChemBioChem 12: 290–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthonisen EH, Berven L, Holm S, Nygard M, Nebb HI, Gronning‐Wang LM (2010) Nuclear receptor liver X receptor is O‐GlcNAc‐modified in response to glucose. J Biol Chem 285: 1607–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariyannur PS, Moffett JR, Madhavarao CN, Arun P, Vishnu N, Jacobowitz DM, Hallows WC, Denu JM, Namboodiri AM (2010) Nuclear‐cytoplasmic localization of acetyl coenzyme a synthetase‐1 in the rat brain. J Comp Neurol 518: 2952–2977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U (2008) SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134: 317–328 [DOI] [PubMed] [Google Scholar]
- Asher G, Schibler U (2011) Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab 13: 125–137 [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Marcacci L, Schibler U (2000) Multiple signaling pathways elicit circadian gene expression in cultured Rat‐1 fibroblasts. Curr Biol 10: 1291–1294 [DOI] [PubMed] [Google Scholar]
- Bare JK (1959) Hunger, deprivation, and the day‐night cycle. J Comp Physiol Psychol 52: 129–131 [DOI] [PubMed] [Google Scholar]
- Bass J (2012) Circadian topology of metabolism. Nature 491: 348–356 [DOI] [PubMed] [Google Scholar]
- Bass J, Takahashi JS (2010) Circadian integration of metabolism and energetics. Science 330: 1349–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg DP, Woods SC (2013) The endocrinology of food intake. Nat Rev Endocrinol 9: 584–597 [DOI] [PubMed] [Google Scholar]
- Berthier A, Vinod M, Porez G, Steenackers A, Alexandre J, Yamakawa N, Gheeraert C, Ploton M, Marechal X, Dubois‐Chevalier J et al (2018) Combinatorial regulation of hepatic cytoplasmic signaling and nuclear transcriptional events by the OGT/REV‐ERBalpha complex. Proc Natl Acad Sci U S A 115: E11033–E11042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindesbøll C, Fan Q, Nørgaard RC, MacPherson L, Ruan H‐B, Wu J, Pedersen TÅ, Steffensen KR, Yang X, Matthews J et al (2015) Liver X receptor regulates hepatic nuclear O‐GlcNAc signaling and carbohydrate responsive element‐binding protein activity. J Lipid Res 56: 771–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray MS, Ratcliffe WF, Grenett MH, Brewer RA, Gamble KL, Young ME (2013) Quantitative analysis of light‐phase restricted feeding reveals metabolic dyssynchrony in mice. Int J Obes 37: 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno NB, de Melo IS, de Oliveira SL, da Rocha AT (2013) Very‐low‐carbohydrate ketogenic diet v. low‐fat diet for long‐term weight loss: a meta‐analysis of randomised controlled trials. Br J Nutr 110: 1178–1187 [DOI] [PubMed] [Google Scholar]
- Buijs RM, Chun SJ, Niijima A, Romijn HJ, Nagai K (2001) Parasympathetic and sympathetic control of the pancreas: a role for the suprachiasmatic nucleus and other hypothalamic centers that are involved in the regulation of food intake. J Comp Neurol 431: 405–423 [DOI] [PubMed] [Google Scholar]
- Canaple L, Rambaud J, Dkhissi‐Benyahya O, Rayet B, Tan NS, Michalik L, Delaunay F, Wahli W, Laudet V (2006) Reciprocal regulation of brain and muscle Arnt‐like protein 1 and peroxisome proliferator‐activated receptor alpha defines a novel positive feedback loop in the rodent liver circadian clock. Mol Endocrinol 20: 1715–1727 [DOI] [PubMed] [Google Scholar]
- Canto C, Gerhart‐Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J (2009) AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458: 1056–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Menzies KJ, Auwerx J (2015) NAD(+) Metabolism and the control of energy homeostasis: a balancing act between mitochondria and the nucleus. Cell Metab 22: 31–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Anderson FE, Jung YJ, Dziema H, Obrietan K (2011) Circadian regulation of mammalian target of rapamycin signaling in the mouse suprachiasmatic nucleus. Neuroscience 181: 79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Li A, Cho HY, Lee B, Obrietan K (2010) Mammalian target of rapamycin signaling modulates photic entrainment of the suprachiasmatic circadian clock. J Neurosci 30: 6302–6314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceriani MF, Hogenesch JB, Yanovsky M, Panda S, Straume M, Kay SA (2002) Genome‐wide expression analysis in Drosophila reveals genes controlling circadian behavior. J Neurosci 22: 9305–9319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai R, Fu H, Zheng Z, Liu T, Ji S, Li G (2017) Resveratrol inhibits proliferation and migration through SIRT1 mediated posttranslational modification of PI3K/AKT signaling in hepatocellular carcinoma cells. Mol Med Rep 16: 8037–8044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaix A, Lin T, Le HD, Chang MW, Panda S (2018) Time‐restricted feeding prevents obesity and metabolic syndrome in mice lacking a circadian clock. Cell Metab 29: 303–319.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaix A, Zarrinpar A, Miu P, Panda S (2014) Time‐restricted feeding is a preventative and therapeutic intervention against diverse nutritional challenges. Cell Metab 20: 991–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapnik N, Rozenblit‐Susan S, Genzer Y, Froy O (2016) Differential effect of fructose on fat metabolism and clock gene expression in hepatocytes vs. myotubes. Int J Biochem Cell Biol 77: 35–40 [DOI] [PubMed] [Google Scholar]
- Chen HH, Xu J, Safarpour F, Stewart AF (2007) LMO4 mRNA stability is regulated by extracellular ATP in F11 cells. Biochem Biophys Res Commun 357: 56–61 [DOI] [PubMed] [Google Scholar]
- Chen S, Feng M, Zhang S, Dong Z, Wang Y, Zhang W, Liu C (2019) Angptl8 mediates food‐driven resetting of hepatic circadian clock in mice. Nat Commun 10: 3518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemenzi MN, Martchenko A, Loganathan N, Tse EK, Brubaker PL, Belsham DD (2020) Analysis of western diet, palmitate and BMAL1 regulation of neuropeptide Y expression in the murine hypothalamus and BMAL1 knockout cell models. Mol Cell Endocrinol 507: 110773 [DOI] [PubMed] [Google Scholar]
- Crane BR, Young MW (2014) Interactive features of proteins composing eukaryotic circadian clocks. Annu Rev Biochem 83: 191–219 [DOI] [PubMed] [Google Scholar]
- Curtis AM, Seo SB, Westgate EJ, Rudic RD, Smyth EM, Chakravarti D, FitzGerald GA, McNamara P (2004) Histone acetyltransferase‐dependent chromatin remodeling and the vascular clock. J Biol Chem 279: 7091–7097 [DOI] [PubMed] [Google Scholar]
- Dallmann R, Viola AU, Tarokh L, Cajochen C, Brown SA (2012) The human circadian metabolome. Proc Natl Acad Sci U S A 109: 2625–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann Benoît, Fleury‐Olela F, Schibler U (2000) Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev 14: 2950–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang F, Sun X, Ma X, Wu R, Zhang D, Chen Y, Xu Q, Wu Y, Liu Y (2016) Insulin post‐transcriptionally modulates Bmal1 protein to affect the hepatic circadian clock. Nat Commun 7: 12696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U (2010) The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol 72: 517–549 [DOI] [PubMed] [Google Scholar]
- Doi M, Hirayama J, Sassone‐Corsi P (2006) Circadian regulator CLOCK is a histone acetyltransferase. Cell 125: 497–508 [DOI] [PubMed] [Google Scholar]
- Donohoe DR, Bultman SJ (2012) Metaboloepigenetics: interrelationships between energy metabolism and epigenetic control of gene expression. J Cell Physiol 227: 3169–3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel‐Mahan KL, Patel VR, de Mateo S, Orozco‐Solis R, Ceglia NJ, Sahar S, Dilag‐Penilla SA, Dyar KA, Baldi P, Sassone‐Corsi P (2013) Reprogramming of the circadian clock by nutritional challenge. Cell 155: 1464–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel‐Mahan K, Sassone‐Corsi P (2013) Metabolism and the circadian clock converge. Physiol Rev 93: 107–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett LJ, Lazar MA (2014) Nuclear receptor Rev‐erbalpha: up, down, and all around. Trends Endocrinol Metab 25: 586–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA (2011) A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331: 1315–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fick LJ, Fick GH, Belsham DD (2010) Rhythmic clock and neuropeptide gene expression in hypothalamic mHypoE‐44 neurons. Mol Cell Endocrinol 323: 298–306 [DOI] [PubMed] [Google Scholar]
- Fick LJ, Fick GH, Belsham DD (2011) Palmitate alters the rhythmic expression of molecular clock genes and orexigenic neuropeptide Y mRNA levels within immortalized, hypothalamic neurons. Biochem Biophys Res Commun 413: 414–419 [DOI] [PubMed] [Google Scholar]
- Foteinou PT, Venkataraman A, Francey LJ, Anafi RC, Hogenesch JB, Doyle FJ 3rd (2018) Computational and experimental insights into the circadian effects of SIRT1. Proc Natl Acad Sci U S A 115: 11643–11648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullerton MD, Galic S, Marcinko K, Sikkema S, Pulinilkunnil T, Chen Z‐P, O'Neill HM, Ford RJ, Palanivel R, O'Brien M et al (2013) Single phosphorylation sites in Acc1 and Acc2 regulate lipid homeostasis and the insulin‐sensitizing effects of metformin. Nat Med 19: 1649–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabel K, Hoddy KK, Haggerty N, Song J, Kroeger CM, Trepanowski JF, Panda S, Varady KA (2018) Effects of 8‐hour time restricted feeding on body weight and metabolic disease risk factors in obese adults: a pilot study. Nutr Healthy Aging 4: 345–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon F, Nagoshi E, Brown SA, Ripperger J, Schibler U (2004) The mammalian circadian timing system: from gene expression to physiology. Chromosoma 113: 103–112 [DOI] [PubMed] [Google Scholar]
- Gatfield D, Schibler U (2007) Physiology. Proteasomes keep the circadian clock ticking. Science 316: 1135–1136 [DOI] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ (1998) Role of the CLOCK protein in the mammalian circadian mechanism. Science 280: 1564–1569 [DOI] [PubMed] [Google Scholar]
- Gilardi F, Migliavacca E, Naldi A, Baruchet M, Canella D, Le Martelot G, Guex N, Desvergne B, Cycli XC (2014) Genome‐wide analysis of SREBP1 activity around the clock reveals its combined dependency on nutrient and circadian signals. PLoS Genet 10: e1004155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S, Le HD, Melkani GC, Panda S (2015) Time‐restricted feeding attenuates age‐related cardiac decline in Drosophila . Science 347: 1265–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco JA, Oosterman JE, Belsham DD (2014) Differential effects of omega‐3 fatty acid docosahexaenoic acid and palmitate on the circadian transcriptional profile of clock genes in immortalized hypothalamic neurons. Am J Physiol Regul Integr Comp Physiol 307: R1049–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillo MA, Colombatto S (2008) S‐adenosylmethionine and its products. Amino Acids 34: 187–193 [DOI] [PubMed] [Google Scholar]
- Guan D, Xiong Y, Borck PC, Jang C, Doulias P‐T, Papazyan R, Fang B, Jiang C, Zhang Y, Briggs ER et al (2018) Diet‐induced circadian enhancer remodeling synchronizes opposing hepatic lipid metabolic processes. Cell 174: 831–842.e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada Y, Sakai M, Kurabayashi N, Hirota T, Fukada Y (2005) Ser‐557‐phosphorylated mCRY2 is degraded upon synergistic phosphorylation by glycogen synthase kinase‐3 beta. J Biol Chem 280: 31714–31721 [DOI] [PubMed] [Google Scholar]
- Hart GW (2019) Nutrient regulation of signaling and transcription. J Biol Chem 294: 2211–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong E, Gill S, Leblanc M, Chaix A, Joens M, Fitzpatrick J et al (2012) Time‐restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high‐fat diet. Cell Metab 15: 848–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley SA, Boudeau J, Reid JL, Mustard KJ, Udd L, Makela TP, Alessi DR, Hardie DG (2003) Complexes between the LKB1 tumor suppressor, STRAD alpha/beta and MO25 alpha/beta are upstream kinases in the AMP‐activated protein kinase cascade. J Biol 2: 28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Nohara K, Park N, Park Y‐S, Guillory B, Zhao Z, Garcia J, Koike N, Lee C, Takahashi J et al (2016) The small molecule nobiletin targets the molecular oscillator to enhance circadian rhythms and protect against metabolic syndrome. Cell Metab 23: 610–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksson E, Huber A‐L, Soto EK, Kriebs A, Vaughan ME, Duglan D, Chan AB, Papp SJ, Nguyen M, Afetian ME et al (2017) The liver circadian clock modulates biochemical and physiological responses to metformin. J Biol Rhythms 32: 345–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibi M, Masumoto A, Naito Y, Kiuchi K, Yoshimoto Y, Matsumoto M, Katashima M, Oka J, Ikemoto S (2013) Nighttime snacking reduces whole body fat oxidation and increases LDL cholesterol in healthy young women. Am J Physiol Regul Integr Comp Physiol 304: R94–R101 [DOI] [PubMed] [Google Scholar]
- Hirao A, Tahara Y, Kimura I, Shibata S (2009) A balanced diet is necessary for proper entrainment signals of the mouse liver clock. PLoS One 4: e6909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Kon N, Itagaki T, Hoshina N, Okano T, Fukada Y (2010) Transcriptional repressor TIEG1 regulates Bmal1 gene through GC box and controls circadian clockwork. Genes Cells 15: 111–121 [DOI] [PubMed] [Google Scholar]
- Hirota T, Okano T, Kokame K, Shirotani‐Ikejima H, Miyata T, Fukada Y (2002) Glucose down‐regulates Per1 and Per2 mRNA levels and induces circadian gene expression in cultured Rat‐1 fibroblasts. J Biol Chem 277: 44244–44251 [DOI] [PubMed] [Google Scholar]
- Hou X, Xu S, Maitland‐Toolan KA, Sato K, Jiang B, Ido Y, Lan F, Walsh K, Wierzbicki M, Verbeuren TJ et al (2008) SIRT1 regulates hepatocyte lipid metabolism through activating AMP‐activated protein kinase. J Biol Chem 283: 20015–20026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang L‐L et al (2003) Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 425: 191–196 [DOI] [PubMed] [Google Scholar]
- Huang Y, He Y, Sun X, He Y, Li Y, Sun C (2014) Maternal high folic acid supplement promotes glucose intolerance and insulin resistance in male mouse offspring fed a high‐fat diet. Int J Mol Sci 15: 6298–6313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Hogenesch JB, Kornacker K (2010) JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome‐scale data sets. J Biol Rhythms 25: 372–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Zhu T, Wu J, Guan KL (2002) TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol 4: 648–657 [DOI] [PubMed] [Google Scholar]
- Inoue M, Senoo N, Sato T, Nishimura Y, Nakagawa T, Miyoshi N, Goda T, Morita A, Miura S (2017) Effects of the dietary carbohydrate‐fat ratio on plasma phosphatidylcholine profiles in human and mouse. J Nutr Biochem 50: 83–94 [DOI] [PubMed] [Google Scholar]
- Joseph SB, Laffitte BA, Patel PH, Watson MA, Matsukuma KE, Walczak R, Collins JL, Osborne TF, Tontonoz P (2002) Direct and indirect mechanisms for regulation of fatty acid synthase gene expression by liver X receptors. J Biol Chem 277: 11019–11025 [DOI] [PubMed] [Google Scholar]
- Kaasik K, Kivimäe S, Allen J, Chalkley R, Huang Y, Baer K, Kissel H, Burlingame A, Shokat K, Ptáček L et al (2013) Glucose sensor O‐GlcNAcylation coordinates with phosphorylation to regulate circadian clock. Cell Metab 17: 291–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katada S, Sassone‐Corsi P (2010) The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat Struct Mol Biol 17: 1414–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy AR, Pissios P, Otu H, Xue B, Asakura K, Furukawa N, Marino FE, Liu F‐F, Kahn BB, Libermann TA et al (2007) A high‐fat, ketogenic diet induces a unique metabolic state in mice. Am J Physiol Endocrinol Metab 292: E1724–E1739 [DOI] [PubMed] [Google Scholar]
- Kettner NM, Mayo SA, Hua J, Lee C, Moore DD, Fu L (2015) Circadian dysfunction induces leptin resistance in mice. Cell Metab 22: 448–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Kim J, Kwon JS, Sandhu J, Tontonoz P, Lee SK, Lee S, Lee JW (2016) Critical roles of the histone methyltransferase MLL4/KMT2D in murine hepatic steatosis directed by ABL1 and PPARgamma2. Cell Rep 17: 1671–1682 [DOI] [PubMed] [Google Scholar]
- Kim EY, Jeong EH, Park S, Jeong HJ, Edery I, Cho JW (2012) A role for O‐GlcNAcylation in setting circadian clock speed. Genes Dev 26: 490–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Jung DY, Nagappan A, Jung MH (2018) Histone H3K9 demethylase JMJD2B induces hepatic steatosis through upregulation of PPARgamma2. Sci Rep 8: 13734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Kwak PB, Weitz CJ (2014) Specificity in circadian clock feedback from targeted reconstitution of the NuRD corepressor. Mol Cell 56: 738–748 [DOI] [PubMed] [Google Scholar]
- Kim K, Boo K, Yu YS, Oh SK, Kim H, Jeon Y, Bhin J, Hwang D, Kim KI, Lee JS et al (2017) RORalpha controls hepatic lipid homeostasis via negative regulation of PPARgamma transcriptional network. Nat Commun 8: 162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinouchi K, Magnan C, Ceglia N, Liu YU, Cervantes M, Pastore N, Huynh T, Ballabio A, Baldi P, Masri S et al (2018) Fasting imparts a switch to alternative daily pathways in liver and muscle. Cell Rep 25: 3299–3314.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, Bass J (2007) High‐fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab 6: 414–421 [DOI] [PubMed] [Google Scholar]
- Koronowski KB, Kinouchi K, Welz P‐S, Smith JG, Zinna VM, Shi J, Samad M, Chen S, Magnan CN, Kinchen JM et al (2019) Defining the independence of the liver circadian clock. Cell 177: 1448–1462.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM (1999) mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98: 193–205 [DOI] [PubMed] [Google Scholar]
- La Fleur SE (2003) Daily rhythms in glucose metabolism: suprachiasmatic nucleus output to peripheral tissue. J Neuroendocrinol 15: 315–322 [DOI] [PubMed] [Google Scholar]
- Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, Vasquez DS, Juguilon H, Panda S, Shaw RJ et al (2009) AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 326: 437–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Cacicedo JM, Ruderman N, Ido Y (2008) SIRT1 modulation of the acetylation status, cytosolic localization, and activity of LKB1. Possible role in AMP‐activated protein kinase activation. J Biol Chem 283: 27628–27635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf D, Tsang AH, Leliavski A, Koch CE, Barclay JL, Drucker DJ, Oster H (2015) Oxyntomodulin regulates resetting of the liver circadian clock by food. eLife 4: e06253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM (2001) Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107: 855–867 [DOI] [PubMed] [Google Scholar]
- Lee JH, Jung JY, Jang EJ, Jegal KH, Moon SY, Ku SK, Kang SH, Cho IJ, Park SJ, Lee JR et al (2015) Combination of honokiol and magnolol inhibits hepatic steatosis through AMPK‐SREBP‐1 c pathway. Exp Biol Med 240: 508–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MD, Ruan HB, Hughes ME, Lee JS, Singh JP, Jones SP, Nitabach MN, Yang X (2013) O‐GlcNAc signaling entrains the circadian clock by inhibiting BMAL1/CLOCK ubiquitination. Cell Metab 17: 303–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Shao SH, Xu Z‐X, Hennessy B, Ding Z, Larrea M, Kondo S, Dumont DJ, Gutterman JU, Walker CL et al (2007) The energy sensing LKB1‐AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat Cell Biol 9: 218–224 [DOI] [PubMed] [Google Scholar]
- Liu S, Brown JD, Stanya KJ, Homan E, Leidl M, Inouye K, Bhargava P, Gangl MR, Dai L, Hatano B et al (2013) A diurnal serum lipid integrates hepatic lipogenesis and peripheral fatty acid use. Nature 502: 550–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockley SW, Brainard GC, Czeisler CA (2003) High sensitivity of the human circadian melatonin rhythm to resetting by short wavelength light. J Clin Endocrinol Metab 88: 4502–4505 [DOI] [PubMed] [Google Scholar]
- Loganathan N, Salehi A, Chalmers JA, Belsham DD (2019) Bisphenol A Alters Bmal1, Per2, and Rev‐Erba mRNA and requires Bmal1 to increase neuropeptide Y expression in hypothalamic neurons. Endocrinology 160: 181–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Zhou Z, Chen Y, Wu Y, Liu Y (2016) RBP4 functions as a hepatokine in the regulation of glucose metabolism by the circadian clock in mice. Diabetologia 59: 354–362 [DOI] [PubMed] [Google Scholar]
- Masri S, Sassone‐Corsi P (2010) Plasticity and specificity of the circadian epigenome. Nat Neurosci 13: 1324–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maury E, Ramsey KM, Bass J (2010) Circadian rhythms and metabolic syndrome: from experimental genetics to human disease. Circ Res 106: 447–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi Y, Qi G, Fan R, Ji X, Liu Z, Liu X (2017) EGCG ameliorates diet‐induced metabolic syndrome associating with the circadian clock. Biochim Biophys Acta Mol Basis Dis 1863: 1575–1589 [DOI] [PubMed] [Google Scholar]
- Minami Y, Kasukawa T, Kakazu Y, Iigo M, Sugimoto M, Ikeda S, Yasui A, van der Horst GTJ, Soga T, Ueda HR (2009) Measurement of internal body time by blood metabolomics. Proc Natl Acad Sci U S A 106: 9890–9895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran‐Salvador E, Lopez‐Parra M, Garcia‐Alonso V, Titos E, Martinez‐Clemente M, Gonzalez‐Periz A, Lopez‐Vicario C, Barak Y, Arroyo V, Claria J (2011) Role for PPARgamma in obesity‐induced hepatic steatosis as determined by hepatocyte‐ and macrophage‐specific conditional knockouts. FASEB J 25: 2538–2550 [DOI] [PubMed] [Google Scholar]
- Morita M, Gravel S‐P, Chénard V, Sikström K, Zheng L, Alain T, Gandin V, Avizonis D, Arguello M, Zakaria C et al (2013) mTORC1 controls mitochondrial activity and biogenesis through 4E‐BP‐dependent translational regulation. Cell Metab 18: 698–711 [DOI] [PubMed] [Google Scholar]
- Morris CJ, Purvis TE, Mistretta J, Scheer FA (2016) Effects of the internal circadian system and circadian misalignment on glucose tolerance in chronic shift workers. J Clin Endocrinol Metab 101: 1066–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherji A, Kobiita A, Chambon P (2015) Shifting the feeding of mice to the rest phase creates metabolic alterations, which, on their own, shift the peripheral circadian clocks by 12 hours. Proc Natl Acad Sci U S A 112: E6683–E6690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Tognini P, Liu Y, Eckel‐Mahan KL, Baldi P, Sassone‐Corsi P (2016) Gut microbiota directs PPARgamma‐driven reprogramming of the liver circadian clock by nutritional challenge. EMBO Rep 17: 1292–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadal A, Marrero PF, Haro D (2002) Down‐regulation of the mitochondrial 3‐hydroxy‐3‐methylglutaryl‐CoA synthase gene by insulin: the role of the forkhead transcription factor FKHRL1. Biochem J 366: 289–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair KS, Short KR (2005) Hormonal and signaling role of branched‐chain amino acids. J Nutr 135: 1547S–1552S [DOI] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone‐Corsi P (2008) The NAD+‐dependent deacetylase SIRT1 modulates CLOCK‐mediated chromatin remodeling and circadian control. Cell 134: 329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone‐Corsi P (2009) Circadian control of the NAD+ salvage pathway by CLOCK‐SIRT1. Science 324: 654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JC, Covarrubias AJ, Zhao M, Yu X, Gut P, Ng CP, Huang Y, Haldar S, Verdin E (2017) Ketogenic diet reduces midlife mortality and improves memory in aging mice. Cell Metab 26: 547–557.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohara K, Mallampalli V, Nemkov T, Wirianto M, Yang J, Ye Y, Sun Y, Han L, Esser KA, Mileykovskaya E et al (2019) Nobiletin fortifies mitochondrial respiration in skeletal muscle to promote healthy aging against metabolic challenge. Nat Commun 10: 3923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noshiro M, Usui E, Kawamoto T, Sato F, Nakashima A, Ueshima T, Honda K, Fujimoto K, Honma S, Honma K et al (2009) Liver X receptors (LXRalpha and LXRbeta) are potent regulators for hepatic Dec1 expression. Genes Cells 14: 29–40 [DOI] [PubMed] [Google Scholar]
- Nymo S, Coutinho SR, Jorgensen J, Rehfeld JF, Truby H, Kulseng B, Martins C (2017) Timeline of changes in appetite during weight loss with a ketogenic diet. Int J Obes 41: 1224–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K, Miyazaki K, Kadota K, Kikuno R, Nagase T, Atsumi G‐I, Ohkura N, Azama T, Mesaki M, Yukimasa S et al (2003) Genome‐wide expression analysis of mouse liver reveals CLOCK‐regulated circadian output genes. J Biol Chem 278: 41519–41527 [DOI] [PubMed] [Google Scholar]
- Oishi K, Shirai H, Ishida N (2005) CLOCK is involved in the circadian transactivation of peroxisome‐proliferator‐activated receptor alpha (PPARalpha) in mice. Biochem J 386: 575–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan K, Robles MS, Westerling T, Weitz CJ (2012) Feedback regulation of transcriptional termination by the mammalian circadian clock PERIOD complex. Science 337: 599–602 [DOI] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB (2002) Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109: 307–320 [DOI] [PubMed] [Google Scholar]
- Park S‐J, Ahmad F, Philp A, Baar K, Williams T, Luo H, Ke H, Rehmann H, Taussig R, Brown A et al (2012) Resveratrol ameliorates aging‐related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell 148: 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partch CL, Green CB, Takahashi JS (2014) Molecular architecture of the mammalian circadian clock. Trends Cell Biol 24: 90–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschos GK, Ibrahim S, Song W‐L, Kunieda T, Grant G, Reyes TM, Bradfield CA, Vaughan CH, Eiden M, Masoodi M et al (2012) Obesity in mice with adipocyte‐specific deletion of clock component Arntl. Nat Med 18: 1768–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschke E, Peschke D, Hammer T, Csernus V (1997) Influence of melatonin and serotonin on glucose‐stimulated insulin release from perifused rat pancreatic islets in vitro . J Pineal Res 23: 156–163 [DOI] [PubMed] [Google Scholar]
- Picinato MC, Haber EP, Cipolla‐Neto J, Curi R, de Oliveira Carvalho CR, Carpinelli AR (2002) Melatonin inhibits insulin secretion and decreases PKA levels without interfering with glucose metabolism in rat pancreatic islets. J Pineal Res 33: 156–160 [DOI] [PubMed] [Google Scholar]
- Puisac B, Ramos M, Arnedo M, Menao S, Gil‐Rodríguez MC, Teresa‐Rodrigo ME, Pié A, de Karam JC, Wesselink J‐J, Giménez I et al (2012) Characterization of splice variants of the genes encoding human mitochondrial HMG‐CoA lyase and HMG‐CoA synthase, the main enzymes of the ketogenesis pathway. Mol Biol Rep 39: 4777–4785 [DOI] [PubMed] [Google Scholar]
- Qi G, Wu W, Mi Y, Shi R, Sun K, Li R, Liu X, Liu X (2018) Tea polyphenols direct Bmal1‐driven ameliorating of the redox imbalance and mitochondrial dysfunction in hepatocytes. Food Chem Toxicol 122: 181–193 [DOI] [PubMed] [Google Scholar]
- Qu M, Duffy T, Hirota T, Kay SA (2018) Nuclear receptor HNF4A transrepresses CLOCK:BMAL1 and modulates tissue‐specific circadian networks. Proc Natl Acad Sci U S A 115: E12305–E12312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadro L, Blaner WS, Salchow DJ, Vogel S, Piantedosi R, Gouras P, Freeman S, Cosma MP, Colantuoni V, Gottesman ME (1999) Impaired retinal function and vitamin A availability in mice lacking retinol‐binding protein. EMBO J 18: 4633–4644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quagliarini F, Mir AA, Balazs K, Wierer M, Dyar KA, Jouffe C, Makris K, Hawe J, Heinig M, Filipp FV et al (2019) Cistromic reprogramming of the diurnal glucocorticoid hormone response by high‐fat diet. Mol Cell 76: 531–545.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan C, Kathale ND, Liu D, Lee C, Freeman DA, Hogenesch JB, Cao R, Liu AC (2018) mTOR signaling regulates central and peripheral circadian clock function. PLoS Genet 14: e1007369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR (2002) Coordination of circadian timing in mammals. Nature 418: 935–941 [DOI] [PubMed] [Google Scholar]
- Rhee J, Inoue Y, Yoon JC, Puigserver P, Fan M, Gonzalez FJ, Spiegelman BM (2003) Regulation of hepatic fasting response by PPARgamma coactivator‐1alpha (PGC‐1): requirement for hepatocyte nuclear factor 4alpha in gluconeogenesis. Proc Natl Acad Sci U S A 100: 4012–4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MN, Wallace MA, Tomilov AA, Zhou Z, Marcotte GR, Tran D, Perez G, Gutierrez‐Casado E, Koike S, Knotts TA et al (2017) A ketogenic diet extends longevity and healthspan in adult mice. Cell Metab 26: 539–546.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruderman NB, Xu XJ, Nelson L, Cacicedo JM, Saha AK, Lan F, Ido Y (2010) AMPK and SIRT1: a long‐standing partnership? Am J Physiol Endocrinol Metab 298: E751–E760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahar S, Sassone‐Corsi P (2012) Regulation of metabolism: the circadian clock dictates the time. Trends Endocrinol Metab 23: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahar S, Zocchi L, Kinoshita C, Borrelli E, Sassone‐Corsi P (2010) Regulation of BMAL1 protein stability and circadian function by GSK3beta‐mediated phosphorylation. PLoS One 5: e8561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini C, Suter DM, Liani A, Gos P, Schibler U (2011) The mammalian circadian timing system: synchronization of peripheral clocks. Cold Spring Harb Symp Quant Biol 76: 39–47 [DOI] [PubMed] [Google Scholar]
- Sato F, Muragaki Y, Zhang Y (2015) DEC1 negatively regulates AMPK activity via LKB1. Biochem Biophys Res Commun 467: 711–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Murakami M, Node K, Matsumura R, Akashi M (2014) The role of the endocrine system in feeding‐induced tissue‐specific circadian entrainment. Cell Rep 8: 393–401 [DOI] [PubMed] [Google Scholar]
- Sato S, Parr EB, Devlin BL, Hawley JA, Sassone‐Corsi P (2018) Human metabolomics reveal daily variations under nutritional challenges specific to serum and skeletal muscle. Mol Metab 16: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Solanas G, Peixoto FO, Bee L, Symeonidi A, Schmidt MS, Brenner C, Masri S, Benitah SA, Sassone‐Corsi P (2017) Circadian reprogramming in the liver identifies metabolic pathways of aging. Cell 170: 664–677.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Watanabe Y, Nishimura Y, Inoue M, Morita A, Miura S (2019) Acute fructose intake suppresses fasting‐induced hepatic gluconeogenesis through the AKT‐FoxO1 pathway. Biochem Biophys Rep 18: 100638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U, Sassone‐Corsi P (2002) A web of circadian pacemakers. Cell 111: 919–922 [DOI] [PubMed] [Google Scholar]
- Schwer B, Verdin E (2008) Conserved metabolic regulatory functions of sirtuins. Cell Metab 7: 104–112 [DOI] [PubMed] [Google Scholar]
- Selhub J, Miller JW (1992) The pathogenesis of homocysteinemia: interruption of the coordinate regulation by S‐adenosylmethionine of the remethylation and transsulfuration of homocysteine. Am J Clin Nutr 55: 131–138 [DOI] [PubMed] [Google Scholar]
- Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GTJ et al (2000) Interacting molecular loops in the mammalian circadian clock. Science 288: 1013–1019 [DOI] [PubMed] [Google Scholar]
- Sherman H, Gutman R, Chapnik N, Meylan J, le Coutre J, Froy O (2011) Caffeine alters circadian rhythms and expression of disease and metabolic markers. Int J Biochem Cell Biol 43: 829–838 [DOI] [PubMed] [Google Scholar]
- Shi SQ, Ansari TS, McGuinness OP, Wasserman DH, Johnson CH (2013) Circadian disruption leads to insulin resistance and obesity. Curr Biol 23: 372–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazu T, Hirschey MD, Newman J, He W, Shirakawa K, Le Moan N, Grueter CA, Lim H, Saunders LR, Stevens RD et al (2013) Suppression of oxidative stress by beta‐hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science 339: 211–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimba S, Ogawa T, Hitosugi S, Ichihashi Y, Nakadaira Y, Kobayashi M, Tezuka M, Kosuge Y, Ishige K, Ito Y et al (2011) Deficient of a clock gene, brain and muscle Arnt‐like protein‐1 (BMAL1), induces dyslipidemia and ectopic fat formation. PLoS One 6: e25231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki A, Misawa K, Ikeda Y, Haraguchi A, Kamagata M, Tahara Y, Shibata S (2017) Potent effects of flavonoid nobiletin on amplitude, period, and phase of the circadian clock rhythm in PER2:LUCIFERASE mouse embryonic fibroblasts. PLoS One 12: e0170904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiwa M, Yoneda M, Okubo H, Ohno H, Kobuke K, Monzen Y, Kishimoto R, Nakatsu Y, Asano T, Kohno N (2015) Distinct time course of the decrease in hepatic AMP‐activated protein kinase and Akt phosphorylation in mice fed a high fat diet. PLoS One 10: e0135554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter KR, Anderson V, Cakora P, Owen A, Lo K, Crossland J, South AC, Felder MR, Vrana PB (2014) Pleiotropic effects of a methyl donor diet in a novel animal model. PLoS One 9: e104942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So AY, Bernal TU, Pillsbury ML, Yamamoto KR, Feldman BJ (2009) Glucocorticoid regulation of the circadian clock modulates glucose homeostasis. Proc Natl Acad Sci U S A 106: 17582–17587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solt LA, Wang Y, Banerjee S, Hughes T, Kojetin DJ, Lundasen T, Shin Y, Liu J, Cameron MD, Noel R et al (2012) Regulation of circadian behaviour and metabolism by synthetic REV‐ERB agonists. Nature 485: 62–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wang Y, Song Y, Cheng XR, Xia S, Rahman MR, Shi Y, Le G (2015) Resveratrol restores the circadian rhythmic disorder of lipid metabolism induced by high‐fat diet in mice. Biochem Biophys Res Commun 458: 86–91 [DOI] [PubMed] [Google Scholar]
- Sun S, Hanzawa F, Umeki M, Ikeda S, Mochizuki S, Oda H (2018) Time‐restricted feeding suppresses excess sucrose‐induced plasma and liver lipid accumulation in rats. PLoS One 13: e0201261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Hanzawa F, Kim D, Umeki M, Nakajima S, Sakai K, Ikeda S, Mochizuki S, Oda H (2019a) Circadian rhythm‐dependent induction of hepatic lipogenic gene expression in rats fed a high‐sucrose diet. J Biol Chem 294: 15206–15217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Hanzawa F, Umeki M, Matsuyama Y, Nishimura N, Ikeda S, Mochizuki S, Oda H (2019b) Impacts of high‐sucrose diet on circadian rhythms in the small intestine of rats. Chronobiol Int 36: 826–837 [DOI] [PubMed] [Google Scholar]
- Sundaram S, Yan L (2016) Time‐restricted feeding reduces adiposity in mice fed a high‐fat diet. Nutr Res 36: 603–611 [DOI] [PubMed] [Google Scholar]
- Sutton EF, Beyl R, Early KS, Cefalu WT, Ravussin E, Peterson CM (2018) Early time‐restricted feeding improves insulin sensitivity, blood pressure, and oxidative stress even without weight loss in men with prediabetes. Cell Metab 27: 1212–1221.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Inoue I, Nakajima Y, Seo M, Nakano T, Yang F, Kumagai M, Komoda T, Awata T, Ikeda M et al (2010) A promoter in the novel exon of hPPARγ directs the circadian expression of PPARγ. J Atheroscler Thromb 17: 73–83 [DOI] [PubMed] [Google Scholar]
- Tognini P, Murakami M, Liu Y, Eckel‐Mahan KL, Newman JC, Verdin E, Baldi P, Sassone‐Corsi P (2017) Distinct circadian signatures in liver and gut clocks revealed by ketogenic diet. Cell Metab 26: 523–538.e5 [DOI] [PubMed] [Google Scholar]
- Torra IP, Tsibulsky V, Delaunay F, Saladin R, Laudet V, Fruchart JC, Kosykh V, Staels B (2000) Circadian and glucocorticoid regulation of Rev‐erbalpha expression in liver. Endocrinology 141: 3799–3806 [DOI] [PubMed] [Google Scholar]
- Ueyama T, Krout KE, Nguyen XV, Karpitskiy V, Kollert A, Mettenleiter TC, Loewy AD (1999) Suprachiasmatic nucleus: a central autonomic clock. Nat Neurosci 2: 1051–1053 [DOI] [PubMed] [Google Scholar]
- Um JH, Pendergast JS, Springer DA, Foretz M, Viollet B, Brown A, Kim MK, Yamazaki S, Chung JH (2011) AMPK regulates circadian rhythms in a tissue‐ and isoform‐specific manner. PLoS One 6: e18450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um JH, Yang S, Yamazaki S, Kang H, Viollet B, Foretz M, Chung JH (2007) Activation of 5'‐AMP‐activated kinase with diabetes drug metformin induces casein kinase Iepsilon (CKIepsilon)‐dependent degradation of clock protein mPer2. J Biol Chem 282: 20794–20798 [DOI] [PubMed] [Google Scholar]
- Verdin E (2015) NAD(+) in aging, metabolism, and neurodegeneration. Science 350: 1208–1213 [DOI] [PubMed] [Google Scholar]
- Vieira E, Marroqui L, Batista TM, Caballero‐Garrido E, Carneiro EM, Boschero AC, Nadal A, Quesada I (2012) The clock gene Rev‐erbalpha regulates pancreatic beta‐cell function: modulation by leptin and high‐fat diet. Endocrinology 153: 592–601 [DOI] [PubMed] [Google Scholar]
- Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S (2009) Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci U S A 106: 21453–21458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan M, Leavens K, Hunter R, Koren S, von Wilamowitz‐Moellendorff A, Lu M, Satapati S, Chu Q, Sakamoto K, Burgess S et al (2013) A noncanonical, GSK3‐independent pathway controls postprandial hepatic glycogen deposition. Cell Metab 18: 99–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LF, Wang XN, Huang CC, Hu L, Xiao YF, Guan XH, Qian YS, Deng KY, Xin HB (2017) Inhibition of NAMPT aggravates high fat diet‐induced hepatic steatosis in mice through regulating Sirt1/AMPKalpha/SREBP1 signaling pathway. Lipids Health Dis 16: 82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Yin Y, Zhang W (2018) Ghrelin restores the disruption of the circadian clock in steatotic liver. Int J Mol Sci 19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzman ED, Fukushima D, Nogeire C, Roffwarg H, Gallagher TF, Hellman L (1971) Twenty‐four hour pattern of the episodic secretion of cortisol in normal subjects. J Clin Endocrinol Metab 33: 14–22 [DOI] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB (2009) ATP‐citrate lyase links cellular metabolism to histone acetylation. Science 324: 1076–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windle RJ, Wood SA, Shanks N, Lightman SL, Ingram CD (1998) Ultradian rhythm of basal corticosterone release in the female rat: dynamic interaction with the response to acute stress. Endocrinology 139: 443–450 [DOI] [PubMed] [Google Scholar]
- Woodie LN, Luo Y, Wayne MJ, Graff EC, Ahmed B, O'Neill AM, Greene MW (2018) Restricted feeding for 9h in the active period partially abrogates the detrimental metabolic effects of a Western diet with liquid sugar consumption in mice. Metab, Clin Exp 82: 1–13 [DOI] [PubMed] [Google Scholar]
- Xie Z, Zhang D, Chung D, Tang Z, Huang H, Dai L, Qi S, Li J, Colak G, Chen Y et al (2016) metabolic regulation of gene expression by histone lysine beta‐hydroxybutyrylation. Mol Cell 62: 194–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamajuku D, Inagaki T, Haruma T, Okubo S, Kataoka Y, Kobayashi S, Ikegami K, Laurent T, Kojima T, Noutomi K et al (2012) Real‐time monitoring in three‐dimensional hepatocytes reveals that insulin acts as a synchronizer for liver clock. Sci Rep 2: 439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Nakahata Y, Tanaka M, Yoshida M, Soma H, Shinohara K, Yasuda A, Mamine T, Takumi T (2005) Acute physical stress elevates mouse period1 mRNA expression in mouse peripheral tissues via a glucocorticoid‐responsive element. J Biol Chem 280: 42036–42043 [DOI] [PubMed] [Google Scholar]
- Yang X, Qian K (2017) Protein O‐GlcNAcylation: emerging mechanisms and functions. Nat Rev Mol Cell Biol 18: 452–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Wang J, Klein PS, Lazar MA (2006) Nuclear receptor Rev‐erbalpha is a critical lithium‐sensitive component of the circadian clock. Science 311: 1002–1005 [DOI] [PubMed] [Google Scholar]
- Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB et al (2007) Rev‐erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science 318: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Zhang D, Tong X, Arthurs B, Guha A, Rui L, Kamath A, Inoki K, Yin L (2014a) Liver clock protein BMAL1 promotes de novo lipogenesis through insulin‐mTORC2‐AKT signaling. J Biol Chem 289: 25925–25935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB (2014b) A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc Natl Acad Sci U S A 111: 16219–16224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Fang B, Emmett MJ, Damle M, Sun Z, Feng D, Armour SM, Remsberg JR, Jager J, Soccio RE et al (2015) Gene regulation. Discrete functions of nuclear receptor Rev‐erbalpha couple metabolism to the clock. Science 348: 1488–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y‐L, Guo H, Zhang C‐S, Lin S‐Y, Yin Z, Peng Y, Luo H, Shi Y, Lian G, Zhang C et al (2013) AMP as a low‐energy charge signal autonomously initiates assembly of AXIN‐AMPK‐LKB1 complex for AMPK activation. Cell Metab 18: 546–555 [DOI] [PubMed] [Google Scholar]
- Zhu Z, Jiang W, McGinley JN, Thompson HJ (2005) 2‐Deoxyglucose as an energy restriction mimetic agent: effects on mammary carcinogenesis and on mammary tumor cell growth in vitro . Cancer Res 65: 7023–7030 [DOI] [PubMed] [Google Scholar]


