Abstract
Some human preimplantation embryos are chromosomally mosaic. For technical reasons, estimates of the overall frequency vary widely from <15 to >90% and the true frequency remains unknown. Aneuploid/diploid and aneuploid/aneuploid mosaics typically arise during early cleavage stages before the embryonic genome is fully activated and when cell cycle checkpoints are not operating normally. Other mosaics include chaotic aneuploid mosaics and mixoploids, some of which arise by abnormal chromosome segregation at the first cleavage division. Chimaeras are similar to mosaics, in having two genetically distinct cell populations, but they arise from more than one zygote and occur less often. After implantation, the frequency of mosaic embryos declines to about 2% and most are trisomic/diploid mosaics, with trisomic cells confined to the placenta. Thus, few babies are born with chromosomal mosaicism. This review discusses the origin of different types of chromosomal mosaics and chimaeras; their fate and the relationship between preimplantation chromosomal mosaicism and confined placental mosaicism in human conceptuses and animal models. Abnormal cells in mosaic embryos may be depleted by cell death, other types of cell selection or cell correction but the most severely affected mosaic embryos probably die. Trisomic cells could become restricted to placental lineages if cell selection or correction is less effective in placental lineages and/or they are preferentially allocated to a placental lineage. However, the relationship between preimplantation mosaicism and confined placental mosaicism may be complex because the specific chromosome(s) involved will influence whether chromosomally abnormal cells survive predominately in the placental trophoblast and/or placental mesenchyme.
Lay summary
Human cells normally have 23 pairs of chromosomes, which carry the genes. During the first few days of development, some human embryos are chromosomal mosaics. These mosaic embryos have both normal cells and cells with an abnormal number of chromosomes, which arise from the same fertilised egg. (More rarely, the different cell populations arise from more than one fertilised egg and these embryos are called chimaeras.) If chromosomally abnormal cells survive to term, they could cause birth defects. However, few abnormal cells survive and those that do are usually confined to the placenta, where they are less likely to cause harm. It is not yet understood how this restriction occurs but the type of chromosomal abnormality influences which placental tissues are affected. This review discusses the origin of different types of chromosomally abnormal cells, their fate and how they might become confined to the placenta in humans and animal models.
Key Words: chromosomal mosaic, chimaera, confined placental mosaicism, aneuploidy, mixoploidy
Introduction
Mosaics are multicellular organisms composed of two or more genetically distinct cell populations that arise from a single zygote (Anderson et al. 1951, Ford 1969). Chromosomal mosaicism arises if a chromosomally different cell population is produced at any stage of development. Mosaicism may affect germ cells, somatic tissues or both and somatic mosaicism may be generalised or confined to a specific tissue or group of tissues. Generalised mosaicism is likely to arise early in development whereas confined mosaicism may arise at any stage. The clinical consequences of mosaicism for a somatic chromosomal abnormality depend on the abnormality; when and where it arises; the tissue distribution and the percentage of abnormal cells overall or in specific tissues (Spinner & Conlin 2014, Papavassiliou et al. 2015).
Chimaeras are similar to mosaics in having two genetically distinct cell populations but they arise in different ways. A chimaera was originally defined as ‘an organism whose cells derive from two or more distinct zygote lineages’ (Anderson et al. 1951). It has also been defined more broadly as ‘any composite animal or plant in which the different cell populations are derived from more than one fertilized egg or the union of more than two gametes” (McLaren 1976). This revised definition includes individuals where the two cell populations are from more than one zygote but not necessarily two whole zygotes.
Chimaeras are often classified as primary chimaeras, which arise very early in development, so all developmental lineages may be affected, or secondary chimaeras, which arise at postimplantation or postnatal stages (Ford 1969). Secondary chimaeras are beyond the scope of this review but include blood chimaeras, arising by placental fusion in dizygotic twin pregnancies (Dunsford et al. 1953); microchimaeras, which have a small number of donor cells from another individual that persist from bidirectional feto-maternal, cross-placental cell trafficking during pregnancy (Boddy et al. 2015) and those produced artificially by tissue transplantation.
Mosaics with numerical chromosome abnormalities contain either aneuploid or polyploid cells. Aneuploidy is when the number of chromosomes is not an exact multiple of the haploid number (n). It includes both chromosome gain (e.g. trisomy; Ts) and chromosome loss (e.g. monosomy; Ms) and may involve multiple chromosomes. Polyploid cells have an exact multiple of the haploid number of chromosomes but more than the diploid (2n) number. Polyploidy usually occurs as triploidy (3n; three haploid sets) or tetraploidy (4n; four haploid sets). Euploidy is when there is an exact multiple of the haploid number of chromosomes and includes haploidy, diploidy and polyploidy.
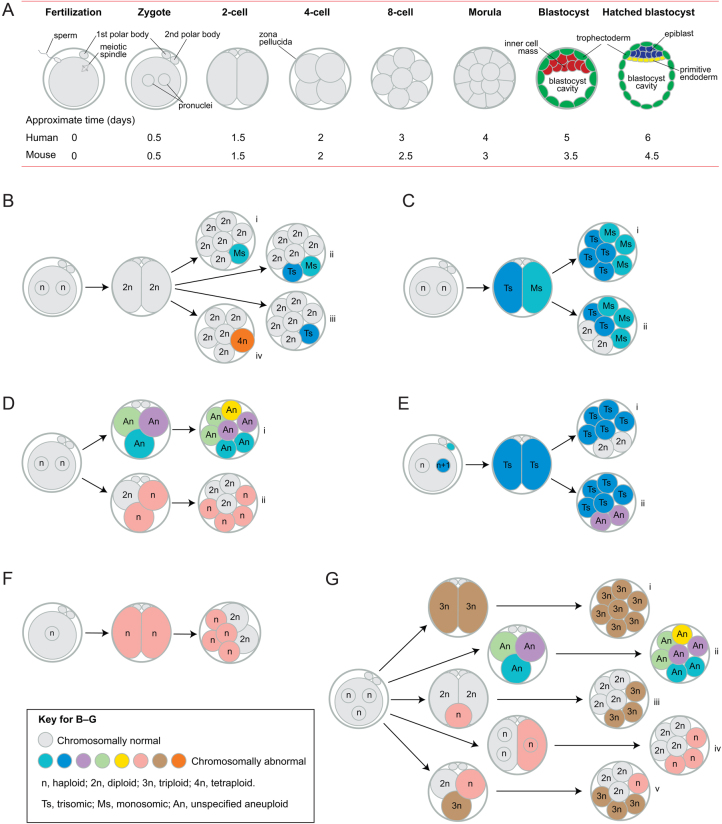
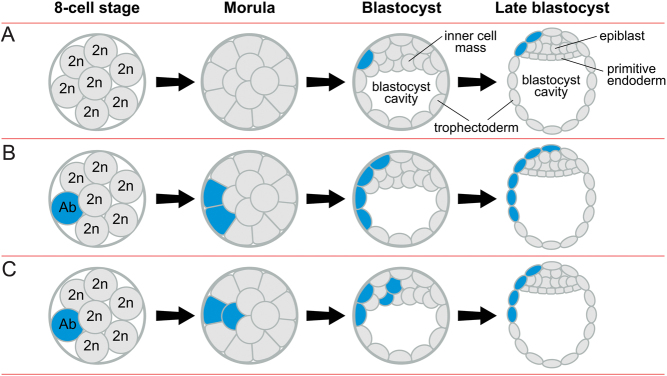
After fertilisation, the preimplantation embryo undergoes three cleavage divisions to the eight-cell stage, then compacts to form a morula, which cavitates to form a blastocyst, with an inner cell mass (ICM) and an outer layer of trophectoderm cells (Fig. 1A). The ICM divides into the epiblast and primitive endoderm (hypoblast) and the human blastocyst implants when it has about 256 cells (Niakan et al. 2012). Some human preimplantation embryos are chromosomal mosaics (Zenzes & Casper 1992, Delhanty et al. 1993) but estimates of their frequency vary widely from <15 to >90% due to the variety and limitations of techniques employed (discussed below) and the true frequency remains unknown. The occurrence of chromosomally mosaic preimplantation embryos has clinical implications for preimplantation genetic testing and this has been widely discussed elsewhere (Popovic et al. 2018).
Figure 1.
Preimplantation development, mosaicism and mixoploidy. (A) After fertilisation, the preimplantation embryo undergoes three cleavage divisions to the eight-cell stage (cleavage divisions are mitotic divisions that increase cell number but not embryo size and cells produced by cleavage division are called blastomeres). The embryo then compacts to form a morula, which cavitates to form a blastocyst, with an outer layer of trophectoderm cells surrounding the inner cell mass (ICM) and the blastocyst cavity. By the late blastocyst stage, the ICM forms the epiblast and primitive endoderm (hypoblast). (B) A normal diploid embryo may produce (i) monosomic/diploid, (ii) monosomic/trisomic/diploid, (iii) trisomic/diploid (origin may be indirect – see text) and (iv) tetraploid/diploid mosaics. (C) Non-disjunction, at the first mitotic division, would produce (i) monosomic/trisomic mosaics or (ii) other types of mosaics, including monosomic/trisomic/diploid mosaics, if further changes, such as trisomic rescue, occurred. (D) Normal bipronuclear zygotes sometimes form tripolar spindles and (i) individual chromosomes may segregate abnormally to produce a chaotic mosaic or (ii) entire haploid sets of chromosomes may segregate to form a haploid/diploid mixoploid. (E) A trisomic zygote could produce (i) trisomic/diploid or (ii) various types of trisomic/aneuploid mosaics. (F) A haploid zygote often produces some diploid cells by endoreplication, so forming a haploid/diploid mosaic. (G) A tripronuclear zygote may (i) produce a non-mosaic, triploid embryo; (ii) form a tripolar spindle and produce a chaotic mosaic which may continue to be unstable, (iii) extrude a small haploid cell which may fuse with a diploid cell to produce a triploid/diploid mixoploid, (iv) undergo atypical early cytokinesis so the pronuclei segregate passively to the two blastomeres or (v) form a tripolar spindle and segregate entire haploid sets of chromosomes to initially form a haploid/triploid/diploid mixoploid. See Fig. 2 for other details.
There are substantially fewer chromosomal mosaics at postimplantation stages and about 2% of chorionic villus samples (CVS) are chromosomally mosaic (Wang et al. 1993, Pittalis et al. 1994, Huang et al. 2009, Malvestiti et al. 2015). Most of these mosaics occur as confined placental mosaicism (CPM) (Malvestiti et al. 2015), which is when chromosomally abnormal cells are present in the placenta but not in the fetus. This was first described by Kalousek and Dill (1983), although Warburton et al. (1978) had previously suggested its occurrence.
This review introduces different types of mosaic preimplantation embryos, involving whole chromosomes, and considers (i) how human preimplantation chromosomal mosaics and primary chimaeras arise; (ii) their fate; (iii) the relationship between preimplantation chromosomal mosaicism and CPM; (iv) some animal models of chromosomal mosaicism.
Literature search
This is a narrative review of a broad topic and the authors decided which articles to include. Relevant articles were identified predominantly using Web of Science and PubMed to search the literature and the weekly PubCrawler alerting service to update them. Other relevant papers cited in these articles were also accessed. Search terms included ‘chimaera’, ‘chimera’, ‘chromosomal mosaic’, ‘confined placental mosaicism’, ‘mixoploidy’, ‘preimplantation genetic screening’ and ‘preimplantation genetic testing for aneuploidy’ in combination with other keywords relevant to the topic.
Identification of mosaic preimplantation embryos
The extent of human preimplantation mosaicism only became apparent after the introduction of in vitro fertilisation (IVF). Initially, classical cytogenetics was used to determine the chromosomal constitution of preimplantation embryos (Angell et al. 1983) and an early review concluded that aneuploid/diploid mosaicism was the most common type of chromosomal abnormality (Zenzes & Casper 1992). However, the paucity of analysable metaphase spreads from preimplantation embryos made conventional cytogenetics inefficient so other methods were evaluated, including in situ hybridisation to specific chromosomes (Angell et al. 1987, West et al. 1987, Penketh et al. 1989). The introduction of multicolour fluorescence in situ hybridisation (FISH), to rapidly identify the copy number of several different chromosomes simultaneously in individual interphase cells of embryos, was a significant advance and provided important evidence for mosaicism among human preimplantation embryos (Griffin et al. 1992, Delhanty et al. 1993, Munné et al. 1993, Coonen et al. 1994, Munné et al. 1994).
FISH was improved to detect 8–12 different chromosomes and became widely used for preimplantation genetic screening for aneuploidy (PGS) for embryos produced by IVF. PGS was replaced by preimplantation genetic testing for aneuploidy (PGT-A) and FISH was superseded by several types of comprehensive chromosome screening (CCS) methods, which can analyse all 24 human chromosomes (Chen et al. 2020, Viotti 2020). CCS platforms that have been used for PGT-A include comparative genomic hybridisation (CGH) to metaphase chromosomes, array-CGH (aCGH), SNP microarrays, quantitative PCR and next-generation sequencing (NGS). NGS has some advantages and became widely used (Fiorentino et al. 2014). Additional powerful new techniques being used to investigate chromosomes in individual cells of embryos include evaluating haplotype and chromosome copy number by haplarithmisis (Destouni et al. 2016) and RNA-seq gene expression profiling (Starostik et al. 2020).
Estimates of the frequency of chromosomally mosaic preimplantation embryos vary. They depend on the detection method, numbers of cells and chromosomes analysed, the source of the cells and the criteria used to classify an embryo as a mosaic. Embryos rejected for transfer after IVF may be atypical and estimates based on a small biopsy may not be representative of the whole embryo. In some studies, an embryo is not classified as mosaic unless the proportion of abnormal cells reaches a specific threshold (van Echten-Arends et al. 2011). Also, blastocysts often contain tetraploid cells but studies differ as to whether they are counted as mosaic or normal.
An early 3–5-chromosome FISH study of embryos not selected for transfer reported 15.2% mosaics at the two to four-cell stage, 49.4% at five to eight cells and 58.1% by the morula stage (Bielanska et al. 2002). van Echten-Arends et al. (2011) reviewed 36 studies, comprising 815 whole embryos analysed by FISH or CCS, and reported mosaicism frequencies from 15 to >90% and a mean of 72% for cleavage stage embryos with at least eight chromosomes analysed. Overall, 22% were diploid, 59% were aneuploid/diploid mosaics, 14% were aneuploid/aneuploid mosaics and 5% had other numerical chromosomal abnormalities.
The high frequencies of mosaicism estimated by FISH are perhaps surprising, given that not all chromosomes are tested. FISH is less efficient with interphase nuclei than metaphase spreads (Ruangvutilert et al. 2000) and a theoretical model of the accuracy of 5-chromosome FISH predicted that it would overestimate the frequency of sporadic aneuploid cells and, therefore, the frequency of mosaicism (Scriven & Bossuyt 2010). Comparison of FISH and SNP-microarray analyses also suggested that FISH might overestimate the frequency of chromosomal mosaicism in cleavage stage embryos (Treff et al. 2010) although another study reported more consistent results for FISH, CGH and aCGH analyses of blastocysts (Fragouli et al. 2011). To what extent overestimation of mosaicism by FISH is offset by not testing all the chromosomes is unclear.
Marin et al. (2017) cited blastocyst mosaicism frequency estimates ranging from 4.8 to 44%, from NGS and other CCS studies of trophectoderm biopsies, with a typical value of approximately 15% for studies with multiple biopsies. An alternative method of estimating the frequency of mosaicism, based on the frequency of discordant CCS results after multiple biopsies, predicted 35.7% of blastocysts were mosaic, including 3.7% with reciprocal aneuploidies, from an analysis of 1124 embryos from 26 studies (Marin et al. 2021).
Starostik et al. (2020) analysed single-cell RNA-seq data from cells of 74 whole embryos on days 4–7 and estimated that, overall, 74% of morulae and blastocysts were mosaics with at least one cell affected by a mitotic error. However, the accuracy of RNA-seq analysis could be hampered by differences in gene expression among cells of early embryos and any differences in expression of maternal and paternal alleles.
Not all CCS platforms can detect mosaicism or polyploidy and results in the normal range are usually cited as euploid rather than diploid. When used in conjunction with SNPs, NGS can detect triploidy (Marin et al. 2018) but not tetraploidy, with equal numbers of maternal and paternal chromosomes. Although, in principle, NGS can detect mosaicism in PGT-A samples, this is not straightforward in a single trophectoderm biopsy of 5–10 cells because of the presence of two cell populations in a putative mosaic sample is inferred indirectly. NGS can produce accurate results with control mixtures of 5–10 aneuploid and diploid cells but is more robust with larger samples (Fragouli et al. 2017, Treff & Franasiak 2017, Biricik et al. 2021).
NGS produces different chromosome copy number profiles for monosomy, disomy and trisomy. Intermediate profiles are consistent with mosaicism but could also result from technical artefacts, so the frequency of mosaicism may be overestimated (Capalbo & Rienzi 2017, Capalbo et al. 2017). Conversely, however, other technical issues would cause the frequency of mosaics to be underestimated. One cell population present in a mosaic blastocyst may often be excluded from a small biopsy. Even if the biopsy is representative, some unbalanced mosaics, with a low proportion of one cell population, will be undetectable and mosaicism may be obscured if a biopsy contains both trisomic and monosomic cells of a reciprocal aneuploidy and is analysed as a single sample (Scott & Galliano 2016, Capalbo & Rienzi 2017, Treff & Franasiak 2017, Gleicher et al. 2021).
These technical limitations cast doubt on the reliability of mosaicism frequency estimates obtained by NGS analysis of small biopsies taken for PGT-A. Some authors suggest that this is likely to overestimate the frequency of mosaicism (Capalbo & Rienzi 2017, Capalbo et al. 2017, Marin et al. 2021, Treff & Marin 2021) but others consider that underestimation is more likely overall (Fragouli et al. 2017, Gleicher et al. 2021). Clinical implications of these technical limitations are discussed elsewhere (Capalbo et al. 2017, Paulson & Treff 2020, Gleicher et al. 2021, Treff & Marin 2021, Viotti et al. 2021a) and are beyond the scope of this review.
Although the combination of cytogenetics, FISH, NGS and RNA-seq analysis provides strong evidence that a substantial proportion of human preimplantation embryos are chromosomally mosaic, the true frequency remains unknown. It has been argued that mosaic preimplantation embryos may be significantly less common than was believed hitherto (Capalbo et al. 2017). This is based, in part, on four sets of data from human blastocysts (Fragouli et al. 2008, Johnson et al. 2010, Northrop et al. 2010, Capalbo et al. 2013), compiled by Capalbo and Rienzi (2017). The focus of this compilation was exclusively on the frequency of aneuploid/euploid mosaic blastocysts, which was estimated to be 6.1% (11/181). However, none of the four individual studies was designed specifically to determine the frequency of mosaicism in a typical cohort of human embryos and if other putative mosaics are included the overall estimated frequency of mosaicism is higher (~15%). Fragouli et al. (2011) also contrasted the total frequency of putative mosaic blastocysts in their study (33%; 17/52) with the lower frequency of putative aneuploid/diploid mosaics (17%; 9/52) and the much lower frequency of putative aneuploid/diploid mosaics with a majority of normal cells (5.8%; 3/52). Although aneuploid/diploid mosaicism may be most relevant to clinical PGT-A, all types of numerical chromosomal mosaicism should be included if the aim is to estimate the total biological frequency of mosaic preimplantation embryos.
Estimates of the total frequency of mosaic embryos are consistently substantially higher among preimplantation embryos than that reported for postimplantation stages. This is typically estimated as only about 2% of CVS (Wang et al. 1993, Pittalis et al. 1994, Huang et al. 2009, Malvestiti et al. 2015). Thus, for the purpose of this review, we make the conventional assumption that the frequency of mosaic embryos declines after implantation (Fragouli et al. 2013). Nevertheless, identification of the true frequency of preimplantation mosaicism would require identification of the chromosomal constitution of every cell from representative embryos at different stages, using an accurate, single-cell method for all the chromosomes.
As all human preimplantation embryos analysed for mosaicism are produced using assisted reproductive technology (ART), such as IVF or intracytoplasmic sperm injection (ICSI), it is not known whether a similar frequency of mosaicism occurs naturally. Some evidence suggests that the mosaicism frequency may be affected by ovarian stimulation protocols (Baart et al. 2007) and in vitro procedures (Swain 2019) and this is supported by several animal studies (discussed later). Nevertheless, postimplantation mosaicism frequencies are similar for natural pregnancies and those conceived by ART (Huang et al. 2009, Zamani Esteki et al. 2019).
Origin of mosaic and chimaeric preimplantation embryos
Chromosomal mosaics are produced by postzygotic mitotic errors in either normal diploid embryos (Fig. 1B, C and D) or chromosomally abnormal embryos (Fig. 1E, F and G). One classification system distinguishes three broad categories: simple mosaics, produced by a single chromosomal error; complex mosaics; resulting from more than one error and chaotic mosaics, containing four or more chromosomally unrelated cell populations (Coonen et al. 2004). Chaotic mosaics may have no normal cells.
Mosaics with aneuploid cells from diploid embryos
Most mitotic errors occur during the first three cleavage divisions (Munné et al. 1994, 2002), when development is still largely dependent on mRNA, protein and mitochondria inherited from the oocyte. (Major waves of human zygotic genome activation occur at 4–8 and 8–16 cells; Jukam et al. 2017.) This vulnerability may underlie why mitotic errors are so frequent during the early cleavage divisions. Inadequate stores of maternal transcripts and proteins could result in dysregulation of mitosis in early embryos, leading to segregation errors and aneuploid/diploid mosaicism (Mantikou et al. 2012).
Anaphase lag and non-disjunction are mitotic segregation errors that occur in early embryos and can arise from faults in cell cycle checkpoints (Vazquez-Diez et al. 2019), mitotic spindles (Chatzimeletiou et al. 2005) or cohesin proteins, which hold the two sister chromatids together (Mantikou et al. 2012). Cell cycle checkpoints G1/S, G2/M and the spindle assembly checkpoint (SAC or metaphase/anaphase checkpoint) each ensure that part of the cell cycle is completed properly before progressing to the next step. If an error occurs, the cell cycle is arrested until the problem is corrected or apoptosis is activated. If chromosomes misalign in early mouse embryos, the SAC components assemble but fail to inhibit the anaphase-promoting complex/cyclosome (APC/C), so anaphase progresses when it should be delayed (Vazquez-Diez et al. 2019). The authors suggest this could be caused by the altered stoichiometry of signalling components during the transition from maternal to embryonic control of gene expression. In human embryos, the SAC is also unable to activate apoptosis, to remove cells with malsegregating chromosomes, until the blastocyst stage (Jacobs et al. 2017).
Anaphase lag is when a chromatid fails to migrate with others at anaphase and can occur when a chromatid attaches to microtubules from both spindle poles or if sister chromatids separate prematurely (perhaps because of cohesin deficiency). The lagging chromatid fails to get incorporated into a nucleus and is often lost, producing an Ms/2n mosaic embryo (Fig. 1Bi). A chromatid can be sequestered into a micronucleus where it may become shattered and rearranged by chromothripsis (Pellestor et al. 2014), which is reminiscent of ‘chromosome demolition’, proposed by Los et al. (1998). Alternatively, the micronucleus may be passively inherited by one daughter cell or form an independent cellular fragment, which may fuse with an adjacent cell (Chavez et al. 2012, Vazquez-Diez et al. 2016, Vazquez-Diez & FitzHarris 2018).
Non-disjunction occurs when sister chromatids fail to separate (possibly from delayed cohesin removal; Mantikou et al. 2012), resulting in reciprocal aneuploidy with one trisomic and one monosomic cell. This produces Ms/Ts/2n mosaics (Fig. 1Bii) or Ms/Ts mosaics if non-disjunction occurs at the first cleavage division (Fig. 1C). Vazquez-Diez and FitzHarris (2018) suggested that anaphase lag could produce an equivalent outcome if a micronucleus, containing the lagging chromosome, is incorporated into a diploid nucleus.
It is unclear how simple Ts/2n mosaics could arise from diploid zygotes (Fig. 1Biii). Some authors have suggested that an individual chromosome may undergo endoreplication to produce a trisomic cell but this seems unlikely because endoreplication is expected to replicate the entire genome (Shu et al. 2018). Some Ts/2n mosaics could be secondary modifications of Ms/Ts/2n mosaics but monosomic cells may not die until apoptosis begins in the morula (Hardy et al. 2001). Ts/2n mosaics can also arise from trisomic zygotes (see below).
Chaotic mosaics
Certain couples, including some with repeated implantation failure, are predisposed to producing complex or chaotic mosaic embryos (Fig. 1Di), suggesting a genetic basis (Delhanty et al. 1997, Mantzouratou et al. 2007, Voullaire et al. 2007). Mutations in genes affecting the SAC or chromatid-spindle attachment are implicated in a rare disease called mosaic variegated aneuploidy syndrome (Yost et al. 2017), which appears similar to complex or chaotic mosaicism in embryos. Comparable mutations might also affect embryos (Schmid et al. 2014), with only the mildest cases surviving postnatally. Evidence suggests that maternal inheritance of variants of the region of chromosome 4 that includes the candidate gene PLK4 (polo-like kinase 4) may cause tripolar spindles in normally fertilised, bipronuclear zygotes, producing abnormal segregation of chromosomes to three daughter cells to form a chaotic mosaic (McCoy et al. 2015b, McCoy et al. 2018). PLK4 is required for regulating centriole duplication (Habedanck et al. 2005) and altered PLK4 expression is associated with repeated implantation failure (Wang & Liu 2020). The production of chaotic mosaics from tripronuclear zygotes that form tripolar spindles is discussed below (Fig. 1Gii). In addition, chaotic mosaics may be produced by fertilisation with sperm retrieved from the seminal tract of some infertile men, particularly those with non-obstructive azoospermia (Magli et al. 2009).
Mosaics from aneuploid embryos
Aneuploid embryos can also produce aneuploid/diploid or aneuploid/aneuploid mosaics (Fig. 1E). For example, a cell in a trisomic embryo may lose one copy of the trisomic chromosome by trisomic rescue (also known as ‘trisomic zygote rescue’), which could involve anaphase lag, non-disjunction or chromosome demolition (Kalousek et al. 1991, Los et al. 1998, Munné et al. 2005, Barbash-Hazan et al. 2009, Vazquez-Diez & FitzHarris 2018). However, unless chromosome loss was somehow targeted to the appropriate chromosome, it would produce new errors. Correction of a trisomic cell by mitotic anaphase lag would produce one corrected disomic cell and one trisomic cell, so a trisomic embryo would become a Ts/2n mosaic. Correction by mitotic non-disjunction would produce a disomic cell and a tetrasomic cell, which are likely to die. In one-third of cases, random loss of one copy of the trisomic chromosome would cause uniparental disomy (UPD, two maternal or paternal copies, instead of one of each). Depending on the chromosome involved, UPD can cause genomic imprinting abnormalities (Kotzot & Utermann 2005, Eggermann et al. 2015). Chromosome duplication for monosomic rescue could occur by non-disjunction (Conlin et al. 2010). This would always produce UPD and a nullisomic cell, which would die. However, although there is evidence for UPD from prenatal and postnatal samples (Eggermann et al. 2015), Gueye et al. (2014) only found UPD in 0.06% of human blastocysts. This suggests that aneuploid cell correction (aneuploid rescue) rarely occurs before implantation, even though mitotic anaphase lag and non-disjunction are likely to be most common during the cleavage stage. Such mitotic errors in aneuploid embryos would probably produce aneuploid/aneuploid mosaics more frequently than aneuploid/diploid mosaics.
Mosaicism for segmental aneuploidy
Although the focus of this review is mosaicism for whole chromosome abnormalities, once CCS methods became available, losses and gains of chromosomal fragments, known as segmental (or partial) aneuploidy, were also identified in preimplantation embryos, either in all cells or in mosaic form (Voullaire et al. 2000, Wells & Delhanty 2000, Vanneste et al. 2009, Rabinowitz et al. 2012, Vera-Rodriguez et al. 2016, Babariya et al. 2017, Escribà et al. 2019). For example, Babariya et al. (2017) used aCGH to estimate that segmental aneuploidy occurred in 10.4% of human oocytes, 24.3% of cleavage stage embryos and 15.6% of blastocysts. Although some segmental errors may be inherited from a carrier of a balanced structural chromosomal abnormality and others may occur during meiosis, most arise during the early cleavage stages (Vera-Rodriguez et al. 2016, Babariya et al. 2017). Babariya et al. (2017) suggested that mosaicism for segmental aneuploidy could arise at cleavage stages if monitoring of DNA damage was dysfunctional before zygotic genome activation was completed and that suboptimal embryo culture conditions might also increase the frequency of double-stranded DNA breaks, leading to segmental aneuploidy. Different ways that segmental aneuploidy may arise are discussed elsewhere (Vanneste et al. 2009, Escribà et al. 2019).
Mixoploidy
Different types of mixoploidy (combinations of polyploidy, haploidy and diploidy) can occur in preimplantation embryos, either alone or in combination with aneuploidy. Bielanska et al. (2002) analysed 216 preimplantation embryos at different stages and reported 2.8% haploid/diploid mixoploids (plus 0.5% haploids) and 14.8% polyploid/diploid mixoploids (plus 5.6% polyploids). The frequency of polyploid/diploid mixoploids was highest at the blastocyst stage (23/33; 69.7%) and most were tetraploid/diploid. Although the presence of tetraploid cells at cleavage stages may be abnormal, they are probably a normal feature of trophectoderm development in blastocysts (Angell et al. 1987, Benkhalifa et al. 1993, Bielanska et al. 2002, de Boer et al. 2004).
Some haploid/diploid mixoploids arise from haploid parthenote embryos, most of which generate diploid cells by the blastocyst stage (Fig. 1F; Leng et al. 2017). This occurs by failed cleavage, whereby the nucleus divides and the cleavage furrow forms but regresses, or endomitosis, a type of endoreplication whereby the nucleus divides without cleavage (Leng et al. 2017). Other haploid/diploid mixoploids may arise by non-canonical divisions that segregate complete haploid sets of chromosomes (Fig. 1Dii; Destouni et al. 2016), as discussed below. Tetraploid/diploid mixoploids may arise from diploid embryos (Fig. 1Biv) by endoreplication (Benkhalifa et al. 1993) or cell fusion (Balakier et al. 2000).
Approximately 2–9% of zygotes produced by conventional IVF are tripronuclear but only 14–25% of these, including most of those with an extra maternal pronucleus, divide into two triploid blastomeres (Fig. 1Gi; Angell et al. 1986, Kola et al. 1987, Pieters et al. 1992, Plachot & Crozet 1992, Palermo et al. 1994, Golubovsky 2003). There are more diandric triploids (extra paternal set of chromosomes) than digynic triploids (extra maternal set of chromosomes) among naturally conceived early abortuses (Zaragoza et al. 2000) and Plachot et al. (1989) found that 86% of preimplantation triploids, produced by conventional IVF, were diandric. However, Marin et al. (2018) reported that 5/5 ICSI-derived triploid blastocysts were digynic. Unlike conventional IVF, ICSI, with a single spermatozoon, precludes chromosomal abnormalities associated with polyspermy, such as diandric triploidy.
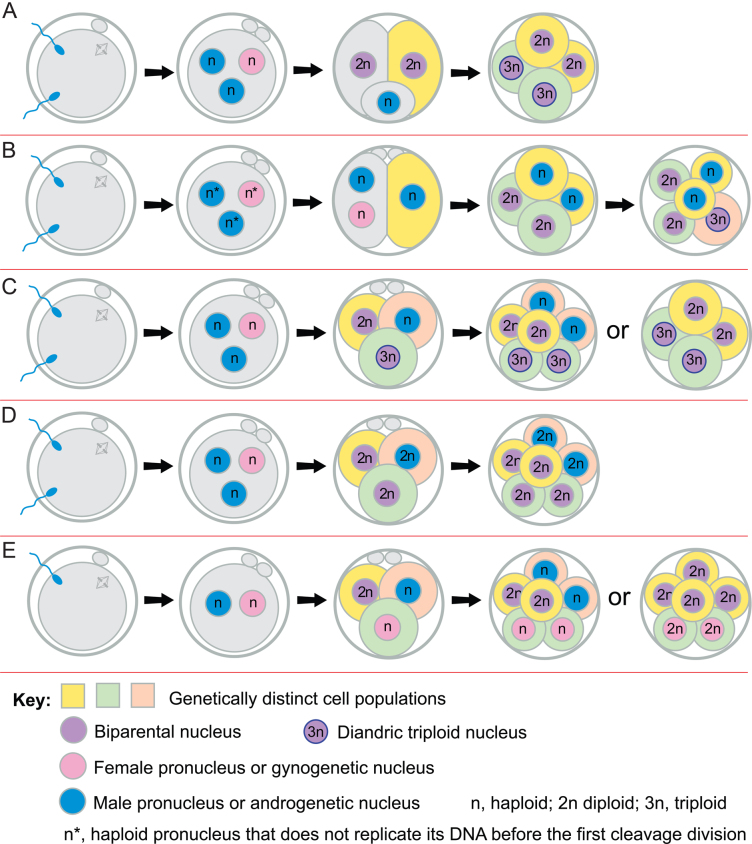
Tripronuclear zygotes that arise by dispermic fertilisation have an extra centriole, as well as the extra paternal pronucleus, because centrioles are paternally inherited in humans (Palermo et al. 1994, Sathananthan et al. 1996). According to Golubovsky (2003) only about 25% of dispermic tripronuclear zygotes become triploid (Fig. 1Gi) and over 50% produce chaotic mosaic embryos because the supernumerary centriole produces a tripolar spindle and chromosomes segregate abnormally (Fig. 1Gii). Golubovsky (2003) further proposed that, in 14–32% of dispermic tripronuclear zygotes, one haploid pronucleus could separate and is either lost, leaving a diploid embryo, or retained, producing different types of mixoploids (Figs 1Giii-iv and 2A, B). Segregation of haploid and diploid sets of chromosome is supported by evidence that, in some tripronuclear zygotes, a complete haploid set of chromosomes is extruded (Fig. 2A) or segregates independently (Fig. 2B) to produce a haploid/diploid mixoploid, which may become 3n/2n by cell fusion and death of any remaining haploid cells (Angell et al. 1986, Kola et al. 1987, Pieters et al. 1992, Plachot & Crozet 1992, Golubovsky 2003, Rosenbusch & Schneider 2009). Mixoploids produced by this type of postzygotic diploidisation of triploids (Golubovsky 2003) could be considered to be chimaeras, by the broader definition given earlier.
Figure 2.
Segregation of complete haploid sets of parental chromosomes. (A, B, C and D) Three pronuclei form after dispermy and may segregate in different ways. (A) One male pronucleus is extruded in a small androgenetic haploid cell and the other two form two normal blastomeres. Later the extruded haploid cell may fuse with one blastomere and both blastomeres divide to form a triploid/diploid mixoploid embryo. (B) The zygote undergoes an atypical early cytokinesis and intact pronuclei are distributed between the two blastomeres. After the next division, there are two biparental diploid cells and two androgenetic haploid cells. In this example (from Supplementary Fig. 4A in Destouni et al. 2016) one diploid and one haploid cell fuse to produce a diandric triploid cell line and other cells divide but various fates are possible. Later the haploid cells may undergo endoreplication, fuse with other cells or die. (C) Entire haploid sets of chromosomes segregate on a tripolar spindle to form a mixoploid embryo with a biparental diploid, diandric triploid and an androgenetic haploid cell. Subsequently, haploid cells may die, fuse or endoreplicate to form androgenetic diploid cells or fuse with other cells. Only two of various possible fates are illustrated. (D) Entire haploid sets of chromosomes segregate on a tripolar spindle to form a chimaera with two biparental diploid cells with different paternal genomes and a diandric diploid cell with two different paternal sets of chromosomes. (E) Two pronuclei form after monospermic fertilisation but, occasionally, entire haploid sets of chromosomes may segregate on a tripolar spindle to form a chimaera with a biparental diploid cell, a gynogenetic haploid and an androgenetic haploid cell. Subsequently, haploid cells may die, endoreplicate to form diploid cells or fuse with other cells. Only two of various possible fates are illustrated.
More recent support for postzygotic diploidisation of triploids was obtained from single-cell haplotyping of tripronuclear bovine embryos by haplarithmisis. This showed that complete haploid, parental sets of chromosomes segregated independently at the first cleavage division in 9/23 (39%) of cleavage stage embryos produced by conventional IVF (Destouni et al. 2016) and this was termed ‘heterogoneic division’ (Greek for different parental origin). Destouni et al. (2016) found evidence for segregation of complete haploid sets of chromosomes in five embryos derived from tripronuclear zygotes, all of which had two paternal genomes, so were dispermic. They proposed that whole sets of chromosomes segregated on tripolar spindles and the chromosome complements could subsequently be modified if haploid cells died, diploidised or fused with other cells. Thus, dispermic, tripronuclear zygotes can produce mixoploidy (Figs 1Gv and 2C) or diploid/diploid chimaeras with androgenetic diploid and two types of biparental diploid cells (Fig. 2D). Destouni et al. (2016) also reported segregation of parental chromosome sets in two apparently normal monospermic, bipronuclear bovine zygotes. Thus, monospermic, bipronuclear zygotes can produce various types of chimaeras with different combinations of biparental, gynogenetic and androgenetic cells (Fig. 2E; Destouni et al. 2016, Destouni & Vermeesch 2017, Masset et al. 2021). Heterogoneic divisions might be facilitated if maternal and paternal chromosomes are kept apart on separate mitotic spindles during the first cleavage division in humans, as in mice (Reichmann et al. 2018), but further work is required to investigate this novel mechanism in humans.
Other primary chimaeras and mosaics
Human chimaeras, including XX/XY chromosomal chimaeras, are much less common than mosaics and are usually only identified after birth, although many will remain undetected (particularly XX/XX and XY/XY chimaeras). Although primary chimaeras have less impact on assisted reproduction, it is appropriate to consider them alongside preimplantation mosaics because they arise at preimplantation stages. Molecular markers, for identifying the number of haploid genomes and their parental origin, have made it easier to distinguish chimaeras (with more than two haploid genomes) from mosaics and predict their possible mode of origin. Madan (2020) listed 50 human XX/XY (or similar) chimaeras (including six with an aneuploid cell line). These were identified because of abnormal sexual development, including hermaphroditism (28), congenital abnormalities (4), patchy skin (1) or by chance (17). Mosaic XX/XY individuals also occur because a 46,XX/46,XY mosaic may be produced from a 47,XXY zygote following two non-disjunction events (Niu et al. 2002).
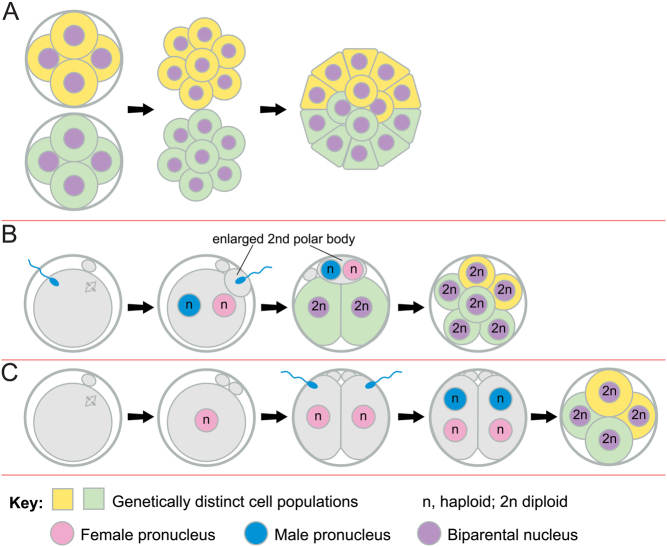
A number of hypothetical mechanisms could produce primary chimaeras (Ford 1969, McLaren 1976) and Fig. 3 shows three that have been commonly invoked (Malan et al. 2006, Madan 2020). First, tetragametic aggregation chimaeras could occur if two preimplantation embryos aggregated after hatching from their zonae pellucidae (Fig. 3A). (The term ‘fusion chimaera’ is sometimes used but no cell fusion occurs.) Some aggregation chimaeras have been conceived in IVF programmes (Strain et al. 1998, Simon-Bouy et al. 2003), raising the possibility that IVF may facilitate embryo aggregation if multiple embryos are transferred to the uterus, particularly if assisted hatching is used. Late aggregation could result in two ICMs within one trophectoderm, which might produce chimaeric or non-chimaeric dizygotic twins within a single chorion (Souter et al. 2003, Miura & Niikawa 2005, Boklage 2006, Peters et al. 2017). Monochorionic dizygotic twins have been produced by IVF and most are chimaeric, at least in the blood (Peters et al. 2017).
Figure 3.
Different types of chimaeras. (A) Two cleavage stage embryos may lose their zonae pellucidae and aggregate to form an aggregation chimaera. (B) Fertilisation of both the egg and the second polar body, which may be enlarged. (C) Parthenogenetic activation of an oocyte produces a two-cell embryo with two haploid nuclei that are then both fertilised by different spermatozoa to produce a chimaera with identical maternal contributions in both cell lines.
Secondly, the egg and second polar body could be fertilised by different spermatozoa (Fig. 3B) and this might occur more readily if the polar body was enlarged, as reported for some aged mouse oocytes (McLaren 1976). Thirdly, diploid/diploid chimaeras, with two different paternal contributions but only one maternal contribution, could be produced if an oocyte is activated parthenogenetically and cleaves into two haploid blastomeres, which are then fertilised by different spermatozoa (Fig. 3C). Souter et al. (2007) reported twin chimaeras (one hermaphrodite and one male) that could have arisen this way if a chimaeric ICM divided to form twins, with different proportions of XX and XY cells.
Triploid/diploid mixoploidy may be produced in several ways other than those discussed earlier. These include aggregation of triploid and diploid embryos (Supplementary Fig. 1A, see section on supplementary materials given at the end of this article; Dewald et al. 1975, Daniel et al. 2003), delayed incorporation of an extra male pronucleus from a second spermatozoon (Supplementary Fig. 1B, C and D; Dewald et al. 1975, Daniel et al. 2003, Quigley et al. 2005) or delayed incorporation of an extra female pronucleus from the second polar body (Supplementary Fig. 1E and F; Ellis et al. 1963, Schmid & Vischer 1967, Müller et al. 1993, van de Laar et al. 2002, Daniel et al. 2003).
Chimaeras and mosaics with an androgenetic, gynogenetic or parthenogenetic diploid cell line plus a biparental diploid cell line may also be produced in several ways (Malan et al. 2006, Madan 2020) and can occur in combination with aneuploidy (Yamazawa et al. 2010). Examples shown in Supplementary Fig. 2 are from Surti et al. (2005) (Supplementary Fig. 2A), Kaiser-Rogers et al. (2006) (Supplementary Fig. 2B), Makrydimas et al. (2002) and Giurgea et al. (2006) (Supplementary Fig. 2C), Robinson et al. (2007) (Supplementary Fig. 2D and E), Sheppard et al. (2019) (Supplementary Fig. 2F) and Strain et al. (1995) (Supplementary Fig. 2G and H). As Destouni et al. (2016) pointed out, some of the types of chimaeras shown in Supplementary Figs 1 and 2 could also be produced by the segregation of complete parental sets of chromosomes at the first division (Fig. 2).
Fate of preimplantation mosaic embryos
The fate of chromosomally abnormal cells in mosaics and chimaeras varies according to the type of abnormality. Haploid cells probably die, fuse with other cells or diploidise before implantation. As noted earlier, tetraploid cells commonly occur in blastocysts and they probably normally contribute to the placental trophoblast (Angell et al. 1987, Benkhalifa et al. 1993, Bielanska et al. 2002, de Boer et al. 2004). Triploid/diploid mixoploids can survive to fetal and postnatal stages (some with genomic imprinting disorders) but sometimes, triploid cells are confined to the placenta (Carson et al. 2018).
Most complex or chaotic mosaic aneuploid embryos die before implantation and some arrest before becoming blastocysts (Bielanska et al. 2002, Santos et al. 2010, McCoy et al. 2015a). The proportion of aneuploid cells in simple aneuploid/diploid mosaics declines between cleavage and blastocyst stages (Bielanska et al. 2002, van Echten-Arends et al. 2011, Fragouli et al. 2013). RNA-seq analysis also indicated that the proportion of aneuploid cells in mosaic embryos declined between days 3 and 6 (Yang et al. 2021) but that low-level mosaicism was still common among morulae and blastocysts, as noted earlier (Starostik et al. 2020). However, reports are contradictory as to whether the frequency of simple aneuploid/diploid mosaics increases or decreases by the blastocyst stage (van Echten-Arends et al. 2011, Fragouli et al. 2019).
Approximately 2% of CVS cases (Wang et al. 1993, Pittalis et al. 1994, Huang et al. 2009, Malvestiti et al. 2015), 0.25% of second-trimester amniocentesis samples (Hsu & Perlis 1984) and 0.02% of live births (Hassold & Jacobs 1984) are mosaic. Over 2700 embryos classified as putative mosaics have been transferred following PGT-A (reviewed by Treff & Marin 2021) but mosaicism has only been observed at term once (Kahraman et al. 2020). However, technical limitations mean that the classification of embryos as euploid, mosaic or aneuploid is inexact and make it difficult to determine the fate of mosaic embryos accurately (Marin et al. 2021, Treff & Marin 2021). Furthermore, chromosomal mosaicism may not be apparent at birth because postnatal cytogenetic investigations are not routine and are often confined to peripheral blood (Belva et al. 2020). Although the fate of mosaic embryos remains unproven, it seems likely that some mosaics, with a high proportion of aneuploid cells, miscarry and most others lose the aneuploid cells from the fetal lineage before term. As noted earlier, for the purpose of this review, we assume that the frequency of mosaicism is higher among preimplantation embryos and declines after implantation.
Relationship between preimplantation chromosomal mosaicism and confined placental mosaicism
Confined placental mosaicism
CPM is present in about 1–2% of viable human pregnancies tested by CVS, at approximately 10–12 weeks (Wang et al. 1993, Pittalis et al. 1994, Huang et al. 2009, Malvestiti et al. 2015). It is more common than true fetal mosaicism (TFM; Table 1) and often persists to term. Amniotic fluid cells, obtained by amniocentesis, are mostly from the fetus (Gosden 1983) and are used to distinguish between TFM and CPM. Different culture conditions for chorionic villus samples enrich the cytotrophoblast or mesenchymal cells and help identify three types of CPM, with abnormal cells in the villus cytotrophoblast (CPM-I), mesenchyme (CPM-II) or both (CPM-III). Studies differ as to whether CPM-I or CPM-II predominates (Pittalis et al. 1994, Malvestiti et al. 2015; Table 1).
Table 1.
Frequencies of different types of mosaicism in two studies of chorionic villi and amniotic fluid cells.
| Type of mosaic | Affected tissues | % of conceptuses | % of mosaics | ||||
|---|---|---|---|---|---|---|---|
| Cytotrophoblast | Mesenchyme | AF cells | Study 1* | 2* | 1* | 2* | |
| Confined placental mosaicism | |||||||
| CPM-I | Abnormal† | Normal | Normal | 0.80 | 0.59 | 53.42 | 35.66 |
| CPM-II | Normal | Abnormal | Normal | 0.31 | 0.68 | 20.55 | 41.26 |
| CPM-III | Abnormal | Abnormal | Normal | 0.16 | 0.17 | 10.96 | 9.99 |
| Total CPM | 1.28 | 1.44 | 84.93 | 86.91 | |||
| Fetal mosaicism | |||||||
| TFM-IV | Abnormal | Normal | Abnormal | 0 | 0.02 | 0 | 1.40 |
| TFM-V | Normal | Abnormal | Abnormal | 0.08 | 0.09 | 5.48 | 5.69 |
| TFM-VI | Abnormal | Abnormal | Abnormal | 0.12 | 0.10 | 8.22 | 5.99 |
| CFM | Normal | Normal | Abnormal | 0.02 | – | 1.37 | – |
| Total FM | 0.23 | 0.22 | 15.07 | 13.09 | |||
*Study 1 data are from Table 2 in Pittalis et al. (1994) and comprised 4,860 conceptuses, 73 of which were mosaics. Study 2 data are from Table 1 in Malvestiti et al. (2015) and comprised 60,347 conceptuses, 1001 of which were mosaics; †Abnormal includes both uniformly abnormal and mosaic tissues.
AF, amniotic fluid. CFM, confined fetal mosaicism; CPM, confined placental mosaicism; FM, fetal mosaicism; TFM, true fetal mosaicism.
Only a subset of numerical chromosomal abnormalities found in preimplantation embryos is identified later at CVS. Most are autosomal trisomies or sex chromosome aneuploidies but triploidy (usually TFM) and tetraploidy (usually CPM) also occur (Pittalis et al. 1994, Malvestiti et al. 2015, Carson et al. 2018). Rare ‘confined placental chimaeras’, where a ‘vanishing twin’ contributes cells to the placenta, also occur (Falik-Borenstein et al. 1994, Surti et al. 2005). Different trisomies predominate in each of the three types of CPM (Lestou & Kalousek 1998, Benn & Grati 2021), which complicates investigations of CPM aetiology.
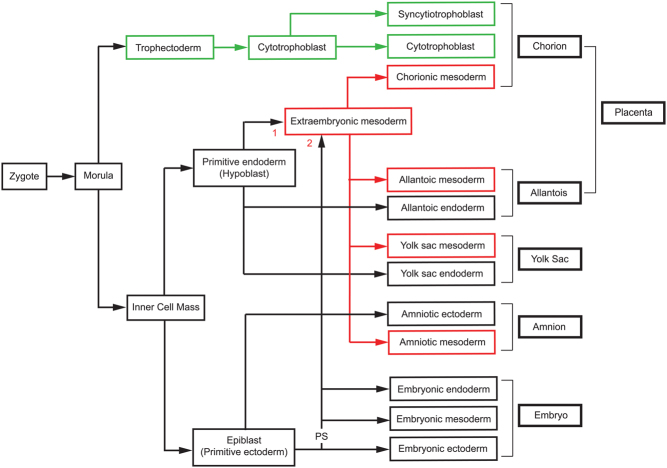
In CPM-III, abnormal (usually trisomic) cells are present in both the ICM and trophectoderm lineages, so they almost certainly exist before these two lineages segregate and are later excluded from the fetal lineage. In CPM-I, abnormal cells become restricted to the placental trophoblast whereas in CPM-II they become restricted to the placental mesenchymal core. This is produced by the extraembryonic mesoderm lineage, which appears to have a dual origin (Fig. 4). The first extraembryonic mesoderm appears before gastrulation and evidence implies this is derived from the primitive endoderm (hypoblast) lineage in both rhesus monkeys (Enders & King 1988) and humans (Bianchi et al. 1993). Later, additional extraembryonic mesoderm is produced from the epiblast lineage via the primitive streak at gastrulation (Enders & King 1988, Robinson et al. 2002).
Figure 4.
Simplified lineage diagram showing the origin of human extraembryonic tissues. The diagram shows the relationship between the embryonic lineage and the two lineages that produce the chorionic villi (trophectoderm and extraembryonic mesoderm). The three germ layers (ectoderm, mesoderm and endoderm) that form the embryo are produced from the epiblast and the embryonic mesoderm and embryonic endoderm emerge from the primitive streak (labelled ‘PS’) during gastrulation. Both the cytotrophoblast and syncytiotrophoblast are produced from the trophectoderm layer of the blastocyst. Evidence suggests that the human extraembryonic mesoderm is produced first by the primitive endoderm (labelled ‘1’) and later from epiblast via the primitive streak (labelled ‘2’). See text for references. For simplicity, both sources of cells are shown feeding into a common pool of extraembryonic mesoderm but they may colonise different tissues.
The type of trisomic CPM is likely to depend on when, where and how (e.g. from a diploid or aneuploid zygote) the mosaicism arises, as well as the chromosome involved. Chance may also play a role because one cell population will be more easily excluded from a lineage founded by few cells and a minor cell population is likely to be more easily excluded. Robinson et al. (1997) proposed that CPM-I and CPM-II mainly arise from diploid zygotes (mitotic aneuploidy) whereas CPM-III is mainly from trisomic zygotes (meiotic aneuploidy) via trisomic rescue, which may cause UPD. CPM, with or without UPD, may affect fetal growth and development (Eggenhuizen et al. 2021).
In principle, four mechanisms could all contribute to the reduction of mosaic embryos and the predominance of CPM among surviving mosaics. These are (i) selective embryonic death (death of the most severely affected embryos), (ii) cell selection, (iii) cell correction and (iv) preferential allocation of abnormal cells to extraembryonic lineages. Preferential allocation would contribute to CPM, as would lineage-biased cell selection and/or lineage-biased cell correction if they were more effective in fetal than extraembryonic lineages. These four potential mechanisms for reducing the frequency of mosaic embryos are also discussed by Coticchio et al. (2021), using different terminology. We use the terms ‘selective embryonic death’ and ‘cell selection’ as previously (Everett & West 1996) and ‘cell correction’ whereas Coticchio et al. (2021) used ‘embryonic mortality’, ‘clonal depletion’ and ‘trisomic/monsomic rescue’, respectively.
Selective embryonic death
As noted earlier, some mosaic embryos die before implantation. Approximately 1.1% of spontaneous abortions and 0.5% of stillbirths are Ts/2n mosaics (Hassold & Jacobs 1984). This suggests that selective embryonic death also occurs after implantation but it is not the main reason why the frequency of Ts/2n mosaics declines after implantation and few survive to term.
In a prospective, non-selection clinical trial, putative euploid and putative low-grade and medium-grade mosaic human blastocysts (estimated to have <50% aneuploid cells in the trophectoderm biopsy by NGS analysis) were made equally available for transfer and transferred to the uterus before NGS classifications were revealed (Capalbo et al. 2021). The putative mosaics were as likely to survive to term as putative euploid embryos but those with higher proportions of aneuploid cells were not transferred. Two retrospective studies suggested that putative mosaics with higher proportions of aneuploid cells and putative complex mosaics were less likely to survive (Munné et al. 2017, Viotti et al. 2021b ). Although this is consistent with selective embryonic death of the most severely affected mosaics, the design of these studies has been criticised (Capalbo et al. 2021) and the difficulty of identifying true mosaic embryos from small biopsies was discussed earlier.
Some studies with animal models, described later, provide evidence for selective embryonic death and suggest that if too many abnormal cells persist in a postimplantation mosaic embryo it is more likely to die.
Cell selection (clonal depletion)
Cell selection probably plays a major role in depleting the number of trisomic cells and restricting their distribution to the placenta but the degree of selection is likely to depend on the nature of the abnormality. Cell selection is thought to begin before implantation because, as noted earlier, the proportion of abnormal cells in mosaic embryos declines between cleavage and blastocyst stages (Bielanska et al. 2002, van Echten-Arends et al. 2011, Fragouli et al. 2013). Selection may involve cell death, loss and/or depletion, by reduced proliferation, but selection pressures are expected to be weak until relevant embryonic genes are activated, probably during the 4–16-cell period (Jukam et al. 2017). Possible mechanisms at preimplantation stages include apoptosis, which begins at the morula stage (Hardy et al. 2001), and exclusion of abnormal cells, particularly during morula compaction (Lagalla et al. 2017). Although direct evidence for selection against trisomic cells in human embryos is lacking, some cells excluded from human blastocysts are chromosomally abnormal (Orvieto et al. 2020). There is also some evidence for postnatal cell selection from longitudinal studies of trisomic/diploid mosaic children and adults but this may be more apparent in blood than tissues with a slower cell turnover (Gravholt et al. 1991, Papavassiliou et al. 2015). In embryos, cell selection against chromosomally abnormal cells might be more stringent in the epiblast or fetal lineage. For example, depletion of heterogeneous aneuploid cells by apoptosis occurred predominantly in the epiblast lineage of mouse chimaeras (Bolton et al. 2016) and the embryonic region of mosaic human ‘gastruloids’, produced with human embryonic stem cells (Yang et al. 2021). In these model systems, heterogeneous chromosomal mosaicism was induced experimentally and the mouse model is discussed later. DNA copy number variation analysis also suggests that the placental trophoblast may tolerate mutant or abnormal cells more readily than other tissues (Coorens et al. 2021).
Cell correction
Aneuploid cells may be corrected by trisomic or monosomic rescue (discussed earlier), which can occur more than once in the same conceptus (Van Opstal et al. 2018). However, it is unclear whether this plays more than a minor role in restricting the distribution of aneuploid cells. As noted earlier, UPD is rare in human blastocysts (Gueye et al. 2014), suggesting that aneuploid cell correction rarely occurs before implantation, but UPD is more common in prenatal and postnatal samples (Eggermann et al. 2015). UPD might arise more frequently (per conceptus) after implantation, simply because many more mitotic divisions have occurred, providing more opportunities for cells with UPD to arise and proliferate. However, detection of a small proportion of UPD cells in a mosaic conceptus might still be challenging unless disomic cells have a selective advantage and outgrow the aneuploid cells.
Preferential allocation to extraembryonic lineages
Preferential allocation of aneuploid cells to placental lineages at various stages could contribute to their restricted distribution. There is no convincing evidence for this in human embryos but studies have failed to allow for the observation that different trisomies predominate in CPM-I, CPM-II and CPM-III (Lestou & Kalousek 1998, Benn & Grati 2021). It, therefore, remains unknown whether any abnormal cell types are preferentially allocated to extraembryonic tissues. Most investigations have considered whether preferential allocation contributes to the restriction of aneuploid cells to the trophectoderm lineage in CPM-I. This restriction could occur if trisomic cells (i) arose preferentially in the trophectoderm or trophoblast, (ii) arose at the cleavage stage and were preferentially allocated to the trophectoderm or (iii) were present in multiple lineages initially but only survived in the trophectoderm/trophoblast (Fig. 5).
Figure 5.
Events in preimplantation embryos that could cause or initiate CPM-I. By the late-blastocyst stage, there are three primary lineages, namely the epiblast, primitive endoderm (or hypoblast) and the trophectoderm, which produces all the placental trophoblast cells (Fig. 4). The outer morula cells form the trophectoderm layer and the inner cells form the ICM of the blastocyst. At least in the mouse, some outer morula cells produce additional inner cells by asymmetrical division and the epiblast and primitive endoderm (hypoblast) precursor cells are initially intermixed in the ICM but they assume their final positions in the late blastocyst (Saiz & Plusa 2013). In reality, restriction of abnormal cells to the trophectoderm/trophoblast lineage in CPM-I is likely to occur gradually and not be completed until after implantation. However, for simplicity, the figure illustrates how complete restriction of abnormal cells to the trophectoderm lineage could occur by the late blastocyst stage. (A) The chromosomally abnormal cell population (shaded blue) could arise exclusively in the trophectoderm at any time after it has separated from the ICM. (B) The abnormal cell population might arise at an early stage but be exclusively or predominantly allocated to the trophectoderm lineage. (C) The abnormal cell population could arise at an early stage and initially be present in both the ICM and trophectoderm but only survive in the trophectoderm lineage. Ab, chromosomally abnormal cell (shaded blue); 2n, normal diploid cell (shaded grey).
A recent NGS analysis of the distribution of putative aneuploid cells, among four trophectoderm samples and the ICM from dissected whole human blastocysts, indicated that they were often confined to one trophectoderm sample in putative aneuploid/diploid mosaics with low proportions of putative aneuploid cells (Capalbo et al. 2021). The authors suggested that aneuploid cells probably arose in the trophectoderm after it separated from the ICM, as shown in Fig. 5A. Aneuploid cells that arose earlier (Fig. 5B and C) might also sometimes produce spatially restricted cell populations if cell mixing was relatively limited before implantation, as in some mouse models (Garner & McLaren 1974, Gardner & Cockroft 1998, Everett et al. 2000).
However, restrictions could also begin later in development and/or involve multiple small steps. For example, abnormal cells might be preferentially (but not exclusively) allocated to the trophectoderm lineage at the blastocyst stage and abnormal cells remaining in other lineages lost after implantation. It is generally assumed that CPM develops from preimplantation mosaicism but, in some cases, trisomic cells may arise de novo in the placenta after implantation.
It is also important to consider how preferential allocation could result in CPM-II. If, the extraembryonic mesoderm of the placenta has a dual origin from the primitive endoderm and the epiblast (Fig. 4), CPM-II could arise if trisomic cells survived in the ICM, epiblast and/or primitive endoderm and later extraembryonic mesoderm lineages but became excluded from the trophectoderm/trophoblast and fetal lineages.
Most recent comparisons of the chromosomal composition of the ICM and trophectoderm have used comprehensive chromosome screening to analyse multicellular samples and categorised them as normal, mosaic or abnormal but none claimed a significant difference (Popovic et al. 2020). A more informative approach is to distinguish or separate ICM and trophectoderm cells and analyse each cell individually. Two such studies, using FISH, provided no evidence for preferential allocation of aneuploid cells to the trophectoderm (Evsikov & Verlinsky 1998, Derhaag et al. 2003). More recently, a study of gene expression profiles, to determine both cell type and chromosome content of individual cells, from day 4 morulae to day 7 blastocysts, showed no difference in the percentage of aneuploid trophectoderm and ICM cells (Starostik et al. 2020). However, aneuploid cells became enriched in the trophectoderm, relative to the epiblast and primitive endoderm, after culture to day 14, but there was no overall change in the aneuploidy frequency (Starostik et al. 2020). To unravel the relationship between preimplantation mosaicism and different types of CPM, it will be necessary to analyse results for different trisomies separately.
Animal models of mosaicism
For ethical reasons, human embryo studies are largely descriptive, so animal models provide an important complementary approach. Indeed, many procedures used for human IVF and embryo culture are based on pioneering animal studies (Johnson 2019). Animal studies of chromosome segregation errors in preimplantation embryos are reviewed elsewhere (Vazquez-Diez & FitzHarris 2018) and some were mentioned earlier. A comprehensive discussion of animal models of chromosome mosaicism is beyond the scope of this review but some selected topics are considered.
Preimplantation mosaicism in vitro and in vivo
Several animal studies suggest that IVF and/or embryo culture might increase mosaicism frequency. Two-colour FISH revealed a higher frequency of mosaicism among mouse blastocysts cultured from the two-cell stage than those developed in vivo (Sabhnani et al. 2011) and a higher frequency of sex-chromosome mosaicism when BALB/cWt strain mouse embryos were produced by IVF and cultured rather than developed in vivo (Bean et al. 2002).
Mixoploidy was reported to be more frequent when bovine, equine and ovine embryos were produced in vitro instead of in vivo (Viuff et al. 1999, Rambags et al. 2005, Coppola et al. 2007) and when bovine embryos were cultured in vitro, in the presence of serum, rather than developed in ewe oviducts in vivo(Lonergan et al. 2004). All four studies used two-colour FISH and reported the abnormal embryos as mixoploid but the scoring criteria precluded the identification of aneuploid/diploid mosaics. A more recent haplarithmisis study showed that both aneuploid and polyploid cells were more common among bovine embryos produced in vitro than in vivo(Tšuiko et al. 2017).
Fate of chromosomally abnormal cells and animal models of CPM
Mice are commonly used to study the postimplantation fate of abnormal cells and lineage relationships are summarised in Supplementary Fig. 3. Early studies established that the mouse epiblast produces the fetus and extraembryonic mesoderm (including that of the amnion and yolk sac), the primitive endoderm produces extraembryonic endoderm, the polar trophectoderm forms placental trophoblast and the mural trophectoderm forms the trophoblast giant cells of the parietal yolk sac (Gardner & Papaioannou 1975, Gardner 1983). Later studies added complexity to the types of trophoblast giant cells (Simmons et al. 2007) and showed that the mouse primitive endoderm also contributes to the embryonic endoderm (Kwon et al. 2008) plus the allantoic and placental extraembryonic mesoderm (Rodriguez & Downs 2017).
Mixoploidy models
The fate of triploid cells has been studied in mouse chimaeras, produced either by aggregating cleavage stage triploid and diploid embryos or by fusing a haploid karyoplast or the second polar body to one cell at the two-cell stage. Triploid cells contributed more often to the trophectoderm than ICM in chimaeric blastocysts (Hino & Tateno 2016 and Azuma et al. 1991b, cited therein) but, in postimplantation chimaeras, they did not contribute consistently more to the placenta than the fetus and other epiblast derivatives (Everett et al. 2007, Hino & Tateno 2016). Instead, they contributed better to the yolk sac (Suwinska et al. 2005) or, specifically, the yolk sac endoderm (Everett et al. 2007). Triploid cells survived in fetal and adult tissues (Azuma et al. 1991a , Suwinska et al. 2005, Hino & Tateno 2016), although their proportion declined with fetal age (Suwinska et al. 2005), possibly because triploid cells divide relatively slowly (Hino & Tateno 2016).
Mouse tetraploid↔diploid (4n↔2n) aggregation chimaeras provided an early CPM model, following the observation that tetraploid cells were more abundant in extraembryonic tissues than the fetus of 4n/2n mouse mosaics (Tarkowski et al. 1977). In chimaeric blastocysts, tetraploid cells initially contributed to the epiblast, primitive endoderm and trophectoderm (Everett & West 1996). By embryonic day 12.5 (E12.5), however, they were excluded or severely depleted in the fetus and other epiblast derivatives but survived in the primitive endoderm and trophectoderm derivatives (James et al. 1995).
Further studies of mouse 4n↔2n chimaeras provided evidence for non-random allocation of tetraploid cells in chimaeric blastocysts, cell selection and selective embryonic death. Non-random cell allocation was suggested by enrichment for tetraploid cells in the trophectoderm, particularly the mural trophectoderm, which was reported in some studies of chimaeric blastocysts (Everett & West 1996, Tang et al. 2000, MacKay & West 2005) but not others (Ishiguro et al. 2005). Experimental manipulation showed that the larger size and increased ploidy of tetraploid cells could each affect their distribution in chimaeric blastocysts (Tang et al. 2000). In contrast, the initial tetraploid:diploid cell ratio had no consistent effect on cell distribution to different lineages (Tang & West 2000) but it affected the overall contribution of tetraploid cells (Goto et al. 2002).
Selection against tetraploid cells occurred during late preimplantation and postimplantation development (Everett & West 1998, Goto et al. 2002, Eakin et al. 2005, Ishiguro et al. 2005, MacKay & West 2005). Proliferation differences may contribute to cell selection (Eakin et al. 2005) but most tetraploid cells are probably eliminated from the epiblast lineage by a p53-dependent, tetraploid checkpoint that triggers apoptosis during epiblast differentiation (Horii et al. 2015). Although tetraploid cells have survived in some tissues of fetal and adult chimaeras (Lu & Markert 1980, Tarkowski et al. 2001, Goto et al. 2002), a high proportion may cause selective embryonic death (James et al. 1995, Tarkowski et al. 2005).
The trophectoderm of bovine embryos contained more putative polyploid cells than the embryonic disc on day 7 but the frequency declined significantly in both lineages by day 12 (Viuff et al. 2002). This provides evidence for cell selection but it is unclear whether the initial distribution resulted from non-random allocation or if more putative polyploid cells arose in the trophectoderm.
Models of mosaicism with aneuploidy
Mouse embryos have relatively low levels of aneuploid/diploid mosaicism but various models have been developed. Two knockout mouse models produce chaotic mosaics. Sycp3−/− female mice have a disrupted synaptonemal complex and produce aneuploid oocytes. About 30% of heterozygous Sycp3+/- embryos, with Sycp3−/− mothers, become chaotic mosaics, undergo p53-independent apoptosis, beginning in the embryonic ectoderm at E7.0, and die by E8.0 (Lightfoot et al. 2006). Homozygous Bub1b−/− embryos have a defective SAC, a high frequency of premature sister chromatid separation, become chaotic mosaics and die between E7.5 and E16.5 (Schmid et al. 2014).
Mouse embryos with non-specific aneuploidies were also produced by transient exposure to the SAC inhibitor, reversine, between the four-cell and eight-cell stages and provided evidence for lineage-biased cell selection and selective embryonic death (Bolton et al. 2016, Singla et al. 2020). Reversine-treated (RT), eight-cell embryos were aggregated with untreated, diploid embryos to produce RT↔diploid chimaeras. Cells from RT embryos (RT cells) were not preferentially allocated to the trophectoderm but were at a selective disadvantage from the blastocyst stage, predominantly in the ICM. This depletion continued after implantation and chimaeras with a high proportion of RT cells were more likely to die (selective embryonic death). Cell selection was mediated by elevated levels of apoptosis and autophagy in RT ICMs (particularly in epiblast cells) and a compensatory increase in proliferation of untreated cells. Apoptosis was less frequent in trophectoderm cells but RT trophectoderm cells divided more slowly than untreated trophectoderm cells by the sixth division. RT cells were not excluded significantly more frequently from the fetus than the placenta in postimplantation chimaeras and survived in some adults.
In the reversine treatment model, each cell of an eight-cell RT embryo can initiate an independent clone of abnormal cells and different clones may have multiple aneuploidies, single aneuploidies or no aneuploidy. Thus, there is heterogeneity among embryos and among cells within embryos. The ideal CPM model should produce simple, identifiable aneuploid/diploid mosaics. The fate of different aneuploidies should then be evaluated separately because some trisomies may model CPM-I and others may model CPM-II or CPM-III.
Mouse embryos with specific aneuploidies can be produced from stocks with Robertsonian translocations (Gropp et al. 1975) and other strategies have been used to produce XXY and XYY trisomic mice (Hirota et al. 2017). Partial trisomies of mouse chromosome 16 (Reeves et al. 1995, Sago et al. 1998) and a trisomic mouse strain carrying human chromosome 21 (O’Doherty et al. 2005) have also been created to model Down syndrome. Several specific aneuploidies have been incorporated into adult mouse chimeras and some were at a selective disadvantage (Epstein 1985). However, as far as we know, the contribution of a specific aneuploidy to the fetus and extraembryonic tissues has only been compared in two Ts3↔2n chimaeras, which did not show CPM (Everett et al. 2007).
Lightfoot et al. (2006) found that 20% of control, wild-type blastocysts were mosaic, suggesting that the frequency of spontaneous chromosomal mosaicism among mouse embryos might be higher than originally thought or vary among strains. Furthermore, culture (with or without IVF) would probably increase this frequency, as discussed earlier. Time-lapse video microscopy of cultured mouse embryos, created by IVF, showed that those with severe chromosome segregation abnormalities often formed micronuclei (Mashiko et al. 2020). This raises the possibility that, even without IVF, the use of time-lapse video microscopy to identify cultured embryos with micronuclei might provide a source of chromosomally mosaic embryos for experimental investigations, without using special stocks or inducing complex mosaicism.
Some limitations of animal models
Care is required when extrapolating from animal models to humans. Mammalian karyotypes have been rearranged during evolution; so specific human aneuploidies have no exact equivalent in model species (Edwards 1994, Ferguson-Smith & Trifonov 2007).
The morphology of early postimplantation human and mouse embryos and the arrangement of the extraembryonic membranes differ (Supplementary Fig. 4) but this may be unimportant if the underlying developmental mechanisms are similar. Developmental differences among mammalian species were discussed by Eakin and Behringer (2004), who suggested that extraembryonic mesoderm has different origins in humans and mice. This would undermine the use of mice to model CPM-II but more recent evidence, discussed above, suggests that both the epiblast and primitive endoderm contribute to extraembryonic mesoderm in both species (Fig. 4 and Supplementary Fig. 3). Nevertheless, it remains unclear whether these different sources of extraembryonic mesoderm contribute comparably to human and mouse placental mesenchyme.
Zygotic genome activation occurs earlier in mouse embryos than in humans (Jukam et al. 2017), so cell cycle checkpoints would function earlier and this could contribute to their lower frequency of mosaicism. In humans and most mammalian species, except rodents, the centrioles are paternally inherited (Sutovsky & Schatten 2000) so dispermy delivers an extra centriole which often causes spindle abnormalities (Golubovsky 2003). However, this does not apply to mice.
Despite these limitations, more is known about the genetics and development of the mouse than other model species and much of this is likely to apply to humans. Undoubtedly, the mouse will continue to be an important model species for human chromosomal mosaicism but other animal models are also required.
Conclusions
The true frequency of human preimplantation aneuploid/diploid mosaicism is unknown but most evidence suggests that it is quite common. Aneuploid/diploid mosaics typically arise during early cleavage stages, before the embryonic genome is activated and when cell cycle checkpoints are not fully functional. Abnormal chromosome segregation at the first cleavage division can also produce chaotic mosaics, mixoploids and some types of chimaeras but chimaeras probably occur relatively infrequently. The frequency of mosaics declines after implantation and mosaicism is usually confined to the placenta. To date, animal models have provided a number of insights into CPM but developmental differences between some animal models and humans need to be considered and might hinder progress. Further work, including separate analysis of different chromosomal abnormalities and the use of more refined animal models, is required to understand the relationship between preimplantation mosaicism and different types of CPM.
Supplementary Material
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of this review.
Funding
This work did not receive any specific grant from any funding agency in the public, commercial, or not-for-profit sector.
Author contribution statement
J D W wrote the first complete draft of the manuscript; C A E wrote drafts of some sections. J D W and C A E reviewed the literature, prepared figures and reviewed and edited the final manuscript.
Acknowledgement
The authors thank Mr Ronnie Grant for professional help with most of the figures.
References
- Anderson D, Billingham RE, Lampkin GH, Medawar PB.1951The use of skin grafting to distinguish between monozygotic and dizygotic twins in cattle. Heredity 5379–397. ( 10.1038/hdy.1951.38) [DOI] [Google Scholar]
- Angell RR, Aitken RJ, Vanlook PFA, Lumsden MA, Templeton AA.1983Chromosome abnormalities in human embryos after in vitro fertilization. Nature 303336–338. ( 10.1038/303336a0) [DOI] [PubMed] [Google Scholar]
- Angell RR, Templeton AA, Messinis IE.1986Consequences of polyspermy in man. Cytogenetics and Cell Genetics 421–7. ( 10.1159/000132242) [DOI] [PubMed] [Google Scholar]
- Angell RR, Sumner AT, West JD, Thatcher SS, Glasier AF, Baird DT.1987Post-fertilization polyploidy in human preimplantation embryos fertilized in vitro. Human Reproduction 2721–727. ( 10.1093/oxfordjournals.humrep.a136621) [DOI] [PubMed] [Google Scholar]
- Azuma S, Fukuda Y, Toyoda Y.1991aStudies on the production of 2n↔3n chimeric mouse embryos. II. Post-implantation development. Japanese Journal of Animal Reproduction 3779–87 (in Japanese). ( 10.1262/jrd1977.37.79) [DOI] [Google Scholar]
- Azuma S, Fukuda Y, Toyoda Y.1991bStudies on the production of 2n↔3n chimeric mouse embryos. I. Preimplantation development. Japanese Journal of Animal Reproduction 3771–77 (in Japanese). ( 10.1262/jrd1977.37.71) [DOI] [Google Scholar]
- Baart EB, Martini E, Eijkemans MJ, Van Opstal D, Beckers NGM, Verhoeff A, Macklon NS, Fauser BC.2007Milder ovarian stimulation for in-vitro fertilization reduces aneuploidy in the human preimplantation embryo: a randomized controlled trial. Human Reproduction 22980–988. ( 10.1093/humrep/del484) [DOI] [PubMed] [Google Scholar]
- Babariya D, Fragouli E, Alfarawati S, Spath K, Wells D.2017The incidence and origin of segmental aneuploidy in human oocytes and preimplantation embryos. Human Reproduction 322549–2560. ( 10.1093/humrep/dex324) [DOI] [PubMed] [Google Scholar]
- Balakier H, Cabaca O, Bouman D, Shewchuk AB, Laskin C, Squire JA.2000Spontaneous blastomere fusion after freezing and thawing of early human embryos leads to polyploidy and chromosomal mosaicism. Human Reproduction 152404–2410. ( 10.1093/humrep/15.11.2404) [DOI] [PubMed] [Google Scholar]
- Barbash-Hazan S, Frumkin T, Malcov M, Yaron Y, Cohen T, Azem F, Amit A, Ben-Yosef D.2009Preimplantation aneuploid embryos undergo self-correction in correlation with their developmental potential. Fertility and Sterility 92890–896. ( 10.1016/j.fertnstert.2008.07.1761) [DOI] [PubMed] [Google Scholar]
- Bean CJ, Hassold TJ, Judis L, Hunt PA.2002Fertilization in vitro increases non-disjunction during early cleavage divisions in a mouse model system. Human Reproduction 172362–2367. ( 10.1093/humrep/17.9.2362) [DOI] [PubMed] [Google Scholar]
- Belva F, Bonduelle M, Buysse A, Van den Bogaert A, Hes F, Roelants M, Verheyen G, Tournaye H, Keymolen K.2020Chromosomal abnormalities after ICSI in relation to semen parameters: results in 1114 fetuses and 1391 neonates from a single center. Human Reproduction 352149–2162. ( 10.1093/humrep/deaa162) [DOI] [PubMed] [Google Scholar]
- Benkhalifa M, Janny L, Vye P, Malet P, Boucher D, Menezo Y.1993Assessment of polyploidy in human morulae and blastocysts using co-culture and fluorescent in-situ hybridization. Human Reproduction 8895–902. ( 10.1093/oxfordjournals.humrep.a138162) [DOI] [PubMed] [Google Scholar]
- Benn P, Grati FR.2021Aneuploidy in first trimester chorionic villi and spontaneous abortions: windows into the origin and fate of aneuploidy through embryonic and fetal development. Prenatal Diagnosis 41519–524. ( 10.1002/pd.5795) [DOI] [PubMed] [Google Scholar]
- Bianchi DW, Wilkinshaug LE, Enders AC, Hay ED.1993Origin of extraembryonic mesoderm in experimental animals – relevance to chorionic mosaicism in humans. American Journal of Medical Genetics 46542–550. ( 10.1002/ajmg.1320460517) [DOI] [PubMed] [Google Scholar]
- Bielanska M, Tan SL, Ao A.2002Chromosomal mosaicism throughout human preimplantation development in vitro: incidence, type, and relevance to embryo outcome. Human Reproduction 17413–419. ( 10.1093/humrep/17.2.413) [DOI] [PubMed] [Google Scholar]
- Biricik A, Cotroneo E, Minasi MG, Greco PF, Bono S, Surdo M, Lecciso F, Sessa M, Fiorentino F, Spinella Fet al. 2021Cross-validation of next-generation sequencing technologies for diagnosis of chromosomal mosaicism and segmental aneuploidies in preimplantation embryos model. Life 11 340. ( 10.3390/life11040340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy AM, Fortunato A, Sayres MW, Aktipis A.2015Fetal microchimerism and maternal health: a review and evolutionary analysis of cooperation and conflict beyond the womb. BioEssays 371106–1118. ( 10.1002/bies.201500059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boklage CE.2006Embryogenesis of chimeras, twins and anterior midline asymmetries. Human Reproduction 21579–591. ( 10.1093/humrep/dei370) [DOI] [PubMed] [Google Scholar]
- Bolton H, Graham SJL, Van der Aa N, Kumar P, Theunis K, Gallardo EF, Voet T, Zernicka-Goetz M.2016Mouse model of chromosome mosaicism reveals lineage-specific depletion of aneuploid cells and normal developmental potential. Nature Communications 7 11165. ( 10.1038/ncomms11165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capalbo A, Rienzi L.2017Mosaicism between trophectoderm and inner cell mass. Fertility and Sterility 1071098–1106. ( 10.1016/j.fertnstert.2017.03.023) [DOI] [PubMed] [Google Scholar]
- Capalbo A, Wright G, Elliott T, Ubaldi FM, Rienzi L, Nagy ZP.2013FISH reanalysis of inner cell mass and trophectoderm samples of previously array-CGH screened blastocysts shows high accuracy of diagnosis and no major diagnostic impact of mosaicism at the blastocyst stage. Human Reproduction 282298–2307. ( 10.1093/humrep/det245) [DOI] [PubMed] [Google Scholar]
- Capalbo A, Ubaldi FM, Rienzi L, Scott R, Treff N.2017Detecting mosaicism in trophectoderm biopsies: current challenges and future possibilities. Human Reproduction 32492–498. ( 10.1093/humrep/dew250) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capalbo A, Poli M, Rienzi L, Girardi L, Patassini C, Fabiani M, Cimadomo D, Benini F, Farcomeni A, Cuzzi Jet al. 2021Mosaic human preimplantation embryos and their developmental potential in a prospective, non-selection clinical trial. American Journal of Human Genetics 1082238–2247. ( 10.1016/j.ajhg.2021.11.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson JC, Hoffner L, Conlin L, Parks WT, Fisher RA, Spinner N, Yatsenko SA, Bonadio J, Surti U.2018Diploid/triploid mixoploidy: a consequence of asymmetric zygotic segregation of parental genomes. American Journal of Medical Genetics: Part A 1762720–2732. ( 10.1002/ajmg.a.40646) [DOI] [PubMed] [Google Scholar]
- Chatzimeletiou K, Morrison EE, Prapas N, Prapas Y, Handyside AH.2005Spindle abnormalities in normally developing and arrested human preimplantation embryos in vitro identified by confocal laser scanning microscopy. Human Reproduction 20672–682. ( 10.1093/humrep/deh652) [DOI] [PubMed] [Google Scholar]
- Chavez SL, Loewke KE, Han JN, Moussavi F, Colls P, Munné S, Behr B, Pera RAR.2012Dynamic blastomere behaviour reflects human embryo ploidy by the four-cell stage. Nature Communications 3 1251. ( 10.1038/ncomms2249) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HF, Chen M, Ho HN.2020An overview of the current and emerging platforms for preimplantation genetic testing for aneuploidies (PGT-A) in in vitro fertilization programs. Taiwanese Journal of Obstetrics and Gynecology 59489–495. ( 10.1016/j.tjog.2020.05.004) [DOI] [PubMed] [Google Scholar]
- Conlin LK, Thiel BD, Bonnemann CG, Medne L, Ernst LM, Zackai EH, Deardorff MA, Krantz ID, Hakonarson H, Spinner NB.2010Mechanisms of mosaicism, chimerism and uniparental disomy identified by single nucleotide polymorphism array analysis. Human Molecular Genetics 191263–1275. ( 10.1093/hmg/ddq003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coonen E, Harper JC, Ramaekers FCS, Delhanty JDA, Hopman AHN, Geraedts JPM, Handyside AH.1994Presence of chromosomal mosaicism in abnormal preimplantation embryos detected by fluorescence in situ hybridisation. Human Genetics 94609–615. ( 10.1007/BF00206952) [DOI] [PubMed] [Google Scholar]
- Coonen E, Derhaag JG, Dumoulin JCM, van Wissen LCP, Bras M, Janssen M, Evers JLH, Geraedts JPM.2004Anaphase lagging mainly explains chromosomal mosaicism in human preimplantation embryos. Human Reproduction 19316–324. ( 10.1093/humrep/deh077) [DOI] [PubMed] [Google Scholar]
- Coorens THH, Oliver TRW, Sanghvi R, Sovio U, Cook E, Vento-Tormo R, Haniffa M, Young MD, Rahbari R, Sebire Net al. 2021Inherent mosaicism and extensive mutation of human placentas. Nature 59280–85. ( 10.1038/s41586-021-03345-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola G, Alexander B, Di Berardino D, St John E, Basrur PK, King WA.2007Use of cross-species in-situ hybridization (ZOO-FISH) to assess chromosome abnormalities in day-6 in-vivo- or in-vitro-produced sheep embryos. Chromosome Research 15399–408. ( 10.1007/s10577-007-1125-2) [DOI] [PubMed] [Google Scholar]
- Coticchio G, Barrie A, Lagalla C, Borini A, Fishel S, Griffin D, Campbell A.2021Plasticity of the human preimplantation embryo: developmental dogmas, variations on themes and self-correction. Human Reproduction Update 27848–865. ( 10.1093/humupd/dmab016) [DOI] [PubMed] [Google Scholar]
- Daniel A, Wu ZH, Darmanian A, Collins F, Jackson J.2003Three different origins for apparent triploid/diploid mosaics. Prenatal Diagnosis 23529–534. ( 10.1002/pd.634) [DOI] [PubMed] [Google Scholar]
- de Boer KA, Catt JW, Jansen RPS, Leigh D, McArthur S.2004Moving to blastocyst biopsy for preimplantation genetic diagnosis and single embryo transfer at Sydney IVF. Fertility and Sterility 82295–298. ( 10.1016/j.fertnstert.2003.11.064) [DOI] [PubMed] [Google Scholar]
- Delhanty JDA, Griffin DK, Handyside AH, Harper J, Atkinson GHG, Pieters MH, Winston RML.1993Detection of aneuploidy and chromosomal mosaicism in human embryos during preimplantation sex determination by fluorescent in-situ hybridization, (FISH). Human Molecular Genetics 21183–1185. ( 10.1093/hmg/2.8.1183) [DOI] [PubMed] [Google Scholar]
- Delhanty JDA, Harper JC, Ao A, Handyside AH, Winston RML.1997Multicolour FISH detects frequent chromosomal mosaicism and chaotic division in normal preimplantation embryos from fertile patients. Human Genetics 99755–760. ( 10.1007/s004390050443) [DOI] [PubMed] [Google Scholar]
- Derhaag JG, Coonen E, Bras M, Janssen JMB, Ignoul-Vanvuchelen R, Geraedts JPM, Evers JLH, Dumoulin JCM.2003Chromosomally abnormal cells are not selected for the extra-embryonic compartment of the human preimplantation embryo at the blastocyst stage. Human Reproduction 182565–2574. ( 10.1093/humrep/deg485) [DOI] [PubMed] [Google Scholar]
- Destouni A, Vermeesch JR.2017How can zygotes segregate entire parental genomes into distinct blastomeres? The zygote metaphase revisited. BioEssays 391600226. ( 10.1002/bies.201600226) [DOI] [PubMed] [Google Scholar]
- Destouni A, Esteki MZ, Catteeuw M, Tsuiko O, Dimitriadou E, Smits K, Kurg A, Salumets A, Van Soom A, Voet Tet al. 2016Zygotes segregate entire parental genomes in distinct blastomere lineages causing cleavage-stage chimerism and mixoploidy. Genome Research 26567–578. ( 10.1101/gr.200527.115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewald G, Alvarez MN, Cloutier MD, Kelalis PP, Gordon H.1975A diploid-triploid human mosaic with cytogenetic evidence of double fertilization. Clinical Genetics 8149–160. ( 10.1111/j.1399-0004.1975.tb04403.x) [DOI] [PubMed] [Google Scholar]
- Dunsford I, Bowley CC, Hutchison AM, Thompson JS, Sanger R, Race RR.1953A human blood-group chimera. BMJ 281. ( 10.1136/bmj.2.4827.81) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eakin GS, Behringer RR.2004Diversity of germ layer and axis formation among mammals. Seminars in Cell and Developmental Biology 15619–629. ( 10.1016/j.semcdb.2004.04.008) [DOI] [PubMed] [Google Scholar]
- Eakin GS, Hadjantonakis AK, Papaioannou VE, Behringer RR.2005Developmental potential and behavior of tetraploid cells in the mouse embryo. Developmental Biology 288150–159. ( 10.1016/j.ydbio.2005.09.028) [DOI] [PubMed] [Google Scholar]
- Edwards JH.1994Comparative genome mapping in mammals. Current Opinion in Genetics and Development 4861–867. ( 10.1016/0959-437x(9490071-x) [DOI] [PubMed] [Google Scholar]
- Eggenhuizen GM, Go A, Koster MPH, Baart EB, Galjaard RJ.2021Confined placental mosaicism and the association with pregnancy outcome and fetal growth: a review of the literature. Human Reproduction Update 27885–903. ( 10.1093/humupd/dmab009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggermann T, Soellner L, Buiting K, Kotzot D.2015Mosaicism and uniparental disomy in prenatal diagnosis. Trends in Molecular Medicine 2177–87. ( 10.1016/j.molmed.2014.11.010) [DOI] [PubMed] [Google Scholar]
- Ellis JR, Marshall R, Normand ICS, Penrose LS.1963A girl with triploid cells. Nature 198 411. ( 10.1038/198411a0) [DOI] [Google Scholar]
- Enders AC, King BF.1988Formation and differentiation of extraembryonic mesoderm in the rhesus monkey. American Journal of Anatomy 181327–340. ( 10.1002/aja.1001810402) [DOI] [PubMed] [Google Scholar]
- Epstein CJ.1985Mouse monosomies and trisomies as experimental systems for studying mammalian aneuploidy. Trends in Genetics 1129–134. ( 10.1016/0168-9525(8590054-X) [DOI] [Google Scholar]
- Escribà MJ, Vendrell X, Peinado V.2019Segmental aneuploidy in human blastocysts: a qualitative and quantitative overview. Reproductive Biology and Endocrinology 17 76. ( 10.1186/s12958-019-0515-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett CA, West JD.1996The influence of ploidy on the distribution of cells in chimaeric mouse blastocysts. Zygote 459–66. ( 10.1017/s0967199400002896) [DOI] [PubMed] [Google Scholar]
- Everett CA, West JD.1998Evidence for selection against tetraploid cells in tetraploid↔diploid mouse chimaeras before the late blastocyst stage. Genetical Research 72225–228. ( 10.1017/s0016672398003498) [DOI] [PubMed] [Google Scholar]
- Everett CA, Stark MH, West JD, Davidson D, Baldock RA.2000Three-dimensional reconstruction of tetraploid↔diploid chimaeric mouse blastocysts. Journal of Anatomy 196341–346. ( 10.1046/j.1469-7580.2000.19630341.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett CA, Keighren MA, Flockhart JH, West JD.2007Evaluation of triploid↔diploid and trisomy-3↔diploid mouse chimeras as models for investigating how lineage restriction occurs in confined placental mosaicism. Reproduction 134799–809. ( 10.1530/REP-07-0285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evsikov S, Verlinsky Y.1998Mosaicism in the inner cell mass of human blastocysts. Human Reproduction 133151–3155. ( 10.1093/humrep/13.11.3151) [DOI] [PubMed] [Google Scholar]
- Falik-Borenstein TC, Korenberg JR, Schreck RR.1994Confined placental chimerism: prenatal and postnatal cytogenetic and molecular analysis, and pregnancy outcome. American Journal of Medical Genetics 5051–56. ( 10.1002/ajmg.1320500112) [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith MA, Trifonov V.2007Mammalian karyotype evolution. Nature Reviews: Genetics 8950–962. ( 10.1038/nrg2199) [DOI] [PubMed] [Google Scholar]
- Fiorentino F, Bono S, Biricik A, Nuccitelli A, Cotroneo E, Cottone G, Kokocinski F, Michel CE, Minasi MG, Greco E.2014Application of next-generation sequencing technology for comprehensive aneuploidy screening of blastocysts in clinical preimplantation genetic screening cycles. Human Reproduction 292802–2813. ( 10.1093/humrep/deu277) [DOI] [PubMed] [Google Scholar]
- Ford CE.1969Mosaics and chimaeras. British Medical Bulletin 25104–109. ( 10.1093/oxfordjournals.bmb.a070658) [DOI] [PubMed] [Google Scholar]
- Fragouli E, Lenzi M, Ross R, Katz-Jaffe M, Schoolcraft WB, Wells D.2008Comprehensive molecular cytogenetic analysis of the human blastocyst stage. Human Reproduction 232596–2608. ( 10.1093/humrep/den287) [DOI] [PubMed] [Google Scholar]
- Fragouli E, Alfarawati S, Daphnis DD, Goodall NN, Mania A, Griffiths T, Gordon A, Wells D.2011Cytogenetic analysis of human blastocysts with the use of FISH, CGH and aCGH: scientific data and technical evaluation. Human Reproduction 26480–490. ( 10.1093/humrep/deq344) [DOI] [PubMed] [Google Scholar]
- Fragouli E, Alfarawati S, Spath K, Jaroudi S, Sarasa J, Enciso M, Wells D.2013The origin and impact of embryonic aneuploidy. Human Genetics 1321001–1013. ( 10.1007/s00439-013-1309-0) [DOI] [PubMed] [Google Scholar]
- Fragouli E, Alfarawati S, Spath K, Babariya D, Tarozzi N, Borini A, Wells D.2017Analysis of implantation and ongoing pregnancy rates following the transfer of mosaic diploid-aneuploid blastocysts. Human Genetics 136805–819. ( 10.1007/s00439-017-1797-4) [DOI] [PubMed] [Google Scholar]
- Fragouli E, Munné S, Wells D.2019The cytogenetic constitution of human blastocysts: insights from comprehensive chromosome screening strategies. Human Reproduction Update 2515–33. ( 10.1093/humupd/dmy036) [DOI] [PubMed] [Google Scholar]
- Gardner RL.1983Origin and differentiation of extraembryonic tissues in the mouse. International Review of Experimental Pathology 2463–133. [PubMed] [Google Scholar]
- Gardner RL, Cockroft DL.1998Complete dissipation of coherent clonal growth occurs before gastrulation in mouse epiblast. Development 1252397–2402. ( 10.1242/dev.125.13.2397) [DOI] [PubMed] [Google Scholar]
- Gardner RL, Papaioannou VE.1975Differentiation in the trophectoderm and inner cell mass. In The Early Development of Mammals. The Second Symposium of the British Society for Developmental Biology, pp. 107–132. Eds Balls M, Wild AE. Cambridge: Cambridge University Press. [Google Scholar]
- Garner W, McLaren A.1974Cell distribution in chimaeric mouse embryos before implantation. Journal of Embryology and Experimental Morphology 32495–503. ( 10.1242/dev.32.2.495) [DOI] [PubMed] [Google Scholar]
- Giurgea I, Sanlaville D, Fournet JC, Sempoux C, Bellanne-Chantelot C, Touati G, Hubert L, Groos MS, Brunelle F, Rahier Jet al. 2006Congenital hyperinsulinism and mosaic abnormalities of the ploidy. Journal of Medical Genetics 43248–254. ( 10.1136/jmg.2005.034116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleicher N, Barad DH, Ben-Rafael Z, Glujovsky D, Mochizuki L, Modi D, Murtinger M, Patrizio P, Orvieto R, Takahashi Set al. 2021Commentary on two recently published formal guidelines on management of ‘mosaic’ embryos after preimplantation genetic testing for aneuploidy (PGT-A). Reproductive Biology and Endocrinology 19 23. ( 10.1186/s12958-021-00716-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubovsky MD.2003Postzygotic diploidization of triploids as a source of unusual cases of mosaicism, chimerism and twinning. Human Reproduction 18236–242. ( 10.1093/humrep/deg060) [DOI] [PubMed] [Google Scholar]
- Gosden CM.1983Amniotic fluid cell types and culture. British Medical Bulletin 39348–354. ( 10.1093/oxfordjournals.bmb.a071847) [DOI] [PubMed] [Google Scholar]
- Goto Y, Matsui J, Takagi N.2002Developmental potential of mouse tetraploid cells in diploid↔tetraploid chimeric embryos. International Journal of Developmental Biology 46741–745. [PubMed] [Google Scholar]
- Gravholt CH, Friedrich U, Nielsen J.1991Chromosomal mosaicism: a follow-up study of 39 unselected children found at birth. Human Genetics 8849–52. ( 10.1007/BF00204928) [DOI] [PubMed] [Google Scholar]
- Griffin DK, Wilton LJ, Handyside AH, Winston RML, Delhanty JDA.1992Dual fluorescent in situ hybridisation for simultaneous detection of X and Y chromosome-specific probes for the sexing of human preimplantation embryonic nuclei. Human Genetics 8918–22. ( 10.1007/BF00207035) [DOI] [PubMed] [Google Scholar]
- Gropp A, Kolbus U, Giers D.1975Systematic approach to study of trisomy in mouse. II. Cytogenetics and Cell Genetics 1442–62. ( 10.1159/000130318) [DOI] [PubMed] [Google Scholar]
- Gueye NA, Devkota B, Taylor D, Pfundt R, Scott RT, Treff NR.2014Uniparental disomy in the human blastocyst is exceedingly rare. Fertility and Sterility 101232–236. ( 10.1016/j.fertnstert.2013.08.051) [DOI] [PubMed] [Google Scholar]
- Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA.2005The Polo kinase Plk4 functions in centriole duplication. Nature Cell Biology 71140–1146. ( 10.1038/ncb1320) [DOI] [PubMed] [Google Scholar]
- Hardy K, Spanos S, Becker D, Iannelli P, Winston RML, Stark J.2001From cell death to embryo arrest: mathematical models of human preimplantation embryo development. PNAS 981655–1660. ( 10.1073/pnas.98.4.1655) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold TJ, Jacobs PA.1984Trisomy in man. Annual Review of Genetics 1869–97. ( 10.1146/annurev.ge.18.120184.000441) [DOI] [PubMed] [Google Scholar]
- Hino T, Tateno H.2016Developmental potential of 2n/3n mixoploid mouse embryos produced by fusion of individual second polar bodies and blastomeres of 2-cell embryos. Reproduction, Fertility, and Development 281982–1989. ( 10.1071/RD15081) [DOI] [PubMed] [Google Scholar]
- Hirota T, Ohta H, Powell BE, Mahadevaiah SK, Ojarikre OA, Saitou M, Turner JMA.2017Fertile offspring from sterile sex chromosome trisomic mice. Science 357932–935. ( 10.1126/science.aam9046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horii T, Yamamoto M, Morita S, Kimura M, Nagao Y, Hatada I.2015p53 suppresses tetraploid development in mice. Scientific Reports 5 8907. ( 10.1038/srep08907) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu LYF, Perlis TE.1984United States survey on chromosome mosaicism and pseudomosaicism in prenatal diagnosis. Prenatal Diagnosis 497–130. ( 10.1002/pd.1970040708) [DOI] [PubMed] [Google Scholar]
- Huang A, Adusumalli J, Patel S, Liem J, Williams J, Pisarska MD.2009Prevalence of chromosomal mosaicism in pregnancies from couples with infertility. Fertility and Sterility 912355–2360. ( 10.1016/j.fertnstert.2008.03.044) [DOI] [PubMed] [Google Scholar]
- Ishiguro N, Kano K, Yamamoto Y, Taniguchi K.2005Tetraploid cells of enhanced green fluorescent protein transgenic mice in tetraploid/diploid-chimeric embryos. Journal of Reproduction and Development 51567–572. ( 10.1262/jrd.17004) [DOI] [PubMed] [Google Scholar]
- Jacobs K, Van de Velde H, De Paepe C, Sermon K, Spits C.2017Mitotic spindle disruption in human preimplantation embryos activates the spindle assembly checkpoint but not apoptosis until day 5 of development. Molecular Human Reproduction 23321–329. ( 10.1093/molehr/gax007) [DOI] [PubMed] [Google Scholar]
- James RM, Klerkx AH, Keighren M, Flockhart JH, West JD.1995Restricted distribution of tetraploid cells in mouse tetraploid↔diploid chimaeras. Developmental Biology 167213–226. ( 10.1006/dbio.1995.1018) [DOI] [PubMed] [Google Scholar]
- Johnson M.2019Human in vitro fertilisation and developmental biology: a mutually influential history. Development 146 dev183145. ( 10.1242/dev.183145) [DOI] [PubMed] [Google Scholar]
- Johnson DS, Cinnioglu C, Ross R, Filby A, Gemelos G, Hill M, Ryan A, Smotrich D, Rabinowitz M, Murray MJ.2010Comprehensive analysis of karyotypic mosaicism between trophectoderm and inner cell mass. Molecular Human Reproduction 16944–949. ( 10.1093/molehr/gaq062) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukam D, Shariati SAM, Skotheim JM.2017Zygotic genome activation in vertebrates. Developmental Cell 42316–332. ( 10.1016/j.devcel.2017.07.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahraman S, Cetinkaya M, Yuksel B, Yesil M, Cetinkaya CP.2020The birth of a baby with mosaicism resulting from a known mosaic embryo transfer: a case report. Human Reproduction 35727–733. ( 10.1093/humrep/dez309) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser-Rogers KA, McFadden DE, Livasy CA, Dansereau J, Jiang R, Knops JF, Lefebvre L, Rao KW, Robinson WP.2006Androgenetic/biparental mosaicism causes placental mesenchymal dysplasia. Journal of Medical Genetics 43187–192. ( 10.1136/jmg.2005.033571) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalousek DK, Dill FJ.1983Chromosome mosaicism confined to the placenta in human conceptions. Science 221665–667. ( 10.1126/science.6867735) [DOI] [PubMed] [Google Scholar]
- Kalousek DK, Howard-Peebles PN, Olson SB, Barrett IJ, Dorfman A, Black SH, Schulman JD, Wilson RD.1991Confirmation of CVS mosaicism in term placentae and high frequency of intrauterine growth retardation association with confined placental mosaicism. Prenatal Diagnosis 11743–750. ( 10.1002/pd.1970111002) [DOI] [PubMed] [Google Scholar]
- Kola I, Trounson A, Dawson G, Rogers P.1987Tripronuclear human oocytes: altered cleavage patterns and subsequent karyotypic analysis of embryos. Biology of Reproduction 37395–401. ( 10.1095/biolreprod37.2.395) [DOI] [PubMed] [Google Scholar]
- Kotzot D, Utermann G.2005Uniparental disomy (UPD) other than 15: phenotypes and bibliography updated. American Journal of Medical Genetics: Part A 136A287–305. ( 10.1002/ajmg.a.30483) [DOI] [PubMed] [Google Scholar]
- Kwon GS, Viotti M, Hadjantonakis AK.2008The endoderm of the mouse embryo arises by dynamic widespread intercalation of embryonic and extraembryonic lineages. Developmental Cell 15509–520. ( 10.1016/j.devcel.2008.07.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagalla C, Tarozzi N, Sciajno R, Wells D, Di Santo M, Nadalini M, Distratis V, Borini A.2017Embryos with morphokinetic abnormalities may develop into euploid blastocysts. Reproductive Biomedicine Online 34137–146. ( 10.1016/j.rbmo.2016.11.008) [DOI] [PubMed] [Google Scholar]
- Leng LZ, Ouyang Q, Kong XY, Gong F, Lu CF, Zhao L, Shi Y, Cheng DH, Hu L, Lu GXet al. 2017Self-diploidization of human haploid parthenogenetic embryos through the Rho pathway regulates endomitosis and failed cytokinesis. Scientific Reports 7 4242. ( 10.1038/s41598-017-04602-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestou VS, Kalousek DK.1998Confined placental mosaicism and intrauterine fetal growth. Archives of Disease in Childhood: Fetal and Neonatal Edition 79F223–F226. ( 10.1136/fn.79.3.f223) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfoot DA, Kouznetsova A, Mahdy E, Wilbertz J, Hoog C.2006The fate of mosaic aneuploid embryos during mouse development. Developmental Biology 289384–394. ( 10.1016/j.ydbio.2005.11.001) [DOI] [PubMed] [Google Scholar]
- Lonergan P, Pedersen HG, Rizos D, Greve T, Thomsen PD, Fair T, Evans A, Boland MP.2004Effect of the post-fertilization culture environment on the incidence of chromosome aberrations in bovine blastocysts. Biology of Reproduction 711096–1100. ( 10.1095/biolreprod.104.030635) [DOI] [PubMed] [Google Scholar]
- Los FJ, Van Opstal D, Van den Berg C, Braat APG, Verhoef S, Wesby-Van Swaay E, Van den Ouweland AMW, Halley DJJ.1998Uniparental disomy with and without confined placental mosaicism: a model for trisomic zygote rescue. Prenatal Diagnosis 18659–668. () [DOI] [PubMed] [Google Scholar]
- Lu TY, Markert CL.1980Manufacture of diploid/tetraploid chimeric mice. PNAS 776012–6016. ( 10.1073/pnas.77.10.6012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay GE, West JD.2005Fate of tetraploid cells in 4n↔2n chimeric mouse blastocysts. Mechanisms of Development 1221266–1281. ( 10.1016/j.mod.2005.09.001) [DOI] [PubMed] [Google Scholar]
- Madan K.2020Natural human chimeras: a review. European Journal of Medical Genetics 63103971. ( 10.1016/j.ejmg.2020.103971) [DOI] [PubMed] [Google Scholar]
- Magli MC, Gianaroli L, Ferraretti AP, Gordts S, Fredericks V, Crippa A.2009Paternal contribution to aneuploidy in preimplantation embryos. Reproductive Biomedicine Online 18536–542. ( 10.1016/s1472-6483(1060131-9) [DOI] [PubMed] [Google Scholar]
- Makrydimas G, Sebire NJ, Thornton SE, Zagorianakou N, Lolis D, Fisher RA.2002Complete hydatidiform mole and normal live birth: a novel case of confined placental mosaicism: case report. Human Reproduction 172459–2463. ( 10.1093/humrep/17.9.2459) [DOI] [PubMed] [Google Scholar]
- Malan V, Vekemans M, Turleau C.2006Chimera and other fertilization errors. Clinical Genetics 70363–373. ( 10.1111/j.1399-0004.2006.00689.x) [DOI] [PubMed] [Google Scholar]
- Malvestiti F, Agrati C, Grimi B, Pompilii E, Izzi C, Martinoni L, Gaetani E, Liuti MR, Trotta A, Maggi Fet al. 2015Interpreting mosaicism in chorionic villi: results of a monocentric series of 1001 mosaics in chorionic villi with follow-up amniocentesis. Prenatal Diagnosis 351117–1127. ( 10.1002/pd.4656) [DOI] [PubMed] [Google Scholar]
- Mantikou E, Wong KM, Repping S, Mastenbroek S.2012Molecular origin of mitotic aneuploidies in preimplantation embryos. Biochimica et Biophysica Acta 18221921–1930. ( 10.1016/j.bbadis.2012.06.013) [DOI] [PubMed] [Google Scholar]
- Mantzouratou A, Mania A, Fragouli E, Xanthopoulou L, Tashkandi S, Fordham K, Ranieri DM, Doshi A, Nuttall S, Harper JCet al. 2007Variable aneuploidy mechanisms in embryos from couples with poor reproductive histories undergoing preimplantation genetic screening. Human Reproduction 221844–1853. ( 10.1093/humrep/dem102) [DOI] [PubMed] [Google Scholar]
- Marin D, Scott RT, Treff NR.2017Preimplantation embryonic mosaicism: origin, consequences and the reliability of comprehensive chromosome screening. Current Opinion in Obstetrics and Gynecology 29168–174. ( 10.1097/GCO.0000000000000358) [DOI] [PubMed] [Google Scholar]
- Marin D, Zimmerman R, Tao X, Zhan Y, Scott Jr RT, Treff NR.2018Validation of a targeted next generation sequencing-based comprehensive chromosome screening platform for detection of triploidy in human blastocysts. Reproductive Biomedicine Online 36388–395. ( 10.1016/j.rbmo.2017.12.015) [DOI] [PubMed] [Google Scholar]
- Marin D, Xu J, Treff NR.2021Preimplantation genetic testing for aneuploidy: a review of published blastocyst reanalysis concordance data. Prenatal Diagnosis 41545–553. ( 10.1002/pd.5828) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiko D, Ikeda Z, Yao T, Tokoro M, Fukunaga N, Asada Y, Yamagata K.2020Chromosome segregation error during early cleavage in mouse pre-implantation embryo does not necessarily cause developmental failure after blastocyst stage. Scientific Reports 10 854. ( 10.1038/s41598-020-57817-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masset H, Tsuiko O, Vermeesch JR.2021Genome-wide abnormalities in embryos: origins and clinical consequences. Prenatal Diagnosis 41554–563. ( 10.1002/pd.5895) [DOI] [PubMed] [Google Scholar]
- McCoy RC, Demko ZP, Ryan A, Banjevic M, Hill M, Sigurjonsson S, Rabinowitz M, Petrov DA.2015aEvidence of selection against complex mitotic-origin aneuploidy during preimplantation development. PLoS Genetics 11 e1005601. ( 10.1371/journal.pgen.1005601) [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy RC, Demko Z, Ryan A, Banjevic M, Hill M, Sigurjonsson S, Rabinowitz M, Fraser HB, Petrov DA.2015bCommon variants spanning PLK4 are associated with mitotic-origin aneuploidy in human embryos. Science 348235–238. ( 10.1126/science.aaa3337) [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy RC, Newnham LJ, Ottolini CS, Hoffmann ER, Chatzimeletiou K, Cornejo OE, Zhan QS, Zaninovic N, Rosenwaks Z, Petrov DAet al. 2018Tripolar chromosome segregation drives the association between maternal genotype at variants spanning PLK4 and aneuploidy in human preimplantation embryos. Human Molecular Genetics 272573–2585. ( 10.1093/hmg/ddy147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren A.1976Mammalian Chimaeras. Cambridge: Cambridge University Press. [Google Scholar]
- Miura K, Niikawa N.2005Do monochorionic dizygotic twins increase after pregnancy by assisted reproductive technology? Journal of Human Genetics 501–6. ( 10.1007/s10038-004-0216-6) [DOI] [PubMed] [Google Scholar]
- Müller U, Weber JL, Berry P, Kupke KG.1993Second polar body incorporation into a blastomere results in 46,XX/69,XXX mixoploidy. Journal of Medical Genetics 30597–600. ( 10.1136/jmg.30.7.597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munné S, Lee A, Rosenwaks Z, Grifo J, Cohen J.1993Diagnosis of major chromosome aneuploidies in human preimplantation embryos. Human Reproduction 82185–2191. ( 10.1093/oxfordjournals.humrep.a138001) [DOI] [PubMed] [Google Scholar]
- Munné S, Weier HUG, Grifo J, Cohen J.1994Chromosome mosaicism in human embryos. Biology of Reproduction 51373–379. ( 10.1095/biolreprod51.3.373) [DOI] [PubMed] [Google Scholar]
- Munné S, Sandalinas M, Escudero T, Márquez C, Cohen J.2002Chromosome mosaicism in cleavage-stage human embryos: evidence of a maternal age effect. Reproductive Biomedicine Online 4223–232. ( 10.1016/s1472-6483(1061810-x) [DOI] [PubMed] [Google Scholar]
- Munné S, Velilla E, Colls P, Bermudez MG, Vemuri MC, Steuerwald N, Garrisi J, Cohen J.2005Self-correction of chromosomally abnormal embryos in culture and implications for stem cell production. Fertility and Sterility 841328–1334. ( 10.1016/j.fertnstert.2005.06.025) [DOI] [PubMed] [Google Scholar]
- Munné S, Blazek J, Large M, Martinez-Ortiz PA, Nisson H, Liu E, Tarozzi N, Borini A, Becker A, Zhang Jet al. 2017Detailed investigation into the cytogenetic constitution and pregnancy outcome of replacing mosaic blastocysts detected with the use of high-resolution next-generation sequencing. Fertility and Sterility 10862–71. ( 10.1016/j.fertnstert.2017.05.002) [DOI] [PubMed] [Google Scholar]
- Niakan KK, Han JN, Pedersen RA, Simon C, Pera RAR.2012Human pre-implantation embryo development. Development 139829–841. ( 10.1242/dev.060426) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu DM, Pan CC, Lin CY, Hwang BT, Chung MY.2002Mosaic or chimera? Revisiting an old hypothesis about the cause of the 46,XX/46,XY hermaphrodite. Journal of Pediatrics 140732–735. ( 10.1067/mpd.2002.124321) [DOI] [PubMed] [Google Scholar]
- Northrop LE, Treff NR, Levy B, Scott RT.2010SNP microarray-based 24 chromosome aneuploidy screening demonstrates that cleavage-stage FISH poorly predicts aneuploidy in embryos that develop to morphologically normal blastocysts. Molecular Human Reproduction 16590–600. ( 10.1093/molehr/gaq037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Doherty A, Ruf S, Mulligan C, Hildreth V, Errington ML, Cooke S, Sesay A, Modino S, Vanes L, Hernandez Det al. 2005An aneuploid mouse strain carrying human chromosome 21 with Down syndrome phenotypes. Science 3092033–2037. ( 10.1126/science.1114535) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvieto R, Shimon C, Rienstein S, Jonish-Grossman A, Shani H, Aizer A.2020Do human embryos have the ability of self-correction? Reproductive Biology and Endocrinology 18 98. ( 10.1186/s12958-020-00650-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palermo G, Munné S, Cohen J.1994The human zygote inherits its mitotic potential from the male gamete. Human Reproduction 91220–1225. ( 10.1093/oxfordjournals.humrep.a138682) [DOI] [PubMed] [Google Scholar]
- Papavassiliou P, Charalsawadi C, Rafferty K, Jackson-Cook C.2015Mosaicism for trisomy 21: a review. American Journal of Medical Genetics: Part A 167A26–39. ( 10.1002/ajmg.a.36861) [DOI] [PubMed] [Google Scholar]
- Paulson RJ, Treff NR.2020Isn’t it time to stop calling preimplantation embryos ‘mosaic’? F&S Reports 1164–165. ( 10.1016/j.xfre.2020.10.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellestor F, Gatinois V, Puechberty J, Genevieve D, Lefort G.2014Chromothripsis: potential origin in gametogenesis and preimplantation cell divisions. A review. Fertility and Sterility 1021785–1796. ( 10.1016/j.fertnstert.2014.09.006) [DOI] [PubMed] [Google Scholar]
- Penketh RJA, Delhanty JDA, Vandenberghe JA, Finklestone EM, Handyside AH, Malcolm S, Winston RML.1989Rapid sexing of human embryos by non-radioactive in situ hybridization: potential for preimplantation diagnosis of X-linked disorders. Prenatal Diagnosis 9489–499. ( 10.1002/pd.1970090706) [DOI] [PubMed] [Google Scholar]
- Peters HE, Konig TE, Verhoeven MO, Schats R, Mijatovic V, Ket JCF, Lambalk CB.2017Unusual twinning resulting in chimerism: a systematic review on monochorionic dizygotic twins. Twin Research and Human Genetics 20161–168. ( 10.1017/thg.2017.4) [DOI] [PubMed] [Google Scholar]
- Pieters MHEC, Dumoulin JCM, Ignoul-Vanvuchelen RCM, Bras M, Evers JLH, Geraedts JPM.1992Triploidy after in vitro fertilization: cytogenetic analysis of human zygotes and embryos. Journal of Assisted Reproduction and Genetics 968–76. ( 10.1007/BF01204118) [DOI] [PubMed] [Google Scholar]
- Pittalis MC, Dalpra L, Torricelli F, Rizzo N, Nocera G, Cariati E, Santarini L, Tibiletti MG, Agosti S, Bovicelli Let al. 1994The predictive value of cytogenetic diagnosis after CVS based on 4860 cases with both direct and culture methods. Prenatal Diagnosis 14267–278. ( 10.1002/pd.1970140406) [DOI] [PubMed] [Google Scholar]
- Plachot M, Crozet N.1992Fertilization abnormalities in human in-vitro fertilization. Human Reproduction 7 (Supplement 1) 89–94. ( 10.1093/humrep/7.suppl_1.89) [DOI] [PubMed] [Google Scholar]
- Plachot M, Mandelbaum J, Junca AM, de Grouchy J, Salat-Baroux J, Cohen J.1989Cytogenetic analysis and developmental capacity of normal and abnormal embryos after IVF. Human Reproduction 4 (Supplement) 99–103. ( 10.1093/humrep/4.suppl_1.99) [DOI] [PubMed] [Google Scholar]
- Popovic M, Dheedene A, Christodoulou C, Taelman J, Dhaenens L, Van Nieuwerburgh F, Deforce D, Van den Abbeel E, De Sutter P, Menten Bet al. 2018Chromosomal mosaicism in human blastocysts: the ultimate challenge of preimplantation genetic testing? Human Reproduction 331342–1354. ( 10.1093/humrep/dey106) [DOI] [PubMed] [Google Scholar]
- Popovic M, Dhaenens L, Boel A, Menten B, Heindryckx B.2020Chromosomal mosaicism in human blastocysts: the ultimate diagnostic dilemma. Human Reproduction Update 26313–334. ( 10.1093/humupd/dmz050) [DOI] [PubMed] [Google Scholar]
- Quigley DI, McDonald MT, Krishnamuthy V, Kishnani PS, Lee MM, Haqq AM, Goodman BK.2005Triploid mosaicism in a 45,X/69,XXY infant. American Journal of Medical Genetics: Part A 138A171–174. ( 10.1002/ajmg.a.30943) [DOI] [PubMed] [Google Scholar]
- Rabinowitz M, Ryan A, Gemelos G, Hill M, Baner J, Cinnioglu C, Banjevic M, Potter D, Petrov DA, Demko Z.2012Origins and rates of aneuploidy in human blastomeres. Fertility and Sterility 97395–401. ( 10.1016/j.fertnstert.2011.11.034) [DOI] [PubMed] [Google Scholar]
- Rambags BPB, Krijtenburg PJ, Van Drie HF, Lazzari G, Galli C, Pearson PL, Colenbrander B, Stout TAE.2005Numerical chromosomal abnormalities in equine embryos produced in vivo and in vitro. Molecular Reproduction and Development 7277–87. ( 10.1002/mrd.20302) [DOI] [PubMed] [Google Scholar]
- Reeves RH, Irving NG, Moran TH, Wohn A, Kitt C, Sisodia SS, Schmidt C, Bronson RT, Davisson MT.1995A mouse model for Down syndrome exhibits learning and behavior deficits. Nature Genetics 11177–184. ( 10.1038/ng1095-177) [DOI] [PubMed] [Google Scholar]
- Reichmann J, Nijmeijer B, Hossain MJ, Eguren M, Schneider I, Politi AZ, Roberti MJ, Hufnagel L, Hiiragi T, Ellenberg J.2018Dual-spindle formation in zygotes keeps parental genomes apart in early mammalian embryos. Science 361189–193. ( 10.1126/science.aar7462) [DOI] [PubMed] [Google Scholar]
- Robinson WP, Barrett IJ, Bernard L, Telenius A, Bernasconi F, Wilson RD, Best RG, Howard-Peebles PN, Langlois S, Kalousek DK.1997Meiotic origin of trisomy in confined placental mosaicism is correlated with presence of fetal uniparental disomy, high levels of trisomy in trophoblast, and increased risk of fetal intrauterine growth restriction. American Journal of Human Genetics 60917–927. [PMC free article] [PubMed] [Google Scholar]
- Robinson WP, McFadden DE, Barrett IJ, Kuchinka B, Penaherrera MS, Bruyere H, Best RG, Pedreira DAL, Langlois S, Kalousek DK.2002Origin of amnion and implications for evaluation of the fetal genotype in cases of mosaicism. Prenatal Diagnosis 221076–1085. ( 10.1002/pd.483) [DOI] [PubMed] [Google Scholar]
- Robinson WP, Lauzon JL, Innes AM, Lim K, Arsovska S, McFadden DE.2007Origin and outcome of pregnancies affected by androgenetic/biparental chimerism. Human Reproduction 221114–1122. ( 10.1093/humrep/del462) [DOI] [PubMed] [Google Scholar]
- Rodriguez AM, Downs KM.2017Visceral endoderm and the primitive streak interact to build the fetal-placental interface of the mouse gastrula. Developmental Biology 43298–124. ( 10.1016/j.ydbio.2017.08.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbusch BE, Schneider M.2009Separation of a pronucleus by premature cytokinesis: a mechanism for immediate diploidization of tripronuclear oocytes? Fertility and Sterility 92394.e5–394.e8. ( 10.1016/j.fertnstert.2009.02.037) [DOI] [PubMed] [Google Scholar]
- Ruangvutilert P, Delhanty JD, Rodeck CH, Harper JC.2000Relative efficiency of FISH on metaphase and interphase nuclei from non-mosaic trisomic or triploid fibroblast cultures. Prenatal Diagnosis 20159–162. () [DOI] [PubMed] [Google Scholar]
- Sabhnani TV, Elaimi A, Sultan H, Alduraihem A, Serhal P, Harper JC.2011Increased incidence of mosaicism detected by FISH in murine blastocyst cultured in vitro. Reproductive Biomedicine Online 22621–631. ( 10.1016/j.rbmo.2011.01.011) [DOI] [PubMed] [Google Scholar]
- Sago H, Carlson EJ, Smith DJ, Kilbridge J, Rubin EM, Mobley WC, Epstein CJ, Huang TT.1998Ts1Cje, a partial trisomy 16 mouse model for Down syndrome, exhibits learning and behavioral abnormalities. PNAS 956256–6261. ( 10.1073/pnas.95.11.6256) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiz N, Plusa B.2013Early cell fate decisions in the mouse embryo. Reproduction 145R65–R80. ( 10.1530/REP-12-0381) [DOI] [PubMed] [Google Scholar]
- Santos MA, Teklenburg G, Macklon NS, Van Opstal D, Schuring-Blom GH, Krijtenburg PJ, de Vreeden-Elbertse J, Fauser BC, Baart EB.2010The fate of the mosaic embryo: chromosomal constitution and development of day 4, 5 and 8 human embryos. Human Reproduction 251916–1926. ( 10.1093/humrep/deq139) [DOI] [PubMed] [Google Scholar]
- Sathananthan AH, Ratnam SS, Ng SC, Tarín JJ, Gianaroli L, Trounson A.1996The sperm centriole: its inheritance, replication and perpetuation in early human embryos. Human Reproduction 11345–356. ( 10.1093/humrep/11.2.345) [DOI] [PubMed] [Google Scholar]
- Schmid W, Vischer D.1967A malformed boy with double aneuploidy and diploid-triploid mosaicism 48,XXYY/71,XXXYY. Cytogenetics 6145–155. ( 10.1159/000129935) [DOI] [PubMed] [Google Scholar]
- Schmid M, Steinlein C, Tian Q, Newell AEH, Gessler M, Olson SB, Rosenwald A, Kneitz B, Fedorov LM.2014Mosaic variegated aneuploidy in mouse BubR1 deficient embryos and pregnancy loss in human. Chromosome Research 22375–392. ( 10.1007/s10577-014-9432-x) [DOI] [PubMed] [Google Scholar]
- Scott RT, Galliano D.2016The challenge of embryonic mosaicism in preimplantation genetic screening. Fertility and Sterility 1051150–1152. ( 10.1016/j.fertnstert.2016.01.007) [DOI] [PubMed] [Google Scholar]
- Scriven PN, Bossuyt PM.2010Diagnostic accuracy: theoretical models for preimplantation genetic testing of a single nucleus using the fluorescence in situ hybridization technique. Human Reproduction 252622–2628. ( 10.1093/humrep/deq196) [DOI] [PubMed] [Google Scholar]
- Sheppard SE, Lalonde E, Adzick NS, Beck AE, Bhatti T, De Leon DD, Duffy KA, Ganguly A, Hathaway E, Ji JLet al. 2019Androgenetic chimerism as an etiology for Beckwith-Wiedemann syndrome: diagnosis and management. Genetics in Medicine 212644–2649. ( 10.1038/s41436-019-0551-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu ZQ, Row S, Deng WM.2018Endoreplication: the good, the bad, and the ugly. Trends in Cell Biology 28465–474. ( 10.1016/j.tcb.2018.02.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons DG, Fortier AL, Cross JC.2007Diverse subtypes and developmental origins of trophoblast giant cells in the mouse placenta. Developmental Biology 304567–578. ( 10.1016/j.ydbio.2007.01.009) [DOI] [PubMed] [Google Scholar]
- Simon-Bouy B, Plachot M, Mokdad A, Lavaud N, Muti C, Bazin A, Vialard F, Belaisch-Allart J.2003Possible human chimera detected prenatally after in vitro fertilization: a case report. Prenatal Diagnosis 23935–937. ( 10.1002/pd.733) [DOI] [PubMed] [Google Scholar]
- Singla S, Iwamoto-Stohl LK, Zhu M, Zernicka-Goetz M.2020Autophagy-mediated apoptosis eliminates aneuploid cells in a mouse model of chromosome mosaicism. Nature Communications 11 2958. ( 10.1038/s41467-020-16796-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souter VL, Kapur RP, Nyholt DR, Skogerboe K, Myerson D, Ton CC, Opheim KE, Easterling TR, Shields LE, Montgomery GWet al. 2003A report of dizygous monochorionic twins. New England Journal of Medicine 349154–158. ( 10.1056/NEJMoa030050) [DOI] [PubMed] [Google Scholar]
- Souter VL, Parisi MA, Nyholt DR, Kapur RP, Henders AK, Opheim KE, Gunther DF, Mitchell ME, Glass IA, Montgomery GW.2007A case of true hermaphroditism reveals an unusual mechanism of twinning. Human Genetics 121179–185. ( 10.1007/s00439-006-0279-x) [DOI] [PubMed] [Google Scholar]
- Spinner NB, Conlin LK.2014Mosaicism and clinical genetics. American Journal of Medical Genetics, Part C 166C397–405. [DOI] [PubMed] [Google Scholar]
- Starostik MR, Sosina OA, McCoy RC.2020Single-cell analysis of human embryos reveals diverse patterns of aneuploidy and mosaicism. Genome Research 30814–825. ( 10.1101/gr.262774.120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strain L, Warner JP, Johnston T, Bonthron DT.1995A human parthenogenetic chimera. Nature Genetics 11164–169. ( 10.1038/ng1095-164) [DOI] [PubMed] [Google Scholar]
- Strain L, Dean JCS, Hamilton MPR, Bonthron DT.1998A true hermaphrodite chimera resulting from embryo amalgamation after in vitro fertilization. New England Journal of Medicine 338166–169. ( 10.1056/NEJM199801153380305) [DOI] [PubMed] [Google Scholar]
- Surti U, Hill LM, Dunn J, Prosen T, Hoffner L.2005Twin pregnancy with a chimeric androgenetic and biparental placenta in one twin displaying placental mesenchymal dysplasia phenotype. Prenatal Diagnosis 251048–1056. ( 10.1002/pd.1255) [DOI] [PubMed] [Google Scholar]
- Sutovsky P, Schatten G.2000Paternal contributions to the mammalian zygote: fertilization after sperm-egg fusion. International Review of Cytology 1951–65. ( 10.1016/s0074-7696(0862703-5) [DOI] [PubMed] [Google Scholar]
- Suwinska A, Ozdzenski W, Waksmundzka M, Tarkowski AK.2005Experimentally produced diploid↔triploid mouse chimaeras develop up to adulthood. Molecular Reproduction and Development 72362–376. ( 10.1002/mrd.20350) [DOI] [PubMed] [Google Scholar]
- Swain JE.2019Controversies in ART: can the IVF laboratory influence preimplantation embryo aneuploidy? Reproductive Biomedicine Online 39599–607. ( 10.1016/j.rbmo.2019.06.009) [DOI] [PubMed] [Google Scholar]
- Tang PC, West JD.2000The effects of embryo stage and cell number on the composition of mouse aggregation chimaeras. Zygote 8235–243. ( 10.1017/s0967199400001039) [DOI] [PubMed] [Google Scholar]
- Tang PC, Ritchie WA, Wilmut I, West JD.2000The effects of cell size and ploidy on cell allocation in mouse chimaeric blastocysts. Zygote 833–43. ( 10.1017/s0967199400000800) [DOI] [PubMed] [Google Scholar]
- Tarkowski AK, Witkowska A, Opas J.1977Development of cytochalasin B-induced tetraploid and diploid/tetraploid mosaic mouse embryos. Journal of Embryology and Experimental Morphology 4147–64. ( 10.1242/dev.41.1.47) [DOI] [PubMed] [Google Scholar]
- Tarkowski AK, Ozdzenski W, Czolowska R.2001Mouse singletons and twins developed from isolated diploid blastomeres supported with tetraploid blastomeres. International Journal of Developmental Biology 45591–596. [PubMed] [Google Scholar]
- Tarkowski AK, Ozdzenski W, Czolowska R.2005Identical triplets and twins developed from isolated blastomeres of 8-and 16-cell mouse embryos supported with tetraploid blastomeres. International Journal of Developmental Biology 49825–832. ( 10.1387/ijdb.052018at) [DOI] [PubMed] [Google Scholar]
- Treff NR, Franasiak JM.2017Detection of segmental aneuploidy and mosaicism in the human preimplantation embryo: technical considerations and limitations. Fertility and Sterility 10727–31. ( 10.1016/j.fertnstert.2016.09.039) [DOI] [PubMed] [Google Scholar]
- Treff NR, Marin D.2021The ‘mosaic’ embryo: misconceptions and misinterpretations in preimplantation genetic testing for aneuploidy. Fertility and Sterility 1161205–1211. ( 10.1016/j.fertnstert.2021.06.027) [DOI] [PubMed] [Google Scholar]
- Treff NR, Levy B, Su J, Northrop LE, Tao X, Scott Jr RT.2010SNP microarray-based 24 chromosome aneuploidy screening is significantly more consistent than FISH. Molecular Human Reproduction 16583–589. ( 10.1093/molehr/gaq039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tšuiko O, Catteeuw M, Zamani Esteki M, Destouni A, Bogado Pascottini O, Besenfelder U, Havlicek V, Smits K, Kurg A, Salumets Aet al. 2017Genome stability of bovine in vivo-conceived cleavage-stage embryos is higher compared to in vitro-produced embryos. Human Reproduction 322348–2357. ( 10.1093/humrep/dex286) [DOI] [PubMed] [Google Scholar]
- van de Laar I, Rabelink G, Hochstenbach R, Tuerlings J, Hoogeboom J, Giltay J.2002Diploid/triploid mosaicism in dysmorphic patients. Clinical Genetics 62376–382. ( 10.1034/j.1399-0004.2002.620504.x) [DOI] [PubMed] [Google Scholar]
- van Echten-Arends J, Mastenbroek S, Sikkema-Raddatz B, Korevaar JC, Heineman MJ, van der Veen F, Repping S.2011Chromosomal mosaicism in human preimplantation embryos: a systematic review. Human Reproduction Update 17620–627. ( 10.1093/humupd/dmr014) [DOI] [PubMed] [Google Scholar]
- Van Opstal D, Diderich KEM, Joosten M, Govaerts LCP, Polak J, Boter M, Saris JJ, Cheung WY, van Veen S, van de Helm Ret al. 2018Unexpected finding of uniparental disomy mosaicism in term placentas: is it a common feature in trisomic placentas? Prenatal Diagnosis 38911–919. ( 10.1002/pd.5354) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste E, Voet T, Le Caignec C, Ampe M, Konings P, Melotte C, Debrock S, Amyere M, Vikkula M, Schuit Fet al. 2009Chromosome instability is common in human cleavage-stage embryos. Nature Medicine 15577–583. ( 10.1038/nm.1924) [DOI] [PubMed] [Google Scholar]
- Vazquez-Diez C, FitzHarris G.2018Causes and consequences of chromosome segregation error in preimplantation embryos. Reproduction 155R63–R76. ( 10.1530/REP-17-0569) [DOI] [PubMed] [Google Scholar]
- Vazquez-Diez C, Yamagata K, Trivedi S, Haverfield J, FitzHarris G.2016Micronucleus formation causes perpetual unilateral chromosome inheritance in mouse embryos. PNAS 113626–631. ( 10.1073/pnas.1517628112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Diez C, Paim LMG, FitzHarris G.2019Cell-size-independent spindle checkpoint failure underlies chromosome segregation error in mouse embryos. Current Biology 29865, .e3–873.e3. ( 10.1016/j.cub.2018.12.042) [DOI] [PubMed] [Google Scholar]
- Vera-Rodriguez M, Michel CE, Mercader A, Bladon AJ, Rodrigo L, Kokocinski F, Mateu E, Al-Asmar N, Blesa D, Simon Cet al. 2016Distribution patterns of segmental aneuploidies in human blastocysts identified by next-generation sequencing. Fertility and Sterility 1051047–1055.e2. ( 10.1016/j.fertnstert.2015.12.022) [DOI] [PubMed] [Google Scholar]
- Viotti M.2020Preimplantation genetic testing for chromosomal abnormalities: aneuploidy, mosaicism, and structural rearrangements. Genes 11 602. ( 10.3390/genes11060602) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viotti M, McCoy RC, Griffin DK, Spinella F, Greco E, Madjunkov M, Madjunkova S, Librach CL, Victor AR, Barnes FLet al. 2021aLet the data do the talking: the need to consider mosaicism during embryo selection. Fertility and Sterility 1161212–1219. ( 10.1016/j.fertnstert.2021.09.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viotti M, Victor AR, Barnes FL, Zouves CG, Besser AG, Grifo JA, Cheng EH, Lee MS, Horcajadas JA, Corti Let al. 2021bUsing outcome data from one thousand mosaic embryo transfers to formulate an embryo ranking system for clinical use. Fertility and Sterility 1151212–1224. ( 10.1016/j.fertnstert.2020.11.041) [DOI] [PubMed] [Google Scholar]
- Viuff D, Rickords L, Offenberg H, Hyttel P, Avery B, Greve T, Olsaker I, Williams JL, Callesen H, Thomsen PD.1999A high proportion of bovine blastocysts produced in vitro are mixoploid. Biology of Reproduction 601273–1278. ( 10.1095/biolreprod60.6.1273) [DOI] [PubMed] [Google Scholar]
- Viuff D, Palsgaard A, Rickords L, Lawson LG, Greve T, Schmidt M, Avery B, Hyttel P, Thomsen PD.2002Bovine embryos contain a higher proportion of polyploid cells in the trophectoderm than in the embryonic disc. Molecular Reproduction and Development 62483–488. ( 10.1002/mrd.90004) [DOI] [PubMed] [Google Scholar]
- Voullaire L, Slater H, Williamson R, Wilton L.2000Chromosome analysis of blastomeres from human embryos by using comparative genomic hybridization. Human Genetics 106210–217. ( 10.1007/s004390051030) [DOI] [PubMed] [Google Scholar]
- Voullaire L, Collins V, Callaghan T, McBain J, Williamson R, Wilton L.2007High incidence of complex chromosome abnormality in cleavage embryos from patients with repeated implantation failure. Fertility and Sterility 871053–1058. ( 10.1016/j.fertnstert.2006.11.043) [DOI] [PubMed] [Google Scholar]
- Wang F, Liu Y.2020Identification of key genes, regulatory factors, and drug target genes of recurrent implantation failure (RIF). Gynecological Endocrinology 36448–455. ( 10.1080/09513590.2019.1680622) [DOI] [PubMed] [Google Scholar]
- Wang BBT, Rubin CH, Williams J.1993Mosaicism in chorionic villus sampling: an analysis of incidence and chromosomes involved in 2612 consecutive cases. Prenatal Diagnosis 13179–190. ( 10.1002/pd.1970130305) [DOI] [PubMed] [Google Scholar]
- Warburton D, Yu CY, Kline J, Stein Z.1978Mosaic autosomal trisomy in cultures from spontaneous abortions. American Journal of Human Genetics 30609–617. [PMC free article] [PubMed] [Google Scholar]
- Wells D, Delhanty JDA.2000Comprehensive chromosomal analysis of human preimplantation embryos using whole genome amplification and single cell comparative genomic hybridization. Molecular Human Reproduction 61055–1062. ( 10.1093/molehr/6.11.1055) [DOI] [PubMed] [Google Scholar]
- West JD, Angell RR, Thatcher SS, Gosden JR, Hastie ND, Glasier AF, Baird DT.1987Sexing the human pre-embryo by DNA-DNA in situ hybridization. Lancet 3291345–1347. ( 10.1016/S0140-6736(8790650-7) [DOI] [PubMed] [Google Scholar]
- Yamazawa K, Nakabayashi K, Kagami M, Sato T, Saitoh S, Horikawa R, Hizuka N, Ogata T.2010Parthenogenetic chimaerism/mosaicism with a Silver-Russell syndrome-like phenotype. Journal of Medical Genetics 47782–785. ( 10.1136/jmg.2010.079343) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Rito T, Metzger J, Naftaly J, Soman R, Hu JJ, Albertini DF, Barad DH, Brivanlou AH, Gleicher N.2021Depletion of aneuploid cells in human embryos and gastruloids. Nature Cell Biology 23314–321. ( 10.1038/s41556-021-00660-7) [DOI] [PubMed] [Google Scholar]
- Yost S, de Wolf B, Hanks S, Zachariou A, Marcozzi C, Clarke M, de Voer R, Etemad B, Uijttewaal E, Ramsay Eet al. 2017Biallelic TRIP13 mutations predispose to Wilms tumor and chromosome missegregation. Nature Genetics 491148–1151. ( 10.1038/ng.3883) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamani Esteki M, Viltrop T, Tsuiko O, Tiirats A, Koel M, Noukas M, Zilina O, Teearu K, Marjonen H, Kahila Het al. 2019In vitro fertilization does not increase the incidence of de novo copy number alterations in fetal and placental lineages. Nature Medicine 251699–1705. ( 10.1038/s41591-019-0620-2) [DOI] [PubMed] [Google Scholar]
- Zaragoza MV, Surti U, Redline RW, Millie E, Chakravarti A, Hassold TJ.2000Parental origin and phenotype of triploidy in spontaneous abortions: predominance of diandry and association with the partial hydatidiform mole. American Journal of Human Genetics 661807–1820. ( 10.1086/302951) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenzes MT, Casper RF.1992Cytogenetics of human oocytes, zygotes, and embryos after in vitro fertilization. Human Genetics 88367–375. ( 10.1007/BF00215667) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



 This work is licensed under a
This work is licensed under a